Assembly of the Gag polyprotein into new viral particles in infected cells is a crucial step in the retroviral replication cycle. Currently, little is known about the onset of assembly in the cytosol. In this paper, we analyzed the cytosolic HIV-1 Gag fraction in real time in live cells using advanced fluctuation imaging methods and thereby provide detailed insights into the complex relationship between cytosolic Gag mobility, stoichiometry, and interactions. We show that Gag diffuses as a monomer on the subsecond timescale with severely reduced mobility. Reduction of mobility is associated with basic residues in its nucleocapsid (NC) domain, whereas capsid (CA) and matrix (MA) domains do not contribute significantly. Strikingly, another diffusive Gag species was observed on the seconds timescale that oligomerized in a concentration-dependent manner. Both NC- and CA-mediated interactions strongly assist this process. Our results reveal potential nucleation steps of cytosolic Gag fractions before membrane-assisted Gag assembly.
Introduction
Assembly of virus particles, from the newly synthesized viral components to the release of virus progeny at the plasma membrane, is an essential part of the replication cycle of retroviruses. The central orchestrator of this process is Gag, the conserved retroviral structural polyprotein (Ganser-Pornillos et al., 2012; Sundquist and Kräusslich, 2012; Bell and Lever, 2013). HIV-1 (Human immunodeficiency virus type 1) Gag consists of three structural domains (matrix [MA], capsid [CA], and nucleocapsid [NC]), the C-terminal domain p6 that acts as an adaptor for several virus and host proteins, and two spacer peptides (SP1 and SP2) separating CA-NC and NC-p6, respectively. HIV-1 Gag is synthesized on cytosolic polysomes and transported to the plasma membrane, where it multimerizes into a hexameric lattice. Gag by itself is sufficient for formation and budding of virus-like particles from the plasma membrane of eukaryotic cells. In virus-producing cells, Gag is also responsible for incorporating all components necessary for creating infectious viruses.
Three major functional interactions of Gag, which have been the subject of extensive studies, govern the assembly process and present targets for antiretroviral drug development (Waheed and Freed, 2012). (1) RNA binding: Virions released from infected cells contain two copies of genomic RNA; selective packaging of the genome is mediated by interaction of Gag with a specific packaging signal (psi) on the viral genome (Kuzembayeva et al., 2014). Particles formed in the absence of RNA carrying the psi sequence incorporate nonspecific RNA in similar amounts, indicating that RNA is an integral structural element of retroviruses (Muriaux et al., 2001; Rulli et al., 2007). Both specific and nonspecific RNA binding critically depend on the NC domain of Gag (Muriaux and Darlix, 2010; Rein et al., 2011): the two zinc fingers in NC are important for specific recognition of the psi sequence, whereas the high proportion of basic residues in NC confers strong nonspecific affinity to RNA in general. Accordingly, mutations that impair RNA binding, or the deletion of NC, affect virus formation. This defect can be partially rescued by overexpression of Gag or replacement of NC with heterologous protein–protein interaction domains. RNA is thus assumed to facilitate assembly by providing a scaffold for concentration of Gag molecules. (2) Protein–protein interactions: Hexameric interactions of the CA domains in the uncleaved Gag polyproteins are the central structural element of the immature Gag lattice, and hexamers of a different geometry are also the main building blocks of the mature cone-shaped CA of HIV (Briggs and Kräusslich, 2011; Ganser-Pornillos et al., 2012). CA thus forms the main building block of both the immature and the mature lattice. The architecture of these two lattices differs significantly (Schur et al., 2015), but the CA dimer interface centered around residues W184 and M185 in the C-terminal domain of CA (CACTD) represents a key feature of both multimeric arrangements. A region comprising CACTD, the adjacent spacer peptide SP1, and the N terminus of NC is critical for immature Gag lattice formation. Mutation of CA residues W184 and M185 to alanine severely impairs protein–protein interactions mediated by the CACTD and thereby interferes with formation of both immature and mature particles (Ganser-Pornillos et al., 2012; Sundquist and Kräusslich, 2012). (3) Membrane binding: Although immature-like virus-like particles assemble in the absence of lipid membranes in vitro, accumulation of Gag at the plasma membrane is required for virus bud formation. Myristoylation of the N-terminal glycine of Gag and a patch of basic residues in the N-terminal MA-domain of Gag confer affinity for the plasma membrane (Chukkapalli and Ono, 2011; Lorizate and Kräusslich, 2011). Specificity of plasma membrane binding appears to be mediated by interaction of the MA domain with PI(4,5)P2, a plasma membrane–specific lipid (Ono et al., 2004; Saad et al., 2008). Mutation of the myristoyl acceptor site prevents membrane association and thereby blocks virus release (Chukkapalli and Ono, 2011; Lorizate and Kräusslich, 2011). However, lack of myristoylation does not prevent assembly of the immature Gag lattice in vitro, and overexpression of nonmyristoylated Gag polyproteins in tissue culture has been shown to lead to assembly of immature-like, nonenveloped spherical particles in the cytosol (Royer et al., 1991; O’Carroll et al., 2012).
All three described interactions are important and contribute to the regulation of Gag assembly in infected cells. A current model proposes that cytosolic Gag is held in a closed conformation through binding of basic regions in both the MA and NC domains to RNA. PI(4,5)P2 binding of Gag induces conformational changes in Gag, leading to insertion of the myristoyl moiety into the inner leaflet of the plasma membrane and exposing the CA-SP1 region required for immature lattice formation.
Contrary to the vast amount of literature on membrane-bound Gag assemblies, little is known about the properties of cytosolic Gag. No clear consensus exists on the stoichiometry of Gag directly after translation and how its different domains contribute to stoichiometry. Data from biochemical and electron microscopy studies indicated the presence of oligomeric Gag complexes (Lee and Yu, 1998; Lee et al., 1999; Nermut et al., 2003). Förster resonance energy transfer experiments on Gag-expressing cells showed Gag–Gag interactions at the membrane, but intracellular Gag punctae were also observed (Derdowski et al., 2004). In the case of Rous sarcoma virus Gag, Förster resonance energy transfer measurements indicated intracellular Gag–Gag interactions for a membrane binding–deficient Gag mutant (Larson et al., 2003). Using fluorescence fluctuation spectroscopy (FFS), cytosolic oligomerization of Gag was observed for membrane binding–defective HIV-1 Gag, but not for wild-type (wt) HIV-1 Gag, nor for the corresponding protein from human T-lymphotropic virus type I (Fogarty et al., 2011a, 2014).
In this paper, we have elucidated the complex cytosolic interactions of HIV-1 Gag in living HeLa cells by separating Gag fractions with different mobilities and stoichiometries using quantitative fluorescence fluctuation imaging. By using point mutants of Gag defective for myristoylation, CA dimerization, and/or RNA binding, we provide clear evidence for the existence of RNA-interacting Gag oligomers in the cytosol, supporting a model in which HIV-1 assembly already initiates in the cytosol.
Results
Cytosolic fluctuation imaging of Gag
For intracellular visualization of Gag, we used an HIV-1 subgenomic plasmid (pKHIV) for transient eukaryotic expression of Venus-tagged Gag (Gag.Venus; Fig. 1 A) in the presence of all HIV-1 proteins except Nef. Immunoblot analyses confirmed expression of Gag.Venus in eukaryotic cells (Fig. S1 A), and immunoblots of virus lysates revealed correct proteolytic processing of the Gag.Venus polyprotein (Fig. S1 B). In agreement with previous studies (Larson et al., 2005; Jouvenet et al., 2008; Ivanchenko et al., 2009), confocal laser scanning microscopy (CLSM) of HeLa cells transfected with this plasmid revealed two phenotypes: (1) diffuse cytoplasmic Gag.Venus or (2) prominent membrane-localized Gag.Venus punctae (Fig. S1 C). Based on live-cell analyses of HIV-1 particle assembly (Ivanchenko et al., 2009; Baumgärtel et al., 2011), phenotype 1 cells can be assigned to the early phase of virus production, in which budding site formation at the plasma membrane is not yet detectable.
Characterization of Gag.Venus. (A) A schematic drawing showing the Gag polyprotein with the Venus fluorescent protein inserted between the MA and CA domains. A frameshift into the +1 reading frame generates the Gag-Pol polyprotein. Arrowheads indicate the PR cleavage sites in Gag. (B) Quantification of membrane-bound fractions in cells. (top right, scheme) A dual-color z stack (gray) is acquired, from which a z plot (cyan) and z image (pink) are calculated. (images) Confocal images of representative cells coexpressing Venus (top row), myristoylated/palmitoylated (MyrPalm)-mYFP (second row), Gag.Venus phenotype 1 (third row), and Gag.Venus phenotype 2 (bottom row), together with mCherry.The z plot (cyan squares) shows the mean intensity as a function of z position. In the z image (pink squares), the ratio of green-to-red (G/R) intensity is calculated per pixel in cytosolic or ventral membrane frames of the stack. The color scaling of the z images is given at the far right. These experiments were performed at least five times for each condition, but only single-cell analyses are shown. Bars, 5 μm for all images. au, arbitrary unit.
Characterization of Gag.Venus. (A) A schematic drawing showing the Gag polyprotein with the Venus fluorescent protein inserted between the MA and CA domains. A frameshift into the +1 reading frame generates the Gag-Pol polyprotein. Arrowheads indicate the PR cleavage sites in Gag. (B) Quantification of membrane-bound fractions in cells. (top right, scheme) A dual-color z stack (gray) is acquired, from which a z plot (cyan) and z image (pink) are calculated. (images) Confocal images of representative cells coexpressing Venus (top row), myristoylated/palmitoylated (MyrPalm)-mYFP (second row), Gag.Venus phenotype 1 (third row), and Gag.Venus phenotype 2 (bottom row), together with mCherry.The z plot (cyan squares) shows the mean intensity as a function of z position. In the z image (pink squares), the ratio of green-to-red (G/R) intensity is calculated per pixel in cytosolic or ventral membrane frames of the stack. The color scaling of the z images is given at the far right. These experiments were performed at least five times for each condition, but only single-cell analyses are shown. Bars, 5 μm for all images. au, arbitrary unit.
It has been shown before that bright oligomers at the plasma membrane can bias FFS experiments in the cytosol, where the thickness of the cell at the position of interest plays a role (Fogarty et al., 2011a). Therefore, we developed a quantitative strategy for imaging membrane-bound fractions inside cells (Materials and methods). The YFP-labeled protein of interest was coexpressed with a freely diffusing RFP (mCherry), and a dual-color z stack was acquired, from which a z plot and z image were calculated (Fig. 1 B). Measurements with Venus were performed as a negative control; a myristoylated and palmitoylated monomeric YFP was used as a positive control for membrane binding (Zacharias et al., 2002). The experiments clearly revealed that the z image, in particular, allows mapping of regions of strong membrane enrichment. Next, we analyzed HeLa cells expressing Gag.Venus and unfused mCherry. From the z images, we conclude that phenotype 1 cells are devoid of membrane punctae and are thus suitable for fluctuation analysis in the cytosol.
Two cytosolic Gag species with profoundly different dynamics
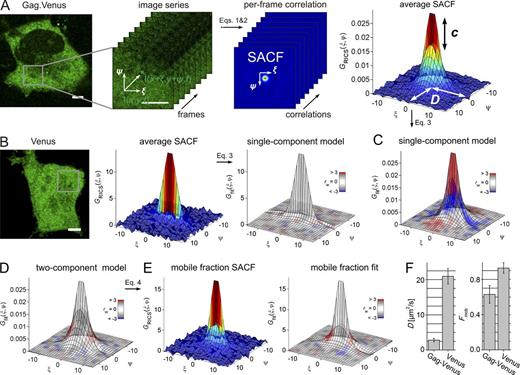
We investigated the mobility of Gag.Venus in the cytosol using raster image correlation spectroscopy (RICS; Digman et al., 2005). Acquisition of a 100-frame image series was performed in HeLa cells expressing Gag.Venus, as illustrated in Fig. 2 A. For each frame in the image series, a spatial autocorrelation function (SACF) was calculated (Eqs. 1 and 2), and all SACFs were averaged (Fig. 2 A). By fitting the SACF, the concentration C and diffusion coefficient D of diffusive Gag.Venus molecules were determined.
RICS analysis of cytosolic Gag. (A) Principle of RICS. (left) A representative HeLa cell expressing Gag.Venus. The highlighted area indicates the region used for RICS (gray square). (middle left) The selected area was consecutively imaged 100 times. (middle right) From each image in the series, an SACF is calculated, and all SACFs are averaged. (right) The mean SACF is depicted in 3D, color coded with the correlation value. From the amplitude and shape of the SACF, the mean concentration over the image and the diffusion coefficient of the sample can be determined. (B) Control RICS analysis of a cell expressing Venus. (left) Confocal image with the region used for RICS analysis indicated as a gray square. (middle) Mean SACF. (right) Single-component fit model to the mean SACF. The fits are color coded according to the value of the goodness-of-fit weighted residuals parameter rw, where gray illustrates a good fit and red–blue indicates regions where the residuals deviate by >3 σ. (C) Single-component fit of the data shown in A. (D) A two-component fit of the data shown in A. (E) Subtracting the immobile fraction determined via fitting with Eq. 4 from the experimental data reveals the SACF and corresponding fit of the mobile fraction. (F) Diffusion coefficient (left) and mobile fraction (right) for Gag.Venus and Venus. Error bars are the SDs from n measurements (see Table S1 for n). Bars, 5 µm.
RICS analysis of cytosolic Gag. (A) Principle of RICS. (left) A representative HeLa cell expressing Gag.Venus. The highlighted area indicates the region used for RICS (gray square). (middle left) The selected area was consecutively imaged 100 times. (middle right) From each image in the series, an SACF is calculated, and all SACFs are averaged. (right) The mean SACF is depicted in 3D, color coded with the correlation value. From the amplitude and shape of the SACF, the mean concentration over the image and the diffusion coefficient of the sample can be determined. (B) Control RICS analysis of a cell expressing Venus. (left) Confocal image with the region used for RICS analysis indicated as a gray square. (middle) Mean SACF. (right) Single-component fit model to the mean SACF. The fits are color coded according to the value of the goodness-of-fit weighted residuals parameter rw, where gray illustrates a good fit and red–blue indicates regions where the residuals deviate by >3 σ. (C) Single-component fit of the data shown in A. (D) A two-component fit of the data shown in A. (E) Subtracting the immobile fraction determined via fitting with Eq. 4 from the experimental data reveals the SACF and corresponding fit of the mobile fraction. (F) Diffusion coefficient (left) and mobile fraction (right) for Gag.Venus and Venus. Error bars are the SDs from n measurements (see Table S1 for n). Bars, 5 µm.
To validate the method and determine a baseline for free cytosolic diffusion of a small protein, cytosolic Venus alone was analyzed by RICS. Venus displayed a D = 21.0 ± 2.2 µm2/s, which could typically be described by a single diffusing species (Fig. 2 B and Table S1). In contrast, an additional second component, representing a species that is static on the subsecond RICS timescale but potentially mobile on a slower timescale (see Gag oligomers are diffusive on the second timescale), was necessary and sufficient to describe the data for Gag.Venus (Fig. 2, C and D). By analyzing many cells displaying low expression levels (with concentrations ranging from 30 to 120 nM; Table S1), we obtained a mean D = 2.8 ± 0.5 µm2/s and a mean mobile fraction of = 0.63 ± 0.1 (Table S1). This D is significantly slower than what we would expect for free diffusion of the Gag.Venus fusion protein. As the effective viscosity of the cytosol depends on the size of the diffusing molecule, we empirically validated the scaling of D with molecular size in the cytosol by RICS analysis of cytosolic tandem fusions of EGFP (Fig. S2 A; Pack et al., 2006). An EGFP tandem trimer (90 kD), which has a similar mass as the Gag.Venus protein (83 kD), displayed a diffusion coefficient of DEGFP3 = 9.5 ± 1.5 µm2/s. Hence, the dynamics of cytosolic Gag are indeed much slower than expected for free cytosolic diffusion of the protein.
NC-mediated interactions contribute to subsecond mobility of cytosolic Gag
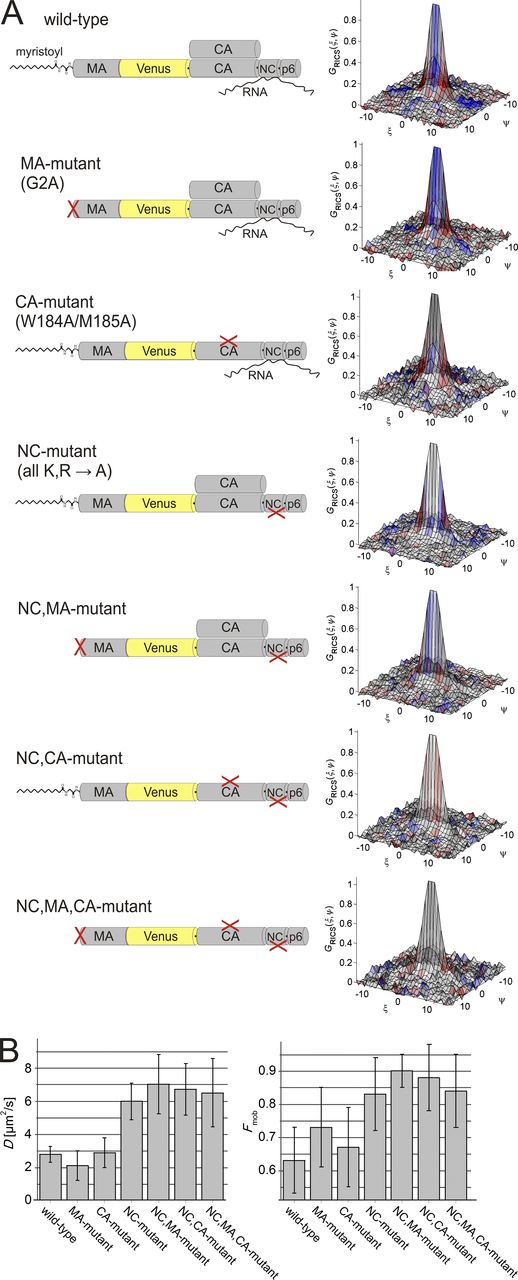
To determine interactions that could give rise to the subsecond mobility of cytosolic Gag, we performed a systematic study of the behavior of a set of Gag mutants (Fig. 3 A). Mutation of the N-terminal glycine to alanine (MA mutant) prevents Gag myristoylation, severely affecting membrane interaction of the protein. The CA mutant contains an exchange of two residues in the dimer interface of the CA domain (W184A and M185A). RNA interactions of the NC domain of Gag are abolished in the NC mutant, in which all 15 lysine and arginine codons in the NC domain were replaced by alanine codons (Cimarelli et al., 2000). We confirmed the known inhibition of HIV-1 particle formation in cells transfected with the respective pKHIVVenus derivatives by immunoblotting (Fig. S1 D).
RICS analysis of the mobility of various Gag mutants. (A) Schematic representation and RICS analysis of the different point mutants of Gag used in this work. Shown on the right are representative, normalized, experimental SACFs of the mobile fraction of wt Gag and given mutants. Data are color coded according to the value of the goodness-of-fit weighted residuals parameter rw, where gray illustrates a good fit and red–blue indicates regions where the residuals deviate by >2.5 σ. The narrower shape of the SACF for the constructs with mutations in the NC domain indicates higher mobility for these mutants. (B) Column graph with the diffusion coefficients (left) and mobile fraction (right) determined from fits to the SACF. Error bars are the SDs of n experiments (see Table S1 for n). For proteins containing NC mutations, the mobile fraction is statistically larger than in experiments with mutants that do not have the NC mutations.
RICS analysis of the mobility of various Gag mutants. (A) Schematic representation and RICS analysis of the different point mutants of Gag used in this work. Shown on the right are representative, normalized, experimental SACFs of the mobile fraction of wt Gag and given mutants. Data are color coded according to the value of the goodness-of-fit weighted residuals parameter rw, where gray illustrates a good fit and red–blue indicates regions where the residuals deviate by >2.5 σ. The narrower shape of the SACF for the constructs with mutations in the NC domain indicates higher mobility for these mutants. (B) Column graph with the diffusion coefficients (left) and mobile fraction (right) determined from fits to the SACF. Error bars are the SDs of n experiments (see Table S1 for n). For proteins containing NC mutations, the mobile fraction is statistically larger than in experiments with mutants that do not have the NC mutations.
RICS analyses revealed that the experimental SACFs of the MA and CA mutant were similar to that of wt Gag with D values of 2.5 ± 1.6 and 2.9 ± 0.9 µm2/s, respectively, whereas the correlation function for the NC mutant revealed faster diffusion (6.0 ± 1.1 µm2/s; Fig. 3 and Table S1). To investigate potential synergistic effects between the different mutations, we combined the NC mutation with the MA mutation, the CA mutation, or both. All variants carrying the NC mutation displayed D values (6–7 µm2/s) and mobile fractions (0.8–0.9) that were both larger in comparison with the wt protein (Fig. 3 and Table S1). The results suggest that the interactions leading to slower cytosolic diffusion of the mobile Gag molecules are directly or indirectly mediated by the NC domain.
Subsecond diffusive Gag is monomeric
To directly investigate oligomerization of the mobile Gag fraction (D = 2.8 ± 0.5 µm2/s), we performed cross-correlation RICS (ccRICS; Digman et al., 2009) using pulsed interleaved excitation (PIE; Müller et al., 2005; Hendrix et al., 2013) on HeLa cells coexpressing Gag.Venus and Gag.mCherry (Fig. 4 A). An immunoblot analysis confirmed the correct intracellular expression and proteolytic cleavage of Gag.mCherry (Fig. S1 A). With ccRICS, the simultaneously recorded image series of two spectrally distinct fluorophores are spatially auto- and cross-correlated (Eqs. 1 and 2 and Fig. 4 B). The ratio of the spatial cross-correlation function (SCCF) amplitude to the Venus SACF amplitude is taken as a measure for the interaction affinity (Fig. 4 B). Either the total data or the “static” component–subtracted data can be used as input for this ratio (CCtotal or CCmob; see Materials and methods).
Investigation of Gag interactions using ccRICS. (A) Schematic drawing of Gag with mCherry inserted between the MA and CA domains. Arrowheads indicate the PR cleavage sites. (B) Principle of ccRICS. (left) Representative HeLa cell coexpressing Gag.Venus (green) and Gag.mCherry (red), with the region used for ccRICS in gray. (middle) Dual-color cross talk–free image series. Bars, 5 µm. (right) The Gag.Venus and Gag.mCherry SACF and spatial cross-correlation function (SCCF) are calculated from each frame using Eq. 2 and averaged. (C) ccRICS analysis of a HeLa cell expressing the tandem heterodimer mVenus-mCherry. The experimental SCCF on the left is color coded according to the correlation value, whereas the color of the two-component SCCF model on the right represents the value of the weighted residuals parameter rw (inset). (D) Data from C, with the static component subtracted. (E) Bar chart summarizing the mean CCtotal (Eq. 6) and CCmob (Eq. 7) parameters obtained after analysis of n cells expressing mVenus-mCherry or Venus + mCherry (as a negative control). The asterisk indicates that no significant cross-correlation was observed. (F) ccRICS analysis of the cell shown in B. Color coding as in C. (G) Data from F, with the static component subtracted. (H and I) Bar chart summarizing the mean CCtotal (Eq. 6) and CCmob (Eq. 7) parameters obtained after analysis of n cells expressing wt Gag (H) or mutants of Gag (I), respectively. Error bars in E, H, and I are the SDs on n experiments (see Table S2 for n). Table S2 also summarizes the most relevant parameters obtained after fitting.
Investigation of Gag interactions using ccRICS. (A) Schematic drawing of Gag with mCherry inserted between the MA and CA domains. Arrowheads indicate the PR cleavage sites. (B) Principle of ccRICS. (left) Representative HeLa cell coexpressing Gag.Venus (green) and Gag.mCherry (red), with the region used for ccRICS in gray. (middle) Dual-color cross talk–free image series. Bars, 5 µm. (right) The Gag.Venus and Gag.mCherry SACF and spatial cross-correlation function (SCCF) are calculated from each frame using Eq. 2 and averaged. (C) ccRICS analysis of a HeLa cell expressing the tandem heterodimer mVenus-mCherry. The experimental SCCF on the left is color coded according to the correlation value, whereas the color of the two-component SCCF model on the right represents the value of the weighted residuals parameter rw (inset). (D) Data from C, with the static component subtracted. (E) Bar chart summarizing the mean CCtotal (Eq. 6) and CCmob (Eq. 7) parameters obtained after analysis of n cells expressing mVenus-mCherry or Venus + mCherry (as a negative control). The asterisk indicates that no significant cross-correlation was observed. (F) ccRICS analysis of the cell shown in B. Color coding as in C. (G) Data from F, with the static component subtracted. (H and I) Bar chart summarizing the mean CCtotal (Eq. 6) and CCmob (Eq. 7) parameters obtained after analysis of n cells expressing wt Gag (H) or mutants of Gag (I), respectively. Error bars in E, H, and I are the SDs on n experiments (see Table S2 for n). Table S2 also summarizes the most relevant parameters obtained after fitting.
For validation, we first analyzed the tandem heterodimer mVenus-mCherry expressed in HeLa cells by ccRICS. The positive cross-correlation (Fig. 4 C, left) indicates that Venus and mCherry are part of the same complex. We fitted the experimental data to a two-component model including a second component that is “static” on the timescale determined by the acquisition parameters (∼1 s for the data presented here; and Fig. 4 C, right). The static component was subtracted (Eq. 8) from the experimental data (Fig. 4 D, left), and what remained is the mobile component (Fig. 4 D, right). We then calculated a CCtotal = 0.414 ± 0.055 (Eq. 6) and CCmob = 0.360 ± 0.046 (Eq. 7) for the total signal and for the mobile component, respectively (Fig. 4 E). We conclude that the mVenus-mCherry tandem fusion is exclusively diffusive on the subsecond timescale. As a negative control, we analyzed cells expressing Venus and mCherry separately. Within experimental error, no cross-correlation was observed (Fig. 4 E), which is expected when PIE excitation is used.
Cells coexpressing Gag.Venus and Gag.mCherry exhibited no detectable cross-correlation after fitting a two-component model to the experimental SCCF and subtracting the static component from the data (Fig. 4, F–H). This result directly shows that Gag molecules that are diffusive on the subsecond timescale are monomeric. We performed an analogous analysis for the point mutants of Gag described in Fig. 3. Interestingly, although no subsecond cross-correlation was observed (CCmob = 0) for any of the investigated mutants (Fig. 4 I, white columns), the total cross-correlation (CCtotal) amplitude appears to depend on the type and number of mutations in Gag (Fig. 4 I, gray columns). This could indicate that the static component is influenced by the mutations. Hence, Gag oligomers with lower mobility may exist, but the ccRICS analysis clearly shows a monomeric subsecond diffusive Gag species.
Gag oligomers are diffusive on the second timescale
To further investigate the possible presence of Gag oligomers, we used temporal image correlation spectroscopy (TICS; Srivastava and Petersen, 1998). Here, fluorescence time traces from individual pixels in a confocal image series are each temporally autocorrelated (Eq. 10 and Fig. 5 A). By fitting a model to the mean temporal autocorrelation function (TACF; Eq. 11), TICS allows quantification of the diffusion coefficient of species that diffuse on the timescale of several seconds, i.e., much slower than the timescales typically accessible with RICS or FFS (microseconds to milliseconds). A 3,000-frame image series was acquired in cells expressing Gag.Venus (Fig. 5 A). TACFs were calculated for the time traces derived from pixels in the image series and averaged. For validation, we first performed TICS analysis of cells expressing freely diffusing Venus. A slow correlation on the order of 10−4 µm2/s was detected that corresponds to microscope drift and/or cellular or organelle motion within the cell. No temporal correlation was present that could be assigned to actual protein diffusion (Fig. 5 B and Table S3). For cells expressing Gag.Venus, on the other hand, a slowly diffusive species with D1 = 0.014 ± 0.002 µm2/s was detected (Table S3).
Investigation of Gag diffusing on the second timescale using (S)TIC(C)S. (A) Principle of TICS. (left) A representative HeLa cell expressing Gag.Venus with the region used for TICS indicated in gray. (middle) Zoom-in on the region that was analyzed. (right) Representative fluorescence time trace of a single pixel after preprocessing (see Materials and methods and Eq. 9). (B) The mean TACFs of all measurements (squares with error bars) for Gag.Venus and Venus. The top graph displays the weighted residuals, rw(τ), for the two-component fits in the bottom graph. The data and corresponding fits are displayed after subtraction of cell or cell-organelle movement. (C) Representative spatiotemporal image correlation spectroscopy (STICS)/iMSD analysis of HeLa cells expressing Gag.Venus illustrating confined diffusion of the Gag complexes. The solid line is a linear fit (Eq. 14) to the first two points yielding values of D = 0.009 µm2/s and ωr = 0.24 µm. In other analyses (four independent experiments), the trends in the data were similar. (D) The principle of TICCS with PIE. (left) A representative HeLa cell coexpressing Gag.Venus and Gag.mCherry is shown with the region used for the TICCS analysis indicated in gray. (right) A dual-color cross talk–free fluorescence time trace calculated on a per pixel basis. (E) Gag-specific TICCS analysis of cells (eight cells) coexpressing Gag.Venus and Gag.mCherry. The data (squares with error bars) represent the mean TCCF from different independent experiments. The data and fit (solid line, Eq. 11) were normalized to GTICCS(τ = 0.32 s) after subtracting the offset y0 and the slow diffusion component representing cell or cell-organelle movement. The top graph shows the reduced residuals, rw(τ). for the different fit functions in the bottom graph. (black) Gag.Venus + mCherry, single-component fit, (green) Gag.Venus + Gag.mCherry, two-component fit. As a control, the mean TCCF from cells coexpressing Gag.Venus and mCherry is displayed. Error bars in B and E are the standard error of the mean from n experiments (see Table S3 for n). Table S3 also summarizes the most relevant parameters obtained from the fits. Bars, 5 µm.
Investigation of Gag diffusing on the second timescale using (S)TIC(C)S. (A) Principle of TICS. (left) A representative HeLa cell expressing Gag.Venus with the region used for TICS indicated in gray. (middle) Zoom-in on the region that was analyzed. (right) Representative fluorescence time trace of a single pixel after preprocessing (see Materials and methods and Eq. 9). (B) The mean TACFs of all measurements (squares with error bars) for Gag.Venus and Venus. The top graph displays the weighted residuals, rw(τ), for the two-component fits in the bottom graph. The data and corresponding fits are displayed after subtraction of cell or cell-organelle movement. (C) Representative spatiotemporal image correlation spectroscopy (STICS)/iMSD analysis of HeLa cells expressing Gag.Venus illustrating confined diffusion of the Gag complexes. The solid line is a linear fit (Eq. 14) to the first two points yielding values of D = 0.009 µm2/s and ωr = 0.24 µm. In other analyses (four independent experiments), the trends in the data were similar. (D) The principle of TICCS with PIE. (left) A representative HeLa cell coexpressing Gag.Venus and Gag.mCherry is shown with the region used for the TICCS analysis indicated in gray. (right) A dual-color cross talk–free fluorescence time trace calculated on a per pixel basis. (E) Gag-specific TICCS analysis of cells (eight cells) coexpressing Gag.Venus and Gag.mCherry. The data (squares with error bars) represent the mean TCCF from different independent experiments. The data and fit (solid line, Eq. 11) were normalized to GTICCS(τ = 0.32 s) after subtracting the offset y0 and the slow diffusion component representing cell or cell-organelle movement. The top graph shows the reduced residuals, rw(τ). for the different fit functions in the bottom graph. (black) Gag.Venus + mCherry, single-component fit, (green) Gag.Venus + Gag.mCherry, two-component fit. As a control, the mean TCCF from cells coexpressing Gag.Venus and mCherry is displayed. Error bars in B and E are the standard error of the mean from n experiments (see Table S3 for n). Table S3 also summarizes the most relevant parameters obtained from the fits. Bars, 5 µm.
We analyzed the space–time relationship of the dynamics of this slow Gag.Venus component by calculating a diffusion law with the recently developed imaging mean squared displacement (iMSD) method (Di Rienzo et al., 2013). The diffusion law calculation for cytosolic Gag.Venus revealed confined rather than Brownian diffusion in the cytosol (Fig. 5 C). This suggests the protein is most likely trapped by binding to a larger interaction partner. As a negative control, an iMSD analysis of cells expressing freely diffusing Venus showed no broadening of the SACF with time (Fig. S3 B).
To investigate the presence of an oligomeric state of the TICS-mobile Gag species, we performed a dual-color cross-correlation TICS (temporal image cross-correlation spectroscopy [TICCS]) analysis (Wiseman et al., 2000). Gag.Venus and Gag.mCherry were coexpressed, and 3,000 frames were recorded with dual-color PIE excitation (Fig. 5 D). If complexes containing both Venus and mCherry fluorophores exist, the amplitude of the mean temporal cross-correlation function (TCCF) will differ from 0. Control cells coexpressing Gag.Venus and free mCherry yielded an amplitude of 0, as expected for noninteracting proteins (Fig. 5 E). For cells coexpressing Gag.Venus and Gag.mCherry, on the other hand, the amplitude was positive (Fig. 5 E), directly proving the existence of diffusive complexes containing heterooligomers of Gag. The minimal model for an adequate fit contained two diffusing components (D1 = 0.017 ± 0.007 µm2/s and D2 = 2.9 [±0.6] × 10−4 µm2/s; Table S3), the faster one (displayed as a green solid line in Fig. 5 E) corresponding to an oligomeric Gag species. The slower component is observed in all TICS experiments and is attributed to microscope drift and/or cellular artifacts (Fig. 5 B).
Concentration-dependent oligomerization of cytosolic Gag
The TICCS data clearly demonstrated cytosolic Gag oligomers. To investigate the stoichiometry of these complexes, we used the number and brightness (N&B) analysis method (Digman et al., 2008). Here, the mean pixel intensity, as well as the variance on this intensity, is determined for each pixel in a confocal image series. The number N of molecules present in the focal volume, as well as the molecular brightness ε (number of photons detected per molecule per second), can be estimated from each pixel, as they contribute differently to the mean and the variance of the signal (Fig. 6 A and Eqs. 16 and 17). For validation, we first investigated cells expressing Venus (Fig. 6 B, left). As expected for a freely diffusing monomeric protein, we observed a broad distribution in N (Fig. 6 B, middle), whereas ε was low and did not depend on the signal intensity (Fig. 6 B, right). Similarly, Gag.Venus at low (∼200 nM) expression levels (Fig. 6 C, left) displayed a broad distribution of N (Fig. 6 C, middle), and relatively constant ε as a function of mean intensity (Fig. 6 C, right). The situation was markedly different in cells with higher overall expression levels (fivefold in the example shown in Fig. 6 D). Whereas the N histogram did not change significantly (Fig. 6 D, middle), many pixels in the image (not only the brighter spots) exhibited a significantly larger ε, and a clear correlation with the mean signal intensity was observed (Fig. 6 D, right). This demonstrates the existence of oligomeric cytosolic Gag complexes in these cells.
Concentration dependence of cytosolic Gag oligomerization investigated using the N&B method. (A, left) A representative HeLa cell expressing Gag.Venus at levels slightly above background (Imean = 27 ± 8 kHz). (middle) The highlighted region (gray square) was imaged 100 times for the N&B analysis. (right) A representative fluorescence time trace of a single pixel after preprocessing (Eq. 15). (B, left) Image of a representative cell expressing Venus along with the corresponding N histogram (middle) and a 2D histogram of ε versus mean intensity (right). (C) Analysis of the cell shown in A. (middle) N histogram (Eq. 16). (right) A 2D histogram of ε (Eq. 17) versus mean intensity. The amplitudes of the 2D histograms are scaled logarithmically. (D, left) A representative image of a cell with higher expression of Gag.Venus (Imean = 150 ± 50 kHz) along with the corresponding N histogram (middle) and 2D histogram of ε versus mean intensity (right). (E) Plot of the mean cytosolic stoichiometry (ε divided by mean εVenus = 5.3 ± 0.3 kHz) versus the total monomer concentration for Venus (black) and Gag.Venus (red). Molar concentrations were obtained from number concentrations (Eq. 19) by dividing the mean image intensity by the mean εVenus. Each data point represents a measurement in a separate cell. Solid lines are linear fits with y intercept = 1. (F) Plot of the mean cytosolic stoichiometry versus the total monomer concentration for the MA mutant (black). The solid red line is the linear fit of the WT data from E. Bars, 5 µm.
Concentration dependence of cytosolic Gag oligomerization investigated using the N&B method. (A, left) A representative HeLa cell expressing Gag.Venus at levels slightly above background (Imean = 27 ± 8 kHz). (middle) The highlighted region (gray square) was imaged 100 times for the N&B analysis. (right) A representative fluorescence time trace of a single pixel after preprocessing (Eq. 15). (B, left) Image of a representative cell expressing Venus along with the corresponding N histogram (middle) and a 2D histogram of ε versus mean intensity (right). (C) Analysis of the cell shown in A. (middle) N histogram (Eq. 16). (right) A 2D histogram of ε (Eq. 17) versus mean intensity. The amplitudes of the 2D histograms are scaled logarithmically. (D, left) A representative image of a cell with higher expression of Gag.Venus (Imean = 150 ± 50 kHz) along with the corresponding N histogram (middle) and 2D histogram of ε versus mean intensity (right). (E) Plot of the mean cytosolic stoichiometry (ε divided by mean εVenus = 5.3 ± 0.3 kHz) versus the total monomer concentration for Venus (black) and Gag.Venus (red). Molar concentrations were obtained from number concentrations (Eq. 19) by dividing the mean image intensity by the mean εVenus. Each data point represents a measurement in a separate cell. Solid lines are linear fits with y intercept = 1. (F) Plot of the mean cytosolic stoichiometry versus the total monomer concentration for the MA mutant (black). The solid red line is the linear fit of the WT data from E. Bars, 5 µm.
We plotted the mean cytosolic stoichiometry as a function of concentration for all independent measurements of cells expressing Gag.Venus or Venus, respectively (Fig. 6 E). For Venus, the stoichiometry did not vary with concentration. In contrast, the mean stoichiometry of cytosolic Gag.Venus (ε divided by mean εVenus = 5.3 ± 0.3 kHz) clearly increased from unity at low concentrations (∼100 nM) to 3–4 at 2.5 µM, indicating that the intrinsic affinity for cytosolic oligomerization of Gag.Venus is in the low micromolar range. As suggested earlier, oligomeric Gag might have a higher affinity for the cell membrane, thereby being effectively removed from the cytosol (Fogarty et al., 2011b). Therefore, we investigated a possible effect of Gag membrane binding on these results by performing a similar N&B analysis on the MA mutant. This mutant exhibited a similar, if not identical, oligomerization behavior as wt Gag (Fig. 6 F). Although this does not exclude that Gag oligomers might have a higher intrinsic affinity for the cell membrane, it is clear that interactions with the cell membrane mediated by myristoylated MA are not required for cytosolic oligomerization. Together, these experiments suggest that the MA domain plays a minor role (if any) in the cytosolic nucleation stage of Gag assembly.
Priming of Gag for cytosolic oligomerization
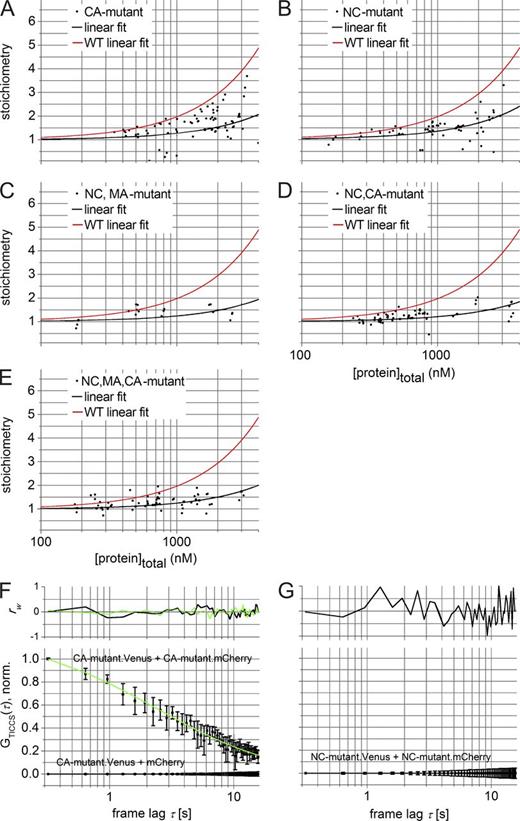
It had previously been suggested that RNA binding primes Gag for oligomerization (Jouvenet et al., 2009; Rein et al., 2011). We set out to investigate this hypothesis using fluctuation imaging. Using the N&B method, we first measured the cytosolic stoichiometry of the described Gag mutants (Fig. 3) as a function of cytosolic Gag concentration. The CA mutant (Fig. 7 A), and, even more so, the NC mutant (Fig. 7 B), displayed a clearly lower propensity for oligomerization than the wt. In agreement with the ccRICS results shown in Fig. 4 I, variants carrying mutations in two or three domains resulted in an even lower tendency to oligomerize, although the additional effect was small relative to the NC mutation alone (Fig. 7, C–E). A TICCS analysis of cells expressing the CA or NC mutants showed detectable cross-correlation for the CA mutant (Fig. 7 F), whereas no cross-correlation was observed for the NC mutant (Fig. 7 G). These results indicate that impairing interactions mediated by the NC or CA domain significantly affects the formation of Gag oligomers that diffuse on the seconds timescale. This is consistent with RNA acting as a primer for the cytosolic assembly of Gag oligomers.
The effect of Gag mutations on cytosolic Gag oligomerization. (A–E) N&B analysis of the different Gag mutants. Plot of the mean cytosolic stoichiometry (ε divided by mean εVenus = 5.3 ± 0.3 kHz) versus the total monomer concentration for the CA mutant (A), NC mutant (B), NC,MA mutant (C), NC,CA mutant (D), and MC,MA,CA mutant (E) of Gag. Molar concentrations are obtained from number concentrations (Eq. 19) by dividing the mean image intensity by the mean εVenus. Each data point represents a measurement in a separate cell. The solid black line is a linear fit with y intercept = 1, and the solid red line is the linear fit of the wt data from Fig. 6 E. (F) Gag-specific TICCS analysis of cells coexpressing the CA mutant.Venus and the CA mutant.mCherry. The data (squares with error bars) represent the mean TCCF from different independent experiments. The data and fit (solid line, Eq. 11) were normalized to GTICCS(τ = 0.32 s) after subtracting the offset y0 and the slow diffusion component representing cell or cell-organelle movement. The top graph shows the reduced residuals, rw(τ), for the different fit functions in the bottom graph. (black) CA mutant.Venus + mCherry, single-component fit; (green) CA-mutant.Venus + CA-mutant.mCherry, two-component fit. (G) Gag-specific TICCS analysis of cells coexpressing Gag.Venus- and Gag.mCherry-tagged versions of the NC mutant Gag. Data (squares with error bars) represent the mean CCF from different independent experiments with the standard error of the mean on n independent experiments (see Table S3 for n). The data and fit (solid line, Eq. 11) were normalized to GTICCS(τ = 0.32 s) after subtracting the offset y0 and the slow diffusion component representing cell or cell organelle movement. The top graph displays the weighted residuals, rw(τ). As a control, the analysis of cells coexpressing the Venus-tagged CA mutant and mCherry are displayed in G. Table S3 summarizes the most relevant parameters obtained after fitting of the TICCS data.
The effect of Gag mutations on cytosolic Gag oligomerization. (A–E) N&B analysis of the different Gag mutants. Plot of the mean cytosolic stoichiometry (ε divided by mean εVenus = 5.3 ± 0.3 kHz) versus the total monomer concentration for the CA mutant (A), NC mutant (B), NC,MA mutant (C), NC,CA mutant (D), and MC,MA,CA mutant (E) of Gag. Molar concentrations are obtained from number concentrations (Eq. 19) by dividing the mean image intensity by the mean εVenus. Each data point represents a measurement in a separate cell. The solid black line is a linear fit with y intercept = 1, and the solid red line is the linear fit of the wt data from Fig. 6 E. (F) Gag-specific TICCS analysis of cells coexpressing the CA mutant.Venus and the CA mutant.mCherry. The data (squares with error bars) represent the mean TCCF from different independent experiments. The data and fit (solid line, Eq. 11) were normalized to GTICCS(τ = 0.32 s) after subtracting the offset y0 and the slow diffusion component representing cell or cell-organelle movement. The top graph shows the reduced residuals, rw(τ), for the different fit functions in the bottom graph. (black) CA mutant.Venus + mCherry, single-component fit; (green) CA-mutant.Venus + CA-mutant.mCherry, two-component fit. (G) Gag-specific TICCS analysis of cells coexpressing Gag.Venus- and Gag.mCherry-tagged versions of the NC mutant Gag. Data (squares with error bars) represent the mean CCF from different independent experiments with the standard error of the mean on n independent experiments (see Table S3 for n). The data and fit (solid line, Eq. 11) were normalized to GTICCS(τ = 0.32 s) after subtracting the offset y0 and the slow diffusion component representing cell or cell organelle movement. The top graph displays the weighted residuals, rw(τ). As a control, the analysis of cells coexpressing the Venus-tagged CA mutant and mCherry are displayed in G. Table S3 summarizes the most relevant parameters obtained after fitting of the TICCS data.
Discussion
In this study, we investigated the cytosolic dynamics of HIV-1 Gag using a comprehensive toolbox based on advanced image correlation spectroscopy techniques. Both the monomeric Gag species, which diffuses on the subsecond timescale, and the slower, oligomeric Gag species, which diffuses on the second timescale, are cytosolic. We presume that both will later contribute to the nucleation and growth of HIV-1 membrane assembly sites during virus replication (Jouvenet et al., 2008; Ivanchenko et al., 2009). We propose a model where a small number of oligomeric Gag complexes bound to viral RNA initiate nucleation of a new assembly site at the membrane to which, as shown by one of our previous studies (Ivanchenko et al., 2009), monomeric Gag is added from a large cytosolic pool to form a complete virus particle (Fig. 8).
Model illustrating the dynamic interplay between different cytosolic Gag species. Cytosolic Gag interacts dynamically with RNA (green box: Species 1a and Species 1b), giving rise to a normally diffusive, yet slow Gag species (Species 1), as well as to a confined diffusive, very slow Gag oligomer (Species 2). Breaking interactions through the NC domain do not render the protein completely freely diffusive, suggesting other cytosolic components still interact with Gag (Species 1b and illustrated by a gray ellipse). The expected diffusion coefficient of D = 9.5 µm2/s for Gag without any interactions is based on RICS measurements of the EGFP tandem trimer having an Mr comparable to free monomeric Gag.Venus. In this model, we do not focus on the actual conformation (bent or extended) of cytosolic Gag but, for simplicity, schematically depicted monomeric Gag in its extended conformation to illustrate the different diffusive species. What species are observed for the different mutants is summarized on the bottom. For simplicity, we only discuss interactions leading to viral assembly. MW, molecular weight; myr, myristoyl.
Model illustrating the dynamic interplay between different cytosolic Gag species. Cytosolic Gag interacts dynamically with RNA (green box: Species 1a and Species 1b), giving rise to a normally diffusive, yet slow Gag species (Species 1), as well as to a confined diffusive, very slow Gag oligomer (Species 2). Breaking interactions through the NC domain do not render the protein completely freely diffusive, suggesting other cytosolic components still interact with Gag (Species 1b and illustrated by a gray ellipse). The expected diffusion coefficient of D = 9.5 µm2/s for Gag without any interactions is based on RICS measurements of the EGFP tandem trimer having an Mr comparable to free monomeric Gag.Venus. In this model, we do not focus on the actual conformation (bent or extended) of cytosolic Gag but, for simplicity, schematically depicted monomeric Gag in its extended conformation to illustrate the different diffusive species. What species are observed for the different mutants is summarized on the bottom. For simplicity, we only discuss interactions leading to viral assembly. MW, molecular weight; myr, myristoyl.
In contrast to fluorescence wide-field studies and to studies that focus on Gag–membrane interactions, we exclusively focus on cytosolic Gag trafficking and oligomerization. With the aid of z plots/images and several control experiments (Fig. 1 and Fig. S4), we could focus on the behavior of cytosolic Gag without being biased by artifacts from (quasi-) immobile cell membrane–bound Gag assemblies. The presence of membrane aggregates can have a significant impact on the diffusion time and diffusion coefficient determined for cytosolic proteins, as was previously shown by Fogarty et al. (2011a) who used two-photon excitation FFS to measure the diffusion of C-terminally EYFP-tagged human T–lymphotropic virus (HTLV-1) and HIV-1 Gag protein in HeLa cells. To minimize this effect, we selected cells that were in the early phase of virus formation and thus devoid of pronounced membrane-associated Gag assemblies. In addition to Gag assembly sites, intracellular vesicles, organelles, and other structures devoid of Gag can also contribute to fluctuations in fluorescence and influence results obtained from image correlation analyses. Therefore, we corrected for slow motion in our analysis, which yielded reliable diffusion values for the different cytosolic Gag species.
The diffusion coefficient of D = 2.8 ± 0.5 µm2/s for faster diffusing, monomeric Gag species (Fig. 8, Species 1) is in good agreement with the Gag mobility study by Larson et al. (2003) who measured a slow diffusive component of D = 3.2 ± 0.6 µm2/s in chicken fibroblast cells expressing C-terminally EGFP-tagged Rous sarcoma virus Gag by two-photon excitation FFS. In contrast to our study, Larson et al. (2003) also observed a fast diffusing subpopulation with a diffusion coefficient of D = 23 ± 5 µm2/s, similar to cytosolic EGFP. This discrepancy could potentially be explained by the fact that the EGFP blinks on the timescale of 100 s of microseconds (Haupts et al., 1998), which could be interpreted as an apparent fast diffusing species when measured with fluorescence correlation spectroscopy (FCS; Hendrix et al., 2011). The diffusion time of 4 ms (corresponding to D = ∼1–3 µm2/s) measured by Fogarty et al. (2011a) for C-terminally EYFP-tagged HTLV-1 Gag protein in HeLa cells is consistent with our observations.
The importance of the NC domain for Gag oligomerization and associated Gag membrane trafficking has been reported in many studies. Deletions of either the complete NC domain (Larson et al., 2003), of only the basic residues (Ottmann et al., 1995; Poon et al., 1996; Cimarelli et al., 2000; Chukkapalli et al., 2010; Datta et al., 2011; Jones et al., 2011) or of the NC zinc finger motifs (Mark-Danieli et al., 2005; Grigorov et al., 2007; El Meshri et al., 2015) strongly reduced Gag–Gag oligomerization, altered Gag membrane trafficking and virus particle formation, and sometimes even leads to intracellular aggregation. This is consistent with our measurements as the diffusion coefficient of monomeric, cytosolic Gag significantly increased from 2.8 ± 0.5 µm2/s to around 6.0 ± 1.2 µm2/s (Fig. 8, Species 1b) when the NC domain was disrupted by exchanging all basic residues (K,R → A). Mutations affecting only the CA dimerization domain (W184A and M185A) or only the myristoylation site (G2A) did not significantly alter the measured mobility of cytosolic Gag. Combined mutations that included alterations to the NC domain displayed increased diffusion but no synergistic effects were observed (Fig. 3 and Table S1).
We attribute the decrease in cytosolic Gag mobility to transient Gag–RNA interactions through the NC domain, which have been observed before by combined membrane flotation and immunoprecipitation assays (Kutluay and Bieniasz, 2010). The observed dynamics of the NC mutant of Gag (D = 6.0 ± 1.2 µm2/s) was still slower than what would be expected for free cytosolic diffusion of monomeric Gag.Venus (Dfree = ∼9.5 µm2/s). This effect cannot be ascribed to the Gag-Pol polyprotein, as it is only present as a minor species (∼5%) in the transfected cells. This suggests that other factors (Fig. 8, gray ellipses) are affecting the cytosolic dynamics of Gag, such as interactions with ABCE1 or Staufen (for an overview of known cytosolic interaction partners of Gag, see Meng and Lever, 2013). One possibility is that Gag–RNA interactions are also mediated by the MA domain, the latter containing a high number of basic residues and showing affinity for RNA (Freed et al., 1994), preferentially tRNA (Kutluay et al., 2014). The interactions need not be long lived; transient interactions can also explain the slower observed mobility of monomeric Gag in the cytosol. In the case of transient interactions with a much slower diffusing biomolecule, the effective diffusion coefficient is given by where and koff are the rate constants for association and dissociation, respectively (Crank, 1975; Hendrix et al., 2011). If transient interactions were the only factor contributing to the decrease in Gag mobility, this would indicate that Gag spends roughly 70% of time bound to other factors (Fig. 8, Species 1a/1b illustrated as rapid on–off equilibrium). In addition, the mobility of proteins is influenced by their conformation. In an earlier study, it has been proposed that Gag can adopt a closed conformation in the cytosol (Datta et al., 2011). A compact conformation would be expected to have higher mobility then an extended conformation and thus cannot be the reason for the slower diffusion. Most likely, the slower diffusion is caused by a combination of multiple effects discussed in this paper.
When we applied the TICS (Fig. 5 and Table S3) analysis, which potentially allows detection of larger complexes, we observed a second cytosolic Gag species that diffuses much more slowly with D = 0.014 ± 0.002 µm2/s (Fig. 8, Species 2). Using dual-color ccRICS (Fig. 4 and Table S2), dual-color TICCS (Fig. 5, Fig. 7 and Table S3), and analysis by N&B (Fig. 6 and Fig. 7), we could confirm that this additional Gag species in the cytosol is indeed oligomeric. Oligomerization of the slow diffusing Gag species was concentration dependent and was not detected at Gag concentrations below ∼200 nM (Fig. 6 E). This observation supports a Gag-dependent interaction mechanism and is consistent with the results from FFS measurements of HIV-1 Gag and HTLV-I Gag alone by Fogarty et al. (2011a). Mutations in the CA dimerization interface severely hamper CA dimerization in vitro (Gamble et al., 1997), impair virus formation, and reduce infectivity (von Schwedler et al., 1998). Accordingly, the CA mutant showed less oligomerization as a function of total Gag concentration in comparison to wt Gag (Fig. 7 A). A similar effect was observed for the NC mutant, suggesting that RNA interactions are also important for formation of these oligomers. The combined CA-NC mutant showed an even stronger impact on oligomerization. Hence, both domains play a significant role in the formation of the cytosolic oligomers. In contrast, mutation of the myristoylation site did not alter the oligomerization behavior of the slow diffusing species with respect to wt Gag.
Recently, Chen et al. (2014) measured the cytosolic diffusion coefficient of Bgl-YFP–labeled HIV-1 RNA in HeLa cells to be in the range of D = 0.07 − 0.3 µm2/s. In their analysis, they also included an additional slower diffusion component (fixed to D1 = 0.01 µm2/s), which was attributed to the mobility of species below their detection limit. The faster diffusing RNA species is distinct from the cytosolic Gag oligomers in our experiments. Upon coexpression with Gag, Chen et al. (2014) neither detected a change in RNA mobility nor cotrafficking with Gag. However, as a result of the much slower mobility of the RNA with respect to Gag, transient interactions of a few monomeric Gag molecules would not be detectable.
Based on our results, we propose a model in which two distinct Gag species, a monomeric and an oligomeric species, exist in the cytosol (Fig. 8). These Gag oligomers are formed independent of plasma membrane interactions because mutation of the myristoylation signal had no impact on cytosolic Gag oligomerization (Fig. 6 F and Fig. 8, MA mutant with species 1 and 2 present). Accordingly, cytosolic oligomerization does not depend on a possible conformational change induced by the interaction of negatively charged PI(4,5)P2 with the positively charged MA domain (Ono et al., 2004; Chukkapalli et al., 2010; Chukkapalli and Ono, 2011; Datta et al., 2011; Rein et al., 2011). This is in agreement with the observation that myristoylation-deficient Gag can form spherical structures in the cytoplasm upon overexpression (O’Carroll et al., 2012).
In contrast, cytosolic oligomerization was critically dependent on basic residues in the NC domain (Fig. 7 B), suggesting that Gag oligomers are bound to RNA. Membrane assembly and release of virus-like particles do not absolutely require an intact NC domain (Ott et al., 2003; O’Carroll et al., 2013), implying that interactions taking place in the cytosol and at the membrane are different at least in part. CA–CA interactions during Gag oligomerization lead to exposure of the myristoyl anchor in the MA domain (Saad et al., 2006, 2007; Li et al., 2007), proposed to promote Gag membrane binding. Thus, we expect that the oligomeric species observed can bind to the plasma membrane. There, they would recruit further Gag molecules to the nascent budding site, where assembly into a multimeric structure is facilitated by PI(4,5)P2 induced plasma membrane binding. This is in agreement with results from Jouvenet et al. (2008) who reported MS2-GFP–bound viral RNA appearing at the membrane several minutes before Gag-mCherry was detected to assemble into virus-like particles. As a result of the detection limit of their method (12 individual Gag molecules), these authors could not observe small Gag oligomers bound to RNA as potential nucleation sites at the membrane. With our methods, we could actually detect the RNA binding–dependent formation of small Gag oligomeric species in the cytosol, which may serve as nucleators for virus assembly. These nucleators are not absolutely essential for formation of virus-like particles but may be functionally important for infectious virus formation.
Conclusions
In this work, we show a clear connection between the cytosolic mobility of the HIV-1 Gag protein, its cytosolic stoichiometry, and its interactions within the cytosol. We provide direct proof for two markedly different Gag species: subsecond-diffusive monomers of Gag undergoing fast interactions with factors in the cytosolic environment mediated by the NC domain and an oligomeric Gag species diffusive on the seconds timescale. Formation of the oligomeric species was dependent both on the integrity of the CA dimer interface and Gag–RNA interactions via the NC domain. The presence of cytosolic Gag oligomers suggests that viral assembly initiates via Gag–RNA and Gag–Gag interactions within the cytosol. Upon binding to the plasma membrane, these oligomers act as a nucleation site for the formation of RNA-containing HIV-1 particles. This study clearly demonstrates that fluctuation imaging can provide crucial quantitative information necessary for understanding complex biological systems such as the onset of HIV-1 Gag assembly in the cytoplasm.
Materials and methods
Plasmids
It has been shown before that assembly and maturation steps of HIV-1 replication can be studied using fluorescence microscopy when an EGFP moiety is inserted between MA and CA subgenes of the gag-pol gene and when expressed as part of a proviral derivative of HIV-1 (pCHIVEGFP) lacking only the long terminal repeat regions and the nef gene (Müller et al., 2004). The latter allows experiments to be performed outside of a P3 facility. To optimize the system for this study, we introduced three modifications to the aforementioned procedure. (1) The HIV-1 coding sequences were expressed from a low copy plasmid backbone. For this, a BgII–XhoI fragment comprising viral protein coding sequences except for nef from proviral plasmid pNL4-3 (position 678–8,893; Adachi et al., 1986) were inserted into the eukaryotic expression vector pKEX-2-XR (cytomegalovirus immediate early promoter/enhancer, simian virus 40 T antigen splice, and polyadenylation signal and hygromycin resistance gene in a pUC19 backbone; Rittner et al., 1991), resulting in plasmid pKHIV (Mergener et al., 1992). The plasmid lacks a eukaryotic origin of replication, resulting in slower accumulation of viral proteins in transfected cells. The egfp gene was replaced by the coding sequence for the EYFP derivative Venus (Nagai et al., 2002; gift of A. Miyawaki, Institute of Physical and Chemical Research, Wako, Japan), whose photochemical properties are preferable for quantitative microscopy (Nagai et al., 2002; Hendrix et al., 2013). For dual-color experiments, the egfp gene was replaced by the coding sequence for mCherry (template pmCherry.N1; Takara Bio Inc.). PCR fragments encoding Venus or mCherry were cloned into a unique ClaI site introduced into a subviral plasmid comprising a BssHII–ApaI fragment from pNL4-3 in a pBluescriptKS backbone followed by transfer of BssHII–SphI fragments comprising part of the Gag coding region into pKHIV, generating pKHIVVenus and pKHIVmCherry, respectively.
The MA mutants were generated by introducing a mutation of codon G2 in MA to A2 through overlap PCR mutagenesis into the respective constructs (primers: forward, 5′-GCTAGAAGGAGAGAGATGGCTGCGAGAGCGTCGGTC-3′; reverse, 5′-GACCGACGCTCTCGCAGCCATCTCTCTCCTTCTAGC-3′; the underlined sequence indicates the mutated codons). The CA mutants were generated by introducing a mutation of codons for W185 and M186 in CA to A185 and A186 through overlap PCR mutagenesis into the respective wt or MA-NC mutant constructs (primers: forward, 5′-CAGGAGGTAAAAAATGCGGCGACAGAAACCTTG-3′; reverse, 5′-CAAGGTTTCTGTCGCCGCATTTTTTACCTCCTG-3′; the underlined sequence indicates the mutated codons). The NC mutants were generated by exchanging an SphI–StuI fragment covering part of Gag and Gag-Pol from the pKHIV-based plasmids and the MA mutant derivatives by the respective fragment from pNL4-3_15A (provided by A. Cimarelli, Ecole Normale Superièure, Lyon, France; Poon et al., 1996; Li et al., 2007). Combinations of mutations were generated by exchange of an SphI–StuI fragment encoding the NC mutation into the respective MA mutant variant and by overlap PCR to introduce the CA mutation into the double mutant. Constructs were verified by sequencing (GATC Biotech).
pMyrPalm.mYFP (Zacharias et al., 2002) was provided by R. Tsien (University of California, San Diego, La Jolla, CA). Plasmid pcsVenus was a gift of A. Miyawaki. The pmVenus-C1 plasmid encoding free monomeric Venus and the pmVenus-mCherry plasmid encoding the cross-correlation control have been described previously (Hendrix et al., 2013). The plasmids encoding the tandem EGFP fusions were provided by M. Kinjo and S. Mikuni (Hokkaido University, Sapporo, Japan).
Cell culture and transfection for microscopy
HeLa cells (National Institutes of Health Reagent program) were cultivated without antibiotics in low-glucose DMEM (Life Technologies) supplemented with 10% heat-inactivated fetal bovine serum (complete medium) at 5% CO2 and 37°C in a humidified atmosphere. For transfection, 3 × 104 cells per well were plated in complete medium in 8-well coverslips (Lab-Tek II Chambered Coverglass; Thermo Fisher Scientific). Plasmid DNA (500 ng in total) was mixed with 1.5 µl XtremeGene 9 transfection reagent (Roche) in 50 µl OptiMEM and incubated for 15–20 min at RT, before addition of 100 ng of pKHIVFP plasmid to cells 6–8 h after seeding. For cotransfections with another pKHIVFP plasmid, a 1:1 plasmid weight ratio was used. For cotransfections with pmCherry-N1 or pVenus encoding the free fluorescent proteins, a 50–100:1 of pKHIVFP/fluorescent protein plasmid weight ratio was used. It has been reported before that only when cotransfecting with equimolar amounts of labeled and unlabeled Gag in assembly experiments, the wt morphology of the virus is maintained (Müller et al., 2004). For the experiments in this paper, however, it was necessary to use only fluorescently labeled constructs. This does not pose a problem as we are investigating the very first steps in Gag oligomerization in the cytosol, rather than the tight packing of Gag in nascent virions.
Virus particle production and immunoblotting
293T cells were transfected with the indicated plasmids using FuGENE 6 (Promega). At 44 h after transfection, cell lysates and culture supernatants were harvested. Viruses were collected from filtered supernatants by ultracentrifugation through a 20% (wt/wt) sucrose cushion. Samples were separated by SDS-PAGE (17.5%; acrylamide/bisacrylamide 200:1), and proteins were transferred to a nitrocellulose membrane by semidry blotting. HIV-1 proteins were stained using polyclonal antisera from sheep (CA) or rabbit (MA), raised against recombinant HIV-1 CA or MA purified from Escherichia coli. Bound antibody was detected by quantitative immunoblotting with an infrared imaging system (Odyssey; LI-COR Biosciences), using secondary antibodies and protocols recommended by the instrument’s manufacturer.
Confocal imaging
At 15–18 h after transfection, cells were washed twice with prewarmed phenol red-free DMEM (Life Technologies) and incubated with the same buffer. To reduce the motion of cells and cellular organelles, all imaging was performed at RT (22–23°C). A home-built PIE fluctuation imaging (PIE-FI) microscope (schematic of the setup is shown in Fig. S5) was used, operated with home-written acquisitioning software (C# language), as described elsewhere (Hendrix et al., 2013). The alignment of the microscope was routinely checked using FCS with PIE (Müller et al., 2005) of a mixture of ATTO565-NHS and ATTO488-CA (D22–23°C = 370 µm2/s; Kapusta, 2010) at 1 µm from the coverslip. In brief, imaging was always performed at the center of the total (60 µm)2 galvanometric mirror scan range, to avoid spherical aberrations introduced by the telescope. In between scans, the focal spot was positioned outside of the scan range to avoid additional photobleaching within the scan region. Images were collected at a distance of 1.5 µm from the coverslip (unless stated otherwise) using one laser (475 nm for Venus/EGFP) or two lasers (475 and 561 nm for mCherry) at a power of ∼0.8 µW per laser in solution. The lasers were synchronized to a master clock of 27.4 MHz but delayed by ∼18 ns with respect to each other for PIE (Müller et al., 2005). The descanned emission was subsequently detected using avalanche photodiodes and recording with time-correlated single-photon counting (Becker et al., 1999). Cross talk–free images were generated by time gating the raw photon data (Lamb et al., 2005), i.e., by selecting only those photons for creating images that were emitted by Venus/EGFP or mCherry after pulsed excitation at 475 or 561 nm, respectively. Image calculation from the raw photon data and subsequent analysis was performed with our Microtime Image Analysis (MIA) software. MIA is a well-documented, user-friendly, stand-alone and freely available program (MATLAB; The MathWorks GmbH) for global, serial, and automated analysis of CLSM images (using continuous-wave excitation or PIE) that can perform (cc)RICS, N&B, and (S)TI(C)CS analyses.
For accurate confocal imaging in the cytosol, the majority of measurements were performed at different z positions of the sample to check for potential biases from the bottom or top membranes. Typically, three z planes at 500-nm intervals were consecutively imaged, one image for each z position, and then repeated 100 times. Consecutive imaging ensured any photobleaching or lateral drift effects were similar in all z planes during the whole experiment. We also used an NA = 1.49 objective (CFI Apochromat TIRF [total internal reflection fluorescence] 100× oil NA 1.49; Nikon) to image thinner optical slices than with a normal NA = 1.2 objective, at higher brightness (and consequently with lower photobleaching; Fig. S4 A and Table S4). With this objective, it was still possible to image cytosolic diffusion constants in the presence of excess membrane-bound fluorescent molecules (Fig. S4 B). Also, we experimentally verified the effect of the distance of the objective focus to the coverslip on the RICS analysis inside cells (Fig. S4 C).
Wide-field imaging
To simplify the localization of cells that have been successfully transfected with all constructs, a wide-field microscope system was integrated into our confocal scanning laser microscope. For the wide-field path, the 488-nm line of a multi-line Ar–Kr laser (Spectra-Physics) and a 561-nm diode-pumped solid-state laser (Jive; Cobolt) were combined and coupled into a single-mode fiber (QPMJ-A3A-A3-488; OZ Optics) that was guided to the microscope. After the collimator (HPUCO-23AF-400; Thorlabs) at the end of the fiber, the beam was expanded 10-fold by two achromatic lenses (Thorlabs) to a diameter of ∼3 cm before being focused onto the back focal plane of the objective (f = 300 mm achromatic lens). A polychroic mirror (LC-470-568-690TBDR; Laser Components GmbH) and a dual-band emission filter (Semrock Brightline HC 523/610; AHF analysentechnik AG) were used to separate the emitted fluorescence from the excitation beam, and then, the fluorescence was guided either to the eyepiece and/or to a charge-coupled device camera for detection (QiCAM; QImaging). Switching from confocal to wide field was possible by flipping a mirror into the confocal path and by switching from a normal to the polychroic mirror in the cube slider of the microscope. Switching was possible without affecting the alignment of the confocal path.
Autofocus
For prolonged imaging without focal drift in the z direction and for acquiring z stacks, a home-built autofocus system was used after a principle similar to what was first published by Hellen and Axelrod (1990). To implement the autofocus system, the mirror that reflected the confocal beam upwards to the objective was replaced with a dichroic mirror (Semrock FF875; AHF analysentechnik AG) that transmits light from a 980-nm laser (CNI-MDL-III-980-100mW; Laser 2000 GmbH). The laser entered the microscope via a side port. The tube lens was removed from behind the objective. The 980-nm beam was focused near the edge of the back aperture of the objective to achieve total internal reflection at the coverslip. The reflected light, and thus axial sample drift, was detected on a position-sensing detector (CONEX-PSD9; Newport Spectra-Physics GmbH) and used to maintain the axial focus position within an accuracy of 50–100 nm via a PID (proportional integral derivative)-type feedback control loop. Focus positioning was physically performed with a piezo-electric sample actuator (P-517.3CL; E-501-00; Physik Instrumente GmbH & Co.), analogously controlled over a PCI (peripheral component interconnect) board (M2i.6034; Spectrum Systementwicklung Microelectronic GmbH) or an objective actuator (MIPOS 100, series NV 40/1; Piezosystems Jena GmbH) digitally controlled over an RS232 serial port.
z profiling
For characterizing membrane binding of Gag, a z stack was recorded with 50-nm intervals between frames, spanning a total distance of 4 µm in the z direction. Individual images were spatially averaged with a Gaussian filter (5 × 5 pixels, σ = 75 nm) to improve data quality. Extracellular regions were omitted from further analysis. The mCherry intensity in each frame of the stack was scaled with a constant to obtain a Venus/mCherry (green to red) intensity ratio of unity in the cytosol. From this normalized stack, a z plot and z image were subsequently calculated. For the z plot, the intensity at a particular (x,y) location in the stack was plotted as a function of z position. For the z image, the green-to-red ratio in the cytosol or ventral cell membrane was represented in a false-color image.
RICS and ccRICS
For RICS and ccRICS, 100 frames per z position were recorded (12.5 × 12.5 µm or 300 × 300 pixels) at a frame time of τf = 1 s, interframe time τfi = ∼0.5 s, line time τl = 3.33 ms, pixel dwell time τp = 11.11 µs, and pixel size δr = 41.7 nm. Slow fluctuations and spatial inhomogeneities were removed by first preprocessing images with a high-pass filter:
where I(x,y,f) is the photon count of pixel (x,y) in frame f,
is the moving average series from frames (f − ΔF) to frame (f + ΔF), and 〈I〉XYF is the mean intensity over all frames F with size X × Y. Unless stated otherwise, a moving average of ΔF = 1 was used for RICS analysis, which corresponded to a 4-s window (3τf + 2τif). Subsequently, for each frame, an SACF and SCCF were calculated as described previously (Petersen et al., 1993) using a two-dimensional discrete Fourier transform algorithm:
where ξ and ψ are the correlation lags (in pixel units) in the fast and slow scanning directions of the galvanometer mirror system, respectively. I1 = I2 for an autocorrelation analysis, I1 ≠ I2 when two different images are used for a cross-correlation analysis, and
is the mean image intensity with X and Y being the respective dimensions of the image, with i as the image channel.
RICS analysis
The mean of the SACF series was analyzed by nonlinear least-squares fitting with a two-component (one mobile and one immobile) model for Brownian diffusion in a three-dimensional Gaussian point spread function (PSF):
where
is the time-dependent component of the correlation function ascribed to fast diffusing molecules. Amob and Aimm are the amplitudes of the mobile and immobile component in the correlation function, respectively, and y0 is a baseline correction term. The fraction of immobile molecules (on the RICS timescale) was described with a symmetrical 2D Gaussian function, where ωimm represents the half-width of the function at 1/e2 of the maximal intensity.
is the average number of molecules in the PSF that are mobile on the RICS timescale, with γ = 2−3/2 as the shape factor for a 3D Gaussian, and is a correction factor needed to calculate absolute concentrations when the moving average subtraction is used. is the ratio of the correlation function amplitude attributed to molecules diffusing on the RICS timescale. This is not the actual fraction of the mobile species, Fmob, as Amob and Aimm scale not only with concentration but also the molecular brightness of the underlying species (Lamb et al., 2000; Rigler and Elson, 2001). Therefore, the values for reported in this work are not to be taken as absolute estimates of the mobile fraction.
After analyzing the individual cells, a mean D parameter was calculated from those analyses that met the following empirical criteria: 95% confidence interval for D smaller than ±D/2, a reduced χ2 goodness-of-fit parameter ≤3, a weighted residuals parameter rw mostly ≤3, and without large systematic deviations and a relative SD, σSACF,rel, <5%. The σSACF,rel is obtained by dividing σSACF, the mean SD on the pixel values in the SACF, by the experimental SACF amplitude, which we approximated by Typical values were σSACF,rel,Venus = 0.5–3% and σSACF,rel,mCherry = 1–4%. Furthermore, both D and could only be determined simultaneously during fitting when In other cases, D was estimated by globally linking between individual SACFs in a large dataset.
The mobile fraction SACF was obtained by subtracting the immobile fit component from the experimental data:
ccRICS analysis
In addition to fitting to the mean SACFs, a cross-correlation model that accounts for a small, lateral shift between the 475- and 561-nm excitation foci was fitted to the mean of the SCCF series:
where
and sx and sy are the offsets in x and y directions between the two images, respectively. Only the time-independent part of this equation is affected by sx and sy, because dual-color images are shifted with respect to each other in space, but not in time. The offset was between 0 and 80 nm, significantly smaller than the PSF and lowers the maximum possible cross-correlation between the two channels (Weidemann et al., 2002). The foci also did not overlap perfectly in the z direction (sz ≈ 200 nm), but because a small axial offset (sz < ωz ≈ 0.6–1 µm) mostly only affects the amplitude of the cross-correlation function and not its shape, this was not taken into account. is the fraction of the CCF amplitude attributed to dual-labeled complexes on the RICS timescale. After fitting, only those data that met the following empirical criteria were included into further analysis: a reduced χ2 goodness-of-fit parameter ≤ 3, a weighted residuals parameter rw mostly ≤ 3, and without large systematic deviations and a relative SD, σSCCF,rel, <15%. The σSCCF,rel is obtained by dividing σSCCF, the mean SD on the pixel values in the SCCF, by the experimental SCCF amplitude, which we approximated by For datasets with σSCCF,rel > 15%, it was no longer possible to distinguish the experimental SCCF from the noise (Fig. S2 B). For these datasets, the amplitude of the cross-correlation function, Gfit,SCCF(0,0), used during analysis was assumed to be 0. Typical values were σSCCF,rel = 4–30%.
Next, an estimate for the cross-correlation between the two channels in the image series was obtained by calculating the normalized total cross-correlation parameter (Saito et al., 2004; Kohl et al., 2005):
The mean CCtotal reported in this work was calculated from those experiments with 0.2 < NVenus/NmCherry < 5, where NVenus and NmCherry are the observed number concentrations of Venus and mCherry carrying molecules, respectively.
An estimate for the cross-correlation signal that is solely caused by diffusion of a dual-labeled protein complex on the subsecond timescale was obtained by calculating the normalized mobile fraction cross-correlation parameter:
The mean CCmob was calculated similarly as for CCtotal. Under the same conditions as above, CCmob approximates the amount of molecules carrying both labels, relative to the total number of molecules carrying the mCherry (or Venus) label.
Analogous to the single-color RICS analysis, the experimental SCCF of the mobile component was obtained by subtracting the immobile fit component from the experimental data:
TICS and TICCS
To obtain adequate counting statistics per pixel, 3,000 frames (5 × 5 µm or 100 × 100 pixels) were recorded for TICS and TICCS analysis at a frame time τf = 0.1 s, interframe time τif = ∼0.22 s, line time τl = 2 ms, pixel dwell time τp = 40 µs, and pixel size δr = 50 nm. To remove overall intensity variations, image series were preprocessed with
where 〈I(f)〉XY is the mean intensity of frame f with size X × Y, and 〈I(f)〉XYF is the mean total intensity from the entire measurement. Next, dim pixels, with 〈ITICS(x,y)〉F < 〈I〉XYF/3, were removed from the analysis. Pixelwise TACF and TCCF were subsequently calculated using a one-dimensional discrete Fourier transform algorithm (Petersen et al., 1993):
where τ is the time lag, I1 = I2 for autocorrelation, and I1 ≠ I2 for dual-color cross-correlation. Next, a one- or two-component model for 3D diffusion was fitted to the data:
where A1 is the correlation amplitude of proteins diffusing on the seconds timescale, and A2 is the correlation amplitude as a result of very slow motion (e.g., cell drift or cell organelle movement). For data recorded with a single detector, the first data point in the TACF (i.e., τ = 0) was omitted from fitting as a result of the autocorrelation of shot noise at τ = 0. To avoid signal end effects during TICS analysis, TACFs and TCCFs were fitted only until a 50-frame lag (i.e., 15.68 s). Slower dynamics (>15.68 s) were too slow to be discerned from cell or cell organelle movement.
STICS, STICCS, and iMSD analysis
For the STICS, STICCS, and iMSD analyses, the same preprocessed data were used as for the TICS/TICCS analysis. Spatio-TACFs and spatio-TCCFs were calculated using a two-dimensional discrete Fourier transform algorithm as described previously (Wiseman et al., 2004):
where time lags τ are equal to 0.33–6.27 s (1–19 image frames). Correlation functions were subsequently fitted to a sum of two 2D Gaussians:
where y0(τ) is an offset, Ai(τ) is the amplitude, and ωi(τ) a measure for the width of Gaussian i. For spatio-TACFs, the lateral offsets sx and sy were 0. Next, a mean squared displacement versus time plot was generated and fit to a straight line to obtain an estimate of the diffusion coefficient of species i (Di Rienzo et al., 2013):
where ωr is on the order of the lateral PSF waist if species i has a physical size smaller than the diffraction limit. For an iMSD simulation, see Fig. S3 A.
Number and brightness
The same data as measured for RICS was used for the N&B analysis. Before preprocessing, images were first corrected for the dead time of the time-correlated single-photon counting system (Hillesheim and Müller, 2003; Hendrix et al., 2013; Becker, 2014). Pixel-specific intensity fluctuations on the >13-s timescale were removed by preprocessing the image series with
where is the spatially averaged image series I(x,y,f), calculated with a pillbox averaging filter of radius R,
is the mean intensity image, and F is the number of recorded frames. For the N&B experiments presented here, R = 5 and ΔF = 4 were used (corresponding to a 13-s time window). Next, pixels with 〈INB(x,y)〉F < 10 kHz were removed from the analysis. Subsequently, the molecular number N(x,y) and brightness ε(x,y) were calculated using (Digman et al., 2008):
and
where
is the temporal variance image over all frames F in the preprocessed image series. For consistency with the number concentration from the RICS analyses, capital N is used throughout this paper rather than n, as in the original publication on N&B (Digman et al., 2008).
In the presence of both mobile and immobile molecules, the measured brightness ε can be used to calculate the fraction of molecules Fimm that is not mobile on the timescale of the N&B analysis (Ossato et al., 2010):
where εcontrol is the measured brightness for a control protein that exhibits a negligible (or no) immobile fraction. In Eq. 18, it is assumed that the brightness of mobile and brightness of immobile molecules are equal. If, on the other hand, the immobile molecules are k-fold brighter, Fimm will be overestimated by
where Nimm and Nmob are the number concentrations of immobile and mobile diffusive molecules, respectively.
Absolute concentrations
Average number concentrations, N, from RICS and N&B were converted to the absolute molar concentration C using (Ivanchenko and Lamb, 2011):
where NA is Avogadro’s number.
Online supplemental material
Fig. S1 shows characterization of the viral particles used. Fig. S2 shows reference RICS measurements on EGFP oligomers and an exemplary ccRICS measurement illustrating the σSCCF,rel. Fig. S3 shows a simulation and reference experiment for STICS/iMSD analysis. Fig. S4 shows different control experiments for cytosolic RICS experiments. Fig. S5 shows a scheme of the microscope setup. Three tables are included that summarize the results of the RICS (Table S1), ccRICS (Table S2), and TI(C)CS (Table S3) analysis. The last table (Table S4) gives a characterization of the objectives used in this study.
Acknowledgments
Anke-Mareil Heuser is gratefully acknowledged for her expert assistance with molecular cloning and biochemistry. Jaroslava Obel’s assistance with cell culture was appreciated. Dipl. Chem. Sigurd Vogler is thanked for assistance during microscopy. Dr. Matthias Höller is acknowledged for preliminary experiments. We thank A. Cimarelli, A. Miyawaki, R. Tsien, M. Kinjo, and S. Mikuni for providing plasmids. We thank M.Sc. Eng. Tomas Dekens (Laboratory for Digital Speech and Signal Processing,Vrije Universiteit Brussel) for fruitful discussions on data analysis.
J. Hendrix is grateful for a postdoctoral fellowship and travel grant from the Research Foundation Flanders. D.C. Lamb wishes to acknowledge financial support of the Deutsche Forschungsgemeinschaft (DFG) through the Excellence Initiatives Nanosystems Initiative Munich, Center for Integrated Protein Science Munich, and SFB 1032 (Project B3) and the Ludwig-Maximilians-Universität München through the Center for Nano Science and the LMUinnovative BioImaging Network. H.-G. Kräusslich and B. Müller acknowledge DFG support through SFB638 (H.-G. Kräusslich) and grant MU885/5-1 (B. Müller). H.-G. Kräusslich and B. Müller are investigators of the CellNetworks Cluster of Excellence. This work was supported by the National Institutes of Health National Institutes of Health grants P41 GM103540 and P50-GM076516 (E. Gratton and M.A. Digman).
The authors declare no competing financial interests.
References
- CA
capsid
- ccRICS
cross-correlation RICS
- CLSM
confocal laser scanning microscopy
- FCS
fluorescence correlation spectroscopy
- FFS
fluorescence fluctuation spectroscopy
- iMSD
imaging mean squared displacement
- MA
matrix
- N&B
number and brightness
- NC
nucleocapsid
- PIE
pulsed interleaved excitation
- PIE-FI
PIE fluctuation imaging
- PSF
point spread function
- RICS
raster image correlation spectroscopy
- SACF
spatial autocorrelation function
- SCCF
spatial cross-correlation function
- STICS
spatio-TICS
- TACF
temporal autocorrelation function
- TCCF
temporal cross-correlation function
- TICS
temporal image correlation spectroscopy
- TICCS
temporal image cross-correlation spectroscopy
- wt
wild type
- STICCS
spatio-TICCS
Author notes
J. Hendrix’s present address is Laboratory for Photochemistry and Spectroscopy, Department of Chemistry, KU Leuven, B-3001 Leuven, Belgium.