TRPC3, a member of the transient receptor potential (TRP) superfamily of cation channels, is a lipid-regulated, Ca2+-permeable channel that mediates essential components of the receptor evoked Ca2+ signal. The modes and mechanisms by which lipids regulate TRPC3 and other members of the TRPC channel family are not well understood. Here, we report that PI(4,5)P2 regulates TRPC3 in three independent modes. PLC-dependent hydrolysis generates diacylglycerol (DAG) that interacts with lipid-binding site 2 in the channel pore. PI(4,5)P2 interacts with lipid site 1 to inhibit TRPC3 opening and regulate access of DAG to the pore lipid site 2. PI(4,5)P2 is required for regulating pore ionic selectivity by receptor stimulation. Notably, the activation and regulation of TRPC3 by PI(4,5)P2 require recruitment of TRPC3 to the ER/PM junctions at a PI(4,5)P2-rich domain. Accordingly, we identified an FFAT site at the TRPC3 N-terminal loop within the linker helices that envelope the C-terminus pole helix. The FFAT site interacts with the ER-resident VAPB to recruit TRPC3 to the ER/PM junctions and control its receptor-mediated activation. The TRPC3’s lipid interacting sites are fully conserved in TRPC6 and TRPC7 and in part in other TRPC channels. These findings inform on multiple modes of regulation of ion channels by lipids that may be relevant to diseases affected by aberrant TRPC channel functions.
Introduction
The transient receptor potential (TRP) superfamily of cation channels includes several families that participate in numerous cellular functions and are mutated in many diseases (Himmel and Cox, 2020). The canonical TRPC family includes six members in humans that function as non-selective, Ca2+ permeable channels. They are activated by G protein–coupled receptors and contribute essentially to the receptor-evoked Ca2+ influx and Ca2+ signaling (see recent reviews in Groschner and Tiapko [2018]; Li et al. [2019]; Wang et al. [2020]). The family is divided into two subgroups based on sequence similarities: TRPC1/4/5 and TRPC3/6/7. All TRPC channels have similar general structures, the details of which were revealed by protein structures solved with cryo-EM of TRPC3 (Fan et al., 2018; Tang et al., 2018), TRPC6 (Tang et al., 2018), TRPC4 (Duan et al., 2018; Vinayagam et al., 2018), and TRPC5 (Duan et al., 2019). All TRPC channels can form homo- or heterotetramers. The cytoplasmic N-terminus is made of four ankyrin repeats and multiple linker helices that are followed by the pre-S1 elbow. The first four of the six transmembrane domains (TMDs), S1–S4, form a voltage-like sensor structure with TMD S5 and S6 forming the channel pore. The cytoplasmic C-terminus starts with the conserved TRP domain and is followed by two connected helices, the rib helix and the pole helix. The N-terminal ankyrin repeats and the four linker helices surround the four cytoplasmic helical bundles composed of four pole helices (Li et al., 2019; Wang et al., 2020).
TRP channels, including TRPC channels, are regulated by lipids. Regulation of TRPC channels by lipids is quite complex. All members of the family can sense lipids. TRPC3/6/7 are directly activated by the lipid diacylglycerol (DAG; Svobodova and Groschner, 2016; Wang et al., 2020) that was identified in the TRPC6 pore (Storch et al., 2017). However, TRPC4/5 can also sense DAG (Imai et al., 2012; Wang et al., 2020), and DAG was located in the pore of TRPC5 when included during the purification process (Song et al., 2021). DAG is generated by Gαq/11-mediated activation of phospholipase C (PLC) that hydrolyzes PI(4,5)P2 to IP3 and DAG. TRPC3/6/7 also responds to PI(4,5)P2 and possibly other phosphoinositides that modulate channel function (Maléth et al., 2014). In addition, PI(4,5)P2 is an essential component of the membrane contact sites (MCS) ER/PM junctions (Lichtenegger et al., 2018; Svobodova et al., 2019). Presence and/or recruitment of TRP channels to the junctions may require recognition of PI(4,5)P2 specifically within the junctions (Maléth et al., 2014).
The structures of the TRPC channels’ lipid interacting sites and their properties are not well understood. Structural modeling suggested that DAG interacts with a fenestration of TRPC3/6/7 pore domain with G652 having an essential role in pore interaction with DAG (Tang et al., 2018). A major advancement has been made with identification of two lipid molecules trapped in the cryo-EM structure of TRPC3 (Fan et al., 2018) and TRPC6 (Lichtenegger et al., 2018; Svobodova et al., 2019). Lipids were found in a cavity formed by the pre-S1 elbow, TMD1, TMD4, and the TMD4–TMD5 linker in both channels (Duan et al., 2019). In the TRPC3 structure, a second lipid was found in a cavity between the P loop and S6 of the neighboring protomer (Fan et al., 2018), where G652 is also located (Lichtenegger et al., 2018; Svobodova et al., 2019), as well as in TRPC6 (Bai et al., 2020) and TRPC5 (Song et al., 2021). This second lipid was identified as DAG. Multiple lipid molecules were observed in the TM surfaces of TRPC5 and TRPC6 that may have structural roles (Bai et al., 2020; Duan et al., 2019; Song et al., 2021). The role of lipid interactions with each site on channel function is not known.
MCS are formed between the ER and all cellular membranes, serve as the site for exchange of materials between organelles, and are the sites for the assembly of signaling complexes (Belardi et al., 2020; Muallem et al., 2017; Prinz et al., 2020). MCS are formed by tether proteins that have an ER localization domain, a domain that spans the space between the ER and target membrane and a domain that interacts with the target membrane (Muallem et al., 2017; Prinz et al., 2020). Well-established tethers are the extended synaptotagmins (E-Syts) that assemble the ER/PM junctions (Giordano et al., 2013; Saheki and De Camilli, 2017). Recently, we identified anoctamin 8 (ANO8) as an important tether of the ER/PM junctions that assembles the entire Ca2+ signaling complex at PI(4,5)P2-dependent ER/PM junctions (Jha et al., 2019). Another protein with tether function is STIM1. STIM1 is the ER Ca2+ content sensor that unfolds in response to ER Ca2+ depletion and is targeted to the ER/PM junctions, where it interacts with and activates the Ca2+ influx channel Orai1 (Yeung et al., 2017). All these tethers—E-Syts, ANO8, and STIM1—have PI(4,5)P2 interacting domains that are essential for their localization and stabilization at the ER/PM junctions by interacting with plasma membrane (PM) PI(4,5)P2 (Jha et al., 2019; Liou et al., 2005; Saheki and De Camilli, 2017; Yuan et al., 2009).
While localization of TRPC channels at MCS has not been examined, let alone the functional significance of such localization, it is known that TRPC channels are activated by STIM1 through mechanisms that are not well understood (Bodnar et al., 2017; Lopez et al., 2020). Moreover, although several studies examined the effect of PI(4,5)P2 on some TRPC channels (Wang et al., 2020), the reported effects are often contradictory and the mechanism and site(s) of PI(4,5)P2 interaction are not known. In the present work, we selected TRPC3 as a model since it has many cellular functions, including contributing to the pathological receptor- and store-mediated Ca2+ influx in diseases such as acute pancreatitis (Kim et al., 2009; Kim et al., 2011), salivary gland damage (Kim et al., 2011), and neuronal (Hartmann and Konnerth, 2015) and cardiac (Nishiyama et al., 2021) functions. In addition, TRPC3 is directly activated by the lipid DAG and by the synthetic activator GSK1702934A (GSK), which interacts with the TRPC3 DAG site (Lichtenegger et al., 2018; Svobodova et al., 2019). Finally, TRPC3 interacts with STIM1, and we previously identified mutants that enhance and reduce interaction of TRPC3 with STIM1 (Lee et al., 2014). Using these mutants and structural guided mutations, we report at least three separable roles of PI(4,5)P2 in regulating TRPC3, and likely other TRPC channels. First, PI(4,5)P2 is essential for recruitment and retention of TRPC3 at the ER/PM junctions, which is mediated by the N-terminal two phenylalanines in an acidic tract (FFAT) motif located in a domain that engulfs the TRPC3 C-terminal pole helix. Second, interaction of pore residues with the PI(4,5)P2 hydrolytic product DAG (site 2) is essential for activation of TRPC3 by receptor stimulation and is affected by localization at the ER/PM junctions. Third, interaction of PI(4,5)P2 with site 1 formed by the pre-S1 elbow, TMD1, TMD4, and the TMD4-TMD5 linker tunes gating of TRPC3 activity and controls interaction of the lipid with site 2. All the TRPC3 PI(4,5)P2 interacting sites are conserved in TRPC6 and TRPC7 and partially in the other TRPC channels. These findings clarify parts of the complex role of lipids in gating TRPC channels that are especially relevant to other TRP channels, as well as regulation of channels by lipids in general.
Results
STIM1 regulates activation of TRPC3 by GPCRs
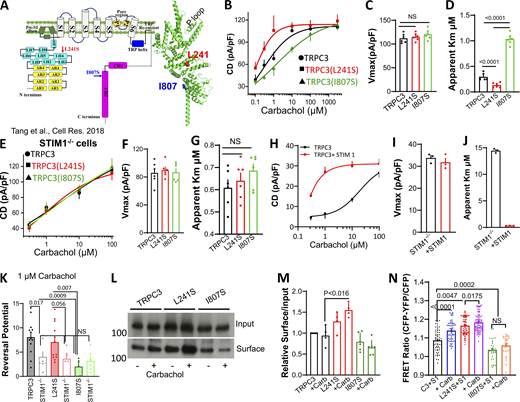
In a previous study, we reported that the TRPC3 L241S mutation enhances while the I807S mutation reduces co-immunoprecipitation (Co-IP) of TRPC3 with STIM1 (Lee et al., 2014; Table S1), which is confirmed in Fig. S1 A. Fig. 1 A shows the position of these residues in the structure of TRPC3 (Tang et al., 2018). Activation of TRPC3 and mutants by STIM1 at low agonist stimulation is shown in Fig. S1, B–J. To better define the effect of STIM1 on channel activation, we determined the dose-response for receptor stimulation of TRPC3 and the mutants. Notably, Fig. 1, B–D show that the main effect of the mutants is on the efficiency of receptor stimulation of TRPC3 (apparent Km), with no effect on maximally stimulated current density (Vmax). Thus, TRPC3(L241S) increased while TRPC3(I807S) reduced the affinity for activation by the M3 receptor (Fig. 1 D).
Effect of STIM1 on TRPC3 current, ER/PM junctions, and Ca2+influx. Current by truncated TRPC3. (A) TRPC3(L241S) enhances Co-IP of TRPC3 with STIM1. (B–J) Time course (B, E, and H), I/V (C, F, and I), and current density (D, G, and J) of TRPC3 (B–D), TRPC3(L241S) (E–G), and TRPC3(I807S) (H–J) in the presence (red) and absence of STIM1 (black) and stimulated with 1 µM carbachol. (K and L) Puncta density (K) and number (L) of MAPPER in wild-type and STIM1−/− cells was measured after treating the cells with CPA. (M) Surface expression of TRPC3 in STIM1−/− resting and stimulated cells. (N and O) Time course (N) and current density (O) of TRPC3 stimulated with 100 µM carbachol in wild-type and in STIM1−/− cells (red). (P) Input and surface of HA-tagged M3Rs expressed together with the indicated TRPC3 mutants and stimulated with or without 100 µM carbachol. This is one of two experiments with similar results. (Q–S) Co-localization of YFP-tagged TRPC3 (Q), TRPC3(L241S) (R), and TRPC3(I807S) (S) with STIM1-CFP in cells treated with 100 µM carbachol and 25 µM CPA. (T) The Pearson’s index for TRPC3, TRPC3(L241S), and TRPC3(I807S) co-localization with STIM1. Source data are available for this figure: SourceData FS1.
Effect of STIM1 on TRPC3 current, ER/PM junctions, and Ca2+influx. Current by truncated TRPC3. (A) TRPC3(L241S) enhances Co-IP of TRPC3 with STIM1. (B–J) Time course (B, E, and H), I/V (C, F, and I), and current density (D, G, and J) of TRPC3 (B–D), TRPC3(L241S) (E–G), and TRPC3(I807S) (H–J) in the presence (red) and absence of STIM1 (black) and stimulated with 1 µM carbachol. (K and L) Puncta density (K) and number (L) of MAPPER in wild-type and STIM1−/− cells was measured after treating the cells with CPA. (M) Surface expression of TRPC3 in STIM1−/− resting and stimulated cells. (N and O) Time course (N) and current density (O) of TRPC3 stimulated with 100 µM carbachol in wild-type and in STIM1−/− cells (red). (P) Input and surface of HA-tagged M3Rs expressed together with the indicated TRPC3 mutants and stimulated with or without 100 µM carbachol. This is one of two experiments with similar results. (Q–S) Co-localization of YFP-tagged TRPC3 (Q), TRPC3(L241S) (R), and TRPC3(I807S) (S) with STIM1-CFP in cells treated with 100 µM carbachol and 25 µM CPA. (T) The Pearson’s index for TRPC3, TRPC3(L241S), and TRPC3(I807S) co-localization with STIM1. Source data are available for this figure: SourceData FS1.
STIM1 controls receptor-mediated stimulation of TRPC3. (A) The domains and structure of a TRPC3 single subunit. The position of L241 and I807 are shown as spheres. (B–D) Concentration dependence of outward current of carbachol-stimulated TRPC3 (black), TRPC3(L241S) (red), and TRPC3(I807S) (green; B). The Vmax (C) and apparent Km (D) were obtained by Hill fitting. (E–G) All experiments were in STIM1−/− cells. Concentration dependence of outward current of carbachol-stimulated TRPC3 (black), TRPC3(L241S) (red), and TRPC3(I807S) (green; E). The Vmax (F) and apparent Km (G) were obtained by Hill fitting. (H–J) Experiments were in STIM1−/− cells transfected with vector (black) or STIM1 (red) depicting the concentration dependence (H), calculated Vmax (I), and apparent Km (J). (K) Reversal potential of TRPC3, TRPC3(L241S), and TRPC3(I807S) measured in wild-type and STIM1−/− cells. (L–M) Example blots and averaged surface expression of TRPC3, TRPC3(L241S), and TRPC3(I807S) in resting cells and cells stimulated with 100 µM carbachol. All blots are measured in kD. (N) FRET ratio between STIM1 and TRPC3, TRPC3(L241S), and TRPC3(I807S) was measured before and after stimulation with 100 µM carbachol. Example time courses and I/V plots are given in Figs. S1 and S2. Source data are available for this figure: SourceData F1.
STIM1 controls receptor-mediated stimulation of TRPC3. (A) The domains and structure of a TRPC3 single subunit. The position of L241 and I807 are shown as spheres. (B–D) Concentration dependence of outward current of carbachol-stimulated TRPC3 (black), TRPC3(L241S) (red), and TRPC3(I807S) (green; B). The Vmax (C) and apparent Km (D) were obtained by Hill fitting. (E–G) All experiments were in STIM1−/− cells. Concentration dependence of outward current of carbachol-stimulated TRPC3 (black), TRPC3(L241S) (red), and TRPC3(I807S) (green; E). The Vmax (F) and apparent Km (G) were obtained by Hill fitting. (H–J) Experiments were in STIM1−/− cells transfected with vector (black) or STIM1 (red) depicting the concentration dependence (H), calculated Vmax (I), and apparent Km (J). (K) Reversal potential of TRPC3, TRPC3(L241S), and TRPC3(I807S) measured in wild-type and STIM1−/− cells. (L–M) Example blots and averaged surface expression of TRPC3, TRPC3(L241S), and TRPC3(I807S) in resting cells and cells stimulated with 100 µM carbachol. All blots are measured in kD. (N) FRET ratio between STIM1 and TRPC3, TRPC3(L241S), and TRPC3(I807S) was measured before and after stimulation with 100 µM carbachol. Example time courses and I/V plots are given in Figs. S1 and S2. Source data are available for this figure: SourceData F1.
Since the mutants affect interaction of TRPC3 with STIM1, these findings imply that a major role of STIM1 is controlling the affinity for receptor activation of TRPC3 and predict that the activation of TRPC3 and the mutants be altered in the absence of STIM1. To test these predictions, we measured the effect of deletion of STIM1 on TRPC3 activation. For this, first we determined the effect of STIM1 deletion on ER/PM junction formation, expression of TRPC3, and the maximal current. The effect of STIM1 on the ER/PM junction was assessed with the ER/PM junction MAPPER probe (Chang et al., 2013). Fig. S1, K and L, show that deletion of STIM1 partially disrupted the junctions by reducing their number and probably their size as shown by the reduced intensity. However, deletion of STIM1 had no effect on the surface expression of TRPC3 (Fig. S1 M) or the maximum current when compared to TRPC3 expressed in wild-type HEK cells (Fig. S1, N and O). Yet, remarkably, the receptor-dependent activation of all three TRPC3 variants collapsed to that of TRPC3(I807S), with low and similar apparent affinity (Fig. 1, E–G). Notably, Fig. 1, H–J, show that the high apparent affinity for receptor stimulation of TRPC3 was rescued by re-expression of STIM1 in STIM1−/− cells. To assess the effect of STIM1 and junctional integrity on channel properties, we determined the reversal potential of TRPC3 and the mutants in wild-type and STIM1−/− cells. Fig. 1 K shows that TRPC3 and TRPC3(L241S) showed higher Na+/Cs+ selectivity than TRPC3(I807S). Disruption of the junctions by deletion of STIM1 increased the Cs+ selectivity of TRPC3 and TRPC3(L241S) and had no effect on that of TRPC3(I807S). Thus, presence in the junctions affects channel pore selectivity.
Measurement of surface expression in Fig. 1, L and M, show that maximally stimulating the type 3 muscarinic receptors (M3Rs) with 100 µM carbachol had no or minimal effect on TRPC3 and the mutants’ surface expression, with only significantly higher surface expression of TRPC3(L241S) in stimulated cells. Similarly, cell stimulation had no apparent effect on surface expression of the M3Rs (Fig. S1 P). Interaction between TRPC3 and STIM1 and the effect of the mutants on the interaction could be demonstrated by Co-IP (Lee et al., 2014) and by confocal microscopy (Fig. S1, Q–T), suggesting reduced colocalization of TRPC3(I807S) with STIM1. However, the limited resolution of confocal microscopy of 200 nm is not sufficient for accurate and quantitative analysis. Therefore, we used Förster resonance energy transfer (FRET) measurement that has a resolution of 2–8 nm. FRET measurement with STIM1-CFP and TRPC3-YFP in Fig. 1 N shows that TRPC3(L241S) increased, while TRPC3(I807S) decreased the basal STIM1-TRPC3 FRET. Cell stimulation increases the STIM1-TRPC3 FRET close to the level measured with basal STIM1-TRPC3(L241S). Cell stimulation further slightly increased STIM1-TRPC3(L241S) FRET, while it had no effect on STIM1-TRPC3(I807S) FRET. Since TRPC3 is at the PM and STIM1 once unfolded by ER Ca2+ depletion associates with PM PI(4,5)P2 at the ER/PM junctions, these findings indicate that cell stimulation primarily enhances the interaction between TRPC3-STIM1 within the ER/PM junctions rather than affecting TRPC3 surface expression and that TRPC3(L241S) is recruited to the junctions, while TRPC3(I807S) cannot access the ER/PM junctions formed by STIM1.
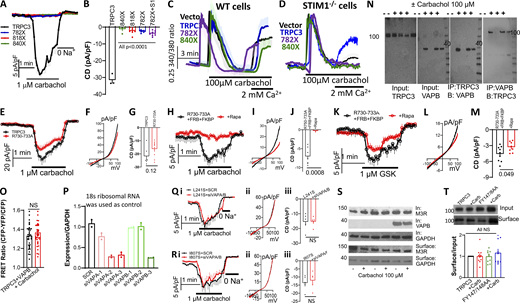
The effects of the TRPC3 mutants on the receptor-mediated stimulation (Fig. 1, B–D) and of STIM1 (Fig. 1, E–J) raised the question as to whether these effects are due to a change in the kinetic properties of the channel or due to a change in TRPC3 response to stimulation by the pore lipid. We addressed this question by characterizing the response of TRPC3 and the mutants to direct activation by GSK. GSK is a substrate that interacts with and activates TRPC3 in the same manner as the pore lipid DAG, but has better cell permeability (Lichtenegger et al., 2018; Svobodova et al., 2019). Fig. 2, A–F, show that TRPC3(L241S) exhibited increased activity in response to GSK, while TRPC3(I807S) had no change. Fig. 2, G–I, show that the kinetic properties of direct activation of the TRPC3 variants by GSK are different than those by receptor stimulation. The TRPC3(L241S) primarily increased the Vmax with a minimal effect on the apparent Km for GSK, while TRPC3(I807S) had no effect on either parameter.
Properties of TRPC3, TRPC3(L241S), and TRPC3(I807S) activation by GSK. (A–F) Time course (A and D), example I/V (B and E), and current density (C and F) of activation of TRPC3 (black), TRPC3(L241S) (red), and TRPC3(I807S) (green) by 0.05 (A–C) and 10 µM GSK (D–F). (G–I) Concentration dependence of GSK stimulation of TRPC3 (black), TRPC3(L241S) (red), and TRPC3(I807S) (green; E). The Vmax (H) and apparent Km (I) were obtained by Hill fitting.
Properties of TRPC3, TRPC3(L241S), and TRPC3(I807S) activation by GSK. (A–F) Time course (A and D), example I/V (B and E), and current density (C and F) of activation of TRPC3 (black), TRPC3(L241S) (red), and TRPC3(I807S) (green) by 0.05 (A–C) and 10 µM GSK (D–F). (G–I) Concentration dependence of GSK stimulation of TRPC3 (black), TRPC3(L241S) (red), and TRPC3(I807S) (green; E). The Vmax (H) and apparent Km (I) were obtained by Hill fitting.
Localization at the ER/PM junctions controls the response of TRPC3 to GPCRs stimulation
In addition to activation of the Orai and TRPC channels (Bodnar et al., 2017; Lopez et al., 2020), STIM1 has tethering function at the ER/PM junctions owing to its ER transmembrane domain, large cytoplasmic domain, and a PM PI(4,5)P2 interacting polybasic domain (Chang et al., 2013; Fig. S1, K and L). To examine further the role of localization in the junctions in the effect of STIM1 on TRPC3, we determine the effect of the other established tethers with critical roles in formation of the junctions E-Syts (Giordano et al., 2013; Saheki and De Camilli, 2017) and ANO8 (Jha et al., 2019). Fig. S2, A and B, show that knockdown or overexpression of E-Syt1 had no effect on TRPC3 surface expression. Fig. 3, A and B, show that knockdown of E-Syt1 significantly inhibited TRPC3 current stimulated by 1 µM carbachol. Interestingly, these effects were specific to E-Syt1. Fig. S3, A–F, show that knockdown of E-Syt2 and E-Syt3 had no effect on receptor stimulation of TRPC3.
Role of ER/PM junctions in regulation of TRPC3 by receptor stimulation. (A and B) Effect of siE-Syt1 (A) and E-Sty1 expression (B) on TRPC3 surface expression. (C and D) Effect of E-Syt1 on TRPC3 and mutant current. C shows example I/V and D the current density. (E–I) Example I/V for TRPC3 and the indicated mutants current as affected (red) by E-Syt1 (E), siANO8 (F), ANO8 (G), PI(4,5)P2 depletion (H and I). (J and K) Surface expression of the indicated TRPC3 mutants. Source data are available for this figure: SourceData FS2.
Role of ER/PM junctions in regulation of TRPC3 by receptor stimulation. (A and B) Effect of siE-Syt1 (A) and E-Sty1 expression (B) on TRPC3 surface expression. (C and D) Effect of E-Syt1 on TRPC3 and mutant current. C shows example I/V and D the current density. (E–I) Example I/V for TRPC3 and the indicated mutants current as affected (red) by E-Syt1 (E), siANO8 (F), ANO8 (G), PI(4,5)P2 depletion (H and I). (J and K) Surface expression of the indicated TRPC3 mutants. Source data are available for this figure: SourceData FS2.
Effect of disruption of the ER/PM junction on activation of TRPC3, TRPC3(L241S), and TRPC3(I807S) by receptor stimulation. (A and B) Cells were treated with scrambled or E-Syt1 siRNA as indicated. (A) Shows the time course of current activated by 1 µM carbachol. (B) Plot the average current densities of experiments as in A of cells treated with scrambled or E-Syt1 siRNA and stimulated with either 0.3, 1, or 10 µM carbachol. (C and D) Cells were transfected with EV or with E-Syt1 as indicated. C shows time course and D the current density. Example I/V and effect of siE-Syt1 and of E-Syt1 on surface TRPC3 are shown in Fig. S2, A–E. (E) FRET ratio in cells expressing TRPC3-YFP and STIM1-CFP was measured in cells expressing vector of E-Syt1 and with or without stimulation with 100 µM carbachol. (F and G) Example images (F) and puncta intensity (G) of TRPC3 in the presence and absence of STIM1 and in cells stimulated with 100 µM carbachol measured by TIRF microscopy. (H) Puncta number/µm2 of TRPC3, TRPC3(L241S), and TRPC3(I807S). (I and J) Cells were treated with scrambled or ANO8 siRNA as indicated. Time course (I) and current density (J) of cells stimulated with 1 µM carbachol and expressing TRPC3 (gray and blue), TRPC3(L241S) (light red and red), and TRPC3(I807S) (light green and green). (K and L) Cells were transfected with EV or with ANO8 as indicated. Time course (K) and current density (L) of cells stimulated with 1 µM carbachol and expressing TRPC3 (gray and blue), TRPC3(L241S) (light red and red), and TRPC3(I807S) (light green and green). EV, empty vector; VC, vector control.
Effect of disruption of the ER/PM junction on activation of TRPC3, TRPC3(L241S), and TRPC3(I807S) by receptor stimulation. (A and B) Cells were treated with scrambled or E-Syt1 siRNA as indicated. (A) Shows the time course of current activated by 1 µM carbachol. (B) Plot the average current densities of experiments as in A of cells treated with scrambled or E-Syt1 siRNA and stimulated with either 0.3, 1, or 10 µM carbachol. (C and D) Cells were transfected with EV or with E-Syt1 as indicated. C shows time course and D the current density. Example I/V and effect of siE-Syt1 and of E-Syt1 on surface TRPC3 are shown in Fig. S2, A–E. (E) FRET ratio in cells expressing TRPC3-YFP and STIM1-CFP was measured in cells expressing vector of E-Syt1 and with or without stimulation with 100 µM carbachol. (F and G) Example images (F) and puncta intensity (G) of TRPC3 in the presence and absence of STIM1 and in cells stimulated with 100 µM carbachol measured by TIRF microscopy. (H) Puncta number/µm2 of TRPC3, TRPC3(L241S), and TRPC3(I807S). (I and J) Cells were treated with scrambled or ANO8 siRNA as indicated. Time course (I) and current density (J) of cells stimulated with 1 µM carbachol and expressing TRPC3 (gray and blue), TRPC3(L241S) (light red and red), and TRPC3(I807S) (light green and green). (K and L) Cells were transfected with EV or with ANO8 as indicated. Time course (K) and current density (L) of cells stimulated with 1 µM carbachol and expressing TRPC3 (gray and blue), TRPC3(L241S) (light red and red), and TRPC3(I807S) (light green and green). EV, empty vector; VC, vector control.
Effect of E-Syt2, E-Syt3, STIM1 deletion, TRPC3 pore and lipid binding mutants on TRPC3 activity stimulated by carbachol of GSK. (A–F) Time course (A and D), example I/V (B and E), and current density (C and F) of TRPC3 expressed in cells treated with scrambled siRNA (SCR; black), siE-Syt2 (red, A–C), or siE-Syt3 (red, D–F) and stimulated with 1 µM carbachol. (G–I) TRPC3 alone (black traces) and TRPC3 + E-Syt1 were expressed in STIM1−/− cells and stimulated with 1 µM carbachol. (J–O) Time course (J and M), example I/V (K and N), and current density (L and O) of TRPC3, TRPC3(E615A), TRPC3(K619A) (J–L) stimulated with 1 µM GSK and TRPC3(I556A) (M and N) stimulated with 3 µM GSK. (P–R) Concentration dependence of GSK stimulation of TRPC3 and TRPC3(N560A) inward current (P). The Vmax (Q) and apparent Km (R) were obtained by Hill fitting.
Effect of E-Syt2, E-Syt3, STIM1 deletion, TRPC3 pore and lipid binding mutants on TRPC3 activity stimulated by carbachol of GSK. (A–F) Time course (A and D), example I/V (B and E), and current density (C and F) of TRPC3 expressed in cells treated with scrambled siRNA (SCR; black), siE-Syt2 (red, A–C), or siE-Syt3 (red, D–F) and stimulated with 1 µM carbachol. (G–I) TRPC3 alone (black traces) and TRPC3 + E-Syt1 were expressed in STIM1−/− cells and stimulated with 1 µM carbachol. (J–O) Time course (J and M), example I/V (K and N), and current density (L and O) of TRPC3, TRPC3(E615A), TRPC3(K619A) (J–L) stimulated with 1 µM GSK and TRPC3(I556A) (M and N) stimulated with 3 µM GSK. (P–R) Concentration dependence of GSK stimulation of TRPC3 and TRPC3(N560A) inward current (P). The Vmax (Q) and apparent Km (R) were obtained by Hill fitting.
Notably, knockdown of E-Syt1 reduced the activity of TRPC3 and TRPC3(L241S) but had no effect on the activity of TRPC3(I807S) (Fig. 3 A). This was the case with all carbachol concentrations tested (Fig. 3 B). In reciprocal experiments, Fig. 3, C and D, show that overexpression of E-Syt1 increased the current measured with TRPC3 and TRPC3(L241S) but not with TRPC3(I807S). Although overexpression of E-Syt1 had no effect on total and surface expression of TRPC3 (Fig. S2 B), the FRET measurements in Fig. 3 E show that E-Syt1 increased the basal and the receptor-stimulated interaction of TRPC3 with STIM1, indicating interaction of STIM1 with TRPC3 takes place at the ER/PM junctions. This was assayed directly with total internal reflection fluorescence (TIRF) microscopy. The sample images and puncta intensity in Fig. 3, F and G, and number of puncta in Fig. 3 H show that TRPC3 is clustered at the TIRF field in unstimulated cells, and STIM1 increased the number of channels at the same clusters (increased intensity). Clustering was not increased further by cell stimulation. The TRPC3(L241S) mutant showed increased clustering (puncta number) relative to TRPC3, and the TRPC3(I807S) mutant showed the same number of puncta as TRPC3. Interestingly, Fig. S3, G–I, show that the effects of E-Syt1 on TRPC3 activity were eliminated in STIM1−/− cells, indicating that the role of E-Syt1 required intact STIM1 ER/PM junctions. Fig. 3, I and J, show that knockdown of ANO8, and Fig. 3, K and L, show that overexpression of ANO8 decreases and increases, respectively, the receptor stimulation of TRPC3 and TRPC3(L241S), but had no effect on TRPC3(I807S), as was found with E-Syt1 and STIM1.
Multiple roles and sites for interaction and regulation of TRPC3 by PI(4,5)P2
The effects of STIM1, E-Syt1, and ANO8 depend on their interaction with PI(4,5)P2 at a PM PI(4,5)P2-rich domain (Giordano et al., 2013; Jha et al., 2019; Liou et al., 2007; Maléth et al., 2014; Saheki and De Camilli, 2017), and TRPC3 is regulated by lipids (Groschner and Tiapko, 2018; Imai et al., 2012; Svobodova and Groschner, 2016). Therefore, we tested the role of PI(4,5)P2 in the regulation of TRPC3 and mutants. PI(4,5)P2 was depleted acutely using the FRB/FKBP-PI5Ptase system (Varnai et al., 2006). Fig. 4, A and B, show that depletion of PI(4,5)P2 reduced the receptor-stimulated TRPC3 and TRPC3(L241S) currents to the level of the TRPC3(I807S) current and had no effect on TRPC3(I807S) current. Measurements of reversal potential in Fig. 4 C show that TRPC3(I807S) has higher Cs+/Na+ selectivity than TRPC3 and TRPC3(L241S). Of note, deletion of PI(4,5)P2 equalized the selectivity of TRPC3, TRPC3(L241S), and TRPC3(I807S) and had no effect on the selectivity of TRPC3(I807S). These findings suggest that PI(4,5)P2 either directly affects the selectivity of the pore or that localization of the channel in a PI(4,5)P2-rich ER/PM junctions affects its selectivity. Further analysis of the effect of PI(4,5)P2 was made by determining the dose-response to carbachol stimulation. Fig. 4, D–F, show that depletion of PI(4,5)P2 reduced TRPC3 Vmax but also increased the apparent Km, combining the effects of both TRPC3(L241S; primarily Vmax) and TRPC3(I807S; primarily Km).
The dependence of TRPC3, TRPC3(L241S), and TRPC3(I807S) activity on plasma membrane PI(4,5)P 2 . (A–C) All cells expressed the indicated TRPC3 channels and the PI(4,5)P2 depletion system FRB-PLCγ-PHD + FKBP-PI5ase. After establishing the whole-cell configuration, cells were treated with vehicle (control) or 0.2 µM rapamycin to 3 min before start of current recording to deplete PI(4,5)P2. Rapamycin was maintained throughout the current recording. The panels show time course (A), current density (B), and reversal potential (C) of TRPC3 (black), TRPC3(L241S) (red), and TRPC3(I807S) (green) treated with vehicle (light colors) or rapamycin (dark colors). (D–F) Concentration dependence of inward current of carbachol-stimulated TRPC3 in control (black) and PI(4,5)P2 depleted cells (purple; D). The Vmax (E) and apparent Km (F) were obtained by Hill fitting. (G and H) Time course (G) and current density (H) of TRPC3(L241S) (red) and TRPC3(I807S) (green) in control (light colors) or PI(4,5)P2 depleted cells (dark colors) and stimulated with 0.3 µM GSK. (I–K) Concentration dependence of outward current of GSK-stimulated TRPC3 in control (black) and PI(4,5)P2 depleted cells (purple; I). The Vmax (J) and apparent Km (K) were obtained by Hill fitting. (L) Effect of PI(4,5)P2 depletion on reversal potential of TRPC3 and mutants stimulated with 0.3 µM GSK.
The dependence of TRPC3, TRPC3(L241S), and TRPC3(I807S) activity on plasma membrane PI(4,5)P 2 . (A–C) All cells expressed the indicated TRPC3 channels and the PI(4,5)P2 depletion system FRB-PLCγ-PHD + FKBP-PI5ase. After establishing the whole-cell configuration, cells were treated with vehicle (control) or 0.2 µM rapamycin to 3 min before start of current recording to deplete PI(4,5)P2. Rapamycin was maintained throughout the current recording. The panels show time course (A), current density (B), and reversal potential (C) of TRPC3 (black), TRPC3(L241S) (red), and TRPC3(I807S) (green) treated with vehicle (light colors) or rapamycin (dark colors). (D–F) Concentration dependence of inward current of carbachol-stimulated TRPC3 in control (black) and PI(4,5)P2 depleted cells (purple; D). The Vmax (E) and apparent Km (F) were obtained by Hill fitting. (G and H) Time course (G) and current density (H) of TRPC3(L241S) (red) and TRPC3(I807S) (green) in control (light colors) or PI(4,5)P2 depleted cells (dark colors) and stimulated with 0.3 µM GSK. (I–K) Concentration dependence of outward current of GSK-stimulated TRPC3 in control (black) and PI(4,5)P2 depleted cells (purple; I). The Vmax (J) and apparent Km (K) were obtained by Hill fitting. (L) Effect of PI(4,5)P2 depletion on reversal potential of TRPC3 and mutants stimulated with 0.3 µM GSK.
To analyze the effect of PI(4,5)P2 on the pore, we determined the effect of PI(4,5)P2 depletion on activation of TRPC3 by GSK. We analyzed primarily the currents measured at +100 mV because of the small current densities stimulated by low concentrations of GSK. Fig. 4, G and H, show that the I807S mutation prevented the dependence of TRPC3 activity on PI(4,5)P2. Surprisingly, the L241S mutation augmented the dependence of TRPC3 activity on PI(4,5)P2, suggesting that PI(4,5)P2 controls access of GSK to the TRPC3(L241S) pore. Indeed, measurement of the GSK dependence of TRPC3 (Fig. 4 I) revealed that PI(4,5)P2 depletion had no effect on Vmax (Fig. 4 J), but markedly increased the apparent Km (Fig. 4 K). Notably, PI(4,5)P2 depletion had no effect on the channel’s reversal potential (Fig. 4 L), which was similar to the reversal potential measured with agonist stimulation in PI(4,5)P2-depleted cells (Fig 4 C). These findings imply multiple effects of PI(4,5)P2: providing a substrate for activation of TRPC3 (DAG), controlling access of DAG to the pore (Km for GSK), and regulating pore architecture when activated by receptor stimulation (change in reversal potential).
The TRPC3 structure (Fan et al., 2018) identified two lipid densities with the periphery labeled as site 1 and the pore as site 2, as shown in Fig. 5 A. Similar sites are present in TRPC6, and the lipid in site 2 was identified as DAG (Bai et al., 2020). We attempted to reveal the role of the two lipid sites in regulation of TRPC3 by PI(4,5)P2. The structure of TRPC3 indicates that E615 and K619 in the pore helix and N645 in S6 of an adjacent subunit make contacts with the pore lipid. Fig. S2 J shows that the E615 and K619 mutations had no effect on total or surface expression of TRPC3, but inhibited TRPC3 current stimulated by carbachol, as shown in Fig. 5, B–D. Surprisingly, the N645A mutation had no effect on TRPC3 current, although the equivalent mutation in TRPC6 inhibited the channel (Bai et al., 2020). The difference may be explained by the assays used. TRPC6 activity was assayed by measurement of Ca2+ influx, while we measured TRPC3 current that more directly reports channel function. The E615A and K619A mutants similarly inhibited activation of TRPC3 by receptor stimulation (Fig. 5 B) and by GSK (Fig. S3, J–L). Mutations of neighboring residues in TRPC6 also inhibited channel activation by DAG (Bai et al., 2020), but their effect on receptor stimulation was not examined. These findings suggest the TRPC3 pore lipid-binding site is likely one of the sites by which PI(4,5)P2 affects channel activity through supply of DAG.
Regulation of TRPC3 activity by lipid sites 1 and 2. (A) The model on the left is of a TRPC3 monomer and doted squares indicate localization of lipid sites 1 (peripheral) and site 2 (pore) in orange. The model on the right shows higher magnification of the site 2 mutated residues as spheres. G652 and N645 in TM6 are shown in the same subunit for simplicity, although G652 and N645 from an adjacent subunit interact with the substrate. Total and surface expression of E615A and K619A are shown in Fig. S2. (B–D) Time course (B), example I/V (C), and current density (D) of TRPC3 (black), TRPC3(E615A) (red), TRPC3(K619A) (blue); and TRPC3(N645A) (purple) are shown. Note that even stimulation with supramaximal concentration of 1 mM carbachol did not activate current of TRPC3(E615A) and TRPC3(K619A). (E) The model shows higher magnification of lipid site 1 in A and the spheres are the residues mutated in this site. Surface expression of W334A, N560A, F563A, and I556A are shown in Fig. S2. (F–J) Time course (F and I), example I/V (G and lower panel in expanded current scale), and current density (H and J) of TRPC3 (black), TRPC3(W334A) (red), TRPC3(Y335A) (blue), TRPC3(I556A) (dark yellow) TRPC3(N560A) (turquoise), and TRPC3(F563A) (purple) are shown. F–H show the inward currents and I and J the outward currents. Note the selective effect of N560A on the inward and of F563A on the outward currents. (K) Reversal potential of TRPC3 and mutants stimulated with 1 µM carbachol.
Regulation of TRPC3 activity by lipid sites 1 and 2. (A) The model on the left is of a TRPC3 monomer and doted squares indicate localization of lipid sites 1 (peripheral) and site 2 (pore) in orange. The model on the right shows higher magnification of the site 2 mutated residues as spheres. G652 and N645 in TM6 are shown in the same subunit for simplicity, although G652 and N645 from an adjacent subunit interact with the substrate. Total and surface expression of E615A and K619A are shown in Fig. S2. (B–D) Time course (B), example I/V (C), and current density (D) of TRPC3 (black), TRPC3(E615A) (red), TRPC3(K619A) (blue); and TRPC3(N645A) (purple) are shown. Note that even stimulation with supramaximal concentration of 1 mM carbachol did not activate current of TRPC3(E615A) and TRPC3(K619A). (E) The model shows higher magnification of lipid site 1 in A and the spheres are the residues mutated in this site. Surface expression of W334A, N560A, F563A, and I556A are shown in Fig. S2. (F–J) Time course (F and I), example I/V (G and lower panel in expanded current scale), and current density (H and J) of TRPC3 (black), TRPC3(W334A) (red), TRPC3(Y335A) (blue), TRPC3(I556A) (dark yellow) TRPC3(N560A) (turquoise), and TRPC3(F563A) (purple) are shown. F–H show the inward currents and I and J the outward currents. Note the selective effect of N560A on the inward and of F563A on the outward currents. (K) Reversal potential of TRPC3 and mutants stimulated with 1 µM carbachol.
To examine the contribution of lipid-binding to site 1, we mutated several residues that contact the lipid, which are shown in Fig. 5 E. We searched for mutants that affected channel activity but retained function. Fig. S2 K shows the effect of the mutants on total and surface expression. The W334A mutant had the only notable effect, reducing total and surface expression. Due to the differential effect of some of the mutants on the inward and outward currents, both currents are shown. Fig. 5, F–H, show the inward currents and Fig. 5, I and J, show the outward currents. Although Y335 makes a hydrogen bond with the lipid head, Y355A had no effect on the current. Fig. 5, F and I, show that the I556A mutation strongly inhibited the current activated by carbachol and Fig. S3, M–O, show the inhibition of the current activated by GSK. This suggests an essential role of I556 in channel function, either by allowing lipid access to this site or by locking the channel in a closed state. Mutation of the nearby F563A had no effect on the inward current but strongly inhibited the outward current, while the N560A mutation had no effect on the outward current while increasing the inward current, as shown in the expanded portion of the I/V in the bottom of Fig. 5 J, suggesting selective effects on the channel pore inner and outer gates. The I556, F563, and N560 residues contact both the lipid and the TRP helix, which regulates TRP channels’ pore opening and voltage dependence (Gees et al., 2012). Hence I556A, F563A, and N560A effects may be mediated by altered TRP helix orientation. The most prominent effect was measured with TRPC3(W334A) that contacts the lipid tail, with the W334A mutant markedly increasing current density. The increased current by TRPC3(W334A) is somewhat underestimated because of the reduced surface expression. Fig. 5 K shows that the changes in channel conductance did not change the pore selectivity, as indicated by the same reversal potential at the Na+ and Cs+ gradients used in these experiments.
The lipid site 1 affects PI(4,5)P2-dependence of TRPC3 and access of lipid to site 2
To gain information on the role of the lipid site 1 in TRPC3 function, we characterized activation of the TRPC3(W334A) and TRPC3(N560A) mutants by receptor stimulation and GSK. Fig. 6, A–F, show the concentration dependence of the inward (Fig. 6, A–C) and outward (Fig. 6, D–F) currents for activation of the mutants by the M3 receptor. Due to the small GSK-activated inward current, more reliable results were obtained with the GSK-activated outward current, and Fig. 6, G–I, show the concentration dependence for activation of the mutants by GSK. The TRPC3(W334A) mutation increased the Vmax for both the receptor-evoked inward and outward currents with minimal reduction of the apparent Km of the outward current. TRPC3(W334A) similarly increased Vmax of the outward currents stimulated by GSK with no effect on apparent Km. Fig. 6, A–C, show that the TRPC3(N560A) mutant increased Vmax for the inward current stimulated by carbachol, as well as by GSK (Fig. S3, P–R) with no effect on the apparent Km. Thus, the primary effect of both the TRPC3(W334A) and TRPC3(N560A) mutants is increased Vmax.
Lipid site 1 modulates TRPC3 activity and substrate access to lipid site 2. (A–F) Concentration dependence of carbachol stimulation of TRPC3 (black), TRPC3(W334A) (red), and TRPC3(N560A) (green; A and D). The Vmax (B and E) and apparent Km (C and F) were obtained by Hill fitting. A–C are results from the inward currents and D–F are results from the outward currents. (G–I) Concentration dependence of outward current of GSK-stimulated TRPC3 (black), TRPC3(W334A) (red), and TRPC3(N560A) (green; G). The Vmax (H) and apparent Km (I) were obtained by Hill fitting.
Lipid site 1 modulates TRPC3 activity and substrate access to lipid site 2. (A–F) Concentration dependence of carbachol stimulation of TRPC3 (black), TRPC3(W334A) (red), and TRPC3(N560A) (green; A and D). The Vmax (B and E) and apparent Km (C and F) were obtained by Hill fitting. A–C are results from the inward currents and D–F are results from the outward currents. (G–I) Concentration dependence of outward current of GSK-stimulated TRPC3 (black), TRPC3(W334A) (red), and TRPC3(N560A) (green; G). The Vmax (H) and apparent Km (I) were obtained by Hill fitting.
In principle, the mutations can increase or decrease PI(4,5)P2 binding to site 1 to affect channel activity and lipid access to pore lipid site 2. To understand the role of lipid site 1 in the effect of PI(4,5)P2 on the channel, we measured the effect of PI(4,5)P2 depletion on the activation of TRPC3, TRPC3(W334A), and TRPC3(N560A) by receptor stimulation and by GSK. Results obtained with the outward currents are shown in Fig. 7, A–K, and with the inward current in Fig. S4, A–J. Deletion of PI(4,5)P2 reduced the TRPC3 current density (Vmax) and increased the apparent Km for carbachol stimulation (outward current, Fig. 7, A, D, and E; and inward current, Fig. 4, D–F), while markedly reducing the affinity for GSK without reducing Vmax (outward current, Fig. 7, F, J, and K; and inward current, Fig. 4, I–K). The TRPC3(W334A) mutation reversed inhibition of receptor-stimulated current by PI(4,5)P2 depletion (Figs. 7 D and S4 C) and markedly increased the apparent affinity for receptor stimulation, as shown in Figs. 7 E and S4 D, while preventing the prominently reduced affinity for GSK observed with TRPC3, as shown in Fig. 7 K and Fig. S4, I and J. PI(4,5)P2 depletion slightly reduced current density and modestly increased the apparent affinity for GSK stimulation of TRPC3(N560A), as shown in Fig. 7, I–K, and Fig. S4, G–J. The different effects of the mutants on stimulation by carbachol and GSK imply multiple roles of PI(4,5)P2. The independence of the increased Vmax for both carbachol and GSK stimulation and on the Km for GSK of PI(4,5)P2 depletion suggests that the TRPC3(W334A) and TRPC3(N560A) inhibit PI(4,5)P2 interaction with site 1. Moreover, PI(4,5)P2 binding to site 1 inhibits TRPC3 activity. The prominent effect of PI(4,5)P2 depletion on TRPC3(W334A) Km for carbachol stimulation suggests an effect specific to receptor stimulation rather than on channel behavior.
Role of PI(4,5)P 2 in the regulation of TRPC3 by lipid sites 1 and 2. (A–E) Concentration dependence of inward current of carbachol-stimulated TRPC3 (A) and TRPC3(W334A) (B) in control (black) and PI(4,5)P2 depleted cells (red). Example I/Vs are in C. The Vmax (D) and apparent Km (E) were obtained by Hill fitting. (F–K) Concentration dependence of outward current of GSK stimulation TRPC3 (F), TRPC3(W334A) (G), and TRPC3(N560A) (I) in control (black) and PI(4,5)P2 depleted cells (red). The Vmax (J) and apparent Km (K) were obtained by Hill fitting. Example I/V are shown in H.
Role of PI(4,5)P 2 in the regulation of TRPC3 by lipid sites 1 and 2. (A–E) Concentration dependence of inward current of carbachol-stimulated TRPC3 (A) and TRPC3(W334A) (B) in control (black) and PI(4,5)P2 depleted cells (red). Example I/Vs are in C. The Vmax (D) and apparent Km (E) were obtained by Hill fitting. (F–K) Concentration dependence of outward current of GSK stimulation TRPC3 (F), TRPC3(W334A) (G), and TRPC3(N560A) (I) in control (black) and PI(4,5)P2 depleted cells (red). The Vmax (J) and apparent Km (K) were obtained by Hill fitting. Example I/V are shown in H.
Effect of PI(4,5)P 2 depletion on carbachol and GSK-stimulated inward current. (A–D) Concentration dependence of carbachol stimulated inward current of TRPC3 (A) and TRPC3(W334A) (B) in control (black) and PI(4,5)P2 depleted cells (red). The Vmax (C) and apparent Km (D) were obtained by Hill fitting. (E–J) Concentration dependence of GSK stimulated inward current of TRPC3 (E), TRPC3(W334A) (F); and TRPC3(N560A) (G) in control (black) and PI(4,5)P2 depleted cells (red). The Vmax (H) and apparent Km (I) were obtained by Hill fitting. J shows the apparent Km with extended scale.
Effect of PI(4,5)P 2 depletion on carbachol and GSK-stimulated inward current. (A–D) Concentration dependence of carbachol stimulated inward current of TRPC3 (A) and TRPC3(W334A) (B) in control (black) and PI(4,5)P2 depleted cells (red). The Vmax (C) and apparent Km (D) were obtained by Hill fitting. (E–J) Concentration dependence of GSK stimulated inward current of TRPC3 (E), TRPC3(W334A) (F); and TRPC3(N560A) (G) in control (black) and PI(4,5)P2 depleted cells (red). The Vmax (H) and apparent Km (I) were obtained by Hill fitting. J shows the apparent Km with extended scale.
The lipid site 1 does not control the PI(4,5)P2-dependent recruitment of TRPC3 to the ER/PM junction
The lack of effect of PI(4,5)P2 depletion on current density and the prominently reduced affinity for carbachol stimulation of the TRPC3(W334A) mutant raised the question if the lipid 1 site affects the recruitment of TRPC3 to the ER/PM junctions, which is required for increased current density, and the high apparent affinity for carbachol stimulation (Figs. 3 and 4). To test this hypothesis directly, we first measured the effect of disrupting the ER/PM junctions on the activity of TRPC3(W334A) and TRPC3(N560A). The ER/PM junctions are disrupted in STIM1−/− cells (Fig. S1, K and L), yet Fig. 8, A–C, show that the increased activity of TRPC3(W334A) and TRPC3(N560A) were maintained in STIM1−/− cells. In addition, Fig. 8, D–F show that the W334A mutation increased Vmax and the apparent affinity for receptor stimulation in STIM1−/− cells. Second, Fig. 8, G and H, show that knockdown of the tethers E-Syt1 and ANO8 reduced the activity of TRPC3(W334A). Third, we examined the effect of the I807S mutation that renders TRPC3 independent of changes in the ER/PM junctions and PI(4,5)P2. Fig. 8, I–K, show that the activity of the double mutant TRPC3(W334A/I807S) was resistant to PI(4,5)P2 depletion. Importantly, Fig. 8, L–N, show that the I807S mutation had minimal effect on the Vmax and prominently increased the apparent Km for receptor stimulation of TRPC3(W334A). Together, these findings indicate that TRPC3(W334A) activity depends on ER/PM junction integrity similar to TRPC3 activity, and thus PI(4,5)P2 at lipid site 1 is not the lipid site mediating interaction of the tethers with PM PI(4,5)P2.
The TRPC3 W334A and N560A mutations do not affect localization at the ER/PM junctions. (A–C) Time course (A), example I/V (B), and current density (C) of TRPC3 (black), TRPC3(W334A) (red), and TRPC3(N560A) expressed in STIM1−/− cells and stimulated with 1 µM carbachol. (D–F) Concentration dependence of inward current of carbachol-stimulated TRPC3 (black) and TRPC3(W334A) (red) expressed in STIM1−/− cells (D). The Vmax (E) and apparent Km (F) were obtained by Hill fitting. (G and H) Time course (G) and current density (H) of TRPC3(W334A) expressed in cells treated with scrambled (black), E-Syt1 (red), and ANO8 (blue) siRNA and stimulated with 1 µM carbachol. (I–K) Time course (I), example I/V (K), and current density (L) of TRPC3(W334A/I807S) expressed in control (black) or PI(4,5)P2 depleted cells and stimulated with 1 µM carbachol. (L–N) Concentration dependence of inward current of carbachol-stimulated TRPC3 (black), TRPC3(I807S) (green), TRPC3(W334A) (red), and TRPC3(W334A/I807S) (blue; L). The Vmax (M) and apparent Km (N) were obtained by Hill fitting.
The TRPC3 W334A and N560A mutations do not affect localization at the ER/PM junctions. (A–C) Time course (A), example I/V (B), and current density (C) of TRPC3 (black), TRPC3(W334A) (red), and TRPC3(N560A) expressed in STIM1−/− cells and stimulated with 1 µM carbachol. (D–F) Concentration dependence of inward current of carbachol-stimulated TRPC3 (black) and TRPC3(W334A) (red) expressed in STIM1−/− cells (D). The Vmax (E) and apparent Km (F) were obtained by Hill fitting. (G and H) Time course (G) and current density (H) of TRPC3(W334A) expressed in cells treated with scrambled (black), E-Syt1 (red), and ANO8 (blue) siRNA and stimulated with 1 µM carbachol. (I–K) Time course (I), example I/V (K), and current density (L) of TRPC3(W334A/I807S) expressed in control (black) or PI(4,5)P2 depleted cells and stimulated with 1 µM carbachol. (L–N) Concentration dependence of inward current of carbachol-stimulated TRPC3 (black), TRPC3(I807S) (green), TRPC3(W334A) (red), and TRPC3(W334A/I807S) (blue; L). The Vmax (M) and apparent Km (N) were obtained by Hill fitting.
The N-terminus FFAT site targets TRPC3 to the ER/PM junction
In a search for potential sites that mediate the recruitment of TRPC3 to the ER/PM junction, we tested truncations around the I807S mutation that affected localization of TRPC3 at the ER/PM junctions. A previous study reported that truncation of TRPC3 at K789 retained nearly normal receptor-stimulated Ca2+ influx (Wedel et al., 2003). However, Fig. S5, A and B, show that similar truncation completely lost activity, even when TRPC3(D782X) was co-expressed with STIM1. Shorter truncations (L818X) and as little as the last eight residues (L840X) eliminated TRPC3 channel activity (Fig. S5, A and B) and receptor-stimulated Ca2+ influx measured in wild-type (Fig. S5 C) or STIM1−/− cells (Fig. S5 D). Moreover, the truncation mutants inhibited the STIM1-independent native receptor-stimulated Ca2+ influx (Fig. S5 D). These findings highlight the importance of the pole helix for TRPC3 activity (and all TRP channels since all TRP channels have pole helices [Li and Fine, 2020]) and did not reveal a role in TRPC3 localization at the ER/PM junctions. Localization of proteins at the ER/PM junctions is stabilized by interaction with PI(4,5)P2 that binds to stretches of positively charged residues (Varnai et al., 2017). The four TRPC3 arginine residues R730–R733 do not make such a motif since TRPC3(R730A-R733A) was activated by receptor stimulation and inhibited by PI(4,5)P2 depletion (Fig. S5, E–M).
TRPC3730RRRR733is not PI(4,5)P2binding motif, VAP siRNA and VAPB-TRPC3 interaction by FRET. (A and B) Time course (A) and current density (B) of the indicated TRPC3 truncation with TRPC3(782X) expressed with empty vector (blue) or with STIM1 (magenta) and stimulated with 1 µM carbachol. (C and D) Carbachol-stimulated Ca2+ influx in wild-type (C) and STIM1−/− cells (D) transfected with vector (black), TRPC3 (blue), TRPC3(782X) (red), or TRPC3(840X) (green). Cells in Ca2+-free media were stimulated with carbachol. Where indicated, Ca2+ influx was measured by addition of Ca2+ to the bath solution. (E–M) Time course (E, H, and K), example I/V (F, I, and L), and current density (G, J, and M). (E–G) Cells transfected with TRPC3 (black) and TRPC3(R730-R733A) (red) were stimulated with 1 µM carbachol. (H–M) TRPC3(R730-R733A) was expressed in control cells (black) or PI(4,5)P2 depleted cells and stimulated with 1 µM carbachol (H–J) or 1 µM GSK (K–M). (N) Cells transfected with TRPC3 and vector or VAPB and were stimulated with 100 µM carbachol as indicated and were used to assay Co-IP of TRPC3 and VAPB. (O) Effect of Carbachol stimulation on TRPC3-YFP/VAPB-mCherry FRET. (P) Effect of VAPA and VAPB siRNA on their mRNA level in HEK cells. siVAPA-2 and siVAPB-3 were used for the experiments. (Q and R) Effect of siVAPA/B on TRPC3 (L) and TRPC3(I807S) (M) current. Shown are time course (i), example I/V (ii), and current density (iii). (S) Effect of VAPB on surface expression of the M3Rs. (T) Surface expression of TRPC3 and TRPC3(F147A/Y148A) with and without carbachol stimulation. Source data are available for this figure: SourceData FS5.
TRPC3730RRRR733is not PI(4,5)P2binding motif, VAP siRNA and VAPB-TRPC3 interaction by FRET. (A and B) Time course (A) and current density (B) of the indicated TRPC3 truncation with TRPC3(782X) expressed with empty vector (blue) or with STIM1 (magenta) and stimulated with 1 µM carbachol. (C and D) Carbachol-stimulated Ca2+ influx in wild-type (C) and STIM1−/− cells (D) transfected with vector (black), TRPC3 (blue), TRPC3(782X) (red), or TRPC3(840X) (green). Cells in Ca2+-free media were stimulated with carbachol. Where indicated, Ca2+ influx was measured by addition of Ca2+ to the bath solution. (E–M) Time course (E, H, and K), example I/V (F, I, and L), and current density (G, J, and M). (E–G) Cells transfected with TRPC3 (black) and TRPC3(R730-R733A) (red) were stimulated with 1 µM carbachol. (H–M) TRPC3(R730-R733A) was expressed in control cells (black) or PI(4,5)P2 depleted cells and stimulated with 1 µM carbachol (H–J) or 1 µM GSK (K–M). (N) Cells transfected with TRPC3 and vector or VAPB and were stimulated with 100 µM carbachol as indicated and were used to assay Co-IP of TRPC3 and VAPB. (O) Effect of Carbachol stimulation on TRPC3-YFP/VAPB-mCherry FRET. (P) Effect of VAPA and VAPB siRNA on their mRNA level in HEK cells. siVAPA-2 and siVAPB-3 were used for the experiments. (Q and R) Effect of siVAPA/B on TRPC3 (L) and TRPC3(I807S) (M) current. Shown are time course (i), example I/V (ii), and current density (iii). (S) Effect of VAPB on surface expression of the M3Rs. (T) Surface expression of TRPC3 and TRPC3(F147A/Y148A) with and without carbachol stimulation. Source data are available for this figure: SourceData FS5.
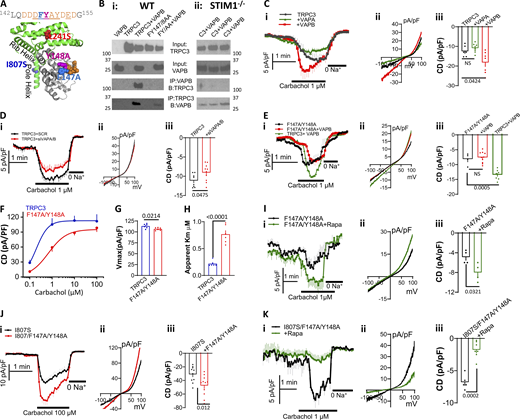
Further inspection of the TRPC3 sequence reveals the presence of a good N-terminal FFAT motif (Slee and Levine, 2019) between residues 142 and 155 within the linker helices (underlined in Fig. 9 A). Although the FFAT motif is not in direct contact with the pole helix where I807 is located, it is in a structure that envelopes the pole helix (Fan et al., 2018; Tang et al., 2018), as illustrated in Fig. 9 A. FFAT motifs interact with the ER located vesicle-associated membrane protein (VAMP)-associated proteins (VAP), VAPA and VAPB, that target them to MCS (Murphy and Levine, 2016; Slee and Levine, 2019). The Co-IP experiments in Fig. 9 B i show that TRPC3 interacts with VAPB, and the Co-IP was prevented by disrupting the FFAT motif in the mutant TRPC3(F147A/Y148A). The interaction of TRPC3 with VAPB was not affected by cell stimulation whether measured by Co-IP (Fig. S5 N) or FRET (Fig. S5 O), but, significantly, disruption of the ER/PM junctions by deletion of STIM1 prominently reduced or prevented the Co-IP as shown in Fig. 9 B ii. To determine whether the TRPC3 FFAT motif is functional, we measured the effect of expressing VAPA and VAPB on TRPC3 current. Fig. 9 C, i–iii, show that VAPB increased the current. Conversely, Fig. 9 D shows that knockdown of VAPA + VAPB reduced the current of TRPC3. The effect of the siRNA on the VAPs mRNA is shown in Fig. S5 P. The L241S mutation recruits TRPC3 to the ER/PM junctions, while the I807S mutation targets TRPC3 to a domain outside the ER/PM junctions (Fig. 1) and is therefore predicted to be independent of VAPA/B knockdown. Fig. S5, Q and R, show that this is indeed the case. The effects of cell stimulation and VAPB on the currents were not due to a change in surface expression of the M3 receptors (Fig. S5 S), and thus are likely due to localization within the ER/PM junctions.
The TRPC3 FFAT motif is requited for TRPC3 regulation by PI(4,5)P2. (A) Shown are the sequence of the FFAT motif and its position within the linker helices (gray) in orange. The mutated FY is shown in spheres. (B, i and ii) Wild-type (Bi) and STIM1−/− (Bii) HEK cells transfected with TRPC3 or TRPC3(F147A/Y148A) and with and without VAPB were used to determine Co-IP of VABP (anti-RFB) and TRPC3 (anti-GFP). (C, i–iii) Cells were transfected with TRPC3 and vector, VAPA or VAPB to measure current time course (i), example I/V (ii), and current density (iii). (D) Cells were treated with scrambled siRNA (black) or siRNA to knock down both VAPA and VAPB (red). Time course (i), example I/V (ii), and current density (iii) of TRPC3. (E, i–iii) Time course (i), example I/V (ii), and current density (iii) are shown for TRPC3 + VAPB (green), TRPC3(F147A/Y148A) (black), and TRPC3(F147A/Y148A) + VAPB (red). (F–H) Concentration dependence of carbachol-stimulated outward current of TRPC3 (black) and TRPC3(F147A/Y148A) (F). The Vmax (G) and apparent Km (H) were obtained by Hill fitting. (I, i–iii) Time course (i), example I/V (ii), and current density (iii) of TRPC3(F147A/Y148A) in control (black) or PI(4,5)P2 depleted cells (red) and stimulated with 1 µM carbachol. (J, i–iii) Time course (i), example I/V (ii), and current density (iii) of TRPC3(I807S) (black) and TRPC3(I807S/F147A/Y148A) stimulated with 100 µM carbachol (red). (Ki–iii) Time course (i), example I/V (ii), and current density (iii) of TRPC3(I807S/F147A/Y148A) in control (black) or PI(4,5)P2 depleted cells (red) and stimulated with 1 µM carbachol. Lack of effect of VAPB on surface M3 receptors and surface expression of TRPC3(F147A/Y148A) are shown in Fig. S2. Source data are available for this figure: SourceData F9.
The TRPC3 FFAT motif is requited for TRPC3 regulation by PI(4,5)P2. (A) Shown are the sequence of the FFAT motif and its position within the linker helices (gray) in orange. The mutated FY is shown in spheres. (B, i and ii) Wild-type (Bi) and STIM1−/− (Bii) HEK cells transfected with TRPC3 or TRPC3(F147A/Y148A) and with and without VAPB were used to determine Co-IP of VABP (anti-RFB) and TRPC3 (anti-GFP). (C, i–iii) Cells were transfected with TRPC3 and vector, VAPA or VAPB to measure current time course (i), example I/V (ii), and current density (iii). (D) Cells were treated with scrambled siRNA (black) or siRNA to knock down both VAPA and VAPB (red). Time course (i), example I/V (ii), and current density (iii) of TRPC3. (E, i–iii) Time course (i), example I/V (ii), and current density (iii) are shown for TRPC3 + VAPB (green), TRPC3(F147A/Y148A) (black), and TRPC3(F147A/Y148A) + VAPB (red). (F–H) Concentration dependence of carbachol-stimulated outward current of TRPC3 (black) and TRPC3(F147A/Y148A) (F). The Vmax (G) and apparent Km (H) were obtained by Hill fitting. (I, i–iii) Time course (i), example I/V (ii), and current density (iii) of TRPC3(F147A/Y148A) in control (black) or PI(4,5)P2 depleted cells (red) and stimulated with 1 µM carbachol. (J, i–iii) Time course (i), example I/V (ii), and current density (iii) of TRPC3(I807S) (black) and TRPC3(I807S/F147A/Y148A) stimulated with 100 µM carbachol (red). (Ki–iii) Time course (i), example I/V (ii), and current density (iii) of TRPC3(I807S/F147A/Y148A) in control (black) or PI(4,5)P2 depleted cells (red) and stimulated with 1 µM carbachol. Lack of effect of VAPB on surface M3 receptors and surface expression of TRPC3(F147A/Y148A) are shown in Fig. S2. Source data are available for this figure: SourceData F9.
In additional experiments, we found that the TRPC3(F147A/Y148A) mutant had no effect on total or surface expression (Fig. S5 T), but eliminated activation by VAPB, as shown in Fig. 9 E, i–iii. Disrupting localization of TRPC3 at the ER/PM junctions causes reduction in the apparent affinity for activation by receptor stimulation (Figs. 1, E–J, 4 D–F, and S4). Fig. 9, F–H, show that mutating the FFAT motif had a small effect on current density, while it prominently reduced the apparent affinity for activation of TRPC3 by carbachol. Disrupted localization at the ER/PM junctions should affect the response of TRPC3 to PI(4,5)P2. Fig. 9 I, i–iii, show that the F147A/Y148A mutations reversed the effect of PI(4,5)P2 depletion on TRPC3 activity, with PI(4,5)P2 depletion increasing, the activity of TRPC3(F147A/Y148A) rather than decreasing activity as previously shown in Fig. 4. This raised the question of whether the F147A/Y148A mutations affect the activity and the PI(4,5)P2 dependence of TRPC3(I807S), the activity of which is independent of PI(4,5)P2 (Fig. 4). Fig. 9 J, i–iii, show that the F147A/Y148A mutation increased, rather than decreased the activity of TRPC3(I807S). Moreover, Fig. 9 K, i–iii, show that the dependence of TRPC3(I807S) on PI(4,5)P2 was restored with strong inhibition of TRPC3(I807S/F147A/Y148A) by PI(4,5)P2 depletion. Together, the findings in Fig. 9 indicate that the TRPC3 FFAT motif is functional and has an important role in the response of TRPC3 to PI(4,5)P2. In addition, the stimulatory and inhibitory effects of PI(4,5)P2 depletion on the TRPC3(F147A/Y148A) and the TRPC3(I807S/F147A/Y148A) mutants illustrate the multiple effects of PI(4,5)P2 on channel function.
Discussion
Both ion channels and transporters are affected and regulated by membrane lipids by virtue of their interaction with them. Lipids were shown to directly gate ion channels, modulate gating by other substrates, and affect their localization in specific membrane domains (Hilgemann, 2020; Thompson and Baenziger, 2020). Lipids can interact with channel pores and/or multiple cytoplasmic lipid binding sites (Bai et al., 2020; Duan et al., 2019; Li et al., 2019; Song et al., 2021; Thompson and Baenziger, 2020). Among the membrane lipids, prominent roles have been described for PI(4,5)P2 in the regulation of numerous ion channels, including many TRP channels (Duncan et al., 2020). Many of the functional studies focused on PI(4,5)P2 and other phosphoinositol lipids because of their role in cell signaling (Balla et al., 2020) and the discovery of their regulation of ion channels and transporters (Hilgemann, 2020). Recent availability of structural information for several channel types identified lipid binding sites, although in most cases, the sites were occupied with the lipids used during the structural studies, rather than by the physiological lipids (Duncan et al., 2020; Thompson and Baenziger, 2020).
TRP channels located at the plasma membrane and in intracellular organelles are regulated by phosphoinositides (Duncan et al., 2020; Hille et al., 2015). They can be regulated by direct interaction of PI(4,5)P2 with a channel lipid-binding site, as was observed in the structure of several TRPV, TRPM, TRPML, and TRPC channels (Duncan et al., 2020; Fine et al., 2020; Li et al., 2019; Wang et al., 2020). In the case of TRPC3/6/7, they are activated by DAG, a hydrolytic product of PI(4,5)P2 (Svobodova and Groschner, 2016; Svobodova et al., 2019; Wang et al., 2020). Another potential mode of regulation is by the localization of the TRP channels to lipid-specific membrane domains at MCS. How lipids regulate TRPC channels is not well understood. Lipid-binding sites were observed in the structure of TRPC4 (Duan et al., 2018; Vinayagam et al., 2018), TRPC5 (Duan et al., 2019; Song et al., 2021), and TRPC3 (Fan et al., 2018) with homologous structures present in TRPC6 (Bai et al., 2020; Tang et al., 2018). Two lipid-binding sites were identified in TRPC3, peripheral site 1, and the pore site 2 (Fan et al., 2018; see Fig. 5).
The key findings of the present work are summarized in the model of Fig. 10. In the resting state, the G protein–coupled receptors and TRPC channels are not in close contact and do not communicate efficiently. Receptor stimulation brings together the receptors and channels in close contact (increased FRET) in a PI(4,5)P2-rich domain within the ER/PM junctions to enhance communication between them and allow receptor-mediated regulation and several modes of regulation of the TRPC channels by PI(4,5)P2. PI(4,5)P2 is shown here to provide the DAG substrate that activates the channel, controls access of DAG to its site of action in the channel pore, and PI(4,5)P2 regulates channel selectivity and localization of the channels at the ER/PM junctions. The dominant tether forming the ER/PM junctions is STIM1, which is required for the roles of other junctional tethers, such as E-Syt1 and the VAPs.
Summary model for regulation of TRPC3 channel at membrane contact site. All structures shown are taken from the literature or were predicted by Robetta. The plasma membrane is shown in purple and the ER in green. The red circles at the plasma membrane stand for PI(4,5)P2. PMCA, plasma membrane Ca2+ ATPase pump; SERCA, sarco/endoplasmic reticulum Ca2+ ATPase pump. In the resting state the core Ca2+ signaling proteins are in a loose communication in their respective membranes and the ER and PM are in a distance. Upon cell stimulation, the core Ca2+ signaling proteins are assembled into complexes at the ER/PM junctions formed by STIM1 that increases their communication and mutual regulation.
Summary model for regulation of TRPC3 channel at membrane contact site. All structures shown are taken from the literature or were predicted by Robetta. The plasma membrane is shown in purple and the ER in green. The red circles at the plasma membrane stand for PI(4,5)P2. PMCA, plasma membrane Ca2+ ATPase pump; SERCA, sarco/endoplasmic reticulum Ca2+ ATPase pump. In the resting state the core Ca2+ signaling proteins are in a loose communication in their respective membranes and the ER and PM are in a distance. Upon cell stimulation, the core Ca2+ signaling proteins are assembled into complexes at the ER/PM junctions formed by STIM1 that increases their communication and mutual regulation.
Several of the present findings point to the importance of lipid-binding to site 2 at the channel pore. Mutation of the TRPC3 P loop E615 and K619 that contact the lipid (Fan et al., 2018) prominently inhibited channel activation both by receptor stimulation and by GSK (Figs. 5 and S3). Moreover, mutation of G652 in TMD 6 that contacts the lipid tail markedly enhanced activation by GSK (Lichtenegger et al., 2018; Svobodova et al., 2019). The DAG site is similar to other TRPC channels (Bai et al., 2020; Song et al., 2021), but it is not identical. For example, the N645A mutation in TRPC3 had no effect on channel function, while the equivalent mutation in TRPC6 N702A abolished DAG-activated Ca2+ influx (Bai et al., 2020), while DAG does not contact this residue in TRPC5 (Song et al., 2021).
We show here that the interaction of the substrate with TRPC3 pore lipid site 2 is modulated by several channel domains. Mutations in residues that form lipid site 1 affect the interaction of GSK with the channel pore. Mutations of I556 and N560 located at the S4–S5 linker at the cytosolic end of the pore either completely prevent TRPC3 activation by GSK or increase inward current density (Fig. S3). The S4–S5 linker contacts the TRP helix (Fan et al., 2018; Tang et al., 2018), and mutations of I556 and N560 may disrupt the position of the TRP helix, which has several roles in all TRP channel functions, including channel gating (Gees et al., 2012). Lipid interaction with site 2 is prominently affected by PI(4,5)P2 interaction with TRPC3. Depletion of PI(4,5)P2 had no effect on maximal current activated by GSK, but reduced the affinity for GSK about 15-fold (Figs. 4 K and 7 K). These effects were more prominent than the effect of PI(4,5)P2 depletion on the affinity for activation of TRPC3 by receptor stimulation, which was reduced by <2-fold (Fig. 4 F and 7 E). Since GSK can fully activate the channel in PI(4,5)P2-depleted cells and PI(4,5)P2 depletion does not affect selectivity of TRPC3 activated by GSK (Fig. 4 L), together the findings indicate interaction of PI(4,5)P2 with TRPC3 lipid-binding site controlling the interaction of the substrate with lipid 2 site. This is likely interaction of PI(4,5)P2 with the peripheral lipid site 1, as suggested by the finding that the W334A mutation completely restored the high affinity for GSK in PI(4,5)P2-depleted cells (Fig. 7 K and S4 I). Thus, lipid binding at site 1 has a prominent role in controlling the binding of the substrate (DAG/GSK) to the pore lipid site 2.
PI(4,5)P2 binding to the lipid site 1 has additional roles in TRPC3 activity. PI(4,5)P2 affected channel selectivity when activated by receptor stimulation (Fig. 4 C), but not when activated directly by GSK (Fig. 4 L). This is likely because when TRPC3 is directly activated by GSK it is not at the ER/PM junctions and the PI(4,5)P2-rich domain and thus is not accessed by PI(4,5)P2, which regulates channel selectivity. The exact nature of this site is not known with certainty. It may not be lipid-binding site 1 since mutations of residues that interact with lipid site 1, W334 and N560, do not change channel selectivity (Fig. 5 K). TRPC channels may have several additional lipid-interacting sites in the periphery of the transmembrane domain sector (Song et al., 2021), one of which may mediate regulation of channel selectivity by PI(4,5)P2.
Mutation of residues interacting with the lipid can completely inhibit (I556A) or increase (W334A) current density that could not be explained by altered total or surface TRPC3 expression (Fig. S2, J and K). The increased activity of TRPC3(W334A) was without any change in the affinity for receptor stimulation or lipid interaction with lipid site 2 (Fig. 6, B and I). The exact effect of PI(4,5)P2 is difficult to determine with certainty since PI(4,5)P2 has both stimulatory and inhibitory effects, as illustrated in Fig. 9, I–K. The simplest explanation for our findings with the W334A mutation is that it prevents PI(4,5)P2 binding to lipid site 1, preventing the inhibitory effect of PI(4,5)P2 on TRPC3 and increasing channel opening. This interpretation is supported by the finding that the W334A mutation increased current stimulated by GSK that was not affected by PI(4,5)P2 depletion (Fig. 7, I and J). Yet, TRPC3(W334A) still responded to PI(4,5)P2 depletion as shown by its reduced affinity for receptor stimulation (Fig. 7 E). This likely reflects a receptor-specific effect of PI(4,5)P2, a role of PI(4,5)P2 in ER/PM junction localization of TRPC3. Binding of PI(4,5)P2 to site 1 appears to also affect channel selectively, as suggested by the selective increases in inward (N560A) and decreases in outward (F563A) currents activated by receptor stimulation (Fig. 5, H–J) and the change in reversal potential of TRPC3 and TRPC3(L241S; Fig. 4 C). All residues—L241, W334, and N560—are in close contact with the TRP helix and their mutation may affect PI(4,5)P2 binding to lipid site 1, thus affecting the shape of the pore and ion access to the external or cytoplasmic sides of the pore.
A role for PI(4,5)P2 in the regulation of any TRP channels that has not yet been examined is regulation by localization at the ER/PM junctions. MCS are zones for assembly of signaling pathways, including Ca2+ and cAMP signaling pathways (Balla et al., 2020; Muallem et al., 2017). Recently, we showed that the tether ANO8 assembles the entire components of the Ca2+ signaling complex at the ER/PM junctions (Jha et al., 2019). In addition, assembly of STIM1–Orai1 complexes at the ER/PM junctions required the tether E-Syt1 and the tethering function of STIM1 that both interact with PM PI(4,5)P2 (Maléth et al., 2014). These tethers and mutations of TRPC3 that affect its interaction with STIM1 (Lee et al., 2014) were used to study the role of localization at the ER/PM junctions in TRPC3 regulation and activity. The present findings show that disruption of the junctions by deletion of STIM1 and the knockdown of E-Syt1 and ANO8 primarily and markedly reduced the apparent affinity for receptor-mediated activation of TRPC3, while overexpression of E-Syt1 and ANO8 increased activation of TRPC3 by receptor stimulation. Targeting of TRPC3 to the junctions is likely mediated by its FFAT site that binds to VAPB since changing expression of VAP proteins and disruption of the TRPC3 FFAT site affected the current and the apparent affinity for receptor stimulation (Fig. 9).
The significance of localization at the ER/PM junctions was further supported using the TRPC3 mutants that affect its interaction with STIM1. The TRPC3(L241S) mutation that increases TRPC3 interaction with STIM1 (Lee et al., 2014) increased the affinity while the TRPC3(I807S) mutation that eliminates TRPC3 interaction with STIM1 (Lee et al., 2014) decreased the affinity for receptor stimulation of TRPC3 (Figs. 1 and 3). The important findings that the TRPC3(L241S) and the TRPC3(I807S) mutants had no effect on the affinity for GSK (Fig. 2) indicate that localization at the ER/PM junctions controls the communication between the receptor complex and TRPC3 rather than access and interaction of the lipid with the channel pore lipid site 1. In this respect, PI(4,5)P2 not only controls lipid interaction with the pore as discussed above, but also affects localization of TRPC3 at the ER/PM junctions because of the role of PI(4,5)P2 in integrity of the junction. Indeed, PI(4,5)P2 controls both the affinity for receptor stimulation and the affinity for GSK (Fig. 4). How efficient communication between TRPC3 and the receptor complex within the ER/PM junctions is achieved is not known at this time and requires further studies.
The present findings with TRPC3 are likely relevant to other TRP channels. The alignment of all TRPC channels in Fig. S6 shows that all elements examined in the present studies are fully conserved in TRPC3/6/7 (highlighted in yellow). Several of the residues that were mutated in TRPC3 and affect lipid binding or regulation by lipids (Table S1) are conserved in other TRPC channels (highlighted in red). Indeed, mutations in these regions in TRPC5 (Song et al., 2021) and TRPC6 (Bai et al., 2020) affected Ca2+ influx. Regulation by PI(4,5)P2 was reported for many TRP channels and is always attributed to direct regulation by PI(4,5)P2 (Duncan et al., 2020). However, part of the regulation can also be due to localization in ER/PM junctions that depends on PI(4,5)P2, as shown here for TRPC3. Clarification of such roles should improve understanding of the regulation of TRP channels by lipids and by localization at MCS.
Alignment of TRPC channels. All residues in red were mutated in TRPC3. The FFAT motif is shown in green. The conserved regions in TRPC3/6/7 in which the residues mutated in TRPC3 are highlighted in yellow.
Alignment of TRPC channels. All residues in red were mutated in TRPC3. The FFAT motif is shown in green. The conserved regions in TRPC3/6/7 in which the residues mutated in TRPC3 are highlighted in yellow.
Materials and methods
Cloning, cell culture, transfection, and siRNA
TRPC3-YFP (Lee et al., 2014) was used as a template for generating TRPC3 mutants using the QuikChange Lightning Site-Directed Mutagenesis Kit (210518; Agilent Technologies). The primers used are shown in Table S1. STIM1, HEK293T, and HEK293T-STIM1−/− cells (generously provided by Dr. Trebak; Emrich et al., 2019), were grown at 37°C with 5% CO2 in a DMEM media supplemented with 10% FBS. Cells were plated in 6-well plates and grown to 50–60% confluency and were transfected with 0.2 μg TRPC3 or mutants and 0.2 μg of the M3R cDNAs 18–24 h before recording. Transfection was with lipofectamine 2000 (catalog 11668019; Life technologies) according to the manufacturer instructions. For PI(4,5)P2 depletion, cells were also transfected with the FRB/FKBP-PI5Ptase system (Varnai et al., 2006) and treated with 0.2 µM rapamycin for 2 min to deplete PI(4,5)P2 prior to start of current measurement. For knockdown of proteins, cells were treated with 40 nM of scrambled RNA as a control or with siRNA targeted against E-Syt1, E-Syt2, E-Syt3 (Maléth et al., 2014), ANO8 (Jha et al., 2019), and VAPA + VAPB as detailed before (Jha et al., 2019; Maléth et al., 2014). After 48 h, the cells were transfected with the desired proteins and were used for current recording 24 h later (total of 72 h treatment with siRNA). The sequences for the E-Syts and ANO8 siRNAs are the same as in (Jha et al., 2019; Maléth et al., 2014). The siVAPA was sequence 2 of ID: hs.Ri.VAPA.13 from IDT and for siVAPB it was sequence 3 of hs.Ri.VAPB.13 from IDT, both from TriFECTa DsiRNA Kit design.
Current recording
TRPC3 current was measured as described before (Lee et al., 2014) 24 h after transfection. Transfected cells were identified by YFP and/or mCherry fluorescence. The pipette solution contained (in mM): 140 CsCl, 2 MgCl2, 1 ATP, 5 EGTA, 1.5 CaCl2 (0.2 free Ca2+), and 10 Hepes (pH 7.2 with CsOH). The bath solution contained (in mM): 140 NaCl, 5 KCl, 0.5 EGTA, 1 MgCl2, and 10 Hepes (pH 7.4 with NaOH). Current was stimulated with the indicated concentrations of the M3R-ligand carbachol or by the direct TRPC3 activator GSK (SML2323; SigmaAldrich). GSK was dissolved in DMSO to prepare a stock solution of 10 mM and was diluted in the bath solution to obtain the desired final concentration. The current was recorded by 400-ms rapid alterations of membrane potential from −100 to +100 mV from a holding potential of 0 mV. The current recorded at −100 and +100 mV was used to calculate current density as pA/pF, and the current recorded in multiple experiments was used to obtain the mean ± SEM and calculate significance. Cells were patched and the whole-cell configuration was obtained in a bath solution containing 2 mM CaCl2 and no EGTA. After 10 sweeps spaced 4 s apart, the solution was changed to the Ca2+-free bath solution, and recording continued for 10 more sweeps. The cells were then stimulated with carbachol or GSK and recording continued for 20 sweeps. Finally, the cells were perfused with Na+-free NMDG solution (bath solution in which NaCl and KCl were replaced with NMDG-Cl) to determine the leak current. For PI(4,5)P2 depletion, 0.2 µM rapamycin was included in the Ca2+-free bath solution and after 2 min (30 sweeps) the cells were stimulated. Longer incubation with rapamycin resulted in increased leak current and thus treatment was restricted to 2 min prior to cell stimulation. Current traces were stored in Origin software (Originlab 2018).
Total and surface expression of TRPC3
As needed, cells were treated with the desired siRNA for 48 h prior to the procedure described below. HEK293T cells grown to 70–80% confluence were transfected with plasmids as specified using Lipofectamine 2000. The growth media was replaced with Opti-MEM before transfection. About 6 h after transfection, the cells were released and replated in poly-L-lysine coated 6-well plates and maintained in DMEM. After 24 h, cells were treated with vehicle or stimulated with 100 µM carbachol for 2 min. The cells were washed with cold PBS and incubated with 1 ml of cold biotinylation solution (0.5 mg/mLSulfo-NHS-SS or -LC biotin in cold PBS) on ice for 30 min with gentle swirling. Biotinylation was terminated by removal of the biotinylation solution and quenching with glycine solution containing 50 mM glycine in cold PBS. After an additional wash with the glycine solution the cells were washed with cold PBS and lysed in 200–600 μl of lysis buffer (1×PBS; 10 mM Na pyrophosphate, 50 mM NaF, 1 mM NaVO3, 5 mM EDTA, and protease inhibitor cocktail tablet [Roche 11836170001]) containing 1% TX-100 by incubation on ice for 15 min. The lysate was transferred to 1.5-ml Eppendorf tubes and sonicated using 2 × 3 pulses (GEX 130 PB at 20% amplitude). The lysates were collected by 20-min centrifugation at 4°C at a speed of 13,000 rpm and stored at −80°C until use (1–2 d). Between 5 and 15 µg proteins were used for input measurement, and between 100 and 200 μg proteins were used to determine surface expression. The biotinylated proteins were isolated using Biosystem: Magnetic beads Neutravidin as detailed by the manufacturer (# 29204; Thermo Fisher Scientific). The amount of protein was analyzed by Western Blots. TRPC3-eGFP was detected with anti-GFP antibodies (A11122; Invitrogen), myc-STIM1 with anti-myc antibodies (2276S; Cell Signalling Technologies), and VAPA and VAPB with anti-RFP antibodies (200-301-379; Rockland). Non-biotinylated Co-IPs were performed using GE HealthCare Protein G Sepharose 4 Fast Flow 17-0618-01. ImageJ was used to quantify bend intensity of all blots. All values are normalized to the surface/input or IP/input of TRPC3 in unstimulated cells.
TIRF microscopy
TIRF imaging was as we described before (Jha et al., 2019). Images were recorded at 37°C with Nikon NIS-Elements paired with the Nikon Elipse Ti with PFS (Perfect Focus System) autofocus capabilities, Nikon N-Storm, Andor iXon Ultra Camera with EMCCD Sensor, D-Eclipse C1, and 60× TIRF objective lens (Nikon), 1.45 Na+ Oil immersion, infinity/0.10–0.22 DIC H. The size and intensity of the puncta were analyzed by ImageJ in imported images recorded by the Nikon NIS software. Background was subtracted from the first image in a sequence and maintained through the time course. The area of the cell from which the puncta were analyzed was determined by the NIS-elements software, and number and intensity were normalized using this area. The results are shown as mean ± SEM, and statistical analysis and figures were made with GraphPad Prism 9.
FRET measurements
Wild-type or STIM1−/− HEK293 cells were plated on glass-bottom dishes (MatTek Corporation) and transfected with TRPC3-YFP, TRPC3(L241S)-YFP, TRPC3(I807S)-YFP (Lee et al., 2014), STIM1-CFP (Maléth et al., 2014), and untagged E-Syt1 using Lipofectamine 2000 (Invitrogen) and incubated for 16–24 h. FRET measurements were with MetaMorph paired with the Olympus IX81 Microscope, Olympus IX2-UCB, and Tripp-Lite Line Conditioner LC2400 connected to the microscope. Elimination to identify transfected cells (Epifluorescence) and FRET measurements were with Air-Therm ATX-H, CoolLED pE-300 LED Illuminator, and ASI MS-2000, Voltran Laser Technology, Inc. Diode Module Stradus Control Box with CDRH ON/OFF key switch and with 405, 445, 488, 515, 561, and 639 nm lasers attached to a Triggerscope V-3B and Laser Aperture. Confocal images were recorded with Yokogawa CSU-X1 (confocal scanner unit) with Filter Wheel Control and shutter, and Photometrics Evolve Delta camera and Olympus UPlanSApo, 60× or 100×, 1.35 NA, infinity/0.13–0.19/FN22 objectives. Cells were perfused using perfusion System from ALA Scientific Instruments VC3-8xP Valve Commander. Data acquisition and processing were with MetaMorph and the intensity values were exported to Excel. The time course intensities were normalized to the 1 min value for each region of interest or cell as appropriate. Traces and intensities were exported to GraphPad Prism 9 for statistical analysis and presentation.
Statistics
All experiments were repeated at least three times and all data were expressed as mean ± SEM. The individual dots in the columns indicate separate cells analyzed. Statistical significance was determined by means of Student’s t test or ANOVA, as appropriate with GraphPad Prism 9 software. P values are listed in the figures, and P values smaller than 0.05 are considered statistically significant.
Online supplemental material
Fig. S1 shows effect of STIM1 on TRPC3 current, ER/PM junctions, and Ca2+ influx, and current by truncated TRPC3. Fig. S2 shows the role of ER/PM junctions in regulation of TRPC3 by receptor stimulation. Fig. S3 shows the effect of E-Syt2, E-Syt3, STIM1 deletion, TRPC3 pore, and lipid binding mutants on TRPC3 activity stimulated by carbachol of GSK. Fig. S4 shows the effect of PI(4,5)P2 depletion on carbachol and GSK-stimulated inward current. Fig. S5 shows that TRPC3 730RRRR733 is not PI(4,5)P2 binding motif, and VAP siRNA and VAPB-TRPC3 interaction by FRET. Fig. S6 shows alignment of TRPC channels. Table S1 lists localization, predicted role, and effect of TRPC3 mutations examined in this study and the G652A examined in Lichtenegger et al., (2018).
Acknowledgments
We thank Dr. Mohamed Trebak (University of Pittsburgh, Pittsburgh, PA) for the STIM1−/− cells, Dr. Jen Liou (University of Texas Southwestern Medical Center, Dallas, TX) for the MAPPER plasmid, and Dr. Pietro De Camilli (Yale School of Medicine, New Haven, CT) for the extended synaptotagmin plasmids.
This work was funded by intramural National Institutes of Health grant NIH/NIDCR DE000735-10.
The authors declare no competing financial interests.
Author contributions: H. Liu, W.-Y. Lin, S.R. Leibow, A.J. Morateck, and M. Ahuja performed experiments; M. Ahuja and S. Muallem directed the studies; S. Muallem drafted the manuscript with input from all authors.
References
Author notes
H. Liu and W.-Y. Lin are co-first authors.