Clathrin-mediated endocytosis depends on polymerization of a branched actin network to provide force for membrane invagination. A key regulator in branched actin network formation is actin capping protein (CP), which binds to the barbed end of actin filaments to prevent the addition or loss of actin subunits. CP was thought to stochastically bind actin filaments, but recent evidence shows CP is regulated by a group of proteins containing CP-interacting (CPI) motifs. Importantly, how CPI motif proteins function together to regulate CP is poorly understood. Here, we show Aim21 and Bsp1 work synergistically to recruit CP to the endocytic actin network in budding yeast through their CPI motifs, which also allosterically modulate capping strength. In contrast, twinfilin works downstream of CP recruitment, regulating the turnover of CP through its CPI motif and a non-allosteric mechanism. Collectively, our findings reveal how three CPI motif proteins work together to regulate CP in a stepwise fashion during endocytosis.
Introduction
Endocytosis is the process by which cells collect cargo along the plasma membrane, invaginate the surrounding membrane, and pinch off the invagination as a vesicle inside of the cell (Doherty and McMahon, 2009). Clathrin-mediated endocytosis (CME) is a major endocytic pathway in all eukaryotic cells (Boettner et al., 2011; McMahon and Boucrot, 2011). CME is highly conserved in protein components, progression, and function from yeast to mammals, making the budding yeast Saccharomyces cerevisiae an attractive model system for the study of CME (Taylor et al., 2011; Boettner et al., 2011). During CME, over 60 endocytic proteins assemble with well-defined timing at endocytic sites along the plasma membrane to drive membrane invagination and vesicle formation (Goode et al., 2015; Kaksonen et al., 2005). An Arp2/3-derived branched actin network polymerizes at late stages of CME, and this aspect of endocytosis has been best studied in yeast (Kaksonen et al., 2003; Galletta et al., 2010). Nearly 30 endocytic factors arrive at CME sites within 5–10 s of each other to nucleate new actin filaments, cap elongating filaments, and turnover the actin network (Goode et al., 2015; Mooren et al., 2012). The result is a dense meshwork of actin filaments (F-actin) averaging ∼50 nm in length or 20 actin subunits (Young et al., 2004). While the function of many endocytic factors involved in the actin polymerization phase of CME is known, the function of many others remains undetermined (Farrell et al., 2015; Burston et al., 2009).
Actin capping protein (CP) is one important component in branched actin network assembly. CP binds the fast-growing barbed ends of F-actin with subnanomolar affinity, preventing addition or loss of actin subunits (Isenberg et al., 1980; Casella et al., 1986; Cooper and Sept, 2008; Carman et al., 2023). CP is a well-conserved heterodimer consisting of an α and β subunit. In budding yeast, the CAP1 and CAP2 genes encode the ∼30 kD α and β subunits, respectively (Amatruda et al., 1990, 1992). Deletion of either CAP1 or CAP2 results in an abnormally large actin network and defects in CME progression (Kaksonen et al., 2005; Amatruda et al., 1990). The CP complex has a characteristic mushroom shape, with a cap region responsible for capping F-actin, and a stalk region, where a diverse group of proteins bind via their CP-interacting (CPI) motifs (Fig. 1 A; Yang et al., 2005; Canton et al., 2005; Bruck et al., 2006, Uruno et al., 2006). Initially described CPI motifs such as those in the CARMIL (CP, Arp2/3, myosin-I linker) family of proteins mediate allosteric regulation of CP, decreasing the affinity of CP for barbed ends and weakening the capping activity of CP in vitro (Hernandez-Valladares et al., 2010; Edwards et al., 2014). As a result, CPI motifs were hypothesized to function downstream of CP binding to barbed ends, working as negative regulators of CP (Hernandez-Valladares et al., 2010). By contrast, subsequent cell biological findings argue that interaction with CPI motif proteins is required for localization of CP to actin networks, suggesting CPI motifs work upstream of CP-binding barbed ends and questioning the negative regulation concept (Edwards et al., 2015; Zhao et al., 2013).
Twinfilin (Twf1) is a well-conserved actin regulator initially identified and characterized in budding yeast (Goode et al., 1998). It is a member of the actin depolymerization factor-homology domain (ADF-H) family of proteins (Lappalainen et al., 1998; Maciver and Hussey, 2002). The physiological function of Twf1 has remained enigmatic due to a multitude of functions displayed in in vitro assays, including as an actin monomer sequestering protein (Goode et al., 1998; Vartiainen et al., 2000; Ojala et al., 2002), a barbed-end CP (Helfer et al., 2006; Paavilainen et al., 2007), and an actin depolymerization factor (Shekhar et al., 2021; Hilton et al., 2018; Johnston et al., 2015). Early experiments in yeast linked Twf1 directly to CP and barbed ends at cortical actin patches (Palmgren et al., 2001; Falck et al., 2004). Recently, the interaction between mammalian Twf1 and CP was shown to occur through a CPI motif in the C-terminal tail of Twf1, which is conserved in yeast Twf1 (Johnston et al., 2018). Unlike other CPI motifs, the CPI motif of Twf1 did not allosterically inhibit CP despite partially sharing a binding site on CP with CARMIL-type CPI motifs (Mwangangi et al., 2021; Johnston et al., 2018; Hakala et al., 2021). Given this, Twf1 was categorized as a pro-capping factor, preventing CARMIL-type CPI motifs from binding to and inhibiting or uncapping CP (Johnson et al., 2018). In contrast, another report suggested that Twf1 is largely responsible for CP turnover (uncapping), with its CPI motif necessary for localization to barbed ends (Hakala et al., 2021). Thus, CPI motif function may be more complex than previously appreciated, and the physiological function of Twf1 remains unclear in both yeast and higher eukaryotes.
Until recently, potential CARMIL-type CPI motif proteins had not been identified in yeast. As such, CP binding to barbed ends was assumed to be an unregulated process in yeast, with CP freely diffusing through the cytosol to cap actin filaments. However, the yeast CME factor Aim21 was recently discovered to contain a CPI motif that facilitates direct interaction with CP, indicating CARMIL-type CPI motif proteins may be conserved in yeast and function in endocytosis (Lamb et al., 2021). Aim21 forms a complex with the endocytic factor Tda2 which together function during the actin polymerization phase of CME (Farrell et al., 2017; Shin et al., 2018). Together with Tda2, Aim21 binds to CP with nanomolar affinity (Lamb et al., 2021). However, mutating the Aim21 CPI motif results in a partial defect in CP recruitment to CME sites, suggesting the CPI motif is not critical for CP localization in yeast. While yeast offers a simpler system to study the CPI motif and CP regulation, our knowledge is very limited.
In this study, we identify the yeast endocytic factor, binding of synaptojanin protein 1 (Bsp1), as a novel CPI motif–containing protein. Bsp1 works synergistically with Aim21 to localize CP to the actin network during CME, and while their CPI motifs allosterically inhibit CP in vitro, the overall in vivo effect is positive. In contrast, Twf1 functions downstream of CP recruitment at CME sites, working to turn over CP from barbed ends through a mechanism that depends on its CPI motif but does not show allosteric inhibition in vitro. Together, our findings shed light on how multiple CPI motifs regulate CP in a stepwise manner during yeast endocytosis.
Results
Aim21, Bsp1, and Twf1 contain CPI motifs
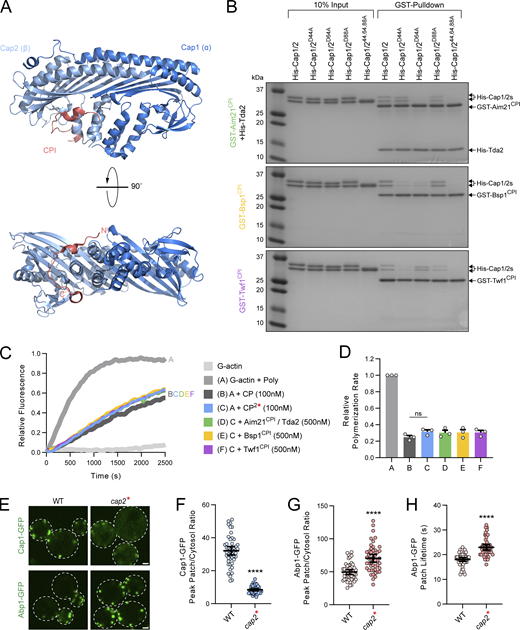
To identify novel CPI motif–containing yeast factors, we performed a sequence alignment of the Aim21 and Twf1 CPI motifs against the S. cerevisiae proteome. While alignment of the Twf1 CPI motif resulted in no clear hits for proteins involved in endocytosis or with the actin network, alignment using the Aim21 CPI motif identified two potential CPI motif–containing proteins involved in CME, Bsp1, and Inp51 (Fig. 1 A). Bsp1 is a little-studied ∼65-kD protein initially identified as a binding partner for the yeast CME factors Inp52 and Inp53 (Wicky et al., 2003). Inositol-polyphosphate 5-phosphatase 1 (Inp51) is a ∼108-kD synaptojanin-like protein important for the regulation of membrane phospholipids during CME (Singer-Krüger et al., 1998; Sun et al., 2007). To test whether these proteins interact with CP, a glutathione-S-transferase (GST) pulldown experiment was performed with GST fused to the potential CPI motifs of Bsp1 and Inp51. In addition, GST was fused to the CPI motifs of the known CP interactors, Aim21 and Twf1. For this experiment and all future in vitro binding experiments, the CPI motif sequence of Aim21 also includes its Tda2-binding region (TBR) located just downstream of its CPI motif, which facilitates interaction with Tda2 and greatly increases the affinity between Aim21 and CP (Lamb et al., 2021). In these cases, 6-histidine–tagged Tda2 (His-Tda2) is included in reactions to allow formation of the Tda2–Aim21 complex. GST fusions were immobilized on glutathione resin and subsequently incubated with 6-histidine–tagged CP (His-Cap1/2). While GST and GST-Inp51 failed to pull down His-Cap1/2, GST-Aim21CPI, GST-Bsp1CPI, and GST-Twf1CPI showed strong interactions with His-Cap1/2 (Fig. 1 B). To determine the binding affinity between CP and the interactors, a series of fluorescence polarization assays were performed using FITC-labeled CPI motif peptides (FITC-Aim21CPI, FITC-Bsp1CPI, and FITC-Twf1CPI). Labeled peptides were titrated with a range of concentrations of His-Cap1/2 (CP), and fluorescence polarization values were measured (Fig. 1, C–E; and Fig. S1, A–C). FITC-Aim21CPI and FITC-Twf1CPI displayed comparable Kd values of 307 ± 23 and 271 ± 23 nM, respectively (Fig. 1 F). Consistent with the GST pulldown experiment, FITC-Bsp1CPI had the most robust interaction with CP, with a Kd of 34 ± 3 nM (Fig. 1, B and F).
Aim21, Bsp1, and Twf1 contain CPI motifs. (A) Top: Crystal structure of the chicken CP complex bound to the CPI motif of human CD2AP (PDB entry 3AA6). The α-subunit (Cap1) is displayed in dark blue, the β-subunit (Cap2) in light blue, and the CPI motif of CD2AP is displayed in red. The N-terminus of the CPI motif is denoted with N’. Bottom: The amino acid sequences of the S. cerevisiae proteins containing potential CPI motifs are shown. For Twf1, * denotes the stop codon. For comparison, the previously described consensus CPI motif sequence (Hernandez-Valladares et al., 2010) is shown above. (B) A GST pulldown assay was performed with GST fused to fragments of Aim21 (491–545), Bsp1 (552–576), Twf1 (308–332), or Inp51 (874–900). Each GST fusion protein was incubated with His-Cap1/2, with Aim21 additionally incubated with His-Tda2 to form the Tda2/Aim21 complex. Bound proteins were analyzed by SDS-PAGE and Coomassie staining. The fragments of Aim21, Bsp1, and Twf1 demonstrated interaction with His-Cap1/2, while the fragment of Inp51 and GST alone did not. (C) A fluorescence polarization assay was performed using 10 nM FITC-labeled Aim21 peptide spanning amino acids 493–540 (FITC-Aim21CPI) and various concentrations of His-Cap1/2 (CP). Reactions included 25 µM His-Tda2 to form the Tda2–Aim21 complex. Data points represent the average and standard deviation from a single experiment performed in three technical replicates fit to a one-site binding isotherm. Note that the error is too small for many data points to produce an error bar larger than the symbol and is therefore not visible. (D) A fluorescence polarization assay was performed as in C using 2.5 nM FITC-labeled Bsp1 peptide spanning amino acids 552–576 (FITC-Bsp1CPI) and various concentrations of His-Cap1/2 (CP). (E) A fluorescence polarization assay was performed as in C using 10 nM FITC-Labeled Twf1 peptide spanning amino acids 308–332 (FITC-Twf1CPI) and various concentrations of His-Cap1/2 (CP). (F) Dissociation constants of CP and Aim21CPI, Bsp1CPI, or Twf1CPI, respectively. Dissociation constants represent the mean ± SEM from three independent experiments each fit to a one-site binding isotherm. (G) Live-cell fluorescence microscopy showing strong colocalization of (top) Aim21-GFP, (middle) Bsp1-GFP, and (bottom) Twf1-GFP with Cap1-mCherry at endocytic patches, respectively (SDY1518, SDY1520, and SDY1522). Endocytic patches used for construction of kymographs to the right are indicated by arrowheads. Scale bars, 1 µm. Source data are available for this figure: SourceData F1.
Aim21, Bsp1, and Twf1 contain CPI motifs. (A) Top: Crystal structure of the chicken CP complex bound to the CPI motif of human CD2AP (PDB entry 3AA6). The α-subunit (Cap1) is displayed in dark blue, the β-subunit (Cap2) in light blue, and the CPI motif of CD2AP is displayed in red. The N-terminus of the CPI motif is denoted with N’. Bottom: The amino acid sequences of the S. cerevisiae proteins containing potential CPI motifs are shown. For Twf1, * denotes the stop codon. For comparison, the previously described consensus CPI motif sequence (Hernandez-Valladares et al., 2010) is shown above. (B) A GST pulldown assay was performed with GST fused to fragments of Aim21 (491–545), Bsp1 (552–576), Twf1 (308–332), or Inp51 (874–900). Each GST fusion protein was incubated with His-Cap1/2, with Aim21 additionally incubated with His-Tda2 to form the Tda2/Aim21 complex. Bound proteins were analyzed by SDS-PAGE and Coomassie staining. The fragments of Aim21, Bsp1, and Twf1 demonstrated interaction with His-Cap1/2, while the fragment of Inp51 and GST alone did not. (C) A fluorescence polarization assay was performed using 10 nM FITC-labeled Aim21 peptide spanning amino acids 493–540 (FITC-Aim21CPI) and various concentrations of His-Cap1/2 (CP). Reactions included 25 µM His-Tda2 to form the Tda2–Aim21 complex. Data points represent the average and standard deviation from a single experiment performed in three technical replicates fit to a one-site binding isotherm. Note that the error is too small for many data points to produce an error bar larger than the symbol and is therefore not visible. (D) A fluorescence polarization assay was performed as in C using 2.5 nM FITC-labeled Bsp1 peptide spanning amino acids 552–576 (FITC-Bsp1CPI) and various concentrations of His-Cap1/2 (CP). (E) A fluorescence polarization assay was performed as in C using 10 nM FITC-Labeled Twf1 peptide spanning amino acids 308–332 (FITC-Twf1CPI) and various concentrations of His-Cap1/2 (CP). (F) Dissociation constants of CP and Aim21CPI, Bsp1CPI, or Twf1CPI, respectively. Dissociation constants represent the mean ± SEM from three independent experiments each fit to a one-site binding isotherm. (G) Live-cell fluorescence microscopy showing strong colocalization of (top) Aim21-GFP, (middle) Bsp1-GFP, and (bottom) Twf1-GFP with Cap1-mCherry at endocytic patches, respectively (SDY1518, SDY1520, and SDY1522). Endocytic patches used for construction of kymographs to the right are indicated by arrowheads. Scale bars, 1 µm. Source data are available for this figure: SourceData F1.
Aim21, Bsp1, and Twf1 contain CPI motifs. (A) Three independent fluorescence polarization assays were performed using 10 nM FITC-labeled Aim21 peptide spanning amino acids 493–540 (FITC-Aim21CPI) and various concentrations of His-Cap1/2 (CP). Reactions included 25 µM His-Tda2 to form the Tda2–Aim21 complex. Each data point represents the average and SD from a single experiment performed in three technical replicates fit to a one-site binding isotherm. Note that the error is too small for many CP concentration data points to produce an error bar larger than the symbol. The dissociation constant determined for individual experiments is listed at the lower right of each graph. (B) Three independent fluorescence polarization assays were performed as in A using 2.5 nM FITC-labeled Bsp1 peptide spanning amino acids 552–576 (FITC-Bsp1CPI) and various concentrations of His-Cap1/2 (CP). (C) Three independent fluorescence polarization assays were performed as in A using 10 nM FITC-labeled Twf1 peptide spanning amino acids 308–332 (FITC-Twf1CPI) and various concentrations of His-Cap1/2 (CP). (D) Fluorescence microscopy images taken with 1-s exposure times of cells expressing Cap1-mCherry along with either Aim21-GFP, Bsp1-GFP, or Twf1-GFP (SDY1518, SDY1520, SDY1522). Aim21-GFP and Twf1-GFP display higher overall fluorescence intensity levels than Bsp1-GFP, likely reflecting higher expression levels. Scale bars, 5 μm.
Aim21, Bsp1, and Twf1 contain CPI motifs. (A) Three independent fluorescence polarization assays were performed using 10 nM FITC-labeled Aim21 peptide spanning amino acids 493–540 (FITC-Aim21CPI) and various concentrations of His-Cap1/2 (CP). Reactions included 25 µM His-Tda2 to form the Tda2–Aim21 complex. Each data point represents the average and SD from a single experiment performed in three technical replicates fit to a one-site binding isotherm. Note that the error is too small for many CP concentration data points to produce an error bar larger than the symbol. The dissociation constant determined for individual experiments is listed at the lower right of each graph. (B) Three independent fluorescence polarization assays were performed as in A using 2.5 nM FITC-labeled Bsp1 peptide spanning amino acids 552–576 (FITC-Bsp1CPI) and various concentrations of His-Cap1/2 (CP). (C) Three independent fluorescence polarization assays were performed as in A using 10 nM FITC-labeled Twf1 peptide spanning amino acids 308–332 (FITC-Twf1CPI) and various concentrations of His-Cap1/2 (CP). (D) Fluorescence microscopy images taken with 1-s exposure times of cells expressing Cap1-mCherry along with either Aim21-GFP, Bsp1-GFP, or Twf1-GFP (SDY1518, SDY1520, SDY1522). Aim21-GFP and Twf1-GFP display higher overall fluorescence intensity levels than Bsp1-GFP, likely reflecting higher expression levels. Scale bars, 5 μm.
To assess the dynamics of the CPI motif–containing proteins during CME, yeast strains were constructed to express Cap1-mCherry together with a GFP-tagged CPI motif protein from their corresponding endogenous loci. Two-color live-cell fluorescence microscopy showed that Aim21-GFP and Cap1-mCherry share nearly identical dynamics during CME, arriving at endocytic patches and leaving in unison (Fig. 1 G, top). Strains expressing Bsp1-GFP or Twf1-GFP, respectively, showed similar dynamics, demonstrating that all three proteins work during the actin polymerization phase of CME along with CP (Fig. 1 G). Still images taken with equal exposure times suggest Aim21-GFP and Twf1-GFP are expressed at similar levels, while Bsp1-GFP is expressed at a lower level (Fig. S1 D). Together, these findings establish Bsp1 as a novel CPI motif–containing protein in yeast and suggest that the three endocytic factors could work together to regulate CP during CME.
CPI motifs are essential for the function of Aim21, Bsp1, and Twf1
CPI motifs are enriched in basic residues, which participate in electrostatic interactions with acidic residues along the CPI motif binding site of CP. We previously abolished the Aim21:CP interaction by mutating three basic residues within the Aim21 CPI motif to glutamic acid (Lamb et al., 2021). Similar to the CPI motif of Aim21, the CPI motifs of Bsp1 and Twf1 are enriched in basic residues (Fig. 2 A). To determine if Bsp1 and Twf1 have a similar reliance on electrostatic interactions for binding to CP, a GST pulldown experiment was performed with GST fused to CPI motifs harboring three basic-to-glutamic acid mutations within the CPI motif (Fig. 2 A). Each wild-type (WT) CPI fusion displayed a robust interaction with His-Cap1/2 (Fig. 2 B). In contrast, each mutant CPI fusion protein, GST-Aim21CPI*, GST-Bsp1CPI*, and GST-Twf1CPI*, displayed no interaction with His-Cap1/2 (Fig. 2 B).
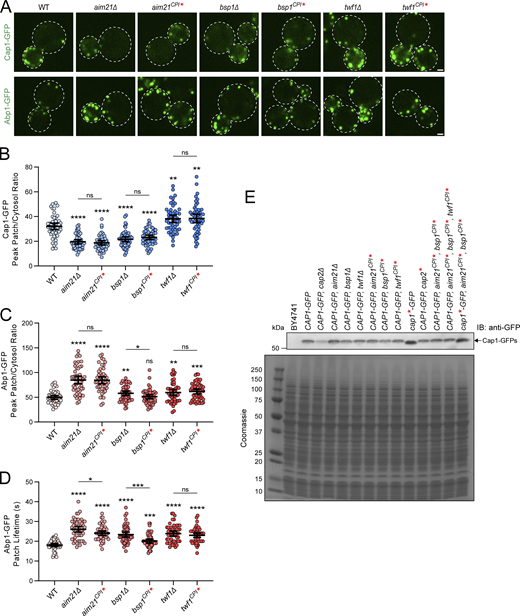
CPI motifs are essential for the function of Aim21, Bsp1, and Twf1. (A) Organization of Aim21, Bsp1, and Twf1 domains. The sequences of the CPI motifs for each of Aim21, Bsp1, and Twf1 are displayed, with the three basic-to-acidic point mutations to the CPI motifs that are used throughout this work shown in red. Aim21CPI* = Aim21504,507,509E. Bsp1CPI* = Bsp1564,566,569E. Twf1CPI* = Twf1322,324,328E. (B) A GST pulldown assay was performed with GST fused to the CPI motifs of Aim21, Bsp1, or Twf1. The GST-CPI fusions either contained no mutations (CPI) or three-point mutations to the CPI motif (CPI*), as indicated in A. Each GST fusion protein was incubated with His-Cap1/2, with Aim21 additionally incubated with His-Tda2 to form the Tda2/Aim21 complex. Bound proteins were analyzed by SDS-PAGE and Coomassie staining. The mutations to each CPI motif abolished the interaction with His-Cap1/2. (C) Top: Live-cell fluorescence microscopy showing reduced recruitment of Cap1-GFP to endocytic sites in cells with the CPI motifs of Aim21 or Bsp1 mutated (SDY1474, SDY1512). In contrast, cells with the CPI motif of Twf1 mutated display an increase in Cap1-GFP patch intensity (SDY1513). Bottom: Live-cell fluorescence microscopy showing increased recruitment of Abp1-GFP to endocytic sites in cells with the CPI motif of Aim21, Bsp1, or Twf1 mutated (SDY1434, SDY1514, SDY1515). Scale bars, 1 µm. (D) Quantification of Cap1-GFP peak fluorescence intensity at endocytic patches in WT, aim21CPI*, bsp1CPI*, and twf1CPI* cells. From left to right, mean peak patch/cytosol ratio = 32.08, 18.73, 23.13, 38.49, and n = 50 for all groups. Error bars, mean with 95% confidence interval (CI). **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001. (E) Quantification of Abp1-GFP peak fluorescence intensity at endocytic patches in WT, aim21CPI*, bsp1CPI*, and twf1CPI* cells. From left to right, the mean peak patch/cytosol ratio = 49.63, 84.36, 51.25, 61.68, and n = 50 for all groups. Error bars, mean with 95% CI. ns = not significant, ***P ≤ 0.001, ****P ≤ 0.0001. (F) Quantification of Abp1-GFP patch lifetime at endocytic patches in WT, aim21CPI*, bsp1CPI*, and twf1CPI* cells. From left to right, mean patch lifetime = 18.06, 24.12, 20.14, 23.02 s, and n = 50 for all groups. Error bars, mean with 95% CI. ***P ≤ 0.001, ****P ≤ 0.0001. (G) Live-cell fluorescence microscopy showing decreased endocytic uptake of Mup1-GFP after 20 min incubation with methionine-rich media in cells with the CPI motif of Aim21, Bsp1, or Twf1 mutated (SDY1471, SDY1749, and SDY1759). Scale bars, 5 µm. (H) Quantification of Mup1-GFP relative plasma membrane fluorescence intensity in WT, aim21CPI*, bsp1CPI*, and twf1CPI* cells. From left to right, mean relative fluorescence intensity = 1.00, 1.00, 1.00, 1.00, 0.35, 0.81, 0.79, 0.66, and n = 52, 55, 73, 62, 53, 55, 64, 65. Error bars, mean with 95% CI. ****P ≤ 0.0001. Source data are available for this figure: SourceData F2.
CPI motifs are essential for the function of Aim21, Bsp1, and Twf1. (A) Organization of Aim21, Bsp1, and Twf1 domains. The sequences of the CPI motifs for each of Aim21, Bsp1, and Twf1 are displayed, with the three basic-to-acidic point mutations to the CPI motifs that are used throughout this work shown in red. Aim21CPI* = Aim21504,507,509E. Bsp1CPI* = Bsp1564,566,569E. Twf1CPI* = Twf1322,324,328E. (B) A GST pulldown assay was performed with GST fused to the CPI motifs of Aim21, Bsp1, or Twf1. The GST-CPI fusions either contained no mutations (CPI) or three-point mutations to the CPI motif (CPI*), as indicated in A. Each GST fusion protein was incubated with His-Cap1/2, with Aim21 additionally incubated with His-Tda2 to form the Tda2/Aim21 complex. Bound proteins were analyzed by SDS-PAGE and Coomassie staining. The mutations to each CPI motif abolished the interaction with His-Cap1/2. (C) Top: Live-cell fluorescence microscopy showing reduced recruitment of Cap1-GFP to endocytic sites in cells with the CPI motifs of Aim21 or Bsp1 mutated (SDY1474, SDY1512). In contrast, cells with the CPI motif of Twf1 mutated display an increase in Cap1-GFP patch intensity (SDY1513). Bottom: Live-cell fluorescence microscopy showing increased recruitment of Abp1-GFP to endocytic sites in cells with the CPI motif of Aim21, Bsp1, or Twf1 mutated (SDY1434, SDY1514, SDY1515). Scale bars, 1 µm. (D) Quantification of Cap1-GFP peak fluorescence intensity at endocytic patches in WT, aim21CPI*, bsp1CPI*, and twf1CPI* cells. From left to right, mean peak patch/cytosol ratio = 32.08, 18.73, 23.13, 38.49, and n = 50 for all groups. Error bars, mean with 95% confidence interval (CI). **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001. (E) Quantification of Abp1-GFP peak fluorescence intensity at endocytic patches in WT, aim21CPI*, bsp1CPI*, and twf1CPI* cells. From left to right, the mean peak patch/cytosol ratio = 49.63, 84.36, 51.25, 61.68, and n = 50 for all groups. Error bars, mean with 95% CI. ns = not significant, ***P ≤ 0.001, ****P ≤ 0.0001. (F) Quantification of Abp1-GFP patch lifetime at endocytic patches in WT, aim21CPI*, bsp1CPI*, and twf1CPI* cells. From left to right, mean patch lifetime = 18.06, 24.12, 20.14, 23.02 s, and n = 50 for all groups. Error bars, mean with 95% CI. ***P ≤ 0.001, ****P ≤ 0.0001. (G) Live-cell fluorescence microscopy showing decreased endocytic uptake of Mup1-GFP after 20 min incubation with methionine-rich media in cells with the CPI motif of Aim21, Bsp1, or Twf1 mutated (SDY1471, SDY1749, and SDY1759). Scale bars, 5 µm. (H) Quantification of Mup1-GFP relative plasma membrane fluorescence intensity in WT, aim21CPI*, bsp1CPI*, and twf1CPI* cells. From left to right, mean relative fluorescence intensity = 1.00, 1.00, 1.00, 1.00, 0.35, 0.81, 0.79, 0.66, and n = 52, 55, 73, 62, 53, 55, 64, 65. Error bars, mean with 95% CI. ****P ≤ 0.0001. Source data are available for this figure: SourceData F2.
Having identified mutations that disrupt each CP:CPI interactions in vitro, we wanted to assess if the CPI motifs were functionally relevant in vivo. First, we assessed the effect of mutating the CPI motifs on recruitment of CP to CME sites. As we reported previously, aim21CPI* cells displayed a reduction in Cap1-GFP recruitment to CME sites compared with WT cells (Fig. 2 C, top, and Fig. 2 D; Lamb et al., 2021). Similar to aim21CPI* cells, bsp1CPI* cells had a defect in Cap1-GFP recruitment, albeit less so than aim21CPI* cells (Fig. 2 C, top, and Fig 2 D). Conversely, twf1CPI* cells showed elevated levels of Cap1-GFP at endocytic sites (Fig. 2 C, top, and Fig. 2 D). Cells carrying a deletion of the AIM21 gene (aim21∆) displayed a comparable reduction in the recruitment of Cap1-GFP to CME sites as aim21CPI* cells (Fig. S2 A, top, and Fig. S2 B). Similarly, bsp1∆ and twf1∆ showed comparable phenotypes to bsp1CPI* and twf1CPI* cells, respectively (Fig. S2 A, top, Fig. S2 B). Importantly, the defect in Cap1-GFP recruitment seen in aim21∆, aim21CPI*, bsp1∆, and bsp1CPI* cells was not due to reduced overall expression of Cap1-GFP (Fig. S2 E).
CPI motifs are essential for the function of Aim21, Bsp1, and Twf1. (A) Top: Live-cell fluorescence microscopy showing reduced recruitment of Cap1-GFP to endocytic sites in cells lacking Aim21 or Bsp1 (SDY1292, SDY1350), or with the CPI motifs of Aim21 and Bsp1 mutated (SDY1474, SDY1512). In contrast, cells lacking Twf1 or with the CPI motif of Twf1 mutated display an increase in Cap1-GFP patch intensity (SDY1351 and SDY1513). Bottom: Live-cell fluorescence microscopy showing increased recruitment of Abp1-GFP to endocytic sites in cells lacking Aim21, Bsp1, or Twf1 (SDY1294, SDY1432, SDY1433), or with the CPI motifs of Aim21, Bsp1, or Twf1 mutated (SDY1434, SDY1514, SDY1515). Scale bars, 1 µm. (B) Quantification of Cap1-GFP peak fluorescence intensity at endocytic patches in WT, aim21∆, aim21CPI*, bsp1∆, bsp1CPI*, twf1∆, and twf1CPI* cells. From left to right, the mean peak patch/cytosol ratio = 32.08, 19.56, 18.73, 21.60, 23.13, 38.11, 38.49, and n = 50 for all groups. Error bars, mean with 95% CI. ns = not significant, **P ≤ 0.01, ****P ≤ 0.0001. (C) Quantification of Abp1-GFP peak fluorescence intensity at endocytic patches in WT, aim21∆, aim21CPI*, bsp1∆, bsp1CPI*, twf1∆, and twf1CPI* cells. From left to right, the mean peak patch/cytosol ratio = 49.63, 84.96, 84.36, 58.10, 51.25, 59.71, 61.68, and n = 50 for all groups. Error bars, mean with 95% CI. ns = not significant, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001. (D) Quantification of Abp1-GFP patch lifetime at endocytic patches in WT, aim21∆, aim21CPI*, bsp1∆, bsp1CPI*, twf1∆, and twf1CPI* cells. From left to right, mean patch lifetime = 18.06, 26.06, 24.12, 22.17, 20.14, 23.02 s, 22.61, and n = 50 for all groups. Error bars, mean with 95% CI. ns = not significant, *P ≤ 0.05, ***P ≤ 0.001, ****P ≤ 0.0001. (E) Cell extracts from Cap1-GFP strains used for fluorescence microscopy (Fig. 2 C, panel A, Fig. 3 A, Fig. 4 D, and Fig. 7 E) were analyzed by immunoblotting (IB) using antibodies to the GFP tag (anti-GFP). The parent strain, BY4741, is shown as a negative control. Bottom: Coomassie-stained gel of extracts show equal loading. The strains display comparable expression of Cap1-GFP except for the cap2∆ strain, which was expected. Source data are available for this figure: SourceData FS2.
CPI motifs are essential for the function of Aim21, Bsp1, and Twf1. (A) Top: Live-cell fluorescence microscopy showing reduced recruitment of Cap1-GFP to endocytic sites in cells lacking Aim21 or Bsp1 (SDY1292, SDY1350), or with the CPI motifs of Aim21 and Bsp1 mutated (SDY1474, SDY1512). In contrast, cells lacking Twf1 or with the CPI motif of Twf1 mutated display an increase in Cap1-GFP patch intensity (SDY1351 and SDY1513). Bottom: Live-cell fluorescence microscopy showing increased recruitment of Abp1-GFP to endocytic sites in cells lacking Aim21, Bsp1, or Twf1 (SDY1294, SDY1432, SDY1433), or with the CPI motifs of Aim21, Bsp1, or Twf1 mutated (SDY1434, SDY1514, SDY1515). Scale bars, 1 µm. (B) Quantification of Cap1-GFP peak fluorescence intensity at endocytic patches in WT, aim21∆, aim21CPI*, bsp1∆, bsp1CPI*, twf1∆, and twf1CPI* cells. From left to right, the mean peak patch/cytosol ratio = 32.08, 19.56, 18.73, 21.60, 23.13, 38.11, 38.49, and n = 50 for all groups. Error bars, mean with 95% CI. ns = not significant, **P ≤ 0.01, ****P ≤ 0.0001. (C) Quantification of Abp1-GFP peak fluorescence intensity at endocytic patches in WT, aim21∆, aim21CPI*, bsp1∆, bsp1CPI*, twf1∆, and twf1CPI* cells. From left to right, the mean peak patch/cytosol ratio = 49.63, 84.96, 84.36, 58.10, 51.25, 59.71, 61.68, and n = 50 for all groups. Error bars, mean with 95% CI. ns = not significant, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001. (D) Quantification of Abp1-GFP patch lifetime at endocytic patches in WT, aim21∆, aim21CPI*, bsp1∆, bsp1CPI*, twf1∆, and twf1CPI* cells. From left to right, mean patch lifetime = 18.06, 26.06, 24.12, 22.17, 20.14, 23.02 s, 22.61, and n = 50 for all groups. Error bars, mean with 95% CI. ns = not significant, *P ≤ 0.05, ***P ≤ 0.001, ****P ≤ 0.0001. (E) Cell extracts from Cap1-GFP strains used for fluorescence microscopy (Fig. 2 C, panel A, Fig. 3 A, Fig. 4 D, and Fig. 7 E) were analyzed by immunoblotting (IB) using antibodies to the GFP tag (anti-GFP). The parent strain, BY4741, is shown as a negative control. Bottom: Coomassie-stained gel of extracts show equal loading. The strains display comparable expression of Cap1-GFP except for the cap2∆ strain, which was expected. Source data are available for this figure: SourceData FS2.
Cells with a defect in CP function show increased levels of the F-actin binding protein Abp1 at CME sites as a result of an overgrown actin network (Kaksonen et al., 2005; Kim et al., 2004). To determine the effect of CPI motif mutations on the actin network at CME sites, the mutant CPI alleles were integrated into cells expressing Abp1-GFP at their endogenous loci. While aim21CPI* cells had significantly increased levels of Abp1-GFP at CME sites, bsp1CPI* cells had levels comparable with WT cells (Fig. 2 C, bottom, and Fig. 2 E). Despite having elevated levels of CP recruitment, twf1CPI* cells also displayed significantly increased levels of Abp1-GFP at CME sites (Fig. 2 C, bottom, and Fig. 2 E). Given the proposed function of Twf1 as an actin depolymerization factor, this phenotype could be explained by a defect in Twf1 function or localization in twf1CPI* cells, resulting in elevated levels of F-actin. While bsp1CPI* cells did not exhibit a significant increase in Abp1-GFP recruitment, each mutant CPI motif allele caused a significant increase in the patch lifetime of Abp1-GFP, indicative of a defect in the actin polymerization phase of CME (Fig. 2 C, bottom, and Fig. 2 F). While aim21CPI* and bsp1CPI* had a noticeable Abp1-GFP phenotype, the phenotypes in aim21∆ and bsp1∆ cells were more severe, suggesting the proteins likely have other functions in addition to their interaction with CP (Fig. S2 A, bottom, and Fig. S2, C and D). Conversely, twf1CPI* and twf1∆ cells had indistinguishable Abp1-GFP and Cap1-GFP phenotypes (Fig. S2, A–D).
We next assessed the importance of each CPI motif on endocytic uptake of cargo. Mup1-GFP, a methionine transporter that localizes to the plasma membrane in cells starved of methionine, was used to track endocytosis. Upon returning cells to methionine-rich media, Mup1-GFP was internalized. A delay in Mup1-GFP internalization was observed in aim21CPI*, bsp1CPI*, and twf1CPI* cells relative to WT cells (Fig. 2, G and H). Thus, the CPI motifs of Aim21, Bsp1, and Twf1 are important for the regulation of CP and efficient endocytosis in vivo, although they likely regulate CP in different ways.
The CPI motifs of Aim21 and Bsp1, but not that of Twif1, are required for proper recruitment of CP to CME sites
Results described above (Fig. 2, B–D; and Fig. S2, A and B) suggest Aim21 and Bsp1 work on CP recruitment to CME sites via CPI motif–CP interaction while Twf1 does not. Given the similar phenotypes seen in aim21CPI* and bsp1CPI* cells regarding CP, we hypothesized that Aim21 and Bsp1 shared synergistic roles in recruiting CP to CME sites. In line with this hypothesis, aim21CPI*/bsp1CPI* cells had a significantly greater Cap1-GFP recruitment defect than either aim21CPI* or bsp1CPI* cells (Fig. 3, A–C). While still present at endocytic sites at very low levels, Cap1-GFP became largely diffuse in the cytosol in aim21CPI*/bsp1CPI* cells (Fig. 3, A–C). Importantly, overall Cap1-GFP expression levels in WT and aim21CPI*/bsp1CPI* cells were comparable (Fig. S2 E). Given the heightened recruitment defect in aim21CPI*/bsp1CPI* cells compared with aim21CPI* cells, we suspected a similar additive effect may be seen in F-actin levels in cells combining the mutations. While Abp1-GFP levels were similar between aim21CPI* and aim21CPI*/bsp1CPI* cells (84.36, 85.68), the patch lifetime of Abp1-GFP had a slight but significant increase in aim21CPI*/bsp1CPI* cells compared with aim21CPI cells with patch lifetimes of 26.02 and 24.12 s, respectively (Fig. 2, C, E, and F; and Fig. S3, A–C). Together these findings point to a shared function of Aim21 and Bsp1 in recruiting CP to endocytic sites.
The CPI motifs of Aim21 and Bsp1 are required for proper recruitment of CP to CME sites. (A) Live-cell fluorescence microscopy showing reduced recruitment of Cap1-GFP to endocytic sites in cells with the CPI motifs of Aim21 or Bsp1 mutated (SDY1474 and SDY1512), or in cells with the CPI mutations combined (SDY1604). (B) Quantification of Cap1-GFP peak fluorescence intensity at endocytic patches in WT, aim21CPI*, bsp1CPI*, and aim21CPI*/bsp1CPI* cells. From left to right, mean peak patch/cytosol ratio = 32.08, 18.73, 23.13, 5.75, and n = 50 for all groups. Error bars, mean with 95% CI. ****P ≤ 0.0001. (C) Top: Live-cell fluorescence microscopy showing strong colocalization of Cap1-GFP with Abp1-mCherry at endocytic patches (SDY1698). Bottom: Live-cell fluorescence microscopy showing reduced recruitment of Cap1-GFP and increased recruitment of Abp1-mCherry to endocytic patches in cells with the CPI motifs of Aim21 and Bsp1 mutated (SDY1702). Endocytic patches used for construction of kymographs to the right are indicated by arrowheads. Scale bars, 1 µm.
The CPI motifs of Aim21 and Bsp1 are required for proper recruitment of CP to CME sites. (A) Live-cell fluorescence microscopy showing reduced recruitment of Cap1-GFP to endocytic sites in cells with the CPI motifs of Aim21 or Bsp1 mutated (SDY1474 and SDY1512), or in cells with the CPI mutations combined (SDY1604). (B) Quantification of Cap1-GFP peak fluorescence intensity at endocytic patches in WT, aim21CPI*, bsp1CPI*, and aim21CPI*/bsp1CPI* cells. From left to right, mean peak patch/cytosol ratio = 32.08, 18.73, 23.13, 5.75, and n = 50 for all groups. Error bars, mean with 95% CI. ****P ≤ 0.0001. (C) Top: Live-cell fluorescence microscopy showing strong colocalization of Cap1-GFP with Abp1-mCherry at endocytic patches (SDY1698). Bottom: Live-cell fluorescence microscopy showing reduced recruitment of Cap1-GFP and increased recruitment of Abp1-mCherry to endocytic patches in cells with the CPI motifs of Aim21 and Bsp1 mutated (SDY1702). Endocytic patches used for construction of kymographs to the right are indicated by arrowheads. Scale bars, 1 µm.
Proper localization and function of CP are required to maintain normal levels of actin at endocytic sites. (A) Live-cell fluorescence microscopy showing increased recruitment of Abp1-GFP to endocytic sites in cells lacking Cap1 (SDY1480), with the α-tentacle of Cap1 mutated (SDY1580), with the CPI motifs of Aim21 and Bsp1 mutated (SDY1610), or with the mutations combined (SDY1634). Scale bar, 1 µm. (B) Quantification of Abp1-GFP peak fluorescence intensity at endocytic patches in WT, cap1∆, cap1*, aim21CPI*/bsp1CPI*, and aim21CPI*/bsp1CPI*/cap1* cells. From left to right, mean peak patch/cytosol ratio = 49.63, 79.03, 80.27, 85.68, 82.70, and n = 50 for all groups. Error bars, mean with 95% CI. ns = not significant, ****P ≤ 0.0001. (C) Quantification of Abp1-GFP patch lifetime at endocytic patches in WT, cap1∆, cap1*, aim21CPI*/bsp1CPI*, and aim21CPI*/bsp1CPI*/cap1* cells. From left to right, mean patch lifetime = 18.06, 33.52, 30.82, 26.02, 30.24 s, and n = 50 for all groups. Error bars, mean with 95% CI. *P ≤ 0.05, ***P ≤ 0.001, ****P ≤ 0.0001. (D) A GST pulldown assay was performed with GST fused to the CPI motifs of Aim21, Bsp1, and Twf1 or a fragment of Inp51. Each GST fusion protein was incubated with either WT CP (His-Cap1/2) or a CP with a mutated His-Cap1 subunit α-tentacle (His-Cap1*/2), as indicated in Fig. 1 B. Aim21 was additionally incubated with His-Tda2 to form the Tda2/Aim21 complex. Bound proteins were analyzed by SDS-PAGE and Coomassie staining. The mutations to His-Cap1* do not affect the interaction between the CPI motifs and His-Cap1*/2. Source data are available for this figure: SourceData FS3.
Proper localization and function of CP are required to maintain normal levels of actin at endocytic sites. (A) Live-cell fluorescence microscopy showing increased recruitment of Abp1-GFP to endocytic sites in cells lacking Cap1 (SDY1480), with the α-tentacle of Cap1 mutated (SDY1580), with the CPI motifs of Aim21 and Bsp1 mutated (SDY1610), or with the mutations combined (SDY1634). Scale bar, 1 µm. (B) Quantification of Abp1-GFP peak fluorescence intensity at endocytic patches in WT, cap1∆, cap1*, aim21CPI*/bsp1CPI*, and aim21CPI*/bsp1CPI*/cap1* cells. From left to right, mean peak patch/cytosol ratio = 49.63, 79.03, 80.27, 85.68, 82.70, and n = 50 for all groups. Error bars, mean with 95% CI. ns = not significant, ****P ≤ 0.0001. (C) Quantification of Abp1-GFP patch lifetime at endocytic patches in WT, cap1∆, cap1*, aim21CPI*/bsp1CPI*, and aim21CPI*/bsp1CPI*/cap1* cells. From left to right, mean patch lifetime = 18.06, 33.52, 30.82, 26.02, 30.24 s, and n = 50 for all groups. Error bars, mean with 95% CI. *P ≤ 0.05, ***P ≤ 0.001, ****P ≤ 0.0001. (D) A GST pulldown assay was performed with GST fused to the CPI motifs of Aim21, Bsp1, and Twf1 or a fragment of Inp51. Each GST fusion protein was incubated with either WT CP (His-Cap1/2) or a CP with a mutated His-Cap1 subunit α-tentacle (His-Cap1*/2), as indicated in Fig. 1 B. Aim21 was additionally incubated with His-Tda2 to form the Tda2/Aim21 complex. Bound proteins were analyzed by SDS-PAGE and Coomassie staining. The mutations to His-Cap1* do not affect the interaction between the CPI motifs and His-Cap1*/2. Source data are available for this figure: SourceData FS3.
CP’s ability to cap barbed ends is required for its localization and function
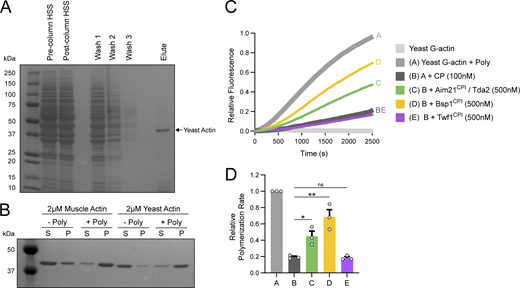
While our data support that the Aim21 and Bsp1 CPI motifs are critical for CP localization to CME sites, a low level of Cap1-GFP was still present at CME sites in aim21CPI*/bsp1CPI* cells. We reasoned the low level of CP still present could be from freely diffusing CP that localizes to CME sites by binding actin filament barbed ends. The capping function of CP is largely dependent on the α-tentacle at the C-terminus of the α-subunit (Cap1; Fig. 4 A). To abolish the capping function of CP, we utilized previously described mutations to the α-tentacle that disrupt the capping function of yeast CP (Kim et al., 2004). Before integrating the mutations into the yeast genome, we wanted to confirm they would produce a large defect in the capping function of CP using pyrene actin polymerization assays. Polymerization of pyrene-labeled actin was induced by addition of polymerization buffer (Poly). In the presence of WT His-Cap1/Cap2 (CP), the polymerization rate of actin was severely reduced, while in the presence of His-Cap1239,240A/Cap2 (CP1*), the rate of polymerization was indistinguishable from a reaction with actin alone (Fig. 4, B and C). Importantly, CP1* was pulled down by GST-CPI fusions with the same efficiency as WT CP in a GST pulldown assay, indicating the mutations did not affect the CP:CPI interactions (Fig. S3 D).
CPs’ ability to cap barbed ends is required for its localization and function. (A) Top: Crystal structure of the chicken CP complex (PDB entry 3AA7) displayed with the α-subunit (Cap1) in dark blue and the β-subunit (Cap2) in light blue. The α-tentacle, which is critical for the capping function of CP, is shown in red. Bottom: Alignment of the S. cerevisiae α-tentacle sequence with homologs from Homo sapiens and Mus musculus. The mutations to Cap1 (Cap1*) that will be used throughout the figure are displayed in red. (B) Pyrene-labeled actin (1 µM, 20% labeled) was polymerized in the absence or presence of His-Cap1/2 (CP, 100 nM) or His-Cap1239,240A/2 (CP1*, 100 nM). CP1* showed no ability to cap actin filaments. (C) Quantification of the relative polymerization rate from three independent pyrene actin polymerization assays in B. From left to right, mean relative polymerization rate = 1.00, 0.25, 0.97. Error bars, mean ± SD. ns = not significant. (D) Live-cell fluorescence microscopy showing reduced recruitment of Cap1-GFP to endocytic sites in cells with the α-tentacle of Cap1 mutated (SDY1582), the CPI motifs of Aim21 and Bsp1 mutated (SDY1604), or with the mutations combined (SDY1608). Scale bar, 1 µm. (E) Quantification of Cap1-GFP peak fluorescence intensity at endocytic patches in WT, cap1*, and aim21CPI*/bsp1CPI* cells. From left to right, mean peak patch/cytosol ratio = 32.08, 11.86, 5.75, and n = 50 for all groups. Error bars, mean with 95% CI. ****P ≤ 0.0001. (F) Live-cell fluorescence microscopy showing no localization of Cap1*-GFP with Abp1-mCherry at endocytic patches in cells with the CPI motifs of Aim21 and Bsp1 mutated (SDY1703). Endocytic patch used for construction of kymograph to the right is indicated by an arrowhead. Scale bar, 1 µm.
CPs’ ability to cap barbed ends is required for its localization and function. (A) Top: Crystal structure of the chicken CP complex (PDB entry 3AA7) displayed with the α-subunit (Cap1) in dark blue and the β-subunit (Cap2) in light blue. The α-tentacle, which is critical for the capping function of CP, is shown in red. Bottom: Alignment of the S. cerevisiae α-tentacle sequence with homologs from Homo sapiens and Mus musculus. The mutations to Cap1 (Cap1*) that will be used throughout the figure are displayed in red. (B) Pyrene-labeled actin (1 µM, 20% labeled) was polymerized in the absence or presence of His-Cap1/2 (CP, 100 nM) or His-Cap1239,240A/2 (CP1*, 100 nM). CP1* showed no ability to cap actin filaments. (C) Quantification of the relative polymerization rate from three independent pyrene actin polymerization assays in B. From left to right, mean relative polymerization rate = 1.00, 0.25, 0.97. Error bars, mean ± SD. ns = not significant. (D) Live-cell fluorescence microscopy showing reduced recruitment of Cap1-GFP to endocytic sites in cells with the α-tentacle of Cap1 mutated (SDY1582), the CPI motifs of Aim21 and Bsp1 mutated (SDY1604), or with the mutations combined (SDY1608). Scale bar, 1 µm. (E) Quantification of Cap1-GFP peak fluorescence intensity at endocytic patches in WT, cap1*, and aim21CPI*/bsp1CPI* cells. From left to right, mean peak patch/cytosol ratio = 32.08, 11.86, 5.75, and n = 50 for all groups. Error bars, mean with 95% CI. ****P ≤ 0.0001. (F) Live-cell fluorescence microscopy showing no localization of Cap1*-GFP with Abp1-mCherry at endocytic patches in cells with the CPI motifs of Aim21 and Bsp1 mutated (SDY1703). Endocytic patch used for construction of kymograph to the right is indicated by an arrowhead. Scale bar, 1 µm.
When the cap1239,240A (cap1*) allele was integrated into the genome of CAP1-GFP cells, there was a large recruitment defect in Cap1*-GFP (Fig. 4, D and E). However, this defect was not as severe as in aim21CPI*/bsp1CPI* cells (Fig. 4, D and E). When the three mutant alleles were combined in aim21CPI*/bsp1CPI*/cap1* cells, Cap1*-GFP was no longer visible at CME sites, becoming completely diffuse in the cytosol (Fig. 4, D–F). To determine if the capping function of CP was essential for its function during CME, Abp1-GFP was imaged in cells lacking the CP complex (cap1∆), cells with defective capping (cap1*), and cells combining the defective recruiting and defective capping mutants (aim21CPI*/bsp1CPI*/cap1*). Indeed, the increase in recruitment of Abp1-GFP reflecting an overgrown actin network was comparable between the cells, and Abp1-GFP persisted at endocytic patches for a similar, increased time period (Fig. 4 F and Fig. S3, A–C). While the patch lifetime of Abp1-GFP in cap1∆ cells was slightly longer than in cap1* or aim21CPI*/bsp1CPI*/cap1* cells, each was increased substantially from WT cells (Fig. S3 C). Thus, while CPI motif–based recruitment is essential for proper CP localization, the ability of CP to bind barbed ends is essential for its function at endocytic sites.
Twf1, but not Aim21 and Bsp1, requires interaction with CP for proper localization to CME sites
Next, we wanted to determine how the CPI motif proteins are recruited to endocytic sites. Aim21 is predicted to be a largely disordered protein. In addition to its CPI motif, it contains a motif for binding Tda2 (TBR) just downstream of the CPI motif and three regions enriched in polyproline (PxxP) motifs (Fig. 5 A). Like Aim21, Bsp1 is predicted to be largely disordered and contains three regions enriched in polyproline motifs (Fig. 5 A). In contrast, Twf1 is a well-folded multidomain protein consisting of twin ADF-H domains followed by a C-terminal tail containing a CPI motif (Fig. 5 A). To determine if CPI motif–containing proteins are reliant on interaction with CP for localization to CME sites, we looked at their recruitment to CME sites in cells with their CPI motifs mutated or in cells lacking CP (cap1∆). When Aim21 CPI motif mutations were integrated into the genome of AIM21-GFP cells (aim21CPI*), Aim21CPI*-GFP was recruited to endocytic sites at higher levels than Aim21-GFP in WT cells (Fig. 5 B, top, and Fig. 5, C and E, top). The corresponding phenotype was seen in cap1∆ cells, but with even higher levels of Aim21-GFP recruitment (Fig. 5 B, top, and Fig. 5 C). Similarly, Bsp1CPI*-GFP and Bsp1-GFP had a significant increase in recruitment to CME sites in bsp1CPI* and cap1∆ cells, respectively (Fig. 5 B, middle, and Fig. 5, D and E, middle). In contrast, Twf1CPI*-GFP was diffuse in the cytosol in twf1CPI* cells and the same phenotype was seen for Twf1-GFP in cap1∆ cells (Fig. 5 B, bottom, and Fig. 5 E, bottom). Importantly, these findings were not due to defects in overall expression, as Aim21-GFP, Bsp1-GFP, and Twf1-GFP expression was comparable with Aim21CPI*-GFP, Bsp1CPI*-GFP, and Twf1CPI*-GFP, respectively (Fig. S4, A–C). These data show that Twf1 is reliant on its CP:CPI interaction for localization to endocytic sites and are consistent with previous findings (Palmgren et al., 2001; Falck et al., 2004). In agreement with the functions of Aim21 and Bsp1 in CP recruitment, Twf1-GFP recruitment levels were decreased in aim21CPI*/bsp1CPI* cells, where CP itself has a severe recruitment defect (Fig. 5, F and G). However, this defect was less severe than the Twf1-GFP recruitment defect in cap1* and aim21CPI*/bsp1CPI*/cap1* cells, demonstrating that the ability for CP to cap actin filaments is needed for normal Twf1 localization (Fig. 5, F and G).
Twf1 but not Aim21 and Bsp1 require interaction with CP for proper localization to CME sites. (A) Organization of Aim21, Bsp1, and Twf1 domains. (B) Top: Live-cell fluorescence microscopy showing increased recruitment of Aim21-GFP to endocytic sites in cells with the CPI motif of Aim21 mutated (SDY1472), or cells lacking Cap1 (SDY1478). Middle: Live-cell fluorescence microscopy showing increased recruitment of Bsp1-GFP to endocytic sites in cells with the CPI motif of Bsp1 mutated (SDY1516), or cells lacking Cap1 (SDY1570). Bottom: Live-cell fluorescence microscopy showing no recruitment of Twf1-GFP to endocytic sites in cells with the CPI motif of Twf1 mutated (SDY1517) or cells lacking Cap1 (SDY1572). Scale bars, 1 µm. (C) Quantification of Aim21-GFP peak fluorescence intensity at endocytic patches in WT, aim21CPI*, and cap1∆ cells. From left to right, mean peak patch/cytosol ratio = 17.61, 30.18, 37.07, and n = 50 for all groups. Error bars, mean with 95% CI. **P ≤ 0.01, ****P ≤ 0.0001. (D) Quantification of Bsp1-GFP peak fluorescence intensity at endocytic patches in WT, bsp1CPI*, and cap1∆ cells. From left to right, mean peak patch/cytosol ratio = 17.50, 25.02, 28.06, and n = 50 for all groups. Error bars, mean with 95% CI. *P ≤ 0.05, ****P ≤ 0.0001. (E) Live-cell fluorescence microscopy of cells coexpressing (top) Aim21CPI*-GFP, (middle) Bsp1CPI*-GFP, and (bottom) Twf1CPI*-GFP with Cap1-mCherry, respectively (SDY1519, SDY1521, SDY1523). Aim21CPI*-GFP and Bsp1CPI*-GFP show strong colocalization with Cap1-mCherry, while Twf1CPI*-GFP shows no colocalization with Cap1-mCherry. Endocytic patches used for construction of kymographs to the right are indicated by arrowheads. Scale bars, 1 µm. (F) Live-cell fluorescence microscopy showing decreased recruitment of Twf1-GFP to endocytic sites in cells with the α-tentacle of Cap1 mutated (SDY1588), the CPI motifs of Aim21 and Bsp1 mutated (SDY1611), or with the mutations combined (SDY1627). Scale bar, 1 µm. (G) Quantification of Twf1-GFP peak fluorescence intensity at endocytic patches in WT, cap1*, aim21CPI*/bsp1CPI*, and aim21CPI*/bsp1CPI*/cap1* cells. From left to right, the mean peak patch/cytosol ratio = 25.06, 5.15, 12.49, 4.43, and n = 50 for all groups. Error bars, mean with 95% CI. ns = not significant, ****P ≤ 0.0001.
Twf1 but not Aim21 and Bsp1 require interaction with CP for proper localization to CME sites. (A) Organization of Aim21, Bsp1, and Twf1 domains. (B) Top: Live-cell fluorescence microscopy showing increased recruitment of Aim21-GFP to endocytic sites in cells with the CPI motif of Aim21 mutated (SDY1472), or cells lacking Cap1 (SDY1478). Middle: Live-cell fluorescence microscopy showing increased recruitment of Bsp1-GFP to endocytic sites in cells with the CPI motif of Bsp1 mutated (SDY1516), or cells lacking Cap1 (SDY1570). Bottom: Live-cell fluorescence microscopy showing no recruitment of Twf1-GFP to endocytic sites in cells with the CPI motif of Twf1 mutated (SDY1517) or cells lacking Cap1 (SDY1572). Scale bars, 1 µm. (C) Quantification of Aim21-GFP peak fluorescence intensity at endocytic patches in WT, aim21CPI*, and cap1∆ cells. From left to right, mean peak patch/cytosol ratio = 17.61, 30.18, 37.07, and n = 50 for all groups. Error bars, mean with 95% CI. **P ≤ 0.01, ****P ≤ 0.0001. (D) Quantification of Bsp1-GFP peak fluorescence intensity at endocytic patches in WT, bsp1CPI*, and cap1∆ cells. From left to right, mean peak patch/cytosol ratio = 17.50, 25.02, 28.06, and n = 50 for all groups. Error bars, mean with 95% CI. *P ≤ 0.05, ****P ≤ 0.0001. (E) Live-cell fluorescence microscopy of cells coexpressing (top) Aim21CPI*-GFP, (middle) Bsp1CPI*-GFP, and (bottom) Twf1CPI*-GFP with Cap1-mCherry, respectively (SDY1519, SDY1521, SDY1523). Aim21CPI*-GFP and Bsp1CPI*-GFP show strong colocalization with Cap1-mCherry, while Twf1CPI*-GFP shows no colocalization with Cap1-mCherry. Endocytic patches used for construction of kymographs to the right are indicated by arrowheads. Scale bars, 1 µm. (F) Live-cell fluorescence microscopy showing decreased recruitment of Twf1-GFP to endocytic sites in cells with the α-tentacle of Cap1 mutated (SDY1588), the CPI motifs of Aim21 and Bsp1 mutated (SDY1611), or with the mutations combined (SDY1627). Scale bar, 1 µm. (G) Quantification of Twf1-GFP peak fluorescence intensity at endocytic patches in WT, cap1*, aim21CPI*/bsp1CPI*, and aim21CPI*/bsp1CPI*/cap1* cells. From left to right, the mean peak patch/cytosol ratio = 25.06, 5.15, 12.49, 4.43, and n = 50 for all groups. Error bars, mean with 95% CI. ns = not significant, ****P ≤ 0.0001.
Aim21 and Bsp1 localize to CME sites through interactions with SH3 domain–containing proteins. (A) Cell extracts from Aim21-GFP strains used for fluorescence microscopy (Fig. 5 B and panel E) were analyzed by immunoblotting (IB) using antibodies to the GFP tag (anti-GFP). The parent strain, BY4741, is shown as a negative control. Bottom: Coomassie-stained gel of extracts shows equal loading. The strains display comparable expression of Aim21-GFP. (B) Cell extracts from Bsp1-GFP strains used for fluorescence microscopy (Fig. 5 B and panel E) were analyzed by immunoblotting (IB) using antibodies to the GFP tag (anti-GFP). The parent strain, BY4741, is shown as a negative control. Bottom: Coomassie-stained gel of extracts shows equal loading. The strains display comparable expression of Bsp1-GFP. (C) Cell extracts from Twf1-GFP strains used for fluorescence microscopy (Fig. 5, B and F) were analyzed by immunoblotting (IB) using antibodies to the GFP tag (anti-GFP). The parent strain, BY4741, is shown as a negative control. Bottom: Coomassie-stained gel of extracts shows equal loading. The strains display comparable expression of Twf1-GFP. (D) Yeast two-hybrid analysis of cells co-transformed with plasmids expressing the GAL4 activation (pGAD424) and binding (pGBT9) domains fused to the SH3 domain of various endocytic factors and polyproline motif (PxxP)–rich regions of Aim21 or Bsp1, respectively. Cells were spotted onto plates containing histidine (+HIS, control) or selective medium lacking histidine (−HIS). Cell growth, indicative of an interaction between proteins, was detected for Aim21 with Abp1 and Bbc1, and for Bsp1 with Abp1, Lsb3, Rvs167, Sla1, and Ysc84. (E) Top: Live-cell fluorescence microscopy showing decreased recruitment of Aim21-GFP to endocytic sites in cells lacking Abp1, Bbc1, or both Abp1 and Bbc1 (SDY1011, SDY1658, SDY1663). Bottom: Live-cell fluorescence microscopy showing decreased recruitment of Bsp1-GFP to endocytic sites in cells lacking Abp1 (SDY1659). In contrast, cells lacking Lsb3, Rvs167, or Ysc84 display an increase in Bsp1-GFP patch intensity (SDY1660, SDY1700, and SDY1661). (F) Quantification of Aim21-GFP peak fluorescence intensity at endocytic patches in WT, abp1∆, bbc1∆, and abp1∆/bbc1∆ cells. From left to right, mean peak patch/cytosol ratio = 17.61, 11.82, 9.70, 4.56, and n = 50 for all groups. Error bars, mean with 95% CI. ****P ≤ 0.0001. (G) Quantification of Bsp1-GFP peak fluorescence intensity at endocytic patches in WT, abp1∆, lsb3∆, rvs167∆, and ysc84∆ cells. From left to right, the mean peak patch/cytosol ratio = 17.50, 7.29, 20.26, 19.24, 21.49, and n = 50 for all groups. Error bars, mean with 95% CI. ns = not significant, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001. Source data are available for this figure: SourceData FS4.
Aim21 and Bsp1 localize to CME sites through interactions with SH3 domain–containing proteins. (A) Cell extracts from Aim21-GFP strains used for fluorescence microscopy (Fig. 5 B and panel E) were analyzed by immunoblotting (IB) using antibodies to the GFP tag (anti-GFP). The parent strain, BY4741, is shown as a negative control. Bottom: Coomassie-stained gel of extracts shows equal loading. The strains display comparable expression of Aim21-GFP. (B) Cell extracts from Bsp1-GFP strains used for fluorescence microscopy (Fig. 5 B and panel E) were analyzed by immunoblotting (IB) using antibodies to the GFP tag (anti-GFP). The parent strain, BY4741, is shown as a negative control. Bottom: Coomassie-stained gel of extracts shows equal loading. The strains display comparable expression of Bsp1-GFP. (C) Cell extracts from Twf1-GFP strains used for fluorescence microscopy (Fig. 5, B and F) were analyzed by immunoblotting (IB) using antibodies to the GFP tag (anti-GFP). The parent strain, BY4741, is shown as a negative control. Bottom: Coomassie-stained gel of extracts shows equal loading. The strains display comparable expression of Twf1-GFP. (D) Yeast two-hybrid analysis of cells co-transformed with plasmids expressing the GAL4 activation (pGAD424) and binding (pGBT9) domains fused to the SH3 domain of various endocytic factors and polyproline motif (PxxP)–rich regions of Aim21 or Bsp1, respectively. Cells were spotted onto plates containing histidine (+HIS, control) or selective medium lacking histidine (−HIS). Cell growth, indicative of an interaction between proteins, was detected for Aim21 with Abp1 and Bbc1, and for Bsp1 with Abp1, Lsb3, Rvs167, Sla1, and Ysc84. (E) Top: Live-cell fluorescence microscopy showing decreased recruitment of Aim21-GFP to endocytic sites in cells lacking Abp1, Bbc1, or both Abp1 and Bbc1 (SDY1011, SDY1658, SDY1663). Bottom: Live-cell fluorescence microscopy showing decreased recruitment of Bsp1-GFP to endocytic sites in cells lacking Abp1 (SDY1659). In contrast, cells lacking Lsb3, Rvs167, or Ysc84 display an increase in Bsp1-GFP patch intensity (SDY1660, SDY1700, and SDY1661). (F) Quantification of Aim21-GFP peak fluorescence intensity at endocytic patches in WT, abp1∆, bbc1∆, and abp1∆/bbc1∆ cells. From left to right, mean peak patch/cytosol ratio = 17.61, 11.82, 9.70, 4.56, and n = 50 for all groups. Error bars, mean with 95% CI. ****P ≤ 0.0001. (G) Quantification of Bsp1-GFP peak fluorescence intensity at endocytic patches in WT, abp1∆, lsb3∆, rvs167∆, and ysc84∆ cells. From left to right, the mean peak patch/cytosol ratio = 17.50, 7.29, 20.26, 19.24, 21.49, and n = 50 for all groups. Error bars, mean with 95% CI. ns = not significant, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001. Source data are available for this figure: SourceData FS4.
Src homology 3 (SH3) domains interact with polyproline motifs and are prevalent among CME factors (Kurochkina and Guha, 2013). To determine if the PxxP motif–rich regions of Aim21 and Bsp1 interacted with an SH3 domain of a CME factor, we used a yeast two-hybrid analysis to test binding of Aim21 and Bsp1 polyproline motif–rich fragments to a library of SH3 domains from CME factors. The PxxP motif–rich fragment of Aim21 spanning amino acids 1–85 interacted with the Bbc1 SH3 domain, while the 367–406 fragment interacted with the Abp1 SH3 domain, and to a lesser extent with the Bbc1 SH3 domain (Fig. S4 D). For Bsp1, the 137–251 fragment interacted with the SH3 domains of Lsb3, Rvs167, Sla1, and Ysc84, and the 295–370 fragment displayed an interaction with the Abp1 SH3 domain (Fig. S4 D). In both abp1∆ and bbc1∆ cells, Aim21-GFP localization was reduced, and in cells combining the gene deletions, there was a drastic recruitment defect, suggesting both proteins are important for localization of Aim21 (Fig. S4 E, top, and Fig. S4 F). Bsp1-GFP only displayed a recruitment defect in abp1∆ cells; however, this recruitment defect was severe (Fig. S4 E, bottom, and Fig. S4 G). Aim21-GFP and Bsp1-GFP were expressed at comparable levels in WT and knockout cells, respectively, suggesting the defect was not a result of reduced expression (Fig. S4, A and B). As sla1∆ cells grew slowly and appeared unhealthy, they were not included in the analysis. Given the reliance of both Aim21 and Bsp1 on interaction with the F-actin binding protein Abp1 for localization, the increased levels of Aim21-GFP and Bsp1-GFP at CME sites seen in cap1∆ cells are consistent with the overgrown actin network phenotype seen in cap1∆ cells (Fig. 5, B–D; and Fig. S3, A and B). Together, the data show that Aim21 and Bsp1 recruitment to CME sites does not depend on CP but is reliant on interaction with SH3 domain proteins, while recruitment of Twf1 depends on CP.
CPI motifs differentially affect the capping function of CP
The CPI motif of several proteins such as CARMIL was shown to bind to and allosterically regulate CP, resulting in a CP with reduced affinity for barbed ends (Takeda et al., 2010; Yang et al., 2005; McConnell et al., 2020). However, the recent discovery of the mammalian Twf1 CPI motif, which does not affect the capping function of CP, has challenged the notion that inhibiting the capping function of CP is a hallmark of CPI motifs (Mwangangi et al., 2021; Johnston et al., 2018; Hakala et al., 2021). While the CPI motif of yeast Twf1 is expected to function like Twf1 in higher eukaryotes, this has not been demonstrated experimentally. To determine how the CPI motifs of Aim21, Bsp1, and Twf1 affect the capping function of CP, a series of pyrene actin polymerization assays were performed. In the presence of CP, the polymerization rate of actin was severely reduced (Fig. 6, A–C). However, when CP was incubated with an unlabeled Aim21 CPI motif peptide (Aim21CPI) prior to actin polymerization, the polymerization rate increased significantly compared with CP alone (Fig. 6 A). Similarly, the addition of Bsp1CPI to CP resulted in a significant increase in the polymerization rate (Fig. 6 B). In contrast, the Twf1CPI peptide had no effect on the capping function of CP, even at high concentrations (Fig. 6 C). While Aim21CPI and Bsp1CPI both had inhibitory effects on CP, this effect was not seen using the mutant peptides Aim21CPI* and Bsp1CPI*, indicating that the inhibitory effect requires the CP:CPI interaction (Fig. 6, A and B). Importantly, the CPI motifs alone had no effect on the polymerization of actin (Fig. 6, A–C). To confirm the inhibitory effects of Aim21CPI and Bsp1CPI on the capping function of CP, actin was purified from budding yeast and utilized in pyrene actin polymerization assays (Fig. S5, A–D). In agreement with our findings using muscle actin (Fig. 6, A–C), Aim21CPI and Bsp1CPI displayed an inhibitory effect on CP while Twf1CPI did not inhibit CP (Fig. S5, C and D). Thus, the Aim21 and Bsp1 CPI motifs function like CARMIL-type CPI motifs, partially inhibiting the capping function of CP while the Twf1 CPI motif does not inhibit capping activity, a behavior analogous to Twf1 CPI motifs from higher eukaryotes.
CPI motifs differentially affect the capping function of CP. (A) Left: Pyrene-labeled actin (1 µM, 20% labeled) was polymerized in the absence or presence of His-Cap1/2 (CP, 100 nM). In addition, reactions were performed with various concentrations of either unlabeled Aim21CPI or Aim21CPI* peptides. His-Tda2 was included to allow for the formation of the Tda2–Aim21 complex. While Aim21CPI partially inhibited the capping function of CP at high concentrations, Aim21CPI* had no effect. Right: Quantification of the relative polymerization rate from three independent pyrene actin polymerization assays. From left to right, mean relative polymerization rate = 1.00, 0.23, 0.98, 0.31, 0.45, 0.54, and 0.22. Error bars, mean ± SEM. *P ≤ 0.05. (B) Left: Pyrene actin polymerization assays were performed as in A with various concentrations of either unlabeled Bsp1CPI or Bsp1CPI* peptides. While Bsp1CPI inhibited the capping function of CP at high concentrations, Bsp1CPI* had no effect. Right: Quantification of the relative polymerization rate from three independent pyrene actin polymerization assays. From left to right, mean relative polymerization rate = 1.00, 0.23, 0.97, 0.37, 0.58, 0.77, and 0.23. Error bars, mean ± SEM. *P ≤ 0.05, **P ≤ 0.01. (C) Left: Pyrene actin polymerization assays were performed as in A with various concentrations of either unlabeled Twf1CPI or Twf1CPI* peptides. Twf1CPI had no effect on the capping function of CP, even at high concentrations. Right: Quantification of the relative polymerization rate from three independent pyrene actin polymerization assays. From left to right, mean relative polymerization rate = 1.00, 0.23, 0.94, 0.24, 0.21, 0.23, and 0.26. Error bars, mean ± SEM.
CPI motifs differentially affect the capping function of CP. (A) Left: Pyrene-labeled actin (1 µM, 20% labeled) was polymerized in the absence or presence of His-Cap1/2 (CP, 100 nM). In addition, reactions were performed with various concentrations of either unlabeled Aim21CPI or Aim21CPI* peptides. His-Tda2 was included to allow for the formation of the Tda2–Aim21 complex. While Aim21CPI partially inhibited the capping function of CP at high concentrations, Aim21CPI* had no effect. Right: Quantification of the relative polymerization rate from three independent pyrene actin polymerization assays. From left to right, mean relative polymerization rate = 1.00, 0.23, 0.98, 0.31, 0.45, 0.54, and 0.22. Error bars, mean ± SEM. *P ≤ 0.05. (B) Left: Pyrene actin polymerization assays were performed as in A with various concentrations of either unlabeled Bsp1CPI or Bsp1CPI* peptides. While Bsp1CPI inhibited the capping function of CP at high concentrations, Bsp1CPI* had no effect. Right: Quantification of the relative polymerization rate from three independent pyrene actin polymerization assays. From left to right, mean relative polymerization rate = 1.00, 0.23, 0.97, 0.37, 0.58, 0.77, and 0.23. Error bars, mean ± SEM. *P ≤ 0.05, **P ≤ 0.01. (C) Left: Pyrene actin polymerization assays were performed as in A with various concentrations of either unlabeled Twf1CPI or Twf1CPI* peptides. Twf1CPI had no effect on the capping function of CP, even at high concentrations. Right: Quantification of the relative polymerization rate from three independent pyrene actin polymerization assays. From left to right, mean relative polymerization rate = 1.00, 0.23, 0.94, 0.24, 0.21, 0.23, and 0.26. Error bars, mean ± SEM.
Effects of CPI motifs on the capping function of CP using yeast actin. (A) Actin was purified from budding yeast using the DNaseI affinity purification method (Goode, 2002). Samples of the high-speed supernantant (HSS), column washes, and column elution were analyzed by SDS-PAGE and Coomassie staining. Yeast actin (∼41 kD) was successfully purified from yeast with a high degree of purity. (B) Actin pelleting experiments were performed using either rabbit skeletal muscle actin or actin purified from yeast (2 µM). Yeast actin was more abundant in the pellet sample (P) than the supernatant sample (S) when mixed with polymerization buffer (+Poly), demonstrating the actin was assembly competent following purification. (C) Yeast actin was combined with pyrene-labeled actin (1 µM total, 20% pyrene-labeled) and polymerized in the absence or presence of His-Cap1/2 (CP, 100 nM). In addition, reactions were performed with 500 nM of unlabeled Aim21CPI, Bsp1CPI, or Twf1CPI peptides. His-Tda2 was included in the Aim21CPI reaction to allow for formation of the Tda2–Aim21 complex. While Aim21CPI and Bsp1CPI partially inhibited the capping function of CP, Twf1CPI had no effect. These results are consistent with the results seen using rabbit skeletal muscle actin. (D) Quantification of the relative polymerization rate from three independent pyrene actin polymerization assays. From left to right, mean relative polymerization rate = 1.00, 0.19, 0.45, 0.69, and 0.18. Error bars, mean ± SEM. ns = not significant, *P ≤ 0.05, **P ≤ 0.01. Source data are available for this figure: SourceData FS5.
Effects of CPI motifs on the capping function of CP using yeast actin. (A) Actin was purified from budding yeast using the DNaseI affinity purification method (Goode, 2002). Samples of the high-speed supernantant (HSS), column washes, and column elution were analyzed by SDS-PAGE and Coomassie staining. Yeast actin (∼41 kD) was successfully purified from yeast with a high degree of purity. (B) Actin pelleting experiments were performed using either rabbit skeletal muscle actin or actin purified from yeast (2 µM). Yeast actin was more abundant in the pellet sample (P) than the supernatant sample (S) when mixed with polymerization buffer (+Poly), demonstrating the actin was assembly competent following purification. (C) Yeast actin was combined with pyrene-labeled actin (1 µM total, 20% pyrene-labeled) and polymerized in the absence or presence of His-Cap1/2 (CP, 100 nM). In addition, reactions were performed with 500 nM of unlabeled Aim21CPI, Bsp1CPI, or Twf1CPI peptides. His-Tda2 was included in the Aim21CPI reaction to allow for formation of the Tda2–Aim21 complex. While Aim21CPI and Bsp1CPI partially inhibited the capping function of CP, Twf1CPI had no effect. These results are consistent with the results seen using rabbit skeletal muscle actin. (D) Quantification of the relative polymerization rate from three independent pyrene actin polymerization assays. From left to right, mean relative polymerization rate = 1.00, 0.19, 0.45, 0.69, and 0.18. Error bars, mean ± SEM. ns = not significant, *P ≤ 0.05, **P ≤ 0.01. Source data are available for this figure: SourceData FS5.
CP:CPI interactions occur through an overlapping binding site needed for normal CP recruitment to CME sites
Numerous crystal structures of CPI motifs in complex with CP have shown that CPI motifs interact with the stalk region of CP, opposite the barbed end binding surface (Fig. 7 A). Of interest, the Cap2 (β) subunit of CP has an extended binding interface with the CPI motif (Fig. 7 A). To determine if yeast CPI motifs interact with CP via a similar mechanism, a series of GST pulldown assays were performed using mutant Cap1/2 complexes. Given the large binding interface on the Cap2 subunit and the reliance of CPI motifs on basic residues for interaction with CP, acidic residues on the stalk region of Cap2 were targeted for mutation. Four CP mutants were tested for interaction with CPI motifs: His-Cap1/2D44A, His-Cap1/2D64A, His-Cap1/2D88A, and a mutant combining the three mutations, His-Cap1/244,64,88A. Each CP mutant displayed different degrees of binding with the CPI motifs (Fig. 7 B). GST-Aim21CPI pulled down normal levels of His-Cap1/2D44A, but less His-Cap1/2D64A and His-Cap1/2D88A than WT CP (Fig. 7 B). GST-Bsp1CPI displayed a weakened interaction with His-Cap1/2D44A and His-Cap1/2D64A but pulled down His-Cap1/2D88A at levels comparable with His-Cap1/2 (Fig. 7 B). GST-Twf1CPI pulled down less His-Cap1/2D44A and His-Cap1/2D88A than WT CP, but normal levels of His-Cap1/2D64A (Fig. 7 B). For each CPI motif fusion, the triple mutant CP, His-Cap1/244,64,88A, abolished the CP:CPI interaction (Fig. 7 B). Together, these results suggest CPI motifs interact with the stalk region of CP, similar to higher eukaryotes.
CP:CPI interactions occur through an overlapping binding site. (A) Top: Crystal structure of chicken CP bound to the CPI motif of human CD2AP (PDB entry 3AA6). The α-subunit (Cap1) is displayed in dark blue, the β-subunit (Cap2) in light blue, and the CPI motif of CD2AP is shown in red. Bottom: The CP complex is rotated 90⁰ for a view of the stalk region of CP. The N-terminus and C-terminus of the CPI motif are labeled N’ and C’, respectively. The CP:CPI interaction occurs largely through a binding interface on the β-subunit. (B) A GST pulldown assay was performed with GST fused to the CPI motifs of Aim21 (top), Bsp1 (middle), or Twf1 (bottom). Each GST fusion protein was incubated with either His-Cap1/2 or with a CP containing a mutated version of the Cap2 subunit potentially affecting interaction with the CPI motifs (His-Cap1/2D44A, His-Cap1/2D64A, His-Cap1/2D88A, and a mutant combining the three mutations, His-Cap1/244,64,88A). Aim21 was additionally incubated with His-Tda2 to form the Tda2/Aim21 complex. Bound proteins were analyzed by SDS-PAGE and Coomassie staining. The mutations to Cap2 affect the interaction with the GST-CPI motif fusions in varying ways and abolish the interaction when combined. (C) Pyrene-labeled actin (1 µM, 20% labeled) was polymerized in the absence or presence of His-Cap1/2 (CP, 100 nM) or His-Cap1/244,64,88A (CP2*, 100 nM). In addition, reactions were performed with either 500 nM of unlabeled Aim21CPI, Bsp1CPI, or Twf1CPI peptides. His-Tda2 was included in the Aim21CPI reaction to allow for formation of the Tda2–Aim21 complex. CP2* displayed an equivalent capping ability to CP, and the capping function of CP2* was not affected by any of the CPI motif peptides. (D) Quantification of the relative polymerization rate from three independent pyrene actin polymerization assays in C. From left to right, mean relative polymerization rate = 1.00, 0.25, 0.32, 0.30, 0.31, and 0.31. Error bars, mean ± SD. ns = not significant. (E) Top: Live-cell fluorescence microscopy showing reduced recruitment of Cap1-GFP to endocytic sites in cells with the CPI motif binding site of Cap2 mutated (SDY1596). Bottom: Live-cell fluorescence microscopy showing increased recruitment of Abp1-GFP to endocytic sites in cells with the CPI motif binding site of Cap2 mutated (SDY1599). Scale bars, 1 µm. (F) Quantification of Cap1-GFP peak fluorescence intensity at endocytic patches in WT and cap2* cells. From left to right, mean peak patch/cytosol ratio = 32.08, 8.44, and n = 50 for all groups. Error bars, mean with 95% CI. ****P ≤ 0.0001. (G) Quantification of Abp1-GFP peak fluorescence intensity at endocytic patches in WT and cap2* cells. From left to right, mean peak patch/cytosol ratio = 49.63, 70.40, and n = 50 for all groups. Error bars, mean with 95% CI. ****P ≤ 0.0001. (H) Quantification of Abp1-GFP patch lifetime at endocytic patches in WT and cap2* cells. From left to right, mean patch lifetime = 18.06, 22.96 s, and n = 50 for all groups. Error bars, mean with 95% CI. ****P ≤ 0.0001. Source data are available for this figure: SourceData F7.
CP:CPI interactions occur through an overlapping binding site. (A) Top: Crystal structure of chicken CP bound to the CPI motif of human CD2AP (PDB entry 3AA6). The α-subunit (Cap1) is displayed in dark blue, the β-subunit (Cap2) in light blue, and the CPI motif of CD2AP is shown in red. Bottom: The CP complex is rotated 90⁰ for a view of the stalk region of CP. The N-terminus and C-terminus of the CPI motif are labeled N’ and C’, respectively. The CP:CPI interaction occurs largely through a binding interface on the β-subunit. (B) A GST pulldown assay was performed with GST fused to the CPI motifs of Aim21 (top), Bsp1 (middle), or Twf1 (bottom). Each GST fusion protein was incubated with either His-Cap1/2 or with a CP containing a mutated version of the Cap2 subunit potentially affecting interaction with the CPI motifs (His-Cap1/2D44A, His-Cap1/2D64A, His-Cap1/2D88A, and a mutant combining the three mutations, His-Cap1/244,64,88A). Aim21 was additionally incubated with His-Tda2 to form the Tda2/Aim21 complex. Bound proteins were analyzed by SDS-PAGE and Coomassie staining. The mutations to Cap2 affect the interaction with the GST-CPI motif fusions in varying ways and abolish the interaction when combined. (C) Pyrene-labeled actin (1 µM, 20% labeled) was polymerized in the absence or presence of His-Cap1/2 (CP, 100 nM) or His-Cap1/244,64,88A (CP2*, 100 nM). In addition, reactions were performed with either 500 nM of unlabeled Aim21CPI, Bsp1CPI, or Twf1CPI peptides. His-Tda2 was included in the Aim21CPI reaction to allow for formation of the Tda2–Aim21 complex. CP2* displayed an equivalent capping ability to CP, and the capping function of CP2* was not affected by any of the CPI motif peptides. (D) Quantification of the relative polymerization rate from three independent pyrene actin polymerization assays in C. From left to right, mean relative polymerization rate = 1.00, 0.25, 0.32, 0.30, 0.31, and 0.31. Error bars, mean ± SD. ns = not significant. (E) Top: Live-cell fluorescence microscopy showing reduced recruitment of Cap1-GFP to endocytic sites in cells with the CPI motif binding site of Cap2 mutated (SDY1596). Bottom: Live-cell fluorescence microscopy showing increased recruitment of Abp1-GFP to endocytic sites in cells with the CPI motif binding site of Cap2 mutated (SDY1599). Scale bars, 1 µm. (F) Quantification of Cap1-GFP peak fluorescence intensity at endocytic patches in WT and cap2* cells. From left to right, mean peak patch/cytosol ratio = 32.08, 8.44, and n = 50 for all groups. Error bars, mean with 95% CI. ****P ≤ 0.0001. (G) Quantification of Abp1-GFP peak fluorescence intensity at endocytic patches in WT and cap2* cells. From left to right, mean peak patch/cytosol ratio = 49.63, 70.40, and n = 50 for all groups. Error bars, mean with 95% CI. ****P ≤ 0.0001. (H) Quantification of Abp1-GFP patch lifetime at endocytic patches in WT and cap2* cells. From left to right, mean patch lifetime = 18.06, 22.96 s, and n = 50 for all groups. Error bars, mean with 95% CI. ****P ≤ 0.0001. Source data are available for this figure: SourceData F7.
Having identified mutations to CP that disrupt interaction with each CPI motif protein, we wanted to assess the endocytic phenotype produced when the mutations were integrated into the yeast genome. Before doing so, we ensured that the CP mutant retained its capping function. When pyrene actin polymerization was induced in the presence of His-Cap1/2 (CP) or the triple mutant His-Cap1/244,64,88A (CP2*), the polymerization rate was reduced to comparable levels (Fig. 7, C and D). Additionally, adding Aim21CPI or Bsp1CPI to CP2* prior to actin polymerization induction did not result in inhibition of CP2* as it did with WT CP, providing further proof that the mutations disrupt the CP:CPI interaction (Fig. 6, A and B; and Fig. 7, C and D).
To determine the endocytic phenotype produced when all three CPI motifs are unable to interact with CP, we integrated the cap244,64,88A (cap2*) allele at the endogenous CAP2 locus, initially in cells expressing Cap1-GFP. Cap1-GFP recruitment was severely impaired in cap2* cells compared with WT cells (Fig. 7 E, top, and Fig. 7 F). This phenotype is similar to that of aim21CPI* and bsp1CPI* cells rather than twf1CPI* cells, suggesting that aim21CPI* and bsp1CPI* work upstream of twf1CPI*. Consistent with synergistic roles of Aim21 and Bsp1 in CP recruitment, the defect in cap2* cells was more severe than in aim21CPI* or bsp1CPI* cells, with peak patch/cytosol ratios of 8.44, 18.73, and 23.13, respectively (Fig. 2 D and Fig. 7 F). The recruitment defect was not due to a decrease in expression of Cap1-GFP, as Cap1-GFP was expressed at comparable levels in WT and cap2* cells (Fig. S2 E). As expected with a defect in CP recruitment, Abp1-GFP levels were significantly elevated in cap2* cells and had an increased patch lifetime, indicative of an enlarged actin network (Fig. 7 E, bottom, and Fig. 7, G and H).
The CPI motif of Bsp1 is required for localization of CP to the contractile actomyosin ring (AMR)
In addition to CPs’ localization to actin patches at endocytic sites, CP was recently found at a structure resembling the AMR at the mother-bud neck (Wirshing et al., 2023). Interestingly, in a subset of cells with large buds, Bsp1-GFP colocalized with Cap1-mCherry to an elongated structure at the mother-bud neck (Fig. 8 A). In agreement with this structure being the AMR, both Bsp1-GFP and Cap1-GFP showed colocalization with the AMR marker Myo1-mCherry in a portion of cells (Fig. 8 B). Both Bsp1-GFP and Cap1-GFP were recruited to the AMR just prior to contraction of Myo1-mCherry, contracted along with Myo1-mCherry, and then disappeared from the AMR (Fig. 8 C, top). In contrast, neither Aim21-GFP nor Twf1-GFP displayed colocalization with the contracting AMR (Fig. 8 C, bottom). While cells with a mutated Bsp1 CPI motif still displayed colocation between Bsp1CPI*-GFP and Myo1-mCherry, Cap1-GFP was no longer seen at the contracting AMR in cells with the bsp1CPI* allele (Fig. 8 D). Consistent with this recruitment defect, Cap1-mCherry did not colocalize with Bsp1CPI*-GFP at the AMR (Fig. 8, E and F). Thus, the CPI motif of Bsp1 has a role in localizing CP to the contractile AMR.
The CPI motif of Bsp1 is required for localization of CP to the AMR. (A) Live-cell fluorescence microscopy showing colocalization of Bsp1-GFP and Cap1-mCherry at the bud neck between the mother and daughter cell (SDY1520). Scale bar, 1 µm. (B) Live-cell fluorescence microscopy showing colocalization of Bsp1-GFP (top) or Cap1-GFP (bottom) with the AMR marker Myo1-mCherry, respectively (SDY1647 and SDY1649). Scale bars, 1 µm. (C) Live-cell fluorescence microscopy of cells expressing Bsp1-GFP, Cap1-GFP, Aim21-GFP, and Twf1-GFP along with the AMR marker Myo1-mCherry, respectively (SDY1647, SDY1649, SDY1654, SDY1655). Bsp1-GFP and Cap1-GFP are recruited to the AMR just prior to contraction. In contrast, Aim21-GFP and Twf1-GFP do not colocalize with Myo1-mCherry at the AMR. AMRs used for construction of kymographs to right are indicated by arrowheads. Arrows indicate the arrival and departure of the GFP-tagged protein. Scale bars, 1 µm. (D) Top: Live-cell fluorescence microscopy showing colocalization of Bsp1CPI*-GFP with the AMR marker Myo1-mCherry (SDY1656). Bottom: Live-cell fluorescence microscopy showing no recruitment of Cap1-GFP to the AMR in cells with the CPI motif of Bsp1 mutated (SDY1657). AMRs used for construction of kymographs to the right are indicated by arrowheads. Arrows indicate the arrival and departure of the GFP-tagged protein. Scale bars, 1 µm. (E) Live-cell fluorescence microscopy of cells expressing Bsp1-GFP (top) or Bsp1CPI*-GFP (bottom) with Cap1-mCherry, respectively (SDY1647, SDY1649). Cap1-mCherry shows strong colocalization with Bsp1-GFP at the AMR. In contrast, Cap1-mCherry does not colocalize with Bsp1CPI*-GFP at the AMR. (F) Quantification of the PCC between Bsp1-GFP or Bsp1CPI*-GFP at the AMR with Cap1-mCherry, respectively. From left to right, mean PCC = 0.544, −0.023, and n = 25, 22. Error bars, mean ± SD.
The CPI motif of Bsp1 is required for localization of CP to the AMR. (A) Live-cell fluorescence microscopy showing colocalization of Bsp1-GFP and Cap1-mCherry at the bud neck between the mother and daughter cell (SDY1520). Scale bar, 1 µm. (B) Live-cell fluorescence microscopy showing colocalization of Bsp1-GFP (top) or Cap1-GFP (bottom) with the AMR marker Myo1-mCherry, respectively (SDY1647 and SDY1649). Scale bars, 1 µm. (C) Live-cell fluorescence microscopy of cells expressing Bsp1-GFP, Cap1-GFP, Aim21-GFP, and Twf1-GFP along with the AMR marker Myo1-mCherry, respectively (SDY1647, SDY1649, SDY1654, SDY1655). Bsp1-GFP and Cap1-GFP are recruited to the AMR just prior to contraction. In contrast, Aim21-GFP and Twf1-GFP do not colocalize with Myo1-mCherry at the AMR. AMRs used for construction of kymographs to right are indicated by arrowheads. Arrows indicate the arrival and departure of the GFP-tagged protein. Scale bars, 1 µm. (D) Top: Live-cell fluorescence microscopy showing colocalization of Bsp1CPI*-GFP with the AMR marker Myo1-mCherry (SDY1656). Bottom: Live-cell fluorescence microscopy showing no recruitment of Cap1-GFP to the AMR in cells with the CPI motif of Bsp1 mutated (SDY1657). AMRs used for construction of kymographs to the right are indicated by arrowheads. Arrows indicate the arrival and departure of the GFP-tagged protein. Scale bars, 1 µm. (E) Live-cell fluorescence microscopy of cells expressing Bsp1-GFP (top) or Bsp1CPI*-GFP (bottom) with Cap1-mCherry, respectively (SDY1647, SDY1649). Cap1-mCherry shows strong colocalization with Bsp1-GFP at the AMR. In contrast, Cap1-mCherry does not colocalize with Bsp1CPI*-GFP at the AMR. (F) Quantification of the PCC between Bsp1-GFP or Bsp1CPI*-GFP at the AMR with Cap1-mCherry, respectively. From left to right, mean PCC = 0.544, −0.023, and n = 25, 22. Error bars, mean ± SD.
Discussion
Until the recent discovery of the Aim21 CPI motif, CP regulators were not believed to be present in yeast. Here, we uncovered a new CPI motif–containing protein, Bsp1, and showed that the Twf1 CPI characterized in mouse is conserved in yeast. Based on our findings, we propose a working model for regulation of CP by CPI motif proteins during CME in Fig. 9. The CPI motifs of Aim21 and Bsp1 mediate interaction with CP and work synergistically to recruit CP to CME sites to bind and cap barbed ends of actin filaments. In addition, a small subset of CP can localize to CME sites by an alternative method, likely freely diffusing through the cytosol to bind barbed ends. Following CP binding to barbed ends of actin, Twf1 is recruited to CP through its CPI motif, which necessitates Aim21 and Bsp1 to dissociate from CP. Twf1 then dissociates CP from barbed ends through its twin ADF-H domains and exposes the barbed ends for efficient depolymerization and recycling for additional rounds of CME. Together, our findings shed light on how multiple CPI motif proteins function at the same subcellular location to regulate CP activity in a sequential manner.
Model of CPI motif regulation of CP during CME. Top: In WT cells, CP is recruited to CME sites through interaction with the CPI motifs of Aim21 and Bsp1, and to a lesser degree through freely binding F-actin barbed ends. Following CP binding to barbed ends, Aim21 and Bsp1 dissociate from CP and allow Twf1 to bind CP through its CPI motif. Twf1 functions to remove CP from barbed ends through its ADF-H domains. The removal of CP and the exposure of aged barbed ends allow for efficient turnover of the branched actin network. Middle: In cells with non-functional Aim21 and Bsp1 motifs, there is a severe CP recruitment defect, leading to reduced recruitment of Twf1. The lack of CP results in an overgrown actin network and elevated levels of Aim21 and Bsp1. The overgrown actin network is turned over at a slower rate due to reduced levels of Twf1. Bottom: In cells with a non-functional Twf1 CPI motif, CP is recruited through Aim21 and Bsp1, but Twf1 is not properly recruited following CP binding to barbed ends. CP is not removed from aged barbed ends by Twf1, and the rate of turnover of the actin network is reduced.
Model of CPI motif regulation of CP during CME. Top: In WT cells, CP is recruited to CME sites through interaction with the CPI motifs of Aim21 and Bsp1, and to a lesser degree through freely binding F-actin barbed ends. Following CP binding to barbed ends, Aim21 and Bsp1 dissociate from CP and allow Twf1 to bind CP through its CPI motif. Twf1 functions to remove CP from barbed ends through its ADF-H domains. The removal of CP and the exposure of aged barbed ends allow for efficient turnover of the branched actin network. Middle: In cells with non-functional Aim21 and Bsp1 motifs, there is a severe CP recruitment defect, leading to reduced recruitment of Twf1. The lack of CP results in an overgrown actin network and elevated levels of Aim21 and Bsp1. The overgrown actin network is turned over at a slower rate due to reduced levels of Twf1. Bottom: In cells with a non-functional Twf1 CPI motif, CP is recruited through Aim21 and Bsp1, but Twf1 is not properly recruited following CP binding to barbed ends. CP is not removed from aged barbed ends by Twf1, and the rate of turnover of the actin network is reduced.
In higher eukaryotes, CP is not constitutively active but is bound in an inactive state by the protein V-1 (Bhattacharya et al., 2006; Taoka et al., 2003). V-1 interacts with the α-tentacle on the cap region of CP with 1:1 stoichiometry and sterically inhibits interaction with barbed ends (Zwolak et al., 2010; Takeda et al., 2010). In addition to partially inhibiting CP in in vitro assays, CARMIL-type CPI motifs can bind to a CP:V-1 complex and induce dissociation of V-1, thus activating CP (McConnell et al., 2020; Fujiwara et al., 2014). Although the CPI motifs of Aim21 and Bsp1 partially inhibited CP in our pyrene actin polymerization assays, they are unlikely to have an analogous CP-activating function because there is no known V-1 homolog in budding yeast. However, we cannot rule out the presence of alternative mechanisms that maintain an inactive cytosolic population of CP in yeast. Additional work is needed to determine if recruitment of a transiently active CP is critical for the proper formation and architecture of branched actin networks, or simply a byproduct of the needs to be activated.
While the status of Twf1 as an actin depolymerization factor is established, the function of its CPI motif and how it may regulate CP function remains a subject of debate. One recent model postulates Twf1 functioning as a pro-capping factor, working to activate CP and protect CP bound to barbed ends from uncapping by CARMIL-type CPI motifs. However, an opposing model suggests CARMIL-type CPI motif proteins are the procapping factors, with Twf1 working downstream of CP binding barbed ends to turn over CP (Hakala et al., 2021). Our cell biological data agree with the latter model as CP is largely diffuse in the cytosol without Aim21 and Bsp1 to localize it to endocytic sites. Moreover, Twf1 is almost completely dependent on CP binding to actin filaments for localization to endocytic sites, demonstrating it works downstream of the “pro-capping” CP recruitment activity of Aim21 and Bsp1. However, aspects of the former model with Twf1 functioning as a pro-capping factor may also be true. The trigger for Twf1-mediated uncapping in our model is unknown, but one possibility is the hydrolysis state of the barbed end actin subunits. Under this hypothesis, Twf1 could function to protect CP from rebinding to Aim21 or Bsp1 in young filaments, preventing potential uncapping. Following a protective role in newly capped filaments, Twf1 could transition to a filament uncapping function as the hydrolysis state of bound ATP changes in older filaments, thereby triggering uncapping through the Twf1 ADF-H domains. Nevertheless, our data indicates the predominant function of Twf1 in cells is to uncap filaments.
In addition to localizing to branched actin networks in mammalian cells, CP also localizes to the AMR where it regulates actin dynamics during cytokinesis (Terry et al., 2018). In agreement with a recent publication that observed CP at an AMR-like structure in budding yeast (Wirshing et al., 2023), our results confirm that CP is a component of the contractile AMR. Additionally, we demonstrated that CP localizes to the AMR in a CPI motif–dependent fashion, requiring interaction with Bsp1 for localization to the AMR. To our knowledge, this is the first example of a CPI motif protein regulating CP at the AMR. Moreover, it underscores the principle that CP requires interactions with CPI motif proteins for proper subcellular localization, even in an organism with a comparatively simple set of actin structures, such as budding yeast. Additional work will be needed to determine the precise function of CP at the AMR and how its regulatory cycle works there in the absence of Twf1.
In conclusion, we used the genetically tractable model system S. cerevisiae to uncover the regulatory cycle of CP during CME. While components of CP regulation may differ between organisms and specific actin networks, our work uncovers themes of CP regulation likely conserved throughout eukaryotes.
Materials and methods
Plasmids and yeast strains
All cloning was performed using the In-fusion HD Cloning System (Takara Bio Inc.). Plasmids for bacterial expression of recombinant GST CPI motif fusion proteins were generated through PCR amplification of the corresponding AIM21, BSP1, TWF1, or INP51 DNA sequence from yeast genomic DNA and cloned into pGEX-5X-1. Generation of plasmids for bacterial expression of recombinant His-Tda2, His-Cap1, and His-Cap2 was described previously (Lamb et al., 2021). Plasmids for yeast two-hybrid analysis of Bsp1 fragments were generated by PCR amplification of the corresponding DNA fragments of BSP1 and cloned into pGBT9. Generation of pGBT9-Aim21 fusions and the pGAD424-SH3 domain fusion library was described previously (Farrell et al., 2017). All plasmids encoding mutant recombinant proteins were engineered by PCR-based mutagenesis using the In-fusion system. All constructs were verified by DNA sequencing.
For plasmids designed to integrate mutations into the S. cerevisiae genome, fragments containing the full ORF of either AIM21, BSP1, CAP1, CAP2, or TWF1 plus 100 base pairs upstream and downstream were generated by PCR amplification from yeast genomic DNA and cloned into pUC18. PCR-based mutagenesis of pUC18-AIM21, pUC18-BSP1, pUC18-CAP1, pUC18-CAP2, and pUC18-TWF1 yielded the templates for genetic manipulation.
The background S. cerevisiae strain BY4741 (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0) was used throughout this study. WT GFP-expressing strains (Aim21-GFP, Abp1-GFP, Bsp1-GFP, Cap1-GFP, Mup1-GFP, and Twf1-GFP) were obtained from the yeast GFP library (Invitrogen). Integration of mutant alleles into the yeast genome was performed using a two-step gene replacement approach described previously (Di Pietro et al., 2010). As an example, SDY1474 (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, CAP1-GFP::HIS3, aim21504,507,509E) was created from the Cap1-GFP library strain (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, CAP1-GFP::HIS3). In step one, URA3 was amplified from pRS316 (Sikorski and Hieter, 1989) using primers that impart 50 base pairs of homology to the sequences directly upstream and downstream of the AIM21 ORF, and then transformed (Ito et al., 1983) into the Cap1-GFP library strain to generate intermediate strain SDY1292 (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, CAP1-GFP::HIS3, aim21Δ::URA3). Second, the intermediate strain SDY1292 was transformed with a DNA fragment containing the mutant allele aim21504,507,509E. Cells were grown overnight on plates containing rich media, then replica-plated onto plates containing 5-fluoroorotic acid (5-FOA). Colonies that grew on plates containing 5-FOA represented cells in which the aim21505,507,509E allele replaced URA3. C-terminal tagging of ABP1, CAP1, and MYO1 ORFs with mCherry was accomplished through PCR amplification of pFA6a-mCherry-KANMX6 and homologous recombination. All integrants were verified by PCR amplification of genomic DNA and DNA sequencing of the purified PCR product. The GFP used to create yeast strains was monomeric. Numerous strains utilized in this study were generated previously (Lamb et al., 2021). All yeast strains and plasmids utilized in this study are listed in Table S1 and Table S2, respectively.
Biochemical methods
Recombinant His and GST fusion proteins were expressed in BL21 codon plus Escherichia coli and purified using HisPur Cobalt Resin (Thermo Fisher Scientific) or Glutathione Sepharose 4 Fast Flow (GE Healthcare) as previously described (Feliciano and Di Pietro, 2012). All proteins were dialyzed in PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4). Protein concentrations were determined using absorbance at 280 nm and the corresponding extinction coefficient.
GST pulldown assays were performed as previously described (Bultema et al., 2014). In short, recombinant GST-tagged proteins (10 μg) were incubated with glutathione Sepharose resin for 30 min at 4°C. Equimolar His-tagged protein was added and incubated for an additional 30 min. Resin was washed three times in PBS containing Triton X-100 (0.1–0.5%) and boiled in Laemmli sample buffer. Bound proteins were analyzed by SDS-PAGE and Coomassie staining. All pulldowns were repeated in an independent experiment to ensure reproducibility.
Total yeast extracts were obtained as previously described (Di Pietro et al., 2010). Immunoblotting of cell extract was performed using mouse anti-GFP antibodies (cat# 11814460001; Sigma-Aldrich). For Aim21-GFP immunoblotting, transfer buffer was supplemented with 0.1% SDS for increased transfer efficiency of Aim21-GFP. For all immunoblots, chemiluminescence was detected on an ImageQuant LAS 500 imager (GE Healthcare).
All peptides utilized in fluorescence polarization and pyrene actin polymerization assays were purchased from GenScript. The sequences of peptides were as follows: (FITC-Aim21CPI) FITC-KTGPLGGTRRGRGPRGRKLPSKVASVEKIEEDDNTNKIEIFNNWNVSS, (Aim21CPI) KTGPLGGTRRGRGPRGRKLPSKVASVEKIEEDDNTNKIEIFNNWNVSS, (Aim21CPI*) KTGPLGGTRRGEGPEGEKLPSKVASVEKIEEDDNTNKIEIFNNWNVSS, (FITC-Bsp1CPI) FITC-KETKPLVHPNKNRTRGPRRKLPTRV, (Bsp1CPI) KETKPLVHPNKNRTRGPRRKLPTRV, (Bsp1CPI*) KETKPLVHPNKNETEGPERKLPTRV, (FITC-Twf1CPI) FITC-SNPDLPNKSNLKFNKPKGPLRKRRT, (Twf1CPI) SNPDLPNKSNLKFNKPKGPLRKRRT, (Twf1CPI*) SNPDLPNKSNLKFNEPEGPLEKRRT. Peptide concentrations were determined using absorbance at 205 nm.
Fluorescence polarization assays
Fluorescence polarization assays were performed as previously described (Feliciano et al., 2015). Briefly, FITC-labeled peptides were titrated with a 24-point dilution series of His-Cap1/2 in experiment buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, 0.01% Triton X-100, and 2 mM DTT). For Aim21 and Twf1, 10 nM of FITC-labeled peptide was utilized, while 2.5 nM of FITC-labeled peptide was used for Bsp1. His-Tda2 was included at a final concentration of 25 μM for all experiments using the Aim21 peptide. Data were collected using 384-well non-binding polystyrene microplates (Greiner Bio-one) in a Victor3 V microplate reader (PerkinElmer) at room temperature following a 30-min incubation. Three technical replicates were performed per experiment, and each experiment was performed independently three times. The dissociation constants for each independent experiment were determined by curve fitting the data to a one-site binding isotherm using GraphPad Prism Software. The reported dissociation constants correspond to the average (and SEM) of values obtained in the three independent experiments.
Actin polymerization assays
Pyrene actin polymerization assays were performed using either rabbit skeletal muscle actin (Cytoskeleton) or yeast actin purified from the yeast strain TVY614 (Feliciano et al., 2011) using the DNaseI affinity purification method (Goode, 2002). Unlabeled actin and pyrene actin were diluted to a concentration of 0.5 mg/ml in G-buffer (5 mM Tris-HCl, pH 8, 0.2 mM CaCl2, 0.2 mM ATP, and 0.5 mM DTT) and left on ice to allow for actin depolymerization. Actin was subsequently centrifuged at 100,000 × g in a TLA100.3 rotor (Beckman Coulter) at 4°C for 30 min. A 2.5 μM stock of 20% pyrene-labeled actin was prepared as described (Doolittle et al., 2013). Reactions were performed with a final volume of 100 μl in 96-well non-binding polystyrene microplates (Greiner Bio-one). 10 μl of exchange buffer (5 mM Tris, 10 mM EGTA, and 1 mM MgCl2) was added to 40 μl of actin stock in a 96-well plate and incubated at room temperature for 10 min. To induce polymerization, a 50 μl mix containing the proteins of interest and 10 μl of 10× polymerization buffer (Final concentration 50 mM KCl, 2 mM MgCl2, 1 mM ATP, and 1 mM EGTA) was added to the 96-well plate. Polymerization was measured over time using a Victor3 V microplate reader (Perkin Elmer-Cetus) with excitation and emission wavelengths of 365 and 406 nm, respectively. Actin polymerization rates were calculated from the slope of the linear portion of assembly curves (25–50% polymerization) and normalized to the polymerization rate of actin with polymerization buffer rate. Statistical significance between groups was determined using an unpaired t test (GraphPad Prism Software).
Actin pelleting assays were performed using rabbit skeletal muscle actin and yeast actin prepared using the previously described method for G-actin preparation. 45 μl of a 2.22 μM G-actin stock (2 μM Final) was mixed with 5 μl of G-buffer (−Poly) or 5 μl of 10× polymerization buffer (+Poly) and incubated at room temperature for 30 min. The reactions were subsequently centrifuged at 100,000 × g in a TLA100.3 rotor (Beckman Coulter) at 24°C for 30 min. Following centrifugation, the 50-μl supernatant fraction was removed to a tube containing 10 μl of 5× Laemmli sample buffer (S fraction) and 60 μl of 1× Laemmli sample buffer was added to the pellet fraction (P fraction). Supernatant and pellet fractions were analyzed by SDS-PAGE and Coomassie staining.
Fluorescence microscopy
Fluorescence microscopy was performed as previously described using an IX81 spinning-disk confocal microscope (Olympus) with an Andor iXon Ultra camera (Oxford Instruments) and a 100× 1.40 NA objective (Tolsma et al., 2018). Briefly, cells grown overnight were diluted in synthetic complete media, grown to early logarithmic phase, and then imaged at room temperature. For imaging of endocytic patches, time-lapse images were generated by collecting an image each second for 90 s for single-color microscopy, and every 2 s for 60 s for two-color microscopy. For imaging of the actomyosin ring, slides were prepared as described (Lewellyn and Miao, 2018) to allow for longer imaging durations, and time-lapse images were generated by collecting images every 2 min for 60 min. Slidebook 6 software (Intelligent Imaging Innovations) was used for acquisition and quantification of endocytic patch intensities, patch lifetimes, intensities of Mup1-GFP cargo at the plasma membrane, and colocalization (Pearson correlation coefficients, PCC). Quantification of peak patch/cytosol ratios and patch lifetimes was performed as previously described (Lamb and Di Pietro, 2022). For quantification of peak patch/cytosol ratios, masks were drawn over individual endocytic sites to track the fluorescence of the patch over time. Patch intensity, average cytosol intensity, and average background intensity were recorded for the frame in which the patch intensity was at its maximum. After subtracting the average background from both values, the values were divided to give the peak patch/cytosol ratio. For quantification of patch lifetimes, masks were drawn over individual endocytic patches to track the maximum fluorescence of each patch over time. Prior to quantification, a threshold fluorescence value was established for each fluorescently tagged protein to establish a start and finish value for the endocytic event. The patch lifetime was determined by the number of frames in which the maximum patch fluorescence was above the threshold value. Mup1-GFP endocytic assays were performed as previously described (Farrell et al., 2017). Briefly, Mup1-GFP cells were grown in minimal media containing methionine to early log phase, moved to minimal media lacking methionine for 2 h, and imaged 20 min after return to methionine-rich media. Fluorescence in the membrane was measured using a mask drawn on the cell periphery and normalized to the background. Individual membrane fluorescence values were normalized by dividing the mean fluorescence intensity of a cell by the mean value of all cells within the group. Statistical significance between groups was determined using an unpaired t test, and distribution of data was tested for normality using the Shapiro–Wilk test for all microscopy data (GraphPad Prism Software). Representative images for groups that were directly compared were displayed with equal brightness and contrast settings.
Yeast two-hybrid assay
AH109 cells were cotransformed with pGBT9 and pGAD424 vectors (Takara Bio Inc.) and grown on synthetic dropout media lacking leucine and tryptophan. Successful cotransformants were grown overnight in synthetic dropout media lacking leucine and tryptophan. The following day, cells were diluted to an O.D. 600 of 0.2 in sterile water. Diluted cells were spotted on synthetic dropout media lacking leucine and tryptophan (control), or lacking leucine, tryptophan, and histidine.
Online supplemental material
Fig. S1 shows fluorescence polarization assays with FITC-labeled peptides corresponding to the CPI motifs of Aim21, Bsp1, and Twf1 demonstrating binding to CP. Fig. S1 also contains fluorescence microscopy images showing that Aim21-GFP and Twf1-GFP display higher overall fluorescence intensity levels than Bsp1-GFP, likely reflecting higher expression levels. Fig. S2 shows that the CPI motifs of Aim21, Bsp1, and Twf1 regulate CP and Abp1 at endocytic sites and that various mutant strains display Cap1-GFP expression levels comparable with WT cells. Fig. S3 shows increased recruitment and patch lifetime of Abp1-GFP at endocytic sites in cells lacking Cap1, with the α-tentacle of Cap1 mutated, with the CPI motifs of Aim21 and Bsp1 mutated, or with the mutations combined. Fig.S3 also shows mutations to the Cap1 α-tentacle do not affect the interaction between the CPI motifs and CP. Fig. S4 shows Aim21 and Bsp1 localize to CME sites through interactions with endocytic SH3 domain–containing proteins and controls demonstrating comparable expression of Aim21-GFP, Bsp1-GFP, and Twf1-GFP in strains harboring various mutations. Fig. S5 shows the effects of CPI motifs of Aim21, Bsp1, and Twf1 on the capping function of CP using yeast actin. Table S1 contains a list of yeast strains utilized in this study. Table S2 contains a list of plasmids utilized in this study. Table S3 contains a list of primers utilized in this study.
Acknowledgments
This work was supported by National Science Foundation grants MCB-1616775 and MCB-2313900 to S.M. Di Pietro.
Author contributions: Conceptualization: A.K. Lamb and S.M. Di Pietro; data curation: A.K. Lamb and S.M. Di Pietro; data curation: A.K. Lamb and S.M. Di Pietro; formal analysis: A.K. Lamb and A.N. Fernandez; funding acquisition: S.M. Di Pietro; investigation: A.K. Lamb, A.N. Fernandez, A. Eadaim, K. Johnson, and S.M. Di Pietro; methodology: A.K. Lamb; project administration: S.M. Di Pietro; resources: S.M. Di Pietro; supervision: A.K. Lamb and S.M. Di Pietro; validation: A.K. Lamb, A.N. Fernandez, and S.M. Di Pietro; visualization: A.K. Lamb and S.M. Di Pietro; writing-original draft preparation: A.K. Lamb and S.M. Di Pietro; writing-review and editing: A.K. Lamb, A.N. Fernandez, and S.M. Di Pietro.
References
Author notes
Disclosures: The authors declare no competing interests exist.
A.N. Fernandez’s current affiliation is Basic Science Division, Fred Hutchinson Cancer Center, Seattle, WA, USA.