Functional membrane proteins in the plasma membrane are suggested to have specific membrane environments that play important roles to maintain and regulate their function. However, the local membrane environments of membrane proteins remain largely unexplored due to the lack of available techniques. We have developed a method to probe the local membrane environment surrounding membrane proteins in the plasma membrane by covalently tethering a solvatochromic, environment-sensitive dye, Nile Red, to a GPI-anchored protein and the insulin receptor through a flexible linker. The fluidity of the membrane environment of the GPI-anchored protein depended upon the saturation of the acyl chains of the lipid anchor. The local environment of the insulin receptor was distinct from the average plasma membrane fluidity and was quite dynamic and heterogeneous. Upon addition of insulin, the local membrane environment surrounding the receptor specifically increased in fluidity in an insulin receptor-kinase dependent manner and on the distance between the dye and the receptor.
Introduction
Cellular membranes contain diverse lipid species with distinct structural and physical properties which are heterogeneously distributed in the cellular membrane, forming dynamic subcompartments with certain sets of lipids and membrane proteins. The function and structure of integral membrane proteins are affected by the physical properties of the local membrane, composition, thickness, and packing of membrane lipids, in which the membrane proteins are exposed. It has been suggested that the activities of several type of ligand-gated ion channels (daCosta and Baenziger, 2009; Kumar et al., 2020), P-type adenosine triphosphatases (Hossain and Clarke, 2019; Lopreiato et al., 2014), and aquaporin (Tong et al., 2012) are altered by cholesterol. Cholesterol regulates the thickness, packing, and fluidity of membranes as do the degree of saturation and length of lipid acyl chains (Harayama and Riezman, 2018), so lipid effects on protein function could come through direct protein lipid interactions or be exerted through biophysical properties.
Membrane lipids provide structural support to maintain the function of membrane proteins, but they also play critical roles in signal transduction. Upon stimulation, receptors often trigger local changes in lipid composition, for example through phosphatidylinositol lipid kinases (Cantley, 2002), which recruit downstream signaling molecules that are currently being investigated as potential therapeutic targets (Prestwich, 2004). However, very little is known about how these pathways, along with the conformational changes of the receptors themselves, affect the local environment of the receptor and potentially its activity. Advanced imaging techniques have been able to demonstrate that there are clusters of specific proteins and lipids in membranes showing that membranes are not completely homogenous laterally (Lingwood and Simons, 2010; Sezgin et al., 2017). For example, single-molecule imaging and fluorescent correlation spectroscopy (FCS) combined with stimulated emission depletion (STED) super-resolution microscopy evaluated the lateral diffusion of molecules of clusters and suggest the existence of nanoscale assemblies of glycosylphosphatidylinositol (GPI)-anchored proteins, cholesterol, and sphingolipids (Eggeling et al., 2009; Honigmann et al., 2014; Kinoshita et al., 2017; Kusumi et al., 2011). Homo-Förster resonance energy transfer (FRET) also revealed the cholesterol-dependent clustering of GPI-anchored proteins (Goswami et al., 2008; Raghupathy et al., 2015). However, these microscopies image the dynamics of each component of lipid-protein clusters separately and it is impossible to know whether they are truly in the same domains. In order to examine the local membrane environment surrounding a specific membrane protein, we need other techniques.
For visualization of the local membrane environment, environment-sensitive dyes are widely utilized in model membranes and in vivo. Environment-sensitive dyes sense water penetration in the membrane, exhibit a red shift in the emission spectrum depending on the local environment polarity, and membrane order is quantified by the ratio of the fluorescent intensity in two fluorescent emission regions as the generalized polarization (GP) value (Bagatolli, 2006; Gaus et al., 2006). Commonly used environment-sensitive dyes such as Laurdan, Di-4ANEPPDHQ, and NR12S, a Nile Red derivative, which has hydrophobic chains and randomly resides within the plasma membrane (Kucherak et al., 2010; Owen et al., 2011).
Previously, we directly linked an environment sensitive dye, Nile Red to target molecules using protein-labeling tags, the SNAP-tag (Keppler et al., 2003), and the acyl carrier protein (ACP)-tag (George et al., 2004) to control environment-sensitive dyes’ distributions in cells, and showed that probe had turn-on fluorescence properties that depended upon attachment to the receptor (Prifti et al., 2014) and we measured membrane potential in cells and in neurons (Sundukova et al., 2019). Nile Red has also been attached to GPCR ligands to investigate lipid microenvironments (Hanser et al., 2021).
Here, we directly visualized the local membrane environment surrounding a GPI-anchored protein and the insulin receptor in living cells upon insulin stimulation by covalently linking Nile Red to the receptor through a flexible tether and the ACP-tag. Nile Red resides in a limited area surrounding the target by our labeling technique and the photo-stability of Nile Red allows a time-lapse observation. Thus, we can garner information about the local membrane environment of the target with high spatial- and temporal-resolution. The membrane environment surrounding the GPI-anchored protein depended upon the saturation of the anchor acyl chains. We show that the membrane fluidity of the local membrane environment surrounding the insulin receptor is increased by insulin. The membrane fluidity change invoked by insulin requires the intrinsic tyrosine kinase activity of the insulin receptor. Meanwhile, changes in the membrane environment of the average plasma membrane due to insulin treatment are negligible. Our findings provide evidence that the local membrane environment of the insulin receptor behaves differently from the bulk plasma membrane.
Results
Development of a method for probing the local membrane environment surrounding the insulin receptor in the plasma membrane
We achieved specific labeling using Nile Red carrying a flexible-PEG linker with the genetically encoded ACP-tagged insulin receptor to monitor the local membrane environment of the insulin receptor. Nile Red (NR) is confined to the region surrounding the receptor by direct linkage to the receptor through a PEG-linker (Fig. 1 A). ACP-tag is a protein-labeling tag which reacts specifically with the coenzyme A (CoA) in the presence of magnesium, ACP synthase, or 4′-phosphopantetheinyl transferase (SFP) synthase. To covalently label the extracellular ACP-tagged insulin receptor (IR), we first synthesized CoA-PEG11-NR. As the membrane localization of Nile Red can be determined by the distance from the ACP-tag on the receptor to the plasma membrane and the PEG-linker length of CoA-PEG-NR (Fig. 1 B), we constructed three insulin receptors with an ACP-tag at distinct positions in the α-subunit, and the ACP was inserted between Met664 and Asp665, Gly677 and Leu678, or Lys730 and Thr731 of the receptor in constructs of PreCT-ACP-IR, 1992-ACP-IR, or 2031-ACP-IR, respectively (Fig. 1 C). Due to the inverted V-shaped structure of the ectodomain of the insulin receptor (Croll et al., 2016), we predicted that PreCT-ACP-IR would be the furthest ACP-tag to the plasma membrane among the three constructs even though the position of ACP-tag is inside the α C-terminal domain near the transmembrane domain (Fig. 1 C).
Strategy for monitoring the local membrane environment surrounding the insulin receptor and validation of membrane localization of the dye. (A) Schematic image of monitoring the local membrane environment surrounding the insulin receptor using ACP-tag and CoA-derivatized Nile Red. The probe fluorescence is extremely low in medium and when covalently attached to the ACP-tag on the receptor, then Nile Red becomes strongly fluorescent (orange). (B) Structure of CoA-PEG-NR with various PEG-lengths, n = 5, 11, 14, and 27. (C) Schematic of constructs used. Domain structures of insulin receptor α chain and the cartoon of whole insulin receptor depicting ACP-tag positions. (D) Specific labeling of ACP-insulin receptor in the plasma membrane with CoA-PEG11-NR. Cyan is DAPI staining in nucleus and magenta is CoA-PEG11-NR staining. Scale bar, 10 μm. (E) Emission spectral shift depending on distance from ACP-tag on the receptor to the plasma membrane. Cells expressing each ACP-IR were stained with CoA-PEG11-NR or NR12S and emission spectrum between 561 and 695 nm was measured from five images for 1992-ACP-IR (n = 59 ROIs), 2031-ACP-IR (n = 37), and PreCT-ACP-IR (n = 69), and from seven images for NR12S (n = 42). Normalized fluorescent intensities were plotted. (F) Sensitivity of emission spectrum to cellular-cholesterol modification. Cell expressing each ACP-IR were treated with 10 mM of cyclodextrin for 15 min or 2.5 mM of cholesterol-cyclodextrin for 1 h at 37°C followed by labeling with CoA-PEG11-NR or NR12S, then the emission spectrum was measured. Data represents the mean ± SEM from 5 images (n = 57–59 ROIs for 1992-ACP-IR, n = 36–37 for 2031-ACP-IR and n = 69–75 for PreCT-ACP-IR), and the mean ± SE from 28 images of NR12S (n = 168 ROIs for each sample).
Strategy for monitoring the local membrane environment surrounding the insulin receptor and validation of membrane localization of the dye. (A) Schematic image of monitoring the local membrane environment surrounding the insulin receptor using ACP-tag and CoA-derivatized Nile Red. The probe fluorescence is extremely low in medium and when covalently attached to the ACP-tag on the receptor, then Nile Red becomes strongly fluorescent (orange). (B) Structure of CoA-PEG-NR with various PEG-lengths, n = 5, 11, 14, and 27. (C) Schematic of constructs used. Domain structures of insulin receptor α chain and the cartoon of whole insulin receptor depicting ACP-tag positions. (D) Specific labeling of ACP-insulin receptor in the plasma membrane with CoA-PEG11-NR. Cyan is DAPI staining in nucleus and magenta is CoA-PEG11-NR staining. Scale bar, 10 μm. (E) Emission spectral shift depending on distance from ACP-tag on the receptor to the plasma membrane. Cells expressing each ACP-IR were stained with CoA-PEG11-NR or NR12S and emission spectrum between 561 and 695 nm was measured from five images for 1992-ACP-IR (n = 59 ROIs), 2031-ACP-IR (n = 37), and PreCT-ACP-IR (n = 69), and from seven images for NR12S (n = 42). Normalized fluorescent intensities were plotted. (F) Sensitivity of emission spectrum to cellular-cholesterol modification. Cell expressing each ACP-IR were treated with 10 mM of cyclodextrin for 15 min or 2.5 mM of cholesterol-cyclodextrin for 1 h at 37°C followed by labeling with CoA-PEG11-NR or NR12S, then the emission spectrum was measured. Data represents the mean ± SEM from 5 images (n = 57–59 ROIs for 1992-ACP-IR, n = 36–37 for 2031-ACP-IR and n = 69–75 for PreCT-ACP-IR), and the mean ± SE from 28 images of NR12S (n = 168 ROIs for each sample).
We first confirmed that plasma membrane staining with CoA-PEG-NR was ACP-IR expression-dependent. The plasma membrane staining with CoA-PEG11-NR was observed in ACP-IR expressing cells, whereas there was no signal of CoA-PEG11-NR detected in parent cells (Fig. 1 D).
It is essential that the Nile Red moiety is inserted into the plasma membrane upon binding to the ACP-IR in order to monitor the local membrane environment of the receptor. We next validated the membrane localization of Nile Red of CoA-PEG11-NR linked to each ACP-IR by fluorescence spectroscopy. Nile Red is a solvatochromic dye which exhibits a red shift depending on water presence in the environment, and we expected that Nile Red could reach to the plasma membrane and emit yellowish fluorescence when the distance between ACP-tag on the receptor and the plasma membrane is shorter than the linker length of CoA-PEG11-NR, whereas Nile Red of CoA-PEG11-NR linked to ACP-insulin receptor whose ACP-tag is far from the plasma membrane would interact somewhere on the receptor and emit reddish fluorescence, which was confirmed by our measurements (Fig. 1 E).
The emission maximum (λmax) of CoA-PEG11-NR linked to the site closest to the membrane, 2031-ACP-IR, was 574 nm which was comparable to that of NR12S, a Nile Red derivative with an amphiphilic anchor exclusively bound to the outer leaflet of the plasma membrane (Kucherak et al., 2010). On the other hand, CoA-PEG11-NR attached to the middle site, 1992-ACP-IR, or the furthest site, PreCT-ACP-IR, showed peaks at 637 or 646 nm, respectively (Fig. 1 E), and these are higher than the emission peak of Nile Red in liquid disordered membranes, around 620 nm (Kucherak et al., 2010). This strongly suggests that Nile Red of CoA-PEG11-NR linked to 1992-ACP- or PreCT-ACP-IR binds somewhere on the receptor rather than being in the membrane.
To further explore the membrane localization of CoA-PEG11-NR connected to ACP-IR, the sensitivity of the emission spectrum to cellular-cholesterol modification was tested. The emission peak at 574 nm was decreased and a shoulder around 620 nm was increased by cholesterol-depletion with cyclodextrin, while the peak at 574 nm was slightly increased and the shoulder peak around 620 nm was lowered by cholesterol-loading when CoA-PEG11-NR was linked to the closest 2031-ACP-IR. Similar but less-pronounced patterns were observed with NR12S. On the other hand, there was no emission spectral shift by cellular-cholesterol modification with CoA-PEG11-NR attached to the furthest PreCT-ACP-IR (Fig. 1 F). These results indicate that Nile Red of CoA-PEG11-NR responds to cholesterol concentration and therefore is in the plasma membrane when the dye is linked to 2031-ACP-IR. Interestingly, the largest changes were obtained with the 1992-ACP-IR construct. Increasing membrane cholesterol seems to increase its insertion into the membrane with a corresponding increase in the peak at 574 nm. Increasing cholesterol may affect lipid packing and membrane thickness, membrane properties which may increase the insertion or stability of insertion of the dye into the membrane under these circumstances where the tether brings the Nile Red chromophore close to the membrane.
The experiment above suggests that some portion of the NR is dynamically close enough to the membrane to insert into the bilayer when it is tethered to the 1992-ACP-IR. In order to test this, we examined the effect of PEG-linker length, using CoA-PEG 5, 14, and 27-NR. The λmax of all CoA-PEG-NR constructs attached to 2031-ACP-IR was 574 nm, consistent with the membrane localization of Nile Red regardless of the PEG-linker length (Fig. 2 A). On the other hand, Nile Red of CoA-PEG-NR linked to the middle 1992-ACP-IR exhibits large emission shifts according to the PEG-linker length. The peak of CoA-PEG5-NR was 646 nm and the peak of CoA-PEG27-NR were 574 nm similar to NR12S, suggesting that Nile Red with the shortest PEG5-linker binds the receptor, while Nile Red with the longest PEG27-linker sits in the membrane (Fig. 2 B). One can see that membrane insertion probably begins with a linker length of 11, but improves as the linker is extended, consistent with the previous explanation concerning membrane properties.
Effect of emission spectrum of CoA-PEG-NR bound ACP-IR to PEG-linker length and sensitivity test to membrane-voltage change. (A and B) Sensitivity of emission spectrum to PEG-linker length. Cells expressing 2031-ACP-IR (A) or 1992-ACP-IR (B) were labeled by CoA-PEGn-NR (n = 5, 11, 14, or 27) or NR12S and emission spectrum was analyzed (each sample: n = 75 from five images). (C) Membrane depolarization led to decreased fluorescent intensity of CoA-PEG11-NR bound 2031-ACP-IR.2031-ACP-IR (n = 9) or 1992-ACP-IR (n = 9) expressing cells labeled by CoA-PEG11-NR and the change in fluorescent intensity induced by membrane depolarization (100 mV) using patch-clump was recorded. Data are means ± SEM. Statistical significance was calculated by a two-sided t test, the change was significant ****, P < 0.0001. (D and E) Correlation between the membrane localization of dye and voltage sensitivity. (D) Cells expressing 1992-ACP-IR were stained with CoA-PEG5-, 11-, or 27-NR and the fluorescent intensity was measured upon membrane depolarization. Data represent means ± SEM. of n = 5 (PEG5), 9 (PEG11), and 18 (PEG27). Statistical significance was calculated by one-way ANOVA; ns, not significant and significant *, P < 0.05. (E) Cells expressing 2031-ACP-IR were stained with CoA-PEG5-, 11-, or 27-NR and the fluorescent intensity was measured upon membrane depolarization. Data represent means ± SEM of n = 12 (PEG5), 9 (PEG11), and 11 (PEG27). Statistical significance; ns.
Effect of emission spectrum of CoA-PEG-NR bound ACP-IR to PEG-linker length and sensitivity test to membrane-voltage change. (A and B) Sensitivity of emission spectrum to PEG-linker length. Cells expressing 2031-ACP-IR (A) or 1992-ACP-IR (B) were labeled by CoA-PEGn-NR (n = 5, 11, 14, or 27) or NR12S and emission spectrum was analyzed (each sample: n = 75 from five images). (C) Membrane depolarization led to decreased fluorescent intensity of CoA-PEG11-NR bound 2031-ACP-IR.2031-ACP-IR (n = 9) or 1992-ACP-IR (n = 9) expressing cells labeled by CoA-PEG11-NR and the change in fluorescent intensity induced by membrane depolarization (100 mV) using patch-clump was recorded. Data are means ± SEM. Statistical significance was calculated by a two-sided t test, the change was significant ****, P < 0.0001. (D and E) Correlation between the membrane localization of dye and voltage sensitivity. (D) Cells expressing 1992-ACP-IR were stained with CoA-PEG5-, 11-, or 27-NR and the fluorescent intensity was measured upon membrane depolarization. Data represent means ± SEM. of n = 5 (PEG5), 9 (PEG11), and 18 (PEG27). Statistical significance was calculated by one-way ANOVA; ns, not significant and significant *, P < 0.05. (E) Cells expressing 2031-ACP-IR were stained with CoA-PEG5-, 11-, or 27-NR and the fluorescent intensity was measured upon membrane depolarization. Data represent means ± SEM of n = 12 (PEG5), 9 (PEG11), and 11 (PEG27). Statistical significance; ns.
Recently, we have demonstrated that NR12S or CoA-PEG5- and 11-NR linked to ACP-tag anchored to the plasma membrane through a GPI were sensitive to membrane voltage change using a patch-clamp technique in HEK cells and neurons (Sundukova et al., 2019). Using this technique, we further confirmed the membrane insertion of Nile Red of CoA-PEG11-NR linked to ACP-IR by the voltage-sensitivity test. The fluorescent intensity of CoA-PEG11-NR attached to 2031-ACP-IR was decreased—3.4 mV ± 0.3% (n = 9) when the membrane voltage was increased to 100 mV; however, CoA-PEG11-NR bound 1992-ACP-IR showed no significant difference (Fig. 2 C). We also tested voltage sensitivity of CoA-Nile Red with various length PEG-linkers in cells expressing 1992-ACP-IR. The fluorescent intensity per 100 mV was decreased with the length of PEG-linker and the delta percentage changes were −0.96 ± 0.16 for PEG5 (n = 5), −1.8 ± 0.2 for PEG11 (n = 9), and −3.5 ± 0.5 for PEG27 (n = 18; Fig. 2 D). Meanwhile, no change was observed among all PEG-linker lengths NR with 2031-ACP-IR (Fig. 2 E). Taken together with the results using fluorescence spectroscopy, there is a correlation between the membrane localization of the dye and voltage sensitivity, showing that Nile Red of CoA-PEG27-NR attached to 1992-ACP-IR is indeed embedded in the membrane, whereas CoA-PEG5 interacts with the receptor itself. Consistent with the results above, Nile Red tethered with 11 PEG may partially insert into membrane.
Our system recognizes the local membrane order change between wild-type ACP-GPI and PGAP2/3 mutant ACP-GPI
Next, we examined that our system senses the membrane order change surrounding a membrane protein at the plasma membrane in physiological conditions using ACP-GPIs. CD59 is a GPI-anchored protein which has two saturated acyl chains when the GPI-lipid remodeling is properly completed. In PGAP2/3 double-mutant cells, CD59 has a saturated acyl chain and an unsaturated acyl chain in the sn1 and sn2 positions, respectively. It was suggested that wild-type (WT) CD59 is associated with ordered membranes, whereas the population of CD59 recruited to membrane ordered domains is greatly reduced in PGAP2/3 double mutant cells (Maeda et al., 2007). We measured the emission spectrum of CoA-PEG11-NR linked to ACP-CD59 in parent cells and PGAP2/3 double-mutant cells. The local membrane environment of WT ACP-GPI is more ordered than that of PGAP2/3 mutant ACP-GPI (Fig. 3 A), whereas the averaged membrane environment of the plasma membrane examined with NR12S in cells expressing WT ACP-GPI or PGAP2/3 mutant ACP-GPI was identical (Fig. 3 B). These results show that the double mutant does not affect overall membrane properties, but that the lipid anchor composition determines the local membrane environment of this GPI-anchored protein, consistent with a lipid-based sorting mechanism. These experiments provide evidence that our system is sensitive enough to monitor changes in the local membrane environment of the targeted membrane protein under physiological conditions.
Emission spectrum of ACP-GPI. (A) Our system recognizes a difference in the local membrane environment between ACP-GPI produced in WT and PGAP2/3 mutant cells. Cells transiently expressing ACP-GPI in WT or PGAP2/3 mutant cells were stained with CoA-PEG11-NR and the emission spectrum between 561 and 695 nm was measured from six images (n = 35 for WT and n = 36 for PGAP2/3 mutant). (B) The emission spectrum of the bulk plasma membrane assessed by NR12S was identical between WT ACP GPI expressing cells and PGAP2/3 mutant ACP-GPI expressing cells. The data represents mean ± SEM from six images (n = 36 ROIs for each sample).
Emission spectrum of ACP-GPI. (A) Our system recognizes a difference in the local membrane environment between ACP-GPI produced in WT and PGAP2/3 mutant cells. Cells transiently expressing ACP-GPI in WT or PGAP2/3 mutant cells were stained with CoA-PEG11-NR and the emission spectrum between 561 and 695 nm was measured from six images (n = 35 for WT and n = 36 for PGAP2/3 mutant). (B) The emission spectrum of the bulk plasma membrane assessed by NR12S was identical between WT ACP GPI expressing cells and PGAP2/3 mutant ACP-GPI expressing cells. The data represents mean ± SEM from six images (n = 36 ROIs for each sample).
The local membrane environment surrounding the insulin receptor becomes more disordered upon insulin stimulation
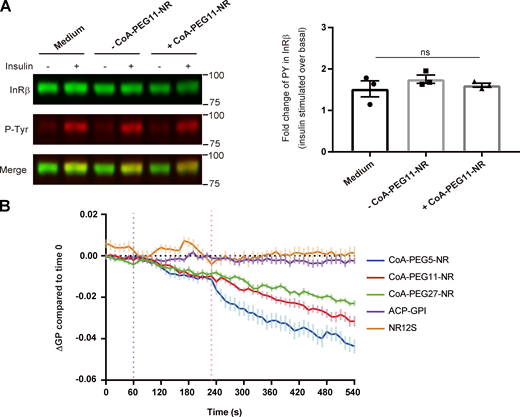
The experiments above confirmed that the Nile Red moiety of CoA-PEG-NR resides in the plasma membrane upon binding to 2031-ACP-IR by fluorescent spectroscopy and voltage-sensitivity tests. Next, we checked the activity of 2031-ACP-IR upon insulin treatment. The basal level of tyrosine phosphorylation of 2031-ACP-IR without insulin addition was similar in the presence or absence of CoA-PEG11-NR. Upon insulin addition, tyrosine phosphorylation was increased on the 2031-ACP-IR, in both unlabeled and CoA-PEG11-NR-labeled forms, showing that the modified receptor is still active and that the attachment of the CoA-PEG-NR with this long linker did not significantly affect the tyrosine kinase activity of 2031-ACP-IR (Fig. 4 A).
CoA-PEG5-NR linked ACP-insulin receptor detects changes in the local membrane environment surrounding the insulin receptor caused by insulin. (A) Tyrosine-phosphorylation of 2031-ACP-IR upon insulin in the presence or absence of attachment of PEG11-NR. 2031-ACP-IR expressing cells were serum-starved for 2 h followed by incubation with serum-free medium, medium including SFP synthase, or medium including SFP synthase and CoA-PEG11-NR for 30 min at room temperatures, then cells were stimulated by 100 nM of insulin at 37°C for 5 min and phosphorylation of 2031-ACP-IR was accessed by Western blotting analysis using α-insulin receptor β antibody (left upper bands) and α-phospho-tyrosine antibody (left middle bands). Molecular weights were indicated in the right side. Tyrosine-phosphorylation levels are presented as the mean fold increase over the basal ±SEM, n = 7–8, for each condition. Statistical significance was calculated by one-way ANOVA, ns, not significant. (B) Time course of mean delta GP change compared to time 0. After 2 h serum starvation, cell stably expressing 2031-ACP-IR were labeled with CoA-PEG5-, 11-, or 27-NR or NR12S and cells transiently expressing ACP-GPI were stained with CoA-PEG11-NR. Spectral imaging was performed every 10 s and the control solution and insulin solution were added at 60 and 240 s, respectively. GP value was calculated with two emission regions, 561–597 and 605–641 nm, and the delta GP compared to the time 0 at each time point was plotted. Data represents the mean ± SEM from 19 time-lapse image sets for CoA-PEG5-NR labeled ACP-IR, 22 for CoA-PEG11-NR-labeled ACP-IR, 17 for CoA-PEG27-NR-labeled ACP-IR, 12 for labeled with NR12S, and 19 for CoA-PEG11-NR-labeled ACP-GPI. Source data are available for this figure: SourceData F4.
CoA-PEG5-NR linked ACP-insulin receptor detects changes in the local membrane environment surrounding the insulin receptor caused by insulin. (A) Tyrosine-phosphorylation of 2031-ACP-IR upon insulin in the presence or absence of attachment of PEG11-NR. 2031-ACP-IR expressing cells were serum-starved for 2 h followed by incubation with serum-free medium, medium including SFP synthase, or medium including SFP synthase and CoA-PEG11-NR for 30 min at room temperatures, then cells were stimulated by 100 nM of insulin at 37°C for 5 min and phosphorylation of 2031-ACP-IR was accessed by Western blotting analysis using α-insulin receptor β antibody (left upper bands) and α-phospho-tyrosine antibody (left middle bands). Molecular weights were indicated in the right side. Tyrosine-phosphorylation levels are presented as the mean fold increase over the basal ±SEM, n = 7–8, for each condition. Statistical significance was calculated by one-way ANOVA, ns, not significant. (B) Time course of mean delta GP change compared to time 0. After 2 h serum starvation, cell stably expressing 2031-ACP-IR were labeled with CoA-PEG5-, 11-, or 27-NR or NR12S and cells transiently expressing ACP-GPI were stained with CoA-PEG11-NR. Spectral imaging was performed every 10 s and the control solution and insulin solution were added at 60 and 240 s, respectively. GP value was calculated with two emission regions, 561–597 and 605–641 nm, and the delta GP compared to the time 0 at each time point was plotted. Data represents the mean ± SEM from 19 time-lapse image sets for CoA-PEG5-NR labeled ACP-IR, 22 for CoA-PEG11-NR-labeled ACP-IR, 17 for CoA-PEG27-NR-labeled ACP-IR, 12 for labeled with NR12S, and 19 for CoA-PEG11-NR-labeled ACP-GPI. Source data are available for this figure: SourceData F4.
Next, we observed the local membrane environment of the insulin receptor upon insulin signaling using 2031-ACP-IR labeled with CoA-PEG5-, 11-, or 27-NR. We also observed the local membrane environment of ACP-GPI linked to CoA-PEG11-NR and the overall properties of the plasma membrane with NR12S. A GP value was calculated to assess the relative membrane order and the delta GP compared to time 0 was plotted at each time point (see Materials and methods). No major changes in membrane order were detected using NR12S upon insulin addition, indicating that the average properties of the plasma membrane did not change upon insulin binding and signaling. Likewise, the local membrane environment surrounding ACP-GPI was unchanged upon on insulin treatment as the GP value was constant through the whole observation. Strikingly, the GP value clearly dropped upon insulin treatment when the 2031-ACP-IR was labeled with CoA-PEG5-NR (Figs. 4 B and S1). This shows that the membrane environment immediately surrounding the insulin receptor became more disordered upon insulin binding and signaling, distinct from the bulk membrane. As the length of the PEG linker was increased allowing the NR to diffuse further away from the receptor, the impact of insulin treatment on the GP value was weaker, but still evident with PEG27 (Fig. 4 B). These results suggest that the changes invoked by insulin treatment happened in the immediate vicinity of the insulin receptor and that the further away the NR was positioned the less apparent the effects. These results further support that our measurements are relevant to the immediate membrane environment of the receptor and suggest that by increasing linker length that we can determine distance effects.
Comparison of emission spectra of CoA-PEG5-NR linked to 2031-ACP-IR in the time-course. Emission spectrum of a representative time-course observation of CoA-PEG5-NR linked to 2031-ACP-IR (Fig. 3 C) at time 0 s, at 230 s, before insulin treatment, and at 540 s, the last time point.
Comparison of emission spectra of CoA-PEG5-NR linked to 2031-ACP-IR in the time-course. Emission spectrum of a representative time-course observation of CoA-PEG5-NR linked to 2031-ACP-IR (Fig. 3 C) at time 0 s, at 230 s, before insulin treatment, and at 540 s, the last time point.
The actual GP change induced by insulin was small, and therefore it was difficult to explore which area of the local membrane environment surrounding the insulin receptor shows significant change after insulin treatment in pseudocolor GP images (Fig. 5). We thus employed zero-mean normalized cross-correlation (ZNCC), a similarity measure metric often used for template-matching, to visualize statistical GP changes between the time-lapse image and the averaged image of control-treatment period from 60 to 230 s. The range of ZNCC value is −1 to 1, and 1 means the perfect matching to the reference image. To take into consideration cell migration and membrane movement through the time-lapse observation, a local region was set to 19 × 19 pixels in each frame image so that the impact of cellular movement can be canceled inside the local region for calculating ZNCC (methods, ZNCC). In a ZNCC image, the pixels with the smaller ZNCC whose GP hardly changes were mapped to blue color, and bigger ZNCC changes whose GP drastically changes to red color. We could clearly see changes in the receptor membrane environment after insulin treatment in ZNCC images. Interestingly, the cellular responses to insulin treatment were quite heterogeneous. There were cells that responded strongly and rapidly to insulin, while some cells showed delayed reactions and a few cells barely responded to insulin (Fig. 6 and Fig. S2). We also noticed that receptors in certain parts of the plasma membrane changed more upon insulin stimulation. To identify receptor environments that responded intensely or hardly to insulin, the top 1,000 pixels showing biggest ZNCC changes or bottom 1,000 pixels showing smallest ZNCC were indicated in all images. The top 1,000 pixels often included regions that exhibited membrane blebbing (Fig. 7 and Video 1). ZNCC kymographs were made for regions containing the top (Fig. 6 B 1, 2, 4, 5, and 6) or bottom 1,000 pixels (Fig. 6 B 3) and the areas of 1, 2, 4 and 6 include region often forms protrusions (Fig. 6 A; and Videos 2 and 3). ZNCC kymograph clearly showed changes were triggered by insulin treatment and the local membrane environment of the insulin receptor was quite heterogeneous. Some areas exhibited significant change just after insulin stimulation (Fig. 6 B 1, 2, 4 and 6), whereas changes were observed late in the position 5 and the position 3 exhibited negligible change upon insulin stimulation (Fig. 6 B).
GP images of the local membrane environment surrounding the insulin receptor. A representative time-lapse data set of pseudocolor GP images filtering the median filter (3 × 3 pixels). The pseudocolor GP images are depicted by logarithmic scale from 10−1–100. The GP value <10−1 is displayed in the same color as 10−1; scale bar, 10 μm. This time-lapse image set was one data set utilized for delta GP change compared to time 0 in Fig. 4 B.
GP images of the local membrane environment surrounding the insulin receptor. A representative time-lapse data set of pseudocolor GP images filtering the median filter (3 × 3 pixels). The pseudocolor GP images are depicted by logarithmic scale from 10−1–100. The GP value <10−1 is displayed in the same color as 10−1; scale bar, 10 μm. This time-lapse image set was one data set utilized for delta GP change compared to time 0 in Fig. 4 B.
ZNCC images reveal that the local membrane environment of the insulin receptor is dynamic, and changes are heterogeneous. (A) A representative time-lapse data set of pseudocolor ZNCC images filtering the median filter (3 × 3 pixels). Arrowheads show areas intensely blebbing and membrane ruffling. Scale bar, 10 μm. The whole time-lapse image used for making the kymographs set is shown in Fig. S2. (B) Kymographs of chosen areas show time-course ZNCC changes. The horizontal dimension of the kymograph represents the vertically averaged ZNCC inside the rectangular area, and the vertical dimension of the kymograph represents the time (from t = 1 to t = 55) change of its averaged ZNCC. Scale bar, 10 μm.
ZNCC images reveal that the local membrane environment of the insulin receptor is dynamic, and changes are heterogeneous. (A) A representative time-lapse data set of pseudocolor ZNCC images filtering the median filter (3 × 3 pixels). Arrowheads show areas intensely blebbing and membrane ruffling. Scale bar, 10 μm. The whole time-lapse image used for making the kymographs set is shown in Fig. S2. (B) Kymographs of chosen areas show time-course ZNCC changes. The horizontal dimension of the kymograph represents the vertically averaged ZNCC inside the rectangular area, and the vertical dimension of the kymograph represents the time (from t = 1 to t = 55) change of its averaged ZNCC. Scale bar, 10 μm.
Top/bottom 1,000 pixel images of the local membrane environment surrounding the insulin receptor. A representative time-lapse data set of top/bottom 1,000-pixel images exhibiting biggest 1,000 pixels of ZNCC changes (red) and smallest 1,000 pixels of ZNCC (blue) in each ZNCC image. Arrowheads show areas of intense blebbing and membrane ruffling. Scale bar, 10 μm.
Top/bottom 1,000 pixel images of the local membrane environment surrounding the insulin receptor. A representative time-lapse data set of top/bottom 1,000-pixel images exhibiting biggest 1,000 pixels of ZNCC changes (red) and smallest 1,000 pixels of ZNCC (blue) in each ZNCC image. Arrowheads show areas of intense blebbing and membrane ruffling. Scale bar, 10 μm.
Time-lapse top/bottom 1,000-pixel images of the local membrane environment surrounding the insulin receptor. The whole time-lapse data set of PEG-Nile-Red labeled CHO cells expressing the ACP insulin receptor showing the top/bottom 1,000-pixel images exhibiting biggest (red) ZNCC changes and smallest (blue) 1,000 pixels of ZNCC in each ZNCC image (Fig. 6 A). Video corresponds to Fig. 7. Arrowheads show areas intensely blebbing and membrane ruffling. Video was acquired at 1 frame/10 s for 540 s and played back at 7 frames/s.
Time-lapse top/bottom 1,000-pixel images of the local membrane environment surrounding the insulin receptor. The whole time-lapse data set of PEG-Nile-Red labeled CHO cells expressing the ACP insulin receptor showing the top/bottom 1,000-pixel images exhibiting biggest (red) ZNCC changes and smallest (blue) 1,000 pixels of ZNCC in each ZNCC image (Fig. 6 A). Video corresponds to Fig. 7. Arrowheads show areas intensely blebbing and membrane ruffling. Video was acquired at 1 frame/10 s for 540 s and played back at 7 frames/s.
Time-lapse ZNCC images of the local membrane environment surrounding the insulin receptor. The whole time-lapse data set of pseudocolor ZNCC images of PEG-Nile Red labeled CHO cells expressing the ACP insulin receptor corresponds to Fig. 6 and Fig. S2. The control solution and insulin solution were added at 60 and 240 s, respectively. Arrowheads show areas intensely blebbing and membrane ruffling. Video was acquired at 1 frame/10 s for 540 s and played back at 7 frames/s.
Time-lapse ZNCC images of the local membrane environment surrounding the insulin receptor. The whole time-lapse data set of pseudocolor ZNCC images of PEG-Nile Red labeled CHO cells expressing the ACP insulin receptor corresponds to Fig. 6 and Fig. S2. The control solution and insulin solution were added at 60 and 240 s, respectively. Arrowheads show areas intensely blebbing and membrane ruffling. Video was acquired at 1 frame/10 s for 540 s and played back at 7 frames/s.
Time-lapse GP images of the local membrane environment surrounding the insulin receptor. The whole time-lapse data set of pseudocolor GP images of PEG-Nile Red labeled CHO cells expressing the ACP insulin receptor corresponds to Fig. 5. The control solution and insulin solution were added at 60 and 240 s, respectively. Arrowheads show areas intensely blebbing and membrane ruffling. Video was acquired at 1 frame/10 s for 540 s and played back at 7 frames/s.
Time-lapse GP images of the local membrane environment surrounding the insulin receptor. The whole time-lapse data set of pseudocolor GP images of PEG-Nile Red labeled CHO cells expressing the ACP insulin receptor corresponds to Fig. 5. The control solution and insulin solution were added at 60 and 240 s, respectively. Arrowheads show areas intensely blebbing and membrane ruffling. Video was acquired at 1 frame/10 s for 540 s and played back at 7 frames/s.
Tyrosine kinase activity is required for GP change in the local membrane environment surrounding the insulin receptor by insulin treatment
We showed the GP change caused by insulin stimulation occurred only in the vicinity of the insulin receptor, whereas neither the GP in the entire plasma membrane nor the local membrane environment of ACP-GPI was altered. These results suggest that the GP change in the immediate membrane environment surrounding the insulin receptor reflects local events catalyzed by the intrinsic tyrosine kinase activity of the receptor. To confirm this possibility, we constructed the kinase-dead (KD) mutant of 2031-ACP-IR with replacement of Lys 1045 in the ATP binding site of the β subunit by Arg (Carpentier et al., 1992; Ebina et al., 1987). Tyrosine-phosphorylation of the mutant insulin receptor was completely blocked upon insulin stimulation (Fig. 8 A). We compared the delta GP change between the WT 2031-ACP-IR and KD 2031-ACP-IR labeled with CoA-PEG5-NR. In this experiment, differential delta GP with insulin-treatment of each sample, WT and KD mutant insulin receptor, against the basal delta GP of each sample with control-treatment set to 0 is plotted. Before the insulin treatment at 120 s, the delta GP of both WT and KD insulin receptor remained at the basal level, whereas it went down immediately after the insulin stimulation in the WT but the KD stayed at pre-insulin levels (Fig. 8 B).
GP changes in the local membrane environment surrounding the insulin receptor caused by insulin require the tyrosine kinase activity of the insulin receptor. (A) Tyrosine-phosphorylation of the KD mutant insulin receptor was inhibited upon insulin. Cells stably expressing the WT 2031-ACP-IR (left) or the KD mutant 2031-ACP-IR (right) were serum-starved for 2 h, then treated with 100 nM of insulin at 37°C for 5 min. Tyrosine-phosphorylation of ACP-IR was analyzed by Western blotting using α-insulin receptor β antibody (upper bands) and α-phospho-tyrosine antibody (lower bands). Molecular weights were indicated in the right side. (B) The differential delta GP suggested that the local membrane environment of the KD mutant insulin receptor showed no GP change by insulin treatment (red), whereas GP in the local membrane environment surrounding the WT insulin receptor dropped after insulin stimulation (blue). Stably expressing the WT 2031-ACP-IR or the KD 2031-ACP-IR cells were serum-starved for 2 h, followed by labeling with CoA-PEG5-NR, then spectral imaging was performed and the control solution or insulin solution were added at 120 s. The differential delta GP with insulin treatment of both WT and KD over the basal control-treated delta GP of each sample was plotted. The basal control-treated delta GP was estimated as the mean delta GP of 11 time-lapse data sets for both WT and KD with. Data represent means ± SEM of 13 time-lapse data sets for WT and 15 for KD. Significant change between WT and KD was statistically analyzed; during 0–120 s, ns, after the insulin treatment at 120 s, the change was significant *, P < 0.05. (C) The differential delta GP was analyzed in cells stably expressing WT 2031-ACP-IR in the presence of DMSO (control), 0.2 μM Latrunculin B, or 50 μM LY294002. The basal control-treated delta GP was calculated as the mean of 8 time-lapse data for control, 11 for Latrunculin B, and 9 for LY294002. Data present means ± of 8 time-lapse data for control, 11 for Latrunculin B, and 10 for LY294002. (D) AKt-phosphorylation upon insulin stimulation in the presence or absence of LY294002. 2031-ACP-IR-expressing cells were serum-starved for 2 h followed by incubation with serum-free medium, medium including 50 μM of LY294002 or DMSO for 10 min at 37°C, then cells were stimulated by 100 nM of insulin at 37°C for 5 min and phosphorylation of Akt was accessed by Western blotting analysis using α-Akt antibody (upper bands) and α phospho-Akt antibody (second upper bands). Actin was used as a loading control (lower bands). (E) Actin cytoskeleton was disrupted by Latrunculin B treatment. Cells plated on glass-bottom dishes were stained with Sir-actin at 37°C for 1 h, followed by washing excess Sir-actin, then cells were treated by DMSO (upper) or 0.2 μM of Latrunculin B (lower) at 37°C for 10 min. Left pictures are Nomarski images and right pictures are images of Sir-actin. Scale bar, 10 μm.
GP changes in the local membrane environment surrounding the insulin receptor caused by insulin require the tyrosine kinase activity of the insulin receptor. (A) Tyrosine-phosphorylation of the KD mutant insulin receptor was inhibited upon insulin. Cells stably expressing the WT 2031-ACP-IR (left) or the KD mutant 2031-ACP-IR (right) were serum-starved for 2 h, then treated with 100 nM of insulin at 37°C for 5 min. Tyrosine-phosphorylation of ACP-IR was analyzed by Western blotting using α-insulin receptor β antibody (upper bands) and α-phospho-tyrosine antibody (lower bands). Molecular weights were indicated in the right side. (B) The differential delta GP suggested that the local membrane environment of the KD mutant insulin receptor showed no GP change by insulin treatment (red), whereas GP in the local membrane environment surrounding the WT insulin receptor dropped after insulin stimulation (blue). Stably expressing the WT 2031-ACP-IR or the KD 2031-ACP-IR cells were serum-starved for 2 h, followed by labeling with CoA-PEG5-NR, then spectral imaging was performed and the control solution or insulin solution were added at 120 s. The differential delta GP with insulin treatment of both WT and KD over the basal control-treated delta GP of each sample was plotted. The basal control-treated delta GP was estimated as the mean delta GP of 11 time-lapse data sets for both WT and KD with. Data represent means ± SEM of 13 time-lapse data sets for WT and 15 for KD. Significant change between WT and KD was statistically analyzed; during 0–120 s, ns, after the insulin treatment at 120 s, the change was significant *, P < 0.05. (C) The differential delta GP was analyzed in cells stably expressing WT 2031-ACP-IR in the presence of DMSO (control), 0.2 μM Latrunculin B, or 50 μM LY294002. The basal control-treated delta GP was calculated as the mean of 8 time-lapse data for control, 11 for Latrunculin B, and 9 for LY294002. Data present means ± of 8 time-lapse data for control, 11 for Latrunculin B, and 10 for LY294002. (D) AKt-phosphorylation upon insulin stimulation in the presence or absence of LY294002. 2031-ACP-IR-expressing cells were serum-starved for 2 h followed by incubation with serum-free medium, medium including 50 μM of LY294002 or DMSO for 10 min at 37°C, then cells were stimulated by 100 nM of insulin at 37°C for 5 min and phosphorylation of Akt was accessed by Western blotting analysis using α-Akt antibody (upper bands) and α phospho-Akt antibody (second upper bands). Actin was used as a loading control (lower bands). (E) Actin cytoskeleton was disrupted by Latrunculin B treatment. Cells plated on glass-bottom dishes were stained with Sir-actin at 37°C for 1 h, followed by washing excess Sir-actin, then cells were treated by DMSO (upper) or 0.2 μM of Latrunculin B (lower) at 37°C for 10 min. Left pictures are Nomarski images and right pictures are images of Sir-actin. Scale bar, 10 μm.
The activation of the insulin receptor tyrosine kinase phosphorylates the insulin receptor and the insulin receptor substrate proteins, leading to the recruitment of phosphatidylinositol 3-kinase (PI3K) to the receptor, then phosphatidylinositol (3,4,5) triphosphate (PIP3) is produced (Haeusler et al., 2018). Therefore, we examined the effect of PIP3 production on the GP change induced by insulin treatment using LY294002, a PI3K inhibitor. The phosphorylation of Akt which is downstream of PI3K was completely blocked by LY294002 treatment (Fig. 8 D); however, the GP change induced by insulin was similar to the untreated control (Fig.8 C). Therefore, it is unlikely that PIP3 production is required for these rapid changes in the local membrane environment.
The actin cytoskeleton is considered to be an important factor that can influence membrane organization (Fritzsche et al., 2017). To explore the involvement of actin cytoskeleton in the GP drop evoked by insulin, we perturbed the actin cytoskeleton using Latrunculin B. The disruption of filamentous actin structures by Latrunculin B (Fig. 8 E) partially prevented the GP decrease in response to insulin (Fig. 8 C), suggesting that actin may play a role in these rapid local membrane environment changes.
Insulin receptor is internalized through the clathrin-dependent endocytic pathway upon insulin binding (Haeusler et al., 2018). To test whether the GP change upon insulin stimulation is caused by transition of the receptor to the clathrin-coated pits, the degree of colocalization between AP2 clathrin adaptor and 2031-ACP-IR was analyzed by the Mander’s correlation coefficient (MCC; Fig. 9, C and D) or Spearman’s rank correlation coefficient (SRCC; Fig. 9 E). As raw data of AP2-mEGFP and 2031-ACP-IR labeled with CoA-Cy3 contain lots of noise (Fig. 9 A), we used segmented images of AP2 and ACP-IR for both colocalization analyses (Fig. 9 B). The value of MCC and SRCC was constant through the observation even after insulin treatment. These results indicate that GP change upon insulin stimulation was not triggered by translocation of the insulin receptor to the clathrin-coated pits by insulin treatment.
Colocalization of clathrin-coated pits and 2031-ACP-IR upon insulin stimulation. (A) 2031-ACP-IR stable expressing cells transfected with AP2-mEGFP plasmid were serum-starved for 2 h and cells were labeled with CoA-Cy3, then time-lapse imaging was performed every 1 min after adding control solution for 3 min and 100 nM insulin solution for 3 min. Left; AP2-mEGFP, middle; 2031-ACP-IR labeled with CoA-Cy3, right; merge. Scale bar, 10 μm. (B) Examples of segmented images of AP2 (left) and ACP-IR (right) in cell1, cell2, and cell3 showed in A). (C–E) Segmented images were used for colocalization analysis in C, D, and E. Scale bar, 10 μm. Colocalization between AP2 clathrin adaptor and 2031-ACP-IR was analyzed by MCC (C and D) or SRCC (E). Data represent the mean ± SEM of six time-lapse data sets (n = 20 cells). Statistical significance was calculated by one-way ANOVA, ns, not significant. (C) The MCC estimated as the ratio of the AP2 colocalized with ACP-IR over the total fluorescent of AP2. (D) The MCC estimated as the ratio of the ACP-IR colocalized with AP2 over the total fluorescent of ACP-IR.
Colocalization of clathrin-coated pits and 2031-ACP-IR upon insulin stimulation. (A) 2031-ACP-IR stable expressing cells transfected with AP2-mEGFP plasmid were serum-starved for 2 h and cells were labeled with CoA-Cy3, then time-lapse imaging was performed every 1 min after adding control solution for 3 min and 100 nM insulin solution for 3 min. Left; AP2-mEGFP, middle; 2031-ACP-IR labeled with CoA-Cy3, right; merge. Scale bar, 10 μm. (B) Examples of segmented images of AP2 (left) and ACP-IR (right) in cell1, cell2, and cell3 showed in A). (C–E) Segmented images were used for colocalization analysis in C, D, and E. Scale bar, 10 μm. Colocalization between AP2 clathrin adaptor and 2031-ACP-IR was analyzed by MCC (C and D) or SRCC (E). Data represent the mean ± SEM of six time-lapse data sets (n = 20 cells). Statistical significance was calculated by one-way ANOVA, ns, not significant. (C) The MCC estimated as the ratio of the AP2 colocalized with ACP-IR over the total fluorescent of AP2. (D) The MCC estimated as the ratio of the ACP-IR colocalized with AP2 over the total fluorescent of ACP-IR.
Discussion
We have established a novel method to visualize the local membrane environment surrounding plasma membrane proteins with a focus on the insulin receptor and have examined its changes due to insulin signaling. As a control protein, we used a GPI-anchored protein and provided evidence that its membrane environment depended upon anchor acyl chain saturation, providing strong evidence for lipid-based sorting of GPI-anchored proteins. In our method, the insulin receptor is covalently tethered to an environment-sensitive dye, Nile Red, using a flexible PEG-linker and the ACP tag. The hydrophilicity of CoA and PEG-linker prevents the dye from crossing the plasma membrane and this is critical for specific labeling of the ACP-IR with CoA-PEG-NR in the plasma membrane (Prifti et al., 2014). Additionally, the enzymatic procedure for attachment of the fluorescent reporter to the ACP tag requires addition of an enzyme, further restricting the labeling to the extracellular face of the plasma membrane. To monitor the local membrane environment surrounding the receptor in the plasma membrane, the Nile Red was linked to ACP-IR through a flexible tether allowing the NR to reside in the plasma membrane. To fulfill this requirement, the ACP-tag on the ectodomain of the receptor needs to be located close enough to the plasma membrane that the length of PEG-linker can be adjusted to bridge this critical distance. We provided several lines of evidence to prove that the NR was embedded in the membrane. First, the λmax of Nile Red was around 574 nm corresponding to the expected values for NR in ordered membranes (580 nm) as opposed to disordered membranes (610 mm; Kucherak et al., 2010) similar to that seen previously and with the membrane probe NR12S. Second, the fluorescence spectrum of the NR was sensitive to changes in amounts of cholesterol. Third, the NR was sensitive to a change in membrane potential using the patch clamp technique. Furthermore, when the probe was not predicted to reach the membrane, with the PreCT-ACP-IR, the fluorescence spectrum peaked at 646 nm indicating that the NR was not membrane-embedded and accordingly was insensitive to cholesterol depletion or addition. It is perhaps a bit surprising that the NR fluorescence was detected using all of our constructs as NR is normally only very weakly fluorescent in aqueous solution. In order to compare the fluorescent intensity of Nile Red between the plasma membrane-inserted state and the receptor-bound state, we further measured the fluorescent intensity of CoA-PEG11-NR linked to 1992-ACP-IR, 2031-ACP-IR, or PreCT-ACP-IR. As the expression level of ACP-IR is different among stably expressing ACP-IR cell lines, ACP-IR was double-stained with CoA-PEG11-NR through the ACP-tag and anti-insulin receptor β antibody, then the fluorescent intensity of Nile Red was normalized to the fluorescence intensity of immunostaining of the insulin receptor. The normalized fluorescent intensity of Nile Red in the receptor-bound state, PreCT-ACP-IR, and the plasma membrane–remaining state, 2031-ACP-IR, was comparable (Figs. S3). Two reasons could explain why the fluorescence labeling intensities of all constructs were similar. It could be that the Nile Red fluorescence increased by non-covalent binding to proteins or it could be increased by the covalent attachment in our system. To test the former, we measured relative quantum yield (QY) of Nile Red in 1-Palmitoyl-2oleoyl-sn-glycero-3-phosphpcholine (POPC) small unilamellar vesicles (SUV), PBS, or NR attached to BSA. The QY of NR in POPC SUV was substantially larger than the QY of NR with BSA (Table S2) consistent with the increase of quantum yield in a hydrophobic environment but showing that non-covalent interaction with BSA does not increase quantum yield. Therefore, we conclude that the increase in fluorescence intensity with these turn-on probes is due to the covalent attachment. One of the proposed reasons for low fluorescence yield in an aqueous environment is dye aggregation. When aggregation is prevented with cyclodextrin, the emission spectrum (Ray et al., 2019) is quite similar to what we see with the PreCT-ACP-IR-bound NR, consistent with the protein bound NR being in an aqueous environment. Therefore, once our probes are attached to the insulin receptor, they cannot aggregate as only one dye can be attached per polypeptide and the difference between insertion into the membrane or not can be judged by examination of the emission spectrum or the other methods that we used to confirm dye insertion. Two limitations of our techniques are the small dynamic range offered by the Nile Red and the difficulty to rule out the effect of changes in protein conformation on the probe, although the latter is probably not applicable to the GPI-anchored protein.
Comparison of the fluorescent intensity of CoA-PEG11-NR linked to 1992-ACP-IR, 2031-ACP-IR, or PreCT-ACP-IP. Top: Images of double staining of PreCT-ACP-IR, 1992-ACP-IR, or 2031-ACP-IR stably expressing cells with CoA-PEG11-NR (left), and with anti-insulin receptor mouse monoclonal antibody and Cy2-conjugated anti mouse secondary antibody (right). Small circles on the plasma membrane are ROIs for measurement of the fluorescent intensity of Cy2 and Nile Red. Bottom: The fluorescent ratio of Nile Red and Cy2 was calculated for PreCT-ACP-IR (n = 169 ROIs), 1992-ACP-IR (n = 53) and 2031-ACP-IR (n = 171). Data represent the mean ± SEM. Statistical significance was calculated by one-way ANOVA, ns, not significant, and ****, P < 0.0001.
Comparison of the fluorescent intensity of CoA-PEG11-NR linked to 1992-ACP-IR, 2031-ACP-IR, or PreCT-ACP-IP. Top: Images of double staining of PreCT-ACP-IR, 1992-ACP-IR, or 2031-ACP-IR stably expressing cells with CoA-PEG11-NR (left), and with anti-insulin receptor mouse monoclonal antibody and Cy2-conjugated anti mouse secondary antibody (right). Small circles on the plasma membrane are ROIs for measurement of the fluorescent intensity of Cy2 and Nile Red. Bottom: The fluorescent ratio of Nile Red and Cy2 was calculated for PreCT-ACP-IR (n = 169 ROIs), 1992-ACP-IR (n = 53) and 2031-ACP-IR (n = 171). Data represent the mean ± SEM. Statistical significance was calculated by one-way ANOVA, ns, not significant, and ****, P < 0.0001.
The effect of insulin on GP decrease is augmented by phospholipase C inhibitor, U73122. Cells stably expressing WT 2031-ACP-IR cultured on glass bottom dishes for 2 d were labeled with CoA-PEG5-NR. The excess dye was washed, 1 ml of 37°C serum-free medium containing 15 mM Hepes was added to the dish, and then the dish was placed on the stage of a Zeiss LSM 780 confocal microscope warmed at 37°C 5 min before the observation. 1 min before observation, 1 μl of DMSO (control) or 1 μl of 1 mM U73122 (final concentration, 1 μM) was added to the dish. Spectrum images in the range of 561–695 nm were captured with 15 channels (step 8.9 nm, 4 times line-average) using lambda mode excited at 514 nm with 1.5% of Argon laser power every 10 s for 7 min, and 100 μl of serum-free medium (control) or insulin in serum-free medium (final concentration, 100 nM) was added to the dish at 120 s. The differential delta GP with insulin treatment of both control and U732122 over the basal control-treated delta GP of each sample was plotted. The basal control-treated delta GP was estimated as the mean delta GP of 8 time-lapse data sets for both control and U732122. Data represent means ± SEM of eight time-lapse data sets for each.
The effect of insulin on GP decrease is augmented by phospholipase C inhibitor, U73122. Cells stably expressing WT 2031-ACP-IR cultured on glass bottom dishes for 2 d were labeled with CoA-PEG5-NR. The excess dye was washed, 1 ml of 37°C serum-free medium containing 15 mM Hepes was added to the dish, and then the dish was placed on the stage of a Zeiss LSM 780 confocal microscope warmed at 37°C 5 min before the observation. 1 min before observation, 1 μl of DMSO (control) or 1 μl of 1 mM U73122 (final concentration, 1 μM) was added to the dish. Spectrum images in the range of 561–695 nm were captured with 15 channels (step 8.9 nm, 4 times line-average) using lambda mode excited at 514 nm with 1.5% of Argon laser power every 10 s for 7 min, and 100 μl of serum-free medium (control) or insulin in serum-free medium (final concentration, 100 nM) was added to the dish at 120 s. The differential delta GP with insulin treatment of both control and U732122 over the basal control-treated delta GP of each sample was plotted. The basal control-treated delta GP was estimated as the mean delta GP of 8 time-lapse data sets for both control and U732122. Data represent means ± SEM of eight time-lapse data sets for each.
The experiments with the 1992-ACP-IR revealed other aspects and a potential use of our technology. At short PEG-linker lengths, the NR is not embedded in the membrane, but as the linker length increases the NR inserts into the plasma membrane and fluoresces in a similar manner as when attached to the 2032-ACP-IR (Fig. 2, A and B). Thus, by varying the linker length, we can bridge the distance between the site of attachment on the protein and the membrane, meaning that our probes can function as molecular rulers to measure the distance between sites on membrane proteins and the membrane. We also saw that with increased linker length the changes in membrane properties due to insulin signaling were reduced (Fig. 4 B), allowing us to measure distance effects with our probes. Interestingly, when the CoA-PEG11-NR was linked to the 1992-ACP-IR we found two emission peaks, a major peak at 637 nm and a minor peak at 574 nm, indicating the majority of Nile Red was bound to the receptor and a part of Nile Red was in the membrane (Fig. 1 E). Interestingly, the two peaks became approximately equal when cholesterol-loading of the membrane was performed. When cholesterol was depleted with cyclodextrin the membrane bound peak was virtually eliminated (Fig.1 F). These results showed that the amount of Nile Red of CoA-PEG11-NR attached to the 1992-ACP-IR that was inserted into the plasma membrane was increased by cholesterol-loading and decreased by cholesterol-depletion. Two explanations are possible to account for this behavior. First, the membrane thickness could have been increased by the cholesterol content in the membrane bringing it closer to the Nile Red. Second, the increased cholesterol content of the membrane could have stabilized the dynamic insertion of the Nile Red into the membrane. In any event, the experiments with the various insertion sites and linker lengths suggest that our probes might also be useful as molecular rulers to map the distance between sites in membrane proteins and the membrane.
Our results observed using the 2031-ACP-IR labeled by CoA-PEG-NR with various PEG-linker length showed that the GP value in the local membrane environment surrounding the insulin receptor was decreased upon insulin stimulation, and the biggest impact of insulin treatment was obtained by CoA-PEG5-NR, which has the shortest PEG linker. Moreover, neither the entire plasma membrane stained by NR12S nor the local membrane environment surrounding the ACP-GPI labeled with CoA-PEG11-NR were altered by insulin (Fig. 4 B). These results suggest that the change of membrane environment caused by insulin occurs only in the vicinity of the insulin receptor. The changes in the local membrane environment induced by insulin were completely blocked in the KD receptor (KD 2031-ACP-IR. Fig. 8 B), demonstrating that the local membrane environment change requires the intrinsic tyrosine kinase upon insulin binding and probably depends upon signaling. This result also supports the notion that the change in fluorescence of the probe upon insulin binding is not solely due to the conformational changes in the receptor, because this should still occur in the kinase-dead mutant. The insulin receptor is a disulphide-linked (αβ)2 homodimer, comprised of heterodimers containing α- and β-subunit, and the insulin receptor undergoes the conformation changes by insulin binding. Detailed conformation changes of the whole insulin receptor driven by insulin have been proposed by single-particle electron microscopy (Gutmann et al., 2018; Scapin et al., 2018). The symmetric ectodomain of the insulin receptor is converted from an inverted U-shaped conformation into a T-shaped conformation upon insulin binding that brings the well-separated two transmembrane domains together, facilitating autophosphorylation of the tyrosine kinase domains in the cytoplasmic tail. To understand the GP decrease in the local membrane environment of the insulin receptor caused by insulin, we compared the emission spectrum of CoA-PEG5-NR linked to the 2031-ACP-IR at time 0, at 230 s, just before the insulin treatment and 540 s, the last time point after insulin treatment. The emission spectra of 0 and 230 s were identical, whereas the peak around 580 nm was decreased in the emission spectrum of 540 s (Fig. S1). This indicates that the local membrane environment surrounding the insulin receptor gets more disordered upon insulin stimulation. The conformational change invoked by insulin may disturb the membrane layer and allow water penetration surrounding the insulin receptor. Certain conformational changes of the insulin receptor are independent of kinase activity, whereas others are dependent on autophosphorylation (Baron et al., 1992). Since the KD mutant could not induce the changes in membrane environment upon insulin binding, either the kinase-dependent conformational change or insulin receptor signaling is required for these changes.
To identify the areas that showed clear GP change in the local membrane environment surrounding the insulin receptor, we employed ZNCC. Due to the local region 19 × 19 pixels, which was slid through each image, our ZNCC analysis succeeded in demonstrating a time-dependent statistical GP-change and ruling out morphological changes like membrane blebbing and ruffling. The plasma membrane in the positions 1 and 2 (Fig. 6 B) showed membrane blebbing and ruffling through the whole observation, but ZNCC changes were observed after insulin pulse at 240 s. Moreover, our top/bottom 1,000 images were particularly striking examples that the level of the response to insulin was spatially heterogeneous in the membrane. ZNCC kymographs revealed that the response of the local membrane environment surrounding the insulin receptor to insulin was temporally and spatially heterogeneous. Certain regions of the plasma membrane including membrane blebs showed clear changes upon insulin stimulation and the timing of response to the insulin varied depending on regions (Figs. 6, 7, and S2). Thus, our technique might be able to resolve local insulin effects in different plasma membrane domains.
Our results using a PI3K inhibitor also demonstrate that the GP change in the local membrane environment of the receptor is not due to PIP3 production surrounding the receptor nor the downstream signaling activated by PI3K after insulin stimulation (Fig. 8 C), but we cannot rule out that the local membrane environment change could be caused by gathering of the substrate, phosphatidyl inositol (4,5) bisphosphate (PI(4,5)P2), to the receptor before conversion to PIP3. Indeed, treatment with U73122, an inhibitor of phospholipase C (PLC) which cleaves PI(4,5)P2 into diacylglycerol and inositol 1,4,5-triphosphate, leading to an increase of the amount of PI(4,5)P2 in the membrane, intensified the GP decrease upon insulin stimulation (Fig. 4; Neacsu et al., 2020). PI(4,5)P2 binds to many actin-binding proteins and regulates actin polymerization (Mandal, 2020). Enhancement of the effect of insulin treatment on GP change by U73122 could be related to actin cytoskeleton dynamics. The actin cytoskeleton was required for the optimal insulin-driven GP change in the local membrane environment of the insulin receptor (Fig. 8 C). It has been reported that the insulin receptor is compartmentalized in plasma membrane domains enriched in cytoskeletal components such as microvilli or the caveolae neck in the plasma membrane and relocates to be internalized upon insulin stimulation (Carpentier, 1993; Carpentier et al., 1992; Foti et al., 2007). The direct interaction of insulin receptor with actin cytoskeleton was observed in 3T3-L1 adipocyte (Foti et al., 2007), meanwhile the indirect interaction through actin-binding protein, filamin, was suggested in HegG2 cells (He et al., 2003). Taken together with these findings, the actin cytoskeleton might be necessary for maintenance and formation of the specific membrane environment of the insulin receptor in insulin signaling. The local membrane environment of the insulin receptor could be mediated by other mechanisms involving actin filaments. The insulin receptor has been suggested to form actin-dependent clusters upon insulin stimulation (Winter et al., 2012) and actin also affects clustering of GPI-anchored proteins (Goswami et al., 2008; Raghupathy et al., 2015). Furthermore, membrane properties are affected by self-organizing actin patterns (Fritzsche et al., 2017). Actin might play multiple roles in the regulation of the local membrane environment of the insulin receptor during signaling.
Strong laser power induces GP decrease and masks the effect of insulin in the local membrane environment of the insulin receptor but not ACP-GPI. Time course of mean delta GP change compared to time 0. (A–C) After 2 h serum starvation, cells stably or transiently expressing 2031-ACP-IR were labeled with CoA-PEG5-NR (A and B), or cells transiently expressing ACP-GPI were labeled with CoA-PEG11-NR (C). Spectral imaging was performed using different laser powers. Control solution or insulin solution were treated at 120 s (A; weaker laser power) or at 180 s (B; stronger laser power). Control solution and insulin solution were added at 60 and 240 s, respectively (C). Data represent the mean ± SEM from 11 time-lapse image sets for control, 13 for insulin of weaker laser powers (A), 10 for both control and insulin of lower laser power (B), and 9 for 1.5% of lower laser power and 8 for 4% laser power (C).
Strong laser power induces GP decrease and masks the effect of insulin in the local membrane environment of the insulin receptor but not ACP-GPI. Time course of mean delta GP change compared to time 0. (A–C) After 2 h serum starvation, cells stably or transiently expressing 2031-ACP-IR were labeled with CoA-PEG5-NR (A and B), or cells transiently expressing ACP-GPI were labeled with CoA-PEG11-NR (C). Spectral imaging was performed using different laser powers. Control solution or insulin solution were treated at 120 s (A; weaker laser power) or at 180 s (B; stronger laser power). Control solution and insulin solution were added at 60 and 240 s, respectively (C). Data represent the mean ± SEM from 11 time-lapse image sets for control, 13 for insulin of weaker laser powers (A), 10 for both control and insulin of lower laser power (B), and 9 for 1.5% of lower laser power and 8 for 4% laser power (C).
Our system should be applicable to examine the local membrane environment of other membrane proteins in the plasma membrane. To observe the local membrane environment of other membrane proteins using our system, consideration should be given to the location of the ACP-tag on the protein and PEG-linker length of CoA-PEG-NR to make sure that the distance to the membrane can be breached. Of course, structural information on the protein of interest would facilitate this strategy, but our technology also has potential to obtain information of protein-membrane distances independent of prior structural information. Care should be taken to ensure that the protein constructs made are functional. In addition, meticulous attention to excitation laser power for time-lapse imaging with our system is required to see clear GP change in the local membrane environment surrounding the insulin receptor upon insulin stimulation. GP was decreased by exposure to excitation laser in a laser-power dependent manner with 2031-ACP-IR labeled by CoA-PEG5-NR (Fig. S3, A and B), but no significant change was observed with ACP-GPI labeled by CoA-PEG11-NR between 1.5% laser power excitation and 4% laser power (Fig. S5 C). Strong laser flash may somehow disorder the local membrane environment of the insulin receptor through the receptor activity.
Materials and methods
Materials
NR12S was a gift from Dr. Klymchenko (Kucherak et al., 2010). SFP synthase was purchased from New England Biolabs. Methyl β cyclodextrin was purchased from CycloLab. Cholesterol-water soluble, human reconstitute insulin, Complete EDTA-free protease inhibitor cocktails, verapamil, dioxane, bovine serum albumin, Nile Red, and Latrunculin B were purchased from Sigma-Aldrich. Pierce BCA Protein Assay Kit, NuPAGE 4–12% Bis-Tris Protein gel and LY294002 was purchased from Thermo Fisher. Phosphatase inhibitor cocktail set V was purchased from Calbiochem. Sir-Actin was purchased from Spirochrome. Anti-insulin receptor β rabbit polyclonal antibody (sc-711) and mouse monoclonal antibody (sc-57342) were purchased from SantaCruz. Anti-Akt mouse monoclonal antibody (Cat. no. 2920), anti-phospho-Akt rabbit monoclonal antibody (Cat. no. 4060) and anti-phospho tyrosine rabbit monoclonal antibody (Cat. no. 8954) were purchased from Cell Signaling Tech. Anti-phospho-tyrosine mouse monoclonal antibody (PY20) was purchased from BD Transduction Lab. IRDye 800CW donkey anti-mouse antibody and IRDye 680RD donkey anti-rabbit antibody were purchased from LI-COR biosciences. Goat-anti-mouse IgG H&L (Cy2; ab6944) was purchased from Abcam. 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphpcholine (POPC; 850457C) was purchased from Avanti Polar Lipids. Methanol was from VWR and phosphate buffered saline (PBS; 10010-015) was from Gibco. U73122 was purchased from Selleck Biotech.
Synthesis of CoA Nile Red compounds
CoA-PEG Nile Red compounds were synthesized by the same procedures as previously (Sundukova et al., 2019). All chemical reagents and anhydrous solvents for synthesis were purchased from commercial suppliers (Sigma-Aldrich, Fluka, Acros) and were used without further purification or distillation. The composition of mixed solvents is given by the volume ratio (vol/vol). 1H magnetic resonance (NMR) spectra were recorded on a Bruker DPX 400 (400 MHz) or Bruker AVANCE III 400 Nanobay (400 MHz) with chemical shifts (δ) reported in ppm relative to the solvent residual signals of CD3OD (3.31 ppm for 1H) or DMSO-d6 (2.50 ppm for 1H). Coupling constants are reported in Hz. High-resolution mass spectra (HRMS) were measured on a Micromass Q-TOF Ultima spectrometer with electrospray ionization (ESI) or LTQ Orbitrap ELITE ETD (Thermo Fisher Scientific). Preparative RP-HPLC was performed on a Dionex system equipped with an UVD 170U UV-Vis detector for product visualization on a Waters SunFire Prep C18 OBD 5 µm 10 × 150 mm Column (buffer A: 0.1% TFA in H2O; buffer B: acetonitrile. Typical gradient was from 0 to 100% B within 30 min with 4 ml/min flow).
Nile Red-PEG5-COOH 2
Fmoc-PEG5-COOH (6.9 mg, 12 µmol) was dissolved in 0.5 ml MeCN/piperidine 9:1 and the solution was stirred for 20 min at room temperature. The solvents were evaporated, the residue was coevaporated three times with MeCN, and dried under high vacuum. Separately, Nile Red-C6-COOH 1 (5.4 mg, 12 µmol) was dissolved in 0.25 ml DMSO, treated with DIPEA (4.1 µl, 24 µmol) and TSTU (3.6 mg, 12 µmol; Jose and Burgess, 2006). The solution was incubated for 30 min at room temperature and was transferred to a vial that contained the above deprotected PEG linker. After 1 h, the reaction was subjected to RP-HPLC and the product was lyophilized. Yield: 4.7 mg (50%). 1H NMR (400 MHz, MeOD) δ 8.09 (d, 1 H, J = 8.8 Hz), 8.00 (d, 1 H, J = 2.5 Hz), 7.63 (d, 1 H, J = 9.2 Hz), 7.18 (dd, 1 H, J = 2.5, 8.8 Hz), 6.92 (dd, 1 H, J = 2.6, 9.3 Hz), 6.64 (d, 1 H, J = 2.6 Hz), 6.27 (s, 1 H), 4.18 (t, 2 H, J = 6.3 Hz), 3.72 (t, 2 H, J = 6.3 Hz), 3.66-3.54 (26 H), 3.39 (t, 2 H, J = 5.4 Hz), 2.55 (t, 2 H, J = 6.3 Hz), 2.30 (t, 2 H, J = 7.3 Hz), 1.92 (m, 2 H), 1.77 (m, 2 H), 1.65-1.56 (m, 2 H), 1.30 (t, 6 H, J = 6.9 Hz). HRMS (ESI/QTOF) m/z: [M + Na]+ calculated for C41H57N3NaO+12 806.3834; Found 806.3834.
Nile Red-PEG11-COOH 3
Fmoc-PEG11-COOH (10.0 mg, 12 µmol) was dissolved in 0.5 ml MeCN/piperidine 9:1 and the solution was stirred for 20 min at room temperature. The solvents were evaporated, the residue was coevaporated three times with MeCN, and dried under high vacuum. Separately, Nile Red-C6-COOH* (5.4 mg, 12 µmol) was dissolved in 0.25 ml DMSO, treated with DIPEA (4.1 µl, 24 µmol) and TSTU (3.6 mg, 12 µmol). The solution was incubated for 30 min at room temperature and was transferred in the vial that contained the above deprotected PEG linker. After 1 h, the reaction was subjected to RP-HPLC and the product was lyophilized. Yield: 4.7 mg (37%). 1H NMR (400 MHz, DMSO) δ 8.04 (d, 1 H, J = 8.7 Hz), 7.94 (d, 1 H, J = 2.4 Hz), 7.88 (t, 1 H, J = 5.6 Hz), 7.64 (d, 1 H, J = 9.1 Hz), 7.26 (dd, 1 H, J = 2.3, 8.7 Hz), 6.83 (dd, 1 H, J = 2.4, 9.1 Hz), 6.66 (d, 1 H, J = 2.3 Hz), 6.20 (s, 1 H), 4.16 (t, 2 H, J = 6.3 Hz), 3.60 (t, 2 H, J = 6.3 Hz), 3.52-3.49 (m, 48 H), 3.40 (t, 2 H, J = 5.9 Hz), 3.20 (q, 2 H, J = 5.6 Hz), 2.44 (t, 2 H, J = 6.3 Hz), 2.12 (t, 2 H, J = 7.2 Hz), 1.79 (m, 2 H), 1.59 (m, 2 H), 1.45 (m, 2 H), 1.17 (m, 6 H). HRMS (ESI/QTOF) m/z: [M + Na]+ calculated for C53H81N3NaO+18 1070.5407; Found 1070.5405.
Nile Red-PEG27-COOH 4
Fmoc-PEG27-COOH (18.5 mg, 12 µmol) was dissolved in 0.5 ml MeCN/piperidine 9:1 and the solution was stirred for 20 min at room temperature. The solvents were evaporated, the residue was coevaporated three times with MeCN, and dried under high vacuum. Separately, Nile Red-C6-COOH* (5.4 mg, 12 µmol) was dissolved in 0.25 ml DMSO, treated with DIPEA (4.1 µl, 24 µmol) and TSTU (3.6 mg, 12 µmol). The solution was incubated for 30 min at room temperature and was transferred in the vial that contained the above deprotected PEG linker. After 1 h, the reaction was subjected to RP-HPLC and the product was lyophilized. Yield: 10.5 mg (50%). 1H NMR (400 MHz, MeOD) δ 8.11 (d, 1 H, J = 8.9 Hz), 8.03 (d, 1 H, J = 2.3 Hz), 7.69 (d, 1 H, J = 9.3 Hz), 7.20 (dd, 1 H, J = 2.5, 8.9 Hz), 7.06 (dd, 1 H, J = 2.6, 9.3 Hz), 6.73 (d, 1 H, J = 2.6 Hz), 6.39 (s, 1 H), 4.19 (m, 2 H), 3.77-3.58 (118 H), 3.55 (t, 2 H, J = 5.4 Hz), 3.38 (t, 2 H, J = 5.4 Hz), 2.55 (t, 2 H, J = 6.3 Hz), 2.29 (t, 2 H, J = 7.3 Hz), 1.91 (m, 2 H), 1.75 (m, 2 H), 1.60 (m, 2 H), 1.34-1.29 (m, 6 H). HRMS (nanochip-ESI/LTQ-Orbitrap) m/z: [M + H]+ calculated for C85H146N3O+34 1752.9782; Found 1,752.9741.
Nile Red-PEG5-C2-maleimide 5
A 12 mM DMSO solution of Nile Red-PEG5-COOH 2 (200 µl, 2.4 µmol) was treated with DIPEA (1.5 µl, 8.4 µmol) and TSTU (0.87 mg, 2.9 µmol). The solution was incubated for 30 min at room temperature with N-(2-aminoethyl)maleimide trifluoroacetate (0.91 mg, 3.6 µmol). After 1 h, the reaction was subjected to RP-HPLC and the product was lyophilized. Yield: 1.6 mg (75%). 1H NMR (400 MHz, MeOD) δ 8.19 (d, 1 H, J = 8.9 Hz), 8.14 (d, 1 H, J = 2.5 Hz), 7.77 (d, 1 H, J = 9.3 Hz), 7.27 (dd, 1 H, J = 8.9, 2.6 Hz), 7.11 (dd, 1 H, J = 9.3, 2.6 Hz), 6.81 (m, 3 H), 6.45 (s, 1 H), 4.24 (t, 2 H, J = 6.3 Hz), 3.70-3.52 (32 H), 3.40-3.36 (m, 4 H), 2.38-2.35 (m, 2 H), 2.30 (t, 2 H, J = 7.3 Hz), 1.97-1.90 (m, 2 H), 1.80-1.73 (m, 2 H), 1.66-1.59 (m, 3 H), 1.32 (m, 6 H). HRMS (ESI/QTOF) m/z: [M + Na]+ calculated for C47H63N5NaO+13 928.4315; Found 928.4316.
Nile Red-PEG11-C2-maleimide 6
A 7.5 mM DMSO solution of Nile Red-PEG11-COOH 3 (400 µl, 3.0 µmol) was treated with DIPEA (1.8 µl, 10.5 µmol) and TSTU (1.1 mg, 3.6 µmol). The solution was incubated for 30 min at room temperature with N-(2-aminoethyl)maleimide trifluoroacetate (1.15 mg, 4.5 µmol). After 1 h, the reaction was subjected to RP-HPLC and the product was lyophilized. Yield: 2.3 mg (65%). 1H NMR (400 MHz, DMSO) δ 8.05 (d, 1 H, J = 8.7 Hz), 7.96-7.94 (m, 2 H), 7.87 (t, 1 H, J = 5.5 Hz), 7.65 (d, 1 H, J = 9.1 Hz), 7.27 (dd, 1 H, J = 2.6, 8.7 Hz), 7.01 (s, 2 H), 6.84 (dd, 1 H, J = 2.6, 9.2 Hz), 6.68 (d, 1 H, J = 2.6 Hz), 6.21 (s, 1 H), 4.17 (t, 2 H, J = 6.3 Hz), 3.56-3.39 (m, 58 H), 3.23-3.18 (m, 4 H), 2.23 (t, 2 H, J = 6.6 Hz), 2.13 (t, 2 H, J = 7.2 Hz), 1.80 (m, 2 H), 1.60 (m, 2 H), 1.45 (m, 2 H), 1.18 (t, 6 H, J = 6.9 Hz). HRMS (ESI/QTOF) m/z: [M + Na]+ calculated for C59H87N5NaO+19 1192.5887; Found 1192.5886.
Nile Red-PEG27-C2-maleimide 7
A 12 mM DMSO solution of Nile Red-PEG27-COOH 4 (200 µl, 2.4 µmol) was treated with DIPEA (1.5 µl, 8.4 µmol) and TSTU (0.87 mg, 2.9 µmol). The solution was incubated for 30 min at room temperature with N-(2-aminoethyl)maleimide trifluoroacetate (0.91 mg, 3.6 µmol). After 1 h, the reaction was subjected to RP-HPLC and the product was lyophilized. Yield: 2.6 mg (58%). 1H NMR (400 MHz, MeOD) δ 8.20 (d, 1 H, J = 8.8 Hz), 8.17 (d, 1 H, J = 2.5 Hz), 7.79 (d, 1 H, J = 9.3 Hz), 7.29 (dd, 1 H, J = 2.6, 8.8 Hz), 7.10 (dd, 1 H, J = 2.7, 9.3 Hz), 6.84 (s, 2 H), 6.81 (d, 1 H, J = 2.6 Hz), 6.45 (s, 1 H), 4.25 (t, 2 H, J = 6.3 Hz), 3.71-3.54 (m, 118 H), 3.41-3.38 (m, 4 H), 2.38 (t, 2 H, J = 6.2 Hz), 2.30 (t, 2 H, J = 7.3 Hz), 1.94 (m, 2 H), 1.77 (m, 2 H), 1.62 (m, 2 H), 1.32 (t, 6 H, J = 6.9 Hz). HRMS (nanochip-ESI/LTQ-Orbitrap) m/z: [M + Na]+ calculated for C91H151N5NaO+35 1897.0082; Found 1897.0065.
Nile Red-PEG5-CoA 8
CoA (1.5 mg, 2 µmol) was dissolved in 44 µl 0.1 M Tris-HCl, pH 7.5, and a 12 mM DMSO solution of Nile Red-PEG5-C2-maleimide 5 (83 µl, 1.0 µmol) was added. After 2 h, the reaction was subjected to RP-HPLC and the product was lyophilized. Yield: 0.7 mg (41%). HRMS (nanochip-ESI/LTQ-Orbitrap) m/z: [M + H2]+2 calculated for C68H101N12O29P3S+2 837.2860; Found 837.2856.
Nile Red-PEG11-CoA 9
CoA (1.5 mg, 2 µmol) was dissolved in 100 µl 0.1 M Tris-HCl, pH 7.5, and a 5 mM DMSO solution of Nile Red-PEG11-C2-maleimide 6 (200 µl, 1.0 µmol) was added. After 2 h, the reaction was subjected to RP-HPLC and the product was lyophilized. Yield: 1.2 mg (60%). MS (ESI) m/z: [M + Na+H]+2 calculated for C68H101N12O29P3S+2 837.2860; Found 837.2856. HRMS (nanochip-ESI/LTQ-Orbitrap) m/z: [M + H2]+2 calculated for C80H125N12O35P3S+2 969.3646; Found 969.3644.
Nile Red-PEG27-CoA 10
CoA (1.5 mg, 2 µmol) was dissolved in 54 µl 0.1 M Tris-HCl, pH 7.5, and a 9.3 mM DMSO solution of Nile Red-PEG27-C2-maleimide 7 (108 µl, 1.0 µmol) was added. After 2 h, the reaction was subjected to RP-HPLC and the product was lyophilized. Yield: 0.9 mg (35%). HRMS (nanochip-ESI/LTQ-Orbitrap) m/z: [M + H2]+2 calculated for C112H189N12O51P3S+2 1321.5744; Found 1321.5730.
Nile Red-PEG14-COOH 11
Fmoc-PEG3-COOH (0.5 mg, 1.2 µmol) was dissolved in 0.1 ml MeCN/piperidine 9:1 and the solution was stirred for 20 min at room temperature. The solvents were evaporated, the residue was coevaporated three times with MeCN, and dried under high vacuum. Separately, a 7.5 mM solution of Nile Red-PEG11-COOH 3 (160 ul, 1.2 µmol) was treated with DIPEA (1 µl, 9.7 µmol) and TSTU (0.36 mg, 1.2 µmol). The solution was incubated for 30 min at room temperature and was transferred to a vial that contained the above deprotected PEG linker. After 1 h, the reaction was subjected to RP-HPLC and the product was lyophilized. Yield: 1.05 µmol (87%).
Nile Red-PEG14-C2-maleimide 12
A 7.5 mM DMSO solution of Nile Red-PEG14-COOH 11 (100 µl, 0.75 µmol) was treated with DIPEA (1 µl, 9.7 µmol) and TSTU (0.27 mg, 0.9 µmol). The solution was incubated for 30 min at room temperature and N-(2-aminoethyl)maleimide trifluoroacetate (0.29 mg, 1.1 µmol) was added. After 1 h, the reaction was subjected to RP-HPLC and the product was lyophilized. Yield: 0.51 µmol (71%).
Nile Red-PEG14-CoA 13
CoA (0.4 mg, 0.42 µmol) was dissolved in 100 µl 0.1 M Tris-HCl, pH 7.5, and a 3.5 mM DMSO solution of Nile Red-PEG14-C2-maleimide 12 (100 µl, 0.35 µmol) was added. After 2 h, the reaction was subjected to RP-HPLC and the product was lyophilized. Yield: 0.13 µmol (31%).
Plasmids
The ACP sequence from the ACP-GPI plasmid (Sundukova et al., 2019) was incorporated to the HIR plasmid previously described (Prifti et al., 2014) by Gibson assembly (New England Biolabs). The HIR (Homo sapiens, NCBI reference sequence NM_001079817) with the HindIII site before the start codon and the XbaI site after the stop codon, was subcloned by replacing the EGFP sequence of pEGFP-N1 (Clontech, Takara; Prifti et al., 2014). The sequences of the HIR, the backbone plasmid, pEGFP-N1 lacking the EGFP sequence, and the ACP were amplified by PCR with PrimeSTAR Max DNA Polymerase (Takara). The primers for PCR were listed in Table S1. The KD 2031-ACP-IR plasmid was generated by a point mutation at Lys1045 to Arg (AAG to AGG). The fragment sequence of HIR, the XhoI site at 3020 to the stop codon, in the plasmid of WT 2031-ACP-IR was replaced to the XhoI-XbaI fragment of HIR sequence including the K1045R mutation with the XbaI site after the stop codon synthesized by GeneArt (Thermo Fisher Scientific). The sequences of plasmids were verified and in the case of plasmids generated by Gibson Assembly all sequences including the plasmid backbone were verified by sequencing by Fasteris SA.
AP2-mGFP plasmid was constructed by replacing mEos4b of AP2-mEos4b plasmid to mEGFP. The sequence of AP2, adaptor-related protein complex2, α subunit transcript variant 2 (NCBI accession no. NM_130787) of the AP2-mEos4b plasmid was obtained from Flexi HaloTag clone AP2A-pFN21A vector (Product ID: FHC03782; Kazusa Genome Technology). The sequence of AP2 and the vector backbone, EGFP-truncated pEGFP-N1, were amplified by PCR with the forward primer; 5′-AGCGGCCGCGACTCTAGAT-3′ and the reverse primer; 5′-GGTGGCGACCGGTGGATC-3′. The mEGFP sequence was amplified by PCR from mGFP-GPI plasmid (Suzuki et al., 2012) with the forward primer; 5′-GGGATCCACCGGTCGCCACCATGGTGAGCAAGGGCGAG-3′ and the reverse primer; 5′-GATCTAGAGTCGCGGCCGCTTTACTTGTACAGCTCGTCCATG-3′. The amplified AP2 with the vector backbone and mEGFP fragment were assembled by Gibson assembly.
Cell culture and stable CHO cell lines expressing ACP-tagged human insulin receptor
Parent CHO-K1 and CHO-K1 cells stably expressing extracellular ACP-tagged human insulin receptors were maintained at 37°C with 5% CO2 in Ham’s F-12 medium (Thermo Fisher Scientific) supplemented 10% fetal bovine serum, 100 U/ml penicillin, and streptomycin. CHO-K1 cells were transfected with ACP-human IR plasmid using Xfect transfection reagent (Takara). 1 d after transfection, the cells were selected in 600 μg mL−1 G418, and stably expressing clones were isolated by limiting dilution method. The clones were screened by staining with CoA-Cy3. Cells stably expressing ACP-IR were maintained in the growth medium supplementing with 600 μg mL−1 G418.
Double staining of CoA-PEG11-NR and DAPI
CHO cells stably expressing ACP-IR were washed with serum-free Ham’s F-12 medium and incubated for 30 min at room temperature in serum-free Ham’s F-12 medium with 1 µM CoA-PEG11-NR, 1 µM SFP synthase, 10 mM MgCl2, and 15 mM Hepes. After washing the excess staining solution with PBS, cells were fixed with 3% paraformaldehyde in PBS and then stained with 300 nM DAPI for 20 min at room temperature. Imaging of the labeled cells was performed under a Zeiss LSM 780 confocal microscope equipped with Plan-Apochromat 63× (1.40 NA) oil immersion objective using Zen software (Zeiss).
Emission spectrum measurement of NR12S or CoA-PEG-NR linked to the ACP-IR
For labeling with NR12S, cells stably expressing ACP-IR cultured on glass bottom (MatTek) dishes for 2 d were labeled with 800 nM of NR12S for 7 min at room temperature in serum-free Ham’s F-12 medium including 15 mM Hepes after washing with serum-free medium. Stably expressing ACP-IR cells were labeled with 2 µM of CoA-PEG-NRs in serum-free medium containing 15 mM Hepes with 1 µM of SFP synthase and 10 mM MgCl2 at room temperature for 20 min. Images were captured using lambda mode under a Zeiss LSM 780 confocal microscope (Zeiss) equipped with plan-apochromat 63× oil immersion objective (1.4 NA) and 32-channel GaAsP detectors, excitation at 514 nm and detection with 15 channels at 561–695 nm (step: 8.9 nm). All images were four-time line averaged. Cellular cholesterol-modification was performed before staining. After cells was washed with serum-free medium, 10 mM of methyl β cyclodextrin was used in serum-free medium at 37°C for 15 min for cholesterol-depletion, and 2.5 mM of cholesterol-water soluble was incubated in serum-free medium at 37°C for 1 h for cholesterol-loading. The emission spectrum was measured in number of regions of interest (ROIs) from 5 images per each sample using ImageJ (National Institutes of Health).
Voltage sensitivity test
Voltage sensitivity of CoA-PEG-NR attached to ACP-IR was measured as described before (Sundukova et al., 2019). Simultaneous optical and electrophysiological recordings were performed at room temperature on a standard patch clamp setup (HEKA, EPC 10Usb) mounted on an inverted Zeiss Axioobserver A1 equipped with an oil-immersion 40× objective (NA 1.3) after cell labeling with NR12S or CoA-PEG-NR. Cells on the microscope stage were perfused with extracellular bath solution (150 mM NaCl, 5 mM KCl, 1 mM MgCl2, 2 mM CaCl2, 10 mM Hepes, and 5 mM glucose, pH 7.4, adjusted with NaOH, 300 mOsm). Pipettes had resistance of 3–7 mΩ when filled with intracellular solution (140 mM of CsCl, 10 mM of Hepes, 10 mM of HEDTA, pH 7.35, adjusted with CsOH, 290 mOsm). After establishing whole cell configuration, cell capacitance and access resistance were routinely compensated. All experiments were performed in whole-cell patch clamp configuration. Membrane potential was changed from −60 to +40 mV and then back to −40 mV. Fluorescence excitation was delivered using a 100 W Mercury short arc lamp (OSRAM) through BP 540–552 nm (RFP) filter or BP360-540 nm (Rhodamine) filters at 28 mW/mm2 power densities measured with an optical power meter at the imaging plane. Fluorescence emission was passed through a LP590 nm (RFP) or BP570-640 nm (Rhodamine) filter and recorded by a Zyla 4.2 Plus (Andor) sCMOS Camera operated with NIS-Elements Ar (Nikon). Fluorescence images were analyzed in NIS-Elements Ar and ImageJ by manually defining the ROI and calculating the mean unweighted mean of pixel values within this region. The cell-free region was used for calculating background fluorescence, which was subtracted from the cell fluorescence. When required, photobleaching was corrected using an exponential decay function. To determine voltage sensitivity, %ΔF/F values were generated by subtracting baseline fluorescence at holding potential from fluorescence during the voltage step and plotted against voltage. Voltage sensitivity of different substrates was then compared as %ΔF/F per 100 mV depolarization from a −60 to +40 mV voltage step. Statistical significance was calculated by a two-sided t test (two-sided; Fig. 2 C) or one-way ANOVA (Fig. 2, D and E) using the Graph Pad Prism 7.
Western blotting
3.0 × 105 of cells stably expressing WT 2031-ACP-IR or KD 2031-ACP-IR were cultured on 6-well plates for 2 d. After 2 h serum-starvation, cells were stimulated with 100 nM of human reconstitute insulin for 5 min at 37°C. The cells were lysed with 120 μl of lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 0.5% TritonX-100) containing protease- and phosphatase-inhibitor cocktails and incubated on ice for 20 min, and then the lysate was centrifuged at 14,000 rpm for 20 min at 4°C. The supernatant was collected, and the protein concentration was determined by BCA protein assay. 20 μg of the lysates was heated at 65°C for 10 min in SDS-sample buffer (6× SDS sample buffer, 375 mM Tris-HCl [pH 6.8], 12% SDS, 40% glycerol, 0.03% bromophenol blue, and 0.6 M DTT) and subjected to SDS-PAGE on NuPAGE 4–12% Bis-Tris protein gel, then the proteins on the gel were transferred to a nitrocellulose membrane. After overnight blocking of the membrane with 10% BSA in TBST (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, and 0.05% Tween-20), the membrane was probed with primary antibodies, anti-insulin receptor β rabbit polyclonal antibody and anti-phospho-tyrosine mouse monoclonal antibody, or anti-insulin receptor beta mouse monoclonal antibody and anti-phospho-tyrosine rabbit polyclonal antibody at 4°C for overnight, followed by secondary antibodies, IRDye 800CW anti-mouse antibody and IRDye 680RD anti-rabbit antibody, then imaged used Fusion FX (VILBER). For experiments to assess the effect of attachment of CoA-PEG11-NR on the receptor tyrosine activity of 2031-ACP-IR, cells were incubated with 1 µM of SFP synthase and 10 mM of MgCl2 in 15 mM Hepes containing medium in the presence and absence of 2 µM CoA-PEG11-NR at room temperatures for 30 min after the serum-starvation. Statistical significance was calculated by one-way ANOVA using the Graph Pad Prism 7 (Fig. 4 A).
Time-lapse imaging of the local membrane environment surrounding the insulin receptor
Cells stably expressing WT 2031-ACP-IR cultured on glass-bottom dishes for 2 d were labeled with 2 µM of CoA-PEG5-NR or CoA-PEG11-NR, 3 µM of CoA-PEG27-NR or 800 nM of NR12S at room temperatures. KD 2031-ACP-IR stably expressing cells plated on glass bottom dishes were labeled with 2 µM of CoA-PEG5-NR. Cells transiently expressing ACP-GPI placed on glass-bottom dishes were stained with 2 µM CoA-PEG11-NR. The excess dye was washed with 37°C serum-free Ham’s F-12 medium containing 15 mM Hepes three times, and 1 ml of 37°C serum-free medium containing 15 mM Hepes was added to the dish, then the dish was placed on the stage top heating chamber (Tokai Hit, INUBTF-WSKM) warmed at 37°C set on the stage of a Zeiss LSM 780 confocal microscope equipped with Plan-Apochromat 63× (1.40 NA) oil immersion objective (Zeiss) for 5 min before the observation. Spectrum images in the range of 561–695 nm were captured with GaAsP detectors (15 channels, step 8.9 nm, 4 times line-average) using lambda mode excited at 514 nm operated by the Zen software every 10 s for 9 min, and 100 μl of serum-free medium (control) and 100 μl of 1 μM human reconstitute insulin in serum-free medium (final concentration, 100 nM) were added to the dish using a syringe with the long needle at 60 and 240 s, respectively, and either of the control or the insulin solution was added at 120 s for 7 min observation. WT 2031-ACP-IR stably expressing cells labeled with CoA-PEG5-NR were used for inhibitor experiments, and cell staining and observation were performed in the presence of DMSO (control), 0.2 μM of Latrunculin B or 50 μM of LY294002. We performed time-lapse imaging by excitation at 514 nm with 2.5% of Argon laser power and we demonstrated the differential delta GP upon insulin stimulation to show the effect of insulin on GP change. GP decrease caused by excitation light was eliminated by subtraction of the basal GP change in control treatment (Fig. 5, B and C). Statistical significance was calculated by one-way ANOVA using the Graph Pad Prism 7 (Fig. 8 B).
Sir-actin staining in living cells
CHO cells stably expressing ACP-2031-IR were incubated with Ham’s F-12 Ham medium containing 3 μM of Sir-actin and 10 μM of verapamil at 37°C for 1 h, followed by washing with Ham’s F-12 medium three times. Imaging of the labeled cells was performed under a Leica SP5 confocal microscope equipped with HCX PL APO CS 63× oil immersion objective (1.4 NA) using LAS AF software (Leica Microsystems).
Staining with CoA-PEG11-NR and immunostaining of the insulin receptor
CHO cells stably expressing ACP-IR were labeled with CoA-PEG11-NR, then immunostaining of the insulin receptor was performed. After washing the excess CoA-PEG11-NR staining solution with PBS, cells were fixed with 3% paraformaldehyde in PBS for 10 min at room temperature, followed by permeabilization with 0.1% TritonX-100 in PBS for 1 min at room temperature and blocking with 5% BSA in PBS for 20 min at room temperature. Cells were incubated with anti-insulin receptor β mouse monoclonal antibody (1:50) for 2 h at room temperature followed by incubation with Cy2-conjugated anti-mouse IgG antibody (1:200) at room temperature for 1 h. After each step, cells were rinsed with PBS three times. Imaging of the labeled cells was performed under an Olympus FV1000-D confocal laser scanning fluorescent microscope equipped with ApoN 60× (1.49 NA) oil immersion objective, Ar laser (488 nm) and LED laser (559 nm). Cy2 was excited at 488 nm with 8% laser power and obtained images at 500–545 nm, and Nile Red was excited at 559 nm with 25% laser power and images were captured in the range of 570–670 nm. Cy2 and Nile Red images were sequentially captured using two photomultipliers. Data analysis was done by Fiji (freely available at https://fiji.sc/).
Determination of relative fluorescence QY of Nile Red
Fluorescence QY of Nile Red (0.1 μM) was measured in dioxane, POPC (75 μg/ml) small unilamellar vesicles (SUV) in PBS, BSA (0.1 mg/ml) in PBS and PBS using a solution of Nile Red in methanol (MeOH) as a reference. A thin film of POPC was dried under vacuum. The film was hydrated with PBS and released from the glass by gentle vortexing. SUVs were made by sonication with a Branson Sonic 450 equipped with a micro-tip at 10% output for 1 min, followed by a 5 min spin at 10,000 rcf. Nile Red was added to the supernatant. Absorbance measurements were performed with a Shimadzu UV-1800 spectrometer using quartz cuvettes with a 1 cm path length from Helma. Water was used as blank. Spectra were acquired from 400 to 800 nm in 1 nm step at medium scan speed, and 5 repeats were averaged. Fluorescence measurements were performed with a Fluorolog-3 (Horiba) equipped with a PPD850 detector. The excitation was set to 520 nm with a slit width of 5 nm. Corrected fluorescence intensities were acquired from 540 to 800 in 1 nm step with a slit width of 5 nm and a detection of 0.1 s/point. Helma quartz cuvettes with 1 cm pathlength were used. Emission intensity was corrected for monochromator and detector sensitivity by the instrument’s software.
Image analysis for time-lapse imaging of the local membrane environment surrounding the insulin receptor
Image segmentation of the receptor-linked environment-sensitive probe
Image analysis was performed on all time-lapse data sets to reveal how the GP value changes during insulin signaling. The captured images, however, usually include information such as image noise that is not relevant to the observation targets. Therefore, image segmentation was performed to extract the target regions, the receptor-linked environment-sensitive probe. The detailed procedures are as follows:
Image sequences of eight spectral channels from 565 to 645 nm (i.e., 565, 575, 585, 595, 615, 625, 635, and 645 nm) were collected throughout the time-lapse observation.
A median filter with a kernel size of W = 3 × 3 × 3 pixels was used to filter each time-lapse image sequence. Median filter was conducted by ImageMagick software version 6.8.9 (https://imagemagick.org/index.php).
A difference of Gaussian (DoG) filter (Marr and Hildreth, 1980) was performed to each median filtered image.
The z-score normalization was performed to the DoG filtered image sequence.
K-means clustering (Kanungo et al., 2002) was performed to the normalized image sequence in order to form two classes (target probe or background noise). K-means was performed by our program and some functions of kmlocal version 1.7.2 (http://www.cs.umd.edu/users/mount/Projects/KMeans/).
The DoG filter enhanced the edges in a digital image and we performed that filter with a standard deviation of 15 and kernel window size W of 25 pixels. As the outputs of K-means clustering, we obtained two-class labeled images, showing to which class belonged each pixel. The class with the higher mean value was identified as the target probe class, and only the pixels belonging to this class were used to calculate the GP value.
The calculation of GP value, delta GP, and differential delta GP
We used GPtheo = −0.735 and −0.728 for NR12S and CoA-PEG-NR, respectively. GPexp was measured and the G factor was calculated every time when the time-lapse observation was performed. Images Ich0 and Ich1 of Eq. 1 are the averaged images of the lower four channels (565–595 nm) and the higher four channels (615–645 nm), respectively. The translated GP values vary between −1.0 (disordered) and 1.0 (ordered). We depict them as pseudocolor GP images (Fig. 5), which reveal the reaction of the target probes surrounding the receptor to the insulin solution.
To compare time-dependent GP changes, the delta GP at each time was estimated by subtraction of the mean GP at time 0 using R scripts and the mean of the delta GP from multiple time-lapse observations was plotted by GraphPad Prism. For 7 min observation of the KD 2031-ACP-IR and inhibitor treatments, the differential delta GP upon insulin stimulation was calculated by subtraction of the basal delta GP with control treatment from the means of delta GP with insulin treatment at each time point. The basal delta GP was the mean of delta GP from control-treated multiple time-lapse data set and the mean of differential delta GP from multiple time-lapse observations was plotted (Fig. 5, B and C).
Dynamic analysis of change in GP using zero-mean normalized cross correlation
Although these pseudocolor GP images provide us with information regarding the range of the GP values, the changes in the GP value are quite small throughout the entire time-lapse observation. As a result, we are not able to visualize the point at which the GP values change before and after the application of insulin solution in these images. Moreover, the GP value of each pixel contains the fluctuation caused by measurement noise or the resolution limit of the microscope.
Colocalization of clathrin-coated pits and 2031-ACP-IR upon insulin stimulation
CHO cells stably expressing ACP-2031-IR were transfected with AP2-mEGFP plasmid using Xfect transfection reagent (Takara). 1 d after transfection, the cells were plated on glass bottom dishes and incubated at 37°C overnight, then cells were stained with 1 µM of CoA-Cy3 in serum-free medium containing 15 mM Hepes with 1 µM of SFP synthase and 10 mM MgCl2 at room temperature for 20 min. The excess dye was washed with 37°C serum-free Ham’s F-12 medium three times, 1 ml of 37°C serum-free medium was added to the dish, and then the dish was placed on the stage of the microscope warmed at 37°C using thermostat incubator (Sankei Co. Ltd). Images were captured under an Olympus FV1000-D confocal laser scanning fluorescent microscope equipped with ApoN 60× (1.49 NA) oil immersion objective and GaAsP detectors every 1 min for 6 min using the FV10 software (Olympus). After taking the image of time 0, 10 μl of serum-free medium (control) was added and 3 min after control solution, 10 μl of 10 μM human reconstitute insulin in serum-free medium (final concentration, 100 nM) were added. GFP and Cy3 were excited at 488 nm with 2% laser power and at 559 nm with 6.9% laser power and images were sequentially captured in the range of 505–540 and 575–675 nm, respectively. The degree of colocalization between AP2 and ACP-IR was analyzed by MCC and SRCC (n = 20 cells for each time point). The MCC was estimated in two ways, as the ratio of the AP2 colocalized with ACP-IR over the total fluorescent of AP2, and the ratio of the ACP-IR colocalized with AP2 over the total fluorescent of ACP-IR. Image segmentation for discriminating regions of AP2 and ACP-IR and to calculate the MCC between GFP and Cy3 was conducted by our program, and the SRCC was conducted by R software version 3.6.2. Statistical significance was calculated by one-way ANOVA using the GraphPad Prism 7.
Online supplemental material
Fig. S1 shows the supplemental material contains a comparison of the emission spectra of PEG5-Nile Red linked to the insulin receptor at the beginning of the time-course before addition of insulin and at the end of the time-course. Fig. S2 shows the complete time-course image set of the ZNCC images. Fig. S3 shows an estimation of the QY of Nile Red linked to the different insulin receptor constructs. Fig. S4 shows that the effects of a phospholipase inhibitor on the local environment of the insulin receptor. Fig. S5 shows the effects of laser power on the GP values of PEG-Nile Red linked to the insulin receptor and ACP-GPI. Table S1 lists the primers used in this study. Table S2 shows the wavelength and quantum yield of Nile Red in solution. Videos 1, 2, and 3 show the full time-course of the experiments displayed as top/bottom 1,000 pixels, ZNCC, or GP, respectively.
Acknowledgments
We thank members of the Riezman lab for their help and comments on the manuscript. We also thank Dr. Christian Bauer and Jerome Bosset at Photonic Bioimaging Center (University of Geneva) and Prof. Fumiyoshi Ishidate at Institute for Integrated Cell-Material Sciences (Kyoto University) for help with imaging. We thank Ms. Shinobu Kawaguchi for preparation of the AP2 plasmid and the ACP-GPI plasmid. We thank Taroh Kinoshita (Osaka University, Osaka, Japan) for providing the PGAP2/PGAP3 cell line.
This work was funded by the NCCR Chemical Biology, the Swiss National Science Foundation (grants 51NF40_185898, 310030, and _184949), and the Leducq Foundation.
Author contributions: M. Umebayashi; conceptualization, methodology, investigation. writing; S. Takemoto, software, formal analysis, writing; L. Reymond; resources, M. Sundukova; investigation, R. Hovius; investigation, A. Bucci; investigation, P.A. Heppenstall; supervision, H. Yokota, software, formal analysis, K. Johnsson; supervision, funding acquisition, H. Riezman; conceptualization, writing, supervision, funding acquisition.
References
Author notes
Disclosures: S. Takemoto reported being partly supported by JSPS KAKENHI grant number JP16K16153. No other disclosures were reported.