Late endosomes (LEs) have characteristic intracellular distributions determined by their interactions with various motor proteins. Motor proteins associated to the dynactin subunit p150Glued bind to LEs via the Rab7 effector Rab7-interacting lysosomal protein (RILP) in association with the oxysterol-binding protein ORP1L. We found that cholesterol levels in LEs are sensed by ORP1L and are lower in peripheral vesicles. Under low cholesterol conditions, ORP1L conformation induces the formation of endoplasmic reticulum (ER)–LE membrane contact sites. At these sites, the ER protein VAP (VAMP [vesicle-associated membrane protein]-associated ER protein) can interact in trans with the Rab7–RILP complex to remove p150Glued and associated motors. LEs then move to the microtubule plus end. Under high cholesterol conditions, as in Niemann-Pick type C disease, this process is prevented, and LEs accumulate at the microtubule minus end as the result of dynein motor activity. These data explain how the ER and cholesterol control the association of LEs with motor proteins and their positioning in cells.
Introduction
Late endosomes (LEs), lysosomes, and lysosome-related organelles are characteristically positioned by motor protein interactions with cytoskeletal elements. The typical steady-state perinuclear positioning of LEs is dictated by the net balance of vectorial movements toward opposite ends of microtubules. The dynein–dynactin motor drives minus end (centripetal) transport, and at least two kinesin motors (KIF5 and KIF3) have been implicated in plus end (centrifugal) transport (Haimo and Rozdzial, 1989; Hollenbeck and Swanson, 1990; Vale et al., 1992; Wubbolts et al., 1999; Brown et al., 2005). Other factors are also involved in maintaining the typical distribution of LEs, including small GTPases, kinases (Pelkmans et al., 2005), motor-associated factors like huntingtin (Colin et al., 2008), and cholesterol (Lebrand et al., 2002; Sugii et al., 2006).
Rab GTPases specify organelle identity (Pfeffer, 2001; Zerial and McBride, 2001) and control selective motor activity on defined compartments (Echard et al., 1998; Jordens et al., 2001, 2005; Hoepfner et al., 2005). The small GTPase Rab7 associates with the LE (Zerial and McBride, 2001), and its effector Rab7-interacting lysosomal protein (RILP) binds the p150Glued subunit of the dynactin protein complex (Jordens et al., 2001; Johansson et al., 2007). The p150Glued protein can bind to dynein heavy chain via the dynein intermediate chain (DIC), but it can also bind to the oppositely directed kinesin II (KIF3A) motor via KAP3 (Wubbolts et al., 1999; Deacon et al., 2003; Brown et al., 2005). Kinesin I (KIF5) also drives plus end transport of LEs (Hollenbeck and Swanson, 1990; Nakata and Hirokawa, 1995) via interactions with other receptors. When p150Glued binding to LEs is inhibited, the kinesin I motor remains active, resulting in net plus end transport (Wubbolts et al., 1999). Although Rab7 and RILP are sufficient for the recruitment of p150Glued-associated motor proteins, the actual translocation of LEs toward the microtubule-organizing center (MTOC) further requires βIII spectrin and the oxysterol-binding protein (OSBP)–related protein 1L (ORP1L; Johansson et al., 2007). ORP1L contains a pleckstrin homology (PH) domain that binds phosphoinositides (Johansson et al., 2005), a protein-interacting FFAT (two phenylalanines [FF] in an acidic tract) motif (Loewen et al., 2003; Loewen and Levine, 2005), and a C-terminal OSBP-related domain (ORD) able to bind 25-hydroxycholesterol (Im et al., 2005; Suchanek et al., 2007). The ER protein OSBP is related to ORP1L and also contains a FFAT motif that interacts with VAMP (vesicle-associated membrane protein)-associated ER protein (VAP) A and VAP-B (Loewen and Levine, 2005). VAP-A and VAP-B are homo- or heterodimeric integral ER proteins (Nishimura et al., 1999; Hamamoto et al., 2005) and participate in ER export of proteins and lipids (Wyles et al., 2002).
Cholesterol has been implicated in LE positioning. Cholesterol-laden LEs in Niemann-Pick type C patients cluster around the MTOC (Mukherjee and Maxfield, 2004). This phenotype is shared with other lysosomal storage diseases like Tangier, Fabry, and Gaucher (Neufeld et al., 2004; Maxfield and Tabas, 2005). Increasing endosomal cholesterol levels with the chemical compound U-18666A causes a similar phenotype (Roff et al., 1991; Koh and Cheung, 2006; Sobo et al., 2007). The LE clustering requires Rab7 and Rab9 (Lebrand et al., 2002; Narita et al., 2005; Chen et al., 2008). Conversely, cholesterol depletion results in LE positioning at the plus end of microtubules (Sugii et al., 2006). This suggests that motor protein activities controlling LE positioning involve Rab GTPases and cholesterol, but the mechanism is unclear.
We show that cholesterol in LEs is sensed by ORP1L, which transmits this information to the Rab7–RILP–p150Glued complex through the formation of ER–LE membrane contact sites (MCSs). At these sites, the ER protein VAP enters the Rab7–RILP complex to control p150Glued binding and positioning of LEs. The characteristic clustering of LEs at the microtubule minus end in Niemann-Pick type C and other lysosomal storage diseases is a result of this process.
Results
Chemical and genetic cholesterol manipulations in LEs and their positioning
Cholesterol is an essential component of cellular membranes and contributes to a host of different processes. To visualize the effects of intracellular cholesterol levels on LE positioning, MelJuSo cells were cultured under control or cholesterol-depleting or -increasing conditions. Cholesterol depletion required culturing cells in lipid-free medium supplemented with a statin to prevent the synthesis of endogenous cholesterol (Sugii et al., 2006). Chemical and genetic cholesterol accumulation in LEs were induced by exposure of MelJuSo cells to U-18666A (Liscum and Faust, 1989; Kobayashi et al., 1999) or by silencing NPC1 to mimic Niemann-Pick type C disease (Carstea et al., 1997).
To visualize the effect of these treatments, cells were fixed and stained with filipin (Sobo et al., 2007) to detect cholesterol (Fig. 1 A and Fig. S1 B) or stained for CD63 to label LEs (Fig. 1 B and Fig. S1 B). Images were made at identical microscope settings and show that these cholesterol-manipulating treatments decreased (statin) or increased (U-18666A or siNPC1) intracellular cholesterol levels (Fig. 1, A and C; and Fig. S1 B).
Chemical and genetic cholesterol manipulations and LEs. (A) Modulation of intracellular cholesterol levels. Mel JuSo cells were cultured in normal medium (FCS), cholesterol-depleted medium supplemented with lovastatin (statin), or normal medium supplemented with U-18666A, as indicated. Fixed cells were stained with filipin to detect cholesterol. A color look up table (LUT) shows fluorescence intensities. n > 100 for each condition. (B) Effects of intracellular cholesterol manipulation (control [CTRL], U-18666A, or statin treatment) on LE positioning in RILP- or ORP1L-silenced Mel JuSo cells. Cells were stained for the LE marker CD63 (red), endogenous ORP1L (green), and actin (blue). The position of the CD63-positive vesicles relative to the nucleus (radial distribution) was determined for the various conditions (>50 cells per condition), binned, and plotted (right). The distributions were statistically different according to the Jonckheere-Terpstra test for control siRNA (P = 1.52 × 10−5 and 7.76 × 10−6 for CD63 and ORP1L, respectively) only. No difference was detected for siRILP (P = 0.20 and 0.54 for CD63 and ORP1L, respectively) and for siORP1L (P = 0.29 for CD63). (C) Dynein motor activity and clustering of LEs in NPC1-silenced cells. Mel JuSo cells were transfected with siRNA for NPC1 for 72 h and, after 48 h, transfected with an expression construct for p50dynamitin to disrupt the dynein–dynactin motor. Fixed cells were stained for CD63 and p50dynamitin to mark the overexpressing cells. n > 100. Bars: (A and C)10 µm; (B) 20 µm.
Chemical and genetic cholesterol manipulations and LEs. (A) Modulation of intracellular cholesterol levels. Mel JuSo cells were cultured in normal medium (FCS), cholesterol-depleted medium supplemented with lovastatin (statin), or normal medium supplemented with U-18666A, as indicated. Fixed cells were stained with filipin to detect cholesterol. A color look up table (LUT) shows fluorescence intensities. n > 100 for each condition. (B) Effects of intracellular cholesterol manipulation (control [CTRL], U-18666A, or statin treatment) on LE positioning in RILP- or ORP1L-silenced Mel JuSo cells. Cells were stained for the LE marker CD63 (red), endogenous ORP1L (green), and actin (blue). The position of the CD63-positive vesicles relative to the nucleus (radial distribution) was determined for the various conditions (>50 cells per condition), binned, and plotted (right). The distributions were statistically different according to the Jonckheere-Terpstra test for control siRNA (P = 1.52 × 10−5 and 7.76 × 10−6 for CD63 and ORP1L, respectively) only. No difference was detected for siRILP (P = 0.20 and 0.54 for CD63 and ORP1L, respectively) and for siORP1L (P = 0.29 for CD63). (C) Dynein motor activity and clustering of LEs in NPC1-silenced cells. Mel JuSo cells were transfected with siRNA for NPC1 for 72 h and, after 48 h, transfected with an expression construct for p50dynamitin to disrupt the dynein–dynactin motor. Fixed cells were stained for CD63 and p50dynamitin to mark the overexpressing cells. n > 100. Bars: (A and C)10 µm; (B) 20 µm.
Does cholesterol affect the positioning of LEs? LysoTracker red–labeled LEs were followed by time-lapse fluorescence microscopy under the different cholesterol-manipulating conditions (Videos 1–3). A fraction of the LEs in cells exposed to low or high cholesterol conditions still moved in a bidirectional and stop and go manner, as in control cells. In response to high cholesterol, more vesicles accumulated near the nucleus and fewer in the cell periphery. This is reversed under low cholesterol conditions (Fig. 1 B). These data suggest that cholesterol influences LE positioning.
Motility of LEs in cholesterol-depleted cells. LysoTracker red was used to label LEs in MelJuSo cells, which were cultured and imaged in cholesterol-depleted medium containing statins overnight (as in Fig. 1, A and B). Vesicle movement is visualized as a ratiometric glow LUT. Cells were imaged using a Leica SP2 system equipped with a heated chamber (37°C with 5% CO2). Frames were taken every 1.6 s. The video is shown at 10 frames/s with 100 total frames.
Motility of LEs in cholesterol-depleted cells. LysoTracker red was used to label LEs in MelJuSo cells, which were cultured and imaged in cholesterol-depleted medium containing statins overnight (as in Fig. 1, A and B). Vesicle movement is visualized as a ratiometric glow LUT. Cells were imaged using a Leica SP2 system equipped with a heated chamber (37°C with 5% CO2). Frames were taken every 1.6 s. The video is shown at 10 frames/s with 100 total frames.
Motility of LEs in cholesterol-enriched cells. LysoTracker red was used to label LEs in MelJuSo cells, which were cultured and imaged in medium containing U-18666A (as in Fig. 1, A and B). Vesicle movement is visualized as a ratiometric glow LUT. Cells were imaged using a Leica SP2 system equipped with a heated chamber (37°C with 5% CO2). Frames were taken every 1.6 s. The video is shown at 10 frames/s with 100 total frames.
Motility of LEs in cholesterol-enriched cells. LysoTracker red was used to label LEs in MelJuSo cells, which were cultured and imaged in medium containing U-18666A (as in Fig. 1, A and B). Vesicle movement is visualized as a ratiometric glow LUT. Cells were imaged using a Leica SP2 system equipped with a heated chamber (37°C with 5% CO2). Frames were taken every 1.6 s. The video is shown at 10 frames/s with 100 total frames.
The effect of cholesterol on motor proteins was then investigated. The Rab7 effectors RILP and ORP1L control dynein motor-mediated transport of LEs. To test their involvement in cholesterol-dependent LE positioning, these proteins were silenced by means of siRNA (efficient silencing was confirmed by quantitative PCR [qPCR]). Cholesterol levels were then manipulated, and LEs were detected by staining for CD63 and ORP1L (Fig. 1 B). Endogenous ORP1L was bound to LEs under all cholesterol-manipulating settings. In addition, cholesterol manipulation did not affect the positioning of LEs when RILP or ORP1L was silenced.
Repositioning of LEs near the MTOC under high LE cholesterol concentrations may require the dynein–dynactin motor. To test this, dynactin's subunit p50dynamitin was overexpressed to physically disrupt the motor complex (Burkhardt et al., 1997) in NPC1-silenced cells, and LEs were detected by CD63 labeling. Overexpression of p50dynamitin abrogated clustering of LEs in NPC1-silenced MelJuSo cells, suggesting that functional dynein–dynactin complexes are involved in clustering NPC1-deficient, cholesterol-laden LEs (Fig. 1 C).
ORP1L controls p150Glued binding to Rab7–RILP
The N-terminal ankyrin repeats of ORP1L bind to Rab7 and are involved in dynein motor transport of LEs (Johansson et al., 2005, 2007). To resolve the ORP1L domains involved in the control of LE transport, a series of monomeric RFP (mRFP)–tagged C-terminal truncations of ORP1L constructs (Fig. 2 A and Fig. S2 A) were coexpressed with GFP-RILP in MelJuSo cells. qPCR data revealed that the ORP1L constructs and RILP were expressed 4- to 10-fold over endogenous expression levels (unpublished data). RILP expression was required to visualize recruitment of endogenous p150Glued and dynein motor binding, but the many motors present on a vesicle (Jordens et al., 2001) rendered localization insensitive to cholesterol manipulation (Fig. S1 C). Vesicles labeled by the mRFP-ORP1L constructs were characterized by immunostaining for LE labeling for CD63, LAMP-1, Rab7, and cholesterol, as detected by filipin (Fig. S2, B and C). Markers for the ER, Golgi, or early endosomes were absent on these vesicles (Fig. S2 D).
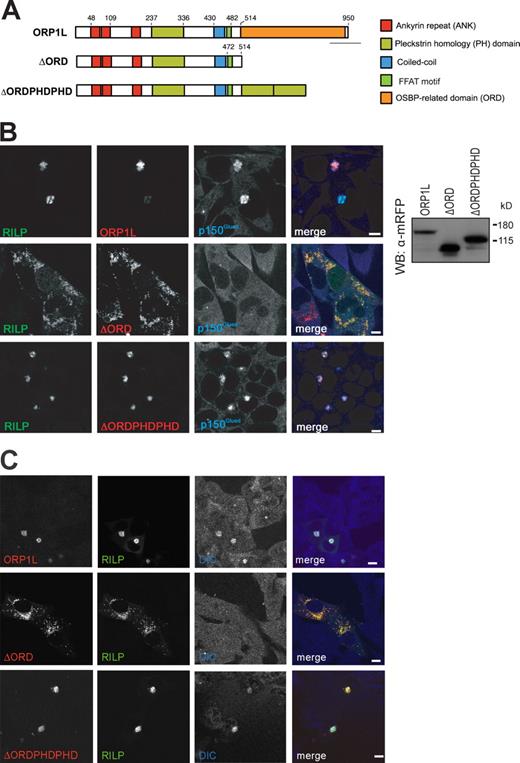
ORP1L controls recruitment of p150Glued to the Rab7–RILP receptor. (A) ORP1L domain structure and constructs. Five domains are predicted in ORP1L. Numbers indicate amino acid residue positions. Constructs were N-terminally tagged with mRFP. The ΔORDPHDPHD chimera had ORD exchanged for a tandem PH domain derived from ORP1L. (B) Effect of ORP1L deletion or chimeric constructs on RILP-mediated p150Glued recruitment. (left) Mel JuSo cells transfected with GFP-RILP and mRFP-ORP1L constructs and stained with anti-p150Glued antibodies before CLSM. For pixel analyses of the images, see Fig. S2 E. n > 200 for each condition. (right) Mel JuSo cells transfected with the indicated mRFP-ORP1L constructs and whole cell lysates analyzed by immunoblotting with anti-mRFP antibodies (WB: α-mRFP). Molecular standards are indicated. WB, Western blot. (C) Effect of ORP1L constructs on RILP recruitment of the p150Glued-associated dynein motor adapter DIC. Mel JuSo cells were transfected with GFP-RILP and the mRFP-ORP1L constructs indicated and stained with anti-DIC antibodies. n > 100 for each condition. Bars, 10 µm.
ORP1L controls recruitment of p150Glued to the Rab7–RILP receptor. (A) ORP1L domain structure and constructs. Five domains are predicted in ORP1L. Numbers indicate amino acid residue positions. Constructs were N-terminally tagged with mRFP. The ΔORDPHDPHD chimera had ORD exchanged for a tandem PH domain derived from ORP1L. (B) Effect of ORP1L deletion or chimeric constructs on RILP-mediated p150Glued recruitment. (left) Mel JuSo cells transfected with GFP-RILP and mRFP-ORP1L constructs and stained with anti-p150Glued antibodies before CLSM. For pixel analyses of the images, see Fig. S2 E. n > 200 for each condition. (right) Mel JuSo cells transfected with the indicated mRFP-ORP1L constructs and whole cell lysates analyzed by immunoblotting with anti-mRFP antibodies (WB: α-mRFP). Molecular standards are indicated. WB, Western blot. (C) Effect of ORP1L constructs on RILP recruitment of the p150Glued-associated dynein motor adapter DIC. Mel JuSo cells were transfected with GFP-RILP and the mRFP-ORP1L constructs indicated and stained with anti-DIC antibodies. n > 100 for each condition. Bars, 10 µm.
To assess the effect of ORP1L and ORP1L mutants on motor recruitment and LE positioning, cells were stained for the dynactin motor subunit p150Glued and DIC. Coexpression of ORP1L and RILP recruited p150Glued (Fig. 2 B) and DIC to LEs (Fig. 2 C) that were clustered near the MTOC. Removal of the C-terminal ORD (ΔORD) prevented p150Glued and DIC recruitment by RILP, resulting in the relocation of RILP-containing compartments to the cell periphery (Fig. 2, B and C).
ORP1L controls cholesterol-dependent LE positioning
OSBP/OSBP-related proteins are involved in nonvesicular transport of sterols, and their ORDs bind and possibly extract sterols from membranes (Ikonen, 2008). The ΔORD mutant of ORP1L should mimic a cholesterol-free conformation of ORP1L. To test whether p150Glued exclusion from Rab7–RILP requires ORD binding to membranes, ORD was exchanged for two ORP1L PH domains in tandem (ΔORDPHDPHD). Tandem PH domains will increase the avidity of binding to phosphoinositides (Lemmon and Ferguson, 2000) and mimic the ORD in a membrane-associated state. The ΔORDPHDPHD chimera allowed p150Glued and DIC recruitment by RILP, resulting in the clustering of RILP-positive compartments near the MTOC (Fig. 2, B and C). Silencing ORP1L with siRNA against ORD before ectopic expression of ΔORD or ΔORDPHDPHD showed no contribution of endogenous ORP1L on the phenotypes caused by the ORP1L mutants (unpublished data). The repositioning of LEs by the different ORP1L variants was confirmed in time-lapse experiments (Videos 4 and 5).
Repositioning of LEs in cells expressing the ORP1L- ΔORD mutant. MelJuSo cells stably expressing CD63-GFP (green) to label LEs were microinjected with vectors encoding mRFP- ΔORD (red), H2B (histone-2B)-GFP (green; nuclear) as an expression control, and 70-kD Texas red dextran as an injection marker in the presence of cycloheximide. The cycloheximide block was washed away 2 h after microinjection. Cells were imaged using a Leica SP2 system equipped with a heated chamber (37°C with 5% CO2). Frames were taken every 10 min. The video is shown at 5 frames/s with 30 total frames.
Repositioning of LEs in cells expressing the ORP1L- ΔORD mutant. MelJuSo cells stably expressing CD63-GFP (green) to label LEs were microinjected with vectors encoding mRFP- ΔORD (red), H2B (histone-2B)-GFP (green; nuclear) as an expression control, and 70-kD Texas red dextran as an injection marker in the presence of cycloheximide. The cycloheximide block was washed away 2 h after microinjection. Cells were imaged using a Leica SP2 system equipped with a heated chamber (37°C with 5% CO2). Frames were taken every 10 min. The video is shown at 5 frames/s with 30 total frames.
Repositioning of LEs in cells expressing the ORP1L- ΔORDPHDPHD mutant. MelJuSo cells stably expressing CD63-GFP (green) to label LEs were microinjected with vectors encoding mRFP- ΔORDPHDPHD (red), H2B (histone-2B)-GFP (green; nuclear) as an expression control, and 70-kD Texas red dextran as an injection marker in the presence of cycloheximide. The cycloheximide block was washed away 2 h after microinjection, immediately before imaging. Cells were imaged using a Leica SP2 system equipped with a heated chamber (37°C with 5% CO2). Frames were taken every 10 min. The video is shown at 5 frames/s with 30 total frames.
Repositioning of LEs in cells expressing the ORP1L- ΔORDPHDPHD mutant. MelJuSo cells stably expressing CD63-GFP (green) to label LEs were microinjected with vectors encoding mRFP- ΔORDPHDPHD (red), H2B (histone-2B)-GFP (green; nuclear) as an expression control, and 70-kD Texas red dextran as an injection marker in the presence of cycloheximide. The cycloheximide block was washed away 2 h after microinjection, immediately before imaging. Cells were imaged using a Leica SP2 system equipped with a heated chamber (37°C with 5% CO2). Frames were taken every 10 min. The video is shown at 5 frames/s with 30 total frames.
To test whether LE positioning requires cholesterol sensing by ORP1L, MelJuSo cells were transfected with mRFP-ORP1L before manipulating cholesterol levels. Cholesterol reduction by statin and lipid-free medium scattered the mRFP-ORP1L–positive vesicles, whereas U-18666A treatment positioned LEs around the MTOC (Fig. 3 A and Fig. S3). Removal of the cholesterol-binding ORD or its replacement by tandem PH domains rendered positioning of LEs insensitive to cholesterol levels. ΔORD induced scattering of LEs under all cholesterol-manipulated conditions, whereas ΔORDPHDPHD always induced dense clustering (Fig. 3 B and Fig. S3). Microscopy analyses revealed that the expression of the ORP1L constructs was increased by six- to 15-fold upon ectopic expression (unpublished data). We conclude that functional ORP1L is required for translating LE cholesterol content into LE positioning.
ORP1L controls cholesterol-dependent LE positioning. (A) Cholesterol-dependent ORP1L vesicle positioning. Mel JuSo cells expressing mRFP-ORP1L were cultured under control conditions (FCS) or conditions causing decreased (statin) or increased (U-18666A) cholesterol levels. Actin was stained with falloidin (green) to mark the cell boundaries before analyses by CLSM. n > 100. (B) ORP1L is dominant in vesicle repositioning as the result of cholesterol manipulations. Mel JuSo cells expressing mRFP-ΔORDPHDPHD or mRFP-ΔORD were treated and imaged as described in A. n > 100. Bars, 10 µm.
ORP1L controls cholesterol-dependent LE positioning. (A) Cholesterol-dependent ORP1L vesicle positioning. Mel JuSo cells expressing mRFP-ORP1L were cultured under control conditions (FCS) or conditions causing decreased (statin) or increased (U-18666A) cholesterol levels. Actin was stained with falloidin (green) to mark the cell boundaries before analyses by CLSM. n > 100. (B) ORP1L is dominant in vesicle repositioning as the result of cholesterol manipulations. Mel JuSo cells expressing mRFP-ΔORDPHDPHD or mRFP-ΔORD were treated and imaged as described in A. n > 100. Bars, 10 µm.
LE cholesterol affects the conformation of ORP1L
To determine whether LE cholesterol content affects the conformation of ORP1L, an mRFP-ORP1L-GFP fusion protein was expressed to monitor intramolecular fluorescence resonance energy transfer (FRET). Energy transfer from a donor fluorophore to a suitable acceptor (Förster, 1948) is dependent on distance and orientation between the two fluorophores (Förster, 1948; Calleja et al., 2003) and was used in this study to reveal conformational changes within mRFP-ORP1L-GFP (Fig. 4 A).
LE cholesterol alters the conformation of ORP1L. (A) Intramolecular FRET for mRFP-ORP1L-GFP. GFP and mRFP are attached to the same molecule, allowing FRET from donor GFP to acceptor mRFP. FRET depends on distance and orientation and thus indicates conformational changes. FRET can be detected by sensitized emission. GFP is excited by 488-nm light, and then, after energy transfer, 582–675-nm light emission by mRFP is detected. (B) Cholesterol effects on ORP1L conformation detected by sensitized emission. Mel JuSo cells expressing mRFP-ORP1L-GFP were cultured under control (FCS) or cholesterol-depleting (statin) or -enhancing (U-18666A) conditions before imaging by CLSM for FRET determination. Panels show the GFP signal, the mRFP signal, calculated FRET, and FRET related to donor fluorophore input: the donor FRET efficiency (ED). The color LUT visualizes the differences in the ED panels. (right) quantification of the donor FRET efficiency detected for mRFP-ORP1L-GFP under the different conditions of cholesterol manipulation. The mean and SD from two experiments (>10 cells analyzed) are shown (*, P = 0.05; **, P = 0.03). (C) ORP1L controls LE positioning in NPC1-silenced cells. Mel JuSo cells were transfected with mRFP-ORP1L, -ΔORD, or -ΔORDPHDPHD and siRNA for NPC1 and analyzed by CLSM. n > 100. (D) Sensitized emission and ORP1L conformation in NPC1-deficient cells. mRFP-ORP1L-GFP–expressing MelJuSo cells were transfected with control (siCTRL) or NPC1 (siNPC1) siRNAs before imaging by confocal FRET. (right) Donor FRET efficiencies determined in >10 control siRNA– or NPC1 siRNA–transfected cells. The mean ± SD is shown (**, P = 5.1 × 10−6). (E) NPC1, cholesterol, and LE clustering. Mel JuSo cells were transfected with control or NPC1 siRNA and then mRFP-ΔORD before staining with filipin for cholesterol. Pixel analyses are shown in Fig. S2 E. n > 50 for each condition. Bars, 10 µm.
LE cholesterol alters the conformation of ORP1L. (A) Intramolecular FRET for mRFP-ORP1L-GFP. GFP and mRFP are attached to the same molecule, allowing FRET from donor GFP to acceptor mRFP. FRET depends on distance and orientation and thus indicates conformational changes. FRET can be detected by sensitized emission. GFP is excited by 488-nm light, and then, after energy transfer, 582–675-nm light emission by mRFP is detected. (B) Cholesterol effects on ORP1L conformation detected by sensitized emission. Mel JuSo cells expressing mRFP-ORP1L-GFP were cultured under control (FCS) or cholesterol-depleting (statin) or -enhancing (U-18666A) conditions before imaging by CLSM for FRET determination. Panels show the GFP signal, the mRFP signal, calculated FRET, and FRET related to donor fluorophore input: the donor FRET efficiency (ED). The color LUT visualizes the differences in the ED panels. (right) quantification of the donor FRET efficiency detected for mRFP-ORP1L-GFP under the different conditions of cholesterol manipulation. The mean and SD from two experiments (>10 cells analyzed) are shown (*, P = 0.05; **, P = 0.03). (C) ORP1L controls LE positioning in NPC1-silenced cells. Mel JuSo cells were transfected with mRFP-ORP1L, -ΔORD, or -ΔORDPHDPHD and siRNA for NPC1 and analyzed by CLSM. n > 100. (D) Sensitized emission and ORP1L conformation in NPC1-deficient cells. mRFP-ORP1L-GFP–expressing MelJuSo cells were transfected with control (siCTRL) or NPC1 (siNPC1) siRNAs before imaging by confocal FRET. (right) Donor FRET efficiencies determined in >10 control siRNA– or NPC1 siRNA–transfected cells. The mean ± SD is shown (**, P = 5.1 × 10−6). (E) NPC1, cholesterol, and LE clustering. Mel JuSo cells were transfected with control or NPC1 siRNA and then mRFP-ΔORD before staining with filipin for cholesterol. Pixel analyses are shown in Fig. S2 E. n > 50 for each condition. Bars, 10 µm.
FRET was determined by sensitized emission using confocal microscopy. GFP was excited with 488-nm light, and the donor mRFP signal was determined. mRFP-ORP1L-GFP–expressing MelJuSo cells were co-cultured with MelJuSo cells expressing only GFP or mRFP for imaging. These latter cells served as internal controls to calculate correction factors for bleed-through of GFP into the mRFP channel and for direct excitation of mRFP by the 488-nm laser (Zwart et al., 2005). FRET and donor FRET efficiency (ED; i.e., FRET normalized to donor fluorophore input) were determined under the various cholesterol-manipulating conditions (Fig. 4 B). Cholesterol depletion decreased FRET efficiency ED, and ED was increased after LE cholesterol accumulation (Fig. 4 B, right), implying that the ORP1L conformational changes were dependent on LE cholesterol content.
To exclude that these differences in ED were caused by the clustering of individual ORP1L molecules under the various conditions rather than conformational changes of each ORP1L molecule, the experiments were repeated with cells expressing both GFP-ORP1L and mRFP-ORP1L (Fig. S4 B). If changes in ED with mRFP-ORP1L-GFP were caused by intermolecular rather than intramolecular FRET, ED with GFP-ORP1L and mRFP-ORP1L would also change. However, ED values were not affected by cholesterol manipulation, implying that the FRET differences result from conformational changes within mRFP-ORP1L-GFP.
Two constructs, mRFP-ΔORD-GFP and mRFP-ΔORDPHDPHD-GFP, were used to test whether cholesterol-dependent conformational changes in ORP1L were mediated by the ORD. These constructs should not respond to variations in cholesterol levels because they lack ORD but may still respond to effects on the PH domain or other domains present in ORP1L. However, no effect of cholesterol manipulation on intramolecular donor FRET efficiency ED was detected, suggesting that cholesterol-induced conformational changes in mRFP-ORP1L-GFP were mediated by the cholesterol-sensing ORD (Fig. S4 A).
FRET efficiencies can also be calculated from differences in donor fluorophore lifetime (Bastiaens and Squire, 1999). The aforementioned FRET experiments were repeated with FRET determination by fluorescence lifetime imaging microscopy (FLIM). The lifetime of GFP is typically 2.7 ns (Pepperkok et al., 1999) but decreases in the case of FRET, when energy is transferred to the acceptor fluorophore mRFP. Cells expressing mRFP-ORP1L-GFP were imaged along with control cells expressing only GFP to determine GFP lifetime under non-FRET conditions. Again, FRET efficiency ED was decreased by cholesterol depletion and increased in response to enhanced cholesterol levels (Fig. S4 C). Cells grown under the various control conditions were also imaged by FLIM and showed no response to cholesterol manipulation, which is fully consistent with the results of FRET experiments measured by sensitized emission (Fig. S4, D and E). This shows that ORP1L senses cholesterol content in LEs through the C-terminal ORD.
ORP1L, cholesterol, and Niemann-Pick type C disease
Silencing NPC1 increases cholesterol in LEs and induces LE clustering. Cholesterol can distribute over the LE-limiting membrane and the internal vesicles by flip-flop mechanisms and/or specific transporters. ORP1L senses cholesterol in the cytosolic leaflet of the LE-limiting membrane before translating this into LE repositioning. To test whether cholesterol sensing by ORP1L is required for LE clustering in Niemann-Pick type C, NPC1-silenced MelJuSo cells (Fig. S1 B) were transfected with different mRFP-ORP1L constructs before analysis by confocal laser-scanning microscopy (CLSM; Fig. 4 C). Whereas ORP1L and ΔORDPHDPHD induced clustering of LEs, ΔORD induced scattering of LEs even when NPC1 is silenced (Fig. S4 F), suggesting that functional ORP1L is required to translate increased cholesterol levels into LE repositioning in NPC1-deficient cells.
To test whether NPC1 deficiency affects the cholesterol levels in the cytosolic leaflet of LEs, intramolecular FRET of mRFP-ORP1L-GFP was measured by sensitized emission in cells transfected with control or NPC1 siRNAs (Fig. 4 D). The FRET efficiency ED increased when NPC1 was silenced (Fig. 4 D). This result was confirmed in FLIM measurements (Fig. S4 G). The donor FRET efficiencies (ED) for mRFP-ORP1L-GFP in NPC1-silenced cells are similar to those observed in cells treated with U-18666A, which mimics NPC1 deficiency (Fig. 4 B; Koh and Cheung, 2006; Sobo et al., 2007).
Cholesterol accumulation in LEs of NPC1-silenced cells may be a cause or a consequence of the LE clustering observed in cells of Niemann-Pick type C patients (Kobayashi et al., 1999). To separate cholesterol accumulation from LE clustering, mRFP-ΔORD was expressed in NPC1-silenced cells. Fixed cells were stained for cholesterol with filipin (Fig. 4 E) and CD63 (Fig. S4 H). The CD63-positive LEs scattered by ΔORD expression still accumulated cholesterol when NPC1 was silenced. This suggests that the characteristic clustering of LEs near the MTOC, which is typical in Niemann-Pick type C, is a consequence of cholesterol accumulation, as sensed by ORP1L (and is abrogated by ΔORD). ORP1L transmits this information to the Rab7–RILP–p150Glued–dynein motor complex for microtubule minus end–driven transport and clustering of LEs.
ORP1L conformation differs between peripheral and perinuclear LEs
If cholesterol levels in LEs determine its intracellular positioning, these levels should be different between peripheral and perinuclear LEs. Cholesterol levels in the cytosolic leaflet of LEs cannot be detected by any currently available probe other than by the mRFP-ORP1L-GFP sensor (Fig. 4). We reanalyzed the images of cells expressing the mRFP-ORP1L-GFP FRET probe (Fig. 5 A) and, as a control, the mRFP-ΔORD-GFP FRET probe (Fig. 5 B). The latter construct lacks the cholesterol-sensing ORD. Donor FRET efficiency ED was determined in five concentric regions spanning the nucleus and cell surface, as indicated (Fig. 5). The results were binned and quantified (Fig. 5, A and B, bottom). ED, as determined with mRFP-ORP1L-GFP, was higher on perinuclear LEs than on peripheral LEs (Fig. 5 A). These differences were sensed by ORD because ED was unaffected with the mRFP-ΔORD-GFP construct (Fig. 5 B). This suggests that the positioning of LEs within one cell correlates with different conformations of ORP1L, as induced by different levels of cholesterol in the cytosolic leaflet of LEs.
ORP1L conformation and LE positioning. (A and B, top) Mel JuSo cells expressing mRFP-ORP1L-GFP (A) or mRFP-ΔORD-GFP (B) were imaged for FRET by CLSM. The GFP image, mRFP image, calculated FRET, and donor FRET efficiency ED images are indicated. ED values are shown in false colors (the LUT shows the corresponding values). The position of the nucleus (N) and cell boundaries are drawn in the GFP channel. The cytosol is divided by five concentric rings, as indicated in the ED panel. (bottom) ED values for the constructs were collected and binned for the different rings. Values are shown for the different rings under the corresponding image. Mean ± SEM of ED values collected for >10 cells analyzed per construct are shown. The asterisks indicate a statistically significant difference in ED of that bin compared to bin 1 (the most internal ring; *, P = 0.024; **, P = 0.026; ***, P = 0.0063). Bars, 10 µm.
ORP1L conformation and LE positioning. (A and B, top) Mel JuSo cells expressing mRFP-ORP1L-GFP (A) or mRFP-ΔORD-GFP (B) were imaged for FRET by CLSM. The GFP image, mRFP image, calculated FRET, and donor FRET efficiency ED images are indicated. ED values are shown in false colors (the LUT shows the corresponding values). The position of the nucleus (N) and cell boundaries are drawn in the GFP channel. The cytosol is divided by five concentric rings, as indicated in the ED panel. (bottom) ED values for the constructs were collected and binned for the different rings. Values are shown for the different rings under the corresponding image. Mean ± SEM of ED values collected for >10 cells analyzed per construct are shown. The asterisks indicate a statistically significant difference in ED of that bin compared to bin 1 (the most internal ring; *, P = 0.024; **, P = 0.026; ***, P = 0.0063). Bars, 10 µm.
ORP1L-dependent recruitment of VAP and removal of the p150Glued dynactin subunit
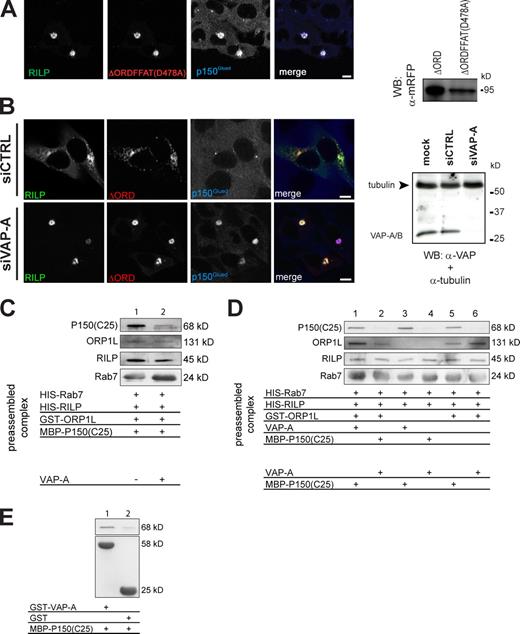
ORP1L allows RILP-induced clustering of LEs. ΔORD relocates LEs to the cell periphery, suggesting the exclusion of p150Glued from RILP (Fig. 2 B). Removal of the 20-kD region preceding the ORD restored RILP-induced clustering and p150Glued binding to RILP (Fig. 2 B). This region contains a predicted coiled-coil region (aa 430–463; Johansson et al., 2007) followed by a FFAT motif (aa 472–482; sequence, SEDEFYDALSD; Fig. 2 A; Loewen et al., 2003) and should contain the information for removal of p150Glued from RILP. To test whether the FFAT motif controlled p150Glued binding to RILP, an inactivating point mutation (D478A) in the FFAT motif (Loewen et al., 2003) was introduced in mRFP-ΔORD. This mutant was coexpressed with GFP-RILP before staining for p150Glued (Fig. 6 A). Whereas ΔORD excludes RILP-mediated recruitment of p150Glued (resulting in LE scattering), the ΔORDFFAT(D478A) point mutant permitted p150Glued binding to RILP, resulting in LE clustering around the MTOC.
ORP1L requires VAP-A to remove p150Glued from Rab7–RILP. (A) Effect of the FFAT motif in ΔORD on RILP-mediated p150Glued recruitment. (left) Mel JuSo cells were transfected with GFP-RILP and mRFP-ΔORD containing a point mutation in its FFAT motif, ΔORDFFAT(D478A). Fixed cells were stained with anti-p150Glued antibodies. n > 200. (right) Mel JuSo cells were transfected with the mRFP-ΔORD or mRFP-ΔORDFFAT(D478A) constructs, and whole cell lysates were analyzed by immunoblotting with anti-mRFP antibodies (WB: α-mRFP). (B) p150Glued exclusion by ΔORD and VAP-A. VAP-A was silenced by siRNA (siVAP-A) in cells expressing GFP-RILP and mRFP-ΔORD. (left) Cells stained for p150Glued. Pixel analyses are shown in Fig. S2 E. n > 100 for each condition. (right) Western blot analysis of cells transfected with transfection reagent (mock), control (siCTRL), or VAP-A siRNA (siVAP-A) and probed for α-tubulin (as loading control) and anti–VAP-A antibodies. (C) VAP-A removes p150Glued from Rab7–RILP. GTP-locked His-Rab7(Q67L) was GTP loaded and coupled to Talon beads before adding purified RILP, ORP1L, and p150Glued(C25) fragments in equimolar amounts to form a preassembled ORP1L–Rab7–RILP–p150Glued(C25) complex. Subsequently, the complex was incubated in the presence or absence of purified VAP-A. After washing, the bead-bound proteins were analyzed by SDS-PAGE and Western blotting. (D) ORP1L requirements for p150Glued removal by VAP-A. GTP-loaded His-Rab7(Q67L) was coupled to Talon Co2+ beads before adding the isolated proteins indicated in equimolar amounts to form a preassembled complex. After washing, these complexes were exposed to isolated VAP-A or the p150Glued(C25) fragment, as indicated, and the effects on the preformed complex were assessed by SDS-PAGE and Western blot analyses with antibodies, as indicated. (E) VAP-A interacts with p150Glued. GST or GST–VAP-A was coupled to beads before exposure to the p150Glued(C25) fragment. After washing, the complexes were analyzed by SDS-PAGE and Western blotting. (A–E) Molecular masses are indicated. WB, Western blot. Bars, 10 µm.
ORP1L requires VAP-A to remove p150Glued from Rab7–RILP. (A) Effect of the FFAT motif in ΔORD on RILP-mediated p150Glued recruitment. (left) Mel JuSo cells were transfected with GFP-RILP and mRFP-ΔORD containing a point mutation in its FFAT motif, ΔORDFFAT(D478A). Fixed cells were stained with anti-p150Glued antibodies. n > 200. (right) Mel JuSo cells were transfected with the mRFP-ΔORD or mRFP-ΔORDFFAT(D478A) constructs, and whole cell lysates were analyzed by immunoblotting with anti-mRFP antibodies (WB: α-mRFP). (B) p150Glued exclusion by ΔORD and VAP-A. VAP-A was silenced by siRNA (siVAP-A) in cells expressing GFP-RILP and mRFP-ΔORD. (left) Cells stained for p150Glued. Pixel analyses are shown in Fig. S2 E. n > 100 for each condition. (right) Western blot analysis of cells transfected with transfection reagent (mock), control (siCTRL), or VAP-A siRNA (siVAP-A) and probed for α-tubulin (as loading control) and anti–VAP-A antibodies. (C) VAP-A removes p150Glued from Rab7–RILP. GTP-locked His-Rab7(Q67L) was GTP loaded and coupled to Talon beads before adding purified RILP, ORP1L, and p150Glued(C25) fragments in equimolar amounts to form a preassembled ORP1L–Rab7–RILP–p150Glued(C25) complex. Subsequently, the complex was incubated in the presence or absence of purified VAP-A. After washing, the bead-bound proteins were analyzed by SDS-PAGE and Western blotting. (D) ORP1L requirements for p150Glued removal by VAP-A. GTP-loaded His-Rab7(Q67L) was coupled to Talon Co2+ beads before adding the isolated proteins indicated in equimolar amounts to form a preassembled complex. After washing, these complexes were exposed to isolated VAP-A or the p150Glued(C25) fragment, as indicated, and the effects on the preformed complex were assessed by SDS-PAGE and Western blot analyses with antibodies, as indicated. (E) VAP-A interacts with p150Glued. GST or GST–VAP-A was coupled to beads before exposure to the p150Glued(C25) fragment. After washing, the complexes were analyzed by SDS-PAGE and Western blotting. (A–E) Molecular masses are indicated. WB, Western blot. Bars, 10 µm.
The FFAT motif in the ER-located ORP1L homologue OSBP interacts with VAP-A and VAP-B, which are involved in protein and lipid export from the ER (Wyles et al., 2002; Loewen and Levine, 2005). VAP is an integral ER protein and not an obvious candidate for controlling p150Glued release from Rab7–RILP on LE membranes. To test whether VAP could be involved in this release, MelJuSo cells were transfected with mRFP-ΔORD and GFP-RILP in combination with siRNA against VAP-A and stained for p150Glued (Fig. 6 B). ΔORD expression again removed p150Glued from RILP in cells transfected with control siRNA, resulting in LE positioning in the cell periphery. When VAP-A was silenced, ΔORD remained associated with p150Glued-RILP, and the LEs clustered around the MTOC. Identical results were obtained when VAP-B was silenced (unpublished data). VAP-A silencing (with four independent siRNAs) reduced VAP-A mRNA levels by >99% and VAP-B levels by 70–80%, as determined by qPCR. Similarly, VAP-B silencing reduced VAP-A expression by 99% (unpublished data). Immunoblotting with VAP-A antibodies confirmed efficient silencing of VAP-A expression (Fig. 6 B).
Purified proteins were used to test whether VAP directly controls p150Glued binding to RILP. The p150Glued(C25) C-terminal domain interacts with the purified Rab7–RILP complex and was used for these experiments (Johansson et al., 2007). The ORP1L–Rab7–RILP–p150Glued complex was first assembled with purified proteins before VAP-A was added (Fig. 6 C). GTP-loaded, His-tagged Rab7 was bound to metal affinity beads before the addition of RILP and ORP1L. The complex was completed by p150Glued(C25), and nonbound proteins were removed by washing after every step during the construction of the complex. Subsequently, purified VAP-A was added to the complex followed again by washing of the beads. Its effect on the ORP1L–Rab7–RILP–p150Glued(C25) complex was determined by Western blotting (Fig. 6 C). Soluble VAP-A removed the C25 fragment of p150Glued from the preassembled Rab7–RILP–ORP1L complex.
To test whether VAP-A prevented binding of p150Glued to the Rab7–RILP complex or whether VAP-A induced the release of p150Glued and to determine the role of ORP1L in this process, various purified Rab7–RILP complexes were assembled in vitro (Fig. 6 D). When VAP-A was included during the assembly of the complex, the p150Glued(C25) fragment still entered the complex (Fig. 6 D, lanes 1 and 2). VAP-A was probably removed by washing during the assembly of the Rab7–RILP complex, which explains why VAP-A only affected C25 binding when it was added to the preassembled Rab7–RILP–C25 complex. The presence or absence of ORP1L in the preassembled Rab7–RILP complex did not affect removal of the C25 fragment of p150Glued by VAP-A (Fig. 6 D, lanes 3 and 4). These data suggest that ORP1L is not directly involved in VAP-A–mediated removal of p150Glued from the Rab7–RILP complex but probably recruits VAP into this complex.
To test whether VAP-A directly interacts with the C25 fragment of p150Glued, GST–VAP-A or GST (as a control) was bound to GST beads, and purified p150Glued(C25) fragment was added. VAP-A interacted with the C-terminal p150Glued(C25) fragment (Fig. 6 E). These in vitro reconstitution experiments reveal a minimal set of proteins controlling dynein motor binding to LEs that includes Rab7, RILP, p150Glued, and the ER protein VAP.
ORP1L and VAP are involved in inducing ER–LE membrane contacts
If the integral ER protein VAP controls motor binding to LEs after recruitment by ORP1L, VAP should somehow contact these vesicles. To visualize this, MelJuSo cells were transfected with the various ORD variants of mRFP-ORP1L and stained for endogenous VAP (Fig. 7 A). Endogenous VAP accumulated on ΔORD- and, although to a lesser extent, on ORP1L-containing vesicles but was excluded from vesicles labeled by ΔORDPHDPHD, indicating that VAP recruitment is dependent on the conformation of ORP1L (Fig. 7 A). These results were confirmed in photoactivation experiments with MelJuSo cells expressing photoactivatable (PA)-GFP–tagged VAP-A and mRFP-ORP1L variants (Fig. S5 A and Videos 6–8) that also showed colocalization of VAP-A and ORP1L on moving vesicles. Accumulation of the ER protein VAP on LEs by ΔORD expression also coincided with the removal of p150Glued from RILP (Fig. S5 B). We then tested whether the ER–LE interactions could be observed by manipulating cholesterol levels in MelJuSo cells in which ORP1L and VAP were not ectopically expressed. TAP1-GFP labeled the ER (Reits et al., 2000). Decreasing cholesterol levels sufficed to induce the arrival of ER markers on LEs, as analyzed by CLSM and quantified by pixel analysis (Fig. 7 B).
VAP-A diffusion in cells expressing ORP1L. MelJuSo cells were transiently transfected with expression vectors for PA-GFP–VAP-A (green) and mRFP-ORP1L (red). PA-GFP–VAP-A was activated by a 405-nm laser at a site where the ER is present but LEs are not. To exclude that the 405-nm laser beam would hit LEs, a z stack was made to locate ER pools of VAP-A away from the mRFP-labeled LEs. Cells were imaged using a Leica AOBS system equipped with a 37°C chamber culture hood with 5% CO2. Frames were taken every 1 min. The video is shown at 3 frames/s with 30 total frames. The still images are presented in Fig. S5 A.
VAP-A diffusion in cells expressing ORP1L. MelJuSo cells were transiently transfected with expression vectors for PA-GFP–VAP-A (green) and mRFP-ORP1L (red). PA-GFP–VAP-A was activated by a 405-nm laser at a site where the ER is present but LEs are not. To exclude that the 405-nm laser beam would hit LEs, a z stack was made to locate ER pools of VAP-A away from the mRFP-labeled LEs. Cells were imaged using a Leica AOBS system equipped with a 37°C chamber culture hood with 5% CO2. Frames were taken every 1 min. The video is shown at 3 frames/s with 30 total frames. The still images are presented in Fig. S5 A.
VAP-A diffusion in cells expressing the ORP1L- ΔORD mutant. MelJuSo cells were transiently transfected with expression vectors for PA-GFP–VAP-A (green) and mRFP- ΔORD (red). PA-GFP–VAP-A was activated by a 405-nm laser at a site where the ER is present but LEs are not. To exclude that the 405-nm laser beam would hit LEs, a z stack was made to locate ER pools of VAP-A away from the mRFP-labeled LEs. Cells were imaged using a Leica AOBS system equipped with a 37°C chamber culture hood with 5% CO2. Frames were taken every 1 min. The video is shown at 3 frames/s with 30 total frames. The still images are presented in Fig. S5 A.
VAP-A diffusion in cells expressing the ORP1L- ΔORD mutant. MelJuSo cells were transiently transfected with expression vectors for PA-GFP–VAP-A (green) and mRFP- ΔORD (red). PA-GFP–VAP-A was activated by a 405-nm laser at a site where the ER is present but LEs are not. To exclude that the 405-nm laser beam would hit LEs, a z stack was made to locate ER pools of VAP-A away from the mRFP-labeled LEs. Cells were imaged using a Leica AOBS system equipped with a 37°C chamber culture hood with 5% CO2. Frames were taken every 1 min. The video is shown at 3 frames/s with 30 total frames. The still images are presented in Fig. S5 A.
VAP-A diffusion in cells expressing the ORP1L- ΔORDPHDPHD mutant. MelJuSo cells were transiently transfected with expression vectors for PA-GFP–VAP-A (green) and mRFP- ΔORDPHDPHD (red). PA-GFP–VAP-A was activated by a 405-nm laser at a site where the ER is present but LEs are not. To exclude that the 405-nm laser beam would hit LEs, a z stack was made to locate ER pools of VAP-A away from the mRFP-labeled LEs. Cells were imaged using a Leica AOBS system equipped with a 37°C chamber culture hood with 5% CO2. Frames were taken every 1 min. The video is shown at 3 frames/s with 30 total frames. The still images are presented in Fig. S5 A.
VAP-A diffusion in cells expressing the ORP1L- ΔORDPHDPHD mutant. MelJuSo cells were transiently transfected with expression vectors for PA-GFP–VAP-A (green) and mRFP- ΔORDPHDPHD (red). PA-GFP–VAP-A was activated by a 405-nm laser at a site where the ER is present but LEs are not. To exclude that the 405-nm laser beam would hit LEs, a z stack was made to locate ER pools of VAP-A away from the mRFP-labeled LEs. Cells were imaged using a Leica AOBS system equipped with a 37°C chamber culture hood with 5% CO2. Frames were taken every 1 min. The video is shown at 3 frames/s with 30 total frames. The still images are presented in Fig. S5 A.
Cholesterol-dependent ORP1L-mediated contacts between LEs and the ER. (A) ΔORD recruits endogenous VAP-A. Mel JuSo cells expressing mRFP-ΔORD, -ORP1L, or -ΔORDPHDPHD were stained for VAP-A. The merge panels show an overlay of the two channels, and the boxes indicate the zoomed-in areas. Pixel analyses of the zoomed-in areas for fluorescence distribution of the mRFP-ORP1L variants and VAP-A are shown. n > 100. (B) Mel JuSo cells expressing TAP1-GFP (red) were exposed to different cholesterol manipulating conditions, as indicated. Cells were stained with antibodies for CD63 (green). Colocalization of the markers was determined by computational pixel analysis and visualized in blue. (right) Quantification of the colocalizing surface of the CD63-positive area. The mean ± SEM for >20 cells analyzed per condition is shown (*, P = 0.068; **, P = 1.55 × 10−6; ***, P = 0.0003). Bars: (A) 10 µm; (B) 5 µm.
Cholesterol-dependent ORP1L-mediated contacts between LEs and the ER. (A) ΔORD recruits endogenous VAP-A. Mel JuSo cells expressing mRFP-ΔORD, -ORP1L, or -ΔORDPHDPHD were stained for VAP-A. The merge panels show an overlay of the two channels, and the boxes indicate the zoomed-in areas. Pixel analyses of the zoomed-in areas for fluorescence distribution of the mRFP-ORP1L variants and VAP-A are shown. n > 100. (B) Mel JuSo cells expressing TAP1-GFP (red) were exposed to different cholesterol manipulating conditions, as indicated. Cells were stained with antibodies for CD63 (green). Colocalization of the markers was determined by computational pixel analysis and visualized in blue. (right) Quantification of the colocalizing surface of the CD63-positive area. The mean ± SEM for >20 cells analyzed per condition is shown (*, P = 0.068; **, P = 1.55 × 10−6; ***, P = 0.0003). Bars: (A) 10 µm; (B) 5 µm.
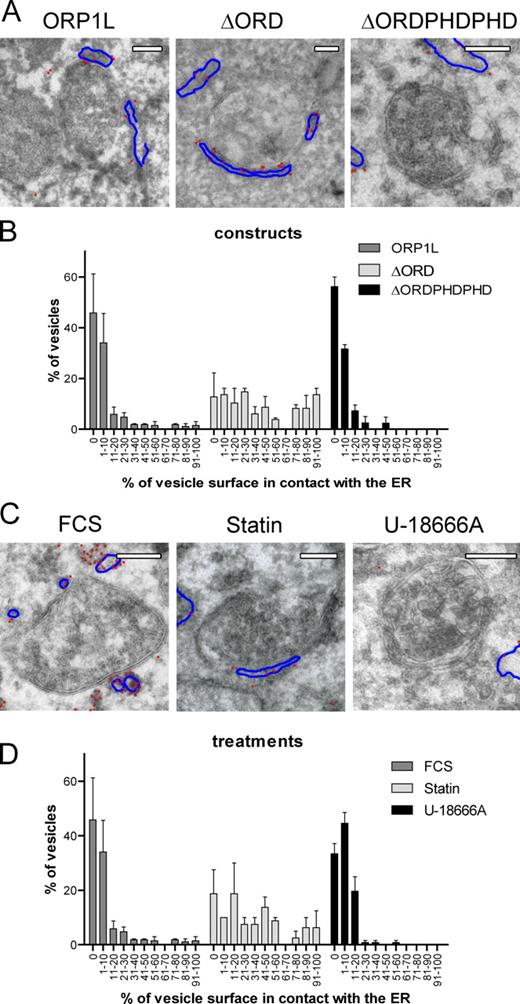
The localization of VAP and ORP1L was subsequently visualized by EM in more detail. EM has a significantly higher spatial resolution than CLSM, which has a maximal resolution of ∼240 nm. MelJuSo cells were transfected with GFP-ΔORD, -ORP1L, or -ΔORDPHDPHD together with HA-tagged VAP-A. After expression of the ORP1L variants was observed (by GFP fluorescence), cells were processed for immuno-EM and stained with HA antibodies to detect VAP-A (Fig. 8 A). The frequency of ER contacts with LEs and the surface of the LEs covered by ER structures was quantified (Fig. 8 B). Note that HA-VAP was never detected at LE membranes, excluding fusion of the LE and ER under these conditions. The ER, as labeled by HA-tagged VAP-A, had to be within 40 nm of the LE-limiting membrane to be considered a contact. Most LEs (>95%) interacted, often over large surface areas, with the ER when ΔORD was expressed. Both the number of ER–LE interactions and the surface covered by the ER decreased when ORP1L was expressed. Additionally, contact sites were almost absent when the high cholesterol conformation of ORP1L (ΔORDPHDPHD) was expressed, suggesting that the cholesterol-dependent conformational state of ORP1L determines ER contact site formation with LEs.
Cholesterol-dependent MCSs between LEs and the ER allowing in trans VAP interactions with the Rab7–RILP complex. (A) Electron micrographs of multivesicular bodies in Mel JuSo cells expressing HA-VAP and different variants of GFP-ORP1L, as indicated. Sections were stained with anti-HA antibodies detected with 15-nm gold particles. The gold particles are highlighted by red dots, and the ER membrane (containing HA-VAP) is indicated by blue lines. The original images are shown in Fig. S5 C. (B) The percentage of multivesicular bodies contacting ER structures and the contact area between the ER and LE membranes was determined in Mel JuSo cells expressing the ORP1L variants as shown in A. Over 50 multivesicular bodies were considered per condition. The data on contact area from two independent quantifications were binned as indicated, and error bars show SEM. (C) Electron micrographs of multivesicular bodies in HA-VAP– and GFP-ORP1L–expressing MelJuSo cells exposed to different cholesterol-manipulating conditions, as indicated. Sections were stained with anti-HA antibodies marked by 15-nm gold particles. The gold particles are highlighted by red dots, and the ER membrane (containing HA-VAP) is indicated by blue lines. The original images are shown in Fig. S5 F. (D) The percentage of multivesicular bodies contacting ER structures and the contact area between the ER and LE membranes from MelJuSo cells expressing ORP1L under various conditions of cholesterol manipulation (as shown in C). Over 50 multivesicular bodies were considered per condition. The data on contact area from two independent quantifications were binned as indicated, and error bars show SEM. Bars, 200 nm.
Cholesterol-dependent MCSs between LEs and the ER allowing in trans VAP interactions with the Rab7–RILP complex. (A) Electron micrographs of multivesicular bodies in Mel JuSo cells expressing HA-VAP and different variants of GFP-ORP1L, as indicated. Sections were stained with anti-HA antibodies detected with 15-nm gold particles. The gold particles are highlighted by red dots, and the ER membrane (containing HA-VAP) is indicated by blue lines. The original images are shown in Fig. S5 C. (B) The percentage of multivesicular bodies contacting ER structures and the contact area between the ER and LE membranes was determined in Mel JuSo cells expressing the ORP1L variants as shown in A. Over 50 multivesicular bodies were considered per condition. The data on contact area from two independent quantifications were binned as indicated, and error bars show SEM. (C) Electron micrographs of multivesicular bodies in HA-VAP– and GFP-ORP1L–expressing MelJuSo cells exposed to different cholesterol-manipulating conditions, as indicated. Sections were stained with anti-HA antibodies marked by 15-nm gold particles. The gold particles are highlighted by red dots, and the ER membrane (containing HA-VAP) is indicated by blue lines. The original images are shown in Fig. S5 F. (D) The percentage of multivesicular bodies contacting ER structures and the contact area between the ER and LE membranes from MelJuSo cells expressing ORP1L under various conditions of cholesterol manipulation (as shown in C). Over 50 multivesicular bodies were considered per condition. The data on contact area from two independent quantifications were binned as indicated, and error bars show SEM. Bars, 200 nm.
To test whether cholesterol levels determine the formation of these contact sites, cells expressing GFP-ORP1L were cultured under control (FCS) or cholesterol-increasing (U-18666A) or -depleting (statin) conditions, and sections were stained for HA-tagged VAP-A (Fig. 8 C). Under control conditions, some ER–LE interactions could be observed, usually over small LE surfaces. Whereas cholesterol accumulation in LEs (as the result of U-18666A) diminished these contacts, cholesterol depletion markedly induced formation of ER–LE contact sites (Fig. 8 D). These data suggest that the conformational state of ORP1L, as controlled by cholesterol levels, determines whether ER contacts the LE. This is required for the ER protein VAP to contact (and manipulate) the Rab7–RILP–p150Glued complex on LEs.
Discussion
Cholesterol is a hydrophobic molecule essential for fluidity and microdomain formation in biomembranes. The transfer of cholesterol to other organelles requires specific cholesterol-binding chaperones. A series of proteins with cholesterol-binding domains like the steroidogenic acute regulatory protein–related lipid transfer domain and ORD have been identified in various compartments (Holthuis and Levine, 2005). Two such proteins, MLN64 and ORP1L, are located at LE membranes (Ikonen, 2008). The transfer of cholesterol over LE membranes requires transporters like ABCA1 and possibly NPC1 (Ikonen, 2008), but flip-flop of cholesterol may be another possibility (Hamilton, 2003). If so, high concentrations of cholesterol in the intraluminal vesicles of LEs should be reflected by high concentrations of cholesterol in their cytosolic leaflet.
High cholesterol content in LEs is typical for a series of lysosomal storage diseases with characteristic LE clustering at the minus end of microtubules. It has been suggested that cholesterol influences LE transport involving Rab GTPases (Holtta-Vuori et al., 2000; Lebrand et al., 2002). Minus end transport of LEs involves the dynein–dynactin motor complex and its receptor complex, Rab7–RILP–ORP1L. Because ORP1L binds both Rab7–RILP (Johansson et al., 2007) and cholesterol (Suchanek et al., 2007), we tested whether ORP1L could transfer information on LE cholesterol content to Rab7–RILP and then to p150Glued-associated motor proteins like the dynein motor. Using intramolecular FRET, we showed that the C-terminal ORD of ORP1L senses LE cholesterol. ORD can be in a membrane-bound state (when cholesterol levels are high) or in a cytosolic, membrane-free conformation (when cholesterol levels are low). Overexpression of ORP1L or one of its mutants did not change cholesterol levels in LEs (unpublished data), which suggests that ORP1L is merely a cholesterol sensor rather than an active contributor to extracting and redistributing cholesterol.
When ORD is in its cytosolic conformation or absent (as in the truncation mutant ΔORD), the FFAT motif of ORP1L prevents access of p150Glued to Rab7–RILP. This does not occur when ORD is in the membrane-associated state (as in the chimeric ΔORDPHDPHD). The ER protein VAP is directly involved in the removal of the dynein–dynactin subunit p150Glued from the Rab7–RILP complex because silencing VAP restores p150Glued binding to ΔORD–Rab7–RILP-containing vesicles. This was confirmed when purified VAP-A removed p150Glued from the RILP–Rab7–ORP1L complex (which also indicated that the minimal set of proteins involved in this process had been identified).
The ER protein VAP would not be an obvious candidate for regulating LE positioning. Light microscopy suggested that VAP localized to LEs when ΔORD was expressed (indicating ER–LE fusion). However, visualization of this process at a higher resolution by EM revealed that the ER (containing VAP) aligned to rather than fused with LEs, creating LE–ER contact sites. The formation of these contact sites is dependent on the conformation of ORP1L. ΔORD or low cholesterol levels in the LEs increased the number and area of contact sites, whereas ΔORDPHDPHD or high cholesterol decreased these interactions. ER–LE contact sites were proposed as sites where lipids like cholesterol could be transferred from LEs to the ER for further distribution (Underwood et al., 1998). However, the ER–LE contact sites are predominantly observed under conditions of low cholesterol and, to a lesser extent, under control conditions. It is possible that they are transiently formed when ORP1L changes conformation to expose the FFAT domain and released when ORP1L adopts the membrane-associated cholesterol-high state. Under low cholesterol conditions when the ER–LE MCSs form, the ER protein VAP can interact in trans with the Rab7–RILP receptor on LEs to release p150Glued. This then results in microtubule plus end transport and scattering of LEs. The ER network is directly linked to the microtubule network that is also used by p150Glued-associated motors on LEs (Vedrenne and Hauri, 2006). The ER–LE contact sites are relatively stable between a dynamic ER and with moving vesicles, as can be appreciated in Videos 5–7. Whether other proteins contribute to stabilizing interactions between these organelles is unclear.
When NPC1 is depleted or mutated, as in Niemann-Pick type C disease (Carstea et al., 1997), or when chemical drugs raise cholesterol levels of LEs (Roff et al., 1991; Sobo et al., 2007), the ORD of ORP1L is membrane associated, preventing the formation of ER–LE MCSs. This allows enduring binding of p150Glued–DIC–dynein motor complexes to Rab7–RILP and transport to the microtubule minus end.
Why cholesterol is selected to dictate LE positioning is unclear. Cholesterol is present in the intraluminal vesicles as well as the cytosolic leaflet of LEs, allowing cytosolic exposure of intraluminal conditions. If cholesterol levels increase with maturation of the LE, the resulting ORP1L-controlled Rab7–RILP–p150Glued–DIC–dynein motor interactions would position the most matured compartments at the extreme minus end of microtubules with “shells” of less matured vesicles surrounding these at more plus end locations. Newly arriving LEs would then first encounter the youngest LE, resulting in spatial maturation before encountering lysosomes. Thus, cholesterol at the LE cytosolic leaflet would control the positioning of these organelles. We have tested this by analyzing the conformation of the cholesterol sensor ORP1L by FRET on LEs located in the perinuclear area versus the periphery. Donor FRET efficiency was lower in the cell periphery, indicating a lower cholesterol concentration.
We have visualized how the Rab7–RILP complex controls intracellular positioning of LEs. This process is controlled by cholesterol levels sensed by ORP1L to induce ER–LE contact sites via interaction with the ER protein VAP. In these contact sites, VAP interacts in trans with the Rab7–RILP receptor, promoting dissociation of p150Glued and its associated motor proteins (Fig. 9). This model explains how cholesterol depletion causes LE scattering and high cholesterol levels induce LE clustering, as observed in Niemann-Pick type C disease. We reveal an unexpected contribution of ER proteins in the control of LE intracellular positioning by motor proteins in a process controlled by cholesterol.
Model showing control of Rab7–RILP–p150Glued motor protein complexes by the ER protein VAP, cholesterol, and the sensor ORP1L. Rab7 recruits the homodimeric effector protein RILP to LEs. RILP binds the p150Glued subunit of the dynein–dynactin motor. ORP1L also binds to the Rab7–RILP complex. ORP1L has a C-terminal cholesterol-sensing ORD that exists in different conformational states determined by cholesterol in LEs. At low cholesterol levels, ORD adopts a conformation in which the adjacent FFAT motif is exposed and which can be detected by the ER protein VAP in ER–LE MCSs. VAP binds in trans to the Rab7–RILP–p150Glued complex and removes p150Glued, thus preventing minus end–directed transport. High cholesterol conditions initiate a different ORP1L conformation, preventing formation of ER–LE MCSs, and VAP fails to interact with the Rab7–RILP–p150Glued complex. Thus, cholesterol levels in LEs determine the conformation of ORP1L and thereby VAP recruitment in ER–LE contact sites. The ER protein VAP then controls p150Glued binding to Rab7–RILP, resulting in the scattering of cholesterol-poor LEs and clustering of cholesterol-laden LEs, as in Niemann-Pick type C disease.
Model showing control of Rab7–RILP–p150Glued motor protein complexes by the ER protein VAP, cholesterol, and the sensor ORP1L. Rab7 recruits the homodimeric effector protein RILP to LEs. RILP binds the p150Glued subunit of the dynein–dynactin motor. ORP1L also binds to the Rab7–RILP complex. ORP1L has a C-terminal cholesterol-sensing ORD that exists in different conformational states determined by cholesterol in LEs. At low cholesterol levels, ORD adopts a conformation in which the adjacent FFAT motif is exposed and which can be detected by the ER protein VAP in ER–LE MCSs. VAP binds in trans to the Rab7–RILP–p150Glued complex and removes p150Glued, thus preventing minus end–directed transport. High cholesterol conditions initiate a different ORP1L conformation, preventing formation of ER–LE MCSs, and VAP fails to interact with the Rab7–RILP–p150Glued complex. Thus, cholesterol levels in LEs determine the conformation of ORP1L and thereby VAP recruitment in ER–LE contact sites. The ER protein VAP then controls p150Glued binding to Rab7–RILP, resulting in the scattering of cholesterol-poor LEs and clustering of cholesterol-laden LEs, as in Niemann-Pick type C disease.
Materials and methods
Reagents
Rabbit anti-GFP and rabbit anti-mRFP antibodies were generated using purified His-mRFP or His-GFP recombinant proteins as immunogen, respectively. Cross-reactivity was excluded by Western blot analyses with various mRFP- or GFP-labeled fusion proteins. Other antibodies used were rabbit polyclonal anti-NPC1 (Novus Biologicals), rabbit anti-ORP1L (gift from V. Olkkonen [National Public Health Institute, Helsinki, Finland] and M. Johansson [Medical Research Council Centre for Developmental Neurobiology, London, England, UK]), rat monoclonal antitubulin antibody (Abcam), rabbit anti–human VAP antiserum (recognizing VAP-A; gift from N. Ridgway, Dalhousie University, Halifax, Nova Scotia, Canada; further analyses showed that this serum recognizes VAP-A only), mouse anti–calnexin AF8 (David et al., 1993), mouse anti-LAMP1 (BD), mouse anti-CD63 (Vennegoor et al., 1985), mouse monoclonal anti-EEA1 (BD), rabbit anti-Rab7 and rabbit anti-HA epitope tag (Santa Cruz Biotechnology, Inc.), rabbit anti–human protein disulfide-isomerase (gift from N. Bulleid, University of Manchester, Manchester, England, UK), mouse monoclonal Golgi-97 (Invitrogen), mouse monoclonal anti–HA epitope tag (12CA5; Roche), and mouse monoclonal anti-DIC (BD). Fluorescent secondary antibodies were obtained from Invitrogen.
Lovastatin (or mevinolin) was purchased from EMD and converted from its inactive prodrug form to its open acid form before use by first dissolving the prodrug in ethanol and heating to 50°C for 2 h in NaOH. Na-phosphate buffer was then added, and this solution was heated to 40°C for 30 min. Finally, the pH was adjusted to 7.3 with concentrated HCl. Mevalonate was prepared by dissolving mevalonolactone (Sigma-Aldrich) in a 0.05-M NaOH solution. The pH was adjusted to 7.0 with 0.1 M HCl. U-18666A (EMD) was dissolved in ethanol to a final concentration of 10 mg/ml. The lipoprotein-deficient serum was prepared as described previously (Goldstein et al., 1983) and was a gift from M. Jauhiainen (National Public Health Institute).
cDNA constructs and vectors
Rab7, RILP, VAP-A, VAP-B, TAP1-GFP, p150Glued(C25), and ORP1L cDNA constructs have been described previously (Reits et al., 2000; Jordens et al., 2001; Wyles et al., 2002; Marsman et al., 2004; Johansson et al., 2007). GST–VAP-A was a gift from M. Hardy and K. Ettayebi (Montana State University, Bozeman, MT), and HA–VAP-A and –VAP-B were gifts from C. Hoogenraad (Erasmus Medical Center, Rotterdam, Netherlands; Teuling et al., 2007). VAP-A was cut with BamHI–XhoI and cloned into both a pAGFP-C1 vector and a pYFP-C1 vector (in the BglII–SalI site). GFP-ANKPHD and mRFP-ANKPHD were generated by subcloning the ANKPHD fragment from pCDNA4-ANKPHD (Johansson et al., 2007) into a pEGFP-C1 vector (BD) or into a custom made pmRFP-C1 vector (Johansson et al., 2007), respectively, using the BamHI and XbaI sites.
ΔORD was generated by PCR using the primers 5′-ACTCGGATCCATGAACACAGAAGCGGAGCAACA-3′ and 5′-AAAGGATCCCTATCTGTGTTTTCTACTGCCCAAATGC-3′ and was inserted into the BamHI site of pcDNA4HisMax (Invitrogen) and the BglII site of pmRFP-C1 vectors (Johansson et al., 2007). To generate mRFP-ΔORDPHDPHD, mRFP-ΔORD was digested with XbaI and EcoRI. The remainder of ΔORD was amplified by PCR using the primers 5′-CTCAAAATGATTAAGGAGTGTGACATGGCTA-3′ and 5′-CCGGAATTCCATTCTGTGTTTTCTACTGCCCAAATGCTC-3′. The amplified fragment was inserted into the XbaI and EcoRI sites of the previously digested mRFP-ΔORD vector. The first PH domain was generated by PCR using the primers 5′-CCGGAATTCCGATATGAGGGCCCTCTCTGGAAGA-3′ and 5′-CGGGGTACCGCTGTAAGCAGAATGTTCTTCTATTGC-3′ and ligated into the EcoRI and KpnI sites. Subsequently, the second PH domain was also generated by PCR using the primers 5′-CGGGGTACCCGATATGAGGGCCCTCTCTGGAAGA-3′ and 5′-CGGGATCCATTCTAGCTGTAAGCAGAATGTTCTTCTATTGC-3′ and inserted using the KpnI and BamHI sites.
mRFP-ΔORDFFAT(D478A) was generated by PCR using mRFP-ΔORD as a template and 5′-CCCAGAATTCTATGCGGCGCTGTCAGATTCCGAGTC-3′ and 5′-AAGCAAGTAAAACCTCTACAAATG-3′ as primers, followed by swapping of the wild-type FFAT motif in mRFP-ΔORD for the mutated FFAT using the EcoRI and BamHI restriction sites. The ORP1L FRET construct (mRFP-ORP1L-GFP) was generated in two steps. First, mRFP-ORP1L was digested with NheI–SacI (internal site in ORP1L), which was subsequently cloned into pEGFP-N1 (Clontech Laboratories, Inc.) to obtain an N-terminal region of ORP1L, which was N-terminally tagged to mRFP and C-terminally tagged to GFP. In the second step, the C-terminal part of ORP1L was produced by PCR with a forward primer (5′-GGAGTGTGACATGGCTAA-3′) and a reverse primer (5′-CGGGATTCCATAAATGTCAGGCAAATTGAAG-3′), mutating the ORP1L stop codon. The PCR product was digested using SacI and BamHI and cloned into the aforementioned intermediate construct.
To obtain mRFP-ΔORDPHDPHD-GFP, mRFP-ΔORDPHD was subcloned into pEGFP-N3 (Clontech Laboratories, Inc.) using the restriction sites NheI and KpnI in mRFP-ΔORDPHDPHD (described above). The second PH domain in the tandem was generated by PCR using the primers 5′-CGGGGTACCCGATATGAGGGCCCTCTCTGGAAGA-3′ and 5′-CGGGGTACCGCTGTAAGCAGAATGTTCTTCTATTGC-3′ and cloned into the KpnI restriction site. To obtain mRFP-ΔORD-GFP, mRFP-ΔORD was cloned into pEGFP-N2 (Clontech Laboratories, Inc.) using the restriction sites NheI and EcoRI in mRFP-ΔORDPHDPHD (described above).
For the generation of mRFP-ORP1L, ORP1L cDNA was cloned into mRFP-C1 using the BglII and BamHI restriction sites. Ectopic expression of RILP or ORP1L resulted in an approximately fourfold or sevenfold increase in mRNA levels (by qPCR), respectively. Expression (analyzed by qPCR) for RILP, ORP1L, and VAP related to actin (set at 100%) was 1.59% (SD, 0.01%), 2.16% (SD, 0.61%), and 1.78% (SD, 1.11%), respectively.
siRNAs for human NPC1 and ORP1L were obtained from Thermo Fisher Scientific (ON-TARGETplus SMARTpool for GenBank/EMBL/DDBJ accession nos. NM_000271 and NM_018030, respectively) and transfected using DharmaFECT 1 transfection reagent (Thermo Fisher Scientific). qPCR showed that RILP and ORP1L were expressed at <1% after siRNA silencing and VAP-A at 0.02%. All constructs were sequence verified. Expressed proteins had the expected molecular weight, as confirmed by SDS-PAGE and Western blot analyses.
Cell culture and drug treatment
Monolayers of MelJuSo cells were maintained in Iscove's modified Dulbecco's medium (IMDM; Invitrogen) supplemented with 8% FCS in 5% CO2 at 37°C. For cholesterol-depleting conditions, MelJuSo cells were washed with PBS and then cultured in IMDM supplemented with 5% lipoprotein-deficient serum, 50 µM lovastatin to inhibit HMG-CoA (3-hydroxy-3-methyl-glutaryl-CoA) reductase, and 230 µM mevalonate to supply essential nonsterol isoprenoids for cell growth and survival (Liscum et al., 1989; Sugii et al., 2006) for 4 h before analyses. For cholesterol enhancement, cells were washed with PBS and cultured in IMDM supplemented with 8% FCS and 3 µg/ml U-18666A for 15 h before analyses.
Microscopy
Transfected cells were fixed at 48 h (DNA or RNAi) or 72 h (RNAi and DNA) after transfection with 4% formaldehyde in PBS for 30 min and permeabilized for 5 min with 0.05% Triton X-100 in PBS at room temperature. Nonspecific binding of antibodies was blocked by 0.5% BSA/PBS for 40 min, after which cells were incubated with primary antibody in 0.5% BSA/PBS for 1 h at room temperature. Bound primary antibodies were visualized with Alexa Fluor secondary antibody conjugates (Invitrogen). Cells were mounted in Vectashield mounting medium (Vector Laboratories). The specimens were analyzed with confocal laser-scanning microscopes (TCS-SP1, TCS-SP2, or AOBS; Leica) equipped with HCX Plan-Apochromat 63× NA 1.32 and HCX Plan-Apochromat lbd.bl 63× NA 1.4 oil-corrected objective lenses (Leica). The acquisition software used was LCS (Leica).
Colocalization analysis for TAP1 and CD63 was performed using Matlab (MathWorks), in which images were stretched to 255 Gy, and colocalization was performed on the pixels with intensities between100 and 255 Gy. The number of colocalizing pixels was related to total CD63 pixels for quantification. The colocalizing pixels were projected on the CD63/TAP1 overlay image for representation.
Photoactivation experiments were performed in MelJuSo cells transfected with PA-GFP tagged to VAP-A and cotransfected with mRFP-tagged ORP1L, ORP1LΔORD, or ORP1LΔORDPDHPDH. Cells were mounted in phenol red–free DME (Invitrogen), limiting autofluorescence of the medium. All measurements were performed on an AOBS system equipped with a 37°C culture chamber and an HCX Plan-Apochromat lbd.bl 63× NA 1.4 lens. Cells were imaged using a 488-nm and 561-nm laser to visualize the activated PA-GFP– and mRFP-tagged proteins, respectively. Point activation with full power from the 405-nm laser was performed for 3 s at the nuclear region, where the ER was activated at the sites below or under the nucleus. This location was devoid of LE structures. To confirm this, a z stack was made before activation.
Quantification of vesicle distribution
A mask was drawn for every cell to be quantified. Within each cell, the center of gravity of the stain to be quantified (e.g., CD63) was determined for each cell with the program CellProfiler (Broad Institute). The cell was divided into six fractions from the defined center to the edge of the cell. The mean fractional intensity at a given radius (fraction of total intensity normalized by fraction of pixels at a given radius) was determined and plotted for each fraction. For the modules and pipelines used, see http://cellprofiler.labstore.nl.
FLIM
FLIM experiments were performed with MelJuSo cells stably expressing mRFP-ORP1L-GFP cultured on Delta T dishes (Bioptechs). Before measurements, cells were mounted in bicarbonate-buffered saline medium (140 mM NaCl, 5 mM KCl, 2 mM MgCl2, 1 mM CaCl2, 23 mM NaHCO3, 10 mM d-glucose, and 10 mM Hepes, pH 7.3) and analyzed at 37°C in a 5% CO2 culture hood surrounding the objective stage of the microscope. Images were taken on an inverted microscope (DM-IRE2; Leica) with an HCX Plan-Apochromat 63× NA 1.35 glycerol-corrected objective lens equipped with a frequency domain lifetime attachment (Lambert Instruments) controlled by the manufacturer's FLIM software (Lambert Instruments). GFP was excited with ∼4 mW of 488-nm light from a light-emitting diode modulated at 40 MHz, and emission was collected at 490–550 nm using an intensified charge-coupled device camera (CoolSNAP HQ; Roper Scientific). To calculate the GFP lifetime, the intensities from 12 phase-shifted images (modulation depth ∼70%) were fitted with a sinus function, and lifetimes were derived from the phase shift between excitation and emission. For internal controls, cells were co-cultured with MelJuSo cells expressing GFP only. Lifetimes were referenced to a 1-µM solution of rhodamine-G6 in saline that was set at a 4.11-ns lifetime. The donor FRET efficiency ED was calculated as ED = 1 − (measured lifetime/GFP lifetime in control cells) (Zwart et al., 2005).
Sensitized emission FRET by confocal microscopy
FRET between GFP and mRFP molecules was determined by calculating the sensitized emission (the mRFP emission after GFP excitation) from separately acquired donor and acceptor images (using an AOBS system equipped with a 37°C culture device at 5% CO2 and an HCX Plan-Apochromat 63× NA 1.32 oil-corrected objective lens). Three images were collected: GFP excited at 488 nm and detected between 500 and 549 nm, indirect mRFP excited at 488 nm and detected between 581 and 675 nm, and direct mRFP excited at 561 nm. Because of GFP-mRFP spectral overlap, indirect mRFP emission was corrected for bleed through of GFP in the 581–675-nm filter and for direct excitation of mRFP by 488-nm light. The correction factors were determined experimentally for each image by co-culturing cells expressing mRFP-ORP1L-GFP (or any of the ORP1L variants) with cells expressing only GFP (to detect the bleed through in the mRFP channel) or mRFP (to detect direct excitation with 488-nm light). FRET and donor FRET efficiency ED were calculated from these data as previously described in detail (van Rheenen et al., 2004; Zwart et al., 2005). For quantification of the intramolecular FRET gradient of mRFP-ORP1L-GFP and mRFP-ΔORD-GFP, ED was measured in five equally sized concentric rings covering the area between the nucleus and the cell surface at the most distant point. The ED in each circle was collected, binned, and expressed per cell analyzed. The mean ED and SEM of the different cells analyzed are calculated for the different rings.
Immuno-EM
Cells were fixed in 2% paraformaldehyde in 0.1 M PHEM buffer (60 mM Pipes, 25 mM Hepes, 2 mM MgCl2, and 10 mM EGTA, pH 6.9) and then processed for ultrathin cryosectioning as previously described (Calafat et al., 1997). 50-nm-thick cryosections were cut at −120°C using diamond knives in a cryoultramicrotome (Leica) and transferred with a mixture of sucrose and methylcellulose onto formvar-coated copper grids. The grids were placed on 35-mm Petri dishes containing 2% gelatin at 37°C. The grids were incubated with the various antibodies at room temperature and then incubated with 10-nm protein A–conjugated colloidal gold (EM Laboratory, Utrecht University) as previously described (Calafat et al., 1997). After immunolabeling, the sections were embedded in a mixture of methylcellulose and 0.4% uranyl acetate and examined with an electron microscope (CM10; Philips). The 40-nm spacing between the ER and LE membranes was derived from both digital and printed electron micrographs.
Filipin stainings
Formaldehyde-fixed cells were stained for 2 h at room temperature with 0.05 mg/ml filipin (Sigma-Aldrich) freshly dissolved in PBS supplemented with 0.5% BSA, and the staining was imaged with a microscope (AxioObserver Z1; Carl Zeiss, Inc.) equipped with a charge-coupled device camera (ORCA-ER; Hamamatsu Photonics) and a 63× NA 1.25 Plan-Neofluar oil-corrected objective lens. Filipin signal was recorded using a 390–415-nm band-pass (BP) excitation filter and with a 450–470-nm BP emission filter. FITC signal was recorded with a 480–510-nm BP excitation filter and with a 515–550-nm BP emission filter. mRFP signal was recorded with a 555–590-nm BP excitation filter and with a 605–655-nm BP emission filter (Carl Zeiss, Inc.). For data acquisition, the manufacturers' software was used.
Protein purification
His-Rab7, His-RILP, and GST-ORP1L were produced as previously described (Johansson et al., 2007). Cells were lysed in 0.5% Triton X-100, 20 mM Hepes, pH 7.5, 200 mM NaCl, 8 mM β-mercaptoethanol, and complete EDTA-free protease inhibitors cocktail (Roche). The lysate was cleared by centrifugation, and the supernatant was incubated with pre-equilibrated Talon Co2+ resin (Clontech Laboratories, Inc.) for 30 min. The resin was then washed extensively with 20 mM Hepes, pH 7.5, 200 mM NaCl, and 8 mM β-mercaptoethanol. Proteins were stored in 8.7% glycerol at −80°C until use in protein reconstitution experiments. GST–VAP-A construct in pGEX was a gift from N. Ridgway (Wyles et al., 2002). GST was removed by cleavage with thrombin by incubation at 30°C for 2 h before inactivation and precipitation by 0.1 mM PMSF. The GST moiety was removed using a glutathione-Sepharose column (GE Healthcare), and the purity of GST-free VAP-A was confirmed by SDS-PAGE and Coomassie staining.
In vitro VAP-A induced p150Glued removal from the Rab7–RILP protein complex
The reconstitution assay was performed in a detergent/salt buffer (20 mM Hepes, pH 7.5, 200 mM NaCl, 4 mM β-mercaptoethanol, 5 mM MgCl2, 0.05% [vol/vol] Triton X-100, and 50 µM GTP). 15 µg His-Rab7(Q67L) was coupled to 3 µl of Talon Co2+ beads and preloaded with GTP. Beads were washed to remove unbound His-Rab7(Q67L), and recombinant purified proteins were added in equimolar concentrations. 3 µg His-RILP, 8.7 µg GST-ORP1L, 2.2 µg VAP-A, and 4.5 µg maltose-binding protein (MBP)–p150Glued(C25) were added to the beads as indicated and incubated in a total volume of 0.4 ml for 2 h at 4°C. Beads were washed extensively, and 4.5 µg MBP-p150Glued(C25) or 2.2 µg VAP-A was added to the beads as indicated. Proteins were subsequently incubated in a total volume of 0.4 ml of detergent/salt buffer for 30 min at 22°C. Beads were washed extensively, and proteins bound to His-Rab7(Q67L) were analyzed by SDS-PAGE and Western blotting. Proteins were detected with the following antibodies: α-His (GE Healthcare), α-MBP (New England Biolabs, Inc.), and α-GST (gift from W. Moolenaar, The Netherlands Cancer Institute, Amsterdam, Netherlands). Direct binding of p150Glued to VAP-A was assayed as follows. 30 µg GST–VAP-A or 15 µg GST was coupled to 15 µl of glutathione–Sepharose 4B beads (GE Healthcare). Beads were washed to remove unbound proteins, and 4.5 µg MBP-p150Glued(C25) was added to the beads in a total volume of 0.4 ml for 30 min at 22°C. Beads were washed extensively, and proteins bound to the glutathione–Sepharose 4B beads were analyzed by SDS-PAGE and Western blotting.
Online supplemental material
Fig. S1 shows cholesterol, NPC1, the ORD, and vesicle distribution. Fig. S2 shows colocalization experiments of RILP and ORP1L variants with compartment markers and dynein motor subunits. Fig. S3 shows the chemical manipulation of cholesterol levels and location of ORP1L variants on LEs. Fig. S4 shows control-sensitized emission FRET and FLIM experiments, the effect of NPC1 silencing on ORP1L conformation, and vesicle distribution of ORP1L mutants. Fig. S5 shows ORP1L-mediated recruitment of VAP-A from the ER and the release of p150Glued from RILP. Videos 1–3 show time-lapse experiments of LysoTracker red–labeled MelJuSo cells when cholesterol levels were manipulated. Videos 4 and 5 show time-lapse experiments of MelJuSo cells in which expression of mRFP-ΔORD and mRFP-ΔORDPHDPHD is induced and the distribution of LEs is followed. Videos 6–8 show time-lapse experiments in MelJuSo cells expressing mRFP-ORP1L, mRFP-ΔORD, and mRFP-ΔORDPHDPHD combined with photoactivation of PA-GFP–tagged VAP-A.
Acknowledgments
We thank K. Jalink for support with sensitized emission and FLIM measurements, P. Peters for support with EM, and V. Olkkonen and M. Johansson for ORP1L reagents and constructs. We also thank N. Ridgway, M. Hardy, K. Ettayebi, and C. Hoogenraad for VAP-A and VAP-B reagents, N. Ong for processing electron micrographs, and M. Hauptmann for help with statistics. We thank H. Pickersgill, R. Michalides, S. Godsave, J. Pierson, and V. Menendez-Benito for critically reading the manuscript and S. Krom for helping with experiments.
N. Rocha was supported by a Fundação para a Ciência ea Tecnologia Fundo Social Europeu PhD scholarship within the Third Framework Program. This work was supported by grants from the Dutch Cancer Society Koningin Wilhelmina Fonds and the Chemical Sciences Section of the Nederlandse Organisatie voor Wetenschappelijk Onderzoek (TOP grant).
References
Abbreviations used in this paper: BP, band pass; CLSM, confocal laser-scanning microscopy; DIC, dynein intermediate chain; FLIM, fluorescence lifetime imaging microscopy; FRET, fluorescence resonance energy transfer; IMDM, Iscove's modified Dulbecco's medium; LE, late endosome; LUT, look up table; MBP, maltose-binding protein; MCS, membrane contact site; mRFP, monomeric RFP; MTOC, microtubule-organizing center; ORD, OSBP-related domain; OSBP, oxysterol-binding protein; PH, pleckstrin homology; qPCR, quantitative PCR; RILP, Rab7-interacting lysosomal protein; VAP, VAMP-associated ER protein.
Author notes
C. Kuijl and R. van der Kant contributed equally to this paper.


![Figure 1. Chemical and genetic cholesterol manipulations and LEs. (A) Modulation of intracellular cholesterol levels. Mel JuSo cells were cultured in normal medium (FCS), cholesterol-depleted medium supplemented with lovastatin (statin), or normal medium supplemented with U-18666A, as indicated. Fixed cells were stained with filipin to detect cholesterol. A color look up table (LUT) shows fluorescence intensities. n > 100 for each condition. (B) Effects of intracellular cholesterol manipulation (control [CTRL], U-18666A, or statin treatment) on LE positioning in RILP- or ORP1L-silenced Mel JuSo cells. Cells were stained for the LE marker CD63 (red), endogenous ORP1L (green), and actin (blue). The position of the CD63-positive vesicles relative to the nucleus (radial distribution) was determined for the various conditions (>50 cells per condition), binned, and plotted (right). The distributions were statistically different according to the Jonckheere-Terpstra test for control siRNA (P = 1.52 × 10−5 and 7.76 × 10−6 for CD63 and ORP1L, respectively) only. No difference was detected for siRILP (P = 0.20 and 0.54 for CD63 and ORP1L, respectively) and for siORP1L (P = 0.29 for CD63). (C) Dynein motor activity and clustering of LEs in NPC1-silenced cells. Mel JuSo cells were transfected with siRNA for NPC1 for 72 h and, after 48 h, transfected with an expression construct for p50dynamitin to disrupt the dynein–dynactin motor. Fixed cells were stained for CD63 and p50dynamitin to mark the overexpressing cells. n > 100. Bars: (A and C)10 µm; (B) 20 µm.](https://cdn.rupress.org/rup/content_public/journal/jcb/185/7/10.1083_jcb.200811005/6/m_jcb_200811005_rgb_fig1.jpeg?Expires=1767738243&Signature=dkycTEEzZAM0uP5v3msa0i9lgpb6ZW5Q5kHG85Lr5pKR06JfjLZMMcjZY9O0E0If8N5VVk~QbVcneW2YORhNMRPDVsq3W9bC3FqjItarzUg8fmjugV8rG3NhhiJIJR7q~kCPboSI4vpsd9IHLcf6mZ~NG~d4wSsjjWKjbkId5RBHFw1vBwpwd~ktvxVTOBw2vbmTSWf0nphURDJ0W2QoYPKoJTO3h4YhOqs7HJR5Ghy9io2SJnRT4wcvM6wkZ2J0hCWVOquf9Iy0H-dSPbznV4SS2WNH7lkwcEYdOG4StDaACEtBLCgg~3WKJzJKRAeH2R608Yay76mAXrB7sWOyog__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)