The actin-rich cortex plays a fundamental role in many cellular processes. Its architecture and molecular composition vary across cell types and physiological states. The full complement of actin assembly factors driving cortex formation and how their activities are spatiotemporally regulated remain to be fully elucidated. Using Dictyostelium as a model for polarized and rapidly migrating cells, we show that GxcM, a RhoGEF localized specifically in the rear of migrating cells, functions together with F-BAR protein Fbp17, a small GTPase RacC, and the actin nucleation-promoting factor WASP to coordinately promote Arp2/3 complex-mediated cortical actin assembly. Overactivation of this signaling cascade leads to excessive actin polymerization in the rear cortex, whereas its disruption causes defects in cortical integrity and function. Therefore, apart from its well-defined role in the formation of the protrusions at the cell front, the Arp2/3 complex-based actin carries out a previously unappreciated function in building the rear cortical subcompartment in rapidly migrating cells.
Introduction
The cell cortex is defined as a thin layer of filamentous actin, myosin motors, and regulatory proteins beneath the plasma membrane (Svitkina, 2020). Assembly and contraction of this actin meshwork generate cortical tension, which enables cells to resist external mechanical stresses, change shape, and exert forces (Chugh et al., 2017; Kelkar et al., 2020). Consequently, the cortex plays a critical role in a variety of cellular processes, including division, migration, and morphogenesis (Heisenberg and Bellaïche, 2013; Salbreux et al., 2012). The mechanical properties of the cortex are key to its physiological function. Changes in cortical mechanics can originate from changes in the architecture of the actin network (Fritzsche et al., 2016; Koenderink and Paluch, 2018; Svitkina, 2020). However, the complete inventory of assembly factors driving the formation of the actin cortex and how their activities are spatiotemporally controlled are not well understood.
Actin polymerization is mainly initiated by two classes of nucleators in the cell: the Arp2/3 complex, which creates branches at the sides of preexisting filaments to generate a dense meshwork, and formins, which nucleate and elongate long and linear actin filaments (Pollard, 2007). Recent biochemical and functional studies have implied that both Arp2/3 and formins are involved in the formation of the cortical actin cytoskeleton, though their relative contributions vary among cell types (Bovellan et al., 2014; Chan et al., 2019; Chugh et al., 2017; Fritzsche et al., 2016; Litschko et al., 2019; Rosa et al., 2015; Severson et al., 2002). For example, the Arp2/3 complex is largely dispensable for the formation of the cell cortex of mitotic epithelial cells within the fly notum and Caenorhabditis elegans embryos (Rosa et al., 2015; Severson et al., 2002). In contrast, in M2 melanoma cells and mitotic HeLa cells, both Arp2/3 and the diaphanous-related formin (DRF) mDia1 contribute to cortical F-actin, though with different effects on cortical integrity and cell behavior (Bovellan et al., 2014). Inhibition of the Arp2/3 complex potentiates the effect of mDia1 depletion, suggesting synergistic activities of the two types of actin nucleators (Bovellan et al., 2014).
The activities of Arp2/3 complex and DRFs are tightly regulated. The DRFs adopt an autoinhibited conformation through intramolecular interactions; they can be activated by binding with Rho family GTPases (Breitsprecher and Goode, 2013; Pollard, 2007). The activation of the Arp2/3 complex relies on nucleation-promoting factors (NPFs) of the Wiskott-Aldrich syndrome protein (WASP) family, which consists of two principal classes of protein: WASPs and WASP family verprolin homologous (WAVE) complex (also called SCAR complex). Both of them contain a C-terminal VCA domain that binds and activates Arp2/3 in response to numerous inputs, including Rho GTPases, phosphoinositide lipids, SH3 domain-containing proteins, kinases, and phosphatases (Pollard, 2007; Pollitt and Insall, 2009). The presence of several Arp2/3 NPFs in the cortex has been shown recently by proteomic analysis and microscope imaging (Bovellan et al., 2014; Cao et al., 2020). Modulating the activities of Arp2/3 and formins may fine-tune the composition, structural organization, and mechanics of the actin cortex.
Dictyostelium discoideum provides a valuable system for studying the architecture of the actin cortex in rapidly moving and polarized cells. Dictyostelium cells exemplify amoeboid migration, which is characterized by weak adhesions, actin-rich protrusions or blebs in the front, and actomyosin-driven contraction in the rear (Devreotes et al., 2017; Lämmermann and Sixt, 2009; Paluch et al., 2016). Previous studies have demonstrated that three DRFs (ForA, ForE, and ForH), which are recruited and activated by the RhoA-like GTPase RacE, act together to safeguard cortical integrity in the rear of migrating cells (Litschko et al., 2019). Evidence suggests that the Arp2/3 complex also takes part in building the rear cortex. First, cryoelectron tomography of peripheral regions in Dictyostelium cells revealed isotropic actin-filament arrays (Medalia et al., 2002). Second, a significant amount of cortical actin meshwork remained in cells with all three DRFs deleted (Litschko et al., 2019). Third, antibodies to the Arp2/3 complex stained around the cortex in related amoeba cells (Machesky et al., 1994; Mullins et al., 1997). However, prominent Arp2/3 activities are mainly found at the leading-edge, including structures of pseudopods and macropinocytic cups (Davidson et al., 2018; Veltman et al., 2012; Veltman et al., 2016; Yang et al., 2021), and whether and how the Arp2/3 complex can be brought to the rear to promote cortical actin generation remains unknown.
In this study, we show that GxcM, a RhoGEF protein localized in the rear of Dictyostelium cells, functions together with F-BAR protein Fbp17, a Rac family GTPase RacC, WASP, and Arp2/3 complex, in a signaling cascade to coordinately regulate the formation of the rear cortical actin subcompartment and maintain cortical integrity.
Results
RhoGEF protein GxcM localizes in the rear of migrating cells
We reasoned that, for Arp2/3-based actin to be involved in the formation of the rear cortex, asymmetric positioning of upstream regulators may be required. Rho family GTPases and their respective guanine nucleotide exchange factors (GEFs) are well-known regulators of Arp2/3-dependent actin polymerization. In a previous screen aiming to identify pleckstrin homology (PH) domain-containing proteins that localize in the rear of cells (Li et al., 2022), we found a putative RhoGEF named GxcM (Fig. 1 A). When expressed as a GFP-fusion protein in cells migrating under agarose along a folic acid gradient, GxcM localized more strongly to the side and rear, resulting in a rear-to-front gradient in its plasma membrane association (Fig. 1, B and C; and Video 1). In randomly moving cells, GxcM-RFP and the well-characterized leading-edge marker PHcrac-GFP (Parent et al., 1998; Yang et al., 2021), a sensor for PIP3/PI(3,4)P2, exhibited an inverse distribution, the latter accumulating in regions where GxcM was depleted (Fig. 1 D). Furthermore, as previously reported for rear proteins (Iijima and Devreotes, 2002; Li et al., 2022; Swaney et al., 2015), GxcM-GFP transiently translocated from the membrane to the cytosol upon global chemoattractant stimulation (Fig. 1 E).
GxcM localizes in the rear of Dictyostelium cells. (A) Schematic representation of GxcM, GxcM4A, and GxcMN790. The star indicates alanine mutations (aa 593–596) in the DH domain. (B) Time-lapse imaging of GxcM-GFP in cells migrating under agarose along a folic acid gradient. The white triangle indicates gradient direction. (C) Left: Schematic of the angle-series plot used to quantify protein distribution. The fluorescent intensity in the cortical region of a cell was plotted along the perimeter, with 0° and +180°/−180° corresponding to the migrating front and rear, respectively. Right: Angle-series plot of GxcM-GFP in under agarose chemotaxing cells as shown in B. Solid line represents the mean and shade represents mean ± SD. n, number of cells analyzed. (D, F, and H) Distribution of PHcrac-GFP and GxcM-RFP (D), GxcMN790-RFP (F), or GxcM4A-RFP (H) in randomly migrating cells. (E, G, and I) Translocation of GxcM-GFP (E), GxcMN790-GFP (G), and GxcM4A-GFP (I) in response to cAMP stimulation added at time 0 (pre: before stimulation). (J, K, N, and O) Distribution of LimEΔcoil-RFP and the GFP control (J), GxcM-GFP (K), GxcMN790-GFP (N), or GxcM4A-GFP (O) in randomly migrating cells. (L, M, P, and Q) Distribution of GFP-ArpC4 and the RFP control (L), GxcM-RFP (M), GxcMN790-RFP (P), or GxcM4A-RFP (Q) in randomly migrating cells. Scale bars, 5 μm.
GxcM localizes in the rear of Dictyostelium cells. (A) Schematic representation of GxcM, GxcM4A, and GxcMN790. The star indicates alanine mutations (aa 593–596) in the DH domain. (B) Time-lapse imaging of GxcM-GFP in cells migrating under agarose along a folic acid gradient. The white triangle indicates gradient direction. (C) Left: Schematic of the angle-series plot used to quantify protein distribution. The fluorescent intensity in the cortical region of a cell was plotted along the perimeter, with 0° and +180°/−180° corresponding to the migrating front and rear, respectively. Right: Angle-series plot of GxcM-GFP in under agarose chemotaxing cells as shown in B. Solid line represents the mean and shade represents mean ± SD. n, number of cells analyzed. (D, F, and H) Distribution of PHcrac-GFP and GxcM-RFP (D), GxcMN790-RFP (F), or GxcM4A-RFP (H) in randomly migrating cells. (E, G, and I) Translocation of GxcM-GFP (E), GxcMN790-GFP (G), and GxcM4A-GFP (I) in response to cAMP stimulation added at time 0 (pre: before stimulation). (J, K, N, and O) Distribution of LimEΔcoil-RFP and the GFP control (J), GxcM-GFP (K), GxcMN790-GFP (N), or GxcM4A-GFP (O) in randomly migrating cells. (L, M, P, and Q) Distribution of GFP-ArpC4 and the RFP control (L), GxcM-RFP (M), GxcMN790-RFP (P), or GxcM4A-RFP (Q) in randomly migrating cells. Scale bars, 5 μm.
Localization of GxcM-GFP in WT cells migrating under agarose along a folic acid gradient. Corresponds to Fig. 1 B. Images were captured at 2-s intervals and played back at 24 frames per second. Scale bar = 5 μm.
Localization of GxcM-GFP in WT cells migrating under agarose along a folic acid gradient. Corresponds to Fig. 1 B. Images were captured at 2-s intervals and played back at 24 frames per second. Scale bar = 5 μm.
GxcM contains a RhoGEF domain (also known as Dbl homology domain or DH domain, 455–638 aa) and a PH domain (626–770 aa) in the central region (Fig. 1 A). Deleting the C-terminus of GxcM (GxcMN790) did not change its mutually exclusive localization with PHcrac or response to stimulation (Fig. 1, F and G), whereas further deletion by removing the DH and PH domains (GxcMN455) caused it to dissociate from the membrane (Fig. S1 A). Though necessary, the DH and PH domains were not sufficient to target GxcM because the truncation containing only the two domains (GxcM455-770) localized in the cytoplasm (Fig. S1 B). Sequence alignment of GxcM with human ECT2, a RhoGEF involved in the regulation of cytokinesis (Tatsumoto et al., 1999), revealed four conserved residues (PVQR, 593–596 aa) within the DH domain (Fig. S1 C). This stretch of residues has been shown to be essential for the GEF activity of ECT2 (Su et al., 2011). Mutating the four residues to alanines (GxcM4A) did not affect the localization or chemoattractant-induced translocation of GxcM (Fig. 1, H and I). These experiments show that GxcM is likely a RhoGEF that localizes selectively in the rear of Dictyostelium cells.
Characterization of GxcM. (A and B) Localization of GxcMN455-GFP and GxcMN456-770-GFP in randomly migrating cells. Scale bars, 5 μm. (C) Sequence alignment of the DH domains of GxcM and human ECT2. The red box indicates the conserved residues shown to be essential for the GEF activity of ECT2. (D) Top: Design of the gxcM knockout construct. A blasticidin-resistant cassette (BSR) was inserted to replace part of the open reading frame of gxcM. Bottom: Targeted clones were confirmed by PCR using the indicated primers. (E) WT and gxcM− cells were plated clonally with bacteria (Klebsiella aerogenes) on standard medium agar for 5 d. Scale bar, 5 mm. (F) Top: WT and gxcM− cells grown for 60 h in shaken suspension were fixed and stained with DAPI to visualize the nuclei. Bottom: Quantification of nuclei in cells. n, number of cells analyzed. Scale bar, 5 μm. (G) Top: Trajectories of randomly migrating cells (n = 38 for WT and 47 for gxcM−). Bottom: Summary of motility parameters. Data were from three independent experiments; mean ± SEM (the average of each biological replicate was used to calculate the mean and SEM). (H) Top: Trajectories of cells migrating under 2% agarose along a folic acid gradient (n = 41 for WT and 41 for gxcM−). Bottom: Summary of chemotaxis parameters. Data were from three independent experiments; mean ± SEM (the average of each biological replicate was used to calculate the mean and SEM). (I) Left: Projection length (Lp) of WT and gxcM− cells determined by micropipette aspiration using a constant pressure of 500 Pa for 5 min. Right: Quantitative analysis of the projection lengths of probed cells. Data were from one representative experiment; the scatter plot shows data points with mean ± SEM; n, number of cells analyzed.
Characterization of GxcM. (A and B) Localization of GxcMN455-GFP and GxcMN456-770-GFP in randomly migrating cells. Scale bars, 5 μm. (C) Sequence alignment of the DH domains of GxcM and human ECT2. The red box indicates the conserved residues shown to be essential for the GEF activity of ECT2. (D) Top: Design of the gxcM knockout construct. A blasticidin-resistant cassette (BSR) was inserted to replace part of the open reading frame of gxcM. Bottom: Targeted clones were confirmed by PCR using the indicated primers. (E) WT and gxcM− cells were plated clonally with bacteria (Klebsiella aerogenes) on standard medium agar for 5 d. Scale bar, 5 mm. (F) Top: WT and gxcM− cells grown for 60 h in shaken suspension were fixed and stained with DAPI to visualize the nuclei. Bottom: Quantification of nuclei in cells. n, number of cells analyzed. Scale bar, 5 μm. (G) Top: Trajectories of randomly migrating cells (n = 38 for WT and 47 for gxcM−). Bottom: Summary of motility parameters. Data were from three independent experiments; mean ± SEM (the average of each biological replicate was used to calculate the mean and SEM). (H) Top: Trajectories of cells migrating under 2% agarose along a folic acid gradient (n = 41 for WT and 41 for gxcM−). Bottom: Summary of chemotaxis parameters. Data were from three independent experiments; mean ± SEM (the average of each biological replicate was used to calculate the mean and SEM). (I) Left: Projection length (Lp) of WT and gxcM− cells determined by micropipette aspiration using a constant pressure of 500 Pa for 5 min. Right: Quantitative analysis of the projection lengths of probed cells. Data were from one representative experiment; the scatter plot shows data points with mean ± SEM; n, number of cells analyzed.
Overexpression of GxcM induces Arp2/3-mediated actin assembly in the rear cortex
To dissect the potential role of GxcM in cortex assembly, we generated gxcM knockout (gxcM−) cells (Fig. S1 D) and examined the cellular activities known to rely on cell shape remodeling and cortex integrity (Litschko et al., 2019; Ramalingam et al., 2015). When Dictyostelium cells are plated clonally with bacteria on an agar plate, they amplify, consume bacteria, and form clear plaques. Feeding cells are located at the outer edges of the plaques, whereas cells within the plaques undergo starvation-induced development to form fruiting bodies (Fey et al., 2007). We found that the development of gxcM− cells on bacterial lawns is indistinguishable from that of WT cells (Fig. S1 E). When assayed in shaken suspension, a condition used to reveal cytokinesis defects in the formin and RacE mutants (Litschko et al., 2019), gxcM− cells exhibited only a mild defect, with the vast majority of gxcM− cells being mono- or binucleated like WT cells, and only ∼7.9% exhibited three or more nuclei (Fig. S1 F). Furthermore, in random motility and under agarose chemotaxis assays, gxcM− cells migrated with speed and directness comparable with WT cells (Fig. S1, G and H). Thus, disruption of gxcM did not appear to markedly impair cell cortex integrity.
Despite the mild phenotypes in gxcM− cells, we observed from the localization experiments (Fig. 1 D) that overexpression of GxcM may alter cell morphology and behavior. Time-lapse imaging further revealed the impact of GxcM overexpression. Cells expressing GxcMN790-RFP, GxcM4A-RFP, or Teep1-RFP, a rear protein characterized previously (Li et al., 2022), exhibited polarized morphology and formed one or two protruding fronts at a given time in the form of pseudopods or macropinocytic cups that were labeled by PHcrac-GFP (Video 2). In contrast, cells expressing GxcM-RFP were less polar and produced rounded GxcM-marked protrusions in the peripheral regions (Video 2). These GxcM-enriched structures did not effectively cause cell displacement or macropinocytosis but generated merely outward bulges of the cell boundary (Video 2). Consistently, GxcM-overexpressing cells exhibited reduced random motility and macropinocytosis (Fig. S2, A and B). Cytokinesis was also weakly impaired in these cells (Fig. S2 C). Similar morphological changes were observed in cells expressing untagged GxcM, ruling out non-specific effects of the engineered fluorescent tag (Fig. S2 D). We further verified that these changes relied on overexpression of GxcM as the expression of GxcM from a single expression cassette integrated into the genome did not seem to cause morphological changes in the cell (Fig. S2 E).
Localization of PHcrac-GFP and RFP-tagged GxcM, GxcMN790, GxcM4A, or Teep1 in WT cells during random migration. Images were captured at 6-s intervals and played back at 12 frames per second. Scale bar = 5 μm.
Localization of PHcrac-GFP and RFP-tagged GxcM, GxcMN790, GxcM4A, or Teep1 in WT cells during random migration. Images were captured at 6-s intervals and played back at 12 frames per second. Scale bar = 5 μm.
Characterization GxcM overexpressing cells. (A) Top: Trajectories of randomly migrating cells (n = 64 for GFP/WT and 50 for GxcM-GFP/WT). Bottom: Summary of motility parameters. Data were from three independent experiments; mean ± SEM. (B) Quantification of TRITC dextran (TD) uptake. Data were from three independent experiments; the scatter plot shows data points with mean ± SEM; n, number of cells analyzed. (C) Left: GFP/WT and GxcM-GFP/WT cells grown on a cell culture plate were fixed and stained with DAPI to visualize the nuclei. Right: Quantification of nuclei. n, number of cells analyzed. (D) Time-lapse imaging of randomly migrating cells expressing GFP-ArpC4 and untagged GxcM. (E) Cells expressing GxcM-GFP from an expression cassette integrated into the genome as a stable single copy via restriction enzyme-mediated integration (REMI) was immunostained with an anti-GFP antibody. (F–I) Localization of GFP-Coronin in randomly migrating cells expressing RFP (F), GxcM-RFP (G), GxcMN790-RFP (H), or GxcM4A-RFP (I). (J and K) Localization of GFP-Coronin and RFP (J) or GxcM-RFP (K) in WT cells migrating under agarose along a folic acid gradient. Angle-series plots on the right show fluorescent intensity distribution of the indicated proteins along the perimeter of the cell, with 0° and +180°/−180° corresponding to the migrating front and rear, respectively. Solid lines represent the mean and shades represent mean ± SD. n, number of cells analyzed. (L) Top: Trajectories of cells migrating under 0.5% agarose along a folic acid gradient (n = 30 for GFP/WT and 29 for GxcM-GFP/WT). Bottom: Summary of chemotaxis parameters. Data were from at least three movies; means ± SEM. Scale bars, 5 μm.
Characterization GxcM overexpressing cells. (A) Top: Trajectories of randomly migrating cells (n = 64 for GFP/WT and 50 for GxcM-GFP/WT). Bottom: Summary of motility parameters. Data were from three independent experiments; mean ± SEM. (B) Quantification of TRITC dextran (TD) uptake. Data were from three independent experiments; the scatter plot shows data points with mean ± SEM; n, number of cells analyzed. (C) Left: GFP/WT and GxcM-GFP/WT cells grown on a cell culture plate were fixed and stained with DAPI to visualize the nuclei. Right: Quantification of nuclei. n, number of cells analyzed. (D) Time-lapse imaging of randomly migrating cells expressing GFP-ArpC4 and untagged GxcM. (E) Cells expressing GxcM-GFP from an expression cassette integrated into the genome as a stable single copy via restriction enzyme-mediated integration (REMI) was immunostained with an anti-GFP antibody. (F–I) Localization of GFP-Coronin in randomly migrating cells expressing RFP (F), GxcM-RFP (G), GxcMN790-RFP (H), or GxcM4A-RFP (I). (J and K) Localization of GFP-Coronin and RFP (J) or GxcM-RFP (K) in WT cells migrating under agarose along a folic acid gradient. Angle-series plots on the right show fluorescent intensity distribution of the indicated proteins along the perimeter of the cell, with 0° and +180°/−180° corresponding to the migrating front and rear, respectively. Solid lines represent the mean and shades represent mean ± SD. n, number of cells analyzed. (L) Top: Trajectories of cells migrating under 0.5% agarose along a folic acid gradient (n = 30 for GFP/WT and 29 for GxcM-GFP/WT). Bottom: Summary of chemotaxis parameters. Data were from at least three movies; means ± SEM. Scale bars, 5 μm.
To examine whether GxcM overexpression altered cell morphology by mediating changes in the actin cytoskeleton, we coexpressed GxcM with LimEΔcoil, a marker of newly polymerized F-actin (Bretschneider et al., 2004). In cells expressing the GFP control, the bulk of the LimEΔcoil-RFP signal was found at the pseudopod or macropinocytic cup region, and less prominent signals were seen at the rear and lateral sides (Fig. 1 J and Video 3). In contrast, in GxcM-GFP-overexpressing cells, the LimEΔcoil signal was highly concentrated at peripheral regions enriched with GxcM (Fig. 1 K and Video 3), suggesting that GxcM expression induces actin polymerization. Further analyses using different actin reporters showed that the GxcM-induced actin assemblies were likely Arp2/3-based because they could be marked by the Arp2/3 complex subunit ArpC4 (Fig. 1, L and M; and Video 3) and Coronin, a central constituent of the Arp2/3-mediated actin network (Fig. S2, F and G). Overexpression of GxcMN790 or GxcM4A did not alter cell morphology or trigger actin polymerization (Fig. 1, N–Q; and Fig. S2, H and I). Therefore, the cortical actin assembly-promoting activity of GxcM seems to require an intact C-terminus, as well as the GEF domain.
Localization of LimEΔcoil-RFP and GFP-ArpC4 in WT cells expressing GxcM-GFP, GxcM-RFP, or a control fluorescent protein during random migration. Corresponds to Fig. 1, J–M. Images were captured at 6-s intervals and played back at 12 frames per second. Scale bar = 5 μm.
Localization of LimEΔcoil-RFP and GFP-ArpC4 in WT cells expressing GxcM-GFP, GxcM-RFP, or a control fluorescent protein during random migration. Corresponds to Fig. 1, J–M. Images were captured at 6-s intervals and played back at 12 frames per second. Scale bar = 5 μm.
We also observed the different actin markers in cells chemotaxing under agarose along a folic acid gradient. In control cells expressing GFP or RFP, the signals of LimEΔcoil, ArpC4, and Coronin were mainly detected in protrusions at the migrating front (Fig. 2, A and C; and Fig. S2 J; and Video 4). In contrast, in cells overexpressing GxcM-GFP or GxcM-RFP, a significant fraction of these reporters were localized to the rear and lateral sides, where intensive GxcM signals were observed (Fig. 2, B and D; and Fig. S2 K; and Video 4). In addition, traveling actin waves were frequently seen in the rear cortical region upon GxcM overexpression (Video 5). The rearrangement of F-actin and Arp2/3 activity from the migrating front to the rear likely interferes with cell function. Although the GxcM-overexpressing cells were still able to orient and move toward the higher concentration of folic acid, they migrated at a considerably slower speed (Fig. S2 L), highlighting the need to fine-tune the level of GxcM. These results suggest that GxcM may function as a cortex assembly factor promoting Arp2/3-based actin polymerization in the rear of cells, and its overexpression likely further boosts such activity, resulting in over-assembly of actin and disruption of cell function.
GxcM overexpression induces cortical actin assembly. Time-lapse imaging of WT cells migrating under agarose along a folic acid gradient. (A) Cells expressing LimEΔcoil-RFP and GFP. (B) Cells expressing LimEΔcoil-RFP and GxcM-GFP. (C) Cells expressing GFP-ArpC4 and RFP. (D) Cells expressing GFP-ArpC4 and GxcM-RFP. Angle-series plots on the right show fluorescent intensity distribution of the indicated proteins along the perimeter of the cell, with 0° and +180°/−180° corresponding to the migrating front and rear, respectively. Solid lines represent the mean and shades represent mean ± SD. n, number of cells analyzed. The white triangles indicate gradient direction. Scale bars, 5 μm.
GxcM overexpression induces cortical actin assembly. Time-lapse imaging of WT cells migrating under agarose along a folic acid gradient. (A) Cells expressing LimEΔcoil-RFP and GFP. (B) Cells expressing LimEΔcoil-RFP and GxcM-GFP. (C) Cells expressing GFP-ArpC4 and RFP. (D) Cells expressing GFP-ArpC4 and GxcM-RFP. Angle-series plots on the right show fluorescent intensity distribution of the indicated proteins along the perimeter of the cell, with 0° and +180°/−180° corresponding to the migrating front and rear, respectively. Solid lines represent the mean and shades represent mean ± SD. n, number of cells analyzed. The white triangles indicate gradient direction. Scale bars, 5 μm.
Localization of LimEΔcoil-RFP and GFP-ArpC4 in WT cells expressing GxcM-GFP, GxcM-RFP, or a control fluorescent protein migrating under agarose along a folic acid gradient. Corresponds to Fig. 2, A–D. Images were captured at 3-s intervals and played back at 6 frames per second. Scale bar = 5 μm.
Localization of LimEΔcoil-RFP and GFP-ArpC4 in WT cells expressing GxcM-GFP, GxcM-RFP, or a control fluorescent protein migrating under agarose along a folic acid gradient. Corresponds to Fig. 2, A–D. Images were captured at 3-s intervals and played back at 6 frames per second. Scale bar = 5 μm.
TIRF microscopy imaging of the localization of GxcM-GFP and LimEΔcoil-RFP in WT cells chemotaxing under agarose. Images were captured at 3-s intervals and played back at 10 frames per second. Scale bar = 5 μm.
TIRF microscopy imaging of the localization of GxcM-GFP and LimEΔcoil-RFP in WT cells chemotaxing under agarose. Images were captured at 3-s intervals and played back at 10 frames per second. Scale bar = 5 μm.
The actin assembly-promoting activity of GxcM relies on interaction with F-BAR protein Fbp17
We noted that the C-terminus (791–1,145 aa) of GxcM is proline-rich (Fig. S3 A). As a diverse array of actin regulators contain proline-binding modules, such as SH3 and EVH1 domains (Ball et al., 2005; Holt and Koffer, 2001), GxcM may function by association with such proteins via its C-terminus. To identify binding partners of GxcM, we immunoprecipitated GxcM-GFP from cell lysates and performed mass spectrometry (MS) analysis. GxcMN790-GFP and Teep1-GFP were immunoprecipitated as controls. We found that an SH3 domain-containing protein (gene ID, DDB_G0271812) was uniquely enriched in the immunocapture of GxcM-GFP (Fig. 3 A and Table S1). Sequence analysis revealed that this protein is a homolog of the FBP17 family of Fes/CIP4 homology-Bin-Amphiphysin-Rvs (F-BAR) proteins (Fig. S3 B). This family includes three closely related proteins, formin-binding protein 17 (FBP17), transactivator of cytoskeletal assembly-1 (TOCA-1), and Cdc42-interacting protein 4 (CIP4), which are all activators of Arp2/3-dependent actin polymerization (Chen et al., 2013; Ho et al., 2004; Takano et al., 2008; Tsujita et al., 2006). DDB_G0271812 shares 21.3% identity and 38.6% similarity to human FBP17 and, similar to FBP17, contains an F-BAR, homology region 1 (HR1), and SH3 domain (Fig. 3 B and Fig. S3 B). Therefore, we named this protein Fbp17.
Characterization of Fbp17. (A) Sequence of the C-terminus of GxcM (aa 791–1,145). Proline residues are highlighted in red. The blue underlines indicate SH3 domain-binding motifs predicted by the LMDIPred web server. (B) Sequence alignment of Fbp17 with human CIP4, FBP17, and TOCA-1. Magenta, green, and blue boxes indicate the F-BAR, homology region 1 (HR1), and SH3 domain, respectively. (C and D) Time-lapse imaging of GFP-Fbp17 in randomly migrating cells (C) or cells moving under agarose along a folic acid gradient (D). Scale bars, 5 μm. (E) Top: Design of the fbp17 knockout construct. Bottom: Targeted clones were confirmed by PCR. (F) The indicated cells were plated clonally with bacteria (Klebsiella aerogenes) on standard medium agar for 5 d. Scale bar, 5 mm. (G and H) Left: Localization of GFP-myosin II in randomly migrating cells (G) or cells migrating under 2% agarose along a folic acid gradient (H). Right: Quantification of the relative cortical localization of GFP-myosin II. Data were from three independent experiments; the scatter plots show data points with mean ± SEM; n, number of cells analyzed. Scale bars, 5 μm.
Characterization of Fbp17. (A) Sequence of the C-terminus of GxcM (aa 791–1,145). Proline residues are highlighted in red. The blue underlines indicate SH3 domain-binding motifs predicted by the LMDIPred web server. (B) Sequence alignment of Fbp17 with human CIP4, FBP17, and TOCA-1. Magenta, green, and blue boxes indicate the F-BAR, homology region 1 (HR1), and SH3 domain, respectively. (C and D) Time-lapse imaging of GFP-Fbp17 in randomly migrating cells (C) or cells moving under agarose along a folic acid gradient (D). Scale bars, 5 μm. (E) Top: Design of the fbp17 knockout construct. Bottom: Targeted clones were confirmed by PCR. (F) The indicated cells were plated clonally with bacteria (Klebsiella aerogenes) on standard medium agar for 5 d. Scale bar, 5 mm. (G and H) Left: Localization of GFP-myosin II in randomly migrating cells (G) or cells migrating under 2% agarose along a folic acid gradient (H). Right: Quantification of the relative cortical localization of GFP-myosin II. Data were from three independent experiments; the scatter plots show data points with mean ± SEM; n, number of cells analyzed. Scale bars, 5 μm.
GxcM interacts with the F-BAR protein Fbp17. (A) Proteomic identification of Fbp17 as a binding partner of GxcM. (B) Schematic representation of full-length Fbp17 and truncation constructs. (C) Co-IP of GxcM-GFP or GxcM4A-GFP with RFP or RFP-Fbp17. IP was performed with RFP-trap and samples were probed with GFP or RFP antibody. (D) Co-IP of the indicated GFP-fusion proteins with RFP-Fbp17. IP was performed with RFP-trap and samples were probed with GFP or RFP antibody. (E) Co-IP of GxcM-GFP with the indicated RFP-fusion proteins. IP was performed with RFP-trap and samples were probed with GFP or RFP antibody. (F) Pull-down of GxcM-GFP from cell lysate with GST or GST-SH3 immobilized on beads. Samples were probed with GFP antibody. The protein-transferred membrane was stained with Ponceau S to show purified GST and GST-SH3. (G) Pull-down of the indicated GFP-fusion proteins from cell lysate with GST-SH3 immobilized on beads. Samples were probed with GFP antibody. The protein-transferred membrane was stained with Ponceau S to show purified GST-SH3. (H) Pull-down of GxcM-GFP from cell lysate with GST-SH3 or MBP-SH3 immobilized on beads. Samples were probed with GFP antibody. The protein-transferred membrane was stained with Ponceau S to show purified GST and MBP fusion proteins. (I) Co-IP of GFP-Fbp17 or GFP-Fbp17F-BAR with RFP or RFP-Fbp17. IP was performed with RFP-trap and samples were probed with GFP or RFP antibody. (J) Co-IP of the indicated GFP-fusion proteins with RFP-Fbp17. IP was performed with RFP-trap and samples were probed with GFP or RFP antibody. Source data are available for this figure: SourceData F3.
GxcM interacts with the F-BAR protein Fbp17. (A) Proteomic identification of Fbp17 as a binding partner of GxcM. (B) Schematic representation of full-length Fbp17 and truncation constructs. (C) Co-IP of GxcM-GFP or GxcM4A-GFP with RFP or RFP-Fbp17. IP was performed with RFP-trap and samples were probed with GFP or RFP antibody. (D) Co-IP of the indicated GFP-fusion proteins with RFP-Fbp17. IP was performed with RFP-trap and samples were probed with GFP or RFP antibody. (E) Co-IP of GxcM-GFP with the indicated RFP-fusion proteins. IP was performed with RFP-trap and samples were probed with GFP or RFP antibody. (F) Pull-down of GxcM-GFP from cell lysate with GST or GST-SH3 immobilized on beads. Samples were probed with GFP antibody. The protein-transferred membrane was stained with Ponceau S to show purified GST and GST-SH3. (G) Pull-down of the indicated GFP-fusion proteins from cell lysate with GST-SH3 immobilized on beads. Samples were probed with GFP antibody. The protein-transferred membrane was stained with Ponceau S to show purified GST-SH3. (H) Pull-down of GxcM-GFP from cell lysate with GST-SH3 or MBP-SH3 immobilized on beads. Samples were probed with GFP antibody. The protein-transferred membrane was stained with Ponceau S to show purified GST and MBP fusion proteins. (I) Co-IP of GFP-Fbp17 or GFP-Fbp17F-BAR with RFP or RFP-Fbp17. IP was performed with RFP-trap and samples were probed with GFP or RFP antibody. (J) Co-IP of the indicated GFP-fusion proteins with RFP-Fbp17. IP was performed with RFP-trap and samples were probed with GFP or RFP antibody. Source data are available for this figure: SourceData F3.
We performed coimmunoprecipitation experiments to verify the interaction between GxcM and Fbp17. Among the GxcM-GFP/RFP-Fbp17, GxcM-GFP/RFP, and Teep1-GFP/RFP-Fbp17 combinations, the interaction was detected only between GxcM-GFP and RFP-Fbp17 (Fig. 3, C and D). A number of additional experiments demonstrated that this interaction is mediated through the C-terminus of GxcM and the SH3 domain of Fbp17. GEF domain mutation (GxcM4A-GFP) did not disrupt the interaction (Fig. 3, C and D), whereas deletion of the C-terminus of GxcM (GxcMN790-GFP) or the SH3 domain of Fbp17 (RFP-Fbp17ΔSH3 or RFP-Fbp17F-BAR) abolished the interaction (Fig. 3, D and E). Moreover, purified recombinant glutathione-S-transferase (GST)-SH3, but not GST, effectively pulled down GxcM-GFP and GxcM4A-GFP, but not GxcMN790-GFP (Fig. 3, F and G). Interestingly, we noted that RFP-fused SH3 (RFP-Fbp17SH3) could not precipitate GxcM-GFP (Fig. 3 E), and purified recombinant maltose-binding protein (MBP)-SH3 also failed to pull down GxcM-GFP (Fig. 3 H). As F-BAR proteins tend to form homodimers (Frost et al., 2008; Shimada et al., 2007), we speculated that the ability to dimerize via either the F-BAR domain or GST may be required for the SH3-GxcM interaction. In support of this notion, we found that Fbp17 self-interacted via the F-BAR region (Fig. 3, I and J).
Consistent with the interaction data, the C-terminus of GxcM and SH3 domain of Fbp17 are required for the colocalization of the two proteins in the cell. Only a small fraction of Fbp17 was detected at the periphery of cells when it was expressed alone as a GFP-fusion protein (Fig. S3, C and D) or coexpressed with RFP (Fig. 4 A). In contrast, coexpression with GxcM-RFP or GxcM4A-RFP strongly recruited GFP-Fbp17 to the cell cortex (Fig. 4, B and C). On the other hand, the expression of GxcMN790-RFP could not recruit GFP-Fbp17 (Fig. 4 D) nor could GxcM-RFP GFP-Fbp17ΔSH3 (Fig. 4 E). Without the potential dimerization via the F-BAR domain, the SH3 domain alone also failed to be recruited by GxcM-RFP (Fig. 4 F).
The actin assembly-promoting activity of GxcM relies on interaction with Fbp17. (A–H) Localization of the indicated fluorescent proteins in randomly migrating cells. (A) WT cells expressing GFP-Fbp17 and RFP. (B) WT cell expressing GFP-Fbp17 and GxcM-RFP. (C) WT cells expressing GFP-Fbp17 and GxcM4A-RFP. (D) WT cell expressing GFP-Fbp17 and GxcMN790-RFP. (E) WT cell expressing GFP-Fbp17ΔSH3 and GxcM-RFP. (F) WT cells expressing GFP-Fbp17SH3 and GxcM-RFP. (G)fbp17– cells expressing GxcM-GFP and LimEΔcoil-RFP. (H)fbp17– cells expressing GxcM-RFP and GFP-ArpC4. (I and J) Time-lapse imaging of fbp17− cells migrating under agarose along a folic acid gradient. (I)fbp17− cells expressing GxcM-GFP and LimEΔcoil-RFP. (J)fbp17− cells expressing GxcM-RFP and GFP-ArpC4. Angle-series plots on the right show fluorescent intensity distribution of the indicated proteins along the perimeter of the cell, with 0° and +180°/−180° corresponding to the migrating front and rear, respectively. Solid lines represent the mean and shades represent mean ± SD. n, number of cells analyzed. Scale bars, 5 μm.
The actin assembly-promoting activity of GxcM relies on interaction with Fbp17. (A–H) Localization of the indicated fluorescent proteins in randomly migrating cells. (A) WT cells expressing GFP-Fbp17 and RFP. (B) WT cell expressing GFP-Fbp17 and GxcM-RFP. (C) WT cells expressing GFP-Fbp17 and GxcM4A-RFP. (D) WT cell expressing GFP-Fbp17 and GxcMN790-RFP. (E) WT cell expressing GFP-Fbp17ΔSH3 and GxcM-RFP. (F) WT cells expressing GFP-Fbp17SH3 and GxcM-RFP. (G)fbp17– cells expressing GxcM-GFP and LimEΔcoil-RFP. (H)fbp17– cells expressing GxcM-RFP and GFP-ArpC4. (I and J) Time-lapse imaging of fbp17− cells migrating under agarose along a folic acid gradient. (I)fbp17− cells expressing GxcM-GFP and LimEΔcoil-RFP. (J)fbp17− cells expressing GxcM-RFP and GFP-ArpC4. Angle-series plots on the right show fluorescent intensity distribution of the indicated proteins along the perimeter of the cell, with 0° and +180°/−180° corresponding to the migrating front and rear, respectively. Solid lines represent the mean and shades represent mean ± SD. n, number of cells analyzed. Scale bars, 5 μm.
To investigate the functional significance of the GxcM-Fbp17 interaction, we generated fbp17 knockout (fbp17−) cells (Fig. S3 E) and examined the effects of GxcM overexpression in these cells. Deletion of fbp17 severely impaired the actin assembly-promoting activity of GxcM. In randomly migrating GxcM/fbp17− cells, LimEΔcoil and ArpC4 signals were no longer concentrated in peripheral regions marked by GxcM (compare Fig. 4, G and H to Fig. 1, L and M). Similarly, in GxcM/fbp17− cells chemotaxing under agarose, the strong rear accumulation of LimEΔcoil and ArpC4 induced by GxcM overexpression was nearly completely abolished, with the bulk of both signals being distributed to the migrating front as in WT cells (compare Fig. 4, I and J to Fig. 2, B and D; compare Video 6 to Video 4). GxcM still exhibited rear localization in fbp17− cells, though in a shallower rear-to-front gradient (compare Fig. 4, I and J to Fig. 2, B and D). These experiments indicate that the actin assembly-promoting activity of GxcM requires association with Fbp17, which likely functions downstream of GxcM to regulate cortical actin generation.
Localization of GxcM-GFP/LimEΔcoil-RFP or GxcM-RFP/GFP-ArpC4 in fbp17−cells migrating under agarose along a folic acid gradient. Corresponds to Fig. 4, I and J. Images were captured at 3-s intervals and played back at 6 frames per second. Scale bar = 5 μm.
Localization of GxcM-GFP/LimEΔcoil-RFP or GxcM-RFP/GFP-ArpC4 in fbp17−cells migrating under agarose along a folic acid gradient. Corresponds to Fig. 4, I and J. Images were captured at 3-s intervals and played back at 6 frames per second. Scale bar = 5 μm.
Fbp17 is required to maintain cortical integrity
We examined whether development, cytokinesis, or migration is affected by fbp17 deletion. On bacterial lawns, fbp17− cells were able to advance through development and form fruiting bodies (Fig. S3 F), but the diameter of the plaques formed by these cells was significantly smaller than those formed by WT cells (mean ± SEM: fbp17−, 6.4 ± 0.8 mm; WT, 9.8 ± 1.2 mm). This defect could be fully rescued by the expression of GFP-Fbp17 (9.3 ± 0.7 mm), but not GFP (5.7 ± 0.8 mm). When assayed for cytokinesis, the majority of WT and rescue (GFP-Fbp17/fbp17−) cells were mono- or binucleated. In contrast, fbp17− cells exhibited increased failure of cytokinesis; merely 33.5% were mononucleated, whereas more than 25% developed three or more nuclei (Fig. 5, A and B). In random motility assays, fbp17− cells exhibited an approximate 40% reduction in speed of movement and a 70% reduction in Euclidean distance, indicating that they migrated more slowly and less persistently (Fig. 5, C–E). When assayed for under agarose chemotaxis, fbp17− cells migrated toward the gradient source at a speed comparable to that of WT cells (Fig. 5, F–H). Despite the recovery of speed, migration persistence and directionality were still significantly impaired in the knockout cells, which was reflected in the more erratic cell tracks and reduced net movement up the gradient (Fig. 5, F–H).
Fbp17 is required to maintain cortical integrity. (A) WT, fbp17−, and GFP-Fbp17/fbp17− (rescue) cells grown for 60 h in shaken suspension were fixed and stained with DAPI to visualize the nuclei. (B) Quantification of nuclei in cells. n, number of cells analyzed. (C) Top: Trajectories of randomly migrating cells (n = 43 for WT and 46 for fbp17−). Bottom: Summary of motility parameters. (D and E) Velocity and Euclidean distance of cells are shown in C. At least 13 cells were quantified per experiment (each experiment shown in a different color); mean ± SEM. (F) Top: Trajectories of cells migrating under 2% agarose along a folic acid gradient (n = 61 for WT and 62 for fbp17−). Bottom: Summary of chemotaxis parameters; FMI, forward migration index. (G and H) Directness and FMI of cells shown in F. At least 17 cells were quantified per experiment (each experiment shown in a different color); mean ± SEM. (I) Localization of GFP-ABD in WT and fbp17− cells. (J) Quantification of the cortex-to-cytoplasm fluorescent intensity ratios of GFP-ABD. (K) WT and fbp17− cells were fixed and stained with Alexa Fluor 555-labeled phalloidin. (L) Quantification of the fluorescence intensity of cortical phalloidin. (M) Projection length (Lp) of WT and fbp17− cells determined by micropipette aspiration using a constant pressure of 500 Pa for 5 min. (N) Quantitative analysis of the projection lengths of probed cells. For C and F, data were from three independent experiments (the average of each biological replicate was used to calculate the mean and SEM). For J, L, and N, data were from three independent experiments; the scatter plots show data points with mean ± SEM; n, number of cells analyzed. Scale bars, 5 μm.
Fbp17 is required to maintain cortical integrity. (A) WT, fbp17−, and GFP-Fbp17/fbp17− (rescue) cells grown for 60 h in shaken suspension were fixed and stained with DAPI to visualize the nuclei. (B) Quantification of nuclei in cells. n, number of cells analyzed. (C) Top: Trajectories of randomly migrating cells (n = 43 for WT and 46 for fbp17−). Bottom: Summary of motility parameters. (D and E) Velocity and Euclidean distance of cells are shown in C. At least 13 cells were quantified per experiment (each experiment shown in a different color); mean ± SEM. (F) Top: Trajectories of cells migrating under 2% agarose along a folic acid gradient (n = 61 for WT and 62 for fbp17−). Bottom: Summary of chemotaxis parameters; FMI, forward migration index. (G and H) Directness and FMI of cells shown in F. At least 17 cells were quantified per experiment (each experiment shown in a different color); mean ± SEM. (I) Localization of GFP-ABD in WT and fbp17− cells. (J) Quantification of the cortex-to-cytoplasm fluorescent intensity ratios of GFP-ABD. (K) WT and fbp17− cells were fixed and stained with Alexa Fluor 555-labeled phalloidin. (L) Quantification of the fluorescence intensity of cortical phalloidin. (M) Projection length (Lp) of WT and fbp17− cells determined by micropipette aspiration using a constant pressure of 500 Pa for 5 min. (N) Quantitative analysis of the projection lengths of probed cells. For C and F, data were from three independent experiments (the average of each biological replicate was used to calculate the mean and SEM). For J, L, and N, data were from three independent experiments; the scatter plots show data points with mean ± SEM; n, number of cells analyzed. Scale bars, 5 μm.
To examine whether these morphogenesis and migration defects were caused by defects in cortex assembly, we visualized the distribution of cortical actin using the ABD120-GFP reporter and phalloidin staining (Pang et al., 1998; Zatulovskiy et al., 2014). In WT cells, the ABD120-GFP signal was more concentrated at the leading-edge protrusions, yet a significant fraction marked the cortical region at the rear and lateral sides (Fig. 5 I). Notably, the deletion of fbp17 did not seem to affect the ABD120 signal at the leading edge but reduced it at the cortical region (Fig. 5 I). Quantification revealed a reduction of ∼20% in the cortical-to-cytosol fluorescent intensity ratio of ABD120-GFP in fbp17− cells compared with WT cells (Fig. 5 J). Similar results were obtained with phalloidin staining, in which fbp17− cells exhibited a decrease of ∼30% in the cortical intensity of phalloidin (Fig. 5, K and L).
As the F-actin shell is considered the main contributor to the cells’ mechanical rigidness (Luo et al., 2014), we examined whether the apparent reduction in cortical F-actin content in fbp17− cells leads to a weakened cortex. To this end, we performed micropipette aspiration (MPA) assays. The initial projection lengths (Lp) of cells captured at a constant pressure of 500 Pa were quantified, which negatively correlate with the cells’ mechanical rigidity (Hochmuth, 2000; Ramalingam et al., 2015). Deletion of fbp17 resulted in significantly longer projection lengths (mean ± SEM: fbp17−, 9.64 ± 0.89 μm WT, 2.23 ± 0.22 µm), implying reduced mechanical rigidity (Fig. 5, M and N). In contrast, the gxcM− cells exhibited equivalent projection lengths as WT cells under this condition (Fig. S1 I), consistent with their lack of defects in cortex-dependent cellular activities. Furthermore, a small fraction of fbp17− cells, but almost none of the WT or gxcM− cells, were aspirated into the micropipette during the experiment. These cells were not included in the analysis, although they likely exhibited more severe defects. Thus, the quantification data likely underestimated the defects in the fbp17− cells. These results substantiate the role of Fbp17 in maintaining cortical integrity.
The actin cortex has been shown to modulate the rear accumulation of myosin II, a driver of cell contractility, so we also compared the distribution of GFP-myosin II in WT and fbp17− cells. Deletion of fbp17 reduced the extent of myosin II accumulation in randomly migrating cells (Fig. S3 G). Interestingly, this defect was alleviated when the fbp17− cells moved under agarose along a chemoattractant gradient (Fig. S3 H). Agar compression, a condition known to relocate myosin II to the cortex to counteract the applied force (Laevsky and Knecht, 2003; Ramalingam et al., 2015), may account for this phenomenon. In support of this notion, we observed increased rear accumulation of myosin II in both WT and fbp17− cells moving under agarose (Fig. S3 H). The differential distribution of myosin II may explain, at least in part, why fbp17− cells exhibited significantly reduced speed during random migration but improved movement under agarose chemotaxis (Fig. 5, C–H).
Fbp17 promotes WASP-mediated actin polymerization
Given the role of Fbp17 in supporting cortical integrity and mediating the downstream effect of GxcM, as well as the roles of its homologs in other organisms, we speculated that Fbp17 may contribute to Arp2/3-dependent actin polymerization by interacting with and stimulating the activities of WASP family NPFs (Ho et al., 2004; Takano et al., 2008; Tsujita et al., 2006). We performed coimmunoprecipitation experiments to examine whether Fbp17 interacts with WASP, which is encoded by a single gene, wasA, in Dictyostelium. GFP-WASP, but not Teep1-GFP, coimmunoprecipitated with RFP-Fbp17 (Fig. 6 A). This interaction depended on the SH3 domain of Fbp17 as RFP-Fbp17ΔSH3 lost the ability to interact with GFP-WASP (Fig. 6 B). Interestingly, as observed for the Fbp17-GxcM interaction, the interaction between Fbp17 and WASP also appeared to require the presence of a potentially dimerized SH3. Purified GST-SH3, but not GST, efficiently pulled down GFP-WASP from cell lysates (Fig. 6, C and D), whereas RFP-Fbp17SH3 or purified MBP-SH3 could not precipitate GFP-WASP (Fig. 6, B and D). Furthermore, consistent with the proposed links among GxcM, Fbp17, and WASP, a fraction of GFP-WASP was recruited to cortical regions with intensive GxcM-RFP signals, and this recruitment was abolished by fbp17 deletion (Fig. 6 E).
Fbp17 promotes WASP-mediated actin polymerization. (A) Co-IP of GFP-WASP and Teep1-GFP with RFP-Fbp17. IP was performed with RFP-trap and samples were probed with GFP or RFP antibody. (B) Co-IP of RFP-Fbp17, -Fbp17ΔSH3, or -Fbp17SH3 with GFP-WASP. IP was performed with RFP-trap and samples were probed with GFP or RFP antibody. (C) Pull-down of GFP-WASP from cell lysate with GST or GST-SH3 immobilized on beads. Samples were probed with GFP antibody. The protein-transferred membrane was stained with Coomassie Brilliant Blue (CBB) to show purified GST and GST-SH3. (D) Pull-down of GFP-WASP from cell lysate with GST-SH3 or MBP-SH3 immobilized on beads. Samples were probed with GFP antibody. The protein-transferred membrane was stained with Ponceau S to show purified GST and MBP fusion proteins. (E) Localization of GxcM-RFP and GFP-WASP in WT and fbp17− cells. Scale bar, 5 μm. (F) GST-SH3, but not SH3 or GST-Fbp17, promotes WASP- and Arp2/3-mediated actin polymerization in pyrene assays. (G) GST-SH3 promotes WASP- and Arp2/3-dependent actin polymerization in a concentration-dependent manner in pyrene assays. The VCA domain of WASP purified as a GST fusion was included as a control. Source data are available for this figure: SourceData F6.
Fbp17 promotes WASP-mediated actin polymerization. (A) Co-IP of GFP-WASP and Teep1-GFP with RFP-Fbp17. IP was performed with RFP-trap and samples were probed with GFP or RFP antibody. (B) Co-IP of RFP-Fbp17, -Fbp17ΔSH3, or -Fbp17SH3 with GFP-WASP. IP was performed with RFP-trap and samples were probed with GFP or RFP antibody. (C) Pull-down of GFP-WASP from cell lysate with GST or GST-SH3 immobilized on beads. Samples were probed with GFP antibody. The protein-transferred membrane was stained with Coomassie Brilliant Blue (CBB) to show purified GST and GST-SH3. (D) Pull-down of GFP-WASP from cell lysate with GST-SH3 or MBP-SH3 immobilized on beads. Samples were probed with GFP antibody. The protein-transferred membrane was stained with Ponceau S to show purified GST and MBP fusion proteins. (E) Localization of GxcM-RFP and GFP-WASP in WT and fbp17− cells. Scale bar, 5 μm. (F) GST-SH3, but not SH3 or GST-Fbp17, promotes WASP- and Arp2/3-mediated actin polymerization in pyrene assays. (G) GST-SH3 promotes WASP- and Arp2/3-dependent actin polymerization in a concentration-dependent manner in pyrene assays. The VCA domain of WASP purified as a GST fusion was included as a control. Source data are available for this figure: SourceData F6.
Via an in vitro pyrene–actin polymerization assay, we measured the activity of recombinant Fbp17 toward WASP. When mixed with actin and the Arp2/3 complex, GST-WASP purified from insect cells that exhibited minimal actin polymerization-promoting activity, indicating that it adopted an autoinhibitory configuration (Fig. 6 F). Adding purified SH3 or GST-Fbp17 did not significantly change the kinetics of the reaction (Fig. 6 F). In contrast, the addition of purified GST-SH3 promoted actin polymerization driven by WASP in a concentration-dependent manner (Fig. 6, F and G), likely through allosteric effects. The VCA domain of Dictyostelium WASP (319–399 aa) was included in the experiment as a positive control (Fig. 6 G). Taken together, these experiments indicate that Fbp17 facilitates WASP activation downstream of GxcM in cortical actin formation.
RacC acts downstream of GxcM to regulate cortex assembly via association with Fbp17 and WASP
Our results described thus far suggest a scenario in which the rear cortex-localized GxcM recruits Fbp17 to promote WASP and Arp2/3-mediated actin assembly. However, as presented earlier, the actin assembly-promoting activity of GxcM also requires its Rho GEF domain, implying the involvement of Rho GTPase(s) in this process. Dictyostelium cells lack canonical Cdc42 and Rho homologs but express 20 Rac proteins, some of which exhibit characteristics of Cdc42 and Rho (Filić et al., 2021). We investigated whether one or more of these Rac proteins act downstream of GxcM. MS of GxcM-associated proteins did not yield a possible candidate. Considering that Rho GTPases are potent regulators of WASP and the proposed mechanism by which FBP17 family proteins function in other organisms involves association with Rho GTPases (Ho et al., 2004; Watson et al., 2016; Watson et al., 2017), we decided to seek the relevant small GTPases by looking for binding partners of WASP and Fbp17.
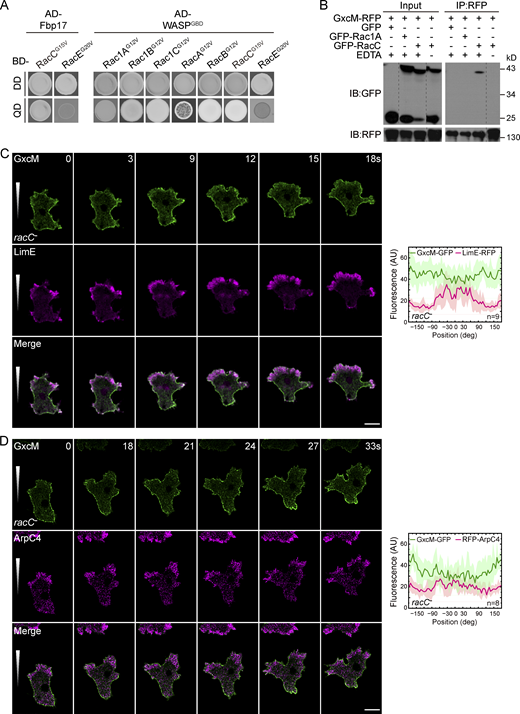
We employed yeast two-hybrid (Y2H) analyses to screen Rac proteins for interaction with the GTPase-binding domain (GBD, aa 126–230) of WASP or full-length Fbp17 (Fig. 7 A and Fig. S4 A). The GBD domain interacted with several Racs (Rac1A, Rac1B, Rac1C, RacA, RacB, and RacC) in their constitutively active (CA) forms, whereas Fbp17 interacted selectively with the CA form of RacC (RacCCA). RacE, which has been shown to regulate cortex assembly via formin proteins (Litschko et al., 2019), did not exhibit an interaction with either GBD or Fbp17. We focused our subsequent investigations on RacC for three reasons. First, in line with the notion that GEFs preferentially bind with target small GTPases in their nucleotide-free forms, we found that RacC, but not Rac1A, exhibited an EDTA-dependent interaction with GxcM (Fig. 7 B). Second, a previous study using a cell-free system showed that GTPγS-charged RacC is capable of stimulating actin polymerization via the activation of WASP (Han et al., 2006). Third, overexpression of RacC has been shown to induce unusual actin-based structures, which somewhat resembles the effect of GxcM overexpression (Seastone et al., 1998).
RacC acts downstream of GxcM to regulate cortical actin assembly. (A) Yeast two-hybrid assay showing the interaction between Fbp17 and the GBD domain of WASP with the constitutively active (CA) forms of the indicated Rac proteins. Yeast was transformed with the indicated constructs and selected for the presence of prey and bait plasmids by growth on double-dropout (DD) agar plate lacking leucine and tryptophan. Interactions were assayed by growth on quadruple-dropout (QD) agar plate additionally lacking histidine and adenine. AD, Gal4-activation domain; BD, Gal4-binding domain. (B) Co-IP of GFP, GFP-RacC, or GFP-Rac1A with GxcM-RFP in the presence or absence of EDTA. IP was performed with RFP-trap and samples were probed with GFP or RFP antibody. (C and D) Time-lapse imaging of racC– cells migrating under agarose along a folic acid gradient. (C)racC− cells expressing GxcM-GFP and LimEΔcoil-RFP. (D)racC− cells expressing GxcM-GFP and RFP-ArpC4. Angle-series plots on the right show fluorescent intensity distribution of the indicated proteins along the perimeter of the cell, with 0° and +180°/−180° corresponding to the migrating front and rear, respectively. Solid lines represent the mean and shades represent mean ± SD. n, number of cells analyzed. Scale bars, 5 μm. Source data are available for this figure: SourceData F7.
RacC acts downstream of GxcM to regulate cortical actin assembly. (A) Yeast two-hybrid assay showing the interaction between Fbp17 and the GBD domain of WASP with the constitutively active (CA) forms of the indicated Rac proteins. Yeast was transformed with the indicated constructs and selected for the presence of prey and bait plasmids by growth on double-dropout (DD) agar plate lacking leucine and tryptophan. Interactions were assayed by growth on quadruple-dropout (QD) agar plate additionally lacking histidine and adenine. AD, Gal4-activation domain; BD, Gal4-binding domain. (B) Co-IP of GFP, GFP-RacC, or GFP-Rac1A with GxcM-RFP in the presence or absence of EDTA. IP was performed with RFP-trap and samples were probed with GFP or RFP antibody. (C and D) Time-lapse imaging of racC– cells migrating under agarose along a folic acid gradient. (C)racC− cells expressing GxcM-GFP and LimEΔcoil-RFP. (D)racC− cells expressing GxcM-GFP and RFP-ArpC4. Angle-series plots on the right show fluorescent intensity distribution of the indicated proteins along the perimeter of the cell, with 0° and +180°/−180° corresponding to the migrating front and rear, respectively. Solid lines represent the mean and shades represent mean ± SD. n, number of cells analyzed. Scale bars, 5 μm. Source data are available for this figure: SourceData F7.
Characterization of RacC. (A) Yeast two-hybrid assay showing interactions between Fbp17 and the GBD domain of WASP with the CA forms of 19 Rac proteins. Yeast was transformed with the indicated constructs and selected for the presence of prey and bait plasmids by growth on double-dropout (DD) agar plate lacking leucine and tryptophan. Interactions were assayed by growth on quadruple-dropout (QD) agar plate additionally lacking histidine and adenine. AD, Gal4-activation domain; BD, Gal4-binding domain. (B) Top: Design of the racC knockout construct. Bottom: Targeted clones were confirmed by PCR. (C) The indicated cells were plated clonally with bacteria (K. aerogenes) on standard medium agar for 5 d. Scale bar, 5 mm. (D–F) Time-lapse imaging of randomly migrating WT cells. (D) Cells expressing GFP-RacCG15V and LimEΔcoil-RFP. (E) Cells expressing GFP-RacCQ64L and LimEΔcoil-RFP. (F) Cells expressing GFP-RacCQ64L and RFP-ArpC4. Scale bars, 5 μm. (G) Left: Localization of GFP-myosin II in WT and racC− cells during random migration. Right: Quantification of the relative cortical localization of GFP-myosin II. Data were from three independent experiments; the scatter plot shows data points with mean ± SEM; n, number of cells analyzed. Scale bars, 5 μm.
Characterization of RacC. (A) Yeast two-hybrid assay showing interactions between Fbp17 and the GBD domain of WASP with the CA forms of 19 Rac proteins. Yeast was transformed with the indicated constructs and selected for the presence of prey and bait plasmids by growth on double-dropout (DD) agar plate lacking leucine and tryptophan. Interactions were assayed by growth on quadruple-dropout (QD) agar plate additionally lacking histidine and adenine. AD, Gal4-activation domain; BD, Gal4-binding domain. (B) Top: Design of the racC knockout construct. Bottom: Targeted clones were confirmed by PCR. (C) The indicated cells were plated clonally with bacteria (K. aerogenes) on standard medium agar for 5 d. Scale bar, 5 mm. (D–F) Time-lapse imaging of randomly migrating WT cells. (D) Cells expressing GFP-RacCG15V and LimEΔcoil-RFP. (E) Cells expressing GFP-RacCQ64L and LimEΔcoil-RFP. (F) Cells expressing GFP-RacCQ64L and RFP-ArpC4. Scale bars, 5 μm. (G) Left: Localization of GFP-myosin II in WT and racC− cells during random migration. Right: Quantification of the relative cortical localization of GFP-myosin II. Data were from three independent experiments; the scatter plot shows data points with mean ± SEM; n, number of cells analyzed. Scale bars, 5 μm.
We investigated the role of RacC in cortex assembly in relation to GxcM, Fbp17, and WASP by generating cells lacking racC or expressing the CA forms of RacC (RacCG15V or RacCQ64L). Deletion of racC (Fig. S4 B) effectively blocked GxcM overexpression-induced rear accumulation of LimEΔcoil or ArpC4 in directionally migrating cells without disrupting the localization of GxcM (Fig. 7, C and D; and Video 7), placing RacC downstream of GxcM. Expression of GFP- or RFP-fused RacCG15V under an inducible promoter generated polarized cells with the RacC signal localized selectively to the rear side. Consistent with the Y2H result, RFP-RacCG15V was capable of recruiting GFP-Fbp17 and GFP-WASP to the rear cortex (Fig. 8, A and B). Moreover, in cells overexpressing GFP-RacCG15V, RFP-ArpC4, and LimEΔcoil-RFP signals were redistributed from the migrating front to where RacC was most concentrated in the rear (Fig. 8 C and Fig. S4 D; and Video 8), similar to what was observed in cells overexpressing GxcM (Fig. 2, B and D). Inducible expression of GFP-RacCQ64L caused a similar redistribution of the actin markers (Fig. S4, E and F). The RacCCA-induced cell polarization and reorganization of the cortical actin network were weakened by fbp17 deletion (Fig. 8 D) and abolished by wasA deletion (Fig. 8 E and Fig. S5 A). Deletion of wasA also suppressed the effect of GxcM overexpression. Though cell polarity was not fully restored, the GxcM-induced intense signals of actin patches were no longer present in wasA− cells (Fig. S5 C, compared to Fig. 1 M). Collectively, these experiments imply that Fbp17 and WASP are needed for RacC activation-dependent cortical actin assembly downstream of GxcM.
Localization of GxcM-GFP with LimEΔcoil-RFP or RFP-ArpC4 in racC–cells migrating under agarose along a folic acid gradient. Corresponds to Fig. 7, C and D. Images were captured at 3-s intervals and played back at 6 frames per second. Scale bar = 5 μm.
Localization of GxcM-GFP with LimEΔcoil-RFP or RFP-ArpC4 in racC–cells migrating under agarose along a folic acid gradient. Corresponds to Fig. 7, C and D. Images were captured at 3-s intervals and played back at 6 frames per second. Scale bar = 5 μm.
RacC regulates cortical actin assembly via interaction with Fbp17 and WASP. (A) Localization of RFP-RacCG15V and GFP-Fbp17 in randomly migrating cells. (B) Localization of RFP-RacCG15V and GFP-WASP in randomly migrating cells. (C–E) Time-lapse imaging of GFP-RacCG15V and RFP-ArpC4 in WT (C), fbp17− (D), and wasA− (E) cells during random migration. Angle-series plots at the bottom show fluorescent intensity distribution of the indicated proteins along the perimeter of the cell, with 0° and +180°/−180° corresponding to the migrating front and rear, respectively. Solid lines represent the mean and shades represent mean ± SD. n, number of cells analyzed. Scale bars, 5 μm.
RacC regulates cortical actin assembly via interaction with Fbp17 and WASP. (A) Localization of RFP-RacCG15V and GFP-Fbp17 in randomly migrating cells. (B) Localization of RFP-RacCG15V and GFP-WASP in randomly migrating cells. (C–E) Time-lapse imaging of GFP-RacCG15V and RFP-ArpC4 in WT (C), fbp17− (D), and wasA− (E) cells during random migration. Angle-series plots at the bottom show fluorescent intensity distribution of the indicated proteins along the perimeter of the cell, with 0° and +180°/−180° corresponding to the migrating front and rear, respectively. Solid lines represent the mean and shades represent mean ± SD. n, number of cells analyzed. Scale bars, 5 μm.
Localization of GFP-RacCG15Vwith RFP-ArpC4 or LimEΔcoil-RFP in WT cells during random migration. Corresponds to Fig. 8 C and Fig. S4 D. Images were captured at 6-s intervals and played back at 12 frames per second. Scale bar = 5 μm.
Characterization of the wasA knockout cells and analyses of clathrin-mediated endocytosis. (A) Top: Design of the wasA knockout construct. Bottom: Targeted clones were confirmed by PCR. (B) The indicated cells were plated clonally with bacteria (K. aerogenes) on standard medium agar for 5 d. Scale bar, 5 mm. (C) Localization of GxcM-RFP and GFP-ArpC4 in wasA− cells. Scale bar, 5 μm. (D) Left: Projection length (Lp) of WT and wasA– cells determined by micropipette aspiration using a constant pressure of 500 Pa for 5 min. Right: Quantitative analysis of the projection lengths of probed cells. Data were from two independent experiments; the scatter plot shows data points with mean ± SEM; n, number of cells analyzed. (E) Left: Localization of GFP-CLC and RFP-ArpC4 in WT, fbp17−, racC−, and wasA− cells imaged by TIRF microscopy; scale bars, 5 μm. Right: Time-lapse imaging of clathrin vesicle internalization; scale bars, 1 μm. Images were acquired every 2–4 s for 8 min. In WT, fbp17−, and racC− cells, recruitment of ArpC4 to clathrin pits coincides with their internalization. In wasA− cells, clathrin pits fail to recruit ArpC4 and persist for hundreds of seconds. (F) Quantification of the lifetime (time between appearance and disappearance from the TIRF field of view) of clathrin-coated pits (CCPs) in WT, fbp17−, and racC− cells. Data were from at least two independent experiments; the scatter plot shows data points with mean ± SEM; n, number of CCP analyzed. In wasA− cells, the lifetime could not be accurately determined because it was often greater than the length of the time-lapse videos.
Characterization of the wasA knockout cells and analyses of clathrin-mediated endocytosis. (A) Top: Design of the wasA knockout construct. Bottom: Targeted clones were confirmed by PCR. (B) The indicated cells were plated clonally with bacteria (K. aerogenes) on standard medium agar for 5 d. Scale bar, 5 mm. (C) Localization of GxcM-RFP and GFP-ArpC4 in wasA− cells. Scale bar, 5 μm. (D) Left: Projection length (Lp) of WT and wasA– cells determined by micropipette aspiration using a constant pressure of 500 Pa for 5 min. Right: Quantitative analysis of the projection lengths of probed cells. Data were from two independent experiments; the scatter plot shows data points with mean ± SEM; n, number of cells analyzed. (E) Left: Localization of GFP-CLC and RFP-ArpC4 in WT, fbp17−, racC−, and wasA− cells imaged by TIRF microscopy; scale bars, 5 μm. Right: Time-lapse imaging of clathrin vesicle internalization; scale bars, 1 μm. Images were acquired every 2–4 s for 8 min. In WT, fbp17−, and racC− cells, recruitment of ArpC4 to clathrin pits coincides with their internalization. In wasA− cells, clathrin pits fail to recruit ArpC4 and persist for hundreds of seconds. (F) Quantification of the lifetime (time between appearance and disappearance from the TIRF field of view) of clathrin-coated pits (CCPs) in WT, fbp17−, and racC− cells. Data were from at least two independent experiments; the scatter plot shows data points with mean ± SEM; n, number of CCP analyzed. In wasA− cells, the lifetime could not be accurately determined because it was often greater than the length of the time-lapse videos.
RacC and WASP are required to maintain cortical integrity
We examined whether deletion of racC or wasA impairs the cortical function of the cell. The racC− cells formed slightly smaller plaques on bacterial lawns (Fig. S4 B), exhibited a severe defect in cytokinesis (Fig. 9, A and B), migrated with significantly reduced speed and directionality (Fig. 9, C–I), and failed to properly localize myosin II during random migration (Fig. S4 G). The cortical F-actin content and mechanical rigidity of the cell (mean projection length ± SEM measured by MPA: racC−, 8.70 ± 0.76 μm; WT, 2.25 ± 0.15 µm) were also considerably reduced in these cells (Fig. 9, J–O). Loss of wasA has been linked to defects in development (Fig. S5 B), cytokinesis, myosin recruitment, and migration (Davidson et al., 2018). In addition, we found that wasA− cells exhibited reduced mechanical rigidity (mean projection length ± SEM measured by MPA: wasA−, 8.39 ± 0.71 μm; WT, 1.96 ± 0.20 µm), similar to racC− and fbp17− cells (Fig. S5 D, compared to Fig. 5 N and Fig. 9 O). The fact that deletion of wasA, racC, or fbp17 leads to similar impairment in the various cortex-dependent cellular activities substantiates their collective involvement in cortex assembly and cortical activity regulation.
RacC is required to maintain cortical integrity. (A) WT, racC−, and GFP-RacC/racC− (rescue) cells grown for 60 h in shaken suspension were fixed and stained with DAPI to visualize the nuclei. (B) Quantification of nuclei in cells. n, number of cells analyzed. (C) Top: Trajectories of randomly migrating cells (n = 65 for WT and 69 for racC−). Bottom: Summary of motility parameters. (D and E) Velocity and Euclidean distance of cells are shown in C. At least 15 cells were quantified per experiment (each experiment shown in a different color); mean ± SEM. (F) Top: Trajectories of cells migrating under 2% agarose along folic acid gradient (n = 76 for WT and 75 for racC−). Bottom: Summary of chemotaxis parameters. (G–I) Speed, FMI, and directness of cells are shown in F. At least 20 cells were quantified per experiment (each experiment shown in a different color); mean ± SEM. (J) Localization of GFP-ABD in WT and racC− cells. (K) Quantification of the cortex-to-cytoplasm fluorescent intensity ratios of GFP-ABD. (L) WT and racC− cells were fixed and stained with Alexa Fluor 555-labeled phalloidin. (M) Quantification of the fluorescence intensity of cortical phalloidin. (N) Projection length (Lp) of WT and racC− cells determined by micropipette aspiration using a constant pressure of 500 Pa for 5 min. (O) Quantitative analysis of the projection lengths of probed cells. For C and F, data were from three independent experiments (the average of each biological replicate was used to calculate the mean and SEM). For K, M, and O, data were from three independent experiments; the scatter plots show data points with mean ± SEM; n, number of cells analyzed. Scale bars, 5 μm.
RacC is required to maintain cortical integrity. (A) WT, racC−, and GFP-RacC/racC− (rescue) cells grown for 60 h in shaken suspension were fixed and stained with DAPI to visualize the nuclei. (B) Quantification of nuclei in cells. n, number of cells analyzed. (C) Top: Trajectories of randomly migrating cells (n = 65 for WT and 69 for racC−). Bottom: Summary of motility parameters. (D and E) Velocity and Euclidean distance of cells are shown in C. At least 15 cells were quantified per experiment (each experiment shown in a different color); mean ± SEM. (F) Top: Trajectories of cells migrating under 2% agarose along folic acid gradient (n = 76 for WT and 75 for racC−). Bottom: Summary of chemotaxis parameters. (G–I) Speed, FMI, and directness of cells are shown in F. At least 20 cells were quantified per experiment (each experiment shown in a different color); mean ± SEM. (J) Localization of GFP-ABD in WT and racC− cells. (K) Quantification of the cortex-to-cytoplasm fluorescent intensity ratios of GFP-ABD. (L) WT and racC− cells were fixed and stained with Alexa Fluor 555-labeled phalloidin. (M) Quantification of the fluorescence intensity of cortical phalloidin. (N) Projection length (Lp) of WT and racC− cells determined by micropipette aspiration using a constant pressure of 500 Pa for 5 min. (O) Quantitative analysis of the projection lengths of probed cells. For C and F, data were from three independent experiments (the average of each biological replicate was used to calculate the mean and SEM). For K, M, and O, data were from three independent experiments; the scatter plots show data points with mean ± SEM; n, number of cells analyzed. Scale bars, 5 μm.
The defects in wasA− cells have previously been attributed to impaired clathrin-mediated endocytosis (Davidson et al., 2018). Considering that homologs of Fbp17 and RacC have also been implicated in this process (Lamaze et al., 1996; Shimada et al., 2007), we finally explored whether a defect in endocytosis could account for the mutant phenotypes. We expressed GFP-fused clathrin light chain (CLC) and RFP-ArpC4 in cells and monitored endocytosis by total internal reflection fluorescence (TIRF) microscopy. In the TIRF view, clathrin-coated pits (CCPs) were seen as transient puncta on the membrane, and the loss of puncta signal, which corresponded with vesicle internalization, was frequently associated with a brief burst of actin polymerization indicated by the recruitment of RFP-ArpC4 (Fig. S5 E). The average lifetime of CCPs was 47.9 ± 2.1 s (mean ± SEM) in WT cells (Fig. S5 F). Deletion of fbp17, racC, or wasA impaired the dynamics, but to different extents. Loss of fbp17 and racC prolonged the lifetime to 67.9 ± 3.0 and 169.6 ± 7.6 s, respectively, whereas loss of wasA resulted in enlarged puncta that persisted for hundreds of seconds with no discernible ArpC4 recruitment (Fig. S5, E and F). The degree of phenotypic defect exhibited by the different knockout cells in endocytosis and other cortex-dependent cellular activities did not consistently correlate, indicating that defects in endocytosis cannot be solely responsible for the mutant phenotypes.
Discussion
In this study, we delineated a signaling cascade that promotes Arp2/3-dependent actin polymerization and maintains cortical integrity in Dictyostelium cells. In this cascade, the cell rear-localized RhoGEF GxcM signals through Fbp17 and RacC, which in turn activates WASP to stimulate actin polymerization (Fig. 10). Prior to our study, sporadic observations hinted at the presence of Arp2/3 and branched F-actin in the rear of Dictyostelium cells. However, because the Arp2/3 complex is a main driver of actin-based protrusions at the leading edge, live cell reporters for Arp2/3 are highly enriched in the front of the cell, likely overshadowing the potential signals in the rear. We were able to visualize the previously unrecognized function of Arp2/3-based actin using cells overexpressing GxcM or activated RacC. Although these were considered non-physiological conditions, the fact that deletion of the respective downstream components abolished or reduced the overexpression effect corroborated the involvement of the signaling cascade in cortex assembly. Defects in the various cortex-dependent cellular activities observed in the fbp17−, racC−, and wasA− cells further substantiated the function of these proteins in maintaining cortical integrity.
The GxcM-Fbp17/RacC-WASP signaling cascade in the formation of a rear cortical subcompartment. GxcM, which localizes selectively in the rear of Dictyostelium cells, signals through Fbp17 and RacC to activate WASP by releasing its VCA tail, which in turn promotes Arp2/3 complex-mediated cortical actin assembly. Over-activation of the signaling cascade triggers excessive actin polymerization in the rear, whereas disruption of the cascade leads to defects in cortical rigidity and function.
The GxcM-Fbp17/RacC-WASP signaling cascade in the formation of a rear cortical subcompartment. GxcM, which localizes selectively in the rear of Dictyostelium cells, signals through Fbp17 and RacC to activate WASP by releasing its VCA tail, which in turn promotes Arp2/3 complex-mediated cortical actin assembly. Over-activation of the signaling cascade triggers excessive actin polymerization in the rear, whereas disruption of the cascade leads to defects in cortical rigidity and function.
Analysis of the GxcM-Fbp17/RacC-WASP signaling cascade revealed crosstalk and redundancy. First, overexpression of GxcM strongly activates the cascade, whereas its deletion causes only subtle defects, suggesting that additional factors, possibly other RhoGEF proteins, may substitute for GxcM. Second, the deletion of wasA only partially suppressed the effect of GxcM overexpression, implying redundancy at the output of the cascade. A potential candidate that could substitute for WASP is the SCAR/WAVE complex. The complex has been linked to FBP17 in Dictyostelium and other systems (Bai and Grant, 2015; Fort et al., 2018). In addition, loss of scrA impaired the mechanical rigidity of Dictyostelium cells (Litschko et al., 2019). Third, GxcM promotes WASP activation by recruiting Fbp17, as well as activating RacC. Both branches are necessary as disruption of either blocks the effect of GxcM overexpression. Fbp17 interacts with activated RacC, indicating that the two branches are not independent but likely function coordinately. Previous studies of mammalian homologs have led to an effector handover model, which explains how FBP17 and Rac family proteins may coordinate for WASP activation (Watson et al., 2016; Watson et al., 2017). In this model, the membrane-bound state of FBP17 is initially favored via protein–lipid interaction and strengthened through homodimerization/oligomerization, which results in sufficient local concentrations to achieve an interaction with Rac, providing that Rac activation has occurred coincidentally. Once colocalized and bound to Rac, FBP17 is poised to activate WASP. The binding of the SH3 domain of FBP17 to the poly-proline region of WASP partially unfolds WASP, allowing interaction between the WASP GBD domain and Rac to occur. The high affinity between GBD and Rac then rapidly pushes the equilibrium in its favor, leading to fully unfolded WASP that is able to trigger Arp2/3 activation and actin nucleation. Our results are in line with the model and offer additional intriguing aspects. In the signaling cascade we outlined, instead of membrane lipids, the rear cortex-localized GxcM likely provides the initial platform for Fbp17 recruitment and activates RacC at the same time. We also provided evidence for the homodimerization of Fbp17 and the potential allosteric effects of the dimerized SH3 on WASP activation. Future studies are needed to address the redundancy issue and fully elucidate the biochemical mechanism by which the signaling cascade mediates the assembly of the cortical branched actin network.
How the GxcM-Fbp17/RacC-WASP signaling cascade is selectively brought to the rear of migrating cells remains an open question. GxcM, which is positioned upstream of the pathway and exhibits dynamic behavior identical to other rear proteins (Iijima and Devreotes, 2002; Swaney et al., 2015), likely plays an important role. We have previously shown that rear-to-front gradients of PI(4,5)P2 and PI(3,5)P2 jointly regulate the rear accumulation of a number of proteins (Li et al., 2022). However, as GxcM does not exhibit apparent PI(4,5)P2- or PI(3,5)P2-binding activity assessed by lipid–protein interaction assays, the molecular mechanism underlying its rear accumulation remains obscure. Intriguingly, activated RacC also selectively targets the rear side of the cell. Furthermore, for both GxcM and activated RacC, their polarized distribution depends on the activities of the respective downstream components. Thus, specific targeting mechanisms and feedback regulation may act together to establish and reinforce the polarity of the signaling cascade.
Our study, together with previous reports showing a critical role of formin proteins in cortex assembly in Dictyostelium (Litschko et al., 2019), indicate that the actin meshwork in the rear cortex of rapidly migrating cells may also be composed of both formin-mediated and Arp2/3-mediated F-actin, similar to what has been observed in relatively non-motile mammalian cells. Different actin nucleators likely differentially affect the length, degree of branching, and density of cortical actin filaments, all of which could influence the emerging physical properties of the cortical network and, hence, cell behavior. The RacE-formin and GxcM-Fbp17/RacC-WASP pathways appear to contribute differently to the cortical function of the cell. Loss of either pathway reduces the cortical F-actin content and cell rigidity. However, the triple formin and racE mutants move with exaggerated fronts and higher speed in unconfined environments and completely lose the ability to migrate in 2D confinement (Litschko et al., 2019), whereas the fbp17 and racC mutants are able to migrate in 2D confinement but move with significantly slower speed in unconfined settings. These observations highlight the functional difference between formin-mediated and Arp2/3-mediated cortical actin structures. For fbp17 and racC mutants, the differential recruitment of myosin II explains, at least in part, their behavioral difference during migration under confined versus unconfined conditions. This result further suggests that the Arp2/3-mediated actin network contributes to the localization and function of myosin II, even though formin-mediated F-actin is generally considered a better substrate. The relative contribution of different actin nucleators and their interplay in cortical function awaits further investigation.
We found that, in addition to the proposed role in building the rear cortical subcompartment and maintaining cortical integrity, components of the signaling cascade, including Fbp17, RacC, and WASP, regulate clathrin-mediated endocytosis similar to their mammalian homologs. Interestingly, the two cellular activities may be spatially coupled, as clathrin-mediated endocytosis tends to occur at the rear in highly motile cells, including Dictyostelium and leukocytes (Damer and O’Halloran, 2000; Davis et al., 1982; Samaniego et al., 2007). Defects in endocytosis may result in reduced membrane tension and misdelivery of membrane proteins (Amato et al., 2019; Djakbarova et al., 2021), which could superimpose on the cortical integrity defects, leading to the various phenotypes observed in the mutant cells. Fbp17, RacC, and WASP family proteins have also been proposed to regulate the formation of front protrusions in migrating cells (Tsujita et al., 2015; Veltman et al., 2012). How the same core actin polymerization machinery drives the formation of different types of cellular structures and, thus, mediates diverse cell functions remains another outstanding question for future investigations.
Materials and methods
Cell culture, transformation, and differentiation
WT cells were derived from Ax2 (Ka) axenic strain provided by Robert Kay laboratory or Ax3 axenic strain provided by Peter Devreotes laboratory. The deletion cell line for wasA was generated in Ax3 and all the other deletion cell lines were generated in Ax2. WT and gene deletion cells were cultured in HL5 medium (cat# HLF3; Formedium) supplemented with antibiotics at 22°C following routine procedures. Cells carrying expression constructs were transformed by electroporation and maintained in HL5 containing G418 (10–40 μg/ml), Hygromycin (50 μg/ml), or both as needed. To induce the expression of constitutively active RacC 20 μg/ml doxycycline was added and incubated for 16 h. For differentiation, cells grown in HL5 were washed with development buffer (DB; 5 mM Na2HPO4, 5 mM KH2PO4, 2 mM MgSO4, and 0.2 mM CaCl2), starved in DB for 1 h, and pulsed with 100 nM cAMP every 6 min for 3–5 h.
Gene disruption and plasmid construction
Plasmids and primers used in this study are listed in Table 1. To make knockout constructs for gxcM, fbp17, racC, and wasA deletion, a blasticidin S resistance (BSR) cassette (Kimmel and Faix, 2006) was inserted into pBlueScript II SK+ to generate pBlueScript-BSR. 5′ and-3′ arms were PCR-amplified from genomic DNA and cloned upstream and downstream of the BSR cassette, respectively. The resulting disruption cassette was amplified by PCR and electroporated into Ax2 or Ax3. Gene disruption was confirmed by resistance to blasticidin (10 μg/ml), PCR, and rescue experiment.
Plasmids and primers used in this study
| Expression in Dictyostelium cells . | |||
|---|---|---|---|
| Usage . | Plasmid backbone . | Sequence, 5′-3′ . | |
| GxcM-GFP or GxcM-RFP | pDM323 or pDM451 | F: 5′-GGAAAGTCGACAAAGCTAGCAGTAAAATAAAAATGAAAATTAAATTTTTTCGATATG-3′ | |
| R: 3′-GTTCTTCTCCTTTACCACTACTAGATTTTCTTGGTGGAATGATTGG-5′ | |||
| GxcMN790-GFP | pDM323 | F: 5′-GGAGCTCAAATAAAAATGAAAATTAAATTTTTTCGATATG-3′ | |
| R: 3′-GGACTAGTACTTCTTTTAATATGATCTAAAAC-5′ | |||
| GxcMN790-RFP | pDM451 | F: 5′-CCGGAGCTCAAATAAAAATGAAAATTAAATTTTTTCGATATG-3′ | |
| R: 3′-CTAGCTAGCACTTCTTTTAATATGATCTAAAACTTTATC-5′ | |||
| GxcM4A-GFP or GxcM4A-RFP | pDM323 or pDM451 | F1: 5′-GAGAGTTTTCTAATTAAAGCAGCAGCAGCACTTTGTAAATATCCATTAC-3′ | |
| R1: 3′-GTAATGGATATTTACAAAGTGCTGCTGCTGCTTTAATTAGAAAACTCTC-5′ | |||
| F: 5′-CCGGAGCTCAAATAAAAATGAAAATTAAATTTTTTCGATATG-3′ | |||
| R: 3′-CTAGCTAGCTTTTCTTGGTGGAATGATTGGTTTTTTAG-5′ | |||
| GxcMN455-GFP | pDM323 | F: 5′-GGAGCTCAAATAAAAATGAAAATTAAATTTTTTCGATATG-3′ | |
| R: 3′-CGGACTAGTCATTTTATTTTCACCTTCTTCTTTCATTAC-5′ | |||
| GxcMN456-770-GFP | pDM323 | F: 5′-GGGGAGCTCATGAGAAACATGGTAATTAATG-3′ | |
| R: 3′-CGGACTAGTTTTAATTAATTCATCCAAATCAG-5′ | |||
| GxcM | pDM358 | F: 5′-CCGGAGCTCAAATAAAAATGAAAATTAAATTTTTTCGATATG-3′ | |
| R: 3′-CTAGCTAGCTTTTCTTGGTGGAATGATTGGTTTTTTAG-5′ | |||
| GFP-Fbp17 or RFP-Fbp17 | pDM317 or pDM449 | F: 5′-CCGGAGCTCATGAGTTATTCAACCGATTTATTAGATGG-3′ | |
| R: 3′-CTAGCTAGCACCATCGATGATGTCAACAAAATTGGCCGGAATG-5′ | |||
| GFP-Fbp17ΔSH3 or RFP-Fbp17ΔSH3 | pDM317 or pDM449 | F: 5′-CCGGAGCTCATGAGTTATTCAACCGATTTATTAGATGG-3′ | |
| R: 3′-CTAGCTAGCCAATTCCTCCAATTGAGCAGTGATAAG-5′ | |||
| GFP-Fbp17SH3 or RFP-Fbp17SH3 | pDM317 or pDM449 | F: 5′-CCGGAGCTCGGTAAAACTAATCTTTCTGGTTCAAG-3′ | |
| R: 3′-CTAGCTAGCACCATCGATGATGTCAACAAAATTGG-5′ | |||
| GFP-Fbp17F-BAR or RFP-Fbp17F-BAR | pDM317 or pDM449 | F: 5′-CCGGAGCTCGGTAAAACTAATCTTTCTGGTTCAAG-3′ | |
| R: 3′-CTAGCTAGCGGTGCCAACTCTAAAGTTTGTTGGAGAG-5′ | |||
| GFP-Fbp17ΔBAR or RFP-Fbp17ΔBAR | pDM317 or pDM449 | F: 5′-CCGGAGCTCCAGATCGACAAGGACGCTGATATCCGTC-3′ | |
| R: 3′-CTAGCTAGCACCATCGATGATGTCAACAAAATTGGCCG-5′ | |||
| GFP-WASP | pDM317 | F: 5′-CTATCTAGAATGGGTAGTCCAACTATTAGTGATC-3′ | |
| R: 3′-CTAGTCGACATAATCTGACCATTCATCATCGTC-5′ | |||
| GFP-ArpC4 | pDM317 | F: 5′-GGGGTACCATGTCCACAGCTCAAGTTCCATATTTAAATTG-3′ | |
| R: 3′-CGAGCGGCCGCAGCAAAGTTCTTTAAGTATTCAGAAGCAAC-5′ | |||
| RFP-ArpC4 | pDM449 | F: 5′-CGGGATCCATGTCCACAGCTCAAGTTCCATATTTAAATTG-3′ | |
| R: 3′-GCTCTAGAAGCAAAGTTCTTTAAGTATTCAGAAGCAAC-5′ | |||
| GFP-Coronin A | pDM317 | F: 5′-GGTCTAGAATGTCTAAAGTAGTCCGTAGTAG-3′ | |
| R: 3′-CCGGCGGCCGCGTTGGTGAGTTCTTTGATTTTGGC-5′ | |||
| GFP-RacC | pDM317 | F: 5′-CCGGAGCTCATGTCAGCAGCAGAAGTTATTAAATTAG-3′ | |
| R: 3′-CTAGCTAGCTTACATAACAATACACTTGGATTTCTTTTTTTTG-5′ | |||
| GFP-RacCG15V | pDM317 | F: 5′-GTTGCTGTAGGTAAAACTTGTTTATTGATTAG-3′ | |
| R: 3′-ATCACCAATAACGACTAATTTAATAACTTC-5′ | |||
| tet on GFP-RacCG15V | pDM371 | F: 5′-CGCGGATCCATGTCAGCAGCAGAAGTTATTAAATTAG-3′ | |
| R: 3′-CGGACTAGTTTACATAACAATACACTTGGATTTCTTTTTTTTG-5′ | |||
| tet on GFP-RacCQ64L | pDM371 | F: 5′-CTTTGGGATACTGCAGGTTTAGAAGAGTACGATAAATTAAG-3′ | |
| R: 3′-CTTAATTTATCGTACTCTTCTAAACCTGCAGTATCCCAAAG-5′ | |||
| tet on RFP-RacCG15V | pDM359 | F: 5′-CGCGGATCCATGGCATCATCAGAAGATGTTATTAAAGAATTTATG-3′ | |
| R: 3′-CGGACTAGTTTACATAACAATACACTTGGATTTCTTTTTTTTG-5′ | |||
| GFP-Rac1A | pDM317 | F: 5′-CCGGAGCTCATGCAAGCAATTAAATGTGTCGTTGTCGGT-3′ | |
| R: 3′-CTAGCTAGCTAAAATGTTGCAACCACCTGAACTTTTCTTCTTTTTGC-5′ | |||
| GFP-ABD | pCV5 | F: 5′-CCGGAGCTCATGGCTGCTGCTCCAAGTGGAAAAAC-3′ | |
| R: 3′-CAGGCTCGAGGGCATCTGAAGTTTCACGACGTTTCTTTTC-5′ | |||
| GFP-Myosin II | pCV5 | F: 5′-CCGGAGCTCAATCCAATTCATGATAGAACTTCAGATTATC-3′ | |
| R: 3′-CAGGCTCGAGTTAAGCTTTGAAACCACCAAAGAAATCGGC-5′ | |||
| GFP-CLC | pDM317 | F: 5′-CAGGAGCTCATGTCAGATCCATTTGGTGAAG-3′ | |
| R: 3′-CTAGCTAGCAACAATTGGTTGATTTTTTAAACG-5′ | |||
| Generation of knockout cells | |||
| gxcM knockout | pBluescript-BSR | Insert 1 F: 5′-CACGGTACCGATACCATCATCGATGATATCAC-3′ | |
| Insert 1 R: 3′-GCCGGAATTCGTGGTGGTATTATTTGATGATG-5′ | |||
| Insert 2 F: 5′-CGGACTAGTGTAAAGTTACAACACCAATTTCTACAC-3′ | |||
| Insert 2 R: 3′-CGGCGGCCGCTTATTTTCTTGGTGGAATGATTGG-5′ | |||
| fbp17 knockout | pBluescript-BSR | Insert 1 F: 5′-CACGGTACCCGTGTATTTCTAAACTATTTTATAC-3′ | |
| Insert 1 R: 3′-GAGAAGCTTCATCTAATAAATCGGTTGAATAACTCAT-5′ | |||
| Insert 2 F: 5′-CGGACTAGTCCAATTTTGTTGACATCATCGATG-3′ | |||
| Insert 2 R: 3′-CGGCGGCCGCGATTAAGGTTGGTATTTTTGAAAG-5′ | |||
| racC knockout | pBluescript-BSR | Insert 1 F: 5′-GACGTCGACCCCCCCTAACATTAAAAGTATTAATAATAG-3′ | |
| Insert 1 R: 3′-GAGAAGCTTCCCTACAGCACCATCACCAATAACG-5′ | |||
| Insert 2 F: 5′-CGGACTAGTGGAATGTTCTGCCAAAACCTCACAAAATCTC-3′ | |||
| Insert 2 R: 3′-CGGCGGCCGCGGTGGCTTCCTAATTTGCCTTTTCTCAAAG-5′ | |||
| wasA knockout | pBluescript-BSR | Insert 1 F: 5′-GACGTCGACGGTATTATATATTGTTCATTTGCATTGGTTT-3′ | |
| Insert 1 R: 3′-GAGAAGCTTCATACAATCTTGCAACAGTAGTTGAATG-5′ | |||
| Insert 2 F: 5′-CGGACTAGTCTATGGCTAATCGTAGAGGTGGTATGAGA-3′ | |||
| Insert 2 R: 3′-CGGCGGCCGCCTGGTAATGTTTTATGACTTGATGTAAATG-5′ | |||
| Expression in bacteria or insect cells | |||
| GST-Fbp17 | pFastBac1-GST | F: 5′-GAGCATATGATGAGTTATTCAACCGATTTATTAGATGGATTTGAAAGATTATATAAAAGAACtTATGGTGTTTTAAAATGTAATC-3′ | |
| R: 3′-TGCTCTAGATTAACCATCGATGATGTCAACAAAATTGGC-5′ | |||
| GST-WASP | pFastBac1-GST | F: 5′-GAGCATATGATGGGTAGTCCAACTATTAGTGATCAAG-3′ | |
| R: 3′-TGCTCTAGAATAATCTGACCATTCATCATCGTCATCATCATC-5′ | |||
| GST-WASPVCA | pGEX-6P-1 | F: 5′-CGCGGATCCCCATCATCAGGAGGTGGTGGTGCC-3′ | |
| R: 3′-TGCTCTAGATTAATAATCTGACCATTCATCATCGTCATC-5′ | |||
| GST-Fbp17SH3 | pGEX-6P-1 | F: 5′-CGCGGATCCGCTGGTGCAACTGGTGGTGCCACTAGAG-3′ | |
| R: 3′-CAGGCTCGAGACCATCGATGATGTCAACAAAATTGGC-5′ | |||
| His-MBP-Fbp17SH3 | pET-MBP-3C | F: 5′-CGCGGATCCGCTGGTGCAACTGGTGGTGCCACTAGAG-3′ | |
| R: 3′-CAGGCTCGAGACCATCGATGATGTCAACAAAATTGGC-5′ | |||
| Expression in yeast cells | |||
| Fbp17 | pGADT7 | F: 5′-GAGCATATGATGAGTTATTCAACCGATTTATTAGATGG-3′ | |
| R: 3′-CGCGGATCCACCATCGATGATGTCAACAAAATTGGCCGG-5′ | |||
| WASPN126-230 | pGADT7 | F: 5′-GAGCATATGATTGGTAAAACTGCTGCAATTAATCCAAATAC-3′ | |
| R: 3′-GG AGATCTACCAATGATATTGACAATGAATTGAGCAG-5′ | |||
| Rac1AG12V | pGBKT7 | F: 5′-GTTGCTGTAGGTAAAACCTGTCTTTTAATTTC-3′ | |
| R: 3′-ATCACCGACAACGACACATTTAATTGCTTG-5′ | |||
| Rac1BG12V | pGBKT7 | F: 5′-GTTGCAGTTGGTAAAACATGTCTTTTAATTTC-3′ | |
| R: 3′-ATCACCAACAACTACACATTTAATTGC-5′ | |||
| Rac1CG12V | pGBKT7 | F: 5′-GTTGCGGTTGGTAAAACATGTCTTTTAATTTC-3′ | |
| R: 3′-ATCACCTACAACTACACATTTAATTGC-5′ | |||
| RacAG12V | pGBKT7 | F: 5′-GTTGCTGTAGGTAAAAGTTGTTTATTAATTGC-3′ R: 3′-ATCACCTACTACTACTAATTTAATTGC-5′ | |
| RacBG12V | pGBKT7 | F: 5′-GTTGCTGTTGGTAAAACTTGTTTATTAATTTC-3′ | |
| R: 3′-ATCACCTACTACCACCAATTTAATTGATTG-5′ | |||
| RacCG15V | pGBKT7 | F: 5′-GTTGCTGTAGGTAAAACTTGTTTATTGATTAG-3′ | |
| R: 3′-ATCACCAATAACGACTAATTTAATAACTTC-5′ | |||
| RacDG17V | pGBKT7 | F: 5′-GTTGCTGTTGGTAAAACTTCACTTTTAATTTTATATAC-3′ | |
| R: 3′-ATCACCTACAACTACTACTTTAACAG-5′ | |||
| RacEG20V | pGBKT7 | F: 5′-GTTGCTGTTGGTAAAACATGTCTTTTAATTTG-3′ | |
| R: 3′-ATCACCGACAACTACTAATTTAACTCTTG-5′ | |||
| RacF1G12V | pGBKT7 | F: 5′-GTTGCAGTTGGTAAAACCTGTATGTTAATTTC-3′ | |
| R: 3′-ATCACCAACAACAACACATTTAATATTTTG-5′ | |||
| RacF2G12V | pGBKT7 | F: 5′-GTTGCAGTTGGTAAAACTTGTATGTTAATTTC-3′ | |
| R: 3′-ATCACCAACAACAACACATTTAATATTTTG-5′ | |||
| RacGG12V | pGBKT7 | F: 5′-GTTGGAATTGGTAAAACATCAATGTTATTAAG-3′ | |
| R: 3′-TTCGCCAACAACACAAACTTTAATACTTTTC-5′ | |||
| RacHM13V | pGBKT7 | F: 5′-GTTAGTGTAGGTAAGACATGTTTACTCATTTC-3′ | |
| R: 3′-ATCACCTACAACCATTACTTTAATATCTTTTAC-5′ | |||
| RacIS14V | pGBKT7 | F: 5′-GTTAAAACTGGAAAAACTACAATGATGATGAC-3′ | |
| R: 3′-ATCACCTAATACTAATAACTTTATATATG-5′ | |||
| RacJD18V | pGBKT7 | F: 5′-GTTGGAGTTGGTAAATCATGTGTAATGAACTC-3′ | |
| R: 3′-ATCTCCAAGACAAAATATTTTAAC-5′ | |||
| RacLG12V | pGBKT7 | F: 5′-GTTGCAGTTGGTAAAACAAGTTTATTAATTGC-3′ | |
| R: 3′-ATCACCACAAATTACCATTTTTATATATTGC-5′ | |||
| RacMY14V | pGBKT7 | F: 5′-GTTGGTGTTGGTAAAACCACCCTATTAATGAC-3′ | |
| R: 3′-ATCGCCAATCGTAACTATTTTAATAG-5′ | |||
| RacNG14V | pGBKT7 | F: 5′-TTAGAGGATTATCCTCGTATAAGACCATTATC-3′ | |
| R: 3′-ACCACCAGTATCCCAACAACCAATTCTC-5′ | |||
| RacOG13V | pGBKT7 | F: 5′-GTTTTAATTGGTAAAACTACATTATTAATGAC-3′ | |
| R: 3′-ATCACCAACTGTCACTATCTTTACTTC-5′ | |||
| RacPG58V | pGBKT7 | F: 5′-GTTTACTGTTGGAAAAACAACACTCTTATTATC-3′ | |
| R: 3′-ATCACCTACAACACCACATTTAATAACCTTTG-5′ | |||
| Other plasmids for expression in Dictyostelium cells | |||
| PHcrac-GFP | Peter Devreotes Laboratory, Johns Hopkins University | ||
| LimEΔcoil-GFP | Peter Devreotes Laboratory, Johns Hopkins University | ||
| LimEΔcoil-RFP | Douglas Robinson Laboratory, Johns Hopkins University | ||
| Expression in Dictyostelium cells . | |||
|---|---|---|---|
| Usage . | Plasmid backbone . | Sequence, 5′-3′ . | |
| GxcM-GFP or GxcM-RFP | pDM323 or pDM451 | F: 5′-GGAAAGTCGACAAAGCTAGCAGTAAAATAAAAATGAAAATTAAATTTTTTCGATATG-3′ | |
| R: 3′-GTTCTTCTCCTTTACCACTACTAGATTTTCTTGGTGGAATGATTGG-5′ | |||
| GxcMN790-GFP | pDM323 | F: 5′-GGAGCTCAAATAAAAATGAAAATTAAATTTTTTCGATATG-3′ | |
| R: 3′-GGACTAGTACTTCTTTTAATATGATCTAAAAC-5′ | |||
| GxcMN790-RFP | pDM451 | F: 5′-CCGGAGCTCAAATAAAAATGAAAATTAAATTTTTTCGATATG-3′ | |
| R: 3′-CTAGCTAGCACTTCTTTTAATATGATCTAAAACTTTATC-5′ | |||
| GxcM4A-GFP or GxcM4A-RFP | pDM323 or pDM451 | F1: 5′-GAGAGTTTTCTAATTAAAGCAGCAGCAGCACTTTGTAAATATCCATTAC-3′ | |
| R1: 3′-GTAATGGATATTTACAAAGTGCTGCTGCTGCTTTAATTAGAAAACTCTC-5′ | |||
| F: 5′-CCGGAGCTCAAATAAAAATGAAAATTAAATTTTTTCGATATG-3′ | |||
| R: 3′-CTAGCTAGCTTTTCTTGGTGGAATGATTGGTTTTTTAG-5′ | |||
| GxcMN455-GFP | pDM323 | F: 5′-GGAGCTCAAATAAAAATGAAAATTAAATTTTTTCGATATG-3′ | |
| R: 3′-CGGACTAGTCATTTTATTTTCACCTTCTTCTTTCATTAC-5′ | |||
| GxcMN456-770-GFP | pDM323 | F: 5′-GGGGAGCTCATGAGAAACATGGTAATTAATG-3′ | |
| R: 3′-CGGACTAGTTTTAATTAATTCATCCAAATCAG-5′ | |||
| GxcM | pDM358 | F: 5′-CCGGAGCTCAAATAAAAATGAAAATTAAATTTTTTCGATATG-3′ | |
| R: 3′-CTAGCTAGCTTTTCTTGGTGGAATGATTGGTTTTTTAG-5′ | |||
| GFP-Fbp17 or RFP-Fbp17 | pDM317 or pDM449 | F: 5′-CCGGAGCTCATGAGTTATTCAACCGATTTATTAGATGG-3′ | |
| R: 3′-CTAGCTAGCACCATCGATGATGTCAACAAAATTGGCCGGAATG-5′ | |||
| GFP-Fbp17ΔSH3 or RFP-Fbp17ΔSH3 | pDM317 or pDM449 | F: 5′-CCGGAGCTCATGAGTTATTCAACCGATTTATTAGATGG-3′ | |
| R: 3′-CTAGCTAGCCAATTCCTCCAATTGAGCAGTGATAAG-5′ | |||
| GFP-Fbp17SH3 or RFP-Fbp17SH3 | pDM317 or pDM449 | F: 5′-CCGGAGCTCGGTAAAACTAATCTTTCTGGTTCAAG-3′ | |
| R: 3′-CTAGCTAGCACCATCGATGATGTCAACAAAATTGG-5′ | |||
| GFP-Fbp17F-BAR or RFP-Fbp17F-BAR | pDM317 or pDM449 | F: 5′-CCGGAGCTCGGTAAAACTAATCTTTCTGGTTCAAG-3′ | |
| R: 3′-CTAGCTAGCGGTGCCAACTCTAAAGTTTGTTGGAGAG-5′ | |||
| GFP-Fbp17ΔBAR or RFP-Fbp17ΔBAR | pDM317 or pDM449 | F: 5′-CCGGAGCTCCAGATCGACAAGGACGCTGATATCCGTC-3′ | |
| R: 3′-CTAGCTAGCACCATCGATGATGTCAACAAAATTGGCCG-5′ | |||
| GFP-WASP | pDM317 | F: 5′-CTATCTAGAATGGGTAGTCCAACTATTAGTGATC-3′ | |
| R: 3′-CTAGTCGACATAATCTGACCATTCATCATCGTC-5′ | |||
| GFP-ArpC4 | pDM317 | F: 5′-GGGGTACCATGTCCACAGCTCAAGTTCCATATTTAAATTG-3′ | |
| R: 3′-CGAGCGGCCGCAGCAAAGTTCTTTAAGTATTCAGAAGCAAC-5′ | |||
| RFP-ArpC4 | pDM449 | F: 5′-CGGGATCCATGTCCACAGCTCAAGTTCCATATTTAAATTG-3′ | |
| R: 3′-GCTCTAGAAGCAAAGTTCTTTAAGTATTCAGAAGCAAC-5′ | |||
| GFP-Coronin A | pDM317 | F: 5′-GGTCTAGAATGTCTAAAGTAGTCCGTAGTAG-3′ | |
| R: 3′-CCGGCGGCCGCGTTGGTGAGTTCTTTGATTTTGGC-5′ | |||
| GFP-RacC | pDM317 | F: 5′-CCGGAGCTCATGTCAGCAGCAGAAGTTATTAAATTAG-3′ | |
| R: 3′-CTAGCTAGCTTACATAACAATACACTTGGATTTCTTTTTTTTG-5′ | |||
| GFP-RacCG15V | pDM317 | F: 5′-GTTGCTGTAGGTAAAACTTGTTTATTGATTAG-3′ | |
| R: 3′-ATCACCAATAACGACTAATTTAATAACTTC-5′ | |||
| tet on GFP-RacCG15V | pDM371 | F: 5′-CGCGGATCCATGTCAGCAGCAGAAGTTATTAAATTAG-3′ | |
| R: 3′-CGGACTAGTTTACATAACAATACACTTGGATTTCTTTTTTTTG-5′ | |||
| tet on GFP-RacCQ64L | pDM371 | F: 5′-CTTTGGGATACTGCAGGTTTAGAAGAGTACGATAAATTAAG-3′ | |
| R: 3′-CTTAATTTATCGTACTCTTCTAAACCTGCAGTATCCCAAAG-5′ | |||
| tet on RFP-RacCG15V | pDM359 | F: 5′-CGCGGATCCATGGCATCATCAGAAGATGTTATTAAAGAATTTATG-3′ | |
| R: 3′-CGGACTAGTTTACATAACAATACACTTGGATTTCTTTTTTTTG-5′ | |||
| GFP-Rac1A | pDM317 | F: 5′-CCGGAGCTCATGCAAGCAATTAAATGTGTCGTTGTCGGT-3′ | |
| R: 3′-CTAGCTAGCTAAAATGTTGCAACCACCTGAACTTTTCTTCTTTTTGC-5′ | |||
| GFP-ABD | pCV5 | F: 5′-CCGGAGCTCATGGCTGCTGCTCCAAGTGGAAAAAC-3′ | |
| R: 3′-CAGGCTCGAGGGCATCTGAAGTTTCACGACGTTTCTTTTC-5′ | |||
| GFP-Myosin II | pCV5 | F: 5′-CCGGAGCTCAATCCAATTCATGATAGAACTTCAGATTATC-3′ | |
| R: 3′-CAGGCTCGAGTTAAGCTTTGAAACCACCAAAGAAATCGGC-5′ | |||
| GFP-CLC | pDM317 | F: 5′-CAGGAGCTCATGTCAGATCCATTTGGTGAAG-3′ | |
| R: 3′-CTAGCTAGCAACAATTGGTTGATTTTTTAAACG-5′ | |||
| Generation of knockout cells | |||
| gxcM knockout | pBluescript-BSR | Insert 1 F: 5′-CACGGTACCGATACCATCATCGATGATATCAC-3′ | |
| Insert 1 R: 3′-GCCGGAATTCGTGGTGGTATTATTTGATGATG-5′ | |||
| Insert 2 F: 5′-CGGACTAGTGTAAAGTTACAACACCAATTTCTACAC-3′ | |||
| Insert 2 R: 3′-CGGCGGCCGCTTATTTTCTTGGTGGAATGATTGG-5′ | |||
| fbp17 knockout | pBluescript-BSR | Insert 1 F: 5′-CACGGTACCCGTGTATTTCTAAACTATTTTATAC-3′ | |
| Insert 1 R: 3′-GAGAAGCTTCATCTAATAAATCGGTTGAATAACTCAT-5′ | |||
| Insert 2 F: 5′-CGGACTAGTCCAATTTTGTTGACATCATCGATG-3′ | |||
| Insert 2 R: 3′-CGGCGGCCGCGATTAAGGTTGGTATTTTTGAAAG-5′ | |||
| racC knockout | pBluescript-BSR | Insert 1 F: 5′-GACGTCGACCCCCCCTAACATTAAAAGTATTAATAATAG-3′ | |
| Insert 1 R: 3′-GAGAAGCTTCCCTACAGCACCATCACCAATAACG-5′ | |||
| Insert 2 F: 5′-CGGACTAGTGGAATGTTCTGCCAAAACCTCACAAAATCTC-3′ | |||
| Insert 2 R: 3′-CGGCGGCCGCGGTGGCTTCCTAATTTGCCTTTTCTCAAAG-5′ | |||
| wasA knockout | pBluescript-BSR | Insert 1 F: 5′-GACGTCGACGGTATTATATATTGTTCATTTGCATTGGTTT-3′ | |
| Insert 1 R: 3′-GAGAAGCTTCATACAATCTTGCAACAGTAGTTGAATG-5′ | |||
| Insert 2 F: 5′-CGGACTAGTCTATGGCTAATCGTAGAGGTGGTATGAGA-3′ | |||
| Insert 2 R: 3′-CGGCGGCCGCCTGGTAATGTTTTATGACTTGATGTAAATG-5′ | |||
| Expression in bacteria or insect cells | |||
| GST-Fbp17 | pFastBac1-GST | F: 5′-GAGCATATGATGAGTTATTCAACCGATTTATTAGATGGATTTGAAAGATTATATAAAAGAACtTATGGTGTTTTAAAATGTAATC-3′ | |
| R: 3′-TGCTCTAGATTAACCATCGATGATGTCAACAAAATTGGC-5′ | |||
| GST-WASP | pFastBac1-GST | F: 5′-GAGCATATGATGGGTAGTCCAACTATTAGTGATCAAG-3′ | |
| R: 3′-TGCTCTAGAATAATCTGACCATTCATCATCGTCATCATCATC-5′ | |||
| GST-WASPVCA | pGEX-6P-1 | F: 5′-CGCGGATCCCCATCATCAGGAGGTGGTGGTGCC-3′ | |
| R: 3′-TGCTCTAGATTAATAATCTGACCATTCATCATCGTCATC-5′ | |||
| GST-Fbp17SH3 | pGEX-6P-1 | F: 5′-CGCGGATCCGCTGGTGCAACTGGTGGTGCCACTAGAG-3′ | |
| R: 3′-CAGGCTCGAGACCATCGATGATGTCAACAAAATTGGC-5′ | |||
| His-MBP-Fbp17SH3 | pET-MBP-3C | F: 5′-CGCGGATCCGCTGGTGCAACTGGTGGTGCCACTAGAG-3′ | |
| R: 3′-CAGGCTCGAGACCATCGATGATGTCAACAAAATTGGC-5′ | |||
| Expression in yeast cells | |||
| Fbp17 | pGADT7 | F: 5′-GAGCATATGATGAGTTATTCAACCGATTTATTAGATGG-3′ | |
| R: 3′-CGCGGATCCACCATCGATGATGTCAACAAAATTGGCCGG-5′ | |||
| WASPN126-230 | pGADT7 | F: 5′-GAGCATATGATTGGTAAAACTGCTGCAATTAATCCAAATAC-3′ | |
| R: 3′-GG AGATCTACCAATGATATTGACAATGAATTGAGCAG-5′ | |||
| Rac1AG12V | pGBKT7 | F: 5′-GTTGCTGTAGGTAAAACCTGTCTTTTAATTTC-3′ | |
| R: 3′-ATCACCGACAACGACACATTTAATTGCTTG-5′ | |||
| Rac1BG12V | pGBKT7 | F: 5′-GTTGCAGTTGGTAAAACATGTCTTTTAATTTC-3′ | |
| R: 3′-ATCACCAACAACTACACATTTAATTGC-5′ | |||
| Rac1CG12V | pGBKT7 | F: 5′-GTTGCGGTTGGTAAAACATGTCTTTTAATTTC-3′ | |
| R: 3′-ATCACCTACAACTACACATTTAATTGC-5′ | |||
| RacAG12V | pGBKT7 | F: 5′-GTTGCTGTAGGTAAAAGTTGTTTATTAATTGC-3′ R: 3′-ATCACCTACTACTACTAATTTAATTGC-5′ | |
| RacBG12V | pGBKT7 | F: 5′-GTTGCTGTTGGTAAAACTTGTTTATTAATTTC-3′ | |
| R: 3′-ATCACCTACTACCACCAATTTAATTGATTG-5′ | |||
| RacCG15V | pGBKT7 | F: 5′-GTTGCTGTAGGTAAAACTTGTTTATTGATTAG-3′ | |
| R: 3′-ATCACCAATAACGACTAATTTAATAACTTC-5′ | |||
| RacDG17V | pGBKT7 | F: 5′-GTTGCTGTTGGTAAAACTTCACTTTTAATTTTATATAC-3′ | |
| R: 3′-ATCACCTACAACTACTACTTTAACAG-5′ | |||
| RacEG20V | pGBKT7 | F: 5′-GTTGCTGTTGGTAAAACATGTCTTTTAATTTG-3′ | |
| R: 3′-ATCACCGACAACTACTAATTTAACTCTTG-5′ | |||
| RacF1G12V | pGBKT7 | F: 5′-GTTGCAGTTGGTAAAACCTGTATGTTAATTTC-3′ | |
| R: 3′-ATCACCAACAACAACACATTTAATATTTTG-5′ | |||
| RacF2G12V | pGBKT7 | F: 5′-GTTGCAGTTGGTAAAACTTGTATGTTAATTTC-3′ | |
| R: 3′-ATCACCAACAACAACACATTTAATATTTTG-5′ | |||
| RacGG12V | pGBKT7 | F: 5′-GTTGGAATTGGTAAAACATCAATGTTATTAAG-3′ | |
| R: 3′-TTCGCCAACAACACAAACTTTAATACTTTTC-5′ | |||
| RacHM13V | pGBKT7 | F: 5′-GTTAGTGTAGGTAAGACATGTTTACTCATTTC-3′ | |
| R: 3′-ATCACCTACAACCATTACTTTAATATCTTTTAC-5′ | |||
| RacIS14V | pGBKT7 | F: 5′-GTTAAAACTGGAAAAACTACAATGATGATGAC-3′ | |
| R: 3′-ATCACCTAATACTAATAACTTTATATATG-5′ | |||
| RacJD18V | pGBKT7 | F: 5′-GTTGGAGTTGGTAAATCATGTGTAATGAACTC-3′ | |
| R: 3′-ATCTCCAAGACAAAATATTTTAAC-5′ | |||
| RacLG12V | pGBKT7 | F: 5′-GTTGCAGTTGGTAAAACAAGTTTATTAATTGC-3′ | |
| R: 3′-ATCACCACAAATTACCATTTTTATATATTGC-5′ | |||
| RacMY14V | pGBKT7 | F: 5′-GTTGGTGTTGGTAAAACCACCCTATTAATGAC-3′ | |
| R: 3′-ATCGCCAATCGTAACTATTTTAATAG-5′ | |||
| RacNG14V | pGBKT7 | F: 5′-TTAGAGGATTATCCTCGTATAAGACCATTATC-3′ | |
| R: 3′-ACCACCAGTATCCCAACAACCAATTCTC-5′ | |||
| RacOG13V | pGBKT7 | F: 5′-GTTTTAATTGGTAAAACTACATTATTAATGAC-3′ | |
| R: 3′-ATCACCAACTGTCACTATCTTTACTTC-5′ | |||
| RacPG58V | pGBKT7 | F: 5′-GTTTACTGTTGGAAAAACAACACTCTTATTATC-3′ | |
| R: 3′-ATCACCTACAACACCACATTTAATAACCTTTG-5′ | |||
| Other plasmids for expression in Dictyostelium cells | |||
| PHcrac-GFP | Peter Devreotes Laboratory, Johns Hopkins University | ||
| LimEΔcoil-GFP | Peter Devreotes Laboratory, Johns Hopkins University | ||
| LimEΔcoil-RFP | Douglas Robinson Laboratory, Johns Hopkins University | ||
To generate constructs expressing GFP- or RFP-fusion proteins, DNA fragments were PCR-amplified from genomic DNA or cDNA and cloned into pDM or pCV5 vectors (Miao et al., 2017; Veltman et al., 2009). To generate GxcM4A, the residues at position 593–596 were mutated to alanines using primers listed in Table 1. For inducible expression of constitutively active (CA) RacC, DNA fragment encoding RacCCA or RFP- RacCCA was cloned into pDM371 or pDM359, respectively.
To generate constructs expressing GST-fusion proteins, cDNA fragment encoding Fbp17SH3 or the VCA domain of WASP was cloned into pGEX-6P-1 vector; cDNA fragment encoding full-length Fbp17 or WASP was cloned into pFastBac1-GST vector using NdeI and XbaI sites. For expression of His-MBP-Fbp17SH3, cDNA fragment encoding the SH3 domain of Fbp17 was cloned into pET-MBP-3C vector using BamHI and XhoI sites.
For yeast two-hybrid assay, cDNA fragment encoding Fbp17 or the GBD domain of WASP (aa 126–230) was PCR-amplified and cloned into pGADT7 prey vector. Constitutively active forms of Dictyostelium Rac GTPases (except for RacQ) were PCR-amplified and cloned into a pGBKT7 bait vector. The CA form mutations were introduced using primers listed in Table 1.
Imaging
To image the localization of fluorescent proteins, cells were plated in coverslip chambers (Lab-Tek; NalgenNunc) filled with HL5 or LoFlo medium (Formedium) and allowed to adhere. Images were acquired on a Zeiss 880 or Zeiss 980 inverted microscope equipped with a 63×/1.4 oil-immersion objective. Chemoattractant stimulations were performed as described previously (Li et al., 2022).
For phalloidin staining, cells were plated in a 8-well coverslip chamber for 2 h in HL5. Cells were fixed for 8 min at room temperature with 2% paraformaldehyde and 0.08% glutaraldehyde in KK2, permeabilized for 8 min with the addition of 0.2% TX-100, quenched in PBS containing 20 mM glycine, and washed with KK2. Cells were then stained with 14 μM Acti-stain 555 phalloidin (cat#PHDH1-A; Cytoskeleton) at room temperature in the dark for 30 min. Images were captured on a Zeiss 880 confocal microscope.
For DAPI staining, cells cultured in suspension for 60 h were gently vortexed, plated on coverslips, and allowed to adhere for 15 min. Cells were fixed with 2 ml ice-cold methanol for 5 min, washed with PBS, and then stained with DAPI containing mounting media (cat#ab104139-20; Abcam). For the experiment presented in Fig. S2 C, cells were grown on 10-cm cell culture plates and collected for DAPI staining. Images were acquired on a Zeiss 880 confocal microscope.
To visualize clathrin-mediated endocytosis, cells expressing GFP-CLC and RFP-ArpC4 were slightly compressed under 0.5% SeaKem GTG agarose and imaged on an in-house Multi-modal SIM system equipped with a Nikon CFI SR HP Apo TIRF 100×/1.49 oil-immersion objective using the TIRF mode. Images were recorded every 2 or 4 s for 8 min.
Image analysis
Most image analyses were performed using Fiji ImageJ 1.53t (National Institutes of Health). For experiments presented in Figs. 5 J, 9 K, S3, G and H; and S4 G, the cortical-to-cytosol fluorescent intensity ratio was determined by dividing the total fluorescent intensity in the rear cortical region by that in the cytosol as described previously (Nguyen et al., 2014). For experiments presented in Fig. 5 L and Fig. 9 M, the total phalloidin intensity in the rear cortical region was measured.
To illustrate protein distribution asymmetry, an angle-series plot was introduced. Image analysis was performed using MATLAB 2021b (MathWorks) and Fiji ImageJ. The fluorescent intensity in the cortical region of a cell was first extracted with custom codes written in MATLAB. During the preprocessing steps, background movement and median filtering were applied to the images. The cell regions were segmented from the background using binarization, open operations, and close operations. The cortex was defined as an area ∼5 to 10 pixels from the cell boundary. The fluorescent intensity data were then plotted along the cell perimeter, with 0° corresponding to the migrating front. The reference point was the centroid of the recognized cell region, and the reference line was the ray pointing in the y direction (0°) starting from the reference point. The angle between the line from a pixel point in the cortex to the reference point and the reference line is the angle coordinate of the pixel point. The values of the angle coordinates are between −180° and +180° (the counterclockwise direction). Finally, the angle coordinates were divided into 72 intervals, and the average fluorescence intensity of pixels in each interval was calculated. Means and standard deviations were calculated from multiple cells. The results were plotted using Origin 2022b (OriginLab). Codes to analyze the data and perform numerical calculations are available in the supplemental material.
Migration assay
For random motility assay, vegetative cells were seeded in a 6-well cell culture plate in HL5 and allowed to adhere for 4 h, except for the experiment presented in Fig. 9 C where cells were allowed to adhere overnight. Before imaging, the medium was replaced with fresh HL5. Images were acquired at 30-s intervals with phase illumination on a Zeiss 880 inverted microscope equipped with a 10×/0.45 objective. To measure the effect of GxcM overexpression on random migration, cells expressing GxcM-GFP or GFP were seeded in a 8-well coverslip chamber and allowed to adhere for 60 min. Time-lapse images were collected at 30-s intervals with a Zeiss 880 microscopy equipped with a 40×/0.95 oil-immersion objective.
Under-agarose folic acid chemotaxis assay was performed as described before (Woznica and Knecht, 2006; Yang et al., 2021). Briefly, 5 ml 0.5% SeaKem GTG agarose melted in LoFlo medium was poured into a 50 mm glass-bottom dish (MatTek Corp). After the setting of agarose, two troughs were cut; one was filled with 1 mM folic acid and the other with vegetative cells resuspended in LoFlo. Cells were allowed to migrate for 5–9 h. For experiments presented in Figs. 5 F, 9 F, S1 H, and S3 H, 5 ml 2% agarose was used. Images were acquired at 20-s intervals with a 10×/0.45 phase objective or 3-s intervals with a 63×/1.4 oil-immersion objective on a Zeiss 880 microscope. In Video 5, cells expressing GxcM-GFP and LimEΔcoil-RPF were exposed to the folic acid gradient under 0.5% agarose and imaged on an in-house Multi-modal SIM system using the TIRF mode. Images were recorded every 3 s.
To quantify migration parameters, including accumulated distance, Euclidean distance, velocity, directness, and forward migration index, cells were tracked using the manual tracking plugin of Fiji ImageJ (https://fiji.sc/) and analyzed using Ibidi chemotaxis tool software.
Micropipette aspiration assay
Micropipette aspiration assay was carried out as described previously with minor modifications (Litschko et al., 2019). Briefly, a hand-made chamber was constructed using two pieces of hydrophobic glass with a space height ∼3 mm. The chamber was filled with PBS buffer and mounted on the stage of a Nikon Eclipse Ti inverted microscope equipped with a 40×/0.75 air objective. 10 μl of cell suspension was injected into the chamber. A bovine serum albumin-coated glass micropipette with an inner diameter of 4.35 ± 0.65 μm was filled with water and positioned into the measurement chamber using a micromanipulator. Aspiration pressure was applied with a height-adjustable water reservoir. The reference pressure (0 Pa) was calibrated by observing the motion of a non-adherent cell in the micropipette. After setting the pressure difference to 500 Pa, cells were aspirated for 5 min and a snapshot was captured at the end. Aspiration length (Lp) was determined by Fiji ImageJ.
Protein purification
Escherichia coli BL21 cells transformed with GST-Fbp17SH3 or GST-WASPVCA were grown until the absorbance at 600 nm of 0.8 and induced with 0.4 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) for 16–18 h at 20°C. The bacterial pellet was resuspended in suspension buffer (50 mM Tris, pH 8.0, 500 mM NaCl, 1 mM EDTA, 1 mM DTT) and lysed by sonication. The cell suspension was centrifuged at 15,000 g for 30 min to pellet the debris. The supernatant was incubated with prewashed glutathione sepharose beads (GE Healthcare) for 1–2 h at 4°C. Beads were washed three times with suspension buffer and once with washing buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 1 mM EDTA, 1 mM DTT). GST-fusion proteins were eluted with elution buffer (50 mM Tris, pH 8.0, 10 mM reduced glutathione) followed by buffer exchange into G buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 1 mM EDTA) using a desalting column (cat#17085101; GE Healthcare). To remove the GST tag, 10 μl PreScission Protease was added into the protein-bead mixture and incubated at 4°C overnight with gentle rotation. Proteins with the GST tag cleaved were collected from the eluate.
GST-Fbp17 and GST-WASP were purified from Sf9 insect cells. Baculovirus packaging was performed according to the manufacturer’s instructions and previous description (Feng et al., 2020). Briefly, pFastBac1-GST-Fbp17 or pFastBac1-GST-WASP were transformed into DH10Bac cells to get recombinant Bacmid, which was then transformed into Sf9 cells using Cellfectin II reagent (cat#10362100; Invitrogen) to produce virus stock. The virus was then amplified by infecting Sf9 cells. After three rounds of infection, GST recombinant proteins were purified following the procedures described above.
His-MBP-Fbp17SH3 was precipitated by amylose beads (cat#E8021L; NEB). After purification, beads were preserved in buffer (25 mM Tris, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, 50% Glycerol) at −20°C.
In vitro actin polymerization assay
Actin nucleation assay was performed as described previously with minor modifications (Diao et al., 2018) Briefly, 3 μM actin (10% pyrene labeled), 20 nM bovine brain Arp2/3 complex (cat#RP01P; Cytoskeleton), and 20 nM GST-WASP were mixed with purified Fbp17SH3, GST- FBP17SH3, or GST-Fbp17 in G buffer and incubated at room temperature. The reaction volume was 135 μl. To initiate actin polymerization, 15 μl 10 × KMEI buffer (500 mM KCl, 10 mM MgCl2, 10 mM EGTA, and 100 mM imidazole-HCl, pH 7.4) was added. Actin assembly was traced by monitoring pyrene fluorescence by a Quanta Master Luminescence QM 3 PH fluorometer (Photo Technology International) with the excitation and emission wavelength set at 365 and 407 nm, respectively. 20 nM GST-WASPVCA was included instead of Fbp17 proteins in one of the reactions as a positive control.
Immunoprecipitation experiments
To identify proteins that interact specifically with GxcM, an immunoprecipitation experiment and mass spectrometry analysis were carried out as described previously with minor modifications (Tu et al., 2022). Briefly, cells expressing GxcM-GFP, GxcMN790-GFP, or Teep1-GFP were starved in DB for 3 h, lysed in ice-cold lysis buffer (10 mM Hepes, pH 7.2, 100 mM NaCl, 0.5% Nonidet P-40, 10% glycerol, 1 mM NaF, 0.5 mM Na3VO4, complete EDTA-free protease inhibitor [Roche], 1 mM DTT) with the addition of 15 mM EDTA, and incubated for 5 min on ice. Lysates were centrifuged at 22,000 × g for 5 min at 4°C. The supernatants were incubated with anti-GFP affinity beads (cat#SA070005; Smart Lifesciences) for 2 h at 4°C. Beads-bound proteins were eluted with SDS loading buffer and subjected to SDS-PAGE. Protein bands were visualized by CBB staining and subjected to in-gel trypsin digestion and mass spectrometry analysis.
For most coimmunoprecipitation experiments, cells expressing GFP- or RFP-fusion proteins were starved in DB for 3 h, lysed in ice-cold lysis buffer, and incubated on ice for 5 min. Lysates were centrifuged at 22,000 × g for 5 min at 4°C. The supernatants were incubated with anti-GFP affinity beads or anti-RFP affinity beads (cat#SA072005; Smart Lifesciences) for 1 h at 4°C. Beads were washed four times in lysis buffer without a protease inhibitor. Samples were eluted with an SDS loading buffer and subjected to SDS-PAGE. For the experiment presented in Fig. 3 E, cells expressing GxcM-GFP and cells expressing RFP-tagged full-length or truncations of Fbp17 were lysed separately in a lysis buffer. Equal volume of lysates containing GxcM or Fbp17 were mixed and then incubated with anti-RFP affinity beads for 1 h at 4°C. Beads were washed with lysis buffer and processed for SDS-PAGE.
To verify the interaction between GxcM and RacC, cells expressing GxcM-RFP and GFP, GFP-Rac1A, or GFP-RacC were lysed in ice-cold Triton lysis buffer (10 mM Hepes, pH 7.2, 100 mM NaCl, 0.1% Triton X-100, 10% glycerol, 1 mM NaF, 0.5 mM Na3VO4, complete EDTA-free protease inhibitor, 1 mM DTT) with or without 15 mM EDTA. Cleared lysates were incubated with anti-RFP affinity beads for 45 min at 4°C. Beads were washed four times with lysis buffer and processed for SDS-PAGE.
Western blotting was performed as described before (Cai et al., 2010). Anti-GFP antibody (1:5,000; WB) was purchased from Roche (cat#11814460001). Anti-DsRed antibody (1:1,000; WB), which was used to detect RFP-fusion proteins, was purchased from Takara (cat#632496).
Pull-down assays
For experiments presented in Fig. 3, F–H and Fig. 6, C and D, cells expressing GFP-fusion proteins were starved, lysed at 5 × 107 cells/ml in lysis buffer, and incubated for 5 min on ice. Lysates were centrifuged at 22,000 × g for 5 min. Supernatants were incubated with beads containing 30 μg purified GST- or MBP-Fbp17SH3 for 1 h at 4°C with gentle agitation. After incubation, beads were washed four times with lysis buffer. Samples were eluted with an SDS loading buffer and subjected to SDS-PAGE.
Yeast two hybrid (Y2H)
To screen Rac GTPases that interact with Fbp17 or WASP, yeast two-hybrid analyses were performed using the Matchmaker GAL4 Two-Hybrid System 3 (Clontech Laboratories). S. cerevisiae strain AH109 was cotransfected with both bait and prey plasmids and grown on double-dropout (DD, lacking leucine and tryptophan) agar plates following the manufacturer’s instructions. Clones were collected, resuspended in 100 μl H2O, and spotted on quadruple-dropout (QD, deficiency in leucine, tryptophan, histidine, and adenine) agar plates. The interactions between tested proteins were analyzed according to the yeast growth on QD agar plates.
Statistics analyses
Statistical analysis was performed using GraphPad Prism. SuperPlots were generated according to Lord et al. (2020). In Fig. S5 F, statistical significance was determined by one-way analysis of variance with Tukey post-test. In all the other figures, statistical significance was determined by a two-tailed unpaired t test. In all figures, *** indicates P < 0.001, **P < 0.01, *P < 0.05, and ns not significant.
Online supplemental material
Fig. S1 shows analyses of the localization of GxcM truncations and characterization of gxcM knockout cells. Fig. S2 shows the characterization of GxcM overexpressing cells. Fig. S3 shows analyses of the sequence and localization of Fbp17 and characterization of fbp17 knockout cells. Fig. S4 shows RacC Y2H and characterization of racC knockout cells and cells overexpressing activated RacC. Fig. S5 shows the characterization of wasA knockout cells and analyses of clathrin-mediated endocytosis. Video 1 shows the localization of GxcM-GFP in WT cells chemotaxing under agarose. Video 2 shows the localization of PHcrac-GFP and RFP-tagged GxcM, GxcMN790, GxcM4A, or Teep1 in WT cells during random migration. Video 3 shows the localization of LimEΔcoil-RFP and GFP-ArpC4 in WT cells expressing GxcM-GFP, GxcM-RFP, or a control fluorescent protein during random migration. Video 4 shows the localization of LimEΔcoil-RFP and GFP-ArpC4 in WT cells expressing GxcM-GFP, GxcM-RFP, or a control fluorescent protein chemotaxing under agarose. Video 5 shows TIRF microscopy imaging of the localization of GxcM-GFP and LimEΔcoil-RFP in WT cells chemotaxing under agarose. Video 6 shows localization of GxcM-GFP/LimEΔcoil-RFP or GxcM-RFP/GFP-ArpC4 in fbp17− cells chemotaxing under agarose. Video 7 shows localization of GxcM-GFP/LimEΔcoil-RFP or GxcM-GFP/RFP-ArpC4 in racC− cells chemotaxing under agarose. Video 8 shows the localization of GFP-RacCG15V with RFP-ArpC4 or LimEΔcoil-RFP in WT cells during random migration. Table S1 shows the proteomic identification of Fbp17 as a binding partner of GxcM. The contour recognition file contains custom codes written for angle-series plot analysis.
Acknowledgments
The authors thank Drs. Hong Zhang (Institute of Biophysics, CAS, Beijing, China), Junjie Hu (Institute of Biophysics, CAS, Beijing, China), Peter Devreotes (Johns Hopkins University, Baltimore, USA), and Robert Kay (MRC Laboratory of Molecular Biology, London, UK) for reagents and cells; Drs. Tianzhi Luo (University of Science and Technology of China, Hefei, China), Yanruo Zhang, and Jianhui Xiao (Institute of Biophysics, CAS, Beijing, China) for helping with the MPA assay and helpful discussions; the proteomics core facilities in Peking University and Tsinghua University for MS analysis; and the Center for Biological Imaging at the Institute of Biophysics for assistance with imaging data collection and analysis.
This work was supported by grants from the Ministry of Science and Technology of China (2021YFA1300301 to H. Cai), the Strategic Priority Research Program of CAS (XDB37020304 to H. Cai), and the National Natural Science Foundation of China (92254303 and 32170701 to H. Cai, 31872828 and 32270743 to Y. Yang, and 11921002 to B. Li).
Author contributions: Conceptualization, H. Cai and D. Li; Methodology, D. Li, Y. Yang, C. Lv, H. Cai, B. Li, P. Gao, and S. Huang; Investigation, D. Li, Y. Yang, C. Li, H. Cai, Y. Wang, X. Chao, J. Huang, S.P. Singh, Y. Yuan, and C. Zhang; Resources, J. Lou; Writing, H. Cai, D. Li, and C. Lv; Funding acquisition, H. Cai, Y. Yang, and B. Li; Supervision, H. Cai.
References
Author notes
D. Li and Y. Yang contributed equally to this paper.
Disclosures: The authors declare no competing interests exist.