Centromeres of higher eukaryotes are epigenetically marked by the centromere-specific CENP-A nucleosome. New CENP-A recruitment requires the CENP-A histone chaperone HJURP. In this paper, we show that a LacI (Lac repressor) fusion of HJURP drove the stable recruitment of CENP-A to a LacO (Lac operon) array at a noncentromeric locus. Ectopically targeted CENP-A chromatin at the LacO array was sufficient to direct the assembly of a functional centromere as indicated by the recruitment of the constitutive centromere-associated network proteins, the microtubule-binding protein NDC80, and the formation of stable kinetochore–microtubule attachments. An amino-terminal fragment of HJURP was able to assemble CENP-A nucleosomes in vitro, demonstrating that HJURP is a chromatin assembly factor. Furthermore, HJURP recruitment to endogenous centromeres required the Mis18 complex. Together, these data suggest that the role of the Mis18 complex in CENP-A deposition is to recruit HJURP and that the CENP-A nucleosome assembly activity of HJURP is responsible for centromeric chromatin assembly to maintain the epigenetic mark.
Introduction
Centromeric chromatin is defined by the incorporation of a unique nucleosome containing the histone H3 variant CENP-A (centromere protein-A), which distinguishes this locus from general chromatin. Centromeric chromatin plays an integral role in organizing and controlling chromosome segregation. The centromere is the site of microtubule attachment and checkpoint signaling during mitosis and also organizes the constitutive centromere components throughout the cell cycle (Cleveland et al., 2003; Musacchio and Salmon, 2007; Cheeseman and Desai, 2008).
The human centromere contains hundreds of thousands to millions of base pairs of DNA arranged in α-satellite higher order repeats (Cleveland et al., 2003; Schueler and Sullivan, 2006; Allshire and Karpen, 2008). Despite the characteristic α-satellite DNA content of centromeres, the existence of stable dicentric chromosomes in which a single centromeric region remains “active” and the formation of neocentromeres at non–α-satellite repeat regions have led to the understanding that centromere specification is an epigenetic process (Marshall et al., 2008). The mechanism by which CENP-A is assembled at preexisting centromeric sites, whether at canonical centromeres or neocentromeres, remains unclear.
The presence of the CENP-A nucleosome within centromeric chromatin directs the recruitment of a large set of proteins present at the centromere throughout the cell cycle (Musacchio and Salmon, 2007). These proteins can be divided into partially overlapping subcomplexes, including the CENP-A nucleosome-associated complex (CENP-ANAC), CENP-A distal components, and CENP-H–I complexes, and are collectively termed the constitutive centromere-associated network (CCAN; Obuse et al., 2004; Foltz et al., 2006; Okada et al., 2009). CENP-N and CENP-C, within the CENP-ANAC, are able to independently discern centromeric CENP-A–containing chromatin from general chromatin by directly recognizing the CENP-A nucleosome (Carroll et al., 2009, 2010).
Several models have been proposed to explain how CENP-A nucleosomes may be differentiated from H3.1 nucleosomes. One model predicts an octameric CENP-A–containing nucleosome with a more compact structure relative to the histone H3.1 nucleosome (Black et al., 2007; Camahort et al., 2009; Sekulic et al., 2010). Also proposed are changes in the CENP-A nucleosome composition that include single copies of each of the four core histones, a lack of histones H2A and H2B, or the inclusion of the nonhistone component Scm3 (Dalal et al., 2007; Mizuguchi et al., 2007; Williams et al., 2009). Finally, it has been suggested that centromeric nucleosomes wrap DNA in a right-handed path around the CENP-A/CenH3-containing histone core, in contrast to the left-handed wrapping of the canonical H3 nucleosome (Furuyama and Henikoff, 2009). All of these distinctions may contribute to the selective assembly of CENP-A nucleosomes into centromeric loci or to the recruitment of a unique set of proteins to the centromeric chromatin.
The redistribution of preexisting CENP-A nucleosomes between newly synthesized sister DNA strands during S phase necessitates the incorporation of new CENP-A nucleosomes during each round of cell division to maintain centromeric identity. Distinct histone chaperones for the different histone H3 variants function to couple the deposition of the appropriate histone variant to a unique site within the genome at distinct times during the cell cycle (Ransom et al., 2010). In the case of vertebrates, new CENP-A is recruited to centromeres in a prenucleosomal complex with HJURP during early G1 (Jansen et al., 2007; Dunleavy et al., 2009; Foltz et al., 2009; Shuaib et al., 2010; Bernad et al., 2011). Point centromeres of Saccharomyces cerevisiae and regional centromeres of Schizosaccharomyces pombe also require the HJURP homologue Scm3 for the recruitment of their respective CENP-A homologues. The interaction between HJURP and CENP-A depends on the centromere-targeting domain (CATD) of CENP-A (Black et al., 2004; Foltz et al., 2009). The CATD is a portion of the histone-fold domain of CENP-A that confers structural changes that alter the shape, surface, and conformational flexibility of the complexes into which CENP-A assembles, relative to its conventional counterpart H3 (Black et al., 2007; Sekulic et al., 2010). This same region dictates the interaction of CENP-A with the Scm3 domain of HJURP as well as CENP-A’s association with the constitutive centromere protein CENP-N (Carroll et al., 2009; Foltz et al., 2009). These data suggest that the structural differences imparted by the CATD mediate the correct localization and incorporation of the variant within the genome and the recruitment of the appropriate proteins required for building the centromere.
New CENP-A nucleosome assembly also requires the human Mis18 complex (Mis18α, Mis18β, and Mis18BP1hsKNL2; also known as M18BP1; Fujita et al., 2007; Maddox et al., 2007). This complex is initially recruited to centromeres during telophase and remains associated with CENP-A–containing chromatin during G1. The S. pombe homologue of Mis18 is required for the localization of Scm3 and CENP-A to centromeres (Camahort et al., 2007; Mizuguchi et al., 2007; Stoler et al., 2007; Pidoux et al., 2009; Williams et al., 2009). In vertebrates, Mis18 is not found as part of the CENP-A–HJURP prenucleosomal complex nor is it associated with isolated CENP-A nucleosomes, but it is contained within CENP-A–containing chromatin (Foltz et al., 2006; Maddox et al., 2007). Because it localizes to centromeres just before new CENP-A loading, it is hypothesized that Mis18 primes the centromere for CENP-A deposition through a mechanism that may involve histone acetylation (Hayashi et al., 2004; Fujita et al., 2007).
Recruitment of HJURP to a noncentromeric LacO (Lac operon) array is sufficient to drive the stable association of CENP-A at the array and to recreate a functional centromere. The HJURP-deposited CENP-A at the array is competent to recruit CCAN proteins and the kinetochore protein NDC80 and form stable microtubule attachments during mitosis. A fragment of HJURP that contains the Scm3 domain is able to specifically assemble CENP-A nucleosomes in vitro. We additionally show the Mis18 complex is required for HJURP recruitment to endogenous centromeres. However, when this recruitment step is bypassed by tethering HJURP to the LacO array, CENP-A deposition can still occur in the absence of Mis18 at this noncentromeric location. Together, our data delineate the roles of Mis18 and HJURP in the recruitment of CENP-A and its assembly into nucleosomes.
Results
HJURP recruitment is sufficient to drive CENP-A deposition into chromatin
HJURP is required for CENP-A deposition and is localized to centromeres during early G1 when new CENP-A is incorporated into centromeres (Jansen et al., 2007; Dunleavy et al., 2009; Foltz et al., 2009). We hypothesized that recruitment of HJURP may couple CENP-A delivery and nucleosome assembly at centromeres by chaperoning CENP-A to centromeres and providing intrinsic CENP-A deposition activity. If true, HJURP recruitment to a noncentromeric site should be sufficient to dictate the site of CENP-A nucleosome deposition. We ectopically localized HJURP to a noncentromeric site within the genome by expressing a LacI (Lac repressor)-HJURP fusion protein in U2OS cells containing 200 copies of a 256×LacO/96×tetracycline-responsive element (TRE) array on chromosome 1 (Janicki et al., 2004). We then determined whether CENP-A deposition occurred at this site.
LacO/TRE U2OS cells were transiently transfected with either mCherry-LacI or the mCherry-LacI-HJURP fusion (hereafter called LacI or LacI-HJURP). GFP-TetR (tet repressor) was cotransfected to independently determine the location of the array. The LacI-HJURP and GFP-TetR fusion proteins successfully localized to and marked the LacO/TRE array (Fig. 1, A and B). When HJURP was localized at the array, endogenous CENP-A was also enriched in 61.3% of transfected cells, 48 h after transfection (Fig. 1, A and C). This was in contrast to 1% of cells showing CENP-A colocalization with the array when the LacI control was expressed, demonstrating the LacO/TRE array did not consistently overlap with an endogenous centromere (Fig. 1, A and C).
HJURP-dependent CENP-A recruitment and incorporation into the LacO/TRE array. (A and B) Recruitment of endogenous CENP-A to the LacO/TRE array in the presence of LacI-HJURP or LacI-HJURPScm3. All LacI constructs have an N-terminal mCherry tag. Representative images of preextracted cells treated with 0 mM IPTG (A) or 10 mM IPTG (B) for 1 h before fixation. Endogenous CENP-A was detected using a monoclonal anti–CENP-A antibody. mCherry-LacI fusions of HJURP or HJURPScm3 and GFP-TetR markers are transiently transfected at equal ratios, and DNA is visualized using DAPI. Cells are fixed at 48 h after transfection. Arrows indicate the array. Insets show magnified views of boxed regions. (C) Quantification of CENP-A staining at the LacO/TRE array. Error bars represent the standard deviations between two experiments. At least 30 cells per condition were analyzed; n = 2. In the case of IPTG treatment, cells in which the residual mCherry signal was still visible at the array were excluded. (D) Quantification of the amount of CENP-A at the array in LacI-HJURP– and LacI-HJURPScm3–transfected cells with and without treatment with IPTG (>28 cells/condition). Middle lines in each box represent the mean integrated intensity for each condition, and whiskers represent the maximum and minimum intensities observed. A.U., arbitrary unit. Bars, 5 µm.
HJURP-dependent CENP-A recruitment and incorporation into the LacO/TRE array. (A and B) Recruitment of endogenous CENP-A to the LacO/TRE array in the presence of LacI-HJURP or LacI-HJURPScm3. All LacI constructs have an N-terminal mCherry tag. Representative images of preextracted cells treated with 0 mM IPTG (A) or 10 mM IPTG (B) for 1 h before fixation. Endogenous CENP-A was detected using a monoclonal anti–CENP-A antibody. mCherry-LacI fusions of HJURP or HJURPScm3 and GFP-TetR markers are transiently transfected at equal ratios, and DNA is visualized using DAPI. Cells are fixed at 48 h after transfection. Arrows indicate the array. Insets show magnified views of boxed regions. (C) Quantification of CENP-A staining at the LacO/TRE array. Error bars represent the standard deviations between two experiments. At least 30 cells per condition were analyzed; n = 2. In the case of IPTG treatment, cells in which the residual mCherry signal was still visible at the array were excluded. (D) Quantification of the amount of CENP-A at the array in LacI-HJURP– and LacI-HJURPScm3–transfected cells with and without treatment with IPTG (>28 cells/condition). Middle lines in each box represent the mean integrated intensity for each condition, and whiskers represent the maximum and minimum intensities observed. A.U., arbitrary unit. Bars, 5 µm.
We disrupted the LacI interaction with the LacO array using IPTG to determine whether CENP-A was stably associated with the underlying chromatin or whether it was tethered at the array solely through its interaction with LacI-HJURP. We expected that the portion of the CENP-A tethered at the array through its binding to LacI-HJURP would be removed by treatment with IPTG. Any remaining CENP-A signal at the array after IPTG treatment would indicate stable association of CENP-A with the DNA. Cells were treated with 10 mM IPTG for 1 h before fixation, 48 h after transfection. Under these conditions, LacI-HJURP was no longer visible at the array in cells counted for CENP-A stability (array marked by GFP-TetR; Fig. 1 B). CENP-A persisted at the array in 51.6% of IPTG-treated cells in which LacI-HJURP was no longer visible. We observed a 46% decrease in the mean intensity of the CENP-A at the arrays in cells treated with IPTG (Fig. 1 D), which is consistent with a subset of CENP-A being tethered at the array through HJURP. The stable association of CENP-A with the LacO DNA after removal of HJURP is consistent with the assembly of CENP-A–containing chromatin at the array (Fig. 1, B and C). Alternatively, the CENP-A observed after removal of HJURP may persist as a prenucleosomal form pending its assembly by other factors, similar to what has been proposed previously in RSF1 knockdowns (Perpelescu et al., 2009).
We used an NIH3T3 cell line containing a LacO-SceI-TRE array to demonstrate that the stable recruitment of CENP-A by LacI-HJURP was not a unique property of the U2OS cells or the genomic location of the array (Soutoglou et al., 2007). Transfected YFP–CENP-A was recruited to the LacO array when LacI-HJURP was expressed and remained stably associated with the array when the cells were treated with IPTG, similar to what we observed in the U2OS cell line (Fig. S1 A).
The extent of homology between the yeast Scm3 proteins and HJURP is contained within a small 52–amino acid stretch in the amino terminus of HJURP (Sanchez-Pulido et al., 2009). A fragment of HJURP that contains the Scm3 homology domain is sufficient to mediate the interaction of CENP-A and HJURP but is not able to direct HJURP to centromeres (Fig. S2; Shuaib et al., 2010). It is reasonable to suppose that HJURP and Scm3 mediate similar functions with respect to CENP-A nucleosome deposition; therefore, we tested whether the region of HJURP that includes the Scm3 homology domain (HJURPScm3), which can interact with CENP-A but is unable to localize to centromeres, is sufficient to direct the stable association of CENP-A at the array. HJURPScm3 was fused to mCherry-LacI (called LacI-HJURPScm3) and transfected into the LacO–containing U20S cells. Targeting LacI-HJURPScm3 resulted in the recruitment of endogenous CENP-A to the array in 39.9% of cells after 48 h in culture, similar to the full-length protein (Fig. 1, A and C). We removed LacI-HJURPScm3 from the array using IPTG, and endogenous CENP-A remained associated with the chromatin in 32.8% of cells, similar to full-length HJURP (Fig. 1, B and C). LacI-HJURPScm3 recruited less CENP-A to the array (Fig. 1 D); however, the proportion of CENP-A retained at the array in LacI-HJURPScm3–transfected, IPTG-treated cells was greater than full-length HJURP. Cells transfected with LacI-HJURPScm3 showed only a 5% decrease in mean CENP-A intensity at the array when treated with IPTG relative to controls.
CENP-A deposition by HJURP recruits CENP-A nucleosome-associated proteins
The recruitment of constitutive centromere proteins is thought to depend on the presence of CENP-A, as indicated by affinity purifications, siRNA, and knockout experiments (Obuse et al., 2004; Régnier et al., 2005; Foltz et al., 2006; Liu et al., 2006). In vitro, CENP-N and CENP-C are able to directly and selectively recognize the CENP-A nucleosome (Carroll et al., 2009, 2010). To test the ability of CENP-A nucleosomes to determine the recruitment of the constitutive centromere, we examined whether components of the CENP-ANAC are recruited to the LacO array after CENP-A was stably associated there using LacI-HJURP (as in Fig. 1). LacO/TRE U2OS cells were cotransfected with LacI-HJURP and GFP-tagged constructs expressing CENP-C, CENP-N, CENP-M, or CENP-T and fixed 72 h later. Only cells with endogenous CENP-A present at the array and a GFP-CENP signal at the centromeres were analyzed. We observed the recruitment of GFP-tagged CENP-C, CENP-N, CENP-M, and CENP-T in 12–28% of cotransfected cells (Fig. 2, A and C). CENP-N was most strongly recruited and selectively interacts with intact CENP-A nucleosomes but does not recognize the prenucleosomal heterotetramer, suggesting CENP-A assembled at the array may be nucleosomal (Carroll et al., 2009). GFP signal was never observed at the array for any of the GFP-tagged CENP-ANAC proteins when cells were cotransfected with the LacI control construct (Fig. 2 B).
HJURP-deposited CENP-A recruits constitutive centromere proteins. (A) LacO/TRE U2OS cells were transiently transfected with LacI-HJURP and constructs expressing LAP (GFP localization and purification)-tagged CENP-C, CENP-M, CENP-N, or CENP-T. Cells were preextracted and fixed 72 h after transfection. The presence of CENP-A was assessed using antibodies against endogenous CENP-A. Insets show the arrays at a higher magnification. (B) LacI control images for LAP-CENP-C–T, indicating that recruitment is never observed in the absence of CENP-A. (C) Graph showing the percentage of doubly transfected (GFP and LacI-HJURP) U2OS LacO/TRE cells with endogenous CENP-A present at the array, which also recruited the indicated constitutive centromere proteins (≥30 cells per condition; error bars represent standard deviation). Bars, 5 µm.
HJURP-deposited CENP-A recruits constitutive centromere proteins. (A) LacO/TRE U2OS cells were transiently transfected with LacI-HJURP and constructs expressing LAP (GFP localization and purification)-tagged CENP-C, CENP-M, CENP-N, or CENP-T. Cells were preextracted and fixed 72 h after transfection. The presence of CENP-A was assessed using antibodies against endogenous CENP-A. Insets show the arrays at a higher magnification. (B) LacI control images for LAP-CENP-C–T, indicating that recruitment is never observed in the absence of CENP-A. (C) Graph showing the percentage of doubly transfected (GFP and LacI-HJURP) U2OS LacO/TRE cells with endogenous CENP-A present at the array, which also recruited the indicated constitutive centromere proteins (≥30 cells per condition; error bars represent standard deviation). Bars, 5 µm.
Ectopic centromeres formed by HJURP act as kinetochores during mitosis
The stable association of CENP-A at the array and the subsequent recruitment of constitutive centromere proteins prompted us to investigate whether mitosis-specific kinetochore proteins would also be recruited to the array. To address this, we immunostained mitotic chromosome spreads for the microtubule-binding kinetochore protein NDC80 and observed its recruitment to the LacO array in 40% of cells transfected with LacI-HJURPScm3 (Fig. 3 A). Endogenous centromeres exhibit a unique morphology in mitotic chromosomes called a constriction. In the LacI-HJURPScm3–transfected cells in which NDC80 is recruited to the array, we observed a second region of constriction at the array in addition to the constriction present at the endogenous centromere consistent with the array acting as a functional kinetochore.
Recruitment of CENP-A by HJURPScm3 mediates kinetochore formation at the LacO array. (A) Mitotic chromosome spreads from U20S-LacO/TRE cells transfected with LacI or LacI-HJURPScm3, arrested in nocodazole, and stained with antibodies for NDC80. 40% of LacI-HJURP arrays recruited NDC80. (B) Monastrol-arrested cells transfected with LacI or LacI-HJURPScm3 and immunostained for centromere marker CENP-T. Radial distribution plots describe the mean centromere position (black circle) in the cells measured (>26 cells per condition) relative to the center of the DNA mass. The array position is diagrammed relative to the center of the DNA mass as blue triangles (LacI) or red diamonds (LacI-HJURPScm3). The gray circle represents one standard deviation from the mean centromere position. The LacI-HJURPScm3 array falls within the centromere region in 69% of transfected cells versus 15% for LacI controls. (C) Selective stabilization of kinetochore-bound microtubules through cold treatment demonstrates the LacI-HJURPScm3 arrays form stable microtubule interactions similar to endogenous centromeres. Insets show magnified views of the boxed region. (D) LacO-SceI-TRE NIH3T3 cells were transfected with YFP–histone H2B and followed by live-cell imaging as they progress through mitosis. Times are given relative to the last frame when cells were in metaphase. Arrows indicate the array, and asterisks indicate nonchromatin-bound unspecific LacI staining. (E) Insets taken from images in D show the behavior of the array (red in merge) and YFP-H2B (green in merge) for (1 and 2) LacI-HJURPScm3 at 6 and 9 min into anaphase, respectively. Bars: (A–D) 5 µm; (E) 2 µm.
Recruitment of CENP-A by HJURPScm3 mediates kinetochore formation at the LacO array. (A) Mitotic chromosome spreads from U20S-LacO/TRE cells transfected with LacI or LacI-HJURPScm3, arrested in nocodazole, and stained with antibodies for NDC80. 40% of LacI-HJURP arrays recruited NDC80. (B) Monastrol-arrested cells transfected with LacI or LacI-HJURPScm3 and immunostained for centromere marker CENP-T. Radial distribution plots describe the mean centromere position (black circle) in the cells measured (>26 cells per condition) relative to the center of the DNA mass. The array position is diagrammed relative to the center of the DNA mass as blue triangles (LacI) or red diamonds (LacI-HJURPScm3). The gray circle represents one standard deviation from the mean centromere position. The LacI-HJURPScm3 array falls within the centromere region in 69% of transfected cells versus 15% for LacI controls. (C) Selective stabilization of kinetochore-bound microtubules through cold treatment demonstrates the LacI-HJURPScm3 arrays form stable microtubule interactions similar to endogenous centromeres. Insets show magnified views of the boxed region. (D) LacO-SceI-TRE NIH3T3 cells were transfected with YFP–histone H2B and followed by live-cell imaging as they progress through mitosis. Times are given relative to the last frame when cells were in metaphase. Arrows indicate the array, and asterisks indicate nonchromatin-bound unspecific LacI staining. (E) Insets taken from images in D show the behavior of the array (red in merge) and YFP-H2B (green in merge) for (1 and 2) LacI-HJURPScm3 at 6 and 9 min into anaphase, respectively. Bars: (A–D) 5 µm; (E) 2 µm.
Cells were treated with the Eg5 inhibitor Monastrol to demonstrate that the array behaves similarly to endogenous centromeres during mitosis. Under these conditions, cells fail to separate their spindle poles during prometaphase and arrest in mitosis with their centromeres oriented toward the central pole and the telomeres toward the periphery. The array of the U2OS cells is incorporated near the telomere of chromosome 1 (Janicki et al., 2004) and is located away from the central cluster of centromeres, as identified by CENP-T staining in control cells (LacI transfected; Fig. 3 B). However, when LacI-HJURPScm3 is expressed, we observe the array closer to the center of the monopolar mitotic structure, consistent with the array acting as a centromere and binding microtubules (LacI-HJURPScm3 transfected; Fig. 3 B).
We predicted that cells in which ectopic centromeres were successfully assembled at the LacO array should form stable microtubule attachments and cause errors in chromosome segregation caused by the presence of two active centromeres on a single chromosome. As expected, cold-stabilized microtubules, characteristic of kinetochore fibers (Brinkley and Cartwright, 1975), were observed to terminate at several LacO arrays assembled by LacI-HJURPScm3 in metaphase cells (Fig. 3 C). Consistent with an interaction between microtubules of the mitotic spindle and the LacO array, cells transfected with LacI-HJURPScm3 exhibited lagging chromosomes during anaphase (Fig. 3 D, Video 1, and Video 2). The lagging chromosomes contained the LacO array (Fig. 3 E), which suggests that the array of each chromosome is interacting with microtubules emanating from both spindle poles in a merotelic-like arrangement. These data support our findings that HJURP-assembled ectopic centromeres at the array can mimic centromeric CENP-A nucleosomes through their ability to recruit CENP-ANAC proteins and assemble a functioning kinetochore.
Chromosome segregation of an NIH3T3-LacO cell expressing LacI. The NIH3T3-LacO YFP–CENP-A stable line was transfected for 48 h with LacI (red) and YFP-H2B (green). Images were analyzed using a DeltaVision deconvolution microscope equipped with a WeatherStation environmental chamber maintained at 37°C. Frames were taken every 3 min for 12 min. Bar, 15 μm.
Chromosome segregation of an NIH3T3-LacO cell expressing LacI. The NIH3T3-LacO YFP–CENP-A stable line was transfected for 48 h with LacI (red) and YFP-H2B (green). Images were analyzed using a DeltaVision deconvolution microscope equipped with a WeatherStation environmental chamber maintained at 37°C. Frames were taken every 3 min for 12 min. Bar, 15 μm.
Chromosome segregation of an NIH3T3-LacO cell expressing LacI-HJURPScm3. NIH3T3-LacO YFP–CENP-A stable line was transfected for 48 h with LacI-HJURPScm3 (red) and YFP-H2B (green). Images were analyzed using a DeltaVision deconvolution microscope equipped with a WeatherStation environmental chamber maintained at 37°C. Frames were taken every 3 min for 12 min. Bar, 15 μm.
Chromosome segregation of an NIH3T3-LacO cell expressing LacI-HJURPScm3. NIH3T3-LacO YFP–CENP-A stable line was transfected for 48 h with LacI-HJURPScm3 (red) and YFP-H2B (green). Images were analyzed using a DeltaVision deconvolution microscope equipped with a WeatherStation environmental chamber maintained at 37°C. Frames were taken every 3 min for 12 min. Bar, 15 μm.
In vitro assembly of CENP-A nucleosomes by HJURPScm3
It is unclear whether HJURP is required only to stabilize and deliver prenucleosomal CENP-A to centromeres or whether HJURP is also actively involved in the assembly of CENP-A nucleosomes at the centromere. Based on the ability of HJURP to drive the stable association of CENP-A at the LacO array, we hypothesized that HJURP plays an active role in the deposition of CENP-A nucleosomes at centromeres. We assessed the ability of HJURP to assemble CENP-A–containing nucleosomes in an in vitro chromatin assembly assay using purified recombinant proteins independent of other assembly factors. The assembly of nucleosomes in this assay is assessed on closed circular plasmid DNA by monitoring the formation of topoisomers (Lusser and Kadonaga, 2004).
Because the Scm3 domain of HJURP is able to bind CENP-A and we demonstrated the stable association of CENP-A at the LacO array in response to LacI-HJURPScm3 expression, we reasoned that this fragment of HJURP should be sufficient to assemble CENP-A nucleosomes. Maltose-binding protein (MBP)–tagged HJURPScm3 containing amino acids 1–208 was expressed in bacteria and purified (Fig. S3 A). As hypothesized, when incubated with CENP-A and histones H4, H2A, and H2B, HJURPScm3 was indeed able to assemble CENP-A nucleosomes as indicated by the accumulation of faster migrating topoisomers (Fig. 4 A). The accumulation of fully supercoiled plasmids increased with increasing the amounts of HJURPScm3 (Fig. 4, A–C). HJURP-assembled CENP-A nucleosomes protected the expected ∼150 bp of DNA after micrococcal nuclease digest (Fig. 4 D).
HJURPScm3 is sufficient to assemble CENP-A nucleosomes in vitro. (A) Plasmid supercoiling assays were conducted using recombinant MBP-tagged HJURPScm3 and recombinant CENP-A octamer (including histones H4, H2A, and H2B) or histone H3.1 octamer to assess the relative ability of HJURP to assemble CENP-A– and H3.1-containing nucleosomes. The relaxed DNA lane contains topoisomerase-treated supercoiled (S.C.) plasmid DNA. HJURPScm3 induced supercoiling more efficiently in the presence of CENP-A relative to H3.1. (B) Line scans across topoisomers within conditions presented in A. Lines indicate the least supercoiled topoisomers. Boxes indicate the location of the maximally assembled topoisomers. (C) Assembly reactions from A containing H3.1 and CENP-A are graphed here as fold intensity over reactions containing no HJURPScm3. Error bars show standard deviations. (D) Assembly reactions in A (using HJURPScm3) or assembly reactions using NPM1 digested with micrococcal nuclease to show DNA protection of the assembled species. A dotted line was drawn to indicate the migration of a 200-bp fragment.
HJURPScm3 is sufficient to assemble CENP-A nucleosomes in vitro. (A) Plasmid supercoiling assays were conducted using recombinant MBP-tagged HJURPScm3 and recombinant CENP-A octamer (including histones H4, H2A, and H2B) or histone H3.1 octamer to assess the relative ability of HJURP to assemble CENP-A– and H3.1-containing nucleosomes. The relaxed DNA lane contains topoisomerase-treated supercoiled (S.C.) plasmid DNA. HJURPScm3 induced supercoiling more efficiently in the presence of CENP-A relative to H3.1. (B) Line scans across topoisomers within conditions presented in A. Lines indicate the least supercoiled topoisomers. Boxes indicate the location of the maximally assembled topoisomers. (C) Assembly reactions from A containing H3.1 and CENP-A are graphed here as fold intensity over reactions containing no HJURPScm3. Error bars show standard deviations. (D) Assembly reactions in A (using HJURPScm3) or assembly reactions using NPM1 digested with micrococcal nuclease to show DNA protection of the assembled species. A dotted line was drawn to indicate the migration of a 200-bp fragment.
General histone chaperones are often promiscuous in their ability to bind and assemble histones into nucleosomes as exemplified by dNAP (Drosophila melanogaster nucleosome assembly protein), which is capable of directly interacting with all four core histones and has been shown to assemble both histone H3 and CENP-A chromatin (Figs. 5 A and S3 B; Yoda et al., 2000; Park and Luger, 2006). We next examined whether the assembly activity of MBP-HJURPScm3 was specific for CENP-A–containing nucleosomes. HJURPScm3 was not able to efficiently assemble nucleosomes when CENP-A was replaced with histone H3.1 (Fig. 4, A–C), which is consistent with the inability of HJURP to bind histone H3–H4 (Foltz et al., 2009) and supporting its role as a CENP-A–specific assembly factor. Although the degree of assembly of CENP-A nucleosomes correlates with the amount of HJURPScm3 present in the reaction, the limited amount of supercoiling observed in H3 assembly reactions did not increase as increasing amounts of HJURPScm3 were titrated into the reactions (Fig. 4 C). This suggests that the limited degree of supercoiling observed with histone H3 does not reflect chaperone-mediated assembly. Together, these data support the hypothesis that HJURP is a CENP-A–specific chromatin assembly factor possessing the intrinsic ability to deposit CENP-A nucleosomes into DNA.
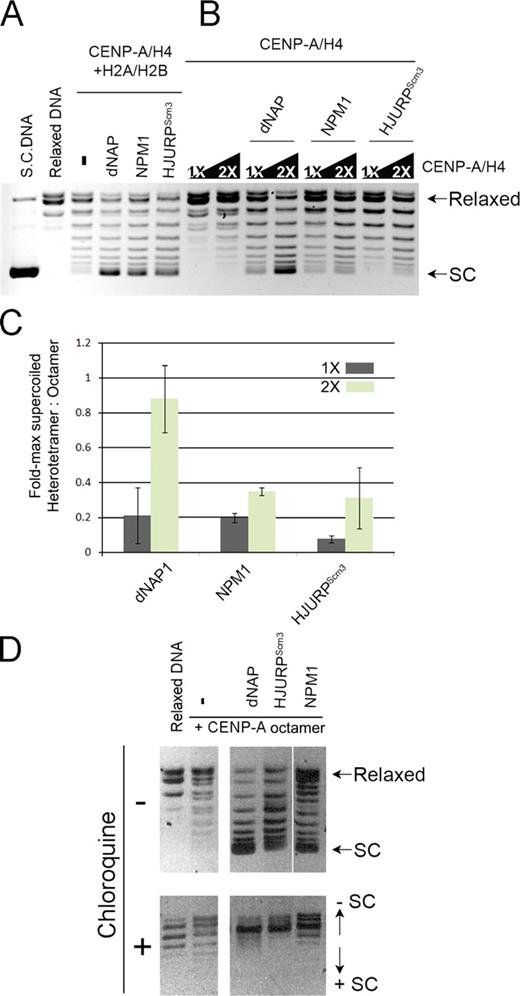
HJURPScm3-assembled CENP-A nucleosomes are negatively supercoiled and contain H2A and H2B. (A and B) Supercoiling assay comparing assembly efficiencies of chaperones dNAP, NPM1, and HJURPScm3 with CENP-A histone octamers (CENP-A–H4 and H2A/H2B) in A or with CENP-A–H4 alone in B. CENP-A–H4 levels added to the reactions were varied from 1 to 2× compared with the amount of CENP-A–H4 present in the reactions in A. Line scans are presented in Fig. S3. SC, supercoiled. (C) Integrated intensities of maximally supercoiled populations were measured from reactions in A and B. Values are graphed as fold-maximally supercoiled heterotetramer to octamer. Error bars show standard deviations. (D) Supercoiling assay showing assembly activities (top) for dNAP, HJURPScm3, and NPM1. Supercoiled DNA was separated by agarose gel electrophoresis with (bottom) or without (top) the DNA intercalating agent chloroquine to distinguish negatively and positively supercoiled DNA. The minus signs indicate no addition of chaperone. The white line indicates that intervening lanes have been spliced out.
HJURPScm3-assembled CENP-A nucleosomes are negatively supercoiled and contain H2A and H2B. (A and B) Supercoiling assay comparing assembly efficiencies of chaperones dNAP, NPM1, and HJURPScm3 with CENP-A histone octamers (CENP-A–H4 and H2A/H2B) in A or with CENP-A–H4 alone in B. CENP-A–H4 levels added to the reactions were varied from 1 to 2× compared with the amount of CENP-A–H4 present in the reactions in A. Line scans are presented in Fig. S3. SC, supercoiled. (C) Integrated intensities of maximally supercoiled populations were measured from reactions in A and B. Values are graphed as fold-maximally supercoiled heterotetramer to octamer. Error bars show standard deviations. (D) Supercoiling assay showing assembly activities (top) for dNAP, HJURPScm3, and NPM1. Supercoiled DNA was separated by agarose gel electrophoresis with (bottom) or without (top) the DNA intercalating agent chloroquine to distinguish negatively and positively supercoiled DNA. The minus signs indicate no addition of chaperone. The white line indicates that intervening lanes have been spliced out.
Because both HJURP and NPM1 (nucleophosmin 1) are consistently copurified with prenucleosomal CENP-A and NPM1 has been shown previously to act as a histone chaperone for histone H3 nucleosomes (Okuwaki et al., 2001), we sought to determine whether NPM1 might contribute to CENP-A nucleosome deposition (Frehlick et al., 2007; Dunleavy et al., 2009; Foltz et al., 2009; Shuaib et al., 2010). NPM1 was expressed and purified from bacteria (Fig. S3 A) and eluted from a size exclusion column as a pentamer (Okuwaki et al., 2001). Like dNAP and HJURPScm3, NPM1 was also able to assemble CENP-A–containing nucleosomes onto plasmid DNA (Figs. 4 D and 5, A and D). When NPM1 and HJURP were both present in the assay, we did not observe an increase in assembly efficiency (unpublished data).
Cse4 in S. cerevisiae is reported to form a centromeric subnucleosomal particle with histone H4 that lacks histones H2A and H2B and may include Scm3 (the homologue of HJURP; Mizuguchi et al., 2004; Stoler et al., 2007; Williams et al., 2009). Subnucleosomal H3–H4 heterotetramer complexes can be deposited onto DNA templates, resulting in supercoiling (Peterson et al., 2007), and recently, Shuaib et al. (2010) demonstrated the ability of HJURP to deposit a single CENP-A–H4 heterotetramer into DNA. We determined whether HJURPScm3 was able to assemble extended arrays of CENP-A–H4 heterotetramers into plasmid DNA and to compare the extent of supercoiling induced by heterotetramer assembly relative to octameric CENP-A assembly. We observed plasmid supercoiling around CENP-A–H4 heterotetramers in response to all three assembly factors tested; although, in each case, the degree of supercoiling was less than that observed with the CENP-A octamers, which included H2A and H2B (Figs. 5, A–C; and S3 C). When the amount of CENP-A–H4 heterotetramer was increased by twofold in the reaction over the amount in the octamer assemblies, dNAP became as efficient as in the presence of all four histones. However, NPM1 and HJURPScm3 continued to yield a lesser degree of plasmid supercoiling around the CENP-A–H4 heterotetramer (Fig. 5, A–C). These observations are consistent with either a lesser activity of HJURPScm3 for the assembly of CENP-A–containing heterotetramers or with a decreased degree of supercoiling per heterotetramer.
It has been reported that CENP-A–containing nucleosomes positively supercoil DNA in Drosophila and S. cerevisiae, in contrast to the negative supercoiling produced by canonical histone H3-containing nucleosomes (Furuyama and Henikoff, 2009). We separated HJURPScm3-induced topoisomers in the presence of chloroquine, an intercalating drug that allows for the detection of both positive and negative supercoiling by a shift in the topoisomers. Positively supercoiled DNA will shift toward faster migrating species in the presence of chloroquine, and negatively supercoiled DNA will shift toward more slowly migrating species (Lusser and Kadonaga, 2004). MBP-HJURPScm3–assembled CENP-A–containing nucleosomes induced negative supercoils as indicated by the upward shift in the gel (Fig. 5 D, bottom), contrary to what has been observed for the Drosophila CID (centromere identifier) and budding yeast Cse4 nucleosomes. These data suggest human CENP-A nucleosomes assembled by their native chaperone include histones H2A and H2B and are wrapped in a left-handed direction, similar to canonical H3.1-containing nucleosomes.
HJURP centromeric localization is dependent on the Mis18 complex at centromeres
HJURP and the Mis18 complex are both required for the recruitment of newly synthesized CENP-A to the centromere; however, the function of the Mis18 complex in CENP-A assembly remains unclear. It was not known whether Mis18 and HJURP are required for independent events in CENP-A deposition or whether the action of the Mis18 complex is required for HJURP recruitment. To address this question, Mis18α, Mis18BP1hsKNL2, or HJURP were knocked down by siRNA treatment in HeLa cell lines that stably express either GFP-tagged Mis18α or GFP-tagged HJURP. The GFP-tagged proteins behave similarly to endogenous proteins, as the proportion of cells with centromeric GFP-Mis18α or GFP-HJURP increases significantly when we enrich for cells in early G1 (Fig. S4).
Knockdown of Mis18α by siRNA reduced GFP-Mis18α protein levels to below 25% of mock-treated levels after 48 h (Fig. 6 A). HJURP siRNA reduced endogenous HJURP to below 25% of normal cellular levels while also substantially decreasing the level of the GFP-tagged protein (Fig. 6 B). Centromeric localization of GFP-tagged Mis18α was abolished after Mis18α siRNA treatment (Fig. 6, C and D). Antibodies that recognize CENP-T were used to identify centromeres. Treating cells with siRNA against Mis18BP1hsKNL2 did not lower the protein level of the exogenous GFP-Mis18α (Fig. 6 A) but did abolish Mis18α localization at centromeres (Fig. 6, C and D) consistent with previous findings (Fujita et al., 2007).
Recruitment of HJURP to centromeres requires the Mis18 complex. (A and B) Cellular extracts from siRNA-treated and control cell lines were analyzed by Western blotting using anti-GFP (A) or anti-HJURP antibodies (B). Each lane contains lysate from 105 cells. Dilution series were generated from mock-treated HeLa GFP-Mis18α (A) or parental HeLa (B) cells. (C) Stable GFP-Mis18α cells lines were treated with siRNA against Mis18α, Mis18BP1hsKNL2, HJURP, or GAPDH (control). Representative images of siRNA-treated GFP-Mis18α cells were selected in which a midbody was clearly present (differential interference contrast [DIC], arrows) to show the cell was in early G1. DAPI staining was overlaid onto the differential interference contrast image. Cells were stained with anti–CENP-T. (D) Mean percentage of GFP-Mis18α centromere-positive nuclei from a population of ≥57 cells in each siRNA treatment from two experiments. (E) Similar image acquisition as in C. Here, stable HeLa GFP-HJURP cells were treated with the same siRNAs. (F) Mean percentage of GFP-HJURP centromere-positive nuclei from a population of ≥135 cells in each siRNA treatment from two experiments. Error bars show standard deviations. Insets show magnified views of boxed regions. Bars, 5 µm.
Recruitment of HJURP to centromeres requires the Mis18 complex. (A and B) Cellular extracts from siRNA-treated and control cell lines were analyzed by Western blotting using anti-GFP (A) or anti-HJURP antibodies (B). Each lane contains lysate from 105 cells. Dilution series were generated from mock-treated HeLa GFP-Mis18α (A) or parental HeLa (B) cells. (C) Stable GFP-Mis18α cells lines were treated with siRNA against Mis18α, Mis18BP1hsKNL2, HJURP, or GAPDH (control). Representative images of siRNA-treated GFP-Mis18α cells were selected in which a midbody was clearly present (differential interference contrast [DIC], arrows) to show the cell was in early G1. DAPI staining was overlaid onto the differential interference contrast image. Cells were stained with anti–CENP-T. (D) Mean percentage of GFP-Mis18α centromere-positive nuclei from a population of ≥57 cells in each siRNA treatment from two experiments. (E) Similar image acquisition as in C. Here, stable HeLa GFP-HJURP cells were treated with the same siRNAs. (F) Mean percentage of GFP-HJURP centromere-positive nuclei from a population of ≥135 cells in each siRNA treatment from two experiments. Error bars show standard deviations. Insets show magnified views of boxed regions. Bars, 5 µm.
HJURP siRNA completely eliminated centromeric localization of the GFP-tagged HJURP protein after 48 h (Fig. 6, E and F). In contrast, siRNA knockdown of HJURP did not alter the localization pattern of GFP-Mis18α to centromeres as compared with the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) control nor did it decrease the protein level of GFP-Mis18α (Fig. 6, A, C, and D). Importantly, siRNA against either Mis18α or Mis18BP1hsKNL2 eliminated HJURP recruitment to centromeres (Fig. 6, E and F) while not affecting the level of endogenous or GFP-tagged HJURP (Fig. 6 B). Therefore, the Mis18 complex is required for HJURP recruitment to centromeres, suggesting that the lack of CENP-A deposition in Mis18 knockdown experiments results from the inability to recruit HJURP (Fujita et al., 2007; Maddox et al., 2007).
To determine whether the Mis18 complex is also required for the function of HJURP in recruiting and stabilizing CENP-A, we performed siRNA knockdown of Mis18BP1hsKNL2 in the LacO-containing U2OS cells. Mis18BP1hsKNL2 protein was reduced to <90% of endogenous levels by siRNA treatment of U20S cells as indicated by immunoblotting and by the loss of centromeric CENP-A (Fig. 7, A and B). The stability of endogenous CENP-A at the LacO array in LacI-HJURPScm3–transfected cells was unaffected by Mis18BP1hsKNL2 depletion after IPTG treatment (Fig. 7 A). Similar numbers of CENP-A–containing arrays were observed in GAPDH and Mis18BP1hsKNL2 siRNA-treated cells after treatment with IPTG (Fig. 7 C). We conclude that the requirement for the Mis18 complex can be bypassed by directly targeting HJURP to DNA.
The Mis18 complex is not required for CENP-A deposition at the LacO/TRE array. (A) Representative images of endogenous CENP-A recruitment in U2OS-LacO cells treated with 15 mM IPTG after 72 h of either GAPDH or Mis18BP1hsKNL2 siRNA treatment. Cells had been transiently transfected with LacI-HJURPScm3 and GFP-TetR 48 h before fixation. Cells were transfected after an initial 24 h siRNA treatment to ensure Mis18BP1hsKNL2 depletion before CENP-A establishment at the array. Insets show magnified views of boxed regions. Bar, 5 µm. (B) Cellular extracts from GAPDH and Mis18BP1hsKNL2 siRNA-treated cells were analyzed by Western blotting using an anti-Mis18BP1hsKNL2 antibody. Each lane contains lysate from 107 cells. (C) Quantification of CENP-A staining at the LacO/TRE array marked by GFP-TetR after 72 h of GAPDH or Mis18BP1hsKNL2 siRNA treatment and 1 h of 15 mM IPTG treatment. At least 30 cells per condition were analyzed; n = 2. Error bars represent the standard deviation between the two experiments. The p-value between GAPDH and Mis18BP1hsKNL2 is 0.3609.
The Mis18 complex is not required for CENP-A deposition at the LacO/TRE array. (A) Representative images of endogenous CENP-A recruitment in U2OS-LacO cells treated with 15 mM IPTG after 72 h of either GAPDH or Mis18BP1hsKNL2 siRNA treatment. Cells had been transiently transfected with LacI-HJURPScm3 and GFP-TetR 48 h before fixation. Cells were transfected after an initial 24 h siRNA treatment to ensure Mis18BP1hsKNL2 depletion before CENP-A establishment at the array. Insets show magnified views of boxed regions. Bar, 5 µm. (B) Cellular extracts from GAPDH and Mis18BP1hsKNL2 siRNA-treated cells were analyzed by Western blotting using an anti-Mis18BP1hsKNL2 antibody. Each lane contains lysate from 107 cells. (C) Quantification of CENP-A staining at the LacO/TRE array marked by GFP-TetR after 72 h of GAPDH or Mis18BP1hsKNL2 siRNA treatment and 1 h of 15 mM IPTG treatment. At least 30 cells per condition were analyzed; n = 2. Error bars represent the standard deviation between the two experiments. The p-value between GAPDH and Mis18BP1hsKNL2 is 0.3609.
Discussion
Determination of the site of centromere assembly is an epigenetic process that depends on the Mis18 complex and the CENP-A–specific chaperone HJURP during early G1. Here, we have demonstrated that recruitment of HJURP to the centromere requires the Mis18 complex. By targeting HJURP to an ectopic location using the LacI/LacO system, we have bypassed the need for Mis18-mediated centromere recruitment of HJURP to assess the role of HJURP in CENP-A deposition independently of the context of the endogenous centromere. Targeting HJURP to the LacO array demonstrates that the recruitment of HJURP is the process that determines the site of CENP-A nucleosome deposition and, subsequently, the assembly of the associated constitutive centromere and mitotic kinetochore (Fig. 8). Furthermore, we have determined that HJURP is directly responsible for CENP-A nucleosome assembly in vitro. Together, these data demonstrate that HJURP recognizes the centromere through the action of the Mis18 complex and that HJURP’s intrinsic ability to assemble CENP-A nucleosomes is an integral step in the epigenetic mechanism by which centromeres are stably propagated.
HJURP recruitment determines centromere position. (A) HJURP is recruited to centromeres through the action of Mis18. During G1, HJURP directly assembles CENP-A nucleosomes at centromeres along with histones H2A and H2B. The recruitment of HJURP is the critical step in determining the site of the centromere. (B) Redirecting HJURP to an integrated LacO array bypasses the requirement for Mis18, results in deposition of CENP-A, and is sufficient to form a functional kinetochore.
HJURP recruitment determines centromere position. (A) HJURP is recruited to centromeres through the action of Mis18. During G1, HJURP directly assembles CENP-A nucleosomes at centromeres along with histones H2A and H2B. The recruitment of HJURP is the critical step in determining the site of the centromere. (B) Redirecting HJURP to an integrated LacO array bypasses the requirement for Mis18, results in deposition of CENP-A, and is sufficient to form a functional kinetochore.
The CENP-A nucleosome is independently recognized by two components of the CENP-ANAC/CCAN, CENP-N and CENP-C (Carroll et al., 2009, 2010), and both of these proteins are recruited to the LacO arrays containing CENP-A. In vitro, CENP-N is unable to bind the CENP-A–H4 heterotetramer but uniquely recognizes the CENP-A–containing nucleosome assembled with histone H4, H2A, and H2B. The recruitment of CENP-N to the LacO array indicates the presence of assembled CENP-A nucleosomes. The CENP-T–W complex is localized in close proximity to the CENP-A nucleosome at centromeres, although it has been proposed to interact directly with histone H3 chromatin at the centromere (Foltz et al., 2006; Hori et al., 2008). We observed CENP-T recruitment to the array, suggesting the incorporation of CENP-A is able to organize the array relative to surrounding H3 nucleosomes in a way that reflects the arrangements present at endogenous centromeres. Recently, it has been demonstrated that recruitment of CENP-T and CENP-C are sufficient to build an ectopic centromere using the LacO system (Gascoigne et al., 2011), which is consistent with our work showing recruitment of CENP-T and CENP-C to the CENP-A–containing arrays that go on to assemble functional kinetochores.
Data examining the ability of CENP-A deposition to drive centromere and kinetochore formation have been somewhat mixed. In human cell lines, overexpression of CENP-A can lead to incorporation of CENP-A throughout the chromatin with an accompanying relocalization of CENP-C; however, these regions do not support kinetochore formation during mitosis (Van Hooser et al., 2001). Recruitment of CENP-A to the LacO array is successful at reconstituting centromere and kinetochore activity. This may be caused by an enrichment of CENP-A at the LacO arrays relative to that deposited into general chromatin by overexpression. Alternatively, the kinetochore activity of the LacO array may reflect a contribution of HJURP-mediated assembly on the CENP-A nucleosomes at the array, as opposed to overexpression-induced chromatin incorporation of CENP-A, which may occur through HJURP-independent mechanisms.
The overexpression of CID in Drosophila was more successful in recapitulating centromere and kinetochore formation than overexpression in human cell lines. In this case, CID overexpression resulted in its accumulation throughout the chromatin and the assembly of centromere-like regions that recruited CENP-C and mediated microtubule attachments. These ectopic centromeres were restricted to a subset of sites within the chromatin (Heun et al., 2006). In our experiments in which CENP-A deposition is restricted to a single location along the chromosome, we observed that not all cells recruited the constitutive centromere proteins and assembled kinetochores at the CENP-A–assembled array. The restricted recruitment of CENP-ANAC proteins to the array may reflect differences in the state of the underlying chromatin. Transcriptional activity or modification state of chromatin can negatively impact centromere formation in human artificial chromosomes (Nakano et al., 2008; Bergmann et al., 2011) and may inhibit the ability of HJURP to deposit CENP-A, acting as a way to avoid spurious CENP-A deposition into noncentromeric loci. Alternatively, a stepwise maturation of the CENP-A–containing region toward a functional centromere may require multiple cell cycles and depend on cell cycle–specific assembly of centromere proteins. Likewise, additional chromatin-remodeling steps after CENP-A nucleosome deposition mediated by RSF1 or MGCRacGAP could influence both long-term stability of CENP-A nucleosomes and the assembly of the constitutive centromere (Perpelescu et al., 2009; Lagana et al., 2010).
Histone H3 variants partner with distinct chaperone complexes to facilitate their different temporal and spatial incorporation within the genome. Histone H3.1 nucleosome incorporation is coupled to DNA synthesis through an interaction between the p150 subunit of the chromatin assembly factor complex (CAF1) and proliferating cell nuclear antigen (Shibahara and Stillman, 1999; Moggs et al., 2000). The assembly of H3.3-containing nucleosomes occurs independently of DNA synthesis and is accomplished through HIRA, ATRX, and DAXX (Tagami et al., 2004; Drané et al., 2010; Goldberg et al., 2010; Wong et al., 2010). Here, we demonstrate the activity of the CENP-A chaperone/assembly factor HJURP is coupled to the centromere through the recruitment by the Mis18 complex. S. pombe contains a single Mis18 protein that directly interacts with Scm3 (Pidoux et al., 2009). However, HJURPScm3, which contains the regions of significant homology between Scm3 and HJURP, does not demonstrate centromeric localization on its own, suggesting the mechanism of HJURP centromeric recruitment in vertebrates is different from Scm3 recruitment in yeast, possibly through a priming mechanism as previously proposed (Fujita et al., 2007).
Several models for the composition of the CENP-A nucleosome have been proposed, including tetrameric forms that contain a single copy of each histone as well as hexameric forms that lack H2A and H2B but incorporate the yeast HJURP homologue Scm3. In human cells, HJURP is present at centromeres during early G1 when new CENP-A nucleosomes are being actively recruited. Similar to a previous study using full-length HJURP (Shuaib et al., 2010), HJURPScm3 is able to assemble CENP-A–H4 heterotetramers into DNA to some degree (Fig. 5); however, the assembly mediated by HJURPScm3 is processive and maximally efficient in the presence of histones H2A and H2B. The assembled structure protects ∼145 bp of DNA. These data suggest human HJURP assembles an octameric nucleosome. Consistent with a centromeric nucleosome containing two copies of CENP-A, disruption of the CENP-A dimerization interface precludes the ability of CENP-A to accumulate at centromeres (Camahort et al., 2009; Sekulic et al., 2010). After deposition by HJURP, further remodeling by additional factors may mediate the conversion of the centromeric nucleosome into the hexameric and/or tetrameric forms observed by others (Dalal et al., 2007; Mizuguchi et al., 2007; Furuyama and Henikoff, 2009; Dimitriadis et al., 2010).
In addition to HJURP, several other chromatin assembly factors have been implicated in the assembly of the human centromeres. NPM1 is also able to assemble CENP-A (Fig. 5) and histone H3 nucleosomes (Okuwaki et al., 2001). NPM1 is associated with the CENP-A–HJURP prenucleosomal complex (Dunleavy et al., 2009; Foltz et al., 2009; Shuaib et al., 2010). The exact role of NPM1 in CENP-A deposition remains somewhat unclear, but it is reasonable to suppose that its assembly activity for CENP-A may play a role in centromere activity given its presence in the CENP-A prenucleosomal complex and at the centromere. In addition, three previously described chromatin-remodeling complexes, FACT (facilitates chromatin transcription), CHD1, and RSF (remodeling and spacing factor) are also associated with the centromere and are important for the stable assembly of CENP-A–containing chromatin (Obuse et al., 2004; Foltz et al., 2006; Okada et al., 2009; Perpelescu et al., 2009). However, CHD1 appears to be dispensable in Drosophila for CID deposition (Podhraski et al., 2010). The activity of RSF1 appears to be restricted to mid-G1 well after new CENP-A nucleosome deposition by HJURP, which immediately follows mitosis (Jansen et al., 2007; Perpelescu et al., 2009). The period of RSF1 recruitment may represent a remodeling event whereby the centromere is reorganized in preparation for the ensuing S phase and mitosis.
Budding yeast and higher eukaryotes use Scm3/HJURP proteins for the common purpose of depositing CENP-A nucleosomes but use disparate mechanisms to determine the site of recruitment (Shivaraju et al., 2011). In the case of budding yeast, the DNA sequence defines the site of CENP-A deposition, whereas in higher eukaryotes, the site of new CENP-A incorporation is influenced by the location of the preexisting CENP-A nucleosomes. Epigenetic inheritance requires the recruitment of HJURP through the activity of the Mis18 complex dictating the site of CENP-A nucleosome deposition.
Materials and methods
siRNA treatment
HeLa cell lines expressing either GFP-Mis18α or GFP-HJURP were plated at 8 × 105 cells in 6-well plates. The next day, cells were transfected with 5 nM Silencer Select siRNAs (Invitrogen) using RNAiMAX (Invitrogen). siRNA sequences were as follows: Mis18α, 5′-GAAGAUGUCUUGAAAGCAUTT-3′; Mis18BP1hsKNL2 (C14orf106), 5′-GGAUAUCCAAAUUAUCUCATT-3′; and HJURP, 5′-CAAGUAUGGAAGUUCGAUATT-3′. The next day, one third of the plating volume of DME with 10% heat-inactivated FBS was added. For Western blot analysis, cells were harvested 48 h after siRNA treatment with PBS + 3 mM EDTA and counted, and whole-cell lysates were made in SDS-PAGE sample buffer. Lysates from 105 cells per lane were separated on 10% SDS-PAGE gel and transferred to nitrocellulose. Blots were incubated in primary anti-GFP or anti-HJURP (Foltz et al., 2009) antibody overnight at 4°C and in secondary (Jackson ImmunoResearch Laboratories, Inc.) for 1 h at 4°C. For U2OS-LacO cells, they were plated at 8 × 104 cells in 24-well plates onto poly-lysine coverslips. They were transfected after 24 h with 5 nM Silencer Select siRNAs (same sequence as mentioned previously in this paragraph for Mis18BP1hsKNL2 siRNA). After 24 h in siRNA, cells were transiently transfected with mCherry-LacI-HJURPScm3 and GFP-TetR using Effectene (QIAGEN) into the media still containing the siRNA. After 8 h of transfection, the media were removed, and cells were retreated with siRNA. Cells were then incubated for 48 h and treated with 15 mM IPTG (Sigma-Aldrich) for 1 h before preextraction, fixation, and staining for endogenous CENP-A as described in the following section.
Cell culture, transfections, and immunocytochemistry
HeLa or U2OS-LacO-TRE (a gift from S. Janicki, Wistar Institute, Philadelphia, PA) cells were plated to poly-lysine–coated coverslips at 1.6 × 105 cells per well in 6-well or 0.8 × 105 cells per well in 24-well plates, respectively. Cells were then transfected 24 h later with 0.2–0.25 µg plasmid DNA (24-well plate) or 1 µg (6-well plate) using GeneJuice (EMD) or Effectene. LacO-SceI-TRE NIH3T3 cells (a gift from T. Misteli, National Cancer Institute, Bethesda, MD) were transfected with Lipofectamine 2000 (Invitrogen). Live-cell imaging was conducted in the NIH3T3-LacO cell line at 37°C in Leibovitz L15 media including 10% FBS after 48 h of transfection. Single-plane images were collected at 1-min intervals on a microscope (Deltavision; Applied Precision) equipped with an environmental chamber (WeatherStation; Applied Precision) maintained at 37°C.
HeLa cells were preextracted with 0.1% Triton X-100 in PBS for 3 min, fixed with 4% formaldehyde in PBS for 10 min, and then quenched by the addition of 100 mM Tris, pH 7.5, for another 10 min at room temperature. HeLa cells were blocked in 2% FBS and 2% BSA in 0.1% Triton X-100–PBS. U2OSLacO/TETr cells were treated with 0 or 10 mM IPTG for 1 h in DME GlutaMAX media before fixation and then preextracted in PHEM buffer (60 mM Pipes, 25 mM Hepes, 10 mM EGTA, 2 mM MgCl2) using 0.1% Triton X-100 for 3 min and finally fixed with 4% paraformaldehyde in PBS for 10 min at room temperature. The cells were quenched for 5 min in 100 mM Tris-HCl followed by a 1-h block in 2% FBS, 2% BSA, and 0.1% Triton X-100 in PBS. Centromeres were visualized with a rabbit polyclonal anti–CENP-T (from D. Cleveland, Ludwig Institute for Cancer Research, San Diego, CA) antibody or monoclonal CENP-A at 1:1,000 dilution (ab13939; Abcam), and DNA was stained with 0.2 mg/ml DAPI. Donkey anti–rabbit Cy5-conjugated (111175003; Jackson ImmunoResearch Laboratories, Inc.) secondary antibodies were used for detection, and coverslips were mounted with ProLong (Invitrogen). GFP-TetR marker detection was enhanced in mitotic spreads and the Mis18BP1hsKNL2 siRNA experiment by staining with a rabbit anti-GFP antibody (1:500) and donkey anti–rabbit FITC-conjugated secondary antibody (711095152; Jackson ImmunoResearch Laboratories, Inc.).
All micrograph images were collected using a 100× oil immersion objective lens (numerical aperture = 1.40; Olympus) on a deconvolution microscope (DeltaVision) using a camera (CoolSNAP HQ2; Photometrics). The acquisition software used was SoftWoRX from Applied Precision. Images were deconvolved and presented as stacked images. Images within cell lines treated with different siRNAs were collected with identical exposure times and scaled equally. Intensities in live-cell and fixed images were analyzed using MetaMorph 7.7 (Molecular Devices). mCherry and YFP signals in live-cell images were subjected to local background subtraction. For determining siRNA knockdown, nuclei with all CENP-T foci colocalized with GFP-HJURP were scored as GFP-HJURP positive. Cells that had GFP-Mis18α at all centromeres with a maximum intensity >7,600 arbitrary units were scored as GFP-Mis18α positive. To assess CENP-A intensity at the LacO/TRE array, images of mCherry-LacI-HJURP– or mCherry-LacI-HJURPScm3–transfected and fixed/immunostained U2OS-LacO cells with or without IPTG treatment 48 h after transfection were taken under identical exposure conditions and analyzed for CENP-A intensity at the array using MetaMorph 7.7. CENP-A signal was subjected to local background subtraction. Intensity measurements were made per a set area, and the integrated intensities over this area for each group of cells were graphed.
Mitotic chromosome spreads
U2OS-LacO cells were arrested overnight in 0.1 ug/ml nocodazole in DME GlutaMAX media 32 h after transfection. Mitotic cells were harvested using a transfer pipette to blow cells off the plate. Cells were spun down, washed in PBS, and resuspended at 106 cells/ml in a hypotonic solution (20 mM Hepes, pH 7.0, 1 mM MgCl2, 0.2 mM CaCl2, 20 mM KCl, LPC [10 µg/ml leupeptin, 10 µg/ml pepstatin A, and 10 µg/ml chymostatin], and 0.5 ug/ml nocodazole/colcemid). After a 10-min incubation in the hypotonic solution, cells were spun onto glass slides using a cytospin (30,000 cells/slide), immediately hydrated with PBS, and then fixed and immunostained as described (see previous section). The anti–mouse NDC80 antibody was used at a 1:500 dilution (GTX70268; GeneTex).
Monastrol arrest
For U2OS-LacO, cells transfected for 48 h with mCherry-LacI-HJURPScm3 or mCherry-LacI, as described above, were arrested overnight in 50 µM Monastrol in DME GlutaMAX media. Mitotic cells were harvested by mitotic blow off, spun down, rinsed with PBS, and then resuspended at 106 cells/ml. Using a cytospin, 30,000 cells/slide were spun onto a glass slide. Cells were immunostained with an anti–rabbit CENP-T antibody (described in the previous section). For the quantification, 26 transfected Monastrol-arrested cells were imaged per condition (LacI or LacI-HJURPScm3). An ellipsoid was drawn to encompass the DNA, and the position of each centromere in each cell was measured relative to the center of this ellipsoid using ImageJ (National Institutes of Health). The black circle represents the mean centromere position over all the cells analyzed. The next gray circle represents one standard deviation from this mean. The blue triangles (LacI) and red diamonds (LacI-HJURPScm3) represent the array positions relative to the center of the ellipsoid in each of the 26 cells measured (Fig. 3).
Cold stable microtubules
Cells were transfected as indicated in the previous section with either mCherry-LacI or mCherry-LacI-HJURPScm3 and synchronized using a double thymidine block. After the second block, cells were released until they were entering mitosis. Cells were then placed on ice and treated with ice-cold media for 10 min. Cells were then cofixed (2% paraformaldehyde and 0.5% Triton X-100 in PHEM buffer) and immunostained for CENP-T and tubulin (using an FITC-conjugated tubulin antibody).
Recombinant protein purification
Recombinant proteins were expressed in the Rosetta (DE3) pLysS bacterial strain. Bacteria expressing His-tagged dNAP1 were sonicated in lysis buffer containing 50 mM sodium phosphate, pH 7.5, 500 mM NaCl, 10 mM imidazole, 10% glycerol, 10 mM β-glycerophosphate, 0.2 mM PMSF, and 0.5 mM benzamidine and purified by Ni–nitrilotriacetic acid (NTA) affinity chromatography. dNAP1 was further purified by size exclusion chromatography on a Superose 6 column in buffer containing 10 mM Hepes/KOH, pH 7.6, 10 mM KCl, 0.1 mM EDTA, 10% glycerol, 0.01% NP-40, 10 mM β-glycerophosphate, 0.2 mM PMSF, and 1 mM DTT followed by anion exchange chromatography on an UnoQ (Bio-Rad Laboratories) column. The topoisomerase I catalytic domain (Shaiu and Hsieh, 1998) was purified as described previously (Fyodorov and Kadonaga, 2003). Bacterial lysates were sonicated in 50 mM sodium phosphate, pH 7.0, 0.5 M NaCl, 15% (vol/vol) glycerol, and 0.1% (vol/vol) NP-40 and purified by Ni-NTA affinity chromatography. A codon-biased human CENP-A was coexpressed with histone H4 from a bicistronic vector (Black et al., 2004). The CENP-A–histone H4 heterotetramer was purified by hydroxyapatite chromatography followed by cation exchange chromatography. Canonical histones (from S. Khorasanizadeh, Burnham Institute, Orlando, FL) were individually expressed and purified by size exclusion chromatography in 7 M guanidinium HCl, 20 mM Tris-HC1, pH 7.5, and 10 mM DTT on a Sephacryl S-200 column followed by cation exchange chromatography in 7 M deionized urea, 20 mM sodium acetate, pH 5.2, 5 mM 2-mercaptoethanol, and 1 mM Na-EDTA as described previously (Luger et al., 1999). His-NPM1 bacterial pellets were resuspended in buffer containing 50 mM Na-phosphate, pH 8.0, 300 mM NaCl, 10 mM imidazole, 10 mM β-glycerophosphate, 1.5 mM MgCl2, and 0.5 mM PMSF and supplemented with 1 mg/ml lysozyme. The mixture was incubated on ice for 30 min and then lysed by using a French press. 1% Triton X-100 was added, and the solution was centrifuged at 26,900 g for 10 min at 4°C. The supernatant was incubated with Ni-NTA agarose beads (QIAGEN) for 1.5 h at 4°C. Beads were washed three times with buffer (50 mM Na-phosphate buffer, pH 8.0, 300 mM NaCl, 20 mM imidazole, 10 mM β-glycerophosphate, 1.5 mM MgCl2, and 0.5 mM PMSF) and finally eluted in elution buffer (50 mM Na-phosphate buffer, pH 8.0, 150 mM NaCl, 250 mM imidazole, 10 mM β-glycerophosphate, 1.5 mM MgCl2, and 1 mM DTT). MBP-HJURPScm3 bacterial pellets were resuspended in lysis buffer (25 mM Tris-Cl, pH 7.2, 200 mM NaCl, 20 mM MgCl2, 10% glycerol, 5 mM β-mercaptoethanol, 10 mM β-glycerophosphate, 0.2 mM PMSF, 1 mM benzamidine, and LPC), stirred on ice for 30 min, and sonicated until no longer viscous. The sample was then centrifuged at 26,900 g for 15 min at 4°C, and the supernatant was incubated with amylose resin (New England Biolabs, Inc.) for 1 h at 4°C. After washing the beads in 10 bead volumes of washing buffer (25 mM Tris-Cl, pH 7.2, 200 mM NaCl, 20 mM MgCl2, 10% glycerol, 5 mM β-mercaptoethanol, 10 mM β-glycerophosphate, 0.5 mM PMSF, 1 mM benzamidine, and LPC), the protein was eluted from beads in elution buffer (25 mM Tris-Cl, pH 7.2, 200 mM NaCl, 20 mM MgCl2, 10% glycerol, 5 mM β-mercaptoethanol, 10 mM β-glycerophosphate, and 10 mM maltose).
Nucleosome assembly
In buffer 1 (20 mM Hepes, pH 7.9, 1 mM EDTA, 10% glycerol, 150 mM NaCl, and 0.1 mg/ml BSA), purified histones (octamer, H3–H4 tetramer, and H2A/H2B dimers) and the purified chaperone were combined in a total volume of 20 µl and incubated at 37°C for 15–30 min (chaperone/histone mix). In a separate reaction, 0.25 µg pUC19 plasmid DNA was added to buffer 2 (20 mM Hepes, pH 7.9, 1 mM EDTA, 10% glycerol, and 150 mM NaCl) containing purified topoisomerase I. This mixture was incubated for 20 min at 37°C and then combined with the chaperone/histone mix followed by an incubation period of 3 h at 37°C. Where indicated, 3 mM CaCl2 was added, and reactions were treated with 3 U micrococcal nuclease (S1 nuclease; Roche) for 10 min at 37°C. The assembly reaction was stopped by addition of EDTA to a final concentration of 50 mM. 100 µl glycogen stop buffer (20 mM EDTA, pH 8.0, 200 mM NaCl, 1% SDS, and 0.25 mg/ml glycogen) and 10 µg proteinase K were added, and the protein digestion was performed at 37°C for 30–60 min. Recovery of DNA was achieved by phenol-chloroform extraction and ethanol precipitation. Topoisomers were separated in 0.8% TBE (89 mM Tris base, 89 mM boric acid, 2 mM EDTA) agarose gels with or without 4.6 µM chloroquine and subsequently stained with ethidium bromide.
Online supplemental material
Fig. S1 demonstrates the recruitment of CENP-A by LacI-HJURP in an NIH3T3 cell line containing a stable LacO-SceI-TRE array as well as live-cell imaging of the CENP-A stability in the NIH3T3 cells after IPTG washout in LacI-HJURP– and LacI-HJURPScm3–transfected cells. Fig. S2 shows that the Scm3 domain of HJURP is not sufficient to target human HJURP to centromeres. Fig. S3 shows the purifications of the recombinant MBP-HJURPScm3, NPM1, and dNAP assembly factors used in the supercoiling assays in Figs. 4 and 5 as well as a supercoiling assay showing the efficiency of dNAP in assembling H3.1- versus CENP-A–containing nucleosomes. Fig. S4 shows the enrichment at centromeres of our GFP-HJURP and GFP-Mis18 constructs upon synchronization of cells in G1 using a double thymidine block. Video 1 shows the control cell from Fig. 3 D undergoing mitotic division. Video 2 shows the cell expressing HJURPScm3 from Fig. 3 D undergoing mitotic division.
Acknowledgments
We give thanks to Sepideh Khorasanizadeh, Tom Misteli, Andrew Holland, Don Cleveland, and Susan Janicki for reagents. We also thank Alexandra Lusser, Jonathan Gaba, and Agnes Sabat for technical assistance and Todd Stukenberg and Dan Burke for comments on this manuscript.
D.R. Foltz was supported by the American Cancer Society. M.C. Barnhart was supported by a Cell and Molecular Biology training grant (National Institutes of Health T32 GM008136). B.E. Black is supported by a research grant (National Institutes of Health GM82989), a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund, and a Rita Allen Foundation Scholar Award. E.A. Bassett has been supported by the Penn Structural Biology Training Grant (National Institutes of Health GM08275) and a predoctoral fellowship from the American Heart Association.
References
Author notes
M.C. Barnhart and P.H.J.L. Kuich contributed equally to this paper.






![Figure 6. Recruitment of HJURP to centromeres requires the Mis18 complex. (A and B) Cellular extracts from siRNA-treated and control cell lines were analyzed by Western blotting using anti-GFP (A) or anti-HJURP antibodies (B). Each lane contains lysate from 105 cells. Dilution series were generated from mock-treated HeLa GFP-Mis18α (A) or parental HeLa (B) cells. (C) Stable GFP-Mis18α cells lines were treated with siRNA against Mis18α, Mis18BP1hsKNL2, HJURP, or GAPDH (control). Representative images of siRNA-treated GFP-Mis18α cells were selected in which a midbody was clearly present (differential interference contrast [DIC], arrows) to show the cell was in early G1. DAPI staining was overlaid onto the differential interference contrast image. Cells were stained with anti–CENP-T. (D) Mean percentage of GFP-Mis18α centromere-positive nuclei from a population of ≥57 cells in each siRNA treatment from two experiments. (E) Similar image acquisition as in C. Here, stable HeLa GFP-HJURP cells were treated with the same siRNAs. (F) Mean percentage of GFP-HJURP centromere-positive nuclei from a population of ≥135 cells in each siRNA treatment from two experiments. Error bars show standard deviations. Insets show magnified views of boxed regions. Bars, 5 µm.](https://cdn.rupress.org/rup/content_public/journal/jcb/194/2/10.1083_jcb.201012017/6/m_jcb_201012017r_rgb_fig6.jpeg?Expires=1771354342&Signature=R3tPwnI2nxSfoRy0K8mJddNFvYqSyturHej4~Om~5KlvJy6K3df910e3gC3YbwQWc~s4-33tjbpT~u5OaCcQllREgFInuFBDNh4D7-ZRR3kZuuPx9YCiYHpOPZsnznf7h8w~SwDSssDXAD6HQ3tsV26lOyKVqt4wvh8Mo~eXpAZlGCcqzZQHDybvI8lXTMfeGG2YG5tXGCKT4E9lSTgotX5AbrPYoBRrihVO7ZGNfqLcKXYniEsQ8Q1Ua30Ftt9PzPbzeqUzCZgTyZkFEzyiAuG4mVY~37eALaQ8R3mbG5VSBbAFP-3HzeaD7FCLMSSDyDej-M4TngJ4T4Yip609dA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)




