The dependency of cancer cells on iron increases their susceptibility to ferroptosis, thus providing new opportunities for patients with treatment-resistant tumors. However, we show that lipid peroxidation, a hallmark of ferroptosis, was found in various areas of patient samples, indicating the potential resistance of ferroptosis. Using whole deubiquitinases (DUBs) sgRNA screening, we found that loss of ZRANB1 confers cancer cell resistance to ferroptosis. Intriguingly, functional studies revealed that ZRANB1 ubiquitinates and represses SLC7A11 expression as an E3 ubiquitin ligase and that ZRANB1 inhibits glutathione (GSH) synthesis through SLC7A11 degradation, leading to elevated lipid peroxidation and ferroptosis. Deletion of the region (residues 463–584) abolishes the E3 activity of ZRANB1. Moreover, we show that ZRANB1 has lower expression in tumors, which is positively correlated with lipid peroxidation. Collectively, our results demonstrate the role of ZRANB1 in ferroptosis resistance and unveil mechanisms involving modulation of E3 ligase activity through an unconventional catalytic domain.
Introduction
Ferroptosis is usually a regulated form of cell death induced by inactivating lipid repair enzymes such as glutathione peroxidase 4 (GPX4) and accumulating lipid reactive oxygen species (ROS) such as lipid hydroperoxides (Yang and Stockwell, 2016). Currently, multiple markers have been developed to detect ferroptotic cells. Among them, lipid peroxidation is the most important marker and must be detected to distinguish ferroptosis from other forms of cell death. To date, there are several means to identify ROS, which are (1) thiobarbituric acid reactive substances assay; (2) liquid chromatography tandem mass spectrometry/mass spectrometry; (3) C11-BODIPY fluorescent probe, and (4) antibodies such as anti-hydroxynonenal or malondialdehyde (MDA) that detect adducts formed by ROS products (Kobayashi et al., 2021). Significant progress has been made in the last decade, particularly in understanding the mechanisms that control ferroptosis. Many researchers have elucidated the specific lipids that drive ferroptosis after oxidation, the intrinsic mechanism of inhibition of ferroptosis, and so on.
The cystine/glutamate antiporter SLC7A11 (also known as xCT) is used to import cystine for glutathione biosynthesis and antioxidant defense and is overexpressed in multiple human cancers. Recent studies have revealed that SLC7A11 can regulate ferroptosis. As one of the most important ferroptosis-defending systems, SLC7A11 normally functions as a strong ferroptotic suppressor together with GPX4 to reduce reactive polyunsaturated fatty acid (PUFA) phospholipid hydroperoxides to nonreactive and nonlethal PUFA phospholipid alcohols. Moreover, GTP cyclohydrolase 1/tetrahydrobiopterin, ferroptosis suppressor protein 1 (FSP1)/CoQ10, and dihydroorotate dehydrogenase (DHODH) have been recently identified to function independent of GPX4 to inhibit ferroptosis. In contrast, as the first identified proferroptotic gene products, ACSL4 and LPCAT3 play roles in facilitating the incorporation of PUFAs into membrane lipids (Stockwell, 2022).
As two important components of the central antioxidant defense system, diverse regulatory mechanisms of SLC7A11 and GPX4 in cancer have been identified. For example, to ensure that SLC7A11 can properly maintain redox homeostasis, its expression is precisely regulated through multiple mechanisms that include transcription factors, epigenetic regulators, and deubiquitinase to orchestrate mRNA and protein turnover (Koppula et al., 2021). In addition, as one of the most promising ferroptosis inducers in vivo, imidazole ketone Erastin (IKE) exerts a promising therapeutic effect on diffuse large B cell lymphoma by blocking SLC7A11-GPX4 signaling (Zhang et al., 2019). Although recent discoveries have suggested that ferroptosis inducers can conquer diverse therapy-resistant cancer cells (Gout et al., 2001; Hangauer et al., 2017; Lang et al., 2019; Lei et al., 2020; Liu et al., 2017; Viswanathan et al., 2017; Wang et al., 2019; Yu et al., 2017), for other solid tumors, such as kidney renal clear cell carcinoma (KIRC), little is known about the antitumor effect of IKE in vivo. In this study, we used a 786-O cell (KIRC) xenograft model and treated them with IKE and found that tumors formed by 786-O cells were significantly resistant to IKE treatment (Fig. S1, A–I). Furthermore, we conducted unbiased screening to investigate which deubiquitinase is involved in conferring ferroptotic resistance to KIRC cells. These findings not only identify the potential mechanisms of ferroptosis resistance but also clarify the relevance between ZRANB1-SLC7A11 axis in tumor samples and ROS-linked ferroptosis resistance.
Screening appropriate tumor models of ferroptosis. (A) Representative phase-contrast cell death images of the indicated 786-O cells induced by 10 μM IKE with or without Liprox and Ferr-1. Scale bars, 50 μm. (B) Histograms showing the viability of the indicated 786-O cells treated as described in A. Error bars are the means ± SEM, n = 3 biological replicates. (C) Diagram of xenograft tumor-loaded mice. Nude mice subcutaneously inoculated with 786-O cells were divided into two groups: the control group was intraperitoneally injected with HBSS solution; the experimental group was intraperitoneally injected with IKE drug. Tumors are marked by red circles. (D) Tumor growth curves of xenograft tumors described in C. Error bars are the means ± SEM, n = 5 mice per group. (E) Weight of tumor xenografts from 786-O cells at the endpoint. Error bars are the means ± SEM, n = 5 independent repeats. (F) Body weight of mice with the indicated genotypes and treatments on different days. Error bars are the means ± SEM, n = 5 independent repeats. (G) Schematic diagram of the sampling tumors from nude mice. (H) Representative images of MDA IHC staining of the tumor xenografts described in C. TS: tumor site. Scale bars, 20 μm. (I) Percentage of MDA-positive stained cells per field. TS, tumor site. Error bars are means ± SEM, n = 5. (E) P values measured using two-tailed unpaired Student’s t test analysis. (B) P values measured using one-way ANOVA. (D, F, and I) P values determined by two-way ANOVA.
Screening appropriate tumor models of ferroptosis. (A) Representative phase-contrast cell death images of the indicated 786-O cells induced by 10 μM IKE with or without Liprox and Ferr-1. Scale bars, 50 μm. (B) Histograms showing the viability of the indicated 786-O cells treated as described in A. Error bars are the means ± SEM, n = 3 biological replicates. (C) Diagram of xenograft tumor-loaded mice. Nude mice subcutaneously inoculated with 786-O cells were divided into two groups: the control group was intraperitoneally injected with HBSS solution; the experimental group was intraperitoneally injected with IKE drug. Tumors are marked by red circles. (D) Tumor growth curves of xenograft tumors described in C. Error bars are the means ± SEM, n = 5 mice per group. (E) Weight of tumor xenografts from 786-O cells at the endpoint. Error bars are the means ± SEM, n = 5 independent repeats. (F) Body weight of mice with the indicated genotypes and treatments on different days. Error bars are the means ± SEM, n = 5 independent repeats. (G) Schematic diagram of the sampling tumors from nude mice. (H) Representative images of MDA IHC staining of the tumor xenografts described in C. TS: tumor site. Scale bars, 20 μm. (I) Percentage of MDA-positive stained cells per field. TS, tumor site. Error bars are means ± SEM, n = 5. (E) P values measured using two-tailed unpaired Student’s t test analysis. (B) P values measured using one-way ANOVA. (D, F, and I) P values determined by two-way ANOVA.
Results
ZRANB1 is linked to ferroptotic resistance
Deubiquitinating enzymes (DUBs) have essential roles in maintaining the stability of the majority of proteins (Amerik and Hochstrasser, 2004) and have been linked to ferroptosis (Lee et al., 2021). Moreover, inactivation of specific deubiquitinases has emerged as an effective anticancer therapy (D’Arcy and Linder, 2014; Pal et al., 2014). To determine which DUB is involved in ferroptosis resistance, we used a CRISPR‒Cas9 screen in cells stably expressing Cas9 (Fig. 1 A) and then individually transfected a gRNA library covering 87 human DUBs, which represent most human DUBs, into these cells. By using all individual DUB stable cell lines, we identified two DUBs that modulate susceptibility to ferroptosis induced by Erastin and cystine deprivation. Specifically, we found that gRNAs targeting ZRANB1 more efficiently promote cell survival under stress than BAP1 depletion, which has been previously reported (Zhang et al., 2018b; Fig. 1, B and C). Taken together, the loss of ZRANB1 may promote resistance to ferroptosis.
ZRANB1 is linked to ferroptosis resistance. (A) Schematic of the CRISPR/Cas9 screening strategy. (B) DUB screen in which 87 DUB KO HeLa cell lines were challenged by cystine starvation. Error bars are means ± SEM, n = 3 biological replicates. (C) DUB screen in which 87 DUB KO HeLa cell lines were treated with Erastin. Error bars are means ± SEM, n = 3 biological replicates. (D) Top panel: Percentage of MDA-positive stained cells per field in samples from 21 patients. Error bars are means ± SEM, n = 5 biological replicates. Bottom panel: Schematic diagram of ROS levels in human KIRC patient samples. (E) IF staining of MDA (green) in human clinical tumor samples. The right images are the overlay of MDA and nuclear DAPI (blue) staining of the same field. Scale bars, 20 μm. (F) IF staining of ZRANB1 (green) and MDA (magenta) in KIRC patients. The right images are the overlay of ZRANB1, MDA, and nuclear DAPI (blue) staining. Scale bars, 20 μm. (G) ZRANB1 staining scores (x axis) positively correlated with MDA staining scores (y axis) in 46 KIRC patients (it should be noted that some circles overlap). The P value was calculated from a linear regression analysis. R is the correlation coefficient. Protein staining score = the percentage of immunopositive cells × immunostaining intensity. (B and C) P value determined by one-way ANOVA.
ZRANB1 is linked to ferroptosis resistance. (A) Schematic of the CRISPR/Cas9 screening strategy. (B) DUB screen in which 87 DUB KO HeLa cell lines were challenged by cystine starvation. Error bars are means ± SEM, n = 3 biological replicates. (C) DUB screen in which 87 DUB KO HeLa cell lines were treated with Erastin. Error bars are means ± SEM, n = 3 biological replicates. (D) Top panel: Percentage of MDA-positive stained cells per field in samples from 21 patients. Error bars are means ± SEM, n = 5 biological replicates. Bottom panel: Schematic diagram of ROS levels in human KIRC patient samples. (E) IF staining of MDA (green) in human clinical tumor samples. The right images are the overlay of MDA and nuclear DAPI (blue) staining of the same field. Scale bars, 20 μm. (F) IF staining of ZRANB1 (green) and MDA (magenta) in KIRC patients. The right images are the overlay of ZRANB1, MDA, and nuclear DAPI (blue) staining. Scale bars, 20 μm. (G) ZRANB1 staining scores (x axis) positively correlated with MDA staining scores (y axis) in 46 KIRC patients (it should be noted that some circles overlap). The P value was calculated from a linear regression analysis. R is the correlation coefficient. Protein staining score = the percentage of immunopositive cells × immunostaining intensity. (B and C) P value determined by one-way ANOVA.
Next, we investigated the correlation between ZRANB1 expression and potential resistance in clinical samples. Heterogeneity provides the fuel for resistance. To first clarify whether ferroptotic resistance is present in clinical samples, we collected 21 samples from KIRC patients, with three to five random sites for each tumor sample. MDA staining (an indicator of lipid peroxidation; Lee et al., 2020) clearly showed that lipid peroxidation (ROS) varied in 62% of the patients. Moreover, ROS in 33% of the patients were low, indicating intrinsic resistance (Fig. 1, D and E). By performing immunofluorescence (IF) staining of ZRANB1 (green) and MDA (magenta) in the KIRC tissue microarrays, we found a highly significant positive correlation (R2 = 0.47, P < 0.0001) between ZRANB1 and MDA (Fig. 1, F and G). These results suggest that ZRANB1 may be involved in tumor-intrinsic resistance to ferroptosis.
ZRANB1 sensitizes cells to ferroptosis
To validate these findings, we generated cell lines stably expressing ZRANB1 using UMRC6 (Fig. S2 A) and 786-O cells (Fig. S2 B) with lower ZRANB1 expression (Fig. S2 C). As expected, overexpression of ZRANB1 significantly promoted Erastin-induced cell death, which could be completely reversed by the ferroptosis inhibitors ferrostatin-1 (Ferr-1), liproxstatin-1 (Liprox), and deferoxamine (DFO) but not the necroptosis inhibitor nec-1s or the apoptosis inhibitor carbobenzoxy-valyl-alanyl-aspartyl-[O-methyl]-fluoromethylketone (Z-VAD-FMK; Fig. 2, A and B; and Fig. S2, D and E). Similarly, we found that treatment with tert-butyl hydroperoxide (TBH), an ROS inducer, resulted in substantial cell death in ZRANB1-overexpressing cells compared with control cells. Consistently, TBH-induced cell death could be largely blocked by Ferr-1, Liprox, and DFO (Fig. 2, C and D; and Fig. S2, F and G).
ZRANB1 promotes ferroptosis in vitro. (A and B) Western blot for ZRANB1 in UMRC6 (A) and 786-O (B) cells transfected with the ZRANB1 expression construct. (C) Immunoblotting analyses of ZRANB1 protein levels in different cancer cell lines. (D and F) Cell viability in the indicated 786-O stable cell lines after treatment with different concentrations of Erastin (D) or TBH (F). (E and G) Bar graphs showing cell viability in 786-O cell lines treated with 10 μM Erastin (E) or 100 μM TBH (G) combined with or without 5 μM Z-VAD, 2 μM Nec-1s, 2 μM Ferr-1, 100 μM DFO, or 10 μM Liprox. (H) Bar graph showing intracellular GSH levels in the indicated 786-O cells. (I) Lipid peroxidation was assessed by flow cytometry after C11-BODIPY staining in the indicated 786-O cells. (J) Cell death measured in the indicated 786-O cells after treatment with 10 μM Erastin together with or without 2 μM Ferr-1 for 24 h. (K) Cell viability of the indicated 786-O cells, measured after culturing under cystine starvation conditions with or without 2 μM Ferr-1 for 48 h. (L and M) Western blot analysis of ZRANB1 expression in ZRANB1-KO RCC4 (L) and ACHN (M) cells. (N) DNA sequencing to validate the gene editing of ZRANB1. (O) Bar graph showing intracellular GSH levels in the indicated ACHN cells. (P and S) Cell viability in the indicated ACHN stable cell lines after treatment with different concentrations of Erastin (P) or TBH (S). (Q) Cell viability measured in the indicated ACHN cells treated with 2 μM Erastin and 1 μM Ferr-1 for 24 h. (R) Cell death measured in the indicated ACHN cells treated with 2 μM Erastin and 1 μM Ferr-1 for 24 h. (T and U) Cell viability of the indicated ACHN cells, measured after treatment with 100 μM TBH and 2 μM Ferr-1 for 24 h (T) or measured after culturing in cystine starvation conditions with or without 2 μM Ferr-1 for 48 h (U). (H) P values measured using two-tailed unpaired Student’s t test analysis. (D–K and O–U) Error bars are the means ± SEM, n = 3 biological replicates. (O) P values measured using one-way ANOVA. (D–G, I–K, and P–U) P values determined by two-way ANOVA. Source data are available for this figure: SourceData FS2.
ZRANB1 promotes ferroptosis in vitro. (A and B) Western blot for ZRANB1 in UMRC6 (A) and 786-O (B) cells transfected with the ZRANB1 expression construct. (C) Immunoblotting analyses of ZRANB1 protein levels in different cancer cell lines. (D and F) Cell viability in the indicated 786-O stable cell lines after treatment with different concentrations of Erastin (D) or TBH (F). (E and G) Bar graphs showing cell viability in 786-O cell lines treated with 10 μM Erastin (E) or 100 μM TBH (G) combined with or without 5 μM Z-VAD, 2 μM Nec-1s, 2 μM Ferr-1, 100 μM DFO, or 10 μM Liprox. (H) Bar graph showing intracellular GSH levels in the indicated 786-O cells. (I) Lipid peroxidation was assessed by flow cytometry after C11-BODIPY staining in the indicated 786-O cells. (J) Cell death measured in the indicated 786-O cells after treatment with 10 μM Erastin together with or without 2 μM Ferr-1 for 24 h. (K) Cell viability of the indicated 786-O cells, measured after culturing under cystine starvation conditions with or without 2 μM Ferr-1 for 48 h. (L and M) Western blot analysis of ZRANB1 expression in ZRANB1-KO RCC4 (L) and ACHN (M) cells. (N) DNA sequencing to validate the gene editing of ZRANB1. (O) Bar graph showing intracellular GSH levels in the indicated ACHN cells. (P and S) Cell viability in the indicated ACHN stable cell lines after treatment with different concentrations of Erastin (P) or TBH (S). (Q) Cell viability measured in the indicated ACHN cells treated with 2 μM Erastin and 1 μM Ferr-1 for 24 h. (R) Cell death measured in the indicated ACHN cells treated with 2 μM Erastin and 1 μM Ferr-1 for 24 h. (T and U) Cell viability of the indicated ACHN cells, measured after treatment with 100 μM TBH and 2 μM Ferr-1 for 24 h (T) or measured after culturing in cystine starvation conditions with or without 2 μM Ferr-1 for 48 h (U). (H) P values measured using two-tailed unpaired Student’s t test analysis. (D–K and O–U) Error bars are the means ± SEM, n = 3 biological replicates. (O) P values measured using one-way ANOVA. (D–G, I–K, and P–U) P values determined by two-way ANOVA. Source data are available for this figure: SourceData FS2.
ZRANB1 promotes ferroptosis in renal cancer cells. (A and C) Cell viability in UMRC6 stable cell lines after treatment with different concentrations of Erastin (A) or TBH (C). (B and D) Bar graphs showing cell viability in UMRC6 cell lines treated with 20 μM Erastin (B) or 100 μM TBH (D) combined with 5 μM Z-VAD-fmk (Z-VAD), 2 μM Necrostatin-1s (Nec-1s), 2 μM Ferr-1, 100 μM DFO, or 10 μM Liprox. (E) Bar graph showing intracellular GSH levels in the indicated UMRC6 cells. (F) Lipid peroxidation was assessed by flow cytometry after C11-BODIPY staining in the indicated UMRC6 cells. (G) Cell death measured in the indicated UMRC6 cells after treatment with 2 μM Ferr-1 and 20 μM Erastin for 24 h. (H) Viability of UMRC6 cells cultured under cystine starvation conditions for the indicated duration. (I) Cell viability of the indicated UMRC6 cells, measured after culturing under cystine starvation conditions with or without 2 μM Ferr-1 for 48 h. (J) Bar graph showing intracellular GSH levels in the indicated RCC4 cells. (K) Lipid peroxidation was assessed by flow cytometry after C11-BODIPY staining in the indicated RCC4 cells. (L and O) Cell viability in RCC4 stable cell lines after treatment with different concentrations of Erastin (L) or TBH (O). (M) Cell viability measured in the indicated RCC4 cells treated with 2 μM Erastin and 2 μM Ferr-1 for 24 h. (N) Cell death measured in the indicated RCC4 cells treated with 2 μM Erastin and 2 μM Ferr-1 for 24 h. (P and Q) Cell viability of the indicated RCC4 cells, measured after treatment with 100 μM TBH and 2 μM Ferr-1 for 24 h (P) or measured after culturing in cystine starvation conditions with or without 2 μM Ferr-1 for 48 h (Q). (R and T) Cell viability in the indicated RCC4 stable cell lines after treatment with different concentrations of Erastin (R) or TBH (T). (S and U) Cell viability measured in the indicated RCC4 cells treated with 2 μM Erastin (S) or 100 μM TBH (U) and 2 μM Ferr-1 for 24 h. (V) Cell viability of the indicated RCC4 cells measured upon culture under cystine starvation conditions for the indicated durations. (W) Cell viability of the indicated RCC4 cells, measured after culturing under cystine starvation conditions with or without 2 μM Ferr-1 for 48 h. (A–W) Error bars are means ± SEM, n = 3 biological replicates. (E) P values measured using two-tailed unpaired Student’s t test analysis. (J) P values measured using one-way ANOVA. (A–D, F–I, and K–W) P values determined by two-way ANOVA.
ZRANB1 promotes ferroptosis in renal cancer cells. (A and C) Cell viability in UMRC6 stable cell lines after treatment with different concentrations of Erastin (A) or TBH (C). (B and D) Bar graphs showing cell viability in UMRC6 cell lines treated with 20 μM Erastin (B) or 100 μM TBH (D) combined with 5 μM Z-VAD-fmk (Z-VAD), 2 μM Necrostatin-1s (Nec-1s), 2 μM Ferr-1, 100 μM DFO, or 10 μM Liprox. (E) Bar graph showing intracellular GSH levels in the indicated UMRC6 cells. (F) Lipid peroxidation was assessed by flow cytometry after C11-BODIPY staining in the indicated UMRC6 cells. (G) Cell death measured in the indicated UMRC6 cells after treatment with 2 μM Ferr-1 and 20 μM Erastin for 24 h. (H) Viability of UMRC6 cells cultured under cystine starvation conditions for the indicated duration. (I) Cell viability of the indicated UMRC6 cells, measured after culturing under cystine starvation conditions with or without 2 μM Ferr-1 for 48 h. (J) Bar graph showing intracellular GSH levels in the indicated RCC4 cells. (K) Lipid peroxidation was assessed by flow cytometry after C11-BODIPY staining in the indicated RCC4 cells. (L and O) Cell viability in RCC4 stable cell lines after treatment with different concentrations of Erastin (L) or TBH (O). (M) Cell viability measured in the indicated RCC4 cells treated with 2 μM Erastin and 2 μM Ferr-1 for 24 h. (N) Cell death measured in the indicated RCC4 cells treated with 2 μM Erastin and 2 μM Ferr-1 for 24 h. (P and Q) Cell viability of the indicated RCC4 cells, measured after treatment with 100 μM TBH and 2 μM Ferr-1 for 24 h (P) or measured after culturing in cystine starvation conditions with or without 2 μM Ferr-1 for 48 h (Q). (R and T) Cell viability in the indicated RCC4 stable cell lines after treatment with different concentrations of Erastin (R) or TBH (T). (S and U) Cell viability measured in the indicated RCC4 cells treated with 2 μM Erastin (S) or 100 μM TBH (U) and 2 μM Ferr-1 for 24 h. (V) Cell viability of the indicated RCC4 cells measured upon culture under cystine starvation conditions for the indicated durations. (W) Cell viability of the indicated RCC4 cells, measured after culturing under cystine starvation conditions with or without 2 μM Ferr-1 for 48 h. (A–W) Error bars are means ± SEM, n = 3 biological replicates. (E) P values measured using two-tailed unpaired Student’s t test analysis. (J) P values measured using one-way ANOVA. (A–D, F–I, and K–W) P values determined by two-way ANOVA.
Glutathione (GSH) biosynthesis originating from cystine could be utilized by GPX4 to detoxify lipid hydroperoxides and protect cells from ferroptosis (Koppula et al., 2018; Lim and Donaldson, 2011; Yang et al., 2014). As expected, we observed that stably expressed ZRANB1 significantly reduced GSH in cells (Fig. 2 E and Fig. S2 H) and increased lipid peroxidation (Fig. 2 F and Fig. S2 I), which eventually led to cell death (Fig. 2 G and Fig. S2 J). Moreover, ZRANB1 overexpression sensitized UMRC6 and 786-O cells to cystine depletion-induced cell death, which could be fully suppressed by Ferr-1 (Fig. 2, H and I; and Fig. S2 K). Conversely, depletion of ZRANB1 in RCC4 and ACHN cells (Fig. S2, L–N) increased GSH in cells (Fig. 2 J and Fig. S2 O), decreased lipid peroxidation (Fig. 2 K), and conferred enhanced resistance to Erastin- (Fig. 2, L–N; and Fig. S2, P–R), ROS- (Fig. 2, O and P; and Fig. S2, S and T), and cystine-depletion-induced ferroptosis (Fig. 2 Q and Fig. S2 U). In addition, we established stable cell lines by restoring ZRANB1 in ZRANB1-deficient cells (Fig. S3, A and B) and found that restoration of ZRANB1 indeed rescued the phenotype induced by ZRANB1 deficiency (Fig. 2 R–W; and Fig. S3 C–H).
Restoration of ZRANB1 resensitized tumor cells to ferroptosis. (A and B) Western blot analysis of ZRANB1 re-expression in ZRANB1-KO RCC4 (A) and ACHN (B) cells. (C and E) Cell viability in the indicated ACHN stable cell lines after treatment with different concentrations of Erastin (C) or TBH (E). (D and F) Cell viability measured in the indicated ACHN cells treated with 2 μM Erastin (D) or 100 μM TBH (F) and 2 μM Ferr-1 for 24 h. (G) Cell viability of the indicated ACHN cells measured upon culture under cystine starvation conditions for the indicated durations. (H) Cell viability of the indicated ACHN cells, measured after culturing under cystine starvation conditions with or without 2 μM Ferr-1 for 48 h. (C–H) Error bars are the means ± SEM, n = 3 biological replicates. (C–H) P values determined by two-way ANOVA. Source data are available for this figure: SourceData FS3.
Restoration of ZRANB1 resensitized tumor cells to ferroptosis. (A and B) Western blot analysis of ZRANB1 re-expression in ZRANB1-KO RCC4 (A) and ACHN (B) cells. (C and E) Cell viability in the indicated ACHN stable cell lines after treatment with different concentrations of Erastin (C) or TBH (E). (D and F) Cell viability measured in the indicated ACHN cells treated with 2 μM Erastin (D) or 100 μM TBH (F) and 2 μM Ferr-1 for 24 h. (G) Cell viability of the indicated ACHN cells measured upon culture under cystine starvation conditions for the indicated durations. (H) Cell viability of the indicated ACHN cells, measured after culturing under cystine starvation conditions with or without 2 μM Ferr-1 for 48 h. (C–H) Error bars are the means ± SEM, n = 3 biological replicates. (C–H) P values determined by two-way ANOVA. Source data are available for this figure: SourceData FS3.
We next sought to investigate the therapeutic potential of ZRANB1 for overcoming resistance to ferroptosis in vivo. First, 786-O cells were subcutaneously inoculated into nude mice, and 46 d after injection, the xenograft tumors were treated with IKE (23 mg kg−1) or Liprox (10 mg kg−1; a ferroptosis inhibitor), as indicated (Fig. 3 A). Although IKE treatment alone did not significantly affect the growth of the 786-O xenograft tumors in mice, ZRANB1 overexpression substantially sensitized the 786-O xenograft tumors to IKE treatment without any side effects; in particular, the cell death of the ZRANB1-overexpressing tumors treated with IKE was largely reversed by Liprox treatment (Fig. 3, B–E). In addition, MDA staining indicated that lipid peroxidation dramatically accumulated in the ZRANB1-overexpressing tumors but not in the mock tumor samples (Fig. 3, F–H). Transmission electron microscopy analysis revealed that cells from the ZRANB1-overexpressing xenograft tumors contained shrunken mitochondria, which is a potential morphological feature of ferroptosis (Fig. 3 I). Moreover, we treated ZRANB1-overexpressing tumors with Liprox and observed that a ferroptosis inhibitor blocked tumor suppression caused by ZRANB1, indicating that ZRANB1 acts as a tumor suppressor partially by regulating intrinsic ferroptosis (Fig. 3, J–M). Together, our results suggest that ZRANB1 determines tumor cell resistance to ferroptosis and that the combination of ZRANB1 with IKE has the potential to treat resistant tumors.
ZRANB1 promotes ferroptosis in vivo. (A) Schematic diagram for evaluating the therapeutic effects of ZRANB1 in a renal cancer xenograft model. (B) Image of xenograft tumors that were inoculated into nude mice with the indicated 786-O cell lines for 60 d. (C) Volumes of xenograft tumors with the indicated genotypes and treatments on different days. Error bars are means ± SEM, n = 5 mice per group. (D) Weight of tumor xenografts as indicated in B at the endpoint. Error bars are means ± SEM, n = 5 independent repeats. (E) Body weight of mice with the indicated genotypes and treatments on different days. Error bars are means ± SEM, n = 5 mice per group. (F and G) Hematoxylin and eosin (HE; F) and IHC staining (MDA; G) of tumor xenografts from the mock and ZRANB1-expressing cell lines. Scale bars, 20 μm. (H) Percentage of MDA-positive stained cells per field as indicated. Error bars are means ± SEM, n = 5. (I) Tumor samples from the mock and ZRANB1-expressing cell lines subjected to transmission electron microscopy. Red arrows indicate mitochondria with obvious cristae, while green arrows represent shrunken mitochondria. Scale bars: right, 500 nm. (J) Image of xenograft tumors that were inoculated into nude mice with the indicated 786-O cell lines for 50 d. (K) Volumes of xenograft tumors with the indicated genotypes and treatments on different days. Error bars are means ± SEM, n = 5 mice per group. (L) Weight of tumor xenografts as indicated in J at the endpoint. Error bars are means ± SEM, n = 5 independent repeats. (M) Body weight of mice with the indicated genotypes and treatments on different days. Error bars are means ± SEM, n = 5 mice per group. (C–E, H, and K–M) P values determined by two-way ANOVA.
ZRANB1 promotes ferroptosis in vivo. (A) Schematic diagram for evaluating the therapeutic effects of ZRANB1 in a renal cancer xenograft model. (B) Image of xenograft tumors that were inoculated into nude mice with the indicated 786-O cell lines for 60 d. (C) Volumes of xenograft tumors with the indicated genotypes and treatments on different days. Error bars are means ± SEM, n = 5 mice per group. (D) Weight of tumor xenografts as indicated in B at the endpoint. Error bars are means ± SEM, n = 5 independent repeats. (E) Body weight of mice with the indicated genotypes and treatments on different days. Error bars are means ± SEM, n = 5 mice per group. (F and G) Hematoxylin and eosin (HE; F) and IHC staining (MDA; G) of tumor xenografts from the mock and ZRANB1-expressing cell lines. Scale bars, 20 μm. (H) Percentage of MDA-positive stained cells per field as indicated. Error bars are means ± SEM, n = 5. (I) Tumor samples from the mock and ZRANB1-expressing cell lines subjected to transmission electron microscopy. Red arrows indicate mitochondria with obvious cristae, while green arrows represent shrunken mitochondria. Scale bars: right, 500 nm. (J) Image of xenograft tumors that were inoculated into nude mice with the indicated 786-O cell lines for 50 d. (K) Volumes of xenograft tumors with the indicated genotypes and treatments on different days. Error bars are means ± SEM, n = 5 mice per group. (L) Weight of tumor xenografts as indicated in J at the endpoint. Error bars are means ± SEM, n = 5 independent repeats. (M) Body weight of mice with the indicated genotypes and treatments on different days. Error bars are means ± SEM, n = 5 mice per group. (C–E, H, and K–M) P values determined by two-way ANOVA.
ZRANB1 associates with and degrades SLC7A11
ZRANB1 (also known as Trabid) is a deubiquitinating enzyme belonging to the ovarian tumor protease (OTU) family with several substrates, such as EZH2 (Zhang et al., 2018a), UVRAG (Feng et al., 2019), Jmjd2d (Jin et al., 2016), Twist (Zhu et al., 2019), VPS34 (Chen et al., 2021), and APC (Tran et al., 2008). As mentioned, recent reports have indicated that there are three major defense mechanisms for ferroptosis. To gain insights into how ZRANB1 regulates ferroptotic resistance, we assessed the expression levels of TFRC, LPCAT3, ACSL4, FSP1, DHODH, GPX4, and SLC7A11 following the overexpression of ZRANB1 and found that ZRANB1 does not regulate the TFRC, LPCAT3, ACSL4, FSP1, DHODH, and GPX4 protein levels (Fig. 4 A and Fig. S4 A). It should be noted that overexpression of ZRANB1 had no effect on ML162- and RAS-selective-lethal-3 (RSL3)–induced cell death (Fig. 4, B–E; and Fig. S4, B–E), implying that this regulation occurs upstream of GPX4. Given that cancer cells predominantly obtain cystine for GSH synthesis through SLC7A11 (Dixon et al., 2012; Sato et al., 1999), whose expression has been reported to dynamically correlate with ferroptosis (Koppula et al., 2021), the findings described in Fig. 1 B prompted us to investigate the mechanistic connection between ZRANB1 and SLC7A11.
ZRANB1 binds and downregulates the SLC7A11 protein. (A) Western blot analysis of TFRC, LPCAT3, ACSL4, FSP1, DHODH, GPX4, and SLC7A11 expression in ZRANB1-overexpressing UMRC6 cells. (B and D) Cell viability in the indicated UMRC6 stable cell lines after treatment with different concentrations of ML162 (B) or RSL3 (D). (C) Cell viability of the indicated UMRC6 cells treated with 2 μM ML162 with or without 2 μM Ferr-1 for 2 h. (E) Cell viability of the indicated UMRC6 cells, measured after culturing in 2 μM RSL3 with or without 2 μM Ferr-1 for 2 h. (F) SLC7A11 expression levels in the indicated UMRC6 cells were measured by western blot. (G) SLC7A11 expression levels in the indicated UMRC6 cells were measured by quantitative RT-PCR. (H) Western blot analysis of ZRANB1 expression in ZRANB1-KO RCC4 cells. (I) Levels of SLC7A11 protein in HeLa-ZRANB1-KO cells re-overexpressing ZRANB1 were measured by western blot. (J) IF staining of SLC7A11 (red) and ZRANB1 (green) in representative human KIRC samples. The right images are the overlay of SLC7A11, ZRANB1, and nuclear DAPI (blue) staining. Scale bars, 20 μm. (K) Correlation between SLC7A11 and ZRANB1 protein levels in KIRC patients. R is the Spearman correlation coefficient. The P value was obtained from a chi-square test. (L–N) Coimmunoprecipitation of endogenous SLC7A11 with ZRANB1. SLC7A11 was immunoprecipitated from HeLa (L), UMRC6 (M), or RCC4 (N) cells, followed by immunoblotting with antibodies against ZRANB1. (O) Western blot analysis of endogenous SLC7A11 after IP of endogenous ZRANB1 from HeLa cells. (P) Left panel: Purified His-ZRANB1 was incubated with purified GST-SLC7A11 and subjected to GST pull-down followed by immunoblotting with the indicated antibodies. Right panel: Purified recombinant His-ZRANB1 and GST-SLC7A11 proteins were analyzed by SDS-PAGE and Coomassie blue staining. (B–E and G) Error bars are means ± SEM, n = 3 biological replicates. (G) P values measured using two-tailed unpaired Student’s t test analysis. (B–E) P values determined by two-way ANOVA. Source data are available for this figure: SourceData F4.
ZRANB1 binds and downregulates the SLC7A11 protein. (A) Western blot analysis of TFRC, LPCAT3, ACSL4, FSP1, DHODH, GPX4, and SLC7A11 expression in ZRANB1-overexpressing UMRC6 cells. (B and D) Cell viability in the indicated UMRC6 stable cell lines after treatment with different concentrations of ML162 (B) or RSL3 (D). (C) Cell viability of the indicated UMRC6 cells treated with 2 μM ML162 with or without 2 μM Ferr-1 for 2 h. (E) Cell viability of the indicated UMRC6 cells, measured after culturing in 2 μM RSL3 with or without 2 μM Ferr-1 for 2 h. (F) SLC7A11 expression levels in the indicated UMRC6 cells were measured by western blot. (G) SLC7A11 expression levels in the indicated UMRC6 cells were measured by quantitative RT-PCR. (H) Western blot analysis of ZRANB1 expression in ZRANB1-KO RCC4 cells. (I) Levels of SLC7A11 protein in HeLa-ZRANB1-KO cells re-overexpressing ZRANB1 were measured by western blot. (J) IF staining of SLC7A11 (red) and ZRANB1 (green) in representative human KIRC samples. The right images are the overlay of SLC7A11, ZRANB1, and nuclear DAPI (blue) staining. Scale bars, 20 μm. (K) Correlation between SLC7A11 and ZRANB1 protein levels in KIRC patients. R is the Spearman correlation coefficient. The P value was obtained from a chi-square test. (L–N) Coimmunoprecipitation of endogenous SLC7A11 with ZRANB1. SLC7A11 was immunoprecipitated from HeLa (L), UMRC6 (M), or RCC4 (N) cells, followed by immunoblotting with antibodies against ZRANB1. (O) Western blot analysis of endogenous SLC7A11 after IP of endogenous ZRANB1 from HeLa cells. (P) Left panel: Purified His-ZRANB1 was incubated with purified GST-SLC7A11 and subjected to GST pull-down followed by immunoblotting with the indicated antibodies. Right panel: Purified recombinant His-ZRANB1 and GST-SLC7A11 proteins were analyzed by SDS-PAGE and Coomassie blue staining. (B–E and G) Error bars are means ± SEM, n = 3 biological replicates. (G) P values measured using two-tailed unpaired Student’s t test analysis. (B–E) P values determined by two-way ANOVA. Source data are available for this figure: SourceData F4.
ZRANB1 interacts with SLC7A11. (A) Western blot analysis of TFRC, LPCAT3, ACSL4, FSP1, DHODH, GPX4, and SLC7A11 expression in ZRANB1-overexpressing 786-O cells. (B and D) Cell viability in the indicated 786-O stable cell lines after treatment with different concentrations of ML162 (B) or RSL3 (D). (C) Cell viability of the indicated 786-O cells treated with 2 μM ML162 with or without 2 μM Ferr-1 for 2 h. (E) Cell viability of the indicated 786-O cells, measured after culturing in 2 μM RSL3 with or without 2 μM Ferr-1 for 2 h. (F) SLC7A11 expression levels in the indicated 786-O cells were measured by western blot. (G) SLC7A11 expression levels in the indicated 786-O cells were measured by quantitative RT-PCR. (H) Western blot analysis of ZRANB1 expression in ZRANB1-KO ACHN cells. (I) Purified S-Tag-SLC7A11 (from HEK293T cells) was incubated with purified His-MYC-ZRANB1, followed by pull-down with nickel beads and immunoblotting with antibodies against SLC7A11 and MYC. (J–L) Coimmunoprecipitation of endogenous SLC7A11 with ZRANB1. SLC7A11 was immunoprecipitated from HEK293T (J), 786-O (K), or ACHN (L) cells, followed by immunoblotting with antibodies against ZRANB1 and SLC7A11. (M) Western blot analysis for SLC7A11 after pull-down with S-protein beads from HEK293T cells cotransfected with MYC-S-Tag-ZRANB1 and FLAG-SLC7A11 individually or together. (N) Western blot analysis of endogenous SLC7A11 after IP of endogenous ZRANB1 from HEK293T cells. (G) P values measured using two-tailed unpaired Student’s t test analysis. (B–E and G) Error bars are the means ± SEM, n = 3 biological replicates. (B–E) P values determined by two-way ANOVA. Source data are available for this figure: SourceData FS4.
ZRANB1 interacts with SLC7A11. (A) Western blot analysis of TFRC, LPCAT3, ACSL4, FSP1, DHODH, GPX4, and SLC7A11 expression in ZRANB1-overexpressing 786-O cells. (B and D) Cell viability in the indicated 786-O stable cell lines after treatment with different concentrations of ML162 (B) or RSL3 (D). (C) Cell viability of the indicated 786-O cells treated with 2 μM ML162 with or without 2 μM Ferr-1 for 2 h. (E) Cell viability of the indicated 786-O cells, measured after culturing in 2 μM RSL3 with or without 2 μM Ferr-1 for 2 h. (F) SLC7A11 expression levels in the indicated 786-O cells were measured by western blot. (G) SLC7A11 expression levels in the indicated 786-O cells were measured by quantitative RT-PCR. (H) Western blot analysis of ZRANB1 expression in ZRANB1-KO ACHN cells. (I) Purified S-Tag-SLC7A11 (from HEK293T cells) was incubated with purified His-MYC-ZRANB1, followed by pull-down with nickel beads and immunoblotting with antibodies against SLC7A11 and MYC. (J–L) Coimmunoprecipitation of endogenous SLC7A11 with ZRANB1. SLC7A11 was immunoprecipitated from HEK293T (J), 786-O (K), or ACHN (L) cells, followed by immunoblotting with antibodies against ZRANB1 and SLC7A11. (M) Western blot analysis for SLC7A11 after pull-down with S-protein beads from HEK293T cells cotransfected with MYC-S-Tag-ZRANB1 and FLAG-SLC7A11 individually or together. (N) Western blot analysis of endogenous SLC7A11 after IP of endogenous ZRANB1 from HEK293T cells. (G) P values measured using two-tailed unpaired Student’s t test analysis. (B–E and G) Error bars are the means ± SEM, n = 3 biological replicates. (B–E) P values determined by two-way ANOVA. Source data are available for this figure: SourceData FS4.
To investigate whether ZRANB1 regulates SLC7A11 protein levels, we overexpressed ZRANB1 in UMRC6 and 786-O cells and found that ZRANB1 significantly downregulated SLC7A11 protein levels (Fig. 4, A and F; and Fig. S4, A and F) without changing the mRNA level of SLC7A11 (Fig. 4 G and Fig. S4 G). Conversely, depleting ZRANB1 expression in RCC4 and ACHN cells (Fig. 4 H and Fig. S4 H) by two independent gRNAs dramatically increased endogenous SLC7A11 protein levels, which could be completely reversed by co-overexpression of ZRANB1 (Fig. 4 I). To further confirm that the regulation of SLC7A11 by ZRANB1 is relevant in human cancer patients, we stained the two proteins on 150 human KIRC tissue microarrays. Consistently, 76% (77/101) of the tumor samples with low ZRANB1 expression exhibited high SLC7A11 expression and 71% (35/49) of the tumor samples with high ZRANB1 expression exhibited low SLC7A11 expression (R = −0.2304, P < 0.0001; Fig. 4, J and K). Coimmunoprecipitation assays confirmed that either exogenous or endogenous ZRANB1 could be detected in SLC7A11 immunoprecipitates (Fig. 4, L–N; and Fig. S4, I–L) and that SLC7A11 was present in ZRANB1 immunoprecipitates (Fig. 4 O and Fig. S4, M and N). Moreover, purified GST-SLC7A11 bound to His-ZRANB1 in a cell-free system (Fig. 4 P), demonstrating a direct interaction between ZRANB1 and SLC7A11.
ZRANB1 ubiquitinates SLC7A11 as an E3 ligase
To determine whether ZRANB1 destabilizes SLC7A11, we treated the cells with cycloheximide (CHX), an inhibitor of protein synthesis, and examined the protein level of endogenous SLC7A11. As expected, overexpression of ZRANB1 markedly shortened the half-life of SLC7A11 (Fig. 5 A and Fig. S5 A); conversely, depletion of ZRANB1 led to a prominent increase in the basal level and half-life of SLC7A11 (Fig. 5 B and Fig. S5, B–G). Based on the data described above, we reasoned that ZRANB1 may indirectly regulate SLC7A11 through its deubiquitinating activity, for example, by directly stabilizing an E3 ligase of SLC7A11. To test this hypothesis, we transiently transfected wild-type (WT) ZRANB1 and its dead mutant lacking DUB activity, C443A ZRANB1, into HEK293T and HeLa cells. Surprisingly, either the WT or dead mutant ZRANB1 abrogated the expression of endogenous SLC7A11 protein (Fig. 5 C and Fig. S5 H), suggesting that ZRANB1 may directly destabilize SLC7A11, which is independent of its DUB enzyme activity.
ZRANB1 atypically degrades SLC7A11. (A) HEK293T cells were cotransfected with V5-SLC7A11, SFB-GFP, and MYC-ZRANB1 expression vectors, treated with 50 μg/ml CHX, collected at different time points and then immunoblotted with MYC, FLAG, and SLC7A11 antibodies. SFB-GFP served as the control for transfection. (B) HeLa-ZRANB1-KO cells were treated with 50 μg/ml CHX, harvested at different time points, and then immunoblotted with antibodies against SLC7A11 and β-actin. (C) Western blot analysis of SLC7A11 and HA-ZRANB1 from ZRANB1-knockdown HEK293T cells cotransfected with SLC7A11 and HA-ZRANB1 (WT and C443A catalytic mutant)-expressing vectors. (D) SLC7A11 expression in ZRANB1-overexpressing cells treated with or without the proteasome inhibitor MG132. (E) ZRANB1-KO HEK293T cells were cotransfected with equal amounts of S-Tag-SLC7A11 and HA-ubiquitin (HA-Ub), together with or without FLAG-ZRANB1, followed by pull-down with S-protein beads and immunoblotting with antibodies against SLC7A11, HA, and FLAG. (F) ZRANB1-KO HEK293T cells were cotransfected with equal amounts of MYC-SLC7A11 and HA-Ub together with or without MYC-ZRANB1, followed by IP with HA beads and immunoblotting with antibodies against SLC7A11, HA, and MYC. (G) In vitro purified SLC7A11 protein was incubated in the presence of purified ZRANB1, ubiquitin-activating enzyme E1, ubiquitin-conjugating enzyme E2, and ubiquitin (Ub) for the in vitro ubiquitination assay. (H) Schematic diagram of full-length ZRANB1 protein and its various truncates. (I) ZRANB1-KO HeLa cells were transfected with SFB-tagged full-length ZRANB1 or its truncates. 48 h after transfection, cells were harvested and immunoblotted with antibodies against SLC7A11, β-actin, and FLAG. (J) In vitro purified SLC7A11 protein was incubated in the presence of purified ZRANB1 or ZRANB1-M3, ubiquitin-activating enzyme E1, ubiquitin-conjugating enzyme E2 and ubiquitin (Ub) for the in vitro ubiquitination assay. (K) Schematic diagram of various ZRANB1 protein truncates (M3, M3-1, and M3-2). (L) HEK293T cells were transfected with MYC-tagged full-length (FL) ZRANB1 or its truncates. 48 h after transfection, cells were harvested and immunoblotted with antibodies against SLC7A11, MYC, and FLAG. (M) Schematic diagram of various ZRANB1 protein truncates (M3-2, M3-2-a, M3-2-b, and M3-2-c). (N) The ZRANB1 OTU domain catalyzes in vitro autopolyubiquitination. ZRANB1 and its truncates were purified from E. coli. (O) Schematic diagram of the full-length ZRANB1 protein and its truncation mutant (ΔM3-2-b). (P) The autoubiquitination ability of ΔM3-2-b in vitro. ZRANB1 and its truncate were purified from E. coli. Source data are available for this figure: SourceData F5.
ZRANB1 atypically degrades SLC7A11. (A) HEK293T cells were cotransfected with V5-SLC7A11, SFB-GFP, and MYC-ZRANB1 expression vectors, treated with 50 μg/ml CHX, collected at different time points and then immunoblotted with MYC, FLAG, and SLC7A11 antibodies. SFB-GFP served as the control for transfection. (B) HeLa-ZRANB1-KO cells were treated with 50 μg/ml CHX, harvested at different time points, and then immunoblotted with antibodies against SLC7A11 and β-actin. (C) Western blot analysis of SLC7A11 and HA-ZRANB1 from ZRANB1-knockdown HEK293T cells cotransfected with SLC7A11 and HA-ZRANB1 (WT and C443A catalytic mutant)-expressing vectors. (D) SLC7A11 expression in ZRANB1-overexpressing cells treated with or without the proteasome inhibitor MG132. (E) ZRANB1-KO HEK293T cells were cotransfected with equal amounts of S-Tag-SLC7A11 and HA-ubiquitin (HA-Ub), together with or without FLAG-ZRANB1, followed by pull-down with S-protein beads and immunoblotting with antibodies against SLC7A11, HA, and FLAG. (F) ZRANB1-KO HEK293T cells were cotransfected with equal amounts of MYC-SLC7A11 and HA-Ub together with or without MYC-ZRANB1, followed by IP with HA beads and immunoblotting with antibodies against SLC7A11, HA, and MYC. (G) In vitro purified SLC7A11 protein was incubated in the presence of purified ZRANB1, ubiquitin-activating enzyme E1, ubiquitin-conjugating enzyme E2, and ubiquitin (Ub) for the in vitro ubiquitination assay. (H) Schematic diagram of full-length ZRANB1 protein and its various truncates. (I) ZRANB1-KO HeLa cells were transfected with SFB-tagged full-length ZRANB1 or its truncates. 48 h after transfection, cells were harvested and immunoblotted with antibodies against SLC7A11, β-actin, and FLAG. (J) In vitro purified SLC7A11 protein was incubated in the presence of purified ZRANB1 or ZRANB1-M3, ubiquitin-activating enzyme E1, ubiquitin-conjugating enzyme E2 and ubiquitin (Ub) for the in vitro ubiquitination assay. (K) Schematic diagram of various ZRANB1 protein truncates (M3, M3-1, and M3-2). (L) HEK293T cells were transfected with MYC-tagged full-length (FL) ZRANB1 or its truncates. 48 h after transfection, cells were harvested and immunoblotted with antibodies against SLC7A11, MYC, and FLAG. (M) Schematic diagram of various ZRANB1 protein truncates (M3-2, M3-2-a, M3-2-b, and M3-2-c). (N) The ZRANB1 OTU domain catalyzes in vitro autopolyubiquitination. ZRANB1 and its truncates were purified from E. coli. (O) Schematic diagram of the full-length ZRANB1 protein and its truncation mutant (ΔM3-2-b). (P) The autoubiquitination ability of ΔM3-2-b in vitro. ZRANB1 and its truncate were purified from E. coli. Source data are available for this figure: SourceData F5.
ZRANB1 regulates SLC7A11 protein stability. (A and C) Quantification of SLC7A11 protein levels (normalized to SFB-GFP) in HEK293T cells corresponding to Fig. 5 A or (normalized to β-actin) in the indicated HeLa cells corresponding to Fig. 5 B. (B) Levels of SLC7A11 protein in HeLa-ZRANB1-Mock/KO cells were measured by western blot. (D and F) RCC4 (D) or ACHN (F) -ZRANB1-KO cells were treated with 50 μg/ml CHX, harvested at different time points, and then immunoblotted with antibodies against SLC7A11 and β-actin. (E and G) Quantification of SLC7A11 protein levels (normalized to β-actin) in the indicated RCC4 (E) or ACHN (G) cells corresponding to D and F. (H) Western blot analysis of SLC7A11 and MYC-ZRANB1 in HeLa cells transfected with MYC-ZRANB1 (WT and C443A) -expressing vectors. (I) Change in the level of SLC7A11 after treatment with the lysosome inhibitor ChlorD. (J) Screening of ubiquitin-conjugating E2 for ubiquitination of SLC7A11. (K) HEK293T cells were cotransfected with MYC-SLC7A11 and FLAG-tagged full-length (FL) ZRANB1 or its truncates, followed by pull-down with anti-S-tag beads and immunoblotting with antibodies against SLC7A11 and FLAG. (L) Purified His-ZRANB1-M3 was incubated with purified GST-SLC7A11 and subjected to GST pull-down followed by immunoblotting. (M) ZRANB1-KO HeLa cells were transfected with MYC-tagged full-length ZRANB1 or its truncates. 48 h after transfection, cells were harvested and immunoblotted with antibodies against SLC7A11, β-actin, and MYC. (N) HEK293T cells were transfected with MYC-tagged full-length ZRANB1 or its truncates. 48 h after transfection, cells were harvested and immunoblotted with antibodies against SLC7A11, MYC, and FLAG. (O) Western blot analysis for SLC7A11 and MYC-ZRANB1 or its truncate from HEK293T cells transfected with MYC-ZRANB1 (full length and ΔM3-2-b) and V5-SLC7A11 expressing vectors. (A, C, E, and G) Error bars are the means ± SEM, n = 3 biological replicates. Source data are available for this figure: SourceData FS5.
ZRANB1 regulates SLC7A11 protein stability. (A and C) Quantification of SLC7A11 protein levels (normalized to SFB-GFP) in HEK293T cells corresponding to Fig. 5 A or (normalized to β-actin) in the indicated HeLa cells corresponding to Fig. 5 B. (B) Levels of SLC7A11 protein in HeLa-ZRANB1-Mock/KO cells were measured by western blot. (D and F) RCC4 (D) or ACHN (F) -ZRANB1-KO cells were treated with 50 μg/ml CHX, harvested at different time points, and then immunoblotted with antibodies against SLC7A11 and β-actin. (E and G) Quantification of SLC7A11 protein levels (normalized to β-actin) in the indicated RCC4 (E) or ACHN (G) cells corresponding to D and F. (H) Western blot analysis of SLC7A11 and MYC-ZRANB1 in HeLa cells transfected with MYC-ZRANB1 (WT and C443A) -expressing vectors. (I) Change in the level of SLC7A11 after treatment with the lysosome inhibitor ChlorD. (J) Screening of ubiquitin-conjugating E2 for ubiquitination of SLC7A11. (K) HEK293T cells were cotransfected with MYC-SLC7A11 and FLAG-tagged full-length (FL) ZRANB1 or its truncates, followed by pull-down with anti-S-tag beads and immunoblotting with antibodies against SLC7A11 and FLAG. (L) Purified His-ZRANB1-M3 was incubated with purified GST-SLC7A11 and subjected to GST pull-down followed by immunoblotting. (M) ZRANB1-KO HeLa cells were transfected with MYC-tagged full-length ZRANB1 or its truncates. 48 h after transfection, cells were harvested and immunoblotted with antibodies against SLC7A11, β-actin, and MYC. (N) HEK293T cells were transfected with MYC-tagged full-length ZRANB1 or its truncates. 48 h after transfection, cells were harvested and immunoblotted with antibodies against SLC7A11, MYC, and FLAG. (O) Western blot analysis for SLC7A11 and MYC-ZRANB1 or its truncate from HEK293T cells transfected with MYC-ZRANB1 (full length and ΔM3-2-b) and V5-SLC7A11 expressing vectors. (A, C, E, and G) Error bars are the means ± SEM, n = 3 biological replicates. Source data are available for this figure: SourceData FS5.
Two major systems have been established for protein turnover in organisms: the ubiquitin‒proteasome pathway (UPP) and the autophagic lysosomal pathway. To clarify which pathway is responsible for regulation, we treated ZRANB1-overexpressing HEK293T cells with either MG132, an inhibitor of the proteasome pathway, or chloroquine (CQ), an inhibitor of the autophagic lysosomal pathway. Interestingly, the downregulation of SLC7A11 expression by ZRANB1 was completely reversed by MG132 (Fig. 5 D) but not CQ treatment (Fig. S5 I), suggesting that ZRANB1 could degrade SLC7A11 in a 26S UPP-dependent manner. Indeed, overexpression of ZRANB1 substantially increased SLC7A11 polyubiquitination under denaturing conditions (Fig. 5, E and F). A20 (an OTU family DUB) has been established as the only protein with DUB activity in the N-terminus and E3 ligase activity in the C-terminus (Wertz et al., 2004). To further determine whether ZRANB1 directly ubiquitinates SLC7A11 as an E3 ligase, we first screened a panel of ubiquitin-conjugating enzymes (E2) for ZRANB1-induced SLC7A11 ubiquitination and found that UBE2G1 is a potential E2 for ZRANB1 (Fig. S5 J). We next confirmed that ZRANB1 ubiquitinates SLC7A11. In vitro purified ubiquitin-activating enzyme (E1), E2, ubiquitin, and ZRANB1 (tentative E3) were incubated with SLC7A11 (substrate) in a cell-free system. Purified ZRANB1 markedly induced SLC7A11 polyubiquitination in vitro (Fig. 5 G), suggesting that SLC7A11 is a direct substrate of ZRANB1, which is a potential E3 ligase.
To further define the functional domain of the ZRANB1 E3 ligase for SLC7A11, we transfected full-length ZRANB1 and three truncated forms (Zhang et al., 2018a) into HEK293T cells (Fig. 5 H) and found that the OTU domain not only interacted with SLC7A11 (Fig. S5, K and L) but was also required for the degradation and ubiquitination of SLC7A11 by ZRANB1 (Fig. 5, I and J). Next, we divided the M3 domain into two truncated domains, namely, M3-1 and M3-2, based on the indicated structural domains (Fig. 5 K). Interestingly, the results showed that the M3-2 region, the OTU structural domain, played a key role in the stabilization of SLC7A11 (Fig. 5 L and Fig. S5 M). We further divided the M3-2 structural domain into three truncates (∼100 amino acids/truncate, M3-2-a, M3-2-b, and M3-2-c; Fig. 5 M). Then, we checked the role of WT-ZRANB1 and each truncation mutant in the stabilization of SLC7A11 and the autoubiquitination ability of ZRANB1. Remarkably, despite the absence of typical ubiquitin transfer domains, such as RING finger and HECT domains, we found that recombinant ZRANB1 from Escherichia coli (E. coli) catalyzed in vitro polyubiquitination and the M3-2-b (residues 463–584) truncation was the functional domain responsible for the activity of the E3 ligase (Fig. 5 N and Fig. S5 N). After deleting this specific region, the ZRANB1 protein lost its E3 ligase activity (Fig. 5, O and P; and Fig. S5 O). Together, ZRANB1 is an atypical E3 ligase for SLC7A11 polyubiquitination and the region (residues 463–584) may be the functional domain.
ZRANB1 promotes ferroptosis via SLC7A11
We next investigated whether SLC7A11 is a functional effector of ZRANB1. Ectopic expression of ZRANB1 in both UMRC6 (Fig. 6 A) and 786-O (Fig. S6 A) cells markedly downregulated SLC7A11 protein expression, inhibited GSH synthesis and promoted ROS-, cystine-depletion-, and Erastin-induced ferroptosis, which could be largely reversed by restoration of SLC7A11 (Fig. 6, B–J; and Fig. S6, B–H). Conversely, depletion of ZRANB1 in both RCC4 (Fig. 6 K) and ACHN (Fig. S6 I) cells resulted in high SLC7A11 protein levels, increased GSH synthesis, and conferred resistance to ROS-, cystine-deprivation-, and Erastin-induced ferroptosis in these two manipulated cell lines. As expected, knockdown of SLC7A11 in ZRANB1 knockout (KO) cells resensitized these cells to ferroptosis (Fig. 6, L–S; and Fig. S6, J–Q), suggesting that acquired ferroptosis resistance in cells is mainly mediated by the ZRANB1-SLC7A11 axis.
ZRANB1 promotes ferroptosis through SLC7A11. (A) Western blot analysis of ZRANB1 and SLC7A11 in the indicated UMRC6 cell lines. (B) Bar graphs showing intracellular GSH levels in the indicated UMRC6 cells. (C and G) Cell viability in UMRC6 stable cell lines after treatment with different concentrations of TBH (C) or Erastin (G). (D, F, and H) Bar graphs showing the viability of the indicated UMRC6 cells treated with 100 μM TBH (D), cultured under cystine starvation conditions for 2 d (F), or treated with 20 μM Erastin (H) with or without 2 μM Ferr-1. (E) Viability of UMRC6 cells cultured under cystine starvation conditions for the indicated durations. (I) Representative phase-contrast images of the indicated UMRC6 cells treated with 20 μM Erastin for 30 h. Scale bars, 50 μm. (J) Bar graphs showing cell death in the indicated UMRC6 cell lines following Erastin treatment. (K) Western blot analysis of SLC7A11 in ZRANB1-KO RCC4 cell lines with or without knockdown of SLC7A11. (L) Bar graphs showing intracellular GSH levels in the indicated RCC4 cells. (M and Q) Cell viability in RCC4 stable cell lines after treatment with different concentrations of TBH (M) or Erastin (Q). (N and P) Bar graphs showing the viability of the indicated RCC4 cells treated with 100 μM TBH (N) or cultured under cystine starvation conditions for 2 d (P) with or without 2 μM Ferr-1. (O) Cell viability of RCC4 cells measured upon culture under cystine starvation conditions for the indicated durations. (R and S) Bar graphs showing the viability (R) or cell death (S) of the indicated RCC4 cells treated with 10 μM Erastin with or without 2 μM Ferr-1. (B–H, J, and L–S) Error bars are means ± SEM, n = 3 biological replicates. (B and L) P values measured using one-way ANOVA. (C–H, J, and M–S) P values determined by two-way ANOVA. Source data are available for this figure: SourceData F6.
ZRANB1 promotes ferroptosis through SLC7A11. (A) Western blot analysis of ZRANB1 and SLC7A11 in the indicated UMRC6 cell lines. (B) Bar graphs showing intracellular GSH levels in the indicated UMRC6 cells. (C and G) Cell viability in UMRC6 stable cell lines after treatment with different concentrations of TBH (C) or Erastin (G). (D, F, and H) Bar graphs showing the viability of the indicated UMRC6 cells treated with 100 μM TBH (D), cultured under cystine starvation conditions for 2 d (F), or treated with 20 μM Erastin (H) with or without 2 μM Ferr-1. (E) Viability of UMRC6 cells cultured under cystine starvation conditions for the indicated durations. (I) Representative phase-contrast images of the indicated UMRC6 cells treated with 20 μM Erastin for 30 h. Scale bars, 50 μm. (J) Bar graphs showing cell death in the indicated UMRC6 cell lines following Erastin treatment. (K) Western blot analysis of SLC7A11 in ZRANB1-KO RCC4 cell lines with or without knockdown of SLC7A11. (L) Bar graphs showing intracellular GSH levels in the indicated RCC4 cells. (M and Q) Cell viability in RCC4 stable cell lines after treatment with different concentrations of TBH (M) or Erastin (Q). (N and P) Bar graphs showing the viability of the indicated RCC4 cells treated with 100 μM TBH (N) or cultured under cystine starvation conditions for 2 d (P) with or without 2 μM Ferr-1. (O) Cell viability of RCC4 cells measured upon culture under cystine starvation conditions for the indicated durations. (R and S) Bar graphs showing the viability (R) or cell death (S) of the indicated RCC4 cells treated with 10 μM Erastin with or without 2 μM Ferr-1. (B–H, J, and L–S) Error bars are means ± SEM, n = 3 biological replicates. (B and L) P values measured using one-way ANOVA. (C–H, J, and M–S) P values determined by two-way ANOVA. Source data are available for this figure: SourceData F6.
ZRANB1 regulates SLC7A11, influencing sensitivity to ferroptosis. (A) Western blot analysis of ZRANB1 and SLC7A11 in the indicated 786-O cell lines. (B) Bar graphs showing intracellular GSH levels in the indicated 786-O cells. (C and E) Cell viability in 786-O stable cell lines after treatment with different concentrations of TBH (C) or culture under cystine starvation conditions for the indicated durations (E). (D and F) Bar graphs showing cell death in the indicated 786-O cells induced by 100 μM TBH with or without 2 μM Ferr-1 (D) or cultured under cystine starvation conditions for 2 d with or without 2 μM Ferr-1 (F). (G) Representative phase-contrast images of the indicated 786-O cells treated with 10 μM Erastin for 30 h. Scale bars, 50 μm. (H) Bar graphs showing cell death in the indicated 786-O cell lines following Erastin treatment. (I) Western blot analysis of SLC7A11 in ZRANB1-KO ACHN cell lines with or without knockdown of SLC7A11. (J) Histograms showing intracellular GSH levels in the indicated ACHN cells. (K and O) Cell viability in ACHN stable cell lines after treatment with different concentrations of TBH (K) or Erastin (O). (L and N) Histograms showing the viability of the indicated ACHN cells treated with 100 μM TBH with or without 2 μM Ferr-1 (L) or cultured under cystine starvation conditions for 2 d with or without 2 μM Ferr-1 (N). (M) Cell viability of ACHN cells measured upon culture under cystine starvation conditions for the indicated durations. (P) Histograms showing the viability of the indicated ACHN cells treated with 10 μM Erastin with or without 2 μM Ferr-1. (Q) Histograms showing cell death in the indicated ACHN cell lines after 10 μM Erastin treatment with or without 10 μM Ferr-1. (B and J) P values measured using one-way ANOVA. (B–F, H, and J–Q) Error bars are the means ± SEM, n = 3 biological replicates. (C–F, H, and K–Q) P values determined by two-way ANOVA. Source data are available for this figure: SourceData FS6.
ZRANB1 regulates SLC7A11, influencing sensitivity to ferroptosis. (A) Western blot analysis of ZRANB1 and SLC7A11 in the indicated 786-O cell lines. (B) Bar graphs showing intracellular GSH levels in the indicated 786-O cells. (C and E) Cell viability in 786-O stable cell lines after treatment with different concentrations of TBH (C) or culture under cystine starvation conditions for the indicated durations (E). (D and F) Bar graphs showing cell death in the indicated 786-O cells induced by 100 μM TBH with or without 2 μM Ferr-1 (D) or cultured under cystine starvation conditions for 2 d with or without 2 μM Ferr-1 (F). (G) Representative phase-contrast images of the indicated 786-O cells treated with 10 μM Erastin for 30 h. Scale bars, 50 μm. (H) Bar graphs showing cell death in the indicated 786-O cell lines following Erastin treatment. (I) Western blot analysis of SLC7A11 in ZRANB1-KO ACHN cell lines with or without knockdown of SLC7A11. (J) Histograms showing intracellular GSH levels in the indicated ACHN cells. (K and O) Cell viability in ACHN stable cell lines after treatment with different concentrations of TBH (K) or Erastin (O). (L and N) Histograms showing the viability of the indicated ACHN cells treated with 100 μM TBH with or without 2 μM Ferr-1 (L) or cultured under cystine starvation conditions for 2 d with or without 2 μM Ferr-1 (N). (M) Cell viability of ACHN cells measured upon culture under cystine starvation conditions for the indicated durations. (P) Histograms showing the viability of the indicated ACHN cells treated with 10 μM Erastin with or without 2 μM Ferr-1. (Q) Histograms showing cell death in the indicated ACHN cell lines after 10 μM Erastin treatment with or without 10 μM Ferr-1. (B and J) P values measured using one-way ANOVA. (B–F, H, and J–Q) Error bars are the means ± SEM, n = 3 biological replicates. (C–F, H, and K–Q) P values determined by two-way ANOVA. Source data are available for this figure: SourceData FS6.
Consistently, we found that overexpression of ZRANB1 alone inhibited xenograft tumor development and that restoration of SLC7A11 in ZRANB1-overexpressing cells partially rescued the phenotype (Fig. 7, A–C; and Fig. S7 A). Conversely, loss of ZRANB1 promoted tumor development, which could be significantly reversed by deletion of SLC7A11 expression (Fig. 7, D–F; and Fig. S7, B–D). Notably, SLC7A11 staining in some areas of tumors was highly enriched compared with that in other areas, which was negatively correlated with ZRANB1 (Fig. 7 G upper panel and Fig. 7 H). Interestingly, staining of SLC7A11 and MDA in different sites of human tumor samples showed a similar correlation. More importantly, most SLC7A11 staining was dramatically attenuated by overexpression of ZRANB1 (Fig. 7 G lower panel and Fig. 7 H), suggesting that ZRANB1 negatively regulated SLC7A11 and led to accumulated lipid peroxidation. In addition, tumors with high ZRANB1 expression also exhibited high MDA staining, which was negatively correlated with SLC7A1 expression in the animal model and human KIRC samples (Fig. 7, I and J), indicating that ZRANB1 inhibits KIRC development through destabilization of SLC7A11 and ferroptosis. Finally, we found that the expression level of ZRANB1 was lower in kidney cancer tissues than in adjacent normal tissues (Fig. 7 K and Fig. S7 E) and negatively correlated with the stage of tumor development (Fig. 7 L), and the prognosis of patients with high ZRANB1 expression was better than that of patients with low ZRANB1 expression (Fig. 7 M and Fig. S7 F). Together, our results suggested that low expression of ZRANB1 in the tumor eliminated the production of lipid peroxidation by enhancing SLC7A11 and eventually led to tumor cell resistance to ferroptosis.
ZRANB1 inhibits tumor development partly through SLC7A11 and ferroptosis. (A) Image of xenograft tumors that were inoculated into nude mice with the indicated 786-O cell lines for 60 d. (B) Volumes of xenograft tumors with the indicated genotypes and treatments on different days. Error bars are the means ± SEM, n = 5 mice per group. (C) Weight of tumor xenografts from the indicated genotypes at the endpoint. Error bars are the means ± SEM, n = 5 independent repeats. (D) Image of xenograft tumors that were inoculated into nude mice with the indicated RCC4 cell lines for 70 d. (E) Volumes of xenograft tumors with the indicated genotypes and treatments on different days corresponding to D. Error bars are means ± SEM, n = 5 mice per group. (F) Weight of tumor xenografts from the indicated genotypes at the endpoint corresponding to D. Error bars are the means ± SEM, n = 5 independent repeats. (G) IF staining of ZRANB1 (green), SLC7A11 (red), and MDA (magenta) in tumor xenografts. The right images are the overlay of ZRANB1, SLC7A11, MDA, and nuclear DAPI (blue) staining. Scale bars, 20 μm. (H) Percentage of ZRANB1/SLC7A11/MDA-positive stained cells per field described in G. Error bars are means ± SEM, n = 5. (I) IF staining of ZRANB1 (green), SLC7A11 (red), and MDA (magenta) in human KIRC samples. The right images are the overlay of ZRANB1, SLC7A11, MDA, and nuclear DAPI (blue) staining. Scale bars, 20 μm. (J) Percentage of ZRANB1-, SLC7A11-, and MDA-positive stained cells per field described in I. Error bars are means ± SEM, n = 5. (K) IF scoring of ZRANB1 staining of KIRC tumor samples and matched adjacent tissues from 30 cancer patients. Error bars are means ± SEM, n = 5 randomly selected magnification fields. Protein staining score = immunostaining intensity × the percentage of immunopositive cells. (L) Correlation between ZRANB1 expression and tumor stage in KIRC (Gene Expression Profiling Interactive Analysis [GEPIA]). (M) Kaplan‒Meier curves of overall survival of KIRC based on ZRANB1 expression (GEPIA). (K) P value measured using two-tailed paired Student’s t test analysis. (B, C, E, F, H–J, and O) P values determined by two-way ANOVA.
ZRANB1 inhibits tumor development partly through SLC7A11 and ferroptosis. (A) Image of xenograft tumors that were inoculated into nude mice with the indicated 786-O cell lines for 60 d. (B) Volumes of xenograft tumors with the indicated genotypes and treatments on different days. Error bars are the means ± SEM, n = 5 mice per group. (C) Weight of tumor xenografts from the indicated genotypes at the endpoint. Error bars are the means ± SEM, n = 5 independent repeats. (D) Image of xenograft tumors that were inoculated into nude mice with the indicated RCC4 cell lines for 70 d. (E) Volumes of xenograft tumors with the indicated genotypes and treatments on different days corresponding to D. Error bars are means ± SEM, n = 5 mice per group. (F) Weight of tumor xenografts from the indicated genotypes at the endpoint corresponding to D. Error bars are the means ± SEM, n = 5 independent repeats. (G) IF staining of ZRANB1 (green), SLC7A11 (red), and MDA (magenta) in tumor xenografts. The right images are the overlay of ZRANB1, SLC7A11, MDA, and nuclear DAPI (blue) staining. Scale bars, 20 μm. (H) Percentage of ZRANB1/SLC7A11/MDA-positive stained cells per field described in G. Error bars are means ± SEM, n = 5. (I) IF staining of ZRANB1 (green), SLC7A11 (red), and MDA (magenta) in human KIRC samples. The right images are the overlay of ZRANB1, SLC7A11, MDA, and nuclear DAPI (blue) staining. Scale bars, 20 μm. (J) Percentage of ZRANB1-, SLC7A11-, and MDA-positive stained cells per field described in I. Error bars are means ± SEM, n = 5. (K) IF scoring of ZRANB1 staining of KIRC tumor samples and matched adjacent tissues from 30 cancer patients. Error bars are means ± SEM, n = 5 randomly selected magnification fields. Protein staining score = immunostaining intensity × the percentage of immunopositive cells. (L) Correlation between ZRANB1 expression and tumor stage in KIRC (Gene Expression Profiling Interactive Analysis [GEPIA]). (M) Kaplan‒Meier curves of overall survival of KIRC based on ZRANB1 expression (GEPIA). (K) P value measured using two-tailed paired Student’s t test analysis. (B, C, E, F, H–J, and O) P values determined by two-way ANOVA.
ZRANB1 negatively correlates with KIRC development. (A and B) Body weight of nude mice with the indicated genotypes and treatments on different days. Error bars are the means ± SEM, n = 5 independent repeats. (C) IHC staining (MDA) of tumor xenografts from the mock and ZRANB1-KO cell lines. Scale bars, 20 μm. (D) Percentage of MDA-positive stained cells per field as indicated. Error bars are means ± SEM, n = 5. (E) Representative hematoxylin and eosin (HE) staining of adjacent and tumor tissues from KIRC patients as indicated in Fig. 4 J. (F) Kaplan‒Meier curves of overall survival of KIRC patients (n = 150), stratified by ZRANB1 protein levels. Protein staining score = immunostaining intensity × the percentage of immunopositive cells.
ZRANB1 negatively correlates with KIRC development. (A and B) Body weight of nude mice with the indicated genotypes and treatments on different days. Error bars are the means ± SEM, n = 5 independent repeats. (C) IHC staining (MDA) of tumor xenografts from the mock and ZRANB1-KO cell lines. Scale bars, 20 μm. (D) Percentage of MDA-positive stained cells per field as indicated. Error bars are means ± SEM, n = 5. (E) Representative hematoxylin and eosin (HE) staining of adjacent and tumor tissues from KIRC patients as indicated in Fig. 4 J. (F) Kaplan‒Meier curves of overall survival of KIRC patients (n = 150), stratified by ZRANB1 protein levels. Protein staining score = immunostaining intensity × the percentage of immunopositive cells.
Discussion
Ferroptosis, a new type of cell death driven by iron-dependent phospholipid peroxidation, is modulated by several cellular metabolic pathways, such as redox homeostasis in vivo, iron handling, mitochondrial activity, amino acids, and lipids (Jiang et al., 2021). Due to the specific characteristics of this type of cell death, some drug-resistant cancer cells, such as mesenchymal-like cells, are highly susceptible to iron-dependent death (Viswanathan et al., 2017). In the past decade, numerous scientists have elucidated the key regulators and mechanisms of ferroptosis from metabolism, ROS biology, and iron biology perspectives. Before that, an HRASV12-selective lethal small molecule, Erastin, was identified to induce nonapoptotic cell death, which was later termed ferroptosis (Dolma et al., 2003). Soon after, another compound, RSL3, was found to similarly induce this type of cell death (Yang and Stockwell, 2008). Erastin and RSL3 represent the first two classes of ferroptosis inducers by inactivating the SLC7A11-GPX4 pathway. Later, other inhibitors, such as ML162, ML210, and FIN56, were invented to target this pathway through different mechanisms. Based on these findings, scientists have attempted to move the conception of ferroptosis to therapeutic applications. IKE, currently the most promising, stable, and effective inhibitor targeting SLC7A11, has been tested in a diffuse large B cell lymphoma xenograft model. However, in our KIRC xenograft model, IKE treatment had little effect due to drug resistance. At present, there are at least three recognized mechanisms by which cancer cells resist ferroptosis: (1) as a major antioxidant transcription factor, NRF2 regulates cellular defense against toxic and oxidative damage through the expression of genes involved in the oxidative stress response and drug detoxification, such as MGST1 in pancreatic cancer cells and ferritin in hepatocarcinoma cells (Kuang et al., 2021; Sun et al., 2021); (2) the transsulfuration pathway generates enough cysteine from methionine, which could compensate for the deficiency of cysteine induced by stress (Hayano et al., 2016); and (3) the mTOR pathway has been recently identified to confer cancer cell resistance to ferroptosis by upregulating the expression of GPX4 at the translational level (Zhang et al., 2021).
Based on our data, we hypothesized that ferroptosis resistance was caused by dysregulation of the SLC7A11 protein. Moreover, we found that diverse expression of ZRANB1 may correlate with ferroptotic resistance through degradation of SLC7A11 in vivo. As a major cystine transporter, SLC7A11 has been reported to be regulated by various stress conditions, such as oxidative and genotoxic stress (Koppula et al., 2021). Through transcriptional activation, ATF4 and NRF2 mediate stress-induced SLC7A11 expression. In contrast, the transcriptional level of SLC7A11 can be repressed by p53. Apart from transcriptional modulation, SLC7A11 can be stabilized by a CD44 variant and a binding partner, OTUB1 (Koppula et al., 2021). However, the underlying mechanisms by which tumor cells maintain high or low levels of SLC7A11 in vivo are still unclear. Our study identified an important posttranslational mechanism linking SLC7A11 regulation to intrinsic ferroptosis resistance. Specifically, we found that a previously established deubiquitinase, ZRANB1, promoted SLC7A11 degradation. As mentioned, ZRANB1 contains an OTU domain that has been known to remove ubiquitin chains from its substrates. In the past, there was only one OTU family deubiquitinase, A20, with both DUB activity and E3 ligase activity in different domains. To our knowledge, the OTU domain has never been reported to play a role as an E3 ligase. Through the in vitro ubiquitinating system, we found that purified full-length ZRANB1 or the truncated ZRANB1 containing the OTU domain efficiently attached ubiquitin to the substrate SLC7A11. E3 ligases are important components of the ubiquitin-proteasome system. By covalently adding ubiquitin to lysine, serine, threonine, or cysteine residues within the substrate, E3 ligases selectively modify proteins. Currently, E3 ligases are divided into two categories: single-subunit E3 ligases, including enzymes from the RING, U-box, HECT, and RBR class E3s, and multisubunit E3 ligases, including enzymes from the Skp-Cul1-Fbox, Elongin-Cul2/5-SOCS box, DCAF-Cul4, and other E3 classes. It will be interesting to test whether the enzyme containing the region (residues 463–584) in the OTU domain represents a new type of E3 ligase. More comprehensive investigations, such as determining the crystal structure, understanding how it recruits E2-ubiquitin, elucidating the mechanisms of priming and positioning ubiquitin and the substrate lysine for transfer, and deciphering how it extends ubiquitin chains to achieve polyubiquitylation, are needed. By degrading the SLC7A11 protein, ZRANB1 dynamically inhibited cystine uptake into cancer cells and rendered them more sensitive to ferroptosis inducers. However, in some cancer cells with low expression of ZRANB1, SLC7A11 was highly enriched, resulting in upregulated cystine uptake and GSH synthesis and eventually leading to ferroptotic resistance. Our study also suggested that the ZRANB1 expression level in KIRC patients may distinguish which patient will benefit from therapy based on ferroptosis.
In summary, from cultured cells to xenograft tumors and human patient samples, we showed that the ZRANB1-SLC7A11 pathway may reshape the lipid peroxidation distribution and further determine the sensitivity of cancer cells to ferroptosis inducers (Fig. S8), whereas the detailed mechanism by which specific characteristic motifs are involved in regulating ZRANB1 activity needs to be further investigated. Moreover, we discovered that heterogeneous MDA distribution in KIRC probably cultivated the ferroptotic plasticity of cancer cells and eventually led to therapeutic resistance. Although other recently discovered antioxidant pathways, such as FSP1 and DHODH (Bersuker et al., 2019; Doll et al., 2019; Jiang et al., 2021; Mao et al., 2021), may be partially involved in this process, targeting ZRANB1 signaling results in ferroptotic death in cancer cells with heterogeneous or low levels of lipid peroxides (Fig. 1 D), presenting a novel strategy to prevent ferroptosis resistance and tumor relapse.
Schematic diagram showing the mechanism. In ZRANB1-deficient tumor cells, the SLC7A11 protein is stable and enriched. Cells with high SLC7A11 expression are quite resistant to ferroptosis inducers such as IKE. However, ectopic overexpression of ZRANB1 in SLC7A11-enriched tumor cells confers tumor cell ferroptosis sensitivity.
Schematic diagram showing the mechanism. In ZRANB1-deficient tumor cells, the SLC7A11 protein is stable and enriched. Cells with high SLC7A11 expression are quite resistant to ferroptosis inducers such as IKE. However, ectopic overexpression of ZRANB1 in SLC7A11-enriched tumor cells confers tumor cell ferroptosis sensitivity.
Materials and methods
Cell lines and culture
HEK293T, HeLa, 786-O, ACHN, BT-549, MDA-MB-231, NCI-H1299, and Caki-1 cells were obtained from the American Type Culture Collection. UMRC6 and RCC4 cells were obtained from Dana-Farber Cancer Institute. OSRC2 cells were obtained from the National Collection of Authenticated Center (Shanghai). SUM159 was obtained from S. Ethier (Medical University of South Carolina, Charleston, SC, USA). All cell lines were free of mycoplasma contamination and cultured under conditions specified by the manufacturer. For cystine deprivation experiments, the culture medium was replaced by DMEM without L-methionine, L-cystine, and L-glutamine (D0422; Sigma-Aldrich).
Reagents
The reagents used for treating cells were Erastin (E7781; Sigma-Aldrich), Ferr-1 (SML0583; Sigma-Aldrich), TBH (458139; Sigma-Aldrich), Z-VAD-FMK (FMK001; R&D Systems), necrostatin-1s (2263; Nec-1, BioVision), MG132 (sc-201270; Santa Cruz Biotechnology), CHX (C7698; Sigma-Aldrich), and chloroquine diphosphate salt (ChlorD, C6628; Sigma-Aldrich). IKE (S8877; Selleckchem) and Liprox (SML1414; Sigma-Aldrich) were used in the xenograft model. All chemicals were dissolved according to the manufacturers’ instructions.
Plasmids
SFB-ZRANB1 and its deletion-mutant-containing expression vectors were described in our previous publication (Zhang et al., 2018a). pCLXSU (GFP)-HA-ZRANB1 and its C443A mutation were gifts from Shao-Cong Sun’s laboratory (Houston, TX, USA) stock as described previously (Jin et al., 2016). Full-length ZRANB1, which was obtained from the HA-ZRANB1 expression vector, was subcloned into the pCDH-MYC-S-tag and pCDH-FLAG vectors, and its C443A mutation was subcloned into the pCDH-MYC-S-tag vector. The SLC7A11-FLAG-HA-pHAGE vector was obtained from Alex Toker’s lab (Boston, MA, USA; Lien et al., 2017). Full-length SLC7A11 was subcloned into the pBabe-SFB and pLenti-V5 vectors using the Gateway system (Invitrogen) or into the pCDH-MYC-S-tag and pHAGE-MYC vectors using the HiFi system (NEB; New England Biolabs). pRK5-HA-ubiquitin was obtained from Addgene (plasmid number: 17608). The SLC7A11 shRNAs were subcloned into pLKO vectors and the sequences were as follows: shSLC-1:5′-CCGGGCTCTCATTTAAGGTTCCCTTCTCGAGAAGGGAACCTTAAATGAGAGCTTTTTG-3′; shSLC-2: 5′-CCGGCCTACATCATCGGTACTTCAACTCGAGTTGAAGTACCGATGATGTAGGTTTTTG-3′.
Cell viability assay
Cell viability was evaluated using methylthiazolyldiphenyl–tetrazolium bromide (MTT, M8180; Solarbio) according to the manufacturer’s instructions. Cells were seeded in 96-well plates (5,000 cells/well) overnight and then treated with DMSO, Erastin, TBH, or cystine starvation for 48 h. The culture medium was replaced with 100 μl fresh medium containing 20 μl of the MTT solution for each well of the plate that was returned to the cell culture incubator for 4 h. Then, the culture medium was discarded, 150 μl DMSO was added, and the plate was incubated at 37°C for 10 min. The absorbance was measured in a microplate reader at 490 nm.
sgRNA screening
An arrayed collection of 344 lentiviral sgRNAs purchased from EdiGene (EdiArrayTM Human CRISPR DeUbiquitin Library: ARR60307) targeting 87 human DUB genes was screened in 96-well plates in HeLa cells. To generate CRISPR KO cells, HeLa cells were first transfected with pLenti-Cas9-2a-mcherry-blasticidin (BSD). After BSD antibiotic selection, we built stable HeLa-cas9 cell lines. 3,000 cells/well were infected in duplicate for 48 h with 2 μl sgRNA-containing viral supernatant and then selected by puromycin (1.5 μg/ml) for 72 h. The sgRNAs targeting GFP served as negative controls. 5,000 HeLa-DUB-KO cells/well were seeded overnight in 96-well plates. The next day, the cells were washed twice with PBS and then treated with DMSO, Erastin (20 μM), or cystine starvation for 48 h. Cell viability was determined by MTT. The final cell viability was measured by the following equation: cell viability % = (ODCys-/Erastin − ODblank)/(ODcontrol − ODblank) × 100.
Cell death and lipid peroxidation assay
To measure cell death, cells were seeded in a 6-well plate 1 d before treatment. After treatment with the indicated drugs, cells were washed twice with PBS, trypsinized, collected in a 1.5 ml tube, resuspended in 0.5 ml staining buffer containing 5 μl propidium iodide (PI, 421301; BioLegend), and incubated for 15 min at 4°C in the dark. Dead cells (PI-positive cells) were analyzed using a Beckman CytoFLEX S flow cytometer. Lipid peroxidation levels were measured as described below. Briefly, cells were seeded in 6-well plates, treated with the indicated drugs, and incubated with fresh culture medium containing 5 μM BODIPY 581/591 dye (D3861; Invitrogen). After incubation for 30 min at 37°C, the cells were trypsinized, collected, resuspended in staining buffer, and then subjected to flow cytometry analysis.
Immunoprecipitation (IP) and western blotting
For IP, cells were washed with ice-cold PBS and lysed in NETN buffer (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA and 0.5% Nonidet P-40) or CHAPS buffer (25 mM Tris-HCl, pH 7.5, 120 mM NaCl, 1 mM EDTA, 0.33% CHAPS) supplemented with the protease and phosphatase inhibitor cocktail (AR1182 and AR1183; BOSTER). For endogenous protein IP, protein extracts lysed in CHAPS buffer were preincubated with magnetic protein-A/G beads (88803; Thermo Fisher Scientific), followed by incubation with a ZRANB1/SLC7A11-specific antibody (ab103417, Rabbit IgG; Abcam and 12691S, Rabbit IgG; Cell Signaling Technology) or control IgG at 4°C for 12 h, and then the magnetic beads were pulled down at room temperature. After washing the beads, the bound proteins were eluted with 2× Laemmli buffer (161-0737; Bio-Rad). For IP of tagged protein, cell extracts lysed in NETN buffer were preincubated with the tag beads as indicated (anti-HA magnetic beads, B26201; Bimake; anti-MYC agarose beads, 20168; Thermo Fisher Scientific; S-protein agarose beads, 69704; Millipore and anti-FLAG antibody, F7425, Rabbit IgG; Sigma-Aldrich) at 4°C overnight. Protein extracts were resolved by SDS-PAGE and transferred to a nitrocellulose membrane according to standard protocols using primary antibodies specific for SLC7A11 (1:5,000, 12691; Cell Signaling Technology), ZRANB1 (1:200, ab103417; Abcam), ubiquitin (1:2,000, sc-271289, Mouse IgG; Santa Cruz Biotechnology), TFRC (1:3,000, 66180-1-Ig, Mouse IgG; Proteintech), LPCAT3 (1:3,000, 67882-1-Ig, Mouse IgG; Proteintech), ACSL4 (1:3,000, 66617-1-Ig, Mouse IgG; Proteintech), FSP1 (1:3,000, 20886-1-AP, Rabbit IgG; Proteintech), DHODH (1:3,000, 14877-1-AP, Rabbit IgG; Proteintech), GPX4 (1:3,000, 67763-1-Ig, Mouse IgG; Proteintech), β-actin (1:5,000, sc-47778, Mouse IgG; Santa Cruz Biotechnology), FLAG (1:5,000, F7425; Sigma-Aldrich), HA (1:2,000, sc-7392, Mouse IgG; Santa Cruz Biotechnology), and MYC (1:2,000, sc-40, Mouse IgG; Santa Cruz Biotechnology). The densitometric analysis of western blots was conducted by the ImageJ program and the quantification values were normalized to the internal control.
Protein purification and GST pull-down assays
The His-ZRANB1-M3/M3-2-a/M3-2-b/ΔM3-2-b protein was overexpressed in the BL21 strain. Bacteria were collected and disrupted by sonication in nondenaturing lysate with a protease inhibitor cocktail (AR1182; Boster). The His-ZRANB1-M3/M3-2-a/M3-2-b/ΔM3-2-b protein was purified according to the manufacturer’s instructions (635713; Takara). For in vitro GST pull-down assays, purified GST-SLC7A11 (H00023657-P01; Abnova) was incubated with purified His-ZRANB1 (E-560-050; R&D Systems) or His-ZRANB1-M3, followed by pull-down with GST-agarose beads (52-2303-00 AK; GE Healthcare). The GST-agarose beads were washed with IP buffer, and the bound proteins were eluted with 2× Laemmli sample buffer (1610737; Bio-Rad).
GSH assay
The relative GSH concentration in cell lysates was detected using a GSH-Glo Glutathione Assay (V6911; Promega) according to the manufacturer’s instructions. Briefly, 5,000 cells/well were seeded in a 96-well plate 1 d before adding drugs. The culture medium from the wells was carefully removed and 100 μl of GSH-Glo Reagent was added and incubated at room temperature for 30 min on a plate shaker. Finally, 100 μl of reconstituted Luciferin Detection Reagent was added to each well, mixed well, and incubated for 15 min. The luminescence was measured by a SYNEGY|H1 microplate reader (BIOTEK).
RNA isolation and real-time PCR
Total RNA was extracted using the HP Total RNA Kit (R6812-02; OMEGA) following the manufacturer’s protocol and was then reverse transcribed with PrimeScript RT Master Mix (RR036A; TaKaRa). The resulting cDNA was used for quantitative PCR using iTaq Universal SYBR Green Supermix (172-5121; Bio-Rad) and the gene expression levels were normalized to β-actin. Real-time PCR and data collection were performed on a Bio-Rad CFX96 Connect instrument and the primer sequences used for PCR were as follows: ZRANB1 forward, 5′-GGCTCTTCCTCAATGCTTGT-3′ and ZRANB1 reverse, 5′-TGTCTCCTCCTGATGACTTG-3′; SLC7A11 forward, 5′-ATGCAGTGGCAGTGACCTTT-3′ and SLC7A11 reverse, 5′-GGCAACAAAGATCGGAACTG-3′; β-actin forward, 5′-CGGAACCGCTCATTGCC-3′ and actin reverse, 5′-ACCCACACTGTGCCCATCTA-3′.
In vitro ubiquitination assay
The experiment was performed according to the manufacturer’s instructions using an in vitro ubiquitination assay kit (K-982; R&D Systems). Briefly, 11 µl of a ubiquitination master mix (including ubiquitin, E1, E3 ubiquitin ligase, substrate, and reaction buffer) was added to 4 μl of E2 in the plate, followed by the addition of 5 μl of 4× ATP and incubation at 37°C for 60 min. 5 μl of 5× SDS-PAGE buffer was added and then analyzed by western blot analysis.
In vitro autoubiquitination assay
The experiment was performed according to the manufacturer’s instructions using an in vitro E3 ligase autoubiquitylation assay kit (ab139469; Abcam). Briefly, assay components were added to 0.5 ml Eppendorf tube(s), including distilled H2O, 10× Ub E3 ligase buffer, 20× ubiquitin activating enzyme solution (E1), 20× UbcH5a, 10× ubiquitin, full-length ZRANB1 and its truncated proteins, 50 mM dithiothreitol, 20× Mg-ATP, and inorganic pyrophosphatase solution. The tube contents were mixed gently. The samples were incubated at 37°C for 60 min. Then, the reactions were quenched by the addition of 50 μl of 2× SDS-PAGE gel loading buffer followed by heating to 95°C for 5 min. Finally, these samples were analyzed by western blot analysis.
Xenograft mouse model
4–6-wk-old athymic NU/NU female nude mice were purchased from Charles River (Beijing). To generate subcutaneous tumors, a total of 5 × 106 786-O cells were mixed with Matrigel (356234; Corning) at a 1:1 ratio (vol/vol) and injected into the left posterior flanks of immunodeficient female nude mice. Mice were fed regular chow and monitored for the progression of tumors by bidimensional tumor measurements once every 4 d until the endpoint. The tumor volume was calculated according to the modified ellipsoidal formula v = length × width2 × 1/2. After 45 d of tumor cell injection, mice were treated with Hank’s Balanced Salt Solution (HBSS; 140250760; Thermo Fisher Scientific) or 23 mg/kg IKE with or without 10 mg/kg Liprox through an intraperitoneal injection every day until the endpoint for drug treatment experiments.
Histology and IF immunohistochemistry
Xenograft tissue samples were collected and immediately fixed in neutral paraformaldehyde fixing solution (G1101; Servicebio) overnight and then submitted to the Servicebio company for embedding and hematoxylin and eosin staining. For IF and immunohistochemical (IHC) staining, tissue sections were processed according to the instructions of Servicebio. Briefly, tissue sections were deparaffinized, rehydrated, and incubated with the corresponding primary and secondary antibodies after antigen repair. The cell nuclei of the tissue sections were restained with DAPI, sealed with blocker, and then photographed by microscopy. The antibodies used for IF were anti-SLC7A11 and anti-ZRANB1, and the antibody used for IHC was anti-MDA (JAI-MMD-030 N; Adipogen, 1:100, Mouse IgG).
Transmission electron microscopy
The tumor tissues were cut into 1–2 mm3 pieces and then the tissue pieces were transferred to a vial with cold fixative solution (G1102; Servicebio), which mainly contained 2.5% glutaraldehyde, using forceps. Notably, when the tissue has more blood and tissue fluid, it should be washed several times with fixative solution before cutting into small pieces for fixation. The tissue pieces can be cut into slightly larger pieces first and then trimmed into 1–2 mm3 pieces while fixing when they cannot be cut into 1–2 mm3 pieces at one time. After fixing, the tissue pieces were submitted to Servicebio company and then examined with a HITACHI HT7700 transmission electron microscope at an accelerating voltage of 80 kV.
Image analysis
Microscopic images were analyzed using Fiji (ImageJ; National Institutes of Health). Background subtraction was performed equally for all images in the same experiment. To observe cell ferroptosis, images of the cell samples were obtained by a Digital Microscope Camera (OD630K USB3.0; SOPTOP) shooting under a normal light microscope (SOPTOP). Images were taken at room temperature with a 10× objective lens and ImageView as the photographing software. For IHC and IF analysis, images were acquired at room temperature with a Pannoramic DESK, Pannoramic MIDI, or Pannoramic 250 FLASH (3DHISTECH) equipped with CaseViewer2.4 (3DHISTECH; 400× magnification). The fluorescence channel type and parameter settings were as follows: DAPI (Ex: 365 nm, Em:420–470 nm), SpGreen (Ex: 484–504 nm, Em:517–537 nm), SpOrange (Ex: 532–554 nm, Em:573–596 nm), and Cy5 (Ex: 608–648 nm, Em:672–712 nm).
Statistical analysis
Data were analyzed by two-sided Student’s t test or one- or two-way ANOVA, depending on the situation. Values are shown as the means ± SEM, and all experiments were repeated at least three times. All statistical analyses were conducted with GraphPad Prism. Data distribution was assumed to be normal, but this was not formally tested. P values < 0.05 were considered significant.
Ethics declarations
This paper describes studies involving human participants, human data, or human tissue, and all the samples were approved by the Ethics Committee of Shanghai Outdo Biotech Company. All animal experiments were approved by the Ethics Committee of Tongji Medical College, Huazhong University of Science and Technology, China.
Online supplemental material
This manuscript is accompanied by eight supplementary figures. Fig. S1 contains data on screening appropriate tumor models of ferroptosis. Fig. S2 contains data supporting Fig. 2. It shows that ZRANB1 promotes ferroptosis in vitro. Fig. S3 contains data supporting Fig. 2. It shows that restoration of ZRANB1 resensitized tumor cells to ferroptosis. Fig. S4 contains data supporting Fig. 4. It also shows that ZRANB1 interacts with SLC7A11. Fig. S5 contains data supporting Fig. 5. It shows that ZRANB1 regulates SLC7A11 protein stability and also contains data on screening E2 for SLC7A11. Fig. S6 contains data supporting Fig. 6. It shows that ZRANB1 regulates SLC7A11, influencing sensitivity to ferroptosis. Fig. S7 contains data supporting Fig. 7. It shows that ZRANB1 negatively correlates with KIRC development. Fig. S8 contains a schematic diagram showing the mechanism. Table S1 is a list of primers used in this study.
Data availability
All information is included in the manuscript (and supplementary information). All data from this study are available from the corresponding author upon reasonable request.
Acknowledgments
We apologize to the colleagues whose relevant work cannot be cited here owing to space limitations. We are grateful to Baoli Hu for their advice and comments. We also thank the Huazhong University of Science and Technology core facility and members of the Zhang lab for discussion.
This research was supported by grants from the National Natural Science Foundation of China (81874116, 82172823, and 81901621), National Key Research and Development Program of China (2021YFA1201200), MOST-Key Program for International S&T Cooperation Projects of China (2017YFE0129100), and startup fund from Huazhong University of Science and Technology. P. Zhang is a scholar in the National Young Talents Program of China.
Author contributions: S. Huang: Data curation; formal analysis; validation; investigation; visualization; writing—original draft. Q. Zhang: Data curation; formal analysis; validation; investigation; visualization. M. Zhao: Formal analysis; methodology. X. Wang: Methodology. Y. Zhang: Resources; methodology. B. Gan: Review and editing. P. Zhang: Conceptualization; resources; formal analysis; supervision; funding acquisition; visualization; writing—original draft; writing—review and editing.
References
Author notes
S. Huang and Q. Zhang contributed equally to this paper.
Disclosures: The authors declare no competing interests exist.






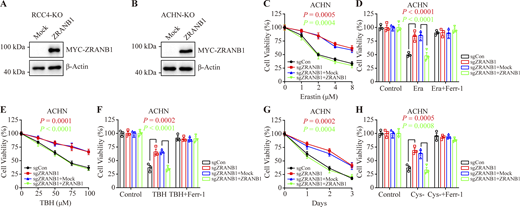

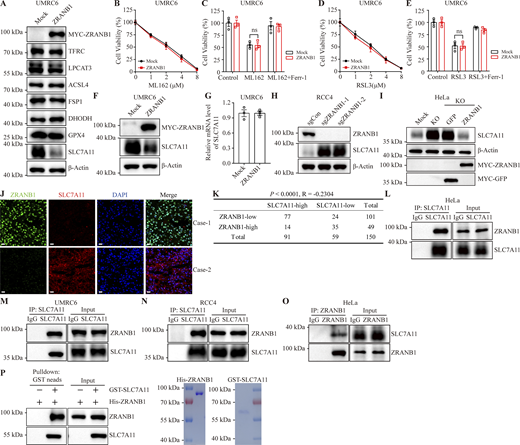


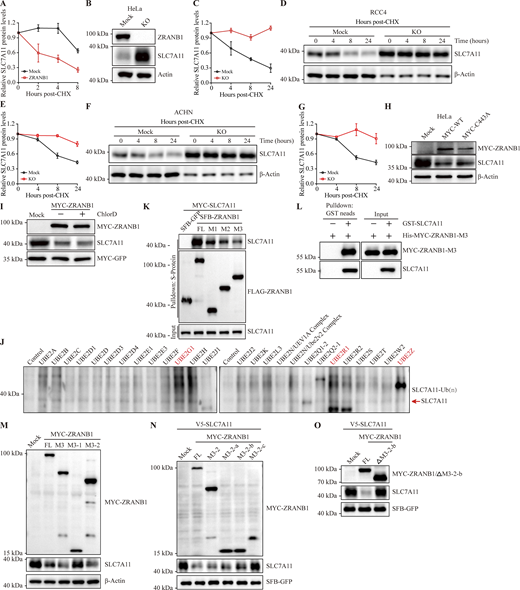
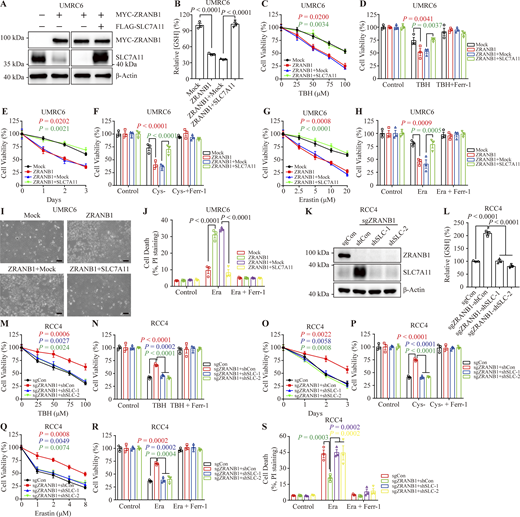

![ZRANB1 inhibits tumor development partly through SLC7A11 and ferroptosis. (A) Image of xenograft tumors that were inoculated into nude mice with the indicated 786-O cell lines for 60 d. (B) Volumes of xenograft tumors with the indicated genotypes and treatments on different days. Error bars are the means ± SEM, n = 5 mice per group. (C) Weight of tumor xenografts from the indicated genotypes at the endpoint. Error bars are the means ± SEM, n = 5 independent repeats. (D) Image of xenograft tumors that were inoculated into nude mice with the indicated RCC4 cell lines for 70 d. (E) Volumes of xenograft tumors with the indicated genotypes and treatments on different days corresponding to D. Error bars are means ± SEM, n = 5 mice per group. (F) Weight of tumor xenografts from the indicated genotypes at the endpoint corresponding to D. Error bars are the means ± SEM, n = 5 independent repeats. (G) IF staining of ZRANB1 (green), SLC7A11 (red), and MDA (magenta) in tumor xenografts. The right images are the overlay of ZRANB1, SLC7A11, MDA, and nuclear DAPI (blue) staining. Scale bars, 20 μm. (H) Percentage of ZRANB1/SLC7A11/MDA-positive stained cells per field described in G. Error bars are means ± SEM, n = 5. (I) IF staining of ZRANB1 (green), SLC7A11 (red), and MDA (magenta) in human KIRC samples. The right images are the overlay of ZRANB1, SLC7A11, MDA, and nuclear DAPI (blue) staining. Scale bars, 20 μm. (J) Percentage of ZRANB1-, SLC7A11-, and MDA-positive stained cells per field described in I. Error bars are means ± SEM, n = 5. (K) IF scoring of ZRANB1 staining of KIRC tumor samples and matched adjacent tissues from 30 cancer patients. Error bars are means ± SEM, n = 5 randomly selected magnification fields. Protein staining score = immunostaining intensity × the percentage of immunopositive cells. (L) Correlation between ZRANB1 expression and tumor stage in KIRC (Gene Expression Profiling Interactive Analysis [GEPIA]). (M) Kaplan‒Meier curves of overall survival of KIRC based on ZRANB1 expression (GEPIA). (K) P value measured using two-tailed paired Student’s t test analysis. (B, C, E, F, H–J, and O) P values determined by two-way ANOVA.](https://cdn.rupress.org/rup/content_public/journal/jcb/222/11/10.1083_jcb.202212072/1/m_jcb_202212072_fig7.png?Expires=1773822992&Signature=THSwu~Dpjd-Del3Wqdj3zE~XbaH5WOibZxNd18VOxv3U5Ib9hxuWI74O6slJbUodoEAPYlnFuPlN6uvl9BqnKMOgLJpWz4LI0LjAsyp50QE1da1HC8267u2jvaASzsoM6urUQ5MrLR6KUsBsdhL3Q~O~hueEjXP8pNEP8MJoRxb4~R2gBQ9vtM1BFps0Y4rAeFe1oM~90Nlri8yeA6UrF-JEC4uE8Ab9FELH1Cm~uMvu5kQMjPWTSE4tNmig6S2uZ-6POHGYI8R4dCeX9OoGZsULZlAph4fuzDjVl8eyFSwBYFCYe3qTzSbL5tP6oUDq9-Qm9uDA0YV-wpe2z-4Jkw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)




