The mitotic segregation apparatus composed of microtubules and chromatin functions to faithfully partition a duplicated genome into two daughter cells. Microtubules exert extensional pulling force on sister chromatids toward opposite poles, whereas pericentric chromatin resists with contractile springlike properties. Tension generated from these opposing forces silences the spindle checkpoint to ensure accurate chromosome segregation. It is unknown how the cell senses tension across multiple microtubule attachment sites, considering the stochastic dynamics of microtubule growth and shortening. In budding yeast, there is one microtubule attachment site per chromosome. By labeling several chromosomes, we find that pericentromeres display coordinated motion and stretching in metaphase. The pericentromeres of different chromosomes exhibit physical linkage dependent on centromere function and structural maintenance of chromosomes complexes. Coordinated motion is dependent on condensin and the kinesin motor Cin8, whereas coordinated stretching is dependent on pericentric cohesin and Cin8. Linking of pericentric chromatin through cohesin, condensin, and kinetochore microtubules functions to coordinate dynamics across multiple attachment sites.
Introduction
The pericentric chromatin along with cohesin and condensin are necessary for the function of a chromatin spring that contributes to faithful chromosome segregation (Ribeiro et al., 2009; Manning et al., 2010; Stephens et al., 2011). It is not known whether chromatin proximal to multiple attachment sites in the yeast spindle or a single mammalian kinetochore behave as separate springs or as one interlinked spring in metaphase.
The structure of point centromeres in Saccharomyces cerevisiae allows precise positioning of cytological labels (lactose operon [LacO]-LacI-GFP) relative to the microtubule attachment, not possible in organisms with regional centromeres. In metaphase, sister pericentric LacO arrays appear as two foci bioriented on the spindle axis (Goshima and Yanagida, 2000; He et al., 2000; Tanaka et al., 2000; Pearson et al., 2001). The arrays are dynamic and can transiently reassociate into one spot. Pericentromere LacO arrays stretch (∼10%) and decompact along the spindle axis (Bachant et al., 2002), and multiply labeled pericentromeres stretch coordinately (Stephens et al., 2013). However, the mechanisms responsible for coordinated stretching or the possibility of correlated motion are not known.
The pericentric chromatin (50 kb surrounding the centromere) is enriched threefold in cohesin and condensin (D’Ambrosio et al., 2008). Cohesin localizes distal to the spindle axis where it radially confines pericentric chromatin, whereas condensin is localized along the spindle axis where it axially compacts the pericentromere (Stephens et al., 2011). Using strains with multiple chromosome labels, we examine correlated chromatin motion and coordinated stretching between pericentromeres of different chromosomes. Our results suggest that a chromatin network comprised of cohesin and condensin exists across multiple microtubule attachment sites in the budding yeast mitotic spindle.
Results and discussion
Pericentromeres of different chromosomes display correlated movement in metaphase
To determine whether pericentromeres of different chromosomes move coordinately during metaphase, we imaged LacO and tetracycline operon (TetO) arrays linked to CEN15 and CEN11, respectively. Movement of each pericentromere was measured relative to its pole (Fig. 1 A, red). The movements of pericentromeres in the same half-spindle were compared using cross-correlation analysis. Correlation increased from G1 to metaphase (G1: 0.15 ± 0.33, n = 80; metaphase: 0.33 ± 0.34, n = 88; P < 0.001; Fig. 1 D). Similar results were found with a different set of labeled chromosomes (CEN3 and CEN11; Fig. S1 A). Thus, pericentromeres show metaphase-dependent correlated movement.
Pericentromeres of different chromosomes display correlated movement in metaphase dependent on condensin and Cin8. (A and B) Compact (foci) pericentromere movements were tracked relative to their respective spindle pole body (Spc29-RFP) in trans (CEN11 and CEN15; A)- and cis (B)-labeled strains (4.5 and 9.4 kb from CEN11). (C) Sister arrays were tracked relative to the midspindle. Each graph depicts a single representative time lapse from n = 50–100. (D) Cross-correlation analysis of trans-, cis-, and sister pericentromere movement in G1 and metaphase (M). t test values are listed above the bars. (E) Time courses were analyzed for which pericentromere label was closest to its respective pole at each time point (trans, n = 742 from 44 experiments; cis, n = 176 from 12 experiments). An equal probability to be closest to the pole suggests that pericentromere (cis) DNA can “flop” over itself (shown on the right). (F–H) Trans (F)-, cis (G)-, and sister (H) pericentromere–correlated motion for chromatin (GalH3, mcm21Δ, and brn1-9) and microtubule motor mutants (cin8Δ and kip1Δ). Asterisks denote significantly different cross-correlation from WT (t test, P < 0.05). Values are listed in Table 1 and Table S1. Error bars represent standard deviations.
Pericentromeres of different chromosomes display correlated movement in metaphase dependent on condensin and Cin8. (A and B) Compact (foci) pericentromere movements were tracked relative to their respective spindle pole body (Spc29-RFP) in trans (CEN11 and CEN15; A)- and cis (B)-labeled strains (4.5 and 9.4 kb from CEN11). (C) Sister arrays were tracked relative to the midspindle. Each graph depicts a single representative time lapse from n = 50–100. (D) Cross-correlation analysis of trans-, cis-, and sister pericentromere movement in G1 and metaphase (M). t test values are listed above the bars. (E) Time courses were analyzed for which pericentromere label was closest to its respective pole at each time point (trans, n = 742 from 44 experiments; cis, n = 176 from 12 experiments). An equal probability to be closest to the pole suggests that pericentromere (cis) DNA can “flop” over itself (shown on the right). (F–H) Trans (F)-, cis (G)-, and sister (H) pericentromere–correlated motion for chromatin (GalH3, mcm21Δ, and brn1-9) and microtubule motor mutants (cin8Δ and kip1Δ). Asterisks denote significantly different cross-correlation from WT (t test, P < 0.05). Values are listed in Table 1 and Table S1. Error bars represent standard deviations.
TetO and LacO arrays in a single pericentromere (cis) provide a measure of maximum correlation. Motion analysis of TetO and LacO arrays (centroids of arrays in CEN11 at 4.5 and 9.4 kb, respectively) revealed similar cross-correlation values for G1 and metaphase (G1: 0.40 ± 0.23, n = 71; metaphase: 0.30 ± 0.36, n = 24; P > 0.2; Fig. 1, B and D). Sister centromere movements, analyzed relative to the midspindle, have a comparable degree of cross-correlation (all metaphase 0.343 ± 0.33, n = 88; P > 0.5; Fig. 1, C and D). The uniformity of motion in metaphase, irrespective of trans-, cis-, or sister markers, is indicative of a multiply cross-linked network rather than independently regulated springs.
To explore the physical state of the chromatin, we examined the spatial position of cis-TetO and -LacO arrays in vivo. Imaging revealed that there is an equal chance for each cis-label to be closer to the pole (Fig. 1 E, Cis). This spatial relationship is similar for labels on different chromosomes as well (Fig. 1 E, Trans). The lack of correspondence in spatial versus physical position reflects the floppiness of the chromatin in vivo (Fig. 1 E, graphic). The apparently low cross-correlation value (∼0.3; Fig. 1 D) across the network is thus limited by the floppiness of the chromatin.
Correlated movement of pericentromeres is dependent on condensin and Cin8
We probed pericentromere dynamics in cohesin and condensin mutants. Correlated motion analysis allows comparison of the behavior of compact arrays. Depletion of condensin (brn1-9; Lavoie et al., 2000) resulted in a significant decrease in correlated movement of different pericentromeres (wild type [WT] = 0.33 to brn1-9 = 0.21 ± 0.35, n = 58; P < 0.05; Fig. 1 F). Deletion of Mcm21, a nonessential kinetochore protein, results in a threefold depletion of pericentric cohesin but maintains arm cohesin (Eckert et al., 2007; Ng et al., 2009). Unlike condensin, depletion of pericentric cohesin did not result in decreased correlated motion (mcm21Δ = 0.36 ± 0.31, n = 54; P > 0.05; Fig. 1 F). Correlated motions of LacO/TetO labels in the same pericentromere or sister pericentromere labels are not altered in either mutant compared with WT (P > 0.05; Fig. 1, G and H). Thus, condensin contributes to correlated motion between pericentromeres of different chromosomes.
Loss of correlated motion between different chromosomes (trans) upon condensin depletion could be caused by chromatin decompaction. Cells depleted of 50% nucleosomes through GalH3 repression (Bouck and Bloom, 2007) display similar pericentromere decompaction as brn1-9 cells (twofold decompaction; Stephens et al., 2011). Trans-correlated motion in GalH3-repressed cells does not decrease relative to WT (0.27 ± 0.35, n = 120; P = 0.15; Fig. 1 F). Interestingly, sister pericentromere–correlated movement is dependent on nucleosome compaction and not on cohesin or condensin (WT = 0.34 vs. GalH3 = 0.24 ± 0.33, n = 120; P = 0.04; Fig. 1 H). The ability to dissect differential mechanisms including nucleosome compaction (histone) versus chromatin compaction (condensin) indicates the complexity of organization within this chromatin network.
Coordinated dynamics may also reflect cross-links in the kinetochore or kinetochore microtubule (kMT). The kinesin 5 motor Cin8 contributes to kinetochore clustering (Tytell and Sorger, 2006; Gardner et al., 2008). To deduce the role of Cin8 in pericentromere dynamics, we tracked pericentromere motion in cin8Δ cells. The correlated movement of trans-labels was reduced in cin8Δ (0.23 ± 0.35, n = 96; P < 0.05; Fig. 1 F). Deletion of KIP1 had an insignificant effect on correlated movement (kip1Δ = 0.29 ± 0.33, n = 36; P > 0.05; Fig. 1 F). Cross-linking of adjacent kMTs would lead to coordinated motion through biasing stochastic microtubule dynamics from individual entities to an ensemble.
Coordinated stretching of pericentromeres
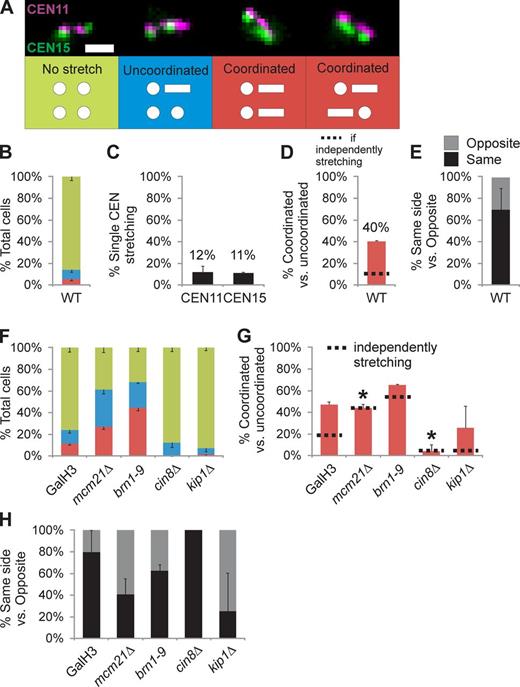
LacO arrays in the pericentromere are observed as compact foci or decompacted linear filaments reflecting the pericentromere chromatin response to force (Bachant et al., 2002; Stephens et al., 2011). Using pericentromeres labeled in trans (CEN11 and CEN15), we investigated the occurrence of coordinated stretching (Fig. 2, A–E). Each pericentromere LacO/TetO displayed similar stretching (CEN11, 12%; CEN15, 11%, n = 267; Fig. 2 C). In cells with a stretched pericentromere, 40% ± 1% of cells exhibit a second stretched pericentromere (n = 37; Fig. 2 D), significantly more than predicted by independent probabilities (11% single stretching dotted line; χ2 < 1 × 10−8; Fig. 2 D). Of the coordinated stretching, events greater than 2/3 displayed stretching on the same side of the spindle (70% same side; Fig. 2 E). Furthermore, both coordinated stretching and bias for the same side of the spindle are reproducible for CEN11 and CEN3 (Fig. S1, B–E). Correlated motion and stretching dynamics between multiple pairs of pericentromeres of different chromosomes is indicative of a cross-linked network.
Coordinated stretching of different pericentromeres is pericentric cohesin and Cin8 dependent. (A and B) Trans-labeled pericentromeres were categorized as no stretch (both arrays compact foci), uncoordinated (stretching in only one of the labeled CEN arrays), or coordinated (both CEN nonsister arrays display stretching). Bar, 1 µm. (C) WT pericentromere stretching frequency for each CEN11 and CEN15. (D) Coordinated stretching occurs in 40 ± 1% of cells that show stretching, higher than predicted by independent stretching frequencies (11% dotted line). (E) Graph of coordinated stretching events that occur on the same or opposite side of the spindle. (F–H) Graphs of categorized stretching (F), coordinated stretching (G), and same versus opposite side coordinated stretching (H) for chromatin (GalH3, mcm21Δ, and brn1-9) and microtubule motor mutants (cin8Δ and kip1Δ). Asterisks denote mutants in which single pericentromere stretching (black lines) and coordinated stretching frequency (red bars) are statistically similar (G, χ2 > 0.4), and thus, stretching is independent. Values are listed in Table 1 and Table S1. Error bars represent standard deviations.
Coordinated stretching of different pericentromeres is pericentric cohesin and Cin8 dependent. (A and B) Trans-labeled pericentromeres were categorized as no stretch (both arrays compact foci), uncoordinated (stretching in only one of the labeled CEN arrays), or coordinated (both CEN nonsister arrays display stretching). Bar, 1 µm. (C) WT pericentromere stretching frequency for each CEN11 and CEN15. (D) Coordinated stretching occurs in 40 ± 1% of cells that show stretching, higher than predicted by independent stretching frequencies (11% dotted line). (E) Graph of coordinated stretching events that occur on the same or opposite side of the spindle. (F–H) Graphs of categorized stretching (F), coordinated stretching (G), and same versus opposite side coordinated stretching (H) for chromatin (GalH3, mcm21Δ, and brn1-9) and microtubule motor mutants (cin8Δ and kip1Δ). Asterisks denote mutants in which single pericentromere stretching (black lines) and coordinated stretching frequency (red bars) are statistically similar (G, χ2 > 0.4), and thus, stretching is independent. Values are listed in Table 1 and Table S1. Error bars represent standard deviations.
Pericentric cohesin and Cin8 coordinate pericentromere stretching of different chromosomes
To determine the components responsible for coordinated stretching, we depleted cells of pericentric cohesin or condensin. Depletion of either results in increased single pericentromere stretching 45–55% (Fig. 2, F and G, black lines; Stephens et al., 2011). Coordinated stretching matches single pericentromere stretching upon depletion of pericentric cohesin (mcm21Δ: single 44% vs. coordinated 44%, χ2 = 1; Fig. 2 G, Table 1, and Table S1). In contrast, condensin mutants maintain a greater than expected coordinated stretching relative to single stretching, similar to WT (brn1-9: single 56% vs. coordinated 65%; χ2 < 0.001; Fig. 2 G and Table S1). If pericentric cohesin is responsible for cross-linking adjacent pericentromeres, coordinated stretching should be reduced on the same side of the spindle. Coordinated stretching on the same side of the spindle decreases in mcm21Δ (41% ± 14%, n = 93) but remains similar to WT in brn1-9 (63% ± 14%, n = 115; Fig. 2 H). Therefore, cohesin is more likely to cross-link pericentromeres in the half-spindle, and upon its depletion, pericentromeres stretch independently.
Summary of WT and mutant correlated motion, coordinated stretching, and interaction between pericentromeres of different chromosomes
| Strain | Trans-correlated movement mean (R) | Single pericentromere stretching mean | Trans-coordinated stretching mean | Pericentromere interaction index mean (4C) |
| % | % | |||
| WT | 0.33 | 11 | 40 | 1.75 |
| GalH3 | 0.27 | 19 | 47 | NA |
| mcm21Δ | 0.36 | 44 | 44* | 1.28** |
| brn1-9 | 0.21* | 56 | 65 | 1.44* |
| cin8Δ | 0.23* | 6 | 4* | 1.66 |
| kip1Δ | 0.29 | 4 | 26 | NA |
| Strain | Trans-correlated movement mean (R) | Single pericentromere stretching mean | Trans-coordinated stretching mean | Pericentromere interaction index mean (4C) |
| % | % | |||
| WT | 0.33 | 11 | 40 | 1.75 |
| GalH3 | 0.27 | 19 | 47 | NA |
| mcm21Δ | 0.36 | 44 | 44* | 1.28** |
| brn1-9 | 0.21* | 56 | 65 | 1.44* |
| cin8Δ | 0.23* | 6 | 4* | 1.66 |
| kip1Δ | 0.29 | 4 | 26 | NA |
Single and double asterisks denote statistically significant change from WT; mutants with different number of asterisks are statistically different. R, mean cross-correlation value; NA, not applicable.
Nucleosome repression via GalH3 results in longer spindle lengths comparable to depletion of pericentric cohesin and condensin (mean spindle length of 2–2.5 µm; Stephens et al., 2011). Single pericentromere stretching increases approximately twofold to 19% in GalH3-repressed cells (n = 233; Fig. 2 G, black lines). GalH3-repressed cells display 47 ± 3% coordinated stretching in cells that stretched (n = 46; Fig. 2 G). Thus, coordinated stretching is not simply a result of chromatin extension; rather, it is dependent on specific cross-linkers such as cohesin (mcm21Δ).
Cells deleted of either CIN8 or KIP1 have shorter spindles (Saunders and Hoyt, 1992), reflecting fewer motors binding to and sliding interpolar microtubules apart. The shorter spindles result in less single stretching of the pericentric chromatin (CEN11 and CEN15, 4–6%; Fig. 2, F and G, black lines). Coordinated and single pericentromere stretching frequency are similar in cin8Δ cells (6% single vs. 4% coordinated; χ2 = 0.46; Fig. 2 G). Oppositely, kip1Δ cells maintained a higher frequency of trans-coordinated stretching compared with single stretching (single 4% vs. coordinated 26%; χ2 < 0.001; Fig. 2 G). Pericentromeres stretch independently in cin8Δ cells, whereas kip1Δ cells display WT-dependent/linked behavior.
Simulations of cross-linking pericentromeres of different chromosomes recapitulate correlated movement and coordinated stretching
We used a mathematical model of the yeast spindle to query the extent that chromatin cross-links could increase correlated motion and stretching in the spindle (Stephens et al., 2013). Addition of cross-linking springs between pericentromeres and their two adjacent neighbors fractionally increases the cross-correlation of kMT plus ends (Fig. 3, A and B, blue). A network in which all pericentromeres were cross-linked to each other significantly increases correlated motion (Fig. 3, A and B, red). Interestingly, simulation of either type of cross-link leads to increased coordinated stretching that matches levels measured in WT cells (Fig. 3 C). Thus, chromatin-based cross-linking of all pericentromeres provides a mechanism for correlated movement and stretching observed in vivo.
Simulation of cross-linking springs between pericentromeres recapitulates correlated movement and stretching. A mathematical model of spindle length force balance, including kMT dynamics and a nonlinear spring, was used to simulate the results of adding cross-linking springs between pericentromeres (Stephens et al., 2013). (A) Springs were added to cross-link neighbors or all pericentromeres into a network. (B and C) Graphs show cross-correlation of kMT plus-end movements (B) or coordinated stretching (C) upon increasing the cross-linking spring constant (kcross) relative to the pericentromere spring constant (kpericentromere; n = 500). All simulations had a 12 ± 2% single pericentromere stretching frequency (black line) similar to in vivo WT. Error bars represent standard deviations.
Simulation of cross-linking springs between pericentromeres recapitulates correlated movement and stretching. A mathematical model of spindle length force balance, including kMT dynamics and a nonlinear spring, was used to simulate the results of adding cross-linking springs between pericentromeres (Stephens et al., 2013). (A) Springs were added to cross-link neighbors or all pericentromeres into a network. (B and C) Graphs show cross-correlation of kMT plus-end movements (B) or coordinated stretching (C) upon increasing the cross-linking spring constant (kcross) relative to the pericentromere spring constant (kpericentromere; n = 500). All simulations had a 12 ± 2% single pericentromere stretching frequency (black line) similar to in vivo WT. Error bars represent standard deviations.
Cohesin and condensin promote physical interaction between pericentromeres
To determine whether pericentromeres are in physical proximity in metaphase, we adapted the 3C (chromosome conformation capture) technique to probe the interaction between two loci on different chromosomes. Inverse primer pairs were used to map the interaction of chromosome 3 and 5 at arm and pericentromere loci (see Materials and methods). Arm loci were used as a control for random interactions between chromosomes 3 and 5. We found that WT pericentromeres interact 1.75 ± 0.05–fold more than random arm interaction (pericentromere/arm, normalized to arm 1.00, n = 10; Fig. 4 A). This recapitulates initial findings of interpericentromere interactions via 3C techniques (cross-linking frequency 1.5; Dekker et al., 2002).
4C reveals interactions between pericentromeres of different chromosomes. (A) Primers were used to assay interactions, via 3C technique, between the pericentromeres and arms of chromosome III and V (see Materials and methods). The interaction index is the ratio of the pericentromere (P) to the arm (A; control for random interactions) PCR product was normalized to 1. Pericentromere interaction index is shown for WT, cin8Δ, brn1-9, mcm21Δ, and GalCEN3 (conditionally off centromere). Values listed in Table 1. Error bars represent standard deviations. Asterisks denote significant difference from WT (P < 0.01). Mutants with a different number of asterisks are significantly different, whereas those with the same numbers are similar. (B) Diagram depicting the half-spindle and a results summary for WT and mutants. kMTs emanate from the spindle pole each bound to a different chromosome at the kinetochore/centromere, whereas interpericentromere interaction is facilitated by a cross-linker. Results of 4C suggest cohesin and condensin act as a cross-linker between different pericentromeres. Plus signs, statistically similar to WT; minus, decreased significantly relative to WT; double minus, decreased significantly relative to single minus; N/A, not applicable.
4C reveals interactions between pericentromeres of different chromosomes. (A) Primers were used to assay interactions, via 3C technique, between the pericentromeres and arms of chromosome III and V (see Materials and methods). The interaction index is the ratio of the pericentromere (P) to the arm (A; control for random interactions) PCR product was normalized to 1. Pericentromere interaction index is shown for WT, cin8Δ, brn1-9, mcm21Δ, and GalCEN3 (conditionally off centromere). Values listed in Table 1. Error bars represent standard deviations. Asterisks denote significant difference from WT (P < 0.01). Mutants with a different number of asterisks are significantly different, whereas those with the same numbers are similar. (B) Diagram depicting the half-spindle and a results summary for WT and mutants. kMTs emanate from the spindle pole each bound to a different chromosome at the kinetochore/centromere, whereas interpericentromere interaction is facilitated by a cross-linker. Results of 4C suggest cohesin and condensin act as a cross-linker between different pericentromeres. Plus signs, statistically similar to WT; minus, decreased significantly relative to WT; double minus, decreased significantly relative to single minus; N/A, not applicable.
The basis for physical interaction could reflect centromere clustering and/or protein-mediated pericentromere interaction. We disrupted the centromere of chromosome III using a conditionally functional centromere (GalCEN3; Hill and Bloom, 1987). Visualization of GalCEN3 LacO/LacI revealed that it becomes unattached from the spindle and does not localize with the cluster of attached centromeres (Fig. S2 and depicted in Fig. 4 B). The intermolecular interaction of pericentromeres is dependent on a functional centromere (GalCEN3 = 1.21 ± 0.02, n = 2; P < 1 × 10−7; Fig. 4 A), resulting in a decreased interaction index close to levels seen by random arm interactions (1.00). Thus, one mechanism for increased interpericentromere physical interaction is the clustering of centromeres attached to kMTs.
Physical interaction between different pericentromeres could also be mediated via chromatin components. Chromatin spring components condensin and cohesin both have the capacity to embrace different chromatin strands (Gruber et al., 2003; Haering et al., 2008; Surcel et al., 2008; Cuylen et al., 2011). Depletion of condensin yields a decrease in pericentromere interaction to 1.44 ± 0.05 (n = 10, P < 0.001; Fig. 4 A). Depletion of pericentric cohesin resulted in a decrease of 4C (chromosome to chromosome conformation capture) interaction to 1.28, similar to GalCEN3 (mcm21Δ = 1.28 ± 0.05, n = 10; P > 0.05; vs. GalCEN3 = 1.21; Fig. 4 A). Cells deleted of CIN8 display kinetochore declustering as well as decreased coordinated interpericentromere dynamics similar to cohesin and condensin mutants (Fig. 1, Fig. 2, and Fig. S3). However, pericentromere interaction via 4C does not significantly decrease in cin8Δ cells (1.66 vs. WT = 1.75, n = 10; P > 0.01; Fig. 4 A). Therefore, decreased interaction between pericentromeres is neither caused by abnormal spindle structure nor altered interpericentromere dynamics (Fig. 4 B). The physical interactions between pericentromeres are dependent on centromere attachment to kMTs as well as condensin- and cohesin-based interpericentromere linkages.
Function of a cross-linked network
What function would a multimicrotubule attachment site gain from cross-links between adjacent microtubules and a cross-linked chromatin network? In a cross-linked network, a single attachment will bias the remaining chromosomes in the same direction. In yeast, pericentric cohesin promotes biorientation (Ng et al., 2009; Sakuno and Watanabe, 2009). Correct orientation is essential to preventing merotelic attachments in mammalian cells in which 16–20 attachments must be bound to kMTs from the same side of the spindle. Mammalian cells depleted of retinoblastoma (pRB) result in pericentromere cohesin and condensin depletion and ensuing merotelic attachments and chromosome missegregation (Manning et al., 2010). Likewise, loss of condensin results in misattached/merotelic chromosomes (Samoshkin et al., 2009; Tada et al., 2011). A second consequence of cross-links is that they could stabilize attachments through a rigid spring or through sharing tension across the network. The addition of cross-links between attachment sites in a mathematical model promoted a stronger spring to the same degree as strengthening individual springs (Stephens et al., 2013). Alternatively, distributing tension through a cross-linked network provides a mechanism to dampen fluctuations resulting from stochastically growing and shortening microtubules. Thus, chromatin cross-links likely serve essential functions in orientation and proper tension-based stabilization of multiple attachment sites in both yeast and mammals.
The resistive properties of the spring likely come from compaction and cross-linking of pericentromeres through condensin and cohesin (Fig. 5; Guacci et al., 1997; Lavoie et al., 2002, 2004; Lam et al., 2006; Heidinger-Pauli et al., 2010; Cuylen et al., 2011; Stephens et al., 2011, 2013). Condensin-dependent chromatin compaction is also critical for tension-sensing mechanisms (Yong-Gonzalez et al., 2007; Uchida et al., 2009). The segregation apparatus allows for a variable number of microtubule attachments by generating an interlinked network in the chromatin, critical for orienting and maintaining bioriented attachments and the kinetochore under tension.
Model of cross-linking in the metaphase spindle apparatus. (A) Diagram of WT metaphase spindle structure and interactions. kMTs (light green) emanating from the spindle pole (dark green) are cross-linked via the kinesin 5 motor Cin8, whereas multiple nonlinear (looped) pericentric chromatin springs (CEN15 and CEN11) are cross-liked via condensin at the base and cohesin radially displaced (Stephens et al., 2011). Cross-links could occur through homotypic interaction or single complex (see Thadani et al. [2012] for condensin and Haering and Jessberger [2012] for cohesin). (B) Condensin functions as an axial cross-linker between compact pericentromeres of different chromosomes to correlate their movement during metaphase. Alternatively, condensin’s contributions to compaction, spring constant, or interactions with topoisomerase II could affect correlated motion. (C) Loss of condensin cross-links results in decreased correlated motion (smaller arrows; Fig. 1 F). (D) Cohesin functions primarily as a distal cross-linker between pericentromeres, resulting in coordinated stretching (double arrows). (E) Loss of pericentric cohesin results in pericentromeres stretching independently (one stretched and one compact; Fig. 2 G).
Model of cross-linking in the metaphase spindle apparatus. (A) Diagram of WT metaphase spindle structure and interactions. kMTs (light green) emanating from the spindle pole (dark green) are cross-linked via the kinesin 5 motor Cin8, whereas multiple nonlinear (looped) pericentric chromatin springs (CEN15 and CEN11) are cross-liked via condensin at the base and cohesin radially displaced (Stephens et al., 2011). Cross-links could occur through homotypic interaction or single complex (see Thadani et al. [2012] for condensin and Haering and Jessberger [2012] for cohesin). (B) Condensin functions as an axial cross-linker between compact pericentromeres of different chromosomes to correlate their movement during metaphase. Alternatively, condensin’s contributions to compaction, spring constant, or interactions with topoisomerase II could affect correlated motion. (C) Loss of condensin cross-links results in decreased correlated motion (smaller arrows; Fig. 1 F). (D) Cohesin functions primarily as a distal cross-linker between pericentromeres, resulting in coordinated stretching (double arrows). (E) Loss of pericentric cohesin results in pericentromeres stretching independently (one stretched and one compact; Fig. 2 G).
Materials and methods
Strain building
To build a strain with pericentric regions labeled on two different chromosomes, we incorporated a 8-kb TetO array 0.5 kb away from CEN11 (at the met14 locus), using the plasmid protocol from the Gasser laboratory (Rohner et al., 2008), into a strain containing a 10-kb LacO array tagged with LacI-GFP 1.8 kb from CEN15 (Goshima and Yanagida, 2000). A target fragment with homology to the met14 locus was transformed into the 1.8-kb LacO strain. The plasmid pSR14 (TetO) with homology to the target fragment was transformed into the strain. We then transformed pDB49 (tetracycline repressor [TetR]-CFP) into the stain to visualize the TetO. Similarly, a strain was created by inserting the 8-kb TetO array at 0.5 kb from CEN11 directed into a strain containing a 10-kb LacO array 3.8 kb from CEN3 (Goshima and Yanagida, 2000). A strain with two arrays on the same chromosome (TetO array 4.5 kb from CEN11 and a LacO array 9.5 kb from CEN11) was constructed by inserting a 8-kb TetO array centromere proximal to a 1.7-kb LacO array (Pearson et al., 2001) using the same protocol.
Cell preparation
WT, cin8Δ, kip1Δ, and mcm21Δ strains were grown at 24°C in synthetic defined–His. Temperature-sensitive allele brn1-9 strains were grown at 24°C and then transferred to restrictive temperatures (37°C) for 3 h before imaging. Cells were grown to log phase as asynchronous cultures and then prepared for imaging. GalH3 strains were α-factor arrested in YPG (yeast/peptone/galactose; 2% galactose), washed, and then released into YPD (yeast/peptone/glucose; 2% glucose) for 3–4 h before viewing, as outlined in Bouck and Bloom (2007).
Microscopy
Images were obtained using a microscope stand (Eclipse TE2000-U; Nikon) with a 100× Plan Apochromat, 1.4 NA digital interference contrast oil immersion lens with a camera (ORCA-ER; Hamamatsu Photonics) at 25°C. Images were acquired using MetaMorph 7.1 (Molecular Devices) and were binned 2 × 2 (pixel size of 130 nm). Images were taken in water on 0.135-mm coverslips. Time-lapse images were obtained in a single z plane at 15- and 30-s intervals with exposure times of 600 ms for CFP, 600 ms for YFP-green filter, 800 ms for RFP, and 250 ms for trans-images. Population images were obtained in z series stacks of 10 images with a step size of 200 nm and similar exposure times as time-lapse images.
Cross-correlation analysis of pericentromere movement
Time-lapse images of the CEN15/CEN11 strain were rotated and aligned relative to the spindle axis using MATLAB (MathWorks, Inc.). Aligned images were used to analyze foci movement relative to the spindle axis (x axis), eliminating movement perpendicular to the spindle (y axis). Correlation was determined in cells with two separated foci for both LacO and TetO arrays. The distance of the foci to their respective pole was measured using MetaMorph 7.1 and logged into Excel (Microsoft) in which cross-correlation analysis was performed using the CORREL function. Cells displaying both arrays separated and maintaining a constant spindle length over the time lapse were considered metaphase.
Coordination of stretching
Pericentromere LacO/TetO array stretching was analyzed in metaphase cells. Cells were considered metaphase if both LacO and TetO arrays displayed separated sisters and spindles did not exhibit anaphase-like linear increases in spindle length. Stretching events were determined as cells with one focus and another fluorescent signal that is linear along the spindle axis (Stephens et al., 2011, 2013). Stretching events are determined by measuring the Gaussian of the fluorescence signal parallel and perpendicular to the spindle axis. Compact pericentromeres will appear as a focus and have an aspect ratio of <1.2 (parallel/perpendicular). Stretched arrays appear as lines and have an aspect ratio of >1.2.
Simulations of movement and stretching
Simulations were run in MATLAB/Simulink (MathWorks, Inc.) using a mathematical model of mitotic force balance in the yeast spindle, including kMT dynamics and a nonlinear spring (Stephens et al., 2013). Cross-links were added to the models as follows: For neighbors, the difference in a pericentromere spring length (L) compared with its two adjacent spring (±1) lengths was calculated, converted into force, and added to pericentromere spring force: Ftotal = −kspring(Li − Lrest) + kcross-link[[Li − (Li + 1)] + [Li − (Li − 1)]]. For a network, the difference between each spring (n) and the other 15 was calculated, converted into force, and added to pericentromere spring force: Ftotal = −kspring(Li − Lrest)+ kcross-link[Σ[Li – (Lj)]], for j ≠ i.
Simulated cross-correlation of kMTs does not match experimentally measured absolute values of chromatin. The movement of the chromatin polymer is not specified in the simulation. A threshold determines the state of a piecewise continuous spring. Below the threshold, a compact spring has a high spring constant, and above the threshold, the stretched spring has a lower spring constant. The cross-linking spring constant (kcross) was increased relative to the pericentromere spring constant (kpericentromere), which was fixed.
4C
Yeast nuclei were prepared and cross-linked with 1% formaldehyde for 10 min at room temperature. The reaction was quenched with the addition of glycine to 0.25 M. Nuclei were washed and resuspended in appropriate restriction digest buffer. 1% SDS was added, and the nuclei were incubated at 65°C for 10 min to remove un–cross-linked proteins. Triton X-100 was added to a final concentration of 1% to remove the SDS and allow for subsequent digestion. 60 U of the restriction enzyme XbaI was added, and the reaction was incubated overnight at 37°C. 10% SDS was added to each tube and incubated at 65°C for 20 min to inactivate XbaI. 800 Weiss units of T4 DNA ligase were added, and the reaction was incubated at 16°C for 2 h to ligate cross-linked DNA. Cross-links were then reversed with the addition of proteinase K and overnight incubation at 65°C. DNA was purified by phenolchloroform extraction and ethanol precipitation. DNA concentration was determined by running of 1% agarose gels and staining with ethidium bromide. All gels were imaged with an imaging system (AlphaImager 2200; Alpha Innotech), and all images were imported into MetaMorph 6.1 for analysis. Gels were analyzed by measurement of the integrated intensity of an area of 5 × 5– and 6 × 6–pixel computer generated boxes centered on each band. The difference in integrated intensity was used to determine the mean background fluorescence per pixel. Integrated intensity was then corrected for background by subtracting the background fluorescence over the 5 × 5 area as described in Joglekar et al. (2006) and Yeh et al. (2008).
Titration PCRs were performed with increasing amounts of input DNA. Input DNA volumes that yielded PCR products that were within the linear range of PCR amplification were then used for 4C analysis. The cross-linking frequencies of regions between chromosomes III and V were compared in the arm and the pericentromere. The centromere 3 primer (pericentromere down) 1,422 bp downstream of CEN3 was paired with a centromere 5 primer (pericentromere up) 1,913 bp upstream of CEN5. The arm region was probed with a chromosome 3 primer (arm down) 75,639 bp from CEN3 and a chromosome 5 primer (arm up) 98,424 bp from CEN5. Ligation products from these regions are detected by PCR, yielding products ∼500–700 bp in size (representing the distance of each primer to the XbaI site). Nonspecific PCR products were not generated in any of the experiments. PCR products from cross-linked DNA were compared with identical products generated from control DNA, which was not cross-linked, allowing all possible ligation products to occur. Analysis of the resultant PCR products showed a mean 75% increase in PCR product for the pericentric region as compared with the region along the arm, indicating a statistically significant increase in physical interaction of the genome at pericentric chromatin versus random in the arm.
Kinetochore declustering
Population images were acquired of strains containing a kinetochore marker (Nuf2, Ndc80, or Ame1) with labeled spindle poles (Spc29). MetaMorph line scans were drawn along the spindle axis (axial) or perpendicular (radial) through the kinetochores to determine whether each sister kinetochore structure remains clustered as one peak or declustered into multiple peaks as outlined in Bouck and Bloom (2007).
Online supplemental material
Fig. S1 shows that a second set of pericentromeres from different chromosomes (LacO 3.8 kb from CEN3 and TetO 0.5 kb from CEN11) display correlated motion and coordinated stretching. Fig. S2 shows that an inactivated centromere loses attachment to the spindle. Fig. S3 shows that pericentric cohesin, condensin, and Cin8 mutants display kinetochore declustering. Table S1 is a comprehensive summary of all coordinated motion and stretching measurements.
Acknowledgments
We thank Will Lewis (Vanderbilt University) for preliminary work to optimize the TetO/TetR insertion protocol. We thank Leandra Vicci, Russell M. Taylor II (University of North Carolina Department of Computer Science), and Michael R. Falvo (University of North Carolina Department of Physics and Astronomy) for insightful discussion. We thank Jolien S. Verdaasdonk and other members of the Bloom laboratory for advice, assistance, and critical readings of the manuscript.
This work was funded by the National Institutes of Health R37 grant GM32238 (K. Bloom).





![Figure 5. Model of cross-linking in the metaphase spindle apparatus. (A) Diagram of WT metaphase spindle structure and interactions. kMTs (light green) emanating from the spindle pole (dark green) are cross-linked via the kinesin 5 motor Cin8, whereas multiple nonlinear (looped) pericentric chromatin springs (CEN15 and CEN11) are cross-liked via condensin at the base and cohesin radially displaced (Stephens et al., 2011). Cross-links could occur through homotypic interaction or single complex (see Thadani et al. [2012] for condensin and Haering and Jessberger [2012] for cohesin). (B) Condensin functions as an axial cross-linker between compact pericentromeres of different chromosomes to correlate their movement during metaphase. Alternatively, condensin’s contributions to compaction, spring constant, or interactions with topoisomerase II could affect correlated motion. (C) Loss of condensin cross-links results in decreased correlated motion (smaller arrows; Fig. 1 F). (D) Cohesin functions primarily as a distal cross-linker between pericentromeres, resulting in coordinated stretching (double arrows). (E) Loss of pericentric cohesin results in pericentromeres stretching independently (one stretched and one compact; Fig. 2 G).](https://cdn.rupress.org/rup/content_public/journal/jcb/203/3/10.1083_jcb.201307104/5/m_jcb_201307104_fig5.jpeg?Expires=1773507971&Signature=moWPRGH9pwVZ2DePQA0AqadEAjNjib-6jqhuQqmitwDEtx~auCy6LENvdNdT4SHIP4usFYd2ooqweHeQCA4g4n4PA1Mt3898EdiOko91JA1jRFyzUQqi7~pOcyIXt8FrX4ydgf6-qaDLno4I5fbMmTdZrBEq-InDJi8uxhYYfVXpi1XohTqZmNW4m7fXBtdzuv4qdwhOTmN2KvjkpMK6YWdJXi9WiJFM27Ud9RZ7lNwjNO1XaYNAjEBuLFrWd9LcxlV7-3cpCYiRhlCc7LK4ZcS~vwRbNpjDkXNmSv8bgD0swnbUcXc~VfyaVpuQY89nY26GhrVrCUBDqnrWdw8RxQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


