Kostmann disease is an inherited severe congenital neutropenia syndrome associated with loss-of-function mutations in an adaptor protein HS1-associated protein X-1 (Hax1). How Hax1 regulates neutrophil function remains largely unknown. In this paper, we use ribonucleic acid interference to deplete Hax1 in the neutrophil-like cell line PLB-985 and identify Hax1 as a negative regulator of integrin-mediated adhesion and chemotaxis. Using microfluidics, we show that depletion of Hax1 impairs neutrophil uropod detachment and directed migration. Hax1-deficient cells also display increased integrin-mediated adhesion and reduced RhoA activity. Moreover, depletion of RhoA induces increased neutrophil adhesion and impaired migration, suggesting that Hax1 regulates neutrophil adhesion and chemotaxis through RhoA. Accordingly, activation of RhoA is sufficient to rescue adhesion of Hax1-deficient neutrophils. Together, our findings identify Hax1 as a novel regulator of neutrophil uropod detachment and chemotaxis through RhoA.
Introduction
Severe congenital neutropenia is a heterogeneous group of inherited disorders characterized by neutropenia and recurrent bacterial infections. Kostmann disease is an autosomal recessive form of severe congenital neutropenia caused by loss-of-function point mutations in the HAX1 gene (Klein et al., 2006). HS1-associated protein X-1 (Hax1) was first identified as a binding partner of the hematopoietic-specific cortactin homologue HS1 (Suzuki et al., 1997) and has been implicated in regulating the actin cytoskeleton and apoptosis. Although it has also been proposed that loss of Hax1 results in neutropenia by affecting neutrophil apoptosis (Klein et al., 2006), a recent study has challenged this role (Jeyaraju et al., 2009).
Hax1 is a ubiquitous protein that regulates the actin cytoskeleton and migration of cancer cells. Hax1 interacts directly with adhesion and cytoskeletal proteins, including cortactin, HS1, Gα13, and β6 integrins. Depletion of endogenous Hax1 using siRNA impairs αvβ6 integrin–mediated migration of squamous cell carcinoma (Ramsay et al., 2007) and reduces migration of NIH3T3 cells (Radhika et al., 2004). However, no previous studies have addressed how Hax1 modulates neutrophil motility. Here, we characterize how Hax1 regulates neutrophil chemotaxis using small hairpin RNA (shRNA) depletion and live imaging with microfluidic gradient generators. We identified a new role for Hax1 as a regulator of neutrophil uropod detachment and chemotaxis through the modulation of integrin-mediated adhesion and Rho GTPase signaling.
Results and discussion
Hax1 localizes to the leading edge during chemotaxis, and its expression is regulated by neutrophil differentiation
Hax1 is a ubiquitously expressed protein that directly interacts with HS1, Gα13, and integrin β6 at its C terminus (Fig. 1 A; Suzuki et al., 1997; Radhika et al., 2004; Ramsay et al., 2007). To characterize Hax1 expression and localization in neutrophils, we used the myeloid leukemia cell line PLB-985, which can be terminally differentiated into neutrophil-like cells (Tucker et al., 1987). To determine whether Hax1 expression is regulated by differentiation, we used immunoblotting to detect endogenous Hax1. Surprisingly, we observed a reduction in Hax1 expression in differentiated PLB-985 cells induced with DMSO compared with undifferentiated cells (Fig. 1 B). This is in contrast to vinculin, HS1, and actin, which were all increased upon differentiation. This finding suggests that Hax1 expression may play a role during neutrophil differentiation.
Hax1 expression in PLB-985 cells and localization during chemotaxis. (A) Schematic of Hax1 modified from (Jeyaraju et al., 2009). Binding sites for HS1 (Suzuki et al., 1997), Gα13 (Radhika et al., 2004), and β6 integrin (Ramsay et al., 2007) are shown. Hax1-GFP constructs are used in C. (B) Hax1 expression is decreased upon differentiation. Vinculin, HS1, and actin were included as controls and show increased expression with neutrophil differentiation. Quantification of expression represents the ratio of differentiated to undifferentiated cells as mean intensity ± SDs from three independent experiments as described in Materials and methods. (C) Still image from time-lapse videos of utrophin-RFP and Hax1-GFP localization in PLB-985 cells. Utrophin-RFP was coexpressed with full-length Hax1-GFP (Video 1), Hax1 1–113–GFP (Video 2), and Hax1Δ113-GFP (Video 3) in differentiated PLB-985 cells and then imaged during chemotaxis on 10 µg/ml fibrinogen in response to a needle containing 1 µM fMLP. Bar, 10 µm.
Hax1 expression in PLB-985 cells and localization during chemotaxis. (A) Schematic of Hax1 modified from (Jeyaraju et al., 2009). Binding sites for HS1 (Suzuki et al., 1997), Gα13 (Radhika et al., 2004), and β6 integrin (Ramsay et al., 2007) are shown. Hax1-GFP constructs are used in C. (B) Hax1 expression is decreased upon differentiation. Vinculin, HS1, and actin were included as controls and show increased expression with neutrophil differentiation. Quantification of expression represents the ratio of differentiated to undifferentiated cells as mean intensity ± SDs from three independent experiments as described in Materials and methods. (C) Still image from time-lapse videos of utrophin-RFP and Hax1-GFP localization in PLB-985 cells. Utrophin-RFP was coexpressed with full-length Hax1-GFP (Video 1), Hax1 1–113–GFP (Video 2), and Hax1Δ113-GFP (Video 3) in differentiated PLB-985 cells and then imaged during chemotaxis on 10 µg/ml fibrinogen in response to a needle containing 1 µM fMLP. Bar, 10 µm.
To characterize the intracellular distribution of Hax1 during neutrophil chemotaxis, we examined the subcellular localization of Hax1 in PLB-985 cells. We coexpressed Hax1-GFP and a marker of the neutrophil uropod that binds to stable F-actin, utrophin-RFP, in PLB-985 cells (Fig. 1 C and Videos 1, 2, and 3; Burkel et al., 2007; Cooper et al., 2008; Yoo et al., 2010). Live imaging demonstrated that Hax1-GFP predominantly localized to the leading edge, although rearward localization was also noted. To characterize the determinants of Hax1 localization, we generated truncation constructs lacking the C-terminal 166 amino acids necessary for binding HS1 and Gα13. Hax1 1–113–GFP no longer showed leading edge localization and concentrated in the middle of the cell body (Fig. 1 C and Video 2). In contrast, Hax1Δ113-GFP, which lacks the N-terminal 113 amino acids, was sufficient to localize to the leading edge of PLB-985 cells (Fig. 1 C and Video 3).
Hax1 1–113–GFP localization in PLB-985 cells during chemotaxis (related to Fig. 1). dPLB-985 cells expressing utrophin-RFP and Hax1 1–113–GFP during chemotaxis on fibrinogen to a needle containing 1 µM fMLP. Video shows a 15-s interval. (top left) DIC. (top right) utrophin-RFP. (bottom left) Hax1 1–113–GFP. (bottom right) RFP-GFP overlay. Time stamp indicates minutes and seconds. Bar, 10 µm.
Hax1 1–113–GFP localization in PLB-985 cells during chemotaxis (related to Fig. 1). dPLB-985 cells expressing utrophin-RFP and Hax1 1–113–GFP during chemotaxis on fibrinogen to a needle containing 1 µM fMLP. Video shows a 15-s interval. (top left) DIC. (top right) utrophin-RFP. (bottom left) Hax1 1–113–GFP. (bottom right) RFP-GFP overlay. Time stamp indicates minutes and seconds. Bar, 10 µm.
Hax1 Δ113-GFP localization in PLB-985 cells during chemotaxis (related to Fig 1). dPLB-985 cells expressing utrophin-RFP and Hax1 Δ113-GFP during chemotaxis on fibrinogen to a needle containing 1 µM fMLP. Video shows a 15-s interval. (top left) DIC. (top right) utrophin-RFP. (bottom left) Hax1 1 Δ113-GFP. (bottom right) RFP-GFP overlay. Time stamp indicates minutes and seconds. Bar, 10 µm.
Hax1 Δ113-GFP localization in PLB-985 cells during chemotaxis (related to Fig 1). dPLB-985 cells expressing utrophin-RFP and Hax1 Δ113-GFP during chemotaxis on fibrinogen to a needle containing 1 µM fMLP. Video shows a 15-s interval. (top left) DIC. (top right) utrophin-RFP. (bottom left) Hax1 1 Δ113-GFP. (bottom right) RFP-GFP overlay. Time stamp indicates minutes and seconds. Bar, 10 µm.
An attractive interacting partner that may mediate this targeting is the actin-binding protein HS1, which stabilizes actin branching at the leading edge (Uruno et al., 2003; Hao et al., 2005). Collectively, our findings identify Hax1 as a leading edge component that is differentially expressed during the differentiation of neutrophil-like PLB-985 cells.
Hax1 is necessary for neutrophil uropod detachment
To characterize Hax1 function, Hax1-deficient PLB-985 cells were generated using lentiviral-mediated shRNA (Fig. 2 A). Kostmann disease patients with loss-of-function Hax1 mutations display increased neutrophil apoptosis in response to TNF-α or H2O2 (Klein et al., 2006). In the presence of H2O2, we observed a mild increase in the apoptosis of Hax1-deficient cells compared with control cells by annexin V staining, suggesting that Hax1 may modulate the sensitivity of PLB-985 cells to proapoptotic stimuli. However, there was no increase in the basal level of apoptosis (Fig. S1 A). Although Hax1 expression is regulated by neutrophil differentiation, we did not observe a defect in the differentiation of Hax1-depleted PLB-985 cells as indicated by normal surface expression of αMβ2 integrin (Fig. S1 B).
Hax1 depletion impairs uropod retraction and reduces RhoA activation. (A) Lentiviral knockdown of Hax1 (Hax1 shRNA A and B) in PLB-985 cells display significant depletion of Hax1 compared with control shRNA. Quantification was normalized to vinculin loading control. ***, P < 0.01; ANOVA with Tukey posttest. (B) Still image from a time-lapse video of control shRNA and Hax1 shRNA A and B PLB-985 cells during chemotaxis on 10 µg/ml fibrinogen (Videos 4 and 5). Arrowheads indicate elongated uropods of Hax1-deficient PLB-985 cells during chemotaxis. Bar, 100 µm. (C) Quantification of uropod length in Hax1-deficient cells compared with control. Control shRNA (n = 279) cells, Hax1 shRNA A (n = 275) cells, and Hax1 shRNA B (n = 241) cells were from three independent experiments. The graph represents means ± 95% confidence interval. ***, P < 0.01; one-way ANOVA with Tukey posttest. (D) GST-RBD pull-down assays show a 10-fold decrease in RhoA activity in Hax1-deficient cells compared with control. ***, P < 0.01; t test. (E) Control and Hax1-deficient PLB-985 cells expressing utrophin-GFP during chemotaxis to 1 µM fMLP on fibrinogen (Videos 6 and 7). For Rho-associated protein kinase inhibition, cells were incubated with 30 µM Y-27632 for 30 min before imaging. Fluorescence intensity from the front to the back of the cell was determined by line scan analysis (yellow lines). Bar, 10 µm. (F) Quantification of line scan analysis of utrophin-GFP localization from E. P-values were obtained by ANOVA with Tukey posttest. Red lines indicate the mean fluorescence ratio from three independent experiments. Error bars are means ± SEM from three independent experiments. A.U., arbitrary unit.
Hax1 depletion impairs uropod retraction and reduces RhoA activation. (A) Lentiviral knockdown of Hax1 (Hax1 shRNA A and B) in PLB-985 cells display significant depletion of Hax1 compared with control shRNA. Quantification was normalized to vinculin loading control. ***, P < 0.01; ANOVA with Tukey posttest. (B) Still image from a time-lapse video of control shRNA and Hax1 shRNA A and B PLB-985 cells during chemotaxis on 10 µg/ml fibrinogen (Videos 4 and 5). Arrowheads indicate elongated uropods of Hax1-deficient PLB-985 cells during chemotaxis. Bar, 100 µm. (C) Quantification of uropod length in Hax1-deficient cells compared with control. Control shRNA (n = 279) cells, Hax1 shRNA A (n = 275) cells, and Hax1 shRNA B (n = 241) cells were from three independent experiments. The graph represents means ± 95% confidence interval. ***, P < 0.01; one-way ANOVA with Tukey posttest. (D) GST-RBD pull-down assays show a 10-fold decrease in RhoA activity in Hax1-deficient cells compared with control. ***, P < 0.01; t test. (E) Control and Hax1-deficient PLB-985 cells expressing utrophin-GFP during chemotaxis to 1 µM fMLP on fibrinogen (Videos 6 and 7). For Rho-associated protein kinase inhibition, cells were incubated with 30 µM Y-27632 for 30 min before imaging. Fluorescence intensity from the front to the back of the cell was determined by line scan analysis (yellow lines). Bar, 10 µm. (F) Quantification of line scan analysis of utrophin-GFP localization from E. P-values were obtained by ANOVA with Tukey posttest. Red lines indicate the mean fluorescence ratio from three independent experiments. Error bars are means ± SEM from three independent experiments. A.U., arbitrary unit.
To determine whether Hax1 is necessary for neutrophil chemotaxis, we performed time-lapse imaging of PLB-985 cells responding to a gradient of N-formyl-l-methionyl-l-leucyl-l-phenylalanine (fMLP) supplied from a micropipette tip. Hax1-deficient cells displayed impaired motility and an elongated uropod during chemotaxis on fibrinogen (Fig. 2, B and C; and Videos 4 and 5). Quantification of uropod length showed a greater than twofold increase in both Hax1-deficient cell lines compared with control (Fig. 2 C). The abnormal uropod morphology and detachment phenotype are similar to what has been reported with Rho inhibition (Alblas et al., 2001; Worthylake and Burridge, 2001; Yoshinaga-Ohara et al., 2002), suggesting that Hax1 may modulate rear retraction by regulating Rho GTPase signaling.
To determine whether Hax1 regulates Rho GTPase activation, RhoA activity assays were performed in control and Hax1-deficient cell lines. Using a pull-down assay, we found that RhoA activation was significantly decreased in Hax1-deficient PLB-985 cells stimulated with fMLP either in suspension (Fig. S1 C) or adherent to fibrinogen (Fig. 2 D). No significant difference in RhoA activity was detected in the absence of fMLP (unpublished data). Quantification of RhoA-GTP demonstrated a >10-fold reduction in RhoA activity in Hax1-deficient cells (Fig. 2 D). In contrast, Rac activation was increased approximately twofold in Hax1 knockdown cells (Fig. S1 D). Collectively, our findings indicate that Hax1 expression is necessary for optimal RhoA activity in neutrophils.
To determine whether Hax1 modulates the localized polarity of F-actin dynamics in motile neutrophils, we examined the localization of a probe for stable F-actin, utrophin-GFP, in control and Hax1-deficient PLB-985 cells. In previous experiments, we have reported that inhibition of the Rho-associated protein kinase with Y-27632 perturbs the uropod localization of utrophin in neutrophils migrating in vivo (Yoo et al., 2010). We also found that Y-27632 impaired the uropod localization of utrophin-GFP (Fig. 2, E and F; and Video 6). To determine whether Hax1 affects the uropod localization of utrophin, we expressed utrophin-GFP in control and Hax1-deficient cells and performed time-lapse imaging during neutrophil chemotaxis. The ratio of fluorescence intensity from the front of the cell to the rear showed a significant reduction of utrophin-GFP at the uropod of Hax1-deficient cells compared with control cells during chemotaxis (Fig. 2, E and F; and Video 7). Collectively, our findings suggest that Hax1 regulates the polarity of F-actin dynamics and rear detachment, likely through RhoA.
Utrophin-GFP localization in PLB-985 cells treated with Y-27632 during chemotaxis (related to Fig. 2). dPLB-985 cells expressing utrophin-GFP exposed to 30 µM Y-27632 for 30 min during chemotaxis on fibrinogen to a needle containing 1 µM fMLP. Video shows a 15-s interval. Time stamp indicates minutes and seconds. Bar, 10 µm.
Utrophin-GFP localization in PLB-985 cells treated with Y-27632 during chemotaxis (related to Fig. 2). dPLB-985 cells expressing utrophin-GFP exposed to 30 µM Y-27632 for 30 min during chemotaxis on fibrinogen to a needle containing 1 µM fMLP. Video shows a 15-s interval. Time stamp indicates minutes and seconds. Bar, 10 µm.
Utrophin-GFP localization in Hax1 shRNA A–expressing PLB-985 cells during chemotaxis (related to Fig. 2). Hax1 shRNA A PLB-985 cells expressing utrophin-GFP during chemotaxis on fibrinogen to a needle containing 1 µM fMLP. Video shows a 15-s interval. Time stamp indicates minutes and seconds. Bar, 10 µm.
Utrophin-GFP localization in Hax1 shRNA A–expressing PLB-985 cells during chemotaxis (related to Fig. 2). Hax1 shRNA A PLB-985 cells expressing utrophin-GFP during chemotaxis on fibrinogen to a needle containing 1 µM fMLP. Video shows a 15-s interval. Time stamp indicates minutes and seconds. Bar, 10 µm.
Inhibition of integrin-mediated adhesion rescues chemotaxis of Hax1-deficient neutrophils
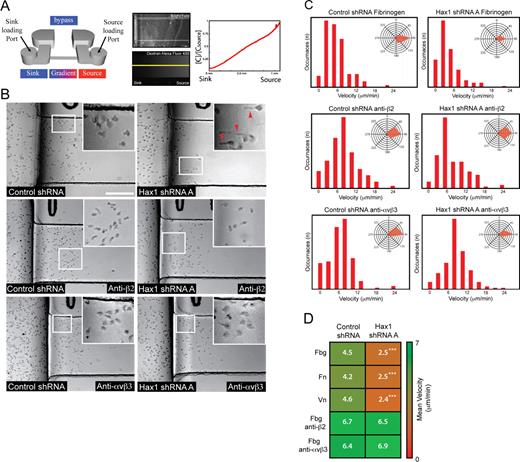
To further characterize how Hax1 regulates neutrophil motility and uropod detachment, live imaging of control and Hax1-deficient cells was performed in microfluidic gradient generators. Static microfluidic gradient generation allows the precise control of chemoattractant gradients over extended durations under flow free conditions and can be used successfully to study neutrophil chemotaxis in a high throughput format (Abhyankar et al., 2008; Berthier et al., 2010). Hax1-deficient PLB-985 cells displayed impaired chemotaxis with a uropod retraction defect in a linear gradient of fMLP on fibrinogen, in accordance with our findings using the micropipette assay (Fig. 3, A and B; and Videos 8 and 9). Quantification by cell tracking revealed a significantly decreased velocity of Hax1-deficient cells compared with control cells on all ligands tested (Fig. 3, C and D). Interestingly, directionality was not impaired in Hax1 knockdown cells, suggesting that directional sensing is intact (Fig. 3 C, insets).
Directed migration is impaired in Hax1-deficient PLB-985 cells using microfluidic gradient generators. (A) Schematic of the microfluidic device showing stable gradient formation with Alexa Fluor 488–conjugated dextran quantified by line scan analysis (yellow line). C/Csource, concentration of the line scan over concentration of the source. (B) Image taken from a time-lapse video of control and Hax1 knockdown cells during chemotaxis on fibrinogen (Videos 8 and 9). Hax1-deficient cells display impaired migration and develop elongated tails (arrowheads). Insets are zoomed regions indicated by the white squares. Bar, 100 µm. (C) Representative velocity and angle (insets) histogram of control and Hax1-deficient cells during chemotaxis on fibrinogen in the absence and presence of β2 or αvβ3 integrin function-blocking antibodies (from one of three independent experiments; n = 10 cells tracked over 30 min during chemotaxis). (D) Mean velocity heat map from three independent experiments in triplicate of control and Hax1 knockdown cells during chemotaxis on fibronectin (Fn), fibrinogen (Fbg), and vitronectin (Vn) and in the presence of integrin function-blocking antibodies on fibrinogen. n = 30 cells per replicate. ***, P < 0.01; t test.
Directed migration is impaired in Hax1-deficient PLB-985 cells using microfluidic gradient generators. (A) Schematic of the microfluidic device showing stable gradient formation with Alexa Fluor 488–conjugated dextran quantified by line scan analysis (yellow line). C/Csource, concentration of the line scan over concentration of the source. (B) Image taken from a time-lapse video of control and Hax1 knockdown cells during chemotaxis on fibrinogen (Videos 8 and 9). Hax1-deficient cells display impaired migration and develop elongated tails (arrowheads). Insets are zoomed regions indicated by the white squares. Bar, 100 µm. (C) Representative velocity and angle (insets) histogram of control and Hax1-deficient cells during chemotaxis on fibrinogen in the absence and presence of β2 or αvβ3 integrin function-blocking antibodies (from one of three independent experiments; n = 10 cells tracked over 30 min during chemotaxis). (D) Mean velocity heat map from three independent experiments in triplicate of control and Hax1 knockdown cells during chemotaxis on fibronectin (Fn), fibrinogen (Fbg), and vitronectin (Vn) and in the presence of integrin function-blocking antibodies on fibrinogen. n = 30 cells per replicate. ***, P < 0.01; t test.
To determine whether increased neutrophil-mediated adhesion was responsible for the impaired migration of Hax1-deficient cells, we characterized the effects of β2 and αvβ3 integrin function-blocking antibodies on the directed migration of Hax1 knockdown cells. Both antibody treatments increased the migration velocity of Hax1-deficient cells to control levels on fibrinogen (Fig. 3, B–D), whereas control IgG antibody had no effect (not depicted), indicating that the impaired migration was caused, at least in part, by increased integrin-mediated adhesion. Collectively, our findings demonstrate that Hax1 is necessary for efficient neutrophil chemotaxis on different integrin ligands by modulating uropod retraction.
Hax1 and RhoA regulate neutrophil adhesion and motility
To determine whether Hax1 regulates global neutrophil adhesion, adhesion of control and Hax1-deficient cells was quantified on different ligands. Hax1-deficient PLB-985 cells treated with fMLP displayed a more than twofold increase in adhesion compared with control cells (Fig. 4 A), which was dependent on ligand density (Fig. S2, A and B). We did not observe differences in the surface expression of αvβ3 integrin, total β2 integrin, or αMβ2 integrin (CD11b) in Hax1-deficient PLB-985 cells compared with control cells (Fig. S1 B and Fig. S2 C). To determine whether the increased adhesion observed in Hax1-deficient cells was mediated by cell surface integrins, we characterized the effects of β2 and αvβ3 integrin function-blocking antibodies on the adhesion of control and Hax1 knockdown cells. Inhibition of either β2 or αvβ3 integrins reduced the maximum adhesion observed (Fig. 4 B), whereas control IgG antibody had no effect (not depicted).
Hax1- and RhoA-deficient PLB-985 cells display increased integrin-mediated adhesion. (A) Adhesion assay of differentiated control and Hax1-deficient cells on fibronectin (Fn), fibrinogen (Fbg), and vitronectin (Vn). (B) Adhesion assay of control and Hax1-deficient cells on 10 µg/ml fibrinogen. For antibody blocking, cells were incubated with β2 or αvβ3 integrin function-blocking antibodies as described in Materials and methods. (C) Lentiviral knockdown of RhoA in PLB-985 cells. (D) Adhesion assay of differentiated control and RhoA-deficient PLB-985 cells on 10 µg/ml fibrinogen. (E) Migration velocities of differentiated control and RhoA-deficient cells during chemotaxis in microfluidic gradient generators. P-values were obtained by ANOVA with Tukey posttest (control shRNA, n = 23 cells and RhoA shRNA 1 and RhoA shRNA 2, n = 22 cells; horizontal lines indicate the mean velocity from three independent experiments). *, P < 0.05. Error bars indicate means ± SEM from three independent experiments.
Hax1- and RhoA-deficient PLB-985 cells display increased integrin-mediated adhesion. (A) Adhesion assay of differentiated control and Hax1-deficient cells on fibronectin (Fn), fibrinogen (Fbg), and vitronectin (Vn). (B) Adhesion assay of control and Hax1-deficient cells on 10 µg/ml fibrinogen. For antibody blocking, cells were incubated with β2 or αvβ3 integrin function-blocking antibodies as described in Materials and methods. (C) Lentiviral knockdown of RhoA in PLB-985 cells. (D) Adhesion assay of differentiated control and RhoA-deficient PLB-985 cells on 10 µg/ml fibrinogen. (E) Migration velocities of differentiated control and RhoA-deficient cells during chemotaxis in microfluidic gradient generators. P-values were obtained by ANOVA with Tukey posttest (control shRNA, n = 23 cells and RhoA shRNA 1 and RhoA shRNA 2, n = 22 cells; horizontal lines indicate the mean velocity from three independent experiments). *, P < 0.05. Error bars indicate means ± SEM from three independent experiments.
To further investigate the relationship between Hax1 and RhoA in the regulation of neutrophil adhesion and motility, RNA interference was used to deplete RhoA from PLB-985 cells (Fig. 4 C). Similar to Hax1 depletion, RhoA-deficient neutrophils displayed a significant increase in adhesion to fibrinogen (Fig. 4 D). Under steep gradients of fMLP using the needle assay, RhoA-deficient neutrophils exhibited impaired migration with multiple protrusions, similar to what has been previously reported (Video 10; Heasman et al., 2010). In microfluidic chambers that produce shallow chemoattractant gradients, RhoA knockdown cells showed impaired migration with elongated uropods and impaired detachment, similar to Hax1-deficient cells (Fig. 4 E and Fig. S2 D). Collectively, our findings demonstrate that Hax1 and RhoA regulate neutrophil chemotaxis through the modulation of integrin-mediated adhesion and uropod detachment.
RhoA-deficient PLB-985 cells during chemotaxis. Time-lapse video demonstrating multiple protrusions and bifurcated pseudopods of RhoA-deficient PLB-985 cells during chemotaxis in a steep fMLP gradient delivered by a micropipette tip. Frames were taken every 15 s. Bar, 10 µm.
RhoA-deficient PLB-985 cells during chemotaxis. Time-lapse video demonstrating multiple protrusions and bifurcated pseudopods of RhoA-deficient PLB-985 cells during chemotaxis in a steep fMLP gradient delivered by a micropipette tip. Frames were taken every 15 s. Bar, 10 µm.
The N-terminal region of Hax1 is sufficient to rescue neutrophil adhesion and motility of Hax1-deficient cells
We next sought to identify the regions of Hax1 that are sufficient to rescue the adhesion and motility defects of Hax1-deficient cells. Stable expression of shRNA-resistant full-length GFP-Hax1 (Fig. S3 A) was sufficient to rescue both neutrophil adhesion and chemotaxis (Fig. 5, A and D). Hax1-deficient PLB-985 cells stably expressing the N-terminal 113 amino acids of Hax1 (GFP–Hax1 1–113), but not the C-terminal 166 amino acids (GFP-Hax1Δ113), rescued neutrophil adhesion and motility (Fig. 5, A and D). Interestingly, there are currently no known interacting partners that bind to the N-terminal 113 amino acids of Hax1.
Constitutively active RhoA and the N-terminal region of Hax1 rescue neutrophil adhesion. (A) Adhesion assays of control cells stably expressing GFP and Hax1 shRNA A cells expressing GFP, or an shRNA-resistant GFP-Hax1, GFP–Hax1 1–113, and GFP-Hax1Δ113 were plated on 10 µg/ml fibrinogen in the presence or absence of 200 nM fMLP. (B) Adhesion assay of control and Hax1-deficient cells plated on 10 µg/ml fibrinogen in the presence or absence of 200 nM fMLP and 10 µM colchicine for 30 min. (C) Adhesion assay of control and Hax1-deficient cells stably expressing GFP or GFP-RhoA (Q63L). (B and C) ***, P < 0.01; one-way ANOVA with Tukey posttest. (D) Migration velocities of control and Hax1-deficient cells stably expressing GFP, GFP-RhoA (Q63L), GFP-Hax1, GFP–Hax1 1–113, and GFP-Hax1Δ113 during chemotaxis to fMLP in microfluidic chemotaxis devices. Horizontal lines indicate the mean velocity. (A and D) Means with common letters indicate P > 0.05 by ANOVA with Tukey posttest.
Constitutively active RhoA and the N-terminal region of Hax1 rescue neutrophil adhesion. (A) Adhesion assays of control cells stably expressing GFP and Hax1 shRNA A cells expressing GFP, or an shRNA-resistant GFP-Hax1, GFP–Hax1 1–113, and GFP-Hax1Δ113 were plated on 10 µg/ml fibrinogen in the presence or absence of 200 nM fMLP. (B) Adhesion assay of control and Hax1-deficient cells plated on 10 µg/ml fibrinogen in the presence or absence of 200 nM fMLP and 10 µM colchicine for 30 min. (C) Adhesion assay of control and Hax1-deficient cells stably expressing GFP or GFP-RhoA (Q63L). (B and C) ***, P < 0.01; one-way ANOVA with Tukey posttest. (D) Migration velocities of control and Hax1-deficient cells stably expressing GFP, GFP-RhoA (Q63L), GFP-Hax1, GFP–Hax1 1–113, and GFP-Hax1Δ113 during chemotaxis to fMLP in microfluidic chemotaxis devices. Horizontal lines indicate the mean velocity. (A and D) Means with common letters indicate P > 0.05 by ANOVA with Tukey posttest.
Constitutively active RhoA rescues adhesion and partially rescues motility of Hax1-deficient neutrophils
To determine whether activation of RhoA is sufficient to rescue the adhesion and motility defects of Hax1-deficient neutrophils, we treated control and Hax1-depleted cells with nocodazole and colchicine or ectopically expressed constitutively active RhoA (Q63L). It has previously been reported that microtubule disruption with either nocodazole or colchicine increases global Rho activity in neutrophils (Niggli, 2003; Xu et al., 2005). Accordingly, we found that treatment with either nocodazole or colchicine rescued the adhesion defect of fMLP-treated Hax1-deficient cells to control levels (Fig. 5 B and Fig. S3 C). Moreover, stable expression of constitutively active GFP-RhoA (Q63L) (Fig. S3 B) in Hax1-deficient cells restored adhesion to control levels (Fig. 5 C). We did not observe any decrease in endogenous RhoA or Rac expression in PLB-985 cells that ectopically express constitutively active GFP-RhoA (Q63L) as has previously been reported for other cell types (Fig. S3 B; Boulter et al., 2010). Ectopic expression of constitutively active RhoA only partially rescued the migration defect of Hax1-deficient cells, suggesting that Hax1 may be necessary for optimum spatiotemporal regulation of RhoA during motility (Fig. 5 D). Collectively, our findings demonstrate that activation of RhoA is sufficient to rescue the adhesion defect of Hax1-deficient cells, indicating that Hax1 negatively regulates neutrophil adhesion through the modulation of RhoA activity.
In summary, we have identified Hax1 as a novel negative regulator of integrin-mediated adhesion that affects uropod detachment and neutrophil chemotaxis. We have also found that Hax1 regulates RhoA activation in neutrophils and that activation of RhoA is sufficient to rescue the adhesion defects of Hax1-deficient cells. Previous studies have suggested that Hax1 loss-of-function mutations mediate the development of congenital neutropenia syndromes through the regulation of neutrophil apoptosis (Klein et al., 2006; Chao et al., 2008). Our findings raise the intriguing alternative hypothesis that Hax1 deficiency may modulate neutrophil function by affecting Rho GTPase signaling and neutrophil adhesion and motility.
It is not clear how Hax1 regulates RhoA activity and neutrophil adhesion. Gα13 and HS1 represent attractive candidates to mediate Hax1 effects. However, we found that the N-terminal region of Hax1, but not the region that interacts with Gα13 and HS1, is sufficient to rescue neutrophil adhesion and motility. Accordingly, the C-terminal region of Hax1 localizes to the leading edge, whereas the N-terminal region of Hax1, which rescues uropod retraction, displays a more rearward localization. Furthermore, we found that ectopic expression of Hax1 and constitutively active RhoA (Q63L) was sufficient to restore the increased adhesion defect in Hax1-deficient cells. Interestingly, RhoA (Q63L) only partially rescued neutrophil motility, suggesting that the proper spatiotemporal regulation of RhoA is critical for efficient chemotaxis. To date, no binding partners have been identified that interact with the N-terminal region of Hax1. Understanding these types of interactions will likely shed light on how Hax1 functions to regulate rear detachment and neutrophil motility.
A final unanswered question is how Hax1 mutations affect neutrophil adhesion and motility to control neutrophil homeostasis in the context of congenital neutropenia syndromes. The finding that Hax1 expression is reduced with neutrophil differentiation is surprising. This raises the intriguing possibility that Hax1 expression has an important function during neutrophil differentiation and that its deficiency may allow the transition to the more adherent phenotype of mature neutrophils. Recent studies have shown that other congenital neutropenia syndromes, including WHIM (warts, hypogammaglobulinemia, infections, and myelokathexis) syndrome, have associated defects in neutrophil retention and motility (Kawai and Malech, 2009; Walters et al., 2010), suggesting that defects in neutrophil adhesion and migration may be key factors that contribute to the pathogenesis of other congenital neutropenia syndromes, such as Kostmann disease. A challenge for future studies will be to establish in vivo systems that will allow these kinds of questions to be answered.
Materials and methods
Antibodies and reagents
Sources for antibodies and reagents used in this study are mouse anti-Hax1 clone 52 and mouse anti-HS1 clone 9 (BD), rabbit anti-Hax1 (Proteintech Group, Inc.), mouse antivinculin clone VIN-11-5 and mouse anti–β-actin clone AC-15 (Sigma-Aldrich), rabbit anti-RhoA (67B9) and rabbit anti-Rac1/2/3 (L129; Cell Signaling Technology), goat anti–rabbit Alexa Fluor 680 and goat anti–mouse Alexa Fluor 800 (Invitrogen), and mouse anti–αvβ3 integrin (clone LM609; Millipore), mouse anti–human CD18 (clone TS1/18), and mouse anti–human Cd11b (clone CBRM1/5; BioLegend). Human fibrinogen, f-Met-Leu-Phe (Sigma-Aldrich), nocodazole (EMD), colchicine (Arcos Organics), and fibronectin were purified from human plasma by affinity chromatography as previously described (Ruoslahti et al., 1982), and human vitronectin was a gift from D. Mosher (University of Wisconsin–Madison, Madison, WI).
Cell culture
PLB-985 cells (gift from C. Parent, National Cancer Institute, National Institutes of Health, Bethesda, MD) were maintained in RPMI 1640 supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 µg/ml streptomycin at a concentration of 0.1–1 × 106 cells/ml. To differentiate, 1.25% DMSO was added to 2 × 105 cells/ml for 6 d. Differentiation was confirmed through FACS analysis of CD11b expression. HEK293T cells (American Type Culture Collection) and Platinum-GP cells (Cell Biolabs, Inc.) were maintained in Dulbecco’s minimal essential medium supplemented with 10% FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin.
DNA constructs, transfections, and viral infections
Hax1 was RT-PCR purified from TRIZOL-extracted total RNA from the neutrophil-like cell line HL-60 using the primers forward, 5′-ATGAGCCTCTTTGATCTCTTCCGGGGCTTTTTCG-3′, and reverse, 5′-CCGGGACCGGAACCAACGTCCCAGGAATAAGT-3′. Full-length Hax1, Hax1 1–113, and Hax1Δ113 sequences were PCR amplified from Hax1 cDNA and cloned into pEGFP-N1 and pEGFP-C1 (Takara Bio Inc.). 2 × 106 PLB-985 cells 6 d after differentiation were transiently transfected with 5 µg DNA using a nucleofector (Cell Line Nucleofector Kit V; Lonza) following the manufacturer’s instructions.
Lentiviral Hax1 and RhoA shRNA targets were purchased from Thermo Fisher Scientific. Targets used in this study were Hax1 shRNA A, 5′-ACAGACACTTCGGGACTCAAT-3′; Hax1 shRNA B, 5′-CCAGAGGCCATTTCATAGGTT-3′; RhoA shRNA 1, 5′-GTACATGGAGTGTTCAGCAAA-3′; RhoA shRNA 2, 5′-TGGAAAGACATGCTTGCTCAT-3′; and control shRNA, 5′-TGTCTCCGAACGTGTCACGTT-3′. HEK293T cells were grown to 70% confluency in a 10-cm tissue-culture dish for each lentiviral target and transfected using 6 µg Hax1 or RhoA shRNA, 0.6 µg VSV-G, and 5.4 µg CMV 8.9.1. A 72-h viral supernatant was collected and concentrated using a lentivirus concentrator (Lenti-X; Takara Bio Inc.) following the manufacturer’s instructions. 106 PLB-985 cells were infected with viral supernatant for 3 d in the presence of 4 µg/ml polybrene. Stable cell lines were generated with 1 µg/ml puromycin selection.
Retroviral transduction of GFP-RhoA (Q63L) and shRNA-resistant GFP-Hax1, GFP–Hax1 1–113, and GFP-Hax1Δ113 was performed as follows: Platinum-GP cells were grown to 70% confluency in a 10-cm tissue-culture dish and transfected using 10 µg retroviral DNA and 3 µg VSV-G. A 72-h viral supernatant was collected and added to 106 PLB-985 cells for 3 d in the presence of 15 µg/ml polybrene. Cells were sorted for equal GFP expression levels using the cell sorter (FACSAria; BD) at the University of Wisconsin flow cytometry facility.
Needle chemotaxis
Stable cell lines expressing shRNA cell lines or transiently transfected cells expressing GFP or RFP fusion proteins were incubated in a 35-mm glass-bottom culture dish that had been coated with 10 µg/ml fibrinogen and blocked with 2% BSA. 106 neutrophils were placed in Gey’s media (20 mM Hepes, 132 mM NaCl, 1 mM MgSO4, 1.2 mM NaH2PO4, 5 mM glucose, 1 mM CaCl2, and 0.5% human serum albumin; American Red Cross Blood Services), and chemotaxis assays were performed using a microinjection system (FemtoJet; Eppendorf) as previously described (Servant et al., 1999; Lokuta et al., 2007). For some experiments, control shRNA cells were incubated with 30 µM Y-27632 for 30 min before chemotaxis. Imaging was performed in a 37°C humidified chamber on an inverted microscope (Eclipse TE300; Nikon) using a 60×, NA of 1.4, or 10×, NA of 0.45, objective. Images were collected using a charge-coupled device camera (CoolSNAP ES2; Photometrics) and captured into MetaVue imaging software (v6.2; Molecular Devices).
Rho GTPase pull-down and immunoblotting
Rho GTPase pull-down assays were performed as previously described (Lokuta et al., 2003). In brief, differentiated PLB-985 cells were serum starved in modified HBSS (150 mM NaCl2, 4 mM KCL, 1 mM MgCl2, 10 mM glucose, 20 mM Hepes, pH 7.4, and 0.2% human serum albumin) and plated on 10 µg/ml fibrinogen or kept in suspension for 10 min and then stimulated with 1 µM fMLP for 5 min, immediately placed on ice, and lysed in radioimmunoprecipitation assay extraction buffer (20 mM Tris-HCL, pH 7.5, 150 mM NaCl2, 2 mM EDTA, 2 mM EGTA, 1% TX-100, 0.5% NP-40, 0.25% sodium deoxycholate, 1 µg/ml pepstatin A, 2 µg/ml aprotinin, 1 µg/ml leupeptin, and 200 nM phenylmethanesulphonyl fluoride). 40 µg of bacterially expressed GST-rhotekin–Rho-binding domain (RBD) or GST-PAK–Rac/Cdc42 (p21)-binding domain (PBD) was incubated with the lysates for 1 h at 4°C. The affinity-precipitated products were run on an SDS-PAGE gel, transferred to nitrocellulose, and immunoblotted for RhoA or Rac. To detect endogenous Rac, we used a Rac1/2/3 antibody. Therefore, the PAK-PBD pull-down does not distinguish between Rac1 and Rac2. Quantification of total RhoA or Rac for all samples was normalized to control shRNA relative to vinculin loading control. The mean ± SEM ratio of RhoA-GTP/RhoA or Rac-GTP/Rac was graphed relative to the control from three independent experiments.
For Hax1 expression analysis, 5 × 106 cells were lysed in radioimmunoprecipitation assay extraction buffer, and 20 µg of total protein was run on SDS-PAGE, transferred to nitrocellulose, and immunoblotted for Hax1, vinculin, HS1, and actin. Western blots were imaged and quantified with an infrared imaging system (Odyssey; LI-COR Biosciences).
Adhesion assays
100,000 Hax1 shRNA A, RhoA shRNA 1 and 2, or control shRNA cells were labeled with calcein AM (Invitrogen) and plated in the presence or absence of 200 nM fMLP for 30 min in quadruplicate in a 96-well black microplate (Greiner Bio-one) coated with fibrinogen, fibronectin, or vitronectin followed by blocking with 2% BSA. In some cases, cells were pretreated with control IgG, β2, or αvβ3 (clone LM609) integrin–blocking antibodies at a concentration of 1:1,000 for 15 min at 4°C before plating. For microtubule disruption, cells were treated with 30 µM nocodazole or 10 µM colchicine for 30 min at 37°C. Fluorescence was measured using a plate reader (Victor3 V; PerkinElmer). Plates were gently washed with modified HBSS in between readings until the remaining control shRNA-fMLP cells were <10% of the original input.
Microfluidic chemotaxis assays
Microfluidic devices were fabricated as previously described (Berthier et al., 2010). In brief, the devices enable the generation of a static gradient of chemoattractant between a source chamber containing fMLP and a sink chamber containing the PLB-985 cells, which were connected by a 1-mm-long, 400-µm-wide thin channel in which the cells migrate. Microfluidic chambers were coated with fibrinogen, fibronectin, or vitronectin for 1 h at room temperature in PBS. 3 µl neutrophil suspension (4 × 106 cells/ml) was added to the device, and 100 nM fMLP was added to the source port. The gradient was allowed to set up and equilibrate in a 37°C humidified chamber for 15 min before imaging. Time-lapse imaging was performed using a 10×, NA of 0.45, objective and motorized stage (Ludl Electronic Products) on an inverted microscope (Eclipse TE300) using a charge-coupled device camera (CoolSNAP ES2) and captured into MetaVue imaging software v6.2. Images were taken every 30 s for 30–45 min with up to eight devices being imaged simultaneously. For integrin-blocking experiments, control IgG and β2 and αvβ3 integrin antibodies were incubated with cells at a concentration of 1:1,000 on ice for 15 min before loading in the microfluidic device. Tracking and velocity measurements were performed using ImageJ analysis software with the manual tracking plugin (National Institutes of Health).
Flow cytometry and apoptosis assay
For integrin αvβ3, integrin β2, and integrin αMβ2 surface staining of differentiated PLB-985 cells, 106 cells were labeled with FITC-conjugated LM609, which binds to the αvβ3 complex, anti-CD18, or anti-CD11b for 30 min on ice. Cells were washed twice with FACS buffer (PBS supplemented with 2% FBS and 0.02% NaN3) and analyzed by flow cytometry. For apoptosis induction of PLB-985 cells, 105 5-d differentiated cells were incubated with 1 mM H2O2 for 30 min in PLB-985 media and then incubated in fresh media for 12 h. Cells were analyzed for apoptosis induction using the Annexin V Apoptosis Detection kit I (BD) following the manufacturer’s instruction and analyzed by flow cytometry.
Statistical analysis
For statistical comparison, unpaired t test or one-way analysis of variance (ANOVA) was used with Tukey posttests with P < 0.05 considered significant.
Online supplemental material
Fig. S1 shows that Hax1-deficient PLB-985 cells have increased sensitivity to H2O2-induced apoptosis, reduced RhoA-GTP, and increased Rac-GTP levels. Fig. S2 shows that Hax1-deficient PLB-985 cells have increased adhesion to vitronectin and fibrinogen, and RhoA-deficient cells display elongated uropods. Fig. S3 shows immunoblotting of Hax1-deficient PLB-985 cells expressing GFP-Hax1, Hax1 mutants, and constitutively active RhoA (Q63L). Video 1 shows Hax1-GFP localization in PLB-985 cells during chemotaxis. Video 2 shows Hax1 1–113–GFP localization in PLB-985 cells during chemotaxis. Video 3 shows Hax1Δ113-GFP localization in PLB-985 cells during chemotaxis. Video 4 shows the chemotaxis of control shRNA PLB-985 cells. Video 5 shows the chemotaxis of Hax1 shRNA A PLB-985 cells. Video 6 shows utrophin-GFP localization in PLB-985 cells treated with Y-27632 during chemotaxis. Video 7 shows utrophin-GFP localization in Hax1 shRNA A–expressing PLB-985 cells during chemotaxis. Video 8 shows control shRNA chemotaxis in microfluidic gradient generators. Video 9 shows Hax1 shRNA A chemotaxis in microfluidic gradient generators. Video 10 shows RhoA-deficient PLB-985 cells during chemotaxis.
Hax1-GFP localization in PLB-985 cells during chemotaxis (related to Fig. 1). dPLB-985 cells expressing utrophin-RFP and Hax1-GFP during chemotaxis on fibrinogen to a needle containing 1 µM fMLP. Video shows a 15-s interval. (top left) differential interference contrast (DIC). (top right) utrophin-RFP. (bottom left) Hax1-GFP. (bottom right) RFP-GFP overlay. Time stamp indicates minutes and seconds. Bar, 10 µm.
Hax1-GFP localization in PLB-985 cells during chemotaxis (related to Fig. 1). dPLB-985 cells expressing utrophin-RFP and Hax1-GFP during chemotaxis on fibrinogen to a needle containing 1 µM fMLP. Video shows a 15-s interval. (top left) differential interference contrast (DIC). (top right) utrophin-RFP. (bottom left) Hax1-GFP. (bottom right) RFP-GFP overlay. Time stamp indicates minutes and seconds. Bar, 10 µm.
Chemotaxis of control shRNA PLB-985 cells (related to Fig. 2). Control shRNA PLB-985 cells during chemotaxis on fibrinogen to a needle containing 1 µM fMLP. Video shows a 15-s interval. Time stamp indicates minutes and seconds. Bar, 100 µm.
Chemotaxis of control shRNA PLB-985 cells (related to Fig. 2). Control shRNA PLB-985 cells during chemotaxis on fibrinogen to a needle containing 1 µM fMLP. Video shows a 15-s interval. Time stamp indicates minutes and seconds. Bar, 100 µm.
Chemotaxis of Hax1 shRNA A PLB-985 cells (related to Fig. 2). Hax1 shRNA A PLB-985 cells during chemotaxis on fibrinogen to a needle containing 1 µM fMLP. Video shows a 15-s interval. Time stamp indicates minutes and seconds. Bar, 100 µm.
Chemotaxis of Hax1 shRNA A PLB-985 cells (related to Fig. 2). Hax1 shRNA A PLB-985 cells during chemotaxis on fibrinogen to a needle containing 1 µM fMLP. Video shows a 15-s interval. Time stamp indicates minutes and seconds. Bar, 100 µm.
Control shRNA chemotaxis in microfluidic gradient generators (related to Fig. 3). Control shRNA PLB-985 cells in microfluidic chemotaxis chambers coated with fibrinogen in a linear gradient of fMLP. Video shows a 15-s interval. Bar, 100 µm.
Control shRNA chemotaxis in microfluidic gradient generators (related to Fig. 3). Control shRNA PLB-985 cells in microfluidic chemotaxis chambers coated with fibrinogen in a linear gradient of fMLP. Video shows a 15-s interval. Bar, 100 µm.
Hax1 shRNA A chemotaxis in microfluidic gradient generators (related to Fig. 3). Hax1 shRNA A PLB-985 cells in microfluidic chemotaxis chambers coated with fibrinogen in a linear gradient of fMLP. Video shows a 15-s interval. Bar, 100 µm.
Hax1 shRNA A chemotaxis in microfluidic gradient generators (related to Fig. 3). Hax1 shRNA A PLB-985 cells in microfluidic chemotaxis chambers coated with fibrinogen in a linear gradient of fMLP. Video shows a 15-s interval. Bar, 100 µm.
Acknowledgments
We would like to thank Carole Parent and Deane Mosher for reagents. We thank Sarah Wernimont and Kathy Schell for their flow cytometry expertise.
This work was supported by National Institutes of Health grants T32ES007015 (to P.J. Cavnar) and GM074827 (to A. Huttenlocher), the American Heart Association grant 10POST3230031 (to P.J. Cavnar), and a Burroughs Wellcome grant (to A. Huttenlocher).