Much of our understanding of actin-driven phenotypes in eukaryotes has come from the “yeast-to-human” opisthokont lineage and the related amoebozoa. Outside of these groups lies the genus Naegleria, which shared a common ancestor with humans >1 billion years ago and includes the “brain-eating amoeba.” Unlike nearly all other known eukaryotic cells, Naegleria amoebae lack interphase microtubules; this suggests that actin alone drives phenotypes like cell crawling and phagocytosis. Naegleria therefore represents a powerful system to probe actin-driven functions in the absence of microtubules, yet surprisingly little is known about its actin cytoskeleton. Using genomic analysis, microscopy, and molecular perturbations, we show that Naegleria encodes conserved actin nucleators and builds Arp2/3–dependent lamellar protrusions. These protrusions correlate with the capacity to migrate and eat bacteria. Because human cells also use Arp2/3–dependent lamellar protrusions for motility and phagocytosis, this work supports an evolutionarily ancient origin for these processes and establishes Naegleria as a natural model system for studying microtubule-independent cytoskeletal phenotypes.
Introduction
Actin is among the most highly expressed proteins in eukaryotic cells and is thought to have evolved before the origin of eukaryotes (Akil and Robinson, 2018; Ettema et al., 2011; Spang et al., 2015; Velle and Fritz-Laylin, 2019). Actin networks, which drive many core cellular functions, are initiated by two main classes of actin nucleators: the Arp/2 complex typically assembles branched actin networks (Mullins et al., 1998), while formins nucleate and elongate linear networks (Breitsprecher and Goode, 2013; Sagot et al., 2002). Actin, the Arp2/3 complex, formins, and many of their regulators are broadly conserved across eukaryotes (Fritz-Laylin et al., 2017a; Kollmar et al., 2012; Pollard and Goldman, 2018; Pruyne, 2017; Velle and Fritz-Laylin, 2019; Veltman and Insall, 2010), but our understanding of cytoskeletal dynamics has come from only a handful of systems, most of which fall within the opisthokonts—the eukaryotic lineage that includes animals and fungi. While studies within this “yeast-to-human” range have made monumental contributions to cell biology, they do not reflect the diversity of eukaryotic cytoskeletal systems. Studying additional, diverse organisms has revealed unique cell biology (Craig et al., 2019; Paredez et al., 2014) and provides opportunities to address questions that are intractable using traditional model organisms (Russell et al., 2017). In this study, we use the amoeba Naegleria—an emerging model system from one of the most evolutionarily distant groups from the opisthokonts (Fig. 1 A)—to explore how a cell undergoes crawling motility and phagocytosis in the absence of microtubules.
Naegleria amoebae employ an actin-based cytoskeletal system, while flagellates use actin and microtubules. (A) This diagram illustrates the evolutionary relationships between Naegleria (top) and other eukaryotes (figure modified from Velle and Fritz-Laylin, 2019). SAR, Stramenopiles, Alveolates, and Rhizaria. (B) A representative amoeba (top) from a growing population and a flagellate (bottom) from a population of differentiated cells were fixed and stained with phalloidin to detect F-actin (green), Tubulin Tracker to visualize tubulin (magenta, γ adjustment of 0.7), and DAPI to label DNA (blue). (C) Changes in actin and microtubule cytoskeletal transcript abundance were calculated using gene expression data collected from amoeba or flagellate populations (also see Fig. S1 A and original data from Fritz-Laylin and Cande, 2010). Positive values indicate higher transcript abundance in amoebae; negative values indicate higher abundance in flagellates (AU = arbitrary units). (D) A subset of data from C highlights the expression pattern of actin assembly factors (also see Fig. S2 B). For graphs in C and D, each bar represents the average relative change in transcript abundance between amoebae and flagellates from three experiments ± SD.
Naegleria amoebae employ an actin-based cytoskeletal system, while flagellates use actin and microtubules. (A) This diagram illustrates the evolutionary relationships between Naegleria (top) and other eukaryotes (figure modified from Velle and Fritz-Laylin, 2019). SAR, Stramenopiles, Alveolates, and Rhizaria. (B) A representative amoeba (top) from a growing population and a flagellate (bottom) from a population of differentiated cells were fixed and stained with phalloidin to detect F-actin (green), Tubulin Tracker to visualize tubulin (magenta, γ adjustment of 0.7), and DAPI to label DNA (blue). (C) Changes in actin and microtubule cytoskeletal transcript abundance were calculated using gene expression data collected from amoeba or flagellate populations (also see Fig. S1 A and original data from Fritz-Laylin and Cande, 2010). Positive values indicate higher transcript abundance in amoebae; negative values indicate higher abundance in flagellates (AU = arbitrary units). (D) A subset of data from C highlights the expression pattern of actin assembly factors (also see Fig. S2 B). For graphs in C and D, each bar represents the average relative change in transcript abundance between amoebae and flagellates from three experiments ± SD.
Decades of research have defined the contributions of actin and microtubules to opisthokont and amoebozoan motility in molecular detail. Outside of these groups, studies investigating actin-driven cell migration in this detail are phylogenetically limited (Frénal et al., 2017; Poulsen et al., 1999). It was therefore unclear if conserved actin-dependent pathways drive motility and phagocytosis in distant eukaryotic lineages. A form of locomotion called “α-motility” relies on highly dynamic, actin-filled pseudopods and low-affinity or nonspecific adhesions to the extracellular environment (Fritz-Laylin et al., 2017a). α-motility is distinct from blebbing motility, where the cell membrane detaches from the underlying actin cortex and blisters outward (Bergert et al., 2012; Yoshida and Soldati, 2006), and from slow mesenchymal motility, in which cells adhere tightly to a 2D surface using focal adhesions (Abercrombie, 1980; Gardel et al., 2010; Paluch et al., 2016). Many of these actin-based motility pathways are also invoked for phagocytosis (Cougoule et al., 2004; Heinrich and Lee, 2011; Jaumouillé et al., 2019). While actin has clear roles in motility and phagocytosis, the contributions of microtubules to these processes vary by cell type (Etienne-Manneville, 2013); microtubules are dispensable for motility in keratocytes (Euteneuer and Schliwa, 1984; Nakashima et al., 2015), while in fibroblasts they are required (Liao et al., 1995; Vasiliev et al., 1970), and in leukocytes they mediate pathfinding and cell shape (Kopf et al., 2020; Renkawitz et al., 2019). Entamoeba histolytica is thought to lack cytoplasmic microtubules (Meza et al., 2006; Vayssié et al., 2004; but see Gómez-Conde et al., 2016), and these amoebae move exclusively by blebbing (Maugis et al., 2010).
While Naegleria spends most of its time as a crawling amoeba, under stressful conditions it can differentiate into a swimming flagellate (Fig. 1 B). This ∼1-h differentiation involves the assembly of an entire microtubule cytoskeleton from scratch (including transcription and translation of tubulin and its regulators) and has been valuable in studying microtubules and de novo centriole assembly (Fritz-Laylin et al., 2010a; Fritz-Laylin and Cande, 2010; Fritz-Laylin et al., 2016; Fulton and Simpson, 1976; Lai et al., 1979). Although these studies form the basis of most of what we know about Naegleria’s cytoskeletal biology, Naegleria usually does not have any microtubules at all (Fulton and Dingle, 1971; Walsh, 2007, 2012). The only microtubules found to date in Naegleria amoebae are found in the mitotic spindle during closed mitosis (Fulton and Dingle, 1971; González-Robles et al., 2009; Walsh, 1984). These mitotic microtubules are built from divergent tubulin that is expressed specifically during mitosis (Chung et al., 2002). This leads to the assumption that interphase Naegleria cells use actin alone to fulfill the cytoskeletal requirements of an amoebic lifestyle, including cell crawling and phagocytosis.
Actin has been implicated in Naegleria’s extremely rapid (>120 µm/min) crawling motility (King et al., 1983; Preston et al., 1990; Preston and King, 2003), as well as in the formation of phagocytic cups in the “brain-eating amoeba,” Naegleria fowleri (Sohn et al., 2010). The genome of a nonpathogenic species, Naegleria gruberi, encodes dozens of actins and actin regulators (Fritz-Laylin et al., 2010b). However, the existing literature lacks a full inventory of Naegleria’s cytoskeletal regulators, quantitative analyses of the actin structures amoebae build, and details of which actin nucleators drive motility and phagocytosis.
Here, we use genomic analyses to show that N. gruberi and the deadly brain-eating N. fowleri each encode an extensive repertoire of actin cytoskeletal regulators, which are mainly expressed in the amoeboid cell type. We also show that N. gruberi amoebae build several distinctive actin-rich structures, including Arp2/3–dependent lamellar ruffles. These ruffles correlate with increased cell motility and phagocytosis. Our findings highlight that despite over a billion years of independent evolutionary history (Betts et al., 2018; Knoll, 2014; Parfrey et al., 2011), Naegleria uses actin-based molecular mechanisms strikingly similar to those of other eukaryotic lineages, supporting an ancient evolutionary origin for α-motility and phagocytosis.
Results and discussion
N. gruberi amoebae lack cytoplasmic microtubules and build diverse actin structures
To define the cytoskeletal differences between Naegleria’s two life stages, we analyzed transcript levels before and after synchronous differentiation from amoebae to flagellates, using data from our previous investigation of centriole assembly (Fritz-Laylin and Cande, 2010). Consistent with our previous findings, actin cytoskeletal transcripts remained at moderate levels in flagellates (Fig. S1), although most were more abundant in amoebae (Fig. 1 C). Meanwhile, Naegleria amoebae had very low levels of any microtubule cytoskeletal element (Fig. S1 A), while these elements were enriched in flagellates (Fig. 1 C). Taken together with the absence of tubulin staining in amoebae (Fig. 1 B), these data support previous reports that interphase amoebae lack microtubules (Fritz-Laylin et al., 2010a; Fulton and Dingle, 1971; Walsh, 2007), while actin is always present.
Actin and microtubule cytoskeletal gene expression differs between Naegleria amoebae and flagellates. (A) The relative transcript abundance for actin cytoskeletal genes (green), microtubule cytoskeletal genes (magenta), and GAPDH (gray) was calculated using expression data collected from amoeba (0 min into differentiation) or flagellate (80 min into differentiation) populations (original data from Fritz-Laylin and Cande, 2010). The relative transcript abundance in flagellates was subtracted from the level in amoebae to generate the graphs shown in Fig. 1, C and D. The microtubule cytoskeletal genes expressed in amoebae include the mitotic tubulins as well as putative spindle components. (B and C) The actin cytoskeletal transcript levels shown in A were organized such that the relative abundances in amoebae (jade green) and flagellates (salmon) were side by side for each of the indicated genes (numbers are in reference to the Joint Genome Institute accession numbers). Actins are shown in the top panel, nucleators and nucleation promoting factors are grouped in the middle panel, and other actin-binding proteins and motors (myosin heavy chains) are shown in the bottom panel. B is set to a linear scale, while C is on a log scale. For all graphs, each bar represents the average relative transcript abundance from three independent experiments ± SD.
Actin and microtubule cytoskeletal gene expression differs between Naegleria amoebae and flagellates. (A) The relative transcript abundance for actin cytoskeletal genes (green), microtubule cytoskeletal genes (magenta), and GAPDH (gray) was calculated using expression data collected from amoeba (0 min into differentiation) or flagellate (80 min into differentiation) populations (original data from Fritz-Laylin and Cande, 2010). The relative transcript abundance in flagellates was subtracted from the level in amoebae to generate the graphs shown in Fig. 1, C and D. The microtubule cytoskeletal genes expressed in amoebae include the mitotic tubulins as well as putative spindle components. (B and C) The actin cytoskeletal transcript levels shown in A were organized such that the relative abundances in amoebae (jade green) and flagellates (salmon) were side by side for each of the indicated genes (numbers are in reference to the Joint Genome Institute accession numbers). Actins are shown in the top panel, nucleators and nucleation promoting factors are grouped in the middle panel, and other actin-binding proteins and motors (myosin heavy chains) are shown in the bottom panel. B is set to a linear scale, while C is on a log scale. For all graphs, each bar represents the average relative transcript abundance from three independent experiments ± SD.
We next looked in depth at specific actin cytoskeletal genes, many of which are conserved in the brain-eating amoeba, N. fowleri (Fig. S2; Herman et al., 2020, Preprint). These Naegleria genomes each encode >20 actins, and N. gruberi possesses 78 total actins and Arps (Fritz-Laylin et al., 2010b). These numbers are similar to those of Dictyostelium discoideum, whose genome encodes 17 identical actins and 41 total actins and Arps (Joseph et al., 2008). This abundance of actin genes likely indicates the importance of actin to an amoeboid lifestyle. Similar to actin, the transcript levels of most actin nucleators and nucleation-promoting factors were elevated in amoebae relative to flagellates (Fig. 1 D). There are, however, a few notable exceptions; one of the two Arp2 paralogs, one of the two WAVEs, two WASH complex subunits, and six formin family proteins were either expressed at similar levels in both populations or were preferentially expressed in flagellates (Fig. 1 D and,Fig. S2). Because actin is still abundant in flagellates (Fig. 1, B and C; and Fig. S1), these could be essential components for the flagellate actin cytoskeleton. Alternatively, because these SCAR/WAVE proteins are not identical, their differential expression may modulate the Arp2/3–activating capacity of the SCAR/WAVE complex and/or could be used for specific phenotypes (Suetsugu et al., 2003). The abundance of the other transcripts in amoebae indicates that the Arp2/3 complex, its activators, and a subset of formins likely facilitate actin-based phenotypes in N. gruberi amoebae.
Naegleria species encode an extensive repertoire of actin cytoskeletal regulators.(A)N. gruberi (N.g.) and N. fowleri (N.f.) genomes each encode a similar cellular complement of actin cytoskeletal proteins. The first column shows the name of the protein family or complex, the second includes the name of the mammalian homolog (with the exception of Class II formins, which are often found in plants, and Diaphanous Like Formins [DLF], which encompass all diaphanous-related formins), and the final columns indicate the number of genes found encoding these proteins in N. gruberi and N. fowleri, respectively. See Table S2 for accession numbers and sequences. We note that N. gruberi also possesses five WH2-domain–containing proteins not listed above, and no homolog of WASP Interacting Protein was identified in either Naegleria genome. (B) The graph from Fig. 1 D is shown (with all actins and Joint Genome Institute accession numbers listed) for comparisons of expression data with the proteins listed in A. Each bar represents the average relative change in transcript abundance between amoebae and flagellates from three experiments ± SD.
Naegleria species encode an extensive repertoire of actin cytoskeletal regulators.(A)N. gruberi (N.g.) and N. fowleri (N.f.) genomes each encode a similar cellular complement of actin cytoskeletal proteins. The first column shows the name of the protein family or complex, the second includes the name of the mammalian homolog (with the exception of Class II formins, which are often found in plants, and Diaphanous Like Formins [DLF], which encompass all diaphanous-related formins), and the final columns indicate the number of genes found encoding these proteins in N. gruberi and N. fowleri, respectively. See Table S2 for accession numbers and sequences. We note that N. gruberi also possesses five WH2-domain–containing proteins not listed above, and no homolog of WASP Interacting Protein was identified in either Naegleria genome. (B) The graph from Fig. 1 D is shown (with all actins and Joint Genome Institute accession numbers listed) for comparisons of expression data with the proteins listed in A. Each bar represents the average relative change in transcript abundance between amoebae and flagellates from three experiments ± SD.
To explore the structures that N. gruberi builds with this extensive actin machinery, we visualized cytoskeletal morphology using structured illumination microscopy (SIM). Amoebae contain a variety of actin structures, including many 0.2–0.8-µm puncta, 3–6-µm hollow actin spheres, a cell cortex, and thin ruffles (Fig. 2 A). Using wide-field fluorescence microscopy to quantify a larger sample size, we found that over 70% of cells had at least 10 actin puncta per cell (Fig. S3), which were present throughout the cell volume. Because the actin spheres were present in less than a third of cells and were more difficult to assess without SIM, we did not conduct further analyses of these structures. The ruffles, however, were clearly present in nearly 70% of cells (Fig. S3 I) and were of particular interest because similar-looking lamellar protrusions drive motility in other eukaryotes (Abercrombie, 1980; Abercrombie et al., 1970; Ingram, 1969). To confirm whether these actin-rich ruffles represented lamellar protrusions, we used scanning electron microscopy (SEM) to visualize the cell surface. We observed obvious lamellar protrusions (Fig. 2 B and Fig. S4) reminiscent of the thin protrusions and pseudopods of animal cells (Abercrombie et al., 1970; Fritz-Laylin et al., 2017a, 2017b; Ingram, 1969). The presence of these ruffles suggests that N. gruberi may move using α-motility, which relies on actin-rich pseudopods, rather than exclusively using blebbing motility like E. histolytica (Maugis et al., 2010).
Naegleria amoebae build lamellar ruffles and other morphologically distinct structures. (A) Amoebae were fixed and stained with phalloidin to detect actin (green, imaged using SIM) and DAPI to label DNA (magenta, imaged with widefield fluorescence). A single Z-slice (top) and maximum intensity projections (bottom) are shown for representative cells (treated with DMSO as part of a larger dataset in Fig. 3). Actin structures defined in the text are indicated. (B) Amoebae were fixed and processed for SEM. Ruffles are indicated in two representative DMSO-treated cells (also see Fig. S4).
Naegleria amoebae build lamellar ruffles and other morphologically distinct structures. (A) Amoebae were fixed and stained with phalloidin to detect actin (green, imaged using SIM) and DAPI to label DNA (magenta, imaged with widefield fluorescence). A single Z-slice (top) and maximum intensity projections (bottom) are shown for representative cells (treated with DMSO as part of a larger dataset in Fig. 3). Actin structures defined in the text are indicated. (B) Amoebae were fixed and processed for SEM. Ruffles are indicated in two representative DMSO-treated cells (also see Fig. S4).
Neither cell morphology nor actin polymer content is affected by jasplakinolide (Jasp), phalloidin (Phall), or cytochalasin (CytoD). (A) Cells were incubated in media ± inhibitors or controls for 10 min, then fixed and stained with Alexa Fluor-488–labeled phalloidin to detect F-actin (green) and DAPI to label DNA (magenta) before imaging using widefield fluorescence microscopy. Representative cells are shown. (B–G) Amoebae treated as in A were stained only with Alexa Fluor-488–labeled phalloidin (with the exception of an unstained control, shown in B) before analysis by flow cytometry. Representative histograms of F-actin staining intensity compare drug treatments with respective controls for one of three biological replicates. (H) Average intensities calculated from E–G were normalized to the stained control (B), which was set to 1 (dashed line). Each point represents the average normalized fluorescence intensity of F-actin staining (experimental replicates are coordinated by shape), and lines indicate the mean of three experimental replicates ± SD. Statistical significance was determined using an ordinary ANOVA followed by Tukey’s multiple comparison test. (I) Images like those shown in A were analyzed with the dataset presented in Fig. 3. Values are averages (Avg) ± SD from three independent replicates, each of which encompassed 50 cells per treatment condition. No statistical differences were found comparing jasplakinolide, phalloidin, or cytochalasin D treatments with controls using an ordinary ANOVA followed by Tukey’s multiple comparison test. *The percentage of cells with actin spheres was calculated for one representative dataset and, due to large variability between control samples, was not quantified for the remaining datasets. AU, arbitrary units; RFU, relative fluorescence units; SM+CK, SMIFH+CK-666 combined treatment.
Neither cell morphology nor actin polymer content is affected by jasplakinolide (Jasp), phalloidin (Phall), or cytochalasin (CytoD). (A) Cells were incubated in media ± inhibitors or controls for 10 min, then fixed and stained with Alexa Fluor-488–labeled phalloidin to detect F-actin (green) and DAPI to label DNA (magenta) before imaging using widefield fluorescence microscopy. Representative cells are shown. (B–G) Amoebae treated as in A were stained only with Alexa Fluor-488–labeled phalloidin (with the exception of an unstained control, shown in B) before analysis by flow cytometry. Representative histograms of F-actin staining intensity compare drug treatments with respective controls for one of three biological replicates. (H) Average intensities calculated from E–G were normalized to the stained control (B), which was set to 1 (dashed line). Each point represents the average normalized fluorescence intensity of F-actin staining (experimental replicates are coordinated by shape), and lines indicate the mean of three experimental replicates ± SD. Statistical significance was determined using an ordinary ANOVA followed by Tukey’s multiple comparison test. (I) Images like those shown in A were analyzed with the dataset presented in Fig. 3. Values are averages (Avg) ± SD from three independent replicates, each of which encompassed 50 cells per treatment condition. No statistical differences were found comparing jasplakinolide, phalloidin, or cytochalasin D treatments with controls using an ordinary ANOVA followed by Tukey’s multiple comparison test. *The percentage of cells with actin spheres was calculated for one representative dataset and, due to large variability between control samples, was not quantified for the remaining datasets. AU, arbitrary units; RFU, relative fluorescence units; SM+CK, SMIFH+CK-666 combined treatment.
Arp2/3 inhibition disrupts the formation of lamellar ruffles. Amoebae were treated with DMSO or CK-666 and then fixed and processed for SEM. Three representative cells for each condition were selected from the same experiment shown in Fig. 2.
Arp2/3 inhibition disrupts the formation of lamellar ruffles. Amoebae were treated with DMSO or CK-666 and then fixed and processed for SEM. Three representative cells for each condition were selected from the same experiment shown in Fig. 2.
Arp2/3 complex activity is required for the assembly of lamellar ruffles
To define how Naegleria regulates diverse actin phenotypes, we employed a panel of small-molecule inhibitors that target actin and/or actin nucleators (Fig. 3 A). To inhibit actin dynamics, we used latrunculin B (LatB), which has a conserved binding site on Naegleria actin (Fig. S5 and Fig. S6) and effectively disrupted total actin polymer content as measured by flow cytometry of cells stained with fluorescent phalloidin (Fig. 3 B). To impair actin nucleation pathways, we used CK-666 to inhibit Arp2/3 complex activity (Hetrick et al., 2013; Nolen et al., 2009) and SMIFH2 to broadly inhibit formin activity (Rizvi et al., 2009). In addition to inhibiting formins, SMIFH2 was also recently shown to inhibit some myosins (Nishimura et al., 2020) and therefore may be better described as an inhibitor of contractile networks. We verified that the target regions for both SMIFH2 and CK-666 were conserved in Naegleria (Fig. S6, C–E), and we found that neither of these inhibitors caused a global disruption in F-actin content (Fig. 3 B and Fig. S3).
Small-molecule inhibitors of the actin cytoskeleton alter Naegleria morphology. (A) Small-molecule inhibitors target actin dynamics. (B) Cells were incubated in media ± inhibitors for 10 min, then fixed and stained with phalloidin to detect F-actin. Total fluorescence was measured using flow cytometry. One representative histogram (left) is shown from a single replicate comparing LatB with its vehicle control (also see Fig. S3). Average F-actin intensities (right) were normalized to the stained control for all conditions. Each point represents the average normalized fluorescence of 20,000 cells from one experimental replicate, with each experimental replicate represented by a different shape. (C) Cells were treated as in B, but were fixed and stained for microscopy using DAPI to label DNA in addition to phalloidin. Cells were analyzed using SIM and widefield fluorescence (Fig. S3). Maximum intensity projections and single Z planes from SIM are shown for representative cells, and filopodia on a SMIFH2-treated cell are magnified in the inset (scale bar, 1 µm). (D–H) 50 cells/condition (imaged using widefield fluorescence) for three independent replicates (150 cells total/treatment) were used to quantify cell area (D) and circularity (E) or the percentage of cells with ≥1 ruffle (F), ≥1 filopodium (G), and ≥10 puncta (H). Small gray symbols represent individual cells, and larger symbols represent the averages for experimental replicates (coordinated by shape). For graphs in B (right) and D–H, black lines represent the means from experimental replicates ± SD, and statistical significance was determined using an ordinary ANOVA and Tukey’s multiple comparison test. Dashed lines indicate the control value. (I) Pixel intensity line scans for F-actin staining were drawn in a single Z-plane bisecting the cell edge, and values were normalized to the average intensity inside the cell, which was set to 1. Curves represent the average relative intensity ± SD for three experimental replicates, each encompassing five cells (also see Fig. S7). AU, arbitrary units; Max, maximum; RFU, relative fluorescence units; SM+CK, SMIFH2+CK-666 combined treatment.
Small-molecule inhibitors of the actin cytoskeleton alter Naegleria morphology. (A) Small-molecule inhibitors target actin dynamics. (B) Cells were incubated in media ± inhibitors for 10 min, then fixed and stained with phalloidin to detect F-actin. Total fluorescence was measured using flow cytometry. One representative histogram (left) is shown from a single replicate comparing LatB with its vehicle control (also see Fig. S3). Average F-actin intensities (right) were normalized to the stained control for all conditions. Each point represents the average normalized fluorescence of 20,000 cells from one experimental replicate, with each experimental replicate represented by a different shape. (C) Cells were treated as in B, but were fixed and stained for microscopy using DAPI to label DNA in addition to phalloidin. Cells were analyzed using SIM and widefield fluorescence (Fig. S3). Maximum intensity projections and single Z planes from SIM are shown for representative cells, and filopodia on a SMIFH2-treated cell are magnified in the inset (scale bar, 1 µm). (D–H) 50 cells/condition (imaged using widefield fluorescence) for three independent replicates (150 cells total/treatment) were used to quantify cell area (D) and circularity (E) or the percentage of cells with ≥1 ruffle (F), ≥1 filopodium (G), and ≥10 puncta (H). Small gray symbols represent individual cells, and larger symbols represent the averages for experimental replicates (coordinated by shape). For graphs in B (right) and D–H, black lines represent the means from experimental replicates ± SD, and statistical significance was determined using an ordinary ANOVA and Tukey’s multiple comparison test. Dashed lines indicate the control value. (I) Pixel intensity line scans for F-actin staining were drawn in a single Z-plane bisecting the cell edge, and values were normalized to the average intensity inside the cell, which was set to 1. Curves represent the average relative intensity ± SD for three experimental replicates, each encompassing five cells (also see Fig. S7). AU, arbitrary units; Max, maximum; RFU, relative fluorescence units; SM+CK, SMIFH2+CK-666 combined treatment.
Naegleria actin retains conserved binding sites for small-molecule inhibitors of actin dynamics. A multiple sequence alignment was generated to compare N. gruberi’s (N.g.’s) most highly expressed actin protein sequence with that of other eukaryotic actins (see Table S3 for sequences and ID numbers). This alignment includes actin sequences from other Discobids, N. fowleri (N.f.) and Trypanosoma cruzi (T.c.); Metamonads, Giardia intestinalis (G.i.) and Trichomonas vaginalis (T.v.); Plants, Chlamydomonas reinhardtii (C.r., including NAP1 [N] and IDA5 [I]) and Ectocarpus siliculosus (E.s.); Stramenopiles, Thalassiosira pseudonana (T.p.); Alveolates, Toxoplasma gondii (T.g.), Plasmodium falciparum (P.f., including actin-1 and actin-2), and Tetrahymena thermophila (T.t.); Amoebozoa, Dictyostelium discoideum (D.d.), Entamoeba histolytica (E.h.), and Acanthamoeba castellanii (A.c.); and Opisthokonts, Saccharomyces cerevisiae (S.c.), Schizosaccharomyces pombe (S.p.), and Oryctolagus cuniculus (O.c.). Sequences were aligned using T-Coffee (Notredame et al., 2000) with defaults (Blosum62 matrix, gap open penalty = −50, gap extension penalty = 0) in Jalview, and residues were colored based on conservation (grayscale) or to highlight differences in amino acids within drug-binding sites (ILVAM: salmon; FWY: tangerine; KRH: blue; DE: red; STNQ: lime; PG: raspberry; and C: yellow). Bars indicate important residues for drug binding, and binding sites are abbreviated as follows: L, latrunculin; P, phalloidin; C, cytochalasin; J, jasplakinolide.
Naegleria actin retains conserved binding sites for small-molecule inhibitors of actin dynamics. A multiple sequence alignment was generated to compare N. gruberi’s (N.g.’s) most highly expressed actin protein sequence with that of other eukaryotic actins (see Table S3 for sequences and ID numbers). This alignment includes actin sequences from other Discobids, N. fowleri (N.f.) and Trypanosoma cruzi (T.c.); Metamonads, Giardia intestinalis (G.i.) and Trichomonas vaginalis (T.v.); Plants, Chlamydomonas reinhardtii (C.r., including NAP1 [N] and IDA5 [I]) and Ectocarpus siliculosus (E.s.); Stramenopiles, Thalassiosira pseudonana (T.p.); Alveolates, Toxoplasma gondii (T.g.), Plasmodium falciparum (P.f., including actin-1 and actin-2), and Tetrahymena thermophila (T.t.); Amoebozoa, Dictyostelium discoideum (D.d.), Entamoeba histolytica (E.h.), and Acanthamoeba castellanii (A.c.); and Opisthokonts, Saccharomyces cerevisiae (S.c.), Schizosaccharomyces pombe (S.p.), and Oryctolagus cuniculus (O.c.). Sequences were aligned using T-Coffee (Notredame et al., 2000) with defaults (Blosum62 matrix, gap open penalty = −50, gap extension penalty = 0) in Jalview, and residues were colored based on conservation (grayscale) or to highlight differences in amino acids within drug-binding sites (ILVAM: salmon; FWY: tangerine; KRH: blue; DE: red; STNQ: lime; PG: raspberry; and C: yellow). Bars indicate important residues for drug binding, and binding sites are abbreviated as follows: L, latrunculin; P, phalloidin; C, cytochalasin; J, jasplakinolide.
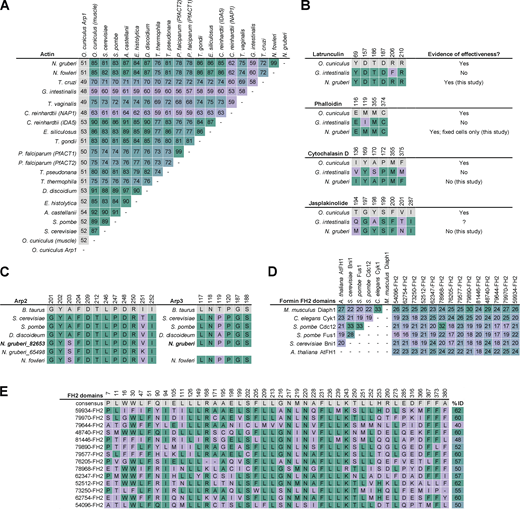
Actin cytoskeletal drugs bind sequences that are conserved in Naegleria proteins.(A) The alignment shown in Fig. S5 was used to calculate the percent identity of actin protein sequences from representative eukaryotes. Values indicating higher conservation are highlighted in green, and values indicating less conservation are highlighted in lilac. Arp1 from rabbit was included for comparison to an actin-related protein (shown in gray). (B) Drug binding sites on mammalian actin (Faulstich et al., 1993; Morton et al., 2000; Nair et al., 2008; Pospich et al., 2017) are shown (gray) in comparison to Giardia actin and Naegleria actin. Identical residues are shown in green, non-identical residues are in lilac, and the numbers above each column indicate the location of the residue in the rabbit actin sequence. Giardia actin is shown as an example due to its low percent identity to mammalian actin, as well as the documented ineffectiveness of actin inhibitors (Paredez et al., 2011). A question mark indicates unknown information. (C) Residues of Arp2 and Arp3 that exist within 5 Å of the CK-666 binding site on Bos taurus (gray) Arp2/3 complex (PDB 3UKR; Baggett et al., 2012) were compared between model organisms and Naegleria. Identical residues are shown in green, non-identical redidues are highlighted in lilac. (D) FH2 domains from six SMIFH2-sensitive formins in diverse model organisms (including Mus musculus and Arabidopsis thaliana) were used to generate an MSA, and the percent identity was calculated in comparison to each other and compared with the FH2 domains of Naegleria’s 14 formins. Higher percent identity values are shown in green, lower values are shown in lilac. (E) SMIFH2-sensitive formins shown in D were used to calculate an FH2 domain consensus sequence (gray). Positions with at least 75% identity among SMIFH2-sensitive FH2 domains are shown, with positions of Naegleria FH2 domains that are identical to the consensus shown in green. The total percent identity to the consensus sequence is shown to the right, with higher percent identities in green, and lower percent identities in lilac. All accession numbers for the sequences used to generate this figure are listed in Table S3.
Actin cytoskeletal drugs bind sequences that are conserved in Naegleria proteins.(A) The alignment shown in Fig. S5 was used to calculate the percent identity of actin protein sequences from representative eukaryotes. Values indicating higher conservation are highlighted in green, and values indicating less conservation are highlighted in lilac. Arp1 from rabbit was included for comparison to an actin-related protein (shown in gray). (B) Drug binding sites on mammalian actin (Faulstich et al., 1993; Morton et al., 2000; Nair et al., 2008; Pospich et al., 2017) are shown (gray) in comparison to Giardia actin and Naegleria actin. Identical residues are shown in green, non-identical residues are in lilac, and the numbers above each column indicate the location of the residue in the rabbit actin sequence. Giardia actin is shown as an example due to its low percent identity to mammalian actin, as well as the documented ineffectiveness of actin inhibitors (Paredez et al., 2011). A question mark indicates unknown information. (C) Residues of Arp2 and Arp3 that exist within 5 Å of the CK-666 binding site on Bos taurus (gray) Arp2/3 complex (PDB 3UKR; Baggett et al., 2012) were compared between model organisms and Naegleria. Identical residues are shown in green, non-identical redidues are highlighted in lilac. (D) FH2 domains from six SMIFH2-sensitive formins in diverse model organisms (including Mus musculus and Arabidopsis thaliana) were used to generate an MSA, and the percent identity was calculated in comparison to each other and compared with the FH2 domains of Naegleria’s 14 formins. Higher percent identity values are shown in green, lower values are shown in lilac. (E) SMIFH2-sensitive formins shown in D were used to calculate an FH2 domain consensus sequence (gray). Positions with at least 75% identity among SMIFH2-sensitive FH2 domains are shown, with positions of Naegleria FH2 domains that are identical to the consensus shown in green. The total percent identity to the consensus sequence is shown to the right, with higher percent identities in green, and lower percent identities in lilac. All accession numbers for the sequences used to generate this figure are listed in Table S3.
To assess whether these inhibitors alter the morphology of Naegleria amoebae, we treated cells for 10 min with inhibitors or controls, fixed and stained cells to detect polymerized actin, and analyzed cells using microscopy (Fig. 3 C and Fig. S3). By SIM, control cells displayed the previously identified actin structures, including ruffles and puncta. Cells treated with LatB, however, were rounded and lacked any distinguishing features, fitting with the defects measured by flow cytometry. While treatment with SMIFH2 did not reliably correlate with any morphological phenotypes we analyzed, CK-666–treated cells had a robust actin cortex and striking actin-rich spikes, reminiscent of filopodia (Fig. 3 C). While control cells and SMIFH2-treated cells also had these filopodia-like protrusions, they were often dimmer and shorter (Fig. 3 C, inset; and Fig. S3 I).
Using widefield fluorescence microscopy to image additional cells, we quantified the cell size (2D area), shape, and presence of actin structures (Fig. 3, D–H). While area did not differ significantly between treatments (Fig. 3 D), cells were more circular following LatB treatment and more elongated following combined treatment with CK-666 and SMIFH2 (Fig. 3 E). Arp2/3 complex inhibition significantly decreased cell ruffles (Fig. 3 F; and Fig. S4), while increasing the percentage of cells with filopodia-like structures (Fig. 3 G). Further, actin puncta were rare in CK-666–treated cells; <1% of treated cells had at least 10 small, actin-rich foci (Fig. 3 H), suggesting these structures are Arp2/3 dependent. Finally, the cell cortex was more intense in CK-666–treated cells (Fig. 3 I; and Fig. S7). Collectively, these results suggest that the Arp2/3 complex is important for forming ruffles and actin puncta, and in the absence of Arp2/3 activity, cells preferentially build filopodia-like structures and may allocate more actin to the cell cortex. Because mammalian leukocytes treated with CK-666 or lacking Arp2/3 complex also show an enrichment in filopodial protrusions (Fritz-Laylin et al., 2017b; Rotty et al., 2017), the increase in these structures in Naegleria upon CK-666 treatment highlights a similarity in actin networks from organisms across the eukaryotic tree.
CK-666 treatment enhances the relative concentration of cortical to intracellular F-actin. (A) 15 amoebae treated with the Arp2/3 complex inhibitor CK-666 from the experiments described in Fig. 3 were imaged using widefield fluorescence microscopy. A line with a width of 50 px, representing 3.25 µm, was drawn perpendicular to the cell edge on a random, relatively straight part of each cell in a Z plane in which the cell edge was in focus. Panels i–iii: examples of these regions on cells are shown, with one example from each of the three biological replicates analyzed. Panel iv: the regions of interest of six additional examples are shown. (B) The pixel intensities along the long axis of each box shown in A were normalized to the average pixel intensity of an area inside the cell, which was set to 1. These normalized intensities were plotted against the distance, with 0 indicating the cell edge (dashed line). The horizontal line indicates the maximum fluorescence intensity at the cell edge. The first three panels (i–iii) correspond to the images in A i–iii, and the rightmost “all” panel depicts all the data used to generate the plot in Fig. 3 J. (C and D) Images mirror A and B, respectively, using the inactive control CK-689. AU, arbitrary units.
CK-666 treatment enhances the relative concentration of cortical to intracellular F-actin. (A) 15 amoebae treated with the Arp2/3 complex inhibitor CK-666 from the experiments described in Fig. 3 were imaged using widefield fluorescence microscopy. A line with a width of 50 px, representing 3.25 µm, was drawn perpendicular to the cell edge on a random, relatively straight part of each cell in a Z plane in which the cell edge was in focus. Panels i–iii: examples of these regions on cells are shown, with one example from each of the three biological replicates analyzed. Panel iv: the regions of interest of six additional examples are shown. (B) The pixel intensities along the long axis of each box shown in A were normalized to the average pixel intensity of an area inside the cell, which was set to 1. These normalized intensities were plotted against the distance, with 0 indicating the cell edge (dashed line). The horizontal line indicates the maximum fluorescence intensity at the cell edge. The first three panels (i–iii) correspond to the images in A i–iii, and the rightmost “all” panel depicts all the data used to generate the plot in Fig. 3 J. (C and D) Images mirror A and B, respectively, using the inactive control CK-689. AU, arbitrary units.
Inhibiting actin nucleation by the Arp2/3 complex reduces Naegleria cell speed, while inhibiting formins impairs directional persistence
To assess whether the actin inhibitors caused defects in cell migration, we introduced inhibitors during live imaging of crawling cells. We then directly compared pre- and post-treatment speeds of individual cells (Fig. 4, A and B; and Video 1). While control cells did not exhibit any robust changes in motility, all cells treated with LatB moved slower after treatment (29.8 ± 4.3 µm/min pretreatment vs. 4.6 ± 2.3 µm/min after treatment). CK-666 treatment reduced speed in 90% of cells, and on average cells moved 13.1 ± 5.5 µm/min slower after CK-666 treatment (Fig. 4, B and C). Together, these data suggest that Arp2/3–dependent actin polymerization is a major contributor to Naegleria’s crawling motility.
Inhibition of actin nucleation pathways impairs Naegleria cell crawling. (A) Crawling cells were imaged using phase/contrast microscopy for 5 min. After 5 min (at t = 0 min), an equal volume of each of the indicated inhibitors or controls was added (diluted in buffer; “control” indicates buffer alone), and imaged for 5 min. Arrowheads indicate the position of a representative cell over time. (B) Cells were treated as in A. 20 randomly selected cells from the center of the field of view at t = 0 were tracked to calculate average speeds before and after treatment. Each point represents the speed of a cell before treatment (x axis) and after treatment (y axis). The dashed line indicates no change. Three experimental replicates are shown using different shapes. Two data points are off the x axis: LatB (79.3, 12.9) and CK-666 (60.8, 23.6). (C) The data collected in B were used to calculate the change in cell speed after treatment (n = 3, 20 cells/trial, 60 cells total/condition). The dashed line is set to zero. (D) Data from B were plotted for 20 cells from one representative experiment. Tracks before treatment (pre-treat; gray) have the −5 min time point at the origin, while tracks after treatment (in color) have the t = 0 time point at the origin (also see Fig. S8). (E) Directional persistence was calculated for each post-treatment cell tracked in B by dividing the maximum displacement from the start by the path length. The dashed line indicates the average from the control. For graphs in C and E, small gray symbols represent the values of single cells, and larger symbols represent experimental averages. Black lines indicate the mean ± SD calculated from three experimental replicates, and statistical significance was determined using an ordinary ANOVA and Tukey’s multiple comparison test. (F) Cells were imaged using differential interference contrast microscopy and treated with CK-666 during imaging. Panels show time lapse images of a representative cell generating a filopodium-like protrusion ∼30 min after treatment. (G) Additional cells treated and imaged as in F were fixed and stained on the microscope after 2 min and 18 s of treatment (top: cell 2, short incubation time) or 53 min (bottom: cell 3, long incubation time). Cells were simultaneously fixed, permeabilized, and stained with DAPI to detect DNA (magenta) and phalloidin to detect F-actin (green). Times after treatment for F and G are in minutes:seconds.
Inhibition of actin nucleation pathways impairs Naegleria cell crawling. (A) Crawling cells were imaged using phase/contrast microscopy for 5 min. After 5 min (at t = 0 min), an equal volume of each of the indicated inhibitors or controls was added (diluted in buffer; “control” indicates buffer alone), and imaged for 5 min. Arrowheads indicate the position of a representative cell over time. (B) Cells were treated as in A. 20 randomly selected cells from the center of the field of view at t = 0 were tracked to calculate average speeds before and after treatment. Each point represents the speed of a cell before treatment (x axis) and after treatment (y axis). The dashed line indicates no change. Three experimental replicates are shown using different shapes. Two data points are off the x axis: LatB (79.3, 12.9) and CK-666 (60.8, 23.6). (C) The data collected in B were used to calculate the change in cell speed after treatment (n = 3, 20 cells/trial, 60 cells total/condition). The dashed line is set to zero. (D) Data from B were plotted for 20 cells from one representative experiment. Tracks before treatment (pre-treat; gray) have the −5 min time point at the origin, while tracks after treatment (in color) have the t = 0 time point at the origin (also see Fig. S8). (E) Directional persistence was calculated for each post-treatment cell tracked in B by dividing the maximum displacement from the start by the path length. The dashed line indicates the average from the control. For graphs in C and E, small gray symbols represent the values of single cells, and larger symbols represent experimental averages. Black lines indicate the mean ± SD calculated from three experimental replicates, and statistical significance was determined using an ordinary ANOVA and Tukey’s multiple comparison test. (F) Cells were imaged using differential interference contrast microscopy and treated with CK-666 during imaging. Panels show time lapse images of a representative cell generating a filopodium-like protrusion ∼30 min after treatment. (G) Additional cells treated and imaged as in F were fixed and stained on the microscope after 2 min and 18 s of treatment (top: cell 2, short incubation time) or 53 min (bottom: cell 3, long incubation time). Cells were simultaneously fixed, permeabilized, and stained with DAPI to detect DNA (magenta) and phalloidin to detect F-actin (green). Times after treatment for F and G are in minutes:seconds.
Naegleria motility is altered by different small-molecule inhibitors. Crawling amoebae were imaged using phase/contrast microscopy at a rate of 5 s/frame. Cells were treated with the indicated inhibitors or controls at the 5-min mark, and imaging proceeded for an additional 5 min.
Naegleria motility is altered by different small-molecule inhibitors. Crawling amoebae were imaged using phase/contrast microscopy at a rate of 5 s/frame. Cells were treated with the indicated inhibitors or controls at the 5-min mark, and imaging proceeded for an additional 5 min.
While SMIFH2 did not result in a statistical difference in cell speed, SMIFH2-treated cells often contacted nearby cells and then moved in circular patterns (Video 1 and,Fig. 4 A). To visualize these behaviors, we used worm plots to display the tracking data for cells before and after treatment, which revealed treated cells do not explore the same area as control cells (Fig. 4 D). SMIFH2-treated cells had an average maximum displacement of 36.2 ± 4.6 µm after treatment compared with 95.2 ± 15.6 µm before treatment. SMIFH2 treatment also reduced the directional persistence of cells; DMSO-treated cells had a persistence of 0.63 ± 0.04, while SMIFH2 decreased this value to 0.41 ± 0.04 (Fig. 4 E). Because SMIFH2 is capable of inhibiting myosins in addition to formins (Nishimura et al., 2020), either effect could disrupt contractile networks that maintain membrane tension, resulting in the loss of polarization (Hind et al., 2016; Houk et al., 2012). Collectively, these data indicate that actin-based motility in N. gruberi is largely dependent on actin nucleation by the Arp2/3 complex, while directional persistence during cell migration may rely on formin-nucleated and/or contractile actin networks.
To address the possibility that the Arp2/3 complex and formins could either cooperate or compensate for each other to drive motility, we treated cells with both CK-666 and SMIFH2. Cells treated with this combination were neither significantly slower nor less directionally persistent than those treated with CK-666 or SMIFH2 alone (Fig. S8, C–E). While these data do not exclude the possibility for some nucleator crosstalk, they are not consistent with synergy between formins and the Arp2/3 complex in this system.
Supplemental motility data. (A) Tracking data show cell movements from the experiments in Fig. 4 (the same SMIFH2 and DMSO graphs are shown in Fig. 4 D and reproduced here for comparison purposes). Cell tracks show movement 5 min before (Pre-treat., gray traces) and 5 min after (colored traces) treatment. (B) In addition to the inhibitors shown in Fig. 4, each experimental replicate also included cells treated with jasplakinolide (Jasp), phalloidin, and cytochalasin D (CytoD). Each small gray symbol represents the change in speed of a single cell after treatment, while larger symbols represent the averages from each experimental replicate, coordinated by shape (circles, squares, and triangles). (C) Cells were imaged as in Fig. 4; after 5 min of imaging in buffer, cells were treated with DMSO, SMIFH2, CK-666, or a cocktail of SMIFH2 and CK-666 (SM+CK) and imaged for an additional 5 min. 20 randomly selected cells from the center of the field of view at the time of treatment were tracked to calculate average speeds before and after treatment. Each point represents the speed of a cell before treatment (plotted on the x axis) and after treatment (plotted on the y axis). Three experimental replicates are indicated using different shapes (squares, triangles, and circles). The dashed line has a slope of 1, indicating where cells would fall if the speed was unchanged. (D) The data collected from experiments in C were used to calculate the change in cell speed after treatment. Each smaller gray symbol represents a single cell, while larger symbols represent the averages from each experimental replicate, with shapes representing independent replicates. (E) Directional persistence was calculated for each post-treatment cell tracked in C by dividing the maximum displacement from the start of the track by the total path length. Each small gray symbol represents the persistence of a single cell, while larger symbols represent experimental averages, coordinated by shape. For B, D, and E, black lines indicate the mean ± SD calculated from the three experimental replicates, and statistical significance was determined using an ordinary ANOVA followed by Tukey’s multiple comparison test.Pre-treat., before treatment.
Supplemental motility data. (A) Tracking data show cell movements from the experiments in Fig. 4 (the same SMIFH2 and DMSO graphs are shown in Fig. 4 D and reproduced here for comparison purposes). Cell tracks show movement 5 min before (Pre-treat., gray traces) and 5 min after (colored traces) treatment. (B) In addition to the inhibitors shown in Fig. 4, each experimental replicate also included cells treated with jasplakinolide (Jasp), phalloidin, and cytochalasin D (CytoD). Each small gray symbol represents the change in speed of a single cell after treatment, while larger symbols represent the averages from each experimental replicate, coordinated by shape (circles, squares, and triangles). (C) Cells were imaged as in Fig. 4; after 5 min of imaging in buffer, cells were treated with DMSO, SMIFH2, CK-666, or a cocktail of SMIFH2 and CK-666 (SM+CK) and imaged for an additional 5 min. 20 randomly selected cells from the center of the field of view at the time of treatment were tracked to calculate average speeds before and after treatment. Each point represents the speed of a cell before treatment (plotted on the x axis) and after treatment (plotted on the y axis). Three experimental replicates are indicated using different shapes (squares, triangles, and circles). The dashed line has a slope of 1, indicating where cells would fall if the speed was unchanged. (D) The data collected from experiments in C were used to calculate the change in cell speed after treatment. Each smaller gray symbol represents a single cell, while larger symbols represent the averages from each experimental replicate, with shapes representing independent replicates. (E) Directional persistence was calculated for each post-treatment cell tracked in C by dividing the maximum displacement from the start of the track by the total path length. Each small gray symbol represents the persistence of a single cell, while larger symbols represent experimental averages, coordinated by shape. For B, D, and E, black lines indicate the mean ± SD calculated from the three experimental replicates, and statistical significance was determined using an ordinary ANOVA followed by Tukey’s multiple comparison test.Pre-treat., before treatment.
Because some cell types of other organisms do not strictly require the Arp2/3 complex for motility (Rotty et al., 2017), we looked more closely at the residual motility of CK-666–treated cells. While a fraction of cells continued to move after treatment, using a mechanism reminiscent of blebbing (Video 1), a large percentage of cells developed filopodia-like protrusions, similar to the structures we previously observed (Fig. 3 C). These structures formed de novo from the leading edge of the cell and grew outward at an average maximum rate of 0.90 ± 0.47 µm/s (maximum growth rate for actin-enriched filopodia over a 3-s window, averaged from four cells; Fig. 4 F). We also imaged cells before and during treatment with CK-666 and fixed and stained these cells for F-actin during the time course (Fig. 4 G and,Video 2). This confirmed that these filopodia-like structures are actin-rich and are likely the same structures we observed in bulk fixed cells. Collectively, these data show that although Naegleria’s main form of cell migration relies on Arp2/3–dependent lamellar protrusions, cells can still migrate, to a degree, under Arp2/3 inhibition.
CK-666 induces actin-rich filopodial protrusions. Cells were imaged using differential interference contrast microscopy at a frame rate of 1 s/frame and treated with CK-666 during imaging. Cells were simultaneously fixed, permeabilized, and stained with DAPI to detect DNA (magenta) and phalloidin to detect F-actin (green). This cell is also shown in Fig. 4 G, cell 2.
CK-666 induces actin-rich filopodial protrusions. Cells were imaged using differential interference contrast microscopy at a frame rate of 1 s/frame and treated with CK-666 during imaging. Cells were simultaneously fixed, permeabilized, and stained with DAPI to detect DNA (magenta) and phalloidin to detect F-actin (green). This cell is also shown in Fig. 4 G, cell 2.
Phagocytosis requires actin assembly and is enhanced by Arp2/3 complex activity
To test the hypothesis that Arp2/3 nucleated actin networks drive phagocytosis in addition to motility in Naegleria, we treated cells with LatB or CK-666 and incubated cells with GFP-expressing bacteria. After fixing cells and visualizing GFP-bacteria by microscopy, we observed that control cells were associated with robust GFP signal, while CK-666–treated cells had considerably less GFP intensity and LatB-treated cells were associated with little-to-no fluorescence (Fig. 5 A).
Phagocytosis is less efficient following Arp2/3 complex inhibition. (A) Amoebae were starved for 1 h, then incubated with controls or inhibitors and GFP-expressing E. coli for 45 min. Cells were then fixed and imaged. GFP intensity is shown using an inverted LUT (lookup table; top) and in magenta (bottom). (B) Cells were treated as in A, then the GFP intensities of 30,000 cells/condition were measured by flow cytometry. Representative histograms display the percentage of cells plotted against GFP intensities for ethanol- and LatB-treated cells (left) or CK-689– and CK-666–treated cells (right). Plots are from one of three experimental replicates (also see Fig. S9). (C) The median fluorescence intensity was calculated from experiments shown in B. For each replicate (coordinated by shape; circles, triangles, and squares), fluorescence intensity of control and treated cells was normalized to a buffer-only control. (D) The population of buffer-only control cells was used to gate a GFP-positive population for each experimental replicate. Each point represents the percentage of cells falling within the GFP-positive gate. For graphs in C and D, lines indicate the mean of three experimental replicates (which are coordinated by shape) ± SD. The dashed lines are set to the average from the control sample. Statistical significance was determined using an ordinary ANOVA and Tukey’s multiple comparison test. AU, arbitrary units; RFU, relative fluorescence units.
Phagocytosis is less efficient following Arp2/3 complex inhibition. (A) Amoebae were starved for 1 h, then incubated with controls or inhibitors and GFP-expressing E. coli for 45 min. Cells were then fixed and imaged. GFP intensity is shown using an inverted LUT (lookup table; top) and in magenta (bottom). (B) Cells were treated as in A, then the GFP intensities of 30,000 cells/condition were measured by flow cytometry. Representative histograms display the percentage of cells plotted against GFP intensities for ethanol- and LatB-treated cells (left) or CK-689– and CK-666–treated cells (right). Plots are from one of three experimental replicates (also see Fig. S9). (C) The median fluorescence intensity was calculated from experiments shown in B. For each replicate (coordinated by shape; circles, triangles, and squares), fluorescence intensity of control and treated cells was normalized to a buffer-only control. (D) The population of buffer-only control cells was used to gate a GFP-positive population for each experimental replicate. Each point represents the percentage of cells falling within the GFP-positive gate. For graphs in C and D, lines indicate the mean of three experimental replicates (which are coordinated by shape) ± SD. The dashed lines are set to the average from the control sample. Statistical significance was determined using an ordinary ANOVA and Tukey’s multiple comparison test. AU, arbitrary units; RFU, relative fluorescence units.
We next measured the levels of cell-associated bacteria using flow cytometry. Compared with the control, the vast majority of LatB-treated cells had virtually no GFP fluorescence (Fig. 5 B). While not as dramatic, CK-666–treated cells also had less GFP signal than CK-689–treated cells (Fig. 5 B and,Fig. S9 A). For LatB, these shifts translated to a significantly lower median intensity compared with ethanol alone and a 50.7% ± 11.9% reduction in the percentage of positive cells (Fig. 5, C and D), indicating that phagocytosis does indeed involve actin polymerization. For cells treated with CK-666, neither the change in GFP intensity nor the percentage of positive cells reached our threshold for statistical significance. However, the three biological replicates had the same trend, suggestive of a defect in phagocytosis despite variability among individual cells (Fig. S9 A).
LatB and CK-666 each impair Naegleria’s phagocytosis. (A) Cells were starved, treated with inhibitors or controls, and fixed as in Fig. 5. Then, the GFP intensities of 30,000 cells per condition were measured by flow cytometry. Cells were initially gated to select only intact, single cells. Then, an untreated control from each replicate (top row) was used to create a gate for GFP+ cells (dashed line). Histograms display the percentage of cells plotted against GFP intensities for ethanol- and LatB-treated cells (middle row) or CK-689– and CK-666–treated cells (bottom row). The histograms from replicate 2 are also shown in Fig. 5, with y axes set to the same scale. (B) Cells were starved and then mixed with GFP-expressing E. coli (shown in magenta) for 1 h. Cells were fixed and stained with Alexa Fluor-568–labeled phalloidin to detect F-actin (green) and imaged using widefield fluorescence microscopy. Maximum intensity projections (max. proj.; top) and single xy, zy, and xz slices are shown intersecting nonintact (white arrowheads) and intact (gray arrowheads) bacteria associated with representative cells. AU, arbitrary units.
LatB and CK-666 each impair Naegleria’s phagocytosis. (A) Cells were starved, treated with inhibitors or controls, and fixed as in Fig. 5. Then, the GFP intensities of 30,000 cells per condition were measured by flow cytometry. Cells were initially gated to select only intact, single cells. Then, an untreated control from each replicate (top row) was used to create a gate for GFP+ cells (dashed line). Histograms display the percentage of cells plotted against GFP intensities for ethanol- and LatB-treated cells (middle row) or CK-689– and CK-666–treated cells (bottom row). The histograms from replicate 2 are also shown in Fig. 5, with y axes set to the same scale. (B) Cells were starved and then mixed with GFP-expressing E. coli (shown in magenta) for 1 h. Cells were fixed and stained with Alexa Fluor-568–labeled phalloidin to detect F-actin (green) and imaged using widefield fluorescence microscopy. Maximum intensity projections (max. proj.; top) and single xy, zy, and xz slices are shown intersecting nonintact (white arrowheads) and intact (gray arrowheads) bacteria associated with representative cells. AU, arbitrary units.
To gain insight into the nature of the residual “bacteria-positive” LatB and CK-666–treated cells, we repeated the experiment and stained cells to visualize F-actin. Cells had two distinct types of GFP staining: intense staining that corresponded to external, intact bacteria as well as weaker staining that appeared to represent internalized bacterial remnants (Fig. S9 B). While both types of staining were often observed in control cells, LatB- and CK-666–treated cells were typically found with only extracellular bacteria. This suggests that the “GFP-positive” LatB and CK-666 cells identified by flow cytometry may have been associated with extracellular bacteria and therefore may not represent bona fide phagocytic events. Collectively, these data support a role for the Arp2/3 complex in phagocytosis as well as motility, suggesting that, as in other organisms, there is some overlap in the pathways responsible for moving and eating.
Concluding remarks
Our work demonstrates that Naegleria has sophisticated actin cytoskeletal machinery, including the Arp2/3 complex, its activators, and formins, which are primarily expressed in amoebae. Using small molecules, we show that cells build Arp2/3–dependent lamellar protrusions and assemble filopodia during Arp2/3 inhibition. We also show that Arp2/3 inhibition impairs cell motility and phagocytosis, while inhibiting formin-derived networks impairs directional persistence. Because Arp2/3–derived networks also promote lamellar protrusions, α-motility, and phagocytosis in opisthokonts and amoebozoans (Davidson et al., 2018; Jaumouillé et al., 2019; Seastone et al., 2001; Veltman et al., 2012; Veltman et al., 2016), our results suggest that the Arp2/3–dependent mechanisms that drive motility and phagocytosis are evolutionarily ancient and can function independently of a microtubule cytoskeleton.
This work also has implications for the brain-eating amoeba, N. fowleri. The lack of effective, reproducible treatment options for N. fowleri infections has resulted in a >90% case fatality rate (Siddiqui et al., 2016). Because motility to and within the brain and the phagocytosis-like mechanism used to consume brain tissue undoubtedly contribute to disease (Siddiqui et al., 2016), defining the molecular underpinnings of these cellular processes may reveal new and much-needed drug targets.
Materials and methods
Cell and bacterial culture
N. gruberi cells (strain NEGM; a gift from Dr. Chandler Fulton, Brandeis University, Waltham, MA) were axenically cultured in M7 Media (see Table S1 for all reagents and recipes) in plug seal tissue culture–treated flasks (CELLTREAT; cat. no. 229330) and grown at 28°C. Cells were split into fresh media every 2–3 d. To induce differentiation (Fig. 1), cells were centrifuged at 1,500 relative centrifugal force (RCF) for 90 s, and pellets were resuspended in ice-cold 2 mM Tris and incubated in a 28°C water bath shaking at 100 rpm for ∼90 min.
For phagocytosis assays, BL21 Escherichia coli expressing superfolder-GFP (Pédelacq et al., 2006) was streaked from frozen glycerol stocks kept at −80°C onto lysogeny broth (LB) plates supplemented with 50 µg/ml kanamycin. Broth cultures of LB + kanamycin were inoculated from single colonies and were grown overnight at 37°C shaking at 175 rpm in a New Brunswick Innova 4330 floor shaker. Cultures were then diluted 1:10 into fresh LB and grown until the OD600 reached 0.4–0.7, at which point IPTG was added to a final concentration of 0.1 mM for 1 h.
Transcript analysis and homolog identification
Microarray data from amoeba and flagellate populations were analyzed from experiments completed in Fritz-Laylin and Cande (2010). Each original biological replicate had been completed in duplicate, so the average for each biological replicate was first calculated. Then, the transcript abundance in flagellates (from the 80-min time point) was subtracted from the transcript abundance in amoebae (from the 0-min time point) for that biological replicate, and the average and SDs from the three replicates were calculated. Only the transcripts corresponding to actin or microtubule cytoskeletal genes were included in this analysis.
To expand upon the original N. gruberi cytoskeletal annotations (Table S5 in Fritz-Laylin et al., 2010b), a database of additional proteins suspected to be present (e.g., additional Arp2/3 complex subunits, additional SCAR/WAVE complex subunits, WASH complex) was generated using protein sequences from human (obtained from PubMed) or D. discoideum homologs (obtained from DictyBase: http://dictybase.org). The resulting database was used as a query to BLAST (Altschul et al., 1990) against the N. gruberi proteome, using default parameters except replacing the scoring matrix with the BLOSUM45 scoring matrix. We further validated protein identities through hmmscan searches using a gathering threshold and Pfam domains using the HMMER website.
To assess the cytoskeletal repertoire of N. fowleri, we compared an N. fowleri (American Type Culture Collection 30863) protein database generated in Augustus (Herman et al., 2020, Preprint) with N. gruberi actin- and microtubule-associated proteins (Fritz-Laylin et al., 2010b) using BLAST with default search parameters. The top N. fowleri hits identified by comparisons to N. gruberi were then compared using BLAST to the full N. gruberi protein library to establish best mutual BLAST hits. See Table S2 for the full list of accession numbers.
Multiple sequence alignments (MSAs) and inhibitor binding sites
Actin sequences from N. gruberi, N. fowleri, and a diverse set of additional eukaryotes were collected (see Table S3) and aligned using T-Coffee (Notredame et al., 2000) with defaults (Blosum62 matrix, gap open penalty = −50, gap extension penalty = 0) in Jalview. Drug binding sites from the literature (Faulstich et al., 1993; Morton et al., 2000; Nair et al., 2008; Pospich et al., 2017) were mapped onto the rabbit muscle actin sequence, using only residues with evidence of direct interactions. It is important to note, however, that additional residues not highlighted in our MSA are likely required for the proper conformation of binding sites.
Cell morphology assays and use of inhibitors
For the experiments shown in Fig. 3, flasks of cells were moved to room temperature on the experiment eve to prevent differentiation to flagellates during the experiment due to temperature fluctuation. 0.5 ml of cells were seeded (at ∼5 × 105 cells/ml) into a 24-well plate preloaded with 0.5 ml fresh M7 medium with inhibitors or controls for final (1×) concentrations as follows: 5 µM LatB, 20 µM cytochalasin D, 140 nM phalloidin, 2 µM jasplakinolide, 50 µM CK-666, 25 µM SMIFH2, 50 µM CK-689, 0.1% DMSO, and 0.09% ethanol. Concentrations were selected based on work in other systems (Bettadapur and Ralston, 2020; Fritz-Laylin et al., 2017b; Jahan and Yumura, 2017; Jasnin et al., 2016; Manich et al., 2018; Olins and Olins, 2004; Ralston et al., 2014; Spector et al., 1983; Velle and Campellone, 2018; Wang et al., 2014; Williams and Kay, 2018; Wilson et al., 2013) and/or were tested at 10-fold dilutions within the range recommended in the company-supplied data sheets in preliminary experiments. From preliminary experiments, the lowest concentration that had an observable phenotype in live cells was chosen, or in the case of cytochalasin D, jasplakinolide, and phalloidin, the highest concentrations we tested were chosen, which still did not produce any obvious phenotypes. While cytochalasin D has generated phenotypes in studies of Naegleria’s transformation into flagellates (Han et al., 1997; Walsh, 2007), the concentrations used in these studies (up to 200 µM) are ∼100 times more concentrated than the amounts used in other systems, so we chose lower, more conservative concentrations. Although we cannot entirely rule out off-target effects, as is generally the case for inhibitor-based studies, we selected short time points (5–10 min) for most experiments to minimize the potential for off-target effects.
After 10 min of incubation with inhibitors or controls, cells were fixed and stained for microscopy following a protocol adapted from Fritz-Laylin et al. (2010a). Medium was aspirated from cells, 1 ml of microscopy fixative (see Table S1 for all recipes and reagents) was added to each well, and 0.5 ml of cells was transferred to each of two coverslips (precoated with 0.1% poly(ethyleneimine)) per condition. Cells were allowed to settle onto the coverslips while fixing. After 15 min of fixation, the fixative was aspirated from the wells and replaced with 0.5 ml per well of permeabilization buffer. After 10 min of incubation, the permeabilization buffer was replaced with staining solution. After staining for 45 min, cells were washed three times in PEMBALG for ∼5 min each, and coverslips were mounted in Prolong Gold antifade reagent and allowed to cure overnight. The next morning, coverslips were sealed with nail polish, and labels were blinded before microscopic analysis.
Images for quantifying cell morphology were acquired using a Nikon Eclipse Ti2 microscope equipped with a pco.panda camera and a Plan Apo λ 100× Oil 1.45 NA objective. Alexa Fluor-488–labeled phalloidin and DAPI were imaged using a pE-300 MultiLaser light-emitting diode light source (excitation: 460 nm or 400 nm; emission: 535 nm or 460 nm). Representative cells were imaged with SIM (shown in Fig. 2 A and Fig. 3 C) using a Nikon A1R-SIMe microscope equipped with an ORCA-Flash4.0 camera and an SR Apo total internal reflection fluorescence 100× 1.49 NA objective. Images were acquired using Nikon NIS Elements software.
Image analysis was performed on blinded datasets using Fiji (Schindelin et al., 2012) to view images as Z-stacks and to generate maximum intensity projections. Phenotypes were defined as follows: Puncta: a cell with at least 10 small (0.2–0.8-µm diameter) F-actin–rich foci in the cell counted as having puncta. Ruffles: Ruffles are actin-rich lamellar protrusions that resemble the ruffled edges of a lasagna noodle. Filopodia-like protrusions: Filopodia were defined as spaghetti-like protrusions from the main body of the cell that are longer than they are wide (<0.75 µm thick, typically 0.2–0.5 µm). Area and shape: The maximum intensity projection was used to draw an outline of the areal footprint of the cell in ImageJ using the “kidney bean” tool. Measurements were taken to include the area and shape descriptors (of which circularity was used). Cortex measurements: The cell cortex was measured by drawing a line (with a thickness of 3.25 µm) perpendicular to the cell edge using the line tool in ImageJ. The center of the line was set to the cell edge so measurements could be normalized. Lines were preferentially drawn on noncurved sections of the cell, and a representative area of the membrane was chosen. The plot profile tool was used to generate pixel intensity plots, which were normalized to an area inside the cell that was set to 1.
Cell migration assays
For the cell motility assays shown in Fig. 4, A–E, flasks of cells were moved to room temperature on the experiment eve to prevent differentiation to flagellates during the experiment. Cells were resuspended by slapping and swirling the flask, and 0.5 ml of cell suspension (∼105 cells/ml) was added to each well of a 24-well plate (with a tissue culture–treated surface). After the cells were allowed to settle for 5 min, medium was aspirated and cells were washed once with 0.5 ml Tris buffer (2 mM Tris, pH 7.2) per well. After the wash was aspirated, a final volume of 0.5 ml Tris buffer was added to the cells in each well. Cells were imaged using phase/contrast microscopy with an Eclipse Ti2 microscope equipped with a pco.panda camera and an Achromat 10× Ph1 apodized dark low 0.25 NA objective, controlled by NIS Elements software. Cells were imaged using an acquisition rate of 5 s/frame. After 5 min of imaging, an equal volume of 2x small-molecule inhibitors or carriers alone (diluted in 2 mM Tris) was added to the wells of live amoebae. Imaging proceeded for an additional 5 min, and the exact time point at which inhibitors or controls were added was noted. File names were blinded, and 20 randomly selected cells from the center of the field of view at the time of treatment were chosen for tracking analysis. Tracked cells were only excluded if they left the field of view or contacted another cell in a way that disrupted tracking (see SMIFH2 treatment; Video 1). Cells were tracked using the mTrackJ plugin (Meijering et al., 2012) in Fiji, and the dataset was divided into “pre” and “post” treatment tracks for each cell with tracking data.
Other fluorescence microscopy
Cells in the experiments in Fig. 1 B were spun down (1,500 RCF/90 s), resuspended in fixative (for 15 min), spun again, resuspended in permeabilization buffer (for 10 min), spun again, and resuspended in staining solution before transfer to a 96-well glass-bottom plate. After 20 min, the staining solution was gently replaced with PBS. The cells in Fig. 5 A were fixed for flow cytometry and imaged in a 96-well glass-bottom plate. The cells in Fig. S9 B were stained following the protocol used for cell morphology assays, except using Alexa Fluor-568 phalloidin. The cells in Fig. 4, F and G were fixed, permeabilized, and stained in one step during imaging in a 35-mm tissue culture–treated glass-bottom MatTek dish. Images in Fig. 1 B; Fig. 4, F and G; Fig. 5 A; and Fig. S9 B were taken using an Eclipse Ti2 microscope equipped with a pco.panda camera and a Plan Apo λ 100× Oil 1.45 NA objective or an Achromat 10× Ph1 ADL 0.25 NA objective (Fig. 5 A only).
Electron microscopy:
NEGM cells were spun down at 1,500 RCF for 90 s, and the cell pellet was resuspended in Tris buffer ± DMSO or CK-666. Cells were seeded onto aclar pieces (coated with 0.1% poly(ethyleneimine)) in a 24-well plate and allowed to settle for ∼10 min before fixing in 2.5% glutaraldehyde + 100 mM cacodylate buffer (pH 7.2). After 20 min of fixation, cells were washed once and dehydrated with a series of 5-min ethanol washes at the following concentrations: 30%, 50%, 70%, 95%, 95%, 100%, and 100%. Samples were then critical point dried, and aclar pieces were mounted onto stubs using carbon tape and sputter coated with 10 nm platinum. Samples were imaged on a Zeiss Supra 40VP scanning electron microscope.
Flow cytometry
Actin polymer content
Our protocol for measuring F-actin by flow cytometry (Kakley et al., 2018) was adapted for Naegleria. NEGM cells were treated with inhibitors for 15 min, spun down, and resuspended in flow cytometry fixative for 10 min. Cells were spun again, and pellets were resuspended in 50 μl PBS before transfer to flow cytometry tubes with permeabilization buffer and phalloidin. An extra ∼1 ml of PBS was added before flow cytometry. 20,000 cells were analyzed for each sample using a Fortessa flow cytometer controlled by FACS DIVA software. Analysis was completed using FlowJo.
Phagocytosis assays
Four T75 tissue culture flasks of ∼60% confluent NEGM cells were transferred into Tris buffer 1 h before the experiment. Cells were then pooled and aliquoted into 125-ml Erlenmeyer flasks (10 ml per flask) preloaded with 5 ml of Tris mixed with inhibitors or controls at 3× the final concentration (so 1× concentration would be reached in the final volume of 15 ml). Then, E. coli induced to express superfolded-GFP (protocol above) were spun down at 3,000 RCF for 4 min, and the pellet was resuspended in an equal volume of Tris to the starting culture. 750 μl of the bacterial suspension was added to each flask. Flasks were incubated while shaking at 28°C for 45 min. Cultures were then transferred to 15-ml conicals, spun at 1,500 RCF/3 min, resuspended in 2 ml Tris buffer, and spun again, and pellets were resuspended in 1 ml flow cytometry fixative and transferred to a 1.5-ml tube. After 15 min of fixing, cells were spun at 1,500 RCF/2 min, and pellets were resuspended in 1 ml PBS and transferred to flow cytometry tubes. 30,000 cells were analyzed for each sample using a Fortessa flow cytometer controlled by FACS DIVA software. Analysis was completed using FlowJo.
Data analysis and statistics
Sample sizes were selected based on balancing experimental robustness with practicality. The samples in Fig. 3, D–H and,Fig. S3 I were blinded before imaging and were only unblinded after analyses on that experimental replicate were completed. Motility data were blinded after the data were acquired but before analysis. Flow cytometry samples were blinded before data acquisition, with the exception of phalloidin-stained and unstained controls (Fig. S3 B), which were always run before the labor-intensive analysis of the remaining controls and samples. All graphs were generated using GraphPad Prism software (version 8.2). Whenever applicable, SuperPlots (Lord et al., 2020) were employed to show data on each individual cell (smaller gray symbols), while also displaying averages (or medians, as in Fig. 5 C, where the underlying data were nonparametric) from each experimental replicate (larger colorful symbols). The experimental replicates were used to determine the mean, SD, and statistical significance (with a one-way ordinary ANOVA and Tukey’s multiple comparison test). We used ordinary ANOVAs because they are a straightforward method to determine significance with a low chance of false positives (Lord et al., 2020) and because cell-to-cell variability was higher than the run-to-run variation. The data distribution of the mean or median values used in determining statistical significance was assumed to be normal, but this was not formally tested. Tukey’s multiple comparison post hoc test was used to allow comparisons between all appropriate samples (e.g., comparing CK-666 with CK-689, DMSO, and untreated populations).
Online supplemental material
Fig. S1 shows the data used to determine the differences in transcript abundance in Fig. 1, B and C. Fig. S2 identifies homologues of actin nucleation factors in N. gruberi and N. fowleri and shows additional data pertaining to Fig. 1 D (on transcript abundance in amoebae vs. flagellates). Fig. S3 contains additional flow cytometry and microscopy data that relate to Fig. 3, including additional actin drugs. Fig. S4 shows additional SEM images of control amoebae, as well as CK-666–treated cells. Fig. S5 is an MSA of diverse eukaryotic actins, with drug-binding sites noted. Fig. S6 shows percent identities for actins based on Fig. S5, as well as MSAs of Arp2, Arp3, and formin FH2 domains, with an analysis of small-molecule target sites. Fig. S7 shows line scans used to generate the plot in Fig. 3 I. Fig. S8 shows motility data for additional drugs and combinations of drugs. Fig. S9 contains flow cytometry histograms used to quantify phagocytosis data in Fig. 5, as well as an analysis of internal versus external bacteria. Video 1 is the live imaging from one experimental replicate in Fig. 4, showing all drug treatments. Video 2 shows the cell in Fig. 4 G before and during treatment, through fixation and staining. Table S1 lists all reagents and recipes, Table S2 lists N. gruberi and N. fowleri actin cytoskeletal gene accession numbers, and Table S3 lists the accession numbers for the actins, Arps, and formin FH2 domains used to create Figs. S5 and S6. Data S1 contains the N. fowleri protein sequences that match the accession numbers in Table S2, and Data S2 contains the eukaryotic actin sequences used to generate the MSA in Fig. S5.
Acknowledgments
We thank Chandler Fulton for NEGM cells and Katherine Dorfman (University of Massachusetts, Amherst, MA) for superfolder-GFP-expressing E. coli. We thank James Chambers for assistance with SIM (performed at the UMass Light Microscopy Facility and Nikon Center of Excellence). We thank Shane Hussey and Amy Burnside for assistance with flow cytometry (performed at the UMass Flow Cytometry Core Facility). We thank Kasia Hammar and the Marine Biological Laboratory for assistance with EM. We thank Simon Shulman for collecting preliminary data. We thank Sam Lord, Pat Wadsworth, Ken Campellone, Edgar Medina, and Meg Titus for providing helpful feedback on this manuscript.
This work was supported by the National Institutes of Health National Institute of Allergy and Infectious Diseases (grant 1R21AI139363), the Pew Charitable Trusts, and an Excellence in Biomedical Research Award from the Richard and Susan Smith Family Foundation.
The authors declare no competing financial interests.
Author contributions: K.B. Velle was responsible for conceptualization, methodology, investigation, writing—original draft, writing—review and editing, visualization, and resources. L.K. Fritz-Laylin was responsible for conceptualization, methodology, investigation, writing—original draft, writing—review and editing, funding acquisition, resources, and supervision.




![Naegleria species encode an extensive repertoire of actin cytoskeletal regulators.(A)N. gruberi (N.g.) and N. fowleri (N.f.) genomes each encode a similar cellular complement of actin cytoskeletal proteins. The first column shows the name of the protein family or complex, the second includes the name of the mammalian homolog (with the exception of Class II formins, which are often found in plants, and Diaphanous Like Formins [DLF], which encompass all diaphanous-related formins), and the final columns indicate the number of genes found encoding these proteins in N. gruberi and N. fowleri, respectively. See Table S2 for accession numbers and sequences. We note that N. gruberi also possesses five WH2-domain–containing proteins not listed above, and no homolog of WASP Interacting Protein was identified in either Naegleria genome. (B) The graph from Fig. 1 D is shown (with all actins and Joint Genome Institute accession numbers listed) for comparisons of expression data with the proteins listed in A. Each bar represents the average relative change in transcript abundance between amoebae and flagellates from three experiments ± SD.](https://cdn.rupress.org/rup/content_public/journal/jcb/219/11/10.1083_jcb.202007158/4/m_jcb_202007158_figs2.png?Expires=1773750043&Signature=RqxT~4~zdb3h0zbbsUTkYkmO0A4F0imRQwKpYpv0FreF5ESjOp1K3M-WxCYnVh4M55jlLznwMjqoHE4fyn6q64YV6iSy-7XMUgFQMJi3EDcYEyV8LhI13jB8DYsnkoSMGIUyAOpk6kllevH~Zm2gahPV2Fg3AP1~NaB4bwaqtwdirDgjZrHTvXgKX-zbIwQp0SgvIq7ZtnyepPqQK~xoGWOcG-dFet3A0tzN2ZVFBlPlf~AjxbI30ecK5PIIKNMVT5f0zvSj47jCWlvLl75Nywkg0c5R7NcmTOHJJqkMiYorqijd7U64Mn1FjtTpF2Vd67Vrs6ynGGSqoxpy6crEwQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)




![Naegleria actin retains conserved binding sites for small-molecule inhibitors of actin dynamics. A multiple sequence alignment was generated to compare N. gruberi’s (N.g.’s) most highly expressed actin protein sequence with that of other eukaryotic actins (see Table S3 for sequences and ID numbers). This alignment includes actin sequences from other Discobids, N. fowleri (N.f.) and Trypanosoma cruzi (T.c.); Metamonads, Giardia intestinalis (G.i.) and Trichomonas vaginalis (T.v.); Plants, Chlamydomonas reinhardtii (C.r., including NAP1 [N] and IDA5 [I]) and Ectocarpus siliculosus (E.s.); Stramenopiles, Thalassiosira pseudonana (T.p.); Alveolates, Toxoplasma gondii (T.g.), Plasmodium falciparum (P.f., including actin-1 and actin-2), and Tetrahymena thermophila (T.t.); Amoebozoa, Dictyostelium discoideum (D.d.), Entamoeba histolytica (E.h.), and Acanthamoeba castellanii (A.c.); and Opisthokonts, Saccharomyces cerevisiae (S.c.), Schizosaccharomyces pombe (S.p.), and Oryctolagus cuniculus (O.c.). Sequences were aligned using T-Coffee (Notredame et al., 2000) with defaults (Blosum62 matrix, gap open penalty = −50, gap extension penalty = 0) in Jalview, and residues were colored based on conservation (grayscale) or to highlight differences in amino acids within drug-binding sites (ILVAM: salmon; FWY: tangerine; KRH: blue; DE: red; STNQ: lime; PG: raspberry; and C: yellow). Bars indicate important residues for drug binding, and binding sites are abbreviated as follows: L, latrunculin; P, phalloidin; C, cytochalasin; J, jasplakinolide.](https://cdn.rupress.org/rup/content_public/journal/jcb/219/11/10.1083_jcb.202007158/4/m_jcb_202007158_figs5.png?Expires=1773750043&Signature=zkbqgKIFxFKokx0DEVArf6hhtrci5HdbLdghUvF0WBKRMJDzfu5O~j6UbEP0RhkTKGTg4hoMFYieamXzFT7O6JCbKYWeMalHqrttzROX0HsNPwqAaFgQuPOGssv16J0zf2bbKpAkgvC7OSlVUz1Mc14phDiZ7BrPqn-yaPf3bFhULaCl2Kcph5I18l6HLGi0U2yJcjJCrRr-6p8PKURBCI1jqRCwD-bieVTKWpGmjI6w4udAltgPEamOK9qHTG9FqoDsV-5l~Iv3LxxF8wXFMd8SSVj34caSyzkhFYZWNnoaaDHWypRZkTqS9ZXMakHd10EYFzdZYXBzkrzClOYW2g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)