Precise chromosome congression and segregation requires the proper assembly of a steady-state metaphase spindle, which is dynamic and maintained by continuous microtubule flux. NuSAP is a microtubule-stabilizing and -bundling protein that promotes chromosome-dependent spindle assembly. However, its function in spindle dynamics remains unclear. Here, we demonstrate that NuSAP regulates the metaphase spindle length control. Mechanistically, NuSAP facilitates kinetochore capture and spindle assembly by promoting Eg5 binding to microtubules. It also prevents excessive microtubule depolymerization through interaction with Kif2A, which reduces Kif2A spindle-pole localization. NuSAP is phosphorylated by Aurora A at Ser-240 during mitosis, and this phosphorylation promotes its interaction with Kif2A on the spindle body and reduces its localization with the spindle poles, thus maintaining proper spindle microtubule flux. NuSAP knockout resulted in the formation of shorter spindles with faster microtubule flux and chromosome misalignment. Taken together, we uncover that NuSAP participates in spindle assembly, dynamics, and metaphase spindle length control through the regulation of microtubule flux and Kif2A localization.
Introduction
The mitotic spindle is a well-organized and evolutionarily conserved microtubule (MT)-based structure that mediates precise chromosome congression and segregation (Karsenti and Vernos, 2001; Mitchison and Salmon, 2001). The spindle microtubules grow and shrink by addition and loss of tubulin subunits at their ends, respectively, allowing the spindle to efficiently probe the cytoplasmic space for biorientation of sister kinetochores and to congress chromosomes to the metaphase plate (Gadde and Heald, 2004; Kline-Smith and Walczak, 2004). Although the spindle microtubules are dynamically unstable and switch between growing and shrinking states, the metaphase spindle will finally reach a balance that maintains a constant length and well-constructed structure (Dumont and Mitchison, 2009; Rogers et al., 2005). In addition, mitotic spindle microtubules display poleward flux (Mitchison, 1989), and a recent study proposed that the flux is driven by four kinesins (Steblyanko et al., 2020). Accordingly, interpolar microtubules are slid apart by microtubule-sliding motors Eg5 and Kif15, sequentially contributed by CENP-E at kinetochores in prometaphase and Kif4A on chromosome arms in metaphase. Kif4A and Kif18A were subsequently identified to regulate kinetochore fiber flux by controlling the length of overlaps between kinetochore fibers and bridging fibers, which promotes kinetochore centering and chromosome alignment (Risteski et al., 2022). When outward sliding apart of interpolar microtubules is balanced by polymerization and depolymerization of the spindle microtubules at the minus-ends and plus-ends, the spindle maintains a steady length (Barisic et al., 2021; Brust-Mascher et al., 2009; Fink et al., 2009; Goshima and Scholey, 2010; Steblyanko et al., 2020; Valdez et al., 2023). This balance is achieved mainly via kinesin-13 family proteins (Brust-Mascher and Scholey, 2002; Ganem and Compton, 2004; Miyamoto et al., 2004; Brust-Mascher et al., 2009; Ems-McClung and Walczak, 2010; Henkin et al., 2023) in concert with other regulators of the spindle microtubule dynamics (Buster et al., 2007; Goshima and Scholey, 2010; Kwok and Kapoor, 2007).
The nucleolar and spindle-associated protein (NuSAP) is a conservative and ∼55 kD protein and is selectively expressed in proliferating cells (Raemaekers et al., 2003). As a microtubule-associated protein (MAP), NuSAP plays an essential role in chromosome-dependent spindle assembly (Raemaekers et al., 2003; Ribbeck et al., 2006, 2007). Previous studies suggested that NuSAP is a mitotic Ran GTPase target that stabilizes and crosslinks microtubules (Ribbeck et al., 2006). During mitosis, NuSAP is dissociated with importins and immobilized on chromatin to induce microtubule attachment to the chromosomes (Ribbeck et al., 2007). Despite the efforts to explore NuSAP roles in mitotic spindle assembly, its function in spindle dynamics remains to be determined.
In this work, we reveal that NuSAP regulates the spindle dynamics to maintain metaphase spindle length by coordinating microtubule depolymerization and microtubule-microtubule sliding. NuSAP is phosphorylated by the kinase Aurora A, which is essential for the proper microtubule flux on the metaphase spindle. NuSAP knockout in cells leads to improperly increasing Kif2A localization onto spindle poles while reducing Eg5 localization on spindle microtubules. Moreover, NuSAP phosphorylation by Aurora A at mitosis enhances its affinity with Kif2A on spindle microtubules, thus preventing the superabundant concentration of Kif2A onto the spindle poles and maintaining the proper metaphase microtubule flux. Collectively, our study reveals a novel mechanism of how NuSAP regulates mitotic spindle assembly, structural dynamics, and metaphase spindle length control for accurate chromosome congression via affecting microtubule flux and Kif2A localization.
Results and discussion
NuSAP deletion results in short spindle formation, whereas its overexpression makes the spindle longer
To understand how cells control spindle assembly and their chromosome congression, we explored the roles of NuSAP in spindle dynamics and function. First, we established NuSAP knockout cell lines with CRISPR-Cas9 in HeLa cell lines. Immunofluorescence microscopy imaging and Western blot analysis showed that we successfully knocked out the expression of NuSAP in HeLa cells (Fig. 1, A and B). Through immunofluorescence microscopy imaging, we observed that NuSAP knockout showed a remarkable effect in generating short bipolar metaphase spindles (Fig. 1 A). The mean distance between spindle poles, as marked by γ-tubulin signals, was reduced to ∼10.07 μm compared with the normal length of ∼11.23 μm (Fig. 1 C). Under conditions that overexpress NuSAP in cells, the metaphase spindle length was significantly longer (∼14.43 μm) than in control cells (∼11.57 μm; Fig. 1, D and E). Notably, overexpression of NuSAP also induced unusual, curved microtubule filaments, extensive bundling of the microtubules, and deficient spindle morphology (Fig. 1 D), as reported before (Raemaekers et al., 2003). These results suggest that the proper amount of NuSAP, not more or less, is essential for a precise metaphase spindle assembly by participating in the metaphase spindle length control.
NuSAP regulates spindle length control and microtubule flux. (A) Control and NuSAP-knockout HeLa cells were stained for γ-tubulin (green), NuSAP (red), and DNA (blue). (B) NuSAP knockout efficiency of HeLa cells was analyzed by Western blotting. (C) Spindle length (indicated by white lines in A) was quantified by γ-tubulin immunofluorescence in control or NuSAP-knockout metaphase cells. Error bars indicate SD. Three independent replicates of 30 cells per replicate were quantified (n = 3). Unpaired two-tailed t test: P = 0.0006. (D) HeLa cells expressing GFP or GFP-NuSAP were stained for γ-tubulin (red), α-tubulin (magenta), and DNA (blue). (E) Spindle length (indicated by white lines) was measured by γ-tubulin immunofluorescence in metaphase cells. Error bars indicate SD. Three independent replicates of 30 cells per replicate were quantified (n = 3). Unpaired two-tailed t test: P = 0.0006. (F and G) NuSAP-knockout HeLa cells were transfected with photoactivatable GFP-tagged α-tubulin (PAGFP-α-tubulin, green). Microtubules were stained with SiR-tubulin (red), and the concentration of SiR-tubulin is 0.04 μM, which does not affect normal mitotic progression. GFP signal in a rectangular region near the microtubule plus ends was activated (time point 0, arrows) and tracked every 10 s. Representative time-course images are shown. (H and I) Corresponding kymograph profiles of the photoactivated regions in F and G (red dotted lines highlight microtubule-flux slopes). Scale bars, 10 s. (J and K) The fluorescence intensity profiles at time points 0 and 120 s in F and G are shown, respectively. (L) The mean velocity was defined by the ratio of the distance that the PAGFP signal travels to 2 min it takes. ΔDistance: the distance between the two points with the strongest PAGFP fluorescence intensity at 0 and 120 s. ΔTime: 2 min. (M) The rates of microtubule flux shown in F and G were measured in control or NuSAP-knockout cells. Error bars indicate SD. Three independent replicates of 14 cells per replicate were quantified (n = 3). Unpaired two-tailed t test: P < 0.0001. ***, P < 0.001; ****, P < 0.0001. n = the number of independent experiments presented. Scale bars, 10 μm. Source data are available for this figure: SourceData F1.
NuSAP regulates spindle length control and microtubule flux. (A) Control and NuSAP-knockout HeLa cells were stained for γ-tubulin (green), NuSAP (red), and DNA (blue). (B) NuSAP knockout efficiency of HeLa cells was analyzed by Western blotting. (C) Spindle length (indicated by white lines in A) was quantified by γ-tubulin immunofluorescence in control or NuSAP-knockout metaphase cells. Error bars indicate SD. Three independent replicates of 30 cells per replicate were quantified (n = 3). Unpaired two-tailed t test: P = 0.0006. (D) HeLa cells expressing GFP or GFP-NuSAP were stained for γ-tubulin (red), α-tubulin (magenta), and DNA (blue). (E) Spindle length (indicated by white lines) was measured by γ-tubulin immunofluorescence in metaphase cells. Error bars indicate SD. Three independent replicates of 30 cells per replicate were quantified (n = 3). Unpaired two-tailed t test: P = 0.0006. (F and G) NuSAP-knockout HeLa cells were transfected with photoactivatable GFP-tagged α-tubulin (PAGFP-α-tubulin, green). Microtubules were stained with SiR-tubulin (red), and the concentration of SiR-tubulin is 0.04 μM, which does not affect normal mitotic progression. GFP signal in a rectangular region near the microtubule plus ends was activated (time point 0, arrows) and tracked every 10 s. Representative time-course images are shown. (H and I) Corresponding kymograph profiles of the photoactivated regions in F and G (red dotted lines highlight microtubule-flux slopes). Scale bars, 10 s. (J and K) The fluorescence intensity profiles at time points 0 and 120 s in F and G are shown, respectively. (L) The mean velocity was defined by the ratio of the distance that the PAGFP signal travels to 2 min it takes. ΔDistance: the distance between the two points with the strongest PAGFP fluorescence intensity at 0 and 120 s. ΔTime: 2 min. (M) The rates of microtubule flux shown in F and G were measured in control or NuSAP-knockout cells. Error bars indicate SD. Three independent replicates of 14 cells per replicate were quantified (n = 3). Unpaired two-tailed t test: P < 0.0001. ***, P < 0.001; ****, P < 0.0001. n = the number of independent experiments presented. Scale bars, 10 μm. Source data are available for this figure: SourceData F1.
Centrosome-independent microtubule nucleation affects the number of microtubules inside the spindle and regulates spindle length (Goshima and Kimura, 2010). To verify whether NuSAP controls the spindle length through mediating the acentrosomal microtubule nucleation, we treated HeLa cells with 500 ng/ml nocodazole for 2 h to disassemble overall microtubules followed by releasing these cells into a fresh medium containing a low concentration of nocodazole (15 ng/ml) to induce the acentrosomal microtubule nucleation (Fu et al., 2013). Through immunostaining of the fixed cells, we observed the acentrosomal microtubule assembly. The results showed that, though the centrosome-based microtubule nucleation was partially inhibited in the presence of a low concentration of nocodazole, the acentrosomal microtubule nucleation was induced, and these nucleated microtubules rapidly assembled into many small acentrosomal asters (Fig. S1 A). We counted the number of acentrosomal microtubule asters 2 min after nocodazole release. The analysis showed that the number of small microtubule asters in NuSAP knockout cells was no different from that in control cells (Fig. S1 B). Thus, the results indicate that NuSAP controlling the metaphase spindle length is not via mediating acentrosomal microtubule nucleation.
NuSAP is not necessary for acentrosomal MT nucleation and is required for kinetochore–microtubule attachment. (A) Control and NuSAP-knockout HeLa cells were treated with 500 ng/ml nocodazole for 2 h. Then, the treated cells were released into a medium containing 15 ng/ml nocodazole and fixed at the indicated time. The cells were stained with anti-α-tubulin (green) and anti-γ-tubulin (red) antibodies. DNA was stained with DAPI (blue). (B) Quantification of the number of small acentrosomal MT asters when cells were released at 2 min (shown in A). Error bars indicate SD. 150 cells were quantified in three independent experiments (n = 3). Unpaired two-tailed t test. P = 0.9233. (C) Detection of the decay rate of PAGFP-α-tubulin signals over time in control and NuSAP KO cells. Three independent replicates of 14 cells per replicate were quantified (n = 3). (D) Control and NuSAP-knockout HeLa cells were placed on ice for 10 min before fixation. Then, the cells were processed for immunofluorescence using anti-Crest (red) and anti-Mad2 (green) antibodies. DNA was stained with DAPI (blue). The box areas were zoomed at the right. Scale bar in zoomed image, 1 μm. (E) Quantification of the percentage of kinetochores with Mad2 signal. Error bars indicate SD. Unpaired two-tailed t test. P = 0.0003. 338 and 340 kinetochores in 100 cells were counted from three independent experiments (n = 3). (F) Control and NuSAP-knockout HeLa cells synchronized to metaphase with MG132 and control HeLa cells synchronized to prometaphase with nocodazole release were stained with anti-α-tubulin (green) and anti-Crest (red) antibodies. DNA was stained with DAPI (blue). The box areas were zoomed at the right. Scale bar in zoomed image, 1 μm. (G) Measurement of interkinetochore (labeled by Crest) distances. Error bars indicate SD. P = 0.0002 for Ctrl/NuSAP KO; P = 0.7585 for NuSAP KO/Prometaphase. Three independent replicates of 100 pairs of kinetochores in 30 cells per replicate were quantified. Unpaired two-tailed t test. ns, not significant; ***, P < 0.001. n = the number of independent experiments presented. Scale bars, 10 μm unless specified otherwise.
NuSAP is not necessary for acentrosomal MT nucleation and is required for kinetochore–microtubule attachment. (A) Control and NuSAP-knockout HeLa cells were treated with 500 ng/ml nocodazole for 2 h. Then, the treated cells were released into a medium containing 15 ng/ml nocodazole and fixed at the indicated time. The cells were stained with anti-α-tubulin (green) and anti-γ-tubulin (red) antibodies. DNA was stained with DAPI (blue). (B) Quantification of the number of small acentrosomal MT asters when cells were released at 2 min (shown in A). Error bars indicate SD. 150 cells were quantified in three independent experiments (n = 3). Unpaired two-tailed t test. P = 0.9233. (C) Detection of the decay rate of PAGFP-α-tubulin signals over time in control and NuSAP KO cells. Three independent replicates of 14 cells per replicate were quantified (n = 3). (D) Control and NuSAP-knockout HeLa cells were placed on ice for 10 min before fixation. Then, the cells were processed for immunofluorescence using anti-Crest (red) and anti-Mad2 (green) antibodies. DNA was stained with DAPI (blue). The box areas were zoomed at the right. Scale bar in zoomed image, 1 μm. (E) Quantification of the percentage of kinetochores with Mad2 signal. Error bars indicate SD. Unpaired two-tailed t test. P = 0.0003. 338 and 340 kinetochores in 100 cells were counted from three independent experiments (n = 3). (F) Control and NuSAP-knockout HeLa cells synchronized to metaphase with MG132 and control HeLa cells synchronized to prometaphase with nocodazole release were stained with anti-α-tubulin (green) and anti-Crest (red) antibodies. DNA was stained with DAPI (blue). The box areas were zoomed at the right. Scale bar in zoomed image, 1 μm. (G) Measurement of interkinetochore (labeled by Crest) distances. Error bars indicate SD. P = 0.0002 for Ctrl/NuSAP KO; P = 0.7585 for NuSAP KO/Prometaphase. Three independent replicates of 100 pairs of kinetochores in 30 cells per replicate were quantified. Unpaired two-tailed t test. ns, not significant; ***, P < 0.001. n = the number of independent experiments presented. Scale bars, 10 μm unless specified otherwise.
NuSAP depletion accelerates metaphase spindle microtubule flux
Microtubule poleward flux can serve as an essential factor in spindle dynamics and controls spindle elongation (Buster et al., 2007; Cimini et al., 2006; Laycock et al., 2006; Maffini et al., 2009; Steblyanko et al., 2020). Based on our observations above, we tested whether NuSAP knockout affects the poleward translocation/flux of tubulin dimers along the spindle microtubules. We transfected control and NuSAP-knockout HeLa cells with photoactivatable (PA) GFP-tagged α-tubulin. Then we used pulse flash of 405 nm laser to activate a rectangular region of PAGFP-α-tubulin in the proximity of chromosomes while time-lapse images were captured every 10 s to measure the velocity at which the activated fluorescent signals approached the pole (Fig. 1, F and G; and Videos 1 and 2). In control metaphase cells, the fluorescent signals approached the spindle pole with a mean velocity of 0.30 μm/min. After NuSAP knockout, the velocity of microtubule flux was significantly increased to 0.44 μm/min (Fig. 1, H–M). In addition, we also detected the decay rate of PAGFP-α-tubulin over time in this experiment. We found that the turnover of tubulins in the spindle is not affected after NuSAP KO, indicating that the effect on the microtubule flux of NuSAP is direct (Fig. S1 C). Thus, these results show that NuSAP depletion shortens the metaphase spindle and accelerates microtubule flux. However, previous study shows that microtubule flux rate and spindle length showed a strong positive correlation overall (Steblyanko et al., 2020). Accordingly, NuSAP deletion may affect other spindle length regulators to counteract spindle elongation caused by microtubule flux rate increase. Collectively, NuSAP regulates metaphase spindle length control and structural dynamics.
MT flux on metaphase spindles in HeLa cells. Control HeLa cells were transfected with photoactivatable GFP-tagged α-tubulin (PAGFP-α-tubulin, green). The cells were treated with thymidine for 17–24 h and then released for 9 h. 405 nm laser was applied to activate the GFP signal in a rectangular region near the MT plus end. Microtubules were stained with SiR-tubulin (red). Confocal images were acquired every 10 s for 2 min using the Spinning disk confocal microscope equipped with a 60×/1.4 NA oil objective lens at 37°C. Reproduced at 2 fps. Selected video stills are shown in Fig. 1 F.
MT flux on metaphase spindles in HeLa cells. Control HeLa cells were transfected with photoactivatable GFP-tagged α-tubulin (PAGFP-α-tubulin, green). The cells were treated with thymidine for 17–24 h and then released for 9 h. 405 nm laser was applied to activate the GFP signal in a rectangular region near the MT plus end. Microtubules were stained with SiR-tubulin (red). Confocal images were acquired every 10 s for 2 min using the Spinning disk confocal microscope equipped with a 60×/1.4 NA oil objective lens at 37°C. Reproduced at 2 fps. Selected video stills are shown in Fig. 1 F.
MT flux on metaphase spindles in HeLa cells with NuSAP knockout. NuSAP-knockout HeLa cells were transfected with photoactivatable GFP-tagged α-tubulin (PAGFP-α-tubulin, green). The cells were treated with thymidine for 17–24 h and then released for 9 h. 405 nm laser was applied to activate the GFP signal in a rectangular region near the MT plus ends. Microtubules were stained with SiR-tubulin (red). Confocal images were acquired every 10 s for 2 min using the Spinning disk confocal microscope equipped with a 60×/1.4 NA oil objective lens at 37°C. Reproduced at 2 fps. Selected video stills are shown in Fig. 1 G.
MT flux on metaphase spindles in HeLa cells with NuSAP knockout. NuSAP-knockout HeLa cells were transfected with photoactivatable GFP-tagged α-tubulin (PAGFP-α-tubulin, green). The cells were treated with thymidine for 17–24 h and then released for 9 h. 405 nm laser was applied to activate the GFP signal in a rectangular region near the MT plus ends. Microtubules were stained with SiR-tubulin (red). Confocal images were acquired every 10 s for 2 min using the Spinning disk confocal microscope equipped with a 60×/1.4 NA oil objective lens at 37°C. Reproduced at 2 fps. Selected video stills are shown in Fig. 1 G.
NuSAP depletion results in kinetochore–microtubule attachment failure
The proper structural dynamics of spindle microtubules are essential for a stable connection between kinetochores and microtubules (Cheeseman and Desai, 2008). We have shown that NuSAP depletion makes spindle microtubules less stable and causes faster poleward movement. Here, we asked whether NuSAP contributes to the kinetochore–microtubule attachment during mitosis. First, mitotic control and NuSAP-knockout HeLa cells were treated with MG132 to achieve fully assembled bipolar spindles and placed on ice for 10 min before fixation. In immunofluorescence experiments, we used checkpoint protein Mad2 to monitor kinetochore–microtubule attachment, whose positive signal revealed unattached kinetochores. We found that NuSAP knockout increased unattached kinetochores (Fig. S1, D and E), suggesting that NuSAP knockout abolished the stable connection between kinetochores and microtubules during mitosis. Consistently, we uncovered that the inter-kinetochore distance of chromosomes aligned at the metaphase plate in NuSAP-knockout cells (∼0.95 μm) was substantially shorter than that in control metaphase cells (∼1.33 μm), but similar to control prometaphase cells (∼0.94 μm), indicating that NuSAP was also required for maintenance of proper inter-kinetochore tension and the tension was lost due to kinetochore detachment from k-fibers (Fig. S1, F and G). It has been reported that NuSAP regulates the dynamics of kinetochore microtubules by attenuating MCAK depolymerization activity (Li et al., 2016), which was a possible explanation for the mechanism of maintaining kinetochore–microtubule attachment of NuSAP. Combining these findings, we conclude that NuSAP-regulated microtubule dynamics contribute to the kinetochore–microtubule attachment during mitotic spindle assembly and chromosome congression.
NuSAP is phosphorylated in mitosis by Aurora A
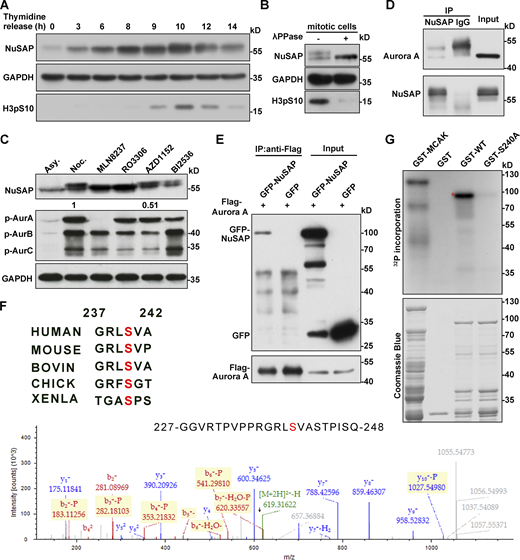
To address how the function of NuSAP is regulated, we examined the levels of the NuSAP protein in different phases of the cell cycle. HeLa cells were arrested at the G1/S boundary by a double-thymidine treatment and then released into fresh medium and collected samples at different time points. Western blot analysis showed NuSAP was a cell cycle–dependent protein with a high expression level in the G2/M phase, followed by a sharp decline in the G1 phase. Moreover, a portion of NuSAP proteins exhibited a motility shift on SDS-PAGE when cells enter mitosis (indicated by Histone H3pS10 antibody; Fig. 2 A), which suggested that NuSAP might be posttranslationally modified during mitosis progression. The mobility change was abolished when the cell mitotic lysates were pretreated with λ-phosphatase (Fig. 2 B), suggesting that NuSAP was phosphorylated during mitosis.
NuSAP is phosphorylated at S240 by Aurora A in mitosis. (A) HeLa cells were blocked at G1/S by double thymidine (Thy.) treatment and then released into fresh medium and harvested at the indicated time point. (B) Mitotic HeLa cell lysates were treated with λ-phosphatase. Samples were analyzed on SDS-PAGE and immunoblotted with the indicated antibodies. (C) HeLa cells were arrested with nocodazole for 17 h and then treated with selected kinase inhibitors (0.25 μM MLN8237 for 30 min, 9 μM RO3306 for 15 min, 0.1 μM AZD1152 for 30 min, 0.1 μM BI2536 for 30 min, respectively). Asy., asynchronized cell samples. The numbers (1.0 and 0.51) indicate the relative gray intensity of p-Aur B. Samples were analyzed on SDS-PAGE and immunoblotted with the indicated antibodies. (D) Mitotic HeLa cells were subjected to an immunoprecipitation (IP) assay with nonspecific IgG or anti-NuSAP antibody followed by Western blotting. (E) Flag-Aurora A was cotransfected with GFP or GFP-NuSAP into HEK293T cells. Lysates of 293T cells were subjected to IP assay with anti-Flag antibody followed by Western blotting. (F) GST-tagged NuSAP protein was incubated with Aurora A kinase at 30°C for 30 min. Samples were processed for MS analysis. Ser-240 site was found to be phosphorylated, consistent with the Aurora A phosphorylation consensus motif. Multiple sequence alignment in the top panel was performed using Uniprot. Red amino acids were conserved. (G) GST-tagged NuSAP proteins with/without point mutation were subjected to Aurora A kinase assay in vitro, followed by autoradiography. GST-tagged MACK was used as positive control. Coomassie Blue staining shows the loading of the GST-tagged NuSAP proteins in the reactions. The red asterisk indicates phosphorylated recombinant NuSAP. Source data are available for this figure: SourceData F2.
NuSAP is phosphorylated at S240 by Aurora A in mitosis. (A) HeLa cells were blocked at G1/S by double thymidine (Thy.) treatment and then released into fresh medium and harvested at the indicated time point. (B) Mitotic HeLa cell lysates were treated with λ-phosphatase. Samples were analyzed on SDS-PAGE and immunoblotted with the indicated antibodies. (C) HeLa cells were arrested with nocodazole for 17 h and then treated with selected kinase inhibitors (0.25 μM MLN8237 for 30 min, 9 μM RO3306 for 15 min, 0.1 μM AZD1152 for 30 min, 0.1 μM BI2536 for 30 min, respectively). Asy., asynchronized cell samples. The numbers (1.0 and 0.51) indicate the relative gray intensity of p-Aur B. Samples were analyzed on SDS-PAGE and immunoblotted with the indicated antibodies. (D) Mitotic HeLa cells were subjected to an immunoprecipitation (IP) assay with nonspecific IgG or anti-NuSAP antibody followed by Western blotting. (E) Flag-Aurora A was cotransfected with GFP or GFP-NuSAP into HEK293T cells. Lysates of 293T cells were subjected to IP assay with anti-Flag antibody followed by Western blotting. (F) GST-tagged NuSAP protein was incubated with Aurora A kinase at 30°C for 30 min. Samples were processed for MS analysis. Ser-240 site was found to be phosphorylated, consistent with the Aurora A phosphorylation consensus motif. Multiple sequence alignment in the top panel was performed using Uniprot. Red amino acids were conserved. (G) GST-tagged NuSAP proteins with/without point mutation were subjected to Aurora A kinase assay in vitro, followed by autoradiography. GST-tagged MACK was used as positive control. Coomassie Blue staining shows the loading of the GST-tagged NuSAP proteins in the reactions. The red asterisk indicates phosphorylated recombinant NuSAP. Source data are available for this figure: SourceData F2.
We then screened the possible mitotic kinases responsible for NuSAP phosphorylation. Through treating mitotic HeLa cells with several mitotic kinase inhibitors, we revealed that treatment with the specific inhibitor MLN8237 for kinase Aurora A and RO3306 for kinase Cdk1 significantly reduced the higher band of NuSAP. In contrast, Aurora B kinase inhibition by AZD1152 or Plk1 kinase inhibition by BI2536 did not down-shift the NuSAP band (Fig. 2 C). A previous report has shown that Cdk1 is responsible for NuSAP phosphorylation at T300 and T338 sites during mitosis, whereas this phosphorylation inhibits NuSAP binding with microtubules (Chou et al., 2011). Thus, we focused on the functions of NuSAP phosphorylation by Aurora A. First, we wanted to know whether NuSAP and Aurora A could bind in vivo. We performed an immunoprecipitation (IP) assay using anti-NuSAP antibodies and found that endogenous Aurora A could be immunoprecipitated from mitotic lysates (Fig. 2 D). In a parallel experiment, we coexpressed Flag-Aurora A with GFP or GFP-NuSAP in HEK293T cells followed by IP assay, and the results also showed that NuSAP interacted with Aurora A (Fig. 2 E). To identify the phosphorylation site(s) of Aurora A on NuSAP, we expressed and purified GST-tagged NuSAP from Escherichia coli and incubated the purified proteins with Aurora A kinase in vitro treated by DMSO or MLN8237 and then prepared the samples for mass spectrum (MS) analysis. The results revealed that Ser-240, which was consistent with the consensus sequence R-X-pS/T-L/V of Aurora A (Ohashi et al., 2006), was phosphorylated (Fig. 2 F, bottom panel). Besides, the sequence alignment showed that the site was conserved in different species (Fig. 2 F, top panel), although it was not in an Aurora A consensus phosphorylation site in Xenopus. To further verify this phosphorylation, we generated non-phosphorylation-mimicking mutant NuSAP-S240A and phosphorylation-mimicking mutant NuSAP-S240D. We also constructed and purified NuSAP mutant protein NuSAP-S240A and performed an in vitro kinase assay. The results showed that NuSAP-WT was phosphorylated by Aurora A kinase, while the mutant NuSAP-S240A could hardly be phosphorylated (Fig. 2 G), indicating that S240 contributed to the proper phosphorylation. This result was also consistent with previous reports (Sardon et al., 2010), whereas the related function was unclear.
NuSAP phosphorylation by Aurora A affects mitotic spindle dynamics
To determine the biological importance of NuSAP phosphorylation by Aurora A, we performed the rescue assay in NuSAP knockout HeLa cells using NuSAP-WT, NuSAP-S240A, or NuSAP-S240D. As we have observed that the NuSAP knockout leads to increased rates of microtubule fluxes, we asked whether the phosphorylation of NuSAP also regulates this process. We cotransfected NuSAP-knockout cells with PAGFP-α-tubulin and different mCherry-tagged NuSAP constructs and performed the photoactivation experiment as before (Fig. 3, A–C; and Videos 3, 4, and 5). By measuring the velocity of the fluorescent signal approaching the spindle pole, we found that in NuSAP knockout metaphase cells, the mean velocity of microtubule flux was 0.28 μm/min when expressing mCherry-NuSAP-WT and was 0.30 μm/min when expressing mCherry-NuSAP-S240D. In contrast, the rate of poleward flux was significantly increased to 0.47 μm/min when expressing NuSAP-S240A (Fig. 3, D–J). Hence, we concluded that the phospho-null NuSAP mutant could not rescue the defects in microtubule flux caused by NuSAP knockout.
Phosphorylation status of NuSAP by Aurora A regulates spindle microtubule flux. (A–C) NuSAP-knockout HeLa cells were transfected with photoactivatable GFP-tagged α-tubulin (PAGFP-α-tubulin, green) and mCherry-tagged NuSAP-WT, -S240A, or -240D (red) followed by live-cell imaging. GFP signal in a rectangular region near the microtubule plus ends was activated (time point 0, arrows) and tracked every 10 s. Representative time-course images are shown. Microtubules were stained with SiR-tubulin (magenta). (D–F) Corresponding kymograph profiles of the photoactivated regions in A–C (red dotted lines highlight microtubule-flux slopes). Scale bars, 10 s. (G–I) The fluorescence intensity profiles at time points 0 and 120 s in A–C are shown, respectively. (J) The rates of microtubule flux shown in A–C were measured (as described in the legend for Fig. 1 L). Error bars indicate SD. Three independent replicates of 13 or 14 cells per replicate were quantified (n = 3). Unpaired two-tailed t test: P < 0.0001 for GFP-NuSAP/GFP-S240A; P < 0.0001 for GFP-S240A/GFP-S240D; P = 0.2511 for GFP-NuSAP/GFP-S240D. (K) NuSAP-knockout HeLa cells were transfected with GFP-tagged NuSAP-WT, -S240A, or -S240D (green) for 48 h, followed by immunofluorescence labeling using anti-α-tubulin antibodies (red) and anti-γ-tubulin antibodies (magenta). (L) Quantification of spindle length in K. Error bars indicate SD. Three independent replicates of 30 cells per replicate were quantified (n = 3). Unpaired two-tailed t test: P = 0.0002 for GFP-NuSAP/GFP-S240A; P = 0.0004 for GFP-S240A/GFP-S240D; P = 0.0550 for GFP-NuSAP/GFP-S240D. ns, not significant; ***, P < 0.001; ****, P < 0.0001. n = the number of independent experiments presented. Scale bars, 10 μm.
Phosphorylation status of NuSAP by Aurora A regulates spindle microtubule flux. (A–C) NuSAP-knockout HeLa cells were transfected with photoactivatable GFP-tagged α-tubulin (PAGFP-α-tubulin, green) and mCherry-tagged NuSAP-WT, -S240A, or -240D (red) followed by live-cell imaging. GFP signal in a rectangular region near the microtubule plus ends was activated (time point 0, arrows) and tracked every 10 s. Representative time-course images are shown. Microtubules were stained with SiR-tubulin (magenta). (D–F) Corresponding kymograph profiles of the photoactivated regions in A–C (red dotted lines highlight microtubule-flux slopes). Scale bars, 10 s. (G–I) The fluorescence intensity profiles at time points 0 and 120 s in A–C are shown, respectively. (J) The rates of microtubule flux shown in A–C were measured (as described in the legend for Fig. 1 L). Error bars indicate SD. Three independent replicates of 13 or 14 cells per replicate were quantified (n = 3). Unpaired two-tailed t test: P < 0.0001 for GFP-NuSAP/GFP-S240A; P < 0.0001 for GFP-S240A/GFP-S240D; P = 0.2511 for GFP-NuSAP/GFP-S240D. (K) NuSAP-knockout HeLa cells were transfected with GFP-tagged NuSAP-WT, -S240A, or -S240D (green) for 48 h, followed by immunofluorescence labeling using anti-α-tubulin antibodies (red) and anti-γ-tubulin antibodies (magenta). (L) Quantification of spindle length in K. Error bars indicate SD. Three independent replicates of 30 cells per replicate were quantified (n = 3). Unpaired two-tailed t test: P = 0.0002 for GFP-NuSAP/GFP-S240A; P = 0.0004 for GFP-S240A/GFP-S240D; P = 0.0550 for GFP-NuSAP/GFP-S240D. ns, not significant; ***, P < 0.001; ****, P < 0.0001. n = the number of independent experiments presented. Scale bars, 10 μm.
MT flux on metaphase spindles in HeLa cells in the background of mCherry-NuSAP-WT. NuSAP-knockout HeLa cells were co-transfected with photoactivatable GFP-tagged α-tubulin (PAGFP-α-tubulin, green) and mCherry-NuSAP-WT (red). The cells were treated with thymidine for 17–24 h and then released for 9 h. 405 nm laser was applied to activate the GFP signal in a rectangular region near the MT plus ends. Microtubules were stained with SiR-tubulin (magenta). Confocal images were acquired every 10 s for 2 min using the spinning disk confocal microscope equipped with a 60×/1.4 NA oil objective lens at 37°C. Reproduced at 2 fps. Selected video stills are shown in Fig. 3 A.
MT flux on metaphase spindles in HeLa cells in the background of mCherry-NuSAP-WT. NuSAP-knockout HeLa cells were co-transfected with photoactivatable GFP-tagged α-tubulin (PAGFP-α-tubulin, green) and mCherry-NuSAP-WT (red). The cells were treated with thymidine for 17–24 h and then released for 9 h. 405 nm laser was applied to activate the GFP signal in a rectangular region near the MT plus ends. Microtubules were stained with SiR-tubulin (magenta). Confocal images were acquired every 10 s for 2 min using the spinning disk confocal microscope equipped with a 60×/1.4 NA oil objective lens at 37°C. Reproduced at 2 fps. Selected video stills are shown in Fig. 3 A.
MT flux on metaphase spindles in HeLa cells in the background of mCherry-NuSAP-S240A. NuSAP-knockout HeLa cells were co-transfected with photoactivatable GFP-tagged α-tubulin (PAGFP-α-tubulin, green) and mCherry-NuSAP-S240A (red). The cells were treated with thymidine for 17–24 h and then released for 9 h. 405 nm laser was applied to activate the GFP signal in a rectangular region near the MT plus ends. Microtubules were stained with SiR-tubulin (magenta). Confocal images were acquired every 10 s for 2 min using the spinning disk confocal microscope equipped with a 60×/1.4 NA oil objective lens at 37°C. Reproduced at 2 fps. Selected video stills are shown in Fig. 3 B.
MT flux on metaphase spindles in HeLa cells in the background of mCherry-NuSAP-S240A. NuSAP-knockout HeLa cells were co-transfected with photoactivatable GFP-tagged α-tubulin (PAGFP-α-tubulin, green) and mCherry-NuSAP-S240A (red). The cells were treated with thymidine for 17–24 h and then released for 9 h. 405 nm laser was applied to activate the GFP signal in a rectangular region near the MT plus ends. Microtubules were stained with SiR-tubulin (magenta). Confocal images were acquired every 10 s for 2 min using the spinning disk confocal microscope equipped with a 60×/1.4 NA oil objective lens at 37°C. Reproduced at 2 fps. Selected video stills are shown in Fig. 3 B.
MT flux on metaphase spindles in HeLa cells in the background of mCherry-NuSAP-S240D. NuSAP-knockout HeLa cells were co-transfected with photoactivatable GFP-tagged α-tubulin (PAGFP-α-tubulin) and mCherry-NuSAP-S240D (red). The cells were treated with thymidine for 17–24 h and then released for 9 h. 405 nm laser was applied to activate the GFP signal in a rectangular region near the MT plus ends. Microtubules were stained with SiR-tubulin (magenta). Confocal images were acquired every 10 s for 2 min using the Spinning disk confocal microscope equipped with a 60×/1.4 NA oil objective lens at 37°C. Reproduced at 2 fps. Selected video stills are shown in Fig. 3 C.
MT flux on metaphase spindles in HeLa cells in the background of mCherry-NuSAP-S240D. NuSAP-knockout HeLa cells were co-transfected with photoactivatable GFP-tagged α-tubulin (PAGFP-α-tubulin) and mCherry-NuSAP-S240D (red). The cells were treated with thymidine for 17–24 h and then released for 9 h. 405 nm laser was applied to activate the GFP signal in a rectangular region near the MT plus ends. Microtubules were stained with SiR-tubulin (magenta). Confocal images were acquired every 10 s for 2 min using the Spinning disk confocal microscope equipped with a 60×/1.4 NA oil objective lens at 37°C. Reproduced at 2 fps. Selected video stills are shown in Fig. 3 C.
Then, we tested how S240 phosphorylation of NuSAP regulates mitotic spindle length control. The results showed that GFP-tagged NuSAP-WT and NuSAP-S240D almost entirely rescued metaphase short spindles resulting from NuSAP knockout, which even caused spindle elongation beyond its normal length due to the overexpression, whereas NuSAP-S240A was unable to rescue this defect (Fig. 3, K and L). Furthermore, we carried out live-cell imaging in NuSAP-knockdown HeLa cells expressing different NuSAP mutants and RFP-H2B and revealed delayed chromosome biorientation and mitosis progression in cells transfected with GFP-NuSAP-S240A. The mean time from NEBD to proper chromosome alignment in cells expressing NuSAP-WT or NuSAP-S240D was 61.40 ± 3.72 and 96.70 ± 7.76 min, respectively, whereas this time was significantly prolonged to 203.4 ± 18.98 min in cells expressing NuSAP-S240A (Fig. S2, A–C; and Videos 6, 7, and 8). Taken together, these results demonstrate that the phosphorylation of NuSAP at Ser-240 by Aurora A regulates the spindle assembly, dynamics, and chromosome congression for normal mitotic progression.
Phospho-null NuSAP resulted in delayed mitosis progression. (A) RFP-H2B (red) expressing HeLa cells with NuSAP siRNA knockdown were processed for rescue experiments with GFP-tagged NuSAP-WT, -S240A, and -S240D (green) followed by live-cell imaging. Images were captured every 3 min with five slices of 8-μm Z-section thickness. (B) Detection of NuSAP RNAi knockdown efficiency in HeLa cells by Western blotting. (C) Quantification of the time from NEBD to chromosome segregation, shown in A. Error bars indicate SD. Three independent replicates of 10 cells per replicate were quantified (n = 3). Unpaired two-tailed t test. P < 0.0001 for WT/S240A. P < 0.0001 for S240A/S240D. ****, P < 0.0001. n = the number of independent experiments presented. Scale bars, 10 μm. Source data are available for this figure: SourceData FS2.
Phospho-null NuSAP resulted in delayed mitosis progression. (A) RFP-H2B (red) expressing HeLa cells with NuSAP siRNA knockdown were processed for rescue experiments with GFP-tagged NuSAP-WT, -S240A, and -S240D (green) followed by live-cell imaging. Images were captured every 3 min with five slices of 8-μm Z-section thickness. (B) Detection of NuSAP RNAi knockdown efficiency in HeLa cells by Western blotting. (C) Quantification of the time from NEBD to chromosome segregation, shown in A. Error bars indicate SD. Three independent replicates of 10 cells per replicate were quantified (n = 3). Unpaired two-tailed t test. P < 0.0001 for WT/S240A. P < 0.0001 for S240A/S240D. ****, P < 0.0001. n = the number of independent experiments presented. Scale bars, 10 μm. Source data are available for this figure: SourceData FS2.
Spindle formation and chromosome alignment in the background of GFP-NuSAP-WT. HeLa cells stably expressing RFP-H2B (red) were depleted of endogenous NuSAP and transiently expressing RNAi-resistant GFP-NuSAP-WT (green). The cells were treated with thymidine for 17–24 h and then released for 9 h. Confocal images were acquired every 3 min for 10–12 h using the DeltaVision system equipped with a 60×/1.42 NA oil objective at 37°C. Reproduced at 10 fps. Selected video stills are shown in Fig. S2 A. Scale bar, 10 μm.
Spindle formation and chromosome alignment in the background of GFP-NuSAP-WT. HeLa cells stably expressing RFP-H2B (red) were depleted of endogenous NuSAP and transiently expressing RNAi-resistant GFP-NuSAP-WT (green). The cells were treated with thymidine for 17–24 h and then released for 9 h. Confocal images were acquired every 3 min for 10–12 h using the DeltaVision system equipped with a 60×/1.42 NA oil objective at 37°C. Reproduced at 10 fps. Selected video stills are shown in Fig. S2 A. Scale bar, 10 μm.
Spindle formation and chromosome alignment in the background of GFP-NuSAP-S240A. HeLa cells stably expressing RFP-H2B (red) were depleted of endogenous NuSAP and transiently expressing RNAi-resistant GFP-NuSAP-S240A (green). The cells were treated with thymidine for 17–24 h and then released for 9 h. Confocal images were acquired every 3 min for 10–12 h using the DeltaVision system equipped with a 60×/1.42 NA oil objective at 37°C. Reproduced at 10 fps. Selected video stills are shown in Fig. S2 A. Scale bar, 10 μm.
Spindle formation and chromosome alignment in the background of GFP-NuSAP-S240A. HeLa cells stably expressing RFP-H2B (red) were depleted of endogenous NuSAP and transiently expressing RNAi-resistant GFP-NuSAP-S240A (green). The cells were treated with thymidine for 17–24 h and then released for 9 h. Confocal images were acquired every 3 min for 10–12 h using the DeltaVision system equipped with a 60×/1.42 NA oil objective at 37°C. Reproduced at 10 fps. Selected video stills are shown in Fig. S2 A. Scale bar, 10 μm.
Spindle formation and chromosome alignment in the background of GFP-NuSAP-S240D. HeLa cells stably expressing RFP-H2B (red) were depleted of endogenous NuSAP and transiently expressing RNAi-resistant GFP-NuSAP-S240D (green). The cells were treated with thymidine for 17–24 h and then released for 9 h. Confocal images were acquired every 3 min for 10–12 h using the DeltaVision system equipped with a 60×/1.42 NA oil objective at 37°C. Reproduced at 10 fps. Selected video stills are shown in Fig. S2 A. Scale bar, 10 μm.
Spindle formation and chromosome alignment in the background of GFP-NuSAP-S240D. HeLa cells stably expressing RFP-H2B (red) were depleted of endogenous NuSAP and transiently expressing RNAi-resistant GFP-NuSAP-S240D (green). The cells were treated with thymidine for 17–24 h and then released for 9 h. Confocal images were acquired every 3 min for 10–12 h using the DeltaVision system equipped with a 60×/1.42 NA oil objective at 37°C. Reproduced at 10 fps. Selected video stills are shown in Fig. S2 A. Scale bar, 10 μm.
Collectively, these results show that phosphorylation of NuSAP Ser-240 maintains normal spindle microtubule flux, while phospho-null NuSAP accelerates the poleward movement of microtubules.
NuSAP negatively regulates the concentration of Kif2A on spindle poles
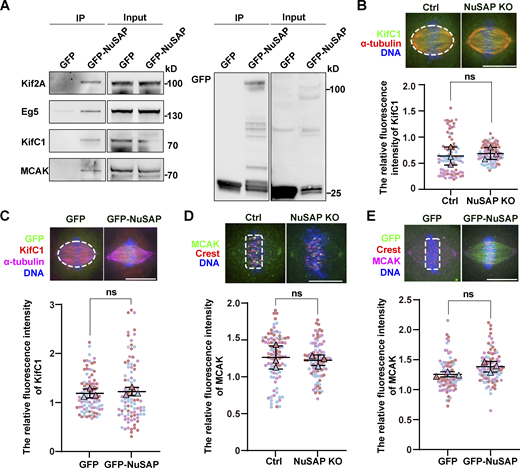
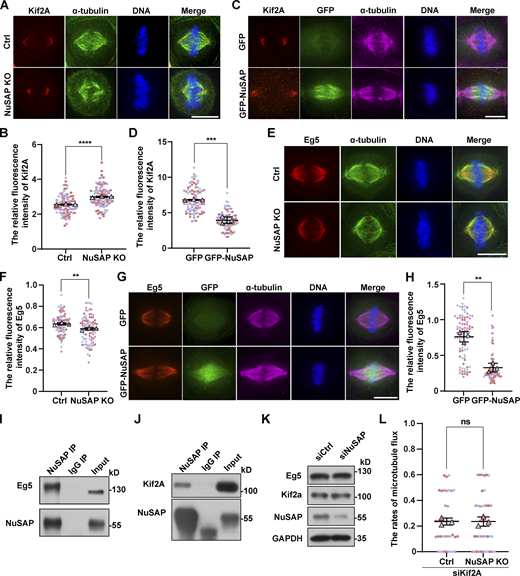
To investigate how NuSAP might affect spindle length, we next identified the cellular targets of NuSAP in mitosis. Firstly, we tested the interaction between NuSAP and several established contributors to microtubule flux and spindle length control, including Kif2A, MCAK, Eg5, and KifC1. Kif2A and MCAK are able to depolymerize microtubules on spindle poles and kinetochores, respectively, and to regulate spindle length (Ganem and Compton, 2004; Henkin et al., 2023). It has been reported that NuSAP regulates the dynamics of kinetochore microtubules by attenuating MCAK depolymerization activity (Li et al., 2016). Eg5 contributes to spindle microtubule flux by mediating the sliding of antiparallel interpolar microtubules, and the changes in spindle length caused by knockdown or overexpression of KifC1 are similar to that of NuSAP (Brust-Mascher et al., 2009; Cai et al., 2009; Miyamoto et al., 2004). Surprisingly, the IP assay showed that NuSAP could interact with all four protein regulators mentioned above (Fig. S3 A). Next, we explored whether NuSAP regulated the localization of these proteins. We analyzed the localization of these regulators after knockout or overexpression of NuSAP. Immunofluorescence statistics showed that NuSAP had no effect on the relative fluorescence intensity of KifC1 on spindle microtubules and that of MCAK at microtubule plus ends (Fig. S3, B–E). In contrast, Kif2A fluorescence intensity on spindle poles was significantly enhanced due to NuSAP knockout and reduced by NuSAP overexpression (Fig. 4, A–D). Moreover, Eg5 was reduced on spindle microtubules in NuSAP-knockout cells compared with the control, whereas it was also reduced by NuSAP overexpression, which was probably caused by spindle elongation (Fig. 4, E–H). To confirm the interaction between NuSAP and the two target proteins, we performed an IP assay using anti-NuSAP antibodies and pulled down endogenous Eg5 or Kif2A from mitotic HeLa lysates. Indeed, the results showed that endogenous NuSAP interacted with endogenous Eg5 and Kif2A (Fig. 4, I and J). Through Western blot analysis, we showed that the protein levels of Eg5 and Kif2A were unaffected by NuSAP knockdown (Fig. 4 K). These results indicate that NuSAP influenced the subcellular localization of Eg5 and Kif2A but not their expression. Furthermore, we codepleted Kif2A and NuSAP in HeLa cells and measured the rates of microtubule flux. We found that the increased rate of microtubule flux caused by NuSAP knockout was rescued by Kif2A depletion, indicating that NuSAP regulated microtubule flux mainly through Kif2A (Fig. 4 L). Collectively, these results indicate that NuSAP negatively regulates the concentration of Kif2A on spindle poles and might promote the localization of Eg5 on spindle microtubules, and so Eg5 and Kif2A might be the main downstream targets of NuSAP.
NuSAP had no effect on the localization of KifC1 and MCAK. (A) NuSAP interacted with Kif2A, Eg5, KifC1, and MCAK. Mitotic HEK293T cells were transfected with GFP or GFP-NuSAP and then immunoprecipitated with GFP-Trap beads. (B and D) Sample images (top) and quantification (bottom) of relative KifC1 (B) and MCAK (D) fluorescence intensity in the control and NuSAP-knockout cells, respectively. Error bars indicate SD. Three independent replicates of 30 cells per replicate were quantified (n = 3). Unpaired two-tailed t test: P = 0.7074 in B; P = 0.7179 in D. The white dotted lines show the areas of measurement. (C and E) Sample images and quantification of relative KifC1 (C) and MCAK (E) fluorescence intensity in HeLa cells transfected with GFP or GFP-NuSAP, respectively. Error bars indicate SD. Three independent replicates of 30 cells per replicate were quantified (n = 3). Unpaired two-tailed t test: P = 0.6413 in C; P = 0.0880 in E. The white dotted lines show the areas of measurement. ns, not significant. n = the number of independent experiments presented. Scale bars, 10 μm. Source data are available for this figure: SourceData FS3.
NuSAP had no effect on the localization of KifC1 and MCAK. (A) NuSAP interacted with Kif2A, Eg5, KifC1, and MCAK. Mitotic HEK293T cells were transfected with GFP or GFP-NuSAP and then immunoprecipitated with GFP-Trap beads. (B and D) Sample images (top) and quantification (bottom) of relative KifC1 (B) and MCAK (D) fluorescence intensity in the control and NuSAP-knockout cells, respectively. Error bars indicate SD. Three independent replicates of 30 cells per replicate were quantified (n = 3). Unpaired two-tailed t test: P = 0.7074 in B; P = 0.7179 in D. The white dotted lines show the areas of measurement. (C and E) Sample images and quantification of relative KifC1 (C) and MCAK (E) fluorescence intensity in HeLa cells transfected with GFP or GFP-NuSAP, respectively. Error bars indicate SD. Three independent replicates of 30 cells per replicate were quantified (n = 3). Unpaired two-tailed t test: P = 0.6413 in C; P = 0.0880 in E. The white dotted lines show the areas of measurement. ns, not significant. n = the number of independent experiments presented. Scale bars, 10 μm. Source data are available for this figure: SourceData FS3.
NuSAP negatively regulates the concentration of Kif2A on spindle poles and promotes the localization of Eg5 on spindle microtubules. (A and E) Control and NuSAP-knockout HeLa cells were stained with anti-Kif2A (red) and anti-α-tubulin (green) antibodies in A and stained with anti-Eg5 (red) and anti-α-tubulin (green) antibodies in E. DNA was stained with DAPI (blue). (B and F) Quantification of relative Kif2A (B) and Eg5 (F) fluorescence intensity on the spindle in A and E, respectively. Error bars indicate SD. Three independent replicates of 30 cells per replicate were quantified (n = 3). Unpaired two-tailed t test: P < 0.0001 in B; P = 0.0080 in F. (C and G) Mitotic HeLa cells were transfected with GFP or GFP-NuSAP (green) and stained with anti-Kif2A (red) and anti-α-tubulin (magenta) antibodies in C, and stained with anti-Eg5 (red) and anti-α-tubulin (magenta) in G. DNA was stained with DAPI (blue). (D and H) Quantification of relative Kif2A (D) and Eg5 (H) fluorescence intensity on the spindle in C and G, respectively. Error bars indicate SD. Three independent replicates of 30 cells per replicate were quantified (n = 3). Unpaired two-tailed t test: P = 0.0004 in D; P = 0.0014 in H. (I and J) Mitotic HeLa cells were subjected to an IP assay using nonspecific IgG or anti-NuSAP antibodies followed by Western blot analysis with anti-NuSAP, anti-Eg5 (I), and anti-Kif2A (J) antibodies. (K) Test the protein level of Kif2A and Eg5 after NuSAP knockdown in HeLa cells. (L) The mean velocity of poleward spindle microtubule flux in Kif2A-depleted and NuSAP/Kif2A-co-depleted cells. Error bars indicate SD. Three independent replicates of 13 or 14 cells per replicate were quantified (n = 3). Unpaired two-tailed t test: P = 0.9884. ns, not significant; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. n = the number of independent experiments presented. Scale bars, 10 μm. Source data are available for this figure: SourceData F4.
NuSAP negatively regulates the concentration of Kif2A on spindle poles and promotes the localization of Eg5 on spindle microtubules. (A and E) Control and NuSAP-knockout HeLa cells were stained with anti-Kif2A (red) and anti-α-tubulin (green) antibodies in A and stained with anti-Eg5 (red) and anti-α-tubulin (green) antibodies in E. DNA was stained with DAPI (blue). (B and F) Quantification of relative Kif2A (B) and Eg5 (F) fluorescence intensity on the spindle in A and E, respectively. Error bars indicate SD. Three independent replicates of 30 cells per replicate were quantified (n = 3). Unpaired two-tailed t test: P < 0.0001 in B; P = 0.0080 in F. (C and G) Mitotic HeLa cells were transfected with GFP or GFP-NuSAP (green) and stained with anti-Kif2A (red) and anti-α-tubulin (magenta) antibodies in C, and stained with anti-Eg5 (red) and anti-α-tubulin (magenta) in G. DNA was stained with DAPI (blue). (D and H) Quantification of relative Kif2A (D) and Eg5 (H) fluorescence intensity on the spindle in C and G, respectively. Error bars indicate SD. Three independent replicates of 30 cells per replicate were quantified (n = 3). Unpaired two-tailed t test: P = 0.0004 in D; P = 0.0014 in H. (I and J) Mitotic HeLa cells were subjected to an IP assay using nonspecific IgG or anti-NuSAP antibodies followed by Western blot analysis with anti-NuSAP, anti-Eg5 (I), and anti-Kif2A (J) antibodies. (K) Test the protein level of Kif2A and Eg5 after NuSAP knockdown in HeLa cells. (L) The mean velocity of poleward spindle microtubule flux in Kif2A-depleted and NuSAP/Kif2A-co-depleted cells. Error bars indicate SD. Three independent replicates of 13 or 14 cells per replicate were quantified (n = 3). Unpaired two-tailed t test: P = 0.9884. ns, not significant; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. n = the number of independent experiments presented. Scale bars, 10 μm. Source data are available for this figure: SourceData F4.
NuSAP controls spindle assembly, dynamics, and metaphase spindle length by negatively regulating Kif2A on spindle poles in an Aurora A–dependent manner
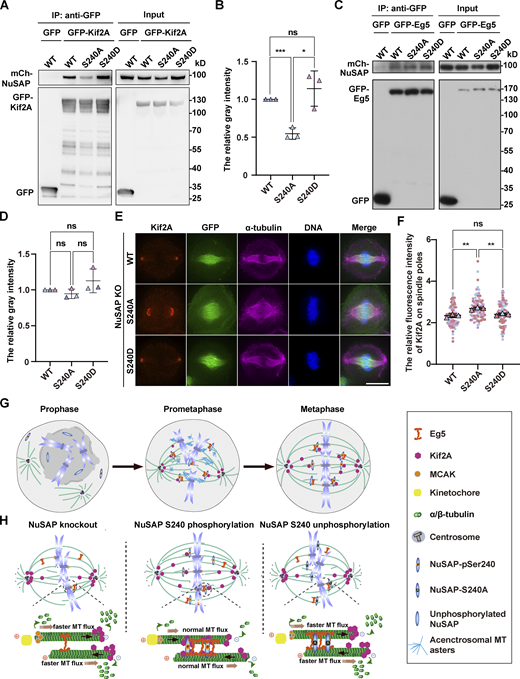
To further test whether phosphorylation of NuSAP at Ser-240 affects these interactions, we cotransfected cells with GFP-Kif2A and different mCherry-tagged NuSAP constructs followed by co-IP assay in mitosis cells. The results showed that Kif2A had a stronger affinity to phosphorylated NuSAP, including WT and S240D, than S240A (Fig. 5, A and B). In the same way, we found that Eg5 bound to NuSAP-WT, -S240A, and -S240D with similar affinity (Fig. 5, C and D). These data show that the interaction between NuSAP and the microtubule depolymerase Kif2A is Aurora A phosphorylation-dependent and that abolishing NuSAP phosphorylation reduces its affinity to Kif2A. Next, through immunostaining for Kif2A in NuSAP knockout-and-rescue cells, we further tested how phosphorylation of NuSAP at Ser-240 influenced Kif2A function in regulating metaphase microtubule flux and spindle assembly. To fully establish the bipolar spindle, we treated synchronized mitotic cells with 10 μM MG132 for 1 h. The results showed that more Kif2A localized on spindle poles in cells expressing NuSAP-S240A than in cells expressing NuSAP-WT and -S240D (Fig. 5, E and F). As reported, Kif2A could depolymerize microtubules at their minus ends (Desai et al., 1999; Ganem et al., 2005). Consequently, an increased concentration of Kif2A on spindle microtubules, caused by NuSAP knockout or NuSAP-240A overexpression, would accelerate the microtubule flux on the spindle. Collectively, these results indicate that NuSAP controls the spindle assembly, structural dynamics, and metaphase length by negatively regulating Kif2A on spindle poles in an Aurora A–dependent manner.
Ser-240 phosphorylation of NuSAP promotes its interaction with Kif2A and reduces the localization of Kif2A on spindle poles. (A and C) Mitotic HEK293T cells cotransfected with the indicated GFP-tagged proteins and mCherry-tagged NuSAP constructs were immunoprecipitated with GFP-Trap beads. (B and D) The quantification of protein–protein interaction discrepancy. Error bars indicate SD. Three independent replicates were quantified (n = 3). Unpaired two-tailed t test: P = 0.0005 for WT/S240A, P = 0.0135 for S240A/S240D, P = 0.3489 for WT/S240D in B; P = 0.2274 for WT/S240A, P = 0.1531 for S240A/S240D, P = 0.2562 for WT/S240D in D. (E) NuSAP-knockout HeLa cells were transfected with GFP-tagged NuSAP-WT, -S240A, or -S240D (green) followed by immunofluorescence labeling using anti-Kif2A (red) and anti-α-tubulin (magenta) antibodies. DNA was stained with DAPI (blue). (F) Quantification of relative Kif2A intensity on spindle poles in E. Error bars indicate SD. Three independent replicates of 30 cells per replicate were quantified (n = 3). Unpaired two-tailed t test: P = 0.0049 for WT/S240A; P = 0.0051 for S240A/S240D; P = 0.3347 for WT/S240D. ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001. n = the number of independent experiments presented. Scale bars, 10 μm. (G) Along with the nuclear envelope broken down during mitotic entry, microtubule asters nucleate around the chromosomes and the centrosomes to form small acentrosomal and large centrosomal asters. These oriented microtubules are bundled and then sorted into the large centrosomal microtubule asters to form a bipolar spindle structure by molecular motors and microtubule-associated proteins till to a well-organized metaphase spindle formation with a dynamically constant length, in which NuSAP plays a crucial role. (H) NuSAP is mainly localized on microtubules near chromosomes and is phosphorylated by the mitotic kinase Aurora A on S240. As a MAP, NuSAP regulates the spindle dynamics to maintain metaphase spindle length through coordinating microtubule depolymerization and microtubule–microtubule sliding. On the one hand, NuSAP interacts with Eg5 on the adjacent microtubules, which allows Eg5 to move along the microtubules and to mediate the sliding of antiparallel interpolar microtubules (shown by the black arrow), which increases spindle length. On the other hand, S240-phosphorylated NuSAP interacts with more Kif2A on the spindle body and then reduces the amount of Kif2A on spindle poles, thus restraining the excess accumulation of Kif2A at the minus ends of microtubules to ensure the proper microtubule depolymerization. At the plus end, NuSAP attenuates MCAK depolymerization activity to stabilize kinetochore microtubules. NuSAP regulates microtubule flux (shown by the orange arrow) and controls metaphase spindle length by coordinating with Eg5, Kif2A, and MCAK. In experimental conditions, NuSAP knockout results in a reduction of Eg5 localization on spindle microtubules, excess accumulation of Kif2A on spindle poles, and high MCAK depolymerization activity. This abnormality leads to faster microtubule flux, kinetochore–microtubule attachment failure, and the formation of a shorter metaphase spindle. Alternatively, overexpression of NuSAP-S240A, a loss-of-function mutant that interacts with less Kif2A on the spindle body, also causes faster microtubule flux and shorter metaphase spindle formation. Source data are available for this figure: SourceData F5.
Ser-240 phosphorylation of NuSAP promotes its interaction with Kif2A and reduces the localization of Kif2A on spindle poles. (A and C) Mitotic HEK293T cells cotransfected with the indicated GFP-tagged proteins and mCherry-tagged NuSAP constructs were immunoprecipitated with GFP-Trap beads. (B and D) The quantification of protein–protein interaction discrepancy. Error bars indicate SD. Three independent replicates were quantified (n = 3). Unpaired two-tailed t test: P = 0.0005 for WT/S240A, P = 0.0135 for S240A/S240D, P = 0.3489 for WT/S240D in B; P = 0.2274 for WT/S240A, P = 0.1531 for S240A/S240D, P = 0.2562 for WT/S240D in D. (E) NuSAP-knockout HeLa cells were transfected with GFP-tagged NuSAP-WT, -S240A, or -S240D (green) followed by immunofluorescence labeling using anti-Kif2A (red) and anti-α-tubulin (magenta) antibodies. DNA was stained with DAPI (blue). (F) Quantification of relative Kif2A intensity on spindle poles in E. Error bars indicate SD. Three independent replicates of 30 cells per replicate were quantified (n = 3). Unpaired two-tailed t test: P = 0.0049 for WT/S240A; P = 0.0051 for S240A/S240D; P = 0.3347 for WT/S240D. ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001. n = the number of independent experiments presented. Scale bars, 10 μm. (G) Along with the nuclear envelope broken down during mitotic entry, microtubule asters nucleate around the chromosomes and the centrosomes to form small acentrosomal and large centrosomal asters. These oriented microtubules are bundled and then sorted into the large centrosomal microtubule asters to form a bipolar spindle structure by molecular motors and microtubule-associated proteins till to a well-organized metaphase spindle formation with a dynamically constant length, in which NuSAP plays a crucial role. (H) NuSAP is mainly localized on microtubules near chromosomes and is phosphorylated by the mitotic kinase Aurora A on S240. As a MAP, NuSAP regulates the spindle dynamics to maintain metaphase spindle length through coordinating microtubule depolymerization and microtubule–microtubule sliding. On the one hand, NuSAP interacts with Eg5 on the adjacent microtubules, which allows Eg5 to move along the microtubules and to mediate the sliding of antiparallel interpolar microtubules (shown by the black arrow), which increases spindle length. On the other hand, S240-phosphorylated NuSAP interacts with more Kif2A on the spindle body and then reduces the amount of Kif2A on spindle poles, thus restraining the excess accumulation of Kif2A at the minus ends of microtubules to ensure the proper microtubule depolymerization. At the plus end, NuSAP attenuates MCAK depolymerization activity to stabilize kinetochore microtubules. NuSAP regulates microtubule flux (shown by the orange arrow) and controls metaphase spindle length by coordinating with Eg5, Kif2A, and MCAK. In experimental conditions, NuSAP knockout results in a reduction of Eg5 localization on spindle microtubules, excess accumulation of Kif2A on spindle poles, and high MCAK depolymerization activity. This abnormality leads to faster microtubule flux, kinetochore–microtubule attachment failure, and the formation of a shorter metaphase spindle. Alternatively, overexpression of NuSAP-S240A, a loss-of-function mutant that interacts with less Kif2A on the spindle body, also causes faster microtubule flux and shorter metaphase spindle formation. Source data are available for this figure: SourceData F5.
Based on our present work and previous reports (Fu et al., 2015; Ma et al., 2010; Zhuo et al., 2015), we propose a working model to illustrate the underlying mechanisms of the regulation of spindle assembly, structural dynamics, and metaphase spindle length control (Fig. 5, G and H). After nuclear envelope breakdown, microtubules nucleate around chromosomes and centrosomes and grow with dynamic instability. These oriented microtubules are bundled with each other and gradually sorted into a bipolar spindle by molecular motors and non-motor proteins. During this process, NuSAP, serving as a MAP, promotes kinetochore–microtubule attachment for k-fiber formation and chromosome congression, which results from affecting microtubule dynamics at the plus end by regulating MCAK (Li et al., 2016). NuSAP coordinates microtubule depolymerization and microtubule–microtubule sliding by interacting with Eg5 and Kif2A on spindle microtubules. First, NuSAP promotes Eg5 to crosslink microtubules, thereby facilitating spindle assembly and mediating the sliding of antiparallel interpolar microtubules to increase metaphase spindle length. Also, more importantly, NuSAP interacts with Kif2A on spindle microtubules in an Aurora A phosphorylation-dependent manner, thereby maintaining a proper amount of Kif2A at spindle poles through a phosphorylation and dephosphorylation cycle. Once the minus-ends of microtubules are incorporated into spindle poles, Eg5 and Kif2A induce the proper microtubule poleward movement. When NuSAP is depleted in an experimental condition, the normal localization of Eg5 on spindle microtubules is reduced and excess Kif2A concentrates on spindle poles, leading to instability of microtubules and increased microtubule flux rates and thus to a short metaphase spindle formation with chromosomal misalignment. This chromosome misalignment after NuSAP depletion may be due to faster kinetochore fiber fluxes, reaching the speed of bridging fiber fluxes, according to the “flux-driven centering model” (Risteski et al., 2021, 2022).
Our results showed Aurora A phosphorylates NuSAP at mitosis at a conserved serine residue S240. The affinity of NuSAP to Kif2A is S240 phospho-dependent, yet this is not the case for the NuSAP-Eg5 interaction. NuSAP is mainly localized on the spindle microtubules near chromosomes. Thus, NuSAP and Kif2A may form complexes on the mitotic spindle microtubules. It is likely that through an unknown mechanism, such as conformational change, phosphorylated NuSAP interacts with more Kif2A on the spindle body and then reduces the amount of Kif2A on spindle poles, thus restraining the excess accumulation of Kif2A at the minus ends of microtubules to ensure the stability of mitotic spindles. Although NuSAP is responsible for recruiting Eg5 onto spindle microtubules, immunofluorescence shows that Eg5 localizes on the whole spindle. NuSAP also interacts with the whole spindle but predominantly concentrates on the central part. Therefore, NuSAP most likely interacted with Eg5 on the spindle, especially on the central part. In addition, the affinity of NuSAP to Eg5 is not S240 phospho-dependent. It has been reported that Eg5 activity is required for flux in Xenopus extract spindles and driving the spindle scale through exerting forces between antiparallel microtubules (Miyamoto et al., 2004; Shirasu-Hiza et al., 2004). Biochemical depletion of Eg5 significantly decreases the flux rate (Miyamoto et al., 2004), which is opposite to our observations after NuSAP knockout. Thus, the increased microtubule rate induced by NuSAP knockout is mainly due to Kif2A but not Eg5. It remains to be determined how NuSAP balances the Kif2A-mediated microtubule depolymerization near the poles and the continuous antiparallel microtubule sliding caused by Eg5. It has been reported in detail that microtubule flux is regulated by four kinesins, Eg5, Kif15, CENP-E, and Kif4A, mainly focused on the interpolar microtubule sliding and the microtubule sliding facilitated by chromosome (Steblyanko et al., 2020). Our results show that NuSAP regulates microtubule flux by coordinating microtubule depolymerization and interpolar microtubule sliding. Eg5 and Kif2A are the main downstream effectors of NuSAP to regulate microtubule dynamics and spindle length.
In conclusion, our findings shed light on the crucial roles of NuSAP in regulating mitotic spindle assembly, structural dynamics, and metaphase spindle length control for proper chromosome congression through modulating microtubule flux, in concert with Eg5, Kif2A, and other regulators of the spindle microtubule dynamics.
Materials and methods
Plasmid construction, protein purification, and antibody preparation
Human NUSAP1 was cloned from a cDNA library (from the Jiahuai Han laboratory, Xiamen University, China) and inserted into pEGFP-C1 (Clontech), pmCherry-C1 (Clontech), or pGEX-4T-1 (Amersham Bioscience) vectors. The NuSAP-S240A and NuSAP-S240D mutants were generated by PCR site-directed mutagenesis and then inserted into pEGFP-C1, pmCherry-C1, or pGEX-4T-1 vectors. Human KIF2A and KIF11 (EG5) were cloned from a cDNA library by RT-PCR and inserted into pEGFP-C1. Photoactivatable GFP-α-tubulin was generated through PCR amplification of the α-tubulin sequence into the PAGFP-C1 (Addgene) vector. GFP was cloned from the pEGFP-C1 vector. His-GFP was generated through PCR amplification of the GFP sequence into pET28a (Addgene). Human AURKA was cloned from a cDNA library by RT-PCR and inserted into pCMV-Tag2B (Addgene). The site-directed mutagenesis PCR primers were as follows:
NuSAP-S240A forward 5′-CCAAGAGGAAGACTCGCTGTGGCTTCTA-3′,
reverse 5′-TAGAAGCCACAGCGAGTCTTCCTCTTGG-3′;
NuSAP-S240D forward 5′-CCAAGAGGAAGACTCGATGTGGCTTCTA-3′,
reverse 5′-TAGAAGCCACATCGAGTCTTCCTCTTGG-3′.
GST-tagged proteins were affinity-purified using glutathione-Sepharose 4B beads (GE Healthcare) according to the manufacturer’s protocols. Anti-NuSAP polyclonal antibody was obtained by immunizing mice (BALB/c) with the full-length recombinant GST-tagged NuSAP. This homemade anti-NuSAP antibody was validated by its disappeared signal (full-length NuSAP) when endogenous NuSAP was knocked out by specific sgRNA. Data are shown in Fig. 1 B. The rabbit anti-GFP antibody used for immunoprecipitation was generated by immunizing rabbits with bacterially expressed recombinant His-tagged GFP and this antibody has been validated in previous papers (Luo et al., 2019; Xu et al., 2017). Other antibodies used in this study were as follows: mouse anti-α-tubulin (T9026; Sigma-Aldrich), mouse anti-γ-tubulin (T6557; Sigma-Aldrich), rabbit anti-γ-tubulin (T3559; Sigma-Aldrich), rabbit anti-NuSAP1 (12024-1-AP; Proteintech), mouse anti-Aurora A (A1231; Sigma-Aldrich), rabbit anti-Aurora A (D3E4Q; Cell Signaling Technology), mouse anti-Flag (M185-3L; MBL), mouse anti-cyclin B1 (ab72; Abcam), mouse anti-GAPDH (60004-1-lg; Proteintech), rabbit anti-H3pS10 (ab5176; Abcam), mouse anti-Eg5 (611186; BD Transduction Laboratories), mouse anti-Kif2A (sc-271471; Santa Cruz Biotechnology), rabbit anti-P-Aurora A/B/C (D13A11; Cell Signaling Technology), mouse anti-mCherry (HX1810; huaxingbio), mouse anti-GFP (M048-3; MBL), mouse anti-IgG (B900620; Proteintech), rabbit anti-IgG (B900610; Proteintech), rabbit anti-IgG Light Chain (A25022; Abbkine), and mouse anti-IgG Light Chain (A25012; Abbkine). The following secondary antibodies were used: Alexa Fluor 488-conjugated anti-mouse IgG (H+L; A-21202; Invitrogen), Alexa Fluor 488-conjugated anti-rabbit IgG (H+L; A-21206; Invitrogen), Alexa Fluor 594-conjugated anti-mouse IgG (H+L; A-21203; Invitrogen), Alexa Fluor 594-conjugated anti-rabbit IgG (H+L; A-21207; Invitrogen), Alexa Fluor 647-conjugated anti-mouse IgG (H+L; A32728; Invitrogen), Alexa Fluor 647-conjugated anti-rabbit IgG (H+L; A-31573; Invitrogen), HRP-conjugated anti-rabbit IgG (323-005-021; Jackson), and HRP-conjugated anti-mouse IgG (223-005-024; Jackson). All animal experiments were performed in the Laboratory Animal Center of Peking University according to guidelines approved by the Institutional Animal Care and Use Committee at Peking University.
Cell culture, synchronization, and transfection
HeLa (ATCC, CCL-2) and HEK293T (CRL-11268; ATCC) cells were cultured in DMEM (CORNING) supplemented with 10% bovine calf serum (CellMax) at 37°C in a 5% CO2 incubator. For the double thymidine block and release experiment, the cells were arrested for 17–20 h with 2.5 mM thymidine (Sigma-Aldrich), released for 9 h, and then arrested for 17–20 h with 2.5 mM thymidine. After release into fresh medium, cells were harvested at the indicated time points. Mitosis-arrested cells were obtained by adding 100 ng/ml nocodazole (Sigma-Aldrich) for 12–15 h after release from the thymidine block. HeLa cells were treated with thymidine for 16–20 h, released for 9 h, and then incubated with 10 μM MG132 for 1 h to fully establish the bipolar spindle. Transient cDNA and siRNA transfections were performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. The following siRNA sequence was used: siNuSAP, 5′-GCACCAAGAAGCUGAGAAUTT-3′ (Raemaekers et al., 2003); siKif2A, 5′-GGCAAAGAGAUUGACCUGG-3′; and siCtrl, 5′-UUCUCCGAACGUGUCACGUTT-3′.
CRISPR/Cas9-mediated editing of NuSAP genes in HeLa cells
The single-guide RNAs (sgRNAs) for targeting human NUSAP genes were predicted by CHOPCHOP (https://chopchop.cbu.uib.no/; Ran et al., 2013). To express sgRNA (5′-CTGCAGGTCACTGTACTTGA-3′) for human NUSAP gene knockout, we annealed the oligonucleotides and cloned them into the BbsI site of dual Cas9 and sgRNA expression vector pSpCas9(BB)-2A-Puromycin (Dr. Feng Zhang laboratory, #48139; Addgene). The plasmids were transfected into HeLa cells using TurboFect Transfection Reagent (Invitrogen). After 48 h, cells were treated with 1 μg/ml puromycin for 2–3 d. Then, the live cells were split individually to make a clonal cell line.
Clones with loss of NuSAP proteins were isolated by immunostaining and confirmed by immunoblotting. The genomic DNA PCR fragments were subcloned into pClone007 vector (pClone007 Blunt simple vector kit; Tsingke) and transformed into the E. coli. Then, several numbers of bacterial colonies were sequenced to confirm the gene disruption. The PCR primers were as follows: forward 5′-ACTCGTTACCTGAACAGGCG-3′ and reverse 5′-TTCGTAATCGACGCCCTGAG-3′.
Immunoprecipitation (IP) and Western blotting
Mitotic cells were washed with cold PBS and lysed in cell lysis buffer (20 mM Tris-HCl, pH = 8.0, 150 mM NaCl, 2 mM EGTA, 0.5 mM EDTA, 0.5% NP-40, 5 mM NaF, 1 mM Na3VO4, 1 mM PMSF, and 500× protease inhibitor cocktail; Calbiochem) for 20 min on ice. Lysates were centrifuged at 13,000 rpm for 15 min and the supernatants were incubated with For NuSAP IP assay. HeLa cell lysates were mixed with mouse anti-NuSAP polyclonal antibody (homemade antibody, as described in “antibody preparation”) or rabbit anti-NuSAP polyclonal antibody (12024-1-AP; Proteintech). For anti-Flag and anti-GFP IP assays, HEK293T cells were transfected with the indicated constructs Flag-Aurora A and GFP or GFP-NuSAP, and the cell lysates were mixed with protein A-Sepharose beads (101041; Thermo Fisher Scientific) conjugated with indicated antibodies for 1 h at 4°C. After five washes with lysis buffer, the beads were suspended in the Laemmli sample buffer. After being resolved on SDS-PAGE gels, the proteins were transferred to nitrocellulose membranes that were then blocked in TTBS (20 mM Tris-HCl, pH = 7.5, 500 mM NaCl, and 0.3% Tween 20) containing 3% nonfat milk at room temperature for 1 h. Then, they were probed with primary antibodies (diluted in nonfat milk) and HRP-conjugated secondary antibodies. The membranes were developed for visualization by enhanced chemiluminescence (Sigma-Aldrich) and X-ray film.
Immunofluorescence and live-cell imaging
Cells were grown on coverslips and fixed in precooled methanol for 5 min on ice, followed by incubation with primary antibodies (diluted in 3% BSA) at 4°C overnight. After washing three times with PBS, the cells were incubated with fluorescence-labeled secondary antibodies for 1 h at room temperature. Coverslips were mounted with Mowiol (Sigma-Aldrich) containing 1 μg/ml DAPI (Sigma-Aldrich) for DNA staining. For live-cell imaging, cells were plated on a glass-bottom dish. Before imaging, the dishes were locked in a heated chamber (37°C) supplemented with 5% CO2.
Microscopy
Some immunofluorescence images were acquired using an imaging system (DeltaVision; Applied Precision) equipped with an inverted microscope (IX-71; Olympus) and a 100×/1.42 NA oil objective at room temperature. Most immunofluorescence images were acquired using PerkinElmer UltraView VoX spinning disk confocal microscope or Andor Dragonfly spinning disk confocal microscope with a 100×/1.4 N.A. oil objective lens of EMCCD at room temperature. Photoactivation images were acquired using a Spinning disk confocal microscope equipped with an inverted microscope (Nikon TiE) and a 60×/1.4 NA oil objective lens at 37°C. Live-cell images were acquired using an imaging system (DeltaVision; Applied Precision) equipped with an inverted microscope (IX-71; Olympus) and a 60×/1.42 NA oil objective at 37°C.
Image acquisition and analysis
Within one experiment, images were acquired under identical imaging settings. The levels of fluorescent signals were sometimes differentially adjusted on final image panels to improve the visibility of dimmer signals. Such adjustments were not performed individually if a quantitative comparison between cells was intended. All immunofluorescence images were captured with a 4-μm Z-section thickness by eight slices and processed for maximum intensity projection. Photoactivation images were acquired every 10 s. Live-cell images were captured with a 6-μm Z-section thickness by six slices and were acquired every 3 min. The images were analyzed using Imaris 9.5 and Volocity 6.1.1.
GFP-α-tubulin photoactivation analysis
For activation studies, mitotic HeLa cells transfected with photoactivatable GFP-α-tubulin and mCherry-tagged constructs were identified by microscopy, and after the acquisition of three preactivation frames, a 405-nm laser was used to activate GFP-α-tubulin in a rectangular region near the microtubule plus ends inside the spindle. For microtubule poleward flux experiments, we measured the distance between the brightest points of the signal at 0 and 120 s, and we defined the mean velocity by the ratio of the above distance to the time of 2 min. To detect the decay rate of the PAGFP-α-tubulin signal, we tracked the brightest point, measured the fluorescence intensity of the PAGFP-α-tubulin signal every 10 s, and normalized the data in the statistics.
λ protein phosphatase (λ-PPase) assay
Mitotic HeLa cells were collected from a 35-mm dish and lysed with lysis buffer, containing 20 mM Tris-HCl, pH = 8.0, 150 mM NaCl, 2 mM EGTA, 0.5% NP-40, and complete EDTA-free protease inhibitors (Roche) on ice for 20 min. Lysates were centrifuged at 13,000 rpm for 15 min and supernatants were incubated with 10× NEBuffer for PMP, 10 × 10 mM MnCl2, and 400 U λ-PPase (New England Biolabs, Inc.) at 30°C for 30 min before being stopped with SDS sample buffer. Then, the reaction system was analyzed by Western blotting analysis.
In vitro kinase assays
2 μg of GST-tagged NuSAP point mutants were incubated with 50 ng human recombinant Aurora A (14-511-M; Sigma-Aldrich) in a water bath at 30°C for 30 min in kinase buffer (50 mM Tris-HCl, pH = 7.5, 10 mM MgCl2, 2 mM EGTA, 5 mM DTT) containing 100 μM ATP and 6,000 Ci/mmol γ-[32P] ATP (GE Healthcare). Reactions were quenched by adding Laemmli sample buffer and analyzed by SDS-PAGE and autoradiography.
Measurement of relative intensity
For fluorescence intensity of Kif2A on spindle poles, four square boxes of 3 × 3 μm were drawn, two on two spindle poles and the other two on their adjacent spindle areas. The mean fluorescence intensity was measured using Velocity software (version 6.1.1). The relative fluorescence intensity was defined by the ratio of the mean intensity on spindle poles and the adjacent spindle areas. The relative fluorescence intensity of Eg5 (or KifC1) was defined by the ratio of the mean fluorescence intensity of Eg5 (or KifC1) and α-tubulin on the entire spindle microtubule area. The relative fluorescence intensity of MCAK was defined by the ratio of the mean fluorescence intensity of MCAK and Crest on the entire chromosome area.
Quantification of Western blots
For mean gray intensities of immunoblotting bands, the same size boxes were drawn on the relevant bands. The intensities of the bands were measured using ImageJ. The relative gray intensity between mCherry-tagged NuSAP constructs and GFP-Kif2A or GFP-Eg5 was defined by the ratio of the mean intensity of interacting mCherry-NuSAP proteins to that of immunoprecipitated GFP-Kif2A or GFP-Eg5. The value of the WT group was set as 1.0 and then the other mutant groups were calculated according to WT.
Statistical analysis
Statistical significance was performed using Graphpad Prism (ver. 6.0c) software. We did not pre-establish exclusion criteria. P values were calculated with a two-tailed t test from the mean values of the indicated data. All statistical data were presented as mean ± SD. Significant difference was marked with asterisks (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, no significant difference).
Online supplemental material
Fig. S1 shows that NuSAP is not necessary for acentrosomal MT nucleation and is required for kinetochore–microtubule attachment. Fig. S2 examines the mitotic progression for rescue experiments with GFP-tagged NuSAP-WT, -S240A, and -S240D in NuSAP siRNA-treated cells. Fig. S3 shows the effect on the localization of NuSAP’s binding partners in NuSAP KO and NuSAP-overexpressed cells. Videos 1 and 2 show NuSAP depletion leads to faster microtubule flux. Videos 3, 4, and 5 show that NuSAP phosphorylation by Aurora A maintains the normal microtubule flux. Videos 6, 7, and 8 show that NuSAP phosphorylation by Aurora A promotes mitotic progression.
Data availability
All pertinent data are provided in the main text and supplemental material. A list of the reagents included in this study is available on request from the corresponding author.
Acknowledgments
We thank Dr. Jiahuai Han (Xiamen University, Xiamen, Fujian, China) for providing NUSAP1 cDNA, and we thank the National Center for Protein Sciences at Peking University (Beijing, China), particularly Drs. Hongxia Lv, Xiaochen Li, Siying Qin, and Dong Liu for technical help. We thank Dr. Zhaoxuan Deng for raising the anti-NuSAP polyclonal antibody in the Zhang lab at Peking University. We thank all other members of our laboratories at Kunming University of Science and Technology and Peking University for their constructive suggestions.
This work was supported by grants from the National Natural Science Foundation of China (32130026, 92254305, and 32070714).
Author contributions: C. Zhang conceived the project; C. Zhang, M. Sun, Y. Wang, G. Xin, and Q. Jiang designed the experiments; M. Sun, Y. Wang, and G. Xin analyzed the data and performed most of the experiments; B. Yang performed some of the experiments; and C. Zhang, M. Sun, Y. Wang, G. Xin, and Q. Jiang wrote the manuscript.
References
Author notes
M. Sun, Y. Wang, and G. Xin contributed equally to this paper.
Disclosures: The authors declare no competing interests exist.