Neuronal migration during development is necessary to form an ordered and functional brain. Postmitotic neurons require microtubules and dynein to move, but the mechanisms by which they contribute to migration are not fully characterized. Using tegmental hindbrain nuclei neurons in zebrafish embryos together with subcellular imaging, optogenetics, and photopharmacology, we show that, in vivo, the centrosome’s position relative to the nucleus is not linked to greatest motility in this cell type. Nevertheless, microtubules, dynein, and kinesin-1 are essential for migration, and we find that interference with endosome formation or the Golgi apparatus impairs migration to a similar extent as disrupting microtubules. In addition, an imbalance in the traffic of the model cargo Cadherin-2 also reduces neuronal migration. These results lead us to propose that microtubules act as cargo carriers to control spatiotemporal protein distribution, which in turn controls motility. This adds crucial insights into the variety of ways that microtubules can support successful neuronal migration in vivo.
Introduction
During brain development, many postmitotic neuronal precursors migrate to organize the brain into regions and layers of distinct populations, so that the correct partners can meet and form a functional network (Hansen et al., 2017). The actin cytoskeleton is crucial for cell motility, as it generates the forces necessary for movement. However, it has been known for some time that several large cell types, such as fibroblasts (Vasiliev et al., 1970) and neurons (Tsai et al., 2007), additionally require microtubules (MTs) for migration, but the mechanisms behind this are not well characterized.
Many migrating neurons exhibit a highly polarized morphology with very long, thin extensions, making the nucleus the greatest obstacle to forward motion (Calero-Cuenca et al., 2018). Pioneering work by Tsai et al. showed that MTs and dynein are required for neuronal migration, and they developed a model in which tip-localized dynein generates pulling forces to position the centrosome in front of the nucleus, while the nucleus is moved toward the leading centrosome by dynein (nucleokinesis; Tsai et al., 2007; Tsai and Gleeson, 2005). There is evidence for this mechanism from rodent studies, specifically from cortical neurons involved in glia-guided radial migration, in which the centrosome clearly precedes the nucleus (Sakakibara et al., 2014; Solecki et al., 2004; Tanaka et al., 2004). However, a number of studies on other neuronal cell types migrating radially and/or tangentially could not confirm that a spatial arrangement of the organelles correlates with forward motion. For example, in cerebellar granule cells in rodents, which can move both radially and tangentially, stabilized MTs are crucial for motility, but the spatial connection between the centrosome and the nucleus seems flexible (Trivedi et al., 2017; Umeshima et al., 2007; Wu et al., 2018). In tangentially migrating zebrafish cerebellar cells, the centrosome can lead the nucleus but can also be overtaken by it, again emphasizing that the arrangement of organelles may not be essential for nucleokinesis and the migration of all neurons (Distel et al., 2010). Retinal ganglion cells exhibit radial migration, but in zebrafish, they are able to complete their migration with a rear-positioned centrosome, and even in the absence of MTs (Icha et al., 2016). This has led to several additional ideas how MTs influence neuronal migration. Among others, it has been suggested that MTs could be anchored by the cells’ actin cortex or sites of adhesion in order to transmit forces to the nucleus or that MTs control the generation of actin-based forces, that they allow nuclear rotation to facilitate efficient nuclear transport, or that MTs are only necessary to fine-tune overall motility or to change the migration type (Hutchins and Wray, 2014; Icha et al., 2016; Jiang et al., 2015; Trivedi et al., 2017; Wu et al., 2018). Neither of these mechanisms address the established role of MTs as cargo-delivery operator in the context of neuronal migration.
Migrating cells need to respond to various extracellular stimuli and to coordinate their response to these cues (e.g., local actin polymerization or cellular adhesion) at sites that can be a hundred microns apart to ensure efficient migration. The dynamic MT array is able to quickly redirect cargo to different parts of the cell, and this property of MTs is thought to coordinate directional changes in large nonneuron cell types and in the axonal growth cone (Dent et al., 2011; Schmoranzer et al., 2003). Targeted cargo delivery is essential for many adhesion proteins. Cadherins are crucial molecules during embryonal development from invertebrates to mammals (Halbleib and Nelson, 2006), as they mediate cell–cell adhesion and participate in cellular signaling (Klezovitch and Vasioukhin, 2015). This makes their subcellular concentration an important factor in emerging cell shape, tissue coherence, and collective cell behavior (Hoffman and Yap, 2015). As transmembrane proteins, cadherins require vesicular transport between different intracellular compartments and the plasma membrane (PM). In mammals, it is performed by MTs and their motors over long distances (Heisler et al., 2014; Mary et al., 2002).
Here, we use tangentially migrating tegmental hindbrain nuclei neurons (THNs) of zebrafish as a model to investigate the contribution of MTs to the migration of neuronal precursors in vivo in the context of a cargo-delivery mechanism. THNs emigrate from their primordial zone, the upper rhombic lip (URL), toward the midbrain–hindbrain boundary (MHB), which they then follow to form clusters at the ventral end of the MHB (Köster and Fraser, 2001; Volkmann et al., 2010). These later develop into the fish homologues of the mammalian parabrachial, parabigeminal, and laterodorsal-pedunculopontine THN (Volkmann et al., 2010). As a model cargo for MT-based transport, we investigate Cadherin-2. Cadherin-2 in zebrafish is highly expressed in neurons, and it is required for the migration of precursors (Lele et al., 2002). Zebrafish THNs move in “steps” of greater velocity, followed by a resting phase, and an accumulation of Cadherin-2 is indicative of forward motion (Rieger et al., 2009). The presence of the repeated cycles of forward movement and resting phases was interpreted as indication that THNs move by MT-mediated nucleokinesis, but the flexible position of the centrosome and the nucleus during forward motion (Distel et al., 2010) left open questions about the significance of MTs and motor proteins for the migration of this neuronal cell type.
By combining THN-specific expression of markers, MT-modifying enzymes and motors, optogenetics, photopharmacological drugs, and quantitative analysis of intracellular traffic, it emerges that in THNs, MTs, and their motors (1) are necessary for migration; (2) may link the centrosome to the nucleus, but this does not correlate with greatest cell motility; (3) are responsible for the long-distance intracellular transport of cargoes such as Cadherin-2; and that (4) imbalanced MT-motor transport reduces motility. These results expand our view about how MTs support neuronal migration in an in vivo setting and reflect the versatility of MTs as a means of regulating a complex process such as cell migration.
Results
MT organization in THNs changes along the migratory route
THNs in zebrafish arise at the URL between 24 h postfertilization (hpf) and 48 hpf (Köster and Fraser, 2001; Volkmann et al., 2010). They emigrate from the URL in a first phase through the cerebellar primordium with bipolar morphology to reach the MHB, which they follow ventrally in a second migratory phase during which they assume a unipolar morphology.
To study the organization of MTs in THNs, we transiently introduced EB3-GFP in a stable transgenic zebrafish line expressing Gal4 from an atonal 1a(atoh1a) promoter, which marks THNs in the developing cerebellum (Fig. 1, A and B; Distel et al., 2010). We first confirmed that upstream activating sequence (UAS)–driven overexpression of the plus-end–binding protein does not affect overall THN migration by observing THN morphology and route (Fig. 1, B and C; and Video 1). THNs in phase 1 exhibited the typical bipolar morphology and followed the normal route. Likewise, THNs in phase 2 possessed a normal unipolar morphology with a long leading process and the nucleus positioned at the rear and continued migrating along the MHB.
MTs reorganize during THN migration. (A) 30-hpf embryo expressing GFP in the atoh1a-Gal4/UAS-system. Anatomical markers are shown. Scale bar, 100 µm. (B) THNs exhibit a bipolar morphology in phase 1 with a long extension contacting the URL (magenta arrow) and a unipolar morphology with a leading process (orange arrow) in phase 2. Dots indicate the nuclei. Trans-light image serves as tissue reference; cb is cerebellum. Scale bar, 10 µm. See also Video 1. (C) Left: Magnified from the magenta region indicated in B, turned by 90°. A phase 1 THN with bipolar morphology positioned between the URL and MHB with EB3-GFP signals emanating from the URL. Right: Magnified from the orange region indicated in B. A phase 2 THN, where EB3 signals originate near the nucleus. In the leading process, individual EB3-GFP comets are difficult to distinguish. Scale bar, 5 µm. (D–F) Cartoon images represent the morphology and tissue position of THNs migrating in different phases; arrow indicates the migration direction. Images of EB3 tracks represent examples from films of ∼2 min (phase 1), ∼17 min (intermediate phase), and ∼30 s (phase 2) duration from one (intermediate, phase 2) or several cells (phase 1). Scale bars, 5 µm. (D) In phase 1, MTs emanate from the distal rear of the THN, and EB3 tracks are mostly unipolar. (E) In intermediate phase, MTs extend in all directions. (F) In phase 2, MTs appear unorganized in the soma, and ∼25% of plus ends are directed into the leading process. (G) MT reorganization correlates with the position of the centrosome at the rear in phase 1 THNs. When the nucleus has reached the MHB, the centrosome moves toward the nucleus together with the retracting rear. Arrows indicate individual examples. Note that the images have been turned by 90°. Scale bar, 10 µm. (H) The centrosome occupies an unstable position near the nucleus in phase 2. Arrows indicate the centrosome. Scale bar, 10 µm. All elapsed time is given in minutes. Where tissue boundaries are indicated, the MHB is marked as a solid line and the URL as a dotted line.
MTs reorganize during THN migration. (A) 30-hpf embryo expressing GFP in the atoh1a-Gal4/UAS-system. Anatomical markers are shown. Scale bar, 100 µm. (B) THNs exhibit a bipolar morphology in phase 1 with a long extension contacting the URL (magenta arrow) and a unipolar morphology with a leading process (orange arrow) in phase 2. Dots indicate the nuclei. Trans-light image serves as tissue reference; cb is cerebellum. Scale bar, 10 µm. See also Video 1. (C) Left: Magnified from the magenta region indicated in B, turned by 90°. A phase 1 THN with bipolar morphology positioned between the URL and MHB with EB3-GFP signals emanating from the URL. Right: Magnified from the orange region indicated in B. A phase 2 THN, where EB3 signals originate near the nucleus. In the leading process, individual EB3-GFP comets are difficult to distinguish. Scale bar, 5 µm. (D–F) Cartoon images represent the morphology and tissue position of THNs migrating in different phases; arrow indicates the migration direction. Images of EB3 tracks represent examples from films of ∼2 min (phase 1), ∼17 min (intermediate phase), and ∼30 s (phase 2) duration from one (intermediate, phase 2) or several cells (phase 1). Scale bars, 5 µm. (D) In phase 1, MTs emanate from the distal rear of the THN, and EB3 tracks are mostly unipolar. (E) In intermediate phase, MTs extend in all directions. (F) In phase 2, MTs appear unorganized in the soma, and ∼25% of plus ends are directed into the leading process. (G) MT reorganization correlates with the position of the centrosome at the rear in phase 1 THNs. When the nucleus has reached the MHB, the centrosome moves toward the nucleus together with the retracting rear. Arrows indicate individual examples. Note that the images have been turned by 90°. Scale bar, 10 µm. (H) The centrosome occupies an unstable position near the nucleus in phase 2. Arrows indicate the centrosome. Scale bar, 10 µm. All elapsed time is given in minutes. Where tissue boundaries are indicated, the MHB is marked as a solid line and the URL as a dotted line.
EB3 and Map4MTB label MTs during THN migration. THNs coexpress H2B-RFP as positional marker in two 34–35-hpf embryos. Expressed markers are labeled in their respective colors. Elapsed time in hours:minutes. Bars: 10 µm. The video is related to Figs. 1 and S1.
MT plus ends in THNs in phase 1 between the URL and the MHB showed unipolar orientation, with the plus ends emanating from the distal rear at the URL and extending toward the MHB (Fig. 1 D). Only at the most distal rear, a small population of short MTs of <3 µm length extending in other directions was observed (11% in the examples given in Fig. 1 D; MT directions are color-coded by their extension toward the front, rear, or sides). At the MHB, some THNs exhibited MT plus ends extending in all directions (Fig. 1 E). Notably, these neurons had not developed a clear leading process, so we referred to these THNs as intermediate phase. In phase 2, MT plus ends in the leading process were oriented toward the leading edge, which was now directed ventrally, but in the soma, the MTs extended outwards from the nuclear region in all directions. In the example in Fig. 1 F, only ∼25% of EB3 signals reached from the nuclear region into the leading process. These observations agree with a previous study which located the centrosome at the distal rear of phase 1 THNs and perinuclear in phase 2 (Distel et al., 2010). Coexpression of Centrin-2 (cetn2), EB3, and Histone-2B (H2B) as nuclear marker confirmed this arrangement (Fig. 1 G). In phase 2, the centrosome is positioned near the nucleus but in an unstable position, shifting from a rear position to a lateral or front position and reverse relative to the nucleus (Fig. 1 H).
Centrosomal shifts occur regularly but do not correlate with THN motility
The original nucleokinesis model (Tsai and Gleeson, 2005) postulates that shifts in centrosome position occur as a consequence of forces pulling on MTs and precede forward motion as forces from the front and dynein at the nucleus organize to transport the organelle forward. Although this model has since been modified by data from additional neuron types and model organisms, a short-lived shift of the centrosome to the front of the nucleus could indicate a buildup of MT-mediated forces, which disperse soon after the nucleus moves (Distel et al., 2010). In this case, a change in relative position of centrosome and nucleus could indicate classic nucleokinesis involving MTs and coincide with greatest forward motion of the nucleus.
To characterize this connection in THNs, we analyzed the centrosome’s position relative to the nucleus and determined its correlation to velocity (Fig. 2 A). In phase 1 THNs, the centrosome is consistently positioned far behind the nucleus; hence, we investigated centrosomal and nuclear positions in phase 2 THNs, where the centrosome is located near the nucleus after the rear has been fully retracted from the ventricular zone. We first recorded THNs coexpressing Centrin-2-YFP and H2B-mRFP for 4–8 h from 31 to 33 hpf on, and we manually tracked the centrosome and the center of the nucleus. We used the nuclear positions to calculate THN speed in 30-min time windows (n = 66 centrosome tracks/10 embryos). Time windows were positioned such that one spanned a front- or rearward shift in the centrosome relative to the nucleus, and one window preceded and another followed this central window for comparison. For control THNs, 45 centrosomal shifts were identified in a total observation time of 6,274 min over all tracks, giving a rate of 7.2 × 10−3 events/min. These included 17 complete profiles with correlated time windows before, during, and after an identified centrosomal shift.
Centrosomal shifts do not correlate with THN migration speed increases. (A) Centrin-2 labeling illustrates the unstable centrosomal position (arrows) in phase 2 THNs relative to the nucleus. Elapsed time in minutes. Scale bar, 10 µm. See also Video 2. (B) Determining THN speed in a 30-min window centered around a centrosomal position shift (labeled “shift”) compared with a time window preceding (“before”) or following (“after”) does not indicate a speed increase correlation with centrosomal position using either Cetn2 or Map4MTB as markers. When THNs were depolarized by ChR2 or ACh, speed increased irrespective of centrosomal position. Note that the speed increase in Map4MTB was not statistically significant (Kruskal–Wallis ANOVA P = 0.126 before/shift, P = 0.075 shift/after for uninduced, P = 0.753 before/shift, P = 0.165 shift/after for ChR2-induced Map4MTB-expressing THNs). n = 10 embryos/66 tracks/45 events for control Cetn2-expressing THNs; n = 11 embryos/87 tracks/53 events for ACh-induced Cetn2-expressing THNs; n = 6 embryos/26 tracks/38 events for control Map4MTB-expressing THNs; n = 7 embryos/40 tracks/23 events for ChR2-induced Map4MTB-expressing THNs. (C) Plotting the data linked for a specific event does not suggest a preference of speed increases around the time of a centrosome shift (red) over other speed profiles (blue) using Cetn2 as marker. n = 10 embryos/66 tracks/17 events. (D) Map4MTB as marker can be used to determine the MTOC position (arrows) relative to the nucleus (dots). Scale bar, 10 µm. (E) No correlation over statistic background was detected in linked THN speed profiles for speed increases around the time of a centrosomal shift using Map4MTB as marker. n = 6 embryos/26 tracks/27 events for control Map4MTB-expressing THNs. Boxes in graphs represent 25–75% of all values and whiskers 1.5 times the quartile. Median is shown as a horizontal bar and mean as a square box.
Centrosomal shifts do not correlate with THN migration speed increases. (A) Centrin-2 labeling illustrates the unstable centrosomal position (arrows) in phase 2 THNs relative to the nucleus. Elapsed time in minutes. Scale bar, 10 µm. See also Video 2. (B) Determining THN speed in a 30-min window centered around a centrosomal position shift (labeled “shift”) compared with a time window preceding (“before”) or following (“after”) does not indicate a speed increase correlation with centrosomal position using either Cetn2 or Map4MTB as markers. When THNs were depolarized by ChR2 or ACh, speed increased irrespective of centrosomal position. Note that the speed increase in Map4MTB was not statistically significant (Kruskal–Wallis ANOVA P = 0.126 before/shift, P = 0.075 shift/after for uninduced, P = 0.753 before/shift, P = 0.165 shift/after for ChR2-induced Map4MTB-expressing THNs). n = 10 embryos/66 tracks/45 events for control Cetn2-expressing THNs; n = 11 embryos/87 tracks/53 events for ACh-induced Cetn2-expressing THNs; n = 6 embryos/26 tracks/38 events for control Map4MTB-expressing THNs; n = 7 embryos/40 tracks/23 events for ChR2-induced Map4MTB-expressing THNs. (C) Plotting the data linked for a specific event does not suggest a preference of speed increases around the time of a centrosome shift (red) over other speed profiles (blue) using Cetn2 as marker. n = 10 embryos/66 tracks/17 events. (D) Map4MTB as marker can be used to determine the MTOC position (arrows) relative to the nucleus (dots). Scale bar, 10 µm. (E) No correlation over statistic background was detected in linked THN speed profiles for speed increases around the time of a centrosomal shift using Map4MTB as marker. n = 6 embryos/26 tracks/27 events for control Map4MTB-expressing THNs. Boxes in graphs represent 25–75% of all values and whiskers 1.5 times the quartile. Median is shown as a horizontal bar and mean as a square box.
When plotting nuclear, and thus THN, velocities for each type of time window, ignoring individual track linkage, the velocity around the time of a centrosomal shift event was not increased over other stages of migration, and statistical analysis did not reveal a significant difference (Fig. 2 B). To see if a weak correlation between centrosomal shift and nuclear migration speed exists in THNs, or whether such a correlation is present in only a subset of tracks, we examined the 17 linked sequences of time windows (Fig. 2 C). In this analysis, only one third of all events exhibited the greatest nuclear velocity around the time of a centrosome shift event, which is expected for a random distribution. Together, these results did not point toward a crucial coupling between centrosomal and nuclear position during normal THN migration.
Next, we extended this analysis to THNs in which motility was enhanced to test if the requirement for greater velocity could induce organelle coupling. We expected that an externally induced increase in cellular migration speed would correlate with either an increase in nuclear movement velocity specifically at the time of a centrosomal shift or an increase in the frequency of observed centrosomal shifts, if centrosome to nucleus positioning was closely linked to the generation of motility.
To this end, we bath applied 5.5 mM acetylcholine (ACh) to the embryos, which is a physiological inducer of cell motility (Theisen et al., 2018). This treatment increased migratory velocity in any stage of migration without revealing a statistically significant connection between centrosome to nuclear position coupling (Fig. 2 B), and contrary to expectations, the frequency of observed centrosome shift events decreased to 4.8 × 10−3 events/min (n = 87 centrosome tracks/11 embryos/53 shift windows, 10,963 min total observation time).
These results are consistent in pointing toward MTs controlling migration in THNs by a mechanism that does not involve positioning the centrosome.
Optogenetic stimulation of neuronal motility does not stabilize MT organizing centers (MTOCs) labeled by Map4MTB
The centrosome likely is the major MTOC in THNs, as indicated by the EB3-GFP tracking in Fig. 1, but other MTOCs could be present in the cells, such as Golgi-based MT nucleation. We were concerned that our previous analysis of centrosome positioning would not detect a potentially significant buildup of MT-mediated forces if they were mediated by such noncentrosomal MTs. EB3 can be used to detect MTOCs, but it would require the recording of cells on two different time scales, one on the order of seconds to visualize the origin of EB3 signals and the other on the order of hours, to measure THN migration. Therefore, we created a new marker protein for MT lattices in order to visualize all MTs in THNs. Map4 is a ubiquitously expressed MT-lattice–binding protein (Chapin and Bulinski, 1991). To avoid MT curving and bundling, we fluorescently tagged only the 269-aa C-terminal Map4MTB fragment, which contains all MT-binding domains (Fig. S1 A; Olson et al., 1995). In THNs, Map4MTB-GFP localized to elongated structures in THNs during phase 1 and in the leading processes of THNs in phase 2 (Fig. S1, B and C), as well as mitotic spindles at the URL (Fig. S1 D and,Video 1). Map4MTB structures were capped by EB3 (Fig. S1 E) and converged near the nucleus. This MTOC could be the centrosome and/or a closely positioned Golgi apparatus (Fig. S1 F). To test whether the C-terminal Map4 fragment affects MT growth, we determined growth speeds for EB3-GFP, Map4MTB-mScarlet, or double-labeled MTs in 28–33-hpf embryos in 1-min-long recordings from neurons in the lower rhombic lip, in which individual or small bundles of MTs could be identified more easily. Growing MT ends at the cortex were either tracked in 2D (Fig. S1, G–I) or growth speeds determined from kymographs (Fig. S1, H–J); both methods revealed that double-labeled MTs did not grow faster than Map4MTB-labeled MTs (2D tracking: mean = 0.116 µm/s [EB3], 0.086 µm/s [Map4MTB], and 0.124 µm/s [both]; kymograph: mean = 0.158 µm/s [EB3], 0.096 µm/s [Map4MTB], and 0.097 µm/s [both]). We detected an MT growth speed difference to EB3-GFP–only expressing neurons, which likely stems from tracking MTs in the cytoplasm, where growth is faster than at the cortex, as MTs lose their EB3 cap (Komarova et al., 2002; Stepanova et al., 2003). In sum, Map4MTB-GFP labels MT lattices in THNs without causing obvious MT defects.
Map4MTB labels MTs in zebrafish (related to Fig. 2 ). (A) Schematic representation of full-length Map4 and Map4MTB fragment. (B and C) In THNs, Map4MTB decorates MTs, which extend from the rear of the cells. Colored arrows indicate bundles in phase 2, arrowheads indicate bundles in phase 1 THNs, and dots mark the corresponding nuclei. Box indicates the region magnified in C. Elapsed time in minutes. Scale bars represent 20 µm in B and 10 µm in C. See also Video 1. (D) THNs coexpressing EB3-GFP and Map4MTB-mKate2 show colocalization in a mitotic spindle. Scale bar, 2 µm. (E) Map4MTB-labeled structures are capped with EB3. Box in overview image shows the position of the magnified region on the right. In the image column on the right, arrowheads point to a growing MT tip. Elapsed time in seconds. Scale bars represent 10 µm in the overview and 2 µm in magnified images. (F) MTOC position (arrow) can be inferred by Map4MTB-mediated MT labeling. Scale bar, 2 µm. (G) Examples of MT growth in hindbrain neurons expressing EB3-GFP, Map4MTB-mScarlet, or both. Arrowheads in images on the left indicate MT tips shown in frame montages on the right. Dotted lines indicate growth of MT tips. Elapsed time in seconds. Scale bars represent 5 µm in overview images and 2 µm in montages. (H) Kymographs of the examples shown in G. Horizontal scale bars, 1 µm; vertical bars, 10 s. (I and J) MT plus ends in THNs expressing EB3-GFP appear to grow two times faster than in Map4MTB-mScarlet overexpressing THNs using 2D tracking (I) or kymograph analysis (J). Mean growth speeds per cell are shown. Box represents 25–75% of all values, the whiskers 1.5 times the quartile. Median is shown as horizontal bar, mean as square box. 2D tracking (I): N (EB3) = 12, mean = 0.116 µm/s, N (Map4MTB) = 14, mean = 0.086 µm/s, P = 0.027 to EB3, F = 5.575; N (both) = 12, mean = 0.124 µm/s, P = 0.124, F = 2.299 to Map4MTB, P = 0.774, F = 0.084 to EB3. Kymograph analysis (J): N (EB3) = 12, mean = 0.158 µm/s, N (Map4MTB) = 14, mean = 0.096 µm/s, P = 0.008 to EB3, F = 8.355; N (both) = 12, P = 0.938, F = 0.006 to Map4MTB, mean = 0.097 µm/s, P = 0.008, F 8.577 to EB3. Significance level in one-way ANOVA: *, P < 0.05; **, P < 0.01; ns, not significant.
Map4MTB labels MTs in zebrafish (related to Fig. 2 ). (A) Schematic representation of full-length Map4 and Map4MTB fragment. (B and C) In THNs, Map4MTB decorates MTs, which extend from the rear of the cells. Colored arrows indicate bundles in phase 2, arrowheads indicate bundles in phase 1 THNs, and dots mark the corresponding nuclei. Box indicates the region magnified in C. Elapsed time in minutes. Scale bars represent 20 µm in B and 10 µm in C. See also Video 1. (D) THNs coexpressing EB3-GFP and Map4MTB-mKate2 show colocalization in a mitotic spindle. Scale bar, 2 µm. (E) Map4MTB-labeled structures are capped with EB3. Box in overview image shows the position of the magnified region on the right. In the image column on the right, arrowheads point to a growing MT tip. Elapsed time in seconds. Scale bars represent 10 µm in the overview and 2 µm in magnified images. (F) MTOC position (arrow) can be inferred by Map4MTB-mediated MT labeling. Scale bar, 2 µm. (G) Examples of MT growth in hindbrain neurons expressing EB3-GFP, Map4MTB-mScarlet, or both. Arrowheads in images on the left indicate MT tips shown in frame montages on the right. Dotted lines indicate growth of MT tips. Elapsed time in seconds. Scale bars represent 5 µm in overview images and 2 µm in montages. (H) Kymographs of the examples shown in G. Horizontal scale bars, 1 µm; vertical bars, 10 s. (I and J) MT plus ends in THNs expressing EB3-GFP appear to grow two times faster than in Map4MTB-mScarlet overexpressing THNs using 2D tracking (I) or kymograph analysis (J). Mean growth speeds per cell are shown. Box represents 25–75% of all values, the whiskers 1.5 times the quartile. Median is shown as horizontal bar, mean as square box. 2D tracking (I): N (EB3) = 12, mean = 0.116 µm/s, N (Map4MTB) = 14, mean = 0.086 µm/s, P = 0.027 to EB3, F = 5.575; N (both) = 12, mean = 0.124 µm/s, P = 0.124, F = 2.299 to Map4MTB, P = 0.774, F = 0.084 to EB3. Kymograph analysis (J): N (EB3) = 12, mean = 0.158 µm/s, N (Map4MTB) = 14, mean = 0.096 µm/s, P = 0.008 to EB3, F = 8.355; N (both) = 12, P = 0.938, F = 0.006 to Map4MTB, mean = 0.097 µm/s, P = 0.008, F 8.577 to EB3. Significance level in one-way ANOVA: *, P < 0.05; **, P < 0.01; ns, not significant.
As Map4MTB thus allowed us to identify the MTOCs in THNs, (Fig. 2 D and,Video 2), we next repeated the MTOC/nuclear correlations in normal THNs by recording phase 2 cells for 4–8 h in 34–38-hpf embryos (n = 6; Fig. 2 D). In total, we observed 38 MTOC shift events relative to the nucleus in an observation time of 4,965 min from 26 tracks, or 7.6 × 10−3 events/min, which closely matches the data from Centrin-2–labeled THNs. Migratory speeds, as determined by nucleus movements around the time of a MTOC shift, were increased when comparing data from all time windows, but again, this was not statistically significant. Plotting track-linked sequences of THN migration speeds before, around, or after the MTOC shift showed that in approximately half of the tracks with full profiles, THN speed increased around an MTOC shift (Fig. 2 E; n = 27 centrosome shift events with full profile). In Centrin-2–labeled THNs, this fraction was only one third (see above), which could hint at a role for Golgi-derived MTs in generating THN motility, but this notion awaits further investigation.
The centrosome changes position relative to the nucleus regularly in control THNs but remains more stable when THN speed is increased by ACh. Left: Control THNs expressing Cetn2-YFP (green) and H2B-mRFP (red). Right: THNs coexpressing the same markers, but with 5.5 mM ACh in bath application. Elapsed time in hours:minutes. Scale bar, 5 µm. The video is related to Fig. 2.
The centrosome changes position relative to the nucleus regularly in control THNs but remains more stable when THN speed is increased by ACh. Left: Control THNs expressing Cetn2-YFP (green) and H2B-mRFP (red). Right: THNs coexpressing the same markers, but with 5.5 mM ACh in bath application. Elapsed time in hours:minutes. Scale bar, 5 µm. The video is related to Fig. 2.
Finally, we used Map4MTB-GFP to track MTOC/nucleus positions when THN motility was induced by activation via the channelrhodopsin ChR2, which is able to efficiently stimulate THN migration in a cell-autonomous way when genetically targeted to THNs (Theisen et al., 2018). The optogenetic activation at 1-min intervals increased velocities across all time windows irrespective of MTOC shift, similar to ACh treatment, and again it did not reveal a statistically significant link between the MTOC and nuclear positions (Fig. 2 B). Instead, the number of MTOC shift events decreased strongly to 23 events in a total observation time of 6,000 min, or 3.8 × 10−3 events/min (n = 7 embryos/40 tracks), a result that is similar to the ACh induction data.
Summarizing the results obtained from these independent approaches, it is clear that MTOC shifts relative to the nucleus can occur at the time of greatest nuclear velocity in THN neurons, yet both movements do not correlate in time, and centrosome shifts therefore do not appear to cause these increases. They may even hinder fast motion, as indicated by the drop in events in response to external stimuli. While this does not rule out that the centrosome and the nucleus are coupled in some still-to-be-characterized form, it implies that the generation of motility in THNs does not rely on localizing the centrosome.
Colchicine treatment affects THN morphology and blocks migration
These findings raise the fundamental question whether MTs are necessary for THNs to migrate in vivo, especially as some neurons in zebrafish are able to use alternative modes of migration in the absence of MTs (Icha et al., 2016). To address this question, we reduced MTs in THNs in three different ways.
First, we disassembled MTs by colchicine and monitored THN movement. Colchicine is readily water soluble and thereby suitable for use in fish, even though zebrafish embryos <5 d postfertilization (dpf) require high doses (Brox et al., 2016; Roche et al., 1994). Accordingly, we initially used 5 mg/ml colchicine on 29-hpf embryos expressing GFP in THNs. After 5 h of exposure to the drug, severe effects were visible: embryos began to develop curved spines (Fig. S2, A and B), and rounded cells were observed at the URL (Fig. S2 C). Although THNs retained their ability to form small, short-lived protrusions (Fig. S2 D), no clustering of THNs at the ventral end of the MHB occurred after 8 h of exposure, similar to a block of actomyosin and thereby migration imposed by 15 µg/ml (−)-blebbistatin (Fig. S2 C). The absence of THN clusters therefore indicated that MTs are important for migration, but the side effects at this high concentration of colchicine did not allow a more detailed analysis. Therefore, we tested lower concentrations of colchicine for their ability to reduce MTs and their effect on THN migration. 1 mg/ml colchicine led to mitotically arrested cells at 8–9 h posttreatment (hpt), with some MTs remaining (Fig. S2 E). At 13 hpt, tissue disintegration began; hence, we determined THN speed in the 9–13-hpt window. We found that the loss of MTs strongly inhibited THN migration. To see if this was dose dependent, we next tested 0.5 mg/ml colchicine. The reduction of migratory velocity was also significant, if weaker, when 0.5 mg/ml colchicine was used (Fig. S2 F; control: median = 4.10 µm/h, n = 145 cells; 1 mg/ml: median = 2.17 µm/h, n = 92 cells, P = 4.58 × 10−8; 0.5 mg/ml: median = 2.97 µm/h, n = 54 cells, P = 0.046).
Colchicine and blebbistatin inhibit THN migration (related to Fig. 3 ). (A) Normal development of 29-hpf embryos. Scale bar, 200 µm. (B) The application of 5 mg/ml colchicine at 29 hpf leads to spine curvature after 5 h. Scale bar, 200 µm. (C) Colchicine induces cell rounding, first apparent in mitotic cells at the URL (arrows). Later, all cells begin to round and the tissue collapses without the formation of THN clusters at the ventral end of the MHB (13 hpt). THN cluster formation is also absent in blebbistatin-treated embryos, although cells with two nuclei (arrowheads) increase over time. Scale bar, 20 µm. (D) THNs treated with colchicine are able to form small, transient protrusions. Colors indicate two examples. Dots label the cell centers, arrowheads point toward protrusions. Elapsed time in minutes. Scale bar, 5 µm. (E) Lower concentrations of colchicine delay the onset of complete MT depolymerization, as indicated by the presence of Map4MTB-positive structures in many THNs after 9 hpt. At 13 hpt, most of these structures have been lost and extensive cell rounding with tissue collapse is apparent. Scale bar, 20 µm. (F) Tracking THNs migrating in the 9–13-hpt window of exposure to lower concentrations of colchicine reveals that incomplete loss of MTs already reduces THN motility. n = 24 embryos/145 tracks for control, n = 4 embryos/92 tracks for 1 mg/ml colchicine, n = 4 embryos/54 tracks for 0.5 mg/ml colchicine; P = 4.58 × 10−8 control/1 mg/ml colchicine, and P = 0.046 control/0.5 mg/ml colchicine. MHB, solid line; URL, dotted line. Boxes in graphs represent 25–75% of all values and whiskers 1.5 times the quartile. Median is shown as a horizontal bar and mean as a square box. Significance level in Kruskal–Wallis ANOVA: *, P < 0.05; ***, P < 0.001.
Colchicine and blebbistatin inhibit THN migration (related to Fig. 3 ). (A) Normal development of 29-hpf embryos. Scale bar, 200 µm. (B) The application of 5 mg/ml colchicine at 29 hpf leads to spine curvature after 5 h. Scale bar, 200 µm. (C) Colchicine induces cell rounding, first apparent in mitotic cells at the URL (arrows). Later, all cells begin to round and the tissue collapses without the formation of THN clusters at the ventral end of the MHB (13 hpt). THN cluster formation is also absent in blebbistatin-treated embryos, although cells with two nuclei (arrowheads) increase over time. Scale bar, 20 µm. (D) THNs treated with colchicine are able to form small, transient protrusions. Colors indicate two examples. Dots label the cell centers, arrowheads point toward protrusions. Elapsed time in minutes. Scale bar, 5 µm. (E) Lower concentrations of colchicine delay the onset of complete MT depolymerization, as indicated by the presence of Map4MTB-positive structures in many THNs after 9 hpt. At 13 hpt, most of these structures have been lost and extensive cell rounding with tissue collapse is apparent. Scale bar, 20 µm. (F) Tracking THNs migrating in the 9–13-hpt window of exposure to lower concentrations of colchicine reveals that incomplete loss of MTs already reduces THN motility. n = 24 embryos/145 tracks for control, n = 4 embryos/92 tracks for 1 mg/ml colchicine, n = 4 embryos/54 tracks for 0.5 mg/ml colchicine; P = 4.58 × 10−8 control/1 mg/ml colchicine, and P = 0.046 control/0.5 mg/ml colchicine. MHB, solid line; URL, dotted line. Boxes in graphs represent 25–75% of all values and whiskers 1.5 times the quartile. Median is shown as a horizontal bar and mean as a square box. Significance level in Kruskal–Wallis ANOVA: *, P < 0.05; ***, P < 0.001.
These experiments suggest that MTs are necessary for the elongated THN morphology as well as migration.
Photopharmacological reduction of MTs separates their functions in THN morphology and migration
To support the colchicine data and gain regional control over MT reduction, we used the photopharmaceutical analogues of colchicine, photostatins (PSTs). PSTs are light-controllable MT inhibitors that exist in an inactive state; they can bind free tubulin only when they are regularly activated by UV illumination, and they revert to their inactive state either under laser illumination at 514–561 nm or with a half-life on the scale of minutes in the dark (Borowiak et al., 2015). At 26–27 hpf, we incubated embryos expressing Map4MTB-GFP and PM-targeted tagRFP-T for 3–4 h in 25 µg/ml PST-1P in the dark. After transfer to the confocal microscope and continuous exposure to 561-nm illumination for 30 min, we tested the efficiency of PST-1P to reduce MTs in the cerebellum. We UV activated the drug at 3-min intervals, and EB3-GFP comets mostly disappeared within 15 min (Fig. 3 A). We did not completely disassemble all MTs in this way, as some Map4MTB-labeled structures persisted (arrows in Fig. 3 A). The nature of these MTs is unknown, but a PST-1P–resistant population has been observed previously (Borowiak et al., 2015; Zenker et al., 2017). Notably, THNs with residual MTs were able to maintain the polarized morphology. Next, regions of interest (ROIs) were placed so that one ROI covered part of the MHB, which was illuminated every 3 min with UV light to activate PST-1P (PST ON), while surrounding ROIs were illuminated by red light to inactivate the drug and record THNs for control (PST OFF; Fig. 3, B and C; region outlined in blue is activated, and region outline in red is control). Many THNs in the activated region showed a largely normal morphology in contrast to the colchicine experiments (Fig. 3 D). Next, we measured the speed of the THNs migrating under MT-depleted and red-light control conditions (Fig. 3 E; n = 63 tracks/8 embryos for UV activated, n = 39 tracks/7 embryos for red-light inactive PST-1P; see Table S1 for statistics on all migration velocities). THNs with fewer MTs moved at only one third of the speed of the red-light illuminated controls (median speed: 2.66 µm/h in activated, 7.42 µm/h in inactivated PST-1P, P = 5.37 × 10−10), comparable to THNs treated with 1 mg/ml colchicine (Fig. S2 F).
Reduction of MTs leads to THN migration decrease. (A) After 10 rounds of PST-1P activation every 3 min, most EB3-GFP signals are lost and do not reappear in subsequent cycles. In contrast, Map4MTB-GFP signal appears weakened, but not completely lost (arrows) after 1 h of PST-1P activation. Elapsed time in minutes. Scale bar, 20 µm. (B) An example of a PST-1P-treated 34-hpf embryo with the activation region outlined in blue and control regions in red. Orange box indicates region magnified in B. (C) Map4MTB-GFP shows that MTs in the activated region are reduced, but some resist depolymerization. Scale bar, 5 µm. (D) Stills from B showing that THNs within the activated region (top row) are impaired in their migration, while control cells (bottom row) are able to move. Dots follow individual examples, and progress is indicated on the right; arrows indicate start position and arrowheads the end point after 3 h. Scale bar, 20 µm. See also Video 3. (E) THNs with reduced MTs generate half the speed of THNs recorded with the same illumination setting without the drug, and one third of the speed of THNs migrating in the control regions of PST-1P-treated embryos. n = 8 embryos/63 tracks for PST-1P activated embryos, n = 7 embryos/39 tracks for PST-1P nonactivated embryos, n = 6 embryos/62 tracks for UV-illuminated embryos; P = 2.59 × 10−6 PST-1P activated/UV only, 5.37 × 10−10 PST-1P activated/nonactivated. (F) Overexpression of Katanin P60 impairs THN migration when combined with aTat1-mediated MT acetylation. n = 17 embryos/103 tracks for Map4MTB + Katanin P60-expressing embryos, n = 20 embryos/101 tracks for aTat1 + Katanin P60-expressing embryos; P = 2.37 × 10−8. (G) THNs tend to escape coexpression of Katanin P60, Map4MTB, and aTat1. Arrows indicate examples of coexpression in phase 2 THNs. Note that these THNs appear to have formed very short leading processes. Scale bar, 20 µm. (H) In examples of embryos coexpressing Katanin P60 with either Map4MTB or aTat1, coexpression is low in aTat1 and Katanin P60-expressing embryos, and significant debris is visible in the tissue. Dots indicate several examples, with orange following a Katanin P60-only expressing THN. Cyan boxes indicate magnified regions. Scale bars, 20 µm. Elapsed time in min. MHB, solid line; URL, dotted line. Boxes in graphs represent 25–75% of all values and whiskers 1.5 times the quartile. Median is shown as a horizontal bar and mean as a square box. Significance level in Kruskal–Wallis ANOVA. ***, P < 0.001.
Reduction of MTs leads to THN migration decrease. (A) After 10 rounds of PST-1P activation every 3 min, most EB3-GFP signals are lost and do not reappear in subsequent cycles. In contrast, Map4MTB-GFP signal appears weakened, but not completely lost (arrows) after 1 h of PST-1P activation. Elapsed time in minutes. Scale bar, 20 µm. (B) An example of a PST-1P-treated 34-hpf embryo with the activation region outlined in blue and control regions in red. Orange box indicates region magnified in B. (C) Map4MTB-GFP shows that MTs in the activated region are reduced, but some resist depolymerization. Scale bar, 5 µm. (D) Stills from B showing that THNs within the activated region (top row) are impaired in their migration, while control cells (bottom row) are able to move. Dots follow individual examples, and progress is indicated on the right; arrows indicate start position and arrowheads the end point after 3 h. Scale bar, 20 µm. See also Video 3. (E) THNs with reduced MTs generate half the speed of THNs recorded with the same illumination setting without the drug, and one third of the speed of THNs migrating in the control regions of PST-1P-treated embryos. n = 8 embryos/63 tracks for PST-1P activated embryos, n = 7 embryos/39 tracks for PST-1P nonactivated embryos, n = 6 embryos/62 tracks for UV-illuminated embryos; P = 2.59 × 10−6 PST-1P activated/UV only, 5.37 × 10−10 PST-1P activated/nonactivated. (F) Overexpression of Katanin P60 impairs THN migration when combined with aTat1-mediated MT acetylation. n = 17 embryos/103 tracks for Map4MTB + Katanin P60-expressing embryos, n = 20 embryos/101 tracks for aTat1 + Katanin P60-expressing embryos; P = 2.37 × 10−8. (G) THNs tend to escape coexpression of Katanin P60, Map4MTB, and aTat1. Arrows indicate examples of coexpression in phase 2 THNs. Note that these THNs appear to have formed very short leading processes. Scale bar, 20 µm. (H) In examples of embryos coexpressing Katanin P60 with either Map4MTB or aTat1, coexpression is low in aTat1 and Katanin P60-expressing embryos, and significant debris is visible in the tissue. Dots indicate several examples, with orange following a Katanin P60-only expressing THN. Cyan boxes indicate magnified regions. Scale bars, 20 µm. Elapsed time in min. MHB, solid line; URL, dotted line. Boxes in graphs represent 25–75% of all values and whiskers 1.5 times the quartile. Median is shown as a horizontal bar and mean as a square box. Significance level in Kruskal–Wallis ANOVA. ***, P < 0.001.
THN speeds naturally decrease along the MHB (Theisen et al., 2018). To rule out that the PST-1P results were the effect of a sampling bias introduced by placing ROIs in the central region of the MHB, we repeated the experiment with the same imaging and ROI setup but without PST-1P for control. These THNs moved at normal velocities (Fig. 3 E and,Video 3; median: 5.14 µm/h, n = 62 tracks/6 embryos, P = 2.59 × 10−6, compare also to control values from other experiments in this article).
25 µM PST-1P reduces MTs in the activated region (green), leading to a decrease in THN migratory speed. THNs migrating under control conditions without UV illumination are visible in the red regions. Colored dots indicate individual examples. THNs coexpressed Map4MTB (green) and a PM marker (red), but due to the different illumination settings to activate PST-1P, only one of the markers may be seen in the activated or control region. Elapsed time in hours:minutes. Scale bar, 10 µm. The video is related to Fig. 3.
25 µM PST-1P reduces MTs in the activated region (green), leading to a decrease in THN migratory speed. THNs migrating under control conditions without UV illumination are visible in the red regions. Colored dots indicate individual examples. THNs coexpressed Map4MTB (green) and a PM marker (red), but due to the different illumination settings to activate PST-1P, only one of the markers may be seen in the activated or control region. Elapsed time in hours:minutes. Scale bar, 10 µm. The video is related to Fig. 3.
In total, this tissue-limited depletion of MT function demonstrates that MTs are crucial for efficient THN migration and that a large number of MTs is required for motion, which appears surprising given that a small population of MTs are able to maintain normal THN morphology. This finding further argues for the ability of MTs to act in different processes simultaneously, such as neuronal progenitor polarity and migration, which can be separated experimentally.
Induced MT severing impairs THN motility
To support these findings, which relied on chemical compounds, we reduced the number of MTs in THNs by genetic means. The catalytic P60 subunit of Katanin severs acetylated MTs (Sudo and Baas, 2010; Toyo-Oka et al., 2005). The overexpression of Katanin P60 together with Map4MTB did not reduce MT numbers or THN speed (median = 4.99 µm/h and n = 103 tracks/17 embryos; Fig. 3, F and G; and Fig. S3 A). As we noticed an overall low level of MT acetylation in THNs (Fig. S3, A and B), which could limit the number of MTs accessible for severing, we increased MT acetylation by coexpressing aTat1 (Fig. S3, A and B; Akella et al., 2010). This resulted in a mosaic distribution of THNs expressing fluorescently tagged Map4MTB, aTat1, and Katanin simultaneously at high levels, as seen by the presence of mostly two fluorescent signals in the cells (Fig. 3 G). Strongly expressing cells typically displayed rounded morphology at the MHB, ultimately ending as debris in the cerebellum at 32–34 hpf. This argues that the efficient removal of MTs in THNs is lethal to the coexpressing cells, similar to the colchicine results (Fig. 3 H, arrows). Tracking of the low coexpressing THNs revealed that these cells migrated at half the speed of cells expressing only Map4MTB and Katanin P60 (median: 2.58 µm/h; n = 101 tracks/20 embryos; Fig. 3, F and G). This is a similar reduction in migratory speed as seen in the PST-1P experiment or 1 mg/ml colchicine treatment, which likewise only reduced, but not completely depolymerized, MTs.
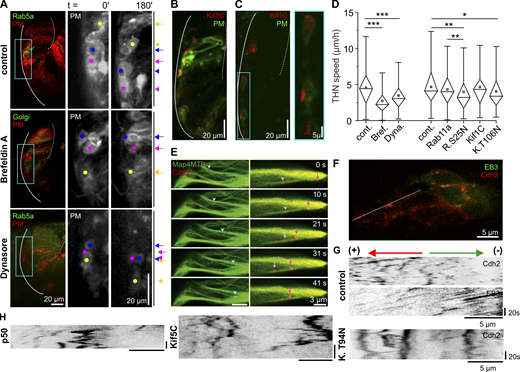
Targeting MT motors or vesicle formation affects THN migration (related to Figs. 4 and 5 ). (A) Endogenous MT acetylation is low in 29-hpf embryos but can strongly be increased by overexpression of aTat1. Scale bar, 10 µm. (B) Quantification of MT acetylation in citrine- or aTat1-expressing THNs. Percentage of MT acetylation–positive cells out of all THNs expressing the genetic marker per embryo is shown. n = 7 embryos (citrine), n = 9 embryos (aTat1). (C) When dynein is inactivated, leading processes are still able to grow along the MHB. Box indicates magnified region on the right. Scale bars, 20 µm. (D) The overexpression of a dominant-negative fragment of Lis1a decreases THN motility. Arrows and arrowheads indicate the start and end points on the right. Scale bars, 20 µm. (E) The reduction in THN motility after quantification is less pronounced than in dynein inactivation by p50. n = 24 embryos/145 tracks for control, n = 14 embryos/191 tracks for Lis1N-expressing embryos, P = 0.027. (F) Brefeldin A treatment disassembles the Golgi apparatus very efficiently within 2 h, while Dynasore blocks endosome formation immediately after injection (G). Blue boxes indicate regions magnified on the right. Scale bars represent 20 µm in the overview and 10 µm in the magnified region. (H) Overexpression of wild-type kinesin-3 Kif1C does not impair THN migration, but the rigor T106N mutation reduces motility. Scale bars, 20 µm. (I) Similarly, the overexpression of WT Rab11a does not interfere with THN motility, but the dominant-negative S25N mutation reduces it. Scale bars, 20 µm. Elapsed time in minutes is given at the top of the respective images. MHB, solid line; URL, dotted line. Colored arrows on the right indicate the start point of an individual THN, and arrowheads of the same color indicate the end point after 180 min. Boxes in graphs represent 25–75% of all values and whiskers 1.5 times the quartile. Median is shown as a horizontal bar and mean as a square box. Significance level in Kruskal–Wallis ANOVA: *, P < 0.05.
Targeting MT motors or vesicle formation affects THN migration (related to Figs. 4 and 5 ). (A) Endogenous MT acetylation is low in 29-hpf embryos but can strongly be increased by overexpression of aTat1. Scale bar, 10 µm. (B) Quantification of MT acetylation in citrine- or aTat1-expressing THNs. Percentage of MT acetylation–positive cells out of all THNs expressing the genetic marker per embryo is shown. n = 7 embryos (citrine), n = 9 embryos (aTat1). (C) When dynein is inactivated, leading processes are still able to grow along the MHB. Box indicates magnified region on the right. Scale bars, 20 µm. (D) The overexpression of a dominant-negative fragment of Lis1a decreases THN motility. Arrows and arrowheads indicate the start and end points on the right. Scale bars, 20 µm. (E) The reduction in THN motility after quantification is less pronounced than in dynein inactivation by p50. n = 24 embryos/145 tracks for control, n = 14 embryos/191 tracks for Lis1N-expressing embryos, P = 0.027. (F) Brefeldin A treatment disassembles the Golgi apparatus very efficiently within 2 h, while Dynasore blocks endosome formation immediately after injection (G). Blue boxes indicate regions magnified on the right. Scale bars represent 20 µm in the overview and 10 µm in the magnified region. (H) Overexpression of wild-type kinesin-3 Kif1C does not impair THN migration, but the rigor T106N mutation reduces motility. Scale bars, 20 µm. (I) Similarly, the overexpression of WT Rab11a does not interfere with THN motility, but the dominant-negative S25N mutation reduces it. Scale bars, 20 µm. Elapsed time in minutes is given at the top of the respective images. MHB, solid line; URL, dotted line. Colored arrows on the right indicate the start point of an individual THN, and arrowheads of the same color indicate the end point after 180 min. Boxes in graphs represent 25–75% of all values and whiskers 1.5 times the quartile. Median is shown as a horizontal bar and mean as a square box. Significance level in Kruskal–Wallis ANOVA: *, P < 0.05.
To summarize, all three independent approaches to MT disruption showed that the complete loss of MTs is lethal for THNs, while a reduction of MTs, although sufficient for survival and overall polarized morphology, impairs active migration. Therefore, different thresholds of MTs are required for MT-dependent homeostasis and migration of neuronal cells. This does not, however, explain how MTs support migration.
The motor proteins dynein and Kif5C are needed for THN migration
As our data suggested that THN motility requires MTs, we next tested the involvement of dynein and kinesin-1 in migration, which has been described for other migrating neuronal cell types (Tsai et al., 2007; Wu et al., 2018). First, we interfered with dynein activation by overexpression of dynamitin/p50, which dissociates the dynactin complex (Melkonian et al., 2007). To avoid adverse effects during early development, we injected the plasmid into the fourth ventricle of 24–28-hpf embryos, which expressed Gal4 from the atoh1a promoter from 17 hpf (Distel et al., 2010), and electroporated the construct into the cerebellar primordium. In THNs, p50 accumulated in punctae after ∼5 h of expression, which were often located near the centrosome at the URL (Fig. 4 A) or perinuclear in phase 2 THNs (Fig. 4 B). As dynein and dynactin often associate with the centrosome (Quintyne and Schroer, 2002), it appears likely that p50 localizes to dynein in THNs. To calculate THN speeds by nucleus tracking, we coexpressed p50 and H2B-mRFP (Fig. 4 C). Under these conditions, p50-expressing THNs migrated at only ∼40% of the speed of controls (median speed: 1.82 µm/h and n = 69 tracks/8 embryos for p50, 4.58 µm/h and n = 85 tracks/7 embryos for control, P = 3.69 × 10−12; Fig. 4 D and,Video 4). Interestingly, and similar to PST-1P–induced MT reduction results, THNs retained their normal morphology upon dynein inactivation, as leading processes continued to grow, while the progress of the somata along the MHB was impaired (Fig. S3 C). To confirm these results, we overexpressed an N-terminal fragment of 87 aa of Lis1a, which has been shown to act as dominant negative to dynein activation (Tai et al., 2002). The effects on THN migratory speeds and morphology were similar yet less pronounced than for p50 overexpression (median speed: 3.63 µm/h and n = 191 tracks/14 embryos, P = 0.027; 4.10 µm/h and n = 145 tracks/24 embryos for control; Fig. S3, D and E). This could be due to a hypomorph function of the dominant-negative fragment, as zebrafish express two homologues of Lis1 in the cerebellar primordium at the MHB at the time of THN migration (Drerup et al., 2010).
MT motor proteins regulate THN speed. (A) Triple-citrine-tagged p50 colocalizes with the centrosome at the URL. Scale bar, 5 µm in overview, 1 µm in the magnified region on the right. Box indicates the magnified region. (B) Similarly, p50 is often found at the origin of Map4MTB-decorated MTs in phase 2 THNs. Scale bars represent 20 µm in the overview and 5 µm in magnified images. Box indicates the magnified region. (C) Blocking either dynein by overexpression of p50 or Kif5C in a rigor mutant (T94N) reduces THN speed. Dots trace individual examples. Cyan boxes indicate the magnified region in the stills. Elapsed time in minutes. Scale bars, 20 µm. See also Video 4. (D) Loss of active dynein approximately halves THN speed, while Kif5C rigor mutant overexpression reduces THN speed by 20–30%. n = 7 embryos/85 tracks for control; n = 8 embryos/69 tracks for p50-expressing embryos; n = 25 embryos/155 tracks for Kif5C-expressing embryos; n = 31 embryos/250 tracks for Kif5C T94N-expressing embryos; P = 3.69 × 10−12 p50/control, P = 0.016 Kif5C/control, P = 1.08 × 10−8 Kif5C/Kif5C T94N, and P = 0.002 control/Kif5C T94N. (E) FISH for kif5c expression in 30-hpf embryos demonstrates that Kif5C is found at the MHB at the time of THN migration. Arrows indicate MHBs and arrowheads URLs. Boxes indicate magnified regions on the right. Scale bars represent 200 µm in the overview and 50 µm in magnified regions on the right. (F) WISH for kif5c confirms the expression of the gene at the MHB at 30 hpf. Top: Antisense. Bottom: Sense for control. Magnified regions on the right are indicated by boxes in overview images. Arrows indicates MHBs and arrowheads URLs. Scale bar represents 100 µm or 50 µm in magnified images. Elapsed time in minutes. MHB, solid line; URL, dotted line. Boxes in graphs represent 25–75% of all values and whiskers 1.5 times the quartile. Median is shown as a horizontal bar and mean as a square box. Significance level in Kruskal–Wallis ANOVA: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
MT motor proteins regulate THN speed. (A) Triple-citrine-tagged p50 colocalizes with the centrosome at the URL. Scale bar, 5 µm in overview, 1 µm in the magnified region on the right. Box indicates the magnified region. (B) Similarly, p50 is often found at the origin of Map4MTB-decorated MTs in phase 2 THNs. Scale bars represent 20 µm in the overview and 5 µm in magnified images. Box indicates the magnified region. (C) Blocking either dynein by overexpression of p50 or Kif5C in a rigor mutant (T94N) reduces THN speed. Dots trace individual examples. Cyan boxes indicate the magnified region in the stills. Elapsed time in minutes. Scale bars, 20 µm. See also Video 4. (D) Loss of active dynein approximately halves THN speed, while Kif5C rigor mutant overexpression reduces THN speed by 20–30%. n = 7 embryos/85 tracks for control; n = 8 embryos/69 tracks for p50-expressing embryos; n = 25 embryos/155 tracks for Kif5C-expressing embryos; n = 31 embryos/250 tracks for Kif5C T94N-expressing embryos; P = 3.69 × 10−12 p50/control, P = 0.016 Kif5C/control, P = 1.08 × 10−8 Kif5C/Kif5C T94N, and P = 0.002 control/Kif5C T94N. (E) FISH for kif5c expression in 30-hpf embryos demonstrates that Kif5C is found at the MHB at the time of THN migration. Arrows indicate MHBs and arrowheads URLs. Boxes indicate magnified regions on the right. Scale bars represent 200 µm in the overview and 50 µm in magnified regions on the right. (F) WISH for kif5c confirms the expression of the gene at the MHB at 30 hpf. Top: Antisense. Bottom: Sense for control. Magnified regions on the right are indicated by boxes in overview images. Arrows indicates MHBs and arrowheads URLs. Scale bar represents 100 µm or 50 µm in magnified images. Elapsed time in minutes. MHB, solid line; URL, dotted line. Boxes in graphs represent 25–75% of all values and whiskers 1.5 times the quartile. Median is shown as a horizontal bar and mean as a square box. Significance level in Kruskal–Wallis ANOVA: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Interfering with the function of dynein by overexpression of p50, Kif5C by overexpression of the rigor mutation T94N, or intracellular transport by expression of the chimera Kif1C-5C reduces THN speed. To improve cell tracking, p50 expression experiments were performed in THNs coexpressing H2B. Expressed markers are indicated at the top. Colored dots indicate individual examples of THNs. Elapsed time in hours:minutes. Bars: 10 µm. The video is related to Figs. 4 and 7.
Interfering with the function of dynein by overexpression of p50, Kif5C by overexpression of the rigor mutation T94N, or intracellular transport by expression of the chimera Kif1C-5C reduces THN speed. To improve cell tracking, p50 expression experiments were performed in THNs coexpressing H2B. Expressed markers are indicated at the top. Colored dots indicate individual examples of THNs. Elapsed time in hours:minutes. Bars: 10 µm. The video is related to Figs. 4 and 7.
While these results do not contradict the idea that MTs and dynein directly transport the nucleus, they are also consistent with a model based on intracellular transport, as dynein is the only retrograde moving motor protein in nonmitotic cells (Allan, 2011). Next, we probed the role of the anterograde motor kinesin-1 in THN migration. There are five members of the kinesin-1 family encoded in the zebrafish genome, of which Kif5C showed a clear expression along the MHB at 30 hpf, similar to previous results (Fig. 4, E and F; Campbell and Marlow, 2013). To test the motor’s contribution to THN migration, we overexpressed the rigor mutation T94N, which stalls the kinesin on MTs (Nakata and Hirokawa, 1995). In the absence of cargo-mediated motor activation, most of the rigor mutant remained cytoplasmic like the WT parent and did not noticeably decorate MTs, arguing against a general roadblock imposed by the mutant stalling on MTs (Fig. 5 C). Compared with WT Kif5C- or citrine-expressing control cells, THNs with the T94N rigor mutation migrated at speeds approximately two thirds that of control cells (4.10 µm/h and n = 145 tracks/24 embryos for citrine control; median: 4.98 µm/h and n = 155 tracks/25 embryos for WT Kif5C; 3.38 µm/h and n = 250 tracks/31 embryos for Kif5C T94N; P = 1.08 × 10−8; Fig. 4, C and D; and Video 4).
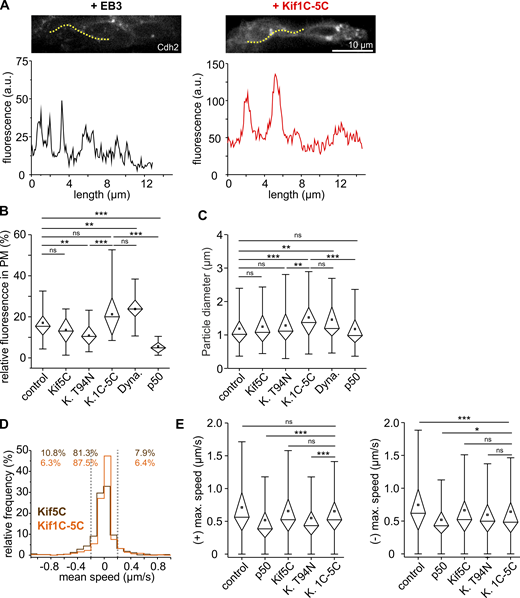
MT motor proteins regulate THN speed.(A) Both Brefeldin A and Dynasore cause a reduction in THN motility. Colored dots follow individual examples. Scale bars represent 20 µm in the overview and 10 µm in magnified images. Elapsed time in minutes. MHB, solid line; URL, dotted line. See also Video 5. (B and C) The overexpression of Kif1C-GFP leads to the accumulation of protein in the tips of phase 1 (B) and phase 2 (C) THNs. Scale bars represent 20 µm in the overview and 5 µm in magnified images. MHB, solid line; URL, dotted line. (D) Quantification of THN speeds confirms the loss of motility when endosomes or Golgi vesicle traffic is targeted. n = 20 embryos/164 tracks for drug control; n = 33 embryos/367 tracks for Brefeldin A; n = 23 embryos/162 tracks for Dynasore; n = 24 embryos/145 tracks for injection control; n = 24 embryos/170 tracks for Rab11a-expressing embryos; n = 25 embryos/252 tracks for Rab11a S25N-expressing embryos; n = 15 embryos/121 tracks for Kif1C-expressing embryos; n = 27 embryos/259 tracks for Kif1C T106N-expressing embryos; P = 9.56 × 10−16 control/Brefeldin A, P = 1.26 × 10−4 control/Dynasore, P = 0.001 control/Rab11a S25N, P = 9.6 × 10−3 Rab11a/Rab11a S25N, and P = 0.018 control/Kif1C T94N T106N. For full statistics, see Table S1. (E) Some Cdh2-positive particles appear to travel along MTs bidirectionally. Arrows indicate anterograde and arrowheads retrograde movement. Scale bar, 3 µm. Elapsed time in seconds on the right. (F) Cdh2 particles in THNs exhibit different motility patterns. Scale bar, 5 µm. Dotted line indicates region for kymograph depicted in G. The unidirectional EB3-GFP signals determine the classification of Cdh2 particles moving into antero- or retrograde direction, indicated above the images. Horizontal scale bars, 5 µm; vertical bars, 20 s. See also Video 6. (H) The kymographs illustrate that Cdh2 particle are less motile when either dynein activation is inhibited (p50) or anterograde vesicle traffic for Cdh2 is blocked (Kif5C T94N). Horizontal scale bars, 5 µm; vertical bars, 20 s. See also Video 6. Boxes in graphs represent 25–75% of all values and whiskers 1.5 times the quartile. Median is shown as a horizontal bar and mean as a square box. Significance level in Kruskal–Wallis ANOVA: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
MT motor proteins regulate THN speed.(A) Both Brefeldin A and Dynasore cause a reduction in THN motility. Colored dots follow individual examples. Scale bars represent 20 µm in the overview and 10 µm in magnified images. Elapsed time in minutes. MHB, solid line; URL, dotted line. See also Video 5. (B and C) The overexpression of Kif1C-GFP leads to the accumulation of protein in the tips of phase 1 (B) and phase 2 (C) THNs. Scale bars represent 20 µm in the overview and 5 µm in magnified images. MHB, solid line; URL, dotted line. (D) Quantification of THN speeds confirms the loss of motility when endosomes or Golgi vesicle traffic is targeted. n = 20 embryos/164 tracks for drug control; n = 33 embryos/367 tracks for Brefeldin A; n = 23 embryos/162 tracks for Dynasore; n = 24 embryos/145 tracks for injection control; n = 24 embryos/170 tracks for Rab11a-expressing embryos; n = 25 embryos/252 tracks for Rab11a S25N-expressing embryos; n = 15 embryos/121 tracks for Kif1C-expressing embryos; n = 27 embryos/259 tracks for Kif1C T106N-expressing embryos; P = 9.56 × 10−16 control/Brefeldin A, P = 1.26 × 10−4 control/Dynasore, P = 0.001 control/Rab11a S25N, P = 9.6 × 10−3 Rab11a/Rab11a S25N, and P = 0.018 control/Kif1C T94N T106N. For full statistics, see Table S1. (E) Some Cdh2-positive particles appear to travel along MTs bidirectionally. Arrows indicate anterograde and arrowheads retrograde movement. Scale bar, 3 µm. Elapsed time in seconds on the right. (F) Cdh2 particles in THNs exhibit different motility patterns. Scale bar, 5 µm. Dotted line indicates region for kymograph depicted in G. The unidirectional EB3-GFP signals determine the classification of Cdh2 particles moving into antero- or retrograde direction, indicated above the images. Horizontal scale bars, 5 µm; vertical bars, 20 s. See also Video 6. (H) The kymographs illustrate that Cdh2 particle are less motile when either dynein activation is inhibited (p50) or anterograde vesicle traffic for Cdh2 is blocked (Kif5C T94N). Horizontal scale bars, 5 µm; vertical bars, 20 s. See also Video 6. Boxes in graphs represent 25–75% of all values and whiskers 1.5 times the quartile. Median is shown as a horizontal bar and mean as a square box. Significance level in Kruskal–Wallis ANOVA: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
These data clearly implicate kinesin-1 in efficient THN migration. While the speed reduction is smaller than upon MT depletion or dynein inactivation, this could be due to the presence of other anterograde kinesins, while dynein is the only retrograde motor. Both dynein and Kif5C are known organelle carriers, so their impact on migration could arise from transporting various cargoes and thereby controlling protein and membrane composition at distinct locations (e.g., via vesicle traffic). This idea has been put forward for migratory nonneuronal cell types (Schmoranzer et al., 2003; Stehbens et al., 2014). Therefore, we next tested to which extent vesicle traffic could be involved in THN migration.
Endosomes and the Golgi apparatus contribute to THN migration to a similar extent as MTs
Both endosomal and Golgi-derived vesicles are typically moved by dynein and kinesins (Granger et al., 2014; Jordens et al., 2005), and we first interfered with intracellular vesicle traffic using pharmacological inhibitors. Dynasore blocks the formation of endosomes at the PM (Macia et al., 2006), while Brefeldin A collapses the Golgi apparatus and thereby prevents the generation of Golgi-derived vesicles and Golgi-derived MTs (Fujiwara et al., 1988). The injection of 2–5 nl of 50 mM Dynasore into the fourth ventricle of 30-hpf embryos or the bath application of 10 µg/ml Brefeldin A led to the disappearance of Rab5-positive endosomes and the dispersal of the Golgi apparatus, respectively (Fig. S3, F and G). In either condition, THN speed was reduced (median: 4.43 µm/h and n = 164 tracks/20 embryos for DMSO control embryos; 3.05 µm/h and n = 162 tracks/23 embryos for Dynasore; P = 1.26 × 10−4; 2.23 µm/h and n = 367 tracks/33 embryos for Brefeldin A; P = 9.56 × 10−16; Fig. 5, A and B; and Video 5), demonstrating that the endocytic pathway and the Golgi apparatus are as important in THN migration as intact MTs. The effect of Brefeldin A on THN motility is slightly greater than Dynasore treatment, which could be due to a reduction of MTs in addition to loss of vesicles. Alternatively, the Golgi-derived supply of components to the PM and other organelles is more important to migration than endocytosis from the PM. The motility of Cadherin-2–positive particles, which is less affected by Brefeldin A treatment than by Dynasore (see below, Fig. 6 and,Fig. S4), argues that the loss of Golgi MTs is not the main reason for slowing THNs, but a future in-depth analysis of Golgi-derived MTs and Golgi-dependent cargo is required to quantify the respective contributions of Golgi–vesicle transport or Golgi MTs to cellular motility.
Golgi dispersal by Brefeldin A or block of endosome formation by Dynasore injection strongly impairs THN migration. Brefeldin A–treated embryos coexpressed a Golgi-targeted YFP and a PM marker, and Dynasore-injected embryos coexpressed GFP-Rab5a and the PM marker. Colored dots indicate individual examples, treatments are indicated at the top. Elapsed time in hours:minutes. Bars: 20 µm. The video is related to Fig. 5.
Golgi dispersal by Brefeldin A or block of endosome formation by Dynasore injection strongly impairs THN migration. Brefeldin A–treated embryos coexpressed a Golgi-targeted YFP and a PM marker, and Dynasore-injected embryos coexpressed GFP-Rab5a and the PM marker. Colored dots indicate individual examples, treatments are indicated at the top. Elapsed time in hours:minutes. Bars: 20 µm. The video is related to Fig. 5.
Cadherin-2 is transported by dynein and Kif5C in THNs.(A–C) When motors or vesicle formation is impaired, Cadherin-2 particles spend less time in motion. For full statistical testing, see Table S2. n = 682 tracks for drug control, n = 711 tracks for Brefeldin A, n = 287 tracks for Dynasore, n = 416 tracks for injection control, n = 308 tracks for p50-expressing embryos, n = 593 tracks for Kif5C-expressing embryos, n = 441 tracks Kif5C T94N-expressing embryos, n = 373 tracks for Rab11a-expressing embryos, and n = 152 tracks for Rab11a S25N-expressing embryos. Instantaneous velocities in the anterograde (B) as well as retrograde (C) direction are reduced when vesicle formation or motor transport is impaired. For full statistical testing, see Table S3 and Table S4. Anterograde: n = 2,728 values for drug control, n = 2,978 values for Brefeldin A, n = 1,451 values for Dynasore, n = 2,059 values for injection control, n = 1,044 values for p50-expressing embryos, n = 2,808 values for Kif5C-expressing embryos, n = 2,055 values Kif5C T94N-expressing embryos, n = 1,815 values for Rab11a-expressing embryos, and n = 837 values for Rab11a S25N-expressing embryos. Retrograde: n = 2,687 values for drug control, n = 3,034 values for Brefeldin A, n = 1,574 values for Dynasore, n = 2,127 values for injection control, n = 1,117 values for p50-expressing embryos, n = 2,791 values for Kif5C-expressing embryos, n = 2,132 values Kif5C T94N-expressing embryos, n = 1,740 values for Rab11a-expressing embryos, and n = 801 values for Rab11a S25N-expressing embryos. (D) Plotting maximal and mean velocities for each of the 416 particle tracks in control THNs illustrates that the majority of Cadherin-2 particles move only at low mean and maximal velocities, with some particles exhibiting higher mobility in both directions. (E) Converting the mean-maximal velocities plots into data density plots reveals that the fraction of slow-moving particles increases when vesicle formation is impaired by drugs. Color code is indicated on the right, where dark red indicates high data density. For comparison of the spread in the datasets, the area of the 0.8 density is outline in gray for each condition, and the area covered by this density level is indicated in the top left corner. The number of particle tracks for each condition is listed in the bottom left corner. Vertical gray lines separate areas without data content. (F) Similar results are obtained when vesicle formation or motor transport of Cadherin-2 particles are genetically targeted. Boxes in graphs represent 25–75% of all values and whiskers 1.5 times the quartile. Median is shown as a horizontal bar and mean as a square box. Significance level in Kruskal–Wallis ANOVA: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant. All data shown in A–F represent n = 13 embryos for drug control, n = 19 embryos for Brefeldin A, n = 8 embryos for Dynasore, n = 13 embryos for injection control, n = 14 p50-expressing embryos, n = 11 Kif5C-expressing embryos, n = 16 Kif5C T94N-expressing embryos, n = 16 Rab11a-expressing embryos, and n = 7 Rab11a S25N-expressing embryos.
Cadherin-2 is transported by dynein and Kif5C in THNs.(A–C) When motors or vesicle formation is impaired, Cadherin-2 particles spend less time in motion. For full statistical testing, see Table S2. n = 682 tracks for drug control, n = 711 tracks for Brefeldin A, n = 287 tracks for Dynasore, n = 416 tracks for injection control, n = 308 tracks for p50-expressing embryos, n = 593 tracks for Kif5C-expressing embryos, n = 441 tracks Kif5C T94N-expressing embryos, n = 373 tracks for Rab11a-expressing embryos, and n = 152 tracks for Rab11a S25N-expressing embryos. Instantaneous velocities in the anterograde (B) as well as retrograde (C) direction are reduced when vesicle formation or motor transport is impaired. For full statistical testing, see Table S3 and Table S4. Anterograde: n = 2,728 values for drug control, n = 2,978 values for Brefeldin A, n = 1,451 values for Dynasore, n = 2,059 values for injection control, n = 1,044 values for p50-expressing embryos, n = 2,808 values for Kif5C-expressing embryos, n = 2,055 values Kif5C T94N-expressing embryos, n = 1,815 values for Rab11a-expressing embryos, and n = 837 values for Rab11a S25N-expressing embryos. Retrograde: n = 2,687 values for drug control, n = 3,034 values for Brefeldin A, n = 1,574 values for Dynasore, n = 2,127 values for injection control, n = 1,117 values for p50-expressing embryos, n = 2,791 values for Kif5C-expressing embryos, n = 2,132 values Kif5C T94N-expressing embryos, n = 1,740 values for Rab11a-expressing embryos, and n = 801 values for Rab11a S25N-expressing embryos. (D) Plotting maximal and mean velocities for each of the 416 particle tracks in control THNs illustrates that the majority of Cadherin-2 particles move only at low mean and maximal velocities, with some particles exhibiting higher mobility in both directions. (E) Converting the mean-maximal velocities plots into data density plots reveals that the fraction of slow-moving particles increases when vesicle formation is impaired by drugs. Color code is indicated on the right, where dark red indicates high data density. For comparison of the spread in the datasets, the area of the 0.8 density is outline in gray for each condition, and the area covered by this density level is indicated in the top left corner. The number of particle tracks for each condition is listed in the bottom left corner. Vertical gray lines separate areas without data content. (F) Similar results are obtained when vesicle formation or motor transport of Cadherin-2 particles are genetically targeted. Boxes in graphs represent 25–75% of all values and whiskers 1.5 times the quartile. Median is shown as a horizontal bar and mean as a square box. Significance level in Kruskal–Wallis ANOVA: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant. All data shown in A–F represent n = 13 embryos for drug control, n = 19 embryos for Brefeldin A, n = 8 embryos for Dynasore, n = 13 embryos for injection control, n = 14 p50-expressing embryos, n = 11 Kif5C-expressing embryos, n = 16 Kif5C T94N-expressing embryos, n = 16 Rab11a-expressing embryos, and n = 7 Rab11a S25N-expressing embryos.
Cadherin-2 particles include stationary and highly motile pools (related to Fig. 6).(A) Mean speeds of all Cadherin-2–positive particles indicate that a large pool is nonmotile, moving either antero- or retrogradely at <0.2 µm/s, with the remaining particles moving at higher speeds in both directions. Interfering with either vesicle formation or MT motors reduces the fast-moving pool and increases the stationary fraction. n = 682 tracks for drug control, n = 711 tracks for Brefeldin A, n = 287 tracks for Dynasore, n = 416 tracks for injection control, n = 308 tracks for p50-expressing embryos, n = 593 tracks for Kif5C-expressing embryos, n = 441 tracks Kif5C T94N-expressing embryos, n = 373 tracks for Rab11a-expressing embryos, and n = 152 tracks for Rab11a S25N-expressing embryos. (B) Similarly, maximal velocities of Cadherin-2 particles are decreased when motor transport is impaired. For full statistical testing, see Table S5 and Table S6. Anterograde: n = 658 tracks for drug control, n = 690 tracks for Brefeldin A, n = 283 tracks for Dynasore, n = 399 tracks for injection control, n = 295 tracks for p50-expressing embryos, n = 579 tracks for Kif5C-expressing embryos, n = 425 tracks for Kif5C T94N-expressing embryos, n = 367 tracks for Rab11a-expressing embryos, n = 150 tracks for Rab11a S25N-expressing embryos. Retrograde: n = 656 tracks for drug control, n = 696 tracks for Brefeldin A, n = 280 tracks for Dynasore, n = 411 tracks for injection control, n = 299 tracks for p50-expressing embryos, n = 580 tracks for Kif5C-expressing embryos, n = 432 tracks for Kif5C T94N-expressing embryos, n = 364 tracks for Rab11a-expressing embryos, and n = 148 tracks for Rab11a S25N-expressing embryos. (C) The modified PST-1P activation protocol is able to deplete MTs. White box indicates PST-1P activation area, which is magnified in the lower row. MHB, solid line; URL, dotted line. Elapsed time in seconds. Scale bar, 10 µm. (D) Kymographs demonstrate that the motility of Cadherin-2 reporter particles is reduced when MTs are depleted. Horizontal scale bars, 5 µm; vertical bars, 20 s. (E) Plotting mean speeds against the respective maximal velocities for each track reveals that, upon MT depletion, many tracks cluster in the low mean-low maximal velocity area. Area of 0.8 density given in the top left corner and number of tracks in the bottom left corner. (F and G) Mean velocities (n = 315 tracks for red light only, control region PST-1P OFF and n = 191 tracks for UV-activated, MT-reduced region PST-1P ON; F) and maximal velocities of Cadherin-2 particles are reduced in the anterograde direction upon MT depletion (G). (anterograde: n = 306 tracks for PST-1P OFF, n = 187 tracks for PST-1P ON, P = 3.51 × 10−4; retrograde: n = 310 tracks for PST-1P OFF, n = 189 tracks for PST-1P ON). (H and I) Pause times (H; n = 315 tracks for PST-1P OFF, n = 191 tracks for PST-1P ON) and instantaneous velocities (I; anterograde: n = 1,751 values for PST-1P OFF, n = 1,000 values for PST-1P ON, P = 0.004; retrograde: n = 1,677 values for PST-1P OFF, n = 1,015 values for PST-1P ON, P = 0.006) show a trend toward lower speed motility/longer stationary times when MTs are depleted. Boxes in graphs represent 25–75% of all values and whiskers 1.5 times the quartile. Median is shown as a horizontal bar and mean as a square box. Significance level in Kruskal–Wallis ANOVA: **, P < 0.01; ***, P < 0.001; ns, not significant. All data shown in A, B, and E–I represent n = 13 embryos for drug control, n = 19 embryos for Brefeldin A, n = 8 embryos for Dynasore, n = 13 embryos for injection control, n = 14 p50-expressing embryos, n = 11 Kif5C-expressing embryos, n = 16 Kif5C T94N-expressing embryos, n = 16 Rab11a-expressing embryos, n = 7 Rab11a S25N-expressing embryos, n = 13 for PST-1P nonactivated (OFF) embryos, and n = 12 for PST-1P activated (ON) embryos.
Cadherin-2 particles include stationary and highly motile pools (related to Fig. 6).(A) Mean speeds of all Cadherin-2–positive particles indicate that a large pool is nonmotile, moving either antero- or retrogradely at <0.2 µm/s, with the remaining particles moving at higher speeds in both directions. Interfering with either vesicle formation or MT motors reduces the fast-moving pool and increases the stationary fraction. n = 682 tracks for drug control, n = 711 tracks for Brefeldin A, n = 287 tracks for Dynasore, n = 416 tracks for injection control, n = 308 tracks for p50-expressing embryos, n = 593 tracks for Kif5C-expressing embryos, n = 441 tracks Kif5C T94N-expressing embryos, n = 373 tracks for Rab11a-expressing embryos, and n = 152 tracks for Rab11a S25N-expressing embryos. (B) Similarly, maximal velocities of Cadherin-2 particles are decreased when motor transport is impaired. For full statistical testing, see Table S5 and Table S6. Anterograde: n = 658 tracks for drug control, n = 690 tracks for Brefeldin A, n = 283 tracks for Dynasore, n = 399 tracks for injection control, n = 295 tracks for p50-expressing embryos, n = 579 tracks for Kif5C-expressing embryos, n = 425 tracks for Kif5C T94N-expressing embryos, n = 367 tracks for Rab11a-expressing embryos, n = 150 tracks for Rab11a S25N-expressing embryos. Retrograde: n = 656 tracks for drug control, n = 696 tracks for Brefeldin A, n = 280 tracks for Dynasore, n = 411 tracks for injection control, n = 299 tracks for p50-expressing embryos, n = 580 tracks for Kif5C-expressing embryos, n = 432 tracks for Kif5C T94N-expressing embryos, n = 364 tracks for Rab11a-expressing embryos, and n = 148 tracks for Rab11a S25N-expressing embryos. (C) The modified PST-1P activation protocol is able to deplete MTs. White box indicates PST-1P activation area, which is magnified in the lower row. MHB, solid line; URL, dotted line. Elapsed time in seconds. Scale bar, 10 µm. (D) Kymographs demonstrate that the motility of Cadherin-2 reporter particles is reduced when MTs are depleted. Horizontal scale bars, 5 µm; vertical bars, 20 s. (E) Plotting mean speeds against the respective maximal velocities for each track reveals that, upon MT depletion, many tracks cluster in the low mean-low maximal velocity area. Area of 0.8 density given in the top left corner and number of tracks in the bottom left corner. (F and G) Mean velocities (n = 315 tracks for red light only, control region PST-1P OFF and n = 191 tracks for UV-activated, MT-reduced region PST-1P ON; F) and maximal velocities of Cadherin-2 particles are reduced in the anterograde direction upon MT depletion (G). (anterograde: n = 306 tracks for PST-1P OFF, n = 187 tracks for PST-1P ON, P = 3.51 × 10−4; retrograde: n = 310 tracks for PST-1P OFF, n = 189 tracks for PST-1P ON). (H and I) Pause times (H; n = 315 tracks for PST-1P OFF, n = 191 tracks for PST-1P ON) and instantaneous velocities (I; anterograde: n = 1,751 values for PST-1P OFF, n = 1,000 values for PST-1P ON, P = 0.004; retrograde: n = 1,677 values for PST-1P OFF, n = 1,015 values for PST-1P ON, P = 0.006) show a trend toward lower speed motility/longer stationary times when MTs are depleted. Boxes in graphs represent 25–75% of all values and whiskers 1.5 times the quartile. Median is shown as a horizontal bar and mean as a square box. Significance level in Kruskal–Wallis ANOVA: **, P < 0.01; ***, P < 0.001; ns, not significant. All data shown in A, B, and E–I represent n = 13 embryos for drug control, n = 19 embryos for Brefeldin A, n = 8 embryos for Dynasore, n = 13 embryos for injection control, n = 14 p50-expressing embryos, n = 11 Kif5C-expressing embryos, n = 16 Kif5C T94N-expressing embryos, n = 16 Rab11a-expressing embryos, n = 7 Rab11a S25N-expressing embryos, n = 13 for PST-1P nonactivated (OFF) embryos, and n = 12 for PST-1P activated (ON) embryos.
To confirm the importance of vesicle transport in THNs excluding side effects on other tissues or numbers of MTs, we generated genetic constructs to interfere with endosomal (Rab11a S25N) or post-Golgi traffic (Kif1C T106N). Rab11a is one of two zebrafish homologues for mammalian Rab11, which regulates protein recycling via endosomes, including Cadherin-2, to ensure migration of cortical neurons (Kawauchi et al., 2010). The T106N mutation is the rigor mutation described for Kif5C introduced into the zebrafish homologue of the kinesin-3 family member Kif1C. Kif1C transports integrin-containing vesicles in epithelial cells (Theisen et al., 2012) and secretory vesicles in neurons (Schlager et al., 2010), and in zebrafish, it has a strong tendency to accumulate at the front of THNs (Fig. 5, B and C). These constructs can therefore be expected to partially block the endocytic or exocytic pathways, respectively. Quantification showed that THNs overexpressing either of the mutated proteins migrated more slowly compared with controls (median: 4.00 µm/h and n = 170 tracks/24 embryos for Rab11a; 3.11 µm/h and n = 252 tracks/25 embryos for Rab11a S25N; P = 0.001; 4.32 µm/h and n = 121 tracks/15 embryos for Kif1C; 3.36 µm/h and n = 259 tracks/27 embryos for Kif1C T106N; P = 0.018; Fig. 5 D and,Fig. S3, H and I).
These experiments demonstrate that interference with MT-based vesicle traffic mimics the effects seen when MTs and motors are inactivated.
Cadherin-2 transport in THNs is in part mediated by dynein and Kif5C
To gain further evidence for a role of MTs and motors in THN migration by distributing cargo, we tested the dependence of Cadherin-2 transport on MTs, motor proteins, and exo/endocytosis in vivo. We chose Cadherin-2 as model cargo, as the protein is regulating coherence and directionality of migrating URL-derived neurons in zebrafish (Lele et al., 2002; Rieger et al., 2009). The protein is dynamically localized during iterative rounds of cellular forward movement, and an accumulation at the front precedes a forward motion (Rieger et al., 2009). While the localization of Cadherin-2 during neuronal migration in fish is less well understood, it is known from mammalian cells that Cadherin-2 is transported by kinesin-1 and dynein (Heisler et al., 2014; Mary et al., 2002) and that its endocytic recycling supports the migration of cortical neurons (Kawauchi et al., 2010).
To see whether Cadherin-2 transport depends on MTs in THNs, we expressed a previously established Cadherin-2 reporter construct (Rieger et al., 2009) together with Map4MTB-GFP and recorded THNs in phase 1 for several minutes at ∼10-s frame intervals. We observed reporter-positive particles of different sizes and motility, several of which appeared to follow MTs in both anterograde (toward the MHB) and retrograde (toward the URL) direction (Fig. 5 E, arrows indicate anterograde and arrowheads retrograde motion). The unipolar orientation of MTs in THNs in phase 1 makes it likely that dynein and a kinesin underlie this transport, which should therefore be altered when the motors are impaired.
We next recorded Cadherin-2 particle motility in THNs for 1 min at 0.9–1.4-s intervals (Fig. 5 F) and observed frequent changes in direction (Fig. 5 G). Interference with dynein or Kif5C appeared to lower particle motility (Fig. 5 H).
To analyze this motility in more detail, we recorded Cadherin-2 reporter particles in a single z-plane for 1 min with a frame interval of ∼1.3 s in THNs in phase 1 under different conditions, altering either organelle generation or impairing MT motors. During this short imaging time, the position of THNs hardly changed (Video 6, note that individual frames were not registered); hence, we tracked all particles which were observed for at least three consecutive frames and calculated motility parameters without tissue displacement correction. We determined relative pause times and mean particle speed as overall motility parameter, maximal velocity, and instantaneous velocity as indicators of processive motor activity. To improve the detection of changes in direction, we introduced a reference axis along the length of the cells onto which particle positions were projected.
Cadherin-2 particles move dynamically in control THNs, but motility is reduced, when motor function or the formation of recycling endosomes is impaired. Top: Control THNs coexpressing EB3-GFP (green) and Cdh2 reporter (red). Panels: Cadherin-2 particles moving under inactivation conditions indicated in the top left corners. Elapsed time in seconds:milliseconds. Scale bar, 5 µm. The video is related to Figs. 5, 7, and S5.
Cadherin-2 particles move dynamically in control THNs, but motility is reduced, when motor function or the formation of recycling endosomes is impaired. Top: Control THNs coexpressing EB3-GFP (green) and Cdh2 reporter (red). Panels: Cadherin-2 particles moving under inactivation conditions indicated in the top left corners. Elapsed time in seconds:milliseconds. Scale bar, 5 µm. The video is related to Figs. 5, 7, and S5.
Stationary times were defined as all steps with a velocity of <0.2 µm/s. At the given frame rate, this meant that a particle moved only 200 nm in a single step, which is close to the resolution of light, or equivalent to 1–2 pixels per step in our setup; hence, we counted such a short-distance movement as paused. We presented the total time paused for each track as percentage of the total observation time for a Cadherin-2 particle. This analysis revealed that, like in mammals, Cadherin-2 traffic relies on endosomes and the Golgi, as well as dynein and Kif5C, as nonmotile phases increased when they were functionally inactivated (Fig. 6 A; see Table S2 for statistics). It also showed that in control THNs, a large percentage of time is spent nonmoving on this short time scale, which may be due to the presence of adherens junctions in the analysis. This is also visible in the mean speeds, which were calculated from the distance between start and end points (Fig. S4 A). Only ∼20% of the particles moved antero- or retrogradely with a mean velocity exceeding 0.2 µm/s. Blocking endocytosis, Golgi, dynein, or Kif5C function again reduced these motile pools and increased the “stationary” fraction of Cadherin-2 particles. These results, however, do not inform about the actual motility of a particle, as low mean speeds can arise from stationary particles that do not move or particles of higher motility in opposite directions that return to their origin.
To further characterize particle motility and their underlying motor transport, we next determined instantaneous velocities. Instantaneous velocities were calculated as sums of consecutive distances traversed in the same direction along the reference axis, divided by the duration of this phase. Dynein and kinesin-1 family members are highly processive motors that can travel for several hundred nanometers (Cai et al., 2007), so interfering with them should reduce instantaneous speeds of Cadherin-2 particles if these motors are directly involved with their transport. Accordingly, instantaneous velocities in both directions were reduced when MT motor function was impaired, although the majority of instantaneous speeds is low, as is expected for a dataset that includes slow-moving particles with many turns such as adherens junctions (Fig. 6, B and C; see Table S3 and Table S4 for statistics). Because such slow, turning particles contribute more values to the instantaneous velocity dataset than fast-moving, unidirectional particles, this could underestimate the fraction of significant motion. Hence, we next analyzed maximal velocities for each track in antero- and retrograde direction. For unidirectional tracks, a single value was included in the respective dataset. Dynein and kinesin-1 are known to move at high velocities (Cai et al., 2007; Encalada et al., 2011), and as expected, targeting the motors directly with p50 or Kif5C T94N produced strong reductions in maximal Cadherin-2 particle velocities (Fig. S4 B; see Table S5 and Table S6 for statistics).
Individually, these parameters demonstrate that high-velocity, directional Cadherin-2 particle transport depends on MT-based mechanisms, but in combination, they can provide more information about how motility patterns are affected by targeting vesicles or motors. For example, plotting the mean speed of each track against the respective maximal velocity values will reveal whether particles become nonmotile or whether they are motile but return to their origin (Fig. 6 B). To facilitate the comparison between different conditions with different number of tracks, the plots were converted into 2D kernel density plots, which were color-coded so that dark-red values indicate higher data density in a given region, and a low level of data density was outlined in Fig. 6, E and F to assess the spread in the dataset. This revealed that, with the exception of the overexpression of WT Kif5C, all treatments reduced Cadherin-2 particle transport, as both maximal and mean velocities decreased simultaneously to result in higher density data areas close to 0, and the data were spread over a smaller area (area indicated in top left corner, number of tracks in bottom left corner in Fig. 6, E and F). This argues that particles become stationary when motor transport of vesicles is impaired rather than particles change their motility to other modalities. It is worth noting that Brefeldin A treatment had a smaller effect on Cadherin-2 transport than Dynasore treatment, which indicates that recycling plays a greater role for Cadherin-2 localization than the delivery of newly synthesized material, although the loss of the Golgi apparatus may decrease the number of MTs as well as block vesicle formation. This also suggests that Golgi-derived MTs are of subordinate importance for Cadherin-2 delivery, but this is likely to be highly cargo specific and needs to be addressed in a study on the role of Golgi-derived MTs in THN migration.
The data condensation near 0 occurred very strongly in the Dynasore and Rab11a S25N datasets, despite a low overall number, indicating that the fraction of stationary particles in the datasets increased. This is particularly interesting for Rab11a S25N, which had not produced a significant difference when comparing individual motility parameters. Rab11a S25N affects only recycling endosomes (Kawauchi et al., 2010), and if a sizable vesicle population unaffected by Rab11a S25N remains, it could explain the lack of effect when pause times and mean and maximal speeds are analyzed separately. The presence of outlying regions in the density plots (Fig. 6 F), which are absent from the Dynasore dataset (Fig. 6 E), indicates that some particles retain the normal motility patterns while others become stationary, arguing that the MT motors are involved in transporting recycling endosomes, as well as other endosomal and anterograde vesicles.
These experiments relied on drugs or genetic constructs that either changed the organelle composition in a THN or targeted the motors. To support our findings, we used PST-1P to target MTs directly. As Cadherin-2 particle transport occurred on a much faster timescale (seconds) than THN migration (hours), we adjusted the PST-1P activating protocol. After the initial 30-min inactivation step at 561 nm, two ROIs were placed on phase 1 THNs expressing the Cadherin-2 reporter-mCherry and EB3-GFP. One ROI was recorded at 405 nm + 488 nm + 561 nm illumination to activate PST-1P and visualize EB3-GFP as depletion control and Cadherin-2 particles (PST ON), whereas the other was recorded at 561 nm only as control (PST OFF) for 5 min continuously at a frame rate of ∼1.3 s. This illumination setup depleted MTs in most THNs after 4 min, although some MTs again remained (Fig. S4 C). THNs that still showed clear EB3 comets at this point were excluded from analysis. Cadherin-2 particles were tracked in the final 1 min of the 5-min recording. Cadherin-2 particles in MT-depleted THNs traveled with reduced mean and maximal velocity and lower instantaneous velocities and had a tendency to spend more time nonmotile (Fig. 6, D–I). Interestingly, in MT-depleted conditions, plus-end–directed transport was more strongly impaired than retrograde transport, while interfering with the motors had reduced transport bidirectionally. This is in line with the idea that dynein is less sensitive to MT subpopulations, while kinesins can be selective for MT surfaces (Sirajuddin et al., 2014; Tas et al., 2017), and that PST-1P treatment could preferentially depolymerize a subset of MTs. Further research into MT modifications and stabilization mechanisms will be necessary to fully explain the differential effects on cargo transport.
In sum, MTs, dynein and Kif5C contribute to Cadherin-2 intracellular transport, likely by moving vesicles, including Cadherin-2–positive (recycling) endosomes toward the cell center and by returning cargo from the recycling compartment and the Golgi to the PM. Many proteins follow similar cycles of import, recycling, and export, and our findings therefore support a model in which MTs and motors regulate neuronal migration by controlling the protein composition at distant locations in a cell.
An imbalance in intracellular Cadherin-2 transport reduces THN motility
This model implies that imbalancing or mistargeting cargo should reduce THN motility. We hypothesized that the overexpression of an artificial motor with high processivity and high affinity for Cadherin-2–positive vesicles could compete against endogenous motors to accumulate the protein in the PM or vesicles, thereby disrupting its transport and consequently THN migration. To test this idea, we constructed a chimeric motor from the motor domain of Kif1C, a leucine zipper and a cargo-binding fragment of Kif5C (Fig. 7 A). Kif1C is a highly processive motor (Siddiqui et al., 2019) that in zebrafish THNs has a strong preference for the front (Fig. 5, C and D). We fused the Kif1C motor domain including the neck coil to a GCN4-derived leucine zipper to ensure dimerization of the chimera. The endogenous coiled coils were removed to avoid heterodimerization with endogenous motors, to disable chimera inactivation, and to limit the binding of adaptor proteins for alternative cargoes. We attached the final 165 aa of zebrafish Kif5C to this construct, which contain binding sites for Cadherin-2 adaptor proteins, while excluding most of the kinesin light chain–binding domain. The light chain connects alternative cargoes such as large organelles to the motor (Wilson and Holzbaur, 2015) and binds mutually exclusive with the adaptors for vesicles containing Cadherin-2 in mammalian cells (Heisler et al., 2014; Twelvetrees et al., 2019). Without a binding site for kinesin light chain, the chimera should not participate in nuclear transport or rotation while enhancing its binding potential for Cadherin-2–positive vesicles.
Imbalanced cargo transport reduces THN motility.(A) Schematic representation of Kif1C-5C chimera. (B and C) In THNs, the chimera localizes to the front of phase 1 (B) and phase 2 (C) cells. Cyan box indicates magnified region on the right. Scale bars represent 20 µm in overview images and 5 µm in magnified images. (D) Kif1C-5C does not colocalize with Cadherin-2 particles, although they are frequently found in the vicinity. Colored boxes indicate magnified regions on the right. Scale bars represent 10 µm in the overview image and 1 µm in magnified images. (E) The expression of Kif1C-5C increases stationary times of Cadherin-2 particles. For full statistical testing, see Table S2; n = 416 tracks for injection control, n = 308 tracks for p50-expressing embryos, n = 593 tracks for Kif5C-expressing embryos, n = 441 tracks Kif5C T94N-expressing embryos, and n = 439 tracks for Kif1C-5C–expressing embryos. (F and G) Kif1C-5C also reduces maximal velocities and mean velocities (F; area of 0.8 data density in top left corner and number of tracks in lower left corner; see also Video 6) as well as instantaneous velocities (G). Note that the retrograde direction is more strongly affected than the anterograde direction, as evidenced by the comparison to other motor-affecting treatments. For full statistical testing, see Table S3 and Table S4. Anterograde: n = 2,059 values for injection control, n = 1,044 values for p50-expressing embryos, n = 2,808 values for Kif5C-expressing embryos, n = 2,055 values Kif5C T94N-expressing embryos, and n = 2,245 values for Kif1C-5C–expressing embryos. Retrograde: n = 2,127 values for injection control, n = 1,117 values for p50-expressing embryos, n = 2,791 values for Kif5C-expressing embryos, n = 2,132 values Kif5C T94N-expressing embryos, and n = 2,187 values for Kif1C-5C–expressing embryos. (H and I) Kif1C-5C reduces overall THN motility. Elapsed time in minutes. Scale bars, 20 µm. n = 24 embryos/145 tracks for control and n = 20 embryos/142 tracks for Kif1C-5C–expressing embryos. Kruskal–Wallis ANOVA: P = 0.007. See also Video 4. (J) Models of how MTs and motors contribute to neuronal migration, with nucleokinesis on the left and the spatiotemporal distribution model on the right. MHB, solid line; URL, dotted line. Boxes in graphs represent 25–75% of all values and whiskers 1.5 times the quartile. Median is shown as a horizontal bar and mean as a square box. Significance level in Kruskal–Wallis ANOVA: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant. All data shown in E–G represent n = 13 embryos for injection control, n = 14 p50-expressing embryos, n = 11 Kif5C-expressing embryos, n = 16 Kif5C T94N-expressing embryos, and n = 9 Kif5C-1C–expressing embryos.
Imbalanced cargo transport reduces THN motility.(A) Schematic representation of Kif1C-5C chimera. (B and C) In THNs, the chimera localizes to the front of phase 1 (B) and phase 2 (C) cells. Cyan box indicates magnified region on the right. Scale bars represent 20 µm in overview images and 5 µm in magnified images. (D) Kif1C-5C does not colocalize with Cadherin-2 particles, although they are frequently found in the vicinity. Colored boxes indicate magnified regions on the right. Scale bars represent 10 µm in the overview image and 1 µm in magnified images. (E) The expression of Kif1C-5C increases stationary times of Cadherin-2 particles. For full statistical testing, see Table S2; n = 416 tracks for injection control, n = 308 tracks for p50-expressing embryos, n = 593 tracks for Kif5C-expressing embryos, n = 441 tracks Kif5C T94N-expressing embryos, and n = 439 tracks for Kif1C-5C–expressing embryos. (F and G) Kif1C-5C also reduces maximal velocities and mean velocities (F; area of 0.8 data density in top left corner and number of tracks in lower left corner; see also Video 6) as well as instantaneous velocities (G). Note that the retrograde direction is more strongly affected than the anterograde direction, as evidenced by the comparison to other motor-affecting treatments. For full statistical testing, see Table S3 and Table S4. Anterograde: n = 2,059 values for injection control, n = 1,044 values for p50-expressing embryos, n = 2,808 values for Kif5C-expressing embryos, n = 2,055 values Kif5C T94N-expressing embryos, and n = 2,245 values for Kif1C-5C–expressing embryos. Retrograde: n = 2,127 values for injection control, n = 1,117 values for p50-expressing embryos, n = 2,791 values for Kif5C-expressing embryos, n = 2,132 values Kif5C T94N-expressing embryos, and n = 2,187 values for Kif1C-5C–expressing embryos. (H and I) Kif1C-5C reduces overall THN motility. Elapsed time in minutes. Scale bars, 20 µm. n = 24 embryos/145 tracks for control and n = 20 embryos/142 tracks for Kif1C-5C–expressing embryos. Kruskal–Wallis ANOVA: P = 0.007. See also Video 4. (J) Models of how MTs and motors contribute to neuronal migration, with nucleokinesis on the left and the spatiotemporal distribution model on the right. MHB, solid line; URL, dotted line. Boxes in graphs represent 25–75% of all values and whiskers 1.5 times the quartile. Median is shown as a horizontal bar and mean as a square box. Significance level in Kruskal–Wallis ANOVA: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant. All data shown in E–G represent n = 13 embryos for injection control, n = 14 p50-expressing embryos, n = 11 Kif5C-expressing embryos, n = 16 Kif5C T94N-expressing embryos, and n = 9 Kif5C-1C–expressing embryos.
To assess the effects of the chimera on intracellular traffic, we first overexpressed the construct in THNs and confirmed that the chimera strongly localized to the front of migrating THNs (Fig. 7, B and C). Cadherin-2 particles were often positioned in close proximity to accumulations of the chimera (Fig. 7 D). Direct colocalization was not expected, given that vesicles at their destination are typically advanced to the PM by actin-based mechanisms and do not remain strongly bound to the MT-based motors. Accordingly, the reporter construct for Cadherin-2 showed diffuse labeling as well as bright, large particles in chimera-expressing THNs. We used line-scan analysis to compare relative levels of the reporter in the PM to the brightest particle by drawing lines along the edge of the cells (Fig. S5, A and B). This revealed the tendency of the chimera to increase diffuse levels of Cadherin-2 at the cell periphery relative to the maximal concentration in particles, as well as an increase in particle size (Fig. S5 C). To evaluate this effect, we repeated this analysis on THNs with blocked endocytosis, which therefore retained Cadherin-2 in the PM, on THNs with excess Kif5C WT for normal delivery of Cadherin-2 or in THNs where delivery to the PM was blocked by Kif5C T94N. Out of these conditions, only the Dynasore-mediated endocytosis block produced a similar effect on increasing Cadherin-2 levels in the PM, albeit with a less pronounced effect on particle size. Taken together, these data imply that the chimera does not interfere with delivery of Cadherin-2 to the PM but rather impairs retrograde transport to cause an imbalance in protein localization.
Kif1C-5C chimera interferes with retrograde traffic (related to Fig. 7).(A) Cadherin-2 reporter particles appear larger and the background increased in THNs overexpressing the Kif1C-5C chimera. Yellow dotted lines indicate the area displayed in the line-scan analysis below the images. Scale bar, 10 µm. See also Video 6. (B) Background fluorescence level relative to the brightest Cadherin-2 reporter particle detected in the line-scan analyses suggest that the expression of Kif1C-5C interferes with endocytosis of the marker. n = 14 embryos/31 values for injection control, n = 13 embryos/25 tracks for p50-expressing embryos, n = 8 embryos/21 values for Kif5C-expressing embryos, 18 embryos/n = 30 values Kif5C T94N-expressing embryos, n = 9 embryos/19 values for Dynasore, and n = 22 values for Kif1C-5C–expressing embryos. Kruskal–Wallis ANOVA: P = 0.004 control/Kif5C T94N, P = 4.7 × 10−3 control/Dynasore, P = 6.4 × 10−8 control/p50, P = 6.10−8 Kif1C-5C/p50, and P = 5.10−4 Kif1C-5C/Kif5C T94N. (C) At the same time, large particles appear in chimera-expressing THNs. n = 14 embryos/164 values for injection control, n = 13 embryos/95 tracks for p50-expressing embryos, n = 8 embryos/124 values for Kif5C-expressing embryos, 18 embryos/n = 121 values Kif5C T94N-expressing embryos, n = 9 embryos/64 values for Dynasore, n = 114 values for Kif1C-5C–expressing embryos. Kruskal–Wallis ANOVA: P = 5.8 × 10−6 control/Kif1C-5C, P = 0.008 Kif5C T94N/Kif1C-5C, P = 0.002 control/Dynasore, P = 7.10−5 for Kif1C-5C/p50, P = 5.10−4 Kif1C-5C/Kif5C T94N. (D) Kif1C-5C reduces retrograde mean speeds of Cadherin-2 reporter particles more strongly than anterogradely moving particles. n = 593 tracks for Kif5C-expressing embryos and n = 439 tracks for Kif1C-5C–expressing embryos. (E) Similarly, maximal velocities in the plus direction are unaffected by chimera expression, while minus-directed velocities are strongly reduced compared with control. For full statistical testing, refer to Table S5 and Table S6. Anterograde: n = 399 tracks for injection control, n = 295 tracks for p50-expressing embryos, n = 579 tracks for Kif5C-expressing embryos, n = 425 tracks Kif5C T94N-expressing embryos, and n = 431 tracks for Kif1C-5C–expressing embryos. Retrograde: Anterograde: n = 411 tracks for injection control, n = 299 tracks for p50-expressing embryos, n = 580 tracks for Kif5C-expressing embryos, n = 432 tracks Kif5C T94N-expressing embryos, and n = 433 tracks for Kif1C-5C–expressing embryos. MHB, solid line; URL, dotted line. Boxes in graphs represent 25–75% of all values and whiskers 1.5 times the quartile. Median is shown as a horizontal bar and mean as a square box. Significance level in Kruskal–Wallis ANOVA: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant. All data shown in D and E represent n = 13 embryos for injection control, n = 14 p50-expressing embryos, n = 11 Kif5C-expressing embryos, n = 16 Kif5C T94N-expressing embryos, n = 8 embryos for Dynasore, and n = 9 Kif5C-1C–expressing embryos.
Kif1C-5C chimera interferes with retrograde traffic (related to Fig. 7).(A) Cadherin-2 reporter particles appear larger and the background increased in THNs overexpressing the Kif1C-5C chimera. Yellow dotted lines indicate the area displayed in the line-scan analysis below the images. Scale bar, 10 µm. See also Video 6. (B) Background fluorescence level relative to the brightest Cadherin-2 reporter particle detected in the line-scan analyses suggest that the expression of Kif1C-5C interferes with endocytosis of the marker. n = 14 embryos/31 values for injection control, n = 13 embryos/25 tracks for p50-expressing embryos, n = 8 embryos/21 values for Kif5C-expressing embryos, 18 embryos/n = 30 values Kif5C T94N-expressing embryos, n = 9 embryos/19 values for Dynasore, and n = 22 values for Kif1C-5C–expressing embryos. Kruskal–Wallis ANOVA: P = 0.004 control/Kif5C T94N, P = 4.7 × 10−3 control/Dynasore, P = 6.4 × 10−8 control/p50, P = 6.10−8 Kif1C-5C/p50, and P = 5.10−4 Kif1C-5C/Kif5C T94N. (C) At the same time, large particles appear in chimera-expressing THNs. n = 14 embryos/164 values for injection control, n = 13 embryos/95 tracks for p50-expressing embryos, n = 8 embryos/124 values for Kif5C-expressing embryos, 18 embryos/n = 121 values Kif5C T94N-expressing embryos, n = 9 embryos/64 values for Dynasore, n = 114 values for Kif1C-5C–expressing embryos. Kruskal–Wallis ANOVA: P = 5.8 × 10−6 control/Kif1C-5C, P = 0.008 Kif5C T94N/Kif1C-5C, P = 0.002 control/Dynasore, P = 7.10−5 for Kif1C-5C/p50, P = 5.10−4 Kif1C-5C/Kif5C T94N. (D) Kif1C-5C reduces retrograde mean speeds of Cadherin-2 reporter particles more strongly than anterogradely moving particles. n = 593 tracks for Kif5C-expressing embryos and n = 439 tracks for Kif1C-5C–expressing embryos. (E) Similarly, maximal velocities in the plus direction are unaffected by chimera expression, while minus-directed velocities are strongly reduced compared with control. For full statistical testing, refer to Table S5 and Table S6. Anterograde: n = 399 tracks for injection control, n = 295 tracks for p50-expressing embryos, n = 579 tracks for Kif5C-expressing embryos, n = 425 tracks Kif5C T94N-expressing embryos, and n = 431 tracks for Kif1C-5C–expressing embryos. Retrograde: Anterograde: n = 411 tracks for injection control, n = 299 tracks for p50-expressing embryos, n = 580 tracks for Kif5C-expressing embryos, n = 432 tracks Kif5C T94N-expressing embryos, and n = 433 tracks for Kif1C-5C–expressing embryos. MHB, solid line; URL, dotted line. Boxes in graphs represent 25–75% of all values and whiskers 1.5 times the quartile. Median is shown as a horizontal bar and mean as a square box. Significance level in Kruskal–Wallis ANOVA: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant. All data shown in D and E represent n = 13 embryos for injection control, n = 14 p50-expressing embryos, n = 11 Kif5C-expressing embryos, n = 16 Kif5C T94N-expressing embryos, n = 8 embryos for Dynasore, and n = 9 Kif5C-1C–expressing embryos.
To corroborate this idea, we repeated the Cadherin-2 particle motility analysis and found that overexpression of the chimera strongly increased stationary phases and reduced mean and maximal velocity of Cadherin-2 particles (Fig. 7, E and F). Interestingly, mean, maximal, and instantaneous velocities in the retrograde direction were strongly reduced by the presence of the chimera (Fig. 7 G and,Fig. S5, D and E), comparable to the effect of p50, while anterograde traffic was largely unaffected. Together with the enhanced PM levels of Cadherin-2, this argues that the chimera is able to retain or return cargo destined for recycling to the PM. Such a transport imbalance should lead to reduced THN motility, as Cadherin-2 traffic is important for efficient migration, so next we analyzed THN speed. THNs expressing Kif1C-5C moved at a median speed of 3.40 µm/h, a clear reduction compared with control levels and similar to the speed decreases measured in Kif5C T94N– or Kif1C T104N–overexpressing THNs (Fig. 7, H and I; n = 142 tracks/20 embryos for Kif1C-5C; P = 0.007). We conclude that the overexpression of the chimera with its limited spectrum of protein interactions supports the notion that MT-based vesicle traffic controls subcellular distribution of crucial factors in migration and that maintaining this fine balance is an important task for MTs to ensure efficient neuronal migration (Fig. 7 J).
Discussion
MTs as versatile neuronal migration regulation tools
It has been known for some time that MTs aid neuronal migration, based on findings in cultured neurons and organotypic slice culture (Tsai et al., 2007; Umeshima et al., 2007). The dependence of neurons on MTs varies, from cerebellar granule neurons, which need MTs in addition to actomyosin during glia-guided radial migration (Solecki et al., 2004; Trivedi et al., 2017; Tsai et al., 2007; Umeshima et al., 2007), to retinal ganglion neurons, which use MTs for improved motility but can migrate without them (Icha et al., 2016). Apart from the cell type, differences in tangential versus radial migration, gliophilic versus neurophilic movement, model organisms, and assay types complicate the evaluation of how much and in which way MTs contribute to neuronal migration.
In this study, we confirm that MTs and dynein are essential to the migration of THNs as model for rhombic lip–derived neurons migrating in vivo in a two-phase manner. In phase 1, the centrosome is positioned far behind the nucleus in these cells. This loosely resembles the situation in migrating zebrafish retinal ganglion cells, which also have a rear-positioned centrosome (Icha et al., 2016). These retinal neurons require nucleokinesis along stabilized MTs from the basal process for efficient migration and can move in the absence of MTs by an actin-based backup mechanism. We observed neither of these phenomena in THNs, where the loss of MTs and their motor proteins cannot be compensated for and MT acetylation levels are low. Specifically, we did not observe an acetylation-stabilized cage- or whisk-like structure of MTs in THNs, which has been reported to be a crucial reinforcement for MTs to transmit pulling forces to the nucleus in migrating rodent cerebellar granule cells and zebrafish retinal ganglion cells (Icha et al., 2016; Umeshima et al., 2007). For phase 2 THNs, we found that centrosomal and nuclear positions relative to one another do not correlate with highest motility, in agreement with a previous study (Distel et al., 2010). This does not rule out that a physical link between MTs and the nucleus exists, and we did not observe a detachment of the centrosome from the nuclear environment after the cell rear was fully retracted at the end of phase 1 of migration. The results on centrosomal position in response to external migration stimulation lead us to conclude that such a link might be structural rather than causative of motility. Whether the external stimulation of THN migration primarily stabilizes MT arrays to increase motility or instead acts on other components (e.g., actomyosin), that in turn produce a more stable MT array, is an open question at this point.
In our study, we have investigated dynein and kinesin-1 in their traditional role of organelle carrier in the context of Cadherin-2 traffic. As dynein is the only retrograde organelle carrier in nonmitotic cells (Allan, 2011), the effects observed in THN migration upon dynein inactivation could therefore derive from deficient organelle transport of the nucleus or other cargo. Our results demonstrate that interference with MTs or motors produced reductions on THN motility very similar to interference with endosomal transport or the Golgi apparatus. The molecular machinery underlying the model cargo Cadherin-2 appears to be very similar to the mammalian situation (Heisler et al., 2014; Mary et al., 2002). It consists of two anterograde delivery pathways, one from the Golgi apparatus and the other from perinuclear recycling compartments, which are served by kinesin-1 and an endosomal pathway, whose retrograde direction is supported by dynein. An imbalance in this transport, which we introduced by the expression of the chimeric motor, produced a reduction of overall THN motility. Together, whether MTs are depleted, motors are inactivated, endo- and exocytosis are disabled, or transport is artificially impaired, THN migration is very similarly impaired. This leads us to conclude that in THNs, the spatiotemporal control of protein composition requires a fine balance for efficient migration and that this task is performed at least in part by MTs.
Such a cargo-distribution model for MTs in neuronal migration can be applied to different kinesins and cargo molecules, and our results on the kinesin-3 family member Kif1C would support this view. Classic examples of asymmetrically distributed proteins in migrating cells are integrins (kinesin-3 cargo) and cadherins (kinesin-1 transport), but they include also proteins involved in the perception of extracellular cues, local translation, and cytoskeletal regulators, among others (Cioni et al., 2018; Kapitein et al., 2010; Salomon et al., 2008). The model may apply to neurons which are not derived from the URL, such as THNs, and may even extend to glia cells supporting radially moving neurons. As was demonstrated by Jaarsma et al. (2014), the radial migration of rodent cerebellar granule cells crucially depends on the presence of the dynein adaptor BICD2 in the Bergmann glial cells guiding them, as extracellular matrix and Cadherin-2 are lost from the Bergmann glia surfaces in its absence, and this consequently reduces the migration of the neurons. It is also possible that cargo-driven neuronal migration support is combined with other ways in which MTs support migration. For example, a cross-talk between MTs and the actin machinery has been uncovered in cultivated cerebellar granule cells, where actin contractility changes upon MT depolymerization (Jiang et al., 2015). One could envision that MTs deliver factors to monitor actin at spatially separated sites to coordinate its activity over long distances in such elongated cells. This presents MTs as versatile tools to control neuronal migration in various ways, depending on the needs of the cell.
Targeting cargo
The maintenance of local protein pools and asymmetries often requires a cycle of internalization, transport, and reexport (Kowalczyk and Nanes, 2012). Where transport to distant locations is required, MTs and motor proteins can provide the necessary means (Stehbens and Wittmann, 2012), but this idea requires that a given cargo is correctly deposited at specific locations. In our study, we used Cadherin-2 as a representative of such a dynamically localized factor with regulatory functions in THN migration (Gärtner et al., 2015; Rieger et al., 2009). The surface amount of Cadherin-2 and its local concentration are critical for cortical neuron migration, and they are regulated by internalization and redistribution (Lai et al., 2015; Peglion et al., 2014). Some Cadherin-2 in THNs may be transported along the cell surface by other mechanisms (Rieger et al., 2009), and in our analysis, only a fraction of particles were involved in long-distance transit within the 1-min observation window. In THNs, GFP-Rab11a predominantly localizes to the URL; hence, it appears likely that recycling endosomes are mostly formed at the extreme cell rear in THNs. A kinesin in phase 1 THNs will be able to deliver the protein from the rear to the leading edge of the cell when necessary in a short time, but many Cadherin-2 particles move bidirectionally in THNs. Such bidirectionality is typical of vesicle transport in vivo (see Hendricks et al., 2010; Wacker et al., 1997; Hancock, 2014 for a review and discussion of potential motor interactions). The uniparallel array of MTs in phase 1 THNs dictates that both dynein and a kinesin must be present on the same particle to allow this motility pattern. Consistent with this, maximal and instantaneous velocities of Cadherin-2 particles are simultaneously reduced for both directions when interfering with dynein or Kif5C. Such codependence could explain the strong effects observed on overall migration, as the absence of either motor would be enhanced by disruption of transport in the other direction, and also why defective dynein activation results in stronger inhibition of motility than the block of one kinesin-1.
If vesicles normally move in both directions, then targeting cargo to a specific location can become complicated, especially where MTs reach from a common MTOC into different cellular compartments, as is the case in THNs in phase 2. In mature neurons, posttranslational modifications of MTs and MT-associated proteins (Maps) that preferentially bind to the MT subpopulations are able to guide cargo to specific cell extensions by differentially activating the motors (Kapitein and Hoogenraad, 2015). Some modifications of MTs appear to exist in THNs, although their nature is at present unknown. PST-1P activation did not depolymerize all MTs, arguing that a stabilized pool of MTs exists in migrating THNs. Such resistant MTs have been observed in other cell types (Borowiak et al., 2015; Zenker et al., 2017); therefore, it appears unlikely that they are artifacts of insufficient PST-1P activation. The axon, which in THNs develops from the leading process, is typically enriched in acetylated MTs (Distel et al., 2010), but this posttranslational modification is not very prominent at this early stage of differentiation. A great number of posttranslational modifications and Maps have been implicated in regulating MT stability (Wloga et al., 2017), making it difficult to identify an obvious alternative candidate for MT stabilization in THNs. In future, the characterization of MT posttranslational modifications and Map decoration could help us understand how different MT populations achieve different tasks, such as maintaining morphology and migration, and provide a hint as to which motors are likely to interact with them to further our understanding of how different motors act together to successfully and efficiently direct specific cargoes.
Materials and methods
Animal model
Adult zebrafish were kept in groups of 20–40 individuals in a fish facility (Aquaneering) maintained at ∼700 mS, pH 6.9–7.1, and 28°C with a 14/10 h light/dark cycle as recommended by common zebrafish handling guidelines (Westerfield, 2007). All experiments used fertilized eggs from brass WT parents (Streisinger et al., 1986), the fish line atoh1a:KalTA (hzm2Tg, ZFIN ID: ZDB-ALT-100624-1; Distel et al., 2010), or crosses of atoh1a:KalTA with 4xUAS-GFP (hzm3Tg, ZFIN ID: ZDB-ALT-100809-1; Distel et al., 2009). The atoh1a:KalTA line expresses a zebrafish-optimized Gal4 in precursors and neurons from an atoh1a promoter, which is able to induce the expression of GFP in the 4xUAS-GFP line, or transgenes behind a UAS element after injection. All eggs were kept in Egg Water (0.03 g/liter sea salt) for 1 h before incubation in 30% Danieau medium (0.12 mM MgSO4, 0.21 mM KCl, 0.18 mM Ca(NO3)2, 17.4 mM NaCl, and 1.5 mM Hepes, pH 7.2) at 28°C. To suppress pigmentation, 150 µM phenylthiourea (PTU) was added to the 30% Danieau medium at 8–12 hpf when needed.
All procedures involving animals were performed according to European Union guidelines and German legislation (EU Directive 2010_63, license number AZ 325.1.53/56.1-TU-BS).
Injection
To achieve tissue-specific expression of transgenes, fertilized eggs from atoh1a:KalTA+/− parents were injected at zygote stage with either 25 ng/µl plasmid DNA or 100 ng/µl plasmid DNA together with 100 ng/µl mRNA encoding for Transposase Tol1 using an Eppendorf Femtojet Express Microinjector. Where two plasmids were coinjected, the plasmids were included in equal amounts, not exceeding the given total DNA amounts. All injection mixes contained 0.05% Phenolred (Sigma-Aldrich) for injection control. Approximately 2 nl was injected per zygote. All mRNA was prepared by in vitro transcription following the manufacturer’s instructions (Sigma-Aldrich). Plasmid DNA was prepared following the manufacturer’s protocol (Macherey Nagel). After injection, eggs were transferred to 30% Danieau and incubated at 28°C. Embryos expressing ChR2 were raised in the dark.
For drug injection, 28–32-hpf embryos were embedded laterally in 1.2% ultra-low-melt agarose (type IX-A; Sigma-Aldrich) and kept in 30% Danieau medium supplemented with PTU and 0.02% Tricaine. A few nanoliters of the injection mix were delivered by a capillary inserted in the fourth ventricle close to the URL. The mixes contained Dynasore stock solution or DMSO and 1:10 dilution of 0.05% Phenolred. Embryos were immediately transferred to the microscope.
Electroporation
To test the effect of p50 on THN migration, a larger number of highly expressing cells was obtained by electroporating the desired plasmid following an established protocol (Hoegler et al., 2011): The 24–28-hpf offspring of atoh1a:KalTA+/− parents were washed once in electroporation buffer (180 mM NaCl, 5 mM KCl, 1.8 mM CaCl2, and 5 mM Hepes, pH 7.2), before embedding laterally in 1.5% ultra-low-melt agarose. The embryos were kept in electroporation buffer supplemented with 0.02% Tricaine. Next, 0.5 µg/µl single-plasmid DNA or 0.45 µg/µl for each plasmid in a two-plasmid coinjection was injected into the fourth ventricle, the anode was positioned behind the head, the cathode was positioned anterior to the forehead, and 10 pulses of 5-ms duration were administered at 75 V and ∼1-s intervals (SD9 Stimulator; Grass Technologies). The embryos were subsequently freed from the agarose and kept at 28°C for 8–10 h before recording THN migration in the plasmid-expressing embryos.
Molecular methods
Plasmid construction
All plasmids from this study carry ampicillin resistance cassettes. Plasmids used to express proteins in zebrafish possess Tol1 integration sites and a 5xUAS element, which is flanked on one or both sides by an E1b promoter. In addition, they usually contain a gene for a fluorescent marker, a β-globin intron, and a termination signal. Constructs that were based on similar backbones from a 5xUAS element to visualize the nucleus in zebrafish embryos via fluorescently tagged Histone 2B (#709; Distel et al., 2010) and the PM via CAAX-targeted YFP (#4014; Theisen et al., 2018) and overexpress 3xCitrine-dynamitin/p50 (#3671; Fei et al., 2018) in THNs have been described previously. All numbers refer to an internal plasmid database and should be quoted when material is requested from the authors.
As control for THNs overexpressing a genetic construct, the vector containing Tol1 sites and 5xUAS-E1b-citrine-p50 was digested with StuI and BsrGI to remove the p50 while retaining citrine, DNaseI/Klenow treated, and religated (#4285).
To visualize the PM, the PM-targeting sequence of Fyn kinase was N-terminally attached to tagRFP-T. Using EcoRI and SalI restriction sites, this fusion protein was inserted into a vector containing inverted repeats for Tol1 genomic integration, a 5xUAS element, and an E1b promoter (#3104).
To visualize EB3-GFP comets together with Cdh2-positive particles, an intermediate vector was constructed in the pBluescript backbone. This intermediate combined the reading frames of EB3-GFP (Distel et al., 2010) and Cdh2-reporter-mCherry (Rieger et al., 2009) bidirectionally behind E1b promoter sequences and a joint 5xUAS sequence (#3811). In a second step, an insert fragment containing all these factors was released by BsrGI digest and introduced in a vector that contained Tol1 integration sites and reading frames for EGFP and tagRFP, which had been opened with BsrGI (#3812).
This resulting vector was digested with BglII and SnaBI to exchange the Cdh2-reporter for H2B-mRFP, obtained from pSKH2B-mRFP:5xUAS:EB3-GFP (Distel et al., 2010) by the same restriction enzymes (#3960).
The resulting vector was subsequently modified by adding 4xUAS-E1b-Centrin2-CFP into the SnaBI restriction site (#3965) to be able to record EB3, the nucleus and the centrosome simultaneously. The 4xUAS-E1b-Centrin2-CFP fragment was obtained by SnaBI and SpeI release and subsequent blunt-ending by DNaseI-Klenow treatment from a vector pB 4xUAS-E1b-Centrin2-CFP (#3963), which had been generated by attaching the ECFP reading frame to the 3′ end of cetn2.
Similarly, the construct coexpressing H2B-mRFP, EB3-GFP, and Centrin2-YFP in THNs (#3966) was created by introducing a 4xUAS-E1b-Centrin2-YFP fragment into the SnaBI site on vector #3960. Again, the insert was obtained by SnaBI and SpeI release with subsequent DNaseI-Klenow treatment from a pB-4xUAS-E1b-Centrin2-YFP (#3964) vector.
The plasmid to visualize Map4MTB was created by amplifying the 3′ end of murine map4 (full-length murine map4 vector provided by A. Straube, University of Warwick, Coventry, UK) containing the MT-binding sites and part of the preceding proline-rich region by PCR (forward primer: 5′-TTAGAATTCAAACATGACTTCCACTCGAGTCAAG-3′, reverse primer: 5′-ATTGAATTCGATGCTTGTCTCCTGGATCTG-3′). The primers introduced a zebrafish-optimized Kozak sequence and removed the stop codon. The PCR product was digested with EcoRI and introduced into a single EcoRI site upstream of EGFP in a vector containing Tol1 repeats, a 5xUAS-E1b promoter element, and a β-globin intron after the EGFP ORF (#3875).
To be able to combine Map4MTB with other fluorescently labeled markers, the EGFP was exchanged for mKate2 by opening the vector with BsrGI and NcoI and inserting mKate2, which had been obtained by PCR with forward primer 5′-TCCACCGGTCGCCACCATGGTGAGCGAGCTGATTAAGGAG-3′ and reverse primer 5′-TAAGGTACCTCAAGATCTGTGCCCCAGTTTGC-3′ to introduce NcoI and Acc65I sites, respectively (#3958).
To create a construct that allows the simultaneous labeling of MTs by EB3-GFP and Map4MTB-mKate2, an intermediate construct was generated by adding a 5xUAS-E1b-Map4MTB-mKate2-containing fragment into the SnaBI site on vector #3960 via XmnI/EcoRV digestion and subsequent DNaseI-Klenow treatment (#4016). This plasmid was next opened by XbaI to remove the E1b-H2B-mRFP fragment (#4485).
Similarly, the GFP-containing EcoRI-BsrGI fragment from #3875 was replaced by an mScarlet-encoding fragment generated with the same enzymes to yield pTol1 5xUAS E1b Map4MTB-mScarlet (#5922).
This plasmid was used to insert the Map4MTB-mScarlet fragment via EcoRI and NotI into an EB3-GFP–containing vector with Tol1 repeats. To enable bidirectional expression of the labeled proteins, an E1b-5xUAS-E1b element was next inserted into the central EcoRI site (#5927).
The plasmid to observe Map4MTB-GFP and the PM was created in a three-fragment ligation by opening a vector containing Tol1 sites and E1b-5xUAS-E1b-FyntagRFP with EcoRI and NotI and introducing Map4MTB-GFP from vector #3875 in two fragments cut with EcoRI and SbfI, SbfI, and NotI (#4486).
The optogenetic experiment was performed using a bidirectional plasmid that contained Tol1 sites, ChR2-YFP, 5xUAS with E1b promoters, and Map4MTB-mKate2. It was generated by cutting #3958 with SacII and SacI and introducing E1b-ChR2-YFP using the same sites from a previously described vector with the same vector backbone and a 5xUAS element (Theisen et al., 2018).
Katanin P60–overexpression experiments were based on a vector containing zebrafish katna1. It was obtained by PCR from cDNA generated from whole 48-hpf zebrafish RNA using forward primer 5′-GAGAATTCAAACATGAGTTTGGGGGAGATCAATG-3′ and reverse primer 5′-GACAATTGGCCGCCGGAGCAGGAGCCAAACTCTG-3′. This product was cloned via EcoRI and MfeI digestion into a vector containing Tol1 sites, 5xUAS-E1b promoter, and EGFP (#4779). From this vector, an intermediate construct was generated, which linked the katna1 to 3xmCherry at its 3′ end under the control of a bidirectional E1b-5xUAS-E1b element, flanked by Tol1 sites. To facilitate the insertion of markers to the other E1b location in front of GFP, the vector was cut by BsrGI and religated to change the orientation of backbone to insert and orient a NotI site toward the open position. This site was subsequently used together with XbaI to insert EB3-GFP (#4782), Map4MTB-GFP (#4781), or aTat1-GFP (#4780).
Overexpression of aTat1-GFP was achieved using a plasmid containing Tol1 sites, 5xUAS-E1b, and atat1 fused at the 3′ end to GFP (#4676). The atat1 ORF was amplified by PCR from cDNA of 72-hpf zebrafish embryos using forward primer 5′-CACGAATTCAAACATGGATTTCCCTTACGACCTG-3′ and reverse primer 5′-GAACAATTGGCCTCCATGGAAGCTAAGCCTAGATC-3′. The product was digested with EcoRI and MfeI and introduced into the EcoRI site before the start of the eGFP ORF. The eGFP was subsequently replaced by mScarlet using PciI and BsrGI (#5544).
This vector was used to combine aTat1-mScarlet expression with Map4MTB-GFP for bidirectional expression from a shared 5xUAS element using SnaBI and XbaI (#5920) and Katanin P60-mCFP from 5xUAS-E1b inserted into the SnaBI site (#5932).
An expression plasmid containing the full-length ORF of kif5c fused to EGFP at the 3′ end was generated by PCR from cDNA of 48-hpf zebrafish embryos using forward primer 5′-AGTCAATTGCAAACATGGTGGACGCGGCTGAAT-3′ and reverse primer 5′-ACGCAATTGGCCGCCTGACTTGCTCCTGTGATGGTAC-3′. The product was cut by EcoRI and MfeI, and introduced in the vector containing Tol1 sites, 5xUAS-E1b and EGFP after an EcoRI site (#4677).
To interfere with dynein activation, the 5′-fragment of pafah1b1b corresponding to the first 87 aa of human Lis1 (Tai et al., 2002) was amplified using forward primer 5′-TGAAGCTTCAATGGTGCTGTCACAGAGGC-3′ and reverse primer 5′-CTGTCGACTTACTGCCCCACGGGGCCTC-3′ on cDNA of 2 dpf embryos. The product was digested with SalI and HindIII, cloned into an intermediate vector, and introduced into a vector behind an 5xUAS-E1b-egfp element via BsrGI and SalI to create an N-terminal GFP-fusion (#5928).
This kif5c-containing vector served as template to introduce the point mutation T94N by site-directed mutagenesis using internal primers 5′-GGGAAGACTCACAATATGGAG-3′ together with outside primers created to obtain the WT ORF. The product was cut by EcoRI and MfeI and introduced in the vector containing Tol1 sites, 5xUAS-E1b, and EGFP after an EcoRI site (#4711). The mutation was confirmed by sequencing.
To generate a probe for the expression of Kif5C, the 3′ end of kif5c was amplified from the full ORF-containing vector using forward primer 5′-CTAGAGAGCACGCAGAGCG-3′ and reverse primer 5′-ACGCAATTGGCCGCCTGACTTGCTCCTGTGATGGTAC-3′. This PCR product was cut with PstI and NsiI and inserted in pBluescript, opened with PstI (#4681). Digoxigenin (DIG)-labeled probes of ∼740-bp lengths were synthesized from T7 and T3 promoters.
To obtain the full-length kif1c, the primers 5′-GAGAATTCAAACATGGCGTCGTCCTCAGTGAAAG-3′ and 5′-TACCCATGGCTACACTACCTCCTCCTGAGGTCTCCATTGGTGTGCTAC-3′ were used to amplify the reading frame from cDNA of 1 dpf zebrafish embryos. After EcoRI/NcoI digestion, the product was inserted before gfp into a vector containing Tol1 sites, 5xUAS-E1b and gfp (#4705), or a bidirectional FyntagRFP-E1b-5xUAS-E1b-gfp element (#5514).
A point mutation to introduce the rigor mutation T106N in this construct was achieved using the internal primer pairs 5′-GGGAAATCTTACAACATGATGG-3′, carrying the desired mutation, and amplified with the primers used in the creation of the full-length plasmid. The product was digested with EcoRI to exchange the sequence on the WT-containing plasmids (#5810 and #5811 for coexpression of FyntagRFP).
The overexpression of Rab11a was achieved by introducing the Rab11a zebrafish ORF, derived from 2 dpf embryos, via digestion with BsrGI and XbaI behind the gfp into the pTol1 5xUAS E1b EGFP vector (#5812). Following the usual point mutation protocol, the S25N mutation was introduced by PCR using primers 5′-GGTGTGGGGAAGAATAACCTGCTG-3′ and the primers used to amplify the WT Rab11a. The resulting fragment was digested with BsrGI and XbaI and introduced into the vector backbone (#5813).
To visualize the Golgi apparatus, the fragment containing the Golgi-targeting sequence fused to mCitrine was released from a previously described plasmid from an earlier study (Distel et al., 2010) by EcoRI and BsrGI digest and inserted into the vector containing pTol1 5xUAS-E1b EGFP (#4679). Using a similar strategy, EGFP-Rab5a was released from a vector containing the full-length ORF by BamHI and BsrGI restriction and introduced into the vector containing pTol1 5xUAS-E1b EGFP (#4682).
To colocalize MTs and Cdh2 reporter, a bidirectional coexpression vector was created by opening plasmid #3875 with EcoRV and introducing Cdh2-reporter-mCherry, which was released from #3812 by SmaI digestion (#3961).
To monitor Cdh2-reporter trafficking when dynein is inhibited, a vector containing Tol1 sites, a bidirectional E1b-5xUAS-E1b module, and Cdh2-reporter-mCherry was opened by BglII and XbaI digestion. 3xCitrine-p50 was introduced by these same sites (#4588).
Similarly, the same vector was opened by XmaI and XbaI digestion to introduce Kif5C-GFP (#4680) or Kif5C T94N-GFP (#4712) and opened with BsrGI and XbaI to introduce Rab11a WT (#5815) or Rab11a S25N (#5814).
To create a Kif1C-Kif5C chimeric construct, the motor domain from Kif1C including the neck linker was amplified from #4705 containing the full-length zebrafish ORF for kif1c with forward primer 5′-GAGAATTCAAACATGGCGTCGTCCTCAGTGAAAG-3′ and reverse primer 5′-GGTAGTTTTTGCTGAGGAGCTCCTCCACTTTGTCCTCGAGCTGTTTCATAGCTCCTCCTGCATGCGTTTTCTCTGCATCAGAGC-3′. The reverse primer contained the N-terminal half of the leucine zipper from yeast GCN4. The remaining half was attached to the cargo-binding domain of the Kif5C heavy chain, which comprised aa 821–985 of zebrafish Kif5C, by primers 5′-GGAGGAGCTCCTCAGCAAAAACTACCACCTCGAGAACGAGGTGGCCAGACTCAAAAAACTCGTGGGAGAGAGAGGAGGAGCTGCATGCGAGAAGATATTCCCAGTTGGAAG-3′ and 5′-ACGCAATTGGCCGCCTGACTTGCTCCTGTGATGGTAC-3′. Both PCR products were fused by PCR, and the combined product was digested with EcoRI and MfeI and inserted in front of eGFP in the vector containing Tol1 sites, 5xUAS, E1b, and egfp to create a C-terminal label (#5921) or in the bidirectional vector for coexpression with Fyn-tagRFP for PM labeling (#5953). To visualize Cadherin-2 particles and the chimera, the Map4MTB-EGFP on vector #3961 was replaced by the chimera-EGFP-containing fragment by XbaI and XmaI restriction. The E1b-5xUAS-E1b cassette was subsequently reintroduced by EcoRI and XbaI (#5957).
Inhibitors
All cytoskeleton inhibitors and ACh were prepared as stock solutions and stored at −20°C in single-use aliquots, with the exception of Brefeldin A, which was purchased as 1,000× stock solution and stored at 4°C. Stock solutions had the following concentrations: Brefeldin A (420601; Biolegend) 5 mg/ml in DMSO; (−)-Blebbistatin (B0560; Sigma-Aldrich) 10 mM in DMSO; colchicine (C9754; Sigma-Aldrich) 25 mg/ml in 30% Danieau; Dynasore (324413; Sigma-Aldrich) 50 mM in DMSO; PST-1P (provided by O. Thorn-Seshold) 5 mM in 30% Danieau; ACh 550 mM in water (A6625; Sigma-Aldrich). Aliquots were dissolved in 30% Danieau medium immediately before use, and embryos were recorded either immediately, after a 2–4-h incubation period for PST-1P or Brefeldin A treatments, or 5 h or longer for colchicine treatments.
Immunohistochemistry
To detect acetylated MTs in situ, embryos were fixed following a protocol by McMenamin et al. (2003). 28–30-hpf dechorionated embryos were incubated in MSB (80 mM Pipes, pH 6.8, 5 mM EGTA, 1 mM MgCl2, 3.7% PFA, 0.25% glutaraldehyde, and 0.2% Triton-X100) for 3–4 h at RT. Subsequently, the solution was removed, and any remaining buffer quenched by the addition of freshly prepared 1 mg/ml NaBH4/PBS and incubation for 40 min at RT. Next, embryos were washed twice with PBS and permeabilized by incubation in PBS containing 0.5% Triton X-100 for 1 h, followed by incubation in −20°C acetone for 20 min. The acetone was removed, and the embryos were washed twice with PBS + 0.1% Triton X-100. Next, unspecific antibody binding was prevented by incubation in PBS + 0.1% Triton X-100 + 10% normal goat serum (NGS) for 2–3 h at RT. After this blocking step, the embryos were incubated in PBS + 0.1% Triton X-100 + 10% NGS containing mouse anti-acetylated α-Tubulin (clone 6–11-B1; T6793; Sigma-Aldrich) at 1:2,000 dilution ON at 4°C. The following day, the embryos were washed six times in PBS + 0.1% Triton X-100 before incubating them in PBS + 0.1% Triton X-100 + 10% NGS, containing goat anti-mouse IgG Cy3 (115–165-003; Research Resource Identifier [RRID]: AB_2338680; Jackson ImmunoResearch) in a dilution of 1:1,000 for 2 h at RT. Next, the embryos were washed three times in PBS before embedding them laterally in 1.2% ultra-low-melt agarose and imaging them immediately.
WISH and FISH
Whole-mount in situ hybridization (WISH) was performed following a previously described method (Theisen et al., 2018). Briefly, WT embryos were fixed at 30 hpf in 4% PFA/PBS ON at 4°C. DIG-labeled probes were created by in vitro transcription from the T7 (sense) and T3 promoters (antisense) with DIG labeling mix (Roche) according to the manufacturers’ instructions. Likewise, Fluorescein-labeled probes were generated using in vitro transcription and Fluorescein RNA Labeling Mix (Sigma-Aldrich).
For WISH, WT embryos were permeabilized with 2% H2O2 and 10 µg/ml Proteinase K. Embryos were prehybridized by incubating them in hybridization buffer (50% formamide, 4× SSC, 50 µg/ml heparin, 50 µg/ml torula RNA, and 0.1% Tween-20) for 6 h at 65°C before adding the probe and further incubation ON. After several washes in 50% formamide in 2× SSC, 2× SSC, and 0.2× SSC at 65°C, the embryos were preadsorbed by adding 10% NGS in PBST, followed by ON incubation with HRP-coupled anti-DIG antibody (11093274910, RRID: AB_2734716; Sigma-Aldrich) at 1:2,000. For detection of the antibody, the embryos were kept in staining buffer (100 mM NaCl, 100 mM Tris, pH 9.5, 50 mM MgCl2, and 1% Tween-20) with 5 µg/µl nitro blue tetrazolium and 3.75 µg/µl BCIP. After the reaction was stopped by washing the embryos several times in PBST, the embryos were transferred to 90% glycerol and imaged after several days.
For FISH, we used custom-made FLUO- and TAMRA-tyramide conjugates, as described previously (Lauter et al., 2011; von Trotha et al., 2014). The atoh1a:KalTA/4xUAS-GFP–positive embryos were treated as described for WISH until hybridization, when they were incubated with a DIG-labeled probe against kif5c and a fluorescein-labeled probe against gfp ON at 65°C in hybridization buffer (50% formamide, 5× SSC, 2.5% dextran sulfate, 50 µg/ml heparin, and 0.5 mg/ml torula RNA). After several washes in hybridization buffer (2× SSC, 2× SSC, 0.2× SSC, and PBST), the embryos were blocked for several hours in maleic acid buffer (0.1 M maleic acid and 0.15 NaCl, pH 7.5) + 10% blocking agent (Roche) before adding anti-FLUO peroxidase (11426346910; Sigma-Aldrich), 1:1,000, and incubation overnight. After several washes in PBST, the antibody was detected by incubation in TAMRA-tyramide staining solution (0.4% TAMRA-tyramide, 0.003% H2O2, and 4% of 50% dextran sulfate in PBST) for 30 min at RT. To stop the reaction, the embryos were washed twice in PBST and 100 mM glycine, pH 2, and several additional washes in PBST. Next, the embryos were again blocked by maleic acid buffer + 10% blocking reagent for 4 h, before adding anti-DIG peroxidase (11207733910; Sigma-Aldrich) at 1:1,000. The reaction was allowed to proceed ON. The detection of the antibody was performed as described above, using a FITC-tyramide staining solution (0.5% FITC-tyramide, 0.003% H2O2, and 4% of 50% dextran sulfate in PBST). Finally, the embryos were washed several times in PBST, embedded, and recorded by confocal imaging.
Image recording
In vivo imaging
As the atoh1a promoter drives expression in URL-derived cells in the cerebellum from 17 hpf (Distel et al., 2010), embryos were selected for imaging from 24 hpf using a stereofluorescence microscope (Leica M205FA or MDG41). These systems consist of various illumination modes for transmitted light and fluorescence imaging and flexible magnification ranging from ∼10× to 120×. They were also used to record the effects of 5 mg/ml colchicine on the embryos using a Leica DCF365 FX camera, controlled by Leica Application Software X software. For image recording, the chorion was removed with forceps. All embryos were sedated for embedding and imaging in 30% Danieau + 150 µM PTU + 0.02% Tricaine for 10 min at RT. Next, up to 14 embryos were embedded in lateral position in 1.2% ultra-low gelling agarose (type IX-A, Sigma-Aldrich) in 30% Danieau. The embryos were transferred to a confocal microscope and kept in 30% Danieau + 150 µM PTU + 0.01% Tricaine for the duration of the experiment (Leica TCS SP8 DMI 6000, Zeiss Airyscan LSM 880). On the confocal, whole embryo images were created by transmitted light and fluorescent imaging on a 10× objective (Plan-Apochromat 10×/0.3 M27) on the Zeiss microscope or a 10× objective on the Leica system (HC PL APO 10×/0.4) and subsequent tile/grid stitching using a plugin for ImageJ (Preibisch et al., 2009). Embryos observed for >6 h were imaged using a 20×/25× oil immersion objective (Leica HC PL APO CS2 20×/0.75 IMM, Zeiss Plan-Apochromat 25×/0.8 IMM Korr DIC M27), shorter experiments such as standard migration assays used a 40× water objective (Leica HC PL APO CS2 40×/1.2 water, Zeiss Plan-Apochromat 40×/1.2 IMM Korr DIC M27), and EB3 visualization was performed with a 63× water objective (Leica HC PL APO CS2 63×/1.2 water, Zeiss LD C-Apochromat 63×/1.15 W Korr M27). Both systems were equipped with a 405-nm diode, an Argon laser to power several lines in the cyan to yellow range, and 561-nm and 633-nm lasers. All experiments were performed at 28°C using a Life Imaging Services heating chamber on the Leica system or an XLmulti S1 incubator on the Zeiss system. Images were recorded at single z-planes for EB3-, Map4MTB-, or Cadherin-2–tracking experiments, and in z-planes of different step-sizes for THN migration assays to cover the cerebellum. Typically, these ranged from 2.5 µm to 5 µm. A standard migration assay recorded THNs over 4 h at 10-min intervals. Image acquisition was controlled by the Leica Application Software X software (for all Leica-based systems) or Zen Black (Zeiss Airyscan LSM 880).
Fixed-sample recording
Embryos stained in WISH produced a purple precipitate which was recorded in 90% glycerol medium using transmitted light on a Leica Leitz DM RBE fluorescence microscope equipped with a 10× objective (Leica HC PL APO 10×/0.4) and a Nikon DS-Vi1 camera, controlled by Nikon NIS-elements software.
Embryos prepared by double-fluorescent in situ hybridization were recorded on the Leica confocal in PBS directly after preparation, using the 20× objective (Leica HC PL APO CS2 20×/0.75 IMM). TAMRA-coupled antibodies against GFP were used to identify atoh1a–positive cells, while FITC-coupled antibodies were directed against the DIG-labeled probe for kif5C. Whole-body images were constructed in ImageJ from individual tiles. Likewise, embryos stained for acetylated MTs were recorded immediately after staining on the same Leica confocal, but using the 40× objective (HC PL APO CS2 40×/1.2 water), keeping the embryos in PBS during recording. Acetylated MTs were detected using antibodies conjugated with Cy3, as expressed constructs for control or aTat1 were tagged with citrine, which was detected in the green/yellow spectrum.
Optogenetics
To investigate centrosome-nucleus position coupling, 28-hpf embryos expressing ChR2-YFP were recorded by confocal light microscopy as described previously (Theisen et al., 2018). Briefly, THNs on the Leica confocal system (HC PL APO CS2 40×/1.2 water) were exposed to 15% of the 476-nm laser line to open the channel to depolarize the cells, which were identified by 0.5–1.5% illumination at 514 nm. This corresponds to an illumination of 6.57 µs/pixel or 1.64 ms/cell of channel opening times for an average-sized THN. The illumination was repeated at 1 min intervals for a total duration of 4 h.
Photopharmacology
For photopharmacological experiments, embryos expressing Map4MTB and Fyn-tagRFP were dechorionated at 26–27 hpf and incubated in 25 µM PST-1P in 30 Danieau + 150 µM PTU for 3–4 h in the dark at 28°C. Next, the embryos were embedded in low-melt agarose and 30% Danieau + PTU + 0.01% Tricaine + 25 µM PST-1P and transferred to the Leica confocal microscope (HC PL APO CS2 40×/1.2 water) in a dark room. As the light during the embedding process may have converted some PST-1P, the cerebellar primordia of these embryos were recorded using only the 561-nm laser continuously for 30 min. PST-1P has a half-life of ∼6 min at 37°C without UV conversion; hence, all active molecules should revert to their inactive state during this time, and cells have time to recover from any damage caused by PST-1P activation during embedding. Morphologically, atypical cell rounding identified by the PM-targeted tagRFP-T disappeared during this step. Next, the cerebellar primordium was recorded in ROIs of different illumination settings, so that one region at the MHB received 405-nm illumination to activate the PST-1P and 488 nm to visualize Map4MTB-GFP, the rest of the tissue only 561 nm illumination to keep PST-1P inactive and to excite Fyn-tagRFP-T for control. For MT polymerization control, EB3-GFP– or Map4MTB-GFP–expressing embryos were recorded with 405-nm and 488-nm illumination only. THNs were recorded for 1 h at 3-min frame intervals for MT depolymerization control, and for 4 h at 3-min intervals for subsequent migration assays. For PST-1P activation and Cadherin-2-reporter imaging, the frame interval was increased to 1.3 s and a 5-min duration, of which only the final 1 min was analyzed, and the recorded region was magnified using the zoom function to improve spatial resolution.
Software for image processing
Images were processed for brightness/contrast, cropped, and annotated using ImageJ/FIJI (Schindelin et al., 2012) and Adobe Photoshop software packages. Line scans and kymographs were generated using standard ImageJ/Fiji plugins. For film presentation in all figures and videos, frames were realigned using the rigid body method, part of the HyperStackReg plugin for ImageJ (DOI 10.5281/zenodo.2252521). Tracking of organelles and cells was performed using either the Manual Tracking plugin or MTrackJ in ImageJ. EB3 tracks in Fig. 1, D–F were visualized using the MTrackJ plugin. Images of whole embryos were reconstructed from tiled images using the Grid/Collection Stitching plugin for ImageJ (Preibisch et al., 2009). Schematic drawings including models were created in CorelDraw. Figures were arranged in CorelDraw.
Quantification and statistical analysis
EB3 and Map4MTB tracking
Embryos expressing EB3-GFP in THNs were assessed by their comet-shape localization at MT tips for adequate expression levels. MT lattice decoration was considered excess levels, and such cells were excluded from analysis. Films were recorded for 2–20 min at 1–3 s frame intervals in single z-planes. The EB3-GFP comets were manually tracked using the MTrackJ plugin for ImageJ. The tracks were plotted and false-colored according to their intracellular growth direction to give the images in Fig. 1, F–H. To determine MT growth speeds, films were recorded for 1 min at 1.2-s frame intervals in single z-planes using the Zeiss Airyscan detector. The EB3-GFP comets as well as growing ends of Map4MTB-mScarlet–decorated MTs at the cell periphery were either manually tracked using the MTrackJ plugin for ImageJ or MT growth speeds were determined from kymographs over the same MT plus ends to give the results depicted in Fig. S1, G–J. Growing MT ends at the cortex were either tracked in 2D (Fig. S1, G and J) or growth speeds were determined from kymographs (Fig. S1, H and I).
Centrosome tracking
To determine the centrosome’s position relative to the nucleus, Map4MTB-GFP or Cetn-2-YFP was expressed in THNs by microinjection at the zygote stage. At 34–38 hpf, the embryos were recorded for 4–8 h at frame rates of 5–10 min, using z-stacks to cover one half of the developing cerebellum in 1–2 µm z-planes. Using the MTrackJ plugin of ImageJ, the position of the centrosome was tracked, as well as the center of the nucleus. Any change in the relative position of the centrosome and the nucleus was marked as “shift” event, and THN speed was calculated in a 30-min window with the event at its center, as well as 30 min preceding and following this time window. A tissue-shift correction was applied (see below). Next, Map4MTB-mKate2 was coexpressed with ChR2-YFP in order to induce a THN migration speed increase by depolarization. The imaging conditions followed the protocol described in (Theisen et al., 2018), relying on 476-nm illumination to open the channel for 1.64 ms per average-sized THN per 1 min. The assay was repeated three times for each condition. The results from this analysis is shown in Fig. 2.
THN tracking
At 27–34 hpf, THN migration was recorded by confocal imaging in z-stacks of typically 5-µm slices and 10-min intervals for 4–8 h. As the embryos continue their normal development during several hours of recording, a displacement of the cerebellar tissue due to natural growth is often observed. This shift is highly individual; hence, tracks from different embryos were corrected for this displacement. Therefore, eight positions at the URL and the MHB were tracked using Manual Tracking in ImageJ and averaged to give a mean displacement of the tissue for each point of the time series. These values were then subtracted from the tracks of THNs, and THN speeds calculated using the bee lines between start and end positions divided by elapsed time. Only THNs which were reliably identified for a minimum of 30 min were included in the analysis.
The assay was repeated three times for each condition investigated, except for the colchicine treatments, which were performed once for each condition for ethical reasons (for statistic parameters, please refer to Table S1). The tracks from all cells are represented in box plots in Figs. 3, 4, 5, 7, S2, and,S3.
Cadherin-2 tracking
Quantitative analysis of Cadherin-2 particle transport was performed in embryos of 27–33 hpf, expressing bidirectional genetic constructs that include a factor to be tested and the Cadherin-2 reporter that has been characterized previously (Rieger et al., 2009). THNs in phase 1 were recorded for 1 min at 0.9–1.4-s intervals. Next, all Cadherin-2 reporter particles in a cell were tracked using MTrackJ of ImageJ, and two marker points were placed on the tissue: one in front of the elongated THNs, representing the (+)-growth direction of MTs, and the other at the URL, the (−) ends of MTs. These marker points were used to create a reference axis in a custom-made software program. Directional movement along this axis was determined for each step. Particle mean speeds for each track were calculated using the distances of end point to start point along the reference axis. Maximal anterograde or retrograde velocities were extracted from all individual step velocities in each direction per track. For unidirectional particles, only one value was included in the dataset. Instantaneous velocities were calculated from distances summing all individual steps in the same direction until the particle either stalled or changed direction along the reference axis. Software developed for this work is available from an online repository (https://github.com/tobiasring/zebrafish-particletracks.git).
Statistical analysis
Boxes in graphs depicting THN migratory speeds or Cadherin-2 particle parameters represent 25–75% of all values and the whiskers 1.5 times the quartile. Median is shown as horizontal bar, mean as square box. All tracking data distributions were analyzed for their Gaussian distribution using a Shapiro-Wilk test, and significance levels calculated using a nonparametric Kruskal–Wallis ANOVA test contained in OriginPro 2019 (OriginLab Corporation) software for all datasets except for MT growth speeds, which were assessed using ANOVA with Bonferroni correction as normal distributions were indicated (Fig. S1, I and J). All conditions were tested against their respective control levels. Significance levels tested are P < 0.05 indicated in figures as *, P < 0.01 as **, or P < 0.001 as ***. Table S1 lists P and χ2 values for THN migration experiments; Tables S2–S6 show values for Cadherin-2 tracking.
Data and software availability
All data supporting the findings of this study are available within the publication and its supplementary information files. All materials generated, including genetic constructs and transgenic fish lines will be made available upon request to the corresponding author.
Online supplemental material
The supplemental figures demonstrate data for the MT lattice marker Map4MTB (Fig. S1), the colchicine treatments (Fig. S2), additional controls and experiments on vesicle traffic in THN migration (Fig. S3), Cadherin-2 tracking parameters and PST-1P in Cadherin-2 tracking (Fig. S4), and Cadherin-2 localization in THNs overexpressing the chimera (Fig. S5). Video 1 shows that EB3 and Map4MTB label MTs during THN migration. Video 2 shows that the centrosome changes position relative to the nucleus regularly in control THNs but remains more stable when THN speed is increased by ACh. Video 3 shows that 25 µM PST-1P reduces MTs in the activated region (green), leading to a decrease in THN migratory speed. Video 4 shows that interfering with the function of dynein by overexpression of p50, Kif5C by overexpression of the rigor mutation T94N, or intracellular transport by expression of the chimera Kif1C-5C reduces THN speed. Video 5 shows that Golgi dispersal by Brefeldin A or block of endosome formation by Dynasore injection strongly impairs THN migration. Video 6 shows that Cadherin-2 particles move dynamically in control THNs, but motility is reduced, and motor function or the formation of recycling endosomes is impaired. In addition, the supplement contains several tables providing information on the number of animals used for the different experimental conditions, the number of tracks analyzed, and the results from statistical testing. Table S1 shows statistic parameters for THN migratory speed assays using Kruskal–Wallis ANOVA. Table S2 shows statistic parameters for Cadherin-2 particle tracking relative pause times using Kruskal–Wallis ANOVA. Table S3 shows statistic parameters for Cadherin-2 particle tracking (+)–directed instantaneous velocities using Kruskal–Wallis ANOVA. Table S4 shows statistic parameters for Cadherin-2 particle tracking (−)–directed instantaneous velocities using Kruskal–Wallis ANOVA. Table S5 shows statistic parameters for Cadherin-2 particle tracking maximal speed in the (+) direction using Kruskal–Wallis ANOVA. Table S6 shows statistic parameters for Cadherin-2 particle tracking maximal speed in the (−) direction using Kruskal–Wallis ANOVA. For improved visualization, six videos are included showing THN migration and Cadherin-2 particle transport under different conditions.
Acknowledgments
The authors would like to thank Anne Straube for the Map4 template, Jakob von Trotha for help with the FISH protocol, Kazuhiko Namikawa for molecular biology advice, and Timo Fritsch for management of the fish lines.
U. Theisen was supported by a European Union Marie Curie Fellowship (FP7, 623612 Cdh2_neuromigration).
The authors declare no competing financial interests.
Author contributions: U. Theisen perceived the project, carried out experiments, analyzed data, and wrote the manuscript. A.U. Ernst and R.L.S. Heyne conducted experiments and performed some data analysis. T.P. Ring analyzed vesicle transport data by programming specific software. O. Thorn-Seshold provided PST and expertise in drug handling and reviewed the manuscript. R.W. Köster intellectually and financially supported the project from the inception and contributed to the discussion and review of the manuscript.