ER tubules form and maintain membrane contact sites (MCSs) with late endosomes/lysosomes (LE/lys). The molecular composition and cellular functions of these MCSs are poorly understood. Here, we find that Tex2, an SMP domain-containing lipid transfer protein conserved in metazoan and yeast, is a tubular ER protein and is recruited to ER–LE/lys MCSs by TMEM55, phosphatases that convert PI(4,5)P2 to PI5P on LE/lys. We show that the Tex2–TMEM55 interaction occurs between an N-terminal region of Tex2 and a catalytic motif in the PTase domain of TMEM55. The Tex2–TMEM55 interaction can be regulated by endosome-resident type 2 PI4K activities. Functionally, Tex2 knockout results in defects in lysosomal trafficking, digestive capacity, and lipid composition of LE/lys membranes. Together, our data identify Tex2 as a tubular ER protein that resides at TMEM55-dependent ER–LE/lys MCSs required for lysosomal functions.
Introduction
The ER is organized into a continuous intracellular membrane network consisting of the nuclear envelope, flattened sheets, and interconnected tubules that spread over the cytosol (Baumann and Walz, 2001; Bian et al., 2011; English and Voeltz, 2013; Levine, 2005b). ER tubules are considered as main sites for the synthesis of a majority of lipids (Borgese et al., 2006). Newly synthesized lipids at the ER are then transferred to other organelles through both vesicular and nonvesicular transport pathways. Although vesicular lipid transport mediates the bulk transport of many lipids, increasing lines of evidence suggest that lipid-transfer protein (LTP)-mediated lipid transport at membrane contact sites (MCSs) is the major transport route for certain lipid types (Holthuis and Levine, 2005; Joshi et al., 2017; Levine, 2004; Levine, 2005a; Prinz et al., 2020; Wong et al., 2017; Wong et al., 2019). MCSs are defined as cytosolic gaps of 10–30 nm between one organelle and the other organelles (Lebiedzinska et al., 2009; Levine, 2004).
The majority of early endosomes and all late endosomes and lysosomes (hereafter referred collectively to as LE/lys) maintain MCSs with ER tubules (Friedman et al., 2013; Zajac et al., 2013), which play critical roles in diverse cellular processes, including modulating endosome maturation and positioning, lipid composition, and fission during cargo sorting (Friedman et al., 2013; Gao et al., 2022; Hoyer et al., 2018; Jongsma et al., 2016; Raiborg et al., 2015; Rocha et al., 2009; Rowland et al., 2014; Wu and Voeltz, 2021). However, the molecular composition and cellular functions of these MCSs are poorly understood.
Tex2 is a synaptotagmin-like mitochondrial-lipid-binding (SMP) domain-containing LTP that resides on the ER. Tex2 is conserved between metazoan and yeast. Nvj2p, the Tex2 homolog in yeast, relocalizes from contacts between the ER and other organelles and increases ER–Golgi contacts upon ER stress. Nvj2p may directly transfer ceramide from the ER to the Golgi complex destined for sphingolipid synthesis and prevents lipotoxicity induced by ceramide accumulation (Liu et al., 2017).
Tex2 is recently reported to cooperate with another SMP-containing protein PDZD8 and the PI(4,5)P2 phosphatases, OCRL-1 and UNC-26/synaptojanin, to regulate endosomal PI(4,5)P2 homeostasis in worms (Jeyasimman et al., 2021). However, whether and how Tex2 is localized to MCSs remains elusive. More importantly, cellular functions of Tex2-mediated MCSs remain largely unknown in mammals. In this study, we found that Tex2 is a tubular ER protein and can be recruited to ER–LE/lys MCSs by TMEM55. The TMEM55-mediated recruitment of Tex2 is regulated by the activities of type 2 PI4Ks, which are resident on endosomal membranes and contribute to the generation of PI4P on endosomal membranes (Balla et al., 2002). Tex2 is required for LE/lys trafficking, lysosomal digestive capacities, and the maintenance of the lipid composition of LE/lys membranes.
Results
Tex2 is a tubular ER protein
E-Syts, a group of SMP domain-containing proteins, transfer a range of glycerophospholipids between the ER and the plasma membrane (PM; Schauder et al., 2014). To identify unknown proteins that may interact with E-Syt1, we performed GFP-Trap assays in HEK293 cells transiently expressing GFP-E-Syt1 followed by mass spectrometry (MS). After the removal of those proteins co-immunoprecipitated (coIPed) by GFP alone, we found an SMP domain-containing protein named testis-expressing protein 2 (Tex2) that strongly interested us owing to its potential lipid transfer activity.
To begin with, we sought to explore the localization of Tex2 and its relation to E-Syt1. We confirmed the Tex2–E-syt1 interaction in GFP-Trap assays using GFP-Tex2 as bait in HEK293 cells (Fig. S1 A). Since E-Syt1 is an integral ER protein that is enriched at ER–PM MSCs upon ER calcium depletion (Bian et al., 2018; Giordano et al., 2013), we asked whether Tex2 colocalized with E-Syt1 at these MCSs by live-cell confocal microscopy. GFP-Tex2 partially colocalized with Halo-E-Syt1 on the ER under normal conditions (Fig. S1, B and D). Halo-E-Syt1 was enriched on the discrete ER microdomains, likely representing the ER–PM MCSs, upon thapsigargin (TG) treatment. However, GFP-Tex2 was not specifically enriched at these sites (Fig. S1 B), suggesting that Tex2 may not function at ER–PM MCSs.
The interaction between Tex2 and E-Syt1, and Genome editing of Tex2. (A) GFP-Trap assays demonstrate an interaction between GFP-Tex2 and Halo-E-Syt1 in COS7 cells. (B) Representative images of a COS7 cell expressing GFP-Tex2 (magenta) and Halo-E-Syt1 (green) before (top panel) and after (bottom panel) TG treatment (2 μM; 30 min). (C) Representative images of a COS7 cell expressing Halo-E-Syt1 (green) and ER-tagRFP (magenta). (D) Pearson’s correlation coefficient of Tex2 vs. ER-resident proteins, including E-Syt1 (13 cells); E-Syt1 upon TG (13 cells); RTN4 (12 cells); Reep5 (12 cells); ARLIP6 (11 cells); and ATL1 (11 cells) in three independent experiments. Ordinary one-way ANOVA with Tukey’s multiple comparisons test. Mean ± SD. (E) CRISPR knock-in of GFP (A206K) Tag at N-terminus of Tex2 locus in HeLa cells. The underlined letters indicate the PAM (TGG) for spCas9; ATG in red in sgRNA showing the start codon of Tex2. (F) Western blots of GFP-Tex2-KI for single cell clone from E. (G) CRISPR knock-out of Tex2 in HeLa cells (Tex2-KO). Two sgRNAs are used with the underlined letters, indicating the PAMs (GGG for sgRNA1 and CGG for sgRNA2) for spCas9. (H) Western blots of two Tex2-KO clones from C. Scale bar, 10 μm in the whole cell images and 2 μm in the insets in B and C. Source data are available for this figure: SourceData FS1.
The interaction between Tex2 and E-Syt1, and Genome editing of Tex2. (A) GFP-Trap assays demonstrate an interaction between GFP-Tex2 and Halo-E-Syt1 in COS7 cells. (B) Representative images of a COS7 cell expressing GFP-Tex2 (magenta) and Halo-E-Syt1 (green) before (top panel) and after (bottom panel) TG treatment (2 μM; 30 min). (C) Representative images of a COS7 cell expressing Halo-E-Syt1 (green) and ER-tagRFP (magenta). (D) Pearson’s correlation coefficient of Tex2 vs. ER-resident proteins, including E-Syt1 (13 cells); E-Syt1 upon TG (13 cells); RTN4 (12 cells); Reep5 (12 cells); ARLIP6 (11 cells); and ATL1 (11 cells) in three independent experiments. Ordinary one-way ANOVA with Tukey’s multiple comparisons test. Mean ± SD. (E) CRISPR knock-in of GFP (A206K) Tag at N-terminus of Tex2 locus in HeLa cells. The underlined letters indicate the PAM (TGG) for spCas9; ATG in red in sgRNA showing the start codon of Tex2. (F) Western blots of GFP-Tex2-KI for single cell clone from E. (G) CRISPR knock-out of Tex2 in HeLa cells (Tex2-KO). Two sgRNAs are used with the underlined letters, indicating the PAMs (GGG for sgRNA1 and CGG for sgRNA2) for spCas9. (H) Western blots of two Tex2-KO clones from C. Scale bar, 10 μm in the whole cell images and 2 μm in the insets in B and C. Source data are available for this figure: SourceData FS1.
One of the most noticeable features was that GFP-Tex2 was exclusively localized to ER tubules (Fig. 1 A), whereas Halo-E-Syt1 was evenly distributed on both tubules and sheets of the ER (Fig. S1, B and C). To avoid potential artifacts of overexpression, we labeled endogenous Tex2 with monomeric GFP (A206K) at its N terminus using Crispr-Cas9 in HeLa cells (GFP-Tex2-KI), in which ∼50% of endogenous Tex2 was labeled by GFP (Fig. S1, E and F). Consistently, endogenous GFP-Tex2 is exclusively localized to ER tubules (Fig. 1 B).
Tex2 is a tubular ER protein. (A) Representative images of a live COS7 cell expressing exogenous GFP-Tex2 (green) and ER-tagRFP (magenta, an ER luminal marker) with insets. (B) Representative images of a live GFP-Tex2-KI HeLa cell expressing endogenous GFP-Tex2 (green) and ER-tagRFP (magenta) with insets. (C) Domain organization of Tex2. (D–H) Representative images of COS7 cells expressing either GFP-Tex2-1-517 (green; D), GFP-Tex2-TM (green; residues 474–517; E), GFP-Tex2-∆1-473 (green; F), GFP-Tex2-(277-517) (green; G), or GFP-Tex2-∆277-473 (green; H), along with ER-tagRFP (magenta) with insets. (I) Pearson’s correlation coefficient of Tex2 proteins vs. ER-tagRFP; GFP-Tex2-TM (22 cells); GFP-Tex2-1-517 (19 cells); GFP-Tex2-∆1-473 (23 cells); GFP-Tex2-(277-517) (26 cells); and GFP-Tex2-∆277-473 (21 cells) in more than three independent experiments. Ordinary one-way ANOVA with Tukey’s multiple comparisons test. Mean ± SD. Scale bar, 10 μm in the whole cell images and 2 μm in the insets in A, B, and D–H.
Tex2 is a tubular ER protein. (A) Representative images of a live COS7 cell expressing exogenous GFP-Tex2 (green) and ER-tagRFP (magenta, an ER luminal marker) with insets. (B) Representative images of a live GFP-Tex2-KI HeLa cell expressing endogenous GFP-Tex2 (green) and ER-tagRFP (magenta) with insets. (C) Domain organization of Tex2. (D–H) Representative images of COS7 cells expressing either GFP-Tex2-1-517 (green; D), GFP-Tex2-TM (green; residues 474–517; E), GFP-Tex2-∆1-473 (green; F), GFP-Tex2-(277-517) (green; G), or GFP-Tex2-∆277-473 (green; H), along with ER-tagRFP (magenta) with insets. (I) Pearson’s correlation coefficient of Tex2 proteins vs. ER-tagRFP; GFP-Tex2-TM (22 cells); GFP-Tex2-1-517 (19 cells); GFP-Tex2-∆1-473 (23 cells); GFP-Tex2-(277-517) (26 cells); and GFP-Tex2-∆277-473 (21 cells) in more than three independent experiments. Ordinary one-way ANOVA with Tukey’s multiple comparisons test. Mean ± SD. Scale bar, 10 μm in the whole cell images and 2 μm in the insets in A, B, and D–H.
The formation of ER tubules is tightly controlled by a group of proteins, including Reep5 (Chen et al., 2021), Reticulon 4 (RTN4; Voeltz et al., 2006), Atlastin-1 (ATL1; Wang et al., 2016), and ARL6IP1 (Yamamoto et al., 2014). We next explored the relationships between Tex2 and these ER-tubule-shaping proteins. Live-cell confocal microscopy showed that GFP-Tex2 colocalized with Reep5 (Fig. S2 A) and RTN4 (Fig. S2 B) on the ER tubules but only partially colocalized with other ER-shaping proteins (Fig. S2, C and D; and Fig. S1 D). The GFP-trap assays showed that Tex2 interacted with Reep5 (Fig. S2 E) and RTN4 (Fig. S2 F), but interacted with ATL-1 (Fig. S2 G) and ARL6IP1 (Fig. S2 H) to a lesser extent. We then asked whether Tex2 was recruited to ER tubules by these ER-shaping proteins. To test this possibility, we suppressed the expression of these ER-shaping proteins by small interfering RNAs (siRNA). However, the tubular ER localization of Tex2 was not disturbed, as revealed by live-cell confocal microscopy (Fig. S2, I and J), suggesting that Tex2 may target tubular ER of its own.
The targeting of Tex2 to tubular ER appears to be independent of ER tubule-shaping proteins. (A–D) Representative images of a COS7 cell expressing GFP-Tex2 (green), ER-tagRFP (blue) and REEP5-Halo (magenta; A), RTN4-Halo (B), Halo-ATL1 (C), and Halo-ARL6IP1 (D) with insets. (E–H) GFP-Trap assays from COS7 cells demonstrate interactions between GFP-Tex2 and Reep5-Halo (E), RTN4-Halo (F), Halo-ATL1 (G), and Halo-ARL6IP1 (H). (I) Representative images of USO2 cells expressing GFP-Tex2 (green), ER-tagRFP (magenta) with insets upon treatments of scrambled, RTN4, REEP5, ARL6IP1, RTN1, and RTN3 siRNAs. (J) qPCR assays showing the efficient of siRNA-mediated suppression in I. Scale bar, 10 μm in the whole cell images and 2 μm in the insets in A–D and I. Source data are available for this figure: SourceData FS2.
The targeting of Tex2 to tubular ER appears to be independent of ER tubule-shaping proteins. (A–D) Representative images of a COS7 cell expressing GFP-Tex2 (green), ER-tagRFP (blue) and REEP5-Halo (magenta; A), RTN4-Halo (B), Halo-ATL1 (C), and Halo-ARL6IP1 (D) with insets. (E–H) GFP-Trap assays from COS7 cells demonstrate interactions between GFP-Tex2 and Reep5-Halo (E), RTN4-Halo (F), Halo-ATL1 (G), and Halo-ARL6IP1 (H). (I) Representative images of USO2 cells expressing GFP-Tex2 (green), ER-tagRFP (magenta) with insets upon treatments of scrambled, RTN4, REEP5, ARL6IP1, RTN1, and RTN3 siRNAs. (J) qPCR assays showing the efficient of siRNA-mediated suppression in I. Scale bar, 10 μm in the whole cell images and 2 μm in the insets in A–D and I. Source data are available for this figure: SourceData FS2.
Therefore, we dissected the Tex2 protein (Fig. 1 C), and found that an N-terminal region containing a TM domain (Tex2-NT; residues 1–517) of Tex2 was sufficient for its targeting to ER tubules (Fig. 1 D), but a smaller region containing the TM domain (Tex2-TM; residues 475–517) failed to exclusively target ER tubules (Fig. 1 E). Consistently, Tex2 without a part of the NT region (Tex2-∆1-473) lost its tubular ER localization (Fig. 1 F), suggesting that the NT region is sufficient and required for Tex2 targeting to ER tubules. Notably, a portion of Tex2-∆1-473 appeared to relocalize to some discrete ER regions, likely ER–PM MCSs (Fig. 1 F). We further found that the residues 277–517 of NT are sufficient to target ER tubules (Fig. 1 G), and Tex2 with a deletion of this region (Tex2-∆277-473) substantially reduced its targeting to tubular ER (Fig. 1, H and I).
Next, we investigated whether Tex2 was able to promote the formation of ER tubules. Overexpression of Climp63-Halo promoted the formation of ER sheets at the cell periphery (Fig. S3, A, B, and E), consistent with a reported role of Climp63 in the formation of ER sheets (Shibata et al., 2010), which was countered upon the coexpression of GFP-Tex2 (Fig. S3, C and E), to a similar extent compared with the effects of RTN4 overexpression (Fig. S3, D and E), a well-studied ER tubule-forming protein (Wang et al., 2016). Interestingly, Crispr–Cas9-mediated Tex2 KO (Fig. S1, G and H) did not substantially affect the tubular ER network at the periphery (Fig. S3, F and G), suggesting a redundant role of these tubular ER-resident proteins in the formation and/or maintenance of the tubular ER network. Collectively, our results suggested that Tex2 exclusively targets tubular ER via the region (residues 277–474) adjacent to the TM.
Supplementary data toFigs. 1 and 2. (A–D). Representative images of COS7 cells expressing the general ER luminal marker ER-tagRFP alone (A), Climp63-Halo (green) and ER-tagRFP (magenta; B), Climp63-Halo (magenta) and GFP-Tex2 (green; C), or Climp63-Halo (magenta) and RTN4-GFP (green; D) with insets at cell periphery. (E) Quantification of ER sheet abundance in A–D. 16 COS7 cells expressing ER-tagRFP, 15 cells expressing Climp63-Halo and ER-tagRFP, 15 cells expressing Climp63-Halo and GFP-Tex2, and 15 cells expressing Climp63-Halo and RTN4-GFP from three independent assays were analyzed. Ordinary one-way ANOVA with Tukey’s multiple comparisons test. Mean ± SD. (F) Representative images of control, or two HeLa Tex2-KO clones expressing ER-tagRFP (magenta) with insets at the cell periphery. (G) Quantification of ER sheet abundance in F. 13 control HeLa cells, 13 Tex2 KO-1 cells, and 14 Tex2 KO-2 cells from three independent assays were analyzed. Ordinary one-way ANOVA with Tukey’s multiple comparisons test. Mean ± SD. (H) Western blots demonstrate the increase in BiP level in control or two Tex2-KO clones upon TG stimulation for 12 h. (I) Representative images of a GFP-Tex2-KI (green) cell expressing a Golgi marker MGAT2-Halo (magenta) upon DMSO and TG (12 h) with insets. (J) Pearson’s correlation coefficient of GFP-Tex2 vs. MGAT2-Halo in DMSO (13 cells) or TG-treated cell (21 cells). Two-tailed unpaired Student’s t test. Mean ± SD. (K) Representative images of a GFP-Tex2-KI (green) cell expressing ER-tagRFP (magenta) upon DMSO, TG (2 h), or TG (12 h), with insets. (L) Pearson’s correlation coefficient of GFP-Tex2 vs. ER-tagRFP in DMSO (12 cells) or TG-treated cell (12 h; 12 cells). Two-tailed unpaired Student’s t test. Mean ± SD. (M) Representative images of a GFP-Tex2-KI (green) cell expressing BFP-TMEM55B (blue) and MGAT2-Halo (magenta) upon DMSO or TG treatment for 6 h with insets. (N) Left: Pearson’s correlation coefficient of GFP-Tex2 vs. Halo-TMEM55B in DMSO (12 cells) or TG-treated cell (13 cells). Right: Pearson’s correlation coefficient of GFP-Tex2 vs. MGAT2-Halo in DMSO (12 cells) or TG-treated cell (13 cells) from more than three independent assays. Two-tailed unpaired Student’s t test. Mean ± SD. Scale bar, 10 μm in the whole cell images and 2 μm in the insets in A–D, F, I, K, and M. Source data are available for this figure: SourceData FS3.
Supplementary data toFigs. 1 and 2. (A–D). Representative images of COS7 cells expressing the general ER luminal marker ER-tagRFP alone (A), Climp63-Halo (green) and ER-tagRFP (magenta; B), Climp63-Halo (magenta) and GFP-Tex2 (green; C), or Climp63-Halo (magenta) and RTN4-GFP (green; D) with insets at cell periphery. (E) Quantification of ER sheet abundance in A–D. 16 COS7 cells expressing ER-tagRFP, 15 cells expressing Climp63-Halo and ER-tagRFP, 15 cells expressing Climp63-Halo and GFP-Tex2, and 15 cells expressing Climp63-Halo and RTN4-GFP from three independent assays were analyzed. Ordinary one-way ANOVA with Tukey’s multiple comparisons test. Mean ± SD. (F) Representative images of control, or two HeLa Tex2-KO clones expressing ER-tagRFP (magenta) with insets at the cell periphery. (G) Quantification of ER sheet abundance in F. 13 control HeLa cells, 13 Tex2 KO-1 cells, and 14 Tex2 KO-2 cells from three independent assays were analyzed. Ordinary one-way ANOVA with Tukey’s multiple comparisons test. Mean ± SD. (H) Western blots demonstrate the increase in BiP level in control or two Tex2-KO clones upon TG stimulation for 12 h. (I) Representative images of a GFP-Tex2-KI (green) cell expressing a Golgi marker MGAT2-Halo (magenta) upon DMSO and TG (12 h) with insets. (J) Pearson’s correlation coefficient of GFP-Tex2 vs. MGAT2-Halo in DMSO (13 cells) or TG-treated cell (21 cells). Two-tailed unpaired Student’s t test. Mean ± SD. (K) Representative images of a GFP-Tex2-KI (green) cell expressing ER-tagRFP (magenta) upon DMSO, TG (2 h), or TG (12 h), with insets. (L) Pearson’s correlation coefficient of GFP-Tex2 vs. ER-tagRFP in DMSO (12 cells) or TG-treated cell (12 h; 12 cells). Two-tailed unpaired Student’s t test. Mean ± SD. (M) Representative images of a GFP-Tex2-KI (green) cell expressing BFP-TMEM55B (blue) and MGAT2-Halo (magenta) upon DMSO or TG treatment for 6 h with insets. (N) Left: Pearson’s correlation coefficient of GFP-Tex2 vs. Halo-TMEM55B in DMSO (12 cells) or TG-treated cell (13 cells). Right: Pearson’s correlation coefficient of GFP-Tex2 vs. MGAT2-Halo in DMSO (12 cells) or TG-treated cell (13 cells) from more than three independent assays. Two-tailed unpaired Student’s t test. Mean ± SD. Scale bar, 10 μm in the whole cell images and 2 μm in the insets in A–D, F, I, K, and M. Source data are available for this figure: SourceData FS3.
TMEM55 recruits Tex2 to ER–LE/Lys MCSs
As a potential LTP resident on ER tubules, Tex2 likely functions at ER-associated MCSs. To identify the type of ER-associated MCSs that Tex2 may function, we sought to identify its adaptor on the other organelle by coIP-MS using GFP-Tex2 as bait. To this end, we identified TMEM55B, a phosphatase that converts PI(4,5)P2 to PI5P on LE/lys as a novel Tex2-interacting protein (Fig. 2 A). Consistently, we found that E-Syt1 and the tubular ER proteins, including RTN4, REEP5, RTN3, RTN1, and ARL6IP1, were identified in our MS analysis (Fig. 2 A). Notably, TMEM55B did not rank at the top of our list. We reason that Tex2 was mainly on ER tubules but was not enriched at ER MCSs under normal conditions in which coIP-MS was performed, suggesting that Tex2–TMEM55B interactions may be subjected to regulation. GFP-trap assays confirmed the interactions between GFP-Tex2 and Halo-TMEM55B (Fig. 2 B). Importantly, live-cell confocal microscopy demonstrated that overexpression of Halo-TMEM55B greatly recruited GFP-Tex2 to another organelle that clustered at perinuclear regions (Fig. 2 C, inset 1), whereas GFP-Tex2 was still localized to the entire ER tubules spreading over the cytosol without TMEM55B overexpression, with a minor enrichment at perinuclear regions (Fig. 2 C, inset 2). As a control, Halo-TMEM55B failed to recruit GFP-E-Syt1 to perinuclear regions (Fig. 2 D), suggesting specific recruitment of Tex2 by TMEM55B.
Identification of TMEM55B as a Tex2 adaptor on LE/Lys. (A) Volcano plot of protein candidates coIPed with Tex2 in HEK293 cells compared with protein candidates coIPed with GFP tag only. Candidates that were considered significant (−log [P value] > 1.3; P < 0.05) were labeled in red (Log2 [fold change] > 0; increased in abundance) or blue (Log2 [fold change] < 0; decreased in abundance). (B) GFP-Trap assays demonstrate an interaction between GFP-Tex2 and Halo-TMEM55B in COS7 cells. (C and D) Representative wide-field images of live COS7 cells expressing either GFP-Tex2 (green; C) or GFP-E-syt1 (green; D) and Halo-TMEM55B with two boxed regions showing at the bottom. (E and F) Representative images of a live COS7 cell expressing GFP-Tex2 (green), Halo-TMEM55B (magenta; E) or Halo-TMEM55A (magenta; F), ER-tagRFP (red), and Lamp1-mCh (blue; a LE/lys marker) with two boxed regions showing at the bottom. (G) Pearson’s correlation coefficient of Tex2 vs. Lamp1 in absence of TMEM55 (11 cells) or upon TMEM55A (10 cells) or TMEM55B (13 cells) overexpression (>3 independent experiments). Ordinary one-way ANOVA with Tukey’s multiple comparisons test. Mean ± SD. (H and I) Representative live-cell images of a GFP-Tex2-KI (green) cell without (H) or with expressing Halo-TMEM55B (I) and Lamp1-mCh (blue) with two boxed regions at the bottom. Yellow arrows denote the specific enrichment of GFP-Tex2-KI at LE/lys extensively contacting the ER. (J) Representative 3D rendering of a COS7 cell expressing GFP-Tex2 (green), Halo-TMEM55B (magenta), and Lamp1-mCh (blue) with y-z projection to the left and x-z projection to the right. (K) COS7 cells expressing GFP-Tex2, Halo-TMEM55B, and stained by DAPI, were fixed and imaged by 3D microscopy (left panel) and processed for TEM (middle panel). Fluorescence and TEM images were correlated (right panel) with two insets showing tight associations between the ER (green) and LE/lys (magenta). Scale bar, 10 μm in the whole cell images and 2 μm in the insets in C, D, E, F, and H–J; 5 μm in the whole cell image, and 1 μm in the insets in K. Source data are available for this figure: SourceData F2.
Identification of TMEM55B as a Tex2 adaptor on LE/Lys. (A) Volcano plot of protein candidates coIPed with Tex2 in HEK293 cells compared with protein candidates coIPed with GFP tag only. Candidates that were considered significant (−log [P value] > 1.3; P < 0.05) were labeled in red (Log2 [fold change] > 0; increased in abundance) or blue (Log2 [fold change] < 0; decreased in abundance). (B) GFP-Trap assays demonstrate an interaction between GFP-Tex2 and Halo-TMEM55B in COS7 cells. (C and D) Representative wide-field images of live COS7 cells expressing either GFP-Tex2 (green; C) or GFP-E-syt1 (green; D) and Halo-TMEM55B with two boxed regions showing at the bottom. (E and F) Representative images of a live COS7 cell expressing GFP-Tex2 (green), Halo-TMEM55B (magenta; E) or Halo-TMEM55A (magenta; F), ER-tagRFP (red), and Lamp1-mCh (blue; a LE/lys marker) with two boxed regions showing at the bottom. (G) Pearson’s correlation coefficient of Tex2 vs. Lamp1 in absence of TMEM55 (11 cells) or upon TMEM55A (10 cells) or TMEM55B (13 cells) overexpression (>3 independent experiments). Ordinary one-way ANOVA with Tukey’s multiple comparisons test. Mean ± SD. (H and I) Representative live-cell images of a GFP-Tex2-KI (green) cell without (H) or with expressing Halo-TMEM55B (I) and Lamp1-mCh (blue) with two boxed regions at the bottom. Yellow arrows denote the specific enrichment of GFP-Tex2-KI at LE/lys extensively contacting the ER. (J) Representative 3D rendering of a COS7 cell expressing GFP-Tex2 (green), Halo-TMEM55B (magenta), and Lamp1-mCh (blue) with y-z projection to the left and x-z projection to the right. (K) COS7 cells expressing GFP-Tex2, Halo-TMEM55B, and stained by DAPI, were fixed and imaged by 3D microscopy (left panel) and processed for TEM (middle panel). Fluorescence and TEM images were correlated (right panel) with two insets showing tight associations between the ER (green) and LE/lys (magenta). Scale bar, 10 μm in the whole cell images and 2 μm in the insets in C, D, E, F, and H–J; 5 μm in the whole cell image, and 1 μm in the insets in K. Source data are available for this figure: SourceData F2.
Next, we sought to identify the type of Tex2-resident ER MCSs by live-cell confocal microscopy. We observed that GFP-Tex2, Halo-TMEM55B, and a LE/lys marker (Lamp1-mCh) were colocalized, with the ER (marked by a luminal ER marker ER-tagRFP) being strongly enriched at these sites (Fig. 2 E), indicating that TMEM55B recruits Tex2 to ER–LE/lys MCSs.
TMEM55A and TMEM55B are phosphoinositide 4-phosphatases that dephosphorylate the D4 position of PI(4,5)P2 mainly on LE/lys membranes, and human TMEM55A and TMEM55B share 51% identity in amino acid sequences (Ungewickell et al., 2005). Both isozymes contain a CX5R motif in their phosphatase domains and two putative TM domains at the C-terminal (Fig. 4 A). Next, we asked whether TMEM55A could also recruit Tex2 by live-cell confocal microscopy. Halo-TMEM55A strongly recruited GFP-Tex2 to TMEM55A-positive LE/lys membranes (Fig. 2 F) to a similar extent as TMEM55B (Fig. 2 G). Since only TMEM55B was identified in our MS analysis, we focused on TMEM55B thereafter in this study.
Next, we confirmed the recruitment of endogenous Tex2 by TMEM55B in GFP-Tex2-KI cells. Live-cell confocal images showed that, though endogenous GFP-Tex2 was mainly distributed over the tubular ER network, a small but significant portion of endogenous GFP-Tex2 was enriched at regions adjacent to LE/lys (Fig. 2 H), likely ER–LE/lys MCSs, in absence of exogenous TMEM55B. Remarkably, upon Halo-TMEM55B expression, endogenous Tex2 was substantially enriched at Halo-TMEM55B-positive LE/lys membranes (Fig. 2 I), suggesting that TMEM55B greatly promoted the recruitment of endogenous GFP-Tex2 to the contacts.
We then used 3D rendering of z-stacks through high-resolution live-cell microscopy to examine the localizations of GFP-Tex2 and Halo-TMEM55B relative to LE/lys. Tex2 was specifically enriched on TMEM55B-positive LE/lys at perinuclear (Fig. 2 J) in reconstructed images, as revealed by colocalization analysis based on x-z and y-z projections of 3D rendering. In addition, we directly examined TMEM55B-mediated recruitment of Tex2-positive ER membranes to LE/lys by correlative light electron microscopy (CLEM). Consistently, transmission electron microscopy showed that the ER was tightly associated with LE/lys at perinuclear regions, where GFP-Tex2 was greatly recruited by Halo-TMEM55B (Fig. 2 K).
Given a specific role of Nvj2p, the yeast homolog of Tex2, at ER–Golgi MCSs upon ER stress, we explored whether Tex2 could be recruited to ER–Golgi MCSs upon TG-induced ER stress by live-cell confocal microscopy. The induction of ER stress was confirmed by an increase in the level of BiP (Fig. S3 H), a marker of ER stress. Upon ER stress, endogenous GFP-Tex2 was not substantially recruited to the Golgi (marked by cis/medial Golgi protein MGAT2; Fig. S3, I and J), and it was mainly localized to the entire ER tubule network upon either 2 h (Fig. S3 K; middle) or 12 h treatment of TG (Fig. S3 K; bottom), similar to untreated conditions (Fig. S3, K and L). In addition, endogenous GFP-Tex2 was still considerably recruited to Halo-TMEM55B-positive LE/lys, but not the Golgi, upon ER stress (Fig. S3, M and N). Together, these results suggest that Tex2 may not be directly involved in TG-induced ER stress. However, it should be noted Tex2 may play important roles in other types of cellular stress, which are not identified in this study.
The N-terminal region of Tex2 is responsible for interacting with TMEM55B
Next, we dissected the Tex2 protein to investigate how Tex2 interacted with TMEM55B. Live-cell microscopy showed that Tex2-ΔTM was mainly cytosolic in absence of TMEM55B (Fig. 3 A; left panel), but a significant portion of this mutant was recruited to Halo-TMEM55B-positive LE/lys (Fig. 3 A; right panel). Another Tex2 mutant with a deletion of both the TM and PH domain (Tex2-ΔTM-ΔPH) was still able to be recruited to TMEM55B-postitive LE/lys, similar to WT Tex2 (Fig. S4 A). Consistently, a Tex2 mutant without the PH domain (Tex2-ΔPH) was recruited to TMEM55B-positive LE/lys, as revealed by the wrapping of Tex2-labeled ER tubules around LE/lys (Fig. S4 B). These results indicated that the PH domain of Tex2 was not required for TMEM55B-mediated recruitment of GFP-Tex2 to ER–LE/lys MCSs. PH domains are often involved in protein targeting to PIs-enriched membranes, including LE/lys membranes (Lemmon, 2007). Notably, PIPs strip assays using the purified PH domain of Tex2 showed that Tex2-PH preferentially bound PI3P and PI4P, but bound, to a less extent, to PI5P, PI3,4P2, PI3,5P2, or PI4,5P2 (Fig. S4 C). Since in this study, the formation of Tex2-mediated MCSs has been examined under TMEM55B overexpression conditions, it is plausible that the PH domain may facilitate the recruitment of Tex2, independent of TMEM55B, to other membranes, for example, the PM, by binding to PI(4,5)P2, under certain conditions.
The N-terminal region of Tex2 is responsible for interacting with TMEM55B. (A) Left: representative images of COS7 cells expressing GFP-Tex2-∆TM (green) alone; middle: representative whole-cell images of a COS7 cell expressing GFP-Tex2-∆TM (green), Halo-TMEM55B (magenta), and Lamp1-mCh (blue); right: two enlarged images from the two boxed regions in the middle panel. (B–E) Representative images of COS7 cells expressing either GFP-Tex2-NT-1 (green; B), GFP-Tex2-NT-2 (green; C), GFP-Tex2 (277-517) (green; D), or GFP-Tex2-∆ (1-276) (green; E), along with Halo-TMEM55B (magenta) and Lamp1-mCh (blue) with insets. (F) Pearson’s correlation coefficient of TMEM55B vs. Tex2 mutants; Tex2-∆TM (10 cells); Tex2-1-540 (10 cells); Tex2-1-517 (10 cells); Tex2-277-517 (10 cells); and Tex2-∆1-276 (10 cells) in more than three independent experiments. Ordinary one-way ANOVA with Tukey’s multiple comparisons test. Mean ± SD. (G) GFP-Trap assays demonstrate interactions between Halo-TMEM55B and Tex2 mutants in COS7 cells. (H) Pulldown assays using GFP-Tex2 bound on GFP-Trap beads and purified GST-TMEM55B demonstrate a direct interaction in a ceramide-independent manner. (I) As in H, pulldown assays using GFP-Tex2 bound on GFP-Trap beads and purified GST-TMEM55B demonstrate a direct interaction in a PS-independent manner. Scale bar, 10 μm in the whole cell images and 2 μm in the insets in A–E. Source data are available for this figure: SourceData F3.
The N-terminal region of Tex2 is responsible for interacting with TMEM55B. (A) Left: representative images of COS7 cells expressing GFP-Tex2-∆TM (green) alone; middle: representative whole-cell images of a COS7 cell expressing GFP-Tex2-∆TM (green), Halo-TMEM55B (magenta), and Lamp1-mCh (blue); right: two enlarged images from the two boxed regions in the middle panel. (B–E) Representative images of COS7 cells expressing either GFP-Tex2-NT-1 (green; B), GFP-Tex2-NT-2 (green; C), GFP-Tex2 (277-517) (green; D), or GFP-Tex2-∆ (1-276) (green; E), along with Halo-TMEM55B (magenta) and Lamp1-mCh (blue) with insets. (F) Pearson’s correlation coefficient of TMEM55B vs. Tex2 mutants; Tex2-∆TM (10 cells); Tex2-1-540 (10 cells); Tex2-1-517 (10 cells); Tex2-277-517 (10 cells); and Tex2-∆1-276 (10 cells) in more than three independent experiments. Ordinary one-way ANOVA with Tukey’s multiple comparisons test. Mean ± SD. (G) GFP-Trap assays demonstrate interactions between Halo-TMEM55B and Tex2 mutants in COS7 cells. (H) Pulldown assays using GFP-Tex2 bound on GFP-Trap beads and purified GST-TMEM55B demonstrate a direct interaction in a ceramide-independent manner. (I) As in H, pulldown assays using GFP-Tex2 bound on GFP-Trap beads and purified GST-TMEM55B demonstrate a direct interaction in a PS-independent manner. Scale bar, 10 μm in the whole cell images and 2 μm in the insets in A–E. Source data are available for this figure: SourceData F3.
Neither the PH-like domain nor the SMP domain is responsible for interacting with TMEM55B. (A and B) Representative images of a COS7 cell expressing either Tex2-ΔTM-ΔPH (green; A), or Tex2-ΔPH (green; B), along with Lamp1-mCh (blue) and Halo-TMEM55B (magenta) with insets. Yellow arrows indicate Tex2 enrichments at TMEM55B-positive LE/lys. (C) PIPs strip assay of purified Tex2-PH. Left: Coomassie blue staining of purified Tex2-PH; right: PIPs strip blots. (D) Representative images of a COS7 cell expressing Tex2-ΔSMP (green) and Lamp1-mCh (blue) and Halo-TMEM55B (magenta) with insets. Yellow arrows indicate Tex2 proteins enrichment at TMEM55B-positive LE/lys. (E) Representative images of a COS7 cell expressing Tex2-SMP alone (gray; left) or Tex2-SMP along with Lamp1-mCh (blue) and Halo-TMEM55B (magenta) with an inset (right). (F and G) Representative images of a COS7 cell expressing Tex2-1-474 (green; F) or Tex2-1-276 (green; G), along with Lamp1-mCh (blue) and Halo-TMEM55B (magenta) with insets. (H) Pearson’s correlation coefficient of either Tex2-ΔTM-ΔPH (10 cells), Tex2-ΔPH (10 cells), Tex2-ΔSMP (11 cells), Tex2-SMP (9 cells), Tex2-1-474 (9 cells), or Tex2-1-276 (11 cells) vs. Halo-TMEM55B in three independent assays. Ordinary one-way ANOVA with Tukey’s multiple comparisons test. Mean ± SD. Scale bar, 10 μm in the whole cell images and 2 μm in the insets in A, B, D, F, and G. Source data are available for this figure: SourceData FS4.
Neither the PH-like domain nor the SMP domain is responsible for interacting with TMEM55B. (A and B) Representative images of a COS7 cell expressing either Tex2-ΔTM-ΔPH (green; A), or Tex2-ΔPH (green; B), along with Lamp1-mCh (blue) and Halo-TMEM55B (magenta) with insets. Yellow arrows indicate Tex2 enrichments at TMEM55B-positive LE/lys. (C) PIPs strip assay of purified Tex2-PH. Left: Coomassie blue staining of purified Tex2-PH; right: PIPs strip blots. (D) Representative images of a COS7 cell expressing Tex2-ΔSMP (green) and Lamp1-mCh (blue) and Halo-TMEM55B (magenta) with insets. Yellow arrows indicate Tex2 proteins enrichment at TMEM55B-positive LE/lys. (E) Representative images of a COS7 cell expressing Tex2-SMP alone (gray; left) or Tex2-SMP along with Lamp1-mCh (blue) and Halo-TMEM55B (magenta) with an inset (right). (F and G) Representative images of a COS7 cell expressing Tex2-1-474 (green; F) or Tex2-1-276 (green; G), along with Lamp1-mCh (blue) and Halo-TMEM55B (magenta) with insets. (H) Pearson’s correlation coefficient of either Tex2-ΔTM-ΔPH (10 cells), Tex2-ΔPH (10 cells), Tex2-ΔSMP (11 cells), Tex2-SMP (9 cells), Tex2-1-474 (9 cells), or Tex2-1-276 (11 cells) vs. Halo-TMEM55B in three independent assays. Ordinary one-way ANOVA with Tukey’s multiple comparisons test. Mean ± SD. Scale bar, 10 μm in the whole cell images and 2 μm in the insets in A, B, D, F, and G. Source data are available for this figure: SourceData FS4.
We also found that Tex2 without the SMP domain (Tex2-ΔSMP) could be recruited to TMEM55B-positive LE/lys, but to a slightly lesser extent compared with WT-Tex2 (Fig. S4, D and H). Consistently, the SMP domain alone (Tex2-SMP) was cytosolic even upon TMEM55B overexpression (Fig. S4 E). These results suggested that the SMP domain may not be required for the Tex2–TMEM55B interaction.
Importantly, live-cell confocal microscopy showed that the residues 1–540 of Tex2 can be substantially recruited by TMEM55B (Fig. 3 B). Further analysis of the NT demonstrated that the Tex2-NT (Tex2-1-517; Fig. 3 C), but not Tex2-277-517 (Fig. 3 D), was sufficient for the recruitment. Consistently, Tex2 with a deletion of the residues 1–276 (Tex2-Δ1-276) failed to be recruited by TMEM55B (Fig. 3 E), suggesting an essential role of the residues 1–276 for Tex2–TMEM55B interactions. Notably, neither residues 1–276 nor residues 1–474 of Tex2-NT could be recruited by TMEM55B (Fig. S4, F and G), suggesting that these two regions were required but not sufficient for the recruitment. Together, these data indicated that the Tex2-NT (1-517) is the minimal functional module for the recruitment of Tex2 by TMEM55B.
We next examined the Tex2–TMEM55B interactions by GFP-Trap assays. In accord with the live-cell microscopy results, GFP-Tex2-NT was strongly coIPed with Halo-TMEM55B, whereas the level of Halo-TMEM55B coIPed by GFP-Tex2-Δ1-276 was greatly reduced (Fig. 3 G), confirming a critical role of Tex2-NT in the Tex2–TMEM55B interactions. Consistently, Tex2-SMP could not be coIPed with Halo-TMEM55B (Fig. 3 G), whereas Tex2-ΔSMP coIPed with Halo-TMEM55B in a similar level as Tex2-NT (Fig. 3 G), confirming that Tex2-SMP was not required for interacting with TMEM55B.
We further asked whether Tex2 directly interacts with TMEM55B by in vitro pulldown assays. At this time, we were unable to produce purified full-length Tex2 or Tex2-NT in sufficient quantities for in vitro pull-down assays. Alternatively, we used GFP-Trap assays to pellet endogenous GFP-Tex2 from HeLa GFP-Tex2-KI using a high-salt (500 mM NaCl) lysis buffer. After rigorous washing to remove proteins that could copellet with GFP-Tex2 under high-salt conditions, GFP-Tex2 beads were incubated with purified Glutathione S-transferase (GST) tag alone or GST-TMEM55B, respectively. Indeed, GFP-Tex2 bound to GST-TMEM55B but not GST tag (Fig. 3 H). In addition, the interaction between GFP-Tex2 and GST-TMEM55B appeared not to be affected by the addition of ceramide or phosphatidylserine (PS; Fig. 3, H and I), two lipid species that were shown to be bound by Tex2 later in this study (Fig. 9).
A catalytic motif of TMEM55B is required for recruiting Tex2
Next, we sought to understand the molecular mechanisms underlying the recruitment of Tex2 by TMEM55B through dissections of the TMEM55B protein (Fig. 4 A). Remarkably, live-cell microscopy showed that a TMEM55B truncation without C-terminal TM domains (TMEM55B-∆TM) was cytosolic (Fig. 4 B), but was greatly recruited to ER tubules upon the expression of GFP-Tex2 (Fig. 4 C), indicating reverse recruitment of cytosolic TMEM55B-∆TM to the ER by GFP-Tex2. Another TMEM55B mutant (TMEM55B-∆NT) still interacted with Tex2 to a similar extent as WT TMEM55B, as revealed by the colocalization between GFP-Tex2 and this mutant on LE/lys (Fig. 4, D and J). However, a truncation mutant containing the TM domain of TMEM55B (TMEM55B ∆1-163) failed to recruit GFP-Tex2 (Fig. 4 E). Importantly, another mutant with the deletion of phosphatase domain (TMEM55B-∆PTase) could target LEs but failed to recruit GFP-Tex2 (Fig. 4 F). These lines of evidence indicated that the PTase domain was critical for the recruitment of Tex2 by TMEM55B. We next investigated whether the recruitment of Tex2 is dependent on the highly conserved catalytic motif CX5R of the PTase domain. Importantly, TMEM55B-∆CX5R completely lost its ability to recruit Tex2 to LE/lys (Fig. 4 G), indicating that the CX5R motif is required for recruiting Tex2. Further, two phosphatase-dead TMEM55B mutants (C140W or C140S) were still able to recruit GFP-Tex2, but to a lesser extent compared with WT TMEM55B (Fig. 4, H, I, and J), suggesting that the Tex2–TMEM55B interaction was not strictly dependent on phosphatase activity of TMEM55B. In addition, Tex2 KO did not significantly affect the localization of TMEM55B, as shown by the colocalization analysis between GFP-TMEM55B and Lamp1-mCh (Fig. 4, K and L). Collectively, our data indicated that the Tex2–TMEM55B interaction is dependent on the CX5R motif in the PTase domain of TMEM55B.
The catalytic motif CX 5 R of TMEM55B is responsible for interacting with Tex2. (A) Domain organization of TMEM55B. (B) Representative images of a COS7 cell expressing Halo-TMEM55B-∆TM (magenta), ER-GFP (green), and Lamp1-mCh (blue) with insets. (C–I) Representative images of COS7 cells expressing either Halo-TMEM55B-∆TM (magenta; C), Halo-TMEM55B-∆NT (75-284aa; D), Halo-TMEM55B-TM (E), Halo-TMEM55B-∆PTase (F), Halo-TMEM55B-∆X5R (G), Halo-PP4P1-C140W (H), and Halo-PP4P1-C140S (I). Yellow arrows indicated TMEM55B-positive LE/lys with GFP-Tex2 enrichments; while red arrows denoted TMEM55B-positive LE/lys without GFP-Tex2 enrichments. (J) Pearson’s correlation coefficient of TMEM55B proteins vs. Tex2; Halo-TMEM55B-∆TM in absence of Tex2 (9 cells); and Halo-TMEM55B-∆TM (9 cells); Halo-TMEM55B-∆1-74 (10 cells); Halo-TMEM55B-∆1-163 (9 cells); Halo-TMEM55B-∆PTase (9 cells); Halo-TMEM55B-∆CX5R (9 cells); Halo-PP4P1-C140W (10 cells); and Halo-PP4P1-C140S (10 cells) upon Tex2 overexpression in three independent experiments. Ordinary one-way ANOVA with Tukey’s multiple comparisons test. Mean ± SD. (K) Representative images of either control (left panel) or Tex2 KO-1 (right panel) HeLa cells expressing GFP-TMEM55B (green) and Lamp1-mCh (magenta) with insets. (L) Pearson’s correlation coefficient of TMEM55B vs. Lamp1 in either control (21 cells) or Tex2 KO-1 (20 cells) HeLa cells in three independent experiments. Two-tailed unpaired Student’s t test. Mean ± SD. Scale bar, 10 μm in the whole cell images and 2 μm in the insets in B–I and K.
The catalytic motif CX 5 R of TMEM55B is responsible for interacting with Tex2. (A) Domain organization of TMEM55B. (B) Representative images of a COS7 cell expressing Halo-TMEM55B-∆TM (magenta), ER-GFP (green), and Lamp1-mCh (blue) with insets. (C–I) Representative images of COS7 cells expressing either Halo-TMEM55B-∆TM (magenta; C), Halo-TMEM55B-∆NT (75-284aa; D), Halo-TMEM55B-TM (E), Halo-TMEM55B-∆PTase (F), Halo-TMEM55B-∆X5R (G), Halo-PP4P1-C140W (H), and Halo-PP4P1-C140S (I). Yellow arrows indicated TMEM55B-positive LE/lys with GFP-Tex2 enrichments; while red arrows denoted TMEM55B-positive LE/lys without GFP-Tex2 enrichments. (J) Pearson’s correlation coefficient of TMEM55B proteins vs. Tex2; Halo-TMEM55B-∆TM in absence of Tex2 (9 cells); and Halo-TMEM55B-∆TM (9 cells); Halo-TMEM55B-∆1-74 (10 cells); Halo-TMEM55B-∆1-163 (9 cells); Halo-TMEM55B-∆PTase (9 cells); Halo-TMEM55B-∆CX5R (9 cells); Halo-PP4P1-C140W (10 cells); and Halo-PP4P1-C140S (10 cells) upon Tex2 overexpression in three independent experiments. Ordinary one-way ANOVA with Tukey’s multiple comparisons test. Mean ± SD. (K) Representative images of either control (left panel) or Tex2 KO-1 (right panel) HeLa cells expressing GFP-TMEM55B (green) and Lamp1-mCh (magenta) with insets. (L) Pearson’s correlation coefficient of TMEM55B vs. Lamp1 in either control (21 cells) or Tex2 KO-1 (20 cells) HeLa cells in three independent experiments. Two-tailed unpaired Student’s t test. Mean ± SD. Scale bar, 10 μm in the whole cell images and 2 μm in the insets in B–I and K.
The regulation of the Tex2–TMEM55B interaction by PI4KII activities
We next tested whether and how the Tex2–TMEM55B interaction was regulated. Importantly, we found that the coexpression of PI4KIIα or PI4KIIβ, PI kinases that convert PI to PI4P on the membranes of LE/lys (Balla et al., 2002), significantly hampered the recruitment of GFP-Tex2 to LE/lys membranes by TMEM55B (Fig. 5, A, B, and F). Yellow arrows indicated diminished Tex2 enrichments on PI4KII-positive/TMEM55B-positive LE/lys; while red arrows denoted Tex2 enrichments on PI4KII-negative/TMEM55B-positive LE/lys. In contrast, coexpression of the kinase-dead mutant PI4KIIα-W359A (Zhou et al., 2014) did not impair the TMEM55B-mediated recruitment of Tex2 (Fig. 5, C and F), indicating that the activities of PI4KIIα play an important role in the regulation of Tex2–TMEM55 interactions. In addition, the colocalization between Tex2 and PI4KIIα-W359A was significantly higher than that of Tex2 and WT PI4KIIα (Fig. 5, C and G), further supporting that the activities of PI4KII inhibit the recruitment of Tex2 to ER–LE/lys MCSs. Moreover, we found that the coexpression of PI4KIIα or PI4KIIβ did not substantially impair the recruitment of Tex2-NT by TMEM55B (Fig. 5, D and F), suggesting a potential regulatory module at the C-terminal region of Tex2. Indeed, the recruitment of GFP-Tex2-ΔPH by TMEM55B was not significantly affected by the co-expression of PI4KIIα or PI4KIIβ (Fig. 5, E and F), suggesting that the PH domain may be involved in the regulation step. Consistently, the colocalization between Tex2-NT or Tex2-ΔPH and PI4KIIs was not substantially altered upon the loss of kinase activities of PI4KIIα (Fig. 5 G). Purified Tex2-PH bound PI4P, PI(3,4)P2, and PI(4,5)P2 in vitro (Fig. S4 C), but this interaction was not strong enough to mediate the ER–LE/lys MCSs (Fig. S4, B, D, and H; data not shown). Therefore, we speculated that the transient binding of these PIPs to the PH domain of Tex2 may hamper the interaction between Tex2-NT and TMEM55B, thus negatively regulating the recruitment of Tex2 by TMEM55B. In addition, siRNA-mediated depletion of PI4KIIα or PI4KIIβ significantly promoted the recruitment of GFP-Tex2 to LE/lys in the absence of exogenous TMEM55B, as revealed by a higher extent of colocalization between GFP-Tex2 and Lamp1-mCh in PI4KII-depleted cells (Fig. 5, H, I, and J). In addition, Tex2 KO did not substantially affect the localization of PI4KIIα (Fig. 5, K and M) or PI4KIIβ (Fig. 5, L and M) relative to LE/lys. Although PI4KIIs are responsible for the generation of PI4P pool on the LE/lys membrane, which can be further converted to other PIPs, such as PI(3,4)P2 and PI(4,5)P2, it is still unclear which PIPs are responsible for the regulation of Tex2–TMEM55 interactions.
The regulation of the Tex2–TMEM55B interaction by PI4KII activities. (A–C) Representative images of COS7 cells expressing either Halo-PI4KIIα (magenta; A), Halo-PI4KIIβ (magenta; B), or kinase-dead PI4KIIα mutant Halo-PI4KIIα-W359A (magenta; C), along with GFP-Tex2 (green), Lamp1-mCh (gray), and BFP-TMEM55B (blue) with two boxed regions shown at the bottom. Yellow arrows indicate reduced Tex2 enrichments at PI4KII-positive/TMEM55B-positive LE/lys while red arrows denote Tex2 enrichments at PI4KII-negative/TMEM55B-positive LE/lys. (D and E) Representative images of COS7 cells expressing either GFP-Tex2-NT (green; D) or GFP-Tex2-ΔPH (green; E) along with Halo-PI4KIIs (magenta), Lamp1-mCh (gray) and BFP-TMEM55B (blue) with insets. (F) Pearson’s correlation coefficient of Tex2 vs. TMEM55B in cells expressing either empty vector (10 cells), Halo-PI4KIIα (13 cells), Halo-PI4KIIβ (14 cells), or Halo-PI4KIIα-W359A (13 cells). Pearson’s correlation coefficient of Tex2-NT vs. TMEM55B in cells expressing either empty vector (14 cells), Halo-PI4KIIα (12 cells), Halo-PI4KIIβ (11 cells), or Halo-PI4KIIα-W359A (13 cells). Pearson’s correlation coefficient of Tex2-ΔPH vs. TMEM55B in cells expressing either empty vector (12 cells), Halo-PI4KIIα (10 cells), Halo-PI4KIIβ (13 cells), or Halo-PI4KIIα-W359A (12 cells) in three independent assays. Ordinary one-way ANOVA with Tukey’s multiple comparisons test. Mean ± SD. (G) Pearson’s correlation coefficient of Tex2 vs. PI4KIIs in cells expressing either Halo-PI4KIIα (13 cells), Halo-PI4KIIβ (14 cells), or Halo-PI4KIIα-W359A (13 cells). Pearson’s correlation coefficient of Tex2-NT vs. TMEM55B in cells expressing either Halo-PI4KIIα (12 cells), Halo-PI4KIIβ (11 cells), or Halo-PI4KIIα-W359A (13 cells). Pearson’s correlation coefficient of Tex2-ΔPH vs. TMEM55B in cells expressing either Halo-PI4KIIα (10 cells), Halo-PI4KIIβ (13 cells), or Halo-PI4KIIα-W359A (12 cells) in three independent assays. Ordinary one-way ANOVA with Tukey’s multiple comparisons test. Mean ± SD. (H) Representative images of COS7 cells expressing GFP-Tex2 (green) and Lamp1-mCh (magenta) upon scrambled (top panel), PI4KIIα (middle panel), or PI4KIIβ siRNAs (bottom panel) with insets. Yellow arrows indicated Tex2 enrichments at LE/lys. (I) qPCR assays showing the efficiency of siRNA-mediated suppression of PI4KIIs. (J) Pearson’s correlation coefficient of Tex2 vs. LE/lys in cells upon scrambled (15 cells), PI4KIIα (18 cells), or PI4KIIβ siRNAs (14 cells). Ordinary one-way ANOVA with Tukey’s multiple comparisons test. Mean ± SD. (K) Representative images of either control (top panel) or Tex2 KO-1(bottom panel) HeLa cells expressing Halo-PI4KIIα (magenta; left panel) or Halo-PI4KIIβ (magenta; right panel) along with Lamp1-mCh (blue) and ER-tagRFP (green) with insets. (L) Pearson’s correlation coefficient of either PI4KIIα or PI4KIIβ vs. Lamp1 in control or Tex2 KO-1 HeLa cells in K and L. For quantification of PI4KIIα, 10 control cells and 10 Tex2 KO-1 cells were analyzed, and for quantification of PI4KIIβ, 10 control cells and 10 Tex2 KO-1 cells were analyzed in three independent experiments. Two-tailed unpaired Student’s t test. Mean ± SD. Scale bar, 10 μm in the whole cell images and 2 μm in the insets in A–E and H.
The regulation of the Tex2–TMEM55B interaction by PI4KII activities. (A–C) Representative images of COS7 cells expressing either Halo-PI4KIIα (magenta; A), Halo-PI4KIIβ (magenta; B), or kinase-dead PI4KIIα mutant Halo-PI4KIIα-W359A (magenta; C), along with GFP-Tex2 (green), Lamp1-mCh (gray), and BFP-TMEM55B (blue) with two boxed regions shown at the bottom. Yellow arrows indicate reduced Tex2 enrichments at PI4KII-positive/TMEM55B-positive LE/lys while red arrows denote Tex2 enrichments at PI4KII-negative/TMEM55B-positive LE/lys. (D and E) Representative images of COS7 cells expressing either GFP-Tex2-NT (green; D) or GFP-Tex2-ΔPH (green; E) along with Halo-PI4KIIs (magenta), Lamp1-mCh (gray) and BFP-TMEM55B (blue) with insets. (F) Pearson’s correlation coefficient of Tex2 vs. TMEM55B in cells expressing either empty vector (10 cells), Halo-PI4KIIα (13 cells), Halo-PI4KIIβ (14 cells), or Halo-PI4KIIα-W359A (13 cells). Pearson’s correlation coefficient of Tex2-NT vs. TMEM55B in cells expressing either empty vector (14 cells), Halo-PI4KIIα (12 cells), Halo-PI4KIIβ (11 cells), or Halo-PI4KIIα-W359A (13 cells). Pearson’s correlation coefficient of Tex2-ΔPH vs. TMEM55B in cells expressing either empty vector (12 cells), Halo-PI4KIIα (10 cells), Halo-PI4KIIβ (13 cells), or Halo-PI4KIIα-W359A (12 cells) in three independent assays. Ordinary one-way ANOVA with Tukey’s multiple comparisons test. Mean ± SD. (G) Pearson’s correlation coefficient of Tex2 vs. PI4KIIs in cells expressing either Halo-PI4KIIα (13 cells), Halo-PI4KIIβ (14 cells), or Halo-PI4KIIα-W359A (13 cells). Pearson’s correlation coefficient of Tex2-NT vs. TMEM55B in cells expressing either Halo-PI4KIIα (12 cells), Halo-PI4KIIβ (11 cells), or Halo-PI4KIIα-W359A (13 cells). Pearson’s correlation coefficient of Tex2-ΔPH vs. TMEM55B in cells expressing either Halo-PI4KIIα (10 cells), Halo-PI4KIIβ (13 cells), or Halo-PI4KIIα-W359A (12 cells) in three independent assays. Ordinary one-way ANOVA with Tukey’s multiple comparisons test. Mean ± SD. (H) Representative images of COS7 cells expressing GFP-Tex2 (green) and Lamp1-mCh (magenta) upon scrambled (top panel), PI4KIIα (middle panel), or PI4KIIβ siRNAs (bottom panel) with insets. Yellow arrows indicated Tex2 enrichments at LE/lys. (I) qPCR assays showing the efficiency of siRNA-mediated suppression of PI4KIIs. (J) Pearson’s correlation coefficient of Tex2 vs. LE/lys in cells upon scrambled (15 cells), PI4KIIα (18 cells), or PI4KIIβ siRNAs (14 cells). Ordinary one-way ANOVA with Tukey’s multiple comparisons test. Mean ± SD. (K) Representative images of either control (top panel) or Tex2 KO-1(bottom panel) HeLa cells expressing Halo-PI4KIIα (magenta; left panel) or Halo-PI4KIIβ (magenta; right panel) along with Lamp1-mCh (blue) and ER-tagRFP (green) with insets. (L) Pearson’s correlation coefficient of either PI4KIIα or PI4KIIβ vs. Lamp1 in control or Tex2 KO-1 HeLa cells in K and L. For quantification of PI4KIIα, 10 control cells and 10 Tex2 KO-1 cells were analyzed, and for quantification of PI4KIIβ, 10 control cells and 10 Tex2 KO-1 cells were analyzed in three independent experiments. Two-tailed unpaired Student’s t test. Mean ± SD. Scale bar, 10 μm in the whole cell images and 2 μm in the insets in A–E and H.
A role of Tex2–TMEM55B interaction in the regulation of LE/lys trafficking
Of note, we observed that LE/lys were substantially confined to perinuclear regions upon TMEM55B overexpression (Fig. 6, A, F, and G), in accord with a reported role of TMEM55B in promoting the retrograde trafficking of LE/lys (Willett et al., 2017). Consistently, siRNA-mediated TMEM55B depletion resulted in a much more dispersed distribution of LE/lys compared with control (Fig. 6, A, B, and G). Interestingly, compared with control, LE/lys were more clustered at the perinuclear region in Tex2 KO cells, similar to the phenotype resulting from PDZD8 suppression (Gao et al., 2022). Importantly, Tex2 KO could significantly rescue the dispersed distribution of LE/lys resulting from TMEM55B depletion (Fig. 6, A, B, and G), suggesting that Tex2 may antagonize the effect of TMEM55B in the retrograde trafficking. In addition, we observed that overexpression of GFP-Tex2 or GFP-Tex2-NT could partially alleviate the clustering of LE/lys at perinuclear regions caused by Halo-TMEM55B overexpression (Fig. 6, C, D, and G). In contrast, coexpression of Tex2 without the NT (Tex2-∆1-276) with TMEM55B had no effect (Fig. 6, E and G), suggesting a role of Tex2-NT in the regulation of TMEM55B-mediated LE/lys trafficking.
A role of Tex2–TMEM55B interaction in retrograde transport of LE/lys. (A) Representative images of live HeLa cell expressing Lamp1-mCh (gray) upon expression of empty vector, GFP-TMEM55B, TMEM55B siRNA, Tex2-KO, and TMEM55B siRNA and Tex2-KO with white dash lines indicating cell shape. (B) Western blots showing the efficiency of siRNA-mediated TMEM55B depletion in control or Tex2 KO HeLa cells. (C–E) Representative images of live HeLa cells expressing Lamp1-mCh (blue), Halo-TMEM55B (magenta), along with either GFP-Tex2 (C), GFP-Tex2-NT (D), or GFP-Tex2-∆1-276 (E) with white dash lines indicating cell shape. (F) Schematic diagram showing the definition of perinuclear regions. (G) Percentage of cells with lysosome clustering at perinuclear region in control (18 cells), TMEM55B overexpression (16 cells), TMEM55B siRNA (14 cells), Tex2-KO (15 cells), TMEM55B siRNA and Tex2-KO (18 cells), TMEM55B and Tex2 (16 cells), TMEM55B and Tex2-NT (1-514) (17 cells), or TMEM55B and Tex2-∆1-276 (16 cells) from more than three independent assays. Ordinary one-way ANOVA with Tukey’s multiple comparisons test. Mean ± SD. (H) Representative images of a live COS7 cell expressing ER-GFP (green) and Lamp1-mCh (magenta) with time-lapse images of a boxed region shown on the right, from more than three independent assays. Yellow arrows denote one LE/lys undergoing retrograde transport with frequent contact with the ER (Video 1; time interval: 1.63 s). (I) Representative images of a live COS7 cell expressing GFP-Tex2 (green), Halo-TMEM55B (magenta), and Lamp1-mCh (blue) with time-lapse images of a boxed region shown on the right, from more than three independent assays. Yellow arrows denote one LE/lys undergoing retrograde transport along with the Tex2-labeled ER (Video 2; time interval: 2.53 s). Scale bar, 10 μm in the whole cell images and 2 μm in the insets in A, C–E, H, and I. Source data are available for this figure: SourceData F6.
A role of Tex2–TMEM55B interaction in retrograde transport of LE/lys. (A) Representative images of live HeLa cell expressing Lamp1-mCh (gray) upon expression of empty vector, GFP-TMEM55B, TMEM55B siRNA, Tex2-KO, and TMEM55B siRNA and Tex2-KO with white dash lines indicating cell shape. (B) Western blots showing the efficiency of siRNA-mediated TMEM55B depletion in control or Tex2 KO HeLa cells. (C–E) Representative images of live HeLa cells expressing Lamp1-mCh (blue), Halo-TMEM55B (magenta), along with either GFP-Tex2 (C), GFP-Tex2-NT (D), or GFP-Tex2-∆1-276 (E) with white dash lines indicating cell shape. (F) Schematic diagram showing the definition of perinuclear regions. (G) Percentage of cells with lysosome clustering at perinuclear region in control (18 cells), TMEM55B overexpression (16 cells), TMEM55B siRNA (14 cells), Tex2-KO (15 cells), TMEM55B siRNA and Tex2-KO (18 cells), TMEM55B and Tex2 (16 cells), TMEM55B and Tex2-NT (1-514) (17 cells), or TMEM55B and Tex2-∆1-276 (16 cells) from more than three independent assays. Ordinary one-way ANOVA with Tukey’s multiple comparisons test. Mean ± SD. (H) Representative images of a live COS7 cell expressing ER-GFP (green) and Lamp1-mCh (magenta) with time-lapse images of a boxed region shown on the right, from more than three independent assays. Yellow arrows denote one LE/lys undergoing retrograde transport with frequent contact with the ER (Video 1; time interval: 1.63 s). (I) Representative images of a live COS7 cell expressing GFP-Tex2 (green), Halo-TMEM55B (magenta), and Lamp1-mCh (blue) with time-lapse images of a boxed region shown on the right, from more than three independent assays. Yellow arrows denote one LE/lys undergoing retrograde transport along with the Tex2-labeled ER (Video 2; time interval: 2.53 s). Scale bar, 10 μm in the whole cell images and 2 μm in the insets in A, C–E, H, and I. Source data are available for this figure: SourceData F6.
We further examined the dynamics of GFP-Tex2 and Halo-TMEM55B over time using live-cell microscopy. Time-lapse video analysis showed that Lamp1-mCh-positive LE/lys frequently contacted but not constantly associated with the ER during retrograde transport in COS7 cells labeled with general ER and LE/lys markers (Fig. 6 H and Video 1). In contrast, GFP-Tex2-labled ER membranes were stably associated with TMEM55B-positive LE/lys during LE/lys transport (Fig. 6 I and Video 2). Collectively, these results indicate that TMEM55B acts as an adaptor on LE/lys membranes to recruit ER-resident Tex2 to ER–LE/lys MCSs, and these MCSs may be involved in the regulation of retrograde trafficking of LE/lys.
Representative time-lapse imaging of a COS7 cell expressing Lamp1-mCh (magenta) and ER-GFP (green) with a lysosome undergoing retrograde transport. Time interval 1.63 s, 5.7 frames per second. Scale bar, 2 μm.
Representative time-lapse imaging of a COS7 cell expressing Lamp1-mCh (magenta) and ER-GFP (green) with a lysosome undergoing retrograde transport. Time interval 1.63 s, 5.7 frames per second. Scale bar, 2 μm.
Representative time-lapse imaging of a COS7 cell expressing GFP-Tex2 (green), Halo-TMEM55B (magenta), and Lamp1-mCh (blue) with a Halo-TMEM55B-positive lysosome tightly associating with GFP-Tex2-labeled ER membranes during intracellular transport. Time interval 2.53 s, 6.25 frames per second. Scale bar, 2 μm.
Representative time-lapse imaging of a COS7 cell expressing GFP-Tex2 (green), Halo-TMEM55B (magenta), and Lamp1-mCh (blue) with a Halo-TMEM55B-positive lysosome tightly associating with GFP-Tex2-labeled ER membranes during intracellular transport. Time interval 2.53 s, 6.25 frames per second. Scale bar, 2 μm.
Tex2 is required for lysosomal functions
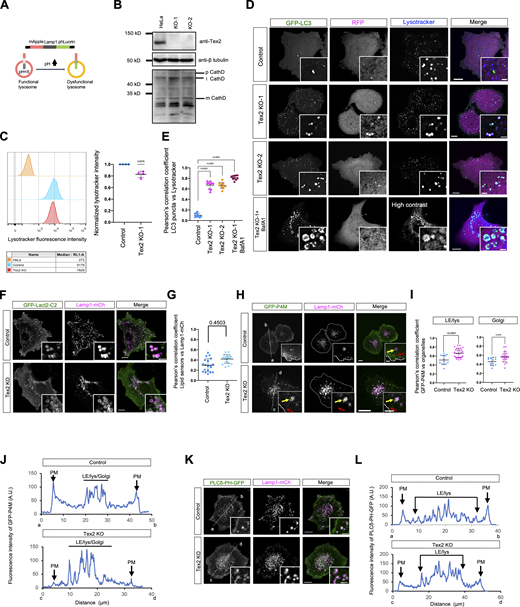
The finding that Tex2 acted at ER–LE/lys MCSs prompts us to explore its roles on these two organelles, in addition to the regulation of LE/lys trafficking. We showed that the localization of Tex2 was not affected by TG-induced ER stress (Fig. S3, H–N). Moreover, the initiation of unfolded protein response induced by ER stress appeared not to be influenced by Tex2 KO as the BiP level remained unchanged upon Tex2 KO (Fig. S3 H). On the other hand, we explored the impacts of Tex2 KO on the functions of LE/lys by a lysosomal function sensor mApple-Lamp1-phLuorin (Fig. S5 A). In control cells, very few LE/lys exhibited phLuorin fluorescence (Fig. 7 A), and the percentage of phLuorin-positive LE/lys strongly increased upon Bafilomycin A1 (BafA1) treatment (Fig. 7 A), a potent v-ATPase inhibitor that blocked lysosomal function. Strikingly, two independent Tex2-KO clones showed a remarkable increase in the percentage of phLuorin-positive LE/lys (Fig. 7 B), suggesting that Tex2 was required for lysosomal functions.
Supplementary data toFigs. 7 and 8. (A) Schematic cartoon of mApple-Lamp1-phLuorin. (B) Western blots show the level of CathepsinD in control or two Tex2-KO clones. (C) Left: representative flow cytometry analysis of either control or Tex2 KO HeLa stained with lysotracker using unstained HeLa cells as a control; right: normalized lysotracker fluorescence intensity of control or Tex2 KO cells as the left panel from four independent assays. One sample t test. Mean ± SD. (D) Representative images of either control (top), two Tex2-KO clones (middle), or Tex2-KO-1 cells treated with BafA1 expressing GFP–LC3–RFP were labeled with Lysotracker (blue) with insets. (E) Pearson’s correlation coefficient of GFP–LC3 vs. Lysotracker in control (10 cells), Tex2 KO-1 (10 cells), Tex2 KO-2 (10 cells), or Tex2 KO-1 treated with BafA1 (10 cells) in three independent assays. Ordinary one-way ANOVA with Tukey’s multiple comparisons test. Mean ± SD. (F) Representative images of either control (top) or Tex2 KO-1 (bottom) cells expressing GFP-Lact2-C2 (green; PS sensor) with insets. (G) Pearson’s correlation coefficient of GFP-Lact2-C2 vs. Lamp1-mCh (18 control; 20 Tex2-KO cells). Two-tailed unpaired Student’s t test. Mean ± SD. (H) Representative images of either control (top) or Tex2 KO-1 (bottom) cells expressing GFP-P4M (PI4P sensor) with insets. (I) Pearson’s correlation coefficient of GFP-P4M vs. either Lamp1-mCh (13 control; 31 Tex2-KO cells) or B4GalT1-TM-BFP (Golgi marker; 13 control; 21 Tex2-KO cells). Two-tailed unpaired Student’s t test. Mean ± SD. (J) Linescan analysis of GFP-P4M at LE/lys/Golgi relative to the PM in control or Tex2 KO-1 HeLa cells, as shown in H. (K) Representative images of either control (top) or Tex2 KO-1 (bottom) cells expressing PI(4,5)P2 sensor PLCδ-PH-GFP with insets. (L) Linescan analysis of PLCδ-PH-GFP at LE/lys relative to the PM in control or Tex2 KO-1 HeLa cells, as shown in K. Two-tailed unpaired Student’s t test. Mean ± SD. Scale bar, 10 μm in the whole cell images and 2 μm in the insets in D, F, H, and K. Source data are available for this figure: SourceData FS5.
Supplementary data toFigs. 7 and 8. (A) Schematic cartoon of mApple-Lamp1-phLuorin. (B) Western blots show the level of CathepsinD in control or two Tex2-KO clones. (C) Left: representative flow cytometry analysis of either control or Tex2 KO HeLa stained with lysotracker using unstained HeLa cells as a control; right: normalized lysotracker fluorescence intensity of control or Tex2 KO cells as the left panel from four independent assays. One sample t test. Mean ± SD. (D) Representative images of either control (top), two Tex2-KO clones (middle), or Tex2-KO-1 cells treated with BafA1 expressing GFP–LC3–RFP were labeled with Lysotracker (blue) with insets. (E) Pearson’s correlation coefficient of GFP–LC3 vs. Lysotracker in control (10 cells), Tex2 KO-1 (10 cells), Tex2 KO-2 (10 cells), or Tex2 KO-1 treated with BafA1 (10 cells) in three independent assays. Ordinary one-way ANOVA with Tukey’s multiple comparisons test. Mean ± SD. (F) Representative images of either control (top) or Tex2 KO-1 (bottom) cells expressing GFP-Lact2-C2 (green; PS sensor) with insets. (G) Pearson’s correlation coefficient of GFP-Lact2-C2 vs. Lamp1-mCh (18 control; 20 Tex2-KO cells). Two-tailed unpaired Student’s t test. Mean ± SD. (H) Representative images of either control (top) or Tex2 KO-1 (bottom) cells expressing GFP-P4M (PI4P sensor) with insets. (I) Pearson’s correlation coefficient of GFP-P4M vs. either Lamp1-mCh (13 control; 31 Tex2-KO cells) or B4GalT1-TM-BFP (Golgi marker; 13 control; 21 Tex2-KO cells). Two-tailed unpaired Student’s t test. Mean ± SD. (J) Linescan analysis of GFP-P4M at LE/lys/Golgi relative to the PM in control or Tex2 KO-1 HeLa cells, as shown in H. (K) Representative images of either control (top) or Tex2 KO-1 (bottom) cells expressing PI(4,5)P2 sensor PLCδ-PH-GFP with insets. (L) Linescan analysis of PLCδ-PH-GFP at LE/lys relative to the PM in control or Tex2 KO-1 HeLa cells, as shown in K. Two-tailed unpaired Student’s t test. Mean ± SD. Scale bar, 10 μm in the whole cell images and 2 μm in the insets in D, F, H, and K. Source data are available for this figure: SourceData FS5.
Tex2 KO affects lysosomal functions. (A and B) Representative images of control, control cells treated with BafA1 (400 nM; 2 h; A), or two HeLa Tex2-KO clones (B), expressing mApple-Lamp1-phLuorin (phLuorin in green; mApple-Lamp1 in magenta) with insets. (C–G) Representative images of two HeLa Tex2-KO clones transiently expressing either Halo-Tex2 (blue; C), Halo-Tex2-∆SMP (D), Halo-Tex2-NT (E), Halo-Tex2-∆1-276 (F), or Halo-Tex2-∆PH (G), and mApple-Lamp1-phLuorin with insets. (H) Percentage of phLuorin-positive lysosomes in control (14 cells), Tex2 KO-1 (11 cells), Tex2 KO-1 rescued by either Halo-Tex2 (13 cells), Halo-Tex2-∆SMP (11 cells), Halo-Tex2-NT (14 cells), Halo-Tex2-∆(1-276) (13 cells), or Halo-Tex2-∆PH (11 cells); and in Tex2 KO-2 (11 cells), Tex2 KO-2 rescued by either Halo-Tex2 (12 cells), Halo-Tex2-∆SMP (13 cells), Halo-Tex2-NT (14 cells), Halo-Tex2-∆(1-276) (12 cells), or Halo-Tex2-∆PH (14 cells) in more than three independent assays. Ordinary one-way ANOVA with Tukey’s multiple comparisons test. Mean ± SD. (I) Western blots demonstrate the level of Halo-Tex2 and Halo-Tex2 mutants in rescue experiments. Scale bar, 10 μm in the whole cell images and 2 μm in the insets in A–G. Source data are available for this figure: SourceData F7.
Tex2 KO affects lysosomal functions. (A and B) Representative images of control, control cells treated with BafA1 (400 nM; 2 h; A), or two HeLa Tex2-KO clones (B), expressing mApple-Lamp1-phLuorin (phLuorin in green; mApple-Lamp1 in magenta) with insets. (C–G) Representative images of two HeLa Tex2-KO clones transiently expressing either Halo-Tex2 (blue; C), Halo-Tex2-∆SMP (D), Halo-Tex2-NT (E), Halo-Tex2-∆1-276 (F), or Halo-Tex2-∆PH (G), and mApple-Lamp1-phLuorin with insets. (H) Percentage of phLuorin-positive lysosomes in control (14 cells), Tex2 KO-1 (11 cells), Tex2 KO-1 rescued by either Halo-Tex2 (13 cells), Halo-Tex2-∆SMP (11 cells), Halo-Tex2-NT (14 cells), Halo-Tex2-∆(1-276) (13 cells), or Halo-Tex2-∆PH (11 cells); and in Tex2 KO-2 (11 cells), Tex2 KO-2 rescued by either Halo-Tex2 (12 cells), Halo-Tex2-∆SMP (13 cells), Halo-Tex2-NT (14 cells), Halo-Tex2-∆(1-276) (12 cells), or Halo-Tex2-∆PH (14 cells) in more than three independent assays. Ordinary one-way ANOVA with Tukey’s multiple comparisons test. Mean ± SD. (I) Western blots demonstrate the level of Halo-Tex2 and Halo-Tex2 mutants in rescue experiments. Scale bar, 10 μm in the whole cell images and 2 μm in the insets in A–G. Source data are available for this figure: SourceData F7.
The lysosomal defect was specific to Tex2 depletion since the transient expression of WT Tex2 could completely rescue the phenotype in these two Tex2-KO clones (Fig. 7, C and H). Importantly, Tex2 with a deletion of lipid transfer domain SMP (Tex2-ΔSMP), a mutant capable of localizing to the MCSs, failed to rescue the lysosomal defect in two Tex2 KO clones (Fig. 7, D and H). Conversely, Tex2-NT could not effectively rescue the phenotype in these two Tex2 KO clones (Fig. 7, E and H), indicating that Tex2-NT, a mutant capable of interacting with TMEM55B, was insufficient for restoring the lysosomal defects. Moreover, the TMEM55B-binding defective mutant, Tex2-Δ(1-276), could restore the lysosomal defects, but to a very limited extent (Fig. 7, F and H), suggesting that Tex2–TMEM55B interaction is required but not sufficient in rescuing the phenotype. In addition, Tex2-ΔPH could partially rescue the phenotypes (Fig. 7, G and H), suggesting that the PH domain was not required but could promote lysosomal function. The striking difference between WT and Tex2 mutants in the rescue experiments was not due to their expression levels, as immunoblot assays showed similar levels of these proteins (Fig. 7 I). Collectively, our results indicated that both the Tex2-NT and Tex2-SMP were required for the Tex2 function with the SMP being the most important one, suggesting that the lipid transfer activity of Tex2 may be essential for the lysosomal function. Notably, since SMP domains typically mediate protein dimerization in addition to lipid transfer (Reinisch and De Camilli, 2016), our results could not distinguish which function of the SMP domain was required for lysosomal function.
The level of Cathepsin D, a lysosomal aspartyl protease in the lysosomal lumen, was examined to explore the effect of Tex2 KO on lysosomal function. The matured form of Cathepsin D was not substantially reduced in Tex2 KO compared with controls (Fig. S5 B), suggesting that Tex2 may not directly regulate the sorting or activities of lysosomal hydrolases. Furthermore, we explored whether Tex2 KO affected the lysosomal pH. Flow cytometry analysis showed that Tex2 KO moderately affected the lysosomal pH, as revealed by ∼15% reduction in mean fluorescence intensity of lysotracker in Tex2 KO than control cells (Fig. S5 C), thus supporting that Tex2 might not directly regulate lysosomal pH.
To gain more insights into Tex2 on the digestive functions of lysosomes, we examined the effects of Tex2 KO on autophagy, a fundamental process closely linked to lysosomal digestive capacity (Dikic and Elazar, 2018). We assessed the basal autophagic flow by a specific sensor RFP–LC3–GFP (Fig. 8 A; Kaizuka et al., 2016). Contrary to the control, in which few autophagosomes labeled by GFP–LC3 puncta were found under normal conditions, the number of autophagosomes was markedly increased in two Tex2 KO clones (Fig. 8, B and E). Importantly, the accumulation of autophagosomes in Tex2 KO could be almost completely rescued by WT–Tex2 other than Tex2–ΔSMP (Fig. 8, C, D, and E). Remarkably, our results showed that the majority of GFP–LC3-labeled autophagosomes were colocalized with Lamp1-labeled LE/lys under either normal or BafA1-treated Tex2-KO cells (Fig. S5, D and E). Therefore, the increase in autophagosome number resulting from Tex2 KO is likely due to the impaired digestive capacity of lysosomes instead of defective autophagosome–lysosome fusion. In addition, the flow of Rapamycin-induced autophagy was also blocked at the autolysosome stage in the two Tex2-KO clones, as revealed by a strong accumulation of autolysosomes labeled by GFP–LC3 and Lysotracker (Fig. 8, F and H), which was substantially rescued by WT–Tex2 other than Tex2–ΔSMP (Fig. 8, G and H), indicating that SMP is required for the proper autophagic flow.
Tex2 depletion results in impaired autophagic flow. (A) Schematic cartoon of GFP–LC3–RFP as an autophagic flow sensor. (B) Representative images of control or two HeLa Tex2-KO clones (middle) expressing GFP–LC3–RFP (GFP–LC3 in green; internal control RFP in magenta). (C) Representative images of two HeLa Tex2-KO clones transiently expressing either Halo-Tex2 (blue) or Halo-Tex2-∆SMP (blue) and GFP–LC3–RFP with insets. (D) Western blots demonstrate the level of Halo-Tex2 and Halo-Tex2-∆SMP in rescue experiments in C. (E) Normalized number of LC3-positive autophagosomes per cell under normal conditions in control (14 cells), Tex2 KO-1 (11 cells), Tex2 KO-1 rescued by Halo-Tex2 (13 cells), Tex2 KO-1 rescued by Halo-Tex2-∆SMP (11 cells), Tex2 KO-2 (12 cells), Tex2 KO-2 rescued by Halo-Tex2 (12 cells), and Tex2 KO-2 rescued by Halo-Tex2-∆SMP (11 cells) in three independent assays. The number of GFP–LC3 dots was normalized to the fluorescence intensity of RFP-LC3∆G. Ordinary one-way ANOVA with Tukey’s multiple comparisons test. Mean ± SD. (F) Representative images of control or two HeLa Tex2-KO clones expressing GFP–LC3–RFP under rapamycin treatments (400 nM; 2 h) with insets. (G) Representative images of two HeLa Tex2-KO clones expressing either Halo-Tex2 (blue) or Halo-Tex2-∆SMP (blue) and GFP–LC3–RFP under rapamycin treatments (400 nM; 2 h) with insets. (H) As in E, the normalized number of LC3-positive autophagosomes per cell under rapamycin stimulation in control (11 cells), Tex2 KO-1 (10 cells), Tex2 KO-1 rescued by Halo-Tex2 (10 cells), Tex2 KO-1 rescued by Halo-Tex2-∆SMP (11 cells), Tex2 KO-2 (13 cells), Tex2 KO-2 rescued by Halo-Tex2 (10 cells), and Tex2 KO-2 rescued by Halo-Tex2-∆SMP (11 cells) in three independent assays. Ordinary one-way ANOVA with Tukey’s multiple comparisons test. Mean ± SD. Scale bar, 10 μm in the whole cell images and 2 μm in the insets in B, C, F, and G. Source data are available for this figure: SourceData F8.
Tex2 depletion results in impaired autophagic flow. (A) Schematic cartoon of GFP–LC3–RFP as an autophagic flow sensor. (B) Representative images of control or two HeLa Tex2-KO clones (middle) expressing GFP–LC3–RFP (GFP–LC3 in green; internal control RFP in magenta). (C) Representative images of two HeLa Tex2-KO clones transiently expressing either Halo-Tex2 (blue) or Halo-Tex2-∆SMP (blue) and GFP–LC3–RFP with insets. (D) Western blots demonstrate the level of Halo-Tex2 and Halo-Tex2-∆SMP in rescue experiments in C. (E) Normalized number of LC3-positive autophagosomes per cell under normal conditions in control (14 cells), Tex2 KO-1 (11 cells), Tex2 KO-1 rescued by Halo-Tex2 (13 cells), Tex2 KO-1 rescued by Halo-Tex2-∆SMP (11 cells), Tex2 KO-2 (12 cells), Tex2 KO-2 rescued by Halo-Tex2 (12 cells), and Tex2 KO-2 rescued by Halo-Tex2-∆SMP (11 cells) in three independent assays. The number of GFP–LC3 dots was normalized to the fluorescence intensity of RFP-LC3∆G. Ordinary one-way ANOVA with Tukey’s multiple comparisons test. Mean ± SD. (F) Representative images of control or two HeLa Tex2-KO clones expressing GFP–LC3–RFP under rapamycin treatments (400 nM; 2 h) with insets. (G) Representative images of two HeLa Tex2-KO clones expressing either Halo-Tex2 (blue) or Halo-Tex2-∆SMP (blue) and GFP–LC3–RFP under rapamycin treatments (400 nM; 2 h) with insets. (H) As in E, the normalized number of LC3-positive autophagosomes per cell under rapamycin stimulation in control (11 cells), Tex2 KO-1 (10 cells), Tex2 KO-1 rescued by Halo-Tex2 (10 cells), Tex2 KO-1 rescued by Halo-Tex2-∆SMP (11 cells), Tex2 KO-2 (13 cells), Tex2 KO-2 rescued by Halo-Tex2 (10 cells), and Tex2 KO-2 rescued by Halo-Tex2-∆SMP (11 cells) in three independent assays. Ordinary one-way ANOVA with Tukey’s multiple comparisons test. Mean ± SD. Scale bar, 10 μm in the whole cell images and 2 μm in the insets in B, C, F, and G. Source data are available for this figure: SourceData F8.
A role of Tex2 in the maintenance of lipid composition of LE/lys membranes
It is intriguing that lysosomal digestive functions are dependent on the potential lipid transfer SMP domain, prompting us to explore whether Tex2 is required for the maintenance of the lipid composition of LE/lys membranes. To begin with, we asked whether the SMP domain of Tex2 might bind lipids by in vitro lipid-binding assays. Purified SMP domain comigrated with nitrobenzoxadiazole (NBD)-labeled glycerophospholipids (phosphatidylcholine [PC], phosphatidylserine [PS], phosphatidylethanolamine [PE]), and sphingolipids (ceramide [Cer]), but not cholesterol, as assessed by native gel electrophoresis (Fig. 9, A and B). Among these lipids, the SMP domain preferentially bound to PC, PS, and ceramide (Fig. 9 C).
A role of Tex2 in the maintenance of lipid composition of LE/lys membranes. (A) Coomassie blue staining of purified 14xHis-Tex2-SMP. (B) In vitro lipid-binding assays for Tex2-SMP. Purified Tex2-SMP was incubated with NBD-tagged lipids and examined by native PAGE. Phospholipids, visualized by their fluorescence, comigrated with protein, visualized by Coomassie blue staining. (C) Ratio of fluorescence of Tex2-SMP-bound lipids to the protein level. Ordinary one-way ANOVA followed by Tukey’s multiple comparisons test. Mean ± SD. (D) Schematic cartoon of non-targeting lipidomic analysis of endogenous GFP-Tex2. (E) Quantification of lipids bound to endogenous GFP-Tex2 from three independent assays. The lipid composition of total membranes in HEK293 cells are shown (Gao et al., 2022). (F) OptiPrep flotation assays showing the distribution of LE/lys membranes (anti-Lamp1) in consecutive 14 fractions from control or Tex2-KO HeLa cells with the eighth fraction being more abundant and specific. (G) Western blots showing the specificity of the fractions enriching ER membranes (anti-Calnexin) using the ER isolation kit from control or Tex2-KO HeLa cells, along with the purified LE/lys membranes from F. (H) Ratio of lipid species of the ER fractions or the LE/lys fractions in Tex2-KO relative to those from control HeLa cells from three independent assays. Mean ± SD. Source data are available for this figure: SourceData F9.
A role of Tex2 in the maintenance of lipid composition of LE/lys membranes. (A) Coomassie blue staining of purified 14xHis-Tex2-SMP. (B) In vitro lipid-binding assays for Tex2-SMP. Purified Tex2-SMP was incubated with NBD-tagged lipids and examined by native PAGE. Phospholipids, visualized by their fluorescence, comigrated with protein, visualized by Coomassie blue staining. (C) Ratio of fluorescence of Tex2-SMP-bound lipids to the protein level. Ordinary one-way ANOVA followed by Tukey’s multiple comparisons test. Mean ± SD. (D) Schematic cartoon of non-targeting lipidomic analysis of endogenous GFP-Tex2. (E) Quantification of lipids bound to endogenous GFP-Tex2 from three independent assays. The lipid composition of total membranes in HEK293 cells are shown (Gao et al., 2022). (F) OptiPrep flotation assays showing the distribution of LE/lys membranes (anti-Lamp1) in consecutive 14 fractions from control or Tex2-KO HeLa cells with the eighth fraction being more abundant and specific. (G) Western blots showing the specificity of the fractions enriching ER membranes (anti-Calnexin) using the ER isolation kit from control or Tex2-KO HeLa cells, along with the purified LE/lys membranes from F. (H) Ratio of lipid species of the ER fractions or the LE/lys fractions in Tex2-KO relative to those from control HeLa cells from three independent assays. Mean ± SD. Source data are available for this figure: SourceData F9.
Next, we sought to identify lipid species bound by full-length GFP-Tex2 in HEK293 cells. Lipid species associated with GFP-Tex2 were assessed by non-targeted lipidomics using liquid chromatography–tandem mass spectrometry (LC–MS/MS) with rigorous washes of the protein before lipid analysis, according to the protocol used in our previous study (Fig. 9 B; Gao et al., 2022). Remarkably, GFP–Tex2 was mainly associated with LPC/PC (∼50%) and sphingolipids (sphingosine [SPH] and Cer) (∼50%; Fig. 9 E). Taking into consideration the lipid composition of total membranes in HEK293 cells, PC accounted for ∼57% of the total phospholipids, whereas sphingolipids only accounted for <15% (Fig. 9 E; Gao et al., 2022). It is plausible that GFP–Tex2 may preferentially bind sphingolipids in cells. It should be noted that Tex2 may also bind phosphatidylinositol phosphates (PIPs) in cells, but the cellular level of PIPs might be too low to be detectable in our assays.
Next, we asked whether Tex2 was required for the proper cellular distribution of these Tex2-associated lipids by live-cell microscopy. We found that the distribution of PS was not substantially affected by Tex2 KO (Fig. S5, F and G), as revealed by GFP-Lact-C2 (Yeung et al., 2008), a PS sensor that specifically marks the PS on the cytoplasmic face of intracellular membranes. In addition, Tex2 KO caused a moderate increase in PI4P at LE/lys membranes, as revealed by a specific PI4P probe GFP-P4M that marked multiple cellular PI4P pools including the Golgi, endosomes, and the PM (Hammond et al., 2014; Fig. S5, H and I). In addition, linescan analysis indicated that Tex2 KO increased the ratio of LE/lys/Golgi PI4P to PM PI4P (Fig. S5 J). Importantly, Tex2 KO resulted in an increase in the ratio of lysosomal PI(4,5)P2 to PI(4,5)P2 on the PM (Fig. S5 K), as shown by the linescan analysis of a PI(4,5)P2 sensor GFP-PLCδ-PH (Stauffer et al., 1998; Fig. S5 L), in accord with a recent study in worms (Jeyasimman et al., 2021).
To get a better understanding of Tex2 in the regulation of lipid compositions of related organelles, we analyzed the abundance of lipid species of the ER and LE/lys membranes by non-targeted lipidomics using LC-MS/MS. The purity of the ER and LE/lys membrane fractions was examined by Western blots (Fig. 9, F and G). Lipidomics results showed that the levels of PG, Cer, and DG of the ER or LE/lys fractions were not substantially affected (Fig. 9 H). In contrast, the levels of PC and SPH of the ER fraction were significantly increased (Fig. 9 H). Importantly, Tex2 KO resulted in a significant increase in the level of SPH in the LE/lys fractions compared with the control, whereas the level of ceramide was not substantially changed upon Tex2 KO. This line of evidence suggests that the aberrant accumulation of SPH might account for the lysosomal dysfunction in Tex2 KO cells. Indeed, accumulation of SPH, which was observed in the lysosomes of Niemann-Pick disease type C patient cells (te Vruchte et al., 2004), was reported to disturb lysosomal calcium homeostasis (Höglinger et al., 2015), a prerequisite for lysosomal functions and autophagy (Tedeschi et al., 2019). Furthermore, the levels of PC and PE were also significantly changed in LE/lys fractions of Tex2 KO cells. In addition, our microscopy results using lipid probes showed that levels of PI(4,5)P2 and PI4P at LE/lys were higher in Tex2 KO than those in control cells. Collectively, these lines of evidence showed that Tex2 depletion resulted in a defect in the lipid composition of LE/lys membranes, which might have accounted for the lysosomal defect in Tex2 KO cells.
Discussion
In this manuscript, we proposed that a putative LTP Tex2 localized exclusively to tubular ER and was recruited to ER–LE/lys MCSs via LE/lys-resident PI phosphatases TMEM55. The recruitment of Tex2 by TMEM55 was negatively regulated by the activities of LE/lys-resident PI4KIIs. The loss of Tex2 at these contacts results in defects in LE/lys trafficking, lysosomal digestive functions, and lipid composition of the LE/lys membranes (Fig. 10).
Working model of Tex2 functions at ER–LE/lys MCSs. (A) Functional dissection of Tex2. Tex2-NT (1-517) containing essential residues 1-277 is required for binding the catalytic motif CX5R of TMEM55. The residues 278-514 is required for targeting Tex2 to tubular ER. The SMP domain is likely responsible for transferring glycerophospholipids and sphingolipids between the ER and LE/lys. The PH domain of Tex2 may be involved in the regulation of Tex2–TMEM55 interactions. (B) Tex2 resides exclusively on tubular ER, is recruited to ER–LE/lys MCSs via LE/lys-resident PI phosphatases TMEM55. The Tex2–TMEM55 interaction is negatively regulated by PI4KII activities. The loss of Tex2 at these contacts resulted in defects in lysosomal digestive functions and impaired autophagic flow, possibly due to the defective lipid composition of LE/lys membranes.
Working model of Tex2 functions at ER–LE/lys MCSs. (A) Functional dissection of Tex2. Tex2-NT (1-517) containing essential residues 1-277 is required for binding the catalytic motif CX5R of TMEM55. The residues 278-514 is required for targeting Tex2 to tubular ER. The SMP domain is likely responsible for transferring glycerophospholipids and sphingolipids between the ER and LE/lys. The PH domain of Tex2 may be involved in the regulation of Tex2–TMEM55 interactions. (B) Tex2 resides exclusively on tubular ER, is recruited to ER–LE/lys MCSs via LE/lys-resident PI phosphatases TMEM55. The Tex2–TMEM55 interaction is negatively regulated by PI4KII activities. The loss of Tex2 at these contacts resulted in defects in lysosomal digestive functions and impaired autophagic flow, possibly due to the defective lipid composition of LE/lys membranes.
ER tubules have evolved to play specific and fundamental roles in mammalian cells. One of such functions is to synthesize the majority of lipids and transfer them to other organelles for organelle biogenesis, trafficking, and alleviation of lipitoxicity (Prinz et al., 2020). Our findings coordinate ER tubule formation, PI4P species homeostasis on LE/lys membranes, and lipid transfer across ER–LE/lys MCSs. To begin with, our findings demonstrate the localization of Tex2 on tubular ER. The RTN proteins localize exclusively to tubular ER via a conserved C-terminal reticulon homology domain (RHD), which forms a characteristic hairpin TM domain that targets RTN proteins to regions of high membrane curvature including tubular ER membranes (Voeltz et al., 2006). We found that Tex2 is exclusively localized to tubular ER via a region (residues 277–474) of the NT adjacent to the TM domain. The question of how the region targets Tex2 to ER tubules remains elusive. We found two potential amphipathic helices (APH) in this region, but disruption of these two APH by point mutations did not block the targeting of Tex2 to ER tubules (unpublished results). Notably, the overexpression of Tex2 could counteract the effects of Climp63 overexpression in the expansion of ER sheets at the cell periphery. However, Tex2 KO did not substantially affect the ER tubule-sheet ratio at the cell periphery. It may suggest that Tex2 is not required for ER tubule formation in cells, which is different from the role of RTN4, the prototype of RTN proteins that generates and maintains the structure of the tubular ER network (Voeltz et al., 2006).
Intracellular LTPs mediate lipid transport between opposing membranes at MCSs through two modes of action—shuttling or bridging. Shuttle transporters typically extract one or two lipid molecules from the membrane of the donor organelle, solubilize it during transport through the cytosol and deposit it in the acceptor organelle membrane, while bridge transporters feature an extended channel, most likely lined with hydrophobic residues that bind tens of lipids at once (Li et al., 2020; Ugur et al., 2020; Wong et al., 2019). In addition to Tex2, a complex of multiple SMP domains is suggested to act as a shuttle transporter for glycerophospholipids and/or ceramides across MCSs in yeast and metazoan, including maintenance of mitochondrial morphology protein 1 (Mmm1) of ERMES (Kornmann et al., 2009) and Nvj2 (Liu et al., 2017) in yeast, and E-Syts (Bian et al., 2018; Giordano et al., 2013), Transmembrane protein 24 (TMEM24; also known as C2CD2L; Lees et al., 2017), and PDZD8 (Gao et al., 2022). To ensure the productivity of lipid shuttling across MCSs, a directional lipid transfer should be fulfilled. Nvj2p, the yeast homolog of Tex2, may mediate a directional ceramide transfer from the ER to the Golgi apparatus for sphingolipid synthesis during ER stress, possibly driven by ceramide gradients (Liu et al., 2017). The direction and driving force of Tex2-mediated lipid transport are unknown. It is plausible that the PI4,5P2 to PI5P conversion catalyzed by TMEM55B may be coupled to the lipid transfer of Tex2 and contributes to a directional lipid transport at ER–LE/lys MCSs in a similar manner as the SMP-containing LTP TMEM24, which directionally transports phosphatidylinositols over other phospholipids at ER-PM MCSs (Lees et al., 2017). Does Tex2 bind these PIPs and other phospholipids (PC or ceramide) via the same sites of SMP? Does Tex2 transfer PIPs, for example, PI5P, over PC or ceramide in cells? These critical questions warrant future investigations.
Tex2 is highly expressed in testes, and our findings highlight the importance of Tex2 in regulating lysosomal digestive function, which raises a question of whether Tex2 is required in the biogenesis of acrosome, a specialized lysosome-like-organelle containing hydrolases in sperm for egg penetration (Moreno and Alvarado, 2006).
Materials and methods
Cell culture, transfection, and RNAi
The African green monkey kidney fibroblast-like COS7 cell line (ATCC), human bone osteosarcoma epithelial U2OS cells (ATCC), and human embryonic kidney 293T (ATCC) were grown in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (Gibco). All the cell lines used in this study were confirmed free of mycoplasma contamination.
Transfection of plasmids and RNAi oligos was carried out with Lipofectamine 2000 and RNAi MAX, respectively. For transfection, cells were seeded at 4 × 105 cells per well in a six-well dish ∼16 h before transfection. Plasmid transfections were performed in OPTI-MEM (Invitrogen) with 2 μl Lipofectamine 2,000 per well for 6 h, followed by trypsinization and replating onto glass-bottom confocal dishes at ∼3.5 × 105 cells per well. Cells were imaged in live-cell medium (DMEM with 10% FBS and 20 mM Hepes with no penicillin or streptomycin) ∼16–24 h after transfection. For all transfection experiments in this study, the following amounts of DNA were used per 3.5-cm well (individually or combined for cotransfection): 1,500 ng for GFP-Tex2 and its mutants; 500 ng for Halo-TMEM55A/B and its mutants, Lamp1-Halo; and 1,500 ng for ER-tagRFP. For siRNA transfections, cells were plated on 3.5-cm dishes at 30–40% density, and 2 μl Lipofectamine RNAimax (Invitrogen) and 50 ng siRNA were used per well. At 48 h after transfection, a second round of transfection was performed with 50 ng siRNAs. Cells were analyzed 24 h after the second transfection for suppression.
Plasmids
Tex2 (NM_018469.5), TMEM55A (NM_018710.3), TMEM55B (NM_001100814.3), ALT1, ARL6IP1, E-Syt1, RTN4, and Reep5 were cloned from HeLa cDNA. The ORFs of Tex2, TMEM55B, TMEM55A, ALT1, PI4KIIα, PI4KII β, and E-syt1 were cloned into mEGFP-C1 between the BglII and SacI or Halo-C1 vector between the SacI and BamHI. The ORFs of Reep5, RTN4, and MGAT2 were cloned between NheI and SacI of Halo-N1. GST-TMEM55B was cloned into PGEX-2T vector between the BamHI and EcoRI. 14xHis-NEDD8-Tex2-SMP was cloned into 14xHis-NEDD8 vector between the BamHI and HindIII. ER-tagRFP and Lamp1-mCh were previously described (Gao et al., 2022; Ji et al., 2017). GFP-Lact-C2 (22852; Addgene), CFP-C1-PLCδ-PH (21262; Addgene), GFP-P4M-SidM (51469; Addgene), mApple-Lamp1-phLuorin-N-8 (54918; Addgene), and PMRX–IP–GFP–LC3–RFP (84573; Addgene) were purchased from Addgene.
CRISPR-Cas9-mediated gene editing
To make Tex2 KO HeLa cell lines, two gRNAs (5′-CCTCTGCACGTGCACTTTAG-3′ and 5′-CAAGTTGGCCATGACCCCGC-3′) were used to delete ∼190 bp from exon 1 of Tex2 gene (Fig. S1 G). Complementary gRNAs were annealed and subcloned into the pSpCas9(BB)-2A-GFP (pX-458) vector (#48138; Addgene) between BbsI endonuclease restriction sites. Upon transfection, HeLa cells were grown in an antibiotic-free medium for 48 h, followed by single-cell sorting by fluorescence-based flow cytometry. Two independent clones were verified by imaging and Western blots (Fig. S1 H). To make GFP-Tex2-KI HeLa cell line, a single gRNA (5′-CCGGCAATGACAAGTCTGTA-3′) was used to target the N-terminus of the Tex2 gene. HeLa cells were transfected with plasmids encoding the gRNA and a donor construct containing super folder GFP (sfGFP) and two homologous arms using Lipofectamine 2000 (Fig. S1 E). 48 h after transfection, single clones were sorted. A positive clone was verified by imaging and Western blots (Fig. S2 F). Table 1 shows all the reagents, including antibodies, drugs, siRNAs, and polar lipids, used in this study. Table 2 shows all the primers used in this study.
Antibodies, drugs, siRNAs, and polar lipids used in this study
| Reagent or Resource . | Source . | Identifier . |
|---|---|---|
| Antibodies and reagents | ||
| Tex2 Rabbit pAb | ABclonal | A14686 |
| GFP antibody | CST | 2555 |
| Anti-Halo Tag pAB | Promega | G928A |
| His Tag Antibody, mAb, Mouse | Genscript | A00186 |
| GST-Tag | Sinobiological | 11213-MM01 |
| Cathepsin D | CST | 2284S |
| Purified Mouse Anti-GM130 | BD Biosciences | 610822 |
| LAMP-1 (H4A3): | Santa cruz | sc-20011 |
| Calnexin Rabbit pAb | ABclonal | A15631 |
| COXIV Polyclonal antibody | Proteintech | 11242-1-AP |
| β-Tubulin mAB(3G6) | Abbkine | A01030 |
| TMEM55B Polyclonal antibody | Proteintech | 23992-1-AP |
| Thapsigargin | Abcam | ab120286 |
| Lysotracker deep red | Thermo Fisher Scientific | L12492 |
| Bafilomycin A1 | MCE | HY-100558 |
| Rapamycin | MCE | HY-10219 |
| siRNA | ||
| siRNA human ARL6IP1#1: 5′-GUACAGUUCAAGUGAAUCUGGAUAA-3′ | (Dong et al., 2018) | N/A |
| siRNA human ARL6IP1#2: 5′-UUAUCCAGAUUCACUUGAACUGUACUA-3′ | (Dong et al., 2018) | N/A |
| siRNA human Reep5: 5′-CGAAGAAAGCUACCGUGAATT-3′ | (Kumar et al., 2019) | N/A |
| siRNA human RTN1: 5′-UAGAUGCGGAAACUGAUGGTT-3′ | (Anderson and Hetzer, 2008) | N/A |
| siRNA human RTN3: 5′-CCUUCUAAUUCUUGCUGAATT-3′ | (Anderson and Hetzer, 2008) | N/A |
| siRNA human RTN4#1: 5′-GATCCAAGCTATCCAGAAA-3′ | Ribobio | N/A |
| siRNA human RTN4#2: 5′-GTGTTGATGTGGGTATTTA-3′ | Ribobio | N/A |
| siRNA human RTN4#3: 5′-GGAATCTGAAGTTGCTATA-3′ | Ribobio | N/A |
| siRNA human scrambled siRNA 5′-CGUUAAUCGCGUAUAAUACGCGUAT-3′ | Ribobio | N/A |
| siRNA human PI4KIIα: 5′-GGUUGGUGGUGCUGGAUUATT-3′ | Ribobio | N/A |
| siRNA human PI4KIIβ: 5′-GGUUCAAGUGGAAGUUACUTT-3′ | Ribobio | N/A |
| siRNA human TMEM55B#1: 5′-GCATCAGCATGTAGTCAAA-3′ | Ribobio | N/A |
| siRNA human TMEM55B#2: 5′-GCCAATCTCTCATCAACGT-3′ | Ribobio | N/A |
| siRNA human TMEM55B#3: 5′-GGGCATTTGTCATCCTGTT-3′ | Ribobio | N/A |
| Polar lipids | ||
| NBD-PC | Avanti | 810133 |
| NBD-PS | Avanti | 810198 |
| NBD-PA | Avanti | 810138 |
| NBD-PE | Avanti | 810144 |
| NBD-Cholesterol | Avanti | 810252 |
| NBD-Ceramide | Avanti | 810211 |
| Reagent or Resource . | Source . | Identifier . |
|---|---|---|
| Antibodies and reagents | ||
| Tex2 Rabbit pAb | ABclonal | A14686 |
| GFP antibody | CST | 2555 |
| Anti-Halo Tag pAB | Promega | G928A |
| His Tag Antibody, mAb, Mouse | Genscript | A00186 |
| GST-Tag | Sinobiological | 11213-MM01 |
| Cathepsin D | CST | 2284S |
| Purified Mouse Anti-GM130 | BD Biosciences | 610822 |
| LAMP-1 (H4A3): | Santa cruz | sc-20011 |
| Calnexin Rabbit pAb | ABclonal | A15631 |
| COXIV Polyclonal antibody | Proteintech | 11242-1-AP |
| β-Tubulin mAB(3G6) | Abbkine | A01030 |
| TMEM55B Polyclonal antibody | Proteintech | 23992-1-AP |
| Thapsigargin | Abcam | ab120286 |
| Lysotracker deep red | Thermo Fisher Scientific | L12492 |
| Bafilomycin A1 | MCE | HY-100558 |
| Rapamycin | MCE | HY-10219 |
| siRNA | ||
| siRNA human ARL6IP1#1: 5′-GUACAGUUCAAGUGAAUCUGGAUAA-3′ | (Dong et al., 2018) | N/A |
| siRNA human ARL6IP1#2: 5′-UUAUCCAGAUUCACUUGAACUGUACUA-3′ | (Dong et al., 2018) | N/A |
| siRNA human Reep5: 5′-CGAAGAAAGCUACCGUGAATT-3′ | (Kumar et al., 2019) | N/A |
| siRNA human RTN1: 5′-UAGAUGCGGAAACUGAUGGTT-3′ | (Anderson and Hetzer, 2008) | N/A |
| siRNA human RTN3: 5′-CCUUCUAAUUCUUGCUGAATT-3′ | (Anderson and Hetzer, 2008) | N/A |
| siRNA human RTN4#1: 5′-GATCCAAGCTATCCAGAAA-3′ | Ribobio | N/A |
| siRNA human RTN4#2: 5′-GTGTTGATGTGGGTATTTA-3′ | Ribobio | N/A |
| siRNA human RTN4#3: 5′-GGAATCTGAAGTTGCTATA-3′ | Ribobio | N/A |
| siRNA human scrambled siRNA 5′-CGUUAAUCGCGUAUAAUACGCGUAT-3′ | Ribobio | N/A |
| siRNA human PI4KIIα: 5′-GGUUGGUGGUGCUGGAUUATT-3′ | Ribobio | N/A |
| siRNA human PI4KIIβ: 5′-GGUUCAAGUGGAAGUUACUTT-3′ | Ribobio | N/A |
| siRNA human TMEM55B#1: 5′-GCATCAGCATGTAGTCAAA-3′ | Ribobio | N/A |
| siRNA human TMEM55B#2: 5′-GCCAATCTCTCATCAACGT-3′ | Ribobio | N/A |
| siRNA human TMEM55B#3: 5′-GGGCATTTGTCATCCTGTT-3′ | Ribobio | N/A |
| Polar lipids | ||
| NBD-PC | Avanti | 810133 |
| NBD-PS | Avanti | 810198 |
| NBD-PA | Avanti | 810138 |
| NBD-PE | Avanti | 810144 |
| NBD-Cholesterol | Avanti | 810252 |
| NBD-Ceramide | Avanti | 810211 |
Primers used in this study
| Primers . | Sequences (5′-3′) . |
|---|---|
| GFP-Tex2-F | 5′-TACAAGTCCGGACTCAGATCTACAAGTCTGTATGGTCGCCATG-3′ |
| GFP-Tex2-R | 5′-CAGAATTCGAAGCTTGAGCTCTCATGGCTGATCAGCAGCCT-3′ |
| GFP-Tex2(1-474)-R | 5′-CAGAATTCGAAGCTTGAGCTCCTACTTCACTGGCAATTCCGGG-3′ |
| GFP-Tex2(1-540)-R | 5′-CAGAATTCGAAGCTTGAGCTCTCAGATATCCAGAGATCTTGTGTTCC-3′ |
| GFP-Tex2(474-1131)-F | 5′-TACAAGTCCGGACTCAGATCTACGTTGGGCTTCTTTATAATGTGTG-3′ |
| GFP-Tex2(1-276)-R | 5′-CAGAATTCGAAGCTTGAGCTCCTAGGAACGAGTATCAGAGGGAGAA-3′ |
| GFP-Tex2(277-517)-F | 5′-TACAAGTCCGGACTCAGATCTCGTTCCTTTTTTAAAGTGCCCG-3′ |
| GFP-Tex2(277-517)-R | 5′-CAGAATTCGAAGCTTGAGCTCTTATGTAAAAAACCAAATCACGCAA-3′ |
| GFP--Tex2(∆ 277-473)-F | 5′-GTTCCTTTACGTTGGGCTTCTTTATAATGTGTG-3′ |
| GFP--Tex2(∆ 277-473)-R | 5′-GCCCAACGTAAAGGAACGAGTATCAGAGGGAGA-3′ |
| Halo-Tex2-F | 5′-CTGGAGATTTCCCTCGAGCTCACAAGTCTGTATGGTCGCCATG-3′ |
| Halo-Tex2-R | 5′-TTATCTAGATCCGGTGGATCCTCATGGCTGATCAGCAGCCT-3′ |
| Tex2(TM)-GFP-F | 5′-TGAACCGTCAGATCCGCTAGCACGTTGGGCTTCTTTATAATGTGTG-3′ |
| Tex2(TM)-GFP-R | 5′-CAGAATTCGAAGCTTGAGCTCTGTAAAAAACCAAATCACGCAAA-3′ |
| GFP-Tex2(∆ TM)-F | 5′-CAGTGAAGCCACCAAGTGCTCATAAATATCACAAG-3′ |
| GFP-Tex2(∆ TM)-R | 5′-ACTTGGTGGCTTCACTGGCAATTCCGGGTGCT-3′ |
| Halo-Tex2(∆ PH)-F | 5′-GAAGAAATCAAGAAGTCATCGGGTGTCTCTGG-3′ |
| Halo-Tex2(∆ PH)-R | 5′-GACTTCTTGATTTCTTCTTTGATATCCAGAGATCTTGTGTT-3′ |
| GFP-Tex2(SMP)-F | 5′-CCACCCTCTATAATGCACTCAGCCATGGACCC-3′ |
| GFP-Tex2(SMP)-R | 5′-TGCATTATAGAGGGTGGAACCTCTGGCAACTT-3′ |
| GFP/Halo-Tex2(∆SMP)-F | 5′-CCACCCTCtATAATGCACTCAGCCATGGACCC-3′ |
| GFP/Halo-Tex2(∆SMP)-R | 5′-TGCATTATaGAGGGTGGAACCTCTGGCAACTT-3′ |
| GFP-Tex2(∆ TM-∆PH)-F | 5′-CAGTGAAGGAGGAGGAAGAACAGGAAGCCTGG-3′ |
| GFP-Tex2(∆ TM-∆PH)-R | 5′-TTCCTCCTCCTTCACTGGCAATTCCGGGTGCT-3′ |
| 14xHis-NEDD8-Tex2(PH)-F | 5′-GGCGCCGGTACCTGCGGATCCCCACCAAGTGCTCATAAATATCACA-3′ |
| 14xHis-NEDD8-Tex2(PH)-R | 5′-GCTCAGCTAATTAAGAAGCTTTCAAGAGGGTGGAACCTCTGG-3′ |
| 14xHis-NEDD8-Tex2(SMP)-F | 5′-GGCGCCGGTACCTGCGGATCCGAGGAGGAAGAACAGGAAGCCT-3′ |
| 14xHis-NEDD8-Tex2(SMP)-R | 5′-GCTCAGCTAATTAAGAAGCTTAGTGATATAAACATCATCCATGTTTGG-3′ |
| Halo-TMEM55A-F | 5′-CTGGAGATTTCCCTCGAGCTCATGGCTGCCGAGGGGGTG-3′ |
| Halo-TMEM55A-R | 5′-GTACCGTCGACTGCAGAATTCCTATGCAAAACTGTGCTCTGGATAA-3′ |
| Halo-TMEM55B-F | 5′-CTGGAGATTTCCCTCGAGCTCATGGCGGCAGATGGAGAGC-3′ |
| Halo-TMEM55B-R | 5′-TTATCTAGATCCGGTGGATCCTCAGGAGAAGTTCTGGACAGGG-3′ |
| BFP-TMEM55B-F | 5′-TGAACCGTCAGATCCGCTAGCGCCACCATGAGCGAGCTGATTAAGGAGAA-3′ |
| BFP-TMEM55B-R | 5′-TCCATCTGCCGCCATGAGCTCATTAAGCTTGTGCCCCAGTTTG-3′ |
| TMEM55B-∆X5R-F | 5′-CCTTATCATTGCATGCCCTCGGCCCTACTGCA-3′ |
| TMEM55B-∆X5R-R | 5′-GGCATGCAATGATAAGGAGACAGTTACAGGGGCA-3′ |
| TMEM55B(∆1-74)-F | 5′-CTCGAGCTCTATTCACCCTTAACTAGCCCGGA-3′ |
| TMEM55B(∆1-74)-R | 5′-GGTGAATAGAGCTCGAGGGAAATCTCCAGAGT-3′ |
| TMEM55B- ∆TM-F | 5′-TATTGGGCGCTGAGGATCCACCGGATCTAGATAAC-3′ |
| TMEM55B- ∆TM-R | 5′-ATCCTCAGCGCCCAATAGATGACACTTTCCTG-3′ |
| TMEM55B(∆1-163)-F | 5′-TTTCCCTCGAGCTCCATCCCGGACCTCTGAGTCC-3′ |
| TMEM55B(∆1-163)-R | 5′-ATGGAGCTCGAGGGAAATCTCCAGAGTAGACA-3′ |
| TMEM55B-∆PTase(73-163)-F | 5′-ACATCCCGGACCTCTGAGTCCAGAACCCCAAC-3′ |
| TMEM55B-∆PTase(73-163)-R | 5′-TCAGAGGTCCGGGATGTGGGTCCTCCCCAGGCAAC-3′ |
| TMEM55B C140W-F | 5′-TCTGGAAAGTGACATCCCAACGGATTGCATGC-3′ |
| TMEM55B C140W-R | 5′-GGATGTCACTTTCCAGATAAGGAGACAGTTACAGGGGCA-3′ |
| TMEM55B C140S-F | 5′-TCTCGAAAGTGACATCCCAACGGATTGCATGC-3′ |
| TMEM55B C140S-R | 5′-GGATGTCACTTTCGAGATAAGGAGACAGTTACAGGGGCA-3′ |
| GST-TMEM55B-F | 5′-GATCTGGTTCCGCGTGGATCCATGGCGGCAGATGGAGAGC-3′ |
| GST-TMEM55B-R | 5′-TCAGTCAGTCACGATGAATTCTCAGGAGAAGTTCTGGACAGGG-3′ |
| Halo-PI4KIIα-F | 5′-CTGGAGATTTCCCTCGAGCTCATGGACGAGACGAGCCCAC-3′ |
| Halo-PI4KIIα-R | 5′-TTATCTAGATCCGGTGGATCCTCACCACCATGAAAAGAAGGGC-3′ |
| Halo-PI4KIIβ-F | 5′-CTGGAGATTTCCCTCGAGCTCATGGAGGATCCCTCCGAGC-3′ |
| Halo-PI4KIIβ-R | 5′-TTATCTAGATCCGGTGGATCCTCACCAGGAGGAAAAAAATGGC-3′ |
| PI4KIIα W359A-F | 5′-ACTCCGCTAGGGCATATCCTTTTTACTGGGCC-3′ |
| PI4KIIα W359A-R | 5′-ATATGCCCTAGCGGAGTCAGGATGCTTCAGTGGG-3′ |
| RTN4-Halo/GFP-F | 5′-TGAACCGTCAGATCCGCTAGCGCCACCATGGAAGACCTGGACCAGTCTCC-3′ |
| RTN4-Halo/GFP-R | 5′-CAGAATTCGAAGCTTGAGCTCTTCAGCTTTGCGCTTCAATCC-3′ |
| Halo-ARL6IP1-F | 5′-CTGGAGATTTCCCTCGAGCTCATGGCGGAGGGAGATAATCG-3′ |
| Halo-ARL6IP1-R | 5′-TTATCTAGATCCGGTGGATCCTCATTCGTTTTTCTTTTCTTTTTGTT-3′ |
| Halo-ATL1-F | 5′-CTGGAGATTTCCCTCGAGCTCATGGCCAAGAACCGCAGG-3′ |
| Halo-ATL1-R | 5′-TTATCTAGATCCGGTGGATCCTTACATTTTTTTCTTTTCTGATTGTTCA-3′ |
| Climp63-halo-F | 5′-TGAACCGTCAGATCCGCTAGCATGCCCTCGGCCAAACAA-3′ |
| Climp63-halo-R | 5′-CGACTGCAGAATTCGAAGCTTGACCTTTTCGTGAATCTTCTCCA-3′ |
| Halo-E-Syt1-F | 5′-CTGGAGATTTCCCTCGAGCTCATGGAGCGATCTCCAGGAGAG-3′ |
| GFP-E-Syt1-F | 5′-TACAAGTCCGGACTCAGATCTATGGAGCGATCTCCAGGAGAG-3′ |
| Halo/GFP-E-Syt1-R | 5′-TTATCTAGATCCGGTGGATCCCTAGGAGCTGCCCTTGTCCTT-3′ |
| Reep5-Halo-F | 5′-CCGGACTCAGATCTCGAGCTCGCCACCATGTCTGCGG-3′ |
| Reep5-Halo-R | 5′-CGGTGGATCCCGGGCCCGCGGGGTGCTCTTCTTTTCTTCACCCA-3′ |
| MGAT2-Halo-F | 5′-TGAACCGTCAGATCCGCTAGCGCCACCATGAGGTTCCGCATCTACAAACG-3′ |
| MGAT2-Halo-R | 5′-CGACTGCAGAATTCGAAGCTTCTGCAGTCTTCTATAACTTTTACAGAGTTC-3′ |
| Tex2 KO sgRNA1-F | 5′-CACCGCCTCTGCACGTGCACTTTAG-3′ |
| Tex2 KO sgRNA1-R | 5′-AAACCTAAAGTGCACGTGCAGAGGC-3′ |
| Tex2 KO sgRNA2-F | 5′-CACCGCAAGTTGGCCATGACCCCGC-3′ |
| Tex2 KO sgRNA2-R | 5′-AAACGCGGGGTCATGGCCAACTTGC-3′ |
| Tex2 KI sgRNA-F | 5′-CACCGCCGGCAATGACAAGTCTGTA-3′ |
| Tex2 KI sgRNA-R | 5′-AAACTACAGACTTGTCATTGCCGGC-3′ |
| Tex2 LHA-F | 5′-GTCGACGGTATCGATAAGCTTAGTCCTTGGTAGAGTTCTTGGTTGG-3′ |
| Tex2 LHA-R | 5′-TTTGCTCATTGCCGGCTTCACAAGGGC-3′ |
| sfGFP-F | 5′-TGAAGCCGGCAATGAGCAAAGGAGAAGAACTTTTCA-3′ |
| sfGFP-R | 5′-GGCGACCATACAGACTTGTTTTGTAGAGCTCATCCATGCCA-3′ |
| Tex2 RHA-F | 5′-AACAAGTCTGTATGGTCGCCATG-3′ |
| Tex2 RHA-R | 5′-CTATAGGGCGAATTGGAGCTCAGTATCAGAGGGAGAAGTCAGTGGG-3′ |
| Primers . | Sequences (5′-3′) . |
|---|---|
| GFP-Tex2-F | 5′-TACAAGTCCGGACTCAGATCTACAAGTCTGTATGGTCGCCATG-3′ |
| GFP-Tex2-R | 5′-CAGAATTCGAAGCTTGAGCTCTCATGGCTGATCAGCAGCCT-3′ |
| GFP-Tex2(1-474)-R | 5′-CAGAATTCGAAGCTTGAGCTCCTACTTCACTGGCAATTCCGGG-3′ |
| GFP-Tex2(1-540)-R | 5′-CAGAATTCGAAGCTTGAGCTCTCAGATATCCAGAGATCTTGTGTTCC-3′ |
| GFP-Tex2(474-1131)-F | 5′-TACAAGTCCGGACTCAGATCTACGTTGGGCTTCTTTATAATGTGTG-3′ |
| GFP-Tex2(1-276)-R | 5′-CAGAATTCGAAGCTTGAGCTCCTAGGAACGAGTATCAGAGGGAGAA-3′ |
| GFP-Tex2(277-517)-F | 5′-TACAAGTCCGGACTCAGATCTCGTTCCTTTTTTAAAGTGCCCG-3′ |
| GFP-Tex2(277-517)-R | 5′-CAGAATTCGAAGCTTGAGCTCTTATGTAAAAAACCAAATCACGCAA-3′ |
| GFP--Tex2(∆ 277-473)-F | 5′-GTTCCTTTACGTTGGGCTTCTTTATAATGTGTG-3′ |
| GFP--Tex2(∆ 277-473)-R | 5′-GCCCAACGTAAAGGAACGAGTATCAGAGGGAGA-3′ |
| Halo-Tex2-F | 5′-CTGGAGATTTCCCTCGAGCTCACAAGTCTGTATGGTCGCCATG-3′ |
| Halo-Tex2-R | 5′-TTATCTAGATCCGGTGGATCCTCATGGCTGATCAGCAGCCT-3′ |
| Tex2(TM)-GFP-F | 5′-TGAACCGTCAGATCCGCTAGCACGTTGGGCTTCTTTATAATGTGTG-3′ |
| Tex2(TM)-GFP-R | 5′-CAGAATTCGAAGCTTGAGCTCTGTAAAAAACCAAATCACGCAAA-3′ |
| GFP-Tex2(∆ TM)-F | 5′-CAGTGAAGCCACCAAGTGCTCATAAATATCACAAG-3′ |
| GFP-Tex2(∆ TM)-R | 5′-ACTTGGTGGCTTCACTGGCAATTCCGGGTGCT-3′ |
| Halo-Tex2(∆ PH)-F | 5′-GAAGAAATCAAGAAGTCATCGGGTGTCTCTGG-3′ |
| Halo-Tex2(∆ PH)-R | 5′-GACTTCTTGATTTCTTCTTTGATATCCAGAGATCTTGTGTT-3′ |
| GFP-Tex2(SMP)-F | 5′-CCACCCTCTATAATGCACTCAGCCATGGACCC-3′ |
| GFP-Tex2(SMP)-R | 5′-TGCATTATAGAGGGTGGAACCTCTGGCAACTT-3′ |
| GFP/Halo-Tex2(∆SMP)-F | 5′-CCACCCTCtATAATGCACTCAGCCATGGACCC-3′ |
| GFP/Halo-Tex2(∆SMP)-R | 5′-TGCATTATaGAGGGTGGAACCTCTGGCAACTT-3′ |
| GFP-Tex2(∆ TM-∆PH)-F | 5′-CAGTGAAGGAGGAGGAAGAACAGGAAGCCTGG-3′ |
| GFP-Tex2(∆ TM-∆PH)-R | 5′-TTCCTCCTCCTTCACTGGCAATTCCGGGTGCT-3′ |
| 14xHis-NEDD8-Tex2(PH)-F | 5′-GGCGCCGGTACCTGCGGATCCCCACCAAGTGCTCATAAATATCACA-3′ |
| 14xHis-NEDD8-Tex2(PH)-R | 5′-GCTCAGCTAATTAAGAAGCTTTCAAGAGGGTGGAACCTCTGG-3′ |
| 14xHis-NEDD8-Tex2(SMP)-F | 5′-GGCGCCGGTACCTGCGGATCCGAGGAGGAAGAACAGGAAGCCT-3′ |
| 14xHis-NEDD8-Tex2(SMP)-R | 5′-GCTCAGCTAATTAAGAAGCTTAGTGATATAAACATCATCCATGTTTGG-3′ |
| Halo-TMEM55A-F | 5′-CTGGAGATTTCCCTCGAGCTCATGGCTGCCGAGGGGGTG-3′ |
| Halo-TMEM55A-R | 5′-GTACCGTCGACTGCAGAATTCCTATGCAAAACTGTGCTCTGGATAA-3′ |
| Halo-TMEM55B-F | 5′-CTGGAGATTTCCCTCGAGCTCATGGCGGCAGATGGAGAGC-3′ |
| Halo-TMEM55B-R | 5′-TTATCTAGATCCGGTGGATCCTCAGGAGAAGTTCTGGACAGGG-3′ |
| BFP-TMEM55B-F | 5′-TGAACCGTCAGATCCGCTAGCGCCACCATGAGCGAGCTGATTAAGGAGAA-3′ |
| BFP-TMEM55B-R | 5′-TCCATCTGCCGCCATGAGCTCATTAAGCTTGTGCCCCAGTTTG-3′ |
| TMEM55B-∆X5R-F | 5′-CCTTATCATTGCATGCCCTCGGCCCTACTGCA-3′ |
| TMEM55B-∆X5R-R | 5′-GGCATGCAATGATAAGGAGACAGTTACAGGGGCA-3′ |
| TMEM55B(∆1-74)-F | 5′-CTCGAGCTCTATTCACCCTTAACTAGCCCGGA-3′ |
| TMEM55B(∆1-74)-R | 5′-GGTGAATAGAGCTCGAGGGAAATCTCCAGAGT-3′ |
| TMEM55B- ∆TM-F | 5′-TATTGGGCGCTGAGGATCCACCGGATCTAGATAAC-3′ |
| TMEM55B- ∆TM-R | 5′-ATCCTCAGCGCCCAATAGATGACACTTTCCTG-3′ |
| TMEM55B(∆1-163)-F | 5′-TTTCCCTCGAGCTCCATCCCGGACCTCTGAGTCC-3′ |
| TMEM55B(∆1-163)-R | 5′-ATGGAGCTCGAGGGAAATCTCCAGAGTAGACA-3′ |
| TMEM55B-∆PTase(73-163)-F | 5′-ACATCCCGGACCTCTGAGTCCAGAACCCCAAC-3′ |
| TMEM55B-∆PTase(73-163)-R | 5′-TCAGAGGTCCGGGATGTGGGTCCTCCCCAGGCAAC-3′ |
| TMEM55B C140W-F | 5′-TCTGGAAAGTGACATCCCAACGGATTGCATGC-3′ |
| TMEM55B C140W-R | 5′-GGATGTCACTTTCCAGATAAGGAGACAGTTACAGGGGCA-3′ |
| TMEM55B C140S-F | 5′-TCTCGAAAGTGACATCCCAACGGATTGCATGC-3′ |
| TMEM55B C140S-R | 5′-GGATGTCACTTTCGAGATAAGGAGACAGTTACAGGGGCA-3′ |
| GST-TMEM55B-F | 5′-GATCTGGTTCCGCGTGGATCCATGGCGGCAGATGGAGAGC-3′ |
| GST-TMEM55B-R | 5′-TCAGTCAGTCACGATGAATTCTCAGGAGAAGTTCTGGACAGGG-3′ |
| Halo-PI4KIIα-F | 5′-CTGGAGATTTCCCTCGAGCTCATGGACGAGACGAGCCCAC-3′ |
| Halo-PI4KIIα-R | 5′-TTATCTAGATCCGGTGGATCCTCACCACCATGAAAAGAAGGGC-3′ |
| Halo-PI4KIIβ-F | 5′-CTGGAGATTTCCCTCGAGCTCATGGAGGATCCCTCCGAGC-3′ |
| Halo-PI4KIIβ-R | 5′-TTATCTAGATCCGGTGGATCCTCACCAGGAGGAAAAAAATGGC-3′ |
| PI4KIIα W359A-F | 5′-ACTCCGCTAGGGCATATCCTTTTTACTGGGCC-3′ |
| PI4KIIα W359A-R | 5′-ATATGCCCTAGCGGAGTCAGGATGCTTCAGTGGG-3′ |
| RTN4-Halo/GFP-F | 5′-TGAACCGTCAGATCCGCTAGCGCCACCATGGAAGACCTGGACCAGTCTCC-3′ |
| RTN4-Halo/GFP-R | 5′-CAGAATTCGAAGCTTGAGCTCTTCAGCTTTGCGCTTCAATCC-3′ |
| Halo-ARL6IP1-F | 5′-CTGGAGATTTCCCTCGAGCTCATGGCGGAGGGAGATAATCG-3′ |
| Halo-ARL6IP1-R | 5′-TTATCTAGATCCGGTGGATCCTCATTCGTTTTTCTTTTCTTTTTGTT-3′ |
| Halo-ATL1-F | 5′-CTGGAGATTTCCCTCGAGCTCATGGCCAAGAACCGCAGG-3′ |
| Halo-ATL1-R | 5′-TTATCTAGATCCGGTGGATCCTTACATTTTTTTCTTTTCTGATTGTTCA-3′ |
| Climp63-halo-F | 5′-TGAACCGTCAGATCCGCTAGCATGCCCTCGGCCAAACAA-3′ |
| Climp63-halo-R | 5′-CGACTGCAGAATTCGAAGCTTGACCTTTTCGTGAATCTTCTCCA-3′ |
| Halo-E-Syt1-F | 5′-CTGGAGATTTCCCTCGAGCTCATGGAGCGATCTCCAGGAGAG-3′ |
| GFP-E-Syt1-F | 5′-TACAAGTCCGGACTCAGATCTATGGAGCGATCTCCAGGAGAG-3′ |
| Halo/GFP-E-Syt1-R | 5′-TTATCTAGATCCGGTGGATCCCTAGGAGCTGCCCTTGTCCTT-3′ |
| Reep5-Halo-F | 5′-CCGGACTCAGATCTCGAGCTCGCCACCATGTCTGCGG-3′ |
| Reep5-Halo-R | 5′-CGGTGGATCCCGGGCCCGCGGGGTGCTCTTCTTTTCTTCACCCA-3′ |
| MGAT2-Halo-F | 5′-TGAACCGTCAGATCCGCTAGCGCCACCATGAGGTTCCGCATCTACAAACG-3′ |
| MGAT2-Halo-R | 5′-CGACTGCAGAATTCGAAGCTTCTGCAGTCTTCTATAACTTTTACAGAGTTC-3′ |
| Tex2 KO sgRNA1-F | 5′-CACCGCCTCTGCACGTGCACTTTAG-3′ |
| Tex2 KO sgRNA1-R | 5′-AAACCTAAAGTGCACGTGCAGAGGC-3′ |
| Tex2 KO sgRNA2-F | 5′-CACCGCAAGTTGGCCATGACCCCGC-3′ |
| Tex2 KO sgRNA2-R | 5′-AAACGCGGGGTCATGGCCAACTTGC-3′ |
| Tex2 KI sgRNA-F | 5′-CACCGCCGGCAATGACAAGTCTGTA-3′ |
| Tex2 KI sgRNA-R | 5′-AAACTACAGACTTGTCATTGCCGGC-3′ |
| Tex2 LHA-F | 5′-GTCGACGGTATCGATAAGCTTAGTCCTTGGTAGAGTTCTTGGTTGG-3′ |
| Tex2 LHA-R | 5′-TTTGCTCATTGCCGGCTTCACAAGGGC-3′ |
| sfGFP-F | 5′-TGAAGCCGGCAATGAGCAAAGGAGAAGAACTTTTCA-3′ |
| sfGFP-R | 5′-GGCGACCATACAGACTTGTTTTGTAGAGCTCATCCATGCCA-3′ |
| Tex2 RHA-F | 5′-AACAAGTCTGTATGGTCGCCATG-3′ |
| Tex2 RHA-R | 5′-CTATAGGGCGAATTGGAGCTCAGTATCAGAGGGAGAAGTCAGTGGG-3′ |
GFP-trap assay
GFP trap (GTA-100; ChromoTek) was used for the detection of protein–protein interactions, and the GFP-Trap assays were performed according to the manufacturer’s protocol. Briefly, after 24 h transfection with the indicated plasmids, cells were lysed in ice-cold lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100 and protease inhibitor cocktail). Lysates were centrifuged at 13,000 rpm for 10 min at 4°C and pellets were removed. Supernatants were incubated with GFP-Trap agarose beads for 1 h at 4°C with gentle rocking. After washing four times with lysis buffer, beads were boiled with SDS sample buffer. Proteins of interest were analyzed by immunoblotting. 5% input was used in GFP traps unless otherwise indicated.
GST Tag and His Tag protein purification
GST and His constructs were transformed into Escherichia coli BL21 (DE3) cells, and the cells were incubated at 37°C until the optical density (OD) at 600 nm reached 0.6–0.8. Subsequently, cells were incubated at 16°C for another hour, followed by induction with 1 mM IPTG overnight at 16°C. Cells were lysed via sonication. GST fusion proteins were purified via the GST-tag Protein Purification kit (C600031-0025; Sangon), and His fusion proteins were purified via the Ni-NTA Sefinose (TM) Resin Purification kit (G600033-0100; Sangon).
In vitro lipid-binding assay
1 μl of either NBD-labeled PS, PE, PC, PA, cholesterol, or ceramide (1 mg/ml in methanol) was incubated with 19 μl purified 14xHis-Tex2-SMP (1 mg/ml) for 2 h at 4°C. Samples were visualized on 10% native PAGE gels. NBD fluorescence was visualized using Bio-Rad ChemiDoc MP, and comigrated proteins were visualized by Coomassie blue staining.
In vitro pull-down assays of GFP-Tex2 and GST-TMEM55B
HEK293 cells transiently transfected with GFP-Tex2 were lysed in high-salt lysis buffer (RIPA buffer containing 500 mM NaCl, proteasome inhibitors, and PMSF). GFP-Trap beads were used to pellet GFP-Tex2 from cell lysates, followed by washing with high-salt lysis buffer 10 times. The GFP-Tex2 beads were incubated with different amounts of ceramide or PS (0, 0.2, 0.4, 0.6, 0.8, and 1.0 μg) for 2 h at 4°C, and then were incubated with purified GST–TMEM55B or GST-only overnight at 4°C, respectively, followed by washing beads with freshly prepared HNM buffer (20 mM Hepes, pH 7.4, 0.1 M NaCl, 5 mM MgCl2, 1 mM DTT, and 0.2% NP-40). GFP-Tex2 beads were resuspended in 100 μl 2 × SDS-sampling buffer. Resuspended beads were boiled for 10 min at 95°C to dissociate protein complexes from beads. Western blotting was performed using anti-GFP, GST, or TMEM55B antibodies. The Coomassie staining was performed for purified GST-TMEM55B.
PIP strip assays
The PIP Strips (P-6001) were blocked by TBS-T + 3% fatty acid-free BSA and then gently agitated for 1 h at room temperature, followed by an incubation with purified His-Tex2-PH (0.5 µg/ml) in TBS-T + 3% fatty acid-free BSA overnight at 4°C. After washing the PIP strips with TBS-T + 3% fatty acid-free BSA three times under gentle agitation for 10 min each time, PIP strips were incubated with the anti-His antibodies overnight at 4°C, followed by repeated washing steps.
Live imaging by high-resolution confocal microscopy
Cells were grown on 35-mm glass-bottom confocal MatTek dishes, and the dishes were loaded to a laser scanning confocal microscope (LSM900; Zeiss) equipped with multiple excitation lasers (405, 458, 488, 514, 561, and 633 nm) and a spectral fluorescence GaAsP array detector. Cells were imaged with the 63 × 1.4 NA iPlan-Apochromat 63× oil objective using the 405 nm laser for BFP; 488 nm for GFP; 561 nm for OFP, tagRFP, or mCherry; and 633 nm for Janilia Fluo 646 HaloTag Ligand.
Mass spectrometry for identification of GFP-Tex2-interacting proteins
The identification of GFP-Tex2-interacting proteins by MS was described in our previous study (Gao et al., 2022). Briefly, the bound proteins were extracted from GFP-Trap agarose beads using SDT lysis buffer (4% SDS, 100 mM DTT, 100 mM Tris-HCl, pH 8.0), followed by sample boiling for 3 min and further ultrasonicated. Undissolved beads were removed by centrifugation at 16,000 g for 15 min. The supernatant, containing proteins, was collected. Protein digestion was performed with the FASP method. Briefly, the detergent, DTT, and IAA in the UA buffer were added to block-reduced cysteine. Finally, the protein suspension was digested with 2 µg trypsin (Promega) overnight at 37°C. The peptide was collected by centrifugation at 16,000 g for 15 min. The peptide was desalted with C18 StageTip for further LC-MS analysis. LC-MS/MS experiments were performed on a Q Exactive Plus mass spectrometer that was coupled to an Easy nLC (Thermo Fisher Scientific). The peptide was first loaded to a trap column (100 µm × 20 mm, 5 µm, C18, Dr. Maisch GmbH) in buffer A (0.1% formic acid in water). Reverse-phase high-performance liquid chromatography (RP-HPLC) separation was performed using a self-packed column (75 µm × 150 mm; 3 µm ReproSil-Pur C18 beads, 120 Å; Dr. Maisch GmbH, Ammerbuch) at a flow rate of 300 nl/min. The RP-HPLC mobile phase A was 0.1% formic acid in water and B was 0.1% formic acid in 95% acetonitrile. The gradient was set as follows: 2–4% buffer B from 0 to 2 min, 4–30% buffer B from 2 to 47 min, 30–45% buffer B from 47 to 52 min, 45–90% buffer B from 52 min and to 54 min, and 90% buffer B kept until to 60 min. MS data were acquired using a data-dependent top20 method dynamically choosing the most abundant precursor ions from the survey scan (350–1,800 m/z) for HCD fragmentation. A lock mass of 445.120025 Da was used as the internal standard for mass calibration. The full MS scans were acquired at a resolution of 70,000 at m/z 200, and 17,500 at m/z 200. The maximum injection time was set to 50 ms for MS and 50 ms for MS/MS. Normalized collision energy was 27 and the isolation window was set to 1.6 Th. Dynamic exclusion duration was 60 s. The MS data were analyzed using MaxQuant software version 1.6.1.0. MS data were searched against the UniProtKB human database (36,080 total entries, downloaded 2019.06.25). Trypsin was selected as the digestion enzyme. A maximum of two missed cleavage sites and a mass tolerance of 4.5 ppm for precursor ions and 20 ppm for fragment ions were defined for database search. Carbamidomethylation of cysteines was defined as a fixed modification, while acetylation of protein N-terminal and the oxidation of methionine were set as variable modifications for database searching. The database search results were filtered and exported with a <1% false discovery rate (FDR) at peptide-spectrum-matched level and protein level, respectively.
Non-targeted lipidomics using LC-MS/MS
The identification of GFP-Tex2-associated lipids in HEK293 cells and the examination of lipid abundance in ER and LE/lys fractions were described in our previous study (Gao et al., 2022). Briefly, to extract lipids, 1 ml methyl tert-butyl ether (MTBE) was added to GFP-Trap agarose beads (Chromoteck) and the samples were shaken for 1 h at room temperature. Next, phase separation was induced by adding 250 μl water, letting it sit for 10 min at room temperature, and centrifuging for 15 min at 14,000 g, 4°C. Due to the low density and high hydrophobicity of MTBE, lipids and lipophilic metabolites are mainly extracted to the upper MTBE-rich phase. The lipid was transferred to fresh tubes and dried with nitrogen. Additionally, to ensure data quality for metabolic profiling, quality control (QC) samples were prepared by pooling aliquots from representative samples for all of the analysis samples and were used for data normalization. QC samples were prepared and analyzed with the same procedure as that for the experiment samples in each batch. Dried extracts were then dissolved in 50% acetonitrile. Each sample was filtered with disposable 0.22 μm cellulose acetate and transferred into 2-ml HPLC vials and stored at −80°C until analysis. For UHPLC-MS/MS analysis, lipid analysis was performed on Q Exactive orbitrap mass spectrometer (Thermo Fisher Scientific) coupled to a UHPLC system Ultimate 3000 (Thermo Fisher Scientific). Samples were separated using a Hypersil GOLD C18 column (100 × 2.1 mm, 1.9 µm; Thermo Fisher Scientific). Mobile phase A was prepared by dissolving 0.77 g of ammonium acetate in 400 ml of HPLC-grade water, followed by adding 600 ml of HPLC-grade acetonitrile. Mobile phase B was prepared by mixing 100 ml of acetonitrile with 900 ml isopropanol. The flow rate was set as 0.3 ml/min. The gradient was 30% B for 0.5 min and was linearly increased to 100% in 10.5 min, maintained at 100% in 2 min, and then reduced to 30% in 0.1 min, with a 4.5-min re-equilibration period employed. Both electrospray ionization (ESI) positive mode and negative mode were applied for MS data acquisition. The positive mode of spray voltage was 3.0 kV and the negative mode was 2.5 kV. The ESI source conditions were set as follows: heater temperature of 300°C, Sheath Gas Flow rate, 45 arb, Aux Gas Flow Rate, 15 arb, Sweep Gas Flow Rate, 1 arb, Capillary Temp, 350°C, and S-Lens RF Level, 50%. The full MS scans were acquired at a resolution of 70,000 at m/z 200 and 17,500 at m/z 200 for MS/MS scans. The maximum injection time was set to 50 ms for MS and 50 ms for MS/MS. MS data were acquired using a data-dependent Top10 method dynamically choosing the most abundant precursor ions from the survey scan (200–1,500 m/z) for HCD fragmentation. Stepped normalized collision energy was set as 15, 25, 35 and the isolation window was set to 1.6 Th. QC samples were prepared by pooling aliquots that were representative of all samples under analysis and used for data normalization. Blank samples (75% acetonitrile in water) and QC samples were injected every six samples during acquisition.
For data preprocessing and filtering, lipids were identified and quantified using LipidSearch 4.1.30 (Thermo Fisher Scientific). Mass tolerance of 5 and 10 ppm were applied for precursor and product ions. A retention time shift of 0.25 min was performed in “alignment.” M-score and chromatographic areas were used to reduce false positives. The lipids with <30% relative standard deviation (RSD) of MS peak area in the QC samples were kept for further data analysis. SIMCAP software (Version 14.0, Umetrics) was used for all multivariate data analyses and modeling. Data were mean-centered using Pareto scaling. Models were built on principal component analysis (PCA), orthogonal partial least-square discriminant analysis (PLS-DA), and partial least-square discriminant analysis (OPLS-DA). All the models evaluated were tested for overfitting with methods of permutation tests. The descriptive performance of the models was determined by R2X (cumulative) [perfect model: R2X (cum) = 1] and R2Y (cumulative) [perfect model: R2Y (cum) = 1] values while their prediction performance was measured by Q2 (cumulative) [perfect model: Q2 (cum) = 1] and a permutation test (n = 200). The permuted model should not be able to predict classes—R2 and Q2 values at the Y-axis intercept must be lower than those of Q2 and the R2 of the non-permuted model. OPLS-DA allowed the determination of discriminating metabolites using the variable importance on projection (VIP). The VIP score value indicates the contribution of a variable to the discrimination between all the classes of samples. Mathematically, these scores are calculated for each variable as a weighted sum of squares of PLS weights. The mean VIP value is 1, and usually, VIP values over 1 are considered significant. A high score is in agreement with a strong discriminatory ability and thus constitutes a criterion for the selection of biomarkers. The discriminating metabolites were obtained using a statistically significant threshold of variable influence on projection (VIP) values obtained from the OPLS-DA model and two-tailed Student’s t test (P value) on the normalized raw data at a univariate analysis level. The P-value was calculated by one-way analysis of variance (ANOVA) for multiple groups analysis. Metabolites with VIP values >1.0 and P value <0.05 were considered to be statistically significant metabolites. Fold change was calculated as the logarithm of the average mass response (area) ratio between two arbitrary classes. On the other side, the identified differential metabolites were used to perform cluster analyses with the R package.
Purification of lysosomal and ER membranes by density gradient centrifugation
OptiPrep flotation assays were performed to enrich lysosomal membrane fractions according to a standard protocol (Uematsu et al., 2017). Briefly, HeLa cells from four confluent 10-cm dishes were collected and resuspended in 2 ml ice-cold homogenization buffer (250 mM sucrose, 20 mM HEPES-KOH, pH 7.4, 1 mM EDTA, 1 mM phenylmethanesulfonyl fluoride, and complete EDTA-free protease inhibitor), followed by the lysis of the cells in a 7-ml Dounce homogenizer with 15–25 strokes. The homogenized cells were centrifuged twice at 3,000 × g for 10 min to remove cell debris and undisrupted cells. The supernatant was diluted with an equal volume of OptiPrep (D1556; Sigma-Aldrich). A discontinuous OptiPrep gradient was generated in an SW41 tube for ultracentrifuge rotors (344059; Beckman Coulter) by overlaying the following OptiPrep solutions all in homogenization buffer: 2.4 ml of the diluted supernatant in 30% OptiPrep, 1.8 ml 20% OptiPrep, 2 ml 15% OptiPrep, 2 ml 10% OptiPrep, 2 ml 5% OptiPrep, and 2 ml 0% OptiPrep. The gradient was centrifuged at 150,200 × g in an SW41Ti rotor (Beckman Coulter) using an Optima XE-90 ultracentrifuge (Beckman Coulter) for 3 h, and subsequently, 14 fractions (0.85 ml each) were collected from the top and analyzed by Western blots using anti-Lamp1 antibody.
ER fractions were enriched using Endoplasmic Reticulum Isolation Kit (ER0100; Sigma-Aldrich) according to the manufacturer's instructions. Briefly, HeLa cells from five confluent 10-cm dishes were collected, followed by centrifugation at 600 × g for 5 min. After washing the cells three times with PBS, the packed cell volume (PCV) was measured and then suspended in a volume of hypotonic extraction buffer (10 mM HEPES, pH 7.8, with 1 mM EGTA and 25 mM potassium chloride) equivalent to three times the PCV. After the incubation of the cells for 20 min at 4°C allowing the cells to swell, the cells were centrifuged at 600 × g for 5 min, followed by the measurement of the “new” PCV. After adding a volume of isotonic extraction buffer (10 mM HEPES, pH 7.8, with 0.25 M sucrose, 1 mM EGTA, and 25 mM potassium chloride) equivalent to two times the “new” PCV, the suspension was then transferred to a 7-ml Dounce homogenizer, followed by the lysis of the cells with 10 strokes and then the centrifugation of the homogenate at 1,000 × g for 10 min at 4°C. After the transfer of the supernatant to another centrifuge tube, the supernatant was centrifuged at 12,000 × g for 15 min at 4°C, followed by another centrifugation for 60 min at 100,000 × g at 4°C. The pellet was the microsomal fraction and further verified by Western blots using anti-Calnexin antibody.
Correlative light electron microscopy
Cells were grown on glass-bottom P35G-2-14-C-Grid dishes (MatTek). The dishes have a high optical quality coverslip with a photo-etched grid and coordinates to facilitate pinpointing the location of individual cells. The cells were fixed with 2% paraformaldehyde (PFA, 16% paraformaldehyde, Ted Pella Co.) in 0.1 M PB buffer pH 7.3 for 30 min at RT. Once the cells of interest were found, their positions on the grid were documented by switching from fluorescence to differential interference contrast (DIC) mode. After fluorescence imaging, the selected areas with positive cells were marked on the bottom of the coverslip under a light microscope to facilitate the processing of EM. After observation with a confocal microscope (Zeiss LSM 980), the samples were fixed in 2.5% glutaraldehyde (25% glutaraldehyde ampules, Ted Pella Co.) and 2% PFA mixture in 0.1 M PB buffer pH 7.3 at 4°C overnight. After washing with PB buffer three times, the samples were stained with 2% OsO4 and 1.5% potassium hexacyanoferrate, followed by sequential washing with PB buffer (three times) and ddH2O (three times). Then, the samples were incubated in 1% TCh and washed with ddH2O 4 times, followed by staining with 2% OsO4 along with another washing step with ddH2O four times. The samples were then stained with 1% UA, followed by washing with ddH2O four times. The samples were stained with a lead aspartate solution and washed with ddH2O five times. Then the samples were dehydrated by incubating with ethanol (30, 50, 70, 95, and 100% X2), followed by incubation with acetone two times. After hydration, the samples were subjected to infiltration and embedding step, in which samples were infiltrated with Epon resin (EMS Corp.) as follows: 3:1, 1:1, 1:3 Acetone: Epon and 100% Epon three times, followed by samples being embedded and polymerized with Epon for 48 h at 60°C. Eventually, the samples were applied to the serial ultrathin sectioning step, and the sections were observed at 80 kV in an FEI Talos 120 kv transmission electron microscopy.
Measurement of the percentage of PHluorin-positive lysosomes in cells
PHluorin-positive lysosomes were manually counted with the assistance of Cell Counter, a plugin of ImageJ (2.1.0/1.53c; National Institutes of Health), and the lysosomes were manually counted in the same way based on Lamp1-mApple fluorescence. The percentage of PHluorin-positive lysosomes in a cell was quantified by the number of PHluorin-positive lysosomes divided by Lamp1-mApple positive lysosomes.
Measurement of ER sheet abundance in cells
Following methods described previously (Shibata et al., 2010) with modifications, ER sheet abundance was measured by calculating the percentage of the areas of ER sheet over the total ER area. In this quantification, the areas of ER sheets and tubules were determined from the fluorescence of Climp63 and RTN4, respectively, after subtraction of the background.
Quantification of the LE/lys positioning
Following the methods described previously (Gao et al., 2022), the number of LE/lys was counted manually with the assistance of ImageJ plugin Cell counter. For quantification of LE/lys positioning, perinuclear regions of cells were defined as shown in Fig. 7 F. Briefly, R was defined by the longer radius of the oval-shaped nucleus, while r was defined by the shorter radius of the oval-shaped nucleus. The length of R or r was measured by ImageJ. The perinuclear region was defined as a region within a distance of 0.5R/r to the nucleus rim. To calculate the percentage of perinuclearly LE/lys, we manually counted the number of total LE/lys and perinuclear LE/lys with the assistance of an ImageJ plugin Cell counter.
Statistical analysis
All statistical analyses and P value determinations were performed in GraphPad Prism6. Data distribution was assumed to be normal, but this was not formally tested. All the error bars represent Mean ± SD. To determine P values, ordinary one-way ANOVA with Tukey’s multiple comparisons test was performed among multiple groups, and a two-tailed unpaired Student’s t test was performed between two groups. One sample Student’s t test was performed for normalized data in Fig. 5 C.
Online supplemental material
Fig. S1 shows the interaction between Tex2 and E-Syt1 by coIP assays and microscopy, and also shows the information about the GFP-Tex2 knock-in and Tex2 knockout cell lines by Crispr-cas9 technology. Fig. S2 provides additional data to support that Tex2 is a tubular ER protein and its targeting to tubular ER is independent of ER tubule-shaping proteins. Fig. S3 contains additional data to show the role of Tex2 in the formation of ER tubules and also provide data to show that Tex2 may not be directly involved in TG-induced ER stress. Fig. S4 provides additional data to show that either the PH domain or the SMP domain of Tex2 is not required for Tex2 recruitment to ER–LE/lys MCSs by TMEM55B. Fig. S5 contains additional data to show the role of Tex2 in lysosomal function and the distribution of PS, PI4P, and PI(4,5)P2. Video 1 shows an example of a lysosome undergoing retrograde transport in a COS7 cell transfected with an ER marker and a LE/lys marker. Video 2 shows an example of a Halo-TMEM55B-positive lysosome tightly associating with GFP-Tex2-labeled ER membranes during intracellular transport in a COS7 cell.
Data availability
All the data and relevant materials, including reagents and primers, that support the findings of this study are available from the corresponding author upon reasonable request.
Acknowledgments
We thank Anbing Shi and Yanling Yan for their insightful suggestions. We thank Mei Yu at Bioimaging Core of Shenzhen Bay Laboratory for providing imaging support. The authors are grateful to Gongzheng Zhao at Multi-Omics Mass Spectrometry Core of Shenzhen Bay Laboratory for assistance with proteomics and lipidomics experiments. We thank Qing Tian and Linfang Yang (Huazhong University of Science and Technology, HUST) for imaging assistance. We thank Jiahao Guo (Cryo-EM Center of Southern University of Science and Technology) for EM assistance.
W. Ji was supported by the National Natural Science Foundation of China (32122025), and the Program for HUST Academic Frontier Youth Team (2018QYTD11). L. Gao was supported by the National Natural Science Foundation of China (8190080864). L. Deng was supported by the National Natural Science Foundation of China (32270779) and the National Key Research and Development Program of China (2022YFA1302800).
Author contributions: Y. Du and W. Ji conceived the project and designed the experiments. Y. Du and L. Gao performed the experiments. Y. Du, W. Chang, L. Deng, and W. Ji analyzed and interpreted the data. W. Ji prepared the manuscript with input and approval from all authors.
References
Author notes
Disclosures: The authors declare no competing interests exist.






![Identification of TMEM55B as a Tex2 adaptor on LE/Lys. (A) Volcano plot of protein candidates coIPed with Tex2 in HEK293 cells compared with protein candidates coIPed with GFP tag only. Candidates that were considered significant (−log [P value] > 1.3; P < 0.05) were labeled in red (Log2 [fold change] > 0; increased in abundance) or blue (Log2 [fold change] < 0; decreased in abundance). (B) GFP-Trap assays demonstrate an interaction between GFP-Tex2 and Halo-TMEM55B in COS7 cells. (C and D) Representative wide-field images of live COS7 cells expressing either GFP-Tex2 (green; C) or GFP-E-syt1 (green; D) and Halo-TMEM55B with two boxed regions showing at the bottom. (E and F) Representative images of a live COS7 cell expressing GFP-Tex2 (green), Halo-TMEM55B (magenta; E) or Halo-TMEM55A (magenta; F), ER-tagRFP (red), and Lamp1-mCh (blue; a LE/lys marker) with two boxed regions showing at the bottom. (G) Pearson’s correlation coefficient of Tex2 vs. Lamp1 in absence of TMEM55 (11 cells) or upon TMEM55A (10 cells) or TMEM55B (13 cells) overexpression (>3 independent experiments). Ordinary one-way ANOVA with Tukey’s multiple comparisons test. Mean ± SD. (H and I) Representative live-cell images of a GFP-Tex2-KI (green) cell without (H) or with expressing Halo-TMEM55B (I) and Lamp1-mCh (blue) with two boxed regions at the bottom. Yellow arrows denote the specific enrichment of GFP-Tex2-KI at LE/lys extensively contacting the ER. (J) Representative 3D rendering of a COS7 cell expressing GFP-Tex2 (green), Halo-TMEM55B (magenta), and Lamp1-mCh (blue) with y-z projection to the left and x-z projection to the right. (K) COS7 cells expressing GFP-Tex2, Halo-TMEM55B, and stained by DAPI, were fixed and imaged by 3D microscopy (left panel) and processed for TEM (middle panel). Fluorescence and TEM images were correlated (right panel) with two insets showing tight associations between the ER (green) and LE/lys (magenta). Scale bar, 10 μm in the whole cell images and 2 μm in the insets in C, D, E, F, and H–J; 5 μm in the whole cell image, and 1 μm in the insets in K. Source data are available for this figure: SourceData F2.](https://cdn.rupress.org/rup/content_public/journal/jcb/222/4/10.1083_jcb.202205133/1/m_jcb_202205133_fig2.png?Expires=1767691610&Signature=kRHrzoWdy~HIDLaDIn2OzUtQSxvn6ZmnJ1P7OE05GgypBo5ToBzMgo-Zv~~DrdAcNNiNqmFfNzCI7JqPPmPUFr3T9gjXvrCrVtfMGw49ESmuw2mIW-cDJLararrrKJDaYWUyrevcOGssvjnTAJYbADhbdSZJ6g86TLCIW~5ayqEVSof2HOsZnNS9L3qJ4ggg9CPoq0BhlRPtQbgcnMDOBE9ns-1v48kpZBOsJbWfET-9H59uaJ2bUtYgfzuhzM2X9XBDvpQyuWd7w2N1DWSRywxst-HRXkmnry370nKvnDAJh088bvLSutmdHI2wkIYmtCdSHCTWzGeLL4j~3p3YSA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)