The central spindle is built during anaphase by coupling antiparallel microtubules (MTs) at a central overlap zone, which provides a signaling scaffold for the regulation of cytokinesis. The mechanisms underlying central spindle morphogenesis are still poorly understood. In this paper, we show that the MT depolymerase Kif2A controls the length and alignment of central spindle MTs through depolymerization at their minus ends. The distribution of Kif2A was limited to the distal ends of the central spindle through Aurora B–dependent phosphorylation and exclusion from the spindle midzone. Overactivation or inhibition of Kif2A affected interchromosomal MT length and disorganized the central spindle, resulting in uncoordinated cell division. Experimental data and model simulations suggest that the steady-state length of the central spindle and its symmetric position between segregating chromosomes are predominantly determined by the Aurora B activity gradient. On the basis of these results, we propose a robust self-organization mechanism for central spindle formation.
Introduction
The central spindle is a microtubule (MT)-based apparatus that is formed between segregating chromosomes and acts as a critical regulator of cytokinesis. It has bipolar organization in which MT bundles of uniform length run parallel to the division axis and interdigitate at the midzone to form antiparallel MT overlaps. The stem body is an electron-dense structure observed with EM at the MT overlapping region (McIntosh and Landis, 1971; Mastronarde et al., 1993). Stem bodies align precisely at the cell equatorial plane and provide scaffolds for the signaling molecules required for contractile ring formation and abscission, which enables a cell to divide at its center (Douglas and Mishima, 2010; Fededa and Gerlich, 2012). Previous studies have revealed that a substantial portion of central spindle MTs is generated de novo at the interchromosomal area during chromosome segregation (Uehara and Goshima, 2010). These MTs, together with MTs that have distinct origins (e.g., centrosomes), become bundled through temporally regulated antiparallel MT-bundling activities (Mishima et al., 2004; Hu et al., 2012). However, the mechanism by which the elaborate central spindle structure is assembled and maintained in between segregating chromosomes is largely unknown.
Phosphoregulation of MT-associated proteins plays an important role in the spatiotemporal organization of the interchromosomal MTs (icMTs). Cdk1-cyclin or Plk1 activity prevents precocious central spindle formation through the inactivation of central spindle-associated factors, such as MKLP1/Kif23 or PRC1, respectively, which enables timely assembly of the central spindle upon anaphase onset (Mishima et al., 2004; Goshima and Vale, 2005; Hu et al., 2012). Aurora B, another crucial mitotic kinase, is involved in cytoskeletal reorganization throughout mitosis as a subunit of the chromosomal passenger complex (CPC). During anaphase, Aurora B forms a characteristic activity gradient that peaks at the central spindle midzone and spreads throughout the interchromosomal area (Fuller et al., 2008; Tan and Kapoor, 2011). Several proteins critical for central spindle formation and/or cytokinesis, such as MKLP1 or RacGAP1, have been identified as targets of Aurora B–mediated phosphoregulation (Carmena et al., 2012; van der Waal et al., 2012). However, these proteins are closely localized to the Aurora B–enriched region (i.e., the midzone). Therefore, the physiological relevance of the Aurora B activity gradient, which spans several micrometers from the midzone, remains unclear.
In this study, we found that MT length control mediated by Kif2A, an MT-depolymerizing kinesin, is key to icMT organization during central spindle formation. Up- or down-regulation of Kif2A affected spindle size and stem body alignment. Our experimental results as well as mathematical modeling support a model in which the Aurora B activity gradient determines the spatial distribution of Kif2A and limits Kif2A-mediated depolymerization in an icMT length-dependent fashion. We propose that MT length control mediated by Aurora B and Kif2A ensures robust self-organization of the characteristic icMT structure, which is important for faithful cell division.
Results
Kif2A-mediated control of MT length is required for proper organization of the central spindle
To investigate mechanisms underlying central spindle formation, we first tested the impact of inhibiting MT dynamics. To monitor the organization of the central spindle, we visualized stem bodies using RacGAP1-GFP. Colocalization of RacGAP1-GFP with the stem body was verified using correlative light microscopy and EM (Fig. S1, A–C). In untreated cells, stem bodies aligned at the equatorial plane within a few minutes after anaphase onset (Fig. S1 D). During cleavage furrow ingression, stem bodies remained aligned at the equator and gradually assembled into a single cluster at the intercellular bridge. However, when cells were acutely treated with 4 µM taxol at anaphase onset, RacGAP1-GFP signals were scattered along the spindle axis, particularly during furrow ingression (Fig. 1, A and B). Thus, dynamic MTs are required for stem body alignment and proper central spindle organization.
Kif2A is required for icMT sizing and organization. (A) Cells expressing RacGAP1-GFP were treated with DMSO (control) or 4 µM taxol for 5–10 min before image acquisition. (B) Frequency of stem body misalignment in DMSO- or taxol-treated cells. The mean ± SE of three independent experiments is shown (≥34 cells were analyzed for each treatment). *, P < 0.01, t test. (C) RacGAP1-GFP dynamics in control (luciferase RNAi) or Kif2A-depleted cells. (middle) Kif2A depletion resulted in stem body misalignment (red arrow) and broadening of the stem body cluster at early telophase (18 min). (bottom) Global misalignment of stem bodies resulted in formation of the perpendicularly tilted division plane. Some of the misaligned stem bodies were separated from the others during telophase (yellow arrowheads). Broken lines indicate cell boundaries. Note that the cell itself did not rotate during division plane rotation. (D) Frequency of stem body misalignment in control or kinesin-depleted cells. The mean ± SE of four (luciferase RNAi) or three (others) independent experiments is shown (≥26 cells were analyzed for each treatment). *, P < 0.01, t test; **, P < 10−4. (E) Immunostaining of MTs and RacGAP1 in control or Kif2A-depleted cells. A buckled MT bundle associated with a misaligned stem body (pink arrow) is highlighted with white arrowheads. (F) Ratio of MT bundle length to chromosome-to-chromosome distance in control or Kif2A-depleted cells. For Kif2A RNAi samples, the MT bundles associated with aligned or misaligned stem bodies were separately analyzed. The mean ratio (±SE) of ≥15 pairs of MT bundles in at least six cells from two independent experiments is shown. *, P < 10−7, t test. Bars, 5 µm.
Kif2A is required for icMT sizing and organization. (A) Cells expressing RacGAP1-GFP were treated with DMSO (control) or 4 µM taxol for 5–10 min before image acquisition. (B) Frequency of stem body misalignment in DMSO- or taxol-treated cells. The mean ± SE of three independent experiments is shown (≥34 cells were analyzed for each treatment). *, P < 0.01, t test. (C) RacGAP1-GFP dynamics in control (luciferase RNAi) or Kif2A-depleted cells. (middle) Kif2A depletion resulted in stem body misalignment (red arrow) and broadening of the stem body cluster at early telophase (18 min). (bottom) Global misalignment of stem bodies resulted in formation of the perpendicularly tilted division plane. Some of the misaligned stem bodies were separated from the others during telophase (yellow arrowheads). Broken lines indicate cell boundaries. Note that the cell itself did not rotate during division plane rotation. (D) Frequency of stem body misalignment in control or kinesin-depleted cells. The mean ± SE of four (luciferase RNAi) or three (others) independent experiments is shown (≥26 cells were analyzed for each treatment). *, P < 0.01, t test; **, P < 10−4. (E) Immunostaining of MTs and RacGAP1 in control or Kif2A-depleted cells. A buckled MT bundle associated with a misaligned stem body (pink arrow) is highlighted with white arrowheads. (F) Ratio of MT bundle length to chromosome-to-chromosome distance in control or Kif2A-depleted cells. For Kif2A RNAi samples, the MT bundles associated with aligned or misaligned stem bodies were separately analyzed. The mean ratio (±SE) of ≥15 pairs of MT bundles in at least six cells from two independent experiments is shown. *, P < 10−7, t test. Bars, 5 µm.
To test whether specific MT-depolymerizing proteins are involved in central spindle organization, we investigated the distribution of stem bodies after knockdown of various MT-depolymerizing kinesins in the kinesin-8 family (Kif18A and Kif18B) and the kinesin-13 family (Kif2A, Kif2B, and Kif2C/mitotic centromere-associated kinesin), which are known to regulate MT dynamics during preanaphase (Gardner et al., 2008; Ems-McClung and Walczak, 2010). Among those candidates, knockdown of Kif2A caused the most prominent and frequent stem body misalignment, as assayed by RacGAP1-GFP localization (Fig. 1, C and D; and Fig. S1, E–L). In severe cases, global misplacement of stem bodies resulted in the formation of a perpendicularly tilted division plane (Fig. 1 C, bottom). The defect in stem body alignment was consistently observed with different siRNA constructs targeting Kif2A (Fig. S1, E and F) and rescued by ectopic expression of the RNAi-resistant version of the Kif2A gene, confirming specificity of the Kif2A RNAi phenotype (Fig. S1, G–I). Immunofluorescence microscopy of Kif2A-depleted cells revealed that MT bundles associated with misaligned stem bodies often buckled (Fig. 1 E, arrowheads) and that they were irregularly positioned in the interchromosomal region. These MT bundles were abnormally long compared with those connected to the aligned stem bodies, and their length exceeded the chromosome to chromosome distance (Fig. 1 F). Thus, Kif2A controls icMT length, which is important for central spindle organization.
Kif2A mediates MT depolymerization at the minus ends of central spindle MTs
We next investigated how Kif2A controls central spindle MT length. Previous studies showed that Kif2A regulates spindle MT dynamics in preanaphase (Gaetz and Kapoor, 2004; Ganem and Compton, 2004; Ganem et al., 2005). However, whether Kif2A plays a role during anaphase is not known. To assay central spindle MT dynamics during anaphase, we imaged live cells expressing EGFP–α-tubulin until anaphase and then added 200 ng/ml nocodazole to induce net MT depolymerization by sequestering tubulin dimers (Fig. 2, A and B). In control cells, the addition of nocodazole resulted in shortening of the central spindle within few minutes, whereas in Kif2A-depleted cells, the nocodazole-induced spindle shortening was completely suppressed (Fig. 2, A and B). This indicates that Kif2A is responsible for MT depolymerization of the central spindle.
Kif2A is involved in the depolymerization of central spindle MTs at their minus ends. (A and B) Shortening of central spindle MTs in control, but not Kif2A-depleted, cells expressing EGFP–α-tubulin after treatment with 200 ng/ml nocodazole (drug was added at time 0). The mean ± SE of at least six samples from two independent experiments is shown in B. (C) Scheme of the photobleaching assay. (D and E) EGFP–α-tubulin–expressing cells were treated with 1 µg/ml nocodazole immediately before image acquisition, and then, slit-shaped photobleach marks were introduced (red arrowheads). A time plot of the central spindle length and distance between two bleach marks. The mean ± SE of seven samples from three independent experiments is shown. Bars, 5 µm.
Kif2A is involved in the depolymerization of central spindle MTs at their minus ends. (A and B) Shortening of central spindle MTs in control, but not Kif2A-depleted, cells expressing EGFP–α-tubulin after treatment with 200 ng/ml nocodazole (drug was added at time 0). The mean ± SE of at least six samples from two independent experiments is shown in B. (C) Scheme of the photobleaching assay. (D and E) EGFP–α-tubulin–expressing cells were treated with 1 µg/ml nocodazole immediately before image acquisition, and then, slit-shaped photobleach marks were introduced (red arrowheads). A time plot of the central spindle length and distance between two bleach marks. The mean ± SE of seven samples from three independent experiments is shown. Bars, 5 µm.
To determine at which end of MTs Kif2A mediates depolymerization in the central spindle, we introduced slit-shaped photobleaching patterns to each half of the central spindle in nocodazole-treated EGFP–α-tubulin cells (Fig. 2, C–E). Because the majority of the plus ends of the antiparallel icMTs are present at the center of the central spindle (Mastronarde et al., 1993), depolymerization of MTs at their plus ends would result in movement of the two slits toward each other during nocodazole-induced spindle shortening. On the other hand, depolymerization at their minus ends, which are mostly distributed at both ends of the central spindle (Mastronarde et al., 1993), would not drive any movement of the bleach marks. As the bleach marks remained immobile during nocodazole-induced central spindle shortening (Fig. 2, D and E), we concluded that Kif2A depolymerizes central spindle MTs predominantly at their minus ends. Consistent with this, immunostaining showed enrichment of endogenous Kif2A at the distal ends of the central spindle (Fig. 3, A and B).
Aurora B controls the size and stability of the central spindle through regulation of Kif2A. (A) Immunostaining of MTs (green) and Kif2A (red) in control or Kif2A-depleted cells treated with or without 5 µM ZM447439 for 15 min. (B) Line profiles of the anti-Kif2A immunostaining intensity along the central spindle in control and ZM447439-treated cells. The mean ± SE of ≥12 cells from three independent experiments is shown. (C) Normalized fluorescence intensity of Kif2A immunostaining on the central spindle. Cells were either treated with 2.5 µM ZM447439 (ZM) for 3 or 10 min during anaphase or cultured without the drug for 30 min (DMSO) before fixation. The mean ± SE of ≥11 samples from two independent experiments is shown. *, P < 10−4, t test. (D) Length of the central spindle after treating cells with 5 µM ZM447439 for 15 min. The mean ± SE of three independent experiments is shown (≥45 cells were analyzed for each treatment). *, P < 0.001, t test. (E) EGFP–α-tubulin–expressing cells depleted of INCENP or codepleted of INCENP and Kif2A. In the absence of INCENP alone, the central spindle (red arrows) oscillated, and the cell later regressed. (F–H) Frequency of central spindle destabilization, furrow regression, or uneven chromosome distribution in control, Kif2A-depleted, INCENP-depleted, or Kif2A/INCENP-codepleted cells. For each sample, ≥47 cells from four independent experiments were analyzed. a.u., arbitrary unit. Bars, 5 µm.
Aurora B controls the size and stability of the central spindle through regulation of Kif2A. (A) Immunostaining of MTs (green) and Kif2A (red) in control or Kif2A-depleted cells treated with or without 5 µM ZM447439 for 15 min. (B) Line profiles of the anti-Kif2A immunostaining intensity along the central spindle in control and ZM447439-treated cells. The mean ± SE of ≥12 cells from three independent experiments is shown. (C) Normalized fluorescence intensity of Kif2A immunostaining on the central spindle. Cells were either treated with 2.5 µM ZM447439 (ZM) for 3 or 10 min during anaphase or cultured without the drug for 30 min (DMSO) before fixation. The mean ± SE of ≥11 samples from two independent experiments is shown. *, P < 10−4, t test. (D) Length of the central spindle after treating cells with 5 µM ZM447439 for 15 min. The mean ± SE of three independent experiments is shown (≥45 cells were analyzed for each treatment). *, P < 0.001, t test. (E) EGFP–α-tubulin–expressing cells depleted of INCENP or codepleted of INCENP and Kif2A. In the absence of INCENP alone, the central spindle (red arrows) oscillated, and the cell later regressed. (F–H) Frequency of central spindle destabilization, furrow regression, or uneven chromosome distribution in control, Kif2A-depleted, INCENP-depleted, or Kif2A/INCENP-codepleted cells. For each sample, ≥47 cells from four independent experiments were analyzed. a.u., arbitrary unit. Bars, 5 µm.
Aurora B controls central spindle length by limiting the accumulation of Kif2A to the distal ends of the structure
How is the Kif2A activity regulated to maintain icMTs at the appropriate length during anaphase? Because kinesin-13 family members are phosphorylated and inactivated by Aurora B (Andrews et al., 2004; Lan et al., 2004; Ohi et al., 2004, 2007; Sampath et al., 2004; Knowlton et al., 2009), we hypothesized that Aurora B controls Kif2A at the central spindle.
To test this, we investigated the effect of an Aurora B kinase inhibitor (ZM447439) on Kif2A localization during anaphase. High concentrations of ZM447439 (50 µM in Fig. S2 A) led to a complete disassembly of central spindle MTs, likely because the centralspindlin complex malfunctioned (Douglas et al., 2010). Lower concentrations of ZM447439 (1.25–5 µM) did not prevent the formation of an antiparallel MT structure in the central spindle, yet this induced abnormal accumulation of Kif2A on the central spindle (Fig. 3, A and B). The fluorescence intensity of Kif2A staining on the central spindle increased around twofold within 10 min after the addition of ZM447439 (Fig. 3 C). The signals were not detected in Kif2A-depleted cells, confirming the specificity of the antibody used for immunostaining (Fig. 3 A).
As another means to inactivate Aurora B, we used RNAi of inner centromere protein (INCENP), another CPC subunit. Similar to high concentrations of ZM447439, use of high concentrations (≥10 nM) of INCENP siRNA caused disassembly of the central spindle (Fig. S2 A). A lower concentration of INCENP siRNA (5 nM), which allowed formation of a central spindle, led to substantial increase of Kif2A at the central spindle (Fig. S2, B and C), similar to Aurora B inhibition by ZM447439.
In the CPC-inhibited cells, the central spindle became significantly shorter than that in untreated cells, which may have been caused by overaccumulation of Kif2A (Fig. 3, A and D; and Fig. S2 D). Consistent with this, the spindle shortening phenotype triggered by ZM447439 or INCENP RNAi treatment was suppressed by codepletion of Kif2A. These data strongly suggest that the CPC controls central spindle length by limiting the access of Kif2A to the distal ends of the structure.
Misregulation of Kif2A impairs coupling of chromosome segregation and cytokinesis
Time-lapse imaging of EGFP–α-tubulin showed that in 31% (n = 104) of the INCENP-depleted cells the central spindle position was unstable during furrow ingression, leading to oscillations along the spindle axis, moving back and forth from one daughter cell to another during furrow ingression (Fig. 3, E and F). During oscillation, each aster stayed in each daughter cell and did not move across the furrow (Fig. S2 E). ZM447439 treatment similarly caused central spindle oscillations during cytokinesis, indicating that Aurora B activity not only controls the morphology but also ensures the stability of the central spindle (Fig. S2, F and G). In 18 out of 33 (54%) INCENP-depleted cells with central spindle oscillations, some or even all chromatids of one of the daughter cells moved across the cleavage furrow to the other daughter cell, resulting in uneven chromosome distribution (Fig. S2 E and Video 1). In 9 out of 33 cells, displaced chromosomes stayed in the furrowing region, which resulted in cytokinesis failure by cleavage furrow regression. Depletion of Kif2A rescued the phenotypes caused by INCENP RNAi or ZM447439 (Fig. 3, E–H; and Fig. S2, F and G). Thus, Aurora B–mediated regulation of Kif2A is essential for spatial coordination of chromosome segregation and cytokinesis.
Uneven chromosome distribution in an INCENP RNAi cell. An anaphase HeLa cell expressing EGFP– α-tubulin (green) and Histone H2B-mRFP (red). The central spindle oscillated between daughter cells, and eventually, two masses of chromatids moved into one daughter cell. Six confocal sections were taken at z intervals of 2 µm every 3 min using TE2000 equipped with a 40×, 1.3 NA Plan Fluor oil immersion objective lens (Nikon), the CSU-X1 confocal unit, and an electron multiplying charge-coupled device camera (ImagEM). Maximum projections of the confocal sections were obtained for every time point using ImageJ. Bar, 5 µm
Uneven chromosome distribution in an INCENP RNAi cell. An anaphase HeLa cell expressing EGFP– α-tubulin (green) and Histone H2B-mRFP (red). The central spindle oscillated between daughter cells, and eventually, two masses of chromatids moved into one daughter cell. Six confocal sections were taken at z intervals of 2 µm every 3 min using TE2000 equipped with a 40×, 1.3 NA Plan Fluor oil immersion objective lens (Nikon), the CSU-X1 confocal unit, and an electron multiplying charge-coupled device camera (ImagEM). Maximum projections of the confocal sections were obtained for every time point using ImageJ. Bar, 5 µm
Central spindle oscillations were also observed in ∼10% of control cells (n = 74). These cells had shorter central spindles than the other cells, suggesting that length control was impaired in the former (Fig. S2 H). In six out of eight cells that showed spindle oscillation, the chromosomes were distributed unevenly. In contrast, spindle oscillation and the accompanying defects were never detected in Kif2A single-depleted cells (n = 47; Fig. 3, F–H). These results suggest the binucleation that sometimes occurs in this cell line is caused, at least in part, by uncoordinated cell division caused by irregular sizing of the central spindle through overfunctioning of Kif2A.
Aurora B–dependent phosphorylation of Kif2A
We next tested whether Kif2A is phosphorylated in an Aurora B–dependent manner during central spindle formation. To monitor the phosphorylation state of Kif2A during mitosis, we used a cell line stably expressing GFP-tagged Kif2A. The subcellular localization of GFP-Kif2A was similar to that of endogenous protein, and GFP-Kif2A overaccumulated at the central spindle upon addition of ZM447439, even in the absence of endogenous protein, suggesting that GFP-Kif2A is regulated by Aurora B in a similar manner to endogenous Kif2A (Fig. S3, A and B). GFP-Kif2A cells were synchronized at interphase, metaphase, or anaphase/telophase, and the phosphorylation state of immunoprecipitated GFP-Kif2A was analyzed by Phos-tag SDS-PAGE followed by immunoblotting (Fig. 4 A; Kinoshita et al., 2006). GFP-Kif2A was detected as a single band in the interphase extract, whereas additional bands with lower mobilities appeared in the samples from metaphase and anaphase/telophase. In the presence of ZM447439, one of the upper bands disappeared (Fig. 4, A and B, arrows), suggesting that Kif2A is phosphorylated in an Aurora B–dependent fashion during mitosis.
Aurora B–dependent phosphorylation of Kif2A during cytokinesis. (A and B) Immunoblotting of GFP-Kif2A wild-type (WT) and mutant proteins that were immunoprecipitated (IP) from cells synchronized at the indicated stages of the cell cycle. Immunoprecipitated samples were subjected to Phos-tag SDS-PAGE followed by immunoblotting using anti-GFP antibody. Note that in the Phos-tag gel, mobility of GFP-Kif2A significantly decreased compared with that in a gel without Phos-tag, and the relative band position to those of molecular markers varied among experiments (see Materials and methods); however, the identical band pattern was reproducibly observed for GFP-Kif2A (n = 2 for each). Arrows indicate upper bands corresponding to phosphorylated GFP-Kif2A. (C) Immunostaining of MTs in cells expressing GFP-Kif2A mutants. (D) Normalized fluorescence intensity of GFP at the central spindle of the cells expressing GFP-Kif2A mutants. (E) Central spindle length in cells expressing GFP-Kif2A mutants. The mean ± SE of at least six cells from two independent experiments is shown in D and E. Asterisks indicate statistically significant differences from the control cells expressing wild-type GFP-Kif2A (P < 10−3, t test), or between the cells expressing GFP-Kif2A (T97A) and GFP-Kif2A (T97A/S157C; P < 0.05). (F and G) Destabilization and oscillation of the central spindle and cytokinesis failure upon GFP-Kif2A (T97A/S157C) or mKate2-Kif2A (T97A/S157C) expression. Arrows and arrowheads indicate the oscillating central spindle and the furrow edge, respectively. (H–J) Frequency of central spindle destabilization, furrow regression, or uneven chromosome distribution in cells expressing GFP-Kif2A mutants. For each mutant, ≥18 cells from seven independent experiments were analyzed. Cells expressing GFP-Kif2A mutants at a comparable level were used for quantification (mean GFP intensity of 100–300 or 100–400 arbitrary units for D and E or H–J, respectively; see also Fig. S3, C and D). Bars, 5 µm.
Aurora B–dependent phosphorylation of Kif2A during cytokinesis. (A and B) Immunoblotting of GFP-Kif2A wild-type (WT) and mutant proteins that were immunoprecipitated (IP) from cells synchronized at the indicated stages of the cell cycle. Immunoprecipitated samples were subjected to Phos-tag SDS-PAGE followed by immunoblotting using anti-GFP antibody. Note that in the Phos-tag gel, mobility of GFP-Kif2A significantly decreased compared with that in a gel without Phos-tag, and the relative band position to those of molecular markers varied among experiments (see Materials and methods); however, the identical band pattern was reproducibly observed for GFP-Kif2A (n = 2 for each). Arrows indicate upper bands corresponding to phosphorylated GFP-Kif2A. (C) Immunostaining of MTs in cells expressing GFP-Kif2A mutants. (D) Normalized fluorescence intensity of GFP at the central spindle of the cells expressing GFP-Kif2A mutants. (E) Central spindle length in cells expressing GFP-Kif2A mutants. The mean ± SE of at least six cells from two independent experiments is shown in D and E. Asterisks indicate statistically significant differences from the control cells expressing wild-type GFP-Kif2A (P < 10−3, t test), or between the cells expressing GFP-Kif2A (T97A) and GFP-Kif2A (T97A/S157C; P < 0.05). (F and G) Destabilization and oscillation of the central spindle and cytokinesis failure upon GFP-Kif2A (T97A/S157C) or mKate2-Kif2A (T97A/S157C) expression. Arrows and arrowheads indicate the oscillating central spindle and the furrow edge, respectively. (H–J) Frequency of central spindle destabilization, furrow regression, or uneven chromosome distribution in cells expressing GFP-Kif2A mutants. For each mutant, ≥18 cells from seven independent experiments were analyzed. Cells expressing GFP-Kif2A mutants at a comparable level were used for quantification (mean GFP intensity of 100–300 or 100–400 arbitrary units for D and E or H–J, respectively; see also Fig. S3, C and D). Bars, 5 µm.
We next searched for physiologically important phosphorylation sites within Kif2A. To this end, we constructed a series of Kif2A mutants in which putative Aurora B phosphorylation sites were substituted with alanine or cysteine and analyzed their phosphorylation states in mitotically arrested cells (Fig. 4 B). Among the mutants tested, T97A eliminated the Aurora B–dependent upper band of GFP-Kif2A in the Phos-tag gel, suggesting that T97 is one of the in vivo Kif2A phosphorylation sites during cell division. We also investigated the subcellular localizations of the mutated Kif2A versions and their influence on central spindle organization (Fig. 4, C–E). The effects of GFP-Kif2A mutants could vary depending on their expression levels; hence, only cells expressing mutant proteins at a comparable level were selected for comparison (Fig. S3, C and D). GFP-Kif2A (T97A) accumulated abnormally on the center of the central spindle (Fig. 4, C and D), leading to significantly shortened central spindles (Fig. 4, C and E), reminiscent of the phenotype of CPC-inhibited cells. All other mutations that did not alter the Phos-tag mobility did not affect GFP-Kif2A localization or central spindle length. These results suggest that phosphorylation on T97 removes Kif2A from the center of the central spindle during anaphase.
It has been reported that phosphorylation on S157 inhibits Kif2A’s MT-depolymerizing activity in vitro (Knowlton et al., 2009). We therefore further tested the possible importance of this site (see Materials and methods). Mutation of S157 alone did not change Kif2A localization, and expression of the mutant protein did not affect central spindle length (Fig. 4, C–E). However, GFP-Kif2A (T97A/S157C) double mutant further decreased central spindle length compared with GFP-Kif2A (T97A) (Fig. 4, C and E). This suggests that S157 also contributes to the regulation of Kif2A in MT depolymerization of central spindle MTs.
We next investigated the stability of the central spindle and the progression of cytokinesis in live cells expressing GFP-Kif2A mutants (Fig. 4, F–J). Cells expressing GFP-Kif2A (T97A) showed elevated incidence of oscillating central spindles, cytokinesis failure, and uneven chromosome distribution. These defects were even more prominent in GFP-Kif2A (T97A/S157C)–expressing cells. During oscillation, disconnection of the central spindle from the cleavage furrow was frequently observed, although anillin and ECT2, which anchor the central spindle to the equatorial cortex (Frenette et al., 2012), localized normally at the cell equator (Fig. 4 G and Fig. S3, E–J). We also observed a delay in the progression of furrowing in these cells (Fig. S3 K). These results suggest that Aurora B–dependent phosphorylation of Kif2A on T97, and possibly on S157, limits Kif2A’s activity to ensure proper sizing of the central spindle and a stable connection between the equatorial cortex and the central spindle for faithful cell division.
Mathematical model for Aurora B– and Kif2A-mediated central spindle length control
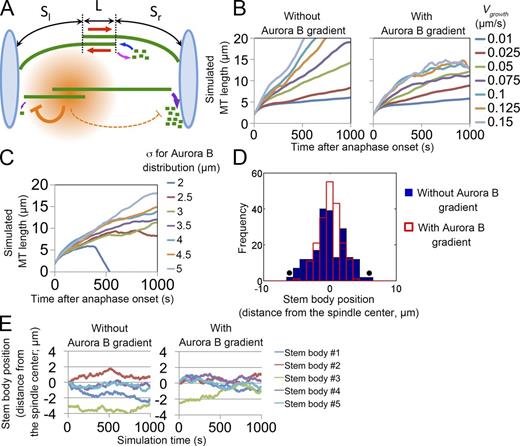
To establish a quantitative framework for Aurora B–dependent Kif2A regulation that ensures proper sizing and organization of icMTs, we performed mathematical modeling on the basis of a previously established theoretical model of the central spindle (see Materials and methods for details; Fig. 5 A; Brust-Mascher et al., 2004). The central spindle is represented by five pairs of antiparallel-coupled MTs with dynamic instability at their plus ends. Sliding motors localized at the MT-overlapping region slide each pair of coupled MTs apart at the rates comparable to those observed in cells (Fig. S4, A and B; Wang et al., 2010b). Distal minus ends are subjected to Kif2A-mediated depolymerization. Aurora B activity was modeled as a gradient centered around the spindle midzone, as previously shown by the use of Aurora B activity biosensors (Fuller et al., 2008; Tan and Kapoor, 2011). Specifically, Aurora B activity was represented as a Gaussian distribution around the antiparallel overlap, which inhibited Kif2A-mediated depolymerization in a signal concentration-dependent manner (Fig. 5 A). Segregated chromosomes were represented as a pair of length-limiting rigid walls at both ends of interchromosomal area, such that icMTs exceeding past this wall are pushed back and buckled as observed in real cells (Fig. 1 E, arrowheads). We simulated icMT elongation and positioning with various values of MT dynamics parameters and compared the outcome with or without the Aurora B activity gradient. In the models without the gradient, icMTs elongated nearly constantly and rarely reached steady states (Fig. 5 B). The kinetics of MT elongation was highly sensitive to changes in the parameter values, showing the unstable nature of the length-controlling system in this model (Fig. 5 B and Fig. S4, C and D). On the other hand, in the models with the gradient, the length of each icMT plateaued within several minutes. Interestingly, the length of icMTs at the steady state was determined predominantly by the size of the gradient (Fig. 5 C). The central spindle failed to form when the size of the gradient was too small, suggesting that an appropriately sized Aurora B signal distribution is necessary for central spindle formation. On the other hand, the length of icMTs was relatively insensitive to changes in other MT dynamics parameters (Fig. 5 B and Fig. S4 C). These results suggest that the dimension of the Aurora B gradient scales icMTs by tuning Kif2A activity and that the gradient provides a robust mechanism for the control of central spindle sizing.
Mathematical modeling of central spindle organization by Aurora B gradient. (A) A schematic image of the central spindle in the 2D model. Pairs of MTs form antiparallel MT overlaps, where they slide apart (red arrows). MT growth and shrinkage occur at plus ends (blue and pink arrow, respectively). The Aurora B gradient (orange) forms around the stem body and inhibits MT depolymerization at minus ends (purple arrows), depending on their distance from the stem bodies. L, length of the antiparallel overlap. Sl and Sr, lengths of nonoverlapping portions. The interchromosomal area is flanked by rigid chromosome walls (light blue), and MTs buckle when their length exceeds the interchromosomal distance (the top MTs). (B and C) Time plots of simulated MT length (sum of L, Sl, and Sr) with various MT growth rates (Vgrowth) or Aurora B gradient sizes (σ). The mean of 25 MT pairs from five independent simulations is shown for each condition. (D) Stem body distributions after 1,000-s simulations. Shown are 200 stem bodies from 40 independent simulations. Closed circles indicate outliers that exceeded the length limit (see Materials and methods). (E) Time plots of simulated stem body positions with or without the Aurora B gradient. A stem body (stem body #3) was misaligned at the beginning of the simulation in both cases. Alignment of stem body #3 was observed only when the Aurora B gradient was present. The mean of five independent simulations is shown for each condition. The parameter values used in each simulation are presented in Table S4.
Mathematical modeling of central spindle organization by Aurora B gradient. (A) A schematic image of the central spindle in the 2D model. Pairs of MTs form antiparallel MT overlaps, where they slide apart (red arrows). MT growth and shrinkage occur at plus ends (blue and pink arrow, respectively). The Aurora B gradient (orange) forms around the stem body and inhibits MT depolymerization at minus ends (purple arrows), depending on their distance from the stem bodies. L, length of the antiparallel overlap. Sl and Sr, lengths of nonoverlapping portions. The interchromosomal area is flanked by rigid chromosome walls (light blue), and MTs buckle when their length exceeds the interchromosomal distance (the top MTs). (B and C) Time plots of simulated MT length (sum of L, Sl, and Sr) with various MT growth rates (Vgrowth) or Aurora B gradient sizes (σ). The mean of 25 MT pairs from five independent simulations is shown for each condition. (D) Stem body distributions after 1,000-s simulations. Shown are 200 stem bodies from 40 independent simulations. Closed circles indicate outliers that exceeded the length limit (see Materials and methods). (E) Time plots of simulated stem body positions with or without the Aurora B gradient. A stem body (stem body #3) was misaligned at the beginning of the simulation in both cases. Alignment of stem body #3 was observed only when the Aurora B gradient was present. The mean of five independent simulations is shown for each condition. The parameter values used in each simulation are presented in Table S4.
In the simulations, the position of each stem body (marked by an asterisk in Video 2) stochastically fluctuated, and deviation of the stem body positions increased over time (Fig. S4 E) because of asymmetric dynamics of each half spindle MT. Interestingly, however, with comparable sets of MT dynamics parameters, the distribution of stem bodies became significantly narrower and more restricted to the center of the central spindle in the model with the gradient, compared with the distribution in the model without the gradient (after 1,000-s simulations; Fig. 5 D and Fig. S4 F). This suggests that the Aurora B activity gradient promotes stem body alignment. To further analyze this result, we placed one or more misaligned stem bodies and simulated their behaviors (Fig. 5 E, green lines; Fig. S4 G; and Video 2). The simulation indicated that the stem bodies, initially distributed at various positions, eventually realigned in an Aurora B gradient-dependent manner, which is reminiscent of the cellular stem body dynamics (Fig. S4 H). In conclusion, the simulations support a model in which the Aurora B activity gradient provides a robust mechanism for the sizing of the central spindle and for alignment of stem bodies.
Simulation of misaligned stem body behavior in the model with or without an Aurora B gradient. The nonoverlap portion of MTs, MT overlap region, stem body positions, and chromosome walls are indicated by green lines, blue lines, red asterisks, and black lines, respectively. Computer programs for simulation and model visualization were written with MATLAB. The simulations were performed with a step size of 1 s. Bar, 2 µm
Simulation of misaligned stem body behavior in the model with or without an Aurora B gradient. The nonoverlap portion of MTs, MT overlap region, stem body positions, and chromosome walls are indicated by green lines, blue lines, red asterisks, and black lines, respectively. Computer programs for simulation and model visualization were written with MATLAB. The simulations were performed with a step size of 1 s. Bar, 2 µm
MKLP2 inhibition causes Kif2A accumulation on the central spindle and icMT disorganization
To experimentally test the effect of the Aurora B localization on the stem body alignment, we investigated the intracellular distribution of Kif2A and the stem body alignment in cells depleted of MKLP2, which is required for the recruitment of CPC to the center of the central spindle and formation of the CPC-mediated phosphogradient (Fig. 6; Gruneberg et al., 2004; Fuller et al., 2008). RNAi of MKLP2 led to the accumulation of Kif2A throughout the spindle, indicating that the localization of CPC is important for the characteristic distribution pattern of Kif2A during cytokinesis (Fig. 6, A and B). MKLP2 depletion also caused a statistically significant reduction in central spindle length (Fig. S5) and misalignment of stem bodies (Fig. 6, C and D). Thus, CPC localization to the center of the central spindle is required for proper sizing of the central spindle and alignment of stem bodies.
MKLP2 is required for proper Kif2A distribution and stem body alignment. (A) Immunostaining of Aurora B (middle) and Kif2A (bottom) in control or MKLP2-depleted cells. (B) Line profiles of anti-Kif2A immunostaining along the central spindle in control and MKLP2-depleted cells. The mean ± SE of ≥36 cells from two independent experiments is shown. (C) Control and MKLP2-depleted cells expressing RacGAP1-GFP. Red arrows indicate misaligned stem bodies. Broken lines indicate cell boundaries. (D) The mean ± SE of three independent experiments is shown (≥62 cells were analyzed in each experiment). *, P < 0.05, t test. a.u., arbitrary unit. Bars, 5 µm.
MKLP2 is required for proper Kif2A distribution and stem body alignment. (A) Immunostaining of Aurora B (middle) and Kif2A (bottom) in control or MKLP2-depleted cells. (B) Line profiles of anti-Kif2A immunostaining along the central spindle in control and MKLP2-depleted cells. The mean ± SE of ≥36 cells from two independent experiments is shown. (C) Control and MKLP2-depleted cells expressing RacGAP1-GFP. Red arrows indicate misaligned stem bodies. Broken lines indicate cell boundaries. (D) The mean ± SE of three independent experiments is shown (≥62 cells were analyzed in each experiment). *, P < 0.05, t test. a.u., arbitrary unit. Bars, 5 µm.
Role of Kif2A-mediated stem body alignment in cytokinesis
We have shown that up-regulation of Kif2A caused an abnormally shortened central spindle and severe cell division defects. On the other hand, down-regulation of Kif2A did not cause cytokinesis defects despite severe misalignment of the stem bodies during anaphase (Fig. 3). We suspect that this may be explained by the merge of misaligned stem bodies into a single cluster at the intercellular bridge during telophase, which supported abscission (Fig. 1 C, middle). We tested whether the Kif2A-dependent mechanism works redundantly with other mechanisms to regulate cytokinesis by codepletion of Kif2A and other cytokinesis regulatory factors. Among these factors, MKLP1 kinesin, a subunit of the centralspindlin complex, plays an essential role in the formation of the central spindle and midbody (Douglas and Mishima, 2010). Although severe cytokinesis failure results when MKLP1 is depleted thoroughly (Matuliene and Kuriyama, 2002), a substantial population of the cells (67%, n = 57) completed cytokinesis when MKLP1 was only partially depleted by suboptimal RNAi conditions. Interestingly, however, codepletion of Kif2A with MKLP1 dramatically increased the frequency of regression during or after furrow ingression (Fig. 7, A and B). These results suggest that the central spindle MT structure is less robust in supporting the progression and completion of cytokinesis in the absence of Kif2A and more susceptible to perturbations of other cytokinesis regulators.
Kif2A-mediated central spindle sizing for cytokinesis. (A) EGFP–α-tubulin cells codepleted of Kif2A and MKLP1. Cells regressed during or after furrow ingression (labeled as early or late regression, respectively). Bar, 5 µm. (B) Frequency of early or late regression in each sample. For each treatment, ≥50 cells from two independent experiments were analyzed. (C) Model for central spindle sizing by Kif2A and Aurora B. The Aurora B activity gradient is formed around stem bodies. In the region where Aurora B concentration is high, Kif2A is inactivated through phosphorylation on T97, whereas it actively depolymerizes MT ends outside the region. This mechanism prevents overshortening of icMTs and guarantees proper sizing of icMTs.
Kif2A-mediated central spindle sizing for cytokinesis. (A) EGFP–α-tubulin cells codepleted of Kif2A and MKLP1. Cells regressed during or after furrow ingression (labeled as early or late regression, respectively). Bar, 5 µm. (B) Frequency of early or late regression in each sample. For each treatment, ≥50 cells from two independent experiments were analyzed. (C) Model for central spindle sizing by Kif2A and Aurora B. The Aurora B activity gradient is formed around stem bodies. In the region where Aurora B concentration is high, Kif2A is inactivated through phosphorylation on T97, whereas it actively depolymerizes MT ends outside the region. This mechanism prevents overshortening of icMTs and guarantees proper sizing of icMTs.
Discussion
Mechanism for controlling central spindle length
Our data support a new regulatory mechanism for central spindle assembly, involving Kif2A depolymerase and Aurora B kinase (Fig. 7 C). We suggest that Kif2A translates the Aurora B activity gradient into a spatial pattern of MT depolymerization activity and thereby into spatial coordination of icMTs. The active form of Kif2A depolymerizes central spindle MTs, but Aurora B–dependent phosphorylation at T97 delocalizes Kif2A from the central spindle and thus inhibits its depolymerization activity. As a result, minus ends further from Aurora B would be more favorably depolymerized by Kif2A, whereas those surrounded by a high concentration of Aurora B would avoid Kif2A-mediated depolymerization. This system allows icMTs to find their steady-state length on the basis of the size of the Aurora B gradient, thus presenting a mechanism of gradient-dependent MT length control. Indeed, our mathematical modeling showed that this simple Kif2A- and Aurora B–mediated mechanism ensures robust regulation of icMT length, which is consistent with all of our experimental results. Notably our model for central spindle assembly does not require additional proposed length-regulating mechanisms (Goshima and Scholey, 2010), such as presetting the balance between MT polymerases and depolymerases (Kinoshita et al., 2001), force-dependent tuning of MT depolymerization activities as proposed for preanaphase spindles (Goshima et al., 2005), or intrinsic length-dependent tuning mechanism of MT depolymerization activities as shown for kinesin-8 (Gupta et al., 2006; Varga et al., 2006).
Spindle length control and stem body alignment
Our results also indicate that Kif2A is required for alignment of the stem bodies during anaphase. Based upon the behavior of the stem body in simulations, we speculate that the symmetric distribution of the Aurora B activity along a coupled MT might maintain the flanking MTs at the same length, resulting in centering of the stem body at the cell equatorial plane (a possible mechanism is depicted in Fig. S4 I). Because the central spindle is crowded with MTs, we cannot observe individual MT dynamics and thereby verify this model in cells. Moreover, our results do not exclude the possibility that other regulatory mechanisms, such as those working at the plus ends of MTs, are present in cells (Bieling et al., 2010; Hu et al., 2011). However, it is noteworthy that our modeling suggests that the Aurora B– and Kif2A-dependent modulation of the minus end dynamics could significantly contribute to the stem body alignment without assuming regulation of plus end dynamics or direct connections between each coupled MT.
Control of Kif2A activity for chromosome segregation and cytokinesis
Our results clarify the importance of controlling Kif2A activity for proper progression of cytokinesis. Overactivation of Kif2A led to central spindle oscillations, uneven chromosome distribution, and cytokinesis failure. One possible cause of spindle oscillation is a weakened connection between the central spindle and the cleavage furrow caused by Kif2A overactivation (Fig. 4 G and Fig. S3, I and J). Excessive MT-depolymerizing activity might make the central spindle structure too small to maintain a stable connection to the equatorial cortex even in the presence of the anchoring proteins such as anillin and ECT2 (Frenette et al., 2012). The observed delay in furrowing may be a result of a loss of integrity in the connection between the central spindle and the equatorial cortex (Gregory et al., 2008). During cytokinesis, blebbing of the polar cortex that accompanies cytoplasmic flow commonly occurs (Charras and Paluch, 2008), and excessive blebbing drives spindle oscillation (Rankin and Wordeman, 2010; Sedzinski et al., 2011). Disconnection of the central spindle from the equatorial cortex and delayed furrowing would make KIF2A-overexpressing cells more susceptible to the mechanical disturbance caused by polar cortical activity, which would increase the chance of spindle oscillation. A subpopulation of untreated HeLa cells also showed uncoordinated cell division accompanied by Kif2A-dependent central spindle shortening and oscillation, suggesting that abnormality in central spindle length control is one of the reasons for genomic instability in this cell line. Interestingly, it has been reported that Kif2A is overexpressed in some cancer cell lines, and this protein is suggested as a target for cancer chemotherapy (Wang et al., 2010a). Our results support this idea.
On the other hand, loss of function of Kif2A did not necessarily cause cytokinesis failure. Despite severe misalignment of the stem bodies during anaphase in Kif2A-depleted cells, cytokinesis completed in these cells, possibly through telophase-specific reassembly of the misaligned stem bodies into a cluster at the intercellular bridge. Because codepletion of Kif2A and MKLP1 had a synergistic effect on the disruption of cytokinesis completion, unknown processes involving this motor protein may comprise a telophase-specific backup mechanism. Future study, combining microscopy and stage-specific gene perturbation techniques, might elucidate such a mechanism. Our mathematical model would serve as a framework to quantitatively describe the process.
Materials and methods
Cell culture, RNAi, and DNA transfection
HeLa Kyoto line, HeLa bacterial artificial chromosome cell lines expressing RacGAP1–localization and affinity purification (LAP) tag or anillin-LAP (a gift from T. Hyman, Max Planck Institute-Dresden, Dresden, Germany; Hutchins et al., 2010), HeLa EGFP–α-tubulin/histone H2B-mRFP line (Steigemann et al., 2009), and HeLa photoactivatable GFP–α-tubulin/histone H2B-mRFP line (a gift from P. Meraldi, University of Geneva, Geneva, Switzerland; Amaro et al., 2010) were cultured in DMEM supplemented with 10% fetal bovine serum and antibiotic antimycotic (Sigma-Aldrich). A HeLa EGFP-Kif2A line was established by transfecting HeLa Kyoto cells with the pEGFP-Kif2A vector and selecting positive cells in the presence of 500 µg/ml G418. siRNA transfection was performed using Lipofectamine RNAiMAX (Invitrogen). The siRNA sequences used in this study are shown in Table S1. DNA transfection was performed using jetPEI (Polyplus Transfection).
Cell culture synchronization
Synchronization of HeLa EGFP-Kif2A cell culture was performed as described previously (Uehara and Goshima, 2010). In brief, interphase cells were obtained by incubating cells in the presence of 2.5 mM thymidine for 24 h. To obtain metaphase-arrested cells, the thymidine-blocked cells were washed three times with DMEM, released for 6 h, treated with 50 ng/ml nocodazole for 4 h, washed again three times with DMEM, and treated with 10 µM MG132 with or without 20 µM ZM447439 for 2 h. To obtain anaphase/telophase cells, metaphase-arrested cells were washed once with DMEM, released for 20 min, and treated with or without 20 µM ZM447439 for 30 min. Prometaphase cells were obtained by treating cells with 50 ng/ml nocodazole for 13 h. These mitotically arrested cells were collected by shaking them off the culture dishes and spun down, and the cell pellets were quickly frozen by liquid nitrogen.
Plasmid construction
Full-length cDNA encoding Kif2A (GenBank accession no. NM_004520.4) was obtained by PCR using a HeLa cell cDNA pool as the template and subcloned into the EcoRI–SalI sites of pEGFP-C1 (Takara Bio Inc.). GFP-Kif2A mutants were also subcloned into pEGFP-C1 using PCR and ligation. Alanine substitution of the S157 residue (corresponding to S132 in Knowlton et al., 2009) disrupted the activity of Kif2A (Knowlton et al., 2009); therefore, we used cysteine substitution instead (Ghosh et al., 2002; Noy et al., 2012). The nontagged Kif2A mutant used for the rescue experiment (Fig. S1, G–I) was made by subcloning a Kif2A gene with RNAi-resistant mutations. All the primers used for PCR are listed in Table S2.
Reagents
ZM447439, taxol (paclitaxel), and nocodazole were purchased from Tocris Bioscience, Molecular Probes, and EMD Millipore, respectively. Rat monoclonal anti–α-tubulin antibody (YOL1/34), mouse monoclonal anti-GFP antibody (JL-8), rabbit polyclonal anti-Kif2A antibody (ab37005), and goat polyclonal antianillin antibody (sc-54859) and rabbit polyclonal anti-ECT2 antibody (sc-1005) were purchased from AbD Serotec, Takara Bio Inc., Abcam, and Santa Cruz Biotechnology, Inc., respectively. Monoclonal antibody against RacGAP1 was obtained by a method described previously (Kimura et al., 1994). In brief, BALB/c mice were immunized using the keyhole limpet hemocyanin-conjugated C-terminal peptide (CTPRFGSKSKSATNLGRQGN) as an antigen. The splenocytes were fused with the myeloma line P3-X63Ag8.653, and the culture media were collected.
Cell imaging
For immunostaining, cells were fixed with 3.2% paraformaldehyde in PBS for 10 min and then permeabilized with 0.5% Triton X-100 in PBS for 10 min at room temperature. For live-cell imaging, cells were cultured at 37°C in phenol red–free DMEM with Hepes (Gibco) for >4 h before and during image acquisition. Fixed and living cells were observed using a microscope (TE2000; Nikon) equipped with a 100×, 1.4 NA Plan Apochromatic, a 60×, 1.4 NA Plan Apochromatic, or a 40×, 1.3 NA Plan Fluor oil immersion objective lens (Nikon), a confocal unit (CSU-X1; Yokogawa Corporation of America), and an electron multiplying charge-coupled device camera (ImagEM; Hamamatsu Photonics). For immunostaining, DyLight 488–, Alexa Fluor 488–, Rhodamine TRITC–, Cy3-, or Cy5-conjugated secondary antibodies against mouse, rat, or rabbit IgG (Jackson ImmunoResearch Laboratories, Inc.) were used. Image acquisition was controlled by µManager software. The photobleaching experiments in Fig. 2 and Fig. S4 A were performed using a confocal microscope (A1R; Nikon) equipped with a 60×, 1.4 NA oil immersion objective lens (Nikon).
Image analysis
Quantification of microscopic data and maximum projection of confocal z stacks were performed using ImageJ software (National Institutes of Health). For line profiling in Fig. 3 B, Fig. 6 B, and Fig. S1 K, we measured the signal intensity along the central spindle and subtracted the cytoplasmic background. For quantification in Fig. 3 C, Fig. 4 D, Fig. S2 C, and Fig. S3 (B, F, and H), we measured the fluorescence intensity of the manually selected regions of interest, subtracted the cytoplasmic background, and multiplied them by the selected area to obtain the total fluorescence intensity. For quantification in Fig. S1 H and Fig. S3 (C and D), we measured the mean signal intensity of the whole cell area and subtracted background outside the cell. For figure preparation, image size and contrast were adjusted using the Photoshop software (Adobe).
Correlative fluorescence and EM
Sample preparation for EM was performed according to a previously described method (Guizetti et al., 2011) with some modifications. RacGAP1-LAP cells were cultured on sapphire discs with grid patterns to identify the position of each cell. Cells were fixed with 2% glutaraldehyde in NaHCa buffer (0.1 M NaCl2, 30 mM Hepes, and 2 mM CaCl2, pH 7.4) for 1.5 h at room temperature and then washed with NaHCa buffer. Z-stack sections of the fixed cells were acquired using the confocal microscope. The fixed samples were then stained with 0.2% tannic acid in NaHCa buffer for 1 min at room temperature, postfixed with 2% osmium tetroxide in NaHCa buffer for 1 h at 4°C, dehydrated, and embedded in Araldite-Epon (Electron Microscopy Sciences). Serial sections (90 nm in thickness) were cut parallel to a substratum using a microtome (UltraCut; Leica) and collected onto Formvar-coated copper grids (special three-slit grid; Nisshin EM Co.). Sections were poststained with 5% uranyl acetate in 60% ethanol and 0.4% lead citrate. Cells identical to those observed with the confocal microscope were imaged using an electron microscope (H7600; Hitachi High-Tech Fielding Corporation) operating at 100 kV. Serially sectioned EM images of anaphase cells were aligned and stacked by ImageJ to examine the correlation with confocal images.
Mathematical modeling
Computer programs were written with MATLAB (MathWorks). We used the version 2012b or later with some toolboxes to run the program. We developed a mathematical model based upon several differential equations. In the model, several pairs of antiparallel MTs, overlapping at each plus end, were aligned at interval of 1 µm in 2D space. The model consisted of four basic elements: asymmetric spindle elongation, catenary function for force balance, dynamic instability of MT plus ends, and MT-depolymerizing activity that can be spatially controlled. As described in the following equations, each element changes its related numerical values following its own rules at each step during a simulation. Thus, integration of these four elements determines polymerization/depolymerization rates at MT ends and stem body positions and thereby simulates dynamic behavior of the model. The model was explored through a computer simulation in which several parameter values were examined (Table S3 and Table S4). We used the qsub command of Portable Batch System for parameter search. The equations were solved numerically using the Runge–Kutta method. All simulations in our study were performed with a step size of 1 s, but we reached identical conclusions with a step size of 0.1 s with several parameters.
Asymmetric model for spindle elongation.
We constructed an asymmetric central spindle model based upon the previously proposed equation for anaphase spindle elongation (Eq. 1 in Brust-Mascher et al., 2004). We divided central spindle MTs into three parts: antiparallel overlap MTs (L) and left and right portions of nonoverlapping MTs (Sl and Sr). In the simulations in Fig. 5 (B and C) and Fig. S4 (C and D), the initial values of L, Sl, or Sr were set to 2, 0, or 0 µm, respectively, whereas in Fig. 5 D and Fig. S4 (B, E, and F), the values were set to 2, 5, or 5 µm, respectively. The dynamics of left and right nonoverlapping MTs were described by the following equations: dSl/dt = Vsliding(t) − Vdepoly,l and dSr/dt = Vsliding(t) − Vdepoly,r, in which Vsliding(t) denotes the sliding rate of overlapping MTs as a function of time and Vdepoly,l and Vdepoly,r denote the mean rate of MT depolymerization at each minus end during anaphase. We assumed that Vsliding(t) is proportional to the number of sliding motors localized at the overlapped region. Therefore, Vsliding(t) was determined according to the following equation: Vsliding(t) = L(t) × Vm, in which Vm denotes the velocity of antiparallel MT sliding generated at a unit antiparallel overlap length, which was set at 0.02/s according to the measured sliding rate (see the following sentence) and a rough estimation of stem body length (∼0.5 µm; Fig. S1 A; McIntosh and Landis, 1971). We found that antiparallel MT sliding occurred at ∼0.01 µm/s in anaphase HeLa cells (Fig. S4 A), which was slower but in the same order as the rate previously reported in Drosophila melanogaster embryonic cells (∼0.05 µm/s; Wang et al., 2010b). Therefore, we tested our model by using different sets of parameters, which reproduced antiparallel sliding at rates covering this physiologically relevant range (0.01–0.05 µm/s). Model behavior was essentially the same within this range of sliding rates, suggesting the generality of our model assumptions. Following the previous model (Brust-Mascher et al., 2004), L was modeled as follows: dL/dt = Vpoly,l + Vpoly,r − Vsliding(t), in which Vpoly,l and Vpoly,r denote the polymerization rate of each plus end.
Catenary model.
Central spindle MTs are pushed back against segregated chromosomes and buckled during anaphase when excessively elongated (Fig. 1 E). To incorporate this behavior into the model, we placed a rigid wall at each side of the simulated interchromosomal area and introduced the catenary equation to model the bending of excessively elongated MTs: y = acosh(x/a), in which the constant a represents curvature of the catenary, and x and y denote each coordinate in 2D space. In our simulation, when the length of a coupled MT exceeded the distance between the two walls, the MT was bent according to the catenary model so that the compression force was balanced. The symbolic math toolbox was used for determination of the catenary constants. On the other hand, when the length of a coupled MT was shorter than the distance between the two walls, it remained straight and parallel to the other MTs. When a coupled MT shorter than the wall to wall distance bumped against a wall, it was pushed back without bending.
Dynamic instability.
The Monte Carlo technique was used to simulate the stochastic nature of MT plus ends undergoing dynamic instability. The probability of the state switching between shrinkage and growth was calculated using the following equation as proposed previously (Sprague et al., 2003): P(switch) = 1 −exp(−kswitch × τstep), in which kswitch is the frequency of catastrophe or rescue event, and τstep denotes the time step of the simulation. In our simulation, a uniformly distributed random number between 0 and 1 was generated at each simulation time step, and a switching event (catastrophe or rescue) occurred when the random value exceeded P(switch). In the growth or shrinkage phase, an MT plus end polymerized or depolymerized at growth velocity Vpoly = Vgrowth or Vpoly = −Vshrink, respectively, in which Vgrowth or Vshrink is the velocity of plus end growth or shrinkage, respectively. Because of asymmetric MT plus end dynamics, the position of each MT overlap stochastically fluctuated. During simulation, because of dramatic MT shrinkage, Sl or Sr sometimes became a negative value, generating outlier stem bodies that localized outside the interchromosomal area (Fig. 5 D, closed circles; and Fig. S4 F). This happened when the degree of disturbance in stem body positioning was beyond the limits of our model.
The Aurora B–mediated signal.
We modeled the Aurora B activity gradient as a Normal distribution, peaking at each stem body where Aurora B is most concentrated in vivo. The concentration of the Aurora B-mediated signal at each minus end C was determined using the equation
if C ≤ 1, or C = 1 if C > 1, in which Iaurora is the Aurora B signal intensity at each stem body, D is the distance between an MT minus end and each stem body, and σ is the standard deviation of each Aurora B distribution. Stem body position was set at the center of antiparallel MT overlap. We incorporated MT depolymerization activity that was anticorrelated with the Aurora B concentration. Therefore, the depolymerization rate at each minus end was determined as Deprate = Vdep,0 + Vdep,max(1 − C), in which Deprate is the depolymerization rate at a minus end, and Vdep,0 and Vdep,max are the minimum and maximum depolymerization rates at the minus end, respectively. Note that we prepared several coupled MTs in our model so that Aurora B on a stem body affected not only the flanking MTs but also other MTs. In the model without the Aurora B gradient, the depolymerization rate was set to constant.
Biochemistry
To analyze the phosphorylation states of the GFP-Kif2A mutants, protein was immunoprecipitated from GFP-Kif2A–expressing cells as described previously (Uehara et al., 2009). In brief, mitotically arrested cells were extracted with the HB100 buffer (50 mM Hepes-KOH, pH 7.6, 100 mM NaCl, 1 mM MgCl2, 1 mM EGTA, 1% Triton X-100, protein inhibitor cocktail [Complete; Roche], phosphatase inhibitor cocktail [PhosSTOP; Roche], and 1 mM DTT) for 10 min on ice and incubated with the anti-GFP–conjugated agarose beads (MBL) for 3 h at 4°C. The immunoprecipitants were washed with HB100 three times and subjected to SDS-PAGE using a 6% acrylamide gel containing 25 µM Phos-tag. Immunoblotting was performed using anti-GFP antibody (JL-8), HRP-conjugated anti–mouse antibody (Jackson ImmunoResearch Laboratories, Inc.), and an ECL Western blotting detection system (GE Healthcare). Precision Plus Protein Dual Color Standards (Bio-Rad Laboratories) were used as the molecular marker in the SDS-PAGE experiments. Note that because of irreproducible mobility of the prestained molecular marker proteins in Phos-tag SDS-PAGE gels (Kinoshita et al., 2012), relative positions of GFP-Kif2A bands to the marker differed among experiments.
Online supplemental material
Fig. S1 shows additional data on stem body misalignment in Kif2A-depleted cells. Fig. S2 shows disorganization of the central spindle by CPC inhibition. Fig. S3 shows localization of GFP-Kif2A upon Aurora B inhibition and defects caused by expression of the Kif2A mutants. Fig. S4 shows mathematical modeling of CPC-mediated central spindle organization. Fig. S5 shows central spindle length in MKLP2-depleted cells. Table S1 and Table S2 show sequences of the siRNAs and primers used in this study, respectively. Table S3 shows parameters used in the simulations. Table S4 lists the parameter values used in each simulation. Video 1 shows uneven chromosome distribution in an INCENP RNAi cell. Video 2 shows simulations of misaligned stem body behavior. A ZIP file is also provided that contains source code files used in the simulation.
Acknowledgments
We are grateful to Drs. Patrick Meraldi and Tony Hyman for the HeLa cell lines, Jiro Usukura (Nagoya University, Nagoya, Japan) for assistance with EM, Shin Ishii and Masataka Yamao (Kyoto University, Kyoto, Japan) for use of the cluster computer to run simulation programs, Shuh Narumiya and Sadanori Watanabe (Kyoto University) for technical support, and Momoko Nishina for technical assistance.
This work was supported by the Next Generation grant (NEXT Program Japan Society for the Promotion of Science) and Human Frontier Science Program (to G. Goshima), the Naito Foundation (to R. Uehara), and Grants-in-Aid for Scientific Research (Ministry of Education, Culture, Sports, Science and Technology; to R. Uehara and G. Goshima). Research in the Gerlich laboratory has received funding from the European Community’s Seventh Framework Programme FP7/2007–2013 under grant agreements no. 241548 (MitoSys) and no. 258068 (Systems Microscopy) and from a European Research Council Starting Grant (agreement no. 281198).
References
Author notes
R. Uehara’s present address is Dept. of Life Sciences, School of Arts and Sciences, The University of Tokyo, Tokyo 153-8902, Japan.