The detyrosination/tyrosination cycle of α-tubulin is critical for proper cell functioning. VASH1–SVBP and VASH2–SVBP are ubiquitous enzymes involved in microtubule detyrosination, whose mode of action is little known. Here, we show in reconstituted systems and cells that VASH1–SVBP and VASH2–SVBP drive the global and local detyrosination of microtubules, respectively. We solved the cryo-electron microscopy structure of VASH2–SVBP bound to microtubules, revealing a different microtubule-binding configuration of its central catalytic region compared to VASH1–SVBP. We show that the divergent mode of detyrosination between the two enzymes is correlated with the microtubule-binding properties of their disordered N- and C-terminal regions. Specifically, the N-terminal region is responsible for a significantly longer residence time of VASH2–SVBP on microtubules compared to VASH1–SVBP. We suggest that this VASH region is critical for microtubule detachment and diffusion of VASH–SVBP enzymes on lattices. Our results suggest a mechanism by which VASH1–SVBP and VASH2–SVBP could generate distinct microtubule subpopulations and confined areas of detyrosinated lattices to drive various microtubule-based cellular functions.
Introduction
Microtubules are key eukaryotic cytoskeletal elements that are regulated by the tubulin code. This code involves the differential expression of α- and β-tubulin isotypes and their post-translational modifications (reviewed in Janke and Magiera, 2020). A prominent tubulin post-translational modification is the cyclic removal and re-addition of the C-terminal tyrosine residue of α-tubulin. This evolutionary conserved cycle, referred to as the “detyrosination/tyrosination cycle,” was shown to be crucial for cell mitosis and differentiation; it is also a key player in the development and functioning of the brain and the heart (Chen et al., 2018; Erck et al., 2005; reviewed in Lopes and Maiato, 2020; Moutin et al., 2021; Sanyal et al., 2021). Accordingly, dysfunctions of the detyrosination/tyrosination cycle lead to cancer, brain disease, and heart failure in humans (Chen et al., 2020; Lafanechere et al., 1998; Peris et al., 2022; Schuldt et al., 2021; reviewed in Lopes and Maiato, 2020; Sanyal et al., 2021).
The known “readers” of the tubulin tyrosine signal emerging from the cycle are kinesin motors and CAP-Gly proteins (reviewed in Sanyal et al., 2021; Steinmetz and Akhmanova, 2008). The activity of the detyrosination/tyrosination cycle regulates cargo trafficking in cells by tuning the microtubule binding of specific kinesins and the dynein–dynactin complex (Dunn et al., 2008; Nirschl et al., 2016; reviewed in Moutin et al., 2021). The cycle also modulates tubulin interactions with regulators of microtubule stability, such as kinesin 13 and CLIP-170 (Chen et al., 2021; Peris et al., 2006; Peris et al., 2009). This modification is thus intimately linked to microtubule dynamics, where the detyrosination signal represents a marker of microtubule stability.
The detyrosination/tyrosination cycle of α-tubulin involves at least four enzymes including the tubulin tyrosine ligase (TTL; Ersfeld et al., 1993) and the three recently discovered detyrosinases comprising the two enzymatic complexes composed of a vasohibin (VASH1 or VASH2) and a small vasohibin-binding protein (SVBP; Aillaud et al., 2017; Nieuwenhuis et al., 2017), and MATCAP (microtubule-associated tubulin carboxypeptidase; Landskron et al., 2022). TTL represents the “writer” of the tyrosine signal, while vasohibins and MATCAP are the “erasers.” SVBP acts as a chaperone and a co-factor for VASH1 and VASH2, which are specific tyrosine/phenylalanine carboxypeptidases (Aillaud et al., 2017; Nieuwenhuis et al., 2017). Mammalian VASH1 and VASH2 proteins share more than 50% overall sequence identity. They belong to the transglutaminase-like cysteine protease superfamily (Sanchez-Pulido and Ponting, 2016) and harbor a Cys–His–Leu catalytic amino acid residue triad conserved among species (Adamopoulos et al., 2019; Wang et al., 2019). Crystal structures of VASH1–SVBP and VASH2–SVBP complexes at high resolution allowed to determine the global organization of the central catalytic regions of the VASHs, which are their most homologous domains (Adamopoulos et al., 2019; Li et al., 2019; Liao et al., 2019; Liu et al., 2019; Wang et al., 2019; Zhou et al., 2019; reviewed in Sanyal et al., 2021). These core domains form a folded, globular structure enclosing the C-terminal region of SVBP. The N- and C-terminal regions of VASHs are less well conserved and are assumed to be disordered as underlined by the lack of structural information in all published structures of VASH–SVBP complexes to date (Adamopoulos et al., 2019; Li et al., 2019; Wang et al., 2019; Zhou et al., 2019).
VASH–SVBP complexes are known to preferentially detyrosinate microtubule-lattice incorporated α-tubulin (Aillaud et al., 2017). We have previously shown that the VASH2–SVBP complex binds microtubules with a 1:1 stoichiometry of VASH2–SVBP:αβ-tubulin heterodimer and with an equilibrium dissociation constant in the low micromolar range (Wang et al., 2019). Mutating the catalytic cysteine residue Cys158 of VASH2 resulted in an enzymatically inactive VASH2 version that was unable to detyrosinate α-tubulin, yet retained the ability to co-localize with microtubules in vitro and in cells. We further identified positively charged VASH2 surface residues located in close proximity to the tyrosine-binding groove. Mutating these VASH2 residues decreased the detyrosination activity of the enzyme but did not completely abrogate microtubule binding in vitro and in cells, leading us to suspect that additional contact sites between VASHs and microtubules remained to be identified. In a recent study, cryo-EM was used to analyze the structure of VASH1–SVBP bound to microtubules. The authors showed that the core domain of VASH1 forms specific contacts with the folded parts of α-tubulin (Li et al., 2020). However, as for the crystal structures obtained on the isolated, free enzyme complex, they did not resolve the flanking regions of the VASH1 core domain, again demonstrating that the N- and C-terminal regions of VASHs are disordered and flexible.
VASH proteins are ubiquitously expressed but at different basal levels, transcripts of VASH1 being in general much higher represented (around 10 times) than transcripts of VASH2 (see the Human Protein Atlas project portal). In cells, detyrosination can occur on microtubules for many purposes and in several places (for review see Roll-Mecak, 2019), and the precise cellular role of each VASH–SVBP complex remains to be understood. VASH1–SVBP was identified as a primary tubulin detyrosinase in neurons and cardiomyocytes (Aillaud et al., 2017; Chen et al., 2020); however, VASH2–SVBP seems to act in cardiac cells as well (Yu et al., 2021). Thus, although a large amount of structural information has been obtained, the mechanism of action of VASH–SVBP enzyme complexes remains poorly understood.
Here, combining single-molecule total internal reflection fluorescence (TIRF) microscopy assays and cryo-EM, we depict the molecular basis of the interaction of VASH1–SVBP and VASH2–SVBP with microtubules in relationship with their detyrosination activities. We show that VASH1–SVBP generates global detyrosination of microtubule while VASH2–SVBP leads to local modifications both in vitro and in cells. The in vitro results obtained with several truncated mutants unveil crucial roles of the disordered N- and C-terminal regions in differentially regulating VASHs’ interactions with microtubules and hence their activities. We reveal that the N-terminal regions, which are highly divergent between the two VASHs, contribute to the difference in the enzymes’ ability to diffuse on the microtubule lattice. Our data further suggest that the disparity in functioning of the two VASH–SVBP enzymes contributes to the generation of different patterns of detyrosinated microtubules and thus to microtubule subpopulations having distinct roles within cells.
Results
VASH1 and VASH2 drive global and local microtubule detyrosination, respectively
We first set up experimental conditions for which both the detyrosinating activity as well as the interaction with microtubules of VASH–SVBP complexes could be studied in parallel using immunofluorescence and TIRF microscopy, respectively. We generated constructs encoding the active full-length human VASH1 or VASH2 tagged with sfGFP (superfolder GFP) at their N-termini and myc-6His at their C-termini. These constructs, together with the plasmid encoding the human SVBP fused to myc and FLAG tags at the C-terminus, were used to prepare full-length sfGFP-VASH1–SVBP and sfGFP-VASH2–SVBP complexes (V1_FL and V2_FL, respectively; Fig. S1, A and B). We immobilized Taxol-stabilized microtubules enriched in tyrosinated HeLa tubulin (Tyr-MTs, containing 80% of tyrosinated-tubulin; see Materials and methods and Fig. S1 C) on the surface of TIRF chambers as previously described (Stoppin-Mellet et al., 2020). Different media were tested in order to find experimental conditions where single molecules of the two enzyme complexes associated with microtubules. In BRB40 (40 mM PIPES-KOH at pH 6.8, 1 mM MgCl2, 1 mM EGTA) supplemented with 50 mM KCl, both enzyme complexes were fully soluble and showed activity and binding events at 50 pM protein concentration, allowing to carry out a comparative study.
Proteins used in the TIRF experiments. (A) Schematic representation of the proteins with color code as in Fig. 5. VASH1 (V1) is represented in blue, VASH2 (V2) in orange, SVBP in purple, sfGFP in green, tags (myc, FLAG, His) in gray. Vertical bars, catalytic triads. FL, full length; CD, core domain; Nt, N-terminal region; Ct, C-terminal region. For most proteins, an inactive protein (dead version, with the catalytic cysteine residue mutated into an alanine residue) was also used. (B) SDS-PAGE and immunoblots of a complete set of VASH1–SVBP protein preparations. Uncropped images are shown in source data. (C) Immunoblot of three separate brains tubulin preparations (1, 2, 3), and of tyrosinated- and detyrosinated-HeLa tubulin preparations (Tyr, deTyr). These samples were co-analyzed with extracts from HEK293T cells transfected with various mCherry α-tubulin variants (Tyr, deTyr, ∆2). Uncropped images are shown in source data. The mean content of the different forms of α-tubulin present in a brain tubulin preparation was estimated after normalization to total α-tubulin levels (and antibody sensitivity (as in Aillaud et al., 2016) to be of 43, 49.5, and 7.5%, for tyrosinated-, detyrosinated-, and ∆2-tubulin respectively. HeLa tubulin was either 100% tyrosinated or 100% detyrosinated. Taxol-stabilized Tyr-MTs and deTyr-MTs were composed of 65% of HeLa tubulin and 35% of brain tubulin (see Materials and methods). Source data are available for this figure: SourceData FS1.
Proteins used in the TIRF experiments. (A) Schematic representation of the proteins with color code as in Fig. 5. VASH1 (V1) is represented in blue, VASH2 (V2) in orange, SVBP in purple, sfGFP in green, tags (myc, FLAG, His) in gray. Vertical bars, catalytic triads. FL, full length; CD, core domain; Nt, N-terminal region; Ct, C-terminal region. For most proteins, an inactive protein (dead version, with the catalytic cysteine residue mutated into an alanine residue) was also used. (B) SDS-PAGE and immunoblots of a complete set of VASH1–SVBP protein preparations. Uncropped images are shown in source data. (C) Immunoblot of three separate brains tubulin preparations (1, 2, 3), and of tyrosinated- and detyrosinated-HeLa tubulin preparations (Tyr, deTyr). These samples were co-analyzed with extracts from HEK293T cells transfected with various mCherry α-tubulin variants (Tyr, deTyr, ∆2). Uncropped images are shown in source data. The mean content of the different forms of α-tubulin present in a brain tubulin preparation was estimated after normalization to total α-tubulin levels (and antibody sensitivity (as in Aillaud et al., 2016) to be of 43, 49.5, and 7.5%, for tyrosinated-, detyrosinated-, and ∆2-tubulin respectively. HeLa tubulin was either 100% tyrosinated or 100% detyrosinated. Taxol-stabilized Tyr-MTs and deTyr-MTs were composed of 65% of HeLa tubulin and 35% of brain tubulin (see Materials and methods). Source data are available for this figure: SourceData FS1.
We first tested the activity of VASH1–SVBP and VASH2–SVBP in vitro using immunofluorescence (Fig. 1, A−C). V1_FL and V2_FL were incubated with Taxol-stabilized Tyr-MTs for 30 min. Both complexes led to a decrease of tyrosinated and an increase of detyrosinated microtubules (Fig. 1 A). Quantification of tyrosination and detyrosination levels showed that the changes were significantly larger for V1_FL than for V2-FL (Fig. 1 B). Interestingly, while the V1_FL induced a global microtubule detyrosination, V2_FL induced the formation of local areas of modifications of the microtubule lattice (Fig. 1, A and C). Extensive regions of microtubules remained unmodified upon incubation with V2_FL over a 30-min period, which was not the case for V1_FL. In order to avoid a contribution of the fusion tags present in both the VASHs on their behavior, we tested enzymatic complexes with no tags on the VASHs and only a 6His tag at the N-terminus of SVBP (Wang et al., 2019). GFP-untagged proteins behaved similarly to their tagged counterparts: the VASH1–SVBP complex induced global detyrosination while VASH2–SVBP led to spots of detyrosination (Fig. S2, A−D).
VASH1–SVBP and VASH2–SVBP complexes exhibit dissimilar microtubule detyrosination activities and very different binding behaviors on microtubules. (A−C) Comparison of the activity of sfGFP-tagged VASH1–SVBP (V1_FL) and VASH2–SVBP (V2_FL) enzyme complexes (50 pM) on Taxol-stabilized Tyr-MTs measured by immunofluorescence in BRB40 supplemented with 50 mM KCl. (A) Representative images of tyrosinated (Tyr, magenta) and detyrosinated (deTyr, green) α-tubulin pools of microtubules after 30 min incubation in the absence (control) or presence of the indicated enzyme complexes. Scale bar, 10 µm. (B) Analysis of tyrosinated- and detyrosinated-tubulin signal intensity. Each point represents a microtubule (at least 300 microtubules were analyzed). Data are represented as the median with the interquartile range. Statistical significance was determined using Kruskal-Wallis test, ****P < 0.0001. (C) Graphs of fluorescence intensity variations of tyrosinated (magenta) and detyrosinated (green) tubulin on selected microtubules in the absence and presence of enzyme. Scale bar, 5 µM. (D−H) TIRF microscopy study of single molecules of sfGFP-tagged VASH1–SVBP (V1_FL and deadV1_FL) and VASH2–SVBP (V2_FL and deadV2_FL) bound to microtubules in the same experimental conditions as in A–C. (D) Representative kymographs. Scale bars: horizontal, 5 µm; vertical, 5 s. Examples of TIRF movies for active VASH1–SVBP and VASH2–SVBP from which kymographs were extracted are presented in supplemental data (Videos 1 and 2). (E−H) Analysis of the binding characteristics. Results for VASH1–SVBP are in blue and for VASH2–SVBP in orange, with plain circles for active enzymes (V1_FL and V2_FL) and empty circles for their catalytically dead versions (deadV1_FL and deadV2_FL). (E) Representation of the microtubule surface covered with VASH–SVBP molecules (white) or not covered (black) during the 45 s of a TIRF movie. Each horizontal line represents a microtubule (at least 19 microtubules were analyzed). The red value corresponds to the mean covering (in %). (F) Cumulative frequency of the residence times measured in TIRF movies taken during the 30 min following addition of enzyme complexes to microtubules. The mean residence time (τ) is obtained by fitting the curve with a mono-exponential function. (G) Analysis of binding frequency. Each point represents a microtubule. (H) Analysis of diffusion. Each point represents a single molecule, the molecules moving on at least 15 microtubules were analyzed. Data are represented as the median with the interquartile range. Statistical significance was determined using Kruskal-Wallis test. *P < 0.01, ***P < 0.005, ****P < 0.0001. Binding characteristics are also summarized in Table 1.
VASH1–SVBP and VASH2–SVBP complexes exhibit dissimilar microtubule detyrosination activities and very different binding behaviors on microtubules. (A−C) Comparison of the activity of sfGFP-tagged VASH1–SVBP (V1_FL) and VASH2–SVBP (V2_FL) enzyme complexes (50 pM) on Taxol-stabilized Tyr-MTs measured by immunofluorescence in BRB40 supplemented with 50 mM KCl. (A) Representative images of tyrosinated (Tyr, magenta) and detyrosinated (deTyr, green) α-tubulin pools of microtubules after 30 min incubation in the absence (control) or presence of the indicated enzyme complexes. Scale bar, 10 µm. (B) Analysis of tyrosinated- and detyrosinated-tubulin signal intensity. Each point represents a microtubule (at least 300 microtubules were analyzed). Data are represented as the median with the interquartile range. Statistical significance was determined using Kruskal-Wallis test, ****P < 0.0001. (C) Graphs of fluorescence intensity variations of tyrosinated (magenta) and detyrosinated (green) tubulin on selected microtubules in the absence and presence of enzyme. Scale bar, 5 µM. (D−H) TIRF microscopy study of single molecules of sfGFP-tagged VASH1–SVBP (V1_FL and deadV1_FL) and VASH2–SVBP (V2_FL and deadV2_FL) bound to microtubules in the same experimental conditions as in A–C. (D) Representative kymographs. Scale bars: horizontal, 5 µm; vertical, 5 s. Examples of TIRF movies for active VASH1–SVBP and VASH2–SVBP from which kymographs were extracted are presented in supplemental data (Videos 1 and 2). (E−H) Analysis of the binding characteristics. Results for VASH1–SVBP are in blue and for VASH2–SVBP in orange, with plain circles for active enzymes (V1_FL and V2_FL) and empty circles for their catalytically dead versions (deadV1_FL and deadV2_FL). (E) Representation of the microtubule surface covered with VASH–SVBP molecules (white) or not covered (black) during the 45 s of a TIRF movie. Each horizontal line represents a microtubule (at least 19 microtubules were analyzed). The red value corresponds to the mean covering (in %). (F) Cumulative frequency of the residence times measured in TIRF movies taken during the 30 min following addition of enzyme complexes to microtubules. The mean residence time (τ) is obtained by fitting the curve with a mono-exponential function. (G) Analysis of binding frequency. Each point represents a microtubule. (H) Analysis of diffusion. Each point represents a single molecule, the molecules moving on at least 15 microtubules were analyzed. Data are represented as the median with the interquartile range. Statistical significance was determined using Kruskal-Wallis test. *P < 0.01, ***P < 0.005, ****P < 0.0001. Binding characteristics are also summarized in Table 1.
Comparison of tubulin detyrosinating activity of sfGFP-tagged and untagged VASH–SVBP complexes , and evolution over the time. VASH1–SVBP and VASH2-SVBP (V1_FL and V2_FL) were produced with or without an sfGFP tag (see Materials and methods) and activity of 50 pM of the different complexes was measured by immunofluorescence in BRB40 with 50 mM KCl on Tyr-MTs. (A and C) Representative images of tyrosinated and detyrosinated α-tubulin pools of microtubules after 30 min incubation in the absence (control) or presence of the indicated enzyme complexes. Scale bar, 10 µm. (B and D) Analysis of tyrosinated-tubulin signals intensities of the experiments presented in A and C, respectively. Each point represents a microtubule (at least 150 microtubules were analyzed). Data are represented as the median with the interquartile range. Kruskal-Wallis test, ****P < 0.0001. (E) Analysis over 30 min (time course of a TIRF experiment) of the decrease in intensity of the tyrosinated-tubulin signals in the presence of active VASH1–SVBP. Each point represents a microtubule (at least 115 microtubules were analyzed). Data are represented as the median with the interquartile range. Statistical significance was determined using Kruskal-Wallis test, ****P < 0.0001. (F) Residence-time analysis of active and dead versions of VASH1–SVBP (V1_FL and deadV1_FL) during a TIRF experiment performed on the same day and in the same experimental conditions as in E. Each point represents the mean residence time of enzymes moving on at least four microtubules (one or two slides were analyzed per indicated time).
Comparison of tubulin detyrosinating activity of sfGFP-tagged and untagged VASH–SVBP complexes , and evolution over the time. VASH1–SVBP and VASH2-SVBP (V1_FL and V2_FL) were produced with or without an sfGFP tag (see Materials and methods) and activity of 50 pM of the different complexes was measured by immunofluorescence in BRB40 with 50 mM KCl on Tyr-MTs. (A and C) Representative images of tyrosinated and detyrosinated α-tubulin pools of microtubules after 30 min incubation in the absence (control) or presence of the indicated enzyme complexes. Scale bar, 10 µm. (B and D) Analysis of tyrosinated-tubulin signals intensities of the experiments presented in A and C, respectively. Each point represents a microtubule (at least 150 microtubules were analyzed). Data are represented as the median with the interquartile range. Kruskal-Wallis test, ****P < 0.0001. (E) Analysis over 30 min (time course of a TIRF experiment) of the decrease in intensity of the tyrosinated-tubulin signals in the presence of active VASH1–SVBP. Each point represents a microtubule (at least 115 microtubules were analyzed). Data are represented as the median with the interquartile range. Statistical significance was determined using Kruskal-Wallis test, ****P < 0.0001. (F) Residence-time analysis of active and dead versions of VASH1–SVBP (V1_FL and deadV1_FL) during a TIRF experiment performed on the same day and in the same experimental conditions as in E. Each point represents the mean residence time of enzymes moving on at least four microtubules (one or two slides were analyzed per indicated time).
Together, these results show that both the VASH1–SVBP and the VASH2–SVBP enzyme complexes effectively detyrosinates microtubules in vitro, however, in a different manner.
Difference in the interaction of VASH1–SVBP and VASH2–SVBP with microtubules
In order to understand the molecular mechanisms involved in the distinct detyrosination patterns, we monitored the interaction of the full-length enzyme complexes, V1_FL and V2_FL, with Taxol-stabilized Tyr-MTs in BRB40 supplemented with 50 mM KCL. We also examined the microtubule-interacting behavior of catalytically inactive enzyme complexes containing a Cys169Ala mutation in VASH1 (deadV1_FL) or a Cys158Ala mutation in VASH2 (deadV2_FL; Aillaud et al., 2017; Wang et al., 2019). Interestingly, as shown in representative kymographs, the binding behavior of the two enzyme complexes on microtubules was very different (Fig. 1 D). Whereas both active and dead versions of VASH1–SVBP exhibited short and frequent binding events, active and dead versions of VASH2–SVBP bound less frequently and for much longer times on microtubules (Videos 1 and 2). As illustrated in Fig. 1 E, the local activity of VASH2–SVBP was correlated with poor covering of microtubules by this enzyme complex in TIRF experiments carried out under the same conditions. In contrast, microtubule covering by VASH1–SVBP enzymes was high (75 and 82% covering for V1_FL and deadV1_FL, respectively, versus 25 and 28% covering for V2_FL and deadV2_FL, respectively).
Interaction of single molecules of sfGFP–VASH1–SVBP (V1_FL) with Taxol-stabilized microtubules enriched in tyrosinated HeLa tubulin studied by single-molecule TIRF microscopy. VASH1 complex (50 pM, green) exhibited short and frequent binding events on microtubules (magenta), and diffused in both directions. Scale bar, 2 μm. Images were collected every 50 ms for 45 s and films are 2.5× accelerated.
Interaction of single molecules of sfGFP–VASH1–SVBP (V1_FL) with Taxol-stabilized microtubules enriched in tyrosinated HeLa tubulin studied by single-molecule TIRF microscopy. VASH1 complex (50 pM, green) exhibited short and frequent binding events on microtubules (magenta), and diffused in both directions. Scale bar, 2 μm. Images were collected every 50 ms for 45 s and films are 2.5× accelerated.
Interaction of single molecules of sfGFP-VASH2–SVBP (V2_FL) with Taxol-stabilized microtubules enriched in tyrosinated HeLa tubulin studied by single-molecule TIRF microscopy. VASH2 complex (50 pM, green) bound less frequently, for much longer times, and diffused significantly less on microtubules (magenta) than VASH1 complex. Scale bar, 2 μm. Images were collected every 50 ms for 45 s and films are 2.5× accelerated.
Interaction of single molecules of sfGFP-VASH2–SVBP (V2_FL) with Taxol-stabilized microtubules enriched in tyrosinated HeLa tubulin studied by single-molecule TIRF microscopy. VASH2 complex (50 pM, green) bound less frequently, for much longer times, and diffused significantly less on microtubules (magenta) than VASH1 complex. Scale bar, 2 μm. Images were collected every 50 ms for 45 s and films are 2.5× accelerated.
We next analyzed the binding parameters (residence-time, binding frequency, diffusion) of the enzyme complexes on microtubule lattices in these experimental conditions (Fig. 1, F−H, and Table 1). Within 30 min after the addition of the enzyme complex, the residence-time distribution of VASH1–SVBP on microtubules was monoexponential with a mean residence-time (τ) of 0.5 and 1.0 s for the active and dead enzyme versions, respectively. The mean residence-time of full-length VASH2–SVBP was estimated higher than 11 s for the active version and higher than 14.6 s for the dead version, and thus at least 15 to 22 times higher than the residence-time of VASH1–SVBP (Fig. 1 F). A precise determination of the residence time for VASH2–SVBP was not possible in the experimental buffer conditions used to study both complexes side by side. Indeed, most VASH2–SVBP molecules were already attached to microtubules at the beginning of the TIRF movies and single-molecule traces disappeared during movie acquisition due to bleaching of the sfGFP fluorophore (see Materials and methods for details). As illustrated in Fig. 1 G, VASH2–SVBP displayed a 6–10-times lower binding frequency than VASH1–SVBP (13.0 events/min.µm.nM for V2-FL and 13.9 events/min.µm.nM for deadV2-FL to be compared to 80.8 events/min.µm.nM for V1_FL and 132.2 events/min.µm.nM for deadV1_FL). Both enzymes diffused along the microtubule lattice, with VASH2–SVBP diffusing more slowly than VASH1–SVBP (1.8 and 1.9 µm.s−1 for V1_FL and deadV1_FL, respectively, to be compared to 1.4 and 1.0 µm.s−1 for V2_FL and deadV2_FL, respectively; Fig. 1 H).
Binding parameters of VASH–SVBP complexes interaction with tyrosinated- and detyrosinated-enriched microtubules in variable ionic strength media
| Experiment (medium) . | Ionic strength (mM) . | Microtubule . | Recombinant protein (concentration) . | Residence time τ (s) . | Binding frequency (min−1.nM−1.µm-1) . |
|---|---|---|---|---|---|
| A (BRB40, 50 mM KCl) | 140 | Tyr-MTs | V1_FL (50 pM) | 0.5 | 80.8 |
| Tyr-MTs | deadV1_FL (50 pM) | 1.0 | 132.2 | ||
| Tyr-MTs | V2_FL (50 pM) | >11 | 13.0 | ||
| Tyr-MTs | deadV2_FL (50 pM) | >14.6 | 13.9 | ||
| B (BRB80) | 173 | Tyr-MTs | V1_FL (50 pM) | 0.8 | 51.6 |
| Tyr-MTs | deadV1_FL (50 pM) | 0.9 | 86.4 | ||
| C (BRB80, 50 mM KCl) | 223 | Tyr-MTs | V1_FL (1 nM) | 0.2 | 3.0 |
| Tyr-MTs | deadV1_FL (1 nM) | 0. 2 | 12.7 | ||
| Tyr-MTs | V2_FL (100 pM) | 3.9 | 20.4 | ||
| Tyr-MTs | deadV2_FL (100 pM) | >7.2 | 13.5 | ||
| D (BRB80, 100 mM KCl) | 273 | Tyr-MTs | V2_FL (50 pM) | 0.9 | 14.9 |
| Tyr-MTs | deadV2_FL (50 pM) | 1.4 | 19.3 | ||
| deTyr-MTs | V2_FL (50 pM) | 0.6 | 13.7 | ||
| deTyr-MTs | deadV2_FL (50 pM) | 0.7 | 41.0 | ||
| E (BRB40, 50 mM KCl) | 140 | Tyr-MTs | V1_FL (50 pM) | 0.8 | 53.4 |
| Tyr-MTs | deadV1_FL (50 pM) | 1.2 | 89.0 | ||
| deTyr-MTs | V1_FL (50 pM) | 0.5 | 42.3 | ||
| deTyr-MTs | deadV1_FL (50 pM) | 0.6 | 66.5 |
| Experiment (medium) . | Ionic strength (mM) . | Microtubule . | Recombinant protein (concentration) . | Residence time τ (s) . | Binding frequency (min−1.nM−1.µm-1) . |
|---|---|---|---|---|---|
| A (BRB40, 50 mM KCl) | 140 | Tyr-MTs | V1_FL (50 pM) | 0.5 | 80.8 |
| Tyr-MTs | deadV1_FL (50 pM) | 1.0 | 132.2 | ||
| Tyr-MTs | V2_FL (50 pM) | >11 | 13.0 | ||
| Tyr-MTs | deadV2_FL (50 pM) | >14.6 | 13.9 | ||
| B (BRB80) | 173 | Tyr-MTs | V1_FL (50 pM) | 0.8 | 51.6 |
| Tyr-MTs | deadV1_FL (50 pM) | 0.9 | 86.4 | ||
| C (BRB80, 50 mM KCl) | 223 | Tyr-MTs | V1_FL (1 nM) | 0.2 | 3.0 |
| Tyr-MTs | deadV1_FL (1 nM) | 0. 2 | 12.7 | ||
| Tyr-MTs | V2_FL (100 pM) | 3.9 | 20.4 | ||
| Tyr-MTs | deadV2_FL (100 pM) | >7.2 | 13.5 | ||
| D (BRB80, 100 mM KCl) | 273 | Tyr-MTs | V2_FL (50 pM) | 0.9 | 14.9 |
| Tyr-MTs | deadV2_FL (50 pM) | 1.4 | 19.3 | ||
| deTyr-MTs | V2_FL (50 pM) | 0.6 | 13.7 | ||
| deTyr-MTs | deadV2_FL (50 pM) | 0.7 | 41.0 | ||
| E (BRB40, 50 mM KCl) | 140 | Tyr-MTs | V1_FL (50 pM) | 0.8 | 53.4 |
| Tyr-MTs | deadV1_FL (50 pM) | 1.2 | 89.0 | ||
| deTyr-MTs | V1_FL (50 pM) | 0.5 | 42.3 | ||
| deTyr-MTs | deadV1_FL (50 pM) | 0.6 | 66.5 |
(A) Related to Fig. 1, D, F, and G. (B) Related to Fig. 2 A. (C) Related to Fig. 2 B. (D) Related to Fig. 2, C, G, and H. (E) related to Fig. 2, E and F. A−E represent experiments performed on different days. At least 15 microtubules were analyzed. See Materials and methods for detailed experimental conditions.
As the binding of microtubule-associated proteins to microtubules often involves electrostatic attractive interactions (Roll-Mecak, 2015; Zhernov et al., 2020), we tested whether an increase in ionic strength affects the interaction of VASH–SVBP with microtubules. This significantly decreased the residence time of both enzyme complexes on microtubules (Fig. 2, A−C, and Table 1) and allowed to determine the residence-time for VASH2–SVBP binding (Fig. 2 C and Table 1 D). Because it does not alter the evaluation of residence time and allows better visibility of traces on microtubules, the concentration of VASH1–SVBP was increased when using high ionic strength buffers. Despite reducing the residence time, VASH2–SVBP still stayed much longer on microtubules than VASH1–SVBP (Table 1 C, τ = 0.2 s for V1_FL versus τ = 3.9 s for V2_FL in BRB80 supplemented with 50 mM KCl, which represents more than 80 mM increase in ionic strength compared to BRB40 supplemented with 50 mM KCl). We next assayed the impact of ionic strength on the enzymatic activity by immunofluorescence. Quantifications of the tyrosinated tubulin signal of images from experiments performed in varying buffers demonstrated that, like the binding behavior, the enzymatic activity of the two VASH–SVBP variants was salt-dependent: increasing the ionic strength led to a significant reduction of the enzymatic activity (Fig. 2 D).
Ionic strength has a strong impact on the interaction of VASH–SVBP complexes with microtubules, whereas tyrosinated-tubulin depletion disrupts less the interaction. (A−D) Effect of ionic strength. Experiments were performed in the indicated buffers. Ionic strengths are indicated in brackets in mM. (A−C) TIRF microscopy study. Representative kymographs of single molecules of sfGFP-tagged VASH1–SVBP and VASH2–SVBP (V1_FL, deadV1_FL, V2_FL, and deadV2_FL) at the indicated concentrations bound to Taxol-stabilized Tyr-MTs. Scale bars: horizontal, 5 µm; vertical, 5 s. Binding characteristics are summarized in Table 1, B–D. (D) Activity measurement by immunofluorescence. Analysis of tyrosinated signals intensities after 30 min incubation in the absence (control) or presence of 50 pM enzymes. Each point represents a microtubule (at least 150 microtubules were analyzed). Data are represented as the median with the interquartile range. (E−H) Effect of decreasing tyrosinated tubulin. TIRF microscopy study of single molecules of sfGFP-tagged VASH1–SVBP (V1_FL and deadV1_FL, 50 pM) and VASH2–SVBP (V2_FL and deadV2_FL, 50 pM) bound to Taxol-stabilized Tyr-MTs or deTyr-MTs. Binding characteristics are summarized in Table 1. (E and G) Representative kymographs. Scale bars: horizontal, 5 µm; vertical, 5 s. (F and H) Analysis of binding frequency. Each point represents a microtubule. At least 15 microtubules were analyzed. Data are represented as the median with the interquartile range. Statistical significance was determined using Kruskal-Wallis test. **P < 0.009, ****P < 0.0001.
Ionic strength has a strong impact on the interaction of VASH–SVBP complexes with microtubules, whereas tyrosinated-tubulin depletion disrupts less the interaction. (A−D) Effect of ionic strength. Experiments were performed in the indicated buffers. Ionic strengths are indicated in brackets in mM. (A−C) TIRF microscopy study. Representative kymographs of single molecules of sfGFP-tagged VASH1–SVBP and VASH2–SVBP (V1_FL, deadV1_FL, V2_FL, and deadV2_FL) at the indicated concentrations bound to Taxol-stabilized Tyr-MTs. Scale bars: horizontal, 5 µm; vertical, 5 s. Binding characteristics are summarized in Table 1, B–D. (D) Activity measurement by immunofluorescence. Analysis of tyrosinated signals intensities after 30 min incubation in the absence (control) or presence of 50 pM enzymes. Each point represents a microtubule (at least 150 microtubules were analyzed). Data are represented as the median with the interquartile range. (E−H) Effect of decreasing tyrosinated tubulin. TIRF microscopy study of single molecules of sfGFP-tagged VASH1–SVBP (V1_FL and deadV1_FL, 50 pM) and VASH2–SVBP (V2_FL and deadV2_FL, 50 pM) bound to Taxol-stabilized Tyr-MTs or deTyr-MTs. Binding characteristics are summarized in Table 1. (E and G) Representative kymographs. Scale bars: horizontal, 5 µm; vertical, 5 s. (F and H) Analysis of binding frequency. Each point represents a microtubule. At least 15 microtubules were analyzed. Data are represented as the median with the interquartile range. Statistical significance was determined using Kruskal-Wallis test. **P < 0.009, ****P < 0.0001.
Together, these results clearly highlight the different binding behaviors of the two VASH–SVBP enzyme complexes and the importance of electrostatic attractive interactions for their microtubule binding.
Tyrosination has a little effect on the interaction of VASH–SVBP complexes with microtubules
We then wondered how VASH–SVBP complexes behave on detyrosinated, non-substrate microtubules. We compared the interaction of both enzyme complexes with microtubules enriched either in tyrosinated or detyrosinated HeLa tubulin (deTyr-MTs; Tyr-MTs contained 80% tyrosinated tubulin, 17% detyrosinated tubulin, and 3% ∆2-tubulin [a variant missing the two C-terminal glutamate residues]; deTyr-MTs contained 15% tyrosinated tubulin, 82% detyrosinated tubulin, and 3% ∆2-tubulin; Fig. S1 C and Materials and methods). We tested both active and inactive enzymes to assay the relevance of detyrosination activity on microtubule binding. To determine the binding characteristics of the two enzymes, a low ionic strength buffer (140 mM, BRB40 supplemented with 50 mM KCl) was used for VASH1–SVBP and a high ionic strength buffer (273 mM, BRB80 supplemented with 100 mM KCl) for VASH2–SVBP. Representative kymographs and binding characteristics are presented in Fig. 2, E−H, and Table 1, D and E.
In low ionic strength buffer, both catalytically active and dead versions of VASH1–SVBP exhibited short residence times on deTyr-MTs (τ = 0.5 s for V1_FL and τ = 0.6 s for deadV1_FL) and stayed longer on Tyr-MTs, with the dead enzyme complex remaining on Tyr-MTs for slightly longer periods of times than its active counterparts (τ = 0.8 s for V1_FL and τ = 1.2 s for deadV1_FL; Table 1 E). The binding frequency of the catalytically dead VASH1–SVBP mutant was significantly higher than the one of its active counterpart on both Tyr-MTs and deTyr-MTs (Fig. 2 F).
In high ionic strength buffer, catalytically active and dead versions of VASH2–SVBP also displayed short residence times on deTyr-MTs (τ = 0.6 s for V2_FL and τ = 0.7 s deadV2_FL) and stayed longer on Tyr-MTs, with the dead enzyme complex staying slightly longer times on Tyr-MTs than its active counterparts (τ = 0.9 s for V2_FL and τ = 1.4 s for deadV2_FL; Table 1 D). The two VASH2–SVBP versions bound Tyr-MTs with similar frequencies (Fig. 2 H). Surprisingly, the binding frequencies of catalytically dead VASH2–SVBP were higher on deTyr-MTs than on Tyr-MTs (Fig. 2, G and H).
Thus, our results reveal slightly longer binding periods of both VASH1–SVBP and VASH2–SVBP enzyme complexes on tyrosinated microtubules than on detyrosinated ones. This was best observed with the catalytically dead mutants that do not modify microtubules (twofold longer binding). With active enzyme complexes, tyrosinated microtubules are progressively detyrosinated which, during the 30-min time of movie acquisition, could progressively reduce the residence time of the enzyme complexes (as shown for the highly efficient VASH1–SVBP enzyme complex; Fig. S2, E and F). In the case of the catalytically dead VASH–SVBP versions, microtubules are not modified over the entire time of the experiment, allowing a detailed analysis of the interaction between enzymes and microtubules. In addition, we found that at an appropriate ionic strength the two dead mutants were able to dissociate from the microtubule lattice, implying that the enzymes’ detyrosination activity is not essential for the release of these enzymes from microtubules. Notably, in such conditions catalytically active and dead versions of the two enzyme complexes showed diffusion in both directions along Taxol-stabilized, tyrosination-enriched microtubules.
Collectively, these results suggest that in vitro, VASH1–SVBP exhibits short and diffusive binding events on microtubules while VASH2–SVBP displays longer and much more static attachments to them. In addition, the catalytic activity of the two enzyme complexes does not appear to be essential for their binding behavior on microtubules.
Cryo-EM structure of VASH2–SVBP in complex with a microtubule
The functional results collected so far suggest that VASH1–SVBP and VASH2–SVBP work very differently on microtubules. We hypothesized that this could be related to a different binding mode of the two enzyme complexes with microtubules. To investigate this idea, we analyzed the structure of VASH2–SVBP in complex with microtubules by cryo-EM. We used the catalytically dead mutant of VASH2–SVBP and GMPCPP (Guanosine-5'-[(α,β)-methyleno]triphosphate)-stabilized HeLa cell microtubules (Fig. 3 A–C, and Fig. S3). Based on a single-particle approach (see Materials and methods), we found that the majority of microtubules contained 14-protofilaments. Downstream extraction and single-particle classification of protofilaments revealed that VASH2–SVBP molecules were present in ∼40% of the total binding sites. Subsequent refinement of the particle subset generated an electron density map of two laterally associated αβ-tubulin heterodimers in complex with VASH2–SVBP (referred to as microtubule–VASH2–SVBP complex from here onwards for simplicity) at a global resolution of 3.1 Å (Fig. 3 B and Fig. S3 B). The resolution calculated for VASH2–SVBP alone was 3.7 Å; however, it varies from 3.3 to 4.8 Å and becomes progressively better in regions located closer to the microtubule (Fig. S3 C and Table 2).
Cryo-EM reconstruction of the microtubule–VASH2–SVBP complex reveals their interaction sites. (A) Electron density map of VASH2–SVBP (catalytically dead version of VASH2 carrying the Cys158Ala mutation) bound to a GMPCPP-stabilized microtubule. A map region of two laterally associated αβ-tubulin dimers from two adjacent protofilaments bound to one VASH2–SVBP complex is shown in two different orientations 90° apart. (B) Overview of the three microtubule–VASH2–SVBP interacting sites from two different orientations 90° apart. Sites 1 and 2 involve the substrate α-tubulin while site 3 involves the α’-tubulin subunit of an adjacent protofilament in the microtubule. All molecules are displayed in cartoon representation. The white arrow close to site 2 in the lower panel indicates the C-terminus of helix αH12 and the black arrow denotes the facing direction of the positively charged groove on VASH2. (C) Surface representation of the VASH2–SVBP structure with the microtubule-binding residues highlighted in orange stick. Insets correspond to VASH2 and tubulin residues implicated in the three interaction sites, displayed in stick representation and labeled in orange and gray, respectively. The α-tubulin, β-tubulin, VASH2, and SVBP molecules are shown in light gray, dark gray, orange, and violet, respectively. (D) Immunoblots of in cellulo tubulin detyrosination assays with WT VASH2 or VASH2 mutants 1 (R134A-R137A), 2 (H257A-F259A), or 3 (R288A-R291A-M292A), and SVBP. Uncropped images are shown in source data. (E) Quantification of the colocalization between VASH2 proteins and microtubules. NIH3T3 cells were cotransfected with plasmids encoding WT or mutant VASH2-eGFP, and SVBP-myc-FLAG. Representative immunofluorescence images are shown on Fig. S4. Each point represents a different cell; at least 8 cells from three independent experiments were analyzed. Bars represent the median and interquartile range. Statistical significance was determined using the Kruskal-Wallis test, ****P < 0.0001. Source data are available for this figure: SourceData F3.
Cryo-EM reconstruction of the microtubule–VASH2–SVBP complex reveals their interaction sites. (A) Electron density map of VASH2–SVBP (catalytically dead version of VASH2 carrying the Cys158Ala mutation) bound to a GMPCPP-stabilized microtubule. A map region of two laterally associated αβ-tubulin dimers from two adjacent protofilaments bound to one VASH2–SVBP complex is shown in two different orientations 90° apart. (B) Overview of the three microtubule–VASH2–SVBP interacting sites from two different orientations 90° apart. Sites 1 and 2 involve the substrate α-tubulin while site 3 involves the α’-tubulin subunit of an adjacent protofilament in the microtubule. All molecules are displayed in cartoon representation. The white arrow close to site 2 in the lower panel indicates the C-terminus of helix αH12 and the black arrow denotes the facing direction of the positively charged groove on VASH2. (C) Surface representation of the VASH2–SVBP structure with the microtubule-binding residues highlighted in orange stick. Insets correspond to VASH2 and tubulin residues implicated in the three interaction sites, displayed in stick representation and labeled in orange and gray, respectively. The α-tubulin, β-tubulin, VASH2, and SVBP molecules are shown in light gray, dark gray, orange, and violet, respectively. (D) Immunoblots of in cellulo tubulin detyrosination assays with WT VASH2 or VASH2 mutants 1 (R134A-R137A), 2 (H257A-F259A), or 3 (R288A-R291A-M292A), and SVBP. Uncropped images are shown in source data. (E) Quantification of the colocalization between VASH2 proteins and microtubules. NIH3T3 cells were cotransfected with plasmids encoding WT or mutant VASH2-eGFP, and SVBP-myc-FLAG. Representative immunofluorescence images are shown on Fig. S4. Each point represents a different cell; at least 8 cells from three independent experiments were analyzed. Bars represent the median and interquartile range. Statistical significance was determined using the Kruskal-Wallis test, ****P < 0.0001. Source data are available for this figure: SourceData F3.
Cryo-EM data collection and processing. (A) Representative cryo-EM micrograph of microtubule–VASH2–SVBP specimens. (B) Map:map FSC resolution plot of the EM map corresponding to the microtubule–VASH2–SVBP complex (mask: two laterally associated αβ-tubulin heterodimers in complex with VASH2–SVBP). (C) Resolution map of the microtubule–VASH2–SVBP complex structure. (D and E) Exerts of the microtubule–VASH2–SVBP electron density map highlighting the quality of representative regions of αβ-tubulin, VASH2, and nucleotides. (F) Model:map FSC plot between the refined structure of the tubulin–VASH2–SVBP and the corresponding EM density map (mask: VASH2–SVBP).
Cryo-EM data collection and processing. (A) Representative cryo-EM micrograph of microtubule–VASH2–SVBP specimens. (B) Map:map FSC resolution plot of the EM map corresponding to the microtubule–VASH2–SVBP complex (mask: two laterally associated αβ-tubulin heterodimers in complex with VASH2–SVBP). (C) Resolution map of the microtubule–VASH2–SVBP complex structure. (D and E) Exerts of the microtubule–VASH2–SVBP electron density map highlighting the quality of representative regions of αβ-tubulin, VASH2, and nucleotides. (F) Model:map FSC plot between the refined structure of the tubulin–VASH2–SVBP and the corresponding EM density map (mask: VASH2–SVBP).
Cryo-EM data collection, refinement, and structure validation
| Data collection . | Microtubule–VASH2–SVBP . |
|---|---|
| Magnification | 47,260× |
| Pixel size (Å) | 1.058 |
| Defocus range (µm) | −0.6 to −2.2 |
| Voltage (kV) | 300 |
| Exposure time (s per frame) | 0.2 |
| Number of frames | 45 |
| Total dose (e/Å2) | 65 |
| Reconstitution (14 protofilamentsmicrotubules) | |
| Box size (pixels) | 600 |
| Inter-box distance (Å) | 82 |
| Asymmetrical units | 1 |
| Micrographs | 3,345 |
| Picked filaments | 8,987 |
| Initial extracted segments | 153,061 |
| Segments after 3D classification | 97,645 |
| Resolution after 3D auto-refine (Å) | 4.2 |
| Final overall resolution (Å) | 3.2 |
| Estimated map sharpening B-factor (Å2) | −61.03 |
| Helical rise (Å) | 8.99 |
| Helical twist (˚) | -25.73 |
| Reconstitution (single protofilament) | |
| Particle after expanding 28× | 2,681,616 |
| Particles after 3D classification | 682,072 |
| Occupancy (%) | 50.87 |
| Resolution map:map FSC = 0.143 (Å) | 3.1 (mask: two laterally associated αβ-tubulin heterodimers in complex with VASH2–SVBP) |
| Estimated map sharpening B-factor (Å2) | −112.03 |
| Resolution map:map FSC = 0.143 (Å) | 3.7 (mask: VASH2–SVBP) |
| Atomic model | |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.005 |
| Bond angles (˚) | 0.835 |
| Validation | |
| Map CC (mask) | 0.80 |
| MolProbity clashscore | 3.36 |
| Overall score | 1.42 |
| Rotamer outliers (%) | 0.21 |
| Resolution map:model FSC= 0.5 (Å) | 3.4 |
| Ramachandran plot | |
| Favored (%) | 96.09 |
| Allowed (%) | 3.91 |
| Outliers (%) | 0.00 |
| Data collection . | Microtubule–VASH2–SVBP . |
|---|---|
| Magnification | 47,260× |
| Pixel size (Å) | 1.058 |
| Defocus range (µm) | −0.6 to −2.2 |
| Voltage (kV) | 300 |
| Exposure time (s per frame) | 0.2 |
| Number of frames | 45 |
| Total dose (e/Å2) | 65 |
| Reconstitution (14 protofilamentsmicrotubules) | |
| Box size (pixels) | 600 |
| Inter-box distance (Å) | 82 |
| Asymmetrical units | 1 |
| Micrographs | 3,345 |
| Picked filaments | 8,987 |
| Initial extracted segments | 153,061 |
| Segments after 3D classification | 97,645 |
| Resolution after 3D auto-refine (Å) | 4.2 |
| Final overall resolution (Å) | 3.2 |
| Estimated map sharpening B-factor (Å2) | −61.03 |
| Helical rise (Å) | 8.99 |
| Helical twist (˚) | -25.73 |
| Reconstitution (single protofilament) | |
| Particle after expanding 28× | 2,681,616 |
| Particles after 3D classification | 682,072 |
| Occupancy (%) | 50.87 |
| Resolution map:map FSC = 0.143 (Å) | 3.1 (mask: two laterally associated αβ-tubulin heterodimers in complex with VASH2–SVBP) |
| Estimated map sharpening B-factor (Å2) | −112.03 |
| Resolution map:map FSC = 0.143 (Å) | 3.7 (mask: VASH2–SVBP) |
| Atomic model | |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.005 |
| Bond angles (˚) | 0.835 |
| Validation | |
| Map CC (mask) | 0.80 |
| MolProbity clashscore | 3.36 |
| Overall score | 1.42 |
| Rotamer outliers (%) | 0.21 |
| Resolution map:model FSC= 0.5 (Å) | 3.4 |
| Ramachandran plot | |
| Favored (%) | 96.09 |
| Allowed (%) | 3.91 |
| Outliers (%) | 0.00 |
In the final map of the microtubule–VASH2–SVBP complex, we could reliably distinguish the α- and β-tubulin subunits by examining their corresponding S9-S10 loop densities (Fig. S3 D), a secondary structure element that differs in length between both subunits. It was straightforward to model unambiguously all regular secondary structure elements of all four protein components, α-tubulin, β-tubulin, VASH2, and SVBP, into the final electron density map, as well as a large number of residue side chains and both the tubulin guanosine nucleotides (Fig. S3, D and E).
Due to their intrinsic flexibility, the densities of the α- and β-tubulin C-terminal tails were not defined beyond the corresponding H12 helices and could thus not be modeled; the same also applied for the N- and C-termini of VASH2 and N-terminus of SVBP. The microtubule–VASH2–SVBP complex structure is very similar to the one of the free enzyme derived by x-ray crystallography (Wang et al., 2019; root-mean-square deviation over 256 Cα atoms of 0.8 Å), suggesting that the core of the enzyme does not undergo global structural rearrangement upon binding. Notably, the observed protofilament-straddling interaction between VASH2–SVBP and the microtubule (Fig. 3, A and B) is not feasible for the free αβ-tubulin heterodimer. Therefore, the characteristic binding mode of VASH2–SVBP with its microtubule-incorporated α-tubulin substrate underlies the mechanism behind the preferential activity of VASH2–SVBP for microtubules compared to free tubulin (Kumar and Flavin, 1981). Table 2 provides a summary of the validation statistics for the final atomic model. The resolution using the model-to-map Fourier Shell Correlation (FSC) curve of the final atomic model and the single particle reconstruction map with FSC = 0.5 was calculated to be 3.4 Å (Fig. S3 F).
The model of the microtubule–VASH2–SVBP complex reveals a specific binding of the enzyme complex on the external surface of the microtubule. The central, positively charged groove of VASH2, encompassing the catalytic amino acid residue triad Cys158, His193, and Ser210 (Aillaud et al., 2017), faces toward the microtubule outer surface and is situated directly above the H12 helix of α-tubulin from which the flexible C-terminal tail emanates (not visible in the electron density; Fig. 3 B). Sites of contacts between VASH2–SVBP and the microtubule outer surface were located primarily on α-tubulin suggestive of specific recognition of the target substrate, the C-terminal tyrosine of α-tubulin. Conversely, the density corresponding to SVBP was distal from the microtubule–VASH2 binding interface and did not appear to contribute to the interaction. Interestingly, the VASH2 part of the enzyme complex binds simultaneously to two α-tubulin subunits: an α-tubulin subunit in close proximity to the VASH2 catalytic site and an α-tubulin subunit on the adjacent protofilament (henceforth denoted α’-tubulin; Fig. 3 B).
In support of our previous mutagenesis results (Wang et al., 2019), inspection of the derived microtubule–VASH2–SVBP complex structural model revealed three primary sites of interaction, denoted site 1, site 2, and site 3, between the folded region of VASH2–SVBP and the ones of α- and α’-tubulin (Fig. 3 C). At site 1, residues Arg134 and Arg137 of VASH2 form electrostatic interactions with the negatively charged residue Glu433 of the H12 helix of α-tubulin. The S4-S5 loop of VASH2 is implicated in site 2 involving residue His257 and Phe259, which form polar interactions with His393 and Glu386 of α-tubulin, respectively. Site 3 engages the α’-tubulin subunit from the adjacent protofilament where a hydrophobic-aromatic residue interaction between Met292 of VASH2 and Tyr108 of α’-tubulin is implicated. In addition, residues Arg288 and Arg291 of VASH2 are likely to form electrostatic interactions with the negatively charged glutamate residues Glu420 and Glu414 of helix H12 of the α’-tubulin subunit, respectively.
To test the functional relevance of the three interfaces, we introduced double or triple mutations into VASH2 to disrupt them, co-transfected the mutants with SVBP in cells and analyzed localization and detyrosinating activity of the mutated complexes. Mutating Arg134 and Arg137 (site 1), or His257 and Phe259 (site 2), or Arg288, Arg291, and Met292 (site 3) to alanines lead to an almost complete loss of detyrosination in cells (Fig. 3 D). Whereas WT VASH2–SVBP showed a strong association with microtubules when expressed in NIH3T3 fibroblasts, all three mutants colocalized weakly with microtubules (Fig. 3 E and Fig. S4), demonstrating that these sites are important for microtubule binding.
VASH2–SVBP mutants associate poorly with microtubules in cells. Representative immunofluorescence images of NIH3T3 fibroblasts cotransfected with plasmids encoding WT or indicated VASH2-eGFP mutants and SVBP-myc-FLAG. Merged images of VASH2-eGFP (green) and α-tubulin (magenta) stainings are displayed in the bottom lane. Insets are magnifications of the white square regions. Scale bars, 25 µM for entire images and 5 µM for insets.
VASH2–SVBP mutants associate poorly with microtubules in cells. Representative immunofluorescence images of NIH3T3 fibroblasts cotransfected with plasmids encoding WT or indicated VASH2-eGFP mutants and SVBP-myc-FLAG. Merged images of VASH2-eGFP (green) and α-tubulin (magenta) stainings are displayed in the bottom lane. Insets are magnifications of the white square regions. Scale bars, 25 µM for entire images and 5 µM for insets.
Taken together, these results show that the structured domain of VASH2–SVBP is capable of binding to three distinctive sites on the microtubule surface. When bound to the microtubule, VASH2–SVBP interacts simultaneously with two adjacent protofilaments by binding both the α-tubulin substrate and the α’-tubulin of a neighboring protofilament. This interaction is driven by three binding sites that are distinct and distal from the catalytic site, and help position the positively charged groove of VASH2 in proximity to the C-terminal tail of α-tubulin. A sequence alignment of vertebrate VASH1 and VASH2 homologues (Fig. S5 A) reveals the high sequence homology for the vasohibin’s core domain, in which the three microtubule-binding interfaces are well conserved. We may thus consider them as substrate recognition elements that confer specificity to the catalytic activity of the vertebrate vasohibin family of tubulin carboxypeptidases.
Sequence alignments of VASHs. (A) ClustalWmultiple alignment of vertebrate VASH1 and VASH2 sequences (only a single VASH gene exists outside vertebrates). Sequences are placental Homo sapiens (human VASH1 NP_055724 and VASH2 NP_001287985), placental Mus musculus (mouse VASH1 NP_796328 and VASH2 NP_659128), monotremata Phascolarctos cinereus (koala VASH1 XP_020841880 and VASH2 XP_020863469), marsupial Ornithorhynchus anatinus (platypus VASH1 XP_001508093 and VASH2 XP_001510897), avian Gallus gallus (chicken VASH1 XP_015143201, XP_015139368), batrachian Xenopus laevis (Western clawed frog VASH1 XP_018085836 and VASH2 XP_018118075), teleost fish Danio rerio (zebrafish VASH1 XP_003200451 and VASH2 XP_005160797), cartilaginous fish Stegostoma fasciatum (zebra shark VASH1 JAFIRC010000009-deduced and VASH2 JAFIRC010000008-deduced), and cyclostomata Petromyzon marinus (sea lamprey VASH1 NC_046101-deduced and VASH2 XP_032818953). Alignment color code: black, dark gray, and gray shading correspond to 100, 80, and 60% of sequence homology, respectively. N-terminal, core, and C-terminal domains are separated by a vertical red line. Sites involved in microtubule binding are indicated. Black triangles located above and under the alignment show residues implicated in microtubule interaction of VASH1 (Li et al., 2020) and VASH2 (Fig. 3), respectively. These residues are red-boxed to highlight the sequence conservation. Red triangle shows the catalytic cysteine. (B) pI analysis of the VASHs domains during evolution. pI values of the N-terminal, core, and C-terminal domains were calculated based on the domain boundaries of the alignment shown in A. Color code is as in Fig. 5 C.
Sequence alignments of VASHs. (A) ClustalWmultiple alignment of vertebrate VASH1 and VASH2 sequences (only a single VASH gene exists outside vertebrates). Sequences are placental Homo sapiens (human VASH1 NP_055724 and VASH2 NP_001287985), placental Mus musculus (mouse VASH1 NP_796328 and VASH2 NP_659128), monotremata Phascolarctos cinereus (koala VASH1 XP_020841880 and VASH2 XP_020863469), marsupial Ornithorhynchus anatinus (platypus VASH1 XP_001508093 and VASH2 XP_001510897), avian Gallus gallus (chicken VASH1 XP_015143201, XP_015139368), batrachian Xenopus laevis (Western clawed frog VASH1 XP_018085836 and VASH2 XP_018118075), teleost fish Danio rerio (zebrafish VASH1 XP_003200451 and VASH2 XP_005160797), cartilaginous fish Stegostoma fasciatum (zebra shark VASH1 JAFIRC010000009-deduced and VASH2 JAFIRC010000008-deduced), and cyclostomata Petromyzon marinus (sea lamprey VASH1 NC_046101-deduced and VASH2 XP_032818953). Alignment color code: black, dark gray, and gray shading correspond to 100, 80, and 60% of sequence homology, respectively. N-terminal, core, and C-terminal domains are separated by a vertical red line. Sites involved in microtubule binding are indicated. Black triangles located above and under the alignment show residues implicated in microtubule interaction of VASH1 (Li et al., 2020) and VASH2 (Fig. 3), respectively. These residues are red-boxed to highlight the sequence conservation. Red triangle shows the catalytic cysteine. (B) pI analysis of the VASHs domains during evolution. pI values of the N-terminal, core, and C-terminal domains were calculated based on the domain boundaries of the alignment shown in A. Color code is as in Fig. 5 C.
Comparison of the microtubule-binding mode of VASH1–SVBP and VASH2–SVBP
A cryo-EM–derived structure of microtubule-bound VASH1–SVBP was recently solved based on a truncated, catalytically inactive version of VASH1 (Cys169 mutated to serine) and recombinant human tubulin (Li et al., 2020; PDB ID 6WSL). Superimposition of the microtubule-bound structures of VASH2–SVBP and VASH1–SVBP revealed minimal structural differences between the two enzyme complexes (root-mean-square deviation over 244 Cα atoms of 1.2 Å). As shown in Fig. 4 A, comparison of the microtubule-binding modes revealed that both enzyme complexes contact two adjacent α-tubulins across neighboring protofilaments simultaneously and position their catalytic site directly above the H12 helix of one of the two α-tubulin subunits. However, they do so at a different relative angle of ∼24° as calculated from a plane defined by the Cα atoms of the three residues Val81, Glu148, and Glu284 of VASH2, and Ala92, Glu159, and Glu295 of VASH1.
Comparison between the microtubule-binding modes of VASH1–SVBP and VASH2–SVBP. (A) Superimposition of the microtubule-VASH1–SVBP (PDB ID 6WSL, blue) and microtubule–VASH2–SVBP (PDB ID 7ZCW, orange) structures reveals a 24° tilt between the two enzyme complexes. (B) Superimposing the VASH2–SVBP structure (orange) onto the microtubule-bound VASH1–SVBP (blue) structure shows a steric clash of Phe259 of the β4-β5 loop of VASH2–SVBP with α-tubulin. On the top, a sequence alignment of the β4-β5 loops of VASH1 and VASH2 is shown with proline residues highlighted in bold. Phe259 of VASH2 and the corresponding Val270 of VASH1 are also highlighted in bold and are underlined. (C) At site 3, the tilt between the VASH1–SVBP (blue) and VASH2–SVBP (orange) enzyme complexes leads to a shift in the position of helix H7 with respect to α’-tubulin.
Comparison between the microtubule-binding modes of VASH1–SVBP and VASH2–SVBP. (A) Superimposition of the microtubule-VASH1–SVBP (PDB ID 6WSL, blue) and microtubule–VASH2–SVBP (PDB ID 7ZCW, orange) structures reveals a 24° tilt between the two enzyme complexes. (B) Superimposing the VASH2–SVBP structure (orange) onto the microtubule-bound VASH1–SVBP (blue) structure shows a steric clash of Phe259 of the β4-β5 loop of VASH2–SVBP with α-tubulin. On the top, a sequence alignment of the β4-β5 loops of VASH1 and VASH2 is shown with proline residues highlighted in bold. Phe259 of VASH2 and the corresponding Val270 of VASH1 are also highlighted in bold and are underlined. (C) At site 3, the tilt between the VASH1–SVBP (blue) and VASH2–SVBP (orange) enzyme complexes leads to a shift in the position of helix H7 with respect to α’-tubulin.
The alignment of vertebrate VASH1 and VASH2 sequences reveals non-identical residues at sites 2 and 3 (Fig. S5 A), which may explain the observed difference in the overall microtubule-binding mode. Indeed, superimposition of the microtubule-bound structure of VASH2–SVBP onto that of VASH1–SVBP revealed that in the S4-S5 loop of VASH2, residue Phe259 (Val270 in VASH1) experiences a steric clash with His209 of the H9-S8 loop of α-tubulin (Fig. 4 B). Inspection of the amino acid sequence of the S4-S5 loop of VASH2 revealed the presence of two additional proline residues (Pro253 and Pro261), which are substituted by a serine and a glutamine, respectively, in VASH1 (Ser264 and Gln272; Fig. 4 B). We think that the additional rigidity imposed by these proline residues might force VASH2–SVBP to adapt a tilted microtubule-binding mode compared to VASH1–SVBP to accommodate the S4-S5 loop of VASH2.
Binding site 3 displays additional differences between the structures of VASH2–SVBP and VASH1–SVBP, which may also account for the changes in the microtubule-binding mode of the two enzyme complexes. The methionine-aromatic interaction of Met292 of VASH2 with Tyr108 of α’-tubulin is not possible in VASH1 as the corresponding residue in VASH1 is a leucine (Leu303; Fig. 4 C). Furthermore, Arg291 and Met292 engage residues Glu414 and Tyr108 of α’-tubulin, respectively. In contrast, the corresponding residues in VASH1 (Arg302 and Leu303) are too far from the α’-tubulin subunit to establish a significant interaction due to a positional shift of the H7 helix (Fig. 4 C).
To determine what effect the conformational difference in microtubule binding may have on the enzymatic activity of the two VASH isoforms, we compared the relative position of the VASH1 and VASH2 catalytic sites (Fig. S6). Using the coordinates of the catalytic triad, we triangulated the centroid point of the three Cα atoms of the corresponding residues as a representative position of the substrate. Compared to VASH2, the centroid point of VASH1 shifts by 6.3 Å in the direction of the plus-end and parallel to the protofilament. This places the catalytic cysteine of VASH1 at a distance of 29.7 Å from the Ser439 residue of the H12 helix of α-tubulin, which is comparable to the distance of 27.3 Å for VASH2. Residue Ser439 was selected, as it is the closest resolved residue of the α-tubulin C-terminus to the processed tyrosine. This analysis suggests that the observed difference in the microtubule-binding mode between VASH1 and VASH2 core domains has only minimal influence in the positioning of the catalytic site relative to the H12 helix of α-tubulin, and that the accessibility of the substrate C-terminal tail is thus only marginally changed between the two vasohibin isoforms.
Relative position of the catalytic site in microtubule–VASH1 and –VASH2 complexes with respect to the C-terminus of helix H12 of α-tubulin. The centroid of the catalytic residue triad is displaced by 6.3 Å in VASH1 (Li et al., 2020) compared to VASH2 due to the observed tilt between the two enzyme complexes (Fig. 4). However, the distance between the catalytic triad centroid of VASH1 and VASH2 to the Cα atom of residue S439 of helix H12 of a-tubulin is very similar (29.7 versus 27.3 Å, respectively).
Relative position of the catalytic site in microtubule–VASH1 and –VASH2 complexes with respect to the C-terminus of helix H12 of α-tubulin. The centroid of the catalytic residue triad is displaced by 6.3 Å in VASH1 (Li et al., 2020) compared to VASH2 due to the observed tilt between the two enzyme complexes (Fig. 4). However, the distance between the catalytic triad centroid of VASH1 and VASH2 to the Cα atom of residue S439 of helix H12 of a-tubulin is very similar (29.7 versus 27.3 Å, respectively).
Taken together, these results show that despite the similarities in the structures of the VASH1–SVBP and VASH2–SVBP core domains, a clear difference in their microtubule-binding pose is observed. These alternate poses are likely driven by the changes in the amino acid sequence of the S4-S5 loop between both enzymes, which forms interactions with site 2 of α-tubulin. Additional contribution may arise from the differences observed in the microtubule-binding residues at site 3 of α’-tubulin. However, this divergence could have only limited effect on the enzyme’s activity since the accessibility of the α-tubulin C-terminal tyrosine is quite similar.
The core domains of VASH1 and VASH2 in complex with SVBP behave similarly
As the cryo-EM derived structures of microtubules in complex with VASH1–SVBP and VASH2–SVBP revealed specific interactions of the enzymatic core domains of VASHs with microtubules, we wondered how truncated versions of the enzymes containing only these central regions would behave on microtubules in vitro.
We first examined the detyrosinating activity of VASH1 and VASH2 core domains in complex with SVBP (V1_CD and V2_CD, respectively; Fig. 5, A and B, and Fig. S1 A). A schematic representation of the two VASH proteins with their different domains is presented in Fig. 5 C. The two VASH core domains were active and, interestingly, they both led to global detyrosination of microtubules (Fig. 5, A and B), as did full-length version V1_FL but not V2_FL (Fig. 1, A−C). Analysis of the overall decrease in tyrosinated tubulin signal showed that the V2_CD was significantly less active than the V1_CD under the buffer conditions used. The latter result could be related to the different binding modes of the two complexes with microtubules as observed by cryo-EM: interactions with α- and α’-tubulin at sites 2 and 3, respectively, could be less favorable for the VASH2 activity than for the VASH1 activity (Figs. 3 and 4).
The N- and C-terminal regions of VASH1 and VASH2 are crucial for their microtubule binding and detyrosination activity. (A and B) Activity of enzyme complexes containing the sfGFP-tagged central core domains of VASH1 and VASH2 with SVBP (V1_CD and V2_CD) bound to Taxol-stabilized Tyr-MTs measured by immunofluorescence in TIRF experimental conditions (50 pM enzyme, BRB80 or BRB40 supplemented with 50 mM KCl, TIRF chamber). (A) Representative images of tyrosinated (Tyr, magenta) and detyrosinated (deTyr, green) α-tubulin pools of microtubules after 30 min in the absence (control) or presence of the enzyme’s core domains. Scale bar, 10 µm. (B) Analysis of tyrosinated and detyrosinated tubulin signals intensity. Each point represents a microtubule (at least 80 microtubules were analyzed). Data are represented as the median with interquartile range. Statistical significance was determined using Kruskal-Wallis test, ****P < 0,001. (C) Schematic representations of human VASH1 (blue) and VASH2 (orange) which are transglutaminase-like cysteine peptidases containing a triad of catalytic residues (Cys, His, and Leu). These proteins share 58% overall sequence identity (74% homology). Identities between core domains (CD, light colored boxes), and between N- and C-terminal regions (Nt and Ct respectively, dark colored boxes) are specified on the diagram. Isoelectric points (pI) of each domain are also provided. The residue numbers are indicated. (D and E) TIRF microscopy study of single molecules of sfGFP-tagged catalytically dead versions of VASH1 and VASH2 full-length versions (dead V1_FL and deadV2_FL), core domains (deadV1_CD and deadV2_CD), and truncated versions missing the flanking regions (deadV1_Nt + CD, deadV1_CD + Ct, deadV2_Nt + CD, deadV2_CD + Ct) bound to Taxol-stabilized Tyr-MTs. Experiments were performed in the presence of 50 pM enzyme in BRB40 supplemented with 50 mM KCl. (D) Representative kymographs. Scale bars: horizontal, 5 µm; vertical, 5 s. (E) Analysis of binding frequency. Each point represents a microtubule. Data are represented as the median with the interquartile range. Statistical significance was determined using Kruskal-Wallis test. **P < 0.002 ***P < 0.0002, ****P < 0.0001. Results for VASH1–SVBP are in blue and for VASH2–SVBP in orange.
The N- and C-terminal regions of VASH1 and VASH2 are crucial for their microtubule binding and detyrosination activity. (A and B) Activity of enzyme complexes containing the sfGFP-tagged central core domains of VASH1 and VASH2 with SVBP (V1_CD and V2_CD) bound to Taxol-stabilized Tyr-MTs measured by immunofluorescence in TIRF experimental conditions (50 pM enzyme, BRB80 or BRB40 supplemented with 50 mM KCl, TIRF chamber). (A) Representative images of tyrosinated (Tyr, magenta) and detyrosinated (deTyr, green) α-tubulin pools of microtubules after 30 min in the absence (control) or presence of the enzyme’s core domains. Scale bar, 10 µm. (B) Analysis of tyrosinated and detyrosinated tubulin signals intensity. Each point represents a microtubule (at least 80 microtubules were analyzed). Data are represented as the median with interquartile range. Statistical significance was determined using Kruskal-Wallis test, ****P < 0,001. (C) Schematic representations of human VASH1 (blue) and VASH2 (orange) which are transglutaminase-like cysteine peptidases containing a triad of catalytic residues (Cys, His, and Leu). These proteins share 58% overall sequence identity (74% homology). Identities between core domains (CD, light colored boxes), and between N- and C-terminal regions (Nt and Ct respectively, dark colored boxes) are specified on the diagram. Isoelectric points (pI) of each domain are also provided. The residue numbers are indicated. (D and E) TIRF microscopy study of single molecules of sfGFP-tagged catalytically dead versions of VASH1 and VASH2 full-length versions (dead V1_FL and deadV2_FL), core domains (deadV1_CD and deadV2_CD), and truncated versions missing the flanking regions (deadV1_Nt + CD, deadV1_CD + Ct, deadV2_Nt + CD, deadV2_CD + Ct) bound to Taxol-stabilized Tyr-MTs. Experiments were performed in the presence of 50 pM enzyme in BRB40 supplemented with 50 mM KCl. (D) Representative kymographs. Scale bars: horizontal, 5 µm; vertical, 5 s. (E) Analysis of binding frequency. Each point represents a microtubule. Data are represented as the median with the interquartile range. Statistical significance was determined using Kruskal-Wallis test. **P < 0.002 ***P < 0.0002, ****P < 0.0001. Results for VASH1–SVBP are in blue and for VASH2–SVBP in orange.
We then used the catalytically dead versions of VASH1 and VASH2 core domains (deadV1_CD and deadV2_CD) in order to avoid evolution of substrate over time and studied the interaction with microtubules in single-molecule TIRF experiments (Fig. 5, D and E). Similar to what we found by probing the activity, deadV1_CD and deadV2_CD exhibited binding characteristics on microtubules similar to those of V1_FL (Fig. 5, D and E, and Table 3 A). They displayed residence times of 1.2–1.4 s, thus close to that of V1_FL and at least 10 times shorter than residence time of V2_FL (Table 3 A). They showed binding frequency close to that of V1_FL and approximately 5 times higher than that of V2_FL (Table 3 A).
Binding parameters on microtubules of full-length, truncated, and chimeric versions of VASH1 and VASH2 enzymes in complex with SVBP
| Experiment (medium) . | Ionic strength (mM) . | Microtubule . | Recombinant protein (concentration) . | Residence time τ (s) . | Binding frequency (min-1.nM−1.µm−1) . |
|---|---|---|---|---|---|
| A (BRB40, 50 mM KCl) | 140 | Tyr-MTs | deadV1_FL (50 pM) | 1.2 | 80.4 |
| deadV1_Nt + CD (50 pM) | - | No binding | |||
| deadV1_CD (50 pM) | 1.4 | 70.7 | |||
| deadV1_CD + Ct (50 pM) | >7.7 | 21.1 | |||
| deadV2_FL (50 pM) | >17.1 | 16.2 | |||
| deadV2_Nt + CD (50 pM) | >4.0 | 35.9 | |||
| deadV2_CD (50 pM) | 1.2 | 85.1 | |||
| deadV2_CD + Ct (50 pM) | >10.4 | 10.4 | |||
| B (BRB40, 50 mM KCl) | 140 | Tyr-MTs | deadV1_FL (50 pM) | 1.2 | 54.1 |
| deadV1_FL(NtV2) (50 pM) | >9.9 | 31.0 | |||
| deadV2_FL (50 pM) | >13.4 | 19.1 | |||
| deadV2_FL(NtV1) (50 pM) | >6.5 | 18.1 | |||
| C (BRB40, 50 mM KCl) | 140 | Tyr-MTs | V1_FL (25 pM) | 1.2 | 96.7 |
| V1_FL(NtV2) (25 pM) | >10.7 | 20.8 | |||
| V2_FL (25 pM) | >12.9 | 22.9 | |||
| V2_FL(NtV1) (25 pM) | >3.9 | 42.4 |
| Experiment (medium) . | Ionic strength (mM) . | Microtubule . | Recombinant protein (concentration) . | Residence time τ (s) . | Binding frequency (min-1.nM−1.µm−1) . |
|---|---|---|---|---|---|
| A (BRB40, 50 mM KCl) | 140 | Tyr-MTs | deadV1_FL (50 pM) | 1.2 | 80.4 |
| deadV1_Nt + CD (50 pM) | - | No binding | |||
| deadV1_CD (50 pM) | 1.4 | 70.7 | |||
| deadV1_CD + Ct (50 pM) | >7.7 | 21.1 | |||
| deadV2_FL (50 pM) | >17.1 | 16.2 | |||
| deadV2_Nt + CD (50 pM) | >4.0 | 35.9 | |||
| deadV2_CD (50 pM) | 1.2 | 85.1 | |||
| deadV2_CD + Ct (50 pM) | >10.4 | 10.4 | |||
| B (BRB40, 50 mM KCl) | 140 | Tyr-MTs | deadV1_FL (50 pM) | 1.2 | 54.1 |
| deadV1_FL(NtV2) (50 pM) | >9.9 | 31.0 | |||
| deadV2_FL (50 pM) | >13.4 | 19.1 | |||
| deadV2_FL(NtV1) (50 pM) | >6.5 | 18.1 | |||
| C (BRB40, 50 mM KCl) | 140 | Tyr-MTs | V1_FL (25 pM) | 1.2 | 96.7 |
| V1_FL(NtV2) (25 pM) | >10.7 | 20.8 | |||
| V2_FL (25 pM) | >12.9 | 22.9 | |||
| V2_FL(NtV1) (25 pM) | >3.9 | 42.4 |
(A) Related to Fig. 5, D and E. (B and C) Related to Fig. 6, A and B. A, B, and C represent experiments performed on different days. At least 10 microtubules were analyzed. See Materials and methods for detailed experimental conditions.
Thus, the catalytic core domains of both VASHs behave quite similarly, which is in contrast to their full-length counterparts. Since the strong divergence between the functioning of the two VASH–SVBP enzymatic complexes did not arise from the microtubule-binding mode of these central regions, we hypothesized that their disordered N- and C-terminal flanking regions could explain the observed differences.
The N- and C-terminal domains of VASHs strongly contribute to the interaction of VASH–SVBP enzyme complexes with microtubules
To better understand the differences in behavior between VASH1–SVBP and VASH2–SVBP, we analyzed the role of their N- and C-terminal disordered core-flanking regions for interaction of the enzymes with microtubules. To this end, we compared the interaction of dead enzymes containing full-length VASH1 and VASH2 (deadV1_FL and deadV2_FL) or their truncated versions lacking either the C- (deadV1_Nt + CD and deadV2_Nt + CD) or N-terminal domains (deadV1_CD + Ct and deadV2_CD + Ct) with Tyr-MTs (Fig. 5, D and E, and Fig. S1 A).
The two mutated enzyme complexes containing the C-terminal region and the catalytic core domain of VASH (CD + Ct proteins) associated strongly to Tyr-MTs, resembling the binding behavior of V2_FL (Fig. 5 D). Their mean residence times on microtubules were >7 s (Table 3 A). In contrast, the two mutant enzymes containing the N-terminal region and the core domain (Nt + CD proteins) behaved very differently: while the VASH1 mutant did not bind to microtubules, the VASH2 mutant showed a long residence-time (Fig. 5 D and Table 3 A). Its residence time was even longer than the complex with only the catalytic core domain of VASH2 (τ = 1.2 s for deadV2_CD and τ > 4.0 s for deadV2_Nt + CD). Intriguingly, high residence-times were usually associated with low binding frequencies, as if long binding events ruled out new ones. This was similarly observed with V2_FL (Fig. 1, D and F; and Table 1 A) and could be due to the presence of fluorescence-bleached molecules that were still attached to the microtubule lattice.
Since ionic strength strongly affected both the binding of enzymes to microtubules as well as their detyrosination activity (Fig. 2, A−D and Table 1), we calculated the isoelectric points of the different regions of VASH1 and VASH2. Interestingly, the C-terminal parts of the VASHs are both very basic, while their N-terminal regions exhibit extremely different isoelectric points. The N-terminal region of VASH1 is acidic as opposed to that of VASH2, which is very basic (Fig. 5 C). Notably, these physicochemical characteristics are conserved among the vertebrates (Fig. S5 B). The basic character of the C-terminal parts of VASHs most probably favors binding of the enzyme complexes to the acidic surface of microtubules composed of stretches of acidic glutamates and aspartates from the α- and β-tubulin subunits (Redeker, 2010; deadV1_CD + CT, deadV2_CD + Ct, Fig. 5 D and Table 3 A). The strong acidic nature of the N-terminal part of VASH1 could explain the lack of binding of the mutant enzyme lacking the basic C-terminal region (deadV1_Nt + CD, Fig. 5 D and Table 3 A). In the case of VASH1–SVBP, its acidic N-terminal part could oppose the interaction of the rest of the protein (core domain and C-terminal region) with microtubules, and thus assist in the release of the enzyme complex from the microtubule. The role of the basic N-terminal part of VASH2 is less clear, but this region might also help the dissociation of the enzyme complex from the microtubule lattice. Indeed, the residence-time of full-length VASH2 is significantly longer than that containing the truncated mutant missing the N-terminal region (τ > 17.1 s for deadV2_FL and τ > 10.4 for deadV2_CD + Ct; Table 3 A).
Overall, these results reveal the important contribution of the N- and C-terminal regions of VASH for the interaction of the two enzyme complexes with microtubules. They further suggest an essential role of the VASHs’ N-terminal regions for their divergent activities in vitro, global versus local detyrosination.
The N-terminal domains of VASHs mediate divergent microtubule-binding and detyrosination activities
To test the hypothesis that the N-terminal regions of VASHs are critical for the distinct behavior of these enzymes, we constructed chimeras by inverting the N-terminal region of one VASH with the one of the other (Fig. S1 A). We tested catalytically active and inactive versions of these chimeras for their interaction with microtubules [V1_FL(NtV2), V2_FL(NtV1), deadV1_FL(NtV2), deadV2_FL(NtV1)] and compared their behavior to the ones of the WT enzymes. Representative kymographs and binding characteristics are presented in Fig. 6, A−F, and Table 3, B and C. As expected, the presence of the basic N-terminal region of VASH2–SVBP significantly changed the binding behavior of VASH1–SVBP (Videos 1 and 3). The residence-times of the chimeras were more than eight times higher than those of the native VASH1–SVBP [τ = 1.2 s for V1_FL; τ = 1.2 s for deadV1_FL; τ > 10.7 s for V1_FL(NtV2); τ > 9.9 s for deadV1_FL(NtV2)]. Their residence times were therefore more similar to the residence time of full-length VASH2–SVBP (τ > 13.4 s for deadV2_FL; τ > 12.9 for V2_FL). The presence of the acidic N-terminal region of VASH1 also significantly changed the binding behavior of VASH2–SVBP (Videos 2 and 4). The residence time of the chimera on microtubules was more than four times shorter than the one of the active WT enzyme (τ > 12.9 for V2_FL; τ > 3.9 for V2_FL(NtV1)), and two times shorter for the dead version (τ > 13.4 for deadV2_FL; τ > 6.5 for deadV2_FL[NtV1]).
The exchange of the N-terminal regions strongly alters binding behavior and activity of VASH–SVBP enzyme complexes. (A−F) Comparison of the interaction with Taxol-stabilized Tyr-MTs of the sfGFP-tagged full length VASH1 and VASH2 enzymes (V1_FL, V2_FL) and chimeras in which the N-termini were exchanged [V1_FL(NtV2), V2_FL(NtV1)]. Both catalytically dead (A, C, and E) and active (B, D, and F) versions were studied. Experiments were performed in the presence of 50 pM enzyme in BRB40 supplemented with 50 mM KCl. (A and B) Representative kymographs. Scale bars: horizontal, 5 µm; vertical, 5 s. Examples of TIRF movies from which kymographs were extracted are presented in supplemental data for the chimeras (Videos 3 and 4). Binding characteristics are summarized in Table 3. (C and D) Analysis of diffusion. Each point represents a single molecule, the molecules moving on at least 17 microtubules were analyzed. (E and F) Representation of the microtubule surface covered with VASH–SVBP molecules (white) or not covered (black) during the 45 s of TIRF movies. Each horizontal line represents a microtubule (at least 17 microtubules were analyzed). The red value corresponds to the mean covering (in %). (G and H) Activity of VASH–SVBP enzyme complexes and chimeras on Tyr-MTs measured by immunofluorescence in the same experimental conditions (50 pM enzyme, BRB40 supplemented with 50 mM KCl, TIRF chamber). (G) Representative images of tyrosinated (Tyr, magenta) and detyrosinated (deTyr, green) α-tubulin pools of microtubules after 30 min incubation in the absence (control) or presence of the indicated enzyme. Scale bar, 10 µM. (H) Analysis of tyrosinated- and detyrosinated-tubulin signals intensities. Each point represents a microtubule (at least 200 microtubules were analyzed). Data are represented as the median. Statistical significance was determined using Kruskal-Wallis test, ****P < 0.0001.
The exchange of the N-terminal regions strongly alters binding behavior and activity of VASH–SVBP enzyme complexes. (A−F) Comparison of the interaction with Taxol-stabilized Tyr-MTs of the sfGFP-tagged full length VASH1 and VASH2 enzymes (V1_FL, V2_FL) and chimeras in which the N-termini were exchanged [V1_FL(NtV2), V2_FL(NtV1)]. Both catalytically dead (A, C, and E) and active (B, D, and F) versions were studied. Experiments were performed in the presence of 50 pM enzyme in BRB40 supplemented with 50 mM KCl. (A and B) Representative kymographs. Scale bars: horizontal, 5 µm; vertical, 5 s. Examples of TIRF movies from which kymographs were extracted are presented in supplemental data for the chimeras (Videos 3 and 4). Binding characteristics are summarized in Table 3. (C and D) Analysis of diffusion. Each point represents a single molecule, the molecules moving on at least 17 microtubules were analyzed. (E and F) Representation of the microtubule surface covered with VASH–SVBP molecules (white) or not covered (black) during the 45 s of TIRF movies. Each horizontal line represents a microtubule (at least 17 microtubules were analyzed). The red value corresponds to the mean covering (in %). (G and H) Activity of VASH–SVBP enzyme complexes and chimeras on Tyr-MTs measured by immunofluorescence in the same experimental conditions (50 pM enzyme, BRB40 supplemented with 50 mM KCl, TIRF chamber). (G) Representative images of tyrosinated (Tyr, magenta) and detyrosinated (deTyr, green) α-tubulin pools of microtubules after 30 min incubation in the absence (control) or presence of the indicated enzyme. Scale bar, 10 µM. (H) Analysis of tyrosinated- and detyrosinated-tubulin signals intensities. Each point represents a microtubule (at least 200 microtubules were analyzed). Data are represented as the median. Statistical significance was determined using Kruskal-Wallis test, ****P < 0.0001.
Interaction of single molecules of chimeric sfGFP-VASH1–SVBP complex bearing the N-terminal region of VASH2 [V1_FL(NtV2)] with Taxol-stabilized microtubules enriched in tyrosinated HeLa tubulin studied by single-molecule TIRF microscopy. The presence of the basic N-terminal region of VASH2–SVBP significantly changed the binding behavior of VASH1–SVBP. The chimeric complex (50 pM, green) showed much higher residence time and diffused poorly along the microtubule (magenta), resembling full length VASH2 complex. Scale bar, 2 μm. Images were collected every 50 ms for 45 s and films are 2.5× accelerated.
Interaction of single molecules of chimeric sfGFP-VASH1–SVBP complex bearing the N-terminal region of VASH2 [V1_FL(NtV2)] with Taxol-stabilized microtubules enriched in tyrosinated HeLa tubulin studied by single-molecule TIRF microscopy. The presence of the basic N-terminal region of VASH2–SVBP significantly changed the binding behavior of VASH1–SVBP. The chimeric complex (50 pM, green) showed much higher residence time and diffused poorly along the microtubule (magenta), resembling full length VASH2 complex. Scale bar, 2 μm. Images were collected every 50 ms for 45 s and films are 2.5× accelerated.
Interaction of single molecules of chimeric sfGFP-VASH2–SVBP complex bearing the N-terminal region of VASH1 [V2_FL(NtV1)] with Taxol-stabilized microtubules enriched in tyrosinated HeLa tubulin studied by single-molecule TIRF microscopy. The presence of the acidic N-terminal region of VASH1 strongly changed the behavior of VASH2–SVBP. The chimeric complex (50 pM, green) showed shorter residence time and recovery of diffusion capacity on microtubules (magenta), resembling full-length VASH1 complex. Scale bar, 2 μm. Images were collected every 50 ms for 45 s and films are 2.5× accelerated.
Interaction of single molecules of chimeric sfGFP-VASH2–SVBP complex bearing the N-terminal region of VASH1 [V2_FL(NtV1)] with Taxol-stabilized microtubules enriched in tyrosinated HeLa tubulin studied by single-molecule TIRF microscopy. The presence of the acidic N-terminal region of VASH1 strongly changed the behavior of VASH2–SVBP. The chimeric complex (50 pM, green) showed shorter residence time and recovery of diffusion capacity on microtubules (magenta), resembling full-length VASH1 complex. Scale bar, 2 μm. Images were collected every 50 ms for 45 s and films are 2.5× accelerated.
The exchange of the N-terminal regions also strongly altered the diffusion of the enzyme complexes on microtubules and their covering of the microtubule lattices (Fig. 6, C−F). The chimeric complex with the N-terminal region of VASH2 and the remainder of VASH1 diffused poorly compared to VASH1–SVBP, and its covering of microtubules was significantly reduced. This was true for both the dead and catalytically active versions of VASH1–SVBP. In contrast, replacement of the N-terminal region of VASH2 with that of VASH1 significantly increased the diffusion and enhanced microtubule covering by VASH2–SVBP. This was particularly evident for the active chimera, which in the presence of the N-terminal region of VASH1 revealed a diffusion capacity equal to that of VASH1–SVBP (Fig. 6 D). The catalytically dead VASH2 chimera, however, retained a significantly lower diffusion capacity than VASH1–SVBP (and of its active counterpart; Fig. 6, C and D). This difference could be due to the absence of a possible structural change occurring during tyrosine cleavage, which could favor VASH2 release from microtubules. This would be consistent with the significantly lower diffusion capacity of the dead VASH2 version compared to its active counterpart (1.4 µm/s for V2_FL and 1.0 for deadV2_FL in BRB40 supplemented with 50 mM KCl; Fig. 1 H; 1.6 µm/s for V2_FL and 1.3 for deadV2_FL in BRB80 supplemented with 100 mM KCl, not shown). The binding frequencies of VASH1 chimeras were strongly reduced compared with those of VASH1 (with a 4.7-fold decrease for active versions and a 1.7-fold decrease for dead versions; Table 3, B and C). The binding frequency was almost unchanged for the dead VASH2 chimera compared to that of the dead VASH2, which was not the case for the active versions, with the VASH2 chimera having a clearly higher binding frequency than VASH2 (1.9-fold increase; Table 3, B and C).
We then examined the microtubule detyrosination activity of the chimeras [V1_FL(NtV2) and V2_FL(NtV1); Fig. 6, G and H] using immunofluorescence. Both chimeras induced global detyrosination, resembling the way VASH1–SVBP acts on microtubules. Thus, the presence of the N-terminal region of VASH1 changed the detyrosinating behavior of VASH2–SVBP from local to global, correlating with the TIRF experiment showing that the chimera is much more diffusive and has a higher binding frequency (Fig. 6 D and Table 3 C). On the other hand, despite inducing an important loss of diffusion capacity (Fig. 6 D), the N-terminal region of VASH2 did not appreciably switch the behavior of VASH1–SVBP (Fig. 6 G). The VASH1 chimera still induced global microtubule modifications. This result could be due to the core domain of VASH1 being significantly more active than the one of VASH2 (Fig. 5 B). Notably, quantification of tyrosinated tubulin levels showed that changes were significantly larger for the VASH1 chimera than for the VASH2 chimera, like for their WT counterparts.
Together, our data reveal the importance of the N-terminal regions of the VASHs in the divergent binding of VASH–SVBP complexes to the microtubule lattice as well as their detyrosination activity. Indeed, by swapping these regions, we were able to switch the microtubule-binding behavior of one enzyme to the other, and change the activity of VASH2–SVBP from local to global, as observed for VASH1–SVBP.
VASH1 and VASH2 give rise to distinct detyrosination patterns in cells
To test whether our in vitro results are relevant in cells, we assayed recombinant VASH–SVBP activities on exposed microtubules of murine embryonic fibroblasts (MEFs). Cultured MEFs were incubated in a buffer containing Triton-X100 for lysis and glycerol to stabilize microtubules (see Material and methods). Fig. 7 shows that native MEFs mainly contain tyrosinated microtubules and that addition of active VASH–SVBP complexes (V1_FL or V2_FL) led to a marked increase of detyrosination of cellular microtubules. The observed increase was greater around nuclei most likely due to the concentration of microtubules. Interestingly and similar to what was observed in vitro, while the VASH1–SVBP complex induced a diffuse and spread microtubule detyrosination, VASH2–SVBP led to the formation of local areas of lattice modifications. Overall, 17.4 and 39.8% of the microtubules surface is detyrosinated by VASH1–SVBP at 50 and 200 nM, respectively, while 2.1 and 3.3% is detyrosinated by VASH2–SVBP at those concentrations (median values; Fig. 7 B). Thus, our data show that the two VASH–SVBP enzymes generate different patterns of microtubule detyrosination both in a reconstituted system and in cells.
VASH1–SVBP and VASH2–SVBP generate different detyrosination profiles in fibroblast cells. (A) Cultured MEFs were permeabilized in BRB80 supplemented with Triton X-100 and glycerol, incubated 30 min with recombinant enzymes (50 or 200 pM, as indicated), fixed in methanol at −20°C, and labeled with anti-tyrosinated (Tyr, magenta) and anti-detyrosinated (deTyr, green) tubulin antibodies to reveal detyrosination/tyrosination levels on microtubules. Scale bar, 10 µm. (B) Quantification of detyrosinated areas of microtubules in fibroblasts treated with VASH–SVBP enzymes as in A. Each point represents a different cell; at least nine cells from three independent experiments were analyzed. Data are represented as the median. Statistical significance was determined using the Kruskal-Wallis test, ****P < 0.0001.
VASH1–SVBP and VASH2–SVBP generate different detyrosination profiles in fibroblast cells. (A) Cultured MEFs were permeabilized in BRB80 supplemented with Triton X-100 and glycerol, incubated 30 min with recombinant enzymes (50 or 200 pM, as indicated), fixed in methanol at −20°C, and labeled with anti-tyrosinated (Tyr, magenta) and anti-detyrosinated (deTyr, green) tubulin antibodies to reveal detyrosination/tyrosination levels on microtubules. Scale bar, 10 µm. (B) Quantification of detyrosinated areas of microtubules in fibroblasts treated with VASH–SVBP enzymes as in A. Each point represents a different cell; at least nine cells from three independent experiments were analyzed. Data are represented as the median. Statistical significance was determined using the Kruskal-Wallis test, ****P < 0.0001.
Discussion
Our in vitro and in cellulo findings reveal divergent activities of the two VASH–SVBP enzymes, with VASH1–SVBP leading to global microtubule detyrosination and VASH2–SVBP generating local areas of microtubule lattice modifications. These different activities are driven by the different microtubule binding behaviors of the two VASH–SVBP enzymes (see graphical abstract).
We found a moderately stable interaction of the two VASH core domains with the microtubule lattice, which includes the specific recognition of two α-tubulin subunits from adjacent protofilaments of the substrate. While the involved interaction sites confer specificity to this family of tubulin carboxypeptidases, the disordered N- and C-terminal regions of VASHs contribute to their microtubule-binding strength and therefore to their mode of action.
The common C-terminal, strongly positively charged region of the two VASHs considerably lengthen the interaction time of both enzyme complexes with microtubules. A reasonable model explaining this observation is that this basic region interacts through electrostatic attractive interactions with the negatively charged outer surface of microtubules (mostly composed of acidic residues stemming from the C-termini of both α- and β-tubulin; Redeker, 2010), allowing to hold an enzyme complex close to the microtubule surface as it moves from one tubulin dimer to another. The C-terminal VASH region could thus maintain an enzyme complex bound to the lattice when its catalytic core domain transiently detaches from it upon cleavage of the C-terminal α-tubulin tyrosine to reach the next substrate. Such a mechanism would increase both the residence-time of a VASH–SVBP complex with microtubule lattices as well as its activity.
In contrast, the N-terminal domain of the two VASHs could be involved in the release mechanism of the enzyme complex from microtubules, and consequently contribute to its diffusion properties on the lattice. This is particularly clear for VASH1–SVBP: its mutant lacking the N-terminal region shows a significantly increased residence time. Moreover, VASH1–SVBP switches from a diffusive to a static binding behavior on microtubules when the N-terminal region is removed. Due to its many negative charges, this domain probably behaves as an electrostatic “repellent,” promoting detachment of the enzyme from the surface of microtubules. This is not the case for the N-terminal region of VASH2, which in contrast to the one of VASH1 is strongly positively charged; it might thus contribute together with its positively charged C-terminal region to the very long residence time of VASH2–SVBP on microtubules.
Our data suggest that the diffusive capacity of VASH1–SVBP, sustained by its N-terminal domain, promotes the global detyrosination of microtubules by this enzyme in vitro. The local detyrosination observed for VASH2–SVBP likely relies on its weak diffusion capacity along microtubules. The two enzymes can nevertheless be considered as processive, i.e., they are capable to catalyze several consecutive reactions without fully detaching from a microtubule. VASH1–SVBP shows higher detyrosination activity than VASH2–SVBP most probably because it has a higher diffusive capability. In addition, VASH1 might have a higher cleavage efficiency than VASH2, as suggested by the increased detyrosination activity of the central domain of VASH1 compared to the one of VASH2 (Fig. 5 B).
Notably, our work reveals that as for molecular motors (Zhernov et al., 2020) electrostatic interactions are key factors in the association between VASH–SVBP complexes and microtubules, making their processive enzymatic functioning possible. Indeed, the disordered flanking regions of the highly structured VASH core domains fine-tune the association through such electrostatic interactions. Interestingly, the N-terminal regions of the VASHs bearing opposite charges give rise to both a divergent microtubule-binding behavior as well as a different activity of the two enzyme complexes in a reconstituted system. Other mechanisms, such as specific post-translational modifications (like, e.g., phosphorylation or polyglutamylation) or association of protein partners (such as MAP4 in cardiomyocytes; Yu et al., 2021) could also positively or negatively modulate these electrostatic interactions in cells, and therefore influence the association between VASH–SVBP and microtubules as well as the enzymes’ detyrosination activity.
In conclusion, our results reveal a novel pathway by which microtubules can be modified differently depending on the detyrosinating enzyme at work. This mechanism could have implications in the diversity of microtubules reported in cells (for reviews see Roll-Mecak, 2019; Roll-Mecak, 2020), which likely contain different compositions of detyrosinating enzymes. With the very recent discovery of the third detyrosinating enzyme MATCAP (Landskron et al., 2022), which uses a fundamentally different catalytic mechanism (being a metalloprotease instead of a cysteine protease) and adopts an alternative microtubule-binding mode and a different way of detyrosinating microtubules, the complexity of microtubule detyrosination in cells may even be greater. These different microtubule detyrosinating enzymes likely encode numerous subpopulations of microtubules bearing different tyrosination signal patterns that, when read by effectors, could fine-tune physiological processes in specific regions of the cell. Of particular interest is that the VASH2–SVBP complex, which is expressed weakly in most cells, can generate highly localized spots of detyrosinated tubulin on microtubule. We speculate that these spots, by generating areas on microtubules unable to bind protein partners such as, for example, CAP-Gly proteins or tyrosine-sensitive molecular motors (Chen et al., 2021; Peris et al., 2006; Peris et al., 2009), may have a role in cytoskeletal organization and/or intracellular trafficking in confined regions of cells. Several physiological processes, including cell division, organization, or migration could require such local signals for proper functioning. In this context, a better understanding of the regulation of tubulin-detyrosinating enzymes as well as their spatial and temporal expression patterns in cells becomes essential.
Material and methods
Constructs and enzymes purification for TIRF and immunofluorescence studies
The cDNA encoding human full-length SVBP (NP_955374, residues 1–66) was cloned into the first multiple cloning site of a modified pETDuET-1 vector, to generate a fusion protein with C-terminal myc and FLAG tags. The cDNAs encoding the human VASH1 (NP_055724) or VASH2 (NP_001287985) were cloned into the second multiple cloning site of the modified vector, to generate fusion proteins with an N-terminal sfGFP followed by a PreScission cleavage site, plus C-terminal myc and His tags (the pI of the sequence added to the C termini was 6.6, which changes very slightly the pI of VASHs and their mutants). The VASH1 and VASH2 constructs used in this study (Fig. S1 A) were: VASH1 full-length (V1_FL, residues 1–365), dead VASH1 full-length, (deadV1_FL, residues 1–365, C169A), VASH1 core domain (V1_CD, residues 57–307), dead VASH1 core domain (deadV1_CD, residues 57–307, C169A), dead VASH1 core domain + N-ter (deadV1_CD + Nt, residues 1–307, C169A), dead VASH1 core domain + C-ter (deadV1_CD + Ct, residues 57–365, C169A), VASH2 full length (V2_FL, residues 1–355), dead VASH2 full-length (deadV2_FL, residues 1–355, C158A), VASH2 core domain (V2_CD, residues 40–296), dead VASH2 core domain (deadV2_CD, residues 40–296, C158A), dead VASH2 core domain + N-ter (deadV2_CD + Nt, residues 1–296, C158A), dead VASH2 core domain + C-ter (deadV2_CD + Ct, residues 40–355, C158A), dead VASH2 with VASH1 N-ter (deadV2_NtV1, VASH1 residues 1–49 and VASH2 residues 40–355, C158A), dead VASH1 with VASH2 N-ter (deadV1_NtV2, VASH2 residues 1–39 and VASH1 residues 51–365, C169A). For the above dead versions, mutations were introduced using a standard PCR procedure. All constructs were verified by DNA sequencing (GENEWIZ). The cloning for non-sfGFP tagged proteins expression were described in Wang et al. (2019).
Protein expression and purification of the various His-tagged VASH1/2–SVBP complexes were performed as previously described (Wang et al., 2019) with slight modifications. Briefly, BL21(DE3) Escherichia coli cells were transformed with the corresponding construction and then cultured in Luria-Bertani medium at 37°C. Protein expression was induced by addition of 0.5 mM IPTG and overnight incubation at 18°C. Cells were collected and resuspended in lysis buffer (50 mM Hepes, pH 8.0, 500 mM NaCl, 10% glycerol, 20 mM imidazole, 2 mM DTT and cOmplete EDTA-free protease inhibitor cocktail tablets [GE, Healthcare]). Following cell lysis by sonication, the extract was clarified by centrifugation at 100,000 g for 30 min at 4°C. Total lysate was loaded in a HisTrap HP Ni2+ Sepharose column (GE Healthcare). Column was then extensively washed using lysis buffer, and enriched VASH1/2–SVBP complexes were eluted from the HisTrap HP Ni2+ Sepharose column using elution buffer (lysis buffer with imidazole concentration raised to 400 mM). Protein fractions were further collected and concentrated, and finally purified with a Superdex 200 10/300GL (GE Healthcare) size exclusion column. The concentration of soluble proteins was measured using Bradford assay with BSA as the standard. All proteins were kept in storage buffer (20 mM Tris-HCl at pH 7.4, 150 mM NaCl, 2 mM DTT) and stored in liquid nitrogen until further use.
Preparation of tubulin and Taxol-stabilized microtubules
Preparation of brain tubulin and its labeling with either biotin or ATTO-565 fluorophore (ATTO-TEC Gmbh) were performed according to Ramirez-Rios et al. (2017). Tubulin proteins from HeLa cells (CCL-2.2; ATCC) used in this study were purified as previously described (Souphron et al., 2019). All tubulin proteins were stored in liquid nitrogen.
Taxol-stabilized microtubules were prepared by polymerizing 45 µM tubulin (composed of 65% of HeLa tyrosinated or detyrosinated tubulin, 30% biotinylated brain tubulin, and 5% ATTO-565–labeled brain tubulin, named Tyr-MTs or deTyr-MTs, respectively) in BRB80 (80 mM Pipes-KOH at pH 6.8, 1 mM MgCl2, 1 mM EGTA) supplemented with 1 mM GTP. Taxol (100 µM) was then added, and microtubules were further incubated for 30 min. Microtubules were then centrifuged for 5 min at 200,000 × g and resuspended in BRB80/10 µM Taxol buffer. All incubations and centrifugations were carried out at 35°C. Using a method described in Aillaud et al. (2016), we estimated that Tyr-MTs contained 80% tyrosinated, 17% detyrosinated, and 3% ∆2-tubulin, while deTyr-MTs contained 82% detyrosinated, 15% tyrosinated, and 3% ∆2-tubulin (see Fig. S1 C).
In vitro binding assays
Experiments were conducted in the media indicated on figures: BRB40 or BRB80 (40 or 80 mM Pipes-KOH at pH 6.8, 1 mM MgCl2, 1 mM EGTA) buffers supplemented or not with the indicated concentrations of KCl. Ionic force of media was calculated as in Thiede et al. (2013) (see Table 1). Flow chambers for TIRF imaging assays were prepared as previously described (Stoppin-Mellet et al., 2020). Perfusion chambers were treated with 30 µl neutravidin (31000, 25 µg/ml; Thermo Fisher Scientifi) incubated for 3 min followed by 50 μl PLL20K-G35-PEG2K (0.1 mg/ml, Jenkem) for 30–45 s. Chambers were then washed 3 times with 100 µl 1% BSA in indicated medium. Subsequently, microtubules enriched in Tyr- or deTyr-MTs prepared as above were diluted to 40 µM and perfused into the chamber. Then, the flow chamber was flushed three times by 100 µl of 1% BSA/10 µM Taxol in medium to remove unbound microtubules. Finally, sfGFP–VASH1/2–SVBP complexes were perfused in an assay-mix solution (medium supplemented with 82 µg/ml catalase, 580 µg/ml glucose oxidase, 1 mg/ml glucose, 4 mM DTT, 0.5 mM Taxol, 0.017% methylcellulose 1,500 cp). All incubations were made at room temperature. The chamber was sealed and then imaged using TIRF microscope.
TIRF in vitro assay
Images were recorded within the first 30 min following addition of the assay-mix solution (see above) on an inverted microscope (Eclipse Ti, Nikon) equipped with a Perfect Focused System, a CFI Apochromat TIRF 100×/1.49 N.A oil immersion objective (Nikon), a warm stage controller (Linkam Scientific) and a Technicoplast chamber to maintain the temperature, an objective heater (OkoLab), an iLas2 TIRF system (Roper Scientific), and a sCMOS camera (Prime95B, Photometrics) controlled by MetaMorph software (version 7.10.3, Molecular Devices). For dual view imaging, an OptoSplit II bypass system (Cairn Research) was used as image splitter and illumination was provided by 488- and 561-nm lasers (150 and 50 mW, respectively). Temperature was maintained at 35°C for all imaging purposes. Acquisition rate was one frame each 50 ms exposure (in streaming acquisition) during 45 s. For each condition at least two slides from two independent experiments using different protein preparations were done.
In vitro assay of detyrosination activity using immunofluorescence
Immunofluorescence in vitro assays were made using exactly the same perfusion chambers and TIRF experiments conditions as above. After addition of the assay-mix solution, incubation was done at 37°C for 30 min followed by three washes with 10 µM Taxol and 1% BSA in indicated medium (wash buffer). Then, incubation with primary antibodies was done for 15 min, followed by three washes with 100 µl of wash buffer, incubation with secondary antibodies for 15 min, and finally three washes with 100 µl of wash buffer. Primary antibodies were rat anti-tyrosinated tubulin (YL1/2 = anti-Tyr, 1:6,000) and rabbit anti-detyrosinated tubulin (anti-deTyr, 1:1,000; see Aillaud et al., 2016) and secondary antibodies were anti-rat coupled to Alexa Fluor 488 (Jackson ImmunoResearch, 712-545-153) and anti-rabbit coupled to Cyanine 3 (711-165-152; Jackson ImmunoResearch), both diluted to 1:500. Images were obtained using a LEICA DMI600/ROPER microscope controlled by Metamorph Video software using the same illumination conditions. For each condition at least three independent experiments with different protein preparations were done.
Assays of detyrosination with low level of recombinant enzymes in cells
MEFs were prepared as previously described (Erck et al., 2005) and were treated with BRB80 supplemented with 0.5% Triton X-100 and 10% glycerol at 35°C for 15 s. Permeabilized cells were washed twice with BRB80 supplemented with 10% glycerol, incubated with 50 or 200 pM of recombinant sfGFP–VASH–SVBP complexes during 30 min at 37°C, and rinsed twice with BRB80 supplemented with 10% glycerol. For each condition at least three independent experiments with different protein preparations were done.
For immunofluorescence, cells were washed in warm PBS supplemented with 0.1% Tween and fixed in freshly prepared −20°C methanol. They were then incubated with primary antibodies (anti-Tyr and anti-deTyr; 1:500), followed by incubation with secondary antibodies (anti-rat conjugated with Alexa Fluor 488 and anti-rabbit conjugated with Cyanine 3, as above). Samples were mounted using DAKO medium supplemented with 1 µg/ml Hoescht 33258 (H3569; Invitrogen). Images were captured with a Zeiss Axiovert 200 M microscope equipped with the acquisition software MetaMorph using the same illumination conditions.
SDS-PAGE, immunoblotting, and antibodies used to analyze purified VASH–SVBP complexes
Equal amounts of purified proteins (1.5 µg) were loaded onto 4–20% gradient polyacrylamide gels (Biorad) and separated by electrophoresis. Proteins were transferred to nitrocellulose membranes and probed as follow: rabbit anti-GFP at 1:10,000 (A6455; Life Technologies), mouse anti-His at 1:10,000 (34660; Qiagen), home-made rabbit anti-VASH1, anti-VASH2, and anti-SVBP (all used at 1:10,000), and mouse anti-α tubulin (αtot, 1:5,000), rat anti-Tyr (YL1/2, 1:5,000), rabbit anti-deTyr (1:20,000), rabbit anti-∆2 tubulin (1:20,000). All anti-tubulin antibodies were described in Aillaud et al. (2016). Anti-VASH1 Gre (against peptide RIRGATDLPKIPIPSVPTFQPTTPV) was described in Chen et al. (2020). Anti-VASH2 Gre and anti-SVBP Gre were produced in a similar manner against peptides AIRNAAFLAKPSIPQVPNYRLSMTI of VASH2 and DPPARKEKSKVKEPAFRVEKAKQKS of SVBP. All peptide sequences were chosen in exposed region of the VASH–SVBP structure (Wang et al., 2019). Specificities of antibodies were tested by immunoblots using extracts of HEK293T cells (ATCC, CRL-3216) expressing recombinant VASH1-eGFP or VASH2-eGFP with SVBP-FLAG (see Wang et al., 2019) and compared to signals obtained with rabbit anti-GFP (A6455; Life Technologies) regarding VASH1 and VASH2, and rabbit anti-FLAG (F7425; Millipore) regarding SVBP. Incubation with primary antibodies were followed by incubation with secondary antibodies conjugated with anti-rabbit horseradish peroxidase (711-035-152; Jackson ImmunoResearch). Blots were finally revealed using Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific) and a ChemiDoc MP Imaging System (BioRad) according to the manufacturer’s protocol.
Constructs and in cellulo assays of WT and mutant VASH2–SVBP
The cDNAs encoding human VASH2 (NP_001287985) with a C-terminal eGFP and human SVBP (NP_955374) with C-terminal myc and FLAG tags are described in Wang et al. (2019). Point mutations were introduced in human VASH2 cDNA by standard PCR procedures and all mutations were verified by DNA sequencing (Eurofins).
For in cellulo tubulin detyrosination assays, HEK293T cells (CRL-3216; ATCC) were transfected using JetPRIME transfection reagent (Polyplus-Transfection) with plasmids encoding WT or mutant VASH2-eGFP and SVBP-myc-FLAG. The cells were collected 24 h after transfection. After washing with PBS medium at 37°C, cells were lysed in BRB80, pH 7.1, 1% Triton X-100 in the presence of a protease inhibitor cocktail (cOmplete EDTA-free, Roche) before being centrifuged for 20 min at 4°C and 12,000 g. Supernatants were then diluted in Laemmli buffer and protein extracts were tested in immunoblot (see above). Primary antibodies were rabbit anti-FLAG (1:10,000) from Millipore (F7425), rabbit anti-GFP (1:5,000) from Life Technologies (A6455), rabbit anti-GAPDH (1:2,500) from Sigma-Aldrich (G9545), and home-made rabbit detyrosinated α-tubulin (1:10,000).
For in cellulo co-localization assays, NIH3T3 cells (laboratory-derived cells, see Wang et al., 2019) were transfected with plasmids encoding WT or mutant VASH2-eGFP and SVBP-myc-FLAG using Lipofectamine or Amaxa (Invitrogen and Lonza Bioscience, respectively). A ratio of 1:1 (µg VASH2:µg SVBP) was used for plasmid co-transfections. After 24 h of transfection, cells were treated with BRB80 supplemented with 0.5% Triton X-100 and 10% glycerol at 37°C for 1 min before fixation in methanol (10 min at −20°C). For immunofluorescence, primary antibodies were home-made mouse anti-α tubulin (1:1,000), rabbit anti-Flag (1:2,000) from Millipore (F7425), chicken anti-GFP (1:2,000) from Millipore (AB16901), and secondary antibodies were anti-chicken coupled to Alexa Fluor 488 (A11039; Invitrogen), anti-rabbit coupled to Cyanine 5 (711-175-152; Jackson ImmunoResearch), and anti-mouse coupled to Cyanine 3 (715-165-150; Jackson ImmunoResearch) all diluted to 1:1,000. Samples were mounted using DAKO medium supplemented with 1 µg/ml Hoescht 33258 (H3569; Invitrogen). Acquisitions of images were performed using a confocal microscope LSM710/Airyscan (Zeiss) equipped with a ×63, numerical aperture 1.4 objective.
Cloning and protein purification of VASH2–SVBP sample for cryo-EM studies
The human cDNAs encoding full-length VASH2 (UniProtKB ID Q86V25) and full-length SVBP (Q8N300) were codon optimized for E. coli expression and synthesized (GENEWIZ). The sequence of VASH2 was modified by mutating Cys158 to alanine resulting in a catalytically inactive protein (Wang et al., 2019). The synthetic cDNAs were then cloned into two different plasmids, VASH2 into pET3d (Novagen) and SVBP into pET28 (EMD Biosciences). When inserted into the pET28 vector, the SVBP sequence was fused to a 6His tag at its C-terminus for purification purposes. Protein expression and purification of the His-tagged VASH2–SVBP complex was performed as previously described (Wang et al., 2019). Final protein samples were snap frozen in liquid nitrogen at 2 mg/ml concentration and stored at −80°C.
GMPCPP-stabilized microtubules were prepared using tubulin purified from HeLa cells (CCL-2.2; ATCC) as described in Souphron et al. (2019). Purified tubulin was resuspended at 4 mg/ml on ice using BRB80 buffer and GMPCPP was added to 0.5 mM final concentration. The tubulin solution was incubated on ice for 5 min then centrifuged for 10 min at 16,000× g at 4°C. The supernatant was transferred to a fresh tube and placed on a 37°C heat block for 40 min. Subsequently, the microtubule solution was kept at 20°C before use.
Cryo-EM grid preparation
Glow discharged Quantifoil Cu300 R2/1 holey carbon EM grids (Electron Microscopy Sciences) were used for specimen preparation. Microtubules were centrifuged for 10 min at 16,000 × g and thoroughly resuspended in BRB80 buffer to 2 mg/ml. 3.5 μl of the microtubule solution was applied to the EM grid and incubated for 1 min at room temperature. The grid was blotted manually and 3.5 μl of a 2 mg/ml V2-SVBP solution was applied to the grid that was then transferred immediately to the chamber of the Vitrobot (Thermo Fisher Scientific) pre-equilibrated at 25°C and 100% relative humidity. After 1 min of incubation, the grid was blotted for 1 s at blot force of 0 and plunge frozen into liquid ethane. Grids prepared this way were stored in liquid nitrogen prior to imaging.
Cryo-EM data collection and image processing
Movies were recorded on a Titan Krios at 300 keV (Thermo Fisher Scientific), with a GIF Quantum LS Imaging filter (20 eV slit width) and a K2 Summit electron counting direct detection camera (Gatan) at a magnification of 47,260 ×, resulting in a pixel size of 1.058 Å, using SerialEM (Mastronarde, 2005). The defocus varied between −0.6 and −2.2 µm. 3,345 movies were recorded with a total dose of 65 e-/Å2 per movie (9 s exposure in total, 0.2 s per frame, 45 frames in total). The dose rate was ∼7.2 e-/Å2 per second (∼1.4 e-/Å2 per frame). For image processing, the software Relion 3.0.8 (Scheres, 2012) was used. The protocols described in Cook et al. (2020) and Debs et al. (2020) were followed with some adaptations. In brief, the movies were drift-corrected and dose-weighted using MotionCor2 (Zheng et al., 2017) and the contrast transfer function parameters were estimated from the motion-corrected electron micrographs using Gctf (Zhang, 2016). Afterwards, microtubule sections were manually picked from the HeLa tubulin data set. Boxes with a size of 600 pixels, re-scaled to 150 pixels with inter-box distance of 82 Å, were extracted. Superparticles were created and processed as previously described (Cook et al., 2020). As the microtubule decoration by VASH2–SVBP was incomplete, we classified the decorated and undecorated protofilaments to improve the signal-to-noise ratio of the VASH2–SVBP density. For this, we expanded the symmetry of the particles depending on the number of protofilaments. We focused on one VASH2–SVBP particle and created a mask including regions of the protofilament and VASH2–SVBP. We subtracted everything outside the mask and used the subtracted particles for 3D classification (Debs et al., 2020). Only particles in the class including VASH2–SVBP were used for further rounds of refinement and to generate the final map.
Model building and refinement
An initial structural model of the microtubule bound VASH2–SVBP complex was generated in Chimera by combining the GMPCPP-stabilized microtubule (Zhang, 2016; PDB ID 3JAL) with the crystal structure of the human VASH2–SVBP complex (PDB ID 6QBY). The models were imported into Chimera (Pettersen et al., 2004), and the Fit-in-Map function was used to perform a rigid-body fit of one VASH2–SVBP complex structure and two laterally aligned αβ-tubulin heterodimers into the final cryo-EM map. The models were then combined and refined in Coot (Emsley and Cowtan, 2004) using “Set Geman-McClure alpha 0.01” while all molecule self-restraints were set to 5.0 in the ProSMART module (Nicholls et al., 2014). The model was then further refined in Coot using the “Chain Refine” command. Finally, the model and map were exported to Phenix (Liebschner et al., 2019) and the “Real-space Refinement” tool was used to complete the refinement. MolProbity (Chen et al., 2010) was used to assess the model quality and generate the values reported in Table 2. Chimera and PyMOL (Schrodinger, 2020) were used for the generation of figures.
Statistical analysis
Data analysis of TIRF
Binding/tracking of single VASH1/2–SVBP molecules for estimation of binding parameters and diffusion were measured on kymographs using FIJI software and a homemade plugin KymoTool (Ramirez-Rios et al., 2016). The tracks were detected from kymographs using “Detection plugin.” Briefly, kymograph images were enhanced by subtracting a blurred image and convolved for enhancement of continuous tracks. After thresholding, the detected tracks were skeletonized and the tracks composed of several molecules were cleaned manually.
Mean residence-time was calculated by fitting the data points to a plateau followed by one-phase decay using GraphPad Prism 7. A precise determination of the residence time for some proteins (such as full-length VASH2–SVBP) was not possible in some experimental conditions and was therefore expressed as “superior to.” Indeed, the molecules were already attached to microtubules at the beginning of the TIRF movies and single-molecule traces disappeared during movie acquisition. This latter observation was due to bleaching of the sfGFP fluorophore, since at lower intensity of the excitation lamp we observed much longer binding events (not shown). However, at these lower lamp intensities, the signal-to-noise ratio was too low for correct analysis of binding characteristics.
Binding frequency calculations were determined by dividing the number of bindings events by the duration of the recording (in min) and by nanomolar VASH1/2–SVBP protein concentration over the microtubule size (in µm).
For diffusion analysis, each track was interpolated with 1 pixel interval and (X, Y) coordinates were collected. The total displacement (sum of dX at each Y) was calculated, and the final displacement (Xfinal − X0) and time of binding (Yfinal − Y0) were calculated. The displacement speed was then estimated as (total displacement in µm)/(binding time in seconds); thus, it included the displacement of the molecule in both microtubule directions.
For quantification of the surface covered by sfGFP (VASH–SVBP complexes) on microtubule, a binary image was produced for each kymograph. The kymograph was resliced to get a stack where different slices represent the time dimension. A maximal projection was generated and the percentage of the image covered by sfGFP was measured. After independently analyzing each kymograph, a scheme was created where each line represents the percentage of sfGFP signal covering on one microtubule.
The significance of difference among multiple groups was tested by non-parametric ANOVA with the Kruskal-Wallis test, and post-hoc pairwise comparisons were tested by the Dunn’s test except for Fig. 1 G and Fig. 7 B where the Conover-Iman test was employed. For the interpretation of the p values, ns means there is no significant difference between the two distributions. * means P value < 0.05, ** means P value < 0.01, *** means P value < 0.001, and **** means P value < 0.0001. Data are generally represented as the median with the interquartile range.
Data analysis of immunofluorescence
For measurement of Tyr and deTyr tubulin intensities, FIJI software was used. A mask was created by addition of the two channels, and each object in the image was saved as a region of interest. The mean intensity of regions of interest was measured on each channel after background subtraction. In each experiment, the values were normalized to the median of the control without enzymes. Data analysis was then performed with GraphPad Prism 7. Statistical analyses were obtained using nonparametric one-way ANOVA assuming non-paired and non-Gaussian distribution (Kruskal–Wallis test).
Quantification of the colocalization of VASH2 proteins with microtubules in cells was performed as in Wang et al. (2019). Briefly, for each cell a cytoplasmic region excluding the perinuclear region was selected. After automatic thresholding, the Pearson’s coefficient of correlation of the two color channels was calculated using the Jacop plugin in Fiji software.
Data are generally represented as the median with the interquartile range. For the interpretation of the P values, ns means there is no significant difference between the two distributions, ** means P value < 0.01, and **** means P value < 0.0001.
Online supplemental material
Fig. S1 shows proteins used in the TIRF experiments. Fig. S2 shows comparison of tubulin detyrosinating activity of sfGFP-tagged and untagged VASH–SVBP complexes, and evolution over the time. Fig. S3 shows cryo-EM data collection and processing. Fig. S4 shows VASH2–SVBP mutants associate poorly with microtubules in cells. Fig. S5 shows sequence alignments of VASHs. Fig. S6 shows relative position of the catalytic site in microtubule–VASH1 and –VASH2 complexes with respect to the C-terminus of helix H12 of α-tubulin. Video 1 shows interaction of single molecules of V1_FL with Taxol-stabilized microtubules enriched in tyrosinated HeLa tubulin studied by single-molecule TIRF microscopy. Video 2 shows interaction of single molecules of V2_FL with Taxol-stabilized microtubules enriched in tyrosinated HeLa tubulin studied by single-molecule TIRF microscopy. Video 3 shows interaction of single molecules of chimeric sfGFP-VASH1–SVBP complex bearing the N-terminal region of VASH2 [V1_FL(NtV2)] with Taxol-stabilized microtubules enriched in tyrosinated HeLa tubulin studied by single-molecule TIRF microscopy. Video 4 shows interaction of single molecules of chimeric sfGFP-VASH2–SVBP complex bearing the N-terminal region of VASH1 [V2_FL(NtV1)] with Taxol-stabilized microtubules enriched in tyrosinated HeLa tubulin studied by single-molecule TIRF microscopy.
Data availability
The cryo-EM electron density map and the structural model of the microtubule–VASH2–SVBP complex have been deposited in the Electron Microscopy Data Bank (EMD-14634) and the Protein Data Bank (ID 7ZCW), respectively.
Acknowledgments
We would like to thank Isabelle Jacquier and Lisa De Macedo of Grenoble Institute Neuroscience (GIN) for technical help in preparation of proteins, Yasmina Saoudi of Photonic Imaging Center of GIN for helping us with TIRF microscopy, and Benoît Decherf for the design of the FIJI software plugin for detection of single-molecule tracks in the kymographs of TIRF experiments. We thank Yves Goldberg for the valuable discussion on statistics. Modified plasmid pETDUet-1 was a generous gift from Sandra Jeudy (IGS, Marseille). We thank the Electron Microscopy Facility of the Paul Scherrer Institute and the BioEM Lab of the Biozentrum, University of Basel, for excellent access to cryo-electron microscopes. We are indebted to Carolyn Moores for initial help with the image analysis procedure.
This work was supported by the Leducq Foundation, research grant n° 20CVD01 (to M.-J. Moutin), by Agence National de la Recherche (ANR), grant SPEED-Y n° ANR-20-CE16-0021 (to M.-J. Moutin) and grant n° ANR-20-CE13-0011 (to C. Janke), by Appel à projets 2019 CNRS/INSERM Défi Santé Numérique (to V. Stoppin-Mellet), by France Alzheimer grant 2023 (to M.M. Magiera), by Institut National de la Santé et de la Recherche Médicale (INSERM), Centre National de la Recherche Scientifique (CNRS), and University Grenoble Alpes, and by the Swiss National Science Foundation, grant n° 310030_192566 (to M.O. Steinmetz). Part of the salaries of C. Sanyal and S. Ramirez-Rios were from a collaborative program between Servier laboratories and M.-J. Moutin team. This work was supported by the Photonic Imaging Center of GIN (University Grenoble Alpes – INSERM U1216), which is part of the Imagerie des Sciences du Vivant core facility and certified by the Infrastructures en Biologie Santé et Agronomie label.
Author contributions. C. Bosc analyzed sequences. B. Blot and C. Bosc designed and performed cloning experiments for the production of sfGFP-tagged proteins. S. Ramirez-Rios, C. Sanyal, and S.R. Choi prepared recombinant proteins. S. Ramirez-Rios and C. Sanyal conceived and performed single-molecule TIRF experiments and assays of activity in reconstituted system and in cells. C. Sanyal generally analyzed binding characteristics (TIRF), and S. Ramirez-Rios enzyme activity (immunofluorescence) and diffusion (TIRF). E. Denarier developed ImageJ macros for TIRF (diffusion) and immunofluorescence analysis, and helped in statistical analysis. F. Krichen started the TIRF project with the help of J.M. Soleilhac. V. Stoppin-Mellet and I. Arnal provided essential advice for single-molecule TIRF all along the project. S. Ramirez-Rios and C. Sanyal produced, purified, and labeled bovine brain tubulin with the help of I. Arnal and V. Stoppin-Mellet. M.M. Magiera and C. Janke generated and purified HeLa tyrosinated and detyrosinated tubulin. S.R. Choi prepared cryo-EM grids. T.B. Blum collected cryo-EM data. T.B. Blum and S.R. Choi processed and refined cryo-EM data. S.R. Choi modeled cryo-EM data. S.R. Choi and M.O. Steinmetz performed structural analyses. M.-J. Moutin supervised the functional experiments. M.O. Steinmetz supervised the structural experiments. M.-J. Moutin prepared Fig. 1, Fig. 2, Fig. 3, D and E, Fig. 5, Fig. 6, Fig. S1, Fig. S2, and Tables 1 and 3 and graphical abstract with the help of S. Ramirez-Rios and C. Sanyal. S.R. Choi prepared Fig. 3, A–C, Fig. 4, Fig. S3, Fig. S6, and Table 2. C. Bosc prepared Fig. S5. S. Ramirez-Rios prepared Fig. 7 A and Fig. S4 and supplemental videos. M.-J. Moutin wrote the manuscript with contributions from all authors.
References
Author notes
S. Ramirez-Rios, S.R. Choi, and C. Sanyal contributed equally to this paper.
Disclosures: M.-J. Moutin reported grants from Servier during the conduct of the study; in addition, M.-J. Moutin had a patent to methods and pharmaceuticals compositions for treating tubulin carboxypeptidases associated diseases licensed “PCT/EP2018/0.” No other disclosures were reported.



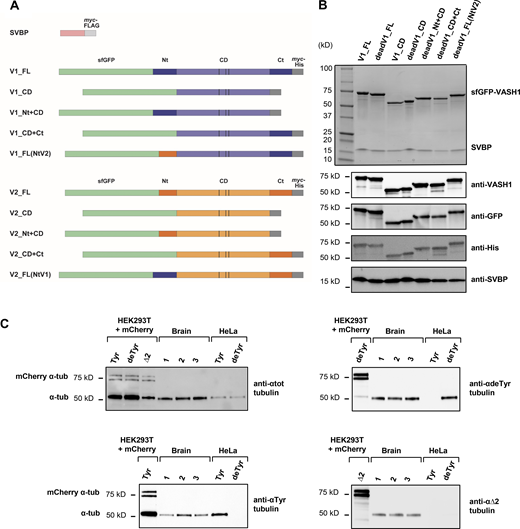



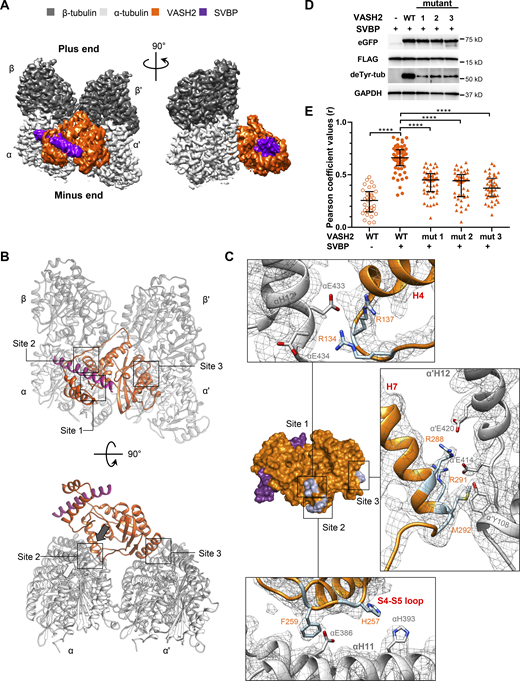






![The exchange of the N-terminal regions strongly alters binding behavior and activity of VASH–SVBP enzyme complexes. (A−F) Comparison of the interaction with Taxol-stabilized Tyr-MTs of the sfGFP-tagged full length VASH1 and VASH2 enzymes (V1_FL, V2_FL) and chimeras in which the N-termini were exchanged [V1_FL(NtV2), V2_FL(NtV1)]. Both catalytically dead (A, C, and E) and active (B, D, and F) versions were studied. Experiments were performed in the presence of 50 pM enzyme in BRB40 supplemented with 50 mM KCl. (A and B) Representative kymographs. Scale bars: horizontal, 5 µm; vertical, 5 s. Examples of TIRF movies from which kymographs were extracted are presented in supplemental data for the chimeras (Videos 3 and 4). Binding characteristics are summarized in Table 3. (C and D) Analysis of diffusion. Each point represents a single molecule, the molecules moving on at least 17 microtubules were analyzed. (E and F) Representation of the microtubule surface covered with VASH–SVBP molecules (white) or not covered (black) during the 45 s of TIRF movies. Each horizontal line represents a microtubule (at least 17 microtubules were analyzed). The red value corresponds to the mean covering (in %). (G and H) Activity of VASH–SVBP enzyme complexes and chimeras on Tyr-MTs measured by immunofluorescence in the same experimental conditions (50 pM enzyme, BRB40 supplemented with 50 mM KCl, TIRF chamber). (G) Representative images of tyrosinated (Tyr, magenta) and detyrosinated (deTyr, green) α-tubulin pools of microtubules after 30 min incubation in the absence (control) or presence of the indicated enzyme. Scale bar, 10 µM. (H) Analysis of tyrosinated- and detyrosinated-tubulin signals intensities. Each point represents a microtubule (at least 200 microtubules were analyzed). Data are represented as the median. Statistical significance was determined using Kruskal-Wallis test, ****P < 0.0001.](https://cdn.rupress.org/rup/content_public/journal/jcb/222/2/10.1083_jcb.202205096/2/m_jcb_202205096_fig6.png?Expires=1772622837&Signature=0fhFWH3YRVm5a~PY09T4~GkQz~2Hc0CoclK1mi4t8EVxhNS4dCOxhiHi6-WEoS1XpIn1vN0QBvucT-MDuYLGZ41Qa4bbmgdWy3z7RIw96OE19WfD4j2dn0~DW~~qMUsfSdFYj6XTv5TYQGucMvpU7awmdCpxlHI3C2sZr2arMFdpJ44rTnjS8TrUzUHOHHB2CnK3cMJ~HIULQmlW7HTXPDYHzTLrhpvmJI5sczRzuyKg9D9l5o0nVWo1QCL26KXdqKUylHKnGR-H~2LbhLA-oyEgT6bT4OyWzAqr8~txCZTEFP68eVleA~KuJzBMWFrEQzA47bDUGJRixIAjh~xG2A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)



