Degradative organelles contain enzymes that function optimally at the acidic pH generated by the V-ATPase. The resulting transmembrane H+ gradient also energizes the secondary transport of several solutes, including Cl−. We report that Cl− influx, driven by the 2Cl−/H+ exchanger ClC-7, is necessary for the resolution of phagolysosomes formed by macrophages. Cl− transported via ClC-7 had been proposed to provide the counterions required for electrogenic H+ pumping. However, we found that deletion of ClC-7 had a negligible effect on phagosomal acidification. Instead, luminal Cl− was found to be required for activation of a wide range of phagosomal hydrolases including proteases, nucleases, and glycosidases. These findings argue that the primary role of ClC-7 is the accumulation of (phago)lysosomal Cl− and that the V-ATPases not only optimize the activity of degradative hydrolases by lowering the pH but, importantly, also play an indirect role in their activation by providing the driving force for accumulation of luminal Cl− that stimulates hydrolase activity allosterically.
Introduction
Phagocytosis plays key roles in immunity and maintaining tissue homeostasis (Flannagan et al., 2012). These functions rely on the ability of immune cells to control the composition of the milieu surrounding the particles they engulf, e.g., the pH, to foster the microbicidal and degradative capacity of the phagosomal compartment (Hackam et al., 1997). Upon scission from the plasma membrane, nascent phagosomes undergo extensive remodeling to acquire pumps and channels from endocytic organelles, including vacuolar H+-pumping ATPases (V-ATPases; Freeman et al., 2023; Lukacs et al., 1990). Once incorporated into the limiting phagosomal membrane, the V-ATPases drive the acidification of the phagosome by converting the energy from ATP hydrolysis into the rotary forces that propel H+ into the phagosomal lumen. Such vectorial transport of H+ generates a transmembrane electrical potential difference (inside positive) that would restrain the activity of the pump if left uncompensated. It has long been appreciated, however, that the charge imbalance generated by the V-ATPase is neutralized by counterion conductance pathways. These are incompletely defined: numerous mechanisms have been proposed but their relative contribution remains controversial (Di et al., 2006; Graves et al., 2008; Kasper et al., 2005; Lange et al., 2006; Steinberg et al., 2010; Weinert et al., 2010).
In lysosomes, putative counterion pathways for the V-ATPase include the parallel influx of Cl− enabled by Cl− channels (Di et al., 2006; Riazanski et al., 2021) or Cl−/H+ exchangers (Graves et al., 2008; Kornak et al., 2001), as well as cation efflux (Lin et al., 2015; Steinberg et al., 2010). As in the case of lysosomes, the generation and maintenance of an acidic steady-state pH in mature phagosomes (phagolysosomes) require the ongoing activity of V-ATPases, which similarly requires a counterion conductance. For phagosomes, which require hours to form, degrade their contents, and ultimately “resolve” (Lancaster et al., 2021; Levin-Konigsberg et al., 2019), such counterion pathways would be predicted to play an essential role.
Lysosomes and mature phagolysosomes continually release (“leak”) H+ equivalents, requiring persistent activity of the V-ATPases even after reaching their steady-state pH. The nature and contribution of the various leak pathways are also incompletely understood. Multiple transporters are seemingly involved: H+ is known to be cotransported with organic solutes including amino acids (Rebsamen et al., 2015) and sugars (Ruivo et al., 2008). The tight coupling of such cotransport, together with the direction of the electrochemical H+ gradient generated by the V-ATPases, drives the net unidirectional efflux of the organic solutes. In addition, H+ leaves acidic organelles in exchange for Cl− via carriers of the CLC family (Jentsch and Pusch, 2018). Five members of this family localize to endomembranes and have been shown to transport two Cl− inward in exchange for one H+ outward, resulting in a net displacement of three charges (negative inside). CLCs are thus poised to act as a counterion pathway as well as an H+ leak pathway (Günther et al., 2003). Still, little is known regarding the exact role of endomembrane CLCs or Cl− in phagosomal maturation, acidification, and resolution, warranting a comprehensive investigation.
In this study, we report that a CLC (ClC-7) is present and largely responsible for Cl− transport in late phagosomes. Importantly, the phagosomal Cl− concentration achieved through rheogenic H+ countertransport exceeds that of the cytosol and is important for activating luminal hydrolases for the degradation of phagocytic targets. Of note, while ClC-7 is required for Cl− accumulation and cargo degradation and resolution, it was found to be dispensable for phagosomal acidification. These observations provide new insights for a role of Cl− in innate immunity and document an indirect role of V-ATPases and luminal acidification in the regulation of hydrolase activity in situ.
Results
The degradation of phagocytic targets requires phagosomal acidification and Cl−
While the formation and maturation of phagosomes have been studied extensively, their subsequent fate, i.e., the process of phagosomal resolution, has been largely neglected. Whether acidification, which is essential for maturation, also plays a role in resolution is not yet known. We, therefore, initiated our studies by establishing a protocol to measure the degradation of ingested biological targets and the resolution of the phagosome in parallel. To that end, we first challenged RAW 264.7 macrophages with IgG-opsonized sheep red blood cells that are effectively internalized via Fc-mediated phagocytosis within minutes. Unbound particles were then washed away and the fate of the engulfed targets was monitored over time. Fluorescent staining of the opsonin coating the target surface enabled us to visualize the red blood cells and estimate their volume (as described in Materials and methods) to track their degradation over time (Fig. 1 A). Independent experiments using fluorescent dextrans indicated that under all conditions studied, the volume calculated based on the distribution of the opsonin was a reliable estimate of the total phagosomal volume (Fig. S1 A). As shown in Fig. 1 B, the initial volume of the phagosomes—which was similar to that of the intact red blood cells (of ∼30 μm3)—decreased steadily as the red cells were broken down. 4 h after internalization, the phagosomes had mostly resolved and the remaining IgG signal (Fig. 1, A and B), as well as that of the soluble dextran marker, were confined into small compartments. Predictably, macrophages treated with the V-ATPase inhibitor, concanamycin A, did not degrade the red blood cells, which remained largely intact over the period of observation (Fig. 1, A and B).
Phagosomal resolution necessitates ongoing activity of the V-ATPase and intracellular Cl − . (A and B) Vehicle control and concanamycin A–treated RAW 264.7 cells challenged with IgG-opsonized sRBCs for 1–4 h and stained for F-actin (cyan) and anti-IgG (red). Concanamycin A (500 nM) was added 30 min after phagocytosis. Representative images shown in A. Mean ± SEM of sRBC phagosome volumes graphed in B. n = 3, each independent experiment counting >50 phagosomes. (C) Representative fluorescence images of vehicle control and concanamycin-treated RAW 264.7 cells challenged with DQ-BSA–coated silica beads for 1 h. (D) Quantification of relative DQ-BSA fluorescence intensity in vehicle control and concanamycin A–treated RAW 264.7 cells. n = 3, each independent experiment counting >30 phagosomes. (E) Colorimetric determinations of intracellular [Cl−] for RAW 264.7 cells incubated in medium containing Cl− or NO−3 (Cl−-depleted) for 1 or 4 h. n = 3. (F) Phagosome resolution in control and Cl−-depleted RAW 264.7 cells challenged with IgG-opsonized sRBCs for 1–4 h. (G) Mean ± SEM volume of sRBC-containing phagosomes. n = 3, each independent experiment counting >50 phagosomes. (H) Representative fluorescence images of control and Cl−-depleted RAW 264.7 cells challenged for 1 h with silica beads precoated with DQ-BSA. (I) Quantification of relative DQ-BSA intensities in control and Cl−-depleted RAW 264.7 cells. n = 5, each independent experiment counting >25 phagosomes.
Phagosomal resolution necessitates ongoing activity of the V-ATPase and intracellular Cl − . (A and B) Vehicle control and concanamycin A–treated RAW 264.7 cells challenged with IgG-opsonized sRBCs for 1–4 h and stained for F-actin (cyan) and anti-IgG (red). Concanamycin A (500 nM) was added 30 min after phagocytosis. Representative images shown in A. Mean ± SEM of sRBC phagosome volumes graphed in B. n = 3, each independent experiment counting >50 phagosomes. (C) Representative fluorescence images of vehicle control and concanamycin-treated RAW 264.7 cells challenged with DQ-BSA–coated silica beads for 1 h. (D) Quantification of relative DQ-BSA fluorescence intensity in vehicle control and concanamycin A–treated RAW 264.7 cells. n = 3, each independent experiment counting >30 phagosomes. (E) Colorimetric determinations of intracellular [Cl−] for RAW 264.7 cells incubated in medium containing Cl− or NO−3 (Cl−-depleted) for 1 or 4 h. n = 3. (F) Phagosome resolution in control and Cl−-depleted RAW 264.7 cells challenged with IgG-opsonized sRBCs for 1–4 h. (G) Mean ± SEM volume of sRBC-containing phagosomes. n = 3, each independent experiment counting >50 phagosomes. (H) Representative fluorescence images of control and Cl−-depleted RAW 264.7 cells challenged for 1 h with silica beads precoated with DQ-BSA. (I) Quantification of relative DQ-BSA intensities in control and Cl−-depleted RAW 264.7 cells. n = 5, each independent experiment counting >25 phagosomes.
Inhibition of protease activity ablates phagosomal resolution. (A) The volume of red blood cells (sRBC) undergoing breakdown closely approximates the total volume of phagosomes at different stages of resolution. RAW 264.7 cells were pulsed overnight with 10 kD tetramethylrhodamine (TMR)-dextran and chased with complete medium 1 h prior to their incubation with FITC-conjugated IgG-opsonized sRBC for 2 or 4 h. Cells were subsequently imaged live. (B) Vehicle control and protease inhibitor-treated RAW 264.7 cells were challenged for 1–4 h with IgG-opsonized sRBC and imaged as in Fig. 1. (C) Volume of sRBC-containing phagosomes as a function of time after internalization. Data are means ± SEM of three experiments, each counting at least 70 phagosomes.
Inhibition of protease activity ablates phagosomal resolution. (A) The volume of red blood cells (sRBC) undergoing breakdown closely approximates the total volume of phagosomes at different stages of resolution. RAW 264.7 cells were pulsed overnight with 10 kD tetramethylrhodamine (TMR)-dextran and chased with complete medium 1 h prior to their incubation with FITC-conjugated IgG-opsonized sRBC for 2 or 4 h. Cells were subsequently imaged live. (B) Vehicle control and protease inhibitor-treated RAW 264.7 cells were challenged for 1–4 h with IgG-opsonized sRBC and imaged as in Fig. 1. (C) Volume of sRBC-containing phagosomes as a function of time after internalization. Data are means ± SEM of three experiments, each counting at least 70 phagosomes.
The H+ gradient established by the V-ATPase is required for the activity of luminal hydrolases. Reduced hydrolase function seemed likely to be the impediment to phagosomal resolution in concanamycin-treated cells. Accordingly, we found that macrophages treated with protease inhibitors failed to degrade red blood cells over 4 h (Fig. S1, B and C). To further assess hydrolytic activity in the presence and absence of the V-ATPase inhibitor, macrophages were challenged with IgG-opsonized silica beads coated with dye-quenched bovine serum albumin (DQ-BSA); proteolytic cleavage of the labeled albumin is indicated by the increased fluorescence that results from dequenching. As expected, phagosomes degraded the DQ-BSA on the surface of the beads in a manner that was dependent on V-ATPase (Fig. 1, C and D).
Phagosomal Cl− is essential for content degradation and resolution
The counterion that offsets charge displacement by the V-ATPase during phagosomal resolution is unknown. To determine if Cl− is required for the resolution of phagosomes, we maintained macrophages in Cl−-free medium for 1–4 h and confirmed that intracellular Cl− was depleted using a colorimetric Cl− assay (Steinberg et al., 2010). By comparing these values to a standard curve and assessing the cell volume electronically, intracellular [Cl−] was determined to be ∼40 mM (Fig. 1 E), a value in reasonable agreement with other measurements (Ince et al., 1987; Ishiguro et al., 2002; Wu et al., 2016), lending confidence to the method used. After 1 h in Cl−-free medium, the intracellular [Cl−] was greatly reduced; by 4 h, intracellular [Cl−] was negligible (Fig. 1 E). As depletion of Cl− may have effects on the metabolic state of cells, we maintained macrophages in Cl−-free medium for 1 h and then assessed their mitochondrial integrity and functionality. To this end, we estimated the potential across the inner mitochondrial membrane using a cationic fluorescent dye. The measurements were validated using the mitochondrial uncoupler carbonyl cyanide-p-trifluoro-methoxyphenyl hydrazone (FCCP), which completely dissipated the potential (Fig. S2, A and B). By comparison, Cl− depletion caused only a modest decrease in dye accumulation, indicative of a slight depolarization of the mitochondrial membrane potential. Nevertheless, the effect was reproducible and was associated with changes in the morphology of the mitochondria, which appeared somewhat smaller/fragmented (Fig. S2, A and B). We, therefore, anticipated that Cl− depletion might affect the ATP content of the cells. Indeed, direct measurements using a luciferase-based assay showed that ATP was significantly reduced (by almost 50%) when Cl− was replaced by NO−3. The partial depletion of ATP, however, had little effect on V-ATPase activity; the lysosomal pH of Cl−-depleted macrophages was only marginally affected (Fig. S2 D). This finding is in good agreement with the known high affinity of the V-ATPase for ATP; its Km is estimated in the µM range (Kishikawa et al., 2022; Nakano et al., 2008), orders of magnitude higher than the cytosolic concentration of ATP remaining in Cl−-depleted cells.
Metabolic consequences of Cl − omission and Clcn7 deletion. (A) RAW 264.7 wildtype and Clcn7 KO macrophages were incubated in either Cl− or NO−3 medium for 1 h prior to addition of rhodamine-123, which accumulates in healthy mitochondria. As a control, FCCP—which collapses the mitochondrial membrane potential—was added to cells 15 min prior to the addition of rhodamine-123. (B) Relative mean rhodamine-123 signal is quantified in B. n = 3 experiments. (C) RAW 264.7 wildtype and Clcn7 KO macrophages were incubated in either Cl− or NO−3 medium for 1 h prior to imaging. Total ATP content was measured by luminescence with the CellTiter Glo kit (Promega). Luminescence signal was normalized by cell count. n = 3 experiments. (D) Lysosomal pH of wildtype and Clcn7 KO macrophages, which were pulsed with FITC-dextran overnight, then incubated in either Cl− or NO−3 medium 1 h prior to imaging as described in Materials and methods. To dissipate the transmembrane pH gradient, cells were treated with 500 nM of concanamycin A 30 min prior to imaging. n = 3 experiments.
Metabolic consequences of Cl − omission and Clcn7 deletion. (A) RAW 264.7 wildtype and Clcn7 KO macrophages were incubated in either Cl− or NO−3 medium for 1 h prior to addition of rhodamine-123, which accumulates in healthy mitochondria. As a control, FCCP—which collapses the mitochondrial membrane potential—was added to cells 15 min prior to the addition of rhodamine-123. (B) Relative mean rhodamine-123 signal is quantified in B. n = 3 experiments. (C) RAW 264.7 wildtype and Clcn7 KO macrophages were incubated in either Cl− or NO−3 medium for 1 h prior to imaging. Total ATP content was measured by luminescence with the CellTiter Glo kit (Promega). Luminescence signal was normalized by cell count. n = 3 experiments. (D) Lysosomal pH of wildtype and Clcn7 KO macrophages, which were pulsed with FITC-dextran overnight, then incubated in either Cl− or NO−3 medium 1 h prior to imaging as described in Materials and methods. To dissipate the transmembrane pH gradient, cells were treated with 500 nM of concanamycin A 30 min prior to imaging. n = 3 experiments.
We, therefore, proceeded to measure the effect of Cl− depletion on the ability of phagosomes to digest and dispose of their contents. These experiments were revealing: despite the minute changes in lysosomal pH, depletion of intracellular Cl− prevented macrophages from breaking down targets and resolving the phagosomes (Fig. 1, F and G). As shown in Fig. 1, H and I, intracellular Cl− was also necessary for the robust proteolytic activity of phagosomes; the signal from DQ-BSA beads was reduced by more than 50% in the absence of Cl−. Thus, both Cl−, presumably transported into phagosomes, and the H+ gradient appeared to be critical determinants of the breakdown of phagocytic targets.
ClC-7 is the Cl− transporter required for phagosomal degradation and resolution
We next sought to determine the source of the phagosomal Cl− required for resolution. We first analyzed mRNA from primary bone marrow–derived macrophages (BMDMs) to determine the relative abundance of transcripts encoding the major Cl− channels and exchangers reported to function in the endocytic pathway (Fig. 2 A; Stauber and Jentsch, 2013). To gauge whether any of these are uniquely required for the phagocytic function of macrophages, their mRNA levels were compared with those of fibroblasts (i.e., non-phagocytic cells; Fig. 2 B). Cystic fibrosis transmembrane conductance regulator (CFTR) had been reported to function in phagocytosis, but we were unable to detect Cftr expression in the macrophages used in our studies. Lrrc8a, which encodes for a conserved subunit of the volume-regulated anion channel (VRAC), was detectable and comparatively more abundant in BMDM than in fibroblasts. However, pharmacological inhibition of VRAC with 4-(2-Butyl-6,7-dichloro-2-cyclopentyl-indan-1-on-5-yl) oxobutyric acid (DCPIB) had no discernible effect on phagosome resolution (Fig. S3, A and B), and the possible contribution of this channel was not pursued further.
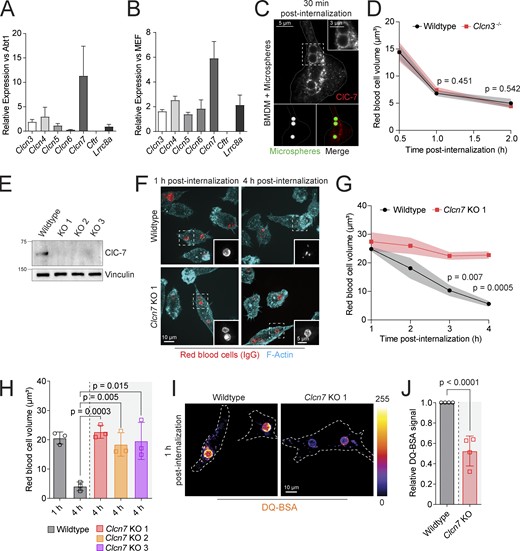
ClC-7 is targeted to the limiting membrane of mature phagosomes and required for phagosomal resolution. (A and B) Determination of Cl− transporter and channel transcript abundance by RT-qPCR. n = 3. (C) Immunostaining of ClC-7 in BMDMs challenged with IgG-opsonized Fluoresbrite YG carboxylate microspheres (green) for 30 min prior to fixation. Inset highlights localization of ClC-7 (red) to phagosomal membrane. (D) Quantification of sRBC-containing phagosomal volumes in wildtype and Clcn3−/− BMDMs. Macrophages were challenged with sRBCs for 0.5–2 h. Data are means ± SEM of five experiments, each counting >50 phagosomes. (E) Lysates of wildtype and Clcn7 KO clones probed with anti–ClC-7 antibody and anti-vinculin antibody used to normalize loading and visualized by SDS-PAGE. Molecular weight markers (kD) are indicated left of the panels. (F and G) Wildtype and Clcn7 KO RAW 264.7 cells challenged for 1–4 h with IgG-opsonized sRBCs and stained for F-actin (cyan) and anti-IgG (red). F shows representative images while G shows mean sRBC-containing phagosome volumes from three independent experiments, each counting >60 phagosomes. (H) Mean red blood cell volumes 1 and 4 h after internalization by RAW 264.7 wildtype or three Clcn7 KO clones (validated in E). (I) Representative fluorescence images of wildtype and Clcn7 KO RAW 264.7 cells challenged with DQ-BSA–coated silica beads for 1 h. (J) Normalized DQ-BSA intensity from four independent experiments, each counting >20 phagosomes. Source data are available for this figure: SourceData F2.
ClC-7 is targeted to the limiting membrane of mature phagosomes and required for phagosomal resolution. (A and B) Determination of Cl− transporter and channel transcript abundance by RT-qPCR. n = 3. (C) Immunostaining of ClC-7 in BMDMs challenged with IgG-opsonized Fluoresbrite YG carboxylate microspheres (green) for 30 min prior to fixation. Inset highlights localization of ClC-7 (red) to phagosomal membrane. (D) Quantification of sRBC-containing phagosomal volumes in wildtype and Clcn3−/− BMDMs. Macrophages were challenged with sRBCs for 0.5–2 h. Data are means ± SEM of five experiments, each counting >50 phagosomes. (E) Lysates of wildtype and Clcn7 KO clones probed with anti–ClC-7 antibody and anti-vinculin antibody used to normalize loading and visualized by SDS-PAGE. Molecular weight markers (kD) are indicated left of the panels. (F and G) Wildtype and Clcn7 KO RAW 264.7 cells challenged for 1–4 h with IgG-opsonized sRBCs and stained for F-actin (cyan) and anti-IgG (red). F shows representative images while G shows mean sRBC-containing phagosome volumes from three independent experiments, each counting >60 phagosomes. (H) Mean red blood cell volumes 1 and 4 h after internalization by RAW 264.7 wildtype or three Clcn7 KO clones (validated in E). (I) Representative fluorescence images of wildtype and Clcn7 KO RAW 264.7 cells challenged with DQ-BSA–coated silica beads for 1 h. (J) Normalized DQ-BSA intensity from four independent experiments, each counting >20 phagosomes. Source data are available for this figure: SourceData F2.
Pharmacological inhibition of VRAC does not affect phagosomal resolution. (A) Vehicle control and DCPIB-treated RAW 264.7 cells challenged for 1–4 h with IgG-opsonized sRBCs, then imaged as in Fig. 1. (B) Determination of the volume of sRBC-containing phagosomes over time. Data are means ± SEM of three experiments, each counting at least 100 phagosomes.
Pharmacological inhibition of VRAC does not affect phagosomal resolution. (A) Vehicle control and DCPIB-treated RAW 264.7 cells challenged for 1–4 h with IgG-opsonized sRBCs, then imaged as in Fig. 1. (B) Determination of the volume of sRBC-containing phagosomes over time. Data are means ± SEM of three experiments, each counting at least 100 phagosomes.
We next quantified Cl− transporters of the CLCN family. Transcripts coding for ClC-3 and ClC-4, which are found in early and late endosomes (Suzuki et al., 2006), were clearly detectable in BMDMs at levels comparatively higher than those in fibroblasts (Fig. 2, A and B). ClC-5 and ClC-6, which can localize to early/recycling and late endosomes, respectively, were expressed at more modest levels yet were nevertheless more abundant in macrophages than in non-phagocytic cells. ClC-7, in contrast, was very prominent in the macrophages, over five times more abundant than in fibroblasts, suggesting a specialized role in phagocytosis (see also Zeziulia et al., 2022). Consistent with this notion, immunostaining revealed ClC-7 to be abundant on the membrane of phagolysosomes (Fig. 2 C and Fig. S4 A), in good agreement with previous proteomic data (Guo et al., 2015; Pauwels et al., 2019). Of note, unlike ClC-3, which was shown to localize to early compartments, ClC-7 was absent from early-stage phagosomes (Fig. S4 A).
ClC-7 localizes to mature phagosomes. (A) BMDMs from wildtype mice were challenged with IgG-opsonized silica beads for 3 min or 1 h, then fixed and immunostained for ClC-7 and LAMP1. (B) Wildtype and Clcn3−/− BMDMs were challenged with IgG-opsonized sRBCs for 1 h and phagocytic index was calculated as described in Materials and methods. Data are means ± SEM of five experiments, each counting at least 30 phagosomes. (C) Wildtype and Clcn7 KO RAW 264.7 macrophages were challenged for 1 h with IgG-opsonized sRBCs and phagocytic index was measured as described in Materials and methods. Data are means ± SEM of three experiments, each counting at least 30 phagosomes.
ClC-7 localizes to mature phagosomes. (A) BMDMs from wildtype mice were challenged with IgG-opsonized silica beads for 3 min or 1 h, then fixed and immunostained for ClC-7 and LAMP1. (B) Wildtype and Clcn3−/− BMDMs were challenged with IgG-opsonized sRBCs for 1 h and phagocytic index was calculated as described in Materials and methods. Data are means ± SEM of five experiments, each counting at least 30 phagosomes. (C) Wildtype and Clcn7 KO RAW 264.7 macrophages were challenged for 1 h with IgG-opsonized sRBCs and phagocytic index was measured as described in Materials and methods. Data are means ± SEM of three experiments, each counting at least 30 phagosomes.
We proceeded to test the functional role of the more abundant ClC isoforms by editing the genes that encode them. Since ClC-3 and ClC-4 can form a heterodimer with functional codependence (Guzman et al., 2017; Weinert et al., 2020), we tested only the effect of deleting ClC-3 on phagosome resolution. We found that BMDMs from Clcn3−/− mice bound and internalized targets as efficiently as their wildtype counterparts (Fig. S4 B), implying that the phagocytic receptors and cytoskeletal remodeling required for phagocytosis were normal in these cells. Importantly, Clcn3−/− BMDMs also degraded IgG-opsonized red blood cells at the same rate as wildtype cells (Fig. 2 D). We concluded that ClC-3 is not responsible for providing the Cl− required for phagosomal cargo degradation.
Clcn7 knockout (KO) RAW 264.7 cells, generated via CRISPR-Cas9 (Fig. 2 E), also bound and internalized targets at normal rates (Fig. S4 C). However, the ClC-7–deficient cells were unable to resolve phagosomes containing red blood cells, which remained largely intact after 4 h (Fig. 2, F–H). Breakdown of DQ-BSA was also impaired in Clcn7 KO macrophages (Fig. 2, I and J), consistent with the notion that proteolytic activity limits the ability of the phagosomes to resolve red cell–containing phagosomes. Importantly, loss of Clcn7 did not affect cell metabolism (Fig. S2, A–D), alleviating possible concerns raised by the metabolic effects of Cl− depletion. Jointly, these findings indicate that, unlike ClC-3, ClC-7 is essential for phagosome resolution.
Phagosome maturation and acidification are independent of ClC-7
Failure to resolve phagosomes could in principle be due to defects in (i) maturation, (ii) acidification, and/or (iii) hydrolase activation. These are not mutually exclusive since an inability to mature would ostensibly prevent the delivery of V-ATPases and hydrolases to the phagosomes. In this regard, loss of ClC-b, the Drosophila equivalent of ClC-7, was reported to impair fusion of phagosomes with lysosomes (Wong et al., 2017). We, therefore, compared the rate of acquisition of the late endosome/lysosome marker LAMP-1 by phagosomes formed by wildtype and Clcn7 KO macrophages. The fraction of phagosomes containing LAMP-1, detected by immunostaining, was indistinguishable between wildtype and Clcn7 KO cells at both early and late time-points (Fig. 3, A and B). In good accordance with these findings, the rate and extent of incorporation of V-ATPases into the phagosomes were also similar in wildtype and Clcn7 KO cells. This was determined using SidK, a bacterial effector that binds selectively to the A subunit of the V-ATPase (Fig. 3, C and D); when conjugated to AlexaFluor-568, SidK can be used as a specific probe to visualize and quantify the H+ pumps (Maxson et al., 2022). Thus, in mammalian cells, the loss of ClC-7 has no appreciable effect on the maturation of the phagosome.
Phagosomal maturation proceeds independently of ClC-7. (A–D) Wildtype and Clcn7 KO RAW 264.7 cells challenged with IgG-opsonized sRBCs for 3 or 20 min. Cells were stained with (A and B) anti-LAMP1 antibodies or incubated with (C and D) SidK-568, a specific probe for the V-ATPase. The percentage of LAMP1-positive phagosomes was quantified in B. The relative fluorescence intensity of SidK-568 was quantified in D. Data are means of three independent experiments, each counting >30 phagosomes.
Phagosomal maturation proceeds independently of ClC-7. (A–D) Wildtype and Clcn7 KO RAW 264.7 cells challenged with IgG-opsonized sRBCs for 3 or 20 min. Cells were stained with (A and B) anti-LAMP1 antibodies or incubated with (C and D) SidK-568, a specific probe for the V-ATPase. The percentage of LAMP1-positive phagosomes was quantified in B. The relative fluorescence intensity of SidK-568 was quantified in D. Data are means of three independent experiments, each counting >30 phagosomes.
While the phagosomes formed in Clcn7 KO macrophages merged with lysosomes and hence incorporated V-ATPases normally, acidification may nevertheless have been affected if, as proposed earlier (Graves et al., 2008), the antiport provides the counterions required for sustained H+ pumping. Impaired acidification could readily explain the observed defects in resolution. To determine the rate and extent of nascent acidification, we challenged macrophages with IgG-opsonized red blood cells conjugated with FITC, a ratiometric pH-sensitive dye. We found that the loss of ClC-7 did not alter the initial development of phagosome acidification (Fig. 4 A). The steady-state pH of red blood cell–containing phagolysosomes, measured 1 h after phagocytosis, was also indistinguishable in wildtype and Clcn7 KO cells (Fig. 4 B). Identical results were obtained when using zymosan as the phagocytic target (Fig. S5 A). As anticipated, deletion of ClC-3, which does not affect phagosome resolution, was also without effect on either the initial rate of or steady-state luminal acidification (Fig. 4 C). Clearly, neither antiport is required for H+ pumping by the V-ATPases. Indeed, replacement of Cl− for NO−3 was similarly without effect on the steady-state phagolysosomal pH (Fig. 4 D). In summary, our data imply that ClC-7 contributes to resolution by means other than enabling maturation and acidification of the phagosome.
Cl− is not required for full acidification of mature phagosomes. (A) Wildtype and Clcn7 KO RAW 264.7 cells were challenged with IgG-opsonized, FITC-conjugated sRBCs, and the fluorescence (ratiometric pH measurements) was recorded for 15 min after internalization. Data are means ± SEM of four experiments, each counting >40 phagosomes. (B)Clcn7 KO RAW 264.7 cells were challenged with IgG-opsonized, FITC-conjugated sRBCs 1 h prior to imaging. The steady-state pH of >60 phagosomes from three independent experiments is shown. (C) The initial rate of acidification was measured in wildtype and Clcn3−/− BMDM as in A, n = 5, each counting >200 phagosomes. (D) Steady-state phagosomal pH was measured as in B in phagosomes from three independent experiments in wildtype RAW 264.7 cells bathed in either Cl−-containing or Cl−-free (NO−3) medium. Cells were depleted of Cl− for 1 h prior to imaging.
Cl− is not required for full acidification of mature phagosomes. (A) Wildtype and Clcn7 KO RAW 264.7 cells were challenged with IgG-opsonized, FITC-conjugated sRBCs, and the fluorescence (ratiometric pH measurements) was recorded for 15 min after internalization. Data are means ± SEM of four experiments, each counting >40 phagosomes. (B)Clcn7 KO RAW 264.7 cells were challenged with IgG-opsonized, FITC-conjugated sRBCs 1 h prior to imaging. The steady-state pH of >60 phagosomes from three independent experiments is shown. (C) The initial rate of acidification was measured in wildtype and Clcn3−/− BMDM as in A, n = 5, each counting >200 phagosomes. (D) Steady-state phagosomal pH was measured as in B in phagosomes from three independent experiments in wildtype RAW 264.7 cells bathed in either Cl−-containing or Cl−-free (NO−3) medium. Cells were depleted of Cl− for 1 h prior to imaging.
Parameters unchanged in macrophages by deletion of Clcn7. (A) Steady-state phagosomal pH of wildtype and Clcn7 KO RAW 264.7 cells. Cells were challenged with IgG-opsonized, FITC-conjugated zymosan particles 1 h prior to imaging. Phagosomal pH was determined by ratiometric imaging as described. Each point represents 1 phagosome; n = 3 independent experiments. (B) Wildtype and Clnc7 KO RAW264.7 macrophages were challenged with IgG-opsonized zymosan-568 for 1 h prior to imaging, and phagocytic index was calculated as described in Materials and methods. (C) Cell volumes of wildtype and Clcn7 KO RAW264.7 macrophages ± IgG-opsonized zymosan-568 were determined with a Coulter counter as described in Materials and methods. Data are means ± SEM of three biological replicates. (D and E) Wildtype and Clcn7 KO RAW 264.7 macrophages were incubated with Magic Red cathepsin B substrate for 30 min according to manufacturer instructions. Relative mean fluorescence signal is quantified in E. n = 3. (F and G) Wildtype and Clcn7 KO RAW 264.7 cells were challenged with IgG-opsonized, FITC-conjugated sRBCs for 1 h prior to imaging. Cells were then acutely treated with 500 nM concanamycin A at time 0 (arrow) and the pH of the phagosome was determined ratiometrically every minute for 10 min. Data are means ± SEM of five experiments, each counting at least 20 phagosomes. Mean instantaneous rates (first derivative) of pH changes at 5 min are quantified in G.
Parameters unchanged in macrophages by deletion of Clcn7. (A) Steady-state phagosomal pH of wildtype and Clcn7 KO RAW 264.7 cells. Cells were challenged with IgG-opsonized, FITC-conjugated zymosan particles 1 h prior to imaging. Phagosomal pH was determined by ratiometric imaging as described. Each point represents 1 phagosome; n = 3 independent experiments. (B) Wildtype and Clnc7 KO RAW264.7 macrophages were challenged with IgG-opsonized zymosan-568 for 1 h prior to imaging, and phagocytic index was calculated as described in Materials and methods. (C) Cell volumes of wildtype and Clcn7 KO RAW264.7 macrophages ± IgG-opsonized zymosan-568 were determined with a Coulter counter as described in Materials and methods. Data are means ± SEM of three biological replicates. (D and E) Wildtype and Clcn7 KO RAW 264.7 macrophages were incubated with Magic Red cathepsin B substrate for 30 min according to manufacturer instructions. Relative mean fluorescence signal is quantified in E. n = 3. (F and G) Wildtype and Clcn7 KO RAW 264.7 cells were challenged with IgG-opsonized, FITC-conjugated sRBCs for 1 h prior to imaging. Cells were then acutely treated with 500 nM concanamycin A at time 0 (arrow) and the pH of the phagosome was determined ratiometrically every minute for 10 min. Data are means ± SEM of five experiments, each counting at least 20 phagosomes. Mean instantaneous rates (first derivative) of pH changes at 5 min are quantified in G.
ClC-7 drives intraphagosomal Cl− accumulation
As ClC-7 is seemingly not required for phagosome maturation or acidification, we reasoned that accumulation of luminal Cl− via the antiporter may instead be the critical determinant of resolution. In this context, it is relevant that early studies demonstrated that cathepsin C, a prototypical lysosomal protease, requires Cl− for optimal activity (Cigic and Pain, 1999; McDonald et al., 1966). To assess whether the intraphagosomal Cl− concentration exceeds that of the cytosol, RAW 264.7 cells were induced to engulf zymosan particles, which are porous and allow permeation of luminal fluid (Fig. 5 A). The ingestion of multiple zymosan particles caused the total volume of the cells to increase; the net volume increase was estimated by electronic cell sizing. We next measured the intracellular Cl− content and used the measured volume to compare the overall intracellular [Cl−] of cells before and after phagocytosis. Ingestion of zymosan was associated with a ∼50% increase in overall intracellular [Cl−] (Fig. 5 B). Assuming that the Cl− content and volume gain are both wholly due to the phagosomes, we calculated the mean intraphagosomal [Cl−] to be 157 ± 78 mM (Fig. 5 C), which is consistent with other reports in the literature (Riazanski et al., 2021; Tan et al., 2013). Importantly, the net Cl− content gain was missing in cells devoid of ClC-7, which nevertheless ingested a comparable number of zymosan particles (Fig. S5 B) and gained an equivalent volume (Fig. S5 C), resulting in a reduced overall Δ[Cl−] that did not attain statistical significance. As a result, the accumulation of Cl− estimated to occur in wildtype phagosomes was absent in the Clcn7 KO phagosomes (Fig. 5 C). These findings are consistent with earlier observations comparing wildtype (Saha et al., 2015) and Clcn7 KO lysosomes (Weinert et al., 2010). The very low [Cl−] calculated for the KO phagosomes may indicate that they are much less permeant to Cl−, but may also result from errors in the calculations, which assume that the cytosolic volume and [Cl−] remain unaltered upon phagocytosis of multiple particles. It is conceivable that plasmalemmal volume-activated Cl− channels may have caused partial loss of cytosolic Cl−, distorting the calculations. In any event, it is apparent that the phagosomal [Cl−] concentration is higher than that of the cytosol, and that ClC-7 is required for the accumulation.
ClC-7 drives intraphagosomal Cl − accumulation and promotes protease activity. (A) Wildtype and Clcn7 KO RAW 264.7 cells were challenged with Alexa-568–conjugated zymosan particles. (B) Total intracellular [Cl−] of wildtype and Clcn7 KO cells with or without zymosan, determined as described in Materials and methods. n = 3. (C) Intraphagosomal [Cl−] in wildtype and Clcn7 KO cells estimated as described in the text based on the cellular Δ[Cl−] and Δ volume between cells with and without zymosan. n = 3.
ClC-7 drives intraphagosomal Cl − accumulation and promotes protease activity. (A) Wildtype and Clcn7 KO RAW 264.7 cells were challenged with Alexa-568–conjugated zymosan particles. (B) Total intracellular [Cl−] of wildtype and Clcn7 KO cells with or without zymosan, determined as described in Materials and methods. n = 3. (C) Intraphagosomal [Cl−] in wildtype and Clcn7 KO cells estimated as described in the text based on the cellular Δ[Cl−] and Δ volume between cells with and without zymosan. n = 3.
Cl− activates select but divergent hydrolases
While cathepsin C has been demonstrated to require Cl− as a cofactor for its optimal activity, it is still unknown whether Cl− is important for the activity of other hydrolases present in phagolysosomes. To address this question, the activity of several other enzymes was tested in vitro as a function of the concentration of Cl−, while keeping the pH at 5.5. Cl− concentration was varied between 0 and 100 mM while maintaining the ionic strength using NO−3 as a substitute. As reported (Cigic and Pain, 1999; McDonald et al., 1966), we confirmed that cathepsin C activity is markedly higher in the presence of 100 mM Cl− than in its absence (Fig. 6 A). Lysozyme, a major antimicrobial glycosidase expressed by macrophages, also displayed an extraordinary dependence on Cl− (Fig. 6 B). DNase II, a nuclease also present in phagolysosomes, similarly showed a profound dependence on Cl− (Fig. 6 C). Not all hydrolases tested were comparably Cl− sensitive: cathepsin B and acid lipase showed similar activity in the presence of 100 mM Cl− (0 mM NO−3) and in its absence (100 mM NO−3; Fig. 6, D and E). To further validate this result, we assayed cathepsin B in cells with a membrane-permeant fluorescent substrate (Magic Red cathepsin B) and found no difference in activity between wildtype and Clcn7 KO macrophages (Fig. S5, D and E).
Cl−activates hydrolases in vitro. (A–E) Enzyme activities were measured in vitro in solutions of pH at 5.5 containing varying concentrations of Cl−, substituted with NO−3. n = 3. RFU: relative fluorescence units. (F) Lysates of wildtype and Clcn7 KO cells were probed with anti–cathepsin C antibody and anti-vinculin antibody used to normalize loading. Molecular weight markers (kD) are indicated left of the panels. (G) Quantification of mature cathepsin C levels relative to the total cathepsin C (mature plus pro-cathepsin) levels from three separate experiments like that shown in F. Source data are available for this figure: SourceData F6.
Cl−activates hydrolases in vitro. (A–E) Enzyme activities were measured in vitro in solutions of pH at 5.5 containing varying concentrations of Cl−, substituted with NO−3. n = 3. RFU: relative fluorescence units. (F) Lysates of wildtype and Clcn7 KO cells were probed with anti–cathepsin C antibody and anti-vinculin antibody used to normalize loading. Molecular weight markers (kD) are indicated left of the panels. (G) Quantification of mature cathepsin C levels relative to the total cathepsin C (mature plus pro-cathepsin) levels from three separate experiments like that shown in F. Source data are available for this figure: SourceData F6.
It should be noted that, in situ, cysteine proteases, including cathepsins, are synthesized as inactive zymogens that are subsequently (auto)proteolytically cleaved to become mature proteases. To determine the role of ClC-7 in the maturation of hydrolases, we analyzed cell lysates of wildtype and Clcn7 KO RAW 264.7 cells using an anti–cathepsin C antibody and found that Clcn7 KO lysates contained markedly reduced levels of mature cathepsin C compared with lysates from wildtype cells (Fig. 6, F and G). Based on these results, we concluded that Cl− and ClC-7 are important for protease activity in vitro and activation in vivo, respectively.
Macrophage activation promotes ClC-7 expression and phagosome resolution
The striking dependence of several antimicrobial enzymes on Cl− prompted us to investigate whether the activation of macrophages influenced the kinetics of phagosomal resolution. To test this, primary BMDMs were treated with lipopolysaccharide (LPS). As shown in Fig. 7 A, activation elevated Clcn7 transcript levels fourfold in BMDMs. Importantly, LPS-activated macrophages degraded internalized red blood cells much faster than untreated counterparts (Fig. 7, B and C). To determine whether this effect was dependent on ClC-7, we primed wildtype and Clcn7 KO macrophages with LPS prior to phagocytosis and found that while wildtype phagosomes showed rapid resolution over 4 h, this was diminished in the Clcn7 KO cells (Fig. 7, D and E). Thus, the ClC-7 pathway that supplies luminal Cl− and fosters an antimicrobial microenvironment is itself boosted by exposure of macrophages to microbial-associated molecular patterns.
Macrophage priming promotes ClC-7 expression and phagosome resolution. (A) Determination of Clcn7 transcript abundance by RT-qPCR. n = 6. (B) Vehicle control and LPS-activated BMDM were challenged with IgG-opsonized sRBCs and imaged after 1–4 h as in Fig. 1. (C) Mean volume of sRBC-containing phagosomes in control and LPS-primed macrophages. n = 3 experiments, each counting >60 phagosomes. (D) Wildtype and Clcn7 KO RAW 264.7 cells treated with LPS and challenged with IgG-opsonized sRBCs for 1–4 h, then imaged as in Fig. 1. (E) Mean volumes of sRBC-containing phagosomes. n = 3 experiments, each counting >80 phagosomes.
Macrophage priming promotes ClC-7 expression and phagosome resolution. (A) Determination of Clcn7 transcript abundance by RT-qPCR. n = 6. (B) Vehicle control and LPS-activated BMDM were challenged with IgG-opsonized sRBCs and imaged after 1–4 h as in Fig. 1. (C) Mean volume of sRBC-containing phagosomes in control and LPS-primed macrophages. n = 3 experiments, each counting >60 phagosomes. (D) Wildtype and Clcn7 KO RAW 264.7 cells treated with LPS and challenged with IgG-opsonized sRBCs for 1–4 h, then imaged as in Fig. 1. (E) Mean volumes of sRBC-containing phagosomes. n = 3 experiments, each counting >80 phagosomes.
Discussion
Inhibiting the activity of the V-ATPase causes gross defects in the degradation of cargo targeted to (phago)lysosomes. While many hydrolases function, often optimally, at neutral pH in vitro, most intracellular degradative enzymes in macrophages depend on the V-ATPase (Pillay et al., 2002). Here, we show that the phagosomal H+ gradient established by the V-ATPase in addition provides the driving force for the secondary accumulation of luminal Cl− via the electrogenic 2Cl−/H+ exchanger, ClC-7, as had been previously demonstrated for lysosomes (Weinert et al., 2010). We found that Cl− was essential for various (phago)lysosomal enzymes to function optimally even at an acidic pH in vitro and was necessary for the destruction of phagocytic targets in situ. Our findings in macrophages are in good agreement with the observations that renal proximal tubule cells of Clcn7−/− mice degrade endocytosed protein more slowly (Wartosch et al., 2009); that loss of clh-6 (the Clcn7 homologue in Caenorhabditis elegans) led to diminished DNA degradation and lysosomal [Cl−] (Chakraborty et al., 2017); and that β-amyloid accumulates in microglia with knocked down expression of ClC-7 by siRNA (Majumdar et al., 2011). The precise manner whereby Cl− fosters the activity of some hydrolases is not known, but two enzymes previously shown to have Cl− dependence, cathepsin C and α-amylase, have Cl− interaction sites at or near their active sites (Aghajari et al., 2002; Cigic and Pain, 1999). Our findings suggest that Cl− may directly bind and act as a co-factor for a number of additional hydrolases, including lysozyme and DNases. Cl− could not only affect the activity of the hydrolases toward their substrates but also the (auto)catalytic conversion of the proforms of hydrolases to their active forms.
We found that a human glycanhydrolase (lysozyme) and a DNase (DNase II) are dependent on Cl− in vitro. Interestingly, these families of enzymes often function extracellularly, where Cl− concentration can exceed 100 mM. Some hydrolases also function critically in compartments generated by close contacts between myeloid cells and their neighboring (target) tissue. Examples include the crowning structures formed around adipocytes by resident macrophages (Sárvári et al., 2015). In a parallel context, ClC-7 plays an essential role in the degradation of bone by osteoclasts that make similarly tight contacts with the underlying bone matrix, generating sealed compartments referred to as lacunae. ClC-7 is targeted to the osteoclast surface facing the lacunae, where it is essential for their resorption: mutations in CLCN7/Clcn7 lead to osteopetrosis in humans and mice (Kornak et al., 2001). The loss of Clcn7 also leads to retinal degeneration in mice (Kornak et al., 2001), suggesting a similar role in juxtacellular target degradation by the retinal pigment epithelium. The striking reliance of lysozyme on high [Cl−] coupled with the intactness of red blood cell membranes 4 h after internalization by Clcn7-null or Cl−-depleted cells points to the potential Cl− dependence of other glycosidases. High [Cl−] may be necessary not only for the degradation of proteins and DNA but also for the protective outer layer (glycocalyx) of many biological targets. The role of high [Cl−] in facilitating the breakdown of the glycocalyx, which would in turn allow proteolytic and lipolytic access to the target, should be investigated further.
Uptake of Cl− via CFTR (Di et al., 2006) or ClC-7 (Graves et al., 2008) has been claimed to provide the counterion pathway that facilitates acidification in the endocytic pathway, notably in lysosomes. However, we found no significant expression of Cftr in primary macrophages, and the deletion of either ClC-3 or ClC-7 was without effect on the acidification of phago(lyso)somes. ClC-5, a 2Cl−/H+ exchanger of the CLC family, was shown to support the acidification of early and recycling endosomes, but has no effect on the acidification of late endosomes (Hara-Chikuma et al., 2005).
Interestingly, full phagosomal acidification was attained also when Cl− was exchanged for NO−3. In this context, it is important to note that ClCs are able to exchange NO−3 for H+, albeit ∼70% as effectively as Cl− (Bergsdorf et al., 2009; Ludwig et al., 2013). However, NO−3 does not support the enzymatic activity of Cl−-sensitive hydrolases. Thus, NO−3 taken up via ClCs may support acidification by serving as a counterion; this would only reinforce the notion that rather than being required for acidification, Cl− is directly required for the efficient degradation of phagocytic targets. Nevertheless, additional counterion transport pathways exist. In this regard, others have reported that cation transport out of phagolysosomes supports their acidification (Lin et al., 2015; Steinberg et al., 2010).
By mediating the efflux of H+ in exchange for Cl−, ClC-7 in principle contributes to the H+ leak from phagolysosomes. This contribution can be assessed by measuring the initial rate of change of the luminal pH when acutely inhibiting the V-ATPases. However, as shown in Fig. S5, F and G, the rate of alkalization of phagolysosomes recorded upon the addition of fully inhibitory concentrations of concanamycin A was not significantly different in cells devoid of ClC-7. This observation implies that ClC-7 may be minimally active once phagolysosomes reach steady-state pH, consistent with our conclusion that the antiporter is not required to either reach or maintain the physiological acidic pH.
While ClC-7 is not ostensibly involved in the establishment of the phagosomal pH, its possible osmotic contribution should be considered. The 2Cl− for 1H+ stoichiometry of the antiporter implies a net osmotic gain with every transport cycle. The osmotic gain is accentuated when considering that the extruded H+ can be replaced by H+ dissociating from luminal buffers. Indeed, gain-of-function mutations in CLCN7 have been reported to cause abnormal lysosomal swelling (Nicoli et al., 2019). Ongoing activity of the antiporter would compound the osmotic gain associated with the degradation of macromolecular cargo like proteins or polysaccharides into its constituent components during phagosome (and lysosome) maturation. Clearly, the activity of ClC-7 must be stringently regulated to avoid undue osmotic swelling that would compromise tubulation and resolution. Phosphoinositides appear to be critical in such regulation. As phagosomes mature, the signaling lipid PI(3)P on the early phagosomal membrane is converted to PI(3,5)P2 by the kinase PIKfyve. Strikingly, inhibition of PIKfyve leads to rapid swelling of endosomal compartments in a variety of cell types, markedly so in macrophages. It is noteworthy that the swelling is dependent on ClC-7 (Gayle et al., 2017) and on V-ATPase activity (Compton et al., 2016). It is also relevant that ClC-7 can directly associate with PI(3)P (Schrecker et al., 2020) and that the Arabidopsis thaliana homolog of ClC-7, ClC-a, has been shown to be inhibited by PI(3,5)P2 (Carpaneto et al., 2017). Recently, it has been demonstrated that PI(3,5)P2 can also directly inhibit ClC-7 activity (Leray et al., 2022). These observations suggest that having accumulated sufficient Cl− to support the activity of the hydrolases, ClC-7 is rendered quiescent by PI(3,5)P2 (and/or by depletion of PI(3)P) to minimize the osmotic burden of mature phagosomes or lysosomes that need to cope with the generation of monomeric products of macromolecule hydrolysis.
Lastly, little is known regarding how the Cl− accumulated in maturing phagolysosomes is cleared when they resorb. Putative efflux pathways include the acid sensitive outward-rectifying anion channel, which was recently demonstrated to serve such a role for nascent macropinosomes (Zeziulia et al., 2022) yet is absent in more mature compartments. Other Cl− channels CLN7 (Wang et al., 2021) and VRAC (Li et al., 2020) were recently reported to localize to the endocytic pathway and may be poised to contribute to the late stages of the resolution process. If so, these channels must be highly regulated to allow for the initial accumulation of luminal [Cl−] required for cargo degradation while enabling the subsequent resolution of the phagolysosomes.
In summary, our findings demonstrate that ClC-7, a 2Cl−/H+ exchanger localized to mature phagosomes, utilizes the H+ gradient established by the V-ATPase to drive the accumulation of intraphagosomal Cl−. This in turn supports the activity of multiple resident hydrolases, which degrade internalized targets and allow for the resolution of the phagosome. We expect that the findings from this study will challenge the long-standing paradigm that lysosomal hydrolases require only low pH to function optimally, highlighting the need for high [Cl−] and contributing to our understanding of the degradative capacity of organelles.
Materials and methods
Cell culture
RAW 264.7 and mouse embryonic fibroblasts were obtained from and authenticated by the American Type Culture Collection. Cell lines were grown in DMEM + 5% FBS at 37°C and under 5% CO2.
Primary BMDMs were isolated from the femurs and tibiae of 6–12-wk-old C57BL/6 wildtype mice, or C57BL/6 and 129/SvJ mixed background Clcn3−/− and Clcn3+/+ mice (Stobrawa et al., 2001), as described (Zeziulia et al., 2022). Briefly, cells were flushed out of bones with PBS, erythrocytes were lysed with red blood cell lysis buffer (ab204733; Abcam), and cells were resuspended in complete DMEM supplemented with 10% FBS, 1% penicillin–streptomycin, and 20 ng/ml recombinant murine macrophage colony-stimulating factor (M-CSF; 315-02; PeproTech). After 3 d in culture, the amount of medium was doubled with complete DMEM containing 20 ng/ml M-CSF. Cells were cultured in a humidified environment at 37°C and 5% CO2 and used 5–10 d after isolation.
Generation of Clcn7 KO clones
RAW 264.7 cells were seeded on 6-well tissue culture plates for 24 h and then transfected with a Guaranteed Predesigned CRISPR gRNA plasmid (Sigma-Aldrich) against Clcn7 (MMPD0000045620) containing Cas9 and GFP to indicate transfection. Cells were then incubated with propidium iodide, and live, transfected cells (GFP-positive and propidium iodide–negative) were sorted by FACS into 96-well tissue culture plates. After expansion, KO clones were verified by immunoblotting.
Reagents
Mammalian expression vectors were obtained from the following sources: OSTM1-OFP (Sino Biological); LAMP1-GFP was described previously (Johnson et al., 2016). Rat ClC-7 sequence was inserted in EGFP-N1 backbone vector with restriction cloning. Primary antibodies were obtained from the following sources: anti-LAMP1 (Developmental Studies Hybridoma Bank; catalog no. H4A3-s), anti-sheep red blood cell (cat#: 55806; Cedarlane Laboratories), anti-GAPDH (EMD Millipore; cat#: MAB374), anti–cathepsin C (cat#: sc-74590; Santa Cruz), and anti-vinculin (cat#: MAB3574; Millipore Sigma). Secondary antibodies conjugated with AlexaFluor-405, -488, -555, -647, or -HRP were obtained from Jackson ImmunoResearch. SidK-568 was prepared according to Maxson et al. (2022). Rabbit anti-ClC-7 was produced as previously described (Kornak et al., 2001).
TaqMan probes for real-time PCR were all obtained from Thermo Fisher Scientific and are cataloged as follows: CLCN3 (Mm01348786); CLCN4 (Mm00441247); CLCN5 (Mm00443851); CLCN6 (Mm00442366); CLCN7 (Mm00442400); LRRC8A (Mm00624918); CFTR (Mm00445197); and ABT1(Mm00803824).
FITC, which was used to label red blood cells and zymosan particles, was from Sigma-Aldrich (cat#: F4274). Other reagents include fluorescently labeled phalloidin (Acti-Stain; cat#: PHDG1-A; Cytoskeleton Inc.), concanamycin A (cat#: C9705; Sigma-Aldrich), nigericin (cat#: ab120494; Abcam), monensin sodium salt (cat#: 475895; Sigma-Aldrich), DCPIB (cat#: 1540; Tocris Bioscience), and 10 kD tetramethylrhodamine dextran (cat#: D1868; Invitrogen). Sheep red blood cells were purchased from MP Biomedicals (cat#: 55876). Paraformaldehyde was obtained from Electron Microscopy Sciences (cat#: 15710). Pepstatin-A (cat#: P5318) and E-64 (cat#: E3132) protease inhibitors were both purchased from Sigma-Aldrich. CellTiter-Glo luminescent viability assay was purchased from Promega (cat#: G7570). Cathepsin B Assay Kit (Magic Red Cathepsin B) was purchased from Abcam (cat#: ab270772). Rhodamine 123 was purchased from Thermo Scientific (cat#: 419091000). Fluoresbrite YG Carboxylate Microspheres were obtained from Tebubio (cat#: 17147-5), and silica beads were obtained from Bangs Laboratories Inc. (cat#: SS05003).
For the in vitro enzyme activity assays, the following enzymes and respective substrates were used: bovine cathepsin C (cat#: C8511; Sigma-Aldrich) and gly-phe-β-naphthylamide (cat#: 14634; Cayman Chemical), human cathepsin B (cat#: C0150; Sigma-Aldrich) and Z-Arg-Arg-MbNA (cat#: 866-31; Echelon Biosciences), human lysozyme (cat#: L1667; Sigma-Aldrich) and Micrococcus lysodeikticus dried cells (cat#: M-0128; Sigma-Aldrich), human lysosomal acid lipase (cat#: CSB-YP012972HU; CusaBio) and LysoLive Lysosomal Acid Lipase assay Kit (cat#: ab253380; Abcam), and bovine Dnase II (cat# D8764; Millipore Sigma) and DNA from fish sperm (cat#: 11467140001; Roche). DTT was obtained from BioBasic Inc. (cat#: DB0058), and ethylenediamine tetraacetic acid was obtained from Fisher Chemical (cat#: SS412-1).
Red blood cell preparation
30 μl of a 10% suspension of sheep red blood cells (sRBCs) was resuspended in 500 μl of PBS. Cells were washed with PBS two times by centrifuging at 2,000 g for 3 min and subsequently labeled with 50 μl of 10 mg/ml FITC in 1.5 ml PBS at 37°C for 2 h, shaking at 700 rpm. sRBCs were then washed three times with PBS and incubated for 30 min with 1.5 μl of anti-sRBC IgG fraction, shaking at 300 rpm at 37°C. Then sRBCs were washed three times with PBS and stored at 4°C for up to 1 wk. Unlabeled sRBCs were prepared in the same manner, omitting the FITC incubation step.
Quantitative RT-PCR (RT-qPCR)
BMDMs and mouse embryonic fibroblasts were plated on 6-well tissue culture plates. Cells were harvested 24 h after and cellular RNA was extracted using the GeneJET RNA purification kit (Thermo Fisher Scientific). Equal RNA from each condition was then used to generate cDNA with the Superscript VILO cDNA Synthesis Kit (Thermo Fisher Scientific). Quantitative real-time PCR was then performed with TaqMan probes (Thermo Fisher Scientific) specific to each assayed gene on a QuantStudio 3 Real-Time PCR System (Applied Biosystems, Thermo Fisher Scientific) in 96-well plates. Readouts for reference (Abt1) and target genes were performed in triplicate. Relative gene expression was calculated based on the 2–∆∆Ct method by comparing the relative quantification of the control gene to each target gene.
Phagocytic index
Phagosome resolution
RAW 264.7 cells and BMDMs were seeded on 18-mm coverslips in 12-well tissue culture plates 24 h prior to experimentation. Cells were then incubated for the indicated duration with sRBCs opsonized with rabbit anti-sheep antibody. In all cases, the cells were washed 20 min after incubation to ensure that all unbound particles were removed. To visualize the macrophages and sRBCs, cells were fixed, permeabilized, and coincubated with fluorescent secondary antibodies against the opsonizing antibody and fluorescent phalloidin to visualize the cells for 30 min prior to imaging.
Transient plasmid DNA transfection
RAW 264.7 cells were seeded on 18-mm coverslips in 12-well tissue culture plates at a concentration of ∼2 × 105 cells a day prior to transfection. Cells were then transiently transfected with desired plasmid DNA using FuGENE HD (Promega) transfection reagent at a ratio of 3:1 (FuGENE to DNA). Medium was replaced 6 h after transfection, and cells were imaged 16–18 h later.
Ratiometric pH measurements
Wildtype and Clcn7 KO RAW 264.7 cells were seeded on 18-mm coverslips in 12-well tissue culture plates 24 h prior to experimentation. Macrophages were incubated with FITC-conjugated IgG opsonized sRBCs for 1 h, then subsequently imaged to determine the steady-state phagosomal pH. To measure the nascent rate of acidification, macrophages were incubated with FITC-conjugated IgG-opsonized sRBCs, spun down at 1,000 rpm for 1 min to synchronize particle binding, and immediately imaged over the course of 15 min. In both cases, the FITC was excited at 490 nm, which emits at 520 nm in a pH-dependent manner, as well as at 440 nm, which emits at 520 nm in a manner that is much less sensitive to pH. Cells were then sequentially bathed in standard buffers (143 mM KCl, 5 mM glucose, 1 mM MgCl2, 1 mM CaCl2, and 20 mM Hepes) at pH 7.5, 6.5, 5.5, and 4.5 also containing 10 μM nigericin and 5 μM monensin for 10 min. The 490 nm/440 nm fluorescence ratio was collected at each pH to construct a calibration curve correlating the mean fluorescence ratio (background subtracted) against the known pH values, fitted by the least squares method. This was ultimately utilized to convert the recorded fluorescence ratio values to pH values.
Immunofluorescence
RAW 264.7 cells and BMDMs were seeded on 18-mm coverslips in 12-well tissue culture plates 24 h prior to experimentation. After phagocytosis was induced, cells were fixed in 3% paraformaldehyde for 5 min, permeabilized in 0.1% Triton-X100 for 5 min, and blocked in 5% BSA for 30 min. Subsequently, the cells were incubated with the indicated primary antibodies in 5% BSA for 1 h, followed by three washes with 1X PBS, and then incubated with appropriate fluorescent secondary antibodies and reagents (e.g., fluorescently labeled phalloidin, SidK) in 5% BSA for 1 h. Cells were then washed with 1X PBS and imaged immediately. All steps were performed at room temperature.
To observe the localization of LAMP1 and the V-ATPase to early and late phagosomes, cells were seeded on 18-mm coverslips in 12-well tissue culture plates 24 h prior to experimentation. Cells were then incubated with IgG-opsonized sRBCs, spun down at 1,000 rpm for 1 min, then allowed to internalize particles for 3 or 20 min, followed by three washes with 1X PBS. Cells were then fixed, permeabilized, and incubated with primary and secondary antibodies (or SidK-568) as described above.
To establish the subcellular localization of ClC-7, BMDMs were seeded on 18-mm coverslips in 12-well tissue culture plates 24 h prior to experimentation. Cells were then subsequently incubated with either Fluoresbrite YG carboxylate microspheres (3.2 μm) for 30 min, or IgG-opsonized silica beads (4.98 μm) for 3 min or 1 h, then fixed, permeabilized, and incubated with primary anti–ClC-7 antibodies followed by secondary antibodies. Samples were then imaged as described above.
Immunoblotting
Wildtype and Clcn7 KO RAW 264.7 cells were grown on 6-well tissue culture plates 24 h prior to harvesting. Cells were subsequently collected on ice and lysed in 100 μl radioimmunoprecipitation assay buffer containing protease and phosphatase inhibitors. Protein concentrations were calculated using BCA standards (Pierce), and samples were diluted in Laemmli buffer containing 10% 2-mercaptoethanol and boiled for 5 min. Samples were then run on 10% SDS-PAGE, transferred onto a polyvinylidene difluoride membrane, and blocked in TBS with 0.05% Tween-20 (TBST) and 5% BSA overnight at 4°C. Primary antibody staining was done in 5% BSA in TBST for 1 h at room temperature at a concentration of 1:1,000 for all antibodies. The membrane was then washed three times with TBST, and subsequently incubated with secondary antibody conjugated to HRP at a concentration of 1:2,000 for 1 h at room temperature before another three washes in TBST. Blots were developed using the ECL Prime Western Blot detection reagent and visualized with a ChemiDoc MP Imaging System (BioRad) and Image Lab software 5.2.1 (BioRad).
DQ-BSA assay
RAW 264.7 cells were seeded on 18-mm coverslips in 12-well tissue culture plates 24 h prior to experimentation. Cells were then incubated with IgG-opsonized DQ-BSA coated silica beads for 1 h prior to imaging. In brief, silica beads were incubated with DQ-BSA red (Molecular Probes) shaking at room temperature at 950 rpm for 2 h. Beads were subsequently washed three times with 1X PBS to remove excess DQ-BSA, then opsonized with human IgG at 37°C, shaking at 950 rpm for 30 min. Beads were then washed three times with excess 1X PBS prior to addition to cells. The fluorescence of the DQ-BSA signal from beads that were never internalized was used as background and was subtracted from intracellular measurements.
Cl− depletion and colorimetric determination of Cl−
Intracellular Cl− depletion and determination
Colorimetric determination of Cl− was adapted from Zall et al. (1956). In brief, cells were either incubated in chloride-containing buffer (130 mM NaCl, 3 mM KCl, 1 mM MgCl2, 1 mM CaCl2, 10 mM glucose, and 20 mM Hepes, pH 7.4), or nitrate buffer (130 mM NaNO3, 3 mM KNO3, 1 mM Mg(NO3)2, 1 mM Ca(NO3)2, 10 mM glucose, and 20 mM Hepes, pH 7.4) for 1 or 4 h, then subsequently washed three times in ice-cold nitrate buffer and lysed in 1% HNO3 overnight. Lysates were then spun down to remove membranes, and the supernatant was mixed in equal parts with a mercury solution (1 part 0.417% mercuric thiocyanate in methanol, 1 part 20.2% ferric nitrate solution, and 13 parts water) in a 96-well plate. The absorbance at 490 nm is indicative of the concentration of Cl− in the sample and was measured using a microplate reader (Biotek, Epoch Microplate Spectrophotometer). By using [Cl−] standards to construct a standard curve, 490 nm absorbance values were correlated to Cl− concentrations. To determine intracellular [Cl−], cell size and number were determined for each sample with a Coulter counter (Beckman Coulter Life Sciences, Multisizer 4) in parallel, which taken together with the [Cl−] of each sample allowed for the calculation of the mean intracellular [Cl−] per cell.
Intraphagosomal [Cl−]
Recombinant hydrolase activity assays
All assays were conducted using 96-well plates and a microplate reader (Varioskan LUX), unless otherwise stated. The activity of cathepsin C was determined with Gly-Phe-β-naphthylamide as substrate. Activity was determined by the appearance of the fluorescent product as a result of proteolytic cleavage, measured at 415 nm with excitation at 360 nm. The activity of human cathepsin B was determined using Z-Arg-Arg-MbNA as substrate, which is cleaved into free 4-methoxy-β-naphthylamine that excites at 350 nm and emits at 440 nm. Cathepsin activity assays were performed in sodium acetate buffer (50 mM sodium acetate, 1 mM DTT, 1 mM EDTA, pH 5.5) with final concentrations of 100 μM of substrate and 0.1 units of enzyme per condition, and then incubated for 1 h at 37°C prior to data collection.
The activity of bovine DNase was determined by adding the recombinant enzyme to solutions of fish DNA and measuring the increase in absorbance at 260 nm, which correlates with the liberation of free nucleotides as the DNA is degraded. The activity assay was performed in sodium acetate buffer (50 mM sodium acetate, 4.2 mM magnesium sulfate, pH 5.5) using a final concentration of 0.003% (wt/vol) of DNA and 300 units of enzyme. DNase activity assays were performed with a 10 mm quartz microcuvette (cat#: 105-QS; Hellma).
The activity of human lysozyme was determined by incubating recombinant enzyme with resuspended Micrococcus lysodeikticus, measuring the loss of absorbance at 450 nm, which correlates with degradation of the bacterium. Activity assays were performed in potassium phosphate buffer (50 mM KH2PO3) with final concentrations of 0.5 μg/ml of bacterium and 30 μg/ml of enzyme per condition at pH 5.5. Absorbance was measured immediately after the addition of enzyme for 5 min, and enzymatic activity was calculated based on the rate of loss of absorbance over time.
The activity of human acid lipase was determined with the LysoLive Lysosomal Acid Lipase Assay Kit. Activity assays were carried out in sodium acetate buffer (50 mM sodium acetate, 1 mM DTT, 1 mM EDTA) with 0.001 unit of LipaGreen reagent and incubated for 8 h at 37°C prior to data collection.
All assays were performed at pH 5.5 in media with varying amounts of NaCl, using sodium nitrate to maintain the final buffer osmolarity at 200 mOsm.
Cathepsin C activation assay
Wildtype and Clcn7 KO RAW 264.7 cell lysates were separated by SDS-PAGE and probed with anti–cathepsin C and anti-vinculin antibodies. The percentage of active cathepsin C was determined by calculating the ratio of the mature cathepsin C (∼23 kD) over the total cathepsin C (sum of mature cathepsin C and procathepsin C band at ∼60 kD). Signals were normalized to the amount of vinculin, used as loading control. Measurements were performed in ImageJ.
Labeling of phagolysosomes with dextran pulse chase
RAW 264.7 cells seeded on 18-mm coverslips in 12-well tissue culture plates were incubated (“pulsed”) with 100 μg/ml 10 kD tetramethylrhodamine conjugated dextran overnight, then washed three times with complete media (“chased”) 1 h prior to experimentation to allow for the accumulation of labeled dextran in lysosomes. Cells were then incubated with IgG-opsonized FITC-conjugated sRBCs, and the volume of phagosomes was visualized via the accumulation of dextran within the sRBC-containing phagosomes.
Determination of intracellular ATP levels
RAW 264.7 wildtype and Clcn7 KO cells were seeded in 96-well tissue culture plates, which were then incubated in Cl−-containing or Cl−-depleted medium 1 h prior to experimentation. Cells were then acutely incubated with CellTiter-Glo reagent according to manufacturer instructions, and luminescence was measured with a Varioskan LUX plate reader.
Determination of H+ leak
RAW 264.7 wildtype and Clcn7 KO cells seeded on 18 mm coverslips in 12-well tissue culture plates were incubated with IgG-opsonized FITC-conjugated sRBCs for 1 h prior to experimentation. Cells were then acutely treated with 500 nM concanamycin A, a potent inhibitor of the V-ATPase, and the pH of the sRBC-containing phagosomes was measured ratiometrically over 10 min.
Microscopy
Confocal microscopy was performed on a spinning-disk system (Quorum Technologies Inc.), which consists of a microscope (Axiovert 200 M, Zeiss), cooled CCD camera (ORCA-Fusion BT, Hamamatsu), five-line laser module (Spectral Applied Research) with 405-, 443-, 491-, 561-, and 655-nm lines, and a filter wheel (MAC5000, Ludl). The microscope is operated using Volocity v6.3 software (Perkin Elmer). Images were acquired with a 63×/1.4 NA oil objective (Zeiss).
Ratiometric pH imaging was performed on a microscopy system that consists of a microscope (Axio Observer, Zeiss), a cooled CCD camera (Evolve 512, Photometrics), and a fluorescent light source (HXP 120V, Zeiss). Ratiometric imaging was performed with two filter cubes: a 470/40 nm and a 436/20 nm. The microscope is operated using ZEN 2 blue edition software (Zeiss), and images were acquired with a 63×/1.4 NA oil objective (Zeiss).
Imaging of Clcn3−/− and wildtype BMDM was performed on a Nikon Ti Eclipse microscope operated with NIS software (version 5.21.03, Build 1481) at 37°C (Okolab incubator) every minute for 15 min on 20 × air objective (Plan Apo NA 0.75, Nikon) with a back-illuminated sCMOS camera (Prime95b, Photometrics). Imaging was done using epifluorescence LED (CoolLED, pE4000) at 435 nm (full width at half maximum: 17 nm) as a normalization wavelength and 490 nm (full width at half maximum: 20 nm) as a pH-sensitive wavelength with excitation filters at 436/20 nm and 480/40 nm; FT510 Dichroic and a 535/30 (Zeiss) were used as emission filters.
Image analysis
Image processing and analyses were performed using Volocity v6.3 (Perkin Elmer). For resolution analyses, the volume of individually labeled sRBCs was determined with the “find objects” function of Volocity. In brief, sRBCs were labeled with fluorescent opsonin, which coated the membrane and created outlines of each particle. These outlines were then detected by “find objects” in each z-stack plane, which when merged together yielded the volume of each sRBC. The mean fluorescence emission at 520 nm of FITC-labeled sRBCs was acquired at 490- and 440-nm with the “find objects” function, and subsequently exported to Microsoft Excel for further data analysis. Pseudocolour representations were created in ImageJ using the “fire” look-up table function and were used to visualize the relative fluorescence intensity of DQ-BSA–coated silica beads contained within phagosomes. Imaging data for ClC3−/− BMDMs were analyzed using custom code written in Python (version 3.8).
General methodology and statistics
Data are presented as mean ± SEM. Statistical significance of data was determined using unpaired Student’s t tests with P < 0.05 considered significant. Statistical analyses were performed in GraphPad Prism 6 and Microsoft Excel. Each experiment was performed at least three times.
Online supplemental material
Fig. S1 shows the validation that phagosomal volume is closely approximated by measuring sheep red blood cell volume and illustrates the defect in phagosomal resolution in RAW 264.7 cells upon protease treatment. Fig. S2 shows the effect of Cl− omission and of Clcn7 deletion on various markers of cellular metabolism and lysosomal pH. Fig. S3 shows the failure of pharmacological inhibition of VRAC to affect phagosomal resolution. Fig. S4 shows that macrophages devoid of ClC-3 or ClC-7 both ingest similar numbers of particles as their wildtype counterparts and that endogenous ClC-7 is localized to mature phagosomes. Fig. S5 shows factors that remain unchanged upon deletion of Clcn7, including phagosomal pH (containing zymosan), phagocytic index, volume gain upon uptake of zymosan, cathepsin B activity, and phagosomal H+ leak.
Data availability
Original data are available from the corresponding authors upon request.
Acknowledgments
Imaging was performed at Hospital for Sick Children Imaging Facility and Leibniz-Forschungsinstitut für Molekulare Pharmakologie Cellular Imaging Facility; we thank Dr. Martin Lehmann for assistance with imaging assays. We thank Mohamad Kabbani for technical assistance with BMDMs preparation.
J.Z. Wu is funded by an Ontario Graduate Scholarship. S.A. Freeman and S. Grinstein are supported by grants PJT-169180 and FDN-143202 from Canadian Institutes of Health Research. S.A. Freeman holds a Canada Research Chair from Canadian Institutes of Health Research. Funded, in part, by grants of the Deutsche Forschungsgemeinschaft (FOR 2652 (Je164/14-1) and under Germany’s Excellence Strategy—EXC-2019-390688087 “Neurocure” to T.J. Jentsch.
Author contribution: J. Wu: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Writing, and Editing. M. Zeziulia: Investigation, Methodology, and Writing. W. Kwon: Investigation. T.J. Jentsch: Resources, Funding acquisition, Writing, Editing, and Supervision. S. Grinstein: Conceptualization, Resources, Funding acquisition, Project administration, Writing, Editing, and Supervision. S.A. Freeman: Conceptualization, Resources, Funding acquisition, Project administration, Writing, Editing, and Supervision.
References
Author notes
Disclosures: The authors declare no competing interests exist.


![Phagosomal resolution necessitates ongoing activity of the V-ATPase and intracellular Cl−. (A and B) Vehicle control and concanamycin A–treated RAW 264.7 cells challenged with IgG-opsonized sRBCs for 1–4 h and stained for F-actin (cyan) and anti-IgG (red). Concanamycin A (500 nM) was added 30 min after phagocytosis. Representative images shown in A. Mean ± SEM of sRBC phagosome volumes graphed in B. n = 3, each independent experiment counting >50 phagosomes. (C) Representative fluorescence images of vehicle control and concanamycin-treated RAW 264.7 cells challenged with DQ-BSA–coated silica beads for 1 h. (D) Quantification of relative DQ-BSA fluorescence intensity in vehicle control and concanamycin A–treated RAW 264.7 cells. n = 3, each independent experiment counting >30 phagosomes. (E) Colorimetric determinations of intracellular [Cl−] for RAW 264.7 cells incubated in medium containing Cl− or NO3− (Cl−-depleted) for 1 or 4 h. n = 3. (F) Phagosome resolution in control and Cl−-depleted RAW 264.7 cells challenged with IgG-opsonized sRBCs for 1–4 h. (G) Mean ± SEM volume of sRBC-containing phagosomes. n = 3, each independent experiment counting >50 phagosomes. (H) Representative fluorescence images of control and Cl−-depleted RAW 264.7 cells challenged for 1 h with silica beads precoated with DQ-BSA. (I) Quantification of relative DQ-BSA intensities in control and Cl−-depleted RAW 264.7 cells. n = 5, each independent experiment counting >25 phagosomes.](https://cdn.rupress.org/rup/content_public/journal/jcb/222/6/10.1083_jcb.202208155/1/m_jcb_202208155_fig1.png?Expires=1773753684&Signature=flFNyfRRwp6Bprxul-yyAVi-ofAsDYd8FH-3pcMSQ~ge-d9-7trYpgH2a8HUzxiuHMaR--XXmaKdYtepVoO3xqoEzvCViAsDcW6uhyCOuRMYKGmQUsbOI59aohWtgLXnhL~yotnrD3~PqJhO7eAauMjKJ4xXwqOJ5DNXQlszJyVWoIGLhWROShyB5ouE7NaX18aP59HFwDGGYKcEo35BX6TF7WXrLyYE3hyN58YVjyN1HsINCH7Dgvo7bbLfyx9taIHRAg6GiqsQ3nkW0P3oPxGqReGYpkPN6fa5zsGx7BCRzyawZBa2viKpZEYni3O49NFCSh~1uDG2PAH8twADFg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)







![ClC-7 drives intraphagosomal Cl−accumulation and promotes protease activity. (A) Wildtype and Clcn7 KO RAW 264.7 cells were challenged with Alexa-568–conjugated zymosan particles. (B) Total intracellular [Cl−] of wildtype and Clcn7 KO cells with or without zymosan, determined as described in Materials and methods. n = 3. (C) Intraphagosomal [Cl−] in wildtype and Clcn7 KO cells estimated as described in the text based on the cellular Δ[Cl−] and Δ volume between cells with and without zymosan. n = 3.](https://cdn.rupress.org/rup/content_public/journal/jcb/222/6/10.1083_jcb.202208155/1/m_jcb_202208155_fig5.png?Expires=1773753684&Signature=PGb0wT7BwCIvA5S34K02EyjHj6kUKXyZKvO8sl~5LJEOH3bgexvnN7vsVTxmMFFqnTwUsjm0vHPzAhhKN9WC5Y7nL~YF6EpGpYHBGRzj96JGY8vWxUswSY3Rf2glnRqTbtxwnaYAzcDa5ht~kfJPbIMIcjBBVTlrQt-~0KwygFrqk9aPA2Y9InNkgB26qIaVkuFK1ofND13anZyIePQ4QZTWy5~zX~BENxEMtK-luj7O~dC6t6Gdk5g6O7NVF~tbka6zEklbmLAbmcU9ljDkR1lVDE5Aopevl5RjKKxOn5fKHJuNsMha~aiN6Uxy0vLRgWiAksDRY0Kx2W7uB495Kw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)




