p120-catenin (p120) binds to the cytoplasmic tails of classical cadherins and inhibits cadherin endocytosis. Although p120 regulation of cadherin internalization is thought to be important for adhesive junction dynamics, the mechanism by which p120 modulates cadherin endocytosis is unknown. In this paper, we identify a dual-function motif in classical cadherins consisting of three highly conserved acidic residues that alternately serve as a p120-binding interface and an endocytic signal. Mutation of this motif resulted in a cadherin variant that was both p120 uncoupled and resistant to endocytosis. In endothelial cells, in which dynamic changes in adhesion are important components of angiogenesis and inflammation, a vascular endothelial cadherin (VE-cadherin) mutant defective in endocytosis assembled normally into cell–cell junctions but potently suppressed cell migration in response to vascular endothelial growth factor. These results reveal the mechanistic basis by which p120 stabilizes cadherins and demonstrate that VE-cadherin endocytosis is crucial for endothelial cell migration in response to an angiogenic growth factor.
Introduction
Dynamic and coordinated changes in cell adhesion are essential for cell migration, tissue patterning, and wound healing. Adherens junctions and their principal cell–cell adhesion molecules, the classical cadherins, are well described (Harris and Tepass, 2010; Saito et al., 2012). However, adherens junction regulation remains poorly understood, limiting our ability to relate static models of cadherin-based adhesion to dynamic biological processes. Cadherins mediate cell adhesion through their extracellular domains, which form calcium-dependent trans-interactions with cadherin molecules on adjacent cells. The cadherin cytoplasmic tail couples cadherins to the actin cytoskeleton and engages in a variety of signaling and membrane trafficking activities. The cytoplasmic tail can be divided into two regions, the catenin-binding domain at the C terminus of the molecule and the more N-terminal juxtamembrane domain. Each of these domains binds to members of the armadillo family of proteins. The catenin-binding domain binds to β-catenin, which links cadherins to the actin cytoskeleton through a mechanism that is not completely understood (Drees et al., 2005; Yamada et al., 2005; Taguchi et al., 2011). The juxtamembrane domain binds to p120-catenin (p120), an important regulator of adherens junction stability. In the absence of p120, cadherins are rapidly internalized from the cell surface and degraded in the lysosome (Davis et al., 2003; Xiao et al., 2003a,b; Miyashita and Ozawa, 2007). Because modulation of cadherin availability at the cell surface has emerged as a key factor determining adhesion strength and because cadherin endocytosis can drive junction disassembly (Troyanovsky et al., 2006), understanding how p120 controls cadherin endocytosis is necessary to understand dynamic regulation of cell adhesion.
In the endothelium, dynamic changes in vascular endothelial cadherin (VE-cadherin)–mediated adhesion are important components of angiogenesis and inflammation, and improper regulation of endothelial cell adhesion can facilitate cancer metastasis (Vincent et al., 2004; Dejana et al., 2008). Similarly to other classical cadherins (Le et al., 1999), VE-cadherin undergoes clathrin-mediated endocytosis (Xiao et al., 2005; Chiasson et al., 2009). In endothelial cells, p120 serves as a master regulator of cadherin expression, balancing cellular levels of VE-cadherin with N-cadherin, which is also expressed in endothelial cells but typically does not contribute significantly to adherens junctions (Ferreri et al., 2008). Endothelial-specific knockout of p120 causes hemorrhaging, defects in vessel patterning, and embryonic lethality, underscoring the importance of p120 regulation of cadherins for endothelial barrier function and vascular development (Oas et al., 2010). Similar results have been reported in a variety of other conditional knockout models (Davis and Reynolds, 2006; Elia et al., 2006; Perez-Moreno et al., 2006; Smalley-Freed et al., 2010; Marciano et al., 2011; Stairs et al., 2011; Chacon-Heszele et al., 2012; Kurley et al., 2012). These findings demonstrate that p120 inhibition of cadherin endocytosis represents a fundamental cellular mechanism that controls cadherin cell surface levels in most cell types.
Although previous studies have highlighted the importance of p120 in regulating cadherin cell surface levels, the precise mechanism by which p120 inhibits cadherin endocytosis has remained elusive (Nanes and Kowalczyk, 2012). This lack of mechanistic insight underlies two significant gaps in our understanding of the role of cadherin endocytosis in development and disease. First, it has been difficult to uncouple p120 binding to the cadherin cytoplasmic tail from control of cadherin endocytosis. Disrupting p120 binding triggers cadherin endocytosis, masking other potential effects. As a result, determining whether cadherins recruit p120 for any purpose other than stabilization of the junction has proven difficult. Second, the contribution of cadherin endocytic trafficking to cell behavior and tissue patterning has thus far been studied through broad perturbations of endocytic pathways (Jarrett et al., 2002; de Beco et al., 2009; Kawauchi et al., 2010; Levayer et al., 2011). These strategies also impact trafficking of other membrane proteins, thereby limiting their specificity and complicating interpretations. Thus, identification of cadherin motifs that mediate endocytosis, allowing for selective perturbation of cadherin internalization and permitting loss-of-function experiments in various model systems, has been an important goal.
The results presented here demonstrate that the core p120-binding region of classical cadherins, which mediates the strongest interactions with p120, comprises a highly conserved endocytic signal and that p120 inhibits cadherin endocytosis by physically occupying this motif. Furthermore, mutation of a three–amino acid acidic cluster uncouples VE-cadherin from p120 while simultaneously preventing endocytosis and stabilizing the cadherin at the cell surface. Functional analysis of this VE-cadherin mutant reveals a critical role for cadherin internalization in endothelial cell migration in response to the angiogenic agent VEGF. These results demonstrate that the core p120-binding region of classical cadherins is a dual-function motif, which mediates both p120 binding and cadherin endocytosis, and reveal a key role for endocytic processing of cadherins in cell migration.
Results
The core p120-binding region of classical cadherins is well conserved
p120 binding to the cadherin juxtamembrane domain regulates cadherin cell surface levels by preventing cadherin endocytosis. This regulatory activity requires p120 association with the cadherin tail but does not require p120 inhibition of RhoA (Ireton et al., 2002; Xiao et al., 2003a, 2005; Miyashita and Ozawa, 2007; Chiasson et al., 2009). These findings support a model in which the cadherin juxtamembrane domain contains an endocytic signal, and p120 binding to the cadherin physically masks that signal (Xiao et al., 2003a; Chiasson et al., 2009). A structural study has revealed that the interaction between E-cadherin and p120 contains both a static binding site, with strong interactions between the cadherin and p120, and a dynamic binding site, with weaker transient interactions (Ishiyama et al., 2010). An important endocytic signal in E-cadherin, a putative AP-2–binding dileucine motif (Miyashita and Ozawa, 2007), falls within the dynamic binding site, suggesting that p120 binding could interfere with this motif, thereby inhibiting E-cadherin endocytosis (Ishiyama et al., 2010). A simulated model of the p120–VE-cadherin complex derived from the p120–E-cadherin crystal structure shows that the interactions of both cadherins with p120 are broadly similar. The important electrostatic and hydrophobic interactions, as well as the general position of the cadherin core p120-binding region along a groove across p120, are conserved in the p120–VE-cadherin model (Fig. 1, A and B). However, VE-cadherin lacks the dileucine endocytic signal present in E-cadherin (Fig. 1 C). VE-cadherin does contain a putative tyrosine-based endocytic signal at Y685, though it is located C terminal to the p120 binding site, making it unlikely to be subject to regulation by p120 binding. Therefore, we hypothesized that the VE-cadherin cytoplasmic tail contains an additional uncharacterized endocytic signal within the p120-binding region.
The core p120-binding region of classical cadherins is well conserved. (A) Schematic illustration of the cadherin–catenin complex. Extracellular cadherin domains mediate adhesion through trans-interactions with cadherins on the adjacent cell. The cadherin juxtamembrane domain binds to p120, and the catenin-binding domain interacts with β-catenin (β-cat). β-Catenin and α-catenin link the cadherin with the actin cytoskeleton (gray) through a mechanism that is not fully understood. CBD, catenin-binding domain; JMD, juxtamembrane domain. (B) Predicted molecular interface between VE-cadherin and p120-catenin. A simulated 3D model of VE-cadherin juxtamembrane domain residues 646–664 bound to the armadillo repeat domain of p120 (surface electrostatic potential: blue, positive; red, negative) was constructed based on the crystal structure of the E-cadherin juxtamembrane domain bound to p120 (Protein Data Bank accession no. 3L6X). Selected residues of VE-cadherin and p120 are labeled in magenta and black, respectively. (C) Multiple sequence alignment of classical cadherins from human (hm), mouse (ms), and Drosophila (dr). Conserved residues are highlighted in green. The core p120-binding region (residues 644–664), which mediates the strongest interactions between the cadherin and p120, is indicated. Other notable features are marked below the alignment: 1, E-cadherin dileucine endocytic signal; 2, E-cadherin Y753 and Y754 Src phosphorylation sites required for Hakai-mediated ubiquitination of E-cadherin; 3–5, VE-cadherin mutations used in Figs. 3, 4, 5, 6, 7, 8, 9, and 10; Δ644 and Δ657, location of VE-cadherin truncation mutations used in Fig. 2.
The core p120-binding region of classical cadherins is well conserved. (A) Schematic illustration of the cadherin–catenin complex. Extracellular cadherin domains mediate adhesion through trans-interactions with cadherins on the adjacent cell. The cadherin juxtamembrane domain binds to p120, and the catenin-binding domain interacts with β-catenin (β-cat). β-Catenin and α-catenin link the cadherin with the actin cytoskeleton (gray) through a mechanism that is not fully understood. CBD, catenin-binding domain; JMD, juxtamembrane domain. (B) Predicted molecular interface between VE-cadherin and p120-catenin. A simulated 3D model of VE-cadherin juxtamembrane domain residues 646–664 bound to the armadillo repeat domain of p120 (surface electrostatic potential: blue, positive; red, negative) was constructed based on the crystal structure of the E-cadherin juxtamembrane domain bound to p120 (Protein Data Bank accession no. 3L6X). Selected residues of VE-cadherin and p120 are labeled in magenta and black, respectively. (C) Multiple sequence alignment of classical cadherins from human (hm), mouse (ms), and Drosophila (dr). Conserved residues are highlighted in green. The core p120-binding region (residues 644–664), which mediates the strongest interactions between the cadherin and p120, is indicated. Other notable features are marked below the alignment: 1, E-cadherin dileucine endocytic signal; 2, E-cadherin Y753 and Y754 Src phosphorylation sites required for Hakai-mediated ubiquitination of E-cadherin; 3–5, VE-cadherin mutations used in Figs. 3, 4, 5, 6, 7, 8, 9, and 10; Δ644 and Δ657, location of VE-cadherin truncation mutations used in Fig. 2.
The core p120-binding region of VE-cadherin harbors an endocytic signal
A gain-of-function approach was adopted to define the roles of different portions of the VE-cadherin cytoplasmic tail in cadherin internalization. Portions of the VE-cadherin cytoplasmic tail were joined to the extracellular and transmembrane domains of IL-2R (interleukin-2 receptor α chain), a transmembrane protein that does not mediate cell adhesion and is not rapidly endocytosed (Xiao et al., 2003a). Sequential deletions from the intracellular (C terminal) end of the resulting chimera allowed us to test the requirements for different portions of the VE-cadherin cytoplasmic tail for catenin binding and internalization. As expected, the VE-cadherin catenin-binding domain was required for coimmunoprecipitation of β-catenin with the IL-2R–VE-cadherin chimera, and the VE-cadherin juxtamembrane domain was required for coimmunoprecipitation of p120. Truncating the VE-cadherin cytoplasmic tail at residues 657 or 644, resulting in the removal of part or all of the core p120-binding region, respectively, prevented p120 coimmunoprecipitation (Fig. 2, A and B).
The core p120-binding region of VE-cadherin functions as an endocytic signal. (A) Portions of the VE-cadherin (VE-cad) cytoplasmic tail (gray) were fused to the extracellular and transmembrane domains of the interleukin-2 receptor α chain (IL-2R; black) to create chimeric proteins. The core p120-binding region is indicated by asterisks. CBD, catenin-binding domain; JMD, juxtamembrane domain; cyto, entire VE-cadherin cytoplasmic tail; Δ657 and Δ644, VE-cadherin cytoplasmic tail truncated at residues 657 and 644. (B) IL-2R–VE-cadherin chimeras were expressed in COS-7 cells and isolated by immunoprecipitation of IL-2R. β-Catenin (β) coprecipitates only with the chimera containing the catenin-binding domain, and p120 coprecipitates only with the chimeras containing the entire juxtamembrane domain. (C) Fluorescence-based internalization assay of IL-2R–VE-cadherin chimeras expressed in COS-7 cells. (top row) Internalized chimera was identified by antibody labeling of surface IL-2R followed by a 10-min incubation to allow internalization and a low pH wash to remove antibody remaining at the cell surface. (bottom row) Cells were then fixed and stained for total IL-2R. (D) Quantification of the ratio of internalized to total chimera. Means ± SEM (n = 8–16 cells per group); *, P < 0.05; **, P < 0.01; ***, P < 0.001 compared with IL-2R and Δ644. (E) The VE-cadherin core p120-binding region (core; asterisk) was fused to IL-2R with a short linker peptide (linker; white oval) used to maintain spacing from the plasma membrane. (F and G) Fluorescence-based internalization assay with a 10-min internalization (intern.) period. Means ± SEM (n = 8–11 cells per group); **, P < 0.01 compared with IL-2R; ◊◊, P < 0.01 compared with linker. IP, immunoprecipitation. Bars, 20 µm.
The core p120-binding region of VE-cadherin functions as an endocytic signal. (A) Portions of the VE-cadherin (VE-cad) cytoplasmic tail (gray) were fused to the extracellular and transmembrane domains of the interleukin-2 receptor α chain (IL-2R; black) to create chimeric proteins. The core p120-binding region is indicated by asterisks. CBD, catenin-binding domain; JMD, juxtamembrane domain; cyto, entire VE-cadherin cytoplasmic tail; Δ657 and Δ644, VE-cadherin cytoplasmic tail truncated at residues 657 and 644. (B) IL-2R–VE-cadherin chimeras were expressed in COS-7 cells and isolated by immunoprecipitation of IL-2R. β-Catenin (β) coprecipitates only with the chimera containing the catenin-binding domain, and p120 coprecipitates only with the chimeras containing the entire juxtamembrane domain. (C) Fluorescence-based internalization assay of IL-2R–VE-cadherin chimeras expressed in COS-7 cells. (top row) Internalized chimera was identified by antibody labeling of surface IL-2R followed by a 10-min incubation to allow internalization and a low pH wash to remove antibody remaining at the cell surface. (bottom row) Cells were then fixed and stained for total IL-2R. (D) Quantification of the ratio of internalized to total chimera. Means ± SEM (n = 8–16 cells per group); *, P < 0.05; **, P < 0.01; ***, P < 0.001 compared with IL-2R and Δ644. (E) The VE-cadherin core p120-binding region (core; asterisk) was fused to IL-2R with a short linker peptide (linker; white oval) used to maintain spacing from the plasma membrane. (F and G) Fluorescence-based internalization assay with a 10-min internalization (intern.) period. Means ± SEM (n = 8–11 cells per group); **, P < 0.01 compared with IL-2R; ◊◊, P < 0.01 compared with linker. IP, immunoprecipitation. Bars, 20 µm.
Using a fluorescence-based internalization assay, we tested which portions of the VE-cadherin cytoplasmic tail were required to mediate endocytosis. The full VE-cadherin cytoplasmic tail and the juxtamembrane domain alone were sufficient to mediate chimera internalization. The 657 truncation chimera was also internalized, even though it lacked the ability to bind p120 (Fig. 2, C and D). In fact, internalization mediated by the 657 truncation was greater than internalization mediated by the juxtamembrane domain or full cytoplasmic tail, consistent with our model that p120 binding masks an endocytic signal to inhibit internalization. In contrast, the 644 truncation chimera was not internalized, indicating that a significant portion, though not all, of the core p120-binding region is necessary for VE-cadherin endocytosis (Fig. 2, C and D).
The VE-cadherin core p120-binding region is not only necessary but also sufficient to mediate endocytosis. Chimeras containing only the VE-cadherin core p120-binding region (residues 644–664) joined to the IL-2R by a short linker peptide were internalized, indicating that the core p120-binding region itself doubles as an endocytic signal (Fig. 2, E–G). To verify that the core p120-binding region mediates endocytosis in a clathrin-dependent manner similar to the full-length VE-cadherin cytoplasmic tail, potassium depletion was used to block clathrin-mediated endocytosis. Internalization mediated by both the full VE-cadherin cytoplasmic tail and the core p120-binding region alone was inhibited (Fig. S1), indicating that the core p120-binding region functions similarly to the full cytoplasmic tail to mediate endocytosis. Together, these results demonstrate that the core p120-binding region of VE-cadherin also functions as an endocytic signal.
p120 binding can be uncoupled from control of cadherin endocytosis
Although there are substantial differences in the juxtamembrane domains of classical cadherins, the core p120-binding regions are well conserved, especially between VE-cadherin residues 644 and 657 (Fig. 1 C). We mutated three consecutive sets of highly conserved amino acids in the core p120-binding region, DEE 646–648, GGG 649–651, and EMD 652–654 to alanines. Each of these mutations prevented coimmunoprecipitation of p120 with the IL-2R–VE-cadherin chimera (Fig. 3 A), consistent with the important roles of these amino acids in the VE-cadherin–p120 binding interaction (Fig. 1 B; Thoreson et al., 2000). However, these three mutations had very different effects on chimera internalization. Mutation of the DEE residues, but not the GGG or EMD residues, almost completely eliminated chimera endocytosis (Fig. 3, B and C). The EMD mutation resulted in a small decrease in internalization, which, while reproducible, did not reach statistical significance, and the GGG mutation had no effect (Fig. 3, B and C). Control experiments confirmed that the steady-state surface expression levels of each chimera were similar (Fig. 3 D).
p120 binding can be uncoupled from control of cadherin endocytosis. Three consecutive sets of amino acids in the core p120-binding region, DEE 646–648, GGG 649–651, and EMD 652–654 (also see Fig. 1 C), were mutated to alanines in IL-2R–VE-cadherin (VE-cad) cytoplasmic tail chimeras and expressed in COS-7 cells. (A) Chimeras were isolated by immunoprecipitation of IL-2R, and coprecipitation of p120 and β-catenin (β) was determined by Western blotting. (B and C) Endocytosis of the chimeras was measured using a fluorescence-based internalization assay with a 10-min internalization period. Although each mutation prevented pull-down of p120, only the DEE mutation significantly inhibited endocytosis. Means ± SEM (n = 15 cells per group); **, P < 0.01; ***, P < 0.001 compared with IL-2R and DEE mutation. (D) Surface expression of the chimeras in COS-7 cells was quantified by antibody labeling of surface IL-2R and immunofluorescence microscopy. Thick line, median (n = 17–21 cells per group); box, interquartile range; whiskers, 90% range. intern., internalization; IP, immunoprecipitation; WT, wild type. Bar, 20 µm.
p120 binding can be uncoupled from control of cadherin endocytosis. Three consecutive sets of amino acids in the core p120-binding region, DEE 646–648, GGG 649–651, and EMD 652–654 (also see Fig. 1 C), were mutated to alanines in IL-2R–VE-cadherin (VE-cad) cytoplasmic tail chimeras and expressed in COS-7 cells. (A) Chimeras were isolated by immunoprecipitation of IL-2R, and coprecipitation of p120 and β-catenin (β) was determined by Western blotting. (B and C) Endocytosis of the chimeras was measured using a fluorescence-based internalization assay with a 10-min internalization period. Although each mutation prevented pull-down of p120, only the DEE mutation significantly inhibited endocytosis. Means ± SEM (n = 15 cells per group); **, P < 0.01; ***, P < 0.001 compared with IL-2R and DEE mutation. (D) Surface expression of the chimeras in COS-7 cells was quantified by antibody labeling of surface IL-2R and immunofluorescence microscopy. Thick line, median (n = 17–21 cells per group); box, interquartile range; whiskers, 90% range. intern., internalization; IP, immunoprecipitation; WT, wild type. Bar, 20 µm.
We next verified these results in the context of full-length VE-cadherin. When expressed in A-431D cells, an epithelial cell line that lacks endogenous cadherin expression, wild-type, DEE mutant, and GGG mutant VE-cadherin all localized to cell–cell contacts. In addition, wild-type VE-cadherin, as well as both mutants, recruited α- and β-catenins to junctions, whereas only wild-type VE-cadherin recruited p120 (Fig. 4, A–C). Neither mutant altered the actin cytoskeleton in A-431D cells (Fig. 4 D) or in primary human microvascular endothelial cells, in which PECAM-1 localization was also unaffected (Fig. 4 E). Furthermore, the DEE mutation almost completely eliminated full-length VE-cadherin internalization, whereas the GGG mutation did not (Fig. 5, A and B). Interestingly, GGG mutant full-length VE-cadherin was internalized more rapidly than wild type (Fig. 5, A and B), consistent with its inability to bind p120. Note that this effect was not observed with the GGG mutant IL-2R–VE-cadherin chimera (Fig. 3, B and C), probably because the chimeras were expressed in excess of available cellular p120, so even wild-type chimera was mostly unbound by p120 and, as a result, was internalized at a rate similar to the p120-uncoupled GGG mutant chimera. Further confirming the ability of the DEE mutation to alter cadherin dynamics, time-lapse imaging of VE-cadherin–RFP expressed in primary human microvascular endothelial cells showed that junctions containing wild-type VE-cadherin were dynamic, with visible endocytic events, whereas junctions containing DEE mutant VE-cadherin lacked visible endocytic events (Fig. 5 C and Videos 1 and 2). Thus, although both the DEE and GGG mutations disrupt p120 binding, only the DEE mutation inhibits internalization, demonstrating that p120 binding to the cadherin can be uncoupled from control of cadherin endocytosis.
p120 binding is not required for adherens junction assembly. (A–D) Wild-type (WT), DEE mutant, or GGG mutant VE-cadherin (VE-cad) with a C-terminal RFP tag was expressed in A-431D cells, which lack endogenous cadherins, using an adenoviral system. Cells were fixed and stained for VE-cadherin and α-catenin (A), β-catenin (β-cat; B), p120 (C), or actin (D). (E) Wild-type or mutant VE-cadherin–RFP was expressed in primary human microvascular endothelial cells. Cells were fixed and stained for RFP (VE-cadherin), actin, and PECAM-1. Bars: (A–D) 5 µm; (E) 20 µm.
p120 binding is not required for adherens junction assembly. (A–D) Wild-type (WT), DEE mutant, or GGG mutant VE-cadherin (VE-cad) with a C-terminal RFP tag was expressed in A-431D cells, which lack endogenous cadherins, using an adenoviral system. Cells were fixed and stained for VE-cadherin and α-catenin (A), β-catenin (β-cat; B), p120 (C), or actin (D). (E) Wild-type or mutant VE-cadherin–RFP was expressed in primary human microvascular endothelial cells. Cells were fixed and stained for RFP (VE-cadherin), actin, and PECAM-1. Bars: (A–D) 5 µm; (E) 20 µm.
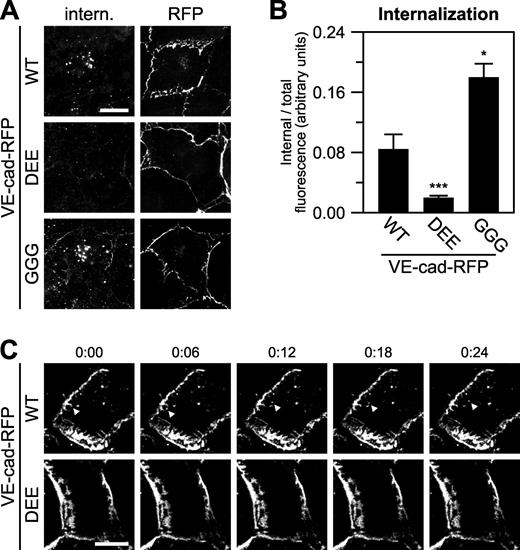
A conserved cluster of three acidic residues is required for full-length VE-cadherin endocytosis. (A and B) Wild-type (WT) or mutant VE-cadherin–RFP was expressed in COS-7 cells, and endocytosis was measured using a fluorescence-based internalization assay with an internalization period of 30 min. Antibody labeling and low pH wash was used to identify internalized (intern.) cadherin (A, left column), and the RFP tag was used to identify total cadherin (A, right column). Means ± SEM (n = 20–24 cells per group); ***, P < 0.001 compared with wild type and GGG mutation; *, P < 0.05 compared with wild type. (C) Time-lapse images of wild-type and DEE mutant VE-cadherin (VE-cad)–RFP expressed in primary human microvascular endothelial cells. Time is shown in minutes and seconds. The arrowheads highlight an endocytic event. See also Videos 1 and 2. Bars: (A) 20 µm; (C) 10 µm.
A conserved cluster of three acidic residues is required for full-length VE-cadherin endocytosis. (A and B) Wild-type (WT) or mutant VE-cadherin–RFP was expressed in COS-7 cells, and endocytosis was measured using a fluorescence-based internalization assay with an internalization period of 30 min. Antibody labeling and low pH wash was used to identify internalized (intern.) cadherin (A, left column), and the RFP tag was used to identify total cadherin (A, right column). Means ± SEM (n = 20–24 cells per group); ***, P < 0.001 compared with wild type and GGG mutation; *, P < 0.05 compared with wild type. (C) Time-lapse images of wild-type and DEE mutant VE-cadherin (VE-cad)–RFP expressed in primary human microvascular endothelial cells. Time is shown in minutes and seconds. The arrowheads highlight an endocytic event. See also Videos 1 and 2. Bars: (A) 20 µm; (C) 10 µm.
p120 occupies the DEE sequence to prevent cadherin endocytosis
The position of the VE-cadherin DEE endocytic signal within the core p120-binding region strongly suggests that p120 binding inhibits cadherin endocytosis by physically occupying the endocytic signal. Indeed, the structural model identifies important electrostatic interactions between p120 and all three side chains of the DEE sequence (Fig. 6, A and B). We therefore tested whether a p120 variant mutated at a critical lysine that interacts with the last residue of the DEE sequence (K444M; Ishiyama et al., 2010) was unable to inhibit cadherin endocytosis. Consistent with our model, increased expression of wild-type p120 significantly inhibited IL-2R–VE-cadherin cytoplasmic tail chimera endocytosis, whereas expression of p120 K444M did not inhibit chimera internalization (Fig. 6, C and D). Furthermore, increased expression of wild-type p120 did not inhibit internalization of the GGG mutant chimera (Fig. 6, C and D), to which p120 is unable to bind (Fig. 3 A). Therefore, p120 occupation of the DEE endocytic signal is required for the inhibition of cadherin endocytosis.
p120 occupies the DEE sequence to prevent cadherin endocytosis. (A and B) Close views of the predicted molecular interface between VE-cadherin and p120. Selected residues of VE-cadherin and p120 are labeled in magenta and black, respectively. Three negatively charged side chains of the DEE sequence are enveloped by positively charged binding pockets (blue) of p120. Hydrogen bonds are indicated by yellow dashes. (C and D) Wild-type (WT) or GGG mutant IL-2R–VE-cadherin cytoplasmic tail chimeras were expressed in COS-7 cells along with wild-type or K444M mutant fluorescently tagged p120. Untransfected cells adjacent to cells transfected with the wild-type p120 construct were used as negative controls (ctrl). Endocytosis of the chimeras was measured using a fluorescence-based internalization (intern.) assay with a 10-min internalization period. Mutation of p120 K444, which is predicted to interact with the last residue of the DEE endocytic signal (A), disrupts p120-mediated inhibition of IL-2R–VE-cadherin chimera internalization, as does mutation of VE-cadherin (VE-cad) GGG 649–651. Means ± SEM (n = 12–15 cells per group); ***, P < 0.001 compared with no exogenous p120 expression and GGG mutant chimera; ◊, P < 0.05 compared with p120 K444M. Bar, 20 µm.
p120 occupies the DEE sequence to prevent cadherin endocytosis. (A and B) Close views of the predicted molecular interface between VE-cadherin and p120. Selected residues of VE-cadherin and p120 are labeled in magenta and black, respectively. Three negatively charged side chains of the DEE sequence are enveloped by positively charged binding pockets (blue) of p120. Hydrogen bonds are indicated by yellow dashes. (C and D) Wild-type (WT) or GGG mutant IL-2R–VE-cadherin cytoplasmic tail chimeras were expressed in COS-7 cells along with wild-type or K444M mutant fluorescently tagged p120. Untransfected cells adjacent to cells transfected with the wild-type p120 construct were used as negative controls (ctrl). Endocytosis of the chimeras was measured using a fluorescence-based internalization (intern.) assay with a 10-min internalization period. Mutation of p120 K444, which is predicted to interact with the last residue of the DEE endocytic signal (A), disrupts p120-mediated inhibition of IL-2R–VE-cadherin chimera internalization, as does mutation of VE-cadherin (VE-cad) GGG 649–651. Means ± SEM (n = 12–15 cells per group); ***, P < 0.001 compared with no exogenous p120 expression and GGG mutant chimera; ◊, P < 0.05 compared with p120 K444M. Bar, 20 µm.
The core p120-binding region is the primary endocytic signal in VE-cadherin
Given the high degree of homology between the core p120-binding regions of classical cadherins, we tested whether the core p120-binding regions of other cadherins also function as endocytic signals. When fused to the IL-2R, the core p120-binding regions of human E-cadherin and N-cadherin, as well as Drosophila melanogaster DE-cadherin, all mediated internalization, although they did so less efficiently than the VE-cadherin core p120-binding region (Fig. 7, A and B). This result suggests that, although this endocytic signal is conserved across a wide variety of classical cadherins, it is particularly important to VE-cadherin. In fact, VE-cadherin does have a putative tyrosine-based endocytic signal at Y685, but substitution of this residue with an alanine inhibited VE-cadherin chimera internalization by only a modest amount (Fig. 7, C and D). Furthermore, this site is distal to the p120-binding region and therefore unlikely to be subject to p120-mediated regulation of VE-cadherin internalization. In contrast, E-cadherin contains a dileucine endocytic signal not present in VE-cadherin. Mutation of these residues in an IL-2R–E-cadherin chimera significantly inhibited internalization, whereas mutation of the DEE sequence in an IL-2R–E-cadherin chimera only modestly inhibited internalization (Fig. 7, E and F). Collectively, these results indicate that the core p120-binding region is a conserved endocytic signal in a variety of classical cadherins, although each cadherin appears to contain flanking sequences that may modulate its function or act as alternative endocytic signals.
The core p120-binding region is the primary endocytic signal in VE-cadherin. Fluorescence-based internalization assays with a 10-min internalization period were used to measure endocytosis of various IL-2R–cadherin chimeras expressed in COS-7 cells. (A and B) Internalization (intern.) of chimeras containing the core p120-binding regions of human VE-cadherin (VE-cad), E-cadherin, and N-cadherin and Drosophila DE-cadherin joined to IL-2R by a linker peptide. Means ± SEM (n = 12–25 cells per group); ***, P < 0.001 compared with IL-2R; ◊, P < 0.05; ◊◊◊, P < 0.001 compared with the linker. (C and D) Internalization of chimeras containing the VE-cadherin cytoplasmic tail, wild type (WT), or with a Y685A mutation joined to IL-2R. Means ± SEM (n = 15 cells per group); ***, P < 0.001 compared with IL-2R. (E and F) Internalization of chimeras containing the E-cadherin cytoplasmic tail, wild type, or with DEE 758–760 (corresponding to the VE-cadherin DEE mutation), EED 664–666 (corresponding the VE-cadherin EMD mutation), or LL 743–744 mutated to alanines joined to IL-2R. Means ± SEM (n = 12 cells per group); *, P < 0.05; ***, P < 0.001 compared with IL-2R and LL mutation. Bars, 20 µm.
The core p120-binding region is the primary endocytic signal in VE-cadherin. Fluorescence-based internalization assays with a 10-min internalization period were used to measure endocytosis of various IL-2R–cadherin chimeras expressed in COS-7 cells. (A and B) Internalization (intern.) of chimeras containing the core p120-binding regions of human VE-cadherin (VE-cad), E-cadherin, and N-cadherin and Drosophila DE-cadherin joined to IL-2R by a linker peptide. Means ± SEM (n = 12–25 cells per group); ***, P < 0.001 compared with IL-2R; ◊, P < 0.05; ◊◊◊, P < 0.001 compared with the linker. (C and D) Internalization of chimeras containing the VE-cadherin cytoplasmic tail, wild type (WT), or with a Y685A mutation joined to IL-2R. Means ± SEM (n = 15 cells per group); ***, P < 0.001 compared with IL-2R. (E and F) Internalization of chimeras containing the E-cadherin cytoplasmic tail, wild type, or with DEE 758–760 (corresponding to the VE-cadherin DEE mutation), EED 664–666 (corresponding the VE-cadherin EMD mutation), or LL 743–744 mutated to alanines joined to IL-2R. Means ± SEM (n = 12 cells per group); *, P < 0.05; ***, P < 0.001 compared with IL-2R and LL mutation. Bars, 20 µm.
VE-cadherin mobility does not require endocytosis
Identification of mutations that uncouple p120 binding from control of cadherin endocytosis allows us to test the effects of inhibiting cadherin endocytosis without affecting the internalization of other membrane proteins. Because a prior study has suggested, based on broad disruption of endocytic pathways with dynamin inhibitors, that cadherin mobility within the plasma membrane results primarily from endocytosis and recycling (de Beco et al., 2009), we used FRAP to test whether selective inhibition of cadherin endocytosis restricted cadherin mobility at cell borders. Surprisingly, neither the endocytosis-defective DEE mutation nor the GGG mutation altered the half-life of VE-cadherin FRAP compared with wild type. In fact, the DEE mutation increased the mobile fraction of VE-cadherin, the proportion of cadherin free to diffuse within the membrane, whereas the GGG mutation slightly decreased the mobile fraction (Fig. 8, A and B; Table 1; and Videos 3, 4, and 5). Recovery rate was not affected by variation in the size of the bleach region, justifying our use of an exponential, rather than diffusion-limited, model (Fig. S2). These observations are incompatible with the hypothesis that cadherin FRAP is facilitated by endocytosis and recycling, and suggest instead that the endocytic signal mediates interactions that restrict VE-cadherin diffusion. We previously reported that the VE-cadherin cytoplasmic tail mediates clustering into clathrin-enriched membrane domains (Chiasson et al., 2009). Therefore, we hypothesized that the DEE mutation might disrupt this interaction. Consistent with this hypothesis, the DEE mutation, but not the GGG mutation, prevented IL-2R–VE-cadherin chimera from co-clustering with clathrin, as determined using both conventional (Fig. 9, A and B) and super resolution (Fig. S3) immunofluorescence microscopy. Thus, the ability of the VE-cadherin cytoplasmic tail to cluster into clathrin-enriched membrane domains limits rather than facilitates lateral mobility of the cadherin within the plasma membrane.
VE-cadherin mobility does not require endocytosis. RFP-tagged wild-type (WT), DEE mutant, or GGG mutant VE-cadherin (VE-cad) was expressed in primary human dermal microvascular endothelial cells and used in FRAP experiments. (A) False-color images of VE-cadherin–RFP at cell junctions before bleaching (left), immediately after bleaching a 5-µm-long section of the junction (indicated by arrowheads), and at 5 and 10 min after bleaching. See also Videos 3, 4, and 5. Bar, 5 µm. (B) Quantification of fluorescence recovery. Mean fluorescence within the bleach area, corrected for image acquisition–related photobleaching, as a fraction of prebleach value. Exponential curves were fit to the data, and their coefficients are given in Table 1. Points, means ± SEM (n = 16–18 sequences per group); lines, exponential models ± 95% confidence interval.
VE-cadherin mobility does not require endocytosis. RFP-tagged wild-type (WT), DEE mutant, or GGG mutant VE-cadherin (VE-cad) was expressed in primary human dermal microvascular endothelial cells and used in FRAP experiments. (A) False-color images of VE-cadherin–RFP at cell junctions before bleaching (left), immediately after bleaching a 5-µm-long section of the junction (indicated by arrowheads), and at 5 and 10 min after bleaching. See also Videos 3, 4, and 5. Bar, 5 µm. (B) Quantification of fluorescence recovery. Mean fluorescence within the bleach area, corrected for image acquisition–related photobleaching, as a fraction of prebleach value. Exponential curves were fit to the data, and their coefficients are given in Table 1. Points, means ± SEM (n = 16–18 sequences per group); lines, exponential models ± 95% confidence interval.
VE-cadherin FRAP model parameters
| Protein | Mobile fraction | Recovery half-life |
| s | ||
| WT VE-cadherin–RFP | 0.465 [0.450–0.481] | 125 [114–137] |
| VE-cadherin–RFP (DEE → AAA) | 0.623 [0.587–0.666] | 132 [113–156] |
| VE-cadherin–RFP (GGG → AAA) | 0.426 [0.409–0.446] | 138 [124–155] |
| Protein | Mobile fraction | Recovery half-life |
| s | ||
| WT VE-cadherin–RFP | 0.465 [0.450–0.481] | 125 [114–137] |
| VE-cadherin–RFP (DEE → AAA) | 0.623 [0.587–0.666] | 132 [113–156] |
| VE-cadherin–RFP (GGG → AAA) | 0.426 [0.409–0.446] | 138 [124–155] |
Square brackets indicate 95% confidence interval. WT, wild type.
Disrupting the endocytic signal prevents VE-cadherin recruitment into clathrin-enriched membrane domains. (A) COS-7 cells expressing wild-type (WT), DEE mutant, or GGG mutant IL-2R–VE-cadherin (VE-cad) cytoplasmic tail chimeras were surface labeled with an antibody against IL-2R. After a 30-min incubation on ice to allow chimera clustering, the cells were fixed and stained for clathrin heavy chain. Boxes show inset areas. Bar, 10 µm. (B) Quantification of IL-2R–VE-cadherin chimera and clathrin colocalization. Pearson’s correlation coefficient; means ± SEM (n = 33–34 cells per group); *, P < 0.05 compared with wild type and GGG mutant.
Disrupting the endocytic signal prevents VE-cadherin recruitment into clathrin-enriched membrane domains. (A) COS-7 cells expressing wild-type (WT), DEE mutant, or GGG mutant IL-2R–VE-cadherin (VE-cad) cytoplasmic tail chimeras were surface labeled with an antibody against IL-2R. After a 30-min incubation on ice to allow chimera clustering, the cells were fixed and stained for clathrin heavy chain. Boxes show inset areas. Bar, 10 µm. (B) Quantification of IL-2R–VE-cadherin chimera and clathrin colocalization. Pearson’s correlation coefficient; means ± SEM (n = 33–34 cells per group); *, P < 0.05 compared with wild type and GGG mutant.
VEGF-induced endothelial cell migration requires cadherin endocytosis
Cadherin endocytosis has been implicated in a variety of developmental events and cellular activities (Nanes and Kowalczyk, 2012). Similarly, p120 association with cadherins has been linked to the regulation of cadherin adhesion and cell migration (Xiao et al., 2007). The ability to selectively uncouple the regulation of endocytosis from p120 binding to cadherins through mutation of the DEE endocytic signal provides an opportunity to define the contribution of these activities of p120 to endothelial cell functions. It is likely that dynamic changes in cell adhesion are necessary for effective cell migration, particularly in the context of angiogenesis and reendothelialization of denuded vessels (Dejana et al., 2009). Using scratch wound assays, we found that expression of endocytosis-defective DEE mutant VE-cadherin in primary human microvascular endothelial cells markedly slowed migration in response to VEGF, whereas expression of wild-type or GGG mutant VE-cadherin had no effect (Fig. 10, A and B). Similar results were obtained when constitutive migration of endothelial cells grown in serum was assessed (Fig. S4, A and B) as well as with cells lacking endogenous cadherin (Fig. S4, C and D). We also confirmed that DEE mutant VE-cadherin was not expressed at a higher level than the other cadherin mutants (Fig. 10 C) and that wild-type, DEE mutant, and GGG mutant VE-cadherin–expressing cells replicated at the same rate as measured by ethynyl-deoxyuridine (EdU) uptake (Fig. 10 D). We performed two experiments to further verify that the observed migration defect was related to cadherin endocytosis. First, we determined that neither wild-type nor mutant VE-cadherin prevented p44/42 MAPK phosphorylation in response to VEGF, indicating that the migration defect caused by endocytosis-defective VE-cadherin was not a result of inhibition of VEGF signaling (Fig. 10 E). Second, we confirmed that inhibition of cadherin endocytosis with a dynamin inhibitor also slowed migration of endothelial cells expressing wild-type or p120-uncoupled VE-cadherin (Fig. S4, E–H). Additionally, because GGG mutant VE-cadherin, which does not bind p120 but undergoes internalization normally, did not affect migration (Fig. 10, A and B), we also conclude that inhibition of migration by the DEE mutant cadherin resulted specifically from inhibition of cadherin endocytosis and not from disruption of p120 recruitment to the adherens junction.
VEGF-induced endothelial cell migration requires cadherin endocytosis. (A–E) Wild-type (WT), DEE mutant, and GGG mutant VE-cadherin (VE-cad) was expressed in monolayers of primary human dermal microvascular endothelial cells using an adenoviral transduction system. Infection with an empty adenovirus (EV) was used as a negative control. (A and B) Endothelial monolayers were serum starved for 1 h and then scratched with a pipette tip. 12 h after the scratch, 100 mg/ml VEGF was added to the medium (indicated by arrowheads). Migration of cells into the wound area was tracked over time. Mean distance closed ± SEM (n = 8 wounds per group); *, P < 0.05 compared with empty adenovirus, wild type, and GGG; ◊, P < 0.05 compared with empty adenovirus and wild type only. (C) VE-cadherin expression was measured by Western blotting. Empty arrowhead, endogenous VE-cadherin; filled arrowhead, exogenously expressed VE-cadherin–RFP. (D) Replication was measured by a thymidine analogue incorporation assay. Confluent monolayers were serum starved for 12 h and then either left untreated or treated with VEGF for 6 h, with incubation in EdU during the final hour. After fixation and labeling, the replication rate was estimated by the fraction of infected cells that were EdU positive. Proportion ± standard error (n = 94–103 cells per group). (E) Activation of VEGF signaling was verified by Western blotting for phosphorylated p44/42 MAPK (top) and total p44/42 MAPK (bottom). Cells were serum starved for 12 h and then treated with VEGF or left untreated for 20 min before harvesting. (F and G) Endocytosis of VE-cadherin in endothelial cells migrating into a scratch wound was measured using a fluorescence-based internalization assay. Confluent monolayers of endothelial cells were serum starved for 45 min and then scratched with a pipette tip and either left untreated (control) or treated with VEGF for 45 min. VE-cadherin endocytosis was measured over a 1-h internalization period in cells near to (75 ± 75 µm) or far from (440 ± 75 µm) the wound edge. Means ± SEM (n = 22–32 cells per group). Bars: (A) 100 µm; (F) 20 µm.
VEGF-induced endothelial cell migration requires cadherin endocytosis. (A–E) Wild-type (WT), DEE mutant, and GGG mutant VE-cadherin (VE-cad) was expressed in monolayers of primary human dermal microvascular endothelial cells using an adenoviral transduction system. Infection with an empty adenovirus (EV) was used as a negative control. (A and B) Endothelial monolayers were serum starved for 1 h and then scratched with a pipette tip. 12 h after the scratch, 100 mg/ml VEGF was added to the medium (indicated by arrowheads). Migration of cells into the wound area was tracked over time. Mean distance closed ± SEM (n = 8 wounds per group); *, P < 0.05 compared with empty adenovirus, wild type, and GGG; ◊, P < 0.05 compared with empty adenovirus and wild type only. (C) VE-cadherin expression was measured by Western blotting. Empty arrowhead, endogenous VE-cadherin; filled arrowhead, exogenously expressed VE-cadherin–RFP. (D) Replication was measured by a thymidine analogue incorporation assay. Confluent monolayers were serum starved for 12 h and then either left untreated or treated with VEGF for 6 h, with incubation in EdU during the final hour. After fixation and labeling, the replication rate was estimated by the fraction of infected cells that were EdU positive. Proportion ± standard error (n = 94–103 cells per group). (E) Activation of VEGF signaling was verified by Western blotting for phosphorylated p44/42 MAPK (top) and total p44/42 MAPK (bottom). Cells were serum starved for 12 h and then treated with VEGF or left untreated for 20 min before harvesting. (F and G) Endocytosis of VE-cadherin in endothelial cells migrating into a scratch wound was measured using a fluorescence-based internalization assay. Confluent monolayers of endothelial cells were serum starved for 45 min and then scratched with a pipette tip and either left untreated (control) or treated with VEGF for 45 min. VE-cadherin endocytosis was measured over a 1-h internalization period in cells near to (75 ± 75 µm) or far from (440 ± 75 µm) the wound edge. Means ± SEM (n = 22–32 cells per group). Bars: (A) 100 µm; (F) 20 µm.
We next attempted to determine whether inhibition of endothelial cell migration by endocytosis-defective VE-cadherin was related to junction formation. Wild-type, DEE mutant, and GGG mutant VE-cadherins exhibited similar distributions at the wound edge in migrating cells (Fig. S5 A). However, migration of sparsely seeded cells, which could not form cell–cell contacts, was apparently unaffected by expression of the DEE mutant (Fig. S5, B–E). To better understand the relationship between junctions, cadherin endocytosis, endothelial cell migration, and VEGF signaling, we monitored VE-cadherin dynamics and internalization in endothelial cells migrating into a scratch wound. Interestingly, we were unable to observe a difference in VE-cadherin endocytosis with and without VEGF treatment and at different distances from the wound edge (Fig. 10, F and G; and Fig. S5, F and G). These results suggest that VE-cadherin internalization is not confined to the wound edge and that VEGF does not induce endothelial migration by stimulating cadherin endocytosis. Rather, efficient endothelial migration appears to require adherens junction plasticity derived from the constitutive endocytosis of VE-cadherin. These findings reveal the importance of cadherin endocytosis to the dynamic changes in cell adhesion necessary for endothelial cell migration and demonstrate that p120 binding and regulation of cadherin endocytosis can be uncoupled during this process.
Discussion
The results presented here reveal that p120 inhibits VE-cadherin endocytosis by binding to and physically masking a cluster of acidic residues in the cadherin cytoplasmic tail (DEE 646–648), which functions as an endocytic signal. This signal is contained within the core p120-binding region, the portion of the cadherin cytoplasmic tail that mediates the strongest interactions with p120 (Fig. 1), and this sequence is both necessary and sufficient to mediate endocytosis (Fig. 2). Thus, when p120 is bound to the cadherin juxtamembrane domain, this endocytic signal is masked, and the cadherin is stabilized. p120 dissociation from the cadherin exposes this signal and triggers cadherin endocytosis. Mutations in either the cadherin or p120, which prevent p120 from masking the endocytic signal, disrupt this regulatory mechanism (Figs. 3, 5, and 6). These findings demonstrate that the core p120-binding region of classical cadherins serves mutually exclusive roles as either a p120 binding site or an endocytic motif. Furthermore, we find that cadherin endocytosis is essential for efficient endothelial cell migration in response to an angiogenic growth factor (Fig. 10), highlighting the importance of this regulatory mechanism in the context of endothelial cell biology.
The DEE acidic cluster of the core p120-binding region is well conserved among classical cadherins, including E-cadherin and N-cadherin (Fig. 1 C). The endocytic function of this cadherin domain is also well conserved, although with varying degrees of efficiency, suggesting that additional endocytic signals may play important roles in other cadherins (Fig. 7, A and B). These additional motifs include the dileucine endocytic signal in E-cadherin and Y685 in VE-cadherin. Interestingly, although the dileucine endocytic signal present in E-cadherin is outside of the core p120-binding region, it is within a region of dynamic binding between p120 and the E-cadherin cytoplasmic tail, so p120 binding may mask this signal as well (Ishiyama et al., 2010). In contrast, VE-cadherin Y685 lies outside of the p120-binding region, and mutation of this tyrosine residue only modestly affects VE-cadherin internalization (Fig. 7, C and D). Furthermore, the Y685 endocytic signal apparently cannot mediate internalization in the absence of the DEE motif (Fig. 3). These findings indicate that the DEE endocytic signal in VE-cadherin is the predominant endocytic motif in this cadherin and that p120 masking of the DEE signal is sufficient to regulate VE-cadherin endocytosis.
Although the endocytic function of the core p120-binding region is conserved in Drosophila DE-cadherin (Fig. 7, A and B), the requirement for p120 binding to maintain cadherin stability is not, as p120-null flies are apparently normal (Myster et al., 2003). p120 also appears to be dispensable in Caenorhabditis elegans (Pettitt et al., 2003). The reason p120 is not an essential gene in invertebrates remains unknown, but corresponding to the mammalian requirement for p120 stabilization of cadherins is a greatly expanded p120 subfamily of catenins, including p0071, δ-catenin/NPRAP, ARVCF, and the plakophilins as well as an expanded repertoire of p120 splicing isoforms (Hatzfeld, 2005). Both the expanded role of p120 and the increased complexity of the p120 subfamily in mammals suggest that vertebrate tissue patterning requires additional pathways for fine tuning cadherin trafficking not needed in simpler organisms.
But if p120 binding to the cadherin juxtamembrane domain is a key mechanism for controlling cadherin endocytosis, what causes p120 to dissociate from the cadherin? One possibility is that Src-mediated phosphorylation of E-cadherin Y753 and Y754 disrupts p120 binding and allows for ubiquitination by the ubiquitin ligase Hakai (Fujita et al., 2002). However, these tyrosine residues are not well conserved, so this mechanism may not be important for all classical cadherins. For example, VE-cadherin and N-cadherin lack the tyrosine residues that are critical for Hakai binding (Fig. 1 C). Furthermore, there is evidence that Hakai-mediated ubiquitination of E-cadherin alters sorting of the cadherin after endocytosis, rather than mediating endocytosis directly (Palacios et al., 2005). In the case of VE-cadherin, VEGF may also induce cadherin endocytosis through Src activation, though the role of p120 in this pathway remains unclear (Gavard and Gutkind, 2006; Gavard et al., 2008; Kidoya et al., 2010; Hashimoto et al., 2011), and Src-mediated phosphorylation alone is apparently insufficient to disrupt p120 binding (Adam et al., 2010). Surprisingly, we were unable to measure any discernible impact of VEGF on VE-cadherin levels or endocytic rates in quiescent or wounded monolayers (Figs. 10 and S5). Nonetheless, it is highly likely that p120 dissociation from the cadherin is a tightly controlled event that is modulated by a variety of humoral pathways and cellular adhesive interactions. Further study of the mechanisms driving p120 dissociation from the cadherin tail is needed to fully understand how cadherin cell surface levels are modulated in various developmental contexts and disease states.
Although the mechanisms regulating p120 binding to the cadherin juxtamembrane domain have not been fully elucidated, the mutations we have identified within the core p120-binding region allow for the functions of cadherin in p120-bound and p120-unbound states to be further explored. For example, the DEE mutation uncouples p120 binding to the cadherin cytoplasmic tail from control of cadherin endocytosis, resulting in a cadherin that is both unbound to p120 and yet also stable at the cell surface. Analysis of this cadherin mutant in a variety of cellular contexts, including p120-null backgrounds, will further our understanding of precisely how p120 binding modulates cadherin function. Furthermore, several recent studies have implicated cadherin endocytosis in the regulation of junction dynamics and in the modulation of cellular activities such as neuronal patterning (Kawauchi et al., 2010) and the establishment of planar polarity during development (Jarrett et al., 2002; Levayer et al., 2011). However, previous work in this area has relied on disruption of the endocytic pathway broadly, rather than selectively inhibiting cadherin endocytosis. For example, previous studies found that preventing endocytosis through dynamin inhibition either modestly increased (Canel et al., 2010) or significantly decreased (de Beco et al., 2009) the rate of E-cadherin FRAP. In contrast, selective inhibition of VE-cadherin endocytosis with the DEE 646–648 mutation does not affect the rate of fluorescence recovery, though it does increase the mobile fraction (Fig. 8 and Table 1). Because specifically blocking cadherin endocytosis does not restrict cadherin mobility, endocytosis and recycling cannot explain cadherin FRAP. Rather, our FRAP results suggest that cadherins can diffuse rapidly in the membrane but that their movement is limited by transient binding interactions, resulting in fluorescence recovery approximated by first-order kinetics (Sprague et al., 2004).
In contrast with our finding that inhibiting cadherin endocytosis does not inhibit FRAP, altering the ability of VE-cadherin to undergo endocytosis has a dramatic effect on endothelial cell migration (Figs. 10 and S4). Mutation of the DEE sequence dramatically stalled endothelial cell migration both in response to VEGF and in cells cultured in the presence of serum. This inhibition was caused by changes in VE-cadherin trafficking, not p120 localization, because GGG mutant VE-cadherin, which undergoes normal internalization but does not bind p120, did not slow migration. Furthermore, because exogenous expression of wild-type VE-cadherin did not slow migration, inhibition is not caused simply by increased cadherin levels. Rather, cadherin endocytosis is apparently essential for the dynamic regulation of cell contacts needed for directed cell migration. Furthermore, our results suggest that VEGF does not stimulate migration by inducing VE-cadherin endocytosis (Figs. 10 and S5). Rather, cell migration appears to require constitutive endocytic cycling of cadherins to impart plasticity to cell–cell contacts, thereby allowing a migratory signal to drive cell movement. Clearly, static models of cell–cell junctions are insufficient to explain important dynamic processes such as cell migration and tissue patterning. Studies of cadherin endocytic mutants in additional model systems will further reveal the biological importance of adherens junction regulation through endocytic mechanisms during a wider variety of developmental and disease processes.
Materials and methods
Cell culture and reagents
Primary cultures of human dermal microvascular endothelial cells were isolated from neonatal foreskin and cultured in Endothelial Growth Medium 2 Microvascular (Lonza). Cells were grown for 24–48 h to 80% confluence on plates or coverslips coated with 0.1% gelatin for most experiments. African green monkey kidney fibroblast-like cell line COS-7 (American Type Culture Collection), human epidermoid carcinoma cell line A-431D (Lewis et al., 1997), human cervical adenocarcinoma cell line HeLa (American Type Culture Collection), and for adenovirus production, human embryonic kidney cell line QBI-293A (MP Biomedicals) were cultured in Dulbecco’s modification of Eagle’s medium with 4.5 g/liter glucose, l-glutamine, and sodium pyruvate (Corning) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific) and 1% antibiotic/antimycotic solution (Corning). Cells were transfected 24 h before the start of experiments using Lipofectamine 2000 (Life Technologies). Dynamin was inhibited using 80 µM Dynasore (Sigma-Aldrich) dissolved in culture medium.
cDNA constructs
Constructs encoding full-length human VE-cadherin in pBluescript (provided by E. Dejana, Italian Foundation for Cancer Research Institute of Molecular Oncology, Milan, Italy; Navarro et al., 1995), the extracellular and transmembrane domains of IL-2R in a cytomegalovirus (CMV) expression vector (provided by S. LaFlamme, Albany Medical College, Albany, NY; LaFlamme et al., 1994), and the IL-2R–VE-cadherin cytoplasmic tail chimera, constructed by PCR and ligation of amino acids 621–784 of VE-cadherin with a C-terminal myc tag into pBluescript followed by subcloning in frame into the IL-2R CMV expression vector were described previously (Venkiteswaran et al., 2002; Xiao et al., 2003a), as were the wild-type and K444M mutant p120-RFP constructs in pcDNA3.1 expression vectors, which were constructed by PCR (Ishiyama et al., 2010). The IL-2R–E-cadherin cytoplasmic tail chimera construct, containing amino acids 728–878 of human E-cadherin ligated in frame into the IL-2R construct using HindIII and XbaI, was provided by C. Niessen (Center for Molecular Medicine, University of Cologne, Cologne, Germany). The IL-2R–VE-cadherin juxtamembrane domain chimera was constructed based on a study mapping the VE-cadherin catenin-binding domain (Navarro et al., 1995). As previously described, amino acids 621–702 of VE-cadherin were isolated by PCR and ligated in frame to the IL-2R construct using HindIII and XhoI restriction sites (Xiao et al., 2003a). The shorter IL-2R–VE-cadherin chimeras were constructed using site-directed mutagenesis to insert tandem stop codons after VE-cadherin residues 657 and 644. Full-length VE-cadherin, IL-2R–VE-cadherin chimera, and IL-2R–E-cadherin chimera point mutants were also constructed using site-directed mutagenesis (primers described in Table S1). Constructs encoding IL-2R linker and IL-2R linker–cadherin core p120-binding region chimeras were constructed by subcloning synthetic DNA plasmids (Table S2; Integrated DNA Technologies) coding for a polyalanine linker with or without residues 644–664 of human VE-cadherin, residues 754–773 of human E-cadherin, residues 772–791 of human N-cadherin, or residues 1,371–1,390 of Drosophila DE-cadherin into the existing IL-2R–VE-cadherin chimera construct using HindIII and XhoI.
Adenovirus production
The full-length VE-cadherin mutants were subcloned into TagRFP-AS-N (Gateway; Evrogen) in frame with a C-terminal TagRFP, a monomeric RFP, and then shuttled into pAd/CMV/V5-DEST using enzymes (Gateway Clonase; Life Technologies). Plasmids were digested with PacI to expose the viral inverted terminal repeats and transfected into QBI-293A cells for production of human adenovirus type 5 packaged with the desired gene. The virus was harvested by concentration and lysis of virus-producing QBI-293A cells. To induce gene expression, cells were infected 24 h before the start of experiments.
Immunoprecipitation and Western blot analysis
For chimera immunoprecipitation experiments, cells were harvested in 0.5% Triton X-100 (Roche) with protease inhibitors (Complete Mini tablets, EDTA free; Roche), 150 mM NaCl, 10 mM Hepes, 1 mM EGTA, and 0.1 mM MgCl2, pH 7.4. After 30-min incubation at 4°C, cell lysates were centrifuged 16,100 g for 10 min, and the soluble fraction was diluted to a final protein concentration of 1 mg/ml. Cell lysate was then incubated with 2 µg antibody against IL-2R (Table S3) conjugated to ferromagnetic beads (Dynabeads; Life Technologies) for 1 h at 4°C. The beads were then washed with 0.1% Triton X-100 and eluted into Laemmli sample buffer (Bio-Rad Laboratories). For other Western blot experiments, cells were harvested directly into Laemmli sample buffer. Samples were analyzed by SDS-PAGE and immunoblotting. Primary antibodies are listed in Table S3. HRP-conjugated secondary antibodies (Bio-Rad Laboratories), a luminol-based detection system (ECL; GE Healthcare), and autoradiography film (Denville Scientific) were used for detection.
Immunofluorescence
Cells cultured on glass coverslips were fixed either in methanol for 5 min or in 4% paraformaldehyde for 10 min followed by 0.1% Triton X-100 for 8 min, depending on the performance of the antibodies used. Primary antibodies are listed in Table S3. Secondary antibodies conjugated to Alexa Fluor dyes (488, 555, or 647 nm; Life Technologies) were used to identify target molecules. Actin was labeled with Alexa Fluor 488–conjugated phalloidin (Life Technologies). Microscopy was performed using either a wide-field fluorescence microscope (DMRXA2; Leica) equipped with 40×/0.65 NA, 63×/1.32 NA, and 100×/1.40 NA oil immersion objectives, narrow band pass filters, and a digital camera (ORCA; Hamamatsu Photonics) or an inverted microscope (DMI 6000B; Leica) equipped with a 63×/1.40 NA oil immersion objective, a 2D array scanner confocal module (VT Infinity; VisiTech International), 561- and 491-nm solid-state lasers, a digital camera (C9100-12; Hamamatsu Photonics), and a temperature-regulated enclosure maintained at 37°C. Images were captured using Simple PCI software (version 6.6; Hamamatsu Photonics). Super resolution microscopy was performed using an N-SIM system (Nikon) in 3D structured illumination microscopy mode on a microscope (Eclipse Ti-E; Nikon) equipped with a 100×/1.49 NA oil immersion objective, 561- and 488-nm solid-state lasers, and an EM charge-coupled device camera (DU-897; Andor Technology). Images were captured and reconstructed using NIS-Elements software with the N-SIM module (version 3.22; Nikon).
Internalization assay
Assays to follow internalization of cadherin or IL-2R–cadherin chimeras were performed as previously described (Xiao et al., 2003a; Chiasson et al., 2009). In brief, cells were incubated with an antibody against the extracellular domain of VE-cadherin or IL-2R dissolved in culture medium at 4°C for 30 min. Unbound antibody was removed by washing with cold PBS. Cells were then incubated in culture medium at 37°C for various time periods to allow internalization to occur. At the end of the internalization period, cells were returned to 4°C and rinsed, and the remaining surface-bound antibody was removed with a low pH wash (PBS with 100 mM glycine, 20 mM magnesium acetate, and 50 mM potassium chloride, pH 2.2). Cells were then rinsed and processed for immunofluorescence, with a second antibody against VE-cadherin or IL-2R, distinguishable based on isotype, or a fluorescent tag used to label the total cadherin or chimera pool. Internalization was quantified by taking the ratio of fluorescence signals corresponding to the internalized and total cadherin or chimera pools. Where indicated, clathrin-mediated endocytosis was inhibited by incubating cells in a potassium depletion buffer (20 mM Hepes, 140 mM NaCl, 1 mM CaCl2, and 1 mM MgCl2) instead of culture medium beginning 30 min before starting the internalization assay.
FRAP
Primary human dermal microvascular endothelial cells expressing fluorescently tagged VE-cadherin were grown in cover glass chambers (Thermo Fisher Scientific). Microscopy was performed using a laser-scanning confocal microscope (A1R; Nikon) equipped with a 60×/1.40 NA oil immersion objective, a 561-nm laser, and a temperature-regulated enclosure maintained at 37°C. Images were captured using NIS-Elements software (version 3.22). Each FRAP sequence consisted of acquisition of two prebleach images, photobleaching of a section of a cell border (3.5–15.3 µm long × 5 µm wide; 33% laser power for 3.5 s), and acquisition of postbleach images at 15-s intervals for 10 min. Images were acquired at 1.5% laser power. A Perfect Focus System (Nikon) was used to maintain focus during the FRAP sequence. Fluorescence recovery was calculated as the ratio of the mean background-subtracted fluorescence intensity within the bleach area to the mean background-subtracted fluorescence intensity of unbleached portions of cell borders to correct for acquisition-related photobleaching, normalized to the prebleach fluorescence intensity. Exponential curves (f(t) = A(1 − e−kt), in which k = ln(2)/t1/2, and A is the mobile fraction) were fit using R (version 2.13; R Foundation for Statistical Computing).
Colocalization analysis
IL-2R–VE-cadherin chimera and clathrin colocalization experiments were designed as modified internalization assays. Chimera-expressing cells were incubated with an antibody against the extracellular domain of IL-2R dissolved in culture medium at 4°C for 30 min, and unbound antibody was removed by washing with cold PBS. Cells were then incubated in culture medium at 4°C for an additional 30 min to allow chimera clustering but not endocytosis. After incubation, cells were processed for immunofluorescence. Colocalization was quantified by computing Pearson’s product–moment correlation coefficient for chimera and clathrin pixel fluorescence intensities in individual cells using an algorithm built on the Commons Math library (version 2.0; Apache Software Foundation).
Migration assays
Migration of cells expressing wild-type or mutant cadherins was measured using scratch wound assays. Cells were grown to confluent monolayers, scratched with a pipette tip, and imaged over time with a bright-field microscope (DM IL; Leica) equipped with a 5×/0.12 NA objective and a camera (DFC420 C; Leica). Images were acquired using FireCam software (version 3.4; Leica). For VEGF-induced migration, endothelial cells were serum starved for 1 h before being scratched, and 100 ng/ml VEGF165 peptide (PeproTech) was added 12 h after wounding. Cell replication rates were measured by incorporation of a thymidine analogue, EdU (Click-iT EdU Imaging kit; Life Technologies). For the single-cell migration assay, cells were imaged at 30-min intervals with an inverted microscope (DMI 6000B) equipped with a 10×/0.30 NA objective, a camera (Retiga EXi; QImaging), and a temperature-regulated enclosure maintained at 37°C. Phase-contrast images were acquired using Simple PCI software (version 6.6). Cell tracking data were extracted using the TrackMate plugin (version 1.2; created by N. Perry, J.-Y. Tinevez, and J. Schindelin) for ImageJ (versions 1.3–1.4; National Institutes of Health).
Image analysis and statistics
ImageJ software (versions 1.3–1.4) was used for all image analysis, and the application of lookup tables was used to produce display images. Custom ImageJ plugins were used to automate quantification. Statistics were computed using R (version 2.13). The Kruskal-Wallis rank sum test with Dunn’s method for multiple comparisons was used to evaluate scaled data, and the χ2 test was used to evaluate count data.
Online supplemental material
Fig. S1 shows that inhibition of clathrin-mediated endocytosis by potassium depletion blocks internalization of IL-2R–VE-cadherin cytoplasmic tail chimeras and IL-2R–VE-cadherin core p120-binding region chimeras. Fig. S2 demonstrates that there is no relationship between the size of the bleach region and VE-cadherin–RFP FRAP, inconsistent with diffusion-limited recovery. Fig. S3 shows super resolution microscopy confirming clustering of IL-2R–VE-cadherin cytoplasmic tail chimeras into clathrin-enriched membrane domains. Fig. S4 shows that constitutive endothelial cell migration requires VE-cadherin endocytosis. Fig. S5 shows that cadherin mutations do not affect distribution at the wound edge or undirected single-cell migration, and cadherin endocytosis is not increased at the wound edge. Videos 1 and 2 show time-lapse images of wild-type (Video 1) and endocytosis-defective (Video 2) fluorescently tagged VE-cadherin expressed in microvascular endothelial cells (see also Fig. 5 C). Videos 3–5 show time-lapse images from FRAP sequences with wild-type (Video 3), DEE mutant (Video 4), and GGG mutant (Video 5) VE-cadherin–RFP (see also Fig. 8 A). Table S1 lists primers used for the creation of mutant cadherin and chimera constructs. Table S2 lists the synthetic genes used for the creation of chimera constructs. Table S3 lists the primary antibodies used for these experiments.
Wild-type VE-cadherin forms dynamic junctions. Time-lapse confocal images of wild-type VE-cadherin–RFP expressed in primary human dermal microvascular endothelial cells. The box highlights an endocytic event. Individual frames are shown in Fig. 5 C. Images were captured by epifluorescence microscopy (inverted DMI 6000B) every 3 s and are 27 µm wide. VE-cad, VE-cadherin.
Wild-type VE-cadherin forms dynamic junctions. Time-lapse confocal images of wild-type VE-cadherin–RFP expressed in primary human dermal microvascular endothelial cells. The box highlights an endocytic event. Individual frames are shown in Fig. 5 C. Images were captured by epifluorescence microscopy (inverted DMI 6000B) every 3 s and are 27 µm wide. VE-cad, VE-cadherin.
Endocytosis-defective VE-cadherin forms static junctions. Time-lapse confocal images of DEE mutant VE-cadherin–RFP expressed in primary human dermal microvascular endothelial cells. Individual frames are shown in Fig. 5 C. Images were captured by epifluorescence microscopy (inverted DMI 6000B) every 3 s and are 27 µm wide. VE-cad, VE-cadherin; mut., mutant.
Endocytosis-defective VE-cadherin forms static junctions. Time-lapse confocal images of DEE mutant VE-cadherin–RFP expressed in primary human dermal microvascular endothelial cells. Individual frames are shown in Fig. 5 C. Images were captured by epifluorescence microscopy (inverted DMI 6000B) every 3 s and are 27 µm wide. VE-cad, VE-cadherin; mut., mutant.
FRAP sequence with wild-type VE-cadherin. False-color time-lapse video of RFP-tagged wild-type VE-cadherin expressed in primary human dermal microvascular endothelial cells. The video begins shortly before bleaching of a 5-µm-long section of the junction (arrow) and continues for 10 min after the bleach. Images were captured by laser-scanning confocal microscopy (A1R) every 15 s and are 31.9 µm wide. Individual frames are shown in Fig. 8 A. VE-cad, VE-cadherin; WT, wild type.
FRAP sequence with wild-type VE-cadherin. False-color time-lapse video of RFP-tagged wild-type VE-cadherin expressed in primary human dermal microvascular endothelial cells. The video begins shortly before bleaching of a 5-µm-long section of the junction (arrow) and continues for 10 min after the bleach. Images were captured by laser-scanning confocal microscopy (A1R) every 15 s and are 31.9 µm wide. Individual frames are shown in Fig. 8 A. VE-cad, VE-cadherin; WT, wild type.
FRAP sequence with DEE mutant VE-cadherin. False-color time-lapse video of RFP-tagged DEE mutant VE-cadherin expressed in primary human dermal microvascular endothelial cells. The video begins shortly before bleaching of a 5-µm-long section of the junction (arrow) and continues for 10 min after the bleach. Images were captured by laser-scanning confocal microscopy (A1R) every 15 s and are 31.9 µm wide. Individual frames are shown in Fig. 8 A. VE-cad, VE-cadherin.
FRAP sequence with DEE mutant VE-cadherin. False-color time-lapse video of RFP-tagged DEE mutant VE-cadherin expressed in primary human dermal microvascular endothelial cells. The video begins shortly before bleaching of a 5-µm-long section of the junction (arrow) and continues for 10 min after the bleach. Images were captured by laser-scanning confocal microscopy (A1R) every 15 s and are 31.9 µm wide. Individual frames are shown in Fig. 8 A. VE-cad, VE-cadherin.
FRAP sequence with GGG mutant VE-cadherin. False color time-lapse video of RFP-tagged GGG mutant VE-cadherin expressed in primary human dermal microvascular endothelial cells. The video begins shortly before bleaching of a 5-µm-long section of the junction (arrow) and continues for 10 min after the bleach. Images were captured by laser-scanning confocal microscopy (A1R) every 15 s and are 31.9 µm wide. Individual frames are shown in Fig. 8 A. VE-cad, VE-cadherin.
FRAP sequence with GGG mutant VE-cadherin. False color time-lapse video of RFP-tagged GGG mutant VE-cadherin expressed in primary human dermal microvascular endothelial cells. The video begins shortly before bleaching of a 5-µm-long section of the junction (arrow) and continues for 10 min after the bleach. Images were captured by laser-scanning confocal microscopy (A1R) every 15 s and are 31.9 µm wide. Individual frames are shown in Fig. 8 A. VE-cad, VE-cadherin.
Acknowledgments
We wish to thank Dr. A. Mattheyses for valuable guidance in developing and analyzing the FRAP experiments, S. Summers for help with adenovirus production and primary cell isolations, Dr. K.J. Green for reviewing the manuscript, and members of the Kowalczyk laboratory for their help and advice.
This work was supported by grants from the National Institutes of Health (R01AR050501 and R01AR048266 to A.P. Kowalczyk), the Integrated Cellular Imaging Microscopy Core of the Emory Neuroscience National Institute of Neurological Disorders and Stroke Core Facilities grant (P30NS055077), and the Canadian Cancer Society (to M. Ikura). B.A. Nanes was supported by fellowships from the American Heart Association and the National Institutes of Health (F30HL110447).
References
Author notes
B.A. Nanes and C. Chiasson-MacKenzie contributed equally to this paper.