In humans, seven evolutionarily conserved genes that cause the cilia-related disorder Bardet-Biedl syndrome (BBS) encode proteins that form a complex termed the BBSome. The function of the BBSome in the cilium is not well understood. We purified a BBSome-like complex from Chlamydomonas reinhardtii flagella and found that it contains at least BBS1, -4, -5, -7, and -8 and undergoes intraflagellar transport (IFT) in association with a subset of IFT particles. C. reinhardtii insertional mutants defective in BBS1, -4, and -7 assemble motile, full-length flagella but lack the ability to phototax. In the bbs4 mutant, the assembly and transport of IFT particles are unaffected, but the flagella abnormally accumulate several signaling proteins that may disrupt phototaxis. We conclude that the BBSome is carried by IFT but is an adapter rather than an integral component of the IFT machinery. C. reinhardtii BBS4 may be required for the export of signaling proteins from the flagellum via IFT.
Introduction
Bardet-Biedl syndrome (BBS; NCBI OMIM #209900) is a rare human disorder causing hypogonadism, rod–cone dystrophy, polydactyly, truncal obesity, and renal abnormalities (Katsanis et al., 2001; Tobin and Beales, 2007; Zaghloul and Katsanis, 2009). This combination of features partially overlaps with the pleiotropic phenotype of disorders caused by defective cilia, which is consistent with the hypothesis that cilia are involved in the etiology of BBS (Rosenbaum and Witman, 2002; Beales, 2005; Blacque and Leroux, 2006). In humans, 12 genes (BBS1–12) have been linked to BBS (Blacque and Leroux, 2006; Stoetzel et al., 2006; Stoetzel et al., 2007); the proteins (BBS1/2/4/5/7/8/9) encoded by 7 of these genes plus BBIP10 form a complex termed the BBSome (Nachury et al., 2007; Loktev et al., 2008).
Experimental data from various species support a role for the BBSome in cilia (Tobin and Beales, 2007). Several BBSome proteins have been localized to basal bodies and cilia in mammalian cells (Ansley et al., 2003; Mykytyn and Sheffield, 2004) and have been shown to undergo intraflagellar transport (IFT) in Caenorhabditis elegans (Blacque et al., 2004). IFT is the bidirectional movement of protein particles consisting of IFT complex A and complex B driven by kinesin 2 (anterograde IFT) and cytoplasmic dynein 1b/2 (retrograde IFT) along the axoneme (Cole et al., 1998; Rosenbaum and Witman, 2002). IFT is required for flagellar assembly, maintenance, and signaling, and the loss of bona fide IFT components results in a failure to assemble cilia, which causes embryonic lethality in mice (Nonaka et al., 1998; Jonassen et al., 2008). In contrast, BBS1/2/4 knockout mice are viable, and cilia are at least initially assembled in the majority of tissues, with sperm flagella being an exception. Thus, the BBSome is dispensable for ciliary assembly in most cell types (Blacque et al., 2004; Kulaga et al., 2004; Mykytyn et al., 2004; Yen et al., 2006; Nachury et al., 2007; Loktev et al., 2008; Mukhopadhyay et al., 2008).
The BBSome subunits are highly conserved and widely distributed among organisms with cilia, suggesting that the function of the BBSome has also been conserved. BBS proteins have been implicated in intracellular transport (Kim et al., 2004; Yen et al., 2006; Gerdes et al., 2007) and protein transport to the cilium and ciliary membrane (Nachury et al., 2007; Berbari et al., 2008; Shah et al., 2008). In C. elegans, BBS7 and -8 are thought to function in controlling IFT by maintaining an association of IFT complexes A and B (Blacque et al., 2004; Ou et al., 2005; Pan et al., 2006). Despite these advances, the functions of the BBSome in cilia remain unclear. This has been caused, in part, by the lack of a genetic model from which BBSome-defective cilia can be isolated and biochemically analyzed.
In this study, we explore the function of the BBSome in Chlamydomonas reinhardtii, which has advantages that allowed us to determine the structural, biochemical, and physiological consequences for cilia when BBSome function is impaired. We show that C. reinhardtii flagella contain a complex, including at least BBS1, -4, -5, -7, and -8, that is orthologous to the mammalian BBSome. Simultaneous tracking of BBS4-GFP and IFT20-mCherry in living cells by total internal reflection fluorescence (TIRF) microscopy (TIRFM) revealed that the BBSome is moved anterogradely and retrogradely in the flagellum in association with a subset of IFT particles. Consistent with this, we find that BBSome proteins are present in flagella at substoichiometric amounts relative to IFT proteins. We identified C. reinhardtii mutants for BBS1, -4, and -7; they have full-length flagella, and IFT is unaffected. Collectively, the results indicate that the BBSome is carried by IFT but is not an integral component of the IFT machinery.
All three BBS mutants are nonphototactic. Biochemical analyses of flagellar fractions of the BBS mutants revealed that several proteins associated with the flagellar membrane and matrix and likely to be involved in signal transduction are abnormally accumulated in the flagella; this apparently impairs the ability of the organelle to carry out phototactic steering because phototaxis is transiently restored to bbs4 cells after the regeneration of new flagella. We propose that the C. reinhardtii BBSome is an IFT adapter required for the turnover or removal of certain proteins from the flagellum via IFT. BBS may be a degenerative disease of the cilium in which certain proteins accumulate over time, leading to progressive ciliary dysfunction.
Results
Defects in BBS1, -4, and -7 impair phototaxis in C. reinhardtii
All eight known subunits of the human BBSome (BBS1/2/4/5/7/8/9) are conserved in C. reinhardtii (Table I). We searched for C. reinhardtii BBSome mutants starting with the BBS4 gene. Genomic DNA from a collection of 350 motility and phototaxis mutants was screened by quantitative RT-PCR using two primer pairs (BBS4N and BBS4C) specific for the C. reinhardtii BBS4 gene (Fig. 1 A). Primer pair BBS4N failed to amplify in four strains, namely the previously characterized nonphototactic mutants ptx5-1, -2, and -3 (Pazour et al., 1995) and the uncharacterized strain F36, which represents the novel allele ptx5-4. Two of these strains (ptx5-1 and -3) also failed to amplify with primer pair BBS4C. The results indicate that all four mutant strains have defects in the BBS4 gene.
C. reinhardtii homologues of BBSome proteins
| Protein | Predicted mass in human | Predicted mass in C. reinhardtii | BLASTP (E value) | Identity |
| kD | kD | % | ||
| BBS1 | 51 | 66 | 7e-111 | 40 |
| BBS2 | 80 | 56 | 1e-67 | 42 |
| BBS4 | 51 | 46 | 4e-106 | 50 |
| BBS5 | 39 | 39 | 1e-103 | 52 |
| BBS7 | 75 | 75 | 22e-109 | 33 |
| BBS8 | 57 | 58 | 1e-156 | 50 |
| BBS9 | 99 | 96 | 1e-139 | 36 |
| BBIP10 | 10 | 9 | 6e-8 | 30 |
| Protein | Predicted mass in human | Predicted mass in C. reinhardtii | BLASTP (E value) | Identity |
| kD | kD | % | ||
| BBS1 | 51 | 66 | 7e-111 | 40 |
| BBS2 | 80 | 56 | 1e-67 | 42 |
| BBS4 | 51 | 46 | 4e-106 | 50 |
| BBS5 | 39 | 39 | 1e-103 | 52 |
| BBS7 | 75 | 75 | 22e-109 | 33 |
| BBS8 | 57 | 58 | 1e-156 | 50 |
| BBS9 | 99 | 96 | 1e-139 | 36 |
| BBIP10 | 10 | 9 | 6e-8 | 30 |
The predicted molecular masses of human and C. reinhardtii BBSome proteins are shown. BLASTP values are shown for the best match to the C. reinhardtii protein in the human reference protein database. All pairs were reciprocal best hits in BLASTP searches. Percent identity is based on the BLASTP alignment.
Identification of C. reinhardtii bbs1, -4, and -7 mutants. (A) Map showing the genomic region near the BBS4 locus. Black arrows indicate PCR primer pairs; plus and minus signs indicate whether or not these primers gave PCR products. bbs4-1 lacks the entire BBS4 gene. The 6,797-bp fragment used to rescue strain bbs4-1 is indicated. JGI protein IDs of the predicted gene products are shown. (B) Map showing the genomic region near the BBS7 locus. Only one of the tested primer pairs failed to amplify in the ptx6-1 strain. Sequencing of this region revealed the insertion of a retrotransposal footprint into the BBS7 gene of ptx6-1, causing a premature stop after 52 codons. (C) Map showing the genomic region near the BBS1 locus. Approximately half of BBS1 is deleted in bbs1-1. Gene orientation is marked by white arrows. wt, wild type.
Identification of C. reinhardtii bbs1, -4, and -7 mutants. (A) Map showing the genomic region near the BBS4 locus. Black arrows indicate PCR primer pairs; plus and minus signs indicate whether or not these primers gave PCR products. bbs4-1 lacks the entire BBS4 gene. The 6,797-bp fragment used to rescue strain bbs4-1 is indicated. JGI protein IDs of the predicted gene products are shown. (B) Map showing the genomic region near the BBS7 locus. Only one of the tested primer pairs failed to amplify in the ptx6-1 strain. Sequencing of this region revealed the insertion of a retrotransposal footprint into the BBS7 gene of ptx6-1, causing a premature stop after 52 codons. (C) Map showing the genomic region near the BBS1 locus. Approximately half of BBS1 is deleted in bbs1-1. Gene orientation is marked by white arrows. wt, wild type.
Mapping of the deleted region in these strains showed that BBS4 and two other potential genes were deleted in ptx5-1 (Fig. 1 A). Transformation of this mutant with a 6,797-bp genomic fragment encompassing only the BBS4 gene (Fig. 1 A) restored the strong negative phototaxis characteristic of the parental strain g1 (Fig. 2 A; Pazour et al., 1995). Therefore, the deletion of the BBS4 gene is responsible for the nonphototactic phenotype of the ptx5-1 mutant (Videos 1–3). For the sake of clarity, the ptx5-1 mutant will be called bbs4-1 in this study.
Time-lapse video showing phototaxis of wild-type C. reinhardtii and the bbs4-1 mutant in a culture dish assay. The direction of incidence of the light is indicated (arrow). Images were recorded at 6 frames/min, and the video is displayed at 24 frames/s.
Time-lapse video showing phototaxis of wild-type C. reinhardtii and the bbs4-1 mutant in a culture dish assay. The direction of incidence of the light is indicated (arrow). Images were recorded at 6 frames/min, and the video is displayed at 24 frames/s.
Phototaxis of wild-type C. reinhardtii cells. Cells were placed in a chamber and illuminated with a stimulus light for ∼5 s before recording cell movements. This video was recorded and is displayed at 30 frames/s.
Phototaxis of wild-type C. reinhardtii cells. Cells were placed in a chamber and illuminated with a stimulus light for ∼5 s before recording cell movements. This video was recorded and is displayed at 30 frames/s.
Lack of phototaxis in C. reinhardtii bbs4-1 mutant cells. Cells were placed in a chamber and illuminated with a stimulus light for ∼5 s before recording cell movements. This video was recorded and is displayed at 30 frames/s.
Lack of phototaxis in C. reinhardtii bbs4-1 mutant cells. Cells were placed in a chamber and illuminated with a stimulus light for ∼5 s before recording cell movements. This video was recorded and is displayed at 30 frames/s.
BBS4 is a flagellar protein required for phototaxis but not IFT. (A) Motion analysis of wild type, bbs4-1, and R26, a strain rescued with a genomic fragment encompassing the BBS4 gene. The direction of light (arrow) is indicated. The radial histograms show the percentage of cells moving in a particular direction relative to the light (24 bins, each 15°). (B) Flagellar length of g1, bbs4-1, bbs7-1, and bbs1-1, as determined by DIC. (C) The velocity of anterograde (a.) and retrograde (r.) IFT particles in wild-type (g1) and bbs4-1 flagella, as determined by DIC. (D) Velocity of KAP-GFP in BBS4 and bbs4-1 flagella, as determined by TIRFM. (C and D) SDs are indicated. (E) Detached flagella from wild type (a–c) or the bbs4-1 mutant (d–f) were stained with anti-IFT46 (a and d) and anti-IFT139 (b and e). (F) Western blot probing proteins of whole cells (a) or isolated flagella (b) with anti-BBS4 (a and b) and anti-IFT139 (a) or anti-IC2, which is specific for an axonemal protein (b), as loading controls. Anti-BBS4 stained a band of ∼41 kD (arrowhead) in whole-cell and flagella samples of wild type and the rescued strain R26; BBS4 was undetectable in bbs4-1. Molecular masses are indicated in kilodaltons. (G) Detached flagella from wild type and the bbs4-1 mutant were stained with anti–acetylated tubulin (ac-tubulin; a and c) and anti-BBS4 (b and d). Bars, 5 µm.
BBS4 is a flagellar protein required for phototaxis but not IFT. (A) Motion analysis of wild type, bbs4-1, and R26, a strain rescued with a genomic fragment encompassing the BBS4 gene. The direction of light (arrow) is indicated. The radial histograms show the percentage of cells moving in a particular direction relative to the light (24 bins, each 15°). (B) Flagellar length of g1, bbs4-1, bbs7-1, and bbs1-1, as determined by DIC. (C) The velocity of anterograde (a.) and retrograde (r.) IFT particles in wild-type (g1) and bbs4-1 flagella, as determined by DIC. (D) Velocity of KAP-GFP in BBS4 and bbs4-1 flagella, as determined by TIRFM. (C and D) SDs are indicated. (E) Detached flagella from wild type (a–c) or the bbs4-1 mutant (d–f) were stained with anti-IFT46 (a and d) and anti-IFT139 (b and e). (F) Western blot probing proteins of whole cells (a) or isolated flagella (b) with anti-BBS4 (a and b) and anti-IFT139 (a) or anti-IC2, which is specific for an axonemal protein (b), as loading controls. Anti-BBS4 stained a band of ∼41 kD (arrowhead) in whole-cell and flagella samples of wild type and the rescued strain R26; BBS4 was undetectable in bbs4-1. Molecular masses are indicated in kilodaltons. (G) Detached flagella from wild type and the bbs4-1 mutant were stained with anti–acetylated tubulin (ac-tubulin; a and c) and anti-BBS4 (b and d). Bars, 5 µm.
To determine whether mutations in other BBS genes might also cause a nonphototactic phenotype in C. reinhardtii, we selected 17 nonphototactic mutants with good motility from our collection and analyzed them by quantitative RT-PCR using 40 primer pairs designed to amplify parts of the BBS genes. Primer pair BBS7-1, amplifying a 230-bp region in the first exon of BBS7, failed to amplify in the previously described strain ptx6-1 (Fig. 1 B; Pazour et al., 1995). Further analysis identified a footprint of the retrotransposon TOC1 (Day et al., 1988) in codon 51 of BBS7 in ptx6-1; this insertion resulted in a premature stop, deleting 77% of the predicted protein (Fig. 1 B). This mutant will be referred to as bbs7-1 in this study. In another nonphototactic strain, RIR7-2, the 5′ half of the BBS1 gene and a gene upstream of BBS1 are deleted; we named this strain bbs1-1 (Fig. 1 C). The results strongly suggest that defects in BBS1 and -7 also impair phototaxis in C. reinhardtii.
Flagellar assembly and IFT are normal without BBS4 in C. reinhardtii
Because defects in BBS proteins have been reported to affect ciliary assembly, IFT particle integrity, and the rate of IFT in other organisms (Blacque et al., 2004; Ou et al., 2005; Nachury et al., 2007; Loktev et al., 2008), we investigated whether the BBSome proteins are required for flagellar assembly and IFT in C. reinhardtii. Swimming velocity is reduced by ∼25% in bbs4-1 and bbs7-1 (Pazour et al., 1995). Flagellar length was unaffected in the bbs mutants (Fig. 2 B), and flagellar regeneration appeared to be normal (not depicted). The ultrastructure of mutant bbs4-1 flagella and IFT particles was indistinguishable from that of wild type by thin-section transmission EM (unpublished data). The velocity of IFT in bbs4-1 was unaffected, as determined by differential interference contrast (DIC; Fig. 2 C) microscopy and by TIRFM of the IFT kinesin motor subunit kinesin-associated protein (KAP)–GFP expressed in bbs4-1/kapts or BBS4/kapts backgrounds (Fig. 2 D; Mueller et al., 2005). In immunofluorescence microscopy (IFM), IFT complex B, visualized by anti-IFT46, and IFT complex A, visualized by anti-IFT139, showed a near perfect colocalization in bbs4-1 flagella (Fig. 2 E, d–f), as in wild-type flagella (Fig. 2 E, a–c). We conclude that flagellar assembly and the integrity and movement of IFT particles in C. reinhardtii are independent of BBS4.
Axonemes of bbs4-1 lack a proper Ca2+ response
Phototactic steering of C. reinhardtii is thought to involve a differential sensitivity of the two flagella to intraflagellar Ca2+ (Witman, 1993). At very low (e.g., 10−9 M) Ca2+, the beating of the cis flagellum (the one closest to the eyespot; Fig. S1 B) will dominate over the trans flagellum, causing the cell to turn away from its eyespot. At higher (e.g., 10−6 M) Ca2+, the trans flagellum is dominant over the cis flagellum, causing the cell to turn toward its eyespot. This can be demonstrated in demembranated cells reactivated in ATP-containing solutions having various concentrations of Ca2+ (Kamiya and Witman, 1984). Pazour et al. (1995) reported that intact bbs4-1 (ptx5-1) cells lacked the ability to respond like wild type in Ca2+-free medium. We confirmed this (Fig. S1 A) and further found that demembranated models of bbs4-1 did not undergo the Ca2+-induced switch in axonemal dominance at high (10−6 M) Ca2+ (Fig. S1 C), indicating that the loss of BBS4 alters the Ca2+ sensitivity of the axoneme.
BBS4 is a component of the flagellar matrix in C. reinhardtii
An antibody (anti-BBS4) was raised against the C-terminal 248 aa of BBS4. Western blots of whole-cell extracts from wild type, bbs4-1, and R26, a strain rescued using genomic BBS4, were probed using anti-BBS4 (Fig. 2 F, a). A band of ∼41 kD, which is close to the expected size of C. reinhardtii BBS4 (46,360 D), was detected in wild type and R26 but not in bbs4-1. When Western blots of isolated flagella of these strains were probed with anti-BBS4, a single band of ∼41 kD was present in wild type and the rescued strain R26; the band was absent in bbs4-1, confirming the loss of BBS4 in this mutant (Fig. 2 F, b). In IFM of detached wild-type flagella, anti-BBS4 gave a punctate staining; as expected, the staining was absent from flagella of bbs4-1 (Fig. 2 G).
Anti-BBS4 was not suitable for localization of BBS4 in whole cells by IFM because in Western blots, it cross reacted with a few other proteins present in whole-cell extracts (Fig. 2 F, a). Therefore, to determine the subcellular distribution of BBS4, bbs4-1 was rescued by expression of BBS4-3xHA (Fig. 3, A and B). Double staining of BBS4HA21 cells (bbs4-1 rescued by BBS4-3xHA) with anti-HA and antitubulin showed that BBS4-3xHA was localized near the two basal bodies and in a spotted fashion along both flagella (Fig. 3 C). This distribution is very similar to that described for IFT particle proteins in C. reinhardtii. Approximately 90% of BBS4-3xHA was present in the cell body, as determined by Western blots comparing deflagellated cell bodies and isolated flagella from BBS4HA21 (Fig. 3 D); similar results were obtained for wild-type BBS4 in wild-type cells (not depicted). A similar distribution was observed for the IFT particle proteins IFT81 and -57. In contrast, only a small cytoplasmic pool was detected for the axonemal protein IC2. Like the IFT particle proteins, BBS4 and BBS4-3xHA were predominately present in the membrane-plus-matrix fraction obtained by extraction of isolated flagella from wild type or BBS4HA21 with 1% NP-40 (Fig. 3 E). Temperature-induced phase partitioning with 1% Triton X-114 (Everberg et al., 2008) was used to separate the membrane-plus-matrix fraction into an aqueous phase (AP) containing proteins of the flagellar matrix and a detergent phase (DP; Fig. S5 B); the latter contained nearly all of the transmembrane protein 183515 (not depicted), indicating that it was enriched in proteins of the flagellar membrane. BBS4 and various IFT particle proteins were enriched in the AP and were not detected in the DP (Fig. 3 F). Thus, both IFM and Western blotting showed that BBS4 is present in the flagella of C. reinhardtii and has a distribution similar to that of IFT particles.
BBS4 has an IFT-like distribution within the cell. (A) Dish phototaxis assay of wild type, bbs4-1, and BBS4HA21 rescued by BBS4-3xHA. (B) Western blot of flagella from the strains shown in C probed with monoclonal anti-HA. (C) BBS4HA21 cells labeled by anti–α-tubulin and anti-HA. BBS4-3xHA is located near the basal bodies (arrowhead) and in dots along both flagella. (D) Western blot of BBS4HA21 to compare the amounts of various proteins in cell bodies (CB) versus isolated flagella (FLA). 1× indicates that approximately two flagella were loaded per cell body, etc. The blot was probed for BBS4-HA, IC2, and IFT57 and -81. BBS4-3xHA is about eight times more abundant in cell bodies than in flagella. (E) Western blot analyzing equivalent amounts of whole flagella, axonemes (AXO), and NP-40–soluble membrane-plus-matrix (M&M) of flagella from BBS4HA21 and wild-type cells. Antibodies were as indicated; those to IC2 and various IFT particle proteins were used as controls. (F) Western blot comparing equivalent amounts of wild-type flagella, axonemes, AP, and DP resulting from Triton X-114 phase partitioning. The blot was probed with the antibodies indicated. (B and D–F) The positions of standard proteins and their molecular masses in kilodaltons are indicated. Bar, 5 µm.
BBS4 has an IFT-like distribution within the cell. (A) Dish phototaxis assay of wild type, bbs4-1, and BBS4HA21 rescued by BBS4-3xHA. (B) Western blot of flagella from the strains shown in C probed with monoclonal anti-HA. (C) BBS4HA21 cells labeled by anti–α-tubulin and anti-HA. BBS4-3xHA is located near the basal bodies (arrowhead) and in dots along both flagella. (D) Western blot of BBS4HA21 to compare the amounts of various proteins in cell bodies (CB) versus isolated flagella (FLA). 1× indicates that approximately two flagella were loaded per cell body, etc. The blot was probed for BBS4-HA, IC2, and IFT57 and -81. BBS4-3xHA is about eight times more abundant in cell bodies than in flagella. (E) Western blot analyzing equivalent amounts of whole flagella, axonemes (AXO), and NP-40–soluble membrane-plus-matrix (M&M) of flagella from BBS4HA21 and wild-type cells. Antibodies were as indicated; those to IC2 and various IFT particle proteins were used as controls. (F) Western blot comparing equivalent amounts of wild-type flagella, axonemes, AP, and DP resulting from Triton X-114 phase partitioning. The blot was probed with the antibodies indicated. (B and D–F) The positions of standard proteins and their molecular masses in kilodaltons are indicated. Bar, 5 µm.
C. reinhardtii BBS4 forms a complex with other BBS proteins
To determine whether a BBSome-like complex is present in the flagellum of C. reinhardtii, detergent extracts from BBS4HA21 and wild-type flagella were fractionated by sucrose gradient centrifugation and analyzed by Western blotting (Fig. 4 A). Both BBS4-3xHA and BBS4 sedimented at ∼12S, suggesting that each was present in a complex. BBS4-3xHA did not cosediment with IFT46 (IFT complex B) or IFT139 (IFT complex A), indicating that it is present in a complex distinct from the IFT complexes. We affinity purified the BBS4-3xHA complex from the BBS4HA21 flagellar membrane-plus-matrix fraction. At least six bands were detected in silver-stained gels of the eluate from BBS4HA21; these proteins were absent in a corresponding eluate from wild type (Fig. 4 B). Analysis by mass spectrometry (MS) identified five of these bands as BBS4-3xHA and BBS1, -5, -7, and -8 (Fig. S2); the sixth band was not identified. We conclude that (a) C. reinhardtii BBS4 is part of a BBSome complex similar to that previously described for vertebrates (Nachury et al., 2007) and (b) the C. reinhardtii BBSome is present in the flagellum.
BBS4 forms a complex with other BBS proteins in the flagellum. (A) Proteins in the flagellar membrane-plus-matrix from BBS4HA21 (top) or wild type (bottom) were separated by sucrose gradient centrifugation, and fractions were analyzed by Western blotting using antibodies to the HA tag or BBS4 and IFT172, -139, and -46. White lines seperate images of the membranes used for this blot. (B) Silver-stained SDS gel of protein complexes affinity purified from the membrane-plus-matrix fraction of BBS4HA21 and wild-type (wt) flagella using anti-HA beads. Proteins identified by MS are indicated. (C) Western blot analysis of isolated flagella or whole-cell extracts from the strains indicated. The blots were probed with antibodies to BBS4 and, as loading controls, IC2 (flagella) or IFT139 (cells). ptx1-1 and ptx7-1 are control nonphototactic strains. (A–C) The positions of standard proteins and their molecular masses in kilodaltons are indicated.
BBS4 forms a complex with other BBS proteins in the flagellum. (A) Proteins in the flagellar membrane-plus-matrix from BBS4HA21 (top) or wild type (bottom) were separated by sucrose gradient centrifugation, and fractions were analyzed by Western blotting using antibodies to the HA tag or BBS4 and IFT172, -139, and -46. White lines seperate images of the membranes used for this blot. (B) Silver-stained SDS gel of protein complexes affinity purified from the membrane-plus-matrix fraction of BBS4HA21 and wild-type (wt) flagella using anti-HA beads. Proteins identified by MS are indicated. (C) Western blot analysis of isolated flagella or whole-cell extracts from the strains indicated. The blots were probed with antibodies to BBS4 and, as loading controls, IC2 (flagella) or IFT139 (cells). ptx1-1 and ptx7-1 are control nonphototactic strains. (A–C) The positions of standard proteins and their molecular masses in kilodaltons are indicated.
To test for genetic interactions between BBS4 and other BBS genes, whole-cell extracts or isolated flagella from bbs7-1 and bbs1-1 were probed with anti-BBS4 (Fig. 4 C). BBS4 was lost from bbs1 cells, suggesting that the expression or stability of BBS4 depends on BBS1. Normal amounts of BBS4 were present in whole cells of bbs7-1, but BBS4 was strongly reduced in bbs7-1 flagella, indicating that the transport of BBS4 into the flagellum depends on BBS7.
BBS proteins are substoichiometric to IFT proteins in C. reinhardtii flagella
The distribution of BBS4 in C. reinhardtii is very similar to that described for IFT particle proteins, and BBS4-3xHA accumulates at the tips of growing flagella, as previously described for IFT particle proteins (Fig. 5 B; Deane et al., 2001). To determine the extent to which BBS4 and IFT proteins colocalize, we performed double IFM with anti-HA and antibodies to various IFT particle proteins. In BBS4HA21 flagella, BBS4-3xHA largely colocalized with IFT139 (Fig. 5 A) and IFT46 (Fig. 5 B), but the number of particles stained by anti-IFT139 or anti-IFT46 exceeded those stained by anti-HA. In wild-type flagella, a majority of the BBS4 particles (95% of 242 BBS4 particles from 35 flagella) colocalized with IFT139 particles, whereas 27% of 314 IFT139 particles lacked a BBS4 signal (Fig. 5 C). In contrast, virtually all IFT B complexes labeled by anti-IFT46 were associated with A complexes labeled by anti-IFT139 and vice versa, as shown in Fig. 2 E (a–c). Therefore, BBS4 is detectable on only a subset of IFT particles in the flagellum. The occasional BBS4 label not colocalized with an IFT label probably represents a BBS particle that transiently dissociated from an IFT particle (Fig. 6, F and G).
BBSomes are less abundant than IFT particles in C. reinhardtii flagella. (A and B) Double immunolabeling of a BBS4HA21 cell with polyclonal anti-HA and anti-IFT139 (A) or anti-IFT46 (B). In B, cells were deflagellated by pH shock and allowed to regrow flagella for ∼10 (a–c), 20 (d–f), and 45 min (g–i). BBS4-HA and the IFT proteins colocalize at the flagellar base, and the signals partially overlap in the flagella. Arrowheads indicate flagellar tips. (C) Detached wild-type flagella were double labeled with anti-BBS4 (a) and anti-IFT139 (b). Closed arrowheads indicate anti-IFT139 signals that did not colocalize with anti-BBS4 signals; the open arrowhead indicates anti-BBS4 signal that did not colocalize with anti-IFT139 staining. (D) Relative abundance of the native and the AQUA peptide LLETLNEDVK of IFT81. The AQUA peptide ion (590.831) and a peptide ion of monoisotopic m/z = 587.323, corresponding to the native LLETLNEDVK peptide ion (theoretical m/z value of 587.322), are marked. For details see Fig. S4. (E) As in D but for the LYVEQTQR peptide of BBS1. Bars: (A and B) 5 µm; (C) 1 µm.
BBSomes are less abundant than IFT particles in C. reinhardtii flagella. (A and B) Double immunolabeling of a BBS4HA21 cell with polyclonal anti-HA and anti-IFT139 (A) or anti-IFT46 (B). In B, cells were deflagellated by pH shock and allowed to regrow flagella for ∼10 (a–c), 20 (d–f), and 45 min (g–i). BBS4-HA and the IFT proteins colocalize at the flagellar base, and the signals partially overlap in the flagella. Arrowheads indicate flagellar tips. (C) Detached wild-type flagella were double labeled with anti-BBS4 (a) and anti-IFT139 (b). Closed arrowheads indicate anti-IFT139 signals that did not colocalize with anti-BBS4 signals; the open arrowhead indicates anti-BBS4 signal that did not colocalize with anti-IFT139 staining. (D) Relative abundance of the native and the AQUA peptide LLETLNEDVK of IFT81. The AQUA peptide ion (590.831) and a peptide ion of monoisotopic m/z = 587.323, corresponding to the native LLETLNEDVK peptide ion (theoretical m/z value of 587.322), are marked. For details see Fig. S4. (E) As in D but for the LYVEQTQR peptide of BBS1. Bars: (A and B) 5 µm; (C) 1 µm.
BBS4 is moved by IFT. (A) BBS4-GFP undergoes IFT. (a) Cell attached to the glass surface by its flagella (arrowheads). (b) TIRFM image of the two flagella of a BBS4-GFP1 cell. (c) Kymogram revealing the anterograde and retrograde movement of BBS4-GFP in the flagella. (B) Western blot of flagella isolated from wild-type control (g1), bbs4-1, IFT20-mCherry, and strain MxG1.3 expressing BBS4-GFP and IFT20-mCherry. The blots were probed as indicated; the positions of the immunoreactive proteins and standard proteins and their molecular masses in kilodaltons are marked. (C) The velocity of fluorescent protein–tagged IFT20 and BBS4 in flagella, as determined by TIRFM. In the BBS4-GFP/IFT20-mCherry strain, only particles with both tags were scored. n, number of particles analyzed. (D) Frequency of BBS4-GFP and fluorescent protein–tagged IFT20, as determined by TIRFM. n, number of flagella analyzed. (C and D) SDs are indicated. (E) Single frame from simultaneous recording of BBS4-GFP and IFT20-mCherry. Flagella are marked by arrowheads, and the cell body is marked by an arrow. (F) Kymograms showing the movement of BBS4-GFP (a) and IFT20-mCherry (b) in one flagellum. The image in c is a merger of a and b. Arrowheads and arrows indicate anterograde and retrograde cotransport of both proteins, respectively. Several tracks representing IFT particles are devoid of BBS4-GFP. Note the fission of a BBS4-GFP signal (arrowhead 1), a part of which is later picked up by a different IFT particle (arrowhead 2). (G) A BBS4-GFP particle falls off (arrowheads) a moving IFT20-mCherry particle (arrows). See Video 7. Bar, 10 µm.
BBS4 is moved by IFT. (A) BBS4-GFP undergoes IFT. (a) Cell attached to the glass surface by its flagella (arrowheads). (b) TIRFM image of the two flagella of a BBS4-GFP1 cell. (c) Kymogram revealing the anterograde and retrograde movement of BBS4-GFP in the flagella. (B) Western blot of flagella isolated from wild-type control (g1), bbs4-1, IFT20-mCherry, and strain MxG1.3 expressing BBS4-GFP and IFT20-mCherry. The blots were probed as indicated; the positions of the immunoreactive proteins and standard proteins and their molecular masses in kilodaltons are marked. (C) The velocity of fluorescent protein–tagged IFT20 and BBS4 in flagella, as determined by TIRFM. In the BBS4-GFP/IFT20-mCherry strain, only particles with both tags were scored. n, number of particles analyzed. (D) Frequency of BBS4-GFP and fluorescent protein–tagged IFT20, as determined by TIRFM. n, number of flagella analyzed. (C and D) SDs are indicated. (E) Single frame from simultaneous recording of BBS4-GFP and IFT20-mCherry. Flagella are marked by arrowheads, and the cell body is marked by an arrow. (F) Kymograms showing the movement of BBS4-GFP (a) and IFT20-mCherry (b) in one flagellum. The image in c is a merger of a and b. Arrowheads and arrows indicate anterograde and retrograde cotransport of both proteins, respectively. Several tracks representing IFT particles are devoid of BBS4-GFP. Note the fission of a BBS4-GFP signal (arrowhead 1), a part of which is later picked up by a different IFT particle (arrowhead 2). (G) A BBS4-GFP particle falls off (arrowheads) a moving IFT20-mCherry particle (arrows). See Video 7. Bar, 10 µm.
The absence of BBS4 from some IFT particles in flagella suggested that BBS proteins are substoichiometric to IFT proteins. To test this, we used the absolute quantification (AQUA) technique (Kirkpatrick et al., 2005) to determine the ratio between BBS1 and IFT81 by MS. Three AQUA peptides containing stable isotopes of carbon (13C) and nitrogen (15N) were synthesized for each protein and used as internal standards to determine the amount of their native peptide counterparts in the wild-type membrane-plus-matrix fraction, which contains virtually all of the flagellar IFT and BBSome proteins (Fig. 3 E). The analyses revealed a ratio of ∼1:6 between BBS1 and IFT81 (Fig. 5, D and E; Fig. S4; and Table II). This result suggests that BBSomes are considerably less abundant than IFT particles in C. reinhardtii flagella.
AQUA of IFT81:BBS1 stoichiometry in the flagellar membrane-plus-matrix by MS
| Protein | Peptide | Femtomole synthetic AQUA peptide/femtomole native peptide | ||
| Analysis 1 | Analysis 2 | |||
| IFT81 | AEV#SENETK | 50:16 | 50:14 | |
| IFT81 | LLETL#NEDVK | 50:18 | 50:18 | |
| IFT81 | TSALEGEVSGL#K | 50:22 | 50:22.5 | |
| BBS1 | ADAGLYSL#GK | 50:4 | 50:3 | |
| BBS1 | LYV#EQTQR | 50:3.5 | 50:3 | |
| BBS1 | LQDAGVVL#QTR | 50:2 | 50:2 | |
| Protein | Peptide | Femtomole synthetic AQUA peptide/femtomole native peptide | ||
| Analysis 1 | Analysis 2 | |||
| IFT81 | AEV#SENETK | 50:16 | 50:14 | |
| IFT81 | LLETL#NEDVK | 50:18 | 50:18 | |
| IFT81 | TSALEGEVSGL#K | 50:22 | 50:22.5 | |
| BBS1 | ADAGLYSL#GK | 50:4 | 50:3 | |
| BBS1 | LYV#EQTQR | 50:3.5 | 50:3 | |
| BBS1 | LQDAGVVL#QTR | 50:2 | 50:2 | |
Membrane-plus-matrix was prepared from isolated wild-type flagella and subjected to AQUA analysis. Data represent duplicate MS analyses, each containing 50 fmol of all six of the indicated synthetic AQUA peptides. # follows residues with 13C- or 15N-stable isotope mass tags. The IFT81 mean ratios for analyses 1 and 2 are 50:18.7 and 50:18.17, respectively. The BBS1 mean ratios for analyses 1 and 2 are 50:3 and 50:2.67, respectively. The ratios of IFT81 to BBS1 for analyses 1 and 2 are 6.23:1 and 6.81:1, respectively. The mean (±SD) of the two IFT81:BBS1 ratio values is 6.52:1 (±0.41).
BBS4 is transported by a subset of IFT particles
We used TIRFM to analyze the movement of BBS4-GFP in flagella of BBS4-GFP1 cells (bbs4-1 rescued by expression of BBS4-GFP; Fig. 6 A). Speckles of BBS4-GFP moved up and down the flagella at velocities typical for IFT in C. reinhardtii (Fig. 6 A, b and c; and Video 4; Kozminski et al., 1993). Anterograde movement of BBS4-GFP progressed at 2.3 µm/s; retrograde transport occurred at 4 µm/s (Fig. 6 C). As expected, the frequency of BBS4-GFP speckles in BBS4-GFP1 flagella was lower than that of IFT complex B protein IFT20 speckles in ift20:IFT20-GFP and ift20:IFT20-mCherry flagella (Fig. 6 D; in ift20, the IFT20 gene is entirely deleted [not depicted]).
TIRFM of BBS4-GFP in C. reinhardtii flagella. Images were recorded and are displayed at 30 frames/s.
TIRFM of BBS4-GFP in C. reinhardtii flagella. Images were recorded and are displayed at 30 frames/s.
To test whether BBS4 is cotransported with IFT particles, we generated a strain (MxG1.3) coexpressing BBS4-GFP and IFT20-mCherry. This strain lacks wild-type BBS4 and IFT20 (Fig. 6 B). Simultaneous observation of both tagged proteins using TIRFM revealed that BBS4-GFP mostly co-migrated with IFT20-mCherry (91% of BBS4-GFP particles; n = 89 particles in 27 flagella; Fig. 6, E and F; and Videos 5 and 6). In contrast, 58% of the IFT particles visualized by IFT20-mCherry did not carry BBS4-GFP (111 of 192 particles). Occasionally, a BBS4-GFP particle was observed to detach from an IFT20-mCherry particle and cease moving (Fig. 6, F and G; and Video 7); conversely, nonmoving BBS4-GFP particles were occasionally picked up and transported by moving IFT20-mCherry particles (Fig. 6 F). The data reveal that a subset of IFT particles transports BBS4-GFP in C. reinhardtii flagella.
TIRFM of BBS4-GFP and IFT20-mCherry in C. reinhardtii flagella. A dual-view system was used to project the BBS4-GFP signal on the top and the IFT20-mCherry signal on the bottom. Images were simultaneously recorded and are displayed at 30 frames/s.
TIRFM of BBS4-GFP and IFT20-mCherry in C. reinhardtii flagella. A dual-view system was used to project the BBS4-GFP signal on the top and the IFT20-mCherry signal on the bottom. Images were simultaneously recorded and are displayed at 30 frames/s.
Color display simultaneously showing BBS4-GFP and IFT20-mCherry in C. reinhardtii flagella. BBS4-GFP is shown in green, and IFT20-mCherry is shown in red. The flagellar tip is oriented to the left, and the base is oriented to the right. Photoshop was used to crop, orient, overlay, and adjust images. TIRFM images were recorded and are displayed at 30 frames/s.
Color display simultaneously showing BBS4-GFP and IFT20-mCherry in C. reinhardtii flagella. BBS4-GFP is shown in green, and IFT20-mCherry is shown in red. The flagellar tip is oriented to the left, and the base is oriented to the right. Photoshop was used to crop, orient, overlay, and adjust images. TIRFM images were recorded and are displayed at 30 frames/s.
Color display simultaneously showing BBS4-GFP and IFT20-mCherry in C. reinhardtii flagella. BBS4-GFP is shown in green, and IFT20-mCherry is shown in red. Note BBS4-GFP falling off the IFT20-mCherry particle, which continues to move toward the tip. The detached BBS4-GFP particle is marked by an arrow. TIRFM images were recorded and are displayed at 30 frames/s. The video corresponds to Fig. 6 G.
Color display simultaneously showing BBS4-GFP and IFT20-mCherry in C. reinhardtii flagella. BBS4-GFP is shown in green, and IFT20-mCherry is shown in red. Note BBS4-GFP falling off the IFT20-mCherry particle, which continues to move toward the tip. The detached BBS4-GFP particle is marked by an arrow. TIRFM images were recorded and are displayed at 30 frames/s. The video corresponds to Fig. 6 G.
bbs4 flagella accumulate putative signaling proteins
To examine the levels of selected proteins in wild-type and bbs4-1 flagella, Western blots were probed with antibodies to IFT particle proteins IFT20, -46, -172, and -139, IFT motor subunits KAP and D1bLIC, and the flagellar membrane proteins polycystin 2 (PKD2), FMG-1 (flagellar membrane glycoprotein-1), FMG-3, and mastigoneme protein. Wild-type and bbs4-1 flagella showed similar amounts of these proteins (Fig. S5 A), suggesting that bbs4-1 has no general defect in the transport of IFT or flagellar membrane proteins.
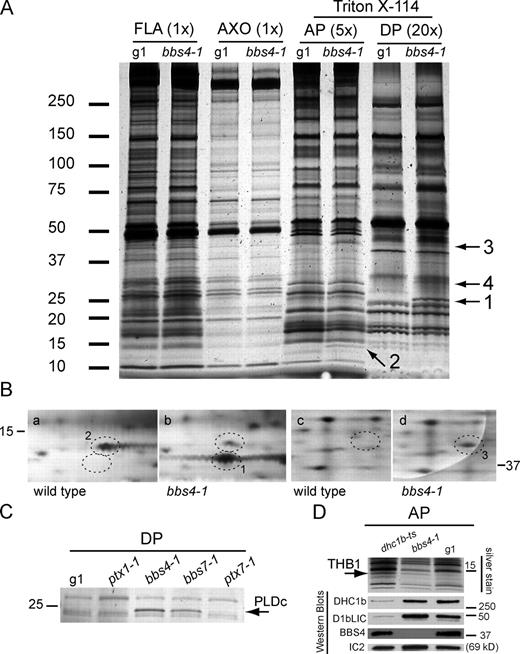
To determine more generally whether the lack of BBS4 affects the protein composition of bbs4-1 flagella, we compared wild-type and mutant flagella and flagellar fractions by 1D and 2D PAGE. No differences were observed between axonemes from wild type and bbs4-1 (Fig. 7 A and Fig. S5 C, a and b). However, SDS-PAGE revealed at least three bands (no. 1, 3, and 4) in the DP and one band (no. 2) in the AP of bbs4-1 that were not detected in the corresponding wild-type fractions (Fig. 7 A). MS identified band 1 as a phospholipase D (PLD) type c (PLDc; Joint Genome Institute [JGI] protein ID 190403), band 2 as a single-domain hemoglobin (THB1; JGI protein ID 81856), band 3 as a serine/threonine protein kinase (STPK; JGI protein ID 193039), and band 4 as JGI protein ID 191821 (a protein of unknown function with putative pleckstrin homology–like domains; Fig. S3). PLDc, STPK, and 191821 are predicted to lack transmembrane domains and to be myristoylated on the second glycine. 2D PAGE (Fig. 7 B) of the AP followed by MS of selected protein spots confirmed the accumulation of THB1 and STPK in bbs4-1 and revealed that OEE3 (oxygen-evolving enhancer 3/PSBQ; JGI protein ID 153656) was reduced in the bbs4-1 mutant flagella. Bands corresponding in size to PLDc (Fig. 7 C), THB1, STPK, and 191821 (not depicted) were also present in bbs1-1 and bbs7-1 flagellar fractions. The enrichment of PLDc and STPK in the DP of bbs7 was confirmed by MS (unpublished data). In contrast, PLDc and 191821 (and THB1 for ptx1-1) were not detected in comparable fractions from the nonphototactic mutants ptx1-1 and ptx7-1 (Fig. 7 C and not depicted; STPK was not tested), which have normal levels of BBS4. Therefore, the accumulation of PLDc, THB1, and STPK is linked to the loss of BBS proteins and is not caused simply by the loss of phototaxis. We conclude that a set of proteins is abnormally enriched in the flagella and specifically in the flagellar membrane-plus-matrix of the bbs mutants.
Biochemical defects in bbs4-1 flagella. (A) Silver-stained 5–15% SDS–polyacrylamide gel of flagella (FLA), axonemes (AXO), AP, and DP from wild type (g1) and bbs4-1. Proteins present in the AP and DP of bbs4-1 (arrows 1–4) but not in the corresponding fractions from wild type were identified by MS analysis as PLDc (band 1), THB1 (band 2), STPK (band 3), and JGI protein ID 191821 (band 4; see Fig. S3). (B) Details of 2D gels (from Fig. S5 C) separating matrix proteins from wild type (a and c) and bbs4-1 flagella (b and d). Protein spots 1 (b) and 3 (d) were enriched in the matrix of bbs4-1 and revealed to be THB1 and STPK, respectively, by MS. Protein spot 2 (a), which was reduced in quantity in the bbs4-1 matrix, was identified as OEE3. (C) Detail of a silver-stained SDS gel of the DP from the strains indicated. The arrow indicates the position of PLDc. (D) Detail of a silver-stained SDS gel and Western blots of the AP of g1, bbs4-1, and the retrograde IFT mutant dhc1bts probed with antibodies to the indicated proteins. The arrow indicates the position of THB1. (A–D) The positions of standard proteins and their molecular masses in kilodaltons are indicated.
Biochemical defects in bbs4-1 flagella. (A) Silver-stained 5–15% SDS–polyacrylamide gel of flagella (FLA), axonemes (AXO), AP, and DP from wild type (g1) and bbs4-1. Proteins present in the AP and DP of bbs4-1 (arrows 1–4) but not in the corresponding fractions from wild type were identified by MS analysis as PLDc (band 1), THB1 (band 2), STPK (band 3), and JGI protein ID 191821 (band 4; see Fig. S3). (B) Details of 2D gels (from Fig. S5 C) separating matrix proteins from wild type (a and c) and bbs4-1 flagella (b and d). Protein spots 1 (b) and 3 (d) were enriched in the matrix of bbs4-1 and revealed to be THB1 and STPK, respectively, by MS. Protein spot 2 (a), which was reduced in quantity in the bbs4-1 matrix, was identified as OEE3. (C) Detail of a silver-stained SDS gel of the DP from the strains indicated. The arrow indicates the position of PLDc. (D) Detail of a silver-stained SDS gel and Western blots of the AP of g1, bbs4-1, and the retrograde IFT mutant dhc1bts probed with antibodies to the indicated proteins. The arrow indicates the position of THB1. (A–D) The positions of standard proteins and their molecular masses in kilodaltons are indicated.
The abnormal accumulation of proteins in the bbs mutants could be caused by failure of a mechanism for excluding nonflagellar proteins from the flagellum or to a failure to remove certain proteins by retrograde IFT. To investigate this, flagellar fractions from the hypomorphic retrograde IFT mutant dhc1bts (unpublished data) were analyzed by SDS-PAGE. Flagella of this strain were assembled at the nonrestrictive temperature used in this study but contained reduced amounts of the retrograde IFT motor dynein 1b (Fig. 7 D). MS showed that THB1 was present in the AP of dhc1bts; PLDc was not detected in the DP of this mutant (unpublished data). Differences in the banding patterns of the mutant versus wild-type DPs precluded assessment of STPK and 191821. The results suggest that at least THB1 normally moves into the flagellum and is removed by retrograde IFT via the BBSome.
Newly assembled flagella of bbs4-1 have transiently restored phototactic steering
The results suggest that the abnormal accumulation of signaling proteins in bbs4-1 flagella somehow impairs the cells’ ability to phototax. If so, freshly formed bbs4-1 flagella should be less severely affected. To test this, the flagella were removed from bbs4-1 by pH shock and, after the cells had regenerated new flagella and recovered motility, their phototactic capability was assessed (Fig. 8). bbs4-1 cells showed improved phototaxis for up to 1 h after the beginning of flagellar regeneration. The results demonstrate that newly formed flagella of bbs4-1 cells are capable of phototactic steering but then lose that ability.
Phototaxis is partially restored in bbs4-1 during flagellar regeneration. Dish phototaxis assays of wild type (g1) and bbs4-1 during flagellar regeneration. Cells were deflagellated by pH shock and allowed to regenerate flagella. Samples were transferred to culture dishes and exposed to directional light (direction indicated by arrows) for ∼10 min. T43, T52, T69, and T120 refer to the time in minutes since deflagellation. Controls were treated identically except that the pH shock was omitted.
Phototaxis is partially restored in bbs4-1 during flagellar regeneration. Dish phototaxis assays of wild type (g1) and bbs4-1 during flagellar regeneration. Cells were deflagellated by pH shock and allowed to regenerate flagella. Samples were transferred to culture dishes and exposed to directional light (direction indicated by arrows) for ∼10 min. T43, T52, T69, and T120 refer to the time in minutes since deflagellation. Controls were treated identically except that the pH shock was omitted.
Discussion
Several C. reinhardtii BBS proteins form an evolutionarily conserved flagellar complex
Nachury et al. (2007) purified, from RPE1 cells and mouse testes, a 12S complex containing seven highly conserved BBS proteins (BBS1/2/4/5/7/8/9); this complex was termed the BBSome. In this study, we demonstrate that C. reinhardtii BBS4 extracted from isolated flagella sediments at ∼12S in sucrose gradients, indicating that it also occurs in a complex. We purified this complex from the membrane-plus-matrix fraction of C. reinhardtii flagella and found it to contain BBS4 and at least four other BBS proteins (BBS1/5/7/8). The presence of a similar complex in C. reinhardtii and mammalian cells indicates that the BBSome is evolutionarily conserved. Although individual BBS proteins previously have been reported to be in cilia (Blacque et al., 2004; Ou et al., 2005; Nachury et al., 2007), this is the first direct evidence that the BBSome complex by itself enters and functions in the flagellum.
C. reinhardtii BBS1, -4, and -7 are not required for flagellar assembly
We identified C. reinhardtii mutants defective in BBS1, -4, and -7. These bbs mutants develop full-length flagella with apparently normal ultrastructure, indicating that these BBSome proteins are not required for flagellar assembly in C. reinhardtii. The loss of BBS proteins has various effects on ciliary assembly in different species and cell types. For example, zebrafish bbs morphants have fewer and shorter cilia in the Kupffer's vesicle (Yen et al., 2006), and Bbs1, -4, and -6 knockout mice are sterile because of a failure to assemble sperm flagella, although other cilia develop more or less normally (Mykytyn et al., 2004; Nishimura et al., 2004). RNAi targeting of BBS5 in C. reinhardtii resulted in the partial or complete loss of flagella (Li et al., 2004). Similarly, the number of ciliated retinal pigment epithelial cells is reduced by siRNAs targeting BBS1 and -5 (Nachury et al., 2007; Loktev et al., 2008). However, cilia are present in human BBS patients and BBS knockout mice as well as in the C. reinhardtii bbs mutants examined in this study, suggesting that loss of the BBS proteins does not cause a general failure of the ciliary assembly machinery. This distinguishes the BBS mutants from IFT loss-of-function mutants, which lack cilia/flagella and are embryonic lethal in mammals (Brazelton et al., 2001; Rosenbaum and Witman, 2002; Hou et al., 2007; Jonassen et al., 2008). It is possible that the effects on ciliary assembly may be an indirect effect of loss of the BBS proteins.
We found that BBS4 was strikingly reduced in the cell body of the bbs1 mutant, indicating that BBS1 is required for the expression or stability of BBS4 and probably of the BBSome. This is consistent with genetic analysis in zebrafish showing that BBS1 has a central role in the interactions between the genes encoding different BBSome proteins (Tayeh et al., 2008). BBS4 was present in the cell body of the C. reinhardtii bbs7 mutant, but only small amounts were transported into the flagella. Thus, flagellar transport of BBS4 requires BBS7. These data provide independent biochemical and genetic evidence that the BBSome proteins interact to form a complex in C. reinhardtii flagella.
C. reinhardtii BBS4 is moved by IFT but is not an integral component of the IFT machinery
During IFT, particles consisting of ∼20 proteins organized into complex A and complex B are transported anterogradely and retrogradely in the flagellum (Rosenbaum and Witman, 2002; Scholey, 2008). In C. elegans cilia, GFP-tagged BBS1, -2, -7, and -8 undergo bidirectional transport similar to that of IFT particles (Blacque et al., 2004). In this study, we report that C. reinhardtii BBS4-GFP is also transported anterogradely and retrogradely in the flagellum at rates identical to that of IFT particles. Using double TIRFM with IFT20-mCherry, we found that BBS4-GFP is indeed carried by IFT particles. This raises the question of whether BBS4, and by extension the BBSome, is an IFT cargo or an integral component of the IFT machinery. In our C. reinhardtii bbs mutants, the assembly of flagella, the amounts of IFT particle and motor proteins in flagella, and the velocity of IFT are normal, and IFT complex A and B colocalize as they do in wild-type flagella. Thus, BBS4 is not an essential component of IFT in C. reinhardtii.
The situation is different in C. elegans. In channel cilia of C. elegans bbs-7 and -8 mutants, IFT particles break down into complex A and B, which then move at distinct velocities, giving rise to the hypothesis that these BBS proteins are required to stabilize the physical interaction between IFT complex A and B (Ou et al., 2005). C. elegans deploys two distinct motors for anterograde transport, kinesin 2 attached to IFT complex A and OSM-3 attached to complex B and solely responsible for IFT in the distal segment of the cilium (Scholey, 2008). Because the two motors have inherently different velocities, their cooperative actions could create tension between the two IFT complexes, and the BBS proteins might be required to prevent complex A and B from separating from each other. In contrast, C. reinhardtii deploys only one anterograde IFT motor and therefore may not require the stabilizing effect of the BBSome.
The C. elegans model for BBS protein function would predict that all IFT particles are associated with a BBSome. However, our double IFM localization of BBS4 and IFT46 (complex B) or IFT139 (complex A) revealed that not all IFT particles carry BBS4 in C. reinhardtii. Consistent with this, double TIRFM showed that many IFT20-mCherry particles were not associated with BBS4-GFP as they moved along the flagellum. We further observed that some BBS4-GFP–labeled particles separated from IFT particles and remained stationary while the IFT particles continued to move along the flagellum. Therefore, in vivo studies indicate that IFT–BBSome interaction differs in C. reinhardtii (this study) versus C. elegans (Blacque et al., 2004).
The C. elegans model would also predict that the IFT particle proteins and BBSome proteins should be present in similar amounts. To test this, we used quantitative MS (Kirkpatrick et al., 2005) to assess the relative amounts of an IFT protein and a BBS protein in C. reinhardtii flagella. The results indicated that IFT81 is approximately six times more abundant than BBS1. Assuming one BBS1 subunit per BBSome (Nachury et al., 2007) and two IFT81 subunits per IFT particle (Lucker et al., 2005), there are at least three times as many IFT particles as BBSomes in the flagellum. Data from the C. reinhardtii flagellar proteome analysis (Pazour et al., 2005) extend this conclusion qualitatively to virtually all of the IFT and BBSome proteins: two (BBS5 and -9) of the eight conserved BBSome proteins were identified by only a single unique peptide each (∼0.2 peptides/10 kD of BBS5 and -9), and the rest were not found at all; in contrast, numerous peptides were found that originated from IFT complex B proteins and IFT motor proteins (2.4 peptides/10 kD and 2.0 peptides/10 kD, respectively), suggesting an ∼1:1 stoichiometry between the motor and complex B proteins and an ∼1:10 stoichiometry between BBS5 and -9 compared with the IFT complex B proteins: Table S4).
In summary, in C. reinhardtii, BBS1, -4, and -7 are not required for IFT, the BBSome proteins are considerably less abundant in flagella than are IFT proteins, and only a subset of IFT particles are involved in BBSome transport. We conclude that the C. reinhardtii BBSome is not an integral part of the IFT machinery. It is more likely to be an adapter that couples specific cargoes to IFT or an IFT cargo that carries a signal. These possibilities are not mutually exclusive.
The BBSome is required for the export of specific proteins from the flagellum
If the BBSome is involved in transport of proteins, cilia and flagella defective in BBSome proteins should have an altered protein composition. Studies of mouse models defective in Bbs genes have revealed a mislocalization of rhodopsin in photoreceptor cells (Nishimura et al., 2004; Abd-El-Barr et al., 2007), and the G protein–coupled receptors Sstr3 and Mchr1 fail to localize to primary cilia of cultured neurons in Bbs2−/− and Bbs4−/− mice (Berbari et al., 2008). This has been interpreted as evidence that the BBSome is involved in transport of at least some flagellar membrane proteins into the cilium. Consistent with this, Nachury et al. (2007) have proposed that the mammalian BBSome interacts with the Rab8 GDP/GTP exchange factor Rabin 8 to promote recruitment of specific post-Golgi membrane vesicles to the base of the cilium followed by entry of membrane proteins into the cilium. Because flagella can be isolated from C. reinhardtii, we were able to assess specific proteins as well as a broad ensemble of flagellar proteins for changes caused by defects in the BBS proteins. In the C. reinhardtii bbs4-1 mutant, the amounts of four selected flagellar membrane proteins were unaltered, as determined by Western blotting, suggesting that bbs mutants do not have a general defect in the delivery of membrane proteins to the flagellum. 1D and 2D PAGE showed that the vast majority of proteins, likewise, were similar in their amounts in wild-type and mutant flagella. However, at least four proteins were abnormally present in bbs flagella, and three of these were in the Triton X-114 DP, which is enriched in proteins of the flagellar membrane. The four proteins together were represented by only one peptide in the flagellar proteome, suggesting that they are not present or present in only small amounts in wild-type flagella.
One possible explanation for the accumulation of specific proteins in the flagella of C. reinhardtii bbs mutants is that the BBSome is required for the export of these proteins from the flagella. To test this hypothesis, we examined the flagella of the retrograde IFT motor mutant dhc1bts and observed an accumulation of THB1 similar to that observed in bbs4 flagella. An accumulation of PLDc was not observed, but this may be because PLDc outcompetes THB1 for binding to the small number of BBSomes presumed to still be trafficking in this hypomorphic mutant or because the mutant is abnormal in other aspects of IFT. Further studies will be necessary to determine whether STPK and protein 191821 accumulate in the flagella of this or other retrograde IFT mutants.
In agreement with a function for the BBSome in the export of flagellar proteins, Shah et al. (2008) observed that membrane vesicles accumulated near the tips of cilia of Bbs1, -2, -4, and -6 knockout mice; they suggested that this phenotype might be caused by a defect in retrograde IFT or loss of cargo from the IFT complex. It also may be relevant that dynein-dependent retrograde transport of melanosomes is defective in zebrafish bbs morphants (Yen et al., 2006).
At least three of the four proteins abnormally accumulated in C. reinhardtii bbs mutants (an STPK, a phospholipase, and a truncated hemoglobin) are potentially involved in intracellular signaling. This raises the possibility that a specific function of the BBSome is to transport signaling proteins. Cilia are involved in various signaling pathways (e.g., phototransduction, olfaction, PDGF-αα, hedgehog, Wnt, etc.), and IFT is thought to be involved in moving receptors within flagella and transmitting some signals (Pazour and Witman, 2003; Pan et al., 2005; Qin et al., 2005; Wang et al., 2006). IFT is also likely to be involved in the turnover of these proteins. For example, polycystin 2 abnormally accumulates in the cilia of mice with defects in Ift88 (Pazour et al., 2002). Similarly, the C. reinhardtii homologue of polycystin 2, PKD2, is transported by IFT and accumulates in the flagella of fla10 mutants when IFT is turned off at the restrictive temperature (Huang et al., 2007). Because no accumulation of PKD2 was observed in flagella of the C. reinhardtii bbs4 mutant, PKD2 may be coupled directly to the IFT particle or to another adapter for its removal from the flagella, whereas the protein kinase, the phospholipase, and the truncated hemoglobin are linked to retrograde IFT via the BBSome. Because the latter proteins are absent or present only in low concentrations in wild-type flagella, it is likely that normally they continually enter and exit the flagella, possibly as part of a signaling system that monitors the flagellum, and build up in the flagella only when export is blocked.
The abnormal accumulation of proteins in bbs flagella is the likely cause of the phototaxis defect
The defects in BBS1, -4, and -7 all cause loss of phototaxis in C. reinhardtii. Phototaxis requires the ability to precisely control the two flagellar axonemes in response to Ca2+-mediated signals induced by photostimulation. bbs4 cells are nonphototactic because their axonemes lack the ability to respond normally to changes in intraflagellar Ca2+. However, BBS proteins are components of the flagellar matrix, and our biochemical analyses have so far failed to identify differences in the axonemes.
The explanation for this conundrum may be that the proteins that accumulate in the bbs mutant flagellar matrix directly or indirectly modify the axoneme and thereby impair a proper Ca2+ response. For example, PLDs generate phosphatidic acid, which acts as a second messenger and can activate or inactivate other proteins (Hancock, 2007). In Chlamydomonas eugametos, a PLD is activated by Ca2+ influx induced by membrane depolarization (Munnik et al., 2000); Ca2+ influx and membrane depolarization are both involved in the photobehavior of C. reinhardtii. The accumulated protein kinase could abnormally phosphorylate axonemal proteins. The I1 inner arm dynein intermediate chain IC138, for example, is hyperphosphorylated in axonemes of nonphototactic mia (modifier of inner arms) mutants (King and Dutcher, 1997; Wirschell et al., 2007). The accumulated THB1 may also disrupt normal signaling. Truncated hemoglobins have been implicated in NO metabolism and are capable of altering redox states (Smagghe et al., 2008). The C. reinhardtii flagellum is rich in flavoproteins and other redox proteins, and at least two, AGG2 and -3, are involved in the regulation of phototaxis (Iomini et al., 2006). Light-dependent changes in redox poise alter the disulfide-based interactions of various components of the outer dynein arms, changing the flagellar beat frequency and the duration of the photophobic response (Wakabayashi and King, 2006). Therefore, the accumulation of one or more of these proteins could easily be responsible for the nonphototactic phenotype of C. reinhardtii bbs mutants. Given the potentially far-reaching effects of these signaling pathways, it is tempting to speculate that a similar disruption of ciliary signaling could be responsible for the diverse and complex phenotypes observed when BBS proteins are defective in other organisms.
Importantly, we found that phototaxis is restored in bbs4-1 cells for a short period of time after the formation of new flagella. This is consistent with a model in which the BBSome continually removes certain proteins from the flagellum and that in the absence of the BBSome, an accumulation of these proteins impairs flagellar function. This experiment also indicates that the functional defect in bbs mutant flagella is not caused by a failure to transport certain proteins into the flagellum; if this were the case, the cells would not have regained the ability to phototax immediately after flagellar regeneration.
The aberrant accumulation of proteins caused by defects in BBSome proteins may lead to progressive degeneration of ciliary function
Because C. reinhardtii disassembles its flagella before mitosis (Cavalier-Smith, 1974), there is only a limited time for proteins to aberrantly accumulate in the flagella and only a limited time for them to damage the flagella. In contrast, many cilia in vertebrates persist for long periods of time, and a failure to remove certain proteins from the cilia could progressively impair their functionality and maintenance. Indeed, the outer segments of rod cells in Bbs knockout mice are initially formed but later degenerate (Nishimura et al., 2004; Abd-El-Barr et al., 2007). Similarly, cilia are progressively lost in the Kupffer's vesicle of zebrafish bbs morphants (Yen et al., 2006). We propose that defects in BBSome proteins cause an aberrant accumulation of specific proteins in the cilia, which, over time, lead to increasingly dysfunctional cilia, ciliary damage, and ciliary degeneration. Therefore, BBS may be a degenerative disease of the cilium.
Materials and methods
Strains and culture conditions
C. reinhardtii strains used in this study are listed in Table S1. Cells were grown in modified minimal medium (Pazour et al., 1995) in 24-well plates, 250-ml Erlenmeyer flasks, or aerated 5-liter diphtheria toxin flasks with a light/dark cycle of 14:10 h at ∼23°C.
Identification and characterization of C. reinhardtii bbs mutants
A genomic DNA library of 350 mutant strains was screened by quantitative RT-PCR using primer pairs BBS4N and BBS4C (see Table S2 for a list of primers used in this work). A total of 40 primer pairs against various parts of the C. reinhardtii BBIP10 and BBS1/2/3/5/7/8/9 genes were tested on DNA isolated from 17 nonphototactic mutants. Primer pair BBS7-1 failed to amplify in strain ptx6-1, and primer pair BBS1-1 failed to amplify in the uncharacterized nonphototactic strain RIR7-2 (provided by C. Dieckmann, University of Arizona, Tucson, AZ). PCR was performed using Quantitect Syber green master mix (QIAGEN), a thermal cycler (Opticon; MJ Research), and the following cycle conditions: 15 min at 94°C; 35 cycles of 30 s at 94°C, 30 s at 54°C, and 30 s at 72°C; followed by 10 min at 72°C. Primer pairs 173322, 148017, 173319, and 190839 were used to map the deletions in the bbs4 mutants. To determine the genetic defect in bbs7/ptx6, primer pairs BBS7FIN1F/BBS7FIN2R and BBS7FIN2F/BBS7FIN1R were used for nested PCR amplification of the affected region from wild type and ptx6; primer BBS7SEQU1F was used for direct sequencing of the PCR products.
Rescue of bbs4-1/ptx5-1
Screening of a bacterial artificial chromosome (BAC) library of C. reinhardtii (Nguyen et al., 2005) using a labeled PCR product generated with primer pair BBS4N identified BAC 1H15 (BAC library CRCCBa, Clemson University Genomics Institute), which contained the entire BBS4 sequence. A 6,797-bp HindIII–BsmI fragment of BAC 1H15 covering BBS4 was subcloned into pBR322. To engineer plasmids encoding epitope-tagged BBS4, the protein-encoding part of the message was amplified from cDNA using primer pair CBBS4 and cloned into FLA14 gene–based expression vector pKL3-3xHA or pKL3-GFP; the former additionally contained the ble selectable marker gene. After transformation or cotransformation together with pSP124S containing the ble resistance gene, transformants were selected on Zeocin Tris-acetate-phosphate plates (Stevens et al., 1996), picked into liquid M medium, and screened for negative phototaxis (Pazour et al., 1995). DNA was isolated from four selected strains rescued with pBR322-BBS4 and tested for the absence of the regions flanking the BBS4 gene and for the presence of the BBS4 gene to verify that they represented transformed bbs4-1/ptx5-1 cells. BBS4HA21 rescued by BBS4-3xHA was mated to strain g2, and random progeny were analyzed for phototactic behavior and for the expression of BBS4-3xHA by indirect IFM. Of 96 strains tested, 50 expressed BBS4-3xHA, and all of these showed phototactic behavior.
Phototaxis assay
To analyze phototactic behavior, cells in a 35 × 10–mm culture dish were illuminated from one side and scored for the ability to accumulate on the other side (Hegemann and Berthold, 2009). Phototaxis was quantitated as described by Moss et al. (1995). Measurements of the Ca2 response of reactivated cells models were performed as described by Kamiya and Witman (1984). All phototaxis experiments were performed 4–8 h after the beginning of the light phase.
Antibody production and epitope tagging
Primers BBS4S3F and BBS4A1R were used to amplify a partial cDNA encoding the C-terminal 248 residues of BBS4; after digestion with EcoRI, the fragment was inserted into the EcoRI site of the bacterial expression vector pMAL-cR1,and the purified fusion protein was used for antibody production in rabbits (Covance). A full-length cDNA encoding BBS4 was cloned into the bacterial expression vector pGEX (GE Healthcare), and the immobilized fusion protein was used to affinity purify the immune serum.
A full-length BBS4 cDNA was cloned into the FLA14-based expression vectors pKL3-3xHA+ble and pKL3-GFP, adding three HA tags or a GFP tag to the C terminus of BBS4 (Lechtreck and Witman, 2007). After transformation into bbs4-1, 2 (BBS4HA21 and BBS4HA24) out of 98 transformants showed restored phototactic behavior and expression of BBS4-3xHA (∼50 kD); BBS4-3xHA was not expressed in 10 other transformants randomly selected for testing. To test whether BBS4 undergoes IFT, we cloned the BBS4 cDNA into a GFP expression vector and used this construct to rescue bbs4-1. 1 out of 78 transformants tested showed restored phototaxis and expression of BBS4-GFP, as detected by Western blots using polyclonal anti-GFP.
Flagellar isolation and fractionation
Flagellar isolation using the dibucaine method and flagellar amputation by pH shock were performed as previously described by Witman (1986) and Lefebvre (1995), respectively. Isolated flagella were extracted with 1% NP-40 Alternative (EMD) for 20 min on ice, and the suspension was separated by centrifugation into a pellet containing the axonemes and a supernatant consisting of the membrane-plus-matrix. For MS quantitation of BBS1 and IFT81 in the membrane-plus-matrix, the AQUA peptide technique was used (see Quantitative MS and Fig. S4). The membrane-plus-matrix was further separated by sucrose gradient centrifugation (36,000 rpm for 12.5 h at 4°C on an SW41 rotor [Beckman Coulter]), and fractions were analyzed by SDS-PAGE and Western blotting. For phase partitioning, isolated flagella were extracted using 1% Triton X-114 for 20 min on ice; after removal of the axonemes by centrifugation (Witman, 1986), the supernatant was incubated briefly at 37°C, and phases were separated by centrifugation (3,300 g for 10 min at RT). The AP was treated with 1% Triton X-114, the DP was diluted with buffer, and the phase separation was repeated to yield the final APs and DPs.
Immunoprecipitation of complexes containing BBS4-3xHA
The membrane-plus-matrix fractions of isolated wild-type and BBS4HA21 flagella were incubated with anti-HA affinity matrix (Roche) in 125 mM NaCl, 30 mM Hepes, 5 mM MgSO4, 0.5 mM EGTA, 25 mM KCl, pH 7.4, and Complete Protease Inhibitor Cocktail (Roche) and incubated overnight at 4°C with agitation. After repeated washes, 1 mg/ml HA peptide (Roche) was added to elute bound proteins. Proteins were separated by 5–15% SDS-PAGE (Bio-Rad Laboratories), excised after silver staining, and analyzed by MS.
IFM
Indirect IFM was performed as described by Lechtreck and Witman (2007). The primary antibodies used in this study are listed in Table S3; secondary antibodies were linked to Alexa Fluor 488, 568, or 594 (Invitrogen). Images were acquired using Axiovision software (Carl Zeiss, Inc.) and a camera (AxioCam MRm; Carl Zeiss, Inc.) on a microscope (Axioskop 2 plus; Carl Zeiss, Inc.) equipped with a 100× NA 1.4 oil DIC Plan-Apochromat objective (Carl Zeiss, Inc.) and epifluorescence. Image brightness and contrast were adjusted using Photoshop 6.0 (Adobe), and figures were assembled using Illustrator 8.0 (Adobe). Capture times and adjustments were similar for images mounted together.
Observation of IFT
IFT in the flagella was observed using DIC or TIRFM at RT. Cells in medium were placed onto poly-l-lysine–treated coverslips and allowed to adhere for ∼0.25–1 min. Coverslips were washed carefully with medium and covered with a second coverslip to form a chamber enclosed by a Vaseline ring. The fluorescence images were captured using a custom-built TIRF microscope based on an inverted microscope (IX71; Olympus) equipped with a Plan-Apochromat 60× NA 1.4 objective (Olympus). Multiline argon/krypton lasers (CVI Melles Griot) provided excitation light at 488 and 568 nm. Both lasers were cleaned up with appropriate MaxLine Laser-line filters (Semrock Inc.). A 488-nm RazorEdge beam splitter (Semrock, Inc.) was used for the GFP signal. An FF498/581 beam splitter (Semrock Inc.) was used for the GFP/mCherry signals. The resulting two-color emission signals were separated using a custom-built dual-view system equipped with an FF562-Di01 dichroic mirror (Semrock Inc.) and 525/50-nm and 630/69-nm emission filters (Semrock Inc.). Signals were recorded using a back-illuminated electron-multiplying charge-coupled device camera (iXon DV860; Andor Technology). Data were analyzed using ImageJ (National Institutes of Health) and Photoshop.
Protein identification by MS
Silver-stained gel bands were excised and digested in gel with trypsin (proteomics grade; Sigma-Aldrich) at 37°C overnight. For purification, eluted peptides were loaded on a µC18 ZipTip (Millipore) equilibrated in 0.1% trifluoroacetic acid. Peptides were deposited directly onto the sample target and allowed to air dry before insertion into the mass spectrometer. Analysis was performed on a matrix–assisted laser desorption/ionization time of flight (MALDI-TOF) mass spectrometer (Kratos Axima QIT; Shimadzu Instruments). Peptides were analyzed in positive ion mode in mid–mass range (700–3,000 D). The instrument was externally calibrated with Angiotensin II (MH+ of 1046.54), P14R (MH+ of 1533.86) and ACTH clip 18–39 (MH+ of 2465.20). Precursors were selected based on signal intensity at a mass resolution width of 250 for collision-induced dissociation fragmentation (MS/MS) using argon as the collision gas. All spectra were peak processed with Mascot Distiller (Matrix Sciences, Ltd.) before database searching. Database searches were performed in house with the Mascot search engine (Matrix Sciences, Ltd.). NMT–The MYR Predictor (http://mendel.imp.ac.at/myristate/SUPLpredictor.htm) and Myristoylator (http://www.expasy.ch/tools/myristoylator/) were used to predict myristoylation sites.
Quantitative MS
The membrane-plus-matrix fraction isolated from wild-type flagella was subjected to SDS-PAGE, and the region from ∼40 to 95 kD (Fig. S4 A) was diced and subjected to in-gel digestion with 6 ng/µl sequencing-grade modified trypsin (Promega) in 50 mM ammonium bicarbonate overnight at 37°C. Peptides were extracted first with 50% MeCN (acetonitrile) and 2.5% formic acid and then with 100% MeCN. Peptides were then dried using a SpeedVac. Peptides were resuspended in 8.5 µl of 2.5% MeCN and 2.5% formic acid containing 12.5 fmol/µl each of the three BBS1 and three IFT81 synthetic AQUA peptides shown in Table II. AQUA peptides with 13C-, 15N-labels were synthesized at Cell Signaling Technology. 4 µl of the samples were shot in duplicate. Mass measurements were made in a mass spectrometer (LTQ-Orbitrap; Thermo Fisher Scientific), which was set up with a liquid chromatography interface essentially as described previously (Ballif et al., 2008). Quantification was performed as outlined in Fig. 5 (D and E) and Fig. S4. Before collection of the measurements reported in Table II, liquid chromatography–MS/MS analyses were conducted on the mixture of six AQUA peptides. For each peptide, the retention time on the reverse-phase C18 column was determined, and MS and MS/MS spectra were collected. Importantly, similar results to those reported in Table II were obtained with a fourfold increase in the concentration of trypsin used in the in-gel digestion, a fourfold increase in the amount of AQUA peptide standards used, or a fivefold increase in the amount of purified flagellar protein.
Online supplemental material
Fig. S1 shows the defective Ca2+ response of bbs4-1. Figs. S2 and S3 show peptides identified by MS. Fig. S4 shows MS data related to the quantification of IFT81. Fig. S5 shows additional data on the fractionation and analysis of bbs4-1 flagella. Video 1 is a time-lapse video showing phototaxis of wild-type C. reinhardtii and the bbs4-1 mutant in a culture dish assay. Video 2 shows phototaxis of wild-type C. reinhardtii cells. Video 3 shows a lack of phototaxis in C. reinhardtii bbs4-1 mutant cells. Video 4 shows TIRFM of BBS4-GFP in C. reinhardtii flagella. Video 5 shows TIRFM of BBS4-GFP and IFT20-mCherry in C. reinhardtii flagella. Videos 6 and 7 depict color display simultaneously showing BBS4-GFP and IFT20-mCherry in C. reinhardtii flagella. Tables S1, S2, and S3 list strains, primers, and antibodies, respectively, used in this study. Table S4 shows that BBS proteins are less abundant than IFT proteins in flagella.
Acknowledgments
We thank Dr. C. Dieckmann for sharing phototaxis mutants, Drs. D. Cole (University of Idaho, Moscow, ID), R. Bloodgood (University of Virginia, Charlottesville, VA), and J. Rosenbaum (Yale University, New Haven, CT) for sharing antibodies, and Dr. J. Brown (University of Massachusetts Medical School [UMMS], Worcester, MA) for analyzing the sedimentation of wild-type BBS4. We are grateful to Drs. G. Hendricks and J. Leszyk of UMMS for expert help with EM and MS, respectively.
Core facilities used in this research were supported by a Diabetes Endocrinology Research Center grant (DK32520). This work was supported by National Institutes of Health grants (GM030626 to G.B. Witman; DC006103, AR048898, and AR048526 to M. Ikebe; and GM060992 to G.J. Pazour), the Vermont Genetics Network through a National Institutes of Health grant (P20 RR16462) from the IDeA Network of Biomedical Research Excellence Program of the National Center for Research Resources (to B.A. Ballif), and the Robert W. Booth Endowment (G.B. Witman).
References
- AP
aqueous phase
- BAC
bacterial artificial chromosome
- BBS
Bardet-Biedl syndrome
- DIC
differential interference contrast
- DP
detergent phase
- IFM
immunofluorescence microscopy
- IFT
intraflagellar transport
- KAP
kinesin-associated protein
- MS
mass spectrometry
- PLD
phospholipase D
- PLDc
PLD type c
- STPK
serine/threonine protein kinase
- TIRF
total internal reflection fluorescence
- TIRFM
TIRF microscopy