Primary cilia function as critical signaling hubs whose absence leads to severe disorders collectively known as ciliopathies; our knowledge of ciliogenesis remains limited. We show that Smo induces ciliogenesis through two distinct yet essential noncanonical Hh pathways in several cell types, including neurons. Surprisingly, ligand activation of Smo induces autophagy via an LKB1-AMPK axis to remove the satellite pool of OFD1. This is required, but not sufficient, for ciliogenesis. Additionally, Smo activates the Gαi-LGN-NuMA-dynein axis, causing accumulation of a portion of OFD1 at centrioles in early ciliogenesis. Both pathways are critical for redistribution of BBS4 from satellites to centrioles, which is also mediated by OFD1 centriolar translocation. Notably, different Smo agonists, which activate Smo distinctly, activate one or the other of these pathways; only in combination they recapitulate the activity of Hh ligand. These studies provide new insight into physiological stimuli (Hh) that activate autophagy and promote ciliogenesis and introduce a novel role for the Gαi-LGN-NuMA-dynein complex in this process.
Introduction
Primary cilia typically form when a cell exits the cell cycle at the G0/G1 stage. At this point, the mother centriole transforms into a basal body, providing a platform for the outgrowth of the ciliary axoneme (Berbari et al., 2009; Choksi et al., 2014; Malicki and Johnson, 2017; Sánchez and Dynlacht, 2016). The absence of primary cilia leads to a variety of severe disorders collectively known as ciliopathies (Reiter and Leroux, 2017). Primary cilia interact with core signaling pathways that play essential roles during development. Among these is the Hedgehog (Hh) pathway, which is crucial for development and regeneration (Sasai and Briscoe, 2012; Briscoe and Thérond, 2013; Bangs and Anderson, 2017; Lee et al., 2016).
In canonical Hh signaling, binding of one of the Hh ligands such as Sonic hedgehog (Shh) to Patched1 (Ptch1) allows Smoothened (Smo; a seven-pass heterotrimeric G-coupled receptor) to accumulate in the primary cilium to ultimately activate Gli transcription factor target genes (Fig. S1 A; Sasai and Briscoe, 2012; Briscoe and Thérond, 2013; Bangs and Anderson, 2017; Lee et al., 2016). In contrast, the noncanonical Hh pathways do not require primary cilia, and in some of these pathways, Smo activates other downstream effectors such as the heterotrimeric G proteins Gαi/o, which activate the small GTPases Rhodamine A and Rac1 (Ras-related C3 botulinum toxin substrate 1) to promote cell migration and dendritic spine formation (Riobo et al., 2006; Jenkins, 2009; Polizio et al., 2011). Interestingly, beyond its role in regulating cAMP levels, Gαi is also part of a conserved protein complex that interacts with the polarity complexes in various organisms to break cell symmetry through spindle orientation and by connecting the plasma membrane to the cytoskeleton. It consists of Gαi, the G protein–signaling modulator 2, also called LGN, Nuclear mitotic apparatus (NuMA), and dynein motor proteins (Kotak et al., 2012; Blumer and Lanier, 2014; di Pietro et al., 2016; Bergstralh et al., 2017). Another Smo effector is the serine-threonine kinase liver kinase B1 (LKB1), which functions in regulating cell metabolism and growth, and whose activation by Smo inhibits adipocyte differentiation (Teperino et al., 2012; Fleury et al., 2016). In other contexts, LKB1 has been shown to form an axis with the key energy sensor AMPK, a protein that can activate autophagy both by inhibiting the mTOR pathway and by phosphorylating the phagophore initiating factor Unc-51-like autophagy activating kinase 1 (ULK1; Shaw et al., 2004; Fan et al., 2015; Mans et al., 2017).
Hh activation induces ciliogenesis in RPE1 cells and primary neurons. (A) Schematic indicating canonical and noncanonical Hh pathways. (B) Immunofluorescence images of RPE1 Ptch1:GFP cells treated with Shh. Ptch1 localized to cilia when Hh pathway was inactive and was removed from cilia upon Hh activation. (C) Immunofluorescence stitched images of RPE1 cells plated at different cell densities indicating a bigger field of view for comparison between different cell densities. (D) Immunofluorescence images of neurons from mouse cortex indicating staining with adenylate cyclase III (AC3) as another ciliary marker. (E) Immunofluorescence images of neurons and “nonneuron” cells in the same culture indicating lack or presence of YAP1 expression in neurons and nonneurons, respectively. Images are stitched to provide a bigger field of view. Yellow and red numbers and arrows indicate the presence or absence of a cilium, respectively.
Hh activation induces ciliogenesis in RPE1 cells and primary neurons. (A) Schematic indicating canonical and noncanonical Hh pathways. (B) Immunofluorescence images of RPE1 Ptch1:GFP cells treated with Shh. Ptch1 localized to cilia when Hh pathway was inactive and was removed from cilia upon Hh activation. (C) Immunofluorescence stitched images of RPE1 cells plated at different cell densities indicating a bigger field of view for comparison between different cell densities. (D) Immunofluorescence images of neurons from mouse cortex indicating staining with adenylate cyclase III (AC3) as another ciliary marker. (E) Immunofluorescence images of neurons and “nonneuron” cells in the same culture indicating lack or presence of YAP1 expression in neurons and nonneurons, respectively. Images are stitched to provide a bigger field of view. Yellow and red numbers and arrows indicate the presence or absence of a cilium, respectively.
Despite the importance of primary cilia function, the upstream regulators that initiate ciliogenesis are poorly understood. Typically, studies on ciliogenesis in tissue culture rely on nutrient starvation to induce autophagy, a process that promotes the degradation and recycling of cytoplasmic proteins (Nakatogawa et al., 2009; Bento et al., 2016). Autophagy induces ciliogenesis by promoting the selective degradation of a ciliopathy protein, oral-facial-digital syndrome 1 (OFD1) from centriolar satellites, the membraneless granules that mediate trafficking of materials to and from centrioles (Pampliega et al., 2013; Tang et al., 2013; Gheiratmand et al., 2019; Odabasi et al., 2019; Quarantotti et al., 2019). In autophagy-deficient cells, an important ciliary protein, Bardet–Biedl syndrome 4 (BBS4), remains in satellites and fails to be recruited to cilia, coinciding with the accumulation of OFD1 on satellites in these cells (Tang et al., 2013). Whether this reflects a direct recruitment of BBS4 by OFD1 is not known. In addition to the satellites, OFD1 also localizes to the distal end of centrioles, which is essential for restraining centriole elongation and promoting ciliogenesis by recruiting distal appendage proteins and the ciliary protein Intraflagellar transport protein 88 (IFT88; Ferrante et al., 2006; Singla et al., 2010).
In physiological conditions where cells are not actively serum starved, high cell density (HCD) is considered one of the main factors that drive ciliogenesis, possibly through Yes-associated protein 1 (YAP1) inactivation (i.e., Hippo pathway activation; Kim et al., 2015). While this could be true for many cells, there are several cell types that contain cilia but exist at low cell density (LCD) in vivo, such as fibroblasts in heart (Villalobos et al., 2019), mesenchymal cells in developing gut (Walton et al., 2012), and neuronal stem cells and neurons (Dahl, 1963; Fuchs and Schwark, 2004; Whitfield, 2004; Arellano et al., 2012). In addition, in some of these cells, such as neurons, YAP1 is not expressed, suggesting that its inactivation is not what drives ciliogenesis in these cells (Huang et al., 2016b; Rojek et al., 2019), but rather that other unknown regulators must be involved.
Here we show that Shh activates two noncanonical pathways downstream of Smo to induce ciliogenesis. Smo triggers the activation of autophagy via the LKB1-AMPK axis even in nutrient-rich conditions. As was previously shown, autophagy promotes the degradation of satellite OFD1. However, our results indicate that activation of autophagy and degradation of OFD1 are not sufficient for cilia formation. In addition, Shh-activated Smo forms an axis with the Gαi-LGN-NuMA-dynein complex, resulting in centriolar accumulation of a portion of the remaining OFD1 at early stages of ciliogenesis. We also show that defects in either of these axes prevent BBS4 accumulation at basal bodies. Hence, the combined activity of these two pathways is necessary to drive Hh-mediated ciliogenesis.
Results
Hh activation induces ciliogenesis at LCD and in the presence of serum
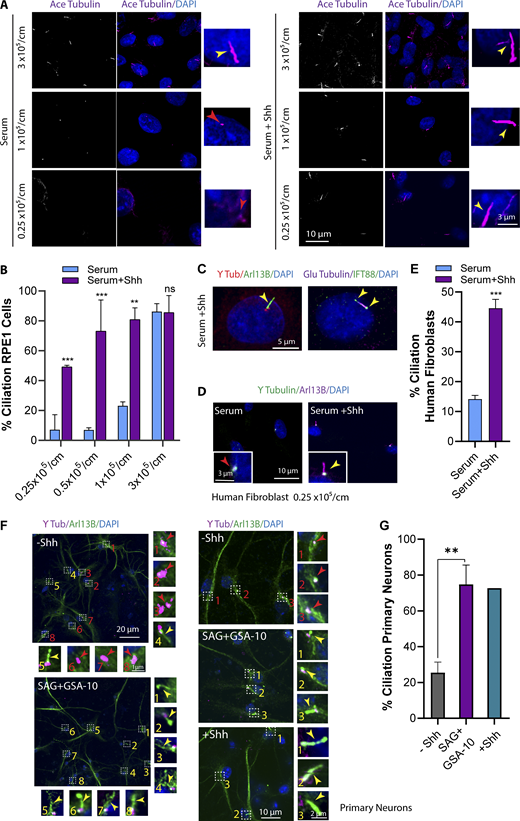
In our search to identify regulators of ciliogenesis, we unexpectedly observed that when retinal pigment epithelial (RPE1) cells growing logarithmically in nutrient-rich conditions (5% FBS) at LCD were treated with Shh conditioned medium (CM), they developed primary cilia. We first confirmed that CM had Shh activity by visualizing Ptch1 removal from cilia using a Ptch:GFP cell line (Fig. S1 B). To confirm that Shh was promoting ciliogenesis, we tested the presence or absence of cilia over a range of cell densities in serum-rich media (Fig. 1, A and B; and Fig. S1 C). At HCD (80–100% confluence), cells formed cilia independent of Shh, consistent with the contribution of Hippo pathway components (YAP1 inactivation) to ciliogenesis (Kim et al., 2015). At medium cell densities (<80% to >30% confluence), even though a population of cells contained cilia, treatment with Shh CM increased the number of ciliated cells up to ∼80%. Surprisingly, however, Hh activation at LCD (20–30% confluence) consistently induced ciliogenesis in cells that otherwise lacked cilia (Fig. 1, A and B; and Fig. S1 C). In addition to ciliary Tubulin markers, these cilia contained bona fide primary cilia proteins such as IFT88 and ADP-ribosylation factor-like protein 13B (Arl13B; Fig. 1 C). We also activated Hh in human foreskin fibroblast cells at LCD in the presence of serum and found that Hh activation induced ciliogenesis in these cells as well (Fig. 1, D and E).
Hh activation induces ciliogenesis at LCD and in the presence of serum. (A) RPE1 cells plated at different cell densities and different conditions as indicated. The number of cells plated/cm2 is indicated. Cells were stained for acetylated (Ace) Tubulin as a ciliary marker. (B) Quantification of ciliated cells at different cell densities. (C) Hh-mediated cilia indicating different ciliary markers localized at cilia. (D) Immunofluorescence images of human fibroblast cells at LCD and treated with Shh (24 h). (E) Quantification of human fibroblast ciliated cells at LCD upon Shh treatment. (F) Immunofluorescence images of neurons from mouse cortex indicating a big field of view (stitches), as well as a higher magnification for ciliogenesis in cells with or without Hh activation. Hh was activated using either Shh CM or dual treatment of SAG plus GSA-10 for 24 h. (G) Quantification of ciliated cells at different conditions as indicated. **, P < 0.001; ***, P < 0.0001 comparing cells with Shh to those without Shh for the same condition. Number of cells (n) used for each experiment is listed in Table S4. Yellow and red numbers and arrows indicate the presence or absence of cilium, respectively.
Hh activation induces ciliogenesis at LCD and in the presence of serum. (A) RPE1 cells plated at different cell densities and different conditions as indicated. The number of cells plated/cm2 is indicated. Cells were stained for acetylated (Ace) Tubulin as a ciliary marker. (B) Quantification of ciliated cells at different cell densities. (C) Hh-mediated cilia indicating different ciliary markers localized at cilia. (D) Immunofluorescence images of human fibroblast cells at LCD and treated with Shh (24 h). (E) Quantification of human fibroblast ciliated cells at LCD upon Shh treatment. (F) Immunofluorescence images of neurons from mouse cortex indicating a big field of view (stitches), as well as a higher magnification for ciliogenesis in cells with or without Hh activation. Hh was activated using either Shh CM or dual treatment of SAG plus GSA-10 for 24 h. (G) Quantification of ciliated cells at different conditions as indicated. **, P < 0.001; ***, P < 0.0001 comparing cells with Shh to those without Shh for the same condition. Number of cells (n) used for each experiment is listed in Table S4. Yellow and red numbers and arrows indicate the presence or absence of cilium, respectively.
Hh activation induces ciliogenesis in neurons
Neurons exist at relatively low cellular density in vivo and contain cilia (Dahl, 1963; Fuchs and Schwark, 2004; Whitfield, 2004; Arellano et al., 2012). In addition, neurons in the neocortex have been shown to lack expression of YAP1 altogether, suggesting that Hippo-mediated translocation of YAP1 from nucleus to cytosol is unlikely to initiate cilia formation (Huang et al., 2016b; Rojek et al., 2019). Neurons contain cilia in vivo but do so poorly in tissue culture, so we tested the possibility that Hh activation may promote their ciliation. To this end, we examined ciliogenesis in neurons obtained from the cortex of mouse brains and activated the Hh pathway. When Hh was activated in these neurons using Shh CM, there was a dramatic increase in the number of ciliated cells in comparison to control (Fig. 1, F and G), however, Shh CM caused many cells to die. For reasons discussed below, we also used a combination of Smo agonists and observed similar results without the associated cell death (Fig. 1, F and G), suggesting that cell death may have been caused by other factors in CM. Additionally, YAP1 expression in cortical neurons was examined using immunofluorescence. Although nonneuronal cells present in the same culture showed expression of YAP1, cortical neurons lacked YAP1 expression, as shown previously (Fig. S1 E; Huang et al., 2016b; Rojek et al., 2019). This suggests that the Hh pathway may induce cilia formation in cells that exist at LCD in vivo, that do not express YAP1, and that inefficiently respond to in vitro techniques such as serum starvation. Cilia formation was also confirmed using a neuronal ciliary marker, adenylate cyclase III (Fig. S1 D).
Noncanonical Hh promotes ciliogenesis through Smo activity
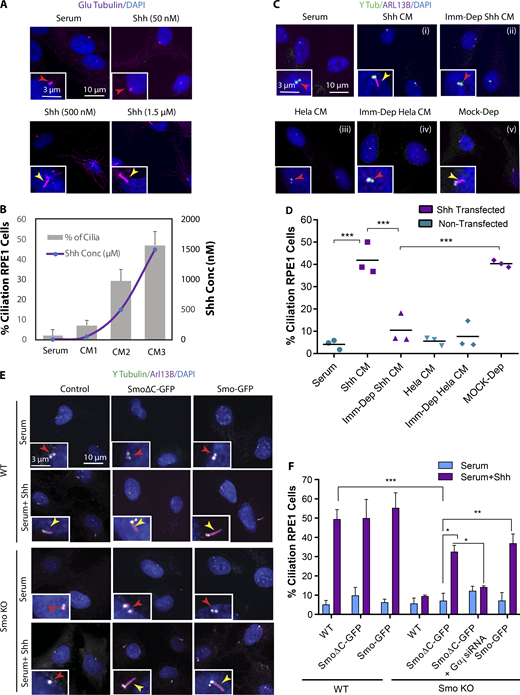
To confirm that the effects were due to Shh ligand in CM, we used two approaches. First, CMs with different concentrations of Shh were produced and quantified by ELISA, and a dose-dependent effect on ciliogenesis was observed (Fig. 2, A and B). In addition, we immunodepleted Shh from CM with antibodies specific for Shh ligand and found that Shh depletion eliminated CM-induced ciliogenesis (Fig. 2, C and D).
Noncanonical Hh promotes ciliogenesis through Smo activity. (A) Immunofluorescence images of RPE1 cells at LCD supplemented with serum and treated with Shh CM (24 h) containing different concentrations of Shh ligand as indicated. (B) Quantification of Shh ligand concentrations and percentage of ciliated cells for each Shh CM as indicated. (C) Immunofluorescence images of RPE1 cells plated at LCD supplemented with serum and treated with Shh CM (i), immunodepleted Shh CM (Imm-Dep Shh CM; ii), CM cultivated from nontransfected HeLa cells (HeLa CM; iii), CM cultivated from nontransfected HeLa cells and immunodepleted for Shh (Imm-Dep HeLa CM; iv), and Shh CM with mock immunodepletion (Mock-Dep), as indicated (v). (D) Quantification of ciliated cells for each condition as indicated. (E) WT and Smo KO RPE1 cells transfected with full-length Smo-GFP and SmoΔC-GFP. Cells were grown in serum, with or without Shh. (F) Quantification of ciliated cells for each condition as indicated. *, P < 0.01; **, P < 0.001; ***, P < 0.0001. Number of cells (n) used for each experiment (three to four repeats) is listed in Table S4. Yellow and red arrows indicate the presence or absence of a cilium, respectively.
Noncanonical Hh promotes ciliogenesis through Smo activity. (A) Immunofluorescence images of RPE1 cells at LCD supplemented with serum and treated with Shh CM (24 h) containing different concentrations of Shh ligand as indicated. (B) Quantification of Shh ligand concentrations and percentage of ciliated cells for each Shh CM as indicated. (C) Immunofluorescence images of RPE1 cells plated at LCD supplemented with serum and treated with Shh CM (i), immunodepleted Shh CM (Imm-Dep Shh CM; ii), CM cultivated from nontransfected HeLa cells (HeLa CM; iii), CM cultivated from nontransfected HeLa cells and immunodepleted for Shh (Imm-Dep HeLa CM; iv), and Shh CM with mock immunodepletion (Mock-Dep), as indicated (v). (D) Quantification of ciliated cells for each condition as indicated. (E) WT and Smo KO RPE1 cells transfected with full-length Smo-GFP and SmoΔC-GFP. Cells were grown in serum, with or without Shh. (F) Quantification of ciliated cells for each condition as indicated. *, P < 0.01; **, P < 0.001; ***, P < 0.0001. Number of cells (n) used for each experiment (three to four repeats) is listed in Table S4. Yellow and red arrows indicate the presence or absence of a cilium, respectively.
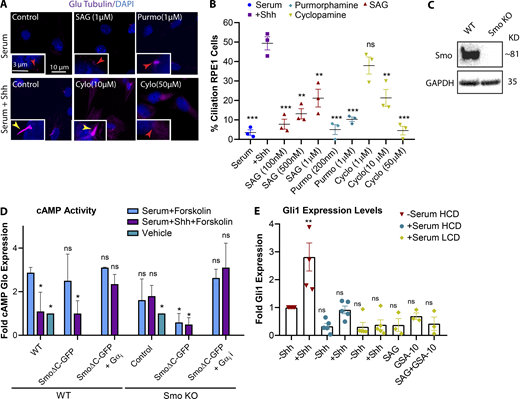
Several Smo agonists and antagonists exist that can activate/inhibit the Hh pathway, but their mechanisms of action depend on where they bind on Smo (Teperino et al., 2012; Gorojankina et al., 2013; Huang et al., 2016a, 2018; Byrne et al., 2018). We used different concentrations of the most commonly used Smo agonists, SAG (Smo agonist) or purmorphamine, to activate Hh pathway, yet even at high concentrations these did not promote cilia formation as efficiently as Shh CM (Fig. S2, A and B). According to recent studies, different Smo agonists activate Smo differently, leading to different downstream outcomes, while Shh ligand may lead to a more complete activation (Teperino et al., 2012, Gorojankina et al., 2013; Huang et al., 2016a, 2018; Byrne et al., 2018). Similarly, different Smo antagonists may target a specific activity of Smo, and some such as cyclopamine can also act as agonists depending on the binding site on Smo (Gorojankina et al., 2013; Huang et al., 2016a, 2018; Byrne et al., 2018). We observed that cyclopamine blocked Hh-mediated ciliogenesis only at very high concentrations (Fig. S2 B). Hence, to confirm that Hh induces ciliogenesis through Smo activity, we used CRISPR to generate a Smo knockout (KO) line in RPE1 cells (Fig. S2 C) and examined cilia formation in the absence of Smo. Unlike the WT cells, Smo KO cells failed to form cilia, indicating that Smo activity is essential for Hh-mediated ciliogenesis (Fig. 2, E and F).
Different Smo agonists and antagonists affect Smo activities, differently. (A) Immunofluorescence images of RPE1 cells plated at LCD and treated with different concentrations of Hh agonists and antagonists as indicated. (B) Quantification of ciliated cells at different conditions as indicated. Statistical significances compare all conditions to control cells with Shh. (C) Western blot probed for Smo indicating Smo KO in CRISPR cell line compared with RPE1 WT. (D) Graph indicating cAMP Glo sensor expression levels as an indicator for cAMP activity for each condition as indicated. (E) Graph indicating Gli1 expression levels for each condition as indicated in comparison to serum-starved ciliated cells. Statistical significance compares all conditions to serum-starved cells at HCD without Shh. *, P < 0.01; **, P < 0.001; ***, P < 0.0001, comparing cells with Shh to those without Shh for the same condition unless otherwise indicated. Number of cells (n) used for each experiment (three repeats) is listed in Table S4. Yellow and red numbers and arrows indicate the presence or absence of a cilium, respectively.
Different Smo agonists and antagonists affect Smo activities, differently. (A) Immunofluorescence images of RPE1 cells plated at LCD and treated with different concentrations of Hh agonists and antagonists as indicated. (B) Quantification of ciliated cells at different conditions as indicated. Statistical significances compare all conditions to control cells with Shh. (C) Western blot probed for Smo indicating Smo KO in CRISPR cell line compared with RPE1 WT. (D) Graph indicating cAMP Glo sensor expression levels as an indicator for cAMP activity for each condition as indicated. (E) Graph indicating Gli1 expression levels for each condition as indicated in comparison to serum-starved ciliated cells. Statistical significance compares all conditions to serum-starved cells at HCD without Shh. *, P < 0.01; **, P < 0.001; ***, P < 0.0001, comparing cells with Shh to those without Shh for the same condition unless otherwise indicated. Number of cells (n) used for each experiment (three repeats) is listed in Table S4. Yellow and red numbers and arrows indicate the presence or absence of a cilium, respectively.
Next, to determine whether Hh-mediated ciliogenesis occurs through the canonical pathway involving Gli activation or one of the noncanonical Hh pathways, we performed a rescue experiment using the Smo KO line. The C-terminus of Smo has been shown to be important for signaling to Gli, and therefore canonical Hh signaling, but is not required for noncanonical Hh signaling (Riobo et al., 2006). To confirm this in our system, given that Gαi activation was previously shown to be a Gli-independent noncanonical Hh pathway, its activation via SmoΔC (Smo1-566; lacking C terminus) at LCD was assessed. To do so, we performed a GloSensor cAMP luciferase-based assay, which allows monitoring of Gαi activity via cAMP levels as a readout. We observed that SmoΔC expression reduced cAMP levels, and that this relies on the presence of Gαi. These results suggest that SmoΔC is sufficient to activate Gαi (Fig. S2 D).
Next, we expressed full-length Smo and SmoΔC constructs in an attempt to rescue the loss of ciliogenesis observed in Smo KO cells. Both full-length Smo and SmoΔC significantly and equivalently rescued the loss of ciliogenesis. We also showed that the ability of SmoΔC to rescue ciliogenesis relies on the presence of Gαi, as SmoΔC failed to rescue ciliogenesis in Smo KO cells lacking Gαi (Fig. 2, E and F). This result indicates that Smo interaction with Gli, and therefore canonical Hh signaling, is not required for its function in cilia formation, and that noncanonical Hh signaling is sufficient to induce ciliogenesis. Moreover, using quantitative RT-PCR, we did not detect significant changes in Gli1 expression levels at LCD in response to Shh (Fig. S2 E).
Hh activates autophagy to promote ciliogenesis
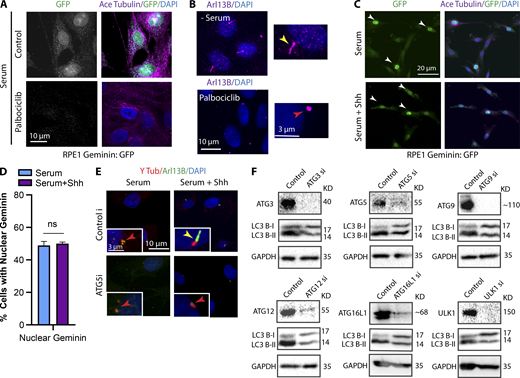
Next, we wanted to determine how noncanonical Hh promotes cilia formation. Ciliogenesis in tissue culture is mainly initiated by serum starvation, which arrests the cell cycle and triggers autophagy; both are thought to be important for ciliogenesis. However, RPE1 cells growing in serum and treated with palbociclib, a CDK4/6 inhibitor that arrests the cells at G0/G1 (Cadoo et al., 2014), did not form cilia, suggesting that cell cycle arrest is not sufficient to initiate cilia formation (Fig. S3, A and B). In addition, using a stable RPE1 cell line expressing a Geminin:GFP reporter, we showed that Shh treatment did not cause cell cycle exit (Fig. S3, C and D). Geminin is a DNA replication factor, and its nuclear expression is an indicator of S-to-G2 progression. The population of cells expressing nuclear Geminin:GFP was similar regardless of presence or absence of Shh, suggesting that Shh did not cause cell cycle exit (Fig. S3, C and D).
Confirmation of Palbociclib activity and depletion ofautophagy proteins by siRNA. (A) Immunofluorescence images of RPE1 Geminin:GFP cells treated with Palbociclib (200 nM) for 24 h to arrest the cell cycle. Geminin, the DNA replication inhibitor, is absent in G1 phase and is highly expressed at the beginning of S phase through G2. Using Palbociclib, cells were Geminin negative showing G0/G1 arrest. (B) Immunofluorescence images of RPE1 cells treated with Palbociclib (200 nM) for 24 h to arrest the cell cycle at G0/G1 phase. Arl13B was used as a ciliary marker. (C) Stitched images of RPE1 Geminin:GFP cells treated with or without Shh at LCD and stained for GFP, indicating cells expressing nuclear GFP signals. White arrows indicate the presence of Geminin in nucleus. (D) Quantification of cells expressing nuclear Geminin at different conditions as indicated. (E) RPE1 cells grown in serum, with or without Shh, after depletion of ATG5 as an example for autophagy genes siRNA screen. (F) Western blots probed for different autophagy proteins and LC3, indicating the efficiency of each KD and its effect on autophagy flux in RPE1 cells. Number of cells (n) used for each experiment (three repeats) is listed in Table S4. Yellow and red arrows indicate the presence or absence of a cilium, respectively.
Confirmation of Palbociclib activity and depletion ofautophagy proteins by siRNA. (A) Immunofluorescence images of RPE1 Geminin:GFP cells treated with Palbociclib (200 nM) for 24 h to arrest the cell cycle. Geminin, the DNA replication inhibitor, is absent in G1 phase and is highly expressed at the beginning of S phase through G2. Using Palbociclib, cells were Geminin negative showing G0/G1 arrest. (B) Immunofluorescence images of RPE1 cells treated with Palbociclib (200 nM) for 24 h to arrest the cell cycle at G0/G1 phase. Arl13B was used as a ciliary marker. (C) Stitched images of RPE1 Geminin:GFP cells treated with or without Shh at LCD and stained for GFP, indicating cells expressing nuclear GFP signals. White arrows indicate the presence of Geminin in nucleus. (D) Quantification of cells expressing nuclear Geminin at different conditions as indicated. (E) RPE1 cells grown in serum, with or without Shh, after depletion of ATG5 as an example for autophagy genes siRNA screen. (F) Western blots probed for different autophagy proteins and LC3, indicating the efficiency of each KD and its effect on autophagy flux in RPE1 cells. Number of cells (n) used for each experiment (three repeats) is listed in Table S4. Yellow and red arrows indicate the presence or absence of a cilium, respectively.
Because autophagy is a positive regulator of cilia formation in serum-starved cells, and Hh was shown to influence the levels of autophagy activation in a cilia-dependent manner (Pampliega et al., 2013), we examined the activation of autophagy via Hh at LCD and in the presence of serum by staining for LC3, an autophagy marker. At HCD, cells exhibited high levels of autophagy and formed cilia independently of Shh. However, at LCD, Shh treatment induced the unexpected activation of autophagy, and cells formed cilia (Fig. 3, A and B). To further confirm this result, autophagy flux, a measure for autophagic degradation, was assessed by Western blot by probing for LC3 and p62. An increase in the LC3-II band is indicative of increased autophagic flux, whereas p62 levels decrease upon activation of autophagy. Consistently, we observed that activation of Hh induced autophagy at LCD even in the presence of serum (Fig. 3, C and D). Next, to see whether the activation of autophagy via Hh is required for Hh-mediated ciliogenesis, several of the proteins important for autophagy were depleted using siRNAs, and ciliogenesis was assessed upon Hh activation. We observed that ciliogenesis was reduced (Fig. 3 E; and Fig. S3, E and F), suggesting that, like serum starvation, Hh-mediated ciliogenesis at least partially depends on activation of autophagy. Interestingly, however, serum-starved cells at LCD were unable to form cilia, even though autophagy was activated (Fig. 3, A–C), suggesting that autophagy is insufficient to induce ciliogenesis. To further confirm this, we treated the cells with Torin1, which mediates the activation of autophagy by inhibiting the mTOR pathway; similarly, Torin1 treatment was not enough to promote ciliogenesis (Fig. 3, A–C). This suggests that while autophagy is required, Hh activates other additional pathways to induce ciliogenesis.
Hh activates autophagy to promote ciliogenesis. (A) RPE1 cells plated at different cell densities and different conditions as indicated. The number of cells plated/cm2 is indicated. Cells were stained for LC3 to assess the activation of autophagy and acetylated (Ace) Tubulin as a ciliary marker. Torin1 concentration is 250 nM. Cells were treated with Torin1 or Shh CM for 24 h. (B) Quantification of average of LC3 puncta/cells for each condition as indicated. Statistical significance compares all conditions to serum-starved cells at HCD. (C) Western blots were probed with LC3 or p62 to assess the activation of autophagy at different conditions as indicated. Cells were treated with Torin1 and bafilomycin A1 (BAF A1; 100 nM) for 4 and 2 h, respectively, followed by an additional 24 h supplemented with Shh CM. Black lines separate nonconsecutive parts of the same sets of Western blots. (D) Quantifications of LC3II/LC3I ratio and p62 levels to measure the autophagy flux for Western blots shown in C. (E) Quantification of ciliated cells for each siRNA depletion as indicated. *, P < 0.01; **, P < 0.001; ***, P < 0.0001 comparing all conditions to control cells with Shh. Number of cells (n) used for each experiment (three to four repeats) is listed in Table S4. Yellow, red, and green arrows point to cilium, no cilium, and LC3 puncta, respectively.
Hh activates autophagy to promote ciliogenesis. (A) RPE1 cells plated at different cell densities and different conditions as indicated. The number of cells plated/cm2 is indicated. Cells were stained for LC3 to assess the activation of autophagy and acetylated (Ace) Tubulin as a ciliary marker. Torin1 concentration is 250 nM. Cells were treated with Torin1 or Shh CM for 24 h. (B) Quantification of average of LC3 puncta/cells for each condition as indicated. Statistical significance compares all conditions to serum-starved cells at HCD. (C) Western blots were probed with LC3 or p62 to assess the activation of autophagy at different conditions as indicated. Cells were treated with Torin1 and bafilomycin A1 (BAF A1; 100 nM) for 4 and 2 h, respectively, followed by an additional 24 h supplemented with Shh CM. Black lines separate nonconsecutive parts of the same sets of Western blots. (D) Quantifications of LC3II/LC3I ratio and p62 levels to measure the autophagy flux for Western blots shown in C. (E) Quantification of ciliated cells for each siRNA depletion as indicated. *, P < 0.01; **, P < 0.001; ***, P < 0.0001 comparing all conditions to control cells with Shh. Number of cells (n) used for each experiment (three to four repeats) is listed in Table S4. Yellow, red, and green arrows point to cilium, no cilium, and LC3 puncta, respectively.
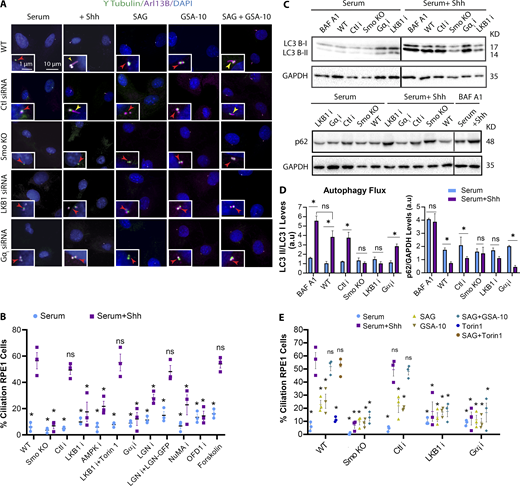
Smo promotes ciliogenesis by activating LKB1-AMPK and Gαi-NuMA axes
To determine the downstream mechanisms driven by Smo that promote ciliogenesis, we investigated the role of two prominent noncanonical Hh pathways, activation of the LKB1-AMPK axis (Teperino et al., 2012; Fleury et al., 2016) and the activation of Gαi via Smo (Riobo et al., 2006; Jenkins, 2009; Polizio et al., 2011). These pathways were investigated because LKB1-AMPK has been linked to the activation of autophagy (Shaw et al., 2004; Fan et al., 2015; Mans et al., 2017), while Gαi, in addition to its role in regulating cAMP levels, can form a conserved complex with LGN, NuMA, and dynein. LGN associates with GDP-bound Gαi via its three GoLoco motifs. When Gαi is GTP-bound, LGN is then released to recruit NuMA to form the Gαi-LGN-NuMA complex, which then regulates dynein motor activity via NuMA-dynein binding (McCudden et al., 2005; Blumer and Lanier, 2014). Dynein has already been implicated as a critical player in the early stages of ciliogenesis (Kotak et al., 2012; Blumer and Lanier, 2014; di Pietro et al., 2016; Bergstralh et al., 2017, Wu et al., 2018). Therefore, to test whether Smo regulates ciliogenesis through either LKB1 or Gαi, we knocked down LKB1, AMPK, Gαi, LGN, and NuMA individually using siRNA and examined ciliogenesis. Knockdown (KD) of each prevented Hh-mediated cilia formation, suggesting that both the LKB1-AMPK and Gαi-LGN-NuMA axes are important for ciliogenesis (Fig. 4, A and B; and Fig. S4, A and B). In support of these data, loss of Hh-mediated ciliogenesis in LGN-depleted cells could be rescued by expression of LGN:GFP (Fig. 4 B). Because Gαi also inhibits adenylyl cyclase, we treated the cell with a cAMP antagonist, Rp-8-bromo-cAMPS, and a cAMP agonist, forskolin, to see whether changes in cAMP could influence Hh-mediated ciliogenesis. Cells treated with Rp-8-bromo cAMPS did not form cilia (Fig. S4 C). Similarly, forskolin treatment did not change the number of ciliated cells in the presence or absence of Hh (Fig. 4 B). These results suggest that Gαi regulation of cAMP levels is unlikely to be a primary cause of Hh-mediated ciliogenesis, whereas its interaction with LGN-NuMA-dynein may play a role in cilia formation.
Smo agonists differentially activate LKB1 or Gαi, and both are required for ciliogenesis. (A) Immunofluorescence images of RPE1 WT, control siRNA, Smo KO, LKB1-depleted, and Gαi (1+2+3)–depleted cells at LCD and treated with Shh or different Smo agonists as indicated. (B) Quantification of ciliated cells at different conditions as indicated. Gαi siRNA represents KD of different isoforms, Gαi (1+2+3), and OFD1 siRNA represents KD of a combination of siRNAs, OFD1(1+2+3). (C) Western blots probed with LC3 or p62 to assess the activation of autophagy at different conditions as indicated. Cells were treated with bafilomycin A1 (BAF A1; 100 nM) for 2 h followed by an additional 6 h supplemented with Shh CM. Black lines separate nonconsecutive parts of the same sets of Western blots. (D) Quantifications of LC3II/LC3I ratio and p62 levels to measure the autophagy flux for Western blots shown in C. (E) Quantification of ciliated cells at different conditions as indicated. *, P < 0.01; **, P < 0.001; ***, P < 0.0001 comparing all conditions to control cells with Shh. Number of cells (n) used for each experiment (three to four repeats) is listed in Table S4. Yellow and red arrows indicate the presence or absence of a cilium, respectively.
Smo agonists differentially activate LKB1 or Gαi, and both are required for ciliogenesis. (A) Immunofluorescence images of RPE1 WT, control siRNA, Smo KO, LKB1-depleted, and Gαi (1+2+3)–depleted cells at LCD and treated with Shh or different Smo agonists as indicated. (B) Quantification of ciliated cells at different conditions as indicated. Gαi siRNA represents KD of different isoforms, Gαi (1+2+3), and OFD1 siRNA represents KD of a combination of siRNAs, OFD1(1+2+3). (C) Western blots probed with LC3 or p62 to assess the activation of autophagy at different conditions as indicated. Cells were treated with bafilomycin A1 (BAF A1; 100 nM) for 2 h followed by an additional 6 h supplemented with Shh CM. Black lines separate nonconsecutive parts of the same sets of Western blots. (D) Quantifications of LC3II/LC3I ratio and p62 levels to measure the autophagy flux for Western blots shown in C. (E) Quantification of ciliated cells at different conditions as indicated. *, P < 0.01; **, P < 0.001; ***, P < 0.0001 comparing all conditions to control cells with Shh. Number of cells (n) used for each experiment (three to four repeats) is listed in Table S4. Yellow and red arrows indicate the presence or absence of a cilium, respectively.
Smo activates autophagy via AMPK, and promotes Gαi activation to induce ciliogenesis. (A) Western blots indicating the efficiency of different KDs in RPE1 cells as indicated. OFD1 siRNAs represent a combination of different OFD1 siRNAs as indicated. (B) Quantification of ciliated cells for depletion of different Gαi isoforms using siRNA in RPE1 cells. (C) Immunofluorescence images of RPE1 cells plated at LCD and treated with Rp-8-Bromo cAMPs in the presence and absence of Shh. (D) Western blot probing for Akt total and phosphorylated to indicate the presence or absence of active mTOR upon Hh activation, respectively. (E) Western blot probing for total and phosphorylated acetyl-CoA carboxylase (ACC) to indicate the activation of AMPK upon Hh activation. (F) Immunofluorescence images of RPE1 cells plated at LCD and treated with SAG or SAG and Torin1 for 24 h.*, P < 0.01; **, P < 0.001, ***, P < 0.0001, comparing all conditions to control cells with Shh. Number of cells (n) used for each experiment (three repeats) is listed in Table S4. Yellow and red arrows indicate the presence or absence of a cilium, respectively.
Smo activates autophagy via AMPK, and promotes Gαi activation to induce ciliogenesis. (A) Western blots indicating the efficiency of different KDs in RPE1 cells as indicated. OFD1 siRNAs represent a combination of different OFD1 siRNAs as indicated. (B) Quantification of ciliated cells for depletion of different Gαi isoforms using siRNA in RPE1 cells. (C) Immunofluorescence images of RPE1 cells plated at LCD and treated with Rp-8-Bromo cAMPs in the presence and absence of Shh. (D) Western blot probing for Akt total and phosphorylated to indicate the presence or absence of active mTOR upon Hh activation, respectively. (E) Western blot probing for total and phosphorylated acetyl-CoA carboxylase (ACC) to indicate the activation of AMPK upon Hh activation. (F) Immunofluorescence images of RPE1 cells plated at LCD and treated with SAG or SAG and Torin1 for 24 h.*, P < 0.01; **, P < 0.001, ***, P < 0.0001, comparing all conditions to control cells with Shh. Number of cells (n) used for each experiment (three repeats) is listed in Table S4. Yellow and red arrows indicate the presence or absence of a cilium, respectively.
Because LKB1-AMPK has previously been linked to autophagy, we hypothesized that Smo can activate autophagy through this axis, whereas other mechanisms, also essential for ciliogenesis, occur through the Gαi-NuMA axis. To confirm this, we first investigated how depletion of LKB1 or Gαi affects the activation of autophagy mediated by Hh, measuring autophagy flux by probing for LC3 or p62 using Western blot. Unlike control cells in which Hh activation promoted the activation of autophagy (as shown by increased autophagic flux), in the absence of Smo or LKB1, the activation of autophagy was interrupted (as shown by the lack of increase in autophagic flux), yet it remained intact in the absence of Gαi (Fig. 4, C and D). These results indicate that Smo can induce the activation of autophagy via LKB1-AMPK, whereas Gαi, along with NuMA, must play other roles in cilia formation.
To further confirm these results, given that LKB1 and AMPK primarily activate autophagy via inactivation of the mTOR pathway, we examined an immediate upstream regulator of mTOR, Akt. Phosphorylation of Akt is expected when mTOR is active, but upon Shh treatment, p-Akt was absent, unlike in control cells (Fig. S4 D). The activation of AMPK via Shh treatment was also confirmed by monitoring phosphorylated acetyl-CoA carboxylase (ACC), a direct substrate of AMPK (Fig. S4 E).
Next, we hypothesized that treating LKB1- or AMPK-depleted cells with Torin1, which activates autophagy, should rescue loss of Hh-mediated ciliogenesis in these cells. Based on our results, unlike LKB1 KD, in which cells were unable to induce cilia formation upon Hh activation, treating cells with Torin1 rescued ciliogenesis upon Hh activation. We also treated AMPK-depleted cells with Torin1 to see whether it rescues ciliogenesis upon Hh activation, but the majority of cells died under this condition, and we were unable to quantify ciliogenesis (Fig. 4 B).
Two Smo agonists differentially activate LKB1 or Gαi, and both are required for ciliogenesis
Previous studies have shown that different Smo agonists can enforce different conformational changes to activate Smo in distinct ways (Teperino et al., 2012, Gorojankina et al., 2013; Huang et al., 2016a, 2018; Byrne et al., 2018). For example, 4-[[(1-hexyl-1,2-dihydro-4-hydroxy-2-oxo-3-quinolinyl)carbonyl]amino]-benzoic acid, propyl ester (GSA-10) does not promote canonical Hh and instead promotes Smo activity toward LKB1 via a noncanonical Hh pathway. Therefore, we hypothesized that Smo could be regulated differentially to activate LKB1 or Gαi. To test this, quantitative RT-PCR was performed first to confirm that GSA-10 does not act through canonical Hh to promote Gli1 expression at LCD (Fig. S2 E). Next, we treated cells with SAG, GSA-10, or a combination of both in the presence or absence of Smo, LKB1, or Gαi and monitored their ability to induce ciliogenesis at LCD in nutrient-rich conditions. Our results indicated that, although WT cells treated with a combination of SAG and GSA-10 formed cilia to the same extent as Shh treatment, SAG or GSA-10 alone could not induce ciliogenesis efficiently (Fig. 4, A and D). In support of these data, when SAG-treated cells were treated with Torin1, which activates autophagy, this combination also promoted ciliogenesis (Figs. 4 D and S4 F). Consistently, in Smo KO and LKB1- or Gαi-depleted cells, cilia did not form regardless of agonist treatment. Taken together, these results suggest that GSA-10 and SAG activate two different pathways downstream of Smo, and although both pathways are necessary, neither on its own is sufficient for Hh-mediated ciliogenesis.
Smo promotes the translocation of OFD1 from satellites to centrioles via Gαi-LGN-NuMA-dynein protein complex
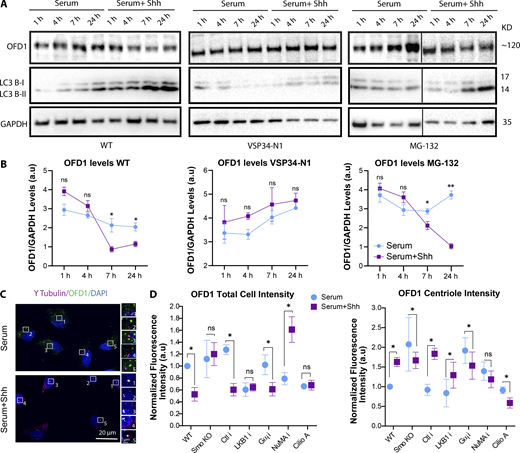
As mentioned, autophagy promotes ciliogenesis by promoting the degradation of OFD1 from centriolar satellites in serum-starved cells. Previous studies had suggested that OFD1 localized at both centrioles and centriolar satellites in cycling cells and is removed from the satellites upon serum starvation (Ferrante et al., 2006; Singla et al., 2010; Tang et al., 2013). Therefore, we sought to see how the levels of OFD1 change upon Hh activation in our conditions over time. To do so, first we performed Western blots to monitor changes in OFD1 levels and observed that OFD1 levels decreased over time upon Hh activation. In contrast, this was interrupted when autophagy was blocked using VSP34-N1 (a PI3K inhibitor that inhibits phagophore formation) but was not affected when the proteasome was blocked using MG-132 (Fig. 5, A and B; and Fig. S5 A).
Smo activation of autophagy via LKB1 promotes the degradation of OFD1. (A) Western blots probed for OFD1 or LC3 to assess degradation of OFD1 or activation of autophagy, respectively, at different conditions as indicated. Cell were treated with or without Shh for different time periods as indicated. Black lines separate nonconsecutive parts of the same sets of Western blots. (B) Quantifications of OFD1 intensity for each Western blot shown in A as indicated. (C) Confocal images (stitched) of RPE1 cells indicating OFD1 distribution in cells plated at LCD and treated with or without Shh for 5 h, stained for OFD1 and γ Tubulin. OFD1 is removed from satellites, while its accumulation increases at centrioles upon Hh activation. Gαi siRNA reparents KD of different isoforms, Gαi (1+2+3). (D) Quantification of OFD1 total cell intensity, and OFD1 centriole intensity at different conditions as indicated. The quantifications were done using images taken with confocal microscopy. *, P < 0.01; **, P < 0.001, comparing Shh treated cells in each condition to those without Shh in the same condition . Number of cells (n) used for each experiment (three to four repeats) is listed in Table S4.
Smo activation of autophagy via LKB1 promotes the degradation of OFD1. (A) Western blots probed for OFD1 or LC3 to assess degradation of OFD1 or activation of autophagy, respectively, at different conditions as indicated. Cell were treated with or without Shh for different time periods as indicated. Black lines separate nonconsecutive parts of the same sets of Western blots. (B) Quantifications of OFD1 intensity for each Western blot shown in A as indicated. (C) Confocal images (stitched) of RPE1 cells indicating OFD1 distribution in cells plated at LCD and treated with or without Shh for 5 h, stained for OFD1 and γ Tubulin. OFD1 is removed from satellites, while its accumulation increases at centrioles upon Hh activation. Gαi siRNA reparents KD of different isoforms, Gαi (1+2+3). (D) Quantification of OFD1 total cell intensity, and OFD1 centriole intensity at different conditions as indicated. The quantifications were done using images taken with confocal microscopy. *, P < 0.01; **, P < 0.001, comparing Shh treated cells in each condition to those without Shh in the same condition . Number of cells (n) used for each experiment (three to four repeats) is listed in Table S4.
OFD1 localization at satellites and centrioles is redistributed upon Hh activation. (A) Confocal stitched images of RPE1 cells to provide a bigger field of view, indicating OFD1 distribution in cells at different conditions as indicated. Cells were plated at LCD, treated with or without Shh for 7 h, and stained for OFD1 and γ Tubulin. OFD1 accumulation decreases at satellites and increases at centrioles upon Hh activation. (B) SIM images indicating OFD1 localization in respect to γ Tubulin as centriole marker in RPE1 WT treated with or without Shh for 5 h; top view from different angles. (C) SIM images indicating OFD1 localization in RPE1 cells at LCD and treated with or without Shh after 24 h when cilium was formed. (D) SIM images indicating BBS4 localization in respect to γ Tubulin as centriole marker in RPE1 WT or Gαi-depleted cells treated with Shh for 5 h; top view from different angles. Unlike WT cells, BBS4 does not localize at centrioles, despite proximity with centrioles.
OFD1 localization at satellites and centrioles is redistributed upon Hh activation. (A) Confocal stitched images of RPE1 cells to provide a bigger field of view, indicating OFD1 distribution in cells at different conditions as indicated. Cells were plated at LCD, treated with or without Shh for 7 h, and stained for OFD1 and γ Tubulin. OFD1 accumulation decreases at satellites and increases at centrioles upon Hh activation. (B) SIM images indicating OFD1 localization in respect to γ Tubulin as centriole marker in RPE1 WT treated with or without Shh for 5 h; top view from different angles. (C) SIM images indicating OFD1 localization in RPE1 cells at LCD and treated with or without Shh after 24 h when cilium was formed. (D) SIM images indicating BBS4 localization in respect to γ Tubulin as centriole marker in RPE1 WT or Gαi-depleted cells treated with Shh for 5 h; top view from different angles. Unlike WT cells, BBS4 does not localize at centrioles, despite proximity with centrioles.
Next, because OFD1 levels started to change within 5–6 h, we monitored changes in OFD1 satellite levels within the cell using confocal microscopy. We observed that within 5 h of Hh activation, the satellite localization of OFD1 was significantly reduced, while OFD1 was concentrated at the centrioles (Fig. 5 C). To confirm that OFD1 is being degraded and not just relocalized to centrioles, we quantified the total cell intensity of OFD1 (which is mainly focused on satellites) versus the portion of OFD1 that colocalizes with the centriole marker γ-Tubulin. Our quantification confirmed that upon Hh activation, the overall intensity of OFD1 in cells was reduced, suggesting that OFD1 is being degraded (Fig. 5 D). In contrast, the centriole localization of OFD1 was increased, suggesting that a portion of OFD1 is also specifically translocated from satellites to centrioles during early ciliogenesis. We also quantified OFD1 levels by interrupting factors downstream of Smo-LKB1 or Smo-Gαi axes. Based on our results, OFD1 degradation from satellites was blocked in Smo KO and LKB1-depleted cells as expected. However, it was intact after Gαi depletion, consistent with our results that autophagy is disrupted in Smo KO and LKB1-depleted cells (Fig. 5 D). Additionally, cells showed defects in OFD1 degradation in the absence of dynein (slight defects) and NuMA (more significant defects), consistent with previous studies indicating roles for dynein and microtubules in autophagosome movement and autophagy (Ravikumar et al., 2005; Nakatogawa et al., 2009; Bento et al., 2016). Surprisingly, we found that in the absence of Smo or Gαi, NuMA, and dynein, OFD1 intensity at centrioles was reduced, suggesting that OFD1 failed to translocate to centrioles (Fig. 5 D). It is noteworthy that the accumulation of OFD1 at centrioles does not simply reflect a redistribution of satellites to the centriole, since the satellite marker PCM1 does not show increased accumulation around the centrioles (top view; Fig. 6 A).
Smo promotes the translocation of OFD1 from satellites to the centrioles via Gαi-LGN-NuMA-dynein protein complex. (A) SIM images (top view) indicating that OFD1 localizes at centriolar satellites using the satellite marker PCM1, and its translocation to distal end of centrioles upon Hh activation using Ninein as subdistal appendage marker. The yellow dashed lines indicate the centriolar satellite area. The cartoon figures indicate how OFD1 looks from different views with respect to other centriolar proteins. (B) SIM images indicating OFD1 localization with respect to γ Tubulin as centriole marker from top view in RPE1 WT, Smo KO, and cells depleted of LKB1, AMPK, Gαi (1+2+3), LGN, and NuMA and treated with Ciliobrevin A (100 nM) at LCD, with or without Shh after 5 h. The cartoon figures are to facilitate better understanding of OFD1 changes in each condition. The red boxes indicate the centriolar satellite area. Yellow and white arrows indicate the presence or absence of OFD1 at centrioles, respectively.
Smo promotes the translocation of OFD1 from satellites to the centrioles via Gαi-LGN-NuMA-dynein protein complex. (A) SIM images (top view) indicating that OFD1 localizes at centriolar satellites using the satellite marker PCM1, and its translocation to distal end of centrioles upon Hh activation using Ninein as subdistal appendage marker. The yellow dashed lines indicate the centriolar satellite area. The cartoon figures indicate how OFD1 looks from different views with respect to other centriolar proteins. (B) SIM images indicating OFD1 localization with respect to γ Tubulin as centriole marker from top view in RPE1 WT, Smo KO, and cells depleted of LKB1, AMPK, Gαi (1+2+3), LGN, and NuMA and treated with Ciliobrevin A (100 nM) at LCD, with or without Shh after 5 h. The cartoon figures are to facilitate better understanding of OFD1 changes in each condition. The red boxes indicate the centriolar satellite area. Yellow and white arrows indicate the presence or absence of OFD1 at centrioles, respectively.
To examine the distribution of OFD1 at the centrioles, we used superresolution structured illumination microscopy (SIM) and examined the changes caused by inhibition of the pathways at different stages. In the majority of interphase cells, OFD1 was focused mainly on satellites and only present in limited amounts at centrioles regardless of which component of the pathway was depleted. Consistent with the quantitation above, within 5 h of Hh activation, the satellite localization of OFD1 was significantly reduced, while a greater portion of OFD1 was concentrated at the centrioles, which remained at basal body after 24 h when cilia were formed (top view; Fig. 6 B; and Fig. S5, B and C; and Videos 1 and 2). Also consistent with our quantifications, the satellite pool of OFD1 failed to be degraded in cells lacking Smo and LKB1, whereas it failed to translocate to centrioles in cells lacking Smo, Gαi, NuMA, and dynein activities. Collectively, these results suggest that following Shh activation, in addition to activation of autophagy via LKB1 resulting in OFD1 degradation from satellites, Smo promotes OFD1 translocation to centrioles by regulating the Gαi-LGN-NuMA-dynein complex; both are essential steps for cilia formation.
Z series of fixed RPE1 WT cell. Cell was imaged using SIM, indicating OFD1 (green) distribution in satellites with respect to centrioles (γ-Tubulin, magenta) as shown in Fig. 6 B. 0.67 Z-stacks (0.11 µm)/frame.
Z series of fixed RPE1 WT cell. Cell was imaged using SIM, indicating OFD1 (green) distribution in satellites with respect to centrioles (γ-Tubulin, magenta) as shown in Fig. 6 B. 0.67 Z-stacks (0.11 µm)/frame.
Z series of fixed RPE1WT cell. Cell was treated with Shh for 5 h and imaged using SIM, indicating OFD1(green) distribution in satellites with respect to centrioles (γ-Tubulin, magenta) as shown in Fig. 6 B. 2 Z-stacks (0.11 µm)/frame.
Z series of fixed RPE1WT cell. Cell was treated with Shh for 5 h and imaged using SIM, indicating OFD1(green) distribution in satellites with respect to centrioles (γ-Tubulin, magenta) as shown in Fig. 6 B. 2 Z-stacks (0.11 µm)/frame.
Both LKB1-AMPK and Gαi-LGN-NuMA-dynein axes are required for BBS4 recruitment to basal body
Previously, it was shown that defects in autophagy interfere with ciliary recruitment of BBS4, a central component of the BBSome and an important ciliary protein (Tang et al., 2013). However, the relationship between OFD1 removal from satellites and accumulation of BBS4 at the centrioles was not clear. One possibility would be that OFD1 retains BBS4 at satellites and its degradation by autophagy is necessary to release it to relocate to the centriole. Alternatively, it is possible that its translocation to centrioles is somehow linked to the translocation of OFD1. To test these possibilities, using SIM, we first monitored BBS4 distribution at satellites and centrioles in the presence or absence of Shh at early stages of ciliogenesis. In the absence of Shh, BBS4 localized to satellites in the vicinity of centrioles (Fig. 7, A and B; and Fig. S5 D; and Videos 3, 4, and 5). However, within 5–6 h of Hh activation, in the majority of cells, BBS4 accumulated at centrioles, similar to what we observed for OFD1. To confirm that this is dependent on OFD1, we then knocked down OFD1 and observed that there was an increased accumulation of BBS4 on satellites upon Hh activation and little if any accumulation at centrioles, suggesting that in the absence of OFD1, its trafficking from satellites and translocation to centrioles is interrupted. Moreover, its retention at satellites argues that OFD1 does not retain BBS4 on satellites (Fig. 7, A and B).
Both LKB1-AMPK and Gαi-LGN-NuMA-dynein axes are required for BBS4 recruitment to basal body. (A) SIM images indicating BBS4 localization with respect to γ Tubulin as a centriole marker in RPE1 WT, Smo KO, and cells depleted of OFD1(1+2+3), LKB1, AMPK, Gαi (1+2+3), LGN, and NuMA and treated with Ciliobrevin A (100 nM) at LCD, with or without Shh for 5 h. Yellow and white arrows point to the presence or absence, respectively, of BBS4 at centrioles. (B) Quantification of BBS4 total cell intensity and BBS4 centriole intensity at different conditions as indicated. The quantifications were done using images taken with confocal microscopy. (C) Graphical abstract indicating how noncanonical Hh induces ciliogenesis via Smo activity. (i and ii) Smo activates LKB1 to promote the activation of autophagy to remove the satellite pool of OFD1 (i) and release BBS4 from satellites (ii). It also activates the Gαi-LGN/NuMA/dynein axis to promote the translocation of a portion of OFD1 to the centrioles and recruitment of BBS4 to basal body and cilia.
Both LKB1-AMPK and Gαi-LGN-NuMA-dynein axes are required for BBS4 recruitment to basal body. (A) SIM images indicating BBS4 localization with respect to γ Tubulin as a centriole marker in RPE1 WT, Smo KO, and cells depleted of OFD1(1+2+3), LKB1, AMPK, Gαi (1+2+3), LGN, and NuMA and treated with Ciliobrevin A (100 nM) at LCD, with or without Shh for 5 h. Yellow and white arrows point to the presence or absence, respectively, of BBS4 at centrioles. (B) Quantification of BBS4 total cell intensity and BBS4 centriole intensity at different conditions as indicated. The quantifications were done using images taken with confocal microscopy. (C) Graphical abstract indicating how noncanonical Hh induces ciliogenesis via Smo activity. (i and ii) Smo activates LKB1 to promote the activation of autophagy to remove the satellite pool of OFD1 (i) and release BBS4 from satellites (ii). It also activates the Gαi-LGN/NuMA/dynein axis to promote the translocation of a portion of OFD1 to the centrioles and recruitment of BBS4 to basal body and cilia.
Z series of fixed RPE1 WT cell. Cell was imaged using SIM, indicating BBS4 (green) distribution in satellites with respect to centrioles (γ-Tubulin, magenta) as shown in Fig. 7 A. 2 Z-stacks (0.11 µm)/frame.
Z series of fixed RPE1 WT cell. Cell was imaged using SIM, indicating BBS4 (green) distribution in satellites with respect to centrioles (γ-Tubulin, magenta) as shown in Fig. 7 A. 2 Z-stacks (0.11 µm)/frame.
Z series of fixed RPE1WT cell. Cell was treated with Shh for 5 h and imaged using SIM, indicating BBS4 (green) distribution in satellites with respect to centrioles (γ-Tubulin, magenta) as shown in Fig. 7 A. 1.33 Z-stacks (0.11 µm)/frame.
Z series of fixed RPE1WT cell. Cell was treated with Shh for 5 h and imaged using SIM, indicating BBS4 (green) distribution in satellites with respect to centrioles (γ-Tubulin, magenta) as shown in Fig. 7 A. 1.33 Z-stacks (0.11 µm)/frame.
Z series of RPE1 Gαi-depleted cells. Cells were treated with Shh, indicating that BBS4 (green) fails to associate with centrioles (γ-Tubulin, magenta). Z series of fixed RPE1 Gαi-depleted cell treated with Shh for 5 h and imaged using SIM, indicating BBS4 (green) distribution in satellites with respect to centrioles (γ-Tubulin, magenta) as shown in Fig. 7 A. 2 Z-stacks (0.11 µm)/frame.
Z series of RPE1 Gαi-depleted cells. Cells were treated with Shh, indicating that BBS4 (green) fails to associate with centrioles (γ-Tubulin, magenta). Z series of fixed RPE1 Gαi-depleted cell treated with Shh for 5 h and imaged using SIM, indicating BBS4 (green) distribution in satellites with respect to centrioles (γ-Tubulin, magenta) as shown in Fig. 7 A. 2 Z-stacks (0.11 µm)/frame.
Interestingly, like OFD1, BBS4 failed to accumulate at centrioles when the Gαi-dynein axis was disrupted, suggesting that its translocation to centrioles coincides with that of OFD1. For example, in Gαi-depleted cells, BBS4 failed to localize to centrioles (Fig. 7, A and B; Fig. S5 D; and Videos 3, 4, and 5). However, in contrast to OFD1, BBS4 also failed to localize to centrioles in most LKB1-depleted cells (Fig. 7, A and B) even though OFD1 still accumulated at centrioles (Figs. 5 D and 6 B). Because OFD1 is unlikely to be retaining BBS4 on satellites, these results suggest that the activation of autophagy and degradation of perhaps another factor other than OFD1 are critical for the release of BBS4 from satellites. Importantly, however, the lack of BBS4 at centrioles in Gαi-depleted cells indicates that its release from satellites is insufficient for its centriolar localization, and this occurs in parallel with the centriolar accumulation of OFD1 via the Gαi-LGN-NuMA-dynein complex. Collectively, these results suggest that after Hh activation, it promotes both the degradation of OFD1 from satellites and its accumulation at centrioles. Shh activation also results in freeing of the satellite pool of BBS4 and the recruitment of BBS4 to centrioles/basal body, possibly via OFD1, all important steps for Hh-mediated cilia formation and function.
Discussion
In this study, we demonstrate that Shh-mediated activation of Smo is a novel regulator of ciliogenesis by activating two separate noncanonical axes: (1) Smo can activate LKB1-AMPK, leading to the activation of autophagy that, as shown previously, is essential for removal of the satellite pool of OFD1, as well as freeing BBS4 from satellites, perhaps by removing other factors; and (2) Smo can also activate the Gαi-LGN-NuMA-dynein axis to promote the translocation of a portion of OFD1 to the centrioles, which in turn mediates the recruitment of BBS4 to the centrioles/basal body (Fig. 7 C). Hh signaling induced ciliogenesis in immortalized retinal pigment epithelial cells, primary human fibroblasts, and primary cortical neurons, all under conditions in which cilia would not be expected, demonstrating the universality of this effect.
We observed that under conditions in which cilia formation or activation of autophagy would not be anticipated, Hh signaling can activate autophagy and induce ciliogenesis. Using a physiological stimulus, the morphogen Shh, we observed the activation of autophagy in cells at low density and in nutrient-rich conditions. Previous studies had shown that Hh activation could increase the levels of autophagic flux in a cilia-dependent manner in cells where autophagy had already been initiated by serum starvation (Pampliega et al., 2013; Tang et al., 2013). Our results, however, indicate that Hh can induce the activation of autophagy independent of cilia and in the presence of serum. Cells lacking LKB1/AMPK, like those lacking Smo, failed to activate autophagy in response to Shh. Given that Smo activates the LKB1-AMPK axis (Teperino et al., 2012; Fleury et al., 2016), and AMPK is a key energy sensor which influences the mTOR pathway, it is likely that Smo activates autophagy through inactivation of mTOR pathway. However, LKB1-AMPK could also contribute to autophagy by phosphorylating one of the essential factors for phagophore nucleation, ULK1 (Shaw et al., 2004; Fan et al., 2015; Mans et al., 2017). We show that Shh activation could activate AMPK and inactivate mTOR. Nonetheless, given that activation of the LKB1-AMPK pathway alone by GSA-10, or inactivation of mTOR via Torin1 were insufficient to induce ciliogenesis, Hh activation must regulate other pathways in addition to autophagy to promote cilia formation. Consistently, activation of autophagy by Torin1 enables SAG to induce ciliogenesis, phenocopying the GSA-10+SAG or Shh CM effects.
In another form of noncanonical Hh, Smo can activate Gαi at the plasma membrane (Riobo et al., 2006; Jenkins, 2009; Polizio et al., 2011). One of the well-known functions of Gαi is regulation of cAMP levels in cells by inhibiting the adenylyl cyclase activity (Blumer and Lanier, 2014). Previously, elevated cAMP levels were shown to promote cilia resorption by activating protein kinase A (PKA) in serum starved cells (Porpora, 2018). However, when we treated cells with a cAMP antagonist that inhibits PKA to phenocopy the effect Gαi on cAMP, cilia formation was not triggered, suggesting that Gαi activation by Smo activates other pathways to promote ciliogenesis.
Dynein function has previously been linked to early stages of ciliogenesis. In particular, a recent paper indicated that at very early stages of ciliogenesis myosin-Va plays a central role in transportation of preciliary vesicles from the Golgi to the mother centriole and their trafficking to the pericentrosomal region depends on dynein activity and microtubules (Ravikumar et al., 2005). Here we show that Gαi promotes the accumulation of OFD1 at centrioles via the Gαi-LGN-NuMA-dynein complex. Given that OFD1 functions in centriole maturation by inhibiting centriole elongation during centriole duplication and promotes distal appendage assembly (Ferrante et al., 2006; Singla et al., 2010), the concentration of OFD1 at centrioles during early stages of ciliogenesis could be a critical step for centriole to basal body transformation. However, centriolar accumulation of OFD1 fails to occur in the absence of Smo, Gαi, NuMA, or dynein. Previously, OFD1 had been reported to localize at both centrioles and centriolar satellites in cycling cells and was removed from the satellites upon serum starvation. However, according to our results its centriole localization is dynamic, and it becomes more concentrated at centrioles during ciliogenesis. Tang et al. (2013) also observed a turnover of the centriolar pool of OFD1, though the authors interpretation was that loss of satellite OFD1 occurs at a significantly faster rate than loss of the centriolar OFD1. However, using super resolution microscopy we found that in the majority of interphase cells, OFD1 was not abundant at centrioles and is, in fact, translocated to the centriole once ciliogenesis has been initiated (e.g., following Hh activation). Therefore, we suggest that in most interphase cells centrioles lack significant levels of OFD1 before ciliogenesis, and OFD1 accumulates at centrioles during ciliogenesis. This is consistent with the fact that OFD1 regulates centriole elongation during the cell cycle and hence before G2, OFD1 may be limited at centrioles to allow for centriole duplication (Ferrante et al., 2006; Singla et al., 2010).
Additionally, as shown previously we observed that BBS4 was associated with satellites in autophagy-deficient cells and was unable to localize to centrioles during early ciliogenesis. We discovered that interrupting either of LKB1 or Gαi axes blocked BBS4 accumulation at centrioles. However, in OFD1 depleted cells we observed an increase in BBS4 satellite pool and lack of its translocation to centrioles. Therefore, in addition to OFD1 degradation from satellites, autophagy seems to release BBS4 from satellites, perhaps through degradation of another factor, while OFD1 translocation to centrioles may be a requisite for BBS4 centriole/basal body accumulation.
Consistent with previous studies showing that the concentration of the Hh morphogen influences the extent of Hh activation (Ribes and Briscoe, 2009; Sasai and Briscoe, 2012), we show that the extent of ciliogenesis also correlates with the concentration of Shh present in CMs. In addition, common Smo agonists such as SAG and puromorphamine were only able to induce ciliogenesis to a limited extent even at high concentrations. Moreover, cyclopamine was unable to inhibit Hh-mediated ciliogenesis to the level we observed in our Smo KO line. These observations are consistent with literature suggesting that different Smo agonists or antagonists could have different impacts on Smo signaling depending on their binding site. Depending on the agonists bound, Smo can adopt distinct conformations, thereby activating specific effectors and leading to different outcomes (Teperino et al., 2012, Gorojankina et al., 2013; Huang et al., 2016a, 2018; Byrne et al., 2018). For example, one Smo agonist, GSA-10, seems to primarily activate LKB1 in a noncanonical Hh pathway, and similar to SAG, it could only induce ciliogenesis to a limited extent. However, combining both SAG and GSA-10 resulted in cilia formation to the same extent as Shh treatment in both RPE1 cells and neurons, suggesting that both Smo activities toward LKB1 (by GSA-10) and Gαi (by SAG) are essential for Hh-mediated ciliogenesis. This result was confirmed when SAG treated cells formed cilia upon treatment with Torin1 (inhibitor of mTOR). Consistently, in Smo KO, as well as in LKB1 or Gαi, depleted cells, cilia did not form regardless of the agonist treatment.
The ability of noncanonical Hh signaling to trigger ciliogenesis could have importance in vivo where the artificial stimuli used to trigger ciliogenesis in vitro, such as serum starvation and Hippo activation, may be lacking. Cells present at low density in certain tissues, such as neuronal stem cells and neurons, may depend on extrinsic signals such as Hh activation, to regulate ciliogenesis. Future studies will be aimed at identifying cell types and conditions where Hh activation may contribute to ciliogenesis in vivo.
Taken together, this study demonstrates novel roles for Smo, acting through noncanonical Hh pathways, to initiate ciliogenesis by indicating: (1) that noncanonical Hh signaling can activate autophagy and promote ciliogenesis without serum starvation; (2) that a novel link exists between the Hh pathway and the Gαi-LGN-NuMA-dynein protein complex; (3) that the Gαi-LGN-NuMA-dynein complex plays an unexpected role in ciliogenesis; (4) that OFD1 is less concentrated at centrioles before ciliogenesis and is translocated to centrioles at early ciliogenesis; and (5) that BBS4 must be released from satellites by autophagy, but that its concentration at centrioles is mediated by OFD1 translocation via the Gαi-LGN-NuMA-dynein complex. These results may provide additional clues about the causes of ciliopathies and the aberrant Hh signaling seen in several cancers.
Materials and methods
Cell culture and transfection
HeLa Tet ON (Clontech), RPE1 WT (CRL-4000; ATCC), and human fibroblasts (generously provided by Dr. Elise Heon, Hospital for Sick Children, Toronto, ON, Canada) cells were grown in DMEM (Wisent), DMEM/F12 (Wisent), and MEM Eagle, alpha modification (Wisent) supplemented with 5–10% FBS (Wisent) and incubated with 5% CO2 at 37°C. For transfection, cells were plated at HCD and transfected using Lipofectamine 2000 (HeLa cells) or Lipofectamine 3000 (RPE1 cells; both Invitrogen), according to the manufacturer’s protocol, except that 3 µl Lipofectamine and 1.5 µg DNA were used per 2 ml of medium. For siRNA transfections, 100 nM siRNAs were used per 1 ml of medium. A list of siRNAs used in this study is provided in Table S1.
For generating Shh CM, HeLa Tet ON cells were transfected with full-length Shh plasmid, and DMEM was replaced by DMEM/F12 at the time of transfection. 24 h after transfection, DMEM/F12 was cultivated from the transfected cells and passed through 0.2-µm filters. Shh CM was stored at 4°C for short storage and −80°C for longer storage (up to 2–3 mo). For LCD studies, cells were transfected at HCD and plated at LCD 5 h after transfection. Hh was activated the next day by replacing Shh CM with DMEM/F12, and cells were fixed and stained 24 h after Hh induction, unless otherwise indicated.
Along with the cell lines mentioned above, the following cell lines were used in this study: RPE1 FlP-IN (kindly provided by Dr. Andrew Wilde, University of Toronto, Toronto, ON, Canada) stably expressing GFP:Ptch1 (generated by Dr. Moshe Kim, Hospital for Sick Children), HeLa Tet ON doxycycline-inducible cells stably expressing full-length mouse Shh plasmid (generated by Dr. Moshe Kim), and RPE1 cells stably expressing Geminin:GFP (generously provided by Dr. Ran Kafri, Hospital for Sick Children).
Small-molecule treatments
The following small molecules were used and dissolved in DMSO, ethanol, or water according to product data sheets as 1,000× stocks: cyclopamine (Cayman Chemical), SAG (Cayman chemical), Pr-8-Bromo cAMPs (sodium salt; Cayman Chemical), purmorphamine (Stemgent), GSA-10 (Tocris), palbociclib (Cayman Chemical), Torin 1, Ciliobrevin A (Tocris), and forskolin (Tocris). Small molecules were added to the cells 2 h before Hh induction and remained in the Shh CM throughout the experiment unless otherwise indicated.
Fixation and immunofluorescence
Cells were fixed for immunofluorescence using 4% (wt/vol) PFA in PBS or 100% cold methanol at 37°C or 4°C, respectively, for 18 min. Before adding the fixing reagent, cells on coverslips were washed with prewarmed (37°C) cytoskeletal buffer (80 mM Pipes, 1 mM MgCl2, and 5 mM EGTA). Cells were then washed three times with 1× TBST buffer (0.15 M NaCl, 0.05 M Tris, pH 7.4, and 0.5% Triton X-100). For BBS4 staining, no cytoskeleton was used, and the three washes were done using PBS. Fixed cells were immunostained for primary antibodies as indicated in Table S2. For staining, coverslips were placed on a piece of parafilm in a chamber with wet paper towels around it to keep the coverslips from drying during the procedure. Cells were blocked using TBST with 5% FBS for 25 min. Then, primary antibodies diluted in TBST with 5% FBS were added to the cells, which were incubated for 1.5 h at RT. Cells were then washed three times with TBST. The secondary antibodies diluted in TBST with 5% FBS were added to the cells and incubated for 1 h at RT. Cells were washed twice with TBST, after which 1:1,000 DAPI (1 mg/ml; Sigma-Aldrich) in TBST was added for 5 min. Cells were washed once with TBST. Lastly, a drop (∼12 µl) of mounting medium (4% n-propyl gallate in 50% glycerol and 0.1 M Tris, pH 8.8) was added to mount each coverslip. The coverslips were placed onto a slide and sealed with nail polish.
Plasmids and molecular biology
The following constructs were used in this study: full-length mouse LGN:GFP, generously provided by Dr. Alisa Piekny (Concordia University, Montreal, QC, Canada); full-length mouse Shh and full-length mouse Smo:GFP plasmids were generously provided by Dr. Chi-chung Hui (Hospital for Sick Children); and SmoΔC:GFP was generated using Gibson Assembly Kit (New England Biolabs) according to the manufacturer’s protocol. Full-length Smo:GFP was used as template, and primers are listed in Table S3. Newly generated plasmid constructs were verified using Sanger sequencing (ACGT Corp.).
SMO CRISPR line
To generate Smo CRISPR KO line in RPE1 cells, gRNA was designed (Table S3) and cloned into Cas-9–enhanced plasmid (71814; Addgene) using a gRNA designing tool and cloning protocol provided at the Zhang laboratory website (MIT, Cambridge, MA). Additionally, a repair plasmid containing Smo homology arms upstream and downstream of the gRNA was generated using the Gibson Assembly Kit. To create the repair plasmid, homology arms were amplified using the primers listed in Table S3 and BAC plasmid clone RP11-595D17 (The Centre for Applied Genomics, Hospital for Sick Children). Next, homology arms 1 and 2 were cloned into pCRISPaint-myc-PuroR plasmid from CRISPaint Gene Tagging Kit (kit 1000000086; Addgene) before and after the Myc-2A-Puro sequence, respectively, using primers in Table S3. Transfection of the repair plasmid along with the Cas-9–enhanced gRNA enabled insertion of the antibiotic resistance gene into to the Smo gene, allowing for selection of the KO cells. Puromycin was used at 10 µM for 10 d, and clonal selection was performed with the remaining cells to obtain a KO clonal line. The selected clones were characterized using immunofluorescence microscopy and Western blotting to ensure the accuracy of the KO. Newly generated plasmid constructs were verified using Sanger sequencing.
ELISA
Shh CMs were diluted with a range of dilutions, and ELISA was performed using mouse ShhN (SHH) ELISA kit (Thermo Fisher Scientific) according to the manufacturer’s instructions.
Immunodepletion
CMs (HeLa or Shh) were cultivated and incubated with Shh or mock antibodies for 4 h at 4°C. Antibodies were added at threefold molar excess. Next, 25 µl of washed protein A beads (Sigma-Aldrich) with ice-cold PBS were incubated with the CM and antibody mix and incubated for another 20–30 min at RT. Beads were pulled down by centrifugation at 4°C for 30 s, and the immunodepleted CMs were collected and sterile filtered before being added to the cells.
cAMP activity assay
cAMP activity levels were measured using cAMP Glo Sensor Technology kit (Promega) according to the manufacturer’s protocol. Cells were transfected with cAMP Glo sensor plasmid along with other constructs or siRNAs depending on the experiment. 5 h after transfection, cells were plated at LCD. The next day, Shh treatment was performed at the end of the day and left overnight. The next day, forskolin was added to cells to activate cAMP for 1 h. Next, cells were incubated with reagent to activate Glo sensor, and cAMP levels were measured according to the manufacturer’s instructions.
Western blot
Cells were lysed using SDS-PAGE sample buffer (50 mM Tris-HCl, pH 6.5, 2% SDS, 10% glycerol, 1% β-mercaptoethanol, 12.5 mM EDTA, and 0.0.2% bromophenol blue). For LCD experiments, 80–90 µl of sample buffer was added to 2-cm wells, and cell lysates from two to three wells were combined. For HCD experiments, 300–400 µl of sample buffer was added per 2-cm wells. Protein lysate containing ∼15–30 µg of total protein (∼20–25 µl of lysed samples) was loaded per well of 4–12% Tris-glycine gels (Invitrogen). Proteins were transferred to polyvinylidene difluoride membrane for 1.5–2 h on an Invitrogen Bolt Minigel Apparatus at 100–120 V and blocked with 5% skim milk overnight at 4°C. Membranes were then incubated with specific antibodies for 1 h at RT. Secondary antibodies conjugated with HRP were used, and blots were developed using the ECL Chemiluminescent Substrate Kit (Invitrogen).
Real-time quantitative PCR
RNA was extracted from cells using TRIzol RNA isolation reagent (Thermo Fisher Scientific). The purified RNA was used for reverse transcription with the SuperScript VILO cDNA Synthesis Kit (Thermo Fisher Scientific). All quantitative PCRs were performed using a CFX Connect Real-Time System (Bio-Rad) with SYBR Green Real-Time PCR Master Mixes (Thermo Fisher Scientific) and 500-nM primer concentration per well. The relative expression of the target genes was normalized to RNA polymerase II levels for each condition and then relative to expression in the HCD sample serum starved without Shh. Primer sequences can be found in Table S3. No-template controls were run for each primer pair to confirm the lack of primer–dimer formation. Five biological replicates were run per condition. Data were analyzed using Excel following Thermo Fisher Scientific’s SYBER Green analysis protocol. All kits were conducted as per the manufacturer’s protocol.
Imaging
Confocal imaging
Fixed cells were imaged using a Quorum Spinning Disk Confocal (Olympus IX81) with 60×/1.35-NA oil-immersion objective and a Hamamatsu C9100-13 electron-multiplying charge-coupled device (EMCCD) camera with Volocity acquisition software (PerkinElmer). Images were acquired as Z-stacks (0.3 or 1 µm for stitches) using the Piezo Z stage (Improvision Piezo). Exposures were set based on control slides 70–95 ms and laser power 30–60. For quantification purposes, stitched images of 4 × 4 were taken and analyzed using Volocity. Image files were exported as TIFFs, which were opened with Java-based software, ImageJ (National Institutes of Health), and converted into maximum-intensity Z-stack projections. Projections and merged color images were then converted into 8-bit images and imported into Illustrator (Adobe).
3D SIM
3D SIM data were collected using an Elyra PS.1 (Carl Zeiss) with a Plan-Apochromat 100×/1.4-NA oil-immersion objective lens with an additional 1.6× optovar. An Andor iXon 885 EMCCD camera was used to acquire images with 101-nm/slice z-stack intervals over a 5–10-µm thickness. The fluorophores were excited with 488 and 555 nm wavelengths, and bandpass 495–550 and 570–620 filters were used to collect the emission wavelengths. Laser powers at the objective focal plane of 52.6 mW in the 5–15% range, exposure time 80–100 ms, and EMCCD camera gain values 10–30 were used during image acquisition. For each image field, grid excitation patterns were collected for five phases and three rotation angles (−75°, −15°, and +45°). The raw data were reconstructed using the SIM module of Zen Black software (v8.1) with noise filter values between 4 and 5.5. Channel alignment was conducted using calibrated files generated from superresolution Tetraspec beads (Carl Zeiss).
Quantification
For quantification, stitched images for each experimental condition were taken. For cilia percentage, the total cell population was determined using the object count function in Volocity using the DAPI channel. Cells positive for ciliary markers were counted using the same function, the percentage of ciliated cells was calculated, and the average of three to four experimental repeats was plotted. For OFD1 measurements, individual cells and OFD1 puncta within each cell were defined using parameters in the find object application in Volocity. A threshold of measurable fluorescence and object size was set in Volocity considering the background, to omit pixels of low signal. An average of the total OFD1 intensity within each cell was determined for each experimental repeat. For OFD1 centriole intensity, OFD1 signals that colocalized with γ-tubulin were determined in each cell, and the average intensity was obtained for each experimental repeat. The average intensities for both total cells and centrioles of OFD1 were then normalized based on serum condition (reported as 1) and plotted on a graph. For all experimental sets, microscope and laser settings remained constant between experiments.
To determine the P values, two-tailed Student’s t test was used assuming unequal values. Error bars represent the SEM for all replicates. For all figures, the following conventions were used: ns, P > 0.01; *, P < 0.01; **, P < 0.001; and ***, P < 0.0001. All the graphs were plotted using GraphPad Prism 8.
Online supplemental material
Fig. S1 shows confocal images of cells at different conditions, supporting involvement of Hh pathway in ciliogenesis. Fig. S2 shows supporting data indicating that Hh induces ciliogenesis through noncanonical pathway. Fig. S3 shows that autophagy is activated and required for noncanonical Hh-mediated ciliogenesis. Fig. S4 shows data supporting that noncanonical Hh activates two distinct pathways via Smo. Fig. S5 shows confocal and SIM images indicating OFD1 and BBS4 distribution in cells treated with or without Shh. Video 1 shows the Z series of RPE1 WT cell indicating OFD accumulation in satellites with respect to centrioles (γ-Tubulin). Video 2 shows the Z series of RPE1 WT cells treated with Shh, indicating OFD1 association with centrioles (γ-Tubulin). Video 3 shows the Z series of RPE1 WT cell indicating BBS4 accumulation in satellites with respect to centrioles (γ-Tubulin). Video 4 shows the Z series of RPE1 WT cells treated with Shh, indicating BBS4 association with centrioles (γ-Tubulin). Video 5 shows the Z series of RPE1 Gαi depleted cells treated with Shh indicating BBS4 fails to associate with centrioles (γ-Tubulin). Table S1 lists the siRNAs used. Table S2 lists the antibodies and dilutions used for each type of experiment. Table S3 lists the primers used. Table S4 lists the number of cells and repeats and experimental conditions for each experiment.
Acknowledgments
We thank Dr. Michael Salter and Janice Hicks at Hospital for Sick Children for generously providing us with primary mouse neurons. We thank Drs. John Brumell, Ran Kafri, Sergio Grinstein, Elise Heon, Chi-chung Hui, and Moshe Kim at the Hospital for Sick Children and Andrew Wilde at the University of Toronto for kindly sharing their cell lines and reagents. We thank Rachel Shannon and Zoe Freidman for preliminary contributions to this work. We thank Kimberly Lau and Paul Paroutis of the Imaging Facility at Hospital for Sick Children for helpful discussions on performing imaging quantifications.
This work was supported by the Canadian Institutes of Health Research (grant PJT-162194 to W.S. Trimble) and a Restracomp Studentship from the Hospital for Sick Children to T. Akhshi. W.S. Trimble is a Canada Research Chair in Molecular Cell Biology.
The authors declare no competing financial interests.
Authors contributions: T. Akhshi and W.S. Trimble designed the study; T. Akhshi performed experiments; T. Akhshi and W.S. Trimble analyzed data; and T. Akhshi and W.S. Trimble wrote the manuscript.
References
Supplementary data
lists the siRNAs used.
lists the primers used.