Haspin phosphorylates histone H3 at threonine-3 (H3T3ph), providing a docking site for the Aurora B complex at centromeres. Aurora B functions to correct improper kinetochore–microtubule attachments and alert the spindle checkpoint to the presence of misaligned chromosomes. We show that Haspin inhibitors decreased H3T3ph, resulting in loss of centromeric Aurora B and reduced phosphorylation of centromere and kinetochore Aurora B substrates. Consequently, metaphase chromosome alignment and spindle checkpoint signaling were compromised. These effects were phenocopied by microinjection of anti-H3T3ph antibodies. Retargeting Aurora B to centromeres partially restored checkpoint signaling and Aurora B–dependent phosphorylation at centromeres and kinetochores, bypassing the need for Haspin activity. Haspin inhibitors did not obviously affect phosphorylation of histone H3 at serine-10 (H3S10ph) by Aurora B on chromosome arms but, in Aurora B reactivation assays, recovery of H3S10ph was delayed. Haspin inhibitors did not block Aurora B localization to the spindle midzone in anaphase or Aurora B function in cytokinesis. Thus, Haspin inhibitors reveal centromeric roles of Aurora B in chromosome movement and spindle checkpoint signaling.
Introduction
The chromosomal passenger complex (CPC), which consists of the kinase Aurora B and the regulatory subunits INCENP, Survivin, and Borealin/Dasra, plays a key role in controlling chromosome segregation and cytokinesis. The CPC was named for its subcellular distribution in mitosis; it localizes on chromosome arms in prophase and, during prometaphase, accumulates at inner centromeres. At the onset of anaphase, the CPC leaves centromeres and transfers to the central spindle. Aurora B phosphorylates multiple substrates, including histone H3 at serine-10 (H3S10ph) on chromatin, mitotic centromere-associated kinesin (MCAK) at inner centromeres, centromere protein A Serine-7, phosphorylated (CENP-AS7ph) at outer centromeres, and KNL1/Mis12 complex/Ndc80 complex (KMN) network proteins at kinetochores (Ruchaud et al., 2007; Welburn et al., 2010).
Aurora B has attracted particular attention because of its functions in regulating kinetochore–microtubule (KT-MT) attachments and spindle checkpoint signaling. If a chromosome attaches to microtubules such that tension is not generated across sister kinetochores, Aurora B acts to destabilize the erroneous attachment. In current models, centromeric Aurora B phosphorylates KMN network proteins at kinetochores, reducing their binding to microtubules (Cheeseman et al., 2006; DeLuca et al., 2006; Liu et al., 2009; Welburn et al., 2010). In this way, Aurora B produces unattached kinetochores that prevent satisfaction of the mitotic spindle checkpoint until all chromosomes establish tension-generating (typically bi-oriented) microtubule attachments (Biggins and Murray, 2001; Tanaka et al., 2002; Hauf et al., 2003; Pinsky et al., 2006; Yang et al., 2009). Emerging evidence suggests that Aurora B also plays a more direct role in spindle checkpoint signaling that is independent of its role in correcting KT-MT attachments (Biggins and Murray, 2001; Kallio et al., 2002; Ditchfield et al., 2003; Hauf et al., 2003; Petersen and Hagan, 2003; King et al., 2007; Vader et al., 2007; Vanoosthuyse and Hardwick, 2009; Maldonado and Kapoor, 2011; Santaguida et al., 2011; Saurin et al., 2011; Matson et al., 2012). However, it remains unclear whether Aurora B must be positioned at inner centromeres to fulfill its function in the spindle checkpoint, particularly because the existence of a kinetochore-bound population of Aurora B has been proposed (DeLuca et al., 2011; Petsalaki et al., 2011).
We and others recently showed that phosphorylation of histone H3 at threonine-3 (H3T3ph), by Haspin creates a chromatin binding site for the BIR domain of Survivin, allowing CPC positioning at inner centromeres in mitosis (Kelly et al., 2010; Wang et al., 2010; Yamagishi et al., 2010). Haspin RNAi, or complementation of Survivin RNAi with Survivin mutants defective in binding to H3T3ph, reduced Aurora B accumulation at centromeres, diminished the Aurora B–dependent centromeric localization of MCAK, and weakened the spindle checkpoint response to the microtubule-stabilizing drug taxol (Wang et al., 2010; Niedzialkowska et al., 2012). However, H3S10ph, CENP-AS7ph, and the spindle checkpoint response to the microtubule-depolymerizing drug nocodazole were relatively unaffected. In addition, although previous work in vitro and using Xenopus laevis egg extracts suggested that H3T3ph contributes to Aurora B activation, either by preventing an inhibitory effect of H3 (Rosasco-Nitcher et al., 2008) or by generating a high local concentration of Aurora B required to allow transactivation on chromatin (Kelly et al., 2007, 2010), this effect was not clear after Haspin RNAi in human cells (Wang et al., 2010). These findings suggested two possibilities. First, some functions of Aurora B might be independent of Haspin and H3T3ph. For example, a Bub1–Sgo1 pathway that also contributes to centromeric CPC localization (Yamagishi et al., 2010; F. Wang et al., 2011) might be sufficient for phosphorylation of some Aurora B substrates, and Survivin BIR domain mutations could alter functions other than H3T3ph binding (Jeyaprakash et al., 2011). Alternatively, the result could be explained if Haspin depletion by RNAi was incomplete in prior studies and different Aurora B substrates require different levels of centromeric Aurora B activity. Because H3T3ph is dependent on the kinase activity of Haspin, small molecule inhibitors of Haspin would provide independent means to address these questions. Compared with RNAi-based approaches, inhibitors offer the potential advantages of selective, rapid, and strong temporal inhibition of kinase activity without depleting the protein itself (Knight and Shokat, 2005), which might have kinase-independent functions in mitosis and roles at other cell cycle stages.
Using high-throughput chemical library screening, we recently identified several Haspin inhibitors (Patnaik et al., 2008). We determined structure-activity relationships for two of these inhibitor classes, and selected one high-potency compound from each, LDN-192960 and LDN-211898, for further studies (Cuny et al., 2010, 2012). A third distinct selective Haspin inhibitor, 5-iodotubercidin, was identified using thermal stability assays (Eswaran et al., 2009; Balzano et al., 2011). Neither LDN-192960 nor LDN-211898 significantly inhibited a range of other mitotic kinases including Cdk1–Cyclin B, Aurora A, Aurora B–INCENP, Aurora C, Nek2, Plk1, and Mps1 (for further selectivity data on >270 kinases, see Cuny et al., 2010, 2012), and 5-iodotubercidin was much less potent against or did not inhibit Cdk1–Cyclin B, Aurora A, Aurora B–INCENP, Nek2, Bub1, Plk1, or Mps1 in vitro (Balzano et al., 2011; for further selectivity data, see Fedorov et al., 2007; and De Antoni et al. in this issue). A recently published study described another inhibitor of Haspin (CHR-6494; Huertas et al., 2012), but the selectivity of this molecule was less well defined, and no analysis of its effects on Aurora B function was reported. Here, we make use of 5-iodotubercidin, LDN-192960, and LDN-211898 to determine the function of Haspin kinase activity in mitotic cells, with emphasis on its role in regulating Aurora B at centromeres.
Results
Three distinct compounds inhibit H3T3 phosphorylation by Haspin in vitro and in cells
We determined IC50 values for inhibition of H3T3 peptide phosphorylation by full-length Haspin of 3 nM for 5-iodotubercidin, 10 nM for LDN-192960, and 100 nM for LDN-211898 (Fig. S1, A–C). These values are consistent with previous studies (Patnaik et al., 2008; Cuny et al., 2010, 2012; Balzano et al., 2011), and demonstrate that these three molecules show a range of potencies for Haspin inhibition in vitro.
To determine compound potency for Haspin inhibition in cells, we arrested HeLa cells in mitosis using nocodazole, then added Haspin inhibitors in the continued presence of nocodazole (and MG132 to counter mitotic exit) for 1 h. Immunoblotting of cell lysates for H3T3ph showed that all three inhibitors strongly inhibited Haspin (Fig. 1 A), with relative potencies that reflected their in vitro activity. In contrast, none of the inhibitors had a detectable effect on the Aurora B product H3S10ph at these doses (but see later in this paper). Immunofluorescence analysis confirmed that all three inhibitors reduced H3T3ph in mitotic U2OS cells. Notably, although 1 µM 5-iodotubercidin or LDN-192960, or 5 µM LDN-211898 caused dramatic reductions in H3T3ph, complete loss of detectable H3T3ph required >3 µM 5-iodotubercidin, >5 µM LDN-192960, or >30 µM LDN-211898 (Fig. 1 B).
Haspin inhibitors reduce H3T3ph and displace Aurora B from centromeres but not the central spindle. (A) HeLa cells were released from double thymidine block and, after 7 h, 5 µM nocodazole was added for 6 h. Mitotic cells collected by “shake-off” were replated in 5 µM nocodazole, 20 µM MG132, and Haspin inhibitors for 1 h. Immunoblots of cell lysates are shown. (B) U2OS cells were released from thymidine block and, after 7 h, 33 nM nocodazole was added. After another 4 h, 0.33 µM nocodazole, 20 µM MG132, and kinase inhibitors were added for 1 h before fixation and immunofluorescence microscopy. To visualize residual H3T3ph, two different red channel exposure times are shown. (C) The ratio of centromere to chromosome arm Aurora B intensity was determined for cells treated as in B (15 centromeres/cell; n = 8 or 9 cells). Means + SD are shown (error bars); ***, P < 0.001 vs. DMSO. (D) Asynchronous U2OS cells were treated with Haspin inhibitors for 2 h. Immunofluorescence microscopy of anaphase cells is shown. Bars, 5 µm.
Haspin inhibitors reduce H3T3ph and displace Aurora B from centromeres but not the central spindle. (A) HeLa cells were released from double thymidine block and, after 7 h, 5 µM nocodazole was added for 6 h. Mitotic cells collected by “shake-off” were replated in 5 µM nocodazole, 20 µM MG132, and Haspin inhibitors for 1 h. Immunoblots of cell lysates are shown. (B) U2OS cells were released from thymidine block and, after 7 h, 33 nM nocodazole was added. After another 4 h, 0.33 µM nocodazole, 20 µM MG132, and kinase inhibitors were added for 1 h before fixation and immunofluorescence microscopy. To visualize residual H3T3ph, two different red channel exposure times are shown. (C) The ratio of centromere to chromosome arm Aurora B intensity was determined for cells treated as in B (15 centromeres/cell; n = 8 or 9 cells). Means + SD are shown (error bars); ***, P < 0.001 vs. DMSO. (D) Asynchronous U2OS cells were treated with Haspin inhibitors for 2 h. Immunofluorescence microscopy of anaphase cells is shown. Bars, 5 µm.
Haspin inhibitors delocalize the CPC from centromeres but not the central spindle
RNAi of Haspin causes premature loss of cohesion in mitosis (Dai et al., 2006). However, centromeres remained paired in numerous immunofluorescence experiments with all three Haspin inhibitors, including when spread mitotic chromosomes were examined (Fig. S1 D). We conclude that the inhibitors allow assessment of kinase-dependent functions of Haspin in the absence of premature sister chromatid separation, which had previously confounded direct analysis of the role of this kinase in error correction and the spindle checkpoint.
Haspin RNAi causes CPC loss from centromeres, but not the central spindle (Wang et al., 2010). Similarly, when added to nocodazole-arrested mitotic cells, all three Haspin inhibitors caused displacement of Aurora B from inner centromeres to a diffuse distribution on chromatin, even when MG132 was included to counter mitotic exit (Fig. 1, B and C; and Fig. 2, A and B). In contrast, although anaphase was disrupted at high doses of Haspin inhibitors (see “Live imaging of cells treated with Haspin inhibitors”), Aurora B was not lost from central spindles (Fig. 1 D), and CPC formation was not affected (Fig. S1 E). Direct comparison of H3T3ph and Aurora B staining suggested that maximal displacement of preaccumulated centromeric CPC required >3 µM 5-iodotubercidin, >10 µM LDN-192960, or >100 µM LDN-211898, which suggested that even low levels of H3T3ph can maintain a significant population of the CPC at centromeres (Figs. 1 B and 2 A). These results provide evidence that the kinase activity of Haspin is required for normal centromeric localization of Aurora B, which is consistent with the notion that H3T3ph provides a docking site for the CPC.
Haspin inhibitors influence the maintenance of Aurora B activity toward centromeric targets. (A) Nocodazole-arrested U2OS cells were obtained as in Fig. 1 B, then kinase inhibitors and 20 µM MG132 were added for 1 h in the presence (MCAK and Aurora B) or absence (CENP-AS7ph and H3S10ph) of 0.33 µM nocodazole. Mitotic Aurora B localization, the centromeric staining intensity of MCAK and CENP-AS7ph, and H3S10ph intensity on chromosomes were classified for at least 100 cells in each condition in one experiment by immunofluorescence microscopy. Similar results were obtained in duplicate experiments. (B and C) Example images of cells treated as in A. Bars, 5 µm. (D) The intensities of CENP-AS7ph and H3S10ph on mitotic chromosomes in each condition were quantified (n = 6–14 cells). Results are expressed as a ratio to centromeric autoantigen staining intensity at the same centromeres. Means + SD are shown (error bars); ***, P < 0.001 vs. DMSO.
Haspin inhibitors influence the maintenance of Aurora B activity toward centromeric targets. (A) Nocodazole-arrested U2OS cells were obtained as in Fig. 1 B, then kinase inhibitors and 20 µM MG132 were added for 1 h in the presence (MCAK and Aurora B) or absence (CENP-AS7ph and H3S10ph) of 0.33 µM nocodazole. Mitotic Aurora B localization, the centromeric staining intensity of MCAK and CENP-AS7ph, and H3S10ph intensity on chromosomes were classified for at least 100 cells in each condition in one experiment by immunofluorescence microscopy. Similar results were obtained in duplicate experiments. (B and C) Example images of cells treated as in A. Bars, 5 µm. (D) The intensities of CENP-AS7ph and H3S10ph on mitotic chromosomes in each condition were quantified (n = 6–14 cells). Results are expressed as a ratio to centromeric autoantigen staining intensity at the same centromeres. Means + SD are shown (error bars); ***, P < 0.001 vs. DMSO.
Haspin inhibitors influence Aurora B activity toward centromeric targets
To determine functional consequences of Haspin inhibition, we conducted additional assays in cells previously arrested in mitosis in nocodazole. This stringent test minimizes indirect effects on other stages of the cell cycle and assesses maintenance of mitotic functions rather than their establishment. Indeed, loss of phosphorylation in these circumstances is likely to be dependent on phosphatase activity. Nevertheless, we observed loss of MCAK from centromeres upon Haspin inhibitor treatment in both U2OS (Fig. 2, A and B) and HeLa cells (Fig. S2 A). The loss of MCAK caused by Haspin inhibition, but not that caused by direct inhibition of Aurora B, could be rescued by artificially restoring Aurora B to centromeres using a CENP-B fusion protein (Liu et al., 2009) containing residues 47 to 920 of INCENP (Fig. 3 A). This confirmed that loss of MCAK caused by Haspin inhibition was likely caused by delocalization of Aurora B, and was unlikely to be caused by direct inhibition of Aurora B.
Artificial retargeting of Aurora B restores MCAK and CENP-AS7ph at centromeres of Haspin-inhibited cells. (A and B) HeLa cells were transfected with CENP-B–EGFP or EGFP–CENP-B–INCENP plasmids between double thymidine treatments. 7 h after release from G1/S, 30 nM nocodazole was added to accumulate mitotic cells. Then, 3.5 h later, medium containing 20 µM MG132 with or without 10 µM Haspin inhibitors or 50 nM Hesperadin, in the presence (A) or absence (B) of 0.33 µM nocodazole, was added for 75 min. Approximately 100 mitotic cells in each condition from one experiment were classified according to the intensity of centromeric MCAK (A) or CENP-AS7ph (B) by immunofluorescence microscopy. Similar results were obtained in a second experiment. Bars, 5 µm.
Artificial retargeting of Aurora B restores MCAK and CENP-AS7ph at centromeres of Haspin-inhibited cells. (A and B) HeLa cells were transfected with CENP-B–EGFP or EGFP–CENP-B–INCENP plasmids between double thymidine treatments. 7 h after release from G1/S, 30 nM nocodazole was added to accumulate mitotic cells. Then, 3.5 h later, medium containing 20 µM MG132 with or without 10 µM Haspin inhibitors or 50 nM Hesperadin, in the presence (A) or absence (B) of 0.33 µM nocodazole, was added for 75 min. Approximately 100 mitotic cells in each condition from one experiment were classified according to the intensity of centromeric MCAK (A) or CENP-AS7ph (B) by immunofluorescence microscopy. Similar results were obtained in a second experiment. Bars, 5 µm.
Previously, we detected only minor changes in phosphorylation of the Aurora B target CENP-AS7 after Haspin RNAi (Wang et al., 2010). However, all three Haspin inhibitors substantially reduced CENP-AS7ph in nocodazole-treated cells (Fig. 2, A and C; and Fig. S2 A), and this loss could be rescued by forced targeting of Aurora B to centromeres with CENP-B–INCENP (Fig. 3 B). A comparison of MCAK and CENP-AS7ph staining in U2OS cells showed that loss of CENP-AS7ph required higher inhibitor doses than loss of MCAK (Fig. 2 A), a finding confirmed by costaining in individual HeLa cells (Fig. S2), and consistent with our previous observation that CENP-AS7ph is less sensitive to loss of Haspin activity than MCAK localization (Wang et al., 2010).
To examine the effect of Haspin inhibition on Aurora B activity beyond centromeres, we used an antibody that recognizes H3S10ph on chromosome arms (Dai et al., 2005; Wang et al., 2010). Immunofluorescence staining showed that H3S10ph was not detectably decreased, even at high concentrations of Haspin inhibitors (Fig. 2, A and C), and in cells in which CENP-AS7ph was reduced (Fig. 2, C and D). Aurora B inhibitors such as Hesperadin caused a strong reduction in H3S10ph, confirming that H3S10ph dephosphorylation can be efficient in these conditions (Fig. 2, A, C, and D). We conclude that in these experimental circumstances, Haspin is required for the full activity of Aurora B toward centromeric targets such as MCAK and CENP-A, but that H3S10 phosphorylation on chromosome arms is significantly less dependent on Haspin.
A role for Haspin in Aurora B activation
Previous studies suggested that H3T3ph contributes to activation of Aurora B (see Introduction). Indeed, Aurora B activation at centromeres is proposed to be crucial for generating a gradient of Aurora B activity emanating from centromeres that can phosphorylate substrates across chromosomes (E. Wang et al., 2011) and along spindle microtubules (Tseng et al., 2010; Tan and Kapoor, 2011). This seems at odds with our finding that H3S10ph is insensitive to Haspin inhibition. However, the studies described so far were performed in cells first blocked in nocodazole, in which Aurora B is strongly active and its substrates phosphorylated before inhibitor addition.
To test if Haspin influences Aurora B activation, we used conditions in which Aurora B is initially inhibited in mitotic cells, but reactivation is then allowed upon removal of Aurora B inhibitor (Fig. 4 A). After treatment with Hesperadin in the absence of Haspin inhibitors, Aurora B was partly delocalized, as expected (F. Wang et al., 2011), but still showed some accumulation at centromeres (Fig. S3, A and B). After removal of Hesperadin, Aurora B resumed a strongly centromeric localization (Fig. S3, A and B), and CENP-AS7ph at centromeres and H3S10ph on chromosome arms returned to near maximal levels within 1 h (Fig. 4, B and C). Notably, Aurora B autophosphorylation at Thr-232 (Aurora B-T232ph; representing an activated form of the kinase; Yasui et al., 2004) recovered more quickly at centromeres than did Aurora B localization (Fig. S3 B), which is consistent with rapid Aurora B activation at centromeres. When we repeated these experiments in the presence of Haspin inhibitors, Aurora B was initially diffuse on chromosomes, and did not recover its centromeric localization upon removal of Hesperadin (Fig. S3, A and B). Aurora B autophosphorylation recovered slowly throughout the chromatin and did not show an accumulation at the centromere (Fig. S3, A and B). Consistent with delayed activation of centromeric Aurora B, phosphorylation of CENP-AS7 was strongly reduced in these conditions (Fig. 4, B and C). In contrast, H3S10ph did recover strongly, showing that strong centromeric accumulation of Aurora B is not essential for H3S10ph generation on arms. However, the kinetics of H3S10ph recovery were delayed by Haspin inhibition (Fig. 4, B and C), which suggests that, in these experimental circumstances, H3T3ph-dependent accumulation of the CPC can contribute to activation of Aurora B and phosphorylation of substrates on chromosome arms. Retargeting of Aurora B to centromeres using CENP-B–INCENP in the presence of Haspin inhibitors caused H3S10ph to increase first at centromeric regions, but also modestly increased the rate at which H3S10ph returned on chromosome arms (Fig. S3, C and D), which is consistent with a report that centromeric activation of Aurora B can enhance phosphorylation of Aurora B targets at distant sites (E. Wang et al., 2011).
Haspin inhibitors delay Aurora B activation. (A) Treatment scheme for Aurora B reactivation assays. (B) Immunofluorescence microscopy of CENP-AS7ph and H3S10ph in HeLa cells treated as in A. Bar, 5 µm. (C) Approximately 100 mitotic cells in each condition from one experiment were classified according to the intensity of CENP-AS7ph or H3S10ph staining. Similar results were obtained in a second experiment using the Aurora B inhibitor ZM447439.
Haspin inhibitors delay Aurora B activation. (A) Treatment scheme for Aurora B reactivation assays. (B) Immunofluorescence microscopy of CENP-AS7ph and H3S10ph in HeLa cells treated as in A. Bar, 5 µm. (C) Approximately 100 mitotic cells in each condition from one experiment were classified according to the intensity of CENP-AS7ph or H3S10ph staining. Similar results were obtained in a second experiment using the Aurora B inhibitor ZM447439.
We then determined if this kinetic difference in Aurora B activation was relevant in a relatively unperturbed mitosis. In cells entering mitosis in the presence of Haspin inhibitors, H3S10 remained strongly phosphorylated, even in cells in which CENP-AS7ph was greatly reduced (Fig. S3 E). Together, these findings indicate that activation of Aurora B for CENP-AS7 phosphorylation at centromeres is more strongly dependent on the correct Haspin-mediated localization of the CPC than H3S10ph on chromosome arms, but that increased centromeric Aurora B localization can contribute to arm substrate phosphorylation in certain experimental situations.
Haspin inhibitors compromise error correction
To determine if the Haspin-dependent population of the CPC is required for KT-MT error correction, we performed monastrol release assays. Monastrol is a kinesin-5/Eg5 inhibitor that prevents centrosome separation during mitotic entry, resulting in the formation of monopolar spindles with erroneously attached chromosomes. Upon removal of monastrol, correction of these attachments is hindered in the presence of Aurora B inhibitors (Lampson et al., 2004). All three Haspin inhibitors compromised the efficiency of chromosome alignment in this assay, with the order of potency expected (Fig. S4 A). As described earlier, we reasoned that the relatively high compound concentrations required might be caused by the presence of already strongly phosphorylated Aurora B substrates at the time of monastrol washout into Haspin inhibitors, allowing substantial error correction before Haspin-dependent Aurora B targets became dephosphorylated. We therefore conducted assays in which Aurora B was initially inhibited but activation was allowed upon monastrol and Hesperadin washout. In this format, all three Haspin inhibitors strongly hindered chromosome alignment at all tested doses (Fig. 5 A). In these assays, we were unable to determine if retargeting Aurora B to centromeres could rescue the defect because expression of CENP-B–INCENP itself disrupts error correction, presumably because the increased local concentration of Aurora B near kinetochores decreases microtubule binding (Liu et al., 2009; Becker et al., 2010). Nevertheless, the results indicate that the CPC population targeted by the Haspin–H3T3ph pathway is required for efficient error correction.
Haspin inhibitors compromise KT-MT attachment correction. (A) HeLa cells were released from thymidine treatment and, after 7 h, 100 µM monastrol was added for 3 h to accumulate cells in mitosis with incorrect KT-MT attachments. Then 50 nM Hesperadin was added to inhibit Aurora B, together with 20 µM MG132. After 1.5 h, monastrol and Hesperadin were removed by washing into fresh medium containing Haspin inhibitors or controls in the continued presence of MG132. Approximately 200 cells were classified in each condition by fluorescence microscopy. Means ± SD are shown (error bars), n = 3. (B) Immunofluorescence microscopy of Dsn1-S109ph during Aurora B reactivation in the presence or absence of Haspin inhibitors in cells treated as in Fig. 4 A. (C) Approximately 100 mitotic cells in each condition from one experiment as in B were classified according to phosphorylated Dsn1 (Dsn1-S109ph) or total Dsn1 (Dsn1) staining intensity at kinetochores. Similar results were obtained in a duplicate experiment. (D) HeLa cells were transfected with plasmids encoding CENP-B–EGFP or EGFP–CENP-B–INCENP between and after double thymidine treatments, then treated as in B. (E) The intensity of Dsn1-S109ph staining in EGFP-positive cells from D was classified in one experiment as in C. Similar results were obtained in a duplicate experiment. Bars, 5 µm.
Haspin inhibitors compromise KT-MT attachment correction. (A) HeLa cells were released from thymidine treatment and, after 7 h, 100 µM monastrol was added for 3 h to accumulate cells in mitosis with incorrect KT-MT attachments. Then 50 nM Hesperadin was added to inhibit Aurora B, together with 20 µM MG132. After 1.5 h, monastrol and Hesperadin were removed by washing into fresh medium containing Haspin inhibitors or controls in the continued presence of MG132. Approximately 200 cells were classified in each condition by fluorescence microscopy. Means ± SD are shown (error bars), n = 3. (B) Immunofluorescence microscopy of Dsn1-S109ph during Aurora B reactivation in the presence or absence of Haspin inhibitors in cells treated as in Fig. 4 A. (C) Approximately 100 mitotic cells in each condition from one experiment as in B were classified according to phosphorylated Dsn1 (Dsn1-S109ph) or total Dsn1 (Dsn1) staining intensity at kinetochores. Similar results were obtained in a duplicate experiment. (D) HeLa cells were transfected with plasmids encoding CENP-B–EGFP or EGFP–CENP-B–INCENP between and after double thymidine treatments, then treated as in B. (E) The intensity of Dsn1-S109ph staining in EGFP-positive cells from D was classified in one experiment as in C. Similar results were obtained in a duplicate experiment. Bars, 5 µm.
Phosphorylation of several KMN network proteins including KNL1, Dsn1, and Hec1/Ndc80 at kinetochores contributes to the regulation of microtubule attachment (Welburn et al., 2010). Consistent with a role of the Haspin-dependent CPC population in error correction, Haspin inhibitors strongly reduced the phosphorylation of Dsn1 at the Aurora B target residue S109 (Dsn1-S109ph) in Aurora B reactivation assays (Fig. 5, B and C; and Fig. S4 B), and Dsn1 phosphorylation could be largely restored by retargeting Aurora B to centromeres using CENP-B–INCENP (Fig. 5, D and E).
Live imaging of cells treated with Haspin inhibitors
To directly observe the effects of Haspin inhibitors on mitosis, we performed time-lapse microscopy of U2OS cells expressing histone H2B-mRFP and γ-tubulin–GFP (Fig. 6 A and Video 1; Dai et al., 2009). All three inhibitors caused a moderate increase in the length of mitosis, defined as the period between nuclear envelope breakdown (NEB) and anaphase onset (Fig. S4 C). This was reminiscent of a similar extension of mitosis reported for cells treated with Aurora B inhibitors (Girdler et al., 2006; Maciejowski et al., 2010; Hégarat et al., 2011). We also noted a dose-dependent decline in the number of cells entering mitosis. This effect was not apparent in prior RNAi studies, and whether it reflects a role for Haspin outside mitosis or off-target effects of the compounds requires further investigation.
Mitosis in vehicle-treated U2OS cells. U2OS cells stably expressing Histone-H2B-mRFP and γ-tubulin–GFP were exposed to 0.1% DMSO (vehicle), and mitotic progression was followed by live laser-scanning confocal fluorescence microscopy (TE-2000; Nikon). Maximum intensity projections of H2B-mRFP (red) and γ-tubulin–GFP (green) fluorescence are shown for 44 frames taken every 5 min. See Fig. 6 A.
Mitosis in vehicle-treated U2OS cells. U2OS cells stably expressing Histone-H2B-mRFP and γ-tubulin–GFP were exposed to 0.1% DMSO (vehicle), and mitotic progression was followed by live laser-scanning confocal fluorescence microscopy (TE-2000; Nikon). Maximum intensity projections of H2B-mRFP (red) and γ-tubulin–GFP (green) fluorescence are shown for 44 frames taken every 5 min. See Fig. 6 A.
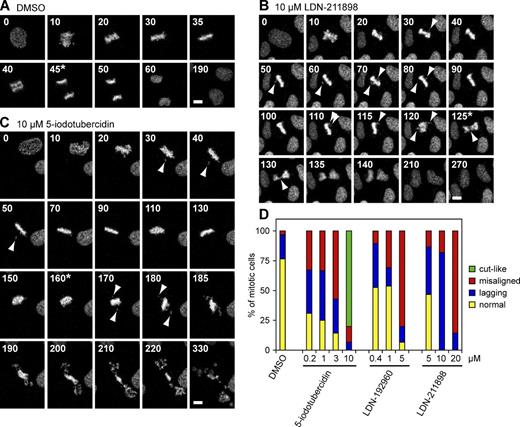
Haspin inhibitors compromise chromosome alignment. (A) U2OS cells expressing Histone-H2B-mRFP and γ-tubulin–GFP were exposed to vehicle alone (DMSO), and mitotic progression was followed by live confocal fluorescence microscopy. Maximum intensity projections of H2B-mRFP fluorescence from selected frames are shown. Video 1 shows complete data including γ-tubulin–GFP fluorescence. (B and C) As above, for a cell treated with 10 µM LDN-211898 (B) or 10 µM 5-iodotubercidin (C). Arrowheads indicate misaligned or lagging chromosomes. Asterisks indicate the first frame in which cytokinetic furrowing was observed. Also see Videos 2 and 3. Bars, 10 µm. (D) Mitotic defects enumerated from live imaging movies. See Fig. S4 C for further details.
Haspin inhibitors compromise chromosome alignment. (A) U2OS cells expressing Histone-H2B-mRFP and γ-tubulin–GFP were exposed to vehicle alone (DMSO), and mitotic progression was followed by live confocal fluorescence microscopy. Maximum intensity projections of H2B-mRFP fluorescence from selected frames are shown. Video 1 shows complete data including γ-tubulin–GFP fluorescence. (B and C) As above, for a cell treated with 10 µM LDN-211898 (B) or 10 µM 5-iodotubercidin (C). Arrowheads indicate misaligned or lagging chromosomes. Asterisks indicate the first frame in which cytokinetic furrowing was observed. Also see Videos 2 and 3. Bars, 10 µm. (D) Mitotic defects enumerated from live imaging movies. See Fig. S4 C for further details.
All three compounds caused a dose-dependent increase in the proportion of defective mitoses. Lagging chromosomes at anaphase were often observed, even at relatively low inhibitor concentrations (Fig. 6, B and D; and Video 2). At higher concentrations, cells that entered anaphase with chromosomes that had not congressed, or that entered anaphase with ill-defined or “loose” metaphase plates, became increasingly apparent (Fig. 6, B–D). At 10 µM of the most potent inhibitor, 5-iodotubercidin, cytokinesis often occurred without obvious chromosome disjunction. In these cases, the cytokinetic furrow impinged upon the chromosome mass, resulting in a “cut-like” phenotype (Fig. 6 C and Video 3) resembling that seen upon microinjection of antibodies against H3T3ph (Wang et al., 2010). Similar mitotic figures, in which central spindle formation was evident in the absence of obvious anaphase chromosome movements, were seen in fixed cells previously treated with 10 µM 5-iodotubercidin or 100 µM LDN-211898 (Fig. 1 D). These results support the conclusion that Haspin inhibition causes defects in error correction, but that it does not affect the central spindle functions of Aurora B or prevent cytokinesis.
Mitosis in LDN-211898-treated U2OS cells. U2OS cells stably expressing Histone-H2B-mRFP and γ-tubulin–GFP were exposed to 10 µM LDN-211898, and mitotic progression was followed by live laser-scanning confocal fluorescence microscopy (TE-2000; Nikon). Maximum intensity projections of H2B-mRFP (red) and γ-tubulin–GFP (green) fluorescence are shown for 60 frames taken every 5 min. See Fig. 6 B.
Mitosis in LDN-211898-treated U2OS cells. U2OS cells stably expressing Histone-H2B-mRFP and γ-tubulin–GFP were exposed to 10 µM LDN-211898, and mitotic progression was followed by live laser-scanning confocal fluorescence microscopy (TE-2000; Nikon). Maximum intensity projections of H2B-mRFP (red) and γ-tubulin–GFP (green) fluorescence are shown for 60 frames taken every 5 min. See Fig. 6 B.
Mitosis in 5-iodotubercidin-treated U2OS cells. U2OS cells stably expressing Histone-H2B-mRFP and γ-tubulin–GFP were exposed to 10 µM 5-iodotubercidin, and mitotic progression was followed by live laser-scanning confocal fluorescence microscopy (TE-2000; Nikon). Maximum intensity projections of H2B-mRFP (red) and γ-tubulin–GFP (green) fluorescence are shown for 73 frames taken every 5 min. See Fig. 6 C.
Mitosis in 5-iodotubercidin-treated U2OS cells. U2OS cells stably expressing Histone-H2B-mRFP and γ-tubulin–GFP were exposed to 10 µM 5-iodotubercidin, and mitotic progression was followed by live laser-scanning confocal fluorescence microscopy (TE-2000; Nikon). Maximum intensity projections of H2B-mRFP (red) and γ-tubulin–GFP (green) fluorescence are shown for 73 frames taken every 5 min. See Fig. 6 C.
Haspin inhibitors compromise maintenance of the spindle checkpoint
The finding that inhibitor-treated cells could exit mitosis before chromosomes were fully aligned suggested either that the spindle checkpoint was satisfied on such spindles, or that a defect in the spindle checkpoint was present. Either of these could result from loss of Haspin-dependent CPC activity because inhibition of Aurora B stabilizes KT-MT attachments and can therefore indirectly promote satisfaction of the spindle checkpoint, and there is also evidence that Aurora B plays a role in the checkpoint that is independent of its function in error correction (see Introduction). To test this second possibility, we monitored the effect of Haspin inhibitors on mitotic exit of HeLa cells previously arrested with high doses of nocodazole (5 µM) that are sufficient to prevent assembly of spindle microtubules detectable by immunofluorescence (Fig. S5 A; Jordan et al., 1992; Brito and Rieder, 2006). 5-Iodotubercidin caused a dose-dependent decrease in mitotic (phospho)-protein monoclonal-2 (MPM-2) phosphoepitopes detected by immunoblotting, indicating that it was able to drive mitotic exit in these conditions (Fig. 7 A). We also found that a dose of the Aurora B inhibitor ZM447439 (1 µM) that did not itself cause detectable mitotic exit was able to lower by ∼10-fold the concentration of 5-iodotubercidin needed to drive exit (Fig. 7 A). Similar findings were made with a second Aurora B inhibitor, Hesperadin (Fig. S5 B).
Haspin inhibitors compromise the spindle checkpoint response to 5 µM nocodazole. (A) HeLa cells were synchronized by thymidine treatment and, 7 h after release, 5 µM nocodazole was added for 7.5 h. Mitotic cells were harvested by mitotic “shake-off” and replated in the continued presence of 5 µM nocodazole together with 5-iodotubercidin and the Aurora B inhibitor ZM447439. After 13.5 h, total cell lysates were analyzed by immunoblotting. (B) HeLa cells were transfected with EGFP–CENP-B or CENP-B–INCENP–EGFP plasmids between and after double thymidine treatments, and then treated essentially as in A, except that cells were replated on coverslips coated with poly-d-lysine. Mitotic indices were determined from ∼100 cells in each condition by DNA staining and fluorescence microscopy (n = 3). Means + SD are shown (error bars); ***, P < 0.001; ND, not detected. (C and D) As for A, but using LDN-192960 (C) or LDN-211898 (D). Black lines indicate that intervening lanes have been spliced out. (E–H) U2OS cells expressing Histone-H2B-mRFP and γ-tubulin–GFP were synchronized and treated with 5 µM nocodazole essentially as described for HeLa cells in A. Mitotic cells were replated in imaging dishes and kinase inhibitors were added in the continued presence of 5 µM nocodazole. Each symbol represents the time at which a cell began to exit from mitosis or die in mitosis, as determined from live imaging series collected over 15 h.
Haspin inhibitors compromise the spindle checkpoint response to 5 µM nocodazole. (A) HeLa cells were synchronized by thymidine treatment and, 7 h after release, 5 µM nocodazole was added for 7.5 h. Mitotic cells were harvested by mitotic “shake-off” and replated in the continued presence of 5 µM nocodazole together with 5-iodotubercidin and the Aurora B inhibitor ZM447439. After 13.5 h, total cell lysates were analyzed by immunoblotting. (B) HeLa cells were transfected with EGFP–CENP-B or CENP-B–INCENP–EGFP plasmids between and after double thymidine treatments, and then treated essentially as in A, except that cells were replated on coverslips coated with poly-d-lysine. Mitotic indices were determined from ∼100 cells in each condition by DNA staining and fluorescence microscopy (n = 3). Means + SD are shown (error bars); ***, P < 0.001; ND, not detected. (C and D) As for A, but using LDN-192960 (C) or LDN-211898 (D). Black lines indicate that intervening lanes have been spliced out. (E–H) U2OS cells expressing Histone-H2B-mRFP and γ-tubulin–GFP were synchronized and treated with 5 µM nocodazole essentially as described for HeLa cells in A. Mitotic cells were replated in imaging dishes and kinase inhibitors were added in the continued presence of 5 µM nocodazole. Each symbol represents the time at which a cell began to exit from mitosis or die in mitosis, as determined from live imaging series collected over 15 h.
To confirm that loss of MPM-2 reactivity reflected exit from mitosis, we repeated similar experiments but examined cells by fluorescence microscopy. Indeed, 5-iodotubercidin caused a dose-dependent increase in the fraction of cells exiting mitosis, as judged by chromosome decondensation and formation of interphase nuclei (Fig. 7 B). Although CENP-B–INCENP does not precisely restore the CPC to its normal location and dynamics at inner centromeres, we determined if targeting Aurora B to centromeres with this fusion protein would rescue the checkpoint response in 5-iodotubercidin–treated cells. We observed a statistically significant increase in the proportion of cells remaining in mitosis in 5 µM nocodazole in the presence of the Haspin inhibitor (Fig. 7 B), confirming that the checkpoint defect is likely to be at least partially caused by delocalization of the CPC.
To corroborate the results in another cell type and to directly visualize mitotic exit, we used U2OS cells expressing histone H2B-mRFP and γ-tubulin–GFP. Mitotic exit was monitored by microscopic imaging of living cells for 15 h. Cells exhibiting membrane ruffling and blebbing characteristic of telophase cells, followed by chromatin decondensation (and often cell spreading), were judged to have exited mitosis. Control cells maintained in 5 µM nocodazole exited mitosis or died at a low and approximately constant rate over this time, as expected (Brito and Rieder, 2009). In contrast, addition of 100 nM of the Aurora B inhibitor Hesperadin caused the majority of cells to exit mitosis within the first 3 h, even in the continued presence of 5 µM nocodazole (P < 0.0001 by log-rank test; Fig. 7 E). 5-Iodotubercidin also caused a dose-dependent increase in the rate of cells exiting mitosis (P < 0.0001 for 1 or 3 µM 5-iodotubercidin vs. DMSO; Fig. 7 F), which suggests that the Haspin-dependent pool of Aurora B is required to maintain full checkpoint activity in cells that are exposed to high doses of nocodazole.
We also tested the ability of the other two Haspin inhibitors to stimulate mitotic exit in 5 µM nocodazole. LDN-192960 alone did not cause detectable exit in the MPM-2 assay in HeLa cells (Fig. 7 C, right). However, we suspected that this might be caused by the occurrence of off-target effects of this compound at doses >∼5 µM. We therefore tested if the dose of LDN-192960 needed to influence mitotic exit could be lowered by combination with Aurora B inhibitors, as for 5-iodotubercidin. Indeed, in the presence of 1 µM ZM447439, even concentrations as low as 0.1 µM LDN-192960 caused substantial loss of MPM-2 epitopes (Fig. 7 C, middle), and similar results were obtained in the presence of Hesperadin (Fig. S5 B). However, 10 µM LDN-192960 did not cause mitotic exit in combination with Aurora B inhibition, which is consistent with off-target effects at this higher dose (Fig. S5 B). In contrast, like 5-iodotubercidin, LDN-211898 was able to drive mitotic exit of HeLa (Fig. 7 D) and U2OS cells (P < 0.0001; Fig. 7 G) in the presence of 5 µM nocodazole, and this could be partly prevented by expression of CENP-B–INCENP (Fig. 7 B). Again, the effects of LDN-211898 were stronger in the presence of low concentrations of ZM447439 or Hesperadin in HeLa (Fig. 7 D and Fig. S5 B) and U2OS cells (P < 0.0001; Fig. 7 H). Therefore, all three Haspin inhibitors can compromise the spindle checkpoint when microtubules are severely disrupted.
Loss of the checkpoint protein BubR1 from kinetochores upon Aurora B inactivation has been widely reported (Ditchfield et al., 2003; Hauf et al., 2003; Emanuele et al., 2008; Becker et al., 2010). In high nocodazole conditions similar to those used in the checkpoint assays, addition of Haspin inhibitors caused a modest reduction in the intensity of BubR1 at kinetochores in a subset of cells (Fig. S5 C). However, in Aurora B reactivation assays (conducted with 5 µM nocodazole), the recovery of BubR1 at kinetochores was substantially delayed in the presence of Haspin inhibitors (Fig. 8, A and B; and Fig. S5 D), and this could be rescued by expression of CENP-B–INCENP (Fig. 8, C and D). Therefore, the failure of delocalized Aurora B to efficiently recruit BubR1 to kinetochores (either by a direct or indirect mechanism) may contribute to the checkpoint deficit seen in Haspin-inactivated cells.
Haspin inhibitors delay BubR1 recruitment to kinetochores. (A) Immunofluorescence microscopy of BubR1 during Aurora B reactivation in the presence or absence of Haspin inhibitors in HeLa cells treated as in Fig. 4 A, but using 5 µM nocodazole throughout. Bar, 5 µm. (B) Approximately 100 mitotic cells in each condition from A were classified according to BubR1 intensity at kinetochores. Means ± SD are shown (error bars), n = 3. (C) HeLa cells were transfected with plasmids encoding EGFP–CENP-B or CENP-B–INCENP–EGFP between and after double thymidine treatments, and then treated essentially as in A. Bars, 5 µm. (D) For cells in C, the intensity of kinetochore BubR1 was classified in at least 100 cells per condition in one experiment. Similar results were obtained in a duplicate experiment.
Haspin inhibitors delay BubR1 recruitment to kinetochores. (A) Immunofluorescence microscopy of BubR1 during Aurora B reactivation in the presence or absence of Haspin inhibitors in HeLa cells treated as in Fig. 4 A, but using 5 µM nocodazole throughout. Bar, 5 µm. (B) Approximately 100 mitotic cells in each condition from A were classified according to BubR1 intensity at kinetochores. Means ± SD are shown (error bars), n = 3. (C) HeLa cells were transfected with plasmids encoding EGFP–CENP-B or CENP-B–INCENP–EGFP between and after double thymidine treatments, and then treated essentially as in A. Bars, 5 µm. (D) For cells in C, the intensity of kinetochore BubR1 was classified in at least 100 cells per condition in one experiment. Similar results were obtained in a duplicate experiment.
Microinjection of H3T3ph antibodies compromises error correction and maintenance of the spindle checkpoint
We previously showed that microinjection of antibodies recognizing H3T3ph into mitotic cells results in displacement of the CPC from centromeres, defects in chromosome alignment, and onset of cytokinesis in the presence of misaligned chromosomes (Wang et al., 2010), all effects that are reminiscent of Haspin inhibition reported here. To test the role of H3T3ph in error correction and maintenance of the spindle checkpoint in a chemical inhibitor–independent manner, we used live imaging to determine the effect of anti-H3T3ph microinjection on LLC-PK cells. Consistent with the results of Haspin inhibition, microinjection of anti-H3T3ph compromised chromosome alignment during release from a kinesin-5/Eg5 inhibitor block (Video 4). To examine mitotic exit, we used LLC-PK cells expressing EGFP–topoisomerase IIα that were previously arrested in mitosis with nocodazole. These cells accumulated efficiently in mitosis at 0.17 µM nocodazole, and exited at a median time of 9 h after live imaging was initiated. In contrast, cells injected with anti-H3T3ph exited mitosis with a median time <4 h (P < 0.0001 by log-rank test; Fig. 9, A and B). This supports the idea that the H3T3ph-dependent CPC population plays a role in the timing of mitotic exit. However, because residual microtubules remain present in LLC-PK cells in these conditions (Centonze and Borisy, 1991), this function could be either indirect through error correction or a more direct one in generating the spindle checkpoint signal itself.
Effects of anti-H3T3ph microinjection in an error correction assay. LLC-PK cells arrested in mitosis with monopolar spindles with 10 µM 5-S-trityl-l-cysteine (a kinesin-5/Eg5 inhibitor) and 25 µM MG132 were injected or not injected with H3T3ph antibodies. Then, 5-S-trityl-l-cysteine was washed out, and the cells were followed for ∼3 h in the presence of 25 µM MG132 to counter mitotic exit. Time-lapse phase-contrast images were collected using a microscope (Axiovert 200M; Carl Zeiss). Times of frame acquisition are shown in hours:minutes:seconds. Of 10 control cells observed for ∼3 h after washout, 60% formed bipolar spindles and then aligned all chromosomes within 20 min, whereas 40% failed to form bipolar spindles and align chromosomes. Of 10 injected cells observed for 3 h, 30% formed bipolar spindles and then aligned all chromosomes within 20 min, whereas 70% failed to align all chromosomes (20% formed bipolar spindles but then did not align all chromosomes for at least 55 min [see video], and 50% failed to form bipolar spindles).
Effects of anti-H3T3ph microinjection in an error correction assay. LLC-PK cells arrested in mitosis with monopolar spindles with 10 µM 5-S-trityl-l-cysteine (a kinesin-5/Eg5 inhibitor) and 25 µM MG132 were injected or not injected with H3T3ph antibodies. Then, 5-S-trityl-l-cysteine was washed out, and the cells were followed for ∼3 h in the presence of 25 µM MG132 to counter mitotic exit. Time-lapse phase-contrast images were collected using a microscope (Axiovert 200M; Carl Zeiss). Times of frame acquisition are shown in hours:minutes:seconds. Of 10 control cells observed for ∼3 h after washout, 60% formed bipolar spindles and then aligned all chromosomes within 20 min, whereas 40% failed to form bipolar spindles and align chromosomes. Of 10 injected cells observed for 3 h, 30% formed bipolar spindles and then aligned all chromosomes within 20 min, whereas 70% failed to align all chromosomes (20% formed bipolar spindles but then did not align all chromosomes for at least 55 min [see video], and 50% failed to form bipolar spindles).
Microinjection of mitotic cells with anti-H3T3ph compromises the spindle checkpoint response in combination with Aurora B inhibition. (A) LLC-PK expressing EGFP-topoisomerase IIα arrested in mitosis with 0.17 µM (left) or 3.3 µM nocodazole (right) were injected with anti-H3T3ph solution containing Dextran Texas red, and time-lapse phase contrast and fluorescence images were collected every 15 min. Note the nuclear reformation at the last time point in each case. Times are in hours:minutes. Bar, 10 µm. (B–E) After treatment with the nocodazole and ZM447439 combinations indicated, exit from mitosis or death in mitosis was enumerated from live imaging series collected over 15 h as in A.
Microinjection of mitotic cells with anti-H3T3ph compromises the spindle checkpoint response in combination with Aurora B inhibition. (A) LLC-PK expressing EGFP-topoisomerase IIα arrested in mitosis with 0.17 µM (left) or 3.3 µM nocodazole (right) were injected with anti-H3T3ph solution containing Dextran Texas red, and time-lapse phase contrast and fluorescence images were collected every 15 min. Note the nuclear reformation at the last time point in each case. Times are in hours:minutes. Bar, 10 µm. (B–E) After treatment with the nocodazole and ZM447439 combinations indicated, exit from mitosis or death in mitosis was enumerated from live imaging series collected over 15 h as in A.
At 3.3 µM nocodazole, a dose that strongly disrupts microtubules in LLC-PK cells (Vandré and Borisy, 1989), anti-H3T3ph–injected cells exited mitosis slightly sooner than control cells, although this difference was not statistically significant (P = 0.1; Fig. 9 C). As with low doses of Haspin inhibitors, we reasoned that antibody injection might not be potent enough to reveal the role of H3T3ph-dependent CPC in the spindle checkpoint. Therefore, we combined anti-H3T3ph microinjection with a dose of Aurora B inhibitor that itself was insufficient to cause mitotic exit in 3.3 µM nocodazole. Upon treatment with 1 µM ZM447439, the median time to mitotic exit was 13 h, similar to controls in the absence of ZM447439. However, coinjection of anti-H3T3ph reduced the median mitotic exit time to 5 h (P < 0.0001; Fig. 9, A and D), whereas microinjection of control antibodies had no significant effect (P = 0.48; Fig. 9 E). Therefore, microinjection of anti-H3T3ph antibodies can compromise the spindle checkpoint even when microtubules are strongly disrupted.
Discussion
Here, we made use of small molecule inhibitors to determine functions of Haspin in mitosis. As expected, Haspin inhibitors strongly reduce phosphorylation of histone H3 at threonine-3 in cells (Dai et al., 2005; Patnaik et al., 2008; Balzano et al., 2011; Huertas et al., 2012). Prior RNAi and microinjection studies in cultured cells (Wang et al., 2010; F. Wang et al., 2011), together with protein depletion in Xenopus extracts (Kelly et al., 2010) and gene deletion in fission yeast (Yamagishi et al., 2010), revealed a role for Haspin in regulating chromatin localization of the CPC in mitosis. Using Haspin inhibitors, we now show that it is the kinase activity of Haspin that is important for the normal positioning of Aurora B on mitotic chromatin, and that this effect is independent of changes in chromosome cohesion. This finding is consistent with the proposed function of H3T3ph to provide a binding site for Survivin on chromatin (Kelly et al., 2010; Wang et al., 2010; Yamagishi et al., 2010; Niedzialkowska et al., 2012).
Although H3T3ph is the only currently known product of Haspin activity, it is possible that other substrates of Haspin exist in cells. Nevertheless, Haspin inhibitors are useful tools to displace H3T3ph-dependent centromeric CPC to examine its functions in mitosis without preventing CPC localization to the central spindle, particularly in combination with artificial retargeting of Aurora B to centromeres. Another study used actinomycin D to delocalize centromeric CPC, but this also compromised midbody localization, and the displacement mechanism and its specificity remain undefined (Becker et al., 2010). Using Haspin inhibitors, we confirmed that the Haspin-dependent CPC pool is required for maintaining centromeric MCAK localization (Wang et al., 2010). In addition, we reveal that centromeric (CENP-AS7) and kinetochore (Dsn1 S109) Aurora B substrates, and its function in error correction, depend on this predominantly centromeric population. This lends support to models that emphasize the role of inner centromeric CPC in controlling the phosphorylation of kinetochore substrates and microtubule attachment stability (Tanaka et al., 2002; Liu et al., 2009).
We also find that Haspin-dependent CPC accumulation increases the rate of Aurora B activation, particularly for centromere and kinetochore substrates. This supports, in cells, suggestions made previously from work in Xenopus extracts and in vitro (Rosasco-Nitcher et al., 2008; Kelly et al., 2010). It is likely that swift concentration and activation is important for feedback regulation of centromeric Aurora B activity on short timescales, such as in response to KT-MT attachment status (Salimian et al., 2011). In contrast, although H3T3ph-dependent localization of Aurora B can increase the rate of H3S10 phosphorylation, this predominantly centromeric Aurora B population may not be strictly necessary for generating H3S10ph on chromosome arms. In fact, when Haspin is inhibited in Aurora B reactivation assays, Aurora B autophosphorylation and H3S10ph return in a diffuse manner that is not first focused at centromeres. This suggests that not all CPC functions require centromeric concentration for activation, nor a soluble gradient of Aurora B activity originating at centromeres. If this were the case, we might expect H3S10ph on arms, at the base of such a gradient, to be particularly sensitive to loss of centromeric CPC, but this is not the case. This suggests that, when largely diffuse on chromatin, the CPC can still reach a concentration sufficient to activate Aurora B for H3S10 phosphorylation. Presumably, the population of Aurora B found prominently on chromosome arms in prophase cells (Ruchaud et al., 2007) contributes directly to H3S10 phosphorylation. It is likely that different target sites phosphorylated by Aurora B have different susceptibilities to Aurora B and opposing phosphatases (Xu et al., 2009; Wang et al., 2010) due both to site-intrinsic features, such as binding affinity, and extrinsic factors, such as substrate abundance. H3S10 appears to be an “easy” substrate for Aurora B to phosphorylate in cells. Thresholds of this type regulate cell cycle events at the cellular level (Coudreuse and Nurse, 2010), but are also likely to be crucial for regional regulation of substrate phosphorylation on a local scale.
Aurora B clearly influences spindle checkpoint responses, although the mechanisms involved have been debated (see Introduction). We find that, like Aurora inhibitors, Haspin inhibitors or microinjection of H3T3ph antibodies compromise maintenance of mitotic arrest when microtubules are severely disrupted. This suggests that the H3T3ph-dependent population of the CPC is required for this activity of Aurora B. This provides support for the idea that Aurora B contributes to generation of the checkpoint response separately from its role in modulating KT-MT attachments, and reduces the concern that off-target effects of Aurora inhibitors were responsible for the effects observed in prior studies. Although we cannot rule out the possibility that Haspin inhibition or anti-H3T3ph microinjection also affects another population of the CPC or another component of the checkpoint pathway (also see De Antoni et al., 2012), we find that the effects of Haspin inhibitors can be partially reversed by retargeting Aurora B to centromeres with CENP-B–INCENP. Our results therefore suggest that the spindle checkpoint involves centromeric CPC. Whether the relevant substrates are within “striking distance” of Aurora B bound to centromeres or depend on a gradient of diffusible Aurora B activity centered on centromeres requires further study. Because the CPC can act as a tension sensor, it remains possible that Aurora B in the checkpoint pathway responds to tension, but it should be noted that our results do not imply that the checkpoint must necessarily be directly responsive to tension.
Previous studies using Haspin RNAi failed to reveal strong effects on CENP-AS7ph or spindle checkpoint responses in nocodazole (Wang et al., 2010), which suggests that Haspin was incompletely depleted in these studies. In contrast to Haspin inhibitors, Haspin RNAi causes a prolonged mitotic delay and premature loss of sister chromatid cohesion in a subset of cells (Dai et al., 2006, 2009). These results suggest that the role of Haspin in cohesion either (a) is independent of its kinase activity or (b) becomes apparent only when Haspin is partially depleted. Indeed, although strong depletion of certain kinetochore proteins compromises the spindle checkpoint, partial depletion of the same proteins can prevent checkpoint satisfaction (McCleland et al., 2003; Meraldi et al., 2004; Meraldi and Sorger, 2005), a condition that might promote “cohesion fatigue” (Daum et al., 2011; Stevens et al., 2011; Logarinho et al., 2012). Further work is required, but we cannot rule out explanation (a) because Haspin overexpression increases arm cohesion (Dai et al., 2006), kinase-deficient mutants of Haspin support cohesion (unpublished data), and we did not observe cohesion loss at intermediate inhibitor concentrations that might mimic partial Haspin depletion.
As with any inhibitor study, we cannot entirely rule out off-target effects of Haspin inhibitors, particularly when high concentrations are used. However, there is a strong theoretical basis for the need to robustly inhibit enzyme activity in cells to cause clear effects, particularly for indirect targets such as the substrates of Aurora B examined here (Knight and Shokat, 2005). Indeed, recent studies highlight the importance of using high Aurora inhibitor concentrations to reveal Aurora B checkpoint functions (Santaguida et al., 2011). Furthermore, we demonstrate that three chemically distinct compounds yield similar phenotypes in cells at relative doses predicted by their ability to inhibit Haspin in vitro and in cells. It seems unlikely that all three inhibitors have a fortuitous off-target activity that would track Haspin inhibition capacity so closely. Furthermore, we used combination treatments with Haspin and Aurora B inhibitors to demonstrate effects at low doses that are less likely to display off-target effects, and we confirmed a role for H3T3ph in error correction and the spindle checkpoint using H3T3ph antibody microinjection experiments that eliminate the use of Haspin inhibitors.
The difficulty in fully inhibiting Aurora B activity in cells by targeting Haspin or Aurora B directly may stem in part from a positive feedback loop between these kinases that drives Aurora B localization in mitosis (F. Wang et al., 2011). Indeed, it is possible that coinhibition of Haspin and Aurora B will provide means to enhance the effects of Aurora B inhibitors currently in clinical trials (Lens et al., 2010), and a compound that inhibits Haspin has shown anti-tumor activity in a mouse xenograft model (Huertas et al., 2012). It seems more certain that the Haspin inhibitors we describe will be useful for further basic studies of chromosome segregation.
Materials and methods
Antibodies
Rabbit polyclonal antibodies used were to H3T3ph (B8634 raised against Histone H3(1–8)T3ph peptide; Dai et al., 2005), γ-tubulin (AK-15; Sigma-Aldrich), Dsn1 no. 110 and Dsn1-S109ph no. 20 (provided by I. Cheeseman, Whitehead Institute, Cambridge, MA; Welburn et al., 2010), Aurora B T232ph (Rockland Immunochemicals), CENP-AS7ph (EMD Millipore), INCENP (I5283; Sigma-Aldrich), and Survivin (NB500-201; Novus Biologicals). Mouse monoclonal antibodies used were to H3S10ph (6G3; Cell Signaling Technology), mitotic phospho-epitopes (MPM-2; EMD Millipore), Aurora B (AIM-1; BD), and α-Tubulin (B-5-1-2; Sigma-Aldrich). Sheep antibodies were to Aurora B and BubR1 (SAB.1 and SBR1.1; S. Taylor, University of Manchester, Manchester, UK; Ditchfield et al., 2003) and MCAK (L. Wordeman, University of Washington, Seattle, WA; Andrews et al., 2004), and human centromere autoantibodies (abbreviated “centromere” in figures) were from ImmunoVision. Secondary antibodies were donkey anti–rabbit or –mouse IgG-HRP; anti–rabbit, –mouse, or –sheep IgG-Cy3; anti–human, –mouse, –sheep, or –rabbit IgG-Cy5 (Jackson ImmunoResearch Laboratories, Inc.); or anti–rabbit, –sheep, or –mouse IgG–Alexa Fluor 488 (Invitrogen).
Cell culture and inhibitors
HeLa and U2OS cells were maintained in 10% FBS/DME at 10% CO2 and 37°C. Cells were arrested at the G1/S boundary by single or double 2 mM thymidine (EMD) treatment, or in prometaphase with nocodazole (Sigma-Aldrich) at stated concentrations. MG132 (Sigma-Aldrich) was used at 20 µM. LDN-211898 (Cuny et al., 2012) was synthesized by Aberjona Laboratories, LDN-192960 was provided by M. Robin (Centre Saint Jérôme, Aix-Marseille Université, Marseille, France; Cuny et al., 2010), and 5-iodotubercidin was from EMD Millipore. ZM447439 (Ditchfield et al., 2003) was obtained from Tocris Bioscience, and Hesperadin (Hauf et al., 2003) was obtained from Selleck Chemicals. For monastrol release experiments (Lampson et al., 2004), cells were treated with 100 µM monastrol (Sigma-Aldrich) for 2 h, followed by washing four times and incubation in medium containing 20 µM MG132. A plasmid encoding a fusion protein of the CENP-B DNA binding domain, CENP-B[1–158] to human INCENP[47–920] and EGFP in pEGFP-N1 (Liu et al., 2009) was provided by M. Lampson (University of Pennsylvania, Philadelphia, PA) and S. Lens (University Medical Center, Utrecht, Netherlands), a plasmid encoding EGFP-CENP-B[1–167] in pEGFP-C1 (Wordeman et al., 2007) was provided by L. Wordeman, and transfections were done with Fugene 6 (Roche).
Immunofluorescence microscopy
In general, cells grown on coverslips were fixed for 10 min with 2% paraformaldehyde in PBS and extracted with 0.5% Triton X-100 in PBS for 5 min, at room temperature. For the error correction assay shown in Figs. 5 A and S4 A, cells were fixed with 4% paraformaldehyde in PBS for 10 min without extraction. For experiments shown in Fig. 5 (B–E), cells were extracted for 5 min in PHEM (60 mM Pipes, 25 mM Hepes, 10 mM EGTA, and 2 mM MgCl2, pH 6.9) plus 1% Triton X-100, and fixed at room temperature for 20 min in PHEM plus 4% formaldehyde. For experiments shown in Fig. S5 A, cells were fixed with 4% paraformaldehyde in PBS for 10 min followed by ice-cold methanol treatment for 5 min. Mitotic chromosome spreads were prepared in hypotonic buffer (75 mM KCl/0.8% sodium citrate/H2O at 1:1:1), attached to glass slides by Cytospin (Thermo Fisher Scientific) at 1,500 rpm for 5 min, fixed with 2% paraformaldehyde in PBS for 20 min at room temperature, and stained with antibodies in 10% FBS, 0.1% Triton X-100, 120 mM KCl, 20 mM, MgCl2, 0.5 mM EDTA, and 10 mM Tris, pH 8.0 (Dai et al., 2006). Blocking was performed in 5% milk in 0.1% or 0.5% Triton X-100 in PBS, and antibodies were diluted in blocking buffer and incubated at room temperature for 1–2 h or overnight at 4°C. DNA was visualized with 1 µM Hoechst 33342 (Thermo Fisher Scientific). Fluorescence microscopy was performed at room temperature using a 60× Plan-Apochromat (NA 1.40) oil immersion objective lens and an inverted microscope (TE2000-U; Nikon) equipped with a SPOT-RT charge-coupled device system and SPOT-RT software (Diagnostic Instruments, Inc.). Photoshop (Adobe) was used to adjust maximum and minimum image brightness using “levels” (equally for all images in a single experiment) and assemble image panels.
Live imaging of U2OS cells
U2OS cells stably expressing γ-tubulin–EGFP (human γ-tubulin fused to the N terminus of EGFP in plasmid pEGFP-N1) and H2B-mRFP (human histone H2B fused to the N terminus of mRFP in plasmid pmRFP-N1) were used (Dai et al., 2009). For imaging, mitotic cells arrested in 5 µM nocodazole for ∼8 h were harvested by “shake-off” and replated in 10% FBS, 25 mM Hepes, and phenol red–free DME (Thermo Fisher Scientific) containing 5 µM nocodazole in a 35-mm single chamber (World Precision Instruments) or 35-mm 4-chamber (Greiner Bio-One) glass-bottom dishes coated with poly-d-lysine. Time-lapse confocal fluorescence imaging was performed using an inverted microscope (TE2000-U; Nikon) equipped with a 40× Plan Fluor (NA 1.30) oil immersion objective lens, a confocal laser scanner (C1 Plus; Nikon), EZ-C1 software (Nikon), and a Proscan II motorized stage (Prior Scientific) in a 37°C heated chamber with CO2 supply. Immediately after kinase inhibitor addition, two-color z stacks with 0.8 µm steps were collected with a 100-nm pinhole every 5 min for 15 h. ImageJ (National Institutes of Health) was used to render maximum intensity projections, adjust brightness, and assemble movies.
Live imaging and microinjection of LLC-PK cells
For assays as in Video 4, asynchronous LLC-PK cells were treated with 10 µM 5-S-trityl-l-cysteine and 25 µM MG132 for 1 h. After mounting in imaging chambers, monopolar mitotic cells were injected using a Burleigh micromanipulator, and microneedles containing 4.5 mg/ml affinity-purified rabbit anti-H3T3ph antibody in PBS. After ∼10 min, 5-S-trityl-l-cysteine was removed by changing to medium containing 25 µM MG132 without 5-S-trityl-l-cysteine. For assays in Fig. 9, a stable LLC-PK cell line expressing human topoisomerase IIα fused to the C terminus of EGFP (in plasmid pEGFP-C3) was used (Tavormina et al., 2002). Cells were injected with 14 mg/ml anti-H3T3ph or rabbit IgG in PBS containing 0.5 mg/ml dextran Texas red. During imaging, 10% FBS in Leibovitz’s L-15 medium supplemented with penicillin and streptomycin and overlaid with mineral oil was used. Time-lapse phase contrast and fluorescence images were collected at 37°C using a 63× objective lens (NA 1.40) and an inverted microscope (Axiovert 200M; Carl Zeiss, Inc.) equipped with a stage heater, air curtain, an ORCA-ER camera (Hamamatsu Photonics), and MetaMorph software (Molecular Devices), or using a 63× (NA 1.40) oil objective and an inverted microscope (AxioObserver; Carl Zeiss, Inc.) equipped with a stage heater, air curtain, an ORCA-ER camera, and Slidebook software (Intelligent Imaging Innovations, Inc.). Images were captured every 2 min (Video 4) or 15 min (Fig. 9). Individual cells were evaluated after a minimum observation period of 3 h (Video 4) or 15 h (Fig. 9). Image panels were assembled using MetaMorph software.
Quantification of immunofluorescence
Quantification of immunofluorescence in fixed mitotic cells was performed using ImageJ using images obtained at identical illumination settings. The total pixel intensity of CENP-AS7ph, H3S10ph, and centromere autoantigen staining within ellipses encompassing individual cells was determined, and the average background pixel intensity was obtained from a smaller ellipse within the cell cytosol. After background subtraction, the ratio of total histone phosphorylation/centromere autoantigen intensity was calculated for each cell. MCAK, CENP-AS7ph, Dsn1, Dsn1-S109ph, and BubR1 intensity levels were quantified at 18 centromeres per cell as follows. Centromeres were defined as regions falling within a 10-pixel diameter circle encompassing paired centromere autoantigen dots. The average pixel intensity within these circles was determined for centromere autoantigen and test staining. After background correction, the ratio of test/centromere autoantigen intensity was calculated for each centromere (Wang et al., 2010). Ratios were normalized to a mean value of 1 in controls.
The ratio of Aurora B at centromeres versus chromosome arms was determined as follows. The intensity of Aurora B and centromere autoantigen staining was determined within 10 × 4 pixel ellipses encompassing paired centromere dots. After background correction, the ratio of Aurora B to centromere autoantigen intensity was calculated independently for each of 15 centromeres per cell. After background subtraction, the average intensity of Aurora B within 10 × 4 pixel ellipses at three locations on chromosome arms was normalized to the mean centromere autoantigen intensity in the same cell. The mean centromere/arm intensity ratio was then calculated for each cell (Niedzialkowska et al., 2012).
Statistical analysis
Statistical analyses of immunofluorescence data were performed by one-way analysis of variance (ANOVA) followed by a Bonferroni’s multiple comparison test, using Instat 2.03 (GraphPad Software). Kaplan-Meier curves and log-rank tests on mitotic exit were performed using Prism 4 (GraphPad Software). The results shown were calculated using datasets from which cells dying in mitosis were excluded, but similar results were obtained if these cells were included.
Immunoprecipitation and immunoblotting
Whole cell lysates for Fig. 1 A were prepared in standard 1× SDS sample buffer with protease inhibitors and phosphatase inhibitors as described previously (Wang et al., 2010). For Fig. S1 E, cells were lysed in buffer P and subjected to immunoprecipitation with ∼3 µg/ml normal sheep IgG (EMD Millipore) or sheep anti–Aurora B antibody followed by immunoblotting as described previously (Wang et al., 2010).
In vitro kinase reactions
Time-resolved fluorescence resonance energy transfer (TR-FRET) assays were performed using recombinant full-length human MBP-Haspin at a near-Km concentration of ATP (200 µM) with 0.1 µM biotinylated H3(1–21) peptide substrate for 10 min at room temperature. Phosphorylation was detected by TR-FRET after addition of Europium-labeled anti-H3T3ph antibody clone JY325 (EMD Millipore) and streptavidin-APC (PerkinElmer) in a final concentration of 25 mM EDTA (Patnaik et al., 2008).
Online supplemental material
Fig. S1 shows Haspin inhibitor structures and IC50s, and that they do not cause cohesion loss or disassemble the CPC. Fig. S2 shows effects of Haspin inhibitors on centromeric MCAK and CENP-AS7ph. Fig. S3 shows effects on H3S10ph and Aurora B localization and autophosphorylation in Aurora B reactivation assays. Fig. S4 shows effects on error correction and the duration of mitosis. Fig. S5 shows disruption of microtubules by nocodazole, the effects of Haspin and Aurora B coinhibition using Hesperadin, and effects of Haspin inhibition on BubR1 localization. Videos 1–3 show the effects of Haspin inhibitors on mitosis by live imaging. Video 4 shows the effects of anti-H3T3ph microinjection on error correction.
Acknowledgments
We thank J. Dai, G. Cuny, M. Glicksman, S. Knapp, R. Stein, and J. Xian for their help during inhibitor development; and S. Santaguida and A. Musacchio for sharing results before publication.
This work was supported by National Institutes of Health (NIH) grant R01CA122608, NIH grant R01GM074210, the Association for International Cancer Research, and Leukemia and Lymphoma Society Scholar Award 1028-12 (to J.M.G. Higgins); and by NIH grant R01GM50412 and the McCasland Foundation (to G.J. Gorbsky).
References
- CENP-AS7ph
centromere protein A Serine-7, phosphorylated
- CPC
chromosomal passenger complex
- H3S10ph
histone H3 at serine-10, phosphorylated
- H3T3ph
histone H3 at threonine-3, phosphorylated
- KMN
KNL1/Mis12 complex/Ndc80 complex
- KT-MT
kinetochore–microtubule
- MCAK
mitotic centromere-associated kinesin
- MPM-2
mitotic (phospho)-protein monoclonal-2
- NEB
nuclear envelope breakdown
- TR-FRET
time-resolved fluorescence resonance energy transfer