TopBP1 and the Rad9–Rad1–Hus1 (9-1-1) complex activate the ataxia telangiectasia mutated and Rad3-related (ATR) protein kinase at stalled replication forks. ATR is recruited to stalled forks through its binding partner, ATR-interacting protein (ATRIP); however, it is unclear how TopBP1 and 9-1-1 are recruited so that they may join ATR–ATRIP and initiate signaling. In this study, we use Xenopus laevis egg extracts to determine the requirements for 9-1-1 loading. We show that TopBP1 is required for the recruitment of both 9-1-1 and DNA polymerase (pol)-α to sites of replication stress. Furthermore, we show that pol-α is also directly required for Rad9 loading. Our study identifies an assembly pathway, which is controlled by TopBP1 and includes pol-α, that mediates the loading of the 9-1-1 complex onto stalled replication forks. These findings clarify early events in the assembly of checkpoint signaling complexes on DNA and identify TopBP1 as a critical sensor of replication stress.
Introduction
Replication stress arises when cellular or template conditions delay replication fork progression during the synthesis (S) phase of the cell cycle. To counter replication stress, eukaryotic cells have evolved a sophisticated replication checkpoint response system that slows cell cycle progression and stabilizes stalled replication forks so that further DNA damage is prevented and so that mitosis is coupled to the completion of S phase. At the heart of this checkpoint are two protein kinases, ataxia telangiectasia mutated and Rad3 related (ATR) and Chk1 (Chen and Sanchez, 2004; Zhang et al., 2006; Paulsen and Cimprich, 2007; Burrows and Elledge, 2008; Cimprich and Cortez, 2008). Stalled replication forks activate ATR, and this allows it to directly phosphorylate and activate Chk1. Because of its central role in organizing the replication stress response, the biochemical mechanism governing ATR activation by stalled replication forks has been an active area of investigation, and important details have begun to emerge.
When a DNA polymerase (pol) stalls, the minichromosome maintenance helicase continues to unwind DNA ahead of the stalled pol (Byun et al., 2005). This produces long tracts of single-stranded DNA (ssDNA), which become coated with replication protein A (RPA). RPA-coated ssDNA is necessary for ATR activation, but it is not sufficient, as at least three additional factors are also required. The first is ATR-interacting protein (ATRIP), an ATR binding partner that interacts directly with RPA to dock the ATR–ATRIP complex onto ssDNA (Cortez et al., 2001; Zou and Elledge, 2003; Unsal-Kacmaz and Sancar, 2004; Ball et al., 2005, 2007; Namiki and Zou, 2006; Burrows and Elledge, 2008). The second is the 9-1-1 complex, a heterotrimeric, ringlike protein composed of the Rad9, Rad1, and Hus1 subunits. 9-1-1 is loaded onto DNA by a clamp-loader protein consisting of Rad17 and four subunits of the replication factor C (RFC) complex (Parrilla-Castellar et al., 2004). The third is TopBP1, an ∼180-kD protein that contains eight copies of the BRCA1 C terminus (BRCT) repeat (Garcia et al., 2005). Located between the BRCT 6 and 7 motifs of TopBP1 is a recently defined ATR activation domain. TopBP1 uses this domain to interact with DNA-bound ATR–ATRIP, and this interaction stimulates ATR kinase activity (Kumagai et al., 2006; Mordes et al., 2008). However, in order for TopBP1 to interact with ATR–ATRIP, it must also interact with the Rad9 component of the 9-1-1 complex (Furuya et al., 2004; Delacroix et al., 2007; Lee et al., 2007). This interaction occurs between the BRCT 1 and 2 domains of TopBP1, and a constitutively phosphorylated Ser residue of Rad9 located in its C terminus (Ser373 in Xenopus laevis) is thought to allow a conformational change in TopBP1 that allows binding to ATR–ATRIP.
The aforementioned results suggest that a core checkpoint complex composed of ATR–ATRIP, TopBP1, and 9-1-1 assembles on RPA-coated ssDNA and that this could be sufficient for ATR activation. If so, the manner in which this complex assembles on ssDNA is of particular interest if we are to understand how ATR is activated. The mechanism for ATR–ATRIP recruitment to ssDNA has been solved; the complex associates with RPA-coated ssDNA by virtue of direct contact between ATRIP and RPA. 9-1-1 is recruited to ssDNA independently of ATR–ATRIP (Kondo et al., 2001; Melo et al., 2001; You et al., 2002; Zou et al., 2002; Lee et al., 2003; Kanoh et al., 2006), but the mechanism that governs 9-1-1 loading is still unclear. In reconstituted systems, Rad17-RFC preferentially loads 9-1-1 onto 5′-DNA junctions in a manner that is stimulated by RPA (Bermudez et al., 2003; Ellison and Stillman, 2003; Zou et al., 2003; Majka et al., 2006). This suggests that the 9-1-1 binding site in vivo contains a 5′-DNA junction; however, how such a structure forms at stalled replication forks is not known.
It is also unclear how TopBP1 is recruited to the checkpoint complex. TopBP1 could join the complex after 9-1-1 has been loaded by virtue of the aforementioned interaction between it and Rad9. However, two previously reported observations suggest that a model whereby Rad9 recruits TopBP1 to stalled forks may be incorrect. First, depletion of the 9-1-1 complex from Xenopus egg extract does not noticeably alter the ability of TopBP1 to associate with replication-stressed chromatin (Lupardus and Cimprich, 2006), as would not be expected if Rad9 recruits TopBP1 to the stalled fork. Second, depletion of TopBP1 prevents the loading of Rad1 onto etoposide-treated chromatin (Parrilla-Castellar and Karnitz, 2003). This result could reflect a role for TopBP1 in organizing the repair of etoposide-induced lesions, or, alternatively, it could indicate that TopBP1 controls the loading of 9-1-1 onto replication forks that stall at etoposide-induced lesions. Given the ambiguity over which factor, TopBP1 or 9-1-1, is responsible for recruiting the other to stalled forks, it is clear that further work is necessary to clarify how these factors arrive at sites of replication stress. This represented one goal of this study.
To date, the known protein requirements for ATR activation include RPA, ATRIP, TopBP1, and the 9-1-1 complex. Work in the Xenopus system has also described the DNA structural requirements for activation of ATR (MacDougall et al., 2007). In these experiments, it was shown that the 5′ end of a primer–template junction is critical for efficient signaling, as blocking the 5′ end of such a structure with biotin-streptavidin abrogated signaling. Thus, this study is consistent with the hypothesis that the 9-1-1 binding site in vivo corresponds to the 5′ end of a primer–template junction. How might such a structure be generated at stalled replication forks? One possibility is that primers synthesized by DNA pol-α are accessed by Rad17-RFC to load the 9-1-1 complex. Pol-α is the only DNA pol that can synthesize DNA on ssDNA templates in the absence of a primed site, and, thus, this pol is a good candidate to generate 5′-DNA junctions at stalled replication forks. Like RPA, pol-α accumulates, or hyperloads, onto chromatin during a replication stress response (Michael et al., 2000; Mimura et al., 2000; Lupardus et al., 2002; Lee et al., 2003). Also linking pol-α to checkpoint signaling are experiments showing that immunodepletion of pol-α from Xenopus egg extracts prevents ATR activation (Michael et al., 2000) and the loading of 9-1-1 onto chromatin (You et al., 2002). However, in these experiments, pol-α was removed from the extract before the addition of DNA templates. Checkpoint activation requires replication forks (Lupardus et al., 2002; Stokes et al., 2002), and because replication fork assembly requires pol-α, it is not yet possible to assign a direct role for pol-α in checkpoint signaling based on these previous studies. Therefore, a second goal of the work in this study was to gain a clearer understanding of the role of pol-α in 9-1-1 loading and ATR activation.
To accomplish our goals, we used Xenopus egg extracts to dissect the assembly of the checkpoint signaling complex, and we focused on two questions. First, which factor, TopBP1 or 9-1-1, initially senses replication stress to recruit the other? Second, does pol-α play a direct role in ATR activation? We report that TopBP1 resides at the top of an ordered assembly pathway that results in the loading of 9-1-1 onto stalled replication forks. TopBP1 senses replication stress, and, in response, it promotes the accumulation of pol-α onto stalled replication forks. Pol-α, in turn, plays a direct role in the recruitment of 9-1-1 to the stalled fork. These results identify TopBP1 as a key sensor of replication stress and provide an explanation as to how 5′-DNA junctions accumulate on stalled forks to allow the loading of 9-1-1 onto the DNA.
Results
TopBP1 controls the loading of 9-1-1 and the hyperloading of pol-α onto stalled replication forks
To study early events in checkpoint signaling, we used Xenopus egg extracts to examine the requirements for the recruitment of TopBP1, Rad9, and pol-α to stalled replication forks. Previous work has shown that ATR–ATRIP is recruited independently of the 9-1-1 complex to stalled forks (Kondo et al., 2001; Melo et al., 2001; You et al. 2002; Zou et al., 2002; Lee et al., 2003; Kanoh et al., 2006). Therefore, we did not examine ATR–ATRIP in our experiments, as our focus was on the 9-1-1 branch of the checkpoint complex assembly pathway. Both TopBP1 and pol-α are required for replication fork assembly (Hubscher et al., 2002; Van Hatten et al., 2002; Hashimoto and Takisawa, 2003), and this demanded an experimental system that would allow bypass of these requirements in replication initiation so that their roles in checkpoint activation could be studied. To accomplish this, we developed a chromatin transfer system whereby sperm chromatin was incubated in egg extract for 30 min to allow initiation and then isolated and transferred to a second extract, where checkpoint activation would then be analyzed (Fig. S1 A). This general approach has been used extensively in the past for studies of DNA replication and checkpoint control (Rowles et al., 1999; Van Hatten et al., 2002; Hashimoto and Takisawa, 2003; Parrilla-Castellar and Karnitz, 2003; Luciani et al., 2004; Trenz et al., 2006; Errico et al., 2007).
Our experimental design is diagrammed in Fig. S1 A. Sperm chromatin is incubated in egg extract for 30 min to allow replication to initiate. The chromatin is then isolated with a high-salt buffer and nuclear isolation buffer containing 250 mM KCl (NIB-250) and transferred to a second extract. A concentration of 250 mM KCl was used to ensure the removal of both TopBP1 and pol-α from the chromatin. To examine protein association with the chromatin in this system, we used a gentle isolation condition involving the low-salt egg lysis buffer (ELB). We characterized the behavior of TopBP1, pol-α, and other replisome components in this system (Fig. S1 B). Sperm chromatin was incubated in egg extract for 30 min and then isolated using either ELB or NIB-250. The ELB chromatin contained TopBP1, pol-α, -δ, and -ε, and proliferating cell nuclear antigen (PCNA; Fig. S1 B, lane 2). In contrast, the NIB-250 chromatin contained pol-δ and -ε as well as PCNA but not TopBP1 or pol-α (Fig. S1 B, lane 3). NIB-250 chromatin was then transferred to a second extract, and, after incubation, the chromatin was isolated again using ELB. This revealed that TopBP1 and pol-α could reassociate with the NIB-250 chromatin after incubation in the second extract (Fig. S1 B, lane 4). Furthermore, aphidicolin-sensitive replication of the NIB-250 chromatin was observed in the second extract (Fig. S1 C). Replication of NIB-250 chromatin was not attenuated by the initiation inhibitor geminin, although geminin blocked the replication of fresh sperm chromatin (Fig. S1 C). Similar results were obtained with the Cdk inhibitor p27 (unpublished data), which shows that replication of NIB-250 chromatin in the second extract is by virtue of the extension of previously initiated replicons and does not require new initiation events.
Next, we examined checkpoint complex assembly and ATR activation in extracts containing NIB-250 chromatin and aphidicolin and observed that aphidicolin induced the hyperloading of pol-α and the recruitment of the Rad9 component of 9-1-1 to the chromatin (Fig. S1 D, lanes 1 and 2). Binding of Orc2 was also examined as a loading control in this and subsequent experiments. To examine ATR activation, we used an antibody that recognizes the Ser344-phosphorylated form of the ATR substrate Chk1 (Guo et al., 2000) and observed that aphidicolin selectively induced ATR activation in extracts containing NIB-250 chromatin. The inclusion of geminin did not suppress pol-α or Rad9 recruitment to stalled replication forks, and it did not inhibit ATR activation (Fig. S1 D, lanes 3 and 4). This shows that, like DNA synthesis, checkpoint activation in extracts containing NIB-250 chromatin occurs independently of new initiation events. The data in Fig. S1 show that the following events occur in our NIB-250 chromatin system. During incubation in the first extract, TopBP1 and pol-α perform their initiation functions, and replication initiates. Isolation of the chromatin removes all detectable TopBP1 and pol-α from the DNA. When NIB-250 chromatin is incubated in a second extract, TopBP1 and pol-α reassociate with the DNA, and DNA replication resumes. If the second extract contains aphidicolin, replication stalls, and the ATR checkpoint is activated.
We used the NIB-250 chromatin system to study the role of TopBP1 in assembly of the checkpoint signaling complex (Fig. 1 A). As shown in Fig. 1 B, in mock-depleted extract, we observed aphidicolin-dependent loading of Rad9 and Rad17, hyperloading of pol-α onto the chromatin, and Chk1 phosphorylation (lane 2). In contrast, in TopBP1-depleted extract, neither Rad9 nor Rad17 accumulated on chromatin in the aphidicolin-treated sample, and pol-α did not hyperload (Fig. 1 B, lane 4). As expected, Chk1 was not phosphorylated in the TopBP1-depleted sample (Fig. 1 B, lane 4). The Rad1 subunit of the 9-1-1 complex also accumulated on chromatin in the control sample that received aphidicolin but not in the TopBP1-depleted extract (Fig. S2). These results suggest that TopBP1 controls the loading of the 9-1-1 complex onto DNA during a replication stress response.
Requirements for the recruitment of TopBP1, pol-α, and 9-1-1 to stalled replication forks. (A) Experimental scheme for B–E. (B) NIB-250 chromatin was incubated in either mock- or TopBP1 (TopBP1−)-depleted extract that optionally contained aphidicolin (+Aph.). After a 45-min incubation, the chromatin-bound fraction was isolated using the ELB procedure. Checkpoint proteins on chromatin and Chk1 phosphorylation in total extract were further examined via Western blotting. (C) NIB-250 chromatin was added to TopBP1-depleted extract supplemented with either blank IVT (no add back) or recombinant IVT TopBP1 (+IVT TopBP1) and, optionally, aphidicolin. Checkpoint proteins on chromatin and Chk1 phosphorylation in extract were examined as in B. (D) Same as B except that pol-α, instead of TopBP1, was depleted in the second egg extract. (E) Either buffer (no add back) or 35 ng/µl purified DNA pol-α (+purified pol-α) was added to pol-α–depleted extract together with NIB-250 chromatin and, optionally, aphidicolin. Rad9 loading onto chromatin and Chk1 phosphorylation in extract were assessed as in B. (F) Experimental scheme for G. (G) Sperm chromatin was combined with mock- or Rad9 (Rad9−)-depleted extract and, where indicated, aphidicolin. Chromatin fractions and total extracts were analyzed as in B.
Requirements for the recruitment of TopBP1, pol-α, and 9-1-1 to stalled replication forks. (A) Experimental scheme for B–E. (B) NIB-250 chromatin was incubated in either mock- or TopBP1 (TopBP1−)-depleted extract that optionally contained aphidicolin (+Aph.). After a 45-min incubation, the chromatin-bound fraction was isolated using the ELB procedure. Checkpoint proteins on chromatin and Chk1 phosphorylation in total extract were further examined via Western blotting. (C) NIB-250 chromatin was added to TopBP1-depleted extract supplemented with either blank IVT (no add back) or recombinant IVT TopBP1 (+IVT TopBP1) and, optionally, aphidicolin. Checkpoint proteins on chromatin and Chk1 phosphorylation in extract were examined as in B. (D) Same as B except that pol-α, instead of TopBP1, was depleted in the second egg extract. (E) Either buffer (no add back) or 35 ng/µl purified DNA pol-α (+purified pol-α) was added to pol-α–depleted extract together with NIB-250 chromatin and, optionally, aphidicolin. Rad9 loading onto chromatin and Chk1 phosphorylation in extract were assessed as in B. (F) Experimental scheme for G. (G) Sperm chromatin was combined with mock- or Rad9 (Rad9−)-depleted extract and, where indicated, aphidicolin. Chromatin fractions and total extracts were analyzed as in B.
To confirm that the failure of 9-1-1 to load in TopBP1-depleted extract was caused by the removal of TopBP1, the experiment was repeated in TopBP1-depleted extract that had been supplemented with recombinant TopBP1. Recombinant TopBP1 was produced by in vitro transcription/translation (IVT) in rabbit reticulocyte lysates, and these lysates were mixed with TopBP1-depleted egg extract at a ratio of 15% (vol/vol). Under these conditions, the amount of IVT-produced TopBP1 present in the reconstituted extract is equivalent to that which is normally present in egg extract (Fig. S3 A, extract panel). Equivalent amounts of TopBP1 were also found in the chromatin-containing fractions from these two extracts (Fig. S3 A, chromatin panel). Furthermore, the IVT-produced TopBP1 restores the capacity for both checkpoint activation and DNA replication to TopBP1-depleted extract (Fig. S3, A and B; Van Hatten et al., 2002; Yan et al., 2006). As shown in Fig. 1 C, inclusion of IVT TopBP1 rescued the loading of Rad9, the hyperloading of pol-α, and the phosphorylation of Chk1 in the TopBP1-depleted extract. These results show that TopBP1 is required for the loading of both Rad17 and 9-1-1 and the hyperloading of pol-α onto replication-stressed chromatin (Fig. 1, B and C). Previous work has clearly shown that TopBP1 is not required for the elongation phase of DNA replication in egg extracts (Van Hatten et al., 2002; Hashimoto and Takisawa, 2003; Parrilla-Castellar and Karnitz, 2003). Therefore, these previous studies alleviated concerns that the effects of TopBP1 on pol-α and 9-1-1 recruitments that we observed are an indirect result of unstable replication forks (Fig. 1, B and C). We conclude that TopBP1 is directly required for the recruitment of pol-α and 9-1-1 to stalled replication forks. This conclusion is in agreement with a previous study, which reported that TopBP1 is required for the recruitment of Rad1 to etoposide-treated chromatin in egg extracts (Parrilla-Castellar and Karnitz, 2003). However, our results extend this previous study because we show that TopBP1-mediated recruitment of 9-1-1 is a general feature of the replication stress response and therefore is not specific to the repair of etoposide-induced DNA damage.
Pol-α plays a direct role in the recruitment of 9-1-1 to stalled replication forks
Next, we examined the requirement for pol-α in checkpoint activation using the NIB-250 chromatin system. Pol-α was immunodepleted from egg extract, and this extract was then combined with NIB-250 chromatin (Fig. 1 A). Control experiments showed that replisome components associated with chromatin (Fig. S4 A) and substantial DNA replication occurred (Fig. S4 B, top) when NIB-250 chromatin was incubated in pol-α–depleted extract. In contrast to NIB-250 chromatin, the replication of fresh sperm chromatin was severely attenuated in pol-α–depleted extract (Fig. S4 B, bottom), which shows that pol-α was efficiently depleted from the extract. These experiments showed that replication forks are active in pol-α–depleted extracts containing NIB-250. However, despite the presence of replication forks, aphidicolin failed to trigger Rad9 loading onto chromatin and Chk1 phosphorylation in the pol-α–depleted extract (Fig. 1 D, lanes 3 and 4). The association of TopBP1 with chromatin was not altered by depletion of pol-α. When purified pol-α (obtained from calf thymus) was added back to pol-α–depleted extract containing aphidicolin, Rad9 loading and Chk1 activation were restored (Fig. 1 E). These data show that pol-α controls the recruitment of 9-1-1 but not TopBP1 to stalled replication forks and plays a direct role in checkpoint activation.
Having established roles for TopBP1 and pol-α in the recruitment of 9-1-1 to stalled forks, we next asked whether Rad9 was important for the recruitment of either TopBP1 or pol-α. For this experiment, either mock- or Rad9-depleted extracts were supplemented with sperm chromatin and, optionally, aphidicolin. After incubation, the chromatin was isolated using ELB, and factor association with the chromatin was determined by Western blotting (Fig. 1 F). We did not use NIB-250 chromatin for this experiment because it has previously been shown that 9-1-1 is not required for DNA replication in Xenopus (Jones et al., 2003). As shown in Fig. 1 G, TopBP1 association with chromatin was not affected by Rad9 depletion and neither was pol-α hyperloading in the sample that received aphidicolin (Fig. 1 G, lane 4). No detectable Rad9 was found on chromatin in the Rad9-depleted sample, and aphidicolin failed to trigger Chk1 phosphorylation, which confirms the fact that Rad9 was functionally depleted from the extract. This result is consistent with a previous study showing that depletion of Rad1 from egg extract did not perturb chromatin association of TopBP1 (Lupardus and Cimprich, 2006); however, pol-α hyperloading was not examined in this previous study. Based on the data in Fig. 1, we conclude that although both TopBP1 and pol-α are required for Rad9 recruitment to stalled replication forks, Rad9 plays no role in the recruitment of either TopBP1 or pol-α.
Recruitment of pol-α and 9-1-1 to stalled replication forks is a novel function of TopBP1
Previous work in Xenopus has shown that TopBP1 is required for the initiation of DNA replication and, independently, to activate ATR at stalled replication forks (Van Hatten et al., 2002; Hashimoto and Takisawa, 2003; Kumagai et al., 2006). The results presented in this study suggest that TopBP1 has an additional, novel function in the replication checkpoint to recruit pol-α and 9-1-1 to stalled replication forks. To determine whether pol-α and 9-1-1 recruitment does indeed represent a distinct TopBP1 function, we asked whether this activity could be mutationally separated from the previously described functions in replication initiation and ATR activation (Van Hatten et al., 2002; Hashimoto and Takisawa, 2003; Kumagai et al., 2006). We used a previously described TopBP1 deletion mutant corresponding to amino acids 1–759 and termed “Mini” (Fig. 2 A; Yan et al., 2006) to explore the relationship between TopBP1 function in replication initiation and in assembling checkpoint complexes on stalled replication forks. Previous work has shown that Mini restores DNA replication to a TopBP1-depleted extract and thus that all of the replication initiation functions are encoded within this fragment (Yan et al., 2006). Therefore, we asked whether Mini could also restore pol-α hyperloading and 9-1-1 loading to a TopBP1-depleted extract. Wild-type (WT) TopBP1 promoted the loading of pol-α and Rad9 onto the chromatin, and it activated ATR (Fig. 2 B, lane 1). In contrast, neither pol-α nor Rad9 accumulated on chromatin in sample containing Mini (Fig. 2 B, lane 2). This shows that although Mini can promote replication initiation, it cannot recruit pol-α or 9-1-1 to stalled replication forks. We conclude that the replication initiation and checkpoint complex assembly functions of TopBP1 are mechanistically distinct.
Recruitment of pol-α and 9-1-1 to stalled replication forks is a novel function of TopBP1. (A) Summary of WT and mutant TopBP1s' functions in replication initiation, pol-α hyperloading, Rad9 loading, and Chk1 phospho-Ser344. A.D., activation domain; a, Yan et al., 2006; b, Kumagai et al., 2006. (B) IVT versions of TopBP1s were added back to TopBP1-depleled extract along with NIB-250 chromatin and aphidicolin. After incubation, the samples were processed for Western blotting as in Fig. 1 C. Additional Western blotting, using an antibody that recognizes the myc tag fused to all recombinant TopBP1s, confirmed that these recombinant forms of TopBP1s were added to extract in equivalent amounts (myc-TopBP1). (C) Blank IVT (−) or recombinant IVT TopBP1s (WT or W1138R) were added to TopBP1-depleted egg extract supplemented with sperm chromatin and aphidicolin. After incubation, the chromatin fractions and total extract were probed via immunoblotting. (D) Same as B except that WT or M5 TopBP1 was added back to TopBP1-depleled extract. (E) Blank IVT (−), WT TopBP1 (WT), or M3 TopBP1 (M3) was added back to TopBP1-depleted egg extract containing AT(70). The AT(70) DNA substrate used for this experiment was prepared as previously described (see Materials and methods; Yan et al., 2006). After incubation, Chk1 phosphorylation and myc-TopBP1 in total extract were determined via Western blotting.
Recruitment of pol-α and 9-1-1 to stalled replication forks is a novel function of TopBP1. (A) Summary of WT and mutant TopBP1s' functions in replication initiation, pol-α hyperloading, Rad9 loading, and Chk1 phospho-Ser344. A.D., activation domain; a, Yan et al., 2006; b, Kumagai et al., 2006. (B) IVT versions of TopBP1s were added back to TopBP1-depleled extract along with NIB-250 chromatin and aphidicolin. After incubation, the samples were processed for Western blotting as in Fig. 1 C. Additional Western blotting, using an antibody that recognizes the myc tag fused to all recombinant TopBP1s, confirmed that these recombinant forms of TopBP1s were added to extract in equivalent amounts (myc-TopBP1). (C) Blank IVT (−) or recombinant IVT TopBP1s (WT or W1138R) were added to TopBP1-depleted egg extract supplemented with sperm chromatin and aphidicolin. After incubation, the chromatin fractions and total extract were probed via immunoblotting. (D) Same as B except that WT or M5 TopBP1 was added back to TopBP1-depleled extract. (E) Blank IVT (−), WT TopBP1 (WT), or M3 TopBP1 (M3) was added back to TopBP1-depleted egg extract containing AT(70). The AT(70) DNA substrate used for this experiment was prepared as previously described (see Materials and methods; Yan et al., 2006). After incubation, Chk1 phosphorylation and myc-TopBP1 in total extract were determined via Western blotting.
Next, we examined the relationship between checkpoint complex assembly and ATR activation for TopBP1. For this, we used a previously described point mutant, W1138R, which cannot stimulate ATR kinase activity (Fig. 2 A; Kumagai et al., 2006). We observed that there was no difference between WT and W1138R TopBP1 in the recruitment of pol-α and Rad9 to chromatin (Fig. 2 C). However, as expected, WT TopBP1 but not W1138R promoted Chk1 phosphorylation in this experiment. This result shows that TopBP1 W1138R can still recruit pol-α and 9-1-1 to stalled forks despite its inability to activate ATR. We conclude that pol-α and 9-1-1 recruitment and ATR activation are mechanistically distinct functions of TopBP1.
To learn more about the TopBP1 sequence determinants that allow it to recruit pol-α and 9-1-1 to stalled replication forks, we performed deletion analysis to determine which N- and C-terminal sequences are required for these functions (the deletion mutants are summarized in Fig. 2 A). We found that deletion of the C-terminal BRCT domains 7 and 8 did not perturb pol-α and 9-1-1 loading (mutants M2 and M3; Fig. 2 B, lanes 3 and 4). However, loss of these domains did attenuate TopBP1's function in double-stranded DNA–induced ATR signaling (Fig. 2 E), as inferred by the failure of the M3 mutant to support ATR signaling in extracts containing 70-nt oligonucleotides composed of homopolymeric A and T annealed to one another (AT(70)). Thus, it appears that BRCT domains 7 and 8 are required for the TopBP1 response to DNA structures that contain double-stranded DNA ends but not for the TopBP1 response to stalled replication forks. Further C-terminal truncation, which removed the sequences separating BRCT domains 6 and 7, eliminated pol-α and 9-1-1–loading activity as well as ATR activation (mutant M4; Fig. 2 B, lane 5). When the N-terminal BRCT 1 and 2 domains were removed from the checkpoint-competent M2 fragment, all TopBP1 replication checkpoint functions were lost (i.e., pol-α and 9-1-1 recruitment and ATR activation; mutant M5; Fig. 2 D). We conclude that both the region containing BRCT domains 1 and 2 as well the region separating BRCT domains 6 and 7 are required for TopBP1 to promote the recruitment of pol-α and 9-1-1 to stalled replication forks (Fig. 2 A).
TopBP1 chromatin association is required for checkpoint complex assembly
Our data suggest that an early event in checkpoint complex assembly is that TopBP1 senses the stalled replication fork and that, in response, it recruits pol-α and the 9-1-1 clamp to the DNA. If so, chromatin binding by TopBP1 is predicted to be a critical event in checkpoint complex assembly. To test this prediction, we searched for mutations in TopBP1 that would attenuate its ability to bind to chromatin to see whether loss of this activity prevented checkpoint complex assembly. We first compared the ability of WT TopBP1 and the M2 and M5 mutants to associate with chromatin (Fig. 3 A). Both WT TopBP1 and the M2 mutant could bind to chromatin regardless of the presence of aphidicolin, whereas the M5 mutant could not (Fig. 3 B). These data show that the BRCT 1 and 2 domains are essential for TopBP1 association with chromatin.
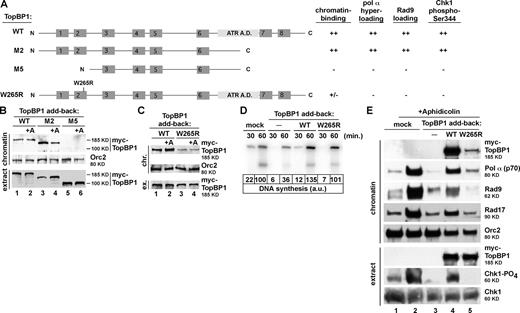
TopBP1 chromatin association is required for pol-α hyperloading and 9-1-1 recruitment to stalled replication forks. (A) Summary of WT and mutant TopBP1s' function in chromatin binding, pol-α hyperloading, Rad9 loading, and Chk1 phospho-Ser344. A.D., activation domain. (B) IVT forms of TopBP1s were added back to TopBP1-depleted egg extract combined with NIB-250 chromatin and, optionally, aphidicolin (+A). The chromatin fraction was isolated and examined for the presence of myc-TopBP1 via Western blotting. (C) Same as B except WT or W265R TopBP1 was added back. (D) Sperm chromatin was added to mock- or TopBP1-depleted (TopBP1 add back) extract optionally containing either blank IVT (−), WT, or W265R TopBP1. DNA synthesis was analyzed at 30 and 60 min as in Fig. S1 C. a.u., arbitrary unit. (E) Sperm chromatin was added to either mock- or TopBP1-depleted (TopBP1 add back) extract supplemented with blank IVT (−), WT, or W265R TopBP1 and, optionally, aphidicolin. Checkpoint proteins on chromatin and Chk1 phosphorylation in extract were assessed by immunoblotting.
TopBP1 chromatin association is required for pol-α hyperloading and 9-1-1 recruitment to stalled replication forks. (A) Summary of WT and mutant TopBP1s' function in chromatin binding, pol-α hyperloading, Rad9 loading, and Chk1 phospho-Ser344. A.D., activation domain. (B) IVT forms of TopBP1s were added back to TopBP1-depleted egg extract combined with NIB-250 chromatin and, optionally, aphidicolin (+A). The chromatin fraction was isolated and examined for the presence of myc-TopBP1 via Western blotting. (C) Same as B except WT or W265R TopBP1 was added back. (D) Sperm chromatin was added to mock- or TopBP1-depleted (TopBP1 add back) extract optionally containing either blank IVT (−), WT, or W265R TopBP1. DNA synthesis was analyzed at 30 and 60 min as in Fig. S1 C. a.u., arbitrary unit. (E) Sperm chromatin was added to either mock- or TopBP1-depleted (TopBP1 add back) extract supplemented with blank IVT (−), WT, or W265R TopBP1 and, optionally, aphidicolin. Checkpoint proteins on chromatin and Chk1 phosphorylation in extract were assessed by immunoblotting.
To pursue these observations further, we generated point mutations in conserved residues of the BRCT 1 and 2 domains and then searched for mutants with an altered ability to bind to chromatin. Various point mutants were produced by IVT and incubated in TopBP1-depleted extract that contained NIB-250 chromatin and, optionally, aphidicolin. The chromatin was then isolated, and association of the IVT proteins with chromatin was assessed by Western blotting. Using this procedure, we identified one mutant, in which Trp265 was changed to Arg (W265R; Fig. 3 A), that bound chromatin less efficiently than WT (Fig. 3 C). To further characterize the W265R mutant, we asked whether it could complement a TopBP1-depleted extract for DNA replication and found that it could (Fig. 3 D). This result shows that W265R does not attenuate the TopBP1 function in initiating DNA replication. We next asked whether the W265R mutant perturbed the checkpoint function of TopBP1. We observed that the W265R mutant failed to efficiently associate with chromatin and, moreover, that neither pol-α hyperloading nor Rad17 or Rad9 recruitment occurred in the extract containing W265R (Fig. 3 E, lane 5). Chk1 phosphorylation also failed in the extract containing W265R TopBP1. These data show that a TopBP1 mutant that fails to associate with chromatin also fails to promote hyperloading of pol-α, recruitment of 9-1-1, and activation of ATR. Based on these data, we conclude that TopBP1 binding to chromatin is an early event in checkpoint signaling and is required for important subsequent steps such as 9-1-1 loading and ATR activation to occur.
Functional organization of the 9-1-1–loading pathway
Our data have described a 9-1-1–loading pathway that requires both TopBP1 and pol-α. TopBP1 acts first to recruit pol-α, and pol-α is required for 9-1-1 to load. To probe this system further, we performed experiments to reveal how the pathway is organized. We reasoned that the pathway may operate in a linear manner, whereby TopBP1 recruits pol-α, pol-α synthesizes a primer, and 9-1-1 is then loaded by Rad17-RFC onto the 5′-DNA junction. In this scenario, the requirements for both TopBP1 and pol-α would be satisfied once the primer has been synthesized. Alternatively, TopBP1 may be required at more than one step: to recruit pol-α and to then participate with Rad17-RFC in the 9-1-1–loading reaction. This second possibility is supported by previous findings showing that TopBP1 interacts with both the 9-1-1 complex and Rad17 in egg extracts (Lee et al., 2007) and our data showing that Rad17 recruitment to stalled replication forks requires TopBP1 (Fig. 1 B).
To dissect how this pathway is organized, we asked whether either pol-α or TopBP1 would need to be present with 9-1-1 during the loading reaction. If the linear pathway model is correct, once a primer has been synthesized, neither TopBP1 nor pol-α would need to be present in the extract when 9-1-1 loads. If the dual-function model is correct, we expect that TopBP1 but not pol-α would have to be present in the extract when 9-1-1 loads. The requirements for pol-α and TopBP1 during the 9-1-1–loading reaction were assessed as follows: Rad9 was depleted from egg extract, and these extracts were then combined with sperm chromatin and aphidicolin. Under these conditions, the replication forks will stall in the presence of TopBP1 and pol-α but in the absence of 9-1-1. The chromatin was then isolated using NIB-250 and transferred to extracts that had been depleted of either pol-α or TopBP1 (Fig. 4 A). The second extracts also contained aphidicolin as well as 9-1-1, and, thus, we determined whether the 9-1-1 complex could load onto the chromatin despite the absence of either pol-α or TopBP1. If either factor (pol-α or TopBP1) can perform its 9-1-1–loading function when 9-1-1 is absent, 9-1-1 will load in the second extract. Conversely, if 9-1-1 requires the presence of a given factor for loading, loading will not occur in the second extract. We examined the requirement for pol-α first. Rad9 could bind to the chromatin, and Chk1 was phosphorylated in a pol-α–depleted second extract (Fig. 4 B). This shows that 9-1-1 does not need to be present for pol-α to perform its 9-1-1–loading function, as would be expected if the role of pol-α in loading 9-1-1 is restricted to primer synthesis. In contrast to the experiment with pol-α, we observed that Rad9 failed to load onto chromatin in a TopBP1-depleted second extract (Fig. 4 C). Control experiments confirmed that the Rad9-, pol-α–, and TopBP1-depleted extracts used in these experiments were functionally depleted of their targets (Fig. S5, B and C) and that the NIB-250 chromatin taken from the Rad9-depleted first extract was devoid of detectable TopBP1 and pol-α (Fig. S5 A). Moreover, these blots show that depletion of any given factor does not noticeably reduce the amount of the other relevant factors in the extract (Fig. S5, B and C). The experiments in Fig. 4 make two important points. First, pol-α can perform its 9-1-1–loading function in a Rad9-depleted extract. This suggests that once primer synthesis has occurred, pol-α is dispensable for 9-1-1 loading. Second, TopBP1 cannot complete its 9-1-1–loading functions in a Rad9-depleted extract. We know that one TopBP1 function, recruitment of pol-α, does not require the presence of 9-1-1, given that the TopBP1-dependent hyperloading of pol-α was observed in Rad9-depleted extract (Fig. 1 G) and that pol-α completes its 9-1-1–loading function in Rad9-depleted extract (Fig. 4 B). Therefore, the failure of 9-1-1 to load in a TopBP1-depleted second extract demonstrates that TopBP1 has at least one additional function in the 9-1-1–loading pathway, which is distinct from pol-α recruitment.
TopBP1 but not pol-α must be present with 9-1-1 for 9-1-1 to load onto stalled replication forks. (A) Experimental design for B and C. (B) NIB-250 chromatin isolated from aphidicolin-containing Rad9-depleted extract was transferred to a second either mock- or pol-α (pol-α−)–depleted extract that also contained aphidicolin. Checkpoint proteins on chromatin and Chk1 phosphorylation in extract were examined as in Fig. 1 B. (C) Same as B except that TopBP1 was depleted in the second extract.
TopBP1 but not pol-α must be present with 9-1-1 for 9-1-1 to load onto stalled replication forks. (A) Experimental design for B and C. (B) NIB-250 chromatin isolated from aphidicolin-containing Rad9-depleted extract was transferred to a second either mock- or pol-α (pol-α−)–depleted extract that also contained aphidicolin. Checkpoint proteins on chromatin and Chk1 phosphorylation in extract were examined as in Fig. 1 B. (C) Same as B except that TopBP1 was depleted in the second extract.
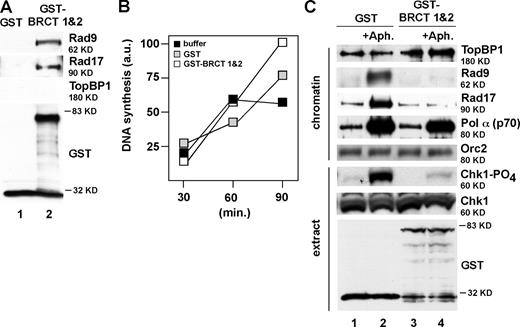
Our data suggest that TopBP1 participates directly in the 9-1-1–loading reaction. Previous work has shown that TopBP1 and Rad9 interact and that the Rad9-binding site on TopBP1 is located within the N terminus of the protein in a region corresponding to BRCT domains 1 and 2 (Lee et al., 2007). If TopBP1 is required to load 9-1-1, we reasoned that perhaps the Rad9–TopBP1 interaction plays a role in 9-1-1 loading. To test this hypothesis, we produced a fusion protein that linked GST to the TopBP1 BRCT 1 and 2 domains (GST–BRCT 1 and 2). Consistent with previous work (Lee et al., 2007), GST pull-down assays show that GST–BRCT 1 and 2, but not GST, could interact with Rad9 and Rad17 in egg extract (Fig. 5 A). Based on this, we reasoned that addition of an excess of GST–BRCT 1 and 2 could dominantly interfere with binding of endogenous TopBP1 to Rad9 and thereby prevent 9-1-1 from loading onto stalled replication forks. Either GST or GST–BRCT 1 and 2 was added to egg extract at a 30-fold molar excess over the concentration of endogenous TopBP1. GST had no effect on the ability of aphidicolin to trigger binding of Rad9, Rad17, TopBP1, or pol-α to the chromatin. In contrast, GST–BRCT 1 and 2 specifically inhibited the binding of Rad9 and Rad17 but not TopBP1 or pol-α. Inclusion of GST–BRCT 1 and 2 also prevented Chk1 phosphorylation (Fig. 5 C). Control experiments showed that, under these conditions, neither GST nor GST–BRCT 1 and 2 inhibited DNA replication in the extract (Fig. 5 B). These data show that GST–BRCT 1 and 2 acts in a dominant-negative manner to block the loading of 9-1-1 onto stalled replication forks and that it does not interfere with the binding of TopBP1 to chromatin or with the hyperloading of pol-α. Based on the data in Figs. 4 and 5, we conclude that TopBP1 plays a direct role in the loading of 9-1-1 onto stalled forks and that interaction between Rad9 and the TopBP1 BRCT 1 and 2 domains is required for 9-1-1 to load.
BRCT 1 and 2 of TopBP1 blocks the loading of 9-1-1 but neither TopBP1 nor pol-α onto stalled replication forks in a dominant-negative manner. (A) Either GST or GST–BRCT 1 and 2 was used for GST pull-down assays as described in Materials and methods. (B) Sperm chromatin was incubated in egg extracts supplemented with buffer, 926 nM GST, or 806 nM GST–BRCT 1 and 2. DNA synthesis products were analyzed at 30, 60, and 90 min as in Fig. S1 C. a.u., arbitrary unit. (C) Either 926 nM GST or 806 nM GST–BRCT 1 and 2 was preincubated with egg extracts, which were then supplemented with sperm chromatin and, optionally, aphidicolin (+Aph.). Checkpoint proteins on chromatin and Chk1 phosphorylation in extract were analyzed as in Fig. 1 B.
BRCT 1 and 2 of TopBP1 blocks the loading of 9-1-1 but neither TopBP1 nor pol-α onto stalled replication forks in a dominant-negative manner. (A) Either GST or GST–BRCT 1 and 2 was used for GST pull-down assays as described in Materials and methods. (B) Sperm chromatin was incubated in egg extracts supplemented with buffer, 926 nM GST, or 806 nM GST–BRCT 1 and 2. DNA synthesis products were analyzed at 30, 60, and 90 min as in Fig. S1 C. a.u., arbitrary unit. (C) Either 926 nM GST or 806 nM GST–BRCT 1 and 2 was preincubated with egg extracts, which were then supplemented with sperm chromatin and, optionally, aphidicolin (+Aph.). Checkpoint proteins on chromatin and Chk1 phosphorylation in extract were analyzed as in Fig. 1 B.
Discussion
In this study, we have combined chromatin-binding assays with the manipulation of Xenopus egg extracts to dissect early events in the assembly of checkpoint signaling complexes on DNA. We report that both TopBP1 and pol-α are required for loading of the 9-1-1 complex onto stalled replication forks. We also report that chromatin binding by TopBP1 triggers assembly of the 9-1-1 complex on DNA. Based on our findings, we propose the following model for loading of the 9-1-1 clamp onto stalled forks (Fig. 6). TopBP1 senses the stalled fork and binds to it (step 1). Pol-α recruitment follows, which allows primer synthesis. This creates either a 5′-RNA junction or, if the RNA portion of the primer is hydrolyzed, a 5′-DNA junction on the DNA (step 2). In our model, TopBP1 also attracts Rad17-RFC and 9-1-1 to the DNA, and this allows Rad17-RFC to load 9-1-1 onto the junction (step 3). The implications of our data and how they support the model in Fig. 6 are discussed in the following sections.
A model for the roles of TopBP1 mediating 9-1-1 loading onto stalled replication forks. See Discussion for details.
A model for the roles of TopBP1 mediating 9-1-1 loading onto stalled replication forks. See Discussion for details.
Sensing of stalled replication forks by TopBP1
An important feature of our model is that TopBP1 initiates 9-1-1 loading by sensing the stalled fork and binding to it (Fig. 6, step 1). We have shown in this study that loss of TopBP1 prevents both pol-α hyperloading and 9-1-1 loading (Fig. 1), which positions TopBP1 at the top of the cascade that leads to 9-1-1 loading. In support of this, we have also shown that the TopBP1 W265R mutant fails to efficiently associate with stalled replication forks and that it cannot support pol-α hyperloading or 9-1-1 loading (Fig. 3). These results identify chromatin binding by TopBP1 as the earliest identifiable step in the 9-1-1–loading pathway and focus attention on the mechanism by which TopBP1 senses stalled forks. We note that, in our experiments, we do not observe a substantial increase in the amount of TopBP1 on chromatin after the induction of replication stress, as might be expected given that TopBP1 binding to stalled forks is required for 9-1-1 loading. Similar results have been reported by others (Hashimoto et al., 2006; Lee et al., 2007). At present, we do not understand why the amount of TopBP1 on chromatin is not substantially increased by replication stress in our experiments. One possibility is that TopBP1 occupies distinct binding sites on chromatin in the absence and presence of replication stress and that when replication forks stall, TopBP1 is transferred from one site to the other. We note that for this possibility to be correct, the same or similar TopBP1 interface would bind to both sites, given that W265R fails to efficiently associate with both unstressed and replication-stressed chromatin. A major goal for future work will be to identify the TopBP1-binding site on stalled replication forks so that the mechanism for how TopBP1 senses stalled forks may be elucidated.
The roles of TopBP1 in 9-1-1 loading
Upon binding to the stalled fork, our data show that TopBP1 recruits pol-α and that pol-α is required for 9-1-1 loading. Therefore, one function of TopBP1 in 9-1-1 loading is the recruitment of pol-α to the stalled fork (Fig. 6, step 2). How might TopBP1 control pol-α activity during a replication stress response? Previous work has clearly shown that, in an unstressed situation, TopBP1 is not required for DNA replication after replication has initiated (Van Hatten et al., 2002; Hashimoto and Takisawa, 2003; Parrilla-Castellar and Karnitz, 2003). This rules out a role for TopBP1 in recruiting pol-α to the lagging strand during unperturbed Okazaki fragment synthesis. However, TopBP1 may be required to stabilize stalled pol-α complexes on the lagging strand during a replication stress response. Alternatively, TopBP1 may play a role in the recruitment of pol-α to the lagging strand specifically during a replication stress response. Recent work has identified MCM10 and And-1 as pol-α–loading factors during unstressed replication (Zhu et al., 2007), and, thus, one possibility is that TopBP1 either replaces or cooperates with these factors to recruit pol-α to the lagging strand during a replication stress response. Finally, as discussed in the following section, TopBP1 and pol-α may play a role on the leading strand during the replication stress response.
In addition to pol-α recruitment, TopBP1 also performs a function in the 9-1-1–loading pathway. We base this conclusion on our observation that, unlike pol-α, TopBP1 cannot complete its 9-1-1–loading function in the absence of 9-1-1. Furthermore, addition of the GST–BRCT 1 and 2 fusion protein to egg extract dominantly interferes with 9-1-1 loading but not pol-α hyperloading. These data show that the TopBP1 function in 9-1-1 loading is not restricted to the recruitment of pol-α. What might this additional function be? Biochemical experiments have shown that Rad17-RFC is sufficient to load 9-1-1 onto RPA-coated 5′-DNA junctions (Ellison and Stillman, 2003; Majka et al., 2006), which shows that TopBP1 is not required for the catalytic activity of Rad17-RFC. One possibility is that, in the context of the cell or a complex extract, TopBP1 is required to concentrate Rad17-RFC and 9-1-1 on or near the DNA so that efficient loading may occur (Fig. 6, step 3). The results presented in this study (Fig. 5) and elsewhere (Lee et al., 2007) show that both Rad17 and Rad9 interact with TopBP1, and these findings support a model whereby TopBP1 increases the local concentration of these factors at the 5′-DNA junction, which could facilitate Rad17-mediated loading of 9-1-1 onto the junction. An important test of this model will be a return to the purified system to see whether the presence of TopBP1 stimulates the 9-1-1–loading reaction or allows 9-1-1 loading when the concentrations of Rad17-RFC and 9-1-1 are otherwise limiting.
The role of pol-α in 9-1-1 loading
Our work shows that pol-α plays a direct role in the recruitment of 9-1-1 to stalled forks. Previous work has shown that 9-1-1 is preferentially loaded onto 5′-DNA junctions (Bermudez et al., 2003; Ellison and Stillman, 2003; Zou et al., 2003; Majka et al., 2006), and, thus, it is likely that the role of pol-α in 9-1-1 loading is to produce this DNA structure on stalled replication forks. Consistent with this, we observed that pol-α can complete its 9-1-1–loading function in the absence of 9-1-1, and, thus, one explanation for this result is that once pol-α has synthesized a primer, it is no longer required for 9-1-1 loading. Where might pol-α–dependent primer synthesis occur on a stalled fork? One possibility is on the lagging strand, where pol-α functions during unperturbed replication to initiate Okazaki fragment synthesis. Another possibility, one that is not mutually exclusive, is that pol-α is recruited by TopBP1 to the leading strand during a replication block. Recent work in budding yeast has shown that large ssDNA gaps form during S phase, on what is likely to be the leading strand, when chromosomes contain irreparable ultraviolet light–induced DNA damage (Lopes et al., 2006). Such gaps can form if replication reinitiates upstream of a stalled leading strand pol, and this type of replication restart reaction would require pol-α–mediated priming. Pol-α–mediated primer synthesis would supply the 5′-DNA junction for 9-1-1 loading in addition to restarting leading strand replication. Therefore, it is possible that TopBP1-mediated recruitment of pol-α to the leading strand during replication stress serves to couple replication restart to assembly of checkpoint complexes at the site of restart.
Materials and methods
Xenopus egg extract methodologies
Xenopus egg extract preparation, sperm chromatin preparation, and DNA synthesis analysis were all performed as described previously (Walter and Newport, 1999). Immunodepletion of TopBP1 and pol-α has been described previously (Yan et al., 2006), and immunodepletion of Rad9 was performed through a similar approach. Unless stated otherwise, aphidicolin and geminin were added to extract at final concentrations of 100 µg/ml and 800 nM, respectively. Chromatin fraction isolation by the ELB procedure for Western blotting was performed as described previously (Stokes et al., 2002). AT(70) used in Fig. 2 E was the duplex preannealed with the oligonucleotides A70 and T70, as described previously (Yan et al., 2006). For the GST pull-down assay (Fig. 5 A), 5 µg GST or GST–BRCT 1 and 2 proteins were incubated with 100-µl egg extracts for 30 min at room temperature. The extract was diluted with 400 µl of immunoprecipitation buffer (100 mM NaCl, 5 mM MgCl2, 10% glycerol, 0.1% NP-40, and 20 mM Tris-HCl, pH 8.0) and supplemented with 20 µl glutathione–Sepharose 4 Fast Flow (GE Healthcare). The mixture was incubated for 1 h with rotation, and the beads were washed three times in immunoprecipitation buffer. The eluted proteins from beads were further analyzed via Western blotting.
NIB-250 chromatin isolation procedure
Sperm chromatin was added to egg extract to 4,000/µl, and the extract was incubated for 30 min at room temperature. The samples were then diluted in NIB-250 (250 mM KCl, 25 mM MgCl2, 25 mM EGTA, 2.5 mM spermidine, 0.75 mM spermine, 2 nM β-mercaptoethanol, 2 µg/ml aprotinin, 2 µg/ml leupeptin, and 50 mM Hepes-KOH, pH 7.6). The mixture was then layered over 15% sucrose/NIB-250 and spun through the cushion in a swinging bucket rotor (TLS-55; Beckman Coulter) for 5 min at 5,000 rpm. After centrifugation, the supernatant was carefully removed, and the pellet was resuspended in sperm dilution buffer (1 mM MgCl2, 100 mM KCl, 150 mM sucrose, and 5 mM Hepes-KOH, pH 7.7). The resuspended suspension (NIB-250 chromatin) was then transferred to a second extract at 10% (vol/vol).
Recombinant proteins
WT and the Mini, M2, M3, M4, and M5 mutants of TopBP1 correspond to TopBP1 nucleotides 1–4,542, 1–2,277, 1–3,540, 1–3,837, 1–2,913, and 1,000–3,540, respectively, and were subcloned into pCS2 + MT. A QuikChange II XL Site-Directed Mutagenesis kit (Agilent Technologies) was used to obtain the point mutants W265R and W1138R of TopBP1. For IVT, an SP6 TNT Quick Master Mix kit (Promega) was used in all cases as described previously (Yan et al., 2006). GST–BRCT 1 and 2 was produced by subcloning TopBP1 nucleotides 1–999 into pGEX-4T-1. Recombinant GST and GST–BRCT 1 and 2 proteins were expressed in Escherichia coli BL21/DE3 and purified by glutathione–Sepharose 4 Fast Flow according to standard protocols. Purified DNA pol-α (calf thymus) was purchased from CHIMERx.
Antibodies
Antibodies against TopBP1, pol-α (p70 subunit), Rad17, and Orc2 have been described previously (Stokes et al., 2002; Van Hatten et al., 2002; Jones et al., 2003; Yan et al., 2006). Antibodies against Rad9, Rad1, and pol-ε (p60 subunit) and -δ (p125 subunit) were gifts from H. Lindsay (Lancaster University, Lancaster, England, UK), K. Cimprich (Stanford University, Stanford, CA), and S. Waga (Osaka University, Osaka, Japan), respectively. Additionally, antibodies against Chk1 (S344-PO4; Cell Signaling Technology), Chk1 (Santa Cruz Biotechnology, Inc.), C-myc (Santa Cruz Biotechnology, Inc.), GST (Santa Cruz Biotechnology, Inc.), MCM5 (Bethyl Laboratories, Inc.), and PCNA (Santa Cruz Biotechnology, Inc.) were purchased from the respective vendors. Peroxidase-conjugated monoclonal mouse anti–rabbit IgG light chain specific (Jackson ImmunoResearch Laboratories) was used to probe Chk1 (S344-PO4) from the depleted extract. Peroxidase-conjugated ImmunoPure goat anti–rabbit IgG (H + L; Thermo Fisher Scientific) and horseradish peroxidase–linked sheep anti–mouse IgG (GE Healthcare) were used when appropriate.
Online supplemental material
Fig. S1 provides characterization of the NIB-250 chromatin isolation and transfer system in Xenopus egg extracts. Fig. S2 shows that TopBP1 is required for the recruitment of the Rad1 subunit of 9-1-1 to stalled DNA replication forks. Fig. S3 demonstrates rescue of DNA replication and Chk1 phosphorylation in TopBP1-depleted egg extract by recombinant TopBP1 produced via IVT. Fig. S4 shows replisome reassembly and activity in pol-α–depleted extract containing NIB-250 chromatin. Fig. S5 shows analysis of NIB-250 chromatin isolated from the Rad9-depleted extract and functional characterization of the immunodepleted (Rad9, pol-α, and TopBP1 depleted) extracts used in Fig. 4.
Acknowledgments
We are grateful to Drs. K. Cimprich, H. Lindsay, and S. Waga for reagents and K. Cimprich and C. Van for informative discussions.
This work was supported by a grant from the National Institute of General Medical Sciences (National Institutes of Health) to W.M. Michael (GM067735).
References
Abbreviations used in this paper: ATR, ataxia telangiectasia mutated and Rad3 related; ATRIP, ATR-interacting protein; BRCT, BRCA1 C terminus; ELB, egg lysis buffer; IVT, in vitro transcription/translation; PCNA, proliferating cell nuclear antigen; pol, polymerase; RFC, replication factor C; RPA, replication protein A; ssDNA, single-stranded DNA; WT, wild type.