Serving as the power plant and signaling hub of a cell, mitochondria contain their own genome which encodes proteins essential for energy metabolism and forms DNA–protein assemblies called nucleoids. Mitochondrial DNA (mtDNA) exists in multiple copies within each cell ranging from hundreds to tens of thousands. Maintaining mtDNA homeostasis is vital for healthy cells, and its dysregulation causes multiple human diseases. However, the players involved in regulating mtDNA maintenance are largely unknown though the core components of its replication machinery have been characterized. Here, we identify C17orf80, a functionally uncharacterized protein, as a critical player in maintaining mtDNA homeostasis. C17orf80 primarily localizes to mitochondrial nucleoid foci and exhibits robust double-stranded DNA binding activity throughout the mitochondrial genome, thus constituting a bona fide new mitochondrial nucleoid protein. It controls mtDNA levels by promoting mtDNA replication and plays important roles in mitochondrial metabolism and cell proliferation. Our findings provide a potential target for therapeutics of human diseases associated with defective mtDNA control.
Introduction
Mitochondria are unique semiautonomous organelles in eukaryotic cells that have their own genome encoding proteins essential for energy metabolism and other mitochondrial functions (Friedman and Nunnari, 2014). The human mitochondrial genome is a circular, 16,569-bp double-stranded DNA (dsDNA) that contains 37 genes encoding 13 proteins involved in the oxidative phosphorylation process and 22 tRNAs and 2 rRNAs required for intramitochondrial protein synthesis (Anderson et al., 1981; Andrews et al., 1999; Falkenberg et al., 2007). As opposed to nuclear DNA (nDNA), mitochondrial DNA (mtDNA) is highly compact and lacks an intron–exon structure. MtDNA is associated with a number of proteins destined to be packaged into macromolecular assemblies called nucleoids, which serve to protect mtDNA and facilitate signaling at the sub-organellar level (Gilkerson, 2009). These nucleoid proteins are particular participants in mtDNA maintenance, for example, mitochondrial single-stranded DNA-binding protein (mtSSB) and transcription factor A of mitochondria (TFAM; Bogenhagen, 2012; Bogenhagen et al., 2008; Garrido et al., 2003; Hensen et al., 2014; Wang and Bogenhagen, 2006). MtDNA polymerase γ (POLγ), mitochondrial RNA polymerase (POLRMT), and mtDNA helicase TWINKLE are present in mitochondrial nucleoids as well (Garrido et al., 2003; Hensen et al., 2014; Wang and Bogenhagen, 2006). Determining the bona fide protein components of the nucleoids has become an important question in mitochondrial nucleoid biology.
Distinct from nDNA, mtDNA exists in multiple copies within each cell, ranging from a few hundreds to many tens of thousands depending on the cell type and metabolic state (Gustafsson et al., 2016). In addition, each mitochondrion may contain several copies of mtDNA. Mitochondria use their own replication machinery whose components are encoded by the nucleus genome to maintain proper mtDNA copy numbers. The core components are POLγ, TWINKLE, mtSSB, and POLRMT (Chouchani et al., 2016; Kazak et al., 2012; Wanrooij and Falkenberg, 2010). Briefly, TWINKLE helicase acts ahead of the mtDNA replication fork to unwind the two strands by translocation on one DNA strand in the 5′–3′ direction. The single-stranded mtDNA loop formed is then coated by tetrameric mtSSB, and POLγ holoenzyme catalyzes the extension of the RNA primer synthesized by POLRMT using the single-stranded mtDNA released by TWINKLE as a template. The primer is then degraded by RNase H1 (Akhmedov and Marín-García, 2015).
Unlike nDNA, mtDNA replicates in both proliferating cells and differentiated cells, such as neuronal and cardiac cells (Magnusson et al., 2003; Ylikallio et al., 2010), indicating that mtDNA actively turns over in postmitotic cells. As such, mtDNA replication is a continuous and indispensable event throughout the life of an organism. Efficient replication to maintain proper mtDNA levels is critical for normal development and healthy life. Defects caused by mutations in proteins involved in mtDNA replication result in a plethora of diseases with impaired mitochondrial functions (Ashley et al., 2007; Moraes et al., 1991; Nishigaki et al., 2003; Viscomi and Zeviani, 2017; Young and Copeland, 2016). For example, about 200 mutations in the POLG (the catalytic POLγ A subunit) and POLG2 (the accessory POLγ B subunit) genes have been detected, and these mutations contribute to the pathogenesis of rare hereditary mitochondrial diseases, including neurodegenerative disorders such as Alpers-Huttenlocher, early childhood hepatocerebral syndromes, ataxia neuropathy, and progressive external ophthalmoplegia (Akhmedov and Marín-García, 2015; Copeland, 2012; Stumpf and Copeland, 2011; Tang et al., 2011; Wong et al., 2008). In addition, over 30 different mutations in the TWINKLE gene have been identified that result in deletions or depletion of mtDNA (Spelbrink et al., 2001; Wanrooij and Falkenberg, 2010) and are associated with neuromuscular disorders (Suomalainen et al., 1997). Moreover, mtDNA abnormalities have been observed in Parkinson’s disease (Bender et al., 2006; Coskun et al., 2012), Alzheimer’s disease (Coskun et al., 2004; Wang et al., 2005), amyotrophic lateral sclerosis (Keeney and Bennett, 2010), and aging (Sondheimer et al., 2011).
It is increasingly appreciated that mtDNA replication is quite complex and finely regulated so that multiple regulatory players can be engaged to coordinate this process. However, it is largely unknown which proteins participate in regulating mtDNA maintenance, though the core constitutes of mtDNA replication machinery have been characterized. Identifying critical regulators not only helps to unravel the mechanisms underlying mitochondrial genome maintenance but also paves the way for understanding the molecular basis of human diseases and aging that are associated with mtDNA abnormalities.
In the current study, we identified C17orf80 as a critical player in regulating mtDNA maintenance. C17orf80 is a functionally uncharacterized protein that has recently been identified as a mitochondrial protein either by a BioID-based proximity-dependent biotinylation assay (Antonicka et al., 2020) or in the human mitochondrial high-confidence proteome (Morgenstern et al., 2021). We show that C17orf80 localizes primarily in the mitochondrial nucleoid foci and is capable of binding mtDNA without sequence specificity. While its upregulation significantly enhances mtDNA copy number, its downregulation shows the opposite effect. Moreover, we demonstrate that C17orf80 plays a crucial role in promoting mtDNA replication. Finally, we show that C17orf80 ablation impairs mitochondrial metabolism and leads to compromised cell proliferation. Our findings indicate that C17orf80 serves as a critical player in maintaining mtDNA copy number via promoting its replication.
Results and discussion
C17orf80 primarily localizes to mitochondrial nucleoids
To hunt for players involved in mtDNA maintenance, we performed immunoprecipitation (IP) in isolated mitochondria from HEK-293T cells using anti-DNA antibody followed by mass spectrometry (MS) identification. Several known mtDNA interactive proteins were found, such as TFAM, MRPS9, and MRPS21 (Fig. 1 A and Fig. S1 A), affirming the reliability of this assay. Interestingly, C17orf80, which is a functionally uncharacterized human protein, also appeared in the mtDNA-interaction protein list (Fig. 1 A and Fig. S1 A). We then selected it for further investigation of its potential role in mtDNA metabolism.
Human C17orf80 and its mouse ortholog D11Wsu47e colocalize with mtDNA. (A) The proteins interacting with mtDNA were screened by IP followed by MS identification. Isolated mitochondria from HEK-293T cells were subjected to IP using an anti-DNA antibody. The score number reflects the levels of protein credibility and abundance. The known mtDNA-interacting proteins (red) and the uncharacterized protein C17orf80 (blue) are marked. (B) Western blot confirmed the interaction between C17orf80 and mtDNA. Anti-DNA antibody was used for IP. TFAM served as the positive control and MCU (the core channel portion of mitochondrial Ca2+ uniporter holocomplex) as the negative control. (C) Western blot showing C17orf80 is highly enriched in mitochondria isolated from HEK-293T cells. ATP5A and β-actin served as the mitochondrial and cytosol markers, respectively. (D) Electron microscopy images of immunogold stained mitochondria in HEK-293T cells not expressing (CON) or stably expressing C17orf80-HA fusion protein (OE). Anti-HA antibody was used for immunogold staining. Left: Western blot showing expression of endogenous and C-terminal HA-tagged C17orf80 in CON and OE cells. Anti-β-actin served as the loading control. Middle: Representative electron microscopy images. Red arrowheads indicate immunogold particles. Scale bars: 100 nm. Right: Quantification of particle numbers. Data are mean ± SEM. n = 25 mitochondria per group. (E and F) Immunofluorescence staining shows that C17orf80 is distributed in a punctate pattern within the mitochondria of HeLa cells (E) and U2OS cells (F). Mitochondria were visualized with anti-ATPB immunofluorescence, and C-terminal Myc-tagged C17orf80 was stained with anti-Myc immunofluorescence. Scale bars are 5 μm in the large-view image and 1 µm in the zoom in image. (G) Colocalization of C17orf80 and mtDNA in HeLa cells. The C-terminal Flag-tagged C17orf80 was indicated by anti-Flag immunofluorescence and mtDNA was stained by Picogreen. Note that nuclear (N, marked by a dotted line) DNA was also stained by Picogreen. The pixel intensity plot of the white dashed line is shown in the right panel. Scale bars: 2 µm. AU, arbitrary unit. (H) Immunofluorescence staining showing colocalization between C17orf80 and mtDNA in HeLa cells. C17orf80-Flag and mtDNA were indicated by anti-Flag and anti-DNA immunofluorescence, respectively. The nucleus (N) was marked by a dotted line. The pixel intensity plot of the white dashed line is shown in the right panel. Scale bars: 2 µm. (I) Colocalization of mouse ortholog D11Wsu47e with mtDNA in MEF cells. D11Wsu47e was indicated by anti-HA immunofluorescence, mtDNA was stained by Picogreen, and mitochondria was indicated by anti-ATPB immunofluorescence. The nucleus (N) was marked by a dotted line. The right panel shows a pixel intensity plot of the white dashed line. Scale bars: 2 µm. (J) Pearson’s correlation coefficients are shown for C17orf80-Flag and Picogreen in G, C17orf80-Flag and mtDNA in H, and D11Wsu47e-HA and Picogreen in I, respectively. Source data are available for this figure: SourceData F1.
Human C17orf80 and its mouse ortholog D11Wsu47e colocalize with mtDNA. (A) The proteins interacting with mtDNA were screened by IP followed by MS identification. Isolated mitochondria from HEK-293T cells were subjected to IP using an anti-DNA antibody. The score number reflects the levels of protein credibility and abundance. The known mtDNA-interacting proteins (red) and the uncharacterized protein C17orf80 (blue) are marked. (B) Western blot confirmed the interaction between C17orf80 and mtDNA. Anti-DNA antibody was used for IP. TFAM served as the positive control and MCU (the core channel portion of mitochondrial Ca2+ uniporter holocomplex) as the negative control. (C) Western blot showing C17orf80 is highly enriched in mitochondria isolated from HEK-293T cells. ATP5A and β-actin served as the mitochondrial and cytosol markers, respectively. (D) Electron microscopy images of immunogold stained mitochondria in HEK-293T cells not expressing (CON) or stably expressing C17orf80-HA fusion protein (OE). Anti-HA antibody was used for immunogold staining. Left: Western blot showing expression of endogenous and C-terminal HA-tagged C17orf80 in CON and OE cells. Anti-β-actin served as the loading control. Middle: Representative electron microscopy images. Red arrowheads indicate immunogold particles. Scale bars: 100 nm. Right: Quantification of particle numbers. Data are mean ± SEM. n = 25 mitochondria per group. (E and F) Immunofluorescence staining shows that C17orf80 is distributed in a punctate pattern within the mitochondria of HeLa cells (E) and U2OS cells (F). Mitochondria were visualized with anti-ATPB immunofluorescence, and C-terminal Myc-tagged C17orf80 was stained with anti-Myc immunofluorescence. Scale bars are 5 μm in the large-view image and 1 µm in the zoom in image. (G) Colocalization of C17orf80 and mtDNA in HeLa cells. The C-terminal Flag-tagged C17orf80 was indicated by anti-Flag immunofluorescence and mtDNA was stained by Picogreen. Note that nuclear (N, marked by a dotted line) DNA was also stained by Picogreen. The pixel intensity plot of the white dashed line is shown in the right panel. Scale bars: 2 µm. AU, arbitrary unit. (H) Immunofluorescence staining showing colocalization between C17orf80 and mtDNA in HeLa cells. C17orf80-Flag and mtDNA were indicated by anti-Flag and anti-DNA immunofluorescence, respectively. The nucleus (N) was marked by a dotted line. The pixel intensity plot of the white dashed line is shown in the right panel. Scale bars: 2 µm. (I) Colocalization of mouse ortholog D11Wsu47e with mtDNA in MEF cells. D11Wsu47e was indicated by anti-HA immunofluorescence, mtDNA was stained by Picogreen, and mitochondria was indicated by anti-ATPB immunofluorescence. The nucleus (N) was marked by a dotted line. The right panel shows a pixel intensity plot of the white dashed line. Scale bars: 2 µm. (J) Pearson’s correlation coefficients are shown for C17orf80-Flag and Picogreen in G, C17orf80-Flag and mtDNA in H, and D11Wsu47e-HA and Picogreen in I, respectively. Source data are available for this figure: SourceData F1.
Mitochondrial nucleoid localization of C17orf80 in HEK-293T cells and sequence alignment between human C17orf80 and mouse D11Wsu47e. (A) The top 18 mtDNA-interacting proteins as shown in Fig. 1 A. (B) Immunofluorescence images showing mitochondrial localization of C17orf80 in HEK-293T cells. C17orf80-Myc was indicated by anti-Myc fluorescence and mitochondria were visualized with anti-ATPB fluorescence. Scale bars are 10 μm in the large-view image and 1 µm in the zoom in image. (C and D) Colocalization of C17orf80 and mtDNA indicated by Picogreen staining (C) or anti-DNA immunofluorescence (D). C17orf80-Flag was indicated by anti-Flag immunofluorescence. Nuclear (N) is marked by a dotted line. The pixel intensity plots of the white dashed lines are shown in the right panels. Scale bars: 2 µm. AU, arbitrary unit. (E) Pearson’s correlation coefficients for C17orf80-Flag and Picogreen in Fig. S1 C and C17orf80-Flag and mtDNA in Fig. S1 D. (F) Full-length sequence alignment of human C17orf80 with its mouse ortholog D11Wsu47e. 58% sequence similarity was observed. (G) Anti-HA Western blot showing expression of C-terminal HA-tagged D11Wsu47e. Anti-ATP5A served as the loading control. Source data are available for this figure: SourceData FS1.
Mitochondrial nucleoid localization of C17orf80 in HEK-293T cells and sequence alignment between human C17orf80 and mouse D11Wsu47e. (A) The top 18 mtDNA-interacting proteins as shown in Fig. 1 A. (B) Immunofluorescence images showing mitochondrial localization of C17orf80 in HEK-293T cells. C17orf80-Myc was indicated by anti-Myc fluorescence and mitochondria were visualized with anti-ATPB fluorescence. Scale bars are 10 μm in the large-view image and 1 µm in the zoom in image. (C and D) Colocalization of C17orf80 and mtDNA indicated by Picogreen staining (C) or anti-DNA immunofluorescence (D). C17orf80-Flag was indicated by anti-Flag immunofluorescence. Nuclear (N) is marked by a dotted line. The pixel intensity plots of the white dashed lines are shown in the right panels. Scale bars: 2 µm. AU, arbitrary unit. (E) Pearson’s correlation coefficients for C17orf80-Flag and Picogreen in Fig. S1 C and C17orf80-Flag and mtDNA in Fig. S1 D. (F) Full-length sequence alignment of human C17orf80 with its mouse ortholog D11Wsu47e. 58% sequence similarity was observed. (G) Anti-HA Western blot showing expression of C-terminal HA-tagged D11Wsu47e. Anti-ATP5A served as the loading control. Source data are available for this figure: SourceData FS1.
Immunoblot confirmed the interaction between mtDNA and C17orf80 (Fig. 1 B). Biochemical analysis revealed that it was highly enriched in isolated mitochondria (Fig. 1 C). Electron microscopy of immunogold-stained C17orf80-HA fusion protein expressed in HEK-293T cells indicated that it was located primarily in the mitochondrial matrix (Fig. 1 D). These three lines of evidence demonstrate that C17orf80 is a mitochondria-localized protein, consistent with previous reports (Antonicka et al., 2020; Morgenstern et al., 2021). To further characterize its sub-organelle location, we expressed the C-terminal Myc-tagged C17orf80 in HeLa, U2OS, or HEK-293T cells and all observed a discrete punctate distribution within the mitochondrial network detected by anti-Myc immunofluorescence (Fig. 1, E and F; and Fig. S1 B). In terms of its interaction with mtDNA, such a punctate distribution prompted us to hypothesize that C17orf80 might localize in mitochondrial nucleoids, which are usually punctiform and distributed throughout the mitochondrial network (Alam et al., 2003; Legros et al., 2004). We then stained mtDNA with Picogreen and observed 79% and 84% overlap between Picogreen staining and Flag-tagged C17orf80 expressed in HeLa and HEK-293T cells, respectively (Fig. 1, G and J; Fig. S1, C and E). In a parallel experiment, exogenously expressed Flag-tagged C17orf80 also appeared to well colocalize with mtDNA stained by anti-DNA immunofluorescence (Fig. 1, H and J; Fig. S1, D and E). In addition, we characterized the mouse ortholog D11Wsu47e, which is also an uncharacterized protein with 58% sequence similarity to human C17orf80 (Fig. S1 F), and found that it displayed 83% overlap with mtDNA in mouse embryonic fibroblasts (MEF; Fig. 1, I and J; and Fig. S1 G). Taken together, these results clearly demonstrate that human C17orf80 and its mouse ortholog D11Wsu47e primarily localize to mitochondrial nucleoid foci.
C17orf80 exhibits non-sequence-specific dsDNA binding activity throughout the mitochondrial genome
We further set to determine whether C17orf80 possessed the ability to bind directly to mtDNA. We exploited chromatin immunoprecipitation combined with quantitative real-time polymerase chain reaction (ChIP-qPCR) and showed that C17orf80 extensively bound mtDNA throughout the mitochondrial genome with a slight enrichment at the QL site, the light (L)-strand origin of replication (Fig. 2 A). Consistent with this result, ChIP-sequencing (ChIP-seq) analysis also revealed abundant C17orf80 binding throughout the mitochondrial genome without obvious sequence specificity (Fig. 2 B). These two experiments demonstrate that C17orf80 displays robust non-specific mtDNA binding activity. The mildly enriched binding at the QL site suggests a possibility that a secondary structure of the mitochondrial genome could affect C17orf80 binding to mtDNA. Further, we characterized its DNA binding activity and specificity using an in vitro electrophoretic mobility shift assay (EMSA). The recombinant human C17orf80 was expressed and purified from E. coli (Fig. 2 C) and identified by MS (Fig. 2 D). The EMSA results showed C17orf80 bound double-stranded DNA (dsDNA) fragments derived from either mtDNA (Fig. 2 E) or nDNA (Fig. 2 F) in a concentration-dependent manner with the Kd values being 46 ± 7 nM and 49 ± 3 nM for mtDNA and nDNA fragments, respectively (Fig. 2 H). However, no obvious single-stranded DNA (ssDNA) binding activity was observed (Fig. 2, G and H). These results provide strong additional evidence to support the non-sequence-specific mtDNA binding activity of C17orf80. All these results indicate that C17orf80 extensively binds mtDNA throughout the mitochondrial genome, constituting a bona fide nucleoid protein.
C17orf80 binds dsDNA throughout the mitochondrial genome without sequence specificity. (A) ChIP-qPCR analysis of mtDNA binding enrichment by C17orf80-HA fusion protein overexpressed in HeLa cells. The primers targeted for different regions of mtDNA for qPCR amplification are indicated in the upper panel. Anti-HA antibody was used for mtDNA-IP. Data are mean ± SEM. n = 3 independent experiments per group. (B) ChIP-seq profile revealing C17orf80 binding mtDNA broadly throughout the mitochondrial genome. HeLa cells expressing C17orf80-HA were subjected to mtDNA-IP using an anti-HA antibody. (C) SDS-PAGE showing the purity of C17orf80 protein expressed and purified in E. coli. (D) MS identification of the purified C17orf80 protein. (E–G) EMSA analyzing C17orf80 binding activity to mitochondrial dsDNA (mito dsDNA, E), nuclear dsDNA (nuc dsDNA, F), and mitochondrial ssDNA (mito ssDNA, G). 10 nM 5′-Cy3 labeled 50 nt dsDNA or ssDNA were incubated with increasing concentrations of C17orf80 protein (0, 200, 400, 600, 800, 1,000, and 1,200 nM). (H) Quantification of bound DNA as a function of C17orf80 concentration. The Kd values for C17orf80 binding to mito dsDNA and nuc dsDNA fragments were calculated. Data are mean ± SEM. n = 3 independent experiments per group. Source data are available for this figure: SourceData F2.
C17orf80 binds dsDNA throughout the mitochondrial genome without sequence specificity. (A) ChIP-qPCR analysis of mtDNA binding enrichment by C17orf80-HA fusion protein overexpressed in HeLa cells. The primers targeted for different regions of mtDNA for qPCR amplification are indicated in the upper panel. Anti-HA antibody was used for mtDNA-IP. Data are mean ± SEM. n = 3 independent experiments per group. (B) ChIP-seq profile revealing C17orf80 binding mtDNA broadly throughout the mitochondrial genome. HeLa cells expressing C17orf80-HA were subjected to mtDNA-IP using an anti-HA antibody. (C) SDS-PAGE showing the purity of C17orf80 protein expressed and purified in E. coli. (D) MS identification of the purified C17orf80 protein. (E–G) EMSA analyzing C17orf80 binding activity to mitochondrial dsDNA (mito dsDNA, E), nuclear dsDNA (nuc dsDNA, F), and mitochondrial ssDNA (mito ssDNA, G). 10 nM 5′-Cy3 labeled 50 nt dsDNA or ssDNA were incubated with increasing concentrations of C17orf80 protein (0, 200, 400, 600, 800, 1,000, and 1,200 nM). (H) Quantification of bound DNA as a function of C17orf80 concentration. The Kd values for C17orf80 binding to mito dsDNA and nuc dsDNA fragments were calculated. Data are mean ± SEM. n = 3 independent experiments per group. Source data are available for this figure: SourceData F2.
C17orf80 plays an essential role in controlling mtDNA copy number
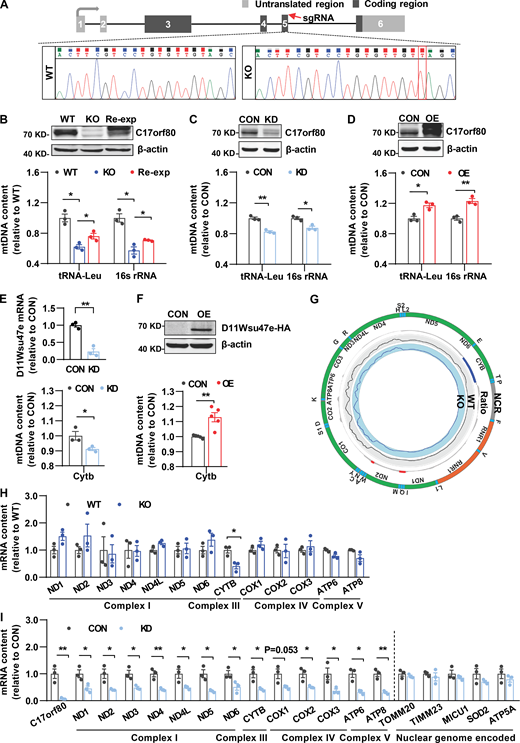
That C17orf80 exhibited robust mtDNA binding activity strongly suggested it could play a potential role in regulating mtDNA levels. We then determined the impact on mtDNA copy number by altering its expression. We generated a stable C17orf80 knockout HEK-293T cell line using the CRISPR/Cas9 system which caused a thymine insertion at the coding region of the C17orf80 gene (Fig. 3 A). The mtDNA levels measured with qPCR decreased by about 40% in the knockout cells (Fig. 3 B), and importantly, C17orf80 reexpression partially rescued the mtDNA contents in the knockout cells (Fig. 3 B). Meanwhile, we also transiently knocked down C17orf80 expression with RNA interference (RNAi) and found that mtDNA levels reduced by 15–20% in either HEK-293T (Fig. 3 C) or HeLa cells (Fig. S2 A). Conversely, C17orf80 overexpression significantly augmented mtDNA levels in both HEK-293T (Fig. 3 D) and HeLa cells (Fig. S2 B). In addition, downregulation and upregulation of the mouse ortholog D11Wsu47e in primary hepatocytes also showed opposing effects on mtDNA levels (Fig. 3, E and F). These results implicate a crucial role of human C17orf80 and its orthologs in controlling mtDNA contents.
Effect of altering C17orf80 and D11Wsu47e expression on mtDNA contents and transcription. (A) Schematic illustrating the C17orf80 knockout strategy in HEK-293T cells. The sgRNA targeted region is indicated (red arrow) and the sequencing result shows a thymine inserted (red square). (B) C17orf80 knockout reduced mtDNA contents that were partially rescued by C17orf80 reexpression in HEK-293T cells. Top: Anti-C17orf80 Western blot showing protein levels in wild type (WT), knockout (KO), and reexpression (Re-exp) cells. Anti-β-actin served as the loading control. Bottom: mtDNA contents in different groups. The tRNA-Leu and 16S rRNA regions were used to indicate mtDNA content. Data are mean ± SEM. *P < 0.05. (C) C17orf80 knockdown reduced mtDNA content in HEK-293T cells. The Western blot in the upper panel shows knockdown efficiency. Anti-β-actin served as the loading control. The lower panel shows mtDNA levels in control (CON) and knockdown (KD) cells. Data are mean ± SEM. *P < 0.05, **P < 0.01. (D) C17orf80 overexpression enhanced mtDNA contents in HEK-293T cells. Top: Anti-C17orf80 Western blot showing protein levels in control (CON) and C17orf80-Myc overexpression (OE) groups. Anti-β-actin served as the loading control. Bottom: mtDNA contents in control and overexpression groups. Data are mean ± SEM. *P < 0.05, **P < 0.01. (E and F) Opposite effects of D11Wsu47e downregulation (E) and upregulation (F) on mtDNA levels in mouse primary hepatocytes. The knockdown efficiency of D11Wsu47e was analyzed by qPCR (E). The D11Wsu47e-HA overexpression was analyzed by anti-HA Western blot (F). The Cytb region was used for indicating mtDNA content. Data are mean ± SEM. *P < 0.05, **P < 0.01. CON, control group; KD, D11Wsu47e knockdown group; OE, D11Wsu47e-HA overexpression group. (G) Few changes in the transcriptome of the mitochondrial genome in C17orf80 knockout HEK-293T cells as determined by RNA-seq coverage on the mtDNA sequence. The ratio was indicated by log2(KO/WT) with increases in red and decreases in blue. n = 2–3 biological repeats. (H) qPCR analysis shows that the vast majority of genes encoded by the mitochondrial genome (12 out of 13) were transcriptionally unchanged in C17orf80 knockout HEK-293T cells. Data are mean ± SEM. *P < 0.05 versus WT group. (I) Transcriptional changes of genes encoded by the mitochondrial genome and nuclear genome in C17orf80 knockdown and control HEK-293T cells. Data are mean ± SEM. *P < 0.05, **P < 0.01. Source data are available for this figure: SourceData F3.
Effect of altering C17orf80 and D11Wsu47e expression on mtDNA contents and transcription. (A) Schematic illustrating the C17orf80 knockout strategy in HEK-293T cells. The sgRNA targeted region is indicated (red arrow) and the sequencing result shows a thymine inserted (red square). (B) C17orf80 knockout reduced mtDNA contents that were partially rescued by C17orf80 reexpression in HEK-293T cells. Top: Anti-C17orf80 Western blot showing protein levels in wild type (WT), knockout (KO), and reexpression (Re-exp) cells. Anti-β-actin served as the loading control. Bottom: mtDNA contents in different groups. The tRNA-Leu and 16S rRNA regions were used to indicate mtDNA content. Data are mean ± SEM. *P < 0.05. (C) C17orf80 knockdown reduced mtDNA content in HEK-293T cells. The Western blot in the upper panel shows knockdown efficiency. Anti-β-actin served as the loading control. The lower panel shows mtDNA levels in control (CON) and knockdown (KD) cells. Data are mean ± SEM. *P < 0.05, **P < 0.01. (D) C17orf80 overexpression enhanced mtDNA contents in HEK-293T cells. Top: Anti-C17orf80 Western blot showing protein levels in control (CON) and C17orf80-Myc overexpression (OE) groups. Anti-β-actin served as the loading control. Bottom: mtDNA contents in control and overexpression groups. Data are mean ± SEM. *P < 0.05, **P < 0.01. (E and F) Opposite effects of D11Wsu47e downregulation (E) and upregulation (F) on mtDNA levels in mouse primary hepatocytes. The knockdown efficiency of D11Wsu47e was analyzed by qPCR (E). The D11Wsu47e-HA overexpression was analyzed by anti-HA Western blot (F). The Cytb region was used for indicating mtDNA content. Data are mean ± SEM. *P < 0.05, **P < 0.01. CON, control group; KD, D11Wsu47e knockdown group; OE, D11Wsu47e-HA overexpression group. (G) Few changes in the transcriptome of the mitochondrial genome in C17orf80 knockout HEK-293T cells as determined by RNA-seq coverage on the mtDNA sequence. The ratio was indicated by log2(KO/WT) with increases in red and decreases in blue. n = 2–3 biological repeats. (H) qPCR analysis shows that the vast majority of genes encoded by the mitochondrial genome (12 out of 13) were transcriptionally unchanged in C17orf80 knockout HEK-293T cells. Data are mean ± SEM. *P < 0.05 versus WT group. (I) Transcriptional changes of genes encoded by the mitochondrial genome and nuclear genome in C17orf80 knockdown and control HEK-293T cells. Data are mean ± SEM. *P < 0.05, **P < 0.01. Source data are available for this figure: SourceData F3.
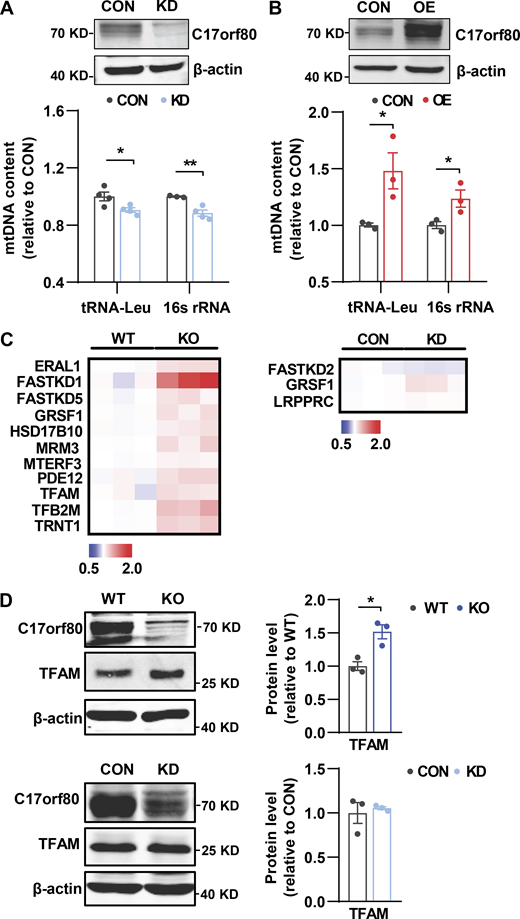
Effect of altering C17orf80 expression on mtDNA contents and mitochondrial transcription-related protein levels. (A and B) Effects of C17orf80 knockdown (KD) and overexpression (OE) on mtDNA levels in HeLa cells. Top: Western blots for C17orf80 knockdown (A) or C17orf80-HA overexpression (B). Anti-β-actin served as the loading control. Bottom: Changes in mtDNA content in different groups. Data are mean ± SEM. *P < 0.05, **P < 0.01. (C) Heatmap of fold changes of the mitochondrial transcription-related proteins analyzed by quantitative proteomics. The protein content in C17orf80 KO or KD cells was normalized to that of WT or CON cells, respectively. The mitochondrial transcription-related proteins with significant changes (P < 0.05) are shown. (D) Western blot showing upregulation of TFAM in the KO HEK-293T cells (upper panels) but no significant changes in the KD cells (lower panels). Anti-β-actin served as the loading control. Left: Representative Western blots. Right: Statistics. Data are mean ± SEM. *P < 0.05. Source data are available for this figure: SourceData FS2.
Effect of altering C17orf80 expression on mtDNA contents and mitochondrial transcription-related protein levels. (A and B) Effects of C17orf80 knockdown (KD) and overexpression (OE) on mtDNA levels in HeLa cells. Top: Western blots for C17orf80 knockdown (A) or C17orf80-HA overexpression (B). Anti-β-actin served as the loading control. Bottom: Changes in mtDNA content in different groups. Data are mean ± SEM. *P < 0.05, **P < 0.01. (C) Heatmap of fold changes of the mitochondrial transcription-related proteins analyzed by quantitative proteomics. The protein content in C17orf80 KO or KD cells was normalized to that of WT or CON cells, respectively. The mitochondrial transcription-related proteins with significant changes (P < 0.05) are shown. (D) Western blot showing upregulation of TFAM in the KO HEK-293T cells (upper panels) but no significant changes in the KD cells (lower panels). Anti-β-actin served as the loading control. Left: Representative Western blots. Right: Statistics. Data are mean ± SEM. *P < 0.05. Source data are available for this figure: SourceData FS2.
Since altering mtDNA content can cause transcriptional perturbance in the mitochondrial genome (Gustafsson et al., 2016), we further assessed mtRNA changes in the C17orf80 knockout cells. Inexplicably, RNA sequencing (RNA-seq) analysis of the stable knockout HEK-293T cells showed that the transcriptome of the mitochondrial genome was largely unaltered (Fig. 3 G). This was confirmed by qPCR analysis showing comparable transcription levels of 12 out of the 13 mitochondrial genome-encoded genes between the knockout and wild-type cells (Fig. 3 H). However, analyzing mtRNA levels in the transient knockdown cells revealed a significant and consistent reduction of all 13 genes encoded by the mitochondrial genome, whereas the transcription of mitochondrial genes encoded by the nuclear genome tested was unaltered (Fig. 3 I). Since both stable knockout and transient knockdown of C17orf80 led to decreased mtDNA levels that should reduce transcription, we looked to see whether the negligible impact of persistent C17orf80 knockout on mtRNA levels might be due to compensatory increases in mitochondrial transcription factors. In support, more significant upregulation of the mtDNA transcription-related players was observed in the knockout cells than in the knockdown cells (Fig. S2, C and D). It is also possible that altered mtRNA stability might contribute to the few mtRNA changes in the knockout cells. Such disparate effects on mitochondrial genome transcription by persistent knockout versus transient knockdown suggest that C17orf80 is not directly involved in the mtDNA transcription process.
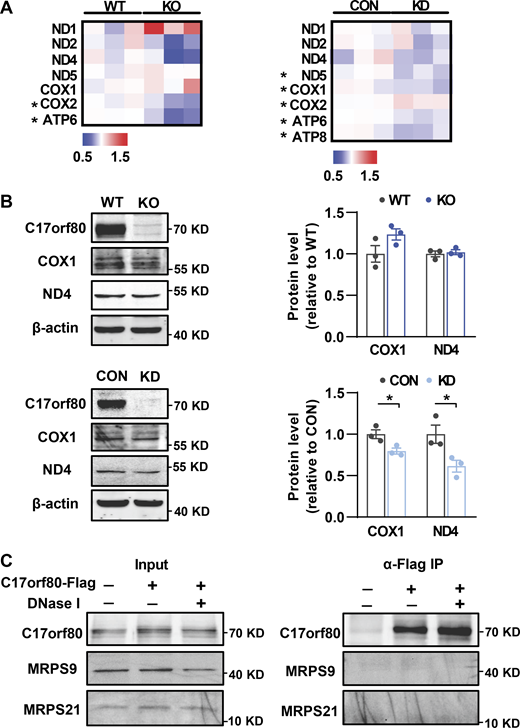
Meanwhile, we determined whether C17orf80 was involved in the mtDNA translation process by assessing the abundance changes of mtDNA-encoded proteins in the knockout and knockdown cells. Quantitative proteomics analysis revealed these mtDNA-encoded proteins detected did not display consistent changes in either C17orf80 knockout or knockdown cells (Fig. S3 A), which was partially confirmed by Western blot (Fig. S3 B). In addition, no apparent interaction between C17orf80 and the mitochondrial ribosome subunits MRPS9 and MRPS21, which were shown to interact with mtDNA (Fig. 1 A and Fig. S1 A), was detected (Fig. S3 C). Though more precise validations are required in the future, these results imply that C17orf80 is not directly involved in the mtDNA translation process.
Effects of C17orf80 ablation on mtDNA translation. (A) Heatmap of fold changes of the mtDNA-encoded proteins analyzed by quantitative proteomics. The protein content in C17orf80 knockout (KO) or knockdown (KD) HEK-293T cells was normalized to that of wild type (WT) or control (CON) cells, respectively. All mtDNA-encoded proteins detected are shown and the proteins with significant changes (P < 0.05) are marked by an asterisk. (B) Western blot showing few changes of COX1 and ND4 in the KO cells (upper panels) and their downregulation in the KD cells (lower panels). Anti-β-actin served as the loading control. Left: Representative Western blot. Right: Statistics. Data are mean ± SEM. *P < 0.05. (C) Co-IP showing C17orf80 exhibits no apparent interaction with MRPS9 or MRPS21. C17orf80-Flag was expressed in HeLa cells and anti-Flag antibody was used for co-IP in the presence or absence of DNase I to degrade mtDNA. Source data are available for this figure: SourceData FS3.
Effects of C17orf80 ablation on mtDNA translation. (A) Heatmap of fold changes of the mtDNA-encoded proteins analyzed by quantitative proteomics. The protein content in C17orf80 knockout (KO) or knockdown (KD) HEK-293T cells was normalized to that of wild type (WT) or control (CON) cells, respectively. All mtDNA-encoded proteins detected are shown and the proteins with significant changes (P < 0.05) are marked by an asterisk. (B) Western blot showing few changes of COX1 and ND4 in the KO cells (upper panels) and their downregulation in the KD cells (lower panels). Anti-β-actin served as the loading control. Left: Representative Western blot. Right: Statistics. Data are mean ± SEM. *P < 0.05. (C) Co-IP showing C17orf80 exhibits no apparent interaction with MRPS9 or MRPS21. C17orf80-Flag was expressed in HeLa cells and anti-Flag antibody was used for co-IP in the presence or absence of DNase I to degrade mtDNA. Source data are available for this figure: SourceData FS3.
C17orf80 acts to promote mtDNA replication
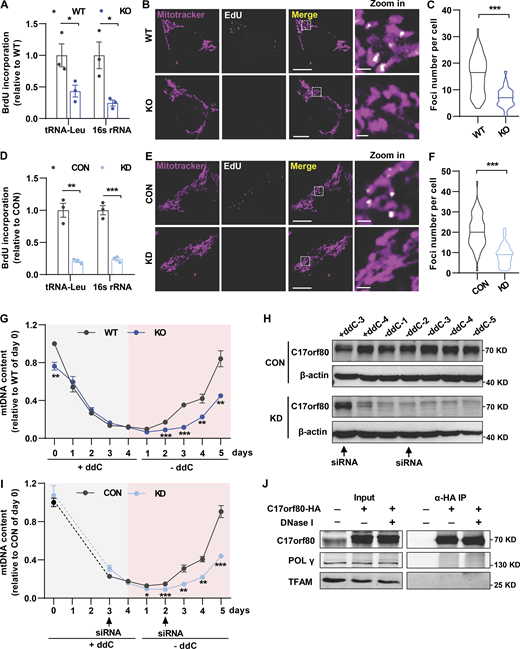
To delineate the underlying mechanism by which C17orf80 maintains mtDNA levels, we determined whether it was involved in the mtDNA replication process. We first used 5-bromo-2′-deoxyuridine (BrdU) to probe the newly synthesized mtDNA, which was then pulled down by IP using anti-BrdU antibody for subsequent qPCR analysis. A dramatic reduction of BrdU labeling was found in both the C17orf80 knockout (Fig. 4 A) and knockdown cells (Fig. 4 D), indicating less mtDNA is engaged in replication. In parallel, 5-ethynyl-2′-deoxyuridine (EdU) immunofluorescence measurement revealed that C17orf80 ablation largely ameliorated the newly synthesized mtDNA in the knockout (Fig. 4, B and C) and knockdown cells (Fig. 4, E and F). Further, we used 2′,3′-dideoxycytidine (ddC), a modified cytosine analog that stalls mtDNA replication via inhibition of POLγ processing and/or nascent strand termination (Brown and Clayton, 2002), to deplete mtDNA content and measured its recovery efficiency after ddC removal. In wild-type cells, the vast majority of mtDNA was depleted after 4 days of ddC treatment followed by a gradual rise to 84% of pretreatment levels after 5-days recovery (Fig. 4 G). However, mtDNA recovery was significantly slowed in the knockout cells (Fig. 4 G). The impaired mtDNA recovery after its depletion by C17orf80 ablation was further corroborated in the knockdown cells in which the siRNA was transfected on day 3 during ddC treatment for the first time and on day 2 during mtDNA recovery for the second time (Fig. 4, H and I). All these three lines of evidence indicate that C17orf80 functions to promote mtDNA replication.
C17orf80 is involved in mtDNA replication. (A) Reduced BrdU incorporation in C17orf80 knockout HEK-293T cells. BrdU-incorporated mtDNA was identified by IP using anti-BrdU antibody and nuclear DNA was used for normalization. Data are mean ± SEM. *P < 0.05. (B) Representative confocal images of EdU labeling in WT and KO HEK-293T cells. Mitochondria were stained with Mitotracker. Scale bars are 10 μm in the large-view image and 1 µm in the zoom-in image. (C) Quantification of the EdU-labeled foci number. n = 50–60 cells per group. ***P < 0.001. (D) As in A, except the experiments were done in control (CON) and knockdown (KD) HEK-293T cells. Data are mean ± SEM. **P < 0.01, ***P < 0.001. (E and F) As in B and C, except the experiments were done in control and knockdown cells. For F, n = 50–60 cells per group. ***P < 0.001. (G) mtDNA copy number changes during induced depletion and repopulation in WT and KO HEK-293T cells. mtDNA depletion was achieved by adding 100 µM ddC to the culture medium for 4 d, and then the cells were cultured for 5 d in the absence of ddC for mtDNA recovery. Data are mean ± SEM. n = 4 independent experiments per group. **P < 0.01, ***P < 0.001. (H) Western blot showing changes of C17orf80 protein level in control and knockdown groups during mtDNA depletion and recovery. The arrows indicate siRNA addition time. Anti-β-actin served as the loading control. (I) As in G, except the experiments were done in control and knockdown HEK-293T cells. Data are mean ± SEM. n = 4 independent experiments per group. *P < 0.05, **P < 0.01, ***P < 0.001. (J) Co-IP shows no obvious interaction between C17orf80 and POLγ or between C17orf80 and TFAM. The C17orf80-HA was expressed in HeLa cells and anti-HA antibody was used for co-IP in the presence or absence of DNase I to degrade mtDNA. Source data are available for this figure: SourceData F4.
C17orf80 is involved in mtDNA replication. (A) Reduced BrdU incorporation in C17orf80 knockout HEK-293T cells. BrdU-incorporated mtDNA was identified by IP using anti-BrdU antibody and nuclear DNA was used for normalization. Data are mean ± SEM. *P < 0.05. (B) Representative confocal images of EdU labeling in WT and KO HEK-293T cells. Mitochondria were stained with Mitotracker. Scale bars are 10 μm in the large-view image and 1 µm in the zoom-in image. (C) Quantification of the EdU-labeled foci number. n = 50–60 cells per group. ***P < 0.001. (D) As in A, except the experiments were done in control (CON) and knockdown (KD) HEK-293T cells. Data are mean ± SEM. **P < 0.01, ***P < 0.001. (E and F) As in B and C, except the experiments were done in control and knockdown cells. For F, n = 50–60 cells per group. ***P < 0.001. (G) mtDNA copy number changes during induced depletion and repopulation in WT and KO HEK-293T cells. mtDNA depletion was achieved by adding 100 µM ddC to the culture medium for 4 d, and then the cells were cultured for 5 d in the absence of ddC for mtDNA recovery. Data are mean ± SEM. n = 4 independent experiments per group. **P < 0.01, ***P < 0.001. (H) Western blot showing changes of C17orf80 protein level in control and knockdown groups during mtDNA depletion and recovery. The arrows indicate siRNA addition time. Anti-β-actin served as the loading control. (I) As in G, except the experiments were done in control and knockdown HEK-293T cells. Data are mean ± SEM. n = 4 independent experiments per group. *P < 0.05, **P < 0.01, ***P < 0.001. (J) Co-IP shows no obvious interaction between C17orf80 and POLγ or between C17orf80 and TFAM. The C17orf80-HA was expressed in HeLa cells and anti-HA antibody was used for co-IP in the presence or absence of DNase I to degrade mtDNA. Source data are available for this figure: SourceData F4.
In terms of C17orf80 acting as a bona fide mitochondrial nucleoid protein, we examined whether it promoted mtDNA replication in cooperation with TFAM and POLγ, the two most abundant mitochondrial nucleoid proteins that play indispensable roles in mtDNA replication (Bogenhagen, 2012; Bogenhagen et al., 2008; Garrido et al., 2003; Hensen et al., 2014; Wang and Bogenhagen, 2006). No obvious physical interaction between C17orf80 and POLγ or between C17orf80 and TFAM was detected by co-IP assay in the presence or absence of mtDNA (Fig. 4 J). This result suggests that C17orf80 promotes mtDNA replication independently by directly interacting with the key component of mtDNA replication machinery POLγ and the most abundant mitochondrial nucleoid protein TFAM. It needs to be determined whether C17orf80 interacts with other components of the mtDNA replication machinery for its action of promoting mtDNA replication.
Importance of C17orf80 in mitochondrial metabolism and cell proliferation
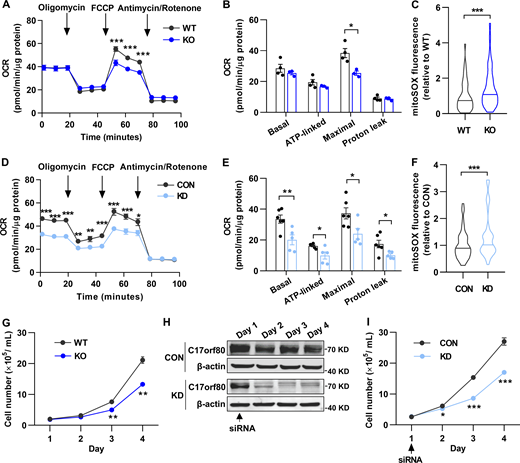
Finally, we examined the functional impact of C17orf80 ablation on mitochondrial function and cell proliferation. Measuring mitochondrial oxygen consumption rate (OCR) showed impaired mitochondrial respiration in both C17orf80 knockout and knockdown cells with decreased maximal respiration in the knockout cells (Fig. 5, A and B) and attenuations of all basal, ATP-linked, maximal, and proton leak-coupled respirations in the knockdown cells (Fig. 5, D and E). The mitochondrial reactive oxygen species (ROS) levels measured by mitoSOX were elevated by C17orf80 ablation (Fig. 5, C and F). These results highlight an important role of C17orf80 in mitochondrial metabolism. Moreover, the knockout cells displayed a significantly compromised proliferation rate compared to wild-type cells (Fig. 5 G). Such defective proliferation was also found in the knockdown cells (Fig. 5, H and I). In agreement with the view that maintaining mtDNA homeostasis is required for cell health (Almannai et al., 2022; El-Hattab et al., 2017; Viscomi and Zeviani, 2017), these results underscore the functional importance of C17orf80 on mtDNA maintenance.
C17orf80 ablation impairs mitochondrial respiration and cell proliferation and elevates mitochondrial ROS levels. (A) OCR changes of wild type (WT) and C17orf80 knockout (KO) HEK-293T cells. 10 mM glucose and 1 mM pyruvate were used as substrates. 1 μM oligomycin, 1 μM FCCP, and 1 μM antimycin/rotenone were sequentially added as indicated by the arrows. Data are mean ± SEM. n = 3–4 independent experiments per group. ***P < 0.001. (B) Statistics of OCR in WT and KO cells. Data are mean ± SEM. *P < 0.05. (C) Increased mitochondrial ROS level in KO HEK-293T cells. n = 50–60 cells per group. ***P <0.001. (D) As in A, except the experiments were done in control (CON) and C17orf80 knockdown (KD) HEK-293T cells. Data are mean ± SEM. n = 5–6 independent experiments per group. *P < 0.05, **P < 0.01, ***P < 0.001. (E) Statistics of D. Data are mean ± SEM. *P < 0.05, **P < 0.01. (F) As in C, except the experiments were done in control and knockdown HEK-293T cells. Data are mean ± SEM. n = 50–60 cells per group. ***P <0.001. (G) Reduced proliferation rate in KO HEK-293T cells. Data are mean ± SEM. n = 3 independent experiments per group. **P < 0.01. (H) Western blot showing changes of C17orf80 protein level in control and knockdown HEK-293T cells. The arrow indicates the time of siRNA addition. Anti-β-actin served as the loading control. (I) Reduced proliferation rate in C17orf80 knockdown HEK-293T cells. Data are mean ± SEM. n = 3–4 independent experiments per group. *P < 0.05, ***P < 0.001. Source data are available for this figure: SourceData F5.
C17orf80 ablation impairs mitochondrial respiration and cell proliferation and elevates mitochondrial ROS levels. (A) OCR changes of wild type (WT) and C17orf80 knockout (KO) HEK-293T cells. 10 mM glucose and 1 mM pyruvate were used as substrates. 1 μM oligomycin, 1 μM FCCP, and 1 μM antimycin/rotenone were sequentially added as indicated by the arrows. Data are mean ± SEM. n = 3–4 independent experiments per group. ***P < 0.001. (B) Statistics of OCR in WT and KO cells. Data are mean ± SEM. *P < 0.05. (C) Increased mitochondrial ROS level in KO HEK-293T cells. n = 50–60 cells per group. ***P <0.001. (D) As in A, except the experiments were done in control (CON) and C17orf80 knockdown (KD) HEK-293T cells. Data are mean ± SEM. n = 5–6 independent experiments per group. *P < 0.05, **P < 0.01, ***P < 0.001. (E) Statistics of D. Data are mean ± SEM. *P < 0.05, **P < 0.01. (F) As in C, except the experiments were done in control and knockdown HEK-293T cells. Data are mean ± SEM. n = 50–60 cells per group. ***P <0.001. (G) Reduced proliferation rate in KO HEK-293T cells. Data are mean ± SEM. n = 3 independent experiments per group. **P < 0.01. (H) Western blot showing changes of C17orf80 protein level in control and knockdown HEK-293T cells. The arrow indicates the time of siRNA addition. Anti-β-actin served as the loading control. (I) Reduced proliferation rate in C17orf80 knockdown HEK-293T cells. Data are mean ± SEM. n = 3–4 independent experiments per group. *P < 0.05, ***P < 0.001. Source data are available for this figure: SourceData F5.
Concluding remarks and future perspectives
In summary, we identified C17orf80, a functionally unknown protein, as a critical regulator of mtDNA abundance. It extensively binds double-stranded mtDNA throughout the mitochondrial genome without sequence specificity, constituting a novel bona fide mitochondrial nucleoid protein. Functionally, it is essentially involved in controlling mtDNA levels by promoting mtDNA replication and plays important roles in mitochondrial metabolism and cell proliferation. Though further study is needed to delineate the molecular mechanism underlying its action in promoting mtDNA replication, our findings underscore C17orf80 as a crucial player in the mtDNA replication process.
Mitochondrial nucleoids are composed of a range of proteins involved in mtDNA maintenance (Bogenhagen, 2012; Bogenhagen et al., 2008; Garrido et al., 2003; Hensen et al., 2014; Wang and Bogenhagen, 2006). For example, TFAM is the most abundant nucleoid protein that binds mtDNA both with and without sequence specificity to regulate mitochondrial transcription initiation and mtDNA copy number, respectively (Campbell et al., 2012). The non-specific mtDNA binding activity enables it to compact mtDNA for replication regulation (Kaufman et al., 2007). Interestingly, like TFAM, C17orf80 also shows genome-wide and non-specific mtDNA binding activity. As such, it is conceivable that C17orf80 might play a role in packaging the mitochondrial genome into a structure that is favorable for the mtDNA replication machinery to proceed. It is also possible that C17orf80 might function to reduce the mtDNA turnover rate by extensively binding it, thus enhancing the steady-state levels of mtDNA.
The mouse ortholog D11Wsu47e not only displayed mitochondrial nucleoid foci distribution but also exerted a similar regulatory role on mtDNA copy number in the mouse cells, as well as what human C17orf80 did. These results highlight a conserved role for C17orf80 and its orthologs in mtDNA abundance regulation. Interestingly, consistent with the 66% sequence similarity between human and mouse mitochondrial genomes, D11Wsu47e shows 58% sequence similarity with C17orf80, suggesting that C17orf80 orthologs might co-evolve with their species-specific mtDNAs for efficient binding.
Serving as the cellular powerhouse and metabolic center, the mitochondrion is both a primary source of ROS production and a primal target of ROS attack (Murphy, 2009; Turrens, 2003; Zorov et al., 2014). The close proximity to the ROS production sites and the lack of protective histones render the mitochondrial genome vulnerable to oxidative damage. Though whether formation of the nucleoid structure has particular specialized functions is unknown, such mtDNA and protein macromolecular assembly could be a way to protect mtDNA against oxidative damage since malfunction of many of the nucleoid proteins can cause damage or loss of mtDNA (Akhmedov and Marín-García, 2015; Almannai et al., 2022; El-Hattab et al., 2017; Gilkerson, 2009). As a new bona fide mitochondrial nucleoid protein, it will be interesting to investigate whether C17orf80 plays a role in protecting mtDNA against oxidative damage.
Functionally, we prove the importance of C17orf80 in mitochondrial metabolism and cell proliferation, underlining the vital role of maintaining mtDNA homeostasis for normal cell life. Disorders in mtDNA maintenance are associated with a wide range of diseases including Parkinson’s disease (Bender et al., 2006; Coskun et al., 2012), Alzheimer’s disease (Coskun et al., 2004; Wang et al., 2005), amyotrophic lateral sclerosis (Keeney and Bennett, 2010), cardiovascular disease (Ide et al., 2001; Karamanlidis et al., 2010), diabetes (Maassen et al., 2004; Simmons et al., 2005), cancer (Fan et al., 2009; Meierhofer et al., 2004), and aging (Sondheimer et al., 2011). A variety of pathogenic mutations in the genes encoding enzymes involving mtDNA replication and repair have been unraveled (Almannai et al., 2022; El-Hattab et al., 2017; Viscomi and Zeviani, 2017). As C17orf80 is a newly identified mtDNA replication regulator and nucleoid protein, it will be important to address its physiological significance and characterize its possible pathogenic mutations that may provide novel targets in mitochondrial biology and medicine.
Materials and methods
Cell culture and C17orf80 knockdown, knockout, and overexpression
Cells were grown in DMEM (Macgene) supplemented with 10% FBS (Gibco), penicillin (100 IU/ml; Macgene), and streptomycin sulfate (0.1 mg/ml; Macgene) at 37°C in 5% CO2. For plasmid transfection, Lipofectamine 3000 (Invitrogen) was used according to the manufacturer’s instructions. For C17orf80 knockdown via RNAi, 150 pmol siRNA were transfected using Lipofectamine RNAi MAX reagent (Invitrogen) according to the manufacturer’s instructions (siRNA sequences listed in Table S1). A C17orf80 knockout HEK-293T cell line was generated using CRISPR/Cas9 system (gRNA sequence: 5′-ACTTCGTCCTGTGTTGTAGC-3′), and 3 μg/ml puromycin was used for selecting monoclonal cells. For generating a stable cell line overexpressing C17orf80-HA fusion protein, a lentivirus carrying the C17orf80-HA gene was constructed to infect HEK-293T cells followed by puromycin selection.
Immunofluorescence and imaging
Cells were fixed with 4% formaldehyde in phosphate-buffered saline (PBS) for 20 min at 37°C and then permeabilized with 0.5% Triton X-100 for 15 min at room temperature (RT). After blocking with 5% BSA in PBS for 2 h at RT, the cells were incubated with the primary antibodies overnight at 4°C, followed by incubation with secondary antibodies for 2 h at RT. After each step, the cells were washed with PBS three times. The primary anti-Myc (Cat#AE070, RRID:AB_2863795; Abclonal), anti-Flag (Cat#AE004, RRID:AB_2771921; Abclonal), anti-HA (Cat#3724S, RRID:AB_1549585; Cell Signaling Technology), anti-DNA (Cat#CBL186, AB_11213573; Sigma-Aldrich), and anti-ATPB (Cat#ab14730, RRID:AB_301438; Abcam) antibodies, and the secondary goat anti-rabbit Alexa Fluor 488 (Cat#A-11008, RRID:AB_143165; Molecular Probes), goat anti-mouse Alexa Fluor 594 (Cat#A-11005, RRID:AB_2534073; Molecular Probes), and goat anti-mouse Alexa Fluor 647 (Cat#A-21235, RRID:AB_2535804; Molecular Probes) antibodies were used. For mtDNA staining, the cells were loaded with 3 µl/ml Picogreen (Invitrogen) for 30 min at RT.
An inverted confocal microscope (980; Zeiss LSM) with a 40×, 1.3 NA oil-immersion objective was used and the images were taken at RT with excitation at 488 nm and emission at 491–586 nm for Alexa Fluor 488 and Picogreen, excitation at 561 nm and emission at 567–646 nm for Alexa Fluor 594, and excitation at 639 nm and emission at 643–756 nm for Alexa Fluor 647, respectively. The images were analyzed by Zen 3.3 software and ImageJ (RRID:SCR_003070).
Mitochondrial isolation
Harvested HEK-293T cells were suspended in an ice-cold mitochondrial isolation medium (210 mM mannitol, 70 mM sucrose, 10 mM Hepes, 1 mM EGTA, 0.1% BSA, pH 7.4, and protease inhibitor cocktail) and transferred to Dounce homogenizer. After gentle homogenization, the cell lysates were centrifuged at 700 g for 10 min, and the supernatants were collected and centrifuged at 12,000 g for 10 min. The pellets were then collected for further assessment.
Anti-mtDNA IP
Crosslink between mtDNA and its binding proteins was performed by treating 3 × 107 HEK-293T cells with 1% formaldehyde for 15 min and then stopped by directly adding glycine to a final concentration of 0.15 M for a 5-min incubation at RT. After washing twice with PBS, the cells were collected and resuspended in a cold mitochondrial isolation solution for mitochondrial isolation as described above. The isolated mitochondria were dissolved in lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl, pH 8, and protease inhibitor cocktail) and sonicated in a Bioruptor (Qsonica) for 5 min at 4°C (15 s at 30% amplitude and 30 s off) to obtain mtDNA fragments between 300 and 800 bp. After sonication, the samples were transferred to a new DNA LoBind Tube (Eppendorf) and centrifuged at 14,000 rpm for 15 min at RT. The supernatants were diluted 1:10 in dilution buffer (1.1% Triton X-100, 0.01% SDS, 1.2 mM EDTA, 167 mM NaCl, 16.7 mM Tris-HCl, pH 8, and protease inhibitor cocktail) and precleaned with Protein L Magnetic Beads (MedChemExpress) for 2 h at 4°C with end-over-end mixing. 5% of the precleaned supernatants were collected as input, and the anti-DNA antibody (Cat#CBL186, AB_11213573; Sigma-Aldrich) was added to the remaining supernatants and incubated at 4°C overnight on a rotator. To precipitate the antibody-DNA/protein complexes, the Protein L Magnetic beads were added to the samples and then incubated for 4–6 h at 4°C. The beads were washed sequentially with low salt immune complex wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 150 mM NaCl, and 20 mM Tris-HCl, pH 8), high salt immune complex wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 500 mM NaCl, and 20 mM Tris-HCl, pH 8), LiCl immune complex wash buffer (0.25 M LiCl, 1% NP-40, 1% deoxycholate sodium, 1 mM EDTA, and 10 mM Tris-HCl, pH 8), and Tris-EDTA (TE) buffer (10 mM Tris-HCl and 1 mM EDTA, pH 8.0). The enriched proteins were finally collected by boiling beads in 1.5× protein loading buffer (TransGen Biotech) for 10 min.
Western blot analysis
The cells were incubated with ice-cold RIPA lysis buffer (C1053+, Applygen) or denaturing lysis buffer (C1052; Applygen) for 30 min on ice and sonicated for 30 s (50% amplitude, 5 s on, and 10 s off). 40–60 μg protein per sample was loaded to 10% or 15% SDS-PAGE, transferred to 0.45 μm PVDF membrane, and blotted with specific antibodies. The blots were visualized using the secondary antibodies conjugated IRDye (926-68070, 925-32211; LI-COR) and an Odyssey imaging system (LI-COR). Monoclonal anti-β-actin (Cat#30101ES60; Yeason) and anti-ATP5A (Cat#ab14748, RRID:AB_301447; Abcam) and polyclonal anti-C17orf80 (Cat#27762-1-AP, RRID:AB_2880964; Proteintech), anti-POLγ (Cat#A1323, RRID:AB_2861682; Abclonal), anti-TFAM (Cat#A3173, RRID:AB_2863025; Abclonal), anti-MCU (Cat#ab272488; Abcam), anti-COX1 (Cat#A7531, RRID:AB_2768058; ABclonal), anti-ND4 (Cat#A9941, RRID:AB_2770457; Abclonal), anti-MRPS9 (Cat#16533-1-AP, RRID:AB_2878272; Proteintech), and anti-MRPS21 (Cat#DF4165, RRID: AB_2836530; Affinity Biosciences) antibodies were used.
Immunogold staining electron microscopy
Cells stably overexpressing C17orf80-HA fusion protein grown on ACLAR 33C Film (Cat#50425; Electron Microscopy Sciences) were fixed with 1% glutaraldehyde in 0.1 M PBS (pH 7.4) for 30 min at RT. After quenching the free aldehyde with 150 mM glycine in 0.1 M PBS for 5 min, cells were washed four times for 6 min each with 0.1 M PBS and post-fixed with 0.1% OSO4 for 30 min avoiding light. Following several washes in ultrafiltered water, cells were dehydrated through increasing concentrations of ethanol in water (30%, 50%, 70%, 85%, 95%, and 100%, 5 min for each) and acetone (2 × 100%, 5 min for each). Samples were infiltrated with EMbed 812 resin (Cat#14120; Electron Microscopy Sciences) mixed 1:3, 1:1, and 3:1 with acetone for 20 min each. Samples were incubated in pure EMbed 812 resin three times (1 h for each) and then placed into molds and polymerized at 65°C for 24 h. After removing ACLAR 33C Film, ultrathin sections (∼75 nm) were taken using a Leica UC7 ultramicrotome and collected on formvar film-coated single slot nickel grids with oval holes. Before immunolabeling, plastic sections were incubated with 1% NaIO4 for 30 min for antigen unmasking. After washing with ultrafiltered water and PBS, sections were blocked with blocking buffer (0.5% BSA and 0.05% Tween in PBS) for 10 min and incubated with primary anti-HA monoclonal antibody (Cat#11867423001, RRID:AB_390918, 1:10; Sigma-Aldrich) for 1 h at RT. Following six washes with PBS (2 min for each), sections were incubated with secondary antibody goat-anti-rat IgG conjugated to 6 nm colloidal gold (Cat#806.055, 1:40; Aurion) for 1 h at RT. After washing with PBS six times, sections were postfixed in 0.1% (vol/vol) glutaraldehyde in PBS for 5 min, washed with ultrafiltered water, and counterstained with 2% uranyl acetate. Nickel grids were observed in the Tecnai G2 Spirit BioTWIN transmission electron microscope (FEI) operating at 120 kV. Images were taken using a Gatan Orius 832 digital camera.
MtDNA copy number quantification
The total DNA from HEK-293T, HeLa, or mouse primary hepatocytes was extracted using Dneasy Blood Tissue Kit (Qiagen) according to the manufacturer’s instructions. For ddC treatment, HEK-293T cells were incubated with 100 μΜ ddC (MedChemExpress) in the culture medium for 4 d and then replaced with a normal culture medium to recover for 5 d. The total DNA was extracted on each day for subsequent quantification. qPCR was used to determine the relative copy number of mtDNA by Trans Start Green qPCR Super Mix (TransGen Biotech) and a BioRad CFX96 Touch RT-PCR Detection System with nDNA content as the reference. The primers specific for nDNA and mtDNA sequences are listed in Table S1. The relative mtDNA copy number was calculated as 2 × 2(Ct (nDNA) − Ct (mtDNA)).
Total RNA extraction and qPCR
Total RNA was extracted using Rneasy Mini Kit (Qiagen) according to the manufacturer’s instructions and then converted to cDNA using EasyScript All-in-One First-Strand cDNA Synthesis SuperMix for qPCR (TransGen Biotech). qPCR reactions were performed using Trans Start Green qPCR Super Mix (TransGen Biotech) with the primer sequences listed in Table S1.
mtDNA ChIP-qPCR
After C17orf80-HA fusion protein was expressed in HeLa cells by adenovirus infection, DNA and protein crosslink were performed by treating 5 × 106 cells with 1% formaldehyde for 15 min and then stopped by directly adding glycine to a final concentration of 0.15 M for a 5 min-incubation at RT. After washing twice with PBS, the cells were collected and resuspended in a cold mitochondrial isolation solution for mitochondrial isolation as described above. The isolated mitochondria were dissolved in lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl, pH 8, and protease inhibitor cocktail) and sonicated in a Bioruptor (Qsonica) for 5 min at 4°C (15 s at 30% amplitude and 30 s off) to obtain DNA fragments between 300 and 800 bp. After sonication, the samples were transferred to a new DNA LoBind Tube (Eppendorf) and centrifuged at 14,000 rpm for 15 min at RT. The supernatants were diluted 1:10 in the dilution buffer (1.1% Triton X-100, 0.01% SDS, 1.2 mM EDTA, 167 mM NaCl, 16.7 mM Tris-HCl, pH 8, and protease inhibitor cocktail) and precleaned with 40 μl Protein A/G Magnetic Beads (MedChemExpress) for 2 h at 4°C with end-over-end mixing. 5% of the precleaned supernatants were collected as input and 4 μg IgG (Cat#AC005, RRID:AB_2771930; ABclonal) or anti-HA antibody (Cat#ab9110, RRID:AB_307019; Abcam) was added to the remaining supernatants and incubated at 4°C overnight on a rotator. Then 60 μl Protein A/G Magnetic Beads were added to precipitate the antibody–DNA/protein complexes with incubation for 4–6 h at 4°C. The beads were washed with low salt immune complex wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 150 mM NaCl, and 20 mM Tris-HCl, pH 8), high salt immune complex wash buffer (0.1% SDS,1% Triton X-100, 2 mM EDTA, 500 mM NaCl, and 20 mM Tris-HCl, pH 8), LiCl immune complex wash buffer (0.25 M LiCl, 1% NP-40, 1% deoxycholate sodium, 1 mM EDTA, and 10 mM Tris-HCl, pH 8), and TE buffer (10 mM Tris-HCl and 1 mM EDTA, pH 8.0), respectively. The crosslink was reversed by incubation with lysis buffer at 70°C for at least 3 h. RNase and proteinase K were then added to remove RNA and proteins, respectively. Finally, the samples were purified using PCI (phenol:chloroform:isoamyl alcohol). The mtDNA ChIP-qPCR was performed using the primers listed in Table S1. The relative fold change was calculated as 2(Ct(IgG) − Ct(HA)).
mtDNA ChIP-seq
The DNA sequencing libraries for input and IP samples were built using NEBNext Ultra II DNA Library Prep Kit for Illumina (New England Biolabs) according to the manufacturer’s instructions. Deep sequencing was performed on an Illumina NovaSeq 6000 platform. The sequencing data were analyzed as described previously (Yang et al., 2022). Briefly, after trimming with a quality drop below a mean of Q15 in a window of five nucleotides and discarding left reads with lengths below 15 bp (RRID:SCR_011848, version: 0.39; Trimmomatic), ChIP-seq reads were aligned to the UCSC hg38 genome using BWA-MEM (Version: 0.7.17-r1188) with the standard default settings. Only reads with mapping quality over 20 were retained. Duplicate reads were further removed using Picard tools (Version: 2.25.0). Mitochondrial coverage profiles were generated with Samtools (RRID:SCR_002105, Version: 1.3.1) and BEDTools (RRID:SCR_006646, Version: 2.30.0). Per base coverage was normalized to total reads mapped to the genome and the circular figures were generated with R library circlize (Version: 0.4.14). The ChIP-seq data have been deposited to the Gene Expression Omnibus (GEO, RRID:SCR_005012) under the accession number GSE224137.
RNA-seq
The total RNA of wild type and C17orf80 knockout HEK-293T cells were extracted using TRIzol reagent (Invitrogen) and cDNA sequencing libraries were built using NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs), according to the manufacturer’s instructions, and sequenced using Illumina NovaSeq 6000 platform. After quality control with FastQC (version: 0.11.9), reads for each library were mapped using HISAT2 (version: 2.2.1) against the human reference genome hg38 with default parameters. Uniquely mapped reads were extracted to calculate the read counts of each gene using the matching gene Annotation from Ensembl with HTSeq (version: 0.12.4). The genes with low expression in all samples (FPKM <0.5) were further filtered. Mitochondrial coverage profiles were generated with Samtools (RRID:SCR_002105, version: 1.3.1) and BEDTools (RRID:SCR_006646, version: 2.30.0). Per base coverage was normalized for each sample, and the circular figures were generated with R library circlize (version: 0.4.14). The RNA-seq data are available in the GEO (RRID:SCR_005012) database under accession number GSE224121.
Tandem mass tag (TMT)–labeled quantitative proteomics
The cells were collected after PBS washing twice, lysed with DB lysis buffer (8 M urea and 100 mM TEBA, pH 8.5), followed by 5 min of ultrasonication on ice. 100 μg proteins per group were transferred into a new microcentrifuge after protein concentration determination. Treated with DTT and iodoacetamide, 3 μl trypsin (1 μg/1 μl) was added, and the samples were digested in 0.1 M TEBA buffer at 37°C overnight with agitation. The procedure of TMT labeling was according to the manufacturer’s instructions (TMT Mass Tagging Kits, Invitrogen) with slight modifications. Briefly, acetonitrile-dissolved TMT labeling reagent was added and mixed with shaking for 2 h at RT. The reaction was quenched by adding 8% ammonia and incubating for 15 min. All labeling samples were mixed with equal volume, desalted, and finally lyophilized. Following this, samples were fractionated by a C18 column (Waters BEH C18, 4.6 × 250 mm, 5 μm) on a Rigol L3000 HPLC system at a flow rate of 1 ml/min. Peptides were combined into 10 fractions, dried under vacuum, reconstituted in 0.1% (vol/vol) formic acid in water, and analyzed by LC-MS/MS.
LC-MS/MS analyses were performed at an EASY-nLC 1200 UHPLC system (Thermo Fisher Scientific) coupled with a Q ExactiveTM HF-X (Thermo Fisher Scientific). 1 μg sample of each fraction was injected into the LC-MS/MS system for analysis. The result spectra of each run were searched against 1685-uniprot-homo-filtered-reviewedyes.fasta (27,392 sequences) by Proteome Discoverer software 2.5 (RRID:SCR_014477; Thermo Fisher Scientific). Database searches were performed with a mass tolerance of 10 ppm for precursor ions and 0.02 D for product ions. Carbamidomethyl was specified as fixed modifications, and oxidation of methionine and iTRAQ/TMT plex were specified as variable modifications. A maximum of two miscleavage sites were allowed. To improve result quality, peptide spectrum matches with a credibility of >99% were identified, and proteins that contained at least one unique peptide with FDR <1% were identified as proteins.
Recombinant expression and purification of C17orf80
For recombinant expression of C17orf80 in E. coli BL21 (nico21), the plasmid pGEX-6P-1 (a gift from Lei Chen’s lab, Peking University, Beijing, China) containing codon-optimized C17orf80 sequence with N-terminally fused with GST was transformed into E. coli. BL21. The cells were cultured in LB media containing ampicillin at 37°C until OD600 reached 0.6–0.8, and then 200 μM isopropyl-β-D-thiogalactoside (IPTG) was added overnight at 16°C to induce the recombinant protein expression. The E. coli cells were then collected, resuspended in lysis buffer (150 mM NaCl, 10 mM DTT, 1 mM PMSF, and 50 mM Tris-HCl, pH 7.5), and sonicated in a Bioruptor (Qsonica) for 20 min at 4°C (5 s at 50% amplitude and 5 s off). After centrifugation at 15,000 rpm for 30 min, the supernatant was transferred to Glutathione Sepharose 4B (GE Healthcare) and washed with wash buffer A (500 mM NaCl, 1 mM DTT, and 50 mM Tris-HCl, pH 7.5) and wash buffer B (150 mM NaCl, 1 mM DTT, and 50 mM Tris-HCl, pH 7.5). For protease digestion, the resin was incubated with 50 U HRV 3C protease (Beyotime) in PreScission buffer (150 mM NaCl, 1 mM EDTA, 1 mM DTT, and 50 mM Tris-HCl, pH 7.5) overnight at 4°C. The recombinant protein was eluted with PreScission buffer and further purified with HiTrap SP HP (GE Healthcare) in ÄKTA pure protein purification system (with a linear gradient from 0 mM NaCl to 1 M NaCl in buffer containing 20 mM Tris, pH 7.5, at 4°C). The protein-containing fractions were collected for SDS-PAGE followed by Coomassie blue staining or for further use.
EMSA
The Cy3-labeled probes of two 50 bp dsDNA and one 50 bp ssDNA were synthesized (sequences are listed in Table S1). The indicated amounts of purified proteins and 10 nM probes were incubated for 30 min at RT in 20 μl binding buffer (100 mM KCl, 1 mM EDTA, 10 mM Tris-Cl, pH 7.5, 0.1 mM DTT, 5% vol/vol glycerol, and 0.01 mg/ml BSA) and then separated on 5.5% native polyacrylamide gel at 4°C and visualized by Cy3 fluorophore.
BrdU IP
The BrdU IP was performed as reported previously with slight modifications (Li et al., 2021). Briefly, 50 μM aphidicolin (Cell Signaling Technology) was added to block nDNA replication for at least 1 h followed by incubation with 50 μM BrdU for 24 h. Total DNA was then extracted and purified using Dneasy Blood Tissue Kit (Qiagen), and ssDNA was obtained by sonication and boiling. The ssDNA was then added to ice-cold BrdU IP buffer (0.0625% [vol/vol] Triton X-100 in PBS) containing tRNA (Roche) and anti-BrdU antibody (Cat#555627, RRID:AB_395993; BD Biosciences) for 2 h at 4°C with rotation in DNA LoBind Tubes (Eppendorf). Prewashed-protein A/G magnetic beads (MedChemExpress) were added to the BrdU IP mix for another 2 h at 4°C with rotation. After washing with BrdU IP buffer and TE buffer (10 mM Tris-HCl and 1 mM EDTA, pH 8.0), EB buffer (10 mM Tris-HCl and 1 mM EDTA, pH 8.0, 1% [wt/vol] SDS) was added and allowed to incubate at 65°C for 15 min to elute the bound single-stranded mtDNA. The eluted ssDNA were further cleaned and concentrated using MinElute PCR purification kit (Qiagen) and detected by qPCR as described above. The input nDNA was used for qPCR normalization.
EdU labeling of mtDNA
EdU incorporation was performed using Click-iT Plus EdU Alexa Fluor 647 Imaging Kit (Invitrogen) according to the manufacturer’s instructions. Briefly, the cells were incubated with 20 μM aphidicolin (Cell Signaling Technology) for 4 h to block nDNA replication followed by incubation with 10 μM EdU for 16 h. The medium containing EdU was removed and 500 nM MitoTracker Red CMXRos (Invitrogen) was added in EdU-free DMEM for 15 min at 37°C. After incubation, the cells were fixed with 3.7% formaldehyde in PBS followed by permeabilization with 0.5% Triton X-100. EdU was detected by Click-iT Alexa Fluor 647 picolyl azide in Click-iT Plus reaction. Images were taken with Zeiss LSM 980 confocal microscopy at RT with excitation at 561 nm and emission at 567–641 nm for MitoTracker, and excitation at 639 nm and emission at 643–756 nm for Alexa Fluor 647. Zen 3.3 and ImageJ (RRID:SCR_003070) were used for image analysis.
Co-IP assay
After C17orf80-HA or C17orf80-Flag fusion protein was expressed in HeLa cells, the cells were collected and resuspended in a cold mitochondrial isolation solution for mitochondrial isolation as described above. The mitochondria were lysed in NP-40–based lysis buffer (50 mM Tris, 150 mM NaCl, 10% vol/vol glycerol, 1 mM EGTA, 1 mM EDTA, and 0.5% vol/vol NP-40, pH 7.40) at 4°C for 45 min, and the supernatants were collected after centrifugation at 15,000 rpm for 15 min. Precleaned anti-HA magnetic beads (Thermo Fisher Scientific) or anti-Flag magnetic beads (Thermo Fisher Scientific) were added to the supernatants and incubated for 6–8 h at 4°C on a rotator. After washing with TBS (20 mM Tris and 150 mM NaCl, pH 7.4) three times, the beads were boiled in protein loading buffer (TransGen Biotech) for 10 min and then the supernatants were collected for Western blot.
OCR measurement
HEK-293T cells were plated in XF24 cell culture microplates at 2.5 × 104 cells/well in DMEM supplemented with 10% FBS and incubated overnight the day before the experiment. OCRs were recorded by Seahorse XF24 Extracellular Flux Analyzer (Agilent Technologies) according to the manufacturer’s instructions. 10 mM glucose and 1 mM pyruvate were used as substrates. 1 µM oligomycin, 1 µM FCCP, and 1 µM rotenone/antimycin A were added sequentially. The basal OCR, ATP production-coupled OCR, proton leak OCR, and maximal OCR were obtained. All data were analyzed using the XF24 software.
Mitochondrial ROS measurement
MitoSOX Red (Invitrogen) was used for detecting mitochondrial ROS levels. Briefly, the cells were incubated with 5 μM MitoSOX in Tyrode’s solution (137 mM NaCl, 5.4 mM KCl, 1.2 mM MgCl2, 1.2 mM NaH2PO4, 20 mM Hepes, 10 mM D-glucose, and 1.8 mM CaCl2, pH 7.35) for 30 min at 37°C, followed by washing three times with Tyrode’s solution. Images were taken with Zeiss LSM 980 confocal microscopy with excitation at 561 nm and emission at 580–740 nm at RT. Zen 3.3 and ImageJ (RRID:SCR_003070) were used for image analysis.
Cell proliferation measurement
To measure cell proliferation, an equal number of cells in different groups were seeded in DMEM for culture and then the cells were collected after trypsin (Macgene) digestion and counted using an automated cell counter (Counterstar) every 24 h.
Statistics
The data are expressed as mean ± SEM. When appropriate, a two-tailed, unpaired Student’s t test was applied to determine statistical significance in GraphPad Prism software 7. P < 0.05 was considered statistically significant.
Online supplemental material
Fig. S1 shows mitochondrial nucleoid localization of C17orf80 in HEK-293T cells and sequence alignment between human C17orf80 and mouse D11Wsu47e. Fig. S2 shows the effect of altering C17orf80 expression on mtDNA contents and mitochondrial transcription-related protein levels. Fig. S3 shows the effects of C17orf80 ablation on mtDNA translation. Table S1 lists primer sequences, siRNA sequences, and probes for EMSA experiments.
Data availability
The data are available from the corresponding author upon reasonable request.
Acknowledgments
We thank Dr. Yingchun Hu for helping perform electron microscopy experiments, Dr. Wen Zhou for technical support with mass spectrometry, Mr. Dong Zhang for technical support of electrophoresis, Ms. Tianyan Liu and Dr. Zhixing Chen for super-resolution imaging, and Drs. Qing Li and Lei Chen for valuable comments.
This work was supported by the National Natural Science Foundation of China (32293211, 92157105, 31971158, 81827809), the National Key Basic Research Program of China (2017YFA0504000), and CAMS Innovation Fund for Medical Sciences (2019-I2M-5-054).
Author contributions: H. Wu and X. Wang conceived and designed the study. H. Wu performed most of the experiments and was involved in all aspects of this study; W. Zhang assisted with the mtDNA replication experiment; X. Liu assisted with the protein purification experiment; F. Xu and Kun Peng assisted with Co-IP and transfection experiments; W. Ding performed the RNA-sequencing data analysis; X. Wang, Q. Ma, and H. Cheng supervised the work. X. Wang, H. Wu, and H. Cheng wrote the manuscript. All authors participated in the discussion and data interpretation and critically reviewed and approved the final version of the manuscript.
References
Author notes
Disclosures: The authors declare no competing interests exist.