In many organisms, the small guanosine triphosphatase RhoA controls assembly and contraction of the actomyosin ring during cytokinesis by activating different effectors. Although the role of some RhoA effectors like formins and Rho kinase is reasonably understood, the functions of another putative effector, Citron kinase (CIT-K), are still debated. In this paper, we show that, contrary to previous models, the Drosophila melanogaster CIT-K orthologue Sticky (Sti) does not require interaction with RhoA to localize to the cleavage site. Instead, RhoA fails to form a compact ring in late cytokinesis after Sti depletion, and this function requires Sti kinase activity. Moreover, we found that the Sti Citron-Nik1 homology domain interacts with RhoA regardless of its status, indicating that Sti is not a canonical RhoA effector. Finally, Sti depletion caused an increase of phosphorylated myosin regulatory light chain at the cleavage site in late cytokinesis. We propose that Sti/CIT-K maintains correct RhoA localization at the cleavage site, which is necessary for proper RhoA activity and contractile ring dynamics.
Introduction
The separation of daughter cells at the end of cell division ensures accurate segregation of genomic and cytoplasmic materials. In animal cells, this process, known as cytokinesis, requires the formation and ingression of a cleavage furrow (CF) that bisects the mother cell after chromosomes reach the spindle poles. In many organisms, the force that drives furrow ingression is the assembly and contraction of actomyosin filaments that often form a contractile ring (CR). The CR is a very dynamic structure in which actomyosin filaments are continuously assembled and disassembled. Compelling evidence indicates that the small GTPase RhoA controls CR assembly and dynamics during cytokinesis (Piekny et al., 2005). This GTPase cycles between an inactive GDP-bound form and an active GTP-bound form, and this RhoA flux seems important for CR dynamics (Miller and Bement, 2008). RhoA can promote profilin-mediated actin polymerization by activating members of the family of formin-related proteins and, in parallel, can control myosin activity through the phosphorylation of the myosin regulatory light chain (MRLC) by Rho kinase (ROK). Another putative RhoA effector, Citron kinase (CIT-K), has also been implicated in cytokinesis in both flies and humans (Madaule et al., 1998; D’Avino et al., 2004; Echard et al., 2004; Naim et al., 2004; Shandala et al., 2004). Initial studies in human cells indicated that CIT-K could also regulate myosin contractility by phosphorylating MRLC (Madaule et al., 2000; Yamashiro et al., 2003). However, this model has been challenged by the finding that the Drosophila melanogaster CIT-K orthologue Sticky (Sti) is not required for furrow ingression and MRLC phosphorylation but rather for the final separation of the daughter cells and proper organization of the CR structure in late cytokinesis (D’Avino et al., 2004; Echard et al., 2004; Naim et al., 2004; Shandala et al., 2004; Dean and Spudich, 2006).
Here, we describe that Sti localizes to the CF via association of a predicted coiled-coil (CC) region with actin and myosin and does not require direct physical interaction with RhoA as previously suggested for CIT-K (Eda et al., 2001). Furthermore, we show that Sti depletion perturbs RhoA localization and causes excessive accumulation of phosphorylated MRLC at the cleavage site in late cytokinesis. Our findings indicate that Sti maintains correct RhoA localization at the cleavage site, which is in turn important for proper CR organization at the end of cytokinesis.
Results and discussion
CR constriction completes successfully in sti mutant neuroblasts
Time-lapse analyses indicated that CF ingression was unaffected in Drosophila S2 cells after sti RNAi (D’Avino et al., 2004; Echard et al., 2004). However, in both Sti-depleted S2 cells and sti mutant tissues, CR components accumulated abnormally at the cleavage site in late cytokinesis (D’Avino et al., 2004; Naim et al., 2004). These results suggest that Sti is not required for CR constriction but for proper CR organization in late cytokinesis. To further address this, we visualized CR dynamics in sti larval neuroblasts expressing the MRLC encoded by the spaghetti squash (sqh) gene tagged with GFP (Royou et al., 2004). We found that CR ingression kinetics in sti mutant neuroblasts was similar to controls, albeit less uniform (Fig. 1 and Videos 1 and 2), indicating that the defects in CR organization observed in the absence of Sti occurred after and not during CF ingression.
CR dynamics in sti mutant neuroblasts. (A and B) Kymographs of time-lapse imaging of sqh-GFP in a control stiGS9053/+ heterozygous (A) or stiGS9053/Df(3L)iro2 hemizygous mutant (B) dividing larval brain neuroblast showing the progression of cytokinesis every 30 s. Bar, 1 µm. (C and D) Equatorial cell diameter during cytokinesis measured during live imaging of control stiGS9053/+ (C; n = 15) or stiGS9053/Df(3L)iro2 mutant (D; n = 14) neuroblast divisions. Time is indicated in seconds after Sqh-GFP concentration at the cell equator. All cells imaged completed contraction within 5 min, with sti mutants showing both faster and slower contractions than the more uniform controls.
CR dynamics in sti mutant neuroblasts. (A and B) Kymographs of time-lapse imaging of sqh-GFP in a control stiGS9053/+ heterozygous (A) or stiGS9053/Df(3L)iro2 hemizygous mutant (B) dividing larval brain neuroblast showing the progression of cytokinesis every 30 s. Bar, 1 µm. (C and D) Equatorial cell diameter during cytokinesis measured during live imaging of control stiGS9053/+ (C; n = 15) or stiGS9053/Df(3L)iro2 mutant (D; n = 14) neuroblast divisions. Time is indicated in seconds after Sqh-GFP concentration at the cell equator. All cells imaged completed contraction within 5 min, with sti mutants showing both faster and slower contractions than the more uniform controls.
Sti localization to the CF requires Rho1 and CR assembly
Previous studies indicated that Sti/CIT-K localization to the CF requires Pebble in flies and RhoA activity in human cells (Eda et al., 2001; Shandala et al., 2004). We confirmed that in Drosophila S2 cells, RNAi depletion of Rho1, the single Drosophila RhoA homologue, prevented accumulation of Sti at the CF (Figs. 2 A and S1 A). The majority (61.5%, n = 104) of Rho1 RNAi cells did not show any localization of Sti to the equatorial cortex, but in 38.5% of cells, we could detect Sti inside the cytoplasm close to microtubules (Fig. 2 A), in agreement with findings in human cells (Eda et al., 2001). These Sti structures associated with microtubules are unlikely to represent remnants of partially ingressed furrows because they do not contain other CR components (Fig. S1 B). We then investigated whether CR assembly, which is regulated by RhoA, was also required for Sti localization. In the large majority (80.8%, n = 120) of cells depleted of the nonmuscle myosin II encoded by the gene zipper, Sti was found to localize either inside the cytoplasm or to only one side of the equatorial cortex, always in proximity of microtubules (Fig. 2 B). The remaining 19.2% of zipper RNAi cells did not show any specific localization of Sti (Fig. 2 B). In contrast, Rho1 was consistently found at the cell’s equatorial cortex after Zipper depletion, even when Sti was absent (Fig. 2 C). Similarly, F-actin depolymerization prevented accumulation of Sti, but not Rho1, at the equatorial cortex (Fig. 2 D). Collectively, these results indicate that CR assembly is essential for Sti localization and raise the possibility that physical interaction with Rho1 might not be directly responsible for Sti recruitment to the CF as previously proposed for CIT-K (Eda et al., 2001).
Sti localization to the CF depends on RhoA and CR assembly. (A) Drosophila S2 cells were treated with dsRNAs directed against the bacterial kanamycin resistance gene (absent in Drosophila and used as a control) or Rho1 for 72 h and then fixed and stained to detect Sti, tubulin, and DNA. (B) S2 cells were treated with dsRNAs directed against kanamycin (control) or zipper for 84 h and then fixed and stained to detect Sti, tubulin, and DNA. Percentages on the left indicate localization of Sti. (C) S2 cells were treated with dsRNAs directed against kanamycin (control) or zipper for 84 h and then fixed and stained to detect Rho1, Sti, tubulin, and DNA (not depicted in the merged images). (D) Drosophila S2 cells were treated with latrunculin A (Lat-A) or its solvent DMSO for 1 h and then fixed and stained to detect Rho1, Sti, tubulin, and DNA (not depicted in the merged images). Bars, 10 µm.
Sti localization to the CF depends on RhoA and CR assembly. (A) Drosophila S2 cells were treated with dsRNAs directed against the bacterial kanamycin resistance gene (absent in Drosophila and used as a control) or Rho1 for 72 h and then fixed and stained to detect Sti, tubulin, and DNA. (B) S2 cells were treated with dsRNAs directed against kanamycin (control) or zipper for 84 h and then fixed and stained to detect Sti, tubulin, and DNA. Percentages on the left indicate localization of Sti. (C) S2 cells were treated with dsRNAs directed against kanamycin (control) or zipper for 84 h and then fixed and stained to detect Rho1, Sti, tubulin, and DNA (not depicted in the merged images). (D) Drosophila S2 cells were treated with latrunculin A (Lat-A) or its solvent DMSO for 1 h and then fixed and stained to detect Rho1, Sti, tubulin, and DNA (not depicted in the merged images). Bars, 10 µm.
Sti localizes to the CF via a CC region that associates with actin and myosin
To identify the region responsible for Sti localization to the CF, we generated cell lines stably expressing several Sti fragments tagged with GFP (Fig. 3 A). Sti contains an N-terminal kinase domain, a central CC region followed by a cysteine-rich (C1) motif, and a C-terminal Citron-Nik1 homology (CNH) domain (Fig. 3 A). All the fragments containing the CC region accumulated at the CF, whereas the kinase domain and the C1-CNH region showed diffuse cytoplasmic localization (Fig. 3, A and B; and not depicted). To further narrow down the region responsible for Sti localization, we initially divided CC into two fragments, CC1 and CC2, on the basis of the Sti CC profile (Fig. S1 C). CC2 localized to the CF (Fig. 3 B), whereas CC1 accumulated at centrosomes in metaphase, weakly localized to central spindle microtubules in anaphase, and then accumulated strongly at the spindle midzone in telophase, where it remained until abscission (Figs. 3 B and S1 D). These results suggest that CC2 is sufficient to direct localization to the CF, whereas CC1 probably interacts with one or more microtubule-associated proteins. The distribution pattern of CC1 could also explain the vicinity of Sti to microtubules observed after depletion of Rho1 and Zipper (Fig. 2, A and B). However, because CC1 is not essential for CF localization, its function remains to be investigated.
Sti localizes to the CF via interaction of its CC region with actin and myosin. (A) Schematic representation illustrating Sti protein domains and the localization of different Sti fragments tagged with GFP. (B) Drosophila S2 cells stably expressing various GFP-tagged Sti fragments (indicated at the top) were fixed and stained to detect GFP, tubulin, and DNA (blue). The arrow in the StiFL image marks a midbody remnant. In the CC1 image, the arrowhead marks the spindle midzone, whereas the arrow indicates CC1 localization to centrosome and astral microtubules. GFP::CC2a localization is shown in early and late telophase and after treatment for 72 h with a dsRNA directed against the CNH region (sti RNAi). Bars, 10 µm. (C) S2 cells stably expressing either the Sti CC2a fragment tagged with PtA or PtA alone were transfected with a Myc::Sqh construct for 48 h, and then protein extracts were used in a PtA pull-down assay. The extracts and pull-downs were analyzed by Western blotting to detect Myc::Sqh, actin, Rho1, and PtA. (D) S2 cells stably expressing PtA::StiCC2a were transfected with a Myc::Sqh construct for 48 h and, 5 h before collection, treated either with Cytochalasin D (Cyto D) or its solvent DMSO. Protein extracts were then subject to PtA pull-down assay, and extracts and pull-downs were analyzed by Western blotting to detect Myc::Sqh, actin, Rho1, and PtA. Numbers on the left indicate the sizes in kilodaltons of the molecular mass markers.
Sti localizes to the CF via interaction of its CC region with actin and myosin. (A) Schematic representation illustrating Sti protein domains and the localization of different Sti fragments tagged with GFP. (B) Drosophila S2 cells stably expressing various GFP-tagged Sti fragments (indicated at the top) were fixed and stained to detect GFP, tubulin, and DNA (blue). The arrow in the StiFL image marks a midbody remnant. In the CC1 image, the arrowhead marks the spindle midzone, whereas the arrow indicates CC1 localization to centrosome and astral microtubules. GFP::CC2a localization is shown in early and late telophase and after treatment for 72 h with a dsRNA directed against the CNH region (sti RNAi). Bars, 10 µm. (C) S2 cells stably expressing either the Sti CC2a fragment tagged with PtA or PtA alone were transfected with a Myc::Sqh construct for 48 h, and then protein extracts were used in a PtA pull-down assay. The extracts and pull-downs were analyzed by Western blotting to detect Myc::Sqh, actin, Rho1, and PtA. (D) S2 cells stably expressing PtA::StiCC2a were transfected with a Myc::Sqh construct for 48 h and, 5 h before collection, treated either with Cytochalasin D (Cyto D) or its solvent DMSO. Protein extracts were then subject to PtA pull-down assay, and extracts and pull-downs were analyzed by Western blotting to detect Myc::Sqh, actin, Rho1, and PtA. Numbers on the left indicate the sizes in kilodaltons of the molecular mass markers.
Alignment of the CIT-K RhoA-binding domain (Madaule et al., 1995) with Sti identified a highly conserved region within CC2-spanning residues 1,235–1,263 that we named Rho-binding homology (RBH; Figs. 3 A and S2 A). A fragment containing RBH, CC2b, showed cytoplasmic localization (Fig. 3 B), whereas the region upstream of RBH, CC2a, showed a distribution identical to full-length Sti even after depletion of the endogenous protein (Fig. 3 B). These results indicate that RBH is not necessary for Sti localization and that CC2a has an independent ability to localize to the CF. To investigate whether CC2a could associate with Rho1 and/or CR components in vivo, we generated cell lines stably expressing either CC2a tagged with two copies of the IgG-binding domain of protein A (PtA) or PtA alone. Because no Sqh antibodies are available, we transiently transfected these cell lines with a transgene expressing Sqh tagged with the Myc epitope and affinity purified PtA-tagged proteins and associated complexes. PtA::CC2a was able to pull down both actin and Myc::Sqh but not Rho1 (Fig. 3 C). Inhibition of actin polymerization did not affect the ability of CC2a to pull down actin and Sqh (Fig. 3 D), suggesting that intact actomyosin is dispensable for this association. Consistent with these results, Sti colocalizes with F-actin (D’Avino et al., 2004) and Sqh at the CF (Fig. S2 B). Collectively, these findings indicate that Sti localizes to the CF via association of its CC2a region with actin and myosin.
Sti is necessary for proper Rho1 localization in late cytokinesis
Because physical interaction with Rho1 was not required for Sti localization to the CF, we asked whether Rho1 localization could instead depend on Sti. To address this question, we visualized Rho1 localization during cytokinesis in sti RNAi cells by immunofluorescence. In the vast majority of control cells (93.7%, n = 205), Rho1 accumulated at the CF in early telophase cells and formed a compact ring around the spindle midzone in mid- and late telophase (Fig. 4 A). After Sti depletion, Rho1 localized normally at the CF at furrowing onset but was found diffuse and did not form a compact ring in 50.3% (n = 151) of mid- and late telophase cells (Fig. 4 A), a phenotype very rarely observed in control cells (2.4%, n = 205). Thus, Sti is necessary for proper Rho1 localization at the cleavage site in late telophase.
Sti is required for proper localization of Rho1 at the CF. (A) Drosophila S2 cells were treated with dsRNAs directed against kanamycin (control) or sti for 72 h and then fixed and stained to detect Rho1, tubulin, and DNA (blue). (B) GST::Rho1 variants were preincubated with GTP and then mixed with Sti1,150–1,331, Sti1,437–1,854, or Rhotekin RBD, translated, and radiolabeled in vitro. The mixtures were then pulled down using glutathione beads, separated on SDS-PAGE gels, transferred onto nitrocellulose membrane, and exposed to x-ray films. The Ponceau S staining of protein loading is shown at the bottom. The numbers on the left indicate the sizes in kilodaltons of the molecular mass marker. (C) S2 cells stably expressing Myc alone or Myc-tagged Sti full length (StiFL), StiΔCNH, or a kinase-dead version of Sti (StiKD) were treated with dsRNAs directed against either kanamycin (kana) or the 3′ UTR of sti for 72 h. The number of multinucleate cells was then counted and plotted. Only Myc-positive cells were counted, and >600 cells were counted in each experiment. (D) Quantification of the cells showing aberrant Rho1 localization from experiments performed as described in C. At least 120 cells were counted in each experiment. (E) Cells expressing the transgenes described in C and depleted of endogenous Sti were stained to detect Rho1, Myc, and tubulin. Bars, 10 µm. n = 4; *, P < 0.05 (Mann–Whitney U test). Error bars indicate standard deviations.
Sti is required for proper localization of Rho1 at the CF. (A) Drosophila S2 cells were treated with dsRNAs directed against kanamycin (control) or sti for 72 h and then fixed and stained to detect Rho1, tubulin, and DNA (blue). (B) GST::Rho1 variants were preincubated with GTP and then mixed with Sti1,150–1,331, Sti1,437–1,854, or Rhotekin RBD, translated, and radiolabeled in vitro. The mixtures were then pulled down using glutathione beads, separated on SDS-PAGE gels, transferred onto nitrocellulose membrane, and exposed to x-ray films. The Ponceau S staining of protein loading is shown at the bottom. The numbers on the left indicate the sizes in kilodaltons of the molecular mass marker. (C) S2 cells stably expressing Myc alone or Myc-tagged Sti full length (StiFL), StiΔCNH, or a kinase-dead version of Sti (StiKD) were treated with dsRNAs directed against either kanamycin (kana) or the 3′ UTR of sti for 72 h. The number of multinucleate cells was then counted and plotted. Only Myc-positive cells were counted, and >600 cells were counted in each experiment. (D) Quantification of the cells showing aberrant Rho1 localization from experiments performed as described in C. At least 120 cells were counted in each experiment. (E) Cells expressing the transgenes described in C and depleted of endogenous Sti were stained to detect Rho1, Myc, and tubulin. Bars, 10 µm. n = 4; *, P < 0.05 (Mann–Whitney U test). Error bars indicate standard deviations.
Shandala et al. (2004) reported that the C-terminal–most 416 amino acids of Sti, which contain the CNH domain, was the only region able to interact with a constitutively active variant of Rho1 in a yeast two-hybrid assay. To confirm and expand these findings, we tested whether two in vitro translated and radiolabeled polypeptides encompassing the CNH (Sti1,437–1,854) or RBH (Sti1,150–1,331; Fig. S2 A) domain could interact with recombinant wild-type (WT), constitutively active (G14V), and dominant-negative (T19N) variants of Rho1 expressed in bacteria. The Rho-binding domain (RBD) of Rhotekin, a known RhoA effector (Reid et al., 1996), was used as a control. In our conditions, Sti1,437–1,854 (containing CNH), but not Sti1,150–1,331 (containing RBH), was pulled down by all three Rho1 variants with similar efficiency (Fig. 4 B). As expected, Rhotekin RBD interacted only with RhoWT and RhoG14V (Fig. 4 B). Our findings support the results of Shandala et al. (2004) that Sti interacts directly with Rho1 through its CNH domain. Because CNH is not required for CF localization (Fig. 3 B), these results further indicate that interaction with Rho1 is not necessary for Sti localization. Finally, the observation that the Sti CNH domain was able to interact with all three Rho1 variants, including dominant-negative RhoT19N (Fig. 4 B), indicates that Sti cannot be considered a canonical RhoA effector.
To investigate whether proper Rho1 localization required Sti kinase activity or interaction with CNH, we tested whether StiΔCNH or a kinase-dead version of Sti (StiKD, containing two lysine to alanine substitutions in the predicted ATP-binding pocket; see Materials and methods) could rescue the cytokinesis defects and aberrant Rho1 localization caused by Sti depletion. We specifically depleted the endogenous Sti protein in cell lines stably expressing one of the following Myc-tagged transgenes: Sti full length (StiFL), StiΔCNH, StiKD, or Myc alone. All these transgenes were expressed and localized properly in Drosophila cells (Fig. S3, A and B). A double-stranded RNA (dsRNA) directed against the sti 3′ untranslated region (UTR), which is absent in all Myc-tagged transgenes, was used to deplete the endogenous Sti. This treatment caused a significant reduction of Sti expression, albeit at lower levels than another dsRNA directed against the Sti CNH region (Fig. S3 C), and this less effective knockdown was mirrored by a less penetrant cytokinesis defect (Fig. S3 D). StiFL fully rescued the cytokinesis failure caused by RNAi of endogenous sti (98.0% rescue; Fig. 4 C), whereas StiKD showed only a partial rescue (47.2%) and StiΔCNH a more significant, although not complete, rescue (79.2%). Moreover, StiFL and StiΔCNH, but not StiKD, were able to rescue Rho1 localization after depletion of endogenous Sti (Fig. 4, D and E). Collectively, these findings indicate that proper Rho1 localization requires Sti kinase activity and not interaction of Rho1 with the CNH domain. However, the CNH domain is necessary for at least some Sti functions because StiΔCNH could not fully rescue the cytokinesis defects caused by sti 3′UTR RNAi (Fig. 4 C).
Sti depletion causes excessive accumulation of mono- and diphosphorylated MRLC at the cleavage site
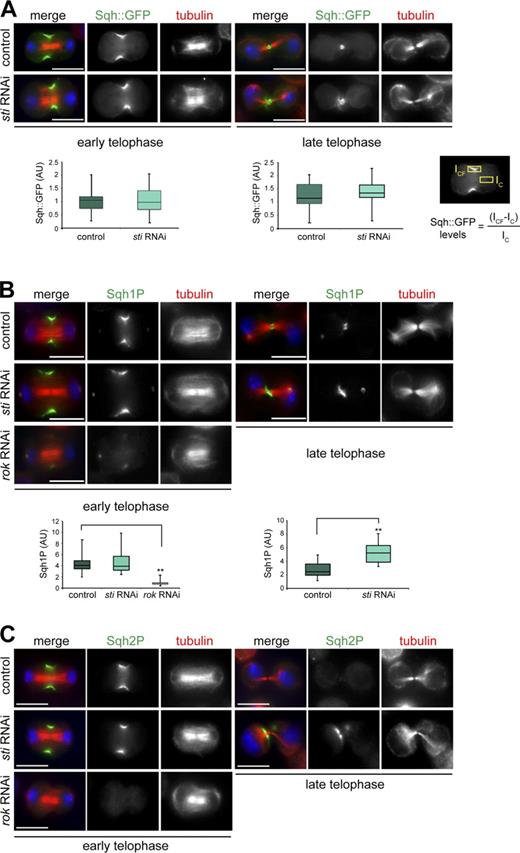
The Rho1 antibody used in our experiment (Fig. 4 A) was not specific for active Rho1, and thus, our results did not reveal whether Rho activation was also affected after sti RNAi. To try to address this issue, we generated stable cell lines expressing two RhoA activity sensors composed of the RBD of either Rhotekin or the formin Diaphanous tagged with GFP. Unfortunately, neither sensor protein localized to the CF in Drosophila cells (unpublished data), and thus, we could not determine whether Sti knockdown affected Rho1 activity at the CF. To overcome this problem, we decided to analyze the level of MRLC phosphorylation as an indirect readout of Rho1 activity. We first determined whether Sti depletion had any effect on the accumulation of Sqh::GFP during cytokinesis. No significant difference in the levels of Sqh::GFP at the CF was observed between control and sti RNAi cells in both early and late telophase (Fig. 5 A). However, Sqh::GFP failed to form a compact ring structure in late telophase sti RNAi cells (Fig. 5 A). This phenotype resembled that observed for other CR components, such as F-Actin and Anillin (D’Avino et al., 2004), and confirmed that Sti is necessary to maintain a compact CR organization in late telophase. We then investigated whether Sti depletion could affect the levels of Sqh phosphorylation at the CF using two phosphospecific antibodies that recognize either the monophosphorylated (Sqh1P, phosphorylated at Ser21 in Drosophila and equivalent to Ser19 in human MRLC) or the diphosphorylated (Sqh2P, phosphorylated at Ser21 and Thr20) form of Sqh (Zhang and Ward, 2011). Sqh1P showed a distribution pattern identical to Sqh::GFP in control cells, whereas sti RNAi cells showed a twofold increase of Sqh1P at the CF in late, but not early, telophase cells (Fig. 5 B). As a control, we also monitored Sqh1P levels after depletion of ROK because it is well established that this kinase phosphorylates Sqh and is required for its recruitment to the CF (Winter et al., 2001; Dean et al., 2005). In agreement with these studies, we found that ROK depletion severely reduced Sqh1P accumulation at the CF (Fig. 5 B). No significant difference in Sqh2P levels at the CF was observed between control and sti RNAi cells in early telophase (Fig. 5 C). However, in late telophase, Sqh2P was detected only in very few control cells (9.0%, n = 199), whereas the majority of sti RNAi cells (67.1%, n = 183) showed a clear Sqh2P signal at the cleavage site in late cytokinesis (Fig. 5 C). Moreover, in these late telophase sti RNAi cells, Sqh2P often appeared to accumulate at one side of the cleavage site, distant from the spindle midzone (Fig. 5 C). These results indicate that Sti depletion causes an increase of mono- and diphosphorylated MRLC at the end of cytokinesis.
Sti depletion causes an increase of phosphorylated Sqh at the CF in late cytokinesis. (A) Drosophila S2 cells stably expressing Sqh::GFP were treated with dsRNAs directed against kanamycin (control) or sti for 72 h and then fixed and stained to detect Sqh::GFP, tubulin, and DNA (blue). The box plots showing the quantification of Sqh::GFP fluorescence levels at the CF in early and late cytokinesis are shown at the bottom. The intensity of Sqh::GFP fluorescence at the CF was calculated using the formula shown, in which ICF is the fluorescence intensity at the CF, and IC represents the background intensity measured within an identical area inside the cytoplasm. At least 30 cells from two separate experiments were analyzed. AU, arbitrary unit. (B) S2 cells were treated with dsRNAs directed against the kanamycin gene (control), sti, or rok for 72 h and then fixed and stained to detect Sqh1P, tubulin, and DNA (blue). Quantification of Sqh1P levels was performed as described in A. Note that the Sqh1P staining on centrosomes is not specific because it was not affected by ROK depletion, and Sqh::GFP never localized to centrosomes (Fig. S2 B). **, P < 0.01 (Student’s t test). (C) S2 cells were treated with dsRNAs directed against kanamycin (control), sti, or rok for 72 h and then fixed and stained to detect Sqh2P, tubulin, and DNA (blue). In each box plot, the box contains the values comprised between the 25th and 75th percentile, and the horizontal line inside the box marks the median. The ends of the whiskers indicate the minimum and maximum values. Bars, 10 µm.
Sti depletion causes an increase of phosphorylated Sqh at the CF in late cytokinesis. (A) Drosophila S2 cells stably expressing Sqh::GFP were treated with dsRNAs directed against kanamycin (control) or sti for 72 h and then fixed and stained to detect Sqh::GFP, tubulin, and DNA (blue). The box plots showing the quantification of Sqh::GFP fluorescence levels at the CF in early and late cytokinesis are shown at the bottom. The intensity of Sqh::GFP fluorescence at the CF was calculated using the formula shown, in which ICF is the fluorescence intensity at the CF, and IC represents the background intensity measured within an identical area inside the cytoplasm. At least 30 cells from two separate experiments were analyzed. AU, arbitrary unit. (B) S2 cells were treated with dsRNAs directed against the kanamycin gene (control), sti, or rok for 72 h and then fixed and stained to detect Sqh1P, tubulin, and DNA (blue). Quantification of Sqh1P levels was performed as described in A. Note that the Sqh1P staining on centrosomes is not specific because it was not affected by ROK depletion, and Sqh::GFP never localized to centrosomes (Fig. S2 B). **, P < 0.01 (Student’s t test). (C) S2 cells were treated with dsRNAs directed against kanamycin (control), sti, or rok for 72 h and then fixed and stained to detect Sqh2P, tubulin, and DNA (blue). In each box plot, the box contains the values comprised between the 25th and 75th percentile, and the horizontal line inside the box marks the median. The ends of the whiskers indicate the minimum and maximum values. Bars, 10 µm.
Closing remarks
Current models propose that in human cells, RhoA recruits and then activates CIT-K, which in turn phosphorylates MRLC at the CF (Eda et al., 2001; Yamashiro et al., 2003). Results from our and another study (Dean and Spudich, 2006) indicate that, at least in Drosophila, this model is incorrect. Indeed, Sti recruitment to the CF does not require interaction with RhoA, and instead, Sti appears necessary to maintain proper RhoA localization at the CF specifically in late cytokinesis (Figs. 3 and 4). In addition, we found that Sti depletion caused an increase and not a decrease of mono- and diphosphorylated MRLC at the cleavage site in late cytokinesis (Fig. 5). Because MRLC phosphorylation is controlled by RhoA signaling (Dean and Spudich, 2006), we propose that aberrant RhoA localization after Sti depletion might cause an increase of phosphorylated MRLC at the CF. MRLC phosphorylation has been described to control actin dynamics (Murthy and Wadsworth, 2005), and thus, this model could well explain the CR disorganization observed after Sti knockdown. Why does Sti depletion cause an increase of MRLC phosphorylation? The simplest hypotheses are that Sti kinase activity could, directly or indirectly, promote the activity of an MRLC phosphatase or inhibit RhoA and/or an MRLC kinase. Identification of Sti targets will be necessary to fully comprehend how this kinase controls RhoA localization during cytokinesis and to distinguish between the aforementioned hypotheses.
Materials and methods
Fly stocks and time-lapse analysis of larval neuroblasts
Flies were raised on a standard cornmeal medium at 25°C. The deficiency Df(3L)iro2 uncovers the sti genomic region (69B–69D6), and the stiGS9053 allele is a P{GSv6} insertion in the 5′UTR of sti as previously described (Shandala et al., 2004). The sqh::GFP stock contains a C-terminal fusion of MRLC with GFP (Royou et al., 2004). The His2Av::GFP stock contains a C-terminal fusion of His2Av with GFP as previously described in Clarkson and Saint (1999). For time-lapse analyses, brains were dissected from sti heterozygous (stiGS9053/+) or sti hemizygous (stiGS9053/Df(3L)iro2) third instar larvae, and neuroblasts were filmed by collecting images every 30 s as previously described (Savoian and Rieder, 2002). In brief, live brains were spread to a monolayer on a coverslip under halocarbon oil (Sigma-Aldrich) using scalpel blades and then mounted using Vaseline as a well. Live imaging was performed at room temperature on a microscope (AX70; Olympus) using a 100× Plan Apochromat objective, NA 1.35. Images were acquired using a camera (CoolSNAP fx; Photometrics) with V++ software (Digital Optics), and kymographs were assembled in Photoshop (Adobe).
Molecular biology, cell culture, DNA, and dsRNA transfections
Gateway technology (Invitrogen) was used in all cloning procedures as previously described (D’Avino et al., 2006). Proteins tagged at the N or C terminus with GFP, Myc, or PtA were expressed in Drosophila cells under the control of either a constitutive Actin5C promoter or an inducible metallothionein promoter using the destination vectors described previously (D’Avino et al., 2006, 2008, 2009). The pDEST15 vector (Invitrogen) was used for bacterial expression of GST-tagged proteins. To generate the Sti kinase-dead mutant, two adjacent lysines (K) located in the predicted ATP-binding pocket of the kinase domain, K142 and K143, were mutated to alanine using a site-directed mutagenesis kit (QuikChange Lightning; Agilent Technologies) following the manufacturer’s instructions. K142 corresponds to K126 in CIT-K, which was shown to be essential for kinase activity (Di Cunto et al., 1998).
The Dmel strain of S2 cells (Invitrogen) was grown in serum-free medium (Express Five; Invitrogen) supplemented with antibiotics. To generate stable cell lines, three million cells were transfected with 3 µg of the appropriate vector along with 0.3 µg pCoBlast plasmid (Invitrogen) carrying a resistance to the antibiotic blasticidin and 15 µl transfection reagent (FuGENE HD; Roche) following the manufacturer’s instructions. Resistant cells were selected by adding 20 µg/ml blasticidin to the media 48 h after transfection. For expression of metallothionein-regulated constructs, cells were incubated in complete medium containing 1 mM CuSO4 for 5–7 h.
dsRNA production and RNAi treatments were performed essentially as described previously (D’Avino et al., 2004). Typically, 1.5 million cells were transfected with 20 µg dsRNA and 20 µl transfection reagent (TransFast; Promega) following the manufacturer’s instructions and incubated for 72 h unless otherwise indicated. The primers used to generate dsRNAs are listed in Table S1. The sti CNH dsRNA was used in all Sti knockdowns with the only exception of the rescue experiments shown in Fig. 3 B. For latrunculin A treatment, cells were plated on coverslips for 2 h and then treated for 1 h with either latrunculin A (Sigma-Aldrich) at a final concentration of 20 µM or an equal volume of its solvent DMSO (Sigma-Aldrich) as a control.
Antibodies and microscopy
The following antibodies were used in this study: mouse monoclonal anti–α-tubulin (clone DM1A; Sigma-Aldrich), chicken polyclonal anti–α-tubulin (ab89984; Abcam), mouse monoclonal antiactin (clone AC-40; Sigma-Aldrich), mouse monoclonal anti-Myc (clone 9E10; Santa Cruz Biotechnology, Inc.), rabbit polyclonal anti-Myc (ab9106; Abcam), rabbit polyclonal anti-Sti (D’Avino et al., 2004), mouse monoclonal anti-Rho1 (clone p1D9; Developmental Studies Hybridoma Bank), rat anti-Sqh2P and guinea pig anti-Sqh1P (provided by R. Ward, University of Kansas, Lawrence, KS; Zhang and Ward, 2011), rabbit polyclonal anti-MRLC S19 (Cell Signaling Technology), peroxidase-ChromPure anti–rabbit IgG (Jackson ImmunoResearch Laboratories, Inc.), and Alexa Fluor 594 phalloidin (Invitrogen). Peroxidase- and Alexa Fluor–conjugated secondary antibodies were purchased from Jackson ImmunoResearch Laboratories, Inc. and Invitrogen.
Dmel cells were grown on 22 × 22–mm coverslips (Menzel-Gläser) and fixed in PHEM buffer (60 mM Pipes, 25 mM Hepes, pH 7, 10 mM EGTA, 4 mM MgCl2, and 3.7% formaldehyde) for 12 min. They were then washed three times for 10 min with PBS and incubated in blocking buffer (PBS, 0.5% Triton X-100, and 3–5% BSA) for 1 h at room temperature. Incubation with primary antibodies was performed overnight at 4°C in PBT (PBS, 0.1% Triton X-100, and 1% BSA). After two washes in PBT, coverslips were incubated with Alexa Fluor–conjugated secondary antibodies (Invitrogen) diluted in PBT for 2 h at room temperature and then washed two times with PBT and once with PBS. Coverslips were finally mounted using mounting medium with DAPI (Vectashield; Vector Laboratories) and visualized on a fluorescence microscope (Axiovert 200; Carl Zeiss) equipped with a 100× objective, NA 1.4. Images were acquired using a camera (CoolSNAP HQ2; Photometrics) with MetaMorph software (Molecular Devices) and assembled and adjusted for contrast using Photoshop CS3. Quantification of Sqh fluorescence was performed using ImageJ software (National Institutes of Health), and fluorescence intensity values were plotted using Excel software (Microsoft).
In vivo pull-down assays
Cells stably expressing either PtA::CC2a or PtA alone were transfected with a plasmid encoding a Myc-tagged Sqh transgene and incubated at 25°C for 42 h, and then the proteasome inhibitor MG132 (Sigma-Aldrich) was added to the medium at a concentration of 25 µM, 5–6 h before harvesting the cells. Cytochalasin D was added 5 h before collection at a final concentration of 10 µM. Cells were then collected and stored at −80°C. The cell pellet was resuspended in 0.5 ml extraction buffer (50 mM Hepes, pH 7.5, 100 mM KAc, 50 mM KCl, 2 mM MgCl2, 2 mM EGTA, 0.1% NP-40, 5 mM DTT, 5% glycerol and protease inhibitors [Complete; Roche]) and homogenized using a high-performance disperser (Thermo Fisher Scientific). Homogenates were centrifuged at 8,000 rpm at 4°C in a centrifuge (5417R; Eppendorf) for 20 min, and supernatants were transferred into new tubes. 50 µl Dynabeads (Invitrogen) conjugated to rabbit IgG (MP Biochemicals) was added to the supernatants and incubated for 2 h on a rotating wheel at 4°C. Beads were then washed five times for 5 min in 1 ml extraction buffer. Proteins were eluted from beads with 0.5 ml elution buffer (0.5 M NH4OH and 0.5 mM EDTA, pH 8.0), lyophilized, and resuspended in Laemmli SDS-PAGE sample buffer (Sigma-Aldrich). Proteins were separated on a SDS-PAGE gel, transferred onto a polyvinylidene fluoride membrane, and probed to detect the antigens shown in Fig. 3 (C and D).
In vitro binding assay
DNA fragments coding for RhoWT, RhoG14V, and RhoT19N were generated by PCR and cloned into pDEST15 (Invitrogen) to express N-terminal GST-tagged polypeptides in Escherichia coli. The GST-tagged products were then purified using glutathione–Sepharose 4B according to manufacturer’s instruction (GE Healthcare). [35S]Methionine-labeled RBH, CNH, and Rhotekin RBD (provided by A. Miller and B. Bement, University of Wisconsin–Madison, Madison, WI) were prepared from corresponding PCR products amplified using primers harboring a T7 promoter and then transcribed and translated in vitro using a transcription/translation system (TnT T7 Quick Coupled; Promega) in the presence of [35S]methionine. Generally, 25 µl glutathione–Sepharose beads containing purified GST-Rho1 proteins (WT, G14V, and T19N) was preincubated in 300 µl NET-N+ buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA, 0.5% NP-40, and a cocktail of protease inhibitors [Complete]) containing 5 mM GTP (Sigma-Aldrich) at 4°C, and after 1 min, 17 mM MgCl2 was added, and the incubation continued for a further 29 min. Then, 10 µl [35S]methionine-labeled polypeptides were added, and the mixture was incubated for 1 h at 4°C with periodic agitation. Samples were then washed five times with 500 µl wash buffer (50 mM Tris-HCl, pH 7.4, 500 mM NaCl, 5 mM EDTA, 5 mM GTP, 0.5% NP-40, and a cocktail of protease inhibitors [Complete]) followed by centrifugation at 500 g for 1 min. Beads were resuspended in 25 µl Laemmli SDS-PAGE sample buffer, and typically 15 µl was loaded on a 4–20% Tris-glycine gel. Proteins were then transferred onto a nitrocellulose membrane using a dry blotting system (iBlot; Invitrogen) and exposed to x-ray films at −80°C with intensifying screens (Sigma-Aldrich). The filters were then stained with Ponceau S to detect GST-tagged proteins.
Online supplemental material
Fig. S1 shows the levels of Rho1 protein and Sti localization after Rho1 RNAi, the predicted CC profile of Sti, and the localization of GFP::CC1 at different mitotic stages. Fig. S2 shows the alignment of the CIT-K Rho-binding region with Sti and the colocalization of Sti and Sqh::GFP. Fig. S3 shows the expression and localization of Myc-tagged Sti proteins, the levels of Sti protein, and the percentage of multinucleate cells after RNAi treatments with sti CNH and sti 3′UTR dsRNAs. Table S1 shows list of the primers used to generate the dsRNAs. Video 1 shows cytokinesis in a control (sti/+) larval neuroblast expressing Sqh::GFP and His2Av::GFP. Video 2 shows cytokinesis in a mutant (sti/Df) larval neuroblast expressing Sqh::GFP and His2Av::GFP.
Cytokinesis in a control sti/+ dividing neuroblast. This video shows cytokinesis in a heterozygous stiGS9053/+ larval neuroblast expressing sqh::GFP and His2Av::GFP transgenes (see Fig. 1 for details). All images were captured at 30-s intervals for 14 min using the 100× objective on a microscope (AX70; Olympus). Playback rate is five frames per second.
Cytokinesis in a control sti/+ dividing neuroblast. This video shows cytokinesis in a heterozygous stiGS9053/+ larval neuroblast expressing sqh::GFP and His2Av::GFP transgenes (see Fig. 1 for details). All images were captured at 30-s intervals for 14 min using the 100× objective on a microscope (AX70; Olympus). Playback rate is five frames per second.
Cytokinesis in a mutant sti dividing neuroblast. This video shows cytokinesis in a hemizygous stiGS9053/Df(3L)iro2 larval neuroblast expressing sqh::GFP and His2Av::GFP transgenes (see Fig. 1 for details). All images were captured at 30-s intervals for 20.5 min using the 100× objective on a microscope (AX70; Olympus). Playback rate is five frames per second.
Cytokinesis in a mutant sti dividing neuroblast. This video shows cytokinesis in a hemizygous stiGS9053/Df(3L)iro2 larval neuroblast expressing sqh::GFP and His2Av::GFP transgenes (see Fig. 1 for details). All images were captured at 30-s intervals for 20.5 min using the 100× objective on a microscope (AX70; Olympus). Playback rate is five frames per second.
Acknowledgments
We would like to thank A. Miller and B. Bement for the Rhotekin RBD plasmid, R. Ward for Sqh1P and Sqh2P antibodies, and T. Takeda for the Sqh::GFP cell line. We are also grateful to M. Savoian for helpful suggestions and T. Takeda for critical reading of the manuscript. The Rho1 antibody was obtained from the Developmental Studies Hybridoma Bank.
This work was supported by a Cancer Research UK grant (C12296/A12541) and a Newton Trust Research grant to P.P. D’Avino. Z.I. Bassi is supported by a Gwynaeth Pretty PhD fellowship of the Department of Pathology of the University of Cambridge. K.J. Verbrugghe and work in the Glover laboratory were supported by a Cancer Research UK program grant.
References
Author notes
K.J. Verbrugghe’s present address is Dept. of Human Genetics, University of Michigan, Ann Arbor, MI 48109.