Pachytene piRNA biogenesis is a hallmark of the germline, distinct from another wave of pre-pachytene piRNA biogenesis with regard to the lack of a secondary amplification process known as the Ping-pong cycle. However, the underlying molecular mechanism and the venue for the suppression of the Ping-pong cycle remain elusive. Here, we showed that a testis-specific protein, ADAD2, interacts with a TDRD family member protein RNF17 and is associated with P-bodies. Importantly, ADAD2 directs RNF17 to repress Ping-pong activity in pachytene piRNA biogenesis. The P-body localization of RNF17 requires the intrinsically disordered domain of ADAD2. Deletion of Adad2 or Rnf17 causes the mislocalization of each other and subsequent Ping-pong activity derepression, secondary piRNAs overproduced, and disruption of P-body integrity at the meiotic stage, thereby leading to spermatogenesis arrested at the round spermatid stage. Collectively, by identifying the ADAD2-dependent mechanism, our study reveals a novel function of P-bodies in suppressing Ping-pong activity in pachytene piRNA biogenesis.
Introduction
In the mammalian germline, it is crucial to restrict transposable elements (TEs) activity to maintain genome integrity. To prevent the deleterious effects of TEs, several mechanisms were evolved to control the activity of TEs in the germline; one of such is the PIWI-interacting RNA (piRNA) pathway, a small RNA-mediated regulatory mechanism (Ernst et al., 2017; Malone and Hannon, 2009). In the piRNA pathway, TE transcripts were forced to fuel the Ping-pong cycle, a powerful positive-feedback amplification process during piRNA biogenesis. Disruption of the Ping-pong cycle in prospermatogonia (also known as gonocytes) leads to the derepression of TEs and male infertility (Shoji et al., 2009). In contrast, ectopic activation of the Ping-pong cycle in meiotic cells is deleterious to the proper expression of protein-coding transcripts (Wasik et al., 2015). Thus, the Ping-pong cycle activity is tightly controlled during spermatogenesis; however, the underlying mechanism and the venue of this control are not well understood.
piRNAs can be categorized into two classes based on the expression periods: pre-pachytene piRNAs and pachytene piRNAs. Pre-pachytene piRNAs are derived from cleavage products of piRNA cluster transcripts termed primary piRNAs, and primary piRNAs generate Ping-pong cycle products termed secondary piRNAs (Aravin et al., 2008; Aravin et al., 2007; Siomi et al., 2011). Pachytene piRNAs are expressed during meiotic prophase, and most pachytene piRNAs are generated from 100 TE-poor genomic loci termed pachytene piRNA clusters (Li et al., 2013; Robine et al., 2009). Pachytene piRNAs are thought not to silence TEs because the TEs located in pachytene piRNA loci are ancient and active copies of these TEs are absent from the mouse genome (Ozata et al., 2019; Reuter et al., 2011; Zheng and Wang, 2012). In addition, recent works demonstrated that the disruption of pachytene piRNA loci (pi6 and/or pi18) did not derepress TE activity in mice (Choi et al., 2021; Wu et al., 2020). Consistent with these observations, Ping-pong cycle is repressed during pachytene piRNA formation (Beyret et al., 2012). In Drosophila, the Tudor-domain protein Qin is required to control the Ping-pong cycle (Zhang et al., 2014; Zhang et al., 2011). The homolog of Qin in mice, RNF17 (also called TDRD4), plays a major role in suppressing the Ping-pong cycles, and the knockout of RNF17 unleashes the Ping-pong cycles and targets protein-coding transcripts improperly, leading to spermatogenesis arrest (Wasik et al., 2015). RNF17 was reported to localize in a novel undefined RNP granule, called RNF17 granules, which is distinguishable from known germ granules or Nuages (Pan et al., 2005), but its interacting partners are unknown. To date, it remains largely unknown how and where RNF17 granules are regulated to suppress the Ping-pong cycles in pachytene piRNA biogenesis.
Germ granules or Nuage are unique electron-dense non-membranous granular ribonucleoprotein (RNP) granules involved with piRNA pathways in the cytoplasm of germ cells, including inter-mitochondrial cement (IMC) and chromatoid bodies (CBs; Aravin et al., 2009; Wang et al., 2020). In addition to these germ granules, germ cells contain processing bodies (P-bodies), the main cytoplasmic RNP granules implicated in post-translational regulation in somatic cells (Voronina et al., 2011). In mouse prospermatogonia, the P-bodies involve the secondary piRNA biogenesis by assisting IMC (also called pi-body) to fuel the Ping-pong cycle, termed piP-body (Aravin et al., 2009). MILI and Nuage core protein TDRD1 are recruited to IMC, while MIWI2 and its partners, such as TDRD9, are recruited to piP-body to modulate the Ping-pong cycle (Ding et al., 2019; Yabuta et al., 2011). CBs are present in late pachytene spermatocytes to early-round spermatids and are enriched with piRNA pathway proteins, including PIWI proteins (MIWI and MILI) and most TDRD family members (Chuma et al., 2006; Hosokawa et al., 2007). Although P-bodies and germ granules are different RNP complexes and share some components, the interplay mechanism between P-bodies and germ granules remains unknown (Gallo et al., 2008; Kotaja et al., 2006; Shoji et al., 2009; Tanaka et al., 2011). A recent study on Bombyx mori (silkworm) indicated that improper exchange of piRNA factors between P-bodies and germ granules leads to massive production of mRNA-derived piRNAs (Chung et al., 2021). However, components of P-bodies and their functions in pachytene piRNA biogenesis remain elusive.
Here, we showed that RNF17 granules are closely associated with P-bodies in mouse spermatocytes, and that spermatogenesis-specific proteins ADAD1 and ADAD2 (ADADs) are novel interacting proteins of RNF17. ADADs are both essential for spermatogenesis and contain a double-stranded RNA binding domain and an adenosine deaminase (AD) domain that has an A-to-I RNA editing activity (Connolly et al., 2005; Snyder et al., 2020). However, neither Adad1 nor Adad2 knockout impairs RNA editing in mouse testes (Snyder et al., 2020). Although ADAD2 interacts with PIWI proteins (MILI and MIWI) and a piRNA pathway factor MAEL (Castañeda et al., 2014; Vagin et al., 2009), ADAD2 does not localize on CBs but forms unique RNA granules and regulates heterochromatin in meiotic male germ cells (Chukrallah et al., 2022; Snyder et al., 2020). In this study, we also found that ADAD2 localizes on P-bodies that overlap with RNF17 granules and the P-body localization of ADAD2 and RNF17 are interdependent. In addition, an Adad2 mutation in mice causes ectopic Ping-pong activation akin to the Rnf17 mutant mice. Further, the loss of ADAD2 or RNF17 disrupts the integrity of P-bodies at the end of the pachytene stage, leading to spermatogenesis arrest at the round spermatid stage. Together, our study demonstrates that both ADAD2 and RNF17 are functional partners in P-bodies, and ADAD2 and RNF17 act in concert to repress the Ping-pong cycle to ensure pachytene piRNA biogenesis.
Results
RNF17 granules are tightly associated with P-bodies in spermatocytes
The Tudor protein Qin/Kumo is the homolog of RNF17 in Drosophila and cooperates with Ago3 and Aub in Nuage (Anand and Kai, 2012; Zhang et al., 2011). However, in mice, RNF17 was reported to localize on a novel undefined RNP granules distinguishable from IMC or CBs, and RNF17 localization has not been fully characterized (Hosokawa et al., 2007; Pan et al., 2005). We thus first characterized the localization of RNF17-granules in detail. Consistent with the previous work, the RNF17-granules initially appeared in early spermatocytes at stage II and formed large prominent dots with the development of the spermatocytes (Fig. S1, A and B). Because BmQIN, the homolog of RNF17 in B. mori, localizes on P-bodies (Chung et al., 2021), we examined whether RNF17 localizes on P-bodies in mammal germ cells. Co-immunofluorescence of RNF17 and EDC3, a P-body marker in mice (Eulalio et al., 2007), showed that RNF17 foci colocalized with EDC3 foci in pachytene spermatocytes from stage II-XI (Fig. 1 A). Co-immunofluorescence of RNF17 and DCP1α, another known component of the P-bodies (Tenekeci et al., 2016), further verified RNF17 localization on P-bodies (Fig. S1 C), suggesting close association of RNF17 granules with P-bodies and a potential role of RNF17 in P-bodies during meiosis. Because, in pro-spermatogonia, P-bodies are associated with germ granules and contain several piRNA factors such as MIWI2, TDRD9, and MAEL (Aravin et al., 2009; Wang et al., 2020), we performed co-immunofluorescence of RNF17 and MILI and examined the relationship between P-bodies and germ granules in meiotic cells. Interestingly, although the RNF17 foci were closely associated with MILI foci initially, these foci developed to form 1–2 distinct foci in the late pachytene/diplotene stage (Fig. 1 B). This result suggests dynamic and functional intercellular communication between P-bodies and germ granules in meiosis and a possible role of P-body proteins in the piRNA pathway.
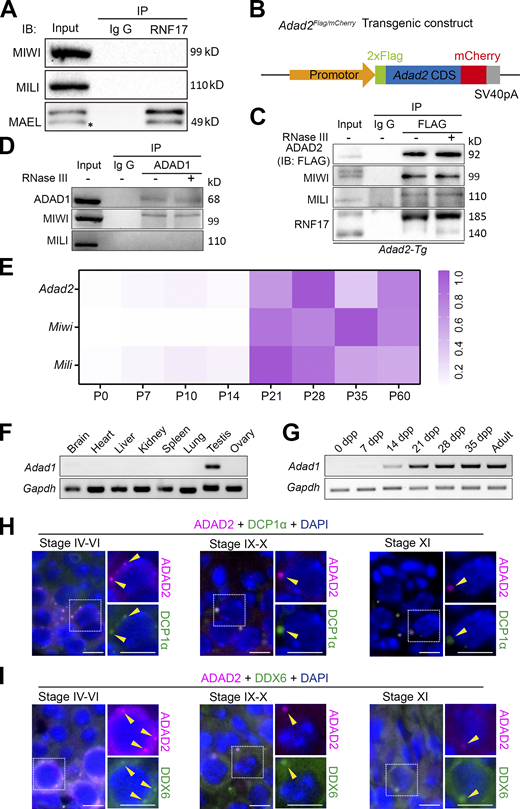
The expression pattern of RNF17 during spermatogenesis. (A) Double immunostaining with RNF17 (red) and GFP-PNA (green) on germ cells from adult WT testis sections are shown. DAPI (blue) marks the nuclei. Scale bars = 20 μm. (B) A schematic summary of the dynamic localizations of RNF17 in adult testis during spermatogenesis. The localization drawing based on the fluorescent signal analyses from three independent experiments. (C) Double immunostaining with DCP1α (red) and RNF17 (green) on spermatocytes from adult WT testis sections at different stages are shown. DAPI (blue) marks the nuclei. Scale bars = 10 μm. P-bodies are marked with arrowheads.
The expression pattern of RNF17 during spermatogenesis. (A) Double immunostaining with RNF17 (red) and GFP-PNA (green) on germ cells from adult WT testis sections are shown. DAPI (blue) marks the nuclei. Scale bars = 20 μm. (B) A schematic summary of the dynamic localizations of RNF17 in adult testis during spermatogenesis. The localization drawing based on the fluorescent signal analyses from three independent experiments. (C) Double immunostaining with DCP1α (red) and RNF17 (green) on spermatocytes from adult WT testis sections at different stages are shown. DAPI (blue) marks the nuclei. Scale bars = 10 μm. P-bodies are marked with arrowheads.
RNF17-granules are associated with P-bodies in spermatocytes. (A) Double immunostaining with RNF17 (green) and EDC3 (magenta) on wild-type (WT) spermatocytes from adult testis sections are shown. DAPI (blue) marks the nuclei. Scale bar = 10 μm. Line graphs showing the immunofluorescence intensity profile along the freely positioned line of RNF17 (green) and EDC3 (magenta). Top-right rectangle is enlarged area from the white box. Scale bar = 5 μm. (B) Double immunostaining with RNF17 (green) and MILI (magenta) on WT spermatocytes from adult testis sections are shown. DAPI (blue) marks the nuclei. Scale bar = 20 μm. Rectangle at the bottom is enlarged area from the white box. Scale bar = 10 μm. Line graphs showing the immunofluorescence intensity profile along the freely positioned line of RNF17 (green) and MILI (magenta). White arrowheads indicate closely associated P-bodies and germ granules, while yellow and purple arrowheads mark P-bodies and germ granules, respectively.
RNF17-granules are associated with P-bodies in spermatocytes. (A) Double immunostaining with RNF17 (green) and EDC3 (magenta) on wild-type (WT) spermatocytes from adult testis sections are shown. DAPI (blue) marks the nuclei. Scale bar = 10 μm. Line graphs showing the immunofluorescence intensity profile along the freely positioned line of RNF17 (green) and EDC3 (magenta). Top-right rectangle is enlarged area from the white box. Scale bar = 5 μm. (B) Double immunostaining with RNF17 (green) and MILI (magenta) on WT spermatocytes from adult testis sections are shown. DAPI (blue) marks the nuclei. Scale bar = 20 μm. Rectangle at the bottom is enlarged area from the white box. Scale bar = 10 μm. Line graphs showing the immunofluorescence intensity profile along the freely positioned line of RNF17 (green) and MILI (magenta). White arrowheads indicate closely associated P-bodies and germ granules, while yellow and purple arrowheads mark P-bodies and germ granules, respectively.
Adenosine deaminase domain-containing proteins are associated with RNF17 in testes
Because the P-body localization of RNF17 is not overlapped with MILI throughout the meiotic stage, we next sought to identify proteins that interact with RNF17 in mouse testes by immunoprecipitation in combination with a mass spectrometric (IP/MS) analysis. We totally identified 97 RNF17-associated proteins (Score > 50); however, only MAEL has been reported to be involved in piRNA biogenesis (Castañeda et al., 2014; Fig. 2 A and Table S1). Surprisingly, the PIWI family protein MIWI was not included in the candidates, which was reported to be associated with RNF17 in mouse testis (Vagin et al., 2009). Further, co-immunoprecipitation using RNF17 antibody in adult mouse testicular lysates confirmed that RNF17 could directly interact with MAEL but not MILI or MIWI (Fig. S2 A). A gene ontology (GO) analysis demonstrated that the proteins involved in RNA processing or gamete generation are enriched in RNF17-associated proteins (Fig. 2 B). Of those, Adenosine deaminase domain-containing proteins (ADAD1 and ADAD2) were highly enriched among RNF17-associated proteins (Fig. 2 A). Combining with the previous proteomic analysis of PIWI proteins (Chen et al., 2009; Vagin et al., 2009), we found that ADAD2 is associated with PIWI proteins (MIWI and MILI) and RNF17 (Fig. 2 C). To confirm the interaction between ADAD2 and PIWI proteins in vivo, we generated transgenic mice expressing Adad2 cDNA with a 2.3 kb sequence upstream of the transcription start site tagged with N-terminal 2× FLAG and C-terminal mCherry (Fig. S2 B). We confirmed specific co-immunoprecipitation of ADAD2 with MIWI and MILI, respectively (Fig. 2 D and Fig. S2 C), which suggests that ADAD2 may function in the piRNA pathway. The interactions between ADAD2, MIWI, and MILI were unaffected by treatment with RNase A or RNase III prior to immunoprecipitation, indicating that the association is direct and RNA-independent (Fig. 2 D and Fig. S2 C). We further verified the RNA-independent interactions between RNF17, ADAD1, and ADAD2 (Fig. 2 E and Fig. S2 C). Interestingly, we detected a weak interaction between ADAD1 and MIWI but not MILI (Fig. S2 D), suggesting a potential role of ADAD1 in the piRNA pathway. Together, these data raised the possibility that ADAD2 functions with RNF17 and PIWI proteins in testes.
Proteomic analysis of endogenous RNF17 in testes. (A) Volcano plot showing enrichment (log2 [mean intensity ratio of RNF17-IP/IgG-IP from WT testes]) and confidence (−log10 [P value of two-sided Student’s t test]) of proteins co-purifying with RNF17 from WT testes lysates (n = 3). The right upper corner of the dotted line area indicates factors with enrichment > fourfold and significance P < 0.05. (B) GO analysis of the most enriched RNF17-associated proteins identified in adult mouse testes. (C) RNF17, MIWI, and MILI protein interaction networks reveal that ADAD2 presents in RNF17, MIWI, and MILI complexes. (D) Co-immunoprecipitation of FLAG-ADAD2-mCherry with MIWI and MILI from lysates generated from WT and Adad2Flag/mCherry mouse testes. Immunoprecipitations were performed in the presence or absence of RNase A. (E) Western blot validation of RNF17 association with ADAD proteins. Source data are available for this figure: SourceData F2.
Proteomic analysis of endogenous RNF17 in testes. (A) Volcano plot showing enrichment (log2 [mean intensity ratio of RNF17-IP/IgG-IP from WT testes]) and confidence (−log10 [P value of two-sided Student’s t test]) of proteins co-purifying with RNF17 from WT testes lysates (n = 3). The right upper corner of the dotted line area indicates factors with enrichment > fourfold and significance P < 0.05. (B) GO analysis of the most enriched RNF17-associated proteins identified in adult mouse testes. (C) RNF17, MIWI, and MILI protein interaction networks reveal that ADAD2 presents in RNF17, MIWI, and MILI complexes. (D) Co-immunoprecipitation of FLAG-ADAD2-mCherry with MIWI and MILI from lysates generated from WT and Adad2Flag/mCherry mouse testes. Immunoprecipitations were performed in the presence or absence of RNase A. (E) Western blot validation of RNF17 association with ADAD proteins. Source data are available for this figure: SourceData F2.
ADAD2 is associated with P-body in male germ cells. (A) Co-immunoprecipitation of RNF17 with MIWI, MILI, and MAEL from lysates isolated from WT testes. Asterisk marks the nonspecific bands. (B) Schematic representation of transgenic cassette expressing FLAG-ADAD2-mCherry cDNA driven by the Adad2 promoter. (C) Co-immunoprecipitation of FLAG with MIWI, MILI, and RNF17 from lysates isolated from Adad2-Tg testes. Immunoprecipitations were performed in the presence or absence of RNase III. (D) Co-immunoprecipitation of ADAD1 with MIWI and MILI from lysates isolated from wild-type testes. Immunoprecipitations were performed in the presence or absence of RNase III. (E) RT-qPCR derived heat maps depicting relative Adad2, Miwi, and Mili transcripts expression throughout testis development. (F) RT-PCR analysis of Adad1 transcript expression in multiple tissues from adult wild-type mice. (G) RT-PCR analysis of Adad1 transcript expression throughout testis development. (H and I) Double immunostaining with ADAD2 (FLAG, red) and DCP1α (green; H) or DDX6 (green; I) on spermatocytes from adult Adad2-Tg mouse testis sections are shown. DAPI (blue) marks the nuclei. Scale bars = 20 μm. P-bodies are marked with arrowheads. Source data are available for this figure: SourceData FS2.
ADAD2 is associated with P-body in male germ cells. (A) Co-immunoprecipitation of RNF17 with MIWI, MILI, and MAEL from lysates isolated from WT testes. Asterisk marks the nonspecific bands. (B) Schematic representation of transgenic cassette expressing FLAG-ADAD2-mCherry cDNA driven by the Adad2 promoter. (C) Co-immunoprecipitation of FLAG with MIWI, MILI, and RNF17 from lysates isolated from Adad2-Tg testes. Immunoprecipitations were performed in the presence or absence of RNase III. (D) Co-immunoprecipitation of ADAD1 with MIWI and MILI from lysates isolated from wild-type testes. Immunoprecipitations were performed in the presence or absence of RNase III. (E) RT-qPCR derived heat maps depicting relative Adad2, Miwi, and Mili transcripts expression throughout testis development. (F) RT-PCR analysis of Adad1 transcript expression in multiple tissues from adult wild-type mice. (G) RT-PCR analysis of Adad1 transcript expression throughout testis development. (H and I) Double immunostaining with ADAD2 (FLAG, red) and DCP1α (green; H) or DDX6 (green; I) on spermatocytes from adult Adad2-Tg mouse testis sections are shown. DAPI (blue) marks the nuclei. Scale bars = 20 μm. P-bodies are marked with arrowheads. Source data are available for this figure: SourceData FS2.
ADAD2 is a testis-specific protein and closely associated with P-bodies in spermatocytes
To understand the physiological function of ADAD2 in testes, we first characterized the tissue expression pattern of ADAD2 in various organs in adult Adad2-transgenic mice. Consistent with a previous study showing a testis-specific expression of ADAD2 (Snyder et al., 2020), ADAD2FLAG/mCherry protein was expressed exclusively in the testes (Fig. 3 A). RT-qPCR analyses revealed that Adad2 expression was dramatically increased after postnatal day 14 (P14) similar to Miwi and Mili (Fig. S2 E). In addition, the expression patterns of Adad1 mRNA in various organs and in developmental testis were similar to those of Adad2 (Fig. S2, F and G). Further RT-qPCR analyses in purified testicular cell populations revealed that Adad2 was predominantly expressed in pachytene spermatocytes similar to Miwi and Mili (Fig. 3 B).
The expression pattern of ADAD2 during spermatogenesis. (A) Western blot analysis of FLAG-ADAD2-mCherry expression in multiple tissues from adult transgenic mice. (B) Relative expression of Adad2, Miwi, and Mili transcripts as measured by RT-qPCR in purified testicular cells. SC, Sertoli cell; Spg, spermatogonia; Spc, spermatocyte; rST, round spermatid; eST, elongating spermatid. Data are presented as mean ± SEM and normalized to the peak expression of each transcript. (C) Double immunostaining with FLAG (magenta) and GFP-PNA (green) on testis sections from adult Adad2 transgenic (Adad2-Tg) mice are shown. DAPI (blue) marks the nuclei. Scale bars = 20 μm. (D) A schematic summary of the dynamic localization of FLAG-ADAD2-mCherry in adult testis during spermatogenesis. The localization drawing is based on the fluorescent signal analyses from three independent experiments. (E) Double immunostaining with ADAD2 (FLAG, magenta) and EDC3 (green) on spermatocytes from adult Adad2-Tg testis sections are shown. DAPI (blue) marks the nuclei. Scale bar = 10 μm. P-bodies are marked with yellow arrowheads. (F) Double immunostaining with ADAD2 (FLAG, magenta) and RNF17 (green) on spermatocytes from adult Adad2-Tg testis sections are shown. DAPI (blue) marks the nuclei. Scale bars = 10 μm. P-bodies are marked with yellow arrowheads. (G) Quantifying the colocalization ratio between ADAD2 and EDC3/RNF17 during spermatogenesis is shown. n = 12 tubules from three mice, error bars are mean ± SEM, unpaired two-tailed Student’s t test. Source data are available for this figure: SourceData F3.
The expression pattern of ADAD2 during spermatogenesis. (A) Western blot analysis of FLAG-ADAD2-mCherry expression in multiple tissues from adult transgenic mice. (B) Relative expression of Adad2, Miwi, and Mili transcripts as measured by RT-qPCR in purified testicular cells. SC, Sertoli cell; Spg, spermatogonia; Spc, spermatocyte; rST, round spermatid; eST, elongating spermatid. Data are presented as mean ± SEM and normalized to the peak expression of each transcript. (C) Double immunostaining with FLAG (magenta) and GFP-PNA (green) on testis sections from adult Adad2 transgenic (Adad2-Tg) mice are shown. DAPI (blue) marks the nuclei. Scale bars = 20 μm. (D) A schematic summary of the dynamic localization of FLAG-ADAD2-mCherry in adult testis during spermatogenesis. The localization drawing is based on the fluorescent signal analyses from three independent experiments. (E) Double immunostaining with ADAD2 (FLAG, magenta) and EDC3 (green) on spermatocytes from adult Adad2-Tg testis sections are shown. DAPI (blue) marks the nuclei. Scale bar = 10 μm. P-bodies are marked with yellow arrowheads. (F) Double immunostaining with ADAD2 (FLAG, magenta) and RNF17 (green) on spermatocytes from adult Adad2-Tg testis sections are shown. DAPI (blue) marks the nuclei. Scale bars = 10 μm. P-bodies are marked with yellow arrowheads. (G) Quantifying the colocalization ratio between ADAD2 and EDC3/RNF17 during spermatogenesis is shown. n = 12 tubules from three mice, error bars are mean ± SEM, unpaired two-tailed Student’s t test. Source data are available for this figure: SourceData F3.
Although the previous study showed that ADAD2 forms cytoplasmic granules in spermatocytes (Snyder et al., 2020), the detailed localization of ADAD2 during meiosis remains to be tested. We performed immunostainings using a FLAG antibody to detect ADAD2FLAG/mCherry protein in testicular sections of adult Adad2-transgenic mice and found that ADAD2FLAG/mCherry protein was mainly expressed in pachytene spermatocytes, but it was absent before early pachytene spermatocytes (stage II), increasingly expressed in mid-pachytene spermatocytes (stages IV–VI), and gradually decreased in late-pachytene spermatocytes (stages VII–X; Fig. 3, C and D). Notably, the subcellular localization of ADAD2 changed dynamically in the pachytene substages, diffused in the cytoplasm of early pachytene spermatocytes, formed several small perinuclear granules, and coalesced into large prominent granules in late pachytene spermatocytes and in diplotene spermatocytes (Fig. 3, C and D). This result raised the possibility that ADAD2 is a novel component of P-bodies in spermatocytes. To test this possibility, ADAD2 was co-stained with a P-body marker EDC3 (Rzeczkowski et al., 2011); more than 50% of the ADAD2-granules were colocalized with EDC3-granules at the pachytene stage (overlapping from ∼55% in stage IV to ∼75% in stage XI) during spermatogenesis, indicating that ADAD2 is associated with P-bodies (Fig. 3, E–G). This result was confirmed by co-staining of ADAD2 with two additional markers of P-bodies, DCP1α and DDX6 (Ayache et al., 2015; Hubstenberger et al., 2017), respectively (Fig. S2, H and I). Consistent with these results, co-immunostaining of ADAD2FLAG/mCherry with RNF17 revealed that more than ∼50% of RNF17 foci overlapped with the ADAD2 foci in pachytene spermatocytes (Fig. 3, F and G), demonstrating that ADAD2 is closely associated with RNF17 in P-bodies.
The association with the P-body of ADAD2 and RNF17 is interdependent in vitro
Many components of P-body or stress granule possess intrinsically disordered regions (IDRs), which can promote condensate formation or liquid–liquid phase separation (Alberti et al., 2019; Fu and Zhuang, 2020). We thus used two algorithms (IUPred3 and VSL3) to analyze the amino acid sequences and secondary structures of ADAD proteins (Mészáros et al., 2018; Peng et al., 2006). We found that the N-terminal parts of ADAD1 and ADAD2 are largely disordered and defined these regions as IDRs (Fig. 4 A and Fig. S3, A and B). We then expressed Flag-tagged ADAD1 or ADAD2 in NIH3T3 cells and analyzed their localization by co-immunostaining with a P-body marker DCP1α. Interestingly, both ADAD1 and ADAD2 displayed a diffused distribution in the cytoplasm but did not colocalize with DCP1α (Fig. 4 B, upper and middle panels). These data indicated that ADAD2 is difficult to localize to P-bodies in NIH3T3 cells, unlike in spermatocytes. Similarly, RNF17 (MYC-RNF17) was incapable of colocalizing with DCP1α as well (Fig. 4 B, lower panel), although the distribution of RNF17 displayed a punctate pattern as previously reported (Pan et al., 2005). Strikingly, when RNF17 was co-expressed with full-length ADAD2 in NIH3T3 or U2OS cells, both RNF17 and ADAD2 concentrated to form granules that overlapped with DCP1α (Fig. 4, C and D and Fig. S3 C), suggesting that RNF17 granules coalesce with P-bodies when ADAD2 is co-expressed in NIH3T3 cells. However, unlike ADAD2, when RNF17 was co-expressed with ADAD1, both RNF17 and ADAD1 showed diffused distribution and could not colocalize with DCP1α (Fig. S3 D). These results suggested that P-body localization of RNF17 requires ADAD2 but not ADAD1 expression.
ADAD2 is essential for the recruitment of RNF17 to DCP1α-positive P-bodies in vitro. (A) Amino acid composition and in silico prediction of IDR in ADAD2 proteins. Residues above the dotted line are predicted to be disordered by IUPred3 and VSL3. (B) Immunofluorescence analysis of FLAG-ADAD1 (green), FLAG-ADAD2(green), and MYC-RNF17(green) with DCP1α (red) on NIH3T3 cells either transfected with Flag-tagged ADAD1 (upper panel), Flag-tagged ADAD2 (middle panel), or Myc-tagged RNF17 (lower panel). Scale bars = 20 μm. Top-right rectangle is enlarged area from the white box in lower panel. DAPI (blue) marks the nuclei. Scale bars = 10 μm. (C) Immunofluorescence analysis of FLAG-ADAD2 or ADAD2 mutants (red), MYC-RNF17 (green) with DCP1α (purple) on NIH3T3 cells co-transfected Myc-tagged RNF17 with Flag-tagged full-length ADAD2 or ADAD2 mutants with deleted fragments. DAPI (blue) marks the nuclei. Scale bar = 20 μm. Right rectangle is enlarged area from the white box. (D) Quantification of the colocalization ratio between ADAD2 (mutants) and RNF17, RNF17 and DCP1α, ADAD2 (mutants) and DCP1α in C, compared with ADAD2 (full length). Each dot represents value from one cell expressing the full-length ADAD2 (n = 16), ΔIDR (n = 16), ΔdsRBD (n = 16), Δ1-238 (n = 15), or Δ292-561 (n = 18) from three independent experiments are shown. Data are presented as mean ± SEM. *P <0.05, **P <0.01, ***P <0.001. Unpaired two-tailed Student’s t test. (E) Co-immunoprecipitation of MYC-RNF17 with FLAG-tagged truncated ADAD2 mutants. HEK293T cells were transfected with indicated plasmids. FLAG-tagged truncated ADAD2 mutants and MYC-RNF17 proteins were detected by western blotting using anti-FLAG and anti-MYC antibodies. (F) Overview of a series of truncated ADAD2 mutants used to map ADAD2 domain(s) responsible for RNF17 binding ability or P-body localization ability. The “+” symbol indicates ability retained. Source data are available for this figure: SourceData F4.
ADAD2 is essential for the recruitment of RNF17 to DCP1α-positive P-bodies in vitro. (A) Amino acid composition and in silico prediction of IDR in ADAD2 proteins. Residues above the dotted line are predicted to be disordered by IUPred3 and VSL3. (B) Immunofluorescence analysis of FLAG-ADAD1 (green), FLAG-ADAD2(green), and MYC-RNF17(green) with DCP1α (red) on NIH3T3 cells either transfected with Flag-tagged ADAD1 (upper panel), Flag-tagged ADAD2 (middle panel), or Myc-tagged RNF17 (lower panel). Scale bars = 20 μm. Top-right rectangle is enlarged area from the white box in lower panel. DAPI (blue) marks the nuclei. Scale bars = 10 μm. (C) Immunofluorescence analysis of FLAG-ADAD2 or ADAD2 mutants (red), MYC-RNF17 (green) with DCP1α (purple) on NIH3T3 cells co-transfected Myc-tagged RNF17 with Flag-tagged full-length ADAD2 or ADAD2 mutants with deleted fragments. DAPI (blue) marks the nuclei. Scale bar = 20 μm. Right rectangle is enlarged area from the white box. (D) Quantification of the colocalization ratio between ADAD2 (mutants) and RNF17, RNF17 and DCP1α, ADAD2 (mutants) and DCP1α in C, compared with ADAD2 (full length). Each dot represents value from one cell expressing the full-length ADAD2 (n = 16), ΔIDR (n = 16), ΔdsRBD (n = 16), Δ1-238 (n = 15), or Δ292-561 (n = 18) from three independent experiments are shown. Data are presented as mean ± SEM. *P <0.05, **P <0.01, ***P <0.001. Unpaired two-tailed Student’s t test. (E) Co-immunoprecipitation of MYC-RNF17 with FLAG-tagged truncated ADAD2 mutants. HEK293T cells were transfected with indicated plasmids. FLAG-tagged truncated ADAD2 mutants and MYC-RNF17 proteins were detected by western blotting using anti-FLAG and anti-MYC antibodies. (F) Overview of a series of truncated ADAD2 mutants used to map ADAD2 domain(s) responsible for RNF17 binding ability or P-body localization ability. The “+” symbol indicates ability retained. Source data are available for this figure: SourceData F4.
ADAD2 but not ADAD1 is required for the recruitment of RNF17 to P-bodies. (A) Homology alignment of ADAD2 protein. (B) Amino acid composition and in silico prediction of IDR in ADAD1 proteins. Residues above the dotted line are predicted to be disordered by IUPred3 and VSL3. (C) Immunofluorescence analysis of FLAG-ADAD2 or ADAD2 mutants(red), MYC-RNF17 (green) with DCP1α (purple) on U2OS cells co-transfected Myc-tagged RNF17 with Flag-tagged full-length ADAD2 or ADAD2 mutants with deleted fragments. DAPI (blue) marks the nuclei. Scale bars = 20 μm. Right rectangle is enlarged area from the white box. (D) Immunofluorescence analysis of DCP1α (red) with FLAG-ADAD1(green) or MYC-RNF17(green) on NIH3T3 cells co-transfected with Myc-tagged RNF17 and Flag-tagged ADAD1. DAPI (blue) marks the nuclei. Scale bars = 20 μm.
ADAD2 but not ADAD1 is required for the recruitment of RNF17 to P-bodies. (A) Homology alignment of ADAD2 protein. (B) Amino acid composition and in silico prediction of IDR in ADAD1 proteins. Residues above the dotted line are predicted to be disordered by IUPred3 and VSL3. (C) Immunofluorescence analysis of FLAG-ADAD2 or ADAD2 mutants(red), MYC-RNF17 (green) with DCP1α (purple) on U2OS cells co-transfected Myc-tagged RNF17 with Flag-tagged full-length ADAD2 or ADAD2 mutants with deleted fragments. DAPI (blue) marks the nuclei. Scale bars = 20 μm. Right rectangle is enlarged area from the white box. (D) Immunofluorescence analysis of DCP1α (red) with FLAG-ADAD1(green) or MYC-RNF17(green) on NIH3T3 cells co-transfected with Myc-tagged RNF17 and Flag-tagged ADAD1. DAPI (blue) marks the nuclei. Scale bars = 20 μm.
To determine the ADAD2 domain which is responsible for the colocalization with RNF17 and the recruitment of RNF17 to P-bodies, we co-expressed a series of FLAG-tagged truncated ADAD2 and MYC-RNF17 in heterologous expression systems using NIH3T3 or U2OS cells. The immunostaining analysis showed that the FLAG-ADAD2-∆IDRs still formed granules and colocalized with RNF17; however, the numbers of FLAG-ADAD2-∆IDRs foci and RNF17 foci that colocalized with DCP1α-foci were significantly decreased (Fig. 4, C and D and Fig. S3 C). This result indicates that the IDRs domain of ADAD2 is not required for the interaction between ADAD2 and RNF17 but is involved in the P-body localization. Of note, we found that the dsRNA binding domain (dsRBD) of ADAD2 is dispensable for the colocalization with RNF17 and the recruitment of RNF17 to P-bodies (Fig. 4, C and D and Fig. S3 C). A recent study revealed that a patient with spermatogenic arrest carried a homozygous mutation in ADAD2, resulting in the production of ADAD2 with the truncated adenosine deaminase (AD) domain (Krausz et al., 2020). Since the AD domain of ADAD2 is highly conserved in humans and mice, we then tested whether AD domain plays a role in the colocalization with RNF17 and the recruitment of RNF17 to P-bodies. The AD domain mutation (Δ292-561) disrupted the ADAD2 colocalization with RNF17 (Fig. 4, C and D and Fig. S3 C), suggesting that the AD domain is required for the interaction with RNF17. Further, IP experiments using truncated proteins of ADAD2 confirmed that the AD domain of ADAD2 is responsible for its interaction with RNF17 (Fig. 4 E). Together, these results demonstrate that the ADAD2 and RNF17 form a functional complex recruited to P-bodies and that the IDR and AD domains of ADAD2 are essential for the recruitment of RNF17 to P-bodies (Fig. 4 F).
ADAD2 is necessary for spermatogenesis but not for germ granule assembly in spermatocytes
To explore the functional roles of Adad2 in spermatogenesis, we utilized the CRISPR/Cas9 gene-editing technology to generate an Adad2 knockout mouse model. Two small guide RNAs (sgRNAs) were designed to target the exon 1 and generated two types of Adad2 mutations; one has a 47 bp deletion on the exon 1 that resulted in premature termination codon mutation (called Adad2Mut1 in Fig. S4 A), and the other has a 24 bp deletion on the exon 1 that resulted in eight amino acids deletion in the IDR domain of ADAD2 (called Adad2Mut2 in Fig. S4 A). Interestingly, Adad2Mut1/Mut1 and Adad2Mut2/Mut2 mice grew healthy without obvious defects compared with control mice (Adad2+/+), whereas both Adad2Mut1/Mut1 and Adad2Mut2/Mut2 males were sterile with a mild but statistically significant decrease in testis weights (Fig. S4, B and C). Histological analysis of testicular sections showed that the seminiferous tubules of Adad2+/+ mice contained various developmental stages of normal spermatogenic cells, including spermatogonia, spermatocytes, round spermatid, and elongated spermatids. In contrast, seminiferous tubules of Adad2Mut1/Mut1 and Adad2Mut2/Mut2 mice were arrested at the round spermatid stage, with the most advanced cells at the step-8 round spermatid stage, and no elongating spermatids were observed (Fig. S4 D). Epididymides from adult Adad2+/+ mice contained normal mature spermatozoa, whereas epididymides of Adad2Mut1/Mut1 and Adad2Mut2/Mut2 mice showed a complete absence of spermatozoa but were filled with degenerating germ cells (Fig. S4 E). This defective spermatogenesis phenotype of Adad2 mutants was consistent with the previous work (Snyder et al., 2020), similar to that in Rnf17, Tdrd7, Tdrd1, and Miwi mutants (Fig. S4 F). Further, periodic acid-Schiff (PAS) staining of Adad2Mut1/Mut1 testes showed that round spermatids failed to undergo cellular elongation at stage IX–XII and exhibited abnormal acrosome shapes (Fig. 5 A). Similarly, immunofluorescence analysis using an acrosome marker ACRV1 showed a cap-shaped acrosome in Adad2+/+ round spermatids, while Adad2Mut1/Mut1 round spermatids occasionally exhibited bi-nuclear, and proacrosomic vesicles failed to flatten (Fig. 5 B), indicating that the acrosome formation was defective in Adad2Mut1/Mut1 round spermatids. Moreover, we found that expression of the transgenic ADAD2 could rescue the defects in spermatogenesis and infertility observed in Adad2Mut1/Mut1 mice (Fig. 5 C), which confirmed the specific targeting of Adad2 in the mutants.
Adad2 is essential for spermatogenesis and male fertility. (A) Schematic representation of the Adad2 locus and design of the two sgRNAs targeting Adad2 exon 1, which harbors the IDR motif. Schematic representation and sequencing trace of the part of Adad2Mut exon 1 carrying the mutation site. The Mut1 site contains 47 bp removed on exon1 and causes a frameshift, while the Mut2 site contains 24 bp removed on exon1 and causes an 8aa lost in IDR motif. (B) The testes weights were reduced in both Adad2Mut1/Mut1 and Adad2Mut2/Mut2 mice at postnatal day 60 (P60). Data presented is the testes/body ratio (n = 6 for each genotype, **P < 0.01, Student’s t test). Error bars are mean ± SEM. (C) Fertility test for adult Adad2Mut1/Mut1and Adad2Mut2/Mut2 males. Each genotype shown was coupled with fertile wild-type mates. N = 3 for each genotype. ***P < 0.001(Student’s t test). (D) PAS-staining of testis sections from Adad2+/+, Adad2Mut1/Mut1, and Adad2Mut2/Mut2 mice at P60. Adad2Mut1/Mut1 and Adad2Mut2/Mut2 seminiferous tubules were devoid of elongating spermatids. Right panels show the high magnifications of the boxed areas on the left. Scale bars = 50 μm. (E) PAS-staining of epididymal tubules from Adad2+/+, Adad2Mut1/Mut1, and Adad2Mut2/Mut2 mice at P60. Adad2+/+ epididymal tubules were full of spermatozoa whereas epididymal tubules from Adad2Mut1/Mut1 and Adad2Mut2/Mut2 mice were devoid of mature sperm but contained degenerating round spermatids. Right panels show the high magnifications of the boxed areas on the left. Scale bars = 50 μm. (F) A diagram representing arrested stages of germ cell development in piRNA pathway associated genes and Adad2 mutants.
Adad2 is essential for spermatogenesis and male fertility. (A) Schematic representation of the Adad2 locus and design of the two sgRNAs targeting Adad2 exon 1, which harbors the IDR motif. Schematic representation and sequencing trace of the part of Adad2Mut exon 1 carrying the mutation site. The Mut1 site contains 47 bp removed on exon1 and causes a frameshift, while the Mut2 site contains 24 bp removed on exon1 and causes an 8aa lost in IDR motif. (B) The testes weights were reduced in both Adad2Mut1/Mut1 and Adad2Mut2/Mut2 mice at postnatal day 60 (P60). Data presented is the testes/body ratio (n = 6 for each genotype, **P < 0.01, Student’s t test). Error bars are mean ± SEM. (C) Fertility test for adult Adad2Mut1/Mut1and Adad2Mut2/Mut2 males. Each genotype shown was coupled with fertile wild-type mates. N = 3 for each genotype. ***P < 0.001(Student’s t test). (D) PAS-staining of testis sections from Adad2+/+, Adad2Mut1/Mut1, and Adad2Mut2/Mut2 mice at P60. Adad2Mut1/Mut1 and Adad2Mut2/Mut2 seminiferous tubules were devoid of elongating spermatids. Right panels show the high magnifications of the boxed areas on the left. Scale bars = 50 μm. (E) PAS-staining of epididymal tubules from Adad2+/+, Adad2Mut1/Mut1, and Adad2Mut2/Mut2 mice at P60. Adad2+/+ epididymal tubules were full of spermatozoa whereas epididymal tubules from Adad2Mut1/Mut1 and Adad2Mut2/Mut2 mice were devoid of mature sperm but contained degenerating round spermatids. Right panels show the high magnifications of the boxed areas on the left. Scale bars = 50 μm. (F) A diagram representing arrested stages of germ cell development in piRNA pathway associated genes and Adad2 mutants.
Loss of ADAD2 impairs spermatogenesis but not germ granule assembly in mice. (A) Representative images of PAS-staining histological testis sections in different stages of the seminiferous cycle from Adad2+/+ and Adad2Mut1/Mut1 mice are shown (n = 3), indicating round spermatid differentiation arrest. Scale bars = 20 μm. (B) Immunofluorescence analysis of proacrosomal vesicles visualized with antibodies to ACRV1 (green) in Adad2+/+ and Adad2Mut1/Mut1 round spermatids. DAPI (blue) marks the nuclei. Scale bar = 20 μm. Red arrowheads indicate defective acrosomes. (C) Representative images of PAS-staining histological testis and epididymis sections of adult Adad2Mut1/Mut1; Adad2Flag/mCherry mice are shown (n = 3). Scale bars = 50 μm. (D and E) Immunofluorescence staining with MIWI (D) and MILI (E) antibodies in seminiferous tubule sections of Adad2+/+ and Adad2Mut1/Mut1 mice. DAPI (blue) marks the nuclei. Scale bars = 50 μm. (F) Western blot analysis of MIWI and MILI in the whole testes of Adad2+/+ and Adad2Mut1/Mut1 mice. GAPDH was shown as a loading control. (G) Western blot analysis of RNF17 in the whole testes of Adad2+/+, Adad2Mut1/Mut1, and Adad2Mut2/Mut2 mice. α-TUBULIN was shown as a loading control. (H) Double immunostaining with DDX6 (green) and MIWI (red) on round spermatids from adult Adad2+/+ and Adad2Mut1/Mut1 testis sections are shown. DAPI (blue) marks the nuclei. Scale bar = 20 μm. Chromatoid bodies are marked with arrowheads. (I) Electron microscopic analysis of testis sections from adult Adad2+/+ and Adad2Mut1/Mut1 mice. Chromatoid bodies are marked with arrowheads. Scale bars = 1 μm. Source data are available for this figure: SourceData F5.
Loss of ADAD2 impairs spermatogenesis but not germ granule assembly in mice. (A) Representative images of PAS-staining histological testis sections in different stages of the seminiferous cycle from Adad2+/+ and Adad2Mut1/Mut1 mice are shown (n = 3), indicating round spermatid differentiation arrest. Scale bars = 20 μm. (B) Immunofluorescence analysis of proacrosomal vesicles visualized with antibodies to ACRV1 (green) in Adad2+/+ and Adad2Mut1/Mut1 round spermatids. DAPI (blue) marks the nuclei. Scale bar = 20 μm. Red arrowheads indicate defective acrosomes. (C) Representative images of PAS-staining histological testis and epididymis sections of adult Adad2Mut1/Mut1; Adad2Flag/mCherry mice are shown (n = 3). Scale bars = 50 μm. (D and E) Immunofluorescence staining with MIWI (D) and MILI (E) antibodies in seminiferous tubule sections of Adad2+/+ and Adad2Mut1/Mut1 mice. DAPI (blue) marks the nuclei. Scale bars = 50 μm. (F) Western blot analysis of MIWI and MILI in the whole testes of Adad2+/+ and Adad2Mut1/Mut1 mice. GAPDH was shown as a loading control. (G) Western blot analysis of RNF17 in the whole testes of Adad2+/+, Adad2Mut1/Mut1, and Adad2Mut2/Mut2 mice. α-TUBULIN was shown as a loading control. (H) Double immunostaining with DDX6 (green) and MIWI (red) on round spermatids from adult Adad2+/+ and Adad2Mut1/Mut1 testis sections are shown. DAPI (blue) marks the nuclei. Scale bar = 20 μm. Chromatoid bodies are marked with arrowheads. (I) Electron microscopic analysis of testis sections from adult Adad2+/+ and Adad2Mut1/Mut1 mice. Chromatoid bodies are marked with arrowheads. Scale bars = 1 μm. Source data are available for this figure: SourceData F5.
Of note, the germ granule assembly was not impaired upon the loss of ADAD2, confirmed by the unaffected MILI and MIWI localization and their protein levels in the Adad2Mut1/Mut1 mouse testes (Fig. 5, D–F). In addition, the protein level of RNF17 was not significantly altered in both Adad2Mut1/Mut1 and Adad2Mut2/Mut2 testes (Fig. 5 G). Since a previous study indicated that the P-bodies and CBs share several components, such as DDX6 and GW182 (Anbazhagan et al., 2022), we thus tested whether the DDX6 localization on CBs is affected by the ADAD2 loss. Co-immunofluorescence of DDX6 and MIWI demonstrated that the DDX6 localization on CBs was unaffected in Adad2Mut1/Mut1 round spermatids (Fig. 5 H). Further, transmission electron microscopy (TEM) visualized the intact CB structure in Adad2Mut1/Mut1 round spermatids (Fig. 5 I). In summary, these data indicate that ADAD2 is required for mouse spermatogenesis but not for CB formation during spermatogenesis.
The interplay between ADAD2 and RNF17 in P-bodies during meiosis
To identify interacting proteins of ADAD2, we performed an IP/MS analysis using FLAG-IP from WT and Adad2Flag/mCherry testes. We identified 37 ADAD2-associated proteins (Score > 50), including RNF17, confirming the interaction between ADAD2 and RNF17 (Fig. 6 A and Table S2). Among these ADAD2-associated proteins, YTHDC2, CNOT1, and RBM25 were reported to be localized on P-bodies (Hubstenberger et al., 2017), and YTHDC2 was reported to be localized on the DCP1α-granules in spermatocytes and required for male fertility (Jain et al., 2018; Liu et al., 2021; Wojtas et al., 2017). Although YTHDC2 signals overlapped with ADAD2-granules in spermatocytes at stage XI, YTHDC2 signals were largely diffused at stages IV–VI (Fig. 6 B), suggesting that YTHDC2 unlikely mediates the connection between ADAD2 and P-body.
The P-body localization of ADAD2 and RNF17 are interdependent in spermatogenesis. (A) Volcano plot showing enrichment (log2(mean intensity ratio of FLAG-IP from WT and Adad2Flag/mCherry testes) and confidence (-log10(FDR)) of proteins co-purifying with FLAG-ADAD2-mCherry from Adad2Flag/mCherry testes lysates (n = 3). Dotted line indicates factors with enrichment > fourfold and significance FDR < 0.1. Red font indicates the known P-body components. (B) Double immunostaining with ADAD2 (FLAG, red) and YTHDC2 (green) on spermatocytes from adult Adad2-Tg testis sections are shown. DAPI (blue) marks the nuclei. Scale bars = 20 μm. (C) Schematic representation of the Rnf17 locus and design of the two sgRNAs targeting Rnf17 exon 1 and exon 10. (D) Western blot analysis of RNF17 in the whole testes of Rnf17+/+ (with or without Adad2-Tg), Rnf17−/− (with or without Adad2-Tg) mice. GAPDH was shown as a loading control. (E) Immunostaining of RNF17 in Rnf17−/−, Adad2-Tg testis are shown. DAPI (blue) marks the nuclei. Scale bar = 50 μm. (F) Double immunostaining with RNF17 (green) and EDC3 (red) on spermatocytes from adult Adad2+/+ and Adad2Mut1/Mut1 testis sections are shown. DAPI (blue) marks the nuclei. Scale bars = 20 μm. Open arrowheads indicate EDC3 positive P-bodies, and solid arrowheads mark RNF17 granules. Quantifying the colocalization ratio between RNF17 and EDC3 in D is shown in the right histogram. n = 15 tubules from three mice, error bars are mean ± SEM., unpaired two-tailed Student’s t test. (G) Double immunostaining with RNF17 (green) and EDC3 (red) on spermatocytes from adult Adad2Mut2/Mut2 mouse testis sections are shown. DAPI (blue) marks the nuclei. Scale bar = 20 μm. (H) Double immunostaining with ADAD2 (red) and EDC3 (green) on spermatocytes from Rnf17−/−, Adad2-Tg mouse testis sections are shown. DAPI (blue) marks the nuclei. Scale bars = 20 μm. (I) Double immunostaining with EDC3/DCP1α (green) and SYCP3 (magenta) on spermatocytes at stage XI from adult WT, Adad2Mut1/Mut1, Adad2Mut2/Mut2, and Rnf17−/− testis sections are shown. DAPI (blue) marks the nuclei. Scale bars = 20 μm. (J) Schematic representation of the interaction between ADAD2/RNF17-granule and P-body in NIH3T3 cells, Adad2 and/or Rnf17 mutant mice based on the immunofluorescence analyses. In NIH3T3 cells, RNF17 granules coalesce with P-bodies once ADAD2 is co-expressed. In Adad2 mutants, RNF17 failed to be enriched on P-bodies instead of forming a few cytoplasmic granules discretely, while ADAD2 failed to form granules in the loss of RNF17. Notably, loss of ADAD2 or RNF17 results in an aberrant disassembly of P-bodies in diplotene spermatocytes. Source data are available for this figure: SourceData F6.
The P-body localization of ADAD2 and RNF17 are interdependent in spermatogenesis. (A) Volcano plot showing enrichment (log2(mean intensity ratio of FLAG-IP from WT and Adad2Flag/mCherry testes) and confidence (-log10(FDR)) of proteins co-purifying with FLAG-ADAD2-mCherry from Adad2Flag/mCherry testes lysates (n = 3). Dotted line indicates factors with enrichment > fourfold and significance FDR < 0.1. Red font indicates the known P-body components. (B) Double immunostaining with ADAD2 (FLAG, red) and YTHDC2 (green) on spermatocytes from adult Adad2-Tg testis sections are shown. DAPI (blue) marks the nuclei. Scale bars = 20 μm. (C) Schematic representation of the Rnf17 locus and design of the two sgRNAs targeting Rnf17 exon 1 and exon 10. (D) Western blot analysis of RNF17 in the whole testes of Rnf17+/+ (with or without Adad2-Tg), Rnf17−/− (with or without Adad2-Tg) mice. GAPDH was shown as a loading control. (E) Immunostaining of RNF17 in Rnf17−/−, Adad2-Tg testis are shown. DAPI (blue) marks the nuclei. Scale bar = 50 μm. (F) Double immunostaining with RNF17 (green) and EDC3 (red) on spermatocytes from adult Adad2+/+ and Adad2Mut1/Mut1 testis sections are shown. DAPI (blue) marks the nuclei. Scale bars = 20 μm. Open arrowheads indicate EDC3 positive P-bodies, and solid arrowheads mark RNF17 granules. Quantifying the colocalization ratio between RNF17 and EDC3 in D is shown in the right histogram. n = 15 tubules from three mice, error bars are mean ± SEM., unpaired two-tailed Student’s t test. (G) Double immunostaining with RNF17 (green) and EDC3 (red) on spermatocytes from adult Adad2Mut2/Mut2 mouse testis sections are shown. DAPI (blue) marks the nuclei. Scale bar = 20 μm. (H) Double immunostaining with ADAD2 (red) and EDC3 (green) on spermatocytes from Rnf17−/−, Adad2-Tg mouse testis sections are shown. DAPI (blue) marks the nuclei. Scale bars = 20 μm. (I) Double immunostaining with EDC3/DCP1α (green) and SYCP3 (magenta) on spermatocytes at stage XI from adult WT, Adad2Mut1/Mut1, Adad2Mut2/Mut2, and Rnf17−/− testis sections are shown. DAPI (blue) marks the nuclei. Scale bars = 20 μm. (J) Schematic representation of the interaction between ADAD2/RNF17-granule and P-body in NIH3T3 cells, Adad2 and/or Rnf17 mutant mice based on the immunofluorescence analyses. In NIH3T3 cells, RNF17 granules coalesce with P-bodies once ADAD2 is co-expressed. In Adad2 mutants, RNF17 failed to be enriched on P-bodies instead of forming a few cytoplasmic granules discretely, while ADAD2 failed to form granules in the loss of RNF17. Notably, loss of ADAD2 or RNF17 results in an aberrant disassembly of P-bodies in diplotene spermatocytes. Source data are available for this figure: SourceData F6.
We next asked whether the P-body localization of ADAD2 and RNF17 is interdependent or not in spermatocytes. To this end, we generated an Rnf17 knockout mouse model. Two sgRNAs were designed to target the exons 2 and 10, which resulted in a 33,223 bp deletion containing a 992 bp coding sequence that causes a premature termination codon (Fig. 6 C). The RNF17 protein was undetectable in Rnf17−/− testes (Fig. 6, D and E). Consistent with the in vitro assays using cell culture systems, the RNF17 formed small granules but failed to localize on the P-bodies in Adad2Mut1/Mut1 and Adad2Mut2/Mut2 spermatocytes (Fig. 6, F and G). Conversely, the FLAG-tagged ADAD2-granules disappeared in the cytoplasm of Rnf17−/−; Adad2-Tg spermatocytes (Fig. 6 H). These data indicate that the P-body localization of ADAD2 and RNF17 is interdependent. Remarkably, cytoplasmic foci of P-body markers (EDC3 and DCP1α) disappeared in Adad2Mut1/Mut1, Adad2Mut2/Mut2, and Rnf17−/− diplotene spermatocytes (Fig. 6 I). Altogether, these results revealed that the integrity of P-bodies during meiosis is defective upon ablation of ADAD2 or RNF17 and that the interplay between ADAD2 and RNF17 is required for the P-body formation (Fig. 6 J).
ADAD2 is required for the suppression of the RNF17-directed ping-pong cycle in pachytene piRNA biogenesis
Since defective spermatogenesis observed in Adad2Mut1/Mut1 (Adad2 mutants) and Adad2Mut2/Mut2 (Adad2 IDR-domain mutants) was akin to the phenotypes of Rnf17−/− and Miwi−/− mice (Fig. S4 F), we thus speculated that the regulation of spermatogenesis by ADAD2 is RNF17-related and ADAD2 may be involved in the piRNA pathway. To elucidate whether ADAD2 is a partner of RNF17 in the regulation of pachytene piRNA biogenesis, we examined the abundance and size of piRNA populations by small RNA sequencing using purified pachytene spermatocytes and round spermatids from adult Adad2+/+ and Adad2Mut1/Mut1 testes. We found that the piRNA profile was not affected with respect to length distribution in both Adad2Mut1/Mut1 pachytene spermatocytes and round spermatids (Fig. 7, A and B and Table S3). This result is consistent with the analysis of published small RNA sequencing on purified wild-type and Rnf17−/− pachytene spermatocytes and round spermatids (Wasik et al., 2015), although the piRNA populations in Rnf17−/− round spermatids showed a slightly decrease (Fig. S5, A and B). This slight decrease is presumably due to RNF17’s association with piRNA precursors. We thus examined the expression levels of four known pachytene piRNAs precursors (piR1, piR2, piR3, and piLR; Bai et al., 2018), and found the expression level of these precursors was not affected (Fig. S5 C). Moreover, we performed an RNA immunoprecipitation (RIP) experiment in Adad2 transgenic mice using an anti-FLAG antibody, followed by RNA isolation and RT-qPCR. The result showed no specific enrichment of piRNA precursors in ADAD2-associated RNA (Fig. S5 D). Interestingly, we observed a peak of 19-nt small RNAs appeared in both Adad2Mut1/Mut1 and Rnf17−/− round spermatids (Fig. 7 B and Fig. S5 B). Based on previous studies (Ichiyanagi et al., 2014; Oey et al., 2011), we suspected that these 19-nt small RNAs are likely the aberrant byproducts of secondary piRNA biogenesis upon depletion of ADAD2 or RNF17.
RNF17-mediated suppression of Ping-pong activity was disrupted in Adad2 mutants. (A and B) Size distribution of small RNA libraries from purified pachytene spermatocytes (A) and round spermatids (B) of Adad2+/+ and Adad2Mut1/Mut1 mice (n = 3). (C and D) Genomic annotation of total piRNA from purified pachytene spermatocytes (C) and round spermatids (D) of Adad2+/+ and Adad2Mut1/Mut1 mice (n = 3). (E and F) Classification of repeat piRNAs from purified pachytene spermatocytes (E) and round spermatids (F) of Adad2+/+ and Adad2Mut1/Mut1 mice (n = 3) based on repeat class. Error bars are mean ± SEM. Two-tailed Student’s t test. (G) Small RNA-seq derived heat maps depicting fold-change of expression relative to wild-type (WT) for the 10 most upregulated LINE TEs in Rnf17−/− testes (Te) and Adad2Mut1/Mut1 pachytene spermatocytes (PS) and round spermatids (RS). (H) Representative images of testis sections from adult Adad2+/+ and Adad2Mut1/Mut1 mice (n = 3) stained for LINE1 ORF1 (green) are shown. DAPI (blue) marks the nuclei. Scale bars = 50 μm. (I) Size profiles of LINE piRNAs in purified pachytene spermatocytes and round spermatids of Adad2+/+ and Adad2Mut1/Mut1 mice (n = 3). Reads were normalized per million unique genomic mappers (RPM). (J and K) Ping-pong analysis of Line1-derived piRNAs in purified pachytene spermatocytes (J) and round spermatids (K) of Adad2+/+ and Adad2Mut1/Mut1 mice (n = 3), showing the relative frequency of the distance between 5′-ends of complementary piRNAs mapping to the LINE1 family. (L) Nucleotide distributions (sequence logos) of the first 10 nt of Line1-derived piRNAs for multi-mappers from purified pachytene spermatocytes (PS) and round spermatids (RS) from adult Adad2+/+ and Adad2Mut1/Mut1 mice (n = 3).
RNF17-mediated suppression of Ping-pong activity was disrupted in Adad2 mutants. (A and B) Size distribution of small RNA libraries from purified pachytene spermatocytes (A) and round spermatids (B) of Adad2+/+ and Adad2Mut1/Mut1 mice (n = 3). (C and D) Genomic annotation of total piRNA from purified pachytene spermatocytes (C) and round spermatids (D) of Adad2+/+ and Adad2Mut1/Mut1 mice (n = 3). (E and F) Classification of repeat piRNAs from purified pachytene spermatocytes (E) and round spermatids (F) of Adad2+/+ and Adad2Mut1/Mut1 mice (n = 3) based on repeat class. Error bars are mean ± SEM. Two-tailed Student’s t test. (G) Small RNA-seq derived heat maps depicting fold-change of expression relative to wild-type (WT) for the 10 most upregulated LINE TEs in Rnf17−/− testes (Te) and Adad2Mut1/Mut1 pachytene spermatocytes (PS) and round spermatids (RS). (H) Representative images of testis sections from adult Adad2+/+ and Adad2Mut1/Mut1 mice (n = 3) stained for LINE1 ORF1 (green) are shown. DAPI (blue) marks the nuclei. Scale bars = 50 μm. (I) Size profiles of LINE piRNAs in purified pachytene spermatocytes and round spermatids of Adad2+/+ and Adad2Mut1/Mut1 mice (n = 3). Reads were normalized per million unique genomic mappers (RPM). (J and K) Ping-pong analysis of Line1-derived piRNAs in purified pachytene spermatocytes (J) and round spermatids (K) of Adad2+/+ and Adad2Mut1/Mut1 mice (n = 3), showing the relative frequency of the distance between 5′-ends of complementary piRNAs mapping to the LINE1 family. (L) Nucleotide distributions (sequence logos) of the first 10 nt of Line1-derived piRNAs for multi-mappers from purified pachytene spermatocytes (PS) and round spermatids (RS) from adult Adad2+/+ and Adad2Mut1/Mut1 mice (n = 3).
Transcriptome and small RNA library analyses of Adad2 Mut1/Mut1 mice. (A and B) Size distribution of small RNA libraries from adult Rnf17+/− and Rnf17−/− pachytene spermatocytes (A) and round spermatids (B). (C) RT-qPCR analysis of the expression of piRNA precursors (piR-1: 9-qC-31469.1, piR-2: 5-qF-14224.1, piR-3: 17-qA3.3–27363.1, piRL: 6-qF3-28913.1) in adult Adad2+/+ and Adad2Mut1/Mut1 testes. Error bars are mean ± SEM. Two-tailed Student’s t test. (D) RT-qPCR analysis of piRNA precursors (piR-1: 9-qC-31469.1, piR-2: 5-qF-14224.1, piR-3: 17-qA3.3–27363.1, piRL: 6-qF3-28913.1) in FLAG-immunoprecipitated RNA from Adad2Flag/mCherry testis lysates. Bars represent the fold enrichment (mean ± SEM, n = 3 independent experiments) of different piRNA precursors isolated by anti-FLAG RIP from Adad2+/+ and Adad2Flag/mCherry testes after normalization to the input, respectively. Two-tailed Student’s t test. (E and F) Genomic annotation of total piRNA from purified pachytene spermatocytes (E) and round spermatids (F) of Rnf17+/− and Rnf17−/− mice. (G and H) Classification of repeat piRNAs from purified pachytene spermatocytes (G) and round spermatids (H) of Rnf17+/− and Rnf17−/− mice based on repeat class. Error bars are mean ± SEM. Two-tailed Student’s t test. (I and J) Bar graph showing the relative frequency of piRNAs mapping to the top 10 IAP-derived piRNA in Adad2Mut1/Mut1 pachytene spermatocytes (PS; I) and round spermatids (RS; J). Error bars are mean ± SEM. Two-tailed Student’s t test. (K) Number of genes exhibiting significantly (fold change > 2, FDR < 0.1) up- or downregulated in Adad2Mut1/Mut1 versus Adad2+/+ testes at P20. N = 3 for each genotype. (L) RNA-seq derived heat-maps depicting genes upregulated or downregulated in Adad2Mut1/Mut1 testes at P20. (M) GO term enrichment analysis of downregulated transcripts in P20 Adad2Mut1/Mut1 testes. (N) RT-qPCR analysis of the expression of downregulated germ cell developmental-associated genes in Adad2Mut1/Mut1 testes compared with Adad2+/+. Data are presented as mean ± SEM, n = 3 independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 using two-tailed Student’s t test.
Transcriptome and small RNA library analyses of Adad2 Mut1/Mut1 mice. (A and B) Size distribution of small RNA libraries from adult Rnf17+/− and Rnf17−/− pachytene spermatocytes (A) and round spermatids (B). (C) RT-qPCR analysis of the expression of piRNA precursors (piR-1: 9-qC-31469.1, piR-2: 5-qF-14224.1, piR-3: 17-qA3.3–27363.1, piRL: 6-qF3-28913.1) in adult Adad2+/+ and Adad2Mut1/Mut1 testes. Error bars are mean ± SEM. Two-tailed Student’s t test. (D) RT-qPCR analysis of piRNA precursors (piR-1: 9-qC-31469.1, piR-2: 5-qF-14224.1, piR-3: 17-qA3.3–27363.1, piRL: 6-qF3-28913.1) in FLAG-immunoprecipitated RNA from Adad2Flag/mCherry testis lysates. Bars represent the fold enrichment (mean ± SEM, n = 3 independent experiments) of different piRNA precursors isolated by anti-FLAG RIP from Adad2+/+ and Adad2Flag/mCherry testes after normalization to the input, respectively. Two-tailed Student’s t test. (E and F) Genomic annotation of total piRNA from purified pachytene spermatocytes (E) and round spermatids (F) of Rnf17+/− and Rnf17−/− mice. (G and H) Classification of repeat piRNAs from purified pachytene spermatocytes (G) and round spermatids (H) of Rnf17+/− and Rnf17−/− mice based on repeat class. Error bars are mean ± SEM. Two-tailed Student’s t test. (I and J) Bar graph showing the relative frequency of piRNAs mapping to the top 10 IAP-derived piRNA in Adad2Mut1/Mut1 pachytene spermatocytes (PS; I) and round spermatids (RS; J). Error bars are mean ± SEM. Two-tailed Student’s t test. (K) Number of genes exhibiting significantly (fold change > 2, FDR < 0.1) up- or downregulated in Adad2Mut1/Mut1 versus Adad2+/+ testes at P20. N = 3 for each genotype. (L) RNA-seq derived heat-maps depicting genes upregulated or downregulated in Adad2Mut1/Mut1 testes at P20. (M) GO term enrichment analysis of downregulated transcripts in P20 Adad2Mut1/Mut1 testes. (N) RT-qPCR analysis of the expression of downregulated germ cell developmental-associated genes in Adad2Mut1/Mut1 testes compared with Adad2+/+. Data are presented as mean ± SEM, n = 3 independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 using two-tailed Student’s t test.
Because, in Rnf17−/− pachytene spermatocytes, secondary piRNA biogenesis is ectopically activated and TE-associated piRNAs are increased (Wasik et al., 2015), we sought to determine whether TE-associated piRNAs was increased by the loss of ADAD2. After mapping to the mouse genome, we detected a significant increase of repeat-derived piRNAs in both Adad2Mut1/Mut1 and Rnf17−/− pachytene spermatocytes and round spermatids, while the pachytene clusters-derived piRNAs were downregulated (Fig. 7, C and D; and Fig. S5, E and F). In addition, both Adad2Mut1/Mut1 pachytene spermatocytes and round spermatids displayed a specific increase in LINE- and IAP-derived piRNAs, which is consistent with Rnf17−/− (Fig. 7, E and F; and Fig. S5, G–J). Of note, further analysis revealed that the piRNAs mapping to evolutionarily young TE subfamilies, such as L1Md_A, L1_Mus, and L1Md_T, were abundant in both Adad2Mut1/Mut1 and Rnf17−/− mice (Fig. 7 G). These evolutionarily young TE subfamilies are deleterious to the genome integrity; thus, they must be appropriately repressed during male germ cell development (Yang et al., 2020). Accordingly, the LINE ORF1 protein was not detected in Adad2Mut1/Mut1 testes (Fig. 7 H), suggesting that the retrotransposition activity was properly silenced in Adad2Mut1/Mut1 testes, similar to the Rnf17−/− testes (Wasik et al., 2015). These results raised the possibility that ADAD2 is a partner of RNF17-directed suppression of the Ping-pong pathway in meiotic cells. To test this possibility, we first examined the source of LINE1 piRNAs in both pachytene spermatocytes and round spermatids from Adad2+/+ and Adad2Mut1/Mut1 testes. As expected, the origins of LINE piRNAs derived from unique loci outside of pachytene clusters and multiple-mapped loci were increased in Adad2Mut1/Mut1 mice (Fig. 7 I). Next, we examined the frequencies of LINE1 piRNAs with the Ping-pong signature (10nt overlap of 5′-ends with opposite orientation) and found that they were elevated in Adad2Mut1/Mut1 pachytene spermatocytes and round spermatids (Fig. 7, J and K). Concomitantly, a large number of LINE1 piRNAs was enriched with A at position 10 in Adad2Mut1/Mut1 pachytene spermatocytes and round spermatids (Fig. 7 L), suggesting the overproduction of secondary piRNAs by cleavage-dependent Ping-pong activity in Adad2Mut1/Mut1 spermatogenesis.
Loss of ADAD2 leads to dramatic transcriptome alterations in postmeiotic germ cells
We next performed RNA-sequencing (RNA-seq) of late juvenile Adad2+/+ and Adad2Mut1/Mut1 testes at P20, a developmental stage with the first appearance of round spermatids. There were no significant changes in the population of germ cells at P20 between Adad2+/+ and Adad2Mut1/Mut1 testes. We only found that, relative to Adad2+/+, 57 genes were downregulated, and 25 genes were upregulated in Adad2Mut1/Mut1 testes (FDR < 0.1, fold change > 2; Fig. S5, K and L and Table S4). Gene ontology (GO) analysis revealed that downregulated transcripts were enriched with the genes involved in the processes of germ cell development (Fig. S5 M). Among these downregulated genes, several essential genes for sperm development were selected and validated by RT-qPCR assays (Fig. S5 N).
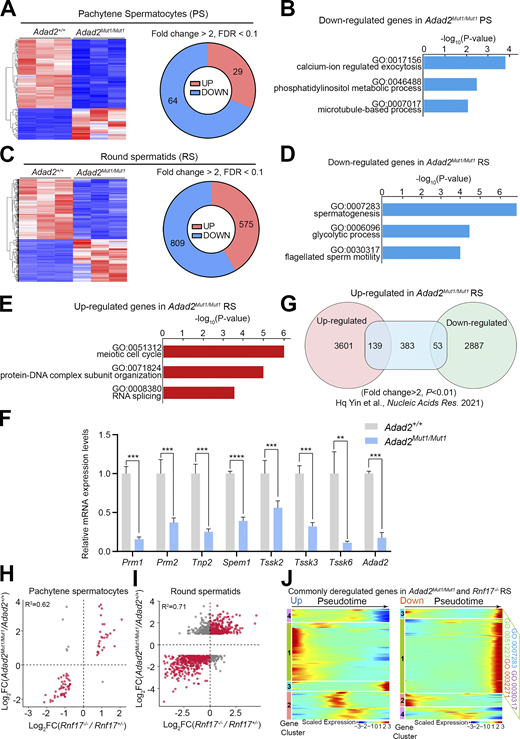
Because these RNA-seq data suggest that the misregulation of transcripts may occur in the round spermatid stage, we then performed RNA-seq using purified pachytene spermatocytes and round spermatids from adult Adad2+/+ and Adad2Mut1/Mut1 testes. We found that only 29 and 64 genes were significantly up- and downregulated, respectively, in Adad2Mut1/Mut1 pachytene spermatocytes compared to controls (Fig. 8 A and Table S5). GO analysis demonstrates that downregulated genes in Adad2Mut1/Mut1 pachytene spermatocytes are enriched with the genes involved in calcium-ion-regulated exocytosis (Fig. 8 B). In contrast, a dramatic alteration was observed in the transcriptome of Adad2Mut1/Mut1 round spermatids, including 575 upregulated and 809 downregulated genes (Fig. 8 C and Table S6). GO analysis of downregulated genes in Adad2Mut1/Mut1 round spermatids revealed enrichment of genes involved in spermatogenesis and flagellated sperm motility (Fig. 8 D). Remarkably, we noticed that upregulated genes in Adad2Mut1/Mut1 round spermatids were enriched with genes involved in the meiotic cell cycle and RNA splicing (Fig. 8 E). Further, among these downregulated genes, several essential genes for sperm development were selected and validated by RT-qPCR assays (Fig. 8 F). Interestingly, by comparing with recently published RNA-seq data (Yin et al., 2021), we found 139 upregulated genes in Adad2Mut1/Mut1 round spermatids are upregulated during normal spermatogenesis from meiotic spermatocytes to round spermatids. In contrast, only 53 upregulated genes in Adad2Mut1/Mut1 round spermatids are downregulated during normal spermatogenesis (Fig. 8 G). These results suggest a role of ADAD2 in promoting the decay of transcripts in round spermatids.
Highly correlated gene expression changes in the absence of either ADAD2 or RNF17. (A) RNA-seq derived heat maps and diagram depicting genes upregulated or downregulated in purified Adad2Mut1/Mut1 pachytene spermatocytes (PS). (B) GO term enrichment analysis for downregulated transcripts in purified Adad2Mut1/Mut1 PS. (C) RNA-seq derived heat maps and diagram depicting genes upregulated or downregulated in purified Adad2Mut1/Mut1 round spermatids (RS). (D) GO term enrichment analysis for downregulated transcripts in purified Adad2Mut1/Mut1 RS. (E) GO term enrichment analysis for upregulated transcripts in purified Adad2Mut1/Mut1 RS. (F) RT-qPCR analysis of the expression of eight selected downregulated germ cell developmental-associated genes in Adad2Mut1/Mut1 RS compared with Adad2+/+. Data are presented as mean ± SEM, n = 3. **P < 0.01, ***P < 0.001 using two-tailed Student’s t test. (G) Overlap of differentially expressed genes during the meiotic spermatocytes-to-postmeiotic round spermatids transition (Hq Yin et al., 2021) with the upregulated genes in Adad2Mut1/Mut1 RS. (H and I) Log2 fold-change (KO/WT) in transcript abundance upon the loss of Adad2 (y-axis) or Rnf17 (x-axis) in purified pachytene spermatocytes (H) and round spermatids (I). Dots represent concordantly (red) or discordantly (gray) misregulated genes. R2 was calculated using Pearson's correlation test. (J) Heatmaps showing the hierarchical relationship among the clusters of commonly deregulated genes in Adad2Mut1/Mut1 and Rnf17−/− round spermatids across pseudotime of spermatogenesis (undifferentiated spermatogonia to round spermatids). Expressions of the commonly upregulated genes (left) and the downregulated genes (right) in round spermatids of Rnf17−/− and Adad2Mut1/Mut1 mice were assessed by reanalyzing scRNA-seq data of spermatogenic cells (GSE109033). Pseudotime (left to right) corresponds to the developmental trajectory of spermatogenesis (undifferentiated spermatogonia to round spermatids). In clusters 1 and 3 of downregulated genes, GO terms (top 4 by P value) with false discovery rate (FDR) <0.01 are shown. GO:0032271: regulation of protein polymerization; GO:0007283: spermatogenesis; GO:0051223: regulation of protein transport; GO:0030317: flagellated sperm motility. See Table S7 for the complete gene list of the GO analyses.
Highly correlated gene expression changes in the absence of either ADAD2 or RNF17. (A) RNA-seq derived heat maps and diagram depicting genes upregulated or downregulated in purified Adad2Mut1/Mut1 pachytene spermatocytes (PS). (B) GO term enrichment analysis for downregulated transcripts in purified Adad2Mut1/Mut1 PS. (C) RNA-seq derived heat maps and diagram depicting genes upregulated or downregulated in purified Adad2Mut1/Mut1 round spermatids (RS). (D) GO term enrichment analysis for downregulated transcripts in purified Adad2Mut1/Mut1 RS. (E) GO term enrichment analysis for upregulated transcripts in purified Adad2Mut1/Mut1 RS. (F) RT-qPCR analysis of the expression of eight selected downregulated germ cell developmental-associated genes in Adad2Mut1/Mut1 RS compared with Adad2+/+. Data are presented as mean ± SEM, n = 3. **P < 0.01, ***P < 0.001 using two-tailed Student’s t test. (G) Overlap of differentially expressed genes during the meiotic spermatocytes-to-postmeiotic round spermatids transition (Hq Yin et al., 2021) with the upregulated genes in Adad2Mut1/Mut1 RS. (H and I) Log2 fold-change (KO/WT) in transcript abundance upon the loss of Adad2 (y-axis) or Rnf17 (x-axis) in purified pachytene spermatocytes (H) and round spermatids (I). Dots represent concordantly (red) or discordantly (gray) misregulated genes. R2 was calculated using Pearson's correlation test. (J) Heatmaps showing the hierarchical relationship among the clusters of commonly deregulated genes in Adad2Mut1/Mut1 and Rnf17−/− round spermatids across pseudotime of spermatogenesis (undifferentiated spermatogonia to round spermatids). Expressions of the commonly upregulated genes (left) and the downregulated genes (right) in round spermatids of Rnf17−/− and Adad2Mut1/Mut1 mice were assessed by reanalyzing scRNA-seq data of spermatogenic cells (GSE109033). Pseudotime (left to right) corresponds to the developmental trajectory of spermatogenesis (undifferentiated spermatogonia to round spermatids). In clusters 1 and 3 of downregulated genes, GO terms (top 4 by P value) with false discovery rate (FDR) <0.01 are shown. GO:0032271: regulation of protein polymerization; GO:0007283: spermatogenesis; GO:0051223: regulation of protein transport; GO:0030317: flagellated sperm motility. See Table S7 for the complete gene list of the GO analyses.
We finally examined the correlation between differential expression genes (DEGs) in pachytene spermatocytes and round spermatids from Adad2Mut1/Mut1 and Rnf17−/− testes. Remarkably, DEGs were highly correlated between Adad2Mut1/Mut1 and Rnf17−/− germ cells (R2 = 0.62 for pachytene spermatocytes and R2 = 0.71 for round spermatids; Fig. 8, H and I). By comparing with previously published single-cell RNA-seq data of spermatogenic cells (Hermann et al., 2018), we found that the commonly downregulated genes in Adad2Mut1/Mut1 and Rnf17−/− round spermatids are expressed in the late stage of pseudotime (Fig. 8 J). Among commonly downregulated genes, 80.1% of the genes belong to clusters 1 and 3, which were enriched with genes involved in spermatogenesis and flagellated sperm motility (Fig. 8 J, right panel, and Table S7), suggesting that ADAD2-RNF17 complex regulates a subset of critical genes required for spermiogenesis. In contrast, many commonly upregulated genes were found in early-to-mid pseudotime (Fig. 8 J, left panel), representing the high expression in spermatogonia or meiotic prophase. Together, we conclude that ADAD2 acts in concert with RNF17 and is required for suppressing the Ping-pong cycle in pachytene piRNA biogenesis.
Discussion
In male germ cells, various types of cytoplasmic RNP granules together make up a regulatory network to ensure the accurate expression of transcripts during spermatogenesis, and P-bodies in mouse spermatocytes share components with germ granules such as CBs (Anbazhagan et al., 2022). In this study, we demonstrated that ADAD2 acts in concert with RNF17 in P-bodies in meiotic cells and is required for suppressing the Ping-pong cycle in pachytene piRNA biogenesis. In addition, the disruption of P-bodies at the end of the pachytene stage in Adad2 and Rnf17 mutants suggest that P-bodies are involved in pachytene piRNA biogenesis. This notion extends the functionalities of P-bodies which are not only involved in pre-pachytene piRNA biogenesis but also in pachytene piRNA biogenesis. We further showed that the loss function of ADAD2 does not affect the CB formation, suggesting distinct functions of ADAD2 in P-bodies and germ granules, including CBs.
Most TDRD family members participate in the assembly of germ granules in mice. Although RNF17 (TDRD4) has long been known as a component of novel granules distinct from germ granules, the exact type of these granules has not been determined yet. In this study, we demonstrated that RNF17 granule is tightly associated with P-bodies in pachytene spermatocytes. In contrast to RNF17, loss of another P-body associated piRNA factor Tdrd9 leads to a meiotic arrest and male infertility primarily due to the secondary piRNA biogenesis defects in prospermatogonia (Shoji et al., 2009; Wenda et al., 2017). In light of these data, our findings clarified the distinct roles of P-bodies between pachytene piRNA biogenesis in meiotic cells and pre-pachytene piRNA biogenesis in prospermatogonia. Specifically, the P-body-localized TDRD9-MIWI2 complex is the driving mechanism to facilitate the production of secondary piRNA by the Ping-pong cycle in prospermatogonia (Aravin et al., 2008; Shoji et al., 2009), while the Ping-pong cycle suppresser ADAD2-RNF17 complex, identified in this study, functions in P-bodies in meiosis, highlighting the functional diversity of P-bodies. Thus, further investigation of P-bodies during meiosis would help uncover molecular underpinings of P-body regulation in piRNA biogenesis.
Deletion of piP-body component genes, such as Tdrd9 and Gtsf1, has no impact on the localization of IMC components (Shoji et al., 2009; Yoshimura et al., 2018). Similarly, we show that the Adad2 deletion did not change the IMC localization of PIWI proteins in spermatocytes. Although P-bodies and germ granules are structurally distinct compartments, two structures merged in round spermatids, as many P-body components appear in CBs (Tanaka et al., 2011). Our data revealed that the defective P-bodies in Adad2 and Rnf17 mutants did not alter the DDX6 recruitment to the CBs in round spermatids, suggesting that P-bodies are dispensable for the structural remodeling of germ granules as spermatogenesis proceeds. In turn, the P-body localization of MAEL, TDRD9, and MIWI2 in prospermatogonia is under the control of an IMC component MILI (Wang et al., 2020). Intriguingly, a recent study reveals that Mael is indispensable for the Qin-positive nuage assembly in B. mori (Namba et al., 2022). In mice, as a partner of RNF17, MAEL is previously reported to associate with IMC (Takebe et al., 2013). Thus, an outstanding question remains whether IMC components regulate the localization of piRNA factors on P-bodies in spermatocytes.
Of note, the Ping-pong cycle is a powerful mechanism that produces secondary piRNAs against TEs in prospermatogonia (Ozata et al., 2019). On the other hand, pachytene spermatocytes suppress Ping-pong cycles presumably due to the following reasons: (1) pachytene piRNAs are primary piRNAs that are transcribed directly from unique piRNA clusters which are transposon-poor (Li et al., 2013); (2) the pachytene piRNAs are unlikely required for the TE silencing in meiotic cells as DNA methylation acts as the primary suppressor of TEs during meiosis; (3) numerous mRNAs required for the late stage of spermiogenesis are transcribed and stored in spermatocytes (Kleene, 2013; Lin et al., 2017), and these mRNAs, such as Rnf168 and Tekt4 (Bohgaki et al., 2013; Roy et al., 2007), are largely devoid of evolutionarily young transposon insertions which are the targets of the Ping-pong mechanism (Molaro et al., 2014; Sookdeo et al., 2013). Considering the common spermatogenic phenotypes between Adad2 mutants and Rnf17−/− mice, the interaction between ADAD2 and RNF17 and their suppression of the Ping-pong cycle are essential in spermatogenesis. Our work raises the possibility that ADAD2 acts as a scaffold to recruit RNF17 and putative piRNA factors to P-bodies to repress the Ping-pong cycle in pachytene piRNA biogenesis.
An interesting finding in this study is that the dsRBD motif of ADAD2 is not required for the interaction between ADAD2 and RNF17 or the P-body localization of RNF17 in cultured cells. We, therefore, proposed that the RNA-binding ability of ADAD2 is not required for the ADAD2-RNF17 complex to suppress Ping-pong activity. Considering that the IDR domain of ADAD2 plays a key role in the P-body localization in vitro, it is possible that phase separation may be driving the recruitment of ADAD2 and RNF17 to P-bodies and facilitating these granules’ formation. This possibility is supported by the notions that the liquid–liquid phase separation is mediated by IDR domains (Martin and Holehouse, 2020) and that the phase separation plays a considerable role in the P-body assembly (Riggs et al., 2020).
In addition to piRNA factors, ADAD2 is also associated with several somatic P-body components such as YTHDC2, a key factor essential for spermatogenesis and male fertility, and YTHDC2 functions in the mRNA decay pathway during spermatogenesis (Bailey et al., 2017). In the current view, a well-known function of the P-bodies is mRNA storage (Parker and Sheth, 2007; Standart and Weil, 2018), which raises the possibility that the mRNA transcripts in spermatocytes may be stored in the P-bodies before they are processed later in the stage of round spermatids during spermatogenesis (Ozturk and Uysal, 2018). Therefore, we assumed that RNF17 and ADAD2 might involve in the protection of mRNAs stored in P-bodies from degradation by the IMC (a center of the Ping-pong cycle). It is reasonable to postulate that IMC and RNF17-/ADAD2-granules are closely related in early-mid pachytene spermatocytes. Additionally, our in vitro cell localization analyses suggest that ADAD2 might act as a scaffold and that the IDR domain of ADAD2 facilitates the connection between RNF17 and P-bodies. Considering the close localization between P-bodies and IMC in spermatocytes before the diplotene stage, we proposed that the Ping-pong suppression function by RNF17 and ADAD2-dependent recruitment of RNF17 to P-bodies are tightly coupled. Therefore, it is reasonable to infer that P-bodies are critical places for mRNA degradation. Additionally, our data also indicated that YTHDC2 is unlikely to involve in recruiting the ADAD2-RNF17 complex to P-bodies. Interestingly, ADAD1 also displayed a diffused distribution in the cytoplasm but did not colocalize with the P-body marker DCP1α. We found that when RNF17 and ADAD1 were ectopically expressed in the cell lines we used, RNF17 localization was impaired by the overexpression of ADAD1 (Fig. S3). However, the localization of RNF17 in vivo may not be guided by the ADAD1 as the existence of other partners of RNF17. This may be explained by the fact that ADAD1 is abundant in the round spermatids rather than spermatocytes, and the phenotype of Adad1 KO mice is also different from Rnf17 KO mice, as elongated spermatids can be detected in Adad1 KO mice (Snyder et al., 2020).
Furthermore, our RNA-seq analyses revealed highly correlated gene expression changes in the absence of either ADAD2 or RNF17, indicating the common function of ADAD2 and RNF17 in gene regulation. This was further supported by small RNA-seq data, which showed that Ping-pong activity was ectopically activated in Adad2 mutants akin to Rnf17 mutants. We thus concluded that RNF17 has two critical roles during pachytene piRNA biogenesis: (1) associating with piRNA precursors to fuel the pachytene piRNA formation because the piRNA abundance in Rnf17 mutant spermatocytes is only half of the wild-type; (2) protecting protein-coding transcripts from Ping-pong cycle. Intriguingly, our data showed that the overall piRNA abundance was not affected in Adad2 mutants, and ADAD2 is not binding to piRNA precursors, suggesting that ADAD2 may not be associated with RNF17-guide piRNA precursors for piRNA processing. Therefore, we hypothesize that ADAD2 selectively functions with RNF17 in pachytene piRNA biogenesis.
In summary, our work reveals a molecular basis underpinning spermatogenic arrest in Adad2 mutants. Improper secondary piRNA machinery induced in the loss of ADAD2 is harmful to meiotic cells, eventually leading to male infertility. Our study sheds light on the regulatory mechanism by which the interaction between ADAD2 and RNF17 in P-bodies represses the Ping-pong cycle to ensure pachytene piRNA biogenesis during meiosis.
Materials and methods
Ethics statement
All the animal procedures were approved by the Institutional Animal Care and Use Committee of Tongji Medical College, Huazhong University of Science and Technology, and the mice were housed in the specific pathogen-free facility of the Laboratory of Animal Center, Huazhong University of Science and Technology. All experiments with mice were conducted ethically according to the Guide for the Care and Use of Laboratory Animal guidelines (Ethics approval number S2795).
Mice
The 2xFLAG-ADAD2-mCherry transgenic mice were generated as previously described (Zhou et al., 2017). Briefly, Adad2 cDNA with a 2× FLAG tag added at the 5′-end and a stop codon removed at the 3′-end was cloned from total RNA isolated from adult mouse testes. The resultant DNA fragment was inserted into the pmCherry-N1 vector (Clontech), with the promoter replaced by a 2.3 kb DNA sequence upstream of the transcription start site of Adad2. A DNA fragment containing Adad2 cDNA was injected into pronuclei of one-cell zygotes for the production of founders. Founders were genotyped by PCR using two sets of primer pairs, which were Tg-Flag-Forward and Tg-Flag-Reverse, which resulted in a 240 bp PCR product, and Tg-mCherry-Forward and Tg-mCherry-Reverse that resulted in a 355 bp PCR product.
Adad2 and Rnf17 mutant mice were generated produced by zygote pronuclear microinjection using CRISPR/Cas9 genome editing as previously described (Li et al., 2021). Two single guide RNAs targeting exon 1 and Cas9 mRNAs were mixed and injected into C57BL/6 zygotes, followed by implanted into pseudo-pregnant C57BL/6 females. Genomic DNA was extracted from founder mice, followed by Sanger sequencing confirmation and PCR analysis. Founder mice were backcrossed to C57BL/6, and F2 generation animals were genotyped for the subsequent experiments. The primers used for genotyping are listed in Table S8.
Vector construction and cell transfections
Full-length and Truncated ADAD2 expression constructs with N-terminal IDR motif absence (aa90-561), dsRNA binding domain absence (aa1–89, 160–561), and C-terminus (aa239–561) were generated by PCR amplification using mouse testis cDNA and cloned into the pCMV-Flag vector that contains an N-terminal FLAG tag. The full-length Adad2 and Rnf17 cDNAs were amplified by PCR using mouse testis cDNA and cloned into the pCMV-Flag and pCMV-Myc vectors, respectively. Each required tagged protein expression construct was transfected in NIH3T3 cells (CRL-1658; ATCC) or U2OS cells (HTB-96; ATCC) with Lipo8000 Transfection Reagent (Beyotime) according to the manufacturer’s protocol. Cells were collected after 48 h transfection, followed by immunofluorescence.
Transmission electron microscopy (TEM)
TEM was performed as our previous study described with minor modifications (Yuan et al., 2015). Briefly, adult testes from Adad2+/+ and Adad2Mut1/Mut1 were fixed in 0.1 M cacodylate buffer (pH = 7.4) supplemented 3% paraformaldehyde and 3% glutaraldehyde plus 0.2% picric acid for 2 h in at room temperature (RT). Following three washes with 0.1 M cacodylate buffer, the samples were post-fixed 0.1 M cacodylate buffer supplemented with 1% osmium tetroxide for 1 h at RT. After washing, the samples were dehydrated in sequentially ethanol solutions (30, 50, 70, 90, and 100%) and embedded in an Eponate mixture (Electron Microscopy Sciences) for polymerization. 70-nm thin sections were cut with a diamond knife. The sections were re-stained with uranyl acetate and lead citrate. Images were taken with a transmission electron microscope (G2 12; FEI Tecnai).
Histology and immunofluorescence
Mouse testes and epididymides were fixed in Bouin’s solution (Lot #SLBJ3855V; Sigma-Aldrich) overnight, dehydrated in ethanol, and embedded in paraffin. 5-μm sections were cut and stained with a periodic acid-Schiff (PAS) kit according to the manufacturer’s instructions. Immunofluorescence was performed on cryosections of testes and cultured cells, respectively. For testes, samples were fixed in 4% paraformaldehyde, dehydrated in sucrose, and embedded in O.C.T (4583; Sakura Finetek). 5-μm sections were cut, and antigen retrieval was performed. For cultured cells, samples were fixed in 4% paraformaldehyde for 15 min, followed by washing with PBS. After three times washing with PBS, the sections or cultured cells were blocked in blocking solution (PBS containing 5% normal donkey serum) for 1 h at RT. Then, tissue sections or cultured cells were incubated with primary antibodies and secondary antibodies and mounted with DAPI (H1200; Vector Laboratories) and photographed. Imaging was performed using a scanning confocal microscope (900; Zeiss) with Zen software. Depending on the experiment, we chose an appropriate lens from an oil-immersion 20 × lens or a water-immersion 40 × lens with an air-conditioned chamber maintained at 25°C was used for all microscopic experiments. Fiji (ImageJ) was used to generate images after data acquisition. Antibodies are listed in Table S9. For all line scan analysis, Fiji (ImageJ) function “Plot Profile” was used. We performed all quantitative colocalization analyses as previously described (Chung et al., 2021). All data for colocalization analysis were obtained as Z-sections with a scanning range of 10.00 and 0.2 μm spacing.
Isolation of spermatogenic cells
The pachytene spermatocytes and round spermatids were isolated by the STA-PUT method described previously with minor modifications (Xiong et al., 2017). In brief, the adult mouse testes were harvested and digested with collagenase IV (1 mg/ml). And then, the dispersed seminiferous tubules were further digested with 0.25% Trypsin containing DNase I (1 mg/ml) and filtered to prepare a single-cell suspension. The single-cell suspension was loaded into a cell separation apparatus (ProScience). After 2 h of sedimentation, cell fractions were harvested and determined according to morphological characteristics, cell diameter, and DAPI staining under a light microscope.
Analysis of RNA-seq data
The previously published RNA-seq datasets (GSE53919) were derived from Rnf17−/− spermatocytes and round spermatids for performing differential expression analyses. To identify all the transcripts, Tophat2 and Cufflinks were used to assemble the sequencing reads based on the UCSC MM10 mouse genome. The differential expression analysis was performed by DEGseq with R. The differential expressed genes were set with the threshold of FDR < 0.1 and fold change ≥ 2. Single-cell RNA-seq data of adult mouse spermatogenic cells was downloaded from NCBI GEO datasets with an accession number of GSE109033. Seurat package was used for cell filtration, normalization of data, reduction of dimensionality reduction, cell cluster, and differentially expressed gene identified (Butler et al., 2018). Dimensionality reduction was constructed by a two-dimensional t-distributed stochastic neighbor embedding (t-SNE) algorithm. Cells types were allocated to each cluster using the known marker genes as described previously (Hermann et al., 2018). All data processing is carried out in the R environment. Gene ontology (GO) analysis was performed by Metascape with a corrected P value cut off of 0.01 (Zhou et al., 2019).
Quantitative RT-PCR assay and RNA-seq
Total RNA was extracted from testes and isolated spermatogenic cells using TRIzol reagent. Total RNA (1 μg) was reverse transcribed using a high-capacity cDNA reverse transcription kit (Vazyme) according to the manufacturer’s instructions. 2 μl diluted cDNA was subjected to real-time PCR using the SYBR Q-PCR master mix (Vazyme). The primers are listed in Table S8. The relative gene expression was quantified using the comparative cycle threshold method, with the Arbp expression used for normalization. For RNA-seq, 1 μg total RNA extracted from testes or isolated spermatogenic cells was used to prepare the mRNA libraries using TruSeq Stranded mRNA Library Preparation Kit Set A (Cat. No. RS-122-2101; Illumina) according to the manufacturers’ instructions. All libraries were sequenced using the Illumina HiSeq 4000 platform. The FASTX-Toolkit was used to remove adaptor sequences, and low-quality reads from the sequencing data. Then, we used Tophat2 and Cufflinks to assemble the sequencing reads based on the UCSC MM10 mouse genome. The differential expression analysis was performed by Cuffdiff. The differential expressed genes were set with the threshold of FDR < 0.1 and fold change ≥ 2.
Immunoprecipitation (IP) and mass spectrometry (MS)
Freshly isolated testes were dissected and lysed in IP buffer (20 mM HEPES, 150 mM NaCl, 0.5% NP-40, 1 mM DTT, pH = 7.3) supplemented with phosphatase inhibitor and protease inhibitor cocktail tablets, followed by centrifugation at 12,000 rpm and the supernatant was collected, and then pre-cleared with protein A or G beads (161-4013; Bio-Rad). The lysate was incubated with primary antibodies overnight at 4°C on a rotator and conjugated with beads for 3 h at 4°C with rotation. The bead complexes were washed three times with IP buffer. For immunoblotting, the protein complex was eluted off the beads into 2 × SDS loading buffer. For the MS assay, the protein complex was eluted off the beads with SDT buffer (4% SDS, 100 mM DTT, 100 mM Tris-HCl). Eluted proteins were trypsin digested as described (Wiśniewski et al., 2009), and the peptide was performed with Q-Exactive Plus mass spectrometer (Thermo Fisher Scientific) for MS analysis.
Immunoblotting
Tissues were rinsed with PBS and lysed in cold NP40 lysis buffer supplemented with phosphatase inhibitor and protease inhibitor cocktail tablets. Tissue lysates were rotated at 4°C for 30 min and centrifuged at 11,000 rpm for 15 min at 4°C. The supernatant was collected, and protein concentration was then determined by Bicinchoninic Acid (BCA) Assay. 20 μg proteins were separated by sodium dodecylsulphate-polyacrylamide gel electrophoresis (SDS-PAGE) on 10% resolving gels. Protein loading was verified by blotting for GAPDH as indicated. The immunoblotting images were detected using the Molecular Imager Gel Doc XR system (Bio-Rad Universal Hood II).
RNA-immunoprecipitation
The testes from Adad2+/+ and Adad2Flag/mCherry mice were collected, de-tunicated, and disrupted by pipetting as previously described with minor modifications (Ding et al., 2018). Briefly, the testicular cell suspension was prepared in ice-cold PBS and subjected to 300 mJ/cm2 UV-C light (1800; Stratagene Stratalinker) three times. Then, the testicular cell suspension was homogenized in IP lysis buffer and incubated at 4°C for 1 h followed by centrifugation at 15,000 rpm for 30 min. Precleared samples were incubated with anti-FLAG antibody overnight at 4°C. After binding with protein G magnetic beads for 3 h, washed three times with wash buffer, and incubated with 1 mg/ml Proteinase K at 55°C for 30 min. RNA extraction, RT, and RT-qPCR were then performed as described above.
Small RNA libraries and bioinformatics
Total RNA extracted from isolated spermatogenic cells was used to construct small RNA libraries. Multiple libraries with different barcodes were pooled and sequenced with the Illumina HiSeq 4000 platform. Fastx_clipper was used to process sequenced reads by clipping the sequencing adapter read-through. Small RNA reads of 24–32 nt for each sample were retrieved for piRNA analyses. These reads were then aligned to piRNA clusters (Li et al., 2013), coding RNAs, non-coding RNAs, Repeats (UCSC RepeatMasker track), and intron based on genomic locations. Reads not mapping to the above sets of sequences were classified as “other.” Alignments were performed with Bowtie (one base mismatch allowed). Repeats included classes of repeats as defined by RepeatMasker.
Statistical analysis
All data are shown as mean ± SEM unless otherwise noted in the figure legends. The Shapiro–Wilk test was used for normality. Significance was tested using the two-tailed unpaired Student's t test (*P < 0.05, **P < 0.01, ***P < 0.001) using Prism 9.0 (GraphPad Software).
Online supplemental material
Fig. S1 shows the expression pattern of RNF17 during spermatogenesis. Fig. S2 shows that ADAD2 is associated with P-body in male germ cells. Fig. S3 shows ADAD2 but not ADAD1 is required to recruit RNF17 to P-bodies. Fig. S4 shows the generation of Adad2 mutants and their phenotype analyses. Fig. S5 shows the data of transcriptome and small RNA-seq in Adad2 mutants. Table S1 shows RNF17-interacting proteins identified from IP-MS. Table S2 shows ADAD2-interacting proteins identified from IP-MS. Table S3 shows the size distribution of small RNA reads. Table S4 contains the differential expression genes in Adad2 mutant testes at P20 determined by RNA-seq. Table S5 contains the differential expression genes in Adad2 mutant pachytene spermatocytes. Table S6 contains the differential expression genes in Adad2 mutant round spermatids. Table S7 contains the gene list of clusters 1 and 3 determined by scRNA-seq data. Table S8 lists the primer sequences used in this study. Table S9 lists the antibody information in this study.
Data availability
All data needed to evaluate the conclusions in the paper are present in the article and/or the supplementary materials. All RNA-seq data are deposited in the NCBI SRA (Sequence Read Achieve) database with the accession number of PRJNA834631. All the raw data of IP-MS are available via the PRIDE repository with identifiers PXD034463 (RNF17) and PXD034437 (ADAD2). All other supporting data of this study are available from the corresponding author upon reasonable request.
Acknowledgments
We are grateful for discussions with colleagues from Huazhong University Science and Technology, China, in the very initial phase of the project.
This work, in part, was supported by grants from the National Natural Science Foundation of China (82171605 and 81971444 to S. Yuan), the Science Technology and Innovation Commission of Shenzhen Municipality (JCYJ20170818160910316 to S. Yuan), and National Institute of Health grants (GM122776 and GM141085 to S.H. Namekawa.).
Author contributions: S. Yuan and M. Xiong conceived and designed the research. M. Xiong, L. Yin, Y. Gui, C. Lv, X. Ma, S. Guo, Y. Wu, S. Feng, X. Fan, S. Zhou, L. Wang, Y. Wen., X. Wang, and Q. Xie performed the bench experiments and data analyses. Y. Gui performed the bioinformatic analysis. S. Guo and X. Fan carried out the pronuclear injection. M. Xiong and S. Yuan wrote the manuscript. S. Yuan supervised the project. S.H. Namekawa revised the manuscript. All authors read and approved the manuscript.
References
Author notes
M. Xiong, L. Yin, Y. Gui, and C. Lv contributed equally to this work.
Disclosures: The authors declare no competing interests exist.




![Proteomic analysis of endogenous RNF17 in testes. (A) Volcano plot showing enrichment (log2 [mean intensity ratio of RNF17-IP/IgG-IP from WT testes]) and confidence (−log10 [P value of two-sided Student’s t test]) of proteins co-purifying with RNF17 from WT testes lysates (n = 3). The right upper corner of the dotted line area indicates factors with enrichment > fourfold and significance P < 0.05. (B) GO analysis of the most enriched RNF17-associated proteins identified in adult mouse testes. (C) RNF17, MIWI, and MILI protein interaction networks reveal that ADAD2 presents in RNF17, MIWI, and MILI complexes. (D) Co-immunoprecipitation of FLAG-ADAD2-mCherry with MIWI and MILI from lysates generated from WT and Adad2Flag/mCherry mouse testes. Immunoprecipitations were performed in the presence or absence of RNase A. (E) Western blot validation of RNF17 association with ADAD proteins. Source data are available for this figure: SourceData F2.](https://cdn.rupress.org/rup/content_public/journal/jcb/222/5/10.1083_jcb.202206067/1/m_jcb_202206067_fig2.png?Expires=1773563220&Signature=cWR7ybnfbAGu3oSX6ij3bohe2w1mc~jZ145wEhl-85igX2H6xyjScXa6tBxQLPYUDdLE7~E~U8HDL40~FdKLbP14zl7Yt1CBXQT8DWO7KE5A~3CISWWAHcCW9lTSsN4KMZZuC2a1KBPJ6qC1pEIDdirBnGO~5Q7i7DfWoW~Kp2raFhrKm-wfaxsiSdwbh5~a3xh1aB9vLeont0RtS0TTl1EsB8b7nmUglNS8FCeajqyQHk1uxMDjWMZ~JOSEFm097ig3EtcA7IctXlaS4A91TaWWg6v6W2yS7W3oTkmW9DuYXtQ18NmE3dHUPcmmHlygFq-7kzVyoK-Kjji0ML9rWw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)