UHRF1 is an epigenetic coordinator bridging DNA methylation and histone modifications. Additionally, UHRF1 regulates DNA replication and cell cycle, and its deletion induces G1/S or G2/M cell cycle arrest. The roles of UHRF1 in the regulation of G2/M transition remain poorly understood. UHRF1 depletion caused chromosome misalignment, thereby inducing cell cycle arrest at mitotic metaphase, and these cells exhibited the defects of spindle geometry, prominently manifested as shorter spindles. Mechanistically, UHRF1 protein directly interacts with EG5, a kinesin motor protein, during mitosis. Furthermore, UHRF1 induced EG5 polyubiquitination at the site of K1034 and further promoted the interaction of EG5 with spindle assembly factor TPX2, thereby ensuring accurate EG5 distribution to the spindles during metaphase. Our study clarifies a novel UHRF1 function as a nuclear protein catalyzing EG5 polyubiquitination for proper spindle architecture and faithful genomic transmission, which is independent of its roles in epigenetic regulation and DNA damage repair inside the nucleus. These findings revealed a previously unknown mechanism of UHRF1 in controlling mitotic spindle architecture and chromosome behavior and provided mechanistic evidence for UHRF1 deletion-mediated G2/M arrest.
Introduction
Mitosis is a highly coordinated process to assure the fidelity of chromosome segregation, which is crucial for genome stability and cellular fitness (Pavin and Tolić, 2016; Nam et al., 2015). Conversely, chromosome missegregation induces aneuploidy, lethality, or malignant transformation (Ben-David and Amon, 2020; Storchova, 2021). The fidelity of chromosome segregation involves interdependent processes at the kinetochore–microtubule interface and the spindle assembly checkpoint (Navarro and Cheeseman, 2021; Jia et al., 2013; Mansfeld et al., 2011). The movement of chromosomes is associated with the structural and dynamic polarity of spindle microtubules, and the directional movement needs force generated by kinesin and dynein, which are microtubule-based molecular motor proteins (Gallisà-Suñé et al., 2023; Sharp et al., 2000).
EG5 is a member of the kinesin motor protein family (Rath and Kozielski, 2012), which exhibits a conserved bipolar homotetramer structure consisting of pairs of motor dimers at opposite ends of an elongated molecule (Acar et al., 2013), allowing the motor dimers to crosslink two adjacent microtubules within the mitotic spindle and to coordinate the antiparallel microtubules sliding during spindle formation, maintenance, and elongation (Kapitein et al., 2005; Blackwell et al., 2017). The precise distribution of EG5 on spindles to perform optimal functions at various mitotic stages is closely related to the maintenance of bipolar spindle architecture and regulated by an intrinsic control system (Mann and Wadsworth, 2019). As a kinesin family member, EG5 has an N-terminal motor domain, a stalk domain that forms a coiled-coil, and a highly conserved C-terminal tail domain that contains a consensus Cdk1 phosphorylation site (Khmelinskii et al., 2009; Goldstein et al., 2017). Cdk1 phosphorylation not only regulates the binding of EG5 to microtubules but also facilitates interactions with other proteins (Hayward et al., 2019; Alfonso-Pérez et al., 2019). Another site in the tail domain is phosphorylated by Nek6/Nek7 kinases, which is necessary for EG5 motor function, localization to the centrosomes, and spindle bipolarity (Eibes et al., 2018; Bertran et al., 2011; Fry et al., 2017). However, in addition to phosphorylation, very little is known about how posttranslational modifications of EG5 determine its intracellular localization and functions.
Ubiquitin-like with PHD and RING finger domains 1 (UHRF1) is an epigenetic coordinator bridging DNA methylation and histone code, and its aberrant overexpression is associated with oncogenic effects. In addition, UHRF1 as a nuclear protein is highly expressed in the proliferating cells and regulates the cell cycle. UHRF1 is essential for the G1/S phase transition since its depletion or downregulation elevated the silenced p53/p21Cip1/WAF1, thereby activating DNA damage response and inducing cell cycle arrest at the G1/S phase transition (Bonapace et al., 2002; Arima et al., 2004). Similarly, another study reported that depletion of UHRF1 activated the DNA damage response and subsequently induced cell cycle arrest at the G2/M phase and caspase 8-dependent apoptosis (Tien et al., 2011). However, the roles of UHRF1 in the regulation of G2/M transition, in particular for the mitotic machinery, remain largely unexplored.
In this study, we first found that depletion of UHRF1 impaired proper spindle architecture and significantly increased the ratio of cells with misaligned chromosomes, suggesting that UHRF1 plays a critical role in the stability of mitotic chromosomes. Mechanistically, we found that the UHRF1 protein directly interacts with the critical mitotic motor EG5 during mitosis. Furthermore, we identified K1034 of the EG5 site as a mitotic target for UHRF1 polyubiquitination. The K1034R mutant disturbed the distribution of EG5 protein during metaphase, thereby resulting in spindle shortening and disordered geometry. Conversely, rescue with wild-type EG5 fixed these spindle defects in EG5-deficient cells. UHRF1 polyubiquitination further promotes its interaction with spindle assembly factor TPX2, thereby ensuring accurate EG5 distribution to the spindles during metaphase. These data established a functional link between nuclear protein UHRF1 and EG5 motor protein coordinating with chromosomal behavior, which is necessary for the successful completion of mitosis and faithful chromosomal transmission.
Results
Depletion of UHRF1 causes chromosome misalignment
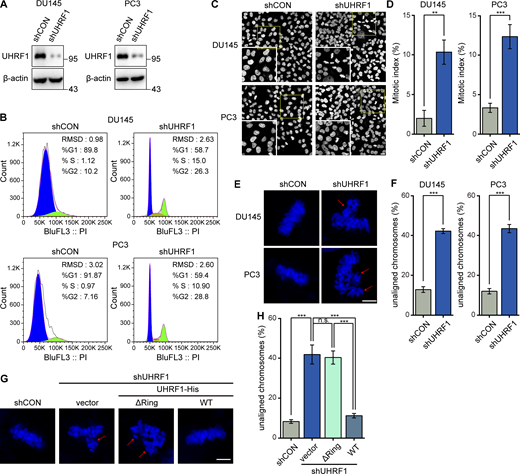
UHRF1 is a nuclear protein and an epigenetic coordinator bridging DNA methylation and histone modifications, thereby regulating gene transcription (Ashraf et al., 2017; Rajakumara et al., 2011). Additionally, UHRF1 is a regulator of DNA replication and the cell cycle, and deletion of UHRF1 induces cell cycle arrest at G1/S or G2/M phase. However, very little is known about how UHRF1 regulates mitosis. Here, we first established DU145 and PC3 cell lines with stable UHRF1 knockdown by infecting cells with lentiviral vectors delivering and expressing UHRF1 shRNA (Fig. 1 A). Consistently in these two cell lines, UHRF1 knockdown significantly increased the proportion of cells at the G2/M phase, indicating cell cycle arrest (Fig. 1 B). To further clarify whether UHRF1 knockdown induces G2/M cell cycle arrest by disturbing mitosis, we observed the nuclear morphology using immunofluorescence and confocal microscopy. As expected, UHRF1 depletion inhibited the process of mitosis, manifesting as prolonged mitotic metaphase, which was judged by whether chromosomes were distributed on the metaphase plate (Fig. 1, C and D). These data suggested that UHRF1 is required for normal progression of mitosis. Prolonged mitosis often causes chromosome misalignment, thereby resulting in mitotic catastrophe. We further observed the chromosome alignment at mitosis when UHRF1 was depleted. As expected, UHRF1 knockdown remarkably increased the frequency of cells with unaligned chromosomes (Fig. 1, E and F).
Depletion of UHRF1 causes chromosome misalignment. (A and B) DU145 and PC3 cells with or without UHRF1 depletion were established by stable transfection with shRNA vectors for UHRF1 or control, and UHRF1 expression was assessed by Western blotting (A). The cell cycle distribution was analyzed by flow cytometry (B). (C–F) The nuclear DNA in DU145 and PC3 cells with or without UHRF1 depletion was stained with DAPI (scale bar, 20 μm; inset scale bar, 10 μm, C). The percent of mitotic cells was assessed according to nuclei morphology. n > 100 cells (D). The chromosomes that failed to congress at the metaphase plate are highlighted by red arrows (scale bar, 5 μm, E). The percentage of cells with unaligned chromosomes was assessed. n > 80 cells (F). (G and H) DU145 cells with UHRF1 depletion were transiently transfected with plasmids expressing UHRF1ΔRING or UHRF1WT, and the nuclear DNA was stained with DAPI (scale bar, 5 μm, G). The percentage of cells with unaligned chromosomes was assessed. n > 80 cells (H). The data for quantification in D, F, and H are from n = 3 independent experiments. Results are represented as mean ± SD (one-way ANOVA test); error bars represent SD. n.s., not significant; **, P < 0.01; ***, P < 0.001. Source data are available for this figure: SourceData F1.
Depletion of UHRF1 causes chromosome misalignment. (A and B) DU145 and PC3 cells with or without UHRF1 depletion were established by stable transfection with shRNA vectors for UHRF1 or control, and UHRF1 expression was assessed by Western blotting (A). The cell cycle distribution was analyzed by flow cytometry (B). (C–F) The nuclear DNA in DU145 and PC3 cells with or without UHRF1 depletion was stained with DAPI (scale bar, 20 μm; inset scale bar, 10 μm, C). The percent of mitotic cells was assessed according to nuclei morphology. n > 100 cells (D). The chromosomes that failed to congress at the metaphase plate are highlighted by red arrows (scale bar, 5 μm, E). The percentage of cells with unaligned chromosomes was assessed. n > 80 cells (F). (G and H) DU145 cells with UHRF1 depletion were transiently transfected with plasmids expressing UHRF1ΔRING or UHRF1WT, and the nuclear DNA was stained with DAPI (scale bar, 5 μm, G). The percentage of cells with unaligned chromosomes was assessed. n > 80 cells (H). The data for quantification in D, F, and H are from n = 3 independent experiments. Results are represented as mean ± SD (one-way ANOVA test); error bars represent SD. n.s., not significant; **, P < 0.01; ***, P < 0.001. Source data are available for this figure: SourceData F1.
It was reported that UHRF1 displays RING-dependent E3 ligase activity through its RING domain (Jenkins et al., 2005), directly participates in the interplay between BRCA1 and 53BP1, and regulates protein ubiquitination such as RIF1 (Zhang et al., 2016). To validate whether the RING domain of UHRF1 plays a critical role in chromosome segregation in mitosis, we constructed a mutant of UHRF1 deleting the RING domain of C-terminus (UHRF1ΔRING) and observed the chromosome morphology at mitosis in DU145 cells. The rescue of full-length UHRF1 (UHRF1WT) in the UHRF1 knockdown cells almost restored the unaligned chromosomes (Fig. 1, G and H). However, the RING domain truncation mutant (UHRF1ΔRING) failed to restore the unaligned chromosomes. These data suggested that UHRF1 is required for proper chromosome congression during mitosis.
Depletion of UHRF1 damages spindle architecture
It is known that chromosome segregation during cell division requires a bipolar mitotic spindle, which is composed of dynamic microtubules (So et al., 2022). It is thus plausible that delayed mitosis of UHRF1-depleted cells may be due to the disruption of mitotic spindle organization. We monitored spindle architecture in mitotic DU145 cells when UHRF1 was knocked down with siRNAs (Fig. S1 A). The distance between two dominant poles was significantly shortened in the UHRF1-depleted cells, manifesting as a remarkable shortening of spindle length (Fig. 2, A and B). The data suggested that UHRF1 depletion impaired mitotic spindle architecture. Furthermore, we observed that the bundles of spindle microtubules at the spindle equator appeared to be distorted or dispersed in some cells, resulting in their disordered connections to chromosomes. We quantified the mitotic cells with abnormal spindle geometry (reduced spindle length or disordered spindle microtubules) and found that the depletion of endogenous UHRF1 caused 80.2% or 80.6% abnormal spindle geometry compared with 9.1% abnormal spindle geometry in the control cells (Fig. 2, A and C). We observed that the asters appeared more prominent due to spindle shortening at metaphase in some UHRF1-depleted cells. We quantified the mean fluorescence intensity of asters and the results showed that UHRF1 knockdown significantly increased the fluorescence intensity of asters (Fig. 2, A and D). To verify the impact of the RING domain on spindle architecture at metaphase, we re-expressed the RING domain truncation mutant (UHRF1ΔRING) in the endogenous UHRF1-deleted cells. Consistent with the above data, UHRF1 depletion significantly shortened spindle length and caused disordered spindle architecture. The rescue of UHRF1WT almost restored the UHRF1 depletion–caused abnormality of spindle architecture to normal spindle length and geometry. However, the rescue of UHRF1ΔRING failed to restore abnormal spindle architecture (Fig. 2, E–H). These data illustrated that UHRF1 maintains proper spindle architecture, which is required for the process of mitosis. Since the spindle length is maintained by balanced outward and inward forces, the UHRF1 deletion–induced shortening of spindle length implies an unbalance of motor forces during mitosis. Therefore, UHRF1 may play a novel role in generating the balanced motor forces to maintain proper metaphase spindles and promote correct cell cycle transition.
In vitro recombinant proteins GST-pull down assay. (A) Knockdown of UHRF1. DU145 cells were transiently transfected with UHRF1 siRNAs and UHRF1 expression was evaluated by Western blotting. (B) GST-pulldown assay of UHRF1 using the indicated GST fusion proteins. EG5 protein was detected. Source data are available for this figure: SourceData FS1.
In vitro recombinant proteins GST-pull down assay. (A) Knockdown of UHRF1. DU145 cells were transiently transfected with UHRF1 siRNAs and UHRF1 expression was evaluated by Western blotting. (B) GST-pulldown assay of UHRF1 using the indicated GST fusion proteins. EG5 protein was detected. Source data are available for this figure: SourceData FS1.
Depletion of UHRF1 damages spindle architecture. (A) DU145 cells were transiently transfected with or without siUHRF1. The mitotic spindles were stained with immunofluorescent anti-α-tubulin antibody (green) and chromosomes were stained with DAPI. Scale bar, 5 μm. (B–D) Spindle pole distance was measured. n = 40 cells (B). The percentage of cells at the metaphases with abnormal spindle geometry was assessed (C) in DU145 cells when transfected with or without siUHRF1. n > 50 cells for each condition. The asters fluorescence intensity of the metaphase spindle was measured. n = 40 cells for each condition. (E) DU145 cells with UHRF1 depletion were transiently transfected with plasmids expressing UHRF1ΔRING or UHRF1WT. The mitotic spindles were stained with immunofluorescent α-tubulin antibody and chromosomes were stained with DAPI. Scale bar, 5 μm. (F–H) Spindle pole distance was measured (n = 40 cells; F) and the percentage of cells at the metaphases with abnormal spindle geometry was assessed (n > 50 cells; G). The mean fluorescence intensity of asters was analyzed by ImageJ. n = 40 cells (H). The data for quantification in C, D, G, and H are from n = 3 independent experiments. Results are represented as mean ± SD; error bars represent SD. Dots represent individual cell samples in B and F; bars are median ± quartile. n.s., not significant; ***, P < 0.001. One-way ANOVA test.
Depletion of UHRF1 damages spindle architecture. (A) DU145 cells were transiently transfected with or without siUHRF1. The mitotic spindles were stained with immunofluorescent anti-α-tubulin antibody (green) and chromosomes were stained with DAPI. Scale bar, 5 μm. (B–D) Spindle pole distance was measured. n = 40 cells (B). The percentage of cells at the metaphases with abnormal spindle geometry was assessed (C) in DU145 cells when transfected with or without siUHRF1. n > 50 cells for each condition. The asters fluorescence intensity of the metaphase spindle was measured. n = 40 cells for each condition. (E) DU145 cells with UHRF1 depletion were transiently transfected with plasmids expressing UHRF1ΔRING or UHRF1WT. The mitotic spindles were stained with immunofluorescent α-tubulin antibody and chromosomes were stained with DAPI. Scale bar, 5 μm. (F–H) Spindle pole distance was measured (n = 40 cells; F) and the percentage of cells at the metaphases with abnormal spindle geometry was assessed (n > 50 cells; G). The mean fluorescence intensity of asters was analyzed by ImageJ. n = 40 cells (H). The data for quantification in C, D, G, and H are from n = 3 independent experiments. Results are represented as mean ± SD; error bars represent SD. Dots represent individual cell samples in B and F; bars are median ± quartile. n.s., not significant; ***, P < 0.001. One-way ANOVA test.
EG5 is a UHRF1-associated mitotic protein
To gain further insight into the UHRF1 mitotic functions, we sought to identify potential coordinators involved in the UHRF1-maintained mitotic spindle architecture. Using a tandem affinity purification with an anti-Flag antibody, followed by high throughput proteomics using a mass spectrometer, we identified EG5 as a potential UHRF1-interacting protein (Fig. 3 A). EG5 plays a key role in the dynamic assembly and function of the mitotic spindle by crosslinking and sliding adjacent microtubules (Mann and Wadsworth, 2019). We validated the affinity purification results by coimmunoprecipitation of His-tagged UHRF1 with Flag-tagged EG5 from HEK293T cells using His or Flag antibody. The interaction between UHRF1-His and EG5-Flag was also detected in DU145 and PC3 cells using coimmunoprecipitation assays (Fig. 3 B). The endogenous protein interaction between UHRF1 and EG5 was also verified in these three cells by coimmunoprecipitation using UHRF1 or EG5 antibody (Fig. 3 C). These results consistently verified that the UHRF1 protein physically interacts with EG5. Moreover, the in vitro GST-pulldown assay using purified recombinant proteins revealed a direct interaction between UHRF1 and EG5 (Fig. S1 B).
EG5 is a UHRF1-interactive mitotic protein. (A) EG5 was identified as a component of UHRF1-interactive protein complexes by immunoprecipitation coupled with mass spectrometry. UHRF1-interacting protein complexes were immunoprecipitated with anti-Flag antibody in HEK293T cells expressing Flag-tagged UHRF1, and then were eluted with Flag peptide. The UHRF1-interacted protein complexes were separated by SDS-PAGE and a 125-kD electrophoretic band was subjected to mass spectrometry analysis. (B) HEK293T, DU145, and PC3 cells were transfected with plasmids expressing UHRF1-His or EG5-Flag, and the interaction between exogenous UHRF1 and EG5 proteins was validated by immunoprecipitation with antibodies against His or Flag, followed by immunoblotting. (C) The interaction between endogenous UHRF1 and EG5 proteins was validated by immunoprecipitation with antibodies against UHRF1 or EG5, followed by immunoblotting in HEK293T, DU145, and PC3 cells. (D) DU145 cells were synchronized in G1/S phase using double thymidine blocking and then released by culture media. The cells were synchronized in G2/M phases at 9 h after release. The interaction between endogenous UHRF1 and EG5 proteins was validated by immunoprecipitation with antibodies against UHRF1, followed by immunoblotting. (E and F) GST-tagged full-length UHRF1 and individual subdomains were constructed for mapping the EG5-binding region. Purified recombinant proteins of GST-tagged individual domains of UHRF1 were incubated with HEK293T cell lysates in vitro as indicated, followed by immunoblotting with anti-EG5 antibody (F). The lysate of HEK293T cells was used for a positive control. (G and H) GST-tagged full-length EG5 and individual subdomains were constructed for mapping the UHRF1-binding region. Purified recombinant proteins of GST-tagged individual domains of EG5 were incubated with HEK293T cell lysates in vitro as indicated, followed by immunoblotting with anti-UHRF1 antibody (H). The lysate of HEK293T cells was used for a positive control. Source data are available for this figure: SourceData F3.
EG5 is a UHRF1-interactive mitotic protein. (A) EG5 was identified as a component of UHRF1-interactive protein complexes by immunoprecipitation coupled with mass spectrometry. UHRF1-interacting protein complexes were immunoprecipitated with anti-Flag antibody in HEK293T cells expressing Flag-tagged UHRF1, and then were eluted with Flag peptide. The UHRF1-interacted protein complexes were separated by SDS-PAGE and a 125-kD electrophoretic band was subjected to mass spectrometry analysis. (B) HEK293T, DU145, and PC3 cells were transfected with plasmids expressing UHRF1-His or EG5-Flag, and the interaction between exogenous UHRF1 and EG5 proteins was validated by immunoprecipitation with antibodies against His or Flag, followed by immunoblotting. (C) The interaction between endogenous UHRF1 and EG5 proteins was validated by immunoprecipitation with antibodies against UHRF1 or EG5, followed by immunoblotting in HEK293T, DU145, and PC3 cells. (D) DU145 cells were synchronized in G1/S phase using double thymidine blocking and then released by culture media. The cells were synchronized in G2/M phases at 9 h after release. The interaction between endogenous UHRF1 and EG5 proteins was validated by immunoprecipitation with antibodies against UHRF1, followed by immunoblotting. (E and F) GST-tagged full-length UHRF1 and individual subdomains were constructed for mapping the EG5-binding region. Purified recombinant proteins of GST-tagged individual domains of UHRF1 were incubated with HEK293T cell lysates in vitro as indicated, followed by immunoblotting with anti-EG5 antibody (F). The lysate of HEK293T cells was used for a positive control. (G and H) GST-tagged full-length EG5 and individual subdomains were constructed for mapping the UHRF1-binding region. Purified recombinant proteins of GST-tagged individual domains of EG5 were incubated with HEK293T cell lysates in vitro as indicated, followed by immunoblotting with anti-UHRF1 antibody (H). The lysate of HEK293T cells was used for a positive control. Source data are available for this figure: SourceData F3.
To verify the mitosis-dependent interaction of UHRF1 and EG5, we synchronized DU145 cells by using double thymidine blocks and then collected them at 0 or 9 h after release into normal medium. The cells were verified at the G1/S or G2/M phase, respectively. The protein interaction was measured by coimmunoprecipitation. The results demonstrated that UHRF1 protein interacts with EG5 in mitotic cells, but not in interphase cells (Fig. 3 D). It has been reported that the RING domain of UHRF1 protein is associated with its E3 ubiquitin ligase activity. We further identified the interactive protein domains of EG5 interact with the SRA and RING domains of UHRF1 by GST-pull down assay (Fig. 3, E and F). The data showed that the SRA and RING domains of UHRF1 interact with the C-terminal tails of EG5 (Fig. 3, G and H).
UHRF1 catalyzes the polyubiquitination of EG5 at the site of K1034
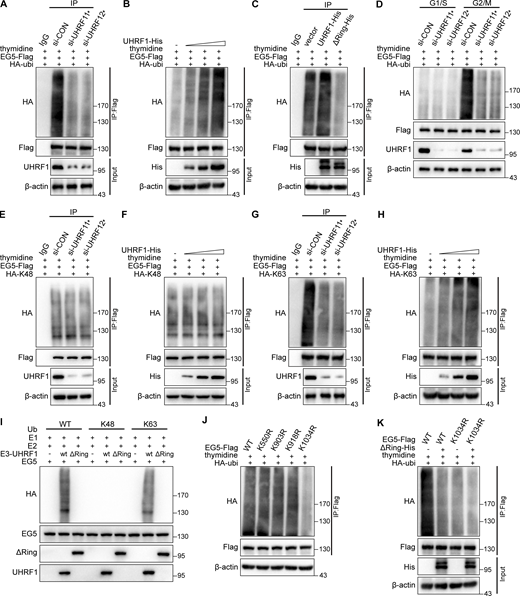
The physical interaction between UHRF1 and EG5 proteins inspired us to investigate whether UHRF1 catalyzes the ubiquitination of EG5 protein. We firstly cotransfected HEK293T cells with plasmids encoding HA-ubiquitin and EG5-Flag, together with siRNA-UHRF1 or UHRF1 expressing plasmids, and then the cells were synchronized at the G2/M phase. EG5 protein was immunoprecipitated with anti-Flag antibodies, and the degree of ubiquitination of EG5 protein was assessed by immunoblotting with antibodies against HA. The knockdown of UHRF1 with siRNA significantly decreased the EG5 ubiquitination levels (Fig. 4 A). Conversely, UHRF1 overexpression steadily increased EG5 ubiquitination levels in a dose-dependent manner (Fig. 4 B). To further clarify whether UHRF1 induced EG5 ubiquitination through the RING domain, we cotransfected HEK293T cells with HA-ubiquitin and EG5-Flag, together with UHRF1WT or UHRF1ΔRING. Compared with UHRF1WT, UHRF1ΔRING failed to induce the ubiquitination of EG5 protein (Fig. 4 C). We further monitored the degree of EG5 ubiquitination in different cell cycle phases when UHRF1 was knocked down. The data showed that decreasing ubiquitination of EG5 only can be detected at 9 h after release into the normal medium by using a double thymidine block (Fig. 4 D). These results suggest that UHRF1 is essential for the ubiquitination of EG5 during mitosis.
UHRF1 catalyzes the polyubiquitination of EG5 at the site of K1034. (A) HEK293T cells were cotransfected with control or UHRF1 siRNAs, together with HA-ubiquitin and EG5-Flag. The cells then were synchronized at the G2/M phase with a double thymidine block. EG5 protein was immunoprecipitated with anti-Flag antibody and the ubiquitination level of EG5 was assessed with HA antibody. (B) HEK293T cells were cotransfected with plasmids expressing UHRF1-His, HA-ubiquitin, and EG5-Flag, and then the cells were synchronized at the G2/M phase. EG5 protein was immunoprecipitated with anti-Flag antibody and the ubiquitination level of EG5 was assessed with an HA antibody. (C) HEK293T cells were cotransfected with plasmids expressing UHRF1-His or UHRF1ΔRING-His, HA-ubiquitin, and EG5-Flag, and then the cells were synchronized at the G2/M phase. EG5 protein was immunoprecipitated with anti-Flag antibody and the ubiquitination level of EG5 was assessed with an HA antibody. (D) HEK293T cells were cotransfected with control or UHRF1 siRNAs together with HA-ubiquitin and EG5-Flag, and then the cells were synchronized in the G1/S phase using double thymidine blocking and released by culture media. The cells were synchronized in G2/M phases at 9 h after release. EG5 protein was immunoprecipitated with an anti-Flag antibody and the ubiquitination level of EG5 was assessed with HA antibody. (E and G) HEK293T cells were cotransfected with control or UHRF1 siRNAs, together with HA-K48 or HA-K63 and EG5-Flag. The cells then were synchronized at the G2/M phase with a double thymidine block. EG5 protein was immunoprecipitated with an anti-Flag antibody, and the ubiquitination level of EG5 was assessed with an HA antibody. (F and H) HEK293T cells were cotransfected with plasmids expressing UHRF1-His, HA-K48/HA-K63, and EG5-Flag, and then the cells were synchronized at the G2/M phase. EG5 protein was immunoprecipitated with an anti-Flag antibody, and the ubiquitination level of EG5 was assessed with an HA antibody. (I) In vitro ubiquitination assay was performed in the presence of Ub (WT, K48, or K63), E1, E2, EG5, and UHRF1 (WT or ΔRING mutant). The ubiquitination of EG5 was examined with HA antibody. (J) HEK293T cells were cotransfected with plasmids expressing HA-ubiquitin and EG5-Flag with four lysine mutations as shown, and then the cells were synchronized at the G2/M phase. EG5 protein was immunoprecipitated with anti-Flag antibody, and the ubiquitination level of EG5 was assessed with an HA antibody. (K) HEK293T cells were cotransfected with plasmids expressing UHRF1ΔRING-His, HA-ubiquitin, and EG5-Flag with wild type or lysine mutation as shown, and then the cells were synchronized at the G2/M phase. EG5 protein was immunoprecipitated with anti-Flag antibody and the ubiquitination level of EG5 was assessed with an HA antibody. Source data are available for this figure: SourceData F4.
UHRF1 catalyzes the polyubiquitination of EG5 at the site of K1034. (A) HEK293T cells were cotransfected with control or UHRF1 siRNAs, together with HA-ubiquitin and EG5-Flag. The cells then were synchronized at the G2/M phase with a double thymidine block. EG5 protein was immunoprecipitated with anti-Flag antibody and the ubiquitination level of EG5 was assessed with HA antibody. (B) HEK293T cells were cotransfected with plasmids expressing UHRF1-His, HA-ubiquitin, and EG5-Flag, and then the cells were synchronized at the G2/M phase. EG5 protein was immunoprecipitated with anti-Flag antibody and the ubiquitination level of EG5 was assessed with an HA antibody. (C) HEK293T cells were cotransfected with plasmids expressing UHRF1-His or UHRF1ΔRING-His, HA-ubiquitin, and EG5-Flag, and then the cells were synchronized at the G2/M phase. EG5 protein was immunoprecipitated with anti-Flag antibody and the ubiquitination level of EG5 was assessed with an HA antibody. (D) HEK293T cells were cotransfected with control or UHRF1 siRNAs together with HA-ubiquitin and EG5-Flag, and then the cells were synchronized in the G1/S phase using double thymidine blocking and released by culture media. The cells were synchronized in G2/M phases at 9 h after release. EG5 protein was immunoprecipitated with an anti-Flag antibody and the ubiquitination level of EG5 was assessed with HA antibody. (E and G) HEK293T cells were cotransfected with control or UHRF1 siRNAs, together with HA-K48 or HA-K63 and EG5-Flag. The cells then were synchronized at the G2/M phase with a double thymidine block. EG5 protein was immunoprecipitated with an anti-Flag antibody, and the ubiquitination level of EG5 was assessed with an HA antibody. (F and H) HEK293T cells were cotransfected with plasmids expressing UHRF1-His, HA-K48/HA-K63, and EG5-Flag, and then the cells were synchronized at the G2/M phase. EG5 protein was immunoprecipitated with an anti-Flag antibody, and the ubiquitination level of EG5 was assessed with an HA antibody. (I) In vitro ubiquitination assay was performed in the presence of Ub (WT, K48, or K63), E1, E2, EG5, and UHRF1 (WT or ΔRING mutant). The ubiquitination of EG5 was examined with HA antibody. (J) HEK293T cells were cotransfected with plasmids expressing HA-ubiquitin and EG5-Flag with four lysine mutations as shown, and then the cells were synchronized at the G2/M phase. EG5 protein was immunoprecipitated with anti-Flag antibody, and the ubiquitination level of EG5 was assessed with an HA antibody. (K) HEK293T cells were cotransfected with plasmids expressing UHRF1ΔRING-His, HA-ubiquitin, and EG5-Flag with wild type or lysine mutation as shown, and then the cells were synchronized at the G2/M phase. EG5 protein was immunoprecipitated with anti-Flag antibody and the ubiquitination level of EG5 was assessed with an HA antibody. Source data are available for this figure: SourceData F4.
We further characterized the types of ubiquitination of EG5 regulated by UHRF1. The synchronized HEK293T cells were cotransfected with plasmids expressing EG5-Flag and HA-K48 or HA-K63 ubiquitin, together with siRNA-UHRF1. The data showed that UHRF1 knockdown reduced K63-linked but not K48-linked ubiquitination chains of EG5 (Fig. 4, E and G). Conversely, stable overexpression of UHRF1 elevated the levels of K63-linked ubiquitination chains of EG5 in a dose-dependent manner but had no impact on K48-linked ubiquitination chains of EG5 (Fig. 4, F and H). The results have been further validated in the UHRF1ΔRING cells. Compared with UHRF1WT, UHRF1ΔRING failed to potentate K63-type ubiquitination of EG5, but K48-type was not significantly different from UHRF1WT (Fig. S2, A and B). These data demonstrated that UHRF1 catalyzed K63-specific protein ubiquitination of EG5 but did not regulate K48-linked ubiquitination chains. To determine whether UHRF1 directly induced EG5 ubiquitination, we reconstituted in vitro EG5 ubiquitination in a ubiquitination reaction buffer containing the recombinant EG5 protein, recombinant UHRF1 protein (WT or ΔRING), and a mixture of E1 and E2 plus ubiquitin (WT, K48 only or K63 only). UHRF1WT induced the ubiquitination of EG5, while UHRF1ΔRING lost the ability to promote EG5 ubiquitination (Fig. 4 I). Furthermore, we found EG5 ubiquitination was mediated by K63-linked ubiquitination chains but not by K48 (Fig. 4 I). Taken together, these data indicated that UHRF1 directly induced the K63-linked ubiquitination of EG5 through its E3 ligase activity.
UHRF1 catalyzes the K63-linked ubiquitination of EG5 at K1034. (A and B) HEK293T cells were cotransfected with plasmids expressing UHRF1-His or UHRF1ΔRING-His, HA-K48/HA-K63, and EG5-Flag, and then the cells were synchronized at the G2/M phase. EG5 protein was immunoprecipitated with an anti-Flag antibody, and the ubiquitination level of EG5 was assessed with an HA antibody. (C) HEK293T cells were cotransfected with plasmids expressing HA-K63 and EG5-Flag with four lysine mutations as shown, and then the cells were synchronized at the G2/M phase. EG5 protein was immunoprecipitated with an anti-Flag antibody, and the ubiquitination level of EG5 was assessed with an HA antibody. (D) HEK293T cells were cotransfected with plasmids expressing UHRF1ΔRING-His, HA-K63, and EG5-Flag with wild-type or lysine mutation as shown, and then the cells were synchronized at the G2/M phase. EG5 protein was immunoprecipitated with anti-Flag antibody and the ubiquitination level of EG5 was assessed with an HA antibody. Source data are available for this figure: SourceData FS2.
UHRF1 catalyzes the K63-linked ubiquitination of EG5 at K1034. (A and B) HEK293T cells were cotransfected with plasmids expressing UHRF1-His or UHRF1ΔRING-His, HA-K48/HA-K63, and EG5-Flag, and then the cells were synchronized at the G2/M phase. EG5 protein was immunoprecipitated with an anti-Flag antibody, and the ubiquitination level of EG5 was assessed with an HA antibody. (C) HEK293T cells were cotransfected with plasmids expressing HA-K63 and EG5-Flag with four lysine mutations as shown, and then the cells were synchronized at the G2/M phase. EG5 protein was immunoprecipitated with an anti-Flag antibody, and the ubiquitination level of EG5 was assessed with an HA antibody. (D) HEK293T cells were cotransfected with plasmids expressing UHRF1ΔRING-His, HA-K63, and EG5-Flag with wild-type or lysine mutation as shown, and then the cells were synchronized at the G2/M phase. EG5 protein was immunoprecipitated with anti-Flag antibody and the ubiquitination level of EG5 was assessed with an HA antibody. Source data are available for this figure: SourceData FS2.
To further identify the potential ubiquitination sites of EG5 protein, we found potential ubiquitination modification sites of lysine (K) between aa 550 and 1057 of EG5 after database analysis. We thus mutated four potential lysine residues within this domain to construct the ubiquitination inactivated mutants. The synchronized HEK293T cells were cotransfected with plasmids expressing EG5 mutants together with HA-ubiquitin or HA-K63, respectively. The data showed that only the K1034R mutant (EG5K1034R) significantly reduced the ubiquitination of EG5 (Fig. 4 J). EG5K1034R but not the other mutants substantially reduced K63-linked ubiquitination of EG5 (Fig. S2 C). In addition, the synchronized HEK293T cells were cotransfected with EG5WT or EG5K1034R, together with UHRF1ΔRING and HA-ubiquitin or HA-K63, respectively. The data showed that the mutations of UHRF1 (UHRF1ΔRING) or/and EG5 (EG5K1034R) impaired the effect of UHRF1 on EG5 ubiquitination (Fig. 4 K and Fig. S2 D). These results indicated that UHRF1 catalyzes the K63-linked ubiquitination of EG5 at K1034.
UHRF1 regulates EG5 interactions with TPX2 to control EG5 localization on the spindle
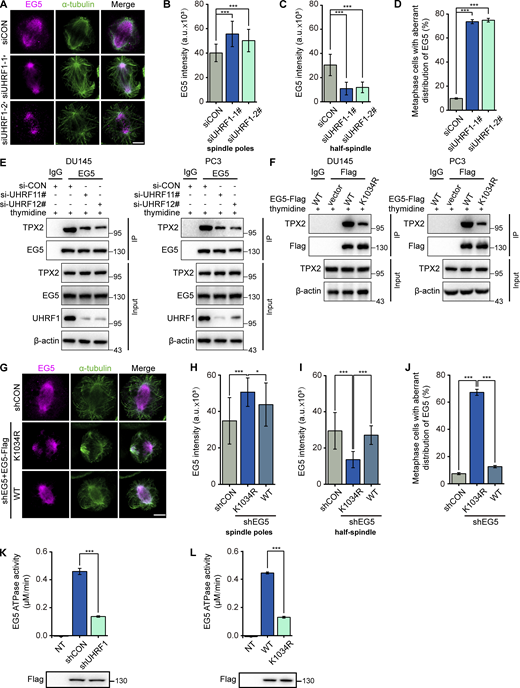
The spatial and temporal dynamics of EG5 ensure its motor function in maintaining spindle architecture (He et al., 2016). Since UHRF1 regulates EG5 ubiquitination, we hypothesized that UHRF1 loss might disrupt the spatial relationship between EG5 and mitotic spindles. Indeed, we observed aberrant EG5 distribution in DU145 cells with UHRF1 knockdown. In the control mitotic cells, EG5 protein uniformly distributed along the spindles when the chromosomes were properly aligned during metaphase. UHRF1 knockdown pushed the EG5 protein clump to both spindle poles and microtubule organizing center but very few on the microtubules of the half-spindle, which is defined as the region between the pole and equator (Fig. 5 A). We quantified the distribution of EG5 protein by measuring the mean intensity in different spindle regions. UHRF1 knockdown significantly reduced the signal intensity in the half-spindle but increased the spindle poles signal intensity. The abnormal distribution of EG5 cells was quantified (Fig. 5, B–D). To rule out the possibility that the reduction of EG5 signal intensity resulted from a decrease in protein levels, we tested whether UHRF1 knockdown reduced the protein level of EG5. The results showed that UHRF1 knockdown faintly changed the EG5 protein level consistently in the three types of tested cells (Fig. S3 A). These results suggested that UHRF1 is necessary for proper protein distribution of EG5 on the mitotic spindles.
UHRF1 regulates EG5 interactions with TPX2 to determine EG5 localization on the spindle. (A) UHRF1 was depleted with siRNAs in DU145 cells. EG5 were stained with immunofluorescent antibodies (red) and the spindle was stained with anti-α-tubulin antibody (green). Scale bar, 5 μm. (B–D) B and C: Corresponding EG5 fluorescence intensity profiles of cells. n = 40 cells for each condition. EG5 fluorescence intensity of the spindle poles (B) or half-spindle (C) was measured, and the percentage of cells at the metaphases with abnormal distribution of EG5 was assessed. n > 50 cells (D). (E) DU145 or PC3 cells were transiently transfected with UHRF1 siRNAs, EG5 protein was immunoprecipitated, and the interacting TPX2 was assessed by immunoblotting. (F) DU145 or PC3 cells were transiently cotransfected with plasmids expressing EG5WT-Flag or EG5K1034R-Flag, EG5 protein was immunoprecipitated with anti-Flag antibody, and the interacting TPX2 was assessed by immunoblotting. (G) DU145 cells with EG5 depletion were transiently transfected with plasmids expressing EG5WT or EG5K1034R. EG5 were stained with immunofluorescent antibodies (red) and the spindle was stained with anti-α-tubulin antibody (green). Scale bar, 5 μm. (H–J) H and I: Corresponding EG5 fluorescence intensity profiles of cells. n = 40 cells for each condition. EG5 fluorescence intensity of the spindle poles (H) or half-spindle (I) was measured, and the percentage of cells at the metaphases with abnormal distribution of EG5 was assessed. n > 50 cells (J). (K and L) DU145 cells with UHRF1 depletion were transiently transfected with plasmids expressing EG5WT-Flag (K), or DU145 cells were transiently transfected with plasmids expressing Flag-tagged wild-type EG5 (WT)/K1034R mutant (L), and immunoprecipitated with a Flag antibody. Half of the immunoprecipitate was used for ATPase assay (bar graph) and the other half was separated by SDS-PAGE and immunoblotted with Flag antibody (gel image). NT, non-transfected. The data for quantification in B–D and H–L are from n = 3 independent experiments. Results are represented as mean ± SD (one-way ANOVA test); error bars represent SD. *, P < 0.05; ***, P < 0.001. Source data are available for this figure: SourceData F5.
UHRF1 regulates EG5 interactions with TPX2 to determine EG5 localization on the spindle. (A) UHRF1 was depleted with siRNAs in DU145 cells. EG5 were stained with immunofluorescent antibodies (red) and the spindle was stained with anti-α-tubulin antibody (green). Scale bar, 5 μm. (B–D) B and C: Corresponding EG5 fluorescence intensity profiles of cells. n = 40 cells for each condition. EG5 fluorescence intensity of the spindle poles (B) or half-spindle (C) was measured, and the percentage of cells at the metaphases with abnormal distribution of EG5 was assessed. n > 50 cells (D). (E) DU145 or PC3 cells were transiently transfected with UHRF1 siRNAs, EG5 protein was immunoprecipitated, and the interacting TPX2 was assessed by immunoblotting. (F) DU145 or PC3 cells were transiently cotransfected with plasmids expressing EG5WT-Flag or EG5K1034R-Flag, EG5 protein was immunoprecipitated with anti-Flag antibody, and the interacting TPX2 was assessed by immunoblotting. (G) DU145 cells with EG5 depletion were transiently transfected with plasmids expressing EG5WT or EG5K1034R. EG5 were stained with immunofluorescent antibodies (red) and the spindle was stained with anti-α-tubulin antibody (green). Scale bar, 5 μm. (H–J) H and I: Corresponding EG5 fluorescence intensity profiles of cells. n = 40 cells for each condition. EG5 fluorescence intensity of the spindle poles (H) or half-spindle (I) was measured, and the percentage of cells at the metaphases with abnormal distribution of EG5 was assessed. n > 50 cells (J). (K and L) DU145 cells with UHRF1 depletion were transiently transfected with plasmids expressing EG5WT-Flag (K), or DU145 cells were transiently transfected with plasmids expressing Flag-tagged wild-type EG5 (WT)/K1034R mutant (L), and immunoprecipitated with a Flag antibody. Half of the immunoprecipitate was used for ATPase assay (bar graph) and the other half was separated by SDS-PAGE and immunoblotted with Flag antibody (gel image). NT, non-transfected. The data for quantification in B–D and H–L are from n = 3 independent experiments. Results are represented as mean ± SD (one-way ANOVA test); error bars represent SD. *, P < 0.05; ***, P < 0.001. Source data are available for this figure: SourceData F5.
EG5 localization and its interaction with TPX2 are affected by UHRF1 mutation. (A) HEK293T, DU145, or PC3 cells were transiently transfected with UHRF1 siRNAs, and UHRF1 or EG5 expression was evaluated by Western blotting. (B) DU145 or PC3 cells with UHRF1 depletion were transiently transfected with plasmids expressing UHRF1ΔRING or UHRF1WT, EG5 protein was immunoprecipitated, and the interacting TPX2 was assessed by immunoblotting. (C) DU145 cells with UHRF1 depletion were transiently transfected with plasmids expressing UHRF1ΔRING or UHRF1WT. EG5 was stained with immunofluorescent antibodies (red), and the spindle was stained with anti-α-tubulin antibody (green). Scale bar, 5 μm. (D and E) Corresponding EG5 fluorescence intensity profiles of cells. n = 40 cells for each condition. EG5 fluorescence intensity of the spindle poles or half-spindle was measured (D), and the percent of cells at the metaphases with abnormal distribution of EG5 was assessed. n > 50 cells (E). (F) DU145 cells with EG5 depletion were transiently transfected with plasmids expressing EG5WT or EG5K1034R. TPX2 were stained with immunofluorescent antibodies (yellow), and the spindle was stained with anti-α-tubulin antibody (green). Scale bar, 5 μm. (G) Corresponding TPX2 fluorescence intensity profiles of cells. n = 40 cells for each condition. TPX2 fluorescence intensity of the spindle poles or half-spindle was measured. The data for quantification in D, E, and G are from n = 3 independent experiments. Results are represented as mean ± SD (one-way ANOVA test); error bars represent SD. n.s., not significant; ***, P < 0.001. Source data are available for this figure: SourceData FS3.
EG5 localization and its interaction with TPX2 are affected by UHRF1 mutation. (A) HEK293T, DU145, or PC3 cells were transiently transfected with UHRF1 siRNAs, and UHRF1 or EG5 expression was evaluated by Western blotting. (B) DU145 or PC3 cells with UHRF1 depletion were transiently transfected with plasmids expressing UHRF1ΔRING or UHRF1WT, EG5 protein was immunoprecipitated, and the interacting TPX2 was assessed by immunoblotting. (C) DU145 cells with UHRF1 depletion were transiently transfected with plasmids expressing UHRF1ΔRING or UHRF1WT. EG5 was stained with immunofluorescent antibodies (red), and the spindle was stained with anti-α-tubulin antibody (green). Scale bar, 5 μm. (D and E) Corresponding EG5 fluorescence intensity profiles of cells. n = 40 cells for each condition. EG5 fluorescence intensity of the spindle poles or half-spindle was measured (D), and the percent of cells at the metaphases with abnormal distribution of EG5 was assessed. n > 50 cells (E). (F) DU145 cells with EG5 depletion were transiently transfected with plasmids expressing EG5WT or EG5K1034R. TPX2 were stained with immunofluorescent antibodies (yellow), and the spindle was stained with anti-α-tubulin antibody (green). Scale bar, 5 μm. (G) Corresponding TPX2 fluorescence intensity profiles of cells. n = 40 cells for each condition. TPX2 fluorescence intensity of the spindle poles or half-spindle was measured. The data for quantification in D, E, and G are from n = 3 independent experiments. Results are represented as mean ± SD (one-way ANOVA test); error bars represent SD. n.s., not significant; ***, P < 0.001. Source data are available for this figure: SourceData FS3.
As is well known, TPX2, a Ran-regulated spindle assembly factor, promotes mitotic spindle formation by enhancing EG5 accumulation on microtubules. Additionally, EG5 requires TPX2 to regulate proper spindle localization in vertebrate cells (Ma et al., 2011). Since UHRF1 knockdown simultaneously disrupted EG5 localization and spindle architecture (Fig. 2), we assessed the impact of UHRF1 knockdown on the EG5–TPX2 protein interaction by coimmunoprecipitation. UHRF1 depletion significantly disrupted the protein interaction of EG5 and TPX2 (Fig. 5 E). The results were validated by using endogenous TPX2 and EG5 proteins in DU145 and PC3 cells. The mutation of the ubiquitination site of EG5 (EG5K1034R) significantly diminished the protein interaction of TPX2 and EG5 (Fig. 5 F). UHRF1 polyubiquitination is required for the interaction of EG5 and TPX2. To clarify the roles of UHRF1 polyubiquitination in the regulation of EG5 protein distribution during mitosis, we monitored the intracellular localization of EG5 protein during mitosis by using immunofluorescence when EG5K1034R was transfected to the EG5-depleted DU145 cells. As expected, EG5K1034R protein accumulated at the spindle poles in prometaphase cells and persisted in the metaphase cells (Fig. 5, G–I). In addition, the metaphase cells with aberrant EG5 distribution significantly increased when UHRF1 polyubiquitination was destroyed (Fig. 5 J). Moreover, we assessed the impact of UHRF1 mutation (UHRF1ΔRING) disrupting the ubiquitination of EG5 on EG5 localization and its interaction with TPX2. We deleted UHRF1 and re-expressed UHRF1ΔRING or UHRF1WT in DU145 cells. Comparing DU145 cells with UHRF1WT, the frequency of abnormal EG5 distribution was significantly increased in the UHRF1ΔRING cells (Fig. S3, C–E). Additionally, UHRF1 mutation significantly diminished the protein interaction of endogenous EG5 and TPX2 in both DU145 and PC3 cells (Fig. S3 B). The results indicated that UHRF1-catalyzed EG5 ubiquitination is required for proper EG5 localization and its interaction with TPX2.
To further clarify whether UHRF1-catalyzed EG5 ubiquitination regulates the localization of TPX2 on spindles, we deleted EG5 and re-expressed wild-type (EG5WT) or mutated EG5 (EG5K1034R) that disrupted the ubiquitination of EG5 in DU145 cells. TPX2 exhibited a pattern of distribution of staining gradients from spindle poles to metaphase plate in the EG5K1034R cells, which was not significantly different from the EG5WT cells (Fig. S3, F and G). The results indicated that the UHRF1-catalyzed EG5 ubiquitination did not control the localization of TPX2 on spindles.
EG5 promotes bipolar spindle formation and maintains spindle architecture through its ATP hydrolysis function. To study the effect of UHRF1 ubiquitination modification on EG5 motor activity, we assessed the ATPase activity of EG5 in DU145 cells with UHRF1 knockdown or EG5K1034R mutant. Following immunoprecipitation with anti-Flag antibodies, the EG5 protein was eluted and subjected to ATPase assays. The ATPase activity of EG5 was significantly reduced when UHRF1 was depleted (Fig. 5 K). Furthermore, we found that the ATPase activity of the K1034R mutant was significantly reduced compared with wild-type EG5 (Fig. 5 L). Therefore, the data indicated that UHRF1-catalyzed EG5 polyubiquitination is required for proper EG5 motor activity and regulates EG5 distribution on spindles during metaphase by promoting the protein interaction of EG5 and TPX2.
UHRF1 ubiquitination is critical for spindle architecture
To further demonstrate the essential roles of the UHRF1–EG5 signaling axis in propagating mitotic spindle architecture, we observed the mitotic spindle architecture when EG5WT or EG5K1034R was rescued in the EG5-depleted DU145 cells. Consistent with previous data, EG5 knockdown significantly increased the number of monopoles. The rescue of EG5WT restored cell cycle arrest in mitosis, while EG5K1034R greatly increased mitotic cells (Fig. 6, A and B). Furthermore, EG5WT, but not EG5K1034R, restored chromosome alignment during mitosis (Fig. 6, C and D). These data demonstrated that UHRF1 protein ubiquitination is required for maintaining normal progression of mitosis and proper chromosome congression. Furthermore, we were surprised to observe that rescue with EG5WT in EG5-depleted cells significantly increased spindle length and restored spindle architecture. In contrast, rescue with EG5K1034R did not restore the spindle shortening or correct the disordered mitotic spindles (Fig. 6, E–G). These data demonstrate the essential roles of UHRF1-induced EG5 ubiquitination in the maintenance of spindle architecture and chromosome stability during mitosis.
UHRF1 ubiquitination is critical for spindle architecture. (A) DU145 cells with EG5 depletion were transiently transfected with plasmids expressing EG5WT or EG5K1034R, and the nuclear DNA was stained with DAPI. Scale bar, 20 μm; inset scale bar, 10 μm. (B) The mitotic cells were identified according to nuclei morphology and the percentage of mitotic cells was assessed. n > 100 cells. (C) Chromosomes that failed to congress at the metaphase plate are highlighted by red arrows. Scale bar, 5 μm. (D) The percentage of DU145 cells with unaligned chromosomes was assessed. n > 50 cells. (E) Spindles were stained with immunofluorescent α-tubulin antibody and chromosomes were stained with DAPI. Scale bar, 5 μm. (F and G) Spindle pole distance was measured. n = 40 cells (F). The percent of cells at the metaphases with abnormal spindle geometry was assessed. n > 50 cells (G). The data for quantification in B, D, and G are from n = 3 independent experiments. Results are represented as mean ± SD; error bars represent SD. Dots represent individual cell samples in F; bars are median ± quartile. **, P < 0.01; ***, P < 0.001. One-way ANOVA test.
UHRF1 ubiquitination is critical for spindle architecture. (A) DU145 cells with EG5 depletion were transiently transfected with plasmids expressing EG5WT or EG5K1034R, and the nuclear DNA was stained with DAPI. Scale bar, 20 μm; inset scale bar, 10 μm. (B) The mitotic cells were identified according to nuclei morphology and the percentage of mitotic cells was assessed. n > 100 cells. (C) Chromosomes that failed to congress at the metaphase plate are highlighted by red arrows. Scale bar, 5 μm. (D) The percentage of DU145 cells with unaligned chromosomes was assessed. n > 50 cells. (E) Spindles were stained with immunofluorescent α-tubulin antibody and chromosomes were stained with DAPI. Scale bar, 5 μm. (F and G) Spindle pole distance was measured. n = 40 cells (F). The percent of cells at the metaphases with abnormal spindle geometry was assessed. n > 50 cells (G). The data for quantification in B, D, and G are from n = 3 independent experiments. Results are represented as mean ± SD; error bars represent SD. Dots represent individual cell samples in F; bars are median ± quartile. **, P < 0.01; ***, P < 0.001. One-way ANOVA test.
Based on the data in the present study, we propose a working model explaining how the UHRF1–EG5 interaction contributes to chromosome congression and spindle assembly during mitosis. First, UHRF1 catalyzes the K63-linked polyubiquitination of EG5 at site K1034 through its RING domain. Next, the ubiquitinated EG5 protein interacts with TPX2, thereby driving EG5 relocalization to spindle microtubules and controlling the distribution of EG5 along microtubules during metaphase. In the cells where UHRF1 is unable to catalyze the ubiquitination of EG5 at K1034, TPX2 is unable to bind EG5 protein, thereby reducing EG5 localization to the antiparallel microtubules on the half-spindle, resulting in the formation of disordered microtubules at the spindle equator and chromosome misalignment (Fig. 7). Altogether, EG5 polyubiquitination regulated by UHRF1 protein is suggested to be a novel regulatory mechanism for spindle assembly.
A schematic model showing the molecular mechanism by which UHRF1 promotes spindle assembly and chromosome congression in mitosis by catalyzing mitotic kinesin motor EG5 polyubiquitination for the interaction with TPX2.
A schematic model showing the molecular mechanism by which UHRF1 promotes spindle assembly and chromosome congression in mitosis by catalyzing mitotic kinesin motor EG5 polyubiquitination for the interaction with TPX2.
Discussion
UHRF1 has been reported to regulate DNA replication (Sharif et al., 2007) and cell cycle progression (Tu et al., 2020; Jeanblanc et al., 2005), while UHRF1 deletion resulted in cell cycle arrest at the G1/S or G2/M phases, suggesting that UHRF1 regulates the process of mitosis. However, the mechanism remains largely elusive. In this present study, we provided direct evidence that UHRF1 regulates EG5-mediated spindle assembly and chromosome congression during the process of mitosis. Our data highlight that the ubiquitin ligase activity of UHRF1 is required for EG5 localization on spindles to perform its functions and avoid subsequent mitotic errors.
UHRF1 is composed of multiple functional domains including the UBL, TUDOR, PHD, SRA, and RING domains. TUDOR, PHD, and SRA domains have been reported to play important roles in epigenetic regulation (Rajakumara et al., 2011; Sharif et al., 2007; Cheng et al., 2013). The RING domain of UHRF1 is relatively less studied. K48-linked polyubiquitination mainly targets proteins for proteasome-dependent protein degradation, whereas K63-linked polyubiquitination serves as a scaffold to recruit the target proteins and facilitates protein/protein interaction, in turn regulating the localization and activity of protein kinase. In addition to the histone, UHRF1 also regulates some non-histone protein ubiquitination. A previous study suggests that the RING domain of UHRF1 is involved in DNMT1 degradation by K48-linked polyubiquitination (Du et al., 2010). UHRF1 also catalyzes the K63-linked polyubiquitination of RIF1, decreasing the interaction between RIF1 and 53BP1 (Zhang et al., 2016). The interplay of UHRF1 with EG5 is critical for the maintenance of genome stability by ensuring faithful chromosome transmission. Furthermore, UHRF1 regulated the protein interaction of TPX2 with EG5 by inducing EG5 K63-linked polyubiquitination at K1034, thereby controlling the localization of EG5 on spindles during mitosis. Importantly, UHRF1 depletion enhanced EG5 accumulation at the spindle poles during metaphase and reduced the ATPase activity of EG5, suggesting that UHRF1 might modulate the motor activity of EG5. To our knowledge, this is the first report that UHRF1 as a nuclear protein is required for the maintenance of chromosome alignment and spindle architecture during mitosis.
The formation of a bipolar mitotic spindle depends on the precise distribution of EG5 on spindles to maintain the spatial accuracy of microtubules and the integrity of spindles. Beyond its functions regulating bipolar spindle formation at the spindle poles in early mitosis (Zheng et al., 2022; Blangy et al., 1995), EG5 is required for metaphase spindle elongation and maintenance of spindle bipolarity by acting on antiparallel microtubules at the spindle midzone during metaphase (Chen et al., 2018; Miyamoto et al., 2004). Moreover, EG5 near the mitotic kinetochore regulates accurate kinetochore-microtubule attachment and chromosome congression (Kajtez et al., 2016; He et al., 2016). Our results showed that EG5 inwardly distributes on the half-spindle at metaphase, which has been confirmed in other studies (Fang et al., 2020). Furthermore, UHRF1 depletion or truncation mutation (UHRF1ΔRING) disrupted the inward distribution of EG5 protein during metaphase and enhanced its spindle poles accumulation. Conversely, the improper distribution of EG5 may thus deregulate microtubule dynamics, resulting in spindle defects and chromosome misalignment.
The localization of EG5 on spindles and its activity at different mitotic stages imply that EG5 distribution on microtubules depends on an intrinsic control system. Spindle localization and activity of EG5 are mainly regulated by phosphorylation modification. EG5 phosphorylation is induced by a series of upstream kinases including NIMA family protein kinase Nek6/Nek7 and Cdk1. Nek9 is activated and directly phosphorylates and activates Nek6/Nek7 through protein interaction. Plk1 controls the phosphorylation of EG5 at the Nek6 site S1033 at centrosomes during early mitosis (Eibes et al., 2018; Rapley et al., 2008). In Homo sapiens, Xenopus laevis, and Drosophila melanogaster, EG5 phosphorylation by Cdk1 at Thr926 promotes its localization on the mitotic spindles and drives bipolar spindle formation (Smith et al., 2011). Most previous studies focused on EG5 phosphorylation, but less on other posttranslational modifications. We for the first time found that UHRF1 directly interacts with EG5 and regulates EG5–TPX2 interactions by inducing EG5 ubiquitination at site K1034. UHRF1 depletion or the K1034R mutation reduced the EG5–TPX2 interaction, resulting in EG5-irregular distribution. EG5 promotes bipolar spindle formation and maintains spindle architecture through its ATP hydrolysis function (Blangy et al., 1995; Kapitein et al., 2008). UHRF1 depletion or the K1034R mutation significantly reduced the ATPase activity of EG5, suggesting UHRF1 regulates EG5 motor activity through ubiquitination modification.
The motility, directionality, or localization of EG5 protein on microtubules was modulated by the interactions with other regulatory proteins, such as TPX2 or dynein (Gable et al., 2012). TPX2, a Ran-regulated spindle assembly factor, binds to microtubules during spindle assembly and subsequently recruits other proteins to the spindles, perhaps by acting as a scaffold (Kufer et al., 2002; Vanneste et al., 2009). Although TPX2 targets EG5 to spindle microtubules, the poleward motion of TPX2 requires EG5, dynein, and microtubule flux (Ma et al., 2010). The data of TPX2 fluorescence distribution showed that the UHRF1-catalyzed EG5 ubiquitination did not control the localization of TPX2 on spindles. UHRF1 depletion impairs mitotic spindle architecture, manifesting as two interrelated yet independent forms of spindle disorganization, shortened spindles, and disordered microtubules at the spindle equator. These data suggested that UHRF1 regulates multiple distinct aspects of mitotic spindle assembly. The mechanism is critical to ensure correct chromosomal behavior in the context of kinetochore–microtubule interactions.
Altogether, we revealed a previously unknown mechanism by which UHRF1 maintains mitotic spindle architecture and chromosome congression to maintain genomic stability by inducing EG5 ubiquitination during mitosis. These functions are independent of its roles in epigenetic regulation as a nuclear protein. Furthermore, the present study provided convincing evidence to explain the mechanism by which UHRF1 deletion induced G2/M cell cycle arrest.
Materials and methods
Cell culture and viral infection
HEK293T (CRL-3216), DU145 (HTB-81), and PC3 (CRL-1435) cells were obtained from the American Type Culture Collection (ATCC). Cells were maintained in DMEM (11995040; Gibco) or 1640 medium (11875093; Gibco) with 10% fetal bovine serum (10270-106; Gibco), 100 U/ml penicillin, and 100 μg/ml streptomycin (14140163; Gibco). Stable cell lines expressing UHRF1 shRNA were generated by transfecting DU145 or PC3 cells with pLKO.1/pLKO.1-UHRF1 shRNA plasmids. Stable DU145 cell lines expressing EG5 shRNA were generated by transfecting with pLKO.1/pLKO.1-EG5 shRNA plasmids and were selected by 1 μg/ml puromycin. All cells were cultured in a humidified incubator at 37°C and 5% CO2.
Plasmids and siRNA transfection
Vectors used in the study and their sources are listed as follows: pGEX-4 T-1 (#40064; Addgene), pcDNA3.1-His (#10958; Addgene), pCMV-Flag (#86099; Addgene), pLKO.1 (#26655; Addgene). pGEX-4 T-1-UHRF1, pGEX-4 T-1-EG5, pcDNA3.1-UHRF1-His, and pCMV–EG5–Flag plasmids were constructed by generating the DNA fragments of UHRF1 and EG5, were amplified from cDNA of HEK293T cells. DNA fragments were inserted into the described vectors. Primers for UHRF1 were 5′-ATGTGGATCCAGGTTCGGACCAT-3′ and 5′-CCGGCCATTGCCGTAGCC-3′; primers for EG5 were 5′-ATGGCGTCGCAGCCAAATTCG-3′ and 5′-TTAAAGGTTGATCTGGGCTCGCAGA-3′. UHRF1 deleting the RING domain mutant and EG5K1034R mutant was generated by the DNA fragments of wild-type and mutated UHRF1 or EG5 by PCR and then subcloned into the above vectors. Primers for UHRF1ΔRing were 5′-ATGTGGATCCAGGTTCGGACCAT-3′ and 5′-CTGGAACGTCTCCTCCACTTT-3′; primers for EG5K1034R were 5′-GGAGAGGTCTAGAGTGGAAGAAACTACAGAGCACTTGG-3′ and 5′-CTTCCACTCTAGACCTCTCCAGTGTGTTAATGCCTC-3′. pLKO.1-UHRF1 shRNA plasmids were generated by subcloning UHRF1 shRNA into pLKO.1 lentiviral vector. (UHRF1 shRNA Sense: 5′-GCGCTGGCTCTCAACTGCT-3′).
siRNA transfection was performed using a Mirus transfection kit according to the manufacturer’s protocol (Mirus). The sequences of UHRF1 siRNA (5′-3′) were as follows: UHRF1 siRNA-1# (5′-GCGCUGGCUCUCAACUGCU-3′) and UHRF1 siRNA-2# (5′-GCAUCUACAAGGUUGUGAA-3′).
Antibodies
The following antibodies were used for immunoblotting: Anti-mouse IgG (H + L; 4410S; 1:5,000), mouse monoclonal α-tubulin antibody (76031; 1:1,000), rabbit monoclonal UHRF1 antibody (87632; 1:1,000), and HA rabbit monoclonal antibody (3724; 1:1,000) were purchased from Cell Signal Transduction (CST). Rabbit polyclonal EG5 antibody (A7907; 1:1,000) and rabbit polyclonal TPX2 antibody (A18327; 1:1,000) were purchased from ABclonal Technology. β-Actin mouse monoclonal antibody (47778; 1:5,000) was purchased from Santa Cruz Biotechnology. Anti-rabbit IgG (H + L; 30000-0-AP; 1:5,000), Flag rabbit polyclonal antibody (20543-1-AP; 1:1,000), and His mouse monoclonal antibody (66005-1-lg; 1:1,000) were purchased from Proteintech.
For immunofluorescence, we employed a mouse monoclonal α-tubulin antibody (76031; 1:100) that was purchased from Cell Signal Transduction (CST). Rabbit polyclonal EG5 antibody (A7907; 1:100) and rabbit polyclonal TPX2 antibody (A18327; 1:100) were purchased from ABclonal Technology. Secondary antibodies labeled with AlexaFluor goat anti-mouse 488 (53-9760-82; 1:500) and goat anti-rabbit 594 (A-21312; 1:500) were purchased from Invitrogen.
Immunoprecipitation
Preparation of extracts for immunoprecipitation used RIPA buffer supplemented with protease inhibitor cocktails (Selleck Chemicals). The extracts were then incubated with 4 μg specified antibodies at 4°C for 12 h on a rotator and then incubated with protein A/G-magnetic beads (MCE) at 4°C for 1 h on a rotator. After that, the beads were washed four times in immunoprecipitation buffer and boiled in 50 μl 1 × loading buffer. IgG was used as a negative control and the extracts were used as a positive control. The precipitate samples were analyzed by Western blot.
Immunoblotting
Cells were lysed and protein was extracted using RIPA buffer supplemented with protease inhibitor cocktails (Selleck Chemicals). Protein concentration was measured with a Bradford assay kit (Thermo Fisher Scientific). The protein was loaded in equal amounts and separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE), with β-actin used as a loading control. The protein samples were run on 10% SDS-PAGE gels and wet transferred to 0.45-μm PVDF membranes. The membranes were blocked in TBST with 5% non-fat dried milk for 1 h at room temperature and then incubated with primary antibodies at 4°C overnight. After three washes with TBST, membranes were incubated with secondary antibodies for 1 h at room temperature. Primary antibodies were used with 1:1,000 dilutions in primary antibody dilution buffer. Secondary antibodies were used with 1:5,000 dilutions in primary antibody dilution buffer. The antigens were detected using ECL Plus Western Blotting Detection Reagents (Advansta).
GST pull-down assay and in vitro ubiquitination assay
According to the manufacturer’s instructions, pGEX-4 T-1-UHRF1 or pGEX-4 T-1-EG5 was constructed by the GST gene fusion system. To produce glutathione S-transferase (GST) and GST-UHRF1 or GST-EG5 fusion proteins, complete BL21 cells were transfected with pGEX-4 T-1 and pGEX-4 T-1-UHRF1 or pGEX-4 T-1-EG5, respectively. A fresh bacterial colony was inoculated into an LB medium containing ampicillin and grown overnight at 37°C. Bacterial cultures were diluted in 200 ml LB with ampicillin and grown for ∼6 h at 37°C until the OD600 value reached 0.6. Then protein expression was induced by 1 mM ispropyl b-D-1-thiogalactopyranoside (IPTG) 8 h at 16°C. GST and GST-UHRF1 or GST-EG5 proteins were purified by binding with glutathione glycan resins (C600031; Sangon Biotech). The resins were incubated with cell lysates of HEK293T cells at 4°C for 12 h on a rotator and then washed four times with the washing buffer. Resin-bound complexes were eluted by boiling in 50 μl 1 × loading buffer and analyzed by Western blot.
Recombinant EG5, recombinant UHRF1 (WT and ΔRING), purified E2 enzyme, and a mixture of E1 plus ubiquitin (WT, K48 only, and K63 only) were prepared. Ubiquitination was analyzed with a ubiquitination kit (K-230; Boston Biochem) following protocols recommended by the manufacturer. The ubiquitination of EG5 was examined with HA antibody.
Cellular immunofluorescence
The cells were synchronized at the G1/S phase by using the double thymidine block and released by culture media. The cells were synchronized in G2/M phases at 9 h after release. Synchronized cells grown on coverslips were fixed in 4% paraformaldehyde solution at room temperature for 15 min. After washing three times, cells were permeabilized with 0.25% Triton X-100 at room temperature for 20 min. Following three PBS washes, cells were blocked with 5% bovine serum albumin for 1 h at room temperature. Then, cells were incubated with primary antibodies in 1% BSA overnight at 4°C, washed three times in PBST, and incubated with the fluorescent-labeled secondary antibodies for 1 h at room temperature in the dark. EG5 and TPX2 incubated with goat anti-rabbit 594 (A-21312; 1:500; Invitrogen) and α-tubulin incubated with goat anti-mouse 488 (53-9760-82; 1:500; Invitrogen) for secondary staining. Primary antibodies were used with 1:100 dilutions in 1% BSA. Secondary antibodies were used with 1:500 dilutions in 1% BSA. Nuclear DNA was stained with 1:1,000 dilutions 4,6-diamidino-2-phenylindole (DAPI; P36935; Thermo Fisher Scientific).
Fixed cells were visualized at 27°C using a Zeiss LSM 980 with Airyscan 2 confocal microscopy equipped with a 100×, 1.4-NA objective in the presence of immersion oil (Carl Zeiss AG) and captured on three detectors (multiakali, GaAsP, and Airyscan). Confocal images were acquired and analyzed with Zeiss Zen 3.7 software (Carl Zeiss AG). The actual length of the spindles was measured using the Zeiss Zen 3.7. To quantify corresponding EG5, TPX2, and asters’ fluorescence intensity, the corresponding areas were outlined manually and the mean fluorescence intensity was analyzed using ImageJ.
ATPase assays
For in vitro ATPase assays, DU145 cells were transfected with Flag-tagged wild-type or K1034R mutant pCMV–EG5 expression plasmids. After the 72 h recovery period, cells were lysed and protein was extracted using RIPA buffer supplemented with protease inhibitor cocktails (Selleck Chemicals), and EG5 was immunoprecipitated using the Flag antibody. Bound Flag-tagged EG5 was eluted using a Flag peptide (B26101; Bimake) by incubating at 4°C for 30 min. Eluted wild-type or K1034R mutant EG5 was used in the ADP-Glo assay, which measures the amount of ADP produced, according to the manufacturer’s instructions (Promega). A reaction containing no immunoprecipitated EG5 was used for background correction.
Flow cytometry
DU145 and PC3 cells with or without UHRF1 depletion were established by stable transfection with shRNA vectors for UHRF1 or control. For cell cycle analysis, DU145 and PC3 cells were first fixed in 70% ethanol at 4°C overnight. Then 0.02 mg/ml propidium iodide containing RNase (30 μg/ml) was used for staining DNA at 37°C for 30 min in the dark. Cell cycle phase distribution was analyzed by flow cytometry (Millipore).
Statistical analysis
All data were analyzed by Origin 8.0 (originlab) and Prism v8 (GraphPad Software). The results of Fig. 1, D, F, and H; Fig. 2, C, D, G, and H; Fig. 5, B, D, H, and L; Fig. 6, B, D, and G; Fig. S3, D, E, and G are presented as mean ± SD; error bars in figures represent SD; for at least triplicate experiments. Dots represent individual cell samples in Fig. 2, B and F; and Fig. 6 F; bars are median ± quartile. The sample size was chosen on the basis of the size of the effect and variance for the different experimental approaches. The data distribution was tested for normality. One-way ANOVA followed by Tukey’s post hoc test was used to analyze the statistical differences among multiple groups. Significance is indicated by asterisks: n.s., not significant; P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001. P values < 0.05 are considered as statistically significant.
Online supplemental material
Fig. S1 shows the Western blotting of UHRF1 knockdown cells and in vitro recombinant proteins by GST-pull-down assay between UHRF1 and EG5. Fig. S2 shows the levels of K48 or K63 ubiquitin-protein of EG5 in HEK293T cells. Fig. S3 shows that EG5 localization and its interaction with TPX2 are affected by UHRF1 mutation and the location of TPX2 on spindles when the motor cannot be modified by UHRF1.
Data availability
Acknowledgments
We thank Dr. Sun Lunquan and the members of the Center of Molecular Medicine, Xiangya Hospital, Central South University for constructive discussion of this study.
This work was supported by grants from the National Natural Science Foundation of China (NSFC) 81572542 and 81874096, National Key Clinical Specialty Construction Project (Clinical Pharmacy), and High-Level Clinical Key Speciality (Clinical Pharmacy) in Guangdong Province.
Author contributions: X. Qi: data collection and analysis, project development, and manuscript writing. Y. Liu, Y. Peng, Y. Fu, Y. Fu, L. Yin: data collection and analysis. X. Li: project administration, data analysis, and manuscript writing. All authors read and approved the final manuscript.
References
Author notes
Disclosures: The authors declare no competing interests exist.