Mechanisms that turn over components of the nucleus and inner nuclear membrane (INM) remain to be fully defined. We explore how components of the INM are selected by a cytosolic autophagy apparatus through a transmembrane nuclear envelope–localized cargo adaptor, Atg39. A split-GFP reporter showed that Atg39 localizes to the outer nuclear membrane (ONM) and thus targets the INM across the nuclear envelope lumen. Consistent with this, sequence elements that confer both nuclear envelope localization and a membrane remodeling activity are mapped to the Atg39 lumenal domain; these lumenal motifs are required for the autophagy-mediated degradation of integral INM proteins. Interestingly, correlative light and electron microscopy shows that the overexpression of Atg39 leads to the expansion of the ONM and the enclosure of a network of INM-derived vesicles in the nuclear envelope lumen. Thus, we propose an outside–in model of nucleophagy where INM is delivered into vesicles in the nuclear envelope lumen, which can be targeted by the autophagosome.
Introduction
The function of the nuclear envelope (NE) is conferred by its biochemical constituents that populate an inner nuclear membrane (INM) with peripherally associated nuclear lamina, an outer nuclear membrane (ONM), and a nuclear pore membrane (Ungricht and Kutay, 2017). The latter defines connections between the INM and ONM where embedded nuclear pore complexes (NPCs) control molecular traffic between the nucleus and cytoplasm (Wente and Rout, 2010; Schmidt and Görlich, 2016). Although we have a considerable understanding of the mechanisms that underly molecular exchange through NPCs, it is less well understood how the NE proteome is turned over under either physiological or pathological conditions.
The need to clear damaged or defective proteins from the nucleus and NE is underscored by the accumulation of nuclear protein aggregates in several human diseases (Bitetto and Di Fonzo, 2020). Furthermore, both NPCs (D’Angelo et al., 2009) and the nuclear lamins accumulate damage with age (Karoutas and Akhtar, 2021) and defects in nuclear transport (Zhang et al., 2015; Chou et al., 2018; Cunningham et al., 2020), and NPC injury may be a cause of certain forms of neurodegeneration, including amyotrophic lateral sclerosis (Coyne et al., 2020, 2021). Interestingly, the protein constituents of NPCs are also characterized by long half-lives in neurons (Savas et al., 2012; Toyama et al., 2013; Ori et al., 2015), which may indicate that they are challenging to productively turn over. Indeed, it is hard to conceptualize how cells might remove these massive macromolecular assemblies without compromising NE integrity. Nonetheless, there is evidence that NPCs may be excised from the NE in both metazoan (Toyama et al., 2019) and yeast model systems (Webster et al., 2014; Lee et al., 2020; Tomioka et al., 2020). While the endosomal sorting complexes required for transport (ESCRTs; Webster et al., 2014; Toyama et al., 2019; Lee et al., 2020) and the macroautophagy (Lee et al., 2020; Tomioka et al., 2020) machinery have been implicated in these events, the molecular and morphological steps in these pathways are just beginning to come to light.
Like NPCs, there is evidence that the lamins can be turned over with several molecular links implicating macroautophagy in this process (Dou et al., 2015; Harhouri et al., 2017; Borroni et al., 2018; Lu and Djabali, 2018). Macroautophagy (hereafter called “autophagy”) is a catabolic mechanism that delivers protein aggregates, lipids, and parts of (and in some cases, whole) organelles to lysosomes for degradation (Gatica et al., 2018). It begins with the formation of a phagophore membrane that is defined by the covalent coupling of the ubiquitin-like protein LC3 (Atg8, in yeast) directly to phosphatidylethanolamine (Melia et al., 2020). The phagophore expands around the cargo, ultimately sealing the cargo inside a closed double-membrane organelle called the autophagosome (Nakatogawa, 2020). The autophagosome fuses with lysosomes (or vacuoles in yeast), where cargo is degraded (Reggiori and Ungermann, 2017).
Interestingly, there is evidence that LC3 can direct a form of nuclear autophagy (nucleophagy) by binding to lamin B1 in the context of oncogene activation (Dou et al., 2015). This interaction plays a part in the selective clearance of lamin B1 from the INM. However, how a cytosolic phagophore selectively targets the INM or nucleoplasm across the double-membraned NE remains unknown. Indeed, in most cases of selective organelle targeting, autophagy cargo adaptors bind to specific proteins and recruit the autophagy machinery to initiate phagophore expansion around themselves (Stolz et al., 2014; Kirkin and Rogov, 2019; Melia et al., 2020). While such a nuclear-specific cargo adaptor has not been identified in metazoans, budding yeast have Atg39 (also known as Esm1; Vevea et al., 2015), a putative type II transmembrane protein (Vevea et al., 2015) that localizes at the NE and is required for the autophagic degradation of both INM and nucleoplasmic proteins (Mochida et al., 2015; Rahman et al., 2018; Mostofa et al., 2018; Mizuno et al., 2020; Otto and Thumm, 2021; Tomioka et al., 2020) but not NPCs (Lee et al., 2020; Tomioka et al., 2020). Thus, Atg39 is the essential cog in the macroautophagic clearance of INM, but how INM is recognized and delivered to the cytoplasm remains unknown.
Here, we explore the mechanism of Atg39-mediated nucleophagy in budding yeast. The data support that Atg39 acts from the ONM and connects to the INM through its lumenal domain. The lumenal domain has functional elements that are required for NE remodeling and the capture of INM cargo into NE blebs that can be targeted by autophagy. By using correlative light and electron microscopy (CLEM) and focused ion beam scanning EM (FIB-SEM) to visualize NE bleb ultrastructure, we observe the capture of INM into vesicles in the NE lumen. We propose a model where nucleophagy proceeds through an outside–in mechanism where putative translumenal interactions coordinate INM and ONM remodeling to ultimately deliver INM cargo to the autophagosome.
Results
Atg39 accumulates at the ONM
Key unknowns to unraveling the nucleophagy mechanism are determining whether Atg39 acts from the ONM or the INM (or both) and whether, like other cargo adaptors (Grumati et al., 2017; Jiang et al., 2020; Mochida et al., 2020), it has any inherent membrane remodeling activity. To address the former, we took advantage of a split-GFP reporter system used to catalog the INM proteome (Smoyer et al., 2016). The system exploits a series of mCherry-tagged reporter proteins that are expressed as fusions to GFP11, a 4-kD fragment of GFP, and are localized in the nucleus (GFP11-mCherry-Pus1; Fig. 1 A) and ER. The two ER reporters differ in that GFP11 either faces the lumen (mCherry-Scs2TM-GFP11) or the cytosol (GFP11-mCherry-Scs2TM; Fig. 1 A). In these backgrounds, the rest of GFP (GFP1–10) is expressed on either the N- or C-terminus of Atg39 (Fig. 1 A) from a galactose-inducible (GAL1) promoter; the reconstitution of a fluorescent GFP provides evidence of physical proximity with which to infer localization.
Atg39 accumulates at the ONM. (A) Schematic of localization and topology of split-GFP constructs. The GFP11–mCherry reporter fusion proteins (i, ii, and iii) are constructed as shown in the key, with localization diagrammed. At the bottom are schematics of the Atg39 GFP1–10 fusions and truncations lacking N- or C-termini. The Atg39 TM domain is depicted as a gray oval. Numbers indicate amino acids. (B) Deconvolved fluorescence micrographs of cells expressing indicated Atg39 fusion proteins and mCherry-reporters (see inset). GFP, mCherry, and merged images are shown. Arrows point to NE blebs. Scale bars are 5 μm. (C) Deconvolved fluorescence micrographs of cells expressing GFP1–10-Atg39 and indicated mCherry-reporter (inset). GFP, mCherry, and merged images are shown. Scale bars are 5 μm. Asterisks indicate vacuolar autofluorescence. (D) Normalized line profiles of GFP (green) and mCherry (magenta) fluorescence bisecting cells as indicated by dotted lines in corresponding top and bottom merge panels from C. Position of NE and cER is indicated by dotted lines. (E) As in C but with cells expressing GFP1–10-atg39-(139–398). (F) As in D but with cells from E. (G) As in C but with cells expressing GFP1–10-atg39-ΔL. Asterisks indicate vacuolar autofluorescence. (H) As in D but with cells from G.
Atg39 accumulates at the ONM. (A) Schematic of localization and topology of split-GFP constructs. The GFP11–mCherry reporter fusion proteins (i, ii, and iii) are constructed as shown in the key, with localization diagrammed. At the bottom are schematics of the Atg39 GFP1–10 fusions and truncations lacking N- or C-termini. The Atg39 TM domain is depicted as a gray oval. Numbers indicate amino acids. (B) Deconvolved fluorescence micrographs of cells expressing indicated Atg39 fusion proteins and mCherry-reporters (see inset). GFP, mCherry, and merged images are shown. Arrows point to NE blebs. Scale bars are 5 μm. (C) Deconvolved fluorescence micrographs of cells expressing GFP1–10-Atg39 and indicated mCherry-reporter (inset). GFP, mCherry, and merged images are shown. Scale bars are 5 μm. Asterisks indicate vacuolar autofluorescence. (D) Normalized line profiles of GFP (green) and mCherry (magenta) fluorescence bisecting cells as indicated by dotted lines in corresponding top and bottom merge panels from C. Position of NE and cER is indicated by dotted lines. (E) As in C but with cells expressing GFP1–10-atg39-(139–398). (F) As in D but with cells from E. (G) As in C but with cells expressing GFP1–10-atg39-ΔL. Asterisks indicate vacuolar autofluorescence. (H) As in D but with cells from G.
Taking advantage of the mCherry-Scs2TM-GFP11 ER lumenal reporter (Fig. 1 A, i), we first confirmed the proposed type II topology of Atg39 (Mochida et al., 2015; Vevea et al., 2015) because only Atg39-GFP1–10, but not GFP1–10-Atg39 resulted in visible GFP fluorescence localized at the NE (Fig. 1 B). We also noted that there were NE extensions, or blebs, at sites of reconstituted GFP fluorescence (Fig. 1 B, arrows), suggesting that Atg39 expression impacted NE morphology, consistent with prior data (Vevea et al., 2015). These blebs were also observed when GFP1–10-Atg39 was expressed alongside the cytosolic-facing ER reporter (Fig. 1 C, top panels, arrow). Consistent with the conclusion that GFP1–10-Atg39 is localized specifically to the NE, the ER reporter itself was distributed throughout the NE and ER, but the GFP fluorescence was only observed at the nuclear periphery and within the blebs extending from the NE. This was particularly evident when line profiles were drawn that bisected the entire cell and nucleus: only the NE, and not the cortical endoplasmic reticulum (cER), peaks of the mCherry fluorescence (magenta) overlapped with the reconstituted GFP-Atg39 fluorescence (Fig. 1 D, green).
Because the ER reporters can access both the ONM and the INM (Fig. 1 A), we tested whether Atg39 could reach the INM using the nucleoplasmic reporter (Fig. 1 A, iii). The extralumenal domain of Atg39 is predicted to be unstructured, with a mol wt of 16 kD. Thus, even with the addition of the GFP1–10, it should, in principle, be able to pass through the peripheral channels along the pore membrane, which are thought to restrict passage of extralumenal domains larger than ∼60 kD (Popken et al., 2015; Smoyer et al., 2016). Interestingly, although we observed low levels of GFP fluorescence when GFP1–10-Atg39 was expressed with the nucleoplasmic reporter (Fig. 1 C, bottom panels), this fluorescence was intranuclear and did not accumulate along the nuclear periphery, even at low levels of expression (see time course of cells treated with galactose; Fig. S1 A). We interpret these data in a model where Atg39 can cross the pore membrane but is then liberated from the INM likely through an INM-associated degradation–type mechanism (Deng and Hochstrasser, 2006; Foresti et al., 2014; Khmelinskii et al., 2014; Koch et al., 2019). Such a model predicts the existence of a degron sequence in Atg39, likely in its N-terminus. Consistent with this idea, deletion of the N-terminus of Atg39 resulted in the accumulation of a reconstituted nuclear rim fluorescence when GFP1–10-atg39-(139–398) was expressed with the nucleoplasmic reporter (Fig. 1 A), even after several hours of growth in galactose (Fig. 1, E and F; and Fig. S1 B), whereas deletion of the C-terminal lumenal domain comprising amino acids 169–398 (GFP1–10-atg39-ΔL; Fig. 1 A) mirrored the full-length protein, albeit with a more visible pool at the nuclear periphery at lower levels (Fig. 1, G and H; and Fig. S1 C). Thus, there are sequence elements in the N-terminus of Atg39 that might trigger its removal from the INM. Taken together, the data are most consistent with a model in which Atg39 localizes at the ONM and may in fact be restricted from accumulating at the INM.
The split-GFP reconstitution of intranuclear GFP fluorescence depends on the N-terminus of Atg39 (supplemental to Fig. 1,).(A–C) Deconvolved fluorescence micrographs of the indicated GFP1–10 constructs coexpressed with the nucleoplasmic split-GFP reporter (see inset and Fig. 1 A). GFP1–10 constructs are expressed behind a GAL1 promoter and are induced by growth in galactose for the indicated times (in hours). GFP (green), mCherry (magenta), and merged images shown. Scale bars are 5 μm.
The split-GFP reconstitution of intranuclear GFP fluorescence depends on the N-terminus of Atg39 (supplemental to Fig. 1,).(A–C) Deconvolved fluorescence micrographs of the indicated GFP1–10 constructs coexpressed with the nucleoplasmic split-GFP reporter (see inset and Fig. 1 A). GFP1–10 constructs are expressed behind a GAL1 promoter and are induced by growth in galactose for the indicated times (in hours). GFP (green), mCherry (magenta), and merged images shown. Scale bars are 5 μm.
The Atg39 lumenal domain is necessary and sufficient for ONM targeting and bleb formation
A model in which Atg39 acts from the outside of the nucleus in predicts physical interactions that connect Atg39 to the INM through the NE lumen/perinuclear space. To explore this, we generated a deletion series of Atg39 to systematically evaluate the sequence elements that conferred ONM localization. We also evaluated the ability of these constructs to induce NE blebs. We first generated C-terminal deletions that sequentially removed lumenal regions that secondary structure prediction suggested were α helical in nature (Fig. 2 A). Deletion of the terminal two α helical segments [GFP-atg39-(1–333)] did not impact NE targeting or the number of NE blebs observed in each cell (Fig. 2, B and E). In contrast, the removal of α2 [GFP-atg39-(1–312)] led to a marked decrease in the number of NE blebs per cell with no obvious impact on NE targeting (Fig. 2, B and E). We confirmed that this result was not simply due to lower expression levels of the GFP-atg39-(1–312) protein (Fig. S2 A) because a direct comparison of cells expressing similar levels (as assessed by total fluorescence) of either GFP-Atg39 or GFP-atg39-(1–312) demonstrated a reduction in NE bleb frequency that was specific for the truncation (Fig. S2 C). Removal of α1 abrogated NE accumulation and abolished NE blebbing as both GFP-atg39-(1–275) and GFP-atg39-ΔL no longer exclusively accumulated at the NE and were found throughout the cER as well (Fig. 2, B and E). Thus, these data support a model in which the lumenal domain of Atg39 has sequence elements required for both NE accumulation and NE remodeling that are contained within the predicted α1 and α2 helices. In analogy to Klarsicht, Anc-1, and Syne homology proteins that accumulate at the ONM by binding to INM Sad1 and Unc-84 proteins (Sosa et al., 2013), we propose that Atg39 accumulates at the ONM by forming a translumenal bridge through direct or indirect interactions with the INM.
The Atg39 lumenal domain is required for NE targeting and bleb formation. (A) Schematic of Atg39 with Atg8-interacting motif (AIM; light orange), Atg11 binding region (blue), predicted TM helix (gray), and predicted α helices 1–4 (colored ovals). Numbers are amino acids. (B) Deconvolved fluorescence micrographs of the indicated GFP fusion proteins. Numbers indicate amino acid position from A. Scale bars are 5 μm. (C) As in B. (D) Deconvolved fluorescence micrographs of GFP-Atg39 in the indicated strains. Scale bars are 5 μm. (E) Bar chart plotting the quantification of the number of NE blebs per cell in the indicated strains from experiments as in B–D. Genotype of strains indicated by bar color as shown in key. At least 50 cells of each genotype were quantified from three independent experiments. Mean and error bars denoting SD are shown. ****, P ≤ 0.0001 by one-way ANOVA.
The Atg39 lumenal domain is required for NE targeting and bleb formation. (A) Schematic of Atg39 with Atg8-interacting motif (AIM; light orange), Atg11 binding region (blue), predicted TM helix (gray), and predicted α helices 1–4 (colored ovals). Numbers are amino acids. (B) Deconvolved fluorescence micrographs of the indicated GFP fusion proteins. Numbers indicate amino acid position from A. Scale bars are 5 μm. (C) As in B. (D) Deconvolved fluorescence micrographs of GFP-Atg39 in the indicated strains. Scale bars are 5 μm. (E) Bar chart plotting the quantification of the number of NE blebs per cell in the indicated strains from experiments as in B–D. Genotype of strains indicated by bar color as shown in key. At least 50 cells of each genotype were quantified from three independent experiments. Mean and error bars denoting SD are shown. ****, P ≤ 0.0001 by one-way ANOVA.
Assessment of total levels of Atg39 fusion proteins and Atg11 (supplemental to Figs. 2, 3, 5,, and 6 ).(AandB) Western blot (WB) of proteins from whole-cell extracts derived from cells expressing the indicated GFP fusions in indicated strains and drug treatments. GFP detected with anti-GFP.1, HRP-conjugated secondary antibodies, and ECL. To assess relative protein loading, a portion of the blots are shown stained with Ponceau S. Position of mol wt standards (in kD) at right. In A, the numbers at the bottom of the blot correspond to the relative amount of each protein normalized to GFP-Atg39 using densitometry. (C) Plot of the percentage of cells with zero, one, or two or more NE blebs in GFP-Atg39 and GFP-atg39-(1–312) cells with similar levels of expression (as assessed by comparing total GFP fluorescence of individual cells). At least 100 cells from three independent replicates were evaluated, and mean and SD are plotted. *, P ≤ 0.05; **, P ≤ 0.01; ****, P ≤ 0.0001 by one-way ANOVA. (D) As in A and B. The numbers at the bottom of the blot correspond to the relative amount of each protein as assessed by densitometry normalized to GFP-Atg39. (E) WB of proteins from whole-cell extracts derived from cells expressing the indicated HA fusions expressed from endogenous ATG39 gene locus after 24 h in SD-N medium. HA detected with anti-HA antibody conjugated to HRP and ECL. To assess relative protein loading, a portion of the blots are shown stained with Ponceau S. Position of mol wt standards (in kD) at right. (F) WB of proteins from whole-cell extracts derived from cells expressing GFP-Atg11 alongside the indicated alleles encoding mCherry fusion proteins treated as indicated with rapamycin. GFP detected with anti-GFP.2, HRP-conjugated secondary antibodies, and ECL. To assess relative protein loading, a portion of the blots are shown stained with Ponceau S. Position of mol wt standards (in kD) at right. (G) As in A and B. Source data are available for this figure: SourceData FS2.
Assessment of total levels of Atg39 fusion proteins and Atg11 (supplemental to Figs. 2, 3, 5,, and 6 ).(AandB) Western blot (WB) of proteins from whole-cell extracts derived from cells expressing the indicated GFP fusions in indicated strains and drug treatments. GFP detected with anti-GFP.1, HRP-conjugated secondary antibodies, and ECL. To assess relative protein loading, a portion of the blots are shown stained with Ponceau S. Position of mol wt standards (in kD) at right. In A, the numbers at the bottom of the blot correspond to the relative amount of each protein normalized to GFP-Atg39 using densitometry. (C) Plot of the percentage of cells with zero, one, or two or more NE blebs in GFP-Atg39 and GFP-atg39-(1–312) cells with similar levels of expression (as assessed by comparing total GFP fluorescence of individual cells). At least 100 cells from three independent replicates were evaluated, and mean and SD are plotted. *, P ≤ 0.05; **, P ≤ 0.01; ****, P ≤ 0.0001 by one-way ANOVA. (D) As in A and B. The numbers at the bottom of the blot correspond to the relative amount of each protein as assessed by densitometry normalized to GFP-Atg39. (E) WB of proteins from whole-cell extracts derived from cells expressing the indicated HA fusions expressed from endogenous ATG39 gene locus after 24 h in SD-N medium. HA detected with anti-HA antibody conjugated to HRP and ECL. To assess relative protein loading, a portion of the blots are shown stained with Ponceau S. Position of mol wt standards (in kD) at right. (F) WB of proteins from whole-cell extracts derived from cells expressing GFP-Atg11 alongside the indicated alleles encoding mCherry fusion proteins treated as indicated with rapamycin. GFP detected with anti-GFP.2, HRP-conjugated secondary antibodies, and ECL. To assess relative protein loading, a portion of the blots are shown stained with Ponceau S. Position of mol wt standards (in kD) at right. (G) As in A and B. Source data are available for this figure: SourceData FS2.
We next asked whether the lumenal domain (with its transmembrane [TM] anchor) was sufficient to confer NE targeting and NE blebbing by making sequential N-terminal deletions. As shown in Fig. 2 C, GFP-atg39-(70–398), which lacks the Atg8 and Atg11 binding regions (Fig. 2 A) showed considerably more NE blebs per cell (mean of 4.3/cell) compared with GFP-Atg39 (Fig. 2 E). Interestingly, these NE blebs were also more elaborate and extended farther from the NE than those induced by GFP-Atg39 (Fig. 2 C). We also observed more numerous NE blebs in cells overexpressing GFP-atg39-(120–398) but not in GFP-atg39-(139–398), which had similar numbers of NE blebs as those induced by the full-length protein. As the first 70 amino acids of Atg39 contain binding motifs for both Atg8 and Atg11, these data suggested that NE remodeling by Atg39 is independent of engaging the autophagy machinery; such a result was further supported by the obvious NE blebbing observed upon overexpression of GFP-Atg39 in atg8Δ, atg11Δ, and atg8Δatg11Δ cells (Fig. 2 D). Although, here too, we consistently counted higher numbers of blebs than those observed when GFP-Atg39 is expressed in WT cells (Fig. 2 E), despite the consistency in the overall levels of GFP-Atg39 between the strains (Fig. S2 D). Cumulatively, these results suggest that Atg8 and Atg11 do not contribute to bleb formation but may play a role in limiting the growth of the NE blebs in the overexpression condition, perhaps as a response to the NE deformation itself.
A model in which Atg11 might attenuate NE bleb growth upon Atg39 overexpression prompted us to examine whether we observed evidence of Atg11 engagement with overexpressed Atg39. We assessed colocalization between GFP-Atg11 and overexpressed mCherry-Atg39 under nutrient-rich conditions where we also tested the impact of induction of autophagy by treating cells with the drug rapamycin. Rapamycin inhibits Tor1 kinase, which induces a starvation-mimicking state (Barbet et al., 1996; Fingar and Blenis, 2004). The latter condition is important because it is typical for cargo adaptor–Atg11 interactions to be controlled by autophagy signaling (Aoki et al., 2011; Farré et al., 2013). As shown in Fig. 3 A, we observed GFP-Atg11 accumulating at the NE colocalized with overexpressed mCherry-Atg39 in ∼55% of carrier-alone (DMSO)–treated cells, which was elevated to ∼75% upon rapamycin treatment (Fig. 3 B). This colocalization likely reflects physical interactions between Atg39 and Atg11 because no such recruitment was observed upon overexpression of mCherry-atg39-(70–398), which lacks the Atg11 binding site (GFP-Atg11 is produced at similar levels in these strains; Fig. S2 F; and Fig. 3, A and B). Thus, overexpressed Atg39 can specifically recruit Atg11 to the NE, even under nutrient-rich conditions. Interestingly, however, simply the exposure of an Atg11 binding site is insufficient to recruit Atg11. For example, the overexpression of atg39-ΔL does not lead to any redistribution of GFP-Atg11 in DMSO-treated conditions (Fig. 3, A and B). Even after addition of rapamycin, we observed colocalization of GFP-Atg11 with mCherry-atg39-ΔL in only ∼10% of cells (Fig. 3 A, inset, and Fig. 3 B). Thus, there must be unique features of the NE environment that promote the specific binding of Atg11 to the Atg11-binding site in Atg39. We suggest a model in which Atg39-mediated NE blebbing provides a platform for Atg11 recruitment, perhaps due to the high local concentration of Atg39 in the blebs, which is not seen with atg39-ΔL. Although Atg11 does not contribute to NE blebbing per se, we cannot rule out that some portion of the NE blebs may be removed or inhibited from forming in this context, which provides an explanation for the more elaborate and abundant NE blebs in the absence of engagement with the autophagy machinery when Atg39 is overexpressed.
Atg39 colocalizes with Atg11. (A) Deconvolved fluorescence micrographs of rapamycin- or carrier-alone (DMSO)–treated cells expressing GFP-Atg11 with the indicated mCherry-fusion proteins. Scale bars are 5 μm. The inset (sixth row, bottom right) shows an example of the ∼10% of cells where mCherry-atg39-ΔL colocalized with GFP-Atg11. The scale bar in the inset is 3 μm. (B) Bar chart plotting the percentage of cells with Atg39–Atg11 colocalization in the indicated strains from experiments as in A. At least 50 cells of each genotype were quantified from three independent experiments. Mean and error bars denoting SD are shown. *, P ≤ 0.05 by one-way ANOVA.
Atg39 colocalizes with Atg11. (A) Deconvolved fluorescence micrographs of rapamycin- or carrier-alone (DMSO)–treated cells expressing GFP-Atg11 with the indicated mCherry-fusion proteins. Scale bars are 5 μm. The inset (sixth row, bottom right) shows an example of the ∼10% of cells where mCherry-atg39-ΔL colocalized with GFP-Atg11. The scale bar in the inset is 3 μm. (B) Bar chart plotting the percentage of cells with Atg39–Atg11 colocalization in the indicated strains from experiments as in A. At least 50 cells of each genotype were quantified from three independent experiments. Mean and error bars denoting SD are shown. *, P ≤ 0.05 by one-way ANOVA.
NE blebs form independently of Atg11 under physiological conditions
As it is typical for cargo adaptor interactions with Atg8 and Atg11 to contribute to membrane deformation in other autophagy contexts (Grumati et al., 2017; Mochida et al., 2020), we wanted to ensure that this autophagy-independent NE blebbing by Atg39 reflects the physiological scenario. We therefore investigated whether NE blebbing is observed with endogenous Atg39-GFP upon induction of autophagy during nitrogen starvation. Under these conditions, we observed the accumulation of endogenous Atg39-GFP in foci that extended from the NE in 20%–30% of WT cells at 4, 8, and 24 h in medium lacking nitrogen (Fig. 4, A and B). Consistent with the conclusion that these NE blebs could form independently of Atg11, we observed more frequent NE blebs in atg11Δ cells, up to 60% over the 24-h time course (Fig. 4 B). Interestingly, in addition to being more frequent, the NE blebs in atg11Δ cells also contained up to four times higher levels of Atg39-GFP (Fig. 4 C). Thus, consistent with the overexpression data, Atg39 can accumulate in NE blebs that form independently of the autophagy machinery.
Endogenous Atg39 accumulates in NE blebs independently of Atg11. (A) Deconvolved fluorescence micrographs of Atg39-GFP in the indicated strains cultured in medium lacking nitrogen (SD-N) for 4, 8, and 24 h. Scale bars are 5 μm. Arrows point to NE blebs. (B) Bar chart plotting the percentage of cells with NE blebs in the indicated strains from experiments in A. At least 50 cells of each genotype were quantified from three independent experiments. Mean and error bars denoting SD are shown. (C) Scatter plot of the quantification of Atg39-GFP fluorescence intensity in NE blebs in the indicated strains from experiments in A. At least 50 cells of each genotype were quantified from three independent experiments. Mean and error bars denoting SD are shown. **, P ≤ 0.01; ****, P ≤ 0.0001 by one-way ANOVA.
Endogenous Atg39 accumulates in NE blebs independently of Atg11. (A) Deconvolved fluorescence micrographs of Atg39-GFP in the indicated strains cultured in medium lacking nitrogen (SD-N) for 4, 8, and 24 h. Scale bars are 5 μm. Arrows point to NE blebs. (B) Bar chart plotting the percentage of cells with NE blebs in the indicated strains from experiments in A. At least 50 cells of each genotype were quantified from three independent experiments. Mean and error bars denoting SD are shown. (C) Scatter plot of the quantification of Atg39-GFP fluorescence intensity in NE blebs in the indicated strains from experiments in A. At least 50 cells of each genotype were quantified from three independent experiments. Mean and error bars denoting SD are shown. **, P ≤ 0.01; ****, P ≤ 0.0001 by one-way ANOVA.
The Atg39 lumenal domain is required for nucleophagy
The data support a model in which Atg39 can accumulate and mediate NE remodeling from the ONM by virtue of sequence motifs in its lumenal domain. To evaluate the importance of these motifs in the nucleophagic clearance of INM proteins, we tested whether they were required for the autophagic degradation of the established integral INM Atg39 cargo Heh1 (also called Src1; Mochida et al., 2015; Mizuno et al., 2020) under conditions of nitrogen starvation. We tested the degradation of Heh1-GFP using a standard autophagy assay that relies on the visualization of a stable fragment of GFP (GFP′) by Western blot, which is liberated from Heh1-GFP by vacuolar proteases. Consistent with published data (Mochida et al., 2015), we observed an ∼65% degradation of the total pool of Heh1-GFP in cells grown in medium lacking nitrogen, which was mitigated in the atg39Δ strain (Fig. 5, A and B). This effect was specific for Heh1-GFP and other model INM cargo. For example, we also observed Atg39-mediated degradation of heh1-ΔL-GFP, a TM-containing fragment of Heh1 that contains its INM-targeting sequences (see Fig. 8 A for schematic). Furthermore, a truncation of Heh1’s paralogue Heh2 (heh2-ΔWH-GFP), which localizes to the INM but, unlike Heh2, does not interact with NPCs (Borah et al., 2021), was sensitive to Atg39-mediated autophagy (Fig. 5, C and E). The latter result suggests that Heh2’s interactions with NPCs may limit its Atg39-mediated degradation. Consistent with this conclusion, neither Heh2-GFP nor Nup82-GFP showed appreciable Atg39-mediated degradation (Fig. 5, D and E). This result was further mirrored by the lack of degradation of Mps3, an integral membrane protein that associates with spindle pole bodies (SPBs; Fig. 5, C and E; Jaspersen et al., 2002; Chalfant et al., 2019). Thus, Atg39 is specifically required for the degradation of INM proteins but not for NE-spanning complexes like NPCs and SPBs.
The autophagic degradation of Heh1-GFP requires the Atg39 lumenal domain.(A) Western blot (WB) of proteins from whole-cell extracts derived from cells expressing Heh1-GFP in strains with the indicated atg39 alleles cultured in medium lacking nitrogen (SD-N) for 24 h. GFP′ is a stable fragment of GFP in vacuoles. GFP detected with anti-GFP.1 antibody, HRP-conjugated secondary antibodies, and ECL. To assess relative protein loading, a portion of the blot is shown stained with Ponceau S. Position of mol wt standards (in kD) at right. (B) Plot of mean and error bars denoting SD of the percent degradation of Heh1-GFP from three independent experiments as in A. (C and D) WBs of proteins from whole-cell extracts derived from cells expressing the indicated GFP fusions in indicated strains cultured in SD-N medium for 24 h. GFP detected with anti-GFP.1 antibody, HRP-conjugated secondary antibodies, and ECL. To assess relative protein loading, a portion of the blots are shown stained with Ponceau S. Position of mol wt standards (in kD) at right. (E) Plot of mean and error bars denoting SD of the percent degradation of GFP-tagged proteins in the indicated strains from three independent experiments as in C and D. ***, P ≤ 0.001; ****, P ≤ 0.0001 by one-way ANOVA with Tukey’s correction. Source data are available for this figure: SourceData F5.
The autophagic degradation of Heh1-GFP requires the Atg39 lumenal domain.(A) Western blot (WB) of proteins from whole-cell extracts derived from cells expressing Heh1-GFP in strains with the indicated atg39 alleles cultured in medium lacking nitrogen (SD-N) for 24 h. GFP′ is a stable fragment of GFP in vacuoles. GFP detected with anti-GFP.1 antibody, HRP-conjugated secondary antibodies, and ECL. To assess relative protein loading, a portion of the blot is shown stained with Ponceau S. Position of mol wt standards (in kD) at right. (B) Plot of mean and error bars denoting SD of the percent degradation of Heh1-GFP from three independent experiments as in A. (C and D) WBs of proteins from whole-cell extracts derived from cells expressing the indicated GFP fusions in indicated strains cultured in SD-N medium for 24 h. GFP detected with anti-GFP.1 antibody, HRP-conjugated secondary antibodies, and ECL. To assess relative protein loading, a portion of the blots are shown stained with Ponceau S. Position of mol wt standards (in kD) at right. (E) Plot of mean and error bars denoting SD of the percent degradation of GFP-tagged proteins in the indicated strains from three independent experiments as in C and D. ***, P ≤ 0.001; ****, P ≤ 0.0001 by one-way ANOVA with Tukey’s correction. Source data are available for this figure: SourceData F5.
To test the role of the Atg39 lumenal domain motifs in nucleophagy, we next examined Heh1-GFP degradation in strains expressing lumenal truncations from the endogenous ATG39 chromosomal locus (i.e., under the control of the ATG39 promoter). Consistent with its importance for executing nucleophagy, the sequential trimming of the C-terminal lumenal domain resulted in a reduction of Heh1-GFP degradation that reflected the NE targeting and remodeling analysis. For example, consistent with the dispensability of the terminal two α helical segments for NE targeting and remodeling, the atg39-(1–333) allele fully complemented the degradation of Heh1-GFP as the WT ATG39 gene (Fig. 5, A and B). In contrast, the sequential removal of the α2 [atg39-(1–312)] and α1 [atg39-(1–275)] coding segments, required for NE remodeling and NE targeting, resulted in a progressive loss of the ability of these alleles to contribute to Heh1-GFP degradation (Fig. 5, A and B). Importantly, except for atg39-(1–275), all truncations were detectable by Western blot with atg39-(1–312) and atg39-(1–333) found at higher levels than Atg39 (Fig. S2 E). Thus, the lumenal sequence motifs that are required for Atg39 NE targeting and blebbing are also needed to effectively execute nucleophagy under conditions of nitrogen starvation.
Atg39-containing NE blebs are delivered to vacuoles by autophagy
Because of the obvious overlap between the requirement of the lumenal motifs for NE targeting/remodeling and nucleophagy, we considered the possibility that the NE blebs formed upon Atg39 overexpression may in fact represent a morphologically relevant intermediate in nucleophagy. Were this to be the case, several criteria needed to be met. First, if the NE blebs were an intermediate in nucleophagy, they would be delivered to vacuoles in a mechanism requiring core autophagy genes. Second, the expectation would be that the NE blebs would contain cargo specific for Atg39-mediated nucleophagy. Third, the NE ultrastructure driven by Atg39 would need to reflect characteristics of protein-mediated membrane remodeling as opposed to simply membrane expansion or the formation of membrane stacks or lamellae that are common artifacts of the overexpression of NE and ER membrane proteins (Wright et al., 1988; Smith and Blobel, 1994; Koning et al., 1996; Yamamoto et al., 1996; Elgersma et al., 1997; Snapp et al., 2003; King et al., 2006; Volkova et al., 2012; Schäfer et al., 2020).
To address these criteria, we first tested whether the NE blebs were delivered to vacuoles. To induce autophagy, we treated Atg39-GFP–expressing cells with rapamycin while arresting Atg39-GFP production by inhibiting the GAL1 promoter with the addition of glucose to the medium; delivery of Atg39-GFP to vacuoles (labeled with FM 4-64 dye) was monitored by fluorescence microscopy at 30-min intervals (Fig. 6 A). To ensure that Atg39-GFP could be visualized in vacuoles, these experiments were performed in a strain lacking Pep4, a vacuolar protease required for activation of vacuolar hydrolases (Ammerer et al., 1986). As shown in Fig. 6 A, we observed internal vacuolar GFP fluorescence in ∼25% of rapamycin-treated cells beginning at the 60-min time point, which progressively increased to ∼95% by the end of the experiment (150 min; Fig. 6, A and B) concomitantly with a reduction in total levels at the NE (Fig. 6 C). To further confirm that overexpressed Atg39-GFP was degraded by autophagy, we also observed the production of GFP′ after treating Atg39-GFP–expressing cells with rapamycin (Fig. 6 D). Consistent with the conclusion that this GFP fragment was the product of a vacuolar protease, it was not observed in a pep4Δ strain (Fig. S2 G). Importantly, this autophagy-dependent degradation of Atg39-GFP also required the Atg39 lumenal domain, as GFP′ was not detected in protein extracts derived from cells expressing any of the lumenal deletions that lacked the α2 [atg39-(1–312)] and α1 [atg39-(1–275) and atg39-ΔL] coding segments upon treatment with rapamycin (Fig. S2 B). We further confirmed that Atg39-GFP is targeted by autophagy by testing the production of GFP′ in both atg8Δ and atg11Δ strains. Deletion of ATG8, and to a lesser extent ATG11, reduced its production (Fig. 6 D). Lastly, in a parallel experiment overexpressing mCherry-Atg39, we observed the coincident accumulation of an INM cargo (heh1ΔL-GFP) colocalized with mCherry-Atg39 in the vacuole (Fig. 6 E). Thus, overexpressed Atg39 alongside INM cargo is delivered to the vacuole through autophagy.
Atg39-containing NE blebs are delivered to vacuoles by autophagy.(A) Deconvolved fluorescence micrographs of a time course (30-min intervals) of rapamycin- or carrier-alone (DMSO)–treated cells expressing Atg39-GFP (green) in a pep4Δ strain; vacuoles are stained with FM 4-64 (magenta). Merged fluorescent images are shown. Image scale in green channel chosen to saturate the bleb fluorescence and allow visualization of NE. Scale bars are 3 μm. (B) Line plot of percentage of cells where Atg39-GFP is visible in vacuoles after treatment with rapamycin (circles and magenta line) or carrier (squares and blue line) over the time points indicated. 75 cells were evaluated each from three independent replicates. SD from the mean percentage is indicated by the shaded area. (C) Violin plot of the fluorescence intensity of Atg39-GFP along the nuclear periphery normalized to background fluorescence in the presence of rapamycin (+) or carrier (−) at the indicated time points. 30 cells each from three independent replicates were evaluated; each replicate was normalized to the mean NE fluorescence at 0 min. Solid line denotes the median; width of the violin plot denotes relative frequency of data points. **, P ≤ 0.01; ****, P ≤ 0.0001 by Brown–Forsythe and Welch ANOVA tests with Games–Howell test for multiple comparisons. (D) Western blot (WB) of proteins from whole-cell extracts derived from rapamycin- or carrier-alone (DMSO)–treated cells expressing Atg39-GFP in the indicated strains. GFP detected with anti-GFP.2 antibody, HRP-conjugated secondary antibodies, and ECL. To assess relative protein loading, a portion of the blots are shown stained with Ponceau S. Position of mol wt standards (in kD) at right. (E) Deconvolved fluorescence micrographs of rapamycin- or carrier-alone (DMSO)–treated cells overexpressing mCherry-Atg39 (magenta) and heh1-ΔL-GFP (green) in a pep4Δ strain; vacuoles are labeled with FM 4-64 (magenta). Merged fluorescent images shown. Asterisks denote the nucleus; arrowheads point to colocalization of GFP and mCherry in vacuole. Scale bars are 5 μm. Source data are available for this figure: SourceData F6.
Atg39-containing NE blebs are delivered to vacuoles by autophagy.(A) Deconvolved fluorescence micrographs of a time course (30-min intervals) of rapamycin- or carrier-alone (DMSO)–treated cells expressing Atg39-GFP (green) in a pep4Δ strain; vacuoles are stained with FM 4-64 (magenta). Merged fluorescent images are shown. Image scale in green channel chosen to saturate the bleb fluorescence and allow visualization of NE. Scale bars are 3 μm. (B) Line plot of percentage of cells where Atg39-GFP is visible in vacuoles after treatment with rapamycin (circles and magenta line) or carrier (squares and blue line) over the time points indicated. 75 cells were evaluated each from three independent replicates. SD from the mean percentage is indicated by the shaded area. (C) Violin plot of the fluorescence intensity of Atg39-GFP along the nuclear periphery normalized to background fluorescence in the presence of rapamycin (+) or carrier (−) at the indicated time points. 30 cells each from three independent replicates were evaluated; each replicate was normalized to the mean NE fluorescence at 0 min. Solid line denotes the median; width of the violin plot denotes relative frequency of data points. **, P ≤ 0.01; ****, P ≤ 0.0001 by Brown–Forsythe and Welch ANOVA tests with Games–Howell test for multiple comparisons. (D) Western blot (WB) of proteins from whole-cell extracts derived from rapamycin- or carrier-alone (DMSO)–treated cells expressing Atg39-GFP in the indicated strains. GFP detected with anti-GFP.2 antibody, HRP-conjugated secondary antibodies, and ECL. To assess relative protein loading, a portion of the blots are shown stained with Ponceau S. Position of mol wt standards (in kD) at right. (E) Deconvolved fluorescence micrographs of rapamycin- or carrier-alone (DMSO)–treated cells overexpressing mCherry-Atg39 (magenta) and heh1-ΔL-GFP (green) in a pep4Δ strain; vacuoles are labeled with FM 4-64 (magenta). Merged fluorescent images shown. Asterisks denote the nucleus; arrowheads point to colocalization of GFP and mCherry in vacuole. Scale bars are 5 μm. Source data are available for this figure: SourceData F6.
To test whether the autophagy machinery selectively targeted the NE blebs that arise from Atg39 overexpression, we tested putative colocalization between endogenous GFP-Atg8 and overexpressed Atg39-mCherry after 2 h of treatment with rapamycin. As shown in Fig. 7, A and B, we observed that most (∼70%) of the GFP-Atg8 foci were found colocalized with the NE blebs. Thus, these data suggest that Atg8 preferentially associates with the NE blebs over the rest of the NE. This conclusion was emphasized when we assessed the dynamics of GFP-Atg8’s interactions with the NE over a 2-h time course. In Video 1, GFP-Atg8 foci colocalized with the Atg39-mCherry NE blebs in waves, which can also be visualized in the still frames shown in Fig. 7 C. When the fluorescence of a region of interest (ROI) encompassing the nucleus was plotted over time, we observed an interesting periodicity of ∼6.5 min between the sequential appearances of GFP-Atg8 at the sites of Atg39-mCherry accumulation (Fig. 7 D). Consistent with the idea that interactions between GFP-Atg8 and Atg39-mCherry result in the removal of Atg39-mCherry, we also observed the continual decline of mCherry fluorescence over the 2-h time course (Fig. 7 D). In the context of the data demonstrating that Atg39 can be degraded by autophagy under analogous conditions, we conclude that the Atg39-induced NE blebs can be directly targeted for removal and degradation by the autophagy machinery.
Atg8 specifically interacts with NE blebs. (A) Deconvolved fluorescence micrographs of GFP-Atg8 and Atg39-mCherry with merge acquired immediately after treatment of cells with rapamycin (0 h) or after 2 h. Scale bars are 5 μm. (B) Stacked bar graph of the percentage of cells with a visible GFP-Atg8 focus and its localization with reference to the NE as in key. 50 cells from three independent replicates were evaluated. (C) Deconvolved fluorescence micrographs of a time course (2-min intervals for 2 h) of cells expressing GFP-Atg8 (green) and Atg39-mCherry (magenta) after treatment with rapamycin for 1 h. Merged fluorescence micrographs are shown. Colocalization events are labeled with Roman numerals. Scale bar is 1 μm. (D) Line plot of normalized fluorescence intensity of GFP-Atg8 (green) and Atg39-mCherry at the nucleus over time from the cell shown in C. GFP peaks correspond to colocalization events labeled with Roman numerals with corresponding labels in C. Average and SD of periodicity (P) of GFP-Atg8/Atg39-mCherry colocalization events was determined from 20 cells, from a total of 324 unique colocalization events.
Atg8 specifically interacts with NE blebs. (A) Deconvolved fluorescence micrographs of GFP-Atg8 and Atg39-mCherry with merge acquired immediately after treatment of cells with rapamycin (0 h) or after 2 h. Scale bars are 5 μm. (B) Stacked bar graph of the percentage of cells with a visible GFP-Atg8 focus and its localization with reference to the NE as in key. 50 cells from three independent replicates were evaluated. (C) Deconvolved fluorescence micrographs of a time course (2-min intervals for 2 h) of cells expressing GFP-Atg8 (green) and Atg39-mCherry (magenta) after treatment with rapamycin for 1 h. Merged fluorescence micrographs are shown. Colocalization events are labeled with Roman numerals. Scale bar is 1 μm. (D) Line plot of normalized fluorescence intensity of GFP-Atg8 (green) and Atg39-mCherry at the nucleus over time from the cell shown in C. GFP peaks correspond to colocalization events labeled with Roman numerals with corresponding labels in C. Average and SD of periodicity (P) of GFP-Atg8/Atg39-mCherry colocalization events was determined from 20 cells, from a total of 324 unique colocalization events.
Time course of deconvolved fluorescence micrographs of GFP-Atg8, Atg39-mCherry, and merged images (related to Fig. 7 C ). Arrow indicates a GFP-Atg8, Atg39-mCherry containing structure. Scale bar is 1 μm and frame rate is 3 frames/s.
Time course of deconvolved fluorescence micrographs of GFP-Atg8, Atg39-mCherry, and merged images (related to Fig. 7 C ). Arrow indicates a GFP-Atg8, Atg39-mCherry containing structure. Scale bar is 1 μm and frame rate is 3 frames/s.
Atg39-derived NE blebs specifically capture the INM
The second criterion that would provide confidence that the NE blebs are a potentially physiological intermediate in nucleophagy would be the selective incorporation of Atg39 cargo into the blebs. To test this, we monitored the distribution of several integral components of the NE (Fig. 8 A) expressed at endogenous levels, including NPCs (Nup85), SPBs (Spc42, Mps3), and integral INM proteins Heh1, Heh2, and Asi1, in the context of mCherry-Atg39 expression. We also tested the model INM cargos heh1-ΔL-GFP and heh2-ΔWH-GFP, which were both degraded by Atg39-dependent autophagy (Fig. 5, C and E). Consistent with the data that NPCs and SPBs (with associated proteins) are not degraded by an Atg39-dependent mechanism (Fig. 5, D and E; and Lee et al., 2020), we did not observe any appreciable accumulation of Nup85-GFP, Heh2-GFP, Mps3-GFP, or Spc42-GFP (Fig. 8 B, right-side GFP panels, and Fig. 8 C) in the NE blebs. In striking contrast, all integral INM proteins and INM reporters colocalized with mCherry-Atg39 within at least 50% of the blebs (Fig. 8 B, arrowheads, and Fig. 8 C). Thus, taken together, Atg39-dependent NE blebs can selectively capture the INM over other elements of the NE, suggesting that the overexpression of Atg39 might be recapitulating key early steps in an Atg39-dependent nucleophagy pathway that targets the INM.
INM is specifically captured in Atg39-containing NE blebs. (A) Cartoon of protein and protein complexes at the NE, including an SPB and NPC. The boxed region is magnified at bottom to show integral INM proteins and truncations of Heh1 and Heh2 that act as INM reporters. Numbers are amino acids. (B) Deconvolved fluorescent micrographs of the indicated GFP fusion proteins under conditions where mCherry-Atg39 expression is repressed (+ glucose [off], left panels) or induced (+ galactose [on], right panels). Merged images of mCherry (magenta) and GFP (green) are also shown. Arrowheads point to NE blebs containing mCherry-Atg39 and INM proteins. Scale bars are 5 μm. (C) Plot of percentage of cells with Atg39 blebs colocalized with indicated NE proteins. Error bars are SD from three independent replicates of 100 cells per replicate. LEM, LAP2/emerin/MAN1.
INM is specifically captured in Atg39-containing NE blebs. (A) Cartoon of protein and protein complexes at the NE, including an SPB and NPC. The boxed region is magnified at bottom to show integral INM proteins and truncations of Heh1 and Heh2 that act as INM reporters. Numbers are amino acids. (B) Deconvolved fluorescent micrographs of the indicated GFP fusion proteins under conditions where mCherry-Atg39 expression is repressed (+ glucose [off], left panels) or induced (+ galactose [on], right panels). Merged images of mCherry (magenta) and GFP (green) are also shown. Arrowheads point to NE blebs containing mCherry-Atg39 and INM proteins. Scale bars are 5 μm. (C) Plot of percentage of cells with Atg39 blebs colocalized with indicated NE proteins. Error bars are SD from three independent replicates of 100 cells per replicate. LEM, LAP2/emerin/MAN1.
NE blebs contain a network of INM-derived vesicles in the NE lumen
Lastly, to evaluate whether Atg39 overexpression leads to changes in NE morphology that might illuminate early steps in nucleophagy, we performed CLEM and tomography. We first examined cells prepared from cultures expressing high levels (∼10 times endogenous levels as assessed by fluorescence microscopy; Fig. S3 A) of Atg39-GFP (Fig. 9 A, Fig. S3 B, and Video 2). Correlation of regions of Atg39-GFP fluorescence (Fig. 9 A; and Fig. S3, B–F, insets) with their position in EMs revealed an impressive proliferation of membranes. These membranes were derived from the NE as direct continuity could be observed with the ONM in single tomographic slices and in 3D reconstructions; ONM ultimately enclosed the entire structure (Fig. 9 A, ii and iv–x, arrowheads point to ONM continuity). Most strikingly, captured within the expanded ONM were additional bilayers (Fig. 9 A, iv, viii, and x; traced in teal), which were most logically derived from the INM. As these membranes were circular within single tomographic slices, we speculated that they were vesicles. Consistent with this, a 3D model of these segmented membranes revealed that they were spherical and similarly sized with a median diameter of ∼115 nm (Fig. 9 A, iii, and Fig. 9 B). Interestingly, a subset of vesicles were connected by membrane constrictions as if undergoing fission (Fig. 9 A, iii, ix, and x, arrows).
NE ultrastructure at sites of Atg39 accumulation (supplemental to Fig. 9 ).(A) Bar plot of the quantification of corrected total cell fluorescence of endogenous Atg39-GFP along with overexpressed Atg39-GFP (high expression) and GFP-Atg39 (low expression). Corrected total cell fluorescence from 75 cells plotted. (B–F) CLEM tomograms from cells expressing low or high levels of Atg39 as indicated. i: Fluorescence micrograph with arrowheads pointing to ROIs similarly annotated in corresponding EM tomogram. ii: Virtual slice from electron tomogram with the location of fluorescence from i indicated by similarly colored arrowheads. iii: Magnification of boxed view in ii. iv: Annotation of virtual slices from iii, with the ONM in blue, the INM in teal, vacuole in green, and mitochondria in orange. Arrow points to continuity of ONM. Asterisks denote nuclear pores. Scale bars are 100 nm.
NE ultrastructure at sites of Atg39 accumulation (supplemental to Fig. 9 ).(A) Bar plot of the quantification of corrected total cell fluorescence of endogenous Atg39-GFP along with overexpressed Atg39-GFP (high expression) and GFP-Atg39 (low expression). Corrected total cell fluorescence from 75 cells plotted. (B–F) CLEM tomograms from cells expressing low or high levels of Atg39 as indicated. i: Fluorescence micrograph with arrowheads pointing to ROIs similarly annotated in corresponding EM tomogram. ii: Virtual slice from electron tomogram with the location of fluorescence from i indicated by similarly colored arrowheads. iii: Magnification of boxed view in ii. iv: Annotation of virtual slices from iii, with the ONM in blue, the INM in teal, vacuole in green, and mitochondria in orange. Arrow points to continuity of ONM. Asterisks denote nuclear pores. Scale bars are 100 nm.
CLEM of Atg39-GFP reveals expanded ONM with INM-derived vesicles in the NE lumen. (A) i: Virtual slice of an electron tomogram of a cell with high expression of Atg39-GFP (fluorescence image overlaying EM in inset). Boxes represent magnified regions shown in the following panels with corresponding colors. ii: 3D model of continuous ONM, with arrowheads pointing to continuities between substructures. iii: 3D model of likely INM-derived vesicles within the ONM compartment, with arrows pointing to constrictions. iv: Example virtual slice of tomogram with ONM and INM traced in blue and teal, respectively. v and vi: Two views of 3D model where ONM and INM are segmented. vii–x: Further magnifications of single tomographic slices (with and without annotation) as defined by surrounding box color with key in i. Arrowheads point to continuity of ONM and arrows to INM constrictions. Scale bars are 200 nm. (B) Quantification of the diameter of INM-derived compartments from cells with high expression (4 cells) and low expression (10 cells) of Atg39 from electron tomography or high expression of Atg39 from FIB-SEM (52 cells). Total number of INM vesicle diameters measured is indicated in the figure. Median and 95% confidence interval are shown. (C) CLEM of low-expressed GFP-Atg39. i: Fluorescence micrograph of GFP-Atg39 with boxed region ultrastructure shown in ii. Arrowhead points to a region of PMN. ii: Virtual slice from electron tomogram acquired at a region indicated by the box in i. iii: Annotation of virtual slice from ii with the ONM in blue and the INM in teal. iv: 3D surface rendering of annotated structures in iii. Scale bars are (i), 2 μm in i and 50 nm in ii–iv. (D) Quantification of indicated morphologies/substructures observed from 26 total GFP-Atg39 NE foci.
CLEM of Atg39-GFP reveals expanded ONM with INM-derived vesicles in the NE lumen. (A) i: Virtual slice of an electron tomogram of a cell with high expression of Atg39-GFP (fluorescence image overlaying EM in inset). Boxes represent magnified regions shown in the following panels with corresponding colors. ii: 3D model of continuous ONM, with arrowheads pointing to continuities between substructures. iii: 3D model of likely INM-derived vesicles within the ONM compartment, with arrows pointing to constrictions. iv: Example virtual slice of tomogram with ONM and INM traced in blue and teal, respectively. v and vi: Two views of 3D model where ONM and INM are segmented. vii–x: Further magnifications of single tomographic slices (with and without annotation) as defined by surrounding box color with key in i. Arrowheads point to continuity of ONM and arrows to INM constrictions. Scale bars are 200 nm. (B) Quantification of the diameter of INM-derived compartments from cells with high expression (4 cells) and low expression (10 cells) of Atg39 from electron tomography or high expression of Atg39 from FIB-SEM (52 cells). Total number of INM vesicle diameters measured is indicated in the figure. Median and 95% confidence interval are shown. (C) CLEM of low-expressed GFP-Atg39. i: Fluorescence micrograph of GFP-Atg39 with boxed region ultrastructure shown in ii. Arrowhead points to a region of PMN. ii: Virtual slice from electron tomogram acquired at a region indicated by the box in i. iii: Annotation of virtual slice from ii with the ONM in blue and the INM in teal. iv: 3D surface rendering of annotated structures in iii. Scale bars are (i), 2 μm in i and 50 nm in ii–iv. (D) Quantification of indicated morphologies/substructures observed from 26 total GFP-Atg39 NE foci.
High expression of Atg39-GFP results in the formation of NE-blebs (related to Fig. 9 A ). Virtual slices from electron tomograph and 3D surface rendering from tracing throughout the tomography with the ONM in blue and INM in teal. Video shown at 10 frames/s. Scale bar is 100 nm.
High expression of Atg39-GFP results in the formation of NE-blebs (related to Fig. 9 A ). Virtual slices from electron tomograph and 3D surface rendering from tracing throughout the tomography with the ONM in blue and INM in teal. Video shown at 10 frames/s. Scale bar is 100 nm.
The vesicle diameter measurements were likely an underestimate as it was not possible to capture many entire vesicle volumes within the ∼200-nm–thick sections. We therefore also performed FIB-SEM on Atg39-GFP–expressing cells. This approach allowed the visualization of 52 whole-cell volumes (Fig. S4 A), revealing cells with expansive networks of NE blebs that emanated from multiple sites on single nuclei (Fig. S4, B and C; and Video 3). In these 3D images, we measured the diameter of 982 INM-derived vesicles, which had a median diameter of ∼164 nm (Fig. 9 B). Interestingly, we also observed lipid droplets associated with virtually all the NE blebs (Fig. S4, B and C). Thus, the overexpression of Atg39 leads to the generation of a network of likely INM-derived vesicles within the NE lumen alongside an expansion of the ONM with associated lipid droplets. While there are certainly many examples of overexpressed membrane proteins driving changes to membrane morphology, the observed Atg39-dependent morphology is most analogous to that observed in NE “budding” or egress pathways (Fradkin and Budnik, 2016).
FIB-SEM of cells expressing Atg39 with model of nucleophagy (supplemental to Fig. 9 ).(A) Total volume of FIB-SEM images of cells expressing Atg39-GFP. (B) Bar chart of the quantification of NE blebs and associated lipid droplets (LDs) observed in a total of 52 cells captured within the FIB-SEM volume shown in A. (C) i: Orthogonal view of a block of FIB-SEM images (from A) of a single cell with isotropic resolution of 7 × 7 × 7 nm voxels. ii: Surface rendering of NE (blue), LDs (yellow), mitochondria (magenta), and vacuole (gray); a scanning EM image of a single z plane is also shown. iii: Top-down view of ii. iv: Side view of surface rendering of just NE and LDs. Arrowheads point to NE blebs. v: Annotated SEM image of a single z-slice with ONM blue and LD in yellow. vi: Top-down view of NE and LDs, with arrowheads pointing to NE blebs. vii and viii: Zoomed view and alternate angles of region containing NE blebs and LDs. Scale bars are 200 nm. (D) A proposed outside–in model of nucleophagy. Atg39 localizes to the ONM and connects (directly or indirectly) to the INM through lumenal motifs. Evagination of the INM and selective capture of INM cargo (Heh1) requires Atg39. INM-derived vesicles form after an INM scission event. Subsequent ONM scission would also be required to liberate the NE bleb before its capture by the autophagosome through interactions with the N-terminus of Atg39.
FIB-SEM of cells expressing Atg39 with model of nucleophagy (supplemental to Fig. 9 ).(A) Total volume of FIB-SEM images of cells expressing Atg39-GFP. (B) Bar chart of the quantification of NE blebs and associated lipid droplets (LDs) observed in a total of 52 cells captured within the FIB-SEM volume shown in A. (C) i: Orthogonal view of a block of FIB-SEM images (from A) of a single cell with isotropic resolution of 7 × 7 × 7 nm voxels. ii: Surface rendering of NE (blue), LDs (yellow), mitochondria (magenta), and vacuole (gray); a scanning EM image of a single z plane is also shown. iii: Top-down view of ii. iv: Side view of surface rendering of just NE and LDs. Arrowheads point to NE blebs. v: Annotated SEM image of a single z-slice with ONM blue and LD in yellow. vi: Top-down view of NE and LDs, with arrowheads pointing to NE blebs. vii and viii: Zoomed view and alternate angles of region containing NE blebs and LDs. Scale bars are 200 nm. (D) A proposed outside–in model of nucleophagy. Atg39 localizes to the ONM and connects (directly or indirectly) to the INM through lumenal motifs. Evagination of the INM and selective capture of INM cargo (Heh1) requires Atg39. INM-derived vesicles form after an INM scission event. Subsequent ONM scission would also be required to liberate the NE bleb before its capture by the autophagosome through interactions with the N-terminus of Atg39.
Visualization of the whole volumes of a cell expressing Atg39-GFP by FIB-SEM with 3D surface rendering of membranes (related to Fig. S4 C ). The NE is shown in blue, lipid droplets in yellow, vacuole in gray, and mitochondria in dark pink. Video shown at 10 frames/s. Scale bar is 200 nm.
Visualization of the whole volumes of a cell expressing Atg39-GFP by FIB-SEM with 3D surface rendering of membranes (related to Fig. S4 C ). The NE is shown in blue, lipid droplets in yellow, vacuole in gray, and mitochondria in dark pink. Video shown at 10 frames/s. Scale bar is 200 nm.
To gain more insight into the biogenesis of the Atg39-induced compartments, we next performed CLEM on cells expressing lower levels of Atg39 (approximately five times endogenous expression; Fig. S3 A). We examined the ultrastructure at NE sites of local GFP-Atg39 accumulation and where emerging blebs were visible by fluorescence microscopy (Fig. 9 C; and Fig. S3, C–F). Ultimately, we examined 26 individual Atg39 focal NE accumulations within 23 cells. Of these, three could not be attributed to any obvious morphology, with two being localized at sites where mitochondria were adjacent to the NE (Fig. 9 D and Fig. S3 C). In nine cells, we could unambiguously correlate the fluorescence to either nucleus–vacuole junctions (NVJs; three of nine) or regions of piecemeal microautophagy of the nucleus (PMN; six of nine; Fig. 9 D and Fig. S3 D), which may be consistent with recent work supporting a role for Atg39 in PMN (Otto and Thumm, 2021). And, in a single cell, we observed likely extensions to the NE that contained NPCs (Fig. 9 D and Fig. S3 E).
The most prevalent morphology (10 of 26; Fig. 9 D) correlated with GFP-Atg39, as presented in Fig. 9 C and Fig. S3 F. As shown in Fig. 9 C, we observed direct continuity between the INM and vesicles within the NE lumen with an ∼25-nm constriction or bud neck at the INM. As in the high-level expression scenario, these vesicles were again similarly sized (median diameter of 139 nm; Fig. 9 B) and were sometimes found in a series where membrane connections could be visualized, segmented, and 3D modeled (Fig. 9 C, iii and iv; and Video 4). Simultaneously, we observed a clear expansion of the ONM, presumably necessary to accommodate the presence of the extra volume occupied by the vesicles in the NE lumen. Thus, we interpret these structures as precursors to the more elaborate compartments observed upon high-level expression of Atg39. These data suggest that Atg39 may have the ability to coordinate membrane remodeling between the INM and ONM and capture components of the INM into vesicles in the NE lumen.
Low expression of GFP-Atg39 leads to formation of INM-derived vesicles in the NE lumen (related to Fig. 9 C ). Virtual slices from an electron tomogram and 3D surface rendering from tracing throughout the tomography with the ONM in blue and INM in teal. Direct continuity between the INM and INM-derived vesicle necks are visible. Video shown at 10 frames/s. Scale bar is 50 nm.
Low expression of GFP-Atg39 leads to formation of INM-derived vesicles in the NE lumen (related to Fig. 9 C ). Virtual slices from an electron tomogram and 3D surface rendering from tracing throughout the tomography with the ONM in blue and INM in teal. Direct continuity between the INM and INM-derived vesicle necks are visible. Video shown at 10 frames/s. Scale bar is 50 nm.
Discussion
Atg39 has recently emerged as a key player in the autophagic degradation of nuclear and INM components (Mochida et al., 2015; Rahman et al., 2018; Mostofa et al., 2018; Chen et al., 2020; Mizuno et al., 2020; Tomioka et al., 2020); however, how nuclear cargo is selectively packaged and delivered to the cytosolic autophagosome, through the double-membraned NE, has remained an open question. Here, we provide a framework for answering this question by proposing an outside–in model of nucleophagy that depends on Atg39 acting from its position at the ONM (see model in Fig. S4 D). Gratifyingly, concurrent work performed by others suggests a similar model (Mochida et al., 2021, Preprint). At the ONM, Atg39 can engage with the cytosolic autophagy machinery through its cytosolically exposed N-terminal domain while connecting to the INM through its lumenal domain. Evidence in support of this model includes the demonstrated importance of the Atg39 lumenal domain not only for NE targeting and NE remodeling (Fig. 2) but also for the nucleophagic degradation of several model INM cargos (Fig. 5). Such a mechanism may prove relevant to other forms of nuclear autophagy as well. For example, PMN requires the generation of an ONM–vacuole contact site by the pairing of Nvj1 and Vac8 (Pan et al., 2000); the close apposition of the INM and ONM at these sites is thought to be mediated by the Nvj1 lumenal domain (Millen et al., 2008). Interestingly, however, Nvj1 overexpression does not lead to any obvious membrane deformation (Pan et al., 2000). Therefore, recent evidence supporting a role for Atg39 in PMN (Otto and Thumm, 2021) might suggest a model in which Atg39 contributes a membrane remodeling activity capable of coevaginating the INM and ONM, which is a prerequisite for both nucleophagy mechanisms.
Perhaps the most exciting element of this model is the proposal that the INM and associated nuclear content are captured within NE lumenal vesicles derived from the INM (Fig. 9, Fig. S3, and Fig. S4). Although we acknowledge that these structures are observed upon Atg39 overexpression, we argue that they are likely bona fide early intermediates in a physiological nucleophagy mechanism. Our confidence with this conclusion is based on several data. First, there is the remarkable ability of the Atg39-driven NE blebs to selectively capture established INM cargo over other components of the NE (Fig. 8). Second, the NE-blebs can be targeted by autophagy provided that the signal, yet to be defined, is supplied (Fig. 6 and Fig. 7). These data also suggest that the interaction between Atg39 and the autophagy apparatus may be regulated in some way, a common theme with selective autophagy cargo adaptors (Aoki et al., 2011; Farré et al., 2013). As Atg39-derived NE blebs can recruit Atg11 outside of triggering autophagy, at least part of this mechanism might respond directly to the morphology of the NE blebs and/or the local concentration of Atg39 (Fig. 3). Third, the overexpression of Atg39 lacking lumenal elements, in particular the predicted α helix 2, does not drive analogous morphologies (Fig. 2), further fortifying that they are a specific consequence of Atg39 and not simply an accumulation of an overexpressed integral NE protein. Lastly, analogous NE blebs (at least at the level of fluorescence microscopy) are observed at endogenous levels under physiological conditions (Fig. 4).
The enclosure of INM within lumenal vesicles is also attractive because it provides a harmonious mechanism for how the INM could be selectively removed without impacting NE integrity. The observed ∼25-nm membrane necks where the vesicle membranes connect with the INM are strongly suggestive of a protein scaffold that would maintain their stability and ultimately drive membrane scission. This observation, in addition to the chain-like concatenation of the INM-derived vesicles, strongly resembles morphologies associated with ESCRT function (McCullough et al., 2018; Vietri et al., 2020). The hypothesis that ESCRT proteins may ultimately be involved in a scission event that would liberate the INM-derived vesicle into the lumen is attractive not only because of the many links between autophagy and ESCRTs (Takahashi et al., 2018; Li et al., 2019; Loi et al., 2019; Zhou et al., 2019; Schäfer et al., 2020; Zhen et al., 2020) but also because Heh1, the only well-established INM protein cargo of Atg39 (Mochida et al., 2015), directly engages with ESCRTs in pathways that ensure NE integrity (Webster et al., 2016; Gu et al., 2017). It is also noteworthy that we could visualize the entire volume of many of the INM-derived vesicles by tomography, suggesting that INM scission had already occurred in the absence of autophagy induction.
Lastly, the removal of INM contents through the proposed mechanism evokes comparisons to the nuclear-to-cytosolic translocation of so-called “mega” RNPs in some Drosophila neurons that proceeds through a vesicular intermediate in the NE lumen (Speese et al., 2012). Similar structures have been observed in yeast upon inhibiting nuclear export (Ding et al., 2017,Preprint) and as part of a stress response (Panagaki et al., 2021). Thus, we anticipate that the removal of intranuclear contents through the NE lumen will prove to be a generalizable principle of protein, and perhaps RNA, quality control that will be relevant beyond the yeast system and that has, in fact, already been hypothesized (Rose and Schlieker, 2012). Ultimately understanding whether this is the case will require the identification of a mammalian functional homologue to Atg39, which so far remains elusive but is the focus of active investigation.
Materials and methods
Yeast strain construction and culturing conditions
All strains used in this study are listed in Table S1. Genomic integration of sequences encoding fluorescent reporter genes, replacement of endogenous gene promoters with GAL1 promoter, and gene deletions were generated using a PCR-based homologous recombination approach using the pFA6a plasmid series (Longtine et al., 1998; Van Driessche et al., 2005), pK-FRB-GFP, or pK-mEGFP as templates.
To generate DTCPL1403, a loxp/Cre-mediated gene disruption was used to integrate the coding sequence for FRB-GFP immediately after the start codon within the endogenous ATG8 gene. Briefly, a fragment containing a kanamycin resistance cassette (KAN-MX6) flanked by loxp sites and followed by FRB-GFP was amplified from pK-FRB-GFP using primers with homology arms flanking the region targeted for genomic insertion. The PCR product was transformed into W303A, and kanamycin-resistant colonies were selected. To generate the GFP-ATG8 allele, a colony containing the KAN-MX6 cassette integrated into the proper location was transformed with NdelI-linearized pTW040 containing the Cre recombinase under the control of the constitutive TDH3 promoter (Terweij et al., 2013) integrating THD3-CRE at the endogenous HIS3 locus. Successful transformants were selected on synthetic complete (SC) media plates lacking histidine. Colonies were screened for loss of KAN-MX6 and acquisition of fluorescence from GFP-Atg8 after growth on yeast extract–peptone–dextrose medium. A similar strategy was used to insert the coding sequence for mEGFP immediately upstream of the start codon of the endogenous ATG11 gene to generate PMCPL638. For this strain, GAL1-CRE was introduced with the pSH47 (Güldener et al., 1996) centromeric plasmid. Successful transformants were selected on plate media lacking uracil and screened for loss of KAN-MX6 as described above.
Yeast were cultured to mid log phase in yeast peptone (YP) media (1% Bacto Yeast Extract [BD Biosciences], 2% Bacto Peptone [BD Biosciences], 0.025% adenine hemisulfate [Sigma-Aldrich]) supplemented with 2% raffinose (R; BD Biosciences), 2% D-galactose (G; Alfa Aesar), and 2% D-glucose (D; Sigma-Aldrich). To maintain selection of plasmids, cells were cultured in SC medium (Sunrise Science) that lacked the appropriate amino acids. All experiments were performed at 30°C. Standard protocols for transformations, mating, sporulation, and dissection were followed (Amberg et al., 2005).
For induction of autophagy using rapamycin, rapamycin (in DMSO; Sigma-Aldrich) or an equivalent volume of DMSO (carrier) was added to mid log phase cultures to a final concentration of 250 ng/ml. Samples collected at time points indicated in the figures were prepared for imaging or immunoblotting as described below.
To induce autophagy by nitrogen starvation, mid log phase cells were pelleted at ∼375 g, washed twice with synthetic defined lacking nitrogen (SD-N) medium (0.17% Difco Yeast Nitrogen Base without amino acids and ammonium sulfate [BD Biosciences], and 2% D), resuspended in SD-N, and returned to a shaking incubator at 30°C for the amount of time indicated in the figure legends.
To assess the localization of Atg39-GFP, GFP-Atg39, mCherry-Atg39, and Atg39-mCherry under the control of the GAL1 promoter, strains (SCCPL39, SCCPL40, SCCPL80, SCCPL95, SCCPL131, SCCPL155, SCCPL157, DTCPL911, PMCPL87, PMCPL112, PMCPL113, PMCPL114, PMCPL115, PMCPL28, PMCPL29, PMCPL390, PMCPL392, PMCPL422, PMCPL424, DTCPL1483, PMCPL471, PMCPL643, PMCPL645, and PMCPL646) were grown in YPR to mid log phase. Expression was induced by the addition of 2% G, and images were acquired at time points indicated in the figures.
To examine the subcellular localization of Atg39 in the split-GFP assay, strains expressing Atg39–split-GFP fusions under the control of the GAL1 promoter (PMCPL21, PMCPL34, PMCPL35, and PMCPL298) were transformed with plasmids containing split-GFP–mCherry reporters for the nucleoplasm (pSJ1321), ONM/ER (pSJ1568), or lumen (pSJ1602). Cells grown overnight in media lacking leucine were diluted into YPR. Expression of Atg39 fusions was induced by the addition of 2% G for 4 h or as otherwise indicated in the figure legends.
To visualize Atg39-GFP within vacuoles upon induction of autophagy, strain SCCPL22 was treated with 2% G to induce Atg39 overexpression. To visualize the vacuole membrane, 10 ml of culture was transferred to a foil-wrapped tube and incubated with FM 4-64 (final concentration of 15 μM in DMSO; Molecular Probes) for 30 min at 30°C. Cells were then pelleted and resuspended in YPG. Atg39 expression was arrested after 4 h with the addition of 2% D, and the samples were split into two. One culture was treated with rapamycin (final concentration 250 ng/ml; Sigma-Aldrich) to induce autophagy and the other with DMSO (carrier).
Plasmid generation
All plasmids used in this study are listed in Table S2. To generate pPM1 (pFA6a-TRP1-GAL1-GFP1–10), an ORF encoding GFP1–10 was PCR amplified (KOD Polymerase; EMD Millipore) from pSJ1256 using primers with PacI and AscI (New England Biolabs) restriction enzyme recognition sites. The amplicon was restriction digested with PacI and AscI (New England Biolabs), gel purified (QIAGEN), and ligated (T4 DNA ligase; Invitrogen) with gel-purified pFA6a-TRP1-GAL1 digested with PacI and AscI (New England Biolabs). Successful subcloning was confirmed by sequencing.
To generate pFA6a-3xHA-mCherry-natMX6, the 3xHA epitope sequence was PCR-amplified with Q5 DNA polymerase using pFA6a-3xHA-his3MX6 (Longtine et al., 1998) as a template. The PCR product was assembled into pFA6a-GFP-his3MX6 (Longtine et al., 1998) digested with SalI and PacI (New England Biolabs) using the Gibson Assembly Master Mix (New England Biolabs).
To generate pK-FRB-GFP, the FRB-GFP sequence was amplified from pFA6a-FRB-GFP using Q5 DNA polymerase (New England Biolabs). pK3F (Zhang et al., 2017) was digested with BamHI and SalI. The FRB-GFP fragment was ligated into linearized pK3F using Gibson Assembly Master Mix (New England Biolabs). Similarly, to generate pK-mEGFP, the mEGFP sequence was amplified from pFA6a-mEGFP using KOD DNA Polymerase. The mEGFP fragment was assembled into pK3F (Zhang et al., 2017) digested with SalI using the Gibson Assembly Master Mix.
To generate pSC8, the sequence of the native promoter and coding sequence for amino acids 1–443 of HEH1 were amplified via PCR (KOD Polymerase) from genomic DNA using primers with the restriction sites BamHI and HindIII. The amplicon was gel purified (QIAGEN) and ligated into pRS415-GFP linearized with BamH1-HF and HindIII-HF using T4 DNA ligase (Invitrogen). The coding sequence for the C-terminal region of Heh1 was subsequently introduced by annealing oligos encoding HEH1 amino acids 449–477 flanked with restriction sites HindIII and SalI followed by ligation using T7 DNA ligase (Invitrogen). The entire fragment GFP-heh1(1–443)-HindIII-heh1(449–477) was excised by restriction digest with BamHI-HF and SalI-HF, purified, and ligated into pRS405 digested with BamHI-HF and SalI-HF.
pSJ1602 (pRS315-NOP1pr-mCherry-SCS2TM-GFP11) was a gift from Sue L. Jaspersen (Stowers Institute for Medical Research, Kansas City, MO; Addgene plasmid # 86417; http://n2t.net/addgene:86417; RRID:Addgene_86417). pSJ1321 (pRS315-NOP1pr-GFP11-mCherry-PUS1) was a gift from Sue L. Jaspersen (Addgene plasmid # 86413; http://n2t.net/addgene:86413; RRID:Addgene_86413). pSJ1568 (pRS315-NOP1pr-GFP11-mCherry-SCS2TM) was a gift from Sue L. Jaspersen (Addgene plasmid # 86416; http://n2t.net/addgene:86416; RRID:Addgene_86416). pSJ1256 (pFA6a-link-yGFP1-10-CaURA3MX) was a gift from Sue L. Jaspersen (Addgene plasmid # 86419; http://n2t.net/addgene:86419; RRID:Addgene_86419).
Microscopy
For all live-cell imaging, mid log phase cells were gently pelleted and washed with SC media containing 2% D and immediately imaged directly on cover glass. All images were acquired on an Applied Precision DeltaVision microscope (GE Healthcare, Life Sciences) equipped with a 100×/1.4 NA oil immersion objective (Olympus), solid state illumination, CoolSNAP HQ2 charge-couple device (CCD; Photometrics) or electron-multiplying CCD (Photometrics) cameras. The microscope stage was maintained at 30°C within an environmental chamber.
Time-lapse imaging of GFP-Atg8 and Atg39-mCherry (DTCPL1683) was performed in microfluidic plates (Y04C; CellASIC) with the ONIX Microfluidic Platform (CellASIC; EMD Millipore). Prior to loading cells in the microfluidic chamber, Atg39-mCherry expression was induced for 2 h with YPG followed by the addition of D to arrest expression. SC with 2% D containing rapamycin (final concentration, 250 ng/ml; Sigma-Aldrich) was perfused through the chamber at 0.25 psi for 1 h before the start of imaging. Z-stacks (0.4-μm sections) were acquired at 2-min intervals for 2 h.
Image processing and analysis
All presented micrographs were deconvolved using an iterative algorithm in softWoRx 6.5.1 software (Applied Precision, GE Healthcare). Micrographs and immunoblots were analyzed in Fiji/ImageJ (Schindelin et al., 2012). Unprocessed images were used for quantification of fluorescence intensity.
Line profiles of fluorescence intensity were generated using the Plot Profile function in Fiji/ImageJ (Schindelin et al., 2012). The minimum and maximum measured values from individual fluorescent channels were normalized to 0 and 1, respectively.
To compare Atg39-GFP levels by fluorescence microscopy (for Fig. S2 C and Fig. S3 A), the fluorescence of individual z-stacks encompassing the whole-cell volume were sum projected, and corrected total cell fluorescence was calculated for the entire cell area in the projection. Corrected total cell fluorescence was calculated as integrated density of the cell minus the product of the area of the selected cell and the mean fluorescence of background. To quantify changes in Atg39-GFP fluorescence intensity at the NE, the integrated density of an ROI at the nuclear periphery comprising 4 pixels was measured and normalized to mean background fluorescence. To quantify the levels of fluorescence of GFP-Atg8 and Atg39-mCherry at the NE throughout the 2-h time course, individual ROIs were drawn in a single z plane. Total fluorescence in each ROI was measured from individual fluorescence channels and normalized between 0 and 1 before plotting as a line graph.
To determine the average period between Atg8–Atg39 colocalization events, the number of observed colocalization events through a 2-h time course was calculated per cell. Within an individual cell, the time of the last observed colocalization event was subtracted from the time of the first observed colocalization event. This total time was divided by one less than the total number of colocalization events observed within that cell through the time course to calculate the average time between colocalization events within that cell. The average and SD for 20 cells was tabulated. To quantify the levels of Atg39-GFP fluorescence intensity at NE blebs, the mean fluorescence of the bleb was measured and normalized to the mean background fluorescence.
Secondary structure prediction
The secondary structure of Atg39’s lumenal domain was predicted by threading the amino acid sequence of Atg39 through Jpred4 (Drozdetskiy et al., 2015).
Statistical methods
Graphs were generated using Prism 9.0 (GraphPad Software). Statistical significance tests were used as indicated in the figure legends. Significance values were calculated within Prism 9.0 and P values are indicated on the graphs or in figure legends as: ns, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; **** P ≤ 0.0001. Error bars are described in figure legends.
CLEM
Correlative microscopy of resin-embedded cells was performed as previously described (Kukulski et al., 2012; Thaller et al., 2019). Expression of Atg39 fusion proteins from cells (DTCPL688 [Atg39-GFP] and PMCPL87 [GFP-Atg39]) cultured in YPR was induced by the addition of 2% G for 3 h. Cells were then collected by centrifugation for 2 min at 350 g. The yeast slurry was transferred to the 200-μm recess of a 1.5-mm wide aluminum platelet (Engineering Office M. Wohlwend) and placed in an HPM100 (Leica Microsystems) for high pressure freezing. Samples were freeze substituted in 0.1% uranyl acetate in acetone and embedded in Lowicryl HM20 (Polysciences) using the automated temperature control of an EM-AFS2 (Leica Microsystems) with manual agitation and solution exchange following the published protocol (Kukulski et al., 2012). The resin was polymerized under UV light, and the resin-embedded cells were cut into 250-nm-thick sections using an ultramicrotome (Leica Artos 3D) equipped with a diamond knife (Diatome) and collected on 200-mesh copper grids with carbon support (Prod. #01840; Ted Pella).
For fluorescence and brightfield micrographs of resin-embedded sections that were generated as described above, several Z-sections were acquired every 200 nm at each grid square of interest, and in-focus planes were selected for CLEM alignment and presentation in figures. Grids selected for tomography were poststained with uranyl acetate and lead citrate. 15-nm protein A–coated gold beads (Cell Microscopy Core, University Medical Center Utrecht) were adhered to the top and bottom surfaces of grids and used as fiducial markers for the alignment and reconstruction of the tilt series. Single-axis (Fig. 9 A; and Fig. S3, B–F) or double-axis (Fig. 9 C) tilt series were collected on an FEI TF20 EM operating at 200 kV using a high-tilt tomography holder (Model 2020; Fichione Instruments) from approximately −65° to +65°, with acquisition at 1° intervals. Images were acquired using SerialEM software (Mastronarde, 2005) at a 2 × 2 binned pixel size of 1.242 nm using a 4k × 4k Eagle CCD (FEI) camera with a 150-μm C2 aperture and a 100-μm objective aperture. Subsequent reconstruction and segmentation were completed in IMOD (Kremer et al., 1996) in an automated fashion (Mastronarde and Held, 2017). For all virtual slices presented, a Gaussian filter in IMOD was applied to reduce noise. Alignment of fluorescence and EM data were completed using the ec-CLEM Plugin (Paul-Gilloteaux et al., 2017) in the ICY imaging suite (de Chaumont et al., 2012). Low-magnification EM was related to fluorescence and brightfield micrographs by selecting approximately six to eight points that corresponded to features visible in both images.
FIB-SEM
For visualization of entire cellular volumes using FIB-SEM, unfixed yeast slurries loaded into a 1.5-mm-wide, 200-μm-deep aluminum platelet (Engineering Office M. Wohlwend) were frozen using a Leica HMP100 at 2,000 psi. The frozen samples were then freeze substituted using a Leica Freeze AFS unit starting at −95°C using 0.1% uranyl acetate in acetone for 50 h to −60°C, rinsed in 100% acetone, and infiltrated over 24 h at −60°C with Lowicryl HM20 resin (Electron Microscopy Science). Samples were placed in gelatin capsules and UV hardened at −45°C for 48 h. The blocks were cured for a further few days before the resin block was trimmed to a rough area of interest and the surface cleanly cut using a Leica UltraCut UC7. The entire block was carefully removed with a fine blade and mounted on an aluminum stub using conductive carbon adhesive and silver paint (Electron Microscopy Sciences) and then sputtered coated with ∼20 nm Pt/Pd (80/20) using Cressington HR equipment (Ted Pella) to reduce charging effects.
The block of embedded samples was loaded into a dual-beam FIB-SEM (CrossBeam 550; Zeiss) equipped with a gallium ion source used for milling and a secondary electron detector (SE2) for image capture. SmartSEM software (Zeiss) was used to set operation parameters and locate the ROI by acquiring initial SEM images at 10 kV, with a 50-μm width and 30-μm height. A protective layer of platinum was subsequently deposited at the ROI with the FIB (30 kV, 3 nA) to protect the structure and reduce charging. Milling was completed at 30 kV, 50 pA, with a carbon deposit of 30 kV, 3 nA. A trench was rough milled at 30 kv, 30 nA, to expose cells followed by polishing at 30 kV, 3 nA. For the final acquisition, the ROI was finely milled at 30 kV, 300 pA, with a 7-nm milling depth. After milling each slice, an image was acquired at a 7-nm pixel size by detecting backscattered electrons of the primary electron beam (acceleration voltage of 1.5 kV, 2 nA, and aperture diameter of 100 μm) with a pixel dwell time of 3 μs. The total dimensions imaged were 19.25 μm × 35.8 μm × 8.9 μm. Atlas5 (Zeiss) was used for preliminary scanning EM stack alignment, and FIB-SEM image stacks were saved as TIFF and MRC files. The image stacks were imported into Dragonfly software (Object Research Systems) for further alignment. Segmentation of structures was done in IMOD (Kremer et al., 1996) approximately every 35 nm.
Lysate preparation
Cells were harvested as previously described (Zhu et al., 2017). Briefly, ∼1.5 × 108 cells were treated with 10% TCA for 1 h on ice and centrifuged at 15,000 g for 10 min at 4°C. The pellet was washed with ice-cold acetone, homogenized by sonication (Bioruptor UCD-200), and pelleted by centrifugation. After two cycles of washing and sonication, the pellet was vacuum dried for 15 min. The dried cell pellet was then mechanically disrupted with 100 μl glass beads (Sigma-Aldrich) and 100 μl urea cracking buffer (50 mM Tris-HCl, pH 7.5, 8 M urea, 2% SDS, and 1 mM EDTA) followed by addition of 100-μl protein sample buffer (Tris-HCl, pH 6.8, 7 M urea, 10% SDS, 24% glycerol, bromophenol blue, and 10% β-mercaptoethanol).
To assess autophagy through GFP-fallout experiments, ∼1.5 × 108 cells were harvested and suspended in 0.2 M NaOH containing 0.1 M DTT, incubated on ice for 10 min, and followed by the addition of 10% TCA with incubation on ice for 15 min. After centrifugation at 15,000 g for 5 min at 4°C, the pellet was washed with acetone, vacuum dried, resuspended in SDS sample buffer (0.1 M Tris-HCl, pH 7.5, 2% SDS, 10% glycerol, 20 mM DTT), and incubated for 10 min at 65°C followed by 95°C for 3 min.
Immunobloting
Proteins from whole-cell extracts were resolved on 4%–20% SDS-PAGE (4561096; Bio-Rad) followed by transfer of the proteins to 0.2-μm nitrocellulose membranes (Bio-Rad). The membranes were blocked in 5% nonfat milk in TBS, Tween 20 for 1 h and immunoblotted with antibodies against GFP (mouse anti-GFP.1, 4°C overnight, 632381 [Takara Bio, Clontech] or rabbit anti-GFP.2, 1 h at room temperature [gift from M.P. Rout; Cristea et al., 2005]), or against HA (anti-HA-peroxidase, 1 h at room temperature, 12013819001; Roche). Blots were incubated with HRP-conjugated secondary antibodies (1 h at room temperature; goat anti-mouse IgG, 31430, or goat anti-rabbit IgG, 31460; Invitrogen) and visualized by ECL (Thermo Fisher Scientific) using a VersaDoc Imaging System (Bio-Rad). Relative protein loading was visualized using Ponceau S Solution (Sigma-Aldrich).
Quantification of autophagic turnover
To calculate the relative percent degradation of GFP fusion proteins, ROIs were drawn around immunoblot bands corresponding to proteolytically cleaved GFP′ and full-length GFP fusion proteins in Fiji/ImageJ (Schindelin et al., 2012), and the total chemiluminescent signal intensity was measured. Measured values for GFP′ were divided by the sum of GFP′ and related GFP-fused full-length protein intensities.
Quantification of relative protein levels by Western blot
To calculate the relative protein levels, ROIs were drawn around the immunoblot band corresponding to the protein of interest in Fiji/ImageJ. The chemiluminescent signal intensity was measured, and the signal from the background was subtracted. To control for loading, the signal in Ponceau S was inverted, an ROI was drawn around two prominent bands, the signal intensity was measured, and the signal from the background was subtracted. The signal intensity from the immunoblot was then normalized to the signal intensity for Ponceau S for each lane to obtain the relative abundances. Each lane was further normalized to the full length (Fig. S2 A) or the WT (Fig. S2 D).
Online supplemental material
Fig. S1 shows a time course of the production of split-GFP by fluorescence microscopy in medium containing galactose. Fig. S2 is a compilation of Western blots assessing total levels of Atg39 fusion proteins in different contexts. Fig. S3 shows quantification of the relative expression of Atg39 fusion proteins and related CLEM micrographs. Fig. S4 shows segmentation and quantification of FIB-SEM of cells expressing Atg39 and a model of nucleophagy. Video 1 is a time course of cells expressing GFP-Atg8 and Atg39-mCherry after treatment with rapamycin (related to Fig. 7 C). Video 2 shows a 3D surface rendering of the ONM, INM, and NE blebs from an electron tomograph of a cell with high expression of Atg39-GFP shown in Fig. 9 A. Video 3 shows the entire cell volume imaged using FIB-SEM of the cell expressing Atg39-GFP shown in Fig. S4 C with 3D surface rendering of the NE, lipid droplets, vacuole, and mitochondria. Video 4 shows a 3D surface render of the ONM and INM from an electron tomograph of a cell with low expression of GFP-Atg39 shown in Fig. 9 C. Table S1 lists the genotypes and origin of all Saccharomyces cerevisiae strains used in the study. Table S2 lists all plasmids used in the study. Table S3 lists all the oligonucleotides used in this study.
Acknowledgments
We thank the members of the Melia and LusKing laboratories for discussion and feedback. We are indebted to Morven Graham and Xinran Liu for invaluable EM expertise. We thank M.P. Rout (The Rockefeller University, New York, NY) and S.L. Jaspersen (Stowers Institute for Medical Research) for reagents.
This work was funded by National Institutes of Health grants R21 AG058033 to C.P. Lusk and T.J. Melia, R01 GM105672 to C.P. Lusk, and F32 GM139285 to N.R. Ader.
The authors declare no competing financial interests.
Author contributions: Conceptualization: S. Chandra, P.J. Mannino, D.J. Thaller, T.J. Melia, and C.P. Lusk. Methodology: S. Chandra, P.J. Mannino, D.J. Thaller, and N.R. Ader. Investigation: S. Chandra, P.J. Mannino, D.J. Thaller, and N.R. Ader. Validation: S. Chandra, P.J. Mannino, D.J. Thaller, and N.R. Ader. Formal analysis: S. Chandra, P.J. Mannino, D.J. Thaller, and N.R. Ader. Writing–original draft: S. Chandra, P.J. Mannino, D.J. Thaller, T.J. Melia, and C.P. Lusk. Writing–review and editing: all authors. Supervision: M.C. King, T.J. Melia, and C.P. Lusk. Funding acquisition: N.R. Ader, T.J. Melia and C.P. Lusk.
References
Author notes
S. Chandra, P.J. Mannino, and D.J. Thaller contributed equally to this paper.





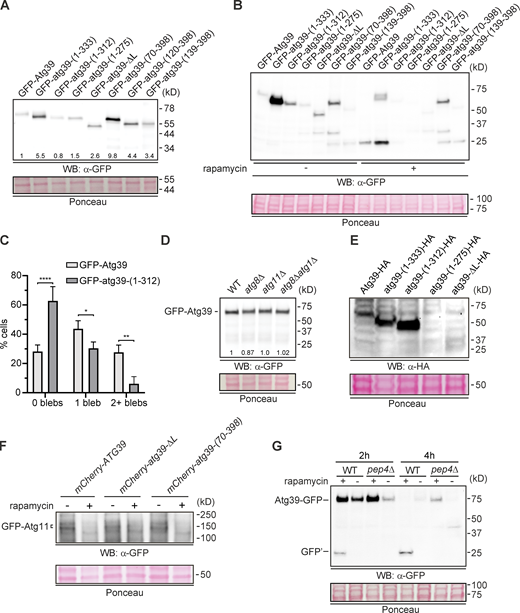



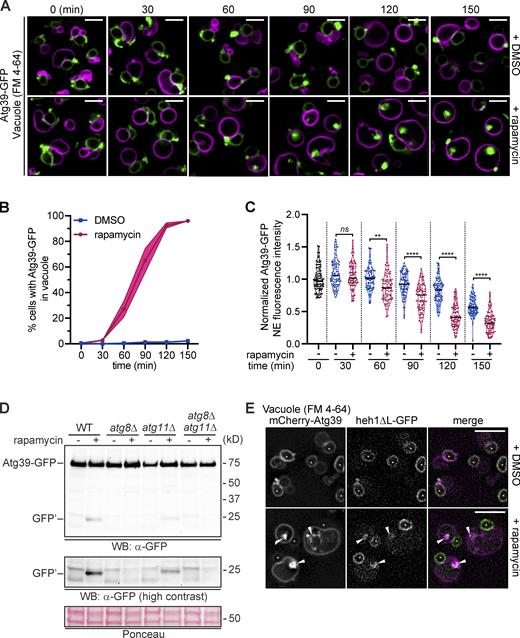

![INM is specifically captured in Atg39-containing NE blebs.(A) Cartoon of protein and protein complexes at the NE, including an SPB and NPC. The boxed region is magnified at bottom to show integral INM proteins and truncations of Heh1 and Heh2 that act as INM reporters. Numbers are amino acids. (B) Deconvolved fluorescent micrographs of the indicated GFP fusion proteins under conditions where mCherry-Atg39 expression is repressed (+ glucose [off], left panels) or induced (+ galactose [on], right panels). Merged images of mCherry (magenta) and GFP (green) are also shown. Arrowheads point to NE blebs containing mCherry-Atg39 and INM proteins. Scale bars are 5 μm. (C) Plot of percentage of cells with Atg39 blebs colocalized with indicated NE proteins. Error bars are SD from three independent replicates of 100 cells per replicate. LEM, LAP2/emerin/MAN1.](https://cdn.rupress.org/rup/content_public/journal/jcb/220/12/10.1083_jcb.202103030/4/m_jcb_202103030_fig8.png?Expires=1773640505&Signature=cuK5aDhsZDmJBPQ3EUhn~VScW~HLB9pJVoDDDLniCFK-Qn9Ak-dIKOG45yPT2QbbrqfUR94wEVnYcLUAlB4j5dXD~aFiTalm~SjzCh40ICT9laKwXufmb-KbEZH42VClgCBLoKeoS5g7aYyqz49dkku7hiLXKFTVKXyozXAfbyAfbTf261kAwjIHzYZkaMp7GFuSFOLte2F7arCl92wtzSdMqsPmy0n8-k246r3CnCyn3oWUph95Uqo8zVdZmOc5fzH-bK2-3BcGvjzExbE7LH2L8cIfcHMbaFFu98IYTHbjyDZO6TEvN2o-K9grMcRSwEVmxbXPe-O1Ht6cLnEyhg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)





