Clathrin ensures mitotic spindle stability and efficient chromosome alignment, independently of its vesicle trafficking function. Although clathrin localizes to the mitotic spindle and kinetochore fiber microtubule bundles, the mechanisms by which clathrin stabilizes microtubules are unclear. We show that clathrin adaptor interaction sites on clathrin heavy chain (CHC) are repurposed during mitosis to directly recruit the microtubule-stabilizing protein GTSE1 to the spindle. Structural analyses reveal that these sites interact directly with clathrin-box motifs on GTSE1. Disruption of this interaction releases GTSE1 from spindles, causing defects in chromosome alignment. Surprisingly, this disruption destabilizes astral microtubules, but not kinetochore-microtubule attachments, and chromosome alignment defects are due to a failure of chromosome congression independent of kinetochore–microtubule attachment stability. GTSE1 recruited to the spindle by clathrin stabilizes microtubules by inhibiting the microtubule depolymerase MCAK. This work uncovers a novel role of clathrin adaptor-type interactions to stabilize nonkinetochore fiber microtubules to support chromosome congression, defining for the first time a repurposing of this endocytic interaction mechanism during mitosis.
Introduction
The precise and differential regulation of the stability of different populations of microtubules (MTs) during mitosis is critical for multiple aspects of cell division, including chromosome alignment and segregation, spindle positioning, and cytokinesis. The congression of chromosomes to the metaphase plate and their stable alignment is facilitated via multiple mechanisms that rely on astral MTs, kinetochore MTs (kMTs), and non-kMT inner-spindle MTs, as well as associated MT motor proteins including dynein, centromere protein E (CENP-E), and chromokinesins (Maiato et al., 2017). Despite their critical importance, the basic mechanisms and regulation by which different MT populations are (de)stabilized over time and space to carry out these and other functions remain poorly understood.
Clathrin plays an integral role in mitotic MT organization/stabilization and chromosome alignment. During mitosis, clathrin localizes to the mitotic spindle and associates with kinetochore fibers (k-fibers), bundles of MTs that connect centrosomes to the kinetochores on chromosomes (Okamoto et al., 2000; Royle et al., 2005; Booth et al., 2011; McDonald et al., 1992; Nixon et al., 2015). Depletion of clathrin from cells leads to loss of MT stability in mitosis, fewer MTs within k-fibers, and defects in spindle integrity and alignment of chromosomes at the metaphase plate (Booth et al., 2011; Royle et al., 2005; Fu et al., 2010; Lin et al., 2010; Cheeseman et al., 2013). Importantly, these mitotic roles of clathrin are independent of its role in endocytosis and membrane trafficking (Royle et al., 2005; Smith and Chircop, 2012; Cheeseman et al., 2013; Royle, 2013). During mitosis, clathrin forms a complex with the proteins TACC3, the MT polymerase ch-Tog, and PI3K-C2α (Hubner et al., 2010; Lin et al., 2010; Fu et al., 2010; Booth et al., 2011; Gulluni et al., 2017). Formation of this complex (hereafter referred to as the CHC/TACC3 complex) and its recruitment to spindles depends on the direct interaction between clathrin heavy chain (CHC) and TACC3 phosphorylated on serine S558 by the Aurora A kinase, thereby restricting the function of this clathrin complex on MTs to mitosis (Hubner et al., 2010; Fu et al., 2010; Lin et al., 2010; Booth et al., 2011; Burgess et al., 2015, 2018; Hood et al., 2013; Gulluni et al., 2017). While the recruitment of the CHC/TACC3 complex during mitosis to spindles is necessary for MT stabilization and chromosome alignment, the mechanisms by which it stabilizes MTs remains unclear.
Despite the initial characterization of TACC3 homologues in Drosophila melanogaster (D-TACC) and Xenopus laevis (XTACC3/Maskin) indicating a role in preferential stabilization of astral/centrosomal MTs (Gergely et al., 2000; Barros et al., 2005; Kinoshita et al., 2005), most analyses of CHC/TACC3 complex function have focused on k-fibers, where several insights have come from EM analysis. Clathrin localizes near electron-dense “bridges” that have been observed connecting MTs within k-fibers (Hepler et al., 1970; Witt et al., 1981; Booth et al., 2011). A more recent EM tomography study has found that these bridges are more akin to a “mesh” that interconnects MTs (Nixon et al., 2015). Depletion of TACC3 or clathrin from cells leads to a reduction in the number of bridges, as well as MTs, within a k-fiber (Booth et al., 2011). These and other observations have contributed to the hypothesis that CHC/TACC3 complexes may perform dual functions within k-fibers to organize and space MTs via physical bridges, as well as to stabilize MTs that make up the k-fiber by lowering catastrophe rates (Booth et al., 2011; Royle, 2012). While a clathrin-based bridging mesh may organize k-fiber MT bundles, it remains unclear how clathrin stabilizes MTs.
Astral MT stability is governed not only by factors tracking the growing plus-ends of astral MTs (e.g., EB1, Kif18B; Rogers et al., 2002; Stout et al., 2011; Tanenbaum et al., 2011a), but also by proteins concentrated around centrosomes (e.g., γ-tubulin, pericentrin [PCNT]; Zimmerman et al., 2004). TACC3 localizes to both MT plus-ends and spindle poles and stabilizes interpolar and astral MTs in mammalian cells (So et al., 2019; Singh et al., 2014; Gutiérrez-Caballero et al., 2015; Nwagbara et al., 2014). Loss of centrosomal MT stability upon XTACC3 depletion has been shown to arise from increased activity of the MT depolymerase, mitotic centromere-associated kinesin (MCAK)/Kif2C, at centrosomes (Kinoshita et al., 2005). MCAK localizes to centromeres, growing MT plus-ends, and spindle poles during mitosis and plays a major role in controlling astral/centrosomal MT stabilization (Lan et al., 2004; Kline-Smith et al., 2004; Srayko et al., 2005; Rizk et al., 2009; Walczak et al., 1996; Wordeman and Mitchison, 1995; Moore et al., 2005). Regulation of MCAK activity, while clearly dependent on Aurora kinase activity, is incompletely understood and involves multiple factors (Andrews et al., 2004; Lan et al., 2004; Tanenbaum et al., 2011b; Bendre et al., 2016; Zhang et al., 2007; Ems-McClung et al., 2013). It was proposed that a function of TACC3 is to antagonize MCAK’s potent activity at centrosomes to stabilize astral MTs, presumably by promoting activity of the MT depolymerase XMAP215/ch-Tog (Kinoshita et al., 2005). However, whether the CHC/TACC3 complex is required for astral MT stabilization has not been addressed.
Another potential functional member of the CHC/TACC3 complex is GTSE1 (pronounced jitsee one; Monte et al., 2000), an intrinsically disordered protein that has been shown by proteomic analysis to interact with the CHC/TACC3 complex in cells (Hubner et al., 2010). Two recent studies showed that GTSE1 localizes to the spindle during mitosis and, like TACC3, is necessary for astral MT stabilization and efficient chromosome alignment (Bendre et al., 2016; Tipton et al., 2017). We showed that GTSE1 stabilizes MTs in mitosis by inhibiting MCAK activity (Bendre et al., 2016). In that study, cancer cells either depleted or knocked out of GTSE1 showed destabilization of both astral MTs and kinetochore–MT attachments. Another study reported that a specific “slow turnover” population of MTs becomes stabilized upon depletion of GTSE1 (Tipton et al., 2017). The reason for these seemingly contradictory results regarding MT stability after GTSE1 depletion is unclear. GTSE1 requires both CHC and TACC3 to localize to spindles, but the interactions and mechanism by which the CHC/TACC3 complex recruits GTSE1 to the spindle is unknown (Hubner et al., 2010; Bendre et al., 2016; Cheeseman et al., 2013).
Clathrin’s function in endocytosis and vesicle trafficking is dependent on interactions between the CHC N-terminal domain (NTD) and clathrin “adaptor” proteins, which facilitate the recruitment of clathrin to membranes (Lemmon and Traub, 2012; Robinson, 2015). The CHC NTD folds into a β-propeller across which four distinct adaptor interaction sites (sites 1 through 4) are distributed. Site 1 of the NTD binds to adaptor proteins containing the clathrin box motif (CBM) consensus sequence LΦXΦ[DE] (where Φ refers to a hydrophobic residue and X to any amino acid; Dell’Angelica et al., 1998; ter Haar et al., 2000). Site 2 binds to the consensus sequence PWxxW, called the “W-box” (Miele et al., 2004). Site 3 has been reported to interact with the “arrestin box” sequence [LI][LI]GxL (Kang et al., 2009), but was recently shown to bind CBM motifs as well (Muenzner et al., 2017). Recently, a fourth site on the CHC NTD was identified (termed the “Royle box”), although no consensus interacting motif has been elucidated (Willox and Royle, 2012; Muenzner et al., 2017). While the NTD of CHC is necessary for its mitotic role, it is not required for interaction with TACC3 (Royle et al., 2005; Royle and Lagnado, 2006; Hood et al., 2013; Smith et al., 2013; Lin et al., 2010). However, to date, no CHC adaptor–site interactions have been implicated in its mitotic function, nor have clathrin adaptor proteins been reported to localize to the metaphase spindle and stabilize MTs (Rao et al., 2016).
Here we show that adaptor interaction sites within the CHC NTD indeed play an important role in clathrin’s ability to stabilize MTs and promote chromosome congression during mitosis. These sites directly recruit GTSE1 to the spindle to inhibit MCAK activity and stabilize MTs. Therefore, in contrast to endocytic adaptor proteins, GTSE1 serves as a MT stability-promoting clathrin “effector” whose function can be determined by clathrin localization. Abolishing the CHC–GTSE1 interaction in cells leads to defects in chromosome congression associated with a loss of centrosomal MT stability, but not kinetochore–MT attachment stability, implicating clathrin in stabilizing non–k-fiber MTs for chromosome congression.
Results
Clathrin directly recruits GTSE1 to the spindle via its adaptor interaction sites
We first confirmed a previous mass spectrometry analysis that indicated an interaction between CHC and GTSE1, by showing that they coimmunoprecipitate in both U2OS and mouse embryonic stem cells (mESCs; Fig. 1, A and B; Hubner et al., 2010). This interaction is direct, as purified GST-CHC pulled down purified GTSE1 (Fig. 1 C). We then used CHC fragment binding analysis to narrow the region of interaction on CHC to the NTD (aa 1–330; Fig. 1 D). To further test whether CHC interacted with GTSE1 via established adaptor binding sites in the CHC NTD, we mutated sites 1–3 individually in GST-CHC(1–330) and assayed for binding to GTSE1. Surprisingly, both site 1 and site 3, but not site 2, were required for interaction with GTSE1 (Fig. 1 E).
Adaptor interaction sites on the CHC NTD directly interact with GTSE1. (A) Immunoblots of cell lysates (input) and IPs of GTSE1 and CHC in U2OS and GTSE1 knockout (GTSE1KO) cells. Immunoblots with anti-GTSE1 or anti-CHC antibody. (B) Immunoblot of cell lysates (input) and IPs of GTSE1-GFP in mESCs. Immunoblots with anti-CHC (top) and anti-CHC and anti-GFP (bottom) antibodies. (C) GST-CHC pulldown of purified recombinant GTSE1. Immunoblot with anti-GTSE1 antibody. Coomassie blue of GST inputs is presented. (D) Pulldown analysis of the interaction between purified GTSE1-GFP and CHC fragments (numbers designate amino acids) fused to GST. Immunoblot with anti-GFP antibody. Coomassie blue of GST inputs is presented. (E) Pulldown analysis of the interaction between purified GTSE1 and CHC(1–330) fused to GST. CHC was mutated at site 1 (S1M: T87A, Q89A, K96E, K98E), site 2 (S2M: Q152L, I154Q), or site 3 (S3M: R188A, Q192A). Immunoblot with anti-GTSE1. Coomassie blue of the GST inputs is presented.
Adaptor interaction sites on the CHC NTD directly interact with GTSE1. (A) Immunoblots of cell lysates (input) and IPs of GTSE1 and CHC in U2OS and GTSE1 knockout (GTSE1KO) cells. Immunoblots with anti-GTSE1 or anti-CHC antibody. (B) Immunoblot of cell lysates (input) and IPs of GTSE1-GFP in mESCs. Immunoblots with anti-CHC (top) and anti-CHC and anti-GFP (bottom) antibodies. (C) GST-CHC pulldown of purified recombinant GTSE1. Immunoblot with anti-GTSE1 antibody. Coomassie blue of GST inputs is presented. (D) Pulldown analysis of the interaction between purified GTSE1-GFP and CHC fragments (numbers designate amino acids) fused to GST. Immunoblot with anti-GFP antibody. Coomassie blue of GST inputs is presented. (E) Pulldown analysis of the interaction between purified GTSE1 and CHC(1–330) fused to GST. CHC was mutated at site 1 (S1M: T87A, Q89A, K96E, K98E), site 2 (S2M: Q152L, I154Q), or site 3 (S3M: R188A, Q192A). Immunoblot with anti-GTSE1. Coomassie blue of the GST inputs is presented.
We next quantitatively analyzed the interdependence of CHC and GTSE1 for their recruitment to the mitotic spindle. RNAi depletion of CHC caused GTSE1-GFP expressed from a bacterial artificial chromosome (BAC) transgene to delocalize from spindles in U2OS cells (Fig. 2, A and B; and Fig. S1, A and B). Similarly, GTSE1 was delocalized from spindles after RNAi depletion of CHC in mESCs (Fig. 2 C and Fig. S1 C). Conversely, CHC remained on spindles in U2OS and mESC cells after either RNAi depletion or stable knockout of GTSE1 (Fig. 2, C–F; and Fig. S1, C and D). Consistently, knockout (U2OS) or RNAi (mESCs) of GTSE1 had no impact on the localization of TACC3 to spindles (Fig. 2, F and G). Thus, while GTSE1 does not impact the CHC/TACC3 complex localization to spindles, clathrin recruits GTSE1 to spindles via a direct interaction with CHC adaptor interaction sites.
Clathrin recruits GTSE1 to the spindle. (A) Immunofluorescence images of U2OS cells stably expressing an RNAi-resistant GTSE1-GFP BAC transgene, stained with antibodies against GFP and α-tubulin. Cells were transfected with control (Ctrl) or CHC siRNA (66 h). All cells were concurrently transfected with GTSE1 siRNA to deplete endogenous GTSE1. (B) Quantification of the spindle recruitment of GTSE1-GFP in cells from A (n = 3 experiments, n ≥ 34 half-spindles from 17 cells per condition and experiment, one experiment presented as a box and whisker plot). (C) Immunofluorescence images of mESCs stained with antibodies against α-tubulin, CHC, and GTSE1, after indicated RNAi. (D) Immunofluorescence images of clonal U2OS cells stably expressing a CHC-GFP BAC transgene after indicated RNAi. Cells were stained for α-tubulin; the GFP signal is from the fluorescing protein. (E) Quantification of CHC-GFP on the spindle of cells from G (n = 3 experiments, n ≥ 44 half-spindles from 22 cells per condition and experiment, one experiment presented as a box and whisker plot). n.s., not significant. (F) Immunofluorescence images of U2OS and GTSE1KO cells stained with antibodies against CHC, TACC3, and α-tubulin. (G) Immunofluorescence images of mESCs stained with antibodies against TACC3 and α-tubulin after indicated siRNA. Scale bars: 5 µm. P values from Wilcoxon test. Numeric data is shown in Table S1.
Clathrin recruits GTSE1 to the spindle. (A) Immunofluorescence images of U2OS cells stably expressing an RNAi-resistant GTSE1-GFP BAC transgene, stained with antibodies against GFP and α-tubulin. Cells were transfected with control (Ctrl) or CHC siRNA (66 h). All cells were concurrently transfected with GTSE1 siRNA to deplete endogenous GTSE1. (B) Quantification of the spindle recruitment of GTSE1-GFP in cells from A (n = 3 experiments, n ≥ 34 half-spindles from 17 cells per condition and experiment, one experiment presented as a box and whisker plot). (C) Immunofluorescence images of mESCs stained with antibodies against α-tubulin, CHC, and GTSE1, after indicated RNAi. (D) Immunofluorescence images of clonal U2OS cells stably expressing a CHC-GFP BAC transgene after indicated RNAi. Cells were stained for α-tubulin; the GFP signal is from the fluorescing protein. (E) Quantification of CHC-GFP on the spindle of cells from G (n = 3 experiments, n ≥ 44 half-spindles from 22 cells per condition and experiment, one experiment presented as a box and whisker plot). n.s., not significant. (F) Immunofluorescence images of U2OS and GTSE1KO cells stained with antibodies against CHC, TACC3, and α-tubulin. (G) Immunofluorescence images of mESCs stained with antibodies against TACC3 and α-tubulin after indicated siRNA. Scale bars: 5 µm. P values from Wilcoxon test. Numeric data is shown in Table S1.
RNAi depletion efficiencies. (A) Immunoblot on the cell lysate of U2OS cells expressing an siRNA-resistant GTSE1-GFP BAC transgene and transfected with control (Ctrl) or CHC siRNA for 66 h. Cells were concomitantly transfected with GTSE1 siRNA to deplete endogenous GTSE1. Immunoblots are with anti-GTSE1, anti-CHC, or anti-α-tubulin antibodies. Anti-CHC blot was performed on same samples run on a different gel. (B) Quantification of CHC and GTSE1-GFP on immunoblot of three siRNA experiments performed as in A. GTSE1-GFP and CHC levels were normalized to α-tubulin and presented in percentage of the control siRNA. Mean percentages (± standard error) are presented (n = 3 experiments). (C) Immunoblot on the cell lysate of mESCs (R1\E) transfected with control, mGTSE1, CHC, or mGTSE1 + CHC siRNA. Immunoblots are with anti-CHC, anti-GTSE1, and anti-α-tubulin antibodies. (D) Immunoblot on the cell lysate of U2OS cells stably expressing a CHC-GFP BAC transgene and transfected with control or GTSE1 siRNA. The immunoblot is with anti-GTSE1 and anti-α-tubulin antibodies. (E) Immunoblot on asynchronous cell lysates of GTSE15xLID cells transfected with control, GTSE1, MCAK, or GTSE1 + MCAK siRNA. Immunoblots are with anti-GTSE1, anti-α-tubulin, or anti-MCAK. (F) Immunoblot on asynchronous cell lysates of U2OS cells transfected with control, CHC, GTSE1, MCAK, CHC + GTSE1, or CHC + MCAK siRNA. Immunoblots are with anti-CHC, anti-GTSE1, anti-MCAK, or anti-α-tubulin.
RNAi depletion efficiencies. (A) Immunoblot on the cell lysate of U2OS cells expressing an siRNA-resistant GTSE1-GFP BAC transgene and transfected with control (Ctrl) or CHC siRNA for 66 h. Cells were concomitantly transfected with GTSE1 siRNA to deplete endogenous GTSE1. Immunoblots are with anti-GTSE1, anti-CHC, or anti-α-tubulin antibodies. Anti-CHC blot was performed on same samples run on a different gel. (B) Quantification of CHC and GTSE1-GFP on immunoblot of three siRNA experiments performed as in A. GTSE1-GFP and CHC levels were normalized to α-tubulin and presented in percentage of the control siRNA. Mean percentages (± standard error) are presented (n = 3 experiments). (C) Immunoblot on the cell lysate of mESCs (R1\E) transfected with control, mGTSE1, CHC, or mGTSE1 + CHC siRNA. Immunoblots are with anti-CHC, anti-GTSE1, and anti-α-tubulin antibodies. (D) Immunoblot on the cell lysate of U2OS cells stably expressing a CHC-GFP BAC transgene and transfected with control or GTSE1 siRNA. The immunoblot is with anti-GTSE1 and anti-α-tubulin antibodies. (E) Immunoblot on asynchronous cell lysates of GTSE15xLID cells transfected with control, GTSE1, MCAK, or GTSE1 + MCAK siRNA. Immunoblots are with anti-GTSE1, anti-α-tubulin, or anti-MCAK. (F) Immunoblot on asynchronous cell lysates of U2OS cells transfected with control, CHC, GTSE1, MCAK, CHC + GTSE1, or CHC + MCAK siRNA. Immunoblots are with anti-CHC, anti-GTSE1, anti-MCAK, or anti-α-tubulin.
GTSE1 binds to CHC in the manner of a clathrin adaptor protein
A search for clathrin-binding sequences in human GTSE1 revealed five potential motifs (denoted A, B, C, D, and E), all located within the last 75 amino acids of GTSE1 (Fig. 3, A and B), that overlap with a minimal C-terminal region of GTSE1(639–739) that binds CHC (Fig. S2 A). To evaluate conservation of these potential motifs, we first performed sensitive sequence searches to identify GTSE1 homologues. This revealed that although a short domain present in the GTSE1 N-terminus can also be found within proteins in some but not all nonvertebrate species (Fig. S3 A, Interpro IPR032768), GTSE1 homologues containing the C-terminal domain that is required for clathrin binding are only conserved in vertebrates, with motifs C, D, and E in these homologues showing the highest conservation (Fig. 3 A and Fig. S3).
Multiple CBMs on GTSE1 and adaptor interaction sites on CHC are required for the GTSE1–CHC interaction. (A) Sequence alignment of GTSE1 C-terminus (aa 661–727 of human GTSE1). Putative clathrin-binding motifs are in green; divergent motifs are italicized. Mutations in the “5xLID” mutant are shown in orange (last row). (B) GTSE1 fragment constructs. Numbers indicate amino acids. Binding of fragments to N-terminal CHC constructs is indicated. (C) Pulldown analysis of the interaction between GST-GTSE1 fragments and CHC(1–642)-6xHis. Immunoblot (anti-His) and Coomassie blue are from separate experiments. (D) AUC analysis of the stoichiometry of the complex between CHC(1–364) and GTSE1(639–739)-6xHis. (E) Structure of GTSE1 motif D interacting with site 3 on CHC. Superimposition (PyMOL) of the structures of CHC(1–364) in complex with GTSE1-C4 (dark green; PDB ID: 6QNN) or GTSE1-C5 (light green; PDB ID: 6QNP). A reference structure (PDB ID: 5M5S) of CHC bound to an amphiphysin peptide is presented in beige. (F) Structure of GTSE1 motif B (light green) interacting with CHC site 1 (PDB ID: 6QNP). A reference structure (PDB ID: 5M5S) of CHC bound to an amphiphysin peptide is presented in beige. (G) Pulldown analysis of the interaction between mutated GST-GTSE1-C3 fragments and CHC(1–642)-6xHis. Immunoblot (anti-His) and Coomassie blue gels are from separate experiments. (H) A model of the interaction between GTSE1 (green) and the CHC NTD. GTSE1 motifs B and D reside in site 1 and site 3 pockets, respectively. The darker green GTSE1 region was modeled manually using Coot.
Multiple CBMs on GTSE1 and adaptor interaction sites on CHC are required for the GTSE1–CHC interaction. (A) Sequence alignment of GTSE1 C-terminus (aa 661–727 of human GTSE1). Putative clathrin-binding motifs are in green; divergent motifs are italicized. Mutations in the “5xLID” mutant are shown in orange (last row). (B) GTSE1 fragment constructs. Numbers indicate amino acids. Binding of fragments to N-terminal CHC constructs is indicated. (C) Pulldown analysis of the interaction between GST-GTSE1 fragments and CHC(1–642)-6xHis. Immunoblot (anti-His) and Coomassie blue are from separate experiments. (D) AUC analysis of the stoichiometry of the complex between CHC(1–364) and GTSE1(639–739)-6xHis. (E) Structure of GTSE1 motif D interacting with site 3 on CHC. Superimposition (PyMOL) of the structures of CHC(1–364) in complex with GTSE1-C4 (dark green; PDB ID: 6QNN) or GTSE1-C5 (light green; PDB ID: 6QNP). A reference structure (PDB ID: 5M5S) of CHC bound to an amphiphysin peptide is presented in beige. (F) Structure of GTSE1 motif B (light green) interacting with CHC site 1 (PDB ID: 6QNP). A reference structure (PDB ID: 5M5S) of CHC bound to an amphiphysin peptide is presented in beige. (G) Pulldown analysis of the interaction between mutated GST-GTSE1-C3 fragments and CHC(1–642)-6xHis. Immunoblot (anti-His) and Coomassie blue gels are from separate experiments. (H) A model of the interaction between GTSE1 (green) and the CHC NTD. GTSE1 motifs B and D reside in site 1 and site 3 pockets, respectively. The darker green GTSE1 region was modeled manually using Coot.
CBMs on the GTSE1 C-terminus interact with adaptor binding sites on the CHC NTD. (A) GST-pulldown analysis of the interaction between GST-CHC(1–364) and GTSE1 fragments fused to a His-tag (GTSE1-His). Numbers indicate amino acids present in the fragments. Immunoblot is with anti-His antibody. (B) Yeast two-hybrid analysis of the interaction between CHC(1–330) fused to activation domain (AD) and different GTSE1(463–739) fragments fused to the DNA binding domain (BD). Letters A–E indicate mutated motifs in GTSE1 (see Fig. 3 A for sequences). Interaction was visualized by yeast growth on −His selection plates. (C) Gel filtration profile (SuperDex200 10 300 GL column) of CHC(1–364) in complex with GTSE1(639–739) fused to 6 His tag. The indicated (pink box) peak protein fraction was run on SDS-PAGE (Coomassie blue stained). (D) Fractions containing CHC(1–364) in complex with GTSE1(639–739)-6xHis were pooled and submitted to AUC-SV. (E) AUC-SV data best-curve-fitting results with root mean square difference (RMSD) value in top. (F) Diagrams showing the amino acids involved in the interactions between CBMs on GTSE1 and sites 1 and 3 on the CHC NTD. The diagrams were designed using Ligplot based on the crystal structures of GTSE1-C4(661–726; upper left panel, PDB ID: 6QNN) and GTSE1-C5(653–719; upper right and lower panels, PDB ID: 6QNP) in complex with CHC(1–364). Site 3 is occupied by motif D in both crystals, while site 1 is occupied by motif E in the GTSE1-C4 crystal and by motif B or motif C in the GTSE1-C5 crystal. Amino acids constituting CBMs are highlighted. (G) Difference electron density maps after refinement of the CHC inthe absence of any atoms of the peptides. The A–E peptide is bound to site 1, and the A–D peptide is shown in site 3.
CBMs on the GTSE1 C-terminus interact with adaptor binding sites on the CHC NTD. (A) GST-pulldown analysis of the interaction between GST-CHC(1–364) and GTSE1 fragments fused to a His-tag (GTSE1-His). Numbers indicate amino acids present in the fragments. Immunoblot is with anti-His antibody. (B) Yeast two-hybrid analysis of the interaction between CHC(1–330) fused to activation domain (AD) and different GTSE1(463–739) fragments fused to the DNA binding domain (BD). Letters A–E indicate mutated motifs in GTSE1 (see Fig. 3 A for sequences). Interaction was visualized by yeast growth on −His selection plates. (C) Gel filtration profile (SuperDex200 10 300 GL column) of CHC(1–364) in complex with GTSE1(639–739) fused to 6 His tag. The indicated (pink box) peak protein fraction was run on SDS-PAGE (Coomassie blue stained). (D) Fractions containing CHC(1–364) in complex with GTSE1(639–739)-6xHis were pooled and submitted to AUC-SV. (E) AUC-SV data best-curve-fitting results with root mean square difference (RMSD) value in top. (F) Diagrams showing the amino acids involved in the interactions between CBMs on GTSE1 and sites 1 and 3 on the CHC NTD. The diagrams were designed using Ligplot based on the crystal structures of GTSE1-C4(661–726; upper left panel, PDB ID: 6QNN) and GTSE1-C5(653–719; upper right and lower panels, PDB ID: 6QNP) in complex with CHC(1–364). Site 3 is occupied by motif D in both crystals, while site 1 is occupied by motif E in the GTSE1-C4 crystal and by motif B or motif C in the GTSE1-C5 crystal. Amino acids constituting CBMs are highlighted. (G) Difference electron density maps after refinement of the CHC inthe absence of any atoms of the peptides. The A–E peptide is bound to site 1, and the A–D peptide is shown in site 3.
Sequence alignments of GTSE1 and TACC3. (A) MUSCLE alignment of the N-terminus (amino acids 1–143 from human GTSE1) and C-terminus (amino acids 679–739 from human GTSE1) of potential GTSE1 homologues. Alignment was performed on full-length genes from the indicated organisms. A conserved N-terminal FDFD motif is labeled. Three putative clathrin binding box motifs (C–E) are indicated in red. Note that some nonvertebrates also have putative clathrin binding box motifs in the C-terminal region (red boxes). (B) MUSCLE alignment of the region of TACC3 that interacts with Aurora A and CHC. Alignment was performed on full-length TACC3 homologues from the indicated organisms using MUSCLE. Only the region encompassing the described interaction sites between TACC3 and Aurora A/CHC is presented. Human residues S558, L559, Y560, F563, L566, and L567 (important for CHC interaction) and F525 and L532 (important for Aurora A interaction) are designated by red and black arrows, respectively. These residues appear conserved in the three nonvertebrate species carefully analyzed (sea squirt, sea urchin, and fruit fly).
Sequence alignments of GTSE1 and TACC3. (A) MUSCLE alignment of the N-terminus (amino acids 1–143 from human GTSE1) and C-terminus (amino acids 679–739 from human GTSE1) of potential GTSE1 homologues. Alignment was performed on full-length genes from the indicated organisms. A conserved N-terminal FDFD motif is labeled. Three putative clathrin binding box motifs (C–E) are indicated in red. Note that some nonvertebrates also have putative clathrin binding box motifs in the C-terminal region (red boxes). (B) MUSCLE alignment of the region of TACC3 that interacts with Aurora A and CHC. Alignment was performed on full-length TACC3 homologues from the indicated organisms using MUSCLE. Only the region encompassing the described interaction sites between TACC3 and Aurora A/CHC is presented. Human residues S558, L559, Y560, F563, L566, and L567 (important for CHC interaction) and F525 and L532 (important for Aurora A interaction) are designated by red and black arrows, respectively. These residues appear conserved in the three nonvertebrate species carefully analyzed (sea squirt, sea urchin, and fruit fly).
To determine the contribution of individual clathrin-binding motifs in GTSE1 to its interaction with the CHC NTD, we first performed a yeast two-hybrid analysis of the CHC–GTSE1 interaction after mutating the first three amino acids of each of the five motifs (A–E) to three alanines. While mutating any single motif lessened the interaction, mutation of motifs A, B, or D had the most dramatic impact (Fig. S2 B). To confirm that multiple motifs on GTSE1 can directly interact with CHC, we tested whether short GTSE1 fragments containing motifs A and B alone (GST-GTSE1-C1), motif C alone (GST-GTSE1-C2), or motifs D and E alone (GST-GTSE1-C3; Fig. 3 B) could independently bind to CHC(1–642). Indeed, the fragment containing motifs A and B (C1), as well as the fragment containing motifs D and E (C3; but not motif C alone [C2]), could efficiently pull down CHC(1–642) (Fig. 3 C). Thus, multiple motifs in GTSE1 and multiple adaptor binding sites on CHC contribute to their interaction.
We next determined the molecular size of the complex formed between the CHC NTD and the GTSE1 C-terminus containing motifs A–E (fragment GTSE1-C) using analytical ultracentrifugation sedimentation velocity (AUC-SV; Fig. 3 D and Fig. S2, C–E). This revealed a 51.4-kD size of the complex, consistent with a heterodimer (theoretical molecular weight of 53.0 kD) and suggesting a 1:1 stoichiometry. Therefore, although there are multiple interaction interfaces between these CHC and GTSE1 proteins, they did not form multimeric complexes. Together, these data suggest a model in which multiple clathrin binding motifs on one GTSE1 molecule bind to multiple adaptor sites on one CHC NTD.
To precisely understand the interaction interface between GTSE1 and CHC, we cocrystallized these regions of the proteins. We determined separate x-ray crystal structures for the complexes formed between the CHC NTD(1–364) and two different GTSE1 C-terminal fragments encompassing either motifs A–E or A–D (GTSE1-C4 and GTSE1-C5, respectively; Fig. 3 B and Table S2 A). In both crystal structures, sites 1 and 3, but not site 2 or 4 on the CHC NTD, were consistently occupied by clear electron densities (Fig. 3, E and F; Fig. S2 F; and Table S2 B), consistent with our pulldown interaction data (Fig. 1 E). In both structures, the electron density present in site 3 could unambiguously be attributed to the region encompassing GTSE1 motif D (Fig. 3 E, Fig. S2 F, and Table S2 B). Of note, the GTSE1 residues (L713, I714, and L716) buried in the hydrophobic pocket of site 3 are those of a conserved CBM. Consistent with an integral role of motif D in binding to CHC, mutation of motif D, but not of motif E, on a GST-GTSE1-C3 fragment abolished its binding to CHC(1–642) (Fig. 3 G). In contrast to what we observed for site 3, site 1 was occupied by three different GTSE1 motifs in the two crystals: motif E in the crystal with GTSE1-C4, and two different motifs, B and C, in different symmetry-related CHC molecules in the crystal with GTSE1-C5 (Fig. 3 F and Table S2 B; see Materials and methods for details). In all cases, bulky hydrophobic residues (i.e., L669, I670, and F672 from motif B; L689, I690, and L692 from motif C; L720, I721, and L723 from motif E) were buried in the hydrophobic cleft formed by blade 1 and 2 of the CHC NTD β propeller, consistent with previously described structures of CBMs interacting with site 1 (Fig. 3 F and Fig. S2 F; amphiphysin peptide bound to CHC is superimposed for comparison; Muenzner et al., 2017; ter Haar et al., 2000; Dell’ Angelica et al., 1998). Mutational analyses indicated a central contribution of motif B in the interaction of GTSE1 with CHC, while motifs C and E contributed the least (Fig. S2 B). Furthermore, the observed motif E interaction with site 1 is likely a crystallization artifact (see Materials and methods). Altogether, these data show that the GTSE1–CHC interaction is reminiscent of the interaction between clathrin and its adaptor proteins in interphase, as CBMs in GTSE1 bind directly to adaptor binding sites 1 and 3 on the CHC NTD. Multiple interaction sites between the two proteins appear to cooperate to facilitate maximum binding of a 1:1 complex, as schematically modeled in Fig. 2 H.
GTSE1 recruitment by the CHC/TACC3 complex is required for efficient chromosome alignment
To determine the importance of the CHC-GTSE1 interaction in mitosis, we generated two stable and clonal U2OS transgenic cell lines expressing either RNAi-resistant WT GTSE1-GFP (GTSE1WT cells) or GTSE1-GFP mutated at all five CBMs (A, B, C, D, and E; denoted GTSE15xLID; see Fig. 3 A) from BACs at levels equivalent to endogenous GTSE1 (Fig. 4 A). Immunoprecipitation (IP) of GTSE15xLID-GFP followed by quantitative mass spectrometry and Western blot confirmed that GTSE15xLID-GFP was no longer able to interact with CHC, nor any other known members of the CHC/TACC3 complex (Fig. 4, B and C; Fig. S4 A; and Table S3). This indicated that GTSE1 associates with the CHC/TACC3 complex critically via adaptor-interaction sites on CHC. Consistently, disruption of the CHC-TACC3 interaction by Aurora A inhibition prevented the association of GTSE1-GFP with TACC3 but not with CHC (Fig. S4 A). Likewise, we could not detect a direct interaction between purified GST-GTSE1 and pTACC3, even after mitotic kinase phosphorylation of GTSE1 (Fig. S4, B and C).
The CHC/TACC3 complex recruits GTSE1 to the spindle to promote chromosome congression. (A) Western blots of U2OS clonal lines expressing RNAi-resistant GTSE1-GFP (GTSE1WT) or GTSE15xLID-GFP (GTSE15xLID) from BAC transgenes. Cells were transfected with control (Ctrl) or GTSE1 siRNA, and lysates were probed for GTSE1 and α-tubulin. (B and C) Mass spectrometry analysis of GTSE1-interacting proteins following anti-GFP IP of mitotic lysates from GTSE1WT or GTSE15xLIDcells, compared with U2OS cells. Curved lines represent significance threshold. (D) Immunofluorescence images of GTSE1WT and GTSE15xLID cells transfected with GTSE1 siRNA. (E) Quantification of the GTSE1-GFP levels on spindles from of GTSE1WT and GTSE15xLID cells depleted for endogenous GTSE1 (n = 3 experiments, n ≥ 26 half spindles from 13 cells per condition and per experiment, one experiment presented as a box and whisker plot, P value from Wilcoxon test). (F) Fraction of cells that have entered anaphase as a function of the time (minutes) after NEB. GTSE15xLID cells have a delayed time of anaphase onset. The mean (± standard error) time between NEB and anaphase onset is indicated in the insets. U2OS, GTSE1WT, and GTSE15xLID cells were transfected with control or GTSE1 siRNA. (n > 223 cells per condition, n = 3 over four experiments; data were pooled for analysis and representation, P values from Wilcoxon test show differences to respective control siRNA). (G) Percentage of fixed metaphase-like cells showing chromosome misalignment. The mean percentage (± standard error) over n = 3 experiments is presented (P values from two-sided, unpaired t test, n ≥ 184 cells per condition). n.s., not significant. Scale bars: 5 µm. Numeric data is shown in Table S1.
The CHC/TACC3 complex recruits GTSE1 to the spindle to promote chromosome congression. (A) Western blots of U2OS clonal lines expressing RNAi-resistant GTSE1-GFP (GTSE1WT) or GTSE15xLID-GFP (GTSE15xLID) from BAC transgenes. Cells were transfected with control (Ctrl) or GTSE1 siRNA, and lysates were probed for GTSE1 and α-tubulin. (B and C) Mass spectrometry analysis of GTSE1-interacting proteins following anti-GFP IP of mitotic lysates from GTSE1WT or GTSE15xLIDcells, compared with U2OS cells. Curved lines represent significance threshold. (D) Immunofluorescence images of GTSE1WT and GTSE15xLID cells transfected with GTSE1 siRNA. (E) Quantification of the GTSE1-GFP levels on spindles from of GTSE1WT and GTSE15xLID cells depleted for endogenous GTSE1 (n = 3 experiments, n ≥ 26 half spindles from 13 cells per condition and per experiment, one experiment presented as a box and whisker plot, P value from Wilcoxon test). (F) Fraction of cells that have entered anaphase as a function of the time (minutes) after NEB. GTSE15xLID cells have a delayed time of anaphase onset. The mean (± standard error) time between NEB and anaphase onset is indicated in the insets. U2OS, GTSE1WT, and GTSE15xLID cells were transfected with control or GTSE1 siRNA. (n > 223 cells per condition, n = 3 over four experiments; data were pooled for analysis and representation, P values from Wilcoxon test show differences to respective control siRNA). (G) Percentage of fixed metaphase-like cells showing chromosome misalignment. The mean percentage (± standard error) over n = 3 experiments is presented (P values from two-sided, unpaired t test, n ≥ 184 cells per condition). n.s., not significant. Scale bars: 5 µm. Numeric data is shown in Table S1.
The CHC–GTSE1 interaction recruits GTSE1 to the CHC/TACC3 complex and the spindle but does not control TACC3 spindle localization. (A) Immunoblot on mitotic lysates of GTSE1WT and GTSE15xLID cells (Input) and IPs of the GFP transgene (GFP IP). MLN8054 was used to inhibit Aurora A. Immunoblots are with anti-GFP, anti-CHC, or anti-TACC3. (B) GST and GST-GTSE1 pulldowns of purified TACC3 phosphorylated by Aurora kinase A. GST-GTSE1 was left unphosphorylated or was phosphorylated by Aurora A (Aur. A), Aurora B (Aur. B), Cdk1, or Aurora A, Aurora B, and Cdk1 (ABCdk1). Phosphorylated TACC3 and GTSE1 were visualized by immunoblot against pTACC3(S558) and GTSE1, respectively. (C) SDS-PAGE stained with Coomassie blue of GST, unphosphorylated GST-GTSE1, and GST-GTSE1 phosphorylated by Aurora A (Aur. A), Aurora B (Aur.B), Cdk1, or Aurora A, Aurora B, and Cdk1 (ABCdk1). Presence of phosphorylated residues was confirmed by Pro-Q staining. (D) Still images from live-cell imaging on GTSE1WT and GTSE15xLID cells transfected with GTSE1 siRNA to deplete the endogenous GTSE1. Cells were treated with siRTubulin before imaging to visualize MTs. (E) Immunofluorescence images showing the localization of TACC3 to the spindle in GTSE1WT and GTSE15xLID cells transfected with the GTSE1 siRNA. TACC3 and α-tubulin staining are shown. (F) Quantification of TACC3 on the spindle of cells from E. Mean TACC3 fluorescence intensity on the spindle was corrected for background and normalized to α-tubulin (n ≥ 51 cells per condition over n = 3 experiments pooled for analysis and presented as a box and whisker plot; P value from ANOVA). n.s., not significant. All scale bars: 5 µm. Numeric data is shown in Table S1.
The CHC–GTSE1 interaction recruits GTSE1 to the CHC/TACC3 complex and the spindle but does not control TACC3 spindle localization. (A) Immunoblot on mitotic lysates of GTSE1WT and GTSE15xLID cells (Input) and IPs of the GFP transgene (GFP IP). MLN8054 was used to inhibit Aurora A. Immunoblots are with anti-GFP, anti-CHC, or anti-TACC3. (B) GST and GST-GTSE1 pulldowns of purified TACC3 phosphorylated by Aurora kinase A. GST-GTSE1 was left unphosphorylated or was phosphorylated by Aurora A (Aur. A), Aurora B (Aur. B), Cdk1, or Aurora A, Aurora B, and Cdk1 (ABCdk1). Phosphorylated TACC3 and GTSE1 were visualized by immunoblot against pTACC3(S558) and GTSE1, respectively. (C) SDS-PAGE stained with Coomassie blue of GST, unphosphorylated GST-GTSE1, and GST-GTSE1 phosphorylated by Aurora A (Aur. A), Aurora B (Aur.B), Cdk1, or Aurora A, Aurora B, and Cdk1 (ABCdk1). Presence of phosphorylated residues was confirmed by Pro-Q staining. (D) Still images from live-cell imaging on GTSE1WT and GTSE15xLID cells transfected with GTSE1 siRNA to deplete the endogenous GTSE1. Cells were treated with siRTubulin before imaging to visualize MTs. (E) Immunofluorescence images showing the localization of TACC3 to the spindle in GTSE1WT and GTSE15xLID cells transfected with the GTSE1 siRNA. TACC3 and α-tubulin staining are shown. (F) Quantification of TACC3 on the spindle of cells from E. Mean TACC3 fluorescence intensity on the spindle was corrected for background and normalized to α-tubulin (n ≥ 51 cells per condition over n = 3 experiments pooled for analysis and presented as a box and whisker plot; P value from ANOVA). n.s., not significant. All scale bars: 5 µm. Numeric data is shown in Table S1.
We next analyzed the importance of the CHC–GTSE1 interaction for the spindle localization of GTSE1 and the CHC/TACC3 complex. GTSE15xLID-GFP was strongly impaired in its localization to the spindle by both fixed and live cell analysis (Fig. 4, D and E; and Fig. S4 D). In contrast, and consistent with GTSE1 depletion data (Fig. 2, D–G; and Fig. S1, C and D), recruitment of the CHC/TACC3 complex to the spindle was not changed in GTSE15xLID mutant cells, as TACC3 remained associated with the spindle (Fig. S4, E and F).
Depletion of GTSE1 leads to a mitotic delay linked to inefficient chromosome alignment (Bendre et al., 2016; Tipton et al., 2017; Fig. 4, F and G). While a WT RNAi-resistant GTSE1-GFP transgene rescued the mitotic delay and chromosome alignment defects in cells depleted for GTSE1, the GTSE15xLID-GFP transgene did not (Fig. 4, F and G). Together, these data indicated that the CHC–GTSE1 interaction is required to recruit GTSE1 to spindles and facilitate chromosome alignment.
CHC interaction with GTSE1 is required for the stabilization of non-kMTs
We next probed for the mechanism by which the CHC-GTSE1 interaction promotes chromosome alignment. CHC is required for the stabilization and organization of MTs within the spindle (Lin et al., 2010; Royle et al., 2005; Booth et al., 2011; Cheeseman et al., 2013), and we previously showed that GTSE1 stabilizes multiple spindle MT subpopulations, including astral MTs and kMTs (Bendre et al., 2016). Spindles in GTSE15xLID cells displayed obvious morphological anomalies and often appeared “wavy,” with half spindles often displaying a concave rather than the normal convex shape and a generally more elongated shape (Fig. 5, A and B). We thus measured MT stability parameters within the spindle. First, we measured MT abundance within the inner spindle. Despite the change in spindle shape, and in contrast to GTSE1-depleted cells, we could not detect a significant change in total inner-spindle tubulin fluorescence or the total spindle volume occupied by MTs in GTSE15xLID cells (Fig. 5, C and D). We next specifically assayed kinetochore–MT attachment stability by treating cells with cold to depolymerize non-kMTs and quantifying the remaining MT intensity (Fig. 5, E and F). While depletion of GTSE1 led to a 50% decrease in the tubulin fluorescence of cold-resistant MTs, this defect was surprisingly not only rescued in GTSE1WT cells but also in the GTSE15xLID cells, indicating that GTSE15xLID-GFP is able to stabilize kinetochore–MT attachment.
Loss of the CHC–GTSE1 interaction impacts spindle shape but not k-fiber stability. (A) Immunofluorescence images of mitotic spindles (α-tubulin staining) from U2OS, GTSE1WT, and GTSE15xLID cells, transfected with control (Ctrl) or GTSE1 siRNA. (B–D) The ratio of spindle width over spindle length (B), the inner-spindle total tubulin fluorescence intensity (C), and the spindle volume (D) were measured in cells from A using 3D objects representing the spindles without astral MTs (n > 60 cells per condition; n = 2 experiments pooled for analysis and presented as box and whisker plots; P values from Wilcoxon test adjusted for multiple comparisons [false discovery rate]). n.s., not significant. (E) Immunofluorescence images showing cold-resistant MTs (α-tubulin) in U2OS, GTSE1WT, and GTSE15xLID cells, transfected with control (Ctrl) or GTSE1 siRNA. G2-synchronized cells (RO3306) were released into mitosis for 55 min before cold treatment. (F) Quantification of the remaining total tubulin fluorescence intensity in cells treated as in E (n = 3, n ≥ 50 cells per condition and per experiment, one experiment presented as a box and whisker plot; P values from Wilcoxon test adjusted for multiple comparisons [false discovery rate]).
Loss of the CHC–GTSE1 interaction impacts spindle shape but not k-fiber stability. (A) Immunofluorescence images of mitotic spindles (α-tubulin staining) from U2OS, GTSE1WT, and GTSE15xLID cells, transfected with control (Ctrl) or GTSE1 siRNA. (B–D) The ratio of spindle width over spindle length (B), the inner-spindle total tubulin fluorescence intensity (C), and the spindle volume (D) were measured in cells from A using 3D objects representing the spindles without astral MTs (n > 60 cells per condition; n = 2 experiments pooled for analysis and presented as box and whisker plots; P values from Wilcoxon test adjusted for multiple comparisons [false discovery rate]). n.s., not significant. (E) Immunofluorescence images showing cold-resistant MTs (α-tubulin) in U2OS, GTSE1WT, and GTSE15xLID cells, transfected with control (Ctrl) or GTSE1 siRNA. G2-synchronized cells (RO3306) were released into mitosis for 55 min before cold treatment. (F) Quantification of the remaining total tubulin fluorescence intensity in cells treated as in E (n = 3, n ≥ 50 cells per condition and per experiment, one experiment presented as a box and whisker plot; P values from Wilcoxon test adjusted for multiple comparisons [false discovery rate]).
Finally, we quantified the stability of all growing MTs by inducing monopolar spindles with the Eg5 kinesin inhibitor S-trityl-l-cysteine (STLC) and determining MT lengths by measuring distances between EB1-stained MT plus-ends and the centrosome. This assay revealed a clear reduction in both the length and abundance of MTs in both GTSE1-depleted U2OS and GTSE15xLID cells (Fig. 6, A–C). Thus, the CHC-GTSE1 interaction is dispensable for the maintenance of metaphase kinetochore–MT attachment stability, but not MT stability in general or spindle morphology.
The CHC–GTSE1 interaction is required for stabilization of nonkinetochore MTs. (A) Immunofluorescence images showing STLC-induced monopolar spindles in indicated cell lines and RNAi conditions. Dashed yellow lines indicate cell borders. (B and C) Mean growing MT length per cell (B) and comet number per cell (C) determined following 3D reconstruction from experiment shown in A. (D) PEF assay. Mean kinetochore (CREST) to monopole (PCNT) distance per cell following 3D reconstruction from A. For B–D, n ≥ 37 cells per condition over n = 3 experiments pooled for analysis and presented as box and whisker plots; P values from Wilcoxon test adjusted for multiple comparisons (false discovery rate). n.s., not significant. Numeric data is shown in Table S1.
The CHC–GTSE1 interaction is required for stabilization of nonkinetochore MTs. (A) Immunofluorescence images showing STLC-induced monopolar spindles in indicated cell lines and RNAi conditions. Dashed yellow lines indicate cell borders. (B and C) Mean growing MT length per cell (B) and comet number per cell (C) determined following 3D reconstruction from experiment shown in A. (D) PEF assay. Mean kinetochore (CREST) to monopole (PCNT) distance per cell following 3D reconstruction from A. For B–D, n ≥ 37 cells per condition over n = 3 experiments pooled for analysis and presented as box and whisker plots; P values from Wilcoxon test adjusted for multiple comparisons (false discovery rate). n.s., not significant. Numeric data is shown in Table S1.
Loss of CHC–GTSE1 interaction destabilizes astral MTs
The above results suggested that the clathrin interaction with GTSE1 facilitates chromosome congression independently of kinetochore–MT attachment stability. How could stabilization of non-kMTs by the CHC–GTSE1 interaction facilitate chromosome congression? To address this question, we imaged both chromosome and spindle dynamics in live GTSE15xLID cells expressing mCherry-β-tubulin and with DNA labeled. Despite forming bipolar spindles in >90% of mitoses, these cells showed an increased mitotic duration and displayed a bipolar spindle with misaligned chromosomes in 42.9 ± 12.8% of mitoses (8.4 ± 2.1% in control cells, average of two experiments ± SD; Fig. S5, A–C). Of the cells with a bipolar spindle and misaligned chromosomes, 92.1 ± 3.0% contained at least one chromosome transiently residing in the vicinity of one of the spindle poles before congression, reminiscent of chromosome behavior when congression is impaired, for example, by loss of dynein-mediated chromosome capture by astral MTs, CENP-E motor activity, or chromokinesin-mediated polar ejection forces (PEFs; Schaar et al., 1997; Li et al., 2007; Wandke et al., 2012).
Analysis of chromosome congression in GTSE15xLID cells and quantification of Aurora B kinase abundance and activity after GTSE1 depletion. (A) Immunoblot on asynchronous cell lysates of GTSE15xLID cells expressing a mCherry-β-tubulin BAC transgene and transfected with control (Ctrl), GTSE1, MCAK, or GTSE1 + MCAK siRNA. Immunoblots are with anti-GTSE1, anti-α-tubulin, or anti-MCAK. (B) Time from NEB to anaphase onset in cells treated as in A. Cells showing a bipolar spindle and misaligned chromosomes are indicated in red (n ≥ 173 cells over n = 2 experiments pooled for analysis and representation). Letters on the plot indicate differences: conditions with the same letter are not statistically different (statistics from Wilcoxon test adjusted for false discovery rate, P < 0.05). (C) Table presenting the average percentage of cells showing the indicated phenotypes in cells from B (the average is presented ± SD). n.d., not determined. (D) Immunoblot on mitotic lysates of U2OS cells transfected with control, GTSE1(B1), GTSE1(B3), or GTSE1(T1) siRNA. Cells were synchronized in mitosis with Nocodazole. Phosphorylation of H3 Serine 10 (H3pS10) is used as a marker of Aurora B activity. Immunoblots are with anti-GTSE1, anti-Aurora B, anti-H3pS10, or anti-α-tubulin antibodies. (E) Quantification of GTSE1 and Aurora kinase B levels in immunoblots of U2OS cells transfected with control or GTSE1 siRNA. Levels were first normalized to α-tubulin level and then to control siRNA. Cells were synchronized in mitosis with Nocodazole. The mean ± SD is presented, n = 2. (F) Immunofluorescence images of metaphase-like U2OS cells transfected with control, GTSE1(B1), GTSE1(B3), or GTSE1(T1) siRNA. DNA and H3pS10 staining are shown. Scale bar: 10 µm. (G) Quantification of the H3pS10 total fluorescence intensity in cells from F. The total H3pS10 fluorescence intensity per cell was measured from 3D reconstructions and corrected for background. Within each experiment, the average total H3pS10 fluorescence intensity per condition was calculated and normalized to the corresponding control siRNA. The mean of these normalized values (± SD) is presented (n ≥ 37 cells per condition over n = 2 experiments). (H) Immunofluorescence images of GTSE15xLID cells transfected with GTSE1 siRNA to deplete the endogenous GTSE1 and showing a misaligned chromosome outside (upper lane) or within the spindle (lower lane). Tubulin, CREST, and MAD1 staining are shown. 4× magnification of the misaligned chromosomes. Positions of the magnified area are indicated. Numeric data is shown in Table S1.
Analysis of chromosome congression in GTSE15xLID cells and quantification of Aurora B kinase abundance and activity after GTSE1 depletion. (A) Immunoblot on asynchronous cell lysates of GTSE15xLID cells expressing a mCherry-β-tubulin BAC transgene and transfected with control (Ctrl), GTSE1, MCAK, or GTSE1 + MCAK siRNA. Immunoblots are with anti-GTSE1, anti-α-tubulin, or anti-MCAK. (B) Time from NEB to anaphase onset in cells treated as in A. Cells showing a bipolar spindle and misaligned chromosomes are indicated in red (n ≥ 173 cells over n = 2 experiments pooled for analysis and representation). Letters on the plot indicate differences: conditions with the same letter are not statistically different (statistics from Wilcoxon test adjusted for false discovery rate, P < 0.05). (C) Table presenting the average percentage of cells showing the indicated phenotypes in cells from B (the average is presented ± SD). n.d., not determined. (D) Immunoblot on mitotic lysates of U2OS cells transfected with control, GTSE1(B1), GTSE1(B3), or GTSE1(T1) siRNA. Cells were synchronized in mitosis with Nocodazole. Phosphorylation of H3 Serine 10 (H3pS10) is used as a marker of Aurora B activity. Immunoblots are with anti-GTSE1, anti-Aurora B, anti-H3pS10, or anti-α-tubulin antibodies. (E) Quantification of GTSE1 and Aurora kinase B levels in immunoblots of U2OS cells transfected with control or GTSE1 siRNA. Levels were first normalized to α-tubulin level and then to control siRNA. Cells were synchronized in mitosis with Nocodazole. The mean ± SD is presented, n = 2. (F) Immunofluorescence images of metaphase-like U2OS cells transfected with control, GTSE1(B1), GTSE1(B3), or GTSE1(T1) siRNA. DNA and H3pS10 staining are shown. Scale bar: 10 µm. (G) Quantification of the H3pS10 total fluorescence intensity in cells from F. The total H3pS10 fluorescence intensity per cell was measured from 3D reconstructions and corrected for background. Within each experiment, the average total H3pS10 fluorescence intensity per condition was calculated and normalized to the corresponding control siRNA. The mean of these normalized values (± SD) is presented (n ≥ 37 cells per condition over n = 2 experiments). (H) Immunofluorescence images of GTSE15xLID cells transfected with GTSE1 siRNA to deplete the endogenous GTSE1 and showing a misaligned chromosome outside (upper lane) or within the spindle (lower lane). Tubulin, CREST, and MAD1 staining are shown. 4× magnification of the misaligned chromosomes. Positions of the magnified area are indicated. Numeric data is shown in Table S1.
One study has suggested that changes in MT dynamics after GTSE1 depletion cause a decrease in Aurora B activity/levels, which in turn leads to weaker PEFs (Tipton et al., 2017). To ask whether the CHC–GTSE1 interaction controls PEFs or CENP-E function, we calculated the distances between kinetochores and poles of STLC-induced monopolar spindles, as has been previously described (Fig. 6 A; Tipton et al., 2017; Barisic et al., 2014, 2015). We did not, however, observe any significant differences in the average kinetochore-to-monopole distances between U2OS control cells, GTSE1-depleted cells, and GTSE1WT or GTSE15xLID cells depleted for GTSE1 (Fig. 6, A and D). We thus analyzed the impact of GTSE1 depletion on Aurora B in our cells. Depletion of GTSE1 did not lead to any detectable decrease in Aurora B levels in mitotic cells (Fig. S5, D and E), nor in its activity as measured by quantifying histone H3 pSer10 on chromosomes (Fig. S5, F and G). Together, these data indicate that defects in Aurora B level/activity, PEFs, or CENP-E function are unlikely to be the cause of the chromosome misalignment observed in U2OS cells following GTSE1 perturbation.
Because astral MTs can promote chromosome congression by capturing chromosomes located outside of the spindle area and facilitating their dynein-mediated poleward transport (Li et al., 2007; Maiato et al., 2017), we specifically quantified the stability of astral MTs in GTSE15xLID cells. Depletion of GTSE1 decreased both the length and abundance of astral MTs (Fig. 7, A–D). While astral MT parameters were restored in GTSE1WT cells, they were clearly defective when the CHC–GTSE1 interaction was impaired in GTSE15xLID cells. This indicated a role of clathrin in stabilizing astral MTs. To confirm this, we measured astral MTs in CHC-depleted U2OS cells. Indeed, CHC depletion alone led to significantly fewer and shorter astral MTs (Fig. 7, E–G). The effect of GTSE1 and CHC on astral MT stability was not specific to transformed cells, as we observed the same effect after depleting them in mESCs (Fig. 7, H–J).
Astral MTs are destabilized upon disruption of the CHC-GTSE1 interaction. (A) Immunofluorescence images illustrating astral length and abundance (α-tubulin and EB1 staining) under labeled conditions. (B) Scheme illustrating the determination of “astral” EB1 comets and quantification method used to measure astral MT length and abundance in 3D reconstruction of cells from A. (C and D) Mean astral MT length (C) and Astral comet number per cell (D) are presented from conditions in A (n ≥ 31 cells per condition over n = 3 experiments pooled for analysis and presented as box and whisker plots; P values from Wilcoxon test adjusted for multiple comparisons [false discovery rate]). (E) Immunofluorescence images of U2OS cells transfected with control (Ctrl) or CHC siRNA (114 h). (F and G) Astral MT length (F) and abundance (G) in cells from E are presented (n ≥ 42 cells per condition over n = 4 experiments pooled for analysis and presented as box and whisker plots; P value from ANOVA and Wilcoxon test, respectively). (H) Immunofluorescence images of mESCs (α-tubulin and EB1 staining) transfected with control, mGTSE1, CHC, or mGTSE1 + CHC siRNA. (I and J) Astral MT length (I) and abundance (J) in cells from H are presented. To account for extreme variation in spindle shape after mGTSE1/CHC depletion in mESCs, the β threshold (see B) was calculated for each spindle based on its geometry (see Materials and methods; n ≥ 24 cells over n = 2 experiments pooled for analysis and presented as box and whisker plots; P values from ANOVA followed by Tukey’s test). n.s., not significant. (K) Stills from live-cell imaging of GTSE15xLID mCherry-β-tubulin U2OS cells with DNA stain, transfected with GTSE1 siRNA. Top and side view from a 3D reconstruction are presented. Scale bar: 10 µm. Letters indicate misaligned chromosomes. Notice misaligned chromosomes remaining outside of inner spindle mass when viewed in 3D. Numeric data is shown in Table S1.
Astral MTs are destabilized upon disruption of the CHC-GTSE1 interaction. (A) Immunofluorescence images illustrating astral length and abundance (α-tubulin and EB1 staining) under labeled conditions. (B) Scheme illustrating the determination of “astral” EB1 comets and quantification method used to measure astral MT length and abundance in 3D reconstruction of cells from A. (C and D) Mean astral MT length (C) and Astral comet number per cell (D) are presented from conditions in A (n ≥ 31 cells per condition over n = 3 experiments pooled for analysis and presented as box and whisker plots; P values from Wilcoxon test adjusted for multiple comparisons [false discovery rate]). (E) Immunofluorescence images of U2OS cells transfected with control (Ctrl) or CHC siRNA (114 h). (F and G) Astral MT length (F) and abundance (G) in cells from E are presented (n ≥ 42 cells per condition over n = 4 experiments pooled for analysis and presented as box and whisker plots; P value from ANOVA and Wilcoxon test, respectively). (H) Immunofluorescence images of mESCs (α-tubulin and EB1 staining) transfected with control, mGTSE1, CHC, or mGTSE1 + CHC siRNA. (I and J) Astral MT length (I) and abundance (J) in cells from H are presented. To account for extreme variation in spindle shape after mGTSE1/CHC depletion in mESCs, the β threshold (see B) was calculated for each spindle based on its geometry (see Materials and methods; n ≥ 24 cells over n = 2 experiments pooled for analysis and presented as box and whisker plots; P values from ANOVA followed by Tukey’s test). n.s., not significant. (K) Stills from live-cell imaging of GTSE15xLID mCherry-β-tubulin U2OS cells with DNA stain, transfected with GTSE1 siRNA. Top and side view from a 3D reconstruction are presented. Scale bar: 10 µm. Letters indicate misaligned chromosomes. Notice misaligned chromosomes remaining outside of inner spindle mass when viewed in 3D. Numeric data is shown in Table S1.
We then asked whether the behavior and attachment status of misaligned chromosomes occurring upon disruption of the CHC–GTSE1 interaction were consistent with a defect in astral MT capture. Closer inspection of misaligned chromosome positions and trajectories in three dimensions in mCherry-β-tubulin GTSE15xLID cells depleted for endogenous GTSE1 revealed that most misaligned chromosomes resided outside of the spindle (i.e., where astral MTs should normally be present) for an extended period of time, before being able to congress to the metaphase plate (Fig. 7 K and Video 1). Closer analysis of misaligned chromosomes in GTSE15xLID cells (39 chromosomes from 16 cells) revealed that most (73%, 14/19) kinetochore pairs located clearly outside of the spindle showed Mad1 staining on both kinetochores, indicating they both lacked MT attachment (Chen et al., 1998; Fig. S5 H, upper panels). In contrast, this was observed for only 22% (2/9) of misaligned chromosome pairs located clearly within the spindle (Fig. S5 H, lower panels), indicating that the majority of these had established a kinetochore–MT attachment and were likely congressing. Together these data show that most misaligned chromosomes in GTSE15xLID cells are located outside of the spindle poles and lack robust kinetochore–MT attachments. This is consistent with a scenario in which astral MTs are too short and sparse to efficiently allow chromosome capture and/or congression. Thus, the CHC-GTSE1 interaction predominantly facilitates chromosome congression through stabilization of nonkinetochore MTs.
Chromosome congression in GTSE15xLID cells expressing mCherry-β-tubulin and transfected with GTSE1 siRNA. Video is based on live-cell imaging of GTSE15xLID cells expressing a mCherry-β-tubulin BAC transgene and transfected with GTSE1 siRNA. Cells were treated with siRDNA to visualize DNA. Videos from top and side view are based on a 3D reconstruction in IMARIS. Scale bar: 10 µm. Frame rates per second = 3.
Chromosome congression in GTSE15xLID cells expressing mCherry-β-tubulin and transfected with GTSE1 siRNA. Video is based on live-cell imaging of GTSE15xLID cells expressing a mCherry-β-tubulin BAC transgene and transfected with GTSE1 siRNA. Cells were treated with siRDNA to visualize DNA. Videos from top and side view are based on a 3D reconstruction in IMARIS. Scale bar: 10 µm. Frame rates per second = 3.
Clathrin promotes chromosome congression and timely mitosis by antagonizing MCAK activity via GTSE1 recruitment
MCAK activity regulates astral MT stability, and we previously showed that GTSE1 binds and inhibits MCAK (Bendre et al., 2016; Srayko et al., 2005; Rizk et al., 2009). Mass spectrometry analysis of GTSE15xLID-GFP interactors (Fig. 4, B and C; and Table S3) indicated a reduced interaction with MCAK, that however did not reach significance. We investigated this more closely under different conditions, and could clearly detect a reduced interaction by Western blot, suggesting that GTSE15xLID that cannot be recruited to spindles fails to inhibit MCAK there (Fig. 8 A). Consistently, disrupting WT GTSE1 localization to spindles by nocodazole treatment also reduced interaction with MCAK (Fig. 8 A). To test whether the phenotypes observed after disruption of the CHC-GTSE1 interaction resulted from excess MCAK activity, we treated both GTSE15xLID cells and mCherry-β-tubulin GTSE15xLID cells with GTSE1, MCAK, or GTSE1 and MCAK siRNA and monitored the time between nuclear envelope breakdown (NEB) and anaphase onset (Fig. 8 B, Fig. S1 E, and Fig. S5, A and B). Strikingly, codepletion of MCAK and endogenous GTSE1 in both cell lines eliminated the increased time to anaphase onset associated with loss of the CHC–GTSE1 interaction. Furthermore, the percentage of mCherry-β-tubulin GTSE15xLID mitotic cells displaying misaligned chromosomes was dramatically reduced, from 42.9 ± 12.8% in GTSE1-depleted cells to 12.1 ± 3.7% in GTSE1 and MCAK codepleted cells (Fig. S5 C). We then asked if the MT stability defect observed upon disruption of the CHC–GTSE1 interaction also depends on MCAK activity. Measurement of MT length in GTSE15xLID cells treated with STLC showed that the decrease in MT length observed upon depletion of GTSE1 was alleviated upon codepletion with MCAK (Fig. 8, C and D). These data are consistent with the MT stability and chromosome congression defects following disruption of the CHC-GTSE1 interaction arising from excess MCAK activity.
Clathrin promotes chromosome congression and timely mitosis by antagonizing MCAK activity via GTSE1 recruitment. (A) Immunoblots of cell lysates (input) and anti-GFP IPs of GTSE1-GFP or GTSE15xLID-GFP under conditions illustrated. Immunoblots with anti-GFP, anti-CHC, or anti-MCAK antibodies. (B) Fraction of cells that entered anaphase as a function of the time (minutes) after NEB in GTSE15xLID cells transfected with control (Ctrl), GTSE1, MCAK, or GTSE1 + MCAK siRNA. The mean time between NEB and anaphase onset is indicated in the inset (± standard error, n ≥ 312 per condition over n = 4 experiments pooled for analysis and representation). In the Stat. columns, conditions with the same letter are not statistically different (statistics from Wilcoxon test adjusted for multiple comparisons [false discovery rate], P < 0.05). (C) Immunofluorescence images (EB1 and tubulin staining) showing STLC-induced monopolar spindles in GTSE15xLID cells after the labeled siRNA transfections. Dashed yellow lines indicate cell borders. (D) Mean growing MT length per cell (EB1 comet to monopole distance) in 3D reconstruction of cells from C (control n = 19, GTSE1 n = 20, GTSE1 + MCAK n = 20, MCAK n = 12, n = 1 experiment, P values from ANOVA followed by Tukey’s test, data presented as box and whisker plots). n.s., not significant. (E) CHC and GTSE1 depletion in U2OS cells lead to similar and nonadditive mitotic delays. Data plotted as described in A. (F) Codepletion of MCAK partially rescues the mitotic delay induced by CHC depletion. Data plotted as in A. Data in E and F were obtained concomitantly (control and CHC siRNAs conditions are shared; n ≥ 255 over n = 3 experiments pooled for analysis and representation). Plots in E and F are cropped on the time abscissa at t = 150 min for representation purposes. In the Stat. columns, conditions with the same letter are not statistically different (statistics from Wilcoxon test adjusted for multiple comparisons [false discovery rate], P < 0.05). Scale bar: 10 µm. Numeric data is shown in Table S1.
Clathrin promotes chromosome congression and timely mitosis by antagonizing MCAK activity via GTSE1 recruitment. (A) Immunoblots of cell lysates (input) and anti-GFP IPs of GTSE1-GFP or GTSE15xLID-GFP under conditions illustrated. Immunoblots with anti-GFP, anti-CHC, or anti-MCAK antibodies. (B) Fraction of cells that entered anaphase as a function of the time (minutes) after NEB in GTSE15xLID cells transfected with control (Ctrl), GTSE1, MCAK, or GTSE1 + MCAK siRNA. The mean time between NEB and anaphase onset is indicated in the inset (± standard error, n ≥ 312 per condition over n = 4 experiments pooled for analysis and representation). In the Stat. columns, conditions with the same letter are not statistically different (statistics from Wilcoxon test adjusted for multiple comparisons [false discovery rate], P < 0.05). (C) Immunofluorescence images (EB1 and tubulin staining) showing STLC-induced monopolar spindles in GTSE15xLID cells after the labeled siRNA transfections. Dashed yellow lines indicate cell borders. (D) Mean growing MT length per cell (EB1 comet to monopole distance) in 3D reconstruction of cells from C (control n = 19, GTSE1 n = 20, GTSE1 + MCAK n = 20, MCAK n = 12, n = 1 experiment, P values from ANOVA followed by Tukey’s test, data presented as box and whisker plots). n.s., not significant. (E) CHC and GTSE1 depletion in U2OS cells lead to similar and nonadditive mitotic delays. Data plotted as described in A. (F) Codepletion of MCAK partially rescues the mitotic delay induced by CHC depletion. Data plotted as in A. Data in E and F were obtained concomitantly (control and CHC siRNAs conditions are shared; n ≥ 255 over n = 3 experiments pooled for analysis and representation). Plots in E and F are cropped on the time abscissa at t = 150 min for representation purposes. In the Stat. columns, conditions with the same letter are not statistically different (statistics from Wilcoxon test adjusted for multiple comparisons [false discovery rate], P < 0.05). Scale bar: 10 µm. Numeric data is shown in Table S1.
Finally, we studied the contribution of GTSE1 to clathrin’s role in astral MT stabilization and chromosome alignment. Depletion of GTSE1 or CHC resulted in very similar defects in astral MT stability (Fig. 7, A–J) and mitotic progression (Fig. 8 E and Fig. S1 F). Clearly, combining loss of GTSE1 and CHC did not lead to further destabilization of astral MTs (Fig. 7, H–J), nor to a greater mitotic delay (Fig. 8 E and Fig. S1 F) than the effect of depletion of either protein alone. These observations are consistent with GTSE1 acting downstream of CHC and significantly contributing to clathrin’s function in this process. We thus asked if the delayed mitosis after loss of CHC was also dependent on MCAK activity. Indeed, U2OS cells codepleted for CHC and MCAK took significantly less time to transit from NEB to anaphase onset than U2OS cells depleted for CHC alone (Fig. 8 F and Fig. S1 F). Altogether, these observations indicate that a substantial part of CHC function in promoting MT stability, chromosome congression, and timely mitosis is to directly recruit the clathrin effector GTSE1 to antagonize MCAK activity.
Discussion
The role of spindle-associated clathrin in mitosis has generally been assumed, based on studies to date, to be to specifically stabilize and organize k-fibers. Here we have shown that clathrin functions to stabilize non-kMTs, such as astral MTs, and that this is an important mechanism by which clathrin promotes chromosome congression. This observation is supported by a recent study showing that clathrin is necessary for spindle positioning via astral MTs (Wolf et al., 2019). We further show here that this function of clathrin is mediated via clathrin adaptor-like interactions of GTSE1 with the CHC NTD, demonstrating for the first time a role for this well-characterized endocytic interaction scheme in mitosis. In contrast to clathrin adaptor proteins that use this interaction to recruit clathrin to sites of coat formation, clathrin on the spindle uses it to recruit GTSE1 and thus stabilize MTs via GTSE1’s ability to inhibit MCAK. Thus, a major mechanism by which clathrin stabilizes MTs is ultimately inhibition of MCAK.
A schematic model of the clathrin/TACC3 complex based on studies to date is illustrated in Fig. 9. GTSE1 does not appear to be required for localization/assembly of CHC and TACC3, and thus likely functions as an “effector” of this complex. Therefore, many of the phenotypes attributed to complex disruption (i.e., depletion of CHC or TACC3) may partly result from delocalization of GTSE1 from spindles and loss of MCAK inhibition. Interestingly, while CHC and TACC3 homologues are conserved among eukaryotes (Peset and Vernos, 2008; Gergely, 2002), published reports of CHC and TACC3 interacting on the mitotic spindle have been limited to vertebrate model systems (human, mouse, chicken, and Xenopus), and it has been proposed that this function of clathrin may have arisen later in evolution (Gulluni et al., 2017; Royle, 2013; Hubner et al., 2010; Lin et al., 2010; Fu et al., 2010; Burgess et al., 2015). Our finding that homologues of human GTSE1 containing the important C-terminal domain exist in only vertebrates raises the question of whether the appearance of GTSE1 (and its interaction with CHC) may be evolutionarily linked to clathrin localization to the spindle and its MT-stabilizing function during mitosis. However, we find that TACC3 residues identified as required for interaction with CHC (Hood et al., 2013; Burgess et al., 2015, 2018) are more distantly conserved, suggesting that a CHC–TACC3 complex on the spindle may have a more ancient origin (Fig. S3). Nevertheless, given that the C-terminal domain of GTSE1 is restricted to vertebrates, the GTSE1-dependent MT stabilization role of the CHC/TACC3 complex likely has a more recent evolutionary origin.
Model of the clathrin/TACC3 complex on a MT. Schematic model of the clathrin/TACC3 complex on a MT based on studies to date. The Aurora A–dependent phosphorylation of S558 on TACC3 is represented as a P. TACC3 is also bound to the MT polymerase ch-TOG. The CHC NTD recruits GTSE1 to inhibit the MT depolymerase MCAK. It is unknown whether PI3K-C2a and GTSE1 can interact with a single CHC on the spindle. Relative protein sizes are roughly conserved.
Model of the clathrin/TACC3 complex on a MT. Schematic model of the clathrin/TACC3 complex on a MT based on studies to date. The Aurora A–dependent phosphorylation of S558 on TACC3 is represented as a P. TACC3 is also bound to the MT polymerase ch-TOG. The CHC NTD recruits GTSE1 to inhibit the MT depolymerase MCAK. It is unknown whether PI3K-C2a and GTSE1 can interact with a single CHC on the spindle. Relative protein sizes are roughly conserved.
With our finding that CHC recruits GTSE1, the clathrin/TACC3 complex becomes a hub incorporating regulation of two major effectors of MT dynamics, ch-Tog/XMAP215 and MCAK, to stabilize MTs (Gard and Kirschner, 1987; Vasquez et al., 1994; Walczak et al., 1996; Desai et al., 1999; Tournebize et al., 2000; Kinoshita et al., 2001). We have shown that GTSE1 inhibition of MCAK provides the major contribution of this complex’s ability to stabilize astral MTs and promote chromosome alignment, as codepletion of GTSE1 and CHC together does not worsen chromosome alignment defects more than either single depletion, and depleting MCAK concurrently with CHC alleviates the severity of the CHC depletion defect (Fig. 8, E and F). Thus, while it has been assumed that the ability of TACC3 to antagonize MCAK activity at spindle poles to regulate astral MT stability is via promoting ch-Tog polymerization activity (Kinoshita et al., 2005), these results indicate an additional, more direct mechanism: TACC3 in complex with CHC directly recruits the MCAK-inhibitor GTSE1. Determining the precise contribution of CHC/TACC3 complex-associated ch-TOG to MT stability has proven difficult to assess, as ch-TOG has multiple roles independent of this complex in spindle assembly, most ch-TOG protein resides at centrosomes and kinetochores independently of CHC/TACC3 complexes, and a TACC3/ch-TOG subcomplex resides on MT plus-ends independent of CHC (Al-Bassam and Chang, 2011; Booth et al., 2011; Gergely et al., 2003; Gutiérrez-Caballero et al., 2015; Lin et al., 2010).
Our results are consistent with multiple mechanistic roles for clathrin operating concurrently during mitosis. One potentially GTSE1-independent role of the CHC/TACC3 complex is the organization of k-fiber MTs via the inter-MT mesh observed in electron micrographs (Nixon et al., 2015). This role has been used to explain observations that loss of clathrin leads to chromosome alignment defects and fewer MTs within k-fibers, and that excess TACC3 disrupts k-fiber MT organization (Booth et al., 2011; Cheeseman et al., 2013; Nixon et al., 2015). Physical bridges/mesh between MTs along the length of k-fibers may explain “stabilizing” MTs within a k-fiber bundle from an organizational perspective. However, it is not clear how they would affect the dynamics/stabilization of kMTs within k-fibers, whose plus-ends are by definition at the kinetochore. In fact, no kinetochore–MT attachment defect was observed upon removal of TACC3/CHC from metaphase spindles or overexpression of TACC3, although k-fiber MT number/organization changed (Cheeseman et al., 2013; Nixon et al., 2015). Perhaps the changes in MT numbers within k-fibers observed after CHC or TACC3 perturbation by EM represent loss of non-kMTs present within the k-fibers, not detectable from fluorescence microscopy assays. The CHC-GTSE1-MCAK pathway may function to stabilize such a MT population, such as augmin-dependent MTs (David et al., 2019), and/or stabilize damaged MTs within k-fibers against catastrophe (Aumeier et al., 2016). Alternatively, or additionally, this could be precisely a role of spindle-localized ch-TOG. Either way, our data support the earlier conjecture that the CHC/TACC3 complex has dual roles on the spindle to organize MTs, as well as to inhibit MT catastrophes (i.e., stabilize MTs; Booth et al., 2011), the latter albeit not only within, but also outside of k-fibers. It will be interesting to see whether loss of GTSE1 affects k-fiber composition of MTs or the mesh, as well as the effect of specifically perturbing spindle associated, CHC/TACC3-recruited ch-TOG.
Our structures of the CHC–GTSE1 interface allow us to compare this interaction to that of CHC-adaptor interactions that function in endocytosis. GTSE1 is able to make multiple contacts with the CHC NTD. Site 3 on the CHC NTD clearly binds to motif D on GTSE1. Interestingly, the GTSE1 residues involved in this interaction with site 3 fit the consensus of a CBM and not the reported site 3–binding motif [LI][LI]GXL (Kang et al., 2009; ter Haar et al., 2000), although a sequence similar to this in GTSE1 (VVGQL) adjacent to motif D makes contacts nearby. The interacting residues of motif D (LIDL) and their position within site 3 are in fact very similar to what has been recently observed with short peptides (Muenzner et al., 2017). Our data thus show that CBMs can indeed interact with more than a single site on the CHC NTD, even in a larger protein domain context, supporting reports using small peptides (Zhuo et al., 2015; Muenzner et al., 2017). Additionally, our work suggests that different CBMs within a polypeptide stretch, commonly observed in adaptor proteins, could interact with a single CHC NTD and enhance binding affinity. The spacing of the GTSE1 motifs would allow a bidentate binding to the CHC NTD with motif D in site 3 and motif B in site 1. While the expanded repertoire of clathrin adaptor proteins and motifs that function in endocytosis results in functional redundancy of CHC adaptor-binding sites for transferrin endocytosis, this was not observed to be the case for the CHC mitotic role, where disruption of site 1 alone, but not site 4, perturbs function (Hood et al., 2013). Thus, there is likely a more limited and/or specific set of interactions with the CHC adaptor-binding sites required on the spindle. Whether our observed GTSE1 binding preferences explain this difference alone, or whether interactions of additional proteins with CHC adaptor sites are also important for mitosis, will be important to establish. Our elucidation of the GTSE1–CHC interaction interface raises the question of whether GTSE1 also functions as a clathrin adaptor in vesicle trafficking. Interestingly, CHC and GTSE1 also interact in interphase (unpublished data), and tethering the C-terminus of GTSE1 to membranes can recruit clathrin and initiate endocytosis, suggesting it may (Wood et al., 2017).
Our results also indicate a separation of function for GTSE1 roles in MT stabilization. While depletion of GTSE1 from U2OS cells leads to defects in the stability of astral MTs and inner-spindle MT and kinetochore–MT attachment, GTSE15xLID that cannot interact with clathrin exhibits defects only in the former, even though apparently completely delocalized from the spindle (Bendre et al., 2016). GTSE1 therefore appears to have the ability, independent of clathrin, to stabilize kinetochore–MT attachment. This could hypothetically function via inhibition of centromere-associated MCAK. However, we have been unable to observe specific localization of GTSE1 to centromeres. Because GTSE1 levels in U2OS and other cancer cell lines are extremely elevated as compared with untransformed cells (Scolz et al., 2012), the observed stabilization of kinetochore–MT attachments by GTSE1 may only occur when GTSE1 is overexpressed, and not represent a normal function of the protein. Consistent with this idea, U2OS cells have an approximately twofold increase in kMT stabilization compared with “normal” RPE-1 cells, while removal of GTSE1 from U2OS cells in our hands results in an approximately twofold decrease in kMT stabilization (Bakhoum et al., 2009; Bendre et al., 2016).
The mechanisms by which clathrin operates in mitosis have been somewhat of an enigma. Here we provide a direct mechanistic link to MT stabilization, whereby adaptor interaction sites in the CHC NTD directly recruit a “clathrin effector,” the MT stabilizing protein GTSE1, to inhibit MCAK activity and stabilize centrosomal MTs. Studies on GTSE1 suggest that its accurate expression and regulation are important not only for chromosome alignment and spindle positioning, but also for avoiding chromosomal instability and potentially tumor progression (Bendre et al., 2016; Tipton et al., 2017; Scolz et al., 2012).
Materials and methods
Cloning and plasmids
The full-length clathrin cDNA was provided by S.J. Royle (Centre for Mechanochemical Cell Biology, Warwick Medical School, Coventry, UK). The full-length GTSE1, clathrin, and TACC3 cDNAs were subcloned into pFastbac vectors with a N-terminal GST tag and a Precision protease cleavage site between the GST and the protein of interest. pGEX4T1-CHC-1-330, 331–1,163, and 996–1,654 were gifts from H.M. Shih (Academia Sinica, Taipei, Taiwan). Clathrin fragments were amplified by PCR and subcloned into pGEX-6P1 vector, and CHC-1-642 was subcloned into pET28a with His tags. Coding sequences for human GTSE1 and its variants were amplified using PCR and cloned into a plasmid vector. GTSE1-C(639–739), GTSE1-C1(652–687), GTSE1-C2(672–708), and GTSE1-C3(693–739) were cloned into pGEX-6P1. GTSE1(1–460), GTSE1(463–739), and GTSE1(639–739) were cloned into pET28a in frame with a His tag. Mutations on clathrin or GTSE1 were generated by site-directed mutagenesis. Clathrin mutants for the adaptor binding sites on the NTD were obtained by introducing the following mutations: T87A, Q89A, K96E, and K98E (S1M), Q152L and I154Q (S2M), and R188A and Q192A (S3M). The CBM on GTSE1 were mutated by introducing the following alanine substitution: LID(664–666) to AAA (motif A), LID(669–671) to AAA (motif B), LID(689–691) to AAA (motif C), LID(713–715) to AAA (motif D), IDL(714–716) to AAA (motif D), LIDL(713–716)AADA (motif D), and E mutant LIQ(720–722) to AAA (motif E). CHC(1–364) and GTSE1(661–726, C4 or 653–719, C5) were subcloned into pGEX-2rbs, a dicistronic derivative of pGEX6P generated in-house, which allowed two genes to be translated from the same transcript. In this construct, clathrin has an N-terminal GST tag while GTSE1 has no tag.
The GTSE1-GFP BAC resistant to the siRNA targeting the sequence 5′-GATTCATACAGGAGUCAAA-3′ is described in Scolz et al. (2012). To obtain the GTSE15xLID-GFP BAC, the point mutations LID(664–666)AAA, LID(669–671)AAA, LID(689–691)AAA, LID(713–715)AAA, and LIQ(720–722)AAA were introduced into the GTSE1-GFP BAC through two successive rounds of counterselection recombineering based on Redβ and Redγ expression (Bird et al., 2011). The mCherry-β-tubulin BAC (MCB6696, HS.E044.J01 backbone), CHC-GFP BAC (MCB4422, RP11-661B17 backbone), and mGTSE1-GFP BAC (MCB5303) were kind gifts from T. Hyman (Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany).
Antibodies
Antibodies used were goat anti-GFP (Max Planck Institute Dresden; described in Poser et al., 2008), rabbit anti-GTSE1 (custom generated; described in Scolz et al., 2012), rabbit anti-TACC3 (sc-22773; Santa Cruz Biotechnology), mouse anti-α-tubulin (DM1α; Sigma-Aldrich), rat anti-tubulin (YL1/2; Santa Cruz Biotechnology), mouse anti-CHC (X22; ab2731, Abcam), goat anti-CHC (N-terminus; LS-C348290; LSBio), rabbit anti-GFP (ab6556, Abcam), rat anti-EB1 (KT-51; Absea Biotechnology), mouse anti-MCAK/Kif2C (1G2; Abnova Corp.), human nuclear antibodies to nuclear antigens centromere autoantibody (CREST; CS1058; Europa Bioproducts), rabbit anti-PCNT (ab4448, Abcam), mouse anti-c-myc (Oncogene EMD Millipore), rabbit anti-Aurora kinase B (ab2254, Abcam), mouse anti-phospho-histone-Ser10 (H3pS10; ab14955, Abcam), mouse anti-His (Penta-His, 34660; Qiagen), rabbit against phospho-TACC3 (gift of K. Kinoshita, RIKEN, Wako, Japan), mouse anti-MAD1 conjugated to Alexa Fluor 488 (gift of A. Musacchio, Max Planck Institute Dortmund, Dortmund, Germany), donkey anti-human antibodies conjugated to Cy5 or Texas red (709-175-149 and 709-075-149; Jackson ImmunoResearch Laboratories), donkey anti-rat Alexa Fluor 488 (A110-337D2; Bethyl), donkey anti-rat Alexa Fluor 594 (A110-337D4; Bethyl), donkey anti-rabbit Alexa Fluor 488 (A-21206; Thermo Fisher Scientific), donkey anti-rabbit Alexa Fluor 594 (A120-208D4; Bethyl), donkey anti-rabbit Alexa Fluor 650 (A120-208D5; Bethyl), donkey anti-mouse Alexa Fluor 488 (A90-337D2; Bethyl), donkey anti-mouse Alexa Fluor 594 (A90-337D4; Bethyl), donkey anti-mouse Alexa Fluor 647 (A31571; Invitrogen), donkey anti-goat Alexa Fluor 488 (705-545-147; Jackson Immunoresearch), donkey anti-goat HRP (SC-2020; Santa Cruz Biotechnology), sheep anti-mouse HRP (NXA931-1ml; Amersham), and donkey anti-rabbit HRP (NXA934-1ml; Amersham).
Protein expression and purification
Full-length GTSE1, clathrin, and TACC3 were produced from Tnao38 insect cells as described previously (Bendre et al., 2016). In brief, cell pellets were harvested 72–96 h after infection and lysed into a buffer containing 50 mM Hepes, pH 8.0, 300 mM NaCl, 5% glycerol, and protease inhibitors. The supernatant was cleared by centrifugation and applied onto a glutathione resin. To remove the GST tags, purified proteins were incubated with GST-Precision protease overnight at 4°C and applied onto a Superose 6 10/300 column. To obtain the clathrin-GTSE1 complex for crystallization, CHC(1–364) and GTSE1 661–726 (C4) or 653–719 (C5) were coexpressed in E. coli BL21 (DE3) cells. Cells were lysed in GST-binding buffer (25 mM Hepes, pH 7.5, 300 mM NaCl, 1 mM EDTA, 5% glycerol, 0.5% Triton X-100, and protease inhibitors [Serva]). The bacterial lysate was first applied on GST affinity column, and the GST-bound fraction was eluted in buffer A pH 8.0 (50 mM Hepes, 300 mM NaCl, 5% glycerol, and 20 mM reduced glutathione) and subjected to size-exclusion chromatography using a Superdex 75 16/600 column. To cleave the GST tag, fractions containing the complex were incubated with GST-PreScission protease overnight at 4°C. GST and GST-PreScission protease were removed by applying the fractions onto a Superdex 75 16/600 column attached to a GST column and equilibrated in a buffer containing 20 mM Tris, pH 7.0, and 50 mM NaCl. The clathrin-GTSE1 complexes were concentrated to ∼30 mg/ml for the crystallization trial. Truncated fragments of clathrin or GTSE1 with GST or His tag were expressed in E. coli BL21 (DE3) cells and further purified onto glutathione, Ni-NTA, or amylose resin. GTSE1-GFP was purified as described in Scolz et al. (2012). pET28 containing N-terminally 6xhis-tagged Aurora kinase A was transformed into Rosetta (DE3) cells with Kanamycin and Chloramphenicol selection. One liter of culture was grown to OD600 1.0 at 37°C and cooled to 18°C. 200 µl of 1-M IPTG was added, and the culture was grown overnight (18 h) at 18°C. Cells were pelleted and resuspended in 30 ml of lysis buffer (50 mM Tris-HCl, pH 7.2, 500 mM NaCl, 0.1% Triton X-100, and 5% glycerol) and frozen in liquid nitrogen. The pellet was then thawed on ice with immediate addition of protease inhibitors (1 complete tablet [Roche], 40 µl peptstatin at 10 mg/ml, and 40 µl PMSF at 10 mg/ml) followed by lysozyme to 0.5 mg/ml final. Suspension was sonicated for 30 s at 50% amplitude on ice. The lysate was cleared by centrifugation at 40,000 rpm for 15 min using an MLA-80 rotor. Imidazole was added to the collected supernatant to 10 mM final. The cleared lysate was loaded onto a 5-ml HisTrap column (GE Life Sciences) and washed with buffer (25 mM Tris-HCl, pH 8.0, 300 mM NaCl, and 20% glycerol) containing 10 mM imidazole (50 ml) and 30 mM imidazole (25 ml). Protein was eluted with the same buffer containing 300 mM imidazole. Peak fractions were pooled and run on a Superdex 200 16/60 column with size exclusion buffer (50 mM Tris, pH 7.5, and 300 mM KCl). Peak fractions were pooled, adjusted to 1 mM DTT and 10% glycerol, aliquoted, and snap frozen in liquid nitrogen.
In vitro protein phosphorylation
Proteins of interest were phosphorylated by Aurora A, Aurora B, or Cdk1 using a molar ratio of 1:100 (kinase:protein) overnight on ice in buffer supplemented with 10 mM MgCl2, 1 mM sodium orthovanadate, and 2 mM ATP. The phosphorylation reaction was stopped by adding 5 µM RO-3306 (Cdk1 inhibitor) or 500 nM MLN8054 (Aurora A inhibitor) for 10 min on ice. Protein phosphorylation was confirmed using Pro-Q Diamond staining (Invitrogen). Purified Aurora B and Cdk1 were gifts from A. Musacchio.
GST pulldown assays
In brief, 1–5 µg GST-tagged proteins were immobilized on glutathione resins and incubated with 2–5 µM proteins of interest in GST-binding buffer (25 mM Hepes, pH 7.5, 300 mM NaCl, 1 mM EDTA, 5% glycerol, and 0.5% Triton X-100) for ∼1 to ∼3 h at 4°C. The reactions were washed with GST binding buffer and analyzed by SDS-PAGE or Western blotting.
Yeast two-hybrid
Clathrin(1–330) was cloned into pGAD vector, and GTSE1(463–739) WT or CBM mutants were in-frame inserted into pGBD vector. Plasmids were transformed into Saccharomyces cerevisiae cells using the lithium acetate, single-strand carrier DNA, polyethylene method (Gietz and Woods, 2002). Cells containing both two-hybrid plasmids were selected on a double dropout plate (SD-ura-leu). A single colony was picked and grown in the double dropout medium till OD0.6. Cells were serially diluted and dropped on double dropout (SD-ura-leu) and triple dropout (SD-ura-leu-his) plates. The interaction strength was examined on the triple dropout plate.
AUC
Bacterial expressed CHC(1–364) was purified as GST fusion protein (using standard protocol and standard buffer, 25 mM Tris, pH 7.5, 100 mM NaCl, 2 mM EDTA, and 1 mM dithioerythritol), and GST tag was cleaved using GST Precision protease. Further tag was removed by performing reverse GST purification followed by gel filtration using a SuperDex 75 column with standard buffer. Bacterial expressed GTSE1(639–739)-6xHis protein was purified as an N-terminal MBP fusion protein (using standard protocol and standard buffer, 25 mM Tris, pH 7.5, 50 mM NaCl, and 2 mM β-mercaptoethanol) and MBP-tag was cleaved using GST-Precision protease followed by Ni-IMAC chromatography using standard protocol (wash and elution buffer containing 30 and 600 mM imidazole, respectively, in standard buffer). Later, buffer was exchanged on a centricon concentrator against standard buffer. For molecular size determination, two proteins were mixed in equal molar ratio in 200-µl volume and injected on a gel filtration (SuperDex200 10 300 GL; GE Healthcare) column (using AUC buffer; 25 mM Tris, pH 7.5, and 50 mM NaCl) to resolve the peak for the complex. Peak fractions were analyzed on 10% SDS-PAGE, and fractions for earlier eluting peaks were pooled together, concentrated on an Amicon Centricon (3-kD cutoff; Millipore), and subjected to molecular size determination by AUC-SV. The AUC-SV run was performed at 42,000 rpm at 20°C in Beckman XL-A ultracentrifuge. The concentrated peak fraction was diluted in AUC buffer to ∼0.8 mg/ml and loaded into standard double-sector centerpieces. The cells were scanned at 280 nm every minute, and >300 scans were recorded. The first 300 image data were processed and analyzed using the program SEDFIT (Schuck, 2000) with the model of continuous c(s) distribution. The partial specific volumes of the proteins (0.742125), buffer density (1.001), and buffer viscosity (0.010142) were estimated using the program SEDNTERP. Data figures were generated using the program GUSSI.
Crystallization
The complex of CHC(1–364) with the GTSE1 peptide C4 (comprising LIDL, motifs A–E) was concentrated to 32 mg/ml after the gel filtration in 20 mM Tris, pH 7.5, and 50 mM NaCl. The sequence of this construct is GPLG-661SQPLIDL…QLSPE726, where GPLGS is a cloning artifact from the PreScission site. Crystals were grown at 20°C with a reservoir solution of 20% PEG 6000 and 0.1 M Tris, pH 8.0, and frozen using PEG300 as cryoprotectant. The complex of clathrin with GTSE1 peptide C5 (encompassing motifs A–D GPLGS-653LAV…SSP719) was purified via gel filtration in 20 mM Tris, pH 7.0, and 50 mM NaCl and concentrated to 30 mg/ml. Crystals grew at 20°C with a reservoir of 12% PEG4000, 0.1 M sodium acetate, and 0.1 M Hepes, pH 7.5, and were frozen using PEG300 as cryoprotectant.
Structure solution
Data were collected at 100K using a Pilatus 6M detector at the X10SA beamline of the SLS in Villigen, Switzerland. All datasets (Table S2 A) were integrated and scaled using XDS and XSCALE (Kabsch, 1993). The structures were solved via molecular replacement with PHASER (Collaborative Computational Project, Number 4, 1994) in space group P43 with a unit cell of 52 52 154 90 90 90 and one molecule per asymmetric unit for the complex with GTSE1-C4 (A–E), or in space group P21 and cell constants 95.1 90.2 100.2 90.0 110.1 90.0 with four molecules per asymmetric unit for the complex with GTSE1-C5 (A–D), using the clathrin moiety of 1UTC or 5M5T as template. The P21 dataset exhibits translational pseudosymmetry corresponding to an approximate doubling of the unit cell in direction of the x axis. This configuration in the crystal leads to every second reflection in this direction being relatively weak, but the processing in the true cell resulted in much clearer electron density compared with refinement trials in the smaller unit cell. Significant differences in the densities and in the positions of the molecules in the larger cell confirmed that refinement in the smaller cell would not be correct.
Example Fo-Fc densities of the A–D and A–E peptides are shown in Fig. S2 G. Refinement with PHENIX (Adams et al., 2010) and building with COOT (Emsley et al., 2010) resulted in models with good Ramachandran geometry and reasonable Rwork/Rfree values (Table S2 A). GTSE1 residues identified in the electron density are listed in Table S2 B.
For the P43 structure with GTSE1-C4 (motifs A–E), data to 2.03 Å was used in spite of the relatively high Rsym values, since this improved the convergence and quality of the refinement, resulting in Rwork/Rfree values of 23.6 and 28.2%, respectively, and 96.8% residues in favored regions of the Ramachandran plot with no outliers. The sequence of the bound peptide could be assigned unambiguously to motif D of GTSE1 (LIDL) in site 3 of the clathrin molecule and to motif E (LIQL) that is fortuitously bound to site 1 of a neighboring, symmetry-related clathrin molecule, with continuous density for the short linker 717SSP719. Indeed, the presence of the peptide leads to a change of the crystal form, since the apo clathrin crystallizes in space group C2 in similar conditions (20% PEG 3350 and 0.2 M sodium formate; not depicted). The motif D interaction seems to be dominant (Fig. 3 G), perhaps enhanced by the preceding residues 706PSPVVGQ712, with well-defined density. Details of the interactions are shown in Fig. S2 F. The central opening of the CHC propeller shows a relatively strong, forked density that cannot be assigned reliably.
For the P21 structure with GTSE1-C5 (motifs A–D), the initial tight noncrystallographic symmetry constraints were gradually relaxed, thus improving the Rwork/Rfree values to 24.1 and 29.1%, respectively. 96.4% of the residues are in favored regions of the Ramachandran plot with no outliers. In all four clathrin monomers in the asymmetric unit, GTSE1-peptide density is visible in sites 1 and 3, with site 3 having the strongest density. Details of the interactions are shown using LigPlot (Laskowski and Swindells, 2011) in Fig. S2 F. It can be clearly seen that motif D is bound in all four sites and three of the four clathrin molecules. Similar to the GTSE1-C4 complex structure, the N-terminal part of GTSE1 motif D (707SPVVGQ712) is clearly visible. The peptides in site 3 of monomers A and B have no crystal contacts, so it is likely that the PVVG motif indeed enhances the affinity of motif D to site 3. The GTSE1 peptide densities in sites 1 and 3 are not connected, so site 1 could harbor any motif of GTSE1. The densities in site 1 of monomers A/B (assigned to motif C 686SRPLIDLM693) are significantly different from those in monomers C/D (most likely motif B 666DLPLIDF672, based on the Phe, Asp, and Pro densities). Motif A (664LIDL667) directly precedes motif B, and Leu664 and Ile665 of the GTSE1 peptides of monomers C/D are actually occupying site 4 of a neighboring CHC, where Leu664 sits in the space of Phe8 of an amphiphysin peptide when superimposed with the 5M5T structure (sequence 3LLDLDFLE10). This crystal packing effect might facilitate an artificial binding of motif B to site 1. Site 4 of monomers C and D is empty, indicating a low intrinsic affinity of the GTSE1 peptide to this region. Thus, site 1 seems to be more promiscuous and binds motifs B or C, depending on the crystal environment.
Cell lines and cell culture
U2OS cells and derivatives were grown at 37°C and 5% CO2 in DMEM (PAN Biotech) supplemented with 10% filtered FBS (Gibco), 2 mM l-glutamine (PAN Biotech), 100 U/ml penicillin, and 0.1 mg/ml streptomycin (PAN Biotech; Pen/Strep mix). R1\E mESCs and derivatives were grown at 37°C and 5% CO2 in DMEM GlutaMAX, high glucose, 4.5 g/liter, and sodium pyruvate (Gibco) supplemented with 17% FBS (PANsera ES, heat-treated 20 min at 56°C), 100 U/ml penicillin, and 0.1 mg/ml streptomycin, 1% non-essential amino acids 100× (Gibco), 0.1% β-mercaptoethanol (Gibco), and 13.34 µg/ml leukemia inhibitory factor (Dortmund Protein Facility, Max Planck Institute Dortmund). At each passage, ≥2.5 × 106 mESCs were seeded into a new 6-cm dish and grown as colonies for 48 h before the next passage. Medium was changed every 24 h.
To obtain U2OS cells expressing GTSE1-GFP, GTSE15xLID-GFP, or CHC-GFP BAC transgenes, the corresponding BACs were transfected into U2OS cells using the Effectene kit (Qiagen) following the manufacturer’s instruction. Clonal lines expressing the BAC transgene were selected with G418 (Sigma-Aldrich). The GTSE1WT and GTSE15xLID clones expressing the GTSE1-GFP and GTSE15xLID-GFP BAC transgenes, respectively, close to endogenous level were selected for phenotypic analysis. The mCherry-β-tubulin GTSE15xLID cells were obtained by transfecting the mCherry-β-tubulin BAC into GTSE15xLID cells and selecting a clonal line expressing both transgenes using G418 (Sigma-Aldrich) and blasticidin selection. GTSE1KO cells were generated in Bendre et al. (2016). To obtain mESCs expressing the mGTSE1-GFP BAC transgene, the corresponding BAC (MCB5303) was transfected in R1\E cells using Effectene Transfect Reagent (Qiagen). Before transfection, cells were seeded into a well coated with cold 0.1% gelatin in ddH2O. Clonal lines expressing the BAC transgene were selected using 200 µg/ml G418 (Sigma-Aldrich).
For the measure of PEFs and MT lengths in monopolar spindles, cells were treated with 10 µM STLC for 2 h before fixation. For the cold-stability assay, cells were synchronized in G2, released for 55 min in normal medium, transferred into ice-cold medium for 17 min, and fixed immediately. Cells were synchronized in G2 32 h after siRNA transfection by a 16-h-long 10-µM RO-3306 (Calibiochem) treatment.
RNAi
The siRNAs 5′-GAUUCAUACAGGAGUCAAATT-3′ (GTSE1(B3)), 5′-GAUCCAACGCAGUAAUGGUTT-3′ (MCAK), and 5′-GGUUGCUCUUGUUACGGAUTT-3′ (CHC) were routinely used to deplete human GTSE1, human MCAK, and mouse and human CHC, respectively. siRNAs 5′-GGAAUUAAAUAAUCCGGUUTT-3′ (GTSE1(B1)) and 5′-GUACAAAGAAGGUCACUUATT-3′ (GTSE1(T1)) were used as extra controls for human GTSE1 depletion in the H3pS10 experiment. The siRNA 5′-GGCGUCUCUCCAGCCUUCCAUUAAU-3′ (mGTSE1) was used to deplete mouse GTSE1. In all experiments, control cells were treated with the Silencer negative control 2 siRNA (AM4637). All siRNAs except GTSE1(T1) (Sigma-Aldrich) and mGTSE1 (Invitrogen) were purchased from Ambion.
GTSE1 and MCAK depletion in human cells was performed using a reverse transfection approach. A transfection mix containing 2.5 µl Oligofectamine (Invitrogen) and siRNA was prepared in OptiMEM (Invitrogen) and incubated 20 min at RT. Meanwhile, cells were seeded at the desired confluence into 24-well plates containing coverslips or 8-well imaging chambers (Ibidi). The transfection mix was added to the cells, and the final volume was adjusted to 500 µl using prewarmed medium. Medium was changed 7–8 h after transfection. Experiments were performed 48 h after transfection unless stated otherwise. GTSE1 and MCAK siRNAs were used at 80 and 12 nM final concentrations, respectively.
CHC depletion in human cells was performed using a forward transfection approach. Cells were seeded into a 3.5-cm dish with coverslips and grown until 75% confluent. Before transfection, medium was exchanged for 1.8 ml OptiMEM supplemented with 100 U/ml penicillin and 0.1 mg/ml streptomycin. A 200-µl transfection mix containing 3 µl Lipofectamine RNAi Max (Invitrogen) and the siRNA was prepared in OptiMEM (Invitrogen) and incubated for 25 min at RT before addition to the cells. Experiments were performed 66 h after transfection. CHC siRNA was used at 50 nM final concentration. When necessary, cells transfected with control or CHC siRNA for 66 h using the forward transfection approach were submitted to a second round of reverse transfection with control, GTSE1 (100 nM), MCAK (12 nM), or CHC (100 nM) siRNAs for an extra 48 h.
GTSE1 and CHC depletion in mouse stem cells was performed using a reverse transfection approach similar to the one described for GTSE1 and MCAK depletion in human cells but with a few variations. The transfection mixed contained 3.2 µl Lipofectamine 2000 (Invitrogen) instead of 2.5 µl Oligofectamine for a final volume of 500 µl in a 24-well plate. Control, mGTSE1, and CHC siRNAs were used at 100 nM final concentration. For microscopy, coverslips were coated overnight at 4°C with BioLaminin 521 (recombinant human Laminin 521; Biolamina) at 5 µg/ml in ice-cold PBS. 105 cells were seeded onto the coated coverslips and grown as a monolayer. For Western blot, the procedure was upscaled to 12-well plates (final volume 750 µl). 2 × 105 cells were seeded directly into uncoated wells and grown as colonies. Experiments were performed 48 h after transfection.
Western blot
Samples in Laemmli buffer were run on SDS-PAGE and transferred onto a nitrocellulose membrane. Membranes were blocked with 5% milk (Carl Roth) in PBS Tween20 0.1% (SERVA electrophoresis) before incubation with the primary antibody diluted in the same blocking solution. For the mouse CHC antibody, the blocking solution was changed to 5% BSA in PBS Tween20 0.1%. Secondary antibodies coupled to HRP were diluted in PBS Tween20 0.1% and milk 5% before incubation on the membrane. For detection, membranes were incubated with ECL reagent (GE Healthcare) before imaging on the ChemiDoc MP imaging system (Bio-Rad) or development onto Hyperfilm (Amersham). Numeric images were obtained by scanning Hyperfilms or using the ImageLab software (Bio-Rad) to generate tif files from the ChemiDoc MP files. Fiji was used to adjust levels, generate 8-bit tif files, and crop images. Western blot quantifications were performed on ChemiDoc MP–generated files showing no saturated pixels and using the volume tools of the ImageLab software. Background-corrected signals were normalized for tubulin intensity and expressed in percentage of the control siRNA condition. If a signal showed a negative value after background correction, the value was set to 0 to avoid problems during normalization. Samples to test RNAi efficiencies were obtained by adding hot Laemmli buffer onto cells from a 12-well plate (reverse transfection procedure scaled up to 750 µl final volume) or by lysing cells from a 3.5-cm dish into 100 µl ice-cold cell lysis buffer (50 mM Na2HPO4, 150 mM NaCl, 10% glycerol, 1% Triton X-100, 1 mM EGTA, 1.5 mM MgCl2, and 50 mM Hepes, pH 7.2, complemented with 2× protease inhibitor mix [SERVA]).
IP and mass spectrometry
To obtain mitotic cells, cells were synchronized with 10 µM STLC for 16 h, harvested by mitotic shake-off, washed with PBS, incubated in medium supplemented with 10 µM MG132 for 45–70 min at 37°C, and harvested by centrifugation. For Aurora kinase A inhibition, 500 nM MLN8054 was applied during MG132 treatment. Asynchronous mESCs were harvested by trypsinization. Cells were lysed for 15 min on ice in ice-cold cell lysis buffer (50 mM Na2HPO4, 150 mM NaCl, 10% glycerol, 1% Triton X-100, 1 mM EGTA, 1.5 mM MgCl2, 50 mM Hepes, pH 7.2, and 1 mM dithioerythritol) supplemented with 2× protease inhibitor mix (SERVA) and PhoStop (EASY Pack, Roche). Debris were removed by centrifugation at 4°C for 15 min at maximum speed in a benchtop centrifuge. 1 ml of cell lysate at 1 or 2.5 mg protein/ml (U2OS cells and mESCs, respectively) was used for IP. 100 µl of cell lysate was saved as Input. Lysates were incubated for 120 or 90 min (U2OS cells and mESCs, respectively) at 4°C with the antibody before addition of 25 µl prewashed Dynabeads (Invitrogen). After 4-h incubation at 4°C, beads were washed three times with cell lysis buffer. For Western blot analysis, proteins were eluted from beads by addition of 2× hot Laemmli buffer. For mass spectrometry analysis, IPs were performed in triplicate to obtain reliable label-free quantitative data and to perform statistical tests. Samples were reduced, alkylated, and digested directly on beads with LysC/Trypsin (Hubner and Mann, 2011). Obtained peptides were desalted and concentrated on C18-reversed phase stage tips (Rappsilber et al., 2007) and then separated with a PepMap100 RSLC C18 nano-HPLC column (2, 100, 75 IDx25 cm, nanoViper, Dionex) on an UltiMat 3000 RSLCnano system (Thermo Fisher Scientific) using a 125-min gradient from 5 to 60% acetonitrile with 0.1% formic acid and then directly sprayed via a nano-electrospray source (Nanospray Flex Ion Source, Thermo Fisher Scientific) in a Q Exactive Plus Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Fisher Scientific). The Q Exactive Plus was operated in data-dependent mode, acquiring one survey scan and subsequently 10 tandem mass spectrometry scans. Resulting raw files were processed with MaxQuant software (v.1.5.2.18) searching against an Uniprot human database (accessed January 2016) using deamidation (NQ), oxidation (M), and acetylation (N-terminus) as variable modifications and carbamidomethylation (C) as fixed modification (Cox and Mann, 2008). A false discovery rate cutoff of 1% was applied at the peptide and protein level. The integrated label-free algorithm was used for relative quantification of the identified proteins (Cox et al., 2014). Quantified proteins were further analyzed with Perseus (v.1.5.1.5; Tyanova et al., 2016). From the list of identified proteins, contaminants and reverse hits were removed. To identify differences between the samples, we required proteins to be quantified in at least two of three replicates for WT or mutant. Missing values were imputated with Perseus at the lower end of the distribution (down shift 1.8, with 0.3). The resulting 1,017 proteins were used for a two-sided t test. The result was visualized with a volcano plot (cutoff: P value 0.01; S0 2).
For the IP comparing the binding of MCAK to GTSE1-GFP and GTSE15xLID-GFP a different procedure was followed. Cells were synchronized in 10 µM STLC for 16 h, harvested by mitotic shake-off, washed with PBS, incubated in medium supplemented with 10 µM MG132 for 90 min at 37°C, and harvested by centrifugation. Alternatively, cells were synchronized with 3.3 µM Nocodazole for 18 h and harvested by mitotic shake-off. Cells were lysed for 15 min on ice in ice-cold lysis buffer 2 (20 mM Tris, pH 8.0, 100 mM KCl, and 1% Triton X-100) complemented with 2× protease inhibitor mix (SERVA) and PhosStop (EASY Pack, Roche). Debris was removed by centrifugation for 15 min at 18,000 relative centrifugal force at 4°C, and 1 ml lysate at 1 mg protein/ml was used for the IP with 1.5 µg goat anti-GFP antibody. Lysates were incubated for 90 min at 4°C with the antibody before addition of 25 µl prewashed Dynabeads (Invitrogen). After 2-h incubation at 4°C, beads were washed three times with ice-cold lysis buffer 2.
Immunofluorescence
Cells seeded onto coverslips were fixed for 10 min at RT with 4% PFA dissolved into a Pipes-based buffer (50 mM Pipes, pH 7.2, 10 mM EGTA, 1 mM MgCl2, and 0.2% Triton X-100). Membranes were permeabilized at RT with 0.25% Triton X-100 for 10 or 5 min (U2OS cells and mESCs, respectively). Cells were blocked at RT for 30 or 45 min (U2OS cells and mESCs, respectively) with 5% BSA (Sigma-Aldrich) dissolved into PBS. Primary antibodies were diluted into 5% BSA in PBS and incubated onto coverslips for 1 h at 37°C in a wet chamber. The same procedure was applied for secondary antibodies. Coverslips were mounted using Prolong Gold antifade mounting with DAPI (Molecular Probes and Thermo Fisher Scientific). Cells were washed three times with PBS between every step. For the assay probing the recruitment of the GTSE1 transgenes to the spindle, PBS was replaced with PBS Tween20 0.1% (SERVA electrophoresis). For the assays measuring the spindle total tubulin fluorescence/volume, astral MTs lengths, or MT lengths/PEF in monopolar spindles, cells were fixed with −20°C methanol for 10 or 15 min (U2OS cells and mESCs, respectively). Cells were then blocked and processed as usual.
A Marianas spinning-disk confocal system (Intelligent Imaging Innovation, 3i) built on an Axio Observer Z1 microscope (Zeiss) with an ORCA-Flash 4.0 camera (Hamamatsu Photonics) was used to acquire images. Images from the MT cold-stability assay were acquired using a 40× 1.4-NA Plan-Apochromat oil objective (ZEISS). Images for the GTSE1 transgene spindle localization, TACC3 localization, astral MT length measurement following CHC depletion, and all mESC experiments were taken using a 100× 1.4-NA Plan-Apochromat oil objective (ZEISS). Other images were taken with a 63× 1.4-NA Plan-Apochromat oil objective (ZEISS).
Live-cell imaging
Culture medium was exchanged for CO2-independent visualization medium (Gibco) 5 h before imaging. To visualize DNA or MTs, 1 h before imaging cells were treated with 1 µM Verapamil and 1 µM siRDNA or 1 µM siRTubulin (Spirochrome), respectively. To measure the time between NEB and anaphase onset, cells were imaged at 37°C overnight. Images of U2OS, GTSE1WT, and GTSE15xLID cells were acquired at 2-min intervals in a series of 2-µm-spaced z-sections. Images of mCherry-β-tubulin GTSE15xLID cells were acquired at 3-min intervals in series of 2.5-µm-spaced z-sections. For the localization of the GTSE1 transgenes in GTSE1WT and GTSE15xLID cells, images were acquired in series of 0.2-µm-spaced z-sections. For the chromosome congression videos, images were acquired at 1.5-min intervals in series of 1-µm-spaced z-sections. All live-cell imaging was performed on a DeltaVision imaging system (GE Healthcare) using a scientific complementary metal–oxide–semiconductor camera (PCO Edge 5.5) and a 40× 1.3-NA UPLFLN 40XO objective (Olympus) or a 60× 1.42-NA Plan Apo N UIS2 objective (Olympus). When required, images were deconvolved using the SoftWoRx software 6.1.l and/or z-projected (maximum intensity) using SoftWoRx software 6.1.1 or Fiji 1.0.
Microscopy image analysis and quantifications
Statistical analysis and plot generation
All statistical analysis, except the mass spectrometry analysis (see IP and mass spectrometry section), were performed in R v.3.4.3. Usually, experiments were analyzed as follows. The normality and homoscedasticity of the data were tested using Shapiro’s test and Bartlett’s test, respectively. If the normality and homoscedasticity of the data was verified, differences were evaluated using ANOVA followed by Tukey’s test. If normality or homoscedasticity were not verified, differences were assessed using a Kruskal-Wallis test followed by direct comparisons using a Wilcoxon test. For multiple comparisons, P values from the Wilcoxon test were adjusted (false discovery rate, Benjamini and Hochberg method). Occasionally, Student’s t test was used for a small dataset, typically n = 3 (see Table S1 for detailed statistical analysis). The ggplot package was used to generate plots that were exported as PDF files. The generated PDF files were imported and edited in Illustrator CS5.1. All box and whisker plots show the median as well as the 25th and 75th quartiles (lower and upper hinge, respectively). The lower whisker is the smallest observation ≥lower hinge − 1.5*interquartile range. The upper whisker is the largest observation ≤upper hinge + 1.5*interquartile range. Values are represented as points.
Evolutionary analysis
To trace the origin of GTSE1 among vertebrates, we first inspected a multiple alignment between the human and many other vertebrate genomes (Sharma and Hiller 2017). This revealed alignments to most vertebrates including the spotted gar and elephant shark, but not the two lamprey species (Petromyzon marinus and Lethenteron camtschaticum) that represent the most basal vertebrate lineage. For the elephant shark, we inspected aligned ESTs and protein sequences, which revealed that the genome contains a full-length GTSE1 gene that aligns over the entire length to GTSE1 of other vertebrates (Fig. S3). Furthermore, the elephant shark GTSE1 is located in a genomic region that contains the same gene order (ALG10, TPRKB, GTSE1, TRMU, and CELSR1) as in the spotted gar, suggesting that this is the ancestral gene order. In mammals, gene order upstream of GTSE1 was changed by genomic rearrangements. BLAST also identified a full-length GTSE1 protein in the whale shark (Fig. 2 B and Fig. S3). To further probe the presence of GTSE1 in lamprey, we queried Lamprey proteins provided by SIMRbase (Smith et al., 2018) with blastp, which revealed that the lamprey possesses a GTSE1 gene (protein identifier PMZ_0002060-RA) that contains the N- and the C-terminal domains. Next, we used the gar and elephant shark GTSE1 protein sequence to perform sensitive searches against the nonvertebrate portion of the NCBI nonredundant protein sequence database using PSI-BLAST with word size 2, gap existence/extension costs of 10 and 3, and the BLOSUM45 matrix.
We used the human TACC3 protein to run PSI-BLAST searches against the NCBI nonredundant protein sequence database and blastp searches against SIMRbase to retrieve homologues in lamprey, ciona, sea urchin, and Drosophila. Multiple alignments of GTSE1 and TACC3 were created with MUSCLE v3.8.31 (Edgar, 2004) and default parameters.
Online supplemental material
Fig. S1 shows RNAi depletion efficiencies for conditions used in the paper. Fig. S2 contains pulldown, yeast two-hybrid, and AUC analyses of the CHC–GTSE1 interaction, as well as graphics showing the electron density maps and interacting amino acids of GTSE1 peptides from crystal structures. Fig. S3 shows GTSE1 and TACC3 sequence alignments. Fig. S4 shows that GTSE15xLID does not interact with CHC/TACC3, GTSE1 does not interact directly with TACC3, and TACC3 localization is not perturbed in GTSE15xLID cells. Fig. S5 shows mitotic timing and chromosome alignment in and GTSE15xLID mCherry-β-tubulin cells and the effect of codepleting MCAK. Also shown is that depletion of GTSE1 does not affect Aurora B stability or activity as measured by H3pS10. Table S1 contains all the numerical data used for plotting and statistics in the paper. Table S2 contains x-ray data collection and refinement statistics for the CHC(1–364)/GTSE1 peptide complexes and GTSE1 residues visible in the electron densities. Table S3 contains mass spectrometry data for the IP of the GTSE1-GFP and GTSE15xLID-GFP transgenes in mitotic lysates. Video 1 shows chromosome congression in GTSE15xLID cells expressing mCherry-β-tubulin and transfected with GTSE1 siRNA.
Acknowledgments
We thank A. Musacchio for purified Aurora B and Cdk1 kinases and anti-MAD1 antibody, H.M. Shih for CHC expression constructs, S.J. Royle for a full-length CHC plasmid, K. Kinoshita for phospho-TACC3 antibody, L. Wordeman for anti-MCAK antibody, and A.A. Hyman for mCherry-β-tubulin, CHC-GFP, and mGTSE1-GFP BACs. We thank S. Chaaban and G. Brouhard for the cartoon MT graphic. We thank G. Vader, R. Cardoso da Silva, M. Pesenti, S. Maffini, and J. Almario for comments on the manuscript. We thank S. Maffini for help with microscopy. We thank the beamline staff of X10SA at the Swiss Light Source Paul Scherrer Institute, Villigen, Switzerland, for support, and our colleagues of Max Planck Institute Dortmund and the TU Dortmund for help with the data collection.
This work was supported by the Max Planck Institute of Molecular Physiology, a Worldwide Cancer Research project grant 16-0093 to A.W. Bird, and the Deutsche Forschungsgemeinschaft (German Research Foundation), Projektnummer 394694869.
The authors declare no competing financial interests.
Author contributions: A. Rondelet, Y-C. Lin, D. Singh, A.T. Porfetye, H.C. Thakur, M. Hiller, I.R. Vetter, and A.W. Bird conceived and designed experiments. A. Rondelet performed most cellular assays, image quantification, analysis, and statistics. Y-C. Lin and P. Brinkert constructed and purified protein fragments and mutants, performed all pull-down assays and Y2H assays, and purified TACC3 and CHC protein constructs. A.T. Porfetye and I.R. Vetter performed crystallization experiments and analysis. D. Singh performed live cell imaging and analyses of cellular Aurora B expression and activity. P.O. Widlund purified Aurora A kinase and GTSE1-GFP proteins. A. Hecker quantified CHC localization. H.C. Thakur performed AUC analysis. M. Hiller performed sequence/evolutionary analyses. N. Schmidt generated cell lines. S. Bendre purified GTSE1 protein. L. Mazul aided in mESC work. F. Müller and T. Bange performed mass spectrometry experiments and analysis. A.R. and A.W. Bird wrote the manuscript.
References
Author notes
A. Rondelet and Y-C. Lin contributed equally to this paper.



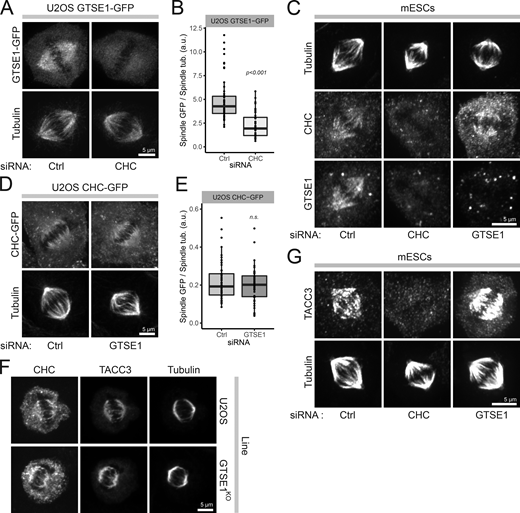




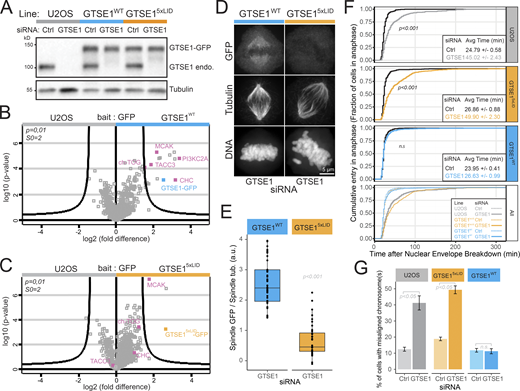
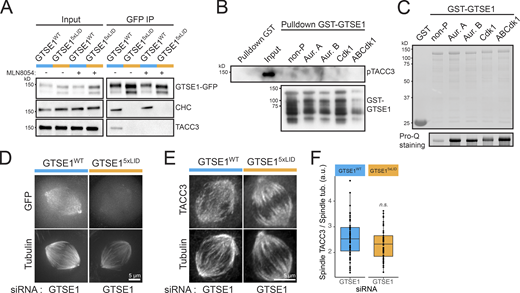
![Loss of the CHC–GTSE1 interaction impacts spindle shape but not k-fiber stability. (A) Immunofluorescence images of mitotic spindles (α-tubulin staining) from U2OS, GTSE1WT, and GTSE15xLID cells, transfected with control (Ctrl) or GTSE1 siRNA. (B–D) The ratio of spindle width over spindle length (B), the inner-spindle total tubulin fluorescence intensity (C), and the spindle volume (D) were measured in cells from A using 3D objects representing the spindles without astral MTs (n > 60 cells per condition; n = 2 experiments pooled for analysis and presented as box and whisker plots; P values from Wilcoxon test adjusted for multiple comparisons [false discovery rate]). n.s., not significant. (E) Immunofluorescence images showing cold-resistant MTs (α-tubulin) in U2OS, GTSE1WT, and GTSE15xLID cells, transfected with control (Ctrl) or GTSE1 siRNA. G2-synchronized cells (RO3306) were released into mitosis for 55 min before cold treatment. (F) Quantification of the remaining total tubulin fluorescence intensity in cells treated as in E (n = 3, n ≥ 50 cells per condition and per experiment, one experiment presented as a box and whisker plot; P values from Wilcoxon test adjusted for multiple comparisons [false discovery rate]).](https://cdn.rupress.org/rup/content_public/journal/jcb/219/2/10.1083_jcb.201907083/6/m_jcb_201907083_fig5.png?Expires=1773662901&Signature=i8zLRchNVHS8REWhz-tF94-lyTmB1b6hA520MD6z2724Gk9R~Oz6J0jvaC4uCn6MVKh9cUOmKy1G2d3FNbK1~L7SnBF3ug-R8IBZ6MLHkoUo6HJFeeEArQ3GOR-kyNBUBi2Ll7wQrZDi3wF4xuTr~Ht9FTTDAgQYfNkcXzGH2DKZqH~vgu-kj7nHPcr4jS0oeLrrYOa4wCtjUS-Jm4sTq5MMiwwCcL9gNUDeWBnzAalaU0UHaelAXrSZk4J1ckN~bOmvlulKhVgbLBo0LCJHXv07XUnMpTEK2ZnltScKq~YChU8dmsroRYSV8hzPz0y9WdLwI14NYywKekL3a62Kaw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

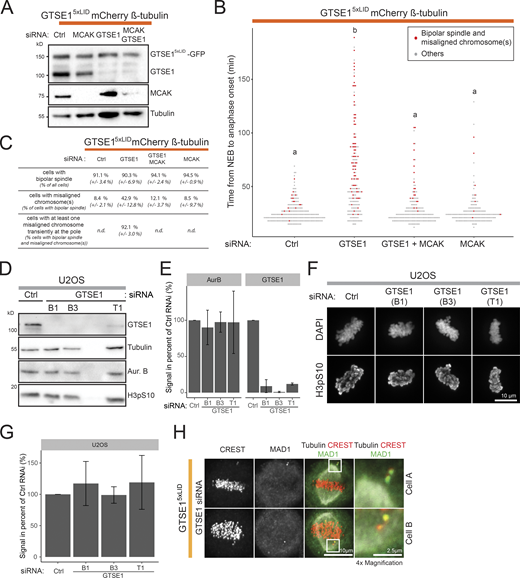
![Astral MTs are destabilized upon disruption of the CHC-GTSE1 interaction. (A) Immunofluorescence images illustrating astral length and abundance (α-tubulin and EB1 staining) under labeled conditions. (B) Scheme illustrating the determination of “astral” EB1 comets and quantification method used to measure astral MT length and abundance in 3D reconstruction of cells from A. (C and D) Mean astral MT length (C) and Astral comet number per cell (D) are presented from conditions in A (n ≥ 31 cells per condition over n = 3 experiments pooled for analysis and presented as box and whisker plots; P values from Wilcoxon test adjusted for multiple comparisons [false discovery rate]). (E) Immunofluorescence images of U2OS cells transfected with control (Ctrl) or CHC siRNA (114 h). (F and G) Astral MT length (F) and abundance (G) in cells from E are presented (n ≥ 42 cells per condition over n = 4 experiments pooled for analysis and presented as box and whisker plots; P value from ANOVA and Wilcoxon test, respectively). (H) Immunofluorescence images of mESCs (α-tubulin and EB1 staining) transfected with control, mGTSE1, CHC, or mGTSE1 + CHC siRNA. (I and J) Astral MT length (I) and abundance (J) in cells from H are presented. To account for extreme variation in spindle shape after mGTSE1/CHC depletion in mESCs, the β threshold (see B) was calculated for each spindle based on its geometry (see Materials and methods; n ≥ 24 cells over n = 2 experiments pooled for analysis and presented as box and whisker plots; P values from ANOVA followed by Tukey’s test). n.s., not significant. (K) Stills from live-cell imaging of GTSE15xLID mCherry-β-tubulin U2OS cells with DNA stain, transfected with GTSE1 siRNA. Top and side view from a 3D reconstruction are presented. Scale bar: 10 µm. Letters indicate misaligned chromosomes. Notice misaligned chromosomes remaining outside of inner spindle mass when viewed in 3D. Numeric data is shown in Table S1.](https://cdn.rupress.org/rup/content_public/journal/jcb/219/2/10.1083_jcb.201907083/6/m_jcb_201907083_fig7.png?Expires=1773662902&Signature=Od0upvffQvq2T-kTuWMbJKwdgPZBOwBrYCUwbVyNQ5Uc5FzuXvgwGWq90iuNnje4UQg2fX3WHvREm2Fu0oUYnEcYEikArVaiOm12Yrn2Mnfcu7W-8jHqbdTZaz11B3U6p~ZiPwagjbh8bAY5AvOiQ~Jwyvw~ED~DB~7r2c00yThQCX5A4kDU5PJt390Y9EIOi9MJZbk5h6Qb6j7AedtYCWNpGWc-Z-JYVYaiUtYGHNttoPiJ~yNL1-hQ7NSCWcT8uJcg9L2quy-hx9xONZ2b6sBhL8d7U4821wsLn06kHkVAHUQ6wN3NvPfVh4lKfY0J9CgNPpXjk4omJaYpp4RaQw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Clathrin promotes chromosome congression and timely mitosis by antagonizing MCAK activity via GTSE1 recruitment. (A) Immunoblots of cell lysates (input) and anti-GFP IPs of GTSE1-GFP or GTSE15xLID-GFP under conditions illustrated. Immunoblots with anti-GFP, anti-CHC, or anti-MCAK antibodies. (B) Fraction of cells that entered anaphase as a function of the time (minutes) after NEB in GTSE15xLID cells transfected with control (Ctrl), GTSE1, MCAK, or GTSE1 + MCAK siRNA. The mean time between NEB and anaphase onset is indicated in the inset (± standard error, n ≥ 312 per condition over n = 4 experiments pooled for analysis and representation). In the Stat. columns, conditions with the same letter are not statistically different (statistics from Wilcoxon test adjusted for multiple comparisons [false discovery rate], P < 0.05). (C) Immunofluorescence images (EB1 and tubulin staining) showing STLC-induced monopolar spindles in GTSE15xLID cells after the labeled siRNA transfections. Dashed yellow lines indicate cell borders. (D) Mean growing MT length per cell (EB1 comet to monopole distance) in 3D reconstruction of cells from C (control n = 19, GTSE1 n = 20, GTSE1 + MCAK n = 20, MCAK n = 12, n = 1 experiment, P values from ANOVA followed by Tukey’s test, data presented as box and whisker plots). n.s., not significant. (E) CHC and GTSE1 depletion in U2OS cells lead to similar and nonadditive mitotic delays. Data plotted as described in A. (F) Codepletion of MCAK partially rescues the mitotic delay induced by CHC depletion. Data plotted as in A. Data in E and F were obtained concomitantly (control and CHC siRNAs conditions are shared; n ≥ 255 over n = 3 experiments pooled for analysis and representation). Plots in E and F are cropped on the time abscissa at t = 150 min for representation purposes. In the Stat. columns, conditions with the same letter are not statistically different (statistics from Wilcoxon test adjusted for multiple comparisons [false discovery rate], P < 0.05). Scale bar: 10 µm. Numeric data is shown in Table S1.](https://cdn.rupress.org/rup/content_public/journal/jcb/219/2/10.1083_jcb.201907083/6/m_jcb_201907083_fig8.png?Expires=1773662902&Signature=OpAi4ecPJi-oXaEwQRHk3ZvdlF~0s3p5d~b4ykqHFTd2dSt5gD-kwiyN2mMWEl47xKqq8B4SOUj-77WSESS7YKCScJCoQ9NrmLn3Vm9KYjdho~298Tiy~kz9fpLOZau5vLyZbh4mE975~FkNOeWXCuCeXFfQ5l0SVZQP2v1kfm6KBsmR-OiV1f2l6HW74u2c2MiWGuymowX1WwV6PyUEDI675nBRM5VbLuVFbZcMWBSuGcH-P47dkZfbq574fdHl2CSgN2hxDEPNWArJQDOOndDR4r5gfbrTHgAQwkG8sNGYIJ~EzD9m7BL4oGv-iwQLOfCvSi4phR1UFcOI9mIMaQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)



