Contractile epithelial tubes are found in various organs, such as lung airways and blood capillaries. Their ability to sense luminal pressure and respond with adequate contractility is essential for their physiology, and its mis-regulation results in diseases such as asthma and hypertension. Here, we describe a mechanoresponsive regulatory pathway downstream of tissue stretching that controls contraction of the C. elegans spermatheca, a tubular structure where fertilization occurs. Using live-imaging, we show that ovulation-induced stretching of spermathecal cells leads to recruitment of the RhoGEF RHGF-1 to stress fibers, which activates RHO-1 and myosin II in a positive feedback loop. Through deletion analysis, we identified the PDZ domain of RHGF-1 as responsible for F-actin binding, and genetic epistasis analysis with the RhoGAP spv-1 demonstrated that tension-dependent recruitment of RHGF-1 to F-actin is required for robust spermathecal contractility. Our study illustrates how mechanosensitive regulators of Rho GTPases provide epithelial tubes the ability to tune their contractility in response to internal pressure.
Introduction
Contractile tubes are found in many tissues, including in the vascular, pulmonary, and reproductive systems (Bernascone et al., 2017). The balance between tube contractility and luminal pressure determines tube diameter and is, therefore, of physiological importance (Sethi et al., 2017). Mis-regulation of tubular contractility can lead to diseases such as hypertension and asthma (Duong-Quy et al., 2013; Tang, 2015). In large tubes, contractility is provided by smooth muscle cells surrounding an inner epithelium, while in small tubes, such as blood capillaries and lung alveoli, contractility is generated by the epithelial cells forming the tube themselves. Smooth muscle and epithelial cell contraction is facilitated by arrays of actin and myosin, and is regulated by two major signaling pathways: calcium and the GTPase RhoA (Szasz and Webb, 2017; Touyz et al., 2018).
In its active, GTP-bound state, RhoA promotes actin polymerization by activating formins and LIM domain Kinase (Geneste et al., 2002), induces myosin filament assembly through activation of Rho Kinase (Yoneda et al., 2005), and facilitates the assembly of actomyosin networks by activating anillin (Piekny and Glotzer, 2008). Since RhoA is such a potent activator of actomyosin, its own activation is tightly regulated in space and time (Heasman and Ridley, 2008; Jaffe and Hall, 2005). Guanine nucleotide exchange factors (GEFs) activate RhoA by catalyzing the exchange of GDP with GTP, while GTPase-activating proteins (GAPs) turn it off by inducing GTP hydrolysis. In its inactive state, GDP-bound RhoA is sequestered in the cytoplasm by guanine nucleotide dissociation inhibitors (Bos et al., 2007; Garcia-Mata et al., 2011). The human genome encodes for 79 RhoGEFs, 64 RhoGAPs, and 2 dual GEFs/GAPs (Müller et al., 2020). They are all multi-domain proteins, each with its own unique interactome and subcellular localization (Müller et al., 2020; Rossman et al., 2005; Tcherkezian and Lamarche-Vane, 2007). The combined action of specific RhoGEFs and RhoGAPs from this pool regulates RhoA activity in various cellular processes, creating a complex network of regulation, whose logic is only beginning to be revealed.
Our model for the study of RhoA regulation in a contractile tube is the Caenorhabditis elegans spermatheca, the tissue where sperm are stored and fertilization takes place (Singaravelu and Singson, 2011). In addition to being visually accessible and genetically tractable, the worm has only 25 RhoGAPs and 20 RhoGEFs, greatly simplifying their investigation.
The spermatheca is composed of 24 epithelial cells organized as a flexible pouch with valves on either end, situated between the oviduct and the uterus (Fig. 1 A; Kimble and Hirsh, 1979). In the ovulation process, an oocyte enters the spermatheca through the distal valve, fertilization takes place, an eggshell is synthesized, and eventually the spermatheca contracts to push the embryo through the spermatheca-uterine (sp-ut) valve into the uterus (Mccarter et al., 1999). Spermatheca contraction is driven by actomyosin contractility of basally located stress fibers within the epithelial cells (Wirshing and Cram, 2017), and is known to depend on both calcium signaling and the C. elegans ortholog of RhoA, RHO-1 (Castaneda et al., 2020; Kelley et al., 2020a; Kovacevic et al., 2013; Tan and Zaidel-Bar, 2015).
Loss of the rhoGEF rhgf-1 rescues spermatheca functions in the RhoGAP spv-1 loss of function mutant. (A) The reproductive system of an adult hermaphrodite C. elegans outlined on stitched DIC microscopy images. Scale bar, 50 µm. (B) Representative stitched DIC images of embryos from wild-type and spv-1(ok1498) loss-of-function worms treated with control (empty vector) or rhgf-1(RNAi). Black arrows point to abnormally shaped embryos. Scale bar, 50 µm. (C) Embryonic lethality of the progeny of wild-type or rhgf-1(ok880) loss-of-function worms treated with control or spv-1(RNAi). Nrepeats ≥ 5, Nmothers ≥ 150, Nembryos ≥ 990. Error bars are ± SEM. Statistical comparisons were performed by Brown-Forsythe ANOVA-Dunnett’s T3 multiple comparisons test. (D) Representative time points from DIC movies of the ovulation process in wild type, spv-1(1498), rhgf-1(ok880), and the double mutant spv-1(1498);rhgf-1(ok880). Time zero (0 s) is the time when the distal valve closes. The spermatheca is pseudo colored pink and the ovulating oocyte/embryo is pseudo colored green. Special phenotypes are labeled. Distal and proximal parts of the spermatheca are labeled. Lines labeled W1 and W2 relate to the measurement of constriction magnitude. Scale bar, 10 µm. Full movies are in Video 1. (E) Dwell time in wild-type, spv-1(1498), rhgf-1(ok880), and spv-1(1498);rhgf-1(ok880) day 1 adults, measured as the duration the oocyte resides in the spermatheca, from distal valve closure until the opening of the sp-ut valve. N ≥ 13. Error bars are mean ± SEM. Statistical comparisons were performed by One-way ANOVA-Tukey’s multiple comparisons test. (F) Constriction magnitude in wild type, spv-1(1498), rhgf-1(ok880), and spv-1(1498);rhgf-1(ok880), measured as the ratio of the width of the distal quarter of the spermatheca (W1) to the width of the proximal quarter of the spermatheca (W2) before the embryo starts to transit out. N ≥ 13. Error bars are mean ± SEM. Statistical comparisons were performed by Brown-Forsythe ANOVA-Dunnett’s T3 multiple comparisons test. (G) Successful embryos transit through the spermatheca of wild-type, spv-1(1498), rhgf-1(ok880), and spv-1(1498);rhgf-1(ok880) worms. N ≥ 14. Statistical comparisons were performed by individuals’ chi-square tests. Stars designate statistical significance (**** P < 0.0001, *** P < 0.001, ** P < 0.01, ns P > 0.05). N, number of samples analyzed.
Loss of the rhoGEF rhgf-1 rescues spermatheca functions in the RhoGAP spv-1 loss of function mutant. (A) The reproductive system of an adult hermaphrodite C. elegans outlined on stitched DIC microscopy images. Scale bar, 50 µm. (B) Representative stitched DIC images of embryos from wild-type and spv-1(ok1498) loss-of-function worms treated with control (empty vector) or rhgf-1(RNAi). Black arrows point to abnormally shaped embryos. Scale bar, 50 µm. (C) Embryonic lethality of the progeny of wild-type or rhgf-1(ok880) loss-of-function worms treated with control or spv-1(RNAi). Nrepeats ≥ 5, Nmothers ≥ 150, Nembryos ≥ 990. Error bars are ± SEM. Statistical comparisons were performed by Brown-Forsythe ANOVA-Dunnett’s T3 multiple comparisons test. (D) Representative time points from DIC movies of the ovulation process in wild type, spv-1(1498), rhgf-1(ok880), and the double mutant spv-1(1498);rhgf-1(ok880). Time zero (0 s) is the time when the distal valve closes. The spermatheca is pseudo colored pink and the ovulating oocyte/embryo is pseudo colored green. Special phenotypes are labeled. Distal and proximal parts of the spermatheca are labeled. Lines labeled W1 and W2 relate to the measurement of constriction magnitude. Scale bar, 10 µm. Full movies are in Video 1. (E) Dwell time in wild-type, spv-1(1498), rhgf-1(ok880), and spv-1(1498);rhgf-1(ok880) day 1 adults, measured as the duration the oocyte resides in the spermatheca, from distal valve closure until the opening of the sp-ut valve. N ≥ 13. Error bars are mean ± SEM. Statistical comparisons were performed by One-way ANOVA-Tukey’s multiple comparisons test. (F) Constriction magnitude in wild type, spv-1(1498), rhgf-1(ok880), and spv-1(1498);rhgf-1(ok880), measured as the ratio of the width of the distal quarter of the spermatheca (W1) to the width of the proximal quarter of the spermatheca (W2) before the embryo starts to transit out. N ≥ 13. Error bars are mean ± SEM. Statistical comparisons were performed by Brown-Forsythe ANOVA-Dunnett’s T3 multiple comparisons test. (G) Successful embryos transit through the spermatheca of wild-type, spv-1(1498), rhgf-1(ok880), and spv-1(1498);rhgf-1(ok880) worms. N ≥ 14. Statistical comparisons were performed by individuals’ chi-square tests. Stars designate statistical significance (**** P < 0.0001, *** P < 0.001, ** P < 0.01, ns P > 0.05). N, number of samples analyzed.
The regulation of spermathecal contraction is critically important for fertility. At first, the spermatheca must be relaxed to allow oocyte entry. Once the oocyte enters the spermatheca, premature contraction before the eggshell is synthesized, will pinch the oocyte, and cause embryonic lethality (Hegsted et al., 2016; Tan and Zaidel-Bar, 2015). On the other hand, delayed or lack of contraction creates a “traffic jam” in the fertility process, with embryos accumulating in the spermatheca and oocytes becoming endomitotic (Kovacevic et al., 2013; McGovern et al., 2018).
We have previously shown that the RhoGAP SPV-1, ortholog of human ARHGAP29, GMIP, and ARHGAP45, functions to inhibit RHO-1 activation in the spermatheca, thus preventing premature spermathecal contractility (Tan and Zaidel-Bar, 2015). SPV-1 binds to the convoluted apical membrane through its F-BAR domain when the spermatheca is empty. Following oocyte entry into the spermatheca—an event that dramatically stretches the tissue—SPV-1 becomes cytoplasmic and inactive (Tan and Zaidel-Bar, 2015). This suggests a mechanobiological feedback pathway by which stretching of the spermatheca shuts off RhoGAP activity. However, the molecular mechanism through which RHO-1 is activated in this system remained unknown.
Here, we identify RHGF-1, ortholog of human ARHGEF11/PDZ-RhoGEF and ARHGEF12/LARG, as the major RhoGEF that activates RHO-1 in the spermatheca and demonstrate that it changes its sub-cellular localization upon oocyte entry. Moreover, we show that RHGF-1 is recruited through its PDZ (post synaptic density protein [PSD95], Drosophila disc large tumor suppressor [Dlg1], and Zonula occludens-1 protein [zo-1]) domain to the basal F-actin stress fibers in a tension-reinforced manner. Thus, RHGF-1 and SPV-1 both regulate RHO-1 in complementary tension-sensitive signaling pathways to induce spermathecal contraction at the right time during ovulation.
Results
Loss of the RhoGEF RHGF-1 rescues the loss of function phenotypes of the RhoGAP SPV-1
To identify RhoGEFs responsible for activating RHO-1 in the C. elegans spermatheca, we performed an RNAi screen with all 20 C. elegans RhoGEFs, using spv-1(ok1498) as a sensitized background. The ok1498 allele is a deletion of the RhoGAP domain, resulting in a hypercontractile spermatheca that often pinched embryos, leading to abnormally shaped embryos on the plate and embryonic lethality (Tan and Zaidel-Bar, 2015). We hypothesized that depletion of the RhoGEF/s that control RHO-1 activation in the spermatheca would alleviate the hypercontractility of spv-1(ok1498) and thus rescue the pinched embryo phenotype. Indeed, our genetic screen revealed that RNAi of the RhoGEF rhgf-1 completely rescued the embryo shape defect of spv-1(ok1498) (Fig. 1 B and Table S1) and partial rescue was achieved with RNAi of unc-73 and Y105E8A.25 (Table S1).
We further investigated the genetic interaction between rhgf-1 and spv-1 by performing the inverse experiment, depleting spv-1 by RNAi in the null mutant rhgf-1(ok880), which has a deletion of its RhoGEF domain. Depletion of spv-1 by RNAi in wild-type worms led to 16 ± 2% (mean ± SEM) embryonic lethality due to embryo pinching. However, depletion of spv-1 in the background of rhgf-1(ok880) resulted in only 7 ± 2% embryonic lethality. The embryonic lethality observed in rhgf-1(ok880) on its own (2.5 ± 0.6%) was not different than wild type (4 ± 1%; Fig. 1 C).
Next, we crossed the RhoGEF and RhoGAP mutants and imaged ovulation transits in wild-type, single mutants, and double-mutant worms, using differential interference contrast (DIC) microscopy (Fig. 1 D and Video 1). We measured three key parameters: dwell time, defined as the duration of time the oocyte resides in the spermatheca before being pushed into the uterus; spermathecal constriction, defined as the ratio between the width of the spermatheca at the distal end (W1) and the width at its proximal end (W2) before the embryo starts to transit out (Fig. 1 D); and the percent of successful embryo transits. In movies with wild-type animals, we measured an average dwell time of 242 ± 6 s, a constriction value of 0.89 ± 0.02, and 100% success rate (Fig. 1, E–G). In spv-1(ok1498), as reported previously (Tan and Zaidel-Bar, 2015), average dwell time significantly decreased to 104 ± 11 s, with a constriction value of 0.58 ± 0.05, indicating a highly constricted spermatheca, and only 56% of the ovulation transits in the spv-1 mutant were successful, due to embryo severing (Fig. 1, E–G). In contrast, the rhgf-1 mutant exhibited an increase in dwell time to 304 ± 12 s, a constriction value of 0.94 ± 0.02, and a 75% success rate due to embryo trapping in the spermatheca, indicating weak or no contraction. The genetic combination of both mutations appeared to largely cancel out each other. The double-mutant rhgf-1(ok880);spv-1(ok1498) had a dwell time of 183 ± 12 s, a constriction value of 0.87 ± 0.02, and importantly, 100% of the embryos successfully transited through the spermatheca (Fig. 1, E–G). Taken together, these experiments suggest that the RhoGEF RHGF-1 and the RhoGAP SPV-1 regulate spermathecal contractility in antagonistic manner.
DIC microscopy time-lapse movies of ovulation and embryo transit in wild-type, spv-1(ok1498), rhgf-1(ok880), and spv-1(ok1498); rhgf-1(ok880) mutant animals. Time interval between frames is 3 s. Frame rate is 7 frames per second (fps). Related to Fig. 1 D.
DIC microscopy time-lapse movies of ovulation and embryo transit in wild-type, spv-1(ok1498), rhgf-1(ok880), and spv-1(ok1498); rhgf-1(ok880) mutant animals. Time interval between frames is 3 s. Frame rate is 7 frames per second (fps). Related to Fig. 1 D.
Following up on the two partial hits from our spv-1 rescue screen, we generated two independent deletion alleles for Y105E8A.25 by CRISPR-Cas9. Measurement of dwell time in these strains did not show any difference from wild type (Fig. S1 A), leading us to conclude that this gene was a false positive in the initial screen. Measurement of dwell time in strains with classical alleles of unc-73 revealed an increase in dwell time in the unc-73(e936) strain that carries a single base pair substitution (Fig. S1 A). Furthermore, spermathecal-specific unc-73(RNAi) in the background of spv-1(ok1498), led to a significant increase in the dwell time, confirming that UNC-73 plays some role in regulating spermathecal contractility (Fig. S1 B).
Dwell time analyses in loss-of-function alleles and RNAi show that the RhoGEF unc-73 regulates spermatheca contractility and Y105E8A.25 does not. (A) Dwell time of wild-type, unc-73(ok322), unc-73(e936), unc-73(ce362), unc-73(ok936), Y105E8A.25(knu182), and Y105E8A.25(knu185) worms. N ≥ 3. Bars are mean ± SEM. Statistical comparisons were performed by One-way ANOVA-Tukey’s multiple comparisons test. (B) Dwell time in spv-1(ok1498) worms treated with spermatheca-specific unc-73(RNAi). N ≥ 7. Bars are mean ± SEM. Statistical comparison was performed by two-tailed unpaired t test. Stars designate statistical significance (*** P <0.001, ** P <0.01). N, number of samples analyzed.
Dwell time analyses in loss-of-function alleles and RNAi show that the RhoGEF unc-73 regulates spermatheca contractility and Y105E8A.25 does not. (A) Dwell time of wild-type, unc-73(ok322), unc-73(e936), unc-73(ce362), unc-73(ok936), Y105E8A.25(knu182), and Y105E8A.25(knu185) worms. N ≥ 3. Bars are mean ± SEM. Statistical comparisons were performed by One-way ANOVA-Tukey’s multiple comparisons test. (B) Dwell time in spv-1(ok1498) worms treated with spermatheca-specific unc-73(RNAi). N ≥ 7. Bars are mean ± SEM. Statistical comparison was performed by two-tailed unpaired t test. Stars designate statistical significance (*** P <0.001, ** P <0.01). N, number of samples analyzed.
RHGF-1 regulates spermathecal RHO-1 levels and does not affect calcium signaling
Both active RHO-1 and calcium signaling are known to initiate spermathecal contractility before embryo expulsion. The RhoGAP SPV-1 directly regulates the activity of RHO-1 (Tan and Zaidel-Bar, 2015), and it was also found to regulate calcium signaling, through its regulation of CDC-42 activity (Bouffard et al., 2019). Moreover, while both human orthologs of RHGF-1, ARHGEF11, and ARHGEF12 were shown to activate RhoA (Müller et al., 2020), it has been reported that ARHGEF11 can gain affinity toward Cdc42 when activated by Gαs (Castillo-Kauil et al., 2020). Therefore, we sought to determine whether RHGF-1 regulates RHO-1 activity or calcium signaling or both.
To examine the effect of RHGF-1 on calcium signaling, we crossed rhgf-1(ok880) with a GCaMP calcium reporter and used widefield fluorescence microscopy to image calcium signaling during embryo transit (Video 2). We used an established image analysis pipeline to quantify calcium dynamics (Bouffard et al., 2019). In all measured parameters, including the number and rate of calcium peaks, and the amount of time required to reach half the maximum or maximum calcium signal, there were no significant differences between rhgf-1(ok880) and the wild-type control (Fig. 2, A–D). Furthermore, we generated kymographs that allowed us to visualize the dynamics of calcium along the entire spermatheca throughout an ovulation transit event in a single image (Fig. 2 E). This analysis revealed that the spatiotemporal dynamics of calcium in rhgf-1(ok880) follow the same pattern as in wild type.
Calcium signaling visualized in rhgf-1(ok880) and control spermatheca with a GCaMP calcium reporter. Time interval between frames is 1 s. Frame rate is 7 fps. Related to Fig. 2 E.
Calcium signaling visualized in rhgf-1(ok880) and control spermatheca with a GCaMP calcium reporter. Time interval between frames is 1 s. Frame rate is 7 fps. Related to Fig. 2 E.
RHGF-1 regulates the activity of RHO-1 and does not affect calcium dynamics. (A–D) Based on GCaMP3 movies, the number of Ca2+ peaks, the number of Ca2+ peaks per second, and the amount of time after oocyte entry required to reach half of the maximum or maximum Ca2+ signal was determined for control and rhgf-1(ok880) Ca2+ traces. N ≥ 12. Error bars are ± SEM. (A, B, and D) Statistical comparisons were performed by two-tailed unpaired t test with Welch’s correction. (C) Statistical comparison was performed by two-tailed unpaired t test. (E) Representative kymographs of control and rhgf-1(ok880) movies are shown with time of entry, distal neck closure, and time the sp-ut valve opens and closes indicated. Kymographs were generated by averaging over the columns of each movie frame (see Materials and methods). Scale bars: vertical scale bar, 1 sec; horizontal scale bar, 10 µm. Time elapsed is indicated on the y-axis. Full movies are in Video 2. (F) Live imaging of spermatheca-specific RHO-1 biosensor (AHPH::GFP) during embryo transit through spermatheca in control and rhgf-1(RNAi) worms. Arrowheads point to the distal end of the spermatheca, where RHO-1 activity is high in the control and low in the rhgf-1(RNAi). Dashed line marks the area where RHO-1 biosensor was quantified. Scale bar, 20 µm. Full movies are in Video 3. (G) Quantification of RHO-1 biosensor fluorescence intensity during ovulation and embryo transit in control and rhgf-1(RNAi) worms. The x-axis (time) was normalized according to distinct events so that all traces begin at the time when the distal valve closes behind the ovulating oocyte (“Oocyte entry complete”) are aligned at the time the sp-ut starts to open (“Embryo exit initiation”) and aligned again when the sp-ut valve closes behind the exiting embryo (“Embryo exit complete”). N = 6. (H) Maximum intensity projections of spermathecae expressing GFP tagged to ACT-1 (actin) in control, rho-1(RNAi), and rhgf-1(RNAi) worms. Arrows point to wavy actin. Scale bar, 10 µm. (I) Tortuosity of actin bundles in the spermathecae of control, rho-1(RNAi), and rhgf-1(RNAi) worms. Tortuosity is defined as the actual length between two ends of an actin bundle divided by the shortest distance between them. N ≥ 12. Error bars are ± SEM. Statistical comparisons were performed by One-way ANOVA-Tukey’s multiple comparisons test. Stars designate statistical significance (*** P < 0.001, ** P < 0.01, ns P > 0.05). N, number of samples analyzed.
RHGF-1 regulates the activity of RHO-1 and does not affect calcium dynamics. (A–D) Based on GCaMP3 movies, the number of Ca2+ peaks, the number of Ca2+ peaks per second, and the amount of time after oocyte entry required to reach half of the maximum or maximum Ca2+ signal was determined for control and rhgf-1(ok880) Ca2+ traces. N ≥ 12. Error bars are ± SEM. (A, B, and D) Statistical comparisons were performed by two-tailed unpaired t test with Welch’s correction. (C) Statistical comparison was performed by two-tailed unpaired t test. (E) Representative kymographs of control and rhgf-1(ok880) movies are shown with time of entry, distal neck closure, and time the sp-ut valve opens and closes indicated. Kymographs were generated by averaging over the columns of each movie frame (see Materials and methods). Scale bars: vertical scale bar, 1 sec; horizontal scale bar, 10 µm. Time elapsed is indicated on the y-axis. Full movies are in Video 2. (F) Live imaging of spermatheca-specific RHO-1 biosensor (AHPH::GFP) during embryo transit through spermatheca in control and rhgf-1(RNAi) worms. Arrowheads point to the distal end of the spermatheca, where RHO-1 activity is high in the control and low in the rhgf-1(RNAi). Dashed line marks the area where RHO-1 biosensor was quantified. Scale bar, 20 µm. Full movies are in Video 3. (G) Quantification of RHO-1 biosensor fluorescence intensity during ovulation and embryo transit in control and rhgf-1(RNAi) worms. The x-axis (time) was normalized according to distinct events so that all traces begin at the time when the distal valve closes behind the ovulating oocyte (“Oocyte entry complete”) are aligned at the time the sp-ut starts to open (“Embryo exit initiation”) and aligned again when the sp-ut valve closes behind the exiting embryo (“Embryo exit complete”). N = 6. (H) Maximum intensity projections of spermathecae expressing GFP tagged to ACT-1 (actin) in control, rho-1(RNAi), and rhgf-1(RNAi) worms. Arrows point to wavy actin. Scale bar, 10 µm. (I) Tortuosity of actin bundles in the spermathecae of control, rho-1(RNAi), and rhgf-1(RNAi) worms. Tortuosity is defined as the actual length between two ends of an actin bundle divided by the shortest distance between them. N ≥ 12. Error bars are ± SEM. Statistical comparisons were performed by One-way ANOVA-Tukey’s multiple comparisons test. Stars designate statistical significance (*** P < 0.001, ** P < 0.01, ns P > 0.05). N, number of samples analyzed.
Next, to investigate the effect of RHGF-1 on RHO-1 activity, we imaged the previously published spermatheca-specific RHO-1 biosensor (Tan and Zaidel-Bar, 2015), using confocal fluorescence microscopy, in control and rhgf-1(RNAi) conditions (Fig. 2 F and Video 3). RHO-1 activity in the sp-ut valve was high throughout the ovulation process. The level of active RHO-1 in the spermatheca bag remained low for the first 2 min after oocyte entry and then it increased, first slowly and then rapidly, prior to and during spermathecal contraction, reaching a maximum intensity 2.2 ± 0.2fold (N = 6) of its value at the time of oocyte entry. In rhgf-1(RNAi) spermathecae, in contrast, RHO-1 activity remained fairly constant until embryo exit initiation, after which it rose moderately, reaching a maximum of 1.5 ± 0.1fold (N = 6) of its value at the time of oocyte entry (Fig. 2, F and G). The lackluster activation of RHO-1 was even more prominent in instances of failed embryo transits (Fig. S2 A). In these cases, the intensity level of the RHO-1 biosensor remained nearly flat for the duration of the movies, up to 26 min after oocyte entry was completed (Fig. S2, A and B; and Video 4), suggesting that RHGF-1 is responsible for activating RHO-1 in the spermatheca.
RHO-1 activity reported by the AHPH::GFP RHO-1 biosensor in control and rhgf-1(RNAi) spermatheca during ovulation and embryo transit. Time interval between frames is 10 s. Frame rate is 3 fps. Related to Fig. 2 F.
RHO-1 activity reported by the AHPH::GFP RHO-1 biosensor in control and rhgf-1(RNAi) spermatheca during ovulation and embryo transit. Time interval between frames is 10 s. Frame rate is 3 fps. Related to Fig. 2 F.
In contrast with RHGF-1, UNC-73 does not appear to regulate RHO-1 in the spermatheca. (A) Snapshots at indicated times from movies of spermatheca-specific RHO-1 biosensor (AHPH::GFP) during ovulation and embryo transit in control, and an rhgf-1(RNAi) spermatheca that trapped the embryo for the duration of the movie. Scale bar, 20 µm. Full movies are in Video 4. (B) Quantification of RHO-1 biosensor fluorescence intensity during ovulation and embryo transit in three controls and three rhgf-1(RNAi) worms that trapped the embryo. (C) Live imaging of spermatheca-specific RHO-1 biosensor (AHPH::GFP) during embryo transit through spermatheca in control and unc-73(RNAi). Scale bar, 20 µm. Full movies are in Video 5. (D) Quantification of RHO-1 biosensor fluorescence intensity during ovulation and embryo transit in control and unc-73(RNAi) worms. The x-axis (time) was normalized according to distinct events so that all traces begin at the time when the distal valve closes behind the ovulating oocyte (“Oocyte entry complete”) are aligned at the time the sp-ut starts to open (“Embryo exit initiation”) and aligned again when the sp-ut valve closes behind the exiting embryo (“Embryo exit complete”). N = 6. (E) Dwell time in rhgf-1(ok880) spermathecae treated with spermatheca-specific unc-73(RNAi). N ≥ 7. Bars are mean ± SEM. Statistical comparison was performed by two-tailed unpaired t test with Welch’s correction. (F) Successful embryos transit through the spermathecae of rhgf-1(ok880) treated with spermatheca-specific unc-73(RNAi). N ≥ 19. Statistical comparison was performed by chi-square test. (G) Fluorescence intensity of UNC-73::GFP treated with unc-73(RNAi), normalize to control fluorescence intensity. N ≥ 27. Bars are mean ± SEM. Statistical comparison was performed by two-tailed unpaired t test with Welch’s correction. Stars designate statistical significance (**** P <0.0001, ns P >0.05). N, number of samples analyzed.
In contrast with RHGF-1, UNC-73 does not appear to regulate RHO-1 in the spermatheca. (A) Snapshots at indicated times from movies of spermatheca-specific RHO-1 biosensor (AHPH::GFP) during ovulation and embryo transit in control, and an rhgf-1(RNAi) spermatheca that trapped the embryo for the duration of the movie. Scale bar, 20 µm. Full movies are in Video 4. (B) Quantification of RHO-1 biosensor fluorescence intensity during ovulation and embryo transit in three controls and three rhgf-1(RNAi) worms that trapped the embryo. (C) Live imaging of spermatheca-specific RHO-1 biosensor (AHPH::GFP) during embryo transit through spermatheca in control and unc-73(RNAi). Scale bar, 20 µm. Full movies are in Video 5. (D) Quantification of RHO-1 biosensor fluorescence intensity during ovulation and embryo transit in control and unc-73(RNAi) worms. The x-axis (time) was normalized according to distinct events so that all traces begin at the time when the distal valve closes behind the ovulating oocyte (“Oocyte entry complete”) are aligned at the time the sp-ut starts to open (“Embryo exit initiation”) and aligned again when the sp-ut valve closes behind the exiting embryo (“Embryo exit complete”). N = 6. (E) Dwell time in rhgf-1(ok880) spermathecae treated with spermatheca-specific unc-73(RNAi). N ≥ 7. Bars are mean ± SEM. Statistical comparison was performed by two-tailed unpaired t test with Welch’s correction. (F) Successful embryos transit through the spermathecae of rhgf-1(ok880) treated with spermatheca-specific unc-73(RNAi). N ≥ 19. Statistical comparison was performed by chi-square test. (G) Fluorescence intensity of UNC-73::GFP treated with unc-73(RNAi), normalize to control fluorescence intensity. N ≥ 27. Bars are mean ± SEM. Statistical comparison was performed by two-tailed unpaired t test with Welch’s correction. Stars designate statistical significance (**** P <0.0001, ns P >0.05). N, number of samples analyzed.
RHO-1 activity reported by the AHPH::GFP RHO-1 biosensor in control and rhgf-1(RNAi) spermatheca during failed embryo transit (“trapping”). Time interval between frames is 10 s. Frame rate is 7 fps. Related to Fig. S2 A.
RHO-1 activity reported by the AHPH::GFP RHO-1 biosensor in control and rhgf-1(RNAi) spermatheca during failed embryo transit (“trapping”). Time interval between frames is 10 s. Frame rate is 7 fps. Related to Fig. S2 A.
As an indirect measure of RHO-1 activity in the spermatheca, we examined the appearance of the actin cytoskeleton. RhoA activity is known to induce stress fiber assembly and contractility (Chrzanowska-Wodnicka and Burridge, 1996). We used a strain expressing fluorescently tagged actin (GFP::ACT-1) to visualize the actin cytoskeleton in control RNAi, rho-1(RNAi), and rhgf-1(RNAi) conditions. In the control, actomyosin stress fibers appeared taut, while reduced RHO-1 activity resulted in loose and tortuous F-actin bundles (Fig. 2 H). Depletion of rhgf-1 affected the actin cytoskeleton similarly to rho-1 depletion, resulting in a wavy appearance of actin bundles as compared to the control. We quantified the tortuosity of actin bundles (actual length divided by distance between both ends) and found a similar increase in both rho-1(RNAi) and rhgf-1(RNAi) compared to the control RNAi (Fig. 2 I).
We then sought to establish whether UNC-73 also regulates RHO-1 in the spermatheca. To this end, we performed unc-73(RNAi) in the spermatheca-specific RHO-1 biosensor strain (Fig. S2 C and Video 5). Quantification of these movies revealed an increase in RHO-1 biosensor fluorescence intensity during embryo transits (1.8 ± 0.1-fold, N = 6) similar to the control movies (1.9 ± 0.2-fold, N = 6; Fig. S2 D), suggesting that UNC-73 does not regulate RHO-1. Furthermore, depletion of unc-73 by RNAi in the rhgf-1(ok880) strain did not affect the dwell time of embryos in the spermatheca, nor did it significantly change the percentage of trapped embryos (Fig. S2, E and F). We confirmed that unc-73(RNAi) reduces UNC-73 in the spermatheca by measuring the fluorescence intensity of endogenously tagged UNC-73::GFP and found it to be reduced to 29 ± 1.3% (N = 27) of wild-type level (Fig. S2 G). Taken together, these results point toward RHGF-1 as the major activator of RHO-1 in the spermatheca.
RHO-1 activity reported by the AHPH::GFP RHO-1 biosensor in control and unc-73(RNAi) spermatheca during ovulation and embryo transit. Time interval between frames is 10 s. Frame rate is 3 fps. Related to Fig. S2 C.
RHO-1 activity reported by the AHPH::GFP RHO-1 biosensor in control and unc-73(RNAi) spermatheca during ovulation and embryo transit. Time interval between frames is 10 s. Frame rate is 3 fps. Related to Fig. S2 C.
RHGF-1 is recruited to the basal side of spermathecal cells following oocyte entry in parallel with RHO-1 activation
Next, we sought to establish the expression pattern and subcellular localization of RHGF-1. To this end, we generated an endogenous RHGF-1::tagRFP strain using CRISPR-Cas9. The embryos in this strain had wild-type–like shape and ovulation movies showed a slightly shorter dwell time compared to control, indicating that tagging RHGF-1 did not perturb its function (Fig. S3, A and B). Endogenous RHGF-1::tagRFP was found to be highly expressed in spermathecal cells and several neurons, consistent with previous reports that used extrachromosomal arrays (Chen et al., 2014; Ziel et al., 2009; Fig. 3 A). We took advantage of the endogenously tagged RHGF-1 to quantify the efficacy of rhgf-1(RNAi) and found that rhgf-1(RNAi) leads to a 95% ± 0.7 (N = 19) reduction in RHGF-1::TagRFP fluorescence in the spermatheca (Fig. S3 C).
Tagging RHGF-1 with tagRFP does not perturb its function and shows that following oocyte entry RHGF-1 relocalizes from the cytoplasm to the basal side. (A and B) Comparison of dwell time and successful embryo transits in wild-type worms versus worm in which genomic RHGF-1 was fused with tagRFP. N ≥ 18. Error bars are mean ± SEM. Statistical comparison for A was performed by two-tailed unpaired t test. Statistical comparison for B was performed by chi-square-Fisher’s exact test. (C) Fluorescence intensity of RHGF-1::tagRFP treated with rhgf-1(RNAi), normalized to control fluorescence intensity. N ≥ 19. Bars are mean ± SEM. Statistical comparison was performed by two-tailed unpaired t test with Welch’s correction. (D) Confocal images from a time-lapse movie of a spermatheca expressing RHGF-1::tagRFP and β-Integrin/PAT-3::GFP as a basal membrane marker. Scale bar, 10 µm. Full movie in Video 6. (E) Representative images demonstrating how we measured the intensity of SPV-1::GFP and RHGF-1::tagRFP in the cytoplasm (yellow) and edge (green), respectively. Region in black box enlarged on right. Scale bars: whole spermatheca, 20 µm; enlargement, 2 µm. Stars designate statistical significance (**** P <0.0001, ** P <0.01, ns P >0.05). N, number of samples analyzed.
Tagging RHGF-1 with tagRFP does not perturb its function and shows that following oocyte entry RHGF-1 relocalizes from the cytoplasm to the basal side. (A and B) Comparison of dwell time and successful embryo transits in wild-type worms versus worm in which genomic RHGF-1 was fused with tagRFP. N ≥ 18. Error bars are mean ± SEM. Statistical comparison for A was performed by two-tailed unpaired t test. Statistical comparison for B was performed by chi-square-Fisher’s exact test. (C) Fluorescence intensity of RHGF-1::tagRFP treated with rhgf-1(RNAi), normalized to control fluorescence intensity. N ≥ 19. Bars are mean ± SEM. Statistical comparison was performed by two-tailed unpaired t test with Welch’s correction. (D) Confocal images from a time-lapse movie of a spermatheca expressing RHGF-1::tagRFP and β-Integrin/PAT-3::GFP as a basal membrane marker. Scale bar, 10 µm. Full movie in Video 6. (E) Representative images demonstrating how we measured the intensity of SPV-1::GFP and RHGF-1::tagRFP in the cytoplasm (yellow) and edge (green), respectively. Region in black box enlarged on right. Scale bars: whole spermatheca, 20 µm; enlargement, 2 µm. Stars designate statistical significance (**** P <0.0001, ** P <0.01, ns P >0.05). N, number of samples analyzed.
RHGF-1 is expressed in the spermatheca and localizes to the basal side following oocyte entry and parallel to RHO-1 activation. (A) RHGF-1::tagRFP expression pattern in the adult hermaphrodite by confocal image (left) and merged with DIC image (right). Arrows point to an occupied spermatheca and arrowheads point to an empty spermatheca. Image is a composite of multiple images stitched in Fiji. Scale bar, 50 µm. (B) Representative confocal images showing the subcellular localization of both RHGF-1::tagRFP and SPV-1::GFP in the same spermatheca from ovulation to the completion of embryo exit. White boxes mark regions enlarged below the images. The graphs show the intensity line profile along the marked lines from the apical side of the cell (“A”) to the basal side (“B”). Dashed lines mark the edges of spermathecal cells. Scale bars: whole spermatheca, 10 µm; enlargement, 2 µm. Full movie in Video 7. (C) Quantification of both RHGF-1::tagRFP and SPV-1::GFP recruitment to the cell edge during ovulation. The x-axis (time) was normalized according to distinct events so that all traces begin at the time when the distal valve closes behind the ovulating oocyte (“Oocyte entry complete”) are aligned at the time the sp-ut starts to open (“Embryo exit initiation”) and aligned again when the sp-ut valve closes behind the exiting embryo (“Embryo exit complete”). N = 3. (D) Representative confocal images of RHGF-1 subcellular localization and RHO-1 biosensor, in the same spermatheca, during oocyte ovulation and embryo transit. White boxes mark regions enlarged below the images. Dashed lines mark the edges of spermathecal cells. Scale bars: whole spermatheca, 20 µm; enlargement, 2 µm. Full movie in Video 8. (E) Quantification of RHGF-1::tagRFP recruitment to the cell edge, and RHO-1 biosensor intensity during ovulation and embryo exit. The x-axis (time) was normalized as in C. N = 6. N, number of samples analyzed.
RHGF-1 is expressed in the spermatheca and localizes to the basal side following oocyte entry and parallel to RHO-1 activation. (A) RHGF-1::tagRFP expression pattern in the adult hermaphrodite by confocal image (left) and merged with DIC image (right). Arrows point to an occupied spermatheca and arrowheads point to an empty spermatheca. Image is a composite of multiple images stitched in Fiji. Scale bar, 50 µm. (B) Representative confocal images showing the subcellular localization of both RHGF-1::tagRFP and SPV-1::GFP in the same spermatheca from ovulation to the completion of embryo exit. White boxes mark regions enlarged below the images. The graphs show the intensity line profile along the marked lines from the apical side of the cell (“A”) to the basal side (“B”). Dashed lines mark the edges of spermathecal cells. Scale bars: whole spermatheca, 10 µm; enlargement, 2 µm. Full movie in Video 7. (C) Quantification of both RHGF-1::tagRFP and SPV-1::GFP recruitment to the cell edge during ovulation. The x-axis (time) was normalized according to distinct events so that all traces begin at the time when the distal valve closes behind the ovulating oocyte (“Oocyte entry complete”) are aligned at the time the sp-ut starts to open (“Embryo exit initiation”) and aligned again when the sp-ut valve closes behind the exiting embryo (“Embryo exit complete”). N = 3. (D) Representative confocal images of RHGF-1 subcellular localization and RHO-1 biosensor, in the same spermatheca, during oocyte ovulation and embryo transit. White boxes mark regions enlarged below the images. Dashed lines mark the edges of spermathecal cells. Scale bars: whole spermatheca, 20 µm; enlargement, 2 µm. Full movie in Video 8. (E) Quantification of RHGF-1::tagRFP recruitment to the cell edge, and RHO-1 biosensor intensity during ovulation and embryo exit. The x-axis (time) was normalized as in C. N = 6. N, number of samples analyzed.
Examination of RHGF-1 subcellular localization within the spermatheca using confocal microscopy revealed a dynamic localization that correlated with the ovulation cycle. RHGF-1 was cytoplasmic in unoccupied spermathecae. However, following oocyte entry and the stretching of the spermatheca, RHGF-1 became increasingly enriched near the basal membrane of spermathecal cells, as marked by β-Integrin/PAT-3::GFP (Fig. S3 D and Video 6). Previously, we showed that SPV-1 behaves in the exact opposite way, localizing to the apical membrane in the unoccupied spermatheca and becoming increasingly cytoplasmic after oocyte entry (Tan and Zaidel-Bar, 2015). To compare the localization dynamics of the RhoGEF and RhoGAP within the same cells over time, we crossed the RHGF-1::tagRFP strain to a strain expressing SPV-1::GFP in the spv-1(ok1498) background and made two-color movies of embryo transits (Fig. 3 B and Video 7). Fluorescence intensity profiles, generated by drawing lines across the cells from the basal to apical side, illustrate their inverse localizations. Following oocyte entry, RHGF-1 was completely cytoplasmic while SPV-1 was enriched at the apical membrane. By the time contraction was initiated, SPV-1 was mostly cytoplasmic and RHGF-1 was primarily at the basal side. To quantify these localization dynamics, we measured the fluorescence intensity of each protein separately at the corresponding cell edge and in the cytoplasm (Fig. S3 E) and calculated their edge/cytoplasm ratios (Fig. 3 C). This quantification showed that while SPV-1 detached from the apical membrane immediately after oocyte entry, RHGF-1 was slowly recruited to the basal side shortly before initiation of embryo exit and its basal enrichment was further enhanced during spermatheca contraction.
Time-lapse movie of a spermatheca expressing RHGF-1::tagRFP and β-Integrin/PAT-3::GFP during ovulation and embryo transit. Time interval between frames is 10 s. Frame rate is 7 fps. Related to Fig. S3 D. Scale bar, 10 μm.
Time-lapse movie of a spermatheca expressing RHGF-1::tagRFP and β-Integrin/PAT-3::GFP during ovulation and embryo transit. Time interval between frames is 10 s. Frame rate is 7 fps. Related to Fig. S3 D. Scale bar, 10 μm.
Time-lapse confocal imaging of spermatheca expressing both RHGF-1::tagRFP and SPV-1::GFP from ovulation to the completion of embryo exit. Time interval between frames is 10 s. Frame rate is 3 fps. Related to Fig. 3 B.
Time-lapse confocal imaging of spermatheca expressing both RHGF-1::tagRFP and SPV-1::GFP from ovulation to the completion of embryo exit. Time interval between frames is 10 s. Frame rate is 3 fps. Related to Fig. 3 B.
To investigate the relationship between RHGF-1 recruitment to the basal side and RHO-1 activation, we crossed the RHO-1 biosensor with the RHGF-1::tagRFP strain and imaged the spermatheca during embryo transit (Fig. 3 D and Video 8). We quantified the recruitment of RHGF-1 to the basal side, as before, and measured the RHO-1 biosensor fluorescence intensity in multiple spermathecae (Fig. 3 E). RHGF-1 recruitment to the basal side and the increase in RHO-1 activity parallel each other, beginning at roughly the same time and proceeding with similar dynamics, consistent with the possibility that they reinforce each other with positive feedback (Fig. 3 E).
Time-lapse confocal imaging of RHGF-1::tagRFP and RHO-1 biosensor, in the same spermatheca, during oocyte ovulation and embryo transit. Time interval between frames is 10 s. Frame rate is 3 fps. Related to Fig. 3 D.
Time-lapse confocal imaging of RHGF-1::tagRFP and RHO-1 biosensor, in the same spermatheca, during oocyte ovulation and embryo transit. Time interval between frames is 10 s. Frame rate is 3 fps. Related to Fig. 3 D.
The PDZ domain of RHGF-1 is responsible for its basal recruitment
To test whether basal recruitment is essential for RHGF-1 function, we first needed to determine which part of the protein is responsible for this localization. RHGF-1 has five protein domains: PDZ, regulator of G protein signaling (RGS), C1, RhoGEF, and pleckstrin homology (PH; Fig. 4 A). To determine which of these domains, if any, is required to drive recruitment of RHGF-1 to the basal side, we generated a plasmid with full-length RHGF-1 cDNA tagged with mKate and a series of derived plasmids with deletions of each domain. Each plasmid was injected into the rhgf-1(ok880) background. We measured the fluorescence intensity of each construct and established that on average they were all expressed at comparable levels (Fig. S4 A). Furthermore, plotting embryo dwell time as a function of fluorescence intensity of all constructs showed that there is no correlation between the two (Fig. S4 B), arguing that any difference in function between the various constructs would be due to their missing domains and not their expression level. Next, we observed the subcellular localization pattern of each construct during ovulation transits (Video 9). Full-length RHGF-1::mKate displayed the same localization dynamics as observed in the RHGF-1::tagRFP strain, starting out as cytoplasmic and then being recruited to the basal side prior to spermatheca contraction (Fig. 4 B). The deletion of the PH, RhoGEF, C1, or RGS domain did not affect the recruitment of RHGF-1 to the basal side (Fig. 4, C–F). Uniquely, deletion of the PDZ domain resulted in a failure of RHGF-1 to be recruited basally and it remained cytoplasmic during the entire ovulation process (Fig. 4 G).
The PDZ domain of RHGF-1 is required and sufficient for its basal recruitment. (A) Schematic presentation of RHGF-1 domains. (B–G) Confocal images showing the subcellular localization of mKate-fused full length RHGF-1 (B), PH domain deletion (C), RhoGEF domain deletion (D), C1 domain deletion (E), RGS domain deletion (F), and PDZ domain deletion (G), during spermatheca contraction. (H and I) Confocal images showing the subcellular localization of mKate-fused PDZ domain, when oocyte entry was completed and during embryo exit, respectively. Region in yellow box enlarged on right. Graphs show the fluorescence intensity profile along the yellow line from the apical side of the cell (“A”) and the basal side (“B”). Arrows point at cytoplasmic peaks and arrowheads point basal side peaks. Scale bars: whole spermatheca, 20 µm; enlargement, 2 µm. Full movies are in Videos 9 and 10.
The PDZ domain of RHGF-1 is required and sufficient for its basal recruitment. (A) Schematic presentation of RHGF-1 domains. (B–G) Confocal images showing the subcellular localization of mKate-fused full length RHGF-1 (B), PH domain deletion (C), RhoGEF domain deletion (D), C1 domain deletion (E), RGS domain deletion (F), and PDZ domain deletion (G), during spermatheca contraction. (H and I) Confocal images showing the subcellular localization of mKate-fused PDZ domain, when oocyte entry was completed and during embryo exit, respectively. Region in yellow box enlarged on right. Graphs show the fluorescence intensity profile along the yellow line from the apical side of the cell (“A”) and the basal side (“B”). Arrows point at cytoplasmic peaks and arrowheads point basal side peaks. Scale bars: whole spermatheca, 20 µm; enlargement, 2 µm. Full movies are in Videos 9 and 10.
Analysis of expression level and dwell time in all RHGF-1 deletion constructs. (A) A comparison of the fluorescence intensity of individual spermathecae in rhgf-1(ok880) worms expressing full-length or various deletion constructs of RHGF-1 fused to mKate. N ≥ 7. Bars are mean ± SEM. Statistical comparisons were performed by One-way ANOVA, P = 0.86 (non-significant). (B) A plot of dwell time as a function of the fluorescence intensity of the RHGF-1 construct expressed. Data includes all the constructs shown in A. Line is simple linear regression. A non-significant negative correlation (r = −0.05) was calculated by correlation assay. N = 43. N, number of samples analyzed.
Analysis of expression level and dwell time in all RHGF-1 deletion constructs. (A) A comparison of the fluorescence intensity of individual spermathecae in rhgf-1(ok880) worms expressing full-length or various deletion constructs of RHGF-1 fused to mKate. N ≥ 7. Bars are mean ± SEM. Statistical comparisons were performed by One-way ANOVA, P = 0.86 (non-significant). (B) A plot of dwell time as a function of the fluorescence intensity of the RHGF-1 construct expressed. Data includes all the constructs shown in A. Line is simple linear regression. A non-significant negative correlation (r = −0.05) was calculated by correlation assay. N = 43. N, number of samples analyzed.
Time-lapse confocal imaging showing the subcellular localization of mKate-fused full length RHGF-1, PH domain deletion, RhoGEF domain deletion, C1 domain deletion, RGS domain deletion, and PDZ domain deletion, from oocyte entry until embryo exit. Time interval between frames is 5 s. Frame rate is 7 fps. Related to Fig. 4, B–G.
Time-lapse confocal imaging showing the subcellular localization of mKate-fused full length RHGF-1, PH domain deletion, RhoGEF domain deletion, C1 domain deletion, RGS domain deletion, and PDZ domain deletion, from oocyte entry until embryo exit. Time interval between frames is 5 s. Frame rate is 7 fps. Related to Fig. 4, B–G.
Then, we tested whether the PDZ domain of RHGF-1 on its own could recapitulate the localization dynamics of full-length RHGF-1 by fusing mKate to the first 240 amino acids of RHGF-1, containing the PDZ domain, and expressing this construct in the rhgf-1(ok880) background. Consistent with the deletion results, the PDZ domain–containing fragment was cytoplasmic upon oocyte entry but was then recruited to the basal side, becoming highly enriched at the basal side by the time the embryo exited (Fig. 4, H and I; and Video 10). Thus, our findings demonstrate that the PDZ domain of RHGF-1 is required for its recruitment to the basal side.
Time-lapse confocal imaging showing the subcellular localization of mKate-fused PDZ domain, from oocyte entry until embryo exit. Time interval between frames is 5 s. Frame rate is 3 fps. Related to Fig. 4, H and I.
Time-lapse confocal imaging showing the subcellular localization of mKate-fused PDZ domain, from oocyte entry until embryo exit. Time interval between frames is 5 s. Frame rate is 3 fps. Related to Fig. 4, H and I.
In a sensitized spv-1(RNAi) background, RHGF-1 lacking its PDZ domain behaves like a RHGF-1 null
Identifying the PDZ domain as required for basal recruitment allowed us to test the hypothesis that basal recruitment of RHGF-1 plays a role in RHO-1 activation. First, we tested the ability of RHGF-1-ΔPDZ to rescue the long oocyte dwell time and 25% trapping phenotypes of the rhgf-1 mutant. To our surprise, expression of RHGF-1-ΔPDZ was able to shorten oocyte dwell time such that its mean dwell time was not significantly different from wild type, and it facilitated 100% successful embryo transits, although the rescue of dwell time was not as robust as full-length RHGF-1 (Fig. 5, A and B). This result suggests that under normal conditions, the basal localization of RHGF-1 is not required for it to activate RHO-1, although basal localization is necessary for its full activity. Importantly, it also indicates that the RhoGEF domain is still functional in RHGF-1-ΔPDZ.
In spv-1(RNAi) conditions, RHGF-1 without its PDZ domain behaves like a complete loss of function. (A and B) Dwell time and successful transits of wild type, rhgf-1(ok880), and rhgf-1(ok880) expressing full-length RHGF-1 or RHGF-1 lacking its PDZ domain (ΔPDZ). N ≥ 8. Error bars in A are ± SEM. Statistical comparisons for A were performed by Brown-Forsythe ANOVA-Dunnett’s T3 multiple comparisons test. Statistical comparisons for B were performed by individuals’ chi-square tests. (C) Representative stitched DIC images of embryos from rhgf-1(ok880) hermaphrodites expressing full-length RHGF-1 or RHGF-1 lacking its PDZ domain (ΔPDZ) treated with control (empty vector) or spv-1(RNAi). Black arrows point to abnormally shaped embryos. Scale bar, 50 µm. (D) Length-to-width ratio of embryos from rhgf-1(ok880) worms expressing full-length RHGF-1 or ΔPDZ treated with control or spv-1(RNAi).N ≥ 19. Box plot from min to max showing the difference in deviation. Statistical comparisons were performed by Brown-Forsythe ANOVA-Dunnett’s T3 multiple comparisons test. (E) Embryonic lethality of the progeny of rhgf-1(ok880) expressing full-length RHGF-1 or ΔPDZ treated with control or spv-1(RNAi). Nrepeats ≥ 4, Nmothers ≥ 120, Nembryos ≥740. Error bars are ± SEM. Statistical comparisons were performed by Brown-Forsythe ANOVA-Dunnett’s T3 multiple comparisons test. (F) Dwell time in rhgf-1(ok880) spermathecae expressing full-length RHGF-1 or ΔPDZ treated with spv-1(RNAi). N = 26. Error bars are mean ± SEM. Statistical comparison was performed by two-tailed unpaired t test. (G) Successful embryos transit through the spermathecae of rhgf-1(ok880) expressing full-length RHGF-1 or ΔPDZ and treated with spv-1(RNAi). N ≥ 40. Statistical comparison was performed by chi-square tests. Stars designate statistical significance (**** P < 0.0001, ** P < 0.01, * P < 0.05, ns P > 0.05). N, number of samples analyzed.
In spv-1(RNAi) conditions, RHGF-1 without its PDZ domain behaves like a complete loss of function. (A and B) Dwell time and successful transits of wild type, rhgf-1(ok880), and rhgf-1(ok880) expressing full-length RHGF-1 or RHGF-1 lacking its PDZ domain (ΔPDZ). N ≥ 8. Error bars in A are ± SEM. Statistical comparisons for A were performed by Brown-Forsythe ANOVA-Dunnett’s T3 multiple comparisons test. Statistical comparisons for B were performed by individuals’ chi-square tests. (C) Representative stitched DIC images of embryos from rhgf-1(ok880) hermaphrodites expressing full-length RHGF-1 or RHGF-1 lacking its PDZ domain (ΔPDZ) treated with control (empty vector) or spv-1(RNAi). Black arrows point to abnormally shaped embryos. Scale bar, 50 µm. (D) Length-to-width ratio of embryos from rhgf-1(ok880) worms expressing full-length RHGF-1 or ΔPDZ treated with control or spv-1(RNAi).N ≥ 19. Box plot from min to max showing the difference in deviation. Statistical comparisons were performed by Brown-Forsythe ANOVA-Dunnett’s T3 multiple comparisons test. (E) Embryonic lethality of the progeny of rhgf-1(ok880) expressing full-length RHGF-1 or ΔPDZ treated with control or spv-1(RNAi). Nrepeats ≥ 4, Nmothers ≥ 120, Nembryos ≥740. Error bars are ± SEM. Statistical comparisons were performed by Brown-Forsythe ANOVA-Dunnett’s T3 multiple comparisons test. (F) Dwell time in rhgf-1(ok880) spermathecae expressing full-length RHGF-1 or ΔPDZ treated with spv-1(RNAi). N = 26. Error bars are mean ± SEM. Statistical comparison was performed by two-tailed unpaired t test. (G) Successful embryos transit through the spermathecae of rhgf-1(ok880) expressing full-length RHGF-1 or ΔPDZ and treated with spv-1(RNAi). N ≥ 40. Statistical comparison was performed by chi-square tests. Stars designate statistical significance (**** P < 0.0001, ** P < 0.01, * P < 0.05, ns P > 0.05). N, number of samples analyzed.
Next, we tested how RHGF-1-ΔPDZ performs when RHO-1 activity is not downregulated by its RhoGAP, by repeating the rescue experiments along with spv-1(RNAi). We quantified four parameters: embryo shape defects, embryonic lethality, dwell time, and percentage of successful transits. Knockdown of spv-1 led to multiple embryos with shape defects in rhgf-1(ok880) worms re-expressing full-length RHGF-1. However, the progeny of rhgf-1(ok880) worms re-expressing RHGF-1-ΔPDZ did not show any shape defect (Fig. 5 C), reminiscent of the cross between spv-1(ok1498) and rhgf-1(ok880). We quantified embryo shape by measuring the ratio between embryo length and width. Depletion of spv-1 led to the appearance of significantly longer embryos in the progeny of rhgf-1(ok880) worms re-expressing full-length RHGF-1, but did not affect the length of embryos of the rhgf-1(ok880) worms re-expressing RHGF-1-ΔPDZ (Fig. 5 D). Note that spv-1(RNAi) was not as severe as the spv-1(ok1498) mutant and did not result in much pinching (round embryos) but rather excessive squeezing (elongated embryos). Abnormal embryo shape leads to embryonic lethality. Accordingly, spv-1(RNAi) in rhgf-1(ok880) worms re-expressing full-length RHGF-1 led to 47 ± 3% embryonic lethality, whilst in the rhgf-1(ok880) worms re-expressing RHGF-1-ΔPDZ it only led to 9 ± 1% embryonic lethality (Fig. 5 E). Similarly, under spv-1(RNAi) conditions, oocyte dwell time was significantly shorter in rhgf-1(ok880) worms re-expressing full-length RHGF-1 compared with rhgf-1(ok880) worms re-expressing RHGF-1-ΔPDZ (Fig. 5 F) and only 64% of embryo transits were successful in the full-length re-expressing worms compared with 93% success rate in the RHGF-1-ΔPDZ re-expressing worms (Fig. 5 G). In all these measures, RHGF-1-ΔPDZ expressing worms behaved similarly to the rhgf-1(ok880) worms, suggesting that RHGF-1-ΔPDZ functions poorly in activating RHO-1.
RHGF-1 localizes along actomyosin stress fibers
After establishing that PDZ domain–mediated basal recruitment of RHGF-1 is required for its full activity, we sought to determine the exact location of recruitment within spermathecal cells. One possibility we considered is the basal plasma membrane. However, deletion of the membrane-binding PH domain did not affect the basal recruitment of RHGF-1 (Fig. 4 C).
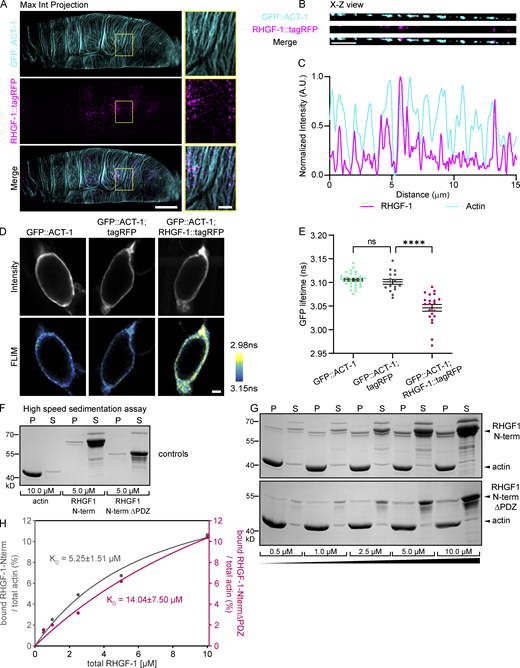
Next, we considered whether RHGF-1 is recruited to the actomyosin bundles present along the basal surface of the spermatheca. We crossed endogenous RHGF-1::tagRFP with a spermatheca-specific actin reporter (GFP::ACT-1) and imaged their relative localizations in occupied spermathecae by live super-resolution confocal microscopy (Fig. 6 A). RHGF-1 appeared in periodic punctate structures along the F-actin bundles in maximum intensity projection views. As illustrated in an X-Z view and its corresponding intensity line profile, RHGF-1 appeared either on top of or adjoining the actin bundles, while in regions without F-actin, RHGF-1 punctae were mostly absent (Fig. 6, B and C). Quantification of the degree of co-localization at the spermatheca surface showed that on average 69 ± 4% (N = 5) of RHGF-1::tagRFP punctae overlapped with GFP::ACT-1 signal.
RHGF-1 localizes along F-actin in basal stress fibers and binds F-actin in vitro. (A) Maximum intensity projection of live super-resolution confocal images of RHGF-1::tagRFP and GFP::ACT-1 in an occupied spermatheca. Region in yellow box enlarged on right. Scale bars: whole spermatheca, 10 µm; enlargement, 2 µm. (B) X-Z view at the surface of the spermatheca. Scale bar, 2 µm. (C) Intensity line profile based on 5-pixel line along X-Z view in B. (D) Representative two-photon images of fluorescence intensity and pseudo-colored FLIM of GFP::ACT-1 alone and co-expressed with cytoplasmic tagRFP or RHGF-1::tagRFP, which acts as acceptor. Scale bar, 20 µm. (E) Comparison of the fluorescence lifetime of GFP::ACT-1 alone and GFP::ACT-1 with cytoplasmic tagRFP or RHGF-1::tagRFP. N ≥ 16. Error bars are mean ± SEM. Statistical comparisons were performed by Brown-Forsythe ANOVA-Dunnett’s T3 multiple comparisons test. Stars designate statistical significance (**** P < 0.0001, ns P > 0.05). N, number of samples analyzed. (F–H) High-speed sedimentation assays demonstrate direct F-actin binding of the N-terminal half of RHGF-1 (RHGF-1-N), weakened by loss of the PDZ domain. (F) In control experiments 10 µM G-actin and 5 µM of either RHGF-1-N or RHGF-1-N-ΔPDZ were incubated alone in polymerization buffer, and the proteins in the pellet (P) and supernatant (S) fractions recovered after centrifugation at 200,000 × g were stained with Coomassie blue to assess spontaneous precipitation of the RHGF-1 fragments in the absence of actin. (G) Co-sedimentation of RHGF-1-N and RHGF-1-N-ΔPDZ with F-actin in high-speed pelleting assays. Increasing concentrations of RHGF-1-N and RHGF-1-N-ΔPDZ as indicated were incubated with 10 µM G-actin in polymerization buffer, and the proteins recovered in the pellet (P) and the supernatant (S) fractions after the centrifugation at 200,000 × g were analyzed as above. (H) Quantification of equilibrium constants of RHGF-1-N and RHGF-1-N-ΔPDZ with F-actin from experiments as shown in G. Solid pink and gray lines represent calculated binding isotherms. Shown values depict calculated KD values and respective standard errors of the fit. Source data are available for this figure: SourceData F6.
RHGF-1 localizes along F-actin in basal stress fibers and binds F-actin in vitro. (A) Maximum intensity projection of live super-resolution confocal images of RHGF-1::tagRFP and GFP::ACT-1 in an occupied spermatheca. Region in yellow box enlarged on right. Scale bars: whole spermatheca, 10 µm; enlargement, 2 µm. (B) X-Z view at the surface of the spermatheca. Scale bar, 2 µm. (C) Intensity line profile based on 5-pixel line along X-Z view in B. (D) Representative two-photon images of fluorescence intensity and pseudo-colored FLIM of GFP::ACT-1 alone and co-expressed with cytoplasmic tagRFP or RHGF-1::tagRFP, which acts as acceptor. Scale bar, 20 µm. (E) Comparison of the fluorescence lifetime of GFP::ACT-1 alone and GFP::ACT-1 with cytoplasmic tagRFP or RHGF-1::tagRFP. N ≥ 16. Error bars are mean ± SEM. Statistical comparisons were performed by Brown-Forsythe ANOVA-Dunnett’s T3 multiple comparisons test. Stars designate statistical significance (**** P < 0.0001, ns P > 0.05). N, number of samples analyzed. (F–H) High-speed sedimentation assays demonstrate direct F-actin binding of the N-terminal half of RHGF-1 (RHGF-1-N), weakened by loss of the PDZ domain. (F) In control experiments 10 µM G-actin and 5 µM of either RHGF-1-N or RHGF-1-N-ΔPDZ were incubated alone in polymerization buffer, and the proteins in the pellet (P) and supernatant (S) fractions recovered after centrifugation at 200,000 × g were stained with Coomassie blue to assess spontaneous precipitation of the RHGF-1 fragments in the absence of actin. (G) Co-sedimentation of RHGF-1-N and RHGF-1-N-ΔPDZ with F-actin in high-speed pelleting assays. Increasing concentrations of RHGF-1-N and RHGF-1-N-ΔPDZ as indicated were incubated with 10 µM G-actin in polymerization buffer, and the proteins recovered in the pellet (P) and the supernatant (S) fractions after the centrifugation at 200,000 × g were analyzed as above. (H) Quantification of equilibrium constants of RHGF-1-N and RHGF-1-N-ΔPDZ with F-actin from experiments as shown in G. Solid pink and gray lines represent calculated binding isotherms. Shown values depict calculated KD values and respective standard errors of the fit. Source data are available for this figure: SourceData F6.
In addition to F-actin, the non-muscle myosin II NMY-1 is a major component of spermathecal stress fibers (Wirshing and Cram, 2017). Therefore, we checked whether RHGF-1 might be recruited to the stress fibers through an interaction with NMY-1. We crossed endogenous RHGF-1::tagRFP with a strain expressing GFP::NMY-1 under a spermatheca-specific promoter, and imaged their relative localizations in occupied spermathecae by live super-resolution confocal microscopy (Fig. S5 A). While they co-localized in some places along the stress fiber, we observed that in some cases RHGF-1 punctae localized independently of NMY-1 (Fig. 5, B and C). To further test the dependence of RHGF-1 stress fiber localization on NMY-1, we repeated the analysis after depletion of the actin cross-linker filamin by RNAi. FLN-1 depletion leads to a separation between the F-actin and NMY-1 networks (Kelley et al., 2020b). Under fln-1(RNAi) conditions, RHGF-1 punctae were aligned in a pattern that appeared separate from the NMY-1 fibers (Fig. S5 D). Quantification of the degree of their co-localization at the spermatheca surface under this condition showed that on average 22 ± 5% (N = 5) of RHGF-1::tagRFP overlapped with GFP::NMY-1 signal, suggesting that RHGF-1 remained associated with F-actin, independently of NMY-1. Thus, it appears that RHGF-1 localizes along stress fibers and this association does not depend on an interaction with myosin II.
Relative subcellular localization of RHGF-1 and NMY-1. (A) Maximum intensity projection of live super-resolution confocal images of RHGF-1::tagRFP and GFP::NMY-1 in an occupied spermatheca. Region in yellow box is enlarged on right. Scale bars: whole spermatheca, 10 µm; enlargement, 2 µm. (B) X-Z view at the surface of the spermatheca. Scale bar, 2 µm. (C) Intensity line profile based on 5-pixel line along X-Z view in B. (D) Maximum intensity projection made from a live super-resolution Z-stack of an occupied spermatheca in a worm expressing endogenous RHGF-1::tagRFP and GFP::NMY-1 under a spermatheca-specific promoter, following fln-1(RNAi). Region in yellow box is enlarged on right. Scale bars: whole spermatheca, 10 µm; enlargement, 2 µm.
Relative subcellular localization of RHGF-1 and NMY-1. (A) Maximum intensity projection of live super-resolution confocal images of RHGF-1::tagRFP and GFP::NMY-1 in an occupied spermatheca. Region in yellow box is enlarged on right. Scale bars: whole spermatheca, 10 µm; enlargement, 2 µm. (B) X-Z view at the surface of the spermatheca. Scale bar, 2 µm. (C) Intensity line profile based on 5-pixel line along X-Z view in B. (D) Maximum intensity projection made from a live super-resolution Z-stack of an occupied spermatheca in a worm expressing endogenous RHGF-1::tagRFP and GFP::NMY-1 under a spermatheca-specific promoter, following fln-1(RNAi). Region in yellow box is enlarged on right. Scale bars: whole spermatheca, 10 µm; enlargement, 2 µm.
RHGF-1 is closely associated with actin in vivo and binds F-actin directly in vitro
To further explore the interaction between RHGF-1 and F-actin within the spermatheca, we employed Förster resonance energy transfer (FRET) and fluorescence lifetime imaging microscopy (FLIM; Yasuda, 2006). Using GFP::ACT-1 as the donor and RHGF-1::tagRFP as the acceptor, a change in the fluorescence lifetime of GFP is expected only if the two fluorescent proteins are at a distance of 10 nm or less, which is the minimal distance for energy transfer to take place (Fig. 6 D). We measured fluorescence lifetime of GFP::ACT-1 in occupied spermathecae, when RHGF-1 is localized to stress fibers. As a control, we also measured the lifetime of GFP::ACT-1 in spermathecae expressing cytoplasmic tagRFP. Compared with a fluorescence lifetime of 3.106 ± 0.003 ns in GFP::ACT-1 alone, we detected a significant decrease in fluorescence lifetime of GFP::ACT-1 to 3.046 ± 0.007 ns in the presence of RHGF-1::tagRFP but not in the presence of cytoplasmic tagRFP (3.101 ± 0.005 ns; Fig. 6 E). Of note, the distribution of GFP lifetime values was wider in the presence of RHGF-1::tagRFP, which may be due to the fact that images were collected at different times after oocyte entry and therefore different amounts of RHGF-1::tagRFP were recruited to F-actin.
To test whether RHGF-1 can bind F-actin directly, we performed an in vitro F-actin co-sedimentation assay using purified recombinant proteins. We were not able to express full-length RHGF-1 in bacteria at large scale, but we were able to express and purify the N-terminus half of RHGF-1 (RHGF-1-N), which contains the PDZ, RGS, and C1 domains. A second recombinant protein we cloned and purified was the N-terminal half of RHGF-1 without the PDZ domain (RHGF-1-N-ΔPDZ). We first performed high-speed sedimentation of each recombinant protein on its own to assess spontaneous precipitation of the RHGF-1 fragments in the absence of actin (Fig. 6 F). Next, we performed high-speed F-actin co-sedimentation with increasing concentrations of RHGF-1-N and RHGF-1-N-ΔPDZ (Fig. 6 G). Quantification of equilibrium constants of RHGF-1-N and RHGF-1-N-ΔPDZ with F-actin from these experiments showed that RHGF-1-N directly interacts with F-actin, albeit weakly (KD = 5.25 ± 1.51 μM), and this interaction is further weakened by deletion of the PDZ domain (KD = 14.04 ± 7.50 μM; Fig. 6 H). Taken together, these data suggest that RHGF-1 can bind directly to F-actin and is in close association with actin in vivo.
RHGF-1 localization to F-actin is increased under tension
We found that RHGF-1 is recruited to F-actin in stress fibers, but only after oocyte entry into the spermatheca, a process that stretches the tissue, and even more so once the stress fibers begin to contract. This led us to hypothesize that tension across the F-actin bundles plays a role in RHGF-1 recruitment. To test this experimentally, we genetically perturbed the actomyosin cytoskeleton to either increase or decrease its contractility and monitored the localization of endogenous RHGF-1::tagRFP under these conditions (Fig. 7 A). We increased the tension in the actin structures by depleting the myosin phosphatase/mel-11 with RNAi and we decreased tension by depleting rho-1, filamin/fln-1, or non-muscle myosin II/nmy-1 by RNAi. We measured actin tortuosity in each of these cases and found the value for mel-11 RNAi to be essentially the same as that of control RNAi, suggesting that stress fibers in normal conditions are already as straight as they can be and therefore an increase in tension would not lead to a decrease in tortuosity. As expected, rho-1, fln-1, and nmy-1 RNAi all resulted in a significant increase in tortuosity of stress fibers (Fig. 7 B). For each of these RNAi depletions, we also measured the ratio of RHGF-1 intensity at the basal side, where F-actin is localized, to that in the cytoplasm at the middle plane of the spermatheca. In the case of mel-11(RNAi), this ratio was 1.71 ± 0.10, similar to control RNAi (1.68 ± 0.10; Fig. 7 C). Thus, RHGF-1 was efficiently recruited to stress fibers upon depletion of mel-11, though it appeared to be concentrated in certain regions overlapping with regions of high actin intensity. In the case of rho-1(RNAi), fln-1(RNAi) and nmy-1(RNAi), we observed very low RHGF-1 fluorescence along the basal side of the cell, despite the presence of large amounts of F-actin. This was reflected in the calculated basal side to cytoplasm intensity ratios [1.08 ± 0.04 for rho-1(RNAi) 1.04 ± 0.02 for fln-1(RNAi) and 1.09 ± 0.03 for nmy-1(RNAi)] that were close to one, demonstrating that RHGF-1 was not recruited to F-actin in the absence of tension. Plotting all the data on one graph, revealed a significant negative correlation between F-actin tortuosity and RHGF-1 recruitment to F-actin, with an R of −0.76 (Fig. 7 D). Thus, the localization of RHGF-1 to stress fibers is reinforced by myosin-induced contractility.
Recruitment of RHGF-1 to F-actin is enhanced by myosin contractility. (A) Maximum intensity projections and middle plane images of GFP::ACT-1 and RHGF-1::tagRFP in spermathecae from control (empty vector), mel-11(RNAi), rho-1(RNAi), fln-1(RNAi), and nmy-1(RNAi) treated worms. Scale bar, 10 µm. (B) Quantification of F-actin bundle tortuosity in each of the conditions shown in A. (C) Quantification of the ratio of RHGF-1 intensity between the basal side of the cell and in the cytoplasm in each of the conditions shown in A. For both B and C: N ≥ 10. Error bars are mean ± SEM. Statistical comparisons were performed by Brown-Forsythe ANOVA-Dunnett’s T3 multiple comparisons test. Stars designate statistical significance (**** P <0.0001, *** P <0.001). (D) Tortuosity as a function of RHGF-1 enrichment at the basal side. Black line is an interpolation using the “Sigmoidal, 4PL, X is log(concentration)” function in GraphPad. Statistical assay was correlation, r = −0.76, P <0.0001. N = 50. N, number of samples analyzed. (E) Schematic model of the mechanosensitive regulation of RHO-1 in the spermatheca, combining the results on RHGF-1 from this study with our previous work on SPV-1 (Tan and Zaidel-Bar, 2015). In the empty spermatheca (left), RHGF-1 is mostly inactive in the cytoplasm, and RHO-1 is mostly inactive due to the GAP activity of SPV-1, which localizes to the apical membrane. Following ovulation, the spermatheca is filled with an oocyte (top) and its membranes and F-actin become stretched. Membrane stretching causes SPV-1 to become cytoplasmic and inactive, while F-actin stretching leads to recruitment of RHGF-1, which activates RHO-1. After fertilization (right), the level of active RHO-1 continues to increase due to a positive feedback loop involving tension-dependent RHGF-1 recruitment, RHO-1 activation, and RHO-1–induced myosin contractility. Finally, when the level of RHO-1 activation and actomyosin contractility reach the level required to squeeze the embryo out of the spermatheca (bottom), emptying of the spermatheca occurs. The loose spermatheca apical membrane folds and recruits SPV-1, which inactives RHO-1, therefore actin becomes under less tension, RHGF-1 detaches, and the cycle is completed.
Recruitment of RHGF-1 to F-actin is enhanced by myosin contractility. (A) Maximum intensity projections and middle plane images of GFP::ACT-1 and RHGF-1::tagRFP in spermathecae from control (empty vector), mel-11(RNAi), rho-1(RNAi), fln-1(RNAi), and nmy-1(RNAi) treated worms. Scale bar, 10 µm. (B) Quantification of F-actin bundle tortuosity in each of the conditions shown in A. (C) Quantification of the ratio of RHGF-1 intensity between the basal side of the cell and in the cytoplasm in each of the conditions shown in A. For both B and C: N ≥ 10. Error bars are mean ± SEM. Statistical comparisons were performed by Brown-Forsythe ANOVA-Dunnett’s T3 multiple comparisons test. Stars designate statistical significance (**** P <0.0001, *** P <0.001). (D) Tortuosity as a function of RHGF-1 enrichment at the basal side. Black line is an interpolation using the “Sigmoidal, 4PL, X is log(concentration)” function in GraphPad. Statistical assay was correlation, r = −0.76, P <0.0001. N = 50. N, number of samples analyzed. (E) Schematic model of the mechanosensitive regulation of RHO-1 in the spermatheca, combining the results on RHGF-1 from this study with our previous work on SPV-1 (Tan and Zaidel-Bar, 2015). In the empty spermatheca (left), RHGF-1 is mostly inactive in the cytoplasm, and RHO-1 is mostly inactive due to the GAP activity of SPV-1, which localizes to the apical membrane. Following ovulation, the spermatheca is filled with an oocyte (top) and its membranes and F-actin become stretched. Membrane stretching causes SPV-1 to become cytoplasmic and inactive, while F-actin stretching leads to recruitment of RHGF-1, which activates RHO-1. After fertilization (right), the level of active RHO-1 continues to increase due to a positive feedback loop involving tension-dependent RHGF-1 recruitment, RHO-1 activation, and RHO-1–induced myosin contractility. Finally, when the level of RHO-1 activation and actomyosin contractility reach the level required to squeeze the embryo out of the spermatheca (bottom), emptying of the spermatheca occurs. The loose spermatheca apical membrane folds and recruits SPV-1, which inactives RHO-1, therefore actin becomes under less tension, RHGF-1 detaches, and the cycle is completed.
Discussion
Understanding how Rho GTPases are spatiotemporally regulated in order to execute specific morphodynamic processes is an ongoing challenge (Hodge and Ridley, 2016; McCormack et al., 2013; Pertz, 2010; Xu et al., 2019; Zegers and Friedl, 2014). Of particular interest, in the case of spermathecal physiology, is the mechanism of timing RHO-1 activation so that it follows eggshell formation. Our previous work identified spermathecal stretching by an incoming oocyte as a mechanical signal that triggered removal from the membrane and inactivation of the RhoGAP SPV-1 (Tan and Zaidel-Bar, 2015). Here we found the RhoGEF RHGF-1 to behave like the mirror image of SPV-1 both in time and space. Spermathecal stretching during ovulation triggered the recruitment of RHGF-1 to basal actin stress fibers and thereby facilitated the activation of RHO-1. Under normal conditions, recruitment of RHGF-1 to stress fibers is not required for it to activate RHO-1, since we could partially rescue the rhfg-1 null allele with RHGF-1-ΔPDZ, which is cytoplasmic. However, in the background of spv-1(RNAi) RHGF-1-ΔPDZ behaves like a null, revealing that when it is not recruited to F-actin its activity is significantly reduced. RHO-1 activates LET-502/Rho Kinase, which in turn leads to the activation of NMY-1, and NMY-1–mediated contraction of stress fibers serves to recruit more RHGF-1, thus forming a positive feedback loop. We propose that the time required for active RHO-1 levels to rise and pass a threshold needed to induce spermathecal contraction, constitutes the dwell time necessary for eggshell formation and thus safe squeezing of the fertilized egg into the uterus. According to our model, mechanical stretching of the spermatheca is sensed by two independent mechanotransduction pathways: one, mediated by membrane tension, serves to switch off the RhoGAP SPV-1, and the second, mediated by actin tension, serves to augment the RhoGEF activity of RHGF-1 (Fig. 7 E).
In C. elegans, RHGF-1 has so far only been studied in neurons, where it was shown to stimulate neurotransmitter release at synapses (Hiley et al., 2006), help guide axon migration and remodeling through inhibition of protrusions and microtubules (Chen et al., 2014; Gujar et al., 2019), and play a role in neuronal regeneration downstream of HIF-1 and serotonin (Alam et al., 2016). The subcellular localization of RHGF-1 in neurons was not examined and therefore cannot be compared to our observation in the spermatheca.
The mammalian orthologs of RHGF-1, ARHGEF11 (a.k.a. PDZ-RhoGEF), and ARHGEF12 (a.k.a. LARG) activate RhoA in a variety of cellular and developmental processes, including cyclic-stretch-induced reorientation of endothelial cells (Abiko et al., 2015), invagination of the lens and otic placodes (Houssin et al., 2020; Sai et al., 2014), contraction of colonic and vascular smooth muscle (Al-Shboul et al., 2014; Ying et al., 2006), and stiffening of endothelial cells to enhance leukocyte transendothelial migration (Lessey-Morillon et al., 2014). Interestingly, ARHGEF12 is associated with hyper-responsiveness of tracheal rings in mouse models and is highly expressed in patients with asthma (Fong et al., 2018). Furthermore, enhanced ARHGEF11 signaling in small-diameter blood vessels was found to lead to vascular hyper-reactivity and result in hypertension (Hilgers et al., 2007). Thus, it appears that RHGF-1 and its orthologs have an evolutionary conserved role in regulating contractility of epithelial tubes.
Similar to RHGF-1, ARHGEF11 was found to bind F-actin, both in cells and in in vitro co-sedimentation assays (Banerjee and Wedegaertner, 2004). Deletion mutagenesis identified in ARHGEF11 a 25–amino acid sequence responsible for actin binding (Banerjee et al., 2009; Banerjee and Wedegaertner, 2004). However, this sequence is not conserved in RHGF-1. Instead, we found that the PDZ domain of RHGF-1 mediates its recruitment to F-actin. Although we found evidence for a direct interaction between RHGF-1 and F-actin in co-sedimentation assays with purified actin and the N-terminal half of RHGF-1, we cannot rule out that another protein is involved in its recruitment to stress fibers in vivo, similar to how the PDZ domain of Enigma recruits it to actin through an interaction with tropomyosin (Guy et al., 1999). However, direct interaction of a PDZ domain with actin has been described for the Drosophila sarcomeric protein Zasp52 (Liao et al., 2020), and our FRET-FLIM results suggest that RHGF-1 is within 10 nm of actin in the spermatheca. Therefore, it is possible that RHGF-1 binds actin directly.
The affinity of RHGF-1 to actin in vitro was rather low and the localization of RHGF-1 along actin bundles in the relaxed spermatheca was minimal. However, upon stretching of the spermathecal cells, the recruitment of RHGF-1 to actin increased, and once the actomyosin stress fibers were tensed, presumably due to rising active RHO-1 levels, the recruitment of RHGF-1 from the cytoplasm further increased. The apparent force-dependent recruitment of RHGF-1 to F-actin was further supported by experiments in which we genetically reduced or increased contractility within the spermatheca. Although it is currently unclear what change in tensed F-actin is being sensed by RHGF-1, it has been postulated that F-actin will change its conformation under tension (Galkin et al., 2012) and recently it was shown that single piconewton forces applied to F-actin enhance binding by αE-catenin (Mei et al., 2020) and several LIM domain proteins (Sun et al., 2020; Winkelman et al., 2020). Force-induced recruitment of ARHGEF12 to focal adhesions following stimulation of integrins with tensional force has been suggested to regulate the response of fibroblasts to force (Guilluy et al., 2011). In migrating neutrophils, ARHGEF11 was found to localize to the cell rear and sides in a myosin II–dependent mechanism (Wong et al., 2007). Thus, RHGF-1 and its mammalian orthologs all appear to form a positive feedback loop between myosin II contractility, RhoGEF localization, and RHO-1/RhoA activation.
In the spermatheca, the positive reinforcement between RHGF-1 and actomyosin contractility culminates in contraction of the spermatheca and expulsion of the embryo. Once emptied, the level of active RHO-1 in the spermatheca must decrease in order to allow another ovulation to take place. This is achieved through the recruitment of SPV-1 to the membrane and detachment of RHGF-1 from the stress fibers (Fig. 7 E). This results in a yin-yang relationship, wherein complementary changes in subcellular localization of a RhoGAP and RhoGEF pair orchestrate a cycle of RhoGTPase activation and inactivation that is externally controlled by tissue stretching. Why SPV-1 localizes to the apical membrane whereas RHGF-1 localizes to basal stress fibers is not clear.
Despite the strong phenotypes of loss of function of either RHGF-1 or SPV-1, their combination cancels out each other and the double mutant has wild-type–like spermatheca function. This result can be explained by the existence of a parallel pathway regulating contractility in the spermatheca, namely, calcium signaling, which also leads to activation of myosin II through MLCK (Kelley et al., 2018). Presumably, the existence of two redundant pathways contributes to robustness of the ovulation process. Our analysis of the RhoGEF UNC-73 in the spermatheca revealed that while it does partially rescue spv-1 it does not appear to regulate RHO-1. RNAi-mediated depletion of unc-73 did not have any effect on RHO-1 activity as measured with the RHO-1 biosensor and it did not increase the severity of the rhgf-1 null mutant. Based on this information, we think that UNC-73 works in parallel to RHGF-1 but not on RHO-1.
It is interesting to note that PDZ-mediated recruitment of RHGF-1 to stress fibers is essential for RHGF-1 activity in the background of spv-1(RNAi), but not for RHGF-1 activity in normal conditions. This probably means that low levels of RHO-1 activation are sufficient for spermatheca contraction. When SPV-1 is not present to keep RHO-1 levels in check, then normal RHGF-1 activity results in too much RHO-1 activity leading to precocious and excessive contractility. However, under the same condition, the RHGF-1 missing its PDZ domain does not lead to excessive RHO-1 activity. From this, we conclude that tension-sensitive recruitment is important to amplify RHGF-1 signaling to RHO-1, which in turn ensures robust generation of cell contraction in each ovulation cycle. This is consistent with a recent experimental and theoretical analysis of a system composed of the RhoGEF GEF-H1, RhoA, and myosin II, in which higher levels of the RhoGEF lead to pulsed contractions that were less sensitive to external inputs (Kamps et al., 2020).
In summary, our study illuminates a complete Rho GTPase regulatory network that controls the cyclic contraction of the C. elegans spermatheca and provides an in vivo example for self-regulation of epithelial tube contractility in response to changing internal pressure, based on mechanosensitive regulators of the actomyosin cytoskeleton.
Materials and methods
C. elegans strains
C. elegans strains were grown on nematode growth medium agar seeded with OP50 Escherichia coli at 20°C. The strains used in this study are listed in Table 1.
C. elegans strains used in this study
| Strain . | Genotype . | Origin . |
|---|---|---|
| N2 | Wild type | |
| RZB25 | spv-1(ok1498) | (Tan and Zaidel-Bar, 2015) |
| RZB241 | rhgf-1(ok880) X outcrossed 2x I3 | RB976 from CGC |
| RZB242 | spv-1(ok1498) line3;rhgf-1(ok880) line I3 double mutant 22 | This study |
| RB595 | unc-73(ok322) I | CGC |
| CB936 | unc-73(e936) I | CGC |
| KG1278 | unc-73(ce362) I | CGC |
| RB1014 | unc-73(ok936) I | CGC |
| COP1113 | Y105E8A.25 (knu182 - 1.7 kb deletion) | This study |
| COP1116 | Y105E8A.25 (knu185 - deletion of 1475 and insertion of 371 bp) | This study |
| RZB333 | xbIs1611[fkh-6P::rde-1, ttx-3::RFP]; WM27 rde-1(ne219);tag-341(ok1498)II | This study |
| UN1108 | xbls1101[fln-1p::GCAMP3, rol-6] | (Kovacevic et al., 2013) |
| RZB246 | rhgf-1(ok880); xbls1101[fln-1p::GCAMP3, rol-6] | This study |
| RZB171 | N2; Ex[psth-1::AHPH::GFP;rol-6(su1006)] | (Tan and Zaidel-Bar, 2015) |
| UN1513 | psth-1:AHPH integrated | (Kelley et al., 2020a) |
| RZB349 | rhgf-1(ok880);xbIs1611[fkh-6P::rde-1, ttx-3::RFP]; WM27 rde-1(ne219) | This study |
| UN1502 | xbIs1502[Y66H1Bp::GFP::Act-1, rol-6] | (Wirshing and Cram, 2017) |
| RZB256 | spv-1(ok1498); Ex[pspv-1::spv-1::GFP;rol-6];rhgf-1::tagRFP | This study |
| RZB395 | Ex[psth-1::AHPH::GFP;rol-6(su1006)]; rhgf-1[ knu148 (tagRFP, loxP::unc-119(+)::loxP)] ; unc-119(ed3) III | This study |
| RZB244 | rhgf-1(ok880); Ex[prhgf-1::rhgf-1 cDNA::mkate] line 6 | This study |
| RZB339 | rhgf-1(ok880); Ex[prhgf-1::rhgf-1∆PH::mKate] | This study |
| RZB338 | rhgf-1(ok880); Ex[prhgf-1::rhgf-1∆DH::mKate] | This study |
| RZB332 | rhgf-1(ok880); Ex[prhgf-1::rhgf-1∆C1::mKate] | This study |
| RZB324 | rhgf-1(ok880); Ex[prhgf-1::rhgf-1∆RGS::mKate] | This study |
| RZB343 | rhgf-1(ok880); Ex[prhgf-1::rhgf-1∆PDZ::mKate] | This study |
| RZB299 | rhgf-1[knu148 (tagRFP, loxP::unc-119(+)::loxP)]; xbIs1502[Y66H1Bp::GFP::Act-1, rol-6] | This study |
| RZB301 | rhgf-1[knu148 (tagRFP, loxP::unc-119(+)::loxP)]; xbIs1527[Fln-1p::GFP::NMY-1::NMY-13′UTR, rol-6] | This study |
| RZB432 | xbIs1502[Y66H1Bp::GFP::Act-1, rol-6];Ex[pMYO-2::dsred;pRHGF-1::tagRFP] | This study |
| COP1071 | rhgf-1[ knu148 (tagRFP, loxP::unc-119(+)::loxP)] ; unc-119(ed3) III | This study |
| RZB289 | pat-3 (knu455 - C-terminal degron::GFP, unc-119(+))/ +; unc-119(ed3); rhgf-1[ knu148 (tagRFP, loxP::unc-119(+)::loxP)] ; unc-119(ed3) | This study |
| RZB434 | rhgf-1(ok880); Ex[prhgf-1::PDZ::mKate] | This study |
| Strain . | Genotype . | Origin . |
|---|---|---|
| N2 | Wild type | |
| RZB25 | spv-1(ok1498) | (Tan and Zaidel-Bar, 2015) |
| RZB241 | rhgf-1(ok880) X outcrossed 2x I3 | RB976 from CGC |
| RZB242 | spv-1(ok1498) line3;rhgf-1(ok880) line I3 double mutant 22 | This study |
| RB595 | unc-73(ok322) I | CGC |
| CB936 | unc-73(e936) I | CGC |
| KG1278 | unc-73(ce362) I | CGC |
| RB1014 | unc-73(ok936) I | CGC |
| COP1113 | Y105E8A.25 (knu182 - 1.7 kb deletion) | This study |
| COP1116 | Y105E8A.25 (knu185 - deletion of 1475 and insertion of 371 bp) | This study |
| RZB333 | xbIs1611[fkh-6P::rde-1, ttx-3::RFP]; WM27 rde-1(ne219);tag-341(ok1498)II | This study |
| UN1108 | xbls1101[fln-1p::GCAMP3, rol-6] | (Kovacevic et al., 2013) |
| RZB246 | rhgf-1(ok880); xbls1101[fln-1p::GCAMP3, rol-6] | This study |
| RZB171 | N2; Ex[psth-1::AHPH::GFP;rol-6(su1006)] | (Tan and Zaidel-Bar, 2015) |
| UN1513 | psth-1:AHPH integrated | (Kelley et al., 2020a) |
| RZB349 | rhgf-1(ok880);xbIs1611[fkh-6P::rde-1, ttx-3::RFP]; WM27 rde-1(ne219) | This study |
| UN1502 | xbIs1502[Y66H1Bp::GFP::Act-1, rol-6] | (Wirshing and Cram, 2017) |
| RZB256 | spv-1(ok1498); Ex[pspv-1::spv-1::GFP;rol-6];rhgf-1::tagRFP | This study |
| RZB395 | Ex[psth-1::AHPH::GFP;rol-6(su1006)]; rhgf-1[ knu148 (tagRFP, loxP::unc-119(+)::loxP)] ; unc-119(ed3) III | This study |
| RZB244 | rhgf-1(ok880); Ex[prhgf-1::rhgf-1 cDNA::mkate] line 6 | This study |
| RZB339 | rhgf-1(ok880); Ex[prhgf-1::rhgf-1∆PH::mKate] | This study |
| RZB338 | rhgf-1(ok880); Ex[prhgf-1::rhgf-1∆DH::mKate] | This study |
| RZB332 | rhgf-1(ok880); Ex[prhgf-1::rhgf-1∆C1::mKate] | This study |
| RZB324 | rhgf-1(ok880); Ex[prhgf-1::rhgf-1∆RGS::mKate] | This study |
| RZB343 | rhgf-1(ok880); Ex[prhgf-1::rhgf-1∆PDZ::mKate] | This study |
| RZB299 | rhgf-1[knu148 (tagRFP, loxP::unc-119(+)::loxP)]; xbIs1502[Y66H1Bp::GFP::Act-1, rol-6] | This study |
| RZB301 | rhgf-1[knu148 (tagRFP, loxP::unc-119(+)::loxP)]; xbIs1527[Fln-1p::GFP::NMY-1::NMY-13′UTR, rol-6] | This study |
| RZB432 | xbIs1502[Y66H1Bp::GFP::Act-1, rol-6];Ex[pMYO-2::dsred;pRHGF-1::tagRFP] | This study |
| COP1071 | rhgf-1[ knu148 (tagRFP, loxP::unc-119(+)::loxP)] ; unc-119(ed3) III | This study |
| RZB289 | pat-3 (knu455 - C-terminal degron::GFP, unc-119(+))/ +; unc-119(ed3); rhgf-1[ knu148 (tagRFP, loxP::unc-119(+)::loxP)] ; unc-119(ed3) | This study |
| RZB434 | rhgf-1(ok880); Ex[prhgf-1::PDZ::mKate] | This study |
Molecular biology and transgenesis
A RHGF-1::mKate expression vector (plasmid pPY43) was generated by cloning rhgf-1 cDNA preceded by 3073 bp of rhgf-1 promoter sequence into the pPD95.75 backbone. RHGF-1 deletion constructs for RGS and C1 were performed by amplifying the entire backbone without the domain to be deleted and then ligating this with PNK ligase. Truncations for PH, DH, and PDZ were performed by overlap PCR and inverse PCR. Briefly, fragments of DNA ∼500 bp on either side of the domain to be deleted were generated by PCR. These fragments were then fused by PCR as they had overhangs for each other. The fused PCR product was then used for inverse PCR with Phusion to loop out the domain to be deleted from the pPY43 plasmid. The deletions involved the following: amino acids 50–125 for ∆PDZ, amino acids 257–385 for ∆RGS, amino acids 485–534 for ∆C1, amino acids 727–961 for ∆GEF, and amino acids 727–961 for ∆PH. The N-terminus with only the PDZ domain (amino acids 1–240) fused with mKate was generated from the pPY43 plasmid by Gibson cloning (New England Biolabs). Cytoplasmic tagRFP driven by the RHGF-1 promoter was generated by assembling tagRFP from pNU758 (used to create RHGF-1::tagRFP) and the pPY43 plasmid without RHGF-1 and mKate, using Gibson assembly. All constructs were checked for deletion by PCR and then confirmed by sequencing before injecting into worms, 50 ng/μl plasmid and 50 ng/μl rol-6 or myo-2::dsRed per injection.
An spv-1 RNAi feeding clone was made by cloning a 580 bp fragment from C. elegans genomic sequence in T444T by overlap PCR, with forward primer 5′-GGCGGCCGCTCTAGAGTTGTTCTGCACAAGCATCTGG-3′ and reverse primer 5′-CCCTCGAGGTCGACGGTGATGTTGTCTGCTCCACAGG-3′.
A rhgf-1 RNAi feeding clone was made by cloning a 558-bp fragment from C. elegans genomic sequence in T444T by Gibson assembly, with forward primer for insert 5′-GAGCTCCACCGCGGTGGCGGCCGCTCTAGACAGAATTATTTCACACGGAGCGG-3′, reverse primer for insert 5′-GGTACCGGGCCCCCCCTCGAGGTCGACGGTGCTCGGTTGTATACTTTGCAATGG-3′, forward primer for vector 5′-TGGAAACCATTGCAAAGTATACAACCGAGCACCGTCGACCTCGAGGGGGG-3′ and reverse primer for vector 5′-CGTGAGTCCGCTCCGTGTGAAATAATTCTGTCTAGAGCGGCCGCCACCGC-3′.
Two unc-73 RNAi feeding clones were made by cloning a 557-bp fragment from C. elegans genomic sequence, with forward primer 5′-GGCGGCCGCTCTAGACGACGAACTCTCCTACACATCG-3′ and reverse primer 5′-CCCTCGAGGTCGACGGTGCGTCATACGGTTCCTTGAGC-3′, and a 780-bp fragment from C. elegans genomic sequence, with forward primer 5′-GGCGGCCGCTCTAGACTACTGAATCGTCAACTTCTGCACC-3′ and reverse primer 5′-CCCTCGAGGTCGACGGTCTCGTTCGGTGAACTTGAGAATATCC-3′ into T444T by overlap PCR.
All the sequences were confirmed by sequencing.
RNAi experiments
RNAi knockdown of gene expression was performed by feeding of HT115 bacteria expressing double-stranded RNA (dsRNA). RNAi feeding clones were obtained from the Ahringer (Source BioScience) or Vidal (GE Dharmacon) RNAi libraries, except for spv-1, rhgf-1, and unc-73 that were cloned into T444T. Bacterial clones with empty L4440 or T444T vectors were used as negative controls. RNAi plates were prepared with nematode growth medium containing 1 mM IPTG and 100 μg/ml of ampicillin. For dsRNA induction, an overnight culture of bacteria was diluted 1:100 and grown at 37°C with shaking for 3–4 h, after which IPTG was added to a final concentration of 1 mM and allowed to grow for an additional 3–4 h. This culture was used to seed the RNAi plates, which were kept overnight at room temperature before placing worms. Adult hermaphrodites of required genotypes were allowed to lay eggs on the RNAi feeding plate and were subsequently removed. The L1 that hatched on the RNAi feeding plate were assayed when they reached adulthood.
For some unc-73 RNAi experiments, we used a spermatheca-specific RNAi approach as described previously (Kelley et al., 2018). Briefly, the essential RNAi machinery protein RDE-1 was re-expressed under the fkh-6 promoter in a rde-1(ne219) background.
Viability and embryo shape assays
Scoring the rescue of embryo shape in the RhoGEF RNAi screen was performed by observing worms under a stereomicroscope as they were cut open with a razor to allow the content of their uterus to spill out. Any worm that had abnormally small or round or pinched embryos (usually accompanied by pieces of severed embryos) was scored as an spv-1 null phenotype (i.e., not rescued). Embryonic viability was measured in the F2 generation, after the F1 fed on RNAi from hatching. 10 day 1 adults (one row of eggs) were allowed to lay eggs for 3 h (50–150 eggs laid), then the adults were removed and the eggs were counted. 24 h later, the L1s and dead eggs were counted. This experiment was performed in triplicates and repeated five times in order to calculate the percent of embryonic viability.
Quantification of embryo shape in ∆PDZ experiments was performed on day 1 F1 adults, which fed on RNAi since hatching. The worms were cut open, and the released embryos were mounted on agar pads on a glass slide and imaged with DIC microscopy at 100×. The length and width of each embryo were measured in FIJI (Schindelin et al., 2012).
Microscopy and imaging
For DIC and fluorescence microscopy imaging of spermatheca dynamics, day 1 adult hermaphrodites with one row of embryos were mounted on glass slides with 10% agarose pads, which served to immobilize them, and 1.38 µl of M9 medium. Image acquisition was performed at 20°C on a Ti-Eclipse inverted microscope (Nikon) equipped with a CSU-X1 spinning-disk confocal head (Yokogawa Electric Corporation), DPSS-Laser (Roper Scientific) of 491- and 561-nm excitation wavelengths and an Evolve electron multiplying charged-coupled device camera (Photometrics). Focus drift during time-lapse acquisition was corrected using Perfect Focus System (Nikon). Acquisition control was performed using MetaMorph (Molecular Devices). Ovulation and embryo transit movies were acquired with a 60× 1.4 NA oil-immersion Plan-Apochromat objective (Nikon). Live super-resolution imaging of protein localization in the spermatheca (Fig. 6, A and B; and Fig. S5) was performed on a Ti-2 Eclipse with a Live-SR module (Gataca systems, France) mounted on the light path of a CSU-W1 spinning disk, a 100× 1.45 NA oil-immersion Plan-Apochromat objective (Nikon, Japan), and a Prime95B SCMOS camera (Photometrics). Actin stress fibers in the spermatheca (Fig. 2 H and Fig. 7 A) and embryos images (Fig. 1 B and Fig. 5 C) were acquired with a 100× 1.45 NA oil-immersion Plan-Apochromat objective (Nikon).
Calcium imaging
All time-lapse GCaMP3 images were acquired as 1,600 × 1,200 pixels for the Spot RT3 OCCD camera and saved as 8-bit tagged image and file format files. All image processing was done using a macro on Image J (Schindelin et al., 2012). All time-lapse images were oriented with the sp-ut valve on the right of the frame and registered to minimize any body movement of the paralyzed animal. An 800 × 400 region of interest encompassing the entire spermatheca was utilized to measure the GCaMP3 signal. The average pixel intensity of each frame was calculated using a custom ImageJ macro (Kovacevic et al., 2013). Calcium pixel intensity (F) was normalized to the average pixel intensity of the first 30 frames prior to the start of ovulation (F0) and plotted against time. Data analysis and graphing were performed using Matlab and GraphPad Prism. Matlab was used to identify peaks in the GCaMP time series. Specifically, data were smoothed using a moving average of five data points through the Matlab “smoothdata” command. Local maxima in the time series with a minimum prominence of 0.1 units and a minimum width of 5 units were then identified using the Matlab “findpeaks” command. These standardized settings were used to analyze all calcium traces. Microsoft Excel was used to quantify the amount of time after oocyte entry required to reach either the half the maximum or maximum calcium signal. Kymograms were generated using an ImageJ macro that calculated the average pixel intensity of each column of a frame and condensing it down to one line per frame of the time-lapse image. Every frame of the time-lapse image was stacked to visualize calcium dynamics of representative ovulations in both space and time. The Fire look-up table color scale was applied to the kymograms using Image J.
Quantifications and image analysis
Quantifications of dwell time, defined as the amount of time from the closure of the distal valve to the opening of the sp-ut valve, were performed from DIC movies.
Constriction magnitude was calculated as described before (Tan and Zaidel-Bar, 2015), briefly, by measuring the width of the spermatheca in two locations, distal (W1) and proximal (W2), and dividing the two (Fig. 1 D).
RHO-1 biosensor activity was quantified by measuring the mean intensity of the middle focal plane of a spermatheca without the sp-ut valve (Fig. 2 F) across time relative to the intensity at the initial time point. Time was normalized to dwell time (where time 0 is closure of distal valve and 1 is opening of sp-ut valve), and the time later on was normalized to 1–1.5 where 1.5 is closing of sp-ut valve. The total amount of RHO-1 biosensor GFP in the spermathecal cells doesn’t change over time, only its localization. The likely explanation for the change in fluorescence intensity that we measured is that much of the cytoplasmic RHO-1 biosensor GFP is below our detection level, whilst RHO-1 biosensor GFP that accumulates at the membrane at high density following RHO-1 activation is detectible and measured.
Fluorescence intensity at the cell edge versus the cytoplasm was quantified by manually tracing a line along the cell edge (1 pixel) or the cytoplasm (4 pixels; Fig. S3 E). Mean intensity of the lines drawn at the cell edge divided by the values obtained for the cytoplasm provided the edge-to-cytoplasm ratio.
Tortuosity of F-actin stress bundles, defined as the ratio of the actual length of the fiber to the shortest distance between the ends of the fiber, was measured with a custom Matlab code. Prior to the analysis, the images were maximum projected and rotated (such that the actin cables are in vertical orientation) in FIJI (Schindelin et al., 2012). Each image was then smoothed with a Gaussian filter, followed by Niblack local thresholding (https://www.mathworks.com/matlabcentral/fileexchange/40849-niblack-local-thresholding). The thresholded image was skeletonized and post-processing was performed to connect neighboring segmented lines as a single line if they do not have drastic angle changes. Finally, the line branches were disconnected to get the final segmentation results for tortuosity measurement.
Co-localization of proteins was calculated by JACoP plugin of FIJI (Schindelin et al., 2012) on maximum intensity projection images.
Quantitative data sets were subjected to statistical analysis using the Prism 9.3.1 software (GraphPad). Statistical tests are specified in the figure legends. All data distributions were assumed to be normal, but this was not formally tested.
Two-photon FLIM
The interaction between GFP::ACT-1 and RHGF-1::tagRFP was measured in day 1 adults using two-photon FLIM. Briefly, GFP was excited with a Ti:sapphire laser (Chameleon, Coherent) at a wavelength of 920 nm and a power of 1.0–2.0 mW. Fluorescence lifetime images were obtained using a Bergamo two-photon microscope (Thorlabs) equipped with a Time-Correlated Single Photon Counting board (Time Harp 260, Picoquant). Emission was collected with a 16 × 0.8 NA objective (Nikon), divided with a 565-nm dichroic mirror (Chroma) and detected with two PhotoMultiplier Tubes with low transfer time spread (H7422-40p, Hamamatsu). Images were collected by 128 × 128 pixels and acquired at 2 ms/line, averaged over 24 frames. The fluorescence lifetime of GFP was measured by curve fitting using custom software written with C# as described previously (Laviv et al., 2020).
Actin co-sedimentation
For the expression of GST-tagged proteins in E. coli, a cDNA fragment encoding the N-terminal half of C. elegans RHGF-1 (residues 1–635) was inserted into the BamHI-EagI sites of pGEX- 6P-1 (GE Healthcare). RHGF-1-N-ΔPDZ lacking the PDZ domain (residues 50–125) was generated by Gibson cloning. The sequences were validated by sequencing.
The N-terminal half of C. elegans RHGF-1 (RHGF-1-N) as well as RHGF-1-N-ΔPDZ tagged with GST were expressed in E. coli host Rosetta 2 (Novagen). Expression was induced with 0.75 mM isopropyl-β-D-thiogalactopyranoside at 21°C for 12 h. The bacteria were harvested and lysed by ultrasonication in lysis buffer containing 30 mM Hepes, pH 7.4, 150 mM NaCl, 2 mM EDTA, 1 mM DTT, 5% (vol/vol) glycerol, 5 mM Benzamidine hydrochloride, 0.5 mM 4-(2-Aminoethyl)benzenesulfonyl fluoride hydrochloride and 2 units/ml Benzonase. The fusion proteins were subsequently purified from bacterial extracts by affinity chromatography using glutathione-conjugated agarose 4B (Macherey-Nagel) using standard procedures. The GST-tag was cleaved off by PreScission protease (GE Healthcare), the tag absorbed on fresh glutathione-conjugated agarose and the proteins in the flow through separated by size-exclusion chromatography with a HiLoad 26/600 Superdex 200 column controlled by an Äkta Purifier System. Protein concentrations were determined by absorption spectroscopy using extinction coefficients predicted from respective amino acid sequences.
Ca2+-ATP G-actin was purified from rabbit skeletal muscle according to Spudich and Watt (1971) and stored in G-buffer (5 mM Tris-HCl, pH 8.0, and 0.2 mM CaCl2, 0.5 mM DTT, 0.2 mM ATP) on ice for the experiments.
Online supplementary material
Table S1 shows RNAi screen of RhoGEFs in spv-1 background scoring abnormal egg shape. Fig. S1 is related to Fig. 1 and shows dwell time analysis in loss of function alleles and RNAi of unc-73 and Y105E8A.25. Fig. S2 is related to Fig. 2 and shows RHO-1 biosensor in control and in trapped rhgf-1(RNAi); RHO-1 biosensor in unc-73(RNAi); analysis of dwell time and successful transits in rhgf-1(ok880) spermathecae treated with spermatheca-specific unc-73(RNAi); and fluorescence intensity of UNC-73::GFP treated with unc-73(RNAi).Fig. S3 is related to Fig. 3 and shows analysis of dwell time and successful embryo transits in RHGF-1::tagRFP; fluorescence intensity of RHGF-1::tagRFP following rhgf-1(RNAi); subcellular localization of RHGF-1::tagRFP and β-Integrin/PAT-3::GFP; and images demonstrating how intensity of SPV-1::GFP and RHGF-1::tagRFP in the cytoplasm and edge was measured. Fig. S4 is related to Fig. 4 and shows analysis of expression level and dwell time in all RHGF-1 deletion constructs. Fig. S5 is related to Fig. 6 and shows relative subcellular localization of RHGF-1 and NMY-1. Video 1 is related to Fig. 1 and shows DIC time-lapse of ovulation and embryo transit in wild-type, spv-1(ok1498), rhgf-1(ok880), and spv-1(ok1498); rhgf-1(ok880) mutant animals. Video 2 is related to Fig. 2 and shows calcium signaling visualized in rhgf-1(ok880) and control spermatheca with a GCaMP calcium reporter. Video 3 is related to Fig. 2 and shows RHO-1 biosensor in control and rhgf-1(RNAi) spermatheca during ovulation and embryo transit. Video 4 is related to Fig. S2 and shows RHO-1 biosensor in control and rhgf-1(RNAi) spermatheca during failed embryo transit. Video 5 is related to Fig. S2 and shows RHO-1 biosensor in control and unc-73(RNAi) spermatheca during ovulation and embryo transit. Video 6 is related to Fig. S3 and shows RHGF-1::tagRFP and β-Integrin/PAT-3::GFP dynamics during ovulation and embryo transit. Video 7 is related to Fig. 3 and shows RHGF-1::tagRFP and SPV-1::GFP dynamics during ovulation and embryo transit. Video 8 is related to Fig. 3 and shows RHGF-1::tagRFP and RHO-1 biosensor dynamics during oocyte ovulation and embryo transit. Video 9 is related to Fig. 4 and shows subcellular localization of mKate-fused full-length RHGF-1 and domain deletions from oocyte entry until embryo exit. Video 10 is related to Fig. 4 and shows subcellular localization of mKate-fused PDZ domain, from oocyte entry until embryo exit.
Acknowledgments
We thank Hanna Grobe for help with image analysis, Jonas Scholz for assistance in protein purification and analyzing the equilibrium constants of RHGF-1 constructs, and Anat Nitzan for drawing the schematic model in Fig. 7 E.
This study was supported by Israel Science Foundation grants 1293/17 and 767/20 awarded to R. Zaidel-Bar, a National Science Foundation (1816640) – U.S. Israel Binational Science Foundation award (2017657) to E.J. Cram and R. Zaidel-Bar, and a grant by the Deutsche Forschungsgemeinschaft Fa330/12-3 awarded to J. Faix. Some strains were provided by the Caenorhabditis Genetics Center (CGC), which is funded by National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). S. Avivi Kela was supported by a travel scholarship from the Joan and Jaime Constantiner Institute for Molecular Genetics.
Author contributions: R. Zaidel-Bar and P.Y. Tan conceived the project. P.Y. Tan, K. Sethi, S. Avivi Kela, and R. Zaidel-Bar designed experiments and analyzed results. P.Y. Tan, K. Sethi, and S. Avivi Kela performed experiments. H.-T. Ong contributed image analysis tools. D. Suresh assisted with cloning. J. Faix performed and analyzed actin co-sedimentation experiments. T. Laviv performed and analyzed FLIM experiments. P.G. Castaneda, M.R. Amin, and E.J. Cram performed calcium analysis. K. Sethi, S. Avivi Kela, and R. Zaidel-Bar prepared the manuscript.
References
Author notes
Disclosures: The authors declare no competing financial interests.