Endocytosis of receptors at the plasma membrane is controlled by a complex mechanism that includes clathrin, adaptors, and actin regulators. Many of these proteins are conserved in yeast yet lack observable mutant phenotypes, which suggests that yeast endocytosis may be subject to different regulatory mechanisms. Here, we have systematically defined genes required for internalization using a quantitative genome-wide screen that monitors localization of the yeast vesicle-associated membrane protein (VAMP)/synaptobrevin homologue Snc1. Genetic interaction mapping was used to place these genes into functional modules containing known and novel endocytic regulators, and cargo selectivity was evaluated by an array-based comparative analysis. We demonstrate that clathrin and the yeast AP180 clathrin adaptor proteins have a cargo-specific role in Snc1 internalization. We additionally identify low dye binding 17 (LDB17) as a novel conserved component of the endocytic machinery. Ldb17 is recruited to cortical actin patches before actin polymerization and regulates normal coat dynamics and actin assembly. Our findings highlight the conserved machinery and reveal novel mechanisms that underlie endocytic internalization.
Introduction
Clathrin-mediated endocytosis selects proteins at the plasma membrane for internalization in membrane-bound vesicles. In mammalian cells, cargo is initially concentrated at endocytic sites by adaptor proteins that promote clathrin recruitment and assembly. A variety of adaptors, such AP2, AP180/clathrin assembly lymphoid myeloid leukemia (CALM), epsins, arrestins, and Dab2, recognize different endocytic motifs and select distinct classes of cargo (Maldonado-Báez and Wendland, 2006). The subsequent recruitment of accessory factors including Wiscott-Aldrich syndrome protein (WASP), dynamin, and amphiphysin stimulates actin polymerization, which results in membrane deformation and vesicle scission (Smythe and Ayscough, 2006).
Studies in yeast have demonstrated that conserved endocytic modules are recruited to sites of internalization in a similar temporal sequence (Kaksonen et al., 2005; Newpher et al., 2005; Perrais and Merrifield, 2005). Clathrin and adaptors are part of the early coat module that establishes the initial site of uptake, whereas the late coat module (Sla1/Sla2/End3/Pan1) couples coat formation to actin polymerization through recruitment and stimulation of yeast WASP (Las17) and Myo5. This activates the Arp2/3 complex to form an actin network that, stabilized by the actin regulatory module (Cap1/2, Sac6, Abp1), promotes rapid inward movement of the internalizing vesicle. Finally, the amphiphysins Rvs161/167 are thought to drive scission of endocytic vesicles from the plasma membrane. Recycling of the endocytic machinery by vesicle uncoating requires additional proteins, including the synaptojanin homologue Inp52.
This sequence of events, defined by live cell imaging studies, suggests that endocytic processes are largely conserved. However, striking differences in the functional requirements for certain regulatory proteins argue that internalization may be differentially regulated in yeast and mammalian cells. For example, the dynamic regulation of actin polymerization is essential for uptake in yeast but not in most mammalian cell types (Engqvist-Goldstein and Drubin, 2003; Smythe and Ayscough, 2006). Conversely, clathrin has an important role in higher cells, yet loss of yeast clathrin causes only a partial reduction in receptor internalization, and yeast homologues of major clathrin adaptor proteins such as AP2 and AP180 are not required for the uptake of known cargo (Wendland and Emr, 1998; Huang et al., 1999). Several other yeast genes are homologous to components of the mammalian endocytic machinery, yet do not lead to observable endocytosis defects when mutated (Engqvist-Goldstein and Drubin, 2003; Kaksonen et al., 2005). Some of these factors may have functionally redundant homologues, or have cargo-specific roles. Additional, unrecognized factors may also regulate cargo uptake in yeast. To better understand the endocytic process in both yeast and mammalian cells, it will be important to identify the complete set of structural and regulatory proteins, and systematically define their specific roles.
The yeast vesicle-associated membrane protein (VAMP)/synaptobrevin homologue Snc1, which regulates the fusion of exocytic vesicles at the cell surface, is a widely studied endocytic cargo protein. After delivery to the plasma membrane of the growing bud, it is rapidly internalized and transported to endosomal and Golgi compartments, where it is incorporated into new secretory vesicles (Lewis et al., 2000). Here, we perform a genome-wide analysis of Snc1 localization in yeast to uncover genes with functional roles in endocytosis, and quantify their relative contribution to this process. We identify functions for known and novel proteins not previously shown to be required for uptake, and demonstrate that the yeast AP180 homologues and clathrin have a cargo-specific role in Snc1 internalization. In addition, we describe an endocytic function for the previously uncharacterized protein low dye binding 17 (Ldb17).
Results
A Snc1-based quantitative assay for endocytic recycling
To quantify defects in endocytosis on a genome-wide scale, we developed an enzymatic assay based on the cell surface localization of a Snc1 reporter. The enzyme invertase (encoded by SUC2) was fused to the extracellular C terminus of Snc1, and GFP was appended to its intracellular N terminus, creating the chimeric protein GSS (GFP-Snc1-Suc2; Fig. 1 A). Cell-surface GSS levels can be determined by a colorimetric invertase activity assay using cell-impermeant reagents (Darsow et al., 2000). By fluorescence microscopy, GSS displayed a polarized distribution at the plasma membrane of small buds similar to Snc1 (Fig. 1 B; Lewis et al., 2000), which has been attributed to a cycle of localized exocytosis followed by rapid endocytosis that prevents diffusion away from the site of new growth (Valdez-Taubas and Pelham, 2003). In support of this model, when the Snc1 endocytosis signal was mutated, the polarization of GSS was lost and the level of invertase activity at the plasma membrane was increased (Fig. 1 B and not depicted). Both Snc1 and GSS were mislocalized to the vacuole in vps51Δ mutants (not depicted), demonstrating that they follow a similar recycling pathway back to the cell surface (Conibear et al., 2003).
Genome-wide screen for endocytic recycling mutants. (A) Schematic of the GFP-Snc1-Suc2 (GSS) reporter. At the plasma membrane (PM), the Suc2 (invertase) portion is accessible to the extracellular space. (B) Localization of GFP-Snc1, GSS, and an endocytosis-defective form of GSS containing snc1V40A,M43A mutations. Bar, 2 µm. (C) Representative portion of the yeast knockout array tested using the invertase activity overlay assay. Mutants defective in Snc1 endocytosis exhibit increased surface invertase activity relative to wild-type cells, and appear dark. Mutants with high (tpm1 and rvs161) or low (snx4 and irs4) levels of cell surface invertase activity are indicated. (D) Relative cell surface invertase activities of the top 1,000 mutants. The red line indicates threshold used to designate top hits.
Genome-wide screen for endocytic recycling mutants. (A) Schematic of the GFP-Snc1-Suc2 (GSS) reporter. At the plasma membrane (PM), the Suc2 (invertase) portion is accessible to the extracellular space. (B) Localization of GFP-Snc1, GSS, and an endocytosis-defective form of GSS containing snc1V40A,M43A mutations. Bar, 2 µm. (C) Representative portion of the yeast knockout array tested using the invertase activity overlay assay. Mutants defective in Snc1 endocytosis exhibit increased surface invertase activity relative to wild-type cells, and appear dark. Mutants with high (tpm1 and rvs161) or low (snx4 and irs4) levels of cell surface invertase activity are indicated. (D) Relative cell surface invertase activities of the top 1,000 mutants. The red line indicates threshold used to designate top hits.
We introduced the GSS reporter into two independent genome-wide collections of viable deletion mutants, and measured its cell surface level for each mutant in parallel using a large-scale plate-based invertase assay (Fig. 1 C). Mutants were ranked according to their median invertase activity score, determined by automated image densitometry (Fig. 1 D and Table S1). Mutants with increased cell-surface GSS included well-characterized endocytosis-defective strains such as rvs161 and vrp1. Other predicted endocytosis genes were enriched in the top 400 hits (Table I), including many not previously shown to cause defects in cargo uptake when deleted, such as yeast homologues of AP180 (yap1801 and yap1802), epsin (ent1), tropomyosin (tpm1), type I myosin (myo5), synaptojanin (inp52), coronin (crn1), and syndapin (bzz1). We generated a “gold standard” list of 33 proteins that have been localized to sites of endocytosis by time-lapse live cell microscopy and that were also represented in the haploid deletion collections we evaluated (Newpher et al., 2005; Maldonado-Báez et al., 2008; Toret et al., 2008; Drubin, D., personal communication). Of these, 23 were found in the top 400 hits, representing a significant degree of enrichment (P < 2.2 × 10−16, Fisher exact test). This set of ∼400 top-scoring mutants was chosen for further analysis. Mutants with cell-surface GSS levels significantly lower than wild type (e.g., snx4 and vps51) were also identified in the screen. This class of mutants, which may have impaired endosomal recycling, will be described in more detail elsewhere.
Predicted yeast endocytic genes identified in the screen
| Previously reported requirement for: | ||||
| Name | Ranka | Homologue | Patch dynamicsb | Internalizationc |
| RVS161 | 1 | Bin3/amphiphysin | Yes | Yes |
| VRP1 | 4 | WIP | Yes | Yes |
| RVS167 | 8 | Bin1/amphiphysin | Yes | Yes |
| SLA1 | 12 | SH3 domain | Yes | Yes |
| ARC18 | 18 | Arp2/3 complex | Yes | Yes |
| YAP1801 | 29 | AP180 | No | No |
| EDE1 | 31 | Eps15 | Yes | Yes |
| YAP1802 | 42 | AP180 | No | No |
| CAP1 | 45 | Actin-capping complex | Yes | No |
| TPM1 | 52 | Tropomyosin | No | No |
| ABP1 | 54 | Abp1 | Yes | No |
| MYO5 | 61 | Type 1 myosin | No | No |
| INP52 | 68 | Synaptojanin | Yes | No |
| YIR003w | 78 | ND | No | ND |
| CAP2 | 127 | Actin-capping complex | Yes | No |
| TWF1 | 155 | Cofilin-like repeats | No | No |
| SAC6 | 209 | Fimbrin | Yes | Yes |
| SYP1 | 216 | ND | No | ND |
| BBC1 | 231 | ND | Yes | No |
| ENT1 | 281 | Epsin | No | No |
| END3 | 315 | ND | Yes | Yes |
| LSB3 | 338 | ND | No | ND |
| CRN1 | 365 | Coronin | No | No |
| AIP1 | 368 | WDR1 | No | ND |
| BZZ1 | 383 | Syndapin | No | No |
| PKH2 | 396 | PDK1 | ND | No |
| Previously reported requirement for: | ||||
| Name | Ranka | Homologue | Patch dynamicsb | Internalizationc |
| RVS161 | 1 | Bin3/amphiphysin | Yes | Yes |
| VRP1 | 4 | WIP | Yes | Yes |
| RVS167 | 8 | Bin1/amphiphysin | Yes | Yes |
| SLA1 | 12 | SH3 domain | Yes | Yes |
| ARC18 | 18 | Arp2/3 complex | Yes | Yes |
| YAP1801 | 29 | AP180 | No | No |
| EDE1 | 31 | Eps15 | Yes | Yes |
| YAP1802 | 42 | AP180 | No | No |
| CAP1 | 45 | Actin-capping complex | Yes | No |
| TPM1 | 52 | Tropomyosin | No | No |
| ABP1 | 54 | Abp1 | Yes | No |
| MYO5 | 61 | Type 1 myosin | No | No |
| INP52 | 68 | Synaptojanin | Yes | No |
| YIR003w | 78 | ND | No | ND |
| CAP2 | 127 | Actin-capping complex | Yes | No |
| TWF1 | 155 | Cofilin-like repeats | No | No |
| SAC6 | 209 | Fimbrin | Yes | Yes |
| SYP1 | 216 | ND | No | ND |
| BBC1 | 231 | ND | Yes | No |
| ENT1 | 281 | Epsin | No | No |
| END3 | 315 | ND | Yes | Yes |
| LSB3 | 338 | ND | No | ND |
| CRN1 | 365 | Coronin | No | No |
| AIP1 | 368 | WDR1 | No | ND |
| BZZ1 | 383 | Syndapin | No | No |
| PKH2 | 396 | PDK1 | ND | No |
ND, not determined.
See Table S1.
Internalization of FM4-64 or cargo proteins. See Engqvist-Goldstein and Drubin (2003), Kaksonen et al. (2005), and Moseley and Goode (2006).
Genetic interaction mapping clusters functionally related genes
The large number of mutants with elevated surface invertase activity suggested that many distinct processes influence GSS reporter distribution. To functionally dissect these processes, we analyzed GSS surface levels in 374 top-scoring deletion mutants in pairwise combination with a panel of 81 deletion mutants that affect a variety of protein trafficking pathways. Genes that cofunction within individual complexes and pathways can be identified in large scale genetic interaction analyses based on two characteristic properties (Schuldiner et al., 2006). When loss of one gene disrupts a pathway or complex, loss of a second gene in the same pathway has no further effect, and the double mutant phenotype is less than would be expected under an additive model of genetic interaction. In addition, genes with related functions share similar patterns of genetic interactions and can be identified by cluster analysis.
In typical applications, such as synthetic lethal analysis, the double mutant array is analyzed using cell viability as the phenotypic readout. Here, we quantified the GSS surface distribution to analyze the ∼30,000 double mutants of our 374 × 81 array. Hierarchical clustering grouped genes according to their pattern of genetic interactions (Figs. 2 A and S1; see Materials and methods for details). Strikingly, many clusters were enriched for genes that act together (Fig. 2 B). Endocytosis genes were found in two distinct clusters, whereas other gene groupings corresponded to distinct processes. We observed that mutants deleted for two different endocytic genes (e.g., vrp1 and inp52) often had lower cell surface levels of GSS than would be expected under an additive model of genetic interaction, which is consistent with a function in the same pathway or complex. In contrast, mutants disrupted for both endocytosis (vrp1 and rvs167) and recycling (snx4 and cvt20/snx42) genes had cell surface GSS levels greater than expected under the additive null model. This is consistent with previous observations that endocytic defects are epistatic to defects in the Snx4-dependent recycling of Snc1 (Lewis et al., 2000).
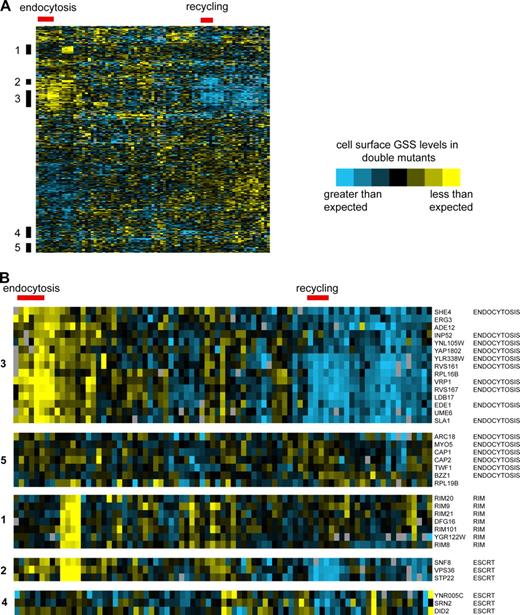
Endocytosis mutants have related genetic interaction profiles. (A) Hierarchical clustering was used to analyze the invertase activity values of double mutant strains generated by crossing top-scoring mutants from the primary screen (y axis) to 81 diverse trafficking mutants (x axis). Yellow indicates lower than expected levels of GSS at the cell surface, whereas blue indicates higher than expected levels (see Methods and materials for details). (B) Detailed view of clusters showing names of genes and highly represented pathways/complexes. Clusters are numbered to indicate their relative position on the heat map in A. Red bar indicates gene clusters on the y axis enriched in endocytosis or recycling genes. See Fig. S1 for a detailed view of gene names and cluster relationships.
Endocytosis mutants have related genetic interaction profiles. (A) Hierarchical clustering was used to analyze the invertase activity values of double mutant strains generated by crossing top-scoring mutants from the primary screen (y axis) to 81 diverse trafficking mutants (x axis). Yellow indicates lower than expected levels of GSS at the cell surface, whereas blue indicates higher than expected levels (see Methods and materials for details). (B) Detailed view of clusters showing names of genes and highly represented pathways/complexes. Clusters are numbered to indicate their relative position on the heat map in A. Red bar indicates gene clusters on the y axis enriched in endocytosis or recycling genes. See Fig. S1 for a detailed view of gene names and cluster relationships.
Integration of genetic and physical interaction data identifies complexes required for Snc1 transport
To comprehensively identify pathways that regulate Snc1 distribution, we integrated our genetic interaction results with genome-wide physical interaction datasets (see Materials and methods for details). In a network analysis, genes are represented as “nodes” (circles) linked by “edges” (lines) that indicate an experimental observation such as a physical interaction. To generate a network from the genetic interaction data, pairwise correlation scores were calculated for all genes in the array, and genes were linked by an edge if their correlation score was greater than a threshold value. The resulting network connects genes that share similar patterns of genetic interaction, and thus may represent components of pathways and complexes. Physical interaction networks were then created for the same set of genes, where edges represent interactions observed in high throughput yeast two-hybrid or mass spectrometry experiments. Highly connected sets of genes in each network were identified and integrated, and significant gene clusters were assigned to subcellular compartments based on the localization of their constituent proteins (Huh et al., 2003).
The resulting map (Fig. 3) provides a visual representation of the processes contributing to GSS localization and an indication of their relative importance. The size of each node is proportional to the phenotype of the corresponding deletion mutant, whereas edges represent either a physical interaction or a genetic correlation extracted from genome-wide data. Four major clusters, containing a total of 20 genes, mapped to the plasma membrane. The largest of these groupings was enriched for genes with known or predicted roles in endocytosis (SLA1, ABP1, INP52, EDE1, YAP1802, RVS161, RVS167, CRN1, and the INP52-overlapping open reading frame [ORF] YNL105W). Additional genes implicated in endocytosis or actin regulation were found in three other clusters at the plasma membrane. These findings highlight the importance of known endocytic regulators in Snc1 trafficking, and predict an endocytic role for uncharacterized genes in these clusters, such as LDB17.
Integration of genome-wide genetic and physical interaction data identifies pathways and complexes. Significant gene clusters contributing to GSS localization were mapped based on integrated physical and genetic interaction networks. Array genes are represented as nodes (where relative size is proportional to GSS surface levels in the corresponding deletion mutant), and are connected by edges that represent either a physical interaction or a genetic correlation extracted from genome-wide data. Clusters were assigned to subcellular compartments based on the localization of their constituent proteins.
Integration of genome-wide genetic and physical interaction data identifies pathways and complexes. Significant gene clusters contributing to GSS localization were mapped based on integrated physical and genetic interaction networks. Array genes are represented as nodes (where relative size is proportional to GSS surface levels in the corresponding deletion mutant), and are connected by edges that represent either a physical interaction or a genetic correlation extracted from genome-wide data. Clusters were assigned to subcellular compartments based on the localization of their constituent proteins.
Several well-characterized protein complexes that function at intracellular organelles or regulate transcription or translation were also identified by our integrative network analysis (Fig. 3). Some have established roles in actin assembly or protein transport, such as the cytosolic prefoldin–GimC complex, which acts as a cochaperone in actin folding (Siegers et al., 1999). ESCRT mutants are known to exhibit increased cell surface levels of receptors, which has been attributed to enhanced recycling from endosomal compartments (Davis et al., 1993). Chromatin remodeling complexes have also been implicated in regulating vesicle transport at endosomes (Schluter et al., 2008), although the mechanism remains unclear. Mutant strains lacking components of the Sin3/Rpd3 histone deacetylation complex, including Dep1 and Ume6, had particularly strong phenotypes. These proteins regulate the expression of phospholipid biosynthesis genes such as OPI3, INO1, and CHO2 (Elkhaimi et al., 2000; Lamping et al., 1994), which were among the top hits in our screen (Table S1). The regulation of plasma membrane lipid composition may also explain the identification of RIM pathway components, which mediate the cellular response to lipid asymmetry and other stimuli (Ikeda et al., 2008).
Clathrin and clathrin assembly proteins are required for Snc1 endocytosis
Our network analysis suggested that 20 genes, in four major clusters, are involved in Snc1 uptake at the plasma membrane. To confirm the results of the large-scale plate-based assay, cell-surface GSS levels in these 20 mutants were quantified by a liquid-based invertase activity assay (Fig. 4 A). Most had elevated GSS levels that correlated well with values derived from the large-scale densitometry data. The significant internalization defect of yap1802 mutants was surprising, as previous studies have not identified a clear role for AP180 homologues in yeast (Huang et al., 1999; Wendland et al., 1999). Yap1802 shares 43% sequence identity with Yap1801, which was also recovered by the genome-wide screen (Table S1). By microscopy, the polarized distribution of Snc1 at the bud was largely maintained in yap1801 or yap1802 single mutants, but Snc1 was evenly distributed at the cell surface of yap1801 yap1802 double mutants (Fig. 4 B). This strong defect in internalization is consistent with a functionally redundant role for the yeast AP180 proteins in Snc1 uptake.
Yeast AP180 homologues have a conserved role in Snc1 internalization. (A) Quantification of cell surface GSS levels in wild-type (WT) and candidate endocytic mutants by the liquid invertase assay. Invertase activity represents relative surface levels of the GSS reporter, quantified as nanomole glucose released per OD600 (mean of at least three experiments ± SD). The red line indicates the baseline invertase activity of the wild-type strain. (B) Localization of GFP-tagged Snc1 and NPF-Sso1 in WT and mutant strains. Bars, 2 µm. (C) Schematic of the GFP-Snc1-Suc2 (GSS) and 3xNPF-GFP-Sso1-Suc2 (NGSS) reporters. (D) Relative cell surface levels of GSS and NGSS reporters measured by liquid invertase assay in mutant strains (mean of three experiments ± SD). Results for each reporter are expressed as the percentage of invertase activity relative to WT. Different endocytosis defective controls (*EN-) were used for each reporter. GSS, endocytosis-defective GSS (snc1V40A M43A) reporter in WT cells; NGSS, NGSS in sla1 mutant strain.
Yeast AP180 homologues have a conserved role in Snc1 internalization. (A) Quantification of cell surface GSS levels in wild-type (WT) and candidate endocytic mutants by the liquid invertase assay. Invertase activity represents relative surface levels of the GSS reporter, quantified as nanomole glucose released per OD600 (mean of at least three experiments ± SD). The red line indicates the baseline invertase activity of the wild-type strain. (B) Localization of GFP-tagged Snc1 and NPF-Sso1 in WT and mutant strains. Bars, 2 µm. (C) Schematic of the GFP-Snc1-Suc2 (GSS) and 3xNPF-GFP-Sso1-Suc2 (NGSS) reporters. (D) Relative cell surface levels of GSS and NGSS reporters measured by liquid invertase assay in mutant strains (mean of three experiments ± SD). Results for each reporter are expressed as the percentage of invertase activity relative to WT. Different endocytosis defective controls (*EN-) were used for each reporter. GSS, endocytosis-defective GSS (snc1V40A M43A) reporter in WT cells; NGSS, NGSS in sla1 mutant strain.
In mammalian cells, the ubiquitous AP180 homologue CALM participates in clathrin-mediated endocytosis, and interacts with the AP-2 adaptor complex (Meyerholz et al., 2005). Deletion of clathrin heavy or light chain (CHC1 or CLC1), but not AP-2 (APL1), also caused a strong Snc1 localization defect (Fig. 4 B and not depicted). Surprisingly, no defect was seen when the major clathrin-binding site in Yap1802 (Wendland and Emr, 1998) was deleted in a strain lacking Yap1801 (Fig. S2 A), which suggests that Yap1802 contains additional clathrin-binding sites, or that additional proteins can bridge the interaction.
In contrast to Snc1 endocytosis, loss of Yap1801 and Yap1802 singly or in combination does not alter ligand-induced down-regulation of the α-factor–Ste2 complex (Huang et al., 1999). We found that loss of Yap1801/2 had little effect on the internalization of a distinct endocytic cargo, Ste6-GFP (Fig. S2 B). Defects may be more apparent for Snc1, which depends on continuous rounds of internalization and recycling to maintain its polarized distribution, compared with Ste6, which is delivered to the vacuole after internalization (Kelm et al., 2004). To determine if Yap1801/2 are generally required for the uptake of proteins subject to endocytic recycling, we appended the well-characterized Sla1-dependent NPFxD endocytosis signal to the t-SNARE GFP-Sso1 to create a different recycling reporter, NPF-Sso1 (Valdez-Taubas and Pelham, 2003). Like Snc1, NPF-Sso1 has a polarized distribution that depends on endocytosis. Consistent with previous results, we found the localization of NPF-Sso1 to be similar to that of Snc1 in wild-type cells, but completely depolarized in sla1 mutants (Fig. 4 B). Its polarized distribution was maintained in yap1801 yap1802 double mutants, which suggests that Yap1801/2 are specifically required for Snc1 internalization.
We created a chimeric version of the NPF-Sso1 reporter by fusing invertase to the NPF-GFP-Sso1 C terminus and adding two additional NPFxD motifs to further enhance internalization (Fig. 4 C), and then measured cell surface levels of the resulting NGSS (3xNPF-GFP-Sso1-Suc2) and GSS reporters (Fig. 4 D). Internalization of GSS was blocked by mutation of the Snc1 endocytic signal, whereas NPF-dependent internalization of NGSS was abolished in a sla1 mutant, as expected. Combined deletion of YAP1801 and YAP1802 dramatically increased levels of GSS at the cell surface, whereas NGSS surface levels were unaffected. Collectively, these results indicate that Yap1801 and Yap1802 act as cargo-specific adaptors for the clathrin-mediated endocytosis of Snc1.
Systematic analysis of cargo specificity
To determine if other genes identified in our genome-wide screen have cargo-specific functions, we reintroduced the GSS and NGSS reporters in parallel into our array of top-scoring deletion mutants. Invertase activity values for each mutant, normalized to the array median, were determined from six independent mutant arrays per reporter and converted into relative sorting scores. The ranked differences between NGSS and GSS sorting scores for each mutant indicated that deletion of YAP1802 and INP52 had the greatest specific effect on GSS (Fig. 5 A). By measuring cell surface reporter levels in liquid culture (Fig. 5 B), we confirmed that inp52 mutants (rank = 2) exhibited a highly significant degree of cargo specificity, whereas a lower-ranking mutant (ldb17) that fell below the 2-SD cutoff did not.
Cargo specificity of genes required for Snc1 endocytosis. (A) Array-based analysis of cargo specificity. The difference (DIFF) in cell surface levels of GSS and NGSS reporters was determined for each mutant by subtracting normalized invertase activity scores measured in parallel analyses of mutant arrays. Mutants with differences >2 SD (P < 0.05) from the mean are shown. Asterisks indicate loss of gene function by deletion of an overlapping ORF. (B) Relative cell surface levels of GSS and NGSS reporters measured by liquid assay in inp52 and ldb17 mutants. Results for each reporter are expressed as the percentage of invertase activity relative to appropriate wild-type (WT) control (mean of three experiments ± SD; P < 0.05). (C) Yap180 and Sla2 are present on internal puncta in inp52 strains. Wild type and inp52 mutants expressing Sla1-GFP, Sla2-GFP, or both Yap1801-GFP and Yap1802-GFP were examined by live cell microscopy. Arrows indicate internal patches. (D) Localization of GFP-tagged Snc1 and NPF-Sso1 in WT, ldb17, and end3 strains. Bars, 2 µm. (E) Ldb17 shares homology with the mammalian SPIN90 and contains a PRD. The conserved PTHR13357 domain is indicated (blue region).
Cargo specificity of genes required for Snc1 endocytosis. (A) Array-based analysis of cargo specificity. The difference (DIFF) in cell surface levels of GSS and NGSS reporters was determined for each mutant by subtracting normalized invertase activity scores measured in parallel analyses of mutant arrays. Mutants with differences >2 SD (P < 0.05) from the mean are shown. Asterisks indicate loss of gene function by deletion of an overlapping ORF. (B) Relative cell surface levels of GSS and NGSS reporters measured by liquid assay in inp52 and ldb17 mutants. Results for each reporter are expressed as the percentage of invertase activity relative to appropriate wild-type (WT) control (mean of three experiments ± SD; P < 0.05). (C) Yap180 and Sla2 are present on internal puncta in inp52 strains. Wild type and inp52 mutants expressing Sla1-GFP, Sla2-GFP, or both Yap1801-GFP and Yap1802-GFP were examined by live cell microscopy. Arrows indicate internal patches. (D) Localization of GFP-tagged Snc1 and NPF-Sso1 in WT, ldb17, and end3 strains. Bars, 2 µm. (E) Ldb17 shares homology with the mammalian SPIN90 and contains a PRD. The conserved PTHR13357 domain is indicated (blue region).
A cargo-specific role for the synaptojanin homologue Inp52, a PIP2 phosphatase required for recycling of coat proteins after vesicle budding is complete (Toret et al., 2008), was unexpected. Because the membrane association of Yap1801/2 is mediated by PIP2-binding AP180 N-terminal homology (ANTH) domains, we considered the possibility that elevated PIP2 levels in inp52 mutants lead to retention of Yap1801/2 on budded vesicles. Consistent with this hypothesis, Yap1801/2-GFP were observed on cytosolic patches in inp52 mutants, but not in wild-type cells (Fig. 5 C). Likewise, loss of Inp52 resulted in cytoplasmic aggregates of the ANTH domain protein Sla2-GFP, but had no effect on Sla1-GFP localization, as described previously (Fig. 5 C; Toret et al., 2008). Prolonged association of Yap1801/2 with budded vesicles would be expected to reduce levels available for GSS internalization, which may explain the apparent cargo specificity of the inp52 deletion.
Ldb17 is a new regulator of yeast endocytosis that is transiently recruited to cortical patches
Mutation of the uncharacterized gene LDB17 (Corbacho et al., 2005) caused a strong GSS localization defect (Fig. 3 and Fig. 4 A) that, in contrast to yap1801 yap1802 and inp52 mutants, was not strongly cargo-specific (Fig. 5 B). We confirmed that both Snc1 and NPF-Sso1 lost their polarized distributions in ldb17 mutants (Fig. 5 D), and a weak internalization defect was also observed for Ste6-GFP (Fig. S2 B), which suggests that Ldb17 is a general component of the endocytic machinery. Although an endocytic role for Ldb17 has not previously been described, it has a PTHR13357 domain also found in the mammalian protein SPIN90 (Fig. 5 E), which has been implicated in clathrin-mediated endocytosis in fibroblasts (Kim et al., 2006, 2007). Most organisms, including yeast and human, contain a single PTHR13357 family member, which indicates that Ldb17 and SPIN90 may have orthologous functions.
To help define the role of Ldb17 in endocytosis, we analyzed its recruitment to sites of internalization using two-color time-lapse fluorescence microscopy. Although difficult to visualize due to its low abundance (Ghaemmaghami et al., 2003), GFP-tagged Ldb17 was observed in faint puncta at the cell surface, where it showed partial overlap with mRFP-tagged forms of the late coat component Sla1, the type I myosin Myo5, and the F-actin–binding protein Abp1 (Fig. 6, A–C), which is consistent with transient colocalization. Kymograph analysis showed that Ldb17 joined preexisting Sla1 patches just before the onset of Sla1 inward movement and Myo5 recruitment (Fig. S3, A–C). Myo5 patches appeared ∼3 s after Ldb17 patches, whereas Ldb17 dissociated from the membrane ∼4 s before Abp1 disassembly. These results allow us to place Ldb17 on the temporal map of the endocytic process (Fig. 6 D); Ldb17 is maximally recruited after assembly of the late coat components, immediately before the recruitment of the myosin module, which precedes the actin-driven inward movement of the invaginating membrane.
Ldb17 is required for proper coat and actin dynamics. (A–C) Colocalization of Ldb17-GFP with Sla1-RFP, Myo5-RFP, and Abp1-RFP. Representative single frames from GFP and RFP channels are shown together with merged images. Arrows indicate cortical patches containing both Ldb17 and RFP-tagged proteins. (D) Schematic diagram, adapted from Kaksonen et al., 2005, illustrating the timing of Ldb17 recruitment to cortical patches relative to markers of the late coat (Sla1), WASP/myo (Myo5), and actin (Abp1) modules, based on kymograph analysis of 10 patches for each GFP/RFP pair (see Fig. S3, A–C). The mean difference in the timing of Ldb17 arrival or disappearance (±SD, in seconds) relative to other markers is indicated, together with the total mean lifetime of Ldb17, Myo5, and Abp1 patches. (E) Localization of Ldb17-GFP and Abp1-RFP in cells treated with DMSO (left), or 200 µM LatA (right) for 30 min at 30°C. (F) Lifetime of GFP-tagged Ldb17 and/or Ldb17ΔPRD at cortical patches in wild type (WT), bzz1, and sla1. Lifetimes were measured for at least 10 patches per strain (mean ± SD). (G) Reduced number and increased brightness of actin patches in ldb17 cells, as assessed by microscopy of Abp1-GFP. (H) Kymograph representation of time-lapse wide-field fluorescence microscopy showing extended lifetime of Sla1-RFP at endocytic patches in ldb17 mutants, as compared with wild-type cells over 120 s. Bars, 2 µm.
Ldb17 is required for proper coat and actin dynamics. (A–C) Colocalization of Ldb17-GFP with Sla1-RFP, Myo5-RFP, and Abp1-RFP. Representative single frames from GFP and RFP channels are shown together with merged images. Arrows indicate cortical patches containing both Ldb17 and RFP-tagged proteins. (D) Schematic diagram, adapted from Kaksonen et al., 2005, illustrating the timing of Ldb17 recruitment to cortical patches relative to markers of the late coat (Sla1), WASP/myo (Myo5), and actin (Abp1) modules, based on kymograph analysis of 10 patches for each GFP/RFP pair (see Fig. S3, A–C). The mean difference in the timing of Ldb17 arrival or disappearance (±SD, in seconds) relative to other markers is indicated, together with the total mean lifetime of Ldb17, Myo5, and Abp1 patches. (E) Localization of Ldb17-GFP and Abp1-RFP in cells treated with DMSO (left), or 200 µM LatA (right) for 30 min at 30°C. (F) Lifetime of GFP-tagged Ldb17 and/or Ldb17ΔPRD at cortical patches in wild type (WT), bzz1, and sla1. Lifetimes were measured for at least 10 patches per strain (mean ± SD). (G) Reduced number and increased brightness of actin patches in ldb17 cells, as assessed by microscopy of Abp1-GFP. (H) Kymograph representation of time-lapse wide-field fluorescence microscopy showing extended lifetime of Sla1-RFP at endocytic patches in ldb17 mutants, as compared with wild-type cells over 120 s. Bars, 2 µm.
The coat module assembles at endocytic sites before the onset of actin polymerization, and its components are recruited before the appearance of F-actin. Treatment with the actin monomer-sequestering drug latrunculin A (LatA) resulted in the expected complete delocalization of Abp1, whereas the membrane localization of Ldb17 was dramatically enhanced (Fig. 6 E). The endocytic coat components Sla1 and Sla2 show similar LatA-resistant cortical localization (Ayscough et al., 1997). This indicates that actin is not required for the membrane recruitment of Ldb17, and is consistent with a role for Ldb17 at late stages of coat assembly.
Sla1 contributes to membrane recruitment of Ldb17
The F-actin–independent localization of Ldb17 suggests that other factors regulate its recruitment to sites of endocytosis. We found that the C-terminal proline-rich domain (PRD) of Ldb17 was important for its localization at the cell periphery (Figs. 6 F and S3 D, left panels). Many endocytic proteins contain SH3 domains that recognize specific PRD sequences and contribute to the assembly of the endocytic network. We confirmed by coimmunoprecipitation that the yeast syndapin homologue Bzz1 binds Ldb17 in a PRD-dependent manner (Fig. S3 E), as predicted in a large-scale study (Tong et al., 2002). Surprisingly, membrane localization of Ldb17-GFP was enhanced in bzz1 mutants (Figs. 6 F and S3 D), and simultaneous loss of Bzz1 and the Ldb17 PRD had additive effects on cell surface GSS levels (Fig. S3 F), which indicates that Bzz1–PRD interactions are not involved in Ldb17 recruitment.
To identify additional PRD binding partners, we used yeast two-hybrid analysis to screen a panel of 16 SH3 domains from proteins implicated in endocytosis or actin regulation (Tong et al., 2002). SH3 domains from Sla1, Lsb3, and Lsb4 all exhibited PRD-dependent interactions with Ldb17 (Fig. S3, G and H). Because Lsb3 and Lsb4 are known binding partners of Sla1 (Dewar et al., 2002; Gavin et al., 2006), their interactions with Ldb17 may be indirect. Interestingly, loss of Sla1 had an effect similar to loss of the PRD; by live-cell microscopy, the lifetime of Ldb17 at the cell surface was reduced from 9 s to 5 s (Fig. 6 F). Collectively, these results suggest that Sla1 binds the Ldb17 PRD to enhance its membrane association at endocytic sites, whereas Bzz1 is important for its release.
Aberrant actin distribution in ldb17 mutants
Based on the timing of its recruitment, we hypothesized that Ldb17 may link coat formation to actin polymerization, as has been suggested for other components of the late coat module. Consistent with this, the actin module protein Abp1 was found in a smaller number of larger, brighter clusters in ldb17 cells compared with wild-type cells (Fig. 6 G), similar to the large actin clumps found in sla1 mutants (Kaksonen et al., 2005). Actin polymerization is required for internalization of the Sla1-containing late coat module; in cells treated with LatA or lacking the Arp2/3 activator Las17, Sla1 patches remain immobile at the cell surface (Ayscough et al., 1997; Sun et al., 2006). Sla1 was similarly stabilized at the membrane in the absence of Ldb17 (Fig. 6 H), which suggests that Ldb17 plays a role in the dynamic regulation of actin polymerization required for the internalization of Sla1-containing endocytic vesicles.
Discussion
We have developed a genome-wide screening method that provides a systematic and quantitative analysis of genes required for the endocytic recycling of Snc1, the yeast VAMP2 homologue. In addition to well-established endocytic regulators, this screen uncovered phenotypes for deletion mutants of predicted endocytosis genes for which no cargo uptake defects had previously been observed. The mammalian homologues of many of these genes also have endocytic roles, underscoring the conserved nature of endocytic processes in eukaryotic cells (Engqvist-Goldstein and Drubin, 2003; Kaksonen et al., 2005; Smythe and Ayscough, 2006).
Collectively, our results highlight the sensitive nature of our reporter assay and reinforce the importance of dynamic actin regulation for the endocytic process. Multiple regulators of the actin cytoskeleton were identified in our screen, including the actin-binding proteins Abp1 and Crn1 (coronin homologue), the actin capping complex (Cap1/2), and the Cap1/2-binding protein twinfilin (Twf1). Lack of sufficiently sensitive assays and study of limited cargo may have prevented detection of endocytic phenotypes for these mutants in previous studies (Kubler and Riezman, 1993). In addition, redundancy of the endocytic machinery may account for the weak but significant phenotypes resulting from loss of individual myosin-I (Myo5) and epsin (Ent1) isoforms (Wendland et al., 1999; Lechler et al., 2000; Maldonado-Báez and Wendland, 2006).
Role for clathrin adaptors in yeast endocytosis
The severe defect in Snc1 uptake caused by loss of Yap1801 and Yap1802, which are homologues of the mammalian clathrin assembly factor AP180/CALM, represents the first demonstration of a unique role for these proteins in yeast endocytosis. Studies in mammalian cells, worms, and flies all support a requirement for AP180 and its homologues in the internalization of VAMP/synaptobrevin. It mediates the selective uptake of this SNARE in worms, and appears to regulate the internalization of additional proteins in flies (Nonet et al., 1999; Bao et al., 2005).
AP180-like proteins may cooperate with other adaptors for the uptake of certain cargo. For example, a redundant role with the epsins in the internalization of the yeast pheromone receptor Ste3 was recently reported (Maldonado-Báez et al., 2008). AP180/CALM contains multiple binding sites for AP2, Eps15, and clathrin (Wendland and Emr, 1998; Maldonado-Báez and Wendland, 2006). A quantitative analysis of synaptobrevin localization in selected C. elegans mutants using the pH-sensitive synaptobrevin-GFP fusion protein synaptopHluorin demonstrated that its surface levels were enhanced in both AP180 and AP2 mutants (Dittman and Kaplan, 2006), which suggests that the interaction between AP180 and AP2 is functionally important in worms. In contrast, our results show that Yap1801/2 does not require AP2 to mediate Snc1 internalization in yeast, which is consistent with a recent study that saw little effect from mutating the AP2-binding site of CALM on synaptobrevin uptake in mammalian cells (Harel et al., 2008).
The specific requirement for Yap1801/2 in Snc1 endocytosis suggests they may act as adaptors to recruit Snc1 into a clathrin-coated pit. Internalization signals recognized by AP180-like monomeric adaptors have yet to be described (Maldonado-Báez and Wendland, 2006), and, like most SNAREs, Snc1 lacks canonical adaptor sorting motifs such as YxxΦ or D/ExxxL/E. Instead, SNARE proteins appear to use specialized sorting signals that differ from those of other cellular cargo (Chidambaram et al., 2004; Miller et al., 2007). The V40A M43A mutations that reduce Snc1 endocytosis could impair a Yap1801/2 recognition site; however, this remains to be determined, as we have not been able to demonstrate a direct physical interaction between Yap1801/2 and Snc1.
Identification of novel components of the yeast endocytic machinery
Our systematic analysis of cargo specificity did not identify additional adaptor proteins that were clearly specific for Snc1. Instead, we discovered that loss of the syntaptojanin homologue Inp52 affected uptake of the Snc1 reporter more strongly than a reporter that used a Sla1-dependent NPFxD signal. This can be explained by the differential recognition of PIP2 by Yap1801/2 and Sla1. High levels of PIP2 resulting from loss of Inp52 prevented efficient recycling of Yap1801/2, which is predicted to reduce its availability for further rounds of internalization. However, no such effect was seen for Sla1, which is consistent with previous studies (Toret et al., 2008).
In contrast to the cargo-specific role of Yap1801/2, we found that the previously uncharacterized gene LDB17 plays a broad role in regulating endocytosis. Ldb17 is transiently recruited to Sla1-containing cortical patches shortly before the onset of actin polymerization, and its membrane association is stabilized by interactions between its PRD and the Sla1 SH3 domain. The aberrant actin assembly and increased lifetime of Sla1 patches in ldb17 mutants suggest that Ldb17 helps trigger the dynamic actin rearrangements that precede coat disassembly.
Ldb17 may link coat assembly to the activation of the Arp2/3 complex through interactions with Sla1 and Arp2/3 activators, or act more directly. The C-terminal region of SPIN90 most similar to Ldb17 is sufficient to promote the WASP-independent activation of Arp2/3-mediated actin polymerization in vitro (Kim et al., 2007). The Arp2/3 and actin-binding motifs present in SPIN90 are only partially conserved in Ldb17, and we did not observe binding of Ldb17 to the Arp2/3 complex and the yeast WASP homologue Las17 in vivo (unpublished data). However, we identified an interaction between the Ldb17 PRD and the Bzz1 SH3 domain that mirrors the PRD-dependent binding of the Ldb17-related protein SPIN90 to the mammalian Bzz1 homologue syndapin (Kim et al., 2006). The conserved interaction of syndapin-like proteins with proline-rich regions in Ldb17 and SPIN90, despite the different placement of the interacting domains, suggests that these proteins may fulfill related roles. Further work will be needed to clarify the function of these conserved proteins in regulating actin polymerization and endocytosis.
Overall, our results suggest that endocytic processes are generally more similar in yeast and higher cells than previously believed, and underscore Snc1 transport in yeast as a model for synaptobrevin recycling at the synapse. Characterization of other genes identified in our screen may yield additional endocytosis factors. Because of the functional redundancy of the endocytic machinery in all eukaryotes, many regulatory proteins are likely to have weak mutant phenotypes that can only be discovered by more sensitive and quantitative approaches. Future application of our genome-wide analysis to different reporter proteins may identify alternative cargo-specific machinery and further define the complexity of endocytic systems.
Materials and methods
Construction of plasmids and yeast strains
Plasmids and yeast strains constructed for this study, and details of their construction, are listed in Table S2. Unless otherwise indicated, plasmids were constructed by homologous recombination after cotransforming yeast with linearized vector and DNA fragments bearing 50–55-bp homology, and recovering recombinant plasmids in Escherichia coli. The MATa yeast strain BY4741 and its gene deletion derivatives were obtained through Thermo Fisher Scientific. Unless otherwise stated, gene disruptions and tagging of genomic ORFs were performed by transformation of a PCR product containing a drug selection marker, flanked by 50–55 bp of homology to the region of interest. Additional strains and plasmids were obtained from C. Boone (University of Toronto, Toronto, Ontario, CAN), S. Fields (University of Washington, Seattle, WA), and S. Michaelis (The Johns Hopkins University, Baltimore, MD; Table S2).
Genome-wide screening
The GFP-Snc1-Suc2 (GSS) reporter was introduced into MATa and MATα knockout collections using the synthetic genetic array procedure (Tong and Boone, 2006). Query strains containing the GSS reporter and a gene deletion marked with NATR were mated to a 384-colony knockout array that contained 374 mutants identified in the genome-wide analysis (top 402 hits minus 28 mitochondrial mutants) and controls (wild type, blank, ent3, and rvs161). Double mutant haploid strains containing the GSS reporter were selected on –LEU –URA +G418 +clonNAT plates.
Manipulation and screening of the resulting colony arrays was performed essentially as described previously (Burston et al., 2008). Colony arrays on 384-array stock plates were pinned four times to YPF plates, creating 1,536 arrays using a Virtek automated colony arrayer (Bio-Rad Laboratories). Digital images of colony arrays subjected to the invertase overlay assay (Darsow et al., 2000) were acquired using a flat-bed scanner (2400; Epson). Local normalization was performed using National Institutes of Health ImageJ software (http://rsb.info.nih.gov/ij/) with the local normalization plugin (bigwww.epfl.ch/sage/soft/localnormalization) to correct for differences in intensity resulting from uneven distribution of reagents. Densitometry on corrected images used the spot-finding program GridGrinder (gridgrinder.sourceforge.net), and values were subjected to background subtraction and filtered to eliminate absent or very slow-growing strains. Densitometry values are inversely correlated with the cell surface levels of the reporter. Therefore, these values were subtracted from the median value for all strains to give a score where a high value represents a high level of the reporter. The invertase overlay assay was performed twice on each knockout collection; mean values were used to generate the final ranking.
Genetic interaction and network analysis
In an additive model of genetic interaction, the phenotype of the double mutant strain (PXY) can be expressed as: PXY = PWT + EX + EY + EGI, where PWT is the wild-type phenotype, EX is the effect of the first mutation, EY is the effect of the second mutation, and EGI is the genetic interaction effect. In a genetic interaction matrix, all double mutants in a given row or column have a mutation in the same “array” gene (row) or “query” gene (column). Assuming genetic interactions are rare, and positive and negative interactions are equally common, dividing double mutant phenotype values (PXY) in each column by the mean of that column removes the effect of the query mutation (Pwt + EY). Subsequently dividing values in each row by the row-wise mean removes the effect of the array mutation (EX), leaving only the genetic interaction effect, EGI.
We applied a modified version of this data preprocessing strategy to the matrix of raw double mutant invertase activity values. Rows with >20% missing observations were removed. Remaining values were divided by row- and column-wise medians, and values in each row and column were scaled so the sum of the squares of the values in each row or column was 1.0. Although these manipulations do not preserve the numerical value of EGI, they optimize clustering of functionally related groups by emphasizing patterns of genetic interactions. Cluster 3.0 software (http://bonsai.ims.u-tokyo.ac.jp/∼mdehoon/software/cluster/software.htm) was used to perform average linkage hierarchical clustering using an uncentered correlation similarity metric on both horizontal and vertical axes, and results were presented using Java Treeview (http://jtreeview.sourceforge.net/), where ordering of genes on both axes reflects the similarity of their genetic interaction profiles, and cluster relationships are indicated by trees. The resulting heat map (Fig. S1) does not provide a true quantitative measure of genetic interaction, but instead approximates the direction of the genetic interaction effect for each double mutant, where yellow is negative (less than expected under the additive null model) and blue is positive (greater than expected).
A network of correlated genetic interaction profiles was generated using the ARACNE algorithm (Margolin et al., 2006) with a threshold of 0.015. The MCODE plugin for Cytoscape (Bader and Hogue, 2003; Shannon et al., 2003) was used to identify densely connected gene clusters in synthetic genetic data and physical interaction networks from Saccharomyces Genome Database (www.yeastgenome.org) and WI-PHI (Kiemer et al., 2007), which were then integrated. The size of each node (gene) was mapped to the strength of the endocytic defect in Cytoscape based on the results from the genome-wide analysis in Table S1.
Liquid invertase assay
The liquid invertase assay was modified from Darsow et al., 2000. Strains expressing GSS or NGSS reporters were grown for 20 h in 2 ml yeast peptone (YP) fructose (OD600 ≈ 10). After dilution to 3 OD600/ml, 5 µl of each culture was mixed with 55 µl of 0.1 M NaOAc buffer, pH 4.9, in a 96-well microtiter plate. Subsequent steps were performed with a multilabel plate reader system (VICTOR3 1420; PerkinElmer). 13 µl of freshly prepared 0.5 M ultra pure sucrose was added to each well, and the plate was incubated for 5 min at 30°C. Next, 100 µl of glucostat reagent (0.1 M K2HPO4, pH 7.0, 347.1 U glucose oxidase, 2.6 ng/ml HRP, 102.6 nM N-ethyl-maleimide, and 0.15 mg/ml O-dianisidine) was added to each well, and the was plate incubated for 5 min at 30°C. Finally, 100 µl of 6 N HCl was added to each well to stop the reaction. The absorbance at 540 nm was used to determine glucose concentration by comparison to a glucose standard curve (which ranges from 5 to 50 nM glucose). Results are reported as nanomolar of glucose produced per 1 OD600 of culture.
Live cell imaging
Yeast cells expressing GFP-tagged proteins were grown to mid-log phase, then viewed directly in minimal selective media at room temperature using a Plan-Apochromat 100× 1.40 NA oil immersion objective lens on an Axioplan 2 fluorescence microscope (both from Carl Zeiss, Inc.). Images were captured with a CoolSNAP camera (Roper Scientific) using MetaMorph 6.1 software (MDS Analytical Technologies) and adjusted using Photoshop CS (Adobe). Alternatively, cells were grown to early log phase on rich medium plates at 30°C and placed in 200 µl of minimal media on concavalin A–coated 8-well Lab-Tek glass-bottom cover dishes (Thermo Fisher Scientific). Images were collected at room temperature with a 3i Marianas microscope (Intelligent Imaging Innovations) equipped with an α-Plan-Fluar 100× 1.46 NA objective lens and a total internal reflection fluorescence slider (both from Carl Zeiss, Inc.). Single-color wide-field GFP images were acquired using 488-nm laser excitation and a GFP dichroic and emission filter (Semrock). Two-color and RFP images were acquired with 488 nm and/or 561 nm laser excitation, with GFP and RFP emission split between two Cascade II 512 cameras with an Optical Insights Dual Cam (Photometrics) equipped with the D520/30m D630/50m splitting optics (MAG Biosystem; Photometrics).
Exposure times varied from 600–750 ms as follows: Ldb17-GFP single experiments, 750 ms; two-color Ldb17/Myo5, 750 ms; Ldb17/Sla1, 750 ms; Lbd17/Abp1, 750 ms for Ldb17 and 650 ms for Abp1. TetraSpeck 100-nm beads (Invitrogen) were used to align the two channels and for subsequent registration in software. Slide-Book 4.2 software (Intelligent Imaging Innovations) was used for image acquisition and dual-channel image registration. Kymographs and montages were created using ImageJ software with kymograph plugins (http://www.embl.de/eamnet/html/kymograph.html).
Live-cell imaging experiments with LatA treatment were performed by incubating cells from early log phase cultures in a final concentration of 200 µM LatA dissolved in DMSO for 30 min at 30°C.
Coimmunoprecipitation
30 OD600 cell pellets made from flash-frozen log phase cells were lysed by vortexing with glass beads in 200 µl of lysis buffer (100 mM NaCl, pH 8.0, 2 mM EDTA, and 0.5% Tween-20 + PMSF) and incubated with mouse α-HA (1:1,000; Applied Biological Materials, Inc.) and 30 µl of protein G–Sepharose (75% slurry in PBS). Coprecipitating proteins were identified by Western blotting with mouse α-GFP (Roche) and HRP-labeled secondary antibodies (Bio-Rad Laboratories). Blots were developed with ECL (Thermo Fisher Scientific), and luminescent images were captured with a Fluor S Max Multi-imager (Bio-Rad Laboratories).
Online supplemental material
Fig. S1 shows full hierarchical clustering of the genetic interaction data. Fig. S2 shows the cargo sorting defects of yap1801 yap1802 mutants. Fig. S3 shows characterization of ldb17 mutants and identification of Ldb17-interacting proteins. Table S1 is a full list of mutants with cell-surface GSS levels greater than or equal to the median value for all strains. Table S2 lists plasmids and yeast strains used in this study.
Acknowledgments
We are grateful to Charlie Boone, Stan Fields, and Susan Michaelis for providing strains and reagents; David Drubin for sharing unpublished data; and Nick Davis and Mara Duncan for critical reading of the manuscript. Chris Fung and Jeffrey Nguyen provided valuable help with genome-wide screens, while Jochen Brumm and Jenny Bryan assisted with data analysis. We thank Michael McCaffery, Ned Perkins, and Michelle Husain of the Integrated Imaging Center at The Johns Hopkins University for help and advice with light microscopy.
This work was supported by Canadian Institutes of Health Research (CIHR) grant FRN No. 64394 (to E. Conibear) and National Institutes of Health (NIH) grants R01 GM-60979 (to B. Wendland) and NIH-NCRR 1S10RR022588-01 (IIC shared instrumentation grant to B. Wendland, who was the co-principle investigator). Additional support was provided by Michael Smith Foundation for Health Research Scholar and CIHR New Investigator Awards (to E. Conibear), Michael Smith Foundation for Health Research doctoral awards (to H.E. Burston and B. Montpetit), Natural Sciences and Engineering Research Council of Canada Postgraduate Scholarship and Canada Graduate Scholarship awards (to H.E. Burston), and a Ford Foundation Diversity Fellowship (to L. Maldonado-Báez).
References
Abbreviations used in this paper: CALM, clathrin assembly lymphoid myeloid leukemia; LatA, latrunculin A; Ldb17, low dye binding 17; ORF, open reading frame; PRD, proline-rich domain; VAMP, vesicle-associated membrane protein; WASP, Wiscott-Aldrich syndrome protein.