In the metazoan replication timing program, clusters of replication origins located in different subchromosomal domains fire at different times during S phase. We have used Xenopus laevis egg extracts to drive an accelerated replication timing program in mammalian nuclei. Although replicative stress caused checkpoint-induced slowing of the timing program, inhibition of checkpoint kinases in an unperturbed S phase did not accelerate it. Lowering cyclin-dependent kinase (Cdk) activity slowed both replication rate and progression through the timing program, whereas raising Cdk activity increased them. Surprisingly, modest alteration of Cdk activity changed the amount of DNA synthesized during different stages of the timing program. This was associated with a change in the number of active replication factories, whereas the distribution of origins within active factories remained relatively normal. The ability of Cdks to differentially effect replication initiation, factory activation, and progression through the timing program provides new insights into the way that chromosomal DNA replication is organized during S phase.
Introduction
The hundreds or thousands of replication origins distributed throughout each eukaryotic genome initiate replication forks at distinct and reproducible times during the course of S phase (Goren and Cedar, 2003; Donaldson, 2005; Zink, 2006). Replication timing profiles of the entire genome of budding yeast (Raghuraman et al., 2001), fission yeast (Heichinger et al., 2006; Eshaghi et al., 2007; Hayashi et al., 2007; Mickle et al., 2007), Drosophila melanogaster (Schübeler et al., 2002; MacAlpine et al., 2004), and mammals (White et al., 2004; Woodfine et al., 2004, 2005; Jeon et al., 2005; Cohen et al., 2006; Farkash-Amar et al., 2008; Hiratani et al., 2008) have been described previously. In general, transcriptionally active euchromatin replicates early in S phase, whereas transcriptionally inactive heterochromatin replicates late. In metazoans, chromosome domains that replicate at similar times are clustered together within the nucleus so that replication occurring at different times in S phase displays distinct intranuclear patterns (Goren and Cedar, 2003; Zink, 2006).
Experiments in yeast suggest that replication origins become programmed for late replication at some stage during G1 (Raghuraman et al., 1997). A similar “timing decision point” was defined in metazoans as a stage in early G1 when tissue culture nuclei acquire the ability to support a normal replication timing program when subsequently driven into S phase by Xenopus laevis egg extracts (Dimitrova and Gilbert, 1999). This represents the establishment of the replication timing program and coincides with movement of early- or late-firing chromosomal regions to appropriate positions within the nucleus, which may involve chromatin modification of the region surrounding replication origins.
Relatively little is known about how the timing program is executed during S phase. Each replication origin is loaded with Mcm2-7 during late mitosis and G1 to form a prereplicative complex that licenses the origin for a single initiation event in the coming S phase (Nishitani and Lygerou, 2004; Blow and Dutta, 2005). During S phase, each prereplicative complex is acted on by Cdks and the Dbf4-dependent kinases to induce initiation. It is likely that Mcm2-7 is the essential substrate for Cdc7 in the initiation of replication. In yeast, the Sld2 and Sld3 proteins have been shown to be essential substrates for Cdks in the initiation of replication, although their vertebrate homologues are currently unknown (Masumoto et al., 2002; Tanaka et al., 2007; Zegerman and Diffley, 2007). Phosphorylation of Sld2 and Sld3 promotes the recruitment of other replisome proteins such as Cdc45 to Mcm2-7 at replication origins.
Replication forks from clusters of adjacent replication origins are organized into replication factories in the nucleus (Jackson and Pombo, 1998; Berezney et al., 2000; Frouin et al., 2003; Kitamura et al., 2006). To account for the number of DNA replication forks generated during S phase, most replication factories must contain multiple replication forks, probably in the range of 5–50 forks per factory (Berezney et al., 2000). Little is known about what causes the clustering of replication origins into factories, although some aspect of chromosomal structure may play a role.
In this study, we examine in detail how Cdk activity drives progression through the replication timing program. We use the experimental system developed by Gilbert et al. (1995), in which replication of mammalian G1 nuclei is driven by incubation in X. laevis egg extracts (Gilbert et al., 1995; Dimitrova and Gilbert, 1999). We provide evidence for a direct function of Cdks in activating replication factories and driving progression through the replication timing program that is distinct from the established function of Cdks in inducing the initiation of replication.
Results
The kinetics of replication in vitro
Nuclei were prepared from CHOC-400 cells released for 4 h into G1 from mitotic synchrony. These had passed the timing decision point and were programmed to replicate according to their normal replication timing program (Dimitrova and Gilbert, 1999). The nuclei were incubated in X. laevis egg extract supplemented with geminin to ensure that only origins licensed in vivo were used (Okuno et al., 2001; Dimitrova et al., 2002). At different times, extracts were pulsed with Cy3-dUTP to label sites of ongoing DNA replication. As reported previously (Dimitrova and Gilbert, 1999), labeling patterns were observed that resembled those seen in vivo. We categorized these as patterns I–V, in accordance with previous nomenclature (Fig. 1 a; O'Keefe et al., 1992). However, it should be noted that the in vitro patterns we observed were not identical to the in vivo ones, particularly at later stages of S phase. In particular, we saw many combined patterns that we designated I/II, II/III, III/IV, and IV/V. The appearance of these combined patterns may be a result of the rapid S phase occurring in vitro (∼2 h compared with ∼12 h in vivo) and is consistent with the idea that the in vitro timing program can progress to later stages before finishing the replication of the earlier stages (see following paragraphs).
Progression of CHOC-400 nuclei through the timing program in vitro. CHOC-400 nuclei were incubated at 10,000 nuclei/µl (∼60 ng DNA/µl) in egg extracts supplemented with geminin. (a) Representative images of CHO nuclei incubated in X. laevis egg extracts and pulse labeled for 5 min with Cy3-dUTP. See Materials and methods for full description of the different labeling patterns. (b) At different times, aliquots were pulse labeled for 5 min with Cy3-dUTP, and the proportion of different replication patterns was assessed. (c) Extract was supplemented with α-[32P]dATP. At different times, total DNA synthesis was measured by TCA precipitation and scintillation counting. The SEM of 14 independent experiments is shown. (d, top) The time of appearance of replication patterns during S phase of CHOC-400 cells as described previously by Dimitrova and Gilbert (1999). (middle) The proportion of time spent by CHOC-400 nuclei replicating specific patterns as calculated in b. (bottom) The amount of DNA synthesis associated with each replication pattern in vitro calculated by scaling the proportions in b by the rate of replication (c).
Progression of CHOC-400 nuclei through the timing program in vitro. CHOC-400 nuclei were incubated at 10,000 nuclei/µl (∼60 ng DNA/µl) in egg extracts supplemented with geminin. (a) Representative images of CHO nuclei incubated in X. laevis egg extracts and pulse labeled for 5 min with Cy3-dUTP. See Materials and methods for full description of the different labeling patterns. (b) At different times, aliquots were pulse labeled for 5 min with Cy3-dUTP, and the proportion of different replication patterns was assessed. (c) Extract was supplemented with α-[32P]dATP. At different times, total DNA synthesis was measured by TCA precipitation and scintillation counting. The SEM of 14 independent experiments is shown. (d, top) The time of appearance of replication patterns during S phase of CHOC-400 cells as described previously by Dimitrova and Gilbert (1999). (middle) The proportion of time spent by CHOC-400 nuclei replicating specific patterns as calculated in b. (bottom) The amount of DNA synthesis associated with each replication pattern in vitro calculated by scaling the proportions in b by the rate of replication (c).
Fig. 1 b shows the proportion of different patterns seen every 10 min during an incubation in vitro. The replication patterns appeared in the same order as in vivo. Although most nuclei reached the late type IV and IV/V patterns after an incubation of 140 min, α-[32P]dATP incorporation suggested that overall replication was inefficient, averaging ∼38% of template DNA replicated (Fig. 1 c). Inefficient replication of somatic nuclei in X. laevis egg extract has been reported previously (Dimitrova and Gilbert, 1998). It is not the result of poor-quality extracts, as CHOC-400 chromosomes and X. laevis sperm nuclei replicated efficiently (unpublished data).
Fig. 1 d shows the total time the egg extract spent replicating the different patterns in vitro compared with the times reported for CHO cells in vivo (Dimitrova and Gilbert, 1999). In vitro, relatively more time is spent replicating the later patterns, likely as a result of the lower rates of replication at later stages (Fig. 1 c). When the pattern proportions are normalized to the rate of replication (Fig. 1 d, bottom), they look roughly similar to the in vivo proportions. However, consistent with Dimitrova and Gilbert (1999), we saw very few pure type V labeling patterns in vivo. Preincubation of X. laevis extracts in the absence of nuclei only slightly altered the appearance of replication patterns once template nuclei were subsequently added (Fig. S1). This suggests that correct progression through the replication timing program requires an interaction between extract and template nuclei.
Initiation times associated with the different patterns
Cdks are required throughout S phase for individual origins to initiate replication. Shortly after addition of Cdk inhibitors such as roscovitine to X. laevis egg extracts, most new initiation events are blocked without affecting forks that have already initiated (Strausfeld et al., 1994, 1996; Luciani et al., 2004). To see how this affected the timing program, we added 1 mM roscovitine to extract at different times after addition of CHO nuclei. Initiation events (as indicated by roscovitine-sensitive DNA replication) took place over a period of >80 min (Fig. 2 a and not depicted). This is significantly longer than the ∼25-min initiation period when X. laevis sperm nuclei replicate in X. laevis egg extract (Luciani et al., 2004). Fig. 2 b shows that roscovitine addition blocked the appearance of new replication patterns as expected of an initiation inhibitor. For example, addition of roscovitine at 20 min prevented the appearance of most type II/III, III, and III/IV patterns, suggesting that few initiation events associated with type III DNA had occurred by 20 min. Fig. 2 c shows a hypothetical time course of initiation events associated with each different replication pattern that would be consistent with our results. There is considerable overlap between the times, which may partly explain the labeling of mixed pattern types (I/II and II/III, etc).
Initiation times associated with different replication patterns. CHOC-400 nuclei were incubated at 10,000 nuclei/µl in egg extracts supplemented with geminin. At different times, aliquots were supplemented with 1 mM roscovitine to block further initiation events. (a) Extract was also supplemented with α-[32P]dATP at the start of the incubation. At different times thereafter, total DNA synthesis was measured by TCA precipitation and scintillation counting. (b) At different times, extract was pulse labeled with Cy3-dUTP for 5 min, and the proportion of nuclei showing different replication patterns was assessed. (c) Data from a and b were combined to provide an estimate of the times over which most initiation events associated with the different replication patterns were occurring.
Initiation times associated with different replication patterns. CHOC-400 nuclei were incubated at 10,000 nuclei/µl in egg extracts supplemented with geminin. At different times, aliquots were supplemented with 1 mM roscovitine to block further initiation events. (a) Extract was also supplemented with α-[32P]dATP at the start of the incubation. At different times thereafter, total DNA synthesis was measured by TCA precipitation and scintillation counting. (b) At different times, extract was pulse labeled with Cy3-dUTP for 5 min, and the proportion of nuclei showing different replication patterns was assessed. (c) Data from a and b were combined to provide an estimate of the times over which most initiation events associated with the different replication patterns were occurring.
Regulation of the timing program by checkpoint kinases
We next supplemented extracts with the DNA polymerase inhibitor aphidicolin at 3 µM, which slows replication forks by two- to three-fold and activates checkpoint kinases in egg extracts (Luciani et al., 2004). Fig. 3 a shows that 3 µM aphidicolin led to a dramatic slowing of the replication timing program, so even after 140 min, most CHO nuclei still showed type II and II/III patterns. This slowing of the replication timing program was at least partly caused by activation of checkpoint kinases because it could be partially reversed by treatment with the ataxia telangiectasia and Rad3–related/ataxia telangiectasia–mutated kinase inhibitor caffeine (Fig. S2, a and b; Blasina et al., 1999; Sarkaria et al., 1999; Luciani et al., 2004; Woodward et al., 2006).
Effect of aphidicolin and caffeine on the timing program. (a and b) CHOC-400 nuclei were incubated at 10,000 nuclei/µl in egg extracts supplemented with geminin ± 3 µM aphidicolin (a) or 5 mM caffeine (b). At different times, extract was pulse labeled with Cy3-dUTP for 5 min, and the proportion of nuclei showing different replication patterns was assessed.
Effect of aphidicolin and caffeine on the timing program. (a and b) CHOC-400 nuclei were incubated at 10,000 nuclei/µl in egg extracts supplemented with geminin ± 3 µM aphidicolin (a) or 5 mM caffeine (b). At different times, extract was pulse labeled with Cy3-dUTP for 5 min, and the proportion of nuclei showing different replication patterns was assessed.
There is some controversy over whether checkpoint kinases play a significant role in slowing the replication timing program during unperturbed S phases. Therefore, we examined the effect of caffeine on the timing of CHO nuclei replicating in egg extract in the absence of any replication inhibitor. Fig. 3 b shows that caffeine did not significantly accelerate the replication timing program; if anything, the program was slightly delayed. This suggests that checkpoint kinases do not normally play a major role in slowing progression through the timing program, although they can do so when fork progression is inhibited. Interestingly, caffeine slightly reduced the rate of replication of CHO nuclei (Fig. S2 c), possibly by reducing the stability of stalled replication forks (Fig. S2 d).
The effect of Cdk activity on the timing program
We next investigated whether, in addition to being required to drive replication initiation, Cdks also play a role in driving the replication timing program. If there was a strict coupling between the rate of initiation and the rate of progression through the timing program, the two would be expected to be reduced together in response to reduction of Cdk activity. However, the replication timing program might be completely independent of both Cdk activity and initiation, and in this case, it would proceed unchanged despite a reduction in the frequency of initiation. Fig. 4 a shows the effect of increasing roscovitine concentrations on total histone H1 kinase activity. The decline in Cdk activity was mirrored by a similar decrease in replication rates (Fig. 4 b), suggesting that Cdk activity is rate limiting for DNA replication (Strausfeld et al., 1994; Moore et al., 2002). Fig. 4 c shows replication patterns at corresponding concentrations. Roscovitine concentrations of 1–10 µM inhibited replication but caused only slight changes in replication timing. However, at 30 µM, roscovitine replication barely reached type III at 140 min, and at 100 µM, replication did not get past type I/II. These results show that the progression of the replication timing program is dependent, either directly or indirectly, on Cdk activity.
Effect of low concentrations of roscovitine on the timing program. (a) Extract was supplemented with histone H1, γ-[32P]ATP, and various concentrations of roscovitine. After incubation for 90 min, protein was separated by SDS-PAGE, and 32P incorporation into H1 was assessed by phosphorimager. (b and c) CHOC-400 nuclei were incubated at 10,000 nuclei/µl in egg extracts supplemented with geminin and different concentrations of roscovitine. Aliquots were also supplemented with α-[32P]dATP at the start of the incubation. (b) At different times thereafter, total DNA synthesis was measured by TCA precipitation and scintillation counting. (c) At different times, extract was pulse labeled with Cy3-dUTP for 5 min, and the proportion of nuclei showing different replication patterns was assessed. No replication patterns were visible in extracts treated with 300 µM roscovitine.
Effect of low concentrations of roscovitine on the timing program. (a) Extract was supplemented with histone H1, γ-[32P]ATP, and various concentrations of roscovitine. After incubation for 90 min, protein was separated by SDS-PAGE, and 32P incorporation into H1 was assessed by phosphorimager. (b and c) CHOC-400 nuclei were incubated at 10,000 nuclei/µl in egg extracts supplemented with geminin and different concentrations of roscovitine. Aliquots were also supplemented with α-[32P]dATP at the start of the incubation. (b) At different times thereafter, total DNA synthesis was measured by TCA precipitation and scintillation counting. (c) At different times, extract was pulse labeled with Cy3-dUTP for 5 min, and the proportion of nuclei showing different replication patterns was assessed. No replication patterns were visible in extracts treated with 300 µM roscovitine.
As shown in Fig. 1, even though during a normal S phase most CHO nuclei reached type IV and IV/V patterns, only ∼40% of the template DNA was typically replicated. Therefore, we wondered whether raising Cdk levels could enhance the rate or extent of replication. We tested this using recombinant cyclin A, which when added to X. laevis egg extracts, binds to Cdk1 and provides an S phase–inducing Cdk activity (Strausfeld et al., 1994, 1996). Fig. 5 (a and b) shows that although recombinant cyclin A barely increased total histone H1 kinase activity, 1 fM–1 nM cyclin A stimulated replication of CHO nuclei with an optimum at ∼1 pM. At 1 µM, cyclin A inhibited replication by forcing entry into a premature mitosis (Strausfeld et al., 1996; unpublished data). Because protein synthesis in our extracts is blocked with cycloheximide, the majority of Cdk activity is supplied by cyclin E–Cdk2 (cyclins A and B having been degraded during mitosis and D-type cyclins not being present at this stage of development; Howe et al., 1995; Rempel et al., 1995; Hartley et al., 1996; Vernon and Philpott, 2003). The ability of cyclin A to stimulate DNA replication without significantly increasing total H1 kinase activity is likely explained by it being 10–100 times more effective at inducing replication initiation than cyclin E (Strausfeld et al., 1996; Moore et al., 2002).
Effect of recombinant cyclin A on the timing program. (a) Extract was supplemented with histone H1 γ-[32P]ATP and various concentrations of cyclin A. After incubation for 90 min, protein was separated by SDS-PAGE and 32P incorporation into H1 assessed by phosphorimager. (b and c) CHOC-400 nuclei were incubated at 10,000 nuclei/µl in egg extracts supplemented with geminin and different concentrations of recombinant cyclin A. (b) Aliquots were also supplemented with α-[32P]dATP at the start of the incubation. At different times afterward, total DNA synthesis was measured by TCA precipitation and scintillation counting. (c) At either 45 or 85 min, extract was pulse labeled with Cy3-dUTP for 5 min, and the proportion of nuclei showing different replication patterns was assessed. (d) Extract was supplemented ± 1 pM cyclin A. At different times, extract was pulse labeled with Cy3-dUTP for 5 min, and the proportion of nuclei showing different replication patterns was assessed.
Effect of recombinant cyclin A on the timing program. (a) Extract was supplemented with histone H1 γ-[32P]ATP and various concentrations of cyclin A. After incubation for 90 min, protein was separated by SDS-PAGE and 32P incorporation into H1 assessed by phosphorimager. (b and c) CHOC-400 nuclei were incubated at 10,000 nuclei/µl in egg extracts supplemented with geminin and different concentrations of recombinant cyclin A. (b) Aliquots were also supplemented with α-[32P]dATP at the start of the incubation. At different times afterward, total DNA synthesis was measured by TCA precipitation and scintillation counting. (c) At either 45 or 85 min, extract was pulse labeled with Cy3-dUTP for 5 min, and the proportion of nuclei showing different replication patterns was assessed. (d) Extract was supplemented ± 1 pM cyclin A. At different times, extract was pulse labeled with Cy3-dUTP for 5 min, and the proportion of nuclei showing different replication patterns was assessed.
Fig. 5 c shows that cyclin A also slightly accelerated the replication timing program. This was most evident at 90 min, when cyclin A induced the appearance of type IV patterns. Fig. 5 d shows that 1 pM cyclin A not only accelerated the timing program but also induced appearance of pure type V patterns, which were very rare in control samples. These experiments show that the replication program can be accelerated by increasing Cdk levels.
The role of Cdks in the activation of replication factories
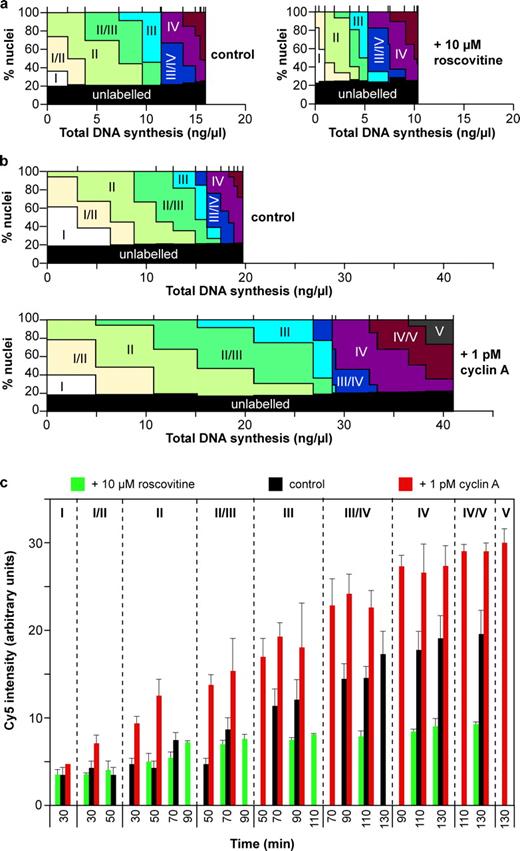
Comparison of Fig. 4 (b and c) suggests that 1–10 µM roscovitine inhibited replication rates more strongly than it inhibited progression through the replication timing program so that nuclei had replicated less DNA than normal when they embarked on later replication patterns. This effect is directly demonstrated in Fig. 6 a. First, a detailed time course was performed with 10 µM roscovitine, with total DNA synthesis and replication patterns measured every 10 min (Fig. S3). The proportion of nuclei showing each pattern was plotted on the vertical axis as previously but with the horizontal axis representing total DNA replication at each time point (Fig. 6 a). This shows that 10 µM roscovitine made the later replication patterns (III, III/IV, IV, and IV) appear at lower levels of DNA replication than in the control. Therefore, lowering Cdk levels decouples initiation from progression through the timing program. The converse effect was seen when extracts were supplemented with 1 pM cyclin A: more DNA replication was seen at each corresponding replication pattern (Fig. 6 b).
Decoupling of replication and the timing program. CHOC-400 nuclei were incubated at 10,000 nuclei/µl in egg extracts supplemented with geminin ± 10 µM roscovitine or 1 pM recombinant cyclin A. (a and b) Data from Fig. 5 and Fig. S2 are shown where the replication patterns at different times are plotted against the total DNA synthesis that had occurred at each time. Vertical ticks above the plot indicate the boundaries of different time points. (c) Extract was also supplemented with Cy5-dCTP at the start of the incubation. At different times, aliquots were pulse labeled with Cy3-dUTP for 5 min, and individual nuclei were analyzed for replication pattern (Cy3 label) or total DNA synthesis (Cy5 label). Error bars indicate mean ± SD.
Decoupling of replication and the timing program. CHOC-400 nuclei were incubated at 10,000 nuclei/µl in egg extracts supplemented with geminin ± 10 µM roscovitine or 1 pM recombinant cyclin A. (a and b) Data from Fig. 5 and Fig. S2 are shown where the replication patterns at different times are plotted against the total DNA synthesis that had occurred at each time. Vertical ticks above the plot indicate the boundaries of different time points. (c) Extract was also supplemented with Cy5-dCTP at the start of the incubation. At different times, aliquots were pulse labeled with Cy3-dUTP for 5 min, and individual nuclei were analyzed for replication pattern (Cy3 label) or total DNA synthesis (Cy5 label). Error bars indicate mean ± SD.
To confirm this, we performed experiments in which total DNA synthesis was measured in individual nuclei. CHO nuclei were continuously labeled with either α-[32P]dATP or Cy5.5-dCTP in extract optionally supplemented with 10 µM roscovitine or 1 pM cyclin A. DNA synthesis in α-[32P]dATP-labeled samples was measured by TCA precipitation and scintillation counting (Fig. S4, a and c). DNA synthesis in the Cy5.5-dCTP–labeled samples was measured by microscopically quantifying Cy5.5 fluorescence in randomly selected nuclei (Fig. S4, b and d). When α-[32P]dATP labeling was plotted against Cy5.5 fluorescence at different times, the result was approximately linear, demonstrating the concordance between the two measurements (Fig. S4, e and f). Cy5.5-dCTP–labeled samples were also pulse labeled with Cy3-dUTP, allowing the determination of replication patterns in nuclei whose total DNA synthesis is known. The results shown in Fig. 6 c reveal two important features. First, in the control sample (Fig. 6 c, black bars), the amount of DNA replication associated with each particular replication pattern fell within fairly narrow confines, as expected of a true replication timing program. Second, 10 µM roscovitine (Fig. 6 c, green bars) significantly lowered the total amount of DNA synthesis associated with each different pattern, whereas cyclin A (Fig. 6 c, red bars) increased it. This shows that altering Cdk activity alters the total rates of replication initiation to a greater degree than progression through the replication timing program and directly demonstrates decoupling of the two processes.
When progression into a new stage of the timing program occurs, new initiation events must occur in newly activated replication factories. Therefore, we investigated which of these aspects were most strongly affected when Cdk activity was varied. CHO nuclei replicating in vitro plus or minus roscovitine or cyclin A were pulsed with Cy3-dUTP in mid– or late S phase (50 or 90 min). Cy3-dUTP labeling revealed the distribution of replication foci, each of which is presumed to consist of one or a small number of replicon clusters (Fig. 7, a–c). We next quantified the number of replication foci present under different Cdk levels and the mean Cy3 intensity of individual foci (which provides an indication of the number of replication forks that they contain). Surprisingly, treatment of extracts with up to 10 µM roscovitine predominantly reduced the total number of foci, leaving their intensity largely unchanged (Fig. 7, d and e). Higher concentrations of roscovitine, which more strongly suppressed both Cdk activity and total DNA replication (Fig. 4, a and b), inhibited both foci number and intensity (Fig. 7, d and e). Conversely, stimulating DNA replication with cyclin A increased the number of replication foci. This effect is highly reproducible (Fig. S5 a) and is also seen when replication factories are visualized with an anti-PCNA antibody (Fig. S5, b–e). Consistent with these results, DNA fiber analysis showed that treatment of extracts with 10 µM roscovitine did not significantly change either replication fork speed or the density of replication forks within active replicon clusters (Fig. 8). These experiments show that modest changes to Cdk activity preferentially alter the activation of replication factories without significantly changing the rate of initiation within active factories.
Replication foci number depends on Cdk levels. CHOC-400 nuclei were incubated in X. laevis egg extracts incubated at 10,000 nuclei/µl in egg extracts supplemented with the indicated concentrations of roscovitine (rosc) or cyclin A (cyc A). Parallel incubations were supplemented with α-[32P]dATP to measure total DNA synthesis. At 50 or 90 min, extract was pulsed for 5 min with Cy3-dUTP. (a–c) Representative images from the 50-min time point are shown. (d and e) The number and mean intensity of Cy3-labeled foci at 50 (d) and 90 (e) min were measured. Error bars indicate SEM between different nuclei.
Replication foci number depends on Cdk levels. CHOC-400 nuclei were incubated in X. laevis egg extracts incubated at 10,000 nuclei/µl in egg extracts supplemented with the indicated concentrations of roscovitine (rosc) or cyclin A (cyc A). Parallel incubations were supplemented with α-[32P]dATP to measure total DNA synthesis. At 50 or 90 min, extract was pulsed for 5 min with Cy3-dUTP. (a–c) Representative images from the 50-min time point are shown. (d and e) The number and mean intensity of Cy3-labeled foci at 50 (d) and 90 (e) min were measured. Error bars indicate SEM between different nuclei.
DNA fiber analysis. (a and b) CHOC-400 nuclei were incubated at 10,000 nuclei/µl in egg extracts supplemented with geminin minus (blue) or plus (red) 10 µM roscovitine (rosc). After incubation for 45 min, extract was pulsed with BrdUTP for 5 min. DNA was isolated, spread on glass slides, and the BrdU-labeled tracks were analyzed. The length of isolated tracks (a) and the distribution of forks within clusters are shown (b).
DNA fiber analysis. (a and b) CHOC-400 nuclei were incubated at 10,000 nuclei/µl in egg extracts supplemented with geminin minus (blue) or plus (red) 10 µM roscovitine (rosc). After incubation for 45 min, extract was pulsed with BrdUTP for 5 min. DNA was isolated, spread on glass slides, and the BrdU-labeled tracks were analyzed. The length of isolated tracks (a) and the distribution of forks within clusters are shown (b).
Discussion
The timing program in X. laevis egg extracts
When X. laevis sperm nuclei are incubated in X. laevis egg extract (mimicking events occurring at fertilization), the sperm DNA is replicated completely in ∼30 min. A rudimentary replication timing program is observed with certain chromosome domains replicating at different stages of this rapid S phase (Labit et al., 2008). In this study, we have examined how the replication timing program is executed in X. laevis egg extracts replicating mammalian CHOC-400 nuclei. As described by Dimitrova and Gilbert (1999), CHO nuclei incubated in X. laevis egg extracts are induced to replicate according to a timing program similar to that occurring normally in CHO cells. In this study, we show that initiation events in the CHO nuclei occurred over 1–2 h in vitro, which is considerably faster than the ∼12 h seen in vivo. Initiation events in vitro associated with the five major labeling patterns occurred in order, but unlike the situation in vivo, they showed considerable overlap.
Given the distribution of forks within origin clusters, it would take >40 min to replicate a typical origin cluster in X. laevis egg extracts, which is similar to the time required in vivo. Because the five stages of the timing program are compressed into ∼2 h in vitro, this makes it inevitable that different replication patterns are seen concurrently. Addition of very high concentrations of roscovitine at different stages through S phase provide further evidence that initiation events associated with different timing stages can occur at the same time. This means that progression from one stage of the timing program to another is not dependent on all of the replication associated with the earlier timing stage having been completed. Indeed, CHO nuclei replicating in vitro progress to late replication patterns with less than half of the template DNA replicated (Dimitrova and Gilbert, 1998; this study).
Progression through the timing program was slowed when fork progression was inhibited by aphidicolin. This slowing of the timing program involved checkpoint kinases and could be partially reversed by coaddition of the checkpoint inhibitor caffeine. When caffeine was added to extract replicating CHO nuclei in the absence of replicative stress, no acceleration of the timing program was seen. This is consistent with work in yeast and mammalian cells suggesting that although checkpoints delay origin firing in response to replicative stress, they do not significantly slow the timing program in unperturbed S phases (Santocanale and Diffley, 1998; Shirahige et al., 1998; Dimitrova and Gilbert, 2000; Feijoo et al., 2001). In contrast, Shechter et al. (2004) showed that under certain circumstances, inhibition of checkpoint kinases in X. laevis egg extracts can accelerate the replication timing program. Importantly, the caffeine-induced acceleration of replication reported by Shechter et al. (2004) was only observed when replication rates had been slowed by the use of very high concentrations of template sperm nuclei. At lower concentrations of nuclei that resemble those seen in the early embryo, inhibition of checkpoint kinases did not greatly accelerate the timing program (Luciani et al., 2004; Shechter et al., 2004; Woodward et al., 2006). This suggests that the high concentrations of sperm nuclei used by Shechter et al. (2004) created replicative stress leading to checkpoint-mediated slowing of the replication timing program.
Cdk activity and the structure of S phase
Cdks are required throughout S phase to induce replication initiation by phosphorylating Sld2 and Sld3 (or their metazoan counterparts) and allowing them to recruit Cdc45 and other replication fork proteins to licensed origins (Masumoto et al., 2002; Tanaka et al., 2007; Zegerman and Diffley, 2007). How then is this initiation function of Cdks coordinated with the timing program so that it drives initiation at origins appropriate for particular stages of S phase? Our results show that Cdk activity also promotes the activation of replication factories (Fig. 9). Reduction of Cdk activity to ∼50% was associated with a reduced number of active replication factories, whereas the number of forks within each factory remained largely unchanged. Conversely, stimulation of Cdk activity increased the number of active replication factories. Treatment of intact U20S cells with roscovitine can also cause a similar reduction in the number of active replication factories (unpublished data). This suggests that the role of Cdks in promoting the activation of new replication factories is conserved throughout higher eukaryotes.
Cdk sensitivity of different levels of replication control. Cartoon showing three different levels of S phase control. The top level shows progression between two different stages of the timing program for a single nucleus. Small green dots represent replication factories, and the black circles represent the nuclear envelope. The middle level shows the activation of a new replication factory (pink dots) next to an existing factory (large green dot). The lower level shows the initiation of a new replication origin (pink circle) on a strand of DNA (green) within an active factory (active replicon cluster).
Cdk sensitivity of different levels of replication control. Cartoon showing three different levels of S phase control. The top level shows progression between two different stages of the timing program for a single nucleus. Small green dots represent replication factories, and the black circles represent the nuclear envelope. The middle level shows the activation of a new replication factory (pink dots) next to an existing factory (large green dot). The lower level shows the initiation of a new replication origin (pink circle) on a strand of DNA (green) within an active factory (active replicon cluster).
Modest changes to Cdk levels changed the number of active replication factories and thus overall replication rate without significantly changing progression through the different replication timing patterns. This changed the amount of total DNA replication associated with each labeling stage of the timing program. However, the timing program was not entirely independent of Cdk activity. Addition of cyclin A caused a modest acceleration of the timing program, whereas high concentrations of roscovitine strongly inhibited progression through the timing program and the appearance of later replication patterns. At present, it is unclear whether the effect of Cdks on the timing program is an indirect consequence of their effect on factory activation and origin initiation or whether Cdks have a separate role in driving the progression from one timing stage to another.
In our experiments, the activation of replication factories showed the highest sensitivity to a change in Cdk levels (Fig. 9). A major effect of Cdks on the activation of replicon clusters has also been reported when sperm nuclei replicate in X. laevis egg extract (Krasinska et al., 2008). This sensitivity to Cdk levels might depend on there being additional Cdk substrates distinct from Cdk substrates required for individual origins to initiate, whose phosphorylation is required to allow the activation of new factories. Alternatively, it might be that the first origin to initiate within a factory requires the highest Cdk activity, but that once this has occurred, initiation of additional origins might be less dependent on high Cdk activity. For example, when initiation occurs in a factory, it might induce a change to the structure of the factory, making it easier for other origins associated with the factory to initiate. Previous work has suggested that the intra–S phase checkpoint may preferentially inhibit the activation of new replication factories, thereby directing new initiation events to inefficient dormant origins within active factories (Woodward et al., 2006; Ge et al., 2007; Blow and Ge, 2009). Our current results suggest that this could be achieved by checkpoint-mediated lowering of Cdk activity (Karlsson-Rosenthal and Millar, 2006; Boutros et al., 2007) or by checkpoint-mediated inhibition of Cdk targets required for factory activation.
Although the mechanisms remain obscure at present, our results show for the first time the multiple ways that Cdk activity can drive progression through S phase, differentially affecting the timing program, factory activation, and replication initiation. Identifying potential Cdk substrates for these transitions is an exciting new goal.
Materials and methods
Cell culture and synchrony
CHOC-400 cells were propagated in DME (Invitrogen) supplemented with nonessential amino acids, 10% fetal calf serum (Perbio), and 10 U/ml streptomycin/penicillin at 37°C in 5% CO2. G1 phase CHOC-400 cells were obtained using mitotic selection as described previously (Gilbert et al., 1995). Essentially, cells were supplemented with fresh medium containing 50 ng/ml nocodazole (Sigma-Aldrich) for 4 h to block cells in metaphase. Mitotic cells were washed in warm medium and released into G1 for 4 h to obtain post–origin decision point cells. Cells were prepared fresh for each experiment.
Preparation of nuclei
Intact nuclei were prepared as described previously (Wu et al., 1997). CHOC-400 cells were washed with ice-cold transport buffer (20 mM Hepes, 110 mM K acetate, 5 mM Na acetate, 2 mM Mg acetate, and 1 mM EGTA, pH 7.6, with KOH), counted, resuspended at 10–15 × 106 cells/ml in transport buffer, and stored on ice. Samples were supplemented with 50 µg/ml digitonin (EMD), incubated on ice for 5 min, and the reaction was stopped by addition of 3% BSA (wt/vol) in transport buffer. Nuclei were recovered by centrifugation at 1,500 rpm for 5 min at 4°C. Cell and nuclear morphology were examined by phase-contrast microscopy. Permeabilization of the plasma and nuclear membranes was verified by staining with 0.1 µg/ml DAPI and 150 kD IgG/FITC exclusion (Dako). Typically, >90% intact nuclei were obtained.
Replication assay
X. laevis egg extract was prepared as described previously (Chong et al., 1997). In brief, unfertilized X. laevis eggs were dejellied and spin crushed at 12,000 g at 4°C for 20 min in a swinging bucket rotor. Cytoplasm was withdrawn, supplemented with 10 µg/ml cytochalasin B and 15% (vol/vol) extract dilution buffer (50 mM KCl, 50 mM Hepes KOH, pH 7.6, 2 mM DTT, 0.4 mM MgCl2, 0.4 mM EGTA, 1 µg/ml each of pepstatin, leupeptin, and aprotinin, and 10% sucrose), and respun at 20,000 g at 4°C for 20 min in a swinging bucket rotor. Cytoplasm was withdrawn and frozen in 20-µl drops in liquid nitrogen. After thawing for use, extracts were supplemented with 250 µg/ml cycloheximide, 25 mM phosphocreatine, 15 µg/ml creatine phosphokinase, and 0.3 mM CaCl2 and incubated for 15 min to promote metaphase exit. They were supplemented with 100 µg/ml geminin and incubated for a further 10 min to prevent further licensing of CHO nuclei. CHOC-400 nuclei were added to extract at 10,000 nuclei/µl. All incubations were performed at 23°C. To measure total DNA synthesis, extracts were supplemented with α-[32P]dATP, and DNA synthesis was measured by TCA precipitation and scintillation counting as described previously (Chong et al., 1997).
Recombinant proteins
The bovine His6-N–cyclin A cDNA plasmid was a provided by T. Hunt (UK London Research Institute, London, England, UK; Brown et al., 1995). Cyclin A was expressed in BL21DE3 induced with 1 mM IPTG for 4 h at 37°C. Bacteria were lysed using Bugbuster (Merck) according to the manufacturer's recommendations. Cyclin A was solubilized in 4 M urea and purified on nickel nitrilotriacetic acid beads (QIAGEN) following the manufacturer's recommendations. Urea was removed by stepwise dialysis against cyclin A solubilization buffer (50 mM glycine, pH 9, 100 mM KCl, 2 mM MgCl2, 5 mM EDTA, 1 mM DTT, 10% [wt/vol] sucrose, and 0.0005% [vol/vol] Tween 20, pH 7.6, with KOH). His-tagged gemininDEL was produced by A. Ferenbach (University of Dundee, Dundee, Scotland, UK) as described previously (Ferenbach et al., 2005).
Replication pattern labeling
Labeling of replication patterns was performed by supplementing X. laevis egg extract with 25 µM Cy3-dUTP (GE Healthcare) for 5 min. For quantification of total DNA synthesis, X. laevis egg was supplemented with 50 µM Cy5.5-dCTP (GE Healthcare). Reactions (20 µl) were stopped by resuspension in 400 µl ice-cold buffer A (60 mM KCl, 15 mM Tris-HCl, pH 7.4, 15 mM NaCl, 1 mM β-mercaptoethanol, 0.5 mM spermidine, and 0.15 mM spermine). The resuspended extract was underlayered with 1 ml buffer A containing 10% sucrose (wt/vol) and was spun at 1,500 rpm in a swinging bucket rotor for 5 min at 4°C. Nuclei were resuspended in 200 µl paraformaldehyde (4%) and incubated for 10 min at 23°C. Meanwhile, 12-mm poly-L-lysine coverslips were placed into 24-well plates and covered with 2 ml TBS (10 mM Tris-HCl, pH 7.5, and 0.15 M NaCl) containing 10% sucrose (wt/vol). After fixation, nuclei were loaded onto the sucrose cushion and spun at 1,500 rpm in a swinging plate rotor for 5 min at 4°C. Coverslips were retrieved and washed three times with TBS plus 0.1% Triton X-100 (vol/vol) followed by three washes with TBS. Nuclei were stained with 1 µg/ml DAPI for 5 min at 23°C. Coverslips were mounted with Vectashield mounting medium, sealed, and dried before visualization.
3D datasets were acquired using a cooled camera (CoolSNAP HQ; Photometrics) on a restoration microscope (DeltaVision Spectris; Applied Precision) built around a stand (lX70; Olympus) with a 60× 1.4 NA Plan Apo lens (Applied Precision). For each nucleus, 22 optical sections were recorded every 0.5 µm, and 3D datasets were deconvolved using the constrained iterative algorithm software (SoftWoRx; Applied Precision). Spatiotemporal patterns could then be visualized and analyzed. The Open Microscopy Environment (OME; Swedlow, 2003; Goldberg et al., 2005) was used for quantitative analysis of images. Quantification of the Cy5.5 intensity was performed using the FindSpots algorithm, and results were exported into an OME XML file for analysis (Platani et al., 2000).
Timing pattern analysis
The following classification of different replication patterns based on previous work (O'Keefe et al., 1992) was used: type I, faintly punctate labeling throughout euchromatic regions; type I/II, intense and diffuse but incomplete labeling of euchromatic regions with distinct lack of nucleolar labeling; type II, complete diffuse labeling of euchromatic regions with lack of nucleolar labeling; type II/III, diffuse labeling of euchromatic regions plus some labeling of the peripheral ring and perinucleolar regions; type III, intense labeling of the peripheral ring, possibly with some perinucleolar labeling; type III/IV, punctate labeling of the peripheral ring plus small-speckled heterochromatic foci within the nuclear interior; type IV, mainly labeling of small-speckled heterochromatic foci within the nuclear interior or at the periphery, with some of the speckled foci forming chain-like structures; type IV/V, labeling of large internal foci plus speckled labeling of small heterochromatic foci within the nuclear interior, which are more punctate than the previous (some peripheral replication may also persist); and type V, predominant labeling of large internal replication foci and at the periphery of the nucleus.
The timing of initiation events associated with different replication patterns was performed as follows: at different times after the start of the in vitro reaction (20, 40, 60, and 80 min), aliquots were taken and supplemented with 1 mM roscovitine. At different times afterward, subaliquots of these were taken, pulsed with 25 µM Cy3-dUTP for 5 min, and the replication patterns were analyzed. If addition of roscovitine at a particular time significantly blocked the subsequent appearance of a particular pattern, the initiation events associated with that pattern were considered to have taken place exclusively after the roscovitine addition time. If addition of roscovitine at a particular time had no significant effect (relative to a control with no added roscovitine) on the appearance of a particular pattern, the initiation events associated with that pattern were considered to have taken place exclusively before the roscovitine addition time. Intermediate cases (in which roscovitine delayed but did not abolish the appearance of a pattern) were considered to indicate that the initiation events associated with that pattern were occurring at the roscovitine addition time.
Analysis of Cy3-labeled foci
Labeling of replication foci was performed at either 50 or 90 min by supplementing X. laevis egg extract with 25 µM Cy3-dUTP for 5 min. Reactions were stopped, fixed, and prepared for microscopy exactly as described for replication pattern labeling. In parallel, total DNA synthesis at either 50 or 90 min was measured in extract supplemented with α-[32P]dATP by TCA precipitation. Datasets were acquired using a camera (Micromax) on a restoration microscope (DeltaVision DV3) built around a stand (Eclipse TE200; Nikon) with a 100× 1.40 NA Plan Apo lens (Nikon). For each nucleus, optical sections were recorded every 0.5 µm, and datasets were deconvolved using the constrained iterative algorithm software (SoftWoRx). For each nucleus, the section with the largest surface area (presumed middle of the nucleus) was selected for quantitation. A 6,400-pixel square (4 × 4 µm) was drawn in the physical center of the section. The number of discernible foci and the total incorporation of label within this area were measured. The mean label incorporated per focus was calculated. Data were generated from 20 nuclei for every condition and time point in each experiment. The OME Remote Objects (OMERO) insight program was used for quantitative analysis (Swedlow, 2003; Goldberg et al., 2005).
Analysis of PCNA foci
For immunodetection of PCNA foci, CHO nuclei incubated in vitro were isolated, fixed, and pelleted onto coverslips as described for replication pattern labeling. Coverslips were washed extensively with PBS plus 0.1% Triton X-100 (vol/vol; PBS-T). Samples were blocked for 1 h in the same buffer plus 3% BSA and incubated for a further 1 h with PC10 anti-PCNA antibody. After extensive washing in PBS-T, cells were labeled with FITC-labeled anti–mouse IgG followed by extensive washing in PBS-T. DNA was stained with 1 µg/ml DAPI for 5 min at 23°C. Coverslips were mounted with Vectashield mounting medium, sealed, and dried before visualization. Datasets were acquired using a camera (Micromax) on a restoration microscope (DeltaVision DV3) built around a stand (Eclipse TE200) with a 100× 1.40 NA Plan Apo lens (Nikon). For each nucleus, optical sections were recorded every 0.5 µm. The OMERO Insight program was used for quantitative analysis (Swedlow, 2003; Goldberg et al., 2005). For each nucleus, the section with the largest surface area (presumed middle of the nucleus) was used for quantitation. A 1,600-pixel square (4 × 4 µm) was drawn in the physical center of each nucleus, and the number of discernible spots was counted. The 10 brightest spots in the whole nucleus were selected and averaged to give mean focus intensity.
DNA fiber labeling
CHO nuclei were incubated in extract supplemented with 100 µM BrdU under the desired conditions (5-min incubation at 45–50 min). Reactions (100 µl) were stopped by resuspension in 400 µl TBS. The resuspended extract was underlayered with 1 ml TBS plus 0.1% Triton X-100 (vol/vol) and 20% sucrose (wt/vol) and was spun at ∼300 g in a swinging bucket rotor for 5 min at 4°C. The supernatant was removed to leave only the sucrose cushion (100 µl; equal to the starting volume), and the cell pellet was resuspended and stored on ice. DNA was spread on glass slides (Superfrost; VWR) according to the following conditions: nuclei were diluted (1:5) in TBS buffer (∼2,000 nuclei/µl). A 1-µl sample was spotted onto the glass slide followed by addition of 9 µl of lysis buffer (0.75% SDS, 200 mM Tris-HCl, pH 7.4, and 50 mM EDTA) and incubated for ∼4 min. DNA fibers were spread by tipping the slides at 36°. After migration down the slide, samples were fixed in methanol/acetic acid (3:1) for >10 min. Slides were rehydrated with H20 and incubated in 2.5 M HCl for 1 h to denature the DNA. Slides were briefly rinsed in TBS and incubated for 1 h in blocking solution containing TBS, 1% (wt/vol) BSA, and 0.1% (vol/vol) Tween 20. Slides were incubated with monoclonal anti-BrdU (BD) at 20 µg/ml in this buffer for 1 h. Slides were washed several times in TBS + Tween, in TBS + Tween + BSA, briefly rinsed in TBS alone, and labeled with 1 µg/ml Alexa Fluor 555 anti–mouse antibody (Invitrogen) in TBS + Tween + BSA for 2 h. Samples were washed extensively in TBS, and DNA was stained with YOYO (Y-3601; diluted at 1:10,000 from a 1 mM stock; Invitrogen) for 10 min. Samples were rinsed five times in TBS and mounted in Vectashield. Random fields were selected using YOYO staining to ensure that only single DNA fibers and not fiber bundles were scored and recorded using a 63× NA 1.32–0.6 oil Plan Apo lens on a microscope (DM IRB; Leica).
Fiber analysis was performed as described previously (Ge et al., 2007). The mean and standard deviation of track lengths were first determined by measuring the length of labeled tracks that were well separated from other tracks (thereby minimizing the risk that they represented fusions between adjacent replicons). Track clusters were selected for the determination of intracluster fork density and origin spacing by the following criteria: clusters (a) consisted of single DNA fibers and not fiber bundles based on YOYO staining, (b) were located in a relatively isolated area, (c) contained at least four consecutive tracks, and (d) each track in the cluster was no longer than the mean track length plus one standard deviation to minimize the risk of including clusters where termination and fusion of neighboring replicons had occurred. For each sample, at least 100 measurements were performed.
Online supplemental material
Fig. S1 shows the effect of preincubation of egg extract on the timing program. Fig. S2 shows that aphidicolin slows the timing program and caffeine causes fork instability. Fig. S3 shows the effect of roscovitine and cyclin A on the timing program. Fig. S4 shows the measurement of DNA synthesis in individual nuclei ± 10 µM roscovitine or 1 pM cyclin A. Fig. S5 shows the effect of roscovitine and cyclin A on replication foci.
Acknowledgments
We thank Silvia Costa and Anatoliy Li for comments on the manuscript and Jason Swedlow and Michael Porter for assistance with the quantitative immunofluorescence.
This work was funded by a Biotechnology and Biological Sciences Research Council studentship to A.M. Thomson and by Cancer Research UK program grants (C303/A3135, C303/A5434, and C303/A7399).


![Figure 1. Progression of CHOC-400 nuclei through the timing program in vitro. CHOC-400 nuclei were incubated at 10,000 nuclei/µl (∼60 ng DNA/µl) in egg extracts supplemented with geminin. (a) Representative images of CHO nuclei incubated in X. laevis egg extracts and pulse labeled for 5 min with Cy3-dUTP. See Materials and methods for full description of the different labeling patterns. (b) At different times, aliquots were pulse labeled for 5 min with Cy3-dUTP, and the proportion of different replication patterns was assessed. (c) Extract was supplemented with α-[32P]dATP. At different times, total DNA synthesis was measured by TCA precipitation and scintillation counting. The SEM of 14 independent experiments is shown. (d, top) The time of appearance of replication patterns during S phase of CHOC-400 cells as described previously by Dimitrova and Gilbert (1999). (middle) The proportion of time spent by CHOC-400 nuclei replicating specific patterns as calculated in b. (bottom) The amount of DNA synthesis associated with each replication pattern in vitro calculated by scaling the proportions in b by the rate of replication (c).](https://cdn.rupress.org/rup/content_public/journal/jcb/188/2/10.1083_jcb.200911037/6/m_jcb_200911037_rgb_fig1.jpeg?Expires=1771366648&Signature=g24rM3ewWKV3ANMkF-Z3uQMRW3s1cTKASziIXm8P1NxjOfu0ajLbE9AWV2l2ZvjBRwnH7ohygGjxSg0vjybXrVRhlTtkPskrE1ByP~cI2KZEh-Zl282fp4EjBBxVktIy45r5OMPhF70Od9hzy26iob04r2XislFYf0HlUlbe~~s7uqRppghggewHp-E2rhaz4ano~nW2T7o74QXxWyU1Exuqz68kAcIU1JD6rdh0EBAZiUDHzALuvKaJjUSYH6iAJmRWz6mSdQHnxVXxULgA4DV7okk5hIWKqNDk2oNrWjnf1LOCVGzCM1DxrOFrUS3~PXfeWK3f~ITXM3~a24sIVQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 2. Initiation times associated with different replication patterns. CHOC-400 nuclei were incubated at 10,000 nuclei/µl in egg extracts supplemented with geminin. At different times, aliquots were supplemented with 1 mM roscovitine to block further initiation events. (a) Extract was also supplemented with α-[32P]dATP at the start of the incubation. At different times thereafter, total DNA synthesis was measured by TCA precipitation and scintillation counting. (b) At different times, extract was pulse labeled with Cy3-dUTP for 5 min, and the proportion of nuclei showing different replication patterns was assessed. (c) Data from a and b were combined to provide an estimate of the times over which most initiation events associated with the different replication patterns were occurring.](https://cdn.rupress.org/rup/content_public/journal/jcb/188/2/10.1083_jcb.200911037/6/m_jcb_200911037_rgb_fig2.jpeg?Expires=1771366648&Signature=e3cAYebvyAj~sB0p8mKR3IbCHkN3WhcZy5Y3NfmEbViRBxBr0mu0JpO5V-8cSWX7iJWDJyvU1Q6t-4RBn0ofVQoL3864Arjqmwbg75wT7MjVmXw~ugV~N5TTt96MYn0svPJ7qTgQiOAzapxjFSAyMBxZj-w3EW3Qo5iz8urHXb3xYNnopfxFw1F1n3dAILmZCe1szV-3ideWmXIRbrjg5Ka8jds67ukpExH83FR0TY57HnnMg7b27gU~afgfreZN1uCE-uDLHaUx05~8CSHc9tuua~exI3L~14N2FVKsYMl~DjwVzSYZhRGGEoh2iUXRSwX-2XhLAOARGNwt-oxH4w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 4. Effect of low concentrations of roscovitine on the timing program. (a) Extract was supplemented with histone H1, γ-[32P]ATP, and various concentrations of roscovitine. After incubation for 90 min, protein was separated by SDS-PAGE, and 32P incorporation into H1 was assessed by phosphorimager. (b and c) CHOC-400 nuclei were incubated at 10,000 nuclei/µl in egg extracts supplemented with geminin and different concentrations of roscovitine. Aliquots were also supplemented with α-[32P]dATP at the start of the incubation. (b) At different times thereafter, total DNA synthesis was measured by TCA precipitation and scintillation counting. (c) At different times, extract was pulse labeled with Cy3-dUTP for 5 min, and the proportion of nuclei showing different replication patterns was assessed. No replication patterns were visible in extracts treated with 300 µM roscovitine.](https://cdn.rupress.org/rup/content_public/journal/jcb/188/2/10.1083_jcb.200911037/6/m_jcb_200911037_rgb_fig4.jpeg?Expires=1771366648&Signature=1Y5PDdZlJSwTDK9jIijDa54spIxfXnsmY2ZE~fIvvbHh58oe1h1Dzpzw3~vPFI2aAxLDFNfLg5ent3w55oDwXfF5u~5Q8bTlMQpAp5Gu4XuGQT3XE-jN~05zrGRsl14w~nIa49Rn4wFLGXh~0-2qNbeijToUyHdX9OjvLxBtVBUY~wQiXdk3wmB0p0TK5pmMICu2jRHLjhRQZBJoPPGARi2SLfwzZhX2u1-~fONc3-VT2Dn8FgycsgNWgHiymsPziGgjPaLbtIKVmdtbU9pa2YQSdE~SA5xBWlJ5NKAsYgFEztCVzv97Dt6gOmeL5qO0TBxiPDgsuuxSctB3zpGyaA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 5. Effect of recombinant cyclin A on the timing program. (a) Extract was supplemented with histone H1 γ-[32P]ATP and various concentrations of cyclin A. After incubation for 90 min, protein was separated by SDS-PAGE and 32P incorporation into H1 assessed by phosphorimager. (b and c) CHOC-400 nuclei were incubated at 10,000 nuclei/µl in egg extracts supplemented with geminin and different concentrations of recombinant cyclin A. (b) Aliquots were also supplemented with α-[32P]dATP at the start of the incubation. At different times afterward, total DNA synthesis was measured by TCA precipitation and scintillation counting. (c) At either 45 or 85 min, extract was pulse labeled with Cy3-dUTP for 5 min, and the proportion of nuclei showing different replication patterns was assessed. (d) Extract was supplemented ± 1 pM cyclin A. At different times, extract was pulse labeled with Cy3-dUTP for 5 min, and the proportion of nuclei showing different replication patterns was assessed.](https://cdn.rupress.org/rup/content_public/journal/jcb/188/2/10.1083_jcb.200911037/6/m_jcb_200911037_rgb_fig5.jpeg?Expires=1771366648&Signature=AxTNT5~LQMGr0vq0nxbakmELYl1isdZDsJNM71lrQCGlnsxr7ad7f29YwzI9uH-2aylLhlfVwQaT4PSvmYxfDRDPrxMAeJST21lo4e62ukZzz8RJqEgmYTP2Zy0vAPYewmKHOpsa~kJha62i4UwBOInLBMISyzCId5UJZDt9wB3dzZx2tNQrwV8kidWgwV2pqpxwghtImleqqHQBENFRip5Q0NvdJkfcckMcVnd4q0442IQjj6M1Qx7rIDcMM3KAKu5mkNtyz6QVtaebJNpRZAfbAGoTXOwVNlF4Dw0nFmSNKDjcv4VDw8xA6cr5O4Aa-~MrTtr9NKBcABk64-muhQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 7. Replication foci number depends on Cdk levels. CHOC-400 nuclei were incubated in X. laevis egg extracts incubated at 10,000 nuclei/µl in egg extracts supplemented with the indicated concentrations of roscovitine (rosc) or cyclin A (cyc A). Parallel incubations were supplemented with α-[32P]dATP to measure total DNA synthesis. At 50 or 90 min, extract was pulsed for 5 min with Cy3-dUTP. (a–c) Representative images from the 50-min time point are shown. (d and e) The number and mean intensity of Cy3-labeled foci at 50 (d) and 90 (e) min were measured. Error bars indicate SEM between different nuclei.](https://cdn.rupress.org/rup/content_public/journal/jcb/188/2/10.1083_jcb.200911037/6/m_jcb_200911037_rgb_fig7.jpeg?Expires=1771366648&Signature=3W~4rNLGHB3uiVXLBGRjf1Bf60BTlAI~9ZS944~EdMjVCaJRVX7T8y2quL1fOHTWvJHziuDzgb7b-4ekf5JrEvs6gIjT4JtKjtuj8Nh~CT8fngvaJT~RfxmOmxBnO496Dq89ytrdv4Dux2Fcb-B2ZPpXnOANzIz-7r73RMu071r22IhRslonNpGm8IfSSRqHHrC7pMSnLFKz~6~NfCLENnU5bq46gesu7E0BJDvYeOkA-WZ6CQlLTNs3EuRCv3S1owMHCUj7x0PhVnKf5nf7FTsF3AcdAyBIFfDhTJ1JMKwWa7YnwHW~Cgo2mhtcduWF8-HaCjx3A9zibfUAOcwgcw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)




