Tim17 is a central, membrane-embedded subunit of the mitochondrial protein import machinery. In this study, we show that Tim17 contains a pair of highly conserved cysteine residues that form a structural disulfide bond exposed to the intermembrane space (IMS). This disulfide bond is critical for efficient protein translocation through the TIM23 complex and for dynamic gating of its preprotein-conducting channel. The disulfide bond in Tim17 is formed during insertion of the protein into the inner membrane. Whereas the import of Tim17 depends on the binding to the IMS protein Mia40, the oxidoreductase activity of Mia40 is surprisingly dispensable for Tim17 oxidation. Our observations suggest that Tim17 can be directly oxidized by the sulfhydryl oxidase Erv1. Thus, import and oxidation of Tim17 are mediated by the mitochondrial disulfide relay, though the mechanism by which the disulfide bond in Tim17 is formed differs considerably from that of soluble IMS proteins.
Introduction
Mitochondria consist of 800–2,000 proteins, most of which are synthesized on cytosolic ribosomes as precursor proteins carrying N-terminal matrix-targeting signals or presequences. These presequences are recognized by receptors of the translocase of the outer membrane of mitochondria and threaded through the protein conducting channels in the outer and inner membrane (Neupert and Herrmann, 2007; Chacinska et al., 2009; Endo et al., 2011; Harbauer et al., 2014; Schulz et al., 2015). Protein translocation is driven by TIM23 (translocase of the inner membrane of mitochondria 23) translocase of the inner membrane that is functionally and physically linked to the ATP-dependent import motor. Although several studies characterized the properties of the import motor, we lack any deeper insight into the structure and function of the two multispanning membrane-embedded subunits of the TIM23 complex, Tim23 and Tim17 (Maarse et al., 1994; Ryan et al., 1998; Alder et al., 2008; Malhotra et al., 2013). Both proteins are structurally related and comprise four transmembrane spans showing characteristic glycine patterns (Demishtein-Zohary et al., 2015). Reconstitution experiments with purified Tim23 as well as patch-clamp analyses of Tim17-depleted inner membranes suggested that Tim23 is able to form a preprotein-responsive channel, although it differs in its behavior from the endogenous TIM23 channel (Truscott et al., 2001; Meinecke et al., 2006; Martinez-Caballero et al., 2007; van der Laan et al., 2007). Tim17 plays an essential role in preprotein translocation, but it is unclear whether Tim17 is part of the TIM23 channel or serves as its regulator.
In addition to this matrix-targeting pathway, alternative import routes are used by certain groups of mitochondrial proteins. Several inner membrane proteins, including members of the mitochondrial carrier family and Tim23, use an alternative inner membrane translocase, the TIM22 complex, for their integration into the inner membrane (Káldi et al., 1998; Rehling et al., 2004). Many proteins of the intermembrane space (IMS) are imported by the Mia40 pathway or mitochondrial disulfide relay (Mesecke et al., 2005; Dabir et al., 2007). Here, the oxidoreductase Mia40 binds to incoming polypeptides via a hydrophobic binding cleft (Naoé et al., 2004; Banci et al., 2009; Kawano et al., 2009; Koch and Schmid, 2014) and introduces disulfide bonds into the preproteins. It was proposed that substrate oxidation is mechanistically linked to protein translocation (folding ratchet; Lutz et al., 2003; Allen et al., 2005; Milenkovic et al., 2007). Recently, two studies presented evidence that the central membrane protein of the TIM22 complex, Tim22, contains a disulfide bond that is formed in a Mia40-dependent fashion (Wrobel et al., 2013; Okamoto et al., 2014). The physiological relevance of this disulfide is not known.
Here, we show that Tim17 contains a structural disulfide bond that is conserved among eukaryotes. The oxidoreductase Mia40 is essential for the import of Tim17, but not for its oxidation. Once formed, the disulfide bond in Tim17 is very stable and serves a structural function that is crucial for TIM23 functionality, particularly at elevated temperatures. Our data are consistent with a role of Tim17 as a central gating element that provides the TIM23 channel with its dynamic properties essential for mitochondrial protein import.
Results
Tim17 contains a conserved disulfide bond
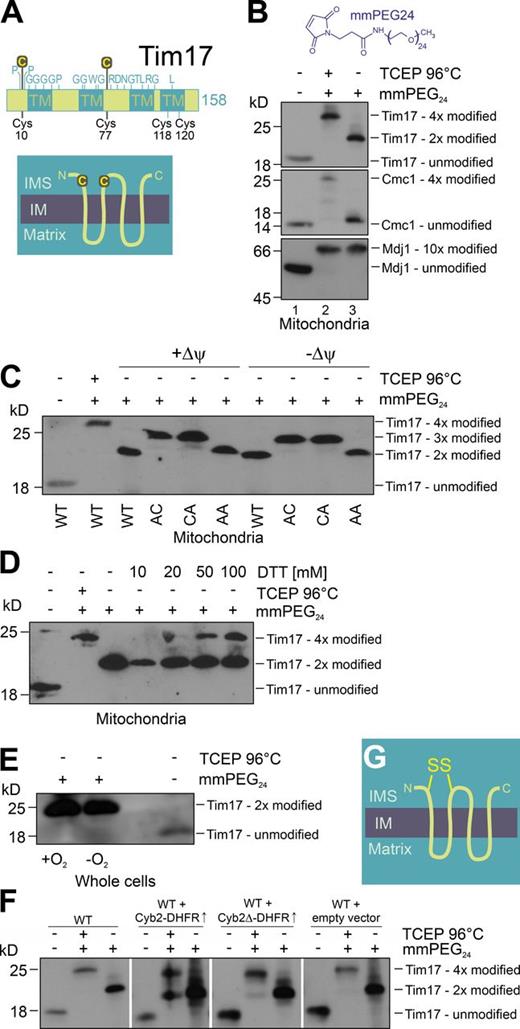
Members of the Tim17 family show a very high degree of sequence conservation, particularly in the N-terminal two thirds of the protein (Fig. S1; Meier et al., 2005). Several amino acid residues are strictly conserved among eukaryotes (Fig. 1 A). These residues comprise glycine patterns in the transmembrane domains that are characteristic for membrane-embedded TIM subunits (Demishtein-Zohary et al., 2015). In addition, two cysteine residues are found in all Tim17 orthologues, one N terminal of the first transmembrane domain and one C terminal of the second transmembrane domain (C10 and C77 in the yeast Tim17, respectively). Both residues are exposed to the IMS and might potentially be in close proximity in the membrane-embedded protein (Fig. 1 A, topology model).
Tim17 contains a structural disulfide bond. (A) Schematic representation of the Tim17 sequence. Invariant residues (see Fig. S1) are indicated above and cysteine residues are indicated below the representation. TM, transmembrane segments. The inset shows the topology of Tim17 and the position of the two conserved cysteine residues. (B) Mitochondrial proteins were TCA precipitated and incubated with the reductant TCEP and/or the thiol-modifying compound mmPEG24. The samples were resolved by SDS-PAGE and used for Western blotting with antibodies against Tim17, Cmc1 (a protein with four oxidized cysteines), or Mdj1 (a protein with 10 reduced cysteines). (C) Mitochondria were isolated from Tim17 mutants in which the indicated cysteines were replaced by alanine residues and used for an mmPEG24 shift experiment as described for B. WT, wild type; AC, Tim17AC; CA, Tim17CA; AA, Tim17AA. (D) To assess the stability of the disulfide bond in Tim17, wild-type mitochondria were pretreated with the indicated concentrations of DTT for 15 min at 28°C. Mitochondrial proteins were TCA precipitated before the redox states of the cysteines in Tim17 were analyzed as described for B. (E) Yeast cells were grown in the presence or absence of molecular oxygen. Cellular proteins were TCA precipitated and the redox state of the cysteines in Tim17 was analyzed. (F) Strains were generated that overexpress Cyb2-dihydrofolate reductase (DHFR) and Cyb2Δ-DHFR from a GAL10 promoter. Mitochondria were isolated from these strains as well as from controls. The redox state of Tim17 was analyzed as described for B. (G) Topology of Tim17 indicating the position of the disulfide bond. IM, inner membrane.
Tim17 contains a structural disulfide bond. (A) Schematic representation of the Tim17 sequence. Invariant residues (see Fig. S1) are indicated above and cysteine residues are indicated below the representation. TM, transmembrane segments. The inset shows the topology of Tim17 and the position of the two conserved cysteine residues. (B) Mitochondrial proteins were TCA precipitated and incubated with the reductant TCEP and/or the thiol-modifying compound mmPEG24. The samples were resolved by SDS-PAGE and used for Western blotting with antibodies against Tim17, Cmc1 (a protein with four oxidized cysteines), or Mdj1 (a protein with 10 reduced cysteines). (C) Mitochondria were isolated from Tim17 mutants in which the indicated cysteines were replaced by alanine residues and used for an mmPEG24 shift experiment as described for B. WT, wild type; AC, Tim17AC; CA, Tim17CA; AA, Tim17AA. (D) To assess the stability of the disulfide bond in Tim17, wild-type mitochondria were pretreated with the indicated concentrations of DTT for 15 min at 28°C. Mitochondrial proteins were TCA precipitated before the redox states of the cysteines in Tim17 were analyzed as described for B. (E) Yeast cells were grown in the presence or absence of molecular oxygen. Cellular proteins were TCA precipitated and the redox state of the cysteines in Tim17 was analyzed. (F) Strains were generated that overexpress Cyb2-dihydrofolate reductase (DHFR) and Cyb2Δ-DHFR from a GAL10 promoter. Mitochondria were isolated from these strains as well as from controls. The redox state of Tim17 was analyzed as described for B. (G) Topology of Tim17 indicating the position of the disulfide bond. IM, inner membrane.
To test whether the two cysteine residues form an intramolecular disulfide bond, we used the alkylating reagent methyl-polyethyleneglycol-maleimide (mmPEG24). This compound reacts with reduced, but not oxidized, thiols, leading to characteristic mass shifts of ∼2 kD per moiety that can be revealed by SDS-PAGE. First, we precipitated mitochondrial proteins with TCA to prevent artificial thiol oxidation. Then, we denatured the proteins with SDS and either directly loaded the extract onto a gel (Fig. 1 B, lane 1) or reduced all cysteine residues with the strong reductant tris-carboxyethyl phosphine (TCEP) after incubation with mmPEG24 (Fig. 1 B, lane 2). The mmPEG24 treatment resulted in an apparent mass shift of the Tim17 signal from ∼19 to 27 kD, consistent with complete alkylation of its four cysteine residues. When the TCEP step was omitted to assess the native redox state of endogenous Tim17, the mass of Tim17 shifted only by ∼4 kD, suggesting that in vivo, only two cysteine residues of Tim17 are present in the reduced state (Fig. 1 B, lane 3). As a control, we used Cmc1 and Mdj1; the IMS protein Cmc1 contains two structural disulfide bonds, and hence its cysteines were inaccessible to mmPEG24, whereas the band of matrix protein Mdj1 was shifted identically in the absence or presence of TCEP, indicating that its cysteines are all reduced in vivo.
To identify which of the four cysteine residues in Tim17 are oxidized, we generated mutants in which either C10 or C77 or both of the conserved cysteine residues were replaced by alanine residues, which we refer to as Tim17AC, Tim17CA, and Tim17AA, respectively. These variants were introduced on single-copy plasmids into a Δtim17 background using a plasmid-shuffling strategy (Meier et al., 2005). In both single mutants, three cysteine residues were modified by mmPEG24, confirming the presence of an intramolecular disulfide bond between C10 and C77 in the endogenous Tim17 protein (Fig. 1 C). This disulfide bond was also present in de-energized mitochondria in which the membrane potential was depleted (Fig. 1 C, −Δψ). We tested for the presence of the Tim17 disulfide bond under many different conditions (log vs. stationary phase, 16–37°C, loss of mitochondrial DNA, presence of respiration inhibitors such as antimycin A, potassium cyanide, or oligomycin). Under all conditions tested, Tim17 contained the disulfide bond between C10 and C77 (unpublished data).
Next, we tested the stability of the disulfide bond in Tim17. To this end, we pretreated isolated mitochondria with increasing amounts of DTT for 15 min at 28°C (Fig. 1 D). However, even under very high concentrations of reductants (100 mM DTT), most Tim17 retained the disulfide bond. The disulfide bond in Tim17 was even present when the cells were grown anaerobically (Fig. 1 E) or when mitochondrial preproteins were highly overexpressed in yeast cells (Fig. 1 F). This extreme stability is reminiscent of what has been observed for other disulfide bonds in the IMS (Curran et al., 2002) and suggests that the disulfide bond in Tim17 plays a structural rather than a regulatory role (Fig. 1 G).
Tim17 is imported in a Mia40-dependent reaction
Next, we tested whether disulfide bond formation in Tim17 is critical for its import into mitochondria and its integration into the inner membrane. We synthesized radiolabeled Tim17 and Tim17AA in reticulocyte lysate and incubated it with purified wild-type mitochondria. After different times, nonimported protein was removed by protease treatment and either directly analyzed or subjected to carbonate extraction to isolate the membrane-inserted protein (Fig. 2, A and B). Tim17 and Tim17AA were imported with comparable efficiency, and roughly half of the protease-protected protein reached a carbonate-resistant location. The carbonate-resistant fraction of Tim17 and Tim17AA was comparable to the fraction that remained protease inaccessible in mitoplasts in which the outer membrane was ruptured by hypotonic swelling (Fig. S2, A and B). Thus, the disulfide bond appears not to be crucial for the import and membrane insertion of Tim17, suggesting that membrane insertion might precede disulfide bond formation (Fig. 2 C).
Tim17 import depends on Mia40. (A and B) 35S-labeled Tim17 or Tim17AA was incubated with wild-type mitochondria at 25°C for the times indicated. Nonimported protein was removed by treatment with proteinase K (PK). Samples were split into two fractions. One was directly subjected to SDS-PAGE and autoradiography (Total), from the other mitochondria were incubated with 0.1 M sodium carbonate for 30 min on ice before carbonate-resistant proteins were pelleted by ultracentrifugation at 100,000 g for 30 min. Western blot signals of the integral membrane protein Oxa1 and the nonmembrane protein Sod1 are shown for control. Quantified signals are shown in B. (C) Model for the import and membrane insertion of Tim17. (D and E) Tim17 or Cmc1, respectively, were incubated with mitochondria from Mia40-containing (Mia40↑) or Mia40-depleted (Mia40↓) cells in the presence or absence of membrane potential (Δψ). The soluble IMS protein Cmc1 is an established Mia40 substrate. (F–H) Import experiments of Tim17 and Tim17AA with wild-type (WT) and mia40-4 mitochondria. (I and J) GAL-Mia40 cells expressing either Mia40 or an SPS mutant or an F315,318E mutant of Mia40 lacking the redox-active cysteines were cultured on glucose for 72 h. Mitochondria were isolated and used for import experiments with radiolabeled Tim17 or Cmc1, respectively.
Tim17 import depends on Mia40. (A and B) 35S-labeled Tim17 or Tim17AA was incubated with wild-type mitochondria at 25°C for the times indicated. Nonimported protein was removed by treatment with proteinase K (PK). Samples were split into two fractions. One was directly subjected to SDS-PAGE and autoradiography (Total), from the other mitochondria were incubated with 0.1 M sodium carbonate for 30 min on ice before carbonate-resistant proteins were pelleted by ultracentrifugation at 100,000 g for 30 min. Western blot signals of the integral membrane protein Oxa1 and the nonmembrane protein Sod1 are shown for control. Quantified signals are shown in B. (C) Model for the import and membrane insertion of Tim17. (D and E) Tim17 or Cmc1, respectively, were incubated with mitochondria from Mia40-containing (Mia40↑) or Mia40-depleted (Mia40↓) cells in the presence or absence of membrane potential (Δψ). The soluble IMS protein Cmc1 is an established Mia40 substrate. (F–H) Import experiments of Tim17 and Tim17AA with wild-type (WT) and mia40-4 mitochondria. (I and J) GAL-Mia40 cells expressing either Mia40 or an SPS mutant or an F315,318E mutant of Mia40 lacking the redox-active cysteines were cultured on glucose for 72 h. Mitochondria were isolated and used for import experiments with radiolabeled Tim17 or Cmc1, respectively.
Does the biogenesis of Tim17 depend on Mia40? Mia40 is an essential protein, and deletion mutants are inviable. We first used mutants in which Mia40 is expressed from a regulatable GAL10 promotor (Fig. S2 C) and isolated from them Mia40-containing (Mia40↑) and Mia40-depleted (Mia40↓) mitochondria which we incubated with radiolabeled Tim17 (Fig. 2, D and E). Tim17 was efficiently imported into Mia40↑ mitochondria. In contrast, Mia40↓ mitochondria were not able to import Tim17, indicating that the presence of Mia40 is, either directly or indirectly, necessary for Tim17 import. In this respect, the import of Tim17 was similar to that of the established Mia40 substrate Cmc1.
A comparable import defect for Tim17 and Tim17AA was observed in temperature-sensitive mia40-4 mitochondria (Chacinska et al., 2004; Mesecke et al., 2005; Sideris and Tokatlidis, 2007; Milenkovic et al., 2009; Sideris et al., 2009) under restrictive conditions (Fig. 2, F–H; and Fig. S2 D), indicating that Mia40 is essential for Tim17 import, whereas the disulfide bond formation per se is not.
Mia40 contains a hydrophobic substrate binding pocket that is flanked by a redox-active cysteine–proline–cysteine motif (Banci et al., 2009; Kawano et al., 2009). Both regions are essential for Mia40 function. To test whether the oxidoreductase activity of Mia40 is required for Tim17 import, we generated a strain in which a Mia40–serine-proline-serine (SPS) variant (or a Mia40 wild-type version for control) was expressed in the Mia40↓ background (Fig. S2 E; Peleh et al., 2016). This mutant lacked any oxidoreductase activity, because both redox-active cysteines are replaced by serines. Mia40 substrates such as the twin Cx9C protein Cmc1 cannot be efficiently imported into Mia40-SPS mitochondria, whereas the import of Tim17 was at least as efficient as with wild-type Mia40 (Fig. 2 I) and more efficient as Mia40-F315,318E (Naoé et al., 2004; Banci et al., 2009; Kawano et al., 2009; Weckbecker et al., 2012; Koch and Schmid, 2014), a Mia40 variant in which two conserved phenylalanine residues of the hydrophobic binding pocket were replaced by glutamates (Fig. 2 J). From this, we conclude that the hydrophobic binding cleft of Mia40 is critical for Tim17 import, whereas the redox-active cysteine pair of Mia40 is dispensable.
In summary, we observed that Mia40 plays an essential (direct or indirect) role in the import of Tim17, for which the oxidoreductase activity of Mia40 was dispensable. We did not observe any critical role of hydrogen peroxide for the import and oxidation of Tim17 (Fig. S2, H–J). Rather, the import of Tim17 tolerated considerable changes in the chemical redox conditions. This supports the notion of a stable structural disulfide bond in Tim17 that is unlikely to be sensitive to intracellular redox conditions.
Mia40 serves as direct binding partner for newly imported Tim17
Next, we tested whether newly imported Tim17 is directly interacting with Mia40. To this end, we incubated isolated wild-type mitochondria with radiolabeled Tim17, Tim17AC, and Tim17AA. Then protein interactions were stabilized by addition of the cleavable cross-linker dithiobis succinimidyl propionate (DSP). Mitochondria were lysed under denaturing conditions with 1% SDS and used for immunoprecipitation with antibodies against Mia40. Thereby radiolabeled Tim17 was efficiently pulled down with Mia40 even in Tim17 mutants lacking the disulfide-forming cysteines (Fig. 3 A, arrowheads). This indicates that Tim17 binds to Mia40 during its import into mitochondria.
Tim17 directly binds to Mia40. (A) Radiolabeled Tim17, Tim17AC, and Tim17AA were imported into wild-type mitochondria for 2 min. The cross-linker DSP was added to the samples for 5 min. Cross-linking was blocked by addition of 100 mM glycine. Mitochondria were reisolated, lysed with 1% SDS, and subjected to immunoprecipitation with antibodies against Mia40 or with preimmune serum (pi). Samples were dissolved in sample buffer containing or lacking 10 mM DTT and analyzed by SDS-PAGE. 10% of the extract used per immunoprecipitation sample was loaded for control (T, total). Arrowheads depict the signals of the Tim17 protein that was pulled down together with Mia40. (B) Tim17 and Tim17AA were imported into Mia40↑ and Mia40↓ mitochondria for 30 min. Nonimported material was removed by protease treatment. Subsequently, the redox state of the imported Tim17 proteins was determined as described for Fig. 1. To show the maximal shift, samples were boiled in SDS and TCEP before incubation with mmPEG24 (lanes 2 and 6). The percent values show the signal intensity of the Mia40-depleted mitochondria relative to those containing Mia40. (C) Tim17 was imported into Mia40↑ and Mia40↓ mitochondria for the indicated time periods. Samples were protease treated and TCA precipitated, and the redox state of the imported Tim17 was analyzed by mmPEG24 treatment as described for Fig. 1 B.
Tim17 directly binds to Mia40. (A) Radiolabeled Tim17, Tim17AC, and Tim17AA were imported into wild-type mitochondria for 2 min. The cross-linker DSP was added to the samples for 5 min. Cross-linking was blocked by addition of 100 mM glycine. Mitochondria were reisolated, lysed with 1% SDS, and subjected to immunoprecipitation with antibodies against Mia40 or with preimmune serum (pi). Samples were dissolved in sample buffer containing or lacking 10 mM DTT and analyzed by SDS-PAGE. 10% of the extract used per immunoprecipitation sample was loaded for control (T, total). Arrowheads depict the signals of the Tim17 protein that was pulled down together with Mia40. (B) Tim17 and Tim17AA were imported into Mia40↑ and Mia40↓ mitochondria for 30 min. Nonimported material was removed by protease treatment. Subsequently, the redox state of the imported Tim17 proteins was determined as described for Fig. 1. To show the maximal shift, samples were boiled in SDS and TCEP before incubation with mmPEG24 (lanes 2 and 6). The percent values show the signal intensity of the Mia40-depleted mitochondria relative to those containing Mia40. (C) Tim17 was imported into Mia40↑ and Mia40↓ mitochondria for the indicated time periods. Samples were protease treated and TCA precipitated, and the redox state of the imported Tim17 was analyzed by mmPEG24 treatment as described for Fig. 1 B.
Because the cysteine residues in Tim17 were obviously not critical for its interaction with Mia40, we next asked whether import and oxidation of Tim17 can be experimentally uncoupled. We imported Tim17 and Tim17AA into mitochondria in which Mia40 was overexpressed or depleted. Nonimported protein was removed by protease treatment before mitochondrial proteins were TCA precipitated. Finally, the redox state of the imported Tim17 was analyzed by mmPEG24 treatment (Fig. 3 B). The import efficiency of Tim17 and Tim17AA were very similar, again suggesting that cysteine oxidation is not critical for Tim17 import. Interestingly, all the imported Tim17 protein contained the disulfide bond, even upon import into the Mia40↓ mitochondria. Here, the amount of the imported Tim17 was lower (23% of that of Mia40↑) but no reduced Tim17 was found in the mitochondria. Even if Tim17 was incubated over prolonged time periods with Mia40↓ mitochondria, no reduced Tim17 was detectable within the mitochondria (Fig. 3 C), suggesting that oxidation of Tim17 can occur independently of Mia40.
Erv1 directly binds to imported Tim17 and is essential for its oxidation
To identify factors critical for the oxidation of Tim17, we imported radiolabeled Tim17 into mitochondria of mutants lacking different redox-relevant factors of the mitochondrial IMS such as Cox17, Cox19, Grx1, Grx2, Pet191, Trx1, Trx2, and Orp1. Tim17 was efficiently imported into all of these mutants (unpublished data). In contrast, no import of Tim17 was observed with mitochondria of a temperature-sensitive mutant of Erv1, the sulfhydryl oxidase of the IMS (Fig. 4 A and Fig. S2 F, erv1-ts). Moreover, the very small amount of radiolabeled Tim17 that was taken up by erv1-ts mitochondria remained fully reduced (Fig. 4 B). Thus, the activity of Erv1 is apparently essential for the import and the oxidation of Tim17 into mitochondria.
Erv1 plays a direct role in the import and oxidation of Tim17. (A) Radiolabeled Tim17 was imported into wild-type (WT) and erv1-ts mitochondria for the times indicated. (B) The redox state of the Tim17 protein imported into erv1-ts mitochondria was analyzed as described for Fig. 3 C. (C) Radiolabeled Tim17, Tim17AC, and Tim17CA were imported into wild-type mitochondria for 2 min. The import was stopped by the addition of cold 1.2 M sorbitol, 20 mM Hepes, pH 7.4, plus 150 mM N-ethyl maleimide. Mitochondria were reisolated, lysed with 1% SDS in the absence or presence of 10 mM DTT and subjected to immunoprecipitation with antibody against Erv1 or with preimmune serum (pi). 10% of the extract used per immunoprecipitation sample was loaded for control (T, total). Arrowheads depict the signals of the Tim17 protein that was pulled down together with Erv1. (D and E) Coimmunoprecipitation experiments of Tim17 after incubation of mitochondria in the presence or absence of membrane potential as indicated and as described for C. (F) Mitochondria were pretreated with 0.5 µM valinomycin to dissipate the membrane potential (Δψ) and incubated with radiolabeled Tim17 for the times indicated. Mitochondria were reisolated, washed, and TCA precipitated. Then, the redox state of Tim17 was analyzed by incubation with mmPEG24. Here, the pattern of the incubated Tim17 was identical to that of Tim17 that was boiled in SDS in the presence of TCEP, indicating that in the absence of membrane potential, all cysteine residues in Tim17 remained reduced. (G) Hypothetical model for the disulfide bond formation in Tim17.
Erv1 plays a direct role in the import and oxidation of Tim17. (A) Radiolabeled Tim17 was imported into wild-type (WT) and erv1-ts mitochondria for the times indicated. (B) The redox state of the Tim17 protein imported into erv1-ts mitochondria was analyzed as described for Fig. 3 C. (C) Radiolabeled Tim17, Tim17AC, and Tim17CA were imported into wild-type mitochondria for 2 min. The import was stopped by the addition of cold 1.2 M sorbitol, 20 mM Hepes, pH 7.4, plus 150 mM N-ethyl maleimide. Mitochondria were reisolated, lysed with 1% SDS in the absence or presence of 10 mM DTT and subjected to immunoprecipitation with antibody against Erv1 or with preimmune serum (pi). 10% of the extract used per immunoprecipitation sample was loaded for control (T, total). Arrowheads depict the signals of the Tim17 protein that was pulled down together with Erv1. (D and E) Coimmunoprecipitation experiments of Tim17 after incubation of mitochondria in the presence or absence of membrane potential as indicated and as described for C. (F) Mitochondria were pretreated with 0.5 µM valinomycin to dissipate the membrane potential (Δψ) and incubated with radiolabeled Tim17 for the times indicated. Mitochondria were reisolated, washed, and TCA precipitated. Then, the redox state of Tim17 was analyzed by incubation with mmPEG24. Here, the pattern of the incubated Tim17 was identical to that of Tim17 that was boiled in SDS in the presence of TCEP, indicating that in the absence of membrane potential, all cysteine residues in Tim17 remained reduced. (G) Hypothetical model for the disulfide bond formation in Tim17.
Next, we tested whether radiolabeled Tim17, during its import into mitochondria, binds directly to Erv1 (Fig. 4 C). To this end, Tim17, Tim17AC, and Tim17CA were incubated with isolated mitochondria before mitochondria were reisolated and lysed under denaturing conditions in SDS buffer. Then, Erv1 was recovered by immunoprecipitation. Surprisingly, Tim17 was efficiently precipitated together with Erv1 and released by addition of DTT, indicating that Erv1 forms a mixed disulfide with newly imported Tim17. A direct interaction of Erv1 was not observed before with other Mia40 substrates such as Atp23 or Cox19 (Fig. S3, A and B).
The direct interaction of Tim17 and Erv1 was also found when the membrane potential was dissipated (Fig. 4 D) and thus established independent of the insertion of Tim17 into the inner membrane (Maarse et al., 1994; Káldi et al., 1998). The SDS-stable interaction depended on the presence of either C10 or C77 in Tim17 because Tim17AA was only recovered with Erv1 from an SDS extract when interactions were stabilized by the cleavable cross-linker DSP (Fig. S3 E). Under native lysis conditions, Tim17 was co-isolated with Mia40 and Erv1, even after incubation in erv1-ts mitochondria in which Erv1 is redox inactive (Fig. S3 D). The binding of imported Tim17 to Erv1 was of a transient nature and was most pronounced after 1 min of the import reaction (Fig. 4 E).
The membrane potential across the inner membrane is a prerequisite for the import of Tim17 (Maarse et al., 1994; Káldi et al., 1998). We observed that, in the absence of membrane potential, no oxidized Tim17 was detectable, even upon prolonged incubation of radiolabeled Tim17 with mitochondria (Fig. 4 F). This suggests that the insertion into the inner membrane is crucial for disulfide bond formation in Tim17, presumably because only in membrane-embedded Tim17 the cysteine residues in position 10 and 77 are close enough to be linked by a disulfide bond.
The data shown here indicate that the mitochondrial disulfide relay system plays a crucial and direct role in the biogenesis of Tim17. Both the oxidoreductase Mia40 and the sulfhydryl oxidase Erv1 are critical players here and directly interact with incoming Tim17 (Fig. 4 G). It is currently unclear whether under physiological conditions Tim17 is predominantly oxidized by Mia40 (Fig. 4 G, steps 4–7) or by Erv1 (Fig. 4 G, steps 4′–6′); however, Tim17 does not have a mitochondria ISS-sorting signal/IMS-targeting signal–like signature like other Mia40 substrates, nor does it form stable intermediates with Mia40. The hydrophobic substrate binding cleft of Mia40 appears to be essential for Tim17 import, presumably in a reaction similar to the translocation of other IMS proteins. However, in contrast to other IMS proteins analyzed so far, Tim17 can be directly bound (and presumably) oxidized by Erv1.
Mutants lacking the structural disulfide bond in Tim17 are temperature sensitive
The strict conservation of the two cysteine residues in Tim17 suggests their critical relevance for TIM23 function. To test the importance of the disulfide bond in Tim17, we constructed yeast mutants in which the endogenous Tim17 was replaced by the Tim17AA variant using a plasmid-shuffling approach (Fig. 5 A). This strain showed an impaired growth on the nonfermentable carbon source glycerol, particularly at higher temperatures (Fig. 5 B). Reduced growth rates of the Tim17AA mutant were also observed upon growth in liquid cultures (Fig. 5 C). The impaired function of the Tim17AA-containing import machinery was very obvious upon overexpression of mitochondrial precursor proteins such as Cox11, which was highly toxic in the Tim17AA background (Fig. S4 A). We did not detect reduced amounts of Tim17 in the cysteine to alanine mutants (Fig. 5 D), but a high percentage of cells showed fragmented mitochondria (Fig. S4 B). Thus, the absence of the disulfide bond in Tim17 leads to reduced cellular fitness, particularly at nonoptimal growing temperatures.
Mutants lacking the structural disulfide bond in Tim17 are temperature sensitive. (A) Plasmid shuffling to generate cysteine to alanine mutants in Tim17. Wild-type (WT) cells or Δtim17 cells containing the wild-type gene of TIM17 on URA3- or HIS3-containing plasmids or the TIM17AA allele on a HIS3-containing plasmid were dropped onto the indicated media. The inability to grow on uracil-deficient medium (SD-URA) and the growth on medium containing 5-fluoro orotic acid indicate the successful loss of the URA3-plasmid containing TIM17 by the shuffling strategy. (B) The indicated strains were grown to log phase and diluted to OD 0.5 to 0.0005, from which 3 µl was spotted onto plates containing glucose or glycerol as carbon sources. (C) Δtim17 mutants expressing either Tim17 or Tim17AA were grown in microtiter plates at 30°C, and the OD was continually measured. (D) The indicated amounts of mitochondrial protein were applied to Western blotting using the indicated antibodies. (E) Radiolabeled Tim17, Tim17AA, and Tim23 were imported for the indicated times into wild-type mitochondria in the presence or absence of membrane potential. Nonimported protein was removed with proteinase K. Samples were solubilized with 1% digitonin and analyzed by BN-PAGE. The asterisk depicts the complex that is found with Tim17AA but barely with Tim17. (F) Isolated mitochondria from the indicated strains were solubilized in buffer containing 1% digitonin and analyzed by BN-PAGE and Western blotting using the indicated antibodies.
Mutants lacking the structural disulfide bond in Tim17 are temperature sensitive. (A) Plasmid shuffling to generate cysteine to alanine mutants in Tim17. Wild-type (WT) cells or Δtim17 cells containing the wild-type gene of TIM17 on URA3- or HIS3-containing plasmids or the TIM17AA allele on a HIS3-containing plasmid were dropped onto the indicated media. The inability to grow on uracil-deficient medium (SD-URA) and the growth on medium containing 5-fluoro orotic acid indicate the successful loss of the URA3-plasmid containing TIM17 by the shuffling strategy. (B) The indicated strains were grown to log phase and diluted to OD 0.5 to 0.0005, from which 3 µl was spotted onto plates containing glucose or glycerol as carbon sources. (C) Δtim17 mutants expressing either Tim17 or Tim17AA were grown in microtiter plates at 30°C, and the OD was continually measured. (D) The indicated amounts of mitochondrial protein were applied to Western blotting using the indicated antibodies. (E) Radiolabeled Tim17, Tim17AA, and Tim23 were imported for the indicated times into wild-type mitochondria in the presence or absence of membrane potential. Nonimported protein was removed with proteinase K. Samples were solubilized with 1% digitonin and analyzed by BN-PAGE. The asterisk depicts the complex that is found with Tim17AA but barely with Tim17. (F) Isolated mitochondria from the indicated strains were solubilized in buffer containing 1% digitonin and analyzed by BN-PAGE and Western blotting using the indicated antibodies.
To assess the assembly of Tim17 and Tim17AA into the TIM23 complex, we resolved protein complexes by blue native (BN) PAGE after in vitro import experiments (Fig. 5 E and Fig. S4 C). Imported Tim17 migrated in a well-characterized protein complex of ∼130 kD, which is referred to as the TIM23CORE complex (van der Laan et al., 2007). This species of the TIM23 complex is associated to the import motor and promotes preprotein translocation into the matrix. In contrast, most Tim17AA migrated in a larger complex of ∼170 kD (Fig. 5 E, asterisk), which presumably represents a TIM23SORT complex. The change in mobility compared with the wild type indicates that the lack of the disulfide bond impairs normal TIM23 assembly. Moreover, when we analyzed the endogenous TIM23 complexes in Tim17 and Tim17AA mitochondria (Fig. 5 F), we observed a considerable depletion of TIM23CORE complexes in the Tim17AA mutant, and all the TIM23 complexes appeared to be locked in the slower-migrating TIM23SORT complex form that additionally contains the Tim21 subunit (Chacinska et al., 2005; van der Laan et al., 2007). In summary, we conclude that in the Tim17AA mutant, the dynamic structure and function of the TIM23 channel is compromised.
The Tim17AA mutant shows a reduced import efficiency for mitochondrial proteins
Next, we analyzed the import efficiency of radiolabeled precursor proteins into Tim17 and Tim17AA mitochondria (Fig. 6, A and B; and Fig. S5, A and B). We chose four proteins which all use the TIM23 complex: Oxa1, an import motor-dependent polytopic inner membrane protein; Mdj1, a soluble matrix protein; cytochrome b2-DHFR, an IMS protein with bipartite presequence that is imported by a stop-transfer mechanism; and cytochrome c1, a stop-transfer–targeted inner membrane protein. In all cases, the proteins were imported into the Tim17AA mitochondria, but the efficiency of the import reaction was between 30 and 60% of that of wild type. In contrast, the carrier protein Aac2, which is imported independently of the TIM23 complex by the TIM22 translocase, was efficiently imported into Tim17AA mitochondria (Fig. 6 C). When Tim17AA mitochondria were pretreated with a heat shock at 37°C before the import experiments, their ability to import proteins was almost completely abolished, whereas wild-type mitochondria were still fully import-competent (Fig. 6, D and E). From this, we conclude that the disulfide bond in Tim17 is important to facilitate efficient protein import of matrix-targeting signal–containing preproteins and essential to maintain the import competence of the TIM23 machinery after heat exposure.
The disulfide bond in Tim17 is critical for mitochondrial protein import. (A–C) The indicated radiolabeled preproteins were incubated with mitochondria at 25°C for the times indicated. Nonimported material was removed by protease treatment. Mitochondria were washed, reisolated, and subjected to SDS-PAGE and autoradiography. Signals of the mature imported proteins were quantified. The dashed lines show the intermediate forms of cytochrome b2. (D and E) As in the above experiments, but the mitochondria were pretreated with import buffer at 37°C before the import reaction at 25°C. (F) The abundance of mitochondrial proteins in Tim17, Tim17CA, and Tim17AA were analyzed by SILAC-based quantitative mass spectrometry. The ratios of the values in the mutants relative to those in the wild type were normalized to the levels of carrier proteins (red dots).
The disulfide bond in Tim17 is critical for mitochondrial protein import. (A–C) The indicated radiolabeled preproteins were incubated with mitochondria at 25°C for the times indicated. Nonimported material was removed by protease treatment. Mitochondria were washed, reisolated, and subjected to SDS-PAGE and autoradiography. Signals of the mature imported proteins were quantified. The dashed lines show the intermediate forms of cytochrome b2. (D and E) As in the above experiments, but the mitochondria were pretreated with import buffer at 37°C before the import reaction at 25°C. (F) The abundance of mitochondrial proteins in Tim17, Tim17CA, and Tim17AA were analyzed by SILAC-based quantitative mass spectrometry. The ratios of the values in the mutants relative to those in the wild type were normalized to the levels of carrier proteins (red dots).
The disulfide bond in Tim17 is of relevance for a broad range of preproteins
The TIM23 complex transports matrix proteins across the inner membrane and stop-transfer–targeted proteins laterally into the inner membrane. Previous studies on tim17 mutants reported about examples that exhibited rather specific defects for proteins that have to be released into the inner membrane or for proteins of the matrix (Chacinska et al., 2005). Because we did not observe a selective import defect for only one of these types of proteins in vitro, we compared the steady-state levels of proteins in Tim17 and Tim17AA mitochondria. Initial experiments by Western blotting did not reveal any considerable differences (Fig. S5 C). To analyze the levels of mitochondrial proteins more generally and quantitatively, we used a SILAC (stable isotope labeling by amino acids in cell culture) labeling strategy followed by mass spectrometry (Fig. S5 D). To this end, we cultured wild-type cells with 13C6,15N4 arginine and 13C6,15N2 lysine, isolated mitochondria, and mixed them in an 1:1 ratio to nonlabeled mitochondria of Tim17, Tim17CA, and Tim17AA, respectively. The intensities of the peptides detected in the mass spectrometer permitted precise determination of the abundance of proteins in these mitochondria. We observed a rather general reduction of the levels of matrix proteins relative to carrier proteins that are imported via the TIM22 pathway (Fig. 6 F, yellow vs. red dots). However, the reduction was rather moderate under the growth conditions tested here (galactose medium at 30°C), and on average, most preproteins were reduced by 10–30% (Table S2). There was no obvious difference between matrix proteins and proteins that need to be laterally sorted to the inner membrane by the TIM23 translocase (Fig. S5, E and F). Moreover, the levels of subunits of the TIM23 complex were not more reduced than other mitochondrial proteins (Fig. S5 G), confirming again that the disulfide bond in Tim17 is not critical for the stable accumulation of TIM23 subunits but is critical for their proper functionality. Thus, we conclude that the disulfide bond in Tim17 plays a general rather than a substrate-specific role for TIM23-mediated protein import into mitochondria.
The disulfide bond in Tim17 is crucial for the dynamic gating behavior of the TIM23 translocase
To identify the molecular defect underlying the reduced import efficiencies into Tim17AA mitochondria, we analyzed the activity of the TIM23 translocase by electrophysiology. This technique has been used very successfully in the past to establish structure–function relationships and link the channel activity of mitochondrial translocases to protein import (Truscott et al., 2001; Grigoriev et al., 2004; Martinez-Caballero et al., 2007). For our analysis, we used a patch-clamp approach for which inner membrane vesicles were isolated from Tim17 and Tim17AA mitochondria and reconstituted into liposome vesicles to form proteoliposomes. Membrane patches were sealed to microelectrodes and excised from the vesicles. Then, the current flowing through single channels was recorded at several voltages between −60 and +60 mV (Fig. 7 A). Representative current traces at either −40 or 40 mV show the highly dynamic gating activity of the TIM23 channel, rapidly fluctuating between three predominant states (open, semiopen, and closed; Fig. 7 B, top traces; and Fig. S4 D), similar to what was reported before for the twin pore structure of the complex (Martinez-Caballero et al., 2007). In the Tim17AA mutant, the flickering or transition rate between the different states was strongly reduced (Fig. 7, A and B; and Fig. S4 D). When we analyzed the population of TIM23 channels in the Tim17AA mutant more closely, two different kinds of behaviors were observed. In some channels (34.5%) the flickering activity was less pronounced compared with the wild type but still clearly recognizable, even though additional smaller substates of various sizes were also noticed (Fig. 7 B, middle traces). We termed this species with damped dynamics “modified.” However, in most cases, the channels were “frozen” and hardly flickered at all. This predominant species apparently lost its ability to rapidly fluctuate between different conductance levels, so that the TIM23 channel lost its gating capacity. Instead, the channels were mostly arrested in its prevailing voltage-induced conformation (Fig. 7, A and B; and Fig. S4 D). However, the peak conductance was similar to that of the twin pore in the wild type, suggesting that the general pore diameter did not generally depend on the disulfide bond.
The disulfide bond in Tim17 is critical for the dynamic gating properties of the TIM23 channel. (A and B) Current–voltage plots (A) and sample current traces (B) are shown to illustrate the behavior and flickering rate of TIM23 channels recorded from wild type and Tim17AA mutant. Single channel containing patches where excised under voltage-clamp conditions, and the voltage was ramped from −40 to 40 mV (A) or held at −40 mV (B). The perfusion chamber and the patch microelectrode contained 150 mM KCl and 5 mM Hepes, pH 7.4. Current was recorded before (Control) and after sequential perfusion of the bath side with 20 µM of the presequence signal peptide yCoxIV(1–13). Dotted lines in current traces (B) correspond to the closed state, and the total amplitude histograms (40–60-s duration) on the side define the channel’s substates registered and their corresponding probability as the percentage of time spent under each current level. Three different behaviors are reported in each case, corresponding to the wild-type channel (Tim17 WT) and the modified and frozen ones of the Tim17AA mutant. (C) Model for the role of the disulfide bond in Tim17 for the stabilization of the TIM23 complex. In the absence of the disulfide bond in Tim17, the gating of the TIM23 complex is severely compromised.
The disulfide bond in Tim17 is critical for the dynamic gating properties of the TIM23 channel. (A and B) Current–voltage plots (A) and sample current traces (B) are shown to illustrate the behavior and flickering rate of TIM23 channels recorded from wild type and Tim17AA mutant. Single channel containing patches where excised under voltage-clamp conditions, and the voltage was ramped from −40 to 40 mV (A) or held at −40 mV (B). The perfusion chamber and the patch microelectrode contained 150 mM KCl and 5 mM Hepes, pH 7.4. Current was recorded before (Control) and after sequential perfusion of the bath side with 20 µM of the presequence signal peptide yCoxIV(1–13). Dotted lines in current traces (B) correspond to the closed state, and the total amplitude histograms (40–60-s duration) on the side define the channel’s substates registered and their corresponding probability as the percentage of time spent under each current level. Three different behaviors are reported in each case, corresponding to the wild-type channel (Tim17 WT) and the modified and frozen ones of the Tim17AA mutant. (C) Model for the role of the disulfide bond in Tim17 for the stabilization of the TIM23 complex. In the absence of the disulfide bond in Tim17, the gating of the TIM23 complex is severely compromised.
Previous analyses showed that the addition of mitochondrial presequence peptides strongly increases the flickering of the TIM23 channel (Meinecke et al., 2006; Martinez-Caballero et al., 2007). In consistence, exposure of patches containing channels to presequence signal peptides greatly boosts their flickering or transition rate between states (Fig. 7, A and B, yCoxIV[1–13]), seemingly because of the transient blockage of the pore. A high affinity interaction of signal peptides with one side of Tim23 has been postulated when this protein, which directly forms at least a part of the translocation pore (Alder et al., 2008; Malhotra et al., 2013), was inserted into bilayers (Truscott et al., 2001). In contrast, in the modified Tim17AA-containing complex, the presequence peptide did not stimulate the flicker rate, and in the frozen Tim17AA complex the signal presequence abruptly and irreversibly blocked the channel. The same behavior was observed regardless of voltage (Fig. 7 A). These results indicate that those species had a severely impaired channel gating.
In summary, the electrophysiology data showed that the TIM23 channel carrying Tim17AA displayed two possible behaviors: (1) a less abounding or modified channel, which in its nature and substrate response largely resembles the wild-type complex but is less dynamic; and (2) a predominant frozen channel, which has lost its ability to gate and does not properly respond to substrates, suggesting that it represents a nonfunctional form. We conclude that the structural disulfide bond in Tim17 is a main component of the TIM23 molecular gating mechanism and thus maintains its critical dynamic properties during preprotein translocation (Fig. 7 C).
Discussion
Import and oxidation of Tim17
Although the core subunits of the TIM23 translocase, Tim17 and Tim23, were identified more than 20 years ago (Dekker et al., 1993; Maarse et al., 1994), their structure and molecular function in protein translocation are poorly understood. It was suggested that Tim23 serves as the pore-forming subunit and that Tim17 is a less important regulator of the TIM23 channel (Truscott et al., 2001; Alder et al., 2008).
In this study, we focused on the relevance of a pair of cysteine residues in Tim17 that is strictly conserved among eukaryotes. We found that these cysteines form a very stable disulfide bond that is exposed toward the IMS and covalently links the first two transmembrane domains of Tim17 (Fig. 7 C). Oxidation of Tim17 is directly coupled to its import, and both processes occur simultaneously. Under the many different conditions tested (anaerobic, presence of oxidants or reductants, fermenting vs. respiring conditions, etc.), we never observed imported Tim17 in the fully reduced state. From this, we conclude that Tim17 oxidation is very robust and not dependent on the physiological conditions, indicating that the disulfide bond does not serve a regulatory role but rather a structural role.
The small Tim complexes of the IMS (particularly the Tim8-Tim13 complex) promote the import of Tim23 into mitochondria, whereas the import of Tim17 was reported not to be affected in mutants of small Tim proteins (Paschen et al., 2000; Davis et al., 2007; Hasson et al., 2010). The severely reduced Tim17 import observed in Mia40-depleted mitochondria can therefore hardly be caused by lower levels of small Tim proteins. Moreover, the binding of Mia40 to Tim17 suggests that Tim17 uses the hydrophobic binding to Mia40 for its import into the IMS in a manner that is comparable to the Tim9-Tim10–driven import of carrier proteins (Koehler et al., 1998; Sirrenberg et al., 1998). Accordingly, we found that a redox-inactive Mia40 mutant (SPS) is still able to efficiently drive the import of Tim17.
Surprisingly, we observed that the oxidoreductase activity of Mia40 was dispensable for Tim17 oxidation. In this respect, Tim17 clearly differs from other Mia40 substrates such as the small Tim proteins or the twin Cx9C proteins (Chacinska et al., 2004; Mesecke et al., 2005; Sideris and Tokatlidis, 2007; Milenkovic et al., 2009; Sideris et al., 2009). Whereas the cysteine-proline-cysteine motif of Mia40 is not required for Tim17 oxidation, the functionality of Erv1 apparently is. Newly imported Tim17 was efficiently recovered with Erv1 in coimmunoprecipitation experiments in an SDS-resistant but DTT-sensitive manner, indicating that Tim17 transiently forms a disulfide-linked import intermediate with Erv1.
Recently, a disulfide bond was identified in Tim22 (Wrobel et al., 2013, 2016; Okamoto et al., 2014). Similar to what we observed for Tim17, the oxidation of Tim22 was found not to depend on Mia40. The observation that Tim17 can be oxidized in a Mia40-independent fashion will certainly inspire studies to identify substrates of Erv1 more comprehensively.
The structural disulfide bond ensures TIM23-mediated translocation
Although the cysteine residues in Tim17 are strictly conserved in evolution, they are not essential for TIM23-mediated protein translocation. However, we found that mutants lacking the disulfide bond in Tim17 are very sensitive to higher temperature, pointing toward strongly reduced thermal stability of the TIM23 complex. The electrophysiological characterization identified two different types of TIM23 channels in the Tim17AA mutants (Fig. 7 B): the modified TIM23 pore, which resembles that of wild type mitochondria but whose gating properties are jeopardized, and a frozen TIM23 pore, which shows a strongly compromised dynamic behavior and presumably is nonfunctional. The presence of a reduced number of functional TIM23 complexes fits very well to the reduced import efficiency observed in the Tim17AA mutant and the reduced levels of presequence-containing proteins detected by quantitative proteomics. This all points to a structural role of the disulfide bond in Tim17, which is crucial for proper gating of the TIM23 channel. Our results together with previous analyses of an N-terminally truncated Tim17 mutant (Meier et al., 2005; Martinez-Caballero et al., 2007) suggest that the highly conserved N-terminal region of Tim17 in conjunction with the IMS-exposed disulfide bond serves as a critical device in the gating of the TIM23 channel.
Thus far, all efforts have failed to recombinantly express and reconstitute Tim17, whereas Tim23 has been successfully studied in liposomes (Truscott et al., 2001). These problems might have in part arisen from the absence of the disulfide bond in the recombinant Tim17. The use of specific bacterial strains optimized for the expression of disulfide-bond–containing proteins (Lobstein et al., 2012) might allow us to tackle this problem in the future.
Materials and methods
Yeast strains and plasmids
Yeast strains used in this study are listed in Table S1. The Δtim17 strain containing the Tim17 wild type or cysteine mutants were derived from the shuffle strain described previously (Meier et al., 2005). The cysteine mutants of Tim17 were generated by plasmid shuffling using a Δtim17 mutant in the BY4741 background as described elsewhere (Meier et al., 2005). For overexpression of mitochondrial preproteins, fusion proteins consisting of the residues 1–167 of cytochrome b2 and DHFR (Cyb2-DHFR) or its Δ19 variant (Cyb2Δ-DHFR; Koll et al., 1992) were cloned into a pYX233 vector. All strains were confirmed by sequencing. The temperature-sensitive mia40-4 mutant was provided by A. Chacinska (International Institute of Molecular and Cell Biology, Warsaw, Poland), the tim17-4 mutant was provided by P. Rehling (University of Gottingen, Gottingen, Germany), and the erv1-ts mutant was provided by T. Lisowski (University of Dusseldorf, Dusseldorf, Germany). Yeast cells were grown as described previously (Peleh et al., 2015). For depletion of Mia40, GAL-Mia40 cells were grown on lactate medium containing 0.2% galactose and shifted to lactate medium containing 0.2% glucose for 72 h.
Mass spectrometry
Pelleted mitochondria were solubilized in 4% SDS and 25 mM NH4HCO3 in a sonication bath to 1 µg/µl protein. Heavy- and light-labeled protein suspensions were mixed 1:1 and precipitated, digested, and desalted as described previously (Mühlhaus et al., 2011). Mass spectrometry analysis was performed as described previously (Bode et al., 2015).
Peptide and protein identification and quantitation were done as described previously (Bode et al., 2015) using MaxQuant software (version 1.2.0.18; Cox and Mann, 2008).
Alkylation shift experiments for redox-state detection
To analyze the redox state of cysteine residues, mitochondrial proteins were TCA precipitated. The pellet was dissolved in modification buffer (80 mM Tris, pH 7.0 [HCl], 10% glycerol, 2% SDS, and bromocresol blue) in the absence or presence of the thiol-free reductant 10 mM TCEP for 20 min at 96°C. Subsequently, 15 mM mmPEG24 was added to modify all reduced cysteines.
Import of radiolabeled proteins into isolated mitochondria
The import reactions were performed as described previously (Mesecke et al., 2005) in the following import buffer: 1 M sorbitol, 100 mM Hepes, pH 7.4, 160 mM KCl, 20 mM magnesium acetate, and 4 mM KH2PO4. In addition, 2 mM ATP, 2 mM NADH, 10 mM creatine phosphate, 100 µg/ml creatine kinase, 2 mM malate, and 2 mM succinate were supplied to energize the mitochondria. To keep the radiolabeled proteins in the reduced unfolded state, 5 mM glutathione and 5 mM EDTA were added to the import mix.
Coimmunoprecipitation
For the immunoprecipitation of import intermediates, mitochondria were reisolated by centrifugation during the import reaction (20,000 g for 20 min at 4°C) and resuspended in lysis buffer I (30 mM Tris/HCl, pH 8, 100 mM NaCl, 1% SDS, and 2 mM PMSF). In some cases, 200 µM DSP was added to the mitochondria before the lysis to stabilize protein interactions by cross-linking. The extract was diluted tenfold in lysis buffer II (30 mM Tris/HCl pH 8, 100 mM NaCl, 1% Triton X-100, and 2 mM PMSF). After incubation for 10 min at 4°C, the extract was cleared by a clarifying spin (20,000 g for 10 min at 4°C). Antibodies were coupled to protein A–Sepharose beads. The beads were incubated with the mitochondrial extract at 4°C, washed three times with the lysis buffer II (2,000 g for 2 min at 4°C), resuspended in SDS-sample buffer, and boiled for 5 min at 96°C. Samples were analyzed by SDS-PAGE and autoradiography. The Tim17 antibodies were provided by D. Mokranjac (University of Munich, Munich, Germany).
Isolation of mitochondria and preparation of proteoliposomes
Wild type and the Tim17AA mutant strain of Saccharomyces cerevisiae were used for the electrophysiological experiments. Cells were cultivated at 30°C on standard SD (synthetic dextrose minimal medium) with 2% glucose in the absence of histidine for 24 h. Mitochondria were isolated from logarithmically growing cells as described previously (Lohret et al., 1997). Homogenization buffer was 0.6 M sorbitol, 10 mM Tris, 1 mM EDTA, 0.2% bovine serum albumin, and 1 mM phenylmethylsulfonyl fluoride, pH 7.4, containing protease inhibitor mixture (P 8215; Sigma-Aldrich). Mitoplasts were prepared from isolated mitochondria by the French press method (Ohba and Schatz, 1987), and the inner membranes were further purified according to Mannella (1982). Membrane purity was routinely assessed, and cross-contamination was typically <10%. Inner membranes were reconstituted into giant proteoliposomes by dehydration–rehydration as described previously (Lohret et al., 1997; Muro et al., 2003) using soybean l-phosphatidylcholine (type IV-S; Sigma-Aldrich).
Patch-clamping techniques
Patch-clamp experiments were performed on reconstituted TIM23 channels of proteoliposomes containing purified mitochondrial inner membranes. In brief, membrane patches were excised from giant proteoliposomes after formation of a gigaseal using microelectrodes with ∼0.4-µm-diameter tips and resistances of 10–30 MΩ. Unless otherwise indicated, the solution in the microelectrode and bath was 150 mM KCl and 5 mM Hepes, pH 7.4. The voltage clamp was established with the inside-out configuration (Hamill et al., 1981) using a List-Medical D-61100 patch-clamp amplifier. Voltages across excised patches were reported as bath potentials. Total amplitude histograms were generated from current traces (40–60-s duration) with WinEDR Software (courtesy of J. Dempster, University of Strathclyde, Strathclyde, Scotland, UK). Filtration was 2 kHz with 5 kHz sampling for all analysis and current traces shown. Permeability ratios were calculated from the reversal potential in the presence of a 150:30 mM KCl gradient as previously described (Muro et al., 2003). Signal peptide yCoxIV(1–13), corresponding to amino acids 1–13 (MLSLRQSIRFFKP) from the N terminus of cytochrome oxidase subunit IV of S. cerevisiae, was introduced by perfusion of the 0.5-ml bath with 3–5 ml of medium containing 20 µM of the peptide. Peptide composition and purity were assessed by high-performance liquid chromatography and mass spectrometry and were typically 90% pure. Flicker rates were determined from current traces (40–60 s) as the number of transition events per second from the open to lower conductance states, with a 50% threshold (∼250 pS) of the predominant event.
Identification of the TIM23 channel activity in the patch-clamp assays
Two large channel activities can be recorded from patches of proteoliposomes containing purified mitochondrial inner membranes, those of TIM23 and TIM22. Routinely, after a gigaseal formation, each patch is electrophysiologically characterized and peak conductance, substate levels, voltage dependence, open probability, and flickering rate are determined. Importantly, the flicker rate (i.e., the transitions frequency between the fully open and the 500 pS substate) is determined in the absence and in the presence of 20 µM of a signal presequence (yCoxIV1–13). Perfusion of patches with this 13–amino acid peptide, corresponding to the N-terminal sequence of subunit IV of cytochrome oxidase, induces a 5–10-fold increase in the flicker rate of TIM23 channels. The channel of TIM22 is not sensitive to N-terminal signal peptides. Even more, the channel of TIM22 is normally silent unless an internal signal sequence is included inside the patch pipette (Peixoto et al., 2007).
Online supplemental material
Fig. S1 shows the sequence conservation of Tim17 proteins. Fig. S2 documents levels of mitochondrial proteins and redox states of Tim17 in different yeast mutants. Fig. S3 presents evidence that Atp23 and Cox19 bind to Mia40, but not to Erv1, and identifies the cysteine residues in position 10 and 77 of Tim17 as essential for its covalent interaction with Mia40 and Erv1. Additional experiments to analyze the phenotypes of the Tim17AA-containing import machinery are shown in Fig. S4. The steady-state levels of proteins in different mitochondria were analyzed by Western blotting and quantitative mass spectrometry as shown in Fig. S5. Table S1 lists the yeast strains used for this study. Table S2 lists the proteins obtained in the SILAC analysis.
Acknowledgments
We thank Sabine Knaus and Lea Düsterwald for technical assistance, Agnieszka Chacinska for the mia40-4 mutant, Peter Rehling for the tim17-4 mutant, Thomas Lisowski for the erv1-ts mutant, Dejana Mokranjac for Tim17 antibodies, Michael Woellhaf and Felix Boos for help with the data analysis, and Jan Riemer and Bruce Morgan for discussion and comments on the manuscript.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (IRTG1830 and SPP1710), the BioComp-Forschungsschwerpunkt Rheinland-Pfalz, the Spanish Ministerio de Ciencia e Innovación (BFU2008-000475), and the Junta de Extremadura-European Social Fund (GR10108).
The authors declare no competing financial interests.