Autophagy is a catabolic pathway required for the recycling of cytoplasmic materials. To define the mechanisms underlying autophagy it is critical to quantitatively characterize the dynamic behavior of autophagy factors in living cells. Using a panel of cell lines expressing HaloTagged autophagy factors from their endogenous loci, we analyzed the abundance, single-molecule dynamics, and autophagosome association kinetics of autophagy proteins involved in autophagosome biogenesis. We demonstrate that autophagosome formation is inefficient and ATG2-mediated tethering to donor membranes is a key commitment step in autophagosome formation. Furthermore, our observations support the model that phagophores are initiated by the accumulation of autophagy factors on mobile ATG9 vesicles, and that the ULK1 complex and PI3-kinase form a positive feedback loop required for autophagosome formation. Finally, we demonstrate that the duration of autophagosome biogenesis is ∼110 s. In total, our work provides quantitative insight into autophagosome biogenesis and establishes an experimental framework to analyze autophagy in human cells.
Introduction
Autophagy is a conserved catabolic process that recycles damaged organelles and protein aggregates or non-specifically degrades cellular material to provide nutrients for cell proliferation, particularly when cells face chemical stress or nutrient starvation (White, 2015; Yu et al., 2018). The hallmark of autophagy is the formation of double-membrane autophagosomes, which sequester cargo and then fuse with lysosomes to trigger the degradation of their contents. Alterations of autophagy have been implicated in the pathology of several human diseases. During the aging of human cells, key autophagy factors are reduced in abundance resulting in the downregulation of autophagic flux, which increases the susceptibility to the two most common neurodegenerative disorders, Alzheimer’s and Parkinson’s disease (Filippone et al., 2022; Lu et al., 2020; Nixon, 2007; Tecalco-Cruz et al., 2022). In contrast, the upregulation of autophagy has been shown to promote a variety of cancers by providing nutrients for rapid tumor proliferation (Gewirtz, 2014). The life cycle of autophagosomes encompasses four distinct steps: phagophore initiation, expansion, and closure forming a mature autophagosome, followed by fusion of the autophagosome to the lysosome leading to its degradation. Autophagosome formation can be initiated non-specifically (non-selective autophagy) or by a target, for instance, a damaged organelle that requires degradation (Kirkin, 2020; Lamb et al., 2013). Non-selective autophagy is induced under starvation conditions or by chemical stress (Lamb et al., 2013). Initially, it was suggested that specific regions of the ER are remodeled into the phagophore (Ge et al., 2017). An alternative hypothesis suggests that the phagophore is formed by ATG9-containing vesicles, which expand to form a mature autophagosome (Chang et al., 2021a; Chang et al., 2021b; Olivas et al., 2022,Preprint; Sawa-Makarska et al., 2020; Tang et al., 2019). ATG9 is the only known autophagy-related protein that contains a transmembrane domain and has recently been shown to have lipid scramblase activity, which facilitates the exchange of phospholipids between the outer to the inner leaflets of the membrane it is embedded in (Ghanbarpour et al., 2021; Maeda et al., 2020; Matoba et al., 2020; Matoba and Noda, 2020; Noda, 2021; Yamamoto et al., 2012). For this reason, ATG9 vesicles are prime candidates for the origin of the phagophore; as ATG2 transfers lipids to the outer leaflets, ATG9 equilibrates lipids across the membrane, growing a vesicle into an autophagosome (Maeda et al., 2019; Maeda et al., 2020; Matoba et al., 2020; Matoba and Noda, 2020; Noda, 2021; Osawa et al., 2019; Valverde et al., 2019). Regardless of the membrane structure autophagosomes originate from, non-selective autophagy is initiated through a complex phosphorylation cascade by the Unc-51-like autophagy activating kinase (ULK1/2) complex. Besides ULK1/2 isoforms, the Ulk1-kinase complex is composed of FIP200, ATG13, and ATG101 (Ganley et al., 2009; Mercer et al., 2009; Shi et al., 2020). The ULK1 substrates include many downstream autophagy factors, and these phosphorylation sites serve as key switches for autophagy protein recruitment to the phagophore (Ganley et al., 2009; Mercer et al., 2018; Mizushima, 2010). One essential step that drives autophagosome biogenesis is the activation of the PI3K complex at the phagophore, which modifies phospho-inositol lipids leading to a local enrichment of PI3P (Mizushima, 2010). The presence of PI3P is sensed by the WIPI3/4 proteins, which subsequently recruit the lipid transferase ATG2 to the growing phagophore (Chowdhury et al., 2018; Dooley et al., 2014; Otomo et al., 2018). In addition to ATG2, WIPI1-4 also recruits the ATG5–ATG12–ATG16 complex (Fracchiolla et al., 2020; Lystad et al., 2019), which acts similarly to ubiquitin ligase complexes but instead conjugates ATG8 family (e.g., LC3 and GABARAP) proteins to phosphatidylethanolamine at the phagophore membrane (Hanada et al., 2007; Kirisako et al., 1999; Shpilka et al., 2011). Once conjugated to the phagophore membrane, LC3 and GABARAP proteins serve as anchors to tether cargo targeted for degradation to the autophagosome (Schaaf et al., 2016). Finally, when expansion and cargo sequestration are complete, the phagophore closes into the double-membrane autophagosome and fuses with the lysosome to trigger the degradation of its contents (Berg et al., 1998; Nakamura and Yoshimori, 2017).
In this study, we establish a collection of genome-edited cell lines that express endogenous HaloTagged autophagy proteins involved in autophagosome biogenesis. This approach maintains the expression levels of the tagged autophagy factors and retains all regulatory mechanisms conferred by the endogenous genomic locus of the respective gene and avoids potential artifacts caused by transgene overexpression. The HaloTag is a versatile protein tag that can be covalently linked to cell-permeable ligands, which facilitates fluorescent labeling, targeted protein degradation, and pulse-chase experiments. Using our collection of cell lines, we systematically quantify the absolute abundance, single-molecule diffusion dynamics, and the recruitment kinetics of these autophagy proteins to the phagophore. The results show that ULK1 and ATG2A are maintained at low expression levels, potentially to prevent uncontrolled autophagosome production, and are key signals in committing the phagophore into developing into an autophagosome. Our live-cell single-molecule imaging experiments reveal that the initiation of autophagosome formation is locally controlled, rather than by cell-wide changes in signaling. Furthermore, we demonstrate that ATG9A is the only autophagy factor that is not locally enriched at sites of autophagosome formation, suggesting that a limited number of ATG9A molecules, potentially those contained in a single ATG9 vesicle, are required in autophagosome formation. Our systematic analysis of autophagy factor foci kinetics reveals that the average duration of autophagosome biogenesis is ∼110 s and that most phagophores do not progress to form mature autophagosomes. In addition, we observe two classes of phagophore foci with distinct diffusion dynamics. Rapidly moving pre-phagophoresare likely mobile ATG9A vesicles that have begun to recruit autophagy factors and phagophores that move more slowly as a result of ATG2A-mediated tethering to lipid donor membranes. Importantly, we show that ATG9A accumulates within the lysosome over time, where it is degraded, suggesting that ATG9A integration into the autophagosome is required for autophagosome expansion. In total, our work supports a model in which the phagophore is initiated by the accumulation of autophagy proteins on mobile ATG9A-containing vesicles that proceed to be tethered to lipid donor membranes by ATG2A, committing the phagophore to maturation into an autophagosome. In addition, we establish a sophisticated experimental framework for the quantitative analysis of autophagy in human cells.
Results
Genomic insertion of HaloTag at endogenous loci of autophagy proteins
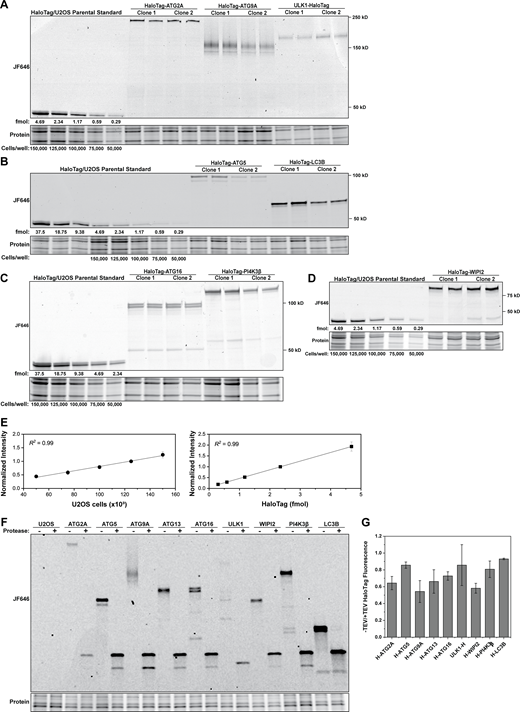
To quantitatively analyze autophagosome formation in human cancer cells (U2OS), we used CRISPR-Cas9-mediated genome editing to introduce the HaloTag into the endogenous loci of autophagy factors that are involved in the phagophore initiation (ULK1, ATG13, PI4K3β), lipid transfer into the growing phagophore (ATG2A and ATG9A), and LC3 conjugation (WIPI2, ATG16, ATG5, LC3; Fig. 1 A). Homozygous insertion of the HaloTag was confirmed by PCR and Sanger sequencing (Fig. S1, A and B), and exclusive expression of the HaloTagged autophagy proteins was validated by Western blot and fluorescent labeling (Fig. 1, B and C). Due to their sequence complexity, we were unable to confirm homozygous editing of WIPI2 and ULK1 loci using PCR amplification. Instead, we validated the specificity of integration and exclusive expression of HaloTagged WIPI2 and ULK1 protein (Fig. S1 C and Fig. 1 B). We attempted to tag DFCP1 and subunits of the PI3K complex, including Beclin-1 and VPS34, but we failed to generate clonal cell lines that exclusively expressed the HaloTagged proteins.
A HaloTag-based platform for quantitative analysis of autophagy in human cells. (A) Model showing autophagy factors and the complexes they form from phagophore initiation, toward phagophore expansion and autophagosome closure. Proteins tagged in this study are indicated in pink. (B) Western blots of autophagy proteins showing size shift of the tagged protein and exclusive expression of the tagged protein in comparison to the parental U2OS cell line. Three concentrations (100, 50, and 25% of initial lysis volume) were loaded on the gel. (C) Fluorescence gel showing gene tagging; two distinct monoclonal lines (C1 and C2) were selected for each edited gene. Cell lines were labeled with saturating amounts of HaloTag ligand JF646 (250 nM, 30 min). Source data are available for this figure: SourceData F1.
A HaloTag-based platform for quantitative analysis of autophagy in human cells. (A) Model showing autophagy factors and the complexes they form from phagophore initiation, toward phagophore expansion and autophagosome closure. Proteins tagged in this study are indicated in pink. (B) Western blots of autophagy proteins showing size shift of the tagged protein and exclusive expression of the tagged protein in comparison to the parental U2OS cell line. Three concentrations (100, 50, and 25% of initial lysis volume) were loaded on the gel. (C) Fluorescence gel showing gene tagging; two distinct monoclonal lines (C1 and C2) were selected for each edited gene. Cell lines were labeled with saturating amounts of HaloTag ligand JF646 (250 nM, 30 min). Source data are available for this figure: SourceData F1.
Validation and functional characterization of genome-edited clones expressing HaloTagged autophagy factors. (A and B) PCR analysis from genomic DNA of genome-edited clones for verifying the correct insertion of the HaloTag at the autophagy loci. For amplifying the insertion, primers outside the homology arms region were designed. The edited clones show an expected shift of ∼2 kb on the PCR product, corresponding to the 3xFlag-HaloTag insert. (C) PCR analysis from genomic DNA of genome-edited clones for verifying the correct insertion of the HaloTag in the high GC-rich ULK1 and WIPI2 gene loci. For amplifying the insertion, primers outside the homology arms and inside the 3xFlag-HaloTag regions were designed. The edited clones show a PCR product, which is absent in the parental U2OS cell line. (D) Western blots for determining the expression levels of the HaloTagged autophagy proteins relative to the wildtype protein before and after removal of HaloTag using the TEV protease. (E) Quantification of the Western blots (A), showing the ratio between HaloTag and parental cell line (N = 3, mean ± SD). (F) Western blot analysis of LC3 levels in parental and genome-edited cell lines in control (Ctr) and upon treatment with rapamycin (R, 100 nM for 2 h), or rapamycin + bafilomycin (R+B, 100 nM each, for 2 h). Source data are available for this figure: SourceData FS1.
Validation and functional characterization of genome-edited clones expressing HaloTagged autophagy factors. (A and B) PCR analysis from genomic DNA of genome-edited clones for verifying the correct insertion of the HaloTag at the autophagy loci. For amplifying the insertion, primers outside the homology arms region were designed. The edited clones show an expected shift of ∼2 kb on the PCR product, corresponding to the 3xFlag-HaloTag insert. (C) PCR analysis from genomic DNA of genome-edited clones for verifying the correct insertion of the HaloTag in the high GC-rich ULK1 and WIPI2 gene loci. For amplifying the insertion, primers outside the homology arms and inside the 3xFlag-HaloTag regions were designed. The edited clones show a PCR product, which is absent in the parental U2OS cell line. (D) Western blots for determining the expression levels of the HaloTagged autophagy proteins relative to the wildtype protein before and after removal of HaloTag using the TEV protease. (E) Quantification of the Western blots (A), showing the ratio between HaloTag and parental cell line (N = 3, mean ± SD). (F) Western blot analysis of LC3 levels in parental and genome-edited cell lines in control (Ctr) and upon treatment with rapamycin (R, 100 nM for 2 h), or rapamycin + bafilomycin (R+B, 100 nM each, for 2 h). Source data are available for this figure: SourceData FS1.
To determine the expression levels of the HaloTagged autophagy proteins relative to the wildtype protein using Western blots, we lysed cells and removed the HaloTag using the Tobacco Etch Virus (TEV) protease before gel electrophoresis. This approach avoids artifacts that we observed caused by the HaloTag affecting Western blot transfer or antibody detection of the autophagy proteins (Fig. S1, D and E). The majority of HaloTagged autophagy factors were expressed at similar levels to their untagged counterparts (Fig. S1, D and E). ULK1-Halo was approximately fourfold overexpressed and Halo-WIPI2 expression was reduced by ∼50% (Fig. S1, D and E). Due to the sequence complexity of the WIPI2 locus, likely not all the WIPI2 alleles were modified causing a reduction in gene dosage. Importantly, Halo-LC3 appeared to be more abundant than wildtype LC3 but the removal of the HaloTag by TEV cleavage eliminated the difference in the Western blot signal (Fig. S1, D and E). This demonstrates that the HaloTag can have a significant impact on the detection of proteins by Western blot. This difference is likely caused by an alteration in Western blot transfer efficiency.
Altogether these observations demonstrate that we have successfully generated a collection of cell lines expressing HaloTagged autophagy proteins from their endogenous loci at or near the levels of their wildtype counterparts.
The HaloTagged autophagy proteins are functional
Our quantitative Western blots shown above demonstrate that our cell lines exclusively express HaloTagged autophagy proteins and allow us to evaluate whether fusion to the HaloTag affected their function in autophagy. To test the functionality of the HaloTagged autophagy proteins, we determined the efficiency of autophagosome formation and degradation by measuring the levels of membrane-conjugated LC3 after autophagy induction by rapamycin with and without the lysosome inhibitor bafilomycin, which prevents autophagosome degradation (Barth et al., 2010; Yamamoto et al., 1998). LC3 Western blots showed that apart from HaloTagged ATG5, where we observed a minor conjugation defect, all cell lines expressing HaloTagged autophagy factors conjugated LC3 to a similar degree as the parental U2OS cells (Fig. S1 F). To verify the colocalization of tagged autophagy proteins with a well-established autophagosome marker, cells were incubated with fluorescent HaloTag-ligand (JF646) alongside baculovirus-mediated transient expression of GFP-LC3. Surprisingly, GFP-LC3 did not form foci indicative of autophagosome formation when combined with N-terminal tags on the conjugation machinery including ATG5, ATG16, and WIPI2 (Fig. S2 A). All the other tagged proteins formed puncta that colocalized with GFP-LC3 foci and responded similarly to the parental cell line when treated with rapamycin and bafilomycin (Fig. S2 A).
Autophagy induced foci formation by HaloTagged autophagy proteins. (A) Live-cell imaging of GFP-LC3 and JF646-labeled HaloTagged proteins in control (Ctr) and upon treatment with rapamycin (Rapa, 100 nM for 2 h) or rapamycin + bafilomycin (Rapa+Baf, 100 nM each, for 2 h). Both treatments show an expected increase in autophagy and LC3 foci. Scale bar = 10 μm. (B) IF with anti-LC3B antibody and HaloTag JF646 labeling for Halo-ATG5, Halo-ATG16, and Halo-WIPI2 cell lines in control (Ctr) and upon treatment with rapamycin (Rapa, 100 nM for 2 h) or rapamycin + bafilomycin (Rapa+Baf, 100 nM each, for 2 h). Both treatments show an expected increase in autophagy and LC3 foci. Scale bar = 10 μm. (C) Representative images of JF646-labeled HaloTagged proteins under control (Ctr) or EBSS starvation media, demonstrating foci-forming ability upon autophagy induction. Scale bar = 10 μm. (D) Quantification of autophagy factor foci from live-cell imaging (A). Data represent the mean ± 1SD of three biological replicates. (E) Quantification of LC3 foci from live-cell imaging (A; N = 3, mean ± SD). (F) Quantification of autophagy factor foci from live-cell imaging (C; N = 3, mean ± SD).
Autophagy induced foci formation by HaloTagged autophagy proteins. (A) Live-cell imaging of GFP-LC3 and JF646-labeled HaloTagged proteins in control (Ctr) and upon treatment with rapamycin (Rapa, 100 nM for 2 h) or rapamycin + bafilomycin (Rapa+Baf, 100 nM each, for 2 h). Both treatments show an expected increase in autophagy and LC3 foci. Scale bar = 10 μm. (B) IF with anti-LC3B antibody and HaloTag JF646 labeling for Halo-ATG5, Halo-ATG16, and Halo-WIPI2 cell lines in control (Ctr) and upon treatment with rapamycin (Rapa, 100 nM for 2 h) or rapamycin + bafilomycin (Rapa+Baf, 100 nM each, for 2 h). Both treatments show an expected increase in autophagy and LC3 foci. Scale bar = 10 μm. (C) Representative images of JF646-labeled HaloTagged proteins under control (Ctr) or EBSS starvation media, demonstrating foci-forming ability upon autophagy induction. Scale bar = 10 μm. (D) Quantification of autophagy factor foci from live-cell imaging (A). Data represent the mean ± 1SD of three biological replicates. (E) Quantification of LC3 foci from live-cell imaging (A; N = 3, mean ± SD). (F) Quantification of autophagy factor foci from live-cell imaging (C; N = 3, mean ± SD).
Since we did not observe any defects in LC3 conjugation by Western blot in Halo-ATG16 and Halo-WIPI2 cells, we hypothesized that the HaloTag sterically interferes with the conjugation of GFP-tagged LC3. To test this hypothesis, we analyzed LC3 foci formation by immunofluorescence (IF) instead of GFP-LC3. Using IF, we detected similar numbers of LC3 foci within the Halo-ATG5, Halo-ATG16, and Halo-WIPI2 cell lines compared with parental U2OS cells, confirming that these HaloTagged autophagy factors can fully support endogenous LC3 conjugation (Fig. S2 B). The formation of foci in response to autophagy induction is a characteristic of autophagy proteins (Karanasios et al., 2013a). In contrast to the LC3 foci number, which increased in response to rapamycin treatment (Fig. S2 E), we did not observe increases in the number of foci formed by other HaloTagged autophagy proteins. HaloTagged ATG2A, ATG13, ULK1, and LC3 foci number was only slightly increased in rapamycin-treated samples while HaloTagged ATG5, ATG9A, and ATG16 showed no increase in foci formation (Fig. S2 D). These observations suggested that inhibition of mTOR with rapamycin did not activate autophagy sufficiently to cause an increase in foci formation in our endogenously tagged cell lines.
To induce autophagy using a more robust approach, we treated cells with Earl’s Balanced Salt Solution (EBSS), which triggers autophagy primarily by amino acid starvation. After treatment with EBSS, the number of foci formed by all HaloTagged autophagy factors was increased compared with cells grown in complete media (Fig. S2 C). Strikingly, while the number of Halo-WIPI2 and ULK1-Halo foci did not change after rapamycin treatment, amino acid starvation using EBSS lead to a fourfold increase in the number of ULK1-Halo and Halo-WIPI2 foci. When imaging Halo-PI4K, we were unable to identify discreet foci but observed an accumulation of Halo-PI4K in the perinuclear region of the cytoplasm (Fig. S2, C and F). This result differed from previous imaging experiments using stable expression cell lines to visualize PI4K (Judith et al., 2019). Notwithstanding, we observed a perinuclear redistribution of Halo-PI4K upon both rapamycin and amino acid starvation, which agrees with previous findings (Judith et al., 2019).
In total, these results demonstrate that the tagged autophagy factors are functional and support autophagosome formation. Only Halo-ATG5 showed a minor defect in the overall level of LC3 conjugation (Fig. S1 F).
Quantification of the absolute abundance of autophagy proteins
To determine the absolute number of protein molecules per cell for each tagged autophagy factor, we expanded upon an established in-gel fluorescence method to quantify the number of HaloTagged molecules per cell (Fig. 2 A; Cattoglio et al., 2019). To avoid the recombinant expression of each HaloTagged autophagy protein to use as a quantification standard, we supplemented U2OS cell lysates of a known number of cells with purified, recombinantly expressed 3xFLAG-HaloTag protein (Fig. 2 A and Fig. S3, A–D). Standard curves for the cell number using total protein levels and HaloTag-fluorescence signal were reproducible across all experiments with R2 values of 0.99 (Fig. S3 E). To account for biases introduced by the banding patterns in fluorescence gels and differences in transfer efficiencies of HaloTagged proteins, we cleaved the fusion proteins using TEV protease. This approach allowed us to calculate correction factors by comparing the signal of the cleaved HaloTag (fluorescence signal) and autophagy factor (Western blot signal) to the HaloTag standard and endogenous autophagy protein signal, respectively (Fig. S1 D and Fig. S3, F and G). The in-gel fluorescence method allowed the quantification of autophagy protein abundance in U2OS cells over a broad range, from low expressed factors (ULK1, ∼3,000 proteins/cell; ATG2A, ∼8,000 proteins/cell) to the highly expressed LC3 (>100,000 proteins/cell). ATG16 and ATG5, which form a complex, were present in a 1:1 ratio, consistent with their constitutive association. WIPI2, a scaffold protein that recruits ATG16-12-5 to the initiation membrane, was present in approximately fourfold excess relative to ATG16. Surprisingly, ATG13 exceeded the abundance of ULK1 ninefold (Fig. 2 C), considering that both proteins are part of a larger kinase complex (the ULK1 complex; Mizushima, 2010). As an orthogonal approach, we determined the relative abundance of HaloTagged proteins in our collection of cell lines using flow cytometry and converted the relative fluorescence into an absolute protein number using Halo-ATG9A C1 as a fiducial point. The absolute protein abundance determined with flow cytometry and in-gel fluorescence were in good agreement, with some differences in the range of the technical error (Fig. 2 D). Collectively, we have determined the absolute protein abundance of key autophagy factors and identified unexpected ratios between the subunits of the ULK1 complex. ULK1 and ATG2A were expressed at substantially lower levels than other autophagy proteins and this may contribute to key regulatory steps that control autophagic flux.
Absolute protein abundance quantification of autophagy factors in human cells. (A) Example in-gel fluorescence containing the quantification standards (HaloTag + cell lysate) and ATG13 protein. (B) Histogram of flow cytometry measurements depicting the relative protein abundances of U2OS (negative control), and two clones of cells expressing Halo-ATG2A and Halo-WIPI2. (C) Corrected protein abundance quantification of the tagged autophagy proteins with in-gel fluorescence and flow cytometry (N = 3, mean ± SD including error propagation). (D) Graph showing the correlation between protein abundance measured by flow cytometry compared to in-gel fluorescence. Source data are available for this figure: SourceData F2.
Absolute protein abundance quantification of autophagy factors in human cells. (A) Example in-gel fluorescence containing the quantification standards (HaloTag + cell lysate) and ATG13 protein. (B) Histogram of flow cytometry measurements depicting the relative protein abundances of U2OS (negative control), and two clones of cells expressing Halo-ATG2A and Halo-WIPI2. (C) Corrected protein abundance quantification of the tagged autophagy proteins with in-gel fluorescence and flow cytometry (N = 3, mean ± SD including error propagation). (D) Graph showing the correlation between protein abundance measured by flow cytometry compared to in-gel fluorescence. Source data are available for this figure: SourceData F2.
Absolute protein abundance quantification of HaloTagged autophagy factors. (A–D) Representative fluorescence gels for the absolute quantification of the autophagy proteins. (E) Standard curve for cell number using stain-free gels (left) and florescent HaloTag protein (right), demonstrating an excellent correlation between intensity and gel loadings. (F) Representative fluorescence gel of HaloTagged proteins in the absence or presence of TEV protease. (G) Ratio of HaloTag fluorescence in the absence and presence of TEV protease (B; N = 3, mean ± SD). Source data are available for this figure: SourceData FS3.
Absolute protein abundance quantification of HaloTagged autophagy factors. (A–D) Representative fluorescence gels for the absolute quantification of the autophagy proteins. (E) Standard curve for cell number using stain-free gels (left) and florescent HaloTag protein (right), demonstrating an excellent correlation between intensity and gel loadings. (F) Representative fluorescence gel of HaloTagged proteins in the absence or presence of TEV protease. (G) Ratio of HaloTag fluorescence in the absence and presence of TEV protease (B; N = 3, mean ± SD). Source data are available for this figure: SourceData FS3.
Single-molecule analysis of the subcellular dynamics of autophagy factors
The dynamic recruitment of autophagy proteins to the sites of autophagosome formation is critical to controlling overall autophagic flux. Autophagosome formation can initiate at cargo-dependent sites such as mitophagy (Dalle Pezze et al., 2021). Alternatively, under starvation conditions, autophagosomes non-specifically engulf cellular material (Lamb et al., 2013). In either case, it is not known how autophagy factors encounter sites of autophagosome formation. Potential mechanisms include 3D diffusion or scanning of existing membrane structures. To define the subcellular distribution of autophagy proteins and the mechanism by which they are recruited to the sites of autophagosome formation, we performed single-molecule live-cell imaging and single-particle tracking of the HaloTagged autophagy factors. Analysis of single-particle trajectories using the SpotOn tool allowed us to define distinct mobility states for each autophagy protein imaged (Fig. 3, A and B; and Videos 1, 2, 3, 4, 5, 6, 7, 8, and 9). In principle, autophagy proteins could exist in three different states: freely diffusing in the cytoplasm, scanning a membrane (through intrinsic phospholipid binding, binding to a protein with intrinsic phospholipid binding, or interacting with transmembrane proteins), or statically bound to a site of autophagosome formation. Lipid vesicles, including ATG9A-containing vesicles, are expected to have diffusion properties closer to membrane-interacting proteins (Rothman et al., 2016; Staufer et al., 2022). To approximate the diffusion parameters for each of these states, we analyzed a HaloTag protein fused to a nuclear export signal (Halo-NES, Video 10), to model freely diffusing proteins, and SNAP-SEC61, as a model membrane-bound protein (Video 10). The Halo-NES diffused rapidly (Dfree = 15 µm2/s, Ffree = 99%) and had a negligible static fraction (Fstatic = 1%; Fig. 3 C). SEC61 particles either slowly diffused (Dslow = 0.75 µm2/s, Fslow = 65%) or were static (Dstatic = 0.08 µm2/s, Fstatic = 35%; Fig. 3 C), which likely represents SEC61 molecules moving freely within the ER membrane or SEC61 molecules that are part of a translocon actively engaged with a ribosome in the process of translation, respectively.
Live-cell single-molecule analysis of autophagy proteins. (A and B) Example of single-particle tracking of (A) ATG9 and (B) ATG16, and the corresponding fitting of the step-size probability distribution with SpotON algorithm. Numbers inside the micrographs indicate the imaging frame associated with the track. Movies were acquired at 6.8 ms per frame, scale bar = 1 μm. (C) Results of diffusive analysis for the HaloTagged autophagy proteins under control and EBSS starvation. Top three panels present the diffusion coefficients of the tracks based on the SpotON analysis. Bottom panel depicts the percentage associated with each fraction. The box indicates confidence interval ± SD, the square indicates the average, and the horizontal line is the median; for each condition, three biological replicates were analyzed, ∼20 cells/replicate.
Live-cell single-molecule analysis of autophagy proteins. (A and B) Example of single-particle tracking of (A) ATG9 and (B) ATG16, and the corresponding fitting of the step-size probability distribution with SpotON algorithm. Numbers inside the micrographs indicate the imaging frame associated with the track. Movies were acquired at 6.8 ms per frame, scale bar = 1 μm. (C) Results of diffusive analysis for the HaloTagged autophagy proteins under control and EBSS starvation. Top three panels present the diffusion coefficients of the tracks based on the SpotON analysis. Bottom panel depicts the percentage associated with each fraction. The box indicates confidence interval ± SD, the square indicates the average, and the horizontal line is the median; for each condition, three biological replicates were analyzed, ∼20 cells/replicate.
Representative live-cell single-molecule imaging movies of control and EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG2A labeled with JFX650. Images were acquired at 146 frames per second. 140 × 140 pixels with a pixel size of 0.16 μm.
Representative live-cell single-molecule imaging movies of control and EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG2A labeled with JFX650. Images were acquired at 146 frames per second. 140 × 140 pixels with a pixel size of 0.16 μm.
Representative live-cell single-molecule imaging movies of control and EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG5 labeled with JFX650. Images were acquired at 146 frames per second. 140 × 140 pixels with a pixel size of 0.16 μm.
Representative live-cell single-molecule imaging movies of control and EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG5 labeled with JFX650. Images were acquired at 146 frames per second. 140 × 140 pixels with a pixel size of 0.16 μm.
Representative live-cell single-molecule imaging movies of control and EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG9A labeled with JFX650. Images were acquired at 146 frames per second. 140 × 140 pixels with a pixel size of 0.16 μm.
Representative live-cell single-molecule imaging movies of control and EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG9A labeled with JFX650. Images were acquired at 146 frames per second. 140 × 140 pixels with a pixel size of 0.16 μm.
Representative live-cell single-molecule imaging movies of control and EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG13 labeled with JFX650. Images were acquired at 146 frames per second. 140 × 140 pixels with a pixel size of 0.16 μm.
Representative live-cell single-molecule imaging movies of control and EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG13 labeled with JFX650. Images were acquired at 146 frames per second. 140 × 140 pixels with a pixel size of 0.16 μm.
Representative live-cell single-molecule imaging movies of control and EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG16 labeled with JFX650. Images were acquired at 146 frames per second. 140 × 140 pixels with a pixel size of 0.16 μm.
Representative live-cell single-molecule imaging movies of control and EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG16 labeled with JFX650. Images were acquired at 146 frames per second. 140 × 140 pixels with a pixel size of 0.16 μm.
Representative live-cell single-molecule imaging movies of control and EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ULK1 labeled with JFX650. Images were acquired at 146 frames per second. 140 × 140 pixels with a pixel size of 0.16 μm.
Representative live-cell single-molecule imaging movies of control and EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ULK1 labeled with JFX650. Images were acquired at 146 frames per second. 140 × 140 pixels with a pixel size of 0.16 μm.
Representative live-cell single-molecule imaging movies of control and EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged WIPI2 labeled with JFX650. Images were acquired at 146 frames per second. 140 × 140 pixels with a pixel size of 0.16 μm.
Representative live-cell single-molecule imaging movies of control and EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged WIPI2 labeled with JFX650. Images were acquired at 146 frames per second. 140 × 140 pixels with a pixel size of 0.16 μm.
Representative live-cell single-molecule imaging movies of control and EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged LC3B labeled with JFX650. Images were acquired at 146 frames per second. 140 × 140 pixels with a pixel size of 0.16 μm.
Representative live-cell single-molecule imaging movies of control and EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged LC3B labeled with JFX650. Images were acquired at 146 frames per second. 140 × 140 pixels with a pixel size of 0.16 μm.
Representative live-cell single-molecule imaging movies of control and EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged PI4K3β labeled with JFX650. Images were acquired at 146 frames per second. 140 × 140 pixels with a pixel size of 0.16 μm.
Representative live-cell single-molecule imaging movies of control and EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged PI4K3β labeled with JFX650. Images were acquired at 146 frames per second. 140 × 140 pixels with a pixel size of 0.16 μm.
Representative live-cell single-molecule imaging movies of control and EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged NES labeled with JFX650 and SNAPTagged SEC61B labeled with JF650. Images were acquired at 146 frames per second. 140 × 140 pixels with a pixel size of 0.16 μm.
Representative live-cell single-molecule imaging movies of control and EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged NES labeled with JFX650 and SNAPTagged SEC61B labeled with JF650. Images were acquired at 146 frames per second. 140 × 140 pixels with a pixel size of 0.16 μm.
To define the diffusion properties of autophagy proteins, cells were imaged under control (control) and starved (EBSS treated) conditions. Except for ATG9A, we used the three-state model described above (freely diffusing, membrane scanning, and statically bound to an autophagosome or other membrane) to fit the step size distributions of all autophagy proteins (Fig. 3 B). Since ATG9A, like SEC61, is a transmembrane protein and resides in lipid vesicles, it does not freely diffuse through the cytoplasm. Diffusion of lipid vesicles within the cytoplasm is characterized by smaller diffusion coefficients than cytosolic proteins (Yamamoto et al., 2012). Lipid vesicles interacting with membrane organelles (i.e., autophagosomes interacting with the ER) are expected to be statically bound. Therefore, a two-state model better describes the expected diffusion properties of ATG9A vesicles and was used to fit the step size distribution of ATG9A trajectories. The step-size distribution for all autophagy factors fits well with a three-state model (or two-state model in the case of ATG9A, Fig. 3 A). For all other autophagy factors analyzed, a large fraction of the particles was freely diffusing (Dfree = 4–6 µm2/s, Ffree = 46–85%; Fig. 3 C). In addition, a significant fraction of molecules for all proteins analyzed moved with a diffusion coefficient comparable with SEC61 (Dslow = 0.5–0.75 µm2/s, Fslow = 12–35%; Fig. 3 C), consistent with these particles representing a membrane-associated population of the autophagy factors. Finally, a small fraction of molecules of all factors imaged were static (Dstatic = 0.02–0.08 µm2/s, Fstatic = 4–22%; Fig. 3 C). Approximately half of the ATG9A molecules diffused slowly (Dslow = 0.3 µm2/s, Fslow = 50%), while the other half of ATG9A particles were static (Dstatic = 0.02 µm2/s, Fstatic = 50%; Fig. 3 C). The diffusion properties of ATG5 and ATG16 were comparable and consistent with ATG5 and ATG16 forming a constitutive complex (Fig. 3 C). Interestingly, the diffusion properties of ATG13 and ULK1 were distinct (Fig. 3 C). ATG13 had a higher fraction of freely diffusing molecules than ULK1 (76% vs. 59%), and the diffusion coefficient for this fraction was significantly higher for ATG13 compared with ULK1 (4.7 µm2/s vs. 3.6 µm2/s, P < 0.001; Fig. 3 C). Together, this suggests that a substantial fraction of ATG13 is not associated with ULK1, which is consistent with our observation that the abundance of ATG13 exceeds the amount of ULK1 by approximately ninefold. Freely diffusing PI4K3β particles displayed the slowest diffusion coefficient of all proteins analyzed, which could be the consequence of transient interactions formed with endosomes and other Golgi-derived organelles (Judith et al., 2019; Waugh, 2019). Finally, more than 50% of LC3 molecules were in the bound or static state. LC3 exists in two primary forms, a lipid-conjugated form (LC3-II) inserted into autophagic membranes and a cytosolic non-conjugated form (LC3-I; Kabeya et al., 2004). Strikingly, the diffusion dynamics of none of the autophagy factors studied significantly changed after exposing cells to starvation conditions (Fig. 3 C). This demonstrates that the dynamic properties of none of the factors studied are globally changed by cell starvation. In addition, the observation that the static populations remain unchanged under starvation conditions shows that only a very small fraction of molecules of a given autophagy factor are actively involved in autophagosome formation. Taken together, our single-molecule analysis of the diffusion dynamics of the tagged autophagy factors suggests that all proteins analyzed exist in a freely diffusing and membrane-associated state. In addition, our data demonstrate that the diffusion properties of the tagged autophagy proteins do not globally change in starvation conditions, indicating that only a small fraction of these proteins actively participates in autophagosome formation.
Quantitative analysis of the autophagosome formation reveals two distinct populations of autophagosomes
We next sought to quantitatively analyze the recruitment of the tagged autophagy factors to autophagosomes, which was not possible using the single-molecule approach described above. The local accumulation of autophagy proteins at sites of autophagosome formation can be visualized as bright cytoplasmic foci and overall autophagic flux can be measured by determining the rate of foci formation using time-lapse microscopy (Dalle Pezze et al., 2021; Itakura and Mizushima, 2010). Autophagy protein foci were automatically identified and tracked using single-particle tracking (Fig. 4, A and B; and Videos 11, 12, 13, 14, 15, 16, 17, and 18; Kuhn et al., 2021). Under control conditions, ULK1, ATG13, ATG5, and ATG16 formed significantly more foci (∼30–50 foci per cell per hour) compared with ATG2A and WIPI2, which formed a limited number of foci in control cells (approximately nine foci per cell per hour, Fig. 4, C and D). Nutrient starvation significantly increased the number of foci formed by all autophagy factors imaged (Fig. 4, C and D). Interestingly, the number of foci formed per cell per hour under starvation conditions was comparable (∼120–150) for ATG5, ATG13, ATG16, and ULK1, which was approximately twofold higher than the number of foci formed by ATG2A (∼60 foci per cell per hour; Fig. 4, C and D). WIPI2 formed an intermediate number of foci (∼90 foci per cell per hour; Fig. 4, C and D). This suggests that ATG2A is only detectably recruited to a subset of phagophores.
High-throughput quantification of autophagy factor foci lifetime and diffusion dynamics. (A) Upon labeling with fluorescent dye (JF646), cells expressing HaloTagged autophagy factors were starved (EBSS) and imaged at four frames per minute for 1 h. TrackIT was used to detect foci based on threshold intensity (left) and connected into tracks using the nearest neighbor algorithm (right). Scale bar = 5 μm. (B) Example images of foci for Halo-ATG2A (upper panel) and Halo-ATG13 (bottom panel). Scale bar = 1 μm. (C) Histograms of foci lifetime for the HaloTagged autophagy proteins in control conditions (Control) and after 1 h nutrient starvation (EBSS). Three biological replicates (20–30 cells per replicate) were performed for each HaloTag cell line. The number of data points (n) is indicated in each graph in the figure panels. (D) Quantification of the number of foci formed per cell by autophagy factors over the course of 1 h imaging in control (Control) and nutrient starvation (EBSS) conditions (N = 3 biological replicates, mean ± SD). A two-tailed t test was used for statistical analysis (*P < 0.05). (E) Histograms of diffusion coefficients of the foci formed by autophagy factors under nutrient starvation. Histograms were fitted with Gaussian curves. For the proteins other than Halo-ATG2A, we fixed the mean of one subpopulation (in red) to match Halo-ATG2A mean. An additional subpopulation (in purple) represents non-ATG2A-like foci. The black line represents the cumulative fitting. (F) Distribution of ATG2A-like and non-ATG2A-like foci diffusion coefficients (N = 3 biological replicates, mean ± SD). (G) Co-localization of ATG13 foci with the ER in control conditions (siCTR) or CHMP2A knock-down (siCHMP2A) cells. The ER was marked with mEmerald-SEC61, and high-resolution images were generated using the CARE algorithm. Halo-ATG13 foci were scaled at low (1×) and high (2.5×) brightness. Scale bar = 5 μm. (H) Histogram of step-size distribution of Halo-ATG2A-positive (H-ATG2A+, red) and Halo-ATG2A-negative (H-ATG2A−, gray) GFP-ATG13 foci. Three biological replicates (20–30 cells per replicate) were performed for each experiment. (I) Fraction of GFP-ATG13 foci showing accumulation of Halo-ATG2A (N = 3 biological replicates, mean ± SD). (J) GFP-ATG13 foci lifetime for Halo-ATG2A+ and Halo-ATG2A− populations (N = 3 biological replicates, mean ± SD).
High-throughput quantification of autophagy factor foci lifetime and diffusion dynamics. (A) Upon labeling with fluorescent dye (JF646), cells expressing HaloTagged autophagy factors were starved (EBSS) and imaged at four frames per minute for 1 h. TrackIT was used to detect foci based on threshold intensity (left) and connected into tracks using the nearest neighbor algorithm (right). Scale bar = 5 μm. (B) Example images of foci for Halo-ATG2A (upper panel) and Halo-ATG13 (bottom panel). Scale bar = 1 μm. (C) Histograms of foci lifetime for the HaloTagged autophagy proteins in control conditions (Control) and after 1 h nutrient starvation (EBSS). Three biological replicates (20–30 cells per replicate) were performed for each HaloTag cell line. The number of data points (n) is indicated in each graph in the figure panels. (D) Quantification of the number of foci formed per cell by autophagy factors over the course of 1 h imaging in control (Control) and nutrient starvation (EBSS) conditions (N = 3 biological replicates, mean ± SD). A two-tailed t test was used for statistical analysis (*P < 0.05). (E) Histograms of diffusion coefficients of the foci formed by autophagy factors under nutrient starvation. Histograms were fitted with Gaussian curves. For the proteins other than Halo-ATG2A, we fixed the mean of one subpopulation (in red) to match Halo-ATG2A mean. An additional subpopulation (in purple) represents non-ATG2A-like foci. The black line represents the cumulative fitting. (F) Distribution of ATG2A-like and non-ATG2A-like foci diffusion coefficients (N = 3 biological replicates, mean ± SD). (G) Co-localization of ATG13 foci with the ER in control conditions (siCTR) or CHMP2A knock-down (siCHMP2A) cells. The ER was marked with mEmerald-SEC61, and high-resolution images were generated using the CARE algorithm. Halo-ATG13 foci were scaled at low (1×) and high (2.5×) brightness. Scale bar = 5 μm. (H) Histogram of step-size distribution of Halo-ATG2A-positive (H-ATG2A+, red) and Halo-ATG2A-negative (H-ATG2A−, gray) GFP-ATG13 foci. Three biological replicates (20–30 cells per replicate) were performed for each experiment. (I) Fraction of GFP-ATG13 foci showing accumulation of Halo-ATG2A (N = 3 biological replicates, mean ± SD). (J) GFP-ATG13 foci lifetime for Halo-ATG2A+ and Halo-ATG2A− populations (N = 3 biological replicates, mean ± SD).
Representative live-cell imaging movies showing the automated foci detection and tracking using TrackIT algorithm. Left: raw movie of Halo-ATG2A cell line in EBSS starvation media; center: automated identification of foci spots; right: compiling of tracks. Cells were labeled with JF646, and images were acquired at four frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell imaging movies showing the automated foci detection and tracking using TrackIT algorithm. Left: raw movie of Halo-ATG2A cell line in EBSS starvation media; center: automated identification of foci spots; right: compiling of tracks. Cells were labeled with JF646, and images were acquired at four frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell foci imaging movies of control and EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG2A labeled with JF646. Images were acquired at four frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell foci imaging movies of control and EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG2A labeled with JF646. Images were acquired at four frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell foci imaging movies of control and EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG5 labeled with JF646. Images were acquired at four frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell foci imaging movies of control and EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG5 labeled with JF646. Images were acquired at four frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell foci imaging movies of control and EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG9A labeled with JF646. Images were acquired at four frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell foci imaging movies of control and EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG9A labeled with JF646. Images were acquired at four frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell foci imaging movies of control and EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG13 labeled with JF646. Images were acquired at four frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell foci imaging movies of control and EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG13 labeled with JF646. Images were acquired at four frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell foci imaging movies of control and EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG16 labeled with JF646. Images were acquired at four frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell foci imaging movies of control and EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG16 labeled with JF646. Images were acquired at four frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell foci imaging movies of control and EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ULK1 labeled with JF646. Images were acquired at four frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell foci imaging movies of control and EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ULK1 labeled with JF646. Images were acquired at four frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell foci imaging movies of control and EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged WIPI2 labeled with JF646. Images were acquired at four frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell foci imaging movies of control and EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged WIPI2 labeled with JF646. Images were acquired at four frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
To further analyze the characteristics of the phagophores detected, we determined the diffusion coefficient of cytoplasmic foci formed by the tagged autophagy factors, which reports on the mobility of the autophagosomes they associate with. The diffusion coefficient distribution of ATG2A-positive autophagosomes revealed a single population with a mean diffusion coefficient of D = 0.002 µm2/s (Fig. 4 E). In contrast, the diffusion coefficient distributions of all other autophagy factors analyzed were clearly made up of two distinct populations, one with a diffusion coefficient comparable with ATG2A-positive autophagosomes and a second population of foci that moved more rapidly (D = 0.01 µm2/s; Fig. 4 E). Importantly, the number of foci formed by ATG5, ATG13, ATG16, ULK1, and WIPI2 (e.g., Fslow,ATG5 = 0.64 * 120 foci per cell per hour = 76 foci per cell per hour) that had a comparable diffusion coefficient to ATG2A foci is in a similar range as the number of ATG2 foci formed (∼60 per cell per hour; Fig. 4 F).
Our observations demonstrate that ATG2-positive foci are less mobile than a large fraction of the foci formed by the other autophagy proteins analyzed. This reduced mobility could be a consequence of ATG2A-mediated tethering of the phagophore to the ER. To dissect the role of ATG2A in tethering autophagosomes to a lipid source, we analyzed the movement dynamics of stalled phagophores by knocking down the autophagosome termination factor CHMP2A in the Halo-ATG13 cell line (Takahashi et al., 2018; Fig. S4 A). Since CHMP2A is required for autophagosome closure (Takahashi et al., 2018), these stalled autophagosomes presumable have undergone expansion by ATG2A-mediated lipid transfer. Depletion of CHMP2A resulted in the accumulation of Halo-ATG13 foci colocalized with the ER, and we observed coordinated movements of ER tubules and Halo-ATG13 foci, indicating that the Halo-ATG13 foci are tethered to the ER (Fig. 4 G and Video 19). We also observed Halo-ATG13 foci that colocalized with the ER in control cells (Fig. 4 G and Video 19).
Quantification of diffusion and kinetic properties of HaloTagged autophagy proteins and autophagy induced foci. (A) Western blot showing partial depletion of CHMP2A in Halo-ATG13 cell line upon siRNA-mediated knockdown. (B) Graphs depicting foci frequency and foci lifetimes of ATG13 in complete media or EBSS treated with VPS34 inhibitor compound 31. Data represent mean ± SD. Letters indicate statistically homogenous groups established by ANOVA (P < 0.05). (C) Distribution of diffusion coefficients for Halo-ATG2A and Halo-ATG2A foci imaged a 3-s frame interval. In Halo-ATG13, Gaussian fitting shows two populations with distinct diffusive properties (solid purple and red lines). Cumulative fitting is shown in a solid black line. (D) Results of diffusive analysis for the parental Halo-ATG13 and ULK1, FIP200, and ATG101 knockout under control and EBSS starvation. Left panel depicts the percentage associated with each fraction. Right panels present the diffusion coefficients of the tracks based on the SpotON analysis. Boxes indicate confidence interval ± SD, the square indicates the average, and the horizontal line is the median; for each condition, three biological replicates were analyzed, ∼20 cells/replicate. (E) Images demonstrating the titration of the virus such that the G120A mutant does not form aggregates (left) and positive control in an ATG9A knockout (right). (F) Bar graph representing the average lifetime of GFP-P62 within ATG-edited cell lines. Data represent mean ± SD of three biological replicates (20–30 cells per replicate). Letters indicate statistically homogenous groups established by ANOVA (P < 0.05). (G) Bar graph depicting the average lifetime of GFP-LC3 within ATG-edited cell lines. Data represent mean ± SD of three biological replicates (20–30 cells per replicate). Letters indicate statistically homogenous groups established by ANOVA (P < 0.05). (H and I) Histograms of the lifetime of Halo-Tagged protein foci that colocalized (green) or did not colocalize (dark gray) with (H) GFP-P62 or (I) GFP-LC3 (light blue). Source data are available for this figure: SourceData FS4.
Quantification of diffusion and kinetic properties of HaloTagged autophagy proteins and autophagy induced foci. (A) Western blot showing partial depletion of CHMP2A in Halo-ATG13 cell line upon siRNA-mediated knockdown. (B) Graphs depicting foci frequency and foci lifetimes of ATG13 in complete media or EBSS treated with VPS34 inhibitor compound 31. Data represent mean ± SD. Letters indicate statistically homogenous groups established by ANOVA (P < 0.05). (C) Distribution of diffusion coefficients for Halo-ATG2A and Halo-ATG2A foci imaged a 3-s frame interval. In Halo-ATG13, Gaussian fitting shows two populations with distinct diffusive properties (solid purple and red lines). Cumulative fitting is shown in a solid black line. (D) Results of diffusive analysis for the parental Halo-ATG13 and ULK1, FIP200, and ATG101 knockout under control and EBSS starvation. Left panel depicts the percentage associated with each fraction. Right panels present the diffusion coefficients of the tracks based on the SpotON analysis. Boxes indicate confidence interval ± SD, the square indicates the average, and the horizontal line is the median; for each condition, three biological replicates were analyzed, ∼20 cells/replicate. (E) Images demonstrating the titration of the virus such that the G120A mutant does not form aggregates (left) and positive control in an ATG9A knockout (right). (F) Bar graph representing the average lifetime of GFP-P62 within ATG-edited cell lines. Data represent mean ± SD of three biological replicates (20–30 cells per replicate). Letters indicate statistically homogenous groups established by ANOVA (P < 0.05). (G) Bar graph depicting the average lifetime of GFP-LC3 within ATG-edited cell lines. Data represent mean ± SD of three biological replicates (20–30 cells per replicate). Letters indicate statistically homogenous groups established by ANOVA (P < 0.05). (H and I) Histograms of the lifetime of Halo-Tagged protein foci that colocalized (green) or did not colocalize (dark gray) with (H) GFP-P62 or (I) GFP-LC3 (light blue). Source data are available for this figure: SourceData FS4.
Representative live-cell foci imaging movies of EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG13, and mEmerald-SEC61 treated with control siRNA. Cells were labeled with JFX650 and images were acquired at one frame per second. mEmerald-SEC61 signal was reconstructed to higher resolution using the CARE algorithm.
Representative live-cell foci imaging movies of EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG13, and mEmerald-SEC61 treated with control siRNA. Cells were labeled with JFX650 and images were acquired at one frame per second. mEmerald-SEC61 signal was reconstructed to higher resolution using the CARE algorithm.
To directly test whether ATG2A is responsible for tethering autophagosomes, we performed dual-color particle tracking of Halo-ATG2A and transiently expressed GFP-ATG13. Only a small fraction (12%) of Halo-ATG2A foci colocalized with GFP-ATG13 foci, and the mobility of GFP-ATG13 foci that colocalized with Halo-ATG2A was substantially lower than GFP-ATG13 foci that lacked Halo-ATG2A signal (Fig. 4, H–J and Video 20). Strikingly, the lifetime GFP-ATG13 foci that colocalized with Halo-ATG2A (100 s) was significantly longer than GFP-ATG13 that did not accumulate Halo-ATG2A (25 s; Fig. 4 L and Video 20), suggesting that foci that recruit both ATG13 and ATG2A likely represent stable phagophores undergoing expansion by lipid transfer and therefore are tether to a lipid donor compartment.
Representative live-cell foci imaging movies of EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG2 and GFP-ATG13. Cells were labeled with JFX650, and images were acquired at one frame per second.
Representative live-cell foci imaging movies of EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG2 and GFP-ATG13. Cells were labeled with JFX650, and images were acquired at one frame per second.
Together these observations demonstrate that ATG5, ATG13, ATG16, and ULK1 are recruited to all phagophores while ATG2A only detectably accumulates at a subset of autophagy-induced foci. In addition, our analysis of the movement dynamics of foci formed by the tagged autophagy factors suggests that ATG2A recruitment triggers the transition of the phagophore to a less mobile state, potentially by forming an anchor point to lipid donor membranes.
Both classes of phagophores identified from tracked populations require ULK1 complex and PI3K activity
The data presented thus far demonstrate that our automated particle tracking approach can quantitatively determine the frequency of formation and biophysical properties of starvation-induced autophagy protein foci. To validate that the tracked foci represent bona fide phagophores that mature into autophagosomes, we dissected the genetic requirements for their initiation by analyzing the contribution of the ULK1 complex and PI3K to foci formation. It is well-established that ULK1 is critical for the initiation of autophagy (Karanasios et al., 2016; Mercer et al., 2009; Mizushima, 2010; Szymańska et al., 2015). As a marker for the ULK1 complex, we used Halo-ATG13. Our analysis of Halo-ATG13 foci dynamics revealed two distinct classes of ATG13-positive structures: a static population with a diffusion coefficient comparable with ATG2A-positive foci and a second population with higher mobility. We hypothesize that these classes of ATG13 foci represent untethered prephagophores (higher mobility), and phagophores tethered to a donor membrane by ATG2A (lower mobility). We, therefore, expected that the formation of both populations of ATG13 foci would require signaling through the ULK1 and PI3K complexes. To dissect the contribution of the ULK1 complex to the formation of both classes of autophagosomes, we knocked out ULK1, FIP200, or ATG101 in the Halo-ATG13 cell line (Fig. 5 A). Clonal knock-out cell lines of ULK1, FIP200, or ATG101 showed a significant defect in LC3 conjugation under starvation conditions and a shifted phospho-P62 band was detected by Western blot in the FIP200 and ATG101 knock-out cells (Fig. 5 B). Interestingly, LC3 conjugation was more strongly inhibited in FIP200 and ATG101 knock-out cells compared with cells lacking ULK1 (Fig. 5, B and C). ULK2 may compensate for the loss of ULK1 activity under these circumstances. We were unable to generate ULK1 and ULK2 double knock-out cells, indicating that loss of ULK1 and ULK2 function may be lethal in U2OS cells. As an alternative approach, we combined the ULK1, FIP200, and ATG101 gene knockouts with ULK-101, a highly potent and specific small molecule inhibitor of both ULK1 and ULK2 (Martin et al., 2018). ULK-101 treatment in ULK1 knock-out cells further decreased LC3 conjugation to the levels observed in cells lacking FIP200 or ATG101, confirming the previously reported role for ULK2 in maintaining LC3 conjugation in the absence of ULK1 (Fig. 5, B and C; Ro et al., 2013). In addition, these observations demonstrate that the knockout of ATG101 and FIP200 leads to a complete loss of ULK1 complex function, which is consistent with previous studies (Itakura and Mizushima, 2010; Kannangara et al., 2021). To determine which step of autophagosome formation is inhibited by the loss of function of ULK1, FIP200, and ATG101, we assessed the formation of ATG13 foci in the knock-out cell lines in both control and starved conditions (Fig. 5 D). As expected, under control conditions, Halo-ATG13 formed a small number of foci in all cell lines (Fig. 5 E and Video 21). After treatment with EBSS, Halo-ATG13 foci formation was increased to ∼200 foci/cell/h in the parental cells (Fig. 5 E). Knockout of FIP200 and ATG101 resulted in a significant reduction in the number of Halo-ATG13 foci formed, from ∼200 to ∼9 and ∼15 foci/cell/h, respectively (Fig. 5 E and Video 21). In ULK1 knock-out cells Halo-ATG13 formed ∼50 foci/cell/h (Fig. 5 E), consistent with the intermediate phenotype observed for LC3 conjugation (Fig. 5, B and C). Together these observations suggest that the tracked foci represent bona fide phagophores or autophagosomes and that their formation is dependent on the assembly of the ULK1 complex.
All tracked populations of autophagy factors foci require ULK1 and PI3K activity. (A) Fluorescence gel and Western blots demonstrating successful knockout of ULK1, FIP200, and ATG101 from the Halo-ATG13 cell line. (B) Western blots demonstrating impaired autophagy when ULK1, FIP200, and ATG101 are individually depleted from the Halo-ATG13 cell line. For the treatment experiments, cells were preincubated in control media with ULK1-101 (1 μM) or without drug for 1 h where indicated. Cells were then switched to their control or EBSS starvation media, with or without bafilomycin (100 nM), for an additional hour. (C) Quantification of the Western blots in B. Data represent mean ± SD over three biological replicates. Phospho-P62 band (red striped bar graph) was detected and quantified only in the FIP200 and ATG101 knock-out cell lines. (D) Histograms of Halo-ATG13 foci lifetimes for the parental Halo-ATG13 cells and ULK1, FIP200, ATG101 knock-out cell lines in control conditions (Control) and after 1 h nutrient starvation (EBSS). Three biological replicates (20–30 cells per replicate) were performed for each cell line. The number of data points (n) is indicated in each graph in the figure panels. (E) Quantification of the number of foci formed per cell by HaloTag-ATG13 over the course of 1 h imaging in control (Control) and nutrient starvation (EBSS) conditions (N = 3, mean ± SD). A two-tailed t test was used for statistical analysis. (F) Histograms of foci lifetimes for the HaloTag-ATG2A (left) and HaloTag-ATG13 (center) in control and nutrient starvation (EBSS, 1 h) conditions with and without wortmannin (1 μM). Cells were pretreated with Wortmannin for 1 h. Right panel presents the quantification of the foci frequency. Data represent mean ± SD over three biological replicates (20–30 cells per replicate). A two-tailed t test was used for statistical analysis (*P < 0.05, ***P < 0.001).
All tracked populations of autophagy factors foci require ULK1 and PI3K activity. (A) Fluorescence gel and Western blots demonstrating successful knockout of ULK1, FIP200, and ATG101 from the Halo-ATG13 cell line. (B) Western blots demonstrating impaired autophagy when ULK1, FIP200, and ATG101 are individually depleted from the Halo-ATG13 cell line. For the treatment experiments, cells were preincubated in control media with ULK1-101 (1 μM) or without drug for 1 h where indicated. Cells were then switched to their control or EBSS starvation media, with or without bafilomycin (100 nM), for an additional hour. (C) Quantification of the Western blots in B. Data represent mean ± SD over three biological replicates. Phospho-P62 band (red striped bar graph) was detected and quantified only in the FIP200 and ATG101 knock-out cell lines. (D) Histograms of Halo-ATG13 foci lifetimes for the parental Halo-ATG13 cells and ULK1, FIP200, ATG101 knock-out cell lines in control conditions (Control) and after 1 h nutrient starvation (EBSS). Three biological replicates (20–30 cells per replicate) were performed for each cell line. The number of data points (n) is indicated in each graph in the figure panels. (E) Quantification of the number of foci formed per cell by HaloTag-ATG13 over the course of 1 h imaging in control (Control) and nutrient starvation (EBSS) conditions (N = 3, mean ± SD). A two-tailed t test was used for statistical analysis. (F) Histograms of foci lifetimes for the HaloTag-ATG2A (left) and HaloTag-ATG13 (center) in control and nutrient starvation (EBSS, 1 h) conditions with and without wortmannin (1 μM). Cells were pretreated with Wortmannin for 1 h. Right panel presents the quantification of the foci frequency. Data represent mean ± SD over three biological replicates (20–30 cells per replicate). A two-tailed t test was used for statistical analysis (*P < 0.05, ***P < 0.001).
Representative live-cell foci imaging movies of EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG13 (top left), and ULK1 (top right), FIP200 (bottom left), and ATG101 (bottom right) knockouts. Cells were labeled with JF646 and acquired at four frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell foci imaging movies of EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG13 (top left), and ULK1 (top right), FIP200 (bottom left), and ATG101 (bottom right) knockouts. Cells were labeled with JF646 and acquired at four frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
A critical consequence of ULK1 activation is thought to be the localized generation of PI3P by VPS34 at the site of autophagosome formation. To determine if PI3P accumulation is downstream of ULK1 complex enrichment at the phagophore, we treated Halo-ATG13 cells with 1 µM Wortmannin, which broadly inhibits the PI3-kinase activity for 1 h prior to and during cell starvation with EBSS and analyzed foci formation by Halo-ATG13. Strikingly, inhibition of PI3P-kinase activity eliminated the induction of Halo-ATG13 foci under starvation conditions (Fig. 5 F and Video 22). Experiments performed with Compound 31 (Pasquier et al., 2015), a specific VPS34 inhibitor, showed a similar reduction of Halo-ATG13 foci (Fig. S4 B). This suggests that PI3P formation by VPS34 is critical for ATG13 accumulation at the phagophore and phagophore initiation. To confirm this finding, we treated Halo-ATG2A cells with Wortmannin and analyzed its impact on ATG2A foci formation. Similar to Halo-ATG13, the number of Halo-ATG2A foci formed in starved cells was reduced to those observed under control conditions by treatment with Wortmannin (Fig. 5 F and Video 23). Importantly, this experiment confirmed that the number of Halo-ATG2A foci formed is approximately half of the number of Halo-ATG13 foci formed in all experimental conditions (Fig. 5 F). Since these experiments were carried out at a higher time resolution compared with the experiments shown in Fig. 4, we also confirmed that the diffusion coefficient distribution of ATG13 foci contained two distinct populations, while that of ATG2A only had a single slowly diffusing population (Fig. S4 B). These observations demonstrate that both populations of ATG13 foci require PI3P formation by VPS34 and ULK1 complex activity and suggest that the two populations of autophagosomes with distinct mobilities represent different stages of autophagosome formation. We will refer to the mobile population as “pre-phagophores” and the static population as “phagophores.”
Representative live-cell foci imaging movies of control and EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG13 with and without Wortmannin. Cells were labeled with JF646, and images were acquired at four frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell foci imaging movies of control and EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG13 with and without Wortmannin. Cells were labeled with JF646, and images were acquired at four frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell foci imaging movies of control and EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG2 with and without Wortmannin. Cells were labeled with JF646, and images were acquired at four frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell foci imaging movies of control and EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG2 with and without Wortmannin. Cells were labeled with JF646, and images were acquired at four frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
It was previously proposed that a positive feedback loop initiated by ULK1 activation and reinforced by additional ULK1 complex recruitment via the formation of PI3P by VPS34 at the phagophore is the critical trigger for autophagosome formation (Ganley et al., 2009; Hosokawa et al., 2009; Jung et al., 2009; Karanasios et al., 2013a; Mercer et al., 2009; Russell et al., 2013). Importantly, ATG13 contains a PI3P/PI4P binding sequence in its N-terminus, which is thought to contribute to the association of the ULK1 complex with the pre-phagophore (Karanasios et al., 2013a). To analyze the dynamic association of ATG13 with cytoplasmic membranes, we took advantage of the live-cell single-molecule imaging approach we developed. Our observations demonstrated that a fraction of Halo-ATG13 particles were highly static and a second subset of Halo-ATG13 molecules moved with a similar diffusion coefficient as SNAP-SEC61. We propose that these slow-moving and static Halo-ATG13 particles are the consequence of the association of Halo-ATG13 with cytoplasmic membrane compartments or phagophores. We first determined whether the diffusion properties of Halo-ATG13 were impacted by the knockout of ULK1, FIP200, or ATG101. Halo-ATG13 diffusion properties were unchanged in cells lacking ULK1, FIP200, or ATG101 under both control and starved conditions (Fig. S4 D and Video 24). These observations suggest that the membrane association of ATG13 does not depend on its association with any of the other ULK1 complex components. Our results demonstrate that the initiation of autophagosome formation requires the ULK1 complex. In addition, ULK1 complex accumulation at the phagophore requires PI3P, which is potentially mediated by the association of ATG13 with PI3P. Altogether, our observations are consistent with a model in which a positive feedback loop initiated by the ULK1 complex and amplified by VPS34 is critical to trigger autophagosome formation.
Representative live-cell single-molecule imaging movies of control (top) and EBSS-treated (bottom) U2OS cells expressing 3xFLAG-HaloTagged ATG13 parental and ULK1, FIP200, and ATG101 knockout. Cells were labeled with JF650, and images were acquired at 146 frames per second. 140 × 140 pixels with a pixel size of 0.16 μm.
Representative live-cell single-molecule imaging movies of control (top) and EBSS-treated (bottom) U2OS cells expressing 3xFLAG-HaloTagged ATG13 parental and ULK1, FIP200, and ATG101 knockout. Cells were labeled with JF650, and images were acquired at 146 frames per second. 140 × 140 pixels with a pixel size of 0.16 μm.
The majority of cytoplasmic foci formed by autophagy proteins do not progress to form mature autophagosomes
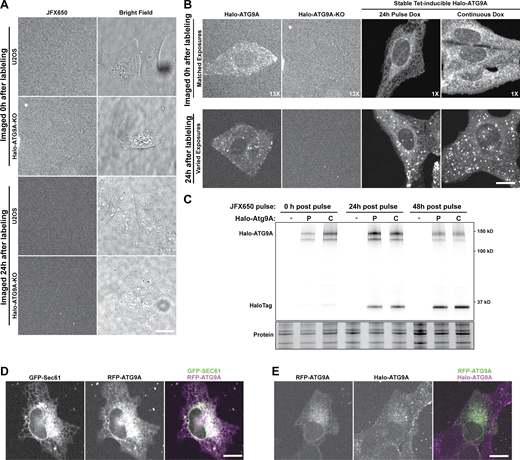
The experiments described thus far focused on the initiation of starvation-induced foci by the tagged autophagy proteins. To analyze the progression of these foci into mature autophagosomes, we performed quantitative dual-color live-cell imaging to assess the recruitment of the cargo adaptor P62 and the ATG8 family protein LC3 to the starvation-induced foci formed by the tagged autophagy factors. To mark growing phagophores and mature autophagosomes, we transduced cells with baculoviruses encoding GFP-LC3 or GFP-P62. To avoid GFP-LC3 aggregation, we optimized the virus concentration using GFP-LC3-G120A, which cannot be conjugated and therefore only forms foci by aggregation. At the viral titers used no GFP-LC3-G120A foci were detected in the Halo-ATG9A cell line, but GFP-LC3-G120A aggregates were observed in ATG9A knock-out cells, which is consistent with the previously observed LC3 aggregates reported in autophagy-deficient cells (Fig. S4 E; Runwal et al., 2019). Using this protocol, cells expressing the HaloTagged autophagy proteins were transduced with GFP-LC3 or GFP-P62 and imaged with high-time resolution (3 s per frame) after cell starvation with EBSS (Fig. 6 A; and Videos 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, and 35). GFP and HaloTag signals were tracked independently, and co-localization was defined as particles whose centroids were <3 pixels apart at any given timepoint (see Materials and methods for details). This approach allowed us to analyze hundreds of autophagosomes in a completely unbiased fashion. A fraction (∼10–20%) of the foci formed by all autophagy factors analyzed colocalized with P62 or LC3 (P62positive, LC3positive), but the majority (∼80–90%) of foci never showed detectable P62 or LC3 accumulation (P62negative, LC3negative, Fig. 6, B–E; and Fig. S4, H and I). The fraction of Halo-ATG2A foci that colocalized with P62 (23%) was approximately double that of all other autophagy proteins tested (10%; Fig. 6 C), while the fraction of autophagy protein foci that colocalized with LC3 was comparable for all proteins tested (Fig. 6 E). Importantly, the overall frequency and lifetime of P62 and LC3 foci was comparable across all cell lines, indicating that the HaloTagged autophagy proteins do not impact the formation of LC3- or P62-positive autophagosomes (Fig. S4, F and G). It is possible that the lack of colocalization of the autophagy proteins analyzed with an autophagy marker was a consequence of the viral transduction, choice of autophagy cargo adaptor, or drift of the autophagy factor foci out of the focal plane of the objective. To rule out these possibilities, we stably expressed GFP-GABARAPL1, GFP-LC3B, and GFP-P62 by integrating a tetracycline-inducible expression cassette into the AAVS1 safe-harbor locus in our Halo-ATG13 cell line. Cells were grown in the absence of tetracycline to limit the expression of the GFP-tagged P62, LC3B, and GABARAPL1. In addition, we increased the time resolution to 1.5 s per frame and imaged three z-sections, allowing us to analyze autophagy protein foci throughout the entire cytosol. The results did not differ from our previous findings: over 80% of ATG13 foci did not colocalize with GABARAPL1, LC3B, and P62 (Fig. 6, F–I).
Analysis of the maturation kinetics of autophagosomes using dual-color imaging. (A) Example images showing formation, growth, and disappearance of colocalized Halo-ATG13 and GFP-p62 foci using dual-color live-cell imaging under EBSS starvation (1 h). Scale bar = 2 μm. (B) Histograms of the lifetimes of Halo-ATG2A and Halo-ATG13 foci that colocalized (green) or did not colocalize (dark gray) with virally transduced GFP-P62. (C) Percentage of HaloTagged autophagy protein foci that colocalized with P62 foci. Data represent mean ± SD of three biological replicates (20–30 cells per replicate). A two-tailed t test was used for statistical analysis. (D) Histograms of the lifetimes of Halo-ATG2A and Halo-ATG13 foci that colocalized (light blue) or did not colocalize (dark gray) with virally transduced GFP-LC3. (E) Percentage of HaloTagged autophagy protein foci that colocalized with LC3 foci. Data represent mean ± SD of three biological replicates (20–30 cells per replicate). A two-tailed t test was used for statistical analysis. (F–H) Histograms of the lifetimes of Halo-ATG13 foci that colocalized or did not colocalize (dark gray) with (F) GFP-GABARAPL1 (yellow), (G) GFP-LC3 (light blue), (H) GFP-p62 (green) stably expressed from an AAVS1 locus insertion. (I) Percentage of HaloTagged autophagy protein foci that colocalized with the adaptors GFP-GABARAPL1, GFP-LC3, and GFP-P62 stably expressed from an AAVS1 locus insertion. Data represent mean ± SD of three biological replicates (20–30 cells per replicate). A two-tailed t test was used for statistical analysis. (J) Quantification of the average HaloTagged autophagy protein foci lifetimes for GFP-P62 positive (green) and negative (dark gray) foci. Data represent mean ± SD of three biological replicates (20–30 cells per replicate). (K) Quantification of the average HaloTagged autophagy protein foci lifetimes for GFP-LC3 positive (light blue) and negative (dark gray) foci. Data represent mean ± SD of three biological replicates (20–30 cells per replicate). (L) Quantification of the average Halo-ATG13 foci lifetimes that are negative (dark gray) or positive for adaptor signals (GABARAPL1, yellow; LC3, light blue; P62, green). Data represent mean ± SD of three biological replicates (20–30 cells per replicate). (M) Quantification of the timing of the three distinct phases of autophagosome formation for the HaloTag proteins under EBSS starvation (1 h). Data represent mean ± SD of three biological replicates (20–30 cells per replicate).
Analysis of the maturation kinetics of autophagosomes using dual-color imaging. (A) Example images showing formation, growth, and disappearance of colocalized Halo-ATG13 and GFP-p62 foci using dual-color live-cell imaging under EBSS starvation (1 h). Scale bar = 2 μm. (B) Histograms of the lifetimes of Halo-ATG2A and Halo-ATG13 foci that colocalized (green) or did not colocalize (dark gray) with virally transduced GFP-P62. (C) Percentage of HaloTagged autophagy protein foci that colocalized with P62 foci. Data represent mean ± SD of three biological replicates (20–30 cells per replicate). A two-tailed t test was used for statistical analysis. (D) Histograms of the lifetimes of Halo-ATG2A and Halo-ATG13 foci that colocalized (light blue) or did not colocalize (dark gray) with virally transduced GFP-LC3. (E) Percentage of HaloTagged autophagy protein foci that colocalized with LC3 foci. Data represent mean ± SD of three biological replicates (20–30 cells per replicate). A two-tailed t test was used for statistical analysis. (F–H) Histograms of the lifetimes of Halo-ATG13 foci that colocalized or did not colocalize (dark gray) with (F) GFP-GABARAPL1 (yellow), (G) GFP-LC3 (light blue), (H) GFP-p62 (green) stably expressed from an AAVS1 locus insertion. (I) Percentage of HaloTagged autophagy protein foci that colocalized with the adaptors GFP-GABARAPL1, GFP-LC3, and GFP-P62 stably expressed from an AAVS1 locus insertion. Data represent mean ± SD of three biological replicates (20–30 cells per replicate). A two-tailed t test was used for statistical analysis. (J) Quantification of the average HaloTagged autophagy protein foci lifetimes for GFP-P62 positive (green) and negative (dark gray) foci. Data represent mean ± SD of three biological replicates (20–30 cells per replicate). (K) Quantification of the average HaloTagged autophagy protein foci lifetimes for GFP-LC3 positive (light blue) and negative (dark gray) foci. Data represent mean ± SD of three biological replicates (20–30 cells per replicate). (L) Quantification of the average Halo-ATG13 foci lifetimes that are negative (dark gray) or positive for adaptor signals (GABARAPL1, yellow; LC3, light blue; P62, green). Data represent mean ± SD of three biological replicates (20–30 cells per replicate). (M) Quantification of the timing of the three distinct phases of autophagosome formation for the HaloTag proteins under EBSS starvation (1 h). Data represent mean ± SD of three biological replicates (20–30 cells per replicate).
Representative live-cell foci imaging movies of EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged LC3 labeled with JF646 and GFP-P62. Right panel is a merge of Halo-LC3 (magenta) and GFP-P62 (green). Images were acquired at 20 frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell foci imaging movies of EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged LC3 labeled with JF646 and GFP-P62. Right panel is a merge of Halo-LC3 (magenta) and GFP-P62 (green). Images were acquired at 20 frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell foci imaging movies of EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG2A labeled with JF646 and GFP-P62. Right panel is a merge of Halo-ATG2A (magenta) and GFP-P62 (green). Images were acquired at 20 frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell foci imaging movies of EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG2A labeled with JF646 and GFP-P62. Right panel is a merge of Halo-ATG2A (magenta) and GFP-P62 (green). Images were acquired at 20 frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell foci imaging movies of EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG5 labeled with JF646 and GFP-P62. Right panel is a merge of Halo-ATG5 (magenta) and GFP-P62 (green). Images were acquired at 20 frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell foci imaging movies of EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG5 labeled with JF646 and GFP-P62. Right panel is a merge of Halo-ATG5 (magenta) and GFP-P62 (green). Images were acquired at 20 frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell foci imaging movies of EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG9A labeled with JF646 and GFP-P62. Right panel is a merge of Halo-ATG9A (magenta) and GFP-P62 (green). Images were acquired at 20 frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell foci imaging movies of EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG9A labeled with JF646 and GFP-P62. Right panel is a merge of Halo-ATG9A (magenta) and GFP-P62 (green). Images were acquired at 20 frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell foci imaging movies of EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG13 labeled with JF646 and GFP-P62. Right panel is a merge of Halo-ATG13 (magenta) and GFP-P62 (green). Images were acquired at 20 frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell foci imaging movies of EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG13 labeled with JF646 and GFP-P62. Right panel is a merge of Halo-ATG13 (magenta) and GFP-P62 (green). Images were acquired at 20 frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell foci imaging movies of EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG16 labeled with JF646 and GFP-P62. Right panel is a merge of Halo-ATG16 (magenta) and GFP-P62 (green). Images were acquired at 20 frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell foci imaging movies of EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG16 labeled with JF646 and GFP-P62. Right panel is a merge of Halo-ATG16 (magenta) and GFP-P62 (green). Images were acquired at 20 frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell foci imaging movies of EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ULK1 labeled with JF646 and GFP-P62. Right panel is a merge of ULK1-Halo (magenta) and GFP-P62 (green). Images were acquired at 20 frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell foci imaging movies of EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ULK1 labeled with JF646 and GFP-P62. Right panel is a merge of ULK1-Halo (magenta) and GFP-P62 (green). Images were acquired at 20 frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell foci imaging movies of EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged WIPI2 labeled with JF646 and GFP-P62. Right panel is a merge of Halo-WIPI2 (magenta) and GFP-P62 (green). Images were acquired at 20 frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell foci imaging movies of EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged WIPI2 labeled with JF646 and GFP-P62. Right panel is a merge of Halo-WIPI2 (magenta) and GFP-P62 (green). Images were acquired at 20 frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell foci imaging movies of EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG2A labeled with JF646 and GFP-LC3. Right panel is a merge of Halo-ATG2A (magenta) and GFP-LC3 (green). Images were acquired at 20 frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell foci imaging movies of EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG2A labeled with JF646 and GFP-LC3. Right panel is a merge of Halo-ATG2A (magenta) and GFP-LC3 (green). Images were acquired at 20 frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell foci imaging movies of EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG13 labeled with JF646 and GFP-LC3. Right panel is a merge of Halo-ATG13 (magenta) and GFP-LC3 (green). Images were acquired at 20 frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell foci imaging movies of EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG13 labeled with JF646 and GFP-LC3. Right panel is a merge of Halo-ATG13 (magenta) and GFP-LC3 (green). Images were acquired at 20 frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell foci imaging movies of EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ULK1 labeled with JF646 and GFP-LC3. Right panel is a merge of ULK1-Halo (magenta) and GFP-LC3 (green). Images were acquired at 20 frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell foci imaging movies of EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ULK1 labeled with JF646 and GFP-LC3. Right panel is a merge of ULK1-Halo (magenta) and GFP-LC3 (green). Images were acquired at 20 frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
To gain further insight into the timeframe of autophagosome formation, we analyzed the kinetics of foci formation and colocalization with P62 or LC3. The lifetime of the P62negative autophagy protein trajectories (mean = 23–41 s) was significantly (P < 0.001) shorter than the P62positive tracks (mean = 91–122 s) for all tagged autophagy factors (Fig. 6 J). Similarly, LC3positive trajectories of Halo-ATG2A, Halo-ATG13, and ULK1-Halo were significantly longer (mean = 110–145 s) than LC3negative tracks of the respective protein (mean = 11–37 s; P < 0.001; Fig. 6 K), and GABARAPL1positive Halo-ATG13 trajectories were significantly longer (mean = 80 s) than Halo-ATG13 tracks that did not colocalize with GABARAPL1 (mean = 25 s; Fig. 6 L). Viral expression and stable integration of GFP-LC3 and GFP-P62 lead to similar results (Fig. 6, J–L). Altogether, these results demonstrate that most autophagosome initiation sites (∼80–90%), marked by the local accumulation of an autophagy protein, rapidly disassemble ∼25 s after their formation without detectably recruiting any of the cargo adaptors tested and therefore do not progress to form mature autophagosomes.
Kinetics of autophagosome formation
To dissect the assembly order and overall formation kinetics of autophagosomes, we analyzed the sequential accumulation of the autophagy factors and P62 or LC3, respectively. To confirm the validity of this approach we first analyzed the accumulation of GFP-P62 at Halo-LC3 foci (Fig. 6 M and Video 25). As expected for LC3-dependent recruitment of P62 to phagophores, GFP-P62 and Halo-LC3 foci appeared nearly simultaneously (Fig. 6 M and Video 25). In addition, the GFP-P62 disappeared when Halo-LC3 foci were still detectable (Fig. 6 M). We were unable to determine the fraction of Halo-LC3 marked autophagosomes that colocalized with GFP-P62 because Halo-LC3 does not get degraded after fusion of the autophagosome with the lysosome, and therefore Halo-LC3 also leads to the accumulation of fluorescent signal in lysosomes. These properties of Halo-LC3 were recently described in detail by Yim and colleagues (Yim et al., 2022). We next analyzed the accumulation kinetics of GFP-LC3 and GFP-P62 at foci formed by the other HaloTagged autophagy factors. On average autophagy protein foci formed ∼40–60 s prior to the recruitment of P62, co-colocalized with P62 for ∼50–85 s, and dissociated from P62 foci ∼30–100 s prior to the disappearance of the P62 signal, which is likely the consequence of autophagosome fusion with the lysosome and quenching of the GFP signal (Fig. 6 M and Videos 26, 27, 28, 29, 30, 31, 32, 33, 34, and 35). Compared to P62, LC3 accumulated at autophagy protein foci more rapidly (∼12–22 s) after their formation (Fig. 6 M), consistent with LC3 conjugation preceding P62 recruitment (Ichimura et al., 2008; Kirkin and Rogov, 2019; Noda et al., 2008; Pankiv et al., 2007). The total lifetime of autophagosomes (ATG signal appearance to P62 or LC3 signal disappearance) did not differ dramatically for all proteins tested (range = 140–226 s, mean = 175 s; Fig. 6 M). In total, these results demonstrate that if the phagophore matures into an autophagosome all autophagy factors analyzed are recruited in close succession, ∼40 s prior to the accumulation of P62, autophagosome biogenesis, from phagophore initiation to dissociation of the autophagy factors, takes ∼110 s, and the total time from phagophore initiation to autophagosome fusion with the lysosomes is ∼175 s.
ATG9A does not detectably accumulate at sites of autophagosome formation
A growing body of evidence suggests that ATG9-containing vesicles are the platform upon which the phagophore is assembled (Chang et al., 2021a; Chang et al., 2021b; Olivas et al., 2022,Preprint; Sawa-Makarska et al., 2020). Live-cell imaging using a transiently expressed RFP-ATG9A suggested that ATG9A foci continuously interact with LC3B-positive phagophores (Orsi et al., 2012). Furthermore, it has been proposed that autophagosomes are initiated at contact sites formed by the ER and ATG9A-positive compartments (Karanasios et al., 2016). When performing dual-color imaging to analyze the kinetics of Halo-ATG9A recruitment to the autophagosome, we were unable to detect the accumulation of endogenous Halo-ATG9A at virally induced GFP-P62–positive foci (Fig. 7 A). To confirm that Halo-ATG9A does not accumulate at sites of autophagosome formation, we edited the Halo-ATG9A cell line to express SNAP-LC3B from its endogenous locus (Fig. 7 B). We confirmed the functionality of SNAP-LC3B by starvation-induced foci formation and LC3 lipid conjugation using a fluorescence gel (Fig. 7, C and D; and Video 36). Similar to GFP-P62 foci, we were unable to detect Halo-ATG9A accumulation at phagophores marked by SNAP-LC3B (Fig. 7 C and Video 36). These observations are at odds with the previous work mentioned above that detected colocalization of ATG9A-positive subcellular structures and LC3 foci (Karanasios et al., 2016; Orsi et al., 2012). To verify the contribution of ATG9A to autophagosome formation in our U2OS cell system, we knocked out ATG9A in the Halo-ATG2A cell line (Fig. 7 E). Compared with the parental Halo-ATG2A cells, we detected a molecular size shift of the P62 band in ATG9A knock-out cells (Fig. 7 E), suggesting aberrant accumulation of P62 due to a defect in autophagy. We then analyzed the foci formation of Halo-ATG2A in the ATG9A knock-out cells. Under control and starvation conditions, the formation of ATG2A foci was significantly reduced in the absence of ATG9A (Fig. 7, G and H; and Video 37). Therefore, while ATG9A accumulation is not detectable at autophagosome initiation sites, it plays a critical role in the formation of ATG2A-positive phagophores. These observations are fully consistent with vesicles containing a small number of ATG9A molecules (undetectable by our imaging approach) forming the platform for autophagosome formation.
ATG9 does not detectably accumulate at the site of autophagosome formation. (A) Example image showing the formation of GFP-P62 spot in the absence of any Halo-ATG9A accumulation. Scale bar = 1 μm. (B) Fluorescent gels showing Halo-ATG9A (JFX650, top) and SNAP-LC3B (JFX650, middle) labeling and Western blot showing probed with an LC3 antibody showing the exclusive expression of SNAP-LC3B in genome-edited cells. (C) Micrographs of cells expressing Halo-ATG9A (JFX650) and SNAP-LC3B (JF503; top, scale bar = 10 µm) and kymograph of a SNAP-LC3B spot showing no accumulation of Halo-ATG9A (one frame per second, frames 79–360). (D) Fluorescent gel of SNAP-LC3B (JFX650) after cell starvation and treatment with bafilomycin, demonstrating lipid conjugation of SNAP-LC3B (bottom band). (E) Fluorescence gel and Western blots demonstrating successful ATG9A gene knockout from cells expressing Halo-ATG2A. The ATG9A knock-out cells accumulate P62, indicating impaired autophagy. (F) Micrographs showing a decrease of Halo-ATG2A foci when ATG9A is knocked out (scale bar = 10 μm). Halo-ATG9A does not form detectable foci under EBSS starvation (right panel). (G) Histograms of Halo-ATG2A foci lifetime in parental and ATG9A knock-out cells (EBSS, 1 h). (H) Quantification of the number of foci formed per cell over the course of an hour by Halo-ATG2A in control and ATG9A knock-out cells. Data represent mean ± SD of three biological replicates (20–30 cells per replicate). A two-tailed t test was used for statistical analysis (*P < 0.05). Source data are available for this figure: SourceData F7.
ATG9 does not detectably accumulate at the site of autophagosome formation. (A) Example image showing the formation of GFP-P62 spot in the absence of any Halo-ATG9A accumulation. Scale bar = 1 μm. (B) Fluorescent gels showing Halo-ATG9A (JFX650, top) and SNAP-LC3B (JFX650, middle) labeling and Western blot showing probed with an LC3 antibody showing the exclusive expression of SNAP-LC3B in genome-edited cells. (C) Micrographs of cells expressing Halo-ATG9A (JFX650) and SNAP-LC3B (JF503; top, scale bar = 10 µm) and kymograph of a SNAP-LC3B spot showing no accumulation of Halo-ATG9A (one frame per second, frames 79–360). (D) Fluorescent gel of SNAP-LC3B (JFX650) after cell starvation and treatment with bafilomycin, demonstrating lipid conjugation of SNAP-LC3B (bottom band). (E) Fluorescence gel and Western blots demonstrating successful ATG9A gene knockout from cells expressing Halo-ATG2A. The ATG9A knock-out cells accumulate P62, indicating impaired autophagy. (F) Micrographs showing a decrease of Halo-ATG2A foci when ATG9A is knocked out (scale bar = 10 μm). Halo-ATG9A does not form detectable foci under EBSS starvation (right panel). (G) Histograms of Halo-ATG2A foci lifetime in parental and ATG9A knock-out cells (EBSS, 1 h). (H) Quantification of the number of foci formed per cell over the course of an hour by Halo-ATG2A in control and ATG9A knock-out cells. Data represent mean ± SD of three biological replicates (20–30 cells per replicate). A two-tailed t test was used for statistical analysis (*P < 0.05). Source data are available for this figure: SourceData F7.
Representative live-cell foci imaging movies of EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG9A and SNAPtag-LC3B. Right panel is a merge of Halo-ATG9A (magenta) and SNAP-LC3B (green). Cells were labeled with JFX650 HaloTag ligand and JF503 SNAPtag ligand, and images were acquired at one frame per second.
Representative live-cell foci imaging movies of EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG9A and SNAPtag-LC3B. Right panel is a merge of Halo-ATG9A (magenta) and SNAP-LC3B (green). Cells were labeled with JFX650 HaloTag ligand and JF503 SNAPtag ligand, and images were acquired at one frame per second.
Representative live-cell foci imaging movies of EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG2 (left), 3xFLAG-HaloTagged ATG2 ATG9A knockout (right). Cells were labeled with JF646 HaloLigand, and images were acquired at four frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
Representative live-cell foci imaging movies of EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG2 (left), 3xFLAG-HaloTagged ATG2 ATG9A knockout (right). Cells were labeled with JF646 HaloLigand, and images were acquired at four frames per minute. 300 × 300 pixels with a pixel size of 0.27 μm.
To resolve the discrepancy in the localization patterns of Halo-ATG9A and RFP-ATG9A, previously used by others (Karanasios et al., 2016; Orsi et al., 2012), we imaged Halo-ATG9A in cells transiently expressing RFP-ATG9A 48 h after transfection. Surprisingly, minimal colocalization of Halo-ATG9A and RFP-ATG9A was observed immediately after labeling with JFX650, whereas RFP-ATG9A formed large structures that colocalized with LAMP1 (Fig. 8, A–E; and Video 38). However, RFP-ATG9A, Halo-ATG9A, and LAMP1 all colocalized 24 h after labeling the HaloTag with JFX650 (Fig. 8, A–E; and Video 38). These observations demonstrate that RFP-ATG9A almost exclusively marks lysosomes. In contrast, the signal derived from Halo-ATG9A initially does not colocalize with lysosomes but accumulates in lysosomes 24 h after HaloTag labeling. There are two possible explanations for this observation. First, at steady state, ATG9A is enriched in lysosomes and the HaloTag ligand fails to react with Halo-ATG9A in this acidic compartment. In this case, the HaloTag signal that accumulates would represent ATG9A molecules that were labeled elsewhere and trafficked to the lysosome over time. Alternatively, little ATG9A is found in lysosomes and the fluorescent signal that accumulates over time results from the buildup of proteolysis-resistant fluorescent HaloTag molecules. Importantly, it was recently demonstrated that Halo-LC3 is partially degraded in lysosomes, causing labeled proteolysis-resistant HaloTag protein to accumulate within the lysosome (Yim et al., 2022). It is also well established that RFP fluorescent signal does not get quenched in lysosomes (Kaizuka et al., 2016; Yim et al., 2022). To analyze the trafficking of ATG9A and to determine whether the HaloTag fused to ATG9A is resistant to degradation, we generated a cell line that expressed Halo-ATG9A under the control of a tetracycline-inducible promotor in an ATG9A knock-out background. This approach allowed us to control both Halo-ATG9A expression and fluorescent labeling. We induced Halo-ATG9A expression for 24 h and imaged Halo-ATG9A immediately after labeling. Under these conditions, Halo-ATG9A localized exclusively to the ER (Fig. S5, A and B). In contrast, 24 h after labeling (48 h after expression induction), Halo-ATG9A localized to large foci resembling lysosomes (Fig. S5 B). Continuous Halo-ATG9A expression led to a similar result, although the enrichment of Halo-ATG9A on the ER immediately after labeling was less pronounced (Fig. S5 B). Importantly, RFP-ATG9A also accumulated in the ER when imaged 24 h after transfection (Fig. S5, D and E). These observations demonstrate that when ATG9A expression is induced it is initially enriched in the ER before trafficking to other subcellular compartments.
ATG9 compartments that co-localize with LC3 foci are lysosomes. (A) Micrographs showing the cellular distribution of transiently expressed RFP-ATG9A, endogenous Halo-ATG9A, and lysosomes, marked with mNeon-LAMP1. Experiments were performed immediately (upper panel) and 24 h (bottom panel) after labeling Halo-ATG9A. Scale bar = 10 μm. (B–E) Quantification of Halo-ATG9A and RFP-ATG9A tracks colocalized with mNeon-LAMP1. The number of cells (n) is indicated in each graph in the figure panels. Top schemes depict which signals were used to calculate the fractions in the corresponding graph. The marker represents mean ± 95% confidence interval. Letters indicate a statistically homogeneous group established by ANOVA (P < 0.05) followed by Bonferroni post-hoc test. (F) Micrographs showing the cellular distribution of endogenous edited SNAP-LC3 and Halo-ATG9A, and lysosomes, marked with LysoTracker. Experiments were performed immediately (upper panel) and 24 h (bottom panel) after Halo-ATG9A labeling. Scale bar = 10 µm. (G) Kymographs showing colocalization of SNAP-LC3/Halo-ATG9A immediately and 24 h after Halo-ATG9A labeling. (H) Number of co-diffusing Halo-ATG9A and SNAP-LC3 foci immediately and 24 h after Halo-ATG9A labeling. The marker represents mean ± 95% confidence interval (*P < 0.05). The number of data points (n) is indicated in each graph in the figure panels. (I) Fraction of colocalized SNAP-LC3 and Halo-ATG9A foci that also colocalized with lysosomes marked with LysoTracker. The marker represents mean ± 95% confidence interval.
ATG9 compartments that co-localize with LC3 foci are lysosomes. (A) Micrographs showing the cellular distribution of transiently expressed RFP-ATG9A, endogenous Halo-ATG9A, and lysosomes, marked with mNeon-LAMP1. Experiments were performed immediately (upper panel) and 24 h (bottom panel) after labeling Halo-ATG9A. Scale bar = 10 μm. (B–E) Quantification of Halo-ATG9A and RFP-ATG9A tracks colocalized with mNeon-LAMP1. The number of cells (n) is indicated in each graph in the figure panels. Top schemes depict which signals were used to calculate the fractions in the corresponding graph. The marker represents mean ± 95% confidence interval. Letters indicate a statistically homogeneous group established by ANOVA (P < 0.05) followed by Bonferroni post-hoc test. (F) Micrographs showing the cellular distribution of endogenous edited SNAP-LC3 and Halo-ATG9A, and lysosomes, marked with LysoTracker. Experiments were performed immediately (upper panel) and 24 h (bottom panel) after Halo-ATG9A labeling. Scale bar = 10 µm. (G) Kymographs showing colocalization of SNAP-LC3/Halo-ATG9A immediately and 24 h after Halo-ATG9A labeling. (H) Number of co-diffusing Halo-ATG9A and SNAP-LC3 foci immediately and 24 h after Halo-ATG9A labeling. The marker represents mean ± 95% confidence interval (*P < 0.05). The number of data points (n) is indicated in each graph in the figure panels. (I) Fraction of colocalized SNAP-LC3 and Halo-ATG9A foci that also colocalized with lysosomes marked with LysoTracker. The marker represents mean ± 95% confidence interval.
Representative live-cell foci imaging movies of EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG9A, RFP-ATG9A, and LAMP-mNeonGreen. Cells were labeled with JFX650 HaloTag ligand, and images were acquired at one frame per second. Green: Halo-ATG9A; red: RFP-ATG9A; blue: Lamp1-mNeon.
Representative live-cell foci imaging movies of EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG9A, RFP-ATG9A, and LAMP-mNeonGreen. Cells were labeled with JFX650 HaloTag ligand, and images were acquired at one frame per second. Green: Halo-ATG9A; red: RFP-ATG9A; blue: Lamp1-mNeon.
Pulse-chase analysis of Halo-ATG9 localization and degradation. (A) Representative micrographs of U2OS and Halo-ATG9A knockout (KO) cells pulse-labeled with JFX650 HaloTag ligand at 0 and 24 h after labeling to demonstrate the level of background fluorescence in the absence of a Halo-ATG9A. Left panel: fluorescence signal; right panel: bright field. (B) Representative micrographs of JFX650 labeled Halo-ATG9A at 0 h (top) and 24 h (bottom) after labeling in the endogenously edited cell line (left) or in the stable tetracycline-inducible Halo-ATG9A add-back in ATG9A knock-out cells (right two panels). ATG9A knock-out cells (same images as in A) were included as a comparison to assess non-specific background signal. (C) Fluorescence gel showing time-dependent cleavage of Halo-ATG9A in cells exposed to a single doxycycline pulse (P) or continuously grown in the presence of doxycycline (C), with progressive accumulation of a lower (∼34 kD) fluorescent band corresponding to the HaloTag protein. (D) Representative micrographs showing the overlap between the endoplasmic reticulum (marked with GFP-SEC61) and RFP-ATG9A. (E) Representative micrographs showing the overlap between Halo-ATG9A and transiently expressed RFP-ATG9A. Source data are available for this figure: SourceData FS5.
Pulse-chase analysis of Halo-ATG9 localization and degradation. (A) Representative micrographs of U2OS and Halo-ATG9A knockout (KO) cells pulse-labeled with JFX650 HaloTag ligand at 0 and 24 h after labeling to demonstrate the level of background fluorescence in the absence of a Halo-ATG9A. Left panel: fluorescence signal; right panel: bright field. (B) Representative micrographs of JFX650 labeled Halo-ATG9A at 0 h (top) and 24 h (bottom) after labeling in the endogenously edited cell line (left) or in the stable tetracycline-inducible Halo-ATG9A add-back in ATG9A knock-out cells (right two panels). ATG9A knock-out cells (same images as in A) were included as a comparison to assess non-specific background signal. (C) Fluorescence gel showing time-dependent cleavage of Halo-ATG9A in cells exposed to a single doxycycline pulse (P) or continuously grown in the presence of doxycycline (C), with progressive accumulation of a lower (∼34 kD) fluorescent band corresponding to the HaloTag protein. (D) Representative micrographs showing the overlap between the endoplasmic reticulum (marked with GFP-SEC61) and RFP-ATG9A. (E) Representative micrographs showing the overlap between Halo-ATG9A and transiently expressed RFP-ATG9A. Source data are available for this figure: SourceData FS5.
To analyze Halo-ATG9A stability, we induced Halo-ATG9A expression with a single doxycycline pulse or grew cells continuously in the presence of doxycycline and collected samples immediately, 24, and 48 h after Halo-ATG9A labeling. We detected a continual accumulation of fluorescently labeled HaloTag in cells stably overexpressing Halo-ATG9A (Fig. S5 C), indicating that Halo-ATG9A, like Halo-LC3, is partially degraded over time releasing fluorescently labeled HaloTag protein. This result strongly suggests that ATG9A is degraded after it localizes to the lysosome and that the fluorescent signal derived from Halo-ATG9A that accumulates in lysosomes over time represents HaloTag protein resistant to proteolysis.
To dissect the relationship between ATG9A- and LC3-positive compartments, and the lysosome, we simultaneously imaged Halo-ATG9A, SNAP-LC3B, and lysosomes marked with LysoTracker (Fig. 8, F and G). Immediately after Halo-ATG9A labeling, we detected a limited number of SNAP-LC3 foci that colocalized with ATG9A-positive compartments (Fig. 8, G and H; and Video 39). In contrast, 24 h after Halo-ATG9A labeling, a large number of LC3 trajectories colocalized with Halo-ATG9A tracks (Fig. 8, G and H; and Video 39). Finally, we determined that >90% of the ATG9A foci that codiffuse with LC3 tracks are also marked by LysoTracker both immediately and 24 h after Halo-ATG9A labeling (Fig. 8 I). Together, these results demonstrate that ATG9A is not detectably enriched at newly formed autophagosomes, and colocalization of ATG9A with LC3B exclusively occurs within the lysosome.
Representative live-cell foci imaging movies of EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG9A, SNAPtag-LC3B, and treated with LysoTracker Deep Red. Cells were labeled with JFX554 HaloTag ligand and JF503 SNAPtag ligand, and images were acquired at one frame per second. Green: Halo-ATG9A; red: SNAP-LC3B; blue: LysoTracker.
Representative live-cell foci imaging movies of EBSS-treated U2OS cells expressing 3xFLAG-HaloTagged ATG9A, SNAPtag-LC3B, and treated with LysoTracker Deep Red. Cells were labeled with JFX554 HaloTag ligand and JF503 SNAPtag ligand, and images were acquired at one frame per second. Green: Halo-ATG9A; red: SNAP-LC3B; blue: LysoTracker.
Discussion
The experiments presented in this study systematically and quantitatively assess the formation of autophagosomes in human cells. Our work provides new mechanistic insights into the initiation and maturation of autophagosomes and the collection of cell lines we have created expressing HaloTagged autophagy factors from their endogenous loci represents a tremendously valuable tool for future studies of autophagosome formation in human cancer cells.
The role of ATG9 vesicles in autophagosome biogenesis
ATG9A/B are the only known transmembrane proteins that are critical for autophagosome formation (Guardia et al., 2020). ATG9A/B resides in lipid vesicles that traffic to and from the TGN and endosomes (Young et al., 2006). Upon starvation, the ULK1/2 complex controls the redistribution of ATG9A vesicles from the TGN to the phagophore site (Orsi et al., 2012). Several recent structural and biochemical studies have demonstrated that ATG9A forms a trimeric membrane pore that functions as a lipid scramblase, passively exchanging lipids between the inner and outer leaflets of lipid bilayers (Guardia et al., 2020; Matoba et al., 2020; Noda, 2021). These observations have led to a model in which ATG9A-containing vesicles constitute the platform upon which the autophagosome is formed via ATG2A-mediated lipid transfer from other membrane sources such as the ER. Work by others has shown that ATG9A-containing proteo-liposomes or immunopurified ATG9A vesicles are sufficient for reconstituted nucleation of the autophagosome (Chang et al., 2021a; Chang et al., 2021b; Olivas et al., 2022,Preprint; Sawa-Makarska et al., 2020). While these in vitro observations are consistent with the hypothesis that ATG9A vesicles are the platform for autophagosome formation, the evidence that ATG9A vesicles transform into autophagosomes in cells is limited.
The work presented here revealed that ATG9A is the only autophagy factor analyzed that does not detectably accumulate at sites of autophagosome biogenesis, yet it is essential for the formation of ATG2A-bound phagophores in U2OS cells. In addition, our results indicate that RFP-ATG9A and Halo-ATG9A accumulate in lysosomes. Therefore, the previously reported association between forming autophagosomes marked by LC3- and ATG9A-positive structures likely represents autophagosomes contacting lysosomes (Bright et al., 2005; Jahreiss et al., 2008). To assess ATG9A localization in the context of autophagosome formation in future experiments, we believe it will be critical to include a lysosomal marker to clearly distinguish lysosomes and autophagosomes. It is important to note that only a small fraction of lysosomes (∼5%, Fig. 8 D) colocalize with Halo-ATG9A, which is consistent with previous observations made using immunofluorescence to detect ATG9A and LAMP1 (Kannangara et al., 2021; Popovic and Dikic, 2014). The trafficking of ATG9A to the lysosome and the accumulation of cleaved, proteolysis-resistant HaloTag protein suggests that ATG9A can be degraded after autophagosomes fuse with the lysosome. Importantly, the lack of accumulation of ATG9A at sites of autophagosome formation is consistent with a lipid vesicle containing a small number of ATG9A molecules (not detectable by conventional fluorescence microscopy) forming the platform for phagophore initiation (Fig. 7 F).
ATG2 recruitment leads to phagophore immobilization
Once an ATG9A vesicle has been specified as a seed for autophagosome formation, it has to expand, engulf cargo, and eventually close into a mature autophagosome. ATG2A, a proposed direct interactor of ATG9A (Gómez-Sánchez et al., 2018; Tang et al., 2019), forms robust foci at autophagosomes, potentially by WIPI3/4-mediated recruitment of ATG2A to the phagophore (Chowdhury et al., 2018; Otomo et al., 2018; Ren et al., 2020). ATG2A is an extended barrel-shaped protein that can form tethers between lipid vesicles and transfer phospholipids between them (Chowdhury et al., 2018; Noda, 2021; Tang et al., 2019; Valverde et al., 2019). Therefore, it is thought that ATG2A is the critical factor for transferring phospholipids from a donor lipid source to the growing phagophore.
Our imaging methodologies allowed us to quantitatively analyze the recruitment of ATG2A to the phagophore. Strikingly, the number of ATG2A foci formed was approximately half of the number formed by all other autophagy factors tested except for WIPI2. In addition, the diffusion coefficient distribution of ATG2A showed a single slowly moving population, while all other factors contained two populations, one which was comparable with ATG2A positive foci and a second more rapidly diffusing population. Importantly, the number of slowly diffusing autophagy factor foci was similar to the number of ATG2A foci observed, suggesting that they reflect the same population of phagophores. These observations demonstrate that ATG2A marks a subset of phagophores with distinct biophysical properties. Consistent with this model, the recruitment of ATG2A to foci formed by the autophagosome initiation factor ATG13 leads to a reduction in the diffusion rate of these structures. We propose that the reduced diffusion coefficient of the ATG2A-positive phagophore population is the result of ATG2A-mediated tethering of the phagophore to donor membranes. The rapidly diffusing population of autophagy factors foci (D = 0.01 µm2/s) is comparable with the diffusion coefficient of static ATG9A vesicles measured in our single-molecule imaging experiments (D = 0.02 µm2/s). Immobilization of an ATG9A vesicle by tethering it to other cellular membranes via ATG2A would be expected to reduce its diffusion coefficient, which is consistent with our observations. For instance, yeast ATG9A vesicles directly involved in the formation of autophagosomes show distinct diffusive behavior than vesicles freely diffusing in the cytosol (Yamamoto et al., 2012). Our results demonstrate that inhibition of phagophore closure by knockdown at CHMP2A leads to the enrichment of ATG13 foci, which were tightly associated with the ER. This observation is consistent with the accumulation of phagophores that have undergone expansion by ATG2A-mediated lipid transfer but remain tethered to the ER due to their inability to close and form mature autophagosomes. In contrast, it is not immediately apparent why ATG2A recruitment should reduce the mobility of the phagophore if it was formed by an alternative ER-remodeling mechanism. In addition, our observations suggest that the ULK1 complex, PI3-kinase, the LC3 lipidation machinery, and to a smaller degree WIPI2 can be recruited to ATG9A vesicles prior to their tethering to donor membranes by ATG2A.
Together these observations demonstrate that the recruitment of ATG2A marks the transition from an ATG9A vesicle into a phagophore that can expand and mature into an autophagosome.
ULK1 is the limiting factor for the assembly of the ULK1 complex
A key open question is how ATG9A vesicles are specifically selected to transform into autophagosomes. The ULK1 complex, composed of ULK1, ATG13, FIP200, and ATG101, is essential in the initiation of autophagy (Dikic and Elazar, 2018; Lin and Hurley, 2016; Yu et al., 2018). Work by others has demonstrated that ATG13 and FIP200 are required for the localization of ULK1 to the phagophore (Chang et al., 2021a; Ganley et al., 2009; Kannangara et al., 2021; Shi et al., 2020). A key function of the ULK1 complex is the activation of the PI3-kinase VPS34 at sites of autophagosome formation to promote the production of PI3P (Mercer et al., 2018; Park et al., 2016; Russell et al., 2013). In addition, it has been shown that ULK1 complex recruitment to the phagophore is reenforced by the association of ATG13 with PI3P (Karanasios et al., 2013a). Therefore, the ULK1 complex and PI3-kinase form a positive feedback loop required to initiate autophagosome formation (Ohashi, 2021).
Our observations demonstrate that ATG13 is ninefold more abundant than ULK1, therefore ULK1 kinase is the limiting factor in ULK1 complex assembly. Despite the differences in protein abundance, similar amounts of ULK1 and ATG13 are recruited to phagophores, suggesting they act in concert during autophagosome biogenesis. Interestingly, clinical studies have shown that ULK1 overexpression is associated with a poor patient prognosis (Lu et al., 2018; Yun et al., 2015), suggesting that ULK1 kinase levels control the overall activity of the ULK1 complex, confirming that ULK1 is a promising clinical target (Martin et al., 2018).
The initiation of autophagosome formation is inefficient
Previous work by others has largely focused on determining the number of cytoplasmic autophagy factor foci as a measurement of autophagosome formation, and a limited number of studies have analyzed the lifetime of foci formed by autophagy proteins (Dalle Pezze et al., 2021; Karanasios et al., 2013a; Karanasios et al., 2013b). Our dual-color imaging of the tagged autophagy proteins and P62, GABARAPL1, or LC3, which mark phagophores that are in the process of cargo sequestration and mature autophagosomes, allowed us to systematically analyze the kinetics and efficiency of phagophore maturation. Surprisingly, ∼90% of the foci formed by ULK1, ATG13, WIPI2, ATG5, and ATG16 do not proceed to accumulate detectable quantities of P62, GABARAPL1, or LC3. In addition, autophagy protein foci that do not colocalize with P62, GABARAPL1, or LC3 are very short-lived (P62negative mean = 33 s; LC3negative mean = 25 s, Halo-ATG13 GABARAPL1negative mean = 24 s) compared with foci that proceed to accumulate P62, GABARAPL1, or LC3 (P62positive mean = 107 s; LC3positive mean = 125 s, Halo-ATG13 GABARAPL1positive mean = 80 s). These observations suggest that these short-lived foci represent aborted autophagosomes that initiate the accumulation of the ULK1 complex, WIPI2, and the LC3 lipidation machinery, but rapidly disassemble rather than maturing into an autophagosome. The use of three distinct markers, the adaptor P62 and ATG8-like family proteins LC3 and GABARAPL1, increases our confidence that these observations are a general feature of autophagosome formation rather than a feature of a specific subset of autophagosomes. It is important to note that our observation that 90% of ULK1, ATG13, WIPI2, ATG5, and ATG16 foci do not proceed to accumulate P62, GABARAPL1, or LC3 is likely an overestimation. Our methodology to track the adaptor protein and autophagy factor foci and to determine their colocalization is very stringent and we likely fail to detect all colocalized trajectories. The close correspondence of the lifetimes of autophagy protein foci that colocalizes with P62 and LC3 strongly suggests that both LC3 and P62 mark the same structure: bona fide autophagosomes.
Strikingly, our analysis of the maturation of ATG2A foci revealed that they are twice as likely to colocalize with P62 compared with the other autophagy factors analyzed (Fig. 6 D). As discussed above, the number of ATG2A foci formed is approximately half of the number formed by ULK1, ATG13, ATG5, and ATG16. Therefore, the total number of foci formed by all autophagy factors analyzed that proceed to colocalize with an adaptor protein is comparable. In addition, the fraction of ATG13 foci that proceed to recruit ATG2A (12%) and adaptor proteins (10–16%) is similar. In addition, the lifetime of ATG13 foci that recruit ATG2A or P62/LC3/GABARAPL1 is comparable (80–100 s), pointing that the colocalization of ATG13 and ATG2A marks phagophores that will complete maturation. Together, these observations demonstrate that the majority of autophagy protein foci do not mature to form autophagosomes and further support our model that the recruitment of ATG2A and the concomitant tethering to donor membranes is a critical step in committing ATG9A vesicles toward maturation into an autophagosome (Fig. 9).
Model for the biogenesis of the autophagosome . Upon autophagy induction, a ULK1 complex–PI3-kinase feedback loop initiates the assembly of autophagy proteins on an untethered ATG9A vesicle. Approximately half of these prephagophores proceed to be tethered to cellular membranes via ATG2A. Only 20% of the ATG2A-positive phagophores mature to a point where the ATG8 family proteins (LC3, GABARAPL1) and the cargo adaptor P62 are detectably recruited, while the majority (80%) of autophagy protein foci disassemble without forming mature autophagosomes. The formation and growth of a full autophagosome take about 90–145 s. Upon closure, the autophagy proteins dissociate, and the mature autophagosome is untethered and delivered to lysosomes (>60 s).
Model for the biogenesis of the autophagosome . Upon autophagy induction, a ULK1 complex–PI3-kinase feedback loop initiates the assembly of autophagy proteins on an untethered ATG9A vesicle. Approximately half of these prephagophores proceed to be tethered to cellular membranes via ATG2A. Only 20% of the ATG2A-positive phagophores mature to a point where the ATG8 family proteins (LC3, GABARAPL1) and the cargo adaptor P62 are detectably recruited, while the majority (80%) of autophagy protein foci disassemble without forming mature autophagosomes. The formation and growth of a full autophagosome take about 90–145 s. Upon closure, the autophagy proteins dissociate, and the mature autophagosome is untethered and delivered to lysosomes (>60 s).
Endogenous tagging combined with automated tracking reveals the maturation kinetics of autophagosomes in human cancer cells
Foci formed by autophagy proteins have been extensively used to infer the lifetime of autophagosomes (Dalle Pezze et al., 2021; Fujita et al., 2008; Karanasios et al., 2013a; Karanasios et al., 2016; Stavoe et al., 2019). Current estimates of autophagosome lifetime rely upon manual identification and analysis of foci formed by stably expressed, fluorescently tagged autophagy protein transgenes (Dalle Pezze et al., 2021; Karanasios et al., 2013a). Using our cell lines that express fluorescently tagged autophagy proteins from their endogenous loci avoids known artifacts due to protein overexpression and provides consistent experimental conditions to directly compare individual autophagy factors. In addition, our observations described above demonstrate that a large fraction of autophagy protein foci do not mature into autophagosomes. To accurately analyze the kinetics of autophagosome formation it is, therefore, necessary to combine the detection of autophagy factor foci with a marker of mature autophagosomes, such as P62, LC3B, or GABARAPL1. In addition, our fully automated detection and particle tracking approach provides an unbiased method that generates a tremendous amount of data.
Our observations demonstrated that all autophagy factors analyzed accumulated ∼40 s prior to P62 recruitment. This suggests that ULK1, ATG13, ATG5, ATG16, WIPI2, and ATG2A are all recruited in rapid succession (Fig. 9). Our current imaging approach does not have the time resolution required to determine a precise recruitment order of the autophagy factors analyzed. In general, we note that previous analysis of foci reports longer-lasting values than our data show. For instance, previous analysis of ATG5 and ATG13 recruitment kinetics showed ATG5 foci lasting 480 s and ATG13 foci, which lasted for 200 s on average (Dalle Pezze et al., 2021; Karanasios et al., 2013a; Koyama-Honda et al., 2013) compared with the average lifetime of 102 and 82 s that we observed in our experiments, respectively. The potential discrepancy may be due to the detection method, expression approach, or cell line differences. Once P62 accumulation is detectable, the autophagy factors remain associated with the phagophore for 50–95 s, and the P62 signal is detected for 40–100 s after the autophagy proteins have dissociated from the autophagosome (Fig. 9). This demonstrates that the autophagy factors we analyzed in this study are recycled and therefore must be largely associated with the cytosol-facing membrane of the autophagosome. This is further supported by the recent observation that the HaloTag protein is resistant to lysosomal degradation (Yim et al., 2022). If the HaloTagged autophagy factors were sequestered inside the autophagosome, we would expect to observe an accumulation of fluorescence signal in the lysosome, which was only observed for Halo-LC3. We assume that the dissociation of the core autophagy factors analyzed in this work is closely correlated with the completion of phagophore expansion and its closure to form a mature autophagosome.
Importantly, we measured similar kinetics when using GFP-LC3, GFP-GABARAPL1, and GFP-P62 expressed by viral transduction and/or stably expressed from a single-genomic insertion as a marker for autophagosomes, which confirms that the P62 signals detected represent autophagosomes rather than protein aggregates marked by P62. LC3 accumulated more rapidly (∼16 s) after autophagy protein foci formation compared with P62 (∼40 s), consistent with the LC3-dependent recruitment of P62 to phagophores (Ichimura et al., 2008; Kirkin and Rogov, 2019; Noda et al., 2008; Pankiv et al., 2007). Overall, similar to autophagy protein foci length, the lifetime of LC3, GABARAPL1, and P62 positive autophagosomes is substantially shorter than previously reported in the literature (Dalle Pezze et al., 2021; Karanasios et al., 2013a; Karanasios et al., 2013b; Fig. 9). We assume that the disappearance of the adaptor or the ATG8 family protein signal is a consequence of the fusion of the autophagosome with the lysosome, which quenches the fluorescent signal of GFP. The quenching of the GFP signal suggests that the average time from the completion of autophagosome formation to autophagosome–lysosome fusion is ∼66 s (ranging from 38 to 101 s). In total, these experiments demonstrate that the average timeframe of autophagosome biogenesis is 110 s (ranging from 90 to 145 s) in U2OS cells (Fig. 9). Importantly, the autophagosome lifetime we determined using automated tracking of thousands of autophagy factor foci was comparable in all HaloTagged autophagy protein cell lines, raising our confidence that we are measuring the lifetime of bona fide phagophores that mature into autophagosomes. The similarity in the overall lifetimes measured also indicates that the autophagosome formation kinetics are not adversely affected by the HaloTag fusion proteins. Finally, our observations demonstrate that all autophagy factors analyzed are removed from autophagosomes prior to the fusion of the autophagosome with the lysosome. This suggests that the signal that leads to the recruitment of these proteins, likely the enrichment of PI3P, is removed from the autophagosome once it has closed and is fully matured (Fig. 9). In total, these experiments precisely define the overall lifetime of autophagosomes, the timing of phagophore maturation, and the time it takes for a mature autophagosome to fuse with a lysosome.
Altogether, our work significantly expands our mechanistic understanding of autophagosome formation and provides a sophisticated experimental framework and toolkit for future quantitative studies of autophagy in human cancer cells.
Materials and methods
Cell lines
All cell lines used in this study were derived from human bone osteosarcoma epithelial cells (U2OS, ATCC HTB-96) and grown in RPMI cell culture media supplemented with 10% FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin at 37°C with 5% CO2.
Plasmid construction and genome editing
The autophagy genes were edited at their endogenous 5′-end, except for ULK1, which was edited at the 3′-end. For the 5′-end editing, the 3XFLAG-HaloTag donor plasmid was generated according to the procedure described by Xi et al. (Schmidt et al., 2016; Xi et al., 2015). Between the HaloTag and the start codon of the tagged gene, a linker sequence including a TEV protease cleavage site was inserted. The homology arms and HaloTag fragments were ligated into pFASTBac linearized with HpaI using Gibson Assembly (NEB). All single-guide RNAs (sgRNAs) were cloned into pX330 Cas9 donor plasmid (Cong et al., 2013). Cells were transfected using the FuGENE HD transfection reagent (Promega Co.); after 72 h of transfection, the cells were selected for 4–6 d with puromycin (1 µg/ml). Following selection with puromycin, cells were transfected with an eGFP-CRE recombinase plasmid. Cells were then labeled with the HaloTag ligand JF646 (Janelia) and sorted with FACS using the eGFP/JF646 positive signals. In single-cell clones, homologous recombination was confirmed by PCR and Sanger sequencing. Halo-NES, SNAP-SEC61 (in the U2OS parental cell line), LAMP1-mNeon (Lamp1-mNeonGreen was a gift from Dorus Gadella; plasmid # 98882; Addgene; http://n2t.net/addgene:98882; RRID:Addgene_98882; Chertkova et al., 2017,Preprint), and the GFP-tagged ATG8 family proteins (LC3, GABARAPL1), and the adaptor P62 (in the genome-edited Halo-ATG13 cell line) were stably expressed by introducing the coding sequence at the AAVS1 safe-harbor locus (PPP1R12C; Qian et al., 2014). All PCR oligonucleotides and sgRNA sequences are listed in Table S1.
Western blotting
The protein samples were separated on 4–15% or 4–20% TGX stain-free polyacrylamide gels (Biorad), followed by standard Western blotting procedures. The primary and secondary antibodies and dilutions used are listed in Table S1. Clarity western ELC Substrate (BioRad) was used to generate a chemiluminescence signal detected with a ChemiDoc imaging system (BioRad). All Western blot quantification was carried out using the ImageQuant software (Cytiva). Expression levels of the edited cell lines were estimated by comparing the chemiluminescence signal with the parental U2OS after normalization with the stain-free signal.
LC3 lipidation assay
Cells were grown in 24-well plates at 60–70% confluency and treated with 100 nM rapamycin and 100 nM rapamycin + 100 nM bafilomycin for 2 h; a control in regular RPMI media was also performed. After treatment, the cells were harvested with 150 μl sample buffer and 20 μl was loaded in a 4–20% Bis-Tris gel. Gels were transferred into polyvinylidene difluoride blot and treated against LC3 antibody according to the protocol supplied by the provider (CellSignaling). The chemiluminescence signal was detected with a ChemiDoc imaging system (BioRad), and the signal corresponding to LC3-I and LC3-II was quantified using the ImageQuant software (Cytiva).
Expression and purification of recombinant His-3XFlag-HaloTag
The His-3XFlag-HaloTag construct was kindly provided by Dr. Youmans and Dr. Cech at the University of Colorado Boulder, Boulder, CO, USA. HaloTag protein was expressed and purified from Escherichia coli BL21(DE3) cells grown in Luria Broth medium. Cells were grown at 37°C and expression was induced at OD ∼0.5–0.8 with 1 ml of IPTG (1M). Upon induction, the temperature was decreased to 18°C and cells were grown overnight (16 h) before harvesting. Cells were centrifugated, the pellet resuspended in wash buffer (50 mM sodium phosphate buffer, pH 8.0, 300 mM sodium chloride, 10 mM imidazole, 5 mM β-mercaptoethanol), and sonicated for 2 min (40% amplitude, 90 s of sonication, 10-s pulses, 20-s pause, Fisherbrand Model 505, 0.5-inch tip). The lysate was then centrifuged (40,000 g and 4°C for 30 min) and the supernatant was loaded to 1 ml of fast flow nickel Sepharose (Cytiva) and incubated under gentle rotation for 1 h at 4°C. Beads were then washed three times with 6 ml of wash buffer, and elution was performed using elution buffer (0 mM sodium phosphate buffer, pH 7.0, 300 mM sodium chloride, 250 mM imidazole, 5 mM β-mercaptoethanol). The eluate was subjected to further purification using size-exclusion chromatography on a Superdex S75 column in isocratic mode (50 mM Tris, pH 7.5, 150 mM potassium chloride, 1 mM DTT). Protein was concentrated to 1 mg/ml, supplemented with 50% glycerol, flash-frozen, and stored at −80°C. To fluorescently label the 6xHIS-3xFLAG-HaloTag, the protein was incubated with a twofold excess of JF646 HaloTag-ligand overnight at room temperature. Excess fluorescent dye was removed by size exclusion chromatography using a Superdex S75 column in isocratic mode (50 mM Tris, pH 7.5, 150 mM potassium chloride, 1 mM DTT). The protein concentration and labeling efficiency were determined by absorption spectroscopy using ε280nm = 41,060 M−1 cm−1 for the 6xHIS-3xFLAG-HaloTag and ε646nm = 152,000 M−1 cm−1 for the JF646 fluorescent dye (Grimm et al., 2015).
In-gel fluorescence absolute protein quantification
His-3XFlag-HaloTag was labeled by combining 15 nmol of JF646 (30 μl from 0.5 mM DMSO resuspended stock) with 0.2 mg recombinant protein in 200 μl of buffer and incubated at RT for 30 min. The labeled protein was then purified by size exclusion on an S75 column. Fractions were combined and concentrated in Vivaspin 10 kD columns and glycerol was added for a final concentration of 20%. Vials were snap-frozen in liquid nitrogen and stored at −80°C. The final concentration was measured using a 40-μl microcuvette in a fluorescence spectrophotometer. Protein concentration was calculated using an extinction coefficient ε646nm = 152,000 M−1 cm−1 (Grimm et al., 2017). The labeled HaloTag protein was then serial diluted in sample buffer and a total of 37.5 fmol down to 0.29 fmol was added to cell lysates that were prepared using direct lysis in sample buffer and diluted from 150,000 cells/ml to 75,000 cells/ml in 25,000 cell/ml increments. The standards were then aliquoted into individual use tubes, boiled, snap-frozen in liquid nitrogen, and stored at −80°C. For the in-gel fluorescence, 80,000 cells were seeded in a 24-well plate. Cells were harvested by adding 30 μl of 1xSDS PAGE sample buffer (Biorad). The protein and the standard were loaded and separated on 4–15% TGX stain-free polyacrylamide gels (Biorad). Fluorescence was detected using the Cy5.5 settings on a ChemiDoc imaging system (BioRad). The gels were quantified using the ImageQuant software (Cytiva). We observed that the band patterns of each protein affected the quantification of both in-gel fluorescence (density of bands) and Western blots (density of bands along with biased transfer). To correct for these biases, we cleaved the HaloTag using TEV protease to compare the fluorescence of the purified HaloTag to cleaved HaloTag and the endogenous proteins with cleaved autophagy proteins (Fig. S1 D and Fig. S3, F and G). For the TEV in‐gel fluorescence corrections, 120,000 cells were labeled for 30 min with 500 nM JF646, washed with PBS, harvested with 5 mM EDTA in PBS, and pelleted. Samples were lysed in 60 μl CHAPS lysis buffer (10 mM TRIS, pH 7.5, 1 mM MgCl2, 1 mM EGTA, 0.5% CHAPS, 10% glycerol) and 20 μl was incubated on ice with five units of TEV protease (P8112; New England Biolabs) for 30 min. Following TEV cleavage, 5 μl 6× SDS sample buffer was added to 25 μl of the digested and undigested samples and boiled for 5 min at 95°C. Samples were then separated by gel electrophoresis. JF646 intensity was measured using the ChemiDoc imaging system (BioRad) and cleaved HaloTag protein was compared to uncleaved full-length HaloTag autophagy factor fusion protein. For TEV-westerns, the protocol followed was similar, but the cells were not labeled and a 50% dilution per sample was added. Gels were transferred using the Trans-Blot Turbo system (with Turbo transfer buffer; BioRad) onto polyvinylidene difluoride or Nitrocellulose. Samples were blocked in 5% milk powder for 1 h, washed in PBS with 0.05% Tween-20, and left overnight in primary antibody. The following day, blots were washed and incubated in goat anti-mouse HRP (1:2,000; 31430; Invitrogen) and goat anti-rabbit HRP (1:2,000; 31460; Invitrogen), and chemiluminescence was measured. Finally, we calculated the cellular protein abundance by first determining the total number of HaloTagged proteins by comparing the in-gel fluorescence signal of the tagged autophagy factor with the HaloTag standard curve. We then measured the total number of cells loaded using the stain-free protein signal compared with the cell number standard curve. Dividing the total number of HaloTag molecules by the number of cells loaded resulted in total protein abundance with the units of molecules per cell. This measurement was then corrected by multiplication with the TEV correction factor (fluorescent signal of the full-length fusion divided by cleaved HaloTag signal) and the Western blot correction factor (Western blot signal of the full-length fusion protein divided by endogenous length protein signal). The protein abundance was computed for two different cell clones; the measurement error was propagated using SD.
Flow cytometry measurement of protein abundance
80,000 cells were plated into a 24-well plate. 24 h after seeding cells were then labeled with JF646 at 500 nM for 30 min in complete media and washed with 5-min incubation in media. Cells were then harvested using 5 mM EDTA in PBS (10 min at 37°C) and transferred to a 2 ml deep 96-well plate using a multichannel pipette. PFA fixation solution was added for a final concentration of 2% PFA in PBS and incubated for 10 min. Samples were then washed once with 1% BSA in PBS and filtered into a 96-well round-bottom plate using a nylon mesh with a 70 µm pore size. Analysis was performed on a Cytek Aurora (Cytek Biosciences) using a 96-well loader and unmixed using the default SpectroFlo software (Cytek Biosciences).
Immunofluorescence to detect LC3 foci in fixed cells
Halo-ATG5, Halo-ATG16, and Halo-WIPI2 cells were plated (200,000 cells) into an ethanol-sterilized 6-well plate containing 22X22 #1.5 glass coverslips and left overnight to adhere. Cells were then labeled with JF646 at 500 nM for 30 min in complete media and washed with 5-min incubation in media. Cells were then treated for 2 h with DMSO control, rapamycin (100 nM), or rapamycin with bafilomycin (100 nM). After 2 h of drug treatment, cells were washed in PBS and fixed in 4% PFA in PBS (10 min at room temperature). The fixed coverslips were then transferred to a humidified chamber and washed three times in PBS for 5 min and simultaneously blocked/permeabilized with 3% BSA in PBS with digitonin (50 μg/ml) for 30 min. Digitonin was added throughout the rest of the experiment for incubation steps. The primary antibody was diluted in 3% BSA in PBS, added at the indicated concentration, and incubated overnight. The samples were then washed three times in PBS and incubated with AlexaFlour conjugated secondary antibody (Invitrogen) for 1 h (1:500) in 3% BSA in PBS. The samples were washed three times in PBS and once in water and then flipped upside down onto microslides containing a drop of antifade mountant (ProLong Diamond, Life Technologies), dried for 10 min, and sealed with nail polish.
Live-cell microscopy
Live-cell imaging experiments were carried out using two distinct microscopes: an Olympus microscope (IX83) with a cellTIRF illuminator with four laser lines (100 mW 405 nm, 200 mW 488 nm, 300 mW 561 nm, and 140 mW 640 nm), and an X-Cite TURBO multiwavelength LED illumination system (Excelitas Technologies). The microscope is equipped with an environmental chamber (cellVivo) to control humidity, temperature, and CO2 level, a 100× TIRF oil-immersion objective (Olympus UApo N, NA = 1.49), a 60× TIRF oil-immersion objective (Olympus UPlanApo, NA = 1.50), a 60× oil immersion objective (Olympus PlanApo N, NA = 1.42), and the appropriate excitation and emission filters. For signal detection, this microscope uses two Andor iXon Ultra 897 EMCCD or two Hamamatsu Orca Fusion BT sCMOS cameras attached to a Twin-cam beamsplitter (Cairn Research). Alternatively, we used an i3 spinning-disk confocal microscope equipped with a CSU-W1 confocal spinning-disk system (Yokogawa), five laser lines (100 mW 445 nm, 150 mW 488 nm, 175 mW 515 nm, 160 mW 561 nm, and 140 mW 638 nm), a Prime 95B sCMOS camera (Photometrics), a 63× oil-immersion objective (Zeiss C Plan-Apo, NA = 1.4), and an incubation chamber to control humidity, temperature, and CO2 level. All live-cell imaging was carried out at 37°C under 5% CO2-containing humidified air.
Autophagy protein and LC3 foci quantification in living cells
Cells (10,000) were grown in 96-well optical plates and after 24 h transduced with GFP-LC3 encoding baculovirus particles using the viral BacMam 2.0 transfection reagent at a concentration of ∼0.25 × 108 viral particles/ml. After 24 h, cells were labeled in 100 nM JF646 in growth media for 10 min, followed by three washes with growth media, and finally a 5 min incubation in growth media at 37°C to remove unincorporated JF646 dye. A nuclear stain (Hoechst dye, diluted 1:10,000) was added during the last wash step. For starvation, cells were washed three times with PBS buffer before the addition of EBSS starvation media. Cells were imaged on the Olympus microscope using the Olympus PlanApo N, NA = 1.42, the Andor iXon 897 Ultra camera, the 630 nm LED at 100% power, the 475 nm LED at 30% power, and the 385 nm LED at 5% power. To avoid signal saturation, for Halo-LC3 the 630 nm LED power was set at 20%. For each cell line, a single frame was taken at 100 ms exposure for the three channels. Images were processed using a home-built foci analysis algorithm written in ICY software (BioImage Analysis Lab; de Chaumont et al., 2012).
Single-molecule live-cell imaging
We performed single-particle tracking analysis for determining diffusion coefficients of HaloTagged autophagy protein under control and EBSS starving conditions. Single-molecule live-cell imaging was carried out on the Olympus microscope using the 640 nm laser (∼25% power) at a HILO angle and the Andor iXon 897 Ultra camera was used for signal detection. Cells (200–300,000) were grown on glass coverslips (170 ± 5 μm, Schott) and imaged 24 h after seeding. Coverslips were cleaned with 1M KOH (1 h in a sonicated water bath), rinsed with double distilled H2O, cleaned with 100% ethanol (1 h in a sonicated water bath), and dried under N2 stream before assembly on a 35-mm diameter imaging dish. Precise determination of diffusion coefficient using single-particle tracking requires sparse labeling of the Halo-tagged protein. Cells were labeled with Halo ligand JFX650 with the following optimized conditions: 1.0 nM, 30 s pulse (ATG9A); 500 pM, 30 s pulse (WIPI2, ULK1, ATG2A, ATG5, ATG13, ATG16L1, PI4K3β); and 50 pM, 30 s pulse (LC3, SEC61, Halo-NES). These labeling conditions allowed a maximum of 10–15 particles per frame throughout the entire imaging experiment (146 frames per second, 3,000 frames, 140 × 512 pixel region of interest). After labeling, the cells were washed three times with fresh media and incubated for an additional 10 min in complete media containing 7-bromophenol (10 μM) to block unreacted HaloTag proteins at 37°C (5% CO2). Before adding EBSS, cells were washed 3× with PBS. EBSS-treated samples were imaged within 1 h of treatment.
Diffusion coefficient determination
For detecting individual single-molecule particles and computing particle tracks, we used the batch parallel-processing version of SLIMFast in MatLab 2020b. This software—kindly provided by Xavier Darzacq (University of California, Berkeley, Berkeley, CA, USA) and Anders Hansen (Massachusetts Institute of Technology, Cambridge, MA, USA)—allows direct import of the TIFF files and uses the multiple-target tracing algorithm for signal detection and particle tracking (Sergé et al., 2008). The following settings were used for all the proteins: exposure time = 6.8 ms, NA = 1.49, PixelSize = 0.16 μm, emission wavelength = 664 nm, Dmax = 15 μm2 s−1, number of gaps allowed = 2, localization error = 10-5, deflation loops = 0. For ATG9A, the Dmax was set at 5 μm2 s−1. From the tracked particles, we determined the diffusion coefficients and the fraction corresponding to distinct particle subpopulations using the MatLab version of SpotOn (kindly provided by Xavier Darzacq and Anders Hansen; Hansen et al., 2018). The following settings were used: TimeGap = 6.8 ms, dZ = 0.700 μm, GapsAllowed = 2, Time-Points = 7, JumpsToConsider = 4, BinWidth = 0.01 μm, PDF-fitting, D_Free1_3State = [2 15], D_Free2_3State = [0.15 2], D_Bound_3State = [0.0001 0.15]. For ATG9A, a two-state model was applied, with the following settings: D_Free2_1State = [0.2 5], D_Bound2_State = [0.0001 0.2]. We carried out three independent biological replicates with at least 20 cells for each cell line for all experiments.
Determination of HaloTag autophagy protein foci lifetime
Autophagy protein foci lifetimes were determined in the HaloTag-edited cell lines by live-cell imaging. Cells were imaged on the Olympus microscope using the Olympus PlanApo N, NA = 1.42, the Andor iXon 897 Ultra camera, using the 630 nm LED at 100% power. Cells (150,000) were grown on glass coverslips (170 ± 5 μm, Schott) and imaged 48 h after seeding. Cells were labeled with Halo ligand 100 nM JF646 in complete media for 10 min, which allows quantitative labeling of HaloTagged proteins. After labeling, the cells were washed three times with fresh media and incubated for an additional 10 min in complete media containing 7-bromophenol (10 μM) to block unreacted HaloTag proteins at 37°C (5% CO2). Before adding EBSS, cells were washed 3× with PBS. EBSS-treated samples were imaged within 1 h of treatment. For each condition, four cell clusters (4–6 cells per cluster) were selected and imaged at 15-s time intervals (50 ms exposure time) for 1 h, corresponding to 240 frames per movie. For ATG2A and ATG13, cells were also imaged at a faster rate (3 s) for over 240 frames. The experiments were run in triplicate, with ∼20–30 cells imaged per cell line for each replicate. Foci were treated as single particles and analyzed using TrackIT (Kuhn et al., 2021) with the following settings: threshold 1.5; tracking radius 6; minimum track length 2; gap frames 2.
Determination of foci colocalization with P62, LC3, and GABARAPL1 foci
Cells were seeded at low confluency (100,000) on glass coverslips (170 ± 5 μm, Schott). After 24 h, cells were transduced with GFP-LC3 or GFP-P62 encoding baculovirus particles using the viral BacMam 2.0 transfection reagent at a concentration of ∼0.25 × 108 viral particles/ml. After an additional 24 h for viral transduction, cells were labeled for foci visualization (100 nM JFX650 for 10 min in complete media) and imaged in control and EBSS starvation media (Grimm et al., 2021). Before adding EBSS, cells were washed 3× with PBS. EBSS-treated samples were imaged within 1 h of treatment. Cells were imaged on the Olympus microscope using the Olympus PlanApo N, NA = 1.42 and the Andor iXon 897 Ultra camera, using the 630 nm LED (100% power) and the 475 nm LED (30% power). For each condition, four-cell clusters (4–6 cells per cluster) were selected and imaged in 3-s time intervals (50 ms exposure time) for 12 min. The experiments were run in triplicate, with ∼20–30 cells imaged per cell line for each replicate. Additional experiments were performed for the Halo-ATG13 cell line stably expressing GFP-tagged adaptors LC3, P62, and GABARAPL1 from the AAVS1 safe harbor locus. Cells were imaged on the same Olympus microscope, with the Olympus TIRF 60× UPlanApo NA = 1.50 objective and the Hamamatsu Orca-Fusion BT sCMOS camera. For each condition, four cell clusters (4–8 cells per cluster) were selected and three z-planes (center plane and ± 2 μm) were imaged at 1.5-s time intervals (50 ms exposure time) for 8 min. The experiments were run in triplicate, with ∼40–60 cells imaged per cell line.
Automated autophagy protein foci tracking and colocalization
Autophagy protein foci were treated as single particles and analyzed using TrackIT (Kuhn et al., 2021) with the following settings: threshold 1.5; tracking radius 6; minimum track length 2; gap frames 6. Tracks were exported and analyzed with a custom MatLab (v. 2022a) algorithm. The algorithm calculated the Euclidean distance between the tracked autophagy factor foci coordinates and the nearest GFP-tagged marker (LC3, P62, or GABARAPL1) signal coordinates. If the calculated distance was ≤3 pixels (0.81 or 0.33 μm for the Andor and Hamamatsu cameras, respectively), the marker signal was considered colocalized with the tracked autophagy protein particle. An autophagy factor track was considered colocalized with a marker track if at least four colocalized events were recorded over the duration of the autophagy factor track. The duration of colocalization between the autophagy factor foci and the marker foci was calculated by determining the difference between the first and last colocalization events.
Step-size analysis was carried out with a home-built algorithm written in MatLab. Diffusion coefficients for the autophagy factors foci were computed using MSDAnalyzer (Tarantino et al., 2014) implemented in MatLab or built in the TrackIt package. In both cases, the algorithm fits the mean-squared displacement curve obtained from the single-particle tracks with a power law equation describing anomalous diffusion. For diffusion analysis, only 60% of the track was fitted.
Live-cell imaging of ATG13 with an ER marker
We visualized ATG13 foci in control cells (siCTR) and upon siRNA knockdown of CHMP2A (siCHMP2A). siRNA experiment was performed using previously validated siRNA pools (Takahashi et al., 2018), and nucleofection was performed according to the supplier’s recommendation (control siRNA [D-001810-10-05] and siCHMP2A [L-020247-01-0005], both supplied by Horizon). mEmerald-SEC61 (a gift from Jennifer Lippincott-Schwartz, plasmid # 90992; Addgene; http://n2t.net/addgene:90992; RRID:Addgene_90992) was added to the siRNA transfection and effective protein depletion was verified at 40 h after siRNA transfection using Western blot using a CHMP2A antibody (Takahashi et al., 2018). The cells were seeded at low confluency (100,000 cells) on glass coverslips (170 ± 5 μm, Schott). The Halo-ATG13 was labeled with JFX650 dye (100 nM in complete media for 10 min) 40 h after transfection and imaged on the Olympus microscope using the Olympus TIRF 60× UPlanApo NA = 1.50, using the 630 nm LED (100% power) and the 475 nm LED (30% power), and the Hamamatsu Orca Fusion BT camera. The ER signal was restored to high resolution using content-aware image reconstruction (CARE), a machine-learning algorithm (Weigert et al., 2018) implemented in ImageJ, referred to as the CSBDeep plugin. The image deconvolution was performed using a pretrained microtubules training set.
Live-cell imaging of ATG9A with LAMP1 lysosomal marker
Halo-ATG9A stably expressing LAMP1-mNeon, a lysosomal marker, was transfected with 2.5 µg of RFP-ATG9A (a gift from Noboru Mizushima; plasmid # 60609; Addgene; http://n2t.net/addgene:60609; RRID:Addgene_60609; Koyama-Honda et al., 2013) using a 4D-Nucleofector unit (Lonza). Nucleofection was performed according to the supplier protocol. The cells were seeded at low confluency (100,000 cells) on glass coverslips (170 ± 5 μm, Schott). Halo-ATG9A was labeled with JFX650 dye (100 nM in complete media for 10 min) 24 and 48 h after transfection to determine if the aging of Halo-ATG9A protein changes its localization with lysosomes. Cells were imaged 48 h after transfection with a 3i spinning-disk confocal microscope with the 488 nm laser (10 ms exposure time, 3% laser power), the 561 nm laser (100 ms exposure time, 100% laser power), and the 640 nm laser (100 ms exposure time, 100% laser power) and imaged at 1 frame per second for 1 min.
Live-cell imaging of Halo-ATG9A and SNAP-LC3B with Deep Red LysoTracker
100,000 double-edited cells expressing Halo-ATG9A and SNAP-LC3B from their endogenous loci were plated in a 35-mm glass bottom dish. The following day, cells were labeled with Halo ligand JFX554 (100 nM in complete media for 10 min) and imaged the same day (0 h experiment) or the subsequent day (24 h experiment). SNAP-LC3B was labeled with JF503 SNAP-ligand (100 nM in complete media) overnight prior to imaging. Samples were then labeled with Deep Red LysoTracker (Invitrogen, 2.5 nM in complete media for 30 min) after labeling the Halo-ATG9A. Cells were washed in complete media and imaged with the 3i spinning-disk confocal microscope with the 488 nm laser (10 ms exposure time, 3% laser power), the 561 nm laser (100 ms exposure time, 100% laser power), and the 640 nm laser (100 ms exposure time, 100% laser power) and imaged at one frame per second for 1 min.
Halo-ATG9A trafficking and stability experiments
ATG9A was knocked out of a Halo-ATG9A cell line using the spCas9 and sgRNAs targeting the 3xFLAG-HaloTag coding sequence and three exons of the ATG9A coding sequence (see Table S1), and knock-out cells were identified by the lack of HaloTag fluorescence. Halo-ATG9A was subsequently added back into the AAVS1 safe locus with a tet-inducible promotor. This Halo-ATG9A add-back cell line was grown with or without 2 µg/ml doxycycline in tetracyclin-free media (Invitrogen). 200,000 cells were then plated in a 6-well dish and doxycycline was added to induce Halo-ATG9A expression in the samples that were grown without doxycyclin (2 µg/ml), and cells were grown in doxycycline-containing media for the remainder of the experiment. The following day, the cells were labeled with JFX650 (100 nM in complete media for 10 min), and the 0 h timepoint was harvested by the addition of 100 μl of 1× SDS PAGE sample buffer. 24 and 48 h after labeling additional samples were harvested. Samples were boiled for 5 min before being separated on 4–15% TGX stain-free polyacrylamide gels (Biorad). Fluorescence was detected using the Cy5.5 setting on the ChemiDoc imaging system (BioRad). For live-cell imaging, the cells were treated as described above, except for the following additions. Endogenously edited Halo-ATG9A was added as a control to compare intensities with doxycycline-induced Halo-ATG9A cells. Additionally, as a control, U2OS and Halo-ATG9A knock-out cells were labeled with JFX650 HaloTag-ligand and imaged at 0 and 24 h after labeling to assess non-specific JFX650 fluorescence signal.
Live-cell imaging of RFP-ATG9A
Plasmids for the expression of RFP-ATG9A (1 ug) and GFP-SEC61 (1 μg) were cotransfected with Lipofectamine 3,000 according to the manufacturer’s instructions in U2OS cells. In an additional experiment, RFP-ATG9A (1 μg) was transfected with Lipofectamine 3,000 according to the manufacturer’s instructions in Halo-ATG9A cells. 24 h after transfection cells were imaged by spinning-disk confocal microscopy as described above.
Quantification and statistical analysis
Data are expressed as ± one SD unless otherwise stated. Data presented as percentages were normalized using a logit function prior to statistical analysis. Statistical differences were evaluated by two-tail t test or one-way ANOVA at P < 0.05 significance level followed by Bonferroni post-hoc test. All the statistical analyses were computed using OriginPro (v. 2023, OriginLab).
Online supplemental material
Fig. S1 shows genomic and protein validation of the genome editing for HaloTag insertion; the figure also presents biochemical validation of the HaloTagged cell lines. Fig. S2 presents additional biochemical validation of the HaloTagged cell lines using immunofluorescence and live-cell imaging. Fig. S3 shows in-gel quantification of HaloTagged autophagy proteins, including the standard curves used for quantification and correction factors upon TEV-cleavage of the HaloTag. Fig. S4 shows additional imaging and biological experiments that accompany Figs. 5 and 6. Fig. S5 contains additional imaging and control experiments for Figs. 7 and 8. Videos 1–10 show representative live-cell single-molecule imaging of control and EBSS-treated U2OS cell lines expressing HaloTagged proteins. Video 11 presents the automated foci detection and tracking method, whereas Videos 12–18 show live-cell foci imaging of control and EBSS-treated U2OS cell lines expressing HaloTagged proteins. Video 19 shows live-cell imaging of the Halo-ATG13 cell line coexpressing an ER marker (mEmerald-SEC61). Video 20 shows live-cell foci imaging of the Halo-ATG2A cell line coexpressing GFP-ATG13. Video 21 shows live-cell foci imaging of the Halo-ATG13 cell line with ULK1-complex unit knockout. Videos 22 and 23 show live-cell foci imaging of Halo-ATG13 and Halo-ATG2A cell lines in the presence of Wortmannin. Video 24 shows live-cell single-molecule imaging for the ULK1-complex unit knockouts. Videos 25–32 show live-cell foci imaging of control and EBSS-treated U2OS cell lines expressing HaloTagged proteins and GFP-P62. Videos 33–35 show live-cell foci imaging of control and EBSS-treated Halo-ATG2A, Halo-ATG13, and ULK1-Halo cell lines coexpressing GFP-LC3. Video 36 shows live-cell foci imaging of the dual-edited Halo-ATG9A/SNAP-LC3 cell line. Video 37 shows live-cell foci imaging of ATG9A knockout in the Halo-ATG2A cell line. Video 38 shows live-cell foci imaging of Halo-ATG9A coexpressing RFP-ATG9A and LAMP1-mNeonGreen. Video 39 shows live-cell foci imaging of Halo-ATG9A coexpressing SNAP-LC3B and stained with LysoTracker DeepRed. Table S1 lists antibodies, chemical reagents, cell lines, oligonucleotides, sgRNA sequences, recombinant DNA, software, algorithms, and miscellaneous reagents used in this study.
Data availability
Data are available in the article itself and its supplementary materials. Live-cell imaging datasets are openly available in a public repository (https://doi.org/10.5061/dryad.866t1g1vh).
Acknowledgments
We thank Dr. Daniel T. Youmans and Dr. Thomas R. Cech (University of Colorado Boulder, Boulder, CO, USA) for providing the plasmid for recombinant production of the 3xFLAG-HaloTag protein, and Luke D. Lavis (Janelia Research Campus, Ashburn, VA, USA) for generously providing the Janelia fluor dyes. We thank Dr. Eric Patrick (Michigan State University, East Lansing, MI, USA) for contributing to the preparation of the recombinant HaloTag protein used in this study. We acknowledge the Flow Cytometry core (Research Technology Support Facility, Michigan State University) and the Confocal Laser Scanning Microscopy core (Center for Advanced Microscopy, Michigan State University) for supporting our cell sorting and microscopy experiments. The order of authors was decided by a randomization process and both authors contributed equally to the paper; co-first authors reserve the right to list themselves first on their curriculum vitae.
This work was supported by a grant from the National Institutes of Health (DP2 GM142307) to J.C. Schmidt. J.C. Schmidt was a Damon Runyon Dale F. Frey Scientist supported (in part) by the Damon Runyon Cancer Research Foundation (DFS-24-17).
Author contributions: Conceptualization: D.G. Broadbent, C. Barnaba, and J.C. Schmidt; Experiments: D.G. Broadbent, C. Barnaba, and G.I. Perez; Data Analysis: D.G. Broadbent and C. Barnaba; Writing—Original Draft: D.G. Broadbent and C. Barnaba; Writing—Review and Editing: D.G. Broadbent, C. Barnaba, and J.C. Schmidt.
References
Author notes
D.G. Broadbent and C. Barnaba contributed equally to this paper.
Disclosures: The authors declare no competing interests exist.