Ataxia telangiectasia (A-T) mutated (ATM) kinase orchestrates deoxyribonucleic acid (DNA) damage responses by phosphorylating numerous substrates implicated in DNA repair and cell cycle checkpoint activation. A-T patients and mouse models that express no ATM protein undergo normal embryonic development but exhibit pleiotropic DNA repair defects. In this paper, we report that mice carrying homozygous kinase-dead mutations in Atm (AtmKD/KD) died during early embryonic development. AtmKD/− cells exhibited proliferation defects and genomic instability, especially chromatid breaks, at levels higher than Atm−/− cells. Despite this increased genomic instability, AtmKD/− lymphocytes progressed through variable, diversity, and joining recombination and immunoglobulin class switch recombination, two events requiring nonhomologous end joining, at levels comparable to Atm−/− lymphocytes. Together, these results reveal an essential function of ATM during embryogenesis and an important function of catalytically inactive ATM protein in DNA repair.
Introduction
Mutations in ataxia telangiectasia (A-T) mutated (ATM) cause the autosomal recessive disorder A-T, a neurodegenerative disease that is often associated with immunodeficiency, genomic instability, and predisposition to cancers (Lavin, 2008). Classical A-T is almost always caused by ATM mutations associated with undetectable ATM protein levels (Gilad et al., 1996; Lakin et al., 1996; Lavin, 2008). Mouse models that express no ATM protein recapitulate most of the human A-T phenotypes with the exception of the spontaneous progressive cerebella atrophy (Barlow et al., 1996; Xu and Baltimore, 1996; Herzog et al., 1998; Borghesani et al., 2000).
ATM kinase exists as inert dimers or higher order oligomers in unstressed cells and is converted to active monomers by agents that induce DNA double-strand breaks (DSBs; Bakkenist and Kastan, 2003; Carson et al., 2003; Uziel et al., 2003; Lee and Paull, 2004, 2005). Although autophosphorylation of ATM at S1981 (corresponding to S1987 in mouse) has been used as a reliable marker of ATM activation, whether S1981 phosphorylation is required for ATM activation is unclear. Expression of human ATM protein containing S1981A mutation fails to restore all ATM function in A-T cells (Bakkenist and Kastan, 2003; Lavin, 2008). However, both purified wild-type (WT) and S1981A mutated human ATM proteins can be activated in vitro (Lee and Paull, 2004). Furthermore, mouse models bearing a homozygous S1987A mutation or S1987A plus two additional autophosphorylation site mutations (corresponding to human S367A and S1893A) have no discernable defects in ATM activation (Pellegrini et al., 2006; Daniel et al., 2008).
As a master regulator of DNA damage responses (Bhatti et al., 2011), ATM has been implicated in both nonhomologous end joining (NHEJ) and homologous recombination (HR), the two principal DNA DSB repair pathways. NHEJ functions throughout the cell cycle. HR is most active in S and G2 phases of the cell cycle, when a homologous template is available. Developing lymphocytes undergo variable, diversity, and joining (V(D)J) recombination and immunoglobulin class switch recombination (CSR), two events that require NHEJ for completion (Rooney et al., 2004). Loss of ATM compromises both chromosomal V(D)J recombination (Borghesani et al., 2000; Bredemeyer et al., 2006; Huang et al., 2007; Callén et al., 2009a; Zha et al., 2011a) and CSR (Lumsden et al., 2004; Reina-San-Martin et al., 2004; Franco et al., 2006), indicating that ATM has an important function in NHEJ. ATM-deficient cells are also hypersensitive to the loss of PARP1/2 function (Ménisser-de Murcia et al., 2001; Huber et al., 2004), suggesting that ATM has an important function in HR. In addition, ATM deficiency is synergistically lethal with several other mutations that compromise the HR pathway (e.g., Nbs1, A-T and Rad3 related [ATR], and FanG; Williams et al., 2002; Kennedy et al., 2007; Murga et al., 2009).
Selective inhibitors targeting ATM kinase activity have been developed and widely used (Hickson et al., 2004; Rainey et al., 2008; White et al., 2008). Although these inhibitors generally recapitulate the effects of ATM protein deficiency (Bredemeyer et al., 2006; Callén et al., 2009b; Zha et al., 2011a), recent studies have identified DNA repair defects in cells treated with ATM kinase inhibitors that are not observed in ATM-null cells, suggesting additional activities of the kinase-inhibited ATM protein (White et al., 2010; Gamper et al., 2012). Here, we show that in contrast to the normal development of ATM-null mice, mice bearing mutations that result in the normal expression of a kinase-dead (KD) ATM (ATM-KD) protein, D2880A/N2885K (corresponding to D2870A/N2875K in humans; Canman et al., 1998), die during early embryonic development.
Results and discussion
AtmKD/KD, but not Atm+/KD mice, die during early embryonic development
To generate mice expressing a KD ATM protein, we introduced the previously described D2880A/N2885K (corresponding to D2870A/N2875K in humans; Canman et al., 1998; Bakkenist and Kastan, 2003) double mutation in the sequence encoding the conserved catalytic loop (Fig. 1 A) of the murine Atm gene along with a floxed neomycin-resistant (NeoR) cassette (referred to as ATM KDN for the presence of NeoR cassette; Fig. 1 B). We selected the D2870A/N2875K double mutation because it was fully characterized for normal protein expression and the absence of kinase activity (Canman et al., 1998; Bakkenist and Kastan, 2003). Six targeted clones were identified by Southern blot analyses (Fig. 1 C), and the mutations were verified in four clones by genomic sequencing. Two independent targeted clones were injected for germline transmission. Atm+/KDN chimeras were then bred with sperm-specific ProtamineCre transgenic mice (O’Gorman et al., 1997) to induce recombination between the loxP sites flanking the NeoR to generate the Atm+/KD mice, in which the KD, AtmKD, allele is transcribed and expressed from the endogenous Atm promoter.
Generation of the AtmKD allele. (A) The corresponding sequence of the catalytic loop of human and mouse ATM kinase are aligned, and the mutated residues are underlined. Gray shade covers the corresponding WT and mutated sequences in human ATM protein. (B) The schematic diagram represents the murine Atm locus (top), targeting vector (second row), targeted allele (AtmKDN, third row), and the neomycin (neo)-deleted mutant allele (AtmKD, bottom). The 5′ probe is marked as a black line. The exons and loxP sites are shown as solid boxes and open triangles, respectively. The exon containing the KD mutation is marked as an open box. Restriction site designations are as follows: X, XhoI; RV, EcoRV; H, HindIII. The map is not drawn to scale. (C) Southern Blot analyses of EcoRV-digested DNA from representative Atm+/+ (WT) and Atm+/KDN ES cells. (D and E) Genotype of embryos and live offspring obtained from timed intercrossing between Atm+/KD mice (D) and Atm+/− mice (E). Compared with the expected Mendelian frequency, the χ2 test p-values are 0.0056 for E9.5-–10.5 AtmKD/KD embryos and 4.01 × 10−8 for AtmKD/KD and 0.16 for Atm−/− mice at birth. Exp, expected.
Generation of the AtmKD allele. (A) The corresponding sequence of the catalytic loop of human and mouse ATM kinase are aligned, and the mutated residues are underlined. Gray shade covers the corresponding WT and mutated sequences in human ATM protein. (B) The schematic diagram represents the murine Atm locus (top), targeting vector (second row), targeted allele (AtmKDN, third row), and the neomycin (neo)-deleted mutant allele (AtmKD, bottom). The 5′ probe is marked as a black line. The exons and loxP sites are shown as solid boxes and open triangles, respectively. The exon containing the KD mutation is marked as an open box. Restriction site designations are as follows: X, XhoI; RV, EcoRV; H, HindIII. The map is not drawn to scale. (C) Southern Blot analyses of EcoRV-digested DNA from representative Atm+/+ (WT) and Atm+/KDN ES cells. (D and E) Genotype of embryos and live offspring obtained from timed intercrossing between Atm+/KD mice (D) and Atm+/− mice (E). Compared with the expected Mendelian frequency, the χ2 test p-values are 0.0056 for E9.5-–10.5 AtmKD/KD embryos and 4.01 × 10−8 for AtmKD/KD and 0.16 for Atm−/− mice at birth. Exp, expected.
Atm +/KD mice are normal in size, fertile, and have no detectable defects in lymphocyte development (Fig. S1, A and B). However, no AtmKD/KD mice were identified in >100 pups generated by breedings between Atm+/KD mice (P = 4.0 × 10−8 for chi-squared test; Fig. 1 D), indicating embryonic lethality. This was unexpected because Atm−/− mice were generated at the roughly Mendelian ratio (P = 0.16 for chi-squared test; Fig. 1 E). Timed heterozygous crosses further revealed that no live AtmKD/KD embryos could be found (P = 0.0056 for chi-squared test; Fig. 1 D) even at embryonic day 9.5–10.5 (E9.5–10.5), indicating early embryonic lethality. We note that the embryonic lethality of KD ATM protein is not caused by a classical dominant-negative mechanism because Atm+/KD mice and cells that express both ATM-WT and ATM-KD proteins have no discernable phenotype.
AtmKD/− embryonic stem (ES) cells have greater genomic instability than Atm−/− cells
To explore the causes of developmental failure in AtmKD/KD mice, we generated AtmKD/− ES cells and control Atm+/+ and Atm−/− cells. To circumvent the embryonic lethality, we first derived AtmKDN/C ES cells. The previously characterized Atm conditional allele (AtmC) behaves as the WT allele and can be efficiently converted to a null (Atm−) allele upon Cre-mediated recombination (Zha et al., 2008, 2011b). We obtained six independent AtmKDN/C clones and three control Atm+/C clones. AtmKD/− ES cells were then generated by infecting AtmKDN/C cells with adenovirus expressing Cre recombinase. Although all 24 Cre-transduced Atm+/C clones were genotyped as Atm+/−, indicating efficient recombination, only 2 of the 96 Cre-transduced AtmKDN/C clones were genotyped as AtmKD/−, implying that AtmKD/− cells have growth and/or survival defects. Western blot analysis verified that ATM-KD protein is expressed at levels comparable with WT endogenous ATM proteins in AtmKD/− ES cells (Fig. 2 A). However, two independently derived AtmKD/− ES cell lines grew significantly slower than both Atm−/− ES and Atm+/+ ES cells, which grew at comparable rates (Fig. 2 B).
Proliferation defects and increased genomic instability in AtmKD/− ES cells. (A) Western blot analysis of total protein (50 µg) for mouse ATM (MAT3; Sigma-Aldrich) and α-tubulin (EMD) in WT, AtmC/KDN, Atm−/−, Atm+/−, and AtmKD/− ES cells. The small amount of protein detectable in the Atm−/− lane likely arises from slight contamination of the ES cells used for this analysis with WT fibroblasts from the feeder layer. (B) Fold increase of cell number relative to day 0, as measured by modified MTT assay (Sigma-Aldrich) in WT, Atm−/−, and AtmKD/− ES cells under normal growth conditions. Three independent experiments were performed in two independently derived AtmKD/− ES cell lines. The graph and the error bars represent standard deviation derived from quadruplicated cultures in one representative experiment. (C) Frequency of metaphases with cytogenetic abnormalities in T-FISH analyses of WT, Atm+/−, and AtmKD/− ES cells. The p-value (0.035) is obtained with χ2 test. (D) The frequency of chromatid and chromosome breaks measured by T-FISH analyses in WT, Atm+/−, and AtmKD/− ES cells. The χ2 test p-value for the frequency of chromatid breaks between Atm+/− and AtmKD/− ES cells is 0.002 and for chromosome breaks is 0.101. (C and D) The error bars represent the standard deviations of multiple independent experiments. (E) Summary of cytogenetic abnormalities in WT, Atm+/−, and AtmKD/− ES cells and controls. Ab, abnormal; MP, metaphase; Chs., chromosome; Cht., chromatid; Br, break; Std.Er, standard error. The data summarize the results from three or more independent experiments using at least two independently derived cell lines of each genotype. 20 metaphases were analyzed per line per experiment.
Proliferation defects and increased genomic instability in AtmKD/− ES cells. (A) Western blot analysis of total protein (50 µg) for mouse ATM (MAT3; Sigma-Aldrich) and α-tubulin (EMD) in WT, AtmC/KDN, Atm−/−, Atm+/−, and AtmKD/− ES cells. The small amount of protein detectable in the Atm−/− lane likely arises from slight contamination of the ES cells used for this analysis with WT fibroblasts from the feeder layer. (B) Fold increase of cell number relative to day 0, as measured by modified MTT assay (Sigma-Aldrich) in WT, Atm−/−, and AtmKD/− ES cells under normal growth conditions. Three independent experiments were performed in two independently derived AtmKD/− ES cell lines. The graph and the error bars represent standard deviation derived from quadruplicated cultures in one representative experiment. (C) Frequency of metaphases with cytogenetic abnormalities in T-FISH analyses of WT, Atm+/−, and AtmKD/− ES cells. The p-value (0.035) is obtained with χ2 test. (D) The frequency of chromatid and chromosome breaks measured by T-FISH analyses in WT, Atm+/−, and AtmKD/− ES cells. The χ2 test p-value for the frequency of chromatid breaks between Atm+/− and AtmKD/− ES cells is 0.002 and for chromosome breaks is 0.101. (C and D) The error bars represent the standard deviations of multiple independent experiments. (E) Summary of cytogenetic abnormalities in WT, Atm+/−, and AtmKD/− ES cells and controls. Ab, abnormal; MP, metaphase; Chs., chromosome; Cht., chromatid; Br, break; Std.Er, standard error. The data summarize the results from three or more independent experiments using at least two independently derived cell lines of each genotype. 20 metaphases were analyzed per line per experiment.
We proceeded to measure spontaneous genomic instability in Atm+/+, AtmKD/−, and Atm−/− ES cells using telomere FISH (T-FISH) assays (Franco et al., 2006; Zha et al., 2008). Under regular growth conditions, ∼30% metaphases derived from AtmKD/− ES cells had spontaneous cytogenetic abnormalities, whereas only 18% of those derived from Atm−/− cells and <2% of those derived from WT cells showed such abnormalities (Fig. 2, C and E). In contrast to the predominant chromosome breaks in Atm−/− ES cells (Fig. 2, D and E; Franco et al., 2006; Zha et al., 2008), the cytogenetic aberrations in AtmKD− ES cells were evenly distributed between chromosome (break involving both sister chromatids) and chromatid (breaks involving one of the two sister chromatids) breaks, with the most dramatic increase observed in chromatid breaks (Fig. 2, D and E; and Fig. S2 C). Although chromosome breaks are derived primarily from DSBs that arise during the G1 phase of the cell cycle, chromatid breaks often result from DSBs in S and G2 phases of the cell cycle, when HR is most active. Together, these results suggest that the physical presence of ATM-KD inhibits DNA repair, likely HR, in a manner that does not occur in the absence of ATM protein. When we expressed mEOS2-ATM in cells in which the endogenous ATM protein was disrupted, both WT and KD ATM are recruited to the site of DNA damage and retained there for ≥5 min (Fig. S2 B). Similar results were obtained in two previous studies (Barone et al., 2009; Davis et al., 2010). Furthermore, previous studies have identified both replication-related DNA repair defects and increased chromatid breaks in cells treated with selective ATM kinase inhibitors (White et al., 2010; Gamper et al., 2012) but not in ATM-null cells. Although it is unclear whether the effects of ATM kinase inhibitors on DNA repair are the same as those observed in AtmKD/− cells, our findings emphasize the need to distinguish the effects of ATM deletion and kinase inhibition in future studies.
The effect of ATM-KD protein on lymphocyte development
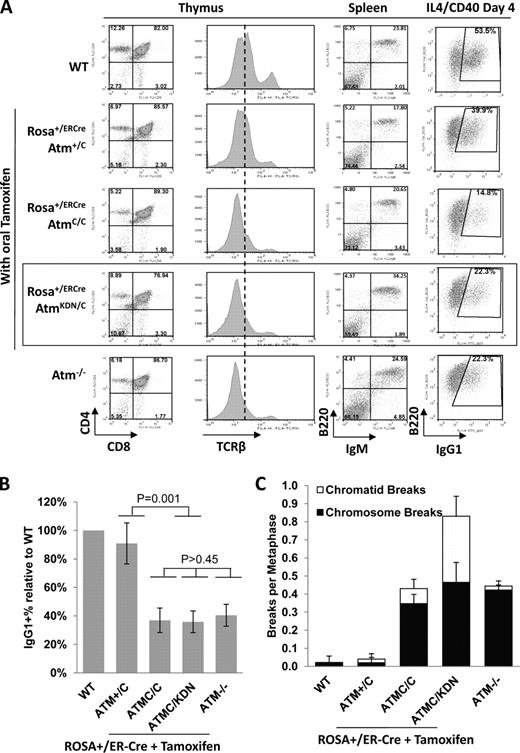
To test the consequences of ATM-KD protein expression in somatic cells, we generated Rosa+/ER-CreAtmC/KDN mice. The ROSA26-ER-Cre (Rosa+/ER-Cre) allele ubiquitously expresses a tamoxifen-inducible fusion protein of estrogen receptor ligand-binding domain and Cre recombinase (ER-Cre) under the endogenous Rosa26 promoter (de Luca et al., 2005; Guo et al., 2007). Lymphocyte development was then analyzed in conditional ATM-KD (Rosa+/ER-CreAtmC/KDN), conditional ATM-null (Rosa+/ER-CreAtmC/C(−)), and control (Rosa+/ER-CreAtmC/+) mice 10 d after oral tamoxifen administration, at which time Cre-mediated recombination of the AtmC and AtmKDN alleles was readily observed in bone marrow, thymus, and spleen (Fig. S3). Western blot of total thymocytes showed that ATM-KD protein is expressed at levels similar to WT protein in thymocytes (Fig. 3 A). Upon irradiation, the levels of phosphorylated KAP-1 and H2AX (γ-H2AX) in thymocytes purified from tamoxifen-treated Rosa+/ER-CreAtmC/KDN mice was comparable to those from Rosa+/ER-CreAtmC/C mice, confirming the loss of ATM kinase activity in AtmKD/− thymocytes (Fig. 3 B). Compared with WT controls (182 ± 27 × 106), the total thymocyte number in tamoxifen-treated Rosa+/ER-CreAtmC/KDN mice (60 ± 17 × 106) was reduced to a level similar to Rosa+/ER-CreAtmC/C (54 ± 12 × 106) and Atm−/− (42 ± 16 × 106) mice (n > 3 for each group). FACS analyses of tamoxifen-treated Rosa+/ER-CreAtmC/KDN mice revealed thymocyte developmental defects, namely decreased TCR-β surface expression and decreased single-positive percentage in the thymus, comparable to those of tamoxifen-treated Rosa+/ER-CreAtmC/C or Atm−/− mice (Fig. 4 A; Borghesani et al., 2000). Together these data suggest that KD ATM protein partially blocks T cell development and, by extension, chromosomal V(D)J recombination to a level comparable with the absence of ATM protein.
ATMKD is catalytically inactive. (A) Western blot for total ATM protein in thymocytes from tamoxifen-treated Rosa+/ER-CreAtm+/C, AtmC/C, or AtmC/KDN mice and control Atm−/− mice. (B) Western blot for phosphorylated H2AX, phosphorylated KAP-1, and total KAP-1 and Actin in irradiated thymocyte (5 Gy) with or without preincubation with 15 µM ATM kinase inhibitor (KU55933; Tocris Bioscience) or 5 µM DNA-PKcs kinase inhibitor (NU7441; Tocris Bioscience). Cell lysates were collected 2 h after irradiation. Primary antibodies were used at the following dilutions: anti-Actin (1:10,000; Sigma-Aldrich), anti-ATM (1:500; MAT3; Sigma-Aldrich), anti–γ-H2AX (1:1,000; EMD Millipore), anti–KAP-1 (1:1,000; Cell Signaling Technology), antiphospho–KAP-1 (S824; 1:1,000; Bethyl Laboratories, Inc.). exp., exposure.
ATMKD is catalytically inactive. (A) Western blot for total ATM protein in thymocytes from tamoxifen-treated Rosa+/ER-CreAtm+/C, AtmC/C, or AtmC/KDN mice and control Atm−/− mice. (B) Western blot for phosphorylated H2AX, phosphorylated KAP-1, and total KAP-1 and Actin in irradiated thymocyte (5 Gy) with or without preincubation with 15 µM ATM kinase inhibitor (KU55933; Tocris Bioscience) or 5 µM DNA-PKcs kinase inhibitor (NU7441; Tocris Bioscience). Cell lysates were collected 2 h after irradiation. Primary antibodies were used at the following dilutions: anti-Actin (1:10,000; Sigma-Aldrich), anti-ATM (1:500; MAT3; Sigma-Aldrich), anti–γ-H2AX (1:1,000; EMD Millipore), anti–KAP-1 (1:1,000; Cell Signaling Technology), antiphospho–KAP-1 (S824; 1:1,000; Bethyl Laboratories, Inc.). exp., exposure.
Analysis of ATM KD lymphocytes. (A) Representative flow cytometric analyses of total thymocytes stained with CD4, CD8, and TCR-β, and total splenocytes stained with B220 and IgM surface markers. For the B220/IgG1 marker staining, CD43− splenocytes were isolated and stimulated in culture with anti-CD40 and IL-4 for 4 d before staining. A total of five independent experiments were performed, and one set of representative FACS analyses is shown. The dotted line shows the median level of surface TCR-β levels in WT thymocytes. The box highlights the analyses of ATMKD/− lymphocytes. (B) Relative frequency of the IgG1+ cells among all B220+ B cells (relative to WT cells in each experiments) in Atm−/− and AtmKD/− B cells. The data represent the mean and standard deviation from at least five experiments. The χ2 test p-values are marked in the graph. (C) The frequency (per metaphase) of chromatid and chromosome breaks measured by T-FISH analyses in stimulated B cells from WT, Atm−/− mice, or tamoxifen-treated Rosa+/ER-CreAtm+/C, AtmC/C, or AtmC/KDN mice. The χ2 test p-value for the frequency of chromatid breaks between Atm+/− and AtmKD/− B cells is 0.007 and for chromosome breaks is 0.816. The table summarized the data obtained from independent experiments performed on two or three mice of each genotype (see also Fig. S3 F).
Analysis of ATM KD lymphocytes. (A) Representative flow cytometric analyses of total thymocytes stained with CD4, CD8, and TCR-β, and total splenocytes stained with B220 and IgM surface markers. For the B220/IgG1 marker staining, CD43− splenocytes were isolated and stimulated in culture with anti-CD40 and IL-4 for 4 d before staining. A total of five independent experiments were performed, and one set of representative FACS analyses is shown. The dotted line shows the median level of surface TCR-β levels in WT thymocytes. The box highlights the analyses of ATMKD/− lymphocytes. (B) Relative frequency of the IgG1+ cells among all B220+ B cells (relative to WT cells in each experiments) in Atm−/− and AtmKD/− B cells. The data represent the mean and standard deviation from at least five experiments. The χ2 test p-values are marked in the graph. (C) The frequency (per metaphase) of chromatid and chromosome breaks measured by T-FISH analyses in stimulated B cells from WT, Atm−/− mice, or tamoxifen-treated Rosa+/ER-CreAtm+/C, AtmC/C, or AtmC/KDN mice. The χ2 test p-value for the frequency of chromatid breaks between Atm+/− and AtmKD/− B cells is 0.007 and for chromosome breaks is 0.816. The table summarized the data obtained from independent experiments performed on two or three mice of each genotype (see also Fig. S3 F).
Similarly, B cell development in tamoxifen-treated Rosa+/ER-CreAtmC/KDN mice was largely comparable with that of Atm−/− mice (Fig. 4 A). To test the effect of ATM-KD protein in CSR, we incubated partially purified (CD43−) AtmKD/− and control splenic B cells with anti-CD40 plus IL-4 (interleukin 4) for 4 d to stimulate CSR to IgG1. PCR and Southern blot analyses confirmed the disappearance of AtmC and AtmKDN alleles and the appearance of the recombined Atm− and AtmKD alleles in purified B cells (Fig. S3, A–C). Western analysis for phosphorylated H2AX (γ-H2AX) in irradiated B cells purified from tamoxifen-treated Rosa+/ER-CreAtmC/KDN and Rosa+/ER-CreAtmC/C mice confirmed the loss of ATM kinase activity in stimulated B cells (Fig. S3 D). Furthermore, B cells from tamoxifen-treated Rosa+/ER-CreAtmC/C(−) mice performed CSR at ∼40% of WT levels with increased genomic instability, similar to those of germline Atm−/− mice (Fig. 4, B and C; and Fig. S3), confirming functional loss of ATM protein in B cells from tamoxifen-treated Rosa+/ER-CreAtmC/C(−) mice. Nevertheless, B cells from tamoxifen-treated Rosa+/ER-CreAtmC/KDN mice underwent CSR at levels comparable with their ATM-null counterparts (Fig. 4 B and Fig. S3). Consistent with our findings on AtmKD/− ES cells, activated B cells from tamoxifen-treated Rosa+/ER-CreAtmC/KDN mice accumulated greater genomic instability, especially chromatid breaks, than Atm−/− B cells upon activation (Fig. 4 C and Fig. S3, D and E). Together, those results support the conclusion that the ATM-KD protein inhibits DNA repair in S and G2 phase of cell cycles.
Although NHEJ plays critical roles in lymphocyte-specific DSB repair events, it is largely dispensable for embryonic development (Lieber, 2010). In contrast, most HR factors are required for embryonic development. In this context, the embryonic lethality of AtmKD/KD, but not Atm−/−, mice may reflect an inhibitory function of ATM-KD protein in a subset of HR. Consistent with this hypothesis, the most dramatic increase of general genomic instability in AtmKD/− cells over ATM-null cells is chromatid breaks, which are typically associated with defects in S and G2 phase DNA repair.
ATM belongs to the family of phosphoinositide 3-kinase–related kinase, which also includes DNA-dependent protein kinase catalytic subunit (PKcs) and ATR. Although ATR is activated by RPA-coated single-strand DNA (Shiotani and Zou, 2009; Liu et al., 2011), ATM and DNA-PKcs are both activated by DNA DSBs, phosphorylate an overlapping pool of substrates (e.g., H2AX, KAP1, and p53; Callén et al., 2009b; Zha et al., 2011a,b), and have redundant functions during embryonic development and DNA repair (Gurley and Kemp, 2001; Sekiguchi et al., 2001; Callén et al., 2009b; Gapud et al., 2011; Zha et al., 2011b). It is possible that the presence of an enzymatically inactive ATM protein might prevent DNA-PKcs, and potentially other phosphoinositide 3-kinase–related kinases, from phosphorylating shared substrates, thereby disrupting DNA repair to a greater extent than complete loss of ATM alone. Loss of both ATM and DNA-PKcs led to CSR defects more severe than loss of either kinase alone (Callén et al., 2009b), but AtmKD/− and Atm−/− B cells performed CSR at similar levels (Fig. 4, A and B), inconsistent with inhibition of DNA-PKcs function in AtmKD/− B cells. Moreover, H2AX and KAP-1, two shared substrates of ATM and DNA-PKcs, are phosphorylated at comparable levels in AtmKD/− and Atm−/− B and T cells (Fig. 3 B and Fig. S3 D). Although phosphorylation of particular substrates of DNA-PKcs or DNA-PKcs itself may be inhibited in AtmKD/− cells, but not ATM−/− cells, our findings are inconsistent with global inhibition of DNA-PKcs activity in AtmKD/− cells. In this context, loss of ATM phosphorylation sites on DNA-PKcs leads to more severe defects in DNA repair than DNA-PKcs–null mice (Zhang et al., 2011).
Past studies of ATM function in DNA repair have focused primarily on its kinase activity. Here, we and others have shown that KD ATM proteins cause early embryonic lethality despite the normal embryonic development in the complete absence of ATM protein (see Daniel et al. in this issue). We further show that AtmKD/− ES and B cells accumulate much higher levels of chromosome instability, particularly chromatid breaks, relative to ATM-null cells. Despite the increased genomic instability, AtmKD/− lymphocytes performed V(D)J recombination and CSR at levels comparable with ATM-null cells. Together, these data suggest that although loss of ATM kinase activity and ATM-mediated phosphorylation of downstream targets compromise both NHEJ and, to a lesser extent, HR function, the ATM-KD protein elicits additional inhibitory effects in postreplication DNA repair, potentially HR. By uncovering unexpected functions for ATM protein in DNA repair and embryonic development, our study raises clinically relevant questions about the mechanism of ATM activation and the application of ATM kinase inhibitors.
Materials and methods
Generation of the ATM-KD allele
The DNA sequence for homologous targeting at the Atm locus was generated via PCR from TC1 ES cell DNA (129 strain). A targeting construct was designed to insert a NeoR gene cassette oriented in the opposite transcriptional direction from the endogenous Atm promoter into intron 57 next to the D2880A/N2885K mutation in exon 58. A 3.5-kb 5′ arm and 5.1-kb 3′ arm were PCR generated separately, cloned into the pBK vector, and sequenced. The 5′ arm was directly subcloned into pLNTK in the desired orientation. The mutation was introduced into the 3′ arm using site-directed mutagenesis and confirmed by sequencing. The mutated 3′ arm was then subcloned into pLNTK. The targeting construct was then electroporated into CSL3 ES cells (129 strain), and successful targeting was determined via Southern blot analyses using EcoRV-digested genomic DNA and a 5′ genomic probe as outlined in Fig. 1 B. The WT band is ∼22 kb, and the targeting introduced an additional EcoRV site and reduced the band to ∼4.7 kb. The corrected clones were confirmed with a 3′ probe with EcoRV digestions. Six independently targeted clones were sequenced for the ATM-KD mutation, and four were verified with correct mutations. Two were injected for germline transmission.
Mice
AtmC/C and Rosa+/ER-Cre mice were previously characterized (de Luca et al., 2005; Guo et al., 2007; Zha et al., 2008, 2011b). To activate Cre recombination, Rosa+/ER-Cre mice were orally fed with tamoxifen (5 mg in 90 µl of sunflower oil and 10 µl ethanol per mouse) daily for a consecutive 2 d. The mice were analyzed 10–14 d after the second treatment. All animal experiments were conducted in a pathogen-free facility and approved by the Institute Animal Care and Use Committee of Columbia University.
Derivation of ES cells from mice
3–4-wk-old female Atm+/KDN mice were superovulated by intraperitoneal injection of 5.0 IU pregnant mare serum gonadotropin followed by intraperitoneal injection of 5.0 IU human chorionic gonadotropin (hCG; Sigma-Aldrich) 46–48 h later. The females were set up with adult male AtmC/C mice (1:1 ratio) immediately after the hCG injection. 4 d after the hCG injection, the mated females were sacrificed, and mature blastocysts were flushed out of the uterus and plated one per well in 96-well plates containing irradiated feeders. After 9–14-d incubation, the cells were expanded to 24-well plates, at which stage, half of the cells were frozen down, and the rest of the cells were used to derive DNA for genotyping. AtmKDN/C ES cells were then identified by PCR.
Cytogenetic analysis for genomic instability
ES cells, not grown on feeders, were plated 24 h before the addition of colcemid (KaryoMAX Colcemid Solution; Gibco) at 100 ng/ml for 1–2 h. ES cells were harvested by trypsinization, and metaphases were prepared as previously described (Zha et al., 2008, 2011b). T-FISH was performed as previously described (Franco et al., 2006). In brief, telomeres were stained with a Cy3-labled (CCCTAA)3 peptide nucleic acid probe (Biosynthesis, Inc.), and DNA was counterstained with DAPI-containing mounting media (Vectashield; Vector Laboratories). Images were acquired using a microscope (Eclipse 80i; Nikon) with remote focus accessory and with a camera unit (CoolSNAP HQ; Photometrics) with the Plan Fluor lens (60 and 100×/1.30 NA oil; Nikon) in room temperature. All images were processed with NIS-Elements AR (version 3.10; Nikon). Chromosome breaks were defined by loss of telomere signal from both sister chromatids. Chromatid breaks were defined by loss of telomere signal from one of the two sister chromatids or a clearly broken DAPI signal in the middle of one chromatid (Fig. S2 C). We note that chromosomal gaps could not be reliably identified with the T-FISH assay; therefore, it is possible that the actual frequency of chromatid or chromosome breaks might be even higher than estimated.
Lymphocyte development and CSR
Lymphocyte populations were analyzed by flow cytometry as previously described (Li et al., 2008). Isolation and activation of splenic B cells and flow cytometric assays were performed as previously described (Li et al., 2008). In brief, CD43− splenic B cells were purified from total spleen using anti–mouse CD43 MACS beads following the manufacturer’s instructions (Miltenyi Biotec). To stimulate the cells for CSR, the cells were incubated with α-CD40 plus IL-4 for 4 d, and the percentage of CSR to IgG1 was determined by surface staining and FACS analysis (Li et al., 2008). To assay for genomic instability, colcemid was added to a fraction of day 4 stimulated B cells for 4 h, and the metaphases were prepared and stained for T-FISH as described for ES cells.
Generation of mEOS2-ATM plasmids
To investigate the accumulation of ATM-WT and ATM-KD at sites of DNA damage in A-T cells, mEOS2-ATM expression constructs were generated. Plasmid pRSETa mEos2 was obtained from Addgene, and mEOS2 (McKinney et al., 2009) was amplified by PCR using primers with NotI linkers. The amplified sequence was inserted into pcDNA3-ATM-WT (Canman et al., 1998) at the NotI site, and plasmids with mEOS2 were inserted in the correct orientation identified by sequencing. mEOS2-ATM-WT Δ3′ untranslated and mEOS2-ATM-KD Δ3′ untranslated were generated by removing the ATM kinase domain and 3′ untranslated sequence in the original pcDNA3-ATM-WT by digestion with BlpI and XhoI and replacing with either the pcDNA3-ATM kinase-inactive sequence containing mutations at D2870A/N2875K (Canman et al., 1998) or the ATM-WT sequence, from which the 3′ end has been deleted, as described previously (Gamper et al., 2012). The expression of recombinant mEOS2-ATM protein is not disrupted by a small hairpin RNA that targets the 3′ untranslated sequence of the endogenous mRNA (Gamper et al., 2012).
Online supplemental material
Fig. S1 shows that the gross weight and lymphocyte development of Atm+/KD mice are comparable with Atm+/+ littermates. Fig. S2 shows that the ATM-KD protein could be normally recruited to the site of DNA damage, and it also shows the representative images of T-FISH analyses. Fig. S3 shows the specific and efficient conversion of AtmC to Atm− alleles in B cells from tamoxifen-treated Rosa+/ER-Cre mice. Table S1 shows the primers used for genotyping the AtmC, Atm−, AtmKDN, and AtmKD alleles.
Acknowledgments
We wish to thank Dr. Richard Baer and Jennifer Crowe for their critical reading and comments on our manuscript. We thank Frances Lee and Chen Li for their technical assistance.
This work was supported by grants CA158073 to S. Zha and CA148644 to C.J. Bakkenist. It is also supported by funding made available by St. Baldrick’s Foundation, Gabrielle Angel Foundation, John Driscoll Jr. Children’s Medical Award, and Leukemia Lymphoma Foundation to S. Zha. K. Yamamoto is partially supported by the graduate program of Pathobiology and Molecular Medicine at Columbia University. W. Jiang is supported in part by the National Institutes of Health/National Cancer Institute training grant T32-CA09503 to Herbert Irving Comprehensive Cancer Research Center at Columbia University.
References
- A-T
ataxia telangiectasia
- ATM
A-T mutated
- ATR
A-T and Rad3 related
- CSR
class switch recombination
- DSB
double-strand break
- ES
embryonic stem
- hCG
human chorionic gonadotropin
- HR
homologous recombination
- KD
kinase dead
- NeoR
neomycin resistant
- NHEJ
nonhomologous end joining
- PE
phycoerythrin
- PKcs
protein kinase catalytic subunit
- T-FISH
telomere FISH
- V(D)J
variable, diversity, and joining
- WT
wild type
Author notes
Thomas Ludwig’s current address is Dept. of Molecular and Cellular Biochemistry, Ohio State University Wexner Medical Center, Columbus, OH 43210.