We report that lysosomal damage is a hitherto unknown inducer of stress granule (SG) formation and that the process termed membrane atg8ylation coordinates SG formation with mTOR inactivation during lysosomal stress. SGs were induced by lysosome-damaging agents including SARS-CoV-2ORF3a, Mycobacterium tuberculosis, and proteopathic tau. During damage, mammalian ATG8s directly interacted with the core SG proteins NUFIP2 and G3BP1. Atg8ylation was needed for their recruitment to damaged lysosomes independently of SG condensates whereupon NUFIP2 contributed to mTOR inactivation via the Ragulator–RagA/B complex. Thus, cells employ membrane atg8ylation to control and coordinate SG and mTOR responses to lysosomal damage.
Introduction
The mammalian autophagy-related (ATG) factors participate in a number of processes including canonical (Morishita and Mizushima, 2019) and noncanonical autophagy (Galluzzi and Green, 2019) with implications in disease and physiology (Levine and Kroemer, 2019). These processes intersect with metabolic regulators, with signals transduced through phosphorylation by mTOR to the ATG apparatus through a module consisting of FIP200-ULK1-ATG13-ATG101 (Jia et al., 2018; Kim et al., 2011). ATGs participate in non-autophagic processes, e.g., ULK1 (mammalian ortholog of yeast Atg1) in glycolysis (Li et al., 2016) and disassembly of stress granules (SGs; Wang et al., 2019). ATGs can play unique roles such as the mammalian ATG8 proteins (mATG8s; Gu et al., 2019; Kumar et al., 2018), which can act even upstream of the lysosomally positioned regulators such as mTOR or TFEB (Kumar et al., 2020; Nakamura et al., 2020). These diverse responses include mATG8s’ conjugation to various stressed or remodeling membranes through mATG8 lipidation or protein modifications (Carosi et al., 2021), collectively termed atg8ylation (Deretic and Lazarou, 2022).
Lysosomal damage elicits mobilization of ESCRT membrane repair systems (Skowyra et al., 2018), mTOR inactivation (Jia et al., 2018), the translocation of TFEB from lysosomes to the nucleus (Chauhan et al., 2016), ubiquitination response (Papadopoulos et al., 2017), AMPK activation (Jia et al., 2020a), lysophagy (Maejima et al., 2013), and lipid changes (Ellison et al., 2020). Since inactivation of mTOR (Shin and Zoncu, 2020) impacts multiple processes such as autophagy and protein translation, it is of interest to consider global changes in transcription and translation during lysosomal damage.
SG formation is a part of global modulation of protein translation (Ivanov et al., 2019; Yang et al., 2020). SGs are cytoplasmic, membraneless liquid–liquid phase separated biomolecular condensates (Alberti et al., 2019) containing ribonucleoprotein particles, translational factors, the 40S ribosomal subunit (Kedersha et al., 2002), and a multitude of other proteins, e.g., G3BP1, TIA1, and NUFIP2 (Ivanov et al., 2019; Yang et al., 2020). Canonical SG formation depends on phosphorylation of eukaryotic translation initiation factor 2α (eIF2α), which blocks the assembly of productive translation preinitiation complexes (Kedersha et al., 1999). Heat shock, oxidative stress, hypoxia, and viral infections are triggers of SG formation and translational arrest (Anderson and Kedersha, 2002). Mammalian eIF2α is phosphorylated by four upstream kinases transducing stress, including PKR (Srivastava et al., 1998), PERK (Patil and Walter, 2001), GCN2 (Kimball, 2001), and HRI (McEwen et al., 2005).
Here, we show that lysosomal damage is a previously unrecognized stressor eliciting canonical SG formation and translation changes. We report that atg8ylation (Deretic and Lazarou, 2022), a process that modifies stressed or remodeling membranes by lipid (Kumar et al., 2021b) or protein (Carosi et al., 2021) conjugation, competes with SG formation. Based on proteomic approaches, we report that individual SG proteins, NUFIP2 and G3BP1, which interact with mATG8s, are recruited to lysosomes and inactivate mTOR through the Ragulator–Rag system. These processes are elicited by lysosomal damaging agents including proteopathic tau, Mycobacterium tuberculosis (Mtb), and SARS-CoV-2ORF3a.
Results
Lysosomal proteome changes during lysosomal damage
To complement our studies of cellular homeostatic responses to lysosomal damage carried out by proximity biotinylation proteomics (Jia et al., 2018; Jia et al., 2020a; Jia et al., 2020b), here we carried out whole organelle proteomic analyses of normative and damaged lysosomes purified by LysoIP (Abu-Remaileh et al., 2017; Jia et al., 2020b; Table S1, Tabs 1 and 2). We chose short pulse for lysosomal damage to capture early events minimizing more advanced degradative processes such as autophagy and other late-stage components of the MERiT response (Jia et al., 2020c). Cells were treated for 30 min with Leu-Leu-O-Me (LLOMe), which is a substrate for reverse peptidase reaction by cathepsin C, resulting in growing polymers in the lysosomal lumen causing membrane damage and permeabilization (Thiele and Lipsky, 1990). Quantitative data-independent acquisition (DIA), mass spectrometry (MS), and proteomic analyses were carried out (Fig. 1 A and Table S1, Tabs 1 and 2 [i]) of damaged vs. undamaged lysosomes in HEK293T cells stably expressing TMEM192-3xHA following a well-established procedure for lysosomal purification (Jia et al., 2020a; Fig. 1 A and Table S1, Tabs 1 and 2). The mass spectrometry data confirmed our prior observations (Jia et al., 2018) that mTOR and Raptor dissociate from lysosomes upon damage (Fig. 1 A) whereas STRING protein interaction network functional analysis (Szklarczyk et al., 2021) revealed enrichment of components of a number of biological processes including several previously not associated with lysosomal damage (Table S1, Tab 2 [ii] and [iii]).
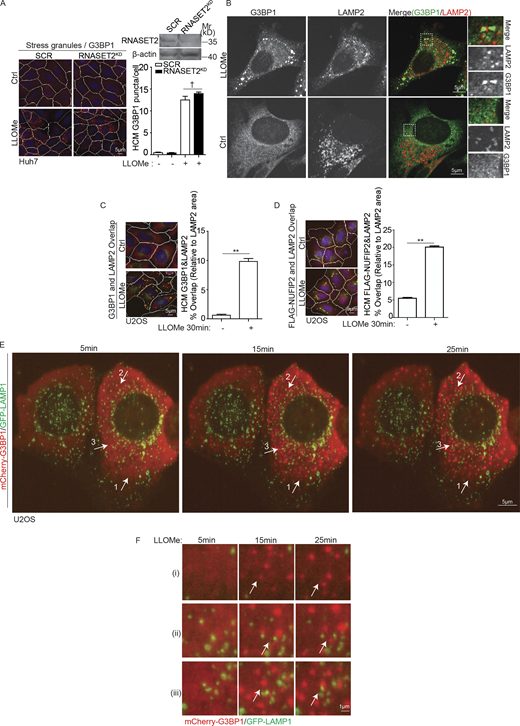
Lysosomal damage induces SG formation. (A) DIA LC/MS/MS quantitative analysis of proteins associated with lysosomes purified by LysoIP (anti-HA IP) from HEK293T cells expressing TMEM192-3xHA untreated or treated with 1 mM LLOMe for 30 min. Three groups of proteins are denoted: ESCRT components, green; autophagy factors, blue; SG components, purple. Scatter (volcano) plot shows log2 fold changes and −Log10 P values; n = 3 (see Table S1, Tab 1). Dashed line, significance cut-off (P < 0.05). (B) Protease accessibility analysis of proteins associated with purified lysosomes (LysoIP). Huh7 cells were treated with 2 mM LLOMe. LysoIP preparations (treated or not with detergent Triton X-100) were digested with 30 µg/ml proteinase K for 30 min and analyzed by immunoblotting. (C) Quantification by HCM of G3BP1 puncta. U2OS cells were treated with EBSS, 4 mM LOMe, 2 mM LLOMe, 200 µM GPN, or 400 µg/ml silica for 30 min. White masks, algorithm-defined cell boundaries (primary objects); red masks, computer-identified G3BP1 puncta (target objects). (D) Fluorescence confocal microscopy imaging of G3BP1. U2OS cells were treated with 2 mM LLOMe for 30 min and immunostained for endogenous G3BP1. Scale bar, 5 µm. (E) Quantification by HCM of G3BP1 puncta in BMM cells treated with 2 mM LLOMe or 100 µM NaAsO2 for 2 h. Green masks, computer-identified G3BP1 puncta. (F) Quantification by HCM of G3BP1 puncta in U2OS cells treated with LLOMe at indicated doses or 100 µM NaAsO2 in the presence or absence of 10 µg/ml cycloheximide (CHX) for 30 min. HCM images in Fig. S1 G. (G) Analysis of proteins associated with purified lysosomes (LysoIP; TMEM192-3xHA) from HEK293T cells treated with 2 mM LLOMe in the presence or absence of 10 µg/ml CHX for 30 min. TMEM192-2xFLAG, control. (H) Immunoblot analysis of eIF2α (S51) phosphorylation in BMM cells treated with 2 mM LLOMe or 100 µM NaAsO2 for 2 h; eIF2α p-S51 quantification, n = 3. (I) Confocal microscopy analysis of G3BP1 (Alexa Fluor 488) and polyA RNA (Cy3-oligo[dT]) by FISH in U2OS cells treated with 2 mM LLOMe for 30 min. Scale bar, 5 µm. (J) HCM analysis of protein synthesis using Click-iT Plus OPP Alexa Fluor 488 Protein Synthesis Assay (Thermo Fisher Scientific) in U2OS cells treated with LLOMe at indicated doses or 100 µM NaAsO2 or 10 µg/ml CHX for 30 min. (K) Immunoblot analysis of ATF4 and phosphorylation of 4EBP1 (S65) and eIF2α (S51) in U2OS cells treated with 2 mM LLOMe for indicated time points; quantification of ATF4 and phosphorylation of 4EBP1 (S65) and eIF2α (S51), n = 3. Ctrl, control (untreated cells). Data, means ± SEM; HCM: n ≥ 3 (each experiment: 500 valid primary objects/cells per well, ≥5 wells/sample). †, P ≥ 0.05 (not significant); *, P < 0.05; **, P < 0.01, ANOVA. See also Fig. S1. Source data are available for this figure: SourceData F1.
Lysosomal damage induces SG formation. (A) DIA LC/MS/MS quantitative analysis of proteins associated with lysosomes purified by LysoIP (anti-HA IP) from HEK293T cells expressing TMEM192-3xHA untreated or treated with 1 mM LLOMe for 30 min. Three groups of proteins are denoted: ESCRT components, green; autophagy factors, blue; SG components, purple. Scatter (volcano) plot shows log2 fold changes and −Log10 P values; n = 3 (see Table S1, Tab 1). Dashed line, significance cut-off (P < 0.05). (B) Protease accessibility analysis of proteins associated with purified lysosomes (LysoIP). Huh7 cells were treated with 2 mM LLOMe. LysoIP preparations (treated or not with detergent Triton X-100) were digested with 30 µg/ml proteinase K for 30 min and analyzed by immunoblotting. (C) Quantification by HCM of G3BP1 puncta. U2OS cells were treated with EBSS, 4 mM LOMe, 2 mM LLOMe, 200 µM GPN, or 400 µg/ml silica for 30 min. White masks, algorithm-defined cell boundaries (primary objects); red masks, computer-identified G3BP1 puncta (target objects). (D) Fluorescence confocal microscopy imaging of G3BP1. U2OS cells were treated with 2 mM LLOMe for 30 min and immunostained for endogenous G3BP1. Scale bar, 5 µm. (E) Quantification by HCM of G3BP1 puncta in BMM cells treated with 2 mM LLOMe or 100 µM NaAsO2 for 2 h. Green masks, computer-identified G3BP1 puncta. (F) Quantification by HCM of G3BP1 puncta in U2OS cells treated with LLOMe at indicated doses or 100 µM NaAsO2 in the presence or absence of 10 µg/ml cycloheximide (CHX) for 30 min. HCM images in Fig. S1 G. (G) Analysis of proteins associated with purified lysosomes (LysoIP; TMEM192-3xHA) from HEK293T cells treated with 2 mM LLOMe in the presence or absence of 10 µg/ml CHX for 30 min. TMEM192-2xFLAG, control. (H) Immunoblot analysis of eIF2α (S51) phosphorylation in BMM cells treated with 2 mM LLOMe or 100 µM NaAsO2 for 2 h; eIF2α p-S51 quantification, n = 3. (I) Confocal microscopy analysis of G3BP1 (Alexa Fluor 488) and polyA RNA (Cy3-oligo[dT]) by FISH in U2OS cells treated with 2 mM LLOMe for 30 min. Scale bar, 5 µm. (J) HCM analysis of protein synthesis using Click-iT Plus OPP Alexa Fluor 488 Protein Synthesis Assay (Thermo Fisher Scientific) in U2OS cells treated with LLOMe at indicated doses or 100 µM NaAsO2 or 10 µg/ml CHX for 30 min. (K) Immunoblot analysis of ATF4 and phosphorylation of 4EBP1 (S65) and eIF2α (S51) in U2OS cells treated with 2 mM LLOMe for indicated time points; quantification of ATF4 and phosphorylation of 4EBP1 (S65) and eIF2α (S51), n = 3. Ctrl, control (untreated cells). Data, means ± SEM; HCM: n ≥ 3 (each experiment: 500 valid primary objects/cells per well, ≥5 wells/sample). †, P ≥ 0.05 (not significant); *, P < 0.05; **, P < 0.01, ANOVA. See also Fig. S1. Source data are available for this figure: SourceData F1.
As an independent control and measure of early cellular response to lysosomal injury, RNA sequencing (RNAseq) analysis was carried out (Fig. 2 A and Table S1, Tab 3). RNAseq data revealed that during early lysosomal damage, several genes were induced including DUSP1 (Fig. 2 A). DUSP1 is a phosphatase inhibiting ERK2 activation (Kirk et al., 2020; Sun et al., 1993), whereas ERK2 is an upstream kinase for TFEB (Napolitano and Ballabio, 2016; Settembre et al., 2011). We tested DUSP1 protein levels and the status of ERK2 and TFEB and found: (i) that DUSP1 was increased (Fig. 2 B); (ii) that ERK2 was dephosphorylated (Fig. 2 B); (iii) that TFEB was dephosphorylated at its Ser142 residue, a known site for phosphorylation by ERK2 (Napolitano and Ballabio, 2016; Settembre et al., 2011; Fig. 2 C); and (iv) that this depended on DUSP1 (Fig. 2 D). Finally, nuclear translocation of TFEB and dephosphorylation at Ser142 in response to lysosomal damage (Chauhan et al., 2016; Settembre et al., 2012) was observed at early time point and was equal in magnitude to the one caused by ERK2 inhibitor AZD6244 (Fig. 2, E and F). We conclude that early lysosomal damage at the chosen time point elicits a relevant cellular response consistent with prior observations (Jia et al., 2020b; Nakamura et al., 2020).
Cellular transcriptional response during lysosomal damage. (A) RNAseq analysis of the change in gene expression (HEK293T cells) in response to treatment with 1 mM LLOMe for 30 min. Scatter (volcano) plot shows log2 fold change and −Log10 P value for the genes identified in three independent experiments (see Table S1, Tab 3). Red dots indicate the genes downregulated; green dots indicate the genes upregulated. Dashed line, significance cut-off (P < 0.05). (B) Immunoblot analysis of DUSP1 expression level and ERK2 (T185/187) phosphorylation in HEK293T cells treated with 1 mM LLOMe for 30 min. (C) Immunoblot analysis of TFEB (S142) phosphorylation in U2OS cells treated with 2 mM LLOMe for 30 min. (D) Immunoblot analysis of ERK2 (T185/187) and TFEB (S142) phosphorylation in Huh7 cells transfected with scrambled siRNA as control (SCR) or DUSP1 siRNA treated with 2 mM LLOMe for 30 min. (E) Quantification by HCM of TFEB nuclear translocation in Huh7 cells treated with or without 530 nM ERK2 inhibitor AZD6244 for 2 h followed by 2 mM LLOMe for 30 min. Blue: nuclei, Hoechst 33342. Red: anti-TFEB antibody, Alexa Fluor 568. White masks, computer-algorithm-defined cell boundaries. Pink masks, computer-identified nuclear TFEB based on the average intensity of Alexa Fluor 568 fluorescence. (F) Immunoblot analysis of ERK2 (T185/187) and TFEB (S142) phosphorylation in Huh7 cells treated with or without 530 nM ERK2 inhibitor AZD6244 for 2 h followed by 2 mM LLOMe for 30 min. Ctrl, control (untreated cells). Data, means ± SEM; HCM: n ≥ 3 (each experiment: 500 valid primary objects/cells per well, ≥5 wells/sample). **, P < 0.01, ANOVA. Source data are available for this figure: SourceData F2.
Cellular transcriptional response during lysosomal damage. (A) RNAseq analysis of the change in gene expression (HEK293T cells) in response to treatment with 1 mM LLOMe for 30 min. Scatter (volcano) plot shows log2 fold change and −Log10 P value for the genes identified in three independent experiments (see Table S1, Tab 3). Red dots indicate the genes downregulated; green dots indicate the genes upregulated. Dashed line, significance cut-off (P < 0.05). (B) Immunoblot analysis of DUSP1 expression level and ERK2 (T185/187) phosphorylation in HEK293T cells treated with 1 mM LLOMe for 30 min. (C) Immunoblot analysis of TFEB (S142) phosphorylation in U2OS cells treated with 2 mM LLOMe for 30 min. (D) Immunoblot analysis of ERK2 (T185/187) and TFEB (S142) phosphorylation in Huh7 cells transfected with scrambled siRNA as control (SCR) or DUSP1 siRNA treated with 2 mM LLOMe for 30 min. (E) Quantification by HCM of TFEB nuclear translocation in Huh7 cells treated with or without 530 nM ERK2 inhibitor AZD6244 for 2 h followed by 2 mM LLOMe for 30 min. Blue: nuclei, Hoechst 33342. Red: anti-TFEB antibody, Alexa Fluor 568. White masks, computer-algorithm-defined cell boundaries. Pink masks, computer-identified nuclear TFEB based on the average intensity of Alexa Fluor 568 fluorescence. (F) Immunoblot analysis of ERK2 (T185/187) and TFEB (S142) phosphorylation in Huh7 cells treated with or without 530 nM ERK2 inhibitor AZD6244 for 2 h followed by 2 mM LLOMe for 30 min. Ctrl, control (untreated cells). Data, means ± SEM; HCM: n ≥ 3 (each experiment: 500 valid primary objects/cells per well, ≥5 wells/sample). **, P < 0.01, ANOVA. Source data are available for this figure: SourceData F2.
An abundance of ESCRT proteins was detected including ALIX (PDCD6IP) and TSG101, shown to contribute to lysosomal damage repair (Jia et al., 2020b; Radulovic et al., 2018; Skowyra et al., 2018) and all ESCRT-III components (Fig. 1 A, green; Table S1, Tab 4). Another category of proteins detected in DIA proteomic analysis of damaged lysosomes were autophagy-associated components (Fig. 1 A, blue; Table S1, Tab 5), with increase in ATG9A, MAP1LC3B, GABARAP, GABARAPL2, and ATG16L1. Thus, the global proteomic analysis was consistent with the ESCRT components being dynamically recruited and participating in repair of damaged lysosomes (Jia et al., 2020b; Skowyra et al., 2018). The evidence of autophagy factors gathering at the damaged lysosomes (Table S1, Tab 5) was consistent with prior studies (Eapen et al., 2021; Jia et al., 2020b; Maejima et al., 2013).
Proteomics of damaged lysosomes reveals connections to SG components
Our proteomic analyses of purified damaged lysosomes revealed abundance of proteins best known for their presence in SGs (Fig. 1 A, purple; Table S1, Tab 6). SGs are canonically induced in response to stressors such as heat shock (Nover et al., 1983), oxidative stress (Kedersha et al., 1999), and viral infection (Srivastava et al., 1998; Williams, 2001); however, lysosomal damage has hitherto not been reported as an inducer of SGs. SG composition is complex and depending upon conditions and complementary genomic vs. proteomic approaches, can include 274–411 proteins (Jain et al., 2016; Yang et al., 2020). These sets of proteins include the proposed core of 36 SG proteins (Yang et al., 2020). Our LysoIP proteomic analysis includes 32 out of the 36 core proteins (Table S1, Tab 6A). Of these, 20 showed statistically significant increase by quantitative DIA analysis (Fig. 1 A and Table S1, Tab 6A). Comparing our LysoIP proteomic data with other summaries of proteins associated with SGs (Ivanov et al., 2019), we detected 13 additional exclusive SG proteins and 10 shared between SGs and P-bodies (Table S1, Tab 6B). Of these, 15 showed statistically significant increase by quantitative proteomics of damaged lysosomes (Fig. 1 A and Table S1, Tab 6B), for a total of 55 SG proteins in LysoIP MS with 27 of those showing increased association with damaged lysosomes. SGs include stalled preinitiation complexes with 40S ribosomal subunit (Ivanov et al., 2019; Riggs et al., 2020). We detected 30 out of 33 human 40S proteins (Nakao et al., 2004) in our proteomic dataset, with 10 of those showing statistically significant increase in association with damaged lysosomes (Fig. 1 A and Table S1, Tab 6C). Thus, our quantitative proteomics analysis detected increased association of SG proteins with damaged lysosomes, including the conventional marker proteins for SGs, G3BP1, and TIA1 (Fig. 1 A; Gilks et al., 2004; Kedersha et al., 2005). Another more recently widely accepted marker of SGs (Yang et al., 2020; Youn et al., 2018), NUFIP2, was prominent in our LysoIP MS, and showed one of the highest enrichments upon lysosomal damage (Fig. 1 A). By LysoIP immunoblotting, we confirmed that NUFIP2, G3BP1, and TIA1 are enriched on damaged lysosomes but not on lysosomes purified from cells treated with arsenite, a conventional inducer of SG formation (Jain et al., 2016; Fig. S1 A). We next tested whether SG proteins associated with lysosomes are present on the surface or within the lysosomal lumen. NUFIP2 and G3BP1, like the regulator of mTOR LAMTOR1, were accessible to and degraded by proteinase K even without detergent treatment of LysoIP preparations (Fig. 1 B). This was in contrast to LAMP2, which is mostly lumenal with only its short C-terminal domain facing the cytosol (Fig. 1 B). Thus, NUFIP2 and G3BP1 were on the surface and not sequestered within the lumen of the lysosomes.
SG formation during lysosomal damage. (A) Immunoblot analysis of proteins associated with purified lysosomes (LysoIP; anti-HA immunoprecipitation TMEM1923xHA) from HEK293T cells treated with 2 mM LLOMe or 100 µM NaAsO2 for 30 min. TMEM1922xFLAG, control. (B) Quantification by HCM of DCP1a and G3BP1 puncta in U2OS cells treated with 2 mM LLOMe for 30 min. PB, P-body. (C) Quantification by HCM of G3BP1 puncta in Huh7 cells treated with 2 mM LLOMe for 30 min. White masks, algorithm-defined cell boundaries (primary objects); Green masks, computer-identified G3BP1 puncta (target objects). (D) Quantification by HCM of Gal3 puncta in BMM cells treated with 2 mM LLOMe or 100 µM NaAsO2 for 2 h. Red masks, computer-identified galectin-3 puncta. (E) Quantification by HCM of TIA1 puncta in U2OS cells treated with 2 mM LLOMe for 30 min. Red masks, computer-identified TIA1 puncta. (F) Quantification by HCM of TIA1 puncta in HeLa cells treated with 4 mM LLOMe for 30 min. Red masks, computer-identified TIA1 puncta. (G) Quantification by HCM of G3BP1 and Gal3 puncta in U2OS cells treated with increasing doses of LLOMe or 100 µM NaAsO2 in the presence or absence of 10 µg/ml cycloheximide (CHX) for 30 min. (i) HCM sample images corresponding to Fig. 1 F. Red masks, computer-identified G3BP1 puncta. (ii and ⅲ) Green masks, computer-identified Gal3 puncta and corresponding quantification in iii. (H) Immunoblot analysis of eIF2α (S51) phosphorylation in U2OS cells treated with 2 mM LLOMe for 30 min and followed by 1 h washout. (I) Quantification by HCM of G3BP1 puncta in U2OS cells treated with 2 mM LLOMe for 30 min and followed by 1 h washout. Red masks, computer-identified G3BP1 puncta. (J) Immunoblot analysis of eIF2α (S51) phosphorylation in HEK293T cells treated with 2 mM LLOMe or 100 µM NaAsO2 for 30 min. (K) Schematic summary of the findings in Fig. 1. (L) Quantification by HCM of Lysotracker Red (LTR) and G3BP1 puncta in parental HeLa WT and Gal3KO cells treated with 4 mM LLOMe for 30 min. Red masks, computer-identified LTR puncta. Green masks, computer-identified G3BP1 puncta. (M) Quantification by HCM of poly(A) RNA (Cy3-oligo[dT]) in U2OS cells transfected with scrambled siRNA as control (SCR) or G3BP1/2 siRNA for single or double knockdown (DKD). Cells were treated with 2 mM LLOMe for 30 min. Red masks, computer-identified poly(A) RNA puncta. Ctrl, control (untreated cells). Data, means ± SEM; HCM: n ≥ 3 (each experiment: 500 valid primary objects/cells per well, ≥5 wells/sample). †, P ≥ 0.05 (not significant); **, P < 0.01, ANOVA. See also Fig. 1. Source data are available for this figure: SourceData FS1.
SG formation during lysosomal damage. (A) Immunoblot analysis of proteins associated with purified lysosomes (LysoIP; anti-HA immunoprecipitation TMEM1923xHA) from HEK293T cells treated with 2 mM LLOMe or 100 µM NaAsO2 for 30 min. TMEM1922xFLAG, control. (B) Quantification by HCM of DCP1a and G3BP1 puncta in U2OS cells treated with 2 mM LLOMe for 30 min. PB, P-body. (C) Quantification by HCM of G3BP1 puncta in Huh7 cells treated with 2 mM LLOMe for 30 min. White masks, algorithm-defined cell boundaries (primary objects); Green masks, computer-identified G3BP1 puncta (target objects). (D) Quantification by HCM of Gal3 puncta in BMM cells treated with 2 mM LLOMe or 100 µM NaAsO2 for 2 h. Red masks, computer-identified galectin-3 puncta. (E) Quantification by HCM of TIA1 puncta in U2OS cells treated with 2 mM LLOMe for 30 min. Red masks, computer-identified TIA1 puncta. (F) Quantification by HCM of TIA1 puncta in HeLa cells treated with 4 mM LLOMe for 30 min. Red masks, computer-identified TIA1 puncta. (G) Quantification by HCM of G3BP1 and Gal3 puncta in U2OS cells treated with increasing doses of LLOMe or 100 µM NaAsO2 in the presence or absence of 10 µg/ml cycloheximide (CHX) for 30 min. (i) HCM sample images corresponding to Fig. 1 F. Red masks, computer-identified G3BP1 puncta. (ii and ⅲ) Green masks, computer-identified Gal3 puncta and corresponding quantification in iii. (H) Immunoblot analysis of eIF2α (S51) phosphorylation in U2OS cells treated with 2 mM LLOMe for 30 min and followed by 1 h washout. (I) Quantification by HCM of G3BP1 puncta in U2OS cells treated with 2 mM LLOMe for 30 min and followed by 1 h washout. Red masks, computer-identified G3BP1 puncta. (J) Immunoblot analysis of eIF2α (S51) phosphorylation in HEK293T cells treated with 2 mM LLOMe or 100 µM NaAsO2 for 30 min. (K) Schematic summary of the findings in Fig. 1. (L) Quantification by HCM of Lysotracker Red (LTR) and G3BP1 puncta in parental HeLa WT and Gal3KO cells treated with 4 mM LLOMe for 30 min. Red masks, computer-identified LTR puncta. Green masks, computer-identified G3BP1 puncta. (M) Quantification by HCM of poly(A) RNA (Cy3-oligo[dT]) in U2OS cells transfected with scrambled siRNA as control (SCR) or G3BP1/2 siRNA for single or double knockdown (DKD). Cells were treated with 2 mM LLOMe for 30 min. Red masks, computer-identified poly(A) RNA puncta. Ctrl, control (untreated cells). Data, means ± SEM; HCM: n ≥ 3 (each experiment: 500 valid primary objects/cells per well, ≥5 wells/sample). †, P ≥ 0.05 (not significant); **, P < 0.01, ANOVA. See also Fig. 1. Source data are available for this figure: SourceData FS1.
Using previously characterized G3BP1-GFP U2OS cells (Mackenzie et al., 2017), we further confirmed by MS that G3BP1 is recruited to lysosomes upon damage but not under arsenite-treatment conditions in our experimental conditions. This is evidenced by G3BP1’s interactions with LAMP1 and LAMP2 almost exclusively under lysosomal damage conditions (Table S1, Tabs 7 and 8). Thus, we conclude that proteins that are primarily known for being components of SGs are recruited to lysosomal membranes upon damage.
Lysosomal damage induces SG formation
We tested whether lysosomal damage induces SG formation using the conventional marker of SGs G3BP1 (Jain et al., 2016; Yang et al., 2020) in cell types amenable to high content microscopy (HCM) analysis (Claude-Taupin et al., 2021; Jia et al., 2018; Jia et al., 2020a; Jia et al., 2020b; Kumar et al., 2021a). In U2OS cells, the human osteosarcoma epithelial cell line that is suitable for HCM, 30 min of LLOMe treatment caused morphologically detectable SGs (Fig. 1, C and D). This was quantified by HCM, indicating a robust SG formation response in cells subjected to lysosomal damage by LLOMe (Fig. 1 C). Unlike SG formation, LLOMe treatment did not elicit P-body formation in U2OS cells, as assessed by the DCP1a marker exclusive to P-bodies (Ivanov et al., 2019; Kedersha et al., 2005; Fig. S1 B). A strong SG formation response was observed with glycyl-N-2-naphthalenyl-L-phenylalaninamide (GPN), another biochemical agent causing lysosomal damage (Berg et al., 1994; Jia et al., 2018) and in cells treated with agents such as silica crystals that physically damage lysosomal membranes (Hornung et al., 2008; Maejima et al., 2013; Fig. 1 C). In contrast, starvation in Earle's buffered salt solution (EBSS), a common method of inducing autophagy or inhibiting mTOR (Deretic and Kroemer, 2021), did not cause SG response (Fig. 1 C) in keeping with a previous report (Prentzell et al., 2021). As another control for LLOMe, we used LOMe, a methoxy esterified leucine (instead of esterified Leu dipeptide; Zoncu et al., 2011), and it did not induce SG formation (Fig. 1 C).
SG response was detected in other cells, including Huh7 cells, the human hepatocyte-derived carcinoma cell line (Fig. S1 C). SGs were detected in murine primary bone marrow-derived macrophages (BMMs) subjected to LLOMe treatment (Fig. 1 E). This response was as robust as a response to canonical SG inducer arsenite (Fig. 1 E). Arsenite, however, did not induce lysosomal damage, monitored by galectin-3 (Gal3) response, a conventional marker for lysosomal damage (Aits et al., 2015; Maejima et al., 2013; Fig. S1 D). We confirmed SG response to lysosomal damage using TIA1, another key immunofluorescence (IF) marker for SGs (Fig. S1, E and F). Further, we tested the effects of cycloheximide, as a known inhibitor of SG formation in response to arsenite treatment, by inhibiting translation elongation and freezing ribosomes on translating mRNAs (Freibaum et al., 2021; Kedersha et al., 2000). Cycloheximide caused similar reduction in SG formation whether cells were treated with LLOMe or with arsenite (Fig. 1 F and Fig. S1 G i), whereas cycloheximide did not affect lysosomal damage monitored by Gal3 response (Fig. S1, G ii and iii). NUFIP2, G3BP1, and TIA1 were recruited to lysosomes independently of SG formation, since their enhanced levels in LysoIP preparations from cells treated with LLOMe were not inhibited by cycloheximide (Fig. 1 G).
Activation of specific protein kinases has been established as a part of SG response, including eIF2α (Kedersha et al., 1999). LLOMe treatment of U2OS cells for 30 min induced phosphorylation of eIF2α on Ser51, whereas a recovery from lysosomal damage during LLOMe washout (Jia et al., 2020b; Maejima et al., 2013) subsided eIF2α pS51 response (Fig. S1 H). This correlated with a reduction in the number of SGs upon LLOMe washout (Fig. S1 I). In BMMs subjected to lysosomal damage, the levels of eIF2α pS51 were similar to those in cells treated with arsenite (Fig. 1 H). A similar increase in eIF2α pS51 in response to LLOMe or arsenite was observed in HEK293T cells used in our proteomic studies (Fig. S1 J). In summary, based on the observed hallmarks of conventional SG response, we conclude that lysosomal damage is a newly identified noncanonical stimulus for induction of canonical SGs (Fig. S1 K). Lysosomal damage was upstream of SG formation, since a knockout of Gal3, in keeping with previous studies (Jia et al., 2020b), sensitized lysosomes to LLOMe-induced damage reflected in increased SG formation in response to the same dose of LLOMe (Fig. S1 L).
The SGs monitored by G3BP1 puncta were authentic SGs as they completely overlapped with polyA RNA probe (Cy3-oligo-dT) detected by fluorescence in situ hybridization (FISH; Fig. 1 I), since functional SGs sequester translationally arrested mRNAs (Anderson and Kedersha, 2006; Ivanov et al., 2019). The detected granules were authentic SGs since individual or double knockdowns of G3BP1 and G3BP2, by far the most critical scaffold factors for various SG inducing conditions, resulted in a reduction of detectable mRNA puncta per cell (Fig. S1 M). SGs contribute to stress-induced translation arrest, which suppresses bulk cap-dependent protein synthesis, but enhances selective translation of ATF4 (Vattem and Wek, 2004), which is a part of integrated stress response (ISR; Costa-Mattioli and Walter, 2020). We first tested whether LLOMe damage causes general translation shutdown using puromycin incorporation assay and found that LLOMe treatment caused general translational shutdown in a dose-dependent fashion (Fig. 1 J). Paralleling this, mTOR was inhibited as measured by 4EBP1 phosphorylation (Fig. 1 K). In contrast, ATF4 expression increased over time (Fig. 1 K). Elevated ATF4 eventually leads to dephosphorylation of eIF2α (Novoa et al., 2003), which was observed later during LLOMe treatment (Fig. 1 K). Thus, lysosomal damage elicits SGs and selective translation.
PKR transmits lysosomal damage signals leading to SG formation
How might lysosomal damage be perceived and relayed to the systems that regulate SG formation? Mammalian eIF2α can be phosphorylated by four kinases (HRI, PKR, PERK, and GCN2) relaying distinct stressors (McCormick and Khaperskyy, 2017; Riggs et al., 2020). In our proteomic analyses of purified damaged lysosomes, only PKR (10 unique peptides) was detected (Fig. 3 A). A trend in PKR increase upon damage was observed by quantitative DIA proteomic analysis of damaged vs. undamaged lysosomes (Fig. 3 A). Thus, we tested whether PKR and other eIF2α kinases were required to transmit lysosomal damage and cause eIF2α phosphorylation. Of the four tested, only a knockdown of PKR abrogated eIF2α phosphorylation in response to lysosomal damage by LLOMe (Fig. 3 B). PKR was activated, as assessed by its phosphorylation at Thr446, in cells subjected to lysosomal damage (Fig. 3 C). An inhibitor of PKR, 2-aminopurine (2-AP; Lu et al., 2012) inhibited eIF2α phosphorylation in cells treated with LLOMe (Fig. 3 C). Thus, PKR is responsible for eIF2α phosphorylation in response to lysosomal damage.
PKR transmits lysosomal damage signals leading to SG formation. (A) Unique PKR peptides and intensity (DIA); LysoIP, n = 3 (see Table S1, Tab 1). Mann-Whitney U test (LLOMe treatment relative to Ctrl). (B) Immunoblot analysis of the phosphorylation of eIF2α (S51) in Huh7 cells transfected with scrambled siRNA as control (SCR) or HRI, PKR, PERK, and GCN2 siRNA for knockdown (KD). Cells were treated with 2 mM LLOMe for 30 min. The level of phosphorylation of eIF2α (S51) was quantified based on three independent experiments. (C) Immunoblot analysis of PKR (T446) and eIF2α (S51) phosphorylation in U2OS cells treated with or without PKR inhibitor 2-AP for 1 h followed by 2 mM LLOMe treatment for 30 min as indicated. (D) Quantification by HCM of G3BP1 puncta in Huh7 cells transfected with scrambled siRNA as control (SCR) or HRI, PKR, PERK, and GCN2 siRNA for knockdown (KD). Cells were treated with 2 mM LLOMe for 30 min. Red masks, computer-identified G3BP1 puncta. (E) Quantification by HCM of G3BP1 puncta in U2OS cells treated with or without PKR inhibitor 2-AP for 1 h followed by 2 mM LLOMe treatment for 30 min as indicated. (F) Quantification by HCM of G3BP1 puncta in U2OS cells treated with or without 210 nM imidazolo-oxindole C16 for 2 h followed by 2 mM LLOMe for 30 min. Red masks, computer-identified G3BP1 puncta. Ctrl, control (untreated cells). Data, means ± SEM; HCM: n ≥ 3 (each experiment: 500 valid primary objects/cells per well, ≥5 wells/sample). *, P < 0.05; **, P < 0.01, ANOVA. Source data are available for this figure: SourceData F3.
PKR transmits lysosomal damage signals leading to SG formation. (A) Unique PKR peptides and intensity (DIA); LysoIP, n = 3 (see Table S1, Tab 1). Mann-Whitney U test (LLOMe treatment relative to Ctrl). (B) Immunoblot analysis of the phosphorylation of eIF2α (S51) in Huh7 cells transfected with scrambled siRNA as control (SCR) or HRI, PKR, PERK, and GCN2 siRNA for knockdown (KD). Cells were treated with 2 mM LLOMe for 30 min. The level of phosphorylation of eIF2α (S51) was quantified based on three independent experiments. (C) Immunoblot analysis of PKR (T446) and eIF2α (S51) phosphorylation in U2OS cells treated with or without PKR inhibitor 2-AP for 1 h followed by 2 mM LLOMe treatment for 30 min as indicated. (D) Quantification by HCM of G3BP1 puncta in Huh7 cells transfected with scrambled siRNA as control (SCR) or HRI, PKR, PERK, and GCN2 siRNA for knockdown (KD). Cells were treated with 2 mM LLOMe for 30 min. Red masks, computer-identified G3BP1 puncta. (E) Quantification by HCM of G3BP1 puncta in U2OS cells treated with or without PKR inhibitor 2-AP for 1 h followed by 2 mM LLOMe treatment for 30 min as indicated. (F) Quantification by HCM of G3BP1 puncta in U2OS cells treated with or without 210 nM imidazolo-oxindole C16 for 2 h followed by 2 mM LLOMe for 30 min. Red masks, computer-identified G3BP1 puncta. Ctrl, control (untreated cells). Data, means ± SEM; HCM: n ≥ 3 (each experiment: 500 valid primary objects/cells per well, ≥5 wells/sample). *, P < 0.05; **, P < 0.01, ANOVA. Source data are available for this figure: SourceData F3.
When we tested the effects of knockdowns of eIF2α kinases on SG formation in response to lysosomal damage, only a knockdown of PKR showed statistically significant reduction in SG formation induced by LLOMe treatment (Fig. 3 D). Furthermore, 2-AP inhibited, in a dose-response fashion, SG formation in response to LLOMe treatment (Fig. 3 E). A more specific inhibitor of PKR, imidazolo-oxindole C16, also reduced SG formation during lysosomal damage (Fig. 3 F). PKR recognizes double stranded RNA during viral infections (Williams, 2001). We thus tested the possibility that RNA potentially released from damaged lysosomes could activate PKR. We knocked down lysosomal RNase RNASET2 (Haud et al., 2011) but did not detect a change in SG formation in response to LLOMe (Fig. S2 A). Whereas the signaling details activating PKR during lysosomal damage remain to be defined, we nevertheless conclude that PKR, an upstream kinase regulating eIF2α and SG formation, associates with lysosomes and that it is important in sensing lysosomal damage and transmitting damage-associated signals to the SG formation systems.
SGs induced by lysosomal damage show limited and dynamic interactions with lysosomes. (A) Quantification by HCM of G3BP1 puncta in Huh7 cells transfected with scrambled siRNA as control (SCR) or RNASET2 siRNA treated with 2 mM LLOMe for 30 min. Red masks, computer-identified G3BP1 puncta. (B) Immunofluorescence confocal microscopy analysis of G3BP1 and LAMP2. U2OS cells were treated with 2 mM LLOMe for 30 min and stained for endogenous G3BP1 and LAMP2. Scale bar, 5 µm. (C) Quantification by HCM of overlaps between G3BP1 and LAMP2 in U2OS cells treated with 2 mM LLOMe for 30 min. White masks, algorithm-defined cell boundaries. Yellow masks, computer-identified overlap of G3BP1 and LAMP2. (D) Quantification by HCM of overlaps between FLAG-NUFIP2 and LAMP2 in U2OS cells expressing FLAG-NUFIP2 treated with 2 mM LLOMe for 30 min. Yellow masks, computer-identified overlap of FLAG-NUFIP2 and LAMP2. (E) Still frames from live-cell fluorescence imaging analysis of mCherry-G3BP1 and GFP-LAMP1. U2OS cells expressing mCherry-G3BP1 and GFP-LAMP1 were incubated with 2 mM LLOMe during live-cell fluorescence imaging. Arrows, the representative regions at indicated timepoint. (F) Zoom views of regions in E. Ctrl, control (untreated cells). Data, means ± SEM; HCM: n ≥ 3 (each experiment: 500 valid primary objects/cells per well, ≥5 wells/sample). †, P ≥ 0.05 (not significant); **, P < 0.01, ANOVA. Source data are available for this figure: SourceData FS2.
SGs induced by lysosomal damage show limited and dynamic interactions with lysosomes. (A) Quantification by HCM of G3BP1 puncta in Huh7 cells transfected with scrambled siRNA as control (SCR) or RNASET2 siRNA treated with 2 mM LLOMe for 30 min. Red masks, computer-identified G3BP1 puncta. (B) Immunofluorescence confocal microscopy analysis of G3BP1 and LAMP2. U2OS cells were treated with 2 mM LLOMe for 30 min and stained for endogenous G3BP1 and LAMP2. Scale bar, 5 µm. (C) Quantification by HCM of overlaps between G3BP1 and LAMP2 in U2OS cells treated with 2 mM LLOMe for 30 min. White masks, algorithm-defined cell boundaries. Yellow masks, computer-identified overlap of G3BP1 and LAMP2. (D) Quantification by HCM of overlaps between FLAG-NUFIP2 and LAMP2 in U2OS cells expressing FLAG-NUFIP2 treated with 2 mM LLOMe for 30 min. Yellow masks, computer-identified overlap of FLAG-NUFIP2 and LAMP2. (E) Still frames from live-cell fluorescence imaging analysis of mCherry-G3BP1 and GFP-LAMP1. U2OS cells expressing mCherry-G3BP1 and GFP-LAMP1 were incubated with 2 mM LLOMe during live-cell fluorescence imaging. Arrows, the representative regions at indicated timepoint. (F) Zoom views of regions in E. Ctrl, control (untreated cells). Data, means ± SEM; HCM: n ≥ 3 (each experiment: 500 valid primary objects/cells per well, ≥5 wells/sample). †, P ≥ 0.05 (not significant); **, P < 0.01, ANOVA. Source data are available for this figure: SourceData FS2.
SGs induced by lysosomal damage show dynamic interactions with lysosomes
The SG core proteins NUFIP2, G3BP1, and TIA1 were recruited to lysosomes independently of SG condensate formation (Fig. 1 G) as described above in experiments where SG formation was inhibited by cycloheximide. Nevertheless, a question remained whether morphologically visible SGs induced by lysosomal damage associated with lysosomes? By confocal fluorescence microscopy, the majority of G3BP1-positive SGs formed during lysosomal damage were either independent of lysosomes or at best juxtaposed to lysosomes (Fig. S2 B). By HCM quantification, only a low number (10–20%) of SGs revealed by G3BP1 antibody or tagged NUFIP2 appeared associated with lysosomes 30 min after exposure to lysosomal damaging agent LLOMe (Fig. S2, C and D). Using live microscopy, we observed that the majority of SGs were forming in locations independent of lysosomes (Video 1 and Fig. S2 E). Overall, the lysosomes and SGs appeared relatively static, albeit there were three types of dynamic events suggesting changing relationships vis-à-vis each other (Video 2): (i) lysosomes and SGs remained independent of each other (Fig. S2 F i); (ii) SGs appeared to be associated with lysosomes initially but then separated (Fig. S2, F ii); and (iii) lysosomes and SGs started separately but then associated (Fig. S2 F iii). Thus, the majority of SGs as morphologically discernible profiles were separate from lysosomes.
Time-lapse confocal microscopy showing of interactions between SGs and lysosomes during lysosomal damage. U2OS cells expressing mCherry-G3BP1 and GFP-LAMP1 were incubated with 2 mM LLOMe during live-cell fluorescence imaging. Red color represents mCherry-G3BP1; green color represents GFP-LAMP1. Image stacks acquired every 30 s. Video plays 6 frames/s; Time is shown in seconds. Related to Fig. S2 E.
Time-lapse confocal microscopy showing of interactions between SGs and lysosomes during lysosomal damage. U2OS cells expressing mCherry-G3BP1 and GFP-LAMP1 were incubated with 2 mM LLOMe during live-cell fluorescence imaging. Red color represents mCherry-G3BP1; green color represents GFP-LAMP1. Image stacks acquired every 30 s. Video plays 6 frames/s; Time is shown in seconds. Related to Fig. S2 E.
Time-lapse confocal microscopy showing three types of dynamic events between SGs and lysosomes during lysosomal damage. Enlarged region of interest in Video 1, shown as snapshots in Fig. S2 F: Video 2 corresponding to Fig. S2 F i, lysosomes and SGs remaining independent of each other. Video 2 corresponding to Fig. S2 F ii, SGs initially associating with lysosomes but then separating. Video 2 corresponding to Fig. S2 F iii, lysosomes and SGs starting separately but then associating. Red color represents mCherry-G3BP1; green color represents GFP-LAMP1. Image stacks acquired every 30 s. Video plays 6 frames/s; Time is shown in seconds.
Time-lapse confocal microscopy showing three types of dynamic events between SGs and lysosomes during lysosomal damage. Enlarged region of interest in Video 1, shown as snapshots in Fig. S2 F: Video 2 corresponding to Fig. S2 F i, lysosomes and SGs remaining independent of each other. Video 2 corresponding to Fig. S2 F ii, SGs initially associating with lysosomes but then separating. Video 2 corresponding to Fig. S2 F iii, lysosomes and SGs starting separately but then associating. Red color represents mCherry-G3BP1; green color represents GFP-LAMP1. Image stacks acquired every 30 s. Video plays 6 frames/s; Time is shown in seconds.
NUFIP2 exits nucleus and localizes to lysosomes upon damage
Despite the separation between lysosomes and SGs as morphologically visualized profiles, our MS data with LysoIP indicated that certain protein components of SGs are enriched on damaged lysosomes. A top hit for this was NUFIP2 (Fig. 1 A), a widely appreciated component of SGs (Yang et al., 2020; Youn et al., 2018). We observed using confocal microscopy that NUFIP2 before LLOMe treatment was mostly in the nucleus of Huh7 cells, separated from the cytosolic G3BP1 (Fig. S3 A). Upon lysosomal damage, NUFIP2 translocated from the nucleus into the cytosol (Fig. S3 A), which was also observed by biochemically analyzing distribution in nuclear vs. postnuclear cell lysate preparations (Fig. S3 C). A bioinformatics analysis of NUFIP2’s primary structure, using consensus/algorithm (Kosugi et al., 2009), revealed a presence of a candidate nuclear localization signal (NLS) in NUFIP2 (Fig. S3 B). When we deleted the putative NLS in NUFIP2, NUFIP2 appeared absent in the nuclear fraction, i.e., was retained in the cytoplasm (Fig. S3 C). Since NUFIP2 WT was found on purified lysosomes only after lysosomal damage (Fig. S1 A), we wondered whether NUFIP2ΔNLS would be by default on lysosomes. However, LysoIP analysis showed that NUFIP2ΔNLS did not partition to lysosomes by default but also required additional signals generated during lysosomal damage to translocate to the lysosomes (Fig. 4 A). Thus, NUFIP2 translocates to lysosomes upon damage.
NUFIP2 exits nucleus and localizes to lysosomes upon damage and cooperates with Gal8 in mTORC1 response to lysosomal damage. (A) Immunofluorescence confocal microscopy analysis of G3BP1 and NUFIP2. Huh7 cells were treated with 2 mM LLOMe for 30 min and stained for endogenous G3BP1 and NUFIP2. Scale bar, 5 µm. (B) The NLS analysis of NUFIP2 by cNLS Mapper. The sequence in red, predicted NLS in NUFIP2, was deleted for generating NUFIP2ΔNLS. (C) Immunoblot analysis of NUFIP2 distribution in nuclear or postnuclear of Huh7 cells transfected with FLAG-NUFIP2 or NUFIP2ΔNLS after the treatment with 2 mM LLOMe for 30 min. (D and E) Confocal microscopy analysis (D) and quantification by HCM (E) of overlaps between mTOR and LAMP2 in U2OS transfected with scrambled siRNA as control (SCR) or NUFIP2 siRNA (NUFIP2KD) treated with 2 mM LLOMe for 30 min. Scale bar, 5 µm. (F) Immunoblot analysis of indicated proteins in U2OS cells transfected with scrambled siRNA as control (SCR) or NUFIP2 siRNA (NUFIP2KD) treated with 2 mM LLOMe for 30 min. The level of phosphorylation of ULK1 (S757), S6K (T389), and 4EBP1 (S65) was quantified based on three independent experiments. (G) Immunoblot validation of NUFIP2-knockout in Huh7 cells. #E7 was used in the following experiments, named as Huh7NUFIP2-KO. (H) HCM images of Fig. 4C. Yellow masks, computer-identified overlap of mTOR and LAMP2. (I) Immunoblot analysis of indicated proteins in Huh7 cells transfected with scrambled siRNA as control (SCR) or TIA1 siRNA (TIA1KD) treated with 2 mM LLOMe for 30 min. (J) Analysis of indicated proteins associated with lysosomes purified by anti-HA immunoprecipitation (LysoIP; TMEM192-3xHA) from HEK293T cells treated with 2 mM LLOMe for 30 min. TMEM192-2xFLAG, control. (K) Immunoblot analysis of the phosphorylation of ULK1 (S757), S6K1 (T389) and 4EBP1 (S65) in parental HeLa (WT) and TSC2-knockout HeLa cells (TSC2KO) treated with 2 mM LLOMe for 30 min. (L) Immunoblot analysis of the phosphorylation of ULK1 (S757), S6K1 (T389), and 4EBP1 (S65) in HEK293T cells or HEK293T cells stably expressing constitutively active RagB GTPase (RagBQ99L) treated with 2 mM LLOMe for 30 min. (M) GST pull-down assay of in vitro translated and radiolabeled Myc-tagged NUFIP2 or G3BP1 with GST or GST-tagged Gal8. AR, autoradiography. Ctrl, control (untreated cells). Data, means ± SEM; HCM: n ≥ 3 (each experiment: 500 valid primary objects/cells per well, ≥5 wells/sample). *, P < 0.05; **, P < 0.01, ANOVA. See also Figs. 4, 5, and 6. Source data are available for this figure: SourceData FS3.
NUFIP2 exits nucleus and localizes to lysosomes upon damage and cooperates with Gal8 in mTORC1 response to lysosomal damage. (A) Immunofluorescence confocal microscopy analysis of G3BP1 and NUFIP2. Huh7 cells were treated with 2 mM LLOMe for 30 min and stained for endogenous G3BP1 and NUFIP2. Scale bar, 5 µm. (B) The NLS analysis of NUFIP2 by cNLS Mapper. The sequence in red, predicted NLS in NUFIP2, was deleted for generating NUFIP2ΔNLS. (C) Immunoblot analysis of NUFIP2 distribution in nuclear or postnuclear of Huh7 cells transfected with FLAG-NUFIP2 or NUFIP2ΔNLS after the treatment with 2 mM LLOMe for 30 min. (D and E) Confocal microscopy analysis (D) and quantification by HCM (E) of overlaps between mTOR and LAMP2 in U2OS transfected with scrambled siRNA as control (SCR) or NUFIP2 siRNA (NUFIP2KD) treated with 2 mM LLOMe for 30 min. Scale bar, 5 µm. (F) Immunoblot analysis of indicated proteins in U2OS cells transfected with scrambled siRNA as control (SCR) or NUFIP2 siRNA (NUFIP2KD) treated with 2 mM LLOMe for 30 min. The level of phosphorylation of ULK1 (S757), S6K (T389), and 4EBP1 (S65) was quantified based on three independent experiments. (G) Immunoblot validation of NUFIP2-knockout in Huh7 cells. #E7 was used in the following experiments, named as Huh7NUFIP2-KO. (H) HCM images of Fig. 4C. Yellow masks, computer-identified overlap of mTOR and LAMP2. (I) Immunoblot analysis of indicated proteins in Huh7 cells transfected with scrambled siRNA as control (SCR) or TIA1 siRNA (TIA1KD) treated with 2 mM LLOMe for 30 min. (J) Analysis of indicated proteins associated with lysosomes purified by anti-HA immunoprecipitation (LysoIP; TMEM192-3xHA) from HEK293T cells treated with 2 mM LLOMe for 30 min. TMEM192-2xFLAG, control. (K) Immunoblot analysis of the phosphorylation of ULK1 (S757), S6K1 (T389) and 4EBP1 (S65) in parental HeLa (WT) and TSC2-knockout HeLa cells (TSC2KO) treated with 2 mM LLOMe for 30 min. (L) Immunoblot analysis of the phosphorylation of ULK1 (S757), S6K1 (T389), and 4EBP1 (S65) in HEK293T cells or HEK293T cells stably expressing constitutively active RagB GTPase (RagBQ99L) treated with 2 mM LLOMe for 30 min. (M) GST pull-down assay of in vitro translated and radiolabeled Myc-tagged NUFIP2 or G3BP1 with GST or GST-tagged Gal8. AR, autoradiography. Ctrl, control (untreated cells). Data, means ± SEM; HCM: n ≥ 3 (each experiment: 500 valid primary objects/cells per well, ≥5 wells/sample). *, P < 0.05; **, P < 0.01, ANOVA. See also Figs. 4, 5, and 6. Source data are available for this figure: SourceData FS3.
NUFIP2 contributes to mTOR inactivation during lysosomal damage. (A) Immunoblot analysis of FLAG-NUFIP2 or FLAG-NUFIP2ΔNLS associated with purified lysosomes (LysoIP; TMEM192-3xHA). Huh7 cells transfected with FLAG-NUFIP2 or FLAG-NUFIP2ΔNLS, treated or not with 2 mM LLOMe for 30 min. TMEM192-2xFLAG, control. (B) Quantification by HCM of G3BP1 puncta in parental Huh7 (WT) and NUFIP2-knockout Huh7 cells (NUFIP2KO) treated with 2 mM LLOMe or 100 µM NaAsO2 for 30 min. Red masks, computer-identified G3BP1 puncta. (C) Quantification by HCM of overlaps and confocal microscopy imaging of mTOR and LAMP2 in parental Huh7 (WT) and NUFIP2-knockout Huh7 cells (NUFIP2KO) treated with 2 mM LLOMe for 30 min. HCM images in Fig. S3 H. Scale bar, 5 µm. (D) Immunoblot analysis of indicated proteins in parental Huh7 (WT) and NUFIP2-knockout Huh7 cells (NUFIP2KO) treated with 2 mM LLOMe for 30 min. The level of phosphorylation of ULK1 (S757), S6K (T389), and 4EBP1 (S65) was quantified based on three independent experiments. (E) Immunoblot analysis of proteins associated with purified lysosomes (LysoIP) from HEK293T cells treated with 1 μM PP242 for 2 h or 2 mM LLOMe for 30 min or HEK293T cells stably expressing constitutively active RagB GTPase (RagBQ99L) treated with 2 mM LLOMe for 30 min. Ctrl, control (untreated cells). Data, means ± SEM; HCM: n ≥ 3 (each experiment: 500 valid primary objects/cells per well, ≥5 wells/sample). †, P ≥ 0.05 (not significant); **, P < 0.01, ANOVA. See also Fig. S3. Source data are available for this figure: SourceData F4.
NUFIP2 contributes to mTOR inactivation during lysosomal damage. (A) Immunoblot analysis of FLAG-NUFIP2 or FLAG-NUFIP2ΔNLS associated with purified lysosomes (LysoIP; TMEM192-3xHA). Huh7 cells transfected with FLAG-NUFIP2 or FLAG-NUFIP2ΔNLS, treated or not with 2 mM LLOMe for 30 min. TMEM192-2xFLAG, control. (B) Quantification by HCM of G3BP1 puncta in parental Huh7 (WT) and NUFIP2-knockout Huh7 cells (NUFIP2KO) treated with 2 mM LLOMe or 100 µM NaAsO2 for 30 min. Red masks, computer-identified G3BP1 puncta. (C) Quantification by HCM of overlaps and confocal microscopy imaging of mTOR and LAMP2 in parental Huh7 (WT) and NUFIP2-knockout Huh7 cells (NUFIP2KO) treated with 2 mM LLOMe for 30 min. HCM images in Fig. S3 H. Scale bar, 5 µm. (D) Immunoblot analysis of indicated proteins in parental Huh7 (WT) and NUFIP2-knockout Huh7 cells (NUFIP2KO) treated with 2 mM LLOMe for 30 min. The level of phosphorylation of ULK1 (S757), S6K (T389), and 4EBP1 (S65) was quantified based on three independent experiments. (E) Immunoblot analysis of proteins associated with purified lysosomes (LysoIP) from HEK293T cells treated with 1 μM PP242 for 2 h or 2 mM LLOMe for 30 min or HEK293T cells stably expressing constitutively active RagB GTPase (RagBQ99L) treated with 2 mM LLOMe for 30 min. Ctrl, control (untreated cells). Data, means ± SEM; HCM: n ≥ 3 (each experiment: 500 valid primary objects/cells per well, ≥5 wells/sample). †, P ≥ 0.05 (not significant); **, P < 0.01, ANOVA. See also Fig. S3. Source data are available for this figure: SourceData F4.
NUFIP2 contributes to mTOR inactivation during lysosomal damage
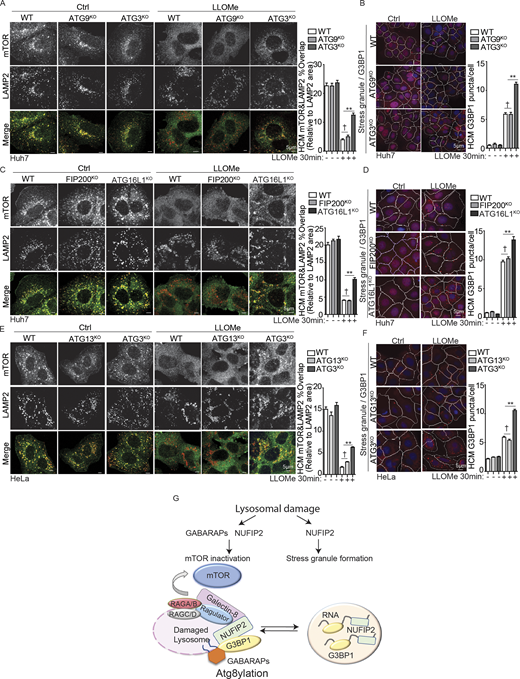
Recent studies have indicated that components of SGs, such as G3BP1 associated in earlier proteomic studies with NUFIP2 (Sowa et al., 2009), can reside on lysosomes, and have additional noncanonical functions outside of the scope of SG formation, including effects on mTOR activity (Prentzell et al., 2021). A knockdown of NUFIP2 reduced mTOR desorption from the lysosomes (Fig. S3, D and E), which serves as a visual proxy for mTOR inactivation in response to various inputs including lysosomal damage (Jia et al., 2018). This was confirmed by testing phosphorylation of mTOR substrates, ULK1 (Ser757), S6K (Thr389), and 4EBP1 (Ser65), which was diminished in cells treated with LLOMe, but less so in cells knocked down for NUFIP2 (Fig. S3 F). We generated a CRISPR knockout of NUFIP2 in Huh7 cells (Huh7NUFIP2-KO; Fig. S3 G). The knockout was validated for its effects on SG formation in response to either canonical inducer arsenite or induction upon lysosomal damage, which were both reduced in Huh7NUFIP2-KO cells (Fig. 4 B). mTOR in Huh7NUFIP2-KO cells resisted inactivation in response to lysosomal damage, quantified by HCM of its desorption from lysosomes (Fig. 4 C and Fig. S3 H). This was also reflected in levels of phospho-ULK1 (Ser757), phospho-S6K (Thr389), and phospho-4EBP1 (Ser65), which resisted reduction in Huh7NUFIP2-KO cells, normally seen upon lysosomal damage (Jia et al., 2018; Fig. 4 D). This effect was specific for NUFIP2 as a knockdown of TIA1 did not affect inhibition of mTOR in response to LLOMe damage (Fig. S3 I). Treatment with a conventional catalytic inhibitor of mTOR, pp242, did not result in NUFIP2 recruitment to lysosomes (Fig. 4 E). NUFIP2 was recruited to damaged lysosomes even in cells expressing constitutively active RagBQ99L, which maintains mTORC1 in active state (Fig. 4 E), indicating that NUFIP2 translocates to damaged lysosomes independently of mTOR activation state and is likely acting upstream of mTOR. Thus, NUFIP2, a key protein classically associated with SGs, acts on lysosomes to inhibit mTOR during lysosomal damage.
Ragulator abundance and activity on damaged lysosomes is controlled by NUFIP2
We have previously shown that mTOR is inactivated during lysosomal damage through Ragulator–RagA/B system by the inactivation of Ragulator’s GEF activity toward RagA/B, which in turn normally keep mTOR active (Jia et al., 2018). Our MS data of purified lysosomes after the damage revealed that four Ragulator components (LAMTOR1, 2, 3, and 5) were elevated on lysosomes (Fig. 1 A and Fig. 5 A), which was confirmed for all Ragulator’s components but was not observed for Rags by LysoIP and Western blot (WB) analyses (Fig. 5 B and Fig. S3 J). The increase in LAMTOR1 and decrease of mTOR on damaged lysosomes appeared abrogated in NUFIP2KO cells (Fig. 5, C i–iii), whereas the total cellular levels of LAMTOR1 did not change (Fig. 5 C iv).
Ragulator abundance and activity on damaged lysosomes is controlled by NUFIP2. (A) Summary of the quantitative changes in relevant proteins of mTORC1 signaling based on DIA LysoIP LC/MS/MS analysis. FC, fold change (see Table S1, Tab 1). (B) Immunoblot analysis of proteins associated with purified lysosomes (LysoIP; TMEM192-3xHA) from HEK293T cells treated with 2 mM LLOMe for 30 min. TMEM192-2xFLAG, control. (C ⅰ–iv) Immunoblot analysis of proteins associated with purified lysosomes (LysoIP) from parental Huh7 WT and NUFIP2-knockout Huh7 cells (NUFIP2KO) treated with 2 mM LLOMe for 30 min (i); quantification (ii–iv), n = 3. (D) HEK293T cells stably expressing FLAG-Metap2 (control) or FLAG-LAMTOR2 transfected with scrambled siRNA (SCR) or NUFIP2 siRNA (NUFIP2KD) were treated with 2 mM LLOMe for 30 min. Cell lysates were immunoprecipitated with anti-FLAG antibody and immunoblotted for indicated proteins. Quantification of interaction between RagA and LAMTOR2, n = 3. (E) HEK293T cells stably expressing FLAG-Metap2 (control) or FLAG-LAMTOR2 transfected with GFP or GFP-NUFIP2 were treated with 2 mM LLOMe for 30 min. Cell lysates were immunoprecipitated with anti-FLAG antibody and immunoblotted for indicated proteins. Quantification of interaction between RagA and LAMTOR2, n = 3. (F) HEK293T cells expressing FLAG (control) or FLAG-NUFIP2 were treated with 2 mM LLOMe for 30 min. Cell lysates were immunoprecipitated with anti-FLAG antibody and immunoblotted for indicated proteins. Data, means ± SEM; †, P ≥ 0.05 (not significant); *, P < 0.05; **, P < 0.01, ANOVA. See also Fig. S3. Source data are available for this figure: SourceData F5.
Ragulator abundance and activity on damaged lysosomes is controlled by NUFIP2. (A) Summary of the quantitative changes in relevant proteins of mTORC1 signaling based on DIA LysoIP LC/MS/MS analysis. FC, fold change (see Table S1, Tab 1). (B) Immunoblot analysis of proteins associated with purified lysosomes (LysoIP; TMEM192-3xHA) from HEK293T cells treated with 2 mM LLOMe for 30 min. TMEM192-2xFLAG, control. (C ⅰ–iv) Immunoblot analysis of proteins associated with purified lysosomes (LysoIP) from parental Huh7 WT and NUFIP2-knockout Huh7 cells (NUFIP2KO) treated with 2 mM LLOMe for 30 min (i); quantification (ii–iv), n = 3. (D) HEK293T cells stably expressing FLAG-Metap2 (control) or FLAG-LAMTOR2 transfected with scrambled siRNA (SCR) or NUFIP2 siRNA (NUFIP2KD) were treated with 2 mM LLOMe for 30 min. Cell lysates were immunoprecipitated with anti-FLAG antibody and immunoblotted for indicated proteins. Quantification of interaction between RagA and LAMTOR2, n = 3. (E) HEK293T cells stably expressing FLAG-Metap2 (control) or FLAG-LAMTOR2 transfected with GFP or GFP-NUFIP2 were treated with 2 mM LLOMe for 30 min. Cell lysates were immunoprecipitated with anti-FLAG antibody and immunoblotted for indicated proteins. Quantification of interaction between RagA and LAMTOR2, n = 3. (F) HEK293T cells expressing FLAG (control) or FLAG-NUFIP2 were treated with 2 mM LLOMe for 30 min. Cell lysates were immunoprecipitated with anti-FLAG antibody and immunoblotted for indicated proteins. Data, means ± SEM; †, P ≥ 0.05 (not significant); *, P < 0.05; **, P < 0.01, ANOVA. See also Fig. S3. Source data are available for this figure: SourceData F5.
Activation state of the Ragulator can be assessed by increased interactions between LAMTOR2 (p14) and RagA when RagA is in its inactive, GDP-bound form (Bar-Peled et al., 2012; Jia et al., 2018). Using this established approach, we quantified complexes between RagA and LAMTOR2, and found them to be increased (reflecting inactive RagA state) during lysosomal damage, in keeping with our prior studies (Jia et al., 2018), but this was reduced in HEK293T cells stably expressing FLAG-LAMTOR2 knocked down for NUFIP2 (Fig. 5 D), indicating that NUFIP2 is required for inactivation of the Ragulator complex. Conversely, overexpression of NUFIP2 further increased the elevated association between FLAG-LAMTOR2 and endogenous RagA during lysosomal damage (Fig. 5 E). Thus, NUFIP2 is required for RagA inactivation. Furthermore, NUFIP2 was in complexes with LAMTOR1 (and other Ragulator components), but only under lysosomal damaging conditions (Fig. 5 F). We conclude that NUFIP2, a functional component of SGs (Yang et al., 2020) is also an important regulator of mTOR through Ragulator during lysosomal damage.
NUFIP2 and galectin-8 (Gal8) cooperate in mTOR response to lysosomal damage
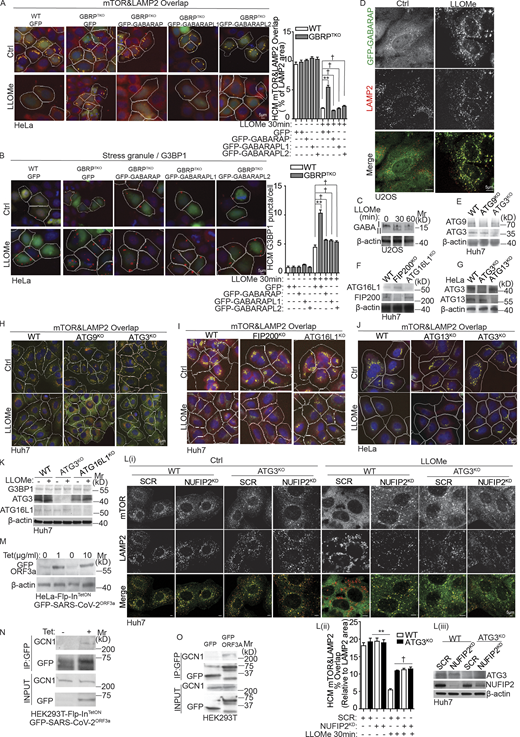
The observation that NUFIP2 is a new regulator of mTOR prompted us to test the previously reported specific regulators of mTOR inactivation during lysosomal damage (Jia et al., 2018). TSC2 did not affect phosphorylation status of mTOR’s substrates (Fig. S3 K), whereas RagB did, as expression of active RagBQ99L (Jia et al., 2018) prevented loss of mTOR activity induced by LLOMe (Fig. S3 L), as previously reported for GPN damage (Jia et al., 2018). We next tested LGALS8 (Gal8), the principal sensor transducing lysosomal damage to inhibit the Ragulator–RagA/B system (Jia et al., 2018). Whereas Gal8 was needed to fully inhibit mTOR based on its retention on lysosomes in Gal8KOHeLa cells exposed to LLOMe (Fig. 6 A), Gal8 had no effect on SG formation. G3BP1 puncta formed as robustly in Gal8KOHeLa cells as in parental WT cells treated with LLOMe (Fig. 6 B).
NUFIP2 and Gal8 cooperate in mTOR response to lysosomal damage. (A) Quantification by HCM of overlaps between mTOR and LAMP2 in Gal8WTHeLa (WT) or Gal8KOHeLa (Gal8KO) cells treated with 2 mM LLOMe for 30 min. Yellow masks, computer-identified overlap of mTOR and LAMP2. (B) Quantification by HCM of G3BP1 puncta in Gal8WTHeLa (WT) or Gal8KOHeLa (Gal8KO) cells treated with 2 mM LLOMe for 30 min. Red masks, computer-identified G3BP1 puncta. (C) HEK293T cells stably expressing FLAG-LAMTOR2 with overexpression of GFP or GFP-NUFIP2 were transfected with scrambled siRNA as control (SCR) or Gal8 siRNA (Gal8KD). Cells were treated with 200 µM GPN for 30 min. Cell lysates were immunoprecipitated with anti-FLAG antibody and immunoblotted for indicated proteins. (D) GST pulldown assay of in vitro translated and radiolabeled Myc-tagged NUFIP2 or G3BP1 with GST or GST-tagged Ragulator or Gal8. (E) GST pull-down assay of in vitro translated and radiolabeled Myc-tagged NUFIP2 with GST or GST-tagged Gal8. Quantification, n = 3. (F) Analysis of indicated proteins associated with lysosomes purified by anti-HA immunoprecipitation (LysoIP; TMEM192-3xHA) from HeLa WT, Gal8KO, GABARAPs knockout (GBRPTKO) or G3BP1 knockdown (G3BP1KD) cells. Cells were treated with 200 µM GPN for 30 min. AR, autoradiography. Ctrl, control (untreated cells). Data, means ± SEM; HCM: n ≥ 3 (each experiment: 500 valid primary objects/cells per well, ≥5 wells/sample). †, P ≥ 0.05 (not significant); **, P < 0.01, ANOVA. See also Fig. S3. Source data are available for this figure: SourceData F6.
NUFIP2 and Gal8 cooperate in mTOR response to lysosomal damage. (A) Quantification by HCM of overlaps between mTOR and LAMP2 in Gal8WTHeLa (WT) or Gal8KOHeLa (Gal8KO) cells treated with 2 mM LLOMe for 30 min. Yellow masks, computer-identified overlap of mTOR and LAMP2. (B) Quantification by HCM of G3BP1 puncta in Gal8WTHeLa (WT) or Gal8KOHeLa (Gal8KO) cells treated with 2 mM LLOMe for 30 min. Red masks, computer-identified G3BP1 puncta. (C) HEK293T cells stably expressing FLAG-LAMTOR2 with overexpression of GFP or GFP-NUFIP2 were transfected with scrambled siRNA as control (SCR) or Gal8 siRNA (Gal8KD). Cells were treated with 200 µM GPN for 30 min. Cell lysates were immunoprecipitated with anti-FLAG antibody and immunoblotted for indicated proteins. (D) GST pulldown assay of in vitro translated and radiolabeled Myc-tagged NUFIP2 or G3BP1 with GST or GST-tagged Ragulator or Gal8. (E) GST pull-down assay of in vitro translated and radiolabeled Myc-tagged NUFIP2 with GST or GST-tagged Gal8. Quantification, n = 3. (F) Analysis of indicated proteins associated with lysosomes purified by anti-HA immunoprecipitation (LysoIP; TMEM192-3xHA) from HeLa WT, Gal8KO, GABARAPs knockout (GBRPTKO) or G3BP1 knockdown (G3BP1KD) cells. Cells were treated with 200 µM GPN for 30 min. AR, autoradiography. Ctrl, control (untreated cells). Data, means ± SEM; HCM: n ≥ 3 (each experiment: 500 valid primary objects/cells per well, ≥5 wells/sample). †, P ≥ 0.05 (not significant); **, P < 0.01, ANOVA. See also Fig. S3. Source data are available for this figure: SourceData F6.
We next examined relationships between NUFIP2, Gal8, and Ragulator components. We found that Gal8 was required for inactivation of RagA by NUFIP2 in response to lysosomal damage by GPN (Fig. 6 C). Whereas the co-immunoprecipitation (co-IP) results (Fig. 5, E and F) indicated that NUFIP2 and all Ragulator components (LAMTOR 1-5) can be in protein complexes, GST pulldowns indicated that these interactions were not direct (Fig. 6 D). Instead, we found that NUFIP2 interacted directly with Gal8 (Fig. 6, D and E; and Fig. S3 M). However, NUFIP2 recruitment to lysosomes surprisingly did not depend on Gal8 (Fig. 6 F), and thus other mechanisms for NUFIP2 translocation to lysosomes must be involved. Nevertheless, a model emerges whereby Gal8, which interacts directly with NUFIP2, transmits the effects of NUFIP2 to the Ragulator–Rag complexes during mTOR inactivation early upon lysosomal damage.
Mammalian Atg8s participate in recruitment of NUFIP2 to damaged lysosomes
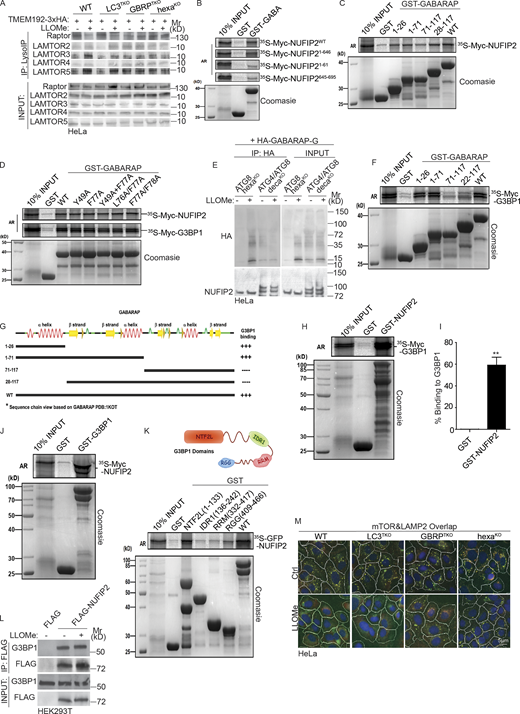
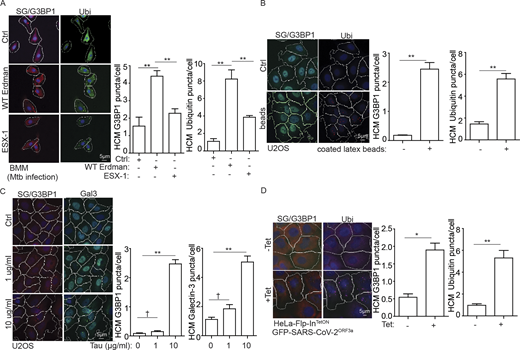
In a previous report (Markmiller et al., 2018) with arsenite-induced SGs, proximity labeling of mATG8 proteins was reported when using G3BP1-APEX2. Thus, we addressed the possibility that mATG8s, usually considered to function primarily in the process of clearance of damaged lysosomes in a process termed lysophagy (Maejima et al., 2013), could play an additional role in recruitment of SG proteins to damaged lysosomes. Our MS analysis revealed enrichment of GABARAP, GABARAPL2, and LC3B on damaged lysosomes (Fig. 7 A and Table S1, Tab 5). Among other autophagy factors increased on purified lysosomes after a short pulse (30 min) of LLOMe-induced damage were ATG16L1 and ATG9A (Fig. 7 A and Table S1, Tab 5), whereas other canonical autophagy factors were not enriched/responsive to lysosomal damage. We next tested whether mATG8s played a role in recruitment of SG proteins to damaged lysosomes. For this we compared the previously characterized (Gu et al., 2019; Nguyen et al., 2016) hexaKO HeLa cell line with inactivated six mATG8s with its parental WT HeLa cell line. These cells expressing TMEM192-3xHA were treated with LLOMe and subjected to unbiased proteomic analysis of purified lysosomes. The volcano plot (Fig. 7 B) and data analyses (Table S1, Tab 9) indicated that NUFIP2 and G3BP1 were the only core SG proteins significantly increased on damaged lysosomes in WT HeLa cells compared to hexaKO HeLa cells. This strongly suggests that mATG8s are the factors responsible for recruitment of these proteins (NUFIP2 and G3BP1) to damaged lysosomes. The uniqueness of NUFIP2 and G3BP1 among SG proteins in proteomic analyses of hexaKO cells is in keeping with their recruitment being independent of SG condensates (Fig. 1 G).
Mammalian ATG8sparticipate in recruitment of NUFIP2 to damaged lysosomes. (A) Summary of the detected autophagy factors and their quantitative changes based on DIA LC/MS/MS analysis of LysoIP in HEK293T cells (see Table S1, Tab 5). FC, fold change. (B) DIA LC/MS/MS analysis of lysosomes purified by anti-HA immunoprecipitation (LysoIP; TMEM192-3xHA) from parental HeLaWT or mATG8 knockout (hexaKO) treated with or without 4 mM LLOMe for 30 min. Scatter (volcano; log2 fold change and −Log10 P values) plot of stress granule core proteins; n = 3 (see Table S1, Tab 9). Dashed line, significance cut-off (P < 0.05). (C) Analysis of indicated proteins associated with purified lysosomes. Lysosomes were purified by anti-HA immunoprecipitation (LysoIP; TMEM192-3xHA) from parental HeLa (WT), LC3TKO, GBRPTKO, and hexaKO cells treated with 4 mM LLOMe for 30 min. (D) GST pulldown assay of in vitro translated and radiolabeled Myc-tagged NUFIP2 with GST or GST-tagged mATG8. GABARAP (GABA); GABARAPL1 (GABAL1); GABARAPL2 (GABAL2). (E) Quantification of D. Data (% binding) represents the percentage of the corresponding protein relative to its input. (F) GST pulldown assay of in vitro translated and radiolabeled Myc-tagged G3BP1 with GST or GST-tagged mATG8. GABARAP (GABA); GABARAPL1 (GABAL1); GABARAPL2 (GABAL2). (G) Quantification of F. Data (% binding) represents the percentage of the corresponding protein relative to its input. (H) Quantification of GST pulldown assay of in vitro translated and radiolabeled Myc-tagged NUFIP2 with GST or GST-tagged GABARAP deletions. Blots in Fig. S4 C. Data (% binding) represents the percentage of the corresponding protein relative to its input. (I) Quantification of GST pull-down assay of in vitro translated and radiolabeled Myc-tagged G3BP1 with GST or GST-tagged GABARAP deletions. Blots in Fig. S4 F. Data (% binding) represents the percentage of the corresponding protein relative to its input. AR, autoradiography. Data, means ± SEM; *, P < 0.05; **, P < 0.01, ANOVA. See also Fig. S4. Source data are available for this figure: SourceData F7.
Mammalian ATG8sparticipate in recruitment of NUFIP2 to damaged lysosomes. (A) Summary of the detected autophagy factors and their quantitative changes based on DIA LC/MS/MS analysis of LysoIP in HEK293T cells (see Table S1, Tab 5). FC, fold change. (B) DIA LC/MS/MS analysis of lysosomes purified by anti-HA immunoprecipitation (LysoIP; TMEM192-3xHA) from parental HeLaWT or mATG8 knockout (hexaKO) treated with or without 4 mM LLOMe for 30 min. Scatter (volcano; log2 fold change and −Log10 P values) plot of stress granule core proteins; n = 3 (see Table S1, Tab 9). Dashed line, significance cut-off (P < 0.05). (C) Analysis of indicated proteins associated with purified lysosomes. Lysosomes were purified by anti-HA immunoprecipitation (LysoIP; TMEM192-3xHA) from parental HeLa (WT), LC3TKO, GBRPTKO, and hexaKO cells treated with 4 mM LLOMe for 30 min. (D) GST pulldown assay of in vitro translated and radiolabeled Myc-tagged NUFIP2 with GST or GST-tagged mATG8. GABARAP (GABA); GABARAPL1 (GABAL1); GABARAPL2 (GABAL2). (E) Quantification of D. Data (% binding) represents the percentage of the corresponding protein relative to its input. (F) GST pulldown assay of in vitro translated and radiolabeled Myc-tagged G3BP1 with GST or GST-tagged mATG8. GABARAP (GABA); GABARAPL1 (GABAL1); GABARAPL2 (GABAL2). (G) Quantification of F. Data (% binding) represents the percentage of the corresponding protein relative to its input. (H) Quantification of GST pulldown assay of in vitro translated and radiolabeled Myc-tagged NUFIP2 with GST or GST-tagged GABARAP deletions. Blots in Fig. S4 C. Data (% binding) represents the percentage of the corresponding protein relative to its input. (I) Quantification of GST pull-down assay of in vitro translated and radiolabeled Myc-tagged G3BP1 with GST or GST-tagged GABARAP deletions. Blots in Fig. S4 F. Data (% binding) represents the percentage of the corresponding protein relative to its input. AR, autoradiography. Data, means ± SEM; *, P < 0.05; **, P < 0.01, ANOVA. See also Fig. S4. Source data are available for this figure: SourceData F7.
Next, we used hexaKO HeLa cells, LC3TKO HeLa cells with inactivated three LC3s and GBRPTKO HeLa cells with inactivated all three GABARAPs (Nguyen et al., 2016). The hexaKO HeLa cells lost ability to recruit NUFIP2 and G3BP1 to damaged lysosomes, as determined by WB analysis of lysosomes purified by LysoIP (Fig. 7 C). The GABARAP subset of mATG8s was responsible for the recruitment of NUFIP2 and G3BP1, since LC3TKO HeLa retained the ability to recruit NUFIP2 and G3BP1 whereas GBRPTKO HeLa cells did not (Fig. 7 C). GABARAPs were also a key for departure of mTOR and Raptor from damaged lysosomes (Fig. 7 C and Fig. S4 A). LAMTOR1 (p18) and LAMTOR complex inversely mirrored mTOR by being enriched on damaged lysosomes (Fig. 7 C and Fig. S4 A). LAMTOR1 enrichment on damaged lysosomes was lost in GBRPTKO HeLa but not in LC3TKO HeLa (Fig. 7 C). Thus, mATG8s, specifically GABARAPs, do not only function in autophagy, but have noncanonical roles in recruitment of NUFIP2 and G3BP1 to the lysosome upon damage.
GABARAPs interact directly with NUFIP2 and G3BP1. (A) Immunoblot analysis of proteins associated with lysosomes purified by anti-HA immunoprecipitation (LysoIP; TMEM192-3xHA) from parental HeLa (WT), LC3TKO, GBRPTKO, and hexaKO cells treated with 4 mM LLOMe for 30 min. (B) GST pulldown assay of in vitro translated and radiolabeled Myc-tagged NUFIP2 deletions with GST or GST-tagged GABARAP (GABA). (C) GST pull-down assay of in vitro translated and radiolabeled Myc-tagged NUFIP2 with GST or GST-tagged GABARAP deletions. (D) GST pulldown assay of in vitro translated and radiolabeled Myc-tagged NUFIP2/G3BP1 with GST or GST-tagged GABARAP mutants. (E) Immunoblot analysis of denaturing HA IP performed on untreated or 4 mM LLOMe treated (30 min) lysates from ATG8 knockout (hexaKO) or ATG4/ATG8 knockout (decaKO) HeLa cells expressing HA-GBRPL1-G. (F) GST pulldown assay of in vitro translated and radiolabeled Myc-tagged G3BP1 with GST or GST-tagged GABARAP deletions. (G) Summary of interactions between GABARAP and G3BP1. (H) GST pulldown assay of in vitro translated and radiolabeled Myc-tagged G3BP1 with GST or GST-tagged NUFIP2. (I) Quantification of Fig. S4 H. Data (% binding) represents the percentage of the corresponding protein relative to its input. (J) GST pulldown assay of in vitro translated and radiolabeled Myc-tagged NUFIP2 with GST or GST-G3BP1. (K) GST pull-down assay of in vitro translated and radiolabeled GFP-tagged NUFIP2 with GST or GST-tagged G3BP1 deletions. (L) Immunoblot analysis of interaction between NUFIP2 and G3BP1 in HEK293T cells transfected with FLAG or FLAG-NUFIP2 with 2 mM LLOMe treatment for 30 min. (M) HCM images corresponding to Fig. 8 C. White masks, algorithm-defined cell boundaries. Yellow masks, computer-identified overlap of mTOR and LAMP2. Ctrl, control (untreated cells). AR, autoradiography. Data, means ± SEM; HCM: n ≥ 3 (each experiment: 500 valid primary objects/cells per well, ≥5 wells/sample). **, P < 0.01, ANOVA. See also Figs. 7 and 8. Source data are available for this figure: SourceData FS4.
GABARAPs interact directly with NUFIP2 and G3BP1. (A) Immunoblot analysis of proteins associated with lysosomes purified by anti-HA immunoprecipitation (LysoIP; TMEM192-3xHA) from parental HeLa (WT), LC3TKO, GBRPTKO, and hexaKO cells treated with 4 mM LLOMe for 30 min. (B) GST pulldown assay of in vitro translated and radiolabeled Myc-tagged NUFIP2 deletions with GST or GST-tagged GABARAP (GABA). (C) GST pull-down assay of in vitro translated and radiolabeled Myc-tagged NUFIP2 with GST or GST-tagged GABARAP deletions. (D) GST pulldown assay of in vitro translated and radiolabeled Myc-tagged NUFIP2/G3BP1 with GST or GST-tagged GABARAP mutants. (E) Immunoblot analysis of denaturing HA IP performed on untreated or 4 mM LLOMe treated (30 min) lysates from ATG8 knockout (hexaKO) or ATG4/ATG8 knockout (decaKO) HeLa cells expressing HA-GBRPL1-G. (F) GST pulldown assay of in vitro translated and radiolabeled Myc-tagged G3BP1 with GST or GST-tagged GABARAP deletions. (G) Summary of interactions between GABARAP and G3BP1. (H) GST pulldown assay of in vitro translated and radiolabeled Myc-tagged G3BP1 with GST or GST-tagged NUFIP2. (I) Quantification of Fig. S4 H. Data (% binding) represents the percentage of the corresponding protein relative to its input. (J) GST pulldown assay of in vitro translated and radiolabeled Myc-tagged NUFIP2 with GST or GST-G3BP1. (K) GST pull-down assay of in vitro translated and radiolabeled GFP-tagged NUFIP2 with GST or GST-tagged G3BP1 deletions. (L) Immunoblot analysis of interaction between NUFIP2 and G3BP1 in HEK293T cells transfected with FLAG or FLAG-NUFIP2 with 2 mM LLOMe treatment for 30 min. (M) HCM images corresponding to Fig. 8 C. White masks, algorithm-defined cell boundaries. Yellow masks, computer-identified overlap of mTOR and LAMP2. Ctrl, control (untreated cells). AR, autoradiography. Data, means ± SEM; HCM: n ≥ 3 (each experiment: 500 valid primary objects/cells per well, ≥5 wells/sample). **, P < 0.01, ANOVA. See also Figs. 7 and 8. Source data are available for this figure: SourceData FS4.
GABARAPs interact directly with NUFIP2 and G3BP1
In GST pulldowns between NUFIP2 and a full panel of mATG8s and in parallel with G3BP1, GABARAP showed strong association with either of the proteins (Fig. 7, D–G). Some appreciable association was also observed with GABARAPL1 (Fig. 7, D–G). We also tested deletion constructs of NUFIP2 for their ability to bind GABARAP (Fig. S4 B). Deletion mapping of GABARAP domains required for interactions with NUFIP2 indicated two binding surfaces, one N-terminally located and another one more centrally located (Fig. 7 H and Fig. S4 C), suggesting that the binding site is not a contiguous linear surface, such as the previously reported LIR docking sites (LDS) or UIM-docking sites (UDS; Johansen and Lamark, 2020; Marshall et al., 2019). Nevertheless, we tested single LDS (Y49A), UDS (F77A), and double LDS/UDS mutant GABARAP for binding to NUFIP2, and none of the mutations in these key residues defining LDS or UDS affected association with NUFIP2 in GST pulldowns (Fig. S4 D). Although the GST pulldown experiments clearly indicate that GABARAP and NUFIP2 noncovalently bind, we tested whether the newly described process of protein atg8ylation (Nguyen et al., 2021) may also take place between these two proteins. Using decaKO HeLa cells lacking all ATG4s delipidating enzymes (which also act as peptidases/isopeptidases), transfected with HA-GABARAP-G (a derivative with the pre-exposed C-terminal Gly residue), we detected HA-GABARAP-atg8ylated form of NUFIP2 (but only in cells lacking ATG4s), as well as the expected non-covalently bound proteins in co-IPs (Fig. S4 E).
G3BP1 has recently been reported to associate with lysosomes (Prentzell et al., 2021) as also seen in our LysoIP preparations (Fig. 7 C). G3BP1 association with damaged lysosomes depended on GABARAPs (Fig. 7 C). We thus tested whether G3BP1 can associate with mATG8s and found in GST pulldown assays that it interacted directly with GABARAP (Fig. 7, F and G). Furthermore, deletion mapping confirmed that the N-terminal region of GABARAP interacts with G3BP1 (Fig. 7, I; and Fig. S4, F and G). Individual or combined LDS and UDS mutants of GABARAP still bound G3BP1 (Fig. S4 D). Finally, NUFIP2 and G3BP1 directly (Fig. S4 H) and very strongly interacted with each other in GST pulldown experiments with 60% of the input [35S] Myc-G3BP1 being bound to its NUFIP2 partner (Fig. S4 I) . This interaction was also observed in reverse pulldown experiments (Fig. S4 J). NUFIP2 strongly interacts with G3BP1 through G3BP1’s N-terminal NTF2L domain (Fig. S4 K). G3BP1 and NUFIP2 constitutively interacted in co-IP experiments (Fig. S4 L). NUFIP2 recruitment to damaged lysosomes was independent of G3BP1 (Fig. 6 F), suggesting that the recruitment through direct interactions with GABARAP is a dominant process for NUFIP2’s translocation to damaged lysosomes.
GABARAPs participate in mTOR inactivation but not in eIF2α phosphorylation in response to lysosomal damage
Based on strong associations in functional and binding experiments between GABARAPs and NUFIP2 and the requirement for NUFIP2 in mTOR inactivation during lysosomal damage, we tested whether NUFIP2’s interactors GABARAPs also played a role in mTOR inactivation. Using ULK1, S6K, and 4EBP1 phosphorylation as a conventional measure of mTOR activity, we detected an expected drop in levels of S6K (Thr389), 4EBP1 (Ser65), and ULK1 (Ser757) phosphorylation upon lysosomal damage with LLOMe, a relationship that was preserved in LC3TKO HeLa cells (Fig. 8, A and B i–iii). In contrast, mTOR inactivation was not observed in hexaKO and GBRPTKO HeLa cells (Fig. 8, A and B i–iii). We next tested by HCM whether mTOR desorption from damaged lysosomes was affected by mATG8s. As with reduction in ULK1, S6K, and 4EBP1 phosphorylation, mTOR association with lysosomes diminished upon lysosomal damage in WT and LC3TKO HeLa cells but not as readily in hexaKO and GBRPTKO HeLa cells (Fig. 8 C and Fig. S4 M).
GABARAPs participate in mTOR inactivation but not in eIF2α phosphorylation in response to lysosomal damage. (A) Immunoblot analysis of the phosphorylation ULK1 (S757), S6K (T389), 4EBP1 (S65), and eIF2α (S51) in parental HeLa (WT), LC3TKO, GBRPTKO, and hexaKO cells treated with 4 mM LLOMe for 30 min. (B ⅰ–ⅳ) Quantification of phosphorylation of ULK1 (S757; i), S6K (T389; ii), 4EBP1 (S65; iii), and eIF2α (S51; iv) in A; Quantification, n = 3. (C) Quantification by HCM and confocal microscopy analysis of overlaps between mTOR and LAMP2 in parental HeLa (WT), LC3TKO, GBRPTKO, and hexaKO cells treated with 4 mM LLOMe for 30 min. HCM images in Fig. S4 M. Scale bar, 5 µm. (D) Quantification by HCM of G3BP1 puncta. Parental HeLa (WT), LC3TKO, GBRPTKO, and hexaKO cells were treated with 4 mM LLOMe for 30 min. Red masks, computer-identified G3BP1 puncta. Data, means ± SEM; HCM: n ≥ 3 (each experiment: 500 valid primary objects/cells per well, ≥5 wells/sample). †, P ≥ 0.05 (not significant); **, P < 0.01, ANOVA. See also Fig. S4. Source data are available for this figure: SourceData F8.
GABARAPs participate in mTOR inactivation but not in eIF2α phosphorylation in response to lysosomal damage. (A) Immunoblot analysis of the phosphorylation ULK1 (S757), S6K (T389), 4EBP1 (S65), and eIF2α (S51) in parental HeLa (WT), LC3TKO, GBRPTKO, and hexaKO cells treated with 4 mM LLOMe for 30 min. (B ⅰ–ⅳ) Quantification of phosphorylation of ULK1 (S757; i), S6K (T389; ii), 4EBP1 (S65; iii), and eIF2α (S51; iv) in A; Quantification, n = 3. (C) Quantification by HCM and confocal microscopy analysis of overlaps between mTOR and LAMP2 in parental HeLa (WT), LC3TKO, GBRPTKO, and hexaKO cells treated with 4 mM LLOMe for 30 min. HCM images in Fig. S4 M. Scale bar, 5 µm. (D) Quantification by HCM of G3BP1 puncta. Parental HeLa (WT), LC3TKO, GBRPTKO, and hexaKO cells were treated with 4 mM LLOMe for 30 min. Red masks, computer-identified G3BP1 puncta. Data, means ± SEM; HCM: n ≥ 3 (each experiment: 500 valid primary objects/cells per well, ≥5 wells/sample). †, P ≥ 0.05 (not significant); **, P < 0.01, ANOVA. See also Fig. S4. Source data are available for this figure: SourceData F8.
We then wondered whether eIF2α phosphorylation, which is a marker of canonical SG formation (Riggs et al., 2020) and is strongly induced by lysosomal damage (Fig. 1, H; and Fig. S1, H and J), might also be affected by GABARAPs. However, contrary to our expectations, the mATG8s subgroup or mATG8s as a whole were not affecting eIF2α response to lysosomal damage (Fig. 8, A and B iv). This suggests a separation of functions of GABARAPs in mTOR inactivation vs. SG formation. In keeping with this interpretation, SG formation in response to lysosomal damage was only increased in hexaKO and GBRPTKO cells (Fig. 8 D). All three GABARAPs individually were capable of suppressing mTOR inactivation and SG formation in GBRPTKO HeLa cells elicited by lysosomal damage (Fig. S5, A and B). In conclusion, GABARAPs control mTOR inactivation and independently affect SG levels by redistributing NUFIP2 to act upon Ragulator and mTOR on damaged lysosomes.
Atg8ylation participates in mTOR inactivation in response to lysosomal damage. (A) Quantification by HCM of overlaps between mTOR and LAMP2 in HeLa (WT), GBRPTKO, and GBRPTKO transfected with GFP-GABARAP/GABARAPL1/GABARAPL2. Cells treated with 4 mM LLOMe for 30 min. White masks, algorithm-defined cell boundaries. Yellow masks, computer-identified overlap of mTOR and LAMP2. (B) Quantification by HCM of G3BP1 puncta in HeLa (WT), GBRPTKO, and GBRPTKO transfected with GFP-GABARAP/GABARAPL1/GABARAPL2. Cells were treated with 4 mM LLOMe for 30 min. Red masks, computer-identified G3BP1 puncta. (C) Immunoblot analysis of GABARAP (GABA) lipidation in U2OS cells treated with 2 mM LLOMe for indicated time points. (D) Immunofluorescence confocal microscopy imaging of GFP-GABARAP and LAMP2. U2OS cells overexpressing GFP-GABARAP were treated with 2 mM LLOMe for 30 min and stained for endogenous LAMP2. Scale bar, 5 µm. (E) WB analysis of ATG9KO, ATG3KO in Huh7 cells. (F) WB analysis of FIP200KO, ATG16L1KO in Huh7 cells. (G) WB analysis of ATG3KO, ATG13KO in HeLa cells. (H) HCM images of Fig. 9 A. Yellow masks, computer-identified overlap of mTOR and LAMP2. (I) HCM images of Fig. 9 C. Yellow masks, computer-identified overlap of mTOR and LAMP2. (J) HCM images of Fig. 9 E. Yellow masks, computer-identified overlap of mTOR and LAMP2. (K) WB analysis of indicated proteins in ATG3KO, ATG16L1KO Huh7 cells. (L) Confocal microscopy imaging (i) and quantification by HCM (ii) of overlaps between mTOR and LAMP2 in parental Huh7 (WT) and ATG3 knockout Huh7 cells (ATG3KO) transfected with scrambled siRNA as control (SCR) or NUFIP2 siRNA (NUFIP2KD). Cells were treated with 2 mM LLOMe for 30 min. WB analysis of indicated protein in iii. (M) WB analysis of the expression of GFP-SARS-CoV-2ORF3a in HeLa Flp-InTetON GFP-SARS-CoV-2ORF3a cells induced by tetracycline (Tet) for 16 h. (N) Immunoblot analysis of interaction between GCN1 and GFP-ORF3a in HEK293T Flp-InTetON GFP-SARS-CoV-2ORF3a cells induced by 1 µg/ml tetracycline (Tet) for 16 h. Cell lysates were immunoprecipitated with anti-GFP antibody and immunoblotted for endogenous GCN1. (O) Immunoblot analysis of interaction between GCN1 and ORF3a in HEK293T cells transfected with GFP or GFP-ORF3a. Cell lysates were immunoprecipitated with anti-GFP antibody and immunoblotted for endogenous GCN1. Ctrl, control (untreated cells). Data, means ± SEM; HCM: n ≥ 3 (each experiment: 500 valid primary objects/cells per well, ≥5 wells/sample). †, P ≥ 0.05 (not significant); **, P < 0.01, ANOVA. See also Figs. 9 and 10. Source data are available for this figure: SourceData FS5.
Atg8ylation participates in mTOR inactivation in response to lysosomal damage. (A) Quantification by HCM of overlaps between mTOR and LAMP2 in HeLa (WT), GBRPTKO, and GBRPTKO transfected with GFP-GABARAP/GABARAPL1/GABARAPL2. Cells treated with 4 mM LLOMe for 30 min. White masks, algorithm-defined cell boundaries. Yellow masks, computer-identified overlap of mTOR and LAMP2. (B) Quantification by HCM of G3BP1 puncta in HeLa (WT), GBRPTKO, and GBRPTKO transfected with GFP-GABARAP/GABARAPL1/GABARAPL2. Cells were treated with 4 mM LLOMe for 30 min. Red masks, computer-identified G3BP1 puncta. (C) Immunoblot analysis of GABARAP (GABA) lipidation in U2OS cells treated with 2 mM LLOMe for indicated time points. (D) Immunofluorescence confocal microscopy imaging of GFP-GABARAP and LAMP2. U2OS cells overexpressing GFP-GABARAP were treated with 2 mM LLOMe for 30 min and stained for endogenous LAMP2. Scale bar, 5 µm. (E) WB analysis of ATG9KO, ATG3KO in Huh7 cells. (F) WB analysis of FIP200KO, ATG16L1KO in Huh7 cells. (G) WB analysis of ATG3KO, ATG13KO in HeLa cells. (H) HCM images of Fig. 9 A. Yellow masks, computer-identified overlap of mTOR and LAMP2. (I) HCM images of Fig. 9 C. Yellow masks, computer-identified overlap of mTOR and LAMP2. (J) HCM images of Fig. 9 E. Yellow masks, computer-identified overlap of mTOR and LAMP2. (K) WB analysis of indicated proteins in ATG3KO, ATG16L1KO Huh7 cells. (L) Confocal microscopy imaging (i) and quantification by HCM (ii) of overlaps between mTOR and LAMP2 in parental Huh7 (WT) and ATG3 knockout Huh7 cells (ATG3KO) transfected with scrambled siRNA as control (SCR) or NUFIP2 siRNA (NUFIP2KD). Cells were treated with 2 mM LLOMe for 30 min. WB analysis of indicated protein in iii. (M) WB analysis of the expression of GFP-SARS-CoV-2ORF3a in HeLa Flp-InTetON GFP-SARS-CoV-2ORF3a cells induced by tetracycline (Tet) for 16 h. (N) Immunoblot analysis of interaction between GCN1 and GFP-ORF3a in HEK293T Flp-InTetON GFP-SARS-CoV-2ORF3a cells induced by 1 µg/ml tetracycline (Tet) for 16 h. Cell lysates were immunoprecipitated with anti-GFP antibody and immunoblotted for endogenous GCN1. (O) Immunoblot analysis of interaction between GCN1 and ORF3a in HEK293T cells transfected with GFP or GFP-ORF3a. Cell lysates were immunoprecipitated with anti-GFP antibody and immunoblotted for endogenous GCN1. Ctrl, control (untreated cells). Data, means ± SEM; HCM: n ≥ 3 (each experiment: 500 valid primary objects/cells per well, ≥5 wells/sample). †, P ≥ 0.05 (not significant); **, P < 0.01, ANOVA. See also Figs. 9 and 10. Source data are available for this figure: SourceData FS5.
Atg8ylation plays a role in mTOR inhibition and competes with SG formation during lysosomal damage
Recently, the concept of atg8ylation has been introduced as a general membrane stress and remodeling response and a unifying mechanism for various roles of mATG8 lipidation in diverse processes beyond their conventional association with canonical autophagy (Deretic and Lazarou, 2022). Thus, we tested whether atg8ylation plays a role in mTOR inactivation by mATG8s, specifically GABARAPs, in conjunction with their binding partner NUFIP2. GABARAP was lipidated during LLOMe treatment (Fig. S5 C). Atg8ylation of damaged lysosomes was further documented by the lipidated form of GABARAP in LysoIP preparations (Fig. 1, G and Fig. 4 E) and by IF (Fig. S5 D). In pairwise comparisons with the atg8ylation (mATG8 lipidation) mutant ATG3KO in Huh7 cells, ATG9A (Claude-Taupin et al., 2021), a canonical autophagy gene, did not affect either mTOR inactivation or SG formation in response to lysosomal damage, whereas ATG3KO (Fig. S5 E) countered mTOR inactivation (quantified by HCM of mTOR desorption from lysosomes) and enhanced SG formation (Fig. 9, A and B; and Fig. S5 H). In another pairwise comparison between the atg8ylation mutant ATG16L1KO and a canonical autophagy factor FIP200KO (Fig. S5 F), ATG16L1KO reduced mTOR inactivation and enhanced SG formation, whereas FIP200KO did not (Fig. 9, C and D; and Fig. S5 I). In the last comparison employed between ATG3KO (atg8ylation mutant) and ATG13KO (canonical autophagy mutant) in HeLa cells (Fig. S5 G), the above relationships held up, i.e., ATG3KO decreased mTOR inactivation and enhanced SG formation in response to lysosomal damage, whereas ATG13KO did not (Fig. 9, E and F; and Fig. S5 J). In ATG3KO and ATG16L1KO cells, G3BP1 protein levels remained unchanged (Fig. S5 K). Knockdown of NUFIP2 in ATG3KO cells did not further enhance the partial rescue of mTOR dissociation from lysosomes in ATG3KO cells (Fig. S5 L), suggesting that atg8ylation and NUFIP2 act along the same pathway.
In conclusion, atg8ylation is important for mTOR inactivation during lysosomal damage, it antagonizes SG formation in response to the same stimulus and competes for factors such as NUFIP2. The competition model (Fig. 9 G) is consistent with the absence of mATG8s’ effects on eIF2α phosphorylation during lysosomal damage.
Atg8ylation plays a role in mTOR inhibition and competes with SG formation during lysosomal damage. (A) Quantification by HCM and confocal microscopy imaging of overlaps between mTOR and LAMP2 in parental Huh7 (WT), ATG9KO, and ATG3KO treated with 2 mM LLOMe for 30 min. HCM images in Fig. S5 H. Scale bar, 5 µm. (B) Quantification by HCM of G3BP1 puncta. Parental Huh7 (WT), ATG9KO, and ATG3KO were treated with 2 mM LLOMe for 30 min. White masks, algorithm-defined cell boundaries. Red masks, computer-identified G3BP1 puncta. (C) Quantification by HCM and confocal microscopy imaging of overlaps between mTOR and LAMP2 in parental Huh7 (WT), FIP200KO, and ATG16L1KO treated with 2 mM LLOMe for 30 min. HCM images in Fig. S5 I. Scale bar, 5 µm. (D) Quantification by HCM of G3BP1 puncta. Parental Huh7 (WT), FIP200KO, and ATG16L1KO were treated with 2 mM LLOMe for 30 min. Red masks, computer-identified G3BP1 puncta. (E) Quantification by HCM and confocal microscopy imaging of overlaps between mTOR and LAMP2 in parental HeLa (WT), ATG13KO, and ATG3KO treated with 4 mM LLOMe for 30 min. HCM images in Fig. S5 J. Scale bar, 5 µm. (F) Quantification by HCM of G3BP1 puncta. Parental HeLa (WT), ATG13KO, and ATG3KO were treated with 4 mM LLOMe for 30 min. Red masks, computer-identified G3BP1 puncta. (G) Schematic summary of the findings in this study. Ctrl, control (untreated cells). Data, means ± SEM; HCM: n ≥ 3 (each experiment: 500 valid primary objects/cells per well, ≥5 wells/sample). †, P ≥ 0.05 (not significant); **, P < 0.01, ANOVA. See also Fig. S5. Source data are available for this figure: SourceData F9.
Atg8ylation plays a role in mTOR inhibition and competes with SG formation during lysosomal damage. (A) Quantification by HCM and confocal microscopy imaging of overlaps between mTOR and LAMP2 in parental Huh7 (WT), ATG9KO, and ATG3KO treated with 2 mM LLOMe for 30 min. HCM images in Fig. S5 H. Scale bar, 5 µm. (B) Quantification by HCM of G3BP1 puncta. Parental Huh7 (WT), ATG9KO, and ATG3KO were treated with 2 mM LLOMe for 30 min. White masks, algorithm-defined cell boundaries. Red masks, computer-identified G3BP1 puncta. (C) Quantification by HCM and confocal microscopy imaging of overlaps between mTOR and LAMP2 in parental Huh7 (WT), FIP200KO, and ATG16L1KO treated with 2 mM LLOMe for 30 min. HCM images in Fig. S5 I. Scale bar, 5 µm. (D) Quantification by HCM of G3BP1 puncta. Parental Huh7 (WT), FIP200KO, and ATG16L1KO were treated with 2 mM LLOMe for 30 min. Red masks, computer-identified G3BP1 puncta. (E) Quantification by HCM and confocal microscopy imaging of overlaps between mTOR and LAMP2 in parental HeLa (WT), ATG13KO, and ATG3KO treated with 4 mM LLOMe for 30 min. HCM images in Fig. S5 J. Scale bar, 5 µm. (F) Quantification by HCM of G3BP1 puncta. Parental HeLa (WT), ATG13KO, and ATG3KO were treated with 4 mM LLOMe for 30 min. Red masks, computer-identified G3BP1 puncta. (G) Schematic summary of the findings in this study. Ctrl, control (untreated cells). Data, means ± SEM; HCM: n ≥ 3 (each experiment: 500 valid primary objects/cells per well, ≥5 wells/sample). †, P ≥ 0.05 (not significant); **, P < 0.01, ANOVA. See also Fig. S5. Source data are available for this figure: SourceData F9.
Diverse pathological agents induce lysosomal damage and SG formation response
We tested whether the above molecular and cell biological processes associated with lysosomal damage are observed in cells affected by agents causing or modeling pathology and disease. Mtb can permeabilize intracellular vacuoles in which it resides, affecting the endolysosomal system in infected macrophages (Manzanillo et al., 2012) and causing lysosomal damage (Chauhan et al., 2016). Hence, we wondered whether virulent Mtb Erdman with its membrane penetrating and lysosome damaging capabilities can induce SGs upon infection of host cells. Murine BMMs were infected with Mtb Erdman WT, and SG formation was quantified after 20 h of infection (Fig. 10 A). As a positive control for endomembrane damage we monitored the ubiquitin response (Chauhan et al., 2016; Yoshida et al., 2017), which paralleled that of SG formation (Fig. 10 A). In contrast, when BMMs were infected with Mtb Erdman mutant in ESX-1, a factor required for permeabilization of endomembranes by Mtb (Manzanillo et al., 2012), both SG formation and ubiquitin puncta formation response were diminished (Fig. 10 A). We further modeled events associated with phagocytosis of membrane permeabilizing bacteria using FuGENE HD–coated latex beads (with FuGENE HD used as a membrane damaging agent), and observed similar SG formation and ubiquitin puncta responses in U2OS cells (Fig. 10 B).
Diverse pathological agents induce lysosomal damage and SG formation response. (A) Quantification by HCM of G3BP1 and ubiquitin (Ubi) puncta in BMM cells infected with Mtb strain Erdman or its ESX-1 mutant at MOI = 10 for 20 h. White masks, algorithm-defined cell boundaries. Red masks, computer-identified G3BP1 puncta. Green masks, computer-identified ubiquitin puncta. (B) Quantification by HCM of G3BP1 and ubiquitin puncta in U2OS cells treated with FuGENE HD-coated latex beads (Polybead Amino Microsphere) for 16 h. Green masks, computer-identified G3BP1 puncta. Red masks, computer-identified ubiquitin puncta. (C) Quantification by HCM of G3BP1 and Gal3 puncta in U2OS cells treated with 1 or 10 µg/ml Tau oligomer overnight. Red masks, computer-identified G3BP1 puncta. Green masks, computer-identified Gal3 puncta. (D) Quantification by HCM of G3BP1 and ubiquitin puncta in the constructed HeLa Flp-InTetON GFP-SARS-CoV-2ORF3a cells induced by 1 µg/ml tetracycline (Tet). Red masks, computer-identified G3BP1 puncta. Pink masks, computer-identified ubiquitin puncta. Data, means ± SEM; HCM: n ≥ 3 (each experiment: 500 valid primary objects/cells per well, ≥5 wells/sample). †, P ≥ 0.05 (not significant); *, P < 0.05; **, P < 0.01, ANOVA. See also Fig. S5. Source data are available for this figure: SourceData F10.
Diverse pathological agents induce lysosomal damage and SG formation response. (A) Quantification by HCM of G3BP1 and ubiquitin (Ubi) puncta in BMM cells infected with Mtb strain Erdman or its ESX-1 mutant at MOI = 10 for 20 h. White masks, algorithm-defined cell boundaries. Red masks, computer-identified G3BP1 puncta. Green masks, computer-identified ubiquitin puncta. (B) Quantification by HCM of G3BP1 and ubiquitin puncta in U2OS cells treated with FuGENE HD-coated latex beads (Polybead Amino Microsphere) for 16 h. Green masks, computer-identified G3BP1 puncta. Red masks, computer-identified ubiquitin puncta. (C) Quantification by HCM of G3BP1 and Gal3 puncta in U2OS cells treated with 1 or 10 µg/ml Tau oligomer overnight. Red masks, computer-identified G3BP1 puncta. Green masks, computer-identified Gal3 puncta. (D) Quantification by HCM of G3BP1 and ubiquitin puncta in the constructed HeLa Flp-InTetON GFP-SARS-CoV-2ORF3a cells induced by 1 µg/ml tetracycline (Tet). Red masks, computer-identified G3BP1 puncta. Pink masks, computer-identified ubiquitin puncta. Data, means ± SEM; HCM: n ≥ 3 (each experiment: 500 valid primary objects/cells per well, ≥5 wells/sample). †, P ≥ 0.05 (not significant); *, P < 0.05; **, P < 0.01, ANOVA. See also Fig. S5. Source data are available for this figure: SourceData F10.
We and others have reported that protopathic tau induces lysosomal damage (Jia et al., 2020b; Papadopoulos et al., 2017). Treatment of U2OS cells with protopathic tau (Jia et al., 2020b) induced both SG formation response and Gal3 puncta formation (Fig. 10 C), the latter being used as a lysosome damage marker (Aits et al., 2015).
Finally, we tested a factor encoded by SARS-CoV-2, ORF3a. The corresponding ORF3a from SARS-CoV (SARS-CoV-1) is known to cause lysosomal damage (Yue et al., 2018) and evidence for reduced acidification of lysosomes with SARS-CoV-2ORF3a has been presented (Ghosh et al., 2020). We generated a stable HeLa cell line with tetracycline controllable with Flp-In GFP-SARS-CoV-2ORF3a (Fig. S5 M). Upon induction of SARS-CoV-2ORF3a expression with tetracycline, we observed SG formation and ubiquitin puncta response, a well-established marker of lysosomal damage (Papadopoulos et al., 2017), consistent with ORF3a causing lysosomal damage and that this in turn induces SG formation (Fig. 10 D). Thus, the relationships presented in this work are of relevance for multiple pathogenic insults of significance for major human diseases.
We carried out ORF3a interactome analysis by constructing HEK293T Flp-InTetON GFP-SARS-CoV-2ORF3a cells and carrying out DIA proteomic analysis with immunopurified GFP-ORF3a (Table S1, Tab 10). The HOPS component VPS39 was observed as one of the enriched ORF3a interactors in our proteomic study (Table S1, Tab 10). VPS39 has been reported in global SARS-CoV-2 interactome studies by others (Gordon et al., 2020; Stukalov et al., 2021). The effects of ORF3a on HOPS have also been validated in the context of lysosomal function within the autophagy pathway (Miao et al., 2021). STRING functional association protein networks analyses indicated 64 and 85 entries assigned to lysosomal membranes and lysosomes (Table S1, Tab 11). Among most abundant proteins found in our DIA proteomic analysis was GCN1, an upstream regulator of GCN2–eIF2α–ATF4 axis during repression of global protein synthesis (Pochopien et al., 2021; Table S1, Tab 10). We validated SARS-CoV2ORF3a interaction with GCN1 in co-IPs (Fig. S5, N and O). These analyses indicate that ORF3a imposes a concerted role on lysosomal function and host cell translational apparatus, which is reflected in the known effects of coronaviruses (Nakagawa et al., 2018) and SARS-CoV-2 (Gordon et al., 2020) on SG formation in host cells.
Discussion
In this study, we report that lysosomal damage induces SG formation as a part of cellular homeostatic responses to stressors. SG formation complements the reported mTOR inactivation during lysosomal damage (Eapen et al., 2021; Goodwin et al., 2021; Jia et al., 2018; Koerver et al., 2019). Together, SG formation and mTOR inhibition cover two key aspects of protein translation during stress (Costa-Mattioli and Walter, 2020; Lu et al., 2004). We found that mTOR inactivation and SG formation are coupled through GABARAPs and atg8ylation (Deretic and Lazarou, 2022).
The elements shared between SGs and mTOR regulations are the individual SG proteins NUFIP2 and G3BP1. They affect mTOR activity independently of SG condensates as demonstrated by cycloheximide experiments whereby SGs are not formed but NUFIP2 and G3BP1 are recruited to damaged lysosomes where they inhibit mTOR. This role of NUFIP2 depends on atg8ylation and interactions with GABARAPs. Both NUFIP2, tested here, and G3BP1 reported elsewhere (Prentzell et al., 2021) play a role in inhibiting mTOR under different conditions through complementary mechanisms (Prentzell et al., 2021). The convergence of NUFIP2’s and Gal8’s actions upon mTOR integrates escalating signals during lysosomal damage, i.e., membrane stress leading to membrane atg8ylation (Deretic and Lazarou, 2022) and overt membrane damage with exposure of lumenal glycans recognized by Gal8 (Jia et al., 2018).
G3BP1 is an essential component acting redundantly with G3BP2 in SG formation (Yang et al., 2020), whereas our quantitative data suggest that NUFIP2 affects how robust the SG formation is. Of significance, G3BP1 and NUFIP2 directly interact as established here in GST pulldowns. Both proteins bind to GABARAPs, which may be essential for their function in mTOR inactivation. GABARAPs do not contribute to SG formation although they have been found in SGs (Markmiller et al., 2018).
The positive role of GABARAPs and atg8ylation in mTOR inactivation and their negative role in SG formation reflect a competition for a limited supply of GABARAPs, balancing shutting down of the cap-dependent translation while favoring selective translation of stress response systems. Considering the canonical (McCormick and Khaperskyy, 2017) and noncanonical (Emara et al., 2012; Fujimura et al., 2012) types of SG responses, they may fit in the continuum of how cells balance mTOR inactivation with the eIF2α phosphorylation.
As shown here, SARS-CoV-2ORF3a induces SGs along with the lysosomal damage. SGs are of significance in viral infections (Lindquist et al., 2010; McCormick and Khaperskyy, 2017; Panas et al., 2012). Viral infections can activate PKR, which phosphorylates eIF2α (Srivastava et al., 1998), and trigger SG formation to inhibit viral translation (Balachandran et al., 2000; Williams, 2001). In our study, PKR was the key upstream kinase affecting eIF2α phosphorylation in response to lysosomal damage and was at least in part on lysosomes. Our finding that virulent Mtb induces SG response in macrophages may be of relevance in pathogenesis of tuberculosis. SGs are a component of a broader ISR (Costa-Mattioli and Walter, 2020; Lu et al., 2004), whereas PKR-dependent eIF2α phosphorylation (Lu et al., 2004) is an aspect of Mtb pathology in necrotic granulomas (Bhattacharya et al., 2021).
How PKR senses lysosomal perturbations remains to be determined. ATG16L1 is a key component of the E3 ligase driving membrane atg8ylation (Kumar et al., 2021b). ATG16L1 is known to interact with V-ATPase (Xu et al., 2019); however, our proteomic data indicate synchronous reduction of V-ATPase subunits on lysosomes during early damage (Table S1, Tab 12). Alternatively, ATG16L1 can bind to ubiquitin (Fujita et al., 2013) and there is a strong ubiquitylation response associated with lysosomal damage (Papadopoulos et al., 2017). Additional work is needed to address the exact mechanism of ATG16L1 increase on damaged lysosomes.
In summary, GABARAPs and atg8ylation balance two important aspects of translational suppression through mTOR and SGs. Atg8ylation and mATG8s play a hitherto unrecognized function in the fine tuning of translational arrest at the interface with the ISR in cells exposed to sources of lysosomal stress in various disease and physiological conditions.
Materials and methods
Antibodies and reagents
Antibodies from Cell Signaling Technology were G3BP1 (1:1,000 for WB; 1:200 for IF), TIA1 (D1Q3K; 1:1,000 for WB; 1:200 for IF), DUSP1 (E6T5S; 1:1,000 for WB), ERK2 (1:1,000 for WB), Phospho-eIF2α (Ser51; 1:1,000 for WB), eIF2α (1:1,000 for WB), Phospho-p70 S6 Kinase (Thr389; 108D2; 1:1,000 for WB), p70 S6 Kinase (49D7; 1:1,000 for WB), Phospho-GCN2 (Thr899; E1V9M; 1:1,000 for WB), GCN2 (1:1,000 for WB), mTOR (7C10; 1:1,000 for WB; 1:400 for IF), Raptor (24C12; 1:1,000 for WB), LAMTOR1 (D11H6; 1:1,000 for WB), Phospho-ULK1 (Ser757; 1:1,000 for WB), ULK1 (D8H5; 1:1,000 for WB), Tuberin/TSC2 (D93F12; 1:1,000 for WB), RagA (D8B5; 1:1,000 for WB), GABARAPL2 (D1W9T; 1:1,000 for WB), Phospho-TFEB (Ser211; E9S8N; 1:1,000 for WB), TFEB (#4240; 1:1,000 for WB; 1:200 for IF), ATG13 (E1Y9V; 1:1,000 for WB), FIP200 (D10D11; 1:1,000 for WB), Atg9A (D4O9D; 1:1,000 for WB), HA-Tag (C29F4; 1:500 for IF), and normal rabbit IgG. Antibodies from Abcam were GFP (ab290; 1:1,000 for WB), GFP(ab38689; for IP) and Anti-β Tubulin (1:1,000 for WB). Antibodies from Sigma Aldrich: FLAG M2 (F1804; for IP and 1:1,000 for WB), phospho TFEB (Ser142; 1:1,000 for WB), anti-ATG7 (1:1,000 for WB), anti-ATG3 (1:1,000 for WB), anti-NUFIP2 (#AV51676; 1:1,000 for WB), anti-NUFIP2 (#HPA067443; 1:100 for IF), and anti-Phospho-ERK2 T185/187 (1:1,000 for WB). Other antibodies used in this study were from the following sources: Gal8 (sc-28254; 1:200 for WB) and β-Actin (C4; 1:1,000 for WB), normal mouse IgG from Santa Cruz Biotechnology; LAMP2 (H4B4; 1:500 for IF) from Developmental Studies Hybridoma Bank of University of Iowa; Anti-Multi Ubiquitin (1:500 for IF) and ATG16L1(PM040; 1:400 for IF) from MBL International. Gal3 (1:1,000 for WB; 1:500 for IF); ALIX (1:200 for IF) from BioLegend. Alexa Fluor 488, 568, 647 (1:500 for IF), G3BP1 (1:1,000 for WB, 1:200 for IF) and secondary antibodies from ThermoFisher Scientific. DMEM, Opti-MEM Reduced Serum Media, and EBSS media from Life Technologies of Thermo Fisher Scientific.
Cells and cell lines
HEK293T, HeLa, and U2OS cells were from American Type Culture Collection. BMMs were isolated from femurs of ATG9fl/fl LysM-Cre–negative mice and cultured in DMEM supplemented with mouse macrophage colony stimulating factor (#5228; CST). TSC2 knockout HeLa cells were from David M. Sabatini (Massachusetts Institute of Technology, Cambridge, MA). HEK293T cells stably expressing FLAG-Metap2/FLAG-p14 and constitutively active RagBQ99L were from Roberto Zoncu (University of California Berkeley, Berkeley, CA). HeLa mATG8 KO cells are from Michael Lazarou (Monash University, Melbourne, Australia). Flp-In cell lines were generated using constructs from Terje Johansen. Huh7 cells are from Rocky Mountain laboratories. Cell lines for LysoIP were generated using constructs obtained from Addgene and the details below. Knockout cell lines were generated by CRISPR/Cas9-mediated knockout system.
Plasmids, siRNAs, and transfection
Plasmids used in this study, such as NUFIP2, cloned into pDONR221 using BP cloning, and expression vectors were made utilizing LR cloning (Gateway, Thermo Fisher Scientific) in appropriate pDEST vectors for immunoprecipitation assay.
NUFIP2 mutants were generated utilizing the QuikChange site-directed mutagenesis kit (Agilent) and confirmed by sequencing (Genewiz). Plasmid transfections were performed using the ProFection Mammalian Transfection System, FuGENE HD Transfection Reagent (Promega), or Lipofectamine 2000 Transfection Reagent (Thermo Fisher Scientific). siRNAs were delivered into cells using either Lipofectamine RNAiMAX (Thermo Fisher Scientific).
Oligonucleotide sequences used in this study are as follows: NUFIP2 Gateway-sense (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGAGGAGAAGCCCGGCCAGCCACAGCC-3′), NUFIP2 Gateway-anti-sense (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTTATTGATCTGGACTATCCATGGCTTC-3′); NUFIP2 ΔNLS sense (5′-GATGGTAGTGGATCTGAGAGCAATAGTGCCAAGGGTTGTGAAAAC-3′); GABARAP-G (C-terminal glycine residue exposed mutant) sense (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCAAGTTCGTGTACAAAGAAG-3′), and GABARAP-G (C-terminal glycine residue exposed mutant) anti-sense (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTCAACCGTAGACACTTTCG-3′).
siRNAs were from Horizon Discovery: siGENOME Non-Targeting Control siRNA (D-001210-01-05); siGENOME human NUFIP2 SMARTpool siRNA (L-021280-01-0005); siGENOME human G3BP1 SMARTpool siRNA (L-012099-00-0005); siGENOME human G3BP2 SMARTpool siRNA (L-015329-01-0005); siGENOME human DUSP1 SMARTpool siRNA (L-003484-02-0005); siGENOME human EIF2AK1(HRI) SMARTpool siRNA (M-005007-00-0005); siGENOME human EIF2AK2 (PKR) SMARTpool siRNA (M-003527-00-0005); siGENOME human EIF2AK3 (PERK) SMARTpool siRNA (M-004883-03-0005); siGENOME human EIF2AK4 (GCN2) SMARTpool siRNA (M-005314-02-0005); siGENOME human RNASET2 SMARTpool siRNA (M-009282-01-0005); and siGENOME human TIA1 SMARTpool siRNA (L-013042-02-0005).
Generation of NUFIP2 CRISPR mutant cells
NUFIP2 knockout cells (Huh7NUFIP2-KO) were generated by CRISPR/Cas9-mediated knockout system. The lentiviral vector lentiCRISPRv2 carrying both Cas9 enzyme and a gRNA targeting NUFIP2 (gRNA1: 5′-AAGTTTGATGATCGGCCCAA-3′/gRNA2: 5′-TAGCCTGAAGCAGACTGTAA-3′; VectorBuilder) was transfected into HEK293T cells together with the packaging plasmids psPAX2 and pCMV-VSV-G at the ratio of 5:3:2. Two days after transfection, the supernatant containing lentiviruses was collected. Huh7 cells were infected by the mixed lentiviruses containing gRNA1 or gRNA 2. 36 h after infection, the cells were selected with puromycin (1 mg/ml) for 1 wk in order to select NUFIP2 knockout cells. NUFIP2 knockout was confirmed by WB. Selection of single clones was performed by dilution in 96-well plates and mutants confirmed by Western blots (Fig. S3 G).
Generating Flp-InTetON GFP-SARS-CoV-2ORF3a cell line
Transfected Hela/HEK293T Flp-In host cells with ORF3a reconstructed plasmid and the pOG44 expression plasmid at ration of 9:1. 24 h after transfection, the cells were washed , and fresh medium added. 48 h after transfection, the cells were split into fresh medium (25% confluency), incubated at 37°C for 2–3 h until they have attached to the culture dish. Then, the medium was removed and added with fresh medium containing hygromycin. The cells were fed with selective medium every 3–4 d until single-cell clone could be identified. Hygromycin-resistant clones were expanded. The clones were incubated in the medium containing 1 or 10 μg/ml tetracycline overnight and examined by WB for the expressing of ORF3a.
Bead transfection assay
Bead transfections were performed as previously reported (Fujita et al., 2013). Transfection reagent-coated beads were prepared by mixing the beads (Polybead Amino Microspheres 0.20 μm; PolySciences, Inc.) with FuGENE HD Transfection Reagent (E2311; Promega), according to the manufacturer’s instructions except that bead suspension was used instead of DNA solution. The resulting bead mixture was mixed with growth medium and further added to cells by replacing the medium at the final concentration of 0.1 μl bead stock/cell. After incubation with the bead mixture overnight at 37°C in a CO2 incubator, the cells were washed twice to remove unattached beads and fixed for the HCM analysis.
Poly(A) RNA FISH analysis
Cells were fixed in 4% paraformaldehyde for 5 min, paraformaldehyde aspirated, and 100% cold methanol added to each well for 10 min. Methanol was replaced with 70% ethanol and incubated for 10 min. The ethanol was aspirated and 1 M Tris, pH 8.0 added to each well for 5 min. After Tris removal, hybridization buffer was added containing the dilution of 5′-labeled Cy3-Oligo-dT(30) stock (GeneLink) for a final concentration of 1 ng/μl. Hybridization was carried out at 37°C for 1 h. After hybridization, samples were washed once with 4 × SSC and then once with 2 × SSC. Incubation with primary antibodies was in 2 × SSC + 0.1% Triton-X-100 for 2 h, washed three times with 2 × SSC, and then incubated with secondary antibodies for 1 h at room temperature. Coverslips were mounted using Prolong Gold Antifade Mountant (Thermo Fisher Scientific).
Nuclear extraction assay
Nuclear extraction followed manufacturer’s instructions (NBP2-29447). Cells (2 × 106) were washed twice with cold 1 × PBS. Cell pellets were resuspended in 200 μl 1 × Hypotonic Buffer (20 mM Tris-HCl [pH 7.4]; 10 mM NaCl; 3 mM MgCl2) on ice for 15 min, after which 10 μl detergent (10% NP40) was added and samples vortexed for 10 s at the highest setting. After centrifugation of the homogenate for 10 min at 3,000 rpm at 4°C, the supernatant was transfered and saveed. This supernatant contains the cytoplasmic fraction. The pellet is the nuclear fraction. The nuclear pellet was resuspended in 20 μl complete Cell Extraction Buffer (10 mM Tris-HCl [pH 7.4]; 100 mM NaCl; 1 mM Na2EDTA; 1 mM EGTA; 10% Glycerol; 0.5% Sodium deoxycholate; 1% Triton X-100; 0.1% SDS; 20 mM Sodium pyrophosphate; 2 mM Na3VO4; 1 mM NaF; 1 mM PMSF) for 30 min on ice with vortexing at 10-min intervals. After centrifugation for 30 min at 14,000× g at 4°C, the supernatant (nuclear fraction) was transfered to a clean microcentrifuge tube, mixed with 2 × Laemmli sample buffer (Bio-Rad), and subjected to immunoblot analysis.
LysoIP assay
Lentiviruses constructs for generating stable LysoIP cells were purchased from Addgene. HEK293T cells were transfected with pLJC5-TMEM192-3xHA or pLJC5-TMEM192-2xFLAG constructs in combination with pCMV-VSV-G and psPAX2 packaging plasmids; 60 h after transfection, the supernatant containing lentiviruses was collected and centrifuged to remove cells and then frozen at −80°C. To establish LysoIP stably expressing cell lines, cells were plated in 10-cm dish in DMEM with 10% FBS and infected with 500 μl of virus-containing media overnight, then add puromycin for selection.
Selected cells in 15-cm plates with 90% confluency were used for each LysoIP. Cells with or without treatment were quickly rinsed twice with PBS and then scraped in 1 ml of KPBS (136 mM KCl, 10 mM KH2PO4, pH 7.25 was adjusted with KOH) and centrifuged at 3,000 rpm for 2 min at 4°C. Pelleted cells were resuspended in 950 μl KPBS and reserved 25 μl for further processing of the whole-cell lysate. The remaining cells were gently homogenized with 20 strokes of a 2 ml homogenizer. The homogenate was then centrifuged at 3,000 rpm for 2 min at 4°C, and the supernatant was incubated with 100 μl of KPBS prewashed anti-HA magnetic beads (Thermo Fisher Scientific) on a gentle rotator shaker for 15 min. Immunoprecipitants were then gently washed three times with KPBS and eluted with 2 × Laemmli sample buffer (Bio-Rad) and subjected to immunoblot analysis.
LysoIP proteinase K protection assay
The lysosomes were isolated by LysoIP assay and immunopurified by anti-HA magnetic beads (ThermoFisher). The protocols of proteinase K protection assay by (Zhang et al., 2015) and (Kimura et al., 2017) were followed. Briefly, immunoprecipitates were gently washed three times with KPBS and resuspended in 30 μl of B88 (20 mM Hepes-KOH, pH 7.2, 250 mM sorbitol, 150 mM potassium acetate, and 5 mM magnesium acetate) or B88 containing 30 μg/ml proteinase K with or without 0.5% Triton X-100, and stored on ice for 30 min. The reactions were stopped by adding 4 × Laemmli sample buffer (Bio-Rad) and then boiled for 10 min for immunoblot analysis.
LysoTracker assay
LysoTracker (LTR) Staining Solution was prepared by freshly diluting 2 μl of LTR stock solution (1 mM LysoTracker Red DND-99; Sigma Aldrich, L7528) in 1 ml of medium. 10 μl of LysoTracker Staining Solution was added to 90 μl of medium in each well in 96 well plates (final volume 100 μl per well, final concentration 0.2 μM LTR) and adherent cells incubated at 37°C for 30 min protected from light. Wells were rinsed gently by 1 × PBS and fixed in 4% paraformaldehyde for 2 min. Wells were washed once in 1 × PBS and nuceli stained with Hoechst 33342 for 2 min before analyzing the plates by HCM.
Protein translation assay
Cells in 96 well plates were subjected to indicated treatment and then stained with O-propargyl-puromycin (OPP) using the Click-iT Plus OPP Alexa Fluor 488 protein synthesis assay kit (Thermo Fisher Scientific) in accordance with the manufacturer’s guidelines. Cells were scanned by HCM, described below. We quantified by HCM the overall fluorescence intensity in cells of fluorescently labeled OPP. OPP is an alkyne analog of puromycin that following incorporation into polypeptides can be fluorescently labeled by Alexa 488 picolyl azide through a chemoselective ligation or “click” reaction, occurring between the picolyl azide dye and the OPP alkyne.
Murine tuberculosis infection assay
Mtb Erdman culture was prepared by thawing frozen stock aliquot and grown in 7H9 Middlebrook liquid medium supplemented with oleic acid, albumin, dextrose, and catalase (Becton Dickinson, Inc.), 0.5% glycerol, and 0.05% Tween 80. Cultures were grown at 37°C. BMMs were infected with Erdman at indicated MOI and incubated with full medium for 18 h. The infected BMMs were lysed and plated on 7H11 agar plates. CFU was enumerated 3–4 wk later.
GST pulldown assay
GST and GST-tagged proteins were produced in SoluBL21 Competent Escherichia coli (C700200; Genlantis) and purified by binding to Glutathionine Sepharose 4 Fast Flow beads (17-5132-01; GE Healthcare) while myc-tagged proteins were in vitro translated using the TNT T7 Reticulocyte Lysate System (14610; Promega) in the presence of 35S-methionine. 10 ml of translated protein were incubated with immobilized GST-tagged protein in NETN buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 1 mM EDTA, 0.5% NP-40) supplemented with cOmplete Mini EDTA-free protease inhibitor cocktail tablets (11836170001; Roche, 1 tablet/10 ml) for 1 h at 4°C followed by five times washing with NETN buffer. 2 × SDS gel loading buffer were added and protein separated by SDS-PAGE. Gels were stained with Coomassie Brilliant Blue R-250 Dye (20278; Thermo Fisher Scientific) to visualize the fusion proteins. Radioactive signals were detected by Fujifilm bioimaging analyzer BAS-5000 and quantified with ScienceLab ImageGuage software (Fujifilm).
HCM
Cells in 96-well plates were fixed in 4% paraformaldehyde for 5 min. Cells were then permeabilized with 0.1% saponin in 3% BSA for 30 min followed by incubation with primary antibodies for 2 h and secondary antibodies for 1 h. Hoechst 33342 staining was performed for 3 min. HCM with automated image acquisition and quantification was carried out using a Cellomics HCS scanner and iDEV software (Thermo Fisher Scientific). Automated epifluorescence image collection was performed for a minimum of 500 cells per well. Epifluorescence images were machine-analyzed using preset scanning parameters and object mask definitions. Hoechst 33342 staining was used for autofocus and to automatically define cellular outlines based on background staining of the cytoplasm. Primary objects were cells, and regions of interest or targets were algorithm-defined by shape/segmentation, maximum/minimum average intensity, total area, and total intensity, etc., to automatically identify puncta or other profiles within valid primary objects. All data collection, processing (object, region of interest, and target mask assignments), and analyses were computer-driven independently of human operators. HCM also provides a continuous variable statistic since it does not rely on parametric reporting cells as positive or negative for a certain marker above or below a puncta number threshold.
co-IP and immunoblotting assays
For co-IP, cells transfected with 8–10 μg of plasmids were lysed in ice-cold NP-40 buffer (Thermo Fisher Scientific) supplemented with protease inhibitor cocktail (11697498001; Roche) and 1 mM PMSF (93482; Sigma-Aldrich) for 30 min on ice. Lysates were centrifuged for 10 min at 10,000 g at 4°C. Supernatants were incubated with (2–3 μg) antibodies overnight at 4°C. The immune complexes were captured with Dynabeads (Thermo Fisher Scientific), followed by three times washing with 1 × PBS. Proteins bound to Dynabeads were eluted with 2 × Laemmli sample buffer (Bio-Rad) and subjected to immunoblot analysis.
For immunoblotting, lysates were centrifuged for 10 min at 10,000× g at 4°C. Supernatants were then separated on 4–20% Mini-PROTEAN TGX Precast Protein Gels (Biorad) and transferred to nitrocellulose membranes. Membranes were blocked in 3% BSA for 1 h at RT and incubated overnight at 4°C with primary antibodies diluted in blocking buffer. They were then incubated with an HRP-conjugated secondary antibody, and proteins were detected using ECL and developed using ChemiDoc Imaging System (Biorad) or x-ray film. Analysis and quantification of bands were performed using ImageJ software.
IF confocal microscopy and analysis
Cells were plated onto coverslips in 6-well plates. After treatment, cells were fixed in 4% paraformaldehyde for 5 min followed by permeabilization with 0.1% saponin in 3% BSA for 30 min. Cells were then incubated with primary antibodies for 2 h and appropriate secondary antibodies Alexa Fluor 488 or 568 (Thermo Fisher Scientific) for 1 h at RT. Coverslips were mounted using Prolong Gold Antifade Mountant (Thermo Fisher Scientific). Images were acquired using a confocal microscope (META; Carl Zeiss) equipped with a 63 3/1.4 NA oil objective, camera (LSM META; Carl Zeiss), and AIM software (Carl Zeiss).
Time-lapse imaging of cultured cells
U2OS cells expressing mCherry-G3BP1 and GFP-LAMP1 were incubated with 2 mM LLOMe for live-cell fluorescence image, which was performed using an inverted microscope (confocal TCS SP5, Leica, LAS AF version 2.6.0), a 63× PlanAPO oil-immersion objective lens (NA 1.4). Two-color time-lapse images were acquired at 340 ms intervals and z-stacks collapsed into 2D projections to generate movies.
RNAseq
HEK293T cells were treated with 1 mM LLOMe or 2 mM LOME for 30 min. Total RNA was extracted using RNeasy Kits (QIAGEN) following the manufacturer’s protocol. The RNAseq service was performed by LC Sciences. Total RNA quantity and purity were analyzed using a Bioanalyzer 2100 and a RNA 6000 Nano LabChip kit (Agilent), with RNA integrity values of >7.0. Poly(A) RNA was purified from total RNA (5 μg) using poly-T oligo-attached magnetic beads using two rounds of purification. Following purification, the mRNA was fragmented into small pieces using divalent cations under elevated temperature. Then, the cleaved RNA fragments were reverse-transcribed to create the final cDNA library in accordance with the protocol for the TruSeq RNA Sample Preparation v.2 (cat. no. RS-122-2001, RS-122-2002; Illumina); the average insert size for the paired-end libraries was 300 bp (±50 bp). The paired-end sequencing was carried out on an Illumina NovaseqTM 6000 at LC Sciences following the manufacturer’s recommended protocol. Using the Illumina paired-end RNAseq approach, the transcriptome was sequenced, generating a total of 2 × 150 million bp paired-end reads. This yielded gigabases of sequence. Before assembly, the low-quality reads (defined as [1] containing sequencing adaptors, [2] reads containing sequencing primer, and [3] nucleotide with a q quality score lower than 20) were removed. Sequencing reads were aligned to the reference genome using HISAT2 package. HISAT allows multiple alignments per read (up to 20 by default) and a maximum of two mismatches when mapping the reads to the reference. HISAT builds a database of potential splice junctions and confirms these by comparing the previously unmapped reads against the database of putative junctions. The mapped reads of each sample were assembled using StringTie. All transcriptomes from samples were merged to reconstruct a comprehensive transcriptome using perl scripts (LC Sciences). After the final transcriptome was generated, StringTie and edgeR were used to estimate the expression levels of all transcripts. StringTie was used to calculate the expression level for mRNAs through the fragments per kilobase million values. Differential gene expression was analyzed using the R package edgeR, which considers dispersions (that is, variations) between biological replicates. P-values were calculated using Fisher’s exact test adapted for overdispersed data; edgeR models read counts with negative binomial distribution64. The differentially expressed mRNAs and genes were selected by R package with log2(fold change) values of ≥1 or log2(fold change) values of ≤−1 and with statistical significance of P < 0.05.
LysoIP-LC/MS/MS DIA
LysoIP-LC/MS/MS data-independent acquisition (DIA) was performed at UC Davis Proteomics Core Facility. Protein samples on magnetic beads were washed four times with 200 μl of 50 mM Triethyl ammonium bicarbonate with a 20-min shake time at 4°C in between each wash. Roughly 2.5 mg of trypsin was added to the beads, and Triethyl ammonium bicarbonate mixture and the samples were digested over night at 800-rpm shake speed. After overnight digestion, the supernatant was removed and the beads were washed once with enough 50 mM ammonium bicarbonate to cover. After 20 min at a gentle shake, the wash is removed and combined with the initial supernatant. The peptide extracts were reduced in volume by vacuum centrifugation and a small portion of the extract was used for fluorometric peptide quantification (Thermo Fisher Scientific Pierce). 1 μg of sample based on the fluorometric peptide assay was loaded for each LC/MS analysis.
Peptides were separated on an Easy-spray 100 mm × 25 cm C18 column using a Dionex Ultimate 3,000 nUPLC. Solvent A = 0.1% formic acid, Solvent B = 100% Acetonitrile 0.1% formic acid; Gradient conditions = 2–50% B over 60 min, followed by a 50–99% B in 6 min and then held for 3 min than 99–2% B in 2 min. Total Run time = 90 min. Thermo Fisher Scientific Fusion Lumos mass spectrometer running in Data independent Analysis mode. Two gas phase fractionated (GPF) injections were made per sample using sequential 4 Da isolation widows. GPF 1 = m/z 362-758, GPF 2 = m/z 758-1158. Tandem mass spectra were acquired using a collision energy of 30, resolution of 30K, maximum inject time of 54 ms, and an automatic gain control target of 50K.
DIA quantification and statistical analysis
DIA data were analyzed using Scaffold DIA (1.3.1). Raw data files were converted to mzML format using ProteoWizard (3.0.11748). Analytic samples were aligned based on retention times and individually searched against Pan human library http://www.swathatlas.org with a peptide mass tolerance of 10.0 ppm and a fragment mass tolerance of 10.0 ppm. Variable modifications considered were: modification on M M and modification on C C. The digestion enzyme was assumed to be Trypsin with a maximum of one missed cleavage site(s) allowed. Only peptides with charges in the range <2..3> and length in the range <6..30> were considered. Peptides identified in each sample were filtered by Percolator (3.01.nightly-13-655e4c7-dirty) to achieve a maximum FDR of 0.01. Individual search results were combined, and peptide identifications were assigned posterior error probabilities and filtered to an FDR threshold of 0.01 by Percolator (3.01.nightly-13-655e4c7-dirty). Peptide quantification was performed by Encyclopedia (0.8.1). For each peptide, the five highest quality fragment ions were selected for quantitation. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis were grouped to satisfy the principles of parsimony. Proteins with a minimum of two identified peptides were thresholded to achieve a protein FDR threshold of 1.0%. Raw data and ScaffoldDIA results were deposited in MassIVE proteomics repository and Proteome Exchange (see Data availablity section).
Quantification and statistical analysis
Data in this study are presented as means ± SEM (n ≥ 3). Data were analyzed with either ANOVA with Tukey’s HSD post-hoc test, or a two-tailed Student’s t test. For HCM, n ≥ 3 includes in each independent experiment: 500 valid primary objects/cells per well, from ≥ 5 wells per plate per sample. Statistical significance was defined as: †, (not significant) P ≥ 0.05; and *, P < 0.05; **, P < 0.01.
Online supplemental material
Fig. S1 shows SG formation during lysosomal damage. Fig. S2 shows the dynamic interactions between SGs and lysosomes during lysosomal damage. Fig. S3 shows NUFIP2 exits nucleus and localizes to lysosomes upon damage and cooperates with Gal8 in mTORC1 response to lysosomal damage. Fig. S4 shows GABARAPs interact directly with NUFIP2 and G3BP1. Fig. S5 shows atg8ylation participates in mTOR inactivation in response to lysosomal damage. Table S1 includes all the raw MS DIA data of this study and corresponding analysis. Video 1 shows dynamic interactions between SGs and lysosomes during lysosomal damage. Video 2 shows the enlarged region of interest in Video 1.
Data availability
Raw MS DIA data of LysoIP in HEK293T cells have been deposited at the MassIVE proteomics repository (MSV000088152) and Proteome Exchange (PXD028745). The rest of raw MS DIA/DDA data in this study have been deposited at the MassIVE proteomics repository (MSV000089622) and Proteome Exchange (PXD034414).
Acknowledgments
We thank K. Bhaskar, University of New Mexico Health Sciences Center, Albuquerque, NM for tau oligomers.
This work was supported by National Institutes of Health grants (R37AI042999 and R01AI111935) and a center grant (P20GM121176) to V. Deretic. The study was in part supported by a pilot award from the AIM Center to J. Jia. Mass spectrometry analysis was supported by a National Institutes of Health shared instrumentation grant S10OD021801 to B. Phinney.
The authors declare no competing financial interests.
Authors contributions: J. Jia and V. Deretic conceptualized the study, designed the experiments, and analyzed the data; J. Jia performed majority of the experiments; Z. Bhujabal and T. Johansen performed and interpreted GST pulldowns; M. Salemi, B. Phinney, and L. Allers performed and interpreted mass spectrometry data; F. Wang, R. Peters, M. Mudd, T. Duque, and R. Javed contributed experiments; V. Deretic wrote the manuscript with J. Jia’s input. All authors read and reviewed the manuscript.



![Lysosomal damage induces SG formation. (A) DIA LC/MS/MS quantitative analysis of proteins associated with lysosomes purified by LysoIP (anti-HA IP) from HEK293T cells expressing TMEM192-3xHA untreated or treated with 1 mM LLOMe for 30 min. Three groups of proteins are denoted: ESCRT components, green; autophagy factors, blue; SG components, purple. Scatter (volcano) plot shows log2 fold changes and −Log10 P values; n = 3 (see Table S1, Tab 1). Dashed line, significance cut-off (P < 0.05). (B) Protease accessibility analysis of proteins associated with purified lysosomes (LysoIP). Huh7 cells were treated with 2 mM LLOMe. LysoIP preparations (treated or not with detergent Triton X-100) were digested with 30 µg/ml proteinase K for 30 min and analyzed by immunoblotting. (C) Quantification by HCM of G3BP1 puncta. U2OS cells were treated with EBSS, 4 mM LOMe, 2 mM LLOMe, 200 µM GPN, or 400 µg/ml silica for 30 min. White masks, algorithm-defined cell boundaries (primary objects); red masks, computer-identified G3BP1 puncta (target objects). (D) Fluorescence confocal microscopy imaging of G3BP1. U2OS cells were treated with 2 mM LLOMe for 30 min and immunostained for endogenous G3BP1. Scale bar, 5 µm. (E) Quantification by HCM of G3BP1 puncta in BMM cells treated with 2 mM LLOMe or 100 µM NaAsO2 for 2 h. Green masks, computer-identified G3BP1 puncta. (F) Quantification by HCM of G3BP1 puncta in U2OS cells treated with LLOMe at indicated doses or 100 µM NaAsO2 in the presence or absence of 10 µg/ml cycloheximide (CHX) for 30 min. HCM images in Fig. S1 G. (G) Analysis of proteins associated with purified lysosomes (LysoIP; TMEM192-3xHA) from HEK293T cells treated with 2 mM LLOMe in the presence or absence of 10 µg/ml CHX for 30 min. TMEM192-2xFLAG, control. (H) Immunoblot analysis of eIF2α (S51) phosphorylation in BMM cells treated with 2 mM LLOMe or 100 µM NaAsO2 for 2 h; eIF2α p-S51 quantification, n = 3. (I) Confocal microscopy analysis of G3BP1 (Alexa Fluor 488) and polyA RNA (Cy3-oligo[dT]) by FISH in U2OS cells treated with 2 mM LLOMe for 30 min. Scale bar, 5 µm. (J) HCM analysis of protein synthesis using Click-iT Plus OPP Alexa Fluor 488 Protein Synthesis Assay (Thermo Fisher Scientific) in U2OS cells treated with LLOMe at indicated doses or 100 µM NaAsO2 or 10 µg/ml CHX for 30 min. (K) Immunoblot analysis of ATF4 and phosphorylation of 4EBP1 (S65) and eIF2α (S51) in U2OS cells treated with 2 mM LLOMe for indicated time points; quantification of ATF4 and phosphorylation of 4EBP1 (S65) and eIF2α (S51), n = 3. Ctrl, control (untreated cells). Data, means ± SEM; HCM: n ≥ 3 (each experiment: 500 valid primary objects/cells per well, ≥5 wells/sample). †, P ≥ 0.05 (not significant); *, P < 0.05; **, P < 0.01, ANOVA. See also Fig. S1. Source data are available for this figure: SourceData F1.](https://cdn.rupress.org/rup/content_public/journal/jcb/221/11/10.1083_jcb.202207091/2/m_jcb_202207091_fig1.png?Expires=1773818234&Signature=IhKt-TaZrOeM2ksIyHd-dwd7C7ytArtg~uXTAq81qO9CqgL-6InupPznT4onceIskzzXQ48QFfHGOLdtL4Aa2c1DwiKljwp6cRhaNBy0qEUUFoN2O2BE92PJeMhWVilvT6bh215CX-7cyTR514kRWUUaHaOJlYSZFZ4A9sMM-jDMCoSYGdbMQNZvWe1TMpsxNUMpWAcU2omkaXHZIXgG3p2czdmvlBIdlMvpOmhYgPeLLiXCUgWe7kkdT9Pjm8OkgOLCGA73JuE1iEb0ddbF6amAWH1gx3lmcXjDJIrmm-rrW0GCjH2cki52Feqo9bL1lLTKp5mXj7otBDhVulU5Pg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![SG formation during lysosomal damage. (A) Immunoblot analysis of proteins associated with purified lysosomes (LysoIP; anti-HA immunoprecipitation TMEM1923xHA) from HEK293T cells treated with 2 mM LLOMe or 100 µM NaAsO2 for 30 min. TMEM1922xFLAG, control. (B) Quantification by HCM of DCP1a and G3BP1 puncta in U2OS cells treated with 2 mM LLOMe for 30 min. PB, P-body. (C) Quantification by HCM of G3BP1 puncta in Huh7 cells treated with 2 mM LLOMe for 30 min. White masks, algorithm-defined cell boundaries (primary objects); Green masks, computer-identified G3BP1 puncta (target objects). (D) Quantification by HCM of Gal3 puncta in BMM cells treated with 2 mM LLOMe or 100 µM NaAsO2 for 2 h. Red masks, computer-identified galectin-3 puncta. (E) Quantification by HCM of TIA1 puncta in U2OS cells treated with 2 mM LLOMe for 30 min. Red masks, computer-identified TIA1 puncta. (F) Quantification by HCM of TIA1 puncta in HeLa cells treated with 4 mM LLOMe for 30 min. Red masks, computer-identified TIA1 puncta. (G) Quantification by HCM of G3BP1 and Gal3 puncta in U2OS cells treated with increasing doses of LLOMe or 100 µM NaAsO2 in the presence or absence of 10 µg/ml cycloheximide (CHX) for 30 min. (i) HCM sample images corresponding to Fig. 1 F. Red masks, computer-identified G3BP1 puncta. (ii and ⅲ) Green masks, computer-identified Gal3 puncta and corresponding quantification in iii. (H) Immunoblot analysis of eIF2α (S51) phosphorylation in U2OS cells treated with 2 mM LLOMe for 30 min and followed by 1 h washout. (I) Quantification by HCM of G3BP1 puncta in U2OS cells treated with 2 mM LLOMe for 30 min and followed by 1 h washout. Red masks, computer-identified G3BP1 puncta. (J) Immunoblot analysis of eIF2α (S51) phosphorylation in HEK293T cells treated with 2 mM LLOMe or 100 µM NaAsO2 for 30 min. (K) Schematic summary of the findings in Fig. 1. (L) Quantification by HCM of Lysotracker Red (LTR) and G3BP1 puncta in parental HeLa WT and Gal3KO cells treated with 4 mM LLOMe for 30 min. Red masks, computer-identified LTR puncta. Green masks, computer-identified G3BP1 puncta. (M) Quantification by HCM of poly(A) RNA (Cy3-oligo[dT]) in U2OS cells transfected with scrambled siRNA as control (SCR) or G3BP1/2 siRNA for single or double knockdown (DKD). Cells were treated with 2 mM LLOMe for 30 min. Red masks, computer-identified poly(A) RNA puncta. Ctrl, control (untreated cells). Data, means ± SEM; HCM: n ≥ 3 (each experiment: 500 valid primary objects/cells per well, ≥5 wells/sample). †, P ≥ 0.05 (not significant); **, P < 0.01, ANOVA. See also Fig. 1. Source data are available for this figure: SourceData FS1.](https://cdn.rupress.org/rup/content_public/journal/jcb/221/11/10.1083_jcb.202207091/2/m_jcb_202207091_figs1.png?Expires=1773818234&Signature=Om8GU1C~SPqSHqebwn5Z6v4-nm-P9fibm8nd40Aqt7RXmm1DCHtS87-y1ZZ8uRH6xKqY59WF6Ot15jrNA5J4GIjNZq8O5n3O7GRD07SQ2dipTFw2VAcjwnS6T3m~9GtP8-V2bMVarmrE8-cNEWmJPhUlKP7Z5aZTQod59F2IPsYueQmH9veHyiJRGtATFOPzGC43PpSDaI1fj8n3g2DqDEv5G4w1S~VG618ww99331wf5CWk49tnH0Kbw05OORx1AxQy5KUSb9Jv1prEYuAr6qegPpvCq2iK3JTDwo6L5VzF7ZRHkGCbFKX4qd2t2OZm9fsUEGhc8HhjC3KJuOOcYQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)