Mechanisms by which microtubule plus ends interact with regions of cell–cell contact during tissue development and morphogenesis are not fully understood. We characterize a previously unreported interaction between the microtubule binding protein end-binding 1 (EB1) and the desmosomal protein desmoplakin (DP), and demonstrate that DP–EB1 interactions enable DP to modify microtubule organization and dynamics near sites of cell–cell contact. EB1 interacts with a region of the DP N terminus containing a hotspot for pathogenic mutations associated with arrhythmogenic cardiomyopathy (AC). We show that a subset of AC mutations, in addition to a mutation associated with skin fragility/woolly hair syndrome, impair gap junction localization and function by misregulating DP–EB1 interactions and altering microtubule dynamics. This work identifies a novel function for a desmosomal protein in regulating microtubules that affect membrane targeting of gap junction components, and elucidates a mechanism by which DP mutations may contribute to the development of cardiac and cutaneous diseases.
Introduction
The ability of tissues to withstand mechanical stress and respond to signaling cues depends on intercellular junctions and their connections to the underlying cytoskeleton (Cowin and Burke, 1996; Jamora and Fuchs, 2002). Cadherin-based adherens junctions and desmosomes are best known for organizing actin and intermediate filaments (IFs) at cell–cell interfaces, respectively (Simpson et al., 2011). However, classic cadherin-associated proteins have also been reported to affect microtubule (MT) dynamics and organization (Chausovsky et al., 2000; Shtutman et al., 2008; Shahbazi et al., 2013). Changes in MT dynamics at cell–cell contacts are in part mediated by interactions of MT plus end–associated proteins with cortical factors that enable local MT plus end capture and stabilization, which influences targeted transport of cargo by MT motor proteins (Gundersen et al., 2004; Lansbergen and Akhmanova, 2006).
The plakin and spectraplakin families comprise versatile proteins that link multiple cytoskeletal components to each other and to plasma membranes (Leung et al., 2002; Suozzi et al., 2012). The modular spectraplakins can associate with actin, IFs, and MTs. The spectraplakin MACF/ACF7 guides MTs along actin toward the cell cortex to promote MT plus end capture (Kodama et al., 2003). Desmoplakin (DP) is a plakin protein best known for tethering IFs to desmosomes through the DP C terminus (Green and Simpson, 2007; Simpson et al., 2011). DP does not associate with MTs directly (Sun et al., 2001), but was shown to mediate MT reorganization during epidermal stratification by redirecting MT minus end proteins including ninein and Lis1 to the cell cortex (Lechler and Fuchs, 2007; Sumigray et al., 2011). Though the MT plus end protein CLIP-170 was reported to localize to desmosomes (Wacker et al., 1992), mechanisms by which DP may regulate MT plus ends are unknown. The discovery that DP regulates MTs suggests that its functions transcend its role in maintaining IF attachment and tissue integrity (Gallicano et al., 1998; Vasioukhin et al., 2001).
Mutations in desmosomal components including DP are associated with epidermal and cardiac diseases such as skin fragility/woolly hair syndrome and arrhythmogenic cardiomyopathy (AC; Delmar and McKenna, 2010; Basso et al., 2011; Simpson et al., 2011). Mechanisms underlying disease pathogenesis are poorly understood and are complicated further by the large spectrum of reported mutations, some of which are nonpathogenic variants. A recent study reported residues 250–604 of the DP N terminus as a “hotspot” for AC mutations with high pathogenicity (Kapplinger et al., 2011). Although the DP N terminus mediates association of DP with other desmosomal proteins, this hotspot is downstream of residues necessary for desmosomal localization (Stappenbeck et al., 1993; Smith and Fuchs, 1998), which suggests that hotspot mutations may act by impairing desmosome-independent functions of the DP N terminus.
Here, we characterize a previously unreported interaction between the DP N terminus and end-binding 1 (EB1), a MT binding protein that regulates MT dynamics and the association of proteins with MT plus ends (Su et al., 1995; Vaughan, 2005; Lansbergen and Akhmanova, 2006). At sites of cell–cell contact, DP regulates the organization and stability of MTs. Using expression constructs harboring cardiac or cutaneous disease mutations in the DP hotspot, we show that DP–EB1 interactions are critical to DP’s regulation of MT dynamics. Impairment of DP–EB1 interactions via expression of a subset of DP disease mutations compromises localization and function of the gap junction protein connexin 43 (Cx43). Together, these findings significantly advance our understanding of mechanisms by which DP mutations may contribute to cardiac and cutaneous diseases involving misregulation of gap junctions.
Results
EB1 is a novel binding partner of the DP N terminus
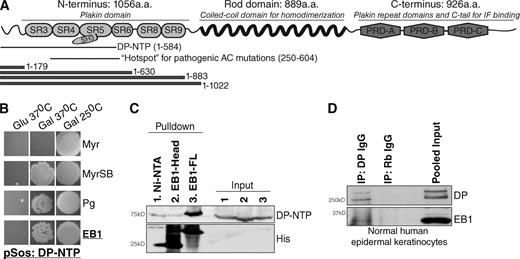
To identify proteins interacting with the DP hotspot for pathogenic AC mutations (residues 250–604; Kapplinger et al., 2011), we conducted a yeast two-hybrid screen using a construct comprising residues 1–584 of the DP N terminus, DP-NTP (DP N-terminal polypeptide; Bornslaeger et al., 1996; Fig. 1 A). A bait DP-NTP construct (pSos-DP-NTP) was incubated with a library of target (pMyr) cDNAs from HeLa cells. Among the targets independently verified to associate with DP-NTP were clones encoding EB1, a MT plus end binding protein (Su et al., 1995; Fig. 1 B). Plakoglobin (pMyr-Pg) was used as a positive control (Ct) for association with DP-NTP (Kowalczyk et al., 1997).
EB1 is a novel binding partner of the DP N terminus. (A) Schematic of DP outlining residues comprising DP-NTP, the DP N-terminal hotspot for pathogenic mutations, and DP N-terminal truncation constructs (see Fig. S1 A). (B) Yeast two-hybrid using DP-NTP as bait. Myr and MyrSB were used as negative and positive controls, respectively, and the known DP-NTP binding partner Pg was used as a positive control. Colony growth on Gal but not Glu plates at 37°C indicates a positive interaction. (C) Pull-down analysis of FLAG-tagged DP-NTP expressed in HEKs with recombinant His-tagged full-length EB1 (EB1-FL), His-tagged EB1 N terminus (residues 1–184), or uncoupled nickel (Ni-NTA) beads. (D) Immunoprecipitation of DP and coimmunoprecipitation of EB1 from NHEKs. Nonspecific rabbit IgG (Rb IgG) was used as a negative control.
EB1 is a novel binding partner of the DP N terminus. (A) Schematic of DP outlining residues comprising DP-NTP, the DP N-terminal hotspot for pathogenic mutations, and DP N-terminal truncation constructs (see Fig. S1 A). (B) Yeast two-hybrid using DP-NTP as bait. Myr and MyrSB were used as negative and positive controls, respectively, and the known DP-NTP binding partner Pg was used as a positive control. Colony growth on Gal but not Glu plates at 37°C indicates a positive interaction. (C) Pull-down analysis of FLAG-tagged DP-NTP expressed in HEKs with recombinant His-tagged full-length EB1 (EB1-FL), His-tagged EB1 N terminus (residues 1–184), or uncoupled nickel (Ni-NTA) beads. (D) Immunoprecipitation of DP and coimmunoprecipitation of EB1 from NHEKs. Nonspecific rabbit IgG (Rb IgG) was used as a negative control.
Using pull-down assays, we mapped the interaction between EB1 and DP-NTP. DP-NTP from human embryonic kidney (HEK) 293 cell lysates associated with recombinant full-length, His-tagged EB1 but did not associate with the EB1 N terminus alone (residues 1–184; Fig. 1 C). Complementary pull-downs using a series of recombinant GST-DP-N-terminal constructs containing spectrin repeats (SRs; Fig. 1 A) showed that although EB1 from normal human epidermal keratinocyte (NHEK) cell lysates associated weakly with DP 1–179 and more so with 1–630, binding was most robust in the presence of all N-terminal SRs (Fig. S1 A). We were not able to stably express a construct ending after SRs 3–4, potentially due to a lack of structural stabilization conferred by the Src homology 3 (SH3) loop of SR5 (Al-Jassar et al., 2011; Choi and Weis, 2011). Coimmunoprecipitation from NHEKs confirmed an interaction between endogenous full-length DP and EB1 (Fig. 1 D).
DP regulates the spatial organization of EB1
Analysis of endogenous DP and EB1 with confocal and structured illumination microscopy (SIM) showed that EB1 comets on MT plus ends came into close proximity and oriented primarily at a perpendicular angle with membrane-localized DP in SCC9 epithelial cells, used here for their robust expression of desmosomal proteins (Fig. 2, A and B). Furthermore, proximity ligation assay (PLA) analysis demonstrated a statistically significant proximity of DP and EB1 in SCC9s (Fig. 2 C). PLA signals were not restricted to sites of cell–cell contact. This result was not unexpected given that PLA of DP with its known binding partner plakophilin 2 (PKP2) is also not restricted to cell–cell contacts (Fig. S1 B), which reflects the presence of DP–PKP2 complexes known to form in the cytoplasm (Godsel et al., 2005; Bass-Zubek et al., 2008). Cytoplasmic signals were reduced by washing cells in buffer containing the detergent saponin, but the signals that remain likely reflect a combination of cortical and noncortical DP–EB1 interactions (Fig. 2 C, DP-EB1 + saponin images). Considered with binding studies (Fig. 1), these data demonstrate an interaction between DP and EB1 in cells.
DP regulates spatial organization of EB1. (A and B) Immunolocalization of endogenous DP and EB1 imaged by confocal microscopy (A) and SIM (B). (C) PLA of DP with EB1 in SCC9s (top) and in SCC9s rinsed with saponin detergent to extract cytoplasmic proteins (bottom), with corresponding immunofluorescence. The graph shows quantification of DP-EB1 PLA, or for PLA of each antigen with nonspecific IgG controls (IgG control images not depicted). (D) Single time frames from time-lapse recordings of EB3-mRFP expressed in cells stably expressing DP-GFP. White lines show representative traces of EB3 paths on MTs (see Videos 1 and 2). (E) Immunolocalization of EB1, DP, Pg, and nuclei (DAPI) in Ct or DP KD SCC9s. Radar chart: EB1 comet angles categorized for frequencies between 0 and 90° relative to cell–cell contacts (broken lines, identified by Pg staining; n > 2,400 comets from three independent experiments). (F) Membrane fluorescence intensities of DP during recruitment to cell–cell contacts (calcium switch) and at steady-state in Ct or EB1 KD cells (see Fig. S2, A and B; n = 20 regions of cell–cell contact for each condition, from one of two independent experiments). All data are from SCC9s. Bars, 10 µm (except for the SIM image, in which they are 5 µm). Error bars indicate SEM.
DP regulates spatial organization of EB1. (A and B) Immunolocalization of endogenous DP and EB1 imaged by confocal microscopy (A) and SIM (B). (C) PLA of DP with EB1 in SCC9s (top) and in SCC9s rinsed with saponin detergent to extract cytoplasmic proteins (bottom), with corresponding immunofluorescence. The graph shows quantification of DP-EB1 PLA, or for PLA of each antigen with nonspecific IgG controls (IgG control images not depicted). (D) Single time frames from time-lapse recordings of EB3-mRFP expressed in cells stably expressing DP-GFP. White lines show representative traces of EB3 paths on MTs (see Videos 1 and 2). (E) Immunolocalization of EB1, DP, Pg, and nuclei (DAPI) in Ct or DP KD SCC9s. Radar chart: EB1 comet angles categorized for frequencies between 0 and 90° relative to cell–cell contacts (broken lines, identified by Pg staining; n > 2,400 comets from three independent experiments). (F) Membrane fluorescence intensities of DP during recruitment to cell–cell contacts (calcium switch) and at steady-state in Ct or EB1 KD cells (see Fig. S2, A and B; n = 20 regions of cell–cell contact for each condition, from one of two independent experiments). All data are from SCC9s. Bars, 10 µm (except for the SIM image, in which they are 5 µm). Error bars indicate SEM.
To determine the potential role of DP in regulating EB1 behavior, we performed time-lapse imaging of exogenous EB3-mRFP in SCC9s stably expressing GFP-tagged DP (Godsel et al., 2005). We used EB3-mRFP as a marker of EB1 based on the ability of these EBs to heterodimerize (Komarova et al., 2009). EB3 comets traveled in a perpendicular vector toward membrane-localized DP-GFP in cells transfected with scrambled Ct siRNA (referred to as Ct knockdown [KD]; Fig. 2 D, Video 1, and Fig. S1 D). In contrast, EB3 traveled along the lateral plane of cell–cell contacts in cells depleted of DP after DP siRNA transfection (DP KD; Fig. 2 D, Video 2, and Fig. S1 D). Analysis of endogenous DP and EB1 in fixed cells showed that the angle of EB1 comets near cell–cell contacts was reduced by ∼21% upon DP KD, which confirms that DP regulates trajectories of EB1-labeled MTs approaching the cortex (Fig. 2 E). DP KD did not compromise membrane localization of E-cadherin or β-catenin (Fig. S1 C). In addition, EB1 reorientation was specific to loss of DP and not caused by loss of adhesion conferred by desmosomes, as KD of desmoglein 2 (Dsg2) did not affect EB1 angles relative to cell–cell contacts (Fig. S1 E). Dsg2 is a desmosomal cadherin whose KD does not affect DP localization or tethering of keratin to junctions (Fig. S1, F and G), but which does compromise adhesive strength (Klessner et al., 2009; Nekrasova et al., 2011).
Time-lapse imaging of EB3-mRFP in Ct KD cells. Time-lapse images of EB3-mRFP (red) in SCC9s stably expressing GFP-tagged DP (green) were acquired after transfection with Ct siRNA. Red and green channels were imaged at 2-s intervals for 30 frames using a microscope (DMI6000; Leica). Corresponding monochrome images of EB3-mRFP comets clarify comet localizations. Particle paths of select EB3 comets (arrowheads) are shown in Fig. 2 D.
Time-lapse imaging of EB3-mRFP in Ct KD cells. Time-lapse images of EB3-mRFP (red) in SCC9s stably expressing GFP-tagged DP (green) were acquired after transfection with Ct siRNA. Red and green channels were imaged at 2-s intervals for 30 frames using a microscope (DMI6000; Leica). Corresponding monochrome images of EB3-mRFP comets clarify comet localizations. Particle paths of select EB3 comets (arrowheads) are shown in Fig. 2 D.
Time-lapse imaging of EB3-mRFP in DP KD cells. Time-lapse images of EB3-mRFP (red) in SCC9s stably expressing GFP-tagged DP (green) were acquired after transfection with DP siRNA. Red and green channels were imaged at 2-s intervals for 30 frames using a microscope (DMI6000; Leica). Corresponding monochrome images of EB3-mRFP comets clarify comet localizations. Particle paths of select EB3 comets (arrowheads) are shown in Fig. 2 D.
Time-lapse imaging of EB3-mRFP in DP KD cells. Time-lapse images of EB3-mRFP (red) in SCC9s stably expressing GFP-tagged DP (green) were acquired after transfection with DP siRNA. Red and green channels were imaged at 2-s intervals for 30 frames using a microscope (DMI6000; Leica). Corresponding monochrome images of EB3-mRFP comets clarify comet localizations. Particle paths of select EB3 comets (arrowheads) are shown in Fig. 2 D.
Although the majority of EB1-binding partners use EB1-mediated MT-based trafficking (Vaughan, 2005; Galjart, 2010), EB1 KD did not affect DP localization in steady-state conditions or in calcium switch assays in which cells are switched from low (0.05 mM) to normal (1.2 mM) calcium to induce formation of calcium-dependent junctions, including desmosomes (Fig. 2 F and Fig. S2, A and B). This result is consistent with reports that DP localization is primarily actin dependent (Godsel et al., 2005). Thus, DP guides the orientation of EB1-labeled MT plus ends, whereas DP localization or expression is not regulated by EB1.
DP alters MT organization and dynamicity
To determine if DP–EB1 interactions contribute to regulation of MT organization and dynamics, we compared the distribution and stability of MTs in Ct and DP KD cells. Similar to EB1, MT ends near cell–cell contacts were often oriented at a perpendicular vector to the contact zone in Ct and Dsg2 KD cells. In contrast, few MT plus ends were oriented perpendicular to the cortex in DP KD cells (Fig. 3 A). To determine if this MT reorganization reflects a change in MT stability, we treated cells with 1 µM demecolcine for 0–25 min and analyzed MT density by quantifying tubulin fluorescence intensity after immunostaining. We observed a significant decrease in demecolcine-resistant MTs in DP KD cells after 10 min of treatment compared with controls (Fig. 3, B and B′). MT alterations in DP KD cells do not appear to be caused by changes in IF organization, as we observed no significant differences in MT resistance to demecolcine in A431 epithelial cells expressing controlled levels of DP-NTP-GFP, which uncouples IFs from desmosomes and results in keratin retraction from cell–cell contacts (Fig. S2 C; Huen et al., 2002). These data suggest that the DP N terminus, which contains the EB1 binding site, is sufficient to stabilize MTs.
DP regulates MT organization and dynamics. (A) Immunostaining of MTs, DP, Pg, and nuclei (DAPI) in Ct, DP, or Dsg2 KD SCC9s. (B) Immunostaining of DP and MTs, with corresponding thresholded MT images, for Ct or DP KD SCC9s treated with 1 µM demecolcine. Lines mark approximate regions of cell–cell contact. (B′) Tubulin intensities during demecolcine treatment (n = 9 image fields). (C) MT polymerization rates and frequency of MT pausing in cytoplasmic and cortical regions of Ct or DP KD SCC9s (n > 119 EB3 tracks for each condition compiled from more than seven cytoplasmic or cortical fields from more than four movies for each condition). All data are from SCC9s. Bars, 10 µm. Error bars indicate SEM.
DP regulates MT organization and dynamics. (A) Immunostaining of MTs, DP, Pg, and nuclei (DAPI) in Ct, DP, or Dsg2 KD SCC9s. (B) Immunostaining of DP and MTs, with corresponding thresholded MT images, for Ct or DP KD SCC9s treated with 1 µM demecolcine. Lines mark approximate regions of cell–cell contact. (B′) Tubulin intensities during demecolcine treatment (n = 9 image fields). (C) MT polymerization rates and frequency of MT pausing in cytoplasmic and cortical regions of Ct or DP KD SCC9s (n > 119 EB3 tracks for each condition compiled from more than seven cytoplasmic or cortical fields from more than four movies for each condition). All data are from SCC9s. Bars, 10 µm. Error bars indicate SEM.
Given the uniform localization of DP–EB1 PLA signals in cells, we next asked if DP regulated MT dynamics in the cytoplasm and at the cortex. We applied MT plus tip tracking algorithms (u-Track 2.0) to time-lapse images of EB3-mRFP (Jaqaman et al., 2008; Applegate et al., 2011). While DP KD dampened MT growth rates in the cytoplasm, the effect was statistically significant near the cortex (Fig. 3 C). More detailed analysis of EB3 tip tracking revealed that DP KD resulted in an increase in the frequency of cortical MT pausing events as well as the overall time in pause (Figs. 3 C and S2 D). These findings are consistent with the reciprocal results observed when EB1 is in excess, which leads to an increase in the MT polymerization rate, a decrease in pause frequency, and an increase in time spent in polymerization (Tirnauer et al., 2002). Increased MT pausing appeared to come at the expense of shrinking MTs, as the frequency of shrinking upon DP KD was reduced, while there was little change in the frequency of growing MTs (Fig. S2 D).
A subset of disease mutations in the DP N terminus interfere with DP–EB1 binding and MT behavior without affecting desmosomal localization or adhesion
Disease mutations have been reported to target all domains of DP (Fig. S3 A), but residues 250–604 of the DP N terminus comprise a mutation hotspot associated with AC (Kapplinger et al., 2011). To determine if mutations in this region interfere with EB1 binding or interfere with DP’s regulation of MTs, we designed constructs harboring mutations associated with skin (N287K) or heart (N458Y, K470E, I533T, and A566T) disease (Whittock et al., 2002; Basso et al., 2006; Rampazzo, A. 2008. European Society of Cardiology meeting. Prevalence of desmosomal protein mutations and clinical features in a large cohort of unrelated consecutive patients affected with ARVC; Xu et al., 2010; Bauce et al., 2011; Cox et al., 2011; Fig. 4 A). These mutations were engineered into DPII, a naturally occurring mRNA splice variant missing two thirds of the DP rod domain (Fig. 4 B). Like DPI, DPII localizes to desmosomes and confers adhesive strength to junctions (Angst et al., 1990; Green et al., 1990; Cabral et al., 2012). Use of DPII constructs allowed us to specifically analyze DP’s regulation of MT plus ends, as DPII is predicted to exclude interactions of the DP rod domain with MT minus end proteins (Lechler and Fuchs, 2007; Sumigray et al., 2011). The reduced size of DPII (∼6.8 kb) also helped circumvent technical challenges associated with expression of DPI (∼8.6 kb). We did confirm, however, that DPI constructs containing these mutations behaved similarly when expressed in more easily transfectable COS cells (Fig. S5 C). Wild-type (WT) DPII-GFP restored keratin attachment and MT stability impaired by DP KD (Fig. S3 B).
Characterization of disease mutations targeting the DP N terminus. (A) Five disease mutations within SRs of the DP N terminus hotspot were selected for analysis. (B) DPII-GFP was created by splicing out base pairs 3,584–5,380 from a construct of DPI-GFP. (C) Immunolocalization of GFP-tagged WT and mutated DPII-GFP constructs expressed by retroviral transduction in SCC9 cells, in the context of endogenous DP. Bar, 10 µm. (D) Densitometric quantification of immunoprecipitated/total GFP for WT DP-NTP-GFP and N287K-DP-NTP-GFP coimmunoprecipitated with plakophilin-1 (PKP1-FLAG) or plakoglobin (Pg-Myc) from HEKs, used for robust protein expression (n = 2 independent experiments for each IP). (E) Dispase assay of adhesive strength for cells expressing WT or mutant DPII-GFP constructs. (E, left) The number of fragments observed. Blots show expression of DP constructs (GFP) in SCC9s. GAPDH was used as a loading control, and EB1 expression levels were equivalent across samples (n = triplicate wells from one of two independent experiments). Error bars indicate SEM.
Characterization of disease mutations targeting the DP N terminus. (A) Five disease mutations within SRs of the DP N terminus hotspot were selected for analysis. (B) DPII-GFP was created by splicing out base pairs 3,584–5,380 from a construct of DPI-GFP. (C) Immunolocalization of GFP-tagged WT and mutated DPII-GFP constructs expressed by retroviral transduction in SCC9 cells, in the context of endogenous DP. Bar, 10 µm. (D) Densitometric quantification of immunoprecipitated/total GFP for WT DP-NTP-GFP and N287K-DP-NTP-GFP coimmunoprecipitated with plakophilin-1 (PKP1-FLAG) or plakoglobin (Pg-Myc) from HEKs, used for robust protein expression (n = 2 independent experiments for each IP). (E) Dispase assay of adhesive strength for cells expressing WT or mutant DPII-GFP constructs. (E, left) The number of fragments observed. Blots show expression of DP constructs (GFP) in SCC9s. GAPDH was used as a loading control, and EB1 expression levels were equivalent across samples (n = triplicate wells from one of two independent experiments). Error bars indicate SEM.
WT and mutated DPII-GFP constructs were retrovirally expressed in SCC9s in the background of endogenous DP, a situation resembling that of patients with one mutant allele. All mutants except for N287K localized prominently in puncta at cell–cell interfaces, which is typical for junctional DP staining and indistinguishable from WT DPII-GFP (Fig. 4 C). N287K exhibited a range of junctional-to-cytoplasmic localization (Figs. 4 C and S5 B), accompanied by varying extents of keratin retraction from cell–cell contacts (see Fig. 6 B). Coimmunoprecipitation analysis revealed that N287K reduced binding to armadillo proteins (Fig. 4 D and Fig. S3 C), which is consistent with its impaired localization. Epithelial sheets expressing N287K exhibited adhesive defects in confluent cell monolayers released with dispase and subjected to mechanical stress (Fig. 4 E; Hudson et al., 2004). Cell sheets expressing WT and junction-associated DP mutants remained intact after mechanical stress, which is consistent with observations that DP-dependent tethering of IFs to cell–cell contacts is important for tissue integrity (Vasioukhin et al., 2001; Huen et al., 2002). None of the mutations detectably altered membrane localization of E-cadherin or β-catenin (Fig. S4 A).
We used PLA analysis to determine whether DP N-terminal mutations interfere with EB1 association. K470E and A566T exhibited PLA signals comparable to WT DPII-GFP. However, N458Y and I533T signals were dramatically reduced in the cell cortex, consistent with a reduction in EB1 association (Fig. 5, A and A′). N287K maintained close proximity to EB1, but because of its increased cytoplasmic localization, signals were distributed throughout the cell.
DP–EB1 interactions are impaired by disease mutations in the DP N terminus. (A) PLA of EB1 with WT and mutated DPII-GFP constructs in SCC9 cells transfected with siRNA targeting the 3′ UTR of endogenous DP. Color panels show PLA dots overlaid with GFP expression. Boxed regions in color overlays are magnified in the panels on the right. PLA values are quantified in the graph (A′; n = 12 image fields from three independent experiments). Bars: (main images) 10 µm; (insets) 5 µm. Error bars indicate SEM. (B) Radar plots: EB1 comet angles categorized for frequencies between 0 and 90° relative to cell–cell contacts in cells expressing WT and mutated DPII-GFP in the background of endogenous DP (n > 250 comets per condition from three independent experiments). Results are compiled in a cumulative frequency plot below the individual graphs, with the x axis representing frequency bins for EB1 angles. All data are from SCC9s.
DP–EB1 interactions are impaired by disease mutations in the DP N terminus. (A) PLA of EB1 with WT and mutated DPII-GFP constructs in SCC9 cells transfected with siRNA targeting the 3′ UTR of endogenous DP. Color panels show PLA dots overlaid with GFP expression. Boxed regions in color overlays are magnified in the panels on the right. PLA values are quantified in the graph (A′; n = 12 image fields from three independent experiments). Bars: (main images) 10 µm; (insets) 5 µm. Error bars indicate SEM. (B) Radar plots: EB1 comet angles categorized for frequencies between 0 and 90° relative to cell–cell contacts in cells expressing WT and mutated DPII-GFP in the background of endogenous DP (n > 250 comets per condition from three independent experiments). Results are compiled in a cumulative frequency plot below the individual graphs, with the x axis representing frequency bins for EB1 angles. All data are from SCC9s.
Next, we analyzed the effects of DP mutations on EB1 organization and MT dynamics. Expression of N458Y or I533T DPII-GFP shifted the angle of EB1 comets relative to cell–cell contacts (Fig. 5 B), corresponding to deficiencies in EB1 binding. EB1 angles were also compromised upon expression of N287K, likely due to the reduced accumulation of this mutant at sites of cell–cell contact, which precluded DP from providing spatial cues for EB1.
As a consequence of altered EB1 organization, there were also significant reductions in demecolcine-resistant MTs in cells expressing N287K, N458Y, or I533T (Fig. 6 A). N458Y and I533T resulted in MT stability defects without altering IFs, though reduced keratin attachment could be observed in cells with cytoplasmic localization of N287K (Fig. 6 B). The reduced sensitivity of MTs to demecolcine in these cells compared with DP KD cells (Fig. 3 B) is likely due to the presence of endogenous DP in the background of expressed DP mutants.
DP mutants increase sensitivity of MTs to demecolcine and decrease MT polymerization rates. (A) DP and MT immunostaining, and thresholded MT images, in demecolcine-treated cells expressing DP mutants. (n > 35 cell fields for each condition from three independent experiments). (A, bottom) Tubulin intensities during demecolcine treatment. (B) Immunolocalization of keratin and GFP in cells expressing DPII-GFP constructs and treated with demecolcine. (C) MT polymerization rates and frequency of pausing events for cells expressing DPII-GFP constructs (n > 127 EB3 tracks for each condition compiled from n > 4 fields from more than four movies for each condition). All data are from SCC9s. Bars, 10 µm. Error bars indicate SEM.
DP mutants increase sensitivity of MTs to demecolcine and decrease MT polymerization rates. (A) DP and MT immunostaining, and thresholded MT images, in demecolcine-treated cells expressing DP mutants. (n > 35 cell fields for each condition from three independent experiments). (A, bottom) Tubulin intensities during demecolcine treatment. (B) Immunolocalization of keratin and GFP in cells expressing DPII-GFP constructs and treated with demecolcine. (C) MT polymerization rates and frequency of pausing events for cells expressing DPII-GFP constructs (n > 127 EB3 tracks for each condition compiled from n > 4 fields from more than four movies for each condition). All data are from SCC9s. Bars, 10 µm. Error bars indicate SEM.
Expression of N287K, N458Y, or I533T resulted in a shift toward slower MT growth rates as compared with WT-DPII-GFP, with the most significant changes in the cortex (Fig. 6 C and Fig. S4 B). These mutants also increased cortical frequencies of pausing events and total time spent in pause, similar to changes observed upon DP KD (Fig. S2 D). Expression of DP-NTP did not affect MT growth rates (Fig. S4 C), which suggests that the observed changes in MT dynamics are unlikely to depend on IF networks. Collectively, these data implicate DP–EB1 interactions in DP-mediated regulation of MT dynamics.
DP promotes membrane localization of Cx43
Trafficking of gap junction proteins to the plasma membrane/sites of cell–cell contact is mediated by MT-based transport (Jordan et al., 1999; Lauf et al., 2002). EB1 was shown to be a key component of the MT-associated machinery involved in localizing Cx43, the predominant connexin isoform in a majority of tissues including the epidermis and myocardium, to the plasma membrane (Laird, 2006; Shaw et al., 2007). However, the significance of EB1 to connexin trafficking in disease has not been determined. In primary NHEKs, which exhibit robust Cx43 localization at the plasma membrane of differentiated cells (Solan and Lampe, 2009), DP or EB1 KD impaired Cx43 membrane localization (Fig. 7 A). DP KD produced a shift in Cx43 Triton solubility (Fig. 7 B), which corresponds to an increase in nonjunctional connexin (Musil and Goodenough, 1991). In contrast, Dsg2 KD did not impair Cx43 membrane localization (Fig. S4 D). Expression of WT DPII-GFP rescued Cx43 membrane localization in cells transfected with DP, but not EB1, siRNA (Fig. S5 A), supporting the idea that DP regulates Cx43 through EB1-dependent MT-based transport.
DP promotes membrane localization of Cx43. (A) Cx43 membrane localization (arrows) in NHEKs electroporated with Ct, DP, or EB1 siRNA. Thresholded Cx43 images are shown to highlight Cx43 membrane localization. Western blot is shown below. (B) Triton X-100 solubility of Cx43 in Ct or DP KD NHEKs, quantified in a graph (n = 3 independent experiments). Lines indicate that intervening lanes have been spliced out. (C) Cx43 membrane localization in Ct or DP KD organotypic raft cultures. Hematoxylin and eosin (H&E) panels demonstrate overall tissue architecture (lines mark dermal-epidermal junctions). Graph: quantification of Cx43 membrane localization (n = 15 image fields from rafts prepared in three independent experiments, with >150 regions of cell–cell contact calculated per condition). A Western blot is shown. (D) Cx43 membrane localization in Ct or DP KD primary NRVCMs. Graph: localization of Cx43 at membranes, marked by Pg (n > 95 regions of cell–cell contact from three independent experiments). Western blot shows protein expression levels. Bars, 10 µm. Error bars indicate SEM.
DP promotes membrane localization of Cx43. (A) Cx43 membrane localization (arrows) in NHEKs electroporated with Ct, DP, or EB1 siRNA. Thresholded Cx43 images are shown to highlight Cx43 membrane localization. Western blot is shown below. (B) Triton X-100 solubility of Cx43 in Ct or DP KD NHEKs, quantified in a graph (n = 3 independent experiments). Lines indicate that intervening lanes have been spliced out. (C) Cx43 membrane localization in Ct or DP KD organotypic raft cultures. Hematoxylin and eosin (H&E) panels demonstrate overall tissue architecture (lines mark dermal-epidermal junctions). Graph: quantification of Cx43 membrane localization (n = 15 image fields from rafts prepared in three independent experiments, with >150 regions of cell–cell contact calculated per condition). A Western blot is shown. (D) Cx43 membrane localization in Ct or DP KD primary NRVCMs. Graph: localization of Cx43 at membranes, marked by Pg (n > 95 regions of cell–cell contact from three independent experiments). Western blot shows protein expression levels. Bars, 10 µm. Error bars indicate SEM.
To study Cx43 localization in a more physiological model, we used organotypic raft cultures grown at an air–liquid interface to induce epidermal stratification and differentiation. We observed a significant reduction in Cx43 membrane localization in DP KD lifted cultures (Fig. 7 C) but not in Dsg1 KD cultures (Fig. S4 E). The lack of Cx43 mislocalization in Dsg1 KD cultures makes it less likely that Cx43 mislocalization in DP KD cultures is due to general defects in tissue architecture caused by desmosome deficiency, though this possibility cannot be fully ruled out. DP KD also impaired Cx43 membrane localization in freshly isolated neonatal rat ventricular cardiac myocytes (NRVCMs; Fig. 7 D). Thus, our data show that DP regulates Cx43 membrane localization in models of skin and heart.
DP–EB1 interactions direct connexin localization and function
Next, we expressed WT and mutant DPII-GFP in NHEKs and in immortalized HL-1 cardiac cells, which retain contractile signaling functions (Claycomb et al., 1998), to determine if DP regulation of Cx43 is EB1 dependent (Fig. 8 A). Using Pg staining to mark areas of cell–cell contact between GFP-labeled cells, we observed a significant reduction in Cx43 membrane localization in cells expressing N287K, N458Y, or I533T (Fig. 8, A and B). Consistent with previous observations, we detected a significant increase in cytoplasmic intensity of N287K DPII-GFP (Fig. 8 C). Cx43 membrane localization was most severely impaired in cells with lower levels of membrane-localized N287K DPII-GFP (Fig. S5 B), which supports the idea that although this mutant retains EB1 binding, its cytoplasmic localization precludes proper EB1-mediated Cx43 membrane localization.
DP disease mutations impair Cx43 membrane localization. (A) Immunostaining of Pg, GFP, and Cx43 in NHEKs expressing WT and mutated DPII-GFP constructs. Pg staining was used to mark regions of cell–cell contact between cells expressing DPII-GFP constructs (white arrows). Regions were overlaid on thresholded Cx43 images for quantification. Bars, 10 µm. (B) Quantification of Cx43 fluorescence intensity at cell–cell contacts in NHEKs (i) and HL-1s (ii; n = 9 image fields from three independent experiments). (C) Quantification of cytoplasmic GFP intensity in NHEKs expressing WT and mutated DPII-GFP constructs (n = 9 image fields from three independent experiments). Error bars indicate SEM.
DP disease mutations impair Cx43 membrane localization. (A) Immunostaining of Pg, GFP, and Cx43 in NHEKs expressing WT and mutated DPII-GFP constructs. Pg staining was used to mark regions of cell–cell contact between cells expressing DPII-GFP constructs (white arrows). Regions were overlaid on thresholded Cx43 images for quantification. Bars, 10 µm. (B) Quantification of Cx43 fluorescence intensity at cell–cell contacts in NHEKs (i) and HL-1s (ii; n = 9 image fields from three independent experiments). (C) Quantification of cytoplasmic GFP intensity in NHEKs expressing WT and mutated DPII-GFP constructs (n = 9 image fields from three independent experiments). Error bars indicate SEM.
We also found that Cx43 was mislocalized when N287K, N458Y, or I533T mutants were expressed in the DPI backbone in COS cells (Fig. S5 C). While used for their efficiency in transient transfections, COS cells did not express DPI-GFP constructs as robustly as retrovirally expressed DPII-GFP in other cell lines. This difference potentially explains the discrepancy in magnitudes of Cx43 mislocalization for mutations expressed in a backbone of DPI versus DPII.
Finally, to determine the functional impact of these changes, we quantified gap junctional intercellular communication (GJIC) of NHEKs expressing DP mutations using a dye-coupling assay. Cells expressing N287K, N458Y, or I533T DPII-GFP demonstrated ∼49%, ∼28%, and ∼38% reductions, respectively, in dye transfer from injected source cells to neighbor cells compared with WT DPII-GFP (Fig. 9). These data support the conclusion that mutations in the DP N terminus impair Cx43 membrane localization and gap junction function by either directly disrupting association with EB1 (N458Y, I533T) or by causing DP mislocalization (N287K) and thus a corresponding mislocalization of EB1 (Fig. 10).
DP mutations impair GJIC. Phase contrast and fluorescence images of NHEKs retrovirally expressing WT and mutated DPII-GFP constructs (in the background of endogenous DP) and injected with Alexa Fluor 568 dye. Arrows mark points of injection in source (s) cells. Graph shows percent dye transfer from source to neighbor (n) cells at 1 min after injection (n > 15 cell pairs for each mutation from three independent experiments). Bars, 10 µm. Error bars indicate SEM.
DP mutations impair GJIC. Phase contrast and fluorescence images of NHEKs retrovirally expressing WT and mutated DPII-GFP constructs (in the background of endogenous DP) and injected with Alexa Fluor 568 dye. Arrows mark points of injection in source (s) cells. Graph shows percent dye transfer from source to neighbor (n) cells at 1 min after injection (n > 15 cell pairs for each mutation from three independent experiments). Bars, 10 µm. Error bars indicate SEM.
Model of DP–EB1 interaction with a summary of mutation pathogenicity. (A) When localized to cell–cell contacts, DP provides spatial cues to regulate EB1 and MT organization. Subsequently, MTs can be used for Cx43 trafficking to adjacent gap junctions (model does not include cytoplasmic DP–EB1 interactions, which may also influence MT dynamics). Upon expression of mutations that impair DP–EB1 binding or cause a cytoplasmic shift in the location of DP–EB1 binding, DP is limited in its ability to regulate EB1 and MT association with cortical regions, resulting in a reduction of Cx43 membrane localization and GJIC. (B) Mutations are summarized based on clinical findings and data obtained in this study.
Model of DP–EB1 interaction with a summary of mutation pathogenicity. (A) When localized to cell–cell contacts, DP provides spatial cues to regulate EB1 and MT organization. Subsequently, MTs can be used for Cx43 trafficking to adjacent gap junctions (model does not include cytoplasmic DP–EB1 interactions, which may also influence MT dynamics). Upon expression of mutations that impair DP–EB1 binding or cause a cytoplasmic shift in the location of DP–EB1 binding, DP is limited in its ability to regulate EB1 and MT association with cortical regions, resulting in a reduction of Cx43 membrane localization and GJIC. (B) Mutations are summarized based on clinical findings and data obtained in this study.
Discussion
Here, we present evidence that cardiac and cutaneous disease mutations in DP result in Cx43 mislocalization by impairing DP–EB1 interactions or by mistargeting DP–EB1 complexes. As a key regulator of MT plus ends, EB1 coordinates the interaction of several plus tip proteins with MTs, thereby mediating association of MTs with various cellular domains for transient stabilization and/or capture (Gundersen et al., 2004; Vaughan, 2005; Galjart, 2010). Many EB1-binding partners contain a conserved SxIP motif facilitating association with EB1 as it tracks MT plus ends (Galjart, 2010). Our data suggest that DP does not require EB1 for proper transport or membrane localization, and that DP–EB1 interactions instead enable DP to regulate EB1 and MT organization at sites of cell–cell contact. Although spectraplakin family proteins BPAG1 and MACF/ACF7 possess distinct MT-binding or EB1-binding domains in their C termini (Yang et al., 1999; Sun et al., 2001; Leung et al., 2002; Subramanian et al., 2003; Slep et al., 2005), the epithelial plakin DP does not contain an SxIP EB1-binding motif nor does it bind directly to MTs (Sun et al., 2001). Thus, DP’s EB1-dependent regulation of MT plus ends through its N terminus expands on mechanisms by which proteins with plakin domains regulate the cytoskeleton.
Our data show that the DP N terminus interacts with the EB1 C terminus (Fig. 1 C). Thus, DP does not compete with EB1 binding to MTs, which is mediated through the EB1 N-terminal calponin-homology domain (Hayashi and Ikura, 2003; Komarova et al., 2009; Galjart, 2010). Our CytoTrap and DP-NTP pull-down data show that residues 1–584 in DP are sufficient for this interaction (Fig. 1, B and C), though pull-down assays suggest that additional regions in the DP N terminus strengthen the interaction with EB1 (Fig. S1 A). The weaker binding exhibited by shorter, bacterially expressed constructs may reflect differences in posttranslational modification states required for optimal binding. It is also plausible that the l-shaped DP N terminus (Al-Jassar et al., 2011) constitutes a binding pocket for EB1, with SR8 and SR9 stabilizing the DP–EB1 interaction. Regardless, data from individual DP-N-terminal point mutants indicates that residues in SR5 (N458, I533) are key in mediating DP–EB1 interactions (Fig. 5 A).
Because EB1 is a master regulator of MT plus end tracking and a core component of plus end capture (Vaughan, 2005; Morrison, 2007), we investigated effects of DP on MT stabilization and dynamics. DP depletion, or expression of DP mutants that do not bind (N458Y, I533T) or likely mislocalize (N287K) EB1, reduces the population of MTs resistant to depolymerization by demecolcine, which indicates a decrease in stable MTs in cells (Fig. 6 A). Analysis of EB3 dynamics on MTs near the cortex also revealed a significant reduction in MT growth rates when DP–EB1 interactions are perturbed (Fig. 6 C). Furthermore, in DP-depleted cells, we observed a >2.5-fold increase in MT pausing near the cortex (Fig. 3 C and Fig. S2 D). Considered together, these findings demonstrate a clear role for DP–EB1 interactions in regulation of MT organization and dynamics. Increased pausing near the cortex may occur if MTs approaching these regions cannot locate or bind membrane-associated DP and thus persist in an attempt to identify cortical attachment cues. Alternatively, in the absence of DP binding, EB1 may instead bind without competition to other cortical factors that also influence MT persistence and organization near the membrane. As indicated by localization of DP–EB1 PLA signals, DP also associates with EB1 in the cytoplasm, where it can influence MT growth rates. Additional studies are needed to fully understand effects of DP on MT dynamics, to determine if DP is a component of cortical MT capture machinery, and to determine how DP–EB1 interactions may relate to previously described interactions of MT plus ends with components of adherens junctions (Ligon and Holzbaur, 2007; Shtutman et al., 2008; Shahbazi et al., 2013).
As a functional outcome of MT plus end interactions with DP at the cell cortex, we investigated Cx43 membrane localization, which is mediated at least in part by EB1-dependent, MT-based trafficking (Shaw et al., 2007). DP KD reduces Cx43 membrane localization in skin and heart cells, which is consistent with in vivo data of gap junction misregulation in DP-knockout mouse models (Gomes et al., 2012; Lyon et al., 2014) and DP-depleted HL-1 cells (Zhang et al., 2013). Here, we show that DP–EB1 interactions are critical for DP’s regulation of gap junctions and act by targeting Cx43 to the membrane. Further analysis will be needed to determine the threshold at which Cx43 mislocalization elicits pathological effects. However, these results suggest a potential mechanism to explain how loss of DP results in improper connexin localization and electrical coupling in epidermal and cardiac tissue (Kaplan et al., 2004; Asimaki et al., 2009; Gomes et al., 2012; Zhang et al., 2013; Lyon et al., 2014), and further provide a molecular basis for observations that in models of cardiac injury, DP relocalizes to lateral membranes along with Cx43 and EB1 (Chkourko et al., 2012).
Our analysis of individual disease mutations provides insight into molecular mechanisms that may lead to observed differences in gap junction function. The N287K mutation was reported in a patient with skin fragility/woolly hair syndrome. Similar to our observations, N287K resulted in cytoplasmic DP localization in patient biopsies (Whittock et al., 2002). N287K targets a region of DP SR4 that is thought to interact with and structurally stabilize the SH3 loop of SR5 (Al-Jassar et al., 2011; Choi and Weis, 2011), which suggests that the mutation could result in conformational instability of the DP N terminus to reduce binding to armadillo proteins and hinder proper membrane localization. AC mutations upstream of N287K—V30M and Q90R—have also been shown to impair association of DP with Pg (Yang et al., 2006). DP mutations that impair armadillo protein binding are all candidates for obstructing DP targeting to desmosomes and/or its ability to confer desmosome-based adhesive strength, as was observed in the case of the N287K mutant. This structural function normally contributes to stabilization of adjacent gap junctions (Saffitz, 2006; Noorman et al., 2009). We show here that mislocalized DP also dampens spatial cues for EB1-directed MT-based trafficking of Cx43 to cell membranes, thus revealing another way that this class of mutants could lead to loss of gap junction communication between cells.
In contrast to the aberrant cytoplasmic DP localization caused by N287K, N458Y and I533T still localize at sites of cell–cell contact. In these cases, impaired Cx43 membrane localization is associated with loss of DP–EB1 binding. N458Y and I533T were reported as single mutations in patients with AC (Rampazzo, A. 2008. European Society of Cardiology meeting. Prevalence of desmosomal protein mutations and clinical features in a large cohort of unrelated consecutive patients affected with ARVC; Xu et al., 2010; Bauce et al., 2011). N458Y targets a residue on the free surface of the SR5 SH3 loop, whereas I533T replaces a core isoleucine residue with a polar threonine residue to potentially introduce a structural kink to SR5 (Choi and Weis, 2011). Because of the close structural proximity of these two mutations, we propose that this region of the DP N terminus SR5 (the junction between the free SH3 loop and the downstream SR5 backbone) constitutes an EB1 binding site. It is important to note that these single point mutations in the DP N terminus do not detectably affect junctional targeting, C-terminal–dependent IF anchorage, membrane localization of adherens junctions components, or cell–cell adhesion. Together, these data support a model wherein EB1 association with DP contributes significantly to regulation of MT stability and gap junction assembly/function, although we cannot rule out the possibility that functions unrelated to EB1 binding may also contribute to these changes.
We did not detect significant changes in EB1 binding or Cx43 membrane localization upon expression of DP K470E or A566T. K470E and A566T were reported as compound heterozygous mutations in a patient with AC (Basso et al., 2006; Bauce et al., 2011; Cox et al., 2011), and it is possible that these mutations are insufficient to produce pathological changes when individually coexpressed with WT DP. It is also possible that stress, e.g., strenuous exercise, may play a role in how these mutations are manifested in disease (Basso et al., 2011); our studies are limited by the absence of these additional antagonistic factors. Alternatively, K470E has been reported to cause local conformational changes in DP while leaving overall protein structure intact (Al-Jassar et al., 2011), which suggests that this mutation could impair interactions with other, unidentified binding partners of the DP N terminus. These possibilities highlight the heterogeneity of AC pathogenesis.
Here, we highlight a role of DP in regulating gap junctions. However, other studies implicate the classical cadherin N-cadherin in directing Cx43 localization (Shaw et al., 2007). This potential functional overlap is of particular interest to consider in the heart, where components of desmosomes and adherens junctions intermingle as a hybrid area composita that stabilizes adjacent gap junctions (Borrmann et al., 2006; Franke et al., 2006). DP localizes to regions of cells resembling classic adherens junctions in cardiac tissue, raising the possibility that multiple components of adhesive junctions cooperate to regulate connexin localization. In agreement with recent studies of DP depletion from epithelial models (Sumigray et al., 2014), our data indicate that DP depletion or mutation do not alter membrane localization of E-cadherin or β-catenin. Nevertheless, we cannot fully rule out some contribution of adherens junctions to Cx43 mislocalization.
In summary, our studies reveal for the first time an interaction between DP and EB1, and elucidate a role for DP in regulating EB1 and MT organization. Importantly, our data provide a mechanistic basis by which mutations in the DP N terminus misregulate gap junctions in cardiac and cutaneous disease: by disrupting DP–EB1 binding or by impairing membrane localization of this interaction. Each of these mechanisms could contribute to development of diseases associated with DP mutations including AC and skin fragility syndromes. These studies highlight and expand on the emerging nontraditional roles for DP and desmosomes in tissue morphogenesis and disease development.
Materials and methods
Cell culture
Human-derived oral squamous cell carcinoma SCC9 cells (a gift from J. Rheinwald, Harvard Medical School, Boston, MA) were maintained in DMEM/F-12 medium (Corning) supplemented with 10% FBS and 1% penicillin/streptomycin. HEK 293 cells, COS monkey kidney cells, and A431 epithelial cell lines were maintained in DMEM (Corning) supplemented with 10% FBS and 1% penicillin/streptomycin. A431 stable cell lines were generated by transfecting parental A431 lines with pTet-On (Takara Bio Inc.), selecting with 400 µg/ml G418 (Corning), and then transfecting with pTRE (Takara Bio Inc.) plasmids expressing DP-NTP-GFP or DP-GFP (Huen et al., 2002; Godsel et al., 2005). Expression of DP-GFP or DP-NTP-GFP in A431 cells was induced by treatment with 4 µg/ml doxycycline for 24 h. Primary NHEKs were cultured in M154 medium (Invitrogen) adjusted to 0.07 mM calcium and supplemented with human keratinocyte growth supplement (HKGS) and gentamicin/amphotericin B. To induce differentiation, NHEKs were switched to media with 1.2 mM calcium in the absence of HKGS. NRVCMs were cultured in M199 medium (Lonza) supplemented with 1% penicillin/streptomycin, heat-inactivated horse serum (Gibco), 15 µM vitamin B12 (Sigma-Aldrich), and 100 µM bromodeoxyuridine (Sigma-Aldrich). Immortalized HL-1 cells derived from mouse cardiac atria (a gift from W. Claycomb, Louisiana State University School of Medicine, New Orleans, LA) were maintained in Claycomb medium (Sigma-Aldrich) with 10% FBS, 1% penicillin/streptomycin, 0.1 mM norepinephrine, and 2 mM l-glutamine.
Antibodies
The following primary antibodies were used in this study: NW161 rabbit anti-DP directed against the first 189 amino acids of DP (Bornslaeger et al., 1996), NW6 rabbit anti-DP directed against the C-terminal domain of DP (Angst et al., 1990), 115F mouse anti-DP directed against the DP C terminus (a gift from D. Garrod, The University of Manchester, Manchester, England, UK), mouse anti-EB1 (610535; BD), rabbit anti-MAPRE1 (ab117821; Abcam), DM1A mouse anti–α-tubulin (Abcam ab7291), 12G10 mouse anti–α-tubulin (Developmental Studies Hybridoma Bank), rabbit anti-Cx43 (AB1728; EMD Millipore), 6D8 mouse anti–desmoglein 2 (a gift from M. Wheelock, University of Nebraska Medical Center, Omaha, NE), JL8 mouse anti-GFP (Living Colors), chicken IgY anti-GFP (Molecular Probes), rabbit anti-GFP (632593; Living Colors), rabbit anti-GFP (A11122; Life Technologies), 1407 chicken anti-Pg (Aves Laboratories), mouse anti–plakophilin 2 (651101; Progen), rabbit anti-GAPDH (Sigma-Aldrich), mouse anti-His (EMD Millipore), rabbit anti-FLAG (Sigma-Aldrich), M2 mouse anti-FLAG (Sigma-Aldrich), mouse anti-cMyc (Life Technologies), KT36 rat anti-EB3 (Abcam), HECD-1 mouse anti–E-cadherin (Abcam), C2206 rabbit anti–β-catenin (Sigma-Aldrich), and KS-B17.2 mouse anti-keratin 18 (Sigma-Aldrich). Western blot analysis included use of peroxidase-conjugated anti–mouse, –rabbit, and –chicken secondary antibodies purchased from the Kirkegaard & Perry Laboratories, Inc. Alexa Fluor 488/568/647–conjugated goat anti–mouse, –rabbit, –rat, and –chicken secondary antibodies (Invitrogen) were used in immunofluorescence studies.
Generation and delivery of DNA constructs
Yeast-two-hybrid (CytoTrap).
The DP-NTP bait construct (pSos-DP-NTP) was generated by cloning a PCR product of DP nucleotides 1–1,752 into the pENTR/D-TOPO Gateway vector (Invitrogen). LR Clonase-mediated recombination was then used to clone the DP-NTP insert into a pSos vector that had been made destination compatible.
EB constructs.
Histidine-tagged EB1 constructs (provided by H. Goodson, University of Notre Dame, Notre Dame, IN) were generated by cloning cDNA of human EB1 (full-length or residues 1–184) into pET15b (Invitrogen; Zhu et al., 2009; Skube et al., 2010). The EB3-mRFP construct (provided by A. Akhmanova, Utrecht University, Utrecht, Netherlands) was generated by substituting mRFP (a gift from R. Tsien, University of California, San Diego, La Jolla, CA) into the GFP-encoding region of a pEGFP vector (Takara Bio Inc.; Grigoriev et al., 2008).
GST-tagged DP constructs.
Truncation constructs of the DP N terminus were generated by purifying PCR products of DP nucleotides 1–537 (aa 1–179), 1–1,890 (aa 1–630), 1–2,649 (aa 1–883), and 1–3,066 (aa 1–1,022) and cloning them into the pDEST15 (N-terminal GST tag) vector (Invitrogen) via pENTR.
Site-specific mutagenesis.
The QuikChange II XL Site-Directed Mutagenesis kit (Agilent Technologies) was used to introduce site-specific mutations to constructs of DP-NTP, DPII, and DPI (Godsel et al., 2005), which were fully verified by sequencing.
Transfections.
Transient transfections were performed using Lipofectamine 2000 (Invitrogen) and 0.5–2 µg/ml DNA.
Retroviral infections.
The phoenix packaging cell line (provided by G. Nolan, Stanford University, Stanford, CA), maintained in DMEM (Mediatech) supplemented with FBS and penicillin/streptomycin, was used to prepare a retrovirus of GFP-tagged DPII constructs. Phoenix cells were transfected with DNA constructs and placed in selection media with 1 µg/ml puromycin 48 h after transfection. Cells were grown at 32°C for 16–24 h to collect viral supernatant, which was then concentrated using Amicon Ultra-15 Centrifugal Filter units (EMD Millipore). NHEKs and HL-1s were infected with virus and 8 µg/ml polybrene for 90–180 min at 32°C, after which cells were washed and returned to growth at 37°C in fresh media. SCC9s were infected with virus and 8 µg/ml polybrene overnight at 37°C. Retrovirus expressing DP-NTP-GFP was provided by J. Broussard (Northwestern University, Evanston, IL).
RNAi-mediated KD
Oligonucleotides against human DP and human EB1/MAPRE1, and scrambled oligonucleotides for controls, were purchased from GE Healthcare. DP and EB1/MAPRE1 siRNAs were each a pool of four siRNA oligonucleotides (target sequences for DP: 5′-GAAGAGAGGUGCAGGCGUA-3′, 5′-GACCGUCACUGAGCUAGUA-3′, 5′-AAACAGAACGCUCCCGAUA-3′, and 5′-CGACAUGAAUCAGUAAGUA-3′; target sequences for EB1/MAPRE1: 5′-GAUGAAGGCUUUGUGAUAC-3′, 5′-GGAAAGCUACGGAACAUUG-3′, 5′-UGACAAAGAUCGAACAGUU-3′, and 5′-GAGAAUGGGUGUUGACAAA-3′). Oligonucleotides targeting the 3′ UTR of human Dsg2 were purchased from IDT (sense sequence: 5′-CCUGGAAGCAGAGACAGUGUGGUCCUU-3′). Additional stealth siRNA oligonucleotides targeting human DP were purchased from Life Technologies (Invitrogen) and used as a pool of three oligonucleotides (sense sequences: 5′-GGAACCUGCCAAGAUGUCAACCAUA-3′, 5′-CAGACCGCUGGCAAAGGAUAGAUAA-3′, and 5′-CAGGGCUCUGUCUUCUGCCUCUGAA-3′). For specific KD of endogenous DP, two oligonucleotides targeting the 3′ UTR of DP (IDT) were used as a mix (sense sequences: 5′-AAUUAAUACCAAACCAAA-3′ and 5′-GCAGUAGAGUGAUAGGA-3′). SCC9 cells were transfected with siRNA oligonucleotides at a final concentration of 20 nM using DharmaFECT (GE Healthcare). NHEKs were electroporated with siRNA oligonucleotides via AMAXA nucleoporation (Lonza) using the Ingenio Electroporation Solution (Mirus) or solution V (Lonza) and program X-001. For DP KD in NRVCMs, the BLOCK-iT PolII miR RNAi Expression Vector kit (Life Technologies) was used to design oligonucleotides targeting the C terminus of rat DP or a scrambled sequence as a control. These constructs were cloned into the pAd/cytomegalovirus (CMV)/V5-DEST vector (Invitrogen) and used to generate virus using the ViraPower Adenoviral Expression System (Invitrogen).
Western blot analysis
For analysis of protein expression levels, cells were washed in PBS and lysed in urea sample buffer (8 M deionized urea, 1% sodium dodecyl sulfate, 10% glycerol, 60 mM Tris, pH 6.8, and 5% β-mercaptoethanol). After equalizing total protein concentrations, samples were run on 7.5–15% SDS-PAGE gels and transferred to polyvinylidene fluoride (PVDF) membranes to be probed with primary and secondary antibodies against proteins of interest.
Immunofluorescence and immunohistochemistry
For analysis of protein localization in cultured cells, coverslips were fixed and permeabilized by submerging in anhydrous methanol for 2–3 min at −20°C. Cells were incubated in solutions with primary and secondary antibodies at 37°C for 60 min and 30 min, respectively, with multiple PBS washes after each step. For analysis of protein localization in tissue sections, paraffin sections of organotypic raft cultures were rehydrated and then heated to 95°C in 0.01 M citrate buffer for antigen retrieval. Sections were incubated in solution with primary antibody at 4°C overnight and in solution with secondary antibody at 37°C for 60 min. Coverslips and tissue sections were mounted using polyvinyl alcohol (Sigma-Aldrich).
Microscope image acquisition
Cells and tissues were visualized using a microscope (model DMR; Leica) fitted with 40× (Plan-Fluotar, NA 1.0) and 63× (Plan-Apochromat, NA 1.32) objective lenses, a confocal microscope (LSM 510 META; Carl Zeiss), or a superresolution microscope (Nikon TiE N-SIM; Nikon). Images on the DMR microscope were acquired using a charge-coupled device (CCD) camera (model C4742-95; Orca 100; Hamamatsu) and analyzed using imaging software (MetaMorph 7.7; Molecular Devices). Images captured on the LSM 510 microscope were taken and analyzed using 63× or 100× objective lenses (Plan Apochromat, NA 1.32 and NA 1.4; Carl Zeiss) and LSM 510 software. SIM images were captured using a 100× objective lens (NA 1.49) and a camera (iXon X3 897; Andor Technology), and reconstructed using Elements version 4 software (Nikon).
Microinjection (dye transfer assay)
Microinjection was performed using a disc scanning unit microscope (IX81 DSU; Olympus; UPlanFL/NA 0.17 Ph1 objective lens) equipped with a FemtoJet injector (Eppendorf). Alexa Fluor 568 Hydrazide (Life Technologies) was injected into GFP-labeled cells at an injection pressure of 10–12 hPa with continuous flow. Injected (source) and neighbor cells were imaged immediately after using phase, FITC, and Cy3 wide-field acquisition channels on a 20× objective lens, with SlideBook imaging software (3i). Fluorescence image intensities were within the linear range of the detector.
Primary cardiac cell isolation and culture
NRVCMs were isolated from day-old Sprague-Dawley pups (Charles River) by excising hearts, dissecting cardiac ventricles, and enzymatically digesting tissue to obtain cell suspensions. After an initial preplating step to allow cardiac fibroblasts to attach to plates, NRVCMs were cultured on plates coated with Collagen IV. For imaging studies, NRVCMs were plated on poly-l-lysine coverslips (BD) that were coated with Collagen IV (Sigma-Aldrich) for 1 h, dried under UV light for 1 h, and washed with PBS. NRVCMs were isolated with approval of the Northwestern University Institutional Animal Care and Use Committee, and all animal care protocols conformed to National Institutes of Health guidelines.
Organotypic raft cultures
3D raft cultures were grown by lifting keratinocytes seeded on collagen plugs to an air–liquid interface (Meyers and Laimins, 1994). Collagen plugs were formed using rat tail collagen type I (BD) seeded with J2 fibroblasts (NIH 3T3). J2 fibroblasts were cultured in DMEM (Corning) supplemented with 10% FBS and 1% penicillin/streptomycin. E-media was made as a 50:50 mix of DMEM/F-12 and DMEM media with 5% FBS, 10 µg/ml gentamicin, 0.4 µg/ml hydrocortisone, 10 ng/ml cholera toxin (Sigma-Aldrich), and 0.25 µg/ml amphotericin B (Corning) mixed with a cocktail of 180 µM adenine, 5 µg/ml human recombinant insulin, 5 µg/ml human apo-transferrin, and 5 µg/ml triiodothyronine (Sigma-Aldrich). After seeding with NHEKs that had been electroporated with Ct or DP siRNA, plugs were grown in E-media with 5 ng/ml EGF. 2 d later, plugs with NHEKs were lifted and placed on a metal grid over E-media without EGF to induce epidermal differentiation. Ct or DP KD organotypic raft cultures were harvested 5 d after the day of lifting. Dsg1-KD rafts were prepared by retroviral transduction of miRNA targeting human lamin A/C or Dsg1, and harvested after 3–10 d (Getsios et al., 2009). Rafts were lysed in urea sample buffer for protein analysis, or embedded in paraffin for histology.
Dispase assay
Dispase assays were used to assess intercellular adhesive strength as described previously (Hudson et al., 2004; Yin et al., 2005). Cells expressing DP mutation constructs were seeded in triplicate onto 60-mm dishes. Upon reaching confluence, cells were switched to growth in 0.7 mM Ca for 4 h, washed twice in Dulbecco’s PBS (DPBS), and incubated in 2.4 U/ml dispase II (Roche) for 30 min at 37°C. Lifted monolayers were subjected to centrifugal force on an orbital shaker and imaged using a dissecting microscope (MZ6; Leica).
Generation of DPII constructs
GFP-tagged DPII was generated by using the QuikChange II XL Site-Directed Mutagenesis kit (Agilent Technologies) to splice out nucleotides 3,584–5,380 of a GFP-tagged construct of full-length human DPI (Godsel et al., 2005). Retroviral DPII LZRS constructs were generated by cloning DPII constructs into the pENTR/D-TOPO Gateway vector (Invitrogen) and then cloning inserts into an LZRS vector that had been made destination compatible.
Yeast two-hybrid screening (CytoTrap)
Yeast two-hybrid screening was performed using the CytoTrap vector kit (Agilent Technologies) as described previously (Harmon et al., 2013). The CytoTrap screen (Agilent Technologies) utilizes the temperature-sensitive cdc25H strain of Saccharomyces cerevisiae, which is deficient in Ras signaling and unable to grow at 37°C. Although yeast grow at the permissive temperature of 25°C, only yeast that are cotransformed with a bait (pSos) construct that binds a membrane-associated myristoylated target (pMyr) protein activate Ras signaling to allow for growth at 37°C. pMyr constructs are expressed under galactose promoters; thus, growth at 37°C on galactose plates without growth at 37°C on glucose plates indicates target–bait interaction. A library of HeLa cell cDNAs expressed in the pMyr vector was cotransformed with pSos-DP-NTP into cdc25H yeast. After obtaining putative positives by identifying clones that were capable of growing on galactose plates at 37°C, pMyr constructs were purified and individually cotransformed with pSos-DP-NTP to confirm specific interactions. pMyr-Pg was used as a positive binding control for DP-NTP, and pMyr and pMyr-SB (Stratagene) were, respectively, used as negative and positive controls for the screen.
GST and Histidine pull-down assays
DP N-terminal truncation constructs in pDEST15 were bacterially expressed by adding l-arabinose to cultured BL21A1 bacteria (Invitrogen). FLAG-tagged DP-NTP was constructed by cloning base pairs 1–1,755 of DP into a p700 CMV promoter-based construct with a C-terminal FLAG tag (Kowalczyk et al., 1999). A negative control of GST in a pGEX vector (GE Healthcare) was expressed by adding IPTG to BL21A1 bacteria. Bacteria were lysed and proteins were purified using glutathione agarose (GE Healthcare). Histidine-tagged EB1 constructs were bacterially expressed by adding IPTG to bacterial cultures of Rosetta 2(DE3) bacteria (EMD Millipore). Histidine-tagged constructs were purified from bacterial lysates via association with nickel-coated Ni-NTA beads (QIAGEN). Beads were incubated with cell lysates at 4°C for 2–3 h, washed with a modified RIPA buffer (500 mM NaCl, 50 mM Tris, pH 7.6, 10 mM MgCl2, 1% Triton X-100, 0.1% SDS, and 0.5% deoxycholate), and eluted in urea sample buffer or Laemmli buffer with 5% β-mercaptoethanol.
Coimmunoprecipitation studies
Cells used for coimmunoprecipitation studies were rinsed twice in PBS and lysed in modified RIPA buffer (500 mM NaCl, 50 mM Tris, pH 7.6, 10 mM MgCl2, 1% Triton X-100, 0.1% SDS, and 0.5% deoxycholate) with complete protease inhibitor cocktail (Roche). After preclearing with GammaBind beads (GE Healthcare), cells were incubated with 0.5–1.5 µg of antibody against the protein of interest overnight at 4°C, followed by conjugation to GammaBind beads. For Pg-Myc and PKP1-FLAG IP studies, lysates were incubated with anti–c-Myc agarose (Sigma-Aldrich) or anti-FLAG M2 agarose (Sigma-Aldrich) for 4 h. Samples were washed and eluted in urea sample buffer or Laemmli buffer with 5% β-mercaptoethanol.
PLA
Reagents used to conduct PLA were purchased from OLink Biosciences through Axxora LLC and used as described previously (Harmon et al., 2013). Cells were rinsed and fixed as described in “Immunofluorescence and immunohistochemistry.” After incubation with primary antibody for 60 min at 37°C, samples were incubated with PLA secondary antibodies conjugated to DNA oligonucleotides for 60 min at 37°C. Samples were then subjected to a 30-min incubation at 37°C for ligation of nucleotides, followed by a 100-min incubation at 37°C for rolling circle polymerization, resulting in the production of red fluorescent dots if the antigens targeted by secondary antibodies were in close proximity (Fredriksson et al., 2002; Söderberg et al., 2006). ImageJ software was used to quantify the number of PLA and DAPI signals per image field (endogenous DP–EB1 interaction), or the intensity of PLA and GFP signals per image field (EB1 interaction with DPII-GFP constructs).
Time-lapse imaging
SCC9 cells stably expressing GFP-tagged DP (Godsel et al., 2005) were plated on Lab-Tek chambered coverglass slides (Nunc) and transfected with EB3-mRFP (Grigoriev et al., 2008). Cells were switched to imaging media (Hanks balanced salt solution, 20 mM Hepes, 1% FBS, 2 mM l-glutamine, 4.5 g/liter glucose, and 1× amino acids) and imaged on a microscope (DMI6000; Leica) housed within a 37°C environmental control system chamber using 100 W mercury halide fiber optic illumination. Red and green channels were captured at 1-s or 2-s intervals with a 100× objective lens (HCX Plan-Apochromat, NA 1.4) using a MAG Dual-Cam adaptor fitted with two ORCA-ER AG cameras (Hamamatsu Photonics). Images were captured using Simple PCI version 6.0 (Hamamatsu Photonics) and processed using Photoshop (Adobe) and MetaMorph 7.7 imaging software. Differential interference contrast (DIC) images were acquired to ensure that cells used for imaging were in contact with neighboring cells. EB3 comet particle paths were compiled using MetaMorph 7.7 imaging software.
For time-lapse imaging of parental SCC9s and SCC9s expressing DP-NTP-GFP, images were acquired on an inverted microscope (TiE; Nikon) equipped with a 7-wavelength Sola light engine (Lumencor Inc.), with perfect focus, using a 60× Plan-Apochromat oil immersion objective lens (1.4 NA). Images were collected using a Neo sCMOS camera (Andor Technology) at 560 MHz for 16-bit images (6.45 µm pixels). Devices were controlled by Elements software (Nikon). Cells were transfected (by Amaxa as described in “RNAi-mediated KD”) with EB3-mRFP and plated onto 25-mm coverslips 1–3 d before image acquisition. Time-lapse images were acquired at 2-s intervals for 3 min.
Quantification of EB1 angles
Cells used for quantification of EB1 angles were costained with anti-Pg antibody to mark the plane of cell–cell contacts. Using MetaMorph 7.7 imaging software, lines were drawn over regions of cell–cell contact and EB1 comets in proximity. The angles of lines over EB1 comets were then normalized to the angle of the relevant cell–cell contact and adjusted to a final value of 0–90°. Subconfluent cell cultures were used to avoid analyzing cells in which MTs had reorganized into cortical arrays, which occurs as cells become confluent (Lechler and Fuchs, 2007).
MT studies
For MT stability studies, cells were treated with 1 µM demecolcine/colcemid (Sigma-Aldrich) at 37°C for the indicated time points. After rinsing cells with 0.01% saponin (Sigma-Aldrich) to extract free tubulin, cells were fixed and stained as described in “Immunofluorescence and immunohistochemistry.” MT intensity was measured using MetaMorph 7.7 imaging software or ImageJ software.
MT growth rates and dynamics
MTs were analyzed by tracking EB3-mRFP comets using tip-tracker software (u-Track 2.0; Jaqaman et al., 2008; Applegate et al., 2011). Defined 7 × 10 µm regions of the cytoplasm and 4 × 10 µm regions of the cortex were used for analysis. MT dynamics were analyzed using u-Track 2.0 software with a maximum closing gap of 5 frames, forward angle of 45°, backward angle of 15°, and a fluctuation radius of 2 pixels.
Cx43 detergent solubility
NHEKs used for detergent solubility analyses were rinsed twice with cold DPBS and lysed in TBS buffer (10 mM Tris-HCl, pH 7.5, 145 mM NaCl, 5 mM EDTA, and 2 mM EGTA) with 1% Triton X-100. Lysates were subjected to three rounds of vortexing for 30 s and incubating on ice for 10 min to extract Triton-soluble fractions. Insoluble pellets (Triton-insoluble fractions) were resuspended in urea sample buffer with 5% β-mercaptoethanol.
Quantification of Cx43 and GFP intensity
Cells used for quantification of Cx43 membrane localization were stained with Pg to mark the areas of cell–cell contacts. MetaMorph 7.7 imaging software or ImageJ software was used to mark regions of cell–cell contacts between cells expressing GFP-tagged constructs. These regions were transferred to corresponding images of GFP and Cx43 staining to quantify membrane localization of GFP and Cx43 for each cell–cell contact. GFP cytoplasmic intensity was measured by quantifying GFP intensity in cytoplasmic regions of cells used for quantification of Cx43 and GFP membrane intensity.
Statistics
P-values were calculated using two-tailed, two-sample unequal variance Student’s t tests. P-values <0.05 were considered statistically significant. For all figures, error bars represent SEM.
Online supplemental material
Figs. S1 and S2 contain further characterization of DP–EB1 interactions, demonstrating that EB1 misregulation is specific to loss of DP. Figs. S3 and S4 include a full schematic of DP disease mutations and show characterization of selected mutations. Figs. S2 and S4 include analysis from u-Track 2.0 software. Fig. S4 also shows Cx43 localization in Dsg KD systems. Fig. S5 shows that WT DPII-GFP can rescue Cx43 localization, characterizes effects of N287K on Cx43 localization, and demonstrates Cx43 localization in cells expressing mutants in DPI. Videos 1 and 2 show time-lapse imaging of EB3-mRFP in Ct and DP KD cells, respectively.
Acknowledgments
Thanks go to members of the Green laboratory. We thank Dr. Holly Goodson (University of Notre Dame) and Dr. Anna Akhmanova for their insightful discussions during development of this manuscript. Thank you to Michael Winding and Rosa Ventrella for assistance with u-Track 2.0 software and MATLAB.
Imaging was performed at the Northwestern University Cell Imaging Facility supported by National Cancer Institute (NCI) grant CCSG P30 CA060553 (Robert H. Lurie Comprehensive Cancer Center). Keratinocytes were obtained from the Keratinocyte Core of the Northwestern University Skin Disease Research Center. Histological analysis was conducted by the Pathology Core of the Northwestern University Skin Disease Research Center (grant 5P30AR057216 02), Chicago, IL, with support from the National Institutes of Health (NIH)/National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS), or by the Mouse Phenotyping Core of the R.H. Lurie Comprehensive Cancer Center supported by the NIH/NCI (grant P30 CA060553-159026). Any opinions, findings, and conclusions or recommendations expressed in this material are of the authors and do not necessarily reflect the views of the Northwestern University Skin Disease Research Center or the NIH/NIAMS/NCI. Sequencing services were performed at the Northwestern University Genomics Core Facility. This work was supported by NIH grants R01 AR041836 and AR43380 (to K.J. Green) and a Leducq Transatlantic Network grant (to principal investigators J. Jalife [University of Michigan, Ann Arbor, MI] and S. Hatem [Pierre-and-Marie-Curie University, Paris, France]) with partial support from grant CA122151 and the J.L. Mayberry endowment. D.M. Patel was supported by a predoctoral institutional training grant (3T32GM008152-26S1) awarded by the National Heart Lung and Blood Institute, and by the Malkin Scholars Program from the Robert H. Lurie Comprehensive Cancer Center of Northwestern University.
The authors declare no competing financial interests.
References
- AC
arrhythmogenic cardiomyopathy
- Ct
control
- Cx43
connexin 43
- DP
desmoplakin
- EB1
end-binding 1
- GJIC
gap junctional intercellular communication
- IF
intermediate filament
- KD
knockdown
- MT
microtubule
- NHEK
normal human epidermal keratinocyte
- NRVCM
neonatal rat ventricular cardiac myocyte
- PLA
proximity ligation assay
- SIM
structured illumination microscopy
- SR
spectrin repeat
- WT
wild type