Canonical WNT signaling stabilizes β-catenin to determine cell fate in many processes from development onwards. One of its main roles in skeletogenesis is to antagonize the chondrogenic transcription factor SOX9. We here identify the SOXC proteins as potent amplifiers of this pathway. The SOXC genes, i.e., Sox4, Sox11, and Sox12, are coexpressed in skeletogenic mesenchyme, including presumptive joints and perichondrium, but not in cartilage. Their inactivation in mouse embryo limb bud caused massive cartilage fusions, as joint and perichondrium cells underwent chondrogenesis. SOXC proteins govern these cells cell autonomously. They replace SOX9 in the adenomatous polyposis coli–Axin destruction complex and therein inhibit phosphorylation of β-catenin by GSK3. This inhibition, a crucial, limiting step in canonical WNT signaling, thus becomes a constitutive event. The resulting SOXC/canonical WNT-mediated synergistic stabilization of β-catenin contributes to efficient repression of Sox9 in presumptive joint and perichondrium cells and thereby ensures proper delineation and articulation of skeletal primordia. This synergy may determine cell fate in many processes besides skeletogenesis.
Introduction
Cell lineage commitment and differentiation are crucial decisions that are continuously made and secured by progenitor/stem and differentiated cells from development onwards. As a pivotal target and transcriptional coactivator of the canonical WNT pathway, β-catenin has a central role in governing such decisions in a plethora of processes (Clevers and Nusse, 2012; Valenta et al., 2012). Its gene is ubiquitously expressed, but in the absence of canonical WNT signaling, newly synthesized cytosolic protein is rapidly recruited into a molecular scaffold known as the adenomatous polyposis coli (APC)–Axin1 destruction complex. Once there, it undergoes a sequence of modifications that lead to its degradation. It is first phosphorylated by CK1 and then by GSK3. It is next ubiquitinated upon interaction with β-TRCP, and it is finally degraded in proteasomes. Upon binding to Frizzled (FZD)–LRP5/6 receptor complexes, canonical WNT ligands initiate a molecular cascade that results in β-catenin stabilization. The mechanisms involved in this cascade are incompletely understood. A paradigm-shifting study has proposed that WNT signaling does not affect β-catenin targeting by CK1 and GSK3 but prevents recruitment of β-TRCP, causing saturation of the destruction complex with phosphorylated, nonubiquitinated β-catenin (Li et al., 2012). In contrast, more recent studies have proposed that recruitment of the destruction complex to the WNT–FZD–LRP5/6 signalosome causes Axin dephosphorylation followed by dissociation of the complex and sequestration of its components into multivesicular bodies (Hernández et al., 2012; Kim et al., 2013; Vinyoles et al., 2014). All models nevertheless agree that canonical WNT signaling allows β-catenin to accumulate in the cytoplasm and to translocate into the nucleus, where it partners with T cell–specific transcription factor (TCF)/lymphoid enhancer-binding factor (LEF) transcription factors to activate genes. Multiple factors have been shown to bind β-catenin and modulate its stability in vitro, but their in vivo contributions are unknown or limited (Kormish et al., 2010; Valenta et al., 2012). Thus, canonical WNT signaling is known today as the leading mechanism for β-catenin stabilization.
Vertebrate skeletogenesis is an archetypal process controlled by β-catenin (Lefebvre and Bhattaram, 2010; Regard et al., 2012; Baron and Kneissel, 2013; Long and Ornitz, 2013). It starts in the embryo when multipotent mesenchymal cells coalesce into skeletogenic masses. Chondrocytes differentiate in the center of condensations and form cartilage primordia. They are surrounded by presumptive joint and perichondrium cells. Cartilage primordia evolve into growth plates, which ensure skeletal elongation before being replaced by bone trabeculae and marrow. Perichondrium evolves into cortical and trabecular bone, and presumptive joints develop into articular cartilage, synovium, and ligaments. Skeletogenesis thus heavily relies on fate decisions made by multipotent mesenchymal cells. The decision to become a chondrocyte is dictated by SOX9, an SRY-related high-mobility group box transcription factor. The Sox9 gene is expressed in multipotent mesenchymal cells and throughout chondrocyte early differentiation but is turned off in cells that commit to nonchondrocytic fates. The latter include presumptive joint and perichondrium cells. Their decision to decline chondrogenesis is driven by β-catenin (Akiyama et al., 2004; Day et al., 2005; Hill et al., 2005, 2006). Although SOX9 promotes β-catenin degradation in chondrocytic cells (Akiyama et al., 2004; Topol et al., 2009), β-catenin effectively represses Sox9 expression in nonchondrocytic cells. Stabilization of β-catenin in perichondrium and joint cells involves multiple canonical WNTs, namely WNT4, WNT9A, and WNT16 (Guo et al., 2004; Später et al., 2006). It is believed that additional WNT ligands remain to be identified, as inactivation of WNT4, WNT9A, and WNT16 results in less severe joint defects than upon β-catenin inactivation. Alternatively, other mechanisms may remain unknown that stabilize β-catenin in presumptive joints. Moreover, although the decision to become a chondrocyte is driven by SOX9 cell autonomously, it remains unknown whether the decision to become a joint or perichondrium cell also depends on cell-autonomous mechanisms that could work at least in part by synergizing with canonical WNT signaling.
The SOX family comprises 20 proteins, most of which determine cell fate and differentiation in discrete lineages (Kamachi and Kondoh, 2013). They are distributed into eight groups, A to H, according to sequence conservation. For instance, SOX9 is a SOXE protein, and its partners in chondrogenesis, SOX5 and SOX6, are SOXD proteins. The SOXC group is composed of SOX4, SOX11, and SOX12. Several studies have started to uncover key roles for these proteins in diverse processes. Their genes overlap in expression in neural, mesenchymal, and other progenitor cell types and are needed, often in redundancy, for proper development of such important organs as the heart, lung, spleen, brain, eye, and spinal cord (Sock et al., 2004; Bhattaram et al., 2010; Potzner et al., 2010; Thein et al., 2010; Paul et al., 2014). SOX4 and SOX11 are up-regulated in multiple types of human tumors, namely, prostate, breast, lymphoma, and colorectal cancer and have been associated with poor patient outcome (Vervoort et al., 2013). A few studies have implicated SOX4 and SOX11 in cell differentiation, as for instance in neuronal cells (Bergsland et al., 2006, 2011), but most studies have pointed to critical cell survival roles. Several mechanisms have been put forward. For instance, SOXC proteins activate the gene for TEAD2, a Hippo pathway transcriptional mediator, in neural and mesenchymal cells in vivo (Bhattaram et al., 2010). SOX4 up-regulates genes involved in the PI3K–AKT and MAPK pathways in acute lymphoblastic leukemia (Ramezani-Rad et al., 2013) and mediates epithelial-to-mesenchymal transition downstream of TGF-β signaling in metastatic cancer cells (Tiwari et al., 2013). In vitro studies have proposed that SOX4 and SOX12 may up-regulate canonical WNT/β-catenin signaling, but in vivo evidence of this action and its consequence and underlying mechanism are lacking (Sinner et al., 2007; Lai et al., 2011; Lee et al., 2011; Saegusa et al., 2012; Duquet et al., 2014). With regard to skeletogenesis, Sox4/11/12-compound mutants feature vertebral column and sternum malformations, caused at least in part by decreased survival of mesenchymal cells, as neither cell proliferation nor cell differentiation were affected (Bhattaram et al., 2010). Sox4+/− adult mice are osteopenic as a result of impaired bone formation (Nissen-Meyer et al., 2007), and Sox11−/− mice are born with an undermineralized skeleton (Sock et al., 2004). In vitro studies have suggested that SOX4 and SOX11 promote osteoblast survival, proliferation, and early differentiation through enhancing expression of osteoblast-specific genes (Nissen-Meyer et al., 2007; Bhattaram et al., 2010; Gadi et al., 2013). SOXC proteins are thus essential regulators of many processes, including skeletogenesis, but further investigations are necessary to decipher the full spectrum of their cellular and molecular actions.
Combining mouse genetic and molecular approaches, we uncover here that SOXC proteins are necessary to ensure proper delineation and articulation of the multiple elements that compose the embryonic skeleton. Their actions are antichondrogenic and involve β-catenin stabilization through cell-autonomous amplification of canonical WNT signaling.
Results
SOXC genes are necessary to delineate and articulate the mouse embryo skeleton
We started this study by testing the importance of SOXC genes in skeletogenesis in mice carrying Sox4 and Sox11 conditional null alleles, Sox12-null alleles, and Prx1Cre, a transgene expressed in limb bud mesenchyme (Logan et al., 2002). Sox4fl/flPrx1Cre, Sox11fl/flPrx1Cre, and Sox12−/− fetuses (Hoser et al., 2008; Bhattaram et al., 2010) looked externally normal at embryonic day 18.5 (E18.5), 1 d before birth (Fig. 1 A). In contrast, Sox4fl/fl11fl/flPrx1Cre fetuses had severe limb malformations, and Sox4fl/fl11fl/fl12−/−Prx1Cre fetuses were even more affected. Double mutants also featured a small head and omphalocele and triple mutants additionally exhibited hematomas (Fig. 1, A and B). Histological analysis showed minor, if any, limb defects in single mutants but severe brachysyndactyly in double and triple mutants (Fig. 1, C–E). Skeletal preparations of triple mutants (referred to as SoxCPrx1Cre) allowed further appreciation of limb defects and revealed underdevelopment of the sternum and skull, other sites expressing Prx1Cre (Fig. S1). These data uncovered that the SOXC genes have critical, redundant roles in the developing appendicular, axial, and craniofacial skeleton. SoxCPrx1Cre limb buds were already shorter and narrower than controls as early as E11.5 (Fig. 1, F and G). Between E12.5 and E14.5, control limbs grew rapidly, developed digital condensations that underwent chondrogenesis, and formed interdigital indentations, but mutant paws remained small, and digital rays were often fused, i.e., lacking joint demarcation and interdigital indentation (Fig. 1, F–I). Proximal elements, such as the tibia, were forming growth plates and quickly elongating in E14.5 control embryos, but cartilage primordia remained small and immature in mutant littermates (Fig. 1 J). SOXC genes are thus necessary from early development to delineate, articulate, and allow maturation of the cartilage primordia of the embryonic skeleton.
Gross and histological analysis of SOXC mutant embryos. (A) Pictures of E18.5 control and SoxC/Prx1Cre mutants. (B) High-magnification pictures of control and triple mutant feet. (C–E) Foot coronal sections stained with Alcian blue (cartilage-specific dye) and nuclear fast red (AB&NFR). Pictures focus on metatarsals and proximal phalanges. Red and black arrows, anterior–posterior and proximal–distal fusions of cartilage primordia, respectively. (F) Pictures of E11.5–13.5 embryo limbs. FL, forelimb; HL, hind limb. (G) Hind limb coronal sections stained with hematoxylin and eosin (E11.5–12.5) or AB&NFR (E13.5). Distal/proximal and anterior/posterior axes are shown. (H–J) Alcian blue staining of E14.5 whole embryos (H) and sections of hind paws (I) and tibias (J). EC, epiphyseal cartilage; PC, proliferating chondrocytes; pHC, prehypertrophic chondrocytes; HC, hypertrophic chondrocytes. All data were verified more than three times. Each panel shows the results of a typical experiment.
Gross and histological analysis of SOXC mutant embryos. (A) Pictures of E18.5 control and SoxC/Prx1Cre mutants. (B) High-magnification pictures of control and triple mutant feet. (C–E) Foot coronal sections stained with Alcian blue (cartilage-specific dye) and nuclear fast red (AB&NFR). Pictures focus on metatarsals and proximal phalanges. Red and black arrows, anterior–posterior and proximal–distal fusions of cartilage primordia, respectively. (F) Pictures of E11.5–13.5 embryo limbs. FL, forelimb; HL, hind limb. (G) Hind limb coronal sections stained with hematoxylin and eosin (E11.5–12.5) or AB&NFR (E13.5). Distal/proximal and anterior/posterior axes are shown. (H–J) Alcian blue staining of E14.5 whole embryos (H) and sections of hind paws (I) and tibias (J). EC, epiphyseal cartilage; PC, proliferating chondrocytes; pHC, prehypertrophic chondrocytes; HC, hypertrophic chondrocytes. All data were verified more than three times. Each panel shows the results of a typical experiment.
SOXC genes are dynamically expressed in skeletogenic cells
To determine where and when SOXC genes are involved in skeletogenesis, we analyzed their expression in the developing mouse limb and compared it to that of SOX9. At E10.5 and E11.5, Sox9 expression was confined to the skeletogenic core mesenchyme, whereas the three SOXC RNAs were detected throughout the mesenchyme and ectoderm of limb buds (Fig. 2 A). At E12.5, Sox9 RNA was marking skeletogenic condensations, whereas the SOXC RNAs were expressed in these condensations and in interdigital mesenchyme. At E13.5 and E14.5, Sox9 RNA remained abundant in developing cartilage but was becoming down-regulated in presumptive joints and perichondrium (Fig. 2, A–C). In contrast, Sox4 RNA was intensely present in presumptive joints and inner layers of perichondrium and was being down-regulated in differentiating chondrocytes. Sox11 RNA was abundant at E13.5 throughout the distal zone of the limb bud and was found at E14.5 primarily in the outer layers of perichondrium and weakly in cartilage epiphyses. Sox12 remained widely expressed but at a low level compared with Sox4 and Sox11. Thus, the SOXC genes are dynamically expressed in the developing skeleton. Their RNAs coexist with Sox9 RNA in skeletogenic mesenchymal cells, but the Sox4 and Sox11 RNAs become mostly restricted to nonchondrocytic cells during overt skeletogenesis, whereas Sox9 RNA specifically labels chondrocytes.
SOXC gene expression in the limb bud. Sections of hind limbs from E10.5–14.5 wild-type embryos were stained with AB&NFR or hybridized with RNA probes (red signals) and counterstained with DAPI (blue dye), as indicated. All SOXC probes had similar length, GC content, and labeling efficiency, allowing comparison of RNA levels. CM, core mesenchyme; DC, digital condensation; DM, distal mesenchyme; E, ectoderm; EC, epiphyseal cartilage; M, mesenchyme; Pc, perichondrium; PJ, presumptive joint. (A) Coronal sections through hind limb buds. E11.5 Sox11 RNA in situ hybridization image and E13.5 Sox9 RNA in situ hybridization images are composite of several pictures. White lines mark the individual image boundaries. (B) High-magnification views of phalangeal regions boxed in A. (C) Proximal region of E14.5 tibia. All data were reproduced more than three times. Each panel shows representative results.
SOXC gene expression in the limb bud. Sections of hind limbs from E10.5–14.5 wild-type embryos were stained with AB&NFR or hybridized with RNA probes (red signals) and counterstained with DAPI (blue dye), as indicated. All SOXC probes had similar length, GC content, and labeling efficiency, allowing comparison of RNA levels. CM, core mesenchyme; DC, digital condensation; DM, distal mesenchyme; E, ectoderm; EC, epiphyseal cartilage; M, mesenchyme; Pc, perichondrium; PJ, presumptive joint. (A) Coronal sections through hind limb buds. E11.5 Sox11 RNA in situ hybridization image and E13.5 Sox9 RNA in situ hybridization images are composite of several pictures. White lines mark the individual image boundaries. (B) High-magnification views of phalangeal regions boxed in A. (C) Proximal region of E14.5 tibia. All data were reproduced more than three times. Each panel shows representative results.
SOXC genes elevate the β-catenin level and repress Sox9 in perichondrium and joints
We previously showed that mesenchymal cells were dying at an abnormally high rate in E10.5–11.5 SoxCPrx1Cre limb buds (Bhattaram et al., 2010). Interestingly, we found here that by E13.5–14.5, SoxCPrx1Cre paws were exhibiting less cell death than controls in interdigital, perichondrium, and joint regions (Fig. 3 A). This suggested that mutant cells in interdigital, joint, and perichondrium regions died earlier or switched to another lineage, such as the chondrocyte lineage, which is protected from cell death by SOX9 (Akiyama et al., 2002). Like control littermates, SoxCPrx1Cre embryos were expressing Msx1 in E13.5 interdigital mesenchyme and Gdf5, Hoxd13, Tgfbr2, Wnt4, and Wnt9a in E14.5 and E18.5 presumptive joints and perichondrium (Fig. 3, B and C; and Fig. S2). Mutant perichondrium and joint cells were thus alive and expressing proper lineage markers. β-Catenin has critical roles in determining the fate of mesenchymal progenitors and presumptive joint and perichondrium cells (Day et al., 2005; Hill et al., 2005). However, inactivation and stabilization of β-catenin in limb bud mesenchyme in Ctnnb1fl/flPrx1CreER and Ctnnb1flEx3/+Prx1CreER embryos, respectively, did not affect Sox4 and Sox11 expression (Fig. S3, A and B), demonstrating that the SOXC genes are expressed independently of β-catenin. Interestingly, the levels of β-catenin protein and activity were substantially reduced in joint and perichondrium cells in E14.5 SoxCPrx1Cre embryos, as assessed by immunostaining (Fig. 3 D) and RNA in hybridization for the canonical WNT signaling targets Lef1 and Ccnd1 (Fig. 3 E). Given the important antagonistic actions of β-catenin and SOX9 in early skeletogenesis, we then analyzed Sox9 expression in SoxCPrx1Cre embryos. At E11.5, control and mutant limb buds were expressing Sox9 throughout most of the mesenchyme (Fig. 3 F). At E12.5–13.5, control limb buds were restricting Sox9 expression to precartilaginous condensations, whereas mutants were still showing wide expression of Sox9. At E14.5 and E18.5, mutant perichondrium and joint cells were still expressing Sox9, but controls were not (Fig. 3 G and Fig. S2). Mutant cells were abundantly surrounded by fibronectin, which is typical of perichondrium, but instead of showing spindle-like organization, they were arranged as a cobblestone, typical of chondrocytes. Western blots of tissue extracts confirmed that SoxCPrx1Cre limb buds contained less β-catenin and more SOX9 than controls (Fig. 3 H). Similarly, SoxCfl/fl primary limb bud cells treated with CRE-encoding adenovirus (AdeCre) to inactivate the SOXC genes contained less nuclear β-catenin and more SOX9 (Fig. 3 I), a finding consistent with SOXC cell-autonomous roles. We concluded that SOXC genes are necessary to increase the β-catenin level and turn off Sox9 and thereby repress chondrogenesis in otherwise properly specified in joint and perichondrium cells.
Cellular and molecular analysis of SoxCPrx1Cre limbs. (A) TUNEL assay (green) in hind limb sections from E13.5–14.5 control and SoxCPrx1Cre littermates. Counterstaining is with DAPI (blue). Arrows, mesenchyme between digits 2 and 3. Arrowheads, presumptive joints. (B and C) RNA in situ hybridization of E13.5–14.5 control and SoxCPrx1Cre hind paw sections with various probes, as indicated. Arrows, presumptive joints. Arrowheads, interdigital and perichondrium cells. (D) E14.5 hind limb sections immunostained for β-catenin (green) and counterstained with DAPI (blue). Magnified images of boxed regions are shown in side panels. j, joint; pC, perichondrium. (E) RNA in situ hybridization of E14.5 control and SoxCPrx1Cre hind paw sections with various probes, as indicated. (F) Sox9 RNA in situ hybridization of hind limb sections from E11.5–13.5 control and SoxCPrx1Cre embryos. (G, left) Coronal section of E14.5 tibias. (right) RNA in situ hybridization and immunofluorescence for SOX9 (red) and fibronectin (green). Pictures were taken at the level of the box shown in the left. (H and I) β-Catenin and SOX9 levels in whole-tissue extracts from control and SoxCPrx1Cre mutant hind limb buds (H) and in membranous/cytoplasmic and nuclear extracts from primary limb bud cells (I). Cells were isolated from E11.5 SoxCfl/fl limb buds and treated in culture with lacZ or CRE adenovirus for 24 h. Representative Western blots are shown. Fold changes in β-catenin and SOX9 levels were normalized to the levels of GAPDH and a nonspecific protein (nsp) in membrane/cytoplasm (mbr/cyt.) and nucleus fractions, respectively. *, P < 0.05; n = 3 embryo littermates (H) and experimental replicates (I). All experiments were repeated at least three times. Each panel shows the results of a representative one.
Cellular and molecular analysis of SoxCPrx1Cre limbs. (A) TUNEL assay (green) in hind limb sections from E13.5–14.5 control and SoxCPrx1Cre littermates. Counterstaining is with DAPI (blue). Arrows, mesenchyme between digits 2 and 3. Arrowheads, presumptive joints. (B and C) RNA in situ hybridization of E13.5–14.5 control and SoxCPrx1Cre hind paw sections with various probes, as indicated. Arrows, presumptive joints. Arrowheads, interdigital and perichondrium cells. (D) E14.5 hind limb sections immunostained for β-catenin (green) and counterstained with DAPI (blue). Magnified images of boxed regions are shown in side panels. j, joint; pC, perichondrium. (E) RNA in situ hybridization of E14.5 control and SoxCPrx1Cre hind paw sections with various probes, as indicated. (F) Sox9 RNA in situ hybridization of hind limb sections from E11.5–13.5 control and SoxCPrx1Cre embryos. (G, left) Coronal section of E14.5 tibias. (right) RNA in situ hybridization and immunofluorescence for SOX9 (red) and fibronectin (green). Pictures were taken at the level of the box shown in the left. (H and I) β-Catenin and SOX9 levels in whole-tissue extracts from control and SoxCPrx1Cre mutant hind limb buds (H) and in membranous/cytoplasmic and nuclear extracts from primary limb bud cells (I). Cells were isolated from E11.5 SoxCfl/fl limb buds and treated in culture with lacZ or CRE adenovirus for 24 h. Representative Western blots are shown. Fold changes in β-catenin and SOX9 levels were normalized to the levels of GAPDH and a nonspecific protein (nsp) in membrane/cytoplasm (mbr/cyt.) and nucleus fractions, respectively. *, P < 0.05; n = 3 embryo littermates (H) and experimental replicates (I). All experiments were repeated at least three times. Each panel shows the results of a representative one.
SOXC and β-catenin genetically interact in perichondrium and joint development
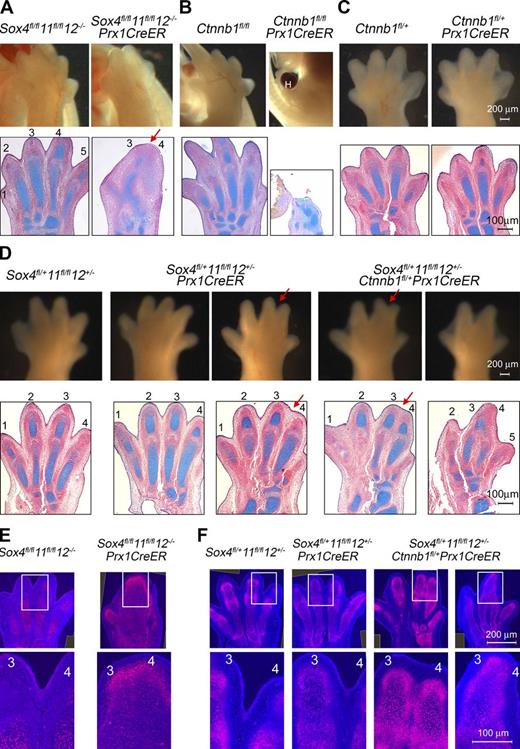
We generated SoxC/Ctnnb1 compound mutants to confirm genetically that SOXC proteins functionally interact with β-catenin during skeletogenesis. We used Prx1CreER (Kawanami et al., 2009) to obtain control and mutant embryos at high frequency and activated Cre recombinase with tamoxifen at E10.5. At E14.5, Sox4fl/fl11fl/fl12−/−Prx1CreER and Ctnnb1fl/flPrx1CreER embryos were similar to respective Prx1Cre mutants (Fig. 4, A and B; Hill et al., 2006). The phenotype of Sox4fl/fl11fl/fl12−/−Prx1CreER limbs was similar to that of Ctnnb1fl/flPrx1CreER embryos, but milder, likely because of incomplete loss of β-catenin. Ctnnbfl/+Prx1CreER and Sox4fl/+11fl/fl12+/−Prx1CreER partial mutant embryos were all normal, except that a few of the latter (2/9; 22%) exhibited soft tissue syndactyly of digits 3 and 4 (Fig. 4, C and D; and Table S1). Importantly, most Sox4fl/+11fl/fl12+/−Ctnnbfl/+Prx1CreER compound mutants (9/11; 82%) exhibited soft-tissue fusions, and many (6/11; 55%) also displayed similar cartilage fusions as SOXC full mutants. SOX9 protein was detected in these fusing regions in Sox4fl/fl11fl/fl12−/−Prx1CreER and Sox4fl/+11fl/fl12+/−Ctnnbfl/+Prx1CreER embryos but not in corresponding regions in Sox4fl/+11fl/fl12+/−Prx1CreER mutants (Fig. 4, E and F). This high frequency and severity of defects in full and compound mutants compared with partial mutants attested that the SOXC and β-catenin genes work in the same pathway to secure the nonchondrocytic fates of perichondrium and joint cells.
Genetic interaction between SoxC and Ctnnb1. (A–D) External and histological aspect of hind paws from E14.5 SoxC/Ctnnb1/Prx1CreER control, partial, and compound mutant embryos. (A) The Sox4fl/fl11fl/fl12−/−Prx1CreER paw displays anterior–posterior (red arrow) fusions of cartilage primordia. (B) The Ctnnb1fl/flPrx1CreER paw is tiny and malformed and has a large hematoma (H). (C) The Ctnnb1fl/+Prx1CreER paw is normal. (D) Most Sox4fl/+11fl/fl12+/−Prx1CreER paws are normal (left), but some develop soft-tissue syndactyly of digits 3 and 4 (red arrow, right). Most Sox4fl/+11fl/fl12+/−Ctnnb1fl/+Prx1CreER paws show soft-tissue fusions without (left) or with (right) cartilage fusions. Also see Table S1. (E and F) SOX9 immunostaining (red) in sections adjacent to those in A–D. Top images are composites of more than one original image. Bottom images are magnified pictures of boxed regions from a single image. They show SOX9 continued expression between digits 3 and 4 in SoxCPrx1CreER (E) and SoxC/Ctnnb1Prx1CreER limbs (F). Data were reproduced in at least three experiments. Each panel shows representative results.
Genetic interaction between SoxC and Ctnnb1. (A–D) External and histological aspect of hind paws from E14.5 SoxC/Ctnnb1/Prx1CreER control, partial, and compound mutant embryos. (A) The Sox4fl/fl11fl/fl12−/−Prx1CreER paw displays anterior–posterior (red arrow) fusions of cartilage primordia. (B) The Ctnnb1fl/flPrx1CreER paw is tiny and malformed and has a large hematoma (H). (C) The Ctnnb1fl/+Prx1CreER paw is normal. (D) Most Sox4fl/+11fl/fl12+/−Prx1CreER paws are normal (left), but some develop soft-tissue syndactyly of digits 3 and 4 (red arrow, right). Most Sox4fl/+11fl/fl12+/−Ctnnb1fl/+Prx1CreER paws show soft-tissue fusions without (left) or with (right) cartilage fusions. Also see Table S1. (E and F) SOX9 immunostaining (red) in sections adjacent to those in A–D. Top images are composites of more than one original image. Bottom images are magnified pictures of boxed regions from a single image. They show SOX9 continued expression between digits 3 and 4 in SoxCPrx1CreER (E) and SoxC/Ctnnb1Prx1CreER limbs (F). Data were reproduced in at least three experiments. Each panel shows representative results.
SOXC proteins stabilize β-catenin
In probing for mechanisms underlying the functional interaction of the SOXC and β-catenin genes and the reduced level of β-catenin in SoxCPrx1Cre limb buds, we first ruled out that β-catenin controls Sox4 and Sox11 at the RNA level (Fig. S3, A and B) and, vice versa, that the SOXC proteins control Ctnnb1 at the RNA level (Fig. S4 A). We ruled out that β-catenin affects the transcriptional activity of SOXC proteins by showing that the ability of SOXC proteins to activate a Tead2 reporter was not affected by treatment of HEK293 cells with WNT3A (Fig. S4 B). Based on these data, we concluded that SOXC proteins likely act on β-catenin at the protein level. 10T1/2 mesenchymal cells transfected with a SOX11 expression plasmid increased their total β-catenin level 1.3-fold and decreased their SOX9 level 2.5-fold in 24 h (Fig. 5 A). Untransfected cells in the same cultures did not show any change in β-catenin and SOX9 levels, indicating that SOX11 acts cell autonomously. These results were consistent with SOXC loss-of-function findings in vivo and thus validated this gain-of-function approach as a suitable in vitro system for our study. In a similar experiment, the analysis of subcellular fractions revealed that SOX11 acted primarily in the nucleus, increasing the β-catenin content of this compartment 3.8-fold and decreasing its SOX9 content fivefold (Fig. 5 B). Overexpression of SOX11 and its group members resulted in increased activation of the β-catenin–dependent TOP-Flash reporter (Fig. 5 C). The amplitude of this effect was similar to that for the β-catenin protein level, inferring that SOXC proteins regulate the amount but not the transcriptional activity of β-catenin. We next tested whether SOXC proteins increase the β-catenin level by stabilizing the protein. To dissociate an effect on protein stability from an effect on RNA translation, we prepared Ctnnb1fl/fl primary osteoblasts and infected them with AdeCre. 1 d later, AdeCre-treated cells were left with a residual amount of Ctnnb1 RNA compared with AdeLacZ-treated control cells (Fig. 5 D, left). We then cotransfected the cells with SOX11- and GFP-encoding plasmids. The next day, GFP-positive/AdeLacZ-treated cells showed 50% more β-catenin in all cell compartments when overexpressing SOX11, as expected (Fig. 5 D, right). GFP-positive/AdeCre-treated cells still showed a low level of β-catenin in the cell membrane (stable pool) but had lost most of β-catenin in the cytoplasm and nucleus (unstable pool), also as expected. Interestingly, GFP-positive/AdeCre-treated cells forced to overexpress SOX11 still exhibited a substantial amount of β-catenin in all cell compartments. Together, these data thus demonstrated that SOXC proteins increase the β-catenin level through protein stabilization.
Stabilization of β-catenin by SOXC proteins. (A and B) β-Catenin and SOX9 protein levels in 10T1/2 cells transfected with GFP and empty (−) or 3FLAG-SOX11 (+) expression plasmids. After 24 h, untransfected and transfected cells were sorted by flow cytometry and whole-cell extracts (A) or membranous/cytoplasmic (mbr/cyt.) and nuclear extracts (B) were made. Western blotting was performed with FLAG antibodies for SOX11 and specific antibodies for other proteins. Numbers indicate protein levels measured in representative blots and normalized with β-actin (A) or GAPDH and HDAC1 (B) levels. Thin vertical white lines added in blot pictures indicate that the order of lanes was rearranged for presentation clarity. (C) TOP-Flash reporter activity in 10T1/2 cells transfected with empty (−) or FLAG-SOXC expression plasmids. Extracts were made after 24 h. Reporter activities are presented for one representative experiment as means of triplicates with standard deviation after normalization for transfection efficiency. The Western blot shows the levels of SOXC and nonspecific (NS) proteins recognized by FLAG antibodies. (D, left) qRT-PCR assay of Ctnnb1 RNA level in Ctnnb1fl/fl osteoblasts infected with lacZ or CRE adenovirus for 24 h and with GFP or SOX11 adenovirus for the next 24 h. Data are shown as means of biological triplicates with standard deviation after normalization with Gapdh mRNA level. Please note that the slight increase in Ctnnb1 mRNA level observed in SOX11-compared with GFP-expressing cells was not statistically significant. (right) β-Catenin level in Ctnnb1fl/fl osteoblasts infected with lacZ or CRE adenovirus for 24 h and then transfected with GFP and empty (−) or SOX11 (+) expression plasmids for another 24 h. GFP-positive/lacZ and GFP-positive/CRE cells were sorted by flow cytometry. Representative Western blots are shown for subcellular fractions. Relative protein levels were quantified using GAPDH, HDAC1, and Na+/K+ ATPase as controls. All data were reproduced in at least three distinct experiments. Each panel shows the results of a representative experiment.
Stabilization of β-catenin by SOXC proteins. (A and B) β-Catenin and SOX9 protein levels in 10T1/2 cells transfected with GFP and empty (−) or 3FLAG-SOX11 (+) expression plasmids. After 24 h, untransfected and transfected cells were sorted by flow cytometry and whole-cell extracts (A) or membranous/cytoplasmic (mbr/cyt.) and nuclear extracts (B) were made. Western blotting was performed with FLAG antibodies for SOX11 and specific antibodies for other proteins. Numbers indicate protein levels measured in representative blots and normalized with β-actin (A) or GAPDH and HDAC1 (B) levels. Thin vertical white lines added in blot pictures indicate that the order of lanes was rearranged for presentation clarity. (C) TOP-Flash reporter activity in 10T1/2 cells transfected with empty (−) or FLAG-SOXC expression plasmids. Extracts were made after 24 h. Reporter activities are presented for one representative experiment as means of triplicates with standard deviation after normalization for transfection efficiency. The Western blot shows the levels of SOXC and nonspecific (NS) proteins recognized by FLAG antibodies. (D, left) qRT-PCR assay of Ctnnb1 RNA level in Ctnnb1fl/fl osteoblasts infected with lacZ or CRE adenovirus for 24 h and with GFP or SOX11 adenovirus for the next 24 h. Data are shown as means of biological triplicates with standard deviation after normalization with Gapdh mRNA level. Please note that the slight increase in Ctnnb1 mRNA level observed in SOX11-compared with GFP-expressing cells was not statistically significant. (right) β-Catenin level in Ctnnb1fl/fl osteoblasts infected with lacZ or CRE adenovirus for 24 h and then transfected with GFP and empty (−) or SOX11 (+) expression plasmids for another 24 h. GFP-positive/lacZ and GFP-positive/CRE cells were sorted by flow cytometry. Representative Western blots are shown for subcellular fractions. Relative protein levels were quantified using GAPDH, HDAC1, and Na+/K+ ATPase as controls. All data were reproduced in at least three distinct experiments. Each panel shows the results of a representative experiment.
SOXC proteins stabilize β-catenin by replacing SOX9 and inhibiting GSK3 activity in the destruction complex
Phosphorylation of β-catenin by the CK1 and GSK3 kinases is a prerequisite for its degradation. When HEK293 cells were cotransfected with plasmids encoding SOX4 and β-catenin, SOX4 increased both the activity of the TOP-Flash reporter and the level of the wild-type form of β-catenin, as expected (Fig. 6, A and B). In contrast, SOX4 was unable to raise the activity and level of a mutant form of β-catenin that was insensitive to CK1 and GSK3. This strongly suggested that SOXC proteins stabilize β-catenin by interfering with its degradation cascade. Both SOX4 and SOX9 were previously shown to physically interact with β-catenin in vitro and SOX9 to destabilize β-catenin by entering the APC–Axin1 complex (Akiyama et al., 2004; Sinner et al., 2007; Topol et al., 2009). We thus asked whether SOXC proteins also enter this complex. Immunoprecipitation (IP) with Axin1 antibodies and extracts from 10T1/2 cells infected with control or SOX11 adenovirus showed that SOX11 was indeed present in the destruction complex (Fig. 6 C). Interestingly, SOX9 was no longer present in the complex in SOX11-overexpressing cells. An in vitro assay with recombinant proteins revealed that prebinding of SOX4 to β-catenin prevented interaction of SOX9 with β-catenin and, vice versa, that prebinding of SOX9 to β-catenin blocked the ability of SOX4 to bind β-catenin (Fig. S5). Together, these data thus provided evidence that SOXC proteins and SOX9 compete with each other to bind to β-catenin. Interestingly, removal of β-catenin from Ctnnb1flfl primary osteoblasts by infection with AdeCre demonstrated that SOX11 was able to down-regulate the level of SOX9 protein and RNA even in the absence of β-catenin (Fig. 6 D). Thus, SOXC proteins may stabilize β-catenin in the destruction complex both by down-regulating Sox9 gene expression and by excluding the SOX9 protein from the destruction complex. We next asked whether SOXC proteins, like SOX9, affect phosphorylation of β-catenin by CK1 and GSK3. As anticipated, Sox9 inactivation in E12.5 Sox9Prx1Cre limb buds increased the total amount of β-catenin but reduced the proportion of the CK1/GSK3-phosphorylated protein (Fig. 6 E). In contrast, E12.5 SoxCPrx1Cre limb buds contained a reduced amount of total β-catenin, and although they had less CK1-phosphorylated β-catenin, they had more GSK3-phosphorylated β-catenin than controls. Consistent with this SOXC loss-of-function result in vivo, 10T1/2 cells overexpressing SOX11 had more CK1-phosphorylated β-catenin and less GSK3-phosphorylated β-catenin (Fig. 6 F). We concluded that SOXC proteins specifically block phosphorylation of β-catenin by GSK3. The lower level of the CK1-phosphorylated β-catenin in SOXC-null limb buds likely reflected the fact that this protein form quickly underwent phosphorylation by GSK3 and then degradation in the absence of the SOXC protein. The higher level of this protein form in SOXC-overexpressing cells likely reflected the inability of GSK3 to process it. The amount of GSK3 protein present in the destruction complex was similar in the presence and absence of SOX11, inferring that SOXC proteins affect the activity of the kinase rather than its recruitment in the complex (Figs. 6 C and S5). Importantly, SOXC proteins were still capable of decreasing the activity of GSK3 on β-catenin in primary limb bud cells deprived of SOX9, showing that SOXC proteins stabilize β-catenin not just by replacing SOX9 in the APC–Axin1 complex but by actively preventing GSK3 phosphorylation of β-catenin (Fig. 6 G). Altogether, these data demonstrated that SOXC proteins stabilize β-catenin by blocking its phosphorylation by GSK3. They perform this action by repressing the Sox9 gene, by replacing SOX9 protein in the destruction complex and by actively repressing the kinase.
SOXC proteins inhibit β-catenin degradation in the destruction complex and repress Sox9 expression. (A) Effect of SOX4 on the activity of wild-type and CK1/GSK3-insensitive (stabilized) β-catenin. HEK293 cells were transfected with TOP-Flash and control reporters, 100 ng empty (−), or 3FLAG-SOX4 expression plasmid, and 0–200 ng of β-catenin expression plasmid. Normalized TOP-Flash activities are presented as mean values with standard deviation of triplicate cultures in a representative experiment. (B) Effect of SOX4 on the levels of wild-type and stabilized β-catenin. Representative blots of membrane/cytoplasm and nuclear extracts from HEK293 cells transfected with 200 ng of wild-type and 20 ng of stabilized β-catenin–FLAG plasmids, respectively. Levels of β-catenin were normalized to HDAC1 and GAPDH levels in nucleus and membrane/cytoplasm (mbr/cyt.) fractions, respectively. *, P < 0.05; n = 3 experimental replicates. (C) Detection of proteins binding to Axin in 10T1/2 cells infected with lacZ or 3FLAG-SOX11 adenovirus for 24 h. Cell lysates (inputs) were immunoprecipitated with nonimmune (IgG-IP) or Axin1 antibodies (Axin-IP). Western blots are shown for a representative experiment. Equal amounts of cell lysates were used for control (lacZ) and tester (SOX11) samples, as shown with GAPDH levels in inputs. SOX11 overexpression was reproducibly found to increase the level of Axin1. (D) β-Catenin–independent repression of Sox9 by SOX11. (left) Ctnnb1fl/fl osteoblasts were infected with lacZ or CRE adenovirus for 44 h and with GFP or SOX11 adenovirus for the next 24 h. Representative blots are shown for whole-cell extracts. (right) qRT-PCR assay in replicate samples. Ctnnb1 and Sox9 RNA levels, normalized to those of Gapdh, are shown as means of triplicates with standard deviation. (E) Levels of total and phosphorylated β-catenin in limb bud extracts from E12.5 SoxCPrx1Cre, Sox9Prx1Cre, and respective control littermates. β-Catenin is phosphorylated at residue S45 by CK1 and at residues S33, S37, and T41 by GSK3. Representative blots are shown. (F) Levels of total and phosphorylated β-catenin in whole extracts of 10T1/2 cells infected with lacZ or 3FLAG-SOX11 adenovirus for 24 h. Representative blots are shown. (G) Levels of SOX9 and total and phosphorylated β-catenin in whole extracts of Sox9fl/fl limb bud cells infected with lacZ or CRE adenovirus for 16 h and then with GFP or 3FLAG-SOX11 adenovirus for 24 h. Representative blots are shown. Levels of total and phosphorylated β-catenin assessed in triplicates were normalized to β-actin levels. *, P < 0.05 between lacZ/GFP controls and other samples; #, P < 0.05 between lacZ/SOX11 and Cre/SOX11 samples. Note that Sox9 depletion has opposite effects on the level of β-catenin phosphorylated by GSK3 in limb bud cells in vivo and in vitro (E and G). The differences may reflect differential effects of Sox9 deletion on cell fate or behavior in the two experimental models. All data were replicated three or more times. Each panel shows typical results.
SOXC proteins inhibit β-catenin degradation in the destruction complex and repress Sox9 expression. (A) Effect of SOX4 on the activity of wild-type and CK1/GSK3-insensitive (stabilized) β-catenin. HEK293 cells were transfected with TOP-Flash and control reporters, 100 ng empty (−), or 3FLAG-SOX4 expression plasmid, and 0–200 ng of β-catenin expression plasmid. Normalized TOP-Flash activities are presented as mean values with standard deviation of triplicate cultures in a representative experiment. (B) Effect of SOX4 on the levels of wild-type and stabilized β-catenin. Representative blots of membrane/cytoplasm and nuclear extracts from HEK293 cells transfected with 200 ng of wild-type and 20 ng of stabilized β-catenin–FLAG plasmids, respectively. Levels of β-catenin were normalized to HDAC1 and GAPDH levels in nucleus and membrane/cytoplasm (mbr/cyt.) fractions, respectively. *, P < 0.05; n = 3 experimental replicates. (C) Detection of proteins binding to Axin in 10T1/2 cells infected with lacZ or 3FLAG-SOX11 adenovirus for 24 h. Cell lysates (inputs) were immunoprecipitated with nonimmune (IgG-IP) or Axin1 antibodies (Axin-IP). Western blots are shown for a representative experiment. Equal amounts of cell lysates were used for control (lacZ) and tester (SOX11) samples, as shown with GAPDH levels in inputs. SOX11 overexpression was reproducibly found to increase the level of Axin1. (D) β-Catenin–independent repression of Sox9 by SOX11. (left) Ctnnb1fl/fl osteoblasts were infected with lacZ or CRE adenovirus for 44 h and with GFP or SOX11 adenovirus for the next 24 h. Representative blots are shown for whole-cell extracts. (right) qRT-PCR assay in replicate samples. Ctnnb1 and Sox9 RNA levels, normalized to those of Gapdh, are shown as means of triplicates with standard deviation. (E) Levels of total and phosphorylated β-catenin in limb bud extracts from E12.5 SoxCPrx1Cre, Sox9Prx1Cre, and respective control littermates. β-Catenin is phosphorylated at residue S45 by CK1 and at residues S33, S37, and T41 by GSK3. Representative blots are shown. (F) Levels of total and phosphorylated β-catenin in whole extracts of 10T1/2 cells infected with lacZ or 3FLAG-SOX11 adenovirus for 24 h. Representative blots are shown. (G) Levels of SOX9 and total and phosphorylated β-catenin in whole extracts of Sox9fl/fl limb bud cells infected with lacZ or CRE adenovirus for 16 h and then with GFP or 3FLAG-SOX11 adenovirus for 24 h. Representative blots are shown. Levels of total and phosphorylated β-catenin assessed in triplicates were normalized to β-actin levels. *, P < 0.05 between lacZ/GFP controls and other samples; #, P < 0.05 between lacZ/SOX11 and Cre/SOX11 samples. Note that Sox9 depletion has opposite effects on the level of β-catenin phosphorylated by GSK3 in limb bud cells in vivo and in vitro (E and G). The differences may reflect differential effects of Sox9 deletion on cell fate or behavior in the two experimental models. All data were replicated three or more times. Each panel shows typical results.
SOXC proteins synergize with canonical WNT signaling
Because β-catenin stabilization typifies canonical WNT signaling, we asked whether SOXC proteins influence this pathway. When forced to overexpress SOX11, HEK293 cells tripled their basal ability to activate TOP-Flash and their ability to respond to WNT3A (Fig. 7 A). Consistent with this gain-of-function effect, SOXC gene inactivation drastically reduced the ability of primary limb bud cells to raise their β-catenin level in response to WNT3A (Fig. 7 B). SOXC proteins thus strongly potentiate canonical WNT signaling. To determine whether SOXC proteins act at the level of the WNT ligand–receptor complex, we treated HEK293 cells with DKK1, a natural inhibitor of WNT ligand binding to LRP5/6 (Semënov et al., 2001). DKK1 virtually abrogated the ability of WNT3A to activate TOP-Flash, as expected, but only partially blocked the effect of SOX11 (Fig. 7 C). Similarly, the ability of SOX4 to raise the β-catenin level produced from a plasmid was only partially blunted by DKK1 (Fig. 7 D). LRP6 phosphorylation, a primary response to the binding of a WNT ligand to the FZD–LRP5/6 complex, was amplified in HEK293 cells treated with WNT3A, as expected, but was not affected by SOX11 overexpression, whether cells were treated or not with WNT3A (Fig. 7 E). These data imply that SOXC proteins do not boost canonical WNT signaling at the level of the ligand–receptor complex and hence strengthen the notion that SOXC proteins act directly at the level of the destruction complex. We therefore asked next whether SOXC proteins amplify canonical WNT signaling through GSK3 inhibition. 10T1/2 cells increased their total β-catenin level in response to WNT3A up to three times faster and two times more efficiently when forced to overexpress SOX11 (Fig. 7, F and G). As described for other cells (Kim et al., 2013), they lowered their amount of GSK3-phosphorylated β-catenin by half within 1 h of WNT3A treatment and returned it to basal level within 4 h. Interestingly, cells overexpressing SOX11 maintained 5–10-fold less GSK3-phosphorylated β-catenin both without and throughout 6 h of WNT3A treatment. This level was lower than achieved with WNT3A treatment alone. We concluded that SOXC proteins boost the response to canonical WNT ligands by making inhibition of β-catenin phosphorylation by GSK3 a constitutive rather than limiting step in the signaling cascade.
SOXC proteins amplify canonical WNT/signaling by inhibiting GSK3-dependent phosphorylation of β-catenin. (A) TOP-Flash reporter activity in HEK293 cells transfected with empty (none) or FLAG-SOX11 expression plasmid and treated with various dilutions of WNT3A medium for the last 6 h of culture. Each dot represents one sample. The equations of linear fits are indicated. (B) β-Catenin level in SoxCfl/fl primary limb bud cells infected with GFP or CRE adenovirus for 24 h. Cells were treated with 20% WNT3A medium for the last 8 h. Representative blots are shown. (C) TOP-Flash reporter activity in 10T1/2 cells transfected for 24 h with empty or 3FLAG-SOX11 expression plasmid. None (−) or DKK1 protein was added 1 h before transfection, and 20% WNT3A medium was added for the last 6 h. Normalized reporter activities are presented as means with standard deviation for triplicates in a typical experiment. Fold increases caused by SOX11 are indicated. *, P < 0.05. (D) β-Catenin level in HEK293 cells transfected for 24 h with β-catenin–FLAG, empty (−), and 3FLAG-SOX4 (+) expression plasmids. DKK1 or solvent (−) was added 1 h before transfection, and 20% WNT3A medium was added for the last 6 h. Cell extracts were subjected to Western blot for β-catenin (FLAG antibody) and β-actin (loading control). Data are shown for a representative experiment. (E) Effect of SOX11 on the level and phosphorylation status of LRP6. HEK293 cells were infected with lacZ (−) or 3FLAG-SOX11 (+) adenovirus for 24 h. WNT3A medium was added for the last 8 h at 0 (−) or 20% (+). Whole-cell extracts were assayed in Western blotting. Data are shown for a representative experiment. (F) Levels of total and GSK3-phosphorylated β-catenin in whole-cell extracts from 10T1/2 cells treated with GFP or 3FLAG-SOX11 adenovirus for 20 h and then with 20% WNT3A medium for the indicated times. Data are shown for a representative experiment. (G) Quantification of protein levels detected in the Western blots shown in F. Total and phosphorylated β-catenin levels were normalized with α-tubulin levels. Each dot represents one sample. Data in each panel are representative of the results of at least three experiments. Thin vertical white lines added in blot pictures indicate that the order of lanes was rearranged for presentation clarity.
SOXC proteins amplify canonical WNT/signaling by inhibiting GSK3-dependent phosphorylation of β-catenin. (A) TOP-Flash reporter activity in HEK293 cells transfected with empty (none) or FLAG-SOX11 expression plasmid and treated with various dilutions of WNT3A medium for the last 6 h of culture. Each dot represents one sample. The equations of linear fits are indicated. (B) β-Catenin level in SoxCfl/fl primary limb bud cells infected with GFP or CRE adenovirus for 24 h. Cells were treated with 20% WNT3A medium for the last 8 h. Representative blots are shown. (C) TOP-Flash reporter activity in 10T1/2 cells transfected for 24 h with empty or 3FLAG-SOX11 expression plasmid. None (−) or DKK1 protein was added 1 h before transfection, and 20% WNT3A medium was added for the last 6 h. Normalized reporter activities are presented as means with standard deviation for triplicates in a typical experiment. Fold increases caused by SOX11 are indicated. *, P < 0.05. (D) β-Catenin level in HEK293 cells transfected for 24 h with β-catenin–FLAG, empty (−), and 3FLAG-SOX4 (+) expression plasmids. DKK1 or solvent (−) was added 1 h before transfection, and 20% WNT3A medium was added for the last 6 h. Cell extracts were subjected to Western blot for β-catenin (FLAG antibody) and β-actin (loading control). Data are shown for a representative experiment. (E) Effect of SOX11 on the level and phosphorylation status of LRP6. HEK293 cells were infected with lacZ (−) or 3FLAG-SOX11 (+) adenovirus for 24 h. WNT3A medium was added for the last 8 h at 0 (−) or 20% (+). Whole-cell extracts were assayed in Western blotting. Data are shown for a representative experiment. (F) Levels of total and GSK3-phosphorylated β-catenin in whole-cell extracts from 10T1/2 cells treated with GFP or 3FLAG-SOX11 adenovirus for 20 h and then with 20% WNT3A medium for the indicated times. Data are shown for a representative experiment. (G) Quantification of protein levels detected in the Western blots shown in F. Total and phosphorylated β-catenin levels were normalized with α-tubulin levels. Each dot represents one sample. Data in each panel are representative of the results of at least three experiments. Thin vertical white lines added in blot pictures indicate that the order of lanes was rearranged for presentation clarity.
Discussion
This study has uncovered that the SOXC genes critically contribute to the patterning of the developing vertebrate skeleton (Fig. 8). Their proteins act cell autonomously in skeletogenic mesenchyme, including presumptive joints and perichondrium. They inhibit GSK3 in the APC–Axin1 complex, and by making this step constitutive rather than limiting, they synergize with canonical WNT signaling to stabilize β-catenin. Through this mechanism and possibly also through β-catenin–independent mechanisms, they robustly repress Sox9 and thereby secure the nonchondrocytic fates of perichondrium and joint cells. The main outcome is proper delineation and articulation of vertebrate skeleton primordia.
Model of the action of SOXC proteins in early skeletogenesis. SOXC proteins cell-autonomously block the activity of GSK3 in perichondrium and presumptive joint mesenchymal cells and thereby synergize with canonical WNT signaling. Enhanced stabilization of β-catenin along with a possible independent action of the SOXC proteins leads to Sox9 repression. SOXC proteins thereby critically contribute to the proper delineation and articulation of the cartilage primordia of the developing vertebrate skeleton.
Model of the action of SOXC proteins in early skeletogenesis. SOXC proteins cell-autonomously block the activity of GSK3 in perichondrium and presumptive joint mesenchymal cells and thereby synergize with canonical WNT signaling. Enhanced stabilization of β-catenin along with a possible independent action of the SOXC proteins leads to Sox9 repression. SOXC proteins thereby critically contribute to the proper delineation and articulation of the cartilage primordia of the developing vertebrate skeleton.
Many pieces of bone and cartilage compose the vertebrate skeleton. Their number, size, and shape define individual morphologies, and their correct articulation ensures adequate locomotion. Their establishment relies on the ability of mesenchymal precursors to develop into distinct cell types. Besides chondrocytes and osteoblasts, whose control mechanisms have been dissected in detail, these cell types also include perichondrium and joint cells, whose regulation is much less understood. SOX9 and β-catenin have been shown to rival with each other to govern the decisions of progenitor cells to become or not to become a chondrocyte, respectively, and hence to specify cartilage, bone, and joint formation. This study has demonstrated that SOXC proteins are authorities in this molecular rivalry. SOX9 and SOXC proteins have overlapping expression patterns and act cell autonomously to drive and repress chondrogenesis, respectively. In contrast, β-catenin is expressed ubiquitously, and its level is controlled through protein stabilization. Secreted canonical WNT ligands have the primacy in stabilizing β-catenin in a wide variety of cell types and act both cell and noncell autonomously. SOX9 antagonizes canonical WNT/β-catenin signaling to secure the fate of chondrocytes, whereas SOXC proteins were shown here to have the opposite function of amplifying this pathway to secure nonchondrocytic cell fates. Thus, as SOX9 and its functional partners SOX5 and SOX6 are called chondrogenic SOX trio (Ikeda et al., 2004), SOXC proteins can now be referred to as antichondrogenic SOX trio. Of note, these designations should not discount the fact that SOXC proteins, like SOX5/6/9, also have key functions in other lineages. Of note too, both trios target β-catenin through nontranscriptional actions but also have chief transcriptional actions. For instance, SOX5/6/9 transactivate major cartilage-specific genes, and SOXC proteins control cell survival by transactivating Tead2 and genes in the PI3K–AKT pathway (Bhattaram et al., 2010; Ramezani-Rad et al., 2013). The two trios might thus also interact at transcriptional levels.
Many SOX proteins, including SOX9, specify both cell lineage fate and differentiation, whereas others, like SOX5/6, only promote cell differentiation. This study has shown that SOXC proteins are necessary to decline chondrogenesis in perichondrium and presumptive joints. The cells in mutant tissues were nevertheless expressing perichondrium and presumptive joint-specific genes, such as Gdf5, and SOX11 was proposed to enhance Gdf5 expression (Kan et al., 2013). Together, these data indicate that SOXC proteins secure, but are not absolutely required for perichondrium and joint cell lineage commitment. Gdf5 and other markers are not expressed in overtly differentiated joint cells and perichondrium-derived osteoblasts, but they remained expressed in SOXC mutants at least until E18.5, by which time they were turned off in control animals. The chondrification of joint and perichondrium cells in SOXC mutants can thus be explained by prolonged maintenance of a SOX9-positive multipotent progenitor status rather than by a complete switch to an overtly differentiated chondrocyte status. This conclusion is in line with data published for β-catenin, as the protein was shown to help joint cells and osteoblasts proceed through differentiation but to be dispensable to activate lineage-specific regulatory and marker genes, including Gdf5 (Hill et al., 2006; Baron and Kneissel, 2013).
SOXC proteins were shown here to amplify canonical WNT signaling. Multiple lines of evidence lead to the conclusion that the mode of functional interaction between SOXC proteins and canonical WNT signaling is synergy. Synergy occurs when two factors/pathways achieve together much more than the sum of their individual actions; their expression/activation occurs independently, and their actions are distinct but directed toward a common goal. SOXC and canonical WNT signaling meet these criteria. SOXC gene expression occurs independently of β-catenin, and vice versa, canonical WNT ligand gene expression is SOX independent. The possibility that SOXC proteins and β-catenin are transcriptional partners, as are TCF/LEF proteins and β-catenin, was excluded. The possibility that SOXC proteins facilitate canonical WNT activation at the receptor level was also excluded. Yet, both SOXC and canonical WNT inactivations (Guo et al., 2004; Später et al., 2006) resulted in chondrification of perichondrium and joints, and compound SoxC/Ctnnb1 mutants exhibited the same phenotype, whereas partial mutants for SoxC or Ctnnb1 did not. Thus, SOXC proteins and canonical WNTs need each other to achieve a key developmental outcome. Their individual actions converge on a focal node, inhibition of β-catenin phosphorylation by GSK3, to most efficiently stabilize β-catenin.
As of today, the mechanisms whereby canonical WNT signaling leads to β-catenin stabilization remain incompletely resolved. This study has proposed that SOXC proteins inhibit β-catenin phosphorylation by GSK3 by binding to β-catenin in the APC–Axin1 complex. SOXC proteins may sterically hinder GSK3 activity or may recruit a factor that neutralizes GSK3. They prevent β-catenin degradation in multiple ways. They exclude SOX9 from the complex by physically preventing SOX9 from entering the complex and by repressing Sox9 through β-catenin–dependent, or possibly, –independent mechanisms. They can also inhibit GSK3 in the absence of SOX9, a mechanism possibly relevant to many cell types. Although many groups view GSK3 inhibition as a key regulated step in canonical WNT signaling (Hernández et al., 2012), others (Li et al., 2012) have proposed that canonical WNT signaling may not affect GSK3 activity. The present study provides a possible resolution to these opposite views. By showing that SOXC proteins transform this limiting step into a constitutive one, this study indeed predicts that this step would be significant in the canonical WNT signaling cascade only in cells that do not have enough SOXC activity to neutralize GSK3. SOXC proteins are believed to be mostly nuclear in vivo. Their role in stabilizing β-catenin may thus be largely confined to the nucleus. Considering that mechanisms have been identified to neutralize or dissociate the destruction complex in the cytoplasm but not in the nucleus and that SOX9 brings this complex and stimulates its activity in the nucleus, SOXC proteins may fulfil a unique function in allowing β-catenin to avoid degradation and exert major actions in the nucleus.
Previous studies, all performed in cancer cell lines in vitro, have suggested several functional links between SOX4 and canonical WNT/β-catenin signaling. Consistent with our findings, SOX4 was proposed to block β-catenin phosphorylation (Sinner et al., 2007). SOX4 was also proposed to stabilize β-catenin by up-regulating CK2 expression (Lee et al., 2011) and to enhance β-catenin transcriptional activity by up-regulating TCF4 expression (Saegusa et al., 2012). In contrast, physical interaction between SOX4 and plakoglobin was suggested to inhibit canonical WNT/β-catenin signaling (Lai et al., 2011). These findings altogether suggest that SOXC proteins may control canonical WNT/β-catenin signaling at multiple levels. Further studies are thus warranted to validate these in vitro findings in vivo and to assess the respective contributions of each type of mechanism in determining cell fate in specific processes.
In conclusion, this study has uncovered that SOXC proteins are critically needed in early skeletogenesis to secure nonchondrocytic cell fates. Their function matches that of canonical WNT signaling, and rightly so, the proteins were demonstrated to synergize with this pathway to stabilize β-catenin and at least partly thereby repress Sox9 in nonchondrocytic skeletal cell types in vivo. Because SOXC proteins, SOX9, and canonical WNT signaling are essential in many cell types, SOXC proteins may amplify the WNT pathway and antagonize SOX9 actions in a myriad of processes in development, physiology, and disease.
Materials and methods
Mice
Mice were used as approved by the Cleveland Clinic Institutional Animal Care and Use Committee. Mice carrying Sox4 and Sox11 conditional null alleles (Sox4fl/fl and Sox11fl/fl, respectively) were previously obtained by flanking the gene’s coding sequences with loxP sites (Penzo-Méndez et al., 2007; Bhattaram et al., 2010). Mice carrying Sox12-null alleles (Sox12−/−) were generated by flanking the SOX12 coding sequence with loxP sites and deleting this sequence through CRE-mediated recombination in the male germline (Bhattaram et al., 2010). Ctnnb1fl/fl mice contained loxP sites flanking exons 2–6 of the β-catenin gene (Brault et al., 2001). Ctnnb1flEx3/+ mice contained loxP sites flanking exon3 of the β-catenin gene (Harada et al., 1999). Sox9fl/fl mice had exons 2 and 3 of the SOX9 gene flanked with loxP sites (Kist et al., 2002). The Prx1Cre and Prx1CreER transgenes featured the CRE and CRE-ER coding sequences, respectively, driven by transcriptional regulatory elements of the Prx1 gene (Logan et al., 2002; Kawanami et al., 2009). CRE recombinase was activated in Prx1CreER embryos by injecting mothers with olive oil containing 0.5 mg tamoxifen and 5 mg progesterone (Sigma-Aldrich) per 10 g of body weight.
In situ analyses
Mouse embryo paraffin sections were generated after embryo fixation in 4% paraformaldehyde. They were deparaffinized with Histo-Clear (National Diagnostics) and rehydrated in graded ethanol solutions. Staining with Lerner-3 Hematoxylin (Lerner Laboratories) and Eosin Y (Polysciences) was performed according to manufacturer’s instructions. Staining with Alcian blue 8GX (Sigma-Aldrich) was performed in 3% (vol/vol) acetic acid and followed by counterstaining using nuclear fast red (Sigma-Aldrich) in aluminum sulfate solution.
RNA in situ hybridization was performed with probes synthesized in the presence of α-[35S]ribo-UTP (PerkinElmer), essentially as previously described (Albrecht et al., 1997). Prehybridization steps included protein digestion with Proteinase K (Roche), postfixation in 4% paraformaldehyde, and blocking of amine groups in triethanolamine-acetic anhydride solution (Sigma-Aldrich). Hybridization was performed overnight at 65°C in 5× SSC, 50% formamide, and 1% SDS solution. Posthybridization washes included digestion of single-stranded RNA with RNases A and T1 (Thermo Fisher Scientific). Slides were coated with emulsion (Kodak NTB-2; Carestream Health) and exposed 1–3 wk in complete darkness. After development and fixation (Kodak), sections were counterstained with Hoechst dye and mounted with Canada balsam (Sigma-Aldrich). In situ probe plasmids were as listed (Table S2).
TUNEL assay was performed using ApopTag kit (EMD Millipore) on sections of embryos frozen in OCT (Sakura). Nucleosome-size apoptotic DNA fragments were detected by incubation with fluorescein-labeled nucleotides for 1 h at room temperature. Sections were counterstained with DAPI-containing mounting medium (Vector Laboratories). Immunostaining was performed on frozen sections. Sections were blocked in normal serum for 1 h and incubated overnight at 4°C with specific antibodies and then with fluorescent secondary antibodies for 1 h in the dark. Antibodies were as described in Table S3.
All imaging was performed at room temperature. Permount (Thermo Fisher Scientific) was used as the mounting medium for histological staining, and Vectashield (Vector Laboratories) was used as the mounting medium for immunostaining. Data were visualized at room temperature with a microscope (DM2500; Leica), captured with a digital camera (MicroPublisher 5.0 Real-Time Viewing; QImaging), using a 10× objective lens for magnification. A 40× objective was used for capturing magnified images in Fig. 3 (D and G). All images were acquired using QCapture Pro 6.0 (QImaging) and processed with Photoshop 7.0 (Adobe) software. Antibodies were used as indicated (Table S3). DAPI (Vector Laboratories) was used to stain nuclei.
Quantitative RT-PCR (qRT-PCR)
Total RNA was prepared using TRIzol (Life Technologies) and RNeasy Mini kit (QIAGEN). cDNA was synthesized using Superscript III First-Strand Synthesis System (Life Technologies) and amplified with primers (Table S4) using SYBR Green PCR Master Mix (Applied Biosystems). PCR conditions were 1 cycle at 95°C for 10 s, 40 cycles of 95°C for 5 s, and 60°C for 30 s. Relative mRNA levels were calculated using the 2−δδCt method.
Cell cultures, adenovirus infection, and transient transfection
HEK293 and 10T1/2 cells were cultured in DMEM with 10% FCS. Primary Ctnnb1fl/fl calvarial osteoblasts were prepared by dissecting from the calvaria from 1-d-old mice under sterile conditions and digesting them in 0.1 mg/ml collagenase P and 0.25% trypsin for 1 h at 37°C. Cells were passed through a 40-µm nylon cell strainer and cultured for 48 h in MEM containing 10% FCS. They were then replated at a density of 3 × 105 cells/10-cm2 dish and maintained in the same medium through the experimental period. Primary limb bud cells were dissociated from E11.5 embryo limb buds in 0.25% trypsin-EDTA for 10 min at 37°C. Cells were passed through a 40-µm nylon cell strainer and cultured at a density of 0.5 × 106 cells per 10-cm2 dish in DMEM containing 10% FCS throughout the experimental period. DKK1 treatment of cells was performed using 100 ng/ml rhDKK1 (R&D Systems). WNT3A medium was produced using LWNT-3A cells (ATCC). For SOX11 adenovirus preparation, the mouse SOX11 coding sequence was cloned in frame with an N-terminal 3FLAG-linker sequence into the pAd5CMVmcsIRESEGFPpA plasmid. This plasmid was used to generate replication-deficient adenoviral particles, in the same way as GFP, lacZ, and CRE adenoviruses (The University of Iowa Gene Transfer Vector Core). Cells were infected with 100 plaque-forming units/cell. For transient transfection, 3 × 105 cells were exposed to mixtures containing 1 µg plasmid and 3 µl FuGENE6 (Roche). Plasmids were as listed (Table S5). For reporter assays, plasmid mixtures contained 200 ng TOP-Flash reporter, 100 ng pSV2βGal (control for normalization), ≤400 ng expression plasmid, and pBluescript ≤1 µg. For cell sorting, plasmid mixtures contained 200 ng pmaxGFP vector (Lonza), 200 ng expression plasmid, and 600 ng pBluescript.
Protein assays
Reporter activities were tested 24 h after transfection using Dual-Light system (Life Technologies) upon cell lysis in Tropix buffer (Applied Biosystems). For sorting, cells were trypsinized, and GFP-positive and GFP-negative cells were separated using a flow cytometer (FACSAria II; BD). Whole-cell extracts were prepared in radioimmunoprecipitation buffer. Cytoplasmic, nuclear, and membranous extracts were prepared using Nuclear Protein Extraction kit or Subcellular Fractionation kit (Thermo Fisher Scientific). Protease and phosphatase inhibitors were included in all lysis buffers (Roche). IP was performed after 10T1/2 cell adenoviral infection for 16 h. Cell lysates were prepared in 20 mM Tris-Cl, pH 8.0, 100 mM NaCl, 1% NP-40, 0.1% Triton X-100, and 10% glycerol and precleared with mouse IgG-bound protein A magnetic beads (Life Technologies) for 6 h at 4°C, as previously described (Li et al., 2012). Precleared lysates were then incubated with 5 µg Axin1 antibody or nonimmune IgG at 4°C overnight followed by immobilization of antibody–protein complexes on protein A magnetic beads for 2 h at 4°C. Beads were washed six times before protein elution in 2× Laemmli buffer at 95°C for 5 min. SDS-PAGE and semidry Western blotting on polyvinylidene fluoride membranes (Bio-Rad Laboratories, Inc.) were performed under standard conditions. Membranes were incubated with primary and HRP-conjugated secondary antibodies (Table S3) diluted in blocking solution containing 5% nonfat dry milk. Signals were detected using ECL Prime Western Blotting Reagent (Life Technologies). Protein levels were quantified using ImageJ software (National Institutes of Health).
Data and statistical analysis
The two-tailed Student’s t test was used to calculate the significance of changes between samples in protein and RNA levels and in reporter activities. The two-tailed Fisher’s exact test was used to calculate the significance of phenotypic changes between control and mutant limbs. P < 0.05 was considered significant in both test types.
Online supplemental material
Fig. S1 shows skeletal preparations of SoxCPrx1Cre fetuses, and Fig. S2 shows Sox9 and Gdf5 expression in fetus joints. Fig. S3 demonstrates that SOXC gene expression is not affected in β-catenin mutant limbs. Fig. S4 illustrates that Ctnnb1 expression is unchanged in SOXC mutant limbs and that SOXC and β-catenin do not affect each other’s transcriptional activities. Fig. S5 shows that SOX4 and SOX9 compete for binding to β-catenin in vitro. Table S1 reports on the frequency of syndactyly in SoxC/Ctnnb1 mutants. Tables S2–S5 list RNA probes, antibodies, primers, and plasmids, respectively.
Acknowledgments
We thank A. Arif, F. Safadi, S. Rao, O. Wessely, and Y. Yang for advice on the project and manuscript.
This work was funded by Arthritis Foundation postdoctoral fellowships to P. Bhattaram and A. Penzo-Méndez, a grant from the Arthritis National Research Foundation to P. Bhattaram, and grants from the National Institutes of Health/National Institutes for Arthritis, Musculoskeletal, and Skin Diseases (AR46249, AR54153, and AR60016) and CARES Foundation to V. Lefebvre.
The authors declare no competing financial interests.