Multicellular organisms can generate and maintain homogenous populations of cells that make up individual tissues. However, cellular processes that can disrupt homogeneity and how organisms overcome such disruption are unknown. We found that ∼100-fold differences in expression from a repetitive DNA transgene can occur between intestinal cells in Caenorhabditis elegans. These differences are caused by gene silencing in some cells and are actively suppressed by parental and zygotic factors such as the conserved exonuclease ERI-1. If unsuppressed, silencing can spread between some cells in embryos but can be repeat specific and independent of other homologous loci within each cell. Silencing can persist through DNA replication and nuclear divisions, disrupting uniform gene expression in developed animals. Analysis at single-cell resolution suggests that differences between cells arise during early cell divisions upon unequal segregation of an initiator of silencing. Our results suggest that organisms with high repetitive DNA content, which include humans, could use similar developmental mechanisms to achieve and maintain tissue homogeneity.
Introduction
Individual tissues in multicellular organisms contain nearly identical cells that continue to behave similarly over time. When a cell divides, however, stochastic variation because of noise (Raj and van Oudenaarden, 2008) or regulated variation because of epigenetic or environmental differences between cells (Snijder and Pelkmans, 2011) can result in nonidentical daughter cells. Such variation is typically reduced during the development of multicellular organisms to ensure robust cell fate determination. The suppression of variation in a process can occur through the use of general mechanisms such as protein chaperones (Hsieh et al., 2013) or of specific mechanisms such as interconnected gene networks (Raj et al., 2010) and regulatory loops (Ji et al., 2013). After cell fate determination, however, variation between cells within a tissue can result in cells that are susceptible to disease (Frank and Rosner, 2012) and drug resistance (Spencer et al., 2009). Yet, in some cases, variation is preserved to generate different cells that together perform a function (e.g., cells that express different photoreceptor proteins that together enable color vision in Drosophila melanogaster; Losick and Desplan, 2008). The mechanisms that generate, preserve, or eliminate variation within a tissue are not well understood, because the large number and unknown developmental lineage of cells that make up a tissue in complex multicellular organisms preclude clear analysis within intact animals.
The worm Caenorhabditis elegans is a tractable model for the analysis of variation between cells within a tissue because it is composed of tissues that develop through a stereotyped series of cell divisions and cell movements (Sulston and Horvitz, 1977; Sulston et al., 1983). The cells within a C. elegans tissue can arise from multiple blastomeres or from a single blastomere. In the case of tissues made from different blastomeres (e.g., body wall muscles from AB, MS, C, and D blastomeres), the different cells that constitute a tissue have different epigenetic histories during development. Observed differences between muscle cells, if any, could thus include differences between blastomeres that arose before tissue specification and persist after tissue specification. In contrast, in the case of tissues made from a single blastomere (e.g., intestine from the E blastomere), any variation between cells must arise after tissue specification. Thus, tissues such as the C. elegans intestine provide an opportunity to examine cell-to-cell variation within a tissue after fate specification.
Cell-to-cell variation in the activity of genes associated with repetitive DNA has been observed in many animals, often between cells of the same tissue. Repetitive DNA can variably effect the expression of nearby genes in different cells in a process called position effect variegation (PEV) in Drosophila (Elgin and Reuter, 2013). An early example showed that the location of the white gene near repetitive DNA results in a variegated expression such that some cells of the Drosophila eye express the white gene but others do not (Muller, 1930). We now know that such repeat-associated gene silencing can occur through RNA-directed mechanisms associated with chromatin modifications and/or DNA methylation (Volpe and Martienssen, 2011; Elgin and Reuter, 2013). However, the origins of the variation between cells and the developmental mechanisms, if any, that control such variation are unclear. Furthermore, despite repetitive sequences constituting an estimated ∼45% (Lander et al., 2001) to ∼69% (de Koning et al., 2011) of the human genome, we do not understand how these large parts of animal genomes are regulated during development.
Studies in C. elegans using repetitive transgenes have provided some insight into expression from repetitive DNA. Genetic screens have identified many conserved factors that promote expression from repetitive DNA through mechanisms that are unclear (Hsieh et al., 1999; Fischer et al., 2013). Insights from the analysis of a few protein factors, however, suggest that expression from repetitive DNA requires the inhibition of RNAi triggered by some form of double-stranded RNA (dsRNA). First, loss of the adenosine deaminases acting on RNA (ADAR) enzymes, which deaminate adenosines in dsRNA, results in the silencing of expression from repetitive DNA (Knight and Bass, 2002) and the recruitment of RNAi on many targets (Wu et al., 2011). Second, loss of the exonuclease ERI-1 (enhancer of RNAi-1), which can trim 3′ overhangs in dsRNA, causes silencing of expression from repetitive DNA (Kennedy et al., 2004). Third, preventing the spread of forms of dsRNA between cells increases the number of cells that show expression from repetitive DNA (Jose et al., 2009). Fourth, silencing observed upon loss of ERI-1 (Kim et al., 2005) or upon loss of ADAR enzymes (Knight and Bass, 2002) can both be relieved by loss of genes required for RNAi. A curious feature of silencing in many genetic backgrounds that lack eri-1 is that it varies from cell to cell (e.g., see Fig. S3 in Kim et al. [2005] and Fig. 1 in Jose et al. [2009]). However, the precise source of dsRNA and the source of cell-to-cell variability are unknown.
Here, we analyze expression from repetitive DNA in the C. elegans intestine at single-cell resolution to uncover a source of cell-to-cell variation and to reveal a developmental mechanism that reduces such variation.
Results
Rearrangements in repetitive DNA generate double-stranded RNA and hairpin RNA
To examine repetitive DNA expression in individual cells without the disruption of cellular function or development in C. elegans, we studied the regulation of the sur-5::gfp repetitive transgene that expresses GFP in all somatic cells, with particularly high levels in intestinal cells. This transgene was generated by transforming worms with a circular plasmid that expresses sur-5::gfp (Fig. S1 A) and integrating the resultant multicopy array into the genome (first used in Winston et al., 2007). Estimations from Illumina sequencing reads suggested that this transgene had 213 ± 26 adjacent copies of the sur-5::gfp plasmid (Figs. 1 A and S1 B). Consistent with early experiments (Stinchcomb et al., 1985), we detected abundant inversions and deletions (Fig. 1 B and Fig. S1, C–E) and a few translocations (Fig. S1, D and E) among the copies of the sur-5::gfp plasmid. The rearrangements were flanked by short sequences with homology (Fig. 1 C), consistent with their generation by recombinases that cause inversions and deletions based on the relative orientation of these sequences (Grindley et al., 2006). These rearrangements, especially inversions, have the potential to generate RNAs that can fold back to form hairpin RNAs or can form dsRNAs with intact mRNA. To examine if such rearranged RNAs are generated from the sur-5::gfp transgene, we performed RNA sequencing (RNA-seq) on polyA-selected RNA isolated from a strain with the sur-5::gfp transgene. We found that RNAs with inversions were present at up to ∼6.5% of the levels of correctly spliced mRNA (Fig. 1 D, blue). The amount of aberrant RNAs detected is likely to be an underestimate, because the library preparation for RNA-seq selected for RNAs with polyA tails. Despite the presence of RNAs expected to trigger RNA-mediated gene silencing (Martienssen and Moazed, 2015), in wild-type animals, GFP fluorescence was reliably detected in all animals and appeared uniform (Fig. 2, A [left] and B [black]). The maximal difference between the brightest and the dimmest intestinal nucleus within a wild-type animal was ∼5-fold, which was only marginally more than the maximum ∼3.5-fold differences that can result from measurement error (Fig. S2, A and B). Thus, although rearrangements within a repetitive transgene generate RNAs that can cause gene silencing, wild-type animals show uniform expression within the intestine.
Analysis of a repetitive transgene reveals inversions and deletions that contribute to the generation of aberrant RNAs. (A) Schematic of the repeat structure of the sur-5::gfp transgene. Repeating units of the ∼12-kb plasmid, which has sur-5 (gray box) and gfp (black box) coding regions, deduced from Illumina sequencing, and units that are not schematized (…) are indicated. Also see Fig. S1. (B) DNA rearrangements in the sur-5::gfp transgene. Inversions (blue) and deletions (orange) observed by DNA sequencing (DNA-seq) are represented on one of the units of the sur-5::gfp repetitive transgene. Percentages reflect reads supporting each rearrangement at a given position compared with those that support no rearrangements at that position. (C) Microhomology-dependent generation of inversions and deletions from a circular plasmid could explain rearrangements found in sur-5::gfp. (top) Schematic illustrating how circular plasmids could undergo inversions (left) or deletions (right) at sites of sequence microhomology (xyz) during the formation of arrays. Dotted lines indicate proposed sites of rearrangements. (bottom) Rearrangements found in sur-5::gfp are associated with regions of microhomology. Identical sequences present near the sites of rearrangement are indicated for inversions (a–r as in B) and deletions (s–w as in B). Dotted lines and sequence colors (brown and blue) are as in the top panel. X indicates nonidentical bases. (D) Rearrangements in RNAs made from the sur-5::gfp transgene. Inversions and deletions (or alternative splicing) observed by RNA sequencing (RNA-seq) and percentages are as in B.
Analysis of a repetitive transgene reveals inversions and deletions that contribute to the generation of aberrant RNAs. (A) Schematic of the repeat structure of the sur-5::gfp transgene. Repeating units of the ∼12-kb plasmid, which has sur-5 (gray box) and gfp (black box) coding regions, deduced from Illumina sequencing, and units that are not schematized (…) are indicated. Also see Fig. S1. (B) DNA rearrangements in the sur-5::gfp transgene. Inversions (blue) and deletions (orange) observed by DNA sequencing (DNA-seq) are represented on one of the units of the sur-5::gfp repetitive transgene. Percentages reflect reads supporting each rearrangement at a given position compared with those that support no rearrangements at that position. (C) Microhomology-dependent generation of inversions and deletions from a circular plasmid could explain rearrangements found in sur-5::gfp. (top) Schematic illustrating how circular plasmids could undergo inversions (left) or deletions (right) at sites of sequence microhomology (xyz) during the formation of arrays. Dotted lines indicate proposed sites of rearrangements. (bottom) Rearrangements found in sur-5::gfp are associated with regions of microhomology. Identical sequences present near the sites of rearrangement are indicated for inversions (a–r as in B) and deletions (s–w as in B). Dotted lines and sequence colors (brown and blue) are as in the top panel. X indicates nonidentical bases. (D) Rearrangements in RNAs made from the sur-5::gfp transgene. Inversions and deletions (or alternative splicing) observed by RNA sequencing (RNA-seq) and percentages are as in B.
Silencing in some cells is suppressed by parental and zygotic ERI-1 and can be partially explained by the canonical RNAi pathway. (A) Loss of the exonuclease ERI-1 silences repetitive DNA in some cells. Representative wild-type (left) or eri-1(−) (right) animals that express nuclear-localized GFP (black) from a repetitive transgene, sur-5::gfp, in intestinal cells (between brackets) are shown. Insets are bright-field images. Bars, 50 µm. (B) Loss of the exonuclease ERI-1 results in bimodal distribution of expression levels. Mean number of intestinal nuclei for each range of relative GFP fluorescence (<0.01, 0.01 to <0.02, 0.02 to <0.03, ..., 0.09 to <0.10, 0.10 to <0.20, 0.20 to <0.30, ..., 0.90 to ≤1.00) in wild-type (black) or eri-1(−) (cyan) animals is shown. Error bars indicate standard error of the mean; n = 20 for wild type L4-staged animals; and n = 60 for eri-1(−) L4-staged animals. (C–E) Parental and zygotic requirements of eri-1 for silencing of a repetitive transgene in intestinal cells. Dosage of the eri-1 gene was varied (+/+, +/−, or −/−) in the hermaphrodite or male, and fractions of cross progeny males with cells that showed >10-fold dimmer GFP fluorescence than the brightest nucleus (fraction of worms with ≥1 dim cell) were determined. Effects on cross progeny males when both parents have the sur-5::gfp (gfp) repetitive DNA (C), when the parental hermaphrodites lack (none) the repetitive DNA (D), or when the parental males lack (none) the repetitive DNA (E) are shown. Error bars indicate 95% confidence intervals, n indicates number of males scored for each genotype, brackets indicate comparisons for maternal and paternal effects. (F) The steps of the RNA silencing pathway (brown) catalyzed by each protein (black) that can eventually cause RNA-directed histone modification (H3K9me3 and/or H3K27me3) are indicated. See text for details. (G) Silencing of repetitive DNA in the absence of ERI-1 occurs through the nuclear RNAi pathway. Fractions of animals that have at least one nucleus with >10-fold lower GFP fluorescence than the brightest nucleus (fraction of worms with ≥1 dim cell) in wild-type, eri-1(−), or eri-1(−) animals that also lack genes of the RNA silencing pathway (rde-4, rde-1, rrf-1, or nrde-3) were determined. Error bars indicate 95% confidence intervals and n indicates the number of animals scored for each genotype. *, P < 0.05. fr., fraction.
Silencing in some cells is suppressed by parental and zygotic ERI-1 and can be partially explained by the canonical RNAi pathway. (A) Loss of the exonuclease ERI-1 silences repetitive DNA in some cells. Representative wild-type (left) or eri-1(−) (right) animals that express nuclear-localized GFP (black) from a repetitive transgene, sur-5::gfp, in intestinal cells (between brackets) are shown. Insets are bright-field images. Bars, 50 µm. (B) Loss of the exonuclease ERI-1 results in bimodal distribution of expression levels. Mean number of intestinal nuclei for each range of relative GFP fluorescence (<0.01, 0.01 to <0.02, 0.02 to <0.03, ..., 0.09 to <0.10, 0.10 to <0.20, 0.20 to <0.30, ..., 0.90 to ≤1.00) in wild-type (black) or eri-1(−) (cyan) animals is shown. Error bars indicate standard error of the mean; n = 20 for wild type L4-staged animals; and n = 60 for eri-1(−) L4-staged animals. (C–E) Parental and zygotic requirements of eri-1 for silencing of a repetitive transgene in intestinal cells. Dosage of the eri-1 gene was varied (+/+, +/−, or −/−) in the hermaphrodite or male, and fractions of cross progeny males with cells that showed >10-fold dimmer GFP fluorescence than the brightest nucleus (fraction of worms with ≥1 dim cell) were determined. Effects on cross progeny males when both parents have the sur-5::gfp (gfp) repetitive DNA (C), when the parental hermaphrodites lack (none) the repetitive DNA (D), or when the parental males lack (none) the repetitive DNA (E) are shown. Error bars indicate 95% confidence intervals, n indicates number of males scored for each genotype, brackets indicate comparisons for maternal and paternal effects. (F) The steps of the RNA silencing pathway (brown) catalyzed by each protein (black) that can eventually cause RNA-directed histone modification (H3K9me3 and/or H3K27me3) are indicated. See text for details. (G) Silencing of repetitive DNA in the absence of ERI-1 occurs through the nuclear RNAi pathway. Fractions of animals that have at least one nucleus with >10-fold lower GFP fluorescence than the brightest nucleus (fraction of worms with ≥1 dim cell) in wild-type, eri-1(−), or eri-1(−) animals that also lack genes of the RNA silencing pathway (rde-4, rde-1, rrf-1, or nrde-3) were determined. Error bars indicate 95% confidence intervals and n indicates the number of animals scored for each genotype. *, P < 0.05. fr., fraction.
Persistence of dsRNA in the absence of ERI-1 silences repetitive DNA in some cells
Unlike the uniform expression observed in wild-type animals, animals that lack ERI-1 showed up to ∼100-fold differences in GFP expression between cells (Fig. 2, A [right] and B [blue]; Jose et al., 2009). The distribution of GFP fluorescence in nuclei was bimodal, dividing nuclei into two classes based on their relative brightness: bright (<10-fold dimmer than the brightest nucleus in an animal) or dim (>10-fold dimmer than the brightest nucleus in an animal; Fig. 2 B). This dramatic enhancement of cell-to-cell variation upon loss of ERI-1 was not observed for GFP expression from single-copy or low-copy transgenes (Fig. S2, C–F), which is consistent with such enhancement being specific for expression from repetitive DNA.
ERI-1 may function by titrating away proteins required for RNA silencing (Lee et al., 2006) and/or by degrading RNA that can silence repetitive DNA (Kennedy et al., 2004; Bühler et al., 2006; Iida et al., 2006). These silencing RNAs derived from the repetitive transgene may be double-stranded RNA (dsRNA; Fire et al., 1998; Hellwig and Bass, 2008) or antisense single-stranded RNA (ssRNA; Tijsterman et al., 2002) that accumulate in the absence of ERI-1. To determine which of these two forms of RNA could explain the observed silencing, we delivered synthetic dsRNA and ssRNA into the embryo by injection into the parent germline and examined silencing of sur-5::gfp (Fig. S2, G and H). Although synthetic dsRNA matching gfp (gfp-dsRNA) caused silencing in wild-type worms and enhanced silencing in eri-1(−) worms, synthetic antisense or sense gfp-ssRNA did not have a detectable effect in wild-type or eri-1(−) worms even when delivered with a strand of complementary phosphorothioate RNA to stabilize the ssRNA in vivo (Fig. S2, G [top] and H [bottom]). Lack of silencing by dsRNA with a phosphorothioate backbone is consistent with a requirement for processing by the endonuclease Dicer, an essential early step of RNAi (Grishok, 2013). Thus, silencing of the repetitive transgene observed in some cells of eri-1(−) animals is likely caused by the presence of dsRNA made from sur-5::gfp.
Both parental and embryonic ERI-1 can enable uniform expression from repetitive DNA
To examine where ERI-1 is required to suppress repetitive DNA silencing by dsRNA, we varied the dosage of maternal, paternal, or embryonic ERI-1 and determined the proportions of animals that showed uniform expression. Animals that lacked uniform expression were identified by the presence of dim nuclei that showed >10-fold reduction in GFP fluorescence in any nucleus compared with the brightest nucleus in that animal (Fig. 2 C). Although embryonic ERI-1 was sufficient to ensure uniform expression from sur-5::gfp in most cases, reduction of paternal ERI-1 (+/− male) in the absence of maternal ERI-1 (−/− hermaphrodite) resulted in loss of uniform expression in some heterozygous eri-1 progeny (Fig. 2 C). Such evidence for paternal contribution was not detectable when the dosage of the transgene was reduced (Fig. 2, D and E), suggesting that paternal ERI-1 is required to suppress cell-to-cell variation only when high levels of dsRNA are made from a repetitive transgene. Furthermore, maternal presence of ERI-1 was sufficient to ensure uniform expression in homozygous mutant progeny, consistent with previous observations of maternal rescue of some eri-1(−) defects (Zhuang and Hunter, 2011). This maternal rescue could be explained by the deposition of the ERI-1 protein or mRNA into embryos because the extent of the maternal effect when both the transgene and ERI-1 were present together in the maternal parent was indistinguishable from that when only ERI-1 was present in the maternal parent (Fig. 2, D and E). In summary, paternal ERI-1 makes a minor contribution to the suppression of cell-to-cell variation compared with zygotic ERI-1 but maternal ERI-1 is sufficient to ensure uniform expression from repetitive DNA.
Silencing of repetitive DNA occurs in part through the canonical RNAi pathway
Many genes required for RNAi can suppress gene silencing that occurs in the absence of ERI-1 (e.g., Kim et al., 2005). Although more than a hundred genes can influence RNAi (Grishok, 2013), the canonical RNAi pathway suggests that dsRNA is processed by the sequential action of the dsRNA-binding protein RDE-4, the primary argonaute RDE-1, the RNA-dependent RNA polymerase RRF-1, and the nuclear argonaute NRDE-3, which directs the deposition of repressive chromatin marks (trimethylation of the histone H3 at lysine 9 or H3K9me3 [Guang et al., 2010] and trimethylation of the histone H3 at lysine 27 or H3K27me3 [Mao et al., 2015]) at loci that produce mRNA of matching sequence (Fig. 2 F; Grishok, 2013). We found that silencing of the repetitive transgene observed in eri-1(−) animals was partially dependent on RDE-4, RDE-1, and RRF-1 such that the number of animals that showed silencing was significantly reduced in the absence of these proteins but entirely dependent on NRDE-3 (Fig. 2 G). These genetic results suggest that silencing of expression from the repetitive transgene in the absence of ERI-1 can occur through RDE-1–dependent and RDE-1–independent mechanisms. The strict requirement for NRDE-3 suggests that both mechanisms converge on NRDE-3–dependent chromatin modifications (H3K9me3 and/or H3K27me3). Because such chromatin modification could be followed by DNA elimination as occurs in ciliates (Mochizuki, 2012), it could also eventually cause deletion of repetitive DNA in somatic cells of C. elegans. Thus, silencing of repetitive DNA in some cells in the absence of ERI-1 occurs in part through the canonical RNAi pathway, likely resulting in the deposition of repressive chromatin marks and possibly including subsequent DNA elimination.
Silencing of repetitive DNA can be repeat specific and independent of homologous loci
Differences between cells in RNA-directed gene silencing may arise either because of inequality between cells in the levels of factors that act through sequence-specific interactions (e.g., dsRNA) or that act independent of nucleotide sequence (e.g., histone-modifying enzymes). If sequence-independent factors were unequal between two intestinal cells, silencing would be expected to co-occur at multiple repeat loci within each cell. To test this possibility, we examined animals that have two different repetitive transgenes that do not share sequence homology: one that expresses GFP and one that expresses DsRed (Fig. 3 A). We found that silencing of GFP could occur without silencing of DsRed within a cell (Fig. 3, B and C), arguing against differences between cells in sequence-independent factors and suggesting that a sequence-dependent factor (likely dsRNA) is different between cells. Consistent with this possibility, a larger number of cells show silencing of the sur-5::gfp repetitive transgene in the presence of dsRNA movement between cells enabled by the dsRNA-selective importer SID-1 than in the absence of such movement (eri-1(−); sid-1(−) versus eri-1(−); sid-1(+) animals in Jose et al., 2009). Collectively, these results suggest that unequal levels of dsRNA that remain despite the spread of dsRNA between cells result in silencing of repetitive DNA within some cells, but not in others.
RNA silencing at one repeat locus can be independent of other repetitive loci or homologous single-copy loci. (A) Schematic of experiment to test if a cell that silences one repeat is likely to also silence another. SUR-5::GFP and nuclear-localized DsRed (nls::DsRED) generated from two repetitive transgenes that lack sequence homology are both expected to localize within intestinal nuclei but be expressed from different promoters (Psur-5 and Psid-2). (B and C) A repeat locus can be silenced independent of another within a cell. Intestinal cells with an integrated repetitive transgene that expresses nuclear-localized GFP (SUR-5::GFP) and an extrachromosomal repetitive transgene that expresses nuclear-localized DsRed (nls::DsRED) were analyzed. (B) Bright-field (top left), GFP fluorescence (top right), DsRed fluorescence (bottom left), and a merged fluorescence image (bottom right) of a representative worm are shown. Arrowhead in merged image indicates a cell that shows silencing of GFP, but not of DsRED. Bars, 100 µm. (C) Frequency of cells with different expression states is consistent with independent silencing of two different repetitive loci. Venn diagram showing number of cells with dim GFP (SUR-5::GFP), dim DsRed (nls::DsRed), and dim DsRed as well as dim GFP. The fraction of cells with dim DsRed as well as dim GFP (30/224) is not significantly different from that expected for chance co-occurrence when dim DsRed is independent of dim GFP (23/224; P = 0.27, n = 224 cells). (D) Schematic of experiment to test if silencing of a repeat locus can spread to another homologous single-copy locus within a cell. SUR-5::GFP and cytoplasmic GFP (GFP) generated from a repetitive transgene and a single-copy transgene that share some sequence homology (gfp) are expected to localize to nuclei and the cytoplasm, respectively, within intestinal cells but be expressed from different promoters (Psur-5 and Peft-3). (E) Silencing of a repeat locus can be independent of a homologous single-copy locus in an eri-1(−) background. Representative bright-field (left) and fluorescent images showing GFP expression (black) in intestinal cells (right) of an animal with a repetitive transgene that expresses nuclear-localized GFP (SUR-5::GFP, top), of an animal with a single-copy transgene that expresses cytoplasmic GFP (GFP, middle), and of an animal with both transgenes (SUR-5::GFP; GFP, bottom) are shown. Arrowhead in animal with both transgenes indicates a cell that shows silencing of SUR-5::GFP, but not of GFP. Bars, 100 µm.
RNA silencing at one repeat locus can be independent of other repetitive loci or homologous single-copy loci. (A) Schematic of experiment to test if a cell that silences one repeat is likely to also silence another. SUR-5::GFP and nuclear-localized DsRed (nls::DsRED) generated from two repetitive transgenes that lack sequence homology are both expected to localize within intestinal nuclei but be expressed from different promoters (Psur-5 and Psid-2). (B and C) A repeat locus can be silenced independent of another within a cell. Intestinal cells with an integrated repetitive transgene that expresses nuclear-localized GFP (SUR-5::GFP) and an extrachromosomal repetitive transgene that expresses nuclear-localized DsRed (nls::DsRED) were analyzed. (B) Bright-field (top left), GFP fluorescence (top right), DsRed fluorescence (bottom left), and a merged fluorescence image (bottom right) of a representative worm are shown. Arrowhead in merged image indicates a cell that shows silencing of GFP, but not of DsRED. Bars, 100 µm. (C) Frequency of cells with different expression states is consistent with independent silencing of two different repetitive loci. Venn diagram showing number of cells with dim GFP (SUR-5::GFP), dim DsRed (nls::DsRed), and dim DsRed as well as dim GFP. The fraction of cells with dim DsRed as well as dim GFP (30/224) is not significantly different from that expected for chance co-occurrence when dim DsRed is independent of dim GFP (23/224; P = 0.27, n = 224 cells). (D) Schematic of experiment to test if silencing of a repeat locus can spread to another homologous single-copy locus within a cell. SUR-5::GFP and cytoplasmic GFP (GFP) generated from a repetitive transgene and a single-copy transgene that share some sequence homology (gfp) are expected to localize to nuclei and the cytoplasm, respectively, within intestinal cells but be expressed from different promoters (Psur-5 and Peft-3). (E) Silencing of a repeat locus can be independent of a homologous single-copy locus in an eri-1(−) background. Representative bright-field (left) and fluorescent images showing GFP expression (black) in intestinal cells (right) of an animal with a repetitive transgene that expresses nuclear-localized GFP (SUR-5::GFP, top), of an animal with a single-copy transgene that expresses cytoplasmic GFP (GFP, middle), and of an animal with both transgenes (SUR-5::GFP; GFP, bottom) are shown. Arrowhead in animal with both transgenes indicates a cell that shows silencing of SUR-5::GFP, but not of GFP. Bars, 100 µm.
The intercellular spread of dsRNA derived from repeat DNA suggests that other single-copy loci of matching sequence could be susceptible to silencing by such dsRNAs. Inversions present within the gfp sequence (d, f, g, m, and p in Fig. 1 B) suggest that dsRNA targeting GFP are made from sur-5::gfp. Therefore, we examined animals with a repetitive transgene (nuclear-localized GFP) and another single-copy transgene (cytosolic GFP) that share ∼900 bp of sequence identity (Fig. 3 D). We found that silencing of nuclear-localized GFP from the repetitive DNA could occur without affecting expression of the unlinked cytosolic GFP from the single-copy transgene (Fig. 3 E). This observation suggests that although forms of dsRNA that match gfp could be transported between cells through SID-1, such dsRNAs can silence matching repetitive DNA, but not single-copy loci.
Together, these results suggest that silencing of repetitive DNA is locus specific but associated with forms of dsRNA that can move between cells. The lack of silencing of homologous single-copy loci suggests that either sufficient amounts of dsRNA are not made to cause such silencing or that other features of the locus (e.g., chromatin modifications present in repetitive DNA, but not at single-copy loci) enhance silencing.
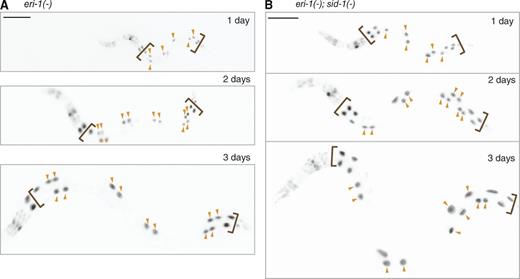
Repetitive DNA is susceptible to apparently stochastic silencing during early development
To determine whether RNA-directed silencing of repetitive DNA is stable once initiated or fluctuates throughout development, we measured GFP fluorescence in the nuclei of individual eri-1(−) worms and of individual eri-1(−); sid-1(−) worms with the sur-5::gfp transgene after 1, 2, and 3 d of development. The measured stability of the GFP protein expressed from the sur-5::gfp transgene in intestinal cells is more than 1 d but less than 1.5 d (Fig. S3, A–C). Thus, the onset of GFP fluorescence from newly synthesized GFP protein as well as loss of GFP fluorescence because of gene silencing can be reliably detected during the period of the experiment (L1 to L4 stage). We observed that the relative fluorescence intensity in most cells did not change more than 10-fold in both eri-1(−) and eri-1(−); sid-1(−) backgrounds (Fig. 4 and Fig. S3, D–G) despite three rounds of endoreduplication of DNA within intestinal cells (Hedgecock and White, 1985) and nuclear divisions in some intestinal cells that occur during this period of development. Thus, silencing or expression of repetitive DNA established in individual intestinal cells during early development is stable despite DNA duplication and nuclear divisions that occur within most intestinal cells.
Pattern of silencing in an animal is established before larval development. (A and B) GFP expression (black) in a representative eri-1(−) (A) or eri-1(−); sid-1(−) (B) animal after 1 (top), 2 (middle), and 3 d (bottom) of development. Arrowheads mark nuclei that continue to show high levels of GFP expression despite nuclear divisions during the 3 d of development. Bars, 100 µm. Also see Fig. S3.
Pattern of silencing in an animal is established before larval development. (A and B) GFP expression (black) in a representative eri-1(−) (A) or eri-1(−); sid-1(−) (B) animal after 1 (top), 2 (middle), and 3 d (bottom) of development. Arrowheads mark nuclei that continue to show high levels of GFP expression despite nuclear divisions during the 3 d of development. Bars, 100 µm. Also see Fig. S3.
To dissect the developmental origin of the variation between cells in the silencing of repetitive DNA, we needed to examine gene silencing within individual intestinal cells and relate it to the lineal origin of each cell. To begin such analyses, we used lineal and morphological information to generate a spatial map of intestinal nuclei (Fig. S4, A and B) that enables unambiguous identification of each nucleus in 16 of the 20 intestinal cells in developed animals in wide-field images. This spatial map was made possible by the known cell divisions and morphogenetic movements of intestinal cells (Sulston and Horvitz, 1977; Sulston et al., 1983; Leung et al., 1999; Hermann et al., 2000) and recent resolution of the resultant helical twist of the intestine in developed animals (Mendenhall et al., 2015; Asan et al., 2016). Measurement of the relative GFP intensity in each nucleus of these 16 intestinal cells in L4-staged animals revealed that no cell showed invariant bright or dim expression from the transgene across all observed animals (Fig. 5 A). The remaining four nuclei are those of the anterior-most cells of the intestine and they are arranged such that fluorescence from two cells located on the right side interferes with fluorescence from the two cells located on the left in wide-field images. Nevertheless, we observed lack of bright GFP expression in all four nuclei in 8 of 60 eri-1(−) animals, suggesting that these cells are also subject to gene silencing. Thus, gene silencing is initiated in some cells before larval development, and none of the intestinal cells are protected from such silencing in all animals.
Spread of silencing between cells is incomplete such that no specific intestinal cell is silenced in all animals that lack ERI-1. (A and B) The heatmap shows relative GFP intensity of each nucleus in 16 intestinal cells for 60 randomly selected eri-1(−) (A) or eri-1(−); sid-1(−) (B) animals (rows). Descendants of the anterior (blue) and posterior (red) daughter of the intestinal blastomere are indicated (A–N; also see Fig. S4). Intensity scale is log10 from maximum relative intensity (black) to ≥100-fold lower in relative intensity (white). When an intestinal cell that could be binucleate did not duplicate its nucleus, the missing nucleus (green) is indicated. (C) Incomplete spread of silencing between cells likely occurs among embryonic cells. Mean relative GFP intensity of each measured intestinal cell in eri-1(−) (gray) and in eri-1(−); sid-1(−) (black) animal is plotted above a lineage diagram of intestinal cells (n = 60 animals). Descendants of the anterior (blue) and posterior (red) daughters of the intestinal blastomere are indicated. Lineal relationships between cells that show significant sid-1–dependent silencing (magenta) suggests that the spread of silencing between cells could begin when the embryo has four daughter cells of the intestinal blastomere (∼60-cell-stage embryo). *, P < 0.05.
Spread of silencing between cells is incomplete such that no specific intestinal cell is silenced in all animals that lack ERI-1. (A and B) The heatmap shows relative GFP intensity of each nucleus in 16 intestinal cells for 60 randomly selected eri-1(−) (A) or eri-1(−); sid-1(−) (B) animals (rows). Descendants of the anterior (blue) and posterior (red) daughter of the intestinal blastomere are indicated (A–N; also see Fig. S4). Intensity scale is log10 from maximum relative intensity (black) to ≥100-fold lower in relative intensity (white). When an intestinal cell that could be binucleate did not duplicate its nucleus, the missing nucleus (green) is indicated. (C) Incomplete spread of silencing between cells likely occurs among embryonic cells. Mean relative GFP intensity of each measured intestinal cell in eri-1(−) (gray) and in eri-1(−); sid-1(−) (black) animal is plotted above a lineage diagram of intestinal cells (n = 60 animals). Descendants of the anterior (blue) and posterior (red) daughters of the intestinal blastomere are indicated. Lineal relationships between cells that show significant sid-1–dependent silencing (magenta) suggests that the spread of silencing between cells could begin when the embryo has four daughter cells of the intestinal blastomere (∼60-cell-stage embryo). *, P < 0.05.
Spread between cells of repetitive DNA silencing could occur during early development
Given that the silencing of repetitive DNA spreads between cells in animals through the dsRNA importer SID-1 (Jose et al., 2009), the persistence of expression through larval development in eri-1(−) animals just as in eri-1(−); sid-1(−) animals suggests that such spread occurs during early development. Identifying the specific cells that show the most SID-1–dependent silencing could provide clues to the earliest stages during development that the transport of dsRNA between cells occurs. Measurement of the relative GFP intensity in each nucleus of the 16 intestinal cells in L4-staged eri-1(−); sid-1(−) animals revealed that no one cell showed invariant bright or dim expression from the transgene across all observed animals (Fig. 5 B) as was the case in eri-1(−) animals (Fig. 5 A). Comparison of the mean relative intensity of GFP expression in each cell of eri-1(−) animals with that in each cell of eri-1(−); sid-1(−) animals revealed significant differences in all cells except in A, B, C, I, and J cells (Fig. 5 C). This observation suggests that all cells except these six were detectably silenced by dsRNAs imported through SID-1. When a set of lineally related cells shows silencing, a parsimonious assumption could be that the silencing was initiated in their common ancestor and persisted through the cell divisions that generated the sister cells. Under such an assumption, these data suggest that the spread of silencing between cells can begin to occur when there are four intestinal cells or at ∼60-cell stage during embryonic development (Fig. 5 C).
Patterns of silencing suggest unequal segregation of initiators of RNA silencing during embryonic cell divisions
Despite the unpredictability of gene silencing in any one cell across multiple animals, how a cell regulates a repetitive transgene may be predictable based on how spatially related or lineally related cells regulate that transgene. To discover whether lineal or spatial relatedness of cells is a better predictor of transgene silencing, we used support vector machines (SVMs; Cortes and Vapnik, 1995) and decision trees (Breiman et al., 1984) to learn classification models of silencing based on different representations of the data. SVMs and decision trees are supervised machine learning algorithms, i.e., they learn a model from labeled training instances that can then be used for classifying new unlabeled instances. In our setup, we learned binary classification models that could classify GFP expression of cells as either bright (0.1 to 1 relative GFP intensity) or dim (0.01 to 0.1 relative GFP intensity). We used three different representations of the data: lineal, spatial, or both. Specifically, we classified GFP expression of cells as either bright or dim based on the relative GFP intensity in lineally related cells (Fig. 6 A), the relative GFP intensity in spatially related cells (Fig. 6 B), or both. We used the relative GFP intensity data of some cells collected from many animals to learn a model for each data representation and then classified the remaining cells using the learned classifier. We found that the accuracy of classification was improved significantly above the baseline of always classifying bright by the lineal model, but not by the spatial model using SVMs (Fig. 6, C and D) or using decision trees (Fig. 6, E and F). Models that use both lineal and spatial information did not improve accuracy more than those using lineal information alone (Fig. 6, D and F).
Silencing of repetitive DNA can be explained by unequal segregation of an initiator of gene silencing soon after tissue specification. (A and B) As illustrated for cell I, SVMs can classify GFP expression on the basis of relative GFP intensity in either lineally or spatially related cells. (A) With the lineal data representation, classification was based on a weighted (w1, w2, w3, and w4) summation of the mean relative GFP intensities of four lineally related sets of cells (l1, l2, l3, and l4) and a constant (c). Also see Fig. S4 A. (B) With the spatial data representation, classification was based on a weighted (w1, w2, and w3) summation of the relative GFP intensities of three spatially adjacent cells (s1, s2, and s3) and a constant (c). Also see Fig. S4 B. (C–F) Two different machine-learning algorithms suggest that lineal models classify transgene expression in a cell better than spatial models. (C) Classifiers learned by SVM using lineal features, spatial features, and both lineal and spatial features are shown. (D) Accuracies of learned SVM models using the three different data representations. The spatial model performed no better than the baseline model of always predicting bright (58% accuracy). The lineal model performed significantly better than the baseline and the spatial model. The model using both lineal and spatial information was no more accurate than that using lineal information alone. Error bars and asterisks are as in Fig. 2 C. (E) The lineal decision tree classifies transgene expression in a cell better than the spatial decision tree. The decision tree learned using the lineal features as in A (left), that learned using spatial features as in B (middle), and that learned using a combination of both lineal and spatial features (right) are shown. The classification of a cell as either bright (B) or dim (D) is determined by traversing the path from the root to the leaf node that applies for that cell. (F) Accuracies of the decision trees that were learned using the three different data representations. Baseline, error bars, and asterisks are as in D. (G–K) Each daughter of the intestinal blastomere had descendants that showed correlated GFP expression but descendants of one daughter showed anti-correlated GFP expression with descendants of the other daughter. Significant (P < 0.05) correlations (orange) and anticorrelations (black) of relative GFP intensity among descendants of the anterior (blue) and posterior (red) daughters of the intestinal blastomere in eri-1(−) (G, n = 60), eri-1(−); sid-1(−) (H, n = 60), eri-1(−); rde-4(−) (I, n = 13), eri-1(−); rde-1(−) (J, n = 29), and eri-1(−); rrf-1(−) (K, n = 39) animals are shown. Also see Fig. S5. (L) Schematic depicting unequal segregation of an initiator of silencing in some cells during early embryonic cell divisions that result in the threshold-dependent initiation of stable silencing or expression at each repeat locus (cyan or magenta).
Silencing of repetitive DNA can be explained by unequal segregation of an initiator of gene silencing soon after tissue specification. (A and B) As illustrated for cell I, SVMs can classify GFP expression on the basis of relative GFP intensity in either lineally or spatially related cells. (A) With the lineal data representation, classification was based on a weighted (w1, w2, w3, and w4) summation of the mean relative GFP intensities of four lineally related sets of cells (l1, l2, l3, and l4) and a constant (c). Also see Fig. S4 A. (B) With the spatial data representation, classification was based on a weighted (w1, w2, and w3) summation of the relative GFP intensities of three spatially adjacent cells (s1, s2, and s3) and a constant (c). Also see Fig. S4 B. (C–F) Two different machine-learning algorithms suggest that lineal models classify transgene expression in a cell better than spatial models. (C) Classifiers learned by SVM using lineal features, spatial features, and both lineal and spatial features are shown. (D) Accuracies of learned SVM models using the three different data representations. The spatial model performed no better than the baseline model of always predicting bright (58% accuracy). The lineal model performed significantly better than the baseline and the spatial model. The model using both lineal and spatial information was no more accurate than that using lineal information alone. Error bars and asterisks are as in Fig. 2 C. (E) The lineal decision tree classifies transgene expression in a cell better than the spatial decision tree. The decision tree learned using the lineal features as in A (left), that learned using spatial features as in B (middle), and that learned using a combination of both lineal and spatial features (right) are shown. The classification of a cell as either bright (B) or dim (D) is determined by traversing the path from the root to the leaf node that applies for that cell. (F) Accuracies of the decision trees that were learned using the three different data representations. Baseline, error bars, and asterisks are as in D. (G–K) Each daughter of the intestinal blastomere had descendants that showed correlated GFP expression but descendants of one daughter showed anti-correlated GFP expression with descendants of the other daughter. Significant (P < 0.05) correlations (orange) and anticorrelations (black) of relative GFP intensity among descendants of the anterior (blue) and posterior (red) daughters of the intestinal blastomere in eri-1(−) (G, n = 60), eri-1(−); sid-1(−) (H, n = 60), eri-1(−); rde-4(−) (I, n = 13), eri-1(−); rde-1(−) (J, n = 29), and eri-1(−); rrf-1(−) (K, n = 39) animals are shown. Also see Fig. S5. (L) Schematic depicting unequal segregation of an initiator of silencing in some cells during early embryonic cell divisions that result in the threshold-dependent initiation of stable silencing or expression at each repeat locus (cyan or magenta).
The lineal machine learning models could have learned from both correlation and anticorrelation of relative GFP intensity between lineally related cells. To identify the cells that show correlated or anticorrelated expression of the repetitive transgene, we compared each cell with every other cell in eri-1(−) animals (Fig. 6 G). We found that a few cells that were descendants of the anterior (Fig. 6 G, blue) or the posterior (Fig. 6 G, red) daughter of the intestinal blastomere showed significant correlation with cells that were also descendants of the same daughter (Fig. S4, A and B). In addition, some descendants of the anterior daughter showed significant anti-correlation with some descendants of the posterior daughter. Because the extent of silencing of sur-5::gfp in eri-1(−) animals can vary upon simply passaging the strain (Devanapally et al., 2015), we examined whether the observed correlations and anticorrelations are reproducible by generating three new isolates of eri-1(−) sur-5::gfp (Fig. S5 A). Although the precise cells that showed correlations and anticorrelations were not reproduced (Fig. S5 A), the general pattern of correlations and anticorrelations in the three new isolates were similar to that observed earlier (29 of 35 relationships agreed with the pattern). We also observed similar patterns of correlation and anticorrelation for the residual silencing that occurs in eri-1(−) animals in the absence of other genes that act in the RNAi pathway. Specifically, measurement of relative GFP intensity in animals that lack both eri-1 and one of the genes of the canonical RNAi pathway (rde-4, rde-1, or rrf-1) or a gene required for the transport of dsRNA between cells (sid-1) revealed that the pattern of residual silencing in each case also varied from animal to animal (Fig. S5 B). Nevertheless, relationships between the descendants of the intestinal blastomere in these double mutants (Fig. 6, H–K; and Fig. S5 B) were similar to those in eri-1(−) single mutants (Fig. 6 G).
The observed anticorrelations suggest the unequal partitioning of a factor (e.g., dsRNA) among the daughters of the first intestinal cell division followed by a few cell divisions when silencing or expression of repetitive DNA is established and subsequently inherited. Consistent with the proposed timing for the origin of differences in silencing between cells, heterochromatin formation and the condensation of repetitive transgene DNA in C. elegans begin at the first intestinal cell division and are accompanied by its positioning at the nuclear periphery (Yuzyuk et al., 2009). This condensation and peripheral positioning of repetitive DNA is dependent on the methylation of histone H3 at lysine 9 (Towbin et al., 2012), which was recently demonstrated to be capable of being maintained independent of the initial RNA trigger in the yeast Schizosaccharomyces pombe (Audergon et al., 2015; Ragunathan et al., 2015). In summary, we propose that, in the absence of the exonuclease ERI-1, the unequal segregation of an initiator of gene silencing (e.g., forms of dsRNA) matching each repeat locus results in the threshold-dependent recruitment of maintainers of gene silencing (e.g., repressive chromatin marks) and subsequent propagation of silencing or expression despite DNA replication and cell divisions (Fig. 6 L).
Discussion
We found that variation in expression from repetitive DNA can arise because of RNA-directed gene silencing that occurs in some cells in the absence of a parentally provided and zygotically expressed exonuclease. Silencing of a repeat locus can be independent of other repeat loci and of single-copy loci with sequence homology, yet the silencing can spread between some cells during early development. Analyses at single-cell resolution and using machine learning suggest that unequal segregation of an initiator of gene silencing (e.g., dsRNA) and threshold-dependent recruitment of a maintenance mechanism (e.g., formation of heterochromatin) can prevent tissue homogeneity.
In vivo analysis of a tissue at single-cell resolution
The measurement of any parameter in live animals across development at single-cell resolution presents considerable challenges. Irregular cellular morphology, complex lineal origins of cells, and movement during morphogenesis can make it difficult to know the precise boundaries and lineal relationships of cells within a tissue. In this study, we benefit from the work of pioneers who have defined all aspects of C. elegans lineage throughout development (Sulston and Horvitz, 1977; Sulston et al., 1983; Leung et al., 1999; Hermann et al., 2000; Mendenhall et al., 2015; Asan et al., 2016) and from nuclear localization, which enables measurement of protein levels despite the irregular shape of intestinal cells. Because cellular behavior is a result of many factors in addition to protein levels, studies in intact animals measuring many parameters (metabolite levels, transcript abundance, chromatin state, signaling activity, etc.) are needed to determine the potentially multifactorial variation between cells within a tissue. Emerging technologies may enable such analyses in the future (Chen et al., 2015; Crosetto et al., 2015). Our studies establish the C. elegans intestine as a model for in vivo analysis across development and complement previous measurements of transcript levels within the intestine during early development in fixed embryos (Raj et al., 2010) and recent measurements in live adult animals (Mendenhall et al., 2015). Despite the clear challenges that lay ahead, in vivo analyses of tissues at single-cell resolution are needed to determine if our understanding of any cellular process is an accurate reflection of the situation in vivo or an artifact of averaging the behavior of many cells (Pelkmans, 2012).
Consequence of repetitive DNA for gene expression
Gene expression requires escape from mechanisms that silence repetitive DNA, especially in the case of mammalian genomes that have large amounts of repetitive sequences (Lander et al., 2001; de Koning et al., 2011). We found that the RNA exonuclease ERI-1 can ensure uniform expression from repetitive DNA by eliminating variation between cells in the early embryo (Fig. 2 A). The conservation of ERI-1 (Thomas et al., 2014), the abundance of repetitive DNA in animals, and the potential for repeats to silence adjacent genes (Elgin and Reuter, 2013) suggest that similar developmental mechanisms exist in other animals to control cell-to-cell variation in the expression of repeats. Our results suggest that cell-to-cell variation within the intestine of animals that lack ERI-1 originates during the first division of the blastomere that generates the C. elegans intestine (Fig. 6). The analysis of PEV in Drosophila similarly suggests that variegation originates during early development (Lu et al., 1996). Our results show that the canonical RNAi pathway and a parallel pathway converge on the nuclear argonaute NRDE-3, which is required for the deposition of H3K9me3 and/or H3K27me3 to cause silencing of repetitive DNA. Both histone modifications have also been implicated in PEV in Drosophila (Elgin and Reuter, 2013). PEV in cultured mammalian cells can affect the expression of ∼900 genes and acts through a protein complex that is not found in Drosophila but nevertheless also requires H3K9me3 (Tchasovnikarova et al., 2015). Collectively, the suppression of H3K9me3 formation at repeats may be an evolutionarily conserved mechanism that is required in organisms with repetitive DNA to ensure uniform expression in cells within a tissue.
Developmental control of tissue homogeneity
Proliferative cell divisions that generate the cells of a tissue likely result in the unequal segregation of many factors between cells (Huh and Paulsson, 2011a,b). Although unequal segregation of factors is used in early development to generate different tissues (Horvitz and Herskowitz, 1992; Osborne Nishimura et al., 2015), unequal segregation after tissue specification could result in disruption of function in some cells within a tissue. RNA silencing at repetitive DNA is one process that can become unequal between cells during proliferative divisions. In the intestinal lineage, the first cell division results in anticorrelated expression of repetitive DNA among daughter cells and subsequent cell divisions result in correlated expression of repetitive DNA among daughter cells (Fig. 6, G–K) despite the spatial separation of lineal sister cells (e.g., the cell pairs E and F, B and C, G and H, and K and L in Fig. 6). This observation suggests that the RNA-directed silencing initiated upon unequal early cell divisions in the intestinal lineage results in persistent silencing in lineal sister cells despite their separation in space during the morphogenesis of the intestine. Developmental mechanisms that reduce the levels of aberrant RNA below the threshold required for maintenance mechanisms (e.g., heterochromatin formation) protect tissues from such dramatic and persistent variation between cells. Variation that escapes such developmental mechanisms may generate defective cells even in the absence of genetic mutations. Loss of tissue homogeneity resulting from this loss of developmental control could predispose a few cells within a tissue to age-related diseases such as cancer (Frank and Rosner, 2012) and potentially play a role in evolution (Feinberg and Irizarry, 2010).
Materials and methods
Worm strains
C. elegans strains were generated using standard genetic crosses and maintained at 15°C using Escherichia coli OP50 as food (Brenner, 1974). The following strains were used in this study: AMJ141 rde-4(ne301) III; eri-1(mg366) nrIs20 (Psur-5::sur-5::gfp) IV, AMJ246 rrf-1(ok589) I; eri-1(mg366) nrIs20 IV (generated by S. Devanapally, University of Maryland, College Park, MD), AMJ259 nrde-3(tm1116) X; eri-1(mg366) nrIs20 IV (generated by S. Devanapally), AMJ284 eri-1(mg366) nrIs20 IV; rde-1(ne219) V (generated by S. Ravikumar, University of Maryland, College Park, MD), AMJ357 oxSi221 ((Peft-3::gfp & unc-119(+)) II; unc-119(ed9)? III; eri-1(mg366) IV, AMJ490 oxSi221 II; unc-119(ed9)? III; eri-1(mg366) nrIs20 IV, AMJ512 jamEx157 (Psid-2::nls::DsRed), AMJ524 jamEx157; eri-1(mg366) nrIs20 IV, EG6070 oxSi221 II; unc-119(ed9) III, AMJ518 (isolate 1) eri-1(mg366) nrIs20 IV, AMJ519 (isolate 2) eri-1(mg366) nrIs20 IV, AMJ520 (isolate 3) eri-1(mg366) nrIs20 IV, AMJ729 eri-1(mg366); unc-119(ed3)?; teIs46 (pRL1417; Pend-1::gfp::H2B + unc-119(+)) AMJ808 stIs10226 (Phis-72::his-24::mCherrry::let-858 3′UTR + unc-119(+)), AMJ811 eri-1(mg366); stIs10226, GR1373 eri-1(mg366) IV, HC195 nrIs20 IV, HC566 nrIs20 IV; sid-1(qt9) V, HC567 eri-1(mg366) nrIs20 IV, HC568 eri-1(mg366) nrIs20 IV; sid-1(qt9) V, N2 wild type, RW10226 unc-119(ed3) III; itIs37 (Ppie-1::mCherry::H2B::pie-1 3′UTR + unc-119(+)) IV, and TX691 unc-119(ed3); teIs46 (pRL1417; Pend-1::gfp::H2B + unc-119(+)).
Oligonucleotides
The following oligonucleotides with a DNA, RNA, or a phosphorothioate-RNA (thio-RNA) backbone (Integrated DNA Technologies) were used in this study: gfp forward RNA and thio-RNA, 5′-ACUGCUCCAAAGAAGAAGCGUAAGGUACCGGUAGAAAAAA-3′; gfp reverse RNA and thio-RNA, 5′-UUUUUUCUACCGGUACCUUACGCUUCUUCUUUGGAGCAGU-3′; unc-22 forward RNA and thio-RNA, 5′-ACAUUCCAGUCAGUGGUGAACCAACUCCAACAAUUACUUG-3′; unc-22 reverse RNA and thio-RNA, 5′-CAAGUAAUUGUUGGAGUUGGUUCACCACUGACUGGAAUGU-3′; P1 DNA, 5′-ATTTGTTGGAGACCAGGCAC-3′; P2 DNA, 5′-CTTCTTCTTTGGAGCAGTCATTTCCTGAAAATATCAGGGTTTTG-3′; P3 DNA, 5′-TCTCAAGGATCTTACCGCTG-3′; P4 DNA, 5′-CAAAACCCTGATATTTTCAGGAAATGACTGCTCCAAAGAAGAAG-3′; P5 DNA, 5′-CTGCCTATTGGGACTCAACG-3′; P5 DNA, 5′-CTGCCTATTGGGACTCAACG-3′; P6 DNA, 5′-ACGCATCTGTGCGGTATTTC-3′; P7 DNA, 5′-CAGACCTCACGATATGTGGAAA-3′; and P8 DNA, 5′-GGAACATATGGGGCATTCG-3′).
Transgenic animals
To express nuclear-localized DsRed in all intestinal cells (Psid-2::nls::DsRed), the promoter for sid-2 (Psid-2) was amplified (Phusion polymerase; New England Biolabs, Inc.) from N2 gDNA using the primers P1 and P2. Nuclear-localized DsRed (nls::DsRed) was amplified (Expand Long Template polymerase; Roche) from pGC306 (a gift from J. Hubbard, New York University, New York, NY; plasmid 19658; Addgene) using the primers P3 and P4. Using these two amplicons as template, Psid-2::nls::DsRed was amplified (Expand Long Template polymerase; Roche) with primers P5 and P6. This final product was purified (QIAquick PCR Purification kit; QIAGEN) and used at a concentration of 40 ng/µl (in 10 mM Tris HCl, pH 8.5) to transform N2 animals by microinjection (Mello et al., 1991). Eight independent transgenic lines were isolated, and the one with the least mosaicism (AMJ512) was used to make the strain shown in Fig. 3 B. Consistent with silencing of DsRed in the eri-1(−) background, fewer cells showed bright DsRed fluorescence in an eri-1(−) background compared with a wild-type background. Because of the mosaicism of the P-sid-2::nls::DsRed transgene, cells that lack bright DsRed fluorescence include cells that have lost the transgene.
Genetic crosses
Male cross progeny were scored for silencing of GFP expressed from nrIs20 (Psur-5::sur-5::gfp) with a fixed magnification on a MVX10 Fluorescence Microscope (Olympus; Fig. 2). Genotypes of scored progeny were confirmed for presence or absence of eri-1 by PCR using primers P7 and P8. Animals with at least one nucleus >10-fold dimmer than the brightest nucleus were scored as silenced, and the proportion of such animals was determined for each genotype (Fig. 2). 95% confidence intervals and p-values for comparison were calculated as described earlier (Jose et al., 2009).
DNA sequencing and RNA-seq
Genomic DNA and total polyA+ RNA of a strain with sur-5::gfp were sequenced using the Illumina sequencing platform. The resultant DNA-sequencing (DNA-seq) and RNA-seq data (available under NCBI GEO accession no. GSE69704) were analyzed using a mix of publicly available bioinformatics tools and custom scripts.
Genomic DNA and total RNA were prepared from liquid cultures of HC566 (E. Traver and P. Raman, University of Maryland, College Park, MD) and used for 101-bp paired-end and 100-bp single-read sequencing of DNA (DNA-seq) or 126-bp single-read sequencing of polyA-selected RNA (RNA-seq). The resulting fastq files were mapped using TopHat2 (Kim et al., 2013) to an inverted tandem copy of pTG96 (linearized after the sequence 5′-AACAACTTGGAAATGAAAT-3′; Fig. S1 C) using default parameters and using the “–fusion search” option. TopHat detects rearrangements that satisfy canonical “splice junctions” (GT-AG, GC-AG, and AT-AC) and thus likely underestimates the number of rearrangements present in sur-5::gfp. The left and right reads of paired-end reads from DNA-Seq were also mapped separately to the template (Fig. S1 B) and the C. elegans genome to estimate the number of copies of pTG96 that were present in the integrated sur-5::gfp transgene. Paired-end reads from DNA-Seq were mapped to two differently linearized versions of pTG96 (Fig. S1, D and E), which were chosen so as to not miss any rearrangements that could be obscured by any one linearization done to allow for mapping. The resultant mapped reads were visualized using Integrative Genomics Viewer (Robinson et al., 2011) after down-sampling the reads, grouping reads based on pair orientation, and coloring read pairs using Illustrator (Adobe).
For both DNA-seq and RNA-seq, the junctions.bed files from Tophat2 were analyzed to identify well-supported inversions and deletions. Well-supported inversions and deletions were determined in two steps: first, inversions with >400 reads and deletions with >100 reads were filtered; and second, only rearrangements (inversions or deletions) supported by >2% of the reads at the site of rearrangement were kept (percentage of reads supporting rearrangement = number of reads supporting rearrangement/[(number of reads at start position of rearrangement + number of reads at end position of rearrangement)/2]) in Fig. 1, B and D). The high frequency of some rearrangements (e.g., ∼21% for “a” in Fig. 1 B) suggest that these rearrangements are present in all cells and occurred during array formation. The independent generation of rearrangements during mitoses, however, cannot be formally excluded.
Fluorescence imaging
Fourth-larval stage (L4) animals in 3 mM tetramisole hydrochloride (Sigma-Aldrich) were individually imaged at a fixed magnification using an AZ100 microscope (Nikon) with a Cool SNAP HQ2 camera (Photometrics). Exposure times (Fig. S2 A) were scaled for each worm to just under saturation based on the most fluorescent intestinal nucleus (owing to GFP expression from sur-5::gfp) in each genetic background tested. Corresponding bright-field images were taken using autoexposure. Worms with evidence of GFP diffusion into the cytoplasm caused by physical distress when the worms were mounted on a slide were not included for the quantitative analysis. Worms assayed for expression of GFP from sur-5::gfp throughout larval development were imaged live on agar plates after 1, 2, and 3 d of development outside the parent worm (Fig. 4, A and B; and Fig. S3, D and E) using stage-specific constant exposure times. Animals with GFP expression from the single-copy transgene oxSi221 (Peft-3::gfp) and from the single-copy or low-copy transgene stIs10226 (Phis-72::his-24::cherry) as well as teIs46 (Pend-1::gfp::H2B) were imaged at a constant exposure time across all compared genetic backgrounds (Fig. S2) and could be silenced using feeding RNAi in a wild-type background. All images were identically adjusted for each figure using Photoshop (Adobe) and Illustrator (Adobe) for display.
Quantitative fluorescence measurements
The intensities of GFP fluorescence from sur-5::gfp expression were determined for each scored nucleus using NIS-Elements (Nikon). GFP intensity for each nucleus was calculated as a product of the area of the nucleus and its mean intensity. Identity of each scored nucleus was determined using expected physical location (Fig. S4, A and B; Sulston and Horvitz, 1977; Sulston et al., 1983; Leung et al., 1999; Hermann et al., 2000; Mendenhall et al., 2015; Asan et al., 2016) and bright-field images. Values of each nucleus were normalized to the brightest nucleus within each animal and the cells were labeled A through N and ordered according to their lineal relationships (Fig. S4, A and B; Sulston and Horvitz, 1977; Sulston et al., 1983; Leung et al., 1999; Hermann et al., 2000; Mendenhall et al., 2015; Asan et al., 2016). When nuclear divisions did not occur in the last four cells (N and J), the missing nucleus was marked green. Cell movements during development position the two most anterior nuclei atop two other nuclei and these four nuclei were not analyzed. When rare abnormal fragments or fusions of nuclei within a cell were observed in some genetic backgrounds, the total intensity value within the cell was divided equally for each expected nucleus. Measurement errors were determined by taking the ratio of GFP expression values between two nuclei within a cell in wild-type animals (Fig. S2 B). Changes in GFP expression as the animal develops were determined by taking the ratio of relative GFP expression values of the same nucleus after 1 d of development to that after 3 d of development. Heatmaps for each strain were generated using Matrix2png (Pavlidis and Noble, 2003) and agglomerative hierarchical clustering using Ward’s method (XLSTAT Pro; Addinsoft). Unless specified, all other statistical analyses were performed using Excel 2011 (Microsoft).
Silencing by injected RNA
For Fig. S2 G and H, forward- and reverse-strand RNA oligos (IDT) against gfp or unc-22 were either injected into one gonad arm of animals at a final concentration of 100 ng/µl individually or after annealing together (cooling at 1°C/min from 95°C to 25°C). The integrity of the injected RNA was checked using nondenaturing polyacrylamide gel electrophoresis. GFP silencing in L4-staged or young adult progeny of each injected worm at 15°C were examined between 4 and 6 d after injection at fixed magnification on a MVX10 fluorescence microscope (Olympus). After scoring for GFP silencing, worms injected with unc-22 dsRNA were scored as silenced if they twitched while suspended in 3 mM tetramisole hydrochloride for at least 30 s.
For Fig. S3 A, the body cavities of L4-staged HC567 animals were injected with either 750 ng/µl in vitro–transcribed gfp-dsRNA (made by J. Marre, University of Maryland, College Park, MD) or 10 mM Tris-HCl, pH 8.5, and the number of brightly fluorescent intestinal nuclei in each injected animal at 15°C was counted after injection at a fixed magnification on a fluorescence microscope (MVX10; Olympus).
Machine learning
The measured fluorescence of nuclei in 14 cells (A–N) of 60 eri-1(−) animals were used. The relative intensity for a cell or pair of cells (in the case of J and N) was calculated as the mean of the relative intensity of all nuclei within that cell. SVM models were implemented using libsvm (Chang and Lin, 2011) from Scikit-learn 14.1 (Pedregosa et al., 2011) with a linear kernel and defaults for the remaining settings. Decision tree models were implemented using the decision tree classifier from Scikit-learn 14.1 with maximum depth set to three and defaults for the remaining settings. The results from both algorithms were validated using 10-fold cross-validation. Specifically, the data were split into ten folds of equal size. Each fold served as a test set once with the remaining nine folds serving as the training data. For each fold, accuracy (number of correctly classified cells/total number of cells) was computed; mean accuracy (over the ten folds) was reported for each of the models; and 95% confidence intervals were computed using Student’s t test.
Correlation analyses
Fluorescence intensity values of nuclei were averaged for each cell and the extents of linear correlation between pairs of cells were computed using the corrcoef function in MATLAB (MathWorks). Heatmaps of correlations were generated using the pcolor function in MATLAB for cells with significant values for Pearson’s r (Fig. S5) and representations of cells with significant values for Pearson’s r (P ≤ 0.05, two-tailed t test) in different genotypes were generated manually using Illustrator (Adobe; Fig. 6, G–K; and Fig. S5, insets).
Online supplemental material
Fig. S1 shows the analyses performed to deduce rearrangements within the sur-5::gfp repetitive transgene. Fig. S2 shows the characteristics of silencing that occurs in the absence of ERI-1. Fig. S3 shows evidence that the patterns of silencing observed in animals that lack ERI-1 are established early in development. Fig. S4 shows the lineal and spatial relationships among intestinal cells in C. elegans. Fig. S5 shows the cells with significant correlated and anticorrelated expression in different genetic backgrounds. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201601050.dv.
Acknowledgments
We thank Jayanth Banavar, Elissa Lei, Norma Andrews, Steve Wolniak, Steve Mount, Katerina Ragkousi, Daniel Damineli, and members of the Jose laboratory for comments; Steven Salzberg, Jeffrey Barrick, and Carl Kingsford for advice on bioinformatics; David Baillie and Andrew Fire for encouragement and advice on determining rearrangements in arrays; Alexander Mendenhall and Roger Brent for explaining the helical twist of the intestine; the Caenorhabditis elegans Genetic Stock Center and the Hunter laboratory (Harvard University) for some worm strains; and Amy Beaven (Cell Biology and Molecular Genetics Imaging Core) for microscopy advice.
This work was supported by a Maryland Summer Scholars grant from the University of Maryland (UMD; H.H. Le), a Howard Hughes Medical Institute grant from UMD (M. Looney), and a Research and Scholarship Awards grant from UMD (A.M. Jose), and in part by National Institutes of Health grants R00GM085200 and R01GM111457 (A.M. Jose).
The authors declare no competing financial interests.
Author contributions: H.H. Le and A.M. Jose initiated the research, and A.M. Jose analyzed DNA-Seq and RNA-Seq data of HC566. H.H. Le, M. Looney, and A.M. Jose designed and performed all other experiments except machine learning, which was designed and performed by M. Bloodgood and B. Strauss. All authors contributed to the writing of the manuscript.