Phenotypic heterogeneity poses a significant hurdle for cancer treatment but is under-characterized in the context of tumor invasion. Amidst the range of phenotypic heterogeneity across solid tumor types, collectively invading cells and single cells have been extensively characterized as independent modes of invasion, but their intercellular interactions have rarely been explored. Here, we isolated collectively invading cells and single cells from the heterogeneous 4T1 cell line and observed extensive transcriptional and epigenetic diversity across these subpopulations. By integrating these datasets, we identified laminin-332 as a protein complex exclusively secreted by collectively invading cells. Live-cell imaging revealed that laminin-332 derived from collectively invading cells increased the velocity and directionality of single cells. Despite collectively invading and single cells having similar expression of the integrin α6β4 dimer, single cells demonstrated higher Rac1 activation upon laminin-332 binding to integrin α6β4. This mechanism suggests a novel commensal relationship between collectively invading and single cells, wherein collectively invading cells promote the invasive potential of single cells through a laminin-332/Rac1 axis.
Introduction
Tumor heterogeneity poses a significant hurdle to cancer treatment (Roulot et al., 2016; Morris et al., 2016; McGranahan and Swanton, 2017; Dagogo-Jack and Shaw, 2018; Dentro et al., 2021). In particular, phenotypic heterogeneity—defined as distinct morphological and behavioral characteristics of subpopulations within a group of cells—plays a crucial role in therapeutic resistance (Marusyk et al., 2020). Identifying and characterizing the subpopulations that emerge as a tumor evolves is crucial to uncovering potential vulnerabilities for multitargeted therapy.
Within this wide range of phenotypic heterogeneity, recent studies have revealed the prominence of invasive heterogeneity across solid tumors (Khalil et al., 2017; Konen et al., 2017; Zhang et al., 2019; Alvarez-Elizondo and Weihs, 2022). Notably, human tissue sections of breast tumors show a coexistence of collectively invading packs alongside single cells at the invasive edge (Khalil et al., 2017). Intravital imaging of murine breast tumors corroborates these invasive modalities and demonstrates the spontaneous emergence of these distinct subpopulations upon mammary fat pad implantation (Ilina et al., 2018). Additionally, primary organoids from a genetically engineered mouse model of triple-negative breast cancer revealed that cells that persistently invade collectively or as single cells could be separated from the same tumor (Henriet et al., 2023). Furthermore, circulating tumor cell analysis in breast and prostate cancer patients identified both circulating tumor clusters and single cells (Aceto et al., 2014), supporting the maintenance of invasive heterogeneity after initial primary tumor invasion.
As separate mechanisms of invasion, collective and single-cell invasion have been extensively characterized (Friedl and Wolf, 2003; Friedl et al., 2012; Te Boekhorst and Friedl, 2016; Lintz et al., 2017; Nagai et al., 2020). Despite these observations, the interactions between invasively distinct subpopulations require further elucidation. Collective invasion depends on the formation of an extensive network of cell–cell junctions to maintain cell–cell adherence during cell motility (Cheung et al., 2013; Loza et al., 2016). Collective “chains” of cells extend outward through highly motile leader cells, which drive directional movement through invadopodial protrusions in 3D and fibronectin micropatterning (Konen et al., 2017; Summerbell et al., 2020). Collective packs of tumor cells have enhanced capabilities to intravasate into the circulation (Chang and Chaudhuri, 2019), override proapoptotic signaling induced by ECM detachment (Zhao et al., 2010), and evade immune attack (Lo et al., 2020). Conversely, single-cell invasion occurs independently of cell–cell junctions, exchanging invasive efficiency for dynamic flexibility. Single-cell invasion is largely split into mesenchymal or amoeboid with the former dependent on filopodia and matrix metalloproteases to move, and the latter being independent of such protrusions and instead relying on physical cellular deformation to navigate porous extracellular matrix environments such as collagen (Friedl and Wolf, 2003; Nagai et al., 2020). The dynamic nature of amoeboid movement allows for cells to exhibit drastically higher velocities than collective packs while simultaneously being highly proliferative, enabling efficacious metastatic seeding into the lung, bones, and nearby lymph nodes (Gao et al., 2017; Graziani et al., 2022). Despite extensive characterization of collective and single-cell motilities in cancer cells, little is known about how subpopulations that adopt these phenotypes interact to drive local cell invasion.
Recent discoveries have pointed to the predisposition for distinct subpopulations to interact to drive invasion and metastasis across multiple cancer models. “Proliferative” and “invasive” subpopulations of zebrafish melanoma cells have been shown to cooperate within metastatic clusters through TFAP2 to drive invasion (Campbell et al., 2021). In human lung adenocarcinoma cell lines, distinct “leader” cells within invasive collective chains used VEGF signaling to bolster the invasive potential of “follower” cells, while follower cells actively secreted growth signals to support leader cell survival (Konen et al., 2017). Leader and follower cell dynamics have also been observed in breast cancer cells, wherein metabolically distinct leader and follower cells spatiotemporally alternate leading collective packs to overcome the energy barrier required for effective invasion through dense collagen matrices (Zhang et al., 2019). Furthermore, ablation and isolation experiments wherein leader or follower cells were deprived of heterotypic interactions demonstrated suppression of overall invasion in a myriad of cancer subtypes (Yang et al., 2019; Zhang et al., 2019; Khalil et al., 2020). Despite these advances, the underlying mechanisms governing the interactions between collectively invading and single-cell subpopulations have yet to be explored.
Here, we sought to isolate the distinct subpopulations observed in triple-negative mammary adenocarcinoma invasion to determine how invasively heterogenous subpopulations interact. We implemented an image-guided technique developed in our laboratory to precisely isolate cells that invade as collective packs or as single cells. RNA sequencing of collectively invading cells and single cells revealed vastly distinct transcriptional programs between these subpopulations. This prompted us to take a multiomic approach involving RNA sequencing, methylation, and proteomic analysis to identify proteins that collectively invading cells secrete to alter single cells, and found laminin-332 components to be highly expressed and secreted in collectively invading cells. We show that laminin-332 increases the velocity and directionality of single cells through live-cell imaging and uncover an enhanced capability for singles to utilize laminin-332 through integrin α6β4 to hyperactivate Rac1. Our work sheds light on the transcriptional and behavioral heterogeneity between collectively invading and single cells and suggests a novel commensal relationship—whereby one subpopulation benefits from another subpopulation that is not being harmed nor benefited—between these subpopulations within a highly heterogeneous triple-negative breast cancer model.
Results
Collectively invading cells and single cells can be isolated as distinct invasive subpopulations
We used Spatiotemporal Genomic and Cellular Analysis (SaGA) to isolate collectively invading cells and single cells from invading 4T1 spheroids (Khatib et al., 2023). Collectively invading cells were identified and isolated as cells invading while maintaining cell–cell junctions. Within a chain, leaders were isolated as the tip-most cell of a chain. Cells posterior to the leader and separated by 2–3 intermediate cells were isolated as followers (Commander et al., 2020). Conversely, singles were identified and isolated as individual cells that were not visually attached to other cells and were separated from the parental spheroid by multiple cell body lengths (Fig. 1, A and B). After sorting, isolated and purified leaders, followers, and singles were maintained separately and their phenotypes were assessed in 2D and 3D cultures. In 2D culture, leaders and followers continued to invade collectively and maintained a high density of E-cadherin positive cell–cell junctions (Fig. 1, C–E; and Fig. S1 A). Singles, in contrast, resembled a rounded amoeboid phenotype with little to no E-cadherin positive cell–cell junctions present between cells (Fig. 1, C–E; and Fig. S1 A). We did not observe expression of other cadherin junction proteins such as N-cadherin or P-cadherin within any of our subpopulations (Fig. 1 D). When embedded in a 3D type I collagen microenvironment, SaGA-isolated leaders and followers exhibited collective movement, wherein cells would aggregate in chains to penetrate the surrounding stroma in a linear, unidirectional manner while singles invaded exclusively as detached, individual cells (Fig. 1 C; and Videos 1 and 2). Surprisingly, these morphological and invasive phenotypes were retained over >40 passages, suggesting that leaders, followers, and singles are not merely cells existing in transient phases, but stable subpopulations with distinct programs that define their cellular behavior. Importantly, we have also observed invasive heterogeneity in the human SUM159 and HCC38 triple-negative breast cancer cell lines. Similar to the 4T1 parental cell line, we observed both cell–cell adherent collectively invading cells and detached single cells within the same spheroid (Fig. S1 B). Taken together, these data support the presence of phenotypic heterogeneity across multiple cancer models, consistent with other published reports (Khalil et al., 2017; Ilina et al., 2018; Henriet et al., 2023).
Subpopulations derived from the invasively heterogeneous 4T1 cell line are morphologically distinct. (A) Schematic detailing the criteria by which leaders, followers, and singles would be identified and isolated via SaGA. (B) Brightfield image (10×) of a 3D 4T1 parental spheroid embedded in 3.0 mg/ml rat-tail collagen type I after 24 h. Select leaders, followers, and singles are encircled with red, blue, and green circles, respectively, and zoomed in. Scale bar, 50 μm. (C) Brightfield images of 4T1 parentals and purified leaders, followers, and singles in 2D (20×) and 3D culture (10×). Scale bar, 50 μm. (D) Protein levels of E-cadherin, N-cadherin, and P-cadherin in whole-cell lysates of 4T1 parentals, leaders, followers, and singles. Actin was used as a loading control. (E) Immunofluorescence images of E-cadherin on 3D spheroids of 4T1 parentals, leaders, followers, and singles at 1.25× zoom (left) and 5.0× zoom (right). Images were acquired at 10× magnification. Yellow in the overlay image denotes nuclei fluorescence emitted from H2B-Dendra2 and red denotes E-cadherin staining. Scale bar, 50 μm. For D and E, three biological replicates were performed. Source data are available for this figure: SourceData F1.
Subpopulations derived from the invasively heterogeneous 4T1 cell line are morphologically distinct. (A) Schematic detailing the criteria by which leaders, followers, and singles would be identified and isolated via SaGA. (B) Brightfield image (10×) of a 3D 4T1 parental spheroid embedded in 3.0 mg/ml rat-tail collagen type I after 24 h. Select leaders, followers, and singles are encircled with red, blue, and green circles, respectively, and zoomed in. Scale bar, 50 μm. (C) Brightfield images of 4T1 parentals and purified leaders, followers, and singles in 2D (20×) and 3D culture (10×). Scale bar, 50 μm. (D) Protein levels of E-cadherin, N-cadherin, and P-cadherin in whole-cell lysates of 4T1 parentals, leaders, followers, and singles. Actin was used as a loading control. (E) Immunofluorescence images of E-cadherin on 3D spheroids of 4T1 parentals, leaders, followers, and singles at 1.25× zoom (left) and 5.0× zoom (right). Images were acquired at 10× magnification. Yellow in the overlay image denotes nuclei fluorescence emitted from H2B-Dendra2 and red denotes E-cadherin staining. Scale bar, 50 μm. For D and E, three biological replicates were performed. Source data are available for this figure: SourceData F1.
Human triple-negative breast cancer cell lines are invasively heterogeneous. (A) High magnification images (20× and 63×) of E-cadherin immunofluorescence on 3D spheroids of 4T1 leaders, followers, and singles. Green in the overlay image denotes nuclei fluorescence emitted from H2B-Dendra2 and red denotes E-cadherin staining. Scale bar, 50 µm. (B) Brightfield image of a SUM159 (10×) and HCC38 (20×) spheroid embedded in rat-tail collagen type I after 24 h invasion. Red circles denote collective chains and green circles denote single cells. Scale bar, 50 µm.
Human triple-negative breast cancer cell lines are invasively heterogeneous. (A) High magnification images (20× and 63×) of E-cadherin immunofluorescence on 3D spheroids of 4T1 leaders, followers, and singles. Green in the overlay image denotes nuclei fluorescence emitted from H2B-Dendra2 and red denotes E-cadherin staining. Scale bar, 50 µm. (B) Brightfield image of a SUM159 (10×) and HCC38 (20×) spheroid embedded in rat-tail collagen type I after 24 h invasion. Red circles denote collective chains and green circles denote single cells. Scale bar, 50 µm.
3D invasion of a collectively invading spheroid. Live-cell imaging of a 4T1 leader spheroid (1,000 cells) invading through 3.0 mg/ml rat-tail collagen type I using confocal microscopy. All cells express nuclear H2B-Dendra2 (green) but only some are shown as fluorescent due to a single z-plane being imaged. Images were acquired every 10 min. Time scale in seconds. Movie displays 3 frames per second.
3D invasion of a collectively invading spheroid. Live-cell imaging of a 4T1 leader spheroid (1,000 cells) invading through 3.0 mg/ml rat-tail collagen type I using confocal microscopy. All cells express nuclear H2B-Dendra2 (green) but only some are shown as fluorescent due to a single z-plane being imaged. Images were acquired every 10 min. Time scale in seconds. Movie displays 3 frames per second.
3D invasion of a singles spheroid. Live-cell imaging of a 4T1 singles spheroid (1,000 cells) invading through 3.0 mg/ml rat-tail collagen type I using confocal microscopy. All cells express nuclear H2B-Dendra2 (green) but only some are shown as fluorescent due to a single z-plane being imaged. Images were acquired every 10 min. Time scale in seconds. Movie displays 3 frames per second.
3D invasion of a singles spheroid. Live-cell imaging of a 4T1 singles spheroid (1,000 cells) invading through 3.0 mg/ml rat-tail collagen type I using confocal microscopy. All cells express nuclear H2B-Dendra2 (green) but only some are shown as fluorescent due to a single z-plane being imaged. Images were acquired every 10 min. Time scale in seconds. Movie displays 3 frames per second.
Collectively invading cells and singles are transcriptionally and epigenetically distinct
To investigate potential underlying drivers governing the invasive phenotypes of the leader, follower, and single subpopulations, we performed bulk RNA sequencing on our purified subpopulations (Fig. 2 A). Principal component analysis revealed that singles have a distinct transcriptional profile when compared with leaders and followers (Fig. 2 B). Leaders and singles exhibited 1,412 differentially expressed genes (DEGs; | log2 fold change | >2, adjusted P value <0.05), while followers and singles exhibited 991 DEGs (Fig. 2 D); importantly, both pair-wise comparisons included E-cadherin (Cdh1) as a highly expressed gene transcript in leaders and followers relative to singles (Fig. 2 C). In addition to Cdh1, leaders and followers also overexpressed Tacstd2, Krt14, Esrp1, Cldn4, and Lama3, all of which exhibited greater than a 7.0 log2 fold change when compared to singles (Fig. 2 E). When Gene Ontology was performed, leaders and followers showed enrichment in multiple gene sets including substrate-dependent cell migration (GO:0006695), cell–cell adhesion (GO:0098609), and tight junction assembly (GO:0120192) when compared with single cells. Conversely, single cells overexpressed Loxl3, Prex1, and Il17rd when compared with collectively invading cells (Fig. 2 C) and showed enrichment in collagen biosynthesis and modifying enzymes (R-HSA-1650814), RAC1 GTPase cycle (R-HSA-9013149), and signaling by receptor tyrosine kinases (R-HSA-9006934) gene sets.
Collectively invading and single cells exhibit distinct transcriptional and epigenetic programs. (A) Heat maps from RNA sequencing data for each pair-wise comparison. Scale denotes z scores from log2-normalized expression counts of most DEGs. (B) Principal component (PC) analysis plot of leaders, followers, and singles based on RNA sequencing data (n = 3). (C) Volcano plots denoting DEGs for each pair-wise comparison. DEGs (small, unbordered red dots) were classified as gene transcripts with −Log10 P values of >1.3 (y-axis) and a log2 fold change difference of >2.0 or less than −2.0 (x-axis). Bordered red dots are select genes overexpressed in collectively invading cells and bordered green dots are select genes overexpressed in single cells. (D) Number of DEGs between each pair-wise comparison. (E) RNA counts for Cdh1, Cldn4, Tacstd2, Esrp1, Lama3, and Krt14 in leaders, followers, and singles (n = 3). (F) Heat map of mouse methylation beta values between leaders and singles and representation of the percentage of hypermethylated regions across all DMRs between leaders and singles. Scale on heat map denotes beta value difference values for each DMR. L-1, L-2, and L-3 denote three replicates of leaders and S-1, S-2, and S-3 denote three replicates of singles. (G) Annotation of DMRs in singles when compared to leaders. (H) Beta value comparison of CpG loci within the promoter region of Cdh1 (n = 7), Cldn4 (n = 4), Tacstd2 (n = 5), Esrp1 (n = 6), Lama3 (n = 7), and Krt14 (n = 3) between leaders and singles. For all panels: mean ± SEM is shown. Unless noted, n.s., no significance, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
Collectively invading and single cells exhibit distinct transcriptional and epigenetic programs. (A) Heat maps from RNA sequencing data for each pair-wise comparison. Scale denotes z scores from log2-normalized expression counts of most DEGs. (B) Principal component (PC) analysis plot of leaders, followers, and singles based on RNA sequencing data (n = 3). (C) Volcano plots denoting DEGs for each pair-wise comparison. DEGs (small, unbordered red dots) were classified as gene transcripts with −Log10 P values of >1.3 (y-axis) and a log2 fold change difference of >2.0 or less than −2.0 (x-axis). Bordered red dots are select genes overexpressed in collectively invading cells and bordered green dots are select genes overexpressed in single cells. (D) Number of DEGs between each pair-wise comparison. (E) RNA counts for Cdh1, Cldn4, Tacstd2, Esrp1, Lama3, and Krt14 in leaders, followers, and singles (n = 3). (F) Heat map of mouse methylation beta values between leaders and singles and representation of the percentage of hypermethylated regions across all DMRs between leaders and singles. Scale on heat map denotes beta value difference values for each DMR. L-1, L-2, and L-3 denote three replicates of leaders and S-1, S-2, and S-3 denote three replicates of singles. (G) Annotation of DMRs in singles when compared to leaders. (H) Beta value comparison of CpG loci within the promoter region of Cdh1 (n = 7), Cldn4 (n = 4), Tacstd2 (n = 5), Esrp1 (n = 6), Lama3 (n = 7), and Krt14 (n = 3) between leaders and singles. For all panels: mean ± SEM is shown. Unless noted, n.s., no significance, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
Interestingly, leaders and followers exhibited only 416 DEGs, suggesting that leaders and followers within the 4T1 cell line are transcriptionally similar subpopulations (Fig. 2 D). This result is consistent with evidence presented previously (Zhang et al., 2019) detailing the fluidity of leader and follower phenotypes in breast cancer models. Due to this phenotypic and transcriptional fluidity, we proceeded to represent collectively invading cells with the leader subpopulation.
To determine whether the transcriptomic differences between leaders and singles have an underlying epigenetic basis, we performed a methylation analysis on leaders and singles. Like the RNA sequencing analysis, we observed stark differences in the methylation patterns of CpG sites across the epigenome of leaders and singles (Fig. 2 F). Leaders and singles exhibited 1,714 differentially methylated regions (DMR; beta difference <−0.2 and >0.2), with singles exhibiting hypermethylation in 95.9% of its DMRs (Fig. 2 G). Among the hypermethylated CpG sites in singles, CpG sites across the gene bodies of Cdh1, Tacstd2, Krt14, Esrp1, Cldn4, and Lama3 were significantly hypermethylated in singles with a mean beta score of 0.17, while being hypomethylated in leaders at a mean beta score of 0.75 (Fig. 2 H). Notably, Cdh1, Tacstd2, Cldn4, and Lama3 each had at least one differentially methylated position (DMP) within the promoter region. These data suggest that the methylation patterns that differentiate leaders and singles parallel the transcriptomic differences found between these subpopulations in the RNA sequencing analysis. These similarities support a methylation-driven transcriptional heterogeneity of invasively distinct subpopulations, with singles notably exhibiting downregulation of key genes due to inherent hypermethylation of CpG sites across its cell genome. Taken together, these analyses show that collectively invading cells and single cells have distinct transcriptional and epigenetic programs that likely underlie their respective morphological and behavioral phenotypes.
Multiomic analyses reveal overexpression of laminin-332 in collectively invading cells
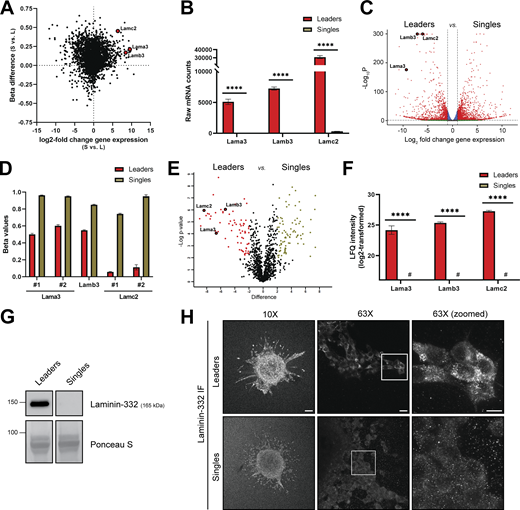
To identify gene transcripts that are both transcriptionally upregulated and hypomethylated at the promoter region of collectively invading cells relative to singles, we integrated RNA sequencing and methylation array data from leaders and singles. Among the nine most overexpressed and hypomethylated gene transcripts in leaders, we identified two components of the laminin-332 complex, laminin α chain-3 (Lama3), and laminin γ chain-2 (Lamc2) (Fig. 3 A). Compared with singles, leaders expressed 673-fold higher mRNA counts of Lama3 and 95-fold higher mRNA counts of Lamc2 (Fig. 3, B and C), while also exhibiting a mean beta difference of 0.23 and 0.45 in the promoter regions of the Lama3 and Lamc2 genes, respectively (Fig. 3 D). Additionally, the third component of the laminin-332 complex, laminin β chain-3 (Lamb3) had 144-fold higher mRNA counts in leaders (Fig. 3 B) while also exhibiting a mean beta difference of 0.30 in the gene promoter (Fig. 3 C). To confirm that this enrichment in laminin subunits in leaders was not due to an overexpression of all laminins, we measured the relative expression of each laminin detected within our RNA sequencing dataset. Surprisingly, only 5 out of the 11 laminin subunits detected within the array were overexpressed in leaders, and the three most overexpressed subunits were Lama3, Lamb3, and Lamc2, confirming the specific upregulation of the laminin-332 complex within leaders (Fig. S2).
Collectively invading cells overexpress and abundantly secrete laminin-332. (A) Integration of RNA sequencing log2 fold change values (x-axis) and mouse methylation array beta difference values (y-axis) for the leaders (L) versus singles (S) pair-wise comparison. Lama3, Lamb3, and Lamc2 are highlighted as red dots among gene transcripts with high mRNA transcription in leaders and significant hypomethylation at the promoter region compared to singles. (B) Raw mRNA counts for gene transcripts of the laminin-332 subunits from RNA sequencing analysis (n = 3, ****P < 0.0001). (C) Volcano plot of the leaders versus singles pair-wise comparison derived from RNA sequencing data highlighting Lama3, Lamb3, and Lamc2 (bordered red dots) as prominent DEGs. (D) Beta value comparison of distinct CpG loci within the promoter regions of Lama3, Lamb3, and Lamc2 between leaders and singles. (E) Volcano plot of differentially secreted peptides extracted via LC-MS/MS from CM from leaders and singles. Red, unbordered dots denote peptides differentially secreted in leaders and green dots denote peptides differentially secreted in singles. (F) Label-free quantification (LFQ) intensity quantification of Lama3, Lamb3, and Lamc2 peptides. # denotes an absence of signal detected (n = 3, ****P < 0.0001). (G) Protein levels of laminin-332 in leaders CM and singles CM. Total protein staining via Ponceau S was used as a loading control. (H) Laminin-332 immunofluorescence (IF) staining on invasive leaders and singles in collagen I 3D culture (10× and 63×). Scale bar: 100 μm for 10× image, 20 μm for 63× image, and 10 μm for 63× zoomed image. For all panels: mean ± SEM is shown. For G and H, three biological replicates were performed. Source data are available for this figure: SourceData F3.
Collectively invading cells overexpress and abundantly secrete laminin-332. (A) Integration of RNA sequencing log2 fold change values (x-axis) and mouse methylation array beta difference values (y-axis) for the leaders (L) versus singles (S) pair-wise comparison. Lama3, Lamb3, and Lamc2 are highlighted as red dots among gene transcripts with high mRNA transcription in leaders and significant hypomethylation at the promoter region compared to singles. (B) Raw mRNA counts for gene transcripts of the laminin-332 subunits from RNA sequencing analysis (n = 3, ****P < 0.0001). (C) Volcano plot of the leaders versus singles pair-wise comparison derived from RNA sequencing data highlighting Lama3, Lamb3, and Lamc2 (bordered red dots) as prominent DEGs. (D) Beta value comparison of distinct CpG loci within the promoter regions of Lama3, Lamb3, and Lamc2 between leaders and singles. (E) Volcano plot of differentially secreted peptides extracted via LC-MS/MS from CM from leaders and singles. Red, unbordered dots denote peptides differentially secreted in leaders and green dots denote peptides differentially secreted in singles. (F) Label-free quantification (LFQ) intensity quantification of Lama3, Lamb3, and Lamc2 peptides. # denotes an absence of signal detected (n = 3, ****P < 0.0001). (G) Protein levels of laminin-332 in leaders CM and singles CM. Total protein staining via Ponceau S was used as a loading control. (H) Laminin-332 immunofluorescence (IF) staining on invasive leaders and singles in collagen I 3D culture (10× and 63×). Scale bar: 100 μm for 10× image, 20 μm for 63× image, and 10 μm for 63× zoomed image. For all panels: mean ± SEM is shown. For G and H, three biological replicates were performed. Source data are available for this figure: SourceData F3.
Fold change difference of laminin gene expression between leaders and singles. Positive values denote higher expression in leaders (L) relative to singles (S) and negative values denote higher expression in singles relative to leaders.
Fold change difference of laminin gene expression between leaders and singles. Positive values denote higher expression in leaders (L) relative to singles (S) and negative values denote higher expression in singles relative to leaders.
To determine if laminin-332 protein was preferentially secreted by leaders, we performed an unbiased proteomic analysis using mass spectrometry on purified protein extracts from conditioned media (CM) of leaders and singles. Among the 17 most differentially secreted proteins present in leader CM, laminin subunits α3 (Lama3), β3 (Lamb3), and γ2 (Lamc2) were all highly abundant exclusively in leaders (Fig. 3 E). In contrast, no protein was detected for Lama3, Lamb3, and Lamc2 in singles, corroborating the binary nature of laminin-332 secretion by leaders (Fig. 3 F). Western blot analysis also revealed that laminin-332 was drastically overabundant in leader CM when compared with singles (Fig. 3 G). 2D and 3D immunofluorescence staining using a laminin-332 antibody revealed higher expression of laminin-332 in leader spheroids than single spheroids (Fig. 3 H). Together, these data demonstrate that components of the laminin-332 complex are not only transcriptionally and epigenetically overrepresented in collectively invading cells, but also exclusively and abundantly secreted as proteins by collectively invading cells.
Laminin-332 promotes the 3D invasion of singles
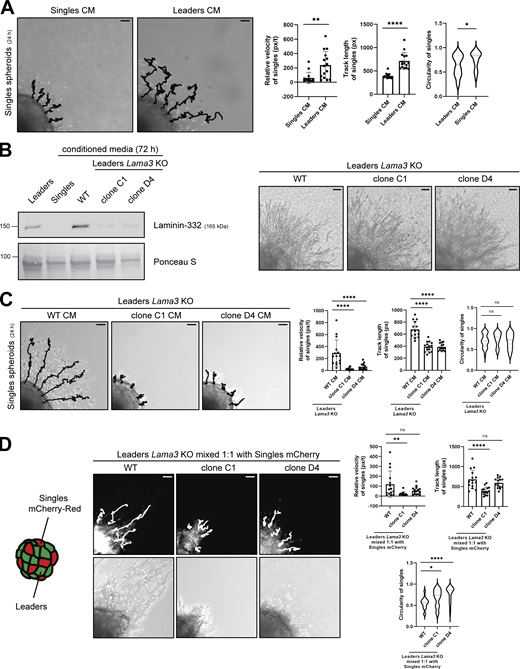
The secretion of laminin-332 complex exclusively by collectively invading cells led us to hypothesize that collectively invading cells could influence the invasive potential of single cells via laminin-332. To test this, we first observed the behavior of singles when treated with laminin-332–high leader CM compared with laminin-332-low single CM in a 3D spheroid model. Within a 24-h time period, singles treated with leader CM exhibited a significantly higher velocity and track length than singles treated with single CM (Fig. 4 A; and Videos 3 and 4). Additionally, singles treated with leader CM displayed a higher potential for directional movement through decreased cellular circularity and a higher meandering index (defined as displacement divided by distance) than singles treated with single CM (Fig. 4 A and Fig. S3 A). To confirm the specific role of laminin-332 in the elevated invasive potential of singles, we treated singles with CM from leaders with a CRISPR/Cas9 knockout (KO) of the Lama3 gene. Western blot analysis of the CM of the KO cells confirmed that Lama3 KO was sufficient for reducing laminin-332 complex assembly and did not hinder the viability or invasive behavior of leaders (Fig. 4 B and Fig. S3 D). Singles treated with CM from two distinct clones of Lama3 KO leaders (clones C1 and D4) showed considerable repression of cell movement, velocity, and directionality when compared with singles treated with CM from leaders with only the Cas9 endonuclease transfected (Lama3 WT cells) (Fig. 4 C; Fig. S3 B; and Videos 5, 6, and 7).
Laminin-332 enhances the invasive potential of singles. (A) Live-cell tracking analysis of singles spheroids after treating with singles CM or leaders CM for 24 h (n = 15). Representative five tracks highlighted for each group. Scale bar, 50 μm. (B) Protein levels of laminin-332 in CM extracted from leaders with WT Lama3, and Lama3 CRISPR/Cas9 KO (clones C1 and D4). Total protein staining via Ponceau S was used as a loading control. Brightfield images were acquired after 24 h. Scale bar, 50 μm. (C) Live-cell tracking analysis of singles spheroids after treating with CM from WT Lama3 leaders and Lama3 KO leaders (clones C1 and D4) (n = 15). Representative five tracks highlighted for each group. Scale bar, 50 μm. (D) Live-cell tracking analysis of mCherry-transfected singles within a spheroid mixed 1:1 with mCherry-transfected singles and either leaders WT, clone C1, or clone D4 (n = 15). Representative five tracks highlighted for each group. Scale bar, 50 μm. For all experiments, three biological replicates were performed. For all panels: mean ± SEM is shown. Px/t stands for pixels/time, and px stands for pixels. Unless noted, n.s., no significance, *P ≤ 0.05, **P ≤ 0.01, ****P ≤ 0.0001. Source data are available for this figure: SourceData F4.
Laminin-332 enhances the invasive potential of singles. (A) Live-cell tracking analysis of singles spheroids after treating with singles CM or leaders CM for 24 h (n = 15). Representative five tracks highlighted for each group. Scale bar, 50 μm. (B) Protein levels of laminin-332 in CM extracted from leaders with WT Lama3, and Lama3 CRISPR/Cas9 KO (clones C1 and D4). Total protein staining via Ponceau S was used as a loading control. Brightfield images were acquired after 24 h. Scale bar, 50 μm. (C) Live-cell tracking analysis of singles spheroids after treating with CM from WT Lama3 leaders and Lama3 KO leaders (clones C1 and D4) (n = 15). Representative five tracks highlighted for each group. Scale bar, 50 μm. (D) Live-cell tracking analysis of mCherry-transfected singles within a spheroid mixed 1:1 with mCherry-transfected singles and either leaders WT, clone C1, or clone D4 (n = 15). Representative five tracks highlighted for each group. Scale bar, 50 μm. For all experiments, three biological replicates were performed. For all panels: mean ± SEM is shown. Px/t stands for pixels/time, and px stands for pixels. Unless noted, n.s., no significance, *P ≤ 0.05, **P ≤ 0.01, ****P ≤ 0.0001. Source data are available for this figure: SourceData F4.
3D invasion of a singles spheroid treated with leaders CM. Live-cell imaging of a 4T1 singles spheroid (1,000 cells) treated with leaders CM using confocal microscopy. Spheroids are embedded in 3.0 mg/ml rat-tail collagen type I. Images were acquired every 20 min. Time scale in seconds. Movie displays 3 frames per second. End-point still of the movie is found in Fig. 4 A.
3D invasion of a singles spheroid treated with leaders CM. Live-cell imaging of a 4T1 singles spheroid (1,000 cells) treated with leaders CM using confocal microscopy. Spheroids are embedded in 3.0 mg/ml rat-tail collagen type I. Images were acquired every 20 min. Time scale in seconds. Movie displays 3 frames per second. End-point still of the movie is found in Fig. 4 A.
3D invasion of a singles spheroid treated with singles CM. Live-cell imaging of a 4T1 singles spheroid (1,000 cells) treated with singles CM using confocal microscopy. Spheroids are embedded in 3.0 mg/ml rat-tail collagen type I. Images were acquired every 20 min. Time scale in seconds. Movie displays 3 frames per second. End-point still of the movie is found in Fig. 4 A.
3D invasion of a singles spheroid treated with singles CM. Live-cell imaging of a 4T1 singles spheroid (1,000 cells) treated with singles CM using confocal microscopy. Spheroids are embedded in 3.0 mg/ml rat-tail collagen type I. Images were acquired every 20 min. Time scale in seconds. Movie displays 3 frames per second. End-point still of the movie is found in Fig. 4 A.
Lama3 KO does not affect leader cell viability and suppresses directional movement of single cells. (A–C) Meandering index quantification for singles (A) treated with leader or singles CM, (B) Lama3 KO leaders CM, and (C) mixed 1:1 with Lama3 KO leaders (n = 15). (D) Quantification of apoptotic events on leaders with Lama3 WT and KO (n = 2). For all panels: mean ± SEM is shown. Unless noted, ns, no significance, *P ≤ 0.05, ***P ≤ 0.001, ****P ≤ 0.0001.
Lama3 KO does not affect leader cell viability and suppresses directional movement of single cells. (A–C) Meandering index quantification for singles (A) treated with leader or singles CM, (B) Lama3 KO leaders CM, and (C) mixed 1:1 with Lama3 KO leaders (n = 15). (D) Quantification of apoptotic events on leaders with Lama3 WT and KO (n = 2). For all panels: mean ± SEM is shown. Unless noted, ns, no significance, *P ≤ 0.05, ***P ≤ 0.001, ****P ≤ 0.0001.
3D invasion of a singles spheroid treated with CM from Lama3 WT leaders. Live-cell imaging of a 4T1 singles spheroid (1,000 cells) treated with CM from Lama3 WT leaders. Spheroids are embedded in 3.0 mg/ml rat-tail collagen type I. Images were acquired every 20 min. Time scale in seconds. Movie displays 3 frames per second. End-point still of the movie is found in Fig. 4 C.
3D invasion of a singles spheroid treated with CM from Lama3 WT leaders. Live-cell imaging of a 4T1 singles spheroid (1,000 cells) treated with CM from Lama3 WT leaders. Spheroids are embedded in 3.0 mg/ml rat-tail collagen type I. Images were acquired every 20 min. Time scale in seconds. Movie displays 3 frames per second. End-point still of the movie is found in Fig. 4 C.
3D invasion of a singles spheroid treated with CM from Lama3 KO leader clone C1. Live-cell imaging of a 4T1 singles spheroid (1,000 cells) treated with CM from Lama3 KO leader clone C1. Spheroids are embedded in 3.0 mg/ml rat-tail collagen type I. Images were acquired every 20 min. Time scale in seconds. Movie displays 3 frames per second. End-point still of the movie is found in Fig. 4 C.
3D invasion of a singles spheroid treated with CM from Lama3 KO leader clone C1. Live-cell imaging of a 4T1 singles spheroid (1,000 cells) treated with CM from Lama3 KO leader clone C1. Spheroids are embedded in 3.0 mg/ml rat-tail collagen type I. Images were acquired every 20 min. Time scale in seconds. Movie displays 3 frames per second. End-point still of the movie is found in Fig. 4 C.
3D invasion of a singles spheroid treated with CM from Lama3 KO leader clone D4. Live-cell imaging of a 4T1 singles spheroid (1,000 cells) treated with CM from Lama3 KO leader clone D4. Spheroids are embedded in 3.0 mg/ml rat-tail collagen type I. Images were acquired every 20 min. Time scale in seconds. Movie displays 3 frames per second. End-point still of the movie is found in Fig. 4 C.
3D invasion of a singles spheroid treated with CM from Lama3 KO leader clone D4. Live-cell imaging of a 4T1 singles spheroid (1,000 cells) treated with CM from Lama3 KO leader clone D4. Spheroids are embedded in 3.0 mg/ml rat-tail collagen type I. Images were acquired every 20 min. Time scale in seconds. Movie displays 3 frames per second. End-point still of the movie is found in Fig. 4 C.
We further validated the role of laminin-332 in stimulating the invasive potential of single cells by co-culturing Lama3-expressing or Lama3-null leaders with singles within a mixed 3D spheroid. Singles were transfected with mCherry-Red and mixed with leaders with Lama3 WT or Lama3 KO at a 1:1 ratio within a single spheroid, and allowed to invade in collagen type I over 48 h (Fig. 4 D). Singles that were mixed with leaders with Lama3 KO invaded significantly less and with less directionality than leaders with Lama3 WT (Fig. 4 D; Fig. 3 C; and Videos 8, 9, and 10), supporting a model whereby local laminin-332 secretion by collectively invading cells promotes directional invasion in single cells within a heterogeneous tumor environment.
3D invasion of a mixed spheroid made up of singles and Lama3 WT leaders. Fluorescence live-cell imaging of a mixed spheroid (1,000 cells) composed of a 1:1 ratio of Lama3 WT leaders and mCherry-tagged singles. Only mCherry-tagged cells are shown. Spheroids are embedded in 3.0 mg/ml rat-tail collagen type I. Images were acquired every 20 min. Time scale in seconds. Movie displays 3 frames per second. End-point still of the movie is found in Fig. 4 D.
3D invasion of a mixed spheroid made up of singles and Lama3 WT leaders. Fluorescence live-cell imaging of a mixed spheroid (1,000 cells) composed of a 1:1 ratio of Lama3 WT leaders and mCherry-tagged singles. Only mCherry-tagged cells are shown. Spheroids are embedded in 3.0 mg/ml rat-tail collagen type I. Images were acquired every 20 min. Time scale in seconds. Movie displays 3 frames per second. End-point still of the movie is found in Fig. 4 D.
3D invasion of a mixed spheroid made up of singles and Lama3 KO clone C1 leaders. Fluorescence live-cell imaging of a mixed spheroid (1,000 cells) composed of a 1:1 ratio of Lama3 KO clone C1 leaders and mCherry-tagged singles. Only mCherry-tagged cells are shown. Spheroids are embedded in 3.0 mg/ml rat-tail collagen type I. Images were acquired every 20 min. Time scale in seconds. Movie displays 3 frames per second. End-point still of the movie is found in Fig. 4 D.
3D invasion of a mixed spheroid made up of singles and Lama3 KO clone C1 leaders. Fluorescence live-cell imaging of a mixed spheroid (1,000 cells) composed of a 1:1 ratio of Lama3 KO clone C1 leaders and mCherry-tagged singles. Only mCherry-tagged cells are shown. Spheroids are embedded in 3.0 mg/ml rat-tail collagen type I. Images were acquired every 20 min. Time scale in seconds. Movie displays 3 frames per second. End-point still of the movie is found in Fig. 4 D.
3D invasion of a mixed spheroid made up of singles and Lama3 KO clone D4 leaders. Fluorescence live-cell imaging of a mixed spheroid (1,000 cells) composed of a 1:1 ratio of Lama3 KO clone D4 leaders and mCherry-tagged singles. Only mCherry-tagged cells are shown. Spheroids are embedded in 3.0 mg/ml rat-tail collagen type I. Images were acquired every 20 min. Time scale in seconds. Movie displays 3 frames per second. End-point still of the movie is found in Fig. 4 D.
3D invasion of a mixed spheroid made up of singles and Lama3 KO clone D4 leaders. Fluorescence live-cell imaging of a mixed spheroid (1,000 cells) composed of a 1:1 ratio of Lama3 KO clone D4 leaders and mCherry-tagged singles. Only mCherry-tagged cells are shown. Spheroids are embedded in 3.0 mg/ml rat-tail collagen type I. Images were acquired every 20 min. Time scale in seconds. Movie displays 3 frames per second. End-point still of the movie is found in Fig. 4 D.
Singles hyperactivate Rac1 upon laminin-332 binding
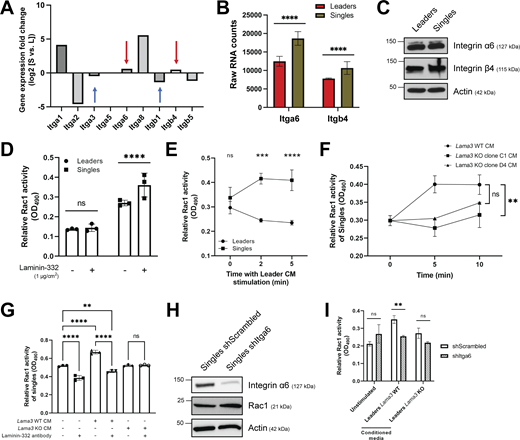
The effect of laminin-332 on the invasive potential of singles led us to investigate the underlying molecular mechanism by which cell movement was being stimulated within singles. We sought to test whether singles overexpress receptors that specifically bind to laminin-332. Canonically, laminin-332 binds to the integrin pairs α6/β4 and α3/β1, each resulting in the downstream activation of distinct small GTPase signaling cascades, Rac1 and Rho, respectively (Marinkovich, 2007). RNA sequencing analysis of integrin expression between leaders and singles revealed that singles only showed significant transcriptional over-expression of integrins α6 (Itga6) and β4 (Itgb4) relative to leaders, while integrins α3 and β1 were overexpressed in leaders relative to singles (Fig. 5, A and B). Interestingly, western blot analysis on whole-cell lysates of leaders and singles revealed no notable difference in total protein expression of integrins α6 and β4 in singles relative to leaders (Fig. 5 C). Furthermore, flow cytometric detection of cell surface integrin α6 expression also showed no significant differences in protein abundance between leaders and singles (Fig. 4 A). Thus, we hypothesized that singles exclusively activate Rac1 via the integrin α6/β4 receptor, despite leaders also expressing the heterodimer.
Singles activate Rac1 activity via binding of integrin α6β4 to leader-derived laminin-332. (A) Log2 fold change of integrin subunits expressed in leaders (L) and singles (S) within the RNA sequencing data. Positive values denote gene transcripts that were upregulated in singles relative to leaders. Red arrows highlight Itga6 and Itgb4 as being upregulated in singles and blue arrows highlight Itga3 and Itgb1 as being downregulated in singles. (B) RNA counts for Itga6 and Itgb4 in leaders and singles (n = 3). (C) Protein levels of integrin α6 and β4 in whole-cell lysates of leaders and singles. Actin was used as a loading control. (D) Relative Rac1 activity of leaders and singles upon direct interaction with 1 μg/cm2 laminin-332 for 5 min (n = 3). (E) Time course measurement of relative Rac1 activity in leaders and singles upon leader CM treatment (n = 3). (F) Time course measurement of relative Rac1 activity in singles upon treatment with CM from leaders Lama3 WT and two distinct leaders with Lama3 KO (clone C1 and clone D4) (n = 3). Statistical annotation only applies for the 10-min time points. (G) Relative Rac1 activity of singles upon treatment with Lama3 WT or Lama3 KO CM with a laminin-332 antibody (1:1,000 dilution) (n = 3). Laminin-332 antibody was mixed in with CM for 30 min prior to treatment. Rac1 activity of cells was measured after 5 min treatment with CM. (H) Protein levels of integrin α6 and total Rac1 upon Itga6 shRNA knockdown in singles. Actin was used as a loading control. (I) Relative Rac1 activity in singles with Itga6 shRNA knockdown (shItga6) upon treatment with CM from leaders Lama3 WT and leaders Lama3 KO. Rac1 activity was measured after 1 h treatment with CM (n = 3). For C–I, three biological replicates were performed. For all panels: mean ± SEM is shown. Unless noted, n.s., no significance, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001. Source data are available for this figure: SourceData F5.
Singles activate Rac1 activity via binding of integrin α6β4 to leader-derived laminin-332. (A) Log2 fold change of integrin subunits expressed in leaders (L) and singles (S) within the RNA sequencing data. Positive values denote gene transcripts that were upregulated in singles relative to leaders. Red arrows highlight Itga6 and Itgb4 as being upregulated in singles and blue arrows highlight Itga3 and Itgb1 as being downregulated in singles. (B) RNA counts for Itga6 and Itgb4 in leaders and singles (n = 3). (C) Protein levels of integrin α6 and β4 in whole-cell lysates of leaders and singles. Actin was used as a loading control. (D) Relative Rac1 activity of leaders and singles upon direct interaction with 1 μg/cm2 laminin-332 for 5 min (n = 3). (E) Time course measurement of relative Rac1 activity in leaders and singles upon leader CM treatment (n = 3). (F) Time course measurement of relative Rac1 activity in singles upon treatment with CM from leaders Lama3 WT and two distinct leaders with Lama3 KO (clone C1 and clone D4) (n = 3). Statistical annotation only applies for the 10-min time points. (G) Relative Rac1 activity of singles upon treatment with Lama3 WT or Lama3 KO CM with a laminin-332 antibody (1:1,000 dilution) (n = 3). Laminin-332 antibody was mixed in with CM for 30 min prior to treatment. Rac1 activity of cells was measured after 5 min treatment with CM. (H) Protein levels of integrin α6 and total Rac1 upon Itga6 shRNA knockdown in singles. Actin was used as a loading control. (I) Relative Rac1 activity in singles with Itga6 shRNA knockdown (shItga6) upon treatment with CM from leaders Lama3 WT and leaders Lama3 KO. Rac1 activity was measured after 1 h treatment with CM (n = 3). For C–I, three biological replicates were performed. For all panels: mean ± SEM is shown. Unless noted, n.s., no significance, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001. Source data are available for this figure: SourceData F5.
To test this, we first evaluated the response of leaders and singles to Rac1 stimulation via EGF treatment. Interestingly, EGF exclusively led to Rac1 activation in singles, while in leaders we observed a gradual deactivation of Rac1 (Fig. S4 B). To more directly assess the effect of laminin-332 on Rac1 activity, we proceeded to measure the Rac1 activity of leaders and singles upon interaction with purified human laminin-332. Surprisingly, laminin-332 only led to Rac1 activation in single cells, while Rac1 activity remained unchanged in leaders (Fig. 5 D). Similar to purified laminin-332, treatment of leaders and singles to laminin-332-rich leader CM only led to activation of Rac1 in singles, while a gradual deactivation of Rac1 was observed in leaders (Fig. 5 E). Furthermore, treatment of singles with CM from Lama3 WT and two Lama3 KO leader clones yielded significantly higher Rac1 activity upon treatment with leader Lama3 WT CM when compared with the leader Lama3 KO CM after 10 min (Fig. 5 F). To confirm that the secretome of Lama3 WT and KO cells are similar (other than Lama3 protein), we performed mass spectrometry–based proteomic analysis of the CM of these cells and found only 33 differentially abundant proteins (|Student’s T test Difference| >1, P < 0.05) out of 783 total proteins [label-free quantification (LFQ) Score <10], including Lama3, which had the lowest P value by a considerable margin (Fig. S4 C). In addition, treatment of singles with Lama3 WT leader CM with a laminin-332 polyclonal antibody led to a reversal of Rac1 activation, further confirming the direct effect of leader-derived laminin-332 on Rac1 activity in single cells among the complex mixture of molecules found in leader CM (Fig. 5 G).
EGF activates Rac1 in single cells and not collectively invading cells. (A) Flow cytometric surface protein quantification of integrin α6 (Itga6) in leaders and singles. Dotted line denotes the unstained sample for each subpopulation. (B) Time course measurement of relative Rac1 activity in leaders and singles upon 50 ng/ml hEGF treatment (n = 3, ***P < 0.001, ****P < 0.0001). Mean ± SEM is shown. (C) Volcano plot of differentially secreted peptides extracted via LC-MS/MS from CM from leaders with Lama3 WT and KO (n = 3). Red circles denote peptides differentially secreted in Lama3 WT leaders and green circles denote peptides differentially secreted in Lama3 KO leaders (|Student’s T test Difference| >1, P value <0.05).
EGF activates Rac1 in single cells and not collectively invading cells. (A) Flow cytometric surface protein quantification of integrin α6 (Itga6) in leaders and singles. Dotted line denotes the unstained sample for each subpopulation. (B) Time course measurement of relative Rac1 activity in leaders and singles upon 50 ng/ml hEGF treatment (n = 3, ***P < 0.001, ****P < 0.0001). Mean ± SEM is shown. (C) Volcano plot of differentially secreted peptides extracted via LC-MS/MS from CM from leaders with Lama3 WT and KO (n = 3). Red circles denote peptides differentially secreted in Lama3 WT leaders and green circles denote peptides differentially secreted in Lama3 KO leaders (|Student’s T test Difference| >1, P value <0.05).
To confirm that laminin-332 binds to the integrin α6/β4 receptor within singles to activate Rac1, we measured the Rac1 activity of singles with Itga6 knockdown (Fig. 5 G). When unstimulated, we observed no difference in Rac1 activity between the vehicle and the knockdown cells (Fig. 5 H). Upon stimulation with Lama3 WT leader CM, however, we found that only the vehicle cells were able to activate Rac1, while the knockdown cells retained the same Rac1 activity as the unstimulated cells (Fig. 5 H). Additionally, we observed a significant decrease in Rac1 activity in shItga6 singles upon treatment leader Lama3 KO CM relative to the leader Lama3 WT CM, confirming the importance of both laminin-332 and integrin α6 for Rac1 activity in single cells (Fig. 5 H). Taken together, these data support a model whereby single cells utilize integrin α6/β4 receptor binding to laminin-332 derived from collectively invading cells for Rac1 activation.
Discussion
Despite the prevalence of phenotypic heterogeneity across multiple solid tumor types, the mechanisms underlying invasively distinct subpopulations remain largely unexplored. Using SaGA, we isolated and analyzed both collectively invading cells (leaders and followers) and individually invading cells (singles) within the 4T1 murine mammary adenocarcinoma model. We performed RNA sequencing on these purified cell subpopulations to investigate transcriptional differences and found that collectively invading and single cells had vastly different transcriptomes that includes upregulation of key epithelial genes such as Cdh1, Krt14, Esrp1, Cldn4, and Tacstd2 in collectively invading cells. E-cadherin (Cdh1) was only expressed in collectively invading cells and not single cells, which has been observed previously (Khalil et al., 2020), and is required for metastasis in multiple breast cancer models (Padmanaban et al., 2019). Similarly, Keratin-14 (Krt14) is also a well-characterized biomarker for breast cancer leader cells within collective packs (Cheung et al., 2013; Hwang et al., 2019) and had significantly higher expression in collectively invading cells compared to singles. Furthermore, keratin-14 knockdown has previously been shown to abrogate the ability of cells to invade collectively and form clustered metastases (Cheung et al., 2013). The relevance of Esrp1, Cldn4, and Tacstd2 within collectively invading cells is less characterized, but they have been shown to drive cell invasion in multiple tumor models (Hwang et al., 2014; Chen et al., 2017; Wu et al., 2017). In contrast to collectively invading cells, cells that exclusively invade as single cells exhibited very low levels of canonically epithelial genes, including Cdh1, Esrp1, and Tacstd2, suggesting that single cells may represent a hybrid amoeboid state within the epithelial-to-mesenchymal transition (EMT) program (Graziani et al., 2022). Additionally, Gene Ontology on single cells showed statistically significant enrichment of multiple EMT-related pathways, including regulation of epithelial-to-mesenchymal transition (GO:0010717) and epithelial-to-mesenchymal transition (GO:0001837). The phenotypic persistence of single cells over multiple passages would suggest that single cells (and collectively invading cells) are not in active transition within this spectrum, but the influence of related pathways to their single-cell phenotype cannot be ruled out.
Subsequent methylation analyses revealed that collectively invading and single-cell subpopulations exhibit significant epigenetic differences. Interestingly, gene transcripts with the highest fold changes in RNA expression between collectively invading and single cells also had prominent differences in methylation patterns at both the promoter regions and gene bodies of the corresponding genes, including Cdh1, Krt14, Esrp1, Cldn4, Tacstd2, and Lama3. An epigenetic basis for the emergence of collectively invading and single cells is supported by the stability we observe in the distinct phenotypes between collectively invading and single-cell subpopulations throughout multiple passages. The stability of these distinct phenotypes could also be attributed to genomic differences between the subpopulations. Although single tandem repeat (STR) analysis of nine genomic markers revealed no prominent differences between these subpopulations (Table S1), we cannot rule out a possible role for an underlying genomic heterogeneity governing these distinct phenotypes. Despite this, our data provide support for collectively invading cells and single cells having distinct epigenetic programs that shape their distinct transcriptional profiles to influence cellular phenotype.
Integration of RNA sequencing and methylation data identified gene transcripts of components of the laminin-332 trimeric complex—Lama3, Lamb3, and Lamc2—as being highly overexpressed and hypomethylated in collectively invading cells relative to single cells. Furthermore, unbiased secretomic analysis confirmed that components of the laminin-332 complex were being exclusively secreted by collectively invading cells. In healthy skin, laminin-332 is specific to the basement membranes of epithelial tissue and is normally produced and secreted by keratinocytes to bolster cell survival and play a role in wound healing (Nguyen et al., 2000; Fujisaki and Hattori, 2002; Manohar et al., 2004). Similarly, cancer cells of epithelial origin have been found to abnormally express laminin-332 to promote anoikis resistance and induce cell motility (Zahir et al., 2003; Carpenter et al., 2009). Recent work established the relevance of laminin-332 within collective invasion, largely focusing on its role in establishing cell–cell contacts within focal adhesions across collective chains (Kumagai et al., 2019). In healthy skin, the integrin α6β4 heterodimer in epithelial cells and laminin-332 in the underlying basement membrane bind to maintain hemidesmosome integrity across highly structured epithelial tissue (Marinkovich, 2007; Te Molder et al., 2021). Interestingly, we show that laminin-332 can also bind to integrin α6β4 as a secreted factor when integrin α6β4 is expressed in a spatially distant cell. Our work corroborates recent discoveries identifying laminin-332 as a factor secreted by collectively invading cells, further clarifying the role of laminin-332 in triple-negative breast cancer invasion (Hwang et al., 2023).
Interestingly, collectively invading cells and single cells had similar protein expression of the integrin α6β4 heterodimer, suggesting that laminin-332 binding to single cells is likely not an exclusively paracrine interaction. Autocrine laminin-332 binding has been reported in skin squamous cell carcinoma and mammary tumor models, wherein laminin-332 binding to syndecan-1 or EGFR leads to further laminin-332 deposition in a positive-feedback loop (Zahir et al., 2003; Okamoto et al., 2003; Baba et al., 2008). Despite this, we showed that one downstream target of integrin α6β4, Rac1, is exclusively activated in singles upon treatment with purified laminin-332 (Fig. 5 D) and laminin-332–rich CM derived from collectively invading cells (Fig. 5 E). Current work supports multiple possible explanations for this duality in Rac1 activation, including mutations or splice variants of either integrin α6 or β4. Instances of such mutations within the ITGB4 genes have been reported in cases of epidermolysis bullosa, wherein the mutations are predicted to reduce integrin α6β4 heterodimer stability (Paine et al., 2022). Additionally, splice variants of integrins α6 and β4 that affect proper dimerization and cell membrane anchoring have been reported with different downstream targets such as ERK and JNK (Chen et al., 2022; Boussaha et al., 2023). Further investigation is required to elucidate the stark differences in Rac1 activity stimulation between collectively invading cells and single cells despite similar membrane-bound integrin α6 and β4 expression.
Our work explores the intricacies of phenotypic heterogeneity within triple-negative cancer and unravels a commensal interaction between phenotypically distinct cancer subpopulations. The attribution of ecological relationships to subpopulation cellular dynamics has been of recent interest, namely in the identification of cooperative interactions between subpopulations of heterogeneous cancer cell lines (Mateo et al., 2014; Konen et al., 2017; Summerbell et al., 2020; Campbell et al., 2021). Thus far, breast cancer cells cooperate through leader–follower dynamics within collectively invading chains to overcome energetic barriers presented by dense collagen matrices through perpetual “shuffling” of leader and follower cells (Zhang et al., 2019). Additionally, lung adenocarcinoma models have profound increases in cell survival and DNA repair within highly dynamic leaders through secreted factors found in follower CM (Konen et al., 2017). More recent evidence has revealed the advantage cooperating clustered cells have as opposed to individual invasive cells in overcoming immune attack from natural killer cells (Lo et al., 2020). Our work describes a commensal relationship, wherein collectively invading cells confer a unilateral invasive benefit unto single cells. To our knowledge, this represents a novel commensal interaction between cancer cell subpopulations, amid extensive work that has characterized commensal interactions between the gut or skin and their respective microbiota (Kumamoto et al., 2020; Zheng et al., 2022; Yang et al., 2023). We speculate that Rac1 activation in singles due to laminin-332 binding could be one aspect of a cooperative effort between collectively invading cells and single cells, but further work is necessary to test the model that actively invasive single cells benefit collectively invading cells to enhance the metastatic potential of a heterogeneous tumor.
In summary, we isolated and characterized collectively invading and single cells within a highly heterogeneous murine triple-negative breast cancer model and discovered the secretion of the laminin-332 complex exclusively by collectively invading cells. We show that laminin-332 derived from collectively invading cells enhances the invasive potential of singles and also stimulates the Rac1 activity of singles. Through this mechanism, we suggest that collectively invading and single cells display a commensal relationship, wherein collectively invading cells unidirectionally benefit single cells. This commensal relationship does not exclude the possibility for additional work revealing a mutualistic relationship wherein both populations benefit one another. Our work contributes toward a more complete understanding of how phenotypic heterogeneity and subpopulation dynamics precede cell invasion and metastatic progression, which we anticipate will be crucial to developing personalized treatments against this ever-evolving heterogeneous disease.
Materials and methods
Cell culture conditions
4T1 cells (American Type Culture Collection; gift from Harold Moses [Vanderbilt University, Nashville, TN, USA]) were cultured in DMEM media treated with penicillin-streptomycin (100 U ml−1) and supplemented with 10% fetal bovine serum (FBS) and maintained at 37°C and 5% CO2. SUM159 cells (gift from Paula Vertino [University of Rochester, Rochester, NY, USA]) were cultured in DMEM/F12 media supplemented with 5% FBS, 5 μg/ml insulin, 1 μg/ml hydrocortisone and treated with penicillin-streptomycin (100 U ml−1) and maintained at 37°C and 5% CO2. HCC38 cells (gift from Jennifer Spangle [Emory University, Atlanta, GA, USA]) were cultured in RPMI media treated with penicillin-streptomycin (100 U ml−1) and supplemented with 10% FBS and maintained at 37°C and 5% CO2. Leader, follower, and single-cell subpopulations were isolated from 4T1 cells transfected with H2B-Dendra2 using SaGA, as previously described (Konen et al., 2017; Khatib et al., 2023). Briefly, 4T1 cells were transfected with nucleus-targeting H2B-Dendra2, a photoconvertible fluorescent protein that facilitates cell visualization during imaging. Prior to photoconversion, all cells emit green fluorescence (maximum excitation, 490 nm; maximum emission, 507 nm) and after excitation with a 405 nm laser, H2B-Dendra2 within selected cells photoconverts to emit red fluorescence (maximum excitation, 553 nm; maximum emission, 573 nm). While cells were invading in 3D collagen type I, individual leader, follower, and single cells were photoconverted separately without fluorescence conversion occurring in nearby cells. After photoconversion, cells were extracted from the collagen type I matrix and sorted using flow cytometry. All isolated subpopulations were authenticated as 4T1 cells via STR analysis by IDEXX Laboratories, Inc., and as such were determined to be free of contamination from another cell line. All cell lines were routinely tested for mycoplasma contamination using a commercially available kit (MycoAlert Mycoplasma Detection Kit; Lonza) according to the manufacturer’s instructions.
Plasmids, transfections, and transductions
4T1 cells were transfected using Lipofectamine 3000 (L3000001; Thermo Fisher Scientific), per the manufacturer’s instructions, or transduced using lentiviral supernatant produced by 293T cells (Barde et al., 2010) with selection based on H2B-Dendra2 expression via flow cytometry. The pLenti.CAG.H2B-Dendra2.W was a gift from Rusty Lansford (plasmid #51005; http://n2t.net/addgene:51005; RRID:Addgene_51005; Addgene). Sleeping Beauty transfections were performed using a 2:1 ratio of transposon to transposase ratio. The pSBbi-Pur-mCherry transposon vector and pCMV(CAT)T7-SB100 transposase vector were gifts from Eric Kowarz (plasmid #60523; http://n2t.net/addgene:60523; RRID:Addgene_60523; Addgene). CRISPR-Cas9 KO of mLama3 on leaders and clonal isolation of KO cells was performed by Synthego (mLama3 gRNA: 5′-CUGCGGGGAGCGGGACCCAG-3′). piggyBac transfections were performed using a 1:1 M ratio of transposon vector to transposase mRNA ratio. mItga6 shRNA knockdown and scrambled vectors (shRNA target sequences: 5′-CGGAAATCCTTTCAAGAGAAA-3′, 5′-CGAGAAGGAAATCAAGACAAA-3′) were designed and purchased from VectorBuilder.
Reagents and antibodies
E-cadherin (CDH1) (24E10; Cell Signaling) was used at 1:10,000 for immunoblotting and 1:500 for immunostaining. Laminin-5 (ab14509; Abcam) was used at 1:200 for immunostaining and 1:1,000 for immunoblotting and experiments requiring inhibition of laminin-332 interactions. Integrin α 6 (ab181551; Abcam) was used at 1:1,000. Integrin α 6-APC (FAB13501A; R&D Systems) was used for flow cytometry. Integrin β 4 (ab236251; Abcam) was used at 1:1,000. Actin (A2066; Sigma-Aldrich) was used at 1:10,000. Peroxidase AffiniPure Goat Anti-Rabbit IgG (H+L) (111-035-144; Jackson ImmunoResearch) was used at 1:10,000. For laminin-332 experiments, human Biolaminin-332 (LN332-0202; BioLamina) was adsorbed per the manufacturer’s instructions. Manganese (II) chlorine solution (M1787; Sigma-Aldrich) was used at 1 mM to activate integrins.
RNA sequencing
RNA sequencing was performed in triplicate on 4T1 leader, follower, and single cells. Cells were grown to 70% confluency and RNA was extracted using an RNeasy Mini Kit (74104; Qiagen). RNA quality control and quantification were performed by the Emory Integrated Genomics Core using a Bioanalyzer and Nanodrop. RNA sequencing library preparation was performed at Novogene utilizing the NEBNext Ultra RNA Library Prep Kit for Illumina (E7530; New England Biolabs) by following the manufacturer’s recommendations. Sequencing libraries were validated on the Agilent 2100 Bioanalyzer System (Agilent Technologies) and quantified using Qubit 2.0 Fluorometer (Invitrogen) as well as by quantitative PCR (Applied Biosystems). The libraries were sequenced to 50 M reads per sample on an Illumina sequencer using a 2 × 150 paired-end (PE) configuration. Raw sequence data (.bcl files) was converted into fastq.gz files and demultiplexed using Illumina’s bcl2fastq software. Compressed, raw sequence data in fastq.gz format were trimmed using Trimmomatic v0.36 (Bolger et al., 2014) to remove Illumina adapter contamination using parameters ILLUMINACLIP:20:10:8:TRUE in PE mode and checked for quality control (QC) using FastQC v0.11.4 and MultiQC v1.12 (Ewels et al., 2016) to generate complete QC reports in HTML format. Trimmed data were also analyzed in FastQC to ensure removal of adapter content and then were aligned to the mm10 mouse reference genome using STAR aligner v2.5.2 (Dobin et al., 2013) with default parameters. HTseq-count v0.13.5 (Anders et al., 2015) was used via the gene-id method to assign counts per gene with the parameters “htseq-count -f bam -m union -r pos -i gene_id -a 10 -s no.” The resulting count tables per sample were merged into an overall counts matrix and analyzed downstream in DESeq2.
DESeq2 was used to determine DEGs between two experimental groups (Love et al., 2014). Raw gene expression counts were used as DESeq2 internally corrects for library size. The expression count table was assumed to follow a negative binomial distribution and the Wald Test was implemented for differential gene expression testing. Raw P values were transformed using the Benjamini–Hochberg correction and a corrected P value of <0.05 was considered for statistical significance (Benjamini and Hochberg, 1995). Heatmaps were generated using the R package pheatmap following the application of a variance stabilizing transformation (Anders et al., 2013). Volcano plots were generated using the R package EnhancedVolcano.
DNA methylation microarray
DNA methylation status was assessed in triplicate on 4T1 leader and single cells. Cells were grown to 70% confluency, trypsinized, and then homogenized using QIAshredder (79654; Qiagen). DNA was isolated using the QIAamp DNA Mini Kit (51304; Qiagen). DNA QC was performed through the Quant-iT Broad-Range dsDNA assay kit (Q33130; Invitrogen) and agarose gel by the Emory Integrated Genomics Core. AKESOgen performed an upfront quantification assay using the Quant-iT PicoGreen dsDNA assay kit (P7589; Invitrogen) to confirm DNA concentration and volume. Bisulfite conversion was performed on the DNA, and the DNA was applied to the Infinium Mouse Methylation BeadChip (20041558; Illumina) and reagents. Data from each sample was first preprocessed by R package “sesame” (Zhou et al., 2018), which includes masking suboptimal probes (e.g., detection P value >0.01, on X and Y chromosomes, non-CpG probes, single nucleotide polymorphism–related probes, and repetitive element), background subtraction, and type-1 probe dye bias. The bias of type-2 probe values was corrected by the method “BMIQ” implemented in R package “wateRmelon” (Pidsley et al., 2013). Batch effect was corrected by “combat” method in R package “ChAMP” (Tian et al., 2017), and between-array normalization was completed by R package “qsmooth” (Hicks et al., 2018). DMPs between group leaders and group singles were detected by function “dmpFinder” implemented in R package “minfi” (Aryee et al., 2014) with criteria absolute beta value difference >0.2 and q-value <0.05. DMRs were identified with function “combp” implemented in R package “ENmix” (Xu et al., 2021) with the following criteria: absolute difference of average betas in a region >0.2, probe number >1, Sidak correction P value <0.1, and minimum region size >50 bp. Gene promoter information was derived from the ENCODE Project.
Liquid chromatography-tandem mass spectrometry (LC-MS/MS)
Proteomic analysis was performed on the CM of adherent cells in 2D plastic. Cells were grown to 50% confluency in complete media, then washed with 1× PBS before replacing with a serum-free defined media supplemented with 1× insulin-transferrin-selenium-ethanolamine, 0.5 μg/ml hydrocortisone, 1 ng/ml cholera toxin, 50 nM O-phosphorylethanolamine, 5 nM triiodothyronine, and 10 ng/ml human EGF. CM was collected over 48 h, centrifuged at top speed using Vivaspin 20, 3 kDa molecular weight cut off (MWCO) polyethersulfone filters (28-9323-58; Cytiva) at 4°C to extract secreted proteins, then flash-frozen. Protein extracts were processed through the Emory Integrated Proteomics Core as previously described (Soucek et al., 2016). Briefly, samples were normalized with 50 mM NH4HCO3, then treated with 5 mM dithiothreitol at RT for 30 min, followed by 10 mM iodoacetimide at RT for 30 min in the dark. Protein digestion was performed with 4 µg of lysyl endopeptidase (101974-094; Fujifilm Wako Chemicals) at RT overnight and proteins were further digested overnight with 4 µg trypsin. The resulting peptides were desalted with an HLB column (186002034; Waters) and then vacuum-dried.
Data acquisition by LC-MS/MS was adapted from a published procedure (Seyfried et al., 2017). Digested peptides were resuspended in 0.1% trifluoroacetic acid and separated on a Water’s Charged Surface Hybrid column (150 µm internal diameter [ID] × 15 cm; particle size: 1.7 µm). An EVOSEP liquid chromatography system was used to run the samples using the preset gradient (88 min). Samples were then monitored on a Q-Exactive Plus Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Fisher Scientific). The mass spectrometer cycle was programmed to collect one full MS scan followed by 20 data-dependent MS/MS scans, and MS scans (400–1,600 m/z range, 3 × 106 automatic gain control [AGC] target, 100 ms maximum ion time) were collected at a resolution of 70,000 at m/z 200 in profile mode. The higher-energy collisional dissociation MS/MS spectra (1.6 m/z isolation width, 28% collision energy, 1 × 105 AGC target, 100 ms maximum ion time) were acquired at a resolution of 17,500 at m/z 200. Dynamic exclusion was set to exclude previously sequenced precursor ions for 30 s. Precursor ions with +1, and +7, +8, or higher charge states were excluded from sequencing.
Label-free quantification analysis was adapted from a published procedure (Seyfried et al., 2017). Andromeda was used to search the spectra, which were subsequently integrated into MaxQuant, against the 2020 Uniprot mouse database (91,439 target sequences). Methionine oxidation (+15.9949 Da), asparagine and glutamine deamidation (+0.9840 Da), and protein N-terminal acetylation (+42.0106 Da) were variable modifications (up to five allowed per peptide); cysteine was assigned as a fixed carbamidomethyl modification (+57.0215 Da). Only fully tryptic peptides were considered with up to 2 missed cleavages in the database search. A precursor mass tolerance of ± 20 ppm was applied prior to mass accuracy calibration and ± 4.5 ppm after internal MaxQuant calibration. The following additional search settings were also included: a maximum peptide mass of 6,000 Da, a minimum peptide length of six residues, 0.05 Da tolerance for orbitrap, and 0.6 Da tolerance for ion trap MS/MS scans. The false discovery rate for peptide spectral matches, proteins, and site decoy fraction were all set to 1%. The following quantification settings were applied: re-quantify with a second peak finding attempt after protein identification has completed; match MS1 peaks between runs; a 0.7-min retention time match window was used after an alignment function was found with a 20-min RT search space. Protein quantification was performed using summed peptide intensities given by MaxQuant. The quantitation method only considered razor plus unique peptides for protein-level quantitation.
3D invasion assays and spheroid microscopy
Spheroids were generated, as previously described (Konen et al., 2016). Cells were seeded in round bottom 96-well plates at a density of 1,000 cells per well and centrifuged at 1500 RPM for 5 min. After incubating the plate at 37°C and 5% CO2 for 72 h, spheroids were embedded in 3 mg/ml rat-tail collagen type I (354249; Corning) and seeded onto glass-bottom culture dishes (P35G; MatTek). The collagen matrix was allowed to polymerize at 37°C and 5% CO2 for 30 min prior to media supplementation.
Images were taken using an Olympus CKX41 microscope with an Infinity 1-3C camera (×4 air, 0.13 numerical aperture [NA], UPlanFL N) at RT using Infinity image acquisition software. For spheroids with mixed subpopulations, cells were seeded together in round-bottom 96-well plates at a 1:1 ratio with 1,000 total cells per spheroid. Images of spheroids that required fluorescent imaging were taken with the Leica TCS SP8 inverted confocal microscope (×10) using 1-mm stack intervals, line scanning (488-nm argon, 561-nm diode-pumped solid-state), 4× line averaging, and both hybrid (HyD) and photomultiplier tube (PMT) detectors. Images were acquired at RT using Leica image acquisition software. Leica software was also used after acquisition to perform maximum projections on multistack acquisitions and to export processed images.
Immunoblotting and immunostaining
For immunoblotting, whole-cell lysate protein expression was assessed by western blotting. Briefly, adherent cells were rinsed with 1× phosphate-buffered saline (PBS) with Ca2+ and Mg2+ and lysed with radioimmunoprecipitation assay buffer (50 mM Tris [pH 7.8], 2 mM EDTA, 150 mM NaCl, 1% Triton X-100, 5% SDS, and 5% sodium deoxycholate) supplemented with Halt Protease and Phosphatase Inhibitor Cocktail (78429; Thermo Fisher Scientific). Lysates were then sonicated briefly to shear the DNA. Protein quantification was performed using a bicinchoninic acid (BSA) protein assay kit (23225; Thermo Fisher Scientific) before SDS–polyacrylamide gel electrophoresis. After electrophoresis, proteins were transferred from the gel to a nitrocellulose membrane for 2 h at 300 mA. The membrane was then blocked using 5% milk diluted in Tris-buffered saline with 0.1% Tween 20 (TBS-T) for 1 h at RT prior to incubating with the primary antibody overnight at 4°C while rocking. After several washes with TBS-T, the membrane was incubated with the secondary antibody at RT for 50 min. Membranes were treated with horseradish peroxidase and developed via film or digital imaging.
For immunostaining (Debnath et al., 2003), cells in 2D or spheroids embedded in rat-tail collagen type I were rinsed with 1× PBS with Ca2+ and Mg2+ and then immediately fixed with freshly prepared 4% paraformaldehyde diluted in 1× PBS with Ca2+ and Mg2+ for 20 min at RT. Spheroids were then rinsed with 100 mM glycine diluted in immunofluorescence buffer (130 mM NaCl, 7 mM Na2HPO4, 3.5 mM NaH2PO4, 0.2% Triton X-100, 0.05% Tween-20) prior to blocking with immunofluorescence buffer supplemented with 10% normal goat serum for 1 h at RT while rocking. Spheroids were then incubated with primary antibody overnight at 4°C while rocking. Spheroids were then washed with immunofluorescence buffer prior to secondary antibody incubation at RT for 50 min. After staining, cells in 2D or 3D spheroids were imaged using the Leica TCS SP8 inverted confocal microscope (×10) using 1-mm stack intervals, line scanning (488-nm argon, 561-nm diode-pumped solid-state), 4× line averaging, and both HyD and PMT detectors.
Flow cytometry
Relative expression of cell surface proteins and detection of cell apoptosis were measured through flow cytometry. For measuring the expression of cell surface proteins, adherent cells were seeded in 10-cm culture plates and grown to 80% confluency prior to dissociation with 1 mM EDTA diluted in 1X PBS. After dissociation, suspended cells were centrifuged into a pellet, washed, and stained with a fluorophore-conjugated antibody diluted in flow buffer (1 mM EDTA, 25 mM HEPES, 1% dialyzed FBS diluted in 1X PBS) for 30 min at 4°C in the dark while agitated. Cells were then washed with flow buffer prior to resuspension for analysis using a BD FACSymphony A3. For the detection of cell apoptosis, the PE Annexin V Apoptosis Detection Kit I (559763; BD Pharmigen) was used, per the manufacturer’s instructions. All data was further analyzed using FlowJo.
Live-cell imaging
Spheroids were embedded in rat-tail collagen type I and plated into cell culture plates with optical glass bottoms as previously described (Konen et al., 2017). Spheroids were then imaged using the Leica TCS SP8 inverted confocal microscope (×10) with a live-cell chamber (37°C and 5% CO2) using 1-mm stack intervals, line scanning using a resonant galvanometric tandem scanner (8 kHz; 488-nm argon, 561-nm diode-pumped solid-state), 4× line averaging, and both HyD and PMT detectors. For CM experiments where no fluorescence was required for image acquisition, images were acquired every 20 min for 24 h. For mixed spheroid experiments, images were acquired every 20 min for 48 h. Leica image acquisition software was used to acquire images and to perform maximum projections on multistack images after acquisition.
Image analysis
For image sequences of 3D spheroids, each 3D image (x, y, and z) was flattened to 2D maximum projections (x and y). Relative velocity and track length of individual cells in 3D spheroids were measured using the MTrackJ plug-in in ImageJ (Meijering et al., 2012). Tracks were drawn by manually selecting a random cell and following the cell’s movement at every time point in the image sequence. Five tracks were drawn per spheroid for three separate spheroids for each condition. Circularity of individual cells was measured at the end of each image sequence by manually outlining the cell border using ImageJ. Five random cells were chosen from a randomized region for three separate spheroids for each condition.
Rac1 G-LISA
Rac1 activity was measured using the colorimetric Rac1 G-LISA Activation Assay Kit (BK128; Cytoskeleton) per manufacturer’s instructions. Cells were grown to 70% confluency with complete media and then serum-starved overnight with serum-free DMEM media. For experiments wherein cells were treated with CM derived from other cells, cells were pretreated with 1 mM Mn2+ diluted in serum-free DMEM for 10 min to activate integrins. Proteins from CM were extracted by centrifugation using Vivaspin 20, 3 kDa MWCO polyethersulfone filters (28-9323-58; Cytiva), and protein concentration was quantified using a BSA protein assay kit (23225; Thermo Fisher Scientific). Protein concentrations were normalized for each experiment and diluted in serum-free DMEM supplemented with 1 mM Mn2+ before stimulating serum-starved cells.
Statistical analysis
Statistical analyses were performed using GraphPad Prism. For all experiments with a single variable and data points acquired from two independent samples, unpaired Student’s T tests were performed to evaluate significance. For all experiments with a single variable and data points acquired from three independent samples, one-way ANOVA tests with multiple comparisons were performed. For all experiments with two variables, two-way ANOVA tests with multiple comparisons were performed. For all ANOVA tests with multiple comparisons, a Tukey correction was applied. For all parametric tests, data distribution was assumed to be normal but this was not formally tested. All bar graph data show the mean, with error bars indicating SEM. For violin plots, thicker segmented lines indicate the mean, while the thinner segmented dots indicate the interquartile range. The figure legends indicate the number of biological replicates (n) for each experiment and all significant denotations.
Online supplemental material
This manuscript is accompanied by four supplementary figures, one supplementary table, and 10 supplemental movies. Fig. S1 shows (A) higher resolution images of E-cadherin immunofluorescence staining on 4T1 subpopulations and (B) examples of invasive heterogeneity in human triple-negative breast cancer cell lines. Fig. S2 shows transcriptional differences in all detectable laminin genes between leaders and singles. Fig. S3 shows (A–C) measurement of the meandering index for single cells for all live-cell experiments performed and (D) quantification of apoptotic events between Lama3 WT and KO leaders. Fig. S4 shows (A) flow cytometric analysis of integrin α6 between leaders and singles, (B) Rac1 activity of leaders and singles after EGF stimulation, and (C) a volcano plot showing the differentially secreted proteins between Lama3 WT and KO leaders from proteomic analysis. Table S1 shows results from STR analysis on 4T1 subpopulations. Videos 1 and 2 show 3D invasion of collectively invading and single-cell subpopulations, respectively. Videos 3 and 4 show 3D invasion of single cells after being treated with leader CM or single CM, respectively. Videos 5, 6, and 7 show 3D invasion of single cells after being treated with CM from Lama3 WT leaders and two Lama3 KO leaders. Videos 8, 9, and 10 show 3D invasion of mCherry-transfected single cells in mixed spheroids comprising of mCherry-transfected single cells and Lama3 WT or KO leaders.
Data availability
RNA sequencing data has been deposited in the Sequence Read Archive (SRA) with the BioProject accession number PRJNA1082274. DNA methylation data has been deposited into the GEO archive with accession number GSE262287. Proteomic data has been deposited into the Synapse archive with accession number syn54010990 (https://www.synapse.org/#!Synapse:syn54010990/files/). All other data, including raw image and video files, are available upon request.
Acknowledgments
We thank H. Moses at Vanderbilt University for the 4T1 cells used to perform SaGA in this study. We also thank P. Vertino at the University of Rochester for the SUM159 cells and J. Spangle at Emory University for the HCC38 cells.
Funding: This work was supported by National Institutes of Health (NIH) National Cancer Institute (NCI) grants R01CA250422 (A.I. Marcus), R01CA247367 (A.I. Marcus), R01CA236369 (A.I. Marcus), PO1CA257906 (A.I. Marcus), and R50CA265345 (J.K. Mouw). This work is also supported by the Brenda Nease Breast Cancer Fund. In addition, research reported in this publication was supported, in part, by the following Shared Resources of Winship Cancer Institute of Emory University and NIH/NCI under award number P30CA138292: the Emory University Integrated Cellular Imaging Microscopy Core (RRID:SCR_023534), the Biostatistics and Bioinformatics Shared Resource (RRID:SCR_023521), the Emory Integrated Genomics Core (RRID:SCR_023529), the Emory Integrated Computational Core (RRID:SCR_023525), the Emory Integrated Proteomics Core (RRID:SCR_023530), and the Pediatrics/Winship Flow Cytometry Core (RRID:SCR_023536). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Author contributions: S.B. Yoon, J.K. Mouw, and A.I. Marcus designed the experiments. J.K. Mouw and N.M. Zohbi performed SaGA to isolate 4T1 subpopulations. S.B. Yoon, J.K. Mouw, I.E. Robinson, and T.O. Khatib performed the experiments. S.B. Yoon, R.A. Arthur, H. Claussen, and L. Chen contributed to the bioinformatics and statistical analysis of the transcriptomics and epigenomics data. S.B. Yoon, R.A. Arthur, H. Claussen, and L. Chen drafted the manuscript. All authors provided input and feedback during manuscript preparation and edited the manuscript.
References
Author notes
Disclosures: The authors declare no competing interests exist.