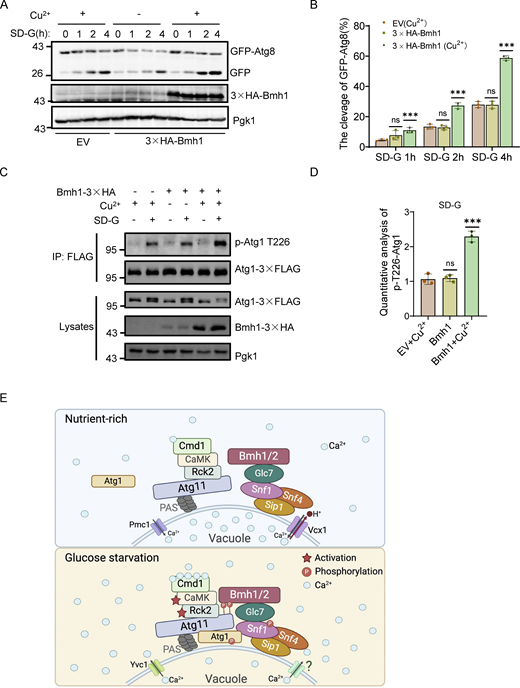
Autophagy is essential for maintaining glucose homeostasis. However, the mechanism by which cells sense and respond to glucose starvation to induce autophagy remains incomplete. Here, we show that calcium serves as a fundamental triggering signal that connects environmental sensing to the formation of the autophagy initiation complex during glucose starvation. Mechanistically, glucose starvation instigates the release of vacuolar calcium into the cytoplasm, thus triggering the activation of Rck2 kinase. In turn, Rck2-mediated Atg11 phosphorylation enhances Atg11 interactions with Bmh1/2 bound to the Snf1–Sip1–Snf4 complex, leading to recruitment of vacuolar membrane-localized Snf1 to the PAS and subsequent Atg1 activation, thereby initiating autophagy. We also identified Glc7, a protein phosphatase-1, as a critical regulator of the association between Bmh1/2 and the Snf1 complex. We thus propose that calcium-triggered Atg11–Bmh1/2–Snf1 complex assembly initiates autophagy by controlling Snf1-mediated Atg1 activation in response to glucose starvation.
Introduction
The ability to adapt to prolonged nutrient deprivation is fundamental for survival in all organisms (Ohsumi, 2014). Autophagy, a highly conserved intracellular degradation process, plays a key role in this adaptation (Kuma et al., 2004; Tsukada and Ohsumi, 1993). Maintaining glucose homeostasis is one of the most important physiological roles connected to autophagy, as evidenced by autophagy-deficient mice suffering fatal hypoglycemia upon fasting (Karsli-Uzunbas et al., 2014). Understanding how cells sense and respond to glucose starvation to induce autophagy is a central question.
Calcium is a ubiquitous signaling molecule essential for many cellular functions (Berridge et al., 2000). In Saccharomyces cerevisiae, cytosolic Ca2+ levels are kept low by various membrane-anchored Ca2+ transporters (Cui et al., 2009). Extracellular Ca2+ enters the cytosol via a high-affinity, low-capacity influx system consisting of Cch1 and Mid1 (Iida et al., 1994; Fischer et al., 1997). Vacuoles store about 95% of the cell’s Ca2+ and serve as the primary Ca2+ reservoir (Cunningham and Fink, 1994). The Ca2+ ATPase Pmc1 and the Ca2+/H+ exchanger Vcx1 import cytosolic Ca2+ into vacuoles (Cunningham and Fink, 1994; Cagnac et al., 2010), while Yvc1 mediates vacuolar Ca2+ efflux into the cytosol (Denis and Cyert, 2002). The endoplasmic reticulum (ER) also helps maintain Ca2+ homeostasis (Samarão et al., 2009), with Csg2 identified as an ER calcium channel involved in Ca2+ transport to the cytosol (Liu et al., 2023). In mammals, Ca2+ signaling is crucial for specifying autophagosome initiation sites (Zheng et al., 2022). Nonetheless, the mechanism by which Ca2+ signaling triggers the autophagy initiation complex assembly in response to stress, particularly in energy deprivation, remains unknown.
Over 40 autophagy-related genes (ATG genes) are identified in yeast (Parzych et al., 2018). Among them, the Atg1 complex comprises six proteins: Atg1, Atg11, Atg13, Atg17, Atg29, and Atg31 (Feng et al., 2014). Atg17 constitutively forms a stable ternary complex with Atg31 and Atg29, crucial for organizing the phagophore assembly site (PAS) and recruiting other ATG proteins (Kabeya et al., 2009). Atg13 is highly phosphorylated under nutrient-rich conditions but dephosphorylates during nitrogen starvation, recruiting to the PAS via Atg17 interaction (Kabeya et al., 2005). Atg11 links autophagy receptors directly with Atg1 for cargo degradation (Feng et al., 2014). Atg1, a Ser/Thr protein kinase, is essential for autophagy, requiring recruitment to the PAS for activation (Kabeya et al., 2005; Yamamoto et al., 2016). Our previous studies have shown that glucose starvation–induced autophagy differs from nitrogen starvation-induced autophagy in both signaling pathways and protein machinery, with mitochondria playing a key role in glucose starvation–induced autophagy (Yi et al., 2017). Atg11 is essential for glucose starvation–induced autophagy, acting as a multifunctional initiation factor for Atg1 activation and PAS recruitment of Atg9 vesicles (Yao et al., 2020). In mammals, activated AMPK phosphorylates and activates ULK1, the Atg1 homolog, for glucose starvation–induced autophagy (Kim et al., 2011). Nonetheless, the mechanisms by which Snf1/AMPK phosphorylates and activates Atg1/ULK1 during glucose starvation remain elusive.
Bmh1 and its paralog Bmh2, sharing 93% amino acid identity, are homologs of mammalian 14-3-3 proteins in S. cerevisiae (Van Heusden et al., 1995). Loss of either BMH1 or BMH2 does not affect cell growth, but deletion of both is lethal (Van Heusden et al., 1995; Van Hemert et al., 2001). The 14-3-3 proteins are phosphor-binding proteins conserved in all eukaryotes, involved in processes like ascospore formation, DNA damage checkpoints, and DNA replication initiation (Cau et al., 2018). In mammals, seven 14-3-3 isoforms exist (Abdrabou et al., 2020). In plants and mammals, 14-3-3 proteins are primarily involved in the negative regulation of nutritional starvation-induced autophagy (Pozuelo-Rubio, 2011; Qi et al., 2022; Xu et al., 2019). However, the specific role(s) of Bmh1/2 in glucose starvation–induced autophagy remain undetermined.
In this study, we show that Ca2+ is a crucial signal linking environmental sensing to autophagy initiation during glucose starvation. Through genetic and biochemical analyses, we reveal a previously undocumented pathway for autophagy initiation in which Ca2+-regulated phosphorylation of Atg11 by Rck2 induces binding of Atg11 with Bmh1/2. This results in vacuolar membrane-localized Snf1 recruitment to the PAS to activate Atg1 in response to glucose deprivation. We thus propose a paradigm in which Ca2+-triggered Atg11–Bmh1/2–Snf1 complex assembly acts as an indispensable prerequisite for Snf1-mediated activation of glucose starvation–induced autophagy by governing the formation of the autophagy initiation complex.
Results
Bmh1/2 are identified as binding partners of Atg11
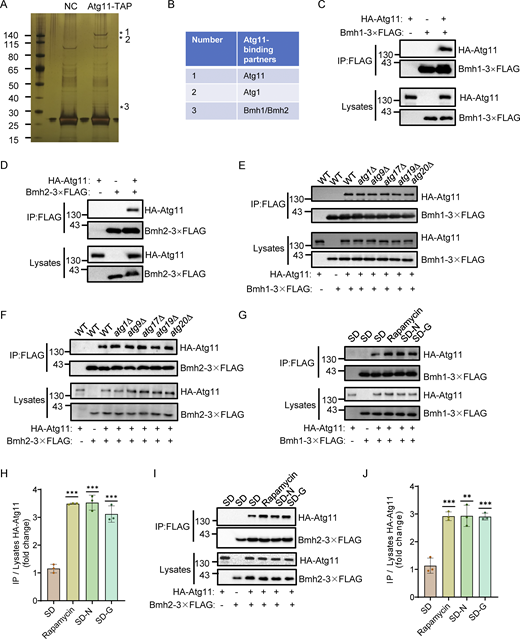
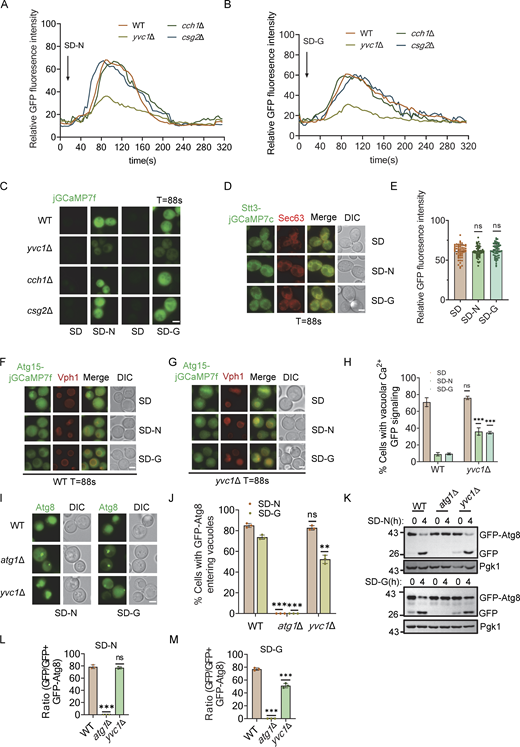
To explore how Atg11 participates in glucose starvation-induced autophagy, we performed affinity purification assays in S. cerevisiae. Atg11-TAP were purified from glucose starvation-treated cells using Rabbit IgG-conjugated Dynabeads. Silver staining revealed that Atg11 interacts with 30–40 kDa proteins, identified as Bmh1 and Bmh2, by mass spectrometry (Fig. 1, A and B). Coimmunoprecipitation (Co-IP) experiments with yeast cells coexpressing HA-Atg11 and Bmh1- or Bmh2-3×FLAG confirmed that Atg11 coprecipitated with both Bmh1 and Bmh2 during glucose starvation (Fig. 1, C and D). Since Atg11 is known to also interact with Atg1, Atg9, Atg17, Atg19, and Atg20 (Yorimitsu and Klionsky, 2005), we next investigated whether the Atg11-Bmh1/2 interaction required any of these other proteins. To this end, we generated ATG1, ATG9, ATG17, ATG19, or ATG20 knockout in yeast cells co-expressing HA-Atg11 with Bmh1-3×FLAG or Bmh2-3×FLAG. Co-IP assays showed that the absence of these proteins did not affect Atg11 binding to Bmh1 or Bmh2, suggesting that Atg11-Bmh1/2 interactions are independent of these proteins (Fig. 1, E and F).
Identification of Bmh1 and Bmh2 as binding partners of Atg11. (A) BY4741 (negative control, NC) or Atg11-3✕FLAG-TEV-ZZ (Atg11-TAP) yeast cells were subjected to glucose starvation for 1 h. Atg11-TAP protein was purified using anti-Rabbit IgG Dynabeads. The samples were separated by 4–12% SDS-PAGE gel, followed by silver staining. Target bands were subjected to LC-MS/MS analysis. (B) Identification of Bmh1/2 proteins by LC-MS/MS analysis. (C and D) Cells co-expressing HA-Atg11 and Bmh1-3✕FLAG (C) or Bmh2-3✕FLAG (D) were treated with glucose starvation for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (E and F) Wild-type (WT), atg1∆, atg9∆, atg17∆, atg19∆, or atg20∆ cells co-expressing HA-Atg11 with Bmh1-3✕FLAG (E) or Bmh2-3✕FLAG (F) were subjected to glucose starvation for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (G and I) Cells co-expressing HA-Atg11 with Bmh1-3✕FLAG (G) or Bmh2-3✕FLAG (I) were cultured in nutrient-rich medium, nitrogen-starvation medium (SD-N), glucose-starvation medium (SD-G), or treated with rapamycin (0.2 μg/ml) for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and analyzed by Western blot using anti-HA antibody. (H and J) The results from G and I were quantified. Data are presented as means ± SD (n = 3). ***P < 0.001; **P < 0.01; two-tailed Student’s t tests were used. Source data are available for this figure: SourceData F1.
Identification of Bmh1 and Bmh2 as binding partners of Atg11. (A) BY4741 (negative control, NC) or Atg11-3✕FLAG-TEV-ZZ (Atg11-TAP) yeast cells were subjected to glucose starvation for 1 h. Atg11-TAP protein was purified using anti-Rabbit IgG Dynabeads. The samples were separated by 4–12% SDS-PAGE gel, followed by silver staining. Target bands were subjected to LC-MS/MS analysis. (B) Identification of Bmh1/2 proteins by LC-MS/MS analysis. (C and D) Cells co-expressing HA-Atg11 and Bmh1-3✕FLAG (C) or Bmh2-3✕FLAG (D) were treated with glucose starvation for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (E and F) Wild-type (WT), atg1∆, atg9∆, atg17∆, atg19∆, or atg20∆ cells co-expressing HA-Atg11 with Bmh1-3✕FLAG (E) or Bmh2-3✕FLAG (F) were subjected to glucose starvation for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (G and I) Cells co-expressing HA-Atg11 with Bmh1-3✕FLAG (G) or Bmh2-3✕FLAG (I) were cultured in nutrient-rich medium, nitrogen-starvation medium (SD-N), glucose-starvation medium (SD-G), or treated with rapamycin (0.2 μg/ml) for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and analyzed by Western blot using anti-HA antibody. (H and J) The results from G and I were quantified. Data are presented as means ± SD (n = 3). ***P < 0.001; **P < 0.01; two-tailed Student’s t tests were used. Source data are available for this figure: SourceData F1.
We next conducted Co-IP assays to determine whether autophagic stimuli regulate the Atg11 association with Bmh1/2. The binding between Atg11 and Bmh1 or Bmh2 was seen to be significantly increased following treatment with rapamycin, nitrogen starvation, or glucose starvation (Fig. 1, G–J). Taken together, these results supported the likelihood that Bmh1/2 are two previously unrecognized binding partners of Atg11, with their interactions influenced by the nutritional status of cells.
Bmh1/2 are required for glucose starvation–induced autophagy by controlling the PAS recruitment and activation of Atg1
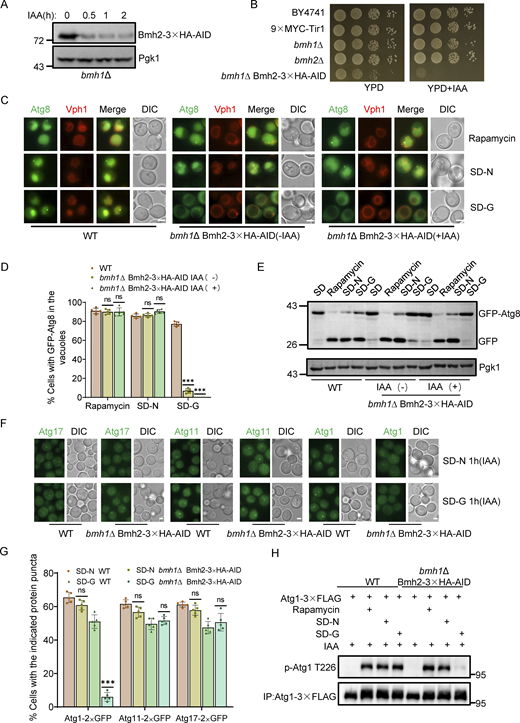
Given the role of Atg11 in glucose starvation–induced autophagy and the Cvt pathway (Yao et al., 2020; Yorimitsu and Klionsky, 2005), we investigated whether Bmh1/2 also participate in these processes. Deleting BMH1 or BMH2 individually had no effect on the generation of free GFP or PrApe1 maturation (Fig. S1, A and B). Since Bmh2 is a paralog of Bmh1, we hypothesized functional redundancy. As BMH1/2 double knockout is lethal (Van Hemert et al., 2001), we generated a BMH2 conditional knockdown line using the auxin-inducible degron (AID) system in a BMH1 deletion strain (Nishimura et al., 2009). Bmh2 was rapidly degraded upon indole-3-acetic acid (IAA) treatment in bmh1∆ cells expressing Bmh2-3✕HA-AID (Fig. 2 A). Spotting assays indicated growth was completely inhibited following the addition of IAA to cultures of bmh1∆ cells expressing Bmh2-3✕HA-AID. Moreover, the growth of this strain was impaired even without IAA treatment, which indicated that 3✕HA-AID tag might negatively affect the normal function of Bmh2 in bmh1∆ cells (Fig. 2 B). Further analysis showed that the growth of bmh1∆ cells expressing Bmh2-3✕HA-AID was compromised on YPD plates with Hydroxyurea (HU) or Methyl methanesulfonate (MMS) (Fig. S1 C). We found that depleting Bmh2 by IAA treatment completely blocked glucose starvation–induced autophagy, but did not affect autophagy induced by rapamycin treatment or nitrogen starvation in the bmh1∆ yeast strain (Fig. 2, C–E). Concurrently, Bmh2-3✕HA-AID depletion by IAA treatment did not affect the PrApe1 maturation process in bmh1∆ cells (Fig. S1 D). These collective results indicated that both Bmh1/2 are specifically required for glucose starvation–induced autophagy.
The deletion of BMH1 or BMH2 had no effect on autophagy or the Cvt pathway, confirming the specific binding of Bmh1/2 with Atg11. (A) Cells co-expressing GFP-Atg8 and Vph1-mCherry in WT, bmh1∆, or bmh2∆ yeast strains were cultured in SD-N, SD-G, or placed under rapamycin treatment, for the indicated time periods. The samples were analyzed by Western blot for the cleavage of GFP-Atg8. Pgk1 served as a loading control. The data are representative of three independent experiments. (B) WT, atg11∆, bmh1∆, or bmh2∆ yeast strains were cultured in a nutrient-rich medium. The samples were analyzed by Western blot for the maturation of PrApe1. Pgk1 served as a loading control. The data are representative of three independent experiments. (C) Growth phenotypes of bmh1∆ Bmh2-3×HA-AID. Serial dilutions of wild-type (BY4741), 9✕MYC-Tir1, bmh1∆, bmh2∆, bmh1∆ Bmh2-3×HA-AID yeast strains were plated in fourfold serial dilution onto YPD plates with or without HU and MMS at the indicated concentrations. The plates were incubated at 30°C or 37°C for 2 days, and spotting assays were performed to assess the growth phenotypes. (D) WT, atg11∆, or bmh1∆ Bmh2-3✕HA-AID yeast strains were grown to the early log-growth phase and then treated with IAA or DMSO for 2 h. The samples were analyzed by Western blot for the maturation of PrApe1. Pgk1 served as a loading control. The data are representative of three independent experiments. (E) Cells expressing3×FLAG-Snf1 in WT or bmh1∆ Bmh2-3✕HA-AID yeast strains were treated with DMSO or IAA for 2 h and then cultured in glucose starvation medium in the absence or presence of IAA for 0.5 h. The activity of Snf1 was detected by immunoblotting with anti-phospho-AMPKα (Thr172) antibody. Pgk1 served as a loading control. The data are representative of three independent experiments. (F) WT, cox6∆ (positive control), bmh1∆, bmh2∆, or bmh1∆ Bmh2-3✕HA-AID yeast strains were treated with DMSO or IAA for 2 h, and then cultured in nutrient-rich medium or glucose starvation medium in the absence or presence of IAA for 0.5 h. Cells were harvested and oxygen consumption was measured using Oroboros O2K. n = 3 independent experiments were quantified. Data are presented as means ± SD. ***P < 0.001; NS, not significant; two-tailed Student’s t tests were used. (G)atg1∆ cells expressing empty vector, Atg1-3✕FLAG, or Atg1-3✕FLAG T226A were grown to the early log-growth phase and then treated with rapamycin for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and analyzed by Western blot using anti-p-T226-Atg1 antibody. The data are representative of three independent experiments. (H) Cells expressing Atg1-3✕FLAG in WT, atg11∆, atg13∆, or atg17∆ yeast strains were grown to the early log-growth phase, and treated with rapamycin or glucose starvation for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and analyzed by Western blot using anti-p-T226-Atg1 antibody. The data are representative of three independent experiments. (I–N) Cells co-expressing Bmh1-3✕FLAG or Bmh2-3✕FLAG with HA-Atg1 (I and J), Atg17-6✕HA (K and L), or Atg13-6✕HA (M and N) were cultured in nutrient-rich medium or SD-G for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (O) Cells co-expressing Atg29-2✕GFP or Atg31-2✕GFP with Bmh1-3✕FLAG or Bmh2-3✕FLAG were cultured in SD-G for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and analyzed by Western blot using anti-GFP antibody. The data are representative of three independent experiments. Source data are available for this figure: SourceData FS1.
The deletion of BMH1 or BMH2 had no effect on autophagy or the Cvt pathway, confirming the specific binding of Bmh1/2 with Atg11. (A) Cells co-expressing GFP-Atg8 and Vph1-mCherry in WT, bmh1∆, or bmh2∆ yeast strains were cultured in SD-N, SD-G, or placed under rapamycin treatment, for the indicated time periods. The samples were analyzed by Western blot for the cleavage of GFP-Atg8. Pgk1 served as a loading control. The data are representative of three independent experiments. (B) WT, atg11∆, bmh1∆, or bmh2∆ yeast strains were cultured in a nutrient-rich medium. The samples were analyzed by Western blot for the maturation of PrApe1. Pgk1 served as a loading control. The data are representative of three independent experiments. (C) Growth phenotypes of bmh1∆ Bmh2-3×HA-AID. Serial dilutions of wild-type (BY4741), 9✕MYC-Tir1, bmh1∆, bmh2∆, bmh1∆ Bmh2-3×HA-AID yeast strains were plated in fourfold serial dilution onto YPD plates with or without HU and MMS at the indicated concentrations. The plates were incubated at 30°C or 37°C for 2 days, and spotting assays were performed to assess the growth phenotypes. (D) WT, atg11∆, or bmh1∆ Bmh2-3✕HA-AID yeast strains were grown to the early log-growth phase and then treated with IAA or DMSO for 2 h. The samples were analyzed by Western blot for the maturation of PrApe1. Pgk1 served as a loading control. The data are representative of three independent experiments. (E) Cells expressing3×FLAG-Snf1 in WT or bmh1∆ Bmh2-3✕HA-AID yeast strains were treated with DMSO or IAA for 2 h and then cultured in glucose starvation medium in the absence or presence of IAA for 0.5 h. The activity of Snf1 was detected by immunoblotting with anti-phospho-AMPKα (Thr172) antibody. Pgk1 served as a loading control. The data are representative of three independent experiments. (F) WT, cox6∆ (positive control), bmh1∆, bmh2∆, or bmh1∆ Bmh2-3✕HA-AID yeast strains were treated with DMSO or IAA for 2 h, and then cultured in nutrient-rich medium or glucose starvation medium in the absence or presence of IAA for 0.5 h. Cells were harvested and oxygen consumption was measured using Oroboros O2K. n = 3 independent experiments were quantified. Data are presented as means ± SD. ***P < 0.001; NS, not significant; two-tailed Student’s t tests were used. (G)atg1∆ cells expressing empty vector, Atg1-3✕FLAG, or Atg1-3✕FLAG T226A were grown to the early log-growth phase and then treated with rapamycin for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and analyzed by Western blot using anti-p-T226-Atg1 antibody. The data are representative of three independent experiments. (H) Cells expressing Atg1-3✕FLAG in WT, atg11∆, atg13∆, or atg17∆ yeast strains were grown to the early log-growth phase, and treated with rapamycin or glucose starvation for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and analyzed by Western blot using anti-p-T226-Atg1 antibody. The data are representative of three independent experiments. (I–N) Cells co-expressing Bmh1-3✕FLAG or Bmh2-3✕FLAG with HA-Atg1 (I and J), Atg17-6✕HA (K and L), or Atg13-6✕HA (M and N) were cultured in nutrient-rich medium or SD-G for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (O) Cells co-expressing Atg29-2✕GFP or Atg31-2✕GFP with Bmh1-3✕FLAG or Bmh2-3✕FLAG were cultured in SD-G for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and analyzed by Western blot using anti-GFP antibody. The data are representative of three independent experiments. Source data are available for this figure: SourceData FS1.
Bmh1/2 regulate glucose starvation–induced autophagy by governing the PAS recruitment and activation of Atg1. (A)bmh1∆ cells expressing Bmh2-3✕HA-AID were treated with 0.5 mM IAA for the indicated periods and the levels of Bmh2 protein were detected by using Anti-HA antibody. Pgk1 served as a loading control. The data are representative of three independent experiments. (B) The indicated yeast strains, either untreated or treated with IAA for 3 days, were plated in fourfold serial dilution onto YPD at 30°C. (C) Cells co-expressing GFP-Atg8 and Vph1-Cherry in wild-type or bmh1∆ Bmh2-3✕HA-AID yeast strains were treated with DMSO or IAA for 2 h and then cultured in SD-N, SD-G, or placed under rapamycin treatment in the absence or presence of IAA for the indicated periods. Images of cells were obtained using an inverted fluorescence microscope. Scale bar, 2 µm. (D) Cells from C were quantified for the vacuolar localization of GFP-Atg8. n = 300 cells were pooled from three independent experiments. Data are presented as means ± SD. ***P < 0.001; ns, no significance; two-tailed Student’s t tests were used. (E) Cells from C were analyzed by Western blot for the cleavage of GFP-Atg8. Pgk1 served as a loading control. The data are representative of three independent experiments. (F) Cells expressing Atg11, Atg17, or Atg1 fused with GFP tag in wild-type or bmh1∆ Bmh2-3✕HA-AID yeast strains were treated with IAA for 2 h and then cultured in nitrogen starvation medium (SD-N) or glucose starvation medium (SD-G) in the presence of IAA for 1 h. Images of cells were obtained using an inverted fluorescence microscope. Scale bar, 2 µm. (G) Cells from F were quantified for the number of cells with the indicated ATG protein puncta. n = 300 cells were pooled from three independent experiments. Data are presented as means ± SD. ***P < 0.001; NS, not significant; two-tailed Student’s t tests were used. (H) Cells expressing Atg1-3✕FLAG in WT or bmh1∆ Bmh2-3✕HA-AID yeast strains were treated with IAA for 2 h and then cultured in SD-N, SD-G, or rapamycin treatment medium in the presence of IAA for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and analyzed by Western blot using anti-p-T226-Atg1 antibody. The data are representative of three independent experiments. Source data are available for this figure: SourceData F2.
Bmh1/2 regulate glucose starvation–induced autophagy by governing the PAS recruitment and activation of Atg1. (A)bmh1∆ cells expressing Bmh2-3✕HA-AID were treated with 0.5 mM IAA for the indicated periods and the levels of Bmh2 protein were detected by using Anti-HA antibody. Pgk1 served as a loading control. The data are representative of three independent experiments. (B) The indicated yeast strains, either untreated or treated with IAA for 3 days, were plated in fourfold serial dilution onto YPD at 30°C. (C) Cells co-expressing GFP-Atg8 and Vph1-Cherry in wild-type or bmh1∆ Bmh2-3✕HA-AID yeast strains were treated with DMSO or IAA for 2 h and then cultured in SD-N, SD-G, or placed under rapamycin treatment in the absence or presence of IAA for the indicated periods. Images of cells were obtained using an inverted fluorescence microscope. Scale bar, 2 µm. (D) Cells from C were quantified for the vacuolar localization of GFP-Atg8. n = 300 cells were pooled from three independent experiments. Data are presented as means ± SD. ***P < 0.001; ns, no significance; two-tailed Student’s t tests were used. (E) Cells from C were analyzed by Western blot for the cleavage of GFP-Atg8. Pgk1 served as a loading control. The data are representative of three independent experiments. (F) Cells expressing Atg11, Atg17, or Atg1 fused with GFP tag in wild-type or bmh1∆ Bmh2-3✕HA-AID yeast strains were treated with IAA for 2 h and then cultured in nitrogen starvation medium (SD-N) or glucose starvation medium (SD-G) in the presence of IAA for 1 h. Images of cells were obtained using an inverted fluorescence microscope. Scale bar, 2 µm. (G) Cells from F were quantified for the number of cells with the indicated ATG protein puncta. n = 300 cells were pooled from three independent experiments. Data are presented as means ± SD. ***P < 0.001; NS, not significant; two-tailed Student’s t tests were used. (H) Cells expressing Atg1-3✕FLAG in WT or bmh1∆ Bmh2-3✕HA-AID yeast strains were treated with IAA for 2 h and then cultured in SD-N, SD-G, or rapamycin treatment medium in the presence of IAA for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and analyzed by Western blot using anti-p-T226-Atg1 antibody. The data are representative of three independent experiments. Source data are available for this figure: SourceData F2.
We next focused on the molecular mechanisms of the Bmh1/2 function in glucose starvation–induced autophagy. Snf1, a yeast homolog of AMPK, is activated by glucose starvation, leading to Atg1 activation by phosphorylation (Yao et al., 2020; Mitchelhill et al., 1994). Using an anti-phos-Thr172-AMPK antibody (Orlova et al., 2008), we found that Snf1 phosphorylation increased under glucose starvation, regardless of Bmh2-3✕HA-AID depletion in bmh1∆ cells (Fig. S1 E). Since mitochondrial respiration is essential for glucose starvation–induced autophagy (Yi et al., 2017), we also examined the mitochondrial respiration rates in wild-type (WT), bmh1∆, bmh2∆, or Bmh2-3✕HA-AID bmh1∆ yeast cells and found that aerobic respiration levels in the mutant yeast strains, with or without IAA treatment, did not show any significant variations from those of the WT under glucose starvation conditions (Fig. S1 F). These data indicate that Bmh1/2 are involved in neither Snf1 activation nor in the maintenance of aerobic respiration under glucose starvation.
Atg proteins are hierarchically recruited to the PAS during autophagy (Suzuki and Ohsumi, 2010). To determine which step in the process of recruiting Atg proteins requires Bmh1/2, GFP-tagged Atg1/11/17 proteins were individually expressed in Bmh2-3✕HA-AID bmh1∆ yeast cells subjected to glucose starvation or nitrogen starvation. Fluorescence image analysis showed that the PAS marker, Atg17-GFP, and Atg11-GFP formed obvious punctate in Bmh2-3✕HA-AID bmh1∆ yeast cells treated with IAA treatment under glucose starvation or nitrogen starvation conditions, indicating that Bmh1/2 are not involved in PAS formation or the PAS recruitment of Atg11 under glucose starvation. In contrast, Atg1-GFP signals appeared well dispersed throughout the cytoplasm in IAA-treated Bmh2-3✕HA-AID bmh1∆ yeast cells under glucose starvation but formed puncta under nitrogen starvation (Fig. 2, F and G), indicating that Bmh1/2 are required specifically for Atg1 recruitment to the PAS during glucose starvation–induced autophagy.
Considering Atg1 recruitment to the PAS is closely linked to its activation (1, 18), we tested whether Bmh1/2 were required for Atg1 activation under glucose starvation. Since the phosphorylation level of Atg1 at site T226 can serve as a marker for the activation status of Atg1 (Yeh et al., 2010), we generated an antibody specifically targeting phosphorylation modifications at Atg1 T226. Western blot analysis indicated that the phosphorylation levels at Atg1 T226 were significantly higher in WT cells under rapamycin treatment compared with nutrient-rich medium (Fig. S1 G). Using this antibody, we found that the phosphorylation of Atg1 T226 was almost completely abolished in IAA-treated Bmh2-3✕HA-AID bmh1∆ cells under glucose starvation but increased under rapamycin treatment (Fig. 1 H). This indicated that Bmh1/2 were specifically involved in Atg1 activation during glucose starvation.
Additionally, we found that both Atg13 and Atg17 were required for Atg1 activation under rapamycin treatment or glucose starvation conditions (Fig. S1 H). To exclude the possibility that Bmh1/2 could bind to other subunits of the Atg1 complex to affect the PAS recruitment and the activation of Atg1, we performed Co-IP assays testing whether Bam1/2 could interact with Atg1, Atg13, Atg17, Atg29, or Atg31. This did not reveal any associations between Bmh1/2 and any of these proteins (Fig. S1, I–O), thus supporting the specificity of Bmh1/2-Atg11 binding. These cumulative results indicate that Bmh1/2 are specifically required for both the recruitment of Atg1 to the PAS and its activation during glucose starvation.
Rck2 is required for the binding of Atg11-Bmh1/2 and glucose starvation–induced autophagy
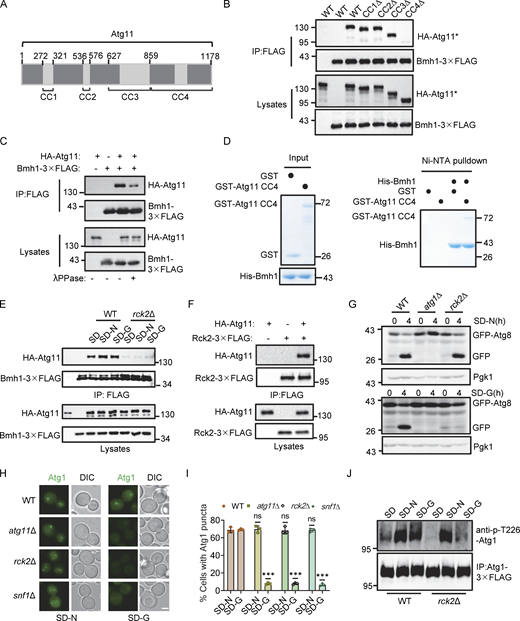
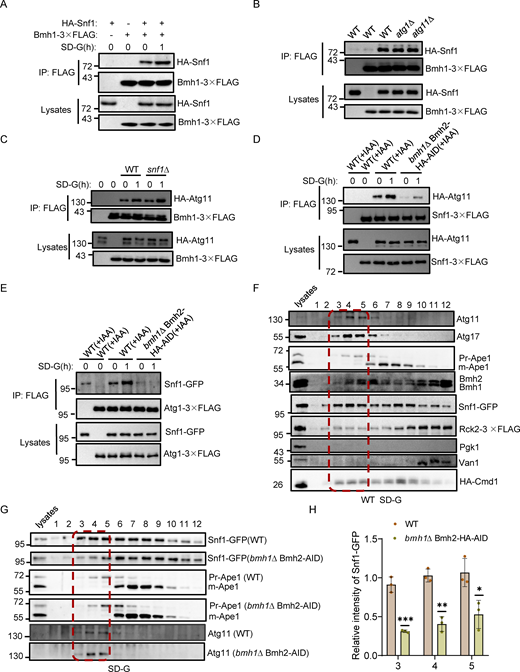
Our previous work demonstrated that ATG11 deletion specifically blocks the PAS recruitment and activation of Atg1 under glucose starvation (Yao et al., 2020). Given our above findings that Bmh1/2 interact with Atg11 and are required for the PAS recruitment and activation of Atg1 during glucose starvation–induced autophagy, we examined the mechanism underlying the Bmh1/2-Atg11 interaction. Atg11 comprises four domains: CC1, CC2, CC3, and CC4 (Fig. 3 A). The CC4 domain is known to mediate the Atg11 interaction with selective autophagy receptors (Yorimitsu and Klionsky, 2005). We constructed deletion mutants lacking each of these domains and expressed them in an atg11∆ yeast strain expressing Bmh1-3✕FLAG or Bmh2-3✕FLAG. Co-IP rescue experiments showed that Bmh1/2 failed to interact with the Atg11 variant lacking the CC4 domain (Fig. 3 B and Fig. S2 A), indicating that the CC4 domain was necessary for the association of Atg11 with Bmh1/2.
Rck2 is required for the binding of Atg11-Bmh1/2 and glucose starvation–induced autophagy. (A) Schematic representation of the Atg11 domains and the deletions. CC1∆, CC2∆, CC3∆, and CC4∆ correspond to the deletion of amino acids at positions 272–321, 536–576, 627–858, and 859–1,178, respectively. (B) Cells co-expressing an empty vector, WT HA-Atg11, HA-Atg11-CC1∆, HA-Atg11-CC2∆, HA-Atg11-CC3∆, or HA-Atg11-CC4∆ with Bmh1-3✕FLAG in the atg11∆ strain were subjected to glucose starvation for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (C) Cells co-expressing WT HA-Atg11 and Bmh1-3✕FLAG in the atg11∆ strains were cultured in SD-G for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads. Bmh1-associated proteins were then treated with or without lambda protein phosphatase (λPPase) and analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (D) In vitro Ni-NTA pulldowns were performed using His-Bmh1with GST-Atg11 CC4 purified from E. coli. Protein samples were separated by SDS-PAGE and detected using Coomassie blue staining. The data are representative of three independent experiments. (E) Cells co-expressing HA-Atg11 and Bmh1-3✕FLAG in the WT or rck2∆ yeast strain were cultured in full medium, SD-N, or SD-G for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (F) Cells co-expressing HA-Atg11 and Rck2-3✕FLAG were subjected to glucose starvation for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (G) Cells expressing GFP-Atg8 and Vph1-mCherry in WT, atg1∆, or rck2∆ yeast strains were cultured in SD-N or SD-G for 4 h. The samples were analyzed by Western blot for the cleavage of GFP-Atg8. Pgk1 served as a loading control. The data are representative of three independent experiments. (H) Cells expressing Atg1-GFP in WT, atg11∆, rck2∆, or snf1∆ yeast strains were cultured in SD-N or SD-G for 1 h. Images of cells were obtained using an inverted fluorescence microscope. Scale bar, 2 µm. (I) Cells from (H) were quantified for the number of cells with Atg1-GFP puncta. n = 300 cells were pooled from three independent experiments. Data are presented as means ± SD. ***P < 0.001; NS, not significant; two-tailed Student’s t tests were used. (J) Cells expressing Atg1-3✕FLAG in WT or rck2∆ yeast strains were were grown to the Log growth phase and then cultured in glucose or nitrogen starvation medium for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and analyzed by Western blot using anti-p-T226-Atg1 antibody. The data are representative of three independent experiments. Source data are available for this figure: SourceData F3.
Rck2 is required for the binding of Atg11-Bmh1/2 and glucose starvation–induced autophagy. (A) Schematic representation of the Atg11 domains and the deletions. CC1∆, CC2∆, CC3∆, and CC4∆ correspond to the deletion of amino acids at positions 272–321, 536–576, 627–858, and 859–1,178, respectively. (B) Cells co-expressing an empty vector, WT HA-Atg11, HA-Atg11-CC1∆, HA-Atg11-CC2∆, HA-Atg11-CC3∆, or HA-Atg11-CC4∆ with Bmh1-3✕FLAG in the atg11∆ strain were subjected to glucose starvation for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (C) Cells co-expressing WT HA-Atg11 and Bmh1-3✕FLAG in the atg11∆ strains were cultured in SD-G for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads. Bmh1-associated proteins were then treated with or without lambda protein phosphatase (λPPase) and analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (D) In vitro Ni-NTA pulldowns were performed using His-Bmh1with GST-Atg11 CC4 purified from E. coli. Protein samples were separated by SDS-PAGE and detected using Coomassie blue staining. The data are representative of three independent experiments. (E) Cells co-expressing HA-Atg11 and Bmh1-3✕FLAG in the WT or rck2∆ yeast strain were cultured in full medium, SD-N, or SD-G for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (F) Cells co-expressing HA-Atg11 and Rck2-3✕FLAG were subjected to glucose starvation for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (G) Cells expressing GFP-Atg8 and Vph1-mCherry in WT, atg1∆, or rck2∆ yeast strains were cultured in SD-N or SD-G for 4 h. The samples were analyzed by Western blot for the cleavage of GFP-Atg8. Pgk1 served as a loading control. The data are representative of three independent experiments. (H) Cells expressing Atg1-GFP in WT, atg11∆, rck2∆, or snf1∆ yeast strains were cultured in SD-N or SD-G for 1 h. Images of cells were obtained using an inverted fluorescence microscope. Scale bar, 2 µm. (I) Cells from (H) were quantified for the number of cells with Atg1-GFP puncta. n = 300 cells were pooled from three independent experiments. Data are presented as means ± SD. ***P < 0.001; NS, not significant; two-tailed Student’s t tests were used. (J) Cells expressing Atg1-3✕FLAG in WT or rck2∆ yeast strains were were grown to the Log growth phase and then cultured in glucose or nitrogen starvation medium for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and analyzed by Western blot using anti-p-T226-Atg1 antibody. The data are representative of three independent experiments. Source data are available for this figure: SourceData F3.
The association of Atg11 with Bmh2 depends on its CC4 domain, and Rck2 is required specifically for glucose starvation–induced autophagy. (A) Cells co-expressing an empty vector, WT HA-Atg11, HA-Atg11 CC1∆, HA-Atg11 CC2∆, HA-Atg11 CC3∆, or HA-Atg11 CC4∆ with Bmh2-3✕FLAG in the atg11∆ strains were subjected to glucose starvation for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (B) Cells co-expressing WT HA-Atg11 and Bmh2-3✕FLAG in the atg11∆ strains were cultured in SD-G for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads. Bmh2-associated proteins were then treated with or without lambda protein phosphatase (λPPase) and analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (C) In vitro Ni-NTA pulldowns were performed using His-Bmh2 with GST-Atg11 CC4 purified from E. coli. Protein samples were separated by SDS-PAGE and then detected using Coomassie blue staining. The data are representative of three independent experiments. (D) Atg11-associated protein kinases were found in the SGD database. (E) Cells co-expressing HA-Atg11 and Bmh2-3✕FLAG in the WT or rck2∆ yeast strains were cultured in nutrient-rich medium, SD-N, or SD-G medium for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (F and G) Cells coexpressing GFP-Atg8 and Vph1-mCherry in WT, atg1∆, or rck2∆ yeast strains were cultured in SD-N or SD-G for 4 h. Images of cells were obtained using an inverted fluorescence microscope. Scale bar, 2 µm. (H) Cells from F and G were quantified for the vacuolar localization of GFP-Atg8. n = 300 cells were pooled from three independent experiments. Data are presented as means ± SD. ***P < 0.001; ns, no significance; two-tailed Student’s t tests were used. (I) Cells expressing HA-Snf1 in WT or rck2∆ yeast strains were grown to the early log-growth phase and then were subjected to glucose starvation for 0.5 h. The activity of Snf1 was detected by immunoblotting with anti-phospho-AMPKα (Thr172) antibody. The data are representative of three independent experiments. (J) WT, atg1∆, or rck2∆ yeast strains were grown to the log-growth phase. The samples were analyzed by Western blot for the maturation of PrApe1. Pgk1 served as a loading control. The data are representative of three independent experiments. Source data are available for this figure: SourceData FS2.
The association of Atg11 with Bmh2 depends on its CC4 domain, and Rck2 is required specifically for glucose starvation–induced autophagy. (A) Cells co-expressing an empty vector, WT HA-Atg11, HA-Atg11 CC1∆, HA-Atg11 CC2∆, HA-Atg11 CC3∆, or HA-Atg11 CC4∆ with Bmh2-3✕FLAG in the atg11∆ strains were subjected to glucose starvation for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (B) Cells co-expressing WT HA-Atg11 and Bmh2-3✕FLAG in the atg11∆ strains were cultured in SD-G for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads. Bmh2-associated proteins were then treated with or without lambda protein phosphatase (λPPase) and analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (C) In vitro Ni-NTA pulldowns were performed using His-Bmh2 with GST-Atg11 CC4 purified from E. coli. Protein samples were separated by SDS-PAGE and then detected using Coomassie blue staining. The data are representative of three independent experiments. (D) Atg11-associated protein kinases were found in the SGD database. (E) Cells co-expressing HA-Atg11 and Bmh2-3✕FLAG in the WT or rck2∆ yeast strains were cultured in nutrient-rich medium, SD-N, or SD-G medium for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (F and G) Cells coexpressing GFP-Atg8 and Vph1-mCherry in WT, atg1∆, or rck2∆ yeast strains were cultured in SD-N or SD-G for 4 h. Images of cells were obtained using an inverted fluorescence microscope. Scale bar, 2 µm. (H) Cells from F and G were quantified for the vacuolar localization of GFP-Atg8. n = 300 cells were pooled from three independent experiments. Data are presented as means ± SD. ***P < 0.001; ns, no significance; two-tailed Student’s t tests were used. (I) Cells expressing HA-Snf1 in WT or rck2∆ yeast strains were grown to the early log-growth phase and then were subjected to glucose starvation for 0.5 h. The activity of Snf1 was detected by immunoblotting with anti-phospho-AMPKα (Thr172) antibody. The data are representative of three independent experiments. (J) WT, atg1∆, or rck2∆ yeast strains were grown to the log-growth phase. The samples were analyzed by Western blot for the maturation of PrApe1. Pgk1 served as a loading control. The data are representative of three independent experiments. Source data are available for this figure: SourceData FS2.
Previous studies have shown that Bmh1/2 binding to many partners is induced by phosphorylation (Trembley et al., 2014). To test whether the Bmh1/2-Atg11 interaction depends on Atg11 phosphorylation, we coexpressed HA-Atg11 with Bmh1-3✕FLAG or Bmh2-3✕FLAG in yeast cells. After Co-IP with anti-FLAG agarose beads and treatment with or without λPPase, we found that Atg11’s binding to Bmh1/2-3✕FLAG decreased after λPPase treatment (Fig. 3 C and Fig. S2 B). Additionally, Ni-NTA pulldown assays showed that purified His-Bmh1/2 had negligible direct interactions with the non-phosphorylated Atg11 CC4 domain in vitro (Fig. 3 D and Fig. S2 C), suggesting that Bmh1/2-Atg11 binding requires both the CC4 domain and Atg11 phosphorylation.
Next, we sought to identify the kinase responsible for Atg11 phosphorylation to induce its association with Bmh1/2. Searching the SGD database for known Atg11 partners, we found seven protein kinases (Fig. S2 D). Individual knockout of each kinase in yeast cells co-expressing Bmh1/2-3✕FLAG and HA-Atg11 revealed that a MAPK-activated protein kinase, Rck2 (Swaminathan et al., 2006), was required for Bmh1/2’s association with Atg11 under nutrient-rich, nitrogen starvation, or glucose starvation conditions (Fig. 3 E and Fig. S2 E). Co-IP assays confirmed Rck2 interaction with Atg11 (Fig. 3 F).
We then tested whether Rck2 participated in glucose starvation–induced autophagy. In rck2∆ cells, the vacuolar localization of GFP-Atg8 was not significantly decreased under nitrogen starvation but was nearly absent under glucose starvation (Fig. S2, F–H). Similarly, free GFP production was almost completely blocked in rck2∆ cells under glucose starvation but not under nitrogen starvation (Fig. 3 G). Given that Bmh1/2 are required for both Atg1 recruitment to the PAS and its activation, and that Rck2 is crucial for the interaction between Bmh1/2 and Atg11, we tested if Rck2 was also necessary for the PAS recruitment and activation of Atg1. Fluorescence microscopy with quantitative image analysis indicated that the numbers of Atg1 puncta were significantly lower in rck2∆ cells than WT cells under glucose starvation conditions, with no difference under nitrogen starvation (Fig. 3, H and I). These results were consistent with the phenotypes of atg11∆ and snf1∆ cells (Fig. 3, H and I). Further examination revealed that Atg1 activation was blocked in rck2∆ cells under glucose starvation but remained unaffected under nitrogen starvation (Fig. 3 J). Snf1 activation and PrApe1 maturation remained unimpaired by RCK2 deletion (Fig. S2, I and J). Cumulatively, these data showed that Rck2 is required for glucose starvation–induced autophagy via regulation of both Atg1 recruitment to the PAS and its activation.
Rck2-mediated Atg11 phosphorylation is required for Atg11-Bmh1/2 interactions and glucose starvation–induced autophagy
Considering the glucose starvation-specific role of Rck2, particularly in the phosphorylation-dependent binding of Bmh1/2 with Atg11, we next confirmed whether Rck2 itself could phosphorylate Atg11. To this end, in vitro kinase assays, using either WT or KD (kinase dead) Rck2-3✕FLAG (purified from glucose-starved yeast) as the kinase and with the Atg11 CC4 domain (purified from E. coli) as a substrate, revealed that the CC4 domain could be phosphorylated by WT, but not KD Rck2 (Fig. 4 A), demonstrating that the Atg11 CC4 domain serves as a direct phosphorylation substrate of Rck2.
Rck2-mediated Atg11 phosphorylation is required for glucose starvation–induced autophagy by regulating Bmh1/2-Atg11 binding. (A) In vitro kinase assays were performed using the Atg11 CC4 domain purified from E. coli as substrates and WT or KD Rck2-3✕FLAG purified from glucose-starved yeast cells as a protein kinase. Phosphorylation of the Atg11 CC4 domain was detected using anti-thioP antibody. The data are representative of three independent experiments. (B) In vitro kinase assays were performed using Atg11 CC4 or Atg11 CC4 2A(T1114A-S1119A) purified from E. coli as substrates with Rck2-3✕FLAG purified from glucose-starved yeast cells as a protein kinase. Phosphorylation of Atg11 CC4 were detected using anti-thioP antibody. The data are representative of three independent experiments. (C)atg11∆ cells co-expressing Bmh1-3✕FLAG with HA-Atg11 WT or 2A were cultured in SD-G for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (D) In vitro GST pulldowns were performed using GST-Bmh1 purified from E. coli with glucose-starved yeast lysates expressing HA-Atg11 WT or 2A. Protein samples were separated by SDS-PAGE and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (E) In vitro phosphorylation assays were performed using GST-Atg11 CC4 WT or 2A purified from E. coli as substrates, with WT or KD Rck2-3✕FLAG purified from glucose-starved yeast cells as a protein kinase. After that, in vitro Ni-NTA pulldowns were performed using His-Bmh1 protein purified from E. coli with the samples of in vitro phosphorylation reaction. Protein samples were separated by SDS-PAGE and then detected using Coomassie blue staining. The data are representative of three independent experiments. (F and G) Cells expressing GFP-Atg8 and Vph1-Cherry in atg11∆, HA-Atg11 WT, HA-Atg11-CC4∆, or HA-Atg11 2A strains were cultured in SD-N(F) or SD-G(G) for 4 h. Autophagic activity was analyzed by Western blot for GFP-Atg8 cleavage. The data are representative of three independent experiments. (H) Cell expressing Atg1-GFP in atg11∆, HA-Atg11 WT, HA-Atg11-CC4∆, or HA-Atg11 2A strains were cultured in SD-N or SD-G for 1 h. Images of cells were obtained using an inverted fluorescence microscope. Scale bar, 2 µm. (I) Cells from H were quantified for the number of cells with Atg1-GFP puncta. n = 300 cells were pooled from three independent experiments. Data are presented as means ± SD. ***P < 0.001; NS, not significant; two-tailed Student’s t tests were used. (J)atg11∆ cells expressing HA-Atg11 WT or 2A with Atg1-3✕FLAG were cultured in SD-G or SD-N for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and analyzed by Western blot using anti-p-T226-Atg1 antibody. (K) the phosphorylation level of Atg1 from J was quantified. Data are presented as means ± SD (n = 3). ***P < 0.001; NS, not significant; two-tailed Student’s t tests were used. Source data are available for this figure: SourceData F4.
Rck2-mediated Atg11 phosphorylation is required for glucose starvation–induced autophagy by regulating Bmh1/2-Atg11 binding. (A) In vitro kinase assays were performed using the Atg11 CC4 domain purified from E. coli as substrates and WT or KD Rck2-3✕FLAG purified from glucose-starved yeast cells as a protein kinase. Phosphorylation of the Atg11 CC4 domain was detected using anti-thioP antibody. The data are representative of three independent experiments. (B) In vitro kinase assays were performed using Atg11 CC4 or Atg11 CC4 2A(T1114A-S1119A) purified from E. coli as substrates with Rck2-3✕FLAG purified from glucose-starved yeast cells as a protein kinase. Phosphorylation of Atg11 CC4 were detected using anti-thioP antibody. The data are representative of three independent experiments. (C)atg11∆ cells co-expressing Bmh1-3✕FLAG with HA-Atg11 WT or 2A were cultured in SD-G for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (D) In vitro GST pulldowns were performed using GST-Bmh1 purified from E. coli with glucose-starved yeast lysates expressing HA-Atg11 WT or 2A. Protein samples were separated by SDS-PAGE and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (E) In vitro phosphorylation assays were performed using GST-Atg11 CC4 WT or 2A purified from E. coli as substrates, with WT or KD Rck2-3✕FLAG purified from glucose-starved yeast cells as a protein kinase. After that, in vitro Ni-NTA pulldowns were performed using His-Bmh1 protein purified from E. coli with the samples of in vitro phosphorylation reaction. Protein samples were separated by SDS-PAGE and then detected using Coomassie blue staining. The data are representative of three independent experiments. (F and G) Cells expressing GFP-Atg8 and Vph1-Cherry in atg11∆, HA-Atg11 WT, HA-Atg11-CC4∆, or HA-Atg11 2A strains were cultured in SD-N(F) or SD-G(G) for 4 h. Autophagic activity was analyzed by Western blot for GFP-Atg8 cleavage. The data are representative of three independent experiments. (H) Cell expressing Atg1-GFP in atg11∆, HA-Atg11 WT, HA-Atg11-CC4∆, or HA-Atg11 2A strains were cultured in SD-N or SD-G for 1 h. Images of cells were obtained using an inverted fluorescence microscope. Scale bar, 2 µm. (I) Cells from H were quantified for the number of cells with Atg1-GFP puncta. n = 300 cells were pooled from three independent experiments. Data are presented as means ± SD. ***P < 0.001; NS, not significant; two-tailed Student’s t tests were used. (J)atg11∆ cells expressing HA-Atg11 WT or 2A with Atg1-3✕FLAG were cultured in SD-G or SD-N for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and analyzed by Western blot using anti-p-T226-Atg1 antibody. (K) the phosphorylation level of Atg1 from J was quantified. Data are presented as means ± SD (n = 3). ***P < 0.001; NS, not significant; two-tailed Student’s t tests were used. Source data are available for this figure: SourceData F4.
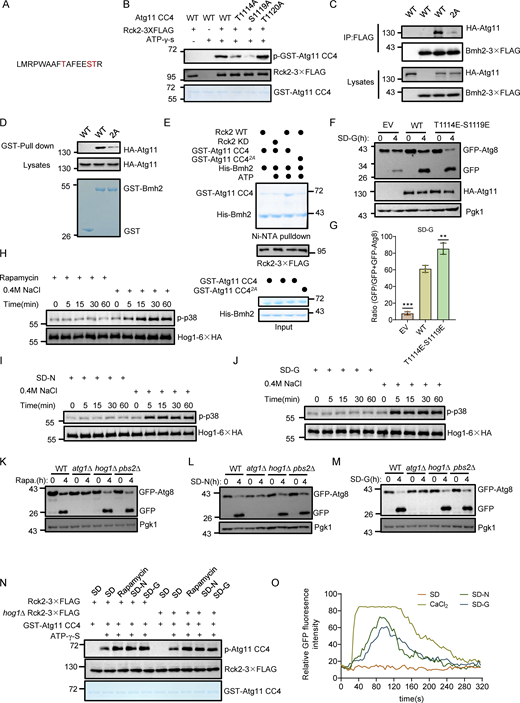
To identify which sites on Atg11 CC4 were phosphorylated by Rck2, we conducted LC-MS analysis of samples from in vitro kinase assays, which revealed a phosphorylated peptide at positions 1106 aa to 1121 aa on Atg11, but the specific phosphorylation site(s) could not be precisely identified (Fig. S3 A). Concurrently, LC-MS identified the same phosphorylated peptide on Atg11-3✕FLAG protein purified from yeast. Narrowing down the potential phosphorylation sites to three serine or threonine residues (T1114, S1119, T1120), we generated three Atg11 CC4 conversion mutants (T1114A, S1119A, or T1120A). Further in vitro kinase assays indicated that the Atg11 CC4 T1114A or S1119A mutants had lower phosphorylation levels than those of Atg11 CC4 T1120A, similar to the Atg11 CC4 results in wild-type controls (Fig. S3 B). We then constructed an Atg11 CC4 T1114A-S1119A(2A) double mutant and found that phosphorylation by Rck2 was completely blocked in the in vitro kinase assays (Fig. 4 B).
Phosphorylation of T1114 and S1119 residues on Atg11 are required for Atg11-Bmh2 binding and the MAPK signaling pathway is not involved in the activation of Rck2 and glucose starvation–induced autophagy. (A) LC-MS/MS identified phosphorylated peptides of Atg11 CC4 by Rck2. (B) In vitro kinase assays were performed using Atg11 CC4 WT, T1114A, S1119A, or T1120A purified from E. coli as substrates with Rck2-3✕FLAG purified from glucose-starved yeast cells as a protein kinase. The phosphorylation levels of Atg11 CC4 and its variants were detected using anti-thioP antibody. The data are representative of three independent experiments. (C)atg11∆ cells co-expressing Bmh2-3✕FLAG with HA-Atg11 WT or 2A were subjected to glucose starvation for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (D) In vitro GST pulldowns were performed using GST- Bmh2 purified from E. coli with glucose-starved yeast lysates expressing HA-Atg11 WT or 2A. Protein samples were separated by SDS-PAGE and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (E) In vitro phosphorylation assays were performed using Atg11 CC4 or 2A purified from E. coli as substrates and with WT or KD Rck2-3✕FLAG purified from glucose-starved yeast cells as a protein kinase. In vitro Ni-NTA pulldowns were then performed using His-Bmh2 purified from E. coli with the samples from the in vitro phosphorylation reaction. Protein samples were separated by SDS-PAGE and then detected using Coomassie blue staining. The data are representative of three independent experiments. (F)atg11∆ yeast cells co-expressing GFP-Atg8 and empty vector, HA-Atg11, or HA-Atg11 T1114E-S1119E were subjected to glucose starvation (SD-G) for 0 or 4 h. The autophagic activity was assessed by Western blotting to detect cleavage of GFP-Atg8, with Pgk1 serving as a loading control. (G) Quantification of the ratio of free GFP/GFP+GFP-Atg8 from F and presented as mean ± SD (n = 3). ***P < 0.001; **P < 0.01; two-tailed Student’s t tests were used. (H–J) Yeast cells expressing Hog1-6✕HA were cultured to early log phase and subsequently subjected to rapamycin treatment (H), nitrogen starvation (SD-N) (I), glucose starvation (SD-G) (J), or 0.4 M NaCl treatment for the indicated time periods. The activation of Hog1 was assessed using anti-phospho-p38 MAPK antibody (#4511; Cell Signaling Technology). The data are representative of three independent experiments. (K–M) Wild-type (WT), atg1∆, hog1∆, or pbs2∆ yeast cells were cultured to early log phase and subsequently subjected to rapamycin treatment (K), nitrogen starvation (SD-N) (L), or glucose starvation (SD-G) (M) for 0 or 4 h. The autophagic activity was detected by Western blots using anti-GFP antibody. Pgk1 served as a loading control. The data are representative of three independent experiments. (N) In vitro kinase assays were performed using GST-Atg11 CC4 purified from E. coli as substrates and Rck2-3✕FLAG purified yeast wild-type or hog1∆ cell cultured under full medium, glucose starvation, nitrogen starvation, or rapamycin-treated conditions as a protein kinase. Phosphorylation levels of GST-Atg11 CC4 were detected using anti-thioP antibody. The data are representative of three independent experiments. (O) Wild-type yeast expressing cytosolic-anchored Ca2+ fluorescence probe jGCaMP7f plasmid were grown to the early log-growth phase, and then subjected to nitrogen starvation (SD-N), glucose starvation (SD-G), or treated with 0.2 M CaCl2. Cells were observed using a fluorescence inverted microscope (Olympus IX83). Images were captured at 4-s intervals using time-lapse microscopy and shown in 24 fps movies. ImageJ software was used to calculate the relative fluorescence intensity of cells to reflect cytoplasmic calcium signaling. The data are representative of three independent experiments. Source data are available for this figure: SourceData FS3.
Phosphorylation of T1114 and S1119 residues on Atg11 are required for Atg11-Bmh2 binding and the MAPK signaling pathway is not involved in the activation of Rck2 and glucose starvation–induced autophagy. (A) LC-MS/MS identified phosphorylated peptides of Atg11 CC4 by Rck2. (B) In vitro kinase assays were performed using Atg11 CC4 WT, T1114A, S1119A, or T1120A purified from E. coli as substrates with Rck2-3✕FLAG purified from glucose-starved yeast cells as a protein kinase. The phosphorylation levels of Atg11 CC4 and its variants were detected using anti-thioP antibody. The data are representative of three independent experiments. (C)atg11∆ cells co-expressing Bmh2-3✕FLAG with HA-Atg11 WT or 2A were subjected to glucose starvation for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (D) In vitro GST pulldowns were performed using GST- Bmh2 purified from E. coli with glucose-starved yeast lysates expressing HA-Atg11 WT or 2A. Protein samples were separated by SDS-PAGE and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (E) In vitro phosphorylation assays were performed using Atg11 CC4 or 2A purified from E. coli as substrates and with WT or KD Rck2-3✕FLAG purified from glucose-starved yeast cells as a protein kinase. In vitro Ni-NTA pulldowns were then performed using His-Bmh2 purified from E. coli with the samples from the in vitro phosphorylation reaction. Protein samples were separated by SDS-PAGE and then detected using Coomassie blue staining. The data are representative of three independent experiments. (F)atg11∆ yeast cells co-expressing GFP-Atg8 and empty vector, HA-Atg11, or HA-Atg11 T1114E-S1119E were subjected to glucose starvation (SD-G) for 0 or 4 h. The autophagic activity was assessed by Western blotting to detect cleavage of GFP-Atg8, with Pgk1 serving as a loading control. (G) Quantification of the ratio of free GFP/GFP+GFP-Atg8 from F and presented as mean ± SD (n = 3). ***P < 0.001; **P < 0.01; two-tailed Student’s t tests were used. (H–J) Yeast cells expressing Hog1-6✕HA were cultured to early log phase and subsequently subjected to rapamycin treatment (H), nitrogen starvation (SD-N) (I), glucose starvation (SD-G) (J), or 0.4 M NaCl treatment for the indicated time periods. The activation of Hog1 was assessed using anti-phospho-p38 MAPK antibody (#4511; Cell Signaling Technology). The data are representative of three independent experiments. (K–M) Wild-type (WT), atg1∆, hog1∆, or pbs2∆ yeast cells were cultured to early log phase and subsequently subjected to rapamycin treatment (K), nitrogen starvation (SD-N) (L), or glucose starvation (SD-G) (M) for 0 or 4 h. The autophagic activity was detected by Western blots using anti-GFP antibody. Pgk1 served as a loading control. The data are representative of three independent experiments. (N) In vitro kinase assays were performed using GST-Atg11 CC4 purified from E. coli as substrates and Rck2-3✕FLAG purified yeast wild-type or hog1∆ cell cultured under full medium, glucose starvation, nitrogen starvation, or rapamycin-treated conditions as a protein kinase. Phosphorylation levels of GST-Atg11 CC4 were detected using anti-thioP antibody. The data are representative of three independent experiments. (O) Wild-type yeast expressing cytosolic-anchored Ca2+ fluorescence probe jGCaMP7f plasmid were grown to the early log-growth phase, and then subjected to nitrogen starvation (SD-N), glucose starvation (SD-G), or treated with 0.2 M CaCl2. Cells were observed using a fluorescence inverted microscope (Olympus IX83). Images were captured at 4-s intervals using time-lapse microscopy and shown in 24 fps movies. ImageJ software was used to calculate the relative fluorescence intensity of cells to reflect cytoplasmic calcium signaling. The data are representative of three independent experiments. Source data are available for this figure: SourceData FS3.
We then investigated whether the phosphorylation of Atg11 was responsible for the observed regulatory effects of Rck2 in the association of Bmh1/2 with Atg11. Co-IP assays showed that expressing the HA-Atg11 T1114A-S1119A (2A) mutant in the atg11∆ background resulted in significantly reduced interactions between Atg11 and Bmh1/2 (Fig. 4 C and Fig. S3 C). In vitro pulldown assays with cell lysates also demonstrated that the association of HA-Atg11 2A with GST-Bmh1/2 purified from E. coli was dramatically decreased compared with WT HA-Atg11 (Fig. 4 D and Fig. S3 D). To validate whether the phosphorylation of Atg11 T1114 and S1119 by Rck2 is essential for the direct association of Atg11 with Bmh1/2, we performed Ni-NTA pulldown experiments immediately following in vitro phosphorylation reactions using either purified WT or kinase-dead (KD) Rck2-3✕FLAG from yeast as the kinase and with WT Atg11 CC4 or Atg11 CC4 2A purified from E. coli as the substrates. Results showed that phosphorylation of Atg11 CC4 by WT Rck2 significantly enhanced the binding between Atg11 CC4 and Bmh1/2, while unphosphorylated Atg11 CC4 (Rck2 KD, Atg11 CC4 2A, or the control groups without ATP) displayed negligible binding with Bmh1/2 (Fig. 4 E and Fig. S3 E). These results provided strong evidence that the phosphorylation of Atg11 residues T1114 and S1119 by Rck2 regulates the binding of Bmh1/2 to Atg11.
We next tested whether the phosphorylation of Atg11 by Rck2 was specifically required for glucose starvation–induced autophagy. To examine this possibility, HA-Atg11, Atg11 CC4∆, Atg11 T1114A-S1119A(2A), or empty vector plasmids were separately introduced into atg11∆ yeast cells coexpressing GFP-Atg8 and Vph1-mCherry. We found that the production of free GFP was almost completely blocked in the HA-Atg11 2A mutant under glucose starvation, despite this mutation showing no effect on GFP-Atg8 cleavage under nitrogen starvation when compared with that in WT cells (Fig. 4, F and G). Subsequently, we investigated whether Atg11 T1114E-S1119E (2E), mimicking Rck2-mediated Atg11 phosphorylation, could enhance autophagy upon glucose starvation. The cleavage of the GFP-Atg8 assay demonstrated that Atg11 2E could promote glucose starvation–induced autophagy (Fig. S3, F and G). Collectively, these results indicated that phosphorylation of Atg11 residues T1114 and S1119 were specifically required for glucose starvation–induced autophagy.
Finally, we next explored the role of Atg11 phosphorylation by Rck2 in glucose starvation–induced autophagy, examining the possibility of this phosphorylation event being integral in the PAS recruitment and activation of Atg1 upon glucose starvation. For this purpose, HA-Atg11, HA-Atg11 2A, or empty vector plasmids, were separately introduced into atg11∆ yeast cells expressing Atg1-GFP. Imaging data revealed that the formation of Atg1-GFP puncta was significantly inhibited in cells expressing the HA-Atg11 2A mutant under glucose starvation conditions, whereas the puncta formed normally in HA-Atg11 2A mutant cells under nitrogen starvation conditions (Fig. 4, H and I). Consistent with the results of RCK2 deletion, expression of the phosphorylation-block Atg11 T1114A-S1119A(2A) mutant significantly impaired the activation of Atg1 under glucose starvation, but not under nitrogen starvation (Fig. 4, J and K). In light of these collective results, we concluded that Rck2-mediated Atg11 phosphorylation is required for glucose starvation–induced autophagy by regulating the association of Atg11 with Bmh1/2.
Glucose starvation triggers the elevation of cytoplasmic calcium and activates Rck2
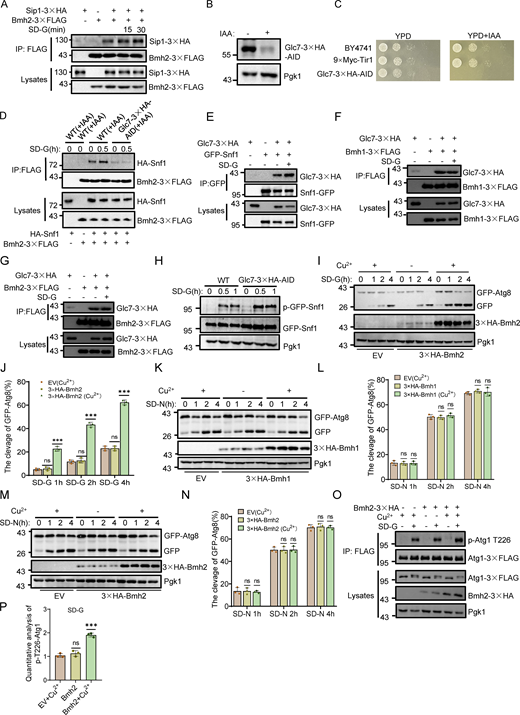
Given that Rck2 and cellular nutritional status regulate Atg11 binding with Bmh1/2, we tested whether Rck2 kinase activity increases during nitrogen or glucose starvation. In vitro kinase assays showed a significant increase in Rck2 kinase activity in response to these autophagic stimuli (Fig. 5 A). As Rck2 is a cytosolic MAPK-activated protein kinase and a direct substrate of Hog1 (Bilsland-Marchesan et al., 2000), we explored whether glucose starvation activates Hog1. Using an anti-phospho-p38 antibody, we observed no Hog1 activation in yeast cells upon rapamycin treatment, nitrogen starvation, or glucose starvation, whereas Hog1 was activated by 0.4 M NaCl (Fig. S3, H–J) (Huang et al., 2020). Subsequently, we examined whether the MAPK signaling pathway is involved in glucose starvation–induced autophagy. Knocking out HOG1 or PBS2, essential for the MAPK signaling pathway (Pelet et al., 2011), did not impair autophagy induced by rapamycin treatment, nitrogen starvation, or glucose starvation (Fig. S3, K–M). We further investigated whether Hog1 regulates Rck2 kinase activity under multiple autophagy-inducing conditions. In in vitro kinase assays, deletion of HOG1 did not impair Atg11 CC4 phosphorylation by Rck2 under any of these conditions (Fig. S3 N), indicating that Hog1 does not activate Rck2 under glucose starvation conditions. These findings suggest that glucose starvation does not activate Hog1, and the MAPK signaling pathway is not involved in autophagy and Rck2-mediated Atg11 phosphorylation in response to glucose starvation.
Glucose starvation triggers the elevation of cytoplasmic calcium and activates Rck2. (A) In vitro phosphorylation assays were performed using Atg11 CC4 purified from E. coli as substrates and Rck2-3✕FLAG purified from nutrient-rich, nitrogen starvation, or glucose starvation-treated yeast cells as a protein kinase. Phosphorylation levels of Atg11 CC4 and its variants were detected using anti-thioP antibody. The data are representative of three independent experiments. (B) Wild-type yeast expressing the cytosolic-anchored Ca2+ fluorescence probe jGCaMP7f plasmid were grown to the early log-growth phase and then subjected to nitrogen starvation, glucose starvation, or treated with 0.2 M CaCl2. Images of cells were obtained using an inverted fluorescence microscope. Scale bar, 2 µm. (C) Cytosolic relative fluorescence intensity from B calculated by ImageJ software. n = 60 cells were pooled from three independent experiments. Data are presented as means ± SD. ***P < 0.001; two-tailed Student’s t tests were used. (D) Cells co-expressing Rck2-3✕FLAG with Atg11-3✕HA were subjected to nitrogen or glucose starvation for 0.5 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-HA antibody. (E) The results from D were quantified. Data are presented as means ± SD (n = 3). **P < 0.01; two-tailed Student’s t tests were used. (F) Cells co-expressing Rck2-GFP, Atg17-2✕mCherry, and Vph1-BFP were grown to the early log-growth phase and then subjected to nitrogen starvation or glucose starvation for 0.5 h. Images of cells were obtained using a spinning disk confocal microscope. Scale bar, 2 µm. (G) Cells from F were quantified for the number of cells in which Rck2-GFP colocalized with Atg17-2×mCherry puncta. n = 300 cells were pooled from three independent experiments. Data are shown as mean ± SD. ***P < 0.001; two-tailed Student’s t tests were used. (H) Yeast cells expressing jGCaMP7f were treated with DMSO, 10 mM EGTA, or 20 mM EGTA for 1 h, and then cultured in nitrogen starvation medium (SD-N) or glucose starvation medium (SD-G) in the presence of DMSO, 10 mM EGTA, or 20 mM EGTA. Images of cells were obtained using an inverted fluorescence microscope. Scale bar, 2 µm. (I and J) Wild-type (WT) or atg1∆ yeast strains expressing GFP-Atg8 were treated with or without 20 mM EGTA for 1 h. Subsequently, they were cultured for 4 h in SD-N medium (I) or SD-G medium (J), with or without 20 mM EGTA. The autophagic activity was assessed by Western blotting to detect cleavage of GFP-Atg8, with Pgk1 serving as a loading control. The data are representative of three independent experiments. (K) Cells expressing GFP-Atg8 in WT, atg1∆, or Cmd1-3✕HA-AID yeast strains were treated with IAA for 2 h and then cultured in SD-N or SD-G in the presence of IAA for 4 h. The autophagic activity was analyzed by Western blot for GFP-Atg8 cleavage. Pgk1 served as a loading control. (L and M) The cleavage of GFP-Atg8 from (k) was quantified and presented as mean ± SD (n = 3). ***P < 0.001; NS, no significance; two-tailed Student’s t tests were used. (N) In vitro kinase assays were performed using GST-Atg11 CC4, purified from E. coli, as substrates, and Rck2-3✕FLAG, purified from yeast cells treated with nutrient-rich, glucose starvation, nitrogen starvation, or rapamycin, with or without 20 mM EGTA treatment, as a protein kinase. Phosphorylation levels of GST-Atg11 CC4 were assessed using an anti-thioP antibody. The data are representative of three independent experiments. Source data are available for this figure: SourceData F5.
Glucose starvation triggers the elevation of cytoplasmic calcium and activates Rck2. (A) In vitro phosphorylation assays were performed using Atg11 CC4 purified from E. coli as substrates and Rck2-3✕FLAG purified from nutrient-rich, nitrogen starvation, or glucose starvation-treated yeast cells as a protein kinase. Phosphorylation levels of Atg11 CC4 and its variants were detected using anti-thioP antibody. The data are representative of three independent experiments. (B) Wild-type yeast expressing the cytosolic-anchored Ca2+ fluorescence probe jGCaMP7f plasmid were grown to the early log-growth phase and then subjected to nitrogen starvation, glucose starvation, or treated with 0.2 M CaCl2. Images of cells were obtained using an inverted fluorescence microscope. Scale bar, 2 µm. (C) Cytosolic relative fluorescence intensity from B calculated by ImageJ software. n = 60 cells were pooled from three independent experiments. Data are presented as means ± SD. ***P < 0.001; two-tailed Student’s t tests were used. (D) Cells co-expressing Rck2-3✕FLAG with Atg11-3✕HA were subjected to nitrogen or glucose starvation for 0.5 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-HA antibody. (E) The results from D were quantified. Data are presented as means ± SD (n = 3). **P < 0.01; two-tailed Student’s t tests were used. (F) Cells co-expressing Rck2-GFP, Atg17-2✕mCherry, and Vph1-BFP were grown to the early log-growth phase and then subjected to nitrogen starvation or glucose starvation for 0.5 h. Images of cells were obtained using a spinning disk confocal microscope. Scale bar, 2 µm. (G) Cells from F were quantified for the number of cells in which Rck2-GFP colocalized with Atg17-2×mCherry puncta. n = 300 cells were pooled from three independent experiments. Data are shown as mean ± SD. ***P < 0.001; two-tailed Student’s t tests were used. (H) Yeast cells expressing jGCaMP7f were treated with DMSO, 10 mM EGTA, or 20 mM EGTA for 1 h, and then cultured in nitrogen starvation medium (SD-N) or glucose starvation medium (SD-G) in the presence of DMSO, 10 mM EGTA, or 20 mM EGTA. Images of cells were obtained using an inverted fluorescence microscope. Scale bar, 2 µm. (I and J) Wild-type (WT) or atg1∆ yeast strains expressing GFP-Atg8 were treated with or without 20 mM EGTA for 1 h. Subsequently, they were cultured for 4 h in SD-N medium (I) or SD-G medium (J), with or without 20 mM EGTA. The autophagic activity was assessed by Western blotting to detect cleavage of GFP-Atg8, with Pgk1 serving as a loading control. The data are representative of three independent experiments. (K) Cells expressing GFP-Atg8 in WT, atg1∆, or Cmd1-3✕HA-AID yeast strains were treated with IAA for 2 h and then cultured in SD-N or SD-G in the presence of IAA for 4 h. The autophagic activity was analyzed by Western blot for GFP-Atg8 cleavage. Pgk1 served as a loading control. (L and M) The cleavage of GFP-Atg8 from (k) was quantified and presented as mean ± SD (n = 3). ***P < 0.001; NS, no significance; two-tailed Student’s t tests were used. (N) In vitro kinase assays were performed using GST-Atg11 CC4, purified from E. coli, as substrates, and Rck2-3✕FLAG, purified from yeast cells treated with nutrient-rich, glucose starvation, nitrogen starvation, or rapamycin, with or without 20 mM EGTA treatment, as a protein kinase. Phosphorylation levels of GST-Atg11 CC4 were assessed using an anti-thioP antibody. The data are representative of three independent experiments. Source data are available for this figure: SourceData F5.
Next, we explored what signals activate Rck2 under glucose starvation. Rck2 has a high similarity to calmodulin-dependent kinases (Teige et al., 2001). However, yeast calmodulin Cmd1 cannot activate Rck2 with or without Ca2+ in vitro (Melcher and Thorner, 1996). Given the structural characteristics of Rck2 and its cytoplasmic localization, coupled with its known involvement in calcium signaling in mammalian cells in autophagy (Zheng et al., 2022), we examined whether cytoplasmic Ca2+ is involved in Rck2 activation under glucose starvation. Imaging data showed a remarkable elevation in cytosolic Ca2+ levels occurred during nitrogen or glucose starvation compared with nutrient-rich conditions (Fig. 5, B and C; and Fig. S3 O and Video 1). Furthermore, nitrogen or glucose starvation enhanced interactions between Rck2 and Atg11 (Fig. 5, D and E). We then tested whether Rck2 colocalized with Atg17 under nitrogen or glucose starvation conditions. Imaging data with statistical analysis showed that only a small proportion of Rck2 colocalized with Atg17 in a nutrient-rich medium, but this proportion significantly increased under nitrogen starvation or glucose starvation conditions (Fig. 5, F and G).
Nitrogen or glucose starvation leads to an increase in cytoplasmic Ca2+levels. (A–D) Wild-type yeast expressing the cytosolic-anchored Ca2+ fluorescence probe jGCaMP7f plasmid were grown to the early log-growth phase (A) and then subjected to 0.2 M CaCl2 treatment (B), nitrogen starvation (C), or glucose starvation (D), and then observed using a fluorescence inverted microscope (Olympus, IX83). Images were captured at 4-s intervals using time-lapse microscopy. The data are representative of three independent experiments. Scale bar, 2 µm. Playback speed is 24 frames per second.
Nitrogen or glucose starvation leads to an increase in cytoplasmic Ca2+levels. (A–D) Wild-type yeast expressing the cytosolic-anchored Ca2+ fluorescence probe jGCaMP7f plasmid were grown to the early log-growth phase (A) and then subjected to 0.2 M CaCl2 treatment (B), nitrogen starvation (C), or glucose starvation (D), and then observed using a fluorescence inverted microscope (Olympus, IX83). Images were captured at 4-s intervals using time-lapse microscopy. The data are representative of three independent experiments. Scale bar, 2 µm. Playback speed is 24 frames per second.
To confirm the involvement of cytosolic Ca2+ in glucose starvation–induced autophagy, we used EGTA, a calcium chelator in yeast cells (Liu et al., 2023), to probe the significance of Ca2+ in this process. The fluorescence signal of jGCaMP7f expressed in the cytoplasm showed that 20 mM EGTA completely inhibited the elevation of cytosolic Ca2+ levels under multiple autophagy-inducing conditions (Fig. 5 H, Fig. S4, A and B; and Video 2). Western blot results indicated that 20 mM EGTA treatment almost completely blocked glucose starvation–induced autophagy, while autophagy induced by treatments with rapamycin or nitrogen starvation remained unaffected (Fig. 5, I and J; and Fig. S4 C). We then assessed whether the Ca2+ sensor, Calmodulin (Cmd1) (Davis et al., 1986), is required for autophagy. Given that CMD1 KO is lethal in yeast (Davis et al., 1986), we constructed a CMD1 conditional KD strain using the AID system. Western blot data showed the rapid degradation of Cmd1 protein after IAA treatment in cells expressing Cmd1-3✕HA-AID (Fig. S4 D). Vacuolar localization and cleavage of GFP-Atg8 assays showed that depletion of Cmd1 by IAA treatment resulted in negligible impairment of autophagy under nitrogen starvation, but completely blocked autophagy during glucose starvation (Fig. 5, K–M; and Fig. S4, E and F). These results suggest that calcium and Cmd1 are specifically required for autophagy induced by glucose starvation.
Calcium signaling is required for glucose starvation–induced autophagy and the interaction between Atg11 and Bmh1/2 and Snf1-Bmh2 interaction is independent of Atg1 or Atg11. (A and B) Yeast cells expressing jGCaMP7f were treated with DMSO, 10 mM EGTA, or 20 mM EGTA for 1 h, and then cultured in nitrogen starvation medium (SD-N) (A) or glucose starvation medium (SD-G) (B) in the presence of DMSO, 10 mM EGTA, or 20 mM EGTA. Cells were observed using a fluorescence-inverted microscope (Olympus IX83). Images were captured at 4-s intervals using time-lapse microscopy and shown in 24 fps movies. ImageJ software was used to calculate the relative fluorescence intensity of cells to reflect cytoplasmic calcium signaling. The data are representative of three independent experiments. (C) Wild-type (WT) or atg1∆ yeast cells expressing GFP-Atg8 were treated with or without 20 mM EGTA for 1 h. Subsequently, they were treated with rapamycin for 4 h, with or without 20 mM EGTA. Autophagic activity was assessed by Western blotting to detect cleavage of GFP-Atg8, with Pgk1 serving as a loading control. The data are representative of three independent experiments. (D) Cells expressing Cmd1-3✕HA-AID were treated with 0.5 mM IAA for 2 h, and the expression levels of Cmd1 protein were detected by using Anti-HA antibody. Pgk1 served as a loading control. The data are representative of three independent experiments. (E) Cells expressing GFP-Atg8 in WT, atg1∆, or Cmd1-3✕HA-AID yeast strains were treated with IAA for 2 h and then cultured in SD-N or SD-G in the presence of IAA for 4 h. Images of cells were obtained using an inverted fluorescence microscope. Scale bar, 2 µm. (F) Cells from E were quantified for the vacuolar localization of GFP-Atg8. n = 300 cells were pooled from three independent experiments. Data are presented as means ± SD. ***P < 0.001; ns, no significance; two-tailed Student’s t tests were used. (G and H) Co-expressing HA-Atg11 and Bmh1-3✕FLAG(G) or Bmh2-3✕FLAG(H) yeast strains were treated with or without 20 mM EGTA for 1 h, and then cultured in nutrient-rich medium, nutrient-rich medium with rapamycin, nitrogen starvation medium, or glucose starvation medium in the presence or absence of EGTA for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (I) Cells co-expressing Bmh2-3✕FLAG and HA-Snf1 were grown to the log-growth phase and then subjected to glucose starvation for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (J) Cells co-expressing Bmh2-3✕FLAG and HA-Atg11 in WT, atg1∆, or atg11∆ yeast strains were cultured in a glucose starvation medium for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (K) Cells co-expressing Bmh2-3✕FLAG and HA-Atg11 in WT or snf1∆ yeast strains were grown to the log-growth phase and then subjected to glucose starvation for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (L) Wild-type (BY4741) or atg11∆ atg17∆ yeast cells expressing Snf1-GFP were cultured in glucose starvation medium for 1 h. Cell lysates were analyzed by gel filtration chromatography using a Superose 6 10/300 GL column. Each fraction was detected by immunoblotting using the indicated antibodies. The red dashed box represents PAS fractions. The data are representative of three independent experiments. Source data are available for this figure: SourceData FS4.
Calcium signaling is required for glucose starvation–induced autophagy and the interaction between Atg11 and Bmh1/2 and Snf1-Bmh2 interaction is independent of Atg1 or Atg11. (A and B) Yeast cells expressing jGCaMP7f were treated with DMSO, 10 mM EGTA, or 20 mM EGTA for 1 h, and then cultured in nitrogen starvation medium (SD-N) (A) or glucose starvation medium (SD-G) (B) in the presence of DMSO, 10 mM EGTA, or 20 mM EGTA. Cells were observed using a fluorescence-inverted microscope (Olympus IX83). Images were captured at 4-s intervals using time-lapse microscopy and shown in 24 fps movies. ImageJ software was used to calculate the relative fluorescence intensity of cells to reflect cytoplasmic calcium signaling. The data are representative of three independent experiments. (C) Wild-type (WT) or atg1∆ yeast cells expressing GFP-Atg8 were treated with or without 20 mM EGTA for 1 h. Subsequently, they were treated with rapamycin for 4 h, with or without 20 mM EGTA. Autophagic activity was assessed by Western blotting to detect cleavage of GFP-Atg8, with Pgk1 serving as a loading control. The data are representative of three independent experiments. (D) Cells expressing Cmd1-3✕HA-AID were treated with 0.5 mM IAA for 2 h, and the expression levels of Cmd1 protein were detected by using Anti-HA antibody. Pgk1 served as a loading control. The data are representative of three independent experiments. (E) Cells expressing GFP-Atg8 in WT, atg1∆, or Cmd1-3✕HA-AID yeast strains were treated with IAA for 2 h and then cultured in SD-N or SD-G in the presence of IAA for 4 h. Images of cells were obtained using an inverted fluorescence microscope. Scale bar, 2 µm. (F) Cells from E were quantified for the vacuolar localization of GFP-Atg8. n = 300 cells were pooled from three independent experiments. Data are presented as means ± SD. ***P < 0.001; ns, no significance; two-tailed Student’s t tests were used. (G and H) Co-expressing HA-Atg11 and Bmh1-3✕FLAG(G) or Bmh2-3✕FLAG(H) yeast strains were treated with or without 20 mM EGTA for 1 h, and then cultured in nutrient-rich medium, nutrient-rich medium with rapamycin, nitrogen starvation medium, or glucose starvation medium in the presence or absence of EGTA for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (I) Cells co-expressing Bmh2-3✕FLAG and HA-Snf1 were grown to the log-growth phase and then subjected to glucose starvation for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (J) Cells co-expressing Bmh2-3✕FLAG and HA-Atg11 in WT, atg1∆, or atg11∆ yeast strains were cultured in a glucose starvation medium for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (K) Cells co-expressing Bmh2-3✕FLAG and HA-Atg11 in WT or snf1∆ yeast strains were grown to the log-growth phase and then subjected to glucose starvation for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (L) Wild-type (BY4741) or atg11∆ atg17∆ yeast cells expressing Snf1-GFP were cultured in glucose starvation medium for 1 h. Cell lysates were analyzed by gel filtration chromatography using a Superose 6 10/300 GL column. Each fraction was detected by immunoblotting using the indicated antibodies. The red dashed box represents PAS fractions. The data are representative of three independent experiments. Source data are available for this figure: SourceData FS4.
20 mM EGTA inhibited an increase in cytoplasmic Ca2+level. (A–F) Yeast cells expressing jGCaMP7f were treated with DMSO, 10 mM EGTA, or 20 mM EGTA for 1 h and then cultured in nitrogen starvation medium (SD-N) (A–C) or glucose starvation medium (SD-G) (D–F) in the presence of DMSO (A and D), 10 mM EGTA (B and E), or 20 mM EGTA (C and F) and observed using a fluorescence inverted microscope (Olympus, IX83). Images were captured at 4-s intervals using time-lapse microscopy. The data are representative of three independent experiments. Scale bar, 2 µm. Playback speed is 24 frames per second.
20 mM EGTA inhibited an increase in cytoplasmic Ca2+level. (A–F) Yeast cells expressing jGCaMP7f were treated with DMSO, 10 mM EGTA, or 20 mM EGTA for 1 h and then cultured in nitrogen starvation medium (SD-N) (A–C) or glucose starvation medium (SD-G) (D–F) in the presence of DMSO (A and D), 10 mM EGTA (B and E), or 20 mM EGTA (C and F) and observed using a fluorescence inverted microscope (Olympus, IX83). Images were captured at 4-s intervals using time-lapse microscopy. The data are representative of three independent experiments. Scale bar, 2 µm. Playback speed is 24 frames per second.
We next investigated the potential role of calcium signaling in the activation of Rck2 in response to glucose starvation. In vitro kinase assays showed that EGTA treatment suppressed Rck2-mediated phosphorylation of Atg11 CC4 under conditions of rapamycin treatment, nitrogen starvation, or glucose starvation (Fig. 5 N), indicating that calcium is required for the activation of Rck2 under autophagy-inducing conditions. We then examined whether Ca2+ was involved in the binding of Bmh1/2-Atg11 under various autophagy-inducing conditions. Co-IP results revealed that the increased binding of Bmh1/2-Atg11during rapamycin treatment, nitrogen starvation, or glucose starvation was almost completely blocked by EGTA treatment (Fig. S4, G and H). Collectively, these findings indicate that calcium is crucial for Rck2 activation during glucose starvation, facilitating the binding of Bmh1/2-Atg11 by phosphorylating Atg11 and initiating glucose starvation–induced autophagy.
Glucose starvation induces the release of vacuolar calcium into the cytoplasm
Next, we investigated the mechanism underlying the increase in cytoplasmic Ca2+ levels during nitrogen or glucose starvation in yeast cells. Previous studies have identified that Ca2+ transporters capable of importing Ca2+ into the cytoplasm primarily include Cch1 on the cell membrane, Yvc1 on the vacuolar membrane, and Csg2 on the ER membrane (Cunningham and Fink, 1994; Liu et al., 2023). To determine which Ca2+ channel contributes to the observed increase in cytosolic Ca2+ under starvation conditions, we individually knocked out these three genes in yeast strains expressing a cytosolic Ca2+ probe (Liu et al., 2023). Imaging these strains after nitrogen or glucose starvation revealed that the cytoplasmic Ca2+ levels were moderately decreased in yvc1∆ cells, whereas knockout of CCH1 or CSG2 had no effect on cytoplasmic Ca2+ levels under autophagy-related stress compared to wild-type cells (Fig. 6, A–C and Video 3), indicating a role for Yvc1 in regulating cytoplasmic Ca2+ during starvation.
Glucose starvation induces the release of vacuolar calcium into the cytoplasm. (A and B) Cells expressing cytoplasmic-anchored jGCaMP7f plasmid in WT, cch1∆, csg2∆, or yvc1∆ yeast strains were subjected to nitrogen starvation or glucose starvation and observed using a fluorescence inverted microscope (Olympus, IX83). Images were captured at 4-s intervals using time-lapse microscopy and shown in 24 fps movies. ImageJ software was used to calculate the relative fluorescence intensity of cells to reflect cytoplasmic calcium signaling. The data are representative of three independent experiments. (C) The image data at 88 s from A and B. Scale bar, 2 µm. (D) Cells co-expressing ER-anchored Stt3-jGCaMP7c (ER Ca2+ probe) and Sec63-mCherry were grown to the early log-growth phase and then subjected to either nitrogen or glucose starvation. Images of cells were obtained using an inverted fluorescence microscope. Scale bar, 2 µm. (E) Relative fluorescence intensity from D calculated by ImageJ software. n = 60 cells were pooled from three independent experiments. Data are presented as means ± SD. ns, no significance; two-tailed Student’s t tests were used. (F and G) Cells co-expressing Vacuolar-localized Atg15-jGCaMP7f (test vacuolar Ca2+ level) and Vph1-mCherry in wild-type (WT) or yvc1∆ yeast strains were grown to the early log-growth phase, and then subjected to nitrogen or glucose starvation. Images of cells were obtained using an inverted fluorescence microscope. Scale bar, 2 µm. (H) Relative fluorescence intensity from F and G, calculated by ImageJ software. n = 60 cells were pooled from three independent experiments. Data are presented as means ± SD. ***P < 0.001; ns, no significance; two-tailed Student’s t tests were used. (I) Cells expressing GFP-Atg8 in WT, atg1∆, or yvc1∆ yeast strains were cultured in either SD-N or SD-G for 4 h. Images of cells were obtained using an inverted fluorescence microscope. Scale bar, 2 µm. (J) Cells from I were quantified for the vacuolar localization of GFP-Atg8. n = 300 cells were pooled from three independent experiments. Data are presented as means ± SD. ***P < 0.001; ns, no significance; two-tailed Student’s t tests were used. (K) Cells expressing GFP-Atg8 in WT, atg1∆, or yvc1∆ yeast strains were cultured in SD-N or SD-G for 4 h. Autophagic activity was analyzed by Western blot for GFP-Atg8 cleavage. (L and M) The cleavage of GFP-Atg8 from K was quantified and presented as mean ± SD (n = 3). ***P < 0.001; NS, no significance; two-tailed Student’s t tests were used. Source data are available for this figure: SourceData F6.
Glucose starvation induces the release of vacuolar calcium into the cytoplasm. (A and B) Cells expressing cytoplasmic-anchored jGCaMP7f plasmid in WT, cch1∆, csg2∆, or yvc1∆ yeast strains were subjected to nitrogen starvation or glucose starvation and observed using a fluorescence inverted microscope (Olympus, IX83). Images were captured at 4-s intervals using time-lapse microscopy and shown in 24 fps movies. ImageJ software was used to calculate the relative fluorescence intensity of cells to reflect cytoplasmic calcium signaling. The data are representative of three independent experiments. (C) The image data at 88 s from A and B. Scale bar, 2 µm. (D) Cells co-expressing ER-anchored Stt3-jGCaMP7c (ER Ca2+ probe) and Sec63-mCherry were grown to the early log-growth phase and then subjected to either nitrogen or glucose starvation. Images of cells were obtained using an inverted fluorescence microscope. Scale bar, 2 µm. (E) Relative fluorescence intensity from D calculated by ImageJ software. n = 60 cells were pooled from three independent experiments. Data are presented as means ± SD. ns, no significance; two-tailed Student’s t tests were used. (F and G) Cells co-expressing Vacuolar-localized Atg15-jGCaMP7f (test vacuolar Ca2+ level) and Vph1-mCherry in wild-type (WT) or yvc1∆ yeast strains were grown to the early log-growth phase, and then subjected to nitrogen or glucose starvation. Images of cells were obtained using an inverted fluorescence microscope. Scale bar, 2 µm. (H) Relative fluorescence intensity from F and G, calculated by ImageJ software. n = 60 cells were pooled from three independent experiments. Data are presented as means ± SD. ***P < 0.001; ns, no significance; two-tailed Student’s t tests were used. (I) Cells expressing GFP-Atg8 in WT, atg1∆, or yvc1∆ yeast strains were cultured in either SD-N or SD-G for 4 h. Images of cells were obtained using an inverted fluorescence microscope. Scale bar, 2 µm. (J) Cells from I were quantified for the vacuolar localization of GFP-Atg8. n = 300 cells were pooled from three independent experiments. Data are presented as means ± SD. ***P < 0.001; ns, no significance; two-tailed Student’s t tests were used. (K) Cells expressing GFP-Atg8 in WT, atg1∆, or yvc1∆ yeast strains were cultured in SD-N or SD-G for 4 h. Autophagic activity was analyzed by Western blot for GFP-Atg8 cleavage. (L and M) The cleavage of GFP-Atg8 from K was quantified and presented as mean ± SD (n = 3). ***P < 0.001; NS, no significance; two-tailed Student’s t tests were used. Source data are available for this figure: SourceData F6.
Yvc1 is involved in the increase of cytoplasmic Ca2+levels under nitrogen or glucose starvation conditions. (A–G) Cells expressing cytoplasmic-anchored jGCaMP7f plasmid in WT (A and E), cch1∆ (B and F), csg2∆ (C and G), or yvc1∆ (D and H) yeast strains were subjected to nitrogen starvation (A–D) or glucose starvation (E–H) and observed using a fluorescence inverted microscope (Olympus, IX83). Images were captured at 4-s intervals using time-lapse microscopy. The data are representative of three independent experiments. Scale bar, 2 µm. Playback speed is 24 frames per second.
Yvc1 is involved in the increase of cytoplasmic Ca2+levels under nitrogen or glucose starvation conditions. (A–G) Cells expressing cytoplasmic-anchored jGCaMP7f plasmid in WT (A and E), cch1∆ (B and F), csg2∆ (C and G), or yvc1∆ (D and H) yeast strains were subjected to nitrogen starvation (A–D) or glucose starvation (E–H) and observed using a fluorescence inverted microscope (Olympus, IX83). Images were captured at 4-s intervals using time-lapse microscopy. The data are representative of three independent experiments. Scale bar, 2 µm. Playback speed is 24 frames per second.
In yeast cells, Ca2+ is stored mainly in vacuoles, with a small proportion residing in the ER (Cunningham and Fink, 1994). Monitoring vacuolar and ER Ca2+ levels using plasmid-based probes during starvation conditions showed stable ER Ca2+ concentrations but a significant decrease in vacuolar Ca2+ levels (Fig. 6, D–F and H) (Liu et al., 2023; Tokumitsu Sakagami, 2022). These results suggest that vacuolar Ca2+ pools likely contribute to the starvation-induced increase in cytoplasmic Ca2+, prompting further investigation into the yvc1∆ yeast strain. Closer scrutiny of Ca2+ flux in these cells revealed that the decrease in vacuolar Ca2+ was partially inhibited by YVC1 knockout (Fig. 6, G and H). This observation hinted at the presence of additional, as-yet-unidentified calcium efflux transporters on the vacuolar membrane, which might participate in the transport of vacuolar Ca2+ into the cytoplasm.
Based on these observations, we assessed the autophagic activity of YVC1 knockout under nitrogen or glucose starvation conditions and found that the deletion of YVC1 partially impaired glucose starvation–induced autophagy but did not affect autophagy induced by nitrogen starvation (Fig. 6, I–M). Collectively, these results imply that the efflux of vacuolar Ca2+ could at least partially account for the starvation-induced increase in cytosolic Ca2+ and highlight the specific requirement of cytoplasmic Ca2+ for glucose starvation–induced autophagy.
Bmh1/2 regulate Snf1 recruitment to the PAS to activate Atg1 upon glucose starvation
Based on our prior evidence that the Atg11 CC4 domain regulates Atg11 binding with Snf1 (Yao et al., 2020), and current results showing that this domain is also required for Atg11 association with Bmh1/2, we investigated whether Bmh1/2 could serve as an adaptor protein to link Atg11 with Snf1. To test this hypothesis, we first determined if Bmh1/2 could associate with Snf1. Co-IP assays showed that Bmh1/2 could indeed bind to Snf1 in a nutrient-rich medium, and this binding increased under glucose starvation (Fig. 7 A and Fig. S4 I). Since Bmh1/2 binds with Atg11 and Snf1 associates with Atg1 and Atg11, we next investigated whether the Bmh1/2 interaction with Snf1 depended on Atg1 or Atg11. As shown in Fig. 7 B and Fig. S4 J, knockout of ATG1 or ATG11 had no effect on the association of Bmh1/2 with Snf1 under glucose starvation, indicating that the binding of Bmh1/2 to Snf1 was independent of both Atg1 and Atg11. We then examined whether Snf1 mediates Atg11-Bmh1/2 interactions. Co-IP assays showed that Atg11 binding with Bmh1/2 was also independent of Snf1 (Fig. 7 C and Fig. S4 K). These data implied that Bmh1/2 may act as an adaptor protein to mediate the interaction of Snf1 with Atg11.
Bmh1/2 is required for Snf1 recruitment to the PAS by regulating the association of Atg11/Atg1 with Snf1 under glucose starvation conditions. (A) Cells co-expressing Bmh1-3✕FLAG and HA-Snf1 were grown to the log-growth phase and then subjected to glucose starvation for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (B) Cells co-expressing Bmh1-3✕FLAG and HA-Snf1 in WT, atg1∆, or atg11∆ yeast strains were cultured in SD-G medium for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (C) Cells co-expressing Bmh1-3✕FLAG and HA-Atg11 in WT or snf1∆ yeast strain were grown to the log-growth phase and then subjected to glucose starvation for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (D) WT and bmh1∆ Bmh2-3✕HA-AID yeast strains were treated with IAA for 2 h, and then cultured in nutrient-rich medium or glucose starvation medium in the presence of IAA for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (E) Cells co-expressing Atg1-3✕FLAG and Snf1-GFP in wild-type and bmh1∆ Bmh2-3✕HA-AID yeast strains were treated with IAA for 2 h, and then cultured in nutrient-rich medium or glucose starvation medium in the presence of IAA for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-GFP antibody. The data are representative of three independent experiments. (F) Gel filtration chromatography experiment using a Superose 6 10/300 GL column for proteins lysed from the indicated yeast strain after 1 h of glucose starvation. Each fraction was detected by immunoblotting using the indicated antibodies. The red dashed box represents PAS fractions. The data are representative of three independent experiments. (G) Wild-type (BY4741) or bmh1∆ Bmh2-3✕HA-AID yeast strains were treated with IAA for 2 h, and then cultured in glucose starvation medium in the presence of IAA for 1 h. Cell lysates were analyzed by gel filtration chromatography using a Superose 6 10/300 GL column. Each fraction was detected by immunoblotting using the indicated antibodies. The red dashed box represents PAS fractions. (H) Snf1 levels in the PAS fractions from G were quantified. Data are presented as means ± SD (n = 3). ***P < 0.001; **P < 0.01; two-tailed Student’s t tests were used. Source data are available for this figure: SourceData F7.
Bmh1/2 is required for Snf1 recruitment to the PAS by regulating the association of Atg11/Atg1 with Snf1 under glucose starvation conditions. (A) Cells co-expressing Bmh1-3✕FLAG and HA-Snf1 were grown to the log-growth phase and then subjected to glucose starvation for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (B) Cells co-expressing Bmh1-3✕FLAG and HA-Snf1 in WT, atg1∆, or atg11∆ yeast strains were cultured in SD-G medium for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (C) Cells co-expressing Bmh1-3✕FLAG and HA-Atg11 in WT or snf1∆ yeast strain were grown to the log-growth phase and then subjected to glucose starvation for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (D) WT and bmh1∆ Bmh2-3✕HA-AID yeast strains were treated with IAA for 2 h, and then cultured in nutrient-rich medium or glucose starvation medium in the presence of IAA for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (E) Cells co-expressing Atg1-3✕FLAG and Snf1-GFP in wild-type and bmh1∆ Bmh2-3✕HA-AID yeast strains were treated with IAA for 2 h, and then cultured in nutrient-rich medium or glucose starvation medium in the presence of IAA for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-GFP antibody. The data are representative of three independent experiments. (F) Gel filtration chromatography experiment using a Superose 6 10/300 GL column for proteins lysed from the indicated yeast strain after 1 h of glucose starvation. Each fraction was detected by immunoblotting using the indicated antibodies. The red dashed box represents PAS fractions. The data are representative of three independent experiments. (G) Wild-type (BY4741) or bmh1∆ Bmh2-3✕HA-AID yeast strains were treated with IAA for 2 h, and then cultured in glucose starvation medium in the presence of IAA for 1 h. Cell lysates were analyzed by gel filtration chromatography using a Superose 6 10/300 GL column. Each fraction was detected by immunoblotting using the indicated antibodies. The red dashed box represents PAS fractions. (H) Snf1 levels in the PAS fractions from G were quantified. Data are presented as means ± SD (n = 3). ***P < 0.001; **P < 0.01; two-tailed Student’s t tests were used. Source data are available for this figure: SourceData F7.
To test this hypothesis, we introduced HA-Atg11 and Snf1-3✕FLAG to the bmh1∆ Bmh2-3✕HA-AID yeast strain. The association of Snf1 with Atg11 was almost abolished after Bmh1/2 depletion by IAA treatment in bmh1∆ Bmh2-3✕HA-AID cells (Fig. 7 D), indicating that Bmh1/2 are crucial for interactions between Atg11 and Snf1. Given that Bmh1/2 are required for Atg11-Snf1 binding and that Atg11 also directly binds to Atg1 (Suzuki and Ohsumi, 2010), we examined whether Snf1-Atg1 interactions require Bmh1/2. Co-IP assays confirmed that Snf1-Atg1 interactions were significantly decreased in bmh1∆ Bmh2-3✕HA-AID cells following Bmh1/2 depletion, indicating that Bmh1/2 are required for the association of Atg1 with Snf1 (Fig. 7 E). Concurrently, gel filtration experiments showed that Bmh1/2, Snf1, Cmd1, and Rck2 appeared simultaneously in the PAS fractions under glucose starvation conditions (Fig. 7 F), while Bmh1/2 depletion significantly decreased Snf1 levels in the PAS fractions under glucose starvation conditions (Fig. 7, G and H). Additionally, we observed that double knockout of ATG11 and ATG17 also reduced Snf1 levels in the PAS fractions upon glucose starvation (Fig. S4 L). Collectively, these data showed that Bmh1/2 are required specifically for glucose starvation–induced autophagy due to its control of both the PAS recruitment of Snf1 and subsequent Snf1-Atg11/Atg1 interactions.
Glc7-mediated vacuolar membrane-localized Snf1 binding with Bmh1/2 is required for glucose starvation–induced autophagy
In S. cerevisiae, Snf1 adopts differing proteins as its β and γ subunits, resulting in three different Snf1 complexes, each corresponding to differing β subunits (Sip1, Sip2, or Gal83), and with such differences effectively regulating cellular localization (vacuolar membrane, cytoplasm, or nucleus) (Yang et al., 1994; Zhang et al., 2010). We explored which Snf1 complex binds to Bmh1/2. To this end, we constructed a yeast strain coexpressing Bmh1-3✕FLAG-TEV-GFP with Snf1-6✕HA and used LC-MS to identify candidate Snf1-binding partners from among Bmh1-associated proteins following two rounds of Co-IP (Fig. 8 A). MS analysis showed that Snf1 binds to Bmh1/2 when found in a complex with Sip1 and Snf4 (Fig. 8 B). We then added a 6✕HA tag to the C-terminus of Sip1, with subsequent Co-IP experiments further confirming this result (Fig. 8 C and Fig. S5 A). Given that Sip1 localizes to the vacuolar membrane (Vincent et al., 2001), this finding suggests that Bmh1/2 likely acts as an adaptor protein to mediate the recruitment of vacuolar membrane-localized Snf1 to the PAS and subsequently activate Atg1 during glucose starvation.
Glc7-mediated Snf1 binding with Bmh1/2 is required for glucose starvation–induced autophagy. (A) Flowchart illustrating the process of identifying candidate Snf1-binding partners among Bmh1-associated proteins under glucose starvation conditions. (B) Representative proteins identified by LC-MS from the experiment described in A. (C) Cells co-expressing Bmh1-3✕FLAG and Sip1-3✕HA were cultured to the log-growth phase and then subjected to glucose starvation for 15 min or 30 min. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (D) Cells co-expressing Bmh1-3✕FLAG and HA-Snf1 in WT and Glc7-3✕HA-AID yeast strains were treated with IAA for 2 h and then cultured in nutrient-rich medium or glucose starvation medium in the presence of IAA for 0.5 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (E) Cells expressing GFP-Atg8 and Vph1-Cherry in wild-type (WT), atg1∆, or Glc7-3✕HA-AID yeast strains were treated with IAA for 2 h and then cultured in SD-N or SD-G in the presence of IAA for 4 h. Autophagic activity was analyzed by Western blot for GFP-Atg8 cleavage. The data are representative of three independent experiments. (F) Wild-type or Glc7-3✕HA-AID yeast strains were treated with IAA for 2 h and then cultured in glucose starvation medium in the presence of IAA for 1 h. Cell lysates were analyzed by gel filtration chromatography using a Superose 6 10/300 GL column. Each fraction was detected by immunoblotting using the indicated antibodies. The red dashed box represents PAS fractions. The data are representative of three independent experiments. (G) Cells expressing Atg1-GFP in WT or Glc7-3✕HA-AID yeast strains were treated with IAA for 2 h and then were cultured in SD-N or SD-G in the presence of IAA for 1 h. Images of cells were obtained using an inverted fluorescence microscope. Scale bar, 2 µm. (H) Cells from G were quantified for the number of cells with Atg1 puncta. n = 300 cells were pooled from three independent experiments. Data are presented as means ± SD. ***P < 0.001; NS, not significant; two-tailed Student’s t tests were used. (I) Cells expressing Atg1-3✕FLAG in WT or Glc7-3✕HA-AID yeast strains were treated with IAA for 2 h and then cultured in glucose or nitrogen starvation medium in the presence of IAA for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and analyzed by Western blot using anti-p-T226-Atg1 antibody. The data are representative of three independent experiments. Source data are available for this figure: SourceData F8.
Glc7-mediated Snf1 binding with Bmh1/2 is required for glucose starvation–induced autophagy. (A) Flowchart illustrating the process of identifying candidate Snf1-binding partners among Bmh1-associated proteins under glucose starvation conditions. (B) Representative proteins identified by LC-MS from the experiment described in A. (C) Cells co-expressing Bmh1-3✕FLAG and Sip1-3✕HA were cultured to the log-growth phase and then subjected to glucose starvation for 15 min or 30 min. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (D) Cells co-expressing Bmh1-3✕FLAG and HA-Snf1 in WT and Glc7-3✕HA-AID yeast strains were treated with IAA for 2 h and then cultured in nutrient-rich medium or glucose starvation medium in the presence of IAA for 0.5 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (E) Cells expressing GFP-Atg8 and Vph1-Cherry in wild-type (WT), atg1∆, or Glc7-3✕HA-AID yeast strains were treated with IAA for 2 h and then cultured in SD-N or SD-G in the presence of IAA for 4 h. Autophagic activity was analyzed by Western blot for GFP-Atg8 cleavage. The data are representative of three independent experiments. (F) Wild-type or Glc7-3✕HA-AID yeast strains were treated with IAA for 2 h and then cultured in glucose starvation medium in the presence of IAA for 1 h. Cell lysates were analyzed by gel filtration chromatography using a Superose 6 10/300 GL column. Each fraction was detected by immunoblotting using the indicated antibodies. The red dashed box represents PAS fractions. The data are representative of three independent experiments. (G) Cells expressing Atg1-GFP in WT or Glc7-3✕HA-AID yeast strains were treated with IAA for 2 h and then were cultured in SD-N or SD-G in the presence of IAA for 1 h. Images of cells were obtained using an inverted fluorescence microscope. Scale bar, 2 µm. (H) Cells from G were quantified for the number of cells with Atg1 puncta. n = 300 cells were pooled from three independent experiments. Data are presented as means ± SD. ***P < 0.001; NS, not significant; two-tailed Student’s t tests were used. (I) Cells expressing Atg1-3✕FLAG in WT or Glc7-3✕HA-AID yeast strains were treated with IAA for 2 h and then cultured in glucose or nitrogen starvation medium in the presence of IAA for 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and analyzed by Western blot using anti-p-T226-Atg1 antibody. The data are representative of three independent experiments. Source data are available for this figure: SourceData F8.
Glc7 is required for the binding Bmh1/2 with Snf1 and Atg1 puncta formation under glucose starvation conditions, and overexpression of Bmh1 or Bmh2 does not affect nitrogen starvation-induced autophagy. (A) Cells co-expressing Bmh2-3✕FLAG and Sip1-3✕HA were grown to the log-growth phase and then subjected to glucose starvation for 15 min or 30 min. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (B) Glc7-3✕HA-AID yeast strains were treated with 0.5 mM IAA for 2 h, and the expression levels of Glc7 protein were detected using anti-HA antibody. Pgk1 served as a loading control. The data are representative of three independent experiments. (C) The indicated yeast strains, either untreated or treated with IAA for 3 days, were plated in fourfold serial dilution onto YPD plates and incubated at 30°C. (D) Cells co-expressing Bmh2-3✕FLAG and HA-Snf1 in WT or Glc7-3✕HA-AID yeast strains were treated with IAA for 2 h and then were cultured in nutrient-rich medium or glucose starvation medium in the presence of IAA for 0.5 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (E) Cells co-expressing Glc7-3✕HA and Snf1-GFP were treated with glucose starvation for 0 h or 1 h. Cell lysates were immunoprecipitated with anti-GFP agarose beads and analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (F and G) Cells co-expressing Glc7-3✕HA and Bmh1-3✕FLAG (F) or Bmh2-3✕FLAG(G) were treated with glucose starvation for 0 h or 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (H) Cells expressing GFP-Snf1 in WT or Glc7-3✕HA-AID yeast strains were treated with DMSO or IAA for 2 h, and then cultured in glucose starvation medium in the absence or presence of IAA for 0.5 h or 1 h. The activity of Snf1 was detected by immunoblotting with anti-phospho-AMPKα (Thr172) antibody. The data are representative of three independent experiments. (I) Cells co-expressing an empty vector or 3✕HA-Bmh2 with the Cup1 promoter and GFP-Atg8 in a wild-type strain were treated with or without CuSO4 for 2 h, and then cultured in glucose starvation medium in the absence or presence of CuSO4 for 0, 1, 2, or 4 h. Autophagic activity were analyzed by Western blot for the cleavage of GFP-Atg8. Pgk1 served as a loading control. (K and M) Cells co-expressing an empty vector or 3✕HA-Bmh1(K)/2(M) with the Cup1 promoter and GFP-Atg8 in a wild-type strain were treated with or without CuSO4 for 2 h, and then cultured in nitrogen starvation medium in the absence or presence of CuSO4 for 0, 1, 2, or 4 h. Autophagic activity were analyzed by Western blot for the cleavage of GFP-Atg8. Pgk1 served as a loading control. (J, L, and N) Quantification of the ratio of free GFP/GFP+GFP-Atg8 from I, K, and M (n = 3). Data are presented as means ± SD. ***P < 0.001; ns, no significance; two-tailed Student’s t tests were used. (O) Atg1-3✕FLAG Cells expressing an empty vector or 3✕HA-Bmh2 with the Cup1 promoter were treated with or without CuSO4 for 2 h and then cultured in glucose starvation medium in the absence or presence of CuSO4 for 0 or 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and analyzed by Western blot using anti-p-T226-Atg1 antibody. (P) The phosphorylation level of Atg1 T226 from O was quantified by calculating the ratio of p-Atg1-T226/FLAG-Atg1 (n = 3). Data are presented as mean ± SD. ***P < 0.001; ns, no significance; two-tailed Student’s t tests were used. Source data are available for this figure: SourceData FS5.
Glc7 is required for the binding Bmh1/2 with Snf1 and Atg1 puncta formation under glucose starvation conditions, and overexpression of Bmh1 or Bmh2 does not affect nitrogen starvation-induced autophagy. (A) Cells co-expressing Bmh2-3✕FLAG and Sip1-3✕HA were grown to the log-growth phase and then subjected to glucose starvation for 15 min or 30 min. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (B) Glc7-3✕HA-AID yeast strains were treated with 0.5 mM IAA for 2 h, and the expression levels of Glc7 protein were detected using anti-HA antibody. Pgk1 served as a loading control. The data are representative of three independent experiments. (C) The indicated yeast strains, either untreated or treated with IAA for 3 days, were plated in fourfold serial dilution onto YPD plates and incubated at 30°C. (D) Cells co-expressing Bmh2-3✕FLAG and HA-Snf1 in WT or Glc7-3✕HA-AID yeast strains were treated with IAA for 2 h and then were cultured in nutrient-rich medium or glucose starvation medium in the presence of IAA for 0.5 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (E) Cells co-expressing Glc7-3✕HA and Snf1-GFP were treated with glucose starvation for 0 h or 1 h. Cell lysates were immunoprecipitated with anti-GFP agarose beads and analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (F and G) Cells co-expressing Glc7-3✕HA and Bmh1-3✕FLAG (F) or Bmh2-3✕FLAG(G) were treated with glucose starvation for 0 h or 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and analyzed by Western blot using anti-HA antibody. The data are representative of three independent experiments. (H) Cells expressing GFP-Snf1 in WT or Glc7-3✕HA-AID yeast strains were treated with DMSO or IAA for 2 h, and then cultured in glucose starvation medium in the absence or presence of IAA for 0.5 h or 1 h. The activity of Snf1 was detected by immunoblotting with anti-phospho-AMPKα (Thr172) antibody. The data are representative of three independent experiments. (I) Cells co-expressing an empty vector or 3✕HA-Bmh2 with the Cup1 promoter and GFP-Atg8 in a wild-type strain were treated with or without CuSO4 for 2 h, and then cultured in glucose starvation medium in the absence or presence of CuSO4 for 0, 1, 2, or 4 h. Autophagic activity were analyzed by Western blot for the cleavage of GFP-Atg8. Pgk1 served as a loading control. (K and M) Cells co-expressing an empty vector or 3✕HA-Bmh1(K)/2(M) with the Cup1 promoter and GFP-Atg8 in a wild-type strain were treated with or without CuSO4 for 2 h, and then cultured in nitrogen starvation medium in the absence or presence of CuSO4 for 0, 1, 2, or 4 h. Autophagic activity were analyzed by Western blot for the cleavage of GFP-Atg8. Pgk1 served as a loading control. (J, L, and N) Quantification of the ratio of free GFP/GFP+GFP-Atg8 from I, K, and M (n = 3). Data are presented as means ± SD. ***P < 0.001; ns, no significance; two-tailed Student’s t tests were used. (O) Atg1-3✕FLAG Cells expressing an empty vector or 3✕HA-Bmh2 with the Cup1 promoter were treated with or without CuSO4 for 2 h and then cultured in glucose starvation medium in the absence or presence of CuSO4 for 0 or 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and analyzed by Western blot using anti-p-T226-Atg1 antibody. (P) The phosphorylation level of Atg1 T226 from O was quantified by calculating the ratio of p-Atg1-T226/FLAG-Atg1 (n = 3). Data are presented as mean ± SD. ***P < 0.001; ns, no significance; two-tailed Student’s t tests were used. Source data are available for this figure: SourceData FS5.
We then explored whether another, unknown protein could mediate Bmh1/2 binding with the Snf1–Sip1–Snf4 complex. Revisiting our above MS data and following two rounds of Co-IP, we found that the degradation of Glc7 by IAA treatment almost completely blocked Bmh1/2 interaction with Snf1 under both nutrient-rich and glucose starvation conditions (Fig. 8 D and Fig. S5, B–D), suggesting Glc7 regulates the binding of Bmh1/2-Snf1. Co-IP results confirmed that Glc7 is associated with both Bmh1/2 and Snf1 under either nutrient-rich medium or glucose starvation conditions (Fig. S5, E–G). However, compared with nutrient-rich conditions, the interaction between Glc7 and Snf1 increases under glucose starvation, while the binding between Glc7 and Bmh1/2 remains unchanged. This indicates that while the association of Glc7-Bmh1/2 is stable, the interaction of Glc7 with Snf1 is regulated by glucose starvation. Glc7, known as the catalytic subunit of protein phosphatase 1, plays a role in glucose repression by dephosphorylating the Snf1 activation loop in S. cerevisiae (Tu and Carlson, 1994). We then investigated whether Glc7-mediated binding between Snf1 and Bmh1/2 is required for glucose starvation–induced autophagy and found that depletion of Glc7 by IAA treatment could completely block glucose starvation–induced autophagy, but not nitrogen starvation–induced autophagy (Fig. 8 E), indicating the specific and vital role of Glc7 in regulating glucose starvation–induced autophagy.
Next, we explored how Glc7 regulates glucose starvation–induced autophagy. Observing that Glc7 is required for Bmh1/2-Snf1 binding, we probed whether Glc7 is involved in Snf1 recruitment to the PAS during glucose starvation. Gel filtration experiments showed that Glc7 conditional KD in yeast substantially reduced Snf1 levels in the PAS fractions following glucose starvation (Fig. 8 F), suggesting that Glc7 plays a pivotal role in facilitating Snf1 recruitment to the PAS in response to glucose starvation. We then examined whether Glc7 degradation blocked Atg1 punctate formation and activation under glucose starvation. For this, Atg1-GFP or Atg1-3✕FLAG plasmids were introduced into wild-type or Glc7-3✕HA-AID yeast strains. Fluorescence microscopy and Western blot detection of Atg1 T226 phosphorylation levels showed that Atg1 puncta formation and activation were almost completely blocked upon Glc7 degradation compared with wild-type cells following glucose starvation (Fig. 8, G–I). However, depletion of Glc7 by IAA did not impair Snf1 activation (Fig. S5 H). Cumulatively, these data indicate that Glc7 plays an essential role in glucose starvation–induced autophagy by controlling Bmh1/2 binding with Snf1, leading to Snf1 recruitment to the PAS and the subsequent activation of Atg1.
Bmh1 or Bmh2 overexpression accelerates glucose starvation–induced autophagy by enhancing Atg1 kinase activity
Since Bmh1/2 control Snf1 binding with Atg1 and is specifically involved in the initiation of glucose starvation–induced autophagy, we examined whether Bmh1 or Bmh2 overexpression would promote this process. For this, we introduced 3✕HA-Bmh1 or 3✕HA-Bmh2 plasmids with Cup1 promoter into a WT yeast strain expressing GFP-Atg8 (Janke et al., 2004). GFP-Atg8 cleavage assays showed that the production of free GFP significantly increased upon the addition of CuSO4 to induce Bmh1 or Bmh2 overexpression under glucose starvation conditions (Fig. 9, A and B; and Fig. S5, I and J), whereas neither Bmh1 nor Bmh2 overexpression had any obvious effect on autophagic activity under nitrogen starvation conditions (Fig. S5, K–N). We then examined whether Bmh1 or Bmh2 overexpression could enhance Atg1 activation in response to glucose starvation. Overexpression of either Bmh1 or Bmh2 resulted in significant promotion of Atg1 activation under glucose starvation conditions (Fig. 9, C and D; and Fig. S5, O and P). Collectively, these data suggest that Bmh1/2 overexpression accelerates glucose starvation–induced autophagy by enhancing Atg1 activation.
Bmh1 or Bmh2 overexpression accelerates glucose starvation–induced autophagy by enhancing Atg1 activation. (A) Cells co-expressing an empty vector and GFP-Atg8, or 3✕HA-Bmh1 with the Cup1 promoter and GFP-Atg8 in the wild-type strain were treated with or without CuSO4 for 2 h, and then cultured in glucose starvation medium in the absence or presence of CuSO4 for 0 h, 1 h, 2 h, or 4 h. Autophagic activity was analyzed by Western blot for the cleavage of GFP-Atg8. Pgk1 served as a loading control. (B) Quantification of the ratio of free-GFP/(GFP+GFP-Atg8) from A (n = 3). Data are presented as means ± SD. ***P < 0.001; ns, no significance; two-tailed Student’s t tests were used. (C) Atg1-3✕FLAG Cells expressing an empty vector or 3✕HA-Bmh1 with the Cup1 promoter were treated with or without CuSO4 for 2 h. The cells were then cultured in glucose starvation medium in the absence or presence of CuSO4 for 0 h or 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and analyzed by Western blot using anti-p-T226-Atg1 antibody. (D) The phosphorylation level of Atg1 T226 from C was quantified by calculating the ratio of p-Atg1-T226/FLAG-Atg1(n = 3). Data are presented as mean ± SD. ***P < 0.001; ns, no significance; two-tailed Student’s t tests were used. (E) Model depicting Ca2+-triggered assembly of the Atg11–Bmh1/2–Snf1 complex governing the initiation of glucose starvation–induced autophagy. CaMK: Calmodulin (CaM)-dependent protein kinase. Source data are available for this figure: SourceData F9.
Bmh1 or Bmh2 overexpression accelerates glucose starvation–induced autophagy by enhancing Atg1 activation. (A) Cells co-expressing an empty vector and GFP-Atg8, or 3✕HA-Bmh1 with the Cup1 promoter and GFP-Atg8 in the wild-type strain were treated with or without CuSO4 for 2 h, and then cultured in glucose starvation medium in the absence or presence of CuSO4 for 0 h, 1 h, 2 h, or 4 h. Autophagic activity was analyzed by Western blot for the cleavage of GFP-Atg8. Pgk1 served as a loading control. (B) Quantification of the ratio of free-GFP/(GFP+GFP-Atg8) from A (n = 3). Data are presented as means ± SD. ***P < 0.001; ns, no significance; two-tailed Student’s t tests were used. (C) Atg1-3✕FLAG Cells expressing an empty vector or 3✕HA-Bmh1 with the Cup1 promoter were treated with or without CuSO4 for 2 h. The cells were then cultured in glucose starvation medium in the absence or presence of CuSO4 for 0 h or 1 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and analyzed by Western blot using anti-p-T226-Atg1 antibody. (D) The phosphorylation level of Atg1 T226 from C was quantified by calculating the ratio of p-Atg1-T226/FLAG-Atg1(n = 3). Data are presented as mean ± SD. ***P < 0.001; ns, no significance; two-tailed Student’s t tests were used. (E) Model depicting Ca2+-triggered assembly of the Atg11–Bmh1/2–Snf1 complex governing the initiation of glucose starvation–induced autophagy. CaMK: Calmodulin (CaM)-dependent protein kinase. Source data are available for this figure: SourceData F9.
Discussion
In this study, we identified a previously undocumented signaling pathway regulating glucose starvation–induced autophagy, in which Ca2+-triggered Atg11–Bmh1/2–Snf1 complex assembly played a pivotal role by regulating the PAS recruitment of vacuolar membrane-localized Snf1 and subsequent Atg1 activation. Mechanistically, glucose starvation induces vacuolar Ca2+ release into the cytoplasm, which in turn activates cytosolic protein kinase Rck2 through calmodulin (CaM)-dependent protein kinase (CaMK). Activated Rck2 then promotes the binding of Atg11 with phosphor-binding proteins Bmh1/2 through phosphorylating the CC4 domain of Atg11. Subsequently, Bmh1/2 recruits vacuolar membrane-localized Snf1–Sip1–Snf4 complex to the PAS through Glc7 and then activates Atg1, thereby initiating glucose starvation–induced autophagy (Fig. 9 E). These results underscore calcium as a critical bridge linking environmental sensing to the formation of the autophagy initiation complex by controlling Snf1-mediated Atg1 activation in response to glucose starvation. Notably, whether Rck2 acts solely as a CaMK during glucose starvation or if other CaMKs are involved remains to be determined.
Calcium transients on the ER surface are known to specify autophagosome initiation sites in mammals (Zheng et al., 2022). However, the calcium-triggered signaling cascade we identified in yeast cells does not affect its formation. Instead, it influences the assembly of the autophagy initiation complex by governing Snf1-mediated Atg1 activation. Additionally, in yeast cells, ER Ca2+ levels remain stable before and after starvation, whereas vacuolar Ca2+ undergoes significant release into the cytoplasm after starvation, contributing to increased cytoplasmic Ca2+. This contrast may arise because vacuoles store ∼95% of cellular Ca2+ in yeast, whereas in mammalian cells, the ER serves as the primary Ca2+ reservoir (Cunningham and Fink, 1994; Berridge, 2002). Furthermore, in mammals, autophagosome initiation sites form at ER–mitochondrial contact sites sensitive to nutrient changes (Hamasaki et al., 2013). By contrast, in yeast, the PAS is situated around the vacuolar membrane, and its formation is almost unaffected by changes in nutrient availability (Ohsumi, 2014). These differences in Ca2+ storage and autophagosome initiation sites across species likely underlie the distinct regulation of autophagy by Ca2+ signaling.
Knockout of vacuolar Ca2+ transporter Yvc1 did not completely inhibit vacuolar Ca2+ release after starvation, suggesting the possible involvement of other vacuolar Ca2+ efflux transporters. This warrants further exploration. Future investigations should focus on identifying unidentified vacuolar Ca2+ efflux transporters that may participate in Ca2+ signaling-mediated regulation of glucose starvation–induced autophagy. Although the molecular mechanism triggering glucose starvation–induced vacuolar Ca2+ release remains unclear, Yvc1, a TRP channel homolog, facilitates vacuolar Ca2+ release in response to hyperosmotic shock (Zhou et al., 2003; Palmer et al., 2001), which glucose starvation can induce (Joyner et al., 2016). Therefore, glucose starvation–induced hyperosmotic shock may trigger vacuolar Ca2+ efflux. Notably, as EGTA acts by chelating extracellular calcium, its effect on abolishing autophagy may indicate that the influx of extracellular calcium is involved in glucose starvation–induced autophagy. More research is needed to understand the source(s) of calcium regulating this autophagic process.
Our observations indicate that Ca2+ signaling is largely uninvolved in autophagy induced by nitrogen starvation. This discrepancy may stem from the fact that nitrogen starvation–induced autophagy does not require Atg11. While Atg11 was previously thought crucial for selective autophagy, our research highlights its vital role in regulating Atg1 activation during glucose starvation. Furthermore, in addition to the role of Atg11 in binding with selective autophagy receptors (Yorimitsu and Klionsky, 2005), we discovered its interaction with Bmh1/2 to recruit vacuolar membrane-localized Snf1 to the PAS. This expands our understanding of Atg11’s function in selective autophagy regulation. Interestingly, nitrogen starvation mirrors glucose starvation in elevating cytoplasmic Ca2+ levels, decreasing vacuolar Ca2+ levels in a partially Yvc1-dependent manner, activating Rck2 toward Atg11, promoting Rck2 colocalization with Atg17, and facilitating Atg11-Bmh1/2 interaction. This suggests that increased Atg11-Bmh1/2 binding due to Rck2 activation under nitrogen starvation conditions may lead to autophagy-mediated degradation of phosphorylated proteins bound to Bmh1/2, a hypothesis to be explored further.
Moreover, our study highlights Bmh1/2’s association with the Snf1–Snf4–Sip1 complex. Given Sip1’s vacuolar membrane localization and the PAS’s proximity to the vacuolar membrane (Ohsumi, 2014; Vincent et al., 2001), this conformation of the Snf1 complex is logically well-suited to function in the phosphorylation and activation of Atg1 in response to glucose starvation. In our previous study, we showed that Atg11 is required for the association of Snf1 with Atg1 under glucose starvation conditions (Yao et al., 2020). Building on this, our study demonstrates that Bmh1/2 involvement is crucial for Snf1 recruitment to the PAS during glucose starvation. Although we identified Glc7 as a regulator of the association between Bmh1/2 and Snf1, further research is required to elucidate the precise mechanisms governing this interaction and its impact on Snf1 recruitment to the PAS upon glucose starvation.
Previously, we proposed a Snf1–Mec1–Atg1 module initiating glucose starvation–induced autophagy on mitochondria (Yi et al., 2017). Recently, we showed that Mec1 regulates the PAS recruitment of Atg13 by directly binding to Atg13 (Yao et al., 2023). In this study, we addressed how Snf1 activates Atg1 on the vacuolar membrane. These findings strongly indicate the existence of a distinct signaling pathway for glucose starvation–induced autophagy. On one hand, glucose starvation increases cellular ROS levels, prompting the recruitment of Mec1 from the nucleus to mitochondria (Wu et al., 2021). Activated Snf1 then phosphorylates Mec1 on the mitochondrial surface. Phosphorylated Mec1 at contact sites interacts with Ggc1 and Atg13, linking mitochondria to autophagosomes, thereby regulating Atg13’s PAS recruitment and Atg1 activation. In this process, mitochondria likely provide energy and membrane sources for glucose starvation–induced autophagosome formation. On the other hand, glucose starvation triggers vacuolar Ca2+ efflux into the cytoplasm, elevating cytoplasmic Ca2+ and activating Rck2. Activated Rck2, in turn, facilitates the binding of Atg11 to Bmh1/2 by phosphorylating Atg11. Bmh1/2 then recruits vacuolar membrane–localized Snf1 to the PAS via Glc7. Direct Atg11-Atg1 interaction allows Snf1 to phosphorylate and activate Atg1. This comprehensive process reveals the coordinated actions of elevated ROS, cytoplasmic Ca2+, mitochondrial respiration, and Snf1 activation in glucose starvation–induced Atg1 activation. Additional factors likely influence Atg1 activation during glucose starvation, warranting further investigation.
In summary, our study unveils a critical regulatory pathway illuminating how cells sense and respond to glucose starvation to induce autophagy. Calcium emerges as a pivotal signaling molecule, bridging environmental sensing with autophagy initiation complex formation, ultimately activating Atg1 and initiating glucose starvation–induced autophagy. These findings suggest a potential parallel mechanism for energy deprivation-induced autophagy in mammals, warranting further investigation. Moreover, the interactions uncovered here may hold implications for autophagy-related pathologies under dysregulated conditions.
Materials and methods
Yeast strains and growth conditions
All yeast strains used in this study are listed in Table S1 and were subjected to verification through polymerase chain reaction (PCR) (P505-d1; Vazyme) or Western blot analysis with the indicated antibodies (Janke et al., 2004). Western blot detection was conducted using the MiniChemi Chemiluminescence imager (SAGECREATION). All plasmids in this study were sequenced or confirmed by Western blotting. Yeast cells were cultured at 30°C in either YPD medium (1% yeast extract, 2% glucose, and 2% peptone) or synthetic medium (SD; 0.17% yeast nitrogen base without amino acids and ammonium sulfate, 0.5% ammonium sulfate, 2% glucose, and corresponding auxotrophic amino acids and vitamins). For autophagy induction, cells were grown to mid-log phase in YPD or SD medium, and then were subjected to glucose starvation medium (SD-G; 0.17% yeast nitrogen base without amino acids and ammonium sulfate, 0.5% ammonium sulfate, and 0.5% casamino acid), nitrogen starvation medium (SD−N; 0.17% yeast nitrogen base without amino acids and ammonium sulfate, 2% glucose), or treated with rapamycin (0.2 μg/ml) for the indicated time periods.
Antibodies
Antibodies were purchased and used in this study at the following dilutions: anti-Pgk1 (1:10,000, NE130/7S; Nordic Immunology), anti-GFP (1:2,500, NE130/7S; Nordic Immunology), anti-FLAG (1:2,500, F1804; Sigma-Aldrich), anti-HA (1:3,000, M20003L; Abmart), anti-p-PRKAA/AMPKα (Thr172) (1:2,000, 2535S; Cell Signaling Technology), anti-Ape1 (1:2,000, GL Biochem Ltd.), anti-thiophosphate ester antibody (ab92570; Abcam), Rabbit anti-p-Atg1(T226) (1:1,000) (produced by Youke); anti-p-P38 MAPK(T180/Y182) (1:1,000) (4511S; Cell Signaling Technology); and goat anti-mouse IgG1, human ads-HRP (1:10,000, 1070-05; SouthernBiotech), goat anti-rabbit, human ads-HRP (1:10,000, 1070-05; SouthernBiotech). The anti-Atg17 antibody was generously provided by Dr. Y. Ohsumi (Tokyo Institute of Technology, Japan).
Fluorescence microscopy
Yeast strains equipped with the indicated fluorescent tags (GFP, mCherry, BFP) were grown to early log phase in nutrient-rich medium and subsequently subjected to glucose starvation, nitrogen starvation, or rapamycin treatment (S1039; Selleck) for the indicated time periods. 200 μl of yeast culture was added to an eight-chambered cover glass (155411; Thermo Fisher Scientific). After the yeast cells settled, fluorescence microscopy was performed at room temperature using either an inverted fluorescence microscope (IX83; Olympus) equipped with a U Plan Super Apochromat objective 100✕/1.4 Oil, (Olympus) or a spinning disk confocal microscope (SpinSR10; Olympus) equipped with UPLSAPO S 60✕/1.30 oil. Images were recorded using their respective microscope systems and then further processed and analyzed using ImageJ software.
Immunoprecipitation
Whole-cell extracts of yeast cells were prepared, and immunoprecipitations were carried out according to previously described methods (Yi et al., 2012). 50 OD600 cells were lysed in 500 μl of ice-cold lysis buffer (50 mM Tris.HCl, pH 7.5, 150 mM NaCl, 0.1% Triton X-100, 10% glycerol, protease inhibitor [Roche], 1 mM PMSF, and 1 mM DTT). An equal volume of glass beads was added to this suspension, followed by vigorous vortexing for 10 min at 4°C to disrupt the cells. The whole-cell extracts were then subjected to two consecutive centrifugations at 12,000 rpm for 15 min at 4°C. The resulting supernatants were incubated with anti-FLAG M2 agarose beads (A2220; Sigma-Aldrich) at 4°C for 2 h. After incubation, the beads were washed three times with 1 ml of lysis buffer and then boiled for 5 min in 50 μl of 1✕Laemmli buffer. The samples were separated by SDS-PAGE, transferred to a nitrocellulose membrane, and probed with the appropriate antibody.
Auxin treatment
The indicated yeast strains were treated with 0.5 mM IAA (indole-3-acetic acid, I2886; Sigma-Aldrich) to induce the degradation of the corresponding proteins, with the addition of an equivalent volume of DMSO to the control strains.
CuSO4-induced the overexpression of Bmh1/2
The indicated yeast strains were treated with 0.1 mM CuSO4 (A603008; Shanghai Sangon Biotech) to induce the overexpression of Bmh1 or Bmh2, while an equivalent volume of H2O was added to the control strains.
In vitro Ni-NTA or GST pulldown assay
For the in vitro Ni-NTA or GST pulldown assay, 5 nM purified protein with either His or GST tag was incubated with 10 μl Ni-NTA or GST beads in 500 μl binding buffer A (50 mM Tris.HCl pH 7.5, 0.5 M NaCl, 1 mM DTT, 1% Triton X-100, 1% PMSF, and 20 mM iminazole; GST-tagged proteins do not require the addition of iminazole) for 2 h on a rotor at 4°C. The beads were then washed once with binding buffer A and twice with binding buffer B (1 mM EDTA, 1 mM EGTA, 150 mM NaCl, 1% Triton X-100 in 1✕PBS). Subsequently, the beads were incubated with Atg11 CC4 or its variants in binding buffer B for 3 h on a rotor at 4°C. Following three washes with binding buffer B, 50 μl 2✕SDS loading buffer was added. Proteins separated by SDS-PAGE were stained using Coomassie brilliant blue dye.
In vitro binding assays of phosphorylated GST-Atg11 CC4 with His-Bmh1/2
rck2∆ cells expressing either WT or KD Rck2-3✕FLAG were subjected to glucose starvation for 1 h. 50 OD600 yeast cells were then harvested and cell lysates were immunoprecipitated using anti-FLAG agarose beads (A2220; Sigma-Aldrich). The enriched WT or KD Rck2-3×FLAG was incubated with 5 µg of purified GST-Atg11 CC4, either WT or 2A variant from E. coli, and 10 µM ATP in Rck2 kinase buffer (30 mM HEPES.KOH pH 7.4, 5 mM NaF, 10 mM MgCl2, 1 mM DTT) at 30°C for 30 min. Concurrently, 10 mM purified His-Bmh1/2 from E. coli was incubated with 10 μl Ni-NTA in 500 μl binding buffer (50 mM Tris pH 7.5, 0.5 M NaCl, 1 mM DTT, 1% Triton X-100, 1% PMSF, and 20 mM) for 2 h on a rotor at 4°C. After incubation, the beads were washed once with binding buffer and then twice with interacting buffer (1 mM EDTA, 1 mM EGTA, 150 mM NaCl, 1% Triton X-100 in 1✕PBS). Subsequently, the beads were incubated with the supernatant of in vitro phosphorylation assays at 4°C for 2 h. Finally, after three washes with interaction buffer, 50 μl 2✕ SDS loading buffer was added to the beads. Proteins separated by SDS-PAGE were then stained with Coomassie brilliant blue dye.
In vitro phosphorylation assay and alkylation protocol
rck2∆ cells expressing either WT or KD Rck2-3✕FLAG plasmids were cultured in a nutrient-rich medium, Rapamycin, SD-N, or SD-G for the indicated time periods. 50 OD600 yeast cells were lysed, and the resulting supernatant was immunoprecipitated using anti-FLAG agarose beads (A2220; Sigma-Aldrich). Purified WT or KD Rck2-3✕FLAG was incubated with 5 µg of purified GST-Atg11 CC4 from E. coli and 1.5 μl 10 mM ATP-γ-S (A1388; Sigma-Aldrich) in Rck2 kinase buffer (30 mM HEPES KOH pH 7.4, 5 mM NaF, 10 mM MgCl2, 1 mM DTT) at 30°C for 30 min. Then, 2 μl of 50 mM p-nitrobenzyl mesylate/PNBM (ab138910; Abcam) was added and left to react for an additional hour at 30°C. The reaction was halted by boiling in protein loading buffer for 5 min. The phosphorylation level of GST-Atg11 CC4 was then assessed using anti-thiophosphate ester antibody (ab92570; Abcam).
Tandem affinity purification of Atg11-3✕FLAG-TEV-ZZ
Yeast cells expressing Atg11-3✕FLAG-TEV-ZZ in 2L SD medium were subjected to glucose starvation for 1 h. The cell pellet was washed with chilled H2O and then broken up with glass beads in a lysis buffer (50 mM Tris.HCl, pH 7.5, 150 mM NaCl, 1% NP-40, 10% glycerol, 10 mM NaF, protease inhibitor). The lysates were obtained by centrifugation at 12,000 rpm for 30 min. Meanwhile, Dynabeads from Invitrogen coupled with rabbit IgG (I5006; Sigma-Aldrich) were firstly washed three times with lysis buffer and then incubated with the lysates overnight at 4°C. The Dynabeads coupled with Rabbit IgG were washed three times with lysis buffer (without protease inhibitor) and then twice with TEV buffer (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.1% NP-40, 1 mM DTT, 1 mM EDTA). Then, 10 μl TEV protease (lab-made) was added to 1 ml TEV cleavage buffer and followed by incubation for 3 h at 4°C. The elutes were concentrated using Amicon Ultra-4 filter units (UFC801096; Millipore). Finally, the protein was resolved by 4–12% SDS-PAGE gel and visualized via silver staining.
Measurement of Ca2+signal intensity in the cytoplasm, ER, or vacuole
Yeast strains expressing cytoplasmic-anchored Ca2+ fluorescent probe jGCaMP7f (D2865; Beyotime) were grown to OD600 = 1.0–1.2 in a nutrient-rich medium and immobilized on the slide with 1 mg/ml Concanavalin A (A804224; Macklin). Fixed cells were carefully washed twice with nitrogen starvation (SD-N) medium, glucose starvation (SD-G) medium, or 0.2 M CaCl2 (A501330; Sangon Biotech) solution and observed at room temperature with an inverted fluorescence microscope (IX83; Olympus) equipped with a U Plan Super Apochromat objective 100✕/1.4 Oil (Olympus). Images were captured at 4-s intervals using time-lapse microscopy. ImageJ software was used to calculate the relative fluorescence intensity of cells to reflect cytoplasmic calcium signaling. Yeast cells expressing cytoplasmic-, ER-, or vacuole-anchored Ca2+ fluorescent probe jGCaMP7 (Beyotime, jGCaMP7c, D2863/jGCaMP7f, D2865) in nutrient-rich medium (SD) or treated with SD-N, SD-G or CaCl2 for 88 s were observed under the consistent exposure conditions. These experiments were conducted three times, each with three independent clones of each yeast strain, and with at least 60 random cells in total assessed. Scale bar: 2 µm.
Quantification and statistical analysis
Quantification of fluorescence microscopy and immunoblotting data were performed using ImageJ software. For all quantitative analyses, the mean values are presented along with the standard deviation (SD), shown as an error bar. Relevant statistical analyses and the number of independent experiments are described in each figure legend. Statistical tests were performed in GraphPad Prism, with all tests using an unpaired two-tailed t test. Significance levels were determined by the indicated P values. Data distribution was assumed to be normal but this was not formally tested.
Online supplemental material
Fig. S1 shows the effects of knocking out the Bmh1 or Bmh2 genes on bulk autophagy and the Cvt pathway, as well as whether the interaction between Bmh1/2 and Atg11 is specific. Fig. S2 shows which domain of Atg11 mediates its interaction with Bmh2 and whether Rck2 is involved in glucose starvation–induced autophagy. Fig. S3 shows whether the phosphorylation of Atg11 regulates the interaction between Atg11 and Bmh2 and whether the MAPK signaling pathway is involved in the activation of Rck2 and autophagy. Fig. S4 shows whether Ca2+ is involved in the binding of Atg11-Bmh1/2 and glucose starvation–induced autophagy, as well as which genes are involved in the regulation of the Snf1–Bmh2 interaction. Fig. S5 shows how Glc7 regulates glucose starvation–induced autophagy and the effect of Bmh1/2 overexpression on nitrogen starvation-induced autophagy. Table S1 lists the yeast strains used in this study. Video 1 is a time-lapse fluorescence video showing the changes in cytosolic Ca2+ levels under nitrogen starvation or glucose starvation conditions. Video 2 is a time-lapse fluorescence video showing the levels of cytosolic Ca2+ under nitrogen starvation or glucose starvation conditions with 10 mM or 20 mM EGTA treatment. Video 3 is a time-lapse fluorescence video showing the changes in cytosolic Ca2+ levels under nitrogen starvation or glucose starvation conditions in WT, cch1∆, csg2∆, or yvc1∆ yeast strains.
Data availability
Data reported in this study are available in the published article and the supplementary material. Further information is available upon reasonable request.
Acknowledgments
We thank Dr. Cheng Ma from the Core facilities, Zhejiang University School of Medicine for their technical support; and the Mass Spectrometry & Metabolomics Core Facility at the Center for Biomedical Research Core Facilities of Westlake University for sample analysis. Fig. 9 E was created by Yuyao Feng using BioRender.
The research was supported by the National Natural Science Foundation of China (32122028, 92254307, and 32070739), and the Zhejiang Provincial Natural Science Foundation of China under grant no. LR21C070001 to C. Yi; National Natural Science Foundation of China (grant no: 32100600) to W. Yao; and the National Key Research and Development Program of China (2021YFC2600104) to L. Zhang.
Author contributions: W. Yao: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing—original draft, Y. Chen: Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Validation, Visualization, Y. Zhang: Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Validation, Visualization, S. Zhong: Data curation, Formal analysis, Investigation, Methodology, Resources, Validation, M. Ye: Formal analysis, Methodology, Software, Yuting Chen: Data curation, Methodology, S. Fan: Validation, M. Ye: Resources, Visualization, H. Yang: Formal analysis, Resources, Y. Li: Validation, C. Wu: Formal analysis, Resources, M. Fan: Data curation, S. Feng: Investigation, Z. He: Methodology, L. Zhou: Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, L. Zhang: Conceptualization, Writing—review & editing, Y. Wang: Investigation, Methodology, Supervision, W. Liu: Writing—review & editing, J. Tong: Data curation, Project administration, Resources, Supervision, D. Feng: Conceptualization, Funding acquisition, Resources, Supervision, Writing—review & editing, C. Yi: Conceptualization, Funding acquisition, Project administration, Supervision, Writing—original draft, Writing—review & editing.
References
Author notes
W. Yao, Y. Chen, Y. Zhang, and S. Zhong share the role of co-first author.
Disclosures: The authors declare no competing interests exist.