Runx1 controls hematopoietic stem cell emergence and hair follicle stem cell (HFSC) activation and proliferation in adult skin. Here we use lineage tracing and mouse genetic manipulation to address the role of Runx1 in the embryonic development of HFSCs. We find Runx1 is expressed in distinct classes of embryonic skin precursors for short-term HF progenitors, adult HFSCs, and mesenchymal progenitors. Runx1 acts in the embryonic epithelium for timely emergence of adult HFSCs and short-term progenitors, but is dispensable for both of them. In contrast, Runx1 is strictly needed in the embryonic mesenchyme for proper adult HFSC differentiation and long-term skin integrity. Our data implicate Runx1 in epithelial cell adhesion and migration and in regulation of paracrine epithelial–mesenchymal cross talk. The latter involves Lef1 and Wnt signaling modulation in opposing directions from two distinct skin compartments. Thus, a master regulator of hematopoiesis also controls HFSC emergence and maintenance via modulation of bidirectional cross talking between nascent stem cells and their niche.
Introduction
The embryonic factors regulating adult-type stem cell (SC) emergence during morphogenesis and the long-term impact of these factors on adult homeostasis are largely obscure (Slack, 2008). The organ rudiments can arise from distinct short-lived “primitive” progenitors before or in parallel with the emergence of long-lived “definitive” adult tissue SCs (Dzierzak and Speck, 2008; Lepper et al., 2009; Messina and Cossu, 2009). Adult blood SCs are set aside during morphogenesis to regenerate the tissue later on in life and their emergence is controlled by a master transcription factor, Runx1 (Dzierzak and Speck, 2008). Hair follicle stem cells (HFSCs) originate in the embryonic hair placodes, and acquire some adult-type characteristics before birth (Levy et al., 2005; Nowak et al., 2008). Here we use Runx1 as an entry point to examine the mechanisms controlling the embryonic development of adult mouse HFSCs.
The skin epithelium—epidermis, HFs, and sebaceous gland (SG)—is made of keratinocytes, whereas the skin mesenchyme (dermis) is made mainly of fibroblasts (Blanpain and Fuchs, 2009). Mouse hair development begins in the embryo and occurs in three waves forming: (a) guard hairs (embryonic day [E] ∼14.5), representing only 2–10% of the mouse pelage; (b) auchene and awl hairs (∼E16.5); and (c) zig-zag hairs (postnatal day [PD] ∼0; Schneider et al., 2009). Due to these waves there is a mix of HF developmental stages (placode, germ, and bulbous peg) in embryonic skin (Fig. S1 A). By birth all HFs are specified and continue to mature up to PD8 (Paus et al., 1999; Schmidt-Ullrich and Paus, 2005). The bulbous peg contains the matrix (M), a class of short-lived HF progenitors (Legué and Nicolas, 2005), which proliferate and differentiate pushing cells upward to generate the inner layers (ILs) of the HF: the hair shaft, and the inner root sheath (IRS). These are surrounded by the outer root sheath (ORS), where adult HFSCs cluster in the hair “bulge” zone (Fig. S1 B; Fuchs, 2009). PD17 marks the end of hair morphogenesis and the start of the first adult hair cycle. This occurs in cyclic and relatively synchronous phases of degeneration and apoptosis (catagen), rest and quiescence (telogen), and growth and proliferation (anagen; Schneider et al., 2009). Signals from the dermal papillae (DP), a mesenchymal hair structure, and the environment activate HFSCs to migrate down and regenerate the matrix (Blanpain and Fuchs, 2009; Zhang et al., 2009).
Several molecular players such as Bmp, Wnt, and Lhx2 regulate both morphogenesis and adult hair cycle (Schneider et al., 2009). Conversely, Sox9, NFATc1, and Stat3 regulate adult HFSCs but not hair morphogenesis (Sano et al., 1999; Vidal et al., 2005; Horsley et al., 2008; Nowak et al., 2008).
Few transcription factors have been shown to regulate both blood and HF-differentiated cell lineages (DasGupta and Fuchs, 1999; Kaufman et al., 2003). Previously we showed Runx1, a blood master regulator, to be important in adult HFSC activation, proliferation, and hair homeostasis (Osorio et al., 2008; Hoi et al., 2010), while others also found it important in the terminal differentiation of the hair shafts (Raveh et al., 2006). Here we find embryonic Runx1 expression in distinct skin compartments essential for proper development and long-term integrity of skin and HFs. Runx1 modulates Lef1 and Wnt signaling in a paracrine fashion and in opposing directions from the epithelial versus mesenchymal skin layers by de-regulating expression of secreted Wnt-regulatory molecules.
Results
Runx1 is dynamically expressed in the skin epithelium and mesenchyme during HF development
Previously, Runx1 was reportedly expressed in mouse skin mesenchyme at E14.5 and E18.5 and in HFs at E18.5 (Raveh et al., 2006). We reexamined in detail Runx1-LacZ embryonic skin (North et al., 2002) and in addition performed antibody staining at E12.5, 13.5, 14.5, 16.5, and 17.5. Not previously reported, at E12.5 we found rare X-Gal+ cells localized to the single layer of ectoderm surrounding the body, whereas later on we detected some X-Gal+ cells in the epithelium of hair placodes and germs (Fig. 1 A). The X-Gal signal was strong in cells of the upper HF and weak and sometimes absent (especially in the placode) in the lower HF. As previously reported (Raveh et al., 2006), the bulbous peg showed strong X-Gal in centrally located cells that were likely the precortex and preIRS (Fig. 1 A, right). From E14.5 to E17.5, we found X-Gal+ cells in the mesenchyme including the dermal condensate and papillae, made of fibroblast (Fig. 1 A), as reported previously (Raveh et al., 2006). LacZ also colocalized rarely in the dermis with markers for blood and endothelial cells (Fig. S3). Immunofluorescence with a Runx1-specific antibody previously generated (Chen et al., 2006) confirmed the Runx1-LacZ skin expression pattern (Fig. 1 B).
Runx1 expression in the epithelial and mesenchymal skin compartments during embryonic HF development. (A) Skin sections from Runx1-LacZ reporter mice at E12.5 and E17.5 stained with X-Gal (blue) and hematoxylin (purple). (B–E) Runx1 immunofluorescence staining of embryonic skin (arrows), Sox9, and NFATc1 expression in the upper follicle (n = 3 WT and 3 Runx1 cKO mice). (D) Serial sections of Runx1 and Sox9 show colocalization of these factors in the HF. *, hair germ; Epi, epidermis; Der, dermis. Bars, 10 µm.
Runx1 expression in the epithelial and mesenchymal skin compartments during embryonic HF development. (A) Skin sections from Runx1-LacZ reporter mice at E12.5 and E17.5 stained with X-Gal (blue) and hematoxylin (purple). (B–E) Runx1 immunofluorescence staining of embryonic skin (arrows), Sox9, and NFATc1 expression in the upper follicle (n = 3 WT and 3 Runx1 cKO mice). (D) Serial sections of Runx1 and Sox9 show colocalization of these factors in the HF. *, hair germ; Epi, epidermis; Der, dermis. Bars, 10 µm.
Runx1 expression in the embryonic epithelium largely resembled (although was generally broader than) that of ORS and bulge markers Sox9 and NFATc1 (Vidal et al., 2005; Horsley et al., 2008; Nowak et al., 2008; Fig. 1, C, D, and D′; and unpublished data). Thus, developing HFs expressed Runx1 highly in the upper zone thought to contain precursors of adult HFSCs, and weakly in the lower follicle likely including the prematrix cells, which generate the first differentiated hair shafts. Moreover, unlike adult skin (Raveh et al., 2006; Osorio et al., 2008), embryonic skin showed Runx1 protein expression in the mesenchyme.
Lineage tracing shows that Runx1-expressing embryonic cells contribute to postnatal mesenchymal skin and distinct epithelial HF lineages
To understand the fate of embryonic Runx1-expressing cells, we performed lineage-tracing experiments during hair morphogenesis. We used Runx1-driven CreER-expressing mice (Samokhvalov et al., 2007) inducible by tamoxifen to turn on the Rosa26 (R26R) reporter (Soriano, 1999) and express stably LacZ in Runx1+ cells and their progeny. Tamoxifen injected daily for 2, 3, and 4 consecutive days in pregnant females resulted in weak X-Gal staining 2 d after last injection. By PD0 the X-Gal+ cells were still scarce in HF epithelium, suggesting rare cells labeling in the hair rudiments (Fig. S2, A and B; and Fig. 2 D). Mice injected 3x (E12.5, 13.5, 14.5; Fig. 2 A) showed X-Gal labeling in many upper dermal cells, DP (10% of HFs), and HF epithelium (7% of HFs; Fig. 2, B–D, n = 3 mice). CreER recombinase reportedly works 6–36 h after tamoxifen injection (Zervas et al., 2004) and most likely labeled HFs of the first hair morphogenetic wave (placodes and germs), and some placodes of the second wave, typically ongoing at ∼E16.5 (Fig. S1 A). All labeled HFs detected at PD0 showed patches of X-Gal+ and X-Gal− cells, supporting the polyclonal HF origin (Fig. 2 D). In summary, Runx1+ hair placode cells marked skin progenitor cells that by birth contributed progeny to the HF epithelium, DP, and upper dermal cells. The Runx1-CreER allele appeared highly inefficient in the HF and DP, but showed considerable efficiency in the upper dermis.
Mesenchymal skin and epithelial HF contribution from embryonic Runx1-expressing cells. (A) Scheme of tamoxifen inductions and sample collection. (B) 50–90-µm skin sections showing efficient X-Gal dermal labeling in mice induced with tamoxifen at E12.4–E14.5 (early). (C) Plot showing the X-Gal labeling efficiency from early and late induction schemes at dates indicated. Minimum of two mice and 100 follicles counted per stage. (D–F) X-gal–stained skin section (25–50 µm) from mice treated as shown in A, sacrificed at different time points. (G) Quantification of patterns in D and E from early labeling show distribution of X-Gal+ HFs in categories labeled exclusively in the outer root sheath (ORS), inner layers and matrix (M/IL), and in both (com, complex). n = 2 mice and 200 X-Gal+ HFs. (H) Same as in E, except shown next to late labeling counts (see A), and without immature HFs (pegs). t test: ***, P ≤ 0.007; **, P < 0.05; *, P ≤ 0.09. Bars, 10–50 µm.
Mesenchymal skin and epithelial HF contribution from embryonic Runx1-expressing cells. (A) Scheme of tamoxifen inductions and sample collection. (B) 50–90-µm skin sections showing efficient X-Gal dermal labeling in mice induced with tamoxifen at E12.4–E14.5 (early). (C) Plot showing the X-Gal labeling efficiency from early and late induction schemes at dates indicated. Minimum of two mice and 100 follicles counted per stage. (D–F) X-gal–stained skin section (25–50 µm) from mice treated as shown in A, sacrificed at different time points. (G) Quantification of patterns in D and E from early labeling show distribution of X-Gal+ HFs in categories labeled exclusively in the outer root sheath (ORS), inner layers and matrix (M/IL), and in both (com, complex). n = 2 mice and 200 X-Gal+ HFs. (H) Same as in E, except shown next to late labeling counts (see A), and without immature HFs (pegs). t test: ***, P ≤ 0.007; **, P < 0.05; *, P ≤ 0.09. Bars, 10–50 µm.
The distribution of the rare X-Gal+ cells in the HF epithelium in thin and thick 90-µm sections at PD0 could be classified in three main labeling patterns: (1) exclusive ORS pattern (35% of all labeled HFs); (2) exclusive matrix and inner layer (M/IL) pattern (18%) with any X-Gal+ cells in the lower bulb and matrix cells, the hair shaft, inner root sheath, inner preSG, and/or hair canal (the inner portion of the infundibulum); and (3) complex pattern (38%), which was a combination of both M/IL and ORS (Fig. 2, D and G). The remaining 10% labeled HF were hair pegs that had not yet developed distinguishable inner layers by this developmental stage (Fig. 2 G and Fig. S2 C).
By PD5 HFs differentiated and produced M/IL with distinct hair shafts (Fig. 2, E and G). The frequency of labeled HFs was 5–9% (Fig. 2 C, n = 2 mice and ∼1,870 HFs). Complex patterns remained roughly the same as for PD0 analysis (∼42% of all labeled HFs), while exclusive ORS patterns increased to ∼56% (Fig. 2 G). This increase was likely due to the contribution of X-Gal+ cells detected in the hair pegs at PD0 to the ORS of the PD5 HFs. In contrast, the frequency of the M/IL-exclusive pattern dropped to ∼2% (Fig. 2 G), likely because many of these lineages appeared to be shed at PD0 into the squames of the epidermis through the hair canal (Fig. 2 D). Moreover, X-Gal+ cells were exclusively present in the hair canal in nearly 3x more HFs at PD5 than at PD0 (3% vs. ∼9%; Fig. S2 D). This was likely because X-Gal+ cells from the M/IL (detected at PD0) had been pushed upward by newly generated X-Gal–negative cells from underneath (by PD5), without concomitant self-renewal of the matrix cells. When counted overall, 98% of labeled HFs contained ORS labeling at PD5 and 56% was exclusive ORS labeling. Exclusive ORS labeling persisted at late stages of HF morphogenesis as shown by X-gal analysis at PD13 (Fig. 2 F, left; and Fig. S2 E). In conclusion, Runx1+ progenitor cells from the early embryonic HF morphogenesis (placode and germ) contributed cells to ORS and/or to M/IL, two lineages considered independent during distinct times of morphogenesis (Legué and Nicolas, 2005; Nowak et al., 2008; and see Discussion). The ORS contribution was most substantial and persisted through late stages of postnatal morphogenesis.
Runx1+ HF cells increased in numbers by late embryogenesis (Fig. 1), and we marked them by tamoxifen injections at E14.5, 15.5, and 16.5 (Fig. 2 A, right). By this time HFs of the second hair growth wave were predominant and reached the placode and germ stages, whereas the few HFs of the first wave were in the early hair peg stage (Fig. S1 A). Because of prolonged Cre activity (see above and Zervas et al., 2004), some advanced pegs and bulbous peg (from ∼E17.5–18.5), including cells of the precortex and preIRS, were also likely labeled.
In PD0 mouse skin from the late induction scheme we detected 18% X-Gal+ HFs (Fig. 2 C, n = 3 mice, 1,300 HFs; Fig. S1 B, bottom). Similar to the early induction, the PD0 dermis also showed X-Gal labeling that was largely lost by PD5 (Fig. S2 F). In the HF epithelium we detected complex labeling pattern (65% of all labeled HFs), M/IL-only (27%), and ORS-only (7%) (Fig. 2 H). When considered together, 93% of all late-labeled HFs showed contribution from the Runx1+ progenitor cells to the M/IL compartments. The distribution of the three HF-labeling patterns remained roughly the same at PD0 and PD5 (Fig. 2 H, red bars). An interesting side observation was the high frequency of HFs with X-Gal+ cells in the hair canal (∼30% of all labeled HFs; Fig. S2 D). In addition, while X-Gal labeling generated by early injection remained strong and relatively constant from E17.5 to PD0 to PD5, the late induction X-Gal labeling was markedly reduced from the dermis and HFs (Fig. 2 C, Fig. S2 F; n = 2 mice, 1,200 HFs), suggesting that the late-embryonic cells were shorter-lived cells (some of which were already part of the precursors of the inner layers) than the early Runx1+ embryonic cells.
In summary, these lineage-tracing experiments can be interpreted as follows: (a) early embryonic Runx1+ epithelial cells were HF progenitors that contributed most substantially to the ORS and were stably maintained during postnatal morphogenesis; (b) Runx1+ epithelial cells from late HF morphogenesis contributed substantially to M/IL, which generated the first hair shaft and inner layers, and were massively lost in postnatal morphogenesis (see Discussion); and (c) early but not late Runx1+ mesenchymal cells showed contribution to the upper dermis and to the DP during postnatal morphogenesis.
Runx1-expressing embryonic cells are precursors of adult HFSCs
To ask if adult HFSCs originate in embryonic Runx1+ cells, we assessed the contribution of Runx1-CreER–marked embryonic cells to the adult skin. We injected mice with tamoxifen during the early (E12.5, 13.5, 14.5) and late (E14.5, 15.5, 16.5) stages of embryonic hair morphogenesis and examined the skin in adulthood up to 8 mo after several adult hair cycles (Fig. 3 A). In both schemes, X-Gal+ dermal cells were much diminished relative to morphogenesis (compare Fig. 3 B with Fig. 2 B; and see Fig. S2 F). X-Gal was also in some DP cells (Fig. 3 C, f), and in rare HF cells (∼7–10% of HFs) both at telogen and anagen (Fig. 3, C and D). In telogen HFs, the X-Gal+ cells were in the bulge ORS (Fig. 3 C, a and c), the inner nonproliferative bulge layer (Fig. 3 C, b), the infundibulum, and the SG (Fig. 3 C, d and e, respectively). In anagen HFs (PD26-PD31), X-Gal+ cells were detectable in the infundibulum, bulge, and bulb (matrix and the differentiated ILs including the newly generated hair shaft; Fig. 3 D). Quantification of these patterns is shown in Fig. 3, F and G. In addition, skin in anagen at ∼PD240 after several hair cycles showed persistent X-Gal labeling in the bulge, ORS, and M/IL, demonstrating the long-term self-renewal ability of the early Runx1+ embryonic cells from the hair placodes (not depicted). As seen before in morphogenesis, early Runx1+ embryonic HF cells or progenitors had a better survival rate into adulthood when compared with the late Runx1+ embryonic HF cells (Fig. 3 E). Taken together, these data demonstrated that emerging precursors of adult HFSCs from the developing hair rudiments expressed Runx1.
Embryonic Runx1-expressing cells contribute to HFSCs and mesenchyme in adult skin. (A) Scheme of tamoxifen inductions and long-term skin analysis. (B) Runx1-expressing cells contribution to the mesenchymal compartment. 50–90-µm skin sections showing X-Gal labeling from mice induced with tamoxifen early and sacrificed at times indicated. (C and D) Skin sections from mice injected early in embryogenesis and sacrificed at time points indicated, stained with X-gal (blue) and hematoxylin (purple). Note X-Gal+ cells in the bulge (Bu), infundibulum (Inf), sebaceous gland (SG), and dermal papilla (DP) and the adult dermis (Der). (E) Fraction of total X-Gal+ HFs in mice treated as shown in A, and sacrificed at postnatal days indicated (PD). n > 2 mice and 1,200 HFs per time point. (F and G) Distribution of X-Gal–labeling patterns in adult HFs at stages indicated. Bars, 10–50 µm.
Embryonic Runx1-expressing cells contribute to HFSCs and mesenchyme in adult skin. (A) Scheme of tamoxifen inductions and long-term skin analysis. (B) Runx1-expressing cells contribution to the mesenchymal compartment. 50–90-µm skin sections showing X-Gal labeling from mice induced with tamoxifen early and sacrificed at times indicated. (C and D) Skin sections from mice injected early in embryogenesis and sacrificed at time points indicated, stained with X-gal (blue) and hematoxylin (purple). Note X-Gal+ cells in the bulge (Bu), infundibulum (Inf), sebaceous gland (SG), and dermal papilla (DP) and the adult dermis (Der). (E) Fraction of total X-Gal+ HFs in mice treated as shown in A, and sacrificed at postnatal days indicated (PD). n > 2 mice and 1,200 HFs per time point. (F and G) Distribution of X-Gal–labeling patterns in adult HFs at stages indicated. Bars, 10–50 µm.
Delayed HF morphogenesis in Runx1 epithelial skin knockout
To test the role of Runx1 in the embryonic epithelial HF cells, we analyzed the embryonic skin phenotype of K14-Cre;Runx1Fl/Fl conditional knock out (cKO) mice (Vasioukhin et al., 1999; Growney et al., 2005). These mice grow hairs by PD9, but showed a prolonged first telogen (Osorio et al., 2008). Embryos and newborn pups looked normal in size and appearance and were born at expected ratios (unpublished data). E12.5 K14-Cre;R26R reporter mice showed targeting in ∼50% of the skin epithelium (unpublished data). Furthermore, K14Cre;Runx1Fl/Fl E16.5 skin showed no Runx1 staining in the fraction mice that was further analyzed (Fig. 4 A).
Runx1 epithelial loss impaired in vivo HF morphogenesis and in vitro cell migration and adhesion. Skin sections from E16.5 Runx1 cKO mice (K14-Cre; Runx1Fl/Fl) and wild-type (WT) littermates immunostained for Runx1 and β4-integrin (A), and for alkaline phosphatase and S-red at E16.5 (B) and PD1 (C). Note fewer HF in cKO skin. (D) Average number of HFs per field of view (FOV); P = 0.02 at E16.5 (n = 4 WT; 630 HFs/130 FOVs and n = 3 cKO; 347 HFs/140 FOVs) and at PD1 (n = 4 WT; 794 HFs/71 FOVs and cKO n = 4; 562 HFs/59 FOVs); P = 0.04. (E) Keratin 5 staining revealed abnormal hair germ morphology in cKO skin (F) Left: an average of ∼13 cells/placode was found in both WT and cKO skin; ∼27 cells/germ in WT, and ∼33 cells/germ in cKO (n = 45 HFs, and n = 3 mice, P = 0.01). Middle: reduced germ size area in cKO skin (P = 0.0004), but no difference in placode. Au: arbitrary unit. Right: cell density plot shows a higher number of cells per area in cKO skin; error bars were calculated using a propagation of uncertainty test. (G and H) Fluorescence image of freshly isolated skin keratinocytes at 24 h after plating onto collagen (G) or mouse embryonic fibroblasts (H), or different ECM factors (I): Col-1, collagen 1; FN, fibronectin; PDL, poly-d-lysine. Note on FN fewer cKO keratinocytes than WT (P = 0.05). (J) Left: keratinocytes that remained round 24 h after plating scored as “not spread.” Right: sub-classification based on surface area (WT: n = 1,299; and cKO: n = 1,352 cells). Runx1 cKO samples have more large cells; *, P < 0.009. (K and L) Phase-contrast images show freshly isolated skin keratinocytes plated for 24 h on collagen, scratch-wounded and quantified for gap size (white bar). P = 0.08, top and 0.04, bottom. MytC, mytomycin C. (M) Skin sections at E16.5 stained with phalloidin to detect actin polymerization status in the hair placode. Note higher signal at the leading edge of placode in WT when compared with cKO. Bars, 20 µm.
Runx1 epithelial loss impaired in vivo HF morphogenesis and in vitro cell migration and adhesion. Skin sections from E16.5 Runx1 cKO mice (K14-Cre; Runx1Fl/Fl) and wild-type (WT) littermates immunostained for Runx1 and β4-integrin (A), and for alkaline phosphatase and S-red at E16.5 (B) and PD1 (C). Note fewer HF in cKO skin. (D) Average number of HFs per field of view (FOV); P = 0.02 at E16.5 (n = 4 WT; 630 HFs/130 FOVs and n = 3 cKO; 347 HFs/140 FOVs) and at PD1 (n = 4 WT; 794 HFs/71 FOVs and cKO n = 4; 562 HFs/59 FOVs); P = 0.04. (E) Keratin 5 staining revealed abnormal hair germ morphology in cKO skin (F) Left: an average of ∼13 cells/placode was found in both WT and cKO skin; ∼27 cells/germ in WT, and ∼33 cells/germ in cKO (n = 45 HFs, and n = 3 mice, P = 0.01). Middle: reduced germ size area in cKO skin (P = 0.0004), but no difference in placode. Au: arbitrary unit. Right: cell density plot shows a higher number of cells per area in cKO skin; error bars were calculated using a propagation of uncertainty test. (G and H) Fluorescence image of freshly isolated skin keratinocytes at 24 h after plating onto collagen (G) or mouse embryonic fibroblasts (H), or different ECM factors (I): Col-1, collagen 1; FN, fibronectin; PDL, poly-d-lysine. Note on FN fewer cKO keratinocytes than WT (P = 0.05). (J) Left: keratinocytes that remained round 24 h after plating scored as “not spread.” Right: sub-classification based on surface area (WT: n = 1,299; and cKO: n = 1,352 cells). Runx1 cKO samples have more large cells; *, P < 0.009. (K and L) Phase-contrast images show freshly isolated skin keratinocytes plated for 24 h on collagen, scratch-wounded and quantified for gap size (white bar). P = 0.08, top and 0.04, bottom. MytC, mytomycin C. (M) Skin sections at E16.5 stained with phalloidin to detect actin polymerization status in the hair placode. Note higher signal at the leading edge of placode in WT when compared with cKO. Bars, 20 µm.
HF counts on skin sections stained with β4-integrin to reveal the basal layer and the ORS (Fig. 4 A), hematoxylin and eosin for contrast (not depicted), or alkaline phosphatase to mark DP and dermal condensates (Fig. 4, B and C) showed fewer follicles in Runx1 cKO than wild-type (WT) control littermate (Fig. 4 D). At E16.5, WT skin had many hair placodes and germs (second hair growth wave) and few hair pegs (first hair growth wave at this age; Schneider et al., 2009). There were 45% fewer follicles per field of view (FOV) in the cKO skin at E16.5 but not at PD0 (Fig. 4 D), indicating a delay in hair placode formation. Moreover, mathematical analysis of these data suggested that HFs were also impaired in their down-growth from placode to hair germ (Fig. S2, G and H). In conclusion, Runx1 loss in the early embryonic epithelium impaired timely hair placode formation and hair germ down-growth, and resulted in delay of hair morphogenesis, largely overcome by birth.
Runx1 loss impairs keratinocyte adherence and migration
Placode formation requires downward migration of epithelial cells, which polarize their cytoskeleton and invade the underlying dermis. Cells in the cKO hair germs appeared more crowded and lacked the regular tiled-pavement WT appearance (Fig. 4, E and M, and quantified in F; see also Materials and methods). This phenotype could not be explained by a potential increased proliferation of cKO cells at E16.5 (Fig. 5). Furthermore, cells in the placodes of cKO stained less with phalloidin, which marks polymerized actin filaments, and this was especially prominent at the placode leading edge (Fig. 4 M). Together, these data suggested potentially impaired downward migration and adhesion of epithelial cells in skin tissue upon Runx1 loss.
Loss of Runx1 in the epithelium causes mild proliferation defects. (A and C) Immunofluorescence image showing embryonic hair germs stained for Ki67 (A) and BrdU (C). (B) Percentage of BrdU+ cells of total sorted from E16.5 skin and spotted on slides showing ∼28% reduction in cKO α6-integrin+ epithelial cells. n > 3 WT and cKO mice and >250 cells counted per population. P ≤ 0.05. (D) Average number of BrDU+ cells per germ in E16.5 skin 3.5 h after BrdU injection. P = 0.01. n > 100 germs WT and cKO. Epi, epidermis; Der, dermis. Bar, 10 µm.
Loss of Runx1 in the epithelium causes mild proliferation defects. (A and C) Immunofluorescence image showing embryonic hair germs stained for Ki67 (A) and BrdU (C). (B) Percentage of BrdU+ cells of total sorted from E16.5 skin and spotted on slides showing ∼28% reduction in cKO α6-integrin+ epithelial cells. n > 3 WT and cKO mice and >250 cells counted per population. P ≤ 0.05. (D) Average number of BrDU+ cells per germ in E16.5 skin 3.5 h after BrdU injection. P = 0.01. n > 100 germs WT and cKO. Epi, epidermis; Der, dermis. Bar, 10 µm.
Runx1 is required for cultured keratinocyte long-term survival and proliferation (Osorio et al., 2008). To determine if Runx1 also played a role in keratinocyte cell adhesion we plated freshly isolated skin cells on different ECM factors: fibronectin (FN), collagen 1 (Col-1), and poly-d-lysine (PDL). By 24 h Runx1 cKO and WT keratinocytes attached similarly on all substrates tested (Fig. 4 I), but cKO cells showed a higher fraction of cells with larger overall area (defined by phalloidin staining; Fig. 4, G and J). This effect was also seen upon plating on mouse embryonic fibroblast layers (Fig. 4 H and unpublished data).
To test cell migration defects we performed in vitro scrape-wound closure assays on monolayers of primary keratinocytes freshly plated onto a collagen matrix. We photographed the wounded area using phase-contrast microscopy at 0, 8, 12, 24, and 31 h after wounding (Fig. 4 K and unpublished data) and found that the distance between edges decreased faster in WT than in KO (n = 4 WT and 4 cKO mice; Fig. 4, K and L, top graph). This was true even in the presence of mitomycin C, a potent inhibitor of cell proliferation (Fig. 4 L, bottom graph). Therefore, Runx1 loss impaired cell spreading and migration in vitro, which correlated with the detected high density of cells in the cKO hair germ in vivo.
De-regulated Lef1 and Wnt signaling pathway in Runx1 epithelial knockout skin
Next we tested the status of HF developmental signaling pathways such as Shh, Bmp/Tgfβ, Wnt, FGFs, and TNFs (Schmidt-Ullrich and Paus, 2005; Schneider et al., 2009). We FACS purified α6-integrinhigh, α6-integrinmedium (HF cells), and α6-integrinnegative (dermal cells) from E16.5 skin (Fig. 1, C and D). QRT-PCR and immunofluorescence of sorted cells confirmed, respectively, the expression of Runx1 mRNA, and a hair specific marker (keratin 17) in these populations (Fig. 6 B and unpublished data). QRT-PCR analysis of HF developmental factors (Nakamura et al., 2001; Andl et al., 2002) revealed significant changes of FGFRII, Wnt10a, and Lef1 in cKO versus WT sorted α6-integrinmedium cells (Fig. 6, A and C). Moreover, immunostaining of skin sections revealed down-regulation of Lef1, a transactivator of Wnt target genes, itself a Wnt-signaling target (Andl et al., 2002; Lowry et al., 2005) in cKO HFs (Fig. 6, D–F, K). These data suggested that Runx1 expression is required directly or indirectly for maintaining high levels of Lef1 in the skin epithelium. Intriguingly, in all four cKO mice analyzed Lef1 immunostaining signal was reduced not only in the skin epithelium, but also in many of the dermal cells adjacent to the HF (Fig. 6, D–F). This suggested a broad paracrine effect of Runx1 on Lef1 protein in both the epithelium and the adjacent mesenchyme. Furthermore, crossing the K14-Cre;Runx1Fl/Fl with reporter BAT-GAL mice, which carry LacZ downstream of an engineered Tcf/Lef enhancer (Maretto et al., 2003), showed broad down-regulation of Wnt signaling in E18.5 cKO skin in HFs at all distinct developmental stages, and this down-regulation extended into the dermis and DP (Fig. 6, G, I, and J). HF and epidermal differentiation lineage markers characteristic to skin were normal at E16.5 and PD9 (Fig. 7), ruling out that lower BAT-GAL signal in the embryo was due to absence of specific hair lineages that normally show high BAT-GAL activity. All these data suggested that Runx1 loss in the epithelium results in a generalized decrease in Lef1 protein and in canonical Wnt signaling in both the epithelial and mesenchymal compartments of the skin, and that Runx1 controls the epithelial–mesenchymal cross talk from the epithelium.
Factors involved in hair morphogenesis are de-regulated in Runx1 cKO skin. (A) Summary of known HF morphogenesis factors (Nakamura et al., 2001) tested in C by QRT-PCR analysis of skin sorted α6-integrinmedium and α6-integrinnegative cells (n = 2 mice per genotype). *, P < 0.06; **, P < 0.006. (B) Same as C but for Runx1 mRNA. Note high expression of Runx1 in the α6-integrinmedium, which indicates enrichment in HF cells over the epidermis. (D–F) Immunofluorescence E16.5 skin images reveals reduction in Lef1 signal in cKO dermal and epidermal skin at all developmental stages analyzed. (G, I, and J) E18.5 BAT-GAL mouse skin showed reduced X-Gal staining in both epithelial and mesenchymal cKO skin when compared with WT. (H) Quantification of G (n = 2 WT and cKO; 300 HFs). (K) Quantification of Lef1 staining from images like D and E (n = 3 WT: 687 HFs and n = 3 cKOs: 248 HFs). **, P ≤ 0.007; *, P = 0.09. Epi, epidermis; Der, dermis. Bars, 10–50 µm.
Factors involved in hair morphogenesis are de-regulated in Runx1 cKO skin. (A) Summary of known HF morphogenesis factors (Nakamura et al., 2001) tested in C by QRT-PCR analysis of skin sorted α6-integrinmedium and α6-integrinnegative cells (n = 2 mice per genotype). *, P < 0.06; **, P < 0.006. (B) Same as C but for Runx1 mRNA. Note high expression of Runx1 in the α6-integrinmedium, which indicates enrichment in HF cells over the epidermis. (D–F) Immunofluorescence E16.5 skin images reveals reduction in Lef1 signal in cKO dermal and epidermal skin at all developmental stages analyzed. (G, I, and J) E18.5 BAT-GAL mouse skin showed reduced X-Gal staining in both epithelial and mesenchymal cKO skin when compared with WT. (H) Quantification of G (n = 2 WT and cKO; 300 HFs). (K) Quantification of Lef1 staining from images like D and E (n = 3 WT: 687 HFs and n = 3 cKOs: 248 HFs). **, P ≤ 0.007; *, P = 0.09. Epi, epidermis; Der, dermis. Bars, 10–50 µm.
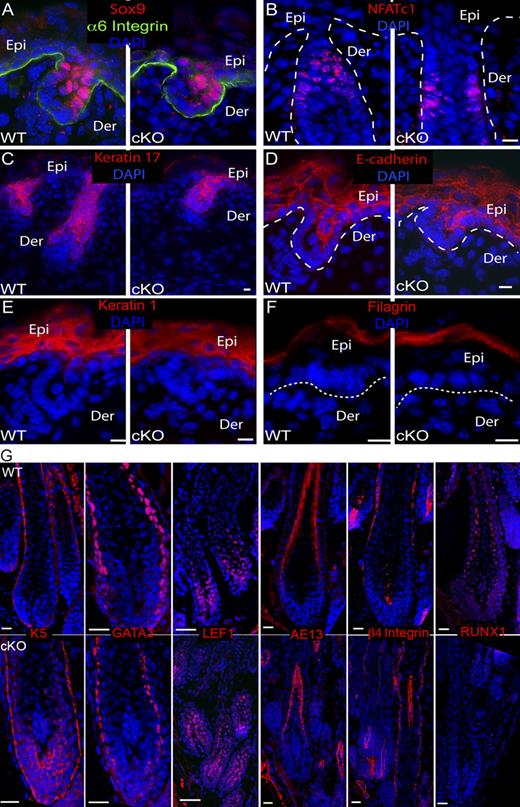
Normal expression of HFSCs and hair markers in Runx1 cKO. (A–F) Skin sections from WT and Runx1 cKO embryos at E16.5 immunostained for indicated markers. n = 3 WT and 3 cKO mice. Epi, epidermis; De, dermis. Bars, 10 µm. (G) PD6-9 skin sections from WT and Runx1 cKO littermates stained for indicated differentiation markers show normal expression. See Fig. S1 B for HF lineage reference. Bars, 10 µm.
Normal expression of HFSCs and hair markers in Runx1 cKO. (A–F) Skin sections from WT and Runx1 cKO embryos at E16.5 immunostained for indicated markers. n = 3 WT and 3 cKO mice. Epi, epidermis; De, dermis. Bars, 10 µm. (G) PD6-9 skin sections from WT and Runx1 cKO littermates stained for indicated differentiation markers show normal expression. See Fig. S1 B for HF lineage reference. Bars, 10 µm.
Runx1CreER targets Runx1 mainly in the skin mesenchyme and impairs adult HF maintenance
Next we asked if Runx1 might also work from the dermis to regulate the epithelial–mesenchymal cross talk. Dermal cells in the HFSC environment work as support or niche cells both in vitro and in vivo (Watt and Hogan, 2000; Fuchs, 2009). Runx1 was highly expressed in the embryo dermis (Fig. 1 A) and was turned off by PD8 (Raveh et al., 2006; Osorio et al., 2008). Runx1CreER embryonic activity was efficient in the skin dermis (Figs. 2 B and 3 B), but highly inefficient in the epithelium (7% of total HFs, which showed mostly rare labeled cells; Fig. 2 C). This allele was designed to result in a nonfunctional truncated Runx1 protein (Samokhvalov et al., 2007). Thus, we reasoned that a Runx1CreER/Fl induced with tamoxifen in embryogenesis (E12.5, 13.5, and 14.5) would generate an essentially mesenchymal Runx1 knockout. As expected, Runx1 staining appeared largely diminished in dermis but not in most HFs at E18.5 (Fig. 8 A). Further analysis of a Runx1CreER/Fl;Rosa26R mouse confirmed these results, and tracked the postnatal and adult fate of the Runx1 mutant skin progenitor cells (see following paragraphs).
Runx1 is essential in the embryonic mesenchyme for long-term maintenance of adult HFs. (A) E18.5 skin largely lacks Runx1 staining in the mesenchyme of Runx1CreER/Fl;R26R mice when compared with control littermate. (B) Runx1CreER/Fl and littermate control mice at PD10. Newborn skin at 14, 22, 37, and 93 post-grafting (PG) onto Nude mice. (C) Skin sections from Runx1CreER/Fl;Rosa26R mice at time points indicated stained with X-Gal and hematoxylin. Note strong X-Gal staining in the upper dermis surrounding the HFs and bent hair shafts and regional loss of HFs by PG93. (D) Oil Red O and hematoxylin-stained skin images from Runx1CreER/Fl and WT littermates at time points indicated. Note enlargement of SG at PD10 and cyst formation with loss of the lower HF structures (bulb/bulge). (E) Quantification of X-Gal+ labeling patterns in distinct HF compartments as indicated. Note increase in SG contribution at the expense of bulb. *, P ≤ 0.004. Inf, infundibulum; SG, sebaceous gland. (F) General increased Lef1 staining in Runx1CreER/Fl (mesenchymal KO) skin at E18.5, PD10, and PG37. Note ectopic Lef1 expression in the bulge at PG37. (G) Increased Wnt signaling in Runx1CreER/Fl mice; BATGAL mice showed in X-gal–stained embryo skin. (H) Phenotype summary of Runx1CreER/Fl induced mice. Bars, 10 µm.
Runx1 is essential in the embryonic mesenchyme for long-term maintenance of adult HFs. (A) E18.5 skin largely lacks Runx1 staining in the mesenchyme of Runx1CreER/Fl;R26R mice when compared with control littermate. (B) Runx1CreER/Fl and littermate control mice at PD10. Newborn skin at 14, 22, 37, and 93 post-grafting (PG) onto Nude mice. (C) Skin sections from Runx1CreER/Fl;Rosa26R mice at time points indicated stained with X-Gal and hematoxylin. Note strong X-Gal staining in the upper dermis surrounding the HFs and bent hair shafts and regional loss of HFs by PG93. (D) Oil Red O and hematoxylin-stained skin images from Runx1CreER/Fl and WT littermates at time points indicated. Note enlargement of SG at PD10 and cyst formation with loss of the lower HF structures (bulb/bulge). (E) Quantification of X-Gal+ labeling patterns in distinct HF compartments as indicated. Note increase in SG contribution at the expense of bulb. *, P ≤ 0.004. Inf, infundibulum; SG, sebaceous gland. (F) General increased Lef1 staining in Runx1CreER/Fl (mesenchymal KO) skin at E18.5, PD10, and PG37. Note ectopic Lef1 expression in the bulge at PG37. (G) Increased Wnt signaling in Runx1CreER/Fl mice; BATGAL mice showed in X-gal–stained embryo skin. (H) Phenotype summary of Runx1CreER/Fl induced mice. Bars, 10 µm.
The Runx1CreER/Fl-induced mice were born, but were generally cannibalized by their mothers, except one who survived to PD10 and showed a marked pigmentation of the skin and delay in hair shaft emergence (Fig. 8 B). To examine the skin phenotype in adulthood we grafted newborn Runx1CreER/Fl;R26R and littermates Runx1Fl/+ (WT) control skin, as well as control Runx1CreER/+;R26R skin induced with tamoxifen in utero (see Materials and methods) onto nude (immune compromised) mice (n = 4 Runx1CreER/Fl; R26R n = 2 control Runx1Fl/+;R26R). We again found a delay in hair shaft emergence and fewer HFs in induced Runx1CreER/Fl relative to control skin at 14 d post-grafting (PG; Fig. 8 B).
Next we analyzed Runx1 mutant and control skin at various stages for the distribution of X-Gal+ cells. As expected, we found many X-Gal+ cells indicative of CreER activity in a large fraction of dermal cells in skin sections from E18.5 and PD10 mice, lost by 90 d PG from both mutant and control mice (Fig. 8 C and unpublished data). In grafted Runx1CreER/Fl;Rosa26R-induced skin the X-Gal+ epithelial HF cells were found in ∼10% of Runx1CreER/Fl and ∼8% Runx1Fl/+ skin at PG37–56 (Fig. 8 E). Thus, skin progenitors tracked here survived into adulthood in the absence of Runx1 and were found in roughly comparable numbers in the different skin compartments. Rare exceptions of over-proliferation, likely attributable to local skin injury and inflammation, are not shown.
The skin and HFs appeared normal at E18.5 in Runx1CreER/Fl but by PD10 the shafts appeared bent in the hair canal, the skin was markedly pigmented, and the SGs enlarged (Fig. 8, B and C). By telogen the HFs of the Runx1CreER/Fl-grafted skin (post-grafting PG39) displayed even larger SGs, which eventually became cyst-like looking structures filled with sebum that lacked the bulb and bulge HF regions (Fig. 8 D and Fig. S4), while some skin regions lacked the HFs altogether (Fig. 8 C, PG93). The preferential differentiation of Runx1-expressing progenitors to SGs was also supported by our counts of X-Gal+ cells in induced Runx1CreER/Fl;R26R-grafted skin during anagen (PG37–39). These counts revealed higher X-gal+ cell numbers (relative to Runx1CreER/+ controls) in the upper HFs (SG and hair canal) at the expense of the hair bulb (lower ORS, matrix, and inner layers; Fig. 8 E).
The conversion of HFs to enlarged cystic SGs in the adult hair cycle (but not in morphogenesis) is reminiscent of the adult phenotype of a transgenic mouse expressing the K14-ΔN-Lef1 allele in the HFSCs from embryogenesis to adulthood (Lowry et al., 2005). Consistent with this observation, Lef1 immunoreactivity was increased in the E18.5 Runx1CreER/Fl skin mesenchyme, as well as in the upper hair epithelial compartment (normally containing the SC compartment with Sox9high, NFATc1high, Runx1high, and Lef1low expression; Fig. 8 F) and remained high in the bulge (a compartment normally negative for Lef1), at PD10 and PG37–39 grafted skin (Fig. 8 F). The generalized up-regulation of Lef1in the HFs (not confined to the 10% HFs lacking Runx1 in the epithelium) pointed to a paracrine effect of Runx1 in the mesenchyme to control Lef1 expression in the epithelium starting in embryogenesis and persisting into adulthood.
Because generalized increase Lef1 expression might impact on Wnt signaling, we generated Runx1CreER/Fl;BAT-GAL Wnt signaling reporter mice and induced Runx1 loss with tamoxifen at E12.5, 13.5, and 14.5. X-Gal signal was strikingly increased in both the mesenchymal and the epithelial compartment (Fig. 8, G and H; n = 2 WT and 2 Runx1CreER/Fl).
Because Runx1 is absent in adult mesenchyme (Raveh et al., 2006; Osorio et al., 2008), it was not surprising that induction of the Runx1CreER/Fl mice with tamoxifen at PD21 resulted in normal-looking skin later on in life (Fig. 8 H and unpublished data). In summary, loss of Runx1 in the skin embryo mesenchyme resulted in preferential differentiation toward SGs over hair bulb lineages and eventually loss of HFs. Given the similarity with previously reported Lef1 and Wnt mutant phenotypes (Gat et al., 1998; Merrill et al., 2001; Andl et al., 2002) and the de-regulation of Lef1 and Wnt signaling described here, it is likely the latter are major factors in mediating the role Runx1 plays in the embryonic skin mesenchyme.
Runx1 interaction with Lef1 and Wnt-signaling pathway
To understand how Runx1 loss induces opposing effects on Wnt signaling from the two skin compartments we performed QRT-PCR (SA-Biosciences) microarrays (n = 2 for each comparison). We compared epidermis or dermis from E18.5 embryos in which Runx1 was deleted either in the epithelium via K14Cre (cKO) or in the mesenchyme via Runx1-CreER (iKO). Data analysis was performed using the SA-Biosciences software, from which scatter plots for each comparison are presented in Fig. 9 A. As expected from the paracrine effect induced by Runx1 loss, we found that both the mesenchymal and the epithelial compartment from the two types of knockout skin showed transcriptional changes (Table S1). Dermis of the mesenchymal KO (iKO) showed mRNA up-regulation of Wnt16, a secreted activating ligand that is likely to elicit a broad effect on neighboring cells. The epidermis seems to respond to the Runx1 loss in the mesenchyme by mRNA up-regulation of several secreted activating molecules such as Wnt11 and Wnt5a, in conjunction with Fzd4 receptor, Wisp1, and Nkd1. All of these changes combined are consistent with the general BAT-GAL up-regulation detected in ikO skin (Fig. 8 G). In the epithelial Runx1 KO (cKO), in which BAT-GAL was down-regulated (Fig. 6), activating molecules such as secreted ligands Wnt11 and Wnt2b were down-regulated along with the Fzd4 receptor. The dermis responded to loss of Runx1 in the epithelium by up-regulating Wnt inhibitory molecules such Sfrp1, Apc, Gsk3β, and Nlk (Ishitani et al., 1999; Chien et al., 2009), and by down-regulating activating Wnt factors Bcl9 (Kramps et al., 2002) and downstream target Foxn1 (Balciunaite et al., 2002). The changes in mRNAs for many Wnt regulatory secreted molecules explain the paracrine effect Runx1 has from either of the two skin compartments. Not all the genes that significantly changed in our QRT-PCR analysis were consistent with the corresponding BAT-GAL signal for each type of Runx1 knockout (Table S1). For example, a few Wnt activators were up-regulated in the iKO and vice versa, some Wnt inhibitors were up-regulated in the cKO. This might suggest that compensation mechanisms could be triggered to counteract the effect of Runx1 KO in the skin, and might explain why some of the phenotypes described here (such as HF developmental delay in cKO) are transient. Alternatively, some of the molecules detected here (such as Wnt or Sfrp) can play opposing effects on Wnt signaling depending on the context (Chien et al., 2009). Clearly, more is to be learned about the interaction of these molecules in skin homeostasis.
Runx1 molecular interaction with the Wnt-signaling pathway. (A) Wnt signaling SA-Bioscience QRT-PCR array on dermis or epidermis from E18.5 embryos with epithelial knockout (cKO) or inducible mesenchymal knockout (iKO) are shown as scattered plots of fold changes normalized to three housekeeping genes for KO vs. WT littermate controls. Red dots are up- and green dots are down-regulated genes. (B) Select list of genes up-regulated on average by greater than twofold by SA-Biosciences software analysis (see Table S1) in each type of KO and skin compartment when compared with the corresponding WT control. (C) Wnt luciferase reporter assay in keratinocytes transfected with DNA constructs indicated at bottom. Note lower activity induced by the Runt domain.
Runx1 molecular interaction with the Wnt-signaling pathway. (A) Wnt signaling SA-Bioscience QRT-PCR array on dermis or epidermis from E18.5 embryos with epithelial knockout (cKO) or inducible mesenchymal knockout (iKO) are shown as scattered plots of fold changes normalized to three housekeeping genes for KO vs. WT littermate controls. Red dots are up- and green dots are down-regulated genes. (B) Select list of genes up-regulated on average by greater than twofold by SA-Biosciences software analysis (see Table S1) in each type of KO and skin compartment when compared with the corresponding WT control. (C) Wnt luciferase reporter assay in keratinocytes transfected with DNA constructs indicated at bottom. Note lower activity induced by the Runt domain.
Finally, we performed luciferase Wnt reporter assay in cultured keratinocytes transfected with a Runx dominant-negative form, Runt-domain (Sakakura et al., 1994; Hoi et al., 2010) fused with myc-tag and GFP and placed in a CMV expression vector (Hoi et al., 2010). Indeed, using previously generated Wnt assay DNA constructs (Zhou et al., 1995; Korinek et al., 1997; Gat et al., 1998) we found that Runt reduced the luciferase signal induced by ΔN-βcatenin and Lef1 action on a TOPFLASH reporter assay (Fig. 9 C). This suggested that appropriate levels of Runx1 activity might be important for amplifying Wnt signaling in conjunction with the core components of the canonical pathway.
Discussion
Here we ask if the master regulator of hematopoietic SC emergence, Runx1 (Dzierzak and Speck, 2008), plays a role in the development and maintenance of adult HFSCs. We show by expression analysis and lineage-tracing experiments that Runx1 is expressed during embryogenesis in: (a) short-term HF progenitors that make the first hair shaft; (b) precursors of adult HFSCs; and (c) skin mesenchyme progenitor cells (Fig. 10 A). Runx1 expression in the skin epithelium modulates development and timely emergence of all the early precursors of hair lineages including those of the adult HFSCs, but is dispensable for the formation of all of them. In contrast, Runx1 expression in the embryonic skin mesenchyme is dispensable for early morphogenesis, but is crucial for specifying competent adult HFSCs that can be properly differentiated and maintained later on in postnatal life. Therefore, Runx1 works both in the SC environment (skin mesenchyme) and within the adult HFSCs precursors to set up the proper development and long-term function of adult HFSCs. Runx1 orchestrates cell adhesion/spreading and migration in epithelial cells and impacts Wnt signaling and Lef1 expression in opposing directions from the two skin compartments (Fig. 10 B).
Model of Runx1 lineage contribution and function during HF morphogenesis. (A) Lineage tracing suggests Runx1-expresing embryonic cells contribute to: (a) the short-term differentiated HF lineages during morphogenesis; (b) the adult HFSC compartment; and (c) the skin mesenchyme. The embryonic mesenchymal population is largely diminished in adulthood. (B) Reciprocal Runx1 targeting in the mesenchyme and epithelium during embryogenesis de-regulates Lef1 and Wnt signaling in opposite directions and has an effect on HFSC differentiation and long-term maintenance in adulthood.
Model of Runx1 lineage contribution and function during HF morphogenesis. (A) Lineage tracing suggests Runx1-expresing embryonic cells contribute to: (a) the short-term differentiated HF lineages during morphogenesis; (b) the adult HFSC compartment; and (c) the skin mesenchyme. The embryonic mesenchymal population is largely diminished in adulthood. (B) Reciprocal Runx1 targeting in the mesenchyme and epithelium during embryogenesis de-regulates Lef1 and Wnt signaling in opposite directions and has an effect on HFSC differentiation and long-term maintenance in adulthood.
Runx1 expression marks distinct precursors of adult HFSCs and short-term progenitors in the earliest stages of embryonic hair development
Previous genetic labeling tracked the fate of epithelial HF cells during morphogenesis, homeostasis, and injury (Legué and Nicolas, 2005; Levy et al., 2005; Nowak et al., 2008; Zhang et al., 2009; Legué et al., 2010). Single-cell lineage tracing in postnatal morphogenesis showed exclusive labeling of either ORS or M/IL cells (Legué and Nicolas, 2005), suggesting that these are independent lineages at discrete stages of morphogenesis. Furthermore, Sox9-Cre marked bulk ORS cells, which did not contribute to the differentiated HF lineages matrix (M) and inner layers (IL) by PD7 (Nowak et al., 2008). In this context, our data in which we detect a large fraction of HFs labeled in early embryonic morphogenesis presenting either ORS or M/IL labeling by PD0, suggested that Runx1 was expressed in the distinct progenitors of these two lineages. In the future, using more specifically expressed factors, such as the ORS marker Sox9 or the early bulge marker NFATc1 (Horsley et al., 2008), would allow better resolution of the two populations likely existent in the hair placode.
Our genetic marking proved that the late Runx1+ embryonic cells have a lower survival rate than the early Runx1+ progenitors, as shown by the drop in total number of X-Gal+ HFs by late postnatal morphogenesis and in adulthood. Thus, most late cells contribute exclusively to hair morphogenesis (Fig. 10 A), resembling the “primitive” short-lived progenitors described for hematopoiesis (Dzierzak and Speck, 2008), whereas most early cells contribute progeny to adult hair homeostasis and long-lived HFSCs. Our detection of ORS-exclusive patterns throughout morphogenesis suggested that at least a fraction of the ORS/bulge cells might remain unused in morphogenesis for later use in adult homeostasis as stem cells.
Adult types of slow-cycling HFSCs were proposed to emerge at ∼E18.5, when HFs are already well formed (Nowak et al., 2008). Our study establishes the existence of a population of Runx1-expressing HFSC precursors largely committed to ORS and adult homeostasis at the earliest stages of hair morphogenesis in the hair placode. This provides another developmental example, in addition to that found in muscle and blood (Dzierzak and Speck, 2008; Messina and Cossu, 2009), in which the adult SCs emerge as independent lineages in the early tissue rudiments (Slack, 2008). Perhaps this developmental mechanism is common among SCs of regenerative tissues.
Embryonic Runx1 works in the epithelium for timely HF development and in the mesenchyme for adult HF integrity
When we delete Runx1 in the HF placode, the emergence of both short-term HF progenitors and precursors of adult HFSCs from the single layer of ectoderm is delayed. This is reminiscent of the role Runx1 plays in adulthood for timely induction of adult HFSC activation and hair regeneration (Osorio et al., 2008; Hoi et al., 2010). Wnt signaling plays important roles in HF morphogenesis and SC activation (Schneider et al., 2009). Thus, the down-regulation of Lef1 and Wnt reporter Bat-Gal in the epithelial cKO Runx1 skin might explain the delay in HF induction.
The skin mesenchyme is an important HFSC niche component (Schmidt-Ullrich and Paus, 2005; Plikus et al., 2008) and Runx1 is expressed there in embryogenesis but not in adulthood (this paper; Raveh et al., 2006; Osorio et al., 2008). Surprisingly, our deletion of Runx1 in the skin embryonic mesenchyme resulted in relatively normal morphogenesis, but induced a generalized defect much later in the adult epithelial skin. This included severe defects of the hair shaft propensity to span the skin surface and preferred differentiation toward SGs at the expense of the hair bulbs. Eventually this resulted in the regional loss of HFs later on in life, which were replaced by sebum-filled cysts. Given the similarity in this phenotype with a Lef1 epithelial transgenic mouse (Merrill et al., 2001), it appears relevant that Lef1 was up-regulated in skin in the mesenchymal Runx1 mutant. As seen in the epithelial knockout, the effect of Runx1 loss on Wnt signaling was not only confined to the Cre-targeted cells, but also spread out in the neighboring cells. In the HF epithelium the ectopic high expression of Lef1 was especially striking in the Runx1highSox9high upper HF compartment where the precursors of adult HFSCs likely reside, which normally express low levels of Lef1. This ectopic expression was maintained in adult HFSCs. Thus, it appears that Runx1 might work in the embryonic mesenchyme to repress noncell-autonomous Lef1 function in the precursors of adult HFSCs for their normal lineage differentiation and maintenance in adulthood (Fig. 10 B).
Our findings show that Runx1 loss perturbed the bi-directional balance of epithelial–mesenchymal interaction in the skin by playing antagonistic and paracrine effects on the Lef1 protein level and Wnt signaling in the two skin compartments. How can such complex function be achieved? Runx1 is known to work as a context-dependent transcriptional activator or repressor (Dzierzak and Speck, 2008). In fact, we found a different set of mRNAs of specific Wnt pathway members changed upon Runx1 loss in the epithelium versus the mesenchyme. Significantly, many of these factors are secreted molecules that can act as either activators or inhibitors on their mother cells and on neighboring cells, explaining the noncell-autonomous role of Runx1 in regulating Wnt signaling. The combined cell-autonomous and noncell-autonomous action of Runx1 appears critical to achieve the appropriate balance of Lef1 levels and Wnt activation in specific regions of the skin. These regional microenvironments orchestrated by Runx1 in the skin are critical for proper regulation of HF development, and for timely HFSC emergence, differentiation, and long-term maintenance.
Materials and methods
Mice
Mouse work was approved by the Cornell University Institutional Animal Care and Use Committee. K14-Cre/Runx1Fl/Fl mice were generated as described previously (Osorio et al., 2008). Runx1+/LacZ mice were maintained in the C57BL6 background. To generate K14-Cre;Runx1Fl/Fl; Bat-gal reporter mice we crossed an F2 generation of Runx1Fl/Fl; Bat gal+ to K14-Cre. Selection of the reporter cKO was based on genotyping for lacZ, K14Cre, and Runx1Fl/Fl (North et al., 1999; Vasioukhin et al., 1999; Growney et al., 2005). Control mice were WT littermates.
Lineage analysis studies and X-Gal staining
Runx1CreER males were mated to R26R females. Day of plug was counted as E0.5. Females received one daily injection for three consecutive days of tamoxifen (16 µg/g body weight) and progesterone (8 µg/g of body weight). At day 19.5 females were sacrificed, and pups were surgically delivered and transferred to foster mothers or sacrificed for tissue analysis. For 5-bromo-4-chloro-3-indoxyl-β-d-galactopyranoside (X-Gal) staining, 10-, 20-, 50-, and 90-µm skin sections were fixed for 1 min in 0.1% glutaraldehyde in PBS, washed 3 × 1 h in cold PBS with 0.01% of NP-40 and 100 mM of sodium dexycholate. After incubation in X-gal solution (North et al., 1999) at 37°C for 12–16 h, slides were rinse in PBS and incubated for 3 h in 1 M NaCO3. Then the slides were washed in PBS, counterstained with hematoxylin, and mounted in 70% glycerol.
Skin grafting
Nu/Nu females were anesthetized with avertin and injected with the pain reliever ketoprofen. Each female received a piece of skin from newborn Runx1CreER/fl;Rosa26R or Runx1CreER/+;Rosa26R mice that were induced with tamoxifen from E12.5 to E14.5. Graft was secured with bandages and gauzes. 2 wk after grafting the bandages were removed.
Histology and immunofluorescence
Immunofluorescence and hematoxylin and eosin staining (H&E) of skin tissue were described previously (Tumbar et al., 2004; Tumbar, 2006). In brief, the skin tissue was collected and embedded in freezing media (OCT), frozen on dry ice, and kept at −80°C for long-term preservation. Frozen skin sections were cut with a Microm HM550 (Richard-Allan Scientific), collected on slides, fixed, and stained. For alkaline phosphatase staining frozen tissue was fixed for 5 min in 2% formaldehyde, washed with 100 mM Tris-Base buffer 9.5, and incubated for 30 min in NCIB/DAB substrate. MOM Basic kit (Vector Laboratories) was used for mouse antibodies. Nuclei were labeled by DAPI. For S-phase labeling, BrdU (Sigma-Aldrich) was injected i.p. at 25 µg/g body weight in saline buffer (PBS) to pregnant dams. Females were sacrificed 3.5 h later and tissue was processed for further analysis (n = 6 cKO and 6 WT). Staining of skin sections was described previously (Tumbar, 2006).
Antibodies for immunofluorescence were from (1) rat: α6- and β4-integrins (1:150), BrdU (1:300; Abcam), α4-integrin (CD49, 1:200; Millipore), VCAM-1 (CD106, 1:200; BD), FITC-conjugated F4/80 (1:50 and 1:62; BioLegend), PECAM-1 (CD31, 1:100; BD), CD34 (1:150; eBioscience), biotin-conjugated CD45 (1:50; BD); (2) rabbit: β-Gal (1:2,000; Cappel), keratin 5 and keratin 17 (1:1,000; Covance), E-cadherin (1:500), LEF1 (1:700; Cell Signaling Technology), Runx1 (1:4,000; T. Jessel, Columbia University, New York, NY), Sox9 (rabbit, 1:500; M. Wegner, Erlangen-Nürnberg University, Erlangen, Germany), Ki67 (1:100; Novocastra), AE13 (1:50; Immunoquest), GATA3 (1:100; Santa Cruz Biotechnology, Inc.), NFATc1 (1:25; Santa Cruz Biotechnology, Inc.), vinculin (1:100), phalloidin-TRed (1:250); (3) mouse: Hsp47 (collagen, 1:50–1:1,000; none worked), SMA (1:50; Thermo Fisher Scientific); and (4) hamster: FITC-conjugated CD11C (1:50; eBioscience). Secondary antibodies coupled to the following fluorophores: FITC, Texas red, streptavidin-conjugated Texas red, or Cy5 were purchased from The Jackson Laboratory. A detailed antibody staining protocol was described in a recently published methods paper (Tumbar, 2006).
Attachment and cell migration studies
Newborn skin was treated with dispase overnight at 4°C. Keratinocytes were isolated by treating the epidermis with 0.1% trypsin for 15 min at room temperature, followed by cell filtration and cell counting using a hemocytometer. For more details see Tumbar (2006). Cells were plated onto collagen 1, fibronectin, and poly-d-lysine–precoated microslides (Cell Signaling Technology). For attachment studies, 8-well microslides were used. In brief, 100,000 live cells were plated per well (n = 5 WT and 5 cKO) and 24 h later cells were fixed in 4% PFA and stained with vinculin, phalloidin, and DAPI. The total number of cells was counted per well in 24 pictures at 10x for each well (2 wells per sample) and analyzed using ImageJ (n = 5 WT and 5 KO).
For migration studies, 4-well collagen-precoated microslides were used. One million cells were plated per well and 36 h after plating a small scratch was made with a pipette tip. Pictures were taken at different time points after the wound. Wound healing was analyzed using ImageJ.
Image acquisition and processing
Image acquisition was performed with a fluorescent Nikon microscope equipped with a Retiga Exi camera (MVI) using the IP-Laboratory software. Transmitted light microscopy images were collected using phase-contrast objectives and rings. When direct comparisons of fluorescence signal levels were needed, wild-type and knockout skin were processed side by side and images were collected the same day, using constant exposure times. Images were imported in Adobe Photoshop, assembled in montages, and enhanced for levels, brightness, and contrast simultaneously to preserve the differences in signal observed in the original data. Images were then cut and transferred in Adobe Illustrator for montage assembly.
QRT-PCR and flow cytometry analysis (FACS)
For flow cytometry cells were isolated from E16.5 back skin stained with phycoerythrin (PE)-labeled α6-integrin (CD49f) antibody (BD), as described previously (Tumbar et al., 2004; Tumbar, 2006). Live cells were those excluding propidium iodide (PI; Sigma-Aldrich). Flow cytometry was performed using BD FACS Aria at Cornell. RNA isolation from sorted cells and RT-PCR of cDNAs were described previously (Tumbar, 2006; Waghmare et al., 2008). RNA for the SA-Biosciences arrays was isolated using Trizol from E18.5 skin that was separated into epidermis and dermis with dispase treatment (Tumbar, 2006). We prepared cDNA using a Superscript III reverse-transcription kit (Invitrogen) and used the equivalent of 1 µg of mRNA per QRT-PCR array plate. QRT-PCR was run with a single-color thermocycler (Bio-Rad Laboratories) and analysis of the Ct values and fold change was performed with the SA-Biosciences proprietary software. Experiments were run in duplicate and three housekeeping genes were used for normalization (Table S1).
Oil Red O staining
10–40-µm-thick frozen tissue sections were incubated for 2 min in hematoxylin and washed in H2O. Slides were incubated in Oil Red working solution (stock solution 0.3% wt/vol diluted in isopropanol, working solution 3:2 ratio of Oil Red [stock solution] to distilled water) for 10 min, rinsed in distilled water, and mounted in 70% glycerol.
Wnt reporter assay
TOPFLASH luciferase assays were done on skin keratinocytes (Merrill et al., 2001). One day after plating in a 24-well dish at 2 x 104 per well, cells received 0.2 µg of DNA transfected via Transfectin (Bio-Rad Laboratories). The following constructs were used for transfection: TOPFLASH, FOPFLASH, K14-ΔNbcat, K14-Lef1 (Zhou et al., 1995; Korinek et al., 1997; Gat et al., 1998; Merrill et al., 2001), CMV-Myc-RUNT-GFP, CMV-GFP-Myc (Hoi et al., 2010), and Renilla. On d 2 cells were cotransfected with either TOPFLASH or FOPFLASH and (1) Renilla; (2) Renilla, K14-ΔNbcat, K14-lef1, and RUNT; or (3) Renilla, K14-ΔNbcat, K14-lef1, and GFP-MYC. 48 h after transfection the cells were harvested and prepared with the Promega Dual Luciferase Reporter Assay System kit for measuring luciferase activity.
Statistical analyses
Data are shown as averages and SDs or SEM. t tests analyses were done with Excel 2008 (Microsoft).
Online supplemental material
Fig. S1 is a schematic of hair morphogenesis and mature hair organization. Fig. S2 shows lineage tracing in morphogenesis and developmental HF delay in Runx1 cKO skin. Fig. S3 shows colocalization of Runx1-expressing cells with dermal markers. Fig. S4 shows phenotypic analysis of Runx1CreER/Fl;Rosa26R during hair morphogenesis and adulthood. Table S1 shows the analysis of Wnt-signaling SA-Biosciences QRT-PCR arrays.
Acknowledgments
We thank Elaine Fuchs, Nancy Speck, Hans Clevers, and Igor Samokhvalov for mice and/or DNA constructs; James Lee Smith for cell sorting; Valerie Horsley for critical reading of the manuscript; and David McDermitt for technical assistance with skin grafting.
This work was funded by grants from NIH/NIAHMS RO1AR053201 and NYSTEM C024354.
The authors declare no competing interest.