Class II formins are key regulators of actin and are essential for polarized plant cell growth. Here, we show that the class II formin N-terminal phosphatase and tensin (PTEN) domain binds phosphoinositide-3,5-bisphosphate (PI(3,5)P2). Replacing the PTEN domain with polypeptides of known lipid-binding specificity, we show that PI(3,5)P2 binding was required for formin-mediated polarized growth. Via PTEN, formin also localized to the cell apex, phragmoplast, and to the cell cortex as dynamic cortical spots. We show that the cortical localization driven by binding to PI(3,5)P2 was required for function. Silencing the kinases that produce PI(3,5)P2 reduced cortical targeting of formin and inhibited polarized growth. We show a subset of cortical formin spots moved in actin-dependent linear trajectories. We observed that the linearly moving subpopulation of cortical formin generated new actin filaments de novo and along preexisting filaments, providing evidence for formin-mediated actin bundling in vivo. Taken together, our data directly link PI(3,5)P2 to generation and remodeling of the cortical actin array.
Introduction
Formins are critical for numerous key actin-based processes ranging from cell polarity and cytokinesis to cell adhesion and migration (Goode and Eck, 2007). In general, formins nucleate and elongate actin filaments to specify where actin arrays are built. Long actin arrays, such as actin cables in budding and fission yeasts, are generated by formins (Feierbach and Chang, 2001; Evangelista et al., 2002; Nakano et al., 2002; Sagot et al., 2002).
In plants, as in other eukaryotes, recent studies have linked formins to a variety of actin-based processes (Banno and Chua, 2000; Cheung and Wu, 2004; Favery et al., 2004; Deeks et al., 2005, 2010; Ingouff et al., 2005; Michelot et al., 2005; Yi et al., 2005; Vidali et al., 2009b; Ye et al., 2009; Cheung et al., 2010; Yang et al., 2011; Zhang et al., 2011). Flowering plants have two classes of formins, typically having at least ten genes in each family (Cvrcková et al., 2004).
Both families share the conserved formin homology (FH) 1 and 2 domains found in all eukaryotic formins; however, the N terminus of plant formins is distinct (Cvrcková et al., 2004). Several lines of evidence suggest that many class I formins are integral membrane proteins (Cvrckova, 2000); most have a putative transmembrane domain and a predicted signal peptide, and several localize to the plasma membrane (Favery et al., 2004; Ingouff et al., 2005; Cheung et al., 2010) or endoplasmic reticulum (Deeks et al., 2010). Membrane association is also suggested for class II formins, albeit with peripheral attachment. Members of this class have an N-terminal domain with high sequence similarity to the phosphatase and tensin (PTEN) homologue thought to mediate lipid binding (Cvrcková et al., 2004; Grunt et al., 2008).
In flowering plants, analysis of formin function has been challenging because of the large size of the gene families. Recently, this challenge was addressed by characterizing formin function in the moss Physcomitrella patens (Vidali et al., 2009b). Like flowering plants, this species has both class I and class II formin families, but with only six genes in the former and two genes in the latter. Additionally the moss has a life-cycle stage in which cells grow by tip growth (Menand et al., 2007), a form of growth where the cell contents are highly polarized to direct and support growth at a restricted region (the tip). In flowering plants, although only a few cell types undergo tip growth, these tip-growing cells accomplish essential processes, including nutrient uptake and fertilization. In all plant taxa characterized to date, whether in algae or angiosperms, the actin cytoskeleton is essential to achieve the polarization required for tip growth.
To dissect the function of formin in moss, RNAi was used taking advantage of silencing constructs that allow targeting of an entire gene family as well as concurrent complementation. This approach revealed that class I formins, rather than contributing to polarized growth, are needed for efficient cytokinesis, and that class II formins are essential for tip growth (Vidali et al., 2009b). Further, the FH1-FH2 domains of class II formins were shown, in vitro, to promote rapid rates of actin elongation, an activity that was argued to underlie their role in tip growth. Class II formins localize near the tip of growing cells and the N-terminal PTEN domain is necessary and sufficient for this localization (Vidali et al., 2009b).
In humans, PTEN is a lipid phosphatase that converts PI(3,4,5)P3 to PI(4,5)P2. PTEN acts as a tumor suppressor by limiting PI(3,4,5)P3 levels, which negatively regulates oncogenic phosphoinositol-3-kinase and AKT signaling pathways, thereby preventing cells from growing and dividing too rapidly (Li et al., 1997; Steck et al., 1997; Maehama and Dixon, 1998; Salmena et al., 2008). In moss, the loss of class II formin function is not complemented by expression of the FH1-FH2 domains alone (Vidali et al., 2009b), suggesting that the formin PTEN domain is required for formin activity in tip growth. This suggestion is consistent with an emerging picture in the formin field that N-terminal formin domains contain key regions, including the small GTPase-binding domains required to regulate actin polymerization (Goode and Eck, 2007). Here we use a combination of genetics, biochemistry, and live-cell imaging to investigate the mechanism of the PTEN domain localization and function with respect to formin-mediated actin polymerization during polarized growth.
Results
Class II formins are recruited to sites of membrane remodeling
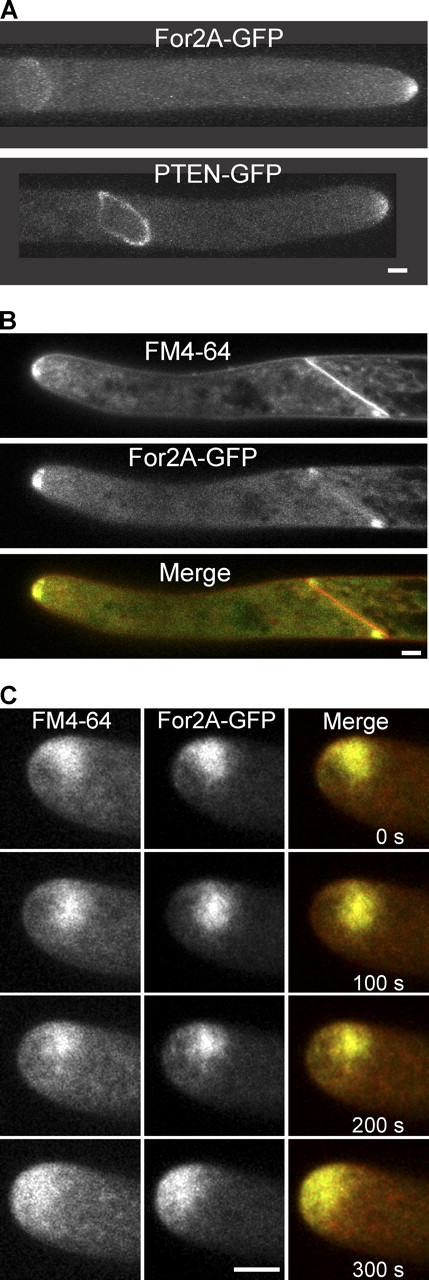
In tip-growing cells, the plasma membrane is actively remodeled at the apex of the cell, where growth is occurring. A combination of active exocytosis of new cell wall material and endocytosis of excess membrane occurs near the cell apex. An additional site of membrane remodeling occurs during cell division as the phragmoplast, the cytokinetic organelle of plants, separates the daughter cells with a new wall. Previously, members of our laboratory localized one of the two functionally redundant class II formins, For2A, to the tip of the cell using a functional GFP fusion protein, For2A-3XmEGFP (referred to hereafter as For2A-GFP; Vidali et al., 2009b). Here, we report that in addition to the cell tip (Fig. 1; Vidali et al., 2009b), For2A-GFP localizes to the phragmoplast (Fig. 1, A and B; Video 1). Furthermore, this localization is reproduced by the For2A PTEN domain alone fused to mEGFP (referred to hereafter as GFP; Fig. 1 A and Video 2). Phragmoplast localization is emphasized by labeling cell membranes with FM4-64. In a medial plane of a cell late in division, FM4-64 labels the cell apex and the entire cell plate membrane. For2A is also at the cell apex, but at the site of cell division it colocalizes with FM4-64 only at the leading edge, coincident with the location of active membrane remodeling within the phragmoplast (Fig. 1 B).
For2A-GFP localizes to the cell tip and phragmoplast during cytokinesis (related to Fig. 1 A). Images are maximum intensity projections of a dividing cell expressing a functionally GFP-tagged copy of For2A. Images were acquired with a laser-scanning confocal microscope (model C1; Nikon). Frames were acquired every 90 s for 30 min. Bar, 5 µm. The video plays at 10 frames per second.
For2A-GFP localizes to the cell tip and phragmoplast during cytokinesis (related to Fig. 1 A). Images are maximum intensity projections of a dividing cell expressing a functionally GFP-tagged copy of For2A. Images were acquired with a laser-scanning confocal microscope (model C1; Nikon). Frames were acquired every 90 s for 30 min. Bar, 5 µm. The video plays at 10 frames per second.
Similar to For2A-GFP, For2A PTEN-GFP is recruited to the phragmoplast during cytokinesis (related to Fig. 1 A). Images are maximum intensity projections of a dividing cell stably expressing PTEN-GFP. Images were acquired with a laser-scanning confocal microscope (model C1; Nikon). Frames were acquired every 90 s for 42 min. The video plays at 10 frames per second. Bar, 5 µm.
Similar to For2A-GFP, For2A PTEN-GFP is recruited to the phragmoplast during cytokinesis (related to Fig. 1 A). Images are maximum intensity projections of a dividing cell stably expressing PTEN-GFP. Images were acquired with a laser-scanning confocal microscope (model C1; Nikon). Frames were acquired every 90 s for 42 min. The video plays at 10 frames per second. Bar, 5 µm.
For2A localizes to sites of dynamic membrane trafficking. (A) For2A-GFP localizes to the tip of the cell and to the phragmoplast. The PTEN domain of For2A alone, fused to GFP also localizes to the tip and the phragmoplast. See also Videos 1 and 2. (B) FM4-64–labeled membranes and For2A colocalize in the phragmoplast, but not the cell plate. Shown is the medial plane of a cell with an almost completed cell division. (C) For2A colocalizes with FM4-64–labeled membranes near the tip. See also Video 3 and Fig. S1. Bars, 5 µm.
For2A localizes to sites of dynamic membrane trafficking. (A) For2A-GFP localizes to the tip of the cell and to the phragmoplast. The PTEN domain of For2A alone, fused to GFP also localizes to the tip and the phragmoplast. See also Videos 1 and 2. (B) FM4-64–labeled membranes and For2A colocalize in the phragmoplast, but not the cell plate. Shown is the medial plane of a cell with an almost completed cell division. (C) For2A colocalizes with FM4-64–labeled membranes near the tip. See also Video 3 and Fig. S1. Bars, 5 µm.
To investigate the colocalization of For2A-GFP and FM4-64 at the cell apex, we observed the dynamics of FM4-64 internalization in cells expressing For2A-GFP using pulse-chase labeling of cells with FM4-64. Interestingly, early internalization of FM4-64, which is likely specific for endocytosis (van Gisbergen et al., 2008), does not colocalize with For2A-GFP (Fig. S1). However, after 30 min, For2A-GFP dynamics and localization coincide with the FM4-64 signal (Fig. 1 C and Video 3), suggesting that the For2A labeling may represent a combination of late endocytic and post-Golgi exocytic membranes. These data together with the phragmoplast localization suggest that For2A-GFP is enriched at sites of active membrane remodeling.
Colocalization of FM4-64–labeled membranes with For2A-GFP at the cell tip (related to Fig. 1 C). For2A-GFP line was pulse-labeled with 20 µM FM4-64 for 5 min and further incubated for 25 min at room temperature. The video was recorded after 30 min incubation. The dynamics of FM4-64–labeled membrane are similar to For2A-GFP at the cell tip. Images are from a single focal plane. Images were acquired with a laser-scanning confocal microscope (model C1; Nikon). Frames were acquired every 10 s for 12.6 min. The video plays at 20 frames per second. Bar, 5 µm.
Colocalization of FM4-64–labeled membranes with For2A-GFP at the cell tip (related to Fig. 1 C). For2A-GFP line was pulse-labeled with 20 µM FM4-64 for 5 min and further incubated for 25 min at room temperature. The video was recorded after 30 min incubation. The dynamics of FM4-64–labeled membrane are similar to For2A-GFP at the cell tip. Images are from a single focal plane. Images were acquired with a laser-scanning confocal microscope (model C1; Nikon). Frames were acquired every 10 s for 12.6 min. The video plays at 20 frames per second. Bar, 5 µm.
Actin polymerization is not directly correlated with all populations of cytoplasmic For2A
To determine the spatial and temporal relationship between For2A and actin, we colocalized actin and formin in tip-growing cells. We generated a line expressing For2A-GFP and Lifeact-mCherry, a validated marker for imaging actin in living protonemal cells (Vidali et al., 2009a). Interestingly, the prominent apical accumulation of For2A-GFP and the Lifeact signal associated only occasionally (Fig. 2 A and Video 4). The image series in Fig. 2 A represents 45 min during which time the cell is actively growing. Both For2A-GFP and Lifeact-mCherry signals are dynamic with changes in position and intensity near the apex of the cell. However, there is a low temporal and spatial correlation between apical accumulation of For2A-GFP and Lifeact-mCherry intensity in this and similar image sequences.
Imaging of For2A-GFP and actin in a growing protonemal cell (related to Fig. 2 A). Images are maximum intensity projections of a For2A-GFP cell stably expressing Lifeact-mCherry. Images were acquired with a spinning confocal microscope (model Ti-E, Nikon; equipped with a spinning disk head [model CSU-X1, Yokogawa Corporation of America]). Frames were acquired every 5 min for 45 min. The video plays at 5 frames per second. Bar, 5 µm.
Imaging of For2A-GFP and actin in a growing protonemal cell (related to Fig. 2 A). Images are maximum intensity projections of a For2A-GFP cell stably expressing Lifeact-mCherry. Images were acquired with a spinning confocal microscope (model Ti-E, Nikon; equipped with a spinning disk head [model CSU-X1, Yokogawa Corporation of America]). Frames were acquired every 5 min for 45 min. The video plays at 5 frames per second. Bar, 5 µm.
Dynamics of For2A and actin in a growing tip cell. (A) For2A-GFP is present in the apex and Lifeact-mCherry is present subapically mostly along the cell cortex. The majority of the time, For2A-GFP and Lifeact-mCherry do not overlap. Occasionally a short overlap followed by quick actin polymerization can be seen (25–35 min). See also Video 4. (B) Away from the tip, bursts of For2A-GFP are observed, followed by actin polymerization near the For2A-GFP enrichment. Here, the For2A-GFP cloud travels through the cell with actin filaments behind it. See also Video 5. Bars, 5 µm.
Dynamics of For2A and actin in a growing tip cell. (A) For2A-GFP is present in the apex and Lifeact-mCherry is present subapically mostly along the cell cortex. The majority of the time, For2A-GFP and Lifeact-mCherry do not overlap. Occasionally a short overlap followed by quick actin polymerization can be seen (25–35 min). See also Video 4. (B) Away from the tip, bursts of For2A-GFP are observed, followed by actin polymerization near the For2A-GFP enrichment. Here, the For2A-GFP cloud travels through the cell with actin filaments behind it. See also Video 5. Bars, 5 µm.
Occasionally though, accumulation of tip-localized For2A-GFP was closely associated with an increase in the Lifeact-mCherry signal (Fig. 2 A, 25–35 min). On average we observed this 2 times in a 50-min time-lapse acquisition (9 cells). In addition to these infrequent events at the tip, For2A-GFP transiently accumulated in other regions of the cell during growth. Time-lapse imaging with higher temporal resolution revealed that these subapical enrichments were always associated with a burst of Lifeact-mCherry signal indicative of actin polymerization (Fig. 2 B; n = 31 accumulations in 9 cells). The formin subapical enrichments were stochastic, from none to as many as 18 occurring in a 60-min time-lapse acquisition. Interestingly, the position of the filaments with respect to For2A-GFP suggested that actin filaments emerged from the formin structure consistent with formin-mediated actin polymerization (Fig. 2 B and Video 5). These data demonstrate that not all cytoplasmic accumulations of For2A actively generate actin filaments.
For2A-GFP generates actin filaments in vivo (related to Fig. 2 B). A burst of For2A-GFP signal, followed by a filamentous Lifeact-mCherry signal, was imaged in the For2A-GFP/LA-mCherry line. Images are from a single focal plane of a subapical region in a protonemal cell. Images were acquired with a spinning confocal microscope (model Ti-E, Nikon; equipped with a spinning disk head [model CSU-X1, Yokogawa Corporation of America]). Frames were acquired every 8 s for 56 s. The video plays at 5 frames per second. Bar, 5 µm.
For2A-GFP generates actin filaments in vivo (related to Fig. 2 B). A burst of For2A-GFP signal, followed by a filamentous Lifeact-mCherry signal, was imaged in the For2A-GFP/LA-mCherry line. Images are from a single focal plane of a subapical region in a protonemal cell. Images were acquired with a spinning confocal microscope (model Ti-E, Nikon; equipped with a spinning disk head [model CSU-X1, Yokogawa Corporation of America]). Frames were acquired every 8 s for 56 s. The video plays at 5 frames per second. Bar, 5 µm.
PTEN homologues fused to the FH1-FH2 domains differentially restore formin-mediated polarized growth
Previous studies demonstrated that neither the FH1-FH2 domains nor the For2A PTEN domain, when expressed alone, complement loss of formin (Vidali et al., 2009b). To further investigate the role of the For2A PTEN domain, we tested whether the PTEN domain could be functionally replaced by PTEN homologues by performing a complementation analysis of the formin RNAi phenotype. As reported previously (Vidali et al., 2009b), RNAi-mediated silencing of both class II formins results in severely stunted plants composed of small round cells (Fig. 3). Silencing is performed using an RNAi construct that contains sequences from the untranslated regions of both For2A and 2B (For2AB-5′UTR). Co-transforming with a construct that expresses the coding sequence of a single formin gene, For2A, ameliorates the growth defect (Fig. 3; Vidali et al., 2009b). These plants are polarized and look like the wild type; however, quantification of area and polarity (via the morphometric parameter, circularity) shows that the plants are modestly smaller and less polarized (Fig. 3 B). Circularity is a ratio of plant area to the square of the perimeter. Wild-type plants have low circularity, resulting from the large perimeter of a highly branched structure. In contrast, the circularity of For2 RNAi plants approaches one because the shape of these plants is far more circular. The incomplete complementation by For2A is due to the method used to generate the For2A construct (as well as all others used here), which introduces eight amino acid insertions between domains, insertions that slightly compromise the protein’s activity (see discussion in Vidali et al., 2009b).
In place of the For2A PTEN domain, PTEN homologues fused to the FH1-FH2 domains of For2A differentially complement formin-mediated polarized growth. (A) Representative chlorophyll autofluorescence images of control RNAi (GUS-RNAi), For2 RNAi (For2AB-5′UTR), and For2 RNAi plants co-transformed with indicated constructs. Bar, 100 µm. (B) Quantification of area and circularity by chlorophyll autofluorescence shows that replacing the For2A PTEN domain with moss PTEN homologues provides full complementation with PTENA, but not with PTEND. Replacement using HsPTEN does not complement the For2 RNAi phenotype. Tagging the formin with an epitope tag (3XFLAG) does not affect its ability to complement the phenotype. Number of plants analyzed is: 101, GUS-RNAi; 101, For2AB-5′UTR; 50, +For2A; 51, +For2A-3XFLAG; 76, +PTENA-FH1FH2; 50, +PTEND-FH1FH2-3XFLAG; 50, +HsPTEN-FH1FH2-3XFLAG. Error bars represent SEM and letters above the bars indicate statistical groups with α = 0.05 from an ANOVA analysis. (C) Alignment of the phosphoinositide-binding regions of human PTEN (HsPTEN) with the For2A and For2B PTEN domains and four P. patens PTEN homologues (PpPTENA-D). The arrow indicates an arginine residue critical for catalytic activity. Note this arginine is absent in the formin PTEN domains.
In place of the For2A PTEN domain, PTEN homologues fused to the FH1-FH2 domains of For2A differentially complement formin-mediated polarized growth. (A) Representative chlorophyll autofluorescence images of control RNAi (GUS-RNAi), For2 RNAi (For2AB-5′UTR), and For2 RNAi plants co-transformed with indicated constructs. Bar, 100 µm. (B) Quantification of area and circularity by chlorophyll autofluorescence shows that replacing the For2A PTEN domain with moss PTEN homologues provides full complementation with PTENA, but not with PTEND. Replacement using HsPTEN does not complement the For2 RNAi phenotype. Tagging the formin with an epitope tag (3XFLAG) does not affect its ability to complement the phenotype. Number of plants analyzed is: 101, GUS-RNAi; 101, For2AB-5′UTR; 50, +For2A; 51, +For2A-3XFLAG; 76, +PTENA-FH1FH2; 50, +PTEND-FH1FH2-3XFLAG; 50, +HsPTEN-FH1FH2-3XFLAG. Error bars represent SEM and letters above the bars indicate statistical groups with α = 0.05 from an ANOVA analysis. (C) Alignment of the phosphoinositide-binding regions of human PTEN (HsPTEN) with the For2A and For2B PTEN domains and four P. patens PTEN homologues (PpPTENA-D). The arrow indicates an arginine residue critical for catalytic activity. Note this arginine is absent in the formin PTEN domains.
To investigate if PTEN homologues could function in place of the For2A PTEN domain, we generated chimeric proteins where the For2A PTEN domain was replaced with human PTEN or moss PTEN. Moss has four PTEN homologues that form two groups based on sequence similarity, with PTENA and B forming one group and PTENC and D the other (Table S1). For complementation studies we chose one from each group. Human PTEN fused to the For2A FH1-FH2 domains (HsPTEN-FH1FH2-3XFLAG) and co-transformed with the formin RNAi construct was unable to rescue either plant area or circularity (Fig. 3), suggesting that some aspect of human PTEN renders this chimera inactive.
Interestingly, the two moss PTEN homologues we investigated showed different degrees of complementation. PTENA-FH1FH2 fully rescues, giving rise to plants whose area and circularity are essentially the same as those given by complementation with For2A itself (Fig. 3). In contrast, we found that PTEND-FH1FH2-3XFLAG does not rescue. Some plants polarize to a limited extent, as seen in the image (Fig. 3 A). But in comparison to formin RNAi, slight improvements in plant area and morphology (Fig. 3 B) are not statistically significant.
Differential rescue might arise from low levels of expression from the complementing plasmid. We have controlled for this in two ways. First, we generate all the expression plasmids similarly, with the same backbone vector and containing the same strong constitutive promoter. Additionally, noncomplementing constructs were generated with an epitope tag (3XFLAG) and expression of the protein in moss cells was confirmed (Fig. S2 A). As a control, For2A was tested with and without the epitope tag and was found to rescue similarly (Fig. 3, A and B). Second, we performed the complementation assay with a range of different plasmid concentrations. Complementation of the formin RNAi phenotype by For2A is optimal using between 5 and 15 µg of the For2A expression plasmid. Below this concentration, we observe a dose-dependent decrease in complementation (Fig. S3). At higher concentrations of For2A, rescue is also diminished (Fig. S3) and it is difficult to recover sufficient numbers of transformants. This is likely a result of toxicity from overexpression of For2A. Similar to For2A, PTENA-FH1FH2 exhibits dose-dependent complementation under the same concentration range (Fig. S3). In contrast, PTEND-FH1FH2 rescues weakly and HsPTEN-FH1FH2 does not restore function at all concentrations tested (Fig. S3). Because the noncomplementing constructs were unable to rescue at the high plasmid concentrations, it should account for lower expression of these fusion proteins (Fig. S2 A). Furthermore, we were unable to recover transformants with high concentrations of HsPTEN-FH1FH2, suggesting that this construct is also toxic at high levels. Taken together, these data indicate that PTENA can functionally replace the For2A PTEN domain, whereas HsPTEN and PTEND cannot.
If PTENA were more similar to the For2A PTEN domain than human PTEN, then that might explain the difference in complementation. However, sequence comparison of human and moss PTENs with the PTEN domains from For2A and For2B does not support this. PTENA is 27% similar to human PTEN, but only 21.6 and 20.4% similar to For2A and For2B PTEN domains, respectively (Table S1). Inspection of the amino acid sequence alignment near the phosphoinositide binding pocket reveals that the phosphoinositide binding region in human PTEN (Lee et al., 1999) is highly similar to the class II formin PTEN domains and the moss PTEN homologues (Fig. 3 C; Grunt et al., 2008). Interestingly, neither of the For2A nor the For2B PTEN domains have a critical arginine required for catalytic activity (Fig. 3 C, arrow; Barford et al., 1994; Lee et al., 1999). Therefore, as has been suggested (Grunt et al., 2008), the formin PTEN domains probably bind phosphoinositides but do not dephosphorylate them. Thus, perhaps the observed differential rescue results from different phosphoinositide binding activities of the tested PTEN homologues.
For2A PTEN domain and moss PTENA homologue bind PI(3,5)P2
Human PTEN not only converts PI(3,4,5)P3 to PI(4,5)P2 (Maehama and Dixon, 1998), it also binds PI(4,5)P2 (Redfern et al., 2008). Because moss PTENA, but not human PTEN, could functionally replace the For2A PTEN domain, we hypothesized that the For2A PTEN domain and moss PTENA might interact with a different phosphoinositide as compared with human PTEN. To test this, we used lipid overlay assays (Dowler et al., 2002) to determine the phosphoinositide binding preferences of the For2A PTEN domain, PTENA, and PTEND. The For2A PTEN domain fused to GST (PTEN-GST) binds phosphoinositides broadly, with the highest specificity to PI(3,5)P2 (Fig. 4 A). GST-PTENA and GST-PTEND appeared more selective, with the former preferring PI(3,5)P2 and the latter both PI(3)P and PI(3,5)P2, although both moss PTEN homologues bound the other phosphoinositides to some extent (Fig. 4 A).
For2A PTEN domain and PTENA specifically bind PI(3,5)P2. (A) Representative lipid-overlay assay shows that the For2A PTEN-GST binds broadly to phosphoinositides, with highest specificity to PI(3,5)P2. PTENA binds PI(3,5)P2 strongly. PTEND also binds PI(3,5)P2, but weaker than PTENA. Additionally, PTEND also binds PI(3)P. GST alone does not interact with phosphoinositides. The numbers indicate amount in picomoles of spotted phosphoinositide. At least three independent experiments were performed for each overlay assay. (B) Immunoblot of For2A PTEN-GST, GST-PTENA, and GST-PTEND using an anti-GST antibody shows that For2A PTEN-GST and GST-PTENA preferentially bind to beads covalently linked to PI(3,5)P2 as compared with control beads. In contrast, GST-PTEND binds similarly to control and PI(3,5)P2 beads. Numbers are molecular weight standards in kD. (C) Immunoblot of For2A PTEN-GST and GST-PTENA using an anti-GST antibody demonstrates that binding to PI(3,5)P2 beads is specifically displaced in the presence of exogenous PI(3,5)P2, but not other phosphoinositides. Numbers are molecular weight standards in kD.
For2A PTEN domain and PTENA specifically bind PI(3,5)P2. (A) Representative lipid-overlay assay shows that the For2A PTEN-GST binds broadly to phosphoinositides, with highest specificity to PI(3,5)P2. PTENA binds PI(3,5)P2 strongly. PTEND also binds PI(3,5)P2, but weaker than PTENA. Additionally, PTEND also binds PI(3)P. GST alone does not interact with phosphoinositides. The numbers indicate amount in picomoles of spotted phosphoinositide. At least three independent experiments were performed for each overlay assay. (B) Immunoblot of For2A PTEN-GST, GST-PTENA, and GST-PTEND using an anti-GST antibody shows that For2A PTEN-GST and GST-PTENA preferentially bind to beads covalently linked to PI(3,5)P2 as compared with control beads. In contrast, GST-PTEND binds similarly to control and PI(3,5)P2 beads. Numbers are molecular weight standards in kD. (C) Immunoblot of For2A PTEN-GST and GST-PTENA using an anti-GST antibody demonstrates that binding to PI(3,5)P2 beads is specifically displaced in the presence of exogenous PI(3,5)P2, but not other phosphoinositides. Numbers are molecular weight standards in kD.
Because lipid overlay assays do not always reliably report a protein’s native phosphoinositide binding specificity, we used an additional approach to test phosphoinositide selectivity. We used beads covalently linked to PI(3,5)P2 to perform a pull-down experiment. We found that both the For2A PTEN domain and PTENA binding to PI(3,5)P2 beads is enhanced as compared with control beads (Fig. 4 B). In contrast, PTEND binds similarly to both control and PI(3,5)P2, suggesting that PTEND does not effectively bind PI(3,5)P2 in solution.
To test for PI(3,5)P2 specificity, we incubated the For2A PTEN-GST or GST-PTENA with PI(3,5)P2 beads in the presence of micellar dispersions of other phosphoinositides. We found that binding to the beads is completely eliminated for For2A PTEN-GST or greatly reduced for GST-PTENA in the presence of PI(3,5)P2, but not other phosphoinositides (Fig. 4 C). Together with the lipid overlay assays, these data strongly suggest that the For2A PTEN domain and moss PTENA interact specifically with PI(3,5)P2. Because the For2A PTEN domain and PTENA rescue formin-mediated polarized growth and specifically interact with PI(3,5)P2, whereas PTEND does not rescue and poorly interacts with PI(3,5)P2, these data argue that PI(3,5)P2 is the critical phosphoinositide.
PI(3,5)P2 binding is sufficient for formin-mediated polarized growth
To confirm that PI(3,5)P2 is critical for formin function in vivo, we replaced the PTEN domain in For2A with a variety of other polypeptides of known phosphoinositide binding specificity. When fused to the For2A FH1FH2 domains, several phosphoinositide binding polypeptides failed to complement the loss-of-function phenotype, including domains that bind PI(3)P (2XFYVE) (Vermeer et al., 2006), PI(3,4)P2 (TAPP1) (Dowler et al., 2000; Thomas et al., 2001; Marshall et al., 2002), and PI(4,5)P2 (PH) (Fig. 5; van Leeuwen et al., 2007). These constructs were epitope tagged and expression in moss cells was verified by isolating proteins from cells transformed with the various constructs. Immunoblots were probed with an antibody to the epitope tag (Fig. S2 B). Additionally, increasing the amount of transformed expression construct did not restore polarized growth; instead, it resulted in very few transformants, suggesting that overexpression of these proteins is toxic (unpublished data). In contrast, complementation is strong from proteins that bind PI(3,5)P2, including the yeast protein ATG18 (Dove et al., 2004; Michell et al., 2006) and the human lipid phosphatase MTM1 (Fig. 5; Schaletzky et al., 2003; Michell et al., 2006). The complementation results are similar to that obtained with For2A, even over a wide range of plasmid concentrations (Fig. S4). We also tested a mutant version of the MTM1 protein where the catalytic Cys was changed to Ser, thereby inactivating the phosphatase activity while maintaining phosphoinositide binding (Taylor et al., 2000). The mutant form of MTM1 (MTM1*) fused to FH1-FH2 also restored tip growth (Fig. 5). Taken together, our data imply that PI(3,5)P2 binding is the key activity conferred on moss class II formins by the PTEN domain.
PI(3,5)P2 binding is essential for For2A function in vivo. (A) Representative chlorophyll autofluorescence images of control RNAi (GUS-RNAi), For2 RNAi (For2AB-5′UTR), and For2 RNAi plants co-transformed with indicated constructs. Bar, 100 µm. (B) Quantification of area and circularity by chlorophyll autofluorescence shows that replacing the For2A PTEN domain with nonhomologous domains from various organisms exclusively rescues For2 RNAi when the domain binds PI(3,5)P2, indicating that PI(3,5)P2 binding is essential for class II formin function in polarized growth. Number of plants analyzed: 175, GUS-RNAi; 176, For2AB-5′UTR; 100, +For2A; 75, +2XFYVE-FH1FH2-3XFLAG; 50, +TAPP1-FH1FH2-3XFLAG; 25, +PH-FH1FH2-3XFLAG; 78, +ATG18-FH1FH2; 75, +MTM1-FH1FH2; 78, +MTM1*-FH1FH2. Error bars represent SEM and letters above the bars indicate statistical groups with α = 0.05 using an ANOVA analysis.
PI(3,5)P2 binding is essential for For2A function in vivo. (A) Representative chlorophyll autofluorescence images of control RNAi (GUS-RNAi), For2 RNAi (For2AB-5′UTR), and For2 RNAi plants co-transformed with indicated constructs. Bar, 100 µm. (B) Quantification of area and circularity by chlorophyll autofluorescence shows that replacing the For2A PTEN domain with nonhomologous domains from various organisms exclusively rescues For2 RNAi when the domain binds PI(3,5)P2, indicating that PI(3,5)P2 binding is essential for class II formin function in polarized growth. Number of plants analyzed: 175, GUS-RNAi; 176, For2AB-5′UTR; 100, +For2A; 75, +2XFYVE-FH1FH2-3XFLAG; 50, +TAPP1-FH1FH2-3XFLAG; 25, +PH-FH1FH2-3XFLAG; 78, +ATG18-FH1FH2; 75, +MTM1-FH1FH2; 78, +MTM1*-FH1FH2. Error bars represent SEM and letters above the bars indicate statistical groups with α = 0.05 using an ANOVA analysis.
PI(3,5)P2 binders localize to the cortex as dynamic spots
Because PTENA and PI(3,5)P2 binders functionally replaced the For2A PTEN domain, we expected that these domains should localize to the tip of the cell, similar to the For2A PTEN domain. In contrast, we predicted that PTEND, which does not rescue formin-mediated polarized growth, should not localize. To test this, we fused PTENA, PTEND, MTM1, and MTM1* to GFP and isolated stable lines expressing these fusion proteins. As expected, PTEND was not enriched at the apex of the cell (Fig. 6 A) or the phragmoplast (not depicted). However, neither were PTENA nor the functional PI(3,5)P2 binders (Fig. 6 A). Instead, these fusion proteins were diffusely cytosolic, similar to GFP alone. To ensure that the expressed fusion proteins were intact, we isolated proteins from plants carrying the various constructs and probed immunoblots with a GFP antibody (Fig. 6 B). Free GFP is undetectable in these protein extracts. Apical accumulation of PTEN-GFP is likely specific to the For2A PTEN domain and not a result of differing expression levels because the average GFP intensity in lines expressing For2A PTEN-GFP, MTM1-GFP, and MTM1*-GFP are similar (Fig. S5 A). Furthermore, a full-length functional MTM1*-FH1FH2-3XGFP (Fig. S5 B) is also not apically enriched when transiently expressed in moss cells (Fig. S5 C). This is in contrast to apical enrichment of transiently expressed For2A PTEN-3XFLAG-GFP (Fig. S5 C), suggesting that the FH1-FH2 domains do not contribute to apical accumulation.
PTENA, PTEND, and the functional PI(3,5)P2 binders fused to GFP are not enriched near the cell tip. (A) Localization of PTENA, PTEND, and the PI(3,5)P2 binders is cytosolic, like GFP alone. The PTEN domain of For2A (second panel) is tip enriched. (B) Immunoblot using an anti-GFP antibody shows that the full-length fusion proteins are expressed. Numbers are molecular weight standards in kD. (C) VAEM images of PTENA, PTEND, and PI(3,5)P2 binders fused to GFP demonstrate that only the fusion proteins that can functionally replace the For2A PTEN domain localize to spots at the cell cortex. In contrast, PTEND-GFP does not appear to have such a specific localization. See also Video 6. (D and E) Density of cortical For2A-GFP spots is reduced in cells overexpressing MTM1*-mCherry. (D) VAEM images of For2A-GFP in a wild-type cell or MTM1*-mCherry–overexpressing cell. (E) Quantification of cortical For2A-GFP density in wild-type or MTM1*-mCherry cells. Number of cells analyzed is: 4, wild type; 9, MTM1*-mCherry. Error bars represent standard SEM. Bars, 5 µm.
PTENA, PTEND, and the functional PI(3,5)P2 binders fused to GFP are not enriched near the cell tip. (A) Localization of PTENA, PTEND, and the PI(3,5)P2 binders is cytosolic, like GFP alone. The PTEN domain of For2A (second panel) is tip enriched. (B) Immunoblot using an anti-GFP antibody shows that the full-length fusion proteins are expressed. Numbers are molecular weight standards in kD. (C) VAEM images of PTENA, PTEND, and PI(3,5)P2 binders fused to GFP demonstrate that only the fusion proteins that can functionally replace the For2A PTEN domain localize to spots at the cell cortex. In contrast, PTEND-GFP does not appear to have such a specific localization. See also Video 6. (D and E) Density of cortical For2A-GFP spots is reduced in cells overexpressing MTM1*-mCherry. (D) VAEM images of For2A-GFP in a wild-type cell or MTM1*-mCherry–overexpressing cell. (E) Quantification of cortical For2A-GFP density in wild-type or MTM1*-mCherry cells. Number of cells analyzed is: 4, wild type; 9, MTM1*-mCherry. Error bars represent standard SEM. Bars, 5 µm.
Although class II formin function in tip growth requires PI(3,5)P2 binding activity, apical enrichment of formin appears dispensable. In protonemal cells, actin filaments are abundant near the plasma membrane and are highly dynamic (Vidali et al., 2009a, 2010). We reasoned that formin activity might be required at the cell cortex. To determine whether formin localizes to the cell cortex, we used variable angle epifluorescence microscopy (VAEM; Konopka and Bednarek, 2008; Staiger et al., 2009). In the For2A-GFP line, formin localizes to discrete spots near the cell membrane (Fig. 6 C), which are highly dynamic (Video 6). Similar to For2A-GFP, the For2A PTEN-GFP also localizes to discrete dynamic spots near the plasma membrane (Fig. 6 C and Video 6), suggesting that the For2A PTEN domain is sufficient to mediate this localization.
For2A-GFP localizes to dynamic cortical dots. The For2A PTEN domain mediates this localization. GFP fusions of PTENA, MTM1, and MTM1*, all proteins that when fused to the FH1-FH2 domains of For2 replace the function of the For2A PTEN domain, also form dynamic cortical dots. In contrast, GFP fusion of PTEND, which is unable to replace the function of the For2A PTEN domain, is diffuse at the cell cortex. These data are related to Fig. 6 C. Images were continuously acquired for at least 30 s with 50-msec exposure time (except for PTEND-GFP, which had 100-msec exposure time) for each frame using VAEM (model Ti-E, Nikon; equipped with TIRF). Video plays at 10 frames per second.
For2A-GFP localizes to dynamic cortical dots. The For2A PTEN domain mediates this localization. GFP fusions of PTENA, MTM1, and MTM1*, all proteins that when fused to the FH1-FH2 domains of For2 replace the function of the For2A PTEN domain, also form dynamic cortical dots. In contrast, GFP fusion of PTEND, which is unable to replace the function of the For2A PTEN domain, is diffuse at the cell cortex. These data are related to Fig. 6 C. Images were continuously acquired for at least 30 s with 50-msec exposure time (except for PTEND-GFP, which had 100-msec exposure time) for each frame using VAEM (model Ti-E, Nikon; equipped with TIRF). Video plays at 10 frames per second.
Because the For2A PTEN domain is required for the localization of For2A-GFP near the plasma membrane in discrete dynamic spots, we hypothesized that this localization is required for formin function in tip growth. If this is the case, then any of the PI(3,5)P2 binders capable of replacing the For2A PTEN domain should likewise form dynamic spots at the cortex. Supporting this hypothesis, PTENA-GFP, MTM1-GFP, and MTM1*-GFP all formed dynamic spots at the cell cortex (Fig. 6 C and Video 6). PTEND, which cannot functionally replace the For2A PTEN domain, localizes diffusely at the cell cortex (Fig. 6 C and Video 6). Interestingly, the spots formed by MTM1-GFP were shorter lived and less discrete then those formed by MTM1*-GFP (Video 6). Given that the former is an active lipid phosphatase whereas the latter is inactive, this suggests that phosphatase activity promotes turnover within the cortex.
To test whether the For2A and MTM1* cortical spots are similar structures, we generated a line that overexpresses MTM1*-mCherry in the background of For2A-GFP. We observed that the density of cortical For2A-GFP spots was reduced in the MTM1*-mCherry line (Fig. 6, D and E), suggesting that For2A-GFP and MTM1*-mCherry are competing for the same sites on the membrane.
Silencing FAB1 kinases impairs polarized growth and results in fewer cortical formin dots
If cortical PI(3,5)P2 sites are critical for formin-mediated polarized growth, then reduction of cellular PI(3,5)P2 should impair polarized growth. We reasoned that silencing the kinases that produce PI(3,5)P2 should lead to a reduction in PI(3,5)P2 levels. Toward this end, we identified three kinases (FAB1-A, FAB1-B, and FAB1-C) that have high sequence similarity with Saccharomyces cerevisiae FAB1. FAB1 is a 1-phosphatidylinositol-3-phosphate 5-kinase, which is responsible for generating PI(3,5)P2 in yeast (Gary et al., 1998). To silence the moss FAB1 homologues, we generated RNAi constructs that target all three FAB1 genes (FAB1-RNAi).
Using RNAi, it is possible to effectively reduce protein levels in moss within 72–96 h after transformation of the RNAi construct (Bezanilla et al., 2003, 2005). However, once the FAB1 kinases are depleted, subsequent dephosphorylation of PI(3,5)P2 must occur to reduce PI(3,5)P2 levels. To allow enough time for these events to occur, we inhibited 4-d-old transformed plants from performing polarized growth by exposing plants to latrunculin B for 3 d. Transformed plants were then transferred to normal growth medium and allowed to grow for an additional 3 d. Using this procedure, we found that plants transformed with the control construct (GUS-RNAi) were able to recover from the latrunculin B treatment and grow normally forming polarized extensions, leading to low levels of circularity (Fig. 7, A and B). As expected, plants transformed with the formin-RNAi construct were unable to recover from the drug treatment, remaining small and composed of spherical cells. Interestingly, plants expressing the FAB1-RNAi constructs were also unable to fully recover from the latrunculin B treatment. They remained small in comparison to controls, but had a higher degree of polarity than the formin-RNAi plants. This is due to the fact that some of the cells in the FAB1-RNAi plants were able to partially polarize and form a limited number of extensions (Fig. 7 A). Additionally, transformation with the FAB1-RNAi constructs consistently yielded significantly fewer transformed plants as compared with the GUS- and formin-RNAi constructs.
Silencing the FAB1 kinases impairs polarized growth and decreases the density of cortical formin dots. (A) Representative chlorophyll autofluorescence images of 10-d-old plants transformed with control (GUS-RNAi), For2-RNAi (For2AB-5′UTR), and FAB1-RNAi constructs. Bar, 100 µm. Plants were grown in the presence of latrunculin B from d 4–7. (B) Quantification of chlorophyll autofluorescence area and circularity shows that FAB1-RNAi has an intermediate phenotype. Number of plants analyzed is: 34, GUS-RNAi; 38, For2AB-5′UTR; 17, FAB1-RNAi. Error bars are SEM and letters above the bars indicate statistical groups with α = 0.05 using an ANOVA analysis. (C) Representative VAEM images of silenced and not silenced FAB1-RNAi plants, stably expressing For2A-GFP. Silenced plants show fewer cortical dots than not silenced plants. See also Video 7. (D) Quantification of the number of cortical dots in silenced and not silenced FAB1-RNAi plants. There is a statistically significant decrease in the number of cortical dots upon FAB1 silencing. Number of cells analyzed is: 5, not silenced; 4, silenced. Error bars are SEM.
Silencing the FAB1 kinases impairs polarized growth and decreases the density of cortical formin dots. (A) Representative chlorophyll autofluorescence images of 10-d-old plants transformed with control (GUS-RNAi), For2-RNAi (For2AB-5′UTR), and FAB1-RNAi constructs. Bar, 100 µm. Plants were grown in the presence of latrunculin B from d 4–7. (B) Quantification of chlorophyll autofluorescence area and circularity shows that FAB1-RNAi has an intermediate phenotype. Number of plants analyzed is: 34, GUS-RNAi; 38, For2AB-5′UTR; 17, FAB1-RNAi. Error bars are SEM and letters above the bars indicate statistical groups with α = 0.05 using an ANOVA analysis. (C) Representative VAEM images of silenced and not silenced FAB1-RNAi plants, stably expressing For2A-GFP. Silenced plants show fewer cortical dots than not silenced plants. See also Video 7. (D) Quantification of the number of cortical dots in silenced and not silenced FAB1-RNAi plants. There is a statistically significant decrease in the number of cortical dots upon FAB1 silencing. Number of cells analyzed is: 5, not silenced; 4, silenced. Error bars are SEM.
To test if reduction of FAB1 kinases led to a concomitant reduction in the cortical targeting of formin, we imaged cells transformed with the FAB1-RNAi constructs. In our RNAi assay, it is possible to identify transformed plants that are actively silencing because these plants have a reduction in a nuclear GFP:GUS reporter (Bezanilla et al., 2005). In any given transformation, a fraction of the transformed plants still express the GFP:GUS reporter and are therefore not silencing the target genes. Taking advantage of this, we were able to use VAEM to image plants that were transformed with the FAB1-RNAi constructs. On the same coverslip and with the same imaging conditions, we identified both silenced and nonsilenced control plants. We found that For2A-GFP was still targeted to the cell cortex in silenced plants (Fig. 7 C and Video 7). However, in comparison to nonsilenced control plants, targeting of For2A-GFP was reduced 2.5-fold (Fig. 7 D). The presence of reduced levels of cortical For2A-GFP may explain the limited extent of polarized outgrowths in the FAB1-RNAi plants. Taken together, these data show that silencing of FAB1 kinases impairs polarized growth and proper targeting of formin to the cell cortex, likely through production of PI(3,5)P2.
Density of For2A-GFP cortical dots is reduced in silenced FAB1-RNAi plants. These data are related to Fig. 7 C. Images were continuously acquired for at least 30 s with 66.7-msec exposure time for each frame using VAEM (model Ti-E, Nikon; equipped with TIRF). Video plays at 10 frames per second.
Density of For2A-GFP cortical dots is reduced in silenced FAB1-RNAi plants. These data are related to Fig. 7 C. Images were continuously acquired for at least 30 s with 66.7-msec exposure time for each frame using VAEM (model Ti-E, Nikon; equipped with TIRF). Video plays at 10 frames per second.
Linear motility of For2A cortical spots is dependent on actin
To determine whether cortical formin generates actin filaments, we investigated the behavior of the cortical formin spots. We analyzed their trajectories using VAEM in For2A-GFP cells expressing Lifeact-mCherry. 29% of cortical formin spots move rapidly in and out of the imaging field, remaining for less than one second on the cortex. The remaining trajectories of cortical For2A-GFP grouped into three main categories: linear, random, and stationary (movement no greater than 0.4 µm from the origin). Linear trajectories can be visualized by displaying the maximum projection of the 30 consecutive frames, which represents 3 s of real time (Fig. 8 A and Video 8). Of the spots that resided on the cortex for more than 1 s, we found that 13.8% displayed linear trajectories (Fig. 8 B; 832 spots from 5 cells). Furthermore, the cortical formin spots that moved in linear trajectories moved at 1.8 ± 0.56 µm/s (51 spots from 4 cells). To determine if these rates were similar to rates of actin polymerization in moss, we analyzed time-lapse series of cells containing Lifeact-mCherry and measured the rate of actin polymerization. We found that new actin filaments grew at 1.93 ± 0.55 µm/s (30 filaments from 7 cells), which is statistically indistinguishable from the rates of formin cortical spots that move in linear trajectories and similar to the rates measured previously for Lifeact-GFP (Augustine et al., 2011). This rate is also consistent with the rate of actin polymerization in Arabidopsis hypocotyl cells (Staiger et al., 2009; Smertenko et al., 2010).
Linear For2A-GFP trajectories are actin dependent. (left) A population of cortical For2A-GFP dots moves along linear trajectories (related to top images in Fig. 8 A). (right) Latrunculin B treatment abolishes the linear population of cortical For2AGFP dots. Data are related to top images in Fig. 8 B. Images were continuously acquired for at least 30 s with 100-msec exposure time for each frame using VAEM (Nikon Ti-E body equipped with TIRF). Video plays at 10 frames per second.
Linear For2A-GFP trajectories are actin dependent. (left) A population of cortical For2A-GFP dots moves along linear trajectories (related to top images in Fig. 8 A). (right) Latrunculin B treatment abolishes the linear population of cortical For2AGFP dots. Data are related to top images in Fig. 8 B. Images were continuously acquired for at least 30 s with 100-msec exposure time for each frame using VAEM (Nikon Ti-E body equipped with TIRF). Video plays at 10 frames per second.
Cortical For2A linear movement is actin dependent. (A) Two representative cells imaged with VAEM show that For2A-GFP localizes to cortical dots (left images) and Lifeact-mCherry labels the cortical F-actin array (center images). For2A linear trajectories are apparent in a maximum projection of 30 frames, which corresponds to three seconds real time (right panels). See also Video 8. (B) Quantification of the percentage of cortical dots found in the three categories of trajectories identified for formin cortical dots that resided on the cortex for more than 1 s. Stationary indicates that no movement greater than 0.4 µm from the origin was observed. (C) Two representative cells imaged with VAEM in the presence of 16 µM latrunculin B show that For2A-GFP localizes to cortical dots (left images) but F-actin has been disrupted, depicted by diffuse Lifeact-mCherry fluorescence (center images). Furthermore, For2A linear trajectories were no longer observed in maximum projections of 30 frames (right panels). See also Video 8. Bars, 5 µm.
Cortical For2A linear movement is actin dependent. (A) Two representative cells imaged with VAEM show that For2A-GFP localizes to cortical dots (left images) and Lifeact-mCherry labels the cortical F-actin array (center images). For2A linear trajectories are apparent in a maximum projection of 30 frames, which corresponds to three seconds real time (right panels). See also Video 8. (B) Quantification of the percentage of cortical dots found in the three categories of trajectories identified for formin cortical dots that resided on the cortex for more than 1 s. Stationary indicates that no movement greater than 0.4 µm from the origin was observed. (C) Two representative cells imaged with VAEM in the presence of 16 µM latrunculin B show that For2A-GFP localizes to cortical dots (left images) but F-actin has been disrupted, depicted by diffuse Lifeact-mCherry fluorescence (center images). Furthermore, For2A linear trajectories were no longer observed in maximum projections of 30 frames (right panels). See also Video 8. Bars, 5 µm.
To test whether cortical formin movement was dependent on actin, we treated cells with latrunculin B and then imaged cortical formin and actin with VAEM. We found that after 25 min in 16 µM latrunculin B, the actin at the cell cortex was completely depolymerized (Fig. 8 C). In the absence of actin, For2A-GFP still formed distinct spots at the cell cortex, indicating that the localization of formin to the cell cortex is independent of actin. This is consistent with the fact that PTEN-GFP and all the proteins that replace the function of PTEN in formin also localize to spots on the membrane. However, cortical formin dynamics was altered in the absence of actin. We did not observe any linear trajectories (Fig. 8 C and Video 8) and the population of random trajectories was greatly diminished, with a concomitant increase in the stable population of formin spots (Fig. 8 B; 245 spots from 5 cells).
Formin generates actin filaments at the cell cortex
We found that formin spots with linear trajectories depend on actin filaments and that their speed is consistent with the rate of actin polymerization in moss. Taken together, these data strongly suggest that the formin spots moving in linear trajectories are generating actin filaments. To test this, we used a variable angle epifluorescence microscope equipped with a dual-view camera, which enabled simultaneous acquisition of For2A-GFP and Lifeact-mCherry. As expected, we found that a large percentage of formin dots did not correlate with actin filaments (Fig. 9, A and B; Videos 9 and 10). However, we did observe that some cortical formin spots coincided with the formation of new filaments (Fig. 9 A, arrowheads). In these cases, the formin spot was found on the end of the new filament and is most clearly visualized in time-lapse (Video 9). We also observed a number of events where formin cortical dots moved along preexisting actin filaments (Fig. 9 B, arrowheads; Video 10). To ensure that these events were more than coincidental, we analyzed 61 linear formin trajectories from 5 cells. We found that 80% of the formin dots that moved in linear trajectories either resided on the end of a new actin filament or coincided with a preexisting filament (Fig. 9 C).
For2A-GFP spot is found at the end of a growing actin filament (related to Fig. 9 A). This longer time-lapse also shows that several formin spots move along preexisting filaments. The three panels are, from left to right, For2A-GFP, Lifeact-mCherry, and merge. In the merge, For2A-GFP and Lifeact-mCherry are false-colored magenta and cyan, respectively. Images were continuously acquired for at least 30 s with 80-msec exposure time for each frame using dual-view VAEM (Nikon Ti-E body equipped with TIRF). Video plays at 10 frames per second.
For2A-GFP spot is found at the end of a growing actin filament (related to Fig. 9 A). This longer time-lapse also shows that several formin spots move along preexisting filaments. The three panels are, from left to right, For2A-GFP, Lifeact-mCherry, and merge. In the merge, For2A-GFP and Lifeact-mCherry are false-colored magenta and cyan, respectively. Images were continuously acquired for at least 30 s with 80-msec exposure time for each frame using dual-view VAEM (Nikon Ti-E body equipped with TIRF). Video plays at 10 frames per second.
For2A-GFP dot glides along a preexisting actin filament (related to Fig. 9 B). The three panels are, from left to right, For2A-GFP, Lifeact-mCherry, and merge. In the merge, For2A-GFP and Lifeact-mCherry are false-colored magenta and cyan, respectively. Images were continuously acquired for at least 30 s with 80-msec exposure time for each frame using dual-view VAEM (Nikon Ti-E body equipped with TIRF). Video plays at 10 frames per second.
For2A-GFP dot glides along a preexisting actin filament (related to Fig. 9 B). The three panels are, from left to right, For2A-GFP, Lifeact-mCherry, and merge. In the merge, For2A-GFP and Lifeact-mCherry are false-colored magenta and cyan, respectively. Images were continuously acquired for at least 30 s with 80-msec exposure time for each frame using dual-view VAEM (Nikon Ti-E body equipped with TIRF). Video plays at 10 frames per second.
For2A-GFP spots generate actin filaments at the cell cortex. (A and B) Simultaneous acquisition of For2A-GFP and Lifeact-mCherry at the cell cortex using VAEM demonstrates that For2A-GFP resides on the ends of newly generated actin filaments (A) and along preexisting filaments (B). See also Videos 9 and 10. In the merge images, For2A-GFP and Lifeact-mCherry are magenta and cyan, respectively. Numbers represent time in milliseconds (ms). Bar, 2 µm. Arrowheads in each frame of the merge indicate the For2A-GFP and the arrow in the last frame indicates the position of the For2A-GFP spot at the beginning of the time-lapse. (C) Quantification of the number of cortical For2A-GFP spots that moved in a linear trajectory and were found at the end of a new actin filament (blue), along a preexisting filament (green), or did not correlate with an actin filament (pink). 61 linear For2A-GFP trajectories were analyzed from 5 cells. (D) Quantification of the speed of For2A-GFP linear movements on new or preexisting filaments. (45 For2A-GFP spots from five cells). (E) Quantification of the change in Lifeact-mCherry fluorescence in the wake of a linearly moving cortical For2A-GFP spot on new or preexisting filaments (37 spots from five cells).
For2A-GFP spots generate actin filaments at the cell cortex. (A and B) Simultaneous acquisition of For2A-GFP and Lifeact-mCherry at the cell cortex using VAEM demonstrates that For2A-GFP resides on the ends of newly generated actin filaments (A) and along preexisting filaments (B). See also Videos 9 and 10. In the merge images, For2A-GFP and Lifeact-mCherry are magenta and cyan, respectively. Numbers represent time in milliseconds (ms). Bar, 2 µm. Arrowheads in each frame of the merge indicate the For2A-GFP and the arrow in the last frame indicates the position of the For2A-GFP spot at the beginning of the time-lapse. (C) Quantification of the number of cortical For2A-GFP spots that moved in a linear trajectory and were found at the end of a new actin filament (blue), along a preexisting filament (green), or did not correlate with an actin filament (pink). 61 linear For2A-GFP trajectories were analyzed from 5 cells. (D) Quantification of the speed of For2A-GFP linear movements on new or preexisting filaments. (45 For2A-GFP spots from five cells). (E) Quantification of the change in Lifeact-mCherry fluorescence in the wake of a linearly moving cortical For2A-GFP spot on new or preexisting filaments (37 spots from five cells).
If these formin spots are generating actin filaments, then we expect that they should move at the rates of actin polymerization. We found that formin associated with the end of a new actin filament moved at 1.80 ± 0.45 µm/sec (27 spots from 5 cells), whereas formin spots associated with preexisting actin filaments moved at 1.98 ± 0.56 µm/sec (18 spots from 5 cells; Fig. 9 D). A student’s t test confirms that these data are not statistically different. Furthermore, if the formin spot generates a new actin filament, then we should observe an increase in Lifeact-mCherry fluorescence, corresponding to the new actin filament. Regardless of whether the formin spot was on the end of a filament or moving along a preexisting filament, we observed a similar change in fluorescence (Fig. 9 E). Taken together, these data strongly suggest that the formin spots that move along linear trajectories are actively polymerizing new actin filaments. The fact that some of these trajectories are along preexisting filaments further suggests that bundling of actin filaments at the cell cortex can occur by addition of new filaments via formin-mediated polymerization along existing actin tracks.
Discussion
Here we show that class II formins interact with PI(3,5)P2 and that this interaction is required for function in vivo. Heretofore understanding of the regulation of plant formins has been elusive because they lack the well-recognized regulatory domains, such as rho GTPase-binding domains found on the N termini of fungal and many animal formins (Goode and Eck, 2007; Grunt et al., 2008; Blanchoin and Staiger, 2010). Interestingly, plant class II formins appear to have acquired a PTEN-like domain (Cvrcková et al., 2004; Grunt et al., 2008) at their N terminus, which interacts with phosphoinositides to promote polarized growth. We find that the PTEN domain of class II formins interacts with PI(3,5)P2 and that this interaction is essential for formin’s role in polarized growth. Furthermore, the For2A PTEN domain is also necessary for class II formin localization to the cell apex, the phragmoplast, and the cell cortex. By analyzing the localization of PI(3,5)P2 binders that functionally replace the formin class II domain, we were surprised to discover that apical localization is not linked to formin’s function in tip growth. Instead, cortical localization appears critical. Our study provides a functional link between membranes containing PI(3,5)P2 and the actin cytoskeleton.
Among PIP2s, PI(3,5)P2 has commanded less attention in comparison to PI(4,5)P2. The latter is enriched on the plasma membrane and is implicated in a large array of signaling pathways in animals, fungi, and plants (Yin and Janmey, 2003; Perera et al., 2006; Sun et al., 2007; Munnik and Vermeer, 2010; Saavedra et al., 2011). In particular, PI(4,5)P2 is known to interact with the actin cytoskeleton through profilin, ADF/cofilin, and activation of the Arp2/3 complex through WAVE and WASP in mammalian systems (Saarikangas et al., 2010; Shewan et al., 2011). Also, PI(4,5)P2 and the PTEN phosphatase are involved in establishing cell polarity (Saarikangas et al., 2010; Shewan et al., 2011). In contrast, much less is known about PI(3,5)P2. In yeast and animals, this phosphoinositide is implicated in endomembrane trafficking (Michell et al., 2006). Although, perhaps due to its low abundance, PI(3,5)P2 has not been directly localized within the endomembrane system. However, in yeast and plants PI(3,5)P2 is generated by the FAB1 kinase, a kinase that is localized to the endomembrane system (Cooke et al., 1998; Gary et al., 1998; Hirano et al., 2011), and when knocked out (or down) results in enlarged vacuoles (Gary et al., 1998; Whitley et al., 2009; Hirano et al., 2011). These results suggest that in plants, as in animals and yeast, PI(3,5)P2 plays a role in membrane trafficking.
To our knowledge, PI(3,5)P2 has not been directly and functionally linked to the actin cytoskeleton in any system. Evidence for a link to actin is indirect. Yeast deficient in the epsin N-terminal homology domain containing protein Ent3p exhibit a defect in actin localization (Eugster et al., 2004). Ent3p is known to bind PI(3,5)P2 and is involved in protein sorting to multivesicular bodies (Friant et al., 2003). Here we show that the For2A PTEN domain binds to PI(3,5)P2 recruiting formin to the cell cortex where it generates actin filaments essential for polarized growth, thereby directly linking PI(3,5)P2 to actin nucleation and filament elongation.
We also find that moss PTENA, in contrast to human PTEN, binds to PI(3,5)P2, not PI(4,5)P2. The binding specificities of vascular plant PTENs are unknown. One of the PTEN homologues in Arabidopsis thaliana catalyzes the conversion of PI(3,4,5)P3 to PI(4,5)P2 in vitro (Gupta et al., 2002), but its ability to bind PI(3,5)P2 was not assessed. Based on our findings, we speculate that PTENs in plants have a different phosphoinositide binding preference, drawing attention to the importance of PI(3,5)P2 in plant signaling. In support of this, a recent study demonstrated that the pleckstrin homology (PH) domain from a rice dynamin-like protein binds PI(3,5)P2, not PI(4,5)P2 (Xiong et al., 2010).
Surprisingly, proteins unrelated in structure to PTEN are functional as chimeras with class II formins, provided that they interact with PI(3,5)P2. This suggests that formin function in tip growth does not rely on the specific structure of the PTEN domain. Rather, the For2A PTEN domain functions to localize formin to sites on the membrane rich in PI(3,5)P2. The PI(3,5)P2 binders that replace the formin PTEN domain do function with respect to polarized growth but are localized diffusely throughout the cytosol. Thus, the apical and phragmoplast enrichment observed for For2A and mediated by the PTEN domain are evidently not required for formin-mediated polarized growth. Thus, we hypothesize that the apical accumulation of For2A represents a pool of inactive formin with respect to polarized growth. These data provide strong evidence unlinking formin apical enrichment from function in polarized growth.
We acquired further evidence for this by imaging live cells containing a functional fluorescent fusion to For2A and a marker for actin, Lifeact-mCherry. We observed variation in the amount of For2A, as well as the position of the enrichment. However, at the cell apex we rarely observed a burst in actin polymerization after accumulation of formin, supporting our hypothesis that the apical enrichment of For2A is a pool of largely inactive formin with respect to actin polymerization.
We propose that active formin is required at the cell cortex. This is supported by the localization of the PI(3,5)P2 binders that functionally replace the formin PTEN domain. Although not accumulating prominently in the cell apex, the functional PI(3,5)P2 binders localize to dynamic spots at the cortex, a localization that is shared by For2A. Also, overexpression of a PI(3,5)P2 binder reduces the density of cortical For2A spots, suggesting that For2A and the PI(3,5)P2 binders are competing for the same sites on the membrane. Furthermore, silencing of the moss FAB1 homologues impairs polarized growth and reduces cortical targeting of formin.
Formin cortical localization is mediated by the PTEN domain and is actin independent. However, detailed analyses of the formin cortical spots demonstrated that a small population of cortical formin moves in linear trajectories and this movement is actin dependent. Simultaneous imaging of formin and actin revealed that the majority of cortical formin spots that move in linear trajectories are either associated with the ends of new actin filaments or along preexisting filaments. Importantly these cortical formin spots move at the rate of actin polymerization in moss cells. Because formin’s actin polymerization activity is required for polarized growth, our data suggest that formin’s essential activity at the cell cortex is to rapidly generate new actin filaments. Furthermore, our data establish a mechanism for bundling in the plant cell cortex. We observed formin-mediated actin polymerization occurring along existing filaments, producing parallel filaments that may be bundled by other factors.
Here, we show that PI(3,5)P2 is essential for formin-mediated polarized growth. The formin PTEN domain, which specifically binds PI(3,5)P2, serves to localize formin to one of four regions in the cell: the cell cortex, an apical cytoplasmic enrichment, stochastic subapical accumulations, and the phragmoplast. However, of these four populations, we show that localization to discrete dynamic cortical spots correlates with formin-mediated polarized growth. Interestingly, only a fraction of formin generates new actin filaments at the cell cortex, suggesting that formin activity is tightly regulated. Thus, we propose that PI(3,5)P2 functions to localize formin to specific cortical sites. Approximately 14% of the time the PI(3,5)P2-containing site is “primed”, perhaps containing a formin activator, and this site stimulates formin’s actin polymerization activity. Interestingly, 30% of the linearly moving For2A-GFP dots were observed to move along existing actin filaments, while polymerizing a new actin filament. These events suggest that formin facilitates bundling of actin filaments by generating new filaments in parallel and in close proximity to existing ones and provide evidence for formin-mediated bundling of actin filaments in vivo. Taken together, our data link PI(3,5)P2 membrane domains to generation and remodeling of the cortical actin array.
Materials and methods
Imaging cell division by confocal laser-scanning microscope (CLSM)
Moss plants were inoculated on top of a thin layer of PpNO3 growth medium (1.03 mM MgSO4, 1.86 mM KH2PO4, 3.3 mM Ca(NO3)2, 45 µM FeSO4, 9.93 µM H3BO3, 220 nM CuSO4, 1.966 µM MnCl2, 231 nM CoCl2, 191 nM ZnSO4, 169 nM KI, and 103 nM Na2MoO4) in a growth chamber with a cover slide glued to the bottom and grown for 7–10 d (Hiwatashi et al., 2008). After 7–10 d, many protonemal cells reach the cover slide and therefore can be imaged with a confocal laser-scanning microscope (CLSM). The chamber was mounted on a confocal microscope (model C1; Nikon) with a 1.45 NA 60x oil immersion objective (Nikon) equipped with photomultiplier tube (PMT) detectors. A 488-nm argon laser with 1% power was used. To image cell division, long protonemal cells were chosen and imaged for several hours at room temperature until cell division was completed. Image acquisition and 3D reconstruction of acquired images were conducted using EZ-C1 3.80 software (Nikon). Subsequent imaging processing, which included smoothing and contrast enhancing, was performed with ImageJ (National Institutes of Health).
FM4-64 treatment and imaging
Moss cells growing in the imaging chamber were incubated with 20 µM FM4-64 (EMD Millipore) diluted in Hoagland’s medium (4 mM KNO3, 2 mM KH2PO4, 1 mM Ca(NO3)2, 89 µM Fe citrate, 300 µM MgSO4, 9.93 µM H3BO3, 220 nM CuSO4, 1.966 µM MnCl2, 231 nM CoCl2, 191 nM ZnSO4, 169 nM KI, 103 nM Na2MoO4, and 1% sucrose) for 5 min and then washed three times with Hoagland’s medium to remove excess FM4-64. The plant was then imaged and processed as described above. The emission was collected with both 515/30-nm and 605/75-nm filter sets for GFP and FM4-64 signals, respectively.
Spinning-disc confocal microscopy
1-wk-old protonemal cells in imaging chambers were mounted on an inverted microscope (model Ti-E; Nikon) equipped with a spinning disk head (model CSU-X1; Yokogawa Corporation of America) and a 512 × 512 electron multiplying CCD camera (iXON; Andor Technology). Images were collected with a 1.4 NA 60x oil immersion objective (Nikon) at room temperature. 30% laser power was used for both 488 and 561 lasers. The electron gain was 300 and exposure time was 200 msec. Image acquisition process was controlled by MetaMorph software (Molecular Devices) and images were further processed with ImageJ as described above.
Plasmid construction
Lifeact-mCherry was constructed by assembling Lifeact-L1L5r (Vidali et al., 2009a) and mCherry-L5L2 into pTHUbi-gate (Vidali et al., 2007). To generate mCherry-L5L2, the mCherry coding sequence was amplified using mEGFP primers (Table S2) with attB5 and attB2 sites and cloned into pDONR221-P5-P2 (Invitrogen) using a BP reaction.
To clone PTENA and PTEND, total RNA was first isolated from 1-wk-old moss protonemal tissue, using the RNeasy plant mini kit (QIAGEN), according to the manufacturer’s recommendations. Total cDNA was generated using oligo(dT) and SuperScript II Reverse transcription (Invitrogen) following the manufacturer’s protocol. Full-length cDNAs of PTENA and PTEND were amplified from total cDNA with primers designed to amplify the open reading frame (Table S2).
For PTEN-GST, GST was amplified from pGEX2tk with primers (Table S2) incorporating EcoRI and XhoI sites and subsequently cloned into pET21, generating pET21-GST. PTEN was amplified from PTEN-pENT with primers (Table S2) incorporating NdeI and EcoRI sites and cloned into pET21-GST. For GST-PTENA and GST-PTEND fusion proteins, cDNAs were first cloned into pENTR-D-Topo (Invitrogen). These entry clones were then transferred to pDEST15 (Invitrogen) using an LR reaction. To generate PTENA-GFP and PTEND-GFP, the coding sequences were amplified from PTENA-pENT and PTEND-pENT, respectively, with primers incorporating specific attB sites (Table S1) and transferred into pDONR221-P1-P5r using a BP clonase reaction. PTENA-L1L5r was assembled with mEGFP-L5L2 (Vidali et al., 2009a) into pTHUbi-gate (Vidali et al., 2007) using LR clonase to generate PTENA-GFP and PTEND-GFP.
All complementation constructs were constructed using Multisite Gateway 3-fragment recombination (Invitrogen). All entry clones were verified by sequencing. Essentially, HsPTEN, MTM1, MTM1*, PTENA, PTEND, PH, TAPP1, 2XFYVE, and ScAtg18 were amplified using primers containing attB1 and attB4 sites (Table S1). MTM1 and MTM1* were amplified from pCDNA3.1(+)-NF-hMTM1 and pCDNA3.1(+)-NF-hMTM1 (C375S) plasmids (Taylor et al., 2000; Robinson and Dixon, 2005). PTENA, and PTEND were amplified from PTENA-pENT and PTEND-pENT, respectively. Human TAPP1 was amplified from pCMV-Taq-nMyc-hTAPP1 (Marshall et al., 2002). Atg18 was amplified from S. cerevisiae genomic DNA. The 2XFYVE and PH sequences were amplified from genomic DNA isolated from A. thaliana lines stably expressing YFP-2XFYVE and YFP-PH (PLCδ1) (Vermeer et al., 2006; van Leeuwen et al., 2007). Human PTEN (HsPTEN) was amplified by RT-PCR from HeLa cells. Total RNA and cDNA of HeLa cells were generated similarly as mentioned above. All PCR products were transferred into pDONR221-P1-P4 using a BP clonase reaction. Resulting entry clones were assembled with 2AFH1-L4L3 and 2AFH2-L3L2 (Vidali et al., 2009b) or 2AFH2-3XFLAG-L3L2 in a 3-fragment recombination reaction into pTHUbi-gate using LR clonase. To generate 2AFH2-3XFLAG-L3L2, three tandem copies of the FLAG epitope were incorporated into 2AFH2-L3L2 using megaprimer PCR (Barik, 1997) with primers listed in Table S2.
To generate MTM1*-FH1FH2-3XGFP, we first removed a BssSI site from the 5′ region of MTM1*-L1R5 using site-directed mutagenesis with primers listed in Table S1. Then we incorporated a BssSI site at the 3′ end of MTM1* using PCR with primers listed in Table S1. Taking advantage of a BssSI site in the vector sequence, the new MTM1* clone was digested with BssSI to remove MTM1* from the L1R5 backbone vector. The full-length coding sequence of For2A in pENTR-D-TOPO (For2A-pENT) (Vidali et al., 2009b) was digested with BssSI removing sequences of the vector and the entire PTEN domain. The BssSI MTM1* fragment was then cloned into For2A-pENT digested with BssSI, generating MTM1*-FH1FH2-pENT. To move this clone into the 2-fragment Multisite Gateway entry clones, the L2 site from pENT had to be replaced with an R5 site. This was done by first generating an FH2-L1R5 clone by PCR amplification and BP clonase reaction. A SmaI and EcoRV fragment from MTM1*-FH1FH2-pENT was replaced with the equivalent fragment from the FH2-L1R5 construct, resulting in replacement of the L2 site with an R5 site and generation of MTM1*-FH1FH2-L1R5. All clones were verified by sequencing. MTM1*-FH1FH2-L1R5 was then recombined with 3XmEGFP-L5L2 using LR clonase to generate MTM1*-FH1FH2-3XGFP.
To generate the FAB1-RNAi constructs, using primers listed in Table S2, we amplified a 400-bp fragment from the first exon of FAB1-A (Pp1S65_215V6) from moss genomic DNA. This fragment has high sequence identity with FAB1-B (Pp1S36_196V6). FAB1-C (Pp1S26_208V6) differs significantly in sequence. FAB1-C is encoded by a gene with no introns, so using primers listed in Table S1 we amplified 400 bp from the 5′ end of the coding sequence from genomic DNA. Amplified fragments were cloned into pENT-D-TOPO and sequenced. Subsequently, the fragments were cloned into pUGGi (Bezanilla et al., 2005) with an LR reaction to generate the RNAi constructs, FAB1-AB-RNAi and FAB1-C-RNAi.
Tissue culture, protoplast transformation, and complementation analysis
All tissue culture and transformations were performed as described previously (Bezanilla et al., 2003, 2005; Vidali et al., 2007) with minor modifications described as follows. Protoplasts were transformed at a concentration of 2 × 106 protoplasts/ml. For isolation of stable transformants, protoplasts were regenerated with top agar (1.03 mM MgSO4, 1.86 mM KH2PO4, 3.3 mM Ca(NO3)2, 2.7 mM (NH4)2-tartrate, 45 µM FeSO4, 9.93 µM H3BO3, 220 nM CuSO4, 1.966 µM MnCl2, 231 nM CoCl2, 191 nM ZnSO4, 169 nM KI, 103 nM Na2MoO4, 6% mannitol, and 10 CaCl2). For transient analysis, protoplasts were plated in 0.5 ml of PpNH4 culture medium (1.03 mM MgSO4, 1.86 mM KH2PO4, 3.3 mM Ca(NO3)2, 2.7 mM (NH4)2-tartrate, 45 µM FeSO4, 9.93 µM H3BO3, 220 nM CuSO4, 1.966 µM MnCl2, 231 nM CoCl2, 191 nM ZnSO4, 169 nM KI, and 103 nM Na2MoO4) supplemented with 8.5% mannitol and 10 mM CaCl2. Transformed plants were selected 4 d after transformation on PpNH4 medium with 0.7% agar containing hygromycin (15 µg/ml) and imaged 7 d after transformation.
For FAB1-RNAi, transformations were performed similarly. To silence all the three FAB1 genes, 30 µg of FAB1-AB-RNAi plus 30 µg of FAB1-C-RNAi were used simultaneously. 4 d after transformation the plants were transferred to PRMB medium containing hygromycin (15 µg/ml) and 13 µM latrunculin B. 7 d after transformation plants were moved to PpNH4 medium containing hygromycin (15 µg/ml) and plants were imaged 10 d after transformation.
Plants were imaged at room temperature with a 1x lens at 63x zoom on a fluorescence stereomicroscope (model MZ16FA; Leica) equipped with a color camera (model DF300FX; Leica) using the GFP2 filter set (Leica). Plant area and morphometric parameters were measured as described previously (Vidali et al., 2007). In brief, a 24-bit RGB image of a 1-wk-old plant was manually cropped, and the red channel corresponding to the chlorophyll autofluorescence was separated. Fluorescence was thresholded using maximum entropy (ImageJ). Total plant area and circularity were determined from the thresholded images. Circularity is the plant area divided by the square of the perimeter. Statistical analyses were performed as described previously (Vidali et al., 2007) except that analysis of variance for multiple comparisons was done on KaleidaGraph (Synergy Software). Area was log transformed to achieve normal distribution. We assessed the possibility of differences between experiments and found none. Pairwise comparisons are corrected for multiple tests using Kramer’s procedure so that the overall α level is 0.05 (Kramer, 1956).
Protein extraction and immunoblotting
To verify expression from constructs that do not rescue the formin-RNAi phenotype, we transformed epitope-tagged constructs into moss protoplasts. After transformation, protoplasts were incubated in 6 ml of plating medium (PpNH4 + 8.5% mannitol) for 3 d. Protoplasts were then harvested and resuspended in 400 µl grinding buffer (100 mM Na3PO4, 10 mM DTT, 20 µg/ml leupeptin, 20% glycerol, and 250 mM PMSF), supplemented with Complete mini EDTA-free protease inhibitor cocktail tablets (1 tablet per 5 ml; Roche). Lysis was done by three freeze–thaw cycles in liquid nitrogen. Cell debris was spun down and the supernatant was precipitated with methanol/chloroform. For immunoblotting, both the pellet and precipitated supernatant were solubilized in sample buffer (175 mM Tris base, 2.5% SDS, 80 mM DTT, and 7.5% glycerol). 80% of the sample was loaded, separated on a 6% SDS-PAGE gel, and transferred to a nitrocellulose membrane.
For protein extraction from stable moss lines, moss protonemal tissue was dried on a paper towel and weighed. For 10 mg of tissue, 100 µl of sample buffer was added and the tissue was ground with a plastic pestle in a microfuge tube. After boiling the sample for 5 min, the tissue debris was removed by centrifugation. The supernatant was collected and the protein concentration was measured by dot-blotting on a nitrocellulose membrane using BSA as the standard. For immunoblotting, 30 µg of protein extract was separated by SDS-PAGE and transferred to a nitrocellulose membrane.
For 3xFLAG immunoblots, membranes were probed with a monoclonal anti-FLAG antibody (Sigma-Aldrich). For GFP immunoblots, the membranes were probed with polyclonal rabbit anti-GFP antibody. The anti-GFP polyclonal rabbit antibody was raised against His-tagged GFP and affinity purified. For immunoblots, the membrane was blocked with 5% nonfat dry milk dissolved in TBS-T (25 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.1% Tween 20) at 25°C for 1 h. After five washes with TBS-T for 5 min, the membrane was incubated with primary antibody (1:5,000 dilution with 1% BSA in TBS-T) at 25°C for 2 h. After five washes with TBS-T for 5 min, the membrane was incubated with horseradish peroxidase–conjugated secondary antibody (1:100,000 dilution for 3xFLAG immunoblots; 1:5,000 dilution for GFP immunoblots) at 25°C for 1 h. The unbound secondary antibody was washed five times for 5 min with TBS-T. For 3xFLAG immunoblots, SuperSignal West Femto (Thermo Fisher Scientific) was used according to the manufacturer’s recommendations. Chemiluminescence emission was detected with a gel dock system equipped for chemiluminescence detection (Bio-Rad Laboratories).
Protein expression and purification
All proteins were expressed in Rosetta (DE3) pLysS strain (EMD Millipore) and induced with the Inducer (KD Medical), an IPTG alternative. Bacteria were grown at 37°C until the optical density of the culture at 600 nm was between 0.6–0.8. The culture was then chilled on ice before adding the Inducer to 2.5 mg/ml. Protein induction was conducted at 20°C overnight.
For GST fusion protein purification, cells were resuspended with column buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, and 1 mM DTT) plus 1 mM final concentration PMSF and 100 µg/ml lysozyme and lysed by sonication. After clearing the cell debris by spinning at 13,000 rpm in a rotor (model SS34; Thermo Fisher Scientific) for 15 min and filtering through a 0.2-µm filter, the supernatant was loaded onto a preequilibrated GST column (EMD Millipore) with column buffer. The column was washed with 15 column volumes of column buffer, followed by 15 column volumes of 0.1% Triton X-100 in column buffer. Excess Triton X-100 was removed by washing with 15 column volumes of column buffer. Protein was eluted with column buffer supplemented with 100 mM reduced glutathione, pH 7.5 (Sigma-Aldrich).
Protein lipid overlay assay
All dipalmitoyl phosphoinositides were purchased from Echelon Biosciences. The assays were conducted following the protocol developed by Dowler et al. (2002). Essentially, lyophilized lipids were reconstituted as 0.5 mM stock with 1:1 methanol and chloroform and stored at −80°C. For the assay, phosphoinositide stocks were diluted with a solution of 2:1:0.8 methanol/chloroform/water. 1 µl of diluted lipids was spotted on Hybond-C extra membrane (GE Healthcare). After the membrane was air-dried at room temperature for 1 h, it was blocked with 2 mg/ml BSA (Thermo Fisher Scientific) dissolved in TBS-T (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.1% Tween 20) for 1 h at room temperature with gentle rocking. The membrane was washed three times with TBS-T, followed by incubation with 10 nM purified protein diluted in the blocking solution (2 mg/ml BSA in TBS-T) at 4°C for 2 h. Then the membrane was washed with TBS-T 10 times for 5 min, followed by standard immunoblotting procedures using anti-GST (Sigma-Aldrich) antibodies.
PI(3,5)P2 bead pull-down and competition assays
PTEN-GST, GST-PTENA, or GST-PTEND (0.06 µM) was incubated with 25 µl of PI(3,5)P2-coupled or control beads (Echelon Biosciences) in pull-down buffer (10 mM Hepes-NaOH, pH 7.5, 1 mM DTT, 150 mM NaCl, and 0.1% Triton X-100) at room temperature for 1 h. Beads were spun down at 1,000 g for 30 s and washed four times with pull-down buffer. Protein was eluted by boiling the beads with SDS-PAGE sample buffer. 1.25% of the input material and 48.8% of the elution was loaded and separated on a 10% SDS-PAGE gel. Immunoblotting was performed with an anti-GST antibody. For the competition assays, incubation of the protein with PI(3,5)P2-coupled beads was performed in the presence of 5-µM micelle dispersions of different phosphoinositides and analyzed similarly.
Variable-angle epifluorescence microscopy (VAEM)
Moss protonemal tissue was grown in PpNH4 medium for 5–6 d. For imaging, protonemal tissue was placed on a 1% agar pad in Hoagland’s medium, covered with a glass coverslip, sealed with VALAP (1:1:1 parts of Vaseline, lanoline, and paraffin), and immediately observed at room temperature. The slide was mounted on an inverted microscope (model Ti-E; Nikon) equipped with a mirror-based T-FL-TIRF illuminator (Nikon) and imaged with a 1.49 NA 100x oil immersion TIRF objective (Nikon). The 1.5x optivar was used for all images to increase magnification. The laser illumination angle was adjusted individually for each sample to achieve the maximum signal-to-noise ratio. GFP was excited with a 488 diode laser and GFP emission from the specimen was captured with a 1024 × 1024 electron-multiplying CCD camera (iXON3; Andor Technology). Dual-view VAEM was acquired on a similar system equipped with a 1.45 NA 60x oil immersion TIRF objective (Nikon) and equipped with a dual-view 512 × 512 EM CCD camera (Andor Technology). GFP and mCherry were simultaneously excited with 488 and 561 argon ion and diode lasers, respectively. The electronic gain was 300 and exposure time was at least 50 msec for both imaging systems. Image acquisition process was controlled by NIS-Elements AR 3.2 software (Nikon) and images were further processed with ImageJ, including background subtraction and enhanced contrast.
Tracking of cortical For2A-GFP dots was performed with MetaMorph Offline 7.0. The original VAEM images were first processed with the software’s “2D Deconvolution” function and then analyzed with the “Track Objects” application. The speed of linear dots was measured manually using ImageJ. Quantification of the density of cortical For2A-GFP was performed in ImageJ, using the “Analyze Particles” function. Images were first corrected for uneven illumination using the background correction plug-in, filtered with an FFT bandpass filter, and then binarized using maximum entropy thresholding. 50–60 slices from a time-lapse acquisition were binarized with this method and particles between 8–80 square pixels were counted. The automated counting method was validated by manually counting the dots in at least five frames from a time-lapse acquisition. The average number of dots was divided by the total area of the VAEM imaging field. Rate of actin polymerization was measured manually using ImageJ. The length of a growing filament was measured. The rate was calculated by dividing the total length the filament grew by the time it took to grow.
Online supplemental material
Fig. S1 shows the kinetics of FM4-64 uptake in the For2A-GFP line. Fig. S2 shows expression of noncomplementing constructs from Figs. 3 and 5. Fig. S3 shows dose-dependent complementation of the formin RNAi phenotype using constructs that replace the PTEN domain with PTEN homologues. Fig. S4 shows dose-dependent complementation of the formin RNAi phenotype using constructs that replace the PTEN domain with unrelated PI(3,5)P2 binding proteins. Fig. S5 shows GFP fluorescence intensity in stable lines expressing various GFP fusion constructs, complementation by MTM1*-FH1-FH2-3XGFP, and localization of MTM1*-FH1-FH2-3XGFP. Video 1 is a time-lapse of For2A-GFP localization at the cell tip and phragmoplast during cytokinesis. Video 2 is a time-lapse of PTEN-GFP localization at the cell tip and phragmoplast during cytokinesis. Video 3 is a time-lapse of FM4-64 labeled For2A-GFP cell. Video 4 is a time-lapse of a growing cell expressing Lifeact-mCherry and For2A-GFP. Video 5 is a time-lapse of a cell expressing Lifeact-mCherry and For2A-GFP showing a cytoplasmic burst of For2A-GFP generating actin filaments. Video 6 is a montage of several videos showing cortical or diffuse localization of a variety of GFP fusion proteins. Video 7 contains two time-lapse series showing cortical For2A-GFP in FAB1-RNAi cells. Video 8 contains two time-lapse series. The left is cortical For2A-GFP without any treatment and the right is cortical For2A-GFP treated with latrunculin B. Videos 9 and 10 show For2A-GFP and Lifeact-mCherry imaged simultaneously with VAEM at the cell cortex. Table S1 shows the sequence comparison between formin PTEN domains and PTEN homologues. Table S2 shows the primers used in this study.
Acknowledgments
We thank Tobias Baskin for extensive editing of the manuscript. We thank Robert Augustine, Caleb Rounds, Graham Burkart, and Peter Hepler for critical reading and helpful suggestions. We thank Jennifer Ross for assistance with simultaneous VAEM imaging of formin and actin. We thank Lawrence Winship for technical advice regarding ImageJ. We also thank members of the Bezanilla laboratory for technical support. We would like to thank Fred Robinson, Jack Dixon, Joop Vermeer, Elison Blancaflor, Teun Munnik, and Aaron Marshall for plasmids encoding phosphoinositide binders; Pat Wadsworth for human HeLa cells; and Wei-Lih Lee for budding yeast genomic DNA.
This work was supported by the National Science Foundation MCB-0747231 (to M. Bezanilla) and the David and Lucille Packard Foundation (to M. Bezanilla). The spinning disc confocal and VAEM microscopes are university facilities funded by an NSF Major Research Instrumentation Development award (0923318).
References
Author notes
P.A.C. van Gisbergen and M. Li contributed equally to this paper.