Dynamic microtubules (MTs) are essential for various intracellular events, such as mitosis. In Drosophila melanogaster S2 cells, three MT tip-localizing proteins, Msps/XMAP215, EB1, and Sentin (an EB1 cargo protein), have been identified as being critical for accelerating MT growth and promoting catastrophe events, thus resulting in the formation of dynamic MTs. However, the molecular activity of each protein and the basis of the modulation of MT dynamics by these three factors are unknown. In this paper, we showed in vitro that XMAP215msps had a potent growth-promoting activity at a wide range of tubulin concentrations, whereas Sentin, when recruited by EB1 to the growing MT tip, accelerated growth and also increased catastrophe frequency. When all three factors were combined, the growth rate was synergistically enhanced, and rescue events were observed most frequently, but frequent catastrophes restrained the lengthening of the MTs. We propose that MT dynamics are promoted by the independent as well as the cooperative action of XMAP215msps polymerase and the EB1–Sentin duo.
Introduction
Microtubules (MTs) are dynamic polymers that consist of α- and β-tubulins. They show dynamic cycles both in vivo and in vitro, during which polymerization and depolymerization are repeated through transition events called catastrophe and rescue (Desai and Mitchison, 1997). A key step toward understanding the mechanism underlying MT polymerization dynamics is to reconstitute the behavior in vitro. Dynamic MTs are generated with a relatively high concentration of tubulin dimers in the presence of GTP in vitro (e.g., Walker et al., 1988). However, these conditions are different from physiological conditions, in which multiple nontubulin proteins are required to generate dynamic MTs (Howard and Hyman, 2007; Akhmanova and Steinmetz, 2008). For example, Xenopus laevis XMAP215 processively adds tubulin onto the MT plus ends, thereby promoting MT growth, whereas it also can promote MT shrinkage by removing tubulin from the end (Kerssemakers et al., 2006; Howard and Hyman, 2007; Brouhard et al., 2008; Slep, 2010). A landmark study on in vitro reconstitution of MT dynamics was published a decade earlier, in which two conserved proteins, XMAP215 and kinesin-13 (MT depolymerase), were mixed with tubulin and reproduced physiological MT dynamics in vitro (Kinoshita et al., 2001). This study gave rise to the concept that balance between an MT polymerizer and a depolymerase is central to MT dynamics in cells. However, other factors have since been identified, the cellular depletion of which also affects MT dynamics. One such factor is the highly conserved protein EB1, which autonomously binds to the growing ends of MTs and recruits many cargo proteins (Akhmanova and Steinmetz, 2008; Slep, 2010). For example, depletion of EB1 in Drosophila melanogaster cells or from Xenopus egg extracts severely reduces the dynamicity of MTs; therefore, EB1 is not a negligible factor with respect to reconstituting plus-end dynamics (Rogers et al., 2002; Tirnauer et al., 2002). Despite our detailed understanding of the mechanism underlying plus-end recognition (Zanic et al., 2009; Maurer et al., 2011, 2012), the molecular activity of EB1 with respect to plus-end dynamics remains unclear (Bieling et al., 2007; Manna et al., 2008; Vitre et al., 2008; Dixit et al., 2009; Komarova et al., 2009; Zhu et al., 2009). Moreover, many of the cargo proteins of EB1, such as cytoplasmic linker-associated protein (CLASP) or kinesins, have MT-binding activity and/or dynamics-modulating activity of their own; this factor has made it challenging to reconstitute EB1-dependent MT dynamics by using purified proteins.
Drosophila has been found to be a good model system for studying MT polymerization dynamics because individual MTs are easily visualized in the living S2 cell line, and the factors governing MT polymerization dynamics have been extensively identified through mutant- or RNAi-based analyses (Rogers et al., 2002; Brittle and Ohkura, 2005; Goshima et al., 2005, 2007; Li et al., 2011). Moreover, these factors are usually present as single genes, unlike mammalian cells that often use multiple paralogues. We have previously identified a plus end–tracking protein, Sentin, the depletion of which phenocopies the deletion of EB1 and XMAP215 (Msps [minispindles] in Drosophila) in S2 cells, in terms of shorter spindle formation and the presence of less dynamic cytoplasmic MTs (i.e., pausing MTs; Li et al., 2011). Sentin is an EB1 cargo, and importantly, when the cargo binding site of EB1 is replaced by the Sentin protein, the fusion protein restores the EB1 RNAi phenotype, despite the fact that recruitment of other cargoes is likely to be absent. This indicates that Sentin is the dominant cargo for EB1 in regulating MT polymerization dynamics in S2 cells (Li et al., 2011). Furthermore, it has been found that Sentin depletion severely reduces plus-end accumulation of XMAP215msps in S2 cells, which led to a model in which Sentin, recruited by EB1, further recruits XMAP215 polymerase to make MTs dynamic in vivo. However, the alternative and mutually nonexclusive model is viable, in which Sentin, independent of XMAP215msps recruitment, functions to modulate MT plus-end dynamics.
This study aimed to understand how XMAP215msps, EB1, and Sentin (the dominant cargo of EB1) regulate MT dynamics. Our data, which were obtained using S2 cells, suggest that Sentin is not a simple recruitment factor for XMAP215msps. The in vitro MT polymerization assay performed using purified proteins showed that Sentin, when recruited by EB1, exhibited growth acceleration activity independent of XMAP215msps as well as catastrophe-promoting activity that was not detected for XMAP215msps. Mixing of the three factors generated the most dynamic MTs, accompanying a synergistic increase in growth rate and the number of rescue events. We suggest that these activities are responsible for the formation of the dynamic MTs in cells.
Results
Additive effect of Sentin and XMAP215msps for interphase MT growth and spindle length control
Sentin phenocopies XMAP215msps in shorter spindle formation, and its RNAi reduces the dynamicity of MTs in S2 cells (Brittle and Ohkura, 2005; Li et al., 2011). To gain insights into the relationship between Sentin and XMAP215msps in cells, we performed double RNAi analyses. We determined the individual MT growth rate in interphase cells by tracing EB1-GFP, which tracks the growing MTs regardless of the presence or absence of XMAP215msps or Sentin (Li et al., 2011). In this experiment, we treated cells with double-strand RNA (dsRNA) for 6 d, so that <1% of residual Sentin was detected by immunoblotting, suggesting that this virtually mimics the Sentin-null condition (Fig. 1 A). XMAP215msps knockdown was much less efficient, and 35% of the residual XMAP215msps was detected after RNAi treatment. Consistent with previous analyses performed using GFP-tubulin, we observed a reduced growth rate after transfection with each RNAi (P < 0.0001; Fig. 1 B; Brittle and Ohkura, 2005; Li et al., 2011). Notably, despite only partial knockdown of XMAP215msps, the growth rate was as severely reduced as seen with Sentin RNAi, suggesting that XMAP215msps contributes more dominantly to growth acceleration in this cell line. Interestingly, double Sentin/XMAP215msps RNAi treatment further reduced the growth rate (P < 0.0001), whereas the knockdown efficiency for each protein was unchanged. These results confirm that Sentin contributes to MT growth acceleration and further indicate that XMAP215msps can function in accelerating growth in cells, even in the absence of Sentin-dependent recruitment.
Sentin functions, at least partially, independent of XMAP215msps in S2 cells. (A) Immunoblotting to determine knockdown efficiency of Sentin and XMAP215msps after RNAi. Sentin RNAi was very efficient, whereas ∼35% residual XMAP215msps was present after RNAi. Tubulin, Aug5 (Dgt5), and the Coomassie staining were used as loading controls. (B) MT growth rate determined by tracing EB1-GFP signals in the kymograph after control (n = 94), Sentin (n = 94), XMAP215msps (n = 47), and double Sentin/XMAP215msps RNAi (n = 49). The error bars represent SEM. (C) The Sentin fragment that lacked the first 230 aa did not restore the normal MT growth rate, although it restored XMAP215msps-GFP accumulation at the tip. Further truncation of the N terminus of Sentin did not rescue either defect. In this experiment, three independent stable cell lines expressing mRFP- or mCherry-tagged Sentin truncations and XMAP215msps-GFP were used, and growing MT ends were identified by mRFP/mCherry signals. Plus-end accumulation of XMAP215msps-GFP was visually determined after kymograph generation (51–121 MTs from two to seven cells). The kymographs and a still cell image of mRFP/mCherry and GFP signals after RNAi knockdown of endogenous Sentin are shown at the top. The XMAP215msps-GFP accumulation frequency and MT growth rate relative to control RNAi for each cell line (±SEM) have been plotted below. (D, left) A monopolar spindle after double Sentin/Klp61F RNAi. A sum projection image of 11 z sections (separated by 0.5 µm) is shown, and the centrosome and astral MT ends are marked. Chromosomes were clustered in the lower side in this cell (not visible in this image). (right) The distance between a centrosome and each astral MT end was measured after sum projection, and the relative value to control RNAi cells has been plotted. Data have been provided for control (n = 187), Sentin (n = 322), XMAP215msps (n = 169), and double Sentin/XMAP215msps RNAi (n = 174) from 22 or 23 cells (±SEM). Bars: (horizontal) 5 µm; (vertical) 1 min.
Sentin functions, at least partially, independent of XMAP215msps in S2 cells. (A) Immunoblotting to determine knockdown efficiency of Sentin and XMAP215msps after RNAi. Sentin RNAi was very efficient, whereas ∼35% residual XMAP215msps was present after RNAi. Tubulin, Aug5 (Dgt5), and the Coomassie staining were used as loading controls. (B) MT growth rate determined by tracing EB1-GFP signals in the kymograph after control (n = 94), Sentin (n = 94), XMAP215msps (n = 47), and double Sentin/XMAP215msps RNAi (n = 49). The error bars represent SEM. (C) The Sentin fragment that lacked the first 230 aa did not restore the normal MT growth rate, although it restored XMAP215msps-GFP accumulation at the tip. Further truncation of the N terminus of Sentin did not rescue either defect. In this experiment, three independent stable cell lines expressing mRFP- or mCherry-tagged Sentin truncations and XMAP215msps-GFP were used, and growing MT ends were identified by mRFP/mCherry signals. Plus-end accumulation of XMAP215msps-GFP was visually determined after kymograph generation (51–121 MTs from two to seven cells). The kymographs and a still cell image of mRFP/mCherry and GFP signals after RNAi knockdown of endogenous Sentin are shown at the top. The XMAP215msps-GFP accumulation frequency and MT growth rate relative to control RNAi for each cell line (±SEM) have been plotted below. (D, left) A monopolar spindle after double Sentin/Klp61F RNAi. A sum projection image of 11 z sections (separated by 0.5 µm) is shown, and the centrosome and astral MT ends are marked. Chromosomes were clustered in the lower side in this cell (not visible in this image). (right) The distance between a centrosome and each astral MT end was measured after sum projection, and the relative value to control RNAi cells has been plotted. Data have been provided for control (n = 187), Sentin (n = 322), XMAP215msps (n = 169), and double Sentin/XMAP215msps RNAi (n = 174) from 22 or 23 cells (±SEM). Bars: (horizontal) 5 µm; (vertical) 1 min.
XMAP215msps recruitment by Sentin may not be sufficient for obtaining fully fast-growing MTs
While evaluating the function of truncated Sentin, we found that the expression of Sentin in which the first 230 aa or more were deleted failed to fully rescue the spindle length phenotype produced by endogenous Sentin RNAi by using dsRNA targeting the UTR, although these fragments still localized to the growing ends of MTs (Fig. S1 A). We next tested whether these fragments could rescue the defects in MT growth speed and XMAP215msps localization during interphase. To this end, we selected cell lines stably expressing XMAP215msps-GFP and mRFP-tagged truncated Sentin. In the rescue experiment, in which endogenous Sentin was specifically knocked down, the fragments that lacked the first 590 aa did not rescue either phenotype. However, the fragment that lacked the first 230 aa restored XMAP215msps-GFP localization, whereas the growth of these GFP-positive MTs remained slower than that of the control MTs (Fig. 1 C). Immunostaining of endogenous XMAP215msps and quantification of the signal intensity showed partial recovery of XMAP215msps accumulation at the tip (Fig. S1 B). Thus, XMAP215msps recruitment by Sentin might not be sufficient to fully rescue the fast-growing MT phenotype.
Distinct effect of Sentin and XMAP215msps on astral MT length control
We have previously shown that astral MTs are less dynamic after Sentin RNAi treatment (Li et al., 2011). However, on performing side-by-side analysis of asters after Sentin RNAi treatment and asters after XMAP215msps RNAi treatment, we noticed that the Sentin asters looked longer. To quantify this difference, monopolar spindles were induced by kinesin-5 (Klp61F) RNAi. The use of kinesin-5 RNAi minimized the possibility that astral MT length was affected by the size of the centrosome; in this cell line, spindles with three or more centrosomes of unequal sizes are frequently observed (Goshima and Vale, 2003). The lengths of the astral MTs that had been generated from singly clustered centrosomes were measured in the absence of Sentin, XMAP215msps, or both (Fig. 1 D). We found that MTs were longest in the absence of Sentin, shortest in the absence of XMAP215msps, and of intermediate length in the double RNAi condition. The appearance of opposite phenotypes after Sentin and XMAP215msps RNAi treatments further supports the notion that Sentin functions in more than merely XMAP215msps recruitment.
Drosophila XMAP215msps has a polymerization-promoting activity, similar to the Xenopus orthologue
We aimed to determine how XMAP215msps, EB1, and Sentin regulate MT dynamics by using the in vitro MT polymerization assay (Video 1). We first purified full-length Drosophila XMAP215msps tagged with HA or GFP and mixed this with tubulin and MT seeds (Fig. 2 A). XMAP215msps-GFP (>20 nM) localized to both ends of MTs throughout the dynamic cycle, stayed at the end of the MT seeds when no MT growth events occurred, and showed occasional MT lattice binding (Fig. 2 B, top). These behaviors are consistent with those of Xenopus XMAP215 in vitro (Brouhard et al., 2008).
In vitro MT polymerization assay with purified tubulin, XMAP215msps, EB1, and Sentin. Images were acquired by total internal reflection fluorescence microscopy (Nikon) every 3 s. EB1, 400 nM. XMAP215msps, 100 nM. GFP-Sentin, 200 nM. Red, rhodamine-labeled tubulin. Green, GFP-Sentin. Bar, 5 µm.
In vitro MT polymerization assay with purified tubulin, XMAP215msps, EB1, and Sentin. Images were acquired by total internal reflection fluorescence microscopy (Nikon) every 3 s. EB1, 400 nM. XMAP215msps, 100 nM. GFP-Sentin, 200 nM. Red, rhodamine-labeled tubulin. Green, GFP-Sentin. Bar, 5 µm.
Drosophila XMAP215 and EB1 promote MT growth and catastrophe, respectively. (A) Drosophila XMAP215msps and EB1 recombinants used in this study. (B, top) Kymographs showing constant MT end localization of XMAP215msps-GFP during growth, shrinkage, and the phase in which no MT growth event took place. Lattice binding was also sometimes observed. (bottom) EB1 exclusively localized to the growing ends of MTs. (C and D) Parameters of MT polymerization dynamics in the presence of 15 µM tubulin and various concentrations of XMAP215msps-HA or EB1 (untagged). Relative values are plotted in these graphs, whereas the actual values are presented in Tables S1, S2, and S3. The mean values of each experiment are marked in gray, whereas the mean values of all the experiments are indicated by black marks. Representative kymographs are shown on the top. See Fig. S2 for the XMAP215msps data using 7.5 µM tubulin. Data from a single experiment are marked with the same shape. Bars: (horizontal) 5 µm; (vertical) 1 min.
Drosophila XMAP215 and EB1 promote MT growth and catastrophe, respectively. (A) Drosophila XMAP215msps and EB1 recombinants used in this study. (B, top) Kymographs showing constant MT end localization of XMAP215msps-GFP during growth, shrinkage, and the phase in which no MT growth event took place. Lattice binding was also sometimes observed. (bottom) EB1 exclusively localized to the growing ends of MTs. (C and D) Parameters of MT polymerization dynamics in the presence of 15 µM tubulin and various concentrations of XMAP215msps-HA or EB1 (untagged). Relative values are plotted in these graphs, whereas the actual values are presented in Tables S1, S2, and S3. The mean values of each experiment are marked in gray, whereas the mean values of all the experiments are indicated by black marks. Representative kymographs are shown on the top. See Fig. S2 for the XMAP215msps data using 7.5 µM tubulin. Data from a single experiment are marked with the same shape. Bars: (horizontal) 5 µm; (vertical) 1 min.
We measured the parameters of MT dynamics under several conditions with different concentrations of XMAP215msps and tubulin. In the presence of 7.5–15 µM tubulin alone, the MTs were dynamic in terms of growth, shrinkage, and catastrophe events, but rescue events were never observed. Consistent with the result of the Xenopus XMAP215 polymerase (Brouhard et al., 2008), the addition of XMAP215msps increased MT growth rate over a wide range of tubulin concentrations (for example, 1.99 ± 0.49–fold, ±SD; P < 0.009 at 100 nM XMAP215msps and 7.5 µM tubulin; n = 4 experiments, 4–117 events from 8–20 MTs were analyzed in each experiment; Figs. 2 C, and S2, A and C; and Tables S1 and S2). MT growth was observed with XMAP215msps, even in the presence of only 3–6 µM tubulin; at these low concentrations, tubulin scarcely polymerized on its own (for example, at a 3-µM concentration of tubulin, none of the 74 seeds showed MT growth, whereas 3 of 30 seeds showed MT growth in the presence of XMAP215msps ; Fig. S2 B). This was also consistent with those obtained in a previous study for Xenopus XMAP215 (Brouhard et al., 2008). In contrast, we could not see a reproducible effect on catastrophe frequency and shrinkage rate by the addition of XMAP215msps (Figs. 2 C and S2 A and Tables S1 and S2). Similar to the tubulin-alone condition, MT rescue was not detected (no clear rescue events were observed in a total of 412 shrinkage events). We conclude that Drosophila XMAP215 has potent activity with respect to promoting and accelerating MT growth.
Drosophila EB1 has weak catastrophe-promoting and growth acceleration activities
The effect of human or yeast EB1 family proteins on MT plus-end dynamics in vitro is very controversial. In some studies, the addition of EB1 accelerates MT growth, whereas in others, this activity has not been detected or the opposite activity has been reported (Bieling et al., 2007; Manna et al., 2008; Vitre et al., 2008; Dixit et al., 2009; Komarova et al., 2009; Zhu et al., 2009). A recent study has suggested that the discrepancy might be caused by the presence or absence of the His tag on EB1, wherein this artificial tag confers growth-promoting activity to EB1 (Zhu et al., 2009). We purified Drosophila EB1 without His tags, mixed it with tubulin, and assessed its molecular activity in four independent experiments (the autonomous plus end–tracking ability of Drosophila EB1 was confirmed by using purified EB1-GFP; Fig. 2, A and B, bottom). Perhaps consistent with the controversial nature of this protein’s activity, some extent of variation was also observed in our study (Fig. 2 D shows a typical kymograph and a complete dataset; also see Table S3). However, in the four independent experiments with 15 µM tubulin, mild growth acceleration was observed (1.19 ± 0.15–fold increase in growth rate, ±SD; P < 0.05 at 800 nM; 10–42 events from 7–20 MTs were analyzed in each experiment). A more prominent effect was generally detected for catastrophe frequency (1.49 ± 0.35–fold increase, ±SD; P < 0.04 at 800 nM; 9–35 events from 7–20 MTs in each experiment), which is consistent with the results of several previous studies performed using yeast or mammalian EB1 orthologues (Bieling et al., 2007; Vitre et al., 2008; Komarova et al., 2009). In contrast, the shrinkage rate did not significantly change in a reproducible manner, which may be consistent with the fact that EB1 is not localized at the tip of shrinking MTs. We conclude that Drosophila EB1 has the ability to promote catastrophe for growing MTs, and it can modestly accelerate growth.
Sentin further increases MT growth rate and catastrophe frequency in the presence of EB1 in vitro
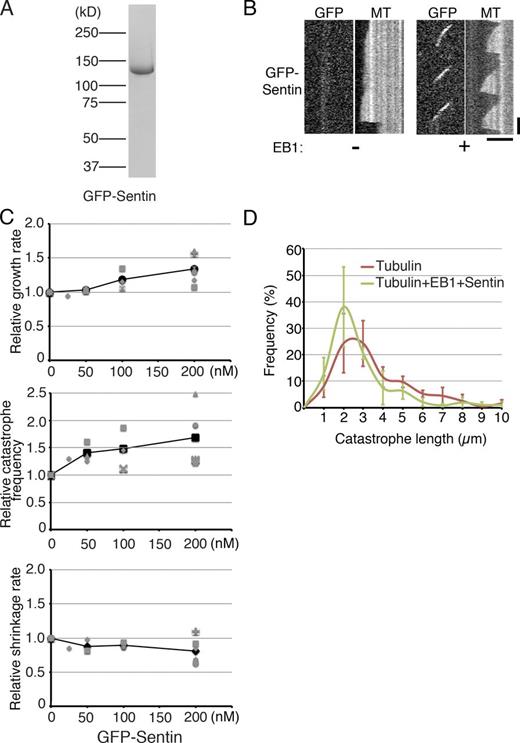
Next, we purified GFP-tagged full-length Sentin, the functionality of which was proven in the rescue experiment in cells (Figs. 3 A and S1 A). We confirmed the EB1-dependent plus end–tracking behavior of full-length GFP-Sentin in vitro (Fig. 3 B). Unlike what was observed with XMAP215msps, the localization of GFP-Sentin was only detected on growing MTs, and no clear lattice binding was observed.
Sentin accelerates MT growth and induces catastrophe in the presence of EB1. (A) Sentin recombinants used in this study. mGFP sequences were added to the N terminus. (B) Kymographs showing GFP-Sentin localization onto the growing MT plus ends in an EB1-dependent manner. Bars: (horizontal) 5 µm; (vertical) 1 min. (C) Parameters of MT polymerization dynamics in the presence of various concentrations of Sentin. Tubulin, 15 µM; EB1, 400 nM. The mean values of each experiment are marked in gray, whereas the mean values of all the experiments are indicated by black marks. The relative values are plotted in these graphs, whereas the actual values are presented in Table S4. Data from a single experiment are marked with the same shape. (D) Catastrophe length, the MT length at the time of catastrophe, was measured in the presence or absence of 400 nM EB1 and 200 nM Sentin. Tubulin was used at a 15-µM concentration. The frequency was plotted with SEM (n = 3 experiments; bin = 1 µm).
Sentin accelerates MT growth and induces catastrophe in the presence of EB1. (A) Sentin recombinants used in this study. mGFP sequences were added to the N terminus. (B) Kymographs showing GFP-Sentin localization onto the growing MT plus ends in an EB1-dependent manner. Bars: (horizontal) 5 µm; (vertical) 1 min. (C) Parameters of MT polymerization dynamics in the presence of various concentrations of Sentin. Tubulin, 15 µM; EB1, 400 nM. The mean values of each experiment are marked in gray, whereas the mean values of all the experiments are indicated by black marks. The relative values are plotted in these graphs, whereas the actual values are presented in Table S4. Data from a single experiment are marked with the same shape. (D) Catastrophe length, the MT length at the time of catastrophe, was measured in the presence or absence of 400 nM EB1 and 200 nM Sentin. Tubulin was used at a 15-µM concentration. The frequency was plotted with SEM (n = 3 experiments; bin = 1 µm).
The parameters of MT plus-end dynamics were determined in the presence or absence of Sentin on the basis of kymographs. When 0–200 nM Sentin was added in the presence of 400 nM EB1 and 15 µM tubulin, the growth rate and the catastrophe frequency increased in a concentration-dependent manner (1.34 ± 0.21– and 1.68 ± 0.51–fold, respectively, ±SD; P < 0.003 and 0.009 at 200 nM; n = 6 experiments, 23–121 events from 10–35 MTs were analyzed in each experiment; Fig. 3 C and Table S4). The growth acceleration activity of EB1–Sentin (∼1.6-fold higher compared with that with 15 µM tubulin alone) was comparable to that of XMAP215msps (∼1.7-fold) at this tubulin concentration. However, unlike what was observed with XMAP215msps, the addition of 400 nM EB1 and 200 nM Sentin could not initiate MT growth at the lower tubulin concentration (Fig. S4 A). Sentin slightly reduced the shrinkage rate (80%); however, the implication of this observation is unclear. These data indicate that Sentin, via EB1 recruitment, accelerates MT growth and increases catastrophe frequency in vitro, independent of XMAP215msps.
A recent study suggested that MT catastrophe is a multistep process and that its frequency depends on the age of the growing MT (Gardner et al., 2011). It was also shown that the potent catastrophe factor mitotic centromere-associated kinesin (MCAK; kinesin-13) eliminates the aging process, whereas Kip3 (kinesin-8) increases the aging rate. We measured MT length at the time of each catastrophe event (catastrophe length), in the presence or absence of EB1–Sentin. Unlike the results reported for MCAK addition, the addition of EB1 and Sentin did not increase the frequency of the appearance of MTs with very short catastrophe length, suggesting that EB1–Sentin does not trigger catastrophe through a mechanism similar to that of MCAK (Fig. 3 D).
Next, by using gel filtration chromatography, we tested whether Sentin binds to free tubulin dimers. In this experiment, neither full-length Sentin nor two truncated Sentin fragments (231–982 and 1–440 aa) clearly co-migrated with tubulin to lower fractions, suggesting that Sentin protein does not have the ability to stably bind to tubulin (Fig. S3).
Combined action of XMAP215msps, EB1, and Sentin maximizes MT dynamicity
The in vitro results thus far indicate that the EB1–Sentin duo and XMAP215msps possess similar (growth acceleration) as well as distinct (catastrophe induction and growth initiation, respectively) activities. We next mixed them with tubulins, which would better mimic the cellular condition, and assessed the effect on MT dynamics and localization dependency.
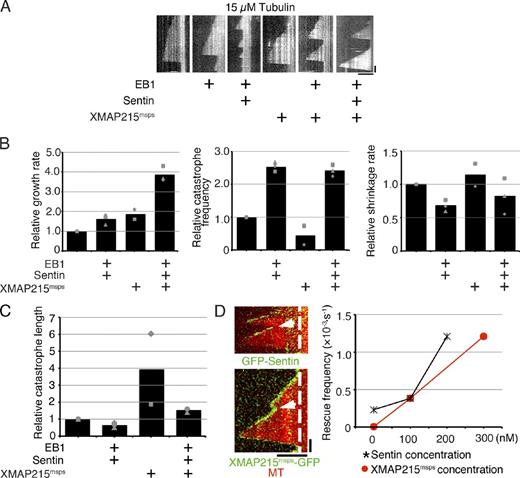
XMAP215msps-HA was mixed with GFP-Sentin, EB1, and 15 µM tubulin. We observed a 3.86 ± 0.40–fold higher growth rate than that obtained under the tubulin-alone condition (±SD; P < 0.0003; n = 3 experiments, 11–85 events from 10–20 MTs were analyzed in each experiment). This value was higher than that for any other combination of proteins, indicating that the presence of each protein is critical to maximize the growth rate (2.4-fold vs. EB1–Sentin addition, 1.6-fold vs. EB1-XMAP215msps addition, and 2.2-fold vs. XMAP215msps addition; Fig. 4, A and B; Video 1; and Table S5). A comparable level of catastrophe frequency was observed, relative to that for the EB1–Sentin-alone condition (∼2.5-fold vs. tubulin alone; n = 3 experiments, 4–80 events from 10–20 MTs were analyzed in each experiment). A similar result was obtained when a lower tubulin concentration (7.5 µM) was used, at which MT growth was rarely seen without XMAP215msps (1.5- and 1.6-fold increase in growth rate and catastrophe frequency vs. XMAP215msps; n = 3 experiments, 6–42 events from 8–20 MTs were analyzed in each experiment; Fig. S4, A and B; and Table S6). Consequently, we found that the catastrophe length in the presence of the three factors was intermediate between those for the XMAP215msps-alone and the EB1–Sentin-alone conditions (Fig. 4 C).
Synergistic growth acceleration and rescue induced by XMAP215msps, EB1, and Sentin. (A) Kymographs showing MT polymerization dynamics with various combinations of EB1, Sentin, and XMAP215msps in the presence of 15 µM tubulin. See also Video 1. (B) Parameters of MT polymerization dynamics. The mean values of each experiment are marked in gray, whereas the mean values of all the experiments have been indicated by black bars. The relative values are shown in these graphs, whereas the actual values are presented in Table S5. 400 nM EB1, 200 nM Sentin, and 100 nM XMAP215msps-HA were added to tubulin. Some of the data (e.g., tubulin + XMAP215msps-HA) overlapped with those in Figs. 2 and 3. (C) Mean catastrophe length in the presence or absence of EB1–Sentin or XMAP215msps. The identical dataset was used as in B. The mean values of each experiment are marked in gray, whereas the mean values of all the experiments are indicated by black bars. The relative values are shown in these graphs, whereas the actual values are presented in Table S5. (D, left) MT rescue (arrows) in the presence of EB1, Sentin, and XMAP215msps. The dashed lines indicate the ends of MT seeds. (right) XMAP215msps- and Sentin-dependent increase in rescue frequency. In the presence of 15 µM tubulin, 400 nM EB1, and 200 nM Sentin (or 300 nM XMAP215msps), various amounts of XMAP215msps (or Sentin) were added. More rescue events were observed with increasing concentrations of Sentin and XMAP215msps. The data shown are from a single representative experiment out of three repeats. Data from a single experiment are marked with the same shape. Bars: (horizontal) 5 µm; (vertical) 1 min.
Synergistic growth acceleration and rescue induced by XMAP215msps, EB1, and Sentin. (A) Kymographs showing MT polymerization dynamics with various combinations of EB1, Sentin, and XMAP215msps in the presence of 15 µM tubulin. See also Video 1. (B) Parameters of MT polymerization dynamics. The mean values of each experiment are marked in gray, whereas the mean values of all the experiments have been indicated by black bars. The relative values are shown in these graphs, whereas the actual values are presented in Table S5. 400 nM EB1, 200 nM Sentin, and 100 nM XMAP215msps-HA were added to tubulin. Some of the data (e.g., tubulin + XMAP215msps-HA) overlapped with those in Figs. 2 and 3. (C) Mean catastrophe length in the presence or absence of EB1–Sentin or XMAP215msps. The identical dataset was used as in B. The mean values of each experiment are marked in gray, whereas the mean values of all the experiments are indicated by black bars. The relative values are shown in these graphs, whereas the actual values are presented in Table S5. (D, left) MT rescue (arrows) in the presence of EB1, Sentin, and XMAP215msps. The dashed lines indicate the ends of MT seeds. (right) XMAP215msps- and Sentin-dependent increase in rescue frequency. In the presence of 15 µM tubulin, 400 nM EB1, and 200 nM Sentin (or 300 nM XMAP215msps), various amounts of XMAP215msps (or Sentin) were added. More rescue events were observed with increasing concentrations of Sentin and XMAP215msps. The data shown are from a single representative experiment out of three repeats. Data from a single experiment are marked with the same shape. Bars: (horizontal) 5 µm; (vertical) 1 min.
In addition to the effects on growth rate and catastrophe frequency, we also recorded a few unexpected observations during these experiments. First, the rescue event was observed to occur much more frequently when XMAP215msps, EB1, and Sentin were added together (eight rescue events were observed against a total of 52 shrinking events, when maximum amounts of Sentin and XMAP215msps were added; Fig. 4 D and Table S7). The occurrence of rescue was considered to be overwhelmingly dependent on the presence of XMAP215msps because no rescue events were observed for shrinking MTs in the presence of only EB1 and Sentin. However, XMAP215msps alone also did not exhibit this activity.
Second, we noticed a very weak but detectable level of MT plus-end accumulation, as well as lattice binding of GFP-Sentin, in the presence of XMAP215msps but in the absence of EB1 (total of 53% of the growing and 14% of the shrinking MTs had GFP-Sentin signals, n = 38 MTs; Fig. 5 A). Because Sentin alone does not show plus-end localization, the data suggest that Sentin is recruited through direct binding to XMAP215msps. In support of this idea, Sentin was coprecipitated with XMAP215msps in the immunoprecipitation experiment using insect cells (Fig. 5 B). However, we could not detect a significant change in MT dynamics parameters, such as growth rate or catastrophe frequency, compared with the case in which XMAP215msps alone was present (Table S5). EB1-GFP was also occasionally (8%; n = 71) detected on the shrinking MTs in the presence of Sentin and XMAP215msps, suggesting the Sentin-mediated connection of EB1 to XMAP215msps (Fig. 5 C).
Interdependence of XMAP215msps and Sentin localization. (A) XMAP215msps recruits Sentin to MT plus ends and the MT lattice independent of EB1. However, GFP-Sentin localization was far weaker in the absence of EB1. Inset shows enlarged view of the boxed area. Arrows in the inset indicate GFP-Sentin signals. (B) Coprecipitation of XMAP215msps and Sentin. XMAP215msps-HA and GFP-Sentin were coexpressed in Sf21 cells, and immunoprecipitation (IP) was performed with anti-HA antibodies. Protein bands after Coomassie staining are displayed (arrow, Sentin; sphere, XMAP215msps). (C) Some shrinking MTs showed EB1-GFP signals in the presence of XMAP215msps and Sentin. (D) Sentin facilitates localization of a low concentration of XMAP215msps (5 nM) to the plus ends. (right) Quantification of the XMAP215msps-GFP intensity at the growing MT tip in the presence or absence of Sentin. The data shown are from a single representative experiment out of two repeats. A.U., arbitrary unit. Bars: (horizontal) 5 µm; (vertical) 1 min.
Interdependence of XMAP215msps and Sentin localization. (A) XMAP215msps recruits Sentin to MT plus ends and the MT lattice independent of EB1. However, GFP-Sentin localization was far weaker in the absence of EB1. Inset shows enlarged view of the boxed area. Arrows in the inset indicate GFP-Sentin signals. (B) Coprecipitation of XMAP215msps and Sentin. XMAP215msps-HA and GFP-Sentin were coexpressed in Sf21 cells, and immunoprecipitation (IP) was performed with anti-HA antibodies. Protein bands after Coomassie staining are displayed (arrow, Sentin; sphere, XMAP215msps). (C) Some shrinking MTs showed EB1-GFP signals in the presence of XMAP215msps and Sentin. (D) Sentin facilitates localization of a low concentration of XMAP215msps (5 nM) to the plus ends. (right) Quantification of the XMAP215msps-GFP intensity at the growing MT tip in the presence or absence of Sentin. The data shown are from a single representative experiment out of two repeats. A.U., arbitrary unit. Bars: (horizontal) 5 µm; (vertical) 1 min.
Finally, when we used a very low concentration of XMAP215msps-GFP (5 nM) with unlabeled EB1 and Sentin, we observed plus-end accumulation of XMAP215msps-GFP more frequently in the presence of EB1 and Sentin; at 5 nM, we detected GFP signals on a total 88% of growing or 35% of shrinking MTs in the presence of Sentin but only 43 or 14% in its absence (n = 38–45 MTs; Fig. 5 D). The analysis of XMAP215msps-GFP intensity at the growing ends confirmed this conclusion (Fig. 5 D, right). This result suggests that Sentin facilitates XMAP215msps accumulation at the plus end, which is similar to the occurrence in vivo.
Interaction between Sentin and XMAP215msps may contribute to the synergistic effect
The dramatic increase in the growth rate and rescue frequency might be caused by the synergistic effect of EB1–Sentin and XMAP215msps because the increase in the growth rate (∼3.9-fold) by EB1–Sentin and XMAP215msps was larger than the sum or product of the increase brought by each protein (1.6-fold by EB1–Sentin and 1.7-fold by XMAP215msps). Moreover, the rescue events were never observed in the presence of EB1–Sentin or XMAP215msps alone. Because Sentin and XMAP215msps facilitate plus-end accumulation of each other, possibly through their direct interaction, one hypothesis is that this interaction contributes to the synergistic effects of EB1–Sentin and XMAP215msps.
To address this hypothesis, we evaluated the activities of two truncated Sentin proteins, GFP-Sentin-N (1–440 aa) and GFP-Sentin-ΔN (231–982 aa; Fig. 6 A). Because the Sentin-N fragment lacked the EB1-binding C-terminal region (Li et al., 2011), it did not track MT ends or accelerate MT growth in the presence of EB1 alone (Fig. 6 B). However, we found that this fragment tracks plus ends of MTs in the presence of XMAP215msps as efficiently as full-length Sentin (total 47%, n = 48 growing MTs), suggesting that this region of Sentin is responsible for interaction with XMAP215msps (Fig. 6 B). Similar to full-length Sentin, however, Sentin-N could not further activate or inhibit XMAP215msps or EB1–Sentin for MT growth acceleration (Fig. S5 A). Interestingly, when Sentin-N was added to the mixture of full-length Sentin, EB1, and XMAP215msps, the synergistic growth acceleration or rescue was significantly suppressed (Figs. 6 C and S5 B). A possible interpretation of this result is that Sentin-N acts as a dominant-negative fragment, interfering with the interaction between EB1-bound, full-length Sentin and XMAP215msps.
The Sentin-N fragment interferes with synergistic growth acceleration and rescue induced by XMAP215msps and EB1–Sentin. (A) GFP-Sentin-N (1–440 aa) and GFP-Sentin-ΔN (231–982 aa) recombinants used in this study. (B) Tracking of Sentin-N or Sentin-ΔN in the presence of EB1 or XMAP215msps. (C) The Sentin-N fragment (600 nM) interfered with growth and rescue induced by 100 nM XMAP215msps (XM), 400 nM EB1 (EB), and 200 nM full-length Sentin (Sen) at 15 µM tubulin. The mean values of each experiment are marked, and the mean values of all the experiments are indicated by black or red bars. For the growth rate, 21–38 events from 10–20 MTs were analyzed in each experiment (n = 2 experiments). None of 35 or 3 of 59 shrinking MTs showed rescue in the presence or absence of GFP-Sentin-N, respectively. (D) 200 nM Sentin-ΔN weakly accelerates growth and promotes rescue in the presence of EB1 and XMAP215msps. Growth rate was 1.2 ± 0.2–fold at 15 µM tubulin (±SD; P < 0.04; n = 6 experiments, 11–43 events from 10–20 MTs were analyzed in each experiment). Rescue occurred more frequently with Sentin-ΔN (3 of 129 or 1 of 134 shrinking MTs showed rescue). (E) Model for the independent as well as cooperative action of XMAP215 polymerase and the EB1–Sentin duo that has growth acceleration and catastrophe promotion activities. N, N terminus; C, C terminus. Data from a single experiment are marked with the same shape. Bars: (horizontal) 5 µm; (vertical) 1 min.
The Sentin-N fragment interferes with synergistic growth acceleration and rescue induced by XMAP215msps and EB1–Sentin. (A) GFP-Sentin-N (1–440 aa) and GFP-Sentin-ΔN (231–982 aa) recombinants used in this study. (B) Tracking of Sentin-N or Sentin-ΔN in the presence of EB1 or XMAP215msps. (C) The Sentin-N fragment (600 nM) interfered with growth and rescue induced by 100 nM XMAP215msps (XM), 400 nM EB1 (EB), and 200 nM full-length Sentin (Sen) at 15 µM tubulin. The mean values of each experiment are marked, and the mean values of all the experiments are indicated by black or red bars. For the growth rate, 21–38 events from 10–20 MTs were analyzed in each experiment (n = 2 experiments). None of 35 or 3 of 59 shrinking MTs showed rescue in the presence or absence of GFP-Sentin-N, respectively. (D) 200 nM Sentin-ΔN weakly accelerates growth and promotes rescue in the presence of EB1 and XMAP215msps. Growth rate was 1.2 ± 0.2–fold at 15 µM tubulin (±SD; P < 0.04; n = 6 experiments, 11–43 events from 10–20 MTs were analyzed in each experiment). Rescue occurred more frequently with Sentin-ΔN (3 of 129 or 1 of 134 shrinking MTs showed rescue). (E) Model for the independent as well as cooperative action of XMAP215 polymerase and the EB1–Sentin duo that has growth acceleration and catastrophe promotion activities. N, N terminus; C, C terminus. Data from a single experiment are marked with the same shape. Bars: (horizontal) 5 µm; (vertical) 1 min.
In contrast, GFP-Sentin-ΔN efficiently tracked plus ends of growing MTs in the presence of EB1 (total 85%, n = 86 growing MTs) and less efficiently in the presence of XMAP215msps alone (13%, n = 57 growing MTs; Fig. 6 B). We found that this fragment does not further accelerate the EB1-dependent growth (i.e., Sentin-ΔN does not possess its own growth acceleration activity; Fig. S5 C). However, it still mildly accelerated growth with XMAP215msps and EB1 and induced more rescue (Fig. 6 D). This result is consistent with the model that the Sentin–XMAP215msps interaction produces the synergistic effect.
Discussion
In this study, we revealed the in vitro growth-accelerating and catastrophe-promoting activities of Drosophila EB1 and Sentin toward growing MTs, in the presence or absence of XMAP215msps, which has been shown to be a potent MT growth promoter. The combination of these two machineries, which likely interact with each other at the MT plus ends, produced MTs with the greatest dynamicity; this observation is consistent with the fact that either of the two machineries is critical for the formation of dynamic MTs in S2 cells (Fig. 6 E). Furthermore, our localization data may reinforce the proposal that Sentin, for which no homologous genes have been identified thus far outside the insect species, might be the functional homologue of the mammalian Slain2 protein that was recently shown to be important for MT growth in cells and was observed to interact with XMAP215 as well as EB1 (Short, 2011; van der Vaart et al., 2011). Thus, this study takes a step forward in understanding the molecular mechanism underlying the regulation of MT dynamics by multiple nontubulin proteins and, moreover, suggests that the mechanism may be widely conserved in animal cells.
Common and distinct activities of Drosophila XMAP215msps, EB1, and Sentin
The growth-promoting activity of XMAP215msps is generally consistent with that of the best-characterized Xenopus orthologue (Brouhard et al., 2008). The approximately twofold increase in MT growth rate detected in this study might be an underestimation, given that pig brain tubulins were used for Drosophila proteins. However, together with the appearance of severe RNAi phenotypes, it is highly probable that XMAP215msps is the major MT polymerase of Drosophila.
EB1 and Sentin had two activities, one that overlaps with the activity of kinesin-13 (catastrophe promotion) and the other with that of XMAP215 (growth acceleration). We believe that these activities would be associated with their in vivo functions because the RNAi of EB1 or Sentin in the S2 cells decreased both growth speed and catastrophe frequency (Rogers et al., 2002; Li et al., 2011). However, our data do not go against the view that XMAP215 and kinesin-13 are the primary growth- and catastrophe-promoting factors, respectively (Kinoshita et al., 2001; Howard and Hyman, 2007). In fact, we propose that the EB1–Sentin machinery would act as a supporter of the core XMAP215–kinesin-13 machinery by accelerating MT growth as well as MT aging for catastrophe and that S2 cells do need both mechanisms to make MTs fully dynamic. This hierarchical relationship might be conserved in vertebrates because MT phenotypes observed after EB1 depletion are rescued by the overexpression of XMAP215 in Xenopus egg extracts (Niethammer et al., 2007).
The growth acceleration on addition of both EB1 and Sentin was comparable with that observed after addition of XMAP215msps. However, we do not consider the EB1–Sentin complex to be a processive polymerase like XMAP215 for a few reasons. First, we did not see growth-promoting activity at a low tubulin concentration in the presence of EB1 and Sentin. Second, unlike the case of XMAP215, we could not obtain data to indicate that Sentin stably binds to soluble tubulins. Third, the known residence time of XMAP215 and EB1 on the MT ends is very different. The average end residence time of a single XMAP215 molecule is 3.8 s, which is long enough to processively add 25 tubulin dimers to the end of each MT protofilament (Brouhard et al., 2008). In contrast, the residence time of yeast and mammalian EB1 family proteins is <0.4 s, which is too short to processively add multiple tubulins (Bieling et al., 2007, 2008; Montenegro Gouveia et al., 2010). A recent structural study showed that EB1 recognizes a structural feature on the MT surface near the plus end and catalyses the transition of tubulin from the recognized state at growing ends to the state tubulin adopts in the lattice distant from the ends, which is assumed to be the basis of the EB1 activity (Maurer et al., 2011, 2012). Sentin bound to EB1 might modulate the plus-end structure for favorable tubulin incorporation, rather than acting as a polymerase. An interesting future experiment would be to determine the structural basis of EB1-recruiting Sentin in this context.
Control of MT dynamics by the combined action of XMAP215msps, EB1, and Sentin
When XMAP215msps, EB1, and Sentin were all combined, we observed the most dynamic MTs thus far; growth rate and catastrophe frequency were maximized, and rescue events were occasionally observed. Interestingly, the dramatic increase in the growth rate and rescue frequency might be because of the synergistic effects of EB1–Sentin and XMAP215msps. On the basis of the experiments using the truncated Sentin fragments, it is tempting to speculate that the physical interaction of Sentin with XMAP215msps facilitates localization of both proteins at the MT ends and modulates their activities, thereby contributing to these synergistic effects. However, other models, including a model in which the XMAP215msps and EB1–Sentin machineries work independently, cannot yet be excluded at this stage.
We could obtain the key features of MT dynamic instability—growth, shrinkage, catastrophe, and rescue—by in vitro combination of three plus-end factors. Our reconstitution, however, still failed to precisely reproduce what happens in vivo. For example, pausing MTs are frequently observed in cells, particularly in interphase (∼40%), but this behavior was rarely seen in any conditions in our in vitro assay. In S2 cells, at least two other factors, kinesin-13 depolymerase and the CLASP rescue factor, are known to significantly affect interphase MT dynamics probably in an EB1-independent manner (Mennella et al., 2005; Sousa et al., 2007; Li et al., 2011). Although CLASP is mostly concentrated at the kinetochore during mitosis and perhaps does not significantly affect the astral MT dynamics, kinesin-13 acts on every MT in every stage of the cell cycle (Lemos et al., 2000; Goshima and Vale, 2005). A next step toward understanding the mechanism controlling MT plus-end dynamics would be to include these two factors in the assay and determine whether they can better reproduce the cellular MT behavior that has, for example, frequent pause events.
Materials and methods
Plasmids, cell culture, and RNAi
Plasmids for GFP, His, or HA fusion proteins expressed in Sf21 or S2 cells were constructed using the Gateway system (Invitrogen), and the plasmids for bacterial protein expression were prepared using conventional DNA ligation (Table S8). Drosophila S2 cell culture, RNAi treatment, transfection, and the rescue experiments were performed using previously described methods (Bettencourt-Dias and Goshima, 2009; Goshima, 2010; Li et al., 2011). In brief, S2 cells were cultured in Schneider’s medium (Invitrogen) supplemented with 10% serum and 1% antibiotics at 23–24°C. Cell lines stably expressing GFP-, mRFP-, or mCherry-tagged proteins were obtained after transfection with the reagent (Cellfectin II; Invitrogen) followed by hygromycin selection. For RNAi treatment, cells cultured in 96-well plates were incubated for 50 min in 50 µl serum-free medium containing 2 µg dsRNA followed by adding back 50 µl medium containing 20% serum and CuSO4 (0–132 µM, depending on the desired expression level). Mitotic arrest for spindle observation was achieved by adding Cdc27 dsRNA during RNAi treatment. dsRNA information has been provided in Table S9. Anti-EB1 antibody is a gift from S. Rogers (University of North Carolina at Chapel Hill, Chapel Hill, NC). The Bac-to-Bac expression system (Invitrogen) was used to construct viral DNA. Spodoptera frugiperda insect cells (Sf9) were cultured in Grace’s insect medium (Invitrogen) with 5% serum and 1% antibiotics at 27°C and were transfected with bacmids by using Cellfectin II. The subsequent baculovirus amplification and protein expression were performed with Sf21 cells that were cultured in Sf900II medium supplemented with 5% serum and 1% antibiotics.
Cell imaging and image analyses
Cells were observed at days 4–6 after RNAi treatment, after being spread on glass-bottom concanavalin A–coated plates. For immunofluorescence microscopy, the cells were fixed with paraformaldehyde and stained with an antitubulin antibody (DM1A; Sigma-Aldrich). When XMAP215msps was stained, methanol fixation was applied instead (antibody was a gift of H. Ohkura, University of Edinburgh, Edinburgh, Scotland, UK). For live imaging of GFP-tagged XMAP215msps, EB1, or Sentin in the cytoplasm, the cells were treated with 10 µg/ml cytochalasin D to eliminate the F-actin network immediately before imaging. The images were collected by a spinning-disk confocal microscope (TE2000 [Nikon]; CSU-X1 [Yokogawa Corporation of America]) equipped with an EM charge-coupled device camera (ImagEM; Hamamatsu Photonics) and a 100×, 1.40 NA lens by using the Micro-Manager software at 24 ± 1°C (Li et al., 2011). The MT growth rate (Fig. 1, B and C) was determined by generating a kymograph in which the signals of EB1-GFP or RFP-Sentin truncations could be traced. For the XMAP215msps-Sentin colocalization experiment (Figs. 1 C and S1 B), the growing plus end was identified by GFP (RFP)-Sentin, and the signal intensity of XMAP215msps at the identical position was measured. Spindle length was defined as the distance between two spindle poles, not centrosomes, and the relative spindle length was calculated by dividing the mean spindle length in each treatment by that of control RNAi-treated cells (Li et al., 2011).
Protein purification and immunoprecipitation
EB1 was purified using the Escherichia coli BL21-AI strain as described previously (Li et al., 2011). In brief, GST-EB1 was purified by using glutathione–Sepharose beads followed by proteolytic cleavage of the GST tag. One modification was that gel filtration chromatography was used as the final step instead of dialysis. EB1-GFP and Sentin-N (1–440-aa fragments) were also purified from E. coli with a similar method. His-mGFP-Sentin, His-Sentin, His-mGFP-Sentin-N (1–440 aa), His-mGFP-Sentin-ΔN (231–982 aa), or XMAP215msps-mGFP (or HA)-His was expressed in insect Sf21 cells. Uninfected Sf21 cells in an Erlenmeyer flask (2 × 106/ml) were treated with virus and incubated with shaking for 3 d (120 rpm at 27°C). The cells were lysed using 5 ml (per 50-ml cell culture) lysis buffer (50 mM K-Hepes, 100 mM NaCl, 1 mM MgCl2, 1 mM EGTA, 1 mM DTT, 30 mM imidazole, and 1% Triton X-100, pH 7.6, with protease inhibitors). After incubation with nickel-coated beads (0.5-ml bed volume) for 1 h, the proteins were eluted using 500 µl of MRB80 buffer (80 mM K-Pipes, pH 6.8, 4 mM MgCl2, and 1 mM EGTA) with 300 mM KCl and 500 mM imidazole followed by gel filtration using the chromatography system (BioLogic DuoFlow; Bio-Rad Laboratories) with a Superdex 200 10/300 GL column (GE Healthcare; equilibrated with MRB80 containing 100 mM KCl and 1 mM DTT). The peak fraction was either flash frozen or kept on ice and used for experiments within 48 h (frozen samples were rarely used for Sentin). Immunoprecipitation was performed by transfecting baculoviruses encoding XMAP215msps-HA and GFP-Sentin followed by anti-HA immunoprecipitation in the lysis buffer (50 mM K-Hepes, pH 7.6, 100 mM NaCl, 1 mM MgCl2, 1 mM EGTA, 1 mM DTT, 1% Triton X-100, and protease inhibitors).
Gel filtration chromatography to evaluate tubulin binding
We mixed full-length or truncated Sentin with tubulin in the binding buffer (80 mM K-Pipes, pH 6.8, 75 mM KCl, 4 mM MgCl2, 1 mM EGTA, and 1 mM GTP) for 20–30 min. After 5× concentration of the solution by centrifugal filters (Amicon Ultra; Millipore) and centrifugation at 106,000 g for 15 min, the supernatant was loaded onto a Superdex 200 10/300 GL column equilibrated with a gel filtration buffer (80 mM K-Pipes, pH 6.8, 75 mM KCl, 4 mM MgCl2, 1 mM EGTA, 0.1 mM GTP, 0.1% Tween 20, and 1 mM DTT). Fractions were collected and analyzed using SDS-PAGE followed by Coomassie staining. The concentration step was necessary to visualize GFP-Sentin or GFP-Sentin-ΔN protein bands by Coomassie staining and also to separate each peak more clearly after fractionation, although the protein concentration increases during this process. The concentration step was skipped for the Sentin-N fragment.
In vitro tracking assay
The end-tracking assay was performed as previously described (Bieling et al., 2010; Gell et al., 2010), with some modifications. Flow chambers (22 mm [width] × 1 mm [height] × 0.15 mm [depth]) were assembled between a coverslip and a precleaned microslide glass with double-sided tape. The silanized coverslip was coated with antibiotin (1% in MRB80; Invitrogen), and the nonspecific surface was blocked with Pluronic F127 (1% in MRB80; Invitrogen). Biotinylated MT seeds (50 µM tubulin mix containing 10% biotinylated tubulin and 10% rhodamine-labeled tubulin with 1 mM guanosine-5′-[(α,β)-methylene] triphosphate) were specifically attached to the functionalized surface by biotinylated tubulin-antibiotin links. After the chamber was washed with MRB80, MT growth was initiated by flowing 3–20 µM tubulin (containing 3.2% rhodamine-labeled tubulin), 0–800 nM EB1, 0–200 nM GFP-Sentin, 0–600 nM GFP-Sentin-N, 0–200 nM GFP-Sentin-ΔN, and/or 0–300 nM XMAP215msps-HA/GFP into the assay buffer (MRB80, 75 mM KCl, 1 mM GTP, 0.5 mg/ml k-casein, and 0.1% methylcellulose; Sigma-Aldrich) and an oxygen scavenger system (50 mM glucose, 400 µg/ml glucose-oxidase, 200 µg/ml catalase, and 4 mM DTT). The samples were sealed with candle wax. During the experiments, the samples were kept at 25 ± 1°C; images were collected every 3 s for 20 min via total internal reflection fluorescence microscopy with a TE2000E or Ti system (Nikon), iXonEM+ 897 (Andor Technology), or Evolve (Roper Scientific), a 100×, 1.49 NA lens, and a 488/561-nm excitation laser. The microscopes were controlled by Micro-Manager. Kymographs were generated for MT seeds using the ImageJ software (National Institutes of Health), and the parameters of MT plus-end dynamics were determined as follows: We first identified the starting points of growth and catastrophe (shrinkage) and the end point of shrinkage in the kymograph. The duration of and change in MT length for each phase were measured, and the rate was determined by calculating the mean velocity between the start and end point of each phase. Catastrophe frequency was determined by dividing the number of shrinkage events by the total time in the growth and shrinkage phases (i.e., we subtracted the time during which dynamic MTs did not appear). The transition from shrinkage to growth was considered a rescue event, and the rescue frequency (for total time) was calculated. Some degrees of variation in MT behaviors were seen on different experiment days. The basis of this variation is unclear; however, differences in protein batches might be responsible. In fact, we noticed that different batches of tubulin could have considerably different abilities to polymerize; therefore, we used 9 µM instead of 7.5 µM in some experiments with low tubulin concentration to equalize the intrinsic polymerization ability and 20 µM instead of 15 µM for high concentration.
Data presentation
In most of the figures that display the results on the in vitro polymerization assay, we have provided the values for each condition relative to the control data acquired on the same day (Figs. 2, C and D; 3 C; 4, B and C; 6, C and D; S2 A; S4, B and C; and S5, A–C). In contrast, the mean values of all the experiments have been provided in Tables S1, S2, S3, S4, S5, S6 and S7. For example, when we obtained growth rates of a1, a2, and a3 (micrometers/minute) in three independent experiments using tubulin alone and b1, b2, and b3 (micrometers/minute) after 400 nM EB1 addition, we presented the relative values b1/a1, b2/a2, and b3/a3 in gray and (b1/a1 + b2/a2 + b3/a3)/3 in black in the figure. In the table, on the other hand, we provided the mean rates (a1 + a2 + a3)/3 and (b1 + b2 + b3)/3 separately with SDs.
Online supplemental material
Fig. S1 shows the effects of truncated Sentin on MT growth and XMAP215msps localization. Fig. S2 shows the effects of XMAP215msps on MT dynamics. Fig. S3 shows the result of the Sentin-tubulin–binding experiments. Fig. S4 shows the effects of EB1, Sentin, and XMAP215msps on MT dynamics. Fig. S5 shows the effects of truncated Sentin on MT dynamics. Table S1 shows kinetic parameters of MT polymerization dynamics in vitro in the presence of various concentrations of XMAP215msps-HA (0−100 nM) at 15 µM tubulin. Table S2 shows kinetic parameters of MT polymerization dynamics in vitro in the presence of various concentrations of XMAP215msps-HA (0–100 nM) at 7.5 µM tubulin. Table S3 shows kinetic parameters of MT polymerization dynamics in vitro in the presence of various concentrations of Drosophila EB1 (0–800 nM) at 15 µM tubulin. Table S4 shows kinetic parameters of MT polymerization dynamics in vitro in the presence of 400 nM EB1and various concentrations of GFP-Sentin (0–200 nM) at 15 µM tubulin. Table S5 shows kinetic parameters of MT polymerization dynamics in vitro in the presence or absence of 400 nM EB1, 200 nM GFP-Sentin, and 100 nM XMAP215msps-HA at 15 µM tubulin. Table S6 shows kinetic parameters of MT polymerization dynamics in vitro in the presence of 100 nM XMAP215msps-HA alone or additionally 400 nM EB1 and 200 nM GFP-Sentin at 7.5 µM tubulin. Table S7 shows kinetic parameters of MT polymerization dynamics in vitro in the presence of 400 nM EB1 and various concentrations of GFP-Sentin (0–200 nM) and XMAP215msps-HA (0–300 nM) at 15 µM tubulin. Table S8 shows plasmids used in this study. Table S9 shows primer sequences for RNAi used in this study. Video 1 shows representative MT behaviors in the presence of XMAP215msps, EB1, and/or Sentin.
Acknowledgments
We are grateful to Ikuko Sugiyama and Marija Zanic for their advice on the in vitro polymerization assay, Hiro Ohkura for the anti-Msps antibody, Steve Rogers for the anti-EB1 antibody, and Tomohiro Miki for technical help.
This work was supported by a Next Generation grant (Japan Society for the Promotion of Science), the Inoue Foundation, and the Human Frontier Science Program (to G. Goshima). W. Li was supported by the Global Centers of Excellence program, the Leading Graduate School program, and the State Scholarship Study Abroad Program of the Chinese Scholarship Council.